- Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs of the PRC, Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, Wuhan, China
Ovate family proteins (OFPs) were firstly identified in tomato as proteins controlling the pear shape of the fruit. Subsequent studies have successively proved that OFPs are a class of negative regulators of plant development, and are involved in the regulation of complex traits in different plants. However, there has been no report about the functions of OFPs in rapeseed growth to date. Here, we identified the OFPs in rapeseed at the genomic level. As a result, a total of 67 members were obtained. We then analyzed the evolution from Arabidopsis thaliana to Brassica napus, illustrated their phylogenetic and syntenic relationships, and compared the gene structure and conserved domains between different copies. We also analyzed their expression patterns in rapeseed, and found significant differences in the expression of different members and in different tissues. Additionally, we performed a GWAS for the number of seeds per silique (NSPS) in a rapeseed population consisting of 204 natural accessions, and identified a new gene BnOFP13_2 significantly associated with NSPS, which was identified as a novel function of OFPs. Haplotype analysis revealed that the accessions with haplotype 3 had a higher NSPS than other accessions, suggesting that BnOFP13_2 is associated with NSPS. Transcript profiling during the five stages of silique development demonstrated that BnOFP13_2 negatively regulates NSPS. These findings provide evidence for functional diversity of OFP gene family and important implications for oilseed rape breeding.
Introduction
Oilseed crops are both an important source of edible vegetable oil and a valuable material of animal feed (Felten et al., 2013; D’Avino et al., 2015). As the second largest source of edible vegetable oil, Brassica napus (B. napus) provides about 13–16% of the total edible vegetable oil for the world (Wang et al., 2018), and the market demand for it is still increasing in recents. Therefore, great efforts have been made to improve its yield. Plant architecture, including main inflorescence length, plant height and branch number, indirectly affects the oilseed rape yield (Reinhardt and Kuhlemeier, 2002; Wang and Li, 2008; Li et al., 2019a), while siliques per plant (SPP), number of seeds per silique (NSPS), and total seed weight (TSW) directly determine the plant yield (Clarke and Simpson, 1978). Similarly, in Arabidopsis, the silique is closely associated with the final yield, and silique-related traits such as silique length (SL) and silique volume (SV) affect the morphology and photosynthetic substances, thus influencing the yield (Ferrándiz et al., 1999). In siliques, NSPS and SL are more important traits. In recent years, many QTLs controlling SL and NSPS have been identified at almost all chromosomes through QTL mapping and genome wide association study (GWAS) (Fu et al., 2015; Wang et al., 2016; Yang et al., 2016, 2017; Luo et al., 2017; Zhu et al., 2020). To date, two genes controlling the SL, BnaA9.ARF18 and BnaA9.CYP78A9, have been cloned in B. napus, which affect SL by regulating cell elongation of the silique wall (Liu et al., 2015a; Shi et al., 2019). Additionally, BnaC9.SMG7B has been cloned as a positive regulator of NSPS, which regulates the formation of normal female gametophyte and finally determines the formation of mature ovules (Li et al., 2015). In recent studies, BnaA08g07940D and BnaA08g07950D were identified as putative candidate genes of a major QTL controlling NSPS by fine mapping (Jiao et al., 2021). Besides, in Brassica juncea, BjCLV1 was found to affect NSPS through the formation of trilocular siliques (Wang et al., 2021). In Arabidopsis, cytokinin and brassinosteroid were found work coordinately to promote ovule initiation and then increase NSPS (Zu et al., 2022). Certainly, the yield is also affected by diseases such as Sclerotinia stem rot, clubroot, blackleg disease and stem canker. Fortunately, many resistance QTLs have been identified. Therefore, integration of these elite QTL alleles controlling different traits into elite cultivars with better plant architecture may be a promising strategy to improve the yield of oilseed rape.
Domestication of fruit-bearing crops involves long-term artificial selection from various wild plant species, and a significant hallmark in this process is the explosive increase in fruit shape variations (Williams, 1965; Hopping et al., 1986). The recessive locus ovate controlling the pear shape of fruit and elongated fruit shape in tomato was identified about one hundred years ago, but the gene was cloned until 20 years ago (Price and Drinkard, 1908; Liu et al., 2002). The ovate proteins were identified as a class of negative regulators in plant development, which contain a C-terminal conserved domain and Von Willebrand factor type C domain, which are conserved in tomato, rice and Arabidopsis (Liu et al., 2002; Wang et al., 2011). Subsequently, studies of plant ovate family proteins (OFPs) have been successively reported. Researchers are increasingly aware of their functions though the exact mechanism remains poorly understood. OFPs play important roles in plant growth and development, and their functions have been studied in both model plants and crops. In Arabidopsis, AtOFP5 affects the cell-fate switch of synergid to egg cell in mature embryo sacs by suppressing the ectopic activity of BELL-KNOX TALE complex (Pagnussat et al., 2007); AtOFP4 and AtOFP1 are involved in secondary cell wall formation; and the Atofp4 mutant exhibited thicker interfascicular fiber cell wall and thinner xylary fiber cell wall (Li et al., 2011; Wang et al., 2011). AtOFP1 interacts with ATH1 to regulate flowering time and stem growth in Arabidopsis (Zhang et al., 2018). Besides, AtOFP1 is involved in the development of male gamete and pollen activity as well as DNA repair (Hackbusch et al., 2005; Wang et al., 2010). In rice (Oryza sativa), overexpression of OsOFP2 led to a series of variations in plant height, leaf morphology, seed shape and abnormity of vascular bundles in stems; OsOFP2 suppresses the expression of GA20ox by modulating the function of KNOX-BELL and inhibits lignin biosynthesis, thereby affecting vasculature development (Schmitz et al., 2015). In radish (Raphanus sativus), RsOFP2.3 is negatively associated with tuberous root elongation and the tuberous root shape (Wang et al., 2020). In peach (Prunus persica), PpOFP1 regulates fruit shape (Cirilli and Rossini, 2021), and another similar study showed that PpOFP1 physically interacts with a ZF-HD_dimer domain protein PpZFHD1 and regulates the salt tolerance of tomato (Tan et al., 2021). In cotton (Gossypium hirsutum), GhOFP4 was found to regulate fiber development (Gong et al., 2014). In Capsicum annuum, CaOFP1 is involved in fruit shaping, and its different expression profiles would result in different shapes via negatively affecting the expression of CaGA20ox1 (Tsaballa et al., 2011), and gene silencing of CaOFP20 increased the fruit length (Borovsky et al., 2022). Ectopic expression of CsOFP12-16c from cucumber (Cucumis sativus) in Arabidopsis affects the silique development and causes blunt and shorter siliques (Han et al., 2022). Previous studies have also suggested that CmOFP13 may control the fruit shape in melon (Cucumis melo) (Monforte et al., 2014; Ma et al., 2021). Also, the expression of MaOFP1 was reported to be negatively associated with fruit ripening in banana (Musa paradisiaca) (Liu et al., 2015b). Although more and more functions of OFPs have been reported in many crops, there are still numerous unknown features remaining to be discovered.
B. napus (2n = 38, AACC) is a polyphyletic polyploidy formed by B. oleracea and B. rapa (Nagaharu, 1935; Allender and King, 2010), and has experienced whole genome duplication (WGD) (Jiao et al., 2011; Chalhoub et al., 2014). Brassica napus napus still retains two sets of chromosomes corresponding to B. oleracea and B. rapa (Chalhoub et al., 2014). Hence, there are many duplicate genes from two sub-genomes or even the earlier progenitor Arabidopsis. Generally, Arabidopsis can serve as an efficient model plant in functional gene research. AtOFPs have been found to have different functions. However, multiple copies of OFP genes have been rarely reported in its closely related species B. napus. In this study, we identified the BnOFP gene family at the genomic level, analyzed its evolution from Arabidopsis to B. napus and compared the difference between copies. A new locus BnOFP13_2 significantly correlated with NSPS was identified, which can be considered as a novel function of OFPs. RNA profiling during silique development suggested that BnOFP13_2 negatively regulates NSPS. Our findings provide evidence for the functional diversity of OFP gene family and important implications for oilseed rape breeding.
Results
Identification and chromosomal distribution of ovate family proteins gene family in Brassica napus
By using the reported 20 AtOFP or AtOFP-like protein sequences as queries, a total of 67 OFP genes were identified through BLAST in B. napus and Arabidopsis databases, which were renamed according to their orthologous genes in Arabidopsis, and their physical and chemical properties were analyzed (Supplementary Table 1). Among the 20 AtOFP genes, AtOFP6 and AtOFP9 had no orthologous gene in B. napus; AtOFP1 had only one orthologous gene BnOFP1_1; while AtOFP2, AtOFP3 and AtOFP5 all had six orthologous genes. The other 13 AtOFP genes, respectively, had two to five orthologous genes in B. napus (Supplementary Table 1). The 67 BnOFP genes were unevenly distributed on the 20 chromosomes of A sub-genome (34 BnOFP genes) and C sub-genome (33 BnOFP genes) (Figure 1). In the A sub-genome, A08 chromosome had no BnOFP gene; chromosome A06 contained one BnOFP gene; and A02, A09, and A10 chromosomes, respectively, harbored five BnOFP genes. On the same chromosome, some BnOFP genes were closely located, while some other genes were far away from each other. For instance, BnOFP17_3 and BnOFP2_5 were closely located on chromosome A04, while BnOFP7_4 and BnOFP14_4 were located on both ends of chromosome A07. In the C sub-genome, chromosome C06 contained no BnOFP member, and eight BnOFP members were located on unknown chromosomes, while other chromosomes, respectively, harbored two to five members (Figure 1). These results suggested the occurrence of genome rearrangement and gene loss during polyploidization.
