- 1Key Laboratory of Fruit Postharvest Biology, Liaoning Province, College of Horticulture, Shenyang Agricultural University, Shenyang, China
- 2General Station of Agricultural Technology Extension, Xinjiang Production and Construction Corps, Urumqi, China
Stone cells are sclerenchyma cells formed by deposition of lignin, which is the most significant factor limiting the quality of pears. Ca2+ was known to inhibit stone cells in pear fruits, but the underlying molecular mechanism remains unclear. Our study revealed that exogenous CaCl2 (Ca2+) treatment of “Nanguo” pear (Pyrus ussuriensis) suppressed the synthesis of lignin and stone cell production. We further analysed the transcriptomes using RNA-seq, identified a transcription factor, PuDof2.5, and its targets gene PuPRX42-like (lignin polymerase gene) expression decreased in CaCl2-treated samples, which are involved in suppressing lignin biosynthesis in pear fruit. PuDof2.5 was found to bind directly to the PuPRX42-like promoter and induced its transcription. Taken together, our results revealed that Ca2+ modulated the key lignin biosynthetic transcription factor PuDof2.5 to suppress stone cell production in pear fruits.
Introduction
The Nanguo pear (Pyrus ussuriensis Maxim) is one of the most popular pears in northern China, and it is deeply loved by consumers for its excellent taste and flavor (Wei et al., 2017). The stone cell content of the Nanguo pear is relatively high, which reduces fruit quality and results in rough flesh texture, lowering its economic value. Therefore, minimizing the stone cell content is essential to improve Nanguo pear quality.
Stone cells are formed from parenchymal cells by deposition of lignin (Rogers and Campbell, 2004). Stone cell production is closely correlated with lignin synthesis, transfer, and deposition (Barros et al., 2015). The biosynthetic pathway of lignin has been studied in pear fruits (Cai et al., 2010). The process starts with the formation of phenylacrylic acid (PA) by the enzyme phenylalanine ammonia lyase (PAL) from L-Phe (L-phenylalanine); cinnamate 4-hydroxylase (C4H) and 4-hydroxycinnamate-CoA ligase (4CL) catalyze the conversion of PA, cinnamyl alcohol dehydrogenase (CAD), and other key enzymes form lignin monomers; finally, the monomers are converted to lignin complexes by peroxidase (PRX) and laccase (LAC).
The key role of PRX genes in plant cell wall lignification has been proved (Shigeto and Tsutsumi, 2016). In Arabidopsis (Arabidopsis thaliana), cell-specific downregulation of the peroxidase AtPRX64 causes a marked delay in the formation of the casparian band, a lignin-based paracellular diffusion barrier in plants (Lee et al., 2019). Overexpression of FaPRX27 in strawberry fruit, supporting peroxidase involved in the root and fruit lignification (Ring et al., 2013). In the loss-of-function Arabidopsis mutant, PRX17 showed lower lignin content in leafs, while overexpression of PRX17 showed the opposite phenotype (Cosio et al., 2017). These findings suggested a broader role of PRXs genes in lignin biosynthesis.
The DNA-binding One Zinc Finger (Dof) is a family of plant-specific transcription factors that bind to the promoter of their targeted genes at the consensus sequence AAAG (Yanagisawa and Schmidt, 1999). These transcription factors also participate in various developmental processes and respond to environmental stimuli in plants (Khaksar et al., 2019). To date, Dof TF-regulated lignin production has been demonstrated in several plants (Rogers et al., 2005; Li et al., 2019; Cao et al., 2020). In Arabidopsis, the double mutants VDof1 and VDof2 showed enhanced lignin deposition, suggesting VDof1 and VDof2 as negative regulators of lignin deposition (Ramachandran et al., 2020). However, PuDof2.5, which regulates the stone cell biosynthesis mechanism in Nanguo pear, is not fully understood.
Calcium is a secondary messenger that participates in cell signaling pathways and is closely related to cell wall development, which plays an important role in regulating fruit quality (Wójcik et al., 2014). Modulation of calcium has been reported in terms of the fruit quality of several pear species (Sajid et al., 2019; Zudaire et al., 2019; Dalzochio et al., 2021). Meanwhile, Ca2+ also regulates the lignin synthesis process. For example, exogenous application of.5% CaCl2 in “Niitaka” pear fruits can decrease PRX enzyme activity to inhibit lignin synthesis (Lee et al., 2007). In “Whangkeumbae” pear, CaCl2 significantly reduced PAL, CAD, and PRX enzyme activities, and the expression levels of PpCAD1 and PpCAD2 were downregulated (Lu et al., 2015). Ca2+ regulates lignin biosynthetic molecular mechanism in “Nanguo” pear remains unclear.
Several studies have indicated that Ca2+ can decrease stone cell content. However, most of them only focused on changes in lignin content and related gene expression. In the present study, we characterized the role of PuDof2.5 in pear fruits. PuDof2.5 was lowly expressed in fruits after Ca2+ treatment. Finally, stone cells were decreased by inhibiting the expression of lignin synthesis gene PuPRX42-like. These findings provide a better understanding of the molecular mechanism by which Ca2+ inhibits stone cell production in pear fruits.
Materials and methods
Plant materials and treatments
Nanguopear (n = 65) trees in the orchard at the Shenyang Agricultural University (Shenyang, China) were used as test materials in this study. The trees were treated with 5 g/L CaCl2, which was sprayed on flowers and fruits at full bloom (FB) and 15 days after full bloom (DAFB) twice. Trees were treated with an equal volume of water as the control. Fruit samples were harvested 20, 35, 50, 65, 80, and 110 DAFB in 2021 and 25, 30, 35, 50, 80, and 125 DAFB in 2020. Fresh fruits were harvested for histochemical analysis and stored at −80°C for further analysis. Three replicates were maintained per treatment with 20 fruits per biological replicate.
Ca2+ localization and paraffin section analysis
Localization of free Ca2+ in Ca2+-treated and control fruits was observed by fluorescence imaging (Wang et al., 2018). The middle flesh was collected with a blade, washed twice with a HEPES buffer, and incubated with Fluo-3/AM at 4°C for 2 h. Then, all the samples were washed with the HEPES buffer at 25°C. Finally, Fluo-3 fluorescence was scanned with a confocal laser scanning microscope (TCS Sp8; Leica, Germany) using a 488-nm laser light to excite the dye and a 525–530-nm long-pass emission filter to read.
Stone cells were observed in paraffin sections by following previous methods (Liu et al., 2021). The pear fruits were fixed with FAA (formaldehyde, acetic acid, and ethanol), followed by safranin and green staining. Finally, the tissue section was mounted with neutral balsam and observed under a light microscope (Nikon, DS-U3).
Analysis of stone cell, lignin content, and lignin histochemistry
The stone cell and lignin contents of the pear fruits were determined 20, 35, 50, 65, 80, and 110 DAFB. The content of stone cells was determined with a previous method (Xue et al., 2019). The calculation method is as follows: stone cell content (mg/g) = stone cell dry weight (mg)/flesh weight (g). Lignin content was determined with a previous method (Anderson et al., 2015) and calculated as follows: lignin content (mg.g−1) = lignin content (mg)/dry weight (g).
Fresh pears were used as the material and were cut and sliced by hand at six developmental stages (20, 35, 50, 65, 80, and 110 DAFB) to confirm the histochemical location of lignin. The sections were placed in 1% phloroglucinol solution and treated with 30% HCl for 5 min. Images were captured with a handheld camera.
