
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 July 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.962253
This article is part of the Research Topic For A Sustainable Future: Novel Insights into Agronomically Important Traits in Cereal Crops View all 11 articles
Lodging in wheat (Triticum aestivum L.) is a complicated phenomenon that is influenced by physiological, genetics, and external factors. It causes a great yield loss and reduces grain quality and mechanical harvesting efficiency. Lodging resistance is contributed by various traits, including increased stem strength. The aim of this study was to map quantitative trait loci (QTL) controlling stem strength-related features (the number of big vascular bundles, stem diameter, stem wall thickness) using a doubled haploid (DH) population derived from a cross between Baiqimai and Neixiang 5. Field experiments were conducted during 2020–2022, and glasshouse experiments were conducted during 2021–2022. Significant genetic variations were observed for all measured traits, and they were all highly heritable. Fifteen QTL for stem strength-related traits were identified on chromosomes 2D, 3A, 3B, 3D, 4B, 5A, 6B, 7A, and 7D, respectively, and 7 QTL for grain yield-related traits were identified on chromosomes 2B, 2D, 3D, 4B, 7A, and 7B, respectively. The superior allele of the major QTL for the number of big vascular bundle (VB) was independent of plant height (PH), making it possible to improve stem strength without a trade-off of PH, thus improving lodging resistance. VB also showed positive correlations with some of the yield components. The result will be useful for molecular marker-assisted selection (MAS) for high stem strength and high yield potential.
Lodging is defined as the permanent displacement of crop stems that can cause devastating agricultural losses such as significant reductions in crop yield and grain quality, as well as harvesting efficiency (Kokubo et al., 1989; Duan et al., 2004; Berry and Berry, 2015; Zhang et al., 2016a,b). Plant stem lodging is attributed to plant height, stem diameter and thickness, upper and lower internodal strength, stem wall thickness, lignin and cellulose accumulation within the stem wall, and spike weight (Zuber et al., 1999; Kong et al., 2013; Shah et al., 2019). Dwarf and semi-dwarf genes have been introduced into wheat breeding programs in recent years (Fischer and Stapper, 1987; Berry et al., 2004), however, positive effects on lodging resistance and grain yield have not always been observed for certain wheat dwarfing genes (Milach and Federizzi, 2001). In addition, 0.7 m of the minimum plant height for optimal grain yield has already been reached in certain genotypes in 1997 (Flintham et al., 1997), so shortening the height would not be the target for further improvement of crop lodging resistance. Furthermore, a recent study indicated that an increase of 28 N mm (the force required to bend a stem) in stem strength could reduce the probability of stem lodging from 0.73 to 0.59 of a crop 130 cm tall yielding 8.9 t ha−1 (Piñera-Chavez et al., 2021).
Wheat (Triticum aestivum L.) is the second most widely grown crop in the world. Its consumption is excepted to account for 19% of the calories in the global human diet (Aksoy and Beghin, 2004). With the vast improvement in wheat grain yield, the weight of stem and spike becomes the main loads for crop stems (Zuber et al., 1999; Kong et al., 2013). In cereals, stem diameter, culm wall thickness, and the number of big and small vascular bundles are related to stem breaking strength and bending strength against lodging (Kokubo et al., 1989; Duan et al., 2004; Wang et al., 2006; Packa et al., 2015). However, Kelbert et al. (2004) found no significant correlations among lodging resistance and other morphological characteristics such as stem diameter, stem wall thickness (Kelbert et al., 2004). The correlations between lodging resistance and the number and size of vascular bundles in cross-section of wheat stem have also been reported but results are inconsistent (Khanna, 1991; Zuber et al., 1999; Kelbert et al., 2004; Wang et al., 2006; Kong et al., 2013).