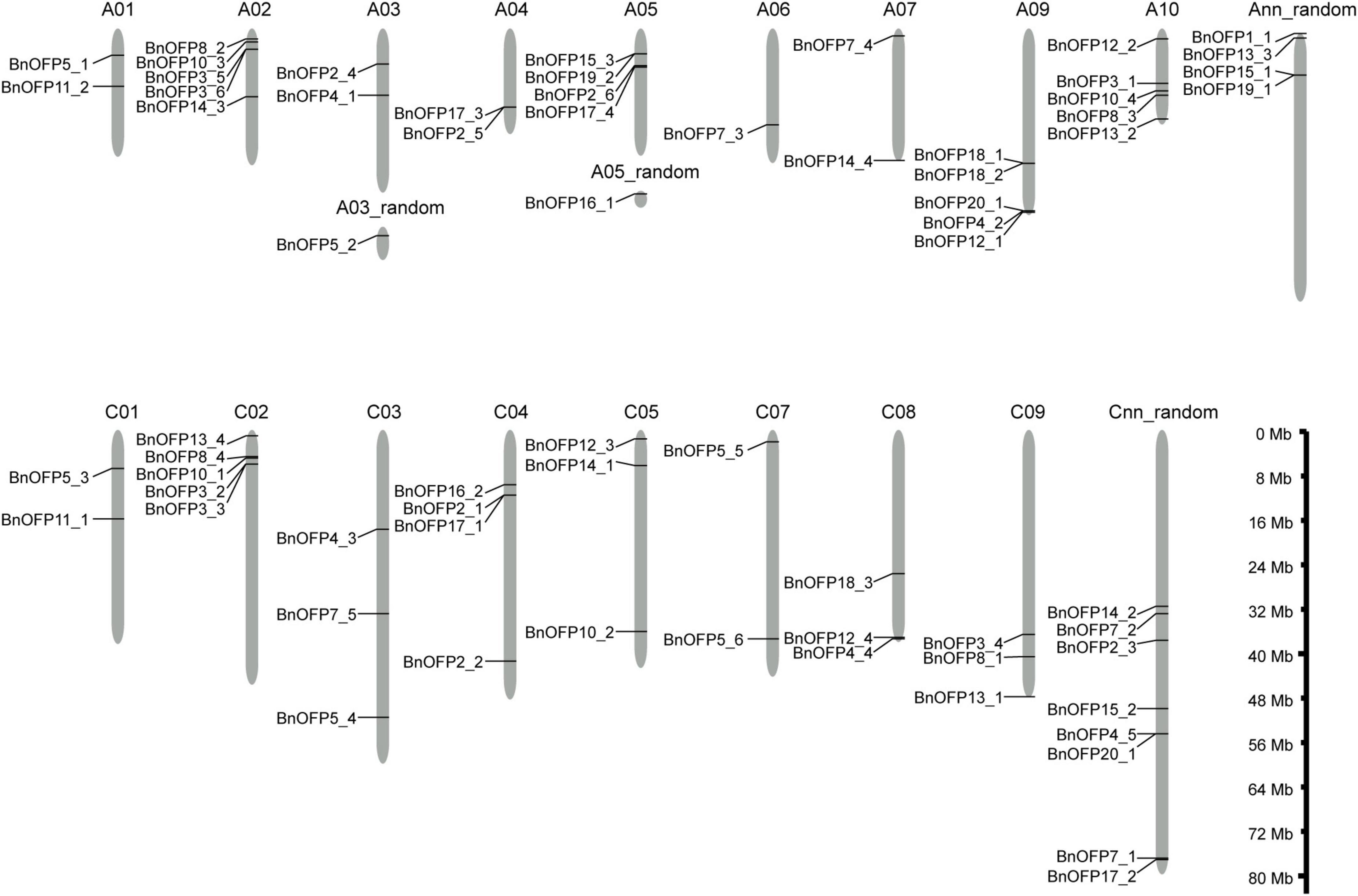
Figure 1. Chromosomal locations of BnOFPs. The 67 BnOFP genes were located on different chromosomes of two sub-genomes. Random chromosome means that the sequence was mapped to corresponding chromosome but the physical location was unknown. Length of chromosomes is also shown in the figure.
Phylogenetic and syntenic relationship of ovate family proteins gene family in Brassica napus
Phylogenetic relationship is an important indicator for gene functional research. The 87 OFP proteins from B. napus and Arabidopsis were used to construct a phylogenetic tree. As a result, five groups were clustered based on the sequence alignment (Figure 2 and Supplementary Table 2). Group 2 was the largest clade (24 proteins), followed by group 5 (19 proteins), group 4 (17 proteins), group 3 (16 proteins), and group 1 (11 proteins). Among the five groups, group 2 and group 3 contained some AtOFPs and all their corresponding B. napus orthologous genes. For example, group 2 included AtOFP5, AtOFP11, AtOFP12, AtOFP15, AtOFP16, AtOFP18 and BnOFP5, BnOFP11, BnOFP12, BnOFP15, BnOFP16, and BnOFP18. On the contrary, some AtOFPs and their orthologous BnOFPs were not clustered together. For example, AtOFP10 was clustered in group 1, but the BnOFP10 orthologs were clustered in group 4 or group 5. Genes with similar functions tended to be clustered in the same group. For instance, AtOFP1 and AtOFP4 were both involved in secondary cell wall formation and thus clustered in group 3 (Li et al., 2011; Wang et al., 2011); AtOFP17 and AtOFP20 were both paralogous genes from segmental duplication blocks, and were also clustered in the same group (Liu et al., 2014; Figure 2). During the polyploidization from Arabidopsis to B. napus, members in different groups had undergone loss-of-function and function divergence. Therefore, the BnOFP gene family members may participate in different biological processes in plant development.
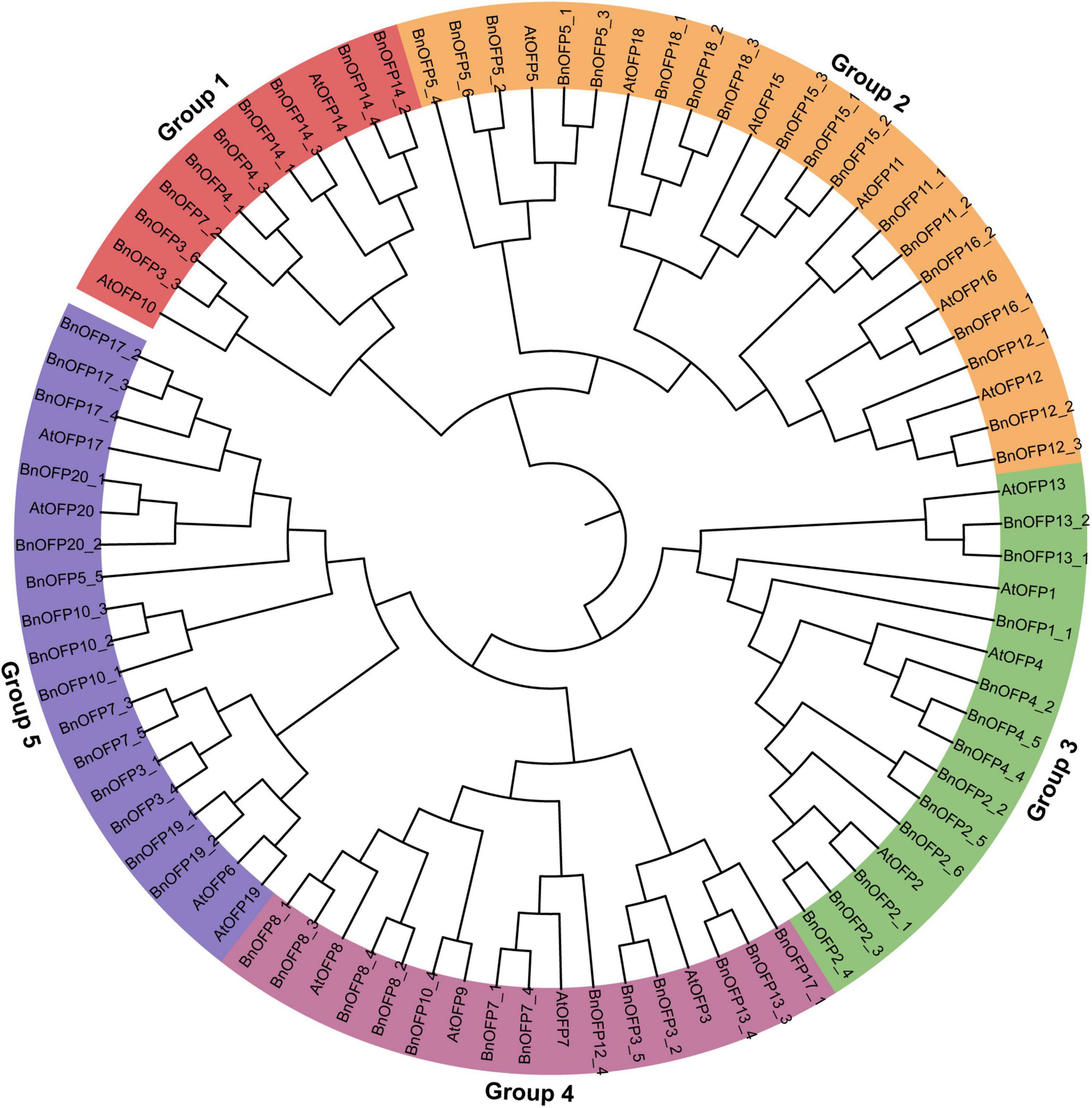
Figure 2. Phylogenetic relationship analysis of ovate family proteins (OFPs) in Arabidopsis and Brassica napus. The 20 AtOFP genes and 67 BnOFP genes were clustered into five groups. Members of each group were represented by different background color.
Since B. napus is of polyphyletic polyploidy with syntenic relationship between the sub-genomes, we analyzed the syntenic relationship between the BnOFPs in A and C sub-genomes (Figure 3). We analyzed the 53 BnOFPs with specific chromosomal locations, and finally identified 19 pairs of syntenic genes. No syntenic gene was detected on chromosome A07 and A08 as well as on C06 and C07, while other chromosomes contained one to five syntenic genes. Chromosome A10 included the most syntenic genes (five genes), with two on chromosome C05 and three on C09. Pairs of syntenic genes were in the same subfamily, such as BnOFP7_3 and BnOFP7_5, and BnOFP2_2 and BnOFP2_5 (Figure 3). Gene phylogenetic and syntenic relationship can be used to explore the functions of unknown genes. Hence, our results may be of great significance for gene functional research in B. napus.
