- Institute of Medical Plant Physiology and Ecology, School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, Guangzhou, China
The perilla anthocyanins have important medicinal and ornamental value, and their contents are significantly affected by light intensity. In view of their molecular mechanisms were not well understood, we integrated the metabolomic and transcriptomic analyses of the light-sensitive perilla variety under different light intensity. The perilla leave color were obviously affected under different treatments. Totally 140 flavonoid metabolites and 2461 genes showed steady change, among which 60 flavonoid metabolites were increased accumulation and 983 genes were upregulated expression under elevated light intensity treatment. Light treatment prominently affected the expression of genes involved in the main anthocyanin metabolites accumulation in perilla leaves. Using WGCNA analysis, we identified 4 key genes in anthocyanin biosynthesis pathway (CHI, DFR, and ANS) and 147 transcription factors (MYB, bHLH, bZIP, ERF, and NAC) involved in malonylshisonin biosynthesis. Among them, 6 MYBs and 4 bZIPs were predicted to play important roles in light regulation of malonylshisonin biosynthesis based on phylogenetic construction, correlation analysis, cis-acting element identification and qPCR verification. The identified key genes and regulatory factors will help us to understand the potential mechanism of photo-regulated anthocyanin accumulation in perilla.
Introduction
Light is essential for plant growth and plays a key environmental factor regulating plant development. The intensity, quality, periodicity, and direction of light directly affect the biosynthesis of metabolites in plants. The influence of light on anthocyanin formation plays an important role in controlling the color of fruits such as grape (Azuma et al., 2012), eggplant (Jiang et al., 2016) and apple (Feng et al., 2013).
Anthocyanins, the largest group of natural water-soluble plant pigments, are widely present in flowers, fruits, leaves, and tubers in the plant kingdom (Chen et al., 2012). Anthocyanins are phenolic compounds containing C6-C3-C6 hydroxyl aromatic rings as a basic skeleton (Kong et al., 2003). The type and final color of anthocyanin are determined by its main backbone and the positions, quantities, and structures of conjugated sugars and covalent modifications (Kocic et al., 2011; Liu Y. et al., 2018). The major anthocyanins include cyanidin, delphinidin, malvidin, pelargonidin, peonidin, and petunidin, as well as their derivatives (Kong et al., 2003; Zhao et al., 2014; Khoo et al., 2017). Anthocyanins are important ingredients of healthy foods, with medicinal value for preventing neurological and cardiovascular diseases, cancer, diabetes, and age-related degenerative diseases (Kong et al., 2003; Zhao et al., 2014; Khoo et al., 2017). Hence, anthocyanins have attracted extensive attention worldwide in recent years.
Perilla frutescens (L.) is an annual herb in the Lamiaceae. Perilla has been widely cultivated in China for more than 2000 years and is currently cultivated in Korea, Japan, and other Asian countries (Nitta et al., 2005a). Perilla is used as traditional herb, oil crops and popular spice. This main medicinal value of perilla is listed in the Chinese Pharmacopeia, including anti-oxidant (Saita et al., 2012), antibacterial (Yamamoto and Ogawa, 2002), anti-inflammatory (Yang et al., 2013; Huang et al., 2014), cough-suppressing (Ahmed, 2019), lipid-lowering, and antitumor properties (Lin et al., 2007). Perilla is divided into red perilla and green perilla according to plant color. Besides its medicinal value, red perilla is also used as an ornamental plant due to its dark red leaves and stems. The red pigment of perilla leaves is also widely used as a food coloring, cosmetic additive, and medicinal compound.
Malonylshisonin [cyanidin 3-O-(6”-O-(E)-p-coumaryl)-β-D-glucopyranoside-5-O- (6”’-O-malonyl)-β-D-glucopyranoside], an unique and major cyanindin-type anthocyanin, was identified in red perilla (Honda et al., 1994; Saito and Yamazaki, 2002; Fujiwara et al., 2018; Jiang et al., 2020). They possessed antioxidant and anti-allergy properties in previous research (Makino et al., 2003; Meng et al., 2006). The biosynthetic pathway of malonylshisonin was recently elucidated. The basic anthocyanin steps involved in 4-coumaroyl-CoA were catalyzed to form cyanidin. The main steps were catalyzed by chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3’-hydroxylase (F3’H), dihydroflavonol 4-reductase (DFR), and anthocyanin synthase (ANS). Whereafter, the cyanidin is converted to malonylshisonin via a process catalyzed by flavonoid 3-glucosyltransferase (3-GT), anthocyanin acyltransferase (ACT), anthocyanin 5-glucosyltransferase (5-GT), and anthocyanin malonyltransferase (MAT) (Saito and Yamazaki, 2002; Jiang et al., 2020; Zhou et al., 2021). As previous research in other plants, anthocyanin biosynthesis is regulated by the MBW ternary transcriptional complex (R2R3-MYB, basic helix-loop-helix [bHLH], and WD40 repeat protein [WDR]) (Holton and Cornish, 1995; Feng et al., 2010; Lloyd et al., 2017), basic leucine zipper (bZIP) (An et al., 2017), and MYC transcription factors.
Light could induce anthocyanin accumulation in perilla leaves. In a previous study of red perilla, anthocyanin accumulation and the expression of anthocyanin biosynthetic genes in leaves were lower under weak vs. strong light conditions (Zheng et al., 2019). However, the molecular mechanisms underlying light-induced anthocyanin biosynthesis in perilla remain unknown. Here, we explored anthocyanin biosynthesis and the mechanism of its photoinduction in red perilla using a light-sensitive cultivated variety. We treated the plants with different light intensities and analyzed the regulatory networks of anthocyanin biosynthesis and accumulation by performing comprehensive metabolomic and transcriptomic analyses. Our findings shed light on the putative molecular regulation of photoinduced anthocyanin accumulation in red perilla.
Materials and methods
Plant materials and treatments
In our previous work, we planted more perilla varieties in light incubator and found that the plant color of ‘PF899’ variety was more sensitive to light intensity. The variety is extremely sensitive to changes in light intensity, with green leaves in weak light and dark red leaves in increased light intensity. Hence, the light-sensitive perilla variety was selected in the study. After germination, the plants were grown in a greenhouse at the Guangzhou University of Chinese Medicine in Guangdong province, China (23°3’N, 113°23’E) under a weak light intensity (WL; 15 μmol/m2) until the three-leaf stage. The plants were transferred to moderate light (ML; 180 μmol/m2) for 6 days, followed by transfer to weak light (15 μmol/m2) for 6 days (recovery light conditions, RL). The plants were grown under a 12 h/12 h light/dark photoperiod and fertilized according to standard practices. Each treatment group was treated with three pots, each pot had five plants. Three biological replicates were performed for each treatment group. The leaves from three treatments were collected, immediately frozen in liquid nitrogen, and stored at –80°C for metabolomic and transcriptomic analysis. And in transcriptome analysis, a group of materials at moderate light (ML; 180 μmol/m2) for 12 h was added.
Measuring total anthocyanin and flavonoid contents
Leaves from different treatment groups were weighed and pulverized. To measure total anthocyanins, each 0.1 g leaf tissue sample was combined with 0.1% hydrochloric acid methanol reagent and incubated at 40°C for 40 min to extract total anthocyanins. The optical densities at 530, 620, and 650 nm were measured with a UV spectrophotometer; 0.1 mol/L hydrochloric acid methanol solution was used as a blank control. Total anthocyanin content was calculated according to Greey’s formula (Lee and Ohnishi, 2003; Nitta et al., 2005b). To measure total flavonoids, perilla leaf powder was combined with an equal volume of 70% ethanol, and 1 mL of the extract and 1 mL of 5% sodium nitrite solution were added to a volumetric bottle. After shaking well for 6 min, 1 mL of 10% aluminum nitrate test solution was added to the flask, which was then mixed for 6 min. After adding 10 mL of sodium hydroxide test solution, the sample was mixed. Finally, 70% ethanol was added to bring the volume to 25 ml. After 15 min, the absorbance of the sample at 500 nm was measured by UV-vis spectrophotometry with 70% ethanol as a blank control. The rutin was used as the standard curve to calculate the content of total flavonoids.
Metabolomics sample preparation and detection
The freeze-dried samples were crushed, and 100 mg of powdered tissue was combined with 1.2 mL of 70% methanol. After centrifuging at 12,000 rpm for 10 min, the extracts were filtered (SCAA-104, 0.22 μm pore size), and the metabolites in the extracts were analyzed used an UPLC-ESI-MS/MS system (UPLC, SHIMADZU Nexera X2; MS, Applied Biosystems 4500 Q TRAP). The conditions were as follows: UPLC: column, Agilent SB-C18 (1.8 μm, 2.1 mm * 100 mm). The mobile phase consisted of solvent A (pure water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). Sample measurements were performed with a gradient program. The column temperature was set to 40°C and the injection volume was 4 μL. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (QTRAP)-MS. LIT, and triple quadrupole (QQQ) scans were acquired on a triple quadrupole-linear ion trap mass spectrometer (Q TRAP), AB4500 Q TRAP UPLC/MS/MS System, equipped with an ESI Turbo Ion-Spray interface, operating in positive and negative ion mode, and controlled by Analyst 1.6.3 software (AB Sciex). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature 550°C; ion spray voltage (IS) 5500 V (positive ion mode)/–4500 V (negative ion mode); ion source gas I (GSI), gas II (GSII), and curtain gas (CUR) were set at 50, 60, and 25.0 psi, respectively; the collision-activated dissociation (CAD) was high. Instrument tuning and mass calibration were performed with 10 and 100 μmol/L polypropylene glycol solution in QQQ and LIT mode, respectively. QQQ scans were acquired as multiple reaction monitoring (MRM) experiments with collision gas (nitrogen) set to moderate. Positive ion mode optimizes cluster removal voltage (DP) and Collision electric (CE) for individual MRM transitions were performed by further DP and CE optimization. A specific set of MRM transitions was monitored for each period based on the metabolites eluted within that period.
RNA sequencing and annotation
A Tiangen RNA Prep Pure Kit for Plants was used to extract total RNA from P. frutescens leaves. A NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States) was used to measure total RNA concentration and quality. RNA samples that met the requirements were sent to Wuhan Bena Technology Service Co., Ltd. (Wuhan, China) for transcriptomic sequencing. RNA-seq libraries (P. frutescens leaves collected at WL, ML12 h, ML6d, and RL × 3 replicates) were constructed and sequenced using the Illumina HiSeq4000 platform, and clean reads were mapped to the perilla reference genome using HISAT with default parameters.
Identification of differential metabolites and differentially expressed genes
The metabolome data in this study was identified with chemical library MWDB (Metware Database), which were established by Metware Metabolism Company. Materials was quantitative based on secondary spectrum information, and isotope signals were removed during analysis, which contained K + ions and Na + ionsRepeated signals of NH4 + ions and fragments of other substances with larger molecular weight were identified. Metabolites with significantly different levels between groups were determined based on Variable Importance in Projection (VIP) ≥ 1 and absolute log2FC (fold change) ≥ 1. VIP values were extracted from the OPLS-DA results, and score plots and permutation plots were generated using the R package MetaboAnalystR. The data were log transformed (log2) and subjected to mean centering before OPLS-DA. To avoid overfitting, a permutation test (200 permutations) was performed. Gene expression levels were calculated as fragments per kilobase of transcript per million mapped reads (FPKM). Differentially expressed genes (DEGs) were identified from normalized read count data using DESeq2.34. Genes with | log2 (fold change) | ≥ 1 and p < 0.01 were considered to be DEGs.
Hierarchical cluster analysis, Pearson correlation coefficients, and principal component analysis
The HCA (hierarchical cluster analysis) results of samples and metabolites were presented as heatmaps with dendrograms, while PCC (Pearson correlation coefficients) between samples were calculated by the core function in R and were presented only as heatmaps. Both HCA and PCC were carried out with the R package pheatmap. For HCA, normalized signal intensities of metabolites (unit variance scaling) were visualized as a color spectrum. Unsupervised PCA (principal component analysis) was performed using the statistics function prcomp within R1. The data were unit variance scaled prior to unsupervised PCA.