RNA-sequencing
Total RNA was isolated from CaCl2-treated and control fruit flesh 35 DAFB (three biological replicates for each treatment), and a library was constructed and sequenced on an Illumina HiSeq 2000 system at LC-Bio Technology Co., Ltd. (Hangzhou, China). We used the fragments per kb per million reads method to calculate differentially expressed genes. Raw RNA-seq data have been deposited on the NCBI Sequence Read Archive (SRA) under accession number PRJNA 797117.
RNA extraction and gene expression analysis
The cetyltrimethylammonium bromide (CTAB) method was used to extract total RNA from CaCl2-treated and untreated fruit flesh (Jaakola et al., 2001), and cDNA synthesis and gene expression analysis were conducted as previously described (Zhang et al., 2019). qRT-PCR was performed on a 7500 Real-time PCR system (Applied Biosystems, Foster City, United States) using SYBR Green Kit (Takara, Tokyo, Japan). The primers used in gene expression analysis are listed in Supplementary Table S1.
PRX enzyme activity
The PRX activity in CaCl2-treated and control fruit flesh was measured as previously described (Alici and Arabaci, 2016). One PRX unit is defined as the amount of enzyme that causes an absorbance change of 0.001 per minute under the test conditions.
Subcellular localization
The PuPRX42-like and PuDof2.5 coding regions were fused with GFP to form the Pro35S:GFP-PuPRX42-like and Pro35S:GFP-PuDof2.5 constructs. Respectively, the construct was co-infiltrated with the mCherry-labeled membrane marker (PM Marker) and nuclear marker (mCherry marker) into onion bulb samples by Agrobacterium tumefaciens-mediated infiltration, the onion bulb samples were cultured in an MS medium for 48 h after infiltration. GFP fluorescence was observed under a confocal microscope (TCS Sp8; Leica, Germany). For green fluorescence observation, the excitation wavelength was 488 nm and the emission wavelength was 520–540 nm. For red fluorescence observation, the excitation wavelength was 561 nm and the emission wavelength was 610–630 nm. Pro35S:GFP was used as a control. All the assays were repeated at least 3 times.
Functional analysis
To overexpress PuDof2.5 and PuPRX42-like in the Nanguo pear fruits, CDS regions were separately cloned into a pRI101 vector to form Pro35S:pRI101-PuDof2.5 and Pro35S: pRI101-PuPRX42-like. These plasmids were transformed into A. tumefaciens strain GV3101, and preparation of infiltration buffer and fruit infiltration were performed as previously described (Li et al., 2016). Briefly, the infiltration buffer was taken with a 1-ml sterile syringe and injected into on-tree Nanguo pear fruits 20 DAFB. For each fruit, one side was used for infiltrating target constructs, and the other side for infiltrating empty pRI101 as control. The infiltrated fruits were harvested 6 day after the infiltration, and the fruit flesh around the infiltrated area was sampled for further use.
GUS analysis
The PuPRX42-like promoter sequence (1,011 bp) was cloned upstream of the GUS gene in the pBI101 vector to obtain the reporter construct. Meanwhile, the PuDof2.5 CDS was introduced into the pRI101 vector to generate the effector construct. The A. tumefaciens strain GV1301 harboring the reporter and effector vectors were infiltrated into N. benthamiana leaves (Li et al., 2017). Histochemical staining was performed with X-gluc, and GUS activity was determined as previously described (Li et al., 2016). Stained leaves were viewed with an optical microscope, the infiltration was repeated three times (three biological replicates).
ChIP-PCR analysis
The PuDof2.5 CDS was cloned into the pRI101-3xflag vector (Yue et al., 2020) and transformed into an A. tumefaciens GV1301 strain. The pear fruits were infected as previously described (Li et al., 2020). ChIP assay was performed using the EpiQuik Plant ChIP Kit (56383; Cell Signaling Technology, Danvers, MA, United States) following the manufacturer′s instructions. Chromatin fragmentation was performed using an Uibra Cell VCX 150PB sonicator (Sonics and Materials Inc., Newtown, CT, United States). The specific for the ChIP-PCR assay was prepared as previously method (Li et al., 2020). Each ChIP assay was repeated three times, and enriched DNA fragments in each ChIP sample were used as one biological replicate for qPCR. Six regions of the PuDof2.5 promoter were analyzed to assess the enrichment; the primers used are listed in Supplementary Table S1.
Yeast one-hybrid assays
The PuDof2.5 CDS sequences were inserted into the pGADT7 vector at the NdeI and EcoRI restriction sites to generate a prey construct. The PuPRX42-like promoter was cloned into the pAbAi vector using the SacI and SmaI restriction sites to produce the bait constructs. The specific operation for the Y1H assay was performed as previously described (Ji et al., 2021). All the primers used in this study are listed in Supplementary Table S1.
Results
Calcium suppresses lignin and stone cell contents in pear fruits
The Nanguo fruits on the tree were treated with 5 g·L−1 CaCl2 in 2020 and 2021. Fruits were harvested 35 days after full bloom (DAFB). The fluo-3/AM staining revealed that flesh cells had higher free Ca2+ in CaCl2-treated fruits than in control fruits (Figure 1A).
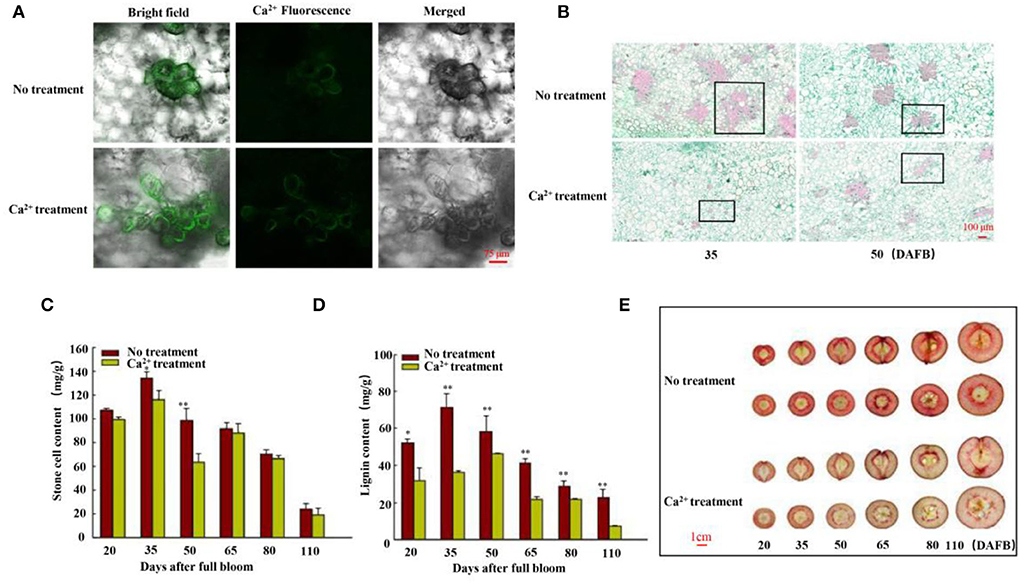
Figure 1. Free Ca2+, stone cell morphology, and lignin and stone cell contents. (A) Free Ca2+ in the flesh cells of fruits was detected using Fluo-3/AM 35 days after full bloom (DAFB). Scale bar 75 μm. (B) Morphology of stone cells was observed using paraffin sections of fruits 35 and 50 DAFB. Scale bar 100 μm. (C) Stone cell production and (D) lignin content of fruits were measured at six stages (20, 35, 50, 65, 80, and 110 DAFB). (E) Staining of sections with phloroglucinol-HCl (scale bar 1 cm). Data are shown as means ± SE (n = 3). Statistical significance was determined by Student's t-test: ** P < 0.01.
