
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 15 September 2022
Sec. Aquatic Photosynthetic Organisms
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.962622
Carbon and nitrogen metabolism are basic, but pivotal metabolic pathways in plants and are tightly coupled. Maintaining the balance of carbon and nitrogen metabolism is critical for plant survival. Comprehensively revealing the metabolic balance of carbon–nitrogen interactions is important and helpful for understanding the adaptation of freshwater plants to CO2 limited aqueous environment. A comprehensive metabolomics analysis combined with physiological measurement was performed in the freshwater plant Ottelia alismoides acclimated to high and low CO2, respectively, for a better understanding of how the carbon and nitrogen metabolic adjustment in freshwater plants respond to carbon limitation. The present results showed that low CO2 acclimated O. alismoides exhibited significant diurnal titratable acidity and malate fluctuations, as well as an opposite diel pattern of starch change and high enzymatic activities required for crassulacean acid metabolism (CAM) photosynthesis, which indicates that CAM was induced under low CO2. Moreover, the metabolomic analysis showed that most intermediates of glycolysis, pentose phosphate pathway (PPP) and tricarboxylic acid (TCA) cycle, were increased under low CO2, indicative of active respiration in low-CO2-treated O. alismoides. Meanwhile, the majority of amino acids involved in pathways of glutamate and arginine metabolism, aspartate metabolism, and the branched-chain amino acids (BCAAs) metabolism were significantly increased under low CO2. Notably, γ-aminobutyric acid (GABA) level was significantly higher in low CO2 conditions, indicating a typical response with GABA shunt compensated for energy deprivation at low CO2. Taken together, we conclude that in low-CO2-stressed O. alismoides, CAM photosynthesis was induced, leading to higher carbon and nitrogen as well as energy requirements. Correspondingly, the respiration was greatly fueled via numerous starch degradation to ensure CO2 fixation in dark, while accompanied by linked promoted N metabolism, presumably to produce energy and alternative carbon sources and nitrogenous substances for supporting the operation of CAM and enhancing tolerance for carbon limitation. This study not only helps to elucidate the regulating interaction between C and N metabolism to adapt to different CO2 but also provides novel insights into the effects of CO2 variation on the metabolic profiling of O. alismoides.
Carbon and nitrogen are the two most abundant nutrient elements in plants, and their metabolisms are the two most basic but pivotal metabolic pathways in plants, and they are tightly coupled (Zhang et al., 2018). Both these two important metabolic processes require carbon skeletons, reducing power, and energy supplied from photosynthetic electron transport or respiration (Sweetlove et al., 2010). The carbon source and energy required for nitrogen metabolism could be provided by carbon metabolism, while nitrogen metabolism could provide photosynthetic pigments and enzymes for carbon metabolism (Nunes-Nesi et al., 2010). Specific to amino acid synthesis, the C skeletons for amino acid synthetic pathways are produced in different sectors of the respiratory pathways. The majority of the photosynthetic fixed C is converted into phosphoenolpyruvate (PEP) via glycolysis and further invested in the synthesis of organic acids through the tricarboxylic acid (TCA) cycle, such as into 2-oxoglutarate and oxaloacetate, which are the main organic acids used for amino acid synthesis (Huppe and Turpin, 1994).
The carbon and nitrogen metabolism involves extensive regulation between the two pathways in plants (Huppe and Turpin, 1994). The active adjustment of carbon and nitrogen metabolism would affect the production and conversion of the photosynthetic products, as well as the synthesis of proteins and absorption of other nutrients. Thus, maintaining the balance of carbon and nitrogen metabolism is critical for plant survival (Krapp and Traong, 2005; Nunes-Nesi et al., 2010). Previous studies have revealed that carbon and nitrogen metabolism in photoautotrophs must closely interact and coordinate when adapting to variable environmental conditions. The treatments that inhibit photosynthetic carbon fixation, such as CO2 deprivation and photosynthetic inhibitors in algae, limit the supply of carbon skeletons and restrict the assimilation of nitrogen into amino acids (Romero et al., 1985). In low CO2 stressed maize seedlings, the assimilation of N was regulated by the capacity of photosynthesis and the availability of stored carbohydrates (Pace et al., 1990). The review of higher terrestrial plants has shown that elevated CO2 improved nitrogen use efficiency and promoted dark respiration, especially in soybean (Leakey et al., 2009).
Macrophytes are important primary producers in productive freshwater ecosystems. The growth of macrophytes is frequently limited by the availability of carbon, as the low rates of CO2 diffusion in the aqueous environment (∼10,000 times lower than in air) and the external boundary layer formed around organisms constrain the uptake of inorganic carbon (Maberly and Madsen, 1998; Iversen et al., 2019). Comprehensively revealing the regulation of carbon and nitrogen metabolism, as well as their interactions are important and helpful for understanding the adaptation of freshwater plants to lower CO2 stress. However, the studies on the photosynthetic carbon and nitrogen metabolism response to CO2 limitation in freshwater plants are fragmented and limited, with the research emphasis frequently being focused on induction or switching of the photosynthetic pathways, as well as the small group of compounds’ metabolism from the induced photosynthetic pathway, such as the crassulacean acid metabolism (CAM) under limited CO2 conditions (Baattrup-Pedersen and Madsen, 1999; Klavsen and Maberly, 2010). CAM could serve to conserve water by minimizing gaseous exchange during the day for terrestrial plants, however, it is also a carbon-conserving mechanism by reducing respiratory carbon loss (Silvera et al., 2010). Recently, it has been shown that O. alismoides (a member of the Hydrocharitaceae) can operate CAM facultatively at low CO2, and at night, the activated phosphoenolpyruvate carboxylase (PEPC) fixes CO2, causing nocturnal malic acid accumulation and concomitant depletion of starch. During the day, this C4 acid is decarboxylated to produce CO2 that is captured by ribulose-bisphosphate carboxylase/oxygenase (Rubisco) into the Calvin–Benson cycle (Shao et al., 2017). Although such studies are essential and pivotal, they could not reveal the coordinating networks of carbon and nitrogen metabolic responses to low CO2 stress. Nevertheless, large metabolite datasets are essential to parameterize flux balance models (Cheung et al., 2014; Shameer et al., 2018), which could be useful for estimating the interaction between C and N metabolism.
Ottelia alismoides generally inhabits shallow waters and forms dense biomass, resulting in large diel fluctuations and low concentrations of CO2 (Shao et al., 2017). To better understand the carbon and nitrogen metabolic adjustment, as well as the carbon–nitrogen interactions of freshwater plant O. alismoides in response to carbon limitation, integrated measurements of diel change of photosynthates, soluble proteins, free amino acids, and photosynthetic enzyme activity, and a comprehensive metabolomics analysis were performed in O. alismoides plants acclimated to high and low CO2, respectively. This work will provide deeper and more reliable insights into the response of O. alismoides to different levels of CO2.
In April 2020, O. alismoides seeds were germinated on sterilized soil covered with sterile tap water (alkalinity ∼2.0 mequiv L–1, TP 1.61 μmol L–1, TN 0.1 mmol L–1) in three plastic pots. They were put in a growth chamber set at 28°C with a 14/10 h photoperiod (120 μmol photons m–2 s–1). About a month and a half later, every two seedlings (∼10 cm height) were transplanted into a plant pot (15 cm diameter and 10 cm height), and a total of eighteen pots were placed in a tank (64 cm high) located on the flat roof of the laboratory for further expanded cultivation. The seedlings in the tank received natural light and were fully submerged during the whole cultivation period.
After ∼8 weeks of growth in the tank, sixteen pots of O. alismoides with similar height (∼30 cm) and lots of oval mature leaves were transferred into eight plastic buckets (25 × 25 × 35 cm), two pots per bucket, used for different CO2 treatments. All the eight buckets were placed in a growth room at 25 ± 2°C and illuminated by white fluorescence tubes with a 14/10 h photoperiod (08:00–22:00 light, ∼130 μmol photons m–2 s–1). Four buckets are for the high CO2 treatment and the other four are for the low CO2 treatment. The O. alismoides plants were cultured with tap water and treated with high and low CO2, respectively. In high CO2 treatment (HC), CO2-saturated tap water was added to the buckets two times each day to produce HC treatment, maintaining the pH at 6.7–7.0. The resultant CO2 over the whole experimental period was between 457 and 864 μmol L–1, with a mean of 649 μmol L–1. In low CO2 treatment (LC), low CO2 was produced by natural photosynthesis of the experimental plants, which consumed the inorganic carbon, and the pH was increased from 8.1 to 10.0, generating a CO2 range of ∼0.03–11.5 μmol L–1 and a mean CO2 of 1.5 μmol L–1 during the experimental period. CO2 concentration was calculated from temperature and pH, and alkalinity was measured by Gran titration based on the equations reported by Maberly (1996). The conditions in low and high CO2 treatments are presented in Supplementary Table 1. After ∼20 days of treatment with different CO2, the newly produced oval mature leaves were sampled at 21:30 (at the end of the light period) and 07:30 (toward the end of the dark period) from different O. alismoides plants from different buckets, respectively, for physiological measurement and metabolomics analyses. Leaf samples per treatment were taken in triplicate from three different plants in different buckets, and samples at both times were collected from the same plant. After collection, all the leaf samples were stored immediately at −80°C before measurement.
The titratable acidity was measured following the previous method with minor modifications (Zhang et al., 2014). A total of 12 ml CO2-free water was added to the known fresh weight of (FW) leaf samples (∼0.2 g) in 15 ml tightly sealed screw-cap plastic tubes and were incubated in a boiling water bath for 60 min. After cooling, the acidity of the samples was measured by titration with 0.01 N NaOH to a pH endpoint of 8.3. The malic acid content was detected as described previously (Han et al., 2020). The leaf samples were extracted with pre-chilled 5% (v/v) perchloric acid and then centrifuged. The pH of the resultant supernatants was adjusted to 3.0–3.5 with saturated K2CO3 solution. After that, the supernatants were centrifuged again and filtered through 0.22 μm filters, and then these solutions were ready for measurement with high-performance liquid chromatography (HPLC). The malic acid concentration was quantitatively determined by analyzing the chromatographic data.
Starch content was determined according to Smith and Zeeman (2006) and Shao et al. (2017). A total of 5 ml of 80% ethanol was added to the frozen leaves (∼0.3 g FW) for homogenization. Then, the homogenate was incubated in a boiling water bath for 5 min and centrifuged at 6,000 × g for 10 min at room temperature. The supernatant was discarded while the pellet was kept for the next step. This ethanol extraction was repeated two times, and then pure water was added to the ethanol-extracted pellets. After a thorough homogenization, the homogenate was incubated in a boiling water bath for 10 min. After the homogenate was cooled to room temperature, 0.2 M Na acetate (pH 5.5), α-amyloglucosidase, and α-amylase were added to the homogenate, and the reaction mixture was incubated at 37°C for 4 h. Then, it was ready for glucose measurements through the amyloglucosidase assay (Smith and Zeeman, 2006).
The content of soluble carbohydrates was determined by the phenol/sulfuric acid method and expressed as mg g–1 FW (Taylor, 1995; Jain et al., 2017). The extraction and assay of soluble proteins were based on the methods described by Bradford (1976) with the bovine serum albumin for calibration. The free amino acids were determined according to the ninhydrin method (Yokoyama and Hiramatsu, 2003; Xian et al., 2020).
