- 1Faculty of Science, Plant Breeding Institute, The University of Sydney, Sydney, NSW, Australia
- 2Faculty of Veterinary and Agricultural Sciences, The University of Melbourne, Parkville, VIC, Australia
Leaf rust of barley causes significant losses in crops of susceptible cultivars. Deploying host resistance is the most cost-effective and eco-sustainable strategy to protect the harvest. However, most known leaf rust resistance genes have been overcome by the pathogen due to the pathogen’s evolution and adaptation. The discovery of novel sources of genetic resistance is vital to keep fighting against pathogen evolution. In this study, we investigated the genetic basis of resistance in barley breeding line GID 5779743 (GID) from ICARDA, found to carry high levels of seedling resistance to prevalent Australian pathotypes of Puccinia hordei. Multipathotype tests, genotyping, and marker-trait associations revealed that the resistance in GID is conferred by two independent genes. The first gene, Rph3, was detected using a linked CAPS marker and QTL analysis. The second gene was detected by QTL analysis and mapped to the same location as that of the Rph5 locus on the telomeric region of chromosome 3HS. The segregating ratio in F2 (conforming to 9 resistant: 7 susceptible genetic ratio; p > 0.8) and F3 (1 resistant: 8 segregating: 7 susceptible; p > 0.19) generations of the GID × Gus population, when challenged with pathotype 5477 P− (virulent on Rph3 and Rph5) suggested the interaction of two genes in a complementary fashion. This study demonstrated that Rph3 interacts with Rph5 or an additional locus closely linked to Rph5 (tentatively designated RphGID) in GID to produce an incompatible response when challenged with a pathotype virulent on Rph3+Rph5.
Introduction
Leaf rust of barley, caused by Puccinia hordei, is the most common and prevalent disease in temperate barley-growing regions (Park et al., 2015). This disease can cause yield losses of up to 62% in susceptible cultivars under epidemic conditions (Clifford, 1985; Cotterill et al., 1992; Griffey et al., 1994; Park et al., 2015). Deploying host resistance has long been considered a cost-effective and environmentally friendly method to protect crop yield from this pathogen (Dinh et al., 2020). So far, 28 loci conferring resistance to leaf rust in barley have been characterized, viz. Rph1 to Rph28 (Mehnaz et al., 2021), of which 25 (Rph1-Rph19, Rph21, Rph22, Rph25, Rph26, Rph27, and Rph28) confer resistance at all growth stages (all stage resistance or ASR), and three (Rph20, Rph23, and Rph24) confer resistance at adult growth stages (adult plant resistance or APR) only. Notably, all the ASR genes conferring resistance to P. hordei identified so far are dominant. Up to date, only four out of 28 Rph genes have been isolated, with three genes encoding proteins of the Nucleotide-Binding Leucine-Rich Repeat (NLR) family (Dracatos et al., 2019; Wang et al., 2019; Chen et al., 2021) and one gene encoding protein of a distinct class (Dinh et al., 2022).
Several ASR Rph genes, including Rph3, Rph7, and Rph9, which were previously known to be effective have been extensively deployed in breeding programs globally (Park et al., 2015). However, the acquisition of virulence matching most of these major Rph genes by P. hordei has hampered their further utilization (Steffenson et al., 1993; Park et al., 2015; Kavanagh et al., 2017), and the deployment of such single major genes in large area can possibly result in serious epidemics (Brooks et al., 2000). Therefore, in parallel with mining new sources of resistance, deploying genes in combinations of two or more has proven to be an effective method of achieving a durable resistance against various pathogens (Kloppers and Pretorius, 1997; Fukuoka et al., 2015; Mundt, 2018). Interestingly, combinations of various “defeated” genes providing residual effects have been reported in multiple crops against different pathogens (Nass et al., 1981; Royer et al., 1984; Brodny et al., 1986; Li et al., 1999). The interaction of an APR and an ASR gene conferring resistance to P. hordei virulent pathotype has also been reported (Singh et al., 2021). When being singly deployed, both the ASR gene Rph5.e and the APR gene Rph20 are ineffective against the pathotype 220 P+ +Rph13 at the seedling stages. However, the Rph5.e + Rph20 combination stayed effective when challenged with the pathotype 220 P+ +Rph13. These findings suggested that the residual effect of “defeated” genes can confer the resistance to P. hordei virulent pathotype.
A barley breeding line GID 5779743 was among germplasm imported to Australia as part of the CAIGE (CIMMYT Australia ICARDA Germplasm Enhancement) project (CAIGE code 67:ZBS15; Australian Grains Genebank accession AGG411915BARL). Tests with the most prevalent pathotype of P. hordei in Australia, viz. 5457 P+, showed that it carried uncharacterized ASR to this pathogen. This study was undertaken to characterize the genetic basis of the resistance to P. hordei in GID 5779743.
Materials and methods
Plant materials and growing conditions
The barley line GID 5779743 (Pedigree: Shenmai No. 3/MSEL) investigated in this study is an accession from the CAIGE barley germplasm collection. The pedigree of Shenmai No. 3 is Shenmai No. 1/Humai No. 10, while the pedigree of MSEL is unknown. Other barley accessions carrying various Rph genes were used as controls (Park et al., 2015). GID 5779743 (here onward referred to as GID) was crossed to a susceptible line (Gus) to develop four F2 populations (POP1–POP4) derived from four F1 seeds. F2 plants of the population POP1 were advanced to the F3 generation for further studies.
Six P. hordei pathotypes designated according to the octal notation proposed by Gilmour (1973) (viz. 200 P− [Plant Breeding Institute culture number 518], 220 P+ +Rph13 [577], 5457 P+ [612], 5477 P− [672], 253 P− [490], and 5652 P+ [561]) were used in this study. The suffix P+/P− added to each octal designation indicated virulence/avirulence for resistance gene Rph19 (Park, 2003). These pathotypes were originally raised from single uredinia on the leaf rust susceptible genotype cv. Gus in the greenhouse and the urediniospores were dried above silica gel for 5–7 days at 12°C before being stored in liquid nitrogen at the Plant Breeding Institute, The University of Sydney, Australia. Details for each pathotype, including pathogenicity on different resistance genes, are listed in the Supplementary Table 1.
Phenotypic analysis
Seedlings were raised in 9-cm-diameter pots. The pots were watered with a soluble fertilizer (Aquasol®, Hortico Pty. Ltd., Revesby, NSW, Australia) at the rate of 35 g in 10 L of water for 100 pots, before sowing. Each F3 family was sown using 25–30 seeds/pot. Seedlings of differential lines and parents were raised by sowing clumps with 8–10 seeds of each.
Greenhouse inoculations were performed following the technique described by Golegaonkar et al. (2009). After sowing, the pots were transferred to rooms maintained at 23 ± 2°C with natural light, and seedlings were raised until ready for inoculations. The inoculations were carried out on 8–10 days-old seedlings with fully expanded first leaves using urediniospores (10 mg of spores per 10 ml of mineral oil per 200 pots) of P. hordei. The inoculated seedlings were placed in the incubation room for 24 h at ambient temperatures in a misted dark room where an ultrasonic humidifier generated the mist. The seedlings were moved to greenhouse chambers at 23 ± 2°C, and disease responses were recorded after 10 days, using a “0”–“4” infection type scale (Park et al., 2015). Infection types (ITs) of “2+” and lower were considered resistant, while “3” or higher indicated susceptibility.