Many quantitative trait loci (QTL) in wheat have been reported for stem strength, culm wall thickness, pith diameter, and stem diameter (Hai et al., 2005; Berry and Berry, 2015). A single solid stem QTL identified on chromosome 3BL contributes to lodging resistance (Kong et al., 2013). From a wheat and spelt cross, a lodging resistance QTL was found to be related to plant height, culm stiffness, leaf width, leaf-growth, days to ear emergence, and culm thickness (Keller et al., 1999). Genes controlling culm diameter and wall thickness at the second basal internode contribute to lodging resistance with culm diameter displaying additive and partial dominant effects (Cui et al., 2002) and culm wall thickness showing both additive and nonadditive effects (Yong et al., 1998). A common genomic region affecting overall stem strength, which included internode material strength, internode diameter, and internode wall width, has been reported in the interval of 278–287 cM on chromosome 3B (Piñera-Chavez et al., 2021). Using 81 chromosome substitution lines between the red hard winter wheat cultivars, Wichita and Cheyenn, the Cheyenne chromosomes 2A and 4D had major effects the number of inner vascular bundles when substituted into Wichita (Al-Qaudhy et al., 1988). QTL for wheat vascular bundle system at 2 cm below the neck of the spike have been identified on chromosomes 1A, 2A, 2D, 5D, 6A, 6D, 7D (Sang et al., 2010).
The vascular bundle is an important structure of source-sink transport system which has a strong impact on the efficiency of photosynthetic production, mineral nutrients uptake, and water transportation (Housley and Peterson, 1982; Lucas et al., 2013). Significant positive correlations were observed between grain yield and the number of vascular bundles in rice (Mohammad et al., 2021), barley, oats (Housley and Peterson, 1982), and wheat (Evans et al., 1970). However, most studies have been focused on the vascular bundles within the upper segment of the plant, including the neck panicles (peduncle; Zhai et al., 2018; Fei et al., 2019), the rachis (Terao et al., 2010; Wolde and Schnurbusch, 2019) not within the stem, especially the basal stem (the third internode), which also contributes to lodging resistance (Duan et al., 2004). In Arabidopsis, genes involved in vascular bundle system, such as MP, PHB, PHV, AtHB15, and REV, have been identified (Hardtke and Berleth, 1998; McConnell et al., 2001; Zhong and Ye, 2004; Du and Wang, 2015). Genes affecting the vascular bundle system have also been reported in rice. Among them, APO1 controls the number of primary rachis branches as well as the vascular bundle formation, DEP1 regulates the number of larger vascular bundles and ABA signaling, and NAL1 affects vein patterning and polar auxin transport (Qi et al., 2008; Terao et al., 2010; Fujita et al., 2013; Fei et al., 2019). WHEAT ORTHOLOG OF APO1 (WAPO1) gene is an orthologue of the rice APO1 gene, affecting spikelet number per spike, but no further evidence has been found for its function on the development of vascular bundle (Kuzay et al., 2019). Restricted genes such as TmBr1 (Deng et al., 2019), an allele of the Q (Qc1) gene (Xu et al., 2018), are involved in the formation of vascular bundles that regulate the efficiency in transporting assimilates in the spikes or the stem strength against lodging in wheat. TaCM, involved in the biosynthesis of lignin, has also been implicated in stem strength (Ma, 2009). However, the functional genes have not yet been fully discovered, and the molecular mechanism of how the vascular bundle system influences crop yield is largely unknown.
Linkage mapping approaches based on individual and multiple populations have become routine in wheat genetic studies to dissect the genetic architecture of complex traits (Wang et al., 2014; Liu et al., 2020; Qu et al., 2021). QTL remains of great importance in identifying the genetic basis of the link between the anatomical and morphophysiological traits of the stem and lodging, and yield potential. In our present study, the number of big vascular bundle (VB), stem diameter (SD, mm), and stem wall thickness (SWT, mm) were examined in a doubled haploid (DH) population of 194 wheat lines under three environmental conditions. The objective of this study was to identify QTL for the vascular bundle system of the third internode along with other stem and panicle traits in wheat. Our results will assist in molecular marker assistant selection to obtain lines with high stem strength and grain-yielding capacity in wheat breeding.
