- 1State Key Laboratory of Rice Biology, China National Rice Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou, China
- 2Hainan Yazhou Bay Seed Laboratory, Sanya, China
- 3Key Laboratory of Herbage and Endemic Crop Biology of Ministry of Education, Inner Mongolia Key Laboratory of Herbage and Endemic Crop Biotechnology, School of Life Sciences, Inner Mongolia University, Hohhot, China
Leaf inclination is a vital agronomic trait and is important for plant architecture that affects photosynthetic efficiency and grain yield. To understand the molecular mechanisms underlying regulation of leaf inclination, we constructed an auxin response factor (arf) rice mutant—osarf4—showing increased leaf inclination using CRISPR/Cas9 gene editing technology. OsARF4 encodes a nuclear protein that is expressed in the lamina joint (LJ) at different developmental stages in rice. Histological analysis indicated that an increase in cell differentiation on the adaxial side resulted in increased leaf inclination in the osarf4 mutants; however, OsARF4-overexpressing lines showed a decrease in leaf inclination, resulting in erect leaves. Additionally, a decrease in the content and distribution of indole-3-acetic acid (IAA) in osarf4 mutant led to a greater leaf inclination, whereas the OsARF4-overexpressing lines showed the opposite phenotype with increased IAA content. RNA-sequencing analysis revealed that the expression of genes related to brassinosteroid (BR) biosynthesis and response was different in the mutants and overexpressing lines, suggesting that OsARF4 participates in the BR signaling pathway. Moreover, BR sensitivity assay revealed that OsARF4-overexpressing lines were more sensitive to exogenous BR treatment than the mutants. In conclusion, OsARF4, a transcription factor in auxin signaling, participates in leaf inclination regulation and links auxin and BR signaling pathways. Our results provide a novel insight into l leaf inclination regulation, and have significant implications for improving rice architecture and grain yield.
Introduction
Plant architecture is governed by a set of complex agronomic traits that determine grain yield and is a pivotal target of artificial selection for rice domestication. Rice tillering pattern, plant height, and leaf inclination are important factors influencing the plant architecture (Van Camp, 2005). Leaf inclination (also called leaf angle), which is the angle between the leaf blade and the culm, is determined by the lamina joint (LJ). LJ, which is composed of a collar, ligules, and auricles, is a unique tissue that connects the leaf blade to leaf sheath in rice, and significantly contributes to the horizontal bending of the blade from the main axis. Division and expansion of the parenchyma and sclerenchyma cells in LJ affect its development, and leaf inclination is influenced by unequal cell division and elongation between the adaxial and abaxial sides of the LJ. Lack of cell expansion and longitudinal elongation of adaxial cells in the LJ results in erect leaves. In contrast, leaf inclination is enhanced by an increase in the expansion of cells on the adaxial surface (Duan et al., 2006). In rice, erect leaves have a higher leaf area index, which can increase the photosynthetic carbon assimilation rate by increasing light capture ability and nitrogen use efficiency to enhance total biomass and grain yield (Sakamoto et al., 2006). Therefore, revealing the molecular mechanism of genes involved in regulation of leaf inclination in rice is urgently required.
Phytohormones play an important role in regulating leaf inclination by changing the cytological structure of LJ. Brassinosteroids (BRs), plant-specific steroid hormones, have a significant effect on the growth and development of the LJ. In rice, studies have shown that BR promotes division and elongation of parenchyma cells at the adaxial side of the LJ and proliferation of sclerenchyma cells at the abaxial side; this unbalanced development between the adaxial and abaxial cells of the LJ leads to leaf bending (Tanabe et al., 2005; Zhang L. Y. et al., 2009; Sun et al., 2015). Several studies have indicated that decreased BR biosynthesis and signal transduction could decrease leaf inclination, whereas excess BR results in increased leaf inclination (Zhang et al., 2014). The mutation of several cytochrome P450 genes involved in BR biosynthesis, such as OsD2, OsD11, and OsDWARF (Hong et al., 2003; Sakamoto et al., 2006; Li et al., 2013), causes shortening of parenchyma cells in the LJ, contributing to reduced leaf inclination and formation of erect leaves. Defective BR signaling genes—BAK1 and BZR1—also result in decreased leaf inclination (Bai et al., 2007; Li et al., 2009). RLA1/SMOS1 (REDUCED LEAF ANGLE1/ SMALL ORGAN SIZE1) acts as an integrator, functioning with OsBZR1 and GSK2, and plays an important role in BR signal transduction and leaf inclination regulation in rice (Qiao et al., 2017). Furthermore, ILI1 (INCLINATION1) and IBH1 (ILI1 binding bHLH) antagonistically regulate the elongation of parenchyma cells in the LJ by interacting with the transcription factor OsBZR1 involved in BR signaling (Zhang L. Y. et al., 2009). In addition, rice U-type cyclin CYCU4; 1 regulates proliferation of sclerenchyma cells on the abaxial side of LJ in the BR-regulated pathway, which plays an important role in promoting the erectness of leaves (Sun et al., 2015). These results suggest that leaf inclination is closely related to BR biosynthesis and signal transduction in rice.
In addition to BR, auxin has been shown to play a negative role in controlling leaf inclination. Studies have shown that reducing auxin levels or signal transduction increases leaf inclination by promoting the elongation of parenchyma cells on the adaxial surface of the LJ (Du et al., 2012; Zhao et al., 2013; Zhang et al., 2015). Furthermore, loss of FIB (Fish Bone) function could reduce the endogenous indole acetic acid (IAA) content and polar auxin transport activity, leading to increased bending of the LJ (Yoshikawa et al., 2014). Auxin early response genes, such as AUX/IAA and GH3 family genes, the auxin receptor TIR1 (TRANSPORT INHIBITOR RESPONSE 1), and auxin response factor (ARF), have been reported to be associated with regulation of leaf inclination. The auxin response factor OsARF1 can inhibit the expression of OsGH3s, which encode IAA amino acid synthases by interacting with the repressor OsIAA1 (AUXIN/INDOLE3-ACETIC ACID 1) in the process of leaf inclination regulation (Wang et al., 2007). Additionally, overexpression of OsIAA1 results in an increased leaf angle (Song et al., 2009). Recent studies have shown that OsIAA6 regulates leaf inclination by inhibiting auxin signaling through interacting with OsARF1 in rice (Xing et al., 2022). The decrease in IAA content promotes elongation of parenchyma cells on the adaxial surface of the LJ, which is consistent with the result that the increase in GH3 expression levels increases leaf inclination. The increase in GH3–5 expression levels and the decrease in free IAA content also lead to increased leaf inclination in OsARF19-overexpressing rice plants (Zhang et al., 2015). LC1 (LEAF INCLINATION 1) encodes OsGH3-1 (an IAA amino synthetase) and promotes cell elongation at the LJ by decreasing the auxin content in rice (Zhao et al., 2013). LC3 (LEAF INCLINATION 3) interacts with LIP1 (LC3-INTERACTING PROTEIN 1) to synergistically suppress auxin signaling, thus controlling leaf inclination (Chen et al., 2018). LPA1 (LOOSE PLANT ARCHITECTURE 1) influences the polar transport of auxin by regulating the expression of auxin transporter OsPIN1a and influences leaf angle regulation (Sun et al., 2019). Inhibition of the expression of auxin receptor OsTIR1 also leads to increased leaf inclination (Bian et al., 2012). In addition, OsARF6 and OsARF17 synergistically control flag leaf angle in response to auxin (Huang et al., 2021).
Both auxin and BR certainly play important roles in regulation of leaf inclination, which involves diverse factors and intricated regulatory networks. In this regard, we identified a novel gene OsARF4 (AUXIN RESPONSE FACTOR 4), mutants of which showed increased leaf inclination, whereas overexpressing lines had erect leaves. Histochemical analysis and scanning electron microscopy (SEM) results showed that the increase in leaf inclination was caused by cell differentiation on the adaxial surface. Moreover, OsARF4 was found to be involved in both auxin and BR signaling in regulation of leaf inclination. These results provide a new potential target for ideal architecture breeding and improving rice yield.
Materials and methods
Plant materials and growth conditions
Wild type variety of rice—Dongjin (Oryza sativa subsp. japonica, WT/DJ)—was used as a control in this study. All seeds were soaked in water in the dark for 3 days at 37°C, and then cultured in a nutrient solution (pH 5.4) and grown in the greenhouse under a 12 h light (30°C) and 12 h dark (24°C) cycle. Thereafter, rice plants were grown in an experimental paddy field under natural conditions of the China National Rice Research Institute in Hangzhou or Hainan Yazhou Bay Seed Laboratory in Sanya, China.
Construction and transformation of binary vectors
The osarf4 mutants were obtained using CRISPR/Cas9 gene editing technology as described by Xie (Xie et al., 2015), and the selected target sites of osarf4 mutants were AGGAGGCATCTCCTTCAGAG and AGTTCCAAAAGGCTTGTTGC. The full-length open reading frame (ORF) of OsARF4 was amplified from WT/DJ cDNA and then cloned into pCAMBIA1300-sGFP binary vector to construct the overexpression vector 35S:OsARF4-GFP. OsARF4 promoter was amplified from WT/DJ genomic DNA and then cloned into the pCAMBIA1301-GUS vector to generate proOsARF4:GUS vector. Agrobacterium strain EHA105 was used for transformation of the vector into WT/DJ. All primers used are shown in Supplementary Table 1.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRA, Kusatsu, Japan) and then reverse transcribed to obtain cDNA using FastKing RT Kit (with gDNase; Tiangen, Beijing, China) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed using the 2*Easy Star Green Fast Mixture (Easy-Do, Zhejiang, China) and a CFX Connect™ Real-time System (Bio-Rad, Hercules, CA, United States). OsACTIN (LOC_Os03g50885) was used as an internal control. Three biological replicates were performed for each experiment. All primers used for qRT-PCR are shown in Supplementary Table 2.
Subcellular localization of OsARF4
High-quality plasmids of 35S:OsARF4-GFP and the nuclear localization marker NSL-mCherry were transiently coexpressed in rice protoplasts using polyethylene glycol as previously described (Qiao et al., 2021). After overnight incubation in the dark at 28°C, fluorescence was observed using a two-photon confocal microscope (Zeiss LSM 710; Carl Zeiss, Oberkochen, Germany).