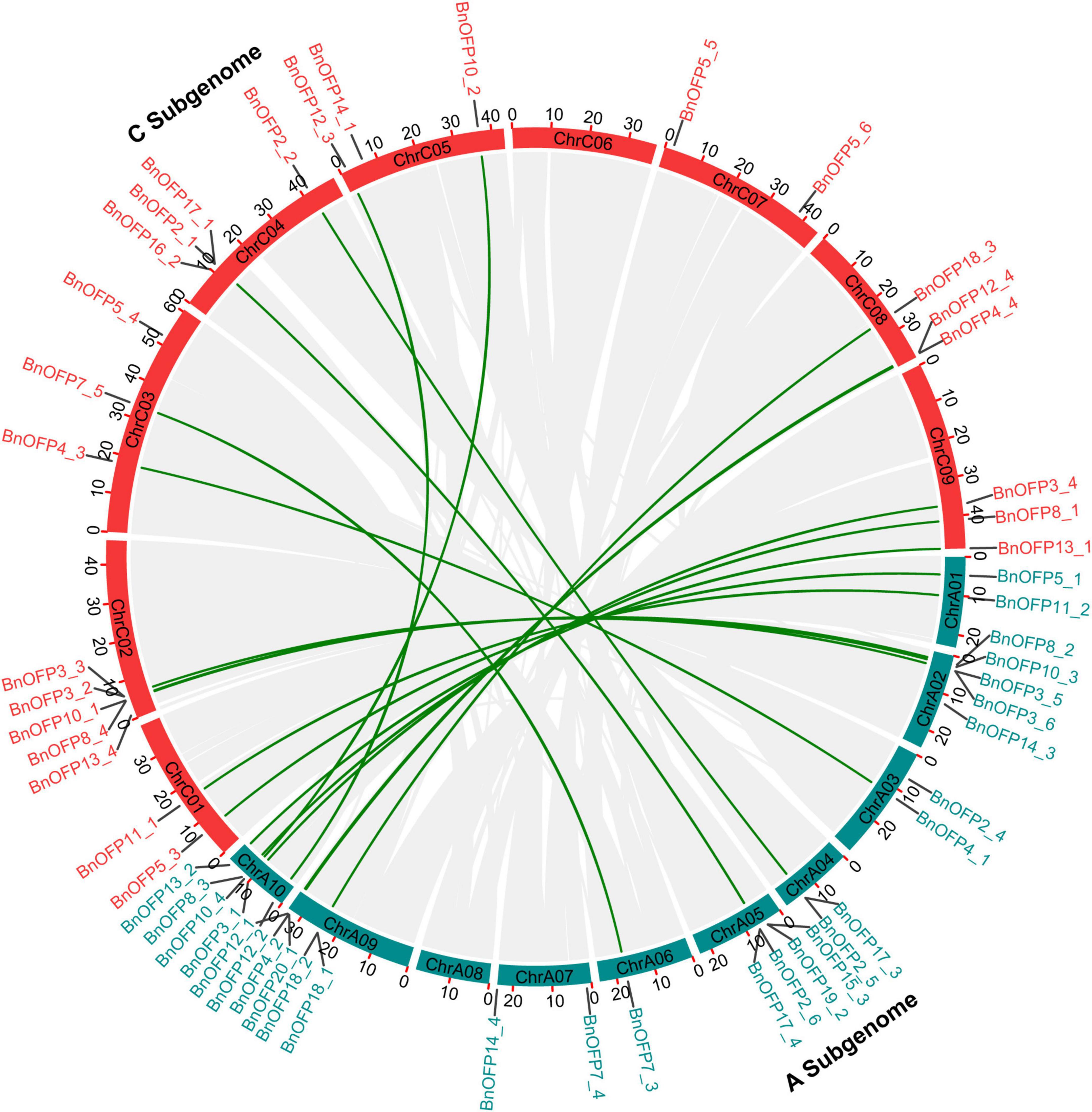
Figure 3. Syntenic relationship between Brassica napus A and C sub-genome. Chromosomes including BnOFP members of two sub-genomes are shown in different color. Gray lines in circle represent the syntenic genes or blocks, and dark green lines represent the syntenic genes in BnOFPs.
Gene structure and conserved domains of BnOFP genes
Gene structure was analyzed to investigate whether there are differences among the 67 BnOFP genes. As expected, large differences were found in gene length and exon number among different members. The gene structure was displayed according to the five clades in the phylogenetic tree (Figure 4A). The gene length ranged from 153 to 4,195 base pairs (bp), and most genes were less than 1,000 bp. The exon number ranged from one to six, and 70.15% (47 out of 67) of the genes had only one exon (Figure 4B). Some genes displayed unique structures. For instance, only seven genes had untranslated region (UTR); BnOFP15_3 and BnOFP5_3 had upstream UTR, BnOFP13_1, BnOFP16_1, BnOFP16_2, and BnOFP2_3 had downstream UTR; while BnOFP19_2 had both upstream and downstream UTR (Figure 3B). This gene family had fewer introns, as well as great differences in intron length. BnOFP17_1, BnOFP13_4, BnOFP13_3, BnOFP5_2, and BnOFP7_2 contained longer introns, and BnOFP5_5 and BnOFP3_2 had shorter introns. Some members derived from the same Arabidopsis gene showed the same gene length and structure, such as the BnOFP8 and BnOFP18 subfamily (Figure 4B).
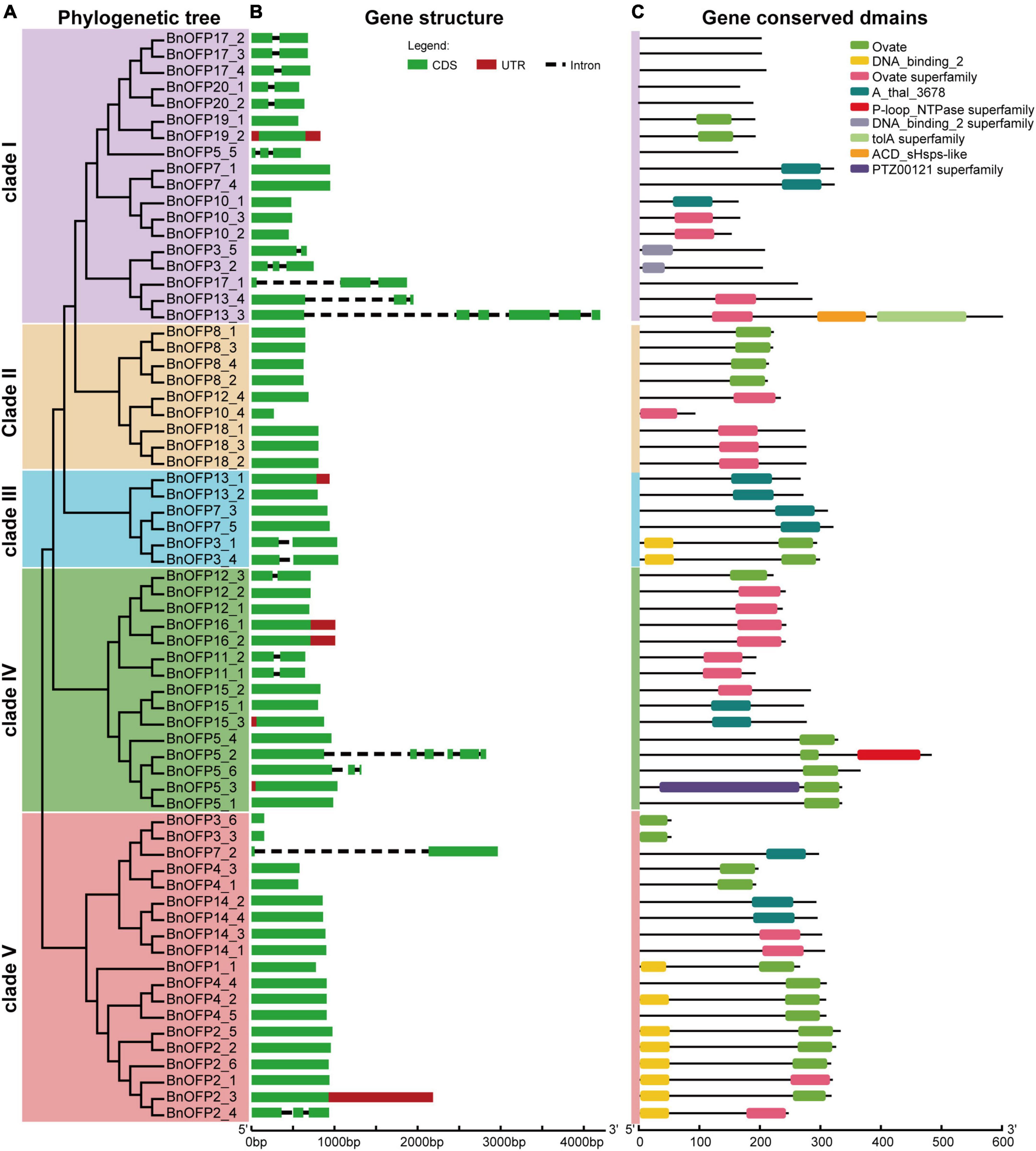
Figure 4. Gene structure and conserved domains of BnOFPs. (A) Phylogenetic tree of BnOFP genes. The 67 BnOFP genes were classified into five clades, each of which is illustrated with different colors. (B) Gene structure of the 67 BnOFP genes. Boxes represent the CDS and UTR section, and the lines indicate introns. (C) Gene conserved domains of the 67 BnOFP genes. Nine conserved domains are represented in different colored boxes, and the line means non-conserved sequence. Length scale is presented at the bottom.
We also analyzed the gene conserved domains, and identified a total of nine conserved domains. Ovate and ovate superfamily domain were detected in 68.66% of the BnOFP members (Figure 4C). Most members contained one domain, and a few members harbored two or three domains. Some domains were rarely detected, such as the P-loop_NTPase superfamily and PTZ00121 superfamily domains, which were only detected in BnOFP5_2 and BnOFP5_3, respectively. However, 31.34% members had no ovate family domains, and seven members in clade I had no conserved domain. This loss of domain may be caused by genome polyploidization. These results may greatly help the research on the functional conservation and divergence of BnOPFs in B. napus evolution.