Then, the lignin and stone cell contents of the fruits were measured at six fruit developmental stages, and the findings were consistent with the paraffin section results (Figures 1B–D, Supplementary Figure S2). We further stained the harvested fruits (25, 30, 35, 50, 80, and 125 DAFB in 2020 and 20, 35, 50, 65, 80, and 110 DAFB in 2021) to confirm the results. Hand-cut sections were prepared by phloroglucinol-HCl staining. The lignin-specific histochemical staining indicated that the level of lignification in the control fruits was higher than in the CaCl2-treated fruits (Figure 1E, Supplementary Figure S1), indicating fewer stone cell primordia in the CaCl2-treated fruits. Thus, the findings collectively indicate that the CaCl2 treatment significantly inhibited lignin biosynthesis and stone cell production in Nanguo pear.
PuPRX42-like gene assists calcium in inhibiting lignin biosynthesis
To explore how CaCl2 inhibits lignin production from 20 to 125 DAFB in Nanguo pear fruits, we measured the PAL, C4H, 4CL, CAD, and PRX activities of the CaCl2-treated and control fruit samples (Supplementary Figure S3). The CaCl2 treatment significantly inhibited PRX activity (Figure 2C), and PRX activity showed the highest correlation with lignin and stone cell contents (Table 1). Subsequently, the expression of lignin biosynthesis genes PuPRX42-like, PuPAL, PuC4H, Pu4CL, and PuCAD was analyzed by qRT-PCR (Figure 2) to identify the key genes that contribute to the lower lignin accumulation in CaCl2-treated fruits. The expression level of PuPRX42-like was lower in the CaCl2-treated fruits than in the control fruits and consistent with PRX activity (Figures 2A,C).
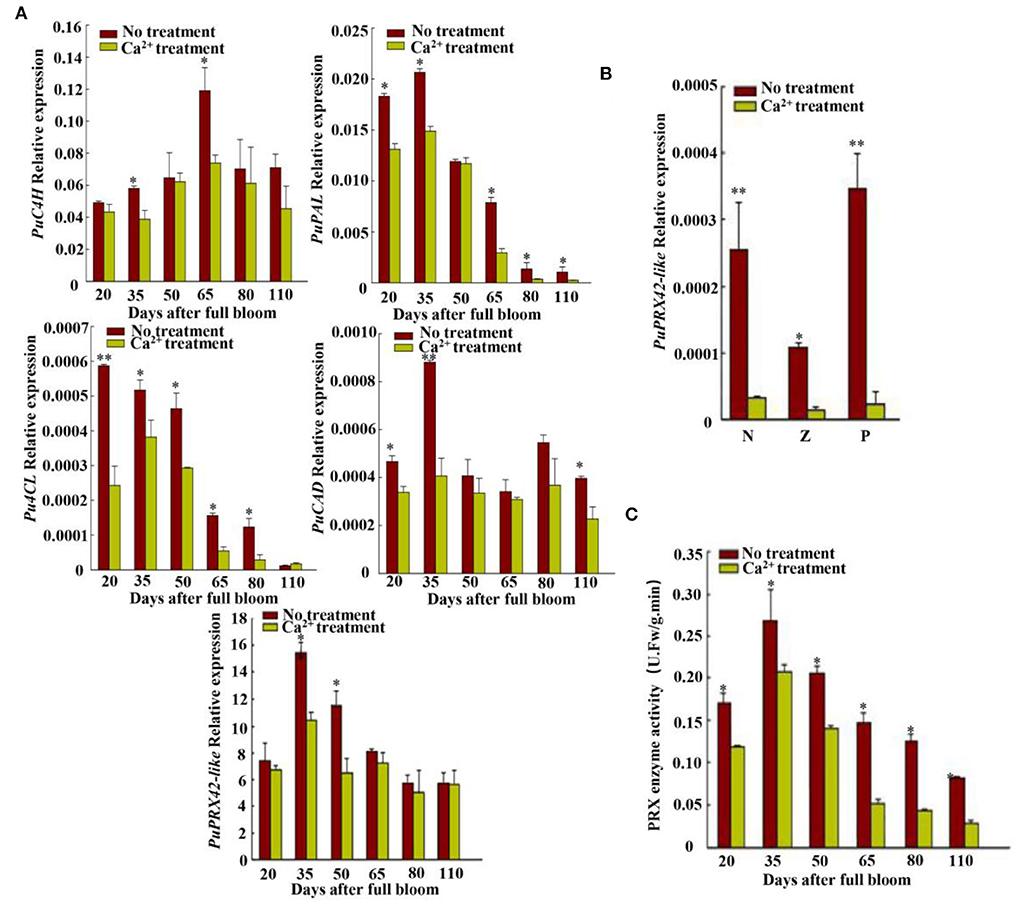
Figure 2. Synthetic lignin relative gene expression and PRX enzyme activity analyses. (A) Expressions of lignin biosynthesis genes PuC4H, PuPAL, Pu4CL, PuCAD, and PuPRX42-like were measured at six fruit developmental stages; PuC4H, cinnamate 4-hydroxylase gene; PuPAL, phenylalanine ammonia lyase gene; Pu4CL, 4-coumarate gene; PuCAD, cinnamyl alcohol dehydrogenase gene; PuPRX42-like, peroxidase gene. (B) PuPRX42-like expression in different pear tissues. N, flesh close to the core; Z, flesh in the middle; P, flesh close to the fruit skin. (C) PRX enzyme activity at six developmental stages. Data are shown as mean ± SE. Statistical significance was determined by Student's t-test: * P < 0.05, ** P < 0.01.
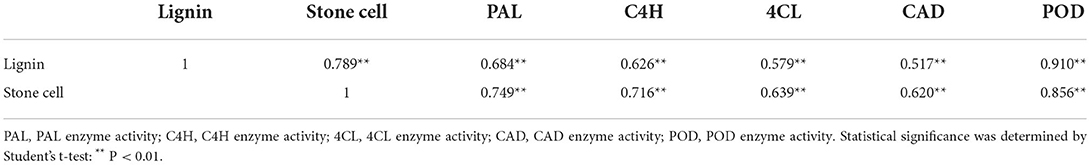
Table 1. Correlation analysis between stone cell, lignin content, and lignin-related enzyme activity.
Furthermore, a transient expression assay was conducted to explore the putative function of PuPRX42-like in lignin production. The recombinant plasmid (35S::PuPRX42-like-OE) was then transiently transformed into Agrobacterium and infiltrated into Nanguo pear fruits on the tree 30 DAFB using the empty vector (pRI101) as the control; The expression of PuPRX42-like was significantly higher in PuPRX42-like-OE fruits than in the control fruits, consistent with lignin content (Figures 3A,C,D,E). These results indicate that PuPRX42-like is important for lignin biosynthesis in pear fruits. PuPRX42-like CDS was tagged with GFP and transformed into onion bulb cells following the Agrobacterium tumefaciens mediation to further explore its subcellular localization. The analysis of onion bulb cells showed that PuPRX42-like was localized on the membranes (Figure 3B).
