- 1State Key Laboratory of Crop Genetics and Germplasm Enhancement, College of Agriculture, Nanjing Agricultural University, Nanjing, China
- 2State Key Laboratory of Cotton Biology, Institute of Cotton Research of Chinese Academy of Agricultural Sciences, Anyang, Henan, China
To date, no ideal effective method for controlling Verticillium wilt in upland cotton (Gossypium hirsutum) has been defined. The purpose of this study was to determine the effects and mechanism through which flagellin C (FLiC) regulates the Gossypium hirsutum cation/proton exchanger 3 gene (GhCAX3), induces plant immunity, and increases resistance to Verticillium wilt. The FLiC gene was cloned from an endophytic bacterium (Pseudomonas) isolated from roots of the upland cotton cultivar Zhongmiansuo 41. The biocontrol effects of FLiC purified in vitro on resistant and susceptible upland cotton cultivars were 47.50 and 32.42%, respectively. FLiC induced a hypersensitive response (HR) in leaves of tobacco and immune responses in upland cotton. Transcriptome data showed that treatment with FLiC significantly enriched the calcium antiporter activity-associated disease-resistant metabolic pathway in seedlings. Moreover, FLiC downregulated GhCAX3 expression to increase intracellular calcium ion (Ca2+) content and stimulate increases in the intracellular hydrogen peroxide (H2O2) and nitric oxide (NO) contents. The coordinated regulation of Ca2+, H2O2, and NO enhanced cotton resistance to Verticillium wilt. Furthermore, transgenic Arabidopsis plants overexpressing FLiC showed significantly improved resistance to Verticillium wilt. FLiC may be used as a resistance gene and a regulator to improve resistance to Verticillium dahliae (VD) in upland cotton.
Highlights
– FLiC induces HRs in tobacco and immune responses in cotton.
– FLiC decreases GhCAX3 expression to increase intracellular Ca2+content.
– Ca2+, H2O2, and NO synergistically regulate the resistance of cotton to VD.
– Transgenic Arabidopsis overexpressing FLiC shows significantly improved resistance to VD.
Introduction
The immune response is closely related to disease resistance in plants. Plants rely mainly on defense pathways such as pathogenic microorganism pattern-triggered immunity (PTI; Nürnberger and Brunner, 2002) and pathogen-secreted effector-triggered immunity (ETI; Thomma et al., 2011; Naveed et al., 2020) to resist pathogen infection. The defense response is achieved by the mutual recognition of and interaction between the recognition receptors of plant cells and the elicitors secreted by pathogenic microorganisms. Systemic disease resistance in plants is ultimately achieved through the transmission and transduction of a series of signals that activate the immune response (Bouizgarne et al., 2006; Jones and Dangl, 2006; Kumar et al., 2020). The early defense response of plants mainly includes the production of early disease resistance signals, such as intracellular calcium ion (Ca2+) influx, reactive oxygen species (ROS) bursts, and nitric oxide (NO) accumulation (Marcec et al., 2019). Ca2+ is a key second messenger and mediates the response of plants to biotic and abiotic stimuli (Gao et al., 2021; Köster et al., 2022). Spatially and temporally defined cytoplasmic Ca2+ signals are shaped through the concerted activity of ion channels, exchangers, and pumps in response to diverse stimuli, and these signals are then decoded through the activity of Ca2+ binding sensor proteins (Köster et al., 2022). In plants, Ca2+ signaling is central to both pattern- and effector-triggered immunity, and the generation of characteristic cytoplasmic Ca2+ elevations in response to potential pathogens are common to both immune reactions (Köster et al., 2022). ROS are widely produced in different cellular compartments under both biotic and abiotic stress conditions (Qi et al., 2017). The plant perception of pathogen-associated molecular patterns triggers a plethora of cellular immune responses, and one of these responses is a rapid and transient burst of ROS mediated by plasma membrane-localized Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases (Sang and Macho, 2017). Numerous studies have demonstrated the importance of NO in the regulation of plant defense against fungal pathogens. NO triggers a reprogramming of defense-related gene expression, the production of secondary metabolites with antimicrobial properties, and the hypersensitive response (HR; Martínez-Medina et al., 2019). These stimulus signals are converted from extracellular signals to intracellular signals and amplified through a cascade reaction to induce downstream defense reactions. These reactions always first occur around the infected tissue and then gradually spread to the surrounding uninfected tissues. The immune system of the entire plant is activated to defend against infection by various pathogens (Dixon et al., 1994; Yano et al., 1998; Buxdorf et al., 2013; Holmes et al., 2021). The resistance of plants is related to NO, hydrogen peroxide (H2O2), and Ca2+. The silencing of the Respiratory Burst Oxidase Homolog Protein D (GhRbohD) genes damage the resistance of cotton to fungi, weaken the generation of ROS, and decrease the content of NO, H2O2, and Ca2+ (Huang et al., 2021). However, few studies have investigated the mutual regulation of Ca2+, H2O2, and NO to mediate the resistance of cotton to Verticillium wilt.
Flagellin isolated from bacteria induces immune responses in arabidopsis, rice, algae, and kelp, but the underlying mechanisms are unclear (Takai et al., 2008; Wang, 2012; Wang et al., 2013; Ma et al., 2017). Plants stimulated by flagellin produce a series of defense responses, including ethylene (ETH) production, callose deposition, ROS bursts, defense-related gene expression, and growth inhibition (Asai et al., 2002; Zipfel et al., 2004). Calcium transporter-related regulatory genes positively and negatively regulate the intracellular Ca2+ levels to participate in plant defense responses (Bi et al., 2021; Liu et al., 2021). Plant Ca2+ gradients, which are found at millimolar levels in the vacuole and molar levels in the cytoplasm, are regulated in part by high-capacity vacuolar cation/H(+) exchangers (Manohar et al., 2011). Therefore, Ca2+ channels are crucial in maintaining Ca2+ levels and regulating plant immune responses (Thor et al., 2020). Cation/proton exchanger 3 (CAX3), a calcium transporter-related regulatory gene, can regulate the Ca2+ levels (Hocking et al., 2017; Li et al., 2021). The mechanism through which flagellin regulates Gossypium hirsutum cation/proton exchanger 3 (GhCAX3) in cotton to induce the plant immune response has not yet been reported. Verticillium wilt caused by Verticillium dahliae (VD) is the most devastating disease of cotton worldwide. No effective measure for controlling Verticillium wilt in upland cotton has been found. This study provides the first demonstration of the effects of flagellin C (FLiC) and the mechanism through which FLiC induces an immune response in upland cotton to increase resistance against VD. In this study, FLiC was cloned from an upland cotton endophytic bacterium (Pseudomonas aeruginosa) and purified in vitro, and the effects of FLiC and the mechanism through which FLiC regulates GhCAX3 to enhance cotton immunity and increase resistance to VD were explored.
Materials and methods
Microbial strains
VD V1070 was generously provided by the Institute of Plant Protection, Jiansu Academy of Agriculture Sciences. Hygromycin B-resistant GFP-labeled VD was maintained on potato dextrose agar (PDA) culture medium at 25°C (Hu, 2012).
Plant materials, culture conditions, and treatments
Seeds of upland cotton (Gossypium hirsutum) cultivars Zhongzhimian 2 and Jimian 11 and Nicotiana benthamiana were sown in soil, and the seedlings were grown in a greenhouse under an 8-h dark/16-h light photoperiod at temperatures of 23°C (dark)/28°C (light) with 60% relative humidity. Seeds of Arabidopsis thaliana were sterilized in 75% ethanol and 3% NaClO for 30 min and then washed three times with sterilized water. The Arabidopsis seeds were sown on 1/2-strength Murashige and Skoog (MS) medium containing 3% sucrose and 0.8% (w/v) agar for 2 weeks. Once they reached the three-leaf stage, the Arabidopsis seedlings were transplanted into nutrient-enriched soil and cultivated in a growth chamber at 22/20°C (night/day) with 60% relative humidity and a 16-h day/8-h night photoperiod. The roots of cotton and Arabidopsis thaliana plants were inoculated via irrigation with spore suspensions adjusted to a concentration of 1 × 107 spores/ml (Feng et al., 2021).
Prokaryotic expression and purification of the FLiC protein
A PGEX-4 T-2 expression vector purchased from Beijing Kinco Xinye Biotechnology Co., Ltd., was used, and a pair of specific primers (FLiC-Pgex-4 T-2F/R) were designed based on the FLiC gene sequence, which is shown in Supplementary Table S2. The recombinant expression plasmid FLiC-PGEX-4 T-2 (verified by PCR amplification, restriction enzyme digestion, and sequencing) was transformed into competent Escherichia coli BL21 (DE3) cells for prokaryotic expression. A single colony of the positive strain was selected, inoculated into 5 ml of fresh LB liquid medium supplemented with 50 mg/l ampicillin, and cultured with shaking at 37°C until the OD600 reached 0.6–0.8. Isopropyl-β-D thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, and the culture solution was shaken at 150 rpm for 5 h at 28°C. FLiC protein was purified using a Beyotime GST-tagged Protein Purification Kit (Shanghai, China). FLiC protein was detected by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE; 12.5%), and liquid that was not inoculated with IPTG was used as a control.
FLiC protein HR assays
Tobacco (Nicotiana benthamiana) leaves were used as the experimental materials; 50 μl of FLiC protein solution (100 μg/ml) was injected into the mesophyll from the abaxial side of the leaves, and H2O was used as a control (Felix et al., 1999; Gómez-Gómez and Boller, 2000). Each experiment was repeated three times.
Plant disease resistance assays
Four-leaf stage seedlings of uniformly growing disease-resistant upland cotton varieties (Zhongzhimian 2) and susceptible varieties (Jimian 11) were selected, and FLiC at a concentration of 100 μg/ml was uniformly applied to the upland cotton leaves; H2O was used as a control. Two days after being sprayed with the FLiC solution, the seedlings were inoculated with a pathogen spore suspension (2 × 107 CFU/ml) using the following method: the upland cotton seedlings were removed, the roots of the experimental plants were soaked in the pathogen spore suspension for 20 min, and the control roots was soaked with H2O. After 15 days, the roots, stems and leaves of the upland cotton plants infected with VD were observed under a microscope (MVX10 MacroView, OLYMPUS, Japan). After 30 days, the disease index of Verticillium wilt was investigated. The disease severity of the seedlings was scored at 30 dpi according to the following criteria: 0 = healthy with no symptoms on the leaves; 1 = one or two cotyledons showing symptoms; 2 = a single true leaf showing symptoms; 3 = more than two leaves showing symptoms; and 4 = plant death. The overall disease index and control efficacy were calculated as follows (Supplementary Figure S1; Liu et al., 2014):
where n0–n4 are the numbers of plants with each of the corresponding disease scores and n is the total number of plants assessed. All experiments were repeated three times, and more than 80 plants were counted each time.
Lignin and callose detection
Forty-eight hours after the cotton stems were sprayed with the FLiC solution, the stems of each group were randomly selected to detect the content of lignin according to the method descried by Xiong et al. (2021). The total lignin content of stem tissue was determined by the acetyl bromide method. Stem samples (100 mg) were rapidly ground into powder in liquid nitrogen, and then washed sequentially with ethanol, 95% ethanol, and deionized water to remove soluble substances. After lyophilization, add 5 ml of acetyl bromide-glacial acetic acid (1:3, vol/vol) to each sample and incubate at 70°C for 30 min. Then 0.9 ml of 2 M NaOH and 3 ml of glacial acetic acid were added, and finally 1 ml of 7.5 M hydroxylamine hydrochloride solution was added to stop the reaction. After centrifugation at 13,000 rpm, the supernatant was collected and the absorbance at 280 nm was measured with a microplate reader SpectraMax iD5. Alkali lignin was used as the standard curve (y = 0.969x + 0.009, R2 = 0.9978). The method described by Millet et al. (2010) was used for callose detection. The amount of callose was quantified using ImageJ software (version 1.48). Three independent biological and technical repeats were performed.