As the key photosynthetic enzymes function in CAM metabolism, the activities of Rubisco, PEPC, and pyruvate phosphate dikinase (PPDK regenerates PEP to provide a substrate for PEPC) need to be determined. The extraction and measurement of Rubisco, PEPC, and PPDK were performed according to the methods described by Zhang et al. (2014) and Shao et al. (2017). Enzyme activities were calculated from the rates of NADH variation at 340 nm.
The low molecular weight metabolites of O. alismoides were extracted and determined by quasi-targeted metabolomics according to the method reported in Want et al. (2012). Leaves (∼0.1 g FW) were grounded with pre-chilled 80% methanol, and then the homogenates were incubated on ice for 5 min. After that, the samples were centrifuged at 15,000 × g at 4°C for 20 min. The resultant supernatant was ready for analysis with an HPLC-MS/MS system. HPLC-MS/MS analyses were performed using a UHPLC chromatography system (ExionLC™ AD, SCIEX, America) equipped with a QTRAP® 6500 + mass spectrometer (SCIEX, America) in Novogene Co., Ltd. (Beijing, China). The extracts were injected into a column (Xselect HSS T3, 2.5 μm, 2.1 × 150 mm, Waters) with a 20 min linear gradient at a 0.4 ml/min flow rate for the positive/negative polarity mode. The samples were detected with multiple reaction monitoring (MRM) and based on Novogene’s self-built database (Luo et al., 2015). The Q1 and Q3, retention time, and declustering potential, as well as collision energy, were used for the identification of metabolites. The data files produced by HPLC-MS/MS were processed for peak integration and correction with SCIEX OS software (Version 1.4). The intensities of the peaks were normalized with metaX software. These metabolites were annotated by applying the KEGG database1, Lipidmaps database2, and HMDB database.3
Analyses of two-way ANOVA were conducted using SPSS 16.0 (SPSS Inc., Chicago, IL, United States) to assess the effect of CO2 concentration and sampling time on the physiological traits of O. alismoides plants, including acidity, the contents of malate, starch, soluble carbohydrates, soluble proteins, and free amino acids, as well as the activities of Rubisco, PEPC, and PPDK, with CO2 concentration and sampling time as factors, and their two-way interactions. Means were significantly different at p < 0.05 based on Duncan’s and Tukey’s post hoc tests.
The effect of different CO2 and sampling times on the metabolic phenotypes of O. alismoides were assessed by running principal component analysis (PCA) with metaX. To further discriminate CO2 and circadian rhythm response, two pairwise comparisons of the metabolome (LC light vs. HC light, LC dark vs. HC dark) were implemented based on the supervised partial least squares discriminant analysis (PLS-DA). The differentially expressed metabolites were selected with the following screening criteria: (1) high confidence (variable importance in the projection, VIP > 1) in discriminations between LC in the light vs. HC in the light, as well as between LC in the dark vs. HC in the dark; (2) mean intensities in LC-treated plants different from those in HC-treated O. alismoides (p-value calculated with an independent t-test was less than 0.05). For clustering heat maps, the data were normalized using z-scores of the intensity areas of differential metabolites and then were produced using the Pheatmap package in R language.
Across the high and low CO2 conditions, the content of acidity varied between 15 and 33 μequiv g–1 FW in the light and 45 and 53 μequiv g–1 FW in the dark (Figure 1A). There was a significant difference in diurnal acidity (∼38 μequiv g–1 FW) in LC-grown O. alismoides (two-way ANOVA, p < 0.05; Figure 1A and Table 1); conversely there was no diurnal acidity variation in HC-treated plants (two-way ANOVA, p > 0.05; Figure 1A and Table 1). The level of malic acid did not fluctuate between light and dark at HC plants (two-way ANOVA, p > 0.05; Figure 1B and Table 1). In contrast, LC-treated plants showed significant light/dark oscillation in malic acid content (two-way ANOVA, p < 0.05; Figure 1B and Table 1). Moreover, the concentration of malic acid in the light was only 19% of that in the dark, showing an obvious depletion of malate during the light period at low CO2 (Figure 1B). However, the malic acid content in the dark did not change with CO2 concentrations (two-way ANOVA, p > 0.05; Figure 1B and Table 1). Similar to the change of malic acid, plants under LC showed significant light/dark fluctuation in starch content, which was also present in HC-treated plants (two-way ANOVA, p < 0.05; Figure 1C and Table 1). The amount of starch was significantly higher (5.6-times) at HC acclimated O. alismoides than that in LC in the light (two-way ANOVA, p < 0.05; Figure 1C). Compared to that in the light, the starch content present in the dark was reduced by 70% for LC-grown leaves, while in HC, this reduction was only 63% (Figure 1C).
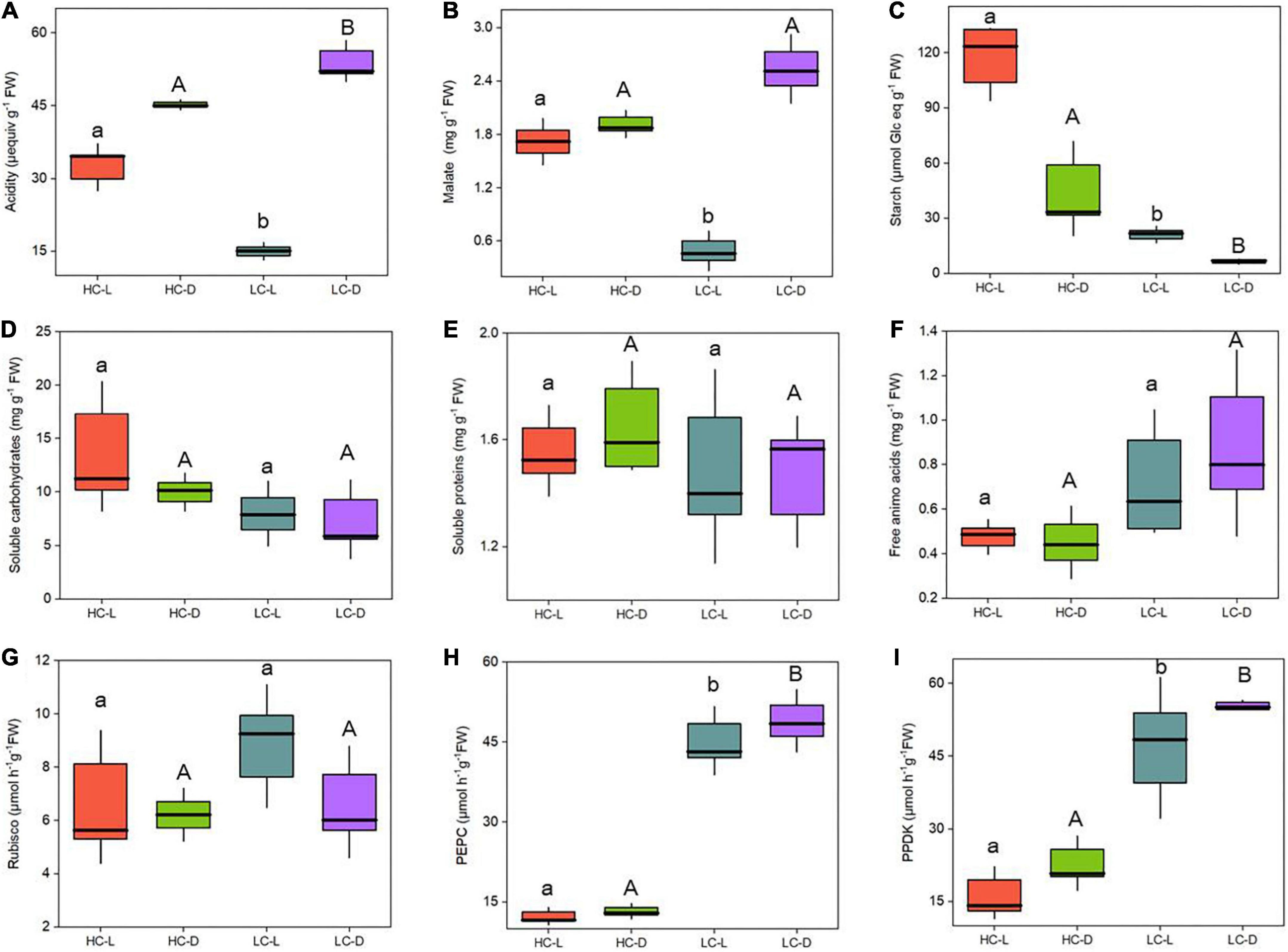
Figure 1. Influence of CO2 concentrations on the physiological parameters from Ottelia alismoides plants sampled at 21:30 (toward the end of the light period) and 07:30 (toward the end of the dark period). (A) Acidity. (B) Malate content. (C) Starch content. (D) Soluble carbohydrates content. (E) Soluble protein content. (F) Free amino acid content. (G) Rubisco activity. (H) PEPC activity. (I) PPDK activity. The enzyme activities (Rubisco, PEPC, and PPDK) were measured from the rate of disappearance of NADH at 340 nm. HC-L, high CO2 in the light; HC-D, high CO2 in the dark; LC-L, low CO2 in the light; LC-D, low CO2 in the dark. Different lowercase letters and uppercase letters indicate significant differences (p < 0.05) between different CO2 treatments in light and dark, respectively.

Table 1. Two-way ANOVA results for physiological parameters in Ottelia alismoides, with CO2 concentration and sampling time as factors.
Growth in low or high CO2 did not have a statistically significant effect on soluble carbohydrates, soluble proteins, and free amino acids of O. alismoides, although the soluble carbohydrates were slightly lower and the free amino acids were slightly higher at low vs. high CO2 (two-way ANOVA, p > 0.05; Figures 1D–F and Table 1).
The Rubisco activity was not statistically significant (two-way ANOVA, p > 0.05; Figure 1G and Table 1) between LC and HC treatments in both light and dark. When compared with high CO2, the PEPC activity was ∼3 times higher at low CO2 in both light and dark (two-way ANOVA, p < 0.05; Figure 1H and Table 1). However, the PEPC activity did not differ significantly between light and dark, regardless of the CO2 treatments (two-way ANOVA, p > 0.05; Figure 1H and Table 1). PPDK displayed a similar variation pattern to PEPC. Compared to HC, LC treatment triggered ∼2 times increase in PPDK activity in O. alismoides (two-way ANOVA, p < 0.05; Figure 1I and Table 1).
To better understand the potential effects of low CO2 on O. alismoides, as well as comprehensively reveal the carbon and nitrogen metabolic adjustment in response to different levels of CO2, LC-MS was performed to identify low-molecular weight metabolites in HC- and LC-acclimated O. alismoides plants. We mainly explored the carbon metabolism (photosynthesis and respiration) and nitrogen assimilation pathways with a metabolomic approach. A total of 790 putative metabolites were detected among all O. alismoides samples (Supplementary Figure 1), and the PCA analysis with an unsupervised pattern was used to evaluate the overall experimental variation, as well as to examine the differences in metabolite profiles among all the O. alismoides samples. The PCA results showed that all samples were distributed into four separate groups (HC dark, HC light, LC dark, and LC light) according to the first two principal components, PC1 and PC2, which represent 31 and 19% of the total variation, respectively (Supplementary Figure 2). This demonstrated that O. alismoides samples in HC and LC groups were clearly separated using the first two components. Furthermore, the PLS-DA score plots displayed a distinct separation between HC and LC groups in the light and dark (Figure 2). The R2Y (represents the interpretation rate of the established model to the Y matrices) and Q2Y (represents the predictive power of the model) were at a high level, which further confirmed that each of the supervised models had a good and valid quality. In addition, the varieties of metabolite profiles including abundant metabolites, visualized through a heat map, were remarkably diverse across all samples (Supplementary Figure 3). Taken together, these results propose that O. alismoides experienced different metabolic processes for adapting to different CO2 conditions.
