- 1All-Russia Research Institute of Agricultural Biotechnology, Kurchatov Genomics Centre – ARRIAB, Moscow, Russia
- 2N.I.Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow, Russia
- 3Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
- 4All-Russian Institute of Plant Genetic Resources (VIR), Department of Wheat Genetic Resources, St. Petersburg, Russia
- 5Agricultural Genetics Institute, Department of Molecular Biology, Hanoi, Vietnam
Aegilops crassa Boiss. is polyploid grass species that grows in the eastern part of the Fertile Crescent, Afghanistan, and Middle Asia. It consists of tetraploid (4x) and hexaploid (6x) cytotypes (2n = 4x = 28, D1D1XcrXcr and 2n = 6x = 42, D1D1XcrXcrD2D2, respectively) that are similar morphologically. Although many Aegilops species were used in wheat breeding, the genetic potential of Ae. crassa has not yet been exploited due to its uncertain origin and significant genome modifications. Tetraploid Ae. crassa is thought to be the oldest polyploid Aegilops species, the subgenomes of which still retain some features of its ancient diploid progenitors. The D1 and D2 subgenomes of Ae. crassa were contributed by Aegilopstauschii (2n = 2x = 14, DD), while the Xcr subgenome donor is still unknown. Owing to its ancient origin, Ae. crassa can serve as model for studying genome evolution. Despite this, Ae. crassa is poorly studied genetically and no genome sequences were available for this species. We performed low-coverage genome sequencing of 4x and 6x cytotypes of Ae. crassa, and four Ae. tauschii accessions belonging to different subspecies; diploid wheatgrass Thinopyrum bessarabicum (Jb genome), which is phylogenetically close to D (sub)genome species, was taken as an outgroup. Subsequent data analysis using the pipeline RepeatExplorer2 allowed us to characterize the repeatomes of these species and identify several satellite sequences. Some of these sequences are novel, while others are found to be homologous to already known satellite sequences of Triticeae species. The copy number of satellite repeats in genomes of different species and their subgenome (D1 or Xcr) affinity in Ae. crassa were assessed by means of comparative bioinformatic analysis combined with quantitative PCR (qPCR). Fluorescence in situ hybridization (FISH) was performed to map newly identified satellite repeats on chromosomes of common wheat, Triticum aestivum, 4x and 6x Ae. crassa, Ae. tauschii, and Th. bessarabicum. The new FISH markers can be used in phylogenetic analyses of the Triticeae for chromosome identification and the assessment of their subgenome affinities and for evaluation of genome/chromosome constitution of wide hybrids or polyploid species.
Introduction
The genus Aegilops L. is closely related to wheat and represents an important gene pool for wheat improvement (Molnár-Láng et al., 2014; Kishii, 2019). Modern taxonomy recognizes 10 diploid and 11 polyploid Aegilops species with various genome compositions (Van Slageren, 1994; Kilian et al., 2011). Six major genomic types – S* (Sitopsis section), U (Ae. umbellulata), C (Ae. markgraphii), M (Ae. comosa), N (Ae. uniaristata), and D (Ae. tauschii) - have been distinguished among diploid Aegilops species (Kimber and Tsunewaki, 1988); they are thought to have occurred approximately 3 million years ago from hybrid populations of the progenitor of Ae. speltoides (S genome) × ancient diploid wheat (A genome) via the mechanism of homoploid hybrid speciation (Marcussen et al., 2014). Polyploid Aegilops emerged from the hybridization of diploid progenitors carrying different genomic types. Despite a broad genome diversity of polyploid Aegilops, the D (sub)genome was detected in four, whereas the U in seven species. Depending on the presence of the D or U genome, which are designated as “pivotal,” all polyploid Aegilops species are divided into the D genome cluster and U genome cluster (Kimber and Feldman, 1987).
Ae. crassa Boiss. is a polyploid grass species belonging to the D genome cluster that naturally grows in the eastern part of the Fertile Crescent, from Turkey on the west to Afghanistan and Middle Asia on the east (Kihara, 1957; Van Slageren, 1994; Kilian et al., 2011). Ae. crassa consists of tetraploid and hexaploid cytotypes [2n = 4x = 28, D1D1XcrXcr and 2n = 6x = 42, D1D1XcrXcrD2D2 respectively] that are similar morphologically. Based on analysis of variation of repeated DNA sequences, Dubkovsky and Dvořák (1995) suggested that 4x Ae. crassa is evolutionary an old species, the subgenomes of which were substantially modified during speciation (Kihara, 1949; Kimber and Zhao, 1983; Rayburn and Gill, 1987; Zhang and Dvořák, 1992; Dubkovsky and Dvořák, 1995; Badaeva et al., 1998, 2002, 2021; Dvořák, 1998; Naghavi et al., 2009; Edet et al., 2018). From another side, it may retain some genomic features of ancient diploid ancestors. One of the Ae. crassa subgenomes, designated D1, is related to the D genome of Ae. tauschii (2n = 2x = 14, DD; Kihara, 1949; Kimber and Zhao, 1983; Zhang and Dvořák, 1992; Badaeva et al., 1998, 2002; Edet et al., 2018), which also served as the cytoplasmic genome donor to this tetraploid species (Terachi et al., 1987; Kimber and Tsunewaki, 1988). The D subgenome is also present in two polyploid Aegilops species and in hexaploid bread wheat Triticum aestivum (2n = 6x = BBAADD). According to molecular analysis of nuclear genome (Dvořák et al., 1998; Luo et al., 2017; Singh et al., 2019) and hybridization pattern of pAs1 probe (Badaeva et al., 1996, 2019a; Zhao et al., 2018; Ebrahimzadegan et al., 2021), the D subgenome of polyploid wheat was inherited from Ae. tauschii subsp. strangulata, while Ae. tauschii subsp. tauschii contributed the D subgenome to polyploid Aegilops species Ae. cylindrica and 6x Ae. crassa (Badaeva et al., 2002). At the same time, the D1 subgenome of Ae. crassa has probably derived from an ancient Ae. tauschii and is distinct from both extant Ae. tauschii subspecies in the distribution of repetitive DNA probes (Rayburn and Gill, 1987; Badaeva et al., 1998, 2002, 2021; Abdolmalaki et al., 2019).
An extinct or still unknown diploid species contributed the second subgenome to Ae. crassa (Dubkovsky and Dvořák, 1995; Dvořák, 1998; Edet et al., 2018). Kihara (1963) proposed that this subgenome could be inherited from Ae. comosa and suggested genomic formula DM for tetraploid and DDM for hexaploid Ae. crassa. Molecular (Zhang and Dvořák, 1992) and meiotic (Kimber and Abu-Bakar, 1981) analyses did not confirm the presence of the M genome in Ae. crassa. Comparison of the restriction profiles of nuclear repeated nucleotide sequences, RNS (Zhang and Dvořák, 1992; Dubkovsky and Dvořák, 1995; Dvořák, 1998), and DArTseq genotyping (Edet et al., 2018) revealed higher similarity of the Xcr subgenome with the S genome of the Sitopsis group, most likely Ae. speltoides, or with the T genome of Ae. mutica (Edet et al., 2018). These observations contradict the result of cytogenetic analysis, which showed correspondence to 5S and 45S rDNA patterns of the Xcr subgenome chromosomes of Ae. crassa and the S* genome chromosomes of the diploid Emarginata species, but not with Ae. speltoides (Badaeva et al., 2002, 2021). Owing to the uncertain origin of the second Ae. crassa subgenome, Dvořák (1998) proposed the genomic formula DXcr for 4x and DDXcr for 6x cytotype.
RFLP analysis showed that evolution of Ae. crassa was associated with significant changes in the fraction of repetitive DNA sequences, in particular, five unique fragments emerged on the restriction profiles of Ae. crassa and its hexaploid derivatives (Zhang and Dvořák, 1992; Dubkovsky and Dvořák, 1995; Dvořák, 1998). Substantial modifications of Ae. crassa subgenomes compared to its putative diploid progenitors were also revealed using chromosome pairing analysis of intraspecific hybrids (Kihara, 1954, 1963; Zhao and Kimber, 1984), C-banding (Badaeva et al., 1998, 2002), and fluorescence in situ hybridization (FISH; Rayburn and Gill, 1987; Badaeva et al., 1998, 2002, 2021; Abdolmalaki et al., 2019).
Despite the significant progress made in genome sequencing, the number of DNA probes employed in FISH analysis of cereal species is still very limited. In addition to pSc119.2 and pAs1 probes that have traditionally been used for chromosome identification and phylogenetic studies of the Triticeae since the middle 80th (Bedbrook et al., 1980; Rayburn and Gill, 1986), several new DNA sequences have been isolated from nuclear DNA and used as probes in FISH analysis of wheat and Aegilops as well as of other grass species (Kato et al., 2004, 2011; Komuro et al., 2013; Chen et al., 2019; Xi et al., 2019).
Progress in whole-genome sequencing and bioinformatics pipelines allows obtaining detailed information on the structure of the repeatome. To date, genome assemblies of Ae. tauschii, Ae. longissima, Ae. speltoides, Ae. sharonensis, and Ae. bicornis have been obtained (Tiwari et al., 2015; Wang et al., 2021; Avni et al., 2022; Li et al., 2022; Yu et al., 2022). However, the genome of Ae. crassa has not yet been sequenced, which certainly limits the possibilities of its comprehensive study. From the other side, even unassembled reads can be used to search for new tandem satellite repeats from which chromosomal markers can be developed (Koo et al., 2016; Du et al., 2017; Liu et al., 2018b; Chen et al., 2019; Kroupin et al., 2019b; Nikitina et al., 2020). In particular, comparative genome analysis has been successfully used to obtain specific chromosomal markers for the detection of alien chromosomes in wheat addition lines (Li et al., 2016; Liu et al., 2018a) and for identification of Y subgenome in Roegneria (Wu et al., 2021). Koo et al. (2016) mapped satellite repeats identified by flow-sorting and sequencing to chromosome 5Mg of Ae. geniculata. However, in most studies of structural genomic diversity of Aegilops, a limited set of “standard” DNA probes based on tandem repeats pTa71, pTa794, pSc119.2, pAs1, and pTa-713, as well as a number of microsatellites, are still being used (Song et al., 2020; Said et al., 2021). Single-gene FISH probes are also employed to compare structural rearrangements of chromosomes in Aegilops species (Danilova et al., 2014, 2017; Tiwari et al., 2015; Said et al., 2021). Despite the informativity of the results obtained using single-gene FISH probes, both flow-sorting and creation of cDNA clones remain cost-and labor-consuming procedures. Owing to modern bioinformatics approaches, tandem repeats can be efficiently selected from even shallow whole-genome sequencing data, and new FISH probes can be obtained by either direct labeling of PCR products or by designing labeled oligonucleotides, which facilitates their transfer between different scientific teams (Kroupin et al., 2019a; Lang et al., 2019a; Xi et al., 2020).
In addition to Ae. crassa, the D subgenome is present in hexaploid wheat and several Aegilops species. According to meiotic, cytogenetic, and molecular analyses, the D subgenome of common wheat is not significantly modified relative to the parental (Kimber and Zhao, 1983; Rayburn and Gill, 1987; Dvořák et al., 1998) and therefore can be used for tracing evolutionary changes in the orthologous chromosomes of other Triticeae species. From another side, the wheatgrass (Thinopyrum) species are genetically related to D subgenome species of wheat and Aegilops (Chen et al., 2001; Guo et al., 2016; Bernhardt et al., 2017, 2020). Introgression of useful genes from wheatgrass to wheat usually occurs between J (wheatgrass) and D (wheat) chromosomes, which might be due to higher homology between them rather than with homoeologues of A or B subgenomes of wheat (Liu et al., 2007; He et al., 2009; Patokar et al., 2015). High syntheny between the Jb genome of Th. bessarabicum and common wheat subgenomes has been detected using a combination of cytogenetic (GISH) and molecular (SNP-mapping) analyses of 12 wheat-Th. bessarabicum introgression lines (Grewal et al., 2018), indicating that the divergence of wheat and Th. bessarabicum genomes was not accompanied with large chromosomal rearrangements, but with alterations of the repeated nucleotide sequences. The comparison of copy number variation of transposable elements between polyploid and diploid Triticeae revealed the similarity between Jb genome of Th. bessarabicum and D genome of Ae. tauschii (Divashuk et al., 2019). Therefore, the comparison of Jb and D (sub)genomes in the abundance and chromosomal localization of repeated DNA elements can be informative not only for repeatome and evolutionary studies, but also may have practical application as a source for increasing genetic diversity of wheat.
The aim of our study was to trace evolutionary changes of Ae. crassa subgenomes (with a special emphasis on the D subgenome) based on a complex approach, which includes low-coverage-sequencing followed by identification of repetitive DNA families using bioinformatics, quantitative assessment of repeats using qPCR, and their physical mapping on chromosomes of Ae. crassa (4x and 6x) in comparison with diploid Ae. tauschii (DD), hexaploid common wheat (BBAADD), and diploid wheatgrass Th. bessarabicum (JbJb) as an outgroup.
Materials and methods
Plant material
The following materials have been used (Table 1). The images of heads, vegetating plants, and spikelets of Ae. crassa accession K-2485 (4x) and IG 131680 (6x) are shown in Supplementary Figure S1.
Sequencing
The fresh young leaves of growing plants were ground in liquid nitrogen, then genomic DNA was extracted using the CTAB protocol (Rogers and Bendich, 1985) and used for whole-genome sequencing, qPCR, and probe preparation for FISH. The quantity and quality of the extracted DNA were checked using a NanoDrop OneC spectrophotometer (Thermo Fisher Scientific) and by electrophoresis in 0.8% agarose gel, respectively. Only genomic DNA samples with OD260/280 value ranging from 1.8 to 2.0 and OD260/230 value ranging from 2.0 to 2.2 were considered as good quality. DNA concentration was measured on a Qubit 4 instrument using Qubit™ dsDNA HS and BR Assay Kits (Thermo Fisher Scientific, Waltham, MA, United States). The shotgun libraries were synthesized using the Swift 2S® Turbo DNA Library Kit (Swift Bioscience, Ann Arbor, MI, USA) according to the manufacturer’s protocol. A test run to check the quality of the libraries was carried out on the MiSeq instrument on MiSeq Reagent Nano Kit v2 (300-cycles). Next, the libraries went through the conversion step and were sequenced on DNBSEQ-G400 on 1 lane. The initial amount of DNA was 25 ng, with the length of fragments around 350 bp and pair-end indexing on Swift 2S Turbo Unique Dual Indexing Kit. The run was performed on the Illumina NextSeq with NextSeq 500/550 Mid Output Kit v2.5 (300 cycles) as described in Illumina protocols for pair-end reads. The length of read was 151 bp, the length of index - 8 bp. The sequencing was performed in Genomed, Ltd. (Moscow, Russia).
Reads preparation and repeat assembly
Adapter sequence and low-quality reads were removed by bbduk.sh from BBMap package (v38.90, github.com/BioInfoTools/BBMap; Bushnell, 2014) at given parameters: ktrim = r k = 20 mink = 10 hdist = 2 maxns = 0 ftl = 19 ftr = 139 minlen = 100 with default bbmap adapters references. After that, they were additionally trimmed from the 3′-end at interleaved = t ftr = 99 for all the reads prepared for assembly to be the same fixed length. We have controlled the quality of resulting reads by using FastQC (v0.11.5, github.com/s-andrews/FastQC; Andrews, 2010) and sampled 2,000,000 paired interlaced reads from them. Interlaced reads were then forwarded to RepeatExplorer2 (v0.3.8., bitbucket.org/petrnovak/repex_tarean; Novák et al., 2013). RepeatExplorer2 output was parsed using custom scripts (github.com/Stathmin/Scripts-for-RepeatExplorer2-parsing).
Repeats alignment and identification
Global alignment is not sensitive enough when applied to repeat consensus monomers because of arbitrary selection of their starts. We took another approach, so each repeat consensus was tripled and aligned with consensuses database by blastn from BLAST+ package (v2.9.0, ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.9.0; Camacho et al., 2009) with –task dc-megablast. All the high-scoring pairs (HSPs) for each aligned pair were analyzed, and total coverages without overlaps of longer sequences by HSPs of shorter ones were calculated with our custom script. We assumed the two repeat consensuses to be related if they produced alignments with e-value <0.05 and their total coverage without overlaps ≥80%. To identify previously known repeats the NCBI Nucleotide database (Coordinators, 2012) was used, and all the repeat-related sequences of Triticeae were downloaded on Oct 18 2021. Each tandem repeat was designated as follows: AC4x_CL##_###nt for 4x Ae. crassa, AC6x_CL##_###nt for 6x Ae. crassa, ATs_CL##_###nt for Ae.tauschii subsp. strangulata and ATt_CL##_###nt for subsp. typica, TB_CL##_###nt for Th. bessarabicum, where CL## is the numerical name of a particular repeat cluster and ###nt is the length of its single monomer. For convenience, we used only CL## designations in subsequent qPCR and FISH experiments. The found repeats were submitted to NCBI GenBank system and acquired the IDs ON872662-ON872692.
Diversity of identified satellites
Acquired repeat consensuses shorter than 200 bp were multiplied until they reached length ≥ 200 bp, which is required for informative mapping with 100 bp paired reads. Samples of 5,000,000 paired reads from each analyzed object, trimmed as described above, were then mapped on acquired repeat consensuses using gsnap (version 2021–12-17, github.com/juliangehring/GMAP-GSNAP; Wu and Nacu, 2010) with flags –max-mismatches 0.2 –min-coverage 40. Produced *.bam mappings were visualized using Jbrowse v1.6.9 (jbrowse.org/jb2/; Buels et al., 2016).
Repeatome structure comparison
RepeatExplorer2 outputs containing proportions of reads by repeat type were parsed. For all the repeat type categories the cumulative proportions, i.e., including subcategories, were summed up and compared between species. For each category the average proportions, standard deviations, and coefficients of variation were obtained.
In silico identification of Xcr subgenome-specific tandem repeats in 4x Aegilops. crassa
Raw reads of 4x Ae. crassa were cleaned up from adapter sequences, and reads were truncated till 100 bp from 3′-end using bbduk from BBTools v38.93 toolkit (sourceforge.net/projects/bbmap/). Trimmed reads were mapped on Ae. tauschii Aet v4.0 genome assembly (Luo et al., 2017) using bwa mem v0.7.17 (github.com/lh3/bwa). For further analysis the reads that perfectly mapped on reference assembly were removed using samtools (Danecek et al., 2021). The resulting 831,000 reads were taken for de novo tandem repeats’ identification using RepeatExplorer2 pipeline (Novák et al., 2017). Novel tandem repeats were identified based on previously obtained consensuses sequences from 4x Ae. crassa using BLAST (Altschul et al., 1990). Primers for identified tandem repeats selected for further estimation of repeat number by qPCR and for probe synthesis for FISH by PCR were designed using Primer3Plus (Untergasser et al., 2012), and their sequences are given in Table 2.
