
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 September 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.977649
 Qingqing Gao1,2†
Qingqing Gao1,2† Wanling Song1,2†
Wanling Song1,2† Xia Li1,2
Xia Li1,2 Chunfan Xiang1,2
Chunfan Xiang1,2 Geng Chen1,2
Geng Chen1,2 Guisheng Xiang1
Guisheng Xiang1 Xiangyu Liu1
Xiangyu Liu1 Guanghui Zhang1
Guanghui Zhang1 Xiaoning Li1
Xiaoning Li1 Shengchao Yang1*
Shengchao Yang1* Chenxi Zhai3*
Chenxi Zhai3* Yan Zhao1,2*
Yan Zhao1,2*Erigeron breviscapus is a Compositae plant, and its rich flavonoids have shown strong preventative and curative effects in the treatment of cardio- and cerebrovascular diseases. bHLH genes play a crucial role in plant growth and development. There are 116 EbbHLH genes in E. breviscapus, and each gene has been named based on its chromosome location. Our phylogenetic analysis divided these genes into 18 subfamilies. To further investigate its function, EbbHLH80 was isolated from E. breviscapus leaves. Next, transcriptomic and metabolomic analyses of tobacco leaves were performed. Among 421 differentially accumulated compounds, 98 flavonoids were identified. In addition, differentially expressed genes were identified using RNA-seq, and further analysis suggested that EbbHLH80-OE could not only regulate the expression of some structural genes in the flavonoid biosynthesis pathway to achieve flavonoid accumulation but also be involved in the regulation of a series of downstream pathways, such as stress response, ABA and ethylene signal transduction, to affect plant growth and development. The results of our analysis provide new insights into the function of EbbHLH80 and lay the foundation for future functional studies on E. breviscapus.
As one of the largest transcription factor families, basic helix-loop-helix (bHLH) proteins are one of the most abundant. In plants, fungi, and animals, these transcription factors (TFs) are widely distributed (Riechmann and Muyldermans, 1999; Carretero-Paulet et al., 2010). A bHLH TF is composed of two connected subregions, a basic region located directly following the helix-loop-helix (HLH) domain and a dimerizing region consisting of 40–50 amino acid residues (Atchley et al., 1999; Carretero-Paulet et al., 2010). bHLHs play an important role in regulating flavonoid biosynthesis and ROS (reactive oxygen species) homeostasis under stress. There has been an increase in the number of bHLH transcription factors found in plant species that regulate flavonoid biosynthesis pathways, and a key function of these transcription factors is to regulate anthocyanin biosynthesis (Jin et al., 2018).
Previous studies have identified several enzyme-coding structural genes involved in phenylpropanoid, flavonoid and anthocyanin biosynthesis (Chen et al., 2019; Sun et al., 2020). It has been demonstrated that the bHLH transcription factor TRANSPARENT TESTA8 (ATT8; AT4G09820) regulates the flavonoid biosynthesis pathway via the transcription of key enzymes BAN (Banyuls) and DFR (dihydroflavonol 4-reductase) (Nesi et al., 2000; Baudry et al., 2004). TT8 is an important transcription factor involved in secondary metabolism as well as the response to stress (Zhai et al., 2020). TT8 loss-of-function lines show altered chromatographic profiles for aglycones of kaempferol and quercetin as well as glycosylated forms (Pelletier et al., 1999; Narasimhan et al., 2003). This complex of R2R3-MYB, bHLH, and WD40 repeat (MBW) proteins directly binds to promoters of flavonoid biosynthesis genes (Dubos et al., 2010; Xu et al., 2014). Previous studies have shown that the flavonoid pathway is regulated in plants by different members of the bHLH family, such as CmbHLH2, which significantly activates CmDFR transcription and triggers anthocyanin accumulation while coexpressed with CmMYB6 in Chrysanthemums (Chrysanthemum morifolium Ramat.) (Xiang et al., 2015). In the inner pericarp of Actinidia chinensis cv. Hongyang, coexpression of AcMYB123 and AcbHLH42 activates AcF3GT1 and AcANS or their homologous gene expression, promoting anthocyanin accumulation (Wang et al., 2019). Moreover, AabHLH1 interacts with AaMYB3 to regulate the accumulation of procyanidin (Li et al., 2019). DhMYB2 was identified in Dendrobium hybrids, interacting with DhbHLH1 to regulate anthocyanin production (Li et al., 2017). Additionally, PPLS1, a bHLH transcription factor, interacts with SiMYB85 to regulate anthocyanin biosynthesis in Setaria italica (Bai et al., 2020). A series of transporters facilitate the transportation of flavonoids to vacuoles after biosynthesis, including glutathione S-transferases (GSTs), multidrug resistance-associated proteins (MRPs), and multidrug and toxic compound extrusion (MATE) proteins (Marrs et al., 1995; Mueller et al., 2000; Zhao and Dixon, 2009).
In the Compositae family, Erigeron breviscapus (E. breviscapus) is an important medicinal plant and is rich in flavonoids in its leaves (Su et al., 2001; Chu et al., 2005). Scutellarin is one of the major active components and is widely used as a prescription drug. E. breviscapus was the first medical plant to undergo an older version genome assembly to accomplish the process from genome sequencing to metabolite biosynthesis, and genome assembly and engineering yeast were used to produce breviscapine (Liu et al., 2018; He et al., 2021). In addition, extensive studies on bHLH gene family members in horticultural and crop plants have increased our understanding of their functions and transcriptional regulatory mechanisms. Moreover, extensive research has been conducted on bHLH genes in horticultural plants, learning more about their regulatory mechanisms. However, it is unclear whether bHLH genes are associated with regulating flavonoid biosynthesis in E. breviscapus. A recent publication of the genome sequence of E. breviscapus provided a way to identify the bHLH gene family (He et al., 2021).
Therefore, whether EbbHLH80 has a similar function as the AtTT8 regulator in regulating the flavonoid pathway, is worthy of further investigation. In previous studies, heterologous overexpression of bHLHs has been shown to regulate the synthesis of flavonoids. Many bHLHs have been found to play a positive role in regulating flavonoid synthesis. The grape VvbHLH1 gene is heterologously overexpressed and increases enzyme activity related to flavonoid synthesis (Wang et al., 2016b). VvbHLH003 and VvbHLH007 were also found to be related to flavonoid synthesis (Wang et al., 2018). Similarly, Arabidopsis PAP2, a MYB transcription factor, is heterologously expressed and increases anthocyanin contents in tomatoes (Li et al., 2018), while the liverwort (Plagiochasma appendiculatum) PabHLH1 is heterogeneously overexpressed in Arabidopsis and induces both flavonoid and anthocyanin synthesis by upregulating structural genes involved in flavonoid synthesis (Zhao et al., 2019). Furthermore, apple MdMYC2 is ectopically overexpressed and significantly upregulates the expression of structural genes in transgenic Arabidopsis (An et al., 2016). In previous studies, TsMYC2 from triticale (Triticum × Secale) and TaMYC1 from wheat (Triticum aestivum) were shown to participate in accumulating anthocyanin and regulate the grain properties associated with the blue aleurone trait (Zong et al., 2019).
Here, we transferred EbbHLH80 into tobacco and observed ectopic flavonoid accumulation. By further integrating metabolome and transcriptome analysis EbbHLH80-OE and Yunyan87 (WT), we identified key metabolites and genes (structural genes or TFs) in leaves to pinpoint key genes controlling metabolite composition in EbbHLH80-OE and Yunyan87. This enabled us to investigate the coordinated regulation of metabolite components and other biological processes directly coregulated with metabolites. Finally, EbbHLH80 was suggested to be an integrator of secondary metabolism TFs that can regulate flavonoid biosynthesis.
Erigeron breviscapus seeds were obtained from Longjin Biotech Co., Ltd. (Xuanwei, Yunnan, China). Nicotiana tabacum (Yunyan 87) seeds were obtained from Yunnan Agricultural University (Kunming, Yunnan, China). The flowers, leaves, roots and stems of E. breviscapus that grew for approximately 8-week-old were collected to quantify EbbHLH80 in the different tissues. Three biological replicates were collected for each sample. The roots, stems, leaves and flowers of E. breviscapus and leaves of tobacco were frozen with liquid nitrogen and then stored at −80°C.
The Hidden Markov Model (HMM) profile of the bHLH DNA-binding domain (PF00010) downloaded from the Pfam database1 was used for the identification of bHLH genes in the E. breviscapus genome (He et al., 2021) using the simple HMM search program TBtools (Chen et al., 2020). The ExPASy proteomics server was used to determine the molecular weights of EbbHLH proteins and their isoelectric points.2 Subcellular localization was predicted by Plant-mPLoc.3
The NCBI Conserved Domain Search4 was used to test for the presence of the bHLH domain. TBtools software was used to visualize NCBI CDD domain patterns (Chen et al., 2020). To analyze the motifs of EbbHLH proteins, the online Multiple Expectation Maximization for Motif Elicitation (MEME)5 was selected and used (Bailey et al., 2006). Furthermore, EbbHLH protein sequences were identified using the MEME program of TBtools. The following parameters were optimized for MEME: AnyNumberOfOccurPerSeq; 15 motifs to be found; and 6–60 residues for each motif. TBtools was also used to visualize MEME results (Chen et al., 2020).