A double haploid (DH) population of 194 lines was generated by anther culture from the cross between two elite wheat cultivars “Baiqimai” (short and thin stem) and “Neixiang 5” (tall and thick stem). Field trials were conducted at Mt. Pleasant Laboratory in Tasmania, Australia (147°08’E, 41°280’S). Fifteen seeds of each line were sown on April 15, 2020 and April 25, 2021 in a row of 0.6 m with a row spacing of 0.3 m, following local farmers’ practices for field management. Under glasshouse condition, five plants of each line were grown in a 2-L pot filled with commercial potting mixture with a distance of 0.2 m between each pot on 7 May 2020. All trials were conducted in a randomized complete block design with three replicates. To evaluate the anatomical properties of the stems, five stems were selected at random from each DH line at post-anthesis and were cross-sectionally cut at the center of every third internode. All samples were collected from main tillers.
Since wheat stems are not as regularly round, we choose the vernier caliper method to measure stem diameter: the stem diameter (SD) = 1/2 (longest diameter (sd1) + shortest diameter (sd2); Figures 1A,B). A transverse loop (less than 0.1 cm in width) in the middle of the third internode was cut by razor blades, observed under a microscope and pictures were taken. The number of the big vascular bundle (VB) was counted. Stem wall thickness was the average of the thickest (swt1) and the thinnest (swt2) stem wall of each cross-section (Figures 1A,B). At maturity, five panicles were collected from each line to count the number of the spikelet (RN; Figure 1C), and measure the total grain weight (PW, g). Plant height (PH, cm) was measured from soil surface to the top of the spike excluding the awns in the field.
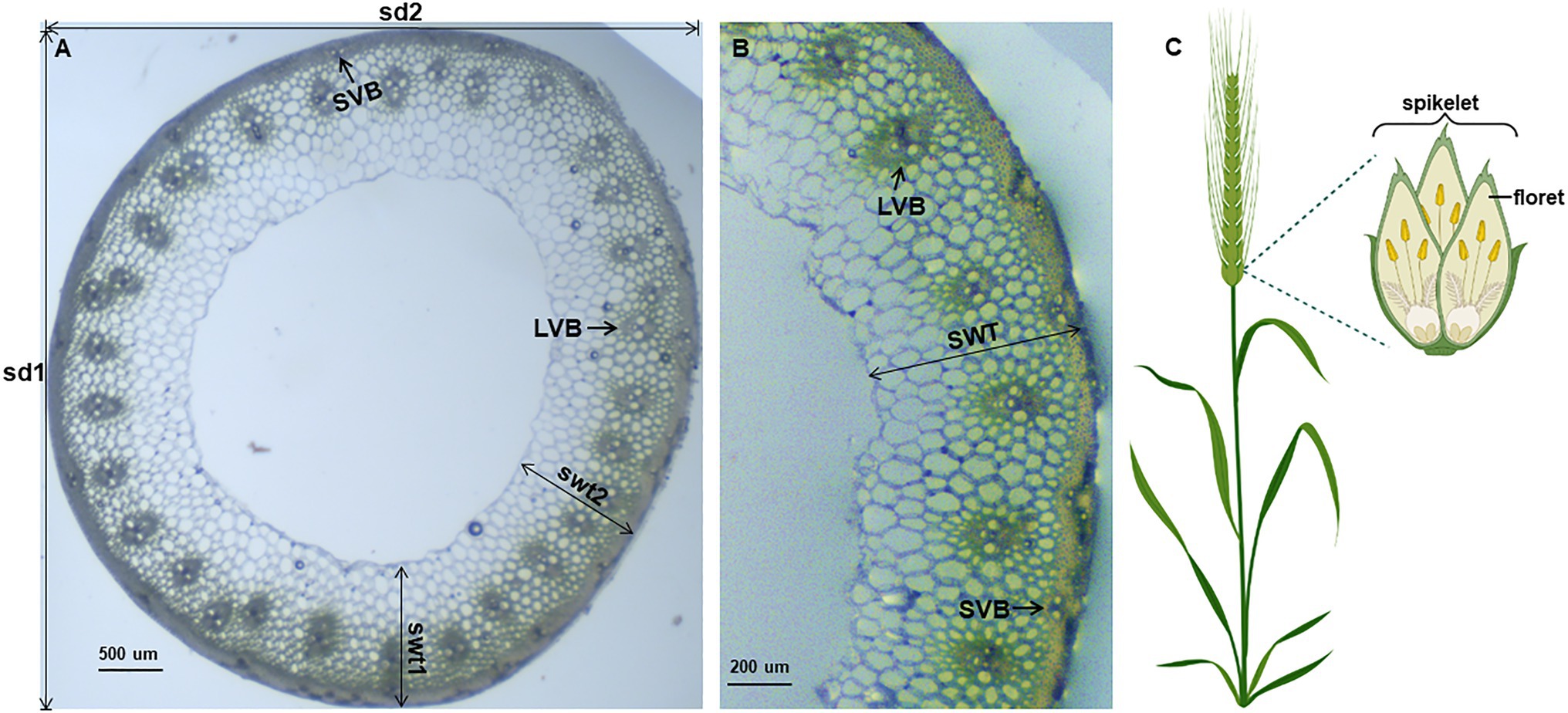
Figure 1. Display of measured wheat characteristics. Anatomical characteristics of wheat stem (A,B). The wheat stems are not that regularly round, so we measured the longest (sd1) and the shortest stem diameter (sd2), and the thickest (swt1) and the thinnest (swt2) stem wall for data collecting. LVB: large vascular bundles; SVB: small vascular bundles; SD: stem diameter; SWT: stem wall thickness. Wheat spikelet architecture (C), created with BioRender.com.