Differential expression of BnOFPs in different tissues
With the rapid development of RNA sequencing technology, transcriptome analysis has been widely used in gene functional research. The expression level of a gene is related to its functional performance. The FPKM (fragments per kilobase of exon model per million mapped fragments) value from RNA sequencing can effectively represent the gene expression levels and be used to analyze the biological processes in plant tissues in different stages and environments. In this study, a transcriptome analysis of twelve tissues in B. napus cultivar line ‘ZS11’ was performed to analyze the expression pattern of BnOFPs. We found that the expression of 67 BnOFPs varied greatly among the twelve tissues (Figure 5A and Supplementary Table 3). The majority of the genes had generally low expression (Supplementary Table 3). BnOFP5_1, BnOFP5_6, BnOFP10_2, BnOFP10_4, BnOFP12_2, and BnOFP12_3 had relatively higher expression in the sepal, while BnOFP14_2, BnOFP16_2, BnOFP7_1, BnOFP18_1, BnOFP14_3, BnOFP15_2, and BnOFP15_3 exhibited relatively higher expression in the bud. BnOFPs were lowly expressed in most tissues. For example, all the genes had low expression in the stem, while in the leaf and silique, only BnOFP17_7 and BnOFP2_1 had relatively high expression (Figure 5A). Ten genes showed no expression in any tissue, probably because they have lost their functions during evolution. A qRT-PCR experiment was then performed to verify the transcriptome data, and the results of five genes in four tissues are presented in Figure 5B. The expression of BnOFP5_4, BnOFP5_6, and BnOFP16_1 was obviously higher in the sepal and lower in the new pistil, while that of BnOFP19_2, BnOFP5_4, and BnOFP5_6 was obviously low in the bud and stem (Figure 5B). The qRT-PCR experimental results were consistent with the transcriptome results, verifying the reliability of the transcriptome data. The expression pattern of BnOFPs suggested the occurrence of functional divergence in this gene family of B. napus during evolution.
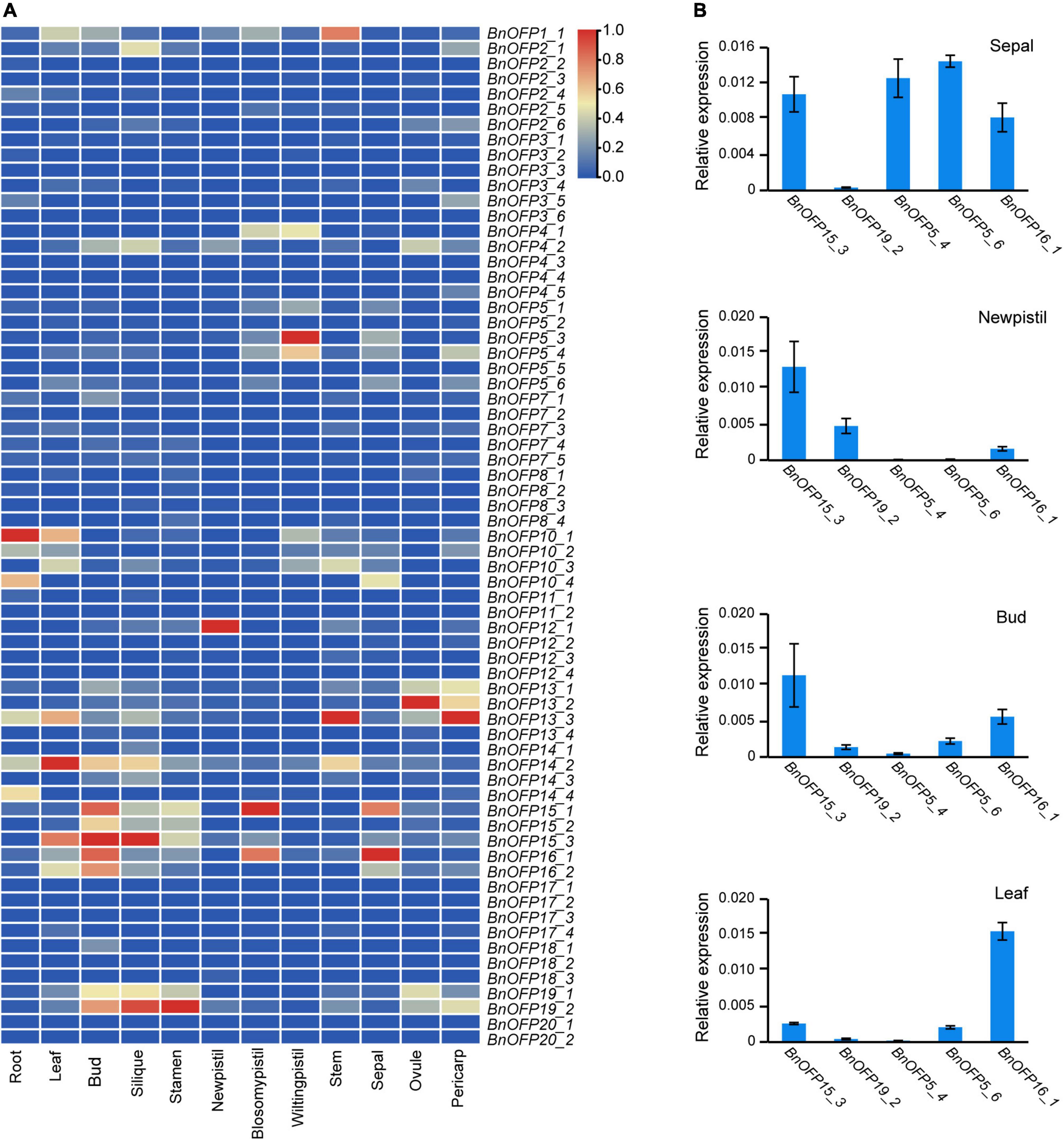
Figure 5. Expression patterns of BnOFP genes. (A) Heatmap of gene expression patterns. Expression levels of 67 BnOFP genes in twelve tissues of ‘ZS11’ obtained from FPKM of transcriptome analysis. (B) Verification of the expression level by qRT-PCR. Five genes and four tissues were used to verify the expression level in transcriptome analysis by qRT-PCR.
Association of BnOFP13_2 with number of seeds per silique
Genome wide association study is a new approach to precisely predict the corresponding genes or QTLs involved in the regulation of complex traits of plants based on linkage disequilibrium (LD) (Nordborg and Weigel, 2008). In this study, GWAS for NSPS was performed with a natural population consisting of 204 accessions. The phenotype value ranged from 7.06 to 24.68, exhibiting large differences among different accessions (Figure 6B). Finally, a significant locus was identified on chromosome A10. The 16.39–16.47 Mb block on A10 contained a BnOFP member, and the most significant SNP was located around BnOFP13_2 (Figure 6A and Supplementary Table 4). Therefore, we speculated that BnOFP13_2 might be associated with NSPS in B. napus, and then analyzed the SNP of BnOFP13_2 in the 204 accessions. Two homozygous non-synonymous SNPs were obtained at the position of +40 and +604 bp of gene sequence. There were three classified haplotypes based on the two SNPs. Haplotype 1 comprised 14 accessions, and haplotype 2 and haplotype 3, respectively, had 141 and 23 accessions (Figure 6C). The accessions in haplotype 1, haplotype 2, and haplotype 3 had the average phenotype value of 16.50, 16.67, and 21.11, respectively, indicating that haplotype 3 had significantly higher NSPS than haplotype 1 and haplotype 2 (Figure 6D). These results suggested that BnOFP13_2 is associated with NSPS.
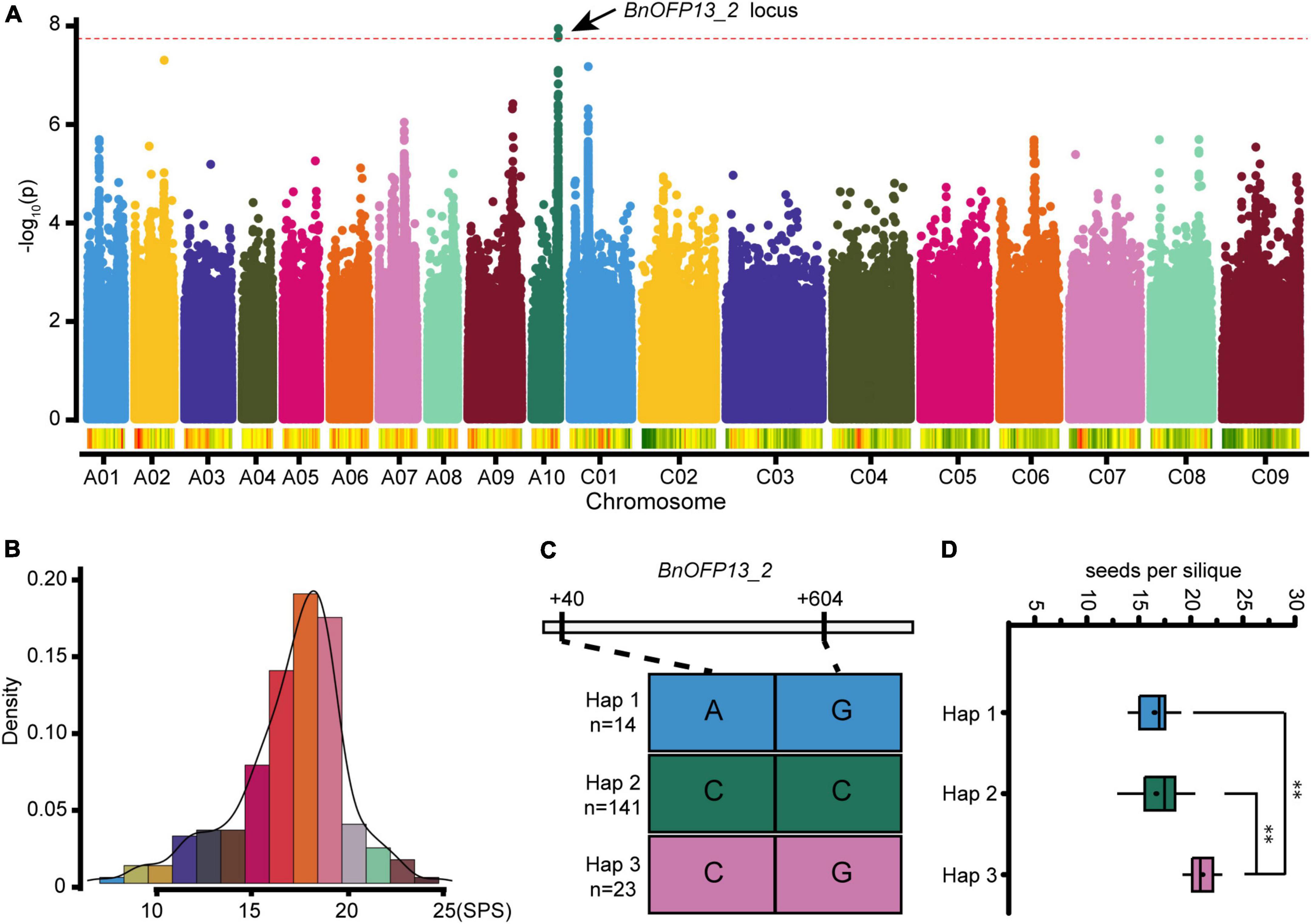
Figure 6. Genome wide association study (GWAS) and haplotype analysis. (A) Manhattan map of GWAS. The red dotted line represents the threshold value 7.72. Dots above the red dotted line indicate the significant loci associated with NSPS. (B) Histogram of frequency distribution. Statistical analysis of NSPS phenotype used in GWAS. Phenotype variations of 204 accessions are presented. (C) Haplotype analysis of BnOFP13_2. All the accessions were classified into three haplotypes based on the two non-synonymous SNPs of BnOFP13_2. (D) Haplotype analysis. Statistical analysis of accessions in each haplotype, and the phenotype value shows significant differences. **p < 0.01 in ANOVA test.