β-Galactosidase (GUS) staining and activity analysis
The auxin reporter DR5:GUS was genetically transformed into WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines. The LJs were treated with the GUS staining solution (10 mM sodium phosphate buffer, pH 7.0; 10 mM Na2EDTA; 1 mM K3[Fe(CN)6]; 1 mM K3[Fe(CN)6]; 0.5% Triton X-100; 20% methyl alcohol, and 0.5 mg/ml X-Gluc) for 30 min at 37°C. Then, chlorophyll was removed by soaking the leaves in 70% ethanol. Images were collected using a stereoscope Nikon SMZ 25 (Nikon Corporation, Tokyo, Japan). The positive pro:OsARF4-GUS transgenic lines were tested by GUS staining, and roots (3-day-old of seedlings), coleoptile (3-day-old seedlings), and 2–10 week old of seedings of pro:OsARF4-GUS were used for expression pattern analysis. GUS activities in the LJs of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines were analyzed using the Plant GUS Elisa Kit1 according to the manufacturer’s instructions.
Quantification of IAA contents
At 10 days after heading, the LJs of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines were ground into a powder using liquid nitrogen. IAA was extracted and its content was measured as previously described (Ljung et al., 2005; Shao et al., 2019; Qiao et al., 2021). Agilent 1,100 high-performance liquid chromatography was performed using a C18 reverse-phase column (250 mm × 4.6 mm, 5 m).
Scanning electron microscopy
Scanning electron microscopy was performed as previously described (Zhao et al., 2010; Wang et al., 2019). Briefly, at 10 days after heading, approximately 1 cm of LJs was excised from the flag leaves of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines. Thereafter, they were dehydrated in graded ethanol series (50–100%), dried in a critical point dryer for 2 h, and subjected to gold sputtering for 1 min. Samples were observed using an S-3000 N scanning electron microscope (Hitachi, Tokyo, Japan).
Leaf inclination measurement and cytological analysis
Leaf inclinations between the sheaths and leaves of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines were photographed and measured at 10 days after heading. For paraffin sectioning, the LJs from flag leaves of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines were taken and fixed using FAA solution (37% formaldehyde, acetic acid, 70% alcohol, 5:5:90, v:v:v), followed by dehydration in a graded ethanol series and embedding in Paraplast Plus (Sigma, United States). Microtome sections (8 μm) were cut to obtain transverse and longitudinal sections using a rotary microtome (RM2245, Leica Microsystems, Hamburg, Germany) and stained with toluidine blue. Sections were observed and photographed using a stereoscope Nikon SMZ 25 (Nikon Corporation, Tokyo, Japan).
RNA sequencing analysis
At 10 days after heading, total RNA was extracted from LJs of flag leaves of WT/DJ, osarf4-1 mutant, and OE-OsARF4-1 overexpression line, and sequenced in BGI with three biological replicates (Shenzhen, China). The analytical methods and software analysis were performed according to previous studies (Zhang et al., 2020). Gene expression levels were expressed as transcript fragments per kilobase (FPKM) per million reads (Florea et al., 2013). Differentially expressed genes (DEGs) were detected using the DESeq 2 package testing at |log2FC| > = 2 and Q value <= 0.05. Additional detailed information is provided on the BGI official website.2 The RNA sequencing data were deposited in the NCBI Sequence Read Archive (SRA) with accession SRR20748275, SRR20748276, SRR20748277, SRR20748278, SRR20748279, SRR20748280, SRR20748281, SRR20748282, and SRR20748283.
BR sensitivity tests
Primary root (PR) inhibition analysis was performed as previously mentioned (Zhang et al., 2015). Seeds of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines were germinated at 37°C and then grown in a normal culture solution supplemented with 0, 0.01, 0.1, 1, or 10 μm 24-epibrassinolide (24-eBL) for 7 days. Then, the PR length was measured. The coleoptile elongation test was performed as previously described (Duan et al., 2006). Seeds of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines were germinated and grown in a normal culture solution containing 0, 0.01, 0.1, 1, or 10 μm 24-eBL for 7 days. Then, a camera (EOS 6D Mark II, Canon, Japan) was used to obtain a photograph to measure the coleoptile length. After 7 days of growth in a normal culture solution, WT/DJ, osarf4, and OsARF4-overexpressing plants were incubated with 24-eBL at different concentrations for 3 days. Thereafter, the leaf inclinations were measured as previously described (Zhang et al., 2015).
Results
OsARF4 is expressed in lamina joint and encodes a nuclear protein
Our previous study has found that the member of the OsARF family, OsARF19, controls rice leaf inclination positively (Zhang et al., 2015). Although OsARF4 and OsARF19 belong to the OsARF family, they exist in different clades. OsARF4 belongs to Class I subfamily, while OsARF19 is from Class II (Wang et al., 2007). OsARF4 is a putative transcriptional repressor with RD (repression domain) in its middle region (MR), while OsARF19 is a putative transcriptional activator with activation domain (AD; Shen et al., 2010). Furthermore, OsARF4 has a negative function in regulating grain size and grain weight in rice (Hu et al., 2018). In that case, whether OsARF4 also takes part in leaf inclination regulation? In order to make this question clearly, the related tests were performed. To determine the functional location of OsARF4, its expression pattern was first investigated. The promoter of OsARF4 was fused with the GUS gene and this construct was then transformed into WT/DJ calluses to obtain the pro:OsARF4-GUS transgenic plants. Pro:OsARF4-GUS transgenic plants were observed at different stages during 2–10 weeks using histochemical staining. The results showed that OsARF4 was expressed in LJ at different development stages, especially at the mature stage (Figure 1A). qRT-PCR further confirmed that OsARF4 was expressed at those stages in LJ (Figure 1B), consistent with the results of GUS staining, implying that OsARF4 may function in LJ in rice.
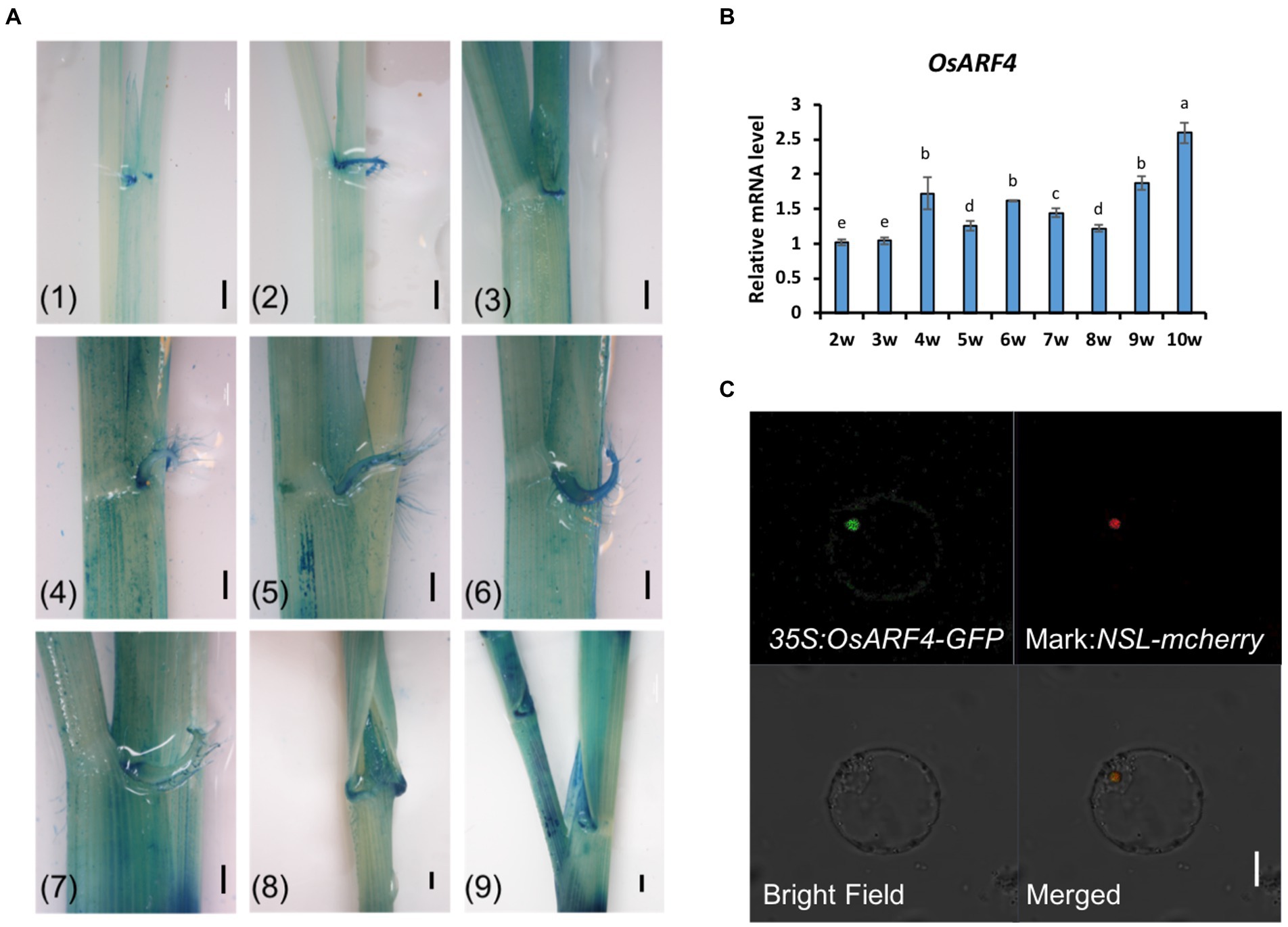
Figure 1. Expression patterns of the OsARF4 gene and subcellular localization of OsARF4. (A) The expression patterns of the OsARF4 gene. (1)–(9) show GUS staining of 2, 3, 4, 5, 6, 7, 8, 9, and 10-week-old seedlings, respectively. Scale bar = 1 mm. (B) qRT-PCR analysis of OsARF4 expression in WT/DJ rice. Three separate biological replicates were used in this experiment. Error bars represent SD (n = 3). Lowercase letters represent significance according to Duncan’s multiple range test (α = 0.05). (C) Subcellular localization of OsARF4 in rice. 35S:OsARF4-GFP and the nuclear localization marker NLS-mCherry were transiently coexpressed in rice protoplasts. Scale bar = 20 mm.
To explore the subcellular localization of OsARF4, the CDS of OsARF4 was fused with GFP and driven by the CaMV35S promoter. Transient co-expression of 35S:OsARF4-GFP and the nuclear marker NLS-mcherry in rice protoplasts showed that OsARF4 was localized in the nucleus (Figure 1C), Furthermore, OsARF4 was also localized in the nuclei of root cells in 35S:OsARF4-GFP transgenic lines (Supplementary Figure 1). These results indicate that OsARF4 encodes a nuclear protein.
