
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 August 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.964604
This article is part of the Research Topic Multi-omics and Computational Biology in Horticultural Plants: From Genotype to Phenotype View all 32 articles
Growth-regulating factors (GRFs) play crucial roles in plant growth and stress response. To date, there have been no reports of the analysis and identification of the GRF transcription factor family in alfalfa. In this study, we identified 27 GRF family members from alfalfa (Medicago sativa L.) “Xinjiang Daye”, and analyzed their physicochemical properties. Based on phylogenetic analysis, these MsGRFs were divided into five subgroups, each with a similar gene structure and conserved motifs. MsGRFs genes are distributed on 23 chromosomes, and all contain QLQ and WRC conserved domains. The results of the collinearity analysis showed that all MsGRFs are involved in gene duplication, including multiple whole-genome duplication or segmental duplication and a set of tandem duplication, indicating that large-scale duplication is important for the expansion of the GRF family in alfalfa. Several hormone-related and stress-related cis-acting elements have been found in the promoter regions of MsGRFs. Some MsGRFs were highly expressed in young leaves and stems, and their expression decreased during development. In addition, the leaf size of different varieties was found to vary, and MsGRF1 to 4, MsGRF18 to 20, and MsGRF22 to 23 were differentially expressed in large and small leaf alfalfa varieties, suggesting that they are critical in the regulation of leaf size. The results of this study can benefit further exploration of the regulatory functions of MsGRFs in growth and development, and can identify candidate genes that control leaf size development.
Growth-regulating factors (GRFs) are plant-specific transcription factors (TFs), that regulate plant growth and development (Kim et al., 2003; Kim and Lee, 2006; Baucher et al., 2013; Omidbakhshfard et al., 2015). The first GRF, named OsGRF1, was discovered 15 years ago in deep-water rice (Oryza sativa). OsGRF1 was identified as the gene responding to gibberellin and differentially expressed in the internode meristems of deep-water rice (van der Knaap et al., 2000). GRFs have two conserved domains: QLQ (Gln-Q, Leu-L, Gln-Q) and WRC (Trp-W, Arg-R, Cys-C), in the N-terminal region. The QLQ domain contains sites for interaction with GRF-interacting factors (GIFs) (Kim and Kende, 2004), while the WRC domain contains a DNA-binding motif and a nuclear localization signal (Choi et al., 2004). The QLQ domains are more conservative than the WRC domains (van der Knaap et al., 2000). All eukaryotes contain QLQ; however, WRC is a plant-specific domain. In the C-terminal region of GRFs, the types and number of amino acid residues vary greatly; thus, the similarity within the family is low. Owing to the diversity of C-terminal domains, GRF proteins have functional diversity. In addition, the length of the C-terminal region determines the protein size.
Initial studies suggested that GRFs only play a role in leaf and stem development. However, recent studies have discovered that GRFs not only regulate flowering, seed, and root development, but also regulate plant longevity and participate in abiotic stress response (Hewezi et al., 2012; Kim et al., 2012; Liang et al., 2013; Debernardi et al., 2014; Liu et al., 2014). In Arabidopsis thaliana, GRF-overexpressed plants have larger leaves than wild-type plants, while the leaves of grf mutant plant are smaller than wild-type plant (Kim et al., 2003). AtGRF1, 2, and 3 control leaf size by regulating cell expansion (Kim et al., 2003), while AtGRF1, 2, 3, 4, 5, and 9 control leaf development through cell proliferation (Horiguchi et al., 2005; Kim and Lee, 2006; Arvidsson et al., 2011; Debernardi et al., 2014). grf1/2/3/4 quadruple mutant plants lack shoot apical meristems (Kim and Lee, 2006). GRF genes are weakly expressed in mature tissues, but are highly expressed in young tissues, such as seeds, shoots, and young leaves (Liang et al., 2013). GRF genes encode transcription factors that bind to sequence-specific DNA, which interacts with the transcriptional cofactor GRF-INTERACTING FACTOR (GIF) to form functional transcriptional complexes that regulate cell proliferation to control leaf size (Kim et al., 2003, 2022; Kim and Kende, 2004; Horiguchi et al., 2005; Lee et al., 2009; Wang et al., 2011; Debernardi et al., 2014; Lee and Kim, 2014; Lu et al., 2020). GRF is negatively regulated by microRNA (miR396), and its expression is suppressed by miR396 after transcription (Wang et al., 2011; Baucher et al., 2013; Debernardi et al., 2014; Liu et al., 2014, 2021; Li et al., 2019; Szczygiel-Sommer and Gaj, 2019; Liebsch and Palatnik, 2020; Beltramino et al., 2021; Lu et al., 2021; Pegler et al., 2021; Kim et al., 2022). GRF/GIF has a universal growth-promoting effect on inflorescences and flower organs of several species. The miR396-GRF/GIF module is involved in the separation of cotyledons and flower organs (Lee et al., 2018), as well as multiple processes of flower organ growth and reproductive development (Nagai et al., 2001; Hewezi et al., 2012; Kim et al., 2012; Baucher et al., 2013). In addition, loss of GRF and GIF function usually leads to varying degrees of sterility in plants, flower organ fusion, and disturbances in the number of flower organs (Wu et al., 2014; Zan et al., 2020). Some GRFs also play an important role in abiotic stress response, including cold and salt stress (Kim et al., 2012; Khatun et al., 2017; Wallace et al., 2017; Shang et al., 2018; Li et al., 2019, 2021; Cao et al., 2020; Pegler et al., 2021). In soybean, the transcription of all GRFs was affected by shading. Under shade stress, almost all expressions of GRFs are significantly downregulated to varying degrees (Chen et al., 2019).
The identification and function of the GRF gene family has been studied in a variety of plants, including Arabidopsis (9) (Kim et al., 2003), soybean (22) (Chen et al., 2019), rice (12) (Choi et al., 2004), apple (16) (Zheng et al., 2018), mulberry (10) (Rukmangada et al., 2018), wheat (8) (Zan et al., 2020), and foxtail millet (Chen and Ge, 2022). However, studies on the GRF gene family in alfalfa are limited. As a widely used forage, alfalfa is a popular feed for livestock and poultry owing to its high yield, good forage quality, and rich nutrition. Leaves and stems, as the main harvest organ, are limiting factors for the yield and quality of alfalfa. Therefore, it is essential to study the control mechanism of leaf size during leaf development to cultivate and select germplasm resources of alfalfa with high quality.
In this study, we identified 27 MsGRFs in the alfalfa genome using hmmscan and verified them with Pfam1 and Conserved Domain Database (CDD) for the presence of QLQ and WRC domains. According to their chromosomal position, they were named MsGRF1- MsGRF27 (Table 1). The coding sequence (CDS) length of MsGRFs varied slightly from 754°bp to 1,932 bp. The shortest GRF proteins were MsGRF20 and MsGRF21, which contained 251 amino acids, whereas the longest, MsGRF1, 2, and 4, had 643 amino acids. Concurrently, the physicochemical properties of MsGRF proteins were predicted. The theoretical molecular weight (MW) of MsGRFs ranged between 28,577.5 Da (MsGRF20 and MsGRF21) and 69,891.83 Da (MsGRF1, MsGRF2, and MsGRF4), and the isoelectric point (pI) ranged from 6.71 to 10.18. MsGRF proteins are rich in basic amino acids, with 92.59% of MsGRFs proteins having an isoelectric point greater than 7 (Table 1).
To clearly demonstrate evolutionary relationships, we constructed a phylogenetic tree with protein sequences of 27 GRFs from alfalfa, 22 GRFs from soybean, and 9 GRFs from Arabidopsis using by MEGA 64 with the Neighbor-Joining (NJ) method. 58 GRFs were divided into six subgroups (I–VI) (Figure 1). Subgroup I contained only one gene, AtGRF9, and subgroups II–VI contained 8, 10, 12, 13, and 14 GRFs, respectively. Twenty-seven MsGRFs were assigned to subgroups II-VI: MsGRFs 8-10 with AtGRF7-8 and GmGRF4-5 were assigned into subgroup II; MsGRFs 1- 4 belonged to subgroup III with AtGRF1-2, GmGRF18-19, and GmGRF21-22; MsGRFs 18-23 comprised subgroup IV along with AtGRF3-4, GmGRF3, 9, 12, and 20; subgroup V only contained MsGRFs11-17 and GmGRF (1 to 2, 8, 10, and 13) proteins; MsGRFs 5, 6, and 24-27 formed subgroup VI with AtGRF 5- 6, GmGRF6, 7, and 14-17. As the functions of many Arabidopsis GRFs have been studied, the function of MsGRF clustered with the GRFs from soybean and Arabidopsis can be inferred from previous research. From the phylogenetic tree, it can be concluded that MsGRFs are more closely related to GmGRFs than to AtGRFs, which may be because both soybean and alfalfa are legumes.