Negative regulation of BnOFP13_2 on number of seeds per silique in early silique development
To investigate how BnOFP13_2 affects NSPS in B. napus, we detected the RNA profiles of BnOFP13_2 at different silique developmental stages in two varieties, respectively, with high NSPS (‘ZC520’) and low NSPS (‘ZC519’). ‘ZC520’ averagely had 24 seeds per silique and ‘ZC519’ only had 18 seeds per silique (Figures 7A,B). The siliques at 7 days after pollination (7 DAP), 14, 21, 28, and 35 DAP were collected to analyze the dynamics of the expression level of BnOFP13_2 (Figure 7C). At 7 and 14 DAP, the expression of BnOFP13_2 was significantly higher in ‘ZC520’ than in ‘ZC519’ (Figure 7D). From 21 to 35 DAP, the expression level of BnOFP13_2 was constantly high without significant difference between the two varieties (Figure 7D). In ‘ZC520’ (high-NSPS variety), the expression of BnOFP13_2 was low in the early developmental stage of silique, which then increased slowly until 21 DAP and remained constant until maturity; while in ‘ZC519’ (low-NSPS variety), BnOFP13_2 showed a high expression level at almost all developmental stages (Figure 7D). These results suggested that the accumulation of BnOFP13_2 transcripts at the early developmental stage of silique is negatively correlated with NSPS. Therefore, we speculated that BnOFP13_2 suppresses the formation or development of ovule, which is similar to the previous finding in Arabidopsis, but the specific molecular mechanism remains to be further studied.
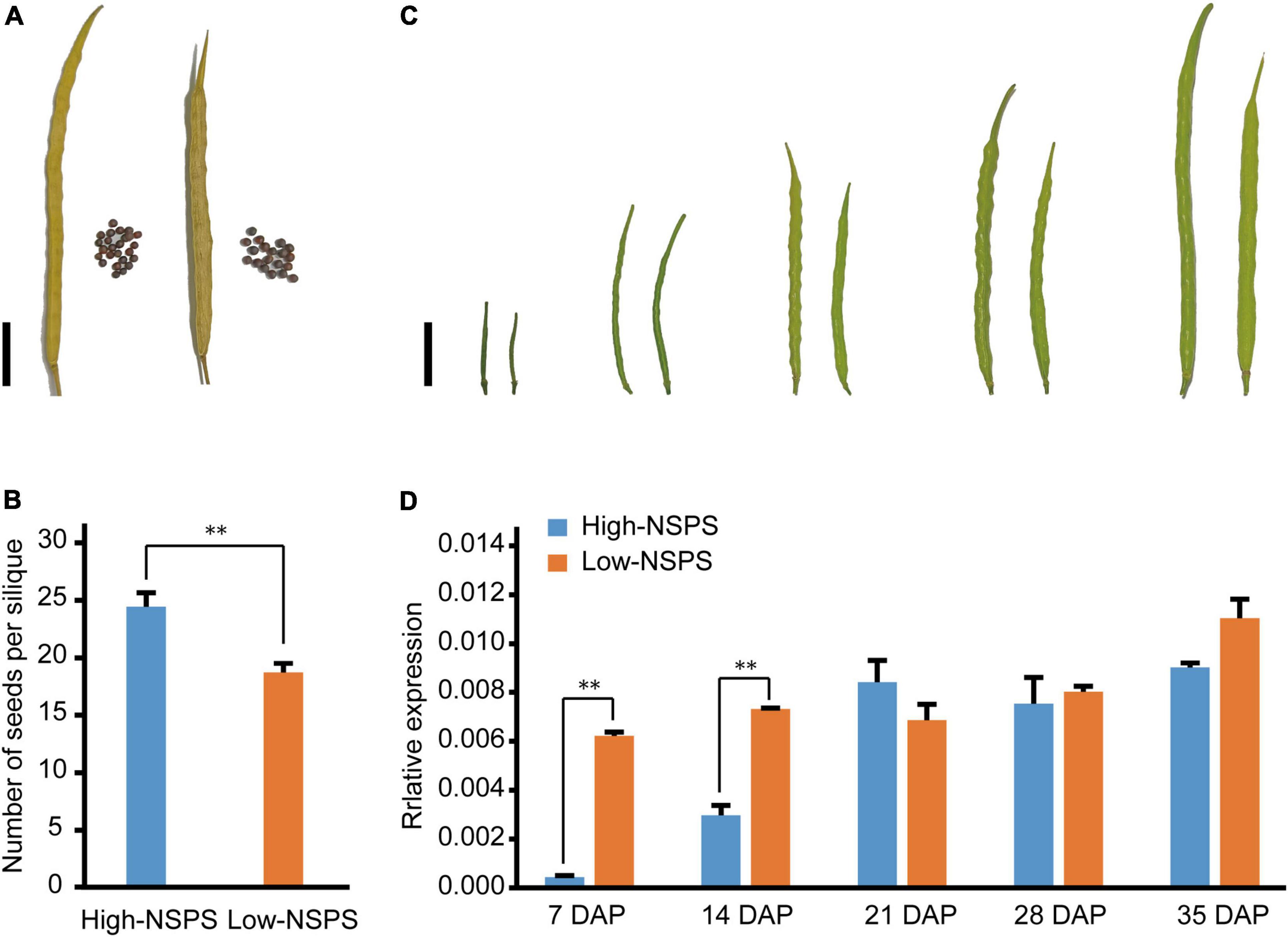
Figure 7. Phenotype of siliques at different developmental stages and expression levels of BnOFP13_2 in high-NSPS variety and low-NSPS variety. (A) Mature silique of high-NSPS variety (left) and low-NSPS variety (right), bar: 2 cm. (B) Statistical analysis of NSPS in high-NSPS variety and low-NSPS variety. (C) Phenotype of high-NSPS variety and low-NSPS variety siliques at 7, 14, 21, 28, and 35 DAP, bar: 2 cm. (D) Relative expression levels of BnOFP13_2 in high-NSPS variety and low-NSPS variety silique at different developmental stages. **p < 0.01.
Discussion
More than 20 years ago, OVATE was a well-known QTL for its effect in controlling the pear shape of tomato fruit, which was then mapped and cloned on chromosome 2 (Ku et al., 2001; Liu et al., 2002). The protein and RNA profiles have suggested that OVATE is a novel class of proteins named as OFPs (ovate family proteins) (Liu et al., 2002). Owing to their effects on fruits, OFPs have been studied in many fruit crops, and the results have indicated that OFPs have great influence on a variety of aspects in plants, including fruit shape (Liu et al., 2002; Tsaballa et al., 2011; Cirilli and Rossini, 2021; Han et al., 2022), tuberous root shape (Wang et al., 2020), ovule development (Pagnussat et al., 2007), secondary cell wall formation (Li et al., 2011; Wang et al., 2011), vasculature development (Schmitz et al., 2015), fruit ripening (Liu et al., 2015b), DNA repair (Wang et al., 2010), floral organs, compound leaf and silique (Liu et al., 2002; Wang et al., 2011). Here, we briefly summarized the functions of OFPs in Arabidopsis and some common crops (Figure 8A), and we believe that more functions of OFPs will be gradually detected in plants. There has been no report about the functions of OFPs in B. napus. In this study, 67 BnOFP members were checked with gene ontology (GO) annotations (Figure 8B). As a result, 63 members were involved in biological regulation, cellular process, metabolic process, negative regulation of biological process and regulation of biological process; 52 members were related to cell part and organelle component; and 12 members had the binding molecular function (Figure 8B). These results suggested that BnOFPs play different roles in plant development. We further identified and characterized the OFPs in B. napus, as well as analyzed their phylogenetic relationship with the orthologs in Arabidopsis and the syntenic relationship between the two sub-genomes. We also analyzed the gene structure, conserved domain and expression pattern, and the differences in the gene structure and expression could provide a theoretical basis for functional research of this gene family. In addition, a novel function of OFPs was identified through GWAS, and haplotype analysis suggested that BnOFP13_2 is significantly associated with NSPS. Further expression profiling at the silique developmental stage indicated that BnOFP13_2 negatively regulates NSPS during early silique development. Our results may provide important information for rapeseed breeding.
