
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 September 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.962973
This article is part of the Research Topic Fungal Wheat Diseases: Etiology, Breeding, and Integrated Management, Volume II View all 13 articles
Transcriptional reprogramming is an essential feature of plant immunity and is governed by transcription factors (TFs) and co-regulatory proteins associated with discrete transcriptional complexes. On the other hand, effector proteins from pathogens have been shown to hijack these vast repertoires of plant TFs. Our current knowledge of host genes' role (including TFs) involved in pathogen colonization is based on research employing model plants such as Arabidopsis and rice with minimal efforts in wheat rust interactions. In this study, we begun the research by identifying wheat genes that benefit rust pathogens during infection and editing those genes to provide wheat with passive resistance to rust. We identified the wheat MYC4 transcription factor (TF) located on chromosome 1B (TaMYC4-1B) as a rust pathogen target. The gene was upregulated only in susceptible lines in the presence of the pathogens. Down-regulation of TaMYC4-1B using barley stripe mosaic virus-induced gene silencing (BSMV-VIGS) in the susceptible cultivar Chinese Spring enhanced its resistance to the stem rust pathogen. Knockout of the TaMYC4-1BL in Cadenza rendered new resistance to races of stem, leaf, and stripe rust pathogens. We developed new germplasm in wheat via modifications of the wheat TaMYC4−1BL transcription factor.
Plants employ a complex network of signaling pathways to defend themselves against pathogen attacks. Signal integration is dictated by transcription factor (TF) regulatory networks. Transcriptional reprogramming is a major component of plant immunity and is administered by TFs and co-regulatory proteins connected within distinct transcriptional complexes (Moore et al., 2011). Over a couple of decades, studies have uncovered numerous TF family members mostly in Arabidopsis thaliana and rice that are critical in regulating proper defense responses when plants are confronted by pathogens. Many of these TFs have been categorized into AP2/ERF, bHLH, bZIP, MYB, NAC and WRKY families. MYC, a basic-helix-loop-helix (bHLH) family TFs were initially discovered from a homology study between an oncogene carried by the Avian virus, Myelocytomatosis (v-MYC) and a human gene overexpressed in different cancers, cellular MYC (c-MYC). They have a DNA binding domain made of 50–60 amino acids, which allows for homo- or heterodimerization to their DNA consensus hexamer sequence CANNTG (Finver et al., 1988). The bHLH TFs have been shown to be key regulators in Jasmonic Acid (JA)-mediated defense responses and in mediating crosstalk with other phytohormones, including salicylic acid (SA), abscisic acid (ABA), gibberellins (GA), and auxin (Kazan and Manners, 2013).
Co-evolved with host defense systems in plants, pathogens are also continually developing counter mechanisms to overcome host defenses. It has become evident that one component of their arsenal is manipulating host cellular processes using effector proteins, including exploiting pathogen susceptible host genes. Efforts in modifying these pathogen targeted host genes to increase resistance against pathogens have become a go-to approach for disease-resistant breeding in model crops such as rice. For example, alteration of rice promoters OsSWEET14 and OsSWEET11 and the OsMPK5 gene enhanced resistance to bacterial blight (caused by Xanthomonas oryzae pv. oryzae), bacterial panicle blight (Burkholderia glumae) and blast (caused by fungus Magnaporthe grisea) of rice (Li et al., 2012; Jiang et al., 2013; Xie and Yang, 2013). Similarly, alteration of wheat TaMLO and TaEDR1 genes (three homeologs) enhanced resistance to powdery mildew (caused by Blumeria graminis f. sp. tritici) (Shan et al., 2013; Wang et al., 2014; Zhang et al., 2017). Identifying host targets of pathogen effectors and modifying these sites to abolish the effector-target interactions would be a quick approach to generate new resistance.
Upon the release of the complete genome sequence of hexaploid wheat (Triticum aestivum L.) by the International Wheat Genome Sequencing Consortium (Appels et al., 2018), it has become apparent that of the over 107,891 high-confidence genes identified, more than 35,000 are transcriptional factors categorized into 40 families and 84 subfamilies (Appels et al., 2018). This genome resource makes gene identification in wheat from a short sequence read possible. In a pilot transcriptomic study on the interaction between wheat and the leaf rust pathogen (Puccinia triticina; Pt) using a pair of near isogenic lines (NILs) of mnr220/MNR220 (the Alpowa background) (Campbell et al., 2012), the expression patterns of some wheat genes were found to be upregulated starting at 5-days post-Pt inoculation (data not shown), a critical rust Pt development stage when the rust has established infection sites in susceptible wheat cultivars. Among those upregulated expressed genes, five of them are transcriptional factors (TFs). Among the five TFs, one of them is 97% similar to a cDNA of TaMYC4-like sequence named TRIUR3_32014 from Triticum urartu (a close wild relative of wheat) after BLAST search in NCBI. MYC2/MYC3/MYC4 were identified as a core of TFs regulating jasmonic acid (JA) and JA-isoleucine accumulation through a positive amplification loop in Arabidopsis (Van Moerkercke et al., 2019). This observation suggested a hypothesis that some rust pathogens upregulate host JA production to suppress SA-mediated defense by manipulating host MYC transcription factors. The wheat MYC4 genes (TaMYC4) are the targets of the rust pathogen during infection. In this study, we have demonstrated that the TaMYC4 homeolog in the long arm of chromosome 1B (TaMYC4−1BL) is upregulated upon pathogen infection and that modification of the TaMYC4−1BL enhanced resistance to wheat against rust pathogens.
Spring wheat cultivar Alpowa (PI 566596) was obtained from the USDA National Plant Germplasm System (NPGS), and Chinese Spring (CS) was obtained from Dr. Evans Lagudah at Commonwealth Scientific and Industrial Research Organization (CSIRO). Cadenza, spring wheat, was obtained from the SeedStor via http://www.seedstor.ac.uk.
The Puccinia graminis f. sp. tritici (Pgt) races QFCSC (isolate 10UML6-1) and TPMKC (isolate 07MT137-2) were provided by Dr. Yue Jin from Cereal Disease Laboratory, USDA-ARS, St. Paul, MN. The P. triticina (Pt) race PBJJG (isolate 09KSAL1-6) was provided by Dr. Robert Bowden (USDA-ARS, Manhattan, KS, USA). A P. striiformis f. sp. tritici (Pst) culture (race and isolate unknown) was collected from the Bozeman Agricultural Research and Teaching Farm of Montana State University (MSU).
For rust screenings, wheat seeds were directly planted into 4-inch small pots (5 seeds/pot) containing SunGro Horticulture Sunshine mix (HeavyGardens Company, Denver, CO). For seed propagation and crosses, wheat seeds were first germinated in Petri dishes on filter paper at room temperature. At root radical emergence, the seeds were transferred to 8-inch pots (one seedling/pot) containing a 1:1 ratio mixture of local soil: Sunshine mix. Growth conditions were set at 22/14°C Day/night temperatures and a 16 h photoperiod in a greenhouse at the Plant Growth Center, MSU. Plants were watered and fertilized every day with Peters General Purpose Plant Food (Scotts-Miracle-Gro Company, Marysville, OH, USA) at a concentration of 150 ppm.
