- Guangdong Key Laboratory of Ornamental Plant Germplasm Innovation and Utilization, Environmental Horticulture Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
Transcription factors (TFs) of the WRKY family play pivotal roles in defense responses and secondary metabolism of plants. Although WRKY TFs are well documented in numerous plant species, no study has performed a genome-wide investigation of the WRKY gene family in Cymbidium sinense. In the present work, we found 64 C. sinense WRKY (CsWRKY) TFs, and they were further divided into eight subgroups. Chromosomal distribution of CsWRKYs revealed that the majority of these genes were localized on 16 chromosomes, especially on Chromosome 2. Syntenic analysis implied that 13 (20.31%) genes were derived from segmental duplication events, and 17 orthologous gene pairs were identified between Arabidopsis thaliana WRKY (AtWRKY) and CsWRKY genes. Moreover, 55 of the 64 CsWRKYs were detectable in different plant tissues in response to exposure to plant hormones. Among them, Group III members were strongly induced in response to various hormone treatments, indicating their potential essential roles in hormone signaling. We subsequently analyzed the function of CsWRKY18 in Group III. The CsWRKY18 was localized in the nucleus. The constitutive expression of CsWRKY18 in Arabidopsis led to enhanced sensitivity to ABA-mediated seed germination and root growth and elevated plant tolerance to abiotic stress within the ABA-dependent pathway. Overall, our study represented the first genome-wide characterization and functional analysis of WRKY TFs in C. sinense, which could provide useful clues about the evolution and functional description of CsWRKY genes.
Introduction
As one of the most prominent flowering plant families, Orchidaceae include 801 genera and more than 30,000 specie (Brown, 2005). In the orchid family, Cymbidium is widely fostered in East Asia, such as China, Japan, and Korea, and Southeast Asia (Liu et al., 2006). Cymbidium has become very prevalent in China, and it is highly desirable in traditional flower markets because of its beauty, fragrant flowers, elegant and upright leaves. However, functional genomics investigations and gene identification of valuable horticultural traits are significantly restricted because of the polyploid genomes and long juvenile phases of the genus Cymbidium. Therefore, it is essential for discovery of functional genes in Cymbidium research. In recent years, next-generation sequencing (NGS) technologies have rapidly developed, offering formidable approaches for high-throughput sequence determination (Leitch et al., 2009; Liu et al., 2012). Furthermore, as RNA-Seq decrease in cost, promoting the identification of new genes by obtaining massive amounts of sequence data with enormous depth and coverage. As a result, a large number of critical regulators related to important agronomic traits and environmental adaptation have been identified in orchid species, including Apostasia (Zhang et al., 2017, 2021a), Cymbidium (Ai et al., 2021; Yang et al., 2021), Dendrobium (Niu et al., 2021; Zhang et al., 2021b), Gastrodia (Xu et al., 2021) Phalaenopsis (Cai et al., 2015; Chao et al., 2018), and Vanilla (Hasing et al., 2020).
The transcription factors (TFs) are critical in controlling plant development and stress response. WRKY, one of the largest families of higher plant TFs, plays essential roles in pathogen defense, abiotic cues, phytohormone signaling, and management of plant development and secondary metabolism. The WRKY domain (WD) is the essential characteristic of the WRKY protein, which is ~60 amino acids in length and consists of the highly conserved signature WRKYGQK, followed by a C2H2-or C2HC-type of zinc-finger motif. The high binding affinity of WRKY TFs to the consensus W-box cis-elements requires both heptapeptide sequence and zinc-finger motif (Maeo et al., 2001). Given these features of the WD, its family members are categorized into three groups (Eulgem et al., 2000). The WRKY Group II can be further divided into five subgroups (IIa, IIb, IIc, IId, and IIe) according to their primary amino acid sequences (Chen et al., 2017). In addition, WRKY preferentially binds to a markedly conservative DNA motif named the W box (T/CTGACC/T; Rushton et al., 2010). Since the first WRKY TF (SPF1) is cloned from sweet potato (Ipomoea batatas; Ishiguro and Nakamura, 1994), an increasing number of WRKY family genes are identified in several species, including Arabidopsis (75; Wu et al., 2005), barley (103; Kan et al., 2021), flax (102; Yuan et al., 2021), ginseng (137; Di et al., 2021), Gossypium (109; Ding et al., 2015), grape (59; Wang et al., 2015), Musa (147; Goel et al., 2016), maize (125; Hu et al., 2021), Taxus (61; Zhang et al., 2018), rice (109; Okay et al., 2014), walnut (103; Hao et al., 2021), and wheat (124; Ye et al., 2021).
In the present study, we analyzed the genome sequences of Cymbidium sinense, a very famous traditional orchid plant in China and Southeast Asia, and identified 64 CsWRKY proteins. Computational analysis was conducted to assess their physicochemical properties, including conserved domain, phylogenetic relationship, motif composition, functional annotation, and protein–protein interaction (PPI). In addition, the expression profiling of CsWRKY genes in various plant tissues in response to hormone treatments abscisic acid (ABA), gibberellic acid, salicylic acid, and methyl jasmonate (MeJA) indicated that most CsWRKY genes were responsive to various plant hormones, and Group III genes appeared to be more actively expressed to cope with the stress response. Collectively, our current findings provided valuable insights into the understanding of WRKY TFs in orchid plants and identified many candidate regulators involved in multiple hormone signaling.
Results
Identification and analysis of WRKY genes in Cymbidium sinense
A total of 64 WRKY TFs were found through the C. sinense genomic database (Supplementary Table S2). Each WRKY gene was consistently named CsWRKY1-CsWRKY64 based on its chromosomal location. The characteristics were analyzed, including the genome location, length of the open reading frame (ORF), and basic information of their encoded proteins, including length, molecular weight (MW), and isoelectric point (pI). Supplementary Table S2 shows that the length of CsWRKY proteins ranged from 67 (CsWRKY38) to 676 (CsWRKY61) amino acids, and their average length was 317 residues. The pI ranged from 4.84 (CsWRKY46) to 10.00 (CsWRKY62), and the MW ranged from 5,929.61 Da (CsWRKY12) to 7,4025.70 Da (CsWRKY61).
Classification of Cymbidium sinense proteins
To explore the phylogenetic relationship among CsWRKY proteins, 275 conserved WDs, including 114 OsWRKY proteins from rice (O. sativa japonica) and 85 AtWRKY proteins from Arabidopsis, were extracted to construct the evolutionary tree using the maximum likelihood (ML) method. As shown in the evolutionary tree (Figure 1), the 64 CsWRKY proteins were categorized into three main groups (I, II, and III), and Group II proteins were further sorted into five subgroups (IIa, IIb, IIc, IId, and IIe). The CsWRKY TFs had two standard motifs, including a WD and a zinc-finger-like domain. The former, typically the WRKYGQK sequence, could combine with the W box cis-element to trigger the expressions of their downstream genes. Besides the WRKYGQK sequence, three variants, WKKYGQK (CsWRKY47) in Group IIa, WRKYGKK (CsWRKY08, 19, and 22) in Group IIb, and WRKYGRK (CsWRKY49, and 62) in Group IIc were also revealed, respectively. The latter, the zinc-finger-like domain, also had two types, namely C2H2 and C2HC.
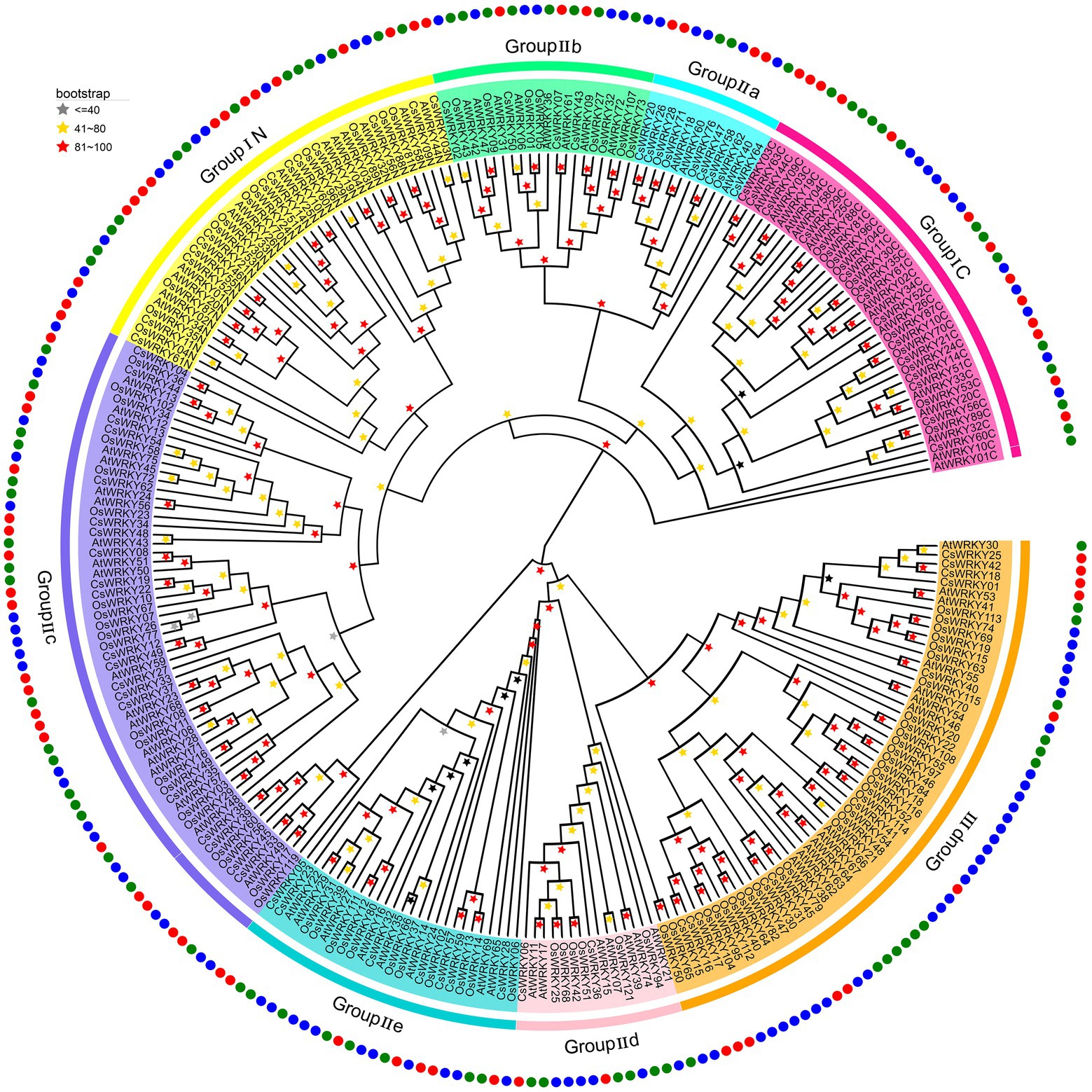
Figure 1. Phylogenetic tree of WRKY genes in Cymbidium sinense (Cs), Arabidopsis thaliana (At), and Oryza sativa (Os). An individual group (I–III) of ancestral relationships were presented in different colors.