Genetic analysis
Four F2 populations (POP1–POP4) derived from four F1 seeds of the cross GID/Gus were subjected to phenotypic assays with four P. hordei pathotypes (POP1 with pt. 5457 P+, POP2 with pt. 220 P+ +Rph13, POP3 with pt. 200 P−, and POP4 with pt. 5477 P−) to examine the inheritance of the resistance against each.
A total of 415 F3 families from POP1 were inoculated with pathotypes 5457 P+ and 5477 P−, while 61 F3 families selected randomly from these 415 families were inoculated with 200 P− and 220 P+ +Rph13. The response to each P. hordei pathotype of these families was recorded 9 days after inoculation when the susceptible control line “Gus” showed the high IT response (“3+”). These F3 families were scored as either non-segregating resistant (NSR), segregating (SEG), or non-segregating susceptible (NSS). The obtained data were statistically analyzed by Chi-squared analyses (χ2) to confirm the goodness-of-fit of observed ratios to theoretical expectations.
DNA isolation and marker analysis
The total DNA from leaf samples was extracted using the SDS method as previously described by Dinh et al. (2022). About 30 mg of the first leaf of each seedling was sampled into a 96-well collection tube (12 × 8 wells) containing two ball bearings and subjected to DNA extraction using an SDS method. To stabilize the DNA, 450 μl of extraction buffer including 0.1 M of Tris–HCl buffer (pH 8.0), 0.005 M EDTA buffer (pH 8.0), 0.5 M NaCl, 2-Mercaptoethanol (70 μl/100 ml buffer), and RNAse (100 μg/ml) were added to each sample before crushing. A TissueLyzer II (Qiagen, Hilden, Germany) at 25 Hz for 2 min was used for crushing the leaf material in the extraction buffer. The final mixture was then added with SDS solution (1.2% final concentration) to solubilize the proteins and lipids at 65°C for 60 min. The remaining proteins were precipitated by adding ammonium acetate 7.5 M to reach a final concentration of 2 M. The mixture was incubated at 4°C for 60 min, followed by centrifuging at 4,800 rpm (4,327 × g) for 10 min to separate debris and the aqueous phase. The upper phase containing genomic DNA was transferred to a new 96 well format plate and pelleted out by adding 100 μl of chilled isopropanol to 100 μl of supernatant. The pellet was washed twice using 100 μl of 70% ethanol before being slowly dissolved in 200 μl TE 0.1x buffer for 6 h for downstream applications.
Primer3Plus software1 was used to design PCR primers that were subsequently synthesized commercially (Sigma-Aldrich, NSW, Australia). Each 10 μl PCR contained 0.2 units of high-fidelity DNA polymerase (MyFi™, Bioline, NSW, Australia), 0.3 μM of each primer, 1x MyFi reaction buffer (Bioline, NSW, Australia), and 20 ng of genomic DNA. Thermocycling conditions consisted of an initial denaturation of 95°C for 10 min followed by 30 cycles of 94°C for 30 s, 55–60°C for 30 s, 72°C for 30 s, followed by a final extension at 72°C for 10 min. PCR products were digested [using a suitable endonuclease when required (Supplementary Table 2)] for 3 h under the recommended temperature. The digested products were monitored by electrophoresis on an agarose gel and visualized by staining with 6x GelRed® (Biotium, California, CA, United States) (1.5 μl/100 ml agarose gel).
Specific markers to detect the presence/absence of Rph3 (Dinh et al., 2022), Rph7 (Dracatos, unpublished), and Rph15 (Chen et al., 2021) were used (Supplementary Table 3).
High throughput genotyping using DArTseq markers and quantitative trait loci analysis
Based on the phenotypic data recorded from F3 families inoculated with P. hordei pathotype 5457 P+, DNA samples extracted from F2 plants corresponding to NSR and NSS families were selected for medium density genotyping. The concentration of extracted DNA was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and adjusted to approximately 80 ng/μl and sent to Diversity Arrays Technology (DArT) Pty. Ltd., Australia for whole-genome profiling. DArTseq silico and DArTseq SNP markers (>23K) were used for genotyping 80 samples (39 samples from each resistant and susceptible group and two parents). DArTseq SNP markers were scored “1” or “0” for the “presence” or “absence” of the marker, while heterozygous genotypes were scored “1/1.” SilicoDArTs are scored in a binary fashion, representing genetically dominant markers with “1” and “0” for the “presence” or “absence” of the restriction fragment with the marker sequence in the genomic representation of the sample. For both sets of markers, the “-” symbol represented missing data.
Genotypic data received from DArTs coupled with the response to pathotype 5457 P+ from 78 F3 families of POP1 were subjected to linkage analysis and mapping. Before analysis, both sets of DArT data consisting of DArTseq silico and DArTseq SNP were curated by removing markers with unknown positions, reproducibility of less than 95%, call rate less than 98%, or markers with more than 20% of missing values. After marker curation, 1,440 DArTseq silico markers and 1,440 DArTseq SNP markers were used separately for QTLs analysis with WinQTLcart software. The phenotypic data used for both analyses were converted from susceptibility and resistance into binary values “0” and “1,” respectively. For both analyses, the QTLs with LOD values higher than the threshold were considered and validated to identify resistance loci.
Developing polymerase chain reaction-based markers for detailed mapping
Based on the physical position of the DArTseq markers, the sequences of annotated high confidence genes listed on the IPK database within the detected locus were used to develop PCR-based markers. Various pairs of primers were designed based on the sequence of Morex to amplify the genomic DNA fragment from both parents of the mapping population (GID 5779743 and Gus). These amplicons were then sequenced using Sanger sequencing (Australian Genome Research Facility Ltd., Victoria, Australia) and aligned to determine the SNP that could be used to nominate suitable restriction enzymes. All markers used were co-dominant and therefore could differentiate heterozygous and homozygous forms. Genomic DNA of all 415 F2 individuals, including 78 samples used for DArT genotyping, was used to construct the detailed map of the detected QTL. The genetic map was constructed using MapChart version 2.32 (Voorrips, 2002). Genetic distances were calculated using the Kosambi mapping function (Kosambi, 2008).
Results
Phenotyping and genotyping of GID
GID was resistant to all P. hordei test pathotypes while the IT responses of various leaf rust differentials varied from resistant to susceptible depending upon the pathotype used (Table 1). Based on multipathotype tests and IT response, the resistance in GID could be conferred by one of the known genes for which all pathotypes used were avirulent, such as Rph7 or Rph15, combination of different resistance genes or a novel gene not reported yet. Genotyping GID with molecular markers linked to Rph3, Rph7, and Rph15 suggested the presence of the Rph3 (Figure 1A), but the absence of Rph7 and Rph15 (Figures 1B,C). Specifically, the genotypic data with the Rph3 linked CAPS marker showed that GID had the same genotype as Bowman + Rph3 NIL (Figure 1A), PCR with the Rph7 linked marker did not generate any product (Figure 1B), and PCR with the Rph15 linked KASP marker showed that GID had the same genotype as Gus and Bowman, which lack Rph15 (Figure 1C). The phenotypic and genotypic analyses provided a strong evidence that GID carries the combination of Rph3 and another resistance gene that was tentatively designated RphGID.