All rust screenings were completed at the two-leaf seedling stage. Rust inoculations were conducted as described in Campbell et al. (2012). In brief, plants were inoculated with rust pathogen urediniospores in Soltrol170 oil suspensions. Inoculated plants were then transferred to a Percival I-60D dew chamber (Percival Scientific Inc., Perry, IA, USA) pre-conditioned to an internal air temperature between 15 and 17°C for Pt and Pst, 19-20°C for Pgt for 24 h. An additional step of 4 h exposure to light prior to removing the plants from a dew chamber for stem rust inoculation. Pgt races QFCSC and TMLKC, Pt race PBJJG and Pst evaluations were conducted at Montana State University in the greenhouse of the Plant Growth Center. Assays with TTKSK (isolate 04KEN156/04) and TKTTF (isolate 13GER10-5) were completed in the Cereal Disease Laboratory, USDA-ARS, St. Paul, MN, following the procedure described by Jin et al. (2007). The tests with other Pst races were conducted at the Wheat Health, Genetics, and Quality Research Unit, USDA-ARS, Pullman, WA, according to the procedure described by Line and Qayoum (1992) and infections types (IT) recorded 18–20 days after inoculation.
Infection types of seedlings to leaf and stem rusts were assessed using the 0-4 IT scale (McIntosh et al., 1995) at 8 and 13–14 days post inoculation (dpi) for Pt and Pgt, respectively, when the symptoms and signs of susceptible controls were fully expressed. Stripe rust was assessed at 14 dpi based 0 (immune)—9 scale (highest susceptible) (Line and Qayoum, 1992).
For RNA extraction, leaf samples were taken from three plants separately per treatment. Each sample was frozen in liquid nitrogen and stored at −80°C. RNA extractions were completed when all the samples at different time points were collected. Sample collection times ranged from 0 to 10 dpi, depending on the experiment. The samples at 0 dpi were taken immediately after inoculation before placing the inoculated plants in a dew chamber. For DNA extraction, leaf samples were taken and immediately used for extraction.
Total RNAs were isolated and treated with DNase I on columns using the Qiagen RNeasy Plant Mini Kit (Qiagen, Valencia, CA) following the manufacturer's instructions. Genomic DNA extraction for conventional PCR was done using the QIAGEN DNeasy Plant Mini Kit (Qiagen Sciences Inc, Germantown, MD, USA). For KASP assays, DNA was extracted from 96 plants using the 96-well plate extraction procedure modified from Holleley and Sutcliffe (2022). The quality and concentration of total RNA/DNA were assessed using 260/280ABS measurements on a NanoDrop 1,000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA). The integrity of DNA or RNA was checked via agarose gel electrophoresis with 2 μl of a sample, 4 μl of water, 1 μl loading buffer (98% formamide, 10 mM EDTA, 0.25% bromophenol blue, and 0.25% xylene cyanol) on a 0.8–1% gel stained by GelRed (Bio-Rad, Hercules, CA) at 125 volts for 25 min.
qRT-PCR was performed using the iScript One-Step RT-PCR Kit with SYBR Green (Bio-Rad, Hercules, CA) on a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA) following the manufacturer's procedure with 100–150 μg sample RNA at annealing temperatures of 56/57°C depending on the primers. ACTB (β-actin) (Kozera and Rapacz, 2013) was used as the housekeeping gene for normalization of transcript abundance (Supplementary Table 1). qRT-PCR was conducted in triplicate.
PCR amplifications were conducted in 20 μl reactions containing 25 mM MgCl2, 10 mM dNTP, 2 μM of each primer (BN1BL primers, Table 2), 50 ng genomic DNA and 1 unit Go Taq Flexi DNA polymerase (Promega, Madison, Wisconsin). Amplifications were performed at 95°C for 7 min, followed by 35 cycles at 95°C for 45 s, 55°C for 45 s, and 68/72°C for 40 s (depending on primers), with a final extension at 68/72°C for 10 min.
KASP genotyping was conducted using the KASP genotyping trial kit following the manufacturer's protocol (Biosearch Technologies Genomic analysis by LGC) using manually designed KASP primers (Supplementary Table 1) on a CFX96 real-time PCR detection system. Products from qRT-PCR, PCR and KASP were checked using gel electrophoresis as described previously.
Gene knockdown was conducted via a BSMV-VIGS assay. The original BSMV vectors were obtained from Dr. Andrew O. Jackson (UC Berkeley, CA, USA). The target fragment for the silencing assay was inserted into the modified γ vector ready for direct PCR cloning as described by Campbell and Huang (2010). BSMV RNA transcripts were synthesized in vitro using the T7 RNA polymerase (New England Biolabs, Ipswich, MA, USA) from linearized α, β, and γ plasmids. The BSMV inoculum was prepared with 3 μL of BSMV RNAs (1:1:1 ratio of α, β, and γ) and 22.5 μL of the inoculation buffer. The inoculum was then rub-inoculated onto the first leaf of two-leaf-stage plants. Leaf tissue was sampled 9 days after virus inoculation to test the silencing efficiency. Stem rust inoculations were done 14 days post virus inoculation when BSMV-induced target gene silencing reached the highest level.
Cadenza mutants were identified from the wheat-tilling database using the sequence of candidate genes as a query. Wheat-tilling is a resource TILLING population consisting of 2,700 individuals developed via ethyl methanesulfonate (EMS) mutagenesis in tetraploid durum cv “Kronos” and the hexaploid wheat cv “Cadenza” backgrounds (Rakszegi et al., 2010). The genome of each mutant has been completely sequenced. Mutations of requested mutants were validated via sequencing of the target regions after PCR amplification from the wild-type Cadenza and mutants using gene-specific primers.
Genetic analysis was conducted to test the genotype-phenotype association using 150 seeds from a self-pollinated heterozygote mutant plant at the selected locus. Also, 96 F4 individuals were used for genotyping and phenotyping via KASP assay using the designed KASP primers. For mutant L683F, the single nucleotide polymorphism (SNP) was a C-T nucleotide change from the wild-type to the L683F mutant. The forward oligos were designed as: Allele 1 with wild-type nucleotide (C) and Allele 2, which has the mutant nucleotide (T). A common reverse primer was designed for both allele oligos. A combination of the three oligos was used in the assay (Supplementary Table 1).
During the time courses of the Pgt TMLKC infections, PR gene expression was assayed at 0, 1, and 2 dpi in both the wild-type Cadenza and mutant. Leaf samples were collected from three biological replicates per dpi for the wild-type and mutant and pretreated under recommended conditions for RNA extraction. Using the corresponding PR gene primers (Supplementary Table 1), PR genes were quantified using extracted RNA via real-time-qPCR as described earlier. The PR gene primers were according to Desmond et al. (2005).
All BLAST and sequence downloads were completed using the International Wheat Genome Sequence Consortium (IWGSC) resources at https://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST. Initially, sequence downloads were completed from IWGSC RefSeq v1.0 (2017-08-07) and later from IWGSC RefSeq v2.1 (2019-02-20). Multiple sequence alignments were conducted using ClustalW Omega at https://www.ebi.ac.uk/Tools/msa/clustalo/. Gene and conserved domain predictions were performed using Softberry at http://www.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind and Pfam at http://pfam.xfam.org/. Primers were designed either manually or using the PrimerQuest® tool at https://www.idtdna.com/pages/tools/primerquest, and primer specificity was assessed by BLAST search of the IWGSC database. RNA-seq data quality was checked using FastQC Version 0.11.6 (Babraham Bioinformatics, Cambridge, CB22 3AT, UK).