VD biomass detection
Two cotyledons were removed from the cotton seedlings of the treatment groups in a sterile environment, weighed and added to 2 ml of sterile water for grinding. After ground into a homogenous mixture, the mixture was diluted according to the gradient dilution method. One hundred microliters of the diluent at different concentrations was spread on red Bengal-resistant medium supplemented with 50 μg/ml hygromycin and 50 μg/ml streptomycin and then placed in an incubator with a constant temperature of 28°C and cultured upside down for 2 days. Afterward, the number of colonies in each Petri dish was counted, and the content of VD per gram of leaf was calculated (Wang, 2014). Three independent biological and technical repeats were performed.
Measurement of chitinase, glucanase, phenylalanine ammonia lyase, polyphenol oxidase, peroxidase, and catalase activities
Chitinase activity was measured using the method described by Emani et al. (2003), and GLU activity was measured using the procedure descried by Abeles and Forrence (1970). PAL activity was measured using the method described by Dickerson et al. (1984), and PPO activity was measured as described by Dickerson et al. (1984). POD activity was measured using the method described by Dong et al. (2003), and CAT activity was measured as described by Plazek and Zur (2003). Three independent biological and technical repeats were performed.
Measurement of resistance-related gene expression by quantitative reverse transcription PCR
Cotton cotyledons were collected, and quantitative reverse transcription PCR (qRT–PCR) analysis was conducted at different time points. RNA extraction and gene expression analysis were performed as previously described (Ren et al., 2013). Each real-time assay was tested via a dissociation protocol to ensure that each amplicon comprised a single product. Sequences of upland cotton defense-related gene primers were used for qRT–PCR (Supplementary Table S1; Han, 2014). The relative expression of genes was calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). Technical replicates of three independent biological samples were performed.
Ca2+, H2O2, and NO detection and quantification
Twelve hours after being sprayed with FLiC protein solution, leaves of each group of plants were randomly selected, and the Ca2+ fluorescence was measured using the method described by Chen et al. (2004). A Calcium Colorimetric Assay Kit (Beyotime Biotechnology Company, Shanghai, China) was used to quantify the Ca2+ concentration. The images were analyzed using Leica Image software. Twelve hours after being sprayed with FLiC protein solution, leaves of each group of plants were randomly selected and stained with Diaminobenzidine (DAB) staining solution. The H2O2 level was measured as described by Kumar et al. (2009). Cotton leaves were washed with distilled water, placed in a Petri dish, mixed with an appropriate amount of DAB (1 mg/ml), and stained under light at 25°C for 8 h. After the staining solution was removed, 95% ethanol solution was added to remove the chlorophyll in the leaves. After boiling for 20 min, the green leaves were removed. The H2O2 deposition in leaves was observed after suspension in clear water. A Hydrogen Peroxide Assay Kit (Beyotime Biotechnology Company, Shanghai, China) was used to quantify the H2O2 concentration, and the NO concentration was measured using the method described by Sun et al. (2012). All the images were visualized by confocal laser scanning microscopy (CLSM; excitation 488 nm; emission 515 nm) and analyzed using Leica Image software. Three independent biological and technical repeats were performed.
RNA sequencing
Two-leaf stage Zhongzhimian 2 plants were used as the materials for transcriptome sequencing. Leaves sprayed with flagellin FLiC were used as the treatment group, and leaves sprayed with water served as the control group. Three replicates of the treatment group and the control group were included, resulting in a total of six transcriptome sequencing samples. Cotton total RNA was extracted with the E.Z.N.A. TM Plant RNA Mini Kit (WEGENE, Shanghai, China). The mRNA with a polyA structure in total RNA was enriched by Oligo(dT) magnetic beads, and the RNA was broken into fragments using the ion breaking method. First-strand cDNA was synthesized using RNA as the template, six-base random primers and reverse transcriptase, and second-strand cDNA was synthesized using first-strand cDNA as the template. After construction of the library, the library fragments were enriched by PCR amplification. The quality of the library was checked using the Agilent 2100 Bioanalyzer, and the total and effective concentrations of the library were then detected. Libraries containing different index sequences were then mixed in proportions based on the effective concentration of the library and the amount of data needed for the library. The pooled libraries were uniformly diluted to 2 nM, and single-stranded libraries were formed by alkali denaturation. After RNA extraction, purification, and library construction, the libraries were paired-end (PE) sequenced based on the Illumina sequencing platform. Based on the RNA sequencing (RNA-seq) results, 12 genes were randomly selected for the design of gene-specific primers using information within the NCBI database and for verification by qRT–PCR (Supplementary Table S2).
GO and KEGG pathway enrichment
Based on the transcriptome screening data, genes that met the following conditions were considered to be differentially expressed: log2|fold change| ≥ 1 and a value of p ≤ 0.05. Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were considered to be significantly enriched in the differentially expressed genes (DEGs) if the p values were <0.05. GO enrichment analysis maps the differentially expressed genes to each term in the database, the number of DEGs in each term is counted, and the GO terms with the same genetic background as the sample DEGs are selected. The DEGs were functionally classified according to the GO annotation results. The enrichment analysis was performed using the phyper function in R software. Biological pathway classification of DEGs was performed based on the KEGG annotation results. Significant enrichment analysis was performed to analyze the physiological pathways and signal transduction pathways involving the DEGs.
Virus-induced gene silencing
The fragments containing part of the coding sequence of GhCAX3 were amplified from the cDNA of Zhongzhimian 2 using the corresponding primer pair GhCAX3-VIGS-F/R and then cloned into a TRV:00 plasmid at the XbaI BamHI sites via T4 DNA Ligase (TaKaRa). The TRV vectors were then transformed into Agrobacterium tumefaciens GV3101. The transformed TRV vectors were agroinfiltrated into two fully expanded cotyledons of Zhongzhimian 2 cotton seedlings. Afterward, the transformed seedlings were transferred to a growth chamber at 25°C with a 16-h light/8-h dark photoperiod. At least 60 plants were inoculated with each construct. A TRV:CLA construct served as a positive marker for evaluating the VIGS efficiency. Fifteen days after inoculation, the expression levels of the target gene were examined, and the successfully silenced plants were subjected to infection with VD V1070. The primers used for this experiment are listed in Supplementary Table S2.
Arabidopsis transformation
The open reading frame (ORF) of FLiC was cloned and inserted into a pBI121 (Cambia) plant binary vector together with the 35S promoter. The resulting pBI121-FLiC plasmid was introduced into Agrobacterium tumefaciens strain GV3101. The primers (pBI121-FLiC F/R) used are listed in Supplementary Table S2. The transformants (T0, T1, and T2 seeds) were screened for survival on half-strength MS medium supplemented with 50 mg/L kanamycin. T3 transgenic lines were identified by RT–PCR.
Statistical analysis
All the experiments used to obtain the data described in this paper were repeated at least three times. The resulting data were subjected to ANOVA using SPSS 19.0 software (SPSS, Inc.). Student’s t-test and one-way ANOVA (p < 0.05) followed by Duncan tests were used for multiple comparisons.
Results
Structure and HR effects of FLiC
The FLiC gene was cloned from an endophytic bacterium (Pseudomonas aeruginosa) isolated from the roots of the upland cotton cultivar Zhongmiansuo 41. The FLiC protein is a type of flagellin with two functional domains (Figure 1A); its three-dimensional structure is shown in Figure 1B. FLiC exhibits 100% homology with Pseudomonas aeruginosa PA103 (Figure 1C). The recombinant plasmid was transformed into Escherichia coli strain BL21 (DE3) and induced by IPTG (0.5 mM) at 28°C for 5 h. The purified FLiC protein was 66 kDa (Figures 1D,E) and induced HR effects and an ROS burst in tobacco leaves (Figures 1F,G).
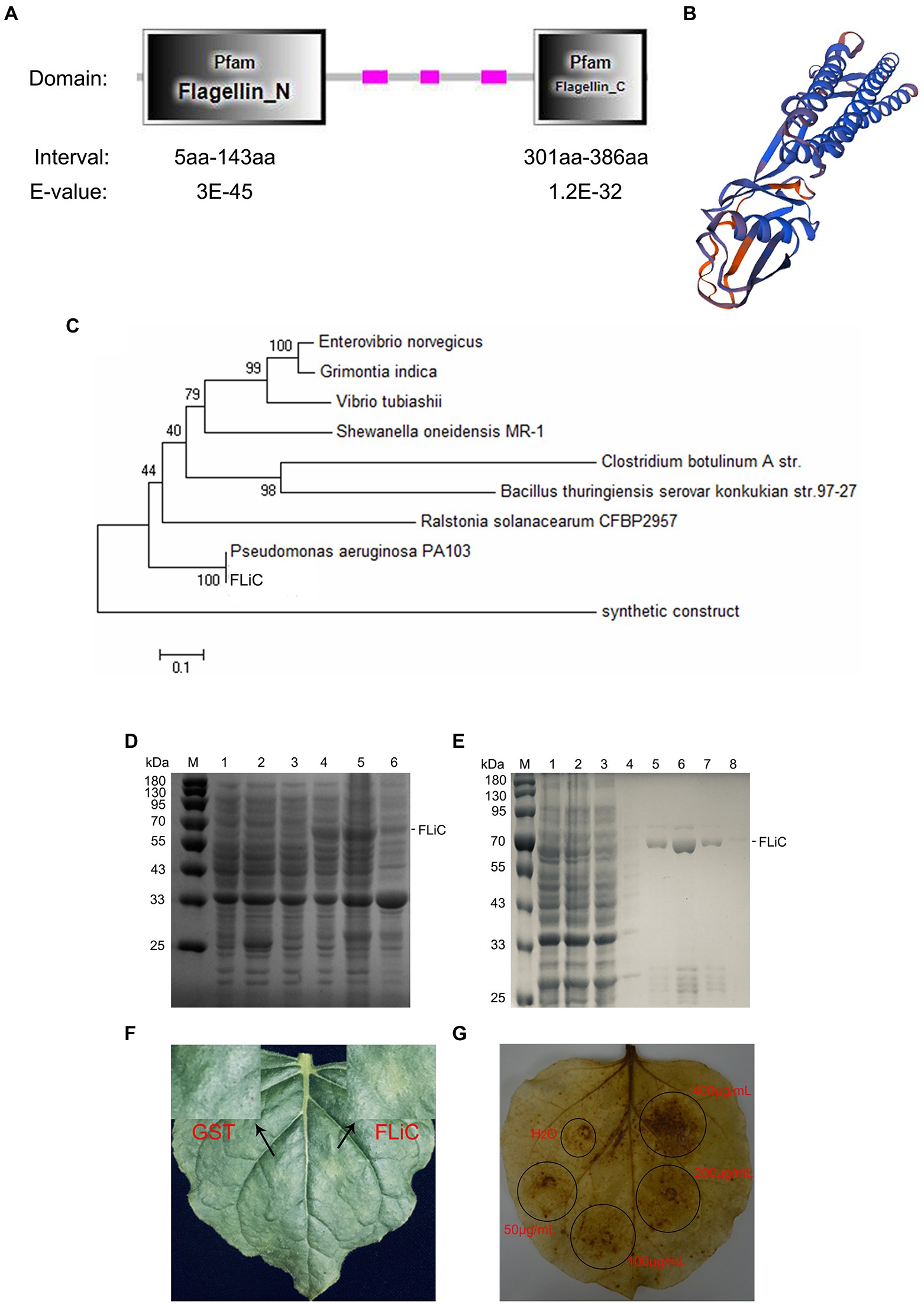
Figure 1. Structure and hypersensitive response (HR) of flagellin C (FLiC). (A) Two functional domains of FLiC protein. (B) Three-dimensional structure diagram of FLiC protein. (C) Evolutionary tree of FLiC proteins. (D) Expression of FLiC recombinant protein in Escherichia coli at 28°C [M protein marker; (1) control containing an empty vector; (2) control containing an empty vector with addition of an inducer (0.5 mM IPTG); (3) whole bacteria with addition of an inducer (0 mM IPTG); (4) whole bacteria with addition of an inducer (0.5 mM IPTG); (5) supernatant after the bacterial cells were sheared; (6) pellet after the bacterial cells were sheared]. (E) Purified FLiC protein (M protein marker; 1, cell lysate; 2, flow-through; 3, wash 1; 4, wash 2; 5, elution 1; 6, elution 2; 7, elution 3; and 8, elution 4). (F) HR of tobacco treated with FLiC. The concentration of FLiC protein was 100 μg/ml; the photograph shows the backside of tobacco 24 h after injection. (G) Hydrogen peroxide (H2O2) generated after treatment with FLiC protein solutions at different concentrations. Diaminobenzidine (DAB) staining was used to visualize H2O2.