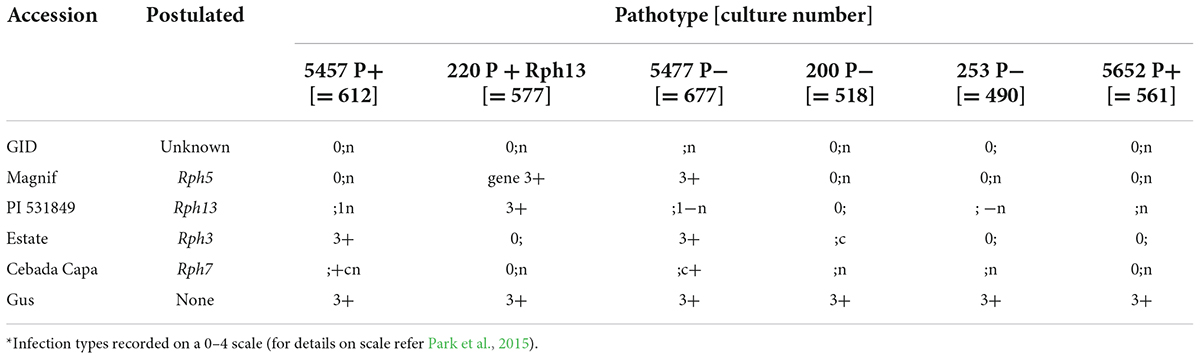
Table 1. Infection type* given by the parents (GID 5779743 and Gus) and controls with six P. hordei pathotypes.
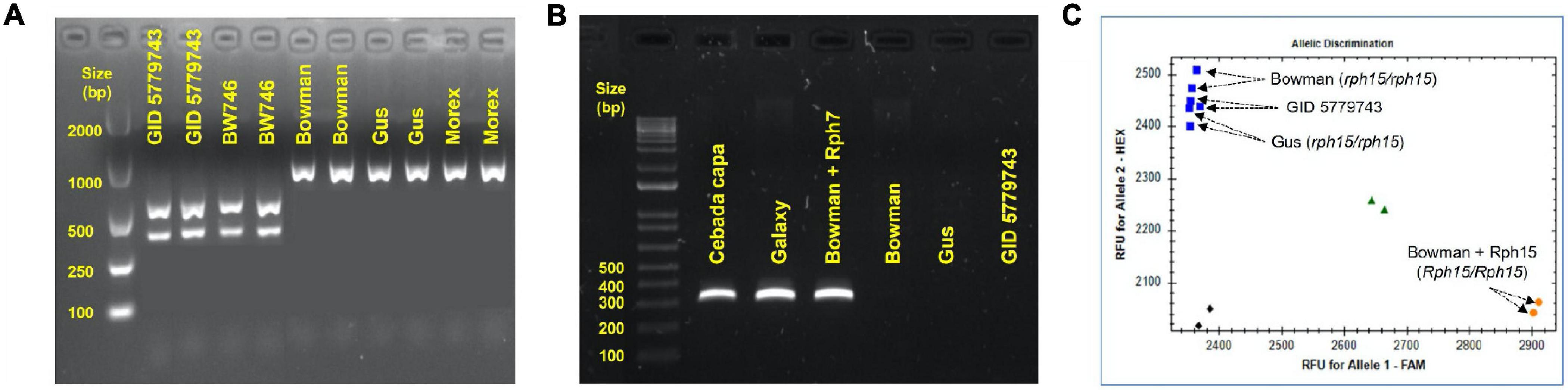
Figure 1. Genotype of GID 5779743 using various closely linked markers with known Rph genes. (A) The barley line BW746 is the NIL of Bowman carry Rph3 that was used as the positive control. The Rph3 gene is absent in the other barley cultivars, namely Bowman, Gus, and Morex, which were used as negative controls. (B) The absence of the resistance gene Rph7 in the barley line GID 5779743 confirmed by marker genotyping. The dominant marker gives the amplicon of 319 bp in length when the gene is present in the barley cv. Cebada Capa, Galaxy, and Bowman+Rph7 while it is absent in three barley cv. Bowman, Gus, and GID 5779743. (C) The absence of the resistance gene Rph15 in the barley line GID 5779743 was confirmed by marker genotyping. KASP marker illustrated the presence of Rph15 in the barley NIL line Bowman+Rph15 by the orange dot in the bottom-right corner, the blue squares in the top-left corner demonstrated the absence of Rph15 in three barley cv. Bowman, Gus, and GID 5779743, the green triangles showed the heterozygous form (Rph15/rph15) while the black diamonds in the bottom-left corner represented the water as no template control. All the PCRs were performed three time and generated similar results.
Genetic inheritance of the resistance in GID
Four F2 populations derived from four F1 seeds of the cross GID/Gus were challenged with various P. hordei pathotypes. The response of four F2 populations (POP1–POP4) revealed that two genes were involved in providing resistance to P. hordei in GID. The response observed in POP1 to pathotype 5457 P+ (virulent on Rph3 and avirulent to Rph5) showed that a single locus (RphGID) conferred the resistance of GID to this pathotype (Tables 2, 3). Similarly, the response of POP2 to pathotype 220 P+ +Rph13 (avirulent on Rph3 and virulent on Rph5) showed a single gene segregation, presumably due to Rph3 (Table 2), and also demonstrated that RphGID is susceptible to this pathotype. The phenotypic data from POP3 challenged with P. hordei pathotype 200 P− (avirulent on Rph3 and Rph5) showed segregation conforming to a digenic inheritance (Table 2) with likely involvement of Rph3 and RphGID. The segregation in the response of POP4 to pathotype 5477 P− (virulent on Rph3 and Rph5) suggested the involvement of two complementary genes (9 resistant: 7 susceptible; Table 2). The segregation ratio in the F3 generation of POP1 against the four pathotypes was consistent with those of the F2 populations (Table 3). The segregation of these four populations against various P. hordei pathotypes demonstrated the presence of two resistance genes, one being Rph3 and the second gene considered RphGID at this stage.

Table 2. The inheritance of resistance to four pathotypes of Puccinia hordei in barley line GID 5779743 based on F2 populations derived from the cross GID/Gus.

Table 3. The inheritance of resistance to four pathotypes of Puccinia hordei in barley line GID 5779743 using F2-derived F3 families from the cross GID/Gus.
Marker trait analysis
QTL conferring resistance to pathotype 220 P+ +Rph13
A total of 1,440 DArTseq Silico markers were retained after data curation. A total of 74 F3 families that were either NSR or NSS against pathotype 220 P+ +Rph13 were selected for QTL analysis. One QTL was detected at the Rph3 locus on the distal region of chromosome 7HL (Figure 2) with the LOD = 7.8.