The RNA sequence data from the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) accession number (PRJEB12497: http://www.ebi.ac.uk/ena/data/view/PRJEB12497) (Dobon et al., 2016) were used to quantify the expression levels of homeologs at different time points via Kallisto software (Bray et al., 2016). The wheat transcriptome downloaded from IWGSC was indexed and reads of RNA-seq were also assessed for quality via FastQC High Throughput Sequence QC Report (version: 0.11.7) in interactive mode. Paired-end sequences were split using Fastq-dump prior to transcript quantification. The paired-end reads were run using default parameters with 100 bootstraps (-b 100). Refer to Kallisto Manual-Pachter Lab (https://pachterlab.github.io/kallisto/manual) for more details to run Kallisto on bulk RNA-seq data.
Data assessment and analysis were conducted in Microsoft Excel and R-studio software version 1.1.453.0. For real-time PCR assays, data were used only if the Ct standard deviation among the triplets was ≤ 0.2 and the mean of the triplet's Ct was used for downstream analysis. Relative expression was calculated using the ΔΔCt method as described in the CFX96 manual (Bio-Rad, Hercules, CA), where fold change = 2−ΔΔCt. Expression measurement of genes was conducted in three technical replicates for each of three biological replicates. Standard deviations were calculated among three biological replicates or using pooled standard deviation formula when comparing two different groups (ΔCt values). Student's t-tests were performed to test whether the expression levels at different time points were significantly different. The p-values were calculated based on an unpaired two-tailed distribution. Expression patterns were graphically represented using averages of the three biological replicates.
To address our question of TaMYC4's possible negative role in wheat plant defense against rust pathogens, we began with further investigation of TaMYC4 conserved domains to relate its structure to function. We identified three copies of the TaMYC4 gene in bread wheat using the cDNA of TaMYC4-like sequence of TRIUR3_32014 from T. urartu to BLAST search the International Wheat Genomic Sequence Consortium (IWGSC) database. TaMYC4-like genes were identified on 1AL, 1BL, and 1DL chromosomes with 97–99% similarities on the DNA level or the amino acid level with an Expect Value of 0.0. The three homeologs were predicted to have conserved basic-helix-loop-helix (bHLH) and helix-loop-helix (HLH) domains at E-values approximately 0.0. A Leucine-zipper domain was predicted on only the 1BL homeolog at an E-value of 0.31. Each of these domains is a classical MYC4 conserved domain (Supplementary Figure 1, Supplementary File 2). The bHLH/HLH and leucine zipper motifs allow binding of MYC proteins to DNA and dimerization with other bHLH TFs (including Max). Based on a ClustalW Omega multiple sequence alignment of the sequences, the percent identity matrix indicated that the three homeologs had over 95% identity at the DNA, mRNA, and protein levels. These insights gave additional validity that the sequence we were interested in is a TF of MYC4.
To further confirm the expression pattern of the TaMYC4 genes observed in the pilot study (data not shown), we conducted a time-course study in Alpowa, the genetic background of the NILs used in the wheat-Pt transcriptomics pilot study. Alpowa was inoculated with Pt race PBJJG and Pgt race QFCSC or buffer Soltrol 170 isoparaffin as a mock inoculated control. An RNA collection time course was set at 0-, 1-, 2-, 3-, 4-, 5-, 8-, and 10-days post-inoculation (dpi). The expression level of TaMYC4 was assayed via qRT-PCR using the BN4RT primers that measured all three homeologs of the TaMYC4 genes (Supplementary Table 1, Supplementary File 1). We did not observe any significant changes in the expression profile of TaMYC4 at any of the time points for the PBJJG-inoculated Alpowa (Supplementary Figure 2). When Alpowa was inoculated with the Pgt race QFCSC, the expression levels of TaMYC4 were similar in samples taken at 0 dpi between Pgt-inoculated Alpowa and the control. However, a significant increase in TaMYC4 transcript abundance was detected at 1 dpi in Pgt infected Alpowa compared to the control plants. The TaMYC4 expression level returned to an undetectable level at 2 dpi and stayed unchanged during the rest of the time points (Figure 1). The expression level of TaMYC4 in buffer inoculated Alpowa showed minor and insignificant changes along from 1 to 3 dpi but showed an increase from 5 to 8 dpi that then declined at 10 dpi. This increase was, however, statistically insignificant. A significantly higher expression of TaMYC4 occurred after rust infection and only in the susceptible host revealed in the pilot study. These observations encouraged us to investigate whether Pgt-induced TaMYC4 upregulation is beneficial to the pathogen during colonization of wheat.
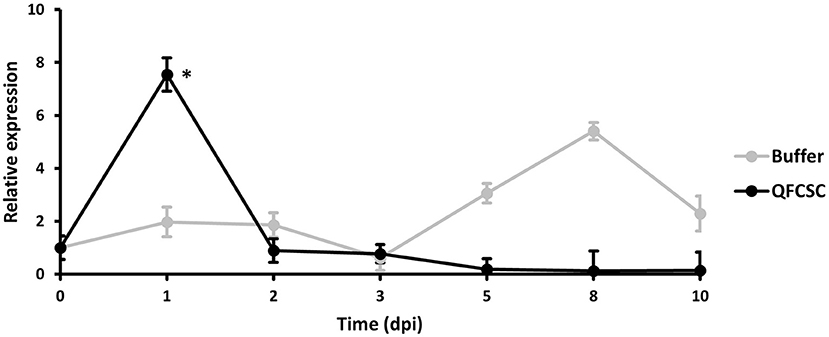
Figure 1. Relative expression of TaMYC4 in wheat cultivar Alpowa inoculated with buffer (control) and the Puccinia graminis f. sp. tritici (Pgt) pathogen (race QFCSC). Alpowa cultivar were inoculated with Pgt urediniospore at 2-leaf stage. RNA samples were extracted from the leaf samples collected at seven time points. Real-time PCR was used to quantify transcript abundances of TaMYC4 genes in both buffer and Pgt inoculated samples. Expression of TaMYC4 genes at each time point was computed relative to the level at 0-dpi. Error bars represent standard deviation computed as the square root of pooled variance between groups and * denote statistical significance at the P ≤ 0.05 levels calculated between each time point compared with 0 dpi.
To quickly determine the function of TaMYC4 regarding stem rust resistance, we downregulated the TaMYC4 genes and examined the infection types on the host. We knocked down all three endogenous copies of TaMYC4 (to avoid functional redundancy) in a rust susceptible wheat cultivar Chinese Spring (CS) using a BSMV-VIGS assay. We used CS instead of Alpowa for the silencing assay for two reasons. One is that BSMV-VIGS is homolog-based, and the sequences of the three TaMYC4 genes obtained from the IWGSC database are from CS. The other reason was to validate our hypothesis in a different cultivar background to study if the pathogen uses the same strategy to infect different cultivars. A construct containing a 247-bp fragment conserved among the gene homeologs after multiple sequence alignment (Supplementary File 1) was obtained via PCR amplification using primers VIGS-F/R (Supplementary Table 1) and used to silence each homoeolog on the three chromosomes (labeled as BSMV:MYC4) (see also Supplementary File 1). A construct carrying only the BSMV genome was used as a no-target control and labeled as BSMV:00. For short, the BSMV-derived construct with no insert was named as γ00, and each BSMV silencing construct was named after the target gene, for example, γMYC4. The concurrent silencing of BSMV inoculum was made by combining the α: β: γ target transcripts in an equal ratio with excess inoculation buffer (FES). In each assay, 20 wheat seedlings were inoculated with γMYC4 or γ00, as a control. At 6 days post BSMV inoculations (dpbi), viral symptoms were visualized on the newly emerged leaves of plants inoculated with BSMV. At nine dpbi, plants inoculated with BSMV constructs showed viral-symptom-free leaf segments, indicating that BSMV induced gene silencing had occurred. Three viral-symptom-free leaf segments were randomly sampled from plants inoculated with γ00 and γMYC4 construct for RT-qPCR. The plants were inoculated with Pgt race QFCSC immediately. Infection type (IT) observed 14 dpi showed enhanced disease resistance in plants that had TaMYC4 silenced. Non-silenced plants were susceptible (Figure 2A. Transcript abundances of TaMYC4 were measured through qRT-PCR using primers BN4RTF/R (Supplementary Table 1), which confirmed a 40% reduction of TaMYC4 in silenced plants relative to the control (Figure 2B). Though the reduction in the expression of the three TaMYC4 homeologous genes resulted in enhanced resistance to Pgt QFCSC, we do not know which homeolog or if all three of them are critical for the rust fungus to colonize the wheat host successfully.