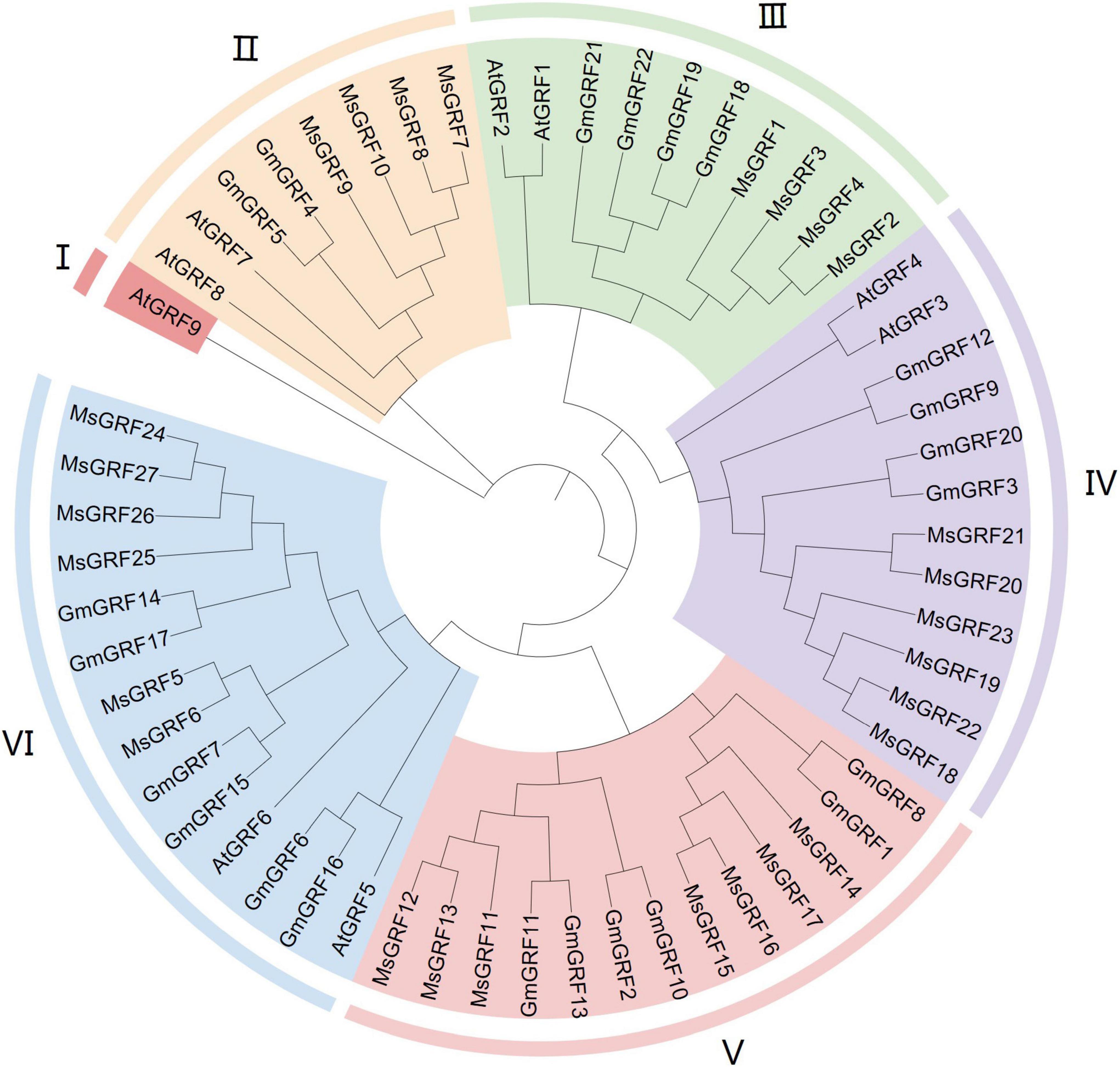
Figure 1. Phylogenetic analysis of growth-regulating factors (GRFs) from Medicago sativa L. (Ms), Glycine max (Gm) and Arabidopsis thaliana (At). MEGA 7.0 software was employed to construct a neighbor-joining phylogenetic tree with 1,000 bootstrap replications. Subgroups are highlighted with different colors.
Gene sequences of MsGRFs showed that the conserved QLQ and WRC domains existed in the N-terminal region of all MsGRFs (Supplementary Figure 1). To further study gene structure and evolutionary relationships, a phylogenetic tree was constructed using the protein sequences of MsGRFs, and their gene structure and motif characteristics were analyzed (Figures 2A–C). The homology of the MsGRF genes was relatively high and the motif distribution was similar, particularly in the same subgroup. Ten conserved motifs were identified using the MEME online program and renamed motifs 1–10 (Supplementary Table 1). All MsGRFs contained different numbers of motifs, ranging from 3 to 10. All MsGRFs had motif 1 and motif 2, annotated by NCBI CDD2 as WRC and QLQ, respectively, which are GRF-specific domains. Four members of subgroup III (MsGRF1, MsGRF2, MsGRF3, and MsGRF4) contained all ten motifs (Figures 1, 2B). Two MsGRFs (MsGRF20 and MsGRF21) contained the least number of motifs. All MsGRFs contained motif 6, however, the distribution on the genes differed (Figure 2B).

Figure 2. Analysis on phylogenetic relationships, motifs, and gene structure of growth-regulating factor genes from Medicago sativa. (A) Phylogenetic tree of 27 MsGRFs in alfalfa. (B) Conserved motif arrangements of MsGRFs. The motifs are indicated in different colored boxes with different numbers. Motifs 1 and 2 represent WRC and QLQ domains, respectively. (C) Exon-intron organizations of MsGRFs. Blue boxes indicate exons; black lines indicate introns.
Exon–intron structures clearly showed that the MsGRFs contained two to four introns (Figure 2C). In all, 18 of the 27 MsGRFs contained three introns; five MsGRFs (MsGRF1, 2, 4, 19, and 23) contained four introns; four genes (MsGRF14, 15, 16, and 17) contained two introns. Genes that were closely related in the phylogenetic tree had approximately the same distribution area of exons and introns. The members of each subgroup in the phylogenetic tree were similar in size and contained similar genetic structures (Figures 1, 2C). All the members of each subgroup contained the same number and similar length of exons. The length of each MsGRF differed depending on the length of the intron. The length of the CDS of MsGRF23 was only 813°bp, while its full length genomic DNA was the longest. In general, motif distribution and gene structure indicate the evolutionary relationship between MsGRFs.
All MsGRFs were unevenly distributed on 23 chromosomes of alfalfa (Figure 3), and were not identified on the nine other chromosomes of alfalfa (2n = 4x = 32). Chromosome 7.4 (chr7.4) contains five MsGRF genes. Only one MsGRF gene was found on chr1.1, chr1.2, chr1.3, chr1.4, chr2.2, chr2.4, chr3.1, chr3.2, chr3.3, chr3.4, chr4.2, chr4.3, chr4.4, chr5.1, chr5.2, chr5.3, chr5.4, chr7.2, chr8.1, chr8.2, chr8.3, and chr8.4. For chromosomes 1, 3, 5, and 8, each allele chromosome had one MsGRF gene, whereas on chromosomes 2 and 7, only allele chromosomes x.2 and x.4 had MsGRF genes. On chromosome 4, only allele chromosome 4.1 had no MsGRF gene. On chromosome 6, the MsGRF gene has not yet been identified in each chromosome allele.