Real-time quantitative PCR
qPCR using primers developed for repeat monomers (Table 2) was performed in triple technical replication with water as negative control and VRN1 as a reference gene (Yaakov et al., 2013) according to the protocol described in Kroupin et al. (2019b). The amplification was performed on a CFX Real-Time PCR Detection System (Bio-Rad) and in Real-Time PCR Mix reaction mixture with Eva Green (Syntol Ltd., Moscow, Russia) according to the manufacturer’s protocol. Primers were synthesized at Syntol Ltd. (Moscow, Russia). The primer concentration was 10 ng/μl, and the DNA concentration was 0.4 ng/μl. The amplification program was as follows: pre-incubation for 10 min at 95°C, then 45 cycles: denaturation for 10 s at 95°C; primer annealing for 30 s at 60°C. The relative quantity (RQ) was calculated using Bio-Rad CFX Manager 3.1 software based on the obtained Ct volumes.
DNA probes for FISH
The following novel repeats identified by means of lowcoverage sequencing followed by bioinformatics analysis were used as FISH probes: (i) derived from 4× Ae. crassa genome: CL3, CL8 (highly homologous to CL16 found in Ae. tauschii subsp. strangulata), CL18, CL60, CL131 (highly homologous to CL149 found in Th. bessarabicum), CL170, CL193, CL209, CL219, CL228, CL232, CL239, CL241, CL244, CL257, CL258, CL261 (highly homologous to CL198 found in Th. bessarabicum); ii) derived from 6x Ae. crassa genome: CL27_232; iii) derived from Th. bessarabicum genome: CL2, CL148. The probes were obtained using PCR with primer sets listed in Table 2. PCR amplification was performed in a 15-μl reaction mixture containing approximately 50 ng genomic DNA, 1.5 μl of 10× PCR buffer, 1.5 mM MgCl2, 0.2 mM of dNTPs, 0.3 μM of each primer, and 0.5 unit of Taq DNA polymerase. The PCR conditions were as follows: an initial denaturation step of 95°C for 5 min, followed by 30 cycles of 94°C for 1 min, annealing at 60°C for 1 min and elongation at 72°C for 1 min with a final extension step at 72°C for 5 min. The obtained amplicons were labeled with either biotin-16-dUTP (CL18, CL60, CL131, CL148, CL170, CL193, CL198, CL219, CL232, CL257, CL258, CL261, CL27_232) or digoxigenin-11-dUTP (CL2, CL3, CL8, CL16, CL18, CL131, CL149, CL170, CL209, CL228, CL232, CL239, CL244) by PCR according to the manufacturer’s instructions (Roche, Germany). Additionally, the probes oligo-pAs1, oligo-pSc119.2, oligo-pTa535 and oligo-pTa71 (Tang et al., 2014), oligo-5S rDNA (Yu et al., 2019), oligo-45 (Tang et al., 2018; Xi et al., 2020), P132 (as homolog to CL241), and P332 derived from Ae. tauschii (Kroupin et al., 2019b), oligo-713 (Tang et al., 2016), and GAAn labeled at the 5′-end with either 6-carboxyfluorescein (FAM; oligo-pAs1, oligo-pSc119.2, oligo-pTa71, oligo-45, P332, P132, and GAAn) or 6-carboxytetramethylrhodamine (TAMRA) or Cy3 (oligo-pAs1, oligo-pTa535, oligo-5S, P332, and oligo-713) were used. For convenience, we further designated the oligo-probes according to the original probe names, i.e., pAs1, pSc119.2, pTa-535, pTa-713, 5S (pTa794), and NOR (pTa71).
Fluorescence in situ hybridization
Chromosomal preparation was carried out according to the protocol published in Badaeva et al. (2017). FISH probes were hybridized to chromosomes of 4x and 6x Ae. crassa, Ae. tauschii (subsp. strangulata and subsp. tauschii), Th. bessarabicum, and common wheat T. aestivum cv. Chinese spring according to protocol in Kuznetsova et al. (2019). Biotinilated probes were detected with streptavidin-Cy3 (Vector laboratories, UK) and digoxigenin-labeled probes were detected using anti-digoxigenin-Fluorescein (Roche, Germany). The slides were stained with DAPI (4′,6-diamidino-2-phenylindole) in Vectashield mounting media (Vector laboratories, Peterborough, UK) and analyzed on Leica DM6 B epifluorescence microscope. Selected images were captured with DFC 9000 GTC (Leica), and the slides were then washed in 2× SSC, and the second hybridization was carried out with “standard” DNA probes (pSc119.2, pAs1, pTa-535, pTa-713, and GGAn) to allow chromosome identification. Probe combination pAs1 + pSc119.2 was used for wheat and Aegilops species and pAs1 + pTa-713 for Th. bessarabicum. The A subgenome chromosomes of wheat were classified using additional probe combination GAAn and pTa-535 according to Komuro et al. (2013); chromosomes of Ae. tauschii were classified as suggested by Badaeva et al. (2002, 2019a) and Zhao et al. (2018), Ae. crassa as in Abdolmalaki et al. (2019), and Badaeva et al. (2021). Th. bessarabicum chromosomes were classified according to Grewal et al. (2018), Badaeva et al. (2019b), and Chen et al. (2019).
Our previous analyses showed that accessions K-2485 and AE 742 (Ae. crassa, 4x), K-112 (Ae. tauschii subsp. strangulata), PI 531711 (Th. bessarabicum), and Chinese Spring have normal karyotypes typical to the respective species (Gill et al., 1991; Badaeva et al., 2019a, 2019b, 2021). At the same time, we have found earlier that hexaploid Ae. crassa, IG 131680 carries a translocation T1D1L:7D1L (T10) with interstitial breakpoints in addition to two species-specific translocations, T1 (Acr:6Xcr) and T2 (4D1S,FcrS; Badaeva et al., 2021).
Results
Assemblies’ characterization
Repeat assemblies for single species (“individual” assemblies) were prepared for tetraploid and hexaploid Ae. crassa, Th. bessarabicum, Ae. tauschii subsp. strangulata, and subsp. typica. The general features of these assemblies are summarized in Supplementary Table S1. In addition, we compiled three comparative assemblies, in which the number of reads for each species were calculated in a direct proportion to ploidy level: 0.5 mln TB + 0.5 mln ATs + 1mln AC4x; 0.5 mln TB + 0.5 mln ATt + 0.5 mln AC4x; 1 mln ATs + 2 mln AC4x. For all repeats extracted from the above assemblies, the extensive summary tables were constructed (Supplementary Table S2). For comparative assembly we have extracted and described clusters containing ≥80% repeats from only one species and, if possible, having no highly similar alignments with clusters from individual assemblies for other species involved in comparative assembly.
Repeats’ characterization
Altogether, 34 repeats were identified in individual assemblies of Ae. crassa 4x (11 high confidence, 8 low confidence satellites and 1 LTR) and Th. bessarabicum (6 high confidence, 6 low confidence satellites and 2 LTRs). The characterization of the identified repeats are shown in Supplementary Tables S3, S4 including consensus sequences, layout of TAREAN graph, estimated proportion of given repeat in the genome, homology to previously found repeats, and repeats found in this study. All repeat consensus sequences (excluding those shorter than 100 bp) together with the putative satellites of 6x Ae. crassa and Ae. tauschii have been clustered after all-to-all blast into 19 groups by their identity and total coverage percentage; based on these data we developed 19 probes. Seventeen of these groups contained sequences from Ae. crassa 4x and were classified into high-probability (№1-№11) and low-probability (№12-№16) repeats, and putative LTR (№17). Additionally, we discriminated an additional group (№18), in which a sequence obtained from 6x Ae. crassa was not paired with a sequence from 4x Ae. crassa. Two other satellites (high confidence №19, and low confidence №20) were found in Th. bessarabicum genome. Based on bioinformatical analysis, all repeats were classified according to copy number as very-low-copy (below 0.29%), low-copy (0.3–0.59%), and common (0.6% and more).
The high-probability repeats, representatives of the groups based on 4x Ae. crassa:
1. AC4x_CL3_339nt is a common repeat, found in all species analyzed. The density of the TAREAN graph layout produced for the given length of repeat monomer (Supplementary Figure S2a) is high, allowing the assumption that there are self-alike features in the monomer influencing the graph. Subsequent analysis with YASS online tool reveals a non-perfect palindromic region 58–176 with 60% identity and e-value of 9.7e−05. Two similar repeats, Ats_CL11_337nt and Ats_CL51_343nt, were identified in the assembly of Ae. tauschii subsp. strangulata by RepeatExplorer2; the latter differed from the analogous Ae. crassa monomer mostly in the given palindromic region. AC4x_CL3_339nt was more dissimilar to P335 repeat from Ae. crassa genome.
2. AC4x_CL131_334nt is a very-low-copy repeat, found in all analyzed species. It is highly similar to P334 found in Ae. tauschii with 90% identity across the 322 bp alignment and highly similar to pTa-465 (FISH-positive repetitive sequence KC290905.1) with 91% identity across 299 bp.
3. AC4x_CL170_369nt is a very-low-copy repeat found in all species analyzed in this study, which directly corresponds to P369 found in Ae. tauschii, and is highly similar to tandem repeat sequence 4P6-14 found in Ae. tauschii (AY249985.1) and ACRI_TR_CL78 satellite sequence found in Agropyron cristatum (MG323512.1).
4. AC4x_CL209_316nt is a very-low-copy repeat, found in all species, except for Th. bessarabicum. It is highly similar to T. aestivum clone p451 (genomic repeat region AF139201.1), and partially more dissimilar from P320 found in Ae. tauschii, aligning for 197 bp with 75% identity.
5. AC4x_CL219_319nt is a very-low-copy repeat, found in all species except for Th. bessarabicum and Ae. tauschii subsp. strangulata. This sequence was found in Ae. tauschii (AH013688.3) in tandem, but was not annotated.
6. AC4x_CL228_312nt is a very-low-copy repeat not yet annotated at NCBI database and found only in the context of T. aestivum BAC libraries and assemblies. It is, however, found in our assemblies of both Ae. tauschii subspecies and 6x Ae. crassa, but is lacking in Th. bessarabicum. Mapping of the AC4x_CL228_312nt monomer with trimmed reads from different sources (Supplementary Figure S2c) shows similar mapping profiles between Ae. crassa 4x and 6x and Ae. tauschii subsp. typica with more notable SNPs in Ae. tauschii subsp. strangulata and lack of given repeat in Th. bessarabicum. AC4x_CL228_312nt is more dissimilar to P321 found in Ae. tauschii, although they produce alignment of 242 bp with 92.6% identity.
7. AC4x_CL232_320nt is a very-low-copy repeat, found in all studied species except for Th. bessarabicum. It is highly similar to T. aestivum clone, to p451 (genomic repeat region AF139201.1), and partially it shows high similarity with P320, aligning for 276 bp with 96% identity.
8. AC4x_CL241_88nt is a very-low-copy repeat, found across all species. This repeat is 97.72% identical to Oligo-44 and Oligo-3A1 found in T. aestivum and highly similar to oligonucleotide BSCL184-2 based on tandem repeat BSCL184 (88 bp long) found in Th. bessarabicum and P132 found in Ae. tauschii.
9. AC4x_CL244_376nt is a very-low-copy repeat, found in all the species except for Ae. tauschii. It is highly similar (with 100% identity) to oligos BSCL1-1 and BSCL1-2 based on putative satellite BSCL1 (376 bp long) found in Th. bessarabicum, and DP4J27982 and DP4J28086 developed based on the sequences of chromosome arm 4JbL of Th. bessarabicum.
10. AC4x_CL257_820nt is a very-low-copy repeat, found exclusively in 4x Ae. crassa and having no similar annotated sequences.
11. AC4x_CL258_1307nt is a very-low-copy repeat, specific to Ae. crassa, and is not annotated either in NCBI database or in analyzed literature.
The low-probability repeats, representatives of the groups based on 4x Ae. crassa:
12. AC4x_CL8_584nt is a common repeat, highly similar to AC6x_CL6_584nt monomer (6x Ae. crassa) and more dissimilar to monomers ATs_CL16_551nt and ATt_CL17_553nt of both Ae. tauschii subspecies. According to gsnap mapping (Supplementary Figure S2b), Th. bessarabicum variant of this repeat differs from the AC4x_CL8_584nt due to the many SNPs. It had no similar annotated sequences either in NCBI database or in analyzed literature.
13. AC4x_CL60_251nt is a low-copy repeat, found exclusively in tetraploid Ae. crassa and being partially more dissimilar to Ae. bicornis RAPD-generated marker sequence (AF120172.1), aligning for 200 bp with 78% identity.
14. AC4x_CL193_504nt is a very-low-copy repeat, found in all species except for Th. bessarabicum. It is somewhat similar to T. aestivum clone pTa-451 (FISH-positive repetitive sequence KC290912.1) with the alignment of 500 bp with 68% identity.
15. AC4x_CL239_178nt is a very-low-copy repeat, found in all species except for Ae. tauschii. This sequence is tandemly organized in Ae. tauschii sequence AH013688.3, but is not annotated either in NCBI database or in analyzed literature.
16. AC4x_CL261_553nt is a very-low-copy repeat, found in all species except for Ae. tauschii. It is more dissimilar to T. aestivum clone CentT550 (satellite sequence MN161206.1), aligning across 540 bp with 83.6% identity.
The putative LTR, representative of the groups based on 4x Ae. crassa:
17. AC4x_CL18_487nt is a common repeat, similar to FAT element.
The high-probability repeats, representative of the groups based on 6x Ae. crassa:
18. AC6x_CL232_145nt is a low-copy repeat, found only in 6x Ae. crassa and in both Ae. tauschii subspecies, being fully identical to P436 found in Ae. tauschii. The FISH probe developed based on AC6x_CL232_145nt is designated CL27_232.
The high-probability repeats, representative of the groups based on Th. bessarabicum:
19. TB_CL2_379nt is a common repeat, found in all species except for Ae. tauschii and somewhat similar to AC4x_CL244_376nt at the 62% identity. It is also highly similar to oligos BSCL1-1, BSCL1-2, DP4J27982, and DP4J28086 with 100% identity (see №9).
The low-probability repeats, representative of the groups based on Th. bessarabicum:
20. TB_CL148_662nt is a low-copy repeat, not found in 6x Ae. crassa and Ae. tauschii subsp. strangulata. It is highly similar to AC4x_CL162_661nt, and to A. cristatum ACRI_TR_CL80 satellite sequence (MG323513.1)
Repeats’ comparative assembly analysis
The comparative analysis allowed us to find repeats unique for 4x Ae. crassa, which are absent in other studied species.
TB + ATs + AC4x_CL82_379nt and TB + ATs + AC4x_CL88_376nt are presumably Th. bessarabicum-specific, even though we get no additional information from this assembly, as the first repeat has a clear analogue in Th. bessarabicum individual assembly, and the second, in contrast with 4x Ae. crassa reads share, aligns well with AC4x known repeat. However, both of them are absent in Ae. tauschii subsp. strangulata. TB + ATs + AC4x_CL217_316nt and TB + ATs + AC4x_CL234_319nt are 4x Ae. crassa-specific, both found in 4x Ae. crassa single species assembly (Supplementary Table S5).
TB + ATt + AC4x_CL87_379nt and TB + ATt + AC4x_CL93_376nt are both absent in Ae. tauschii subsp. typica and are equivalent to the first pair of repeats described above for the previous assembly. TB + ATt + AC4x_CL204_316nt and TB + ATt + AC4x_CL220_319nt are 4x Ae. crassa-specific and are equivalent to the last pair of repeats described above for the previous assembly (Supplementary Table S6).
ATs + AC4x_CL203_316nt, ATs + AC4x_CL213_319nt, ATs + AC4x_CL247_1319nt, ATs + AC4x_CL224_178nt, and Ats + AC4x_CL240_553nt are 4x Ae. crassa-specific, and all of them have a clear analogue in 4x Ae. crassa single species assembly (Supplementary Table S7).
In silico identification of Xcr subgenome-specific tandem repeats in 4x Aegilops crassa
Four novel tandem repeats were identified in RepeatExplorer2 output among filtered reads (Supplementary Table S3). Homology search showed that they have no similarity to previously identified tandem repeats of 4x Ae. crassa. Biased filtering (removing reads perfectly mapping on Ae. tauschii D genome) probably causes an increase of fraction of these reads of interest and makes it possible to assemble and classify assembled consensuses as tandem repeats. BLASTn search against the entire Nucleotide database showed numerous hits with other satellite and transposon sequences from other grass species for CL162 and CL244 and no hits against repeat-related sequences for CL257 and CL262.
Assessment of copy numbers of repetitive DNA clusters using qPCR
The values of threshold cycle (Ct) were directly determined using qPCR for each repeat and for each species and relative copy number normalized against single-copy gene VRN1, which was calculated for each repeat (Supplementary Table S8). Decimal logarithm of relative quantity (RQ) was provided for copy number comparison (Table 3), and all repeats were conventionally (with highly probability) divided into high-copy (log10RQ > 4), medium-copy (2 < log10RQ < 4), and low-copy (log10RQ < 2). In 4x and 6x Ae. crassa CL3, CL18, CL209, CL170, CL8, and CL239 represented high-copy repeats; CL131 and CL2 were low-copy repeats, while others were classified as medium-copy repeats. In Ae. tauschii genome CL3, CL18, and CL170 were highly abundant, CL148, CL60, CL8, CL232, CL193, CL261, CL228, and CL27_232 - medium abundant, while others were found to be low-copy repeats. In T. aestivum, the high-copy repeats were CL3, CL18, the low-copy repeats were CL228, CL239, CL131, CL258, CL2, and CL257, while others were classified as medium-copy. In Th. bessarabicum CL244 and CL3 demonstrated high abundance, CL148, CL170, CL239, CL60, CL193, CL8, CL261, CL18, and CL27_232 were medium-copy, while others were classified as low-copy. Based on our findings, the following groups of repeats were distinguished:
i. repeats that occur predominantly in genomes of Ae. crassa (4x and 6x): CL257 and CL258;
ii. repeats found only in genomes of wheat and Ae. crassa: CL209 and CL219;
iii. repeats abundant in genomes of Ae. tauschii, wheat, Ae. crassa: CL232 and CL228;
iv. repeats with similarly high abundance in all studied species: CL261, CL193, CL60, CL148, CL8, CL170, CL18, CL3, CL27_232;
v. repeats not detected in genome of Ae. tauschii, but abundant in other species: CL244;
vi. repeats with high copy number in Th. bessarabicum and Ae. crassa: CL239;
vii. repeats with low copy number in all studied species: CL2 and CL131.