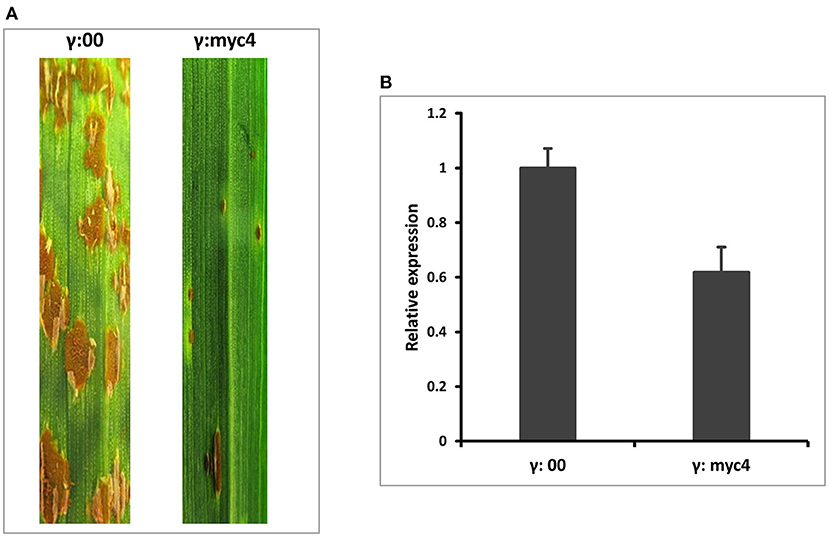
Figure 2. BSMV-VIGS of TaMYC4 in wheat cultivar Chinese Spring (CS). The first leaf of each plant was rub inoculated with the indicated BSMV constructs at the two-leaf stage and then spray inoculated with Pgt race QFCSC at 10 days after BSMV inoculation. The disease was assessed and photographed at 14 dpi. CS without any viral inoculation, used as a rust inoculation control; γ:00, CS inoculated with BSMV: MYC4; γ:myc4. RNA was extracted from viral-free leaf samples taken from γ:00 and γ:myc4 plants prior to pathogen inoculation. Transcript abundance was quantified via RT-qPCR. Error bars represent standard deviation among three biological reps. (A) Infection types of MYC4-silenced plants (B) Relative expression of TaMYC4 in γ:00 and γ:myc4 silenced plants.
We explored whether all three TaMYC4 homeologs are negatively involved in the plant's defense mechanism based on their expressions. We quantified each TaMYC4 homeolog via Kallisto software using the RNA sequence data generated from a previous study (Dobon et al., 2016) involving Avocet Yr5 (resistant) and Vuka (susceptible) inoculated with Pst pathogen at 0, 1, 2, 3, and 5 dpi and accessed from the NCBI. We used just the Pst-wheat data set since we could not find readily available Pgt-wheat RNAseq data for the specific time-course study of our interest. Also, the Pst-wheat study helped us to assess the TaMYC4 expression pattern in the wheat-Pst interaction, which we already examined beforehand in Pgt/Pt-wheat (Supplementary Figures 2; Figure 1). FastQC is a quality control tool for high throughput sequence data. Overall, FastQC checks on the RNA-seq data indicated good quality features such as per base sequence quality and overrepresented sequences. The RNA-seq was pseudo-aligned and quantified via Kallisto software (Bray et al., 2016). The output of transcript abundance was recorded in transcript per million (tpm). The transcript abundances of the genes of interest (1AL, 1BL, and 1DL) were imported using the gene IDs; TRIAE_CS42_1AL_TGACv1_000298_AA0008240.1, TRIAE_CS42_1BL_TGACv1_726352_AA2170300.1, TRIAE_CS42_1DL_TGACv1_061684_AA0201690.1, respectively. Pre-examination of the transcript data for abundance satisfied normality requirement in the R studio. A graph of expression of the gene was plotted using the averages of biological replicates at each time point.
Homeolog TaMYC4-1BL was significantly upregulated in the susceptible cultivar at 1 dpi compared with the resistant cultivar. It was also the most expressed of the three homeologs (Figure 3). This result led us to the search for permanent alteration (mutants) of TaMYC4-1BL.
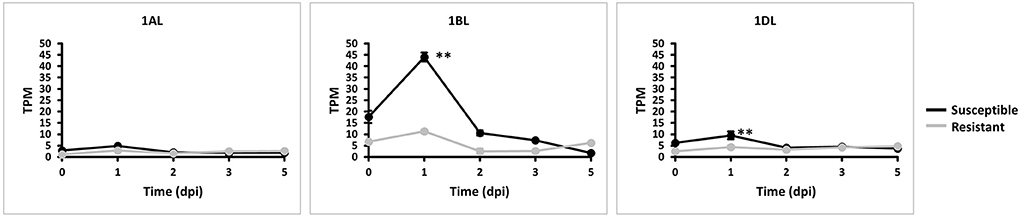
Figure 3. Expression of TaMYC4 (1AL, 1BL and 1DL) homeologs during stripe rust–Avocet Yr5 (resistant) and Vuka (susceptible) interactions. TPM is transcript per million. Error bars represent standard deviation among three biological reps and ** denote statistical significance at the P ≤ 0.01 levels calculated between each time point and 0 dpi.
Using the TaMYC4-1BL cDNA sequence as a query, a BLAST search for matches in the database of wheat-tilling mutant lines revealed more than 40 lines carrying a mutation on the TaMYC4-1BL gene in the wheat cv Cadenza background. We selected three mutants that had a Sorting Intolerant from Tolerant (SIFT) score of 0.0. Two of the three mutations were confirmed after genotyping with TaMYC4−1BL specific primers (Supplementary Table 1). One is a homozygote with a missense mutation at the protein position 683, changing amino acid L to F, hereafter identified as L683F-MYC4−1BL or L683F. The other is a heterozygote with a missense mutation from M to I at position 635, designated as M635I-MYC4-1BL or M635I (Table 1).