According to the positions assigned in genome annotations of E. breviscapus, EbbHLH genes were located on its chromosomes using TBtools software (Chen et al., 2020). For the phylogenetic analysis, the multiple sequence alignments of the bHLH proteins from E. breviscapus and Arabidopsis were aligned by the ClustalW program and adjusted manually. The phylogenetic tree was constructed by the neighbor-joining method in MEGA 7.0 with 1,000 bootstrap replications (Kumar et al., 2016). The same method was adopted to construct phylogenetic trees of bHLH proteins from E. breviscapus. The phylogenetic trees of all EbbHLH and 120 AtbHLH were constructed using the same method described above, and the AtbHLH sequences were downloaded from TAIR (The Arabidopsis Information Resource).6 According to the phylogenetic tree, certain bHLH proteins were predicted to have certain biological functions.
The CDS of EbbHLH80 was cloned from leaves of 8-week-old E. breviscapus. The PC1300-35S-EbbHLH80 construct was generated by subcloning the target fragments between the BamHI and XbaI sites of the PC1300-35S vector using the primers listed in Supplementary Table 1. Then, the plasmids were transformed into Agrobacterium tumefaciens GV3101, which was then used in tobacco cultivar Yunyan87 (Nicotiana tabacum) transformation with the leaf disc methods described by Huang et al. (2016). Regeneration shoots and healthy resistant shoots were selected on the selective shooting medium and rooting medium, both containing 50 mg ⋅ L–1 kanamycin and 250 mg ⋅ L–1 carbenicillin. Well-developed rooted plants were transferred to soil and then grown in a growth room at 25 ± 2°C with 65–70% relative humidity.
The leaves samples were obtained from 7-week-old Yunyan87 (wild-type, WT) and homozygous T2 lines of transgenic tobacco plants (EbbHLH80-OE), were harvested for transcriptome and metabolome sequencing. Liquid nitrogen was used to freeze all samples immediately, and then they were stored at -80°C. Three biological replicates were used in this study, each comprising leaves from six individual plants.
To quantify the total flavonoids content, the leaves were harvested from six different 7-week-old tobacco plants. Total flavonoids were extracted and quantified using the method described by Wang et al. (2016a). A spectrophotometer (UV-1800, Shimadzu) was used to measure the absorbance of the samples at 535 nm. Methanol containing 1% HCl was used as a blank control. Content of flavonoids was calculated by Zhao et al. (2022), and data was expressed as mg/g FW. For each sample, at least three biological replicates were performed.
Wild-type (WT) and EbbHLH80-OE metabolite profiling was performed using a widely targeted metabolome method with three biological replicates in each group (Chen et al., 2013). Quality control (QC) analysis was conducted before the data analysis. Analyst 1.6.1 was used to filter data, detect peaks, align, and perform all other calculations. A variable importance in projection (VIP) score of the (O)PLS model was applied to rank the most clearly distinguished metabolites between groups. Metabolites with significant differences in content were defined as having a variable importance in the project (VIP) ≥ 1 and T-test P < 0.05.
The leaves were obtained from 7-week-old WT and EbbHLH80-OE tobacco for transcriptome analysis with three biological replicates in each group. The total RNA was extracted for reverse-transcribing into cDNA to construct a cDNA library. To perform the quality control, the raw data was controlled by fastp. RNA purity and integrity were determined with 1% agarose gel, Nanophotometer spectrophotometer, and Agilent 2100 Bioanalyzer. These data were subsequently used to analyze base composition and mass distribution to confirm the quality of this set of data. HISAT2 software was used for alignment analysis with the reference genome, the transcripts of the new genes were assembled with Stringtie, the gene expression level was analyzed with FPKM, the significantly differentially accumulated genes were screened according to FDR < 0.05 and log2FC > |1|, Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were enriched with TBtools software (Chen et al., 2020) and the KEGG database.
The RNA extraction kit (Magen, China) was used to extract total RNA from the samples. RNA was reverse-transcribed into cDNA using a reverse transcription kit (Takara, China). In accordance with qPCR SYBR Green Master Mix, a 20 μL reaction system was constructed (Vazyme, China). PCR was performed on a QuantStudio 5 (ABI) instrument (Thermo Fisher Scientific, Singapore) with the primers listed in Supplementary Table 2. The NtActin gene was used as an internal control, thus obtaining true differences in gene of interest specific expression. The quantitative real-time PCR (RT-qPCR) cycling conditions were as follows: 95°C for 30 s, 40 cycles of 95°C for 10 s and 60°C for 30 s, followed by 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. After amplification, the melting curves were used to verify the PCR products. Gene expression levels were analyzed by the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Statistical analysis was conducted with one-way analysis of variance. Values of P < 0.05 or P < 0.01 were considered to be statistically significant. Double and single asterisks indicate significant differences between comparing samples at 0.01 and 0.05, respectively.
A total of 159 bHLH TFs were identified. After removing redundant proteins by BLASTP with NCBI Batch CD-Search tools, 116 bHLH TFs remained (Supplementary Table 3). All bHLHs were named EbbHLH1–EbbHLH116 according to their chromosomal distribution, with details including the renaming, physical position, isoelectric point (pI), molecular weight and length of the coding protein (Supplementary Table 4).
In addition, analysis of EbbHLH physicochemical properties showed that the shortest EbbHLH is EbbHLH050, with 89 amino acids, and the longest is EbbHLH113, with 870 amino acids. The molecular weight (MW) of the proteins ranged from 10.0 (EbbHLH050) to 100.1 kDa (EbbHLH113), and their isoelectric points (PI) ranged from 4.8 (EbbHLH039) to 10.11 (EbbHLH014). Subcellular localization predicted that 115 EbbHLH proteins were located in the nucleus. One EbbHLH protein was found in the mitochondrion (Supplementary Table 5). The large-scale gene expression required for the biogenesis and functional operation of mitochondria is controlled by a related network of TFs and regulators that bind to nuclear DNA.
The chromosomal location analyses revealed that the 116 EbbHLH genes were randomly distributed on 9 chromosomes (Figure 1). The results showed that 28 EbbHLH genes were found on chromosome 1, followed by 15 EbbHLH genes located on chromosome 2. In addition, 8 EbbHLH genes were located on chromosome 9. There were no significant correlations between chromosome length and the number of EbbHLH genes.
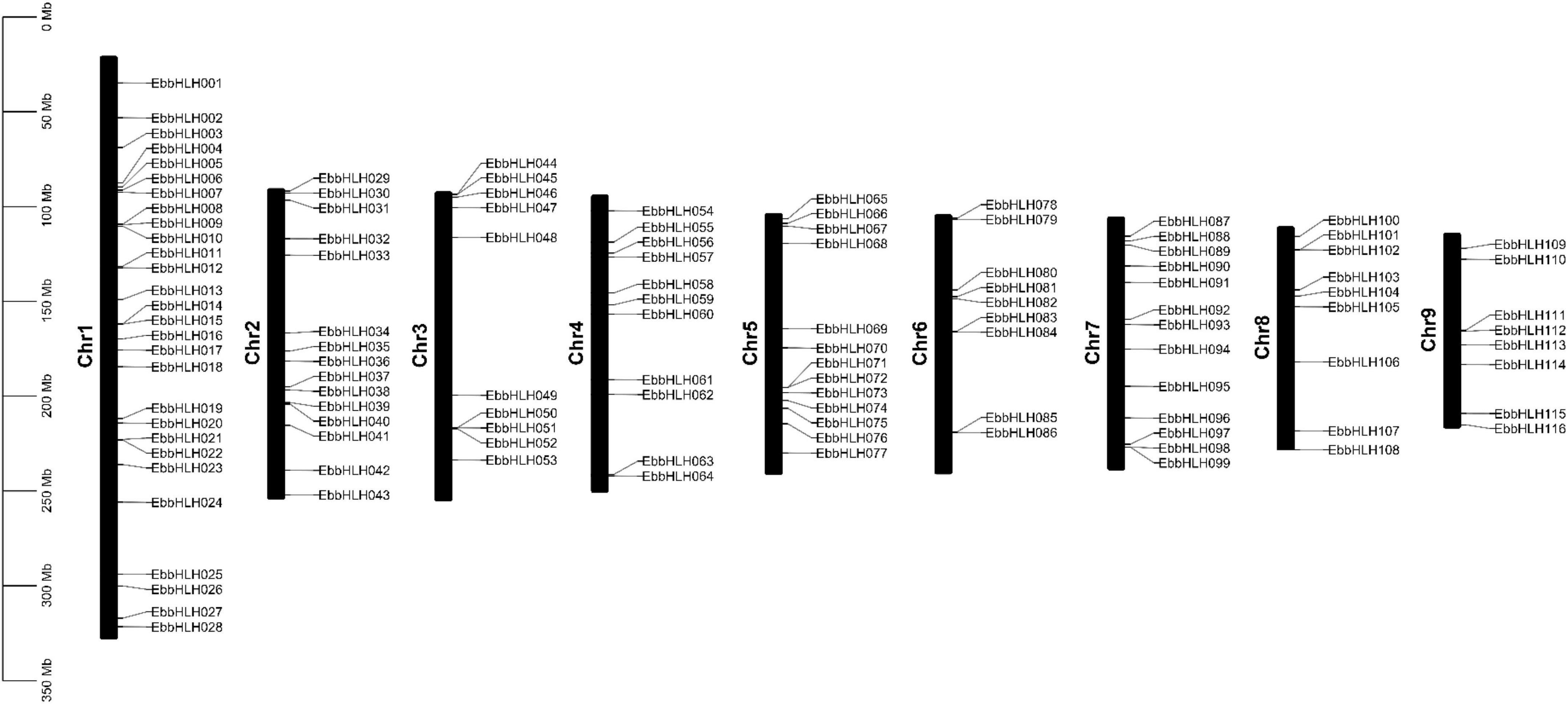
Figure 1. Chromosome distribution of EbbHLHs gene. The chromosome number is shown in the middle of each chromosome.