It is worth noting that Group I was composed of 15 CsWRKY proteins, 11 of which contained two WDs. The remaining CsWRKY63 had a single intact WD in its C terminus, and the other three members (CsWRKY03, 46, and 55) had a single WD at the N terminus (Supplementary Table S2; Supplementary Figure S1), implying that they most likely experienced domain loss or acquisition events in the evolutionary process (Ross et al., 2007). In addition, the zinc-finger motifs of the CsWRKYs in Group I belonged to the C2H2 type with a C-X4-C-X22–23-H-X1-H motif (Supplementary Table S2). There were 38 members in Group II, while 75 contained the motif of C-X4–5-6-C-X23–25-29-H-X1-H, and three members, CsWRKY01, 38, and 44, lacked a typical zinc-finger-like motif. According to their phylogenetic relationship, all 38 Group II members were further categorized into five subgroups as follows: Group IIa (five proteins), Group IIb (four proteins), Group IIc (20 proteins), Group IId (two proteins), and Group IIe (seven proteins). The zinc-finger motifs of Group III members (11) belonged to the C2HC type, with the C-X7-C-X23–24-26-H-X1-C motif (Supplementary Table S2), except for CsWRKY17 and 41 only containing fragmentary WRKY structure.
Analyses of chromosomal location, gene duplication, and genome synteny
A total of 64 candidate CsWRKY genes were mapped to 16 of the 20 chromosomes of C. sinense with an irregular arrangement (Figure 2). Chromosome 2 harbored the most significant number of CsWRKYs (10 genes), followed by chromosome 8 (eight genes). With only one gene, the least number of CsWRKYs was found on Chromosome 10. Three chromosomes contained the members from all three groups, 10 chromosomes contained the members from two groups, and the other three only had the members from one group.
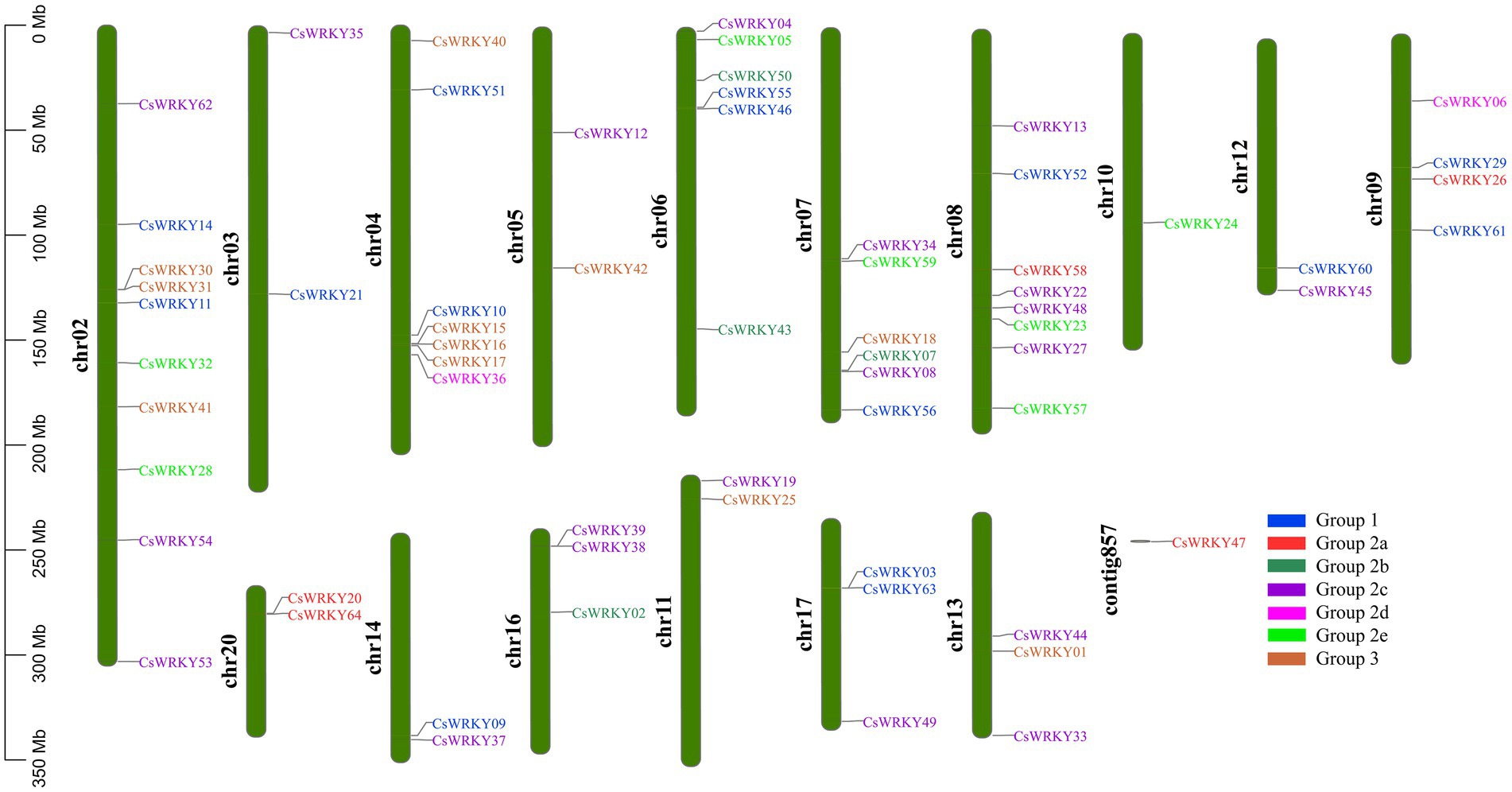
Figure 2. The chromosomal distribution and positioning of CsWRKYs across all seven chromosomes of C. sinense. A total of 16 chromosomes and one contig with varying lengths are shown in Mb (million base pair) scale on the left, where individual chromosomes (bars) are labeled with respective CsWRKY genes. The subfamily members are presented in various colors.
To further assess the origin and evolution of the CsWRKY gene family, we constructed two comparative syntenic maps between C. sinense and A. thaliana, O. sativa, V. vinifera, M. cuminata, or Z. mays at the genome-wide level. Supplementary Figure S2 shows that we finally identified 27, 28, and 29 orthologous gene pairs between C. sinense and O. sativae, V. vinifera, or M. cuminata, respectively. However, only 12 and 19 orthologous gene pairs between C. sinense and Z. mays or A. thaliana, were identified, respectively. More details about these orthologous gene pairs are given in Supplementary Table S5. There were far more orthologous genes between C. sinense and O. sativae than those between C. sinense and A. thaliana, which probably resulted from the nearer phylogenetic relationship between C. sinense and O. sativae.
To assess the selection pressure on various duplicated WRKY genes, we calculated the Ka and Ks substitution rates and the Ka/Ks ratios for each repeat CsWRKY gene pair. Typically, a Ka/Ks ratio of 1, >1, or <1 indicates neutral selection, adaptive evolution with positive selection, and negative or purifying selection, respectively (Nekrutenko et al., 2002). The Ka/Ks ratio for all seven segmentally duplicated gene pairs was <1 (Supplementary Table S4; Supplementary Figure S3), showing the high conservation of CsWRKY genes during evolution.
Gene structure and conserved motif analysis of CsWRKY genes
To gain more in-depth insight into the development of the WRKY gene family in C. sinense, we mapped the genetic structure of each CsWRKY gene. Based on the full-length CsWRKY proteins (Figure 3), we constructed a phylogenetic tree to clarify the gene structure and conserved motifs better. The number of introns in CsWRKY genes ranged from 0 to 10, similar to rice from 0 to 8 (Xie et al., 2005). Most CsWRKY genes harbored one to four introns, with 23 members containing two introns, 13 containing three introns, 10 containing one intron, and nine containing four introns. The other two CsWRKY genes contained five introns, and three CsWRKY genes contained seven, eight, and 10 introns. Additionally, the remaining four CsWRKY genes contained no intron. Figure 3 reveals that most CsWRKY genes within the same group or subgroup had a similar gene organization, showing the functional similarity among members.
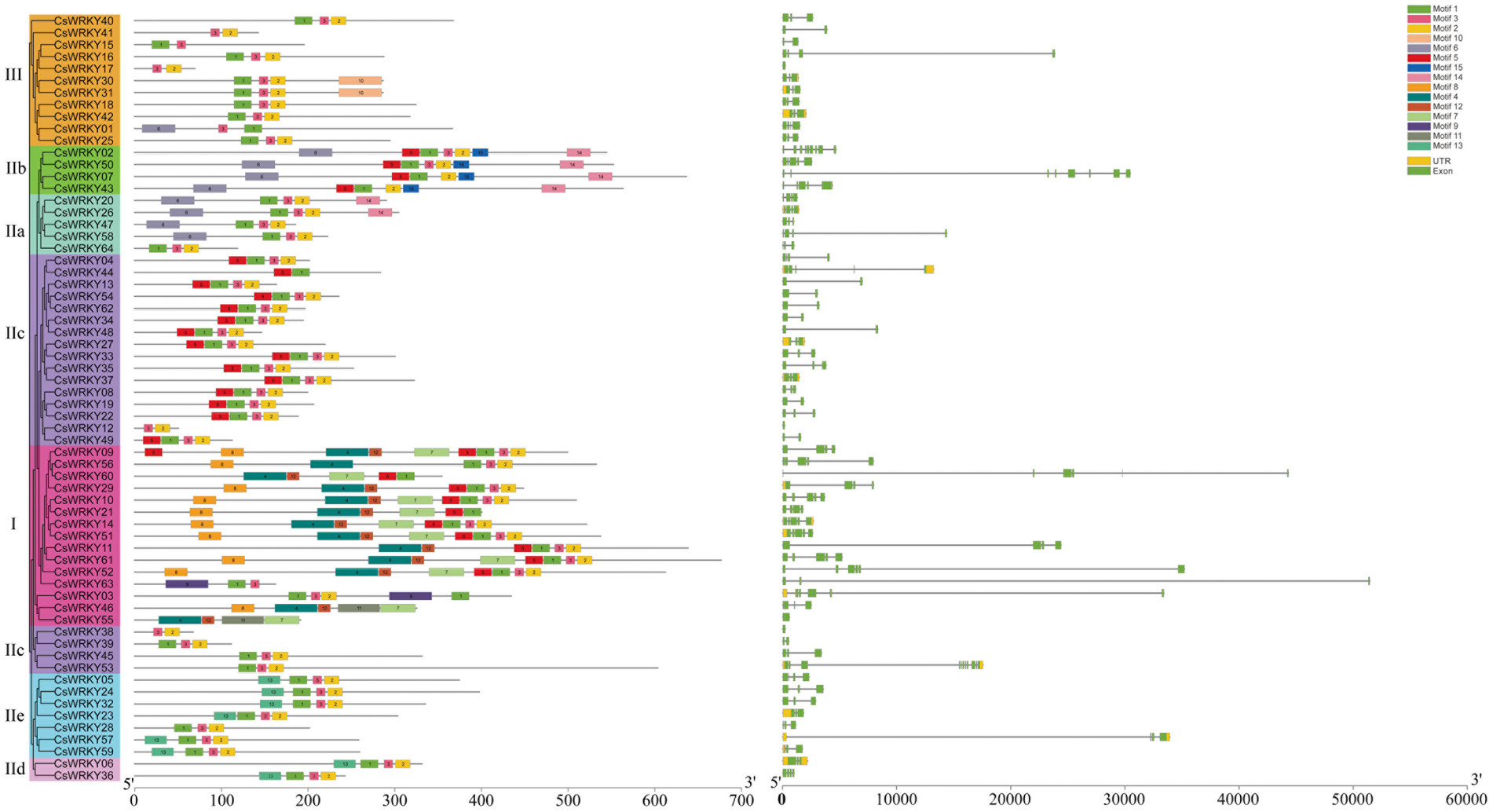
Figure 3. Phylogenetic relationships, gene structure, and architecture of conserved protein motifs in WRKY genes from C. sinense.