The Cadenza L683F mutant showed enhanced resistance to Pgt race TPMKC, Pt race PBJJG, and one field-collected race of Pst (Figure 4; Table 2). The L683F mutant was then tested using additional races of Pgt and Pst (Table 2). The mutant showed higher levels of resistance to Pgt QFCSC and Pst race PSTv-4. However, the mutant was as susceptible as Cadenza to Pgt races TTKSK and TRTTF, and Pst races PSTv-37, PSTv-41 and PSTv-47. The rust screening results indicated the resistance conferred by the mutation L683F in TaMYC4-1B is race-specific.

Figure 4. Reactions of the wild-type Cadenza and the TaMYC4 mutant L683F after inoculation with Puccinia graminis f. sp. tritici (Pgt), P. triticina (Pt) and P. striiformis f. sp. tritici (Pst) inoculation. Infection types (IT) of Cadenza: L683F in each pair of images were, Pgt (4: 1+), Pt (3: 2-;) and Pst (4: 3c) Photos of Pgt and Pst inoculated plants were taken 14 days post inoculation. Photos of Pt inoculated plants were taken 9 days post inoculation.
To confirm the new rust resistance in the mutant due to the mutation in TaMYC4-1B and not due to other mutations in the background, we crossed the mutant L683F with a susceptible cultivar Alpowa. F1 plants were self-pollinated to produce F2 segregating populations. Out of the 150 individuals in the first genetics analysis, infection types of 148 plants with clear phenotypes were scored. A resistant to susceptible ratio of 35:113 was observed, fitting the 1:3 ratio, [χ2 (1, N = 148) = 0.14, p = 0.70], indicating that resistance was a recessive phenotype. The genotyping results from 12 susceptible and 3 resistant plants confirmed the resistant plants had the mutant nucleotide (T/A) and the susceptible plants had the (C/G) base. The second step of confirmation was to design a KASP assay to detect the SNP for mutant L683F using the design primers (Supplementary Table 1) based on the TaMYC4-1BL scaffold sequence (Supplementary File 1). The result of the KASP assay from 94 F2 individuals and the two parental lines showed that all 35 resistant plants had a monomorphic SNP marker of L683F. Among the 58 susceptible plants, 17 were heterozygotes of the SNP, and 42 were homozygotes of the wild-type SNP.
To understand the genetic mechanism of the new rust resistance, we tested the expressions of four PR genes, including SA-dependent PR2 and PR5 and JA-dependent PR3 and PR10 (Van Loon and Van Strien, 1999) in a time-course study in the wild-type Cadenza and mutant L683F with Pgt TPMKC infection. The level of each PR gene was monitored at three-time points with qRT-PCR using PR gene-specific primers (Supplementary Table 1). The basal expression of the PR genes at 0 dpi was at a similar level between the wild-type Cadenza and mutant L683F (Figure 5), about 0-0.5 relative to the expression of the reference gene actin (data not shown). The result suggested that these four PR genes had minimal expression in the absence of Pgt. However, PR5 was highly upregulated (12-fold) at 1 dpi in L683F relative to its expression in the wild-type Cadenza (Figure 5), suggesting that the mutation in L683F permitted an elevated level of an SA-mediated PR gene. Meanwhile, PR2, PR3, and PR10 had no significant differences compared to the wild-type Cadenza across the three-time points. This result suggested that the mutation in L683F did not affect the two JA-mediated PR genes in the defense response to Pgt TPMKC infection.
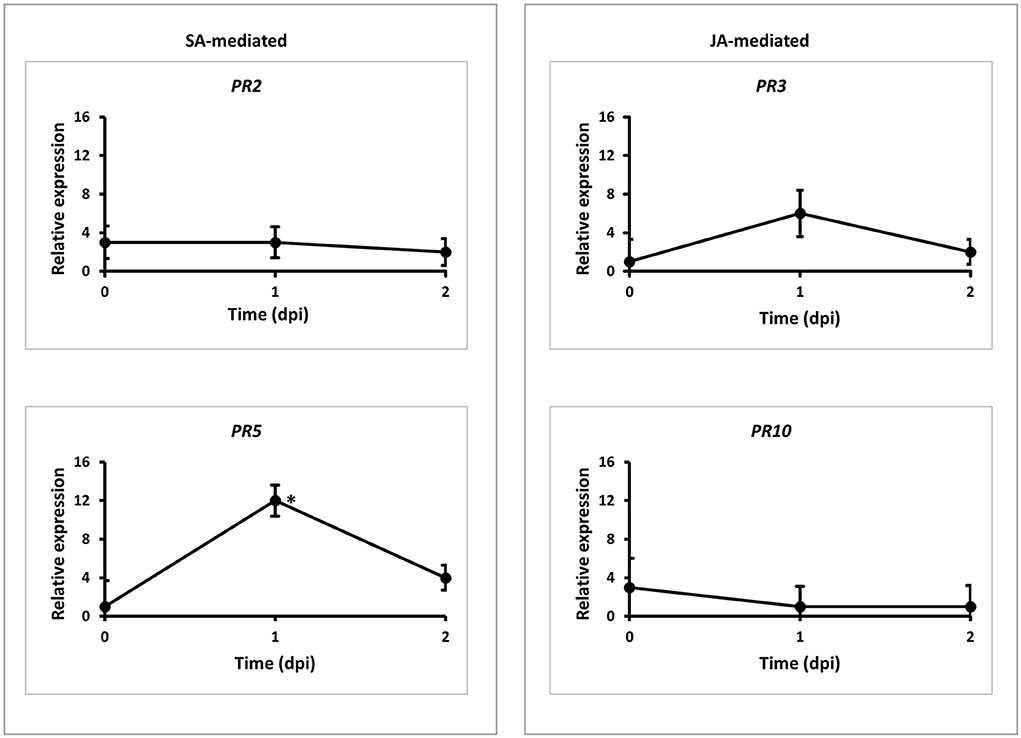
Figure 5. Expressions of pathogen related (PR) genes responding to Pgt (race TMLKC) in the wild-type and mutant wheat lines. Wild-type Cadenza and mutant L683F-MYC4-1BL were inoculated with the Pgt pathogen at 2-leaf stage. RNA samples were extracted from the leaf samples collected at three time points. Real-time PCR was used to quantify transcript abundances of the four PR genes. Expression of PR genes at each time point was expressed in the mutant relative to the level in the wild-type. * denote statistical significance at the P ≤ 0.05 levels compared between wildtype and mutant at each time point. Error bars represent standard deviation computedas the square root of pooled variance between groups.
The first evidence that supports the hypothesis that the TaMYC4 genes are targets of the pathogen for assistance in colonization is the upregulation of TaMYC4 only in rust inoculated but not in the mock-inoculated wheat plants. This provided initial evidence that differentially expressed TaMYC4 TF was due to the presence of rust pathogens (Figure 1). Secondly, the upregulation of TaMYC4-1B occurred only in a susceptible host (Figure 3). The expression trajectory indicated that the targeting of the TaMYC4-1B gene happened at the early stage of pathogen colonization. Also, down-regulation of all three copies of TaMYC4 reduced the susceptibility of a susceptible host to Pgt race QFCSC (Figure 2A). The relative reduction in TaMYC4 transcript abundance in the silenced plants (Figure 2B) indicated that the observed enhanced resistance was attributable to the TaMYC4 knockdown. The silencing assay suggested that the effect of the TaMYC4 upregulation was a benefit to the pathogen and negative to the wheat host. Arabidopsis MYC4 has been shown positively involved in JA and JA-isoleucine accumulation (Van Moerkercke et al., 2019). JA and ABA are positive regulators of stomatal closure (Sarwat and Tuteja, 2017). Bacterial pathogen Pseudomonas syringae secret coronatine (COR), a structural and functional analog of the active form of JA, to open host stomata during infection (Zhou et al., 2015). We hypothesize that rust pathogens upregulate TaMYC4-1B to increase the JA level to open up host stomata because rust fungal germ tubes enter the host through the stomata. Additional support of this hypothesis is a study of PR gene expression during rust pathogen infections. JA-mediated PR gene expressions were low in resistant lines, suggesting a low JA level at the early time point post-inoculation (Zhang et al., 2018). Finally, a recessive loss-of-function mutation in TaMYC4−1BL conferred resistance to rust pathogens (Figure 4), supporting our claim that the TaMYC4-1B gene facilitated infection by rust pathogens in a compatible wheat host.