Further analysis of the gene structure and the conserved domains in EbbHLHs and their exon and intron structures were obtained by comparing the corresponding genomic files (Figure 2C). A comparison of the number and position of the exons and introns revealed that the 116 EbbHLH genes have different numbers of exons, varying from 2 to 11. In addition, 18 genes (16%) contained 2 exons, and the remaining genes had 3 or more exons. The largest proportion of EbbHLH genes (n = 21) had 3 introns. Furthermore, members of subfamilies 8, 10, and 12 contained only 1 intron, and the remaining protein sequences had more than 2 introns. EbbHLH089 had the largest number of 10 introns. Both EbbHLH013 and EbbHLH033 had 9 introns. In general, all the members of different subfamilies had similar numbers of CDS-intron junctions.
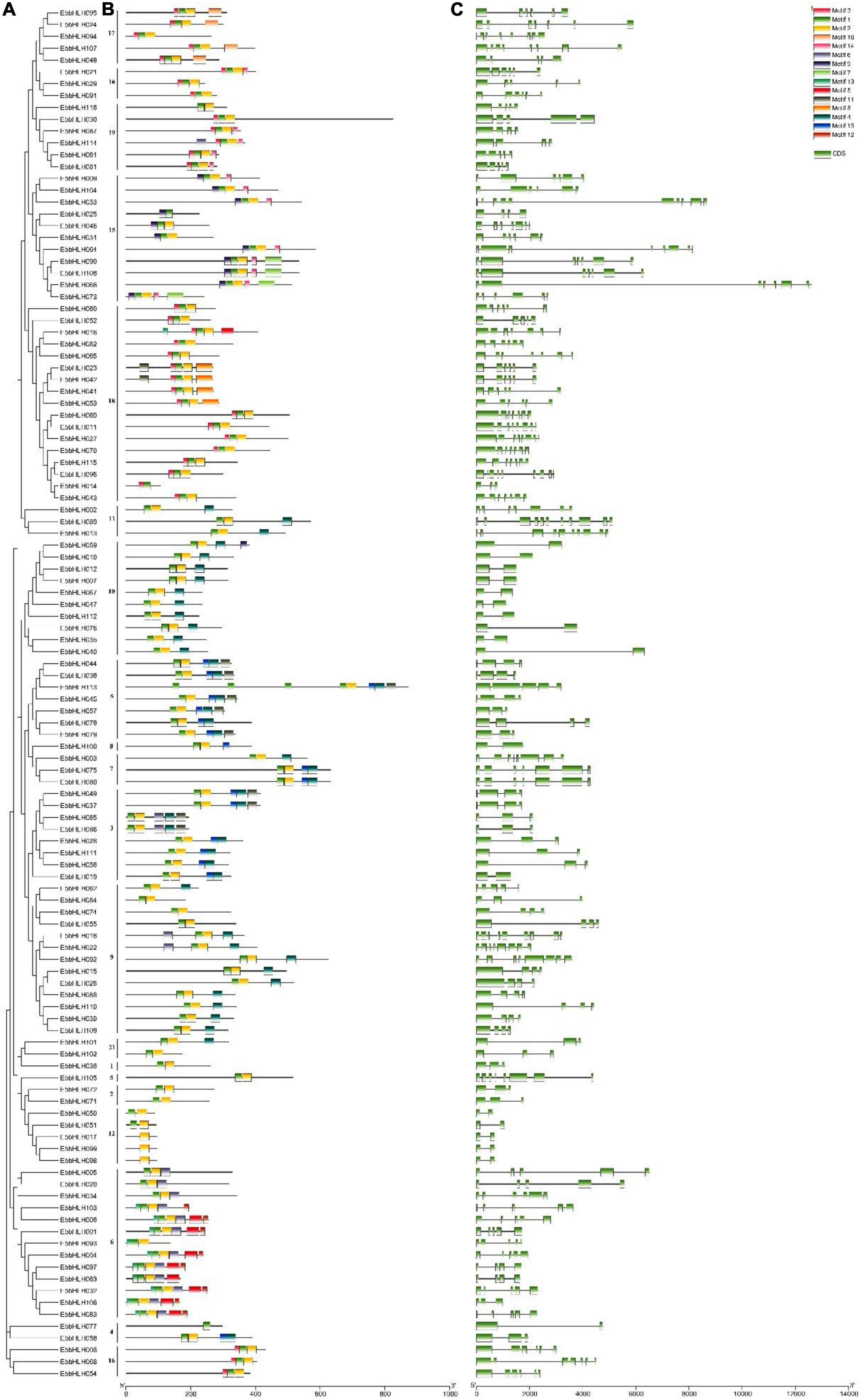
Figure 2. Phylogenetic analysis, gene structure and motif distribution analysis of the bHLH gene family of E. breviscapus. (A) Phylogenetic trees were constructed by the NJ method, repeated 1,000 times at each node. (B) Fifteen amino acid motif in the EbbHLH protein is indicated by a colored box. (C) Exons and introns are indicated by green rectangles and gray lines, respectively.
Then, 15 conserved motifs were identified in E. breviscapus bHLH proteins (Figure 2B). Eighty-six percent of the EbbHLH protein sequences contained both motifs 1 and 2. Each bHLH protein had a conserved motif position. In addition to the highly conserved bHLH domain, EbbHLH members within the same subgroups usually shared similar motif compositions, while high variance was observed among different subgroups, indicating conserved protein architectures within a specific subgroup. Moreover, some subgroup-specific motifs were detected, which might be required for subgroup-specific functions. For example, motifs 1, 2, 4, and 15 were found in all members of subgroup 5, while motifs 1, 2, and 4 were found in all members of subgroup 9. However, some motifs were only found in specific subgroups. For example, motif 12 was only found in subgroup 6, and motif 10 was unique to subgroup 17, whereas motif 9 was detected in subgroups 10 and 15.
To investigate the evolutionary relationships of EbbHLH genes in E. breviscapus, a neighbor-joining method was constructed with amino acid sequences of 116 EbbHLHs from E. breviscapus and 120 AtbHLHs from Arabidopsis (Figure 3). In phylogenetic analysis, EbbHLH proteins were divided into 18 subfamilies (Figure 2A), and their regulatory roles were predicted based on AtbHLH classification. Of these, 17 EbbHLHs were assigned to subfamily 18; 13 EbbHLHs were clustered into subfamily 6 and subfamily 9. However, only 1 EbbHLH was assigned to subfamily 1, subfamily 5, and subfamily 8. Additionally, none of the EbbHLHs were assigned to subfamilies 13, 14, or 20. Moreover, the majority of the EbbHLH gene members of subfamily 7 might be regulatory genes of flavonoid biosynthesis. Of all 116 EbbHLH genes, EbbHLH80 was most similar to AtbHLH042, which is known to be associated with flavonoid biosynthesis. Due to the high homology with AtbHLH042 (AtTT8), it is worth investigating whether EbbHLH80 serves a similar function.