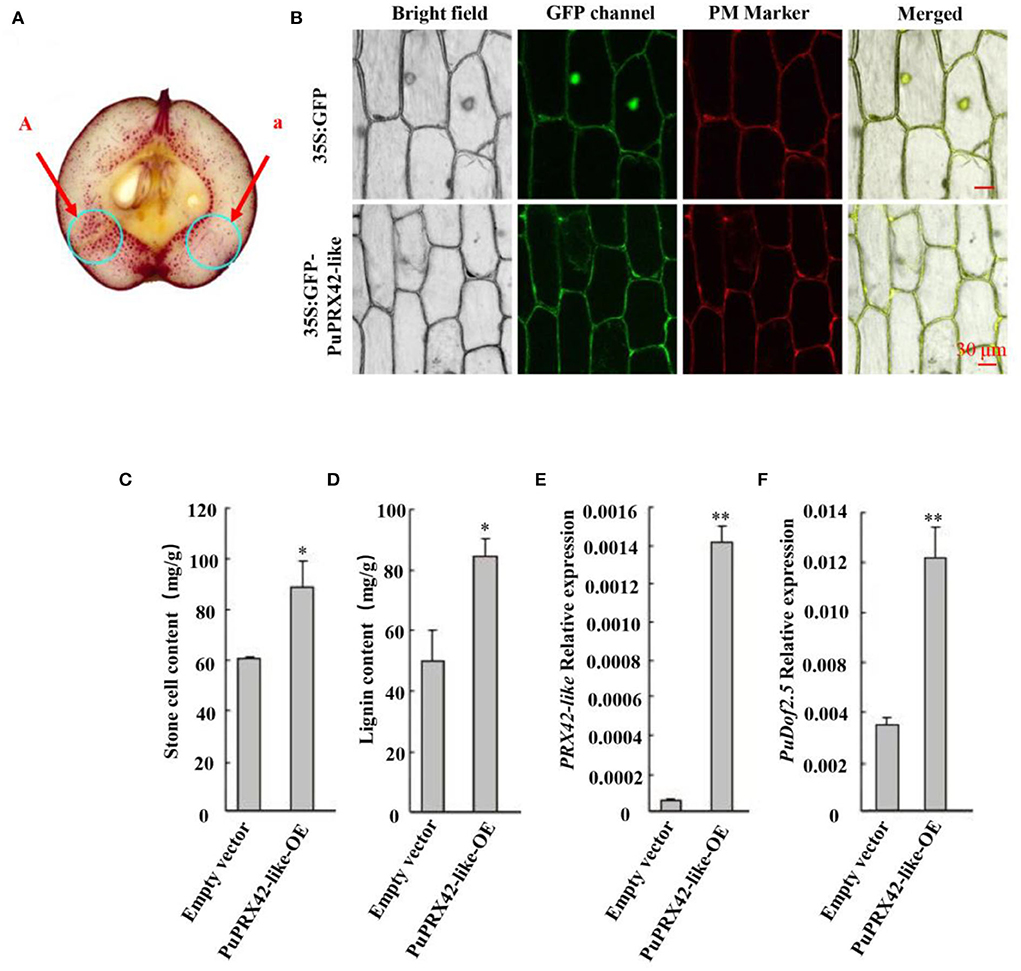
Figure 3. Functional analysis of PuPRX42-like. PuPRX42-like was transiently expressed in pear fruits (PuPRX42-like-OE) 30 DAFB following Agrobacterium-mediated transient transformation, The transformed pear fruits were harvested 7 days after infiltration to be analyzed by (A) staining the sections with phloroglucinol-HCl (A, pRI101-PuPRX42-like; a, pRI101). (B) Subcellular localization of PuPRX42-like was determined by transiently overexpressing PuPRX42-like, 35Spro::GFP, and 35Spro::PuPRX42-like-GFP constructs in onion bulb cells; PM marker served as a membrane marker (scale bar 30 μm). (C) Stone cell production and (D) lignin content were measured. (E) PuPRX42-like and (F) PuDof2.5 expression levels in PuPRX42-like-OE fruits were determined by qRT-PCR. Data are shown as mean ± SE (n = 3). Statistical significance was determined by Student's t-test: ** P < 0.01, * P < 0.05.
PuDof2.5 was the key transcription factor after calcium-treated pear fruits
We compared the transcriptomes of CaCl2−treated and control fruits harvested 35 DAFB, and RNA-seq data had different transcription factors, PuDof2.5, PuWRKY2, PuNAC1, PuERF1, and PuMYB1 (Figure 4). Among them, all genes expressions were downregulated after Ca2+ treatment, but only PuDof2.5 expression trend was the same as stone cell change at six fruit developmental stages; these were confirmed by qRT-PCR. We also analyzed PuDof2.5 expression in different tissues and found a high expression level in flesh close to the core. Furthermore, we identified cis-elements (AAAG) of Dof transcription factor in the promoter of PuPRX42-like (1,500 bp) and hypothesized the role of PuDof2.5 in regulating PuPRX42-like expression.
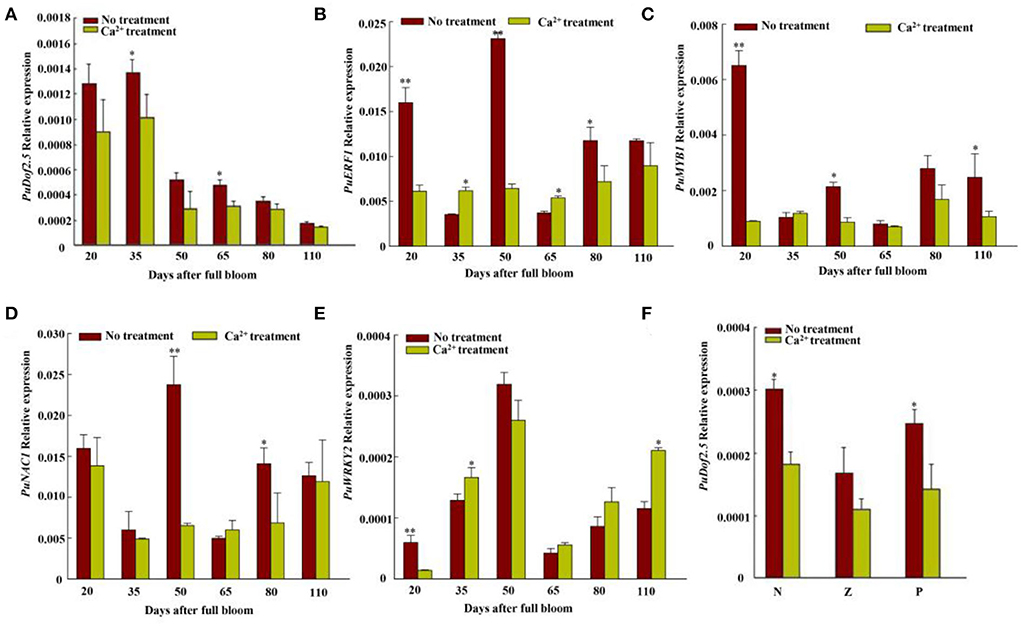
Figure 4. Transcriptome-filtered transcription factor expression level analyses of Nanguo pear fruits. (A) Expression level of PuDof2.5 at six fruit developmental stages after Ca2+ treatment. (B) Expression level of PuERF1 at six fruit developmental stages after Ca2+ treatment. (C) Expression level of PuMYB1 at six fruit developmental stages after Ca2+ treatment. (D) Expression level of PuNAC1 at six fruit developmental stages after Ca2+ treatment. (E) Expression level of PuWRKY2 at six fruit developmental stages after Ca2+ treatment. (F) PuDof2.5 expression in different pear tissues. N, flesh close to the pit; Z, flesh in the middle; P, flesh close to the fruit skin. Data are shown as mean ± SE (n = 3). Statistical significance was determined by Student's t-test: * P < 0.05, ** P < 0.01.
