
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 10 August 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.967607
This article is part of the Research Topic The Role of Stress Proteins in Plants Under Abiotic Stress View all 5 articles
Plants dynamically manipulate their gene expression in acclimation to the challenging environment. Hereinto, the histone methylation tunes the gene transcription via modulation of the chromatin accessibility to transcription machinery. Osmotic stress, which is caused by water deprivation or high concentration of ions, can trigger remarkable changes in histone methylation landscape and genome-wide reprogramming of transcription. However, the dynamic regulation of genes, especially how stress-inducible genes are timely epi-regulated by histone methylation remains largely unclear. In this review, recent findings on the interaction between histone (de)methylation and osmotic stress were summarized, with emphasis on the effects on histone methylation profiles imposed by stress and how histone methylation works to optimize the performance of plants under stress.
Post-translational modification (PTM) is a critical regulatory mechanism of the cellular organisms, which governs all aspects of the genomic functionality from DNA (deoxyribonucleic acid) replication, to mRNA (messenger ribonucleic acid) production, and to protein turnover, enabling them to dynamically respond to developmental cues and environmental fluctuations (Ytterberg and Jensen, 2010; Hashiguchi and Komatsu, 2017; Kosová et al., 2021). In eukaryotic cells, the octameric nucleosome consists of two molecules of H2A, H2B, H3, and H4 each, which are further wrapped by 147 base pairs of DNA. The occupation of histones and modifications on the core histones primarily determines the nucleosome density and chromatin configuration, which further controls the accessibility to a repertoire of transcription factors (Jenuwein and Allis, 2001; Shahbazian and Grunstein, 2007; Deal and Henikoff, 2011; Klemm et al., 2019). Methylation on the N-terminal of histones is an important covalent modification that dictates the gene transcription programs to combat the environmental stresses (Kim et al., 2008, 2010, 2015; Yuan et al., 2013; Van Oosten et al., 2014; Asensi-Fabado et al., 2017; Wong et al., 2017). Furthermore, the methyl-groups are dynamically added to or removed from the histones, allowing precise transcriptional manipulation (Pfluger and Wagner, 2007; Kim et al., 2008; Deal and Henikoff, 2011; Xiao et al., 2016; Park et al., 2018).
Histones can be methylated at the lysine residues and the arginine residues, which are catalyzed by histone lysine methyltransferases (HKMT) and histone arginine methyltransferases (HRMT), respectively. Histone lysine methylation is specifically catalyzed by the SET-Domain Group proteins (SDGs), which can be divided into seven subfamilies including E(z), Ash, Trx, Suv, PRDM, SMYD, and SETD (Springer et al., 2003; Zhou et al., 2020). E(z), Ash, Trx, and Suv are four major subfamilies involved in the methylation of histones (Springer et al., 2003). In plants, most characterized histone lysine methylation occurs at H3K4, H3K9, H3K27 and H3K36 while the methylation on H3K79 is rare. The histone arginine methylation primarily occurs on H3R2, H3R8, H3R17, H3R26, and H4R3 (Feng et al., 2002; Niu et al., 2007; Pfluger and Wagner, 2007; Wu et al., 2009; Liu et al., 2010; Berr et al., 2011; Huang et al., 2014, 2015; Zacarias and Casas-Mollano, 2021). Histone methylation can serve either positive or negative functions in gene transcription. Methylation on H3K9, H3K27, and H4R3 marks transcriptional repression. By contrast, methylation on H3K4 and H3K36 is labelled with transcriptional activation (Yu et al., 2009; Wang et al., 2015a; Zacarias and Casas-Mollano, 2021). The histone lysine methylation and arginine methylation together build histone hallmarks that can be interpreted into transcription activation or repression. The histone methylation is a reversible process where methyl groups can be added onto specific residues by histone methyltransferases/methylases and be removed by histone demethylases, which plays vital roles in maintaining the homeostasis of histone methylation (Klose and Zhang, 2007; Liu et al., 2010). In higher plants, there are two distinct histone demethylation mechanisms: amine oxidation by LSD1 (lysine-specific demethylase1) which specifically demethylates H3K4 and hydroxylation by JmjC proteins (Jumonji domain-containing) with broader specificity (Shi et al., 2004; Tsukada et al., 2006; Williams et al., 2014; Qian et al., 2015).
Plants have sessile lifestyle and their vitality is stringently dependent on their growth conditions (Frolov et al., 2017). Osmotic stresses (OS), as a major common abiotic stress limiting the crop yield, are most likely to occur periodically (Ault, 2020). Drought and salinity are two representatives of osmotic stress, which are non-ionic and ionic, respectively, (Munns et al., 2010). Drought is one of the most devastating abiotic stresses in agronomy and caused by water deficit (Ault, 2020). Several other abiotic environmental factors, such as high salinity and reduced or elevated temperatures, also decrease water availability, leading to the onset of osmotic response (Verslues et al., 2006; Frolov et al., 2017). The molecular basis underpinning response to osmotic stress has been a hot topic in plant stress biology. In certain ecosystems, plants adapt to environmental stresses by different mechanism including epigenetic regulation (Granot et al., 2009; Kim et al., 2010; Balao et al., 2018). As the drought stress occurs, plants correspondingly shape their histone methylome including H3K4me2, H3K9me2/3 and H3K27me2/3. Moreover, this epigenetic mechanism seems to be associated with their living surroundings and species-specific (Granot et al., 2009). These findings evidence that plants can respond to the environment on the chromatin level. Herein we summarized recent studies on the histone methylation regulation upon osmotic stress (Table 1), discussed the underlying mechanism (Figure 1), pinpointed several questions to be addressed and postulated the perspectives of study on histone methylation.
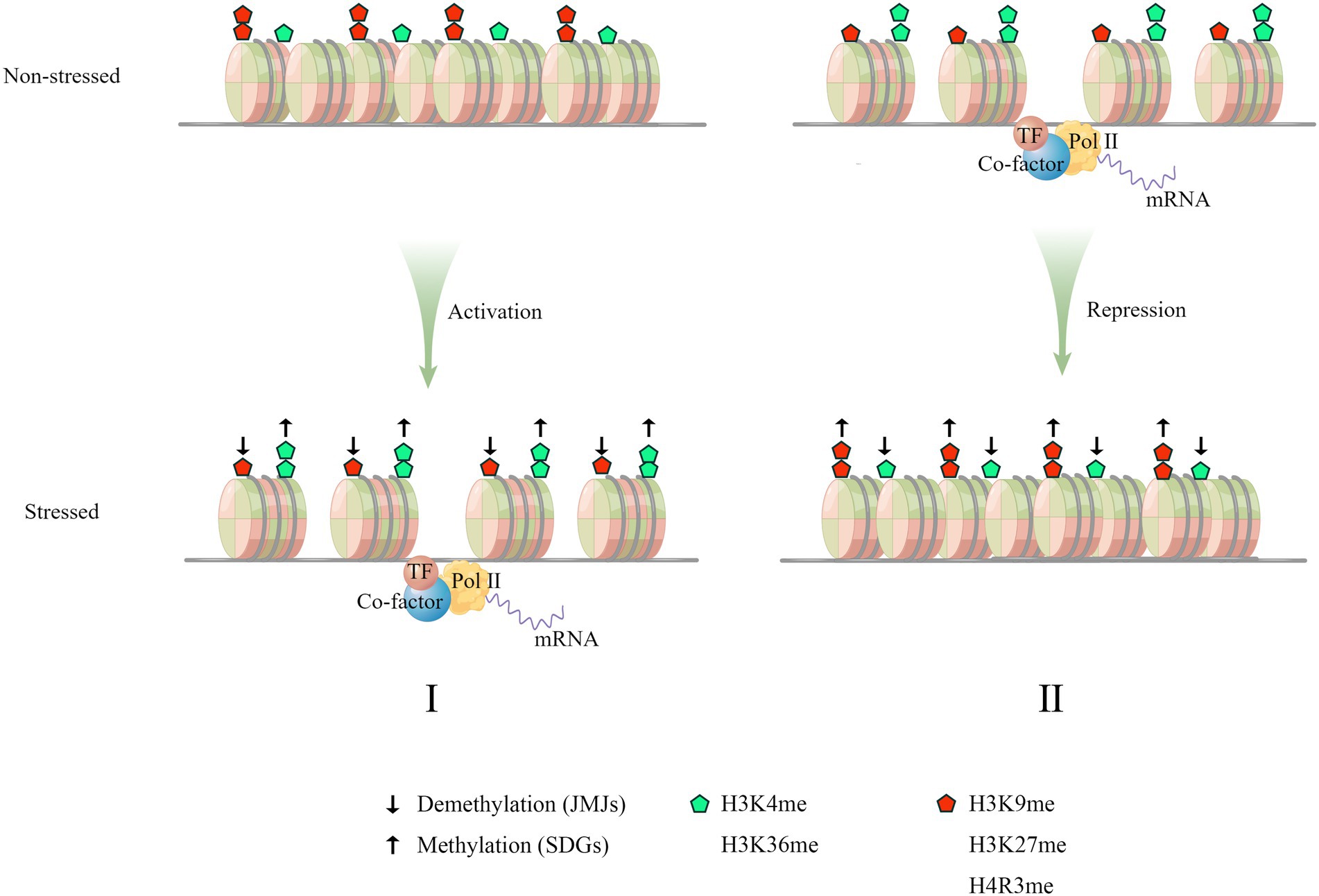
Figure 1. A paradigm of stress-induced histone hallmarks and gene transcription. Osmotic stress induces both transcriptional activation (I) and repression of stress-responsive genes (II). These two mechanisms collaboratively orchestrate the stress response by modulating gene transcription. (I) Under non-stressful conditions, the chromatin region of genes positively regulating stress-response is enriched in repressive histone methylations (the red pentagon) and has higher nucleosome occupancy, which decreases the chromatin accessibility to the transcription machinery. Upon stress, the chromatin region is marked with more active histone methylations (the green pentagon) and bears fewer nucleosomes, leading to increased accessibility and transcriptional activation. (II) As for the genes negatively regulating stress response, the corresponding genic loci are marked with higher active histone methylations and lower repressive histone methylations. The active transcription helps to repress inappropriate stress-response. Upon stress, the deposition of active markers is reduced while the repressive markers are deposited, which leads to transcriptional repression. This negative regulation counteracts the repression of stress response imposed by negative genes in stress signaling pathway. The depletion of histone methylations is mediated by JMJ demethylase (↓) and the increase of histone methylations is mediated by histone methylases/methyl-transferases (↑), which represents two biological processes termed histone demethylation and histone methylation, respectively. In both cases (I, II), the histone methylation and histone demethylation might be simultaneously involved.