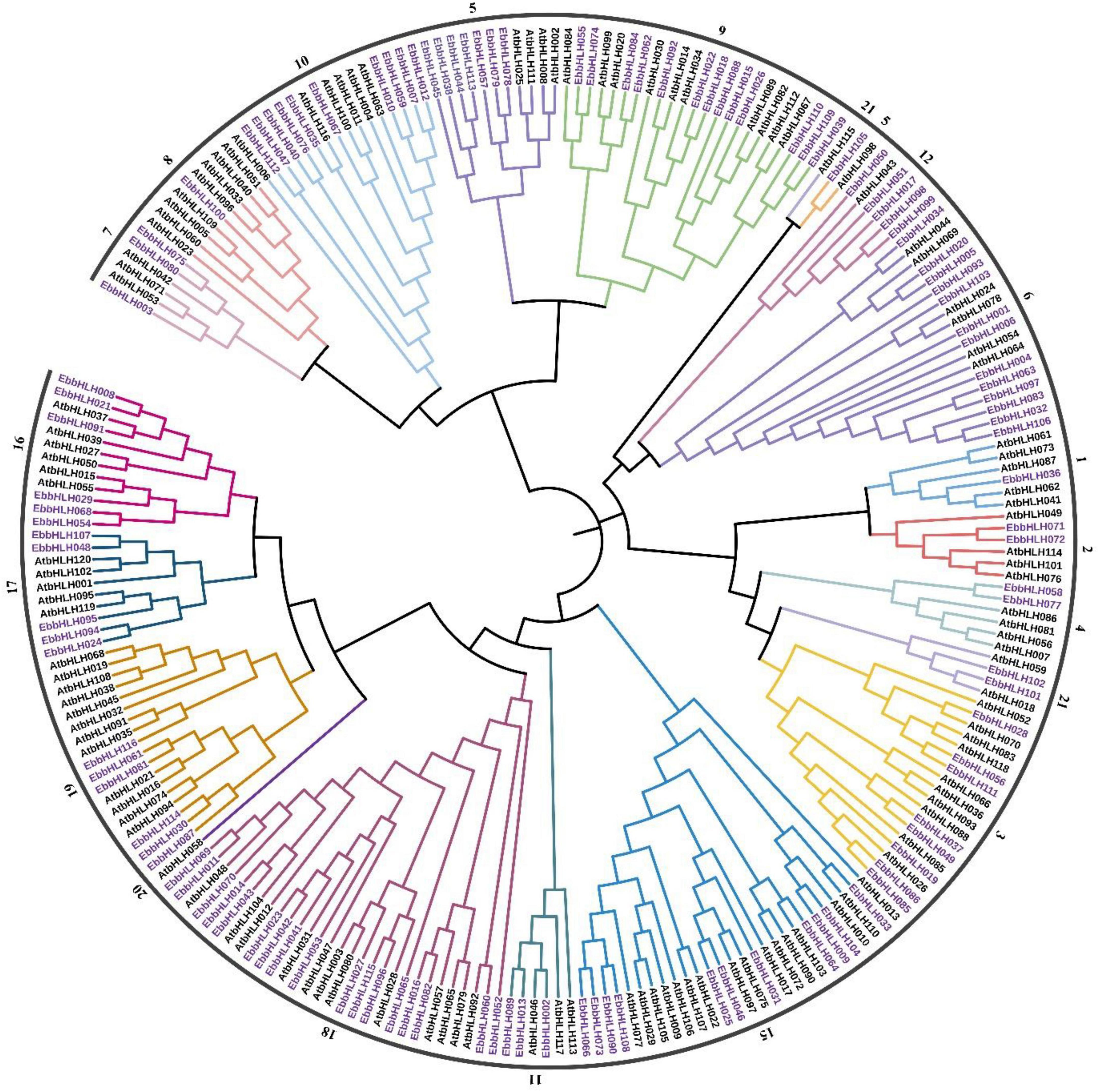
Figure 3. Phylogenetic tree between E. breviscapus and Arabidopsis thaliana. The phylogenetic tree was obtained using the NJ method in MEGA7. The tree shows 21 phylogenetic subfamilies, each subfamily represented by different colors. bHLH protein from Arabidopsis is labeled with the prefix “At.”
In our previous study, EbPAL, EbCHS, and EbCHI genes exhibited high expression levels in leaves, while the main component scutellarin, is rich in the leaves, followed by flowers, stems, and roots (Zhao et al., 2022). To analyze the expression of EbbHLH80, the results showed that the highest amount of EbbHLH80 was found in the leaves of E. breviscapus (Figure 4), which was used in subsequent experiments. To investigate the possible function of EbbHLH80 in flavonoid biosynthesis, the complete ORF of EbbHLH80 (1,896 bp) was isolated and obtained, and then the 35S:EbbHLH80 construct ectopically expressed EbbHLH80 in tobacco (Figure 5A). Several independent 35S:EbbHLH80 transgenic tobacco lines were screened, and three transgenic lines (OE1, 5, and 9) showing higher EbbHLH80 expression levels than other lines were selected for subsequent experiments (Figures 5B,D). The transcription levels of the EbbHLH80 genes in the WT and OE lines were analyzed. Compared with WT, the expression levels of EbbHLH80 in OE lines were significantly increased. Next, we quantified the level of total flavonoids in transgenic and WT plants. The total flavonoid content of OE1, OE5, and OE9 transgenic lines was significantly higher by 1. 41-, 1. 49-, and 1.45-fold, respectively, than that of the WT (P < 0.01; Figure 5C). Therefore, OE5 was chosen and used for further transcriptome and metabolome analysis.
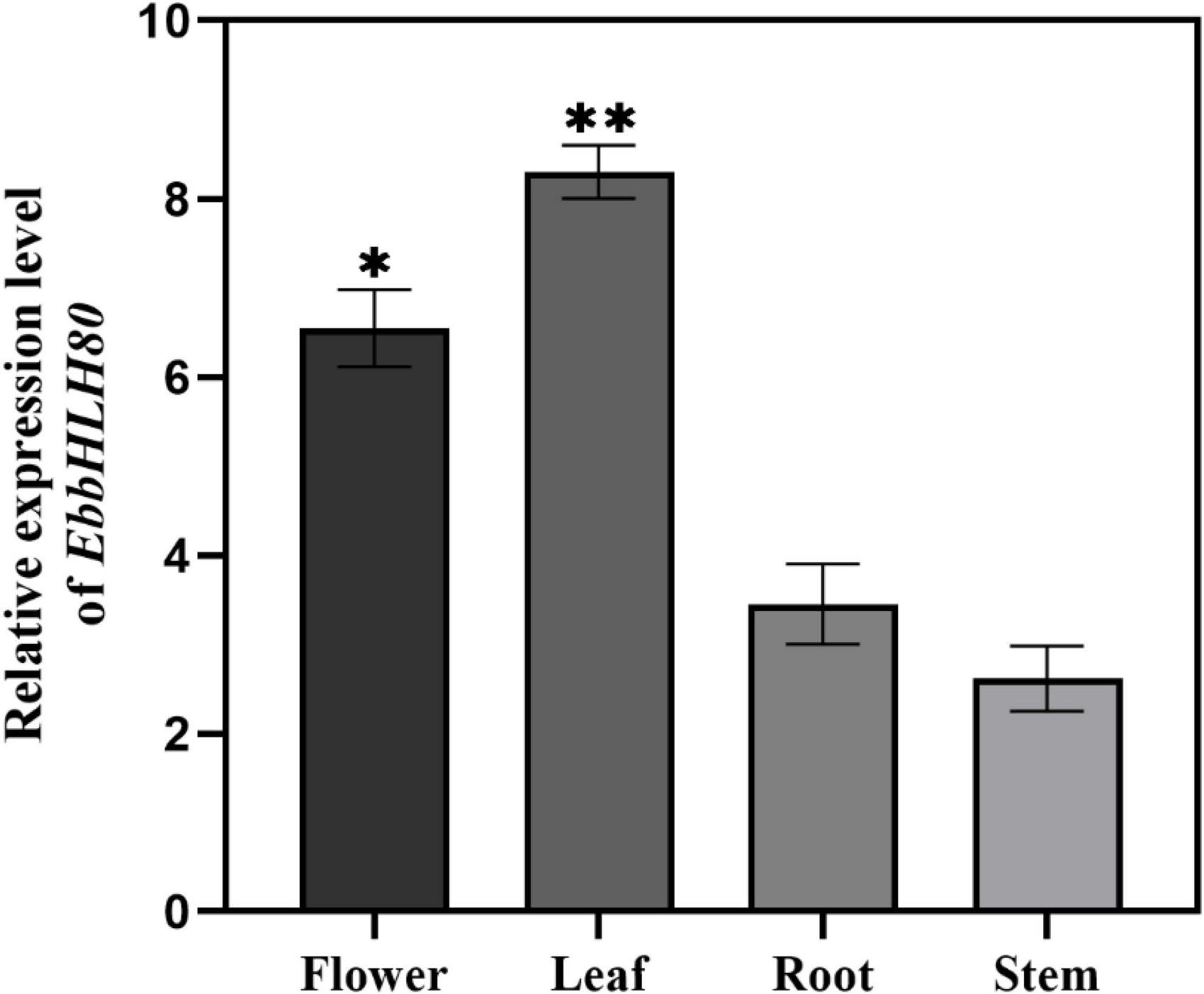
Figure 4. Relative expression of EbbHLH80 in the different tissues of E. breviscapus. Data represent mean ± SD of three biological replicates and the one-way analysis of variance was used for statistical analyses (*P < 0.05; **P < 0.01).