Furthermore, the PuDof2.5 CDS was cloned into the pRI101 vector under the control of the 35S promoter to build a 35S: PuDof2.5 overexpression construct (PuDof2.5-OE). The vector was transformed into Agrobacterium and infiltrated into on-tree pear fruits. pRI101 alone was used as control, and qRT-PCR confirmed higher expression of PuDof2.5 in PuDof2.5-OE fruits than in control fruits (Figure 5). We detected that the stone cell and lignin contents in PuDof2.5-OE fruits were significantly increased. Notably, the expression level of PuPRX42-like was also high in the PuDof2.5-OE fruits, suggesting that PuDof2.5 might play a role in suppressing stone cells by regulating the expression of PuPRX42-like.
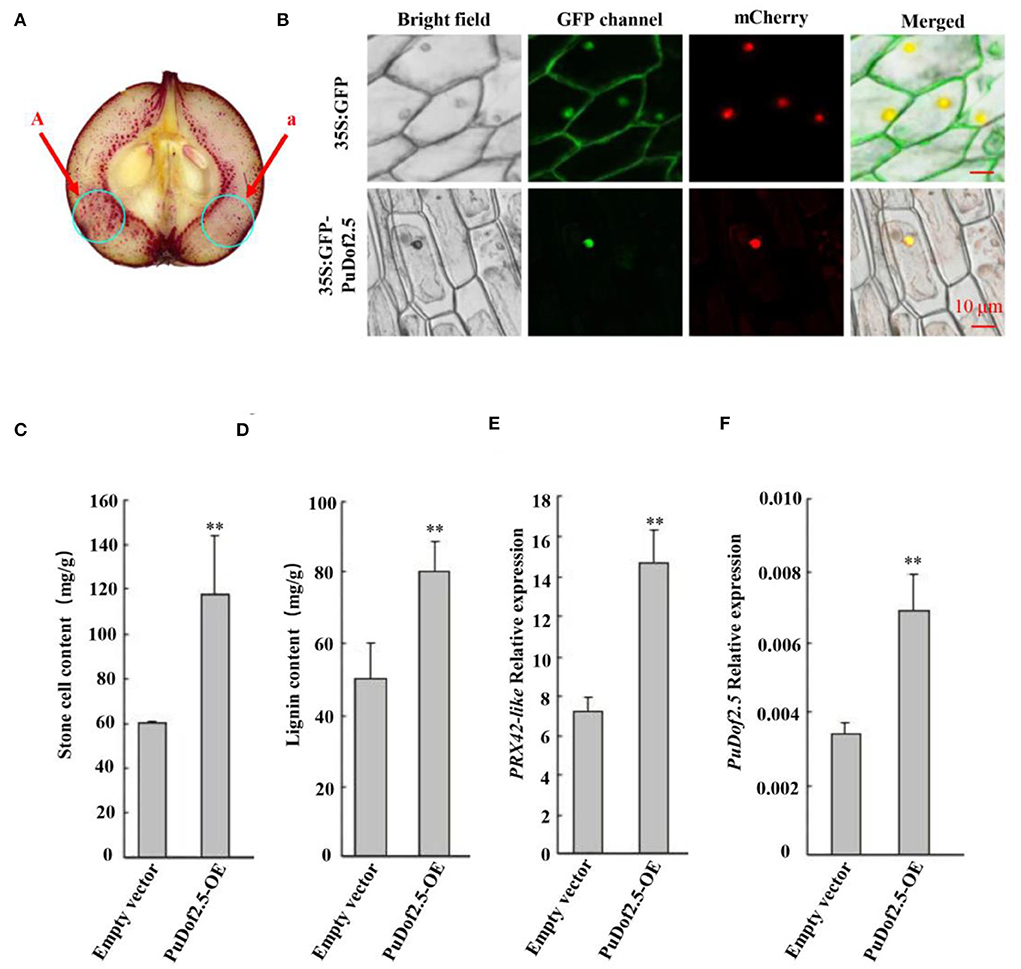
Figure 5. Functional analysis of PuDof2.5. (A) staining the sections with phloroglucinol-HCl (A, pRI101-PuDof2.5; a, pRI101). (B) Subcellular localization of PuDof2.5 was determined by transiently overexpressing PuDof2.5, 35Spro::GFP, and 35Spro:: PuDof2.5-GFP constructs in onion bulb cells; PM marker served as a membrane marker (scale bar 10 μm). (C) Stone cell content and (D) lignin content were measured. (E) PuPRX42-like and (F) PuDof2.5 expression levels in PuDof2.5-OE fruits were determined by qRT-PCR. Data are shown as mean ± SE (n = 3). Statistical significance was determined by Student's t-test: ** P < 0.01.
PuDof2.5 binds to the promoter of PuPRX42-like
Furthermore, a yeast one-hybrid (Y1H) assay was conducted to investigate the binding of PuDof2.5. The results showed that PuDof2.5 bound to the PuPRX42-like promoter (Figure 6A). To investigate whether PuPRX42-like is a direct target of PuDof2.5, ChIP-qPCR assays were performed. The analysis confirmed that PuDof2.5 could bind to regions of PuPRX42-like promoters (Figure 6B). Next, we investigated the PuDof2.5 regulation to the PuPRX42-like promoter by β-glucuronidase (GUS) transactivation assay. There was significantly increased activity of the PuPRX42-like promoter compared to the empty vector (Figures 6C,D), suggesting that PuDof2.5 is a transcriptional activator of PuPRX42-like. Taken together, the results suggested that PuDof2.5 directly binds to the PuPRX42-like promoter to activate its expression.