Gene ontology enrichment and Kyoto encyclopedia of genes and genomes enrichment analysis
The identified metabolites were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Compound database2. Gene function annotation was performed using four databases: National Center for Biotechnology Information (NCBI3), non-redundant protein sequences (NR4), the Swiss-Prot protein sequence database, and Gene Ontology (GO5). The annotated metabolites and genes were then mapped to the KEGG Pathway database6. Pathways with significantly regulated metabolites were subjected to MSEA (metabolite sets enrichment analysis), and their significance was determined based on p-values from hypergeometric tests. Finally, the DEGs in the WL, ML12h, ML6d, and RL groups were subjected to GO and KEGG pathway enrichment analysis.
Weighted gene co-expression network analysis of metabolome and transcriptomic data
The weighted gene co-expression network analysis (WGCNA) package was used to generate co-expression network modules between metabolites and genes. Using the automatic network construction function (blockwise Modules) with default parameters, co-expression modules were obtained based on TOM (topological overlap measure). The initial clusters were merged on eigengenes. The eigengene value was calculated for each module and used to search for associations with key anthocyanin substances. The transcriptional regulatory networks were generated by combining the Pearson correlation coefficient (PCC > 0.85) between genes and transcription factors, and the cis-element binding sites were predicted in the promoter regions of key anthocyanin genes in the same module. The networks were visualized by CYTOSCAPE (v.3.7.2, United States) (Kohl et al., 2011).
Phylogenetic analysis
The conservative domain of the candidate MYBs and bZIPs from perilla and other plants were used to construct phylogenetic. Accession numbers of MYBs and bZIPs from other plants are shown in Supplementary Table S8. The neighbor-joining (NJ) tree was constructed using the poison model with MEGA-X with 1000 bootstrap replicates. The phylogenetic trees were visualized using the iTOL web tool7.
Quantitative reverse-transcription PCR
Quantitative reverse-transcription PCR was performed as previously described (Livak and Schmittgen, 2001). Total RNA was isolated from the samples using a MAGEN RNA Extraction Kit (MAGEN, United States), and reverse transcription was performed using an Evo M-WLV RT Kit (Accurate, Hunan, China). Primers for each gene (ANS, CHI, DFR, and so on) were designed using Primer 5.0; the primers are listed in Supplementary Table S9. The specificity of the primers was verified by agarose gel electrophoresis. PCR amplification was performed using a LightCycler 480 II REAL-TIME PCR system (Roche, Basel, Switzerland). The qPCR cycling conditions were 1 cycle of pre-denaturation at 95°C, 30 s; 40 cycles of 95°C for 5 s and 60°C for 30 s; dissolution at 95°C for 5 s; 60°C for 1 min. All genes were amplified with three replicates, and the perilla Actin gene was used as an internal reference gene.
Results
Various anthocyanin content of perilla under different light intensity treatments
To identify the leaves color and major pigment components under different light, we observed and measured the total anthocyanin and total flavonoid contents during different light intensity treatments. The perilla leaves color under different light intensities were obviously affected. The leaves and seedlings of the light-sensitive red perilla variety were green when cultured under weak light intensity (15 μmol/m2, WL). After transferring to moderate light intensity (180 μmol/m2, ML) for 6 days, the leaves and whole plants turned dark red. When returned to weak light intensity for 6 days (15 μmol/m2, RL), the leaf color gradually returned to partially green (Figure 1A). Like the changes of the leaf color, total anthocyanin and total flavone levels significantly increased under ML treatment and then decreased in RL compared with WL (Figures 1B,C). These results demonstrate that light intensity significantly affects the leaf color and the accumulation of total anthocyanins and total flavones in leaves of the light-sensitive perilla variety.
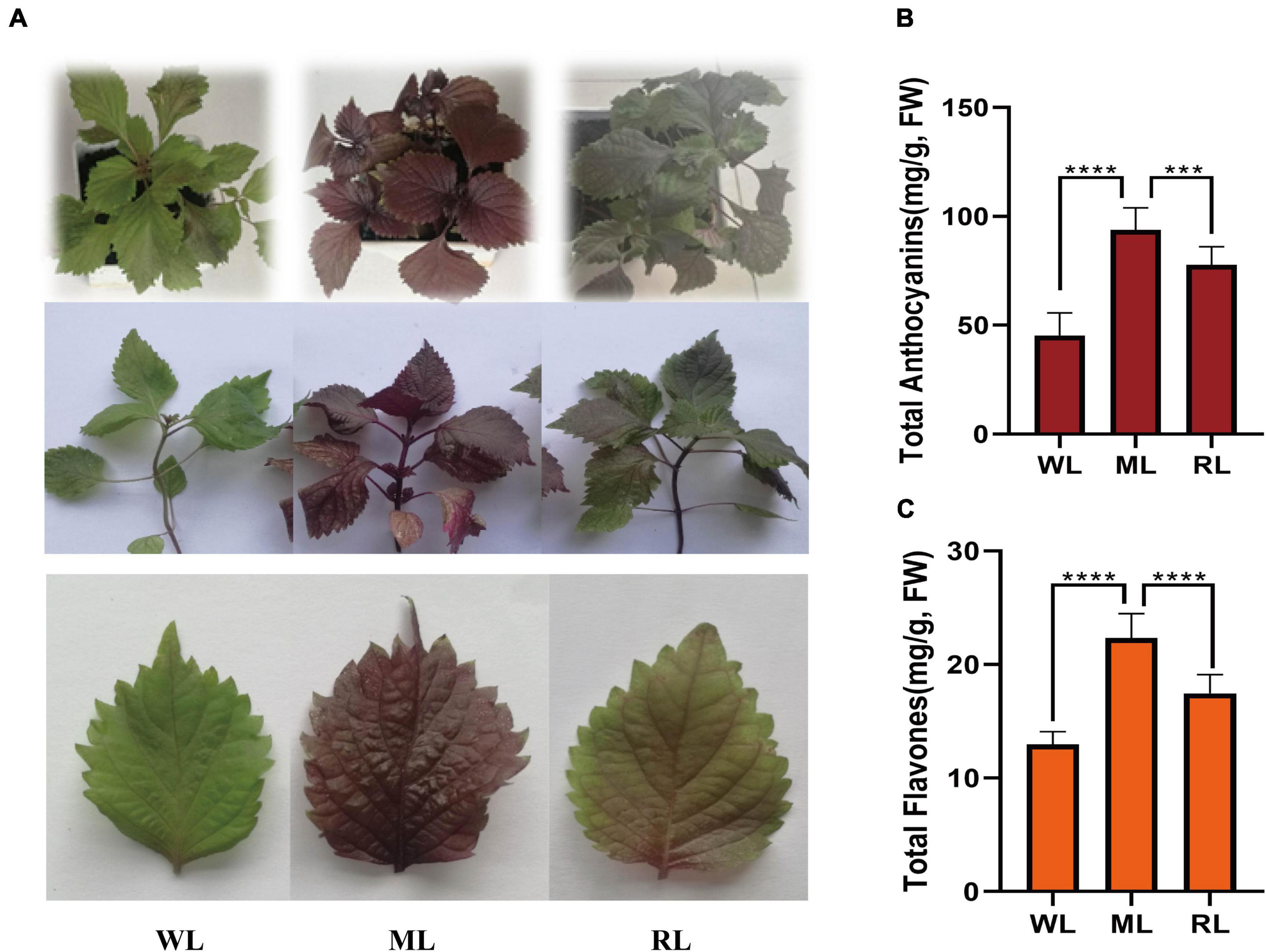
Figure 1. Leaf color, anthocyanin, and flavone contents of perilla under different light intensity treatments. (A) Phenotypes of perilla. (B) Total anthocyanin contents of perilla leaves. (C) Total flavone contents of perilla leaves under WL, weak light; ML, moderate light for 6 days; RL, recovered weak light for 6 days. **** Indicates P value < 0.0001.
Metabolome detection under different light intensity treatments
To assess anthocyanin components under different light treatments, we performed metabolomic analysis using UPLC-ESI-MS/MS. The samples were clearly clustered by PCA (Supplementary Figure S1). In total, 293 flavonoid metabolites were identified, including 103 flavonols, 72 flavones, 34 anthocyanins, and 84 other derivatives (Figure 2A; Supplementary Table S1). There were 90, 55, and 81 differentially expressed metabolites (DEM) in WL vs. ML, ML vs. RL, and WL vs. RL, respectively (Figure 2B).
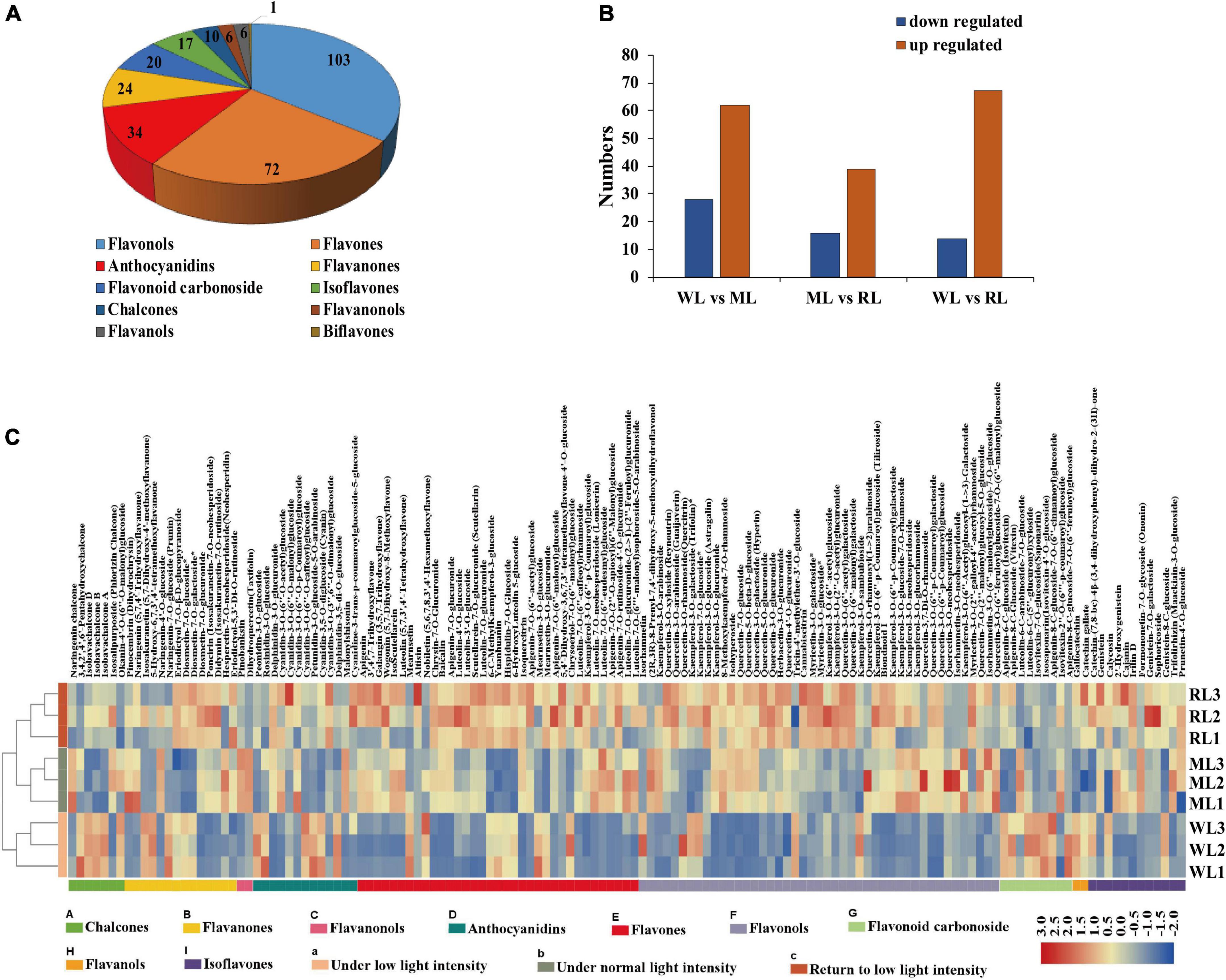
Figure 2. The flavonoid metabolites of perilla leaves under different light intensity treatments. (A) Types of flavonoid metabolites detected. (B) Differentially expressed metabolites (DEMs) in WL vs. ML6d, ML 6d vs. RL, and WL vs. RL. (C) Contents of flavonoid metabolites in perilla leaves under different light intensity treatments.