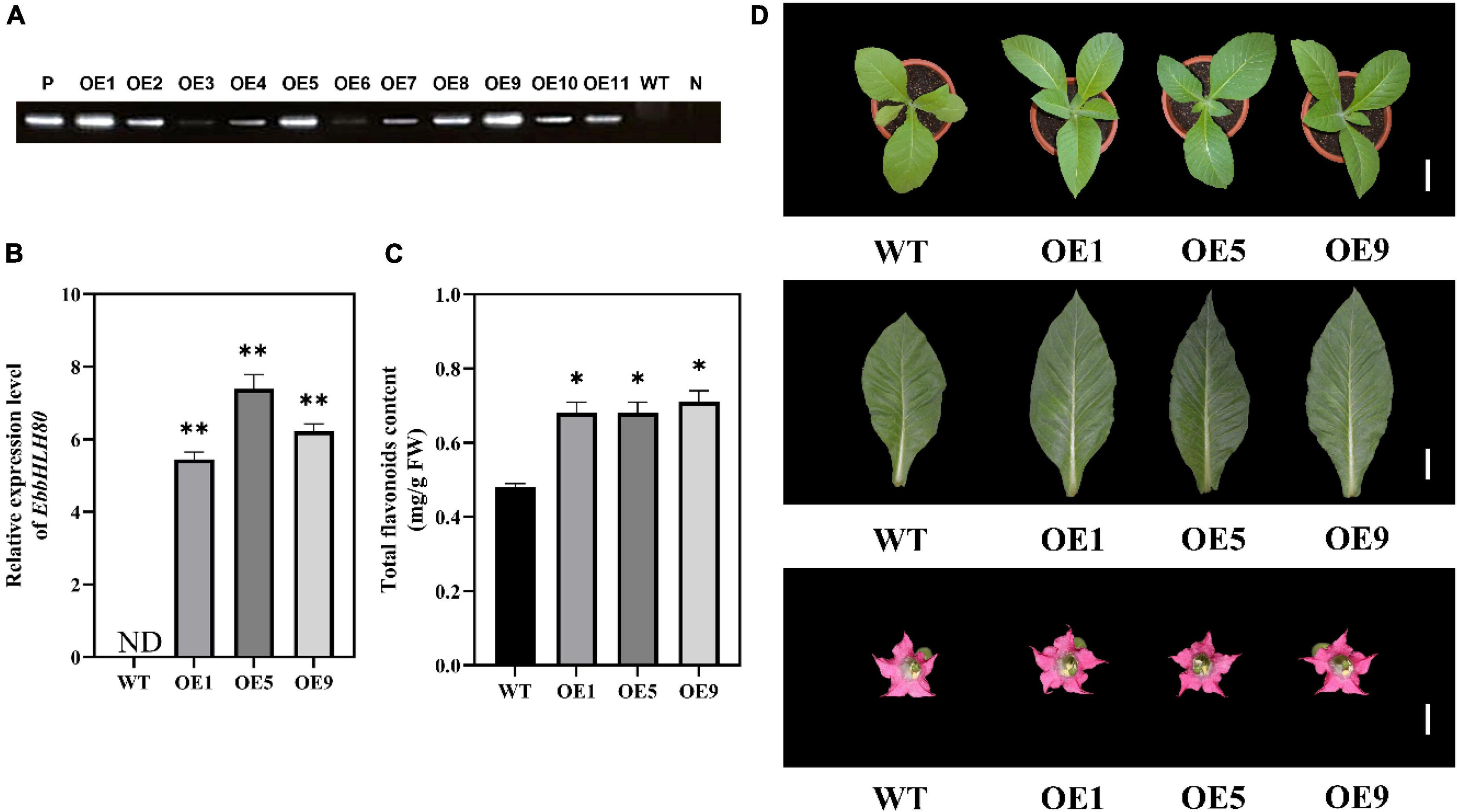
Figure 5. Characterization of transgenic tobacco lines overexpression EbbHLH80. (A) PCR amplification of EbbHLH80 in transgenic lines (OE1-11) and WT plants. P, positive control. N, negative control (double-distilled water, ddH2O). (B) Relative expression level of EbbHLH80 in WT and EbbHLH80–OE (OE1, OE5, and OE9) tobacco transgenic plants. (C) Total Flavonoids content in leaves from 7-week-old plants from WT as well as EbbHLH80–OE (OE1, OE5, and OE9) tobacco transgenic plants. (D) The phenotypes of OE1, OE5, and OE9 tobacco transgenic plants and WT. Bar = 1 cm. Data represent mean ± SD of three biological replicates and the one-way analysis of variance was used for statistical analysis (*P < 0.05 and **P < 0.01).
To compare metabolite accumulation, leaf samples were analyzed using UPLC−ESI−MS/MS. The metabolite profiles of EbbHLH80-OE and WT leaves showed significantly differences. Using the metabolite database, a total of 421 differentially accumulated metabolites were identified. Furthermore, 98 of the metabolites were identified as flavonoid-related, constructing a hierarchical heatmap clustering (Figure 6), including 4 anthocyanins, 2 chalcones, 5 dihydroflavonoids, 3 dihydroflavonols, 3 flavanols, 29 flavonoids, 7 flavonoid carbonosides, 41 flavonols, and 4 isoflavones (Figure 6 and Supplementary Table 6). A clear separation could be observed between EbbHLH80-OE and WT, suggesting distinct flavonoid profiles.
Differentially accumulated metabolites (DAMs) were selected using VIP ≥ 1 and T-test P < 0.05. 214 DAMs were significantly different between the compared samples, and 18 flavonoid metabolites were present at significantly higher levels in the EbbHLH80-OE line than in the WT (Supplementary Table 7). Based on the results, the EbbHLH80 may play a crucial role in regulating flavonoid biosynthesis.
Global gene expression was further profiled in the leaf samples of EbbHLH80-OE and WT. High-quality libraries (with Q30 values higher than 91%) were constructed. The libraries contained 46,694,552–63,472,428 clean reads that mapped successfully to the tobacco genome, with matching rates in the range of 94.03–96.50%. Consequently, a total of 9,377 differentially expressed genes (DEGs) were identified by an absolute log2FC > |1| with FDR < 0.05 between EbbHLH80-OE and WT, including 4,262 upregulated genes and 5,115 downregulated genes (Supplementary Table 8).
GO annotations and KEGG pathway enrichment analysis were used to predict the functions of DEGs. GO annotations assigned these DEGs to 45 functional terms. Twenty-one were categorized into biological processes, 10 into molecular functions, and 14 into cellular components (Figure 7). The genes clustered in the biological processes group were mainly related to metabolic processes (1,379 sequences), cellular processes (1,107 sequences) and single tissue processes (904 sequences). The molecular function terms were related to catalytic activity (660 sequences), binding (778 sequences) and transporter activity (113 sequences). The majority of genes involved in cellular components were located in the cell (760 sequences), cell part (760 sequences) and organelle (561 sequences). Moreover, KEGG pathway enrichment analysis showed that DEGs were significantly enriched in biosynthesis of secondary metabolites, ribosome plant hormone signal transduction and the MAPK signaling pathway (Figure 8 and Supplementary Table 9).
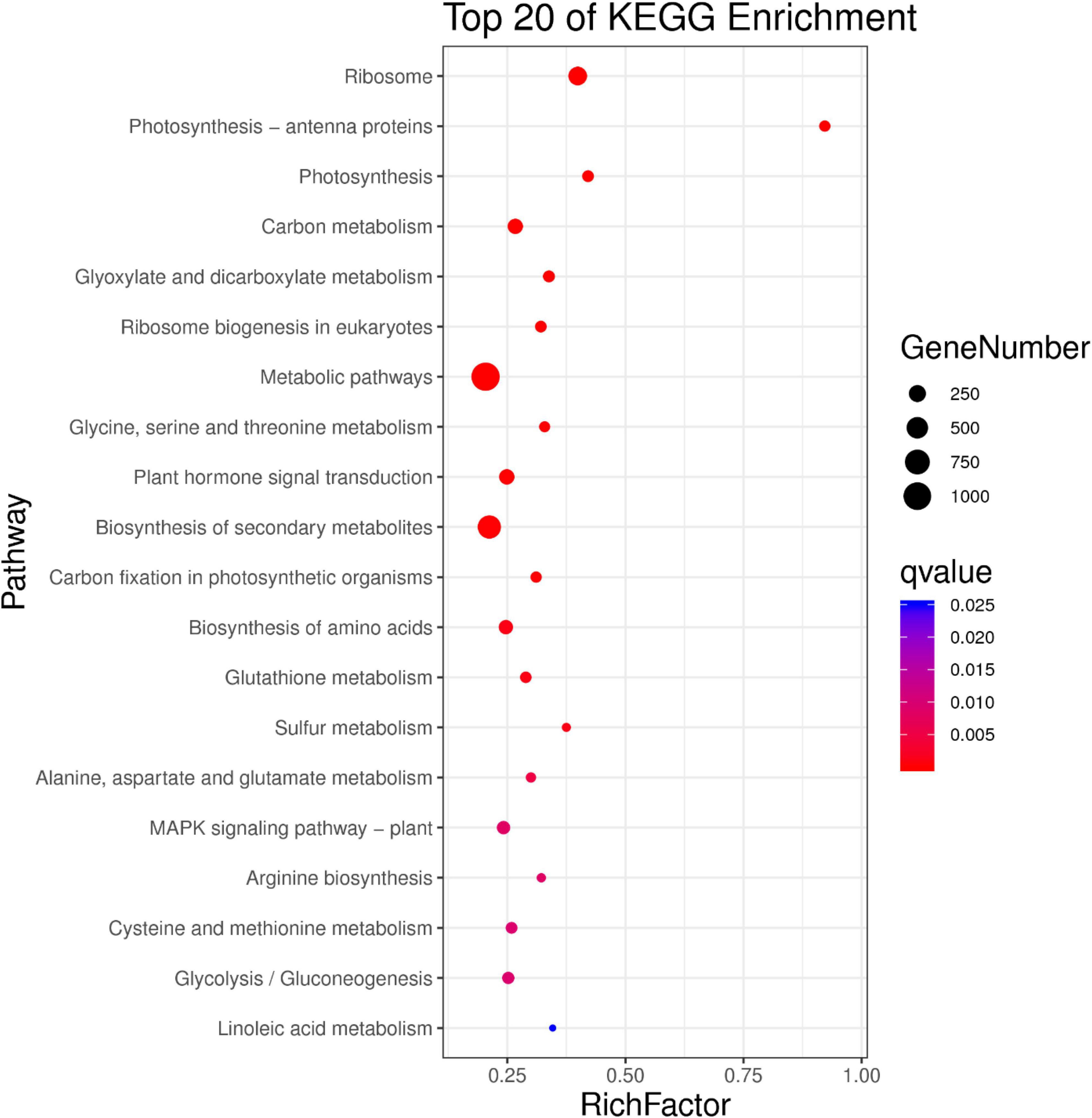
Figure 8. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs between WT and OE5. The X-axis represents the enrichment factor, and the Y-axis represents the enrichment gene. The size of the dot represents the number of differentially enriched genes.