Osmotic stress and stress-triggered abscisic acid (ABA) signaling cast vast effects on the global landscape of histones. In the Arabidopsis genome, about 80% of the annotated genes bear H3K4me1, 84% bear H3K4me2 marks, and 62% bear H3K4me3 mark. In total, up to about 90% of annotated genes are methylated on H3K4 to different extent and a train of dehydration/ABA inducible genes marked by H3K4me3. Though the H3K4me3 peaks around the transcription start site (TSS) of most transcribed genes, dehydration and ABA can induce wider distribution of H3K4me3 profiles. Moreover, H3K4me1 and H3K4me2 are less sensitive to dehydration stress compared to H3K4me3 (van Dijk et al., 2010). In maize (Zea mays) seedlings stressed by salinity and drought, the active methylation mark H3K4me2 remarkably increases while the repressive methylation marks on H3K9 and H3K27 drop, implying a genome-wide gene activation through chromatin state might be involved in osmotic stress (Kamal et al., 2021). In soybean (Glycine max), salt stress induces de novo establishment of H3K27me3 to deactivate gene transcription (Sun et al., 2019). In the tea plants (Camellia sinensis), drought stress decreases the methylation of histone H3K4 (Chen et al., 2021c). These results pinpoint the correlation between stress-induced histone methylation and genome-wide transcription reprogramming.
Stress signals not only induce global changes in histone methylome but also specifically affects the methylation on stress-responsive genes. In Arabidopsis, MYB44 is a highly active gene and a potent repressor of the salt-induced expression of PROTEIN PHOSPHATASE TYPE2 C (PP2C) genes. ABA lowers the nucleosome occupancy and mannitol induces a remarkable increase in H3K4me3 and H3ac both in the promoter and the gene body of AtMYB44 (Nguyen and Cheong, 2018). The ABA-signaling components including PP2C genes (e.g., ABI1, ABI2, and HAI1) are upregulated under salt stress, accompanied with increased active markers H3ac and H3K4me3, and decreased nucleosome density. Under non-stress conditions, a functional SWI2/SNF2 chromatin remodeling ATPase BRAHMA (BRM) is required for the repression of PP2C genes (Nguyen et al., 2019). NCED3 (Nine Cis-Epoxycarotenoid Dioxygenase 3) encodes a rate-limiting enzyme in the pathway of ABA biosynthesis and its transcription is fine-tuned to maintain proper stress response (Qin and Zeevaart, 1999; Iuchi et al., 2001; Tan et al., 2018). In response to osmotic stress, NCED3 expression is dramatically upregulated and induces the ABA biosynthesis. In Arabidopsis, Drought decreases nucleosome occupancy in the RD29A (RESPONSIVE TO DESICCATION 29A) region, and H3K4me3 and H3K9ac deposition at RD20 (RESPONSIVE TO DESICCATION 20) and RD29A to upregulate their expression (Kim et al., 2008). Arabidopsis TRX proteins (ARABIDOPSIS HOMOLOG OF TRITHORAX) including ATX1, ATX3, ATX4, and ATX5, are involved in deposition of H3K4me3 which is an active histone mark (de la Paz Sanchez et al., 2015; Pu and Sung, 2015; Chen et al., 2017; Song et al., 2021). ATX1 positively regulates the expression of NCED3 through trimethylation of H3K4. The expression of RD29A/B is impaired in atx1 mutants. Meanwhile, the stallment of RNA Polymerase II which is a core component of the transcription machinery mirrors the decrease of H3K4me3 (Ding et al., 2011, 2012). Collectively, ATX1 probably plays a multipurpose role in drought response through regulation of genes involved in both ABA biosynthesis and the signaling cascade. Other TRX members, ATX4 and ATX5 function redundantly in drought response. Following the perception of ABA signal, both ATX4 and ATX5 are directed to AHG3 (ABA-HYPERSENSITIVEGERMINATION 3). The association of ATX4 and ATX5 to AHG3 locus increases H3K4me3 deposition and stallment of RNA polymerase II, leading to enhanced expression of AHG3, which finally decreases ABA-sensitivity (Liu et al., 2018b). Thus, ATX homologs might confer opposite stress response through shared biochemical activity but distinctive loci-specificity. These studies together illustrate a H3K4me3-based mechanism by which the ABA/drought response is fine-tuned through modulation of ABA-biosynthesis and ABA signaling (Ding et al., 2011; Liu et al., 2018b; Nguyen et al., 2019).
In addition to active marks including H3K4 methylation, repressive methylation marks on H3K27 and H4R3 also participate in osmotic stress. LHP1 (LIKE HETEROCHROMATIN PROTEIN 1), a component of PRC1 (POLYCOMB REPRESSIVE COMPLEX 1), mediates the H3K27me3 deposition on ABA-responsive genes to exert the transcription repression (Ramirez-Prado et al., 2019). CAU1(CALCIUM UNDERACCUMULATION 1), a H4R3sme2 methylase, binds to the CAS (CALCIUM SENSING RECEPTOR) promoter for H4R3sme2 methylation and consequently represses the expression of CAS to decrease stomatal closure and drought tolerance. However, this process is highly dynamic as elevated Ca2+ induces dissociation of CAU1 from the CAS promoter, further relieving the epigenetic repression effects (Fu et al., 2013). In another study, drought stress downregulates the expression of CAU1, which results in decreased H4R3sme2 in the promoter region and elevated expression of ANAC055(ATAF-LIKE NAC DOMAIN CONTAINING PROTEIN 55). Upregulated ANAC055 promotes proline accumulation through elevated expression of P5CS1(DELTA1-PYRROLINE-5-CARBOXYLATE SYNTHASE 1; Fu et al., 2017). These two studies have uncovered a negative role of H4R3 methylation in drought response through CAS for stomatal aperture and ANAC055 for proline decumulation.
There is mounting evidence that histone methylation is also an important regulatory mechanism of stress response in other plant species. The salt-responsive gene OsBZ8 encoding a bZIP transcription factor bears distinctive H3K27me3 and H3K4me3 profiles both under resting conditions and stressful conditions, which might in part explain the different salt-tolerance across rice varieties (Paul et al., 2017). The rice ARGONAUTE2 (AGO2) is associated with BIG GRAIN3(BG3) locus and increase the H3K4me3 but decrease the H3K27me3. The activation of BG3 transcription leads to enhanced salt response (Yin et al., 2020). OsSDG708, encoding a putative H3K36 methyltransferase, positively regulates drought tolerance by directly targeting and activating the crucial ABA biosynthesis genes OsNCED3 and OsNCED5 in rice. Overexpression of OsSDG708 enhances the drought tolerance and increases the grain yield either, highlighting its potential role epigenetic regulation (Chen et al., 2021b). In dehydrated maize plants, higher active methylation (H3K36me3 and H3K4me3) and lower repressive methylation (H3K27me3) are deposited in the loci of antisense transcripts, indicating biogenesis of small RNAs might be regulated by histone methylation in drought response (Xu et al., 2017). In tomato (Solanum lycopersicum), two repressive histone methylation marks exhibit distinctive response to dehydration. The H3K27me3 barely changes while the H3K9me2 rapidly drops after dehydration (González et al., 2013). The pathogen- and drought-inducible SlSDG33 and SlSDG34, which are homologous to Arabidopsis SDG8, negatively regulate drought tolerance by promoting the H3K4me3 and H3K36me3 deposition on a set of genes with negative regulatory roles in stress tolerance. SlSDG33 and SlSDG35 might act additively in drought response as the double mutant showed superior tolerance to the single mutants (Zong et al., 2022). During the postharvest desiccation process in tea plants, the ABA-biosynthesis genes are activated with elevated histone acetylation and decreased H3K9me2, leading to ABA accumulation (Gu et al., 2021). In salt-stressed castor beans (Ricinus communis), H3K4me3 and H3K27me3 showed a correlation with expression of the stress-regulated genes. RSM1, a MYB-like transcription factor, was under dynamic bivalent control of H3K4me3 (active) and H3K27me3 (repressive) by salt stress (Han et al., 2020). In bread wheat (Triticum aestivum L.), 166 SDG members are identified, 36 of which had incomplete SET domain. During the seedling stage, 30 out of the 166 TaSDGs are downregulated by heat and drought, suggesting they are involved in stress response (Batra et al., 2020). In a Chinese medicinal monocot, the Dendrobium catenatum, 5 out of 42 SDG genes showed altered expression level during drought-recovery (Chen et al., 2020). Taken together, the histone methylation in osmotic stress responses might be a common regulatory mechanism across the plant species.