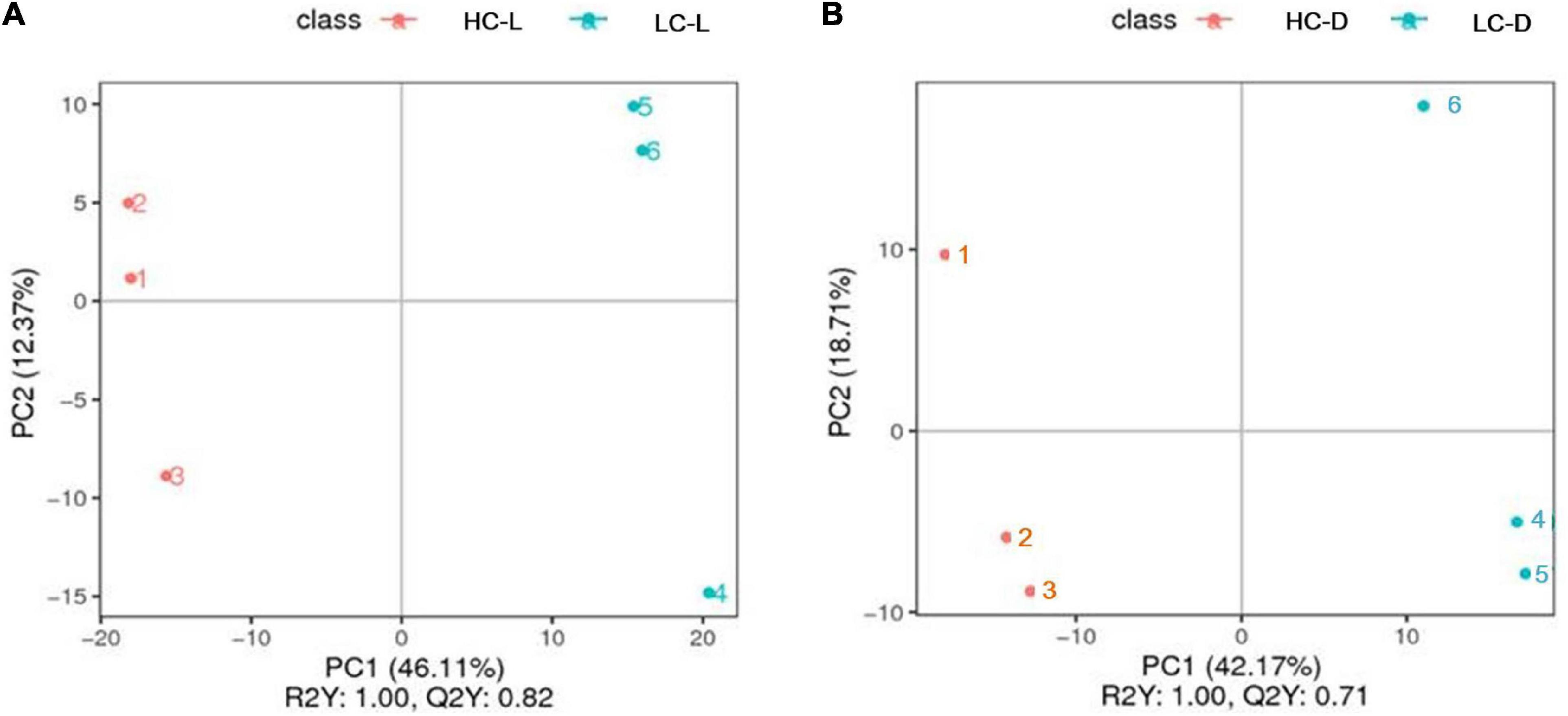
Figure 2. Partial least-squares discriminate analysis (PLS-DA) score plots of Ottelia alismoides metabolic profiles for different pairwise comparisons. (A) Low CO2 in the light vs. high CO2 in the light, (B) low CO2 in the dark vs high CO2 in the dark. HC-L, high CO2 in the light; HC-D, high CO2 in the dark; LC-L, low CO2 in the light; LC-D, low CO2 in the dark.
A p-value < 0.05 and a VIP > 1 were utilized to identify the important differently expressed metabolites associated with CO2 treatment and circadian rhythm conditions. Compared with high CO2, there were 156 differently expressed metabolites at low CO2 when in the light, including 102 increased and 54 decreased, whereas when in the dark, there were 107 differently expressed metabolites, including 82 increased and 25 decreased (Figure 3A). The differentially changed metabolites in different CO2-treated O. alismoides plants mainly included amino acids, organic acids, carbohydrates, nucleotides, phospholipids, and phytohormones (Figures 3B,C).
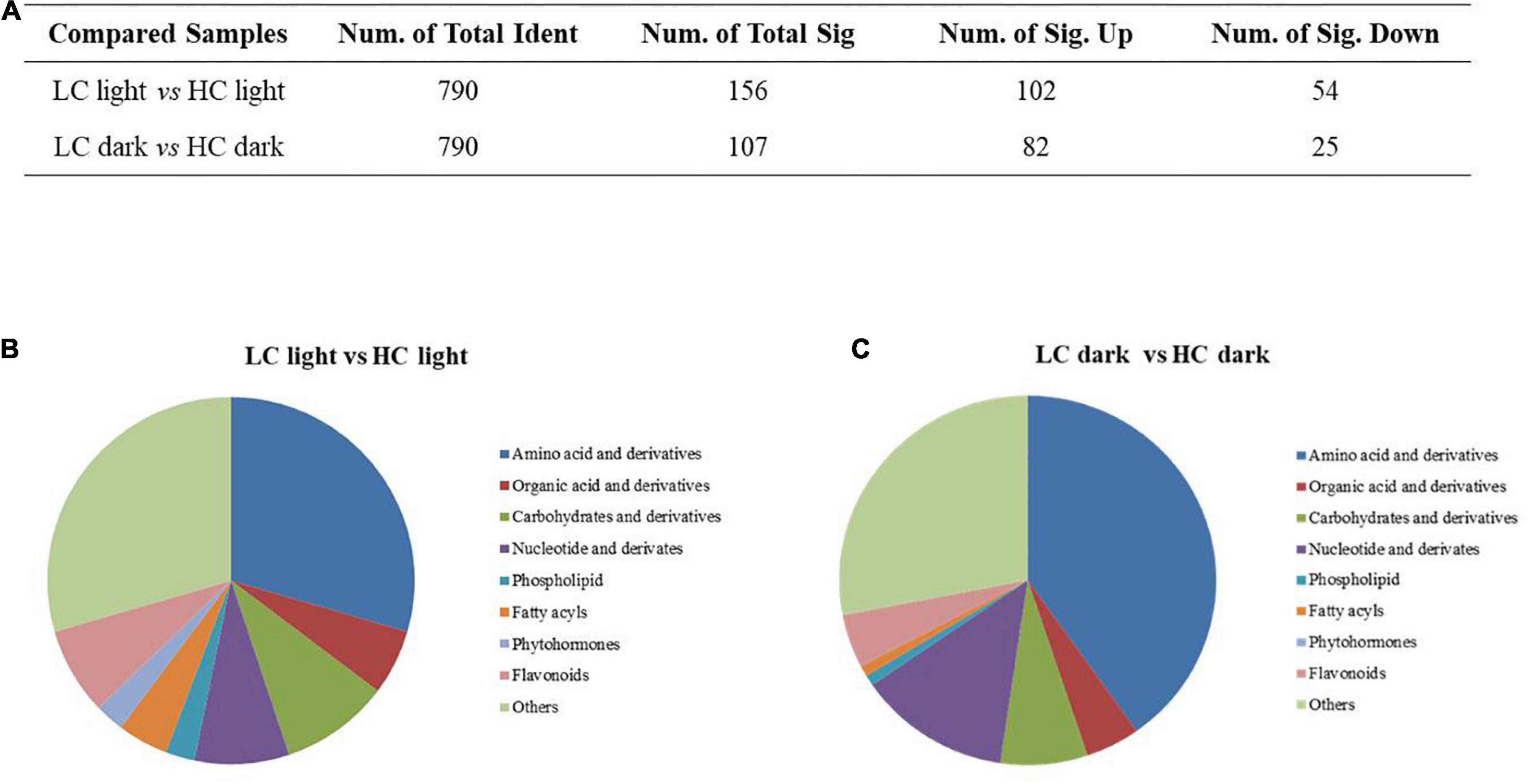
Figure 3. Effects of different CO2 on the metabolome of Ottelia alismoides. (A) Statistics of differently accumulated metabolites in each pairwise comparison. (B) Classification of differently accumulated metabolites between low and high CO2 treated O. alismoides sampled in the light. (C) Classification of differently accumulated metabolites between low and high CO2 treated O. alismoides sampled in the dark.
A total of 55 differentially expressed metabolites were selected, including organic acids, amino acids, and carbohydrates, as shown in Table 2. The corresponding up- or downregulated tendency, indicative of the log2 (fold change), showed how these metabolites varied at LC-treated O. alismoides plants compared to HC. Among the organic acids, a significant increase was observed in D-galactonic acid (independent t-test, p < 0.05; Table 2) in light under low CO2. As for the amino acids, significantly increased metabolites include glutamate (independent t-test, p < 0.001 in the light and p < 0.05 in the dark; Table 2), glutamine (independent t-test, p < 0.05 in the light and p < 0.01 in the dark; Table 2), γ-aminobutyric acid (GABA) (independent t-test, p < 0.01 in the light and p < 0.05 in the dark; Table 2), lysine (independent t-test, p < 0.05 in the light and p < 0.01 in the dark; Table 2), homoserine, and threonine (independent t-test, p < 0.05 in both light and dark; Table 2), except the succinic acid, which only increased in light (independent t-test, p < 0.05; Table 2). On the contrary, tryptophan, serine, and histidine significantly declined under light (independent t-test, p < 0.05; Table 2). In response to low CO2, the upregulated carbohydrates included L-arabinose (3.33-fold in the light, independent t-test, p < 0.001 and 2.51-fold in the dark, independent t-test, p < 0.05, respectively; Table 2), 3-phosphoglyceric acid (1.63-fold in the light, independent t-test, p < 0.05; Table 2), D-xylulose (3.41-fold in the light, independent t-test, p < 0.05 and 2.81-fold in the dark, independent t-test, p < 0.05, respectively; Table 2), and D-glucose 1-phosphate (0.94-fold in the light, independent t-test, p < 0.05 and 0.70-fold in the dark, independent t-test, p < 0.05, respectively; Table 2). On the contrary, glucose (−0.56-fold in the dark, independent t-test, p < 0.05; Table 2), fructose (−0.49-fold in the dark, independent t-test, p < 0.05; Table 2), D-melezitose (−0.97-fold in the light, independent t-test, p < 0.05; Table 2), and 2-deoxy-D-galactose (−1.18-fold in the light, independent t-test, p < 0.05 and −0.84-fold in the dark, independent t-test, p < 0.05, respectively; Table 2) were significantly decreased. Among the phytohormones, 3-indolebutyric acid (IBA) was downregulated considerably (−1.43-fold in the light, independent t-test, p < 0.05; Table 2) in LC-treated plants in comparison to HC.