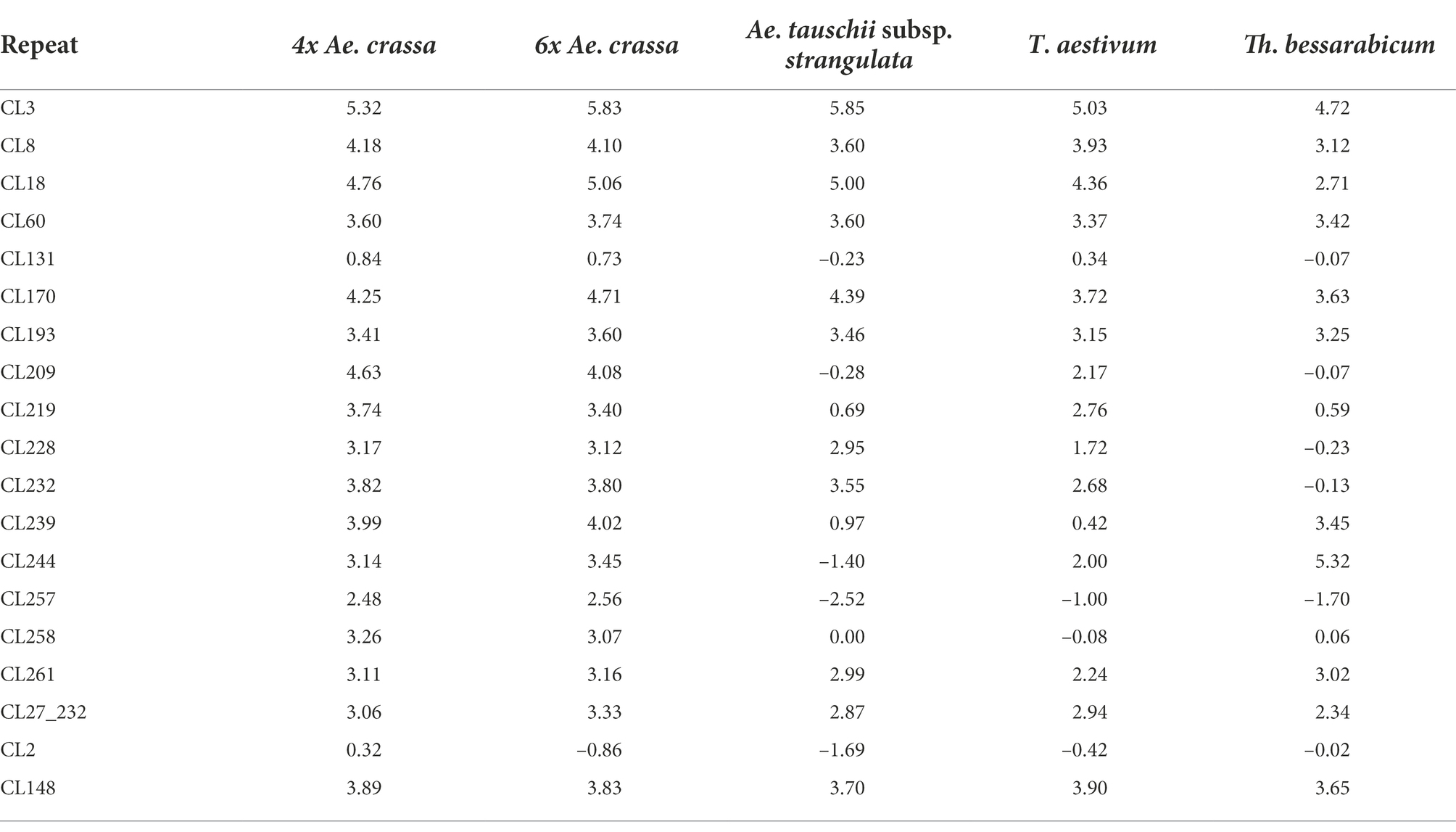
Table 3. Relative quantity (expressed in decimal logarithm) of the tandem repeats found in this study.
Fluorescence in situ hybridization
FISH analysis of repeated DNA sequences currently isolated from 4x Ae. crassa (AE 742) and previously identified repeats showed that individual repeats differ in organization (dispersed vs. discrete signals), species and subgenome specificity, the number and chromosome position(s), and signal intensities (Supplementary Figures S3–S7). Five repeats were found to be homologous to already known DNA sequences: CL3 is homologous to pAs1, CL18 to FAT, CL131 to pTa-465, and CL241 to Oligo-3A1. Repeats CL2, CL8, CL60, CL148, CL170, CL193, CL209, CL219, CL228, CL232, CL239, CL244, CL257, CL258, CL261 (and its homolog CL198), and CL27_232 are considered as novel FISH probes.
Two Ae. crassa repeats, CL8 and CL60, and Th. bessarabicum repeat CL148 are distributed evenly along the entire lengths of all chromosomes irrespective of their (sub)genome affinities (Supplementary Figures S3, S7). Owing to the usefulness of these repeats for genome or chromosome identification, we excluded them from further analyses.
Other DNA probes containing already known and novel repeated sequences demonstrated discrete signals. Three of them are species-specific and produce signals on one chromosome (pair) each. Thus, CL257 and CL258 occur only in Ae. crassa (4x and 6x). Small CL257 signals appear in subterminal region of 5D1S arm, and CL258 signals distally on the 1D1S arm (Figures 1A,I,M; Supplementary Figures S3, S4). Although 6x Ae. crassa possesses two copies of the D* subgenome, only one chromosome pair belonging to D1 subgenome carries the signals of each of the two abovementioned probes; signals are a little more intense for CL258 than for CL257 (Figures 1A,I).
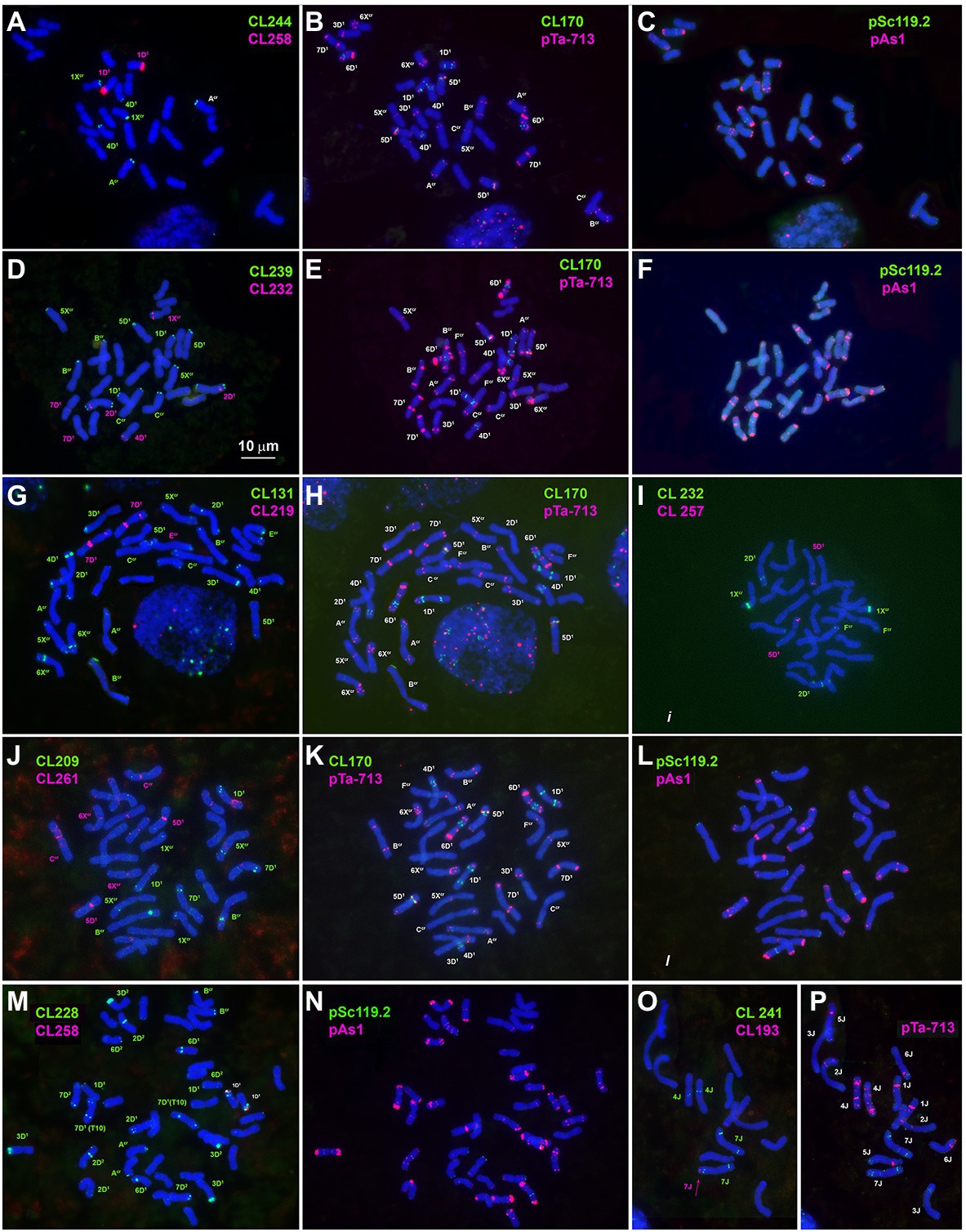
Figure 1. Metaphase cells of Ae. crassa, 4x (A–L) and 6x (M,N), and of Th. bessarabicum (O,P) after FISH with different probe combinations; (A–C) the cell of 4x Ae. crassa AE 742 hybridized consecutively with CL244 + CL258 followed by CL170 + pTa-713 and pAs1 + pSc119.2; (D–F) - the cell of 4x Ae. crassa AE 742 hybridized consecutively with CL239 + CL232 followed by CL170 + pTa-713 and pAs1 + pSc119.2; (G,H) - the cell of 4x Ae. crassa AE 742 hybridized consecutively with CL131 + CL219 followed by CL170 + pTa-713; (I)- the cell of 4x Ae. crassa AE 742 hybridized with CL232 and CL257; (J–L) - the cell of 4x Ae. crassa AE 742 hybridized consecutively with CL209 + CL261 followed by CL170 + pTa-713 and pAs1 + pSc119.2; (M,N) – the cell of 6x Ae. crassa AE 131680 hybridized consecutively with CL228 + CL258 followed by pAs1 + pSc119.2; (O,P) – the cell of Th. bessarabicum PI 531711 hybridized with CL193 + CL241 followed by pTa-713. Probe combinations are given on the top of the respective images; probe color corresponds to signal color.
CL193 probe produces a single hybridization site on the tip of the short arm of one of the two Th. bessarabicum homologous chromosomes 7 J (Figure 1O). FISH fails to detect the CL193 on Ae. crassa, Ae. tauschii, or common wheat chromosomes.
Large signals of CL27_232 probe are found in the proximal region of 3D1 short arms and faint signals in a distal part of 5Xcr long arms in both 4x and 6x cytotypes of Ae. crassa (Figures 2A; Supplementary Figures S3, S4). Hexaploid Ae. crassa possesses additional CL27_232 signals in the short arm of 3D2 and in the terminus of short arm of 4D1, which was transferred from Fcr short arm as a result of species-specific translocation T2 (Figure 2A; Supplementary Figure S4). Only one pair of clear CL27_232 signals is detected on chromosome 3D of Ae. tauschii (Figure 2C) and 3D of common wheat (Figure 2J).
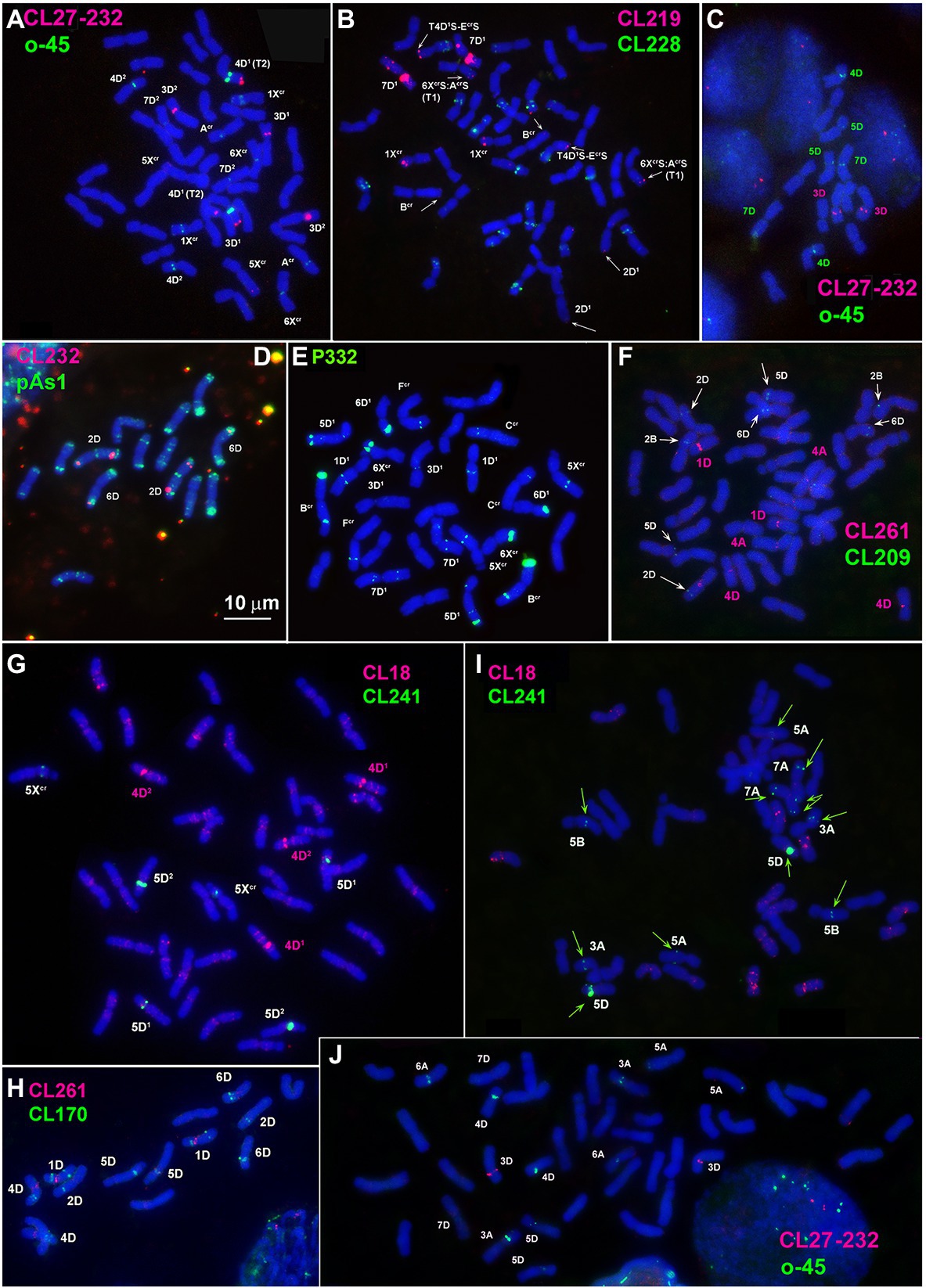
Figure 2. Localization of: (A,C,J) CL27_232 (red) + o45 (green), (B) CL219 (red) + CL228(green); (D) CL232 (red) + pAs1 (green); (E) P332 (green); (F) CL261 (red) + CL209 (green); (G,I) CL18 (red) + CL241 (green); (H) CL261 (red) + CL170 (green) on chromosomes of Ae. crassa, 4x (E), 6x (A,B,F,G), Ae. tauschii (C,D,H) and common wheat (I,J).
CL244 probe displays relatively poor hybridization to Ae. crassa chromosomes. Thus, three pairs of very faint signals appear in the terminal regions of 1XcrL, AcrL, and 4D1S of 4x Ae. crassa (Figures 1A, 3A,B). In karyotype of 6x Ae. crassa the largest CL244 signals appear on 5D1S, and smaller signals - on chromosomes AcrL and FcrS. As Fcr was modified following species-specific translocation T2, this site is probably derived from 4D1S (Figure 3C). Common wheat possesses three pairs of very small CL244 signals on chromosomes 2AS, 1BS, and 4BL (Figures 3E,F), and no hybridization is detected in Ae. tauschii. On the contrary, most Th. bessarabicum chromosomes carry intense hybridization sites in either the short or long arm (Figure 3D). Signals appear on both homologous chromosomes 1JS, 5JS, and 7JS, but only on one homolog of 3JL and 6JS pairs.
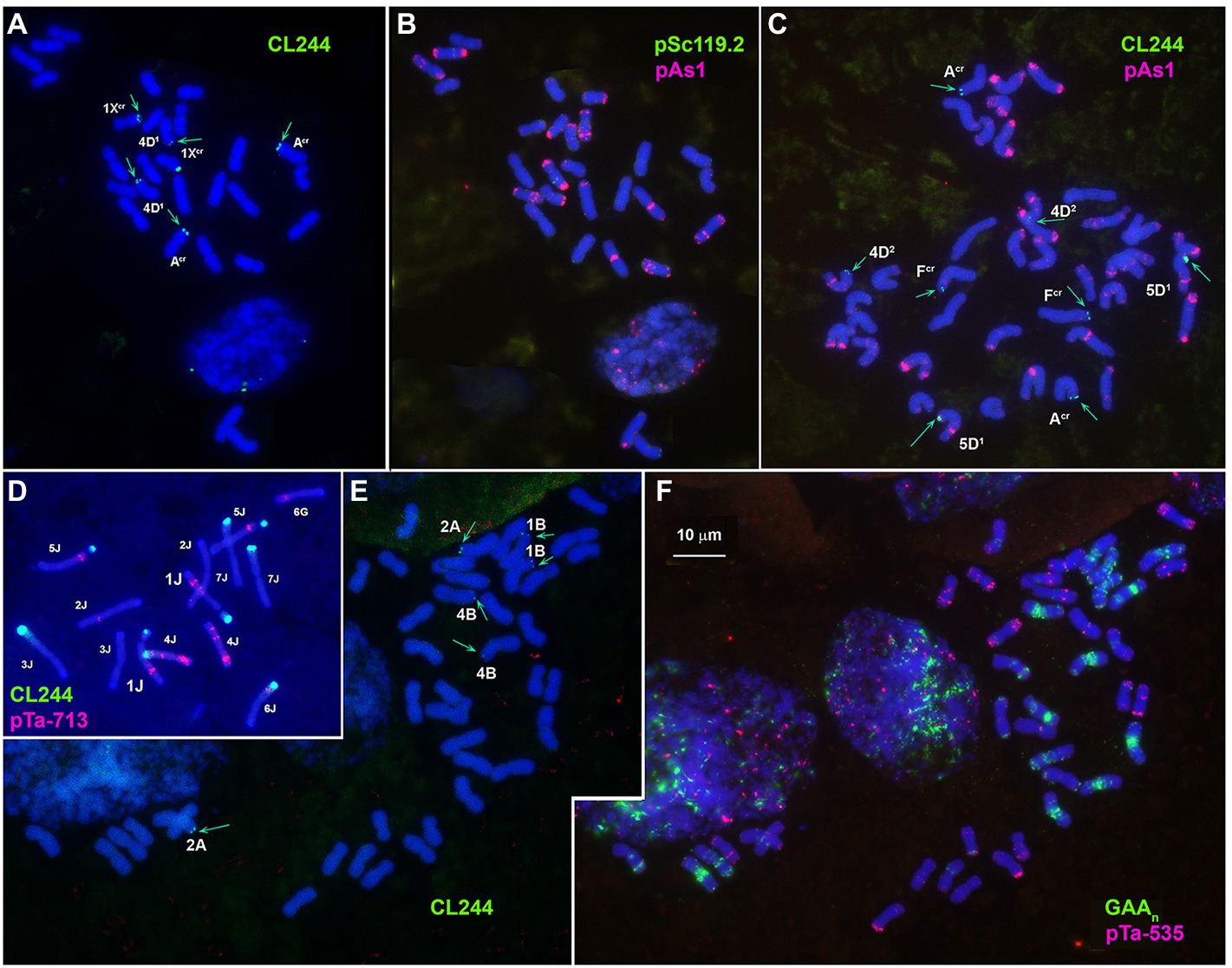
Figure 3. Localization of CL244 (green) repeat on Ae. crassa, 4x (A,B), Ae. crassa, 6x (C), Th. bessarabicum (D), and common wheat (E,F) chromosomes. Chromosomes are identified using pAs1 (B,C, red), pTa-713 (D, red) or GAAn (F, green) and pTa-535 (F, red). Chromosomes carrying CL244 signals are indicated. Scale bar, 10 mcm.