Data processing for QTL mapping was described in a previous study (Niu et al., 2021). Mean phenotypic values across replicates in each environment and best linear unbiased estimation (BLUE) values across multiple environments were then generated for statistical analysis. The mean values of BLUEs of each lines were used for QTL mapping for measured traits (Supplementary Table S1). Pearson’s correlation coefficients were analyzed using GraphPad Software, San Diego, California USA.1 Plots for the distribution of phenotypes were conducted on the data visualization web server “ImageGP” (Chen et al., 2022).
DNA was isolated from young leaves of each line, including the parents, using the CTAB method (Murray and Thompson, 1980). Whole genome diversity array technology (DArT) and single nucleotide polymorphism (SNP) genotyping based on the reference genome of the bread wheat variety Chinese Spring (IWGSC RefSeq v2.0, International Wheat Genome Sequencing Consortium)2 assembly were conducted by Diversity Arrays Technology (Canberra, Australia).3 The genetic map was constructed in JoinMap 4.0 (Ooijen et al., 2006) using 2,518 polymorphism markers (χ2-test, p < 0.05) with <10% missing data (Supplementary Table S2). The genetic and physical positions of the markers were aligned with the Chinese Spring wheat reference genome assembly (Alaux et al., 2018), the order of the markers on each chromosome in the linkage map was compared with the order of the physical map of each chromosome (Supplementary Figure S1). QTL analysis was conducted with MapQTL 6.0 (Van Ooijen, 2009). Digenic interactions analysis between non-allelic QTL were similar to the previously reported (Fan et al., 2015; Gill et al., 2017). The R package LinkageMapView (Ouellette et al., 2017) was used to visualize the constructed map. MapChart 2.2 (Voorrips, 2002) was used for the plotting linkage groups and QTL locations.
The genetic map was generated from 451 high-quality polymorphic SNP markers and 2067 DArT markers covering a total map distance of 4996.1 cM in 21 linkage groups corresponding to the 21 wheat chromosomes, with chromosome sizes ranging from 73.7 cM (4D) to 325.5 cM (2D). On average, each chromosome contained 120 markers, ranging from 19 (4D) to 184 on (5B), and the total marker density was 1.87 cM, ranging from 0.65 cM on 6B to 6.52 cM on 5D (Table 1; Supplementary Tables S1–S3).