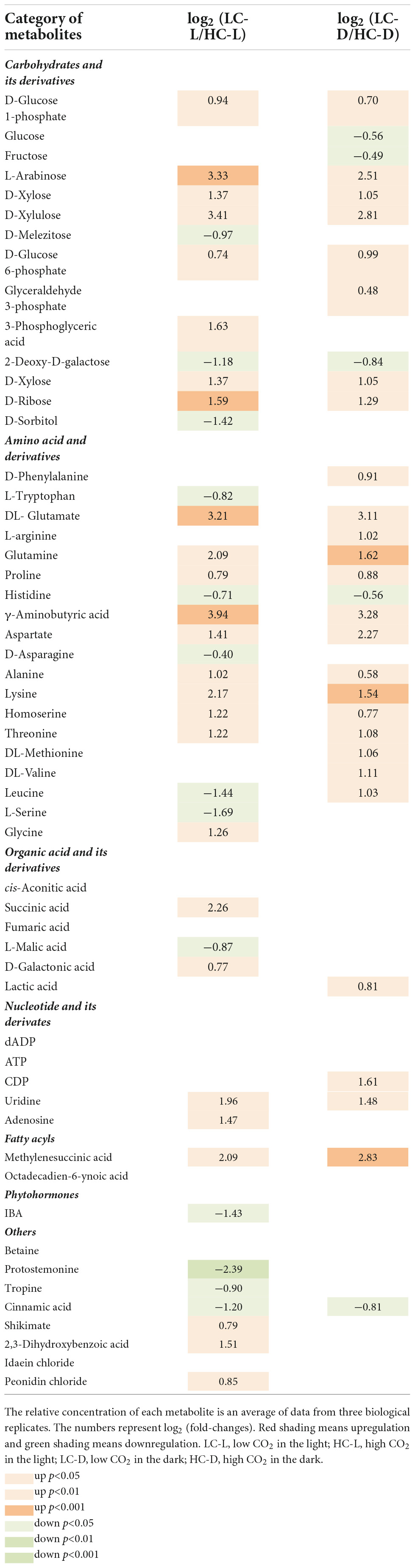
Table 2. List of significantly affected metabolites in low CO2 in the light compared to high CO2 in the light, and low CO2 in the dark compared to high CO2 in the dark.
These above-mentioned metabolites found to be involved with carbon metabolism, assimilation of amino acids, and biosynthesis of secondary metabolites were assorted into different groups based on their metabolic functions, according to the analysis of KEGG pathways, which is the major pathway-related public database that includes genes and metabolites. Based on these metabolomics results, we primarily focused on the key metabolites participating in the vital carbon and nitrogen metabolic pathways in O. alismoides plants, such as glycolysis, pentose phosphate pathway (PPP), TCA cycle, and amino acid metabolism.
Metabolites associated with energy metabolism including glycolysis, PPP, and TCA cycle intermediates were altered significantly following low CO2 stress in O. alismoides plants (Figure 4). The relative abundance of glucose-6-phosphate (G-6-P, 0.74-fold in the light, independent t-test, p < 0.05 and 0.99-fold in the dark, independent t-test, p < 0.05, respectively), glyceraldehyde 3-phosphate (G-3-P, 0.48-fold in the dark, independent t-test, p < 0.05), and 3-phosphoglyceric acid (3-PG, 1.63-fold in the light, independent t-test, p < 0.05) significantly increased in LC-treated O. alismoides plants (Figure 4A and Table 2). Pentose phosphate pathway, an alternative branch of glycolysis to produce sugars, appeared to be highly activated in O. alismoides under low CO2 based on the metabolites present in this pathway. The levels of D-ribose (1.59-fold in the light, independent t-test, p < 0.001 and 1.29-fold in the dark, independent t-test, p < 0.05, respectively), D-galactonic acid (0.77-fold in the light, independent t-test, p < 0.05), and D-xylose (1.37-fold in the light, independent t-test, p < 0.05 and 1.05-fold in the dark, independent t-test, p < 0.05, respectively), significantly increased in LC-treated O. alismoides, respectively (Figure 4B and Table 2). Figure 4C shows the content changes of six organic acids involved in the TCA cycle in light and dark, including citric acid, cis-aconitic acid, isocitric acid, succinic acid, fumaric acid, and malic acid in low and high CO2 conditions, respectively. Malic acid, the key nocturnal carbon storage metabolite for CAM, cycled as expected with the lowest level in the light and with the highest level in the dark, when under low CO2 (independent t-test, p < 0.05). However, when under HC, the malic acid levels did not differ between light and dark (independent t-test, p > 0.05). In addition, we observed significantly elevated levels of succinic acid in LC-treated O. alismoides in light (2.26-fold, independent t-test, p < 0.05; Figure 4C and Table 2). The other metabolites in the TCA cycle (citric acid, cis-aconitic acid, fumaric acid, and isocitric acid) have no significant alterations under LC when compared to HC (independent t-test, p > 0.05; Figure 4C and Table 2).
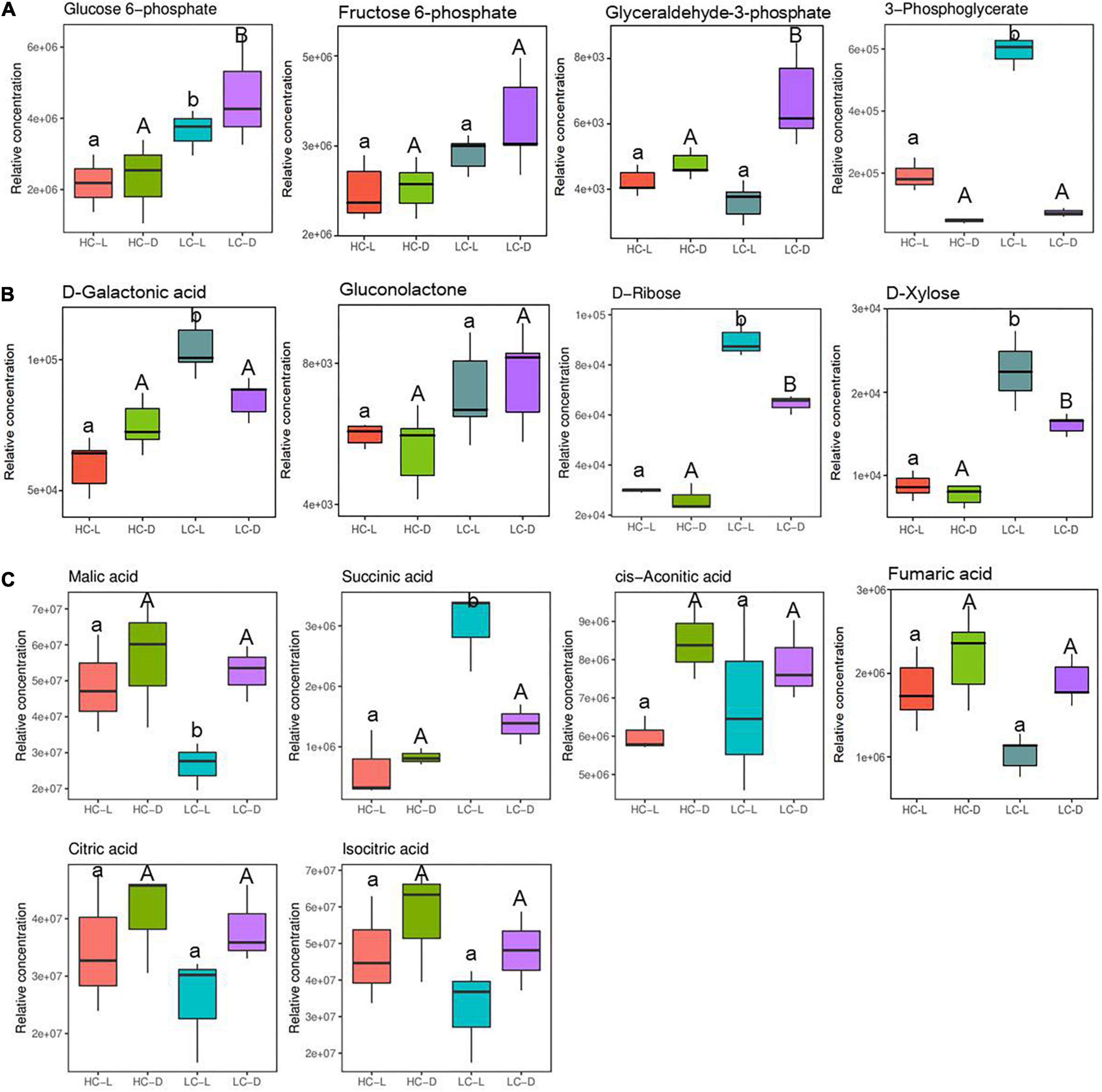
Figure 4. Box plots of relative abundance of the metabolites related to the C metabolism pathway in Ottelia alismoides, based on the analysis of KEGG pathways. (A) Glycolytic pathway; (B) PPP; (C) TCA cycle. HC-L, high CO2 in the light; HC-D, high CO2 in the dark; LC-L, low CO2 in the light; LC-D, low CO2 in the dark. Different lowercase letters and uppercase letters indicate significant differences (p < 0.05) between different CO2 treatments in the light and dark, respectively.