FLiC induces resistance to VD in upland cotton plants
Jimian 11 (a susceptible cultivar) and Zhongzhimian 2 (a resistant cultivar) were sprayed with FLiC protein solution, and the amount of VD infection in the roots, stems, and leaves of these plants were significantly lower than those found in the control group under the microscope (Figures 2B,C), indicating that FLiC can induce resistance to VD infection (Figure 2). Moreover, according to the classification standard of cotton verticillium wilt disease (Supplementary Figure S1), the incidence of infection in Zhongzhimian 2 was lower than that in Jimian 11. Thirty days after FLiC treatment, the disease index of Zhongzhimian 2 (29) was lower than that of Jimian 11 (46.23). The relative biocontrol effects of FLiC on Zhongzhimian 2 and Jimian 11 were 47.50 and 32.42, respectively (Table 1). Therefore, FLiC induces systemic disease resistance in upland cotton, and the induction effect in the resistant cultivars is stronger than that in the susceptible cultivars.
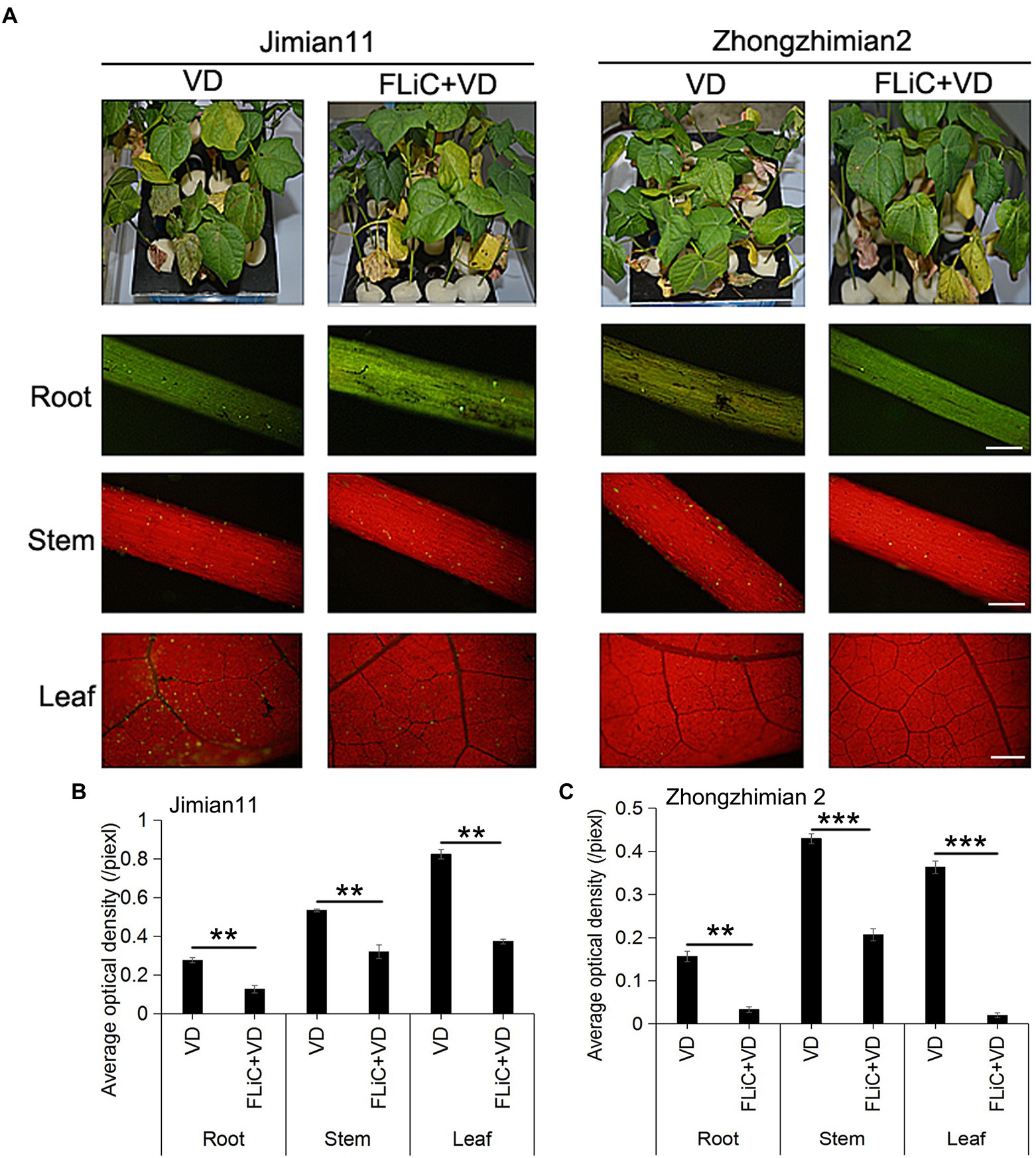
Figure 2. Effect of FLiC on infection with VD in Jimian 11 and Zhongzhimian 2. (A) Differences in phenotype and spore colonization of the roots, stems, and leaves between the disease-resistant variety Zhongzhimian 2 and the susceptible variety Jimian 11. (B) Statistics of the average fluorescence density of spores in the roots, stems, and leaves of Jimian 11. (C) Statistics of the average fluorescence density of spores in the roots, stems, and leaves of Zhongzhimian 2. The data represent the means ± SDs; n = 3 (**p < 0.01; ***p < 0.001; t test).Bars = 500 μm.
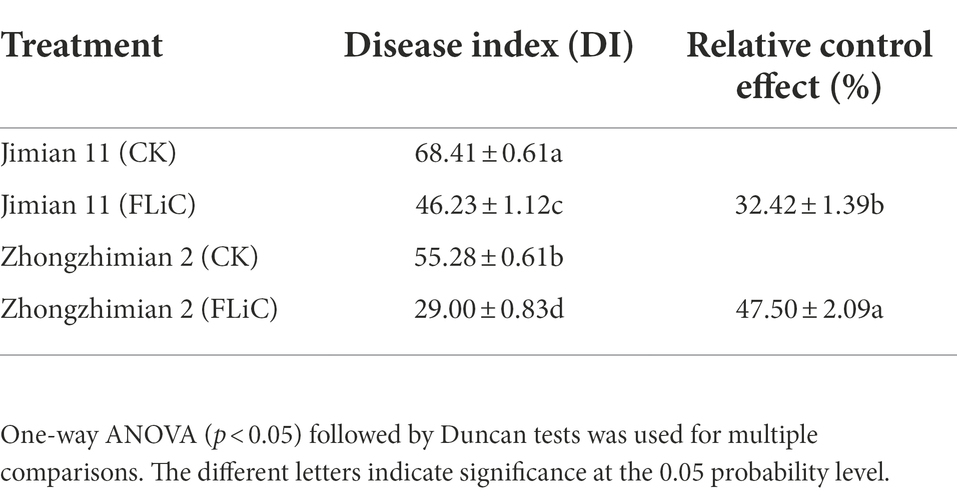
Table 1. FLiC induces VD disease resistance in 30-day seedlings of different upland cotton varieties.
FLiC induces deposition of callose in leaves and lignin in stems
Many studies have identified the accumulation of lignin as a general and basal defense reaction in plant immunity and cotton resistance to VD (Hu et al., 2021). Forty-eight hours after cotton leaves were sprayed with FLiC solution, obvious callose deposits were observed in the leaves of Zhongzhimian 2, and the lignin content in the stems was higher than the control level (Figures 3A–C). To further confirm that the callose and lignin induced by FLiC are involved in enhancing resistance to VD, solutions of 2-deoxy-D-glucose (2-DDG; callose synthesis inhibitor) and α-aminooxyacetic acid-β-phenylpropionic acid (AOPP; Lignin synthesis inhibitor) were sprayed onto cotton leaves, and this spraying decreased the amount of callose and lignin deposition and significantly increased the amount of VD spores on the roots, which decreased the FLiC-induced resistance (Figures 3D,E). Therefore, the results demonstrate that FLiC can induce a strong immune response in cotton and enhance resistance to VD through the production of callose and lignin.
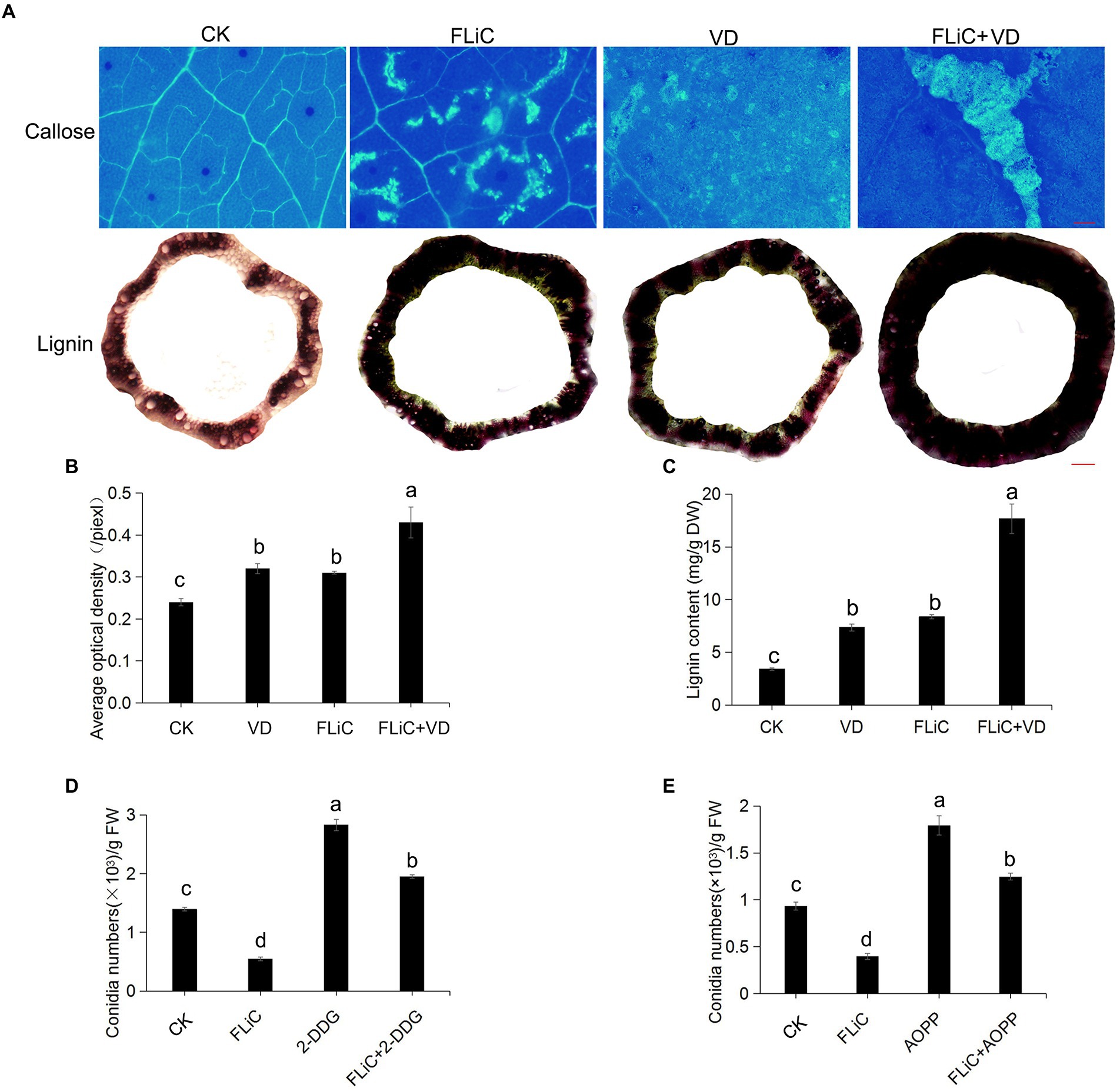
Figure 3. FLiC induces the deposition of lignin and callose in upland cotton. (A) FLiC induced the deposition of leaf callose and stem lignin in cotton (callose bars = 1,000 μm; lignin bars = 2,000 μm). The concentration of FLiC was 100 μg/ml, and the cotton leaves were treated for 48 h. (B) Statistics of the average optical density of callose. (C) Determination of the lignin content. (D) Statistics of the bacterial mass after the clearance of callose with 2-DDG. (E) Statistics of the bacterial mass after the clearance of lignin with AOPP. The data represent the means ± SDs; n = 3. One-way ANOVA (p < 0.05) followed by Duncan’s test was used for multiple comparisons. The different letters indicate significance at the 0.05 probability level.