Quantitative analysis showed that approximately 140 (48%) flavonoid metabolites were affected under different light intensity treatments in perilla leaves. Among these, 62 metabolites were significantly induced under increased light intensity, including 23 flavonols, 19 flavones, and 5 anthocyanins (Supplementary Table S2). Among them, the contents of malonylshisonin increased 1.7-fold under ML treatment compared with WL and decreased 0.8-fold under RL treatment compared with ML (Supplementary Table S3). Naringin chalcone and dihydroquercetin (early intermediates in the anthocyanin biosynthesis pathway), as well as cyanidin-3,5-O-diglucoside, cyanidin-3-O-(6”-O-p-coumaryl) glucoside-5-O-glucoside, and cyanidin-3-O-(6”-O-p-coumaryl) glucoside (important precursors of malonylshisonin) were all detected in this study. Interestingly, the levels of naringin chalcone, dihydroquercetin, and cyanidin-3-O-(6”-O-p-coumaryl) glucoside increased by 4.3-, 5.8-, and 3.2-fold (Supplementary Table S3), respectively, under ML treatment compared with WL. By contrast, the cyanidin-3,5-O-diglucoside contents decreased by 0.6-fold under ML treatment compared with WL. These results suggest that light intensity affects accumulation of the metabolites in anthocyanin biosynthesis pathway in perilla.
According to the change trends of flavonoid metabolites under different treatments, they were divided into eight categories. The levels of subclass 4 and subclass 7 members initially increased, then decreased, possessed consistent pattern with total anthocyanin contents. Both malonylshisonin and naringin chalcone belong to subclass 7. The levels of subclass 2 and subclass 6 members continuously increased under ML and RL treatment (Supplementary Figure S2). The cyanidin-3-O-(6”-O-P-coumaryl) glucoside-5-O-glucoside and dihydroquercetin belong to subclass 2 and subclass 4, respectively. The four subclasses attracted our main attention.
Transcriptomic analysis under different light intensity treatments
To explore the molecular regulatory mechanism of the perilla anthocyanin metabolites in response to light intensity, we performed transcriptomic sequence. Considering the rapid response of transcriptomic level to light treatment, we added a short treatment (12 h) under 180 μmol/m2 of light (ML12h). After RNA sequencing and data filtering, 701.64 million clean reads with 105 Gb clean data were obtained. The Q20 reached 97%, with an average GC content of 48%. When we mapped the sequencing reads to perilla reference genomes, we obtained an average unique mapping ratio of 82.94% and a multiple mapping ratio of 10.19% (Supplementary Table S4).
Subsequently, we identified DEGs among all samples. And most of DEGs were responsed to light intensity showing to the results of GO analysis (Supplementary Figure S3). Compared to the WL group, 3290 and 3026 genes were upregulated in ML12h and ML6d, respectively. Compared to the RL group, 4043 and 3219 genes were upregulated in ML12h and ML6d, respectively (Figure 3A). Among the upregulated genes under ML treatment, 983 genes showed increased expression under short (12 h) and long (6 days) treatments compared with WL (Figure 3B). These DEGs are mainly involved in metabolic pathways, phenylalanine and tyrosine tryptophan biosynthesis and phenylalanine metabolism in the KEGG enrichment (Figure 3C). By contrast, 1478 genes were downregulated in RL compared with ML (Figure 3D). The down-regulated genes were enriched in biosynthesis of secondary metabolites, phenylalanine and tyrosine tryptophan biosynthesis, tyrosine metabolism, phenylalanine metabolism, and flavonoid biosynthesis in the KEGG enrichments (Figure 3E). Based on the expression pattern, we divided all DEGs into 26 categories (Supplementary Figure S4). The 983 and 1478 DEGs most belong to Profile 20 (upregulated in ML12h and ML6d compared with WL) and Profile 23 (downregulated in RL compared with ML12h and ML6d), respectively. These photo-response genes attracted our main attention.
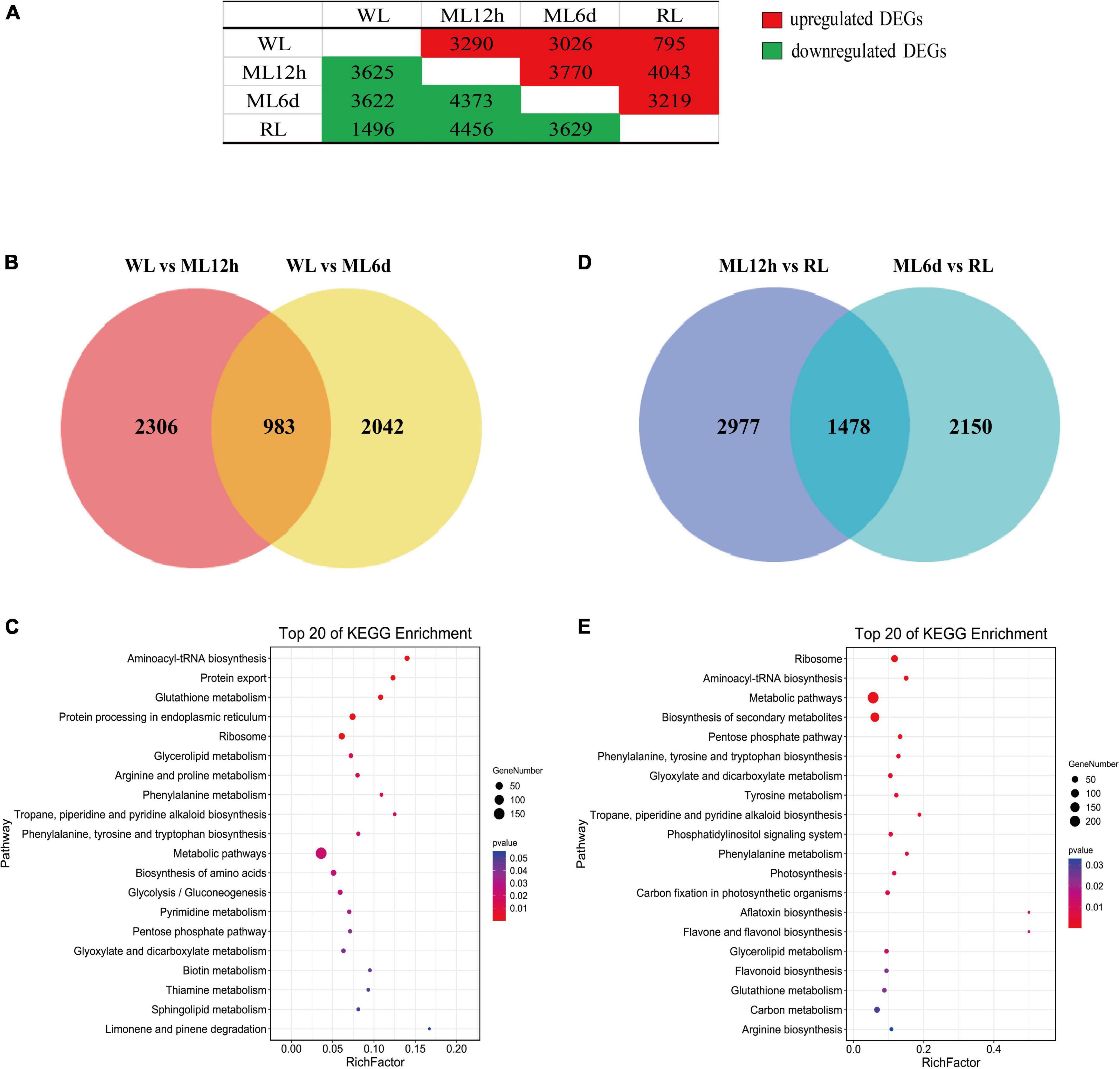
Figure 3. Venn diagram and KEGG pathways of the DEGs under different light intensity treatments. (A) Number of upregulated and downregulated differentially expressed genes (DEGs) in pair-wise comparisons. (B) Venn diagram and (C) top 20 KEGG pathways of upregulated genes of the ML12h and ML6d groups compared with WL. (D) Venn diagram and (E) top 20 KEGG pathways of downregulated genes in the RL group compared with ML12h and ML6d.
The metabolites and key genes in the anthocyanin biosynthesis pathway
We identified metabolites and genes involved in the anthocyanin and flavones biosynthetic pathways (Figure 4). Malonylshisonin biosynthesis involves 10 catalytic reactions, containing the initial cyanidin biosynthesis and the latter cyanidin derivatization pathway (Saito and Yamazaki, 2002; Jiang et al., 2020). Naringin chalcone, dihydroquercetin, cyanidin-3,5-O-diglucoside, cyanidin-3-O-(6”-O-p-Coumaroyl) glucoside, cyanidin-3-O-(6”-O-p-Coumaroyl) glucoside-5-O-diglucoside, and malonylshisonin were identified in the metabolomic analysis. Except for cyanidin 3,5-O-glucoside, these metabolites obviously accumulated under increased light intensity. Then, we identified 95 genes encoding the 10 anthocyanin synthases in the malonyshisonin biosynthetic pathway (Figure 4 and Supplementary Table S5). Furthermore, 32 genes were upregulated after light treatment, which encode the key rate-limiting enzymes in the basic anthocyanin biosynthetic pathway. Among these, 18 genes (including PAL, 4CL, CHS, F3’H, ANS, 5-GT, and MAT) were more strongly upregulated under 12 h ML treatment, and five genes (including F3H, 3-GT, ACT, and CHI) had higher expression levels under 6 days ML treatment. Only one C4H gene was expressed at high levels under both 12 h and 6 days ML treatments. These results suggest that light intensity remarkably influences the gene expression and metabolite accumulation involving in anthocyanin biosynthesis in perilla leaves.
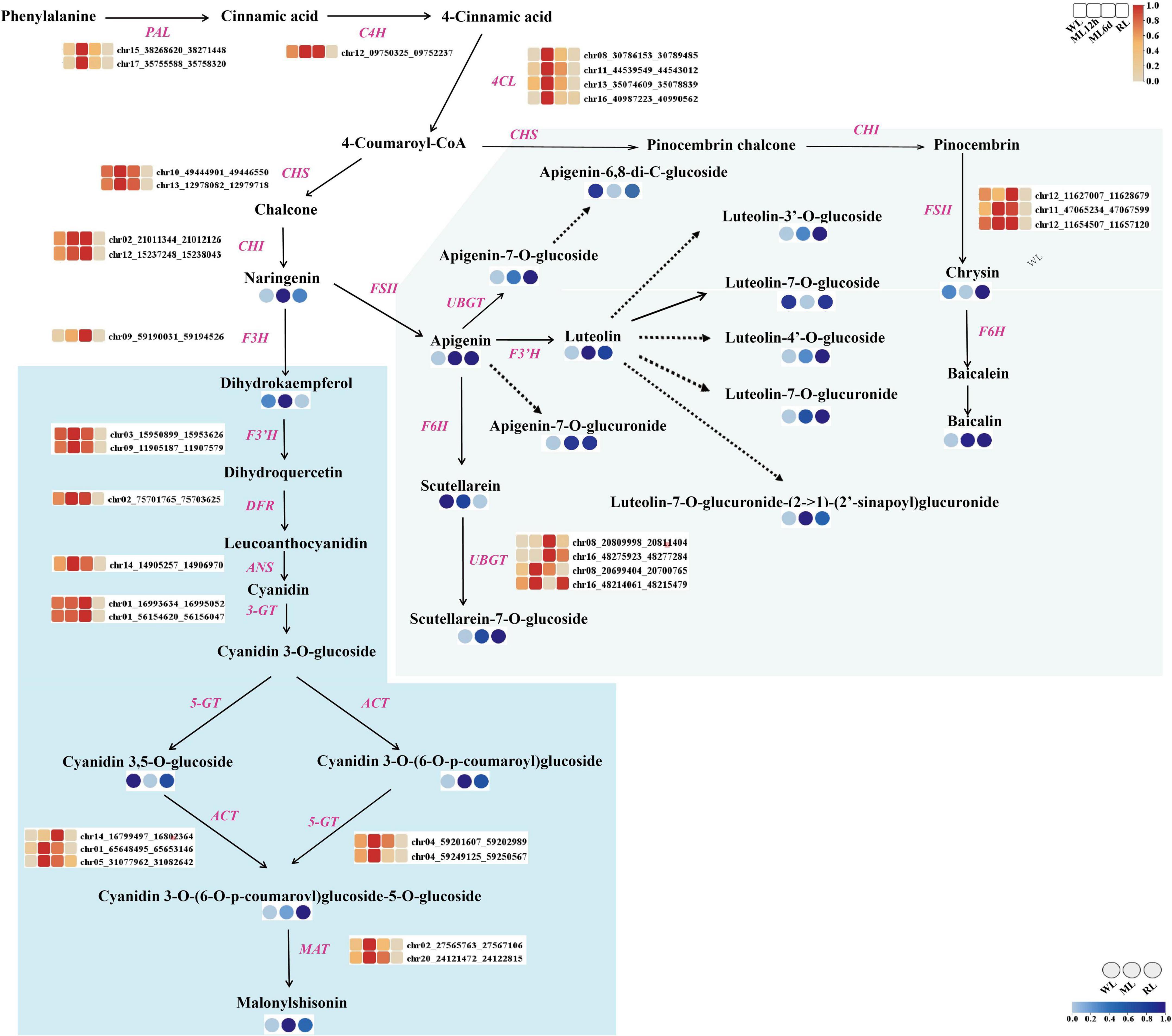
Figure 4. The metabolites and genes involving in the anthocyanin biosynthesis pathway under different light intensity treatments in perilla leaves. The blue background indicates the major malonylshisonin biosynthetic pathway. The green background indicates the major flavonoid biosynthetic pathway. The blue color in the circle indicates the level of the metabolite under WL, ML, and RL treatment. The red color in the blocks represents the relative expression levels of genes under WL, ML12h, ML6d, and RL treatments. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3’H, flavonoid 3’-hydroxylase; DFR, dihydroflavonol reductase; ANS, anthocyanin synthase (leucoanthocyanin dioxgenase); 3-GT, flavonoid 3-glucosyltransferase; ACT, anthocyanin acyltransferase; 5-GT, anthocyanin 5-glucosyltransferase; MAT, anthocyanin malonyltransferase.