The three homeologs of TaMYC4 have bHLH/HLH conserved domains characteristic of MYC transcription factors. Nonetheless, an LZ domain is only found in the TaMYC4-1B protein (Supplementary Figure 1), suggesting a possible non-redundancy function under different conditions. Most literature have provided evidence of the positive contribution of the leucine zipper TFs to biotic (Milligan et al., 1998; Ballvora et al., 2002; Alves et al., 2013) and abiotic stresses (Yu et al., 2020). Generally, LZ TFs heterodimerize with other proteins involved in cell proliferation, survival, and metabolism (Adhikary and Eilers, 2005). These functions could be hijacked by pathogens for their benefit with enhanced host susceptibility, as observed with the mutation of TaMYC4-1B resulting in enhanced resistance to the rust pathogens. More research is however needed to arrive at this conclusion. Indeed, several studies (Shitsukawa et al., 2007; Hovav et al., 2008; Chaudhary et al., 2009; Chen et al., 2011) have revealed transcriptional divergence among homeologs. However, non-redundancy can also arise due to different expression patterns, not due to the protein sequence. Therefore, we suggest that the TaMYC4−1BL gene acts as a rust pathogen susceptibility gene such that it acts as a factor needed by the pathogens to colonize the host.
The segregation ratio of resistant to susceptible phenotypes confirmed that TaMYC4−1BL confers a recessive phenotype for resistance or a dominant phenotype for susceptibility. At 24 h post-inoculation, development of haustorial mother cells of a wheat rust fungus commences (Serfling et al., 2016). Haustoria are known to play a vital role in cellular communication between pathogen and host (Heath, 1997), nutrient acquisition (Hahn and Mendgen, 2001), manipulation of host metabolism, and the suppression of host defenses (Voegele and Mendgen, 2003). This knowledge further strengthens our claim that TaMYC4-1BL was a target to manipulate host defense to benefit the pathogen at the early pathogenesis stage.
Indeed, the higher level of TaMYC4−1BL transcripts at 1-day post Pgt inoculation (Figure 1) resulted in lower PR5 expression in Cadenza L683F mutant (Figure 5), suggesting that PR5 expression was hampered when the TF was increased in the presence of Pgt. PR genes are involved in host defense under different wheat-pathogen race interactions (Zhang et al., 2018). PR5 is an SA-dependent thaumatin-like protein (Van Loon, 1982) and has been shown to inhibit the growth of various fungi (Muthukrishnan et al., 2001). Strategies of bacteria hijacking plant hormones to manipulate host defense has been well studied. For example, Agrobacterium tumefaciens uses its T-DNA to facilitate production of host auxin and cytokinin hormones in the formation of crown galls. Various strains of P. syringae produces coronatine (COR) phytotoxin to manipulate host hormones to enhance bacterial growth and symptom development (Mittal and Davis, 1995). It was later shown that COR is structurally similar to JA isoleucine hence functioning in antipathy to the SA pathway, which plays a crucial role in defense against this bacterial (Brooks et al., 2005; Browse, 2009). Zheng et al. (2012) demonstrated that COR targets host NAC TFs to case stomata reopening and systemically induced susceptibility. We suspect that PR5 might have been suppressed by rust pathogen effectors using TaMYC4−1BL as a host target gene. The insignificant expression of PR10 (the other SA-related proteins) in this interaction (Figure 5) suggested that for specific signal transduction, separate PR proteins may be involved during different plant-pathogen interactions. Also, it has been established that a cross-communication between SA- and JA-dependent defense pathways exists (Felton and Korth, 2000; Pieterse et al., 2001). The relatively lower and insignificant expression levels of the JA-pathway associated PR proteins (PR2 and PR3) at 1 dpi (Figure 5) allude to this crosstalk that enables plants to fine-tune their defense reactions depending on the type of stress they encounter.
The bHLH superfamily of transcription factors, including TaMYC4, have essential regulatory components in transcriptional networks of many developmental pathways (Atchley and Fitch, 1997). In Arabidopsis, MYC4 TFs are known to bind to the G-box of promoters and are involved in JA gene regulation (Niu et al., 2011). Collectively, MYC4, MYC2 and MYC3 were shown to control JA-dependent responses (Fernández-Calvo et al., 2011). It was demonstrated that MYC4 could form complexes with glucosinolate-related MYBs to regulate glucosinolate biosynthesis (Schweizer et al., 2013). A recent study unraveled that an MYC2/MYC3/MYC4-controlled positive-feedback loop transcriptionally regulated spray-induced jasmonate accumulation (Van Moerkercke et al., 2019). These activities of MYC4 in Arabidopsis give additional credence to our hypothesis that TaMYC4 is beneficial to pathogens in suppressing host defense in the early stages of wheat rust interaction. However, contrary to its implication in JA-pathways in Arabidopsis, we found its negative function in the SA-dependent pathway implicated in PR5 suppression. This emphasizes our knowledge that a gene could function in different pathways in different species and under varying conditions. At this point, this study has not ascertained if this mechanism is similar for all the three rust pathogens and has also not uncovered the detailed mode of action of TaMYC4-1BL during the compatible wheat-rust interactions. Hence further study is necessary to elucidate these unknowns.
The Cadenza L683F mutant showed resistance to Pt race PBJJG, Pgt race TPMKC, but not to Pgt races TTKSK (Ug99) and TRTTF. Also, the mutant was susceptible to all the Pst races tested except for a field-collected uncharacterized Pst isolate and race PSTv-4 (Table 2). The race-specific resistance was shown by hypersensitive reaction and moderate resistance at the seedling stage during their interaction with the pathogens. The resulting infection types range from 1 to 3 in seedlings. The wild-type Cadenza had a moderate resistance level to Pgt race QFCSC, but the mutant L683F had enhanced resistance to the pathogen. Furthermore, the same mutation enhanced resistance to three rusts, suggesting a common target in a host used by the rust pathogens as a strategy.
We also noticed that down-regulation of the TaMYC4 genes in wheat cv. CS enhanced resistance to Pgt race TPMKC, and mutated TaMYC4-1BL in different wheat cultivars could enhance resistance to the same rust pathogen race, suggesting a rust pathogen race used the same strategy targeting the same genes in different genetic backgrounds of the same host.
Over the years, wheat rust resistance breeding has been focused on using adult-plant resistance (APR) and all-stage resistance (R) genes from wheat and related species. At the same time, this is very resource-consuming because these resistance genes are continually overcome by evolving virulent races of rust pathogens, mainly because most of these genes confer race-specific resistance. There have been considerations for the building resistance gene cassettes to confer efficient resistances against different rust pathogens or races. However, this effort requires the availability of effective resistance genes as resources.