FLiC-induced resistance depends on changes in defensive enzyme activity
To further verify that FLiC can induce changes in enzyme activity in upland cotton to enhance the defense response, we evaluated the changes in the activities of six enzymes in the leaves of Zhongzhimian 2 sprayed with the FLiC solution only. After the plants were sprayed with FLiC solution, the activities of defense-related enzymes changed to varying degrees (Supplementary Figure S2). Specifically, CAT activity in these plants was significantly higher than that in the control plants on the tenth day, and the activities of PPO, POD, and PAL on the fourth day were significantly higher than those of the control; on the eighth day, the activities were comparable to those of the control; in addition, on the tenth day, all the enzymes except CHI and GLU showed significant increases in activity compared with the activity levels in the control. Moreover, CHI activity showed a significant increase in the treated plants compared with that in the control on the second and fourth days and then began to decrease to the control level. GLU activity was significantly higher on the second day compared with that of the control and then began to decrease. These results showed that the activity of defense-related enzymes exhibits differential changes at different times after inoculation to improve cotton resistance to VD.
FLiC induces the expression of VD resistance-related genes
Twenty-four hours after the seedlings were sprayed with FLiC solution, defense-related genes such as pathogenesis-related proteins 1 (PR1), CHI, GLU, POD4, ERF5, PR10, lipoxygenase (LOX), and CDN1 were induced to varying degrees (Supplementary Figure S2). Six hours after inoculation, the expression levels of the MAPK2, MAPK6, and MAPK7 genes in the roots of seedlings belonging to the treatment group were lower than those in the control roots, and the expression levels of the MAPK16 gene were higher than the control levels. However, 12 and 24 h after inoculation, the expression levels of the MAPK2, MAPK6, MAPK7, and MAPK16 genes were basically the same as those in the control plants. These findings indicated that the MAPK16 gene may be involved in the early immune response induced by FLiC. Six hours after inoculation with VD, the expression levels of the WRKY5 and WRKY6 genes were higher than the control levels, whereas those of WRKY2, WRKY3, and WRKY4 were not. At 12 and 24 h after inoculation, the expression levels of WRKY2, WRKY3, WRKY4, WRKY5, and WRKY6 were not significantly different from the control levels. Thus, the WRKY5 and WRKY6 transcription factors may be involved in the early immune response induced by FLiC (Supplementary Figure S3).
FLiC induces an increase in intracellular Ca2+
To verify that FLiC protein-induced resistance to VD is related to an increase in intracellular Ca2+ content and causes a downstream immune response, epidermal cells were loaded with the Ca2+-sensitive fluorescent probe Fluo-3/AM; afterward, solutions of FLiC protein at a concentration of 100 μg/ml were applied, and the effects were observed by LCSM. The results showed that FLiC proteins can cause a significant increase in the fluorescence intensity of epidermal cells (Figure 4). After the seedlings were sprayed with the Ca2+ chelating agent ethylene glycol-bis(2-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) and a Ca2+ channel blocker (LaCl3), the fluorescence intensity induced by FLiC was significantly reduced. Therefore, FLiC can induce an increase in the Ca2+ concentration in upland cotton epidermal cells.
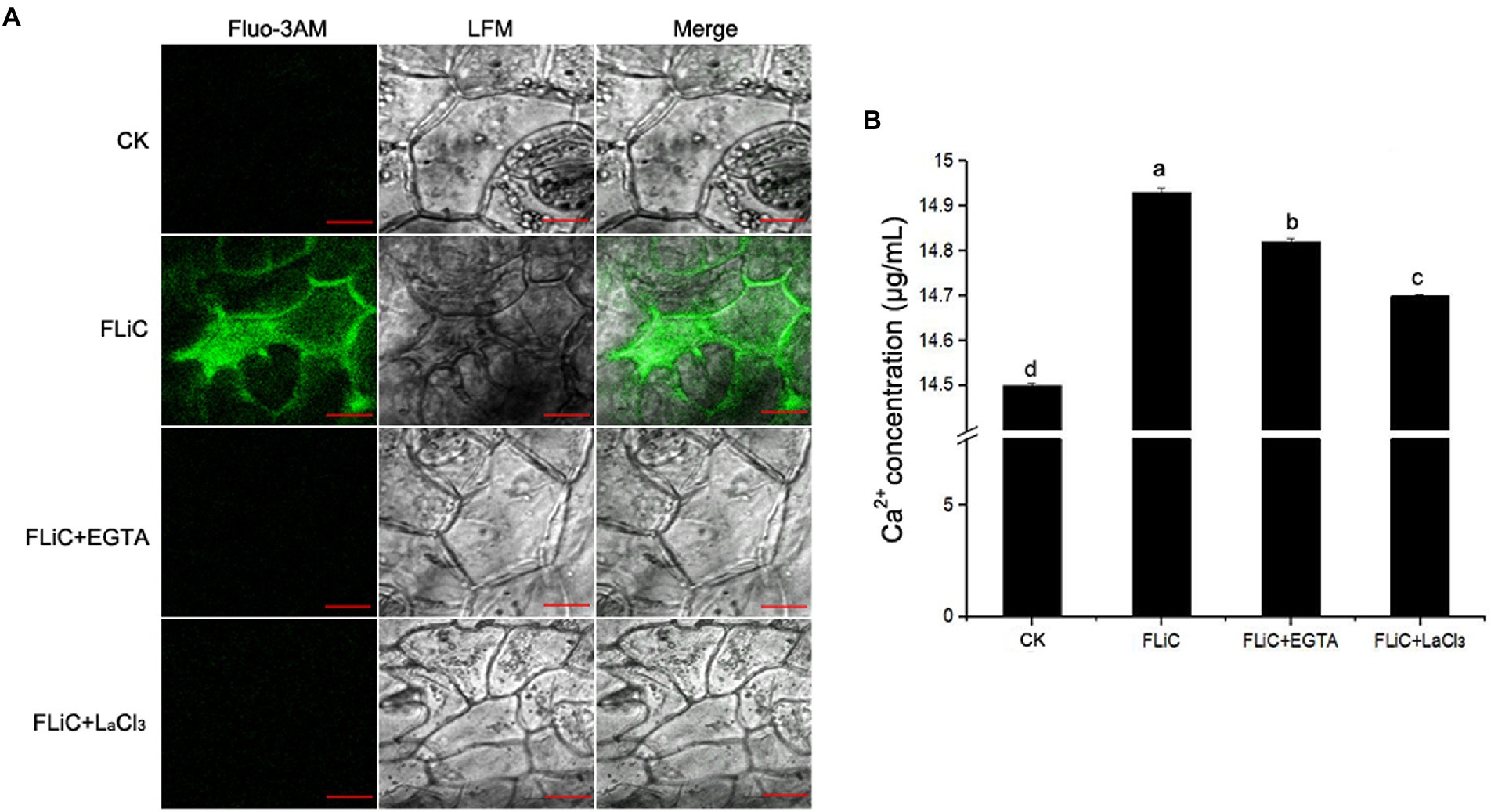
Figure 4. FLiC induces Ca2+ production in upland cotton epidermal cells. (A) FLiC induces increased intracellular Ca2+ content. (B) Intracellular Ca2+ quantification. The data represent the means ± SDs; n = 3. One-way ANOVA (p < 0.05) followed by Duncan’s test was used for multiple comparisons. The different letters indicate significant differences at the 0.05 probability level. Bars = 50 μm.
The regulatory relationship between Ca2+ and H2O2 was clarified
Twelve hours after cotton leaves were sprayed with FLiC solution, an obvious ROS burst was observed in the leaves (Supplementary Figure S4 A). To determine the relationship between Ca2+ and H2O2, the seedlings were sprayed with CAT (After H2O2 is synthesized, it will first accumulate outside the cell membrane, and CAT will remove these formed H2O2 molecules), EGTA (After the Ca2+ chelator EGTA enters plant cells, it will chelate Ca2+ and reduce the intracellular Ca2+ concentration) and LaCl3(the Ca2+ channel inhibitor LaCl3 blocks the influx of extracellular Ca2+ into the cell and reduces the intracellular Ca2+ concentration) solutions, and this spraying significantly decreased the H2O2 content in the cotton leaves to the control level. These results indicated that Ca2+ acts upstream of H2O2 and affects the synthesis of H2O2. Moreover, 5–30 days after the cotton leaves were sprayed with the FLiC solution, the H2O2 content in the leaves of the seedlings continued to be significantly higher than that of the control. Until the 30th day, the H2O2 content began to decrease, but the level was still higher than that of the control (Supplementary Figure S4 C). Therefore, FLiC first induces Ca2+ increase and then affects the contents of intracellular H2O2 such that the immune response improves over a long period of time.
The induction of NO by FLiC improves resistance to VD
Forty-eight hours after inoculation with VD, the number of pathogens colonizing the roots of seedlings sprayed with FLiC solution was significantly lower than that of the control (Supplementary Figure S5 A). To verify that the NO generated after spraying FLiC hinders the colonization of pathogens. The spraying of carboxy-2-phenyl-4,4,5,5-tetramethylimidazoline-3-oxide-1-oxyl (C-PTIO; a scavenger of NO) onto cotton leaves increased the number of colonized pathogens, and the difference between this number and the control was significant (Supplementary Figure S5 D). Five days after the leaves were inoculated with the VD spore solution, lesions were observed on the leaf surface (Supplementary Figure S5 B). The area of damaged leaves in the seedlings sprayed with FLiC solution was significantly lower than that of the control. The spraying of C-PTIO increased the area of damaged leaves in the seedlings of both the FLiC and control groups. These results further indicate that the accumulation of NO induced by FLiC participates in the resistance of upland cotton to VD.
The relationship among ROS, Ca2+, and NO enhance resistance to VD
To further illuminate the relationship among ROS, Ca2+, and NO, strong fluorescence was observed around the epidermal cells and stomata of cotton sprayed with the FLiC solution, which indicated that FLiC can induce the production of NO in cotton epidermal cells of seedlings (Figures 5A,E). The fluorescence intensity of the seedlings was strongest 1 h after being sprayed with 400 μg/ml FLiC solution (Figures 5B,C and D), indicating that the production of NO induced by FLiC depends on the concentration of FLiC and the time after treatment. The spraying of solutions containing the NO scavenger C-PTIO and the NOS pathway inhibitor nitro-L-arginine methyl ester (L-NAME) onto cotton leaves significantly decreased the fluorescence intensity (Figures 5A,E), which indicated that C-PTIO and L-NAME exert an inhibitory effect on FLiC-induced NO production in epidermal cells. The nitrate reductase (NR) pathway inhibitor tungstate had only a weak inhibitory effect on FLiC-induced NO production in epidermal cells. These results indicated that FLiC induces the production of NO in epidermal cells; NO is synthesized mainly through the nitric oxide synthase (NOS) pathway. The spraying of a solution containing CAT, EGTA, and LaCl3 onto cotton leaves significantly decreased the NO content. These results showed that Ca2+ and H2O2 are involved in the production of NO. After the seedlings were sprayed with C-PTIO and L-NAME solutions, FLiC could still induce increased H2O2 production. These results showed that the production of H2O2 occurs independently of the NO content. However, excessive amounts of NO exert an inhibitory effect on FLiC-induced H2O2 production (Supplementary Figure S4 B). The spraying of the seedlings with EGTA and LaCl3 solutions significantly decreased the content of H2O2 (Supplementary Figures S4A,B). Therefore, in the immune response induced by FLiC, Ca2+ induces H2O2 and participates in the production of H2O2; H2O2 subsequently stimulates the production of NO; and excessive NO inhibits the production of H2O2.