The probe CL241 hybridized to the proximal part of the short arms of all group 5 chromosomes of 4x and 6x Ae. crassa (Figure 2G; Supplementary Figures S3, S4) and of Ae. tauschii. In common wheat, the CL241 sites on 5A and 5B are located in the long, but not in the short arms, while additional CL241 loci are found on 3A (long arm) and both arms of 7A (Figure 2I; Supplementary Figure S6). Chromosome 7J of Th. bessarabicum carries two CL241 sites in opposite arms (Figure 1O; Supplementary Figure S7). Additional CL241 sites are observed in the middle of 4JL, but there are no signals on group 5 chromosomes.
The CL209 signals appear mainly on the Xcr subgenome chromosomes of Ae. crassa. Small CL209-sites are present on chromosomes Ccr (middle of the short arm), 5Xcr (distal third of the long arm), and on a distal part of 7D1L arm (Figure 1J; Supplementary Figures S3, S4). Hexaploid Ae. crassa has additional large subtelomeric CL209 clusters on chromosomes 1XcrS and in a distal part of 6D2L (Figure 2F; Supplementary Figure S4). CL209 is absent in Ae. tauschii and Th. bessarabicum, but produces very weak signals on common wheat chromosomes 2BS, 2DL, and 6DS (Supplementary Figure S6).
CL219 sequence is present in abundance on Ae. crassa (4x and 6x) chromosome 7D1, which contains two prominent clusters located in a proximal half of the short arm in a distal third of an opposite arm (Figures 1G, 2B; Supplementary Figures S3, S4). In addition, small signals occur in a distal part of 5D1S and in the terminus of FcrS. In 6x cytotype the site from chromosome Fcr is transferred onto chromosome 4D1S following species-specific translocation T2. Hexaploid Ae. crassa also contains clear CL219 signals on three pairs of Xcr subgenome chromosomes: in the short arm of 1Xcr, the long arm of 6Xcr, and in the end of BcrS (Supplementary Figure S4). Common wheat possesses distinct CL219 sites in the terminal part of 2BS chromosomes and this repeat is absent in Ae. tauschii and Th. bessarabicum (Supplementary Figures S5–S7).
FISH reveals large signal of CL232 probe in a distal part of the long arm of chromosome 2D (or its derivative) in all wheat and Aegilops species (Figure 4, Supplementary Figures S3–S6), while a smaller site in the short arm of 2D is present only in Ae. tauschii, common wheat, and the D2 subgenome of Ae. crassa (6x). Prominent subtelomeric CL232 signals are detected in the long arm of 1Xcr in 4x (Figures 1D,I) as well as in 6x Ae. crassa, which also possesses a smaller signal terminally in the short arm (Figure 4). Hexaploid Ae. crassa has smaller signals of CL232 in a terminal part of 4D1S and distal quarters of 7D1L and 6D2L. Small CL232 site in 7DL appears in diploid Ae. tauschii (Figure 2D), and this sequence is absent in Th. bessarabicum genome.
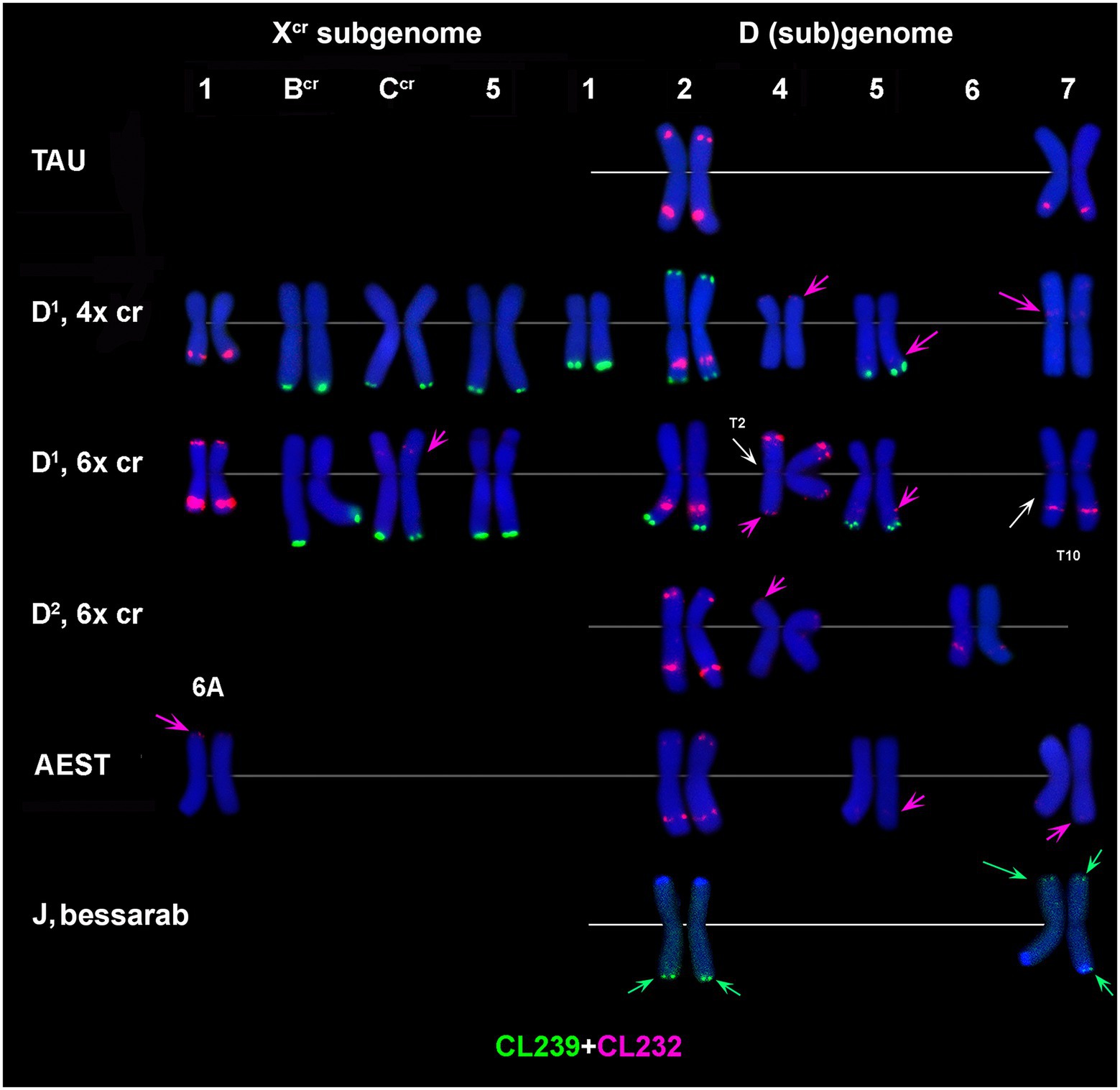
Figure 4. Distribution of CL239 (green) and CL232 (red) sequences on chromosomes of different cereal species: TAU – Ae. tauschii ssp. strangulata; D1, 4x cr – D1 subgenome of tetraploid Ae. crassa; D1, 6x cr, D2, 6x cr – D1 and D2 subgenomes of hexaploid Ae. crassa; AEST – A and D subgenomes of T. aestivum, J, bessarab – Jb genome of Th. bessarabicum. 1–7 – homoeologous groups. Pink arrows point to minor CL232 sites.
CL239 is not detected in Ae. tauschii and common wheat; it is also absent in the D2 subgenome chromosomes of 6x Ae. crassa (Figure 4). Both Ae. crassa cytotypes contain large CL239 clusters in subterminal regions of several Xcr and D1 chromosomes (Figure 1D). Among them, five sites are common (BcrL, CcrL, 5XcrL, 2D1L, and 3D1), but tetraploid form contains two additional loci on 1D1L and 2D1S. Two pairwise CL239 signals and one odd signal are detected terminally on Th. bessarabicum chromosomes 2JL and 7JS + L (Supplementary Figure S7).
The largest signals of oligo-45 (o-45) probe occur in a proximal region of chromosome 4D in Ae. tauschii (Figure 2C; Supplementary Figure S5), 4x and 6x Ae. crassa (Figure 2A, Supplementary Figures S3, S4), and common wheat (Figure 2J; Supplementary Figures S6). Additional, smaller signals are found in pericentromeric regions of 5D and 7D and (Ae. tauschii and common wheat), proximal part of 3AS, subterminal region of 5AL, and pericentromeric part of 6A (common wheat, Figure 2J). Ae. crassa contain several o-45 sites located in the middle of 6D1S; on the chromosome AcrS, close to the centromere; interstitially and terminally in 1XcrL; in the middle of 5XcrL; and in pericentromere of 6Xcr (4x and 6x Ae. crassa) and 7D2 (6x Ae. crassa; Supplementary Figures S3, S4).
All studied species possess the CL261(=CL198) sequence. According to FISH, it localizes in pericentromeric regions of most chromosomes and signal intensity varies between chromosomes and between species (Figures 1J, 2F,H; Supplementary Figures S3–S7).
According to FISH, CL170 sequence is present in D (sub)genome(s) of Triticum and Aegilops species and in the Jb genome of Th. bessarabicum (Figure 2H; Supplementary Figures S3–S7). Distribution of CL170 sites along the D (sub)genome chromosomes is highly conserved across species (Figures 5). Two clear signals are located interstitially in the opposite arms of 1D; two signals in the middle 4DL are interrupted by a small pTa-713 site; large signal is present proximally in 5D short arm alongside a distinct pTa-713 site, and large double signals in the proximal half of 6DS (Figures 1B,E,H,K). A distinct CL170 site occurs in the middle of 2D chromosome of Ae. tauschii (Figures 2H, 5, TAU) and 2D of common wheat (Figure 5, D AEST). Two small CL170 sites are detected on opposite arms of chromosome Acr of the tetraploid and in the long arms of Acr and 6Xcr chromosomes of the hexaploid Ae. crassa cytotypes; the split of two CL170 sites between two different chromosomes is due to species-specific translocation T1. Hexaploid Ae. crassa contains CL170 sites in both, the long (subterminal) and short (double distal) arms of chromosome 7D2, which are absent on the orthologous chromosomes of other species.
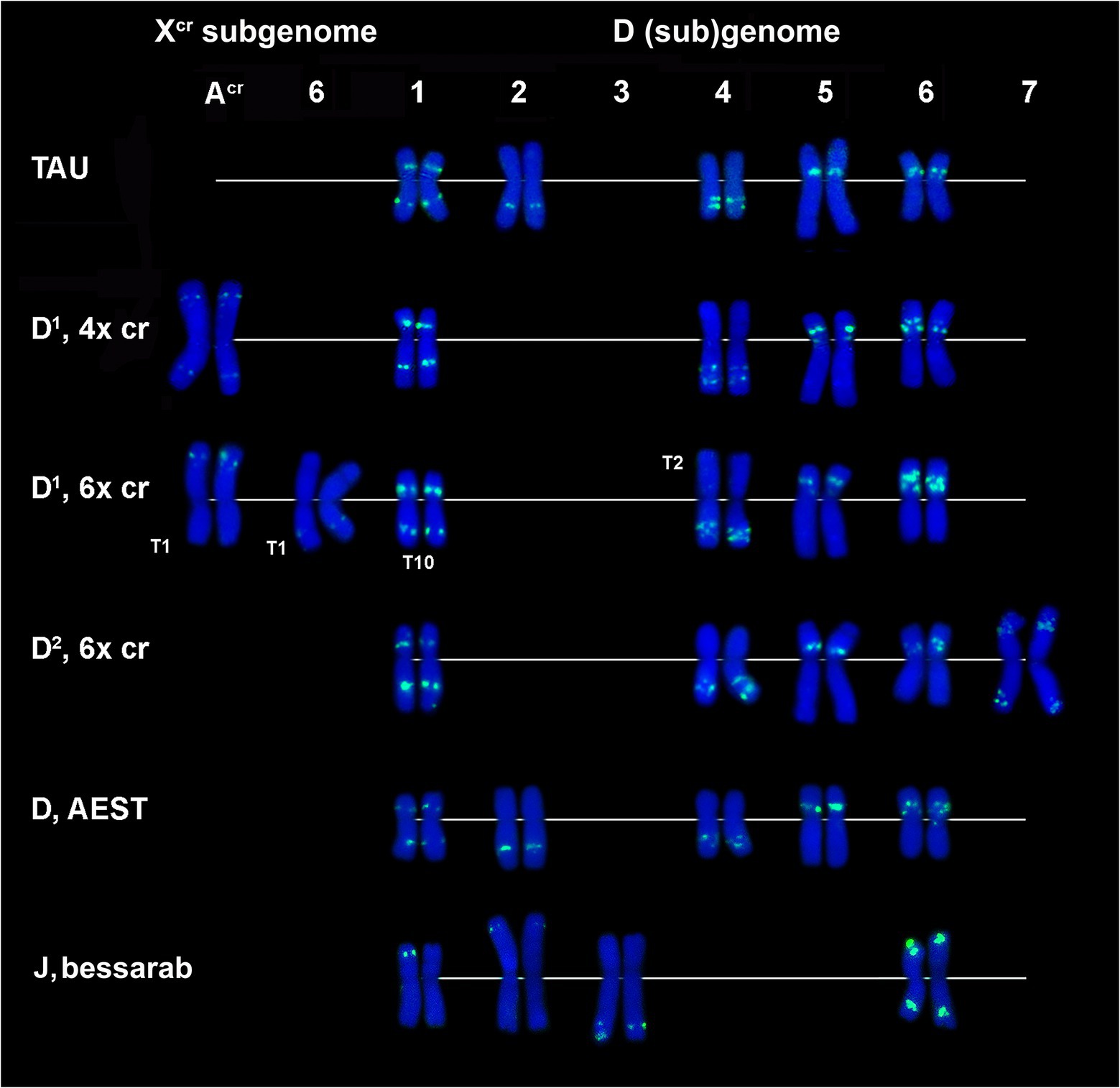
Figure 5. Comparison of CL170 patterns on chromosomes of different cereal species: TAU – Ae. tauschii ssp. strangulata; D1, 4x cr – D1 subgenome of tetraploid Ae. crassa; D1, 6x cr, D2, 6x cr – D1 and D2 subgenomes of hexaploid Ae. crassa; AEST – D subgenome of T. aestivum, J, bessarab – Jb genome of Th. bessarabicum. 1–7 – homoeologous groups. T1, T2, T10 – translocated chromosome of Ae. crassa. T10 causes small length reduction of chromosome region distal to CL170 site in the long arm of 1D1.
CL228 is located predominantly on the D1 subgenome chromosomes of 4x (Supplementary Figure S3) and 6x Ae. crassa (Figures 1M, 2B; Supplementary Figure S4). Two clear signals are detected in the terminus and in the middle part of 1D1 and 6D1 short arms; a prominent, probably double signal is observed in the terminal part of 3D1L, and one or a pair of small signals are present in opposite arms of 2D1 and 7D1 (Figure 6). Distinct signals are found on chromosomes BcrL and AcrL (6x Ae. crassa)/ 6XcrL (4x Ae. crassa), in the latter case the signal exchange is due to species-specific translocation T1.
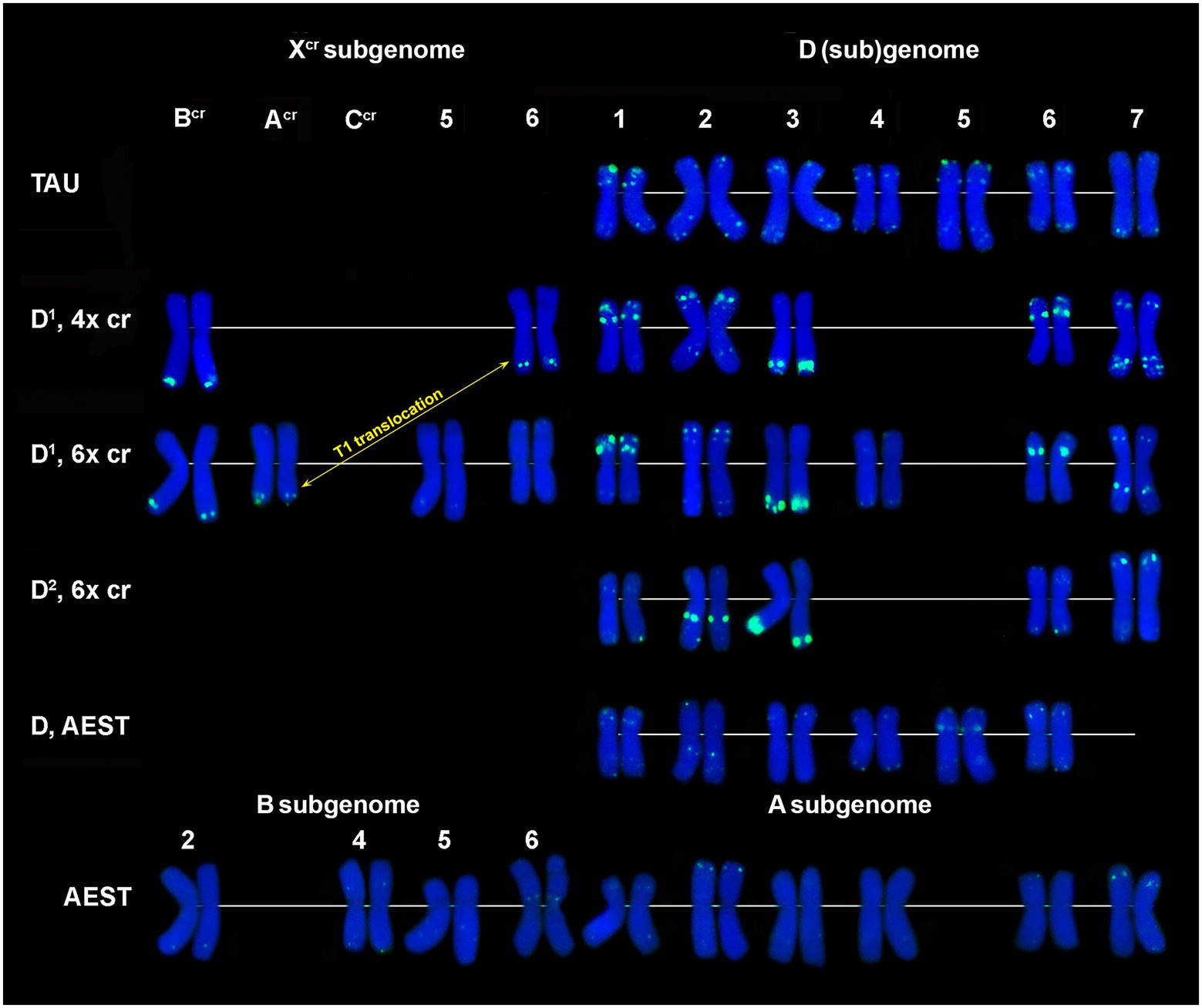
Figure 6. Comparison of CL228 labeling patterns on chromosomes of different cereal species: TAU – Ae. tauschii ssp. strangulata; D1, 4x cr – D1 subgenome of tetraploid Ae. crassa; D1, 6x cr, D2, 6x cr – D1 and D2 subgenomes of hexaploid Ae. crassa; AEST – A, B, and D subgenomes of T. aestivum. 1–7 – homoeologous groups. White arrowheads show minor CL228 sites detected on the Xcr, A, B, and D subgenome chromosomes.