The marker positions on each chromosome in this map were similar to the published genetic maps (Maccaferri et al., 2015; Wen et al., 2017; Qu et al., 2021). In comparison with genomes A and B, genome D was the shortest and contained much fewer markers and more gaps (Figure 2), suggesting that fewer crossing-over events occurred on the D genome. In general, a high collinearity at the genome-wide level was observed between the genetic and published Chinese Spring consensus map. Lower collinearities were observed in some chromosomal regions due to low marker densities (Supplementary Figure S1), which has also been reported previously (Wingen et al., 2017; Qu et al., 2021). Recombination happened much more frequently in distal chromosomal regions, while recombination near the centromeres tended to be suppressed, consistent with previous studies (Sourdille et al., 2004). The longer map length was due to (1) increased recombination events and map resolution with an higher number of markers and density (Ferreira et al., 2006; Wingen et al., 2017), and (2) differences in chromosomal structure in different mapping populations and application of different ordering algorithms (Ferreira et al., 2006; Qu et al., 2021).
The frequency distribution of each measured trait showed a continuous distribution, with all measured traits in the DH lines across all environments exhibiting significant differences between genotypes (Figure 3; Supplementary Figure S2). VB was significantly positively correlated not only with SD but also with yield-related traits RN and PW, while the correlations with SWT and PH did not reach the level of significance (Figure 4). The correlation coefficients for SD and other traits ranged from 0.13 to 0.51 (p < 0.05), and the correlation coefficients for PW with the rest of the measured traits ranged from 0.30 to 0.39 (p < 0.001). SWT was significantly associated with PW with a correlation coefficient of 0.30 (p < 0.001). In addition, data from the three harvests in 2020 and 2021 showed high correlation coefficients for all measured traits (Supplementary Figure S3). All five traits showed very high heritability ranging from 0.78 for SD to 0.92 for RN across different environments, except PW that had only 0.28 (Table 2; Supplementary Figure S3).
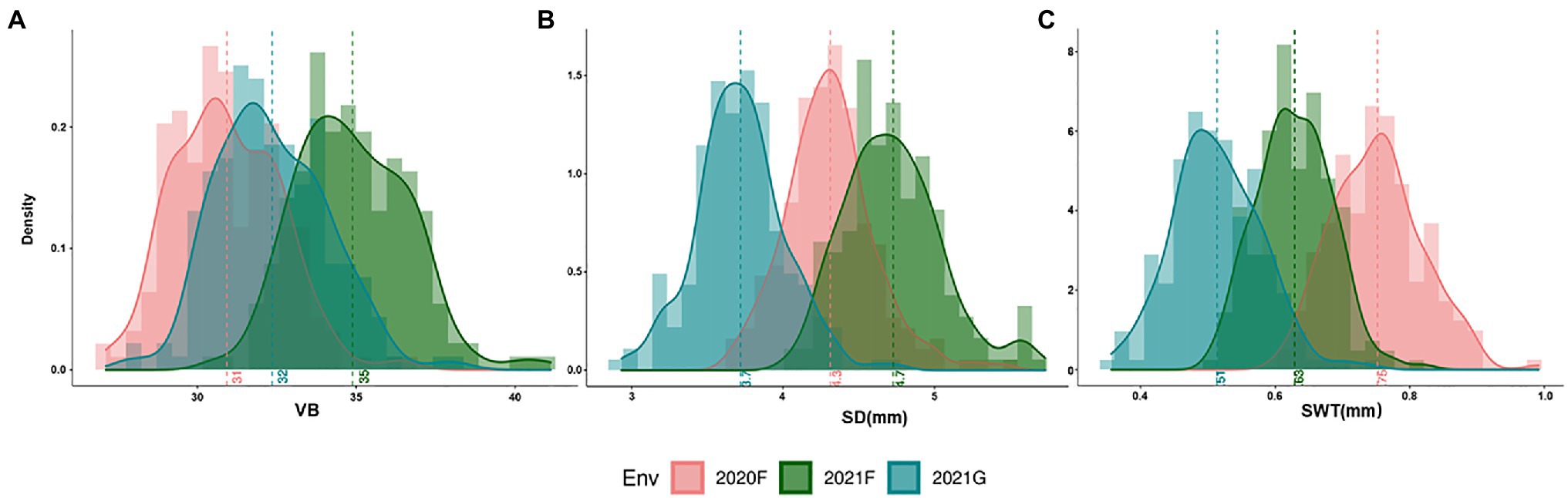
Figure 3. Phenotypic distributions of VB (A), SD (B) and SWT (C) in three environments. VB: number of big vascular bundle; SD: stem diameter; SWT: stem wall thickness.