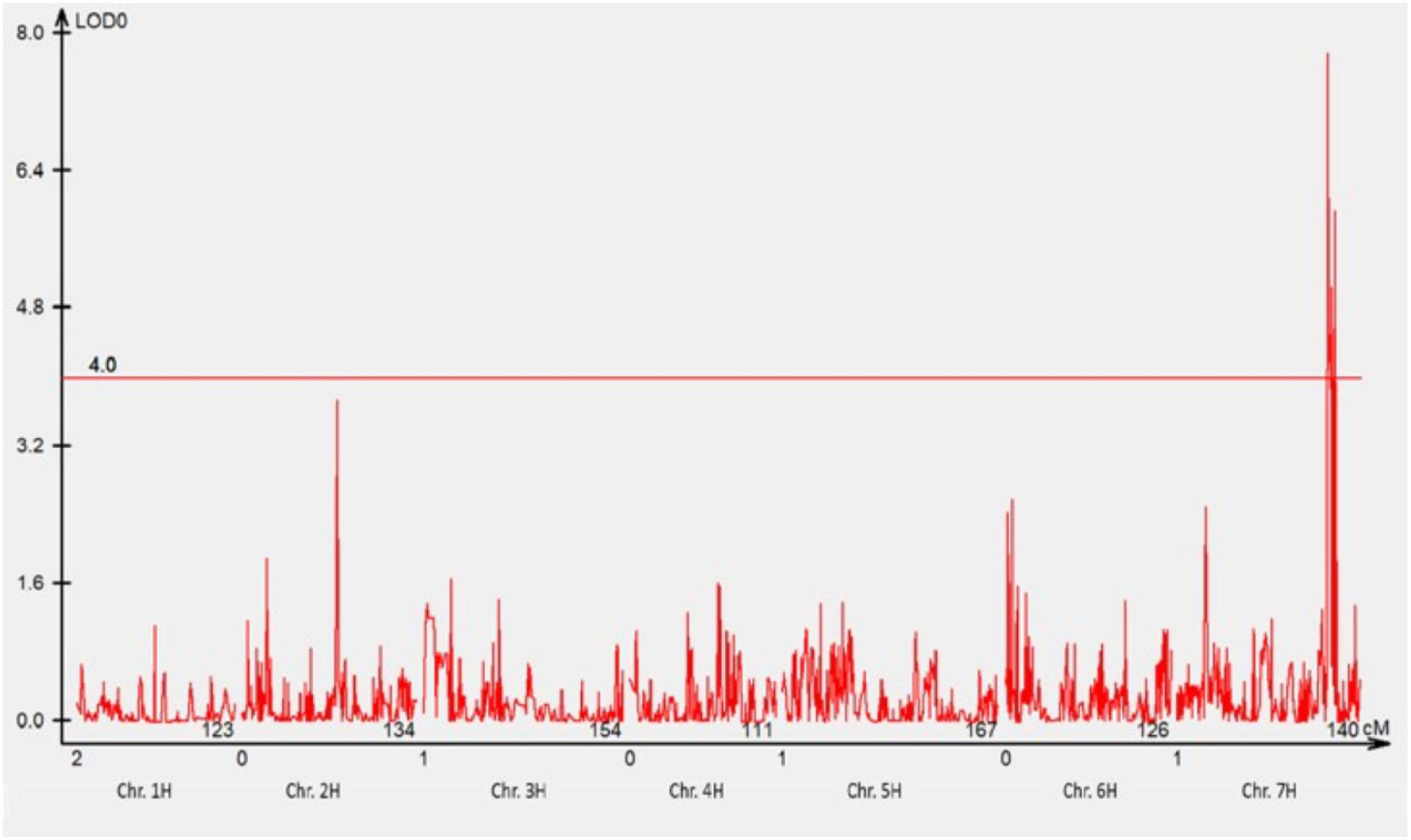
Figure 2. The most significant QTL conferring resistance to pathotype 220 P+ +Rph13 was detected using genotypic data of DArTseq Silico markers. One QTL was detected on the long arm of chromosome 7H. The threshold value was set at 4.0.
QTL conferring resistance to pathotype 5457 P+
The mapping population comprising of 78 F3 families was genotyped using 23,309 DArTseq silico and 11,458 DArTseq SNP markers covering the whole genome. After the data curation, 1,440 DArTseq SNP markers and 1,440 DArTseq silico markers were used together with the phenotypic data generated in phenotypic assay with pt. 5457 P+ for analyses. One locus on chromosome 3HS (RphGID) was detected with the LOD = 19, and 18 linked markers (Figure 3).
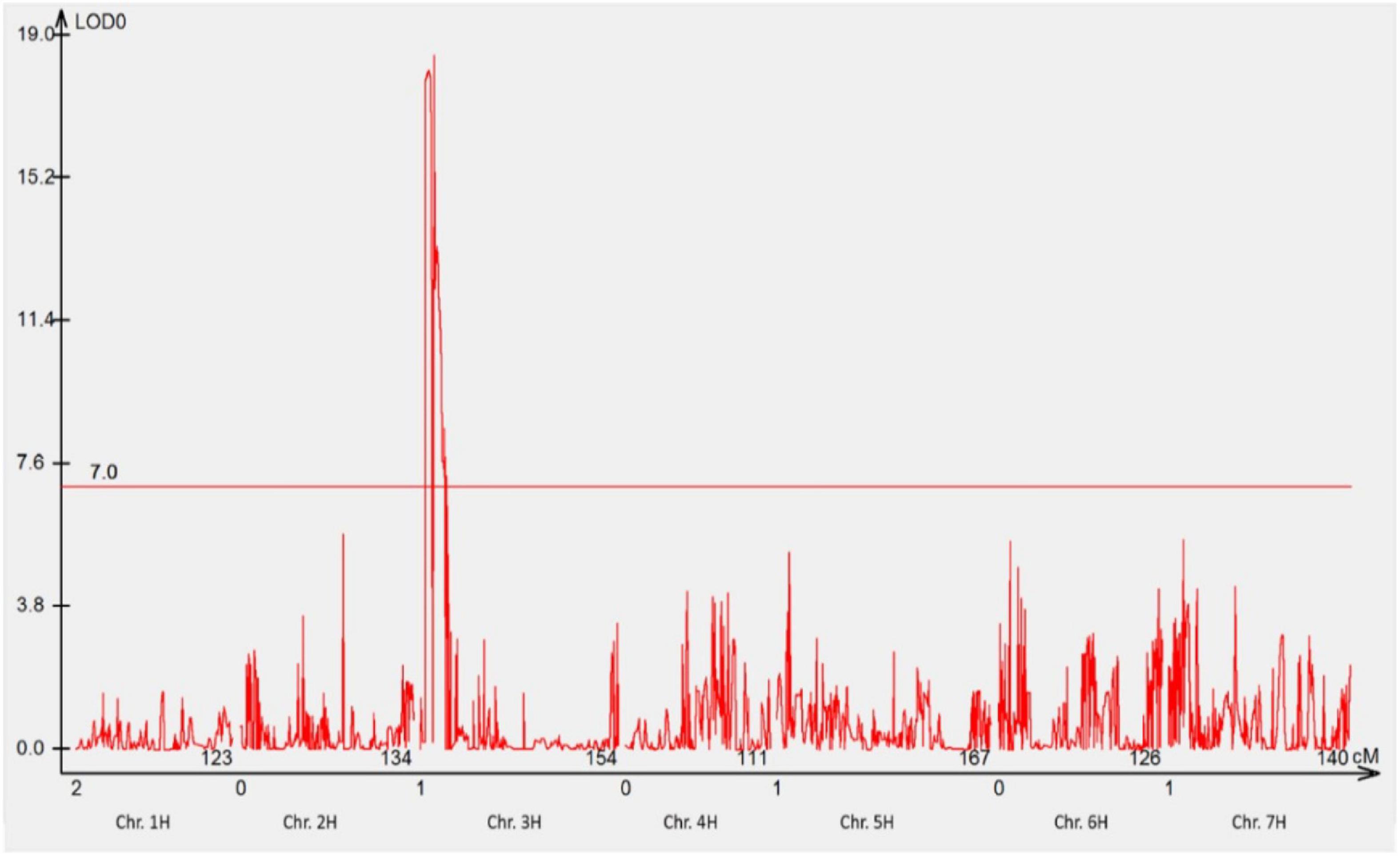
Figure 3. The most significant QTL conferring resistance to pathotype 5457 P+ detected using genotypic data of DArTseq SNP markers. One QTL was detected on the distal region of the short arm of chromosome 3H. The threshold value was set at 7.0.