Moreover, we identified 15 conserved motifs within proteins by the MEME program. MEME motif study showed that each CsWRKY protein had its specific conserved motifs (Supplementary Table S3). Figure 3 depicts that the WRKY family members with similar motif structures could be categorized into the same group. Almost all CsWRKYs had the conserved heptapeptides WRKYGQK (Motif 1 or Motif 4) and harbored at least one motif. Besides, Motif 2 was composed of the C2H2 motif, while Motif 12 consisted of the C2HC motif. Furthermore, the conserved motifs were explicitly present in various groups. For instance, Group IIa members harbored four conserved motifs (Motifs 1, 2, 3, and 6), Group IIb members contained five conserved motifs (Motifs 1, 2, 5, 6, and 15), and 14 Group IIc members contained four conserved motifs (Motifs 1, 2, 3, and 5). Clearly, a few motifs existed in one or more groups and subgroups. For example, Motifs 4, 8, 7, and 12 existed in Group I members, and Motif 10 existed only in Group III, while Motif 15 was mainly present in Group IIb. This result suggested that these groups were endowed various functions during evolution. However, even if these members belong to the same clade, the functions of family members vary greatly (Yang et al., 2019). These WRKY proteins may have different functions though classified in the same group. Despite CsWRKY28 belonged to group IIe, it had the similar conserved motifs with the group IIc.
The cis-elements in the promoters of Cymbidium sinense WRKY genes
Cis-elements in promoter region are important for gene expression (Rombauts et al., 1999). To verify the potential function of CsWRKY genes in response to abiotic stress, we extracted the 2,000-bp promoter sequences of the CsWRKY genes and analyzed them for cis-elements using the PlantCARE database. Supplementary Figure S4 depicts that 20 types of cis-acting elements related to stresses and phytohormone responses were identified in the promoters of CsWRKY genes, including four defense-and stress-responsive elements (W box and TC-rich repeats) and seven hormone-related elements (ABRE, AuxRE, As-1, TGA-element, CGTCA-motif, and TGACG-motif). Supplementary Figure S4 shows that MYC and MYB were the most numerous elements in the promoter regions of all 64 CsWRKYs, with 61 genes containing these two elements. Box 4 and G box were two types of light-response elements, ARE was involved in anaerobic induction, and ABRE (ABA-responsive elements), CGTCA-motif, and TGACG-motif (MeJA-responsive elements) also appeared frequently in promoter regions of CsWRKY genes, which were found in 59, 49, 49, 47, 42, and 42 promoters, respectively. Many W-boxes were identified in 37 CsWRKY gene promoters, indicating that these genes were modulated by other WRKY TFs or themselves. LTR and MBS elements that respond to low temperature and drought stresses were identified in the promoters of 17 and 28 CsWRKY genes, respectively. Moreover, 25 promoters showed a TC-rich repeats element (cis-acting element participated in defense and stress response), and only 15 promoters had a GARE-motif (gibberellin-responsive element). Thus, the cis-element analysis showed that the expressions of CsWRKY genes in C. sinense might be related to various environmental factors.
Interaction analysis of specific CsWRKY proteins
A PPI network of CsWRKY proteins was established using STRING 10.5 software based on the Arabidopsis association model. Our results showed that there were close interaction networks among 43 CsWRKYs (Supplementary Figure S5). Among them, 14 proteins belonged to Group I, 22 proteins belonged to Group II, and seven proteins belonged to Group III. The extensive WRKY-WRKY interactions manifested a mechanism for functional cooperation and antagonism among WRKY proteins for dynamic regulation of target genes in C. sinense. In addition, CsWRKY20, CsWRKY26, CsWRKY47, CsWRKY58, and CsWRKY64 belonging to Group IIa, CsWRKY10, CsWRKY11, CsWRKY14, CsWRKY21, and CsWRKY51 belonging to Group I, and CsWRKY01, CsWRKY25, CsWRKY42 and CsWRKY15 belonging to Group III were also involved in a more robust interaction network with other proteins. For instance, MAP kinase 4 (MPK4), functioning as a regulator of pathogen defense responses (Rasmussen et al., 2012; Bazin et al., 2020), and SIB1 are negative regulators of ABA-mediated leaf senescence (Zhang et al., 2022a), indicating that these genes played a kernel role in the biotic and abiotic stress response.
Analysis of expression profiles of Cymbidium sinense WRKY genes in different tissues
Based on RNA-seq and expression profiling analysis, 55 CsWRKYs were detectable in different plant tissues (Figure 4; Supplementary Table S6). However, the other nine transcripts (CsWRKY10, 12, 15, 17, 38, 41, 47, 59, and 64) were not detected, showing that these genes might be pseudogenes or have particular temporal and spatial expression patterns. Most of the 55 CsWRKY transcripts investigated were expressed in all tissues with low expression levels, suggesting that these TFs worked with other proteins synergistically or interactively during plant growth and development. Moreover, many genes had preferential expression in various tissues. For example, 19 CsWRKY transcripts (CsWRKY21, 55, 56, 60, and 61 from Group I, CsWRKY07, 43, and 50 from Group IIb, CsWRKY19, 22, 33, 34, 48, and 54 from Group IIc, CsWRKY25 from Group IId, CsWRKY05, 24, and 57 from Group IIe, and CsWRKY42 from Group III) exhibited high expression levels in root particularly. By contrast, 2, 10, 3, and 2 exhibited the highest transcript abundances in the stem, leaf, flower, and fruit, respectively. These genes might play special roles in specific tissues. Additionally, several gene pairs with a close relationship, such as CsWRKY03/63 and CsWRKY19/33, indicated similar expression profiles, suggesting that the functionality of these genes was redundant.
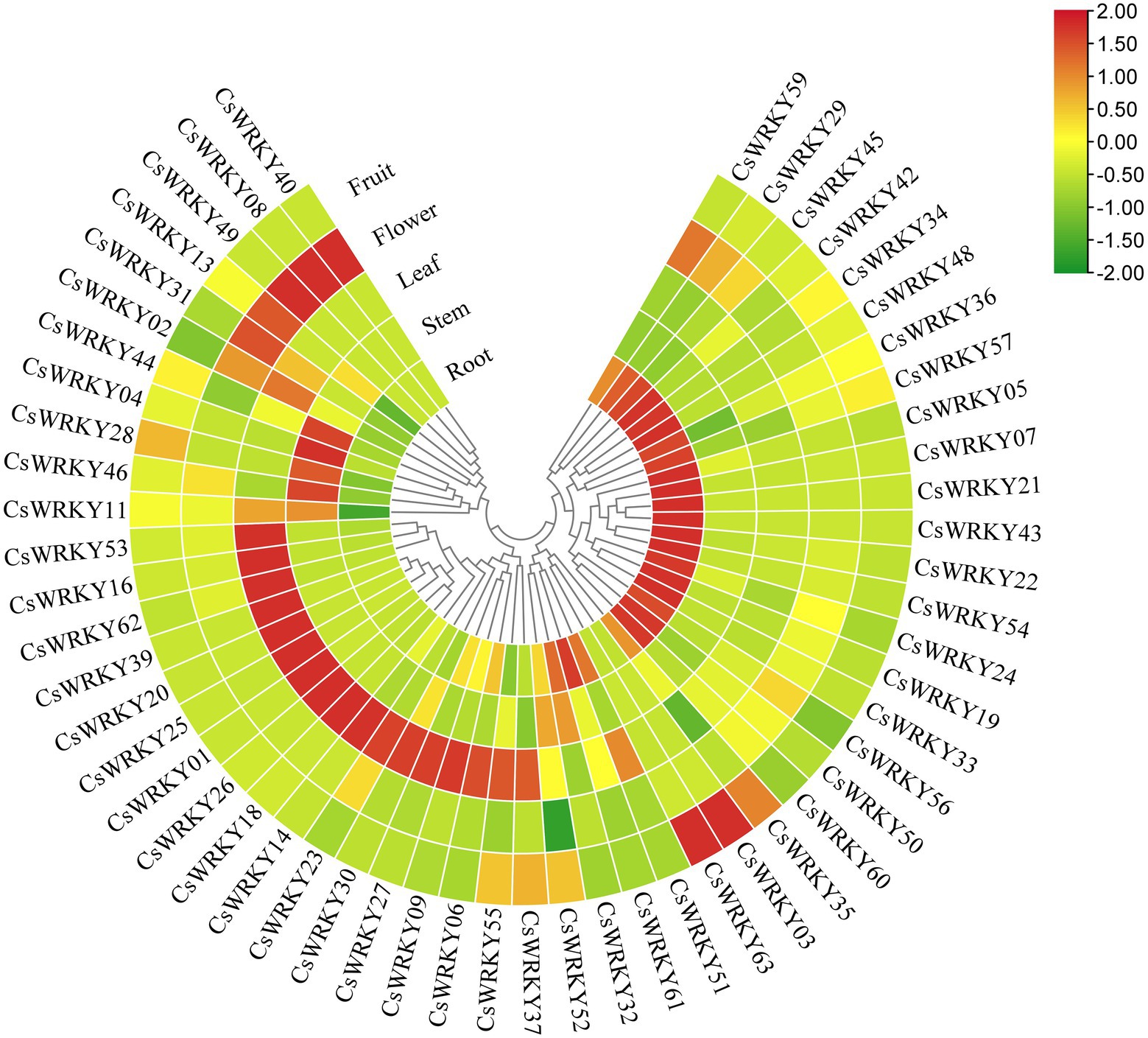
Figure 4. Heat map representation and hierarchical clustering of the CsWRKY gene expression profiles in five tissues.
Expression profiles of Cymbidium sinense WRKY genes with hormone treatment
WRKY are widely involved in plant abiotic stress regulation (Jiang et al., 2017). To further certify the hormone response of CsWRKY genes, quantitative real-time PCR (qRT-PCR) was accomplished for 21 randomly chosen CsWRKY genes, including four Group I members, 11 Group II members, and six Group III members. We first optimized the treatment conditions, and the seedling leaves were sampled after exposure to 100 μmol GA, ABA, SA, IAA, JA, or ACC for 2 h.