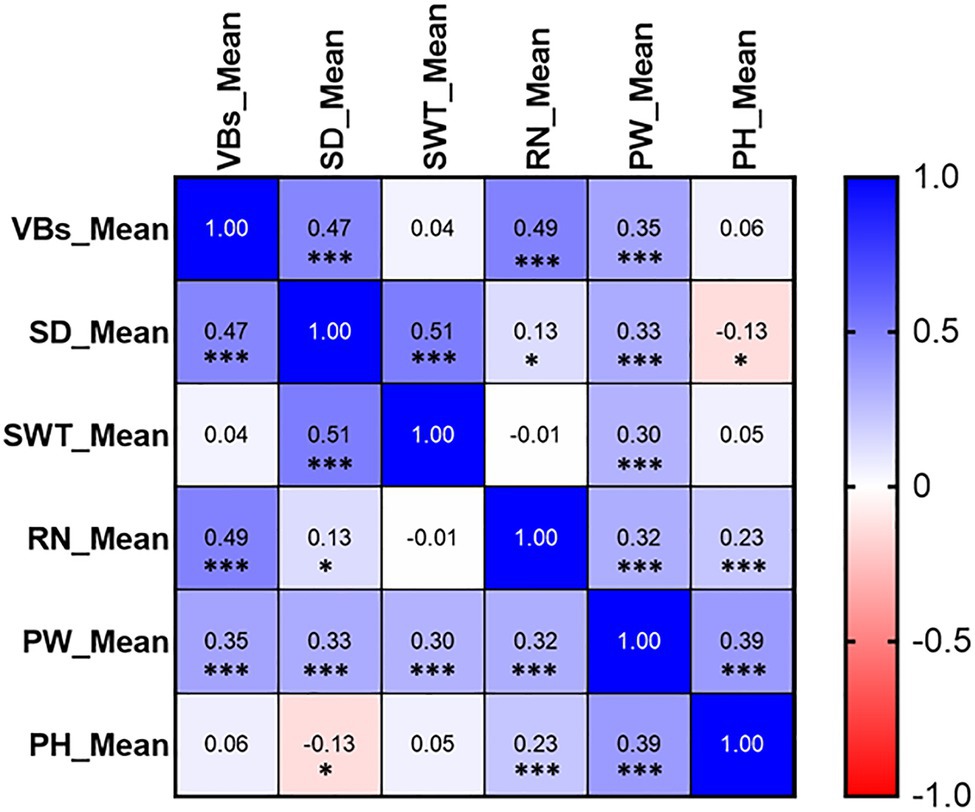
Figure 4. Correlations between stem- and yield-related traits. The number in the middle of the cell is the correlation coefficient; “*” and “***” refer to significant correlations (p < 0.05 and p < 0.001). VB: number of big vascular bundle; SD: stem diameter; SWT: stem wall thickness; RN: the number of the spikelet; PW: panicle weight; PH: plant height.