Five pairs of the D2 subgenome chromosomes of 6x Ae. crassa possess CL228 sites. The largest signal occurs on 3D2L, similarly to 3D1L, and other intense sites are present in the proximal third of 2D2L and a distal part of 7D2S. Signals detected on chromosomes 1D2 and 6D2 are very weak, however their location is similar to that observed on the orthologous D1 subgenome chromosomes (Figure 6).
CL228 probe hybridized to all chromosomes of a diploid Ae. tauschii subsp. strangulata, however hybridization is much weaker compared to Ae. crassa (Figure 6; Supplementary Figure S5). Labeling patterns of the D subgenome chromosomes of common wheat are similar to that of Ae. tauschii, except for the lack of signals on 7D and less intense sites on 1D. The CL228 sequence hybridized to some A and B subgenome chromosomes of common wheat. Small but distinct signals are present on 6BS close to the centromere, in distal parts of 2AS and 7AS. Several very weak but consistent signals appear on 2B, 4B, 5B, 1A, 3A, 4A, and 6A chromosomes (Figure 6).
CL131 is homologous to T. aestivum clone pTa-465. CL131 hybridization patterns obtained in our study on common wheat chromosomes (Figure 7, AEST; Supplementary Figure S6) corresponds to previously reported patterns for pTa-465, thus confirming that these sequences are homologous. In common wheat and Ae. tauschii karyotypes only four out of seven D (sub)genome chromosomes carry small CL131 signals; these are 1D, 2D, 6D, and 7D in Aegilops and 2D, 5D, 6D, and 7D in wheat. Both Ae. crassa cytotypes carry numerous CL131 sites (Figure 1G), which have higher intensities and appear on both D1 and Xcr chromosomes (Figure 7). Most prominent sites appear on chromosome 2D1; subterminal signals on 3D1L and 3D2L are also large. Prominent CL131 signals are observed in the perinucleolar region of 6Xcr and subtelomeric parts of 7CcrL chromosomes; hexaploid accession also contains a clear signal on CcrS (Figure 7; Supplementary Figures S3, S4).
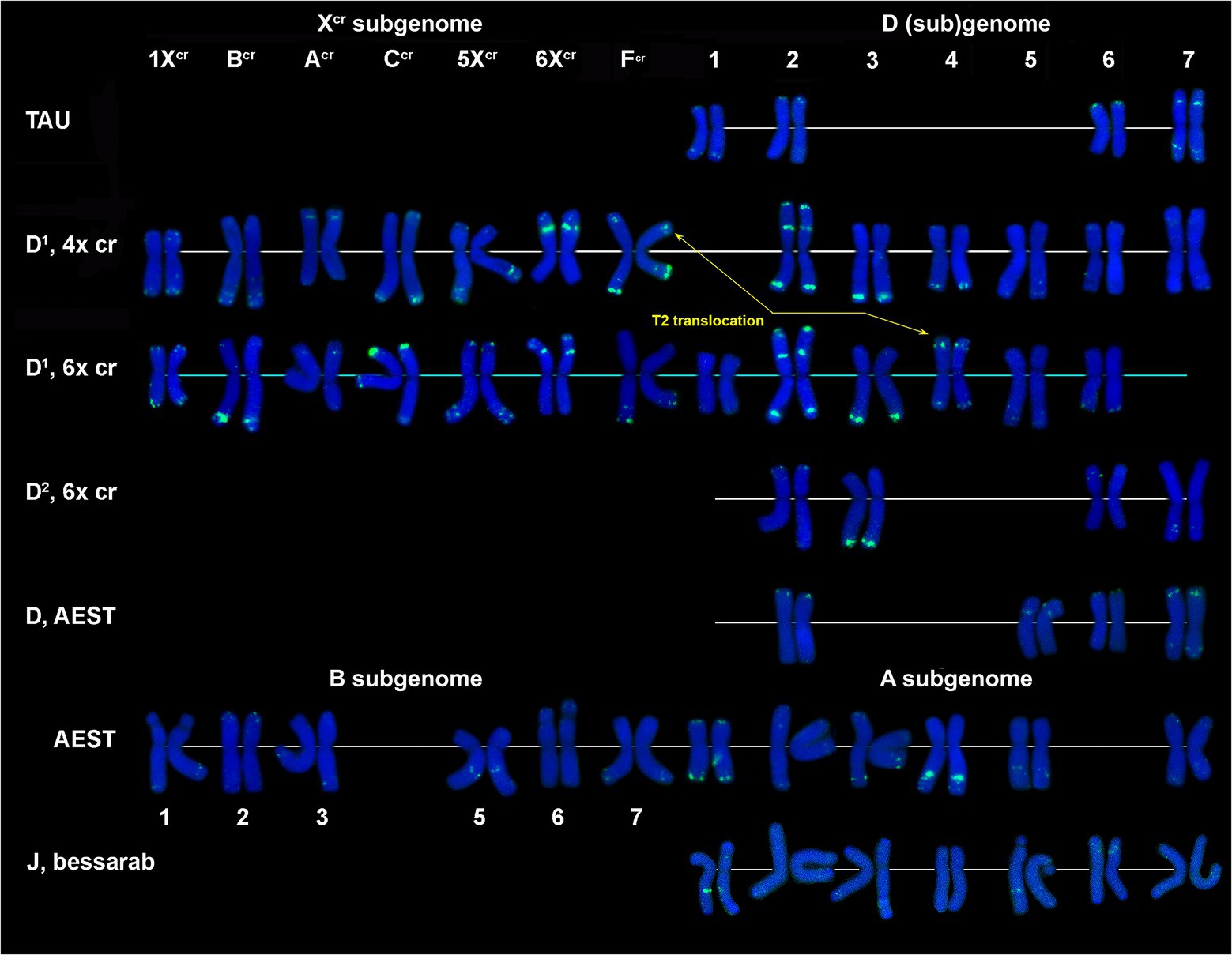
Figure 7. Comparison of CL131 labeling patterns on chromosomes of different cereal species: TAU – Ae. tauschii ssp. strangulata; D1, 4x cr – D1 subgenome of tetraploid Ae. crassa; D1, 6x cr, D2, 6x cr – D1 and D2 subgenomes of hexaploid Ae. crassa; AEST – A, B, and D subgenomes of T. aestivum; J, bessarab – Jb genome of Th. bessarabicum. 1–7 – homoeologous groups.
The P332 is homologous to pTa-k566 sequence and is similar to it in the distribution pattern on common wheat chromosomes (Figure 8). FISH detected P332 signals of variable sizes on most Ae. crassa (4x and 6x) and Ae. tauschii chromosomes (Figures 2E, 8; Supplementary Figures S3–S6), but not in Th. bessarabicum. No intraspecific variation of labeling patterns has been observed in Ae. crassa, and 4x and 6x cytotypes differ only in the size of 332 site on the chromosome Ccr (Figure 8). The D1 subgenome differs from D2 in labeling patterns of 2D, to a lesser extent of 5D and 7D chromosomes, and D2 subgenome shows high similarity with the D (sub)genomes of Ae. tauschii and common wheat.
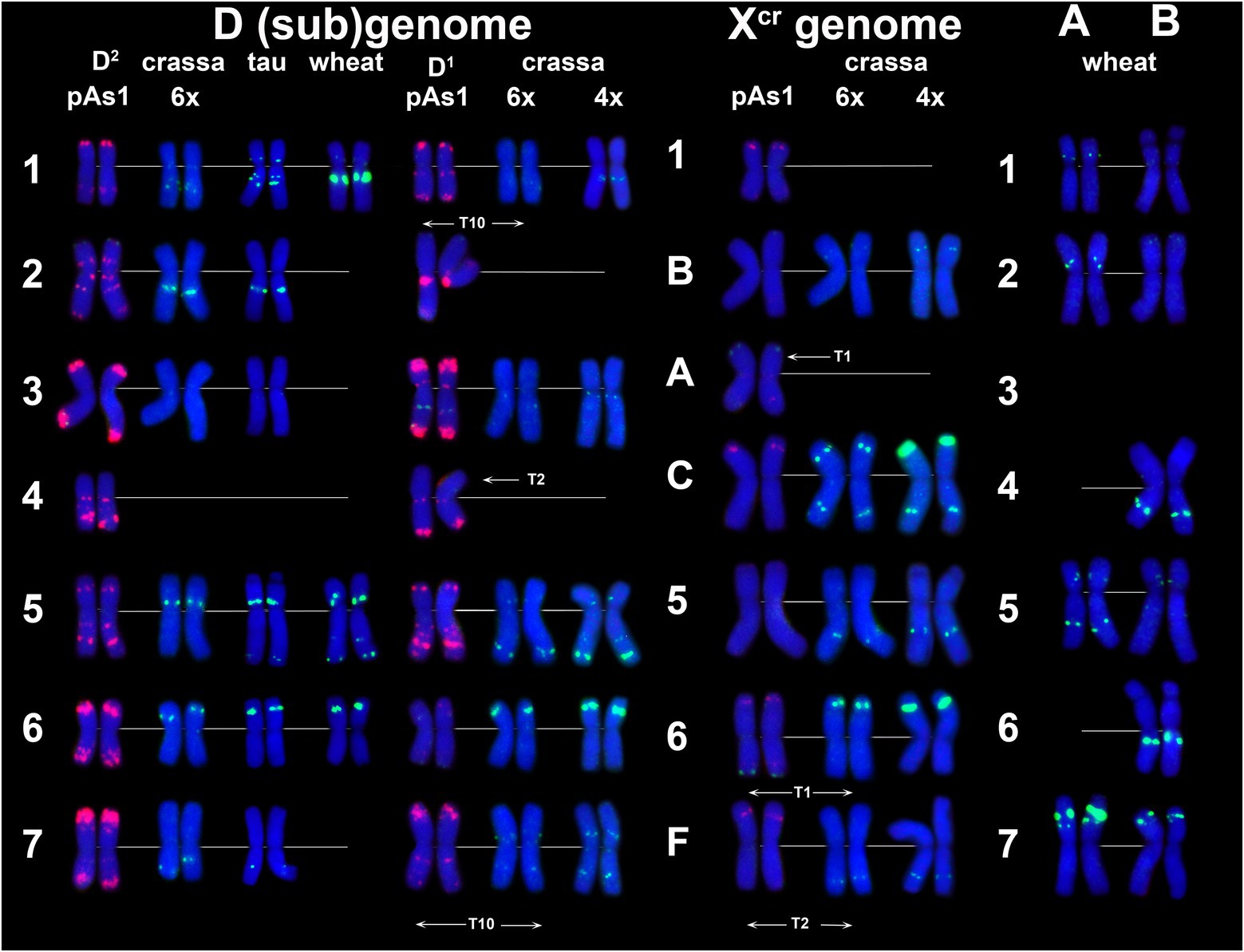
Figure 8. Comparison of P332 labeling patterns on chromosomes of different cereal species. D (sub)genome: D2 – subgenome of 6x Ae. crassa; TAU – Ae. tauschii ssp. tauschii; wheat – Chinese spring; D1 – subgenomes of tetraploid (4x) and hexaploid (6x) Ae. crassa; Xcr – subgenomes of tetraploid (4x) and hexaploid (6x) Ae. crassa; A and B – subgenomes of wheat (Chinese spring). 1–7 – homoeologous groups. A, B, C, F – chromosomes of Ae. crassa with unknown homoeologous group.
CL18 is homologous to FAT repeat and is similar to it in chromosomal distribution. Thus, CL18 signals localize unevenly along the length of all chromosomes in all species studied. It shows more intense labeling in proximal chromosome regions (Figures 2G,I; Supplementary Figures S3–S6) being especially abundant in the chromosome(s) 4DS. The sequence CL3 is homologous to pAs1 clone and CL187 to pSc119.2. Labeling patterns of these probes are almost identical to those reported earlier for the respective sequences and are not described in this paper.
Six new repeats identified in Ae. crassa, CL8 = CL16, CL131 = CL149, CL170, CL239, CL241, and Th. bessarabicum-derived repeat CL148, were shared by Aegilops and Thinopyrum species. Two Th. bessarabicum sequences, CL16 (homologous to CL8) and CL148, are dispersed along all chromosomes; of them CL16 is more abundant in proximal, while CL148 in distal chromosome regions (Supplementary Figure S7). The probe CL198 (related to CL261) hybridizes to pericentromeric chromosome regions, while CL2, CL193, CL239, and CL244 to subterminal regions of one to several Th. bessarabicum chromosomes. A clustered pattern of CL2 is observed in terminal regions of the Jb genome chromosomes. Three probes, CL149 (=CL131), CL241, and CL170, hybridize to interstitial regions of Th. bessarabicum chromosomes. Signals obtained with CL131 are very faint, inconsistent, and therefore not secure for chromosome identification. Together, CL241 and CL170 can serve as reliable chromosomal markers for Th. bessarabicum (Supplementary Figure S7). For all these repeats heteromorphisms of homologous chromosomes in signal presence and/ or size is often observed.
Discussion
Repeated nucleotide sequences are a major component of plant genome and play an important role in evolution (Salina et al., 2004b; Sharma and Raina, 2005; Dvořák, 2009; Mehrotra and Goyal, 2014; Macas et al., 2015; Liu et al., 2019). Divergence of diploid species or formation of new species via polyploidization are often associated with alterations in a fraction of repetitive DNA manifested in the emergence of new repeated DNA families, amplifications/eliminations of repeats, or their re-distribution between chromosomes (Zhao et al., 1998; Hemleben et al., 2007; Macas et al., 2015; Liu et al., 2019; Kuo et al., 2021; Waminal et al., 2021). Repetitive DNAs are located in structurally and functionally important chromosome regions (Heslop-Harrison et al., 2003; Shapiro and von Sternberg, 2005; Sharma and Raina, 2005; Heslop-Harrison and Schwarzacher, 2011; Mehrotra and Goyal, 2014; Garrido-Ramos, 2015) and are essential for maintenance of chromosome and genome integrity. From another side, chromosomal breaks causing chromosomal rearrangements often occur at sites enriched with repetitive DNA (Raskina et al., 2008; Molnár et al., 2011; Murat et al., 2017; Pollak et al., 2018). Owing to this, many researchers studying genome evolution and speciation in plants were focused on analysis repetitive DNA (Liu et al., 2019; Song et al., 2020; Ebrahimzadegan et al., 2021; Waminal et al., 2021). Fluorescence in situ hybridization is one of the most broadly exploited approaches in this field.
Development of new markers for genome studies is an important task in molecular biology and cytogenetics (Du et al., 2017; Meng et al., 2018; Said et al., 2018; Liu et al., 2019; Nikitina et al., 2020; Xi et al., 2020; Zagorski et al., 2020; Li et al., 2021; Liu and Zhang, 2021; Singh et al., 2021). Current progress in plant genome sequencing and bioinformatic analysis has opened broad perspectives for discovering new DNA sequences that can potentially be used as FISH probes, and the number of such markers rapidly increases. In our current study, as in some other publications (Liu et al., 2019; Zagorski et al., 2020; Waminal et al., 2021), we combine the benefits of molecular biology (in vitro), bioinformatics (in silico), and molecular cytogenetics (in situ) to get deeper insight on genome organization and karyotype evolution of Ae. crassa. The integration of different methods allowed us to identify several new repetitive DNA families, to assess their genome abundance, and map them on chromosomes by FISH. We reveal complicated genome organization of Ae. crassa and trace changes in the pattern of repetitive DNA over the course of evolution.
Novel markers for chromosome and genome identification
The results obtained in a current study provided us more detailed information on genome and chromosome organization of Ae. crassa and the related species. We found that some repetitive sequences are widespread, whereas other sequences are restricted to particular species, subgenome(s), homoeologous group, or even a single chromosome of particular species. Thus, eight repeats isolated from Ae. crassa (CL8, CL18, CL131, CL170, CL239, CL241, CL261 = CL198, and CL244) and one Th. bessarabicum (CL148) repeats were found to be common between wheat, Aegilops, and Th. bessarabicum; the results of qPCR and FISH assays correlates to each other rather strongly. Importantly, at least two of these sequences, CL241 and CL170, exhibited unique labeling patterns, which differed from other FISH probes suggested for cytogenetic analysis of this grass (Du et al., 2017; Grewal et al., 2018; Chen et al., 2019; Badaeva et al., 2019b). The probes CL2 and CL244 produced large signals overlapping with the position of C-bands on Th. bessarabicum chromosomes (Mirzaghaderi et al., 2010); these sequences may be important constituents of heterochromatin blocks in this species. Taken together with CL244 and CL2 probes, the sequences CL241 and CL170 can be used as supplementary probes in FISH analysis of wheatgrass and its hybrids with common wheat.
Our analyses showed that individual Ae. crassa chromosomes differ in the number and combination of repeated families, their abundance, and physical distribution. Chromosome 5D1, comprising nearly all known repetitive DNA families, showed the highest diversity, while only few variants were recorded for 2D1 and 7Xcr.
Five variants of repetitive DNAs hybridized exceptionally to terminal chromosome regions – CL239, CL244, CL257, CL258, and CL2 (only in Th. bessarabicum). One of the newly discovered Ae. crassa repeats, CL261, was localized in pericentromeric regions of several chromosome pairs of wheat, Ae. crassa, and Ae. tauschii. Bioinformatically, CL261 sequence was assigned to satellites (low probability) with the length of 553 nt (Supplementary Table S3). Satellite repeats are found in pericentromeric chromosome regions of many plant and animal species (Miller et al., 1998; Hudakova et al., 2001; Kishii et al., 2001; Sharma et al., 2013; Teo et al., 2013; Su et al., 2019) and CL261 could also belong to centromere-specific repeat family. CL261 showed homology to Th. bessarabicum repeat CL198, which hybridizes to pericentromeric regions of Th. ponticum chromosomes (Nikitina et al., 2020) and centromere-specific repeat CentT550 identified in common wheat (Su et al., 2019).