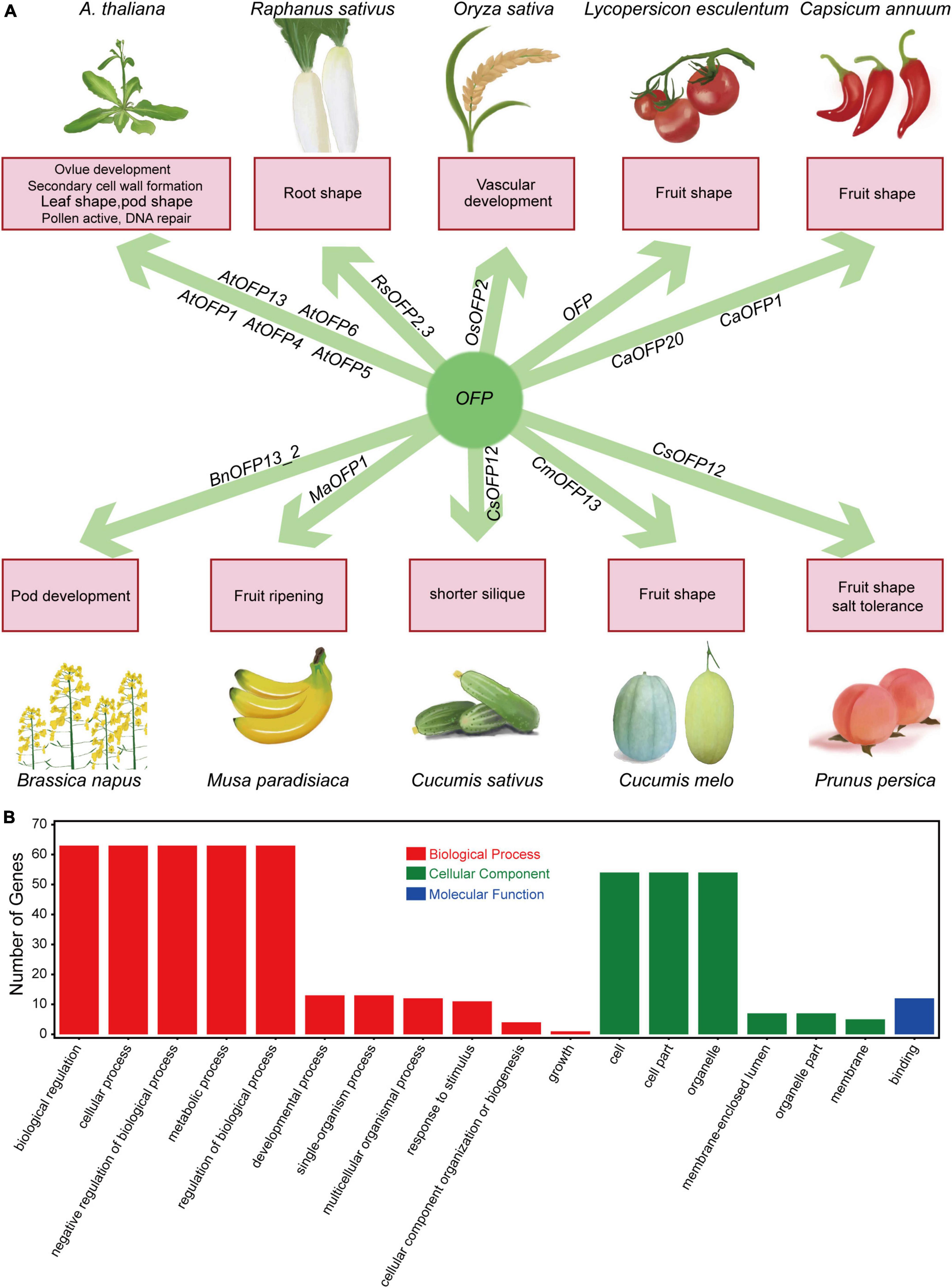
Figure 8. Brief summary of OFP functions and GO annotations of the 67 BnOFP genes. (A) Function summary of OFPs in common crops. The crop pictures are painted. (B) GO annotations of OFP genes in B. napus. Gene numbers and annotations in biological processes, cellular components and molecular functions are shown in the figure.
In Arabidopsis, AtOFP1 was firstly identified as a transcriptional repressor of cell elongation in many organs. An increase in the expression of AtOFP1 reduced the length of hypocotyl, inflorescence stem, cauline leaf, rosette leaf, floral organ, and silique (Wang et al., 2007). Overexpression of AtOFP2, AtOFP4, and AtOFP7 led to similar phenotypes to overexpression of AtOFP1 such as round and curled leaves, implying that these three genes are also transcriptional repressors of cell elongation (Wang et al., 2007; Li et al., 2011). Overexpression of AtOFP6 and AtOFP8 resulted in thick and cyan rosette leaves in plants, while that of AtOFP13, AtOFP15, and AtOFP16 led to short and blunt-ended siliques (Wang et al., 2011). In this study, high expression of BnOFP13_2 was detected at early developmental stage of siliques in low-NSPS variety. Therefore, it can be speculated that BnOFP13_2 negatively regulates NSPS by reducing silique length and inhibiting ovule development at early developmental stage.
Since OFPs are negative regulators of plant growth and development, the underlying mechanisms have received great attention. Although the mechanisms remain largely unknown, there has been some evidence implying that OFPs may directly regulate the expression of the target genes. In Arabidopsis, AtOFP1 contains a putative NLS domain and is located in the nucleus, and a yeast one-hybrid experiment indicated that AtOFP1 represses the expression of the reporter gene. These results suggest that AtOFP1 is a transcription repressor directly regulating gene expression (Wang et al., 2007, 2011). In addition, AtOFP1 also contains the LxLxL domain, which was also found in ERF transcription factors and is necessary for transcriptional repression (Hiratsu et al., 2003). On the other hand, OFPs were found to form a complex to perform their functions via interacting with other proteins. For instance, AtOFP4 and AtOFP1 interact with KNAT7 and BLH6 to regulate secondary cell wall formation (Li et al., 2011); AtOFP1 coordinates with ATH1 to affect flowering time and stem growth (Zhang et al., 2018); AtOFP1 has interaction with BLH3 to regulate the transition from vegetative phase to reproductive phase (Zhang et al., 2016); AtOFP5 cooperates with KANT2 and BLH1 in regulating female gametophyte development (Pagnussat et al., 2007); MaOFP1 interacts with MuMADS1 to regulate fruit ripening (Liu et al., 2015b); GhOFP4 coordinates with GhKNL1 in mediating secondary cell wall formation (Gong et al., 2014); and OsOFP2 interacts with OsKNAT7 and BLH6-like1 to mediate vascular development (Schmitz et al., 2015). With a better understanding of these negative regulators, OFPs can be better used for crop improvement via the gene editing technology.
In plants, polyploidization occurs at high frequency. It has been demonstrated that about 25–30% of the existing flowering plants are polyploids and have not been diploidized yet (Wood et al., 2009; Scarpino et al., 2014). Polyploids are generally classified into autopolyploids and allopolyploids, with the former being formed through doubling of one chromosome set, while the latter being formed via hybridization or merging of different chromosome sets in different species (Ramsey and Schemske, 1998; Barker et al., 2016; Van de Peer et al., 2017). Allopolyploidy is involved in epigenetic remodeling and changes in gene expression, which will contribute to a higher adaptive potential (Yoo et al., 2014; Hu et al., 2015b; Van de Peer et al., 2017). It is widely accepted that polyploids have increased the mutational robustness and adaptability compared with diploids. Polyploids can overcome sympatrically speciate and cytotype minority exclusion for rapid adaptation, which is also owing to their higher genomic plasticity than diploids (te Beest et al., 2012; Schoenfelder and Fox, 2015). Gene duplication generally has four modes, including the tetraploid, segmental, tandem, and transpositional modes, with each mode retaining genes in a biased method (Neufeld et al., 1991; Freeling, 2009). After genome or segment duplication, some duplicated genes are often lost (Freeling, 2009). Polyploidization or WGD is an important driving force for evolution in animals and plants (Abdel-haleem, 2007; Edger and Pires, 2009), and the success of angiosperms is partially attributed to WGD (De Bodt et al., 2005). In Arabidopsis, OFPs are involved in different functions. After polyploidization into B. napus, the OFP members have experienced gene doubling, loss and functional divergence, which may lead to the appearance of novel gene structure, conserved domains and functions. In this study, seven BnOFP members were detected to have no ovate or related domains (Figure 4C), which might have been caused by loss-of-function during polyploidization.
Conclusion
In this study, we identified 67 BnOFP genes at the genome level, analyzed their evolution from Arabidopsis to B. napus and compared the gene structure and conserved domains between copies. We also identified a new potential locus significantly correlated with NSPS, which is a novel function of OFPs. RNA profiling in different stages of silique development suggested that BnOFP13_2 negatively regulates the NSPS by decreasing the silique length and inhibiting the ovule development at early developmental stage. Our findings provide evidence for the functional diversity of OFP gene family and new implications for oilseed rape breeding.
Materials and methods
Plant materials and phenotype identification
The 204 natural accessions and ‘ZS11’ used for GWAS analysis and qRT-PCR experiment were obtained from the Key Laboratory of Biology and Genetic Improvement of Oil Crops at OCRI and planted in our experimental field in WuHan, HuBei province, China. These materials used for GWAS were collected from all over the world, including 52 spring accessions, 40 winter accessions and 112 semi-winter accessions, and were cultivated under natural growing conditions. The phenotype data were collected in 1 year (2017–2018). Planting was conducted with twelve plants in a row and a row spacing of 30 cm. All varieties were self-bred for many generations and were pure inbred lines. Ten siliques from different parts of the main inflorescence were collected to evaluate the NSPS. The phenotype of each variety was represented by the average of three biological replicates.