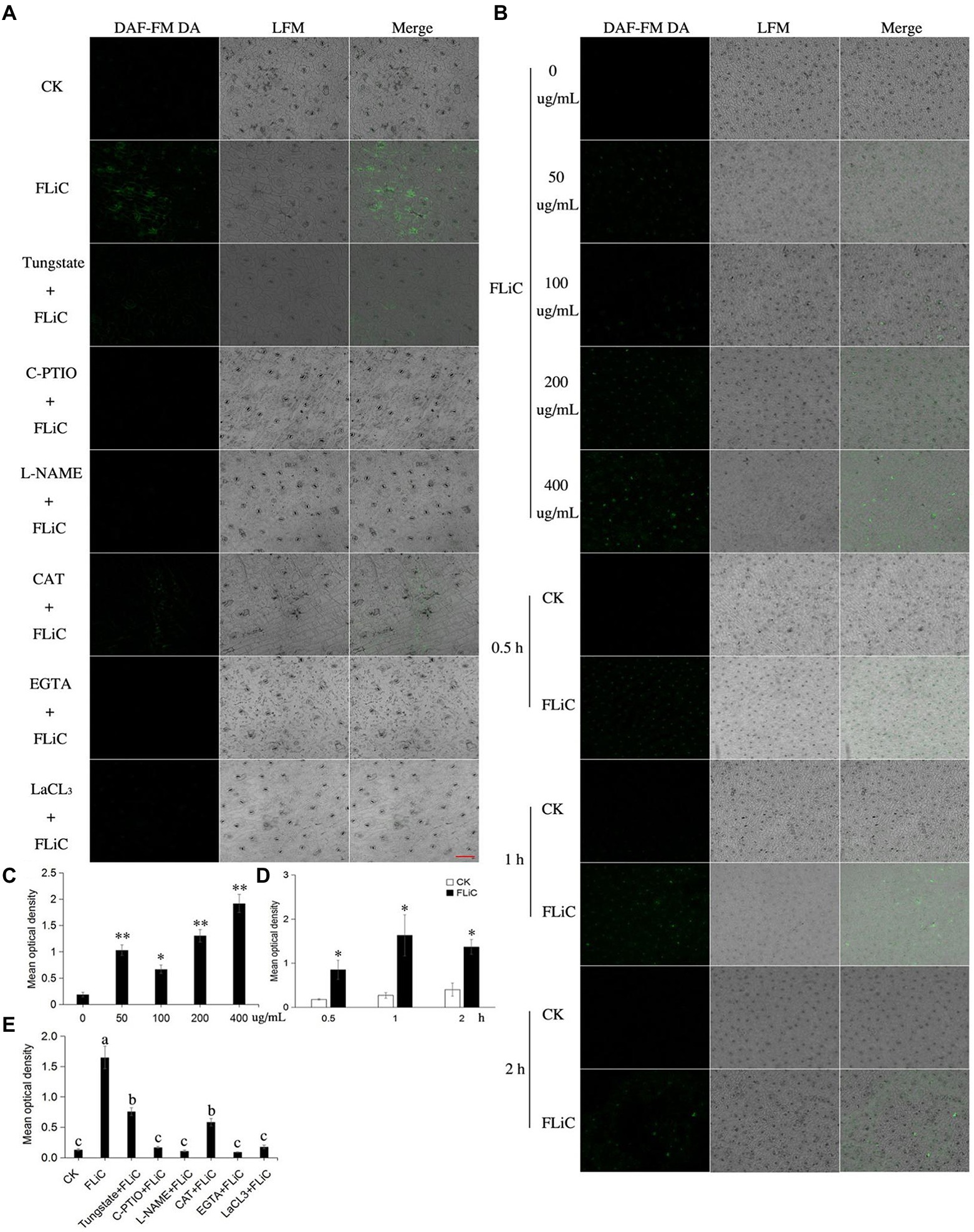
Figure 5. NO production in upland cotton leaves in response to FLiC treatment. (A) Fluorescence of upland cotton epidermal cells in response to different treatments (200 μM C-PTIO, 200 μM L-NAME, 100 μM tungstate, 100 unit/ml CAT, 5 mM EGTA, and 200 μM LaCl3). (B) Fluorescence of upland cotton epidermal cells at different time points after treatment with solutions of different protein concentrations. (C) Average fluorescence density of NO in upland cotton epidermal cells treated with solutions of different protein concentrations. (D) Average fluorescence density of NO in upland cotton epidermal cells at different time points after treatment. (E) Average fluorescence density of NO in upland cotton epidermal cells after different treatments. The data represent the means ± SDs; n = 3 (*p < 0.05; **p < 0.01; t test). One-way ANOVA (p < 0.05) followed by Duncan’s test was used for multiple comparisons. The different letters indicate that the difference between the treatments is significant at the 0.05 probability level. Bars = 20 μm.
Significant metabolic pathways enriched in transcriptome results
To analyze which metabolic pathways were associated with the transcriptome results, we performed GO and KEGG enrichment analyses. The three most significant GO enrichment pathways were metal ion transmembrane transporter activity, cellular potassium ion transport, and potassium ion transmembrane transport. The differentially expressed genes were enriched in the following pathways (in descending order): oxidoreductase activity (22 DEGs), metal ion transport (seven DEGs), metal ion transmembrane transporter activity (seven DEGs), and monovalent inorganic cation transmembrane transporter activity (six DEGs). In addition, significantly enriched to calcium antiporter activity. The three most significant pathways identified in the KEGG enrichment analysis were glycolysis/gluconeogenesis, carbon fixation in photosynthetic organisms, and diterpenoid biosynthesis. The genes were enriched in the following pathways (in descending order): glycolysis/gluconeogenesis (five DEGs), phenylpropanoid biosynthesis (four DEGs), and carbon fixation in photosynthetic organisms (three DEGs). These results suggest that the resistance induced by FLiC in cotton is related to calcium transporter activity.
The cation/proton exchangers gene family negatively regulates cotton plant resistance to VD
The above-described studies confirmed that FLiC induces the immune response in cotton due to increased intracellular Ca2+ content, and the coordinated regulation of Ca2+, H2O2, and NO enhances the resistance to VD. To clarify which metabolic pathways regulate intracellular Ca2+ content, we analyzed the transcriptome results. The RNA-seq results showed that 87 genes were upregulated and 90 genes were downregulated. Twelve genes were randomly selected from the RNA-seq results to verify the transcriptome results by qRT–PCR. The results showed that the expression trends of 11 of the 12 genes were the same as those identified from the transcriptome data, with an accuracy rate of 91.67%. This finding shows that the transcriptome results are credible (Supplementary Figure S6). The significantly enriched DEGs were classified and clustered based on the results (Figures 6A,B). The DEGs were significantly enriched in related disease-resistant metabolic pathways, such as those involving calcium antiporter activity, potassium ions, diterpenoid biosynthesis, and phenylpropanoid biosynthesis (Figures 6C,D). Further analysis revealed that calcium antiporter activity is regulated by the cation/proton exchangers gene family. The expression level of this gene family in the plants sprayed with FLiC solution was lower than the control level. Therefore, this gene family may negatively regulate cotton plant resistance to VD.
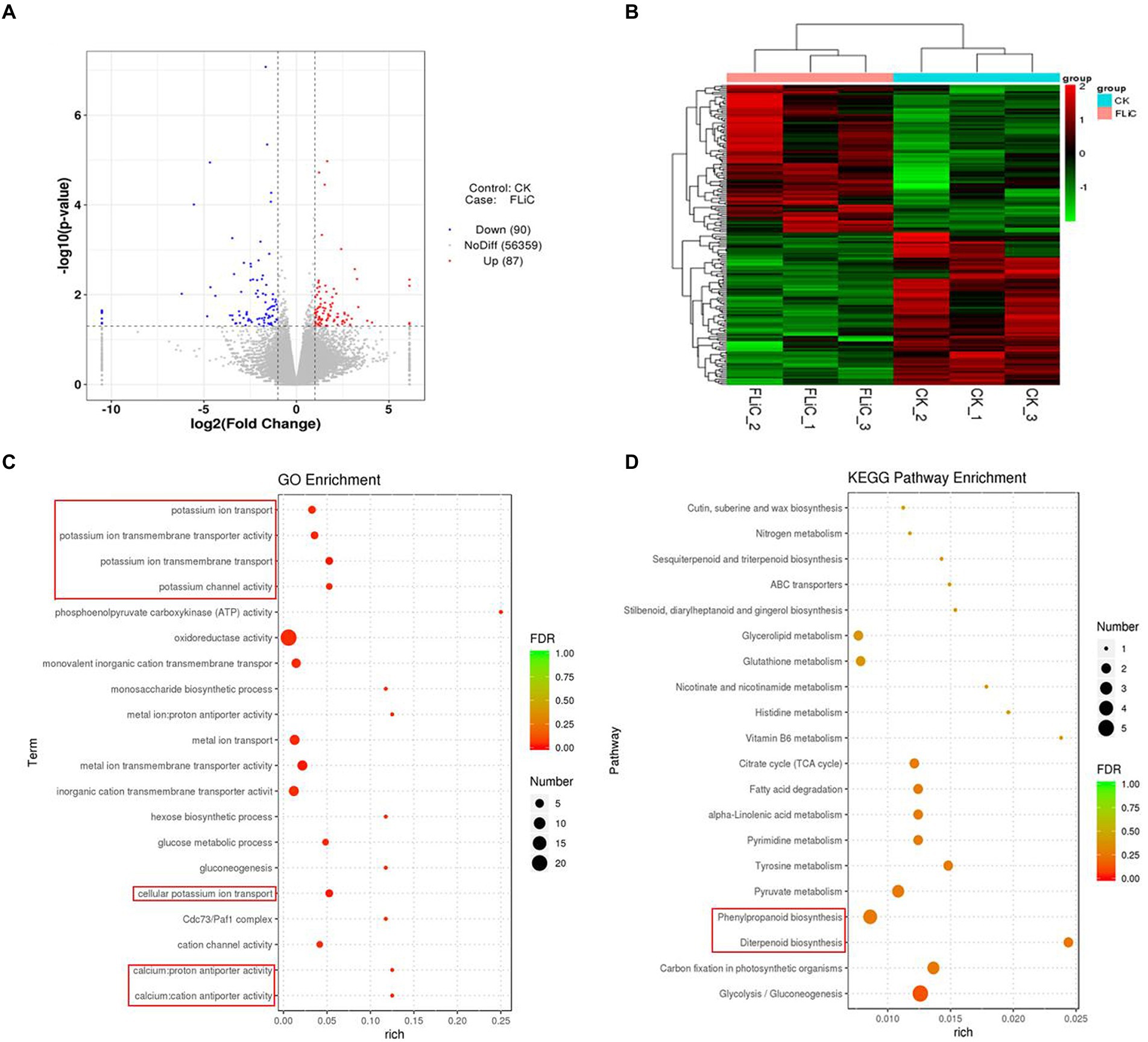
Figure 6. Pathways in which the DEGs are significantly enriched according to the transcriptomic data. (A) Volcano plot of DEGs. (B) DEG cluster analysis. (C) Bubble chart of the top 20 metabolic pathways enriched in GO terms. (D) Bubble chart of the top 20 metabolic pathways enriched in KEGG pathways.
The immune response is enhanced after GhCAX3 silencing
Pathogen induced Ca2+ signaling and homeostasis is mediated by two factors, the Ca2+ influx channels and the Ca2+ efflux pumps/exchangers (Ca2+-ATPase or CAX type Ca2+/H+ exchangers). To verify that the cation/proton exchanger 3 (GhCAX3; CAX type Ca2+/H+ exchangers) gene negatively regulates cotton resistance to VD, the GhCAX3 gene was silenced. After true leaves of the positive control became albino, the silencing efficiency of GhCAX3 was determined to have reached 62% (Figures 7A,B). Twenty-five days after inoculation with VD, the disease index was determined, and the TRV2::GhCAX3 group exhibited a significant lower index than the control TRV2::00 group (Figures 7I,J). After the GhCAX3 gene was silenced, the intracellular Ca2+ and NO contents in the epidermis of leaves of plants sprayed with FLiC solution were significantly higher than those of wild-type (WT) plants and plants transformed with an empty vector (TRV2::00; Figures 7C,D). The levels of ROS, callose and lignin om the sprayed plants were significantly higher than those in WT and TRV2::00-transformed plants (Figures 7E–H). Obviously, silencing of GhCAX3 through VIGS decreases the Ca2+ extrusion from the cytosol and thus causing the enhanced Ca2+ concentration as well as Ca2+-dependent ROS and NO signaling. Therefore, FLiC negatively regulates the expression of the GhCAX3 gene to enhance the resistance of seedlings to VD.