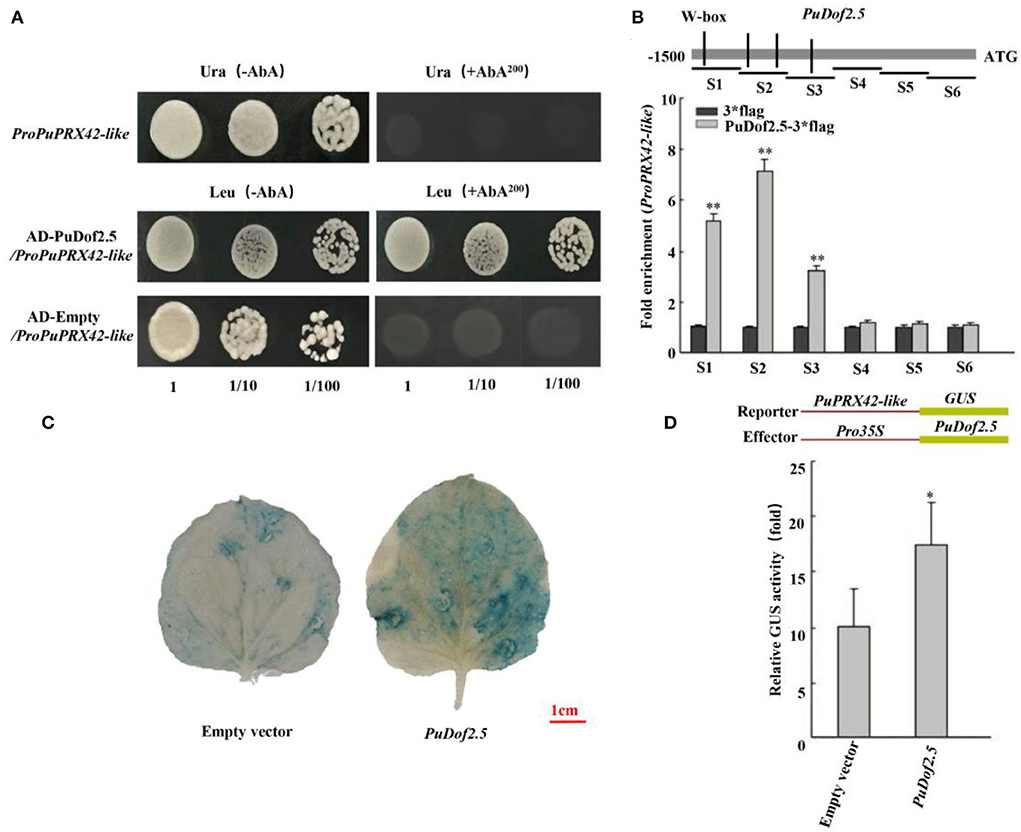
Figure 6. PuDof2.5 regulates PuPRX42-like transcription by promoter binding. (A) Y1H analysis of PuDof2.5 binding to the promoter of PuPRX42-like. AbA is a yeast growth inhibitor, and the basal concentration was 200 ng/ml. The empty vector and the PuPRX42-like promoter were used as negative controls. (B) ChIP-qPCR analysis to determine how PuDof2.5 directly binds to the PuPRX42-like promoter. Eluted DNA was used in the quantitative PCR (qPCR) performed to analyze the six fragments (S1–S6). Immunoprecipitate from cells transiently expressing the vector alone was used as a negative control. (C) Histochemical GUS (β-glucosidase) staining. (D) GUS activity analysis to determine how PuDof2.5 promotes the activity of the PuPRX42-like promoter. Data are shown as mean ± SE (n = 3). Statistical significance was determined by Student's t-test: * P < 0.05, ** P < 0.01.
Discussion
Stone cells are a restrictive factor of Nanguo pear fruit quality (Yan et al., 2014). The formation of stone cells is closely related to the deposition of lignin in pear fruits (Li et al., 2007). However, although previous studies have described that Ca2+ induces the biosynthesis of lignin in fruit species. CaCl2 significantly inhibited lignin biosynthesis enzyme and PAL activities to decrease lignin content in apple fruits (Zhao and Wang, 2015). The studies only investigated changes in stone cell production after exogenous Ca2+ treatment and the expression profile of genes involved in lignin biosynthesis and signal transduction. The detailed mechanism by which Ca2+ regulates lignin biosynthesis to suppress stone cell production in pear fruits was unclear. Here, we determined that Ca2+-inhibited PuDof2.5 induces the expression of a lignin biosynthetic gene (PuPRX42-like) by directly binding to its promoter. Our findings suggest that Ca2+ has an essential role in reducing stone cell accumulation in pear fruits.
Calcium reduces stone cell production by reducing lignin accumulation
The texture of pear fruits depends on stone cell mass density and content (Wang et al., 2021). In this study, stone cells developed as clusters, presenting a radial shape throughout the fruit flesh in control fruits, which is consistent with earlier reports (Li et al., 2017). However, Ca2+ significantly suppressed stone cell mass content and density (Figure 1B). Higher free Ca2+ levels were observed in the Ca2+-treated fruits, whereas the levels were lower in the control pear fruits (Figure 1A). The phloroglucinol staining revealed lignin deposition in the flesh of pear fruits and lignified stone cells; the detailed analysis revealed that lignin accumulated rapidly from 20 to 50 DAFB (Figure 1D). Meanwhile, the control fruits were stained darker than the Ca2+-treated fruits (Figure 1E), consistent with stone cell content (Figure 1C). A similar result was reported in “Whangkeumbae” (Pyrus pyrifolia) pear (Lu et al., 2015). Based on these findings, we proposed that Ca2+ regulates lignin accumulation by inhibiting the expression of lignin biosynthesis genes. Together, the studies provided detailed information that Ca2+ decreased stone cell content by inhibiting lignin accumulation in pear fruits.
PuPRX42-like is important for Ca2+-induced lignin biosynthesis
Studies have proven the role of various enzymes in lignin synthesis. Among PAL, C4H, CAD, 4CL and PRX (Tao et al., 2009), PRX is the key enzyme to catalyze monolignol polymerization into lignin (Dean and Eriksson, 1994; Cheng et al., 2017). The present study analyzed the activities of PRX, PAL, C4H, CAD, and 4CL related to lignin biosynthesis after Ca2+ treatment (Supplementary Figure S3). Interestingly, the Ca2+ treatment only reduced PRX enzyme activity, and it is consistent with lignin and stone cell contents (Figures 1C,D, 2C, Supplementary Figure S3). An earlier finding with the same results in “Niitaka” pear fruit (Lee et al., 2007) suggested that the PRX enzyme acts a key factor in lignin accumulation to form stone cell. The correlation analysis between lignin synthesis enzyme (PRX, PAL, C4H, CAD, and 4CL) with lignin and stone cell contents also supported this observation (Table 1).
Previous studies have revealed that PuPRXs regulated PRX activity and lignin biosynthesis in pear, and that the high expression level of PuPRX2 promotes lignin accumulation in PuRBOHF-overexpressing pear calli (Wang et al., 2021). In our present study, PuPRX42-like expression levels were consistent with PRX activity changes (Figures 2A,C). Meanwhile, the overexpression of PuPRX42-like significantly accelerated lignin accumulation and stone cell production. PuPRX42-like was localized on the plasma membrane, including sites where lignification occurred. To sum up, the results indicated that PuPRX42-like was a key point in the regulation of lignin biosynthesis.
PuDof2.5 binds to PuPRX42-like promoter and regulates lignin biosynthesis
Dofs are plant-specific transcription factors that act as either transcriptional activators or repressors to influence plant developmental processes (Licausi et al., 2013), i.e., OsDof15 promoted rice root elongation response to salt stress, and overexpression of GhDof1 resulted in significant cold tolerance (Su et al., 2017). Meanwhile, overexpression of AtDof2.1 promoted rice leaf senescence (Zhou et al., 2021). Nonetheless, the role of Dof in regulating lignin biosynthesis in pear fruits is not well-known. We observed that Ca2+ significantly suppressed the expression of PuDof2.5 (Figure 4A). Moreover, PuDof2.5 is a transcriptional activator that binds to the lignin biosynthetic gene PuPRX42-like promoter, directly upregulating its expression (Figures 5A,B,D). Overexpression of PuDof2.5 increased lignin and stone cell contents and upregulated the expression level of PuPRX42-like. Thus, our findings propose that PuDof2.5 acted as a useful marker to respond to Ca2+ treatment.
Our results indicated that Ca2+ induced stone production in pear fruits (Figure 7). When endogenous Ca2+ levels are high, PuDof2.5 expression declines, and PuDof2.5 suppresses the transcription of lignin biosynthetic gene PuPRX42-like by direct promoter binding; thus, PRX enzyme activity is reduced, lignin biosynthesis is inhibited, and stone cell production is decreased. When endogenous Ca2+ levels decline, PuDof2.5 expression is enhanced, which accelerates PRX enzyme activity and the transcription of PuPRX42-like, leading to a burst of stone cells. Taken together, the results provide a novel insight into the mechanism by which Ca2+ regulates stone cell accumulation to improve pear quality.