OsARF4 negatively affects leaf inclination
Studies have shown that OsARF gene family members OsARF19, OsARF6, and OsARF1 were involved in regulating leaf inclination in rice (Zhang et al., 2015; Huang et al., 2021; Xing et al., 2022). To explore whether OsARF4 functions to control leaf inclination, two osarf4 mutants were constructed using CRISPR/Cas9 gene editing technology (Xie et al., 2015), and two OsARF4-overexpressing lines were obtained (Figures 2A,B). Compared with WT/DJ, osarf4-1 showed a 71-bp deletion in the eighth exon in the ORF (1834–1904 bp) of OsARF4, which caused premature termination of protein translation, resulting in the formation of 627 amino acid protein instead of the 808 amino acid protein in osarf4-1. osarf4-2 showed a 56-bp deletion in the seventh exon in the ORF (1631–1,684 bp), which caused premature termination of protein translation resulting in the formation of a truncated protein with 564 amino acids instead of 808 amino acids. The protein structure in both mutants was changed (Figure 2A). The expression of OsARF4 was upregulated four and nine folds in OsARF4-1 and OsARF4-7-overexpressing lines, respectively, compared with that in WT/DJ (Supplementary Figure 2). Phenotypic observation showed that at the heading stage, leaf angles of osarf4 mutants (osarf4-1 and osarf4-2) were much larger than that of WT/DJ, and the OsARF4-overexpressing lines (OE-OsARF4-1 and OE-OsARF4-7) showed a greater number of erect leaves than inclined leaves (Figure 2C). This was more evident in the flag leaves (reaching to ~85° in osarf4 mutants and ~ 18° in OsARF4-overexpressing lines compared with 28° in WT/DJ; Figures 2D,E).
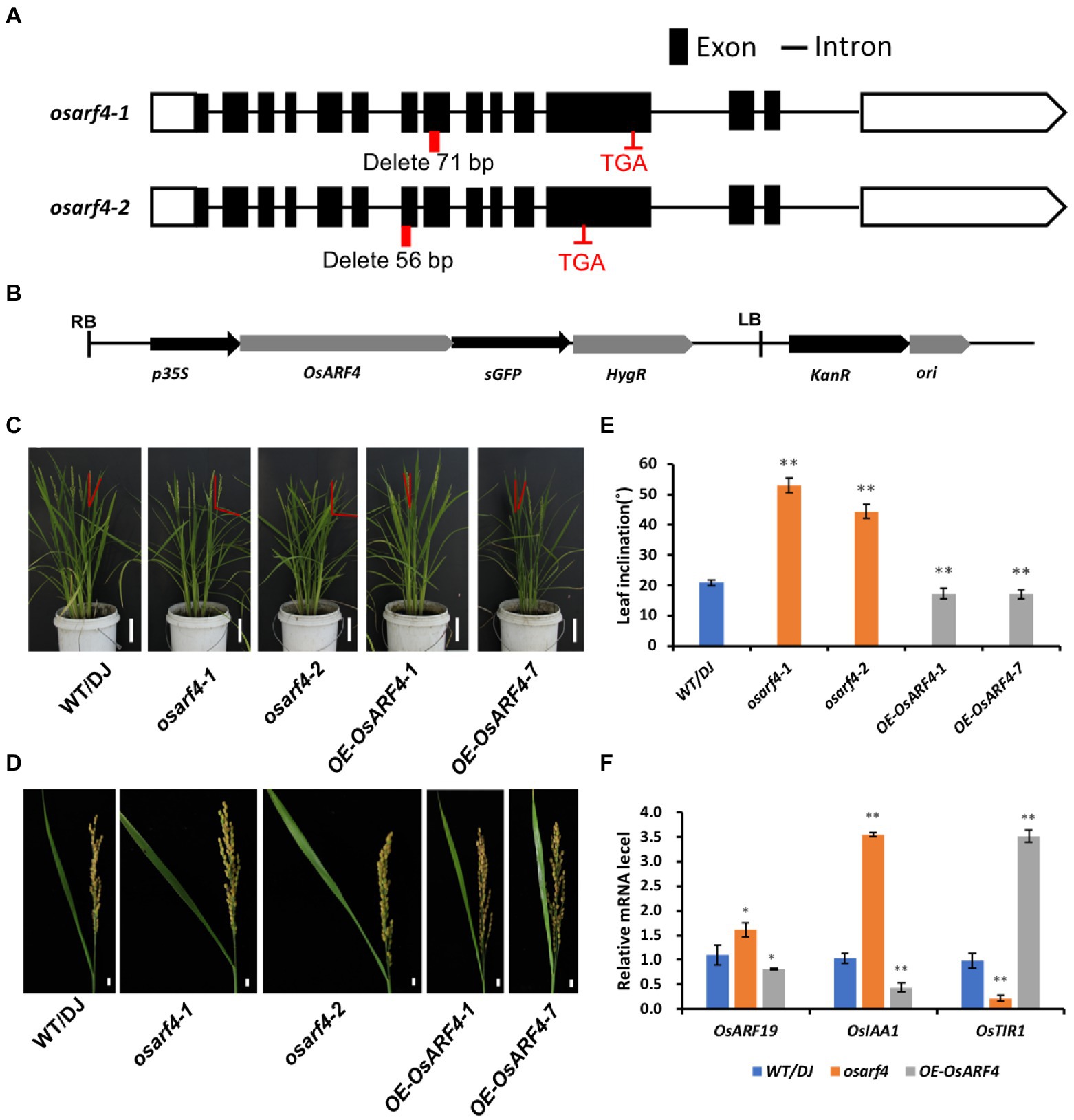
Figure 2. Phenotypic characterization of OsARF4. (A) Construction of two osarf4 mutants using CRISPR/Cas9 technology. Black lines, black boxes, and red boxes indicate introns, exons, and editing sites, respectively. TGA is a stop codon. (B) Map of the OsARF4-overexpressing vector. (C) Phenotypes of 3-month-old seedlings of WT/DJ, osarf4 mutants (osarf4-1 and osarf4-2), and OsARF4-overexpressing lines (OE-OsARF4-1 and OE-OsARF4-7). Bars = 10 cm. (D) Flag leaf angles of WT/DJ, osarf4 mutants, and OsARF4-overexpressing seedlings at mature stages. Bars = 3 cm. (E) Quantification of flag leaf angles at mature stages. Ten biological replicates were taken for each experiment. ** indicates a significant difference at p < 0.01. (F) Relative expression levels of leaf inclination-related genes in WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines. Three individual biological replicates were taken for each experiment. *p < 0.05 and **p < 0.01 indicate significant differences compared with WT/DJ using Student’s t-test.
To further verify the function of OsARF4 in controlling leaf inclination, we analyzed the expression of genes related to regulation of leaf inclination in the LJs of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines (Figure 2F). The transcriptional abundances of the genes OsARF19 and OsIAA1 were induced in the osarf4 mutants, whereas they were reduced in OsARF4-overexpressing lines compared with those in WT/DJ. In addition, OsTIR1 showed a opposite trend compared with OsIAA1. These results are consistent with those reported previously that OsARF19 and OsIAA1 positively regulate leaf inclination, and OsTIR1 negatively regulates leaf inclination (Song et al., 2009; Bian et al., 2012; Zhang et al., 2015), implying that OsARF4 might play a negative role in regulating leaf inclination.
Increased cell proliferation on the adaxial surface leads to increased leaf inclination in osarf4
Previous studies have shown that the LJ significantly contributes to the formation of leaf inclination, and the increased leaf inclination may cause alterations in LJ development (Yamamuro et al., 2000). Therefore, the collar length of the LJ at adaxial and abaxial epidermis in WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines was measured. As shown in Figures 3A,B, the collar length of the LJ at adaxial surface of osarf4 mutants increased by 60%, whereas it decreased by 45% in OsARF4-overexpressing lines compared with that in WT/DJ. In contrast, there were no significant differences in the length at the abaxial surface between WT/DJ and osarf4 mutants or OsARF4-overexpressing lines. These results indicate that the main difference in leaf inclination in those lines was due to the imbalanced elongation between the adaxial and abaxial surfaces of LJ.
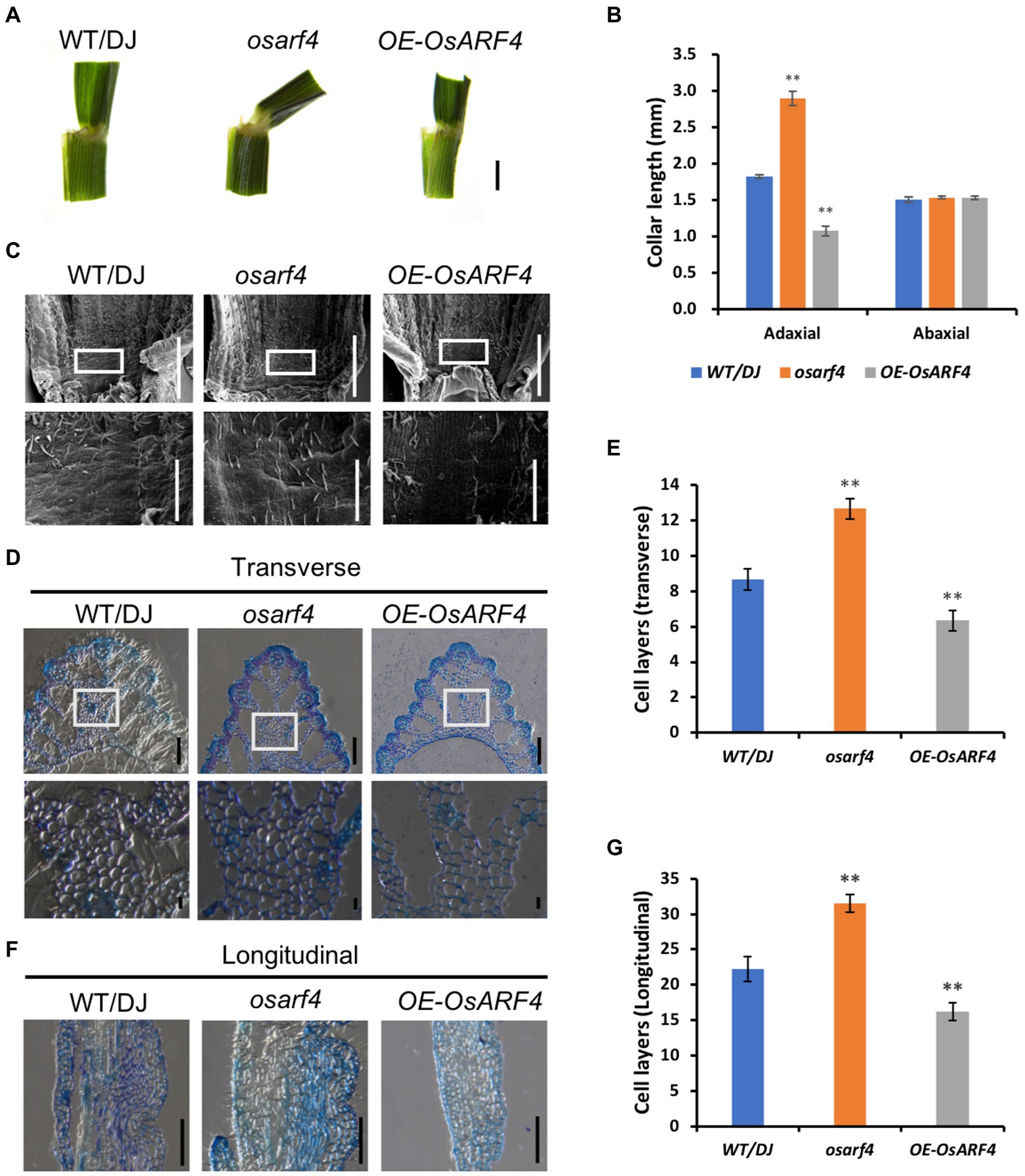
Figure 3. Microstructure analysis of LJs in WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines. (A) Comparison of LJs of flag leaves in 3-month-old seedlings of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines. (B) The collar length of the flag leaves at adaxial and abaxial surfaces in the above-mentioned lines. (C) SEM images of the adaxial surface of the LJ of the flag leaves. (D) Transverse (cross) section of LJs of flag leaves in the above-mentioned lines. Upper panel, bar = 200 μm; lower panel shows the magnified parts of the upper panel, bar = 20 μm. (E) Cell layers in transverse sections. Ten biological replicates were considered; **p < 0.01 represents significant differences using Student’s t-test. (F) Longitudinal section of LJs of flag leaves in the above-mentioned lines. Bar = 200 μm. (G) Cell layers in longitudinal section of LJs. Ten biological replicates were considered for each test. **p < 0.01 indicates a significant difference compared with WT/DJ using Student’s t-test.