Moreover, we identified 14 light-regulated flavones based on analysis of the flavonoid biosynthesis pathway. Most of these compounds accumulated under light treatment. Specifically, the contents of luteolin, apigenin, and baicalin increased by 5.0-, 5.4-, and 11.6-fold, respectively, under ML treatment compared with WL. Comparatively, the levels of apigenin-6,8-di-C-glucoside, luteolin-7-O-glucoside, and chrysin decreased under ML treatment. Because many genes involved in the accumulation of these substances are unknown, we only detected FSII, F6H, and UBGT in the flavone biosynthetic pathway (Supplementary Table S5). Like the malonylshisonin biosynthesis genes, both FSII and UBGT were upregulated under ML treatment compared with WL. In view of luteolin, apigenin and more flavones possess pharmacological effects (Lei et al., 2020; Liang et al., 2020; Rizzo et al., 2021), the results show that strongly accumulated flavones under elevated light treatment, with provided potential application for further research.
Integrated transcriptomic and metabolomic analysis
To gain further insight into the regulation of anthocyanin biosynthesis in response to light intensity, we performed WGCNA to identify co-expression networks between anthocyanin metabolites and gene expression levels. Ten co-expression modules were identified based on their similar expression patterns (Figure 5A). The turquoise module is positively correlated with anthocyanins, malonylshisonin, and other metabolites (Figure 5B). We therefore focused on those genes in the turquoise module. We identified anthocyanin biosynthesis and transcription factor genes in this module and used gene pairs with correlation coefficients more than 0.85 to construct a regulatory network (Figure 5C and Supplementary Table S7). Five important genes involved in anthocyanin biosynthesis were identified in the turquoise module, including ANS, 4CL, DFR, and CHI, which were positively correlated with total anthocyanin and malonylshisonin contents. The 147 transcription factors were also identified, mainly encoding MYB, bHLH, bZIP, ERF, and NAC in the module. Notably, MYB5, MYB12, bZIP2, bZIP6, bHLH1, bHLH7, and bHLH9 had high connectivity with malonylshisonin and five anthocyanin biosynthesis genes. The MYB_related2, MYB_related4 had high positive correlation with malonylshisonin. Moreover, the expression level of most identified MYB and bZIP genes were strongly upregulated under ML treatment compared with WL and downregulated under RL treatment compared with ML (Figure 5D). By contrast, MYB22, bHLH1, bHLH7, and bHLH9 were negatively correlated with malonylshisonin content. MYB22 and bHLH1 were also negatively correlated with total anthocyanin contents (Supplementary Table S6). Hence, these transcription factors were speculated to play important roles in regulating malonylshisonin accumulation under different light intensities.
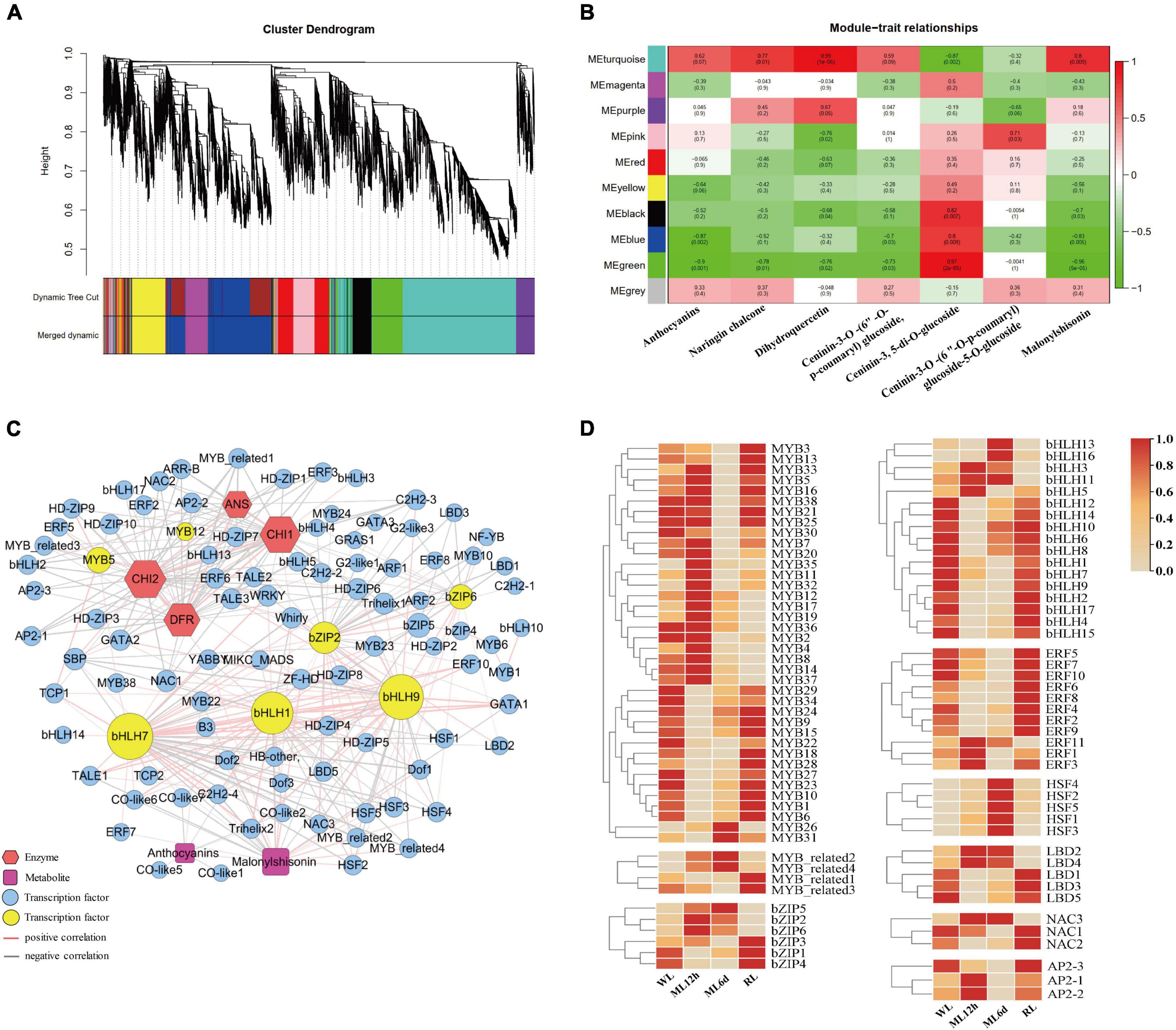
Figure 5. Weighted correlation network analysis (WGCNA) of the anthocyanin biosynthetic pathway. (A) Dendrogram of the co-expression modules (clusters). The major tree branches constitute 10 modules labeled with different colors. (B) Heat map showing the correlations between modules and anthocyanin compounds. Red indicates a positive correlation, and green indicates a negative correlation between the cluster and the sample. (C) Co-expression network of the genes and metabolites with correlation coefficients greater than 0.85. (D) Heat maps of the expression levels of genes encoding MYBs, MYB-related, bZIPs, bHLHs, ERFs, HSFs, LBDs, NACs, and AP2s based on the transcriptomic data.
Candidate MYB genes involved in anthocyanin biosynthesis
MYB transcription factors are major members of MBW ternary transcriptional complex, which is an important regulator of anthocyanin metabolism in many plants (Lloyd et al., 2017; Li et al., 2020; Wang et al., 2020; Zhang et al., 2020; Ning et al., 2021). In WGCNA analysis, we identified 38 MYB transcription factors, containing 37 R2R3-MYB subfamily members and one 1R-MYB (MYB10). These MYBs showed high correlation coefficient with malonylshisonin and five anthocyanin biosynthesis genes (Supplementary Figure S5). Among them, 22 R2R3-MYB members were highly induced under ML treatment, while 15 were downregulated in response to light treatment (Figure 5D).
To examine the MYB transcription factors involved in regulating anthocyanin accumulation, we constructed a phylogenetic tree combined the MYBs that we identified in perilla and were previously reported to regulate anthocyanin accumulation in other plants. The phylogenetic tree was divided into three subclasses (Figure 6A). Subclass I contains perilla MYB12, MYB2 and key anthocyanin-regulated MYBs from Arabidopsis and apple. Subclass II includes 11 perilla MYBs and three Arabidopsis R2R3-MYB family members. Among them, MYB8 and MYB14 showed peak expression under ML12h treatment. Subclass III includes 25 MYBs from perilla and IbMYB340. Among them, MYB4 and MYB37 were expressed at high levels and responded to light treatment.