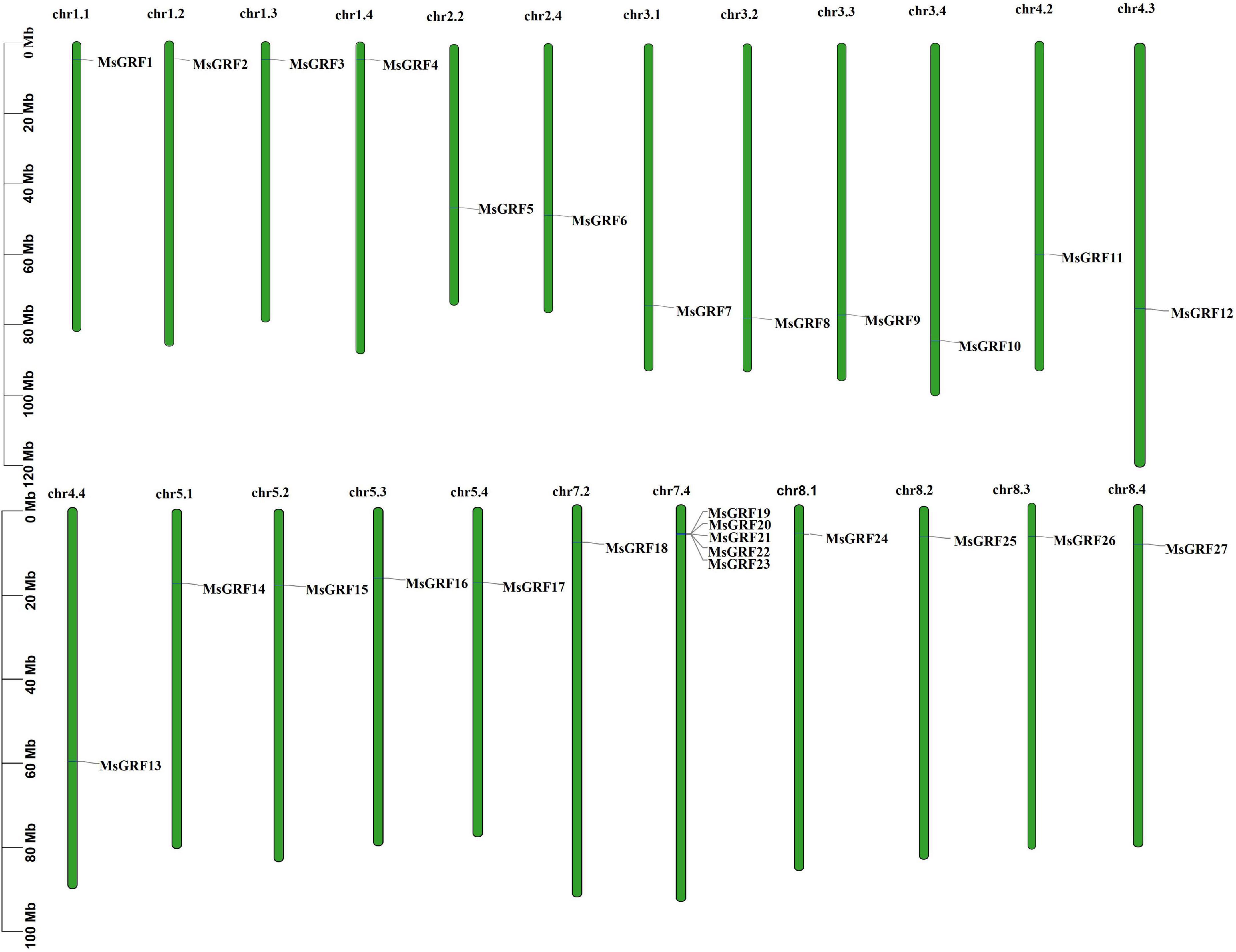
Figure 3. The distribution of MsGRFs on alfalfa chromosomes. The green bars represent each chromosome, and the black lines label the position of each MsGRF gene.
Gene duplication is considered as one of the primary driving forces in the evolution of genomes and genetic systems. To study the gene duplication relationship of the alfalfa GRF family, collinearity analysis of MsGRFs was performed using Tbtools (Figure 4). All MsGRFs are involved in the duplication process, including tandem duplication, whole-genome duplication (WGD) or segment duplication. MsGRF19-MsGRF23 is a set of tandem duplications located on chr7.4. Other duplicated gene pairs are genome-wide duplication or segment duplications. The non-synonymous substitution rates (Ka) and synonymous substitution rates (Ks) for each duplicated gene pair were calculated (Supplementary Table 2). The Ka/Ks values of all gene pairs were less than 1, indicating that the MsGRF gene family is subject to purifying selection.
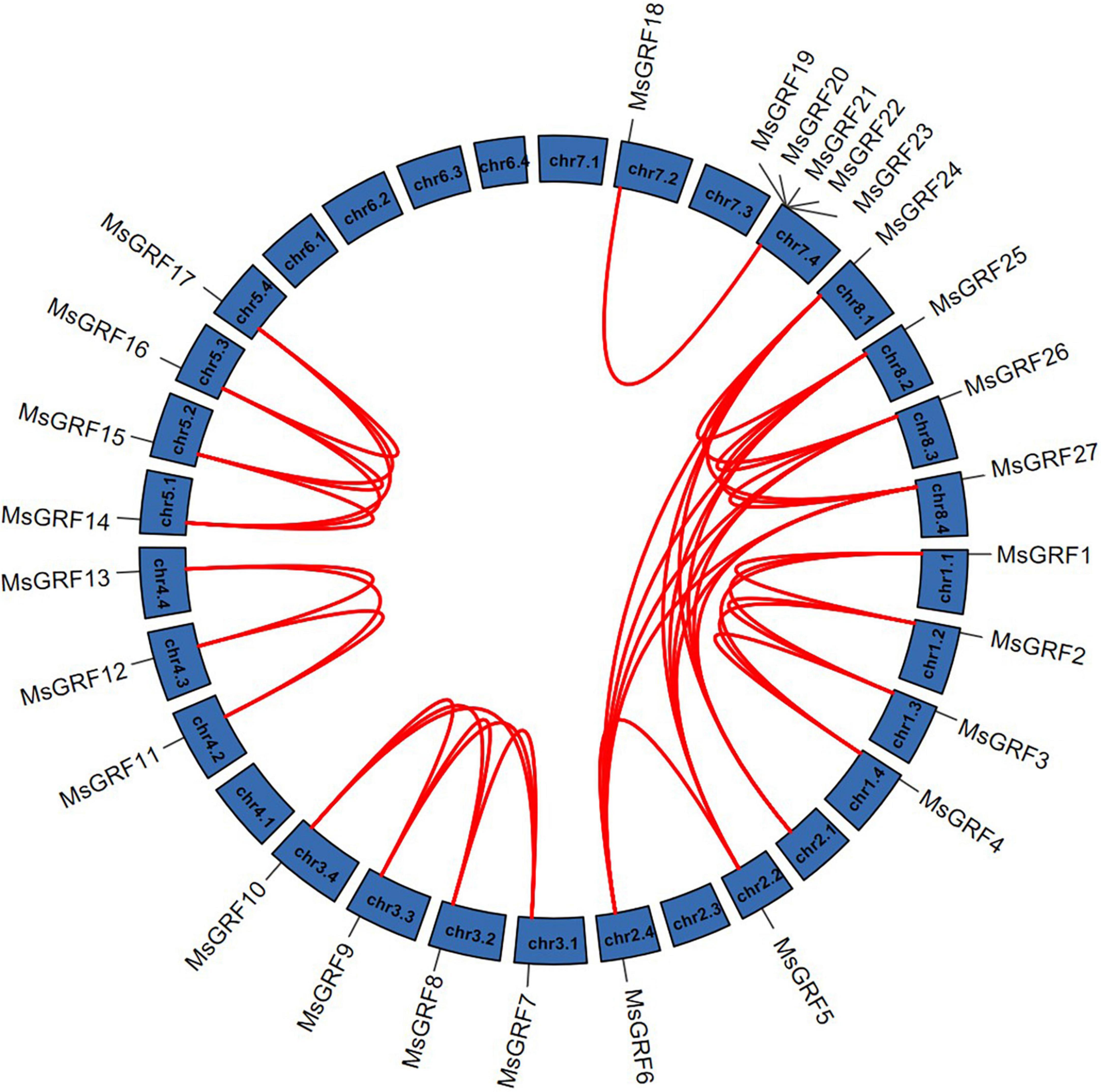
Figure 4. Synteny analysis of MsGRFs genes in alfalfa. Red lines indicate the replicated MsGRFs gene pairs in alfalfa.
The online cis-element database PlantCARE was used to analyze the promoter sequences (upstream 2,000 bp) of MsGRFs. Conserved core elements TATA-box and enhancement elements CAAT-box in the promoter sequences were observed, which conformed to the basic structural characteristics of eukaryotic gene promoters. The promoter sequence also contained many elements related to hormonal and abiotic stress responses (Figure 5). Hormone-responsive elements include jasmonic acid-responsive elements (CGTCA-motif and TGACG-motif), salicylic acid cis-acting element (TCA-element), gibberellin-responsive elements (GARE motif, P-box and TATC-box), abscisic acid-responsive elements (ABRE), and auxin-responsive elements (AuxRR-core, TGA-element). Abiotic stress response elements include the anaerobic inducible element (ARE), disease resistance and stress response element (TC-rich repeats), low temperature responsive cis-acting element (LTR), and MYB binding site involved in drought-inducibility (MBS). In addition, we found certain unique cis-acting elements in the promoter sequence: CAT-box (cis-acting regulatory element related to meristem expression), MSA-like (cis-acting element involved in cell cycle regulation), and HD-Zip 1 (element involved in differentiation of the palisade mesophyll cells). Each MsGRF contains at least one hormone-related cis-element and one stress-related cis-acting element, however, the types vary.