Figure 5 shows that the expressions of CsWRKY01, CsWRKY18, CsWRKY20, CsWRKY25, CsWRKY30, CsWRKY31, CsWRKY37, and CsWRKY42 were significantly up-regulated by over 5-fold under the GA treatment (Figures 5A,B). Under the ABA treatment, all of CsWRKYs were significantly induced, except that CsWRKY45 was significantly down-regulated (Figure 5). Under the SA treatment, CsWRKY20, CsWRKY25, CsWRKY30, and CsWRKY31 were significantly induced by over 15-fold (Figures 5B,C). The expression of CsWRKY20 was up-regulated by over 5-fold under the IAA treatment, which was induced by over 6-fold under the JA treatment and over 125-fold under the ACC treatment (Figure 5B). More interestingly, Group III genes sensitive to responses to various hormones played important roles in responses to abiotic stress in C. sinense.
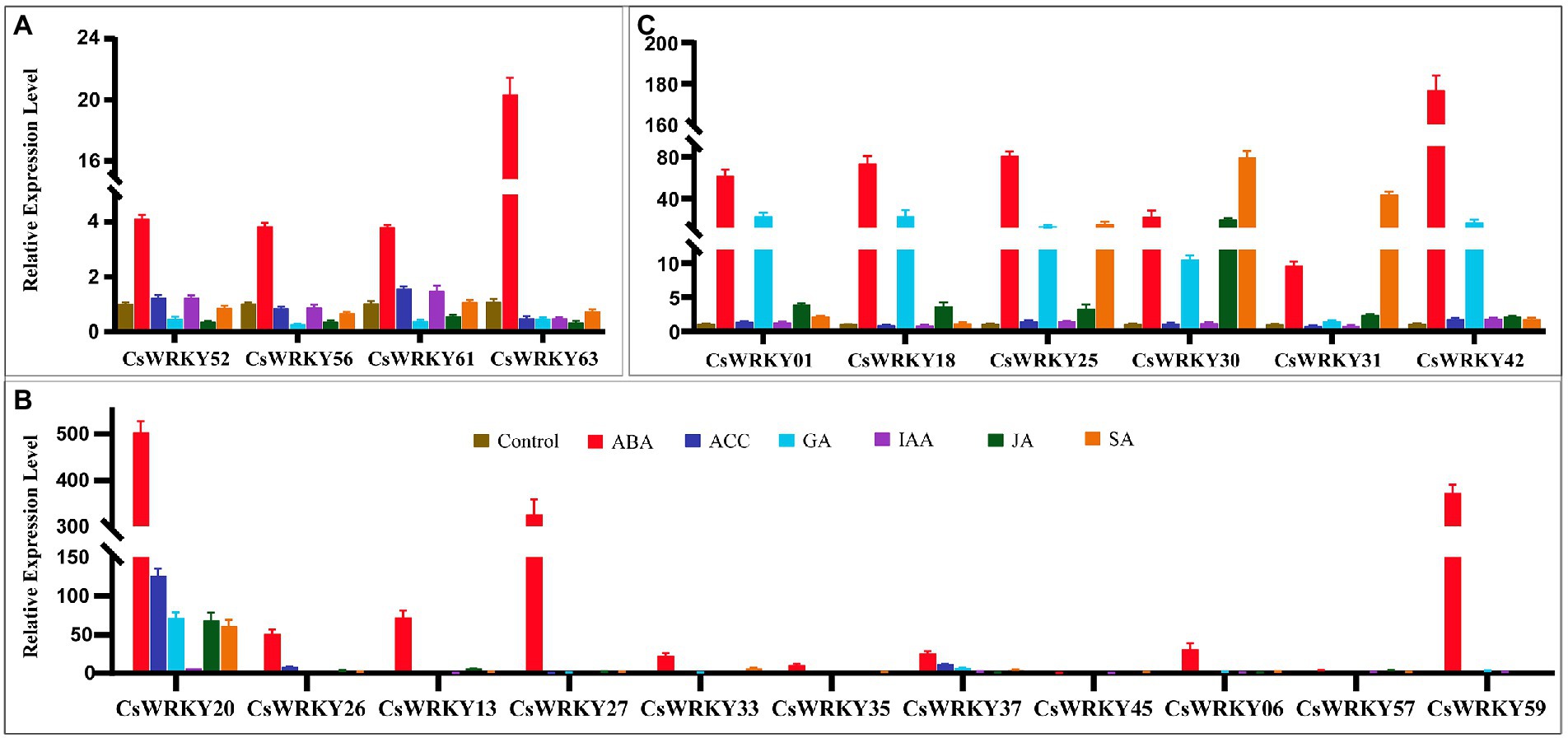
Figure 5. Expression patterns of 21 selected CsWRKY genes in C. sinense leaves under ABA, ACC, GA, IAA, JA, and SA stresses (A. Group I, B. Group II, and C. Group III). qRT-PCR data were normalized using the Cymbidium actin gene. X-axes represent various treatments (CK, normal condition). Different genes and y-axes are scales of relative expression level. Error bars result from three biological replicates.
Subcellular localization of CsWRKY18
Since subcellular location information can provide some clues for protein function research, we used the online software Wolf PSORT to predict the subcellular locations of CsWRKY proteins in this study (Supplementary Table S7). Almost all CsWRKY proteins were mainly located in the nucleus, except for seven genes located in various organelles, such as cytoplasm, chloroplast, mitochondria, and peroxisome. CsWRKY18 protein was chosen for subcellular localization verification. The full-length cDNA without the termination codon was fused in-frame to the 5′-end of the GFP gene under the control of the CaMV 35S promoter (Figure 6A). The recombined vector pAN580-CsWRKY18-GFP was transfected into leaf base protoplasts of Cymbidium. Confocal microscopy revealed that the fusion protein CsWRKY18-GFP was detected specifically in the nucleus, while GFP control displayed ubiquitous distribution in the whole cell. Furthermore, DAPI (4′,6-diamidino-2-phenylindole) staining showed that CsWRKY was clearly localized to the nucleus (Figure 6B).
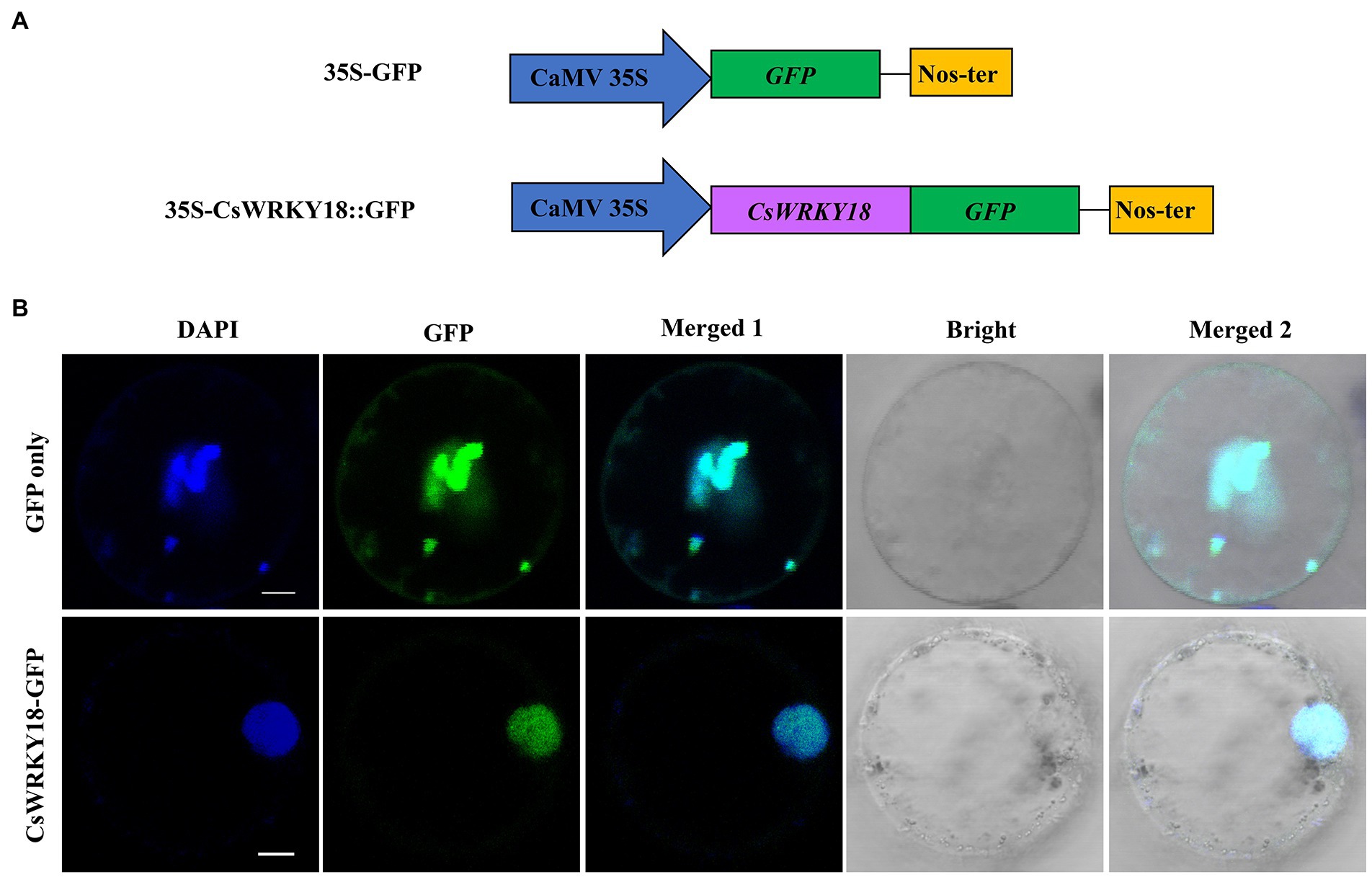
Figure 6. Subcellular localization of Group III CsWRKY18 protein. (A) Schematic illustration of the constructs. (B) Subcellular location of free GFP and CsWRKY18-GFP protein in C. sinense leaves.
Overexpression of the CsWRKY18 gene increases ABA sensitivity
Figure 7A shows that under normal conditions, no significant difference in germination between WT plants and three transgenic lines was found. However, when 5 μM exogenous ABA was given, the transgenic Arabidopsis lines showed dramatically lower cotyledon greening rates than WT plants. Moreover, during the post-germinative growth stage, 4-day-old seedlings grown on 0.5× MS medium were transferred to vertical agar plates containing 1 μM or 5 μM ABA. We found that the root growths of WT and transgenic lines were both markedly suppressed. However, the root lengths of transgenic plants were remarkably shorter compared with WT plants (Figure. 7C).