All the eukaryotic cells have similar chromatin structure which is subject to a set of shared epigenetic modifications. It is reasonable that those modifications are likely conserved to some extent. For instance, ectopic expression of AtHUB2, encoding an Arabidopsis histone mono-ubiquitination E3 ligase, significantly improves the yield of transgenic cotton (Gossypium hirsutum) plants under drought stress conditions and concomitantly enhances the drought tolerance. By contrast, RNAi knockdown of GhHUB2 genes decreased the drought resistance. The ectopically expressed AtHUB2 directly interacts with the endogenous GhH2B to deposit H2Bub1 at a stress-responsive gene GhDREB (DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN), resulting in increased H3K4me3 and gene activation (Chen et al., 2019). Considering the cross-species activity of AtHUB2, the role of H2Bub1 in drought tolerance is conserved in Arabidopsis and cotton plants. Moreover, this epigenetic regulation has promising future as the agronomic traits of transgenic cotton under drought condition were dramatically improved. Ectopic expression of CsSDG36 from the tea plants, which is homologous to ATX4 of Arabidopsis, downregulates stomatal development-related genes, increases stomatal density, and consequently decreases drought tolerance in Arabidopsis (Chen et al., 2021c). As aforementioned, both genes involved in stress-signaling and catalytic enzymes of stress signaling are subject to various histone methylations (Ding et al., 2011; Liu et al., 2018b; Nguyen et al., 2019). Though there are extensive studies revealing the role of histone methylation, their regulatory targets seem to be unpredictable based on the existing literature (as summarized in Table 1). Such phenomenon is likely due to variations in the genomic context and different evolutionary trajectories across the species. Meanwhile, all the epigenetic codes must have their genetic basis which is encoded by the DNA sequence. Development of new methodologies in high-throughput sequencing and advances in bio-informatics of learning the genome-epigenome-phenome relationship will greatly benefit epigenetic studies in crops as well as provide insights into the nucleic acid foundation of histone codes (Li et al., 2022).
Since the aforementioned histone methylation can either be active or repressive, the demethylation also plays different roles in gene transcription activity depending on their target sites and the chromatin context. The SnRKs (SNF1-RELATED PROTEIN KINASE) are important signaling component of osmotic stress. In Arabidopsis, H3K4me3 in the regions of promoter and gene body of SnRK2.6 can be demethylated by JMJ17 (Jumonji Domain-Containing Protein 17). Functional loss of JMJ17 leads to hyper-expression of SnRK2.6, increased ABA-sensitivity, reduced stomatal aperture, and enhanced the tolerance to drought stress (Huang et al., 2019). ABI3 activates the transcription of JMJ30 at post-germination stage. Then JMJ32 and the upregulated JMJ30 together control the ABA-mediated post-germination growth arrest through H3K27me3 demethylation and activation of SnRK2.8 (Wu et al., 2019a,b). JMJ15 preferentially represses the genes marked by H3K4me2 and H3K4me3 probably through histone demethylation and enhances salt tolerance (Shen et al., 2014). Hyper-expression of JMJ15 down-regulates a consortium of stress-responsive genes encoding transcription factors including ZAT10, WRKY33, WRKY44, CBF2, ERF6, and ERF 10. Meanwhile, the over-presented function of JMJ15 leads to higher expression level of RD29A, RD22, and COR15. However, this study failed to identify the direct targets of JMJ15, which most likely underpin the salt tolerance. Arabidopsis histone demethylases LDL1 and LDL2 (LYSINE-SPECIFIC DEMETHYLASE LIKE 1 and 2) act redundantly in repressing the seed dormancy, and their function is genetically dependent on seed-specific gene DOG1 (DELAY OF GERMINATION1), ABA-biosynthesis gene ABA2 (ABA DEFICIENT 2) and ABA-signaling gene ABI3 (ABA-INSENSITIVE3; Zhao et al., 2015). Thus, DOG1, ABA2, and ABI3 are potential targets of LDL1/2.
In non-model plants, there is mounting evidence of histone demethylation involved in osmotic stress. OsJMJ710 targets to MYB48-1 and demethylates H3K36me2, and dowregulates the expression of MYB48-1, leading to repressed drought tolerance in rice (Zhao et al., 2022). OsJMJ703 negatively regulates both the floral development and the drought tolerance. Though its targets have not been identified, its potential role as a demethylase of H3K4me3 is proposed due to the observation of increased global H3K4me3 in knock-down transgenic lines (Song et al., 2018). Tomato SlJMJ4 positively regulates dark- and ABA-induced leaf senescence by two partially overlapping mechanisms. In dark, SlJMJ4 is directed to the loci of SlORE1, SlNAP2, SlSAG113, and SlSAG12. In response to ABA, SlJMJ4 is targeted to SlABI5 and SlNCED3, in addition to SlORE1, SlNAP2, SlSAG113, SlSAG12. SlJMJ4 activates the transcription through removal of H3K27me3 in both processes (Ding et al., 2022). Considering the importance of ABI5 in ABA signaling and NCED3 in ABA biosynthesis, SlJMJ4 might also be involved in regulation of ABA-dependent drought tolerance. Moreover, there is masssive evidence showing that histone demethylation is responsive to osmotic stress though the mechanism has not been fully specified. Pan-genomic analysis in 11 rice species has identified 151 JmjC genes which suggests JmjC family underwent duplication and diversification during evolution. Some of the rice JmjC genes are involved in response to salt (Chowrasia et al., 2018). There are 64 JmjC genes in the genome of allotetraploid Brassica napus, of which 29 are from Brassica rapa and 23 from the Brassica oleracea. The BnaKDM5 (lysine demethylase 5) subfamily genes respond to stresses including salt, drought and high temperature (He et al., 2021). In cotton species, JmjC genes are divided into five subfamilies. Gossypium raimondii has 25 JmjC genes, Gossypium arboreum has 26, Gossypium hirsutum has 52, and Gossypium barbadense has 53. Several GhJMJs respond to salt and PEG treatment. Ectopic expression of GhJMJ34/40 imparts osmotic tolerance Saccharomyces cerevisiae (Sun et al., 2021). In birch, a total of 21 JmjC domain-containing histone demethylase proteins (JHDMs) are identified and classified into five subfamilies. In their promoter regions, cis-elements associated with hormone and abiotic stress responses are overrepresented. Their expression profiles also support their role in abiotic stress (Chen et al., 2021a). Thus, the genes encoding histone demethylases themselves are regulated by osmotic stress. In conclusion, JMJ- or LSD1-mediated histone demethylation allows regulatory plasticity of gene transcription under stress.
Histone methylation extensively cross-talks with other mechanisms both directly and indirectly to orchestrate the transcription of stress-related genes. Trithorax (TRX) group proteins are evolutionarily conserved activators with putative methylation activity on H3K4. The Arabidopsis TRX group consists of 12 members including ATX1–5 and ATX-RELATED (ATXR) 1–7 in (Alvarez-Venegas and Avramova, 2002; Veerappan et al., 2008; Chen et al., 2017). ATX5 deposits H3K4me3 at HY1 locus, activating its transcription to represses the transcription of ABI4 (ABA INSENSITIVE4). By contrast, glucose deactivates ATX5 thus diminishing H3K4me3 on HY1 locus, which further leads to a mitigation of HY1-repressed ABI4 expression (Liu et al., 2018a). The ATX1-HY1-ABI4 module epigenetically couples glucose signaling with transcription. Since the ABI4 is an important ABA-signal transducer in plants, this module might also be involved in ABA-signaling. Stomatal movement plays a critical role in drought tolerance as the stomata controls the transpiration rate. Light induces stomatal opening through H3K4me3 deposition at SOC1(SUPPRESSOR OF OVEREXPRESSION OF CO 1) gene, which is dependent of FT (FLOWERING LOCUS T; Aoki et al., 2019). Therefore, H3K4me3 might integrate light signaling into flowering and drought response.
Though Arabidopsis SDG26 (SET DOMAIN GROUP 26) which belongs to the ASH family has been characterized as a histone methyltranseferase with H3K36 specificity, which marks gene activation (Berr et al., 2015). Paradoxically, loss of functional FLD/LD/SDG26 leads to over-accumulation of H3K4me1/2 and H3K36me3, which hints SDG26 might be a versatile modifier involved both in gene activation and repression (Fang et al., 2020). Later in another study, the authors found that FLD/SDG26/LD interacts with R-loop to slow both the transcription initiation and the elongation (Xu et al., 2021). Since the ASH-group HKMTs are involved in drought tolerance (Chen et al., 2021b), the output when multiple epigenetic mechanisms are combinatorially involved needs to be reconsidered.
The PcG proteins and TrxG proteins represent two groups of histone modifiers with contrasting roles in gene transcription. PcG proteins maintain gene repression through deposition of repressive histone marks including H3K27me3 and H2Aub1 while TrxG proteins can function oppositely by catalyzing active histone marks such as H3K4me3. A well-characterized antagonistic model is the PcG-TxG counteraction in flowering. However, the ATX1 (TrxG)-ULT1 (TrxG)-EMF1 (PcG) module can also function synergistically and additively during seed germination (Xu et al., 2018). Moreover, a novel hypothesis was raised that PcG and TrxG complexes could function in concert to deposit bivalent chromatin marks as the H3K27me3 and H3K4me3 level in clf and atx1 double mutant was partially restored at the silent AG locus (Saleh et al., 2007). There is crosstalk in different PcG groups. Generally, Histone ubiquitination by PRC1 requires H3K27me3 deposited by PRC2 in Arabidopsis. On the contrary, PRC1 seems to function independently of PRC2 and is required for PRC2’s function in other reports (Merini and Calonje, 2015; Zhou et al., 2017; Wang and Shen, 2018). Histone ubiquitination also participates in interplay with histone methylation. In Arabidopsis, UBC1/2 (UBIQUITIN CARRIER 1/2) encode two E2 conjugated enzymes which are invoved in Histone H2B monoubiquitination (H2Bub1). Both UBC1 and UBC2 positively regulate salt response through the MPK4-MYB42-SOS2 module. H2B monoubiquitination mediated by UBC1/2 enhances H3K4me3 at both MYB42 and MPK4 and reinforces their expression (Sun et al., 2020).