To identify the cellular mechanism underlying the regulation of leaf inclination by OsARF4, SEM analysis was conducted to observe microstructure morphology. In osarf4 mutant, there was much larger and formed a bulge in the adaxial surface of the LJ, whereas it was smoother in OsARF4-overexpressing lines compared to the smooth adaxial surface in WT/DJ (Figure 3C). These results further confirmed that the development of the LJ in osarf4 was altered and resulted in increased leaf inclination.
Previous studies have shown that aberrant cellular development, such as lack of the longitudinal cell elongation or increase in cell expansion at the adaxial sides in the collar, can lead to changes in leaf inclination (Duan et al., 2006; Zhao et al., 2010; Zhang et al., 2015; Guo et al., 2021; Tian et al., 2021). To further reveal the reason for the changed leaf inclination in the mutants and overexpression lines of OsARF4, we prepared paraffin sections with both transverse and longitudinal sections of the LJs of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines. The results indicate that the number of sclerenchyma cell layers on the adaxial surface increased in both transverse and longitudinal sections in osarf4 mutants and decreased in the OsARF4-overexpressing lines, compared with that in WT/DJ (Figures 3D–G), which illustrates that cell division, but not cell elongation, in the LJ resulted in increased leaf inclination in osarf4.
OsARF4 regulates leaf inclination by controlling auxin distribution and content in lamina joints
Auxin is an important plant hormone that plays a negative role in controlling leaf inclination (Du et al., 2012; Zhou et al., 2017). Studies have shown that OsARF regulates leaf angles induced by auxin (Zhang et al., 2015). To explore the role of auxin in OsARF4-mediated leaf inclination pathway, the relative expression level of OsARF4 was detected under auxin treatment. It was found that auxin treatment could induce OsARF4 expression (Supplementary Figure 3), indicating that the function of OsARF4 might be positively regulated by auxin.
As the auxin response reporter, DR5:GUS has been widely used to study the distribution of endogenous auxin (Ulmasov et al., 1997; Qiao et al., 2021). To determine whether the changes in leaf inclination of OsARF4 are influenced by the changes in auxin distribution, DR5:GUS was transformed into the osarf4 mutants and OsARF4-overexpressing lines. GUS staining in LJs indicated that DR5:GUS expression in osarf4 mutants was markedly lower than that in WT/DJ but more intense in OsARF4-overexpressing lines (Figure 4A). Moreover, GUS activity was lower in osarf4 but higher in OsARF4-overexpressing lines, compared with that in WT/DJ (Figure 4B). Furthermore, the free auxin content in LJs of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines was measured. The results showed that the auxin content was lower and higher in osarf4 mutants and OsARF4-overexpressing lines, respectively, compared with that in WT/DJ (Figure 4C), implying that OsARF4 regulates leaf inclination by controlling local auxin levels in LJs.
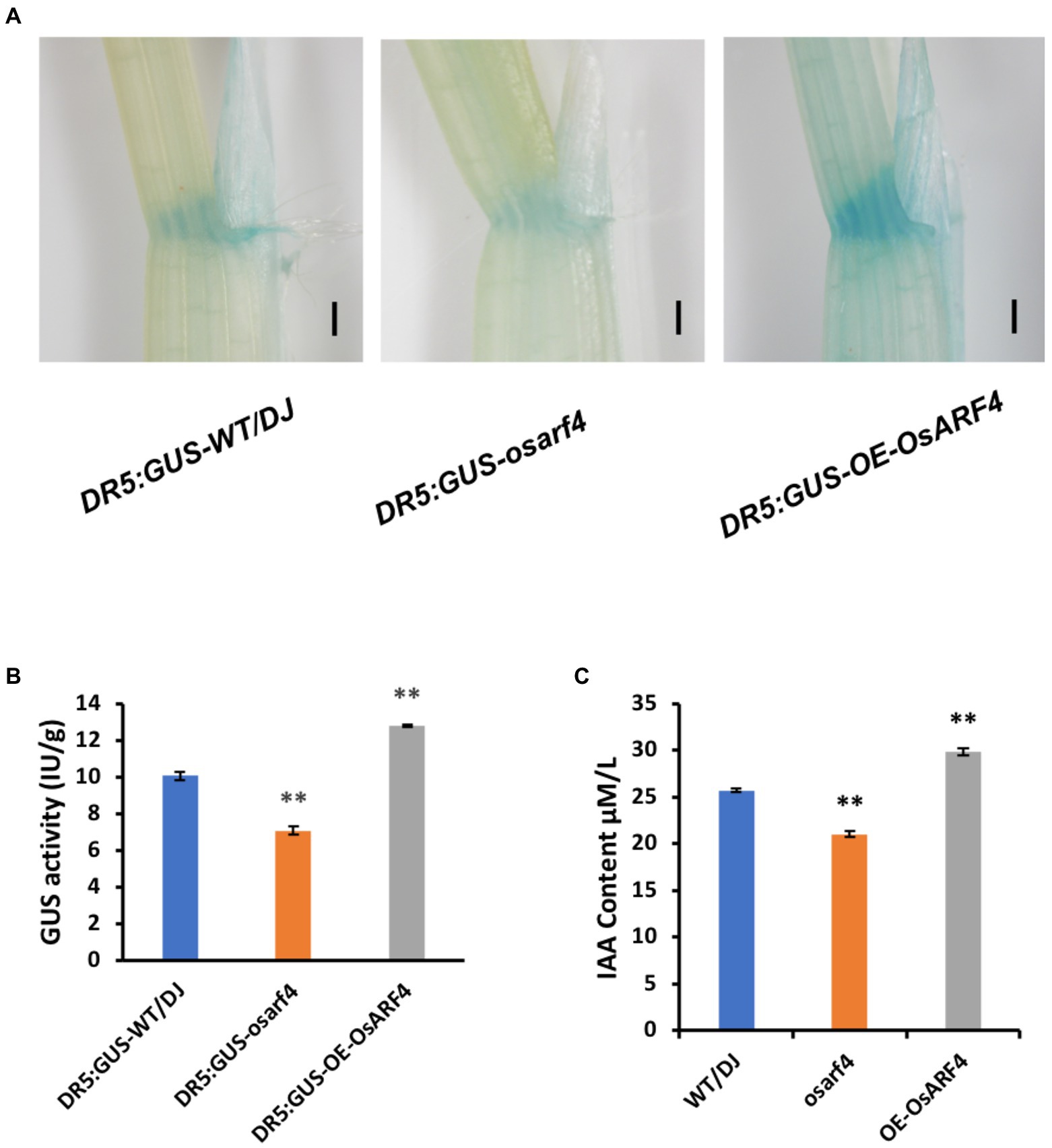
Figure 4. Analysis of auxin distribution and content in LJs in WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines. (A) GUS staining of DR5:GUS-WT/DJ, DR5:GUS-osarf4, and DR5:GUS-OE-OsARF4 transgenic lines in the LJs. Bars = 500 μm. (B) Quantification of GUS activity in DR5:GUS-WT/DJ, DR5:GUS-osarf4, and DR5:GUS-OE-OsARF4 transgenic lines. Ten biological replicates were evaluated. **p < 0.01 indicates a significant difference in means ± SD compared with DR5:GUS-WT/DJ based on Student’s t-test. (C) Determination of auxin contents in LJs of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines. Five separate biological replicates were used in each test. **p < 0.01 indicates significant differences from WT/DJ based on Student’s t-test.
RNA-seq analysis of WT/DJ, mutant and overexpression line of OsARF4
To analyze genes of OsARF4-mediated pathway that participate in the regulation of leaf inclination, transcriptome of the flag LJs from WT/DJ, osarf4 mutant, and OsARF4-overexpressing line was sequenced. A total of 32,938 genes were obtained in the RNA-seq analysis. Among those differentially expressed genes (DEGs), 70 genes were upregulated and 130 genes were downregulated between WT and osarf4 mutant; 1,229 genes were upregulated and 1,452 genes were downregulated between the WT and OsARF4-overexpressing lines; and 12 genes were upregulated and 11 genes were downregulated between the osarf4 mutant and OsARF4-overexpressing line (Figures 5A,B). The results of gene ontology (GO) enrichment analysis demonstrated that genes related to auxin signal transduction and BR biosynthesis were significantly overrepresented (Figure 5C), which implies that OsARF4 might be involved in auxin signal transduction and BR biosynthesis. In previous studies, genes related to regulation of leaf inclination, including OsBZR2 (Min et al., 2019), OsREM4.1 (Li et al., 2021), OsNCED5 (Zhu et al., 2009), OsBZR1 (Bai et al., 2007), OsDWARF (Hong et al., 2002), OsD11 (Tanabe et al., 2005), OsD2 (Hong et al., 2003) OsGSR1 (Wang et al., 2009), OsBRI1 (Brassinazole-resistant 1; Yamamuro et al., 2000), and OsLPA1 (Wu et al., 2013), have been reported, and these genes showed variable expression in LJs of osarf4 mutant and OsARF4-overexpressing line compared with that in WT/DJ (Figure 5D and Supplementary Figure 4). These results indicate the importance of OsARF4 in BR signaling. Furthermore, as shown in Figure 5D, the RNA-seq results indicated that the expression of OsBZR1 is upregulated in osarf4 mutant and downregulated in OE-OsARF4 line. On the contrary, the expression of OsD2 is downregulated in osarf4 mutant and upregulated in OE-OsARF4 line. To confirm the results, qRT-PCR was performed in the flag leaf of WT/DJ, osarf4 mutants, and OE-OsARF4 lines. The results were consistent with the RNA-seq analysis (Figures 5D,E). OsBZR1 is a famous gene associated with brassinosteroid signal transduction in rice (Bai et al., 2007). OsD2 encodes a cytochrome P450 BR synthesis enzyme in the late BR biosynthesis pathway (Hong et al., 2003). Overall, whether OsBZR1 and OsD2 are the potential target genes of OsARF4 needs to be further explored.