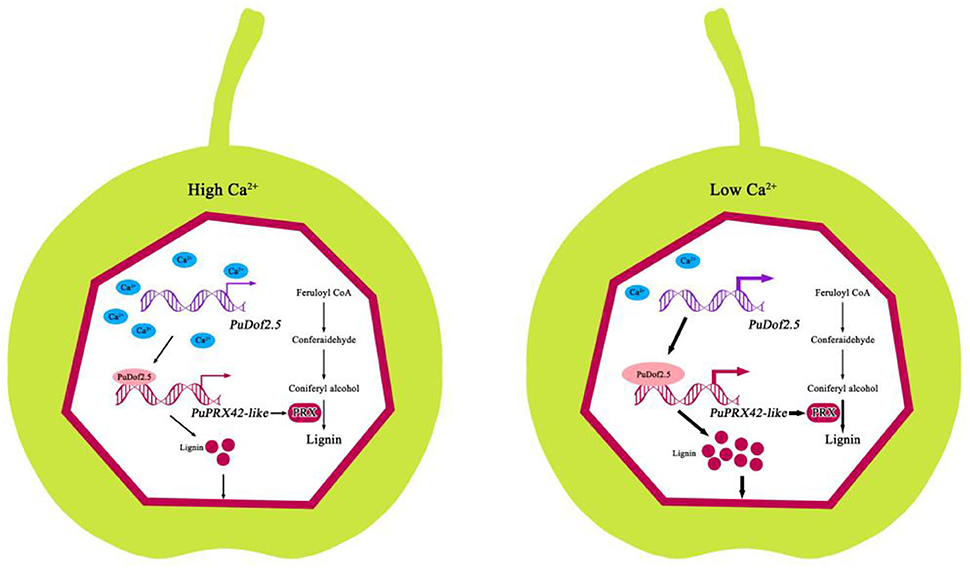
Figure 7. Model of the mechanism by which Ca2+ suppresses stone cell production in pear fruits. At high levels of endogenous Ca2+, PuDof2.5 downregulated the transcription of lignin biosynthetic gene PuPRX42-like by directly binding to the promoter, decreasing lignin accumulation and stone cell production. At low levels of endogenous Ca2+, PuDof2.5 upregulated the transcription of lignin biosynthetic s PuPRX42-like, increasing lignin accumulation and stone cell production.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
XL and GD designed this study. HZ, TW, SG, and MX performed the experiments. XL, GD, and HZ wrote the manuscript. All the authors read and approved the final version of the manuscript.
Funding
This study was supported by the National Pear Industry Technology System Project (CARS-28-15), Liaoning Provincial National Natural Science Foundation of China (2019-ZD-0716), Education Department of Liaoning Province Serves Local Projects (LSNFW202009), and Inter-university Cooperation Project of Education Department of Liaoning Province.
Acknowledgments
The authors would like to thank Prof. Zhifu Guo and Prof. Aide Wang for providing the vectors. We would also like to thank topedit (https://topeditsci.com) for editing the language of a draft of this manuscript.
Conflict of interest
Author TW was employed by Xinjiang Production and Construction Corps.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.976977/full#supplementary-material
References
Alici, E. H., and Arabaci, G. (2016). Determination of SOD, POD, PPO and CAT enzyme activities in Rumex obtusifolius L. Annu. Rev. Med. 11, 1–7. doi: 10.9734/ARRB/2016/29809
Anderson, N. A., Tobimatsu, Y., Ciesielski, P. N., Ximenes, E., Ralph, J., Donohoe, B. S., et al. (2015). Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27, 2195–2209. doi: 10.1105/tpc.15.00373
Barros, J., Serk, H., Granlund, I., and Pesquet, E. (2015). The cell biology of lignification in higher plants. Ann. Bot. 115, 1053–1074. doi: 10.1093/aob/mcv046
Cai, Y. P., Li, G. Q., Nie, J. Q., Lin, Y., Nie, F., Zhang, J. Y., et al. (2010). Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 125, 374–379. doi: 10.1016/j.scienta.2010.04.029
Cao, Y. P., Meng, D. D., Li, X. X., Wang, L. H., Cai, Y. P., and Jiang, L (2020). A Chinese white pear (Pyrus bretschneideri) BZR gene PbBZR1 act as a transcriptional repressor of lignin biosynthetic genes in fruits. Front. Plant Sci. 11, 1087. doi: 10.3389/fpls.2020.01087
Cheng, X., Li, M. L., Li, D. H., Zhang, J. Y., Jin, Q., Sheng, L. L., Cai, Y. P., et al. (2017). Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biology 6, 1602–1613. doi: 10.1242/bio.026997
Cosio, C., Ranocha, P., Francoz, E., Burlat, V., Zheng, Y. M., Perry, E. S., et al. (2017). The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 213, 50–263. doi: 10.1111/nph.14127
Dalzochio, O. Â., Silvestre, W. P., and Pauletti, G. F. (2021). Effect of the application of prohexadione-calcium on the growth of ‘Packham's Triumph' and ‘Hosui' pears (Pyrus communis L.). Res. Soc. Dev. 10, e3110816801. doi: 10.33448/rsd-v10i8.16801
Dean, J. F., and Eriksson, K. E. L. (1994). Laccase and the deposition of lignin in vascular plants. Holzforschung 48, 21–33. doi: 10.1515/hfsg.1994.48.s1.21
Jaakola, L., Pirttila, A. M., Halonen, M., and Hohtola, A. (2001). Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 19, 201–203. doi: 10.1385/MB:19:2:201
Ji, Y. L., Qu, Y., Jiang, Z. Y., Yan, J. J., Chu, J. F., Xu, M. Y., et al. (2021). The mechanism for brassinosteroids suppressing climacteric fruit ripening. Plant Physiol. 185, 1875–1893. doi: 10.1093/plphys/kiab013
Khaksar, G., Sangchay, W., Pinsorn, P., Sanqpong, L., and Sirikantaramas, S. (2019). Genome-wide analysis of the Dof gene family in durian reveals fruit ripening-associated and cultivar-dependent Dof transcription factors. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-019-48601-7
Lee, M. H., Jeon, H. S., Kim, S. H., Chung, J. H., Roppolo, D., Lee, H. J., et al. (2019). Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 38, e101948. doi: 10.15252/embj.2019101948
Lee, S. H., Choi, J. H., Kim, W. S., Park, Y. S., and Gemma, H. (2007). Effects of calcium chloride spray on peroxidase activity and stone cell development in pear fruit (Pyrus pyrifolia ‘Niitaka'). J. Jpn. Soc. Hortic. Sci. 76, 191–196. doi: 10.2503/jjshs.76.191
Li, M. T., Cheng, C. X., Zhang, X. F., Zhou, S. P., Wang, C. H., Ma, C. H., et al. (2019). PpNAC187 enhances lignin synthesis in ‘Whangkeumbae' pear (Pyrus pyrifolia) ‘hard-end' fruit. Molecules. 24, 4338. doi: 10.3390/molecules24234338
Li, S. H., Schneider, B., and Gershenzon, J. (2007). Microchemical analysis of laser-microdissected stone cells of Norway spruce by cryogenic nuclear magnetic resonance spectroscopy. Planta 225, 771–779. doi: 10.1007/s00425-006-0376-z
Li, T., Jiang, Z. Y., Zhang, L. C., Tan, D. M., Wei, Y., Yuan, H., et al. (2016). Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 88, 735–748. doi: 10.1111/tpj.13289
Li, T., Xu, Y. X., Zhang, L. C., Ji, Y. L., Tan, D. M., Yuan, H., et al. (2017). The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29, 1316–1334. doi: 10.1105/tpc.17.00349
Li, X. Y., Guo, W., Li, J. C., Yue, P. T., Bu, H. D., Jiang, J., et al. (2020). Histone acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in pear fruit. Plant Physiol. 182, 2035–2046. doi: 10.1104/pp.20.00002