The following sequences occurred in subterminal and/or interstitial positions of several Ae. crassa chromosomes: CL131, CL170, CL209, CL219, CL228, CL232, CL131, CL241, and CL27_232. Particular sites, for example, proximal parts of 4D1S, 5D1S, and 7D1S, were especially enriched with satellite DNA and may contain several different variants of repeats. Thus, large signals of GAAn (Abdolmalaki et al., 2019; Badaeva et al., 2021), CL170, CL241, and CL18 overlapping with faint signals of pTa-713 and pTa-k566 probes are found in the short arm of 5D1. C-banding analysis (Badaeva et al., 1998, 2002) revealed two distinct heterochromatin blocks in this chromosomal region. Another satellite-rich cluster was detected in the proximal part of chromosome 7D1S, which possessed an abundant quantity of CL219 and pTa-713 repeats and much lesser amounts of CL232, CL18, and pTa-k566 sequences. As in a previous case, a large Giemsa band was observed in the respective chromosome region (Badaeva et al., 1998, 2002).
Repeats CL27_232 and CL241 found in Triticum-Aegilops species occupied a similar position on the orthologous chromosomes. CL27_232 clusters appeared in the short arm of 3D of common wheat, Ae. crassa, and Ae. tauschii. Although few additional minor signals have been detected on chromosomes 4D1 (T2) and 5Xcr of 6x Ae. crassa, CL27_232 repeat can be used as a marker of the short arm of 3D chromosome. Such chromosome-specific markers based on single-copy genes (Danilova et al., 2014) or pooled oligo-probes (Li et al., 2021) have been developed for wheat and found broad application in phylogenetic studies of the Triticeae (Danilova et al., 2017). Repeat CL241 was found in the short arms of all homoeologous group 5 chromosomes of Ae. crassa and Ae. tauschii. This syntheny however was disturbed in wheat and Thinopyrum species, which possessed clear CL241 sites on A, B, and Jb (sub)genome chromosomes, but in different positions.
Two sequences, CL170 and CL228, occurred mainly in the D1 subgenome. Hybridization pattern of CL170 probe was highly conserved across the D (sub)genomes of wheat and Aegilops species (Figure 5), whereas more intense hybridization sites of CL228 on Ae. crassa chromosomes compared to Ae. tauschii or common wheat (Figure 6) suggested sequence amplification following speciation of this amphiploid. Interestingly, labeling patterns of CL170, CL244 probes on Ae. tauschii subsp. strangulata and D subgenome chromosomes of common wheat differed from that in the D1 and D2 chromosomes, suggesting that these differences may exist in the genome of diploid progenitor. This assumption is legitimate, as many molecular (Luo et al., 2007; Wang et al., 2013; Li et al., 2018; Singh et al., 2019; Gaurav et al., 2021) and cytogenetic (Majka et al., 2017; Zhao et al., 2018; Badaeva et al., 2019a; Ebrahimzadegan et al., 2021) data supported significant genome divergence between two Ae. tauschii subspecies.
Two repeats were more abundant in the Xcr subgenome, CL131 homologous to pTa-465 and P332 homologous to pTa-k566 (Komuro et al., 2013), but occurred also in D1 and D2 subgenomes (Figures 7; 8, Supplementary Figures S3, S4). Both probes hybridized to subtelomeric and interstitial chromosome regions and showed different labeling patterns between D genomes of Ae. tauschii subsp. strangulata and tauschii vs. D1 and D2 subgenome chromosomes of Ae. crassa. The CL131 is, according to our current results, more abundant in the Xcr rather than D1 subgenome of Ae. crassa (Figure 7). Thus, three prominent CL131 clusters were detected on 2D1 and another one, overlapping with CL228, on 3D1L. Such prominent CL131 (pTa-465) clusters were not found in wheat (Komuro et al., 2013) or in subsp. strangulata (accession K-112), but smaller pTa-465 sites in similar positions were recorded in several Ae. tauschii accessions by Majka et al. (2017). Unfortunately, these authors did not provide full taxonomic description on the material they used.
Most CL131 sites on Xcr chromosomes were small and only 6Xcr possessed large signals comparable in size with signals on chromosome 4A of wheat. We found differences between 4x and 6x accessions in the size of CL131 sites on chromosomes CcrS and FcrL, which are probably not related to the formation of hexaploid form, whereas modifications of labeling patterns of FcrS and 4D1S chromosomes were likely caused by species-specific translocation T2. No traces of CL131 repeat were recorded on Th. bessarabicum chromosomes by FISH, which corresponds to qPCR results.
According to bioinformatics and qPCR the novel CL239 repeat is absent in the Ae. tauschii genomes. Indeed, FISH detected CL239 sites on most Xcr chromosomes, but only on two D1 subgenome chromosomes. In addition to Ae. crassa, CL239 was discovered in Th. bessarabicum, but it was absent in the D (sub)genomes of wheat and Ae. tauschii, and the D2 subgenome of 6x Ae. crassa, in agreement with results obtained by other methods. Based on these observations we suggest that CL239 sequence was probably contributed to Ae. crassa by the putative progenitor of Xcr subgenome, and it spread to the D1 subgenome chromosomes following species evolution. Alternatively, this sequence could present in a putative progenitor of the D1 subgenome, but after formation of primary allopolyploid was amplified in polyploid descendant, but eliminated from diploid ancestor. Amplification of CL131 and CL239 repeats in Ae. crassa can be suggested based on the lack of this sequence in Ae. tauschii genome. Comparison with 6x Ae. crassa however showed that this is true only for CL239, because most D2 subgenome chromosomes had a CL131 hybridization pattern similar with the D1 chromosomes.
Although CL232 repeat hybridized only on a few Ae. crassa chromosomes, it helped us to shed some light on the structure of chromosome 2D1, the origin of which is still highly speculative. Labeling patterns of all repeats used in our current and in all previous studies (Badaeva et al., 1998, 2002, 2021; Abdolmalaki et al., 2019) showed that most drastic changes occurred in this Ae. crassa chromosome as compared to Ae. tauschii. Chromosome 2D1 was assigned to the D subgenome because i) it showed distinct hybridization with the D genome-specific probe pAs1; and ii) labeling pattern of CL232 and CL239 probes on the long arm of 2D1 were almost identical to the long arm of 2D orthologs of Ae. tauschii, the D2 subgenome of 6x Ae. crassa, and D subgenome of common wheat. At the same time, 2D1 lacks hybridization sites of CL170, CL228, and pTa-k566, and acquires three CL131 clusters, which point to significant structural rearrangement of Ae. crassa chromosome 2D1.
Comparison of the results of the preliminary estimation of the repeats abundance using qPCR and the localization of the identified repeats to the chromosomes of the studied species generally showed the collinearity of the results obtained by these methods. Thus, CL8, CL18, CL60, and CL148 demonstrated a high copy number and noticeable dispersed signals on the chromosomes in all studied species. CL3, CL261, CL27_232, and CL170 also showed high copy number in all studied species according to qPCR results, and discrete pattern of hybridization on chromosomes. It can also be noted that the species specificity revealed by the qPCR results was also observed when analyzing the FISH results: CL257 and CL258 are found only in Ae. crassa, CL209 and CL219 in Ae. crassa and T. aestivum, CL228 and CL232 in Ae. crassa, Ae. tauschii and less in wheat, absence of CL239 in wheat and Ae. tauschii, absence of CL244 in Ae. tauschii, and high abundance and bright signals on the Th. bessarabicum chromosomes.
However, we also revealed differences between qPCR data and localization of repeats on chromosomes using FISH for CL193, CL2, and CL131. Although CL193 showed a high copy number in all species in the qPCR experiment, it hybridized to one pair of Th. bessarabicum chromosomes only. CL193 is homologous to the dispersed-clustered FAT and P631 repeats, as well as to the microsatellite-related FISH-positive repetitive sequence pTa-451 (Supplementary Table S9). We can assume that the CL193 repeat is quite strongly dispersed throughout the chromosomes without cluster localization, so that FISH is not able to identify its presence. On the other hand, qPCR in this case could give false positive results due to primer annealing and non-specific amplification on other repeating elements. CL2, homologous to terminal repeats of various Triticeae species (Supplementary Table S9), in our experiments, was absent in the studied species by qPCR, but showed bright terminal signals on the Th. bessarabicum chromosomes. Similarly, CL131 also showed extremely low abundance in qPCR, but showed clear local signals on the chromosomes of all species studied. In these cases, qPCR showed false-negative results, which can be explained by the low efficiency of the selected primers or difficult amplification regions for the polymerase. Nevertheless, despite individual cases, in general, the qPCR method has demonstrated its suitability for preliminary screening of novel DNA repeats for their application as chromosomal markers.
Evolutionary changes of repetitive DNA families in Aegilops crassa genome
The five repeats we identified, CL2, CL239, CL244, CL257, and CL258, are distinguished by their conservative subtelomeric localization. Sequence analysis showed that CL2, which was found in Th. bessarabicum genome, is homologous to terminal/subterminal repeats found in Th. bessarabicum, Leymus racemosus, Dasypyrum villosum, and Secale cereale (Wilkes et al., 1995; Francki et al., 1997; Kishii et al., 1999; Pace et al., 2011; Du et al., 2017; Chen et al., 2019), as well as to CL244; the CL244 repeat itself showed homology to terminal repeats of Th. bessarabicum, Ae. speltoides (Spelt52.1), and S. cereale (including pSc200; Appels et al., 1981; Vershinin et al., 1995; Salina et al., 2004a; Du et al., 2017; Chen et al., 2019), whereas CL239 is homologous to Spelt1-like repeat Tri-MS-6 (EF469549.1; Supplementary Table S9). Subtelomeric repeats are localized in terminal heterochromatic blocks and their copy number may vary between accessions and between species and can change during evolution. Moreover, even homologous FISH probes may give different signals (or no signals at all) in the same species. For example, oligo-probes DP4J20764 and DP4J30938, which are homologous to CL2, gave signals on chromosomes of Th. bessarabicum and wheat, while the DP4J31304 was found only in chromosomes of Th. bessarabicum (Du et al., 2017). CL2 hybridized only to Th. bessarabicum chromosomes, while CL244 and CL239 showed hybridization to chromosomes of Th. bessarabicum, as well as the Xcr and D1 subgenome chromosomes of Ae. crassa, but absent in Ae. tauschii. Judging from the wide range of genomes in which the terminal repeats CL2, CL239, and CL244 (V, R, J, S) occur, we can propose their antiquity and even possible common origin of CL2 and CL244. The CL2, CL239, and CL244 repeats can probably be amplified during Th. bessarabicum speciation, but CL2 and CL239 were totally eliminated from Aegilops and Triticum, while CL244 was retained in the S, R, Xcr genomes and the putative ancestor of wheat B subgenome. Signals on D and A subgenome chromosomes of common wheat may appear due to the transfer of CL244 repeat from B to the A subgenome chromosomes, while CL239 and CL244 may be transferred from Xcr to the D1 subgenome chromosomes of Ae. crassa after polyploidization, as a result of coevolution of subgenomes, as was supposedly for terminal repeats Spelt1 and Spelt52 (Zoshchuk et al., 2007; Raskina et al., 2011).
Subtelomeric repeats play an important role in recognition of homologous chromosomes during meiosis (Corredor et al., 2007; Aguilar and Prieto, 2020). We found CL257 and CL258 repeats only in the D1 subgenome of the Ae. crassa (4x and 6x) on chromosomes belonging to genetic groups 1 and 5, respectively, which may indirectly indicate their role in the recognition of these chromosomes during meiosis and promote their differentiation from homoeologues. In turn, CL244 forms signals of varying intensity in termini of several Xcr and D1 chromosomes, which may also indirectly indicate their putative involvement in chromosome recognition in meiosis.
CL244, which was filtered bioinformatically as the putative Xcr subgenome-specific repeat, may also exhibit partial sequence elimination in Ae. crassa. Only a few faint FISH signals were observed on Ae. crassa and common wheat chromosomes, and it was totally absent in Ae. tauschii, which agrees with qPCR results. Despite some polymorphisms in CL244 location, the signals were always found in subtelomeric regions of wheat and Aegilops chromosomes, and most chromosomes carrying CL244 sites belong to subgenomes other than D/ D1. According to both FISH and qPCR analyses, CL244 is highly abundant in Th. bessarabicum, which presumed that this repeat was inherited from an ancient grass ancestor and remained mainly in Jb and Xcr (sub)genomes.
Only two repetitive DNA families analyzed in our study, CL257 and CL258, were found to be species-specific. Each of them was mapped to a single Ae. crassa chromosome, 5D1 and 1D1, respectively, and in both cases hybridization signals had terminal location. This is not surprising because many pieces of experimental evidence prove that species-specific repeats are often accumulated in subtelomeric regions of plant chromosome, which comprise rapidly evolving families of satellite repeats (Anamthawat-Jonsson and Heslop-Harrison, 1993; Salina et al., 2004b; Lim et al., 2005; Sharma and Raina, 2005; González-García et al., 2006). Bioinformatically, CL257 and CL258 were classified as “true satellites,” which occur in genome of Ae. crassa with the same frequency of 0.019% (Supplementary Table S3). Real-time qPCR confirmed their presence in both 4x and 6x Ae. crassa and absence in other species (Supplementary Table S8). None of the methods we used here revealed these repeats in other species, and no hits have been found in NCBI database. Based on these observations we assume that these two repeats emerged de novo in the D1 subgenome at the stage of formation of primary amphiploids or during subsequent evolution of 4x Ae. crassa. Probably owing to this novelty, bioinformatics attributed the CL257 repeat to the Xcr subgenome, although physically it localizes on the D1 chromosome that emphasize the necessity of complex approaches in repeatome studies. The emergence of five novel Ae. crassa-specific repetitive DNA sequences was earlier documented by Dubkovsky and Dvořák (1995), however, the relationships of these repeats with CL257 and CL258 remain not known. No differences in CL257 and CL258 signal intensities have been observed between 4x and 6x Ae. crassa, suggesting that hexaploidization did not cause significant changes in their content.
Formation of 4x Ae. crassa was associated with massive amplification of certain satellite repeats that may already pre-exist in the genome of the putative progenitor species. This mechanism was probably responsible for the emergence of two huge CL219 clusters on 7D1 in both 4x and 6x Ae. crassa. According to FISH, this sequence is lacking on the orthologous chromosomes of common wheat, Ae. tauschii, and on 7D2 of Ae. crassa. On the other side, CL219 was absent in Th. bessarabicum and Ae. tauschii, but occurred on chromosomes 2BS of common wheat and 1Xcr, 6Xcr, and 7Xcr of Ae. crassa. All these chromosomes belong to subgenomes other than D, and all sites were only found in subtelomeric regions. Based in these observations, we assumed that CL219 satellite was present in minor quantities in the putative Xcr subgenome progenitor. It could probably be transferred to the proximal region of 7D, which was highly enriched with other satellite sequences (e.g., pTa-713, pTa-k566, CL18, and CL232) via the mechanism of ectopic pairing (Schubert and Lysak, 2011; Waminal et al., 2021) soon after formation of primary Ae. crassa amphiploid. Genomic shock might cause massive amplification and spread of the repeat to other chromosomal sites, leading to the emergence of prominent CL219 clusters in proximal (7D1S) and distal (7D1L) chromosome regions.
We identified two repeats, CL261 and CL198, localized in the pericentromeric region, which are found to be homologous to the centromeric repeats CentT550 and 17–202 (Supplementary Table S9). Centromeres play an important role in the precise segregation of sister chromatids in mitosis and meiosis, mediated by the centromere-specific histone protein CENH3. Localization of this protein coincides with the centromeric arrangement of CentT550 repeats, which is characteristic mainly for the D subgenome of wheat (and Ae. tauschii), and CentT566, which is characteristic mainly for the B subgenome of wheat (and Ae. speltoides) and, probably, served as a source of these repeats in other subgenomes of bread wheat after polyploidization (Mach, 2019). Repeat 17–202 was found in the genome of Th. pontium and is localized to chromosomes of both wheat and wheatgrass (Nikitina et al., 2020). The CL261 repeat is localized mainly on the D1 chromosomes of 4x and 6x Ae. crassa and on the D subgenome chromosomes of common wheat. Its analogue, CL198, is localized on five pairs of Th. bessarabicum chromosomes. Thus, most likely we identified a new centromeric repeat that has a common origin with CentT550 and passed to both the J and D (sub)genomes of wheatgrass and Aegilops during evolution. Considering the importance of this repeat in ensuring proper meiosis, we can assume that the simultaneous presence of CentT550-like repeats in wheat and wheatgrass chromosomes of intergeneric hybrids may explain predominant introgressions of the J genome chromatin of Thinopyrum to the D subgenome of wheat.
We also identified a few chromosome-or group-specific repeats. Thus, CL241, which is homologous to Oligo-3A1 (Lang et al., 2018; Lang et al., 2019b) and Oligo-44 (Tang et al., 2018; Supplementary Table S9), was mapped to the same wheat or Ae. tauschii chromosomal regions as was described in literature. Their signals are predominantly found on group 5 chromosomes, either in the short [(sub)genomes D, Sl, Ss] or long [(sub)genomes S, B, A] arms. In Th. bessarabicum, we detected signals on two chromosome pairs (with one and two hybridization sites), while Lang et al. (2019b) identified two and four chromosomes carrying a single Oligo-3A1 site each in Th. ponticum and Th. intermedium, respectively. We did not find this repeat in the Xcr subgenome, Lang et al. (2019b) did not detect it in barley and Dasypyrum breviaristatum. Thus, we can assume that the Xcr subgenome donor diverged from a common ancestor after barley and D. breviaristatum, but prior to active amplification of this repeat in Triticum/Aegilops/Thinopyrum/D. villosum.
We can reach a similar conclusion considering that the CL170 repeat is absent in Xcr genome, but is abundant in the D (sub)genome of wheat and Ae. tauschii, Ae. crassa, and Th. bessarabicum. The homologous sequences were detected on chromosomes of Th. ponticum, Th. bessarabicum, Th. intermedium, A. cristatum, but they were absent in D. villosum and Pseudoroegneria spicata chromosomes (Supplementary Table S9). Probably, the putative Xcr genome progenitor diverged from a common ancestor after separation of the V and St genomes, and prior to the start of massive amplification of the CL170-like repeat in Triticum/Aegilops/Thinopyrum/A. cristatum.
Results obtained in our study showed that evolution of Ae. crassa was associated not only with amplification, but also with elimination of repetitive DNA sequences, as was described for other plant species (Dvořák and Zhang, 1992; Salina et al., 2004b; Adams and Wendel, 2005; Feldman and Levy, 2005; Kumar et al., 2010). For example, bioinformatic analysis failed to detect sequence AC4x_CL193_504nt in Th. bessarabicum Jb genome; qPCR showed its high abundance in genomes of all analyzed species, while FISH detected a single CL193 signal on one of the two homologs 7 J, and no signals were found in Ae. crassa. Probably, this sequence was present in the genome of the putative progenitor(s) of Ae. crassa, but it was eliminated during evolution of Aegilops species. Ae. crassa, being the most ancient polyploid species in the genus Aegilops, may retain a tracing amount of this repeat, which cannot be detected at chromosomal level by FISH.