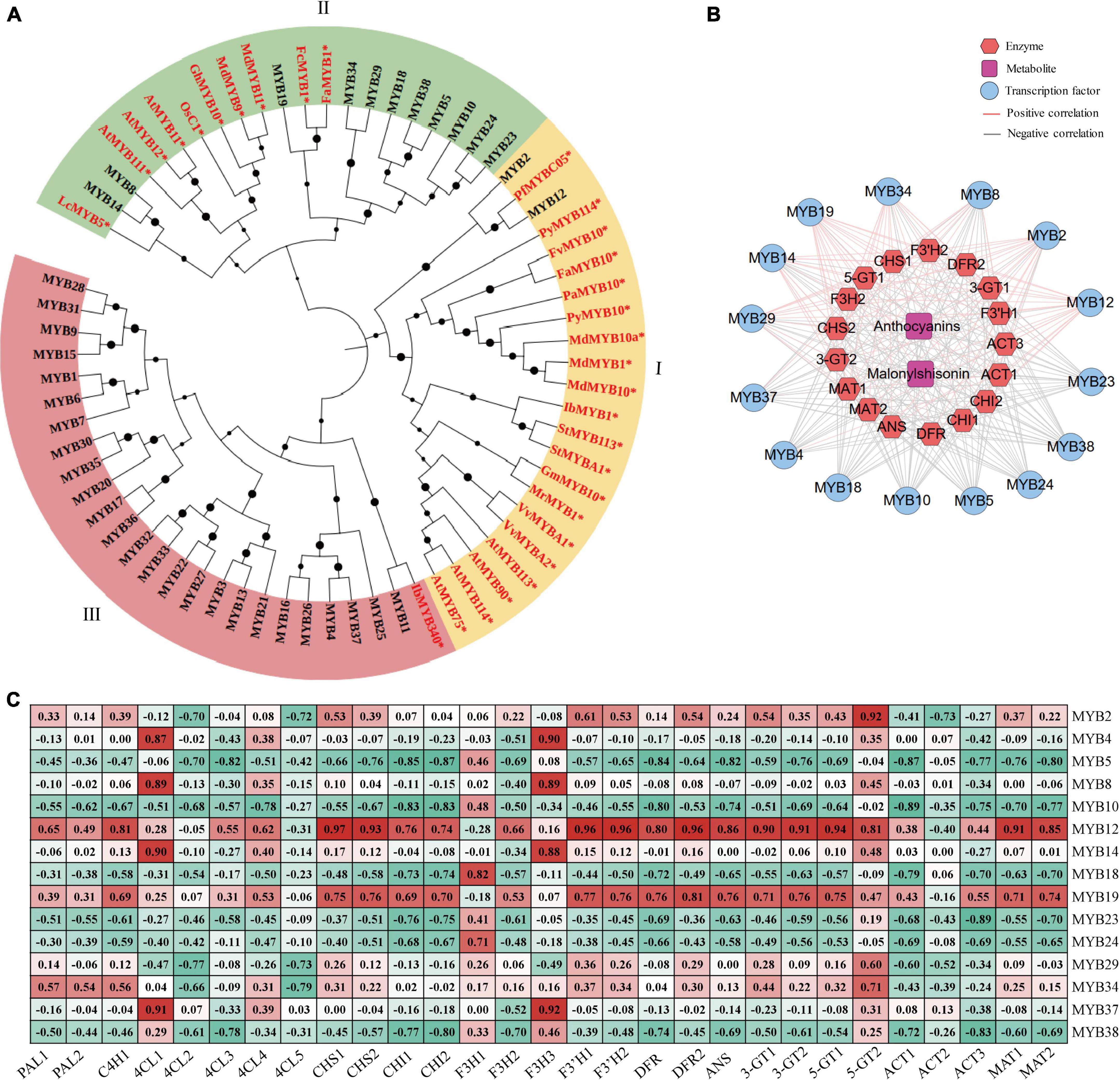
Figure 6. The MYB transcription factors involved in anthocyanin biosynthesis in perilla. (A) Phylogenetic relationships of MYBs between perilla and other plants. Yellow, green, and pink background indicate three subclasses in the evolutionary tree. The dots next to the nodes represent bootstrap values of 1000 replicates. The red font and symbol * indicate that these objects have been reported. (B) Co-expression networks between candidate MYBs. (C) Correlation coefficients between candidate MYB transcription factors and key anthocyanin biosynthesis genes. At, Arabidopsis thaliana; Vv, Vitis vinifera; St, Solanum tuberosum; Pb, Pyrus x bretschneideri; Os, Oryza sativa Japonica; Pf, Perilla frutescens var. crispa; Pa, Prunus avium; Py, Pyrus pyrifolia var. culta; Mr, Morella rubra; Md, Malus domestica; Lc, Litchi chinensis; Ib, Ipomoea batatas; Gh, Gossypium hirsutum; Gm, Garcinia mangostana; Fa, Fragaria x ananassa; Fv, Fragaria vesca; Fc, Fragaria chiloensis.
For further explore the relationships between the MYBs and anthocyanin biosynthesis, we performed correlation analysis of 15 selected MYBs with anthocyanin metabolites and 17 key anthocyanin biosynthesis genes. Among these, 9 MYBs showed positive correlations with anthocyanin metabolites and 6 showed positive correlations with key anthocyanin biosynthesis genes (Figure 6B). Interestingly, MYB12 and MYB19 showed significantly positive correlations with most anthocyanin biosynthesis genes (Figure 6C). The MYB12 had been reported in malonylshisonin formation, which was found in responsive to light treatment in the study.
Furthermore, the cis-acting elements of four key anthocyanin biosynthesis genes, including DFR, CHI1, CHI2, and ANS, were analyzed. Several light-response elements and hormone-response elements were identified in the four anthocyanin genes (Supplementary Figure S7). Interestingly, we identified CCAAT-box and MYB-recognition element (MRE) cis-acting elements in the promoter regions of perilla CHI1 and ANS, respectively (Supplementary Figure S7). Combining WGCNA analysis, phylogenetic analysis, and cis-acting element analysis, more MYB transcription factors were predicted to regulate malonylshisonin accumulation under the light treatment in perilla.
Candidate bZIP genes involved in anthocyanin biosynthesis
The bZIP transcription factors were recently reported to regulate anthocyanin biosynthesis (Hu et al., 2019; Liu et al., 2019; Wang et al., 2020). In the turquoise module, we also identified 6 bZIP transcription factors (Supplementary Figure S6). These perilla bZIPs were divided into two subclasses in the evolutionary tree (Figure 7A). Subclass I consist of bZIP4 and bZIP3 from perilla, along with HY5 transcription factors from other plants. Subclass II consists of 4 bZIP transcription factors from perilla, along with MdbZIP44, LcABF3, and MdABI5. Co-expression analysis showed that bZIP2, bZIP5, and bZIP6 share positive regulatory relationships with key anthocyanin biosynthesis genes (CHI1, CHI2, ANS, and DFR), while bZIP1, bZIP3, and bZIP4 share negative regulatory relationships with these genes (Figure 7B). Interestingly, bZIP2, bZIP5, and bZIP6 showed significant positive correlations with total anthocyanin and malonylshisonin contents and most anthocyanin biosynthesis genes (Figure 7C). The promoter sequences of CHI1, CHI2, ANS, and DFR all contain G-boxes, a light-responsive binding site that is recognized by bZIP transcription factors (Supplementary Figure S7). The bZIP2 and bZIP6 were identified to encode G-box binding factor 3 in perilla, suggesting that they may play key roles in the light response and the anthocyanin regulation.
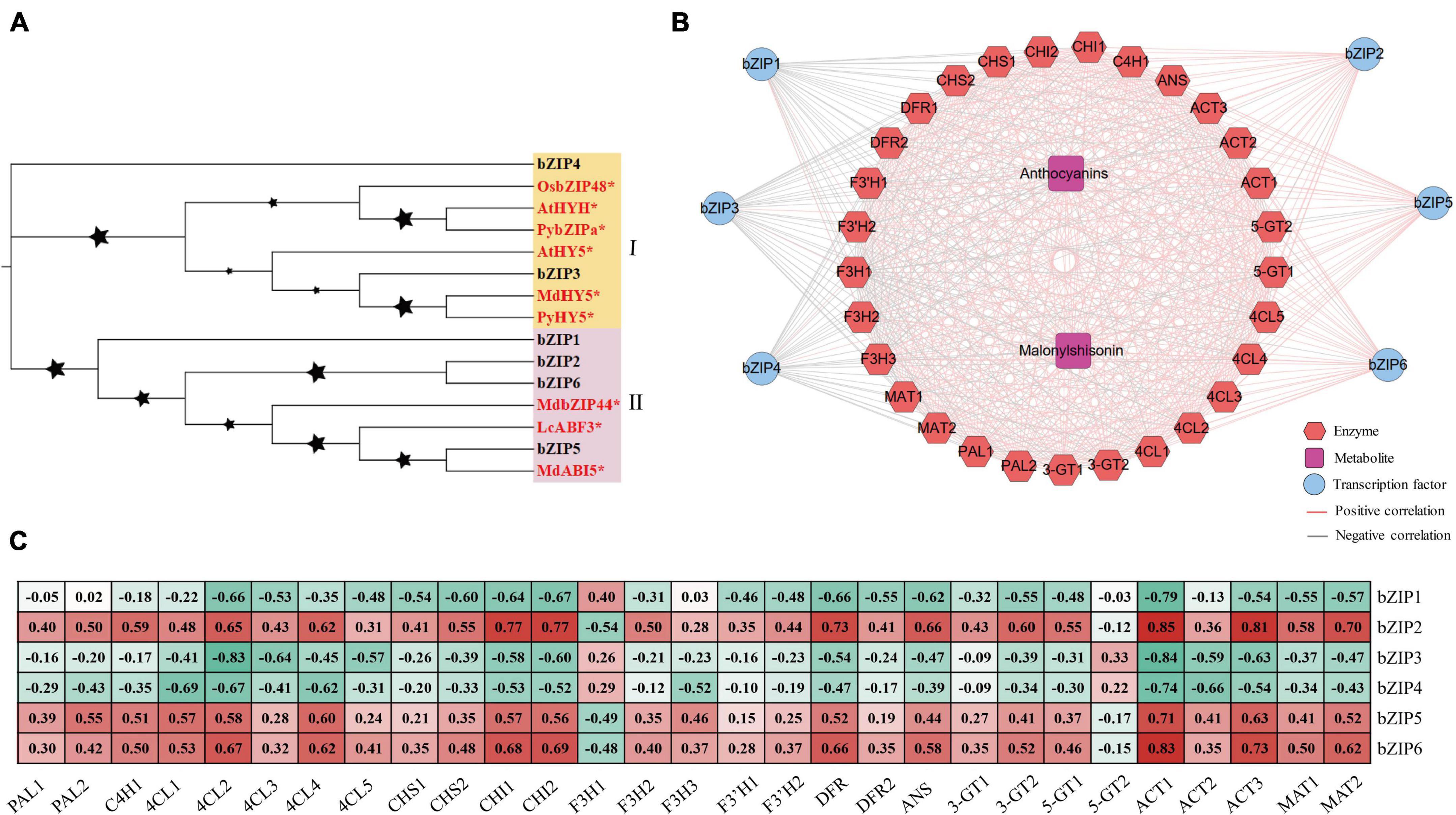
Figure 7. The bZIP transcription factors involved in the light response and anthocyanin regulation in perilla. (A) Phylogenetic relationships of bZIP transcription factors. Yellow and pink background indicate two subclasses in the evolutionary tree. The stars next to the nodes represent bootstrap values of 1000 replicates. The red font and symbol * indicate that these objects have been reported. (B) Co-expression networks between candidate bZIPs. (C) Correlation coefficients between candidate bZIP transcription factors and key anthocyanin biosynthesis genes. At, Arabidopsis thaliana; Py, Pyrus pyrifolia; Os, Oryza sativa Japonica; Md, Malus domestica; Lc, Litchi chinensis.
Quantitative RT-PCR analysis of the candidate genes involved in anthocyanin biosynthesis in Perilla frutescens
We measured the expression levels of anthocyanin biosynthesis genes (CHI1, CHI2, ANS, and DFR) and important transcription factor genes (MYB, bZIP, and bHLH) involved in anthocyanin regulation by qRT-PCR (Figure 8). Combined the transcriptomic and qRT-PCR data, the candidate genes showed similar expression patterns. The anthocyanin biosynthesis genes (CHI1, CHI2, ANS, and DFR), MYB genes, and bZIP genes were upregulated under increased light intensity. These genes were all upregulated after a short (12 h) moderate light intensity treatment and downregulated after a long treatment (6 days), and their expression continued to decline under low light intensity. The response of the anthocyanin biosynthesis genes to light intensity treatment might be the direct cause of accumulation of anthocyanin metabolites. The MYB transcription factor genes were more sensitive to short vs. long light treatment, pointing to their important regulatory functions in anthocyanin metabolite accumulation. The bZIP2 and bZIP6 are homologous to the important light-response transcription factor gene HY5. Both were upregulated after a short light treatment. We also identified bHLH1 and bHLH7 transcription factor genes in perilla. Interestingly, both genes were downregulated under light treatment. The bHLH1 and bHLH7 encode PIF3 and PIF7, respectively. The PIFs were negatively regulation factors in response to light in other plants. The downregulated trend of bHLH1 and bHLH7 under light treatment is consistent with their functions. The expression patterns of key anthocyanin biosynthesis genes and transcription factors in response to light points to their important roles in regulating anthocyanin biosynthesis.