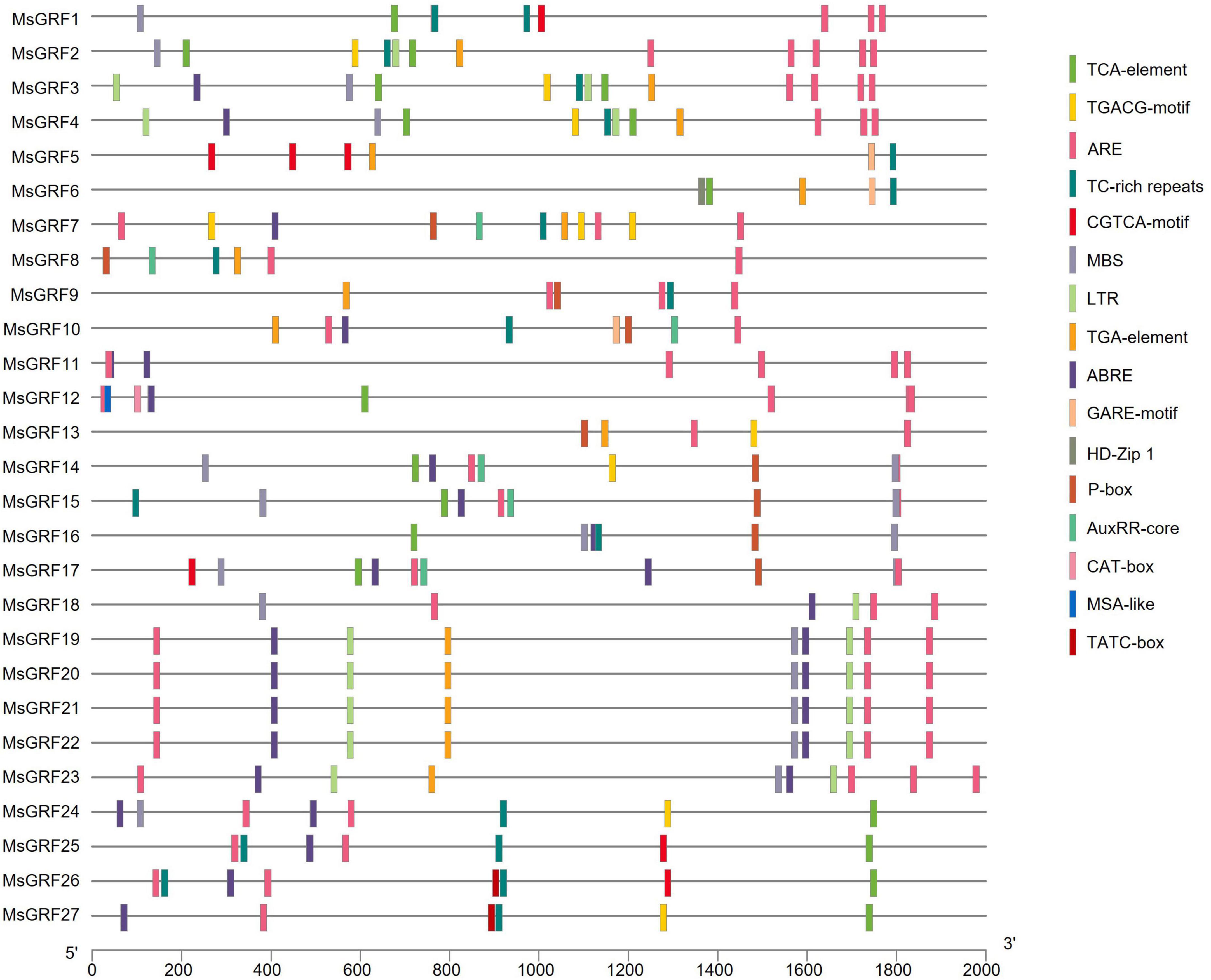
Figure 5. The cis-acting element contained in the 2 kb promoter sequence of the MsGRF gene. Different cis-elements are indicated by different colored rectangles and placed in the matching position on the promoter.
To analyze the expression of MsGRFs at different developmental stages of stems and leaves, the expression levels of 27 genes at different growth and developmental stages were verified by qRT-PCR, and the results were visualized as heatmaps (Figures 6A,B and Supplementary Table 3). Each stem internode, from the apex to base of the stem, is used as a developmental stage, labeled as S1 to S8. The first leaf that has not fully unfolded is regarded as the first stage of leaf development (L1), and is then divided into L1 to S4 according to leaf position (Supplementary Figure 2).
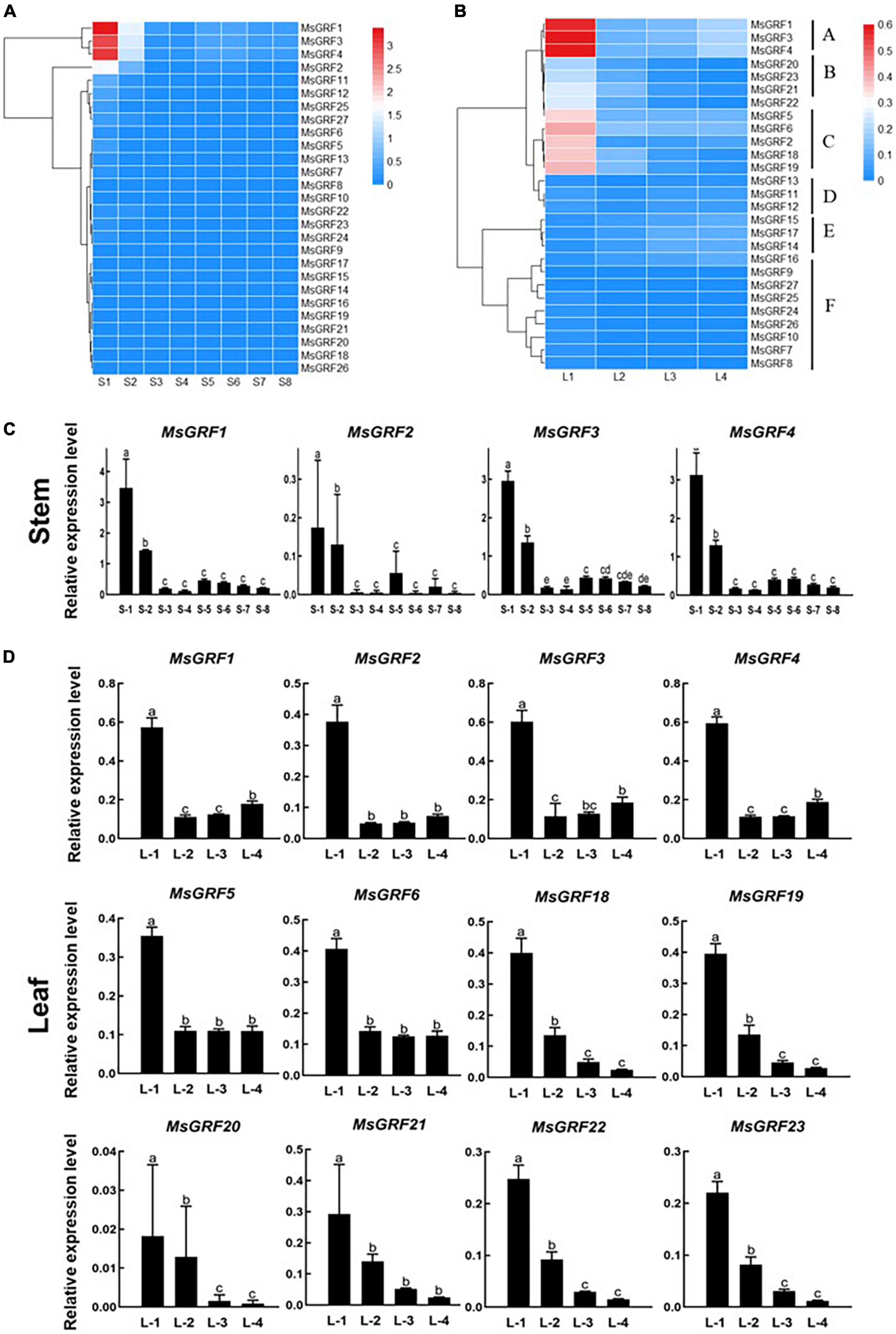
Figure 6. Expression profiling of MsGRFs. (A) Expression profiles of MsGRFs at different developmental stages of stem internodes. Each stem internode starting from the apex is used as a developmental stage, labeled as Stem-1 to 8. (B) Expression profiles of MsGRFs at different developmental stages of leaves clustered into A to F six clades. L1 to L4 indicates the leaf development according to leaf position, and the first leaf that has not fully unfolded is regarded as the first stage of leaf development (L1). (C,D) qRT-PCR quantification of gene expression levels of selected MsGRF genes from (A,B) displayed in a column chart. The different letters (a, b, c, etc.) indicate the significant difference at P < 0.05 by Student’s t-test analysis.
The expression of MsGRFs in stems is shown in the Figure 6A. According to the expression patterns at different developmental stages of the stem, most of the GRF family genes were weakly expressed. Compared with other MsGRF genes, the expression levels of MsGRF1 to 4 were significantly higher. These four genes were most strongly expressed in the S1 stage, followed by the S2 stage, and weakly expressed in the stems at other developmental stages. The qRT-PCR results in column chart showed clearly that the expression levels of these four MsGRFs decreased dramatically from the first period, increased a little in the S5 period, and then decreased gradually (Figure 6C). Overall, the expression of the four MsGRFs gradually decreased during stem growth and development. In conclusion, it is speculated that MsGRFs1-4 plays an important role in regulating stem development.