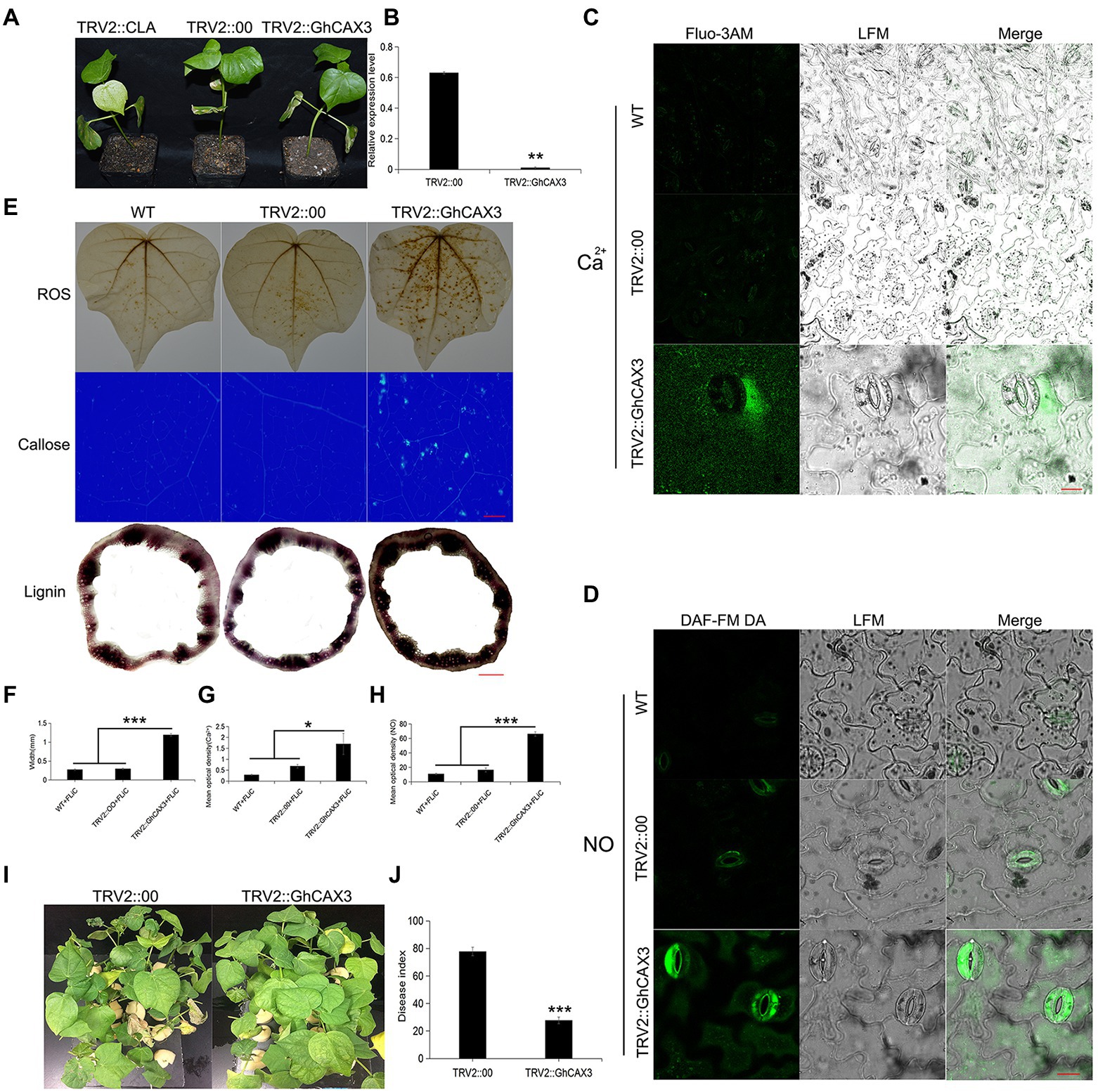
Figure 7. Anti-disease substances increase after the silencing of GhCAX3. (A) Imaging of plants transformed with TRV2::GhCAX3 at 10 days after gene silencing; TRV2::CLA served as the positive control, and TRV2::00 was the negative control. (B) GhCAX3 gene silencing efficiency. (C) Intracellular Ca2+ content of cotton after GhCAX3 gene silencing. (D) Intracellular NO content of cotton after GhCAX3 gene silencing. (E) ROS, callose, and lignin contents after GhCAX3 gene silencing. (F) Determination of the lignin content. (G) Quantification of intracellular Ca2+ in cotton after GhCAX3 gene silencing. (H) Intracellular NO quantification in cotton after GhCAX3 gene silencing. (I) Detection of disease resistance after GhCAX3 gene silencing. (J) Investigation of disease index after GhCAX3 gene silencing. The data represent the means ± SDs; n = 3 (*p < 0.05; **p < 0.01; ***p < 0.001; t test). Bars (C,D) = 50 μm. Bars (E) = 2000 μm.
Transgenic Arabidopsis overexpressing the FLiC gene exhibits increased resistance to VD and increased expression of VD resistance genes
Arabidopsis plants overexpressing the FLiC gene exhibited enhanced disease resistance to VD compared with WT Arabidopsis plants (Supplementary Figure S7 A). The expression of the FLiC gene was significantly increased 24 h after inoculation (Supplementary Figure S7 B). Therefore, overexpression of the FLiC gene in Arabidopsis might improve resistance to VD. To verify that Arabidopsis plants overexpressing the FLiC gene exhibit enhanced resistance to VD, the expression levels of LOX, PAL, PR1, and vegetative storage protein (VSP) genes were measured, and the results showed that the expression of the four disease resistance genes were significantly increased. Therefore, overexpression of the FLiC gene in Arabidopsis can effectively increase the expression of disease resistance genes related to the salicylic acid (SA) and jasmonic acid (JA) signaling pathways and enhance resistance to VD. The expression level of the glutathione peroxidase (GPX7) gene was similar to or lower than that of the control; POD expression more obvious in the FLiC-overexpressing plants exposed to VD than in the control plants exposed to VD; and the glutathione S-transferase (GSTU3) gene showed significantly increased expression (Supplementary Figure S7). Therefore, in transformed Arabidopsis plants, the FLiC gene can effectively upregulate the expression of ROS and NO signal-related genes in roots and can enhance resistance to VD.
Discussion
Ca2+, NO, and H2O2 synergistically enhance upland cotton resistance to VD
The relationships among Ca2+, NO, and H2O2 in plants and the regulatory mechanisms through which plant disease resistance can improve remain unknown. We found that NO participates in the signal transduction of plants in response to various biotic and abiotic stresses (Yan et al., 2007; Sang et al., 2008; Martínez-Medina et al., 2019). Induction with the elicitor Ca2+ increases the production of NO in tobacco and grape, and NO can in turn cause an increase in the intracellular Ca2+ concentrations (Lamotte et al., 2004; Vandelle et al., 2006; Besson-Bard et al., 2008). These investigations indicate an interaction between Ca2+ and NO signals. An increase in the Ca2+ concentration in mesophyll cells was found to be needed for the cells to produce ROS after pathogen infection (Qiao et al., 2015). Exogenous H2O2 application to Arabidopsis thaliana root epidermis results in dose-dependent transient increases in the net Ca2+ influx (Demidchik et al., 2007). These results indicate an interaction between Ca2+ and H2O2 signals. The synergistic effect of NO and H2O2 triggers the death of hypersensitive cells, but the absence of either one of these factors prevents the induction of cell death (Tada et al., 2004). By affecting the activity of NO synthase, H2O2 can induce the synthesis and accumulation of NO (Zhang et al., 2007). Similarly, NO can regulate the H2O2 levels, and H2O2 is involved in mediating the NO-induced resistance of tomato to Rhizopus nigricans (Fan et al., 2008). These results showed that transient changes in the contents of NO and H2O2 can activate a series of physiological responses in plants and that these compounds can interact to regulate the same or related signaling pathways to enhance a certain response. Ca2+ and H2O2 act upstream of NO production to induce the death of hypersensitive cells (Qiao et al., 2015). Our results showed that Ca2+ and H2O2 also act upstream of NO. However, whether the content of NO negatively alters the content of H2O2 is unclear. Our results suggest that FLiC binds to membrane receptors and negatively regulates GhCAX3 to increase the intracellular content of Ca2+ and induce the production of H2O2 and NO; that H2O2 induces the production of NO, whereas NO inhibits the synthesis of H2O2; and that H2O2 and NO can induce the production of defense response-related compounds. These findings show that Ca2+, NO, and H2O2 synergistically regulate the resistance of upland cotton plants to VD, which is induced by FLiC (Figure 8). Our results provide the first demonstration that FLiC negatively regulates GhCAX3 to enhance the resistance of upland cotton to VD.
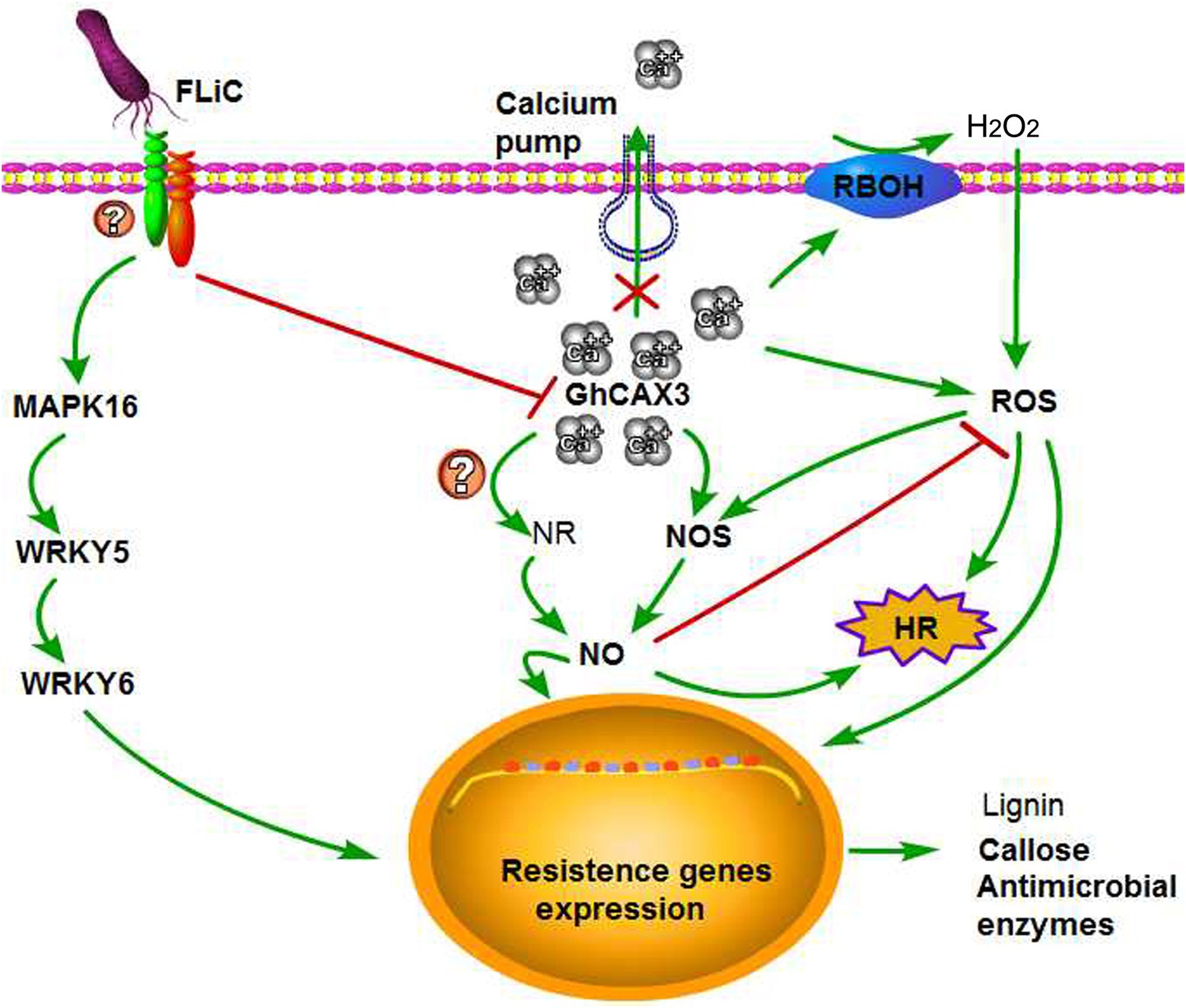
Figure 8. Possible explanation of the FLiC-induced resistance signaling pathway. FLiC increases the intracellular calcium ion content by inhibiting the expression of the calcium ion pump-regulating gene GhCAX3, which in turn induces increases in the H2O2 and NO contents and thereby improves the resistance of cotton to Verticillium wilt.