Predicted coding genes associated with transport were identified by transcriptome data analysis (Supplementary Table 10), including 38 glutathione S–transferase (GST), 24 multidrug and toxic compound extrusion (MATE), and 48 ATP-binding cassette (ABC) transporters. Among them, 20 ABCs, 3 GSTs, and 7 MATEs were significantly upregulated in EbbHLH80-OE (FDR < 0.05 and log2FC > |1|), indicating that they may be key flavonoid transport genes.
Three hundred and fifty-five DEGs were predicted to encode TFs that regulate flavonoid synthesis (Supplementary Table 11). Of these DEGs, there were 78 ethylene response factors (ERFs), 22 MYBs, 24 bHLHs, 45 WRKYs, and 3 NACs. Genes annotated as MYB35 (LOC107815451), MYB86 (LOC107802100, LOC107762209), MYB48 (LOC107814331), MYB32 (LOC107807690), and MYB44-like (LOC107816351) were upregulated transcription factors. A significant change in expression was also observed for TFs annotated as bHLH, WRKY, NAC, MYB, and ERF family members (Supplementary Tables 12–16). In addition, these genes are not only involved in plant growth and development, but also involved in responses to ethylene signaling pathways, biosynthesis of flavonoids and anthocyanins, regulation of the expression of ABA inducible genes, and involvement in resistance to multiple stress responses. Moreover, 1 DEG was identified that was involved in the light signal transduction pathway. Genes associated with GATA transcription factor 9 were annotated as GATA transcription factors (Supplementary Table 17).
An analysis of the expression levels of nine DEGs involved in flavonoid biosynthesis in WT and EbbHLH80-OE plants was conducted to further validate the RNA-seq results. The results confirmed that flavonoid biosynthesis and regulatory genes, including PAL, C4H, 4CL, CHS, CHI, FLS2, F3H, DFR, and ANS, were upregulated in EbbHLH80-OE (Figure 9). A complete correlation was observed between the RT-qPCR and RNA-seq results, which demonstrated the reliability of the RNA-seq data and DEGs analysis.
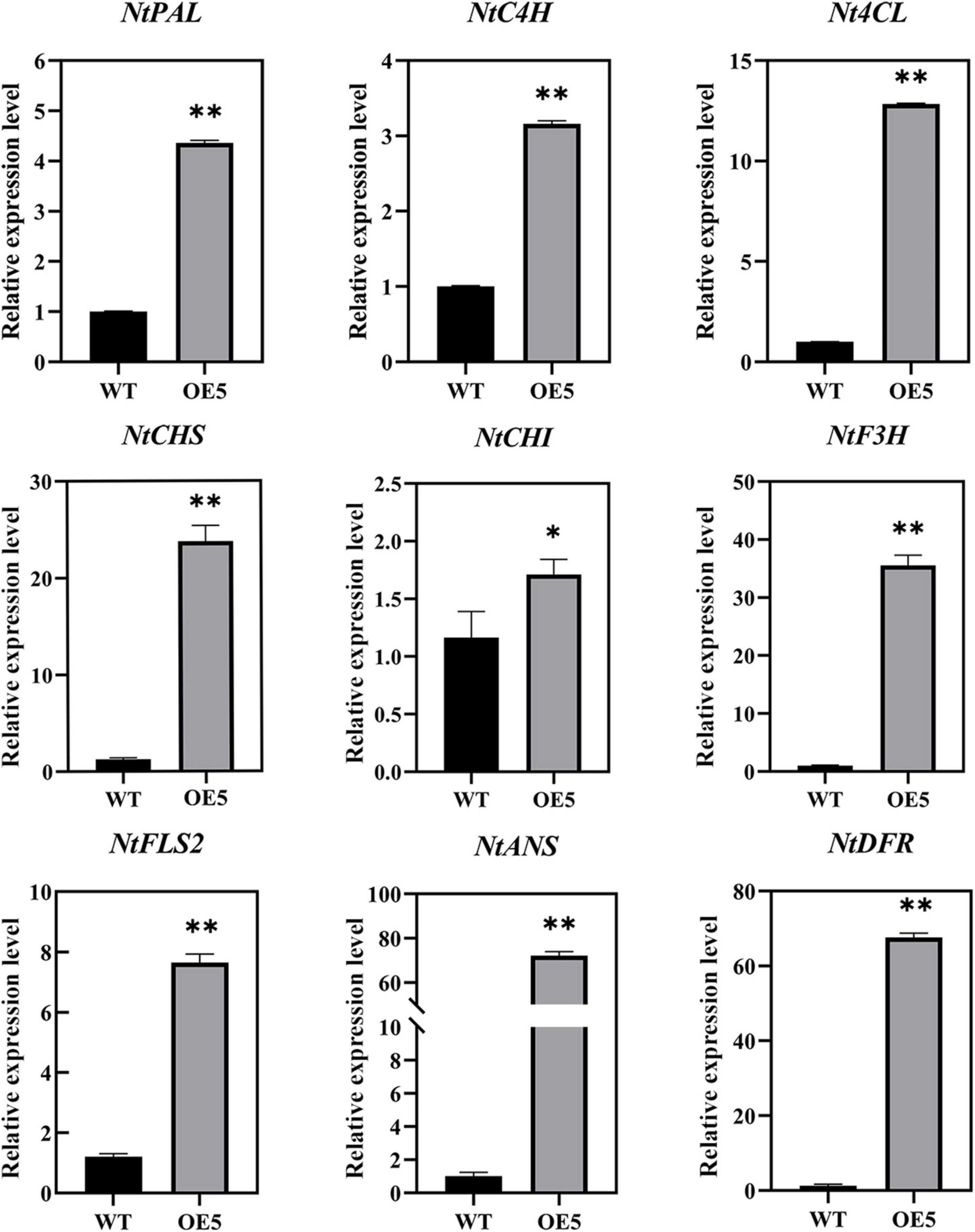
Figure 9. Expression analysis of flavonoid biosynthesis genes between WT and OE5. The characters on the X-axis indicate the WT and OE5. The Y-axis represents the relative level of gene expression. Data represent mean ± SD of three biological replicates and the one-way analysis of variance was used for statistical analysis (*P < 0.05; **P < 0.01).
Since the full genome sequence of E. breviscapus was completed (He et al., 2021), we have been able to identify and characterize all members of the bHLH family. The E. breviscapus genome contains 116 bHLH genes whose predominant nuclear location is consistent with their function as transcription factors (Supplementary Table 5). The number of EbbHLH genes was smaller than that in Arabidopsis (162) (Bailey et al., 2003), rice (167) (Li et al., 2006), tomato (152) (Wang et al., 2015), cucumber (142) (Li et al., 2020), pepper (122) (Zhang et al., 2020), and pear (197) (Dong et al., 2021). The reason for this may be the difference in genome size between the plants or evolutionary divergence. A previous study showed that the Arabidopsis bHLH gene family contained 21 subgroups (Toledo-Ortiz et al., 2003). The phylogenetic analysis showed that the E. breviscapus bHLH gene family contained 18 subgroups (Figure 3) with wildly varied gene numbers from 2 (subgroups 2, subgroups 4, and subgroups 21) to 17 (subgroups 18) and three one-gene subgroups 1, subgroups 5, and subgroups 8. Most EbbHLH proteins clustered in the same group share similar motifs, suggesting that these conserved motifs play crucial roles in the group functions. The gene and protein structure analysis showed that the EbbHLH family also has a broad diversity in intron/exon organizations as well the protein motif patterns.