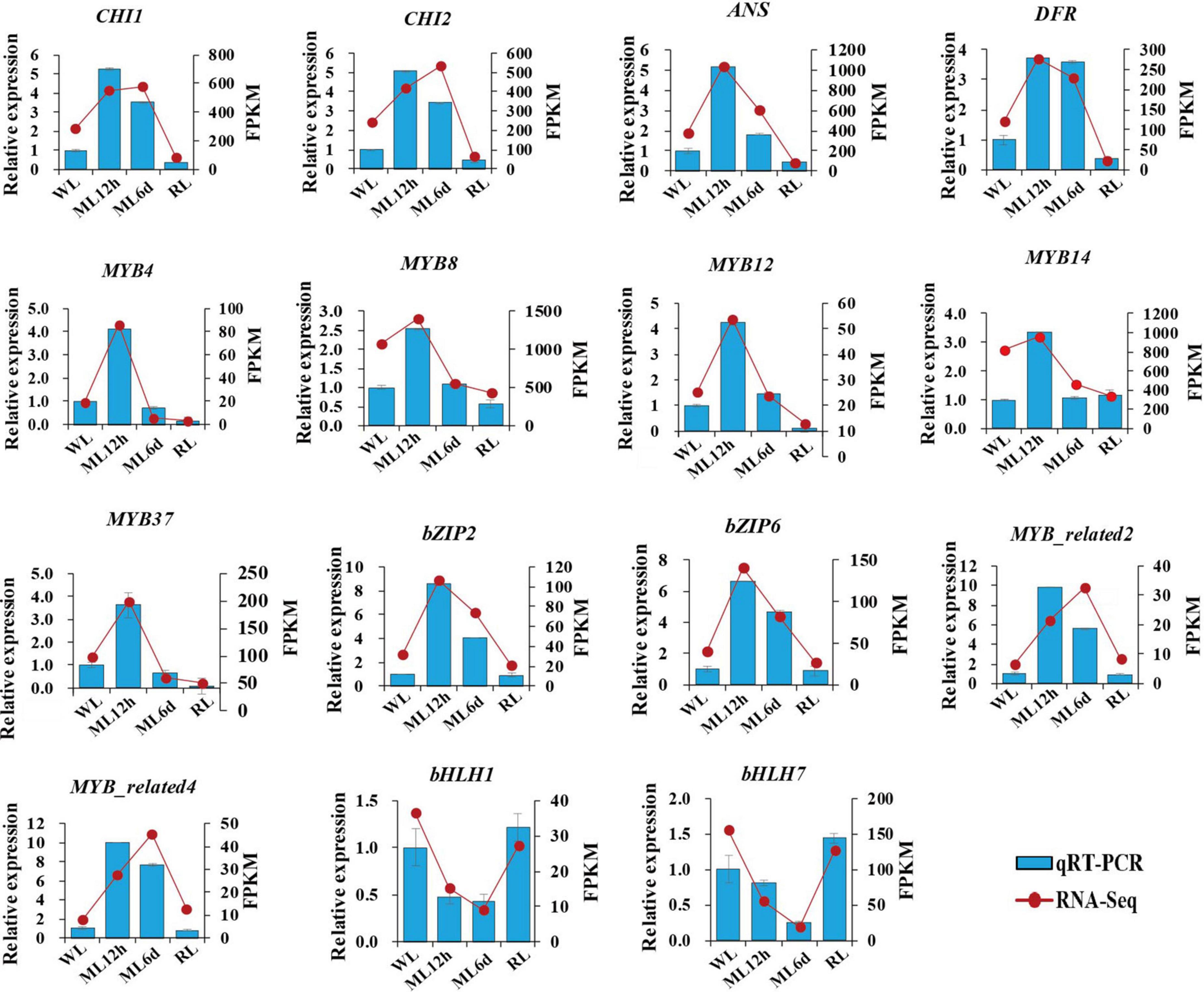
Figure 8. Relative expression levels of candidate genes. The histogram indicates gene expression data form qRT-PCR. The line chart indicates FPKM from transcriptomic sequence. Data are presented as means ± standard deviation (n = 3).
Discussion
Leaf color and anthocyanin accumulation are important traits for the medicinal and ornamental value of perilla (Ishikura, 1981; Yu et al., 2017). Perilla leaves color is highly sensitive to light conditions, providing an opportunity to study the molecule mechanism of anthocyanin biosynthesis and regulation (Mastropasqua et al., 2020).
In this study, we performed metabolomic and transcriptomic analyses of the light-sensitive perilla variety under different light intensity treatments. The metabolomic analysis identified 293 flavonoid compounds. Focus on malonylshisonin, the major anthocyanin metabolites in perilla, obviously increased in response to increased light treatment. Malonylshisonin biosynthesis involves 10 catalytic reactions, containing the initial cyanidin biosynthesis and the latter cyanidin derivatization pathway. In the latter steps, cyanidin-3-O-glucoside is converted to cyanidin-3-O-(6”-O-p-coumaryl) glucoside and cyanidin 3,5-O-glucoside (Springob et al., 2003). Under ML treatment, the content of cyanidin 3,5-O-glucoside was decreased, whereas the content of cyanidin-3-O-(6”-O-P-coumaryl) glucoside was increased, suggesting that the metabolic flow favored the formation of cyanidin-3-O-(6”-O-P-coumaryl) glucoside under light treatment. Next, we investigated the biosynthesis genes of malonylshisonin accumulation under different light treatments. Previous work found that nearly all the genes encoding the enzymes (except for CHI) showed higher expression in red perilla than in green perilla (Jiang et al., 2020). In the current study, the genes encoding CHS, CHI, F3H, DFR, and ANS, the rate-limiting enzymes in the anthocyanin biosynthetic pathway (Takos et al., 2006), were obviously responded to light treatment. These genes may play important roles in malonylshisonin accumulation in perilla leaves under light treatment.
In the WGCNA and correlation analysis, ANS, CHI, and DFR were positively correlated with malonylshisonin content. The homologous proteins of ANS, CHI, and DFR were responded to light treatment in other plants (Britsch, 1990; Menssen et al., 1990; Saito et al., 1999). CHI is the rate-limiting enzyme for flavonoid biosynthesis (Bednar and Hadcock, 1988; McKhann et al., 1998; Shimada et al., 2003). The expression of CHI increases in rice in response to high light levels and UV irradiation (Albert et al., 2009; Park et al., 2021). DFR catalyzes the conversion of dihydroquercetin to leucoanthocyanidin, which is recognized as a pivotal step in anthocyanin biosynthesis (Xie et al., 2004; Miyagawa et al., 2015; Ni et al., 2020). The expression of DFR is downregulated in the shade in Brassica oleracea (Liu et al., 2020). ANS catalyzes the oxidation of leucoanthocyanidin (Turnbull et al., 2000, 2003), and ANS is upregulated by high-light treatment in Arabidopsis thaliana (Zheng et al., 2019, 2021). In the current study, the expression levels of the perilla genes CHI1, CHI2, DFR, and ANS all increased under ML treatment. These results indicate that these genes play a major role in determining malonylshisonin biosynthesis in different light intensities.
Plants respond to light via photoreceptors and employ multiple transcription factors to regulate anthocyanin metabolites formation (Zhang et al., 2021). To further identify the key regulation factors, we used WGCNA method to comprehensively analyze metabolites and key genes in perilla. In this study, the 147 transcription factors were also identified based on WGCNA analysis. Among them, R2R3-MYBs have been widely researched. There are four Arabidopsis thaliana R2R3-MYB family members (AtMYB113, AtMYB90, AtMYB114, and AtMYB75) were reported control anthocyanin biosynthesis in vegetative tissues (Gonzalez et al., 2008). AtMYB75 (also called PAP1) activates the transcription of CHS, DFR, and ANS (Shi and Xie, 2010, 2011) and participates in the regulation of anthocyanin biosynthesis by HY5 via the transcriptional activation of its promoter (Shin et al., 2013). The MdMYB10 and MdMYB1 are key anthocyanin-related transcription factors in apple (Takos et al., 2006; Espley et al., 2007). In apple, the expression of MdMYB1 and its allele MdMYB10 is induced by sunlight (Takos et al., 2006; Feng et al., 2013). The expression of MdMYB10 is also regulated by HY5, which binds to the G-box in the MdMYB10 promoter in a light-responsive manner, thereby affecting the expression of downstream structural genes and anthocyanin accumulation in apple (An et al., 2017). Interestingly, PfMYB12 were identified shares complete sequence identity with MYBC05. The MYBC05 was previously reported to promote the production of anthocyanins, which is thought to bind to myC-F3G1 and PFWD to form the MBW complex, which promotes the production of anthocyanins in red perilla (Yamazaki et al., 2003; Yamazaki and Saito, 2011). Hence, the PfMYB12 and PfMYB2 are clustered with above MYBs, which are thought to important function in anthocyanin light-responsive accumulation. Next, The PfMYB8 and PfMYB14 were clustered with three AtMYBs (AtMYB11, AtMYB12, AtMYB111), which targeted several flavonoid biosynthesis genes, including CHS, CHI, F3H, and FLS1 in Arabidopsis (Stracke et al., 2007). AtMYB12 and AtMYB111 also control flavonol accumulation in roots in a light-dependent manner (Stracke et al., 2017). PfMYB4 and PfMYB37 were clustered with IbMYB340, which interacted with IbERF71 and IbbHLH2 to regulate anthocyanin biosynthesis in sweet potato (Ning et al., 2021). Moreover, MYB-binding sites, including MBS, MRE, and CCAAT-box elements, were identified in the promotors of CHI1, CHI2, and ANS. The CCAAT-box is the binding site of the HvMYB1 transcription factor in barley (Alexander et al., 2019). The MRE cis-acting elements is recognized and bound by BrPAP1 transcription factor, which regulates anthocyanin biosynthesis in Brassica rapa (Yang et al., 2021). These findings suggest that these MYBs play important roles in regulating anthocyanin accumulation in perilla in response to different light intensities. To sum up, the six PfMYB (PfMYB2, PfMYB12, PfMYB8, PfMYB14, PfMYB4, and PfMYB37) were predicted to possess important function in photosensitive anthocyanin regulation in perilla.
The bZIP family, one of the largest transcription factor families in plants, is involved in physiological processes such as plant development, environmental signaling, and stress responses (Djamei et al., 2007; Liu C. C. et al., 2018; Xiao et al., 2021). The bZIP transcription factor HY5 is an important transcription factor involved in light signal transduction and plant pigment accumulation in response to light (Holm et al., 2002; Stracke et al., 2010; Shin et al., 2013; Gangappa and Botto, 2016). HY5 also promotes flavonoid accumulation in response to both visible and UV-B light. Using WGCNA, we identified six bZIP transcription factors in the co-expression modules. Perilla bZIP3 and bZIP4 clustered with HY5s from other plants. In the previous research, HY5 could responds to receptors for blue, red/far-red, and ultraviolet light (Oyama et al., 1997; Ang et al., 1998; Chattopadhyay et al., 1998; Brown and Jenkins, 2008) and directly binds to G-box or ACE-box elements in the promoters of MYB genes to promote their expression to promote flavonoid biosynthesis (Holm et al., 2002; Stracke et al., 2010; Shin et al., 2013). Perilla bZIP3 and bZIP4 were both induced by light but showed negative regulatory relationships with anthocyanin pathway genes in the current study. By contrast, bZIP2 and bZIP6 showed positive regulatory relationships with most anthocyanin biosynthesis genes and possess light-regulated traits in perilla. bZIP2 and bZIP6 encode G-box binding protein 3 (GBF3) transcription factors. In Arabidopsis, GBF3 has the same gene expression pattern as HY5, and its binding sites showed a 86.7% overlap with those of HY5 under changing light conditions, suggesting that GBF3 might interfere with the function of HY5 (Kurihara et al., 2020). bZIP2 and bZIP6 also clustered with MdbZIP44 and LcABF3. MdbZIP44 promotes anthocyanin accumulation in response to abscisic acid by enhancing the binding of MdMYB1 to the promoters of downstream target genes in apple (An et al., 2018). LcABF3 activates the promoter region of LcMYB1 and structural genes such as LcF3’H and LcDFR to modulate anthocyanin biosynthesis in lychee (Hu et al., 2019). Interestingly, bZIP2 and bZIP6 had the highest correlations with CHI1 and CHI2 in WGCNA. The CHI1 and CHI2 promoters contain G-box cis-acting elements, which can be recognized and combined by bZIPs. That means bZIP2 and bZIP6 perhaps together with MYBs, activated the transcription of the key candidate anthocyanin biosynthesis genes and promote the accumulation of malonylshisonin under moderate light intensity. Moreover, the bHLH1 and bHLH7 encode PIF3 and PIF7. Phytochrome-interacting factor (PIF) genes are downregulated in the presence of light. These bHLH transcription factors function as primary partners of the red/far-red light receptors phytochromes in light signaling (Nozue et al., 2007). Our results indicate that bHLH1 and bHLH7 negatively regulate anthocyanin biosynthesis in the light, which is consistent with the function of PIFs in the light response.