In the expression profile of leaves (Figure 6B), MsGRFs can be clustered into six clades, denoted by A-F. The MsGRF genes of cluster F were negligibly expressed. Cluster D and E expression was weak, but the expression level of cluster E increased slightly at the L3 and L4 stage. In contrast, cluster A was strongly expressed in leaves, especially at the L1 stage. The expression levels of clusters B and C were higher in the early stages of leaf development, and differed significantly from those in the other stages (Figure 6B). The expression of MsGRFs1-6 was the strongest in the L1 stage, fluctuated in the L2, L3, and L4 stages, and showed a downward trend in general, revealed by qRT-PCR analysis in column chart (Figure 6D). The expression of MsGRFs18 to 23 reached the summit at the L1 stage, and then decreased significantly during leaf development (Figure 6D). In summary, the expression of MsGRFs was high in the early stages of leaf development and weak in mature leaves. These results indicate that MsGRF1-6 and MsGRF18-23 play important roles in the leaf development.
To analyze GRF function on leaf development, the small leaf, and large leaf alfalfa varieties was investigated. The leaves of “Xinjiang Daye” were larger than those of the “Nei 1 × Nei 2” varieties (Figure 7A). To clarify whether the development of alfalfa leaf size was regulated by cell proliferation or expansion, the number of lower epidermal cells of the L4 stage leaves from the two varieties under a microscope and the average cell area of a single epidermal cell was investigated (Figures 7A,B). The average area of a single lower epidermal cell in “Xinjiang Daye” is larger than “Nei 1 × Nei 2” (Figure 7C). The average cell number of a single leaf in “Xinjiang Daye” is much higher than that in “Nei 1 × Nei 2” (Figure 7D). These results implied that leaf size is regulated by both cell proliferation and expansion. According to the expression of MsGRFs at different developmental stages of leaves (Figure 6D), we selected MsGRFs with high expression at L1 stage for qRT-PCR analysis to verify their expression in large leaves (“Xinjiang Daye”) and small leaves (“Nei 1 × Nei 2”) alfalfa varieties. The selected MsGRFs were highly expressed in these two varieties, but they had differences in the expression levels. It was found that the expression levels of MsGRF1 to 4, MsGRF18 to 20, and MsGRF22 to 23 were much higher in “Xinjiang Daye” than that in “Nei 1 × Nei 2” (Figure 8).
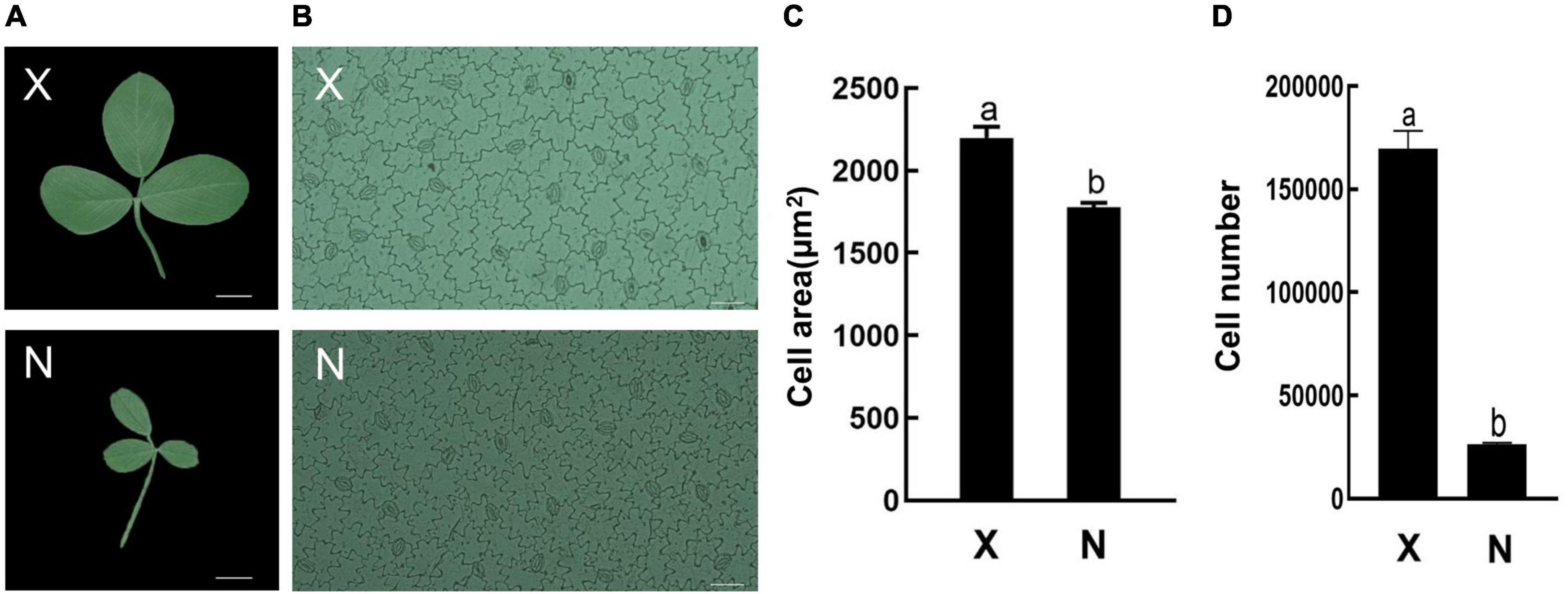
Figure 7. Leaf morphology and lower epidermal cells observed under microscope. (A) Morphological observation of large (X, “Xinjiang Daye”) and small (N, “Nei 1 × Nei 2”) leaves. Scale bar, 1 cm. (B) Epidermal cells of large (X, “Xinjiang Daye”) and small (N, “Nei 1 × Nei 2”) leaves under microscope. Scale bar, 50 μm. (C) The average area of a single epidermal cell in the leaf of “Xinjiang Daye” and “Nei 1 × Nei 2”. (D) Estimates of the average total cells number in a single leaf of “Xinjiang Daye” and “Nei 1 × Nei 2”. The letters (a, b) indicate the significant difference at P < 0.05 by Student’s t-test. The L4 stage leaves were used for the observation.
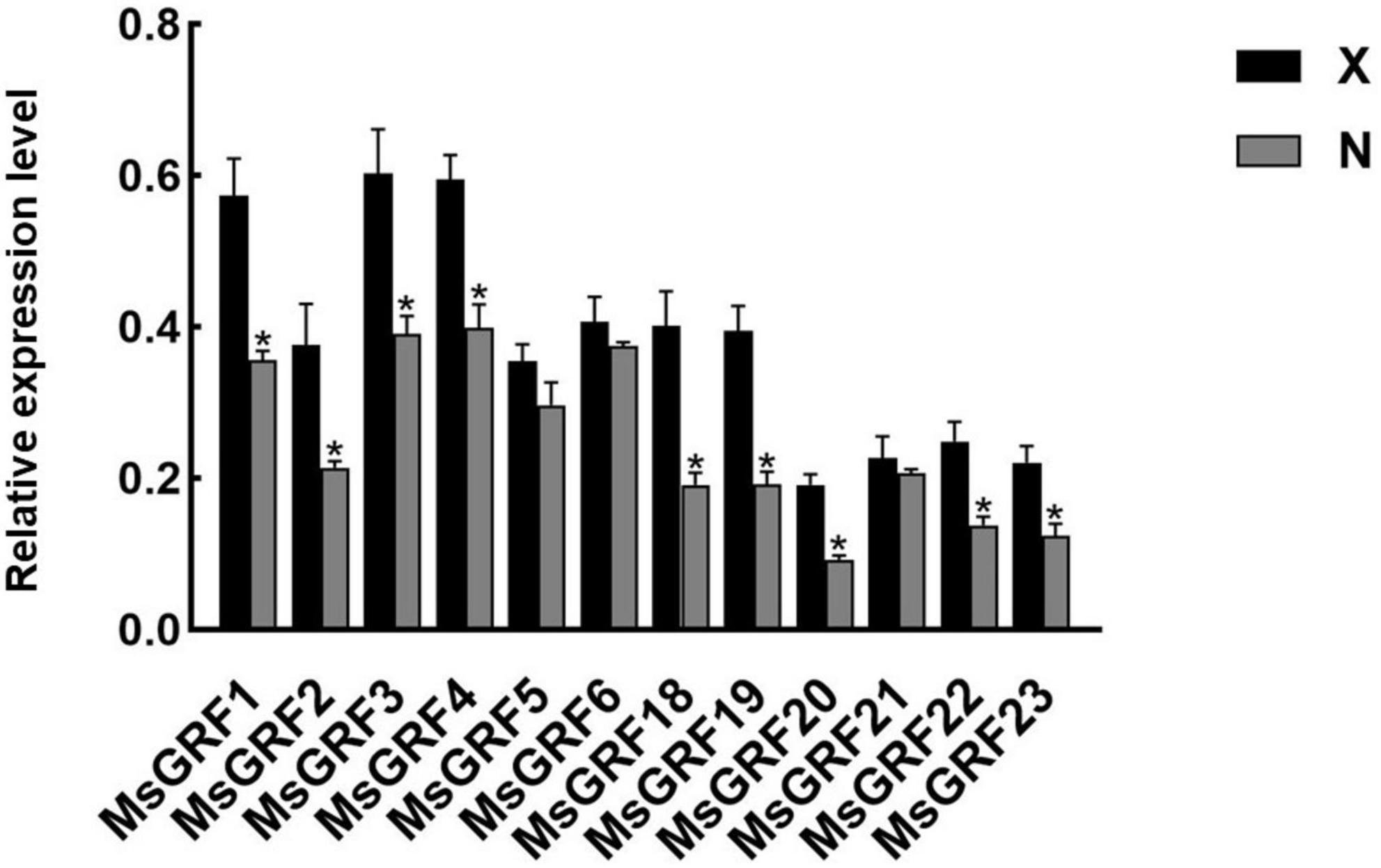
Figure 8. Quantification of the expression levels of selected MsGRFs in the leaves of X, “Xinjiang Daye” and N, “Nei 1 × Nei 2” using qRT-PCR. Vertical bars indicate standard deviation. The asterisk (*) indicates the significant difference at P < 0.05 by Student’s t-test.