Detailed map of RphGID
Pt. 5457 P+
The whole F2 population consisting of 415 individuals was phenotyped with pt. 5457 P+ and genotyped using eight CAPS markers to construct a detailed map of the QTL RphGID (Supplementary Table 3). All of these markers resided on one side of the RphGID locus on chromosome 3H. The genetic distance from the closest marker ZG_13 to RphGID was 1.8 cM (Figure 4). Due to the telomeric location of the locus, the marker ZG_13 delimited locus RphGID in a physical window of 33 kb in length based on the reference genome of Morex v2.0 (2019). Notably, the RphGID was mapped to the same position as the Rph5 locus, on the extreme telomeric region of chromosome 3HS.
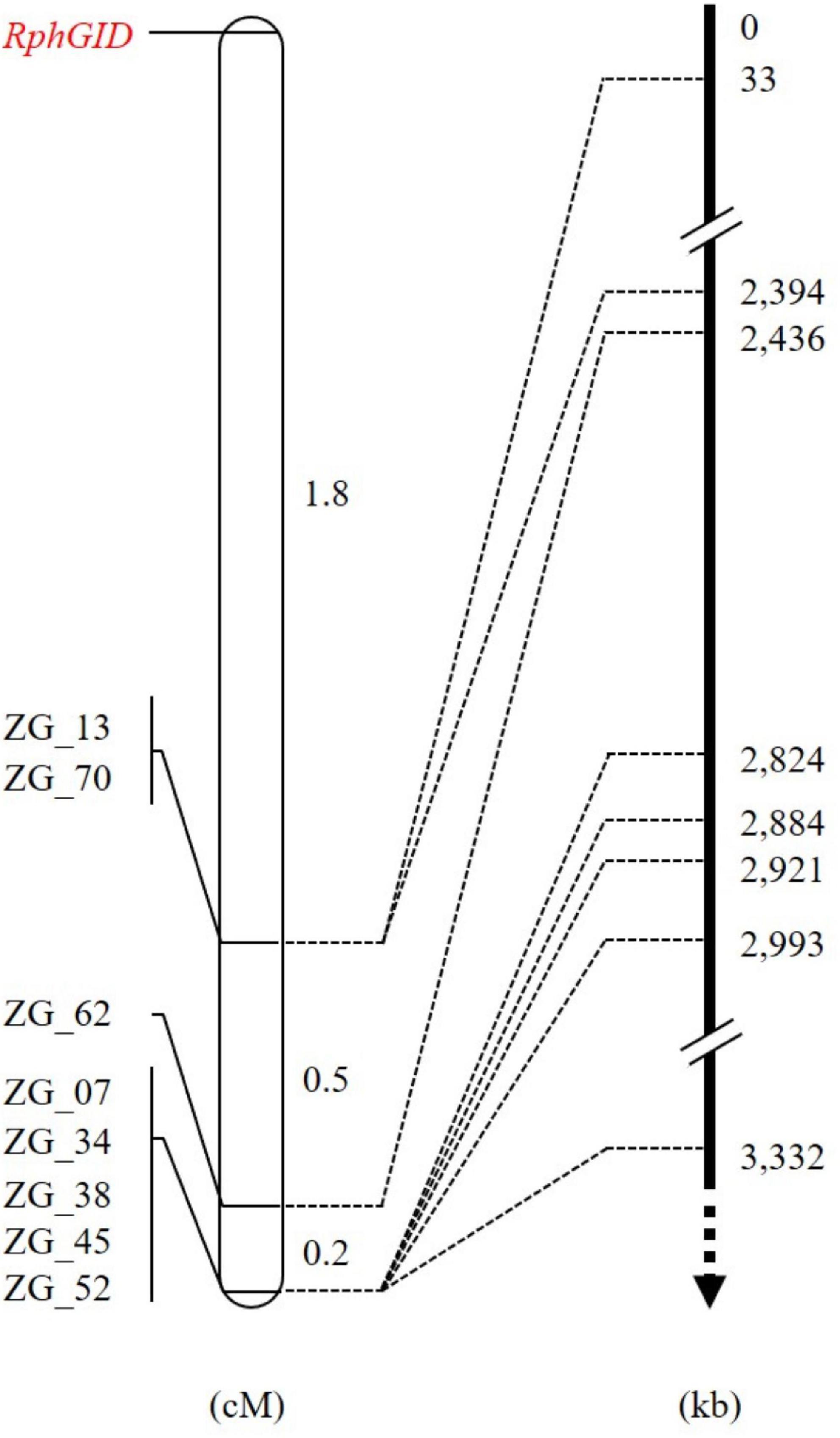
Figure 4. The detailed map of the RphGID locus. ZG_13 is the closest marker to the RphGID locus. The arrow is toward the centromere of chromosome 3H.
Pt. 5477 P−
The response of POP4 and 411 F3 families derived from POP1 to P. hordei pathotype 5477 P− (Tables 2, 3) revealed the involvement of two genes interacting with each other in a complementary manner. Fifty-six lines showing either NSR or NSS responses to pt. 5477 P− were selected for QTL analysis. The analysis detected two major QTLs (on chromosomes 3HS and 7HL) involved in the resistance of GID to pt. 5477 P− (Figure 5). The position of linked DArT markers aligned with RphGID (3HS) and Rph3 (7HL). Furthermore, the homozygous form of the Rph3 gene was detected in all 26 F2 plants whose F3 families showed NSR responses to pt. 5477 P−. These results demonstrated the presence of the Rph3 gene in GID and illustrated the involvement of this gene in the observed complementary interactions.