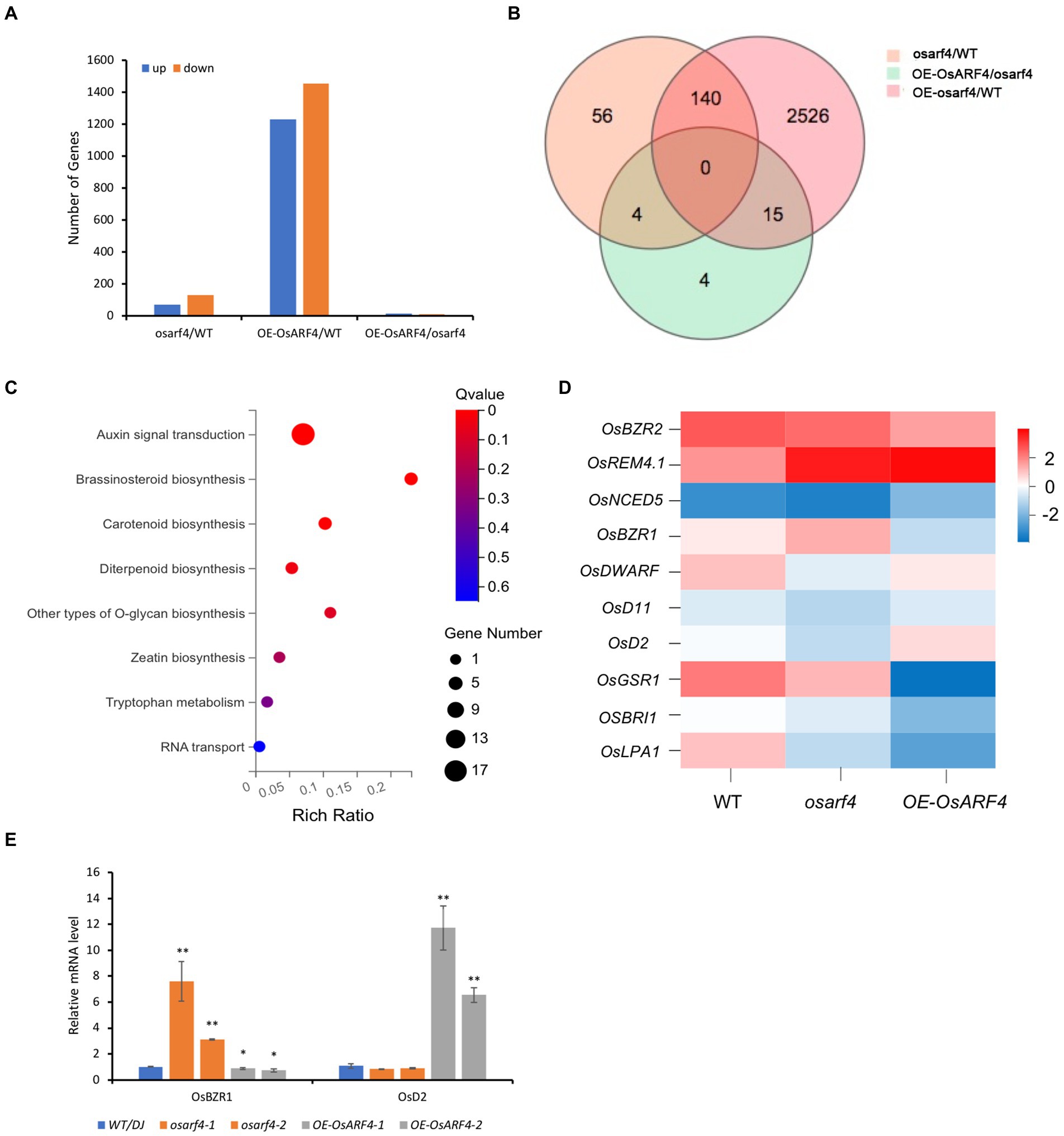
Figure 5. RNA-seq analysis of OsARF4-modulated genes in rice LJs. (A) Numbers of DEGs in the LJs of WT/DJ versus osarf4, WT/DJ versus OE-OsARF4, and osarf4 versus OE-OsARF4. Blue columns represent up-regulated genes, and orange columns represent downregulated genes. (B) Venn diagram showing an overlap of DEGs in LJs in WT/DJ versus osarf4, WT/DJ versus OE-OsARF4, and osarf4 versus OE-OsARF4 in LJs. (C) GO enrichment functional clustering of OsARF4-modulated genes that were differentially expressed in the transcriptome. X-axis represents enrichment ratio. The size of bubbles represents the number of DEGs annotated to a certain GO term. The color represents enrichment significance value. (D) Heat map of the RNA-seq analysis revealed the expression patterns of selected genes during BR biosynthesis and signaling in WT/DJ, osarf4 mutant, and OsARF4-overexpressing lines. (E) Relative expression of OsBZR1 and OsD2 in two osarf4 mutants and two OE-OsARF4 lines. Three individual biological replicates were taken for each experiment. *p < 0.05 and **p < 0.01 indicate significant differences compared with WT/DJ using Student’s t-test.
OsARF4 is sensitive to exogenous BR
In rice, BR has been proved to play a pivotal role in the determination of leaf inclination (Sakamoto et al., 2013; Zhang et al., 2015). To further confirm whether OsARF4 is involved in BR signaling in regulation of leaf inclination, the three classical BR sensitivity experiments were performed. In addition to LJs, OsARF4 was also expressed in primary root, lateral root, and coleoptile (Figure 1A; Supplementary Figure S5). Therefore, BR sensitivity experiments, PR elongation, coleoptile elongation, and the degree of leaf inclination tests were performed under treatments with different concentrations of 24-eBL (Figure 6). The PR growth was inhibited by BR treatments and this inhibition increased with the increased in BR concentration. Compared to the WT/DJ, osarf4 mutants were insensitive to exogenous BR treatments, whereas OsARF4-overexpressing lines were relatively more sensitive (Figures 6A,D). After BR treatments, the elongation of coleoptile increased in OsARF4-overexpressing lines and became more pronounced with the increase in BR concentration, whereas osarf4 mutants showed slight increase compared with that in WT/DJ (Figures 6B,D). The leaf inclination in OsARF4-overexpressing lines and osarf4 mutants increased by 8–80% and 3–20%, respectively, under treatments with different concentrations of 24-eBL (Figures 6C,D). Taken together, the OsARF4-overexpressing lines were more sensitive to exogenous BR treatments, whereas the osarf4 mutants were insensitive, compared with WT/DJ.
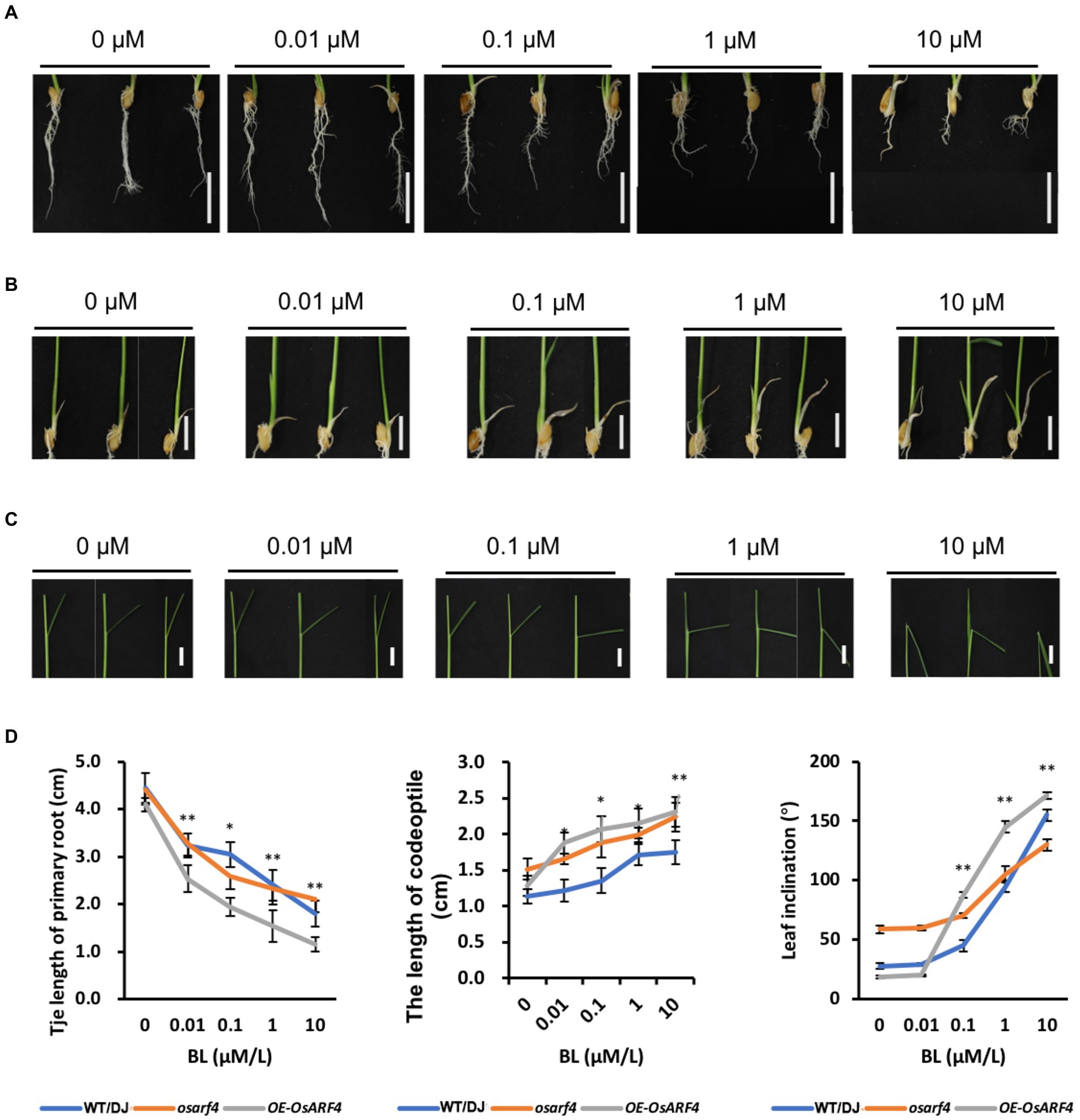
Figure 6. OsARF4 mediates the morphological response of rice to 24-epibrassinolide (24-eBL). (A–C) primary root (PR; A), coleoptile elongation (B), and Leaf inclination (C) response to 24-eBL treatments. Panels from left to right show roots of seedlings of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines grown for 7 days (A,B) or 3 days (C) under 0, 0.01, 0.1, 1, and 10 μm 24-eBL treatment. Bars = 2 cm in A and 1 cm in B/C. (D) Statistical analyses of PR length, coleoptile, and leaf inclination in WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines under above indicated 24-eBL treatments (n = 10). *p < 0.05 and **p < 0.01 based on Student’s t-test.