The GRF family, as a class of plant-specific transcription factors, plays an important role in plant growth and development (Kim et al., 2003; Kim and Lee, 2006; Baucher et al., 2013; Omidbakhshfard et al., 2015). The GRF gene family has been identified and studied in many species, but has not been reported in alfalfa. Existing studies have confirmed that the GRF family can regulate the development of roots, stems, leaves, flowers, and fruits, the maintenance of shoot apical meristems, regulate plant longevity and respond to abiotic stresses (van der Knaap et al., 2000; Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Kim and Lee, 2006; Marcotrigiano, 2010; Wang et al., 2011; Hewezi et al., 2012; Baucher et al., 2013; Debernardi et al., 2014; Lee and Kim, 2014; Wu et al., 2014; Tao et al., 2016; Beltramino et al., 2018; Lee et al., 2018; Lockhart, 2018; Zhang D. et al., 2018; Lu et al., 2020; Kim et al., 2022). In Arabidopsis, GRF genes regulate leaf size via cell proliferation and expansion (Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Wang et al., 2011; Debernardi et al., 2014; Beltramino et al., 2018; Shimano et al., 2018). The leaves are the main site of photosynthesis (Tsukaya, 2014), and the main harvesting organ in alfalfa. Therefore, it is essential to study the regulatory mechanisms of the GRF gene family in the development of alfalfa leaf size. In this study, we performed bioinformatic analysis of 27 GRF gene family members identified in alfalfa variety “Xinjiang Daye”, and predicted the physicochemical properties and cis-acting elements of MsGRFs. To clarify the evolutionary relationship of the GRF gene family, a phylogenetic tree was constructed and its gene structure and motif distribution were studied. The replication relationship between genes was analyzed using chromosomal location and collinearity analysis. The expression patterns of MsGRFs at different growth and developmental stages of the leaves and stems were quantitatively analyzed by qRT-PCR.
The GRF family contains QLQ and WRC conserved domains at the N-terminus, and the WRC region contains DNA-binding motifs and nuclear localization signal regions, which can combine with the cis-acting regions of downstream genes to regulate the expression of such genes (Kim and Kende, 2004). The QLQ domain can combine with GIF to form a transcriptional activator and play a regulatory role (Choi et al., 2004). These 27 genes were verified to contain QLQ and WRC domains (Supplementary Figure 1); therefore, they were finally identified as GRF family members. The protein lengths of MsGRFs ranged from 251 to 643 amino acids. Subsequently, the physicochemical properties of the MsGRFs were predicted, including isoelectric point and molecular weight. The theoretical MW of MsGRFs was between 28,577.5 Da and 69,891.83 Da, and the isoelectric point (pI) was between 6.71 and 10.18 (Table 1).
Phylogenetic analysis showed that 27 MsGRFs were divided into six subgroups according to their phylogenetic relationships (Figure 1). AtGRF1, 2, 3 have been shown to regulate leaf size through cell proliferation and expansion (Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Hewezi et al., 2012; Beltramino et al., 2018), therefore, the function of MsGRFs in the same subgroup can be inferred according to clustering in the evolutionary tree, which provides a foundation for future studies on the mechanism of GRFs that control leaf size development. The gene structure and motif distribution were consistent with the phylogenetic results, which confirmed the phylogenetic relationship among MsGRF genes (Figure 2). Members of the same subgroup contain similar gene structures and conserved motifs. Studies have shown that most genes in the GRF family contain three introns (Wang et al., 2014; Shang et al., 2018; Zhang J. et al., 2018), which is consistent with our results of gene structure analysis (Figure 2C).
Gene duplication is considered to be one of the primary driving forces in the evolution of genomes and genetic systems. Tandem duplication events and large-segment duplication events are considered the main reasons for the expansion of gene families in the genome (Levasseur and Pontarotti, 2011). MsGRFs were distributed on 23 chromosomes, and no GRF family members were identified in any of the copies of chr 6.1–6.4 (Figure 3). According to sequence alignment, the sequences of the MsGRFs on each chromosomal copy were highly homologous. The results of the collinearity analysis showed that the MsGRF gene family was expanded by large segment duplication. The MsGRFs in alfalfa were involved in gene duplication events (Figure 4). Chromosome 7.4 contains five genes identified as tandem repeats of MsGRFs, which are arranged in neighboring positions, and form a gene cluster with similar sequences. In other species, such as soybean, wheat, and foxtail millet, duplication events of the GRF gene family have also been demonstrated, (Chen et al., 2019; Zan et al., 2020; Chen and Ge, 2022). The Ka/Ks of all replicating gene pairs was less than 1 (Supplementary Table 2), and most of the non-synonymous substitutions were harmful, indicating that the environmental selection pressure during the evolution process was negative, and the MsGRF genes were selected for purification.
Cis-acting elements are DNA sequences that exist upstream of a gene and participate in the regulation its expression. They do not encode any protein but only provide a binding site for action (Hernandez-Garcia and Finer, 2014). In this study, we predicted that cis-acting elements located 2,000 bp upstream of the promoter using PlantCARE (Figure 5). The promoter sequences of MsGRFs contain hormone-related cis-acting elements and stress-related cis-acting elements, among which ARE is the most widely distributed, followed by ABRE. In addition, only MsGRF6 contained HD-Zip 1 (an element involved in differentiation of the palisade mesophyll cells), and MsGRF12 contained CAT-box (cis-acting regulatory element related to meristem expression) and MSA-like (cis-acting element involved in cell cycle regulation) element. Each MsGRF contained abiotic stress-related cis-acting elements, indicating that these genes responded to different stresses. Based on these results, candidate genes are provided for studies related to abiotic stress.
The expression of MsGRFs in various tissues plays an important role in growth and development. It has been demonstrated in previous studies that the GRF gene family is strongly expressed in young tissues and weakly expressed in mature tissues (Zhang et al., 2008; Khatun et al., 2017; Zheng et al., 2018; Zhou et al., 2018; Zan et al., 2020; Tang et al., 2021). In this study, the expression patterns of the MsGRF family were similar in leaves and stems, with high expression in young stems and leaves, which decreased with growth and development (Figure 7). MsGRFs1-4 were significantly expressed in leaves and stems, indicating that these genes play an important role in regulating their growth and development. By observing the lower epidermal cells of different sizes of leaves from different varieties, we found that the size of alfalfa leaves was controlled by cell proliferation and expansion (Figure 7). The expression of several MsGRF genes was significantly different in large and small leaf alfalfa varieties, such as MsGRF1 to 4, MsGRF18 to 20, and MsGRF22 to 23 (Figure 8), which may be related to the regulation of leaf size. As the main site of photosynthesis, the leaves are also the main harvesting organs of alfalfa. Studying the control mechanism of leaf size is crucial to understanding the ecology and increasing production of alfalfa (Liu et al., 2012, 2021). According to the expression of MsGRF genes in different developing leaves, many candidate genes have been identified, and the key genes controlling leaf size need to be further investigated.
In this study, 27 GRF family members in alfalfa were identified and their basic characteristics and functions were subjected to preliminarily analysis. QLQ and WRC are two domains unique to the GRF gene family that helped us identify MsGRFs in alfalfa. To study the evolutionary relationships between GRFs, a phylogenetic tree was constructed and divided into six subgroups. Members of the same subgroup have similar gene structures and motif distributions. In alfalfa, there are 23 chromosomes with GRF family genes, among which chr7.4 contains five genes and the other chromosomes only contain one gene. All MsGRFs are involved in gene duplication events including tandem duplication, whole-genome duplication, and segment duplication. The results of the collinear analysis showed that gene duplication facilitated the expansion of MsGRFs. The upstream regions of the promoters of MsGRFs all contain one or more hormone or stress-related cis-acting elements. MsGRF1-4 were strongly expressed in young stems and leaves, whereas MsGRF5, 6 and MsGRF18-23 were only highly expressed in young leaves and not in stem. The expression of MsGRF1-4, MsGRF18-20, and MsGRF22-23 were significantly different in large and small leaf alfalfa varieties. In conclusion, these results lay a foundation for us to further study the function and regulatory mechanism of the alfalfa GRF gene family in leaf development and screen the key genes for controlling leaf size.