The histone modifiers are also recruited to specific genomic loci through assembly into protein complexes containing DNA-binding subunits. The PhD-finger protein VIL1 (VIN3-LIKE1) can directly target to ABI3, ABI4, and ABI5, which encodes important transcription factors required for ABA signaling. Further, VIL1 recruits PRC2 complex to trimethylate H3K27me3 and repress the transcription of ABI3/4/5. Loss-of-VIL1 leads to hypersensitivity to ABA and enhanced drought tolerance (Zong et al., 2022). GmPHD6 (PLANT HOMEODOMAIN 6) interacts with the GmLHP1/2 coactivators through the PHD domain to form a transcriptional activation complex. GmPHD6 recognizes the G-rich elements in target gene promoters and is recruited by non-methylated (H3K4me0) and low methylated histone (H3K4me1,2) but not high methylated histone (H3K4me3). Once recruited, GmPHD6 further recruits GmLHP1 to activate ABA-responsive genes including CYP75B1 and CYP82C4. Therefore, GmPHD6 improves the salt tolerance in a GmLHP1-dependent manner (Wei et al., 2017). In most studies, LHP1 has been proposed as a subunit of PRC1 (Polycomb Repressive Complex 1) to repress gene transcription through mono-ubiquitination of H2A (Xu and Shen, 2008; Bratzel et al., 2010; Wang et al., 2014), and an accessory protein of PRC2 (Polycomb Repressive Complex2) to repress gene transcription through H3K27me3 (Zhang et al., 2007; Yuan et al., 2016). Therefore, contradiction of how GmLHP1/2 and GmPHD6 are coordinated to activate gene transcription is yet to be addressed. On the other hand, the plant homeodomain (PHD) finger proteins can recognize and bind to modified histone H3 and function as histone code readers (Shi et al., 2007; Bürglin and Affolter, 2016; Wei et al., 2017), the recognition of existing histone modification might also be instrumental to subsequent modifications for further gene regulation in response to salt stress. Besides the existing histone modifications, the cis-elements on the genome also bear important information for histone modifications. Such is the case that the cis-elements in the genic region of MsMYB4, are required for its expression and the local H3K4me3 and H3K9ac induced by salt in alfafa (Medicago sativa; Dong et al., 2020). In another way, some chromatin-associated factors can modulate histone methylation through physical interaction with histone modifiers. BPC (BASIC PENTACYSTEINE) proteins binds to ABI4 promoter and physically recruit the PRC2 to deposit H3K27me3, further leading to repressed ABI4 expression (Mu et al., 2017). Histone readers VP1/ABI3-LIKE 1/2 can recognize a cis-regulatory element at the FLC locus and recruits HDA9 for locus-specific H3K27 deacetylation. Deacetylated H3K27 in the FLC region is subsequently marked with H3K27me3 by PRC2 (Zeng et al., 2019). This hierarchy between histone deacetylation and H3K27me3 might enable plants to flower smartly in acclimation the environment because the HDA9 is involved in various stress response (Chen and Wu, 2010; Zheng et al., 2016, 2020; Shen et al., 2019; Van Der Woude et al., 2019; Baek et al., 2020; De Rooij et al., 2020). Other histone deacetylases including HD2C, HDA6 and HDA19 can regulate the histone methylation at the ABA-responsive genes including ABI1 and ABI2 (Luo et al., 2012).
The noncoding RNAs (ncRNAs) are functional ribonucleic acids that do not encode proteins or function independently of their peptide products. The role of noncoding RNAs including miRNA and lncRNAs in mediating the chromatin state has revealed (Böhmdorfer and Wierzbicki, 2015). Hereby, the histone methylation mediated by non-coding RNAs is discussed.
microRNAs (miRNAs) play an important role in plant stress responses. The Arabidopsis miR778 targets to the SUVH6 which encoding a histone H3K9 methyltransferase. The downregulation of SUVH6 mediated by miR778 causes dramatic upregulation of Pi deficiency-responsive genes. Though the direct evidence of H3K9 methylation at these loci is missing, it is likely that miR778 regulates the Pi-deficiency responsive genes transcription through regulation of SUVH6-mediated H3K9 methylation (Wang et al., 2015b). In a recent study, the miR778 negatively regulate both SUVH5 and SUVH6, thus activating the nematode-responsive genes (Bennett et al., 2022). These two studies support the multifaceted role of miR778 both in abiotic and biotic stress by targeting the histone methyltransferase genes to reduce the deposition of H3K9 methylation. However, the role of miRNAs in osmotic stress-induced histone (de)methylation remains elusive.
The lncRNAs probably serve both shared and distinctive roles the in plant stress responses (Di et al., 2014). The expression of DROUGHT INDUCED lncRNA (DRIR) can be dramatically upregulated. Hyper-expression of DRIR increases tolerance to drought and salt stress in an ABA-dependent manner. A consortium of genes involved in ABA signaling, water transport, and other stress response are misregulated by hyperexpression of DRIR (Qin et al., 2017). lncRNAs regulate histone methylation by recruiting histone modifiers to regulate gene transcription. COOLAIR and COOLAIR are two distinct lncRNAs transcribed from the 3′ end and the first intron of FLC. COOLAIR recruits CLF which is a component of PRC2 to FLC to establish H3K27me3 for gene repression (Heo and Sung, 2011). The COOLAIR antisense transcripts are targeted to FLC locus. The biogenesis of COOLAIR transcripts precedes the deposition of H3K27me3 at FLC locus by prolonged cold, which mediated the replacement of H3K36 methylation with H3K27me3 in the intragenic region. Interestingly, the COOLAIR lncRNA mediates a cold-induced synchronization from H3K36 methylation to H3K27 methylation, thus repressing the transcriptional activity of FLC. In the study, the researchers proposed that COOLAIR worked independently of the polycomb complex (Csorba et al., 2014). Likewise, MAS4 is the antisense transcript of MAF4 (MADS AFFECTING FLOWERING4). MAS4 is associated with WDR5a and recruits it to MAF4 locus, which further promotes the H3K4me3 and activate the transcription of MAF4 (Zhao et al., 2018). A short splice variant of COOLAIR affects the recruitment of the histone demethylase FLD (FLOWERING LOCUS D) to FLC, leading to demethylation of H3K4 at FLC (Liu et al., 2007; Marquardt et al., 2014). The FLC gene is a hot topic and represents a recognized platform to study epigenetic basis in the model plants (Whittaker and Dean, 2017). However, the direct evidence of lncRNA-regulated histone methylation in osmotic response is still missing. In other model species, recent studies might better our understanding of the relationship between lncRNA and histone methylation as the epigenetic basis is often highly conserved in eukaryotes. In mice, transcription factor OCT4 activates the expression of lncRNA Suv39h1as which targets to the Suv39h1 and down regulates its expression. As the Suv39h1 is a major methyltransferase catalyzing the di- and tri-methylation of H3K9, Suv39h1as implements negative regulation on the H3K9me2 and H3K9me during the differentiation of embryonic cells. Thus, the lncRNA-regulated can also be indirect (Bernard et al., 2022).
Similar to the interactive networks entailing histone methylation, the cross-talks sponsored by histone demethylation also play an unneglectable role in response to osmotic stress. In Arabidopsis thaliana, WRKY40 recruits histone demethylase JMJ17 to the ABI5 chromatin to deplete the deposition of H3K4me3. The decreased H3K4me3 level consequently deactivates the transcription of ABI5. Meanwhile, WRKY40 represses the transcription of HY5 to downregulate ABI5 expression. Upon stress, WRKY40 and JMJ17 are dissociated from ABI5 loci to withhold the transcriptional repression. The elevated ABI5 protein forms heteromeric dimer with HY5, and reinforces its own expression. Thus under resting conditions, this mechanism forms a double-safety clutch to repress constitutive ABA-response. Under stressful conditions, the concomitant release of WRKY40-JMJ17 and WRKY40-HY5 can boost the expression of ABI5 by increasing both the chromatin accessibility and activity of transcription factor (Wang et al., 2021b). This model represents a multilayer crosstalk between transcription factors and histone methylation status. Under non-stressful conditions, RPN1a (REGULATORY PARTICLE NON-ATPASE 1a) interacts with JMJ27 and mediates its degradation through the 26S proteasome pathway. Drought stress diminishes RPN1a abundance and indirectly elevates the protein level of JMJ27 to reinforce the activation of GOLS2 and RD20 through demethylation of H3K9me2 (Wang et al., 2021a). The proteasome-mediate turnover of histone demethylases provides another layer of regulation via the enzyme homeostasis. The enzymatic activity of JmjC proteins requires cofactors including ferrous iron and alpha-ketoglutarate in the oxidative demethylation reaction. Thus, it is convincing that disruption in the biochemical pathway required for demethylation can interfere with the demethylation process. Blocking the alpha-ketoglutarate biogenesis pathway concomitantly impairs the function of JMJ14, JMJ15, and JMJ18, leading to a global increase in H3K4me3 which further affects the thermosensory response (Cui et al., 2021). This results represent a metabolic regulation of histone methylation. An antagonistic model has been proposed where the PRC2 complex deposits H3K27me3 to repress gene transcription but JmjC proteins erase methylation from H3K27me3 to activate gene transcription (Crevillén, 2020). It is noteworthy that plant histone demethylases counteract the gene repression through erasure of the repressive histone methyl marks in most reported cases, which establish a de-repression mechanism upon stress.
Though a multitude of evidence show that histone methylation status is closely associated with the gene transcription activity, there are some exceptions. For instance, H3K4me3 enrichment on 4,837 rice genes in the genome were altered by drought stress while only a small proportion of them showed altered mRNA level (Zong et al., 2013), hinting a stress-memory imposed by drought through H3K4me3. In Arabidopsis, previous transient hyperosmotic stress imposes a long somatic memory by shaping the epigenomic landscape. Stress-induced H3K27me3 seems to be stable at least for 10 days before retreated to the normal level. By contrast, the H3K4me2, H3K4me3, and H3K9me2 islands are barely changed (Sani et al., 2013). The methylation on H3K4 might persist during stress-recovery stage (Ding et al., 2011, 2012). Likewise, H3K4me3 mediated by SDG25 and ATX1 plays an important role in heat-responsive gene expression, and is required for establishment of the heat stress memory (Song et al., 2021). However, functional loss of the H3K4me3 catalyzer ATX1 has no obvious impact on drought stress-memory in Arabidopsis (Ding et al., 2012). The genic loci of some drought-responsive transcription factors are less enriched in H3K27me3 in Arabidopsis, which might contribute to more efficient response to recurring drought (Ramirez-Prado et al., 2019). JMJ-mediated demethylation of H3K27me3 plays an indispensable role in heat acclimation with an imprinted stress memory at HSP22 and HSP17.6C loci, which potentiates their expression when heat stress reoccurs (Yamaguchi et al., 2021). These studies have suggested that different memory mechanism might be involved depending on the stress type. APX2 (ASCORBATE PEROXIDASE2) gene shows stress memory after heat stress, correlating with persisting transcription and H3K4 hyper-methylation. Using a heat-inducible dCas9 to target a JMJ demethylase domain required for methylation of H3K4 significantly reduces the stress-induced H3K4me3 and swipes off the transcriptional memory (Oberkofler and Bäurle, 2022). Collectively, stress-memory established by histone methylation is possibly a common mechanism allowing plants to combat the environmental challenges (Ding et al., 2012; Sani et al., 2013; Kinoshita and Seki, 2014; Avramova, 2015; Liu et al., 2018b; Zhang et al., 2020; Song et al., 2021; Yamaguchi et al., 2021). Moreover, editing the epigenome might not only be a powerful tool for studying chromatin basis of stress memory but also at the frontline of utilization of epigenetics in field crops.