Moreover, OsARF4 expression was induced by BR treatments (Figure 7A). When osarf4 mutants and OsARF4-overexpressing lines were treated with 0.1 μM 24-eBL (the optimum concentration selected from the above gradient treatment) for 3 days simultaneously, the leaf inclination of WT/DJ, osarf4 mutants, and OsARF4-overexpressing lines increased by 96, 38, and 390%, respectively (Figures 7B–C). Hence, OsARF4-overexpressing lines were more sensitive, whereas osarf4 mutants were insensitive to BR treatments, compared with WT/DJ. These results suggest that OsARF4 is related to regulation of BR signaling-mediated leaf inclination in rice.
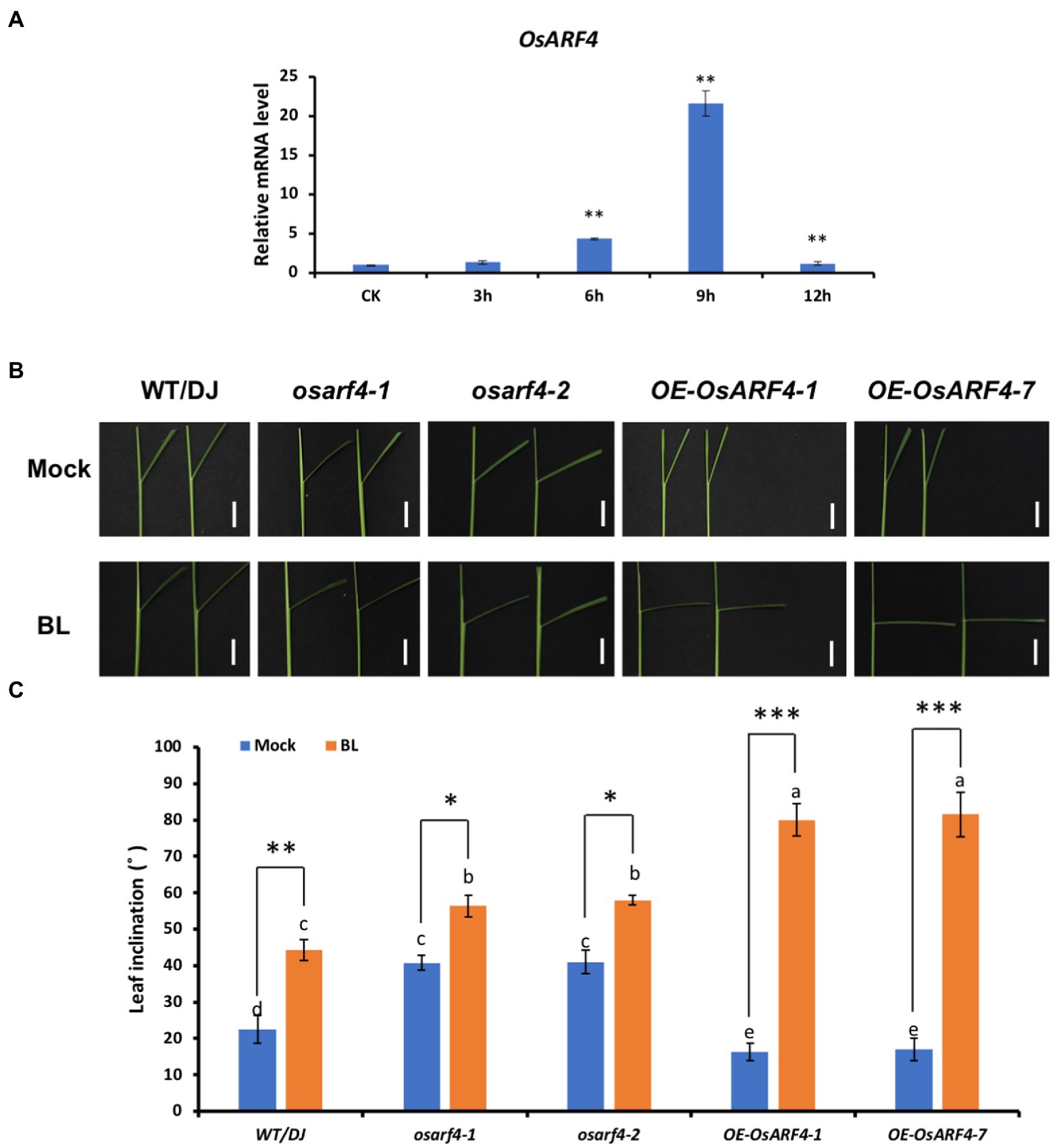
Figure 7. OsARF4 is sensitive to low concentration of BR in increasing leaf inclination of rice seedling. (A) The expression of OsARF4 under 0.1 μm 24-eBL treatment at different time intervals. (B) Leaf inclination response of 7 days seedlings of WT/DJ, osarf4-1, osarf4-2, OE-OsARF4-1, and OE-OsARF4-7 to 0.1 μm 24-eBL treatment for 3 days. (C) Measurement of leaf inclination in response to 0.1 μm 24-eBL treatment for 3 days as shown in B. Fifteen rice lines were measured for each treatment. Error bars indicate SD (n = 3). Lowercase letters represent significance based on Duncan’s multiple range test (α = 0.05). *p < 0.05, **p < 0.01 and ***p < 0.001 indicate significant differences of means ± SD compared with control based on t-test.
Discussion
OsARF4 functions in regulating leaf inclination
As plant-specific B3-type transcription factors, ARFs play a role in activating or repressing the auxin response genes by specifically binding to auxin response element (AuxRE; Chandler, 2016). There are 25 ARFs in rice, many of which have been reported to execute multiple functions (Wang et al., 2007). For instance, OsARF12 and OsARF16 are associated with root development and phosphate homeostasis (Qi et al., 2012; Shen et al., 2013, 2014; Wang et al., 2014). OsARF25 is involved in controlling grain size, and its T-DNA insertion mutant has shorter grains (Zhang et al., 2018). OsmiR160 negatively regulates the expression of OsARF18 and improves rice growth and development via auxin signaling pathway (Huang et al., 2016a). OsARF6 participates in the miR167a-OsARF6-OsAUX3 module to regulate grain length and grain weight in rice (Qiao et al., 2021). OsARF6 and OsARF17 bind to the promoter of ILA1 to determine flag leaf angle (Huang et al., 2021). OsARF1 interacts with OsIAA6 to regulate leaf inclination in rice through auxin and BR pathway (Xing et al., 2022). In a previous study, the research of our laboratory have found that OsARF19 is involved in positive regulation of lamina inclination regulation by binding to the promoters of OsGH3-5 and OsBRI1, through auxin and BR pathway (Zhang et al., 2015). In this study, we generated two osarf4 mutants using CRISPR/Cas9 gene editing technology, and two OsARF4-overexpressing lines using genetic transformation. Phenotype analysis showed that the flag leaf inclination increased by approximately 150% in osarf4 mutants and decreased by 44% in OsARF4-overexpressing lines after heading in rice (Figure 2; Supplementary Figure 2). Leaf inclination is mainly determined by the LJ, which is a unique tissue connecting the leaf blade and sheath in rice (Wang et al., 2020). GUS staining and qRT-PCR results indicated that OsARF4 was expressed in LJ at different developmental stages (Figures 1A,B), thus confirming that OsARF4 is involved in leaf inclination regulation. Interestingly, both OsARF4 and OsARF19 belong to the OsARF family, but they play converse roles in the regulation of leaf inclination in rice, because they were from different clades, namely, OsARF4 belongs to Class I with transcript repression domain while OsARF19 belongs to Class II with transcript activation domain (Wang et al., 2007; Shen et al., 2010), lead to they have different functions in leaf inclination regulation.
From a cytological perspective, the change in leaf inclination is attributed to the imbalance in the cell development of the adaxial and abaxial sides in LJs (Duan et al., 2006). In previous studies, most of the identified rice mutants with altered leaf inclination showed abnormal division and expansion of adaxial cells in the leaf collar (Nakamura et al., 2009; Zhang S. W. et al., 2009; Zhao et al., 2010; Ning et al., 2011). The leaf inclination is caused by the bending of the blade. From the perspective of a cytological structure, the most obvious change is the cell division and expansion at the adaxial side of the LJ (Wang et al., 2020). To illustrate the cytological structures of mutants and overexpression lines of OsARF4, the collar lengths in the adaxial and abaxial sides were measured. The results demonstrated that the asymmetric collar length in LJs alters the leaf inclination in the mutants and overexpression lines of OsARF4. Furthermore, according to the cytological structural analysis of LJs, it was found that OsARF4 regulates leaf inclination through modulating adaxial cell division rather than cell expansion of the collar in LJs (Figure 3). The increase of leaf inclination in osarf4 is achieved by promoting the differentiation of the adaxial side parenchyma cells, which is mainly to promote the synthesis of genes related to cell wall such as the cyclin gene family members. The cell cycle-related genes OsCYCU4;1, OsCYCD2;1, OsCYCD6;1, and OsCYCD1;2 also changed differently in the RNA-seq analyses of WT/DJ, osarf4 mutant, and OsARF4-overexpressing line in LJs (Supplementary Figure 6). In addition, whether auxin has polar distribution in the adaxial side and abaxial side of LJs needs to be further explored.