The “Xinjiang Daye” genome used in this article is publicly available in the NCBI database under project PRJNA540215, and the genome assembly files are available at https://figshare.com/projects/whole_genome_sequencing_and_assembly_of_Medicago_sativa/66380 (Chen et al., 2020). Alfalfa protein sequences and genome annotations were downloaded from the Alfalfa Breeder’s Toolbox.3 Transcription factor prediction and blast analysis were performed using Majorbio Cloud Platform.4 Analysis of transcription factors was performed using hmmscan with parameter E-value ≤ 1e–5 (Lozano et al., 2015). Finally, sequences were verified with Pfam(see text footnote 1) and CDD.5 The genes containing the WRC and QLQ domains were considered to be MsGRFs. The ExPASy proteomics server6 was used to predict the physicochemical properties of each MsGRF protein, including the molecular weight (MW) and theoretical isoelectric point (pI) (Wilkins et al., 1999).
Full-length amino acid sequences of GRF in alfalfa were aligned using MEGA 7.0 (Kumar et al., 2016). Conserved motifs for predicted MsGRFs protein sequences were identified using the MEME online program7 with default settings, except that the motif number was set as 10 (Bailey et al., 2009). Gene structure and motif distribution were visualized using the TBtools software (Chen et al., 2018). Nine Arabidopsis GRF sequences from The Arabidopsis Information Resource (TAIR)8 and 22 GRF sequences of soybean from Phytozome v139 were used to construct a phylogenetic tree (Supplementary Table 5). MEGA 7.0 was used to construct phylogenetic trees using the neighbor-joining method with Poisson model, pairwise deletion, and 1,000 bootstrap replications (Kumar et al., 2016). The cis-acting elements in the 2,000 bp upstream sequences of the promoter of MsGRFs were predicted using PlantCARE10 (Lescot et al., 2002), and TBtools was used to visualize the cis-acting elements.
The chromosomal location of the alfalfa GRF gene was obtained from the genome assembly files,11 and the chromosomal distribution was mapped using TBtools. Collinearity analysis of 27 MsGRFs gene was performed using TBtools software to detect gene duplication events. Based on the results of the collinearity analysis, calculation of non-synonymous (Ka) and synonymous (Ks) substitutions for each pair of duplicated genes were made using TBtools. The ratio of Ka/Ks was used to do the analysis of selection pressure.
To investigate the expression patterns of MsGRFs, total RNA from different tissues were used for qRT-PCR (Schmittgen and Livak, 2008). Total RNA was extracted using the Takara MiniBEST Plant RNA Extraction Kit (Takara Bio Inc., Kusatsu, Japan), and the RNAs were reverse transcribed into cDNAs using HiScript III® RT SuperMix for qRT-PCR (+ gDNA wiper) (Vazyme Biotech Co., Ltd., Nanjing, China). ChamQ SYBR Color qRT-PCR MasterMix (Vazyme Biotech Co., Ltd., Nanjing, China) was used for qRT-PCR, and the MsUBC Q-2F gene was used as a reference gene, each of which had three technical replicates. Beacon Designer 7.9 was used to design real-time quantitative primers, and the sequences of the primers used for qRT-PCR was listed in Supplementary Table 4.
In this experiment, we used cultivated “Xinjiang Daye” and “Nei 1 × Nei 2” alfalfa as plant material. The alfalfa seeds were placed in a petri dish containing H2O and then placed in a germination bag. After seven days, the germinated seedlings were transferred to 1/2 Hoagland’s nutrient solution for growth and cultivation, during which the nutrient solution was changed every 3 days. Plants were placed in an artificial climate incubator with a 16-hour photoperiod, day and night temperature of 25°C/22°C, and relative humidity of 60–70%. The plant materials used for the morphological observation were “Nei 1 × Nei 2” and “Xinjiang Daye” alfalfa cultivated in the same environment.
To analyze the expression patterns in different growth stages, the leaves and stems of 30-day-old alfalfa were selected. The leaves were divided into four developmental stages (L1, L2, L3, and L4), and the stems of the same plant were divided into eight developmental stages (S1 to S8) according to the order of stem nodes from the apex to base (Supplementary Figure 2). All samples were immediately frozen in liquid nitrogen and stored at −80° until use.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material
MC, YS, and HL designed and planned the experiments. HL, YS, JW, and WT performed the experiments. YS and HL analyzed the data and wrote the manuscript. MC, LC, HX, and Z-YW revised the manuscript. All authors read, revised, and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31972958), the Start Up Funds for High Level Talents of Qingdao Agricultural University (663-1120001), and the First Class Grassland Science Discipline Program of Shandong Province, China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.964604/full#supplementary-material
Supplementary Figure 1. Protein sequence alignment of 27 MsGRFs and conserved domains of QLQ and WRC.
Supplementary Figure 2. Different developmental stages of stems and leaves in alfalfa. L1 represents the first leaf that is not fully expanded, then L2, L3, and L4 are defined according to leaf position. From top to bottom in the stem, each stem node is used as a developmental stage, represented by S1–8.
Arvidsson, S., Pérez-Rodríguez, P., and Mueller-Roeber, B. (2011). A growth phenotyping pipeline for Arabidopsis thaliana integrating image analysis and rosette area modeling for robust quantification of genotype effects. New Phytol. 191, 895–907. doi: 10.1111/j.1469-8137.2011.03756.x
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Baucher, M., Moussawi, J., Vandeputte, O. M., Monteyne, D., Mol, A., Perez-Morga, D., et al. (2013). A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol. 15, 892–898. doi: 10.1111/j.1438-8677.2012.00696.x
Beltramino, M., Debernardi, J. M., Ferela, A., and Palatnik, J. F. (2021). ARF2 represses expression of plant GRF transcription factors in a complementary mechanism to microRNA miR396. Plant Physiol. 185, 1798–1812. doi: 10.1093/plphys/kiab014
Beltramino, M., Ercoli, M. F., Debernardi, J. M., Goldy, C., Rojas, A. M. L., Nota, F., et al. (2018). Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions. Sci. Rep. 8:13447. doi: 10.1038/s41598-018-29859-9
Cao, J. F., Huang, J. Q., Liu, X., Huang, C. C., Zheng, Z. S., Zhang, X. F., et al. (2020). Genome-wide characterization of the GRF family and their roles in response to salt stress in Gossypium. BMC Genom. 21:575. doi: 10.1186/s12864-020-06986-0
Chen, C., Xia, R., Chen, H., and He, Y. (2018). TBtools, a Toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv [Preprint]. doi: 10.1101/289660
Chen, F., Yang, Y., Luo, X., Zhou, W., Dai, Y., Zheng, C., et al. (2019). Genome-wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biol. 19:269. doi: 10.1186/s12870-019-1861-4
Chen, H. L., and Ge, W. A. (2022). Identification, molecular characteristics, and evolution of GRF gene family in foxtail millet (Setaria italica L.). Front. Genet. 12:727674. doi: 10.3389/fgene.2021.727674
Chen, H., Zeng, Y., Yang, Y., Huang, L., Tang, B., Zhang, H., et al. (2020). Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 11:2494. doi: 10.1038/s41467-020-16338-x
Choi, D., Kim, J. H., and Kende, H. (2004). Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 45, 897–904. doi: 10.1093/pcp/pch098
Debernardi, J. M., Mecchia, M. A., Vercruyssen, L., Smaczniak, C., Kaufmann, K., Inze, D., et al. (2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79, 413–426. doi: 10.1111/tpj.12567
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217-218, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hewezi, T., Maier, T. R., Nettleton, D., and Baum, T. J. (2012). The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant physiol. 159, 321–335. doi: 10.1104/pp.112.193649
Horiguchi, G., Kim, G. T., and Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365-313X.2005.02429.x
Khatun, K., Robin, A. H. K., Park, J. I., Nath, U. K., Kim, C. K., Lim, K. B., et al. (2017). Molecular characterization and expression profiling of tomato GRF Transcription factor family genes in response to abiotic stresses and phytohormones. Int. J. Mol. Sci. 18:1056. doi: 10.3390/ijms18051056
Kim, J. H., and Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 13374–13379. doi: 10.1073/pnas.0405450101
Kim, J. H., and Lee, B. H. (2006). Growth-regulating factor4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49, 463–468. doi: 10.1007/BF03031127
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313x.2003.01862.x