The trans-cis interaction between transcription factors and target DNA sequences has been well documented and the input of stress signals determines the output of gene transcription. Nonetheless, there remains gaps between the gene transcription and the anchoring of transcription factors, wherein chromatin accessibility might play a critical role. Unlike DNA methylation or histone acetylation, which marks gene repression and activation respectively, histone methylation is one of the most functionally diversified epigenetic mechanism as it can either exert activation or repression on target genes depending on the site of lysine residues. Moreover, the histone lysine can be methylated to different extent enabling plasticity in gene regulation. Here in this review, recent advances in the histone methylation and demethylation which establish histone methylome and orchestrate gene transcription in response to osmotic stress have been summarized and discussed. Apart from the conserved biochemical activity of histone methylation writers and erasers, their specificity is yet to be elucidated. Another unneglectable aspect is the cross-talks between histone methylation and other regulatory mechanisms including histone acetylation, DNA methylation, histone ubiquitination, transcription factors and small RNAs, etc. In some cases, well-recognized antagonistic regulators can work synergistically, which further hinders our in-depth understanding of the role of loci-specific histone methylation. Though the complexity of histone methylation and gene transcription has not been fully detangled in lab, the potential of histone methylation in field comes into sight as a set of studies on stress memory (also known as “priming”) have revealed that stress can impose both somatic retention and transgenerational inheritance accompanied with multiple epigenetic modifications, avoiding growth penalty caused by constitutive expression of stress response (Figure 2). Conclusively, we propose three directions as follows, toward which the study on histone methylation might be greatly advanced:
1. The loci-specificity of histone methylation;
2. The interplay and hierarchy between histone methylation and other mechanisms;
3. The formation, maintenance, and erasure of histone methylation-based stress memory.
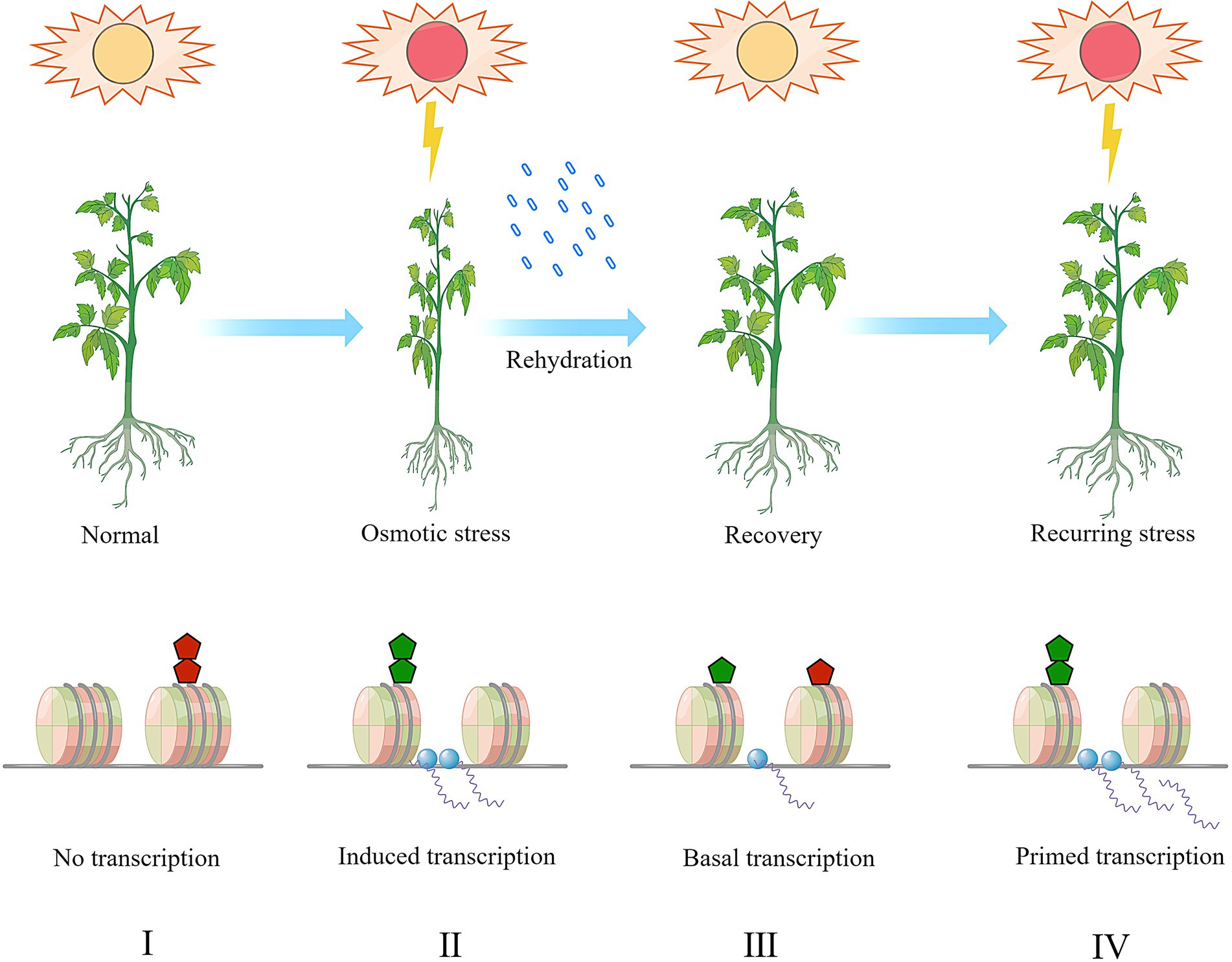
Figure 2. A hypothetic model of stress-memory and histone (de)methylation in plants combating recurring osmotic stress. The histone methylation dynamics at stress responsive genes and the transcriptional activation was exemplified and presented in this model. Under normal condition (I), repressive histone methylation (the red pentagon, e.g., H3K9me1/2/3, H3K27me1/2/3) is enriched and the chromatin is inaccessible to the transcription machinery (the blue dots). The low accessibility of chromatin results in transcriptional repression. Upon a primary drought response (II), the enrichment of repressive histone methylation is depleted the and active histone methylation (the green pentagon, e.g., H3K4me1/2/3, H3K36me1/2/3) is deposited to form an open chromatin status. Correspondingly, the stress responsive genes are transcribed upon induction. After recovery (III), the histone methylation pattern persists for a certain period and then both the active and repressive methylation marks gradually retreat to the normal level. The stress memory is imprinted during the transition from the stage (II) to the stage (III). During this stage, the transcription activity might be maintained in a lower basal level. When a secondary stress occurs (IV), the plants combat the stress more efficiently as the stress-responsive genes have been primed, and the active histone methylation and stallment of transcription machinery allow plants with quicker or more robust response, and finally reduces the plant vulnerability to stress. During the stage (IV), the stress memory stored on the histones is re-accessed and translated into gene transcription activity.
MX conceived this review and drafted the manuscript. JW participated in the figure configuration. FX conducted this work and gave insightful viewpoints to the manuscript. All authors contributed to the article and approved the submitted version.