In the research, we also identified 14 important flavone metabolites in perilla leaves, including luteolin, apigenin, baicalein, scutellarein, and their glycoside derivatives. Luteolin can attenuate allergic nasal inflammation (Jeon et al., 2014; Liang et al., 2020). Apigenin and apigenin-7-diglucuronide can mitigate oxidative stress and possesses anti-inflammatory activity (Bian et al., 2017; Kasiri et al., 2018). Baicalin represses C/EBP beta via redox homeostasis, representing a potentially effective treatment for Parkinson’s disease (Lei et al., 2020). They are all possess a certain of pharmacological functions and got more attention. The accumulation of 10 flavones increased in perilla leaves in response to moderate light treatment. This increase in the levels of pharmacologically important flavones indicates that the value of perilla can be increased under the proper light conditions. The flavonoid biosynthesis and its regulation require further study in perilla.
In summary, the regulation of light intensity signal in plants is a complex process. We mainly focused on the red perilla malonylshisonin synthesis pathway. In addition, the function of each candidate gene needs to be confirmed by different methods. Therefore, further work is required to elucidate the mechanism of light signal in perilla plants.
Conclusion
This study integrated the metabolomic and transcriptomic analyses of the light-sensitive perilla variety under different light intensity. The light intensity significantly affects the color, metabolite accumulation and gene expression involved in the main anthocyanin biosynthesis in perilla leaves. Based on WGCNA analysis, key genes and transcription factors were identified. What’s more, 6 MYBs and 4 bZIPs were predicted to play important roles in light-regulated anthocyanin biosynthesis. The identified key genes and regulatory factors will help us to understand the potential mechanism of photo-regulated anthocyanin accumulation in perilla.
Statistical analysis
Statistical analyses were performed by Student’s t-test and one-way ANOVA using SPSS 23.0 (SPSS Inc., Chicago, IL, United States). Least significant difference (LSD) was used to compare treatment means, and p = 0.05 was considered as statistically significant.
Data availability statement
The datasets presented in this study are deposited in the Genome Sequence Archive in National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences. The accession number is PRJNA837648.
Author contributions
GX: writing – original draft, conceptualization, methodology, formal analysis, investigation, and data curation. HW: supervision, project administration, and funding acquisition. QS: planned and designed the research, methodology, data curation, and writing – review and editing. XZ and ZL: performed the experiments. HJ: supervision. DW: data analysis. JH and KX: plants cultivation. All authors have read and approved the final version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation for regional fund (31860391), the National Natural Science Foundation of China Grant (31970261), and the Talent Support Project of Guangdong (2019TQ05N182).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.976449/full#supplementary-material
Footnotes
- ^ www.r-project.org
- ^ http://www.kegg.jp/kegg/compound/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://www.ncbi.nlm.nih.gov/refseq/about/nonredundantproteins
- ^ http://www.geneontology.org/
- ^ http://www.kegg.jp/kegg/pathway.html
- ^ http://itol.embl.de/index.shtml
References
Ahmed, H. M. (2019). Ethnomedicinal, Phytochemical and Pharmacological Investigations of Perilla frutescens (L.) Britt. Molecules 24:102. doi: 10.3390/molecules24010102
Albert, N. W., Lewis, D. H., Zhang, H., Irving, L. J., Jameson, P. E., and Davies, K. M. (2009). Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 60, 2191–2202.
Alexander, R. D., Wendelboe-Nelson, C., and Morris, P. C. (2019). The barley transcription factor HvMYB1 is a positive regulator of drought tolerance. Plant Physiol. Biochem. 142, 246–253. doi: 10.1016/j.plaphy.2019.07.014
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4:17023. doi: 10.1038/hortres.2017.56
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., and Hao, Y. J. (2018). Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 41, 2678–2692. doi: 10.1111/pce.13393
Ang, L. H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., et al. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. doi: 10.1016/S1097-2765(00)80022-2
Azuma, A., Yakushiji, H., Koshita, Y., and Kobayashi, S. (2012). Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 236, 1067–1080. doi: 10.1007/s00425-012-1650-x
Bednar, R. A., and Hadcock, J. R. (1988). Purification and characterization of chalcone isomerase from soybeans. J. Biol. Chem. 263, 9582–9588. doi: 10.1016/S0021-9258(19)81556-9
Bian, M., Zhang, Y., Du, X., Xu, J., Cui, J., Gu, J., et al. (2017). Apigenin-7-diglucuronide protects retinas against bright light-induced photoreceptor degeneration through the inhibition of retinal oxidative stress and inflammation. Brain Res. 1663, 141–150. doi: 10.1016/j.brainres.2017.03.019
Britsch, L. (1990). Purification and characterization of flavone synthase I, a 2-oxoglutarate-dependent desaturase. Arch Biochem. Biophys. 282, 152–160. doi: 10.1016/0003-9861(90)90099-k
Brown, B. A., and Jenkins, G. I. (2008). UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8. HY5, and HYH. Plant Physiol. 146, 576–588. doi: 10.1104/pp.107.108456
Chattopadhyay, S., Ang, L. H., Puente, P., Deng, X. W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. doi: 10.1105/tpc.10.5.673
Chen, X. Q., Nagao, N., Itani, T., and Irifune, K. (2012). Anti-oxidative analysis, and identification and quantification of anthocyanin pigments in different coloured rice. Food Chem. 135, 2783–2788. doi: 10.1016/j.foodchem.2012.06.098
Djamei, A., Pitzschke, A., Nakagami, H., Rajh, I., and Hirt, H. (2007). Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 318, 453–456. doi: 10.1126/science.1148110
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Feng, F., Li, M., Ma, F., and Cheng, L. (2013). Phenylpropanoid metabolites and expression of key genes involved in anthocyanin biosynthesis in the shaded peel of apple fruit in response to sun exposure. Plant Physiol. Biochem. 69, 54–61. doi: 10.1016/j.plaphy.2013.04.020
Feng, S., Wang, Y., Yang, S., Xu, Y., and Chen, X. (2010). Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232, 245–255. doi: 10.1007/s00425-010-1170-5
Fujiwara, Y., Kono, M., Ito, A., and Ito, M. (2018). Anthocyanins in perilla plants and dried leaves. Phytochemistry 147, 158–166. doi: 10.1016/j.phytochem.2018.01.003
Gangappa, S. N., and Botto, J. F. (2016). The Multifaceted Roles of HY5 in Plant Growth and Development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Holm, M., Ma, L. G., Qu, L. J., and Deng, X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. doi: 10.1101/gad.969702
Holton, T. A., and Cornish, E. C. (1995). Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 7, 1071–1083. doi: 10.2307/3870058
Honda, G., Yuba, A., Nishizawa, A., and Tabata, M. (1994). Genetic control of geranial formation in Perilla frutescens. Biochem. Genet. 32, 155–159. doi: 10.1007/BF00554619
Hu, B., Lai, B., Wang, D., Li, J., Chen, L., Qin, Y., et al. (2019). Three LcABFs are Involved in the Regulation of Chlorophyll Degradation and Anthocyanin Biosynthesis During Fruit Ripening in Litchi chinensis. Plant Cell Physiol. 60, 448–461. doi: 10.1093/pcp/pcy219
Huang, B., Lin, C., Chen, Y., and Kao, S. (2014). Anti-inflammatory effects of Perilla frutescens leaf extract on lipopolysaccharide-stimulated RAW264.7 cells. Mol. Med. Rep. 10, 1077–1083. doi: 10.3892/mmr.2014.2298
Ishikura, N. (1981). Anthocyanins and Flavones in Leaves and Seeds of Perilla Plant. Agric. Biol. Chem. 45, 1855–1860. doi: 10.1271/bbb1961.45.1855
Jeon, I. H., Kim, H. S., Kang, H. J., Lee, H., Jeong, S. I., Kim, S. J., et al. (2014). Anti-Inflammatory and Antipruritic Effects of Luteolin from Perilla (P. frutescens L.) Leaves. Molecules 19, 6941–6951. doi: 10.3390/molecules19066941
Jiang, M., Ren, L., Lian, H., Liu, Y., and Chen, H. (2016). Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (Solanum melongena L.). Plant Sci. 249, 46–58. doi: 10.1016/j.plantsci.2016.04.001
Jiang, T., Guo, K., Liu, L., Tian, W., Xie, X., Wen, S., et al. (2020). Integrated transcriptomic and metabolomic data reveal the flavonoid biosynthesis metabolic pathway in Perilla frutescens (L.) leaves. Sci. Rep. 10:16207. doi: 10.1038/s41598-020-73274-y
Kasiri, N., Rahmati, M., Ahmadi, L., and Eskandari, N. (2018). The significant impact of apigenin on different aspects of autoimmune disease. Inflammopharmacology 26, 1359–1373. doi: 10.1007/s10787-018-0531-8
Khoo, H. E., Azlan, A., Tang, S. T., and Lim, S. M. (2017). Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 61:1361779. doi: 10.1080/16546628.2017.1361779
Kocic, B., Filipovic, S., Nikolic, M., and Petrovic, B. (2011). Effects of anthocyanins and anthocyanin-rich extracts on the risk for cancers of the gastrointestinal tract. J. Buon. 16, 602–608.
Kohl, M., Wiese, S., and Warscheid, B. (2011). Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303.