Five QTL for VB were identified on chromosomes 2D, 3A, 4B, 5A, and 7A and designated Qvb-2D, Qvb-3A, Qvb-4B, Qvb-5A, and Qvb-7A, respectively (Figure 5; Table 3). Qvb-5A with 5357328 as the nearest marker explained 15.4% of phenotypic variation. The other four QTL explained a total of 24.3% of the phenotypic variation. The closest markers for these four QTL were D1179294-2D21.4, 7487752—0, 1120870-4B43.8, and D1107943-7A1150, respectively. Six QTL for SD were detected on 2D (Qsd-2D), 3A (Qsd-3A), 3B (Qsd-3B), 5A (Qsd-5A), 6B (Qsd-6B), and 7A (Qsd-7A), with the two major ones, Qsd-3A and Qsd-5A determining 12.6 and 11.5% of the phenotypic variation, respectively. Four QTL for SWT were mapped to chromosomes 2D, 3B, 3D, and 7D, respectively. The major QTL, Qswt-3B with the nearest marker of D3942570-2B81.7 determined 13.4% of the phenotypic variation. Qvb-2D and Qsd-2D were located at the same position (38.09 cM) and Qvb-3A overlapped the region of Qsd-3A (Figure 5; Table 3), confirming the significant correlation between VB and SD (Figure 4).
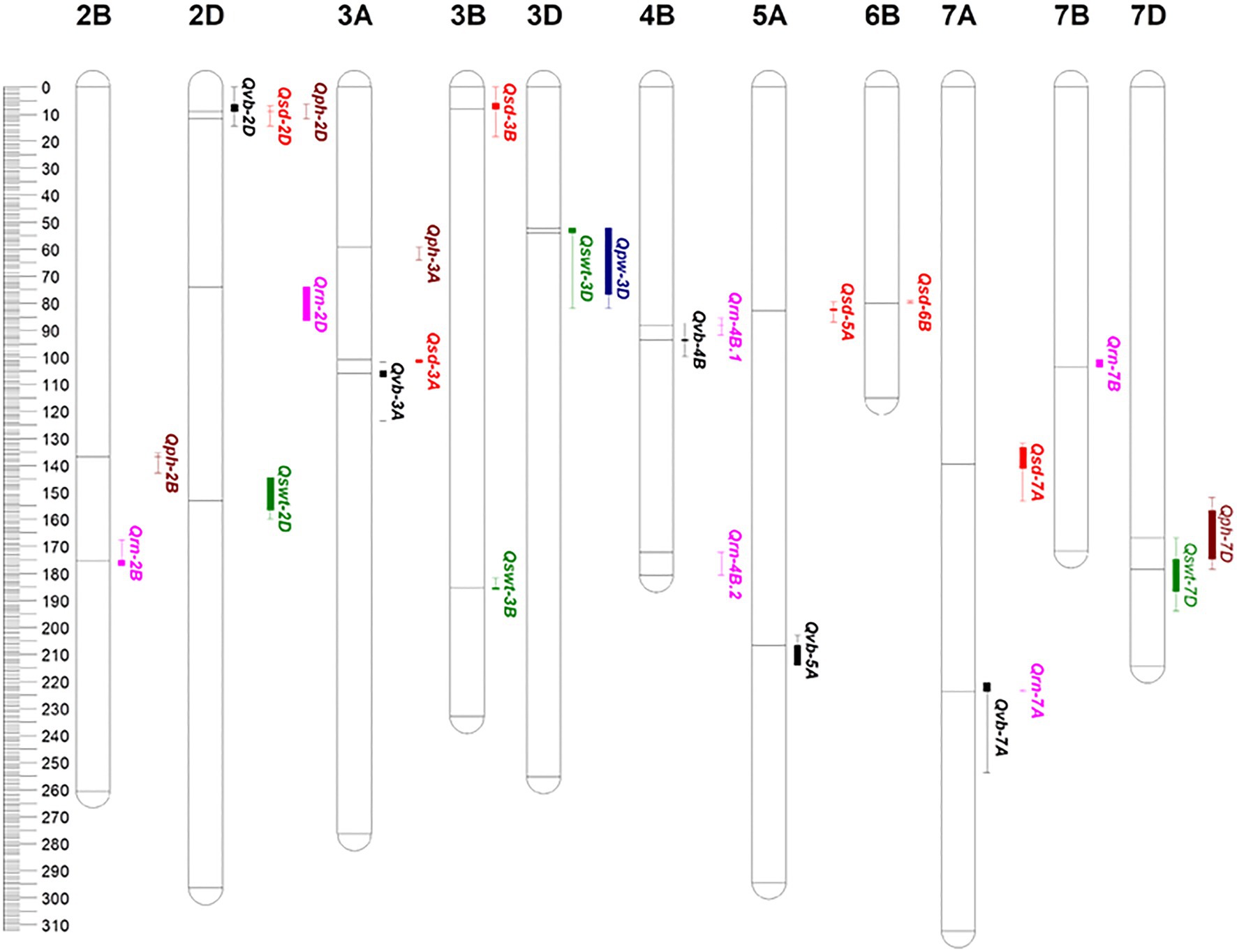
Figure 5. QTL identified for stem- and yield-related traits. The solid line near each QTL within each chromosome indicate the nearest marker of individual QTL, respectively. vb: number of big vascular bundle; sd: stem diameter; swt: stem wall thickness; rn: the number of the spikelet; pw: panicle weight; ph: plant height. Black color for VB; red color for SD; green color for SWT; purple color for RN; blue color for PW; brown color for PH.