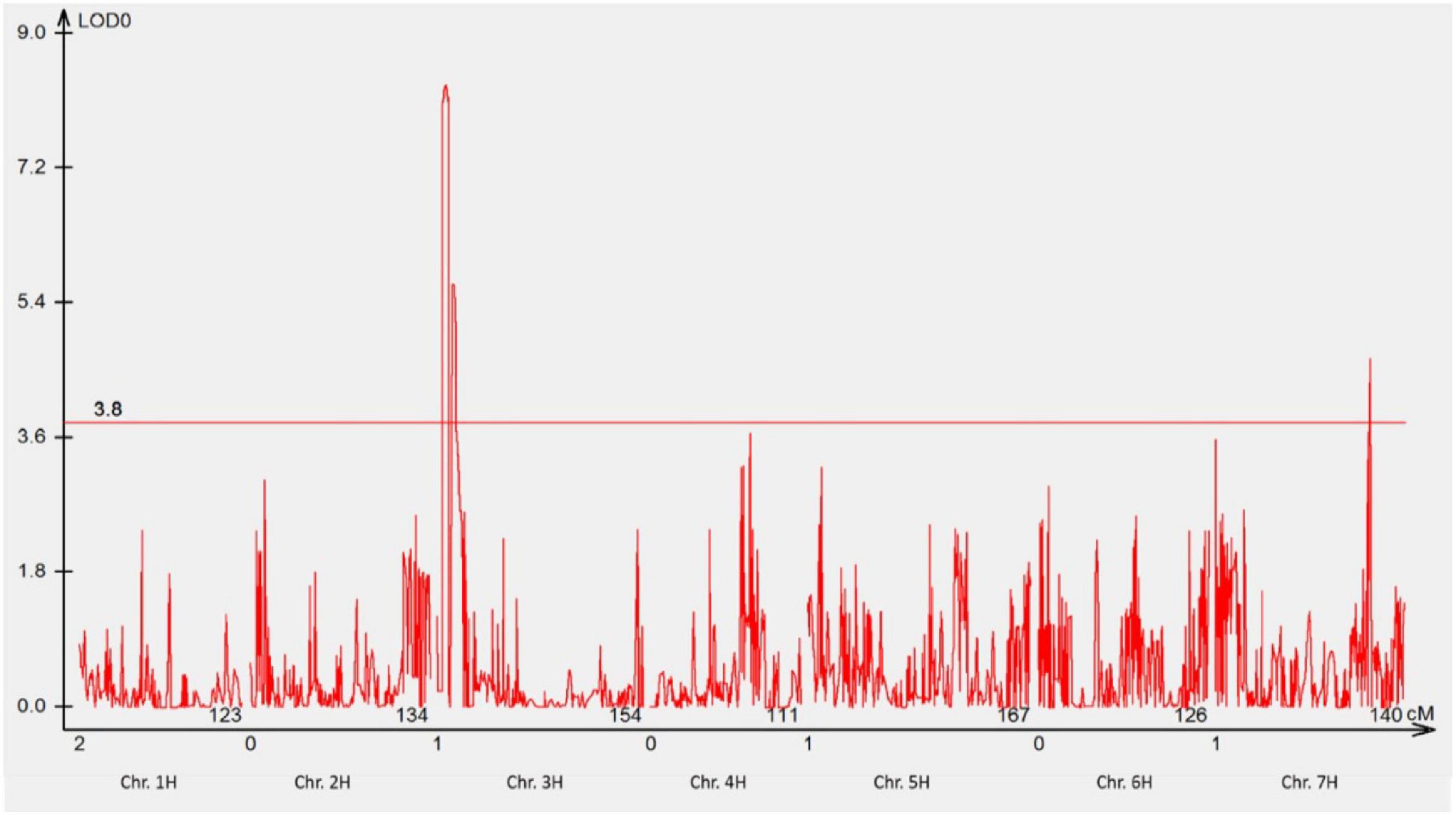
Figure 5. Manhattan plot showing the most significant QTL conferring resistance to pathotype 5477 P– using genotypic data of DArTseq silico markers. Two QTLs were detected, one QTL was located on the distal region of the short arm of chromosome 3H, and another one was located on the long arm of chromosome 7H. The threshold value was set at 3.8.
Gene annotation for the RphGID locus
Most of the 33-kb delimited physical window sequence in the reference genome (cv. Morex) consisted of repetitive elements, and the FGENESH prediction tool showed five putative genes designated as GID_ORF1 to GID_ORF5 (Table 4). Among them, GID_ORF1 encodes an unknown function protein without any conserved domains. Two conserved domains found in protein GID_ORF2, DEAD-like helicase and Helitron-like, were reported to be involved in ATP-dependent RNA or DNA unwinding. The MDN1 domain of GID_ORF3 was involved in ribosome maturation. The KOW motif of GID_ORF4 was known as an RNA-binding domain shared by some ribosomal proteins. The protein GID_ORF5 contains two conserved domains, namely RNase_HI_RT_Ty1 and RVT_2. The genomic DNA sequences of these five putative genes have been deposited in NCBI with the accession numbers OP021633, OP021634, OP021635, OP021636, and OP021637, respectively.
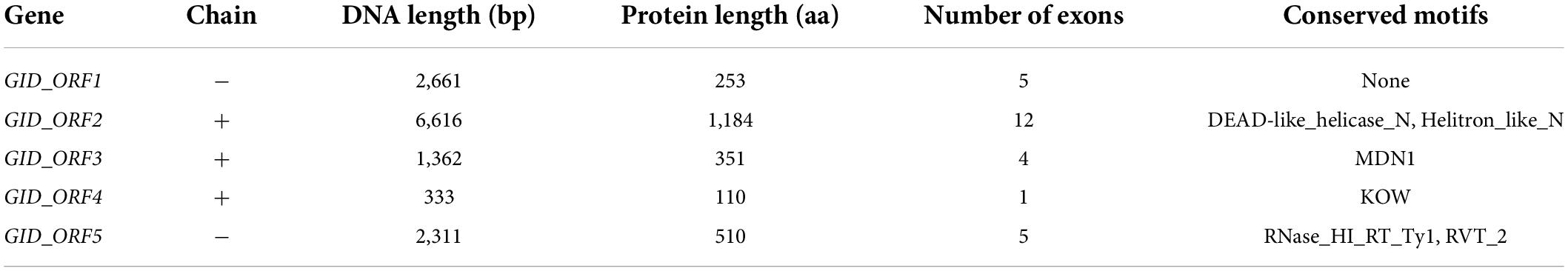
Table 4. Five putative genes were identified within the first 33 kb sequence of barley cv. Morex on the short arm of chromosome 3H.
Discussion
This study combined multi-pathotype rust tests, marker genotyping, genetic analysis, and marker-trait associations to characterize leaf rust resistance in the ICARDA barley breeding line GID 5779743. The line was found to be seedling resistant to the most common pathotypes of P. hordei in Australia. Genotyping with closely linked markers demonstrated the presence of Rph3 and the absence of Rph7 and Rph15 in this line. The genetic inheritance of resistance in four populations derived from a cross of GID/Gus revealed the involvement of two resistance genes. QTL analysis revealed that one of these two genes is Rph3, and the second (RphGID) is located in the extreme telomeric region of the short arm of chromosome 3H in the same genomic region as previously reported for Rph5 (Mammadov et al., 2003). The low infection type produced by GID in response to pathotype 5457 P+ (avirulent on Rph5) was very similar to the typical low infection type (;+N) reported for gene Rph5 (Park et al., 2015). The infection type patterns with multiple pathotypes, QTL analysis, and the high-resolution map of RphGID strongly suggested that RphGID is Rph5.