Figure 5A shows the content changes of glutamate and its derivatives including arginine, proline, glutamine, and GABA in light and dark. Glutamate was observed to increase (3.21-fold in the light, independent t-test, p < 0.001 and 3.11-fold in the dark, independent t-test, p < 0.05, respectively) in LC-treated O. alismoides plants (Figure 5A and Table 2). Consistent with the glutamate increases, arginine (1.02-fold in the dark, independent t-test, p < 0.05), proline (0.79-fold in the light, independent t-test, p < 0.05 and 0.88-fold in the dark, independent t-test, p < 0.05, respectively), glutamine (2.09-fold in the light, independent t-test, p < 0.05 and 1.62-fold in the dark, independent t-test, p < 0.01, respectively), and GABA (3.94-fold in the light, independent t-test, p < 0.01 and 3.28-fold in the dark, independent t-test, p < 0.05, respectively) also increased in LC-treated O. alismoides plants. Contrary, the abundance of histidine (−0.71-fold in the light, independent t-test, p < 0.05 and −0.56-fold in the dark, independent t-test, p < 0.05, respectively) decreased under LC (Figure 5A and Table 2). The amino-acid pathway derived from aspartate is in charge of the distribution of carbon flux from aspartate into the synthesis of lysine, threonine, and methionine (Curien et al., 2009). Low CO2 caused a significant increase in metabolites produced by aspartate metabolism, including aspartate (1.41-fold in the light, independent t-test, p < 0.05 and 2.27-fold in the dark, independent t-test, p < 0.05, respectively), alanine (1.02-fold in the light, independent t-test, p < 0.05 and 0.58-fold in the dark, independent t-test, p < 0.05, respectively), homoserine (1.22-fold in the light, independent t-test, p < 0.05 and 0.77-fold in the dark, independent t-test, p < 0.05, respectively), threonine (1.22-fold in the light, independent t-test, p < 0.05 and 1.08-fold in the dark, independent t-test, p < 0.05, respectively), lysine (2.17-fold in the light, independent t-test, p < 0.05 and 1.54-fold in the dark, independent t-test, p < 0.01, respectively), and DL-methionine (1.06-fold in the dark, independent t-test, p < 0.05) in O. alismoides (Figure 5B and Table 2). However, the asparagine content (−0.40-fold in the light, independent t-test, p < 0.05) decreased in low-CO2-treated O. alismoides (Figure 5B and Table 2). The shikimate pathway provides carbon sources for the synthesis of aromatic amino acids, such as tryptophan, phenylalanine, and tyrosine; approximately 30% of the fixed carbon is fluxed into this pathway in plants (Maeda and Dudareva, 2012). In the present study, the content of shikimate (0.79-fold in the light, independent t-test, p < 0.05) and phenylalanine (0.91-fold in the dark, independent t-test, p < 0.05) were significantly increased under LC treatment. In contrast, tryptophan was significantly decreased (−0.82-fold in the light, independent t-test, p < 0.05) under LC. In addition, the metabolites that were derived from phenylalanine in the shikimate pathway, cinnamic acid (−1.20-fold in the light, independent t-test, p < 0.05 and −0.81-fold in the dark, independent t-test, p < 0.05, respectively) also significantly decreased in abundance under LC (Figure 5C and Table 2). No significant difference was observed in tyrosine and 4-hydrocinnamic acid levels (independent t-test, p > 0.05; Figure 5C).
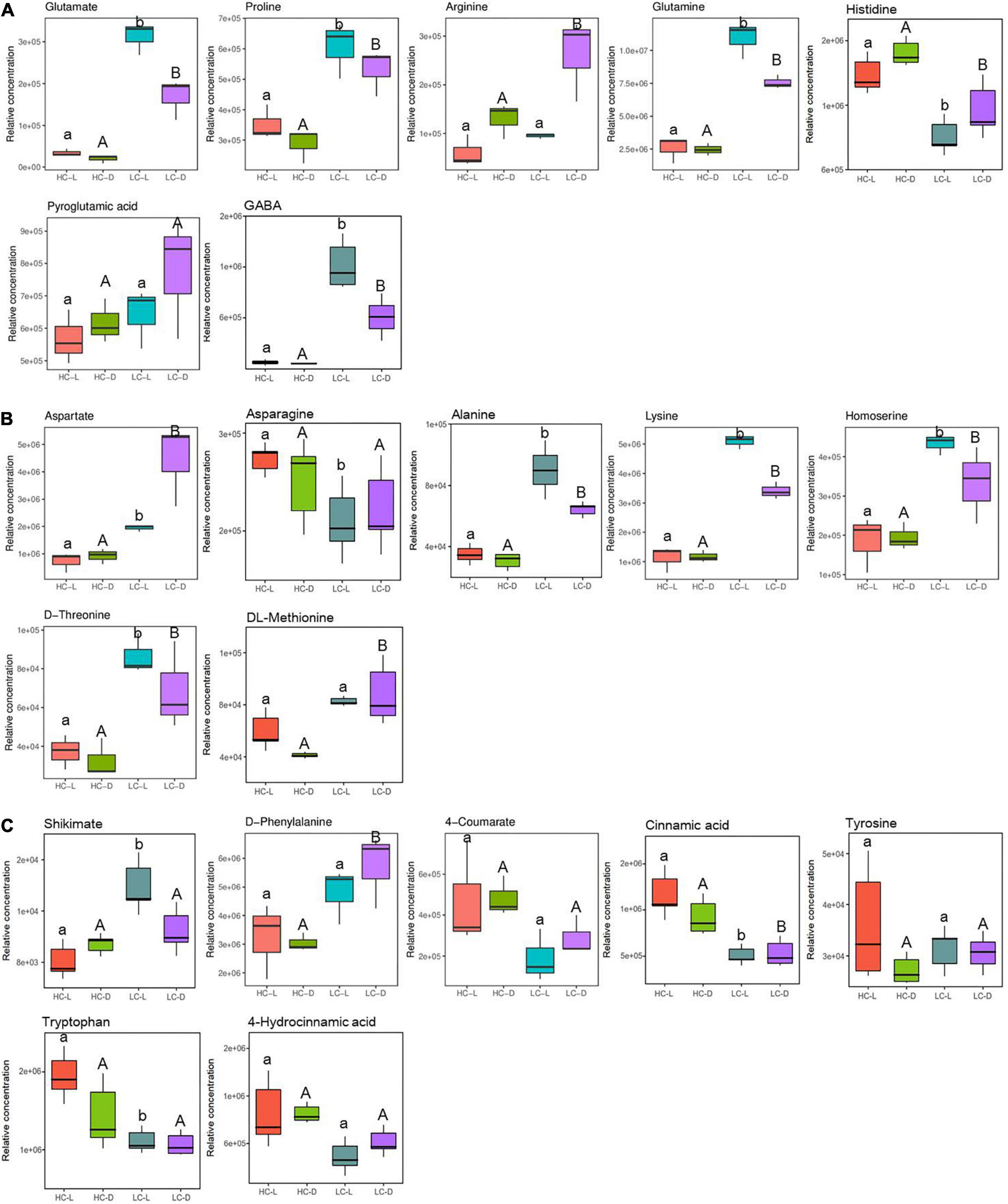
Figure 5. Box plots of relative abundance of the metabolites related to N metabolism pathway in Ottelia alismoides, based on the analysis of KEGG pathways. (A) Arginine and proline metabolism; (B) aspartate metabolism; (C) shikimate–phenylpropanoid metabolism. HC-L, high CO2 in the light; HC-D, high CO2 in the dark; LC-L, low CO2 in the light; LC-D, low CO2 in the dark. Different lowercase letters and uppercase letters indicate significant differences (p < 0.05) between different CO2 treatments in the light and dark, respectively.
The synergistic regulation of carbon and nitrogen metabolism plays a crucial role in the adaptation of plants to all kinds of stresses. For freshwater plants, the availability of CO2 is often one of the growth-limiting factors. To understand how the freshwater plant O. alismoides regulates carbon and nitrogen metabolism for better adapting to environmental low CO2 limitations, physiological measurements combined with a comprehensive metabolomics analysis were performed in O. alismoides acclimated to high and low CO2, respectively. In this section, the physiological and biochemical relevance of the present data, and the differential roles of metabolites involved in CAM metabolism, respiratory metabolism, and amino acid metabolism were discussed to reveal how the carbon and nitrogen metabolism were co-regulated when O. alismoides acclimated to low CO2. To the best of our knowledge, this is the first study to reveal the metabolic response of C and N metabolism to low CO2 stress in the freshwater plant O. alismoides with metabolome.
In aquatic ecosystems, less than 10% of the tested aquatic species have been discovered to operate CAM photosynthesis, which has been considered to be a carbon-conservation strategy that effectively increases carbon utilization (Maberly and Gontero, 2017). The features of CAM generally include the following: (1) dark fixation of CO2 and nocturnal accumulation of malic acid, (2) diurnal fluctuations in acidity/malate and starch content, and the opposite diel pattern of acidity/malate and starch, (3) high activity of PEPC at night that allows the production of malate, and (4) high ratio of PEPC:rubisco activity (Keeley, 1998; Igamberdiev and Eprintsev, 2016; Maberly and Gontero, 2017). In the present study, only low-CO2-acclimated O. alismoides exhibited significant diurnal titratable acidity and malate fluctuations, and an opposite diel pattern of starch change, consistent with the expectation and characteristics of CAM metabolism. Furthermore, based on the enzymatic activity analysis (Rubisco, PEPC, and PPDK), when O. alismoides were treated with low CO2, the activities of PEPC and PPDK (regenerates PEP to ensure the substrate requirements for PEPC) were very high in the dark and were much higher than that of HC plants, in accordance with nocturnal carboxylation due to CAM photosynthesis. The increased enzyme activity of PEPC and PPDK induced by low CO2 was consistent with the previous reports for O. alismoides (Zhang et al., 2014; Shao et al., 2017) and was consistent with that in low-CO2-grown terrestrial CAM species Opuntia ficus-indica (Israel and Nobel, 1994) and the very low-CO2-treated microalgae Nannochloropsis oceanica (Wei et al., 2019). Overall, these patterns of variation of enzyme activities, as well as acidity and starch content, confirm that CAM photosynthesis was induced in LC-stressed O. alismoides, which confirmed our previous results (Huang et al., 2018; Han et al., 2020).
It has been pointed out that an active CAM would suffer energy costs for investing in the machinery and running of CAM (Raven and Lucas, 1985). Glycolysis, PPP, and TCA cycle are primary metabolic pathways in plants and could provide energy and carbon skeletons for other metabolic pathways. According to the metabolomic analyses, a metabolic network (Figure 6) and a summary of metabolic changes in carbon and nitrogen metabolism (Figure 7) were reconstructed for O. alismoides under different CO2 conditions. Glucose-6-phosphate is linked to the start of both glycolysis and PPP pathways, and these are critical for the plants to respond to abiotic stress (Aranda et al, 2020). Glycolysis is the predominant pathway to fuel plant respiration; in addition, it is directly involved in many biochemical adaptations of plants to environmental stresses, such as nutrient limitation, as well as osmotic, drought, and cold stresses, which are crucial in plants (Plaxton, 1996). In the present study, the upregulation of the metabolites (G-6-P, G-3-P, and 3-PG) involved in glycolysis under LC indicates that there was a higher accumulation of intermediates of the glycolytic pathway, which presumably reflects some adjustment in the pattern of carbon flux responding to reduced photosynthesis and higher energy demand in low-CO2-acclimated O. alismoides. In addition, the changed carbohydrates were not only involved in glycolysis but also related to PPP, as revealed in this study. In detail, increased levels of D-ribose, D-xylose, and D-galactonic acid derived from the PPP pathway were observed in LC-treated plants, reflecting increased PPP metabolism in these plants. Correspondingly, the general decrease of glucose and fructose under low CO2 was most likely caused by increased glycolysis and PPP pathways (Hasler-Sheetal et al., 2016). The biological pathway analysis further confirms that the PPP pathway was altered by LC treatment (Supplementary Figure 4). A similar promotion of glycolysis and PPP has also been observed previously in other species, such as wheat cultivars under drought stress (Bowne et al., 2012). Pyruvate, the final metabolite of glycolysis, failed to be detected through metabolomic analysis in the present study, however, the amino acids derived from pyruvate, alanine, valine, and leucine were significantly upregulated in LC-treated O. alismoides. These results imply that when O. alismoides plants were treated with LC, they increased glycolytic flux and consumed carbohydrate pools to meet the requirements of energy and intermediate substance for the biosynthesis of other metabolites. As the core of respiratory machinery, the improved activity of the TCA cycle indicates the elevated energy demand and metabolic rate (Zhang et al., 2021). As revealed by the metabolomic data, six metabolites were identified in the TCA cycle in the present study. The succinic acid was found to be significantly increased in LC-treated O. alismoides plants. This finding is consistent with the result from Hasler-Sheetal et al. (2015), who reported that succinic acid was significantly increased in anoxic-stressed seagrass Zostera marina. The elevation of TCA pathway intermediates probably indicates the activation of the TCA cycle in LC-treated O. alismoides, which was in accordance with the report of Tcherkez et al. (2008), who clarified that the TCA cycle activity and glutamate production are promoted under a low CO2 environment. Wei et al. (2019) also recently reported that the TCA cycle genes were enriched in low-CO2-grown microalgae Nannochloropsis oceanica by tracking the transcriptomic profiles.