Metabolic pathways through which FLiC protein induces an immune response
The mechanisms through which full-length FLiC protein affects the plant immune response have not yet been reported. We cloned a newly identified flagellin gene, FLiC, which encodes the same amino acid sequence as Flg22. In cotton, FLiC can induce immune responses, such as MAPK cascade reactions, and upregulation of key genes involved in disease resistance pathways, such as those involving SA, JA, and ETH (Supplementary Figure S4). According to a transcriptome analysis, the DEGs identified after application of the protein to cotton (by spraying of solutions) were significantly enriched in pathways involving potassium ions, Ca2+, diterpenoid synthesis, phenylpropanoid biosynthesis, lignin biosynthesis, nitrogen metabolism, and other disease resistance-related metabolic pathways. Therefore, the disease resistance induced by FLiC protein in cotton is related to the activation of these pathways. Moreover, the CAX3 gene has not been found to regulate the Ca2+ levels or participate in the disease resistance of cotton. In this study, we found that GhCAX3 negatively regulated the Ca2+ levels and thus hampered the VD resistance of cotton plants sprayed with FLiC solution. Plant Ca2+ signals are involved in a wide array of intracellular signaling pathways following pathogen invasion (Kanchiswamy et al., 2013; Aldon et al., 2018; Wang et al., 2019). Ca2+-dependent protein kinases have been predicted to mediate signaling following Ca2+ influx after pathogen infection (Kanchiswamy et al., 2013; Ma et al., 2013; Bredow and Monaghan, 2019). The pathogen-associated molecular patterns involving bacterial flagellin are the most intensively studied PAMPs (Wang, 2012). After the treatment of Arabidopsis thaliana with Flg22, Ca2+ channels and their involvement in the activation of the stomatal closure mechanism in the process of immune signal transduction indicated the specificity of the Ca2+ influx mechanism in response to different stresses (Thor et al., 2020). Moreover, after treatment with Flg22, Arabidopsis thaliana double-knockout plants (in which the genes encoding the tonoplast-targeted pump aca 4/11 were knocked out) showed higher basal Ca2+ levels and greater Ca2+ signals than WT plants (Hilleary et al., 2020). The HopZ-activated resistance 1 resistosome acts as a calcium-permeable cation channel to trigger plant immunity and cell death (Bi et al., 2021). Taken together, these findings show that calcium transporters can negatively regulate intracellular Ca2+content. However, the GhCAX3 gene has not been previously found to regulate the Ca2+ levels or participate in the disease resistance of cotton. Through transcriptome analysis, we found that GhCAX3 negatively enhanced upland cotton resistance to VD. Many studies have investigated the ability of flagellin to induce plant immunity, but whether flagellin regulates CAX3 to induce plant immunity has not been reported. We cloned a new type of flagellin gene, FLiC, which has the same encoded amino acid sequence as that of Flg22. Whether Flg22 regulates CAX3 to induce plant immunity has not been reported, and how Flg22 regulates the relationship among Ca2+, NO, and H2O2 to improve disease resistance has not yet been elucidated. We found that the silencing of GhCAX3 in cotton increased the intracellular Ca2+ content and thereby induced increases in the contents of H2O2, NO, and disease resistance-related substances after the plants were sprayed with FLiC solution. These results showed that FLiC inhibits the expression of GhCAX3, which increases the intracellular Ca2+ content and induces increases in the H2O2 and NO contents to enhance upland cotton resistance to VD.
FLiC-transformed Arabidopsis exhibits increased resistance to VD
The flagellin gene of Bacillus subtilis has been successfully transferred into rice, which results in increased resistance to rice blast and bacterial streaks (Wang et al., 2014, 2015). The inoculation of Arabidopsis plants overexpressing FLiC with VD significantly increased the relative expression levels of the SA and JA defense signal-related genes LOX, PAL, PR1, and VSP, and the expression levels of both LOX and PR1 increased 40-fold. Moreover, the expression of the GSTU3, GPX7, and POD genes, which are related to the ROS and NO signaling pathways, changed to varying degrees. Therefore, the FLiC transgene in Arabidopsis can induce the expression of key genes involved in the SA, JA, ROS, and NO signaling pathways to improve plant disease resistance. Therefore, our research not only revealed a possible use of a new full-length flagellin gene but also used upland cotton as a research material to explore a new approach for inducing an immune response in plants and provide a new protein that can improve the resistance of upland cotton to VD.
Conclusion
Flagellin C induced HRs in tobacco and immune responses in cotton. FLiC reduces the expression of GhCAX3 to increase the intracellular Ca2+ content and then stimulate the increase of intracellular H2O2 and NO content. The coordinated regulation of Ca2+, H2O2, and NO enhanced the resistance to VD, and transgenic Arabidopsis plants overexpressing FLiC exhibited significant resistance to VD.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: NCBI, PRJNA847674.
Author contributions
CT: conceptualization, supervision, and funding acquisition. HZ, YW, and YZ: data curation. HZ and YW: investigation. HZ and HN: formal analysis and writing–original draft. HZ, YX, and CT: writing–review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Key R&D Program of China (2016YFD0102105) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX20_0582).
Acknowledgments
We thank Delin Hong (College of Agriculture, Nanjing Agricultural University) for kindly assisting with the experiments and for critically reading the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.969506/full#supplementary-material
References
Abeles, F. B., and Forrence, L. E. (1970). Temporal and hormonal control of beta-1, 3-glucanase in Phaseolus vulgaris L. Plant Physiol. 45, 395–400. doi: 10.1104/pp.45.4.395
Aldon, D., Mbengue, M., Mazars, C., and Galaud, J. P. (2018). Calcium Signalling in plant biotic interactions. Int. J. Mol. Sci. 19:665. doi: 10.3390/ijms19030665
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Besson-Bard, A., Courtois, C., Gauthier, A., Dahan, J., Dobrowolska, G., Jeandroz, S., et al. (2008). Nitric oxide in plants: production and cross-talk with Ca2+ signaling. Mol. Plant 1, 218–228. doi: 10.1093/mp/ssm016
Bi, G., Su, M., Li, N., Liang, Y., Dang, S., Xu, J., et al. (2021). The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cells 184, 3528–3541.e12. doi: 10.1016/j.cell.2021.05.003
Bouizgarne, B., El-Maarouf-Bouteau, H., Frankart, C., Reboutier, D., Madiona, K., Pennarun, A. M., et al. (2006). Early physiological responses of Arabidopsis thaliana cells to fusaric acid: toxic and signalling effects. New Phytol. 169, 209–218. doi: 10.1111/j.1469-8137.2005.01561.x
Bredow, M., and Monaghan, J. (2019). Regulation of plant immune signaling by calcium-dependent protein kinases. Mol. Plant-Microbe Interact. 32, 6–19. doi: 10.1094/MPMI-09-18-0267-FI
Buxdorf, K., Rahat, I., Gafni, A., and Levy, M. (2013). The epiphytic fungus Pseudozyma aphidis induces jasmonic acid- and salicylic acid/nonexpressor of PR1-independent local and systemic resistance. Plant Physiol. 161, 2014–2022. doi: 10.1104/pp.112.212969
Chen, Y. L., Huang, R., Xiao, Y. M., Lü, P., Chen, J., and Wang, X. C. (2004). Extracellular calmodulin-induced stomatal closure is mediated by heterotrimeric G protein and H2O2. Plant Physiol. 136, 4096–4103. doi: 10.1104/pp.104.047837
Demidchik, V., Shabala, S. N., and Davies, J. M. (2007). Spatial variation in H2O2 response of Arabidopsis thaliana root epidermal Ca2+ flux and plasma membrane Ca2+ channels. Plant J. 49, 377–386. doi: 10.1111/j.1365-313X.2006.02971.x
Dickerson, D. P., Pascholati, S. F., Hagerman, A. E., Butler, L. G., and Nicholson, R. L. (1984). Phenylalanine ammonialyase and hydroxy cinnamate: CoA ligase in maize mesocotyls inoculated with Helminthosporium maydis or Helminthosporium carbonum. Physiol. Plant Pathol. 25, 111–123. doi: 10.1016/0048-4059(84)90050-X
Dixon, R. A., Harrison, M. J., and Lamb, C. J. (1994). Early events in the activation of plant defense responses. Annu. Rev. Phytopathol. 32, 479–501. doi: 10.1146/annurev.py.32.090194.002403
Dong, H., Li, W., Zhang, D., and Wei, T. (2003). Differential expression of induced resistance by an aqueous extract of killed Penicillium chrysogenum against Verticillium wilt of cotton. Crop Prot. 22, 129–134. doi: 10.1016/S0261-2194(02)00122-9
Emani, C., Garcia, J. M., Lopata-Finch, E., Pozo, M. J., Uribe, P., Kim, D. J., et al. (2003). Enhanced fungal resistance in transgenic cotton expressing an endochitinase gene from Trichoderm virens. Plant Biotechnol. J. 1, 321–336. doi: 10.1046/j.1467-7652.2003.00029.x
Fan, B., Shen, L., Liu, K., Zhao, D., Yu, M., and Sheng, J. (2008). Interaction between nitric oxidenand hydrogen peroxide in postharvest tomato resistance response to Rhizopus Nigricans. J. Sci. Food Agric. 88, 1238–1244. doi: 10.1002/jsfa.3212
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313x.1999.00265.x
Feng, H., Li, C., Zhou, J., Yuan, Y., Feng, Z., Shi, Y., et al. (2021). A cotton WAKL protein interacted with a Dna J protein and was involved in defense against Verticillium dahliae. Int. J. Biol. Macromol. 167, 633–643. doi: 10.1016/j.ijbiomac.2020.11.191
Gao, M., He, Y., Yin, X., Zhong, X., Yan, B., Wu, Y., et al. (2021). Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cells 184, 5391–5404.e17. doi: 10.1016/j.cell.2021.09.009
Gómez-Gómez, L., and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/s1097-2765(00)80265-8
Han, Q. (2014). Screening and identification of cotton anti-VD biocontrol bacteria and research on disease resistance mechanism. Dissertation/doctor’s thesis. Nanjing Agricultural University, Nanjing (Jiangsu).
Hilleary, R., Paez-Valencia, J., Vens, C., Toyota, M., Palmgren, M., and Gilroy, S. (2020). Tonoplast-localized Ca2+ pumps regulate Ca2+ signals during pattern-triggered immunity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 117, 18849–18857. doi: 10.1073/pnas.2004183117
Hocking, B., Conn, S. J., Manohar, M., Xu, B., Athman, A., Stancombe, M. A., et al. (2017). Heterodimerization of Arabidopsis calcium/proton exchangers contributes to regulation of guard cell dynamics and plant defense responses. J. Exp. Bot. 68, 4171–4183. doi: 10.1093/jxb/erx209
Holmes, E. C., Chen, Y. C., Mudgett, M. B., and Sattely, E. S. (2021). Arabidopsis UGT76B1 glycosylates N-hydroxy-pipecolic acid and inactivates systemic acquired resistance in tomato. Plant Cell 33, 750–765. doi: 10.1093/plcell/koaa052
Hu, H. L. (2012). Construction of transgenic VD with GFP gene and study on the process of infecting cotton. Dissertation/master’s thesis. Nanjing Agricultural University, Nanjing (Jiangsu).