The change in auxin level in lamina joint might alter leaf inclination
Auxin responses play a crucial role in plant growth and development by forming local concentration gradients. Auxin induces rapid transcriptional responses, incorporating a series of auxin early response genes (SAUR, Aux/IAA, and GH3) and ARFs. It is known that auxin synthesis and signal transduction are related to the regulation of leaf inclination (Xu et al., 2021). Moreover, during early LJ development, high local auxin concentrations were formed due to auxin synthesis and polar transport, which will stimulate cell division. During LJ development, a decrease in IAA content will promote the elongation of parenchyma cells on the adaxial side of the LJ, which is consistent with the increase in GH3 expression, thus increasing leaf inclination (Wang et al., 2007; Chen et al., 2018). The auxin signaling pathway is normally mediated by ARFs that regulate transcription in response to auxin in plants (Guilfoyle and Hagen, 2007). In this study, we have shown that OsARF4 regulated leaf inclination via altering the auxin distribution and content in LJ. As shown in Figure 4, the auxin distribution and GUS activity decreased in osarf4 mutants, they significantly increased in OsARF4-overexpressing lines compared with those in WT/DJ. The free auxin levels in LJs were consistent with this result, implying that the alteration of auxin level in LJ regulated by OsARF4 results in changes in leaf inclination. The RNA-seq analyses also revealed that some auxin-catabolism and auxin signaling-related genes such as OsGH3-1, OsGH3-2, OsGH3-5, OsAFB2, OsARF1, and OsIAA1 showed different expression levels in the LJs of WT/DJ, osarf4 mutant, and OsARF4-overexpressing line (Supplementary Figure 6).
OsARF4 mediates auxin-BR crosstalk during regulation of leaf inclination
Auxin and BR are two vital phytohormones that regulate leaf inclination in rice (Tong and Chu, 2018). Studies have shown that lacking of BR leads to an erect leaf phenotype because of the inhibition of elongation of parenchyma cells and division of sclerenchyma cells on the abaxial side of the LJ (Zhang L. Y. et al., 2009; Sun et al., 2015). Most BR-deficient and BR-insensitive mutants show altered BR sensitivity by altering auxin synthesis or signal transduction, and many auxin-related mutants also show altered BR responses by changing leaf inclination, thus suggesting crosstalk between BR and auxin during LJ development (Song et al., 2009; Zhao et al., 2013; Zhang et al., 2015). The signaling pathways of auxin and BR share multiple genes, which play important roles in plant growth and development (Vert et al., 2008). In rice, OsARF19 regulates leaf inclination by binding to the promoter region of OsBRI1 to connect auxin and BR signaling pathways (Zhang et al., 2015). The ds1 mutant shows decreased leaf angle owing to lower BR sensitivity, and DS1 participates in plant architecture regulation through regulating the expression of OsBRI1 via interacting with OsARF11 in rice (Liu et al., 2018). Therefore, BR and auxin genes act synergistically to co-regulate the development of adaxial cells in LJ (Liu et al., 2016). In this study, both auxin and BR induced the expression of OsARF4 (Figure 7A; Supplementary Figure 3). DR5:GUS staining and measurement of free IAA content showed that OsARF4 is involved in auxin signaling (Figure 4). RNA-seq analysis indicated that OsARF4 is related to BR biosynthesis and signaling (Figure 5). Furthermore, OsARF4-overexpressing lines were sensitive to BR treatments and had increased leaf inclination (Figures 6, 7B–D). These results indicate that OsARF4 acts as a bridge between auxin and BR signaling during the regulation of leaf inclination.
In addition, a previous study has reported that OsARF4 interacts with the rice GSK3-like kinase, OsGSK5/OsSK41, to negatively regulate grain size and weight in rice. And, osarf4 mutant was no effect on BR sensitivity in terms of leaf lamina inclination at the seedling stage (Hu et al., 2018). In this study, we observed that osarf4-1 and osarf4-2 had enlarged leaf inclination at the flag leaf of mature period (Figures 2C–D). As for BR sensitivity, the BR sensitivity experiments were treated with different concentrations 0, 0.01, 0.1, 1, and 10 μm of 24-eBL. The above tests presented that osarf4 mutants are insensitive to exogenous BR treatments, while OsARF4-overexpressing lines are more sensitive not only in the degree of leaf inclination but also in PR elongation and coleoptile growth (Figure 6). Besides, the different editing sites and background plants from previous research might result in a little difference of regulatory mechanism (Hu et al., 2018).
Taken together, the semi-dwarf phenotype, characterized by erect leaves and shorter panicles, is an ideal phenotype for improving grain yield in high-density planting in rice (Morinaka et al., 2006; Sakamoto et al., 2006; Wu et al., 2008). Therefore, the regulation mechanism of leaf inclination can provide a theoretical basis for plant architecture breeding that would help develop highly productive rice varieties. In our study, evidence from genetic analysis, cellular biological observation, molecular biological analysis, and physiological experiments support the finding that OsARF4 regulates leaf inclination via auxin and BR signaling pathways in rice. Improving plant architecture is an important goal for breeding of high-yield rice varieties. Erect leaves can increase planting density, improve photosynthetic capacity, increase accumulation of photosynthetic products, and ultimately increase the grain yield of rice (Huang et al., 2016b). Our research uncovers a normal regulatory factor of leaf inclination and shows that a molecular bridge exists between auxin and BR signaling. These findings may help optimize plant architecture and increase yield of rice.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA862303.
Author contributions
YQ and QQ designed the experiments. JQ, YZ, SH, and SC performed experiments. YQ, QQ, ZG, and JQ analyzed the data. YQ, QQ, and JQ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by grants from the National Natural Science Foundation of China (32060451), the Zhejiang Provincial Natural Science Foundation of China (grant no. LZ19C020001), Natural Science Foundation of Inner Mongolia (2022ZD11), and Applied Technology Research and Development Foundation of Inner Mongolia (2021PT0001) to YHQ, the Special Fund of Hainan Yazhou Bay Seed Laboratory to JYQ (B21Y10202), and China Postdoctoral Science Foundation (2022T150714).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.979033/full#supplementary-material
SUPPLEMENTARY FIGURE 1 | Nuclear localization of OsARF4 in root cells. 35S:OsARF4-GFP was transformed into WT/DJ. Green fluorescence represents the nucleus of the root tip cell; bar scale = 20 μm.
SUPPLEMENTARY FIGURE 2 | Relative expression of OsARF4 in OsARF4-overexpressing lines. qRT-PCR was performed using the LJs of each line for three individual biological replicates. *P < 0.05 and **P < 0.01, t-test.
SUPPLEMENTARY FIGURE 3 | OsARF4 expression under auxin treatments. Seven-day-old of WT/DJ seedlings were grown in a solution containing 10 μm NAA and 1 μm 2,4-D for 0, 3, 6, 9, and 12 h. (n = 3) **P < 0.01 indicates significant differences compared with 0 h using Student’s t-test.
SUPPLEMENTARY FIGURE 4 | Expression profiles of hormone-related genes in RNA-seq in LJs of WT/DJ, osarf4-1 and OE-OsARF4-1 line.
SUPPLEMENTARY FIGURE 5 | OsARF4 is expressed in primary root, lateral root and coleoptile of pro:OsARF4-GUS transgenic seedlings. (A) GUS staining of 3-d-old primary root, lateral root and coleoptile of pro:OsARF4-GUS transgenic seedlings. Bar scale = 500 μm. (B) qRT-PCR analysis of OsARF4 expression in 3-d-old primary root, lateral root and coleoptile. Error bars represent SD (n = 3). Lowercase letters represent significance according to Duncan’s multiple range test (α = 0.05).
SUPPLEMENTARY FIGURE 6 | Heat map of the RNA-seq analysis of genes involved in cell division and auxin signaling in LJs.
SUPPLEMENTARY TABLE 1 | Primers used for vector construction.
SUPPLEMENTARY TABLE 2 | Primers used for qRT-PCR.