The work was supported by Taishan Scholars program of Shandong Province (tsqn201812018), Natural Science Foundation of Shandong Province (ZR2019ZD16), and Project for Scientific Research Innovation Team of Young Scholar in Colleges and Universities of Shandong Province (2020KJE002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alvarez-Venegas, R., and Avramova, Z. (2002). SET-domain proteins of the Su(var)3-9, E(z) and Trithorax families. Gene 285, 25–37. doi: 10.1016/S0378-1119(02)00401-8
Aoki, S., Toh, S., Nakamichi, N., Hayashi, Y., Wang, Y., Suzuki, T., et al. (2019). Regulation of stomatal opening and histone modification by photoperiod in Arabidopsis thaliana. Sci. Rep. 9:10054. doi: 10.1038/s41598-019-46440-0
Asensi-Fabado, M.-A., Amtmann, A., and Perrella, G. (2017). Plant responses to abiotic stress: the chromatin context of transcriptional regulation. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1860, 106–122. doi: 10.1016/j.bbagrm.2016.07.015
Ault, T. R. (2020). On the essentials of drought in a changing climate. Science 368, 256–260. doi: 10.1126/science.aaz5492
Avramova, Z. (2015). Transcriptional ‘memory’ of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 83, 149–159. doi: 10.1111/tpj.12832
Baek, D., Shin, G., Kim, M. C., Shen, M., Lee, S. Y., and Yun, D.-J. (2020). Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front. Plant Sci. 11:143. doi: 10.3389/fpls.2020.00143
Balao, F., Paun, O., and Alonso, C. (2018). Uncovering the contribution of epigenetics to plant phenotypic variation in Mediterranean ecosystems. Plant Biol. 20, 38–49. doi: 10.1111/plb.12594
Batra, R., Gautam, T., Pal, S., Chaturvedi, D., Rakhi, J. I., et al. (2020). Identification and characterization of SET domain family genes in bread wheat (Triticum aestivum L.). Sci. Rep. 10:14624. doi: 10.1038/s41598-020-71526-5
Bennett, M., Piya, S., Baum, T. J., and Hewezi, T. (2022). miR778 mediates gene expression, histone modification, and DNA methylation during cyst nematode parasitism. Plant physiol. 189, 2432–2453. doi: 10.1093/plphys/kiac228
Bernard, L. D., Dubois, A., Heurtier, V., Fischer, V., Gonzalez, I., Chervova, A., et al. (2022). OCT4 activates a Suv39h1-repressive antisense lncRNA to couple histone H3 lysine 9 methylation to pluripotency. Nucleic Acids Res. 50, 7367–7379. doi: 10.1093/nar/gkac550
Berr, A., Shafiq, S., Pinon, V., Dong, A., and Shen, W.-H. (2015). The trxG family histone methyltransferase SET DOMAIN GROUP 26 promotes flowering via a distinctive genetic pathway. Plant J. 81, 316–328. doi: 10.1111/tpj.12729
Berr, A., Shafiq, S., and Shen, W.-H. (2011). Histone modifications in transcriptional activation during plant development. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1809, 567–576. doi: 10.1016/j.bbagrm.2011.07.001
Böhmdorfer, G., and Wierzbicki, A. T. (2015). Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 25, 623–632. doi: 10.1016/j.tcb.2015.07.002
Bratzel, F., López-Torrejón, G., Koch, M., Del Pozo, J. C., and Calonje, M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A Monoubiquitination. Curr. Biol. 20, 1853–1859. doi: 10.1016/j.cub.2010.09.046
Bürglin, T. R., and Affolter, M. (2016). Homeodomain proteins: an update. Chromosoma 125, 497–521. doi: 10.1007/s00412-015-0543-8
Chen, B., Ali, S., Zhang, X., Zhang, Y., Wang, M., Zhang, Q., et al. (2021a). Genome-wide identification, classification, and expression analysis of the JmjC domain-containing histone demethylase gene family in birch. BMC Genomics 22:772. doi: 10.1186/s12864-021-08063-6
Chen, K., Du, K., Shi, Y., Yin, L., Shen, W.-H., Yu, Y., et al. (2021b). H3K36 methyltransferase SDG708 enhances drought tolerance by promoting abscisic acid biosynthesis in rice. New Phytol. 230, 1967–1984. doi: 10.1111/nph.17290
Chen, H., Feng, H., Zhang, X., Zhang, C., Wang, T., and Dong, J. (2019). An Arabidopsis E3 ligase HUB2 increases histone H2B monoubiquitination and enhances drought tolerance in transgenic cotton. Plant Biotechnol. J. 17, 556–568. doi: 10.1111/pbi.12998
Chen, Q., Guo, L., Yuan, Y., Hu, S., Guo, F., Zhao, H., et al. (2021c). Ectopic Overexpression of histone H3K4 methyltransferase CsSDG36 from tea plant decreases hyperosmotic stress tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 22:5064. doi: 10.3390/ijms22105064
Chen, L.-Q., Luo, J.-H., Cui, Z.-H., Xue, M., Wang, L., Zhang, X.-Y., et al. (2017). ATX3, ATX4, and ATX5 encode putative H3K4 methyltransferases and are critical for plant development. Plant Physiol. 174, 1795–1806. doi: 10.1104/pp.16.01944
Chen, D.-H., Qiu, H.-L., Huang, Y., Zhang, L., and Si, J.-P. (2020). Genome-wide identification and expression profiling of SET DOMAIN GROUP family in Dendrobium catenatum. BMC Plant Biol. 20:40. doi: 10.1186/s12870-020-2244-6
Chen, L.-T., and Wu, K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5, 1318–1320. doi: 10.4161/psb.5.10.13168
Chowrasia, S., Panda, A. K., Rawal, H. C., Kaur, H., and Mondal, T. K. (2018). Identification of jumonjiC domain containing gene family among the Oryza species and their expression analysis in FL478, a salt tolerant rice genotype. Plant Physiol. Biochem. 130, 43–53. doi: 10.1016/j.plaphy.2018.06.031
Crevillén, P. (2020). Histone demethylases as counterbalance to H3K27me3 silencing in plants. iScience 23:101715. doi: 10.1016/j.isci.2020.101715
Csorba, T., Questa, J. I., Sun, Q., and Dean, C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. 111, 16160–16165. doi: 10.1073/pnas.1419030111
Cui, X., Zheng, Y., Lu, Y., Issakidis-Bourguet, E., and Zhou, D.-X. (2021). Metabolic control of histone demethylase activity involved in plant response to high temperature. Plant Physiol. 185, 1813–1828. doi: 10.1093/plphys/kiab020
de la Paz Sanchez, M., Aceves-García, P., Petrone, E., Steckenborn, S., Vega-León, R., Álvarez-Buylla, E. R., et al. (2015). The impact of Polycomb group (PcG) and Trithorax group (TrxG) epigenetic factors in plant plasticity. New Phytol. 208, 684–694. doi: 10.1111/nph.13486
De Rooij, P. G., Perrella, G., Kaiserli, E., and Van Zanten, M. (2020). The diverse and unanticipated roles of histone deacetylase 9 in coordinating plant development and environmental acclimation. J. Exp. Bot. 71, 6211–6225. doi: 10.1093/jxb/eraa335
Deal, R. B., and Henikoff, S. (2011). Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 14, 116–122. doi: 10.1016/j.pbi.2010.11.005
Di, C., Yuan, J., Wu, Y., Li, J., Lin, H., Hu, L., et al. (2014). Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 80, 848–861. doi: 10.1111/tpj.12679
Ding, Y., Avramova, Z., and Fromm, M. (2011). The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 66, 735–744. doi: 10.1111/j.1365-313X.2011.04534.x
Ding, Y., Fromm, M., and Avramova, Z. (2012). Multiple exposures to drought 'train' transcriptional responses in Arabidopsis. Nat. Commun. 3:740. doi: 10.1038/ncomms1732
Ding, X., Zhang, D., Gu, D., Li, Z., Liang, H., Zhu, H., et al. (2022). Histone H3K27 demethylase SlJMJ4 promotes dark- and ABA- induced leaf senescence in tomato. Horticul. Res. 9:77. doi: 10.1093/hr/uhab077
Dong, W., Gao, T., Wang, Q., Chen, J., Lv, J., and Song, Y. (2020). Salinity stress induces epigenetic alterations to the promoter of MsMYB4 encoding a salt-induced MYB transcription factor. Plant Physiol. Biochem. 155, 709–715. doi: 10.1016/j.plaphy.2020.08.015
Fang, X., Wu, Z., Raitskin, O., Webb, K., Voigt, P., Lu, T., et al. (2020). The 3′ processing of antisense RNAs physically links to chromatin-based transcriptional control. Proc. Natl. Acad. Sci. 117, 15316–15321. doi: 10.1073/pnas.2007268117
Feng, Q., Wang, H., Ng, H. H., Erdjument-Bromage, H., Tempst, P., Struhl, K., et al. (2002). Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058. doi: 10.1016/S0960-9822(02)00901-6
Frolov, A., Bilova, T., Paudel, G., Berger, R., Balcke, G. U., Birkemeyer, C., et al. (2017). Early responses of mature Arabidopsis thaliana plants to reduced water potential in the agar-based polyethylene glycol infusion drought model. J. Plant Physiol. 208, 70–83. doi: 10.1016/j.jplph.2016.09.013
Fu, Y., Ma, H., Chen, S., Gu, T., and Gong, J. (2017). Control of proline accumulation under drought via a novel pathway comprising the histone methylase CAU1 and the transcription factor ANAC055. J. Exp. Bot. 69, 579–588. doi: 10.1093/jxb/erx419
Fu, Y.-L., Zhang, G.-B., Lv, X.-F., Guan, Y., Yi, H.-Y., and Gong, J.-M. (2013). Arabidopsis histone methylase CAU1/PRMT5/SKB1 acts as an epigenetic suppressor of the calcium signaling gene CAS to mediate stomatal closure in response to extracellular calcium. Plant Cell 25, 2878–2891. doi: 10.1105/tpc.113.113886
González, R. M., Ricardi, M. M., and Iusem, N. D. (2013). Epigenetic marks in an adaptive water stress-responsive gene in tomato roots under normal and drought conditions. Epigenetics 8, 864–872. doi: 10.4161/epi.25524
Granot, G., Sikron-Persi, N., Gaspan, O., Florentin, A., Talwara, S., Paul, L. K., et al. (2009). Histone modifications associated with drought tolerance in the desert plant Zygophyllum dumosum Boiss. Planta 231, 27–34. doi: 10.1007/s00425-009-1026-z
Gu, D., Yang, J., Wu, S., Liao, Y., Zeng, L., and Yang, Z. (2021). Epigenetic regulation of the phytohormone abscisic acid accumulation under dehydration stress during postharvest processing of tea (Camellia sinensis). J. Agric. Food Chem. 69, 1039–1048. doi: 10.1021/acs.jafc.0c07220
Han, B., Xu, W., Ahmed, N., Yu, A., Wang, Z., and Liu, A. (2020). Changes and associations of genomic transcription and histone methylation with salt stress in Castor bean. Plant Cell Physiol. 61, 1120–1133. doi: 10.1093/pcp/pcaa037
Hashiguchi, A., and Komatsu, S. (2017). “Chapter Six - Posttranslational Modifications and Plant–Environment Interaction,” in Methods in enzymology. ed. A. K. Shukla (Cambridge, MA: Academic Press), 97–113.