In plants, flavonoids play an important role in pigmentation (Lai et al., 2014). Flavonoids are found in six main groups: anthocyanins, flavan-3-ols, flavanonols, flavanols, flavones, and phenolic acids. Flavonoids are related to plant stress resistance, as they can improve the ability of plants to resist multiple stresses, thereby increasing plant resistance to damage and protecting the plant from disease (Panche et al., 2016). According to recent research, flavonoid structural genes are regulated by bHLH transcription factors (Bai et al., 2011). There have been many studies on flavonoid metabolism and accumulation in plants based on transcriptome and targeted metabolome analyses. Analyses of TT8 inducible overexpression and loss-of-function lines revealed that TT8 coordinates glycosylation of nucleotides and flavonoids, suggesting that TT8 controls nucleotide sugar synthesis (Rai et al., 2016). In TT8 of the B. napus cultivar QinYou NO. 7, TT8 is primarily expressed during seed development. BnTT8 can produce yellow seed coats, higher fatty acid concentrations, and lower storage protein concentrations in tt8-4 mutants (Qi et al., 2017). Through the metabolome analysis of E. breviscapus, 214 DAMs were identified, 18 of which were flavonoid metabolites. In EbbHLH80-OE lines, the contents of flavonoid metabolites are significantly higher compared with WT, and these findings will provide a more comprehensive approach to EbbHLH80-mediated regulation of flavonoid biosynthesis.
EbbHLH80-OE leaves accumulate significantly more flavonoids than WT, suggesting that this is a direct result of the higher expression of flavonoid biosynthesis genes (PAL, C4H, 4CL, CHI, CHS, F3H, FLS2, DFR, and ANS). The correlations between key DEGs and metabolites associated with flavonoid biosynthesis in EbbHLH80-OE tobacco leaves were analyzed. Of these, LOC107800382, LOC107781346, LOC107781089, and LOC107824957 showed significant positive correlations with nine types of flavonoid metabolites (Supplementary Figure 1). Previous studies showed that anthocyanin content was positively correlated with the expression of early or late biosynthesis genes (Rahim et al., 2014). The coexpression of flavonoid-related genes in synthesis and metabolism may be a universal regulatory mechanism by which the flavonoid pathway controls structural gene expression. Anthocyanins are transferred from the endoplasmic reticulum to the vacuole by GSTs in plants (Mueller et al., 2000). In Arabidopsis, GSTs such as TT19 were found to correlate positively with anthocyanin accumulation (Kitamura et al., 2004). Compared to the WT, we found that the transcription of 3 GST genes increased significantly in EbbHLH80-OE tobacco leaves, corresponding to higher anthocyanin levels. Thus, upregulated GSTs are likely to facilitate anthocyanin transport and increase flavonoid levels within the vacuoles. Additionally, anthocyanin is transported and accumulated by ABC transporters and MATE family members (Goodman et al., 2004; Gomez et al., 2011). In this study, 3 GSTs, 7 MATEs, and 20 ABCs were positively correlated with flavonoid accumulation.
It has been shown that MYB alone, coexpression of MYB and bHLH, or the MYB-bHLH-WD40 complex is sufficient to induce flavonoid accumulation in plants (Xie et al., 2016). Several studies have shown that flavonoid biosynthesis is also controlled by TFs, such as the WRKY and ERF families, as well as by lncRNAs and miRNAs (Yao et al., 2017; Yang et al., 2019; An et al., 2020). According to functional assays, ERFs play a role in fruit peel degreening, fruit ripening, and hormone signal transduction (Zhou et al., 2016; Han et al., 2018). Additionally, ERFs have recently been discovered to be involved in anthocyanin biosynthesis (An et al., 2020). In this study, 49 ERFs were upregulated in EbbHLH80-OE compared to WT (Supplementary Table 16). In a previous study, the ethylene response factor (ERF) protein MdERF109 was found to be related to anthocyanin biosynthesis, promoting coloration by directly binding to the promoters of anthocyanin-related genes and simultaneously interacting with anthocyanin biosynthesis genes (Ma et al., 2021).
Taken together, these results suggest that EbbHLH80 activates ERF109 TFs, activating genes involved in flavonoid biosynthesis pathways and increasing flavonoid concentrations. This indicated that the highly accumulated flavonoid in EbbHLH80-OE might be attributed to the activation of ERF genes. However, there will be a need to investigate how the spontaneously changed genes in EbbHLH80-OE and evaluate how it could cause the accumulations in the expression levels of these transcription factors and their structural genes.
The present study provided a comprehensive analysis of the bHLH gene family from a genome-wide perspective. In E. breviscapus, 116 EbbHLH genes are found, divided into 18 subfamilies and distributed unevenly on 9 chromosomes. A phylogenetic analysis of these genes was further supported by their similar exon–intron structures and conserved motif compositions. Combining similar gene expression patterns with integrated metabolomic and transcriptomic analysis, EbbHLH80-OE tobacco leaves contain higher flavonoids and have altered gene expression patterns compared with Yunyan87 tobacco leaves. Several key genes involved in flavonoid synthesis were upregulated in the transgenic EbbHLH80-OE lines, resulting in significantly higher levels of flavonoid.
The original contributions presented in this study are publicly available. This data can be found here: NCBI, SRR19135011–SRR19135016.
YZ, CZ, and SY conceived and designed the experiments. QG, WS, XL, and CX performed the experiments. GC, GX, XYL, GZ, and XNL analyzed the data. YZ, QG, and CZ wrote the article. All authors contributed to the article and approved the submitted version.
This work was supported by the Major Science and Technique Programs in Yunnan Province (No. 2019ZF011-1) and the National Natural Science Foundation of China (No. 81760694).
We are grateful to Hannah Dahl for reading and commenting on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.977649/full#supplementary-material
An, J. P., Li, H. H., Song, L. Q., Su, L., Liu, X., You, C. X., et al. (2016). The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol. Biochem. 108, 24–31. doi: 10.1016/j.plaphy.2016.06.032
An, J. P., Zhang, X. W., Bi, S. Q., You, C. X., Wang, X. F., and Hao, Y. J. (2020). The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 101, 573–589. doi: 10.1111/tpj.14555
Atchley, W. R., Terhalle, W., and Dress, A. (1999). Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 48, 501–516. doi: 10.1007/pl00006494
Bai, H., Song, Z., Zhang, Y., Li, Z., Wang, Y., Liu, X., et al. (2020). The bHLH transcription factor PPLS1 regulates the color of pulvinus and leaf sheath in foxtail millet (Setaria italica). Theor. Appl. Genet. 133, 1911–1926. doi: 10.1007/s00122-020-03566-4
Bai, Y., Pattanaik, S., Patra, B., Werkman, J. R., Xie, C. H., and Yuan, L. (2011). Flavonoid-related basic helix-loop-helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 234, 363–375. doi: 10.1007/s00425-011-1407-y
Bailey, P. C., Martin, C., Toledo-Ortiz, G., Quail, P. H., Huq, E., Heim, M. A., et al. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15, 2497–2502. doi: 10.1105/tpc.151140
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Hu, B., Qin, Y., Hu, G., and Zhao, J. (2019). Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 136, 178–187. doi: 10.1016/j.plaphy.2019.01.024
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Chu, Q., Wu, T., Fu, L., and Ye, J. (2005). Simultaneous determination of active ingredients in Erigeron breviscapus (Vant.) Hand-Mazz. by capillary electrophoresis with electrochemical detection. J. Pharmaceutical Biomed. Anal. 37, 535–541. doi: 10.1016/j.jpba.2004.11.018
Dong, H., Chen, Q., Dai, Y., Hu, W., Zhang, S., and Huang, X. (2021). Genome-wide identification of PbrbHLH family genes, and expression analysis in response to drought and cold stresses in pear (Pyrus bretschneideri). BMC Plant Biol. 21:86. doi: 10.1186/s12870-021-02862-5
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Gomez, C., Conejero, G., Torregrosa, L., Cheynier, V., Terrier, N., and Ageorges, A. (2011). In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 67, 960–970. doi: 10.1111/j.1365-313X.2011.04648.x
Goodman, C. D., Casati, P., and Walbot, V. (2004). A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16, 1812–1826. doi: 10.1105/tpc.022574