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA 2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, X. F., Li, S. R., Feng, X. X., and Li, L. L. (2021). Study on cell wall composition, fruit quality and tissue structure of hardened ‘Suli' pears (Pyrus bretschneideri Rehd). J. Plant Growth Regul. 40, 2007–2016. doi: 10.1007/s00344-020-10248-4
Lu, G. L., Li, Z. J., Zhang, X. F., Wang, R., and Yang, S. L. (2015). Expression analysis of lignin-associated genes in hard end pear (Pyrus pyrifolia Whangkeumbae) and its response to calcium chloride treatment conditions. J. Plant Growth Regul. 34, 251–262. doi: 10.1007/s00344-014-9461-x
Ramachandran, V, Tobimatsu, Y., Masaomi, Y., Sano, R., Umezawa, E., Demura, T., et al. (2020). Plant-specific Dof transcription factors vascular-related DOF1 and vascular-related DOF2 regulate vascular cell differentiation and lignin biosynthesis in Arabidopsis. Plant Mol. Biol. Rep. 104, 263–281. doi: 10.1007/s11103-020-01040-9
Ring, L., Yeh, S. Y., Hücherig, S., Hoffmann, T., Blanco-Portales, R., Fouche, M., et al. (2013). Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiol. 163, 43–60. doi: 10.1104/pp.113.222778
Rogers, L. A., and Campbell, M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164, 17–30. doi: 10.1111/j.1469-8137.2004.01143
Rogers, L. A., Dubos, C., Surman, C., Willment, J., Cullis, L., Mansfield, S. D., et al. (2005). Comparison of lignin deposition in three ectopic lignification mutants. New Phytol. 168, 123–140. doi: 10.1111/j.1469-8137.2005.01496.x
Sajid, M., Basit, A., Ullah, I., Tareen, J., Asif, M., Khan, S., et al. (2019). Efficiency of calcium chloride (CaCl2) treatment on post-harvest performance of pear (Pyrus communis L.). Pure Appl. Biol. 8, 1111–1125. doi: 10.19045/bspab.2019.80053
Shigeto, J., and Tsutsumi, Y. (2016). Diverse functions and reactions of class III peroxidases. New Phytol. 209, 1395–1402. doi: 10.1111/nph.13738
Su, Y., Liang, W., Liu, Z. J., Wang, Y. M., Zhao, Y. P., Ijaz, B., et al. (2017). Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J. Plant Physiol. 218, 222–234. doi: 10.1016/j.jplph.2017.07.017
Tao, S. T., Khanizadeh, S., Zhang, H., and Zhang, S. L. (2009). Anatomy, ultrastructure and lignin distribution of stone cells in two Pyrus species. Plant Sci. 176, 413–419. doi: 10.1016/j.plantsci.2008.12.011
Wang, X. Q., Liu, S. Q., Sun, H. L., Liu, C. Y., Li, X. Y., Liu, Y., et al. (2021). Production of reactive oxygen species by PuRBOHF is critical for stone cell development in pear fruit. Hortic. Res. 8, 1–11. doi: 10.1038/s41438-021-00674-0
Wang, Y. L., Zhang, X. F., Wang, Y. Z., Yang, S. L., and Qu, H. Y. (2018). The changes of intracellular calcium concentration and distribution in the hard end pear (Pyrus pyrifolia cv. ‘Whangkeumbae') fruit. Cell Calcium 71, 15–23. doi: 10.1016/j.ceca.2017.11.002
Wei, S. W., Qin, G. H., Zhang, H. P., Tao, S. T., Wu, J., Wang, S. M., et al. (2017). Calcium treatments promote the aroma volatiles emission of pear (Pyrus ussuriensis ‘Nanguoli') fruit during post-harvest ripening process. Sci. Hortic. 215, 102–111. doi: 10.1016/j.scienta.2016.12.008
Wójcik, P., Skorupińska, A., and Filipczak, J. (2014). Impacts of preharvest fall sprays of calcium chloride at high rates on quality and ‘Conference' pear storability. Sci. Hortic. 168, 51–57. doi: 10.1016/j.scienta.2014.01.017
Xue, C., Yao, J. L., Qin, M. F., Zhang, M. Y., Allan, A. C., Wang, D. F., et al. (2019). PbrmiR397a regulates lignification during stone cell development in pear fruit. Plant Biotechnol. J. 17, 103–117. doi: 10.1111/pbi.12950
Yan, C. C., Yin, M., Zhang, N., Jin, Q., Fang, Z., Lin, Y., et al. (2014). Stone cell distribution and lignin structure in various pear varieties. Sci. Hortic. 174, 142–150. doi: 10.1016/j.scienta.2014.05.018
Yanagisawa, S., and Schmidt, R. J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214. doi: 10.1046/j.1365-313X.1999.00363.x
Yue, P. T., Lu, Q., Liu, Z., Lv, T. X., Li, X. Y., Liu, H. D., et al. (2020). Auxin-activated MdARF5 induces the expression of ethylene biosynthetic genes to initiate apple fruit ripening. New Phytol. 226, 1781–1795. doi: 10.1111/nph.16500
Zhang, H. Z., Yang, J. L., Li, W. L., Chen, Y. X., Lu, H., Zhao, S. C., et al. (2019). PuHSFA4a enhances tolerance to excess zinc by regulating reactive oxygen species production and root development in Populus. Plant Physiol. 180, 2254–2271. doi: 10.1104/pp.18.01495
Zhao, Y., and Wang, C. (2015). Effect of calcium chloride in combination with salicylic acid on post-harvest freshness of apples. Food Sci. Biotechnol. 24, 1139–1146. doi: 10.1007/s10068-015-0145-5
Zhou, S. L., Li, X., Liu, Q., Zhao, Y., Jiang, W., Wu, A. Q., et al. (2021). DNA demethylases remodel DNA methylation in rice gametes and zygote and are required for reproduction. Mol. Plant 14, 1569–1583. doi: 10.1016/j.molp.2021.06.006
Keywords: pear, Ca2+, PuDof2.5, PuPRX42-like, lignin, stone cell
Citation: Zhang H, Gao S, Wang T, Xu M, Li X and Du G (2022) Ca2+ mediates transcription factor PuDof2.5 and suppresses stone cell production in pear fruits. Front. Plant Sci. 13:976977. doi: 10.3389/fpls.2022.976977
Received: 24 June 2022; Accepted: 02 August 2022;
Published: 24 August 2022.
Edited by:
Karel Dolezal, Academy of Sciences of the Czech Republic, CzechiaReviewed by:
Zhang Zongying, Shandong Agricultural University, ChinaYe Han, Gansu Agricultural University, China
Copyright © 2022 Zhang, Gao, Wang, Xu, Li and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinyue Li, MjAyMDUwMDA1MUBzeWF1LmVkdS5jbg==; Guodong Du, Z3VvZG9uZ2R1QHN5YXUuZWR1LmNu
 He Zhang
He Zhang Siyang Gao1
Siyang Gao1