He, X., Wang, Q., Pan, J., Liu, B., Ruan, Y., and Huang, Y. (2021). Systematic analysis of JmjC gene family and stress-response expression of KDM5 subfamily genes in Brassica napus. PeerJ 9:e11137. doi: 10.7717/peerj.11137
Heo, J. B., and Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Huang, H., Lin, S., Garcia, B. A., and Zhao, Y. (2015). Quantitative proteomic analysis of histone modifications. Chem. Rev. 115, 2376–2418. doi: 10.1021/cr500491u
Huang, H., Sabari, B. R., Garcia, B. A., Allis, C. D., and Zhao, Y. (2014). SnapShot: histone modifications. Cell 159:458. doi: 10.1016/j.cell.2014.09.037
Huang, S., Zhang, A., Jin, J. B., Zhao, B., Wang, T.-J., Wu, Y., et al. (2019). Arabidopsis histone H3K4 demethylase JMJ17 functions in dehydration stress response. New Phytol. 223, 1372–1387. doi: 10.1111/nph.15874
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jenuwein, T., and Allis, C. D. (2001). Translating the histone code. Science 293, 1074–1080. doi: 10.1126/science.1063127
Kamal, K. Y., Khodaeiaminjan, M., Yahya, G., El-Tantawy, A. A., Abdel El-Moneim, D., El-Esawi, M. A., et al. (2021). Modulation of cell cycle progression and chromatin dynamic as tolerance mechanisms to salinity and drought stress in maize. Physiol. Plant. 172, 684–695. doi: 10.1111/ppl.13260
Kim, J. M. T. K., Nishioka, T., and Seki, M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33, 604–611. doi: 10.1111/j.1365-3040.2009.02076.x
Kim, J.-M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Kim, J.-M., To, T. K., Ishida, J., Morosawa, T., Kawashima, M., Matsui, A., et al. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49, 1580–1588. doi: 10.1093/pcp/pcn133
Kinoshita, T., and Seki, M. (2014). Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 55, 1859–1863. doi: 10.1093/pcp/pcu125
Klemm, S. L., Shipony, Z., and Greenleaf, W. J. (2019). Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220. doi: 10.1038/s41576-018-0089-8
Klose, R. J., and Zhang, Y. (2007). Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8, 307–318. doi: 10.1038/nrm2143
Kosová, K., Vítámvás, P., Prášil, I. T., Klíma, M., and Renaut, J. (2021). Plant Proteoforms Under environmental stress: functional proteins arising From a single gene. Front. Plant Sci. 12:793113. doi: 10.3389/fpls.2021.793113
Li, Z., Li, D., Li, Y., Guo, X., and Yang, R. (2022). Deciphering the regulatory code of histone modifications in plants. J. Genet. Genom. 003. doi: 10.1016/j.jgg.2022.07.003 (in press).
Liu, C., Lu, F., Cui, X., and Cao, X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Liu, F., Quesada, V., Crevillén, P., Bäurle, I., Swiezewski, S., and Dean, C. (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 Homolog to downregulate FLC. Mol. Cell 28, 398–407. doi: 10.1016/j.molcel.2007.10.018
Liu, Y., Wang, J., Yin, H., Zhang, A., Huang, S., Wang, T.-J., et al. (2018a). Trithorax-group protein ATX5 mediates the glucose response via impacting the HY1-ABI4 signaling module. Plant Mol. Biol. 98, 495–506. doi: 10.1007/s11103-018-0791-0
Liu, Y., Zhang, A., Yin, H., Meng, Q., Yu, X., Huang, S., et al. (2018b). Trithorax-group proteins ARABIDOPSIS TRITHORAX4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol. 217, 1582–1597. doi: 10.1111/nph.14933
Luo, M., Wang, Y.-Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Marquardt, S., Raitskin, O., Wu, Z., Liu, F., Sun, Q., and Dean, C. (2014). Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54, 156–165. doi: 10.1016/j.molcel.2014.03.026
Merini, W., and Calonje, M. (2015). PRC1 is taking the lead in PcG repression. Plant J. 83, 110–120. doi: 10.1111/tpj.12818
Mu, Y., Zou, M., Sun, X., He, B., Xu, X., Liu, Y., et al. (2017). BASIC PENTACYSTEINE proteins repress ABSCISIC ACID INSENSITIVE4 expression via direct recruitment of the Polycomb-repressive complex 2 in Arabidopsis root development. Plant Cell Physiol. 58, 607–621. doi: 10.1093/pcp/pcx0006
Munns, R., James, R. A., Sirault, X. R., Furbank, R. T., and Jones, H. G. (2010). New phenotyping methods for screening wheat and barley for beneficial responses to water deficit. J. Exp. Bot. 61, 3499–3507. doi: 10.1093/jxb/erq199
Nguyen, N. H., and Cheong, J.-J. (2018). The AtMYB44 promoter is accessible to signals that induce different chromatin modifications for gene transcription. Plant Physiol. Biochem. 130, 14–19. doi: 10.1016/j.plaphy.2018.06.030
Nguyen, N. H., Jung, C., and Cheong, J.-J. (2019). Chromatin remodeling for the transcription of type 2C protein phosphatase genes in response to salt stress. Plant Physiol. Biochem. 141, 325–331. doi: 10.1016/j.plaphy.2019.06.012
Niu, L., Lu, F., Pei, Y., Liu, C., and Cao, X. (2007). Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 8, 1190–1195. doi: 10.1038/sj.embor.7401111
Oberkofler, V., and Bäurle, I. (2022). Inducible epigenome editing probes for the role of histone H3K4 methylation in Arabidopsis heat stress memory. Plant Physiol. 189, 703–714. doi: 10.1093/plphys/kiac113
Park, J., Lim, C. J., Shen, M., Park, H. J., Cha, J.-Y., Iniesto, E., et al. (2018). Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. 115, E5400–E5409. doi: 10.1073/pnas.1721241115
Paul, A., Dasgupta, P., Roy, D., and Chaudhuri, S. (2017). Comparative analysis of histone modifications and DNA methylation at OsBZ8 locus under salinity stress in IR64 and Nonabokra rice varieties. Plant Mol. Biol. 95, 63–88. doi: 10.1007/s11103-017-0636-2
Pfluger, J., and Wagner, D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10, 645–652. doi: 10.1016/j.pbi.2007.07.013
Pu, L., and Sung, Z. R. (2015). PcG and trxG in plants – friends or foes. Trends Genet. 31, 252–262. doi: 10.1016/j.tig.2015.03.004
Qian, S., Wang, Y., Ma, H., and Zhang, L. (2015). Expansion and functional divergence of Jumonji C-containing histone demethylases: significance of duplications in ancestral angiosperms and vertebrates. Plant Physiol. 168, 1321–1337. doi: 10.1104/pp.15.00520
Qin, X., and Zeevaart, J. A. D. (1999). The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. 96, 15354–15361. doi: 10.1073/pnas.96.26.15354
Qin, T., Zhao, H., Cui, P., Albesher, N., and Xiong, L. (2017). A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 175, 1321–1336. doi: 10.1104/pp.17.00574
Ramirez-Prado, J. S., Latrasse, D., Rodriguez-Granados, N. Y., Huang, Y., Manza-Mianza, D., Brik-Chaouche, R., et al. (2019). The Polycomb protein LHP1 regulates Arabidopsis thaliana stress responses through the repression of the MYC2-dependent branch of immunity. Plant J. 100, 1118–1131. doi: 10.1111/tpj.14502
Saleh, A., Al-Abdallat, A., Ndamukong, I., Alvarez-Venegas, R., and Avramova, Z. (2007). The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 35, 6290–6296. doi: 10.1093/nar/gkm464
Sani, E., Herzyk, P., Perrella, G., Colot, V., and Amtmann, A. (2013). Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 14:R59. doi: 10.1186/gb-2013-14-6-r59
Shahbazian, M. D., and Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100. doi: 10.1146/annurev.biochem.76.052705.162114
Shen, Y., Conde Silva, N., Audonnet, L., Servet, C., Wei, W., and Zhou, D.-X. (2014). Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 5:290. doi: 10.3389/fpls.2014.00290
Shen, Y., Lei, T., Cui, X., Liu, X., Zhou, S., Zheng, Y., et al. (2019). Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J. 100, 991–1006. doi: 10.1111/tpj.14492
Shi, X., Kachirskaia, I., Walter, K. L., Kuo, J.-H. A., Lake, A., Davrazou, F., et al. (2007). Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at Either lysine 4 or lysine 36. J. Biol. Chem. 282, 2450–2455. doi: 10.1074/jbc.C600286200
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase Homolog LSD1. Cell 119, 941–953. doi: 10.1016/j.cell.2004.12.012
Song, Z.-T., Zhang, L.-L., Han, J.-J., Zhou, M., and Liu, J.-X. (2021). Histone H3K4 methyltransferases SDG25 and ATX1 maintain heat-stress gene expression during recovery in Arabidopsis. Plant J. 105, 1326–1338. doi: 10.1111/tpj.15114
Song, T., Zhang, Q., Wang, H., Han, J., Xu, Z., Yan, S., et al. (2018). OsJMJ703, a rice histone demethylase gene, plays key roles in plant development and responds to drought stress. Plant Physiol. Biochem. 132, 183–188. doi: 10.1016/j.plaphy.2018.09.007
Springer, N. M., Napoli, C. A., Selinger, D. A., Pandey, R., Cone, K. C., Chandler, V. L., et al. (2003). Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 132, 907–925. doi: 10.1104/pp.102.013722
Sun, L., Song, G., Guo, W., Wang, W., Zhao, H., Gao, T., et al. (2019). Dynamic changes in genome-wide Histone3 Lysine27 trimethylation and gene expression of soybean roots in response to salt stress. Front. Plant Sci. 10:1031. doi: 10.3389/fpls.2019.01031
Sun, Z., Wang, X., Qiao, K., Fan, S., and Ma, Q. (2021). Genome-wide analysis of JMJ-C histone demethylase family involved in salt-tolerance in Gossypium hirsutum L. Plant Physiol. Biochem. 158, 420–433. doi: 10.1016/j.plaphy.2020.11.029
Sun, Y., Zhao, J., Li, X., and Li, Y. (2020). E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis. New Phytol. 227, 455–472. doi: 10.1111/nph.16538