Interestingly, GID maintained its resistance when challenged with pathotype 5477 P− virulent on Rph3 and Rph5. The genetic analysis of the F2 population and F3 families based on tests with this pathotype showed the involvement of two complementary genes mapped to the Rph3 (7HL) and a locus on 3HS that can be Rph5 or closely linked to Rph5. In addition, the detection of the Rph3 gene in homozygous form in all 26 F2 plants whose F3 families showed no segregation responses to pt. 5477 P− demonstrated that the presence of Rph3 in GID may be playing a role in conferring the resistance of GID to this pathotype. It is possible that Rph3 interacts with another resistance gene, possibly Rph5 (or another locus in the same genomic region) in a complementary fashion to produce an incompatible resistance response.
Combinations of some defeated genes can provide resistance to pathotypes with matching individual virulences, and these residual effects have been reported in multiple crops against different pathogens (Nass et al., 1981; Royer et al., 1984; Brodny et al., 1986; Li et al., 1999). In wheat, the interaction between Yr73 and Yr74 in a complementary fashion confers resistance to Puccinia striiformis f. sp. tritici (Dracatos et al., 2016), while complementary action between Lr27 and Lr31 provided resistance to Puccinia triticina in wheat (Singh et al., 1999). The complementary action conferring APR to P. hordei was also reported on the barley cv. Mecknes Maroc (Elmansour et al., 2017). Very recently, Singh et al. (2021) reported on the interaction of an APR (Rph20) and an ASR (Rph5.e) gene conferring resistance to P. hordei virulent pathotype. In soybean, the resistance of accession JS 95-60 to the fungus Colletotrichum truncatum was also regulated by the interactions of two major genes in complementary fashion (Nataraj et al., 2020). In the current study, we hypothesized that two defeated genes (Rph3 and RphGID/Rph5) interact in a complementary fashion to produce resistance against a pathotype virulent on each gene individually. Allelic studies are recommended to further resolve the genetic basis of resistance in GID.
Most plant genes conferring resistance to pathogens encode proteins of the NLR family (Dodds and Rathjen, 2010; Zipfel, 2014). None of five putative genes identified by the FGENESH tool within 33-kb of the RphGID locus encodes a protein of the NLR family, and none were reported to be associated with the resistance to any plant diseases. Notably, the putative genes were identified based on the genome of barley cv. Morex, while Morex is susceptible to P. hordei pathotype 5457 P+, the RphGID locus may not exist in this accession. Many disease resistance genes isolated so far such as the genes conferring resistance to Xanthomonas bacteria in rice and pepper (Yoshimura et al., 1998; Wu et al., 2008; Wang et al., 2015, 2018), and especially the barley leaf rust resistance gene Rph3 (Dinh et al., 2022) encode other types of protein without any conserved domains. In our study, we were not able to pinpoint whether all the identified genes in ORF region play role in providing resistance against all the six pathotypes used in this study and therefore further studies on validation of these genes is highly recommended.
In conclusion, the current study demonstrates that the barley line GID carries two independent loci, Rph3 and RphGID. This study also illustrated that the interactions between Rph3 and RphGID in a complementary manner confers the resistance to the P. hordei pathotype 5477 P−, which is virulent on Rph3 when this gene is singly deployed. Based on the responses to various P. hordei pathotypes and the detailed genetic map, we suggest that RphGID can be Rph5 or an allele of this gene. A test of allelism is required to validate this hypothesis. Besides, more efforts are needed to fine-map toward identifying the genetic sequences behind the resistance to P. hordei in the GID 5779743 line. The interactions between defeated genes to confer resistance to pathogens is also a prospect due to the limited number of identified resistance genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
DS and RP conceived the project, designed multi-pathotype tests, oversaw all rust phenotyping, and performed the gene postulation. DS provided the materials. RP provided all rust isolates and information on pathogenicities. HD performed multi-pathotype tests, genotyped the materials, and performed the QTL analysis. HD and MP designed the markers, analyzed the genotypic data, and constructed the high-resolution map. HD and DS wrote the manuscript. All authors reviewed the manuscript and approved the submitted version.
Acknowledgments
Seeds of line GID 5779743 provided by the Australian Grains Genebank, Horsham, Australia and International Center for Agricultural Research in the Dry Areas (ICARDA) is gratefully acknowledged. We are thankful to Peter Dracatos for providing Rph7-linked molecular markers, C. Chen for helping us to test the presence/absence of Rph15, M. Williams for technical support with plant growth facilities, and Australian Awards Scholarship for providing financial support to HD.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.988322/full#supplementary-material
Footnotes
References
Brodny, U., Nelson, R., and Gregory, L. (1986). Residual and interactive expressions of “Defeated” wheat stem rust resistance genes. Phytopathology 76, 546–549. doi: 10.1094/Phyto-76-546
Brooks, W. S., Griffey, C., Steffenson, B., and Vivar, H. (2000). Genes governing resistance to Puccinia hordei in thirteen spring barley accessions. Phytopathology 90, 1131–1136. doi: 10.1094/PHYTO.2000.90.10.1131
Chen, C., Jost, M., Clark, B., Martin, M., Matny, O., Steffenson, B. J., et al. (2021). BED domain-containing NLR from wild barley confers resistance to leaf rust. Plant Biotechnol. J. 19, 1206–1215. doi: 10.1111/pbi.13542
Clifford, B. (1985). “Barley leaf rust,” in The cereal rusts Vol. II: Diseases, distribution, epidemiology and control’, eds A. P. Roelfs and W. R. Bushnell (Amsterdam: Elsevier), 173–205. doi: 10.1016/B978-0-12-148402-6.50014-6
Cotterill, P., Rees, R., Platz, G., and Dill-Macky, R. (1992). Effects of leaf rust on selected Australian barleys. Aust. J. Exp. Agric. 32, 747–751. doi: 10.1071/EA9920747
Dinh, H. X., Singh, D., Gomez de la Cruz, D., Hensel, G., Kumlehn, J., Mascher, M., et al. (2022). The barley leaf rust resistance gene Rph3 encodes a predicted membrane protein and is induced upon infection by avirulent pathotypes of Puccinia hordei. Nat. Commun. 13, 1–13. doi: 10.1038/s41467-022-29840-1
Dinh, H. X., Singh, D., Periyannan, S., Park, R. F., and Pourkheirandish, M. (2020). Molecular genetics of leaf rust resistance in wheat and barley. Theor. Appl. Genet. 133, 2035–2050. doi: 10.1007/s00122-020-03570-8
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dracatos, P. M., Barto<, J., Elmansour, H., Singh, D., Karafiátová, M., Zhang, P., et al. (2019). The coiled-coil NLR Rph1, confers leaf rust resistance in barley cultivar Sudan. Plant physiol. 179, 1362–1372. doi: 10.1104/pp.18.01052
Dracatos, P. M., Zhang, P., Park, R. F., McIntosh, R. A., and Wellings, C. R. (2016). Complementary resistance genes in wheat selection ‘Avocet R’confer resistance to stripe rust. Theor. Appl. Genet. 129, 65–76. doi: 10.1007/s00122-015-2609-7
Elmansour, H., Singh, D., Dracatos, P. M., and Park, R. F. (2017). Identification and characterization of seedling and adult plant resistance to Puccinia hordei in selected African barley germplasm. Euphytica 213:119. doi: 10.1007/s10681-017-1902-8
Fukuoka, S., Saka, N., Mizukami, Y., Koga, H., Yamanouchi, U., Yoshioka, Y., et al. (2015). Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 5, 1–7. doi: 10.1038/srep07773
Gilmour, J. (1973). Octal notation for designating physiologic races of plant pathogens. Nature 242, 620. doi: 10.1038/242620a0
Golegaonkar, P. G., Singh, D., and Park, R. F. (2009). Evaluation of seedling and adult plant resistance to Puccinia hordei in barley. Euphytica 166, 183–197. doi: 10.1007/s10681-008-9814-2
Griffey, C., Das, M., Baldwin, R., and Waldenmaier, C. (1994). Yield losses in winter barley resulting from a new race of Puccinia hordei in North America. Plant Dis. (USA) 78, 256–260. doi: 10.1094/PD-78-0256
Kavanagh, P. J., Singh, D., Bansal, U. K., and Park, R. F. (2017). Inheritance and characterization of the new and rare gene Rph25 conferring seedling resistance in Hordeum vulgare against Puccinia hordei. Plant Breed. 136, 908–912. doi: 10.1111/pbr.12535
Kloppers, F., and Pretorius, Z. (1997). Effects of combinations amongst genes Lr13, Lr34 and Lr37 on components of resistance in wheat to leaf rust. Plant Pathol. 46, 737–750. doi: 10.1046/j.1365-3059.1997.d01-58.x
Kosambi, D. D. (2008). “Kosambi’s function,” in Encyclopedia of genetics, genomics, proteomics and informatics, ed. G. P. Rédei (Dordrecht: Springer), 1066–1066.