Kong, J. M., Chia, L. S., Goh, N. K., Chia, T. F., and Brouillard, R. (2003). Analysis and biological activities of anthocyanins. Phytochemistry 64, 923–933. doi: 10.1016/S0031-9422(03)00438-2
Kurihara, Y., Makita, Y., Shimohira, H., and Matsui, M. (2020). Time-Course Transcriptomics Study Reveals Mode of bZIP Transcription Factors on Light Exposure in Arabidopsis. Int. J. Mol. Sci. 21:1993. doi: 10.3390/ijms21061993
Lee, J. K., and Ohnishi, O. (2003). Genetic relationships among cultivated types of Perilla frutescens and their weedy types in East Asia revealed by AFLP markers. Genet. Resour. Crop Evol. 50, 65–74. doi: 10.1023/A:1022951002271
Lei, K., Shen, Y., He, Y., Zhang, L., Zhang, J., Tong, W., et al. (2020). Baicalin Represses C/EBP beta via Its Antioxidative Effect in Parkinson’s Disease. Oxid. Med. Cell Longev. 2020:8951907. doi: 10.1155/2020/8951907
Li, C., Wu, J., Hu, K. D., Wei, S. W., Sun, H. Y., Hu, L. Y., et al. (2020). PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 7:37. doi: 10.1038/s41438-020-0254-z
Liang, K., Yu, S., Huang, W., and Yen, H. (2020). Luteolin Attenuates Allergic Nasal Inflammation via Inhibition of Interleukin-4 in an Allergic Rhinitis Mouse Model and Peripheral Blood from Human Subjects with Allergic Rhinitis. Front. Pharmacol. 11:291. doi: 10.3389/fphar.2020.00291
Lin, C., Kuo, C., Wang, J., Cheng, J., Huang, Z., and Chen, C. (2007). Growth inhibitory and apoptosis inducing effect of Perilla frutescens extract on human hepatoma HepG2 cells. J. Ethnopharmacol. 112, 557–567. doi: 10.1016/j.jep.2007.05.008
Liu, C., Yao, X., Li, G., Huang, L., and Xie, Z. (2020). Transcriptomic profiling of purple broccoli reveals light-induced anthocyanin biosynthetic signaling and structural genes. PeerJ 8:e8870. doi: 10.7717/peerj.8870
Liu, C. C., Chi, C., Jin, L. J., Zhu, J., Yu, J. Q., and Zhou, Y. H. (2018). The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 41, 1762–1775. doi: 10.1111/pce.13171
Liu, H., Su, J., Zhu, Y., Yao, G., Allan, A. C., Ampomah-Dwamena, C., et al. (2019). The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 6:134. doi: 10.1038/s41438-019-0217-4
Liu, Y., Tikunov, Y., Schouten, R. E., Marcelis, L., Visser, R., and Bovy, A. (2018). Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 6:52. doi: 10.3389/fchem.2018.00052
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB-bHLH-WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Makino, T., Furuta, Y., Wakushima, H., Fujii, H., Saito, K., and Kano, Y. (2003). Anti-allergic effect of Perilla frutescens and its active constituents. Phytother. Res. 17, 240–243. doi: 10.1002/ptr.1115
Mastropasqua, L., Dipierro, N., and Paciolla, C. (2020). Effects of Darkness and Light Spectra on Nutrients and Pigments in Radish, Soybean, Mung Bean and Pumpkin Sprouts. Antioxidants 9:558. doi: 10.3390/antiox9060558
McKhann, H. I., Paiva, N. L., Dixon, R. A., and Hirsch, A. M. (1998). Expression of genes for enzymes of the flavonoid biosynthetic pathway in the early stages of the Rhizobium-legume symbiosis. Adv. Exp. Med. Biol. 439, 45–54. doi: 10.1007/978-1-4615-5335-9_4
Meng, L., Lozano, Y., Bombarda, I., Gaydou, E., and Li, B. (2006). Anthocyanin and flavonoid production from Perilla frutescens: Pilot plant scale processing including cross-flow microfiltration and reverse osmosis. J. Agric. Food Chem. 54, 4297–4303. doi: 10.1021/jf0604079
Menssen, A., Höhmann, S., Martin, W., Schnable, P. S., Peterson, P. A., Saedler, H., et al. (1990). The En/Spm transposable element of Zea mays contains splice sites at the termini generating a novel intron from a dSpm element in the A2 gene. EMBO J. 9, 3051–3057. doi: 10.1002/j.1460-2075.1990.tb07501.x
Miyagawa, N., Miyahara, T., Okamoto, M., Hirose, Y., Sakaguchi, K., Hatano, S., et al. (2015). Dihydroflavonol 4-reductase activity is associated with the intensity of flower colors in delphinium. Plant Biotechnol. 32, 249–255. doi: 10.5511/plantbiotechnology.15.0702b
Ni, J., Ruan, R., Wang, L., Jiang, Z., Gu, X., Chen, L., et al. (2020). Functional and correlation analyses of dihydroflavonol-4-reductase genes indicate their roles in regulating anthocyanin changes in Ginkgo biloba. Ind. Crop Prod. 152:112546. doi: 10.1016/j.indcrop.2020.112546
Ning, Z., Hu, K., Zhou, Z., Zhao, D., Tang, J., Wang, H., et al. (2021). IbERF71, with IbMYB340 and IbbHLH2, coregulates anthocyanin accumulation by binding to the IbANS1 promoter in purple-fleshed sweet potato (Ipomoea batatas L.). Plant Cell Rep. 40, 157–169. doi: 10.1007/s00299-020-02621-0
Nitta, M., Lee, J. K., Kang, C. W., Katsuta, M., Yasumoto, S., Liu, D. J., et al. (2005a). The distribution of Perilla species. Genet. Resour. Crop Evolution 52, 797–804. doi: 10.1007/s10722-003-6017-5
Nitta, M., Lee, J. K., Kobayashi, H., Liu, D., and Nagamine, T. (2005b). Diversification of Multipurpose Plant Perilla Frutescens. Genet. Resour. Crop Evolution 52, 663–670. doi: 10.1007/s10722-003-6013-9
Nozue, K., Covington, M. F., Duek, P. D., Lorrain, S., Fankhauser, C., Harmer, S. L., et al. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361. doi: 10.1038/nature05946
Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes. Dev. 11, 2983–2995. doi: 10.1101/gad.11.22.2983
Park, S. I., Park, H. L., Bhoo, S. H., Lee, S. W., and Cho, M. H. (2021). Biochemical and molecular characterization of the rice chalcone isomerase family. Plants (Basel) 10:2064. doi: 10.3390/plants10102064
Rizzo, V., Ferlazzo, N., Curro, M., Isola, G., Matarese, M., Bertuccio, M. P., et al. (2021). Baicalin-Induced Autophagy Preserved LPS-Stimulated Intestin22(5):2315.al Cells from Inflammation and Alterations of Paracellular Permeability. Int. J. Mol. Sci. 22:2315. doi: 10.3390/ijms22052315
Saita, E., Kishimoto, Y., Tani, M., Iizuka, M., Toyozaki, M., Sugihara, N., et al. (2012). Antioxidant Activities of Perilla frutescens against Low-Density Lipoprotein Oxidation in Vitro and in Human Subjects. J. Oleo. Sci. 61, 113–120. doi: 10.5650/jos.61.113
Saito, K., Kobayashi, M., Gong, Z., Tanaka, Y., and Yamazaki, M. (1999). Direct evidence for anthocyanidin synthase as a 2-oxoglutarate-dependent oxygenase: Molecular cloning and functional expression of cDNA from a red forma of Perilla frutescens. Plant J. 17, 181–189. doi: 10.1046/j.1365-313x.1999.00365.x
Saito, K., and Yamazaki, M. (2002). Biochemistry and molecular biology of the late-stage of biosynthesis of anthocyanin: Lessons from Perilla frutescens as a model plant. New Phytol. 155, 9–23. doi: 10.1046/j.1469-8137.2002.00440.x
Shi, M. Z., and Xie, D. Y. (2010). Features of anthocyanin biosynthesis in pap1-D and wild-type Arabidopsis thaliana plants grown in different light intensity and culture media conditions. Planta 231, 1385–1400. doi: 10.1007/s00425-010-1142-9
Shi, M. Z., and Xie, D. Y. (2011). Engineering of red cells of Arabidopsis thaliana and comparative genome-wide gene expression analysis of red cells versus wild-type cells. Planta 233, 787–805. doi: 10.1007/s00425-010-1335-2
Shimada, N., Aoki, T., Sato, S., Nakamura, Y., Tabata, S., and Ayabe, S. (2003). A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol. 131, 941–951. doi: 10.1104/pp.004820
Shin, D. H., Choi, M., Kim, K., Bang, G., Cho, M., Choi, S. B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. Febs. Lett. 587, 1543–1547. doi: 10.1016/j.febslet.2013.03.037
Springob, K., Nakajima, J., Yamazaki, M., and Saito, K. (2003). Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 20, 288–303. doi: 10.1039/b109542k
Stracke, R., Favory, J. J., Gruber, H., Bartelniewoehner, L., Bartels, S., Binkert, M., et al. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33, 88–103. doi: 10.1111/j.1365-3040.2009.02061.x
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Turgut-Kara, N., and Weisshaar, B. (2017). The AtMYB12 activation domain maps to a short C-terminal region of the transcription factor. Z. Naturforsch. C. J. Biosci. 72, 251–257. doi: 10.1515/znc-2016-0221
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Turnbull, J., Sobey, W., Aplin, R., Hassan, A., Schofield, C., Firmin, J., et al. (2000). ChemInform Abstract: Are Anthocyanidins the Immediate Products of Anthocyanidin Synthase? Chem. Commun. 32, 2473–2474. doi: 10.1039/b007594i
Turnbull, J. J., Nagle, M. J., Seibel, J. F., Welford, R. W., Grant, G. H., and Schofield, C. J. (2003). The C-4 stereochemistry of leucocyanidin substrates for anthocyanidin synthase affects product selectivity. Bioorg. Med. Chem. Lett. 13, 3853–3857. doi: 10.1016/S0960-894X(03)00711-X
Wang, Y., Zhang, X., Zhao, Y., Yang, J., He, Y., Li, G., et al. (2020). Transcription factor PyHY5 binds to the promoters of PyWD40 and PyMYB10 and regulates its expression in red pear ‘Yunhongli No. 1’. Plant Physiol. Biochem. 154, 665–674. doi: 10.1016/j.plaphy.2020.07.008
Xiao, S., Liu, Y., Wang, A., Liu, Y., Li, X., Liu, Z., et al. (2021). The response of tartary buckwheat and 19 bZIP genes to abscisic acid (ABA). Mol. Biol. Rep. 48, 4341–4350. doi: 10.1007/s11033-021-06449-z
Xie, D. Y., Jackson, L. A., Cooper, J. D., Ferreira, D., and Paiva, N. L. (2004). Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol. 134, 979–994. doi: 10.1104/pp.103.030221
Yamamoto, H., and Ogawa, T. (2002). Antimicrobial activity of perilla seed polyphenols against oral pathogenic bacteria. Biosci. Biotech. Bioch. 66, 921–924. doi: 10.1271/bbb.66.921
Yamazaki, M., Makita, Y., Fester, K., and Saito, K. (2003). Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perilla frutescens var. crispa. Biochem. Eng. J. 14, 191–197. doi: 10.1016/S1369-703X(02)00222-X
Yamazaki, M., and Saito, K. (2011). Molecular genetic study on the anthocyanin chemotypes of Perilla frutescens var. crispa. Nat. Prod. Commun. 6, 423–427. doi: 10.1177/1934578X1100600322
Yang, E., Ku, S., Lee, W., Lee, S., Lee, T., Song, K., et al. (2013). Barrier protective effects of rosmarinic acid on HMGB1-induced inflammatory responses in vitro and in vivo. J. Cell Physiol. 228, 975–982. doi: 10.1002/jcp.24243
Yang, J., Song, H. D., Chen, Y., Chen, B., Kim, M., Kim, P., et al. (2021). A single amino acid substitution in the R2R3 conserved domain of the BrPAP1a transcription factor impairs anthocyanin production in turnip (Brassica rapa subsp. rapa). Plant Physiol. Biochem. 162, 124–136. doi: 10.1016/j.plaphy.2021.02.011
Yu, H., Qiu, J. F., Ma, L. J., Hu, Y. J., Li, P., and Wan, J. B. (2017). Phytochemical and phytopharmacological review of Perilla frutescens L. (Labiatae), a traditional edible-medicinal herb in China. Food Chem. Toxicol. 108, 375–391. doi: 10.1016/j.fct.2016.11.023
Zhang, S., Zhang, L., Zou, H., Qiu, L., Zheng, Y., Yang, D., et al. (2021). Effects of Light on Secondary Metabolite Biosynthesis in Medicinal Plants. Front. Plant Sci. 12:781236. doi: 10.3389/fpls.2021.781236
Zhang, Z., Shi, Y., Ma, Y., Yang, X., Yin, X., Zhang, Y., et al. (2020). The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 18, 2267–2279. doi: 10.1111/pbi.13382
Zhao, C. L., Chen, Z. J., Bai, X. S., Ding, C., Long, T. J., Wei, F. G., et al. (2014). Structure-activity relationships of anthocyanidin glycosylation. Mol. Divers 18, 687–700. doi: 10.1007/s11030-014-9520-z
Zheng, X. T., Chen, Y. L., Zhang, X. H., Cai, M. L., Yu, Z. C., and Peng, C. L. (2019). ANS-deficient Arabidopsis is sensitive to high light due to impaired anthocyanin photoprotection. Funct. Plant Biol. 46, 756–765. doi: 10.1071/FP19042
Zheng, X. T., Yu, Z. C., Tang, J. W., Cai, M. L., Chen, Y. L., Yang, C. W., et al. (2021). The major photoprotective role of anthocyanins in leaves of Arabidopsis thaliana under long-term high light treatment: Antioxidant or light attenuator? Photosynth. Res. 149, 25–40.
Keywords: Perilla frutescens, metabolomic, transcriptomic, photoinduced, weighted gene co-expression network analysis (WGCNA), transcription factor
Citation: Xie G, Zou X, Liang Z, Wu D, He J, Xie K, Jin H, Wang H and Shen Q (2022) Integrated metabolomic and transcriptomic analyses reveal molecular response of anthocyanins biosynthesis in perilla to light intensity. Front. Plant Sci. 13:976449. doi: 10.3389/fpls.2022.976449
Received: 23 June 2022; Accepted: 22 August 2022;
Published: 23 September 2022.
Edited by:
Jordi Moreno-Romero, Universitat Autònoma de Barcelona, SpainCopyright © 2022 Xie, Zou, Liang, Wu, He, Xie, Jin, Wang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbin Wang, d2FuZ2hvbmdiaW5nQGd6dWNtLmVkdS5jbg==; Qi Shen, c2hlbnFpQGd6dWNtLmVkdS5jbg==
 Guanwen Xie
Guanwen Xie Xiuzai Zou
Xiuzai Zou Zishan Liang
Zishan Liang Duan Wu
Duan Wu