Footnotes
References
Bai, M. Y., Zhang, L. Y., Gampala, S. S., Zhu, S. W., Song, W. Y., Chong, K., et al. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. U. S. A. 104, 13839–13844. doi: 10.1073/pnas.0706386104
Bian, H., Xie, Y., Guo, F., Han, N., Ma, S., Zeng, Z., et al. (2012). Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 196, 149–161. doi: 10.1111/j.1469-8137.2012.04248.x
Chandler, J. W. (2016). Auxin response factors. Plant Cell Environ. 39, 1014–1028. doi: 10.1111/pce.12662
Chen, S. H., Zhou, L. J., Xu, P., and Xue, H. W. (2018). SPOC domain-containing protein leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PLoS Genet. 14:e1007829. doi: 10.1371/journal.pgen.1007829
Du, H., Wu, N., Fu, J., Wang, S., Li, X., Xiao, J., et al. (2012). A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 63, 6467–6480. doi: 10.1093/jxb/ers300
Duan, K., Li, L., Hu, P., Xu, S. P., Xu, Z. H., and Xue, H. W. (2006). A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 47, 519–531. doi: 10.1111/j.1365-313X.2006.02804.x
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res 2:188. doi: 10.12688/f1000research.2-188.v2
Guilfoyle, T. J., and Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Guo, J., Li, W., Shang, L., Wang, Y., Yan, P., Bai, Y., et al. (2021). OsbHLH98 regulates leaf angle in rice through transcriptional repression of OsBUL1. New Phytol. 230, 1953–1966. doi: 10.1111/nph.17303
Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada, Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. doi: 10.1046/j.1365-313x.2002.01438.x
Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910. doi: 10.1105/tpc.014712
Hu, Z., Lu, S. J., Wang, M. J., He, H., Sun, L., Wang, H., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11, 736–749. doi: 10.1016/j.molp.2018.03.005
Huang, G., Hu, H., van de Meene, A., Zhang, J., Dong, L., Zheng, S., et al. (2021). AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell 33, 3120–3133. doi: 10.1093/plcell/koab175
Huang, J., Li, Z., and Zhao, D. (2016a). Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in Rice. Sci. Rep. 6:29938. doi: 10.1038/srep29938
Huang, X., Yang, S., Gong, J., Zhao, Q., Feng, Q., Zhan, Q., et al. (2016b). Genomic architecture of heterosis for yield traits in rice. Nature 537, 629–633. doi: 10.1038/nature19760
Li, H., Jiang, L., Youn, J. H., Sun, W., Cheng, Z., Jin, T., et al. (2013). A comprehensive genetic study reveals a crucial role of CYP90D2/D2 in regulating plant architecture in rice (Oryza sativa). New Phytol. 200, 1076–1088. doi: 10.1111/nph.12427
Li, D., Wang, L., Wang, M., Xu, Y. Y., Luo, W., Liu, Y. J., et al. (2009). Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 7, 791–806. doi: 10.1111/j.1467-7652.2009.00444.x
Li, Q., Xu, F., Chen, Z., Teng, Z., Sun, K., Li, X., et al. (2021). Synergistic interplay of ABA and BR signal in regulating plant growth and adaptation. Nat. Plants 7, 1108–1118. doi: 10.1038/s41477-021-00959-1
Liu, J. M., Park, S. J., Huang, J., Lee, E. J., Xuan, Y. H., Je, B. I., et al. (2016). Loose plant Architecture1 (LPA1) determines lamina joint bending by suppressing auxin signalling that interacts with C-22-hydroxylated and 6-deoxo brassinosteroids in rice. J. Exp. Bot. 67, 1883–1895. doi: 10.1093/jxb/erw002
Liu, X., Yang, C. Y., Miao, R., Zhou, C. L., Cao, P. H., Lan, J., et al. (2018). DS1/OsEMF1 interacts with OsARF11 to control rice architecture by regulation of brassinosteroid signaling. Rice 11:46. doi: 10.1186/s12284-018-0239-9
Ljung, K., Hull, A. K., Celenza, J., Yamada, M., Estelle, M., Normanly, J., et al. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17, 1090–1104. doi: 10.1105/tpc.104.029272
Min, H. J., Cui, L. H., Oh, T. R., Kim, J. H., Kim, T. W., and Kim, W. T. (2019). OsBZR1 turnover mediated by OsSK22-regulated U-box E3 ligase OsPUB24 in rice BR response. Plant J. 99, 426–438. doi: 10.1111/tpj.14332
Morinaka, Y., Sakamoto, T., Inukai, Y., Agetsuma, M., Kitano, H., Ashikari, M., et al. (2006). Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141, 924–931. doi: 10.1104/pp.106.077081
Nakamura, A., Fujioka, S., Takatsuto, S., Tsujimoto, M., Kitano, H., Yoshida, S., et al. (2009). Involvement of C-22-hydroxylated brassinosteroids in auxin-induced lamina joint bending in rice. Plant Cell Physiol. 50, 1627–1635. doi: 10.1093/pcp/pcp106
Ning, J., Zhang, B., Wang, N., Zhou, Y., and Xiong, L. (2011). Increased leaf Angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the lamina joint of Rice. Plant Cell 23, 4334–4347. doi: 10.1105/tpc.111.093419
Qi, Y., Wang, S., Shen, C., Zhang, S., Chen, Y., Xu, Y., et al. (2012). OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 193, 109–120. doi: 10.1111/j.1469-8137.2011.03910.x
Qiao, J., Jiang, H., Lin, Y., Shang, L., Wang, M., Li, D., et al. (2021). A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 14, 1683–1698. doi: 10.1016/j.molp.2021.06.023
Qiao, S., Sun, S., Wang, L., Wu, Z., Li, C., Li, X., et al. (2017). The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate Brassinosteroid signaling and Rice architecture. Plant Cell 29, 292–309. doi: 10.1105/tpc.16.00611
Sakamoto, T., Morinaka, Y., Inukai, Y., Kitano, H., and Fujioka, S. (2013). Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 73, 676–688. doi: 10.1111/tpj.12071
Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S., Ueguchi-Tanaka, M., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24, 105–109. doi: 10.1038/nbt1173
Shao, Y., Zhou, H. Z., Wu, Y., Zhang, H., Lin, J., Jiang, X., et al. (2019). OsSPL3, an SBP-domain protein, regulates crown root development in Rice. Plant Cell 31, 1257–1275. doi: 10.1105/tpc.19.00038
Shen, C., Wang, S., Bai, Y., Wu, Y., Zhang, S., Chen, M., et al. (2010). Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). J. Exp. Bot. 61, 3971–3981. doi: 10.1093/jxb/erq208
Shen, C., Wang, S., Zhang, S., Xu, Y., Qian, Q., Qi, Y., et al. (2013). OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 36, 607–620. doi: 10.1111/pce.12001
Shen, C., Yue, R., Yang, Y., Zhang, L., Sun, T., Tie, S., et al. (2014). OsARF16 is involved in cytokinin-mediated inhibition of phosphate transport and phosphate signaling in rice (Oryza sativa L.). PLoS One 9:e112906. doi: 10.1371/journal.pone.0112906
Song, Y., You, J., and Xiong, L. (2009). Characterization of OsIAA1 gene, a member of rice aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol. Biol. 70, 297–309. doi: 10.1007/s11103-009-9474-1
Sun, S., Chen, D., Li, X., Qiao, S., Shi, C., Li, C., et al. (2015). Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev. Cell 34, 220–228. doi: 10.1016/j.devcel.2015.05.019
Sun, Q., Li, T. Y., Li, D. D., Wang, Z. Y., Li, S., Li, D. P., et al. (2019). Overexpression of Loose plant architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN-FORMED 1a in rice. Plant Biotechnol. J. 17, 855–857. doi: 10.1111/pbi.13072
Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17, 776–790. doi: 10.1105/tpc.104.024950
Tian, Q., Luan, J., Guo, C., Shi, X., Deng, P., Zhou, Z., et al. (2021). A bHLH protein, OsBIM1, positively regulates rice leaf angle by promoting brassinosteroid signaling. Biochem. Biophys. Res. Commun. 578, 129–135. doi: 10.1016/j.bbrc.2021.09.035
Tong, H., and Chu, C. (2018). Functional specificities of Brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 23, 1016–1028. doi: 10.1016/j.tplants.2018.08.007
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. doi: 10.1105/tpc.9.11.1963
Van Camp, W. (2005). Yield enhancement genes: seeds for growth. Curr. Opin. Biotechnol. 16, 147–153. doi: 10.1016/j.copbio.2005.03.002
Vert, G., Walcher, C. L., Chory, J., and Nemhauser, J. L. (2008). Integration of auxin and brassinosteroid pathways by Auxin response factor 2. Proc. Natl. Acad. Sci. U. S. A. 105, 9829–9834. doi: 10.1073/pnas.0803996105
Wang, A., Hou, Q., Si, L., Huang, X., Luo, J., Lu, D., et al. (2019). The PLATZ transcription factor GL6 affects grain length and number in rice. Plant Physiol. 180, 2077–2090. doi: 10.1104/pp.18.01574
Wang, R., Liu, C., Li, Q., Chen, Z., Sun, S., and Wang, X. (2020). Spatiotemporal resolved leaf angle establishment improves rice grain yield via controlling population density. iScience 23:101489. doi: 10.1016/j.isci.2020.101489
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, L., Wang, Z., Xu, Y., Joo, S. H., Kim, S. K., Xue, Z., et al. (2009). OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 57, 498–510. doi: 10.1111/j.1365-313X.2008.03707.x
Wang, S., Zhang, S., Sun, C., Xu, Y., Chen, Y., Yu, C., et al. (2014). Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 201, 91–103. doi: 10.1111/nph.12499
Wu, X., Tang, D., Li, M., Wang, K., and Cheng, Z. (2013). Loose plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 161, 317–329. doi: 10.1104/pp.112.208496
Wu, C. Y., Trieu, A., Radhakrishnan, P., Kwok, S. F., Harris, S., Zhang, K., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20, 2130–2145. doi: 10.1105/tpc.107.055087
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U. S. A. 112, 3570–3575. doi: 10.1073/pnas.1420294112
Xing, M., Wang, W., Fang, X., and Xue, H. (2022). Rice OsIAA6 interacts with OsARF1 and regulates leaf inclination. Crop J. doi: 10.1016/j.cj.2022.02.010
Xu, J., Wang, J. J., Xue, H. W., and Zhang, G. H. (2021). Leaf direction: Lamina joint development and environmental responses. Plant Cell Environ. 44, 2441–2454. doi: 10.1111/pce.14065
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1605. doi: 10.1105/tpc.12.9.1591
Yoshikawa, T., Ito, M., Sumikura, T., Nakayama, A., Nishimura, T., Kitano, H., et al. (2014). The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 78, 927–936. doi: 10.1111/tpj.12517
Zhang, C., Bai, M. Y., and Chong, K. (2014). Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 33, 683–696. doi: 10.1007/s00299-014-1578-7
Zhang, L. Y., Bai, M. Y., Wu, J., Zhu, J. Y., Wang, H., Zhang, Z., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780. doi: 10.1105/tpc.109.070441
Zhang, S. W., Li, C. H., Cao, J., Zhang, Y. C., Zhang, S. Q., Xia, Y. F., et al. (2009). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151, 1889–1901. doi: 10.1104/pp.109.146803
Zhang, Z., Li, J., Tang, Z., Sun, X., Zhang, H., Yu, J., et al. (2018). Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3-OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J. Exp. Bot. 69, 4723–4737. doi: 10.1093/jxb/ery256
Zhang, S., Wang, S., Xu, Y., Yu, C., Shen, C., Qian, Q., et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654. doi: 10.1111/pce.12397
Zhang, X., Wang, Y., Zhu, X., Wang, X., Zhu, Z., Li, Y., et al. (2020). Curled flag leaf 2, encoding a cytochrome P450 protein, regulated by the transcription factor Roc5, influences flag leaf development in Rice. Front. Plant Sci. 11:616977. doi: 10.3389/fpls.2020.616977
Zhao, S. Q., Hu, J., Guo, L. B., Qian, Q., and Xue, H. W. (2010). Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 20, 935–947. doi: 10.1038/cr.2010.109
Zhao, S. Q., Xiang, J. J., and Xue, H. W. (2013). Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in LEAF INCLINATION control. Mol. Plant 6, 174–187. doi: 10.1093/mp/sss064
Zhou, L. J., Xiao, L. T., and Xue, H. W. (2017). Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol. 174, 1728–1746. doi: 10.1104/pp.17.00413
Keywords: leaf inclination, auxin, OsARF4, rice, brassinosteroid
Citation: Qiao J, Zhang Y, Han S, Chang S, Gao Z, Qi Y and Qian Q (2022) OsARF4 regulates leaf inclination via auxin and brassinosteroid pathways in rice. Front. Plant Sci. 13:979033. doi: 10.3389/fpls.2022.979033
Edited by:
Yule Liu, Tsinghua University, ChinaReviewed by:
Ning Ning Wang, Nankai University, ChinaJianming Li, University of Michigan, United States
Haodong Chen, Tsinghua University, China
Copyright © 2022 Qiao, Zhang, Han, Chang, Gao, Qi and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhua Qi, cXloanBAemp1LmVkdS5jbg==; Qian Qian, cWlhbnFpYW4xODhAaG90bWFpbC5jb20=
 Jiyue Qiao1,2
Jiyue Qiao1,2 Zhenyu Gao
Zhenyu Gao Yanhua Qi
Yanhua Qi Qian Qian
Qian Qian