Kim, J.-S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012). Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Kim, Y., Takahashi, S., and Miyao, M. (2022). Relationship between reduction in rice (Nipponbare) leaf blade size under elevated CO2 and miR396-GRF module. Plant Signal. Behav. 17:2041280. doi: 10.1080/15592324.2022.2041280
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, B. H., and Kim, J. H. (2014). Spatio-temporal distribution patterns of GRF-interacting factor expression and leaf size control. Plant Signal. Behav. 9:e29697. doi: 10.4161/psb.29697
Lee, B. H., Ko, J. H., Lee, S., Lee, Y., Pak, J. H., and Kim, J. H. (2009). The Arabidopsis GRF-interacting factor gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151, 655–668. doi: 10.1104/pp.109.141838
Lee, S. J., Lee, B. H., Jung, J. H., Park, S. K., Song, J. T., and Kim, J. H. (2018). Growth-regulating factor and GRF-interacting factor specify meristematic cells of gynoecia and anthers. Plant Physiol. 176, 717–729. doi: 10.1104/pp.17.00960
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Levasseur, A., and Pontarotti, P. (2011). The role of duplications in the evolution of genomes highlights the need for evolutionary-based approaches in comparative genomics. Biol. Direct 6, 11–11. doi: 10.1186/1745-6150-6-11
Li, A. L., Wen, Z., Yang, K., and Wen, X. P. (2019). Conserved miR396b-GRF regulation is involved in abiotic stress responses in Pitaya (Hylocereus polyrhizus) (vol 20, 2501, 2019). Int. J. Mol. Sci. 20:4795. doi: 10.3390/ijms20194795
Li, Z. Q., Xie, Q., Yan, J. H., Chen, J. Q., and Chen, Q. X. (2021). Genome-wide identification and characterization of the abiotic-stress-responsive GRF gene family in diploid woodland strawberry (Fragaria vesca). Plants Basel 10:1916. doi: 10.3390/plants10091916
Liang, G., He, H., Li, Y., Wang, F., and Yu, D. (2013). Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 164, 249–258. doi: 10.1104/pp.113.225144
Liebsch, D., and Palatnik, J. F. (2020). MicroRNA miR396, GRF transcription factors and GIF co-regulators: A conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 53, 31–42. doi: 10.1016/j.pbi.2019.09.008
Liu, H., Guo, S., Xu, Y., Li, C., Zhang, Z., Zhang, D., et al. (2014). OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 165, 160–174. doi: 10.1104/pp.114.235564
Liu, J., Hua, W., Yang, H. L., Zhan, G. M., Li, R. J., Deng, L. B., et al. (2012). The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 63, 3727–3740. doi: 10.1093/jxb/ers066
Liu, Y., Yan, J., Wang, K., Li, D., Yang, R., Luo, H., et al. (2021). MiR396-GRF module associates with switchgrass biomass yield and feedstock quality. Plant Biotechnol. J. 19, 1523–1536. doi: 10.1111/pbi.13567
Lockhart, J. (2018). Design stars: How GRF-interacting factors help determine the layout of the root tip. Plant Cell 30, 265–266. doi: 10.1105/tpc.18.00059
Lozano, R., Hamblin, M. T., Prochnik, S., and Jannink, J. L. (2015). Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 16:360. doi: 10.1186/s12864-015-1554-9
Lu, Y., Meng, Y., Zeng, J., Luo, Y., Feng, Z., Bian, L., et al. (2020). Coordination between growth-regulating factor1 and GRF-interacting factor1 plays a key role in regulating leaf growth in rice. BMC Plant Biol. 20:200. doi: 10.1186/s12870-020-02417-0
Lu, Y., Zeng, J., and Liu, Q. (2021). The rice miR396-GRF-GIF-SWI/SNF module: A player in GA signaling. Front. Plant Sci. 12:786641. doi: 10.3389/fpls.2021.786641
Marcotrigiano, M. (2010). A role for leaf epidermis in the control of leaf size and the rate and extent of mesophyll cell division. Am. J. Bot. 97, 224–233. doi: 10.3732/ajb.0900102
Nagai, M., Tanaka, S., Tsuda, M., Endo, S., Kato, H., Sonobe, H., et al. (2001). Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc. Natl. Acad. Sci. U.S.A. 98, 3843–3848. doi: 10.1073/pnas.061036798
Omidbakhshfard, M. A., Proost, S., Fujikura, U., and Mueller-Roeber, B. (2015). Growth-Regulating Factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010. doi: 10.1016/j.molp.2015.01.013
Pegler, J. L., Nguyen, D. Q., Oultram, J. M. J., Grof, C. P. L., and Eamens, A. L. (2021). Molecular manipulation of the MiR396/GRF expression module alters the salt stress response of Arabidopsis thaliana. Agron. Basel 11:1751. doi: 10.3390/agronomy11091751
Rukmangada, M. S., Sumathy, R., Sivaprasad, V., and Naik, V. G. (2018). Genome-wide identification and characterization of growth-regulating factors in mulberry (Morus spp.). Trees 32, 1695–1705. doi: 10.1007/s00468-018-1744-6
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shang, S., Wu, C., Huang, C., Tie, W., Yan, Y., Ding, Z., et al. (2018). Genome-wide analysis of the GRF family reveals their involvement in abiotic stress response in Cassava. Genes (Basel) 9:110. doi: 10.3390/genes9020110
Shimano, S., Hibara, K. I., Furuya, T., Arimura, S. I., Tsukaya, H., and Itoh, J. I. (2018). Conserved functional control, but distinct regulation, of cell proliferation in rice and Arabidopsis leaves revealed by comparative analysis of GRF-interacting factor 1 orthologs. Development 145:dev.159624. doi: 10.1242/dev.159624
Szczygiel-Sommer, A., and Gaj, M. D. (2019). The miR396-GRF regulatory module controls the embryogenic response in Arabidopsis via an auxin-related pathway. Int. J. Mol. Sci. 20:5221. doi: 10.3390/ijms20205221
Tang, Y., Cheng, W., Li, S., Li, Y., Wang, X., Xie, J., et al. (2021). Genome-wide identification and expression analysis of the growth regulating factor (GRF) family in Jatropha curcas. PLoS One 16:e0254711. doi: 10.1371/journal.pone.0254711
Tao, W. J., Zhang, J. Y., Li, G. Y., Liu, T., Liu, F. P., Yi, J. G., et al. (2016). A wearable sensor system for lower-limb rehabilitation evaluation using the GRF and CoP distributions. Meas. Sci. Technol. 27:025701. doi: 10.1088/0957-0233/27/2/025701
Tsukaya, H. (2014). Comparative leaf development in angiosperms. Curr. Opin. Plant Biol. 17, 103–109. doi: 10.1016/j.pbi.2013.11.012
van der Knaap, E., Kim, J. H., and Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704. doi: 10.1104/pp.122.3.695
Wallace, B. D., Berman, Z., Mueller, G. A., Lin, Y. F., Chang, T., Andres, S. N., et al. (2017). APE2 Zf-GRF facilitates 3 ‘-5 ‘ resection of DNA damage following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 114, 304–309. doi: 10.1073/pnas.1610011114
Wang, F., Qiu, N., Ding, Q., Li, J., Zhang, Y., Li, H., et al. (2014). Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 15:807. doi: 10.1186/1471-2164-15-807
Wang, L., Gu, X., Xu, D., Wang, W., Wang, H., Zeng, M., et al. (2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 62, 761–773. doi: 10.1093/jxb/erq307
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. 58, 531 doi: 10.1385/1-59259-
Wu, L., Zhang, D., Xue, M., Qian, J., He, Y., and Wang, S. (2014). Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 56, 1053–1063. doi: 10.1111/jipb.12220
Zan, T., Zhang, L., Xie, T. T., and Li, L. Q. (2020). Genome-wide identification and analysis of the Growth-Regulating Factor (GRF) gene family and GRF-interacting factor family in Triticum aestivum L. Biochem. Genet. 58, 705–724.
Zhang, D., Sun, W., Singh, R., Zheng, Y., Cao, Z., Li, M., et al. (2018). GRF-interacting factor1 regulates shoot architecture and meristem determinacy in maize. Plant Cell 30, 360–374. doi: 10.1105/tpc.17.00791
Zhang, J., Li, Z., Jin, J., Xie, X., Zhang, H., Chen, Q., et al. (2018). Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum). Gene 639, 117–127. doi: 10.1016/j.gene.2017.09.070
Zhang, D.-F., Li, B., Jia, G.-Q., Zhang, T.-F., Dai, J.-R., Li, J.-S., et al. (2008). Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.). Plant Sci. 175, 809–817.
Zheng, L. W., Ma, J. J., Song, C. H., Zhang, L. Z., Gao, C., Zhang, D., et al. (2018). Genome-wide identification and expression analysis of GRF genes regulating apple tree architecture. Tree Genet. Genomes 14:54. doi: 10.1007/s11295-018-1267-8
Keywords: gene family, growth-regulating factor, leaf size, alfalfa, expression profile
Citation: Sun Y, Li H, Wu J, Zhang K, Tang W, Cong L, Xie H, Wang Z-Y and Chai M (2022) Genome-wide identification of growth-regulating factor transcription factor family related to leaf and stem development in alfalfa. Front. Plant Sci. 13:964604. doi: 10.3389/fpls.2022.964604
Received: 08 June 2022; Accepted: 05 August 2022;
Published: 23 August 2022.
Edited by:
Suvendu Mondal, Bhabha Atomic Research Centre, IndiaCopyright © 2022 Sun, Li, Wu, Zhang, Tang, Cong, Xie, Wang and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maofeng Chai, Y2hpbXUyMTAwQDEyNi5jb20=
†These authors have contributed equally to this work
‡ORCID: Maofeng Chai, orcid/0000-0001-9915-0321
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.