Identification of BnOFPs in Brassica napus
The OFP gene protein sequences of Arabidopsis were obtained from the TAIR database1, and used to search for the OFP genes in B. napus. The annotation and genome information of B. napus cultivar ‘Darmor-bzh’ and corresponding orthologous genes in Arabidopsis were obtained from the BRAD (Brassicaceae Database) database2 (Chalhoub et al., 2014). The HMMER3.03 was used to search for OFP genes in B. napus (E value was set to 1e-5).
Gene structure and conserved domain analysis
Gene structure files of BnOFPs were downloaded from EnsemblPlants database4, and illustrated through GSDS2.0 (Gene Structure Display Server) online tools5 (Hu et al., 2015a). Gene conserved domains were analyzed through CD-search in NCBI (National Center for Biotechnology Information) database6 using the amino acid sequence, and illustrated through TBtools (Chen et al., 2020).
Phylogenetic and syntenic relationship analysis
The protein sequences of OFPs in Arabidopsis and B. napus were used to construct a phylogenetic tree using the MEGA software. The beautification of phylogenetic tree was conducted with iTOL online tools.7 The syntenic relationship of gene or block between two sub-genomes were obtained from the BRAD database (Chalhoub et al., 2014). The syntenic relationship figure was drawn through TBtools (Chen et al., 2020).
Transcriptome analysis and qRT-PCR analysis
The transcriptome data of twelve tissues in ‘ZS11’ used for the expression pattern analysis were obtained from our lab and have been already published (SRA accession: PRJNA474576) (Li et al., 2019b). The heatmap was drawn through TBtools (Chen et al., 2020). The sepal, leaf, bud, and newpistil samples used for RNA extraction were collected in experimental field. The silique samples were collected at 7, 14, 21, 28, and 35 DAP. All the samples had three biological replicates. The TRIzol reagent (Invitrogen, Carlsbad, CA, United States) was used for total RNA extraction. About 2 μg RNA was used to reverse transcribe using the PrimeScript™ RT reagent Kit (TaKaRa Co., Ltd., Beijing, China). The B. napus gene BnaA10g22340D was used as the reference gene. The relative expression was evaluated using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Genome wide association study analysis in natural population
The genomic DNA of all plants was extracted from tender rosette leaves using the modified CTAB method (Allen et al., 2006). The SNP data were obtained from 7 × re-sequencing data mapped to the reference genome ‘Darmor-bzh’. Re-sequencing was performed by the commercial Illumina HiSeq XTen service (BGI-Shenzhen, China). The SNPs were finalized under the minor allele frequency <0.05, and 2611513 valid SNPs were obtained at last, the original SNPs were obtained from published data of our lab (Ding et al., 2020). GWAS for NSPS was performed with the R package using the general linear model (Yin et al., 2021). The threshold value was set to p < -log10(0.05/N), where N represents the number of used SNPs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JL, XhC, and JH designed the research. SL supervised the research. JL and YW performed the experiments and wrote the manuscript. YW, XbC, and XZ collected the data. MX helped analyze the data. LL and YL provided the plant materials. XhC and JH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Key Research and Development Program of China (2021YFD1600500), the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ZDRW202105 and CAAS-ASTIP-2013-OCRI), Central Public-interest Scientific Institution Basal Research Fund (Y2020YJ03 and 1610172021001), and China Agriculture Research System of MOF and MARA (CARS-12).
Acknowledgments
Thanks to all the laboratory members for investigating the phenotype in GWAS analysis, thanks to Minqiang Tang for providing the SNP data, thanks to XhC and YL for organizing the phenotype investigation, thanks to Luyao Fan for painting the crop pictures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.962592/full#supplementary-material
Footnotes
- ^ https://www.arabidopsis.org/
- ^ http://brassicadb.cn/#/
- ^ http://www.hmmer.org/
- ^ http://plants.ensembl.org/index.html
- ^ http://gsds.gao-lab.org/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://itol.embl.de/
References
Abdel-haleem, H. (2007). The origins of genome architecture. J. Hered. 98, 633–634. doi: 10.1093/jhered/esm073
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Allender, C. J., and King, G. J. (2010). Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10:54. doi: 10.1186/1471-2229-10-54
Barker, M. S., Arrigo, N., Baniaga, A. E., Zheng, L., and Levin, D. A. (2016). On the relative abundance of autopolyploids and allopolyploids. New Phytol. 210, 391–398. doi: 10.1111/nph.13698
Borovsky, Y., Raz, A., Doron-Faigenboim, A., Zemach, H., Karavani, E., and Paran, I. (2022). Pepper fruit elongation is controlled by capsicum annuum ovate family protein 20. Front. Plant Sci. 12:815589. doi: 10.3389/fpls.2021.815589
Chalhoub, B., Denoeud, F., Liu, S., and Parkin, I. A. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cirilli, M., and Rossini, L. (2021). Many candidates for a single chair: A critical review of the genetic determinant of flat fruit shape trait in peach (Prunus persica L. Batsch). Tree Genet. Genom. 17:34. doi: 10.1007/s11295-021-01515-w
Clarke, J. M., and Simpson, G. M. (1978). Influence of irrigation and seeding rates on yield and yield components of Brassica napus cv. tower. Can. J. Plant Sci. 58, 731–737. doi: 10.4141/cjps78-108
D’Avino, L., Dainelli, R., Lazzeri, L., and Spugnoli, P. (2015). The role of co-products in biorefinery sustainability: Energy allocation versus substitution method in rapeseed and carinata biodiesel chains. J. Clean. Prod. 94, 108–115. doi: 10.1016/j.jclepro.2015.01.088
De Bodt, S., Maere, S., and Van de Peer, Y. (2005). Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 20, 591–597. doi: 10.1016/j.tree.2005.07.008
Ding, L.-N., Li, M., Guo, X.-J., Tang, M.-Q., Cao, J., Wang, Z., et al. (2020). Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus. Plant Biotechnol. J. 18, 1255–1270. doi: 10.1111/pbi.13289
Edger, P. P., and Pires, J. C. (2009). Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosom. Res. 17:699. doi: 10.1007/s10577-009-9055-9
Felten, D., Fröba, N., Fries, J., and Emmerling, C. (2013). Energy balances and greenhouse gas-mitigation potentials of bioenergy cropping systems (Miscanthus, rapeseed, and maize) based on farming conditions in Western Germany. Renew. Energy 55, 160–174. doi: 10.1016/j.renene.2012.12.004
Ferrándiz, C., Pelaz, S., and Yanofsky, M. F. (1999). Control of carpel and fruit development in Arabidopsis. Annu. Rev. Biochem. 68, 321–354. doi: 10.1146/annurev.biochem.68.1.321
Freeling, M. (2009). Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-Genome, Segmental, or by Transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Fu, Y., Wei, D., Dong, H., He, Y., Cui, Y., Mei, J., et al. (2015). Comparative quantitative trait loci for silique length and seed weight in Brassica napus. Sci. Rep. 5:14407. doi: 10.1038/srep14407
Gong, S. Y., Huang, G. Q., Sun, X., Qin, L. X., Li, Y., Zhou, L., et al. (2014). Cotton KNL1, encoding a class II KNOX transcription factor, is involved in regulation of fibre development. J. Exp. Bot. 65, 4133–4147. doi: 10.1093/jxb/eru182
Hackbusch, J., Richter, K., Müller, J., Salamini, F., and Uhrig, J. F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 4908–4912. doi: 10.1073/pnas.0501181102
Han, L.-j, Song, X.-f, Wang, Z.-y, Liu, X.-f, Yan, L.-y, Han, D.-g, et al. (2022). enome-wide analysis of OVATE family proteins in cucumber (Cucumis sativus L.). J. Integr. Agric. 21, 1321–1331. doi: 10.1016/s2095-3119(21)63788-7
Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. doi: 10.1046/j.1365-313x.2003.01759.x
Hopping, M. E., Monselise, S. P., Monselise, S., and Hopping, M. E. (1986). CRC handbook of fruit set and development. Boca Raton, FL: CRC Press Inc. doi: 10.1201/9781351073042
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015a). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, G., Koh, J., Yoo, M. J., Chen, S., and Wendel, J. F. (2015b). Gene-expression novelty in allopolyploid cotton: A proteomic perspective. Genetics 200, 91–104. doi: 10.1534/genetics.115.174367
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi: 10.1038/nature09916
Jiao, Y., Zhang, K., Cai, G., Yu, K., Amoo, O., Han, S., et al. (2021). Fine mapping and candidate gene analysis of a major locus controlling ovule abortion and seed number per silique in Brassica napus L. Theor. Appl. Genet. 134, 2517–2530. doi: 10.1007/s00122-021-03839-6
Ku, H. M., Liu, J., Doganlar, S., and Tanksley, S. D. (2001). Exploitation of Arabidopsis-tomato synteny to construct a high-resolution map of the ovatecontaining region in tomato chromosome 2. Genome 44, 470–475. doi: 10.1139/g01-024
Li, E., Wang, S., Liu, Y., Chen, J. G., and Douglas, C. J. (2011). OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 67, 328–341. doi: 10.1111/j.1365-313X.2011.04595.x
Li, S., Chen, L., Zhang, L., Li, X., Liu, Y., Wu, Z., et al. (2015). BnaC9.SMG7b Functions as a Positive Regulator of the Number of Seeds per Silique in Brassica napus by Regulating the Formation of Functional Female Gametophytes. Plant Physiol. 169, 2744–2760. doi: 10.1104/pp.15.01040
Li, H., Li, J., Song, J., Zhao, B., Guo, C., Wang, B., et al. (2019a). An auxin signaling gene BnaA3.IAA7 contributes to improved plant architecture and yield heterosis in rapeseed. New Phytol. 222, 837–851. doi: 10.1111/nph.15632
Li, Y., Dong, C., Hu, M., Bai, Z., Tong, C., Zuo, R., et al. (2019b). Identification of Flower-Specific Promoters through Comparative Transcriptome Analysis in Brassica napus. Intl. J. Mol. Sci. 20:5949. doi: 10.3390/ijms20235949
Liu, D., Sun, W., Yuan, Y., Zhang, N., Hayward, A., Liu, Y., et al. (2014). Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann. Bot. 113, 1219–1233. doi: 10.1093/aob/mcu061
Liu, J., Van Eck, J., Cong, B., and Tanksley, S. D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 99, 13302–13306. doi: 10.1073/pnas.162485999
Liu, J., Hua, W., Hu, Z., Yang, H., Zhang, L., Li, R., et al. (2015a). Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc. Natl. Acad. Sci. U.S.A. 112, E5123–E5132. doi: 10.1073/pnas.1502160112
Liu, J., Zhang, J., Hu, W., Miao, H., Zhang, J., Jia, C., et al. (2015b). Banana Ovate family protein MaOFP1 and MADS-box protein MuMADS1 antagonistically regulated banana fruit ripening. PLoS One 10:e0123870. doi: 10.1371/journal.pone.0123870
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, Z., Wang, M., Long, Y., Huang, Y., Shi, L., Zhang, C., et al. (2017). Incorporating pleiotropic quantitative trait loci in dissection of complex traits: Seed yield in rapeseed as an example. Theor. Appl. Genet. 130, 1569–1585. doi: 10.1007/s00122-017-2911-7
Ma, J., Li, C., Zong, M., Qiu, Y., Liu, Y., Huang, Y., et al. (2021). CmFSI8/CmOFP13 encoding an OVATE family protein controls fruit shape in melon. J. Exp. Bot. 73, 1370–1384. doi: 10.1093/jxb/erab510
Monforte, A. J., Diaz, A., Cano-Delgado, A., and van der Knaap, E. (2014). The genetic basis of fruit morphology in horticultural crops: Lessons from tomato and melon. J. Exp. Bot. 65, 4625–4637. doi: 10.1093/jxb/eru017
Nagaharu, U. (1935). Genome analysis in Brassica carinata with special reference to the experimental formation of Brassica napus, a peculiar mode of fertilization. Japanese J. Bot. 7, 389–452.