Hu, Q., Xiao, S., Wang, X., Ao, C., Zhang, X., and Zhu, L. (2021). GhWRKY1-like enhances cotton resistance to Verticillium dahliae via an increase in defense-induced lignification and S monolignol content. Plant Sci. 305:110833. doi: 10.1016/j.plantsci.2021.110833
Huang, W., Zhang, Y., Zhou, J., Wei, F., Feng, Z., Zhao, L., et al. (2021). The respiratory burst oxidase homolog protein D (GhRbohD) positively regulates the cotton resistance to Verticillium dahliae. Int. J. Mol. Sci. 22:13041. doi: 10.3390/ijms222313041
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kanchiswamy, C. N., Mohanta, T. K., Capuzzo, A., Occhipinti, A., Verrillo, F., Maffei, M. E., et al. (2013). Differential expression of CPKs and cytosolic Ca2+ variation in resistant and susceptible apple cultivars (Malus x domestica) in response to the pathogen Erwinia amylovora and mechanical wounding. BMC Genomics 14:760. doi: 10.1186/1471-2164-14-760
Köster, P., DeFalco, T. A., and Zipfel, C. (2022). Ca2+ signals in plant immunity. EMBO J. 41:e110741. doi: 10.15252/embj.2022110741
Kumar, R., Barua, P., Chakrabort, Y. N., and Nandi, A. K. (2020). Systemic acquired resistance specific proteome of Arabidopsis thaliana. Plant Cell Rep. 39, 1549–1563. doi: 10.1007/s00299-020-02583-3
Kumar, V., Parkhi, V., Kenerley, C. M., and Rathore, K. S. (2009). Defense-related gene expression and enzyme activities in transgenic cotton plants expressing an endochitinase gene from Trichoderma virens in response to interaction with Rhizoctonia solani. Planta 230, 277–291. doi: 10.1007/s00425-009-0937-z
Lamotte, O., Gould, K., Lecourieux, D., Sequeira-Legrand, A., Lebrun-Garcia, A., Durner, J., et al. (2004). Analysis of nitric oxide signaling functions in tobacco cells challenged by the elicitor cryptogein. Plant Physiol. 135, 516–529. doi: 10.1104/pp.104.038968
Li, H., Guo, Y., Lan, Z., Zhang, Z., Ahammed, G. J., Chang, J., et al. (2021). Melatonin antagonizes ABA action to promote seed germination by regulating Ca2+ efflux and H2O2 accumulation. Plant Sci. 303:110761. doi: 10.1016/j.plantsci.2020.110761
Liu, Q., Ding, Y., Shi, Y., Ma, L., Wang, Y., Song, C., et al. (2021). The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 40:e104559. doi: 10.15252/embj.2020104559
Liu, T., Song, T., Zhang, X., Yuan, H., Su, L., Li, W., et al. (2014). Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5:4686. doi: 10.1038/ncomms5686
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Y., Zhao, Y., and Berkowitz, G. A. (2017). Intracellular Ca2+ is important for flagellin-triggered defense in Arabidopsis and involves inositol polyphosphate signaling. J. Exp. Bot. 68, 3617–3628. doi: 10.1093/jxb/erx176
Ma, Y., Zhao, Y., Walker, R. K., and Berkowitz, G. A. (2013). Molecular steps in the immune signaling pathway evoked by plant elicitor peptides: Ca2+-dependent protein kinases, nitric oxide, and reactive oxygen species are downstream from the early Ca2+ signal. Plant Physiol. 163, 1459–1471. doi: 10.1104/pp.113.226068
Manohar, M., Shigaki, T., Mei, H., Park, S., Marshall, J., Aguilar, J., et al. (2011). Characterization of Arabidopsis Ca2+/H+ exchanger CAX3. Biochemistry 50, 6189–6195. doi: 10.1021/bi2003839
Marcec, M. J., Gilroy, S., Poovaiah, B. W., and Tanaka, K. (2019). Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 283, 343–354. doi: 10.1016/j.plantsci.2019.03.004
Martínez-Medina, A., Pescador, L., Terrón-Camero, L. C., Pozo, M. J., and Romero-Puertas, M. C. (2019). Nitric oxide in plant-fungal interactions. J. Exp. Bot. 70, 4489–4503. doi: 10.1093/jxb/erz289
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990. doi: 10.1105/tpc.109.069658
Naveed, Z. A., Wei, X., Chen, J., Mubeen, H., and Ali, G. S. (2020). The PTI to ETI continuum in Phytophthora-plant interactions. Front. Plant Sci. 11:593965. doi: 10.3389/fpls.2020.593905
Nürnberger, T., and Brunner, F. (2002). Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5, 318–324. doi: 10.1016/s1369-5266(02)00265-0
Plazek, A., and Zur, I. (2003). Cold-induced plant resistance to necrotrophic pathogens and antioxidant enzyme activities and cell membrane permeability. Plant Sci. 164, 1019–1028. doi: 10.1016/S0168-9452(03)00089-X
Qi, J., Wang, J., Gong, Z., and Zhou, J. M. (2017). Apoplastic ros signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. doi: 10.1016/j.pbi.2017.04.022
Qiao, M., Sun, J., Liu, N., Sun, T., Liu, G., Han, S., et al. (2015). Changes of nitric oxide and its relationship with H2O2 and Ca2+ in defense interactions between wheat and Puccinia Triticina. PLoS One 10:e0132265. doi: 10.1371/journal.pone.0132265
Ren, A., Li, M. J., Shi, L., Mu, D. S., Jiang, A. L., Han, Q., et al. (2013). Profiling and quantifying differential gene transcription provide insights into ganoderic acid biosynthesis in Ganoderma lucidum in response to methyl jasmonate. PLoS One 8:e65027. doi: 10.1371/journal.pone.0065027
Sang, J., Jiang, M., Lin, F., Xu, S., Zhang, A., and Tan, M. (2008). Nitric oxide reduces hydrogen peroxide accumulation involved in water stress-induced subcellular antioxidant defense in maize plants. J. Integr. Plant Biol. 50, 231–243. doi: 10.1111/j.1744-7909.2007.00594.x
Sang, Y., and Macho, A. P. (2017). Analysis of PAMP-triggered ROS burst in plant immunity. Methods Mol. Biol. 1578, 143–153. doi: 10.1007/978-1-4939-6859-6_11
Sun, A., Nie, S., and Xing, D. (2012). Nitric oxide-mediated maintenance of redox homeostasis contributes to NPR1-dependent plant innate immunity triggered by lipopolysaccharides. Plant Physiol. 160, 1081–1096. doi: 10.1104/pp.112.201798
Tada, Y., Mori, T., Shinogi, T., Yao, N., Takahashi, S., Betsuyaku, S., et al. (2004). Nitrie oxide and reactive oxygen species do not elicit hypersensitive cell death but induce apoptosis in the adjacent cells during the defense response of oat. Mol. Plant Microb. Interaet. 17, 245–253. doi: 10.1094/MPMI.2004.17.3.245
Takai, R., Isogai, A., Takayama, S., and Che, F. S. (2008). Analysis of flagellin perception mediated by Flg 22 receptor OsFLS2 in rice. Mol. Plant-Microbe Interact. 21, 1635–1642. doi: 10.1094/MPMI-21-12-1635
Thomma, B. P., Nurnberger, T., and Joosten, M. H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15. doi: 10.1105/tpc.110.082602
Thor, K., Jiang, S., Michard, E., George, J., Scherzer, S., Huang, S., et al. (2020). The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 585, 569–573. doi: 10.1038/s41586-020-2702-1
Vandelle, E., Poinssot, B., Wendehenne, D., Bentejac, M., and Pugin, A. (2006). Integrated signalling network involving calcium, nitric oxide, and active oxygen species but not mitogen - activated protein kinases in BcPG1-elicited grapevine defenses. Mol. Plant-Microbe Interact. 19, 429–440. doi: 10.1094/MPMI-19-0429
Wang, S. S. (2012). A preliminary study on the defense response of kelp induced by Flg 22. Dissertation/master’s thesis. Qingdao (Shangdong), Ocean University of China.
Wang, X. N. (2014). Study on the infection of cotton leaves and monocots by VD labeled with green fluorescent protein. Dissertation/master’s thesis. Nanjing Agricultural University, Nanjing (Jiangsu).
Wang, X. Y., Chen, Z. Y., Liu, W. Z., Luo, C. P., Zhang, R. S., and Zhou, H. F. (2014). Resistance to bacterial leaf streak of transgenic Rice with Flagellin gene (Chinese abstract). Acta Bot. Northwest. 34, 1534–1539. doi: 10.7606/j.issn.1000-4025.2014.08.1534
Wang, X. Y., Chen, Z. Y., Liu, W. Z., Luo, C. P., Zhang, R. S., and Zhou, H. F. (2015). Breeding of fla transgenic rice and analysis of disease resistance spectrum (Chinese abstract). Mol. Plant Breed. 13, 61–65. doi: 10.13271/j.mpb.013.000061
Wang, J., Liu, X., Zhang, A., Ren, Y., Wu, F., Wang, G., et al. (2019). A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 29, 820–831. doi: 10.1038/s41422-019-0219-7
Wang, S., Zhao, F., Wei, X., Lu, B., Duan, D., and Wang, G. (2013). Preliminary study on Flg 22-induced defense response in female gametophytes in Saccharina japonica (Phaeophyta). J. Appl. Phycol. 25, 1215–1223. doi: 10.1007/s10811-012-9911-4
Xiong, X. P., Sun, S. C., Zhu, Q. H., Zhang, X. Y., Li, Y. J., Liu, F., et al. (2021). The cotton lignin biosynthetic gene Gh4CL30 regulates lignification and phenolic content and contributes to verticillium wilt resistance. Mol. Plant-Microbe Interact. 34, 240–254. doi: 10.1094/MPMI-03-20-0071-R
Yan, J., Tsuichihara, N., Etoh, T., and Iwai, S. (2007). Reactive oxygen species and nitric oxide are involved in ABA inhibition of stomatal opening. Plant Cell Environ. 30, 1320–1325. doi: 10.1111/j.1365-3040.2007.01711.x
Yano, A., Suzuki, K., Uchimiya, H., and Shinshi, H. (1998). Induction of hypersensitive cell death by a fungal protein in cultures of tobacco cells. Mol. Plant-Microbe Interact. 11, 115–123. doi: 10.1094/MPMI.1998.11.2.115
Zhang, A., Jiang, M., Zhang, J., Ding, H., Xu, S. H., and Hu, X. (2007). Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of mitogen-aetivated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol. 175, 36–50. doi: 10.1111/j.1469-8137.2007.02071.x
Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E. J., Jones, J. D., Felix, G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. doi: 10.1038/nature02485
Glossary
Keywords: flagellin, upland cotton, RNA-seq, immune response, Verticillium dahliae
Citation: Zhou H, Wang Y, Zhang Y, Xie Y, Nadeem H and Tang C (2022) Flagellin C decreases the expression of the Gossypium hirsutum cation/proton exchanger 3 gene to promote calcium ion, hydrogen peroxide, and nitric oxide and synergistically regulate the resistance of cotton to Verticillium wilt. Front. Plant Sci. 13:969506. doi: 10.3389/fpls.2022.969506
Edited by:
Wolfgang Moeder, University of Toronto, CanadaReviewed by:
Jian Sun, Jiangsu Normal University, ChinaXianpeng Xiong, Agricultural Genomics Institute at Shenzhen (CAAS), China
Yijie Dong, Chinese Academy of Agricultural Sciences, China
Xing Fen Wang, Hebei Agricultural University, China
Copyright © 2022 Zhou, Wang, Zhang, Xie, Nadeem and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Canming Tang, dGFuZ2NtQG5qYXUuZWR1LmNu
 Heng Zhou
Heng Zhou Yi Wang1
Yi Wang1 Yihao Zhang
Yihao Zhang