Han, Z., Hu, Y., Lv, Y., Rose, J. K. C., Sun, Y., Shen, F., et al. (2018). Natural variation underlies differences in ETHYLENE RESPONSE FACTOR17 activity in fruit peel degreening. Plant Physiol. 176, 2292–2304. doi: 10.1104/pp.17.01320
He, S., Dong, X., Zhang, G., Fan, W., Duan, S., Shi, H., et al. (2021). High quality genome of Erigeron breviscapus provides a reference for herbal plants in Asteraceae. Mol Ecol. Resour. 21, 153–169. doi: 10.1111/1755-0998.13257
Huang, W., Khaldun, A. B. M., Chen, J., Zhang, C., Lv, H., Yuan, L., et al. (2016). A R2R3-MYB transcription factor regulates the flavonol biosynthetic pathway in a traditional Chinese medicinal plant, Epimedium sagittatum. Front. Plant Sci. 7:1089. doi: 10.3389/fpls.2016.01089
Jin, S. W., Rahim, M. A., Kim, H. T., Park, J. I., Kang, J. G., and Nou, I. S. (2018). Molecular analysis of anthocyanin-related genes in ornamental cabbage. Genome 61, 111–120. doi: 10.1139/gen-2017-0098
Kitamura, S., Shikazono, N., and Tanaka, A. (2004). TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 37, 104–114. doi: 10.1046/j.1365-313x.2003.01943.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lai, B., Li, X. J., Hu, B., Qin, Y. H., Huang, X. M., Wang, H. C., et al. (2014). LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis. PLoS One 9:e86293. doi: 10.1371/journal.pone.0086293
Li, C., Qiu, J., Ding, L., Huang, M., Huang, S., Yang, G., et al. (2017). Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiol. Biochem. 112, 335–345. doi: 10.1016/j.plaphy.2017.01.019
Li, J., Wang, T., Han, J., and Ren, Z. (2020). Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol. 20:272. doi: 10.1186/s12870-020-02440-1
Li, L., Hao, X., Liu, H., Wang, W., Fu, X., Ma, Y., et al. (2019). Jasmonic acid-responsive AabHLH1 positively regulates artemisinin biosynthesis in Artemisia annua. Biotechnol. Appl. Biochem. 66, 369–375. doi: 10.1002/bab.1733
Li, N., Wu, H., Ding, Q., Li, H., Li, Z., Ding, J., et al. (2018). The heterologous expression of Arabidopsis PAP2 induces anthocyanin accumulation and inhibits plant growth in tomato. Funct. Integr. Genomics 18, 341–353. doi: 10.1007/s10142-018-0590-3
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Liu, X., Cheng, J., Zhang, G., Ding, W., Duan, L., Yang, J., et al. (2018). Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 9:448. doi: 10.1038/s41467-018-02883-z
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, H., Yang, T., Li, Y., Zhang, J., Wu, T., Song, T., et al. (2021). The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell 33, 3309–3330. doi: 10.1093/plcell/koab188
Marrs, K. A., Alfenito, M. R., Lloyd, A. M., and Walbot, V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. doi: 10.1038/375397a0
Mueller, L. A., Goodman, C. D., Silady, R. A., and Walbot, V. (2000). AN9, a petunia glutathione S-Transferase required for anthocyanin sequestration, is a flavonoid-binding protein1. Plant Physiol. 123, 1561–1570. doi: 10.1104/pp.123.4.1561
Narasimhan, K., Basheer, C., Bajic, V. B., and Swarup, S. (2003). Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol. 132, 146–153. doi: 10.1104/pp.102.016295
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Panche, A. N., Diwan, A. D., and Chandra, S. R. (2016). Flavonoids: an overview. J. Nutritional Sci. 5:e47. doi: 10.1017/jns.2016.41
Pelletier, L. G., Dion, S. C., Tuson, K. M., and Green-Demers, I. (1999). Why do people fail to adopt environmental protective behaviors? Toward Taxonomy Environ. Amotivation 29, 2481–2504. doi: 10.1111/j.1559-1816.1999.tb00122.x
Qi, S., Liu, K., Gao, C., Li, D., Jin, C., Duan, S., et al. (2017). The effect of BnTT8 on accumulation of seed storage reserves and tolerance to abiotic stresses during Arabidopsis seedling establishment. Plant Growth Regulation 82, 271–280. doi: 10.1007/s10725-017-0257-4
Rahim, M. A., Busatto, N., and Trainotti, L. (2014). Regulation of anthocyanin biosynthesis in peach fruits. Planta 240, 913–929. doi: 10.1007/s00425-014-2078-2
Rai, A., Umashankar, S., Rai, M., Kiat, L. B., Bing, J. A., and Swarup, S. (2016). Coordinate regulation of metabolite glycosylation and stress hormone biosynthesis by TT8 in Arabidopsis. Plant Physiol. 171, 2499–2515. doi: 10.1104/pp.16.00421
Riechmann, L., and Muyldermans, S. (1999). Single domain antibodies: comparison of camel VH and camelised human VH domains. J. Immunol. Methods 231, 25–38. doi: 10.1016/s0022-1759(99)00138-6
Su, W., Lu, J., Zhang, G., and Wang, C. (2001). Ecological and biological analysis of total flavonoids in Erigeron breviscapus. Chinese Traditional Herbal Drugs 32, 1119–1121.
Sun, X., Li, L., Pei, J., Liu, C., and Huang, L. F. (2020). Metabolome and transcriptome profiling reveals quality variation and underlying regulation of three ecotypes for Cistanche deserticola. Plant Mol. Biol. 102, 253–269. doi: 10.1007/s11103-019-00944-5
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Wang, F., Zhu, H., Chen, D., Li, Z., Peng, R. H., and Yao, Q. (2016b). A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Culture (PCTOC) 125, 387–398. doi: 10.1007/s11240-016-0953-1
Wang, F., Kong, W., Wong, G., Fu, L., Peng, R., Li, Z., et al. (2016a). AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genomics 291, 1545–1559. doi: 10.1007/s00438-016-1203-2
Wang, J., Hu, Z., Zhao, T., Yang, Y., Chen, T., Yang, M., et al. (2015). Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genomics 16:39. doi: 10.1186/s12864-015-1249-2
Wang, L., Tang, W., Hu, Y., Zhang, Y., Sun, J., Guo, X., et al. (2019). A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 99, 359–378. doi: 10.1111/tpj.14330
Wang, P., Su, L., Gao, H., Jiang, X., Wu, X., Li, Y., et al. (2018). Genome-Wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis. Front. Plant Sci. 9:64. doi: 10.3389/fpls.2018.00064
Xiang, L., Liu, X., Li, X., Yin, X., Grierson, D., Li, F., et al. (2015). A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS One 10:e0143892. doi: 10.1371/journal.pone.0143892
Xie, Y., Tan, H., Ma, Z., and Huang, J. (2016). DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant 9, 711–721. doi: 10.1016/j.molp.2016.01.014
Xu, W., Grain, D., Bobet, S., Le Gourrierec, J., Thévenin, J., Kelemen, Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 202, 132–144. doi: 10.1111/nph.12620
Yang, T., Ma, H., Zhang, J., Wu, T., Song, T., Tian, J., et al. (2019). Systematic identification of long noncoding RNAs expressed during light-induced anthocyanin accumulation in apple fruit. Plant J. 100, 572–590. doi: 10.1111/tpj.14470
Yao, G., Ming, M., Allan, A. C., Gu, C., Li, L., Wu, X., et al. (2017). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92, 437–451. doi: 10.1111/tpj.13666
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18, 1153–1168. doi: 10.1111/pbi.13281
Zhang, Z., Chen, J., Liang, C., Liu, F., Hou, X., and Zou, X. (2020). Genome-Wide identification and characterization of the bHLH transcription factor family in pepper (Capsicum annuum L.). Front. Genet. 11:570156. doi: 10.3389/fgene.2020.570156
Zhao, J., and Dixon, R. A. (2009). MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21, 2323–2340. doi: 10.1105/tpc.109.067819
Zhao, Y., Zhang, G., Tang, Q., Song, W., Gao, Q., Xiang, G., et al. (2022). EbMYBP1, a R2R3-MYB transcription factor, promotes flavonoid biosynthesis in Erigeron breviscapus. Front. Plant Sci. 13:946827. doi: 10.3389/fpls.2022.946827
Zhao, Y., Zhang, Y., Liu, H., Zhang, X., Ni, R., Wang, P., et al. (2019). Functional characterization of a liverworts bHLH transcription factor involved in the regulation of bisbibenzyls and flavonoids biosynthesis. BMC Plant Biol. 19:497. doi: 10.1186/s12870-019-2109-z
Zhou, X., Zhang, Z. L., Park, J., Tyler, L., Yusuke, J., Qiu, K., et al. (2016). The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol. 171, 2760–2770. doi: 10.1104/pp.16.00154
Keywords: EbbHLH80, flavonoids, bHLH transcription factors, anthocyanins, Erigeron breviscapus
Citation: Gao Q, Song W, Li X, Xiang C, Chen G, Xiang G, Liu X, Zhang G, Li X, Yang S, Zhai C and Zhao Y (2022) Genome-wide identification of bHLH transcription factors: Discovery of a candidate regulator related to flavonoid biosynthesis in Erigeron breviscapus. Front. Plant Sci. 13:977649. doi: 10.3389/fpls.2022.977649
Received: 24 June 2022; Accepted: 15 August 2022;
Published: 14 September 2022.
Edited by:
Chunzhao Liu, Qingdao University, ChinaReviewed by:
Feng Xu, Yangtze University, ChinaCopyright © 2022 Gao, Song, Li, Xiang, Chen, Xiang, Liu, Zhang, Li, Yang, Zhai and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengchao Yang, c2hlbmdjaGFveWFuZ0AxNjMuY29t; Chenxi Zhai, Y2hlbnhpemhhaUBvdXRsb29rLmNvbQ==; Yan Zhao, emhhb3lhbmttQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.