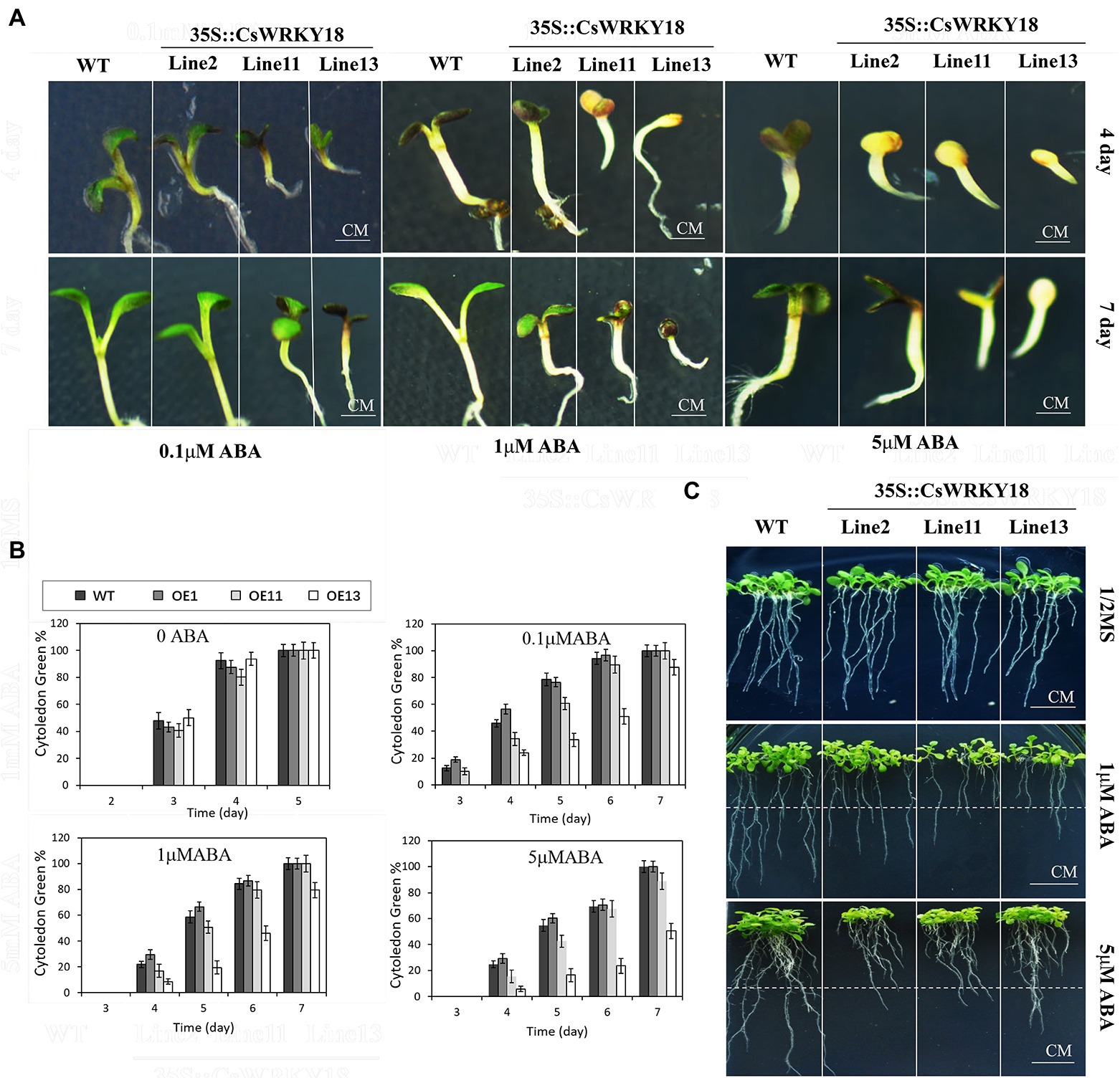
Figure 7. 35S::CsWRKY18 seedlings development under ABA treatment. (A) Seedling development of wild-type and 35S::CsWRKY18 treated with 0.1, 1, and 5 μM ABA. (B) Cytoledon green ratio of seedlings under ABA treatment. (C) Root elongation of wild-type and 35S::CsWRKY18.
Overexpression of CsWRKY18 enhances plant tolerance to abiotic stress
We further compared the root morphology of the WT and transgenic plants grown on 1/2 MS medium containing 0 mM, 100 mM, 150 mM, and 200 mM NaCl (Figure 8). Three transgenic lines exhibited longer roots compared with WT plants growing on all culture medium (Figure 8). In parallel, leaves of 4-week-old seedlings were treated with 200 mg/L PEG6000. After 12 h, dehydration and shrinkage of the WT leaves were observed. In contrast, the leaves of transgenic plants remained fresh and upright (Supplementary Figure S6).
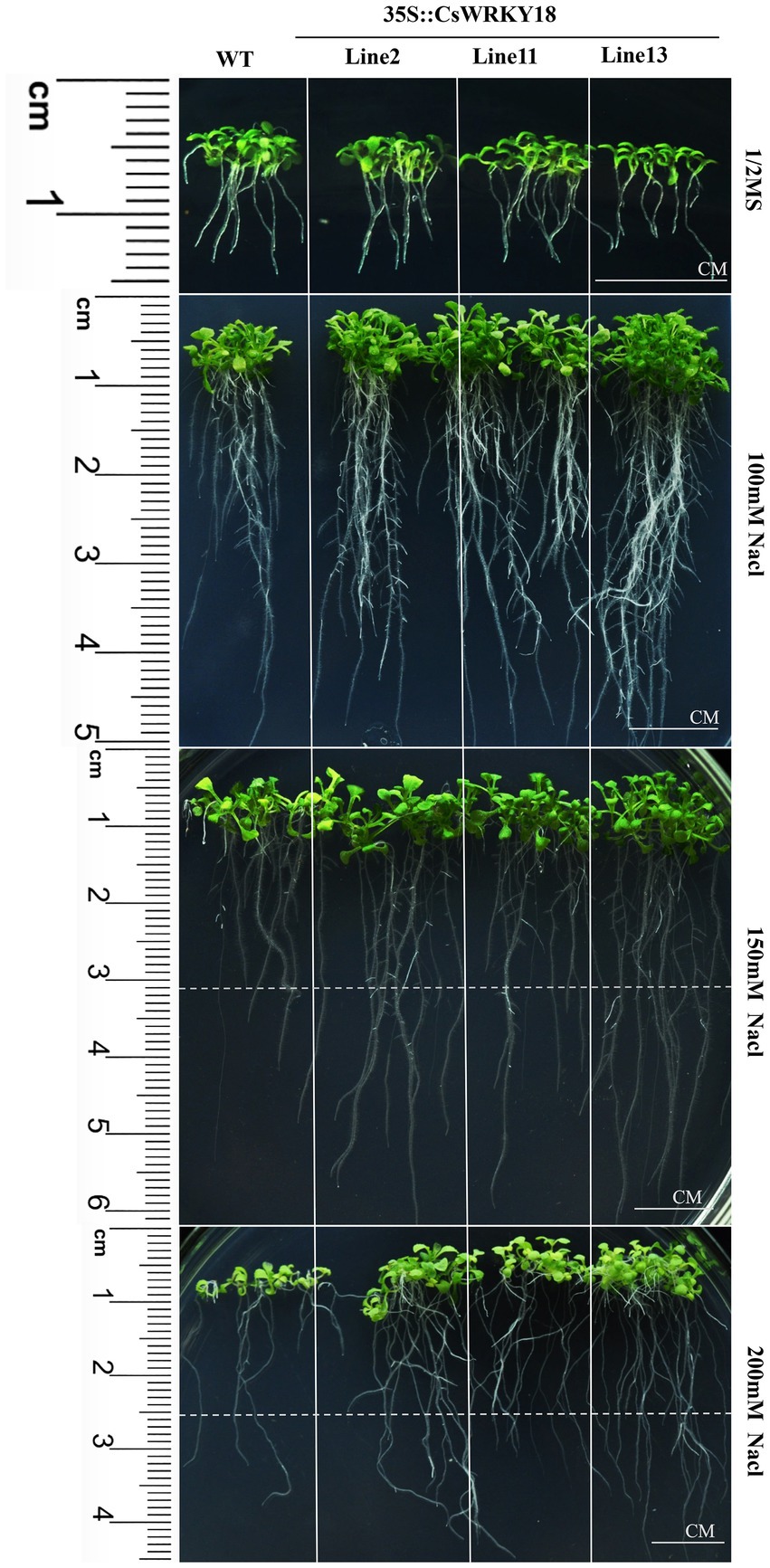
Figure 8. Root elongation of wild-type and 35S::CsWRKY18 under NaCl treatment in 100, 150, and 200 mM.
To explore the mechanism of CsWRKY18 in response to salt and osmotic stress, Tu, a well-known ABA biosynthesis inhibitor, was applied. When 0.75 mM Tu was supplemented to a medium containing 150 mM NaCl buffer, the salt and osmotic tolerance phenotypes in the transgenic lines were obviously weakened (Supplementary Figure S7). These results suggested that the CsWRKY18 transgenic plants were tolerant to salt and osmotic stress, indicating a potential role of CsWRKY18 in the ABA pathway.
Materials and methods
Plant materials and treatment
In the present study, the plants of C. sinense’ Dharma’ were grown in a greenhouse under controlled conditions (26°C/23°C, day/night, 16 h/8 h light/dark photoperiod with relative humidity 80%) at Environmental Horticulture Research Institute, Guangdong Academy of Agricultural Sciences (China). For tissue/organ-specific expression analysis, various plant parts, such as roots, leaves, stems, and flowers, were harvested from a 2-year-old plant. For expression in fruit tissues, 1/2/3/cm, mature green fruits, breaker fruits, and 10-day breaker fruits were harvested. For phytohormone treatment, 2-year-old plants in flowerpots were used, and each group (n = 3) was sprayed with 100 μM ABA, gibberellin (GA3), auxin (IAA), and MeJA. The samples were collected at 0, 0.5, 1, 2, 3, and 4 h after exposure from nine individual plants. For different organ samples, roots, stems, and leaves were collected from three plants grown for 45 days. All materials were immediately frozen in liquid nitrogen and preserved at −80°C for further analysis of WRKY genes. Considering the sensitive response of Group III to hormone signals, we randomly selected CsWRKY18 for further functional analysis. We generated transgenic A. thaliana plants overexpressing the CsWRKY18 gene under the CaMV 35S promoter, according to the general method (Clough and Bent, 1998; Feng et al., 2021) and we established 23 homozygous transgenic lines using hygromycin selection and PCR. Three T2-independent lines, L2, L11, and L13, were chosen for further phenotypic analysis according to the PCR analysis (Figure 7B). Seeds from transgenic and WT plants were incubated for germination on 1/2 MS medium supplemented with 1 μM ABA.