Li, Z.-K., Luo, L., Mei, H., Paterson, A., Zhao, X., Zhong, D., et al. (1999). A “defeated” rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol. Gen. Genet. 261, 58–63. doi: 10.1007/s004380050941
Mammadov, J., Zwonitzer, J., Biyashev, R., Griffey, C., Jin, Y., Steffenson, B., et al. (2003). Molecular mapping of leaf rust resistance gene Rph5 in barley. Crop Sci. 43, 388–393. doi: 10.1094/PHYTO.2003.93.5.604
Mehnaz, M., Dracatos, P., Pham, A., March, T., Maurer, A., Pillen, K., et al. (2021). Discovery and fine mapping of Rph28: A new gene conferring resistance to Puccinia hordei from wild barley. Theor. Appl. Genet. 134, 2167–2179. doi: 10.1007/s00122-021-03814-1
Mundt, C. C. (2018). Pyramiding for resistance durability: Theory and practice. Phytopathology 108, 792–802. doi: 10.1094/PHYTO-12-17-0426-RVW
Nass, H., Pedersen, W., MacKenzie, D., and Nelson, R. (1981). The residual effects of some “Defeated” powdery mildew resistance genes. Phytopathology 71, 1315–1318. doi: 10.1094/PHYTO.1999.89.7.533
Nataraj, V., Maranna, S., Kumawat, G., Gupta, S., Rajput, L. S., Kumar, S., et al. (2020). Genetic inheritance and identification of germplasm sources for anthracnose resistance in soybean [Glycine max (L.) Merr.]. Genet. Resour. Crop Evol. 67, 1449–1456. doi: 10.1007/s10722-020-00917-4
Park, R. F. (2003). Pathogenic specialization and pathotype distribution of Puccinia hordei in Australia, 1992 to 2001. Plant Dis. 87, 1311–1316. doi: 10.1094/PDIS.2003.87.11.1311
Park, R. F., Golegaonkar, P. G., Derevnina, L., Sandhu, K. S., Karaoglu, H., Elmansour, H. M., et al. (2015). Leaf rust of cultivated barley: Pathology and control. Annu. Rev. Phytopathol. 53, 565–589. doi: 10.1146/annurev-phyto-080614-120324
Royer, M., Nelson, R., MacKenzie, D., and Diehle, D. (1984). Partial resistance of near-isogenic wheat lines compatible with Erysiphe graminis f. sp. tritici. Phytopathology 74, 1001–1006. doi: 10.1094/Phyto-74-1001
Singh, D., Park, R. F., and McIntosh, R. (1999). Genetic relationship between the adult plant resistance gene Lr12 and the complementary gene Lr31 for seedling resistance to leaf rust in common wheat. Plant Pathol. 48, 567–573. doi: 10.1046/j.1365-3059.1999.00391.x
Singh, L., Park, R. F., Dracatos, P., Ziems, L., and Singh, D. (2021). Understanding the expression and interaction of Rph genes conferring seedling and adult plant resistance to Puccinia hordei in barley. Can. J. Plant Pathol. 43(suppl. 2), S218–S226. doi: 10.1080/07060661.2021.1936649
Steffenson, B., Jin, Y., and Griffey, C. (1993). Pathotypes of Puccinia hordei with virulence for the barley leaf rust resistance gene Rph7 in the United States. Plant Dis. 77, 867–869. doi: 10.1094/PDIS-08-12-0785-PDN
Voorrips, R. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Heredity 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2015). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 8, 290–302. doi: 10.1016/j.molp.2014.10.010
Wang, J., Zeng, X., Tian, D., Yang, X., Wang, L., and Yin, Z. (2018). The pepper Bs4C proteins are localized to the endoplasmic reticulum (ER) membrane and confer disease resistance to bacterial blight in transgenic rice. Mol. Plant Pathol. 19, 2025–2035. doi: 10.1111/mpp.12684
Wang, Y., Subedi, S., de Vries, H., Doornenbal, P., Vels, A., Hensel, G., et al. (2019). Orthologous receptor kinases quantitatively affect the host status of barley to leaf rust fungi. Nat. Plants 5, 1129–1135. doi: 10.1038/s41477-019-0545-2
Wu, L., Goh, M. L., Sreekala, C., and Yin, Z. (2008). XA27 depends on an amino-terminal signal-anchor-like sequence to localize to the apoplast for resistance to Xanthomonas oryzae pv oryzae. Plant physiol. 148, 1497–1509. doi: 10.1104/pp.108.123356
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. U.S.A. 95, 1663–1668. doi: 10.1073/pnas.95.4.1663
Keywords: barley (Hordeum vulgare), Puccinia hordei, leaf rust, complementary genes, interaction, disease resistance, mapping, QTL
Citation: Dinh HX, Pourkheirandish M, Park RF and Singh D (2022) The genetic basis and interaction of genes conferring resistance to Puccinia hordei in an ICARDA barley breeding line GID 5779743. Front. Plant Sci. 13:988322. doi: 10.3389/fpls.2022.988322
Received: 07 July 2022; Accepted: 27 July 2022;
Published: 16 August 2022.
Edited by:
Satish Kumar, Indian Council of Agricultural Research (ICAR), IndiaReviewed by:
Sripada M. Udupa, International Center for Agricultural Research in the Dry Areas (ICARDA), MoroccoPrem Lal Kashyap, Indian Institute of Wheat and Barley Research (ICAR), India
Copyright © 2022 Dinh, Pourkheirandish, Park and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Davinder Singh, ZGF2aW5kZXIuc2luZ2hAc3lkbmV5LmVkdS5hdQ==; orcid.org/0000-0003-1411-9291
 Hoan X. Dinh
Hoan X. Dinh Mohammad Pourkheirandish
Mohammad Pourkheirandish Robert F. Park
Robert F. Park Davinder Singh
Davinder Singh