Tan, W., Zhang, D., Zhou, H., Zheng, T., Yin, Y., and Lin, H. (2018). Transcription factor HAT1 is a substrate of SnRK2. 3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 14:e1007336. doi: 10.1371/journal.pgen.1007336
Tsukada, Y.-I., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P., et al. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. doi: 10.1038/nature04433
Van Der Woude, L. C., Perrella, G., Snoek, B. L., Van Hoogdalem, M., Novák, O., Van Verk, M. C., et al. (2019). HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A Z depletion. Proceed. Nat. Acad.Sci. 116, 25343–25354. doi: 10.1073/pnas.1911694116
van Dijk, K., Ding, Y., Malkaram, S., Riethoven, J.-J. M., Liu, R., Yang, J., et al. (2010). Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 10:238. doi: 10.1186/1471-2229-10-238
Van Oosten, M. J., Bressan, R. A., Zhu, J.-K., Bohnert, H. J., and Chinnusamy, V. (2014). The role of the epigenome in gene expression control and the epimark changes in response to the environment. Crit. Rev. Plant Sci. 33, 64–87. doi: 10.1080/07352689.2014.852920
Veerappan, C. S., Avramova, Z., and Moriyama, E. N. (2008). Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi. BMC Evol. Biol. 8:190. doi: 10.1186/1471-2148-8-190
Verslues, P. E., Agarwal, M., Katiyar-Agarwal, S., Zhu, J., and Zhu, J. K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45, 523–539. doi: 10.1111/j.1365-313X.2005.02593.x
Wang, Y., Gu, X., Yuan, W., Schmitz, R. J., and He, Y. (2014). Photoperiodic control of the floral transition through a distinct Polycomb repressive complex. Dev. Cell 28, 727–736. doi: 10.1016/j.devcel.2014.01.029
Wang, T.-J., Huang, S., Zhang, A., Guo, P., Liu, Y., Xu, C., et al. (2021b). JMJ17–WRKY40 and HY5–ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytol. 230, 567–584. doi: 10.1111/nph.17177
Wang, Q., Liu, P., Jing, H., Zhou, X. F., Zhao, B., Li, Y., et al. (2021a). JMJ27-mediated histone H3K9 demethylation positively regulates drought-stress responses in Arabidopsis. New Phytol. 232, 221–236. doi: 10.1111/nph.17593
Wang, J., Meng, X., Yuan, C., Harrison, A. P., and Chen, M. (2015a). The roles of cross-talk epigenetic patterns in Arabidopsis thaliana. Brief. Funct. Genom. 15, 278–287. doi: 10.1093/bfgp/elv025
Wang, Q., and Shen, W.-H. (2018). Chromatin modulation and gene regulation in plants: insight about PRC1 function. Biochem. Soc. Trans. 46, 957–966. doi: 10.1042/BST20170576
Wang, L., ZengJ, H. Q., Song, J., Feng, S. J., and Yang, Z. M. (2015b). miRNA778 and SUVH6 are involved in phosphate homeostasis in Arabidopsis. Plant Sci. 238, 273–285. doi: 10.1016/j.plantsci.2015.06.020
Wei, W., Tao, J.-J., Chen, H.-W., Li, Q.-T., Zhang, W.-K., Ma, B., et al. (2017). A histone code reader and a transcriptional activator interact to regulate genes for salt tolerance. Plant Physiol. 175, 1304–1320. doi: 10.1104/pp.16.01764
Whittaker, C., and Dean, C. (2017). The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Williams, S. T., Walport, L. J., Hopkinson, R. J., Madden, S. K., Chowdhury, R., Schofield, C. J., et al. (2014). Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates. Epigenetics 9, 1596–1603. doi: 10.4161/15592294.2014.983381
Wong, M. M., Chong, G. L., and Verslues, P. E. (2017). “Epigenetics and RNA processing: connections to drought, salt, and ABA?” in Plant Stress Tolerance. ed. R. Sunkar (Berlin: Springer), 3–21.
Wu, J., Ichihashi, Y., Suzuki, T., Shibata, A., Shirasu, K., Yamaguchi, N., et al. (2019a). Abscisic acid-dependent histone demethylation during postgermination growth arrest in Arabidopsis. Plant Cell Environ. 42, 2198–2214. doi: 10.1111/pce.13547
Wu, J., Yamaguchi, N., and Ito, T. (2019b). Histone demethylases control root elongation in response to stress-signaling hormone abscisic acid. Plant Signal. Behav. 14:1604019. doi: 10.1080/15592324.2019.1604019
Wu, T., Yuan, T., Tsai, S.-N., Wang, C., Sun, S.-M., Lam, H.-M., et al. (2009). Mass spectrometry analysis of the variants of histone H3 and H4 of soybean and their post-translational modifications. BMC Plant Biol. 9, 1–15. doi: 10.1186/1471-2229-9-98
Xiao, J., Lee, U.-S., and Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi: 10.1016/j.pbi.2016.08.002
Xu, C., Fang, X., Lu, T., and Dean, C. (2021). Antagonistic cotranscriptional regulation through ARGONAUTE1 and the THO/TREX complex orchestrates FLC transcriptional output. Proc. Natl. Acad. Sci. 118:e2113757118. doi: 10.1073/pnas.2113757118
Xu, F., Kuo, T., Rosli, Y., Liu, M.-S., Wu, L., Chen, L.-F. O., et al. (2018). Trithorax group proteins act together with a Polycomb group protein to maintain chromatin integrity for epigenetic silencing during seed germination in Arabidopsis. Mol. Plant 11, 659–677. doi: 10.1016/j.molp.2018.01.010
Xu, L., and Shen, W.-H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18, 1966–1971. doi: 10.1016/j.cub.2008.11.019
Xu, J., Wang, Q., Freeling, M., Zhang, X., Xu, Y., Mao, Y., et al. (2017). Natural antisense transcripts are significantly involved in regulation of drought stress in maize. Nucleic Acids Res. 45, 5126–5141. doi: 10.1093/nar/gkx085
Yamaguchi, N., Matsubara, S., Yoshimizu, K., Seki, M., Hamada, K., Kamitani, M., et al. (2021). H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nature. Communications 12:3480. doi: 10.1038/s41467-021-23766-w
Yin, W., Xiao, Y., Niu, M., Meng, W., Li, L., Zhang, X., et al. (2020). ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in Rice. Plant Cell 32, 2292–2306. doi: 10.1105/tpc.19.00542
Ytterberg, A. J., and Jensen, O. N. (2010). Modification-specific proteomics in plant biology. J. Proteome 73, 2249–2266. doi: 10.1016/j.jprot.2010.06.002
Yu, Y., Bu, Z., Shen, W.-H., and Dong, A. (2009). An update on histone lysine methylation in plants. Prog. Nat. Sci. 19, 407–413. doi: 10.1016/j.pnsc.2008.07.015
Yuan, L., Liu, X., Luo, M., Yang, S., and Wu, K. (2013). Involvement of histone modifications in plant abiotic stress responses. J. Integr. Plant Biol. 55, 892–901. doi: 10.1111/jipb.12060
Yuan, W., Luo, X., Li, Z., Yang, W., Wang, Y., Liu, R., et al. (2016). A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 48, 1527–1534. doi: 10.1038/ng.3712
Zacarias, E., and Casas-Mollano, J. A. (2021). “Advances in plant omics and systems biology approaches,” in Cataloging Posttranslational Modifications in Plant Histones. ed. F. V. Winck (Berlin: Springer), 131–154.
Zeng, X., Gao, Z., Jiang, C., Yang, Y., Liu, R., and He, Y. (2019). HISTONE DEACETYLASE 9 functions with Polycomb silencing to repress FLOWERING LOCUS C Expression1. Plant Physiol. 182, 555–565. doi: 10.1104/pp.19.00793
Zhang, X., Germann, S., Blus, B. J., Khorasanizadeh, S., Gaudin, V., and Jacobsen, S. E. (2007). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14, 869–871. doi: 10.1038/nsmb1283
Zhang, X., Ménard, R., Li, Y., Coruzzi, G. M., Heitz, T., Shen, W.-H., et al. (2020). Arabidopsis SDG8 potentiates the sustainable transcriptional induction of the pathogenesis-related genes PR1 and PR2 During plant Defense response. Front. Plant Sci. 11:277. doi: 10.3389/fpls.2020.00277
Zhao, X., Li, J., Lian, B., Gu, H., Li, Y., and Qi, Y. (2018). Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 9:5056. doi: 10.1038/s41467-018-07500-7
Zhao, W., Wang, X., Zhang, Q., Zheng, Q., Yao, H., Gu, X., et al. (2022). H3K36 demethylase JMJ710 negatively regulates drought tolerance by suppressing MYB48-1 expression in rice. Plant Physiol. 189, 1050–1064. doi: 10.1093/plphys/kiac095
Zhao, M., Yang, S., Liu, X., and Wu, K. (2015). Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front. Plant Sci. 6:159. doi: 10.3389/fpls.2015.00159
Zheng, Y., Ding, Y., Sun, X., Xie, S., Wang, D., Liu, X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713. doi: 10.1093/jxb/erv562
Zheng, Y., Ge, J., Bao, C., Chang, W., Liu, J., Shao, J., et al. (2020). Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant 13, 598–611. doi: 10.1016/j.molp.2019.12.011
Zhou, H., Liu, Y., Liang, Y., Zhou, D., Li, S., Lin, S., et al. (2020). The function of histone lysine methylation related SET domain group proteins in plants. Protein Sci. 29, 1120–1137. doi: 10.1002/pro.3849
Zhou, Y., Romero-Campero, F. J., Gómez-Zambrano, Á., Turck, F., and Calonje, M. (2017). H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 18:69. doi: 10.1186/s13059-017-1197-z
Zong, W., Kim, J., Bordiya, Y., Qiao, H., and Sung, S. (2022). Abscisic acid negatively regulates the Polycomb-mediated H3K27me3 through the PHD-finger protein, VIL1. New Phytologist n/a(n/a) 235, 1057–1069. doi: 10.1111/nph.18156
Keywords: osmotic stress, histone methylation, histone demethylation, gene transcription, stress memory
Citation: Xiao M, Wang J and Xu F (2022) Methylation hallmarks on the histone tail as a linker of osmotic stress and gene transcription. Front. Plant Sci. 13:967607. doi: 10.3389/fpls.2022.967607
Received: 13 June 2022; Accepted: 25 July 2022;
Published: 10 August 2022.
Edited by:
Longxing Hu, Hunan Agricultural University, ChinaReviewed by:
Juncheng Lin, Xiamen University, ChinaCopyright © 2022 Xiao, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Xu, Znh1QHNkdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.