Due to the limitations of natural existing resistance genes in wheat or its relatives, scientists are looking for strategies to generate new resistance genes via mutagenesis or genome editing. Decades of studies on plant-pathogen interactions revealed that when pathogens successfully colonize host plants, some of the host genes are reprogrammed by the pathogens. Those host genes could be transcriptionally activated by the pathogen transcription activator-like effectors (TALEs), interact with the pathogen effectors to suppress the plant defense response (Fukuoka et al., 2009), or redirect the host nutrient sinks to the host-pathogen interfaces (Chen et al., 2010). In nature, a single mutation of one of those host genes could happen and make the host less desirable to the pathogen, therefore less severity of the disease. For example, rice xa13-mediated resistance is due to a mutation of the effector binding site (EBE) in the promoter of the host glucose transporter gene OsSWEET11 (Xa13). The mutation leads to the loss of the bacterial pathogen Xanthomonas oryzae pathovar oryzae (Xoo) strain PXO99 PthXo1-mediated induction of the SWEET gene and reduced bacterial growth (Chen et al., 2010). In wheat, changes of only two amino acids in a conserved region of a hexose transporter at the Lr67 locus between lr67 and Lr67 reduce the growth of multiple biotrophic pathogen species (Moore et al., 2015). Understanding the specificity of pathogen targeting DNA sequences has enabled the development of new host resistance in rice (Hummel et al., 2012; Zeng et al., 2015), pepper (Romer et al., 2009), and resistance against Ralstonia solanacearum in a broad host range (de Lange et al., 2013).
Our study demonstrated an effective scheme of using a transcriptomic-guided approach in wheat to select candidates of pathogen targets, a BSMV-induced gene silencing assay to identify and confirm the targets, and phenotype and genetic analysis of mutations on selected targets to develop new resistant germplasm. TaMYC4-1BL was selected because the upregulation of the gene was only detected in susceptible lines in the presence of rust pathogens. A reverse genetic approach using BSMV-induced gene silencing revealed that the host demonstrated improved resistance when the TaMYC4-1BL was downregulated. With the known DNA sequence of TaMYC4-1BL and an EMS mutagenized population, we were able to identify mutations in the gene. Phenotype and genetic analysis of the L683F mutation in TaMYC4-1BL of Cadenza confirmed a new resistance to rust pathogens in wheat. This approach provides a means of navigating the challenges associated with germplasm creation and studying gene function in wheat, low efficiency in wheat transformation, and concerns with detrimental effects of a host gene mutation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Conceptualization and funding acquisition: LH. Methodology: BN, LH, HZ, MR, MW, and XC. Investigation and formal analysis: BN. Writing—original draft: BN and LH. Writing—review and editing: BN, LH, MR, MW, and XC. All authors contributed to the article and approved the submitted version.
This research was supported by the NSF BREAD program (Grant No. IOS-0965429).
Our sincere gratitude to Dr. Evans Lagudah, for providing us with Chinese Spring wheat; to Rothamsted Research and JIC for providing the Cadenza mutants, to Dr. Yue Jin, for providing Pgt races QFCSC (isolate 10UML6-1) and TPMKC (isolate 07MT137-2); to Dr. Robert Bowden for providing Pt race PBJJG (isolate 09KSAL1-6) and the USDA National Plant Germplasm System (NPGS) for spring wheat cultivar Alpowa (PI 566596).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.962973/full#supplementary-material
Adhikary, S., and Eilers, M. (2005). Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell. Biol. 6, 635–645. doi: 10.1038/nrm1703
Alves, M. S., Dadalto, S. P., Gonçalves, A. B., De Souza, G. B., Barros, V. A., and Fietto, L. G. (2013). Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14, 7815–7828. doi: 10.3390/ijms14047815
Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Atchley, W. R., and Fitch, W. M. (1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. U. S. A. 94, 5172–5176. doi: 10.1073/pnas.94.10.5172
Ballvora, A., Ercolano, M. R., Weiß, J., Meksem, K., Bormann, C. A., Oberhagemann, P., et al. (2002). The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 30, 361–371. doi: 10.1046/j.1365-313X.2001.01292.x
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Brooks, D. M., Bender, C. L., and Kunkel, B. N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant. Pathol. 6, 629–639. doi: 10.1111/j.1364-3703.2005.00311.x
Browse, J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Ann. Rev. Plant Biol. 60, 183–205. doi: 10.1146/annurev.arplant.043008.092007
Campbell, J., and Huang, L. (2010). Silencing of multiple genes in wheat using Barley stripe mosaic virus. J. Biotech. Res. 2, 12
Campbell, J., Zhang, H., Giroux, M. J., Feiz, L., Jin, Y., Wang, M., et al. (2012). A mutagenesis-derived broad-spectrum disease resistance locus in wheat. Theor. Appl. Genet. 125, 391–404. doi: 10.1007/s00122-012-1841-7
Chaudhary, B., Flagel, L., Stupar, R. M., Udall, J. A., Verma, N., Springer, N. M., et al. (2009). Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182, 503–517. doi: 10.1534/genetics.109.102608
Chen, C., Costa, M. G., Yu, Q., Moore, G. A., and Gmitter, F. G. (2010). Identification of novel members in sweet orange carotenoid biosynthesis gene families. Tree Genet. Genomes 6, 905–914. doi: 10.1007/s11295-010-0300-3
Chen, X., Truksa, M., Snyder, C. L., El-Mezawy, A., Shah, S., and Weselake, R. J. (2011). Three homologous genes encoding sn-glycerol-3-phosphate acyltransferase 4 exhibit different expression patterns and functional divergence in Brassica napus. Plant Physiol. 155, 851–865. doi: 10.1104/pp.110.169482
de Lange, O., Schreiber, T., Schandry, N., Radeck, J., Braun, K. H., Koszinowski, J., et al. (2013). Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytologist 199, 773–786. doi: 10.1111/nph.12324
Desmond, O. J., Edgar, C. I., Manners, J. M., Maclean, D. J., Schenk, P. M., and Kazan, K. (2005). Methyl jasmonate induced gene expression in wheat delays symptom development by the crown rot pathogen Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 67, 171–179. doi: 10.1016/j.pmpp.2005.12.007
Dobon, A., Bunting, D. C. E., Cabrera-Quio, L. E., Uauy, C., and Saunders, D. G. O. (2016). The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genomics 17, 380–380. doi: 10.1186/s12864-016-2684-4
Felton, G. W., and Korth, K. L. (2000). Trade-offs between pathogen and herbivore resistance. Curr. Opin. Plant Biol. 3, 309–314. doi: 10.1016/S1369-5266(00)00086-8
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J. -M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701. doi: 10.1105/tpc.110.080788
Finver, S. N., Nishikura, K., Finger, L. R., Haluska, F. G., Finan, J., Nowell, P. C., et al. (1988). Sequence analysis of the MYC oncogene involved in the t chromosome translocation in a human leukemia T-cell line indicates that putative regulatory regions are not altered. Proc. Natl. Acad. Sci. 85, 3052–3056. doi: 10.1073/pnas.85.9.3052
Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., et al. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325, 998–1001. doi: 10.1126/science.1175550
Hahn, M., and Mendgen, K. (2001). Signal and nutrient exchange at biotrophic plant–fungus interfaces. Curr. Opin. Plant Biol. 4, 322–327. doi: 10.1016/S1369-5266(00)00180-1
Heath, M. C. (1997). Signalling between pathogenic rust fungi and resistant or susceptible host plants. Ann. Botany 80, 713–720. doi: 10.1006/anbo.1997.0507
Holleley, C., and Sutcliffe, A. (2022). 3.12 96 Well DNA Extraction Protocol. Available online at: https://www.researchgate.net/profile/Clare-Holleley-2/publication/242744494_96_Well_DNA_Extraction_Protocol/links/0f317531d650ba76a1000000/96-Well-DNA-Extraction-Protocol.pdf
Hovav, R., Udall, J. A., Chaudhary, B., Rapp, R., Flagel, L., and Wendel, J. F. (2008). Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. 105, 6191–6195. doi: 10.1073/pnas.0711569105
Hummel, A. W., Doyle, E. L., and Bogdanove, A. J. (2012). Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytologist 195, 883–893. doi: 10.1111/j.1469-8137.2012.04216.x
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic. Acids Res. 41, e188–e188. doi: 10.1093/nar/gkt780
Jin, Y., Singh, R., Ward, R., Wanyera, R., Kinyua, M., Njau, P., et al. (2007). Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 91, 1096–1099. doi: 10.1094/PDIS-91-9-1096
Kazan, K., and Manners, J. M. (2013). MYC2: The master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Kozera, B., and Rapacz, M. (2013). Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406. doi: 10.1007/s13353-013-0173-x
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390. doi: 10.1038/nbt.2199
Line, R. F., and Qayoum, A. (1992). Virulence, Aggressiveness, Evolution, and Distribution of Races of Puccinia Striiformis (the cause of stripe rust of wheat) in North America, 1968-87. Technical bulletin-United States Department of Agriculture (1788)
McIntosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat Rusts: An Atlas of Resistance Genes. Kluwer Academic Publishers. doi: 10.1071/9780643101463
Milligan, S. B., Bodeau, J., Yaghoobi, J., Kaloshian, I., Zabel, P., and Williamson, V. M. (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10, 1307–1319. doi: 10.1105/tpc.10.8.1307
Mittal, S., and Davis, K. R. (1995). Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol. Plant Microbe Interact. 8, 165–171. doi: 10.1094/MPMI-8-0165
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Moore, J. W., Loake, G. J., and Spoel, S. H. (2011). Transcription dynamics in plant immunity. Plant Cell 23, 2809–2820. doi: 10.1105/tpc.111.087346
Muthukrishnan, S., Liang, G. H., Trick, H. N., and Gill, B. S. (2001). Pathogenesis-related proteins and their genes in cereals. Plant Cell Tissue Organ Culture 64, 93. doi: 10.1023/A:1010763506802
Niu, Y., Figueroa, P., and Browse, J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154. doi: 10.1093/jxb/erq408
Pieterse, C. M., Ton, J., and Van Loon, L. (2001). Cross-talk between plant signalling pathways: boost or burden. Ag. Biotech. Net. 3, 1–8.
Rakszegi, M., Kisgyörgy, B., Tearall, K., Shewry, P., Láng, L., Phillips, A., et al. (2010). Diversity of agronomic and morphological traits in a mutant population of bread wheat studied in the Healthgrain program. Euphytica 174, 409–421. doi: 10.1007/s10681-010-0149-4
Romer, P., Strauss, T., Hahn, S., Scholze, H., Morbitzer, R., Grau, J., et al. (2009). Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 150, 1697–1712. doi: 10.1104/pp.109.139931
Sarwat, M., and Tuteja, N. K. (2017). Hormonal signaling to control stomatal movement during drought stress. Plant Genet. 11, 143–153. doi: 10.1016/j.plgene.2017.07.007
Schweizer, F., Fernández-Calvo, P., Zander, M., Diez-Diaz, M., Fonseca, S., Glauser, G., et al. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117. doi: 10.1105/tpc.113.115139
Serfling, A., Templer, S. E., Winter, P., and Ordon, F. (2016). Microscopic and molecular characterization of the prehaustorial resistance against wheat leaf rust (Puccinia triticina) in einkorn (Triticum monococcum). Front. Plant Sci. 7, 1668. doi: 10.3389/fpls.2016.01668
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686doi: 10.1038/nbt.2650
Shitsukawa, N., Tahira, C., Kassai, K. -i., Hirabayashi, C., Shimizu, T., Takumi, S., et al. (2007). Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19, 1723–1737. doi: 10.1105/tpc.107.051813
Van Loon, L. (1982). Regulation of changes in proteins and enzymes associated with active defence against virus infection. In Active defense mechanisms in plants. Springer, 247–273. doi: 10.1007/978-1-4615-8309-7_14
Van Loon, L. C., and Van Strien, E. A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. doi: 10.1006/pmpp.1999.0213
Van Moerkercke, A., Duncan, O., Zander, M., Šimura, J., Broda, M., Vanden Bossche, R., et al. (2019). A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. 116, 23345. doi: 10.1073/pnas.1911758116
Voegele, R. T., and Mendgen, K. (2003). Rust haustoria: nutrient uptake and beyond. New Phytol. 159, 93–100. doi: 10.1046/j.1469-8137.2003.00761.x
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947. doi: 10.1038/nbt.2969
Xie, K., and Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 6, 1975–1983. doi: 10.1093/mp/sst119
Yu, Y., Qian, Y., Jiang, M., Xu, J., Yang, J., Zhang, T., et al. (2020). Regulation mechanisms of plant basic leucine zippers to various abiotic stresses. Front. Plant Sci. 11, 1258. doi: 10.3389/fpls.2020.01258
Zeng, X., Tian, D., Gu, K., Zhou, Z., Yang, X., Luo, Y., et al. (2015). Genetic engineering of the Xa10 promoter for broad-spectrum and durable resistance to Xanthomonas oryzae pv. oryzae. Plant Biotechnol. J. 13, 993–1001. doi: 10.1111/pbi.12342
Zhang, H., Qiu, Y., Yuan, C., Chen, X., and Huang, L. (2018). Fine-tuning of PR genes in wheat responding to different Puccinia rust species. J. Plant Physiol. Pathol. 06, 178. doi: 10.4172/2329-955X.1000178
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Zheng, X.-Y., Spivey, N. W., Zeng, W., Liu, P.-P., Fu, Z. Q., Klessig, D. F., et al. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11, 587–596. doi: 10.1016/j.chom.2012.04.014
Keywords: MYC4 Transcription factor, Triticum aestivum (bread wheat), rust, (Puccinia), BSMV-VIGS, kallisto program, wheat-tilling, enhanced resistance
Citation: Nyamesorto B, Zhang H, Rouse M, Wang M, Chen X and Huang L (2022) A transcriptomic-guided strategy used in identification of a wheat rust pathogen target and modification of the target enhanced host resistance to rust pathogens. Front. Plant Sci. 13:962973. doi: 10.3389/fpls.2022.962973
Received: 07 June 2022; Accepted: 15 August 2022;
Published: 02 September 2022.
Edited by:
Maria Rosa Simon, National University of La Plata, ArgentinaReviewed by:
Yulin Cheng, Chongqing University, ChinaCopyright © 2022 Nyamesorto, Zhang, Rouse, Wang, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bernard Nyamesorto, YmVybmFyZC5ueWFtZXNvcnRvQG1vbnRhbmEuZWR1; Li Huang, bGh1YW5nQG1vbnRhbmEuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.