Identification of WRKY genes from Cymbidium sinense
The genome sequences of C. sinense, which were sampled in Guangzhou, Guangdong Province, China, were established by our lab (Yang et al., 2021), and the HMM file (PF03106) of the WD was obtained from the Pfam database.1 To comprehensively identify the CsWRKYs, HMMER 3.02 software was adopted to search against the C. sinense protein sequences with default parameters (E-value cut-off <1E-5). The incorrect and redundant predicted sequences were manually removed, and then all putative CsWRKY genes were further verified using NCBI’s Conserved Domain Database.3 The MW and PI were evaluated using ExPASy-ProtParam online software.4 Meanwhile, the protein sequences of A. thaliana and O. sativa were downloaded from TAIR5 and Rice Genome Annotation Project,6 respectively.
Phylogenetic analysis of Cymbidium sinense WRKY TFs
Multiple sequence alignment of the WDs and full-length proteins was conducted by CLUSTAL (gap opening penalty: 10 and gap extension penalty: 0.2; Larkin et al., 2007). The phylogenetic tree was reconstructed from 1,000 ultrafast bootstrap ML tree replicates using IQ-TREE v1.6.12 (Nguyen et al., 2015) with best-fit model selection (PMB + I + G4) by ModelFinder (Kalyaanamoorthy et al., 2017). The ML phylogenetic tree was visualized using Evolview v3 (Subramanian et al., 2019).
Characterization of the structure and motif of CsWRKYs
The available information of exons and introns was retrieved from the C. sinense genome sequences and then visualized by the TBtools v1.098725 (Chen et al., 2020) with coding sequences and genomic sequences. The MW and pI of the putative WRKY proteins were calculated by the ExPASy proteomics server.7 The motifs of each deduced CsWRKY protein were analyzed by MEME suite software8 (Bailey et al., 2009) with parameters as follows: the maximum number of motifs, 10. The upstream 2-Kb sequences of WRKY genes were extracted to determine the cis-elements by using the PlantCARE database.9
Comparative genome synteny analysis
The alignment was performed by LASTZ using CDS sequences of C. sinense and five representative species (A. thaliana, Oryza sativa, Vitis vinifera, Musa acuminate, and Zea mays). The syntenic block map was established by MCscan with cscore = 0.7. Each orthologous gene pair was then further investigated with PAL2NAL10 (Suyama et al., 2006) to determine the Ks (synonymous substitution rate) and Ka (non-synonymous substitution rate).
Transcriptomic data sets to analyze the expression patterns of WRKYs
To evaluate gene expression profiles of CsWRKYs, the expression patterns of CsWRKY genes in five different tissues were identified, such as roots, stems, leaves, flowers, and fruits. The transcriptome data of all tissues were came from our previous report (Yang et al., 2021). The expression values were calculated by log2 (FPKM) and displayed as a heat map generated using TBtools.
Subcellular localization of CsWRKY proteins
The subcellular localization was predicted using the online software WOLF PSORT.11
RNA extraction, cDNA synthesis, and qRT-PCR analysis
The above leaf samples were ground into a fine powder. Total RNA of these samples was extracted using Trizol Reagent (Invitrogen), and then 1 μg RNA was used to synthesize cDNA in a 20-μl reaction system according to the instructions in PrimeScript™ RT reagent Kit with gDNA Eraser (TAKARA). qRT-PCR was carried out on a Bio-Rad iCycler Real-Time PCR Detection System (United States) by TaKaRa SYBR Premix Ex Taq™ (Tli RNaseH Plus) with three biological replications. Briefly, the reaction was conducted in a 20-μl reaction system containing 10 μl SYBR Premix (2×), 1 μl cDNA, 1 μl sense and anti-sense primer (10 μM), and 7 μl ddH2O. After an initial denaturation step at 95°C for 1 min, the amplifications were carried out with 40 cycles at a melting temperature of 95°C for 10 s, an annealing temperature of 56°C for 30 s, and an extension temperature of 72°C for 30 s. Primer pairs were designed using Primer Premier 5.0, and the NCBI blast program was used to identify the specificity of all primers (Supplementary Table S1). The β-actin gene (Mol013347) in C.sinense was used as an internal reference, and the relative expression levels were calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001).
CsWRKY PPI analysis
A functional protein association network was constructed in the STRING program based on the Arabidopsis association model with the confidence parameter of 0.15 and the number of interactions of 5.
Discussion
Genes of the WRKY superfamily, one of the largest groups of TFs in plants, are also present in other eukaryotic lineages, such as flagellated protozoan (Giardia lamblia) and soil-living amoeba-like slime mold (Dictyostelium discoideum; Zhang and Wang, 2005; Phukan et al., 2016). WRKY family plays a vital functional Progress in gene identification and functional evaluation of WRKY family in plant germination to senescence or seed production in the whole plant’s life cycle. WRKY TFs can enhance tolerance against heat, freezing, drought, salt, cadmium stress, UV radiation, and disease-causing organisms in different crop species, including Arabidopsis, Chrysanthemum, cucumber, peanut, pepper, rice, tobacco, tomato, and wheat (Dang et al., 2013; Liang et al., 2017; Chakraborty et al., 2018; Gao et al., 2018; Han et al., 2018, 2019; Hong et al., 2018; Khan et al., 2018; Kiranmai et al., 2018; Liu et al., 2018; Luan et al., 2019; Hussain et al., 2021).
However, compared with other crops, knowledge of the WRKY gene family has remained relatively scarce in the orchid field due to the relatively backward release of a complete and high-quality genome. In the present study, we identified 64 WRKY genes from the C. sinense genome. All CsWRKY were also classified into seven groups (groups I, IIa, IIb, IIc, IId, IIe, and III). Possibly since the green alga Chlamydomonas reinhardtii contains only one Group I WRKY gene (Zhang and Wang, 2005), Group I WRKY genes are considered the ancestors of other WRKY genes. In C. sinense, as many monocotyledonous species occur domain loss events (Ross et al., 2007; Wei et al., 2012; Brand et al., 2013), the 15 members of Group I contained 27 WDs. Subgroup II genes exist early in the evolution of plants (Hori et al., 2014). Interestingly, CsWRKY47 in Group IIa had unique sequences in the WD (WKKYGQK) except for CsWRKY03 and CsWRKY63 in Group I. The WD sequence variation may influence the normal interactions and binding specificities with downstream target genes (van Verk et al., 2008; Zhou et al., 2008). Recently, a great quantity of Group I and Group II functions have been identified. For example, CitWRKY28 (Group I) and CitNAC029 can promote the accumulation of cuticular wax in Arabidopsis leaves (Yang et al., 2022). In Dioscorea composita, the expression of DcWRKY3 (Group I) is strongly affected by salt stress (Yu et al., 2022). The transcription level of CsWRKY25 (Group I) is up-regulated in Penicillium digitatum-infected citrus peels (Wang et al., 2022b). PcWRKY11 (Group II WRKY from Polygonum cuspidatum) significantly increases the tolerance to salt stress in transgenic A. thaliana (Wang et al., 2022a).
Compared with Group I and II WRKY TFs, Group III TFs alter C2H2 to C2HC zinc-finger motif C-X7-C-X26-HTC. It has been considered the most adaptable and most advanced in monocot evolution (Eulgem et al., 2000; Zhang and Wang, 2005). In addition, Group III is the youngest in the WRKY family (Song and Gao, 2014; Wu et al., 2016). Therefore, in numerous species, Group III is the key to determining the number of WRKY genes. Nevertheless, Group III WRKY genes are considered the most vital group in gene family evolution and seemingly play an important role in the adaptation and evolution of plants (Zhang and Wang, 2005; Goel et al., 2016; Shan et al., 2021; Khan et al., 2022). In C. sinense, 11 CsWRKY gene members belonged to Group III, and such a number is fewer than the corresponding number in most other plants, such as 13 in Arabidopsis (Kalde et al., 2003), 28 in rice (Wang et al., 2015), 35 in maize (Hu et al., 2021), 62 in Dendrobium catenatum (Zhang et al., 2022b), 57 in Phalaenopsis, and 43 in Apostasia (unpublished). These findings suggested the functional conservation of WRKY genes during orchid evolution.
We analyzed the function of CsWRKY18 and indicated its role in the ABA pathway and abiotic stress. Different abiotic stresses can induce AtWRKY30. Meanwhile, the overexpression of AtWRKY30 greatly enhances the resistance of Arabidopsis in response to salt stress (Scarpeci et al., 2013) and improves heat and drought stress tolerance in wheat (El-Esawi et al., 2019). Moreover, AtWRKY30 expression is induced by oxidative stress treatment, fungal elicitor, mosaic virus, JA, auxin, SA, and ABA (Jiang et al., 2017; Zou et al., 2019). Evolutionary processes including duplication events and chromosomal set changes (polyploidy) could extend the members of a gene family in plants (Faraji et al., 2021; Heidari et al., 2021). Besides, it seems that WRKY gene family has been subjected to more evolutionary pressures and extended. However, evolutionary events could increase the members of gene family, but point mutations in coding site regions and upstream/downstream site of new members can affect the function of new members and cause diversity of members expression (Abdullah et al., 2021; Musavizadeh et al., 2021). Tandem duplication event as a 200 kb chromosome region containing two or more genes (Holub, 2001). All the 13 CsWRKY gene pairs among 64 WRKY genes were identified as segmental duplication, but no tandem duplication, indicating that tandem duplication events might have not articipated in the amplification of CsWRKY gene family. More functions of CsWRKYs should be further studied.
Furthermore, six CsWRKYs were subdivided into one subclass IIIa, and five CsWRKYs were classified into another subclass IIIb. OsWRKY45 (Group IIIa) is an essential positive player in the resistance against the rice blast fungus M. oryzae (Shimono et al., 2007). Various abiotic stresses can induce AtWRKY30 (Group IIIb). Meanwhile, the overexpression of AtWRKY30 significantly improves the resistance of Arabidopsis in response to salt stress (Scarpeci et al., 2013). These years, WRKY-c311842 TFs play key roles in berberine accumulation in Coptis chinensis (Liu et al., 2022). As an essential plant hormone in plants, ABA plays a vital role in development, physiological processes, and stress responses, and the ABA signaling pathway is an important component of the stress regulation network (Li et al., 2017, 2018; Sun et al., 2018). Several WRKY TFs have been reported to be positive regulators of ABA-mediated stomatal closure, while some are negative regulators of seed germination and can also indirectly control flowering (Rushton et al., 2012). The ectopic expressions of TaWRKY75-A and ZmWRKY79 in Arabidopsis improve the survival rate under drought stress by regulating ABA biosynthesis (Gulzar et al., 2021; Ye et al., 2021). OsWRKY50 enhances salt stress tolerance via an ABA-independent pathway (Huang et al., 2021). In order to explore the CsWRKY Group III gene family, we elucidated the function of CsWRKY18 in drought stress through regulating ABA biosynthesis.
In summary, we identified 64 WRKY genes in C. sinense genome and analyzed their expression patterns in response to GA, ABA, SA, IAA, JA, and ACC treatments and their expression profiles in the leaves of C. sinense. The ectopic expression of CsWRKY18 in Arabidopsis improved the survival rate under drought stress, suggesting that CsWRKY18 was a regulator in ABA biosynthesis.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found at: NCBI, PRJNA743748.