Thus, if the full-length genome assemblies are unavailable, identification of new repetitive DNA sequences in particular species based on results of low-coverage NGS sequencing makes it possible to broaden the pool of data and get closer to their analysis at the level of “omics” technologies with lesser time and financial expenses.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, ON872662–ON872692.
Author contributions
EB, GK, and MD conceived and designed the experiments, analyzed data, interpreted results, wrote and edited the manuscript. EB, VS, TK, and AY designed and conducted the cytogenetic experiments and analyzed chromosome images. NC and MB provided and botanically verified plant material for analysis. SS designed and synthesized oligo-probes for FISH analyses. EN, DU, and AE performed bioinformatics analysis. AK performed and analyzed qPCR experiments. EB, OR, and PK wrote the first manuscript version. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Russian Science Foundation, grant number 21-16-00123.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.980764/full#supplementary-material
References
Abdolmalaki, Z., Mirzaghaderi, G., Mason, A. S., and Badaeva, E. D. (2019). Molecular cytogenetic analysis reveals evolutionary relationships between polyploid Aegilops species. Plant Syst. Evol. 305, 459–475. doi: 10.1007/s00606-019-01585-3
Adams, K. L., and Wendel, J. F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141. doi: 10.1016/j.pbi.2005.01.001
Aguilar, M., and Prieto, P. (2020). Sequence analysis of wheat subtelomeres reveals a high polymorphism among homoeologous chromosomes. Plant Genome 13:e2006. doi: 10.1002/tpg2.20065
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Anamthawat-Jonsson, K., and Heslop-Harrison, J. S. (1993). Isolation and characterisation of genome-specific DNA sequences in Triticeae species. Mol. Gen. Genet. 240, 151–158. doi: 10.1007/BF00277052
Andrews, S. (2010). "FASTQC. A quality control tool for high throughput equence data". 08.01.19 ed. (Babraham Institute, U.K.: Babraham Bioinformatics).
Appels, R., Dennis, E. S., Smyth, D. R., and Peacock, W. J. (1981). Two repeated DNA sequences from the heterochromatic regions of rye (Secale cereale) chromosomes. Chromosoma 84, 265–277. doi: 10.1007/bf00399137
Avni, R., Lux, T., Minz-Dub, A., Millet, E., Sela, H., Distelfeld, A., et al. (2022). Genome sequences of three Aegilops species of the section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J. 110, 179–192. doi: 10.1111/tpj.15664
Badaeva, E. D., Ruban, A. S., Alieva-Schnorr, L., Municio, C., Hesse, S., and Houben, A. (2017). “In situ hybridization to plant chromosomes” in Fluorescence in situ hybridization (FISH) – Application guide. ed. T. Liehr . 2nd ed (Berlin: Springer), 477–494. doi: 10.1007/978-3-662-52959-1
Badaeva, E., Zoshchuk, S., Paux, E., Gay, G., Zoshchuk, N., Roger, D., et al. (2010). Fat element—a new marker for chromosome and genome analysis in the Triticeae. Chromosom. Res. 18, 697–709. doi: 10.1007/s10577-010-9151-x
Badaeva, E. D., Amosova, A. V., Muravenko, O. V., Samatadze, T. E., Chikida, N. N., Zelenin, A. V., et al. (2002). Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant Syst. Evol. 231, 163–190. doi: 10.1007/s006060200018
Badaeva, E. D., Chikida, N. N., Belousova, M. K., Ruban, A. S., Surzhikov, S. A., and Zoshchuk, S. A. (2021). A new insight on the evolution of polyploid Aegilops species from the complex crassa: molecular-cytogenetic analysis. Plant Syst. Evol. 307:3. doi: 10.1007/s00606-020-01731-2
Badaeva, E. D., Fisenko, A. V., Surzhikov, S. A., Yankovskaya, A. A., Chikida, N. N., Zoshchuk, S. A., et al. (2019a). Genetic heterogeneity of a diploid grass Ae. tauschii revealed by chromosome banding methods and electrophoretic analysis of the seed storage proteins (gliadins). Russ. J. Genet. 55, 1315–1329. doi: 10.1134/S1022795419110024
Badaeva, E. D., Friebe, B., and Gill, B. S. (1996). Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome 39, 293–306. doi: 10.1139/g96-040
Badaeva, E. D., Friebe, B., Zoshchuk, S. A., Zelenin, A. V., and Gill, B. S. (1998). Molecular cytogenetic analysis of tetraploid and hexaploid Ae. crassa. Chromosom. Res. 6, 629–637. doi: 10.1023/A:1009257527391
Badaeva, E. D., Surzhikov, S. A., and Agafonov, A. V. (2019b). Molecular-cytogenetic analysis of diploid wheatgrass Th. bessarabicum (Savul and Rayss). A. Löve. Comp Cytogenet. 13, 389–402. doi: 10.3897/CompCytogen.v13i4.36879
Bedbrook, R. J., Jones, J., O'Dell, M., Thompson, R. J., and Flavell, R. B. (1980). A molecular description of telomeric heterochromatin in Secale species. Cells 19, 545–560. doi: 10.1016/0092-8674(80)90529-2
Bernhardt, N., Brassac, J., Dong, X., Willing, E.-M., Poskar, C. H., Kilian, B., et al. (2020). Genome-wide sequence information reveals recurrent hybridization among diploid wheat wild relatives. Plant J. 102, 493–506. doi: 10.1111/tpj.14641
Bernhardt, N., Brassac, J., Kilian, B., and Blattner, F. R. (2017). Dated tribe-wide whole chloroplast genome phylogeny indicates recurrent hybridizations within Triticeae. BMC Evol. Biol. 17:141. doi: 10.1186/s12862-017-0989-9
Buels, R., Yao, E., Diesh, C. M., Hayes, R. D., Munoz-Torres, M., Helt, G., et al. (2016). JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 17:66. doi: 10.1186/s13059-016-0924-1
Bushnell, B. (2014). "BBMap: a fast, accurate, splice-aware aligner". Lawrence Berkeley National Laboratory United States. Department of Energy. Office of Scientific Technical Information: United States. Department of Energy. Office of Science.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformat. 10:421. doi: 10.1186/1471-2105-10-421
Chen, J., Tang, Y., Yao, L., Wu, H., Tu, X., Zhuang, L., et al. (2019). Cytological and molecular characterization of Th. bessarabicum chromosomes and structural rearrangements introgressed in wheat. Mol. Breed. 39:146. doi: 10.1007/s11032-019-1054-8
Chen, Q., Conner, R., Laroche, A., and Ahmad, F. (2001). Molecular cytogenetic evidence for a high level of chromosome pairing among different genomes in Triticum aestivum–Thinopyrum intermedium hybrids. Theor. Appl. Genet. 102, 847–852. doi: 10.1007/s001220000496
Coordinators, N. R. (2012). Database resources of the National Center for biotechnology information. Nucl. Ac. Res. 41, D8–D20. doi: 10.1093/nar/gks1189
Corredor, E., Lukaszewski, A. J., Pachon, P., Allen, D. C., and Naranjo, T. (2007). Terminal regions of wheat chromosomes select their pairing partners in meiosis. Genetics 177, 699–706. doi: 10.1534/genetics.107.078121
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008
Danilova, T. V., Akhunova, A. R., Akhunov, E. D., Friebe, B., and Gill, B. S. (2017). Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Plant J. 92, 317–330. doi: 10.1111/tpj.13657
Danilova, T. V., Friebe, B., and Gill, B. S. (2014). Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor. Appl. Genet. 127, 715–730. doi: 10.1007/s00122-013-2253-z
Divashuk, M., Karlov, G., and Kroupin, P. (2019). Copy number variation of transposable elements in Thinopyrum intermedium and its diploid relative species. Plan. Theory 9:15. doi: 10.3390/plants9010015
Du, P., Zhuang, L. F., Wang, Y. Z., Yuan, L., Wang, Q., Wang, D. R., et al. (2017). Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Th. bessarabicum chromosomes. Genome 60, 93–103. doi: 10.1139/gen-2016-0095
Dubkovsky, J., and Dvořák, J. (1995). Genome identification of the Triticum crassum complex (Poaceae) with the restriction patterns of repeated nucleotide sequences. Am. J. Bot. 182, 131–140.
Dvořák, J. (2009). “Triticeae genome structure and evolution,” in Genetics and genomics of the Triticeae. 1st Edn. eds. G. J. Muehlbauer and C. Feuillet (New York: Springer), 685–711.
Dvořák, J. (1998). “Genome analysis in the Triticum-Aegilops alliance.” in: Proceedings of the 9th international wheat genetics symposium, 2–7 August 1998, ed. A.E. Slinkard: Printcrafters Inc.), 8–11.
Dvořák, J., and Zhang, H. B. (1992). Application of molecular tools for study of the phylogeny of diploid and polyploid taxa in Triticeae. Hereditas 116, 37–42.
Dvořák, J., Luo, M. C., Yang, Z. L., and Zhang, H. B. (1998). The structure of the Ae. tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 97, 657–670.
Ebrahimzadegan, R., Orooji, F., Ma, P., and Mirzaghaderi, G. (2021). Differentially amplified repetitive sequences among Ae. tauschii subspecies and genotypes. Front. Plant Sci. 12:716750. doi: 10.3389/fpls.2021.716750
Edet, O. U., Gorafi, Y. S. A., Nasuda, S., and Tsujimoto, H. (2018). DArTseq-based analysis of genomic relationships among species of tribe Triticeae. Sci. Rep. 8:16397. doi: 10.1038/s41598-018-34811-y
Feldman, M., and Levy, A. A. (2005). Allopolyploidy - a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109, 250–258. doi: 10.1159/000082407
Francki, M. G., Crasta, O. R., Sharma, H. C., Ohm, H. W., and Anderson, J. M. (1997). Structural organization of an alien Thinopyrum intermedium group 7 chromosome in US soft red winter wheat (Triticum aestivum L.). Genome 40, 716–722. doi: 10.1139/g97-794
Garrido-Ramos, M. A. (2015). Satellite DNA in plants: more than just rubbish. Cytogenet. Genome Res. 146, 153–170. doi: 10.1159/000437008
Gaurav, K., Arora, S., Silva, P., Sánchez-Martín, J., Horsnell, R., Gao, L., et al. (2021). Population genomic analysis of Ae. tauschii identifies targets for bread wheat improvement. Nat. Biotech 40, 422–431. doi: 10.1038/s41587-021-01058-4
Gill, B. S., Friebe, B., and Endo, T. R. (1991). Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34, 830–839. doi: https://doi.org/10.1139/g95-030
González-García, M., González-Sánchez, M., and Puertas, M. J. (2006). The high variability of subtelomeric heterochromatin and connections between nonhomologous chromosomes, suggest frequent ectopic recombination in rye meiocytes. Cytogenet. Genome Res. 115, 179–185. doi: 10.1159/000095240
Grewal, S., Yang, C., Edwards, S. H., Scholefield, D., Ashling, S., Burridge, A. J., et al. (2018). Characterisation of Th. bessarabicum chromosomes through genome-wide introgressions into wheat. Theor. Appl. Genet. 131, 389–406. doi: 10.1007/s00122-017-3009-y
Guo, J., Yu, X., Yin, H., Liu, G., Li, A., Wang, H., et al. (2016). Phylogenetic relationships of Thinopyrum and Triticum species revealed by SCoT and CDDP markers. Plant Syst. Evol. 302, 1301–1309. doi: 10.1007/s00606-016-1332-4
He, R., Chang, Z., Yang, Z., Yuan, Z., Zhan, H., Zhang, X., et al. (2009). Inheritance and mapping of powdery mildew resistance gene Pm43 introgressed from Thinopyrum intermedium into wheat. Theor. Appl. Genet. 118, 1173–1180. doi: 10.1007/s00122-009-0971-z
Hemleben, V., Kovarik, A., Torres-Ruiz, R. A., Volkov, R. A., and Beridze, T. (2007). Plant highly repeated satellite DNA: molecular evolution, distribution and use for identification of hybrids. Syst. Biodivers. 5, 277–289. doi: 10.1017/S147720000700240X
Heslop-Harrison, J. S., and Schwarzacher, T. (2011). Organisation of the plant genome in chromosomes. Plant J. 66, 18–33. doi: 10.1111/j.1365-313X.2011.04544.x
Heslop-Harrison, J. S., Brandes, A., and Schwarzacher, T. (2003). Tandemly repeated DNA sequences and centromeric chromosomal regions of Arabidopsis species. Chromosom. Res. 11, 241–253. doi: 10.1023/A:1022998709969
Hudakova, S., Michalek, W., Presting, G. G., Hoopen, R. T., Santos, K. D., Jasencakova, Z., et al. (2001). Sequence organization of barley centromeres. Nucleic Acids Res. 29, 5029–5035. doi: 10.1093/nar/29.24.5029
Kato, A., Lamb, J. C., Albert, P. S., Danilova, T., Han, F., Gao, Z., et al. (2011). “Chromosome painting for plant biotechnology,” in Plant chromosome engineering. Methods in molecular biology. ed. J. A. Birchler (Humana Totowa, NJ, United States: Springer Protocols), 67–96.
Kato, A., Lamb, J. C., and Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. U. S. A. 101, 13554–13559. doi: 10.1073/pnas.0403659101
Kihara, H. (1949). Genomanalyse bei Triticum und Aegilops. IX. Systematischer Aufbau der Gattung Aegilops auf genomanalytischer Grundlage. Cytologia 14, 135–144.
Kihara, H. (1954). Considerations on the evolution and distribution of Aegilops species based on the analyser-method. Cytologia 19, 336–357.
Kihara, H. (1957). Completion of genome-analysis of three 6x species of Aegilops. Wheat Inf. Serv. 6:11.
Kihara, H. (1963). Nucleus and chromosome substitution in wheat and Aegilops. II. Chromosome substitution. Seiken Ziho 15, 13–23.
Kilian, B., Mammen, K., Millet, E., Sharma, R., Graner, A., Salamini, F., et al. (2011). “Aegilops,” in Wild crop relatives: Genomics and breeding resources. Cereals. ed. C. Kole (Berlin Heidelberg: Springer), 1–76.
Kimber, G., and Abu-Bakar, M. (1981). The genome relationships of Triticum dichasians and T. umbellulatum. Z. Pflanzenzucht. 87, 265–273.
Kimber, G., and Feldman, M. (1987). Wild wheat, an introduction. Columbia, Mo, U.S.A.: College of Agriculture University of Missouri.
Kimber, G., and Tsunewaki, K. 1988. “Genome symbols and plasma types in the wheat group”, in: Proceedings of the 7th international wheat genetics symposium, 13-19-July 1988. eds. T.E. Miller & R.M.D. Koebner: Bath Press, Avon, 1209–1210.
Kimber, G., and Zhao, Y. H. (1983). The D genome of the Triticeae. Can. J. Genet. Cytol. 25, 581–589.
Kishii, M. (2019). An update of recent use of Aegilops species in wheat breeding. Front. Plant Sci. 10:585. doi: 10.3389/fpls.2019.00585
Kishii, M., Nagaki, K., and Tsujimoto, H. (2001). A tandem repetitive sequence located in the centromeric region of common wheat (Triticum aestivum) chromosomes. Chromosom. Res. 9, 417–428. doi: 10.1023/A:1016739719421
Kishii, M., Tsujimoto, H., and Sasakuma, T. (1999). Exclusive localization of tandem repetitive sequences in subtelomeric heterochromatin regions of Leymus racemosus (Poaceae, Triticeae). Chromosom. Res. 7, 519–529. doi: 10.1023/A:1009285311247
Komuro, S., Endo, R., Shikata, K., and Kato, A. (2013). Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56, 131–137. doi: 10.1139/gen-2013-0003
Koo, D. H., Tiwari, V. K., Hřibová, E., Doležel, J., Friebe, B., and Gill, B. S. (2016). Molecular cytogenetic mapping of satellite DNA sequences in Aegilops geniculata and wheat. Cytogenet. Genome Res. 148, 314–321. doi: 10.1159/000447471
Kroupin, P., Kuznetsova, V., Romanov, D., Kocheshkova, A., Karlov, G., Dang, T., et al. (2019a). Pipeline for the rapid development of cytogenetic markers using genomic data of related species. Genes 10:113. doi: 10.3390/genes10020113
Kroupin, P. Y., Kuznetsova, V. M., Nikitina, E. A., Martirosyan, Y. T., Karlov, G. I., and Divashuk, M. G. (2019b). Development of new cytogenetic markers for Thinopyrum ponticum (Podp.) Z.-W. Liu & R.-C. Wang. Comp. Cytogenet. 13, 231–243. doi: 10.3897/CompCytogen.v13i3.36112
Kumar, S., Friebe, B., and Gill, B. S. (2010). Fate of Aegilops speltoides-derived, repetitive DNA sequences in diploid Aegilops species, wheat-Aegilops amphiploids and derived chromosome addition lines. Cytogenet. Genome Res. 129, 47–54. doi: 10.1159/000314552
Kuo, Y.-T., Ishii, T., Fuchs, J., Hsieh, W.-H., Houben, A., and Lin, Y.-R. (2021). The evolutionary dynamics of repetitive DNA and its impact on the genome diversification in the genus Sorghum. Front. Plant Sci. 12:729734. doi: 10.3389/fpls.2021.729734
Kuznetsova, V., Razumova, O., Karlov, G., Dang, T., Kroupin, P., and Divashuk, M. (2019). Some peculiarities in application of denaturating and non-denaturating in situ hybridization on chromosomes of cereals. Moscow Univer. Biol. Sci. Bul. 74, 75–80. doi: 10.3103/S0096392519020056
Lang, T., La, S., Li, B., Yu, Z., Chen, Q., Li, J., et al. (2018). Precise identification of wheat–Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 61, 177–185. doi: 10.1139/gen-2017-0229
Lang, T., Li, G., Wang, H., Yu, Z., Chen, Q., Yang, E., et al. (2019a). Physical location of tandem repeats in the wheat genome and application for chromosome identification. Planta 249, 663–675. doi: 10.1007/s00425-018-3033-4
Lang, T., Li, G., Yu, Z., Ma, J., Chen, Q., Yang, E., et al. (2019b). Genome-wide distribution of novel Ta-3A1 mini-satellite repeats and its use for chromosome identification in wheat and related species. Agronomy 9:60. doi: 10.3390/agronomy9020060
Li, C., Sun, X., Conover, J. L., Zhang, Z., Wang, J., Wang, X., et al. (2018). Cytonuclear coevolution following homoploid hybrid speciation in ae. tauschii. Mol. Biol. Evol. 36, 341–349. doi: 10.1093/molbev/msy215
Li, G., Wang, H., Lang, T., Li, J., La, S., Yang, E., et al. (2016). New molecular markers and cytogenetic probes enable chromosome identification of wheat-Thinopyrum intermedium introgression lines for improving protein and gluten contents. Planta 244, 865–876. doi: 10.1007/s00425-016-2554-y
Li, G., Zhang, T., Yu, Z., Wang, H., Yang, E., and Yang, Z. (2021). An efficient oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J. 105, 978–993. doi: 10.1111/tpj.15081
Li, L.-F., Zhang, Z.-B., Wang, Z.-H., Li, N., Sha, Y., Wang, X.-F., et al. (2022). Genome sequences of five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol. Plant 15, 488–503. doi: 10.1016/j.molp.2021.12.019
Lim, K. Y., Matyasek, R., Kovarik, A., Fulnecek, J., and Leitch, A. R. (2005). Molecular cytogenetics and tandem repeat sequence evolution in the allopolyploid Nicotiana rustica compared with diploid progenitors N. paniculata and N. undulata. Cytogenet. Genome Res. 109, 298–309. doi: 10.1159/000082413
Liu, G., and Zhang, T. (2021). Single copy oligonucleotide fluorescence in situ hybridization probe design platforms: development, application and evaluation. Int. J. Mol. Sci. 22:7124. doi: 10.3390/ijms22137124
Liu, L., Luo, Q., Li, H., Li, B., Li, Z., and Zheng, Q. (2018a). Physical mapping of the blue-grained gene from Thinopyrum ponticum chromosome 4Ag and development of blue-grain-related molecular markers and a FISH probe based on SLAF-seq technology. Theor. Appl. Genet. 131, 2359–2370. doi: 10.1007/s00122-018-3158-7
Liu, L., Luo, Q., Teng, W., Li, B., Li, H., Li, Y., et al. (2018b). Development of Thinopyrum ponticum-specific molecular markers and FISH probes based on SLAF-seq technology. Planta 247, 1099–1108. doi: 10.1007/s00425-018-2845-6
Liu, Q., Li, X., Zhou, X., Li, M., Zhang, F., Schwarzacher, T., et al. (2019). The repetitive DNA landscape in Avena (Poaceae): chromosome and genome evolution defined by major repeat classes in whole-genome sequence reads. BMC Plant Biol. 19:226. doi: 10.1186/s12870-019-1769-z
Liu, Z., Li, D., and Zhang, X. (2007). Genetic relationships among five basic genomes St, E, A, B and D in Triticeae revealed by genomic southern and in situ hybridization. J. Integr. Plant Biol. 49, 1080–1086. doi: 10.1111/j.1672-9072.2007.00462.x
Luo, M.-C., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Ae. tauschii. Nature 551:498. doi: 10.1038/nature24486
Luo, M. C., Yang, Z. L., You, F. M., Kawahara, T., Waines, J. G., and Dvořák, J. (2007). The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet. 114, 947–959. doi: 10.1007/s00122-006-0474-0
Macas, J., Novák, P., Pellicer, J., Čížková, J., Koblížková, A., Neumann, P., et al. (2015). In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS One 10:e0143424. doi: 10.1371/journal.pone.0143424
Mach, J. (2019). Polyploid pairing problems: how centromere repeat divergence helps wheat sort it all out. Plant Cell 31, 1938–1939. doi: 10.1105/tpc.19.00622
Majka, M., Kwiatek, M. T., Majka, J., and Wisniewska, H. (2017). Ae. tauschii accessions with geographically diverse origin show differences in chromosome organization and polymorphism of molecular markers linked to leaf rust and powdery mildew resistance genes. Front. Plant Sci. 8:1149. doi: 10.3389/fpls.2017.01149
Marcussen, T., Sandve, S. R., Heier, L., Spannagl, M., Pfeifer, M., Jakobsen, K. S., et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092:10.1126/science.1250092
Mehrotra, S., and Goyal, V. (2014). Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genom. Proteom. Bioinformat. 12, 164–171. doi: 10.1016/j.gpb.2014.07.003
Meng, Z., Zhang, Z., Yan, T., Lin, Q., Wang, Y., Huang, W., et al. (2018). Comprehensively characterizing the cytological features of Saccharum spontaneum by the development of a complete set of chromosome-specific oligo probes. Front. Plant Sci. 9:1624. doi: 10.3389/fpls.2018.01624
Miller, J. T., Jackson, S. A., Nasuda, S., Gill, B. S., Wing, R. A., and Jiang, J. (1998). Cloning and characterization of a centromere-specific repetitive DNA element from Sorghum bicolor. Theor. Appl. Genet. 96, 832–839.