Neufeld, T. P., Carthew, R. W., and Rubin, G. M. (1991). Evolution of gene position: Chromosomal arrangement and sequence comparison of the Drosophila melanogaster and Drosophila virilis sina and Rh4 genes. Proc. Natl. Acad. Sci. U.S.A. 88, 10203–10207. doi: 10.1073/pnas.88.22.10203
Nordborg, M., and Weigel, D. (2008). Next-generation genetics in plants. Nature 456, 720–723. doi: 10.1038/nature07629
Pagnussat, G. C., Yu, H. J., and Sundaresan, V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19, 3578–3592. doi: 10.1105/tpc.107.054890
Price, H. L., and Drinkard, A. W. (1908). Inheritance in tomato hybrids. Zeitschrift für induktive Abstammungs- und Vererbungslehre 1, 402–403. doi: 10.1007/BF01990617
Ramsey, J., and Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. doi: 10.1146/annurev.ecolsys.29.1.467
Reinhardt, D., and Kuhlemeier, C. (2002). Plant architecture. EMBO Rep. 3, 846–851. doi: 10.1093/embo-reports/kvf177
Scarpino, S. V., Levin, D. A., and Meyers, L. A. (2014). Polyploid formation shapes flowering plant diversity. Am. Nat. 184, 456–465. doi: 10.1086/677752
Schmitz, A. J., Begcy, K., Sarath, G., and Walia, H. (2015). Rice Ovate Family Protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci. 241, 177–188. doi: 10.1016/j.plantsci.2015.10.011
Schoenfelder, K. P., and Fox, D. T. (2015). The expanding implications of polyploidy. J. Cell Biol. 209, 485–491. doi: 10.1083/jcb.201502016
Shi, L., Song, J., Guo, C., Wang, B., Guan, Z., Yang, P., et al. (2019). A CACTA-like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 98, 524–539. doi: 10.1111/tpj.14236
Tan, Q., Jiang, S., Wang, N., Liu, X., Zhang, X., Wen, B., et al. (2021). OVATE Family Protein PpOFP1 Physically Interacts With PpZFHD1 and Confers Salt Tolerance to Tomato and Yeast. Front. Plant Sci. 12:759955. doi: 10.3389/fpls.2021.759955
te Beest, M., Le Roux, J. J., Richardson, D. M., Brysting, A. K., Suda, J., Kubesová, M., et al. (2012). The more the better? The role of polyploidy in facilitating plant invasions. Anna. Bot. 109, 19–45. doi: 10.1093/aob/mcr277
Tsaballa, A., Pasentsis, K., Darzentas, N., and Tsaftaris, A. S. (2011). Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 11:46. doi: 10.1186/1471-2229-11-46
Van de Peer, Y., Mizrachi, E., and Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. doi: 10.1038/nrg.2017.26
Wang, B., Wu, Z., Li, Z., Zhang, Q., Hu, J., Xiao, Y., et al. (2018). Dissection of the genetic architecture of three seed-quality traits and consequences for breeding in Brassica napus. Plant Biotechnol. J. 16, 1336–1348. doi: 10.1111/pbi.12873
Wang, G., Zhang, X., Huang, W., Xu, P., Lv, Z., Zhao, L., et al. (2021). Increased seed number per silique in Brassica juncea by deleting cis-regulatory region affecting BjCLV1 expression in carpel margin meristem. Plant Biotechnol. J. 19, 2333–2348. doi: 10.1111/pbi.13664
Wang, S., Chang, Y., Guo, J., and Chen, J. G. (2007). arabidopsis ovate family protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50, 858–872. doi: 10.1111/j.1365-313X.2007.03096.x
Wang, S., Chang, Y., Guo, J., Zeng, Q., Ellis, B. E., and Chen, J. G. (2011). Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS One 6:e23896. doi: 10.1371/journal.pone.0023896
Wang, X., Chen, L., Wang, A., Wang, H., Tian, J., Zhao, X., et al. (2016). Quantitative trait loci analysis and genome-wide comparison for silique related traits in Brassica napus. BMC Plant Biol. 16:71. doi: 10.1186/s12870-016-0759-7
Wang, Y. K., Chang, W. C., Liu, P. F., Hsiao, M. K., Lin, C. T., Lin, S. M., et al. (2010). Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair. Plant. Mol. Biol. 74, 453–466. doi: 10.1007/s11103-010-9685-5
Wang, Y., and Li, J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. doi: 10.1146/annurev.arplant.59.032607.092902
Wang, Y., Wang, Q., Hao, W., Sun, H., and Zhang, L. (2020). Characterization of the OFP gene family and its putative involvement of tuberous root shape in radish. Int. J. Mol. Sci. 21:1293. doi: 10.3390/ijms21041293
Wood, T. E., Takebayashi, N., Barker, M. S., Mayrose, I., Greenspoon, P. B., and Rieseberg, L. H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. U.S.A. 106, 13875–13879. doi: 10.1073/pnas.0811575106
Yang, Y., Shen, Y., Li, S., Ge, X., and Li, Z. (2017). High density linkage map construction and QTL detection for three silique-related traits in orychophragmus violaceus derived brassica napus population. Front. Plant Sci. 8:1512. doi: 10.3389/fpls.2017.01512
Yang, Y., Shi, J., Wang, X., Liu, G., and Wang, H. (2016). Genetic architecture and mechanism of seed number per pod in rapeseed: Elucidated through linkage and near-isogenic line analysis. Sci. Rep. 6:24124. doi: 10.1038/srep24124
Yin, L., Zhang, H., Tang, Z., Xu, J., Yin, D., Zhang, Z., et al. (2021). rMVP: A Memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinform. 19, 619–628. doi: 10.1016/j.gpb.2020.10.007
Yoo, M.-J., Liu, X., Pires, J. C., Soltis, P. S., and Soltis, D. E. (2014). Nonadditive gene expression in polyploids. Annu. Rev. Genet. 48, 485–517. doi: 10.1146/annurev-genet-120213-092159
Zhang, L., Sun, L., Zhang, X., Zhang, S., Xie, D., Liang, C., et al. (2018). OFP1 Interaction with ATH1 regulates stem growth, flowering time and flower basal boundary formation in Arabidopsis. Genes (Basel) 9, 399. doi: 10.3390/genes9080399
Zhang, L., Zhang, X., Ju, H., Chen, J., Wang, S., Wang, H., et al. (2016). Ovate family protein1 interaction with BLH3 regulates transition timing from vegetative to reproductive phase in Arabidopsis. Biochem. Biophys. Res. Commun. 470, 492–497. doi: 10.1016/j.bbrc.2016.01.135
Zhu, Y., Ye, J., Zhan, J., Zheng, X., Zhang, J., Shi, J., et al. (2020). Validation and characterization of a seed number per silique quantitative trait locus qSN.A7 in rapeseed (Brassica napus L.). Front. Plant Sci. 11:68. doi: 10.3389/fpls.2020.00068
Keywords: ovate family protein, Brassica napus, GWAS, yield traits, negative regulator, silique development
Citation: Liu J, Wu Y, Cui X, Zhang X, Xie M, Liu L, Liu Y, Huang J, Cheng X and Liu S (2022) Genome-wide characterization of ovate family protein gene family associated with number of seeds per silique in Brassica napus. Front. Plant Sci. 13:962592. doi: 10.3389/fpls.2022.962592
Received: 06 June 2022; Accepted: 23 August 2022;
Published: 14 September 2022.
Edited by:
Sarvajeet Singh Gill, Maharshi Dayanand University, IndiaReviewed by:
Mingli Yan, Hunan Academy of Agricultural Sciences, ChinaWei Qian, Southwest University, China
Copyright © 2022 Liu, Wu, Cui, Zhang, Xie, Liu, Liu, Huang, Cheng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohui Cheng, Y2hlbmd4aWFvaHVpQGNhYXMuY24=; Junyan Huang, aHVhbmdqeUBvaWxjcm9wcy5jbg==
†These authors have contributed equally to this work and share first authorship
 Jie Liu
Jie Liu Yupo Wu
Yupo Wu Xiaobo Cui
Xiaobo Cui Junyan Huang
Junyan Huang