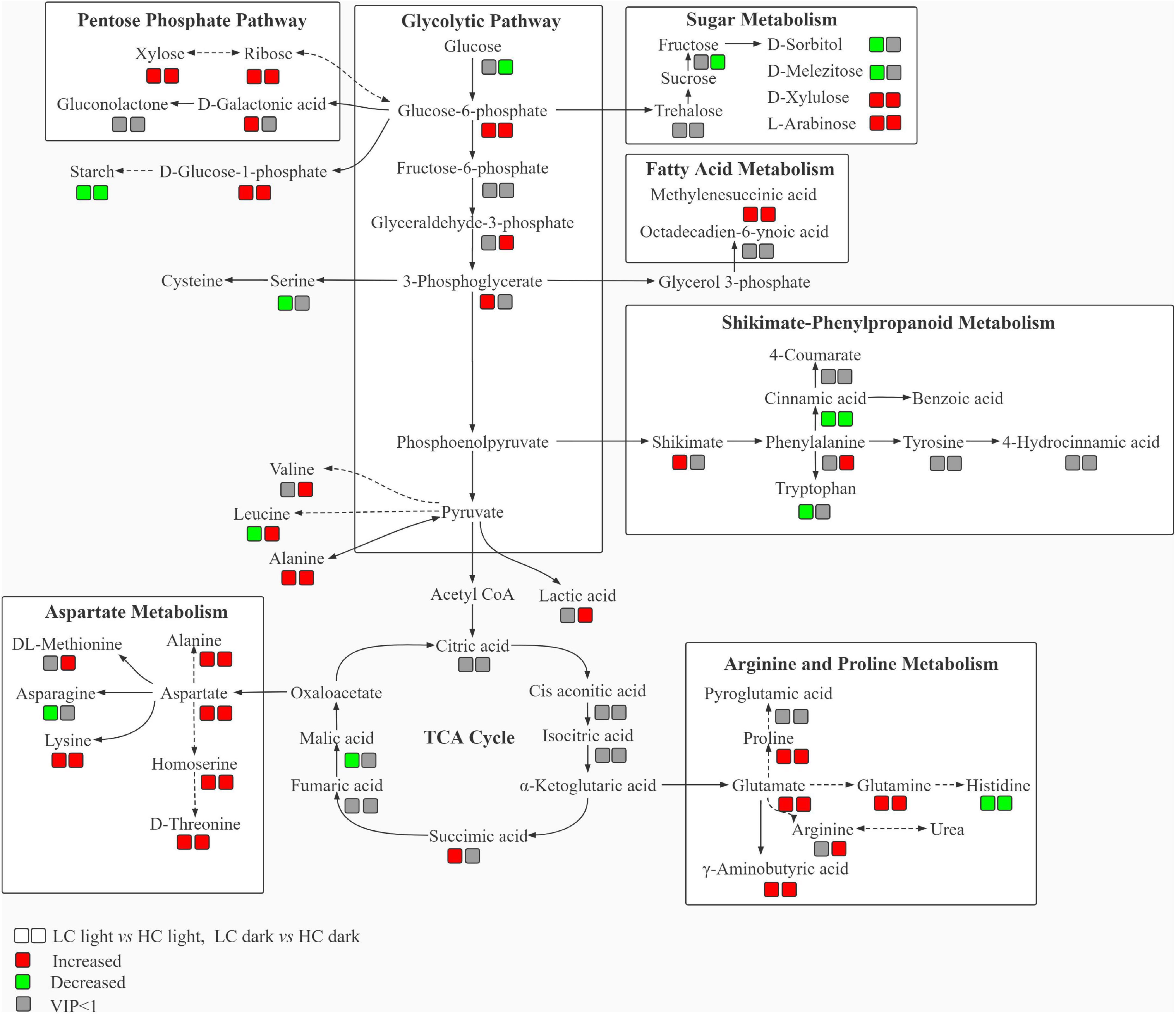
Figure 6. Schematic diagram of the proposed metabolic network in Ottelia alismoides responding to low and high CO2. Results represent fluctuations performing pairwise comparisons; the symbols underneath the metabolite names correspond to the result of the pairwise comparison that is indicated in the legend. Dashed lines symbolize multi-step or not fully elucidated pathway sections and solid lines symbolize one-step consecutive metabolites in a biosynthetic pathway.
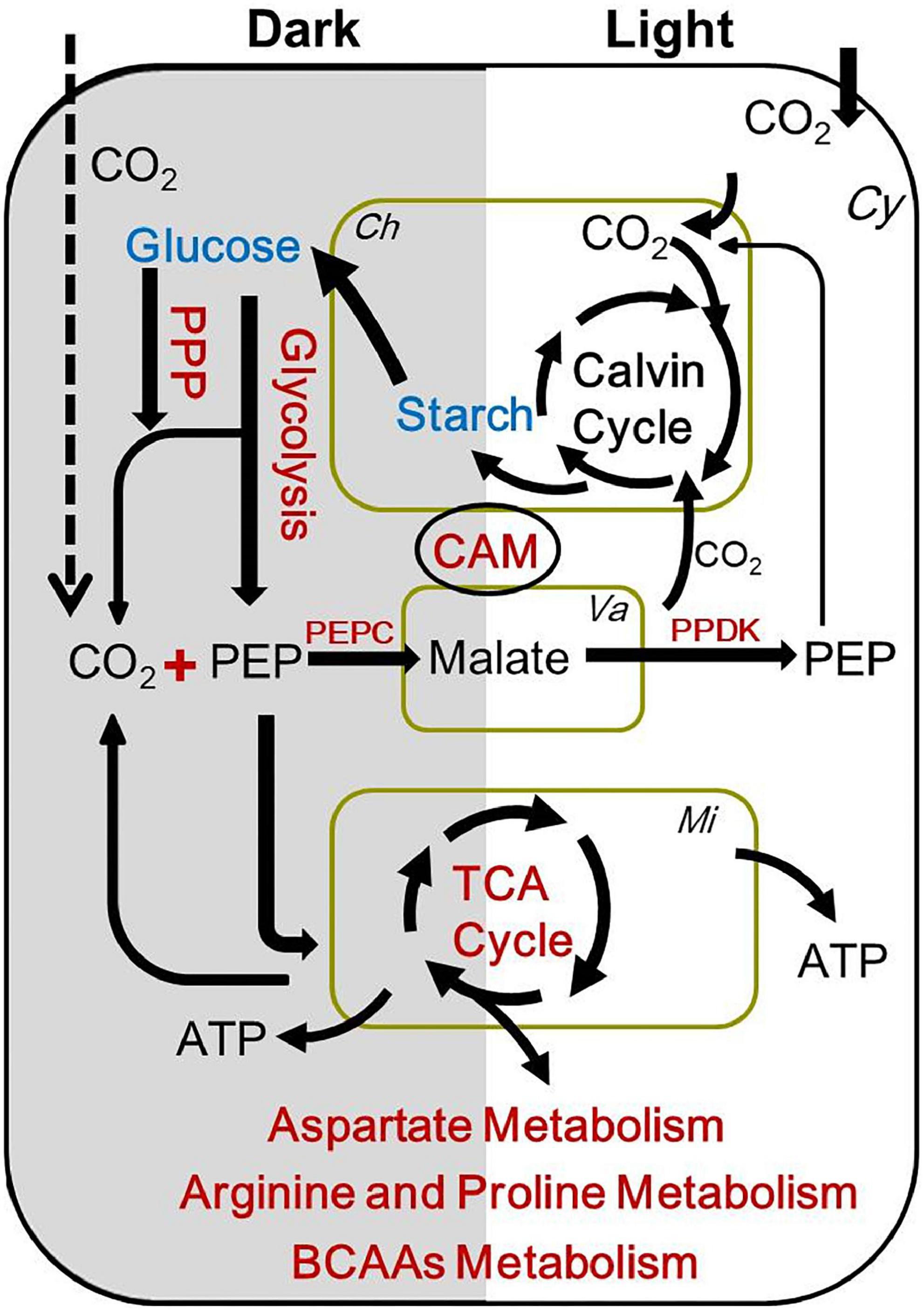
Figure 7. A summary of the metabolic changes in carbon and nitrogen metabolism in low CO2 compared to high CO2 grown Ottelia alismoides. Red indicates an increased metabolite content/upregulated reaction/induced metabolism and blue indicates a decreased metabolite content as compared with high CO2 grown plants. The dotted line indicates that the CO2 in solution perhaps enters the plant cell in dark for CAM carbon fixation. Ch, chloroplast; Cy, cytoplasm; Mi, mitochondria; Va, vacuole; PEP, phosphoenolpyruvate; BCAAs, branched-chain amino acids; PPP, pentose phosphate pathway.
Taken together, these observations indicate that the respiratory metabolism was upregulated in LC-treated O. alismoides, which was consistent with the previous study (Tcherkez et al., 2008). As we mentioned above, CAM photosynthesis was induced in O. alismoides under low CO2. Aquatic CAM photosynthesis is a CO2 concentrating mechanism that evolved in response to carbon limitation in water (Keeley, 1998). During the operation of CAM, the aquatic CAM plants are capable of dark fixation of CO2 and storage of malic acid, and daytime decarboxylation of malic acid to release CO2, which reduces carbon limitation, but with a certain investment of energy. It is therefore likely that when O. alismoides was stressed by LC, CAM photosynthesis was induced to adapt to the carbon stress, and the respiration was correspondingly upregulated to provide more energy for sustaining CAM photosynthesis.
Nitrogen metabolism regulation, which is closely linked with carbon metabolism in basic biochemical pathways in plants, is crucial for plant stress tolerance (Lawlor, 2002; Zhong et al., 2017). The carbon skeletons for amino acid synthetic pathways are generated in different sectors of the respiration process. Furthermore, ATP and reductant required for amino acid biosynthesis are also from respiration. Thus, nitrogen assimilation inevitably closely interacts with respiration, and the C produced by photosynthetic CO2 assimilation is a building block for amino acid synthesis (Stitt et al., 2002). In the present study, low CO2 treatment significantly affected the nitrogen metabolism and had a great impact on the amino acids pool in O. alismoides. Most of the identified amino acids, as well as a number of N-containing molecules derived from glutamine and glutamate, such as GABA, were strongly induced under low CO2 stress. These differentially changed amino acids were mainly involved in pathways of glutamate and arginine metabolism, aspartate metabolism, and branched-chain amino acids (BCAAs) metabolism.