Author contributions
G-FZ and F-XY designed the experiments and edited the manuscript. Y-LW, J-PJ, and JG executed the experiments and assembled the figures. C-QL, DL, JL, and QX conducted the qRT-PCR and PPI. F-XY, J-PJ, and Y-LW wrote the paper with inputs from other authors. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Natural Science Foundation of Guangdong province (2017A030312004), grants from the National Key R&D Program (2018YFD1000404 and 2019YFD1001003), the National Natural Science Foundation of China (31902065), the Science and Technology Project of Guangdong Province (2019B030316033), Guangzhou Science and Technology Project (201707010307 and 202002030016), Innovation Team of Modern Agriculture Industry Technology System in Guangdong Province (2021KJ121), and Guangdong Academy of Agricultural Sciences Discipline Team Construction Project (202127TD and BZ202006).
Acknowledgments
We thank the engineers of Nextomics Biosciences Co., Ltd. and PubBio Co., Ltd. for technical assistance in data processing and bioinformatics analysis and the Yuandong Chinese Orchid Corporation for plant cultivation. We are grateful to Da Luo of Sun Yat-sen University and Di-Qiu Yu of Yunnan University for their help during the preparation of the article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.969010/full#supplementary-material
Abbreviations
ABA, Abscisic acid; qRT-PCR, Quantitative RT-PCR; WGCNA, Weighted gene co-expression network analysis; WD, WRKY domain; GO, Gene ontology; PPI, Protein–protein interaction; ET, Ethylene; JA, Jasmonic acid; HMM, Hidden Markov Model; NJ, Neighbor-joining; Ks, Synonymous substitution rate; Ka, Non-synonymous substitution rate.
Footnotes
1. ^http://pfam.xfam.org/ (Accessed December 4, 2021).
3. ^https://www.ncbi.nlm.nih.gov/Structure/cdd/ (Accessed December 3, 2021).
4. ^http://web.expasy.org/protparam/ (Accessed January 3, 2022).
5. ^https://www.arabidopsis.org/
6. ^http://rice.plantbiology.msu.edu/ (Accessed November 11, 2021).
7. ^https://web.expasy.org/protparam/ (Accessed January 3, 2022).
8. ^http://meme-suite.org/tools/meme (Accessed January 12, 2022).
9. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (Accessed February 2, 2022).
References
Abdullah,, Faraji, S., Mehmood, F., Malik, H. M. T., Ahmed, I., Heidari, P., et al. (2021). The GASA gene family in cacao (Theobroma cacao, Malvaceae): genome wide identification and expression analysis. Agronomy 11:1425. doi: 10.3390/agronomy11071425
Ai, Y., Li, Z., Sun, W. H., Chen, J., Zhang, D. Y., Ma, L., et al. (2021). The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 8:264. doi: 10.1038/s41438-021-00709-6
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bazin, J., Mariappan, K., Jiang, Y., Blein, T., Voelz, R., Crespi, M., et al. (2020). Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis. PLoS Pathog. 16:e1008401. doi: 10.1371/journal.ppat.1008401
Brand, L. H., Fischer, N. M., Harter, K., Kohlbacher, O., and Wanke, D. (2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41, 9764–9778. doi: 10.1093/nar/gkt732
Brown, P. M. (2005). Wild Orchids of Florida: With References to the Atlantic and Gulf Coastal Plains. Gainesville: University Press of Florida.
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W. C., Liu, K. W., et al. (2015). The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72. doi: 10.1038/ng.3149
Chakraborty, J., Ghosh, P., Sen, S., and Das, S. (2018). Epigenetic and transcriptional control of chickpea WRKY40 promoter activity under Fusarium stress and its heterologous expression in Arabidopsis leads to enhanced resistance against bacterial pathogen. Plant Sci. 276, 250–267. doi: 10.1016/j.plantsci.2018.07.014
Chao, Y. T., Chen, W. C., Chen, C. Y., Ho, H. Y., Yeh, C. H., Kuo, Y. T., et al. (2018). Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 16, 2027–2041. doi: 10.1111/pbi.12936
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Hu, Y., Vannozzi, A., Wu, K. C., Cai, H. Y., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Clough, S. J., and Bent, A. F. (1998). Floral dip:a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343
Dang, F. F., Wang, Y. N., Yu, L., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Di, P., Wang, P., Yan, M., Han, P., Huang, X. Y., Yin, L., et al. (2021). Genome-wide characterization and analysis of WRKY transcription factors in Panax ginseng. BMC Genomics 22:834. doi: 10.1186/s12864-021-08145-5
Ding, M., Chen, J., Jiang, Y., Lin, L., Cao, Y., Wang, M., et al. (2015). Genome-wide investigation and transcriptome analysis of the WRKY gene family in Gossypium. Mol. Gen. Genomics. 290, 151–171. doi: 10.1007/s00438-014-0904-7
El-Esawi, M. A., Al-Ghamdi, A. A., Ali, H. M., and Ahmad, M. (2019). Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.). Genes 10:13. doi: 10.3390/genes10020163
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Faraji, S., Heidari, P., Amouei, H., Filiz, E., Abdullah,, and Poczai, P. (2021). Investigation and computational analysis of the Sulfotransferase (SOT) gene family in potato (Solanum tuberosum): insights into sulfur adjustment for proper development and stimuli responses. Plan. Theory 10:2597. doi: 10.3390/plants10122597
Feng, C., He, C., Wang, Y., Xu, H., Xu, K., Zhao, Y., et al. (2021). Genome-wide identification of soybean shaker K+ channel gene family and functional characterization of GmAKT1 in transgenic Arabidopsis thaliana under salt and drought stress. J. Plant Physiol. 266:153529. doi: 10.1016/j.jplph.2021.153529
Gao, H. M., Wang, Y. F., Xu, P., and Zhang, Z. B. (2018). Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 9:997. doi: 10.3389/fpls.2018.00997
Goel, R., Pandey, A., Trivedi, P. K., and Asif, M. H. (2016). Genome-wide analysis of the Musa WRKY gene family: evolution and differential expression during development and stress. Front. Plant Sci. 7:299. doi: 10.3389/fpls.2016.00299
Gulzar, F., Fu, J. Y., Zhu, C. Y., Yan, J., Li, X. L., Meraj, T. A., et al. (2021). Maize WRKY Transcription factor ZmWRKY79 positively regulates drought tolerance through elevating ABA biosynthesis. Int. J. Mol. Sci. 22:19. doi: 10.3390/ijms221810080
Han, D. G., Ding, H. B., Chai, L. J., Liu, W., Zhang, Z. Y., Hou, Y. J., et al. (2018). Isolation and characterization of MbWRKY1, a WRKY transcription factor gene from Malus baccata (L.) Borkh involved in drought tolerance. Can. J. Plant Sci. 98, 1023–1034. doi: 10.1139/cjps-2017-0355
Han, Y. Y., Fan, T. T., Zhu, X. Y., Wu, X., Ouyang, J., Jiang, L., et al. (2019). WRKY12 represses GSH1 expression to negatively regulate cadmium tolerance in Arabidopsis. Plant Mol. Biol. 99, 149–159. doi: 10.1007/s11103-018-0809-7
Hao, F., Yang, G., Zhou, H. J., Yao, J. J., Liu, D. R. L., Zhao, P., et al. (2021). Genome-wide identification and transcriptional expression profiles of transcription factor WRKY in common walnut (Juglans regia L.). Genes 12:19. doi: 10.3390/genes12091444
Hasing, T., Tang, H. B., Brym, M., Khazi, F., Huang, T. F., and Chambers, A. H. (2020). A phased Vanilla planifolia genome enables genetic improvement of flavour and production. Nature Food 1, 811–819. doi: 10.1038/s43016-020-00197-2
Heidari, P., Abdullah,, Faraji, S., and Poczai, P. (2021). Magnesium transporter gene family: genome-wide identification and characterization in Theobroma cacao, Corchorus capsularis, and Gossypium hirsutum of family Malvaceae. Agronomy 11:1651. doi: 10.3390/agronomy11081651
Holub, E. B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. doi: 10.1038/35080508
Hong, Y. H., Cui, J., Liu, Z., and Luan, Y. S. (2018). SpWRKY6 acts as a positive regulator during tomato resistance to Phytophthora infestans infection. Biochem. Biophys. Res. Commun. 506, 787–792. doi: 10.1016/j.bbrc.2018.10.155
Hori, K., Maruyama, F., Fujisawa, T., Togashi, T., Yamamoto, N., Seo, M., et al. (2014). Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 5:3978. doi: 10.1038/ncomms4978
Hu, W., Ren, Q., Chen, Y., Xu, G., and Qian, Y. (2021). Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 21:427. doi: 10.1186/s12870-021-03206-z
Huang, S. Z., Hu, L. J., Zhang, S. H., Zhang, M. X., Jiang, W. Z., Wu, T., et al. (2021). Rice OsWRKY50 mediates ABA-dependent seed germination and seedling growth, and ABA-independent salt stress tolerance. Int. J. Mol. Sci. 22:12. doi: 10.3390/ijms22168625
Hussain, A., Khan, M. I., Albaqami, M., Mahpara, S., Noorka, I. R., Ahmed, M. A. A., et al. (2021). CaWRKY30 positively regulates pepper immunity by targeting CaWRKY40 against Ralstonia solanacearum inoculation through modulating defense-related genes. Int. J. Mol. Sci. 22:18. doi: 10.3390/ijms222112091
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/bf00282746
Jiang, J. J., Ma, S. H., Ye, N. H., Jiang, M., Cao, J. S., and Zhang, J. H. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Kalde, M., Barth, M., Somssich, I. E., and Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant-Microbe Interact. 16, 295–305. doi: 10.1094/MPMI.2003.16.4.295
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kan, J. H., Gao, G. Q., He, Q., Gao, Q., Jiang, C. C., Ahmar, S., et al. (2021). Genome-wide characterization of WRKY transcription factors revealed gene duplication and diversification in populations of wild to domesticated barley. Int. J. Mol. Sci. 22:17. doi: 10.3390/ijms22105354
Khan, M. A., Kang, D. R., Wu, Y. F., Wang, Y., Ai, P. H., and Wang, Z. C. (2022). Characterization of WRKY gene family in whole-genome and exploration of flowering improvement genes in Chrysanthemum lavandulifolium. Front. Plant Sci. 13:861193. doi: 10.3389/fpls.2022.861193
Khan, M. I., Zhang, Y. W., Liu, Z. Q., Hu, J., Liu, C. L., Yang, S., et al. (2018). CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 19:17. doi: 10.3390/ijms19051403
Kiranmai, K., Rao, G. L., Pandurangaiah, M., Nareshkumar, A., Reddy, V. A., Lokesh, U., et al. (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 9:346. doi: 10.3389/fpls.2018.00346
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leitch, I. J., Kahandawala, I., Suda, J., Hanson, L., Ingrouille, M. J., Chase, M. W., et al. (2009). Genome size diversity in orchids: consequences and evolution. Ann. Bot. 104, 469–481. doi: 10.1093/aob/mcp003
Li, W., De Ollas, C., and Dodd, I. C. (2018). Long-distance ABA transport can mediate distal tissue responses by affecting local ABA concentrations. J. Integr. Plant Biol. 60, 16–33. doi: 10.1111/jipb.12605
Li, K., Yang, F., Miao, Y., and Song, C. P. (2017). Abscisic acid signaling is involved in regulating the mitogen-activated protein kinase cascade module, AIK1-MKK5-MPK6. Plant Signal. Behav. 12:e1321188. doi: 10.1080/15592324.2017.1321188
Liang, Q. Y., Wu, Y. H., Wang, K., Bai, Z. Y., Liu, Q. L., Pan, Y. Z., et al. (2017). Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci. Rep. 7:4799. doi: 10.1038/s41598-017-05170-x
Liu, Z., Chen, S., Chen, S., and Chen, S. (2006). The Genus CYMBIDIUM in China. The Genus Cymbidium in China: Science Press.
Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., et al. (2012). Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012:251364. doi: 10.1155/2012/251364
Liu, Q., Li, X., Yan, S. J., Yu, T., Yang, J. Y., Dong, J. F., et al. (2018). OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice. BMC Plant Biol. 18:257. doi: 10.1186/s12870-018-1479-y
Liu, X. M., Tan, J. P., Cheng, S. Y., Chen, Z. X., Ye, J. B., Zheng, J. R., et al. (2022). Comparative transcriptome analysis provides novel insights into the molecular mechanism of berberine biosynthesis in Coptis chinensis. Sci. Hortic. 291:110585. doi: 10.1016/j.scienta.2021.110585
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, Q. Q., Chen, C. H., Liu, M. Y., Li, Q., Wang, L. N., and Ren, Z. (2019). CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci. 279, 59–69. doi: 10.1016/j.plantsci.2018.11.002
Maeo, K., Hayashi, S., Kojima-Suzuki, H., Morikami, A., and Nakamura, K. (2001). Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem. 65, 2428–2436. doi: 10.1271/bbb.65.2428
Musavizadeh, Z., Najafi-Zarrini, H., Kazemitabar, S. K., Hashemi, S. H., Faraji, S., Barcaccia, G., et al. (2021). Genome-wide analysis of potassium channel genes in rice: expression of the OsAKT and OsKAT genes under salt stress. Genes 12:784. doi: 10.3390/genes12050784
Nekrutenko, A., Makova, K. D., and Li, W. H. (2002). The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 12, 198–202. doi: 10.1101/gr.200901
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Niu, Z. T., Zhu, F., Fan, Y. J., Li, C., Zhang, B. H., Zhu, S. Y., et al. (2021). The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm. Sin. B 11, 2080–2092. doi: 10.1016/j.apsb.2021.01.019
Okay, S., Derelli, E., and Unver, T. (2014). Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol. Gen. Genomics. 289, 765–781. doi: 10.1007/s00438-014-0849-x
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Rasmussen, M. W., Roux, M., Petersen, M., and Mundy, J. (2012). MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 3:169. doi: 10.3389/fpls.2012.00169
Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Ross, C. A., Liu, Y., and Shen, Q. J. (2007). The WRKY gene family in Rice (Oryza sativa). J. Integr. Plant Biol. 49, 827–842. doi: 10.1111/j.1744-7909.2007.00504.x
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Scarpeci, T. E., Zanor, M. I., Mueller-Roeber, B., and Valle, E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 83, 265–277. doi: 10.1007/s11103-013-0090-8
Shan, N., Xiang, Z. J., Sun, J. Y., Zhu, Q. L., Xiao, Y., Wang, P. T., et al. (2021). Genome-wide analysis of valine-glutamine motif-containing proteins related to abiotic stress response in cucumber (Cucumis sativus L.). BMC Plant Biol. 21:492. doi: 10.1186/s12870-021-03242-9
Shimono, M., Sugano, S., Nakayama, A., Jiang, C. J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
Song, Y., and Gao, J. (2014). Genome-wide analysis of WRKY gene family in Arabidopsis lyrata and comparison with Arabidopsis thaliana and Populus trichocarpa. Chin. Sci. Bull. 59, 754–765. doi: 10.1007/s11434-013-0057-9
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Sun, L. R., Wang, Y. B., He, S. B., and Hao, F. S. (2018). Mechanisms for Abscisic acid inhibition of primary root growth. Plant Signal. Behav. 13:e1500069. doi: 10.1080/15592324.2018.1500069
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi: 10.1093/nar/gkl315
van Verk, M. C., Pappaioannou, D., Neeleman, L., Bol, J. F., and Linthorst, H. J. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. doi: 10.1104/pp.107.112789
Wang, Y., Feng, L., Zhu, Y., Li, Y., Yan, H., and Xiang, Y. (2015). Comparative genomic analysis of the WRKY III gene family in populus, grape, arabidopsis and rice. Biol. Direct 10:48. doi: 10.1186/s13062-015-0076-3
Wang, W., Li, T., Chen, Q., Yao, S., Deng, L., and Zeng, K. (2022b). CsWRKY25 improves resistance of Citrus fruit to Penicillium digitatum via modulating reactive oxygen species production. Front. Plant Sci. 12:818198. doi: 10.3389/fpls.2021.818198
Wang, G., Wang, X., Ma, H., Fan, H., Lin, F., Chen, J., et al. (2022a). PcWRKY11, an II-d WRKY transcription factor from Polygonum cuspidatum, enhances salt tolerance in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23:4357. doi: 10.3390/ijms23084357
Wei, K. F., Chen, J., Chen, Y. F., Wu, L. J., and Xie, D. X. (2012). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19, 153–164. doi: 10.1093/dnares/dsr048
Wu, J., Chen, J. B., Wang, L. F., and Wang, S. M. (2016). Genome-wide investigation of WRKY transcription factors involved in terminal drought stress response in common bean. Front. Plant Sci. 8:380. doi: 10.3389/fpls.2017.00380
Wu, K. L., Guo, Z. J., Wang, H. H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Xie, Z., Zhang, Z. L., Zou, X. L., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Xu, Y. X., Lei, Y. T., Su, Z. X., Zhao, M., Zhang, J. X., Shen, G. J., et al. (2021). A chromosome-scale Gastrodia elata genome and large-scale comparative genomic analysis indicate convergent evolution by gene loss in mycoheterotrophic and parasitic plants. Plant J. 108, 1609–1623. doi: 10.1111/tpj.15528
Yang, F. X., Gao, J., Wei, Y. L., Ren, R., Zhang, G. Q., Lu, C. Q., et al. (2021). The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 19, 2501–2516. doi: 10.1111/pbi.13676
Yang, K., Li, Y., Wang, S., Xu, X., Sun, H., Zhao, H., et al. (2019). Genome-wide identification and expression analysis of the MYB transcription factor in moso bamboo (Phyllostachys edulis). PeerJ 6:e6242. doi: 10.7717/peerj.6242
Yang, H., Zhu, Z., Zhang, M., Li, X., Xu, R., Zhu, F., et al. (2022). CitWRKY28 and CitNAC029 promote the synthesis of cuticular wax by activating CitKCS gene expression in citrus fruit. Plant Cell Rep. 41, 905–920. doi: 10.1007/s00299-021-02826-x
Ye, H., Qiao, L., Guo, H., Guo, L., Ren, F., Bai, J., et al. (2021). Genome-wide identification of wheat WRKY gene family reveals That TaWRKY75-A is referred to drought and salt resistances. Front. Plant Sci. 12:663118. doi: 10.3389/fpls.2021.663118
Yu, S. J., Lan, X., Zhou, J. C., Gao, K. X., Zhong, C. M., and Xie, J. (2022). Dioscorea composita WRKY3 positively regulates salt-stress tolerance in transgenic Arabidopsis thaliana. J. Plant Physiol. 269:153592. doi: 10.1016/j.jplph.2021.153592
Yuan, H. M., Guo, W. D., Zhao, L. J., Yu, Y., Chen, S., Tao, L., et al. (2021). Genome-wide identification and expression analysis of the WRKY transcription factor family in flax (Linum usitatissimum L.). BMC Genomics 22:375. doi: 10.1186/s12864-021-07697-w
Zhang, M., Chen, Y., Nie, L., Jin, X., Liao, W., Zhao, S., et al. (2018). Transcriptome-wide identification and screening of WRKY factors involved in the regulation of taxol biosynthesis in Taxus chinensis. Sci. Rep. 8:5197. doi: 10.1038/s41598-018-23558-1
Zhang, G. Q., Liu, K. W., Li, Z., Lohaus, R., Hsiao, Y. Y., Niu, S. C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549, 379–383. doi: 10.1038/nature23897
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. doi: 10.1186/1471-2148-5-1
Zhang, T. T., Xu, Y., Ding, Y. D., Yu, W. A., Wang, J., Lai, H. G., et al. (2022b). Identification and expression analysis of WRKY gene family in response to abiotic stress in Dendrobium catenatum. Front. Genet. 13:800019. doi: 10.3389/fgene.2022.800019
Zhang, H., Zhang, L., Ji, Y., Jing, Y., Li, L., Chen, Y., et al. (2022a). Arabidopsis SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2 inhibit WRKY75 function in abscisic acid-mediated leaf senescence and seed germination. J. Exp. Bot. 73, 182–196. doi: 10.1093/jxb/erab391
Zhang, W. X., Zhang, G. Q., Zeng, P., Zhang, Y. Q., Hu, H., Liu, Z. J., et al. (2021a). Genome sequence of Apostasia ramifera provides insights into the adaptive evolution in orchids. BMC Genomics 22:536. doi: 10.1186/s12864-021-07852-3
Zhang, Y., Zhang, G. Q., Zhang, D., Liu, X. D., Xu, X. Y., Sun, W. H., et al. (2021b). Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic Res 8:183. doi: 10.1038/s41438-021-00621-z
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Keywords: Cymbidium sinense, genome-wide, WRKY, expression pattern, abiotic stress
Citation: Wei Y-L, Jin J-P, Liang D, Gao J, Li J, Xie Q, Lu C-Q, Yang F-X and Zhu G-F (2022) Genome-wide identification of Cymbidium sinense WRKY gene family and the importance of its Group III members in response to abiotic stress. Front. Plant Sci. 13:969010. doi: 10.3389/fpls.2022.969010
Edited by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanReviewed by:
Yavar Vafaee, University of Kurdistan, IranParviz Heidari, Shahrood University of Technology, Iran
Copyright © 2022 Wei, Jin, Liang, Gao, Li, Xie, Lu, Yang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gen-Fa Zhu, Z2VuZmF6aHVAMTYzLmNvbQ==; Feng-Xi Yang, ZmVuZ3hpX3dlaUBzaW5hLmNvbQ==
 Yong-Lu Wei
Yong-Lu Wei Jian-Peng Jin
Jian-Peng Jin Di Liang
Di Liang Jie Gao
Jie Gao Jie Li
Jie Li Feng-Xi Yang
Feng-Xi Yang Gen-Fa Zhu
Gen-Fa Zhu