Mirzaghaderi, G., Shahsevand Hassani, H., and Karimzadeh, G. (2010). C-banded karyotype of Th. bessarabicum and identification of its chromosomes in wheat background. Genet. Resour. Crop. Evol. 57, 319–324. doi: 10.1007/s10722-009-9509-0
Molnár, I., Cifuentes, M., Schneider, A., Benavente, E., and Molnár-Lang, M. (2011). Association between simple sequence repeat-rich chromosome regions and intergenomic translocation breakpoints in natural populations of allopolyploid wild wheats. Ann. Bot. 107, 65–76. doi: 10.1093/aob/mcq215
Molnár-Láng, M., Molnár, I., Szakács, É., Linc, G., and Bedö, Z. (2014). “Production and molecular cytogenetic identification of wheat-alien hybrids and introgression lines,” in Genomics of plant genetic resources. Volume 1. Managing, sequencing and mining genetic resources. eds. R. Tuberosa, A. Graner, and E. Frison (New York Heidelberg Dordrecht London: Springer), 255–284.
Murat, F., Armero, A., Pont, C., Klopp, C., and Salse, J. (2017). Reconstructing the genome of the most recent common ancestor of flowering plants. Nat. Genet. 49, 490–496. doi: 10.1038/ng.3813
Naghavi, M. R., Ranjbar, Z. A., Aghaei, M., Mardi, M., and Pirseyedi, S. M. (2009). Genetic diversity of Ae. crassa and its relationship with Ae. tauschii and the D genome of wheat. Cereal Res. Commun. 37, 159–167. doi: 10.1556/CRC.37.2009.2.2
Nikitina, E., Kuznetsova, V., Kroupin, P., Karlov, G. I., and Divashuk, M. G. (2020). Development of specific Thinopyrum cytogenetic markers for wheat-wheatgrass hybrids using sequencing and qPCR data. Int. J. Mol. Sci. 21:4495. doi: 10.3390/ijms21124495
Novák, P., Ávila Robledillo, L., Koblížková, A., Vrbová, I., Neumann, P., and Macas, J. (2017). TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 45:e111. doi: 10.1093/nar/gkx257
Novák, P., Neumann, P., Pech, J., Steinhaisl, J., and Macas, J. (2013). Repeat explorer: a galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29, 792–793. doi: 10.1093/bioinformatics/btt054
Pace, C. D., Vaccino, P., Cionini, P. G., Pasquini, M., Bizzarri, M., and Qualset, C. O. (2011). Dasypyrum. In wild crop relatives: Genomic and breeding resources (pp. 185–292). Springer, Berlin, Heidelberg.
Patokar, C., Sepsi, A., Schwarzacher, T., Kishii, M., and Heslop-Harrison, J. (2015). Molecular cytogenetic characterization of novel wheat-Th. bessarabicum recombinant lines carrying intercalary translocations. Chromosoma 125, 163–172. doi: 10.1007/s00412-015-0537-6
Pollak, Y., Zelinger, E., and Raskina, O. (2018). Repetitive DNA in the architecture, repatterning, and diversification of the genome of Aegilops speltoides Tausch (Poaceae, Triticeae). Front. Plant Sci. 9:1779. doi: 10.3389/fpls.2018.01779
Raskina, O., Barber, J. C., Nevo, E., and Belyayev, A. (2008). Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet. Genome Res. 120, 351–357. doi: 10.1159/000121084
Raskina, O., Brodsky, L., and Belyayev, A. (2011). Tandem repeats on an eco-geographical scale: outcomes from the genome of Aegilops speltoides. Chromosom. Res. 19, 607–623. doi: 10.1007/s10577-011-9220-9
Rayburn, A. L., and Gill, B. S. (1986). Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol. Biol. Report. 4, 102–109. doi: 10.1007/BF02732107
Rayburn, A. L., and Gill, B. S. (1987). Molecular analysis of the D-genome of the Triticeae. Theor. Appl. Genet. 73, 385–388.
Rogers, S., and Bendich, A. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76.
Said, M., Holušová, K., Farkas, A., Ivanizs, L., Gaál, E., Cápal, P., et al. (2021). Development of DNA markers from physically mapped loci in Aegilops comosa and Aegilops umbellulata using single-gene FISH and chromosome sequences. Front. Plant Sci. 12:689031. doi: 10.3389/fpls.2021.689031
Said, M., Hřibová, E., Danilova, T. V., Karafiátová, M., Čížková, J., Friebe, B., et al. (2018). The Agropyron cristatum karyotype, chromosome structure and cross-genome homoeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes. Theor. Appl. Genet. 131, 2213–2227. doi: 10.1007/s00122-018-3148-9
Salina, E. A., Adonina, I. G., Vatolina, T. Y., and Kurata, N. (2004a). A comparative analysis of the composition and organization of two subtelomeric repeat families in Aegilops speltoides Tausch. And related species. Genetica 122, 227–237. doi: 10.1007/s10709-004-5602-7
Salina, E. A., Numerova, O. M., Ozkan, H., and Feldman, M. (2004b). Alterations in subtelomeric tandem repeats during early stages of allopolyploidy in wheat. Genome 47, 860–867. doi: 10.1139/g04-044
Schubert, I., and Lysak, M. A. (2011). Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27, 207–216. doi: 10.1016/j.tig.2011.03.004
Shapiro, J. A., and von Sternberg, R. (2005). Why repetitive DNA is essential to genome function. Biol. Rev. 80, 227–250. doi: 10.1017/S1464793104006657
Sharma, A., Wolfgruber, T. K., and Presting, G. G. (2013). Tandem repeats derived from centromeric retrotransposons. BMC Genomics 14:142. doi: 10.1186/1471-2164-14-142
Sharma, S., and Raina, S. N. (2005). Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet. Genome Res. 109, 15–26. doi: 10.1159/000082377
Singh, A. K., Zhang, P., Dong, C., Li, J., Singh, S., Trethowan, R., et al. (2021). Generation and molecular marker and cytological characterization of wheat - Secale strictum subsp. anatolicum derivatives. Genome 64, 29–38. doi: 10.1139/gen-2020-0060
Singh, N., Wu, S., Tiwari, V., Sehgal, S., Raupp, J., Wilson, D., et al. (2019). Genomic analysis confirms population structure and identifies inter-lineage hybrids in Ae. tauschii. Front. Plant Sci. 10.9. doi: 10.3389/fpls.2019.00009
Song, Z., Dai, S., Bao, T., Zuo, Y., Xiang, Q., Li, J., et al. (2020). Analysis of structural genomic diversity in Aegilops umbellulata, Ae. markgrafii, Ae. comosa, and Ae. uniaristata by fluorescence in situ hybridization karyotyping. Front. Plant Sci. 11, 710–710. doi: 10.3389/fpls.2020.00710
Su, H., Liu, Y., Liu, C., Shi, Q., Huang, Y., and Han, F. (2019). Centromere satellite repeats have undergone rapid changes in polyploid wheat subgenomes. Plant Cell 31, 2035–2051. doi: 10.1105/tpc.19.00133
Tang, S., Qiu, L., Xiao, Z., Fu, S., and Tang, Z. (2016). New oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 7:118. doi: 10.3390/genes7120118
Tang, S., Tang, Z., Qiu, L., Yang, Z., Li, G., Lang, T., et al. (2018). Developing new oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Front. Plant Sci. 9:1104. doi: 10.3389/fpls.2018.01104
Tang, Z., Yang, Z., and Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Teo, C., Lermontova, I., Houben, A., Mette, M., and Schubert, I. (2013). De novo generation of plant centromeres at tandem repeats. Chromosoma 122, 233–241. doi: 10.1007/s00412-013-0406-0
Terachi, T., Ogihara, Y., and Tsunewaki, K. (1987). The molecular basis of genetic diversity among cytoplasms of Triticum and Aegilops. VI. Complete nucleotide sequences of the rbc L genes encoding H-and L-type rubisco large subunits in common wheat and Ae. crassa 4x. Jpn. J. Genet. 62, 375–387.
Tiwari, V. K., Wang, S., Danilova, T., Koo, D. H., Vrána, J., Kubaláková, M., et al. (2015). Exploring the tertiary gene pool of bread wheat: sequence assembly and analysis of chromosome 5Mg of Aegilops geniculata. Plant J. 84, 733–746. doi: 10.1111/tpj.13036
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer 3--new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Van Slageren, M.W. (1994). Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. Et Spach) Eig (Poaceae). Wageningen: Wageningen Agricultural University, Wageningen and ICARDA, Aleppo, Syria.
Vershinin, A. V., Schwarzacher, T., and Heslop-Harrison, J. S. (1995). The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell 7, 1823–1833. doi: 10.1105/tpc.7.11.1823
Waminal, N. E., Pellerin, R. J., Kang, S.-H., and Kim, H. H. (2021). Chromosomal mapping of tandem repeats revealed massive chromosomal rearrangements and insights into Senna tora dysploidy. Front. Plant Sci. 12:629898. doi: 10.3389/fpls.2021.629898
Wang, J., Luo, M.-C., Chen, Z., You, F. M., Wei, Y., Zheng, Y., et al. (2013). Ae. tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 198, 925–937. doi: 10.1111/nph.12164
Wang, L., Zhu, T., Rodriguez, J. C., Deal, K. R., Dubcovsky, J., McGuire, P. E., et al. (2021). Ae. tauschii genome assembly Aet v5.0 features greater sequence contiguity and improved annotation. G3 11:jkab325. doi: 10.1093/g3journal/jkab325
Wilkes, T. M., Francki, M. G., Langridge, P., Karp, A., Jones, R. N., and Forster, J. W. (1995). Analysis of rye B-chromosome structure using fluorescence in situ hybridization (FISH). Chromosom. Res. 3, 466–472. doi: 10.1007/BF00713960
Wu, D., Zhu, X., Tan, L., Zhang, H., Sha, L., Fan, X., et al. (2021). Characterization of each St and Y genome chromosome of Roegneria grandis based on newly developed FISH markers. Cytogenet. Genome Res. 161, 213–222. doi: 10.1159/000515623
Wu, T. D., and Nacu, S. (2010). Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881. doi: 10.1093/bioinformatics/btq057
Xi, W., Tang, S., Du, H., Luo, J., Tang, Z., and Fu, S. (2020). ND-FISH-positive oligonucleotide probes for detecting specific segments of rye (Secale cereale L.) chromosomes and new tandem repeats in rye. Crop J. 8, 171–181. doi: 10.1016/j.cj.2019.10.003
Xi, W., Tang, Z., Tang, S., Yang, Z., Luo, J., and Fu, S. (2019). New ND-FISH-positive oligo probes for identifying Thinopyrum chromosomes in wheat backgrounds. Int. J. Mol. Sci. 20:2031. doi: 10.3390/ijms20082031
Yaakov, B., Meyer, K., Ben-David, S., and Kashkush, K. (2013). Copy number variation of transposable elements in Triticum–Aegilops genus suggests evolutionary and revolutionary dynamics following allopolyploidization. Plant Cell Rep. 32, 1615–1624. doi: 10.1007/s00299-013-1472-8
Yu, G., Matny, O., Champouret, N., Steuernagel, B., Moscou, M. J., Hernández-Pinzón, I., et al. (2022). Aegilops sharonensis genome-assisted identification of stem rust resistance gene Sr62. Nat. Commun. 13:1607. doi: 10.1038/s41467-022-29132-8
Yu, Z., Wang, H., Xu, Y., Li, Y., Lang, T., Yang, Z., et al. (2019). Characterization of chromosomal rearrangement in new wheat—Thinopyrum intermedium addition lines carrying Thinopyrum—specific grain hardness genes. Agronomy 9:18. doi: 10.3390/agronomy9010018
Zagorski, D., Hartmann, M., Bertrand, Y. J. K., Paštová, L., Slavíková, R., Josefiová, J., et al. (2020). Characterization and dynamics of repeatomes in closely related species of Hieracium (Asteraceae) and their synthetic and apomictic hybrids. Front. Plant Sci. 11:591053. doi: 10.3389/fpls.2020.591053
Zhang, H. B., and Dvořák, J. (1992). The genome origin and evolution of hexaploid Triticum crassum and Triticum syriacum determined from variation in repeated nucleotide sequences. Genome 35, 806–814.
Zhao, L., Ning, S., Yi, Y., Zhang, L., Yuan, Z., Wang, J., et al. (2018). Fluorescence in situ hybridization karyotyping reveals the presence of two distinct genomes in the taxon Ae. tauschii. BMC Genomics 19:3. doi: 10.1186/s12864-017-4384-0
Zhao, X. P., Si, Y., Hanson, R. E., Crane, C. F., Price, H. J., Stelly, D. M., et al. (1998). Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Res. 8, 479–492. doi: 10.1101/gr.8.5.479
Zhao, Y. H., and Kimber, G. (1984). New hybrids with D genome wheat relatives. Genetics 106, 509–515. doi: 10.1093/genetics/106.3.509
Zoshchuk, S. A., Badaeva, E. D., Zoshchuk, N. V., Adonina, I. G., Shcherban, A. B., and Salina, E. A. (2007). Intraspecific divergence in wheats of the Timopheevi group as revealed by in situ hybridization with tandem repeats of the spelt 1 and spelt 52 families. Russ. J. Genet. 43, 636–645. doi: 10.1134/S1022795407060063
Keywords: Aegilops, shallow whole-genome sequencing, fluorescence in situ hybridization, chromosomes, satellite repeats, repeatome
Citation: Kroupin PY, Badaeva ED, Sokolova VM, Chikida NN, Belousova MK, Surzhikov SA, Nikitina EA, Kocheshkova AA, Ulyanov DS, Ermolaev AS, Khuat TML, Razumova OV, Yurkina AI, Karlov GI and Divashuk MG (2022) Aegilops crassa Boiss. repeatome characterized using low-coverage NGS as a source of new FISH markers: Application in phylogenetic studies of the Triticeae. Front. Plant Sci. 13:980764. doi: 10.3389/fpls.2022.980764
Edited by:
Vijay Kumar Tiwari, University of Maryland, College Park, College Park, United StatesReviewed by:
Zujun Yang, University of Electronic Science and Technology of China, ChinaMaría-Dolores Rey, University of Cordoba, Spain
Copyright © 2022 Kroupin, Badaeva, Sokolova, Chikida, Belousova, Surzhikov, Nikitina, Kocheshkova, Ulyanov, Ermolaev, Khuat, Razumova, Yurkina, Karlov and Divashuk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pavel Yu. Kroupin, cGF2ZWxrcm91cGluMTk4NUBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Pavel Yu. Kroupin
Pavel Yu. Kroupin Ekaterina D. Badaeva
Ekaterina D. Badaeva Victoria M. Sokolova
Victoria M. Sokolova Nadezhda N. Chikida4
Nadezhda N. Chikida4 Ekaterina A. Nikitina
Ekaterina A. Nikitina Alina A. Kocheshkova
Alina A. Kocheshkova Daniil S. Ulyanov
Daniil S. Ulyanov Aleksey S. Ermolaev
Aleksey S. Ermolaev Olga V. Razumova
Olga V. Razumova Anna I. Yurkina
Anna I. Yurkina Gennady I. Karlov
Gennady I. Karlov Mikhail G. Divashuk
Mikhail G. Divashuk