The amino acids involved in glutamate and arginine metabolism, including glutamate, glutamine, proline, GABA, and arginine (only in the dark), were derived from TCA cycle intermediate α-ketoglutaric acid and were all significantly promoted under LC. Both glutamate and glutamine have an important effect on nitrogen metabolism (Zhao et al., 2018). The increase in glutamate and glutamine content under LC was in line with a previous report in Arabidopsis that glutamine level was upregulated when subjected to combined stress with drought and heat (Rizhsky et al., 2004). It is also reported that low N induced significant upregulation of amino acids in plants, including glutamate and glutamate-derived amino acids (Kusano et al., 2011). The gene glutamate dehydrogenase 2 (GDH2), which was involved in the synthesis of glutamate, was upregulated by drought stress in wild species of tomato (Egea et al., 2018). The amino acid GABA is mainly synthesized from glutamate through the catalyzation of glutamate decarboxylase (Michaeli et al., 2011). This metabolite always accumulates rapidly under adverse environmental conditions; moreover, genes involved in the GABA metabolism, glutamate decarboxylase 1 (GAD1) and γ-aminobutyrate transaminase (GABA-T), were specifically induced by drought stress in the wild relative of tomato, Solanum pennellii (Egea et al., 2018). The high level of GABA is tightly correlated with the enhanced resistance to stressful conditions (Kinnersley and Turano, 2000; Renault et al., 2010; Podlešáková et al., 2018). Numerous metabolic processes are regulated by GABA, such as the TCA cycle, nitrogen metabolism, as well as metabolic responses to oxidative stress (Bouché and Fromm, 2004). It has been suggested that seagrass could compensate for the deprivation of energy by utilizing the carbon derived from glutamine via the GABA shunt (Hasler-Sheetal et al., 2015). In this study, the significantly increased GABA throughout the diel cycle in low CO2 treated plants is in agreement with the previous studies within the low-light-stressed seagrass Zostera marina (Hasler-Sheetal et al., 2016). The enhanced GABA perhaps implies a GABA shunt compensated for energy deprivation caused by low carbon limitation in O. alismoides.
Concerning other reactions of N metabolism, low CO2 also promoted the aspartate-family pathway in O. alismoides, revealed by the significantly increased concentration of aspartate, lysine, methionine, threonine, alanine, and homoserine. Lysine, methionine, and threonine, as the essential amino acids, are commonly referred to as aspartate (Asp)-family amino acids; they are generated from a common precursor Asp via complicated pathways (Kirma et al., 2012). These Asp-family amino acids originated from TCA intermediates oxaloacetate and contribute to nitrogen storage and utilization. Plenty of evidence has revealed that the Asp-family metabolism is linked with the TCA cycle, which plays an important role in regulating the physiological response to energy deprivation caused by various abiotic stresses in plants (Kirma et al., 2012). Taking lysine as an example, it not only acted as an essential constituent for proteins but also catabolized in the TCA cycle for generating energy (Galili, 2011). The promotion in the Asp-family pathway observed in LC stressed O. alismoides suggests that the metabolic response caused by carbon limitation is similar to that found for many other abiotic stresses. In the metabolomic study of a model plant Lemna minor L., the amino acids of the Asp families were also upregulated in herbicides in glyphosate-treated plants (Kostopoulou et al., 2020). Interestingly, in this study, asparagine was found to be significantly decreased under low CO2 in light. Generally, when under C limitation, N is channeled to asparagine as a transient storage pool through the transcriptionally activated asparagine synthetase (Weigelt et al., 2008). However, the reason for asparagine reduction in this study is unknown.
In addition, alanine, valine, and leucine, which are associated with pyruvate metabolism, were observed to be increased in low-CO2-treated O. alismoides. Alanine is the major free amino acid and the latter two (valine and leucine) belong to the BCAAs (Pernollet et al., 1986), which are jointly regulated in their biosynthesis due to sharing common enzymes (Joshi et al., 2010). It has been proposed that BCAAs could provide alternative carbon sources for stressed plants. Previous studies have reported that BCAAs’ content was significantly increased in sugar-starved or drought-stressed Arabidopsis (Rizhsky et al., 2004; Taylor et al., 2004). Urano et al. (2009) also found the increased accumulation of BCAAs under dehydration stress and BCAAs were transcriptionally regulated. Egea et al. (2018) reported that the gene involved in BCAAs biosynthesis, the branched-chain-amino-acid transaminase 5 (BCAT5), was induced in drought-stressed Solanum pennellii. As such, the increased accumulation of BCAAs in low-CO2-stressed O. alismoides might be associated with the adaptation of plants to carbon deficiency to fulfill the demands of growing in C-limited conditions.
Collectively, these observations indicate that the changes in amino acids caused by low CO2 are closely associated with the regulation of intermediates involved in glycolysis, PPP, and TCA cycle. Low CO2 promoted the biosynthesis of amino acids in O. alismoides through the upregulation of respiration. Wei et al. (2019) have also reported that in low-CO2 grown Nannochloropsis oceanica, the carbon flow was switched from protein synthesis to other pathways (e.g., gluconeogenesis and secondary metabolite biosynthesis). The enhanced respiration process under low CO2 could produce more energy and precursors (such as α-ketoglutaric acid and oxaloacetate) for amino acid synthesis. Moreover, the operation of CAM induced by low CO2 is inevitable to increase the demand for energy and other resources such as proteins (enzymes) (Raven and Lucas, 1985; Maberly and Gontero, 2017). Therefore, the promoted N metabolism in low CO2, including glutamate and arginine metabolism, aspartate metabolism, and BCAAs metabolism, and these in turn presumably, could be used to support extra energy and alternative carbon sources, as well as to provide components for the construction of CAM machines for better adaptation of O. alismoides to carbon limitation conditions.
In conclusion, this work provides a valuable insight into the metabolic response of O. alismoides to variable CO2 based on a conjoint analysis of physiology and metabolomics. Comparative metabolomic analysis revealed significant changes in the metabolite profiles, especially the regulation of C and N metabolism, responding to the variable CO2 in O. alismoides. Nevertheless, more investigation is required to understand the relevance and contributions of these metabolic processes mediated by variable CO2.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
WL and WH designed the experiments. SH and WH performed the experiments and collected the data. WH, SH, and LW analyzed the data. WH prepared the manuscript. All authors contributed and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31970368, 32170390, and 31870346) and the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB31000000).
We thank the reviewers and editors for their valuable suggestions on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.962622/full#supplementary-material
Aranda, I., Cadahía, E., and Fernández de Simón, B. (2020). Leaf ecophysiological and metabolic response in Quercus pyrenaica willd seedlings to moderate drought under enriched CO2 atmosphere. J. Plant Physiol. 244:153083. doi: 10.1016/j.jplph.2019.153083
Baattrup-Pedersen, A., and Madsen, T. V. (1999). Interdependence of CO2 and inorganic nitrogen on crassulacean acid metabolism and efficiency of nitrogen use by Littorella uniflora (L.) aschers. Plant Cell Environ. 22, 535–542. doi: 10.1046/j.1365-3040.1999.00413.x
Bouché, N., and Fromm, H. (2004). GABA in plants: just a metabolite? Trends Plant Sci. 9, 110–115. doi: 10.1016/j.tplants.2004.01.006
Bowne, J. B., Erwin, T. A., Juttner, J., Schnurbusch, T., Langridge, P., Bacic, A., et al. (2012). Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol. Plant 5, 418–429. doi: 10.1093/mp/ssr114
Bradford, M. M. (1976). Arapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cheung, C. Y., Poolman, M. G., Fell, D. A., Ratcliffe, R. G., and Sweetlove, L. J. (2014). A diel flux balance model captures interactions between light and dark metabolism during day-night cycles in C3 and crassulacean acid metabolism leaves. Plant Physiol. 165, 917–929. doi: 10.1104/pp.113.234468
Curien, G., Bastien, O., Robert-Genthon, M., Cornish-Bowden, A., Cárdenas, M. L., and Dumas, R. (2009). Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol. Syst. Biol. 5:271. doi: 10.1038/msb.2009.29
Egea, I., Albaladejo, I., Meco, V., Morales, B., Sevilla, A., Bolarin, M. C., et al. (2018). The drought-tolerant Solanum pennellii regulates leaf water loss and induces genes involved in amino acid and ethylene/jasmonate metabolism under dehydration. Sci. Rep. 8:2791. doi: 10.1038/s41598-018-21187-2
Galili, G. (2011). The aspartate-family pathway of plants: Linking production of essential amino acids with energy and stress regulation. Plant Signal. Behav. 6, 192–195. doi: 10.4161/psb.6.2.14425
Han, S. J., Xing, Z. F., Li, W., and Huang, W. M. (2020). Response of anatomy and CO2-concentrating mechanisms to variable CO2 in linear juvenile leaves of heterophyllous Ottelia alismoides: Comparisons with other leaf types. Environ. Exp. Bot. 179:104194. doi: 10.1016/j.envexpbot.2020.104194
Hasler-Sheetal, H., Castorani, M. C. N., Glud, R. N., Canfield, D. E., and Holmer, M. (2016). Metabolomics reveals cryptic interactive effects of species interactions and environmental stress on nitrogen and sulfur metabolism in seagrass. Environ. Sci. Technol. 50, 11602–11609. doi: 10.1021/acs.est.6b04647
Hasler-Sheetal, H., Fragner, L., Holmer, M., and Weckwerth, W. (2015). Diurnal effects of anoxia on the metabolome of the seagrass Zostera marina. Metabolomics 11, 1–11. doi: 10.1007/s11306-015-0776-9
Huang, W. M., Shao, H., Zhou, S. N., Zhou, Q., Fu, W. L., Zhang, T., et al. (2018). Different CO2 acclimation strategies in juvenile and mature leaves of Ottelia alismoides. Photosynth. Res. 138, 219–232. doi: 10.1007/s11120-018-0568-y
Huppe, H. C., and Turpin, D. H. (1994). Integration of carbon and nitrogen metabolism in plant and algal cells. Annu. Rev. Plant Physiol. Plant Mol. Bioi. 45, 577–607. doi: 10.1146/annurev.pp.45.060194.003045
Igamberdiev, A. U., and Eprintsev, A. T. (2016). Organic acids: The pools of fixed carbon involved in redox regulation and energy balance in higher plants. Front. Plant Sci. 7:1042. doi: 10.3389/fpls.2016.01042
Israel, A. A., and Nobel, P. S. (1994). Activities of carboxylating enzymes in the CAM species Opuntia ficus-indica grown under current and elevated CO2 concentrations. Photosynth. Res. 40, 223–229. doi: 10.1007/BF00034772
Iversen, L. L., Winkel, A., and Baastrup-Spohr, L. (2019). Catchment properties and the photosynthetic trait composition of freshwater plant communities. Science 366, 878–881. doi: 10.1126/science.aay5945
Jain, V. M., Karibasappa, G. N., Dodamani, A. S., and Mali, G. V. (2017). Estimating the carbohydrate content of various forms of tobacco by phenol-sulfuric acid method. J. Educ. Health Promot. 6:90. doi: 10.4103/jehp.jehp_41_17
Joshi, V., Joung, J. G., Fei, Z., and Jander, G. (2010). Interdependence of threonine, methionine and isoleucine metabolism in plants: Accumulation and transcriptional regulation under abiotic stress. Amino Acids 39, 933–947. doi: 10.1007/s00726-010-0505-7
Keeley, J. E. (1998). CAM photosynthesis in submerged aquatic plants. Bot. Rev. 64, 121–175. doi: 10.1007/BF02856581
Kinnersley, A. M., and Turano, F. J. (2000). Gamma aminobutyric acid (GABA) and plant responses to stress. Crit. Rev. Plant Sci. 19, 479–509. doi: 10.1080/07352680091139277
Kirma, M., Araújo, W. L., Fernie, A. R., and Galili, G. (2012). The multifaceted role of aspartate-family amino acids in plant metabolism. J. Exp. Bot. 63, 4995–5001. doi: 10.1093/jxb/ers119
Klavsen, S. K., and Maberly, S. C. (2010). Effect of light and CO2 on inorganic carbon uptake in the invasive aquatic CAM plant Crassula helmsii. Funct. Plant Biol. 37, 737–747. doi: 10.1071/FP09281
Kostopoulou, S., Ntatsi, G., Arapis, G., and Aliferis, K. A. (2020). Assessment of the effects of metribuzin, glyphosate, and their mixtures on the metabolism of the model plant Lemna minor L. applying metabolomics. Chemosphere 239, 124582. doi: 10.1016/j.chemosphere.2019.124582
Krapp, A., and Traong, H. N. (2005). Regulation of C/N interaction in model plant species. J. Crop Improv. 15, 127–173. doi: 10.1300/J411v15n02_05
Kusano, M., Fukushima, A., Redestig, H., and Saito, K. (2011). Metabolomic approaches toward understanding nitrogen metabolism in plants. J. Exp. Bot. 62, 1439–1453. doi: 10.1093/jxb/erq417
Lawlor, D. W. (2002). Carbon and nitrogen assimilation in relation to yield: mechanisms are the key to understanding production systems. J. Exp. Bot. 53, 773–787. doi: 10.1093/jexbot/53.370.773
Leakey, A. D. B., Ainsworth, E. A., Bernacchi, C. J., Rogers, A., Long, S. P., and Ort, D. R. (2009). Elevated CO2 effects on plant carbon, nitrogen, and water relations: Six important lessons from FACE. J. Exp. Bot. 60, 2859–2876. doi: 10.1093/jxb/erp096
Luo, P., Dai, W. D., Yin, P. Y., Zeng, Z. D., Kong, H. W., Zhou, L. N., et al. (2015). MRM-ion pair finder: A systematic approach to transform non-targeted mode to pseudotargeted mode for metabolomics study based on liquid chromatography-mass spectrometry. Anal. Chem. 87, 5050–5055. doi: 10.1021/acs.analchem.5b00615
Maberly, S. C. (1996). Diel, episodic and seasonal changes in pH and concentrations of inorganic carbon in a productive lake. Fresh. Biol. 35, 579–598. doi: 10.1111/j.1365-2427.1996.tb01770.x
Maberly, S. C., and Gontero, B. (2017). Ecological imperatives for aquatic CO2-concentrating mechanisms. J. Exp. Bot. 68, 3797–3814. doi: 10.1093/jxb/erx201
Maberly, S. C., and Madsen, T. V. (1998). Affinity for CO2 in relation to the ability of freshwater macrophytes to use HCO3. Funct. Ecol. 12, 99–106. doi: 10.1046/j.1365-2435.1998.00172.x
Maeda, H., and Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Michaeli, S., Fait, A., Lagor, K., Nunes-Nesi, A., Grillich, N., Yellin, A., et al. (2011). A mitochondrial GABA permease connects the GABA shunt and the TCA cycle, and is essential for normal carbon metabolism. Plant J. 67, 485–498. doi: 10.1111/j.1365-313X.2011.04612.x
Nunes-Nesi, A., Fernie, A. R., and Stitt, M. (2010). Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 3, 973–996. doi: 10.1093/mp/ssq049
Pace, G. M., Volk, R. J., and Jackson, W. A. (1990). Nitrate reduction in response to CO2-limited photosynthesis. relationship to carbohydrate supply and nitrate reductase activity in maize seedlings. Plant Physiol. 92, 286–292. doi: 10.1104/pp.92.2.286
Pernollet, J. C., Huet, J. C., Moutot, F., and Morot Gaudry, J. F. (1986). Relationship between photosynthesis and protein synthesis in maize. II. interconversions of the photoassimilated carbon in the ear leaf and in the intermediary organs to synthesize the seed storage proteins and starch. Plant Physiol. 80, 216–222. doi: 10.1104/pp.80.1.216
Plaxton, W. C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 185–214. doi: 10.1146/annurev.arplant.47.1.185
Podlešáková, K., Ugena, L., Spíchal, L., Doležal, K., and De Diego, N. (2018). Phytohormones and polyamines regulate plant stress responses by altering GABA pathway. Nat. Biotechnol. 48, 53–65. doi: 10.1016/j.nbt.2018.07.003
Raven, J. A., and Lucas, W. J. (1985). “Energy cost of carbon acquisition,” in Inorganic Carbon Uptake By Aquatic Photosynthetic Organisms, eds W. J. Lucas and J. A. Berry (Rockville, MD: American Society of Plant Physiologists), 305–324.
Renault, H., Roussel, V., El Amrani, A., Arzel, M., Renault, D., Bouchereau, A., et al. (2010). The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 10:20. doi: 10.1186/1471-2229-10-20
Rizhsky, L., Liang, H., Shuman, J., Shulaev, V., Davletova, S., and Mittler, R. (2004). When defense pathways collide: The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134, 1683–1696. doi: 10.1104/pp.103.033431
Romero, J. M., Lara, C., and Guerrero, M. G. (1985). Dependence of nitrate utilization upon active CO2 fixation in Anacystis nidulans: A regulatory aspect of the interaction between photosynthetic carbon and nitrogen metabolism. Arch. Biochem. Biophys. 237, 396–401. doi: 10.1016/0003-9861(85)90291-7
Shameer, S., Baghalian, K., Cheung, C. Y. M., Ratcliffe, R. G., and Sweetlove, L. J. (2018). Computational analysis of the productivity potential of CAM. Nat. Plants 4, 165–171. doi: 10.1038/s41477-018-0112-2
Shao, H., Gontero, B., Maberly, S. C., Jiang, H. S., Cao, Y., Li, W., et al. (2017). Responses of Ottelia alismoides, an aquatic plant with three CCMs, to variable CO2 and light. J. Exp. Bot. 68, 3985–3995. doi: 10.1093/jxb/erx064
Silvera, K., Neubig, K. M., Whitten, W. M., Williams, N. H., Winter, K., and Cushman, J. C. (2010). Evolution along the crassulacean acid metabolism continuum. Funct. Plant Biol. 37, 995–1010. doi: 10.1071/FP10084
Smith, A. M., and Zeeman, S. C. (2006). Quantification of starch in plant tissues. Nat. Protoc. 1, 1342–1345. doi: 10.1038/nprot.2006.232
Stitt, M., Muller, C., Matt, P., Gibon, Y., Carillo, P., Morcuende, R., et al. (2002). Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 53, 959–970. doi: 10.1093/jexbot/53.370.959
Sweetlove, L. J., Beard, K. F., Nunesnesi, A., Fernie, A. R., and Ratcliffe, R. G. (2010). Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 15, 462–470. doi: 10.1016/j.tplants.2010.05.006
Taylor, K. A. C. C. (1995). A modification of the phenol/sulfuric acid assay for total carbohydrates giving more comparable absorbances. Appl. Biochem. Biotechnol. 53, 207–214. doi: 10.1007/BF02783496
Taylor, N. L., Heazlewood, J. L., Day, D. A., and Millar, A. H. (2004). Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol. 134, 838–848. doi: 10.1104/pp.103.035675
Tcherkez, G., Bligny, R., Gout, E., Mahe, A., Hodges, M., and Cornic, G. (2008). Respiratory metabolism of illuminated leaves depends on CO2 and O2 conditions. Proc. Natl. Acad. Sci. U.S.A. 105, 797–802. doi: 10.1073/pnas.0708947105
Urano, K., Maruyama, K., Ogata, Y., Morishita, Y., Takeda, M., Sakurai, N., et al. (2009). Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 57, 1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x
Want, E. J., Masson, P., Michopoulos, F., Wilson, I. D., Theodoridis, G., Plumb, R. S., et al. (2012). Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 8, 17–32. doi: 10.1038/nprot.2012.135
Wei, L., El Hajjami, M., Shen, C., You, W. X., Lu, Y. D., Li, J., et al. (2019). Transcriptomic and proteomic responses to very low CO2 suggest multiple carbon concentrating mechanisms in Nannochloropsis oceanica. Biotechnol. Biofuels 12:168. doi: 10.1186/s13068-019-1506-8
Weigelt, K., Küster, H., Radchuk, R., Müller, M., Weichert, H., Fait, A., et al. (2008). Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J. 55, 909–926. doi: 10.1111/j.1365-313X.2008.03560.x
Xian, L., Zhang, Y., Cao, Y., Wan, T., Gong, Y., Dai, C., et al. (2020). Glutamate dehydrogenase plays an important role in ammonium detoxification by submerged macrophytes. Sci. Total Environ. 722:137859. doi: 10.1016/j.scitotenv.2020.137859
Yokoyama, S., and Hiramatsu, J. (2003). A modified ninhydrin reagent using ascorbic acid instead of potassium cyanide. J. Biosci. Bioeng. 95, 204–205. doi: 10.1016/S1389-1723(03)80131-7
Zhang, C. C., Zhou, C. Z., Burnap, R. L., and Peng, L. (2018). Carbon/nitrogen metabolic balance: Lessons from cyanobacteria. Trends Plant Sci. 23, 1116–1130. doi: 10.1016/j.tplants.2018.09.008
Zhang, Y. Z., Yin, L. Y., Jiang, H. S., Li, W., Gontero, B., and Maberly, S. C. (2014). Biochemical and biophysical CO2 concentrating mechanisms in two species of freshwater macrophyte within the genus ottelia (hydrocharitaceae). Photosynth. Res. 121, 285–297. doi: 10.1007/s11120-013-9950-y
Zhang, Z. Q., Zhou, C., Fan, K. P., Zhang, L., Liu, Y., and Liu, P. F. (2021). Metabolomics analysis of the effects of temperature on the growth and development of juvenile European seabass (Dicentrarchus labrax). Sci. Total Environ. 769:145155. doi: 10.1016/j.scitotenv.2021.145155
Zhao, L., Huang, Y., and Keller, A. A. (2018). Comparative metabolic response between cucumber (Cucumis sativus) and corn (Zea mays) to a Cu(OH)2 nanopesticide. J. Agr. Food Chem. 66, 6628–6636. doi: 10.1021/acs.jafc.7b01306
Keywords: carbon metabolism, glycolysis, low CO2 stress, metabolic profiling, N metabolism, TCA cycle
Citation: Huang W, Han S, Wang L and Li W (2022) Carbon and nitrogen metabolic regulation in freshwater plant Ottelia alismoides in response to carbon limitation: A metabolite perspective. Front. Plant Sci. 13:962622. doi: 10.3389/fpls.2022.962622
Received: 06 June 2022; Accepted: 15 August 2022;
Published: 15 September 2022.
Edited by:
Gwenael Piganeau, UMR 7232 Biologie Intégrative des Organismes Marins (BIOM), FranceReviewed by:
Shengqing Shi, Chinese Academy of Forestry, ChinaCopyright © 2022 Huang, Han, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Li, bGl3ZWlAd2JnY2FzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.