QTL were also mapped for PW, RN and PH. The major QTL for PH (Qph-2D) was overlapped with only one minor QTL for VB (Qvb-2D) and one for SD (Qsd-2D), Qph-7D on chromosome 7D was overlapped with only one minor QTL for SWT (Qswt-7D), confirming the less significant correlations between PH and these three stem traits. One major QTL for RN was identified on chromosome 7A (Qrn-7A) which is co-located with (Qrn-7A). Another minor QTL on chromosome 4B (Qrn-4B.1) is co-located with Qvb-4B, indicating potential relationship between RN and VB (Figure 4). Only one minor QTL was identified for PW (Qpw-3D) due to its low heritability (Table 2) and this QTL was at a similar position to a QTL for SWT on chromosome 3D (Qswt-3D; Table 3; Figure 5).
Plant height is one of the most important features of plants’ architecture affecting plant lodging resistance under harsh environmental conditions (Berry and Berry, 2015). However, as shown in Figure 4, only SD exhibited a negatively close association with PH (−0.13, p < 0.05), and the major QTL for stem-related traits were totally different with that for PH. To further confirm the relationships between PH and those stem-related traits in wheat, QTL analyses were performed using PH as a covariate. Of the five QTL for VB, all QTL showed increased R2 with the percentage of the phenotypic variation determined by the major QTL Qvb-5A being from 15.4 to 18.5% (Tables 3, 4), while the remaining four minor QTL for the trait showed decreased R2 (Tables 3, 4), when using PH as a covariate. In contrast, when PH was used as a covariate, Qsd-2D and Qsd-3B became insignificant, Qsd-3A showed a slight reduction in R2, Qsd-5A and Qsd-7A showed a slight increase in R2, and Qsd-6B remained the same (Table 4). The R2 of Qswt-2D for SWT showed a slight increase (from 8.2 to 10.2%), and the R2 of all other QTL for SWT had a slight decrease when using PH as a covariate (Table 4). In general, PH was independent of VB, although Qvb-2D and Qph-2D were co-located at the same position on 2D (Table 3; Figure 5).
Table 4. Changes in the significance and percentage variation determined by the QTL after PH or SWT/VB were used as covariates.
VB was also strongly correlated with RN and PW in the current study (Figure 4). No significant changes to the QTL for PW when using VB as a covariate. However, four out of six QTL for RN became insignificant when using VB as a covariate and the R2 of the major QTL for RN on 7A (Qrn-7A) showed a slight decrease from 27.3 to 20.9% (Table 4).
Several genes have been reported to affect VB formation in Arapidopsis and rice (Hardtke and Berleth, 1998; McConnell et al., 2001; Zhong and Ye, 2004; Qi et al., 2008; Terao et al., 2010; Fujita et al., 2013; Du and Wang, 2015; Fei et al., 2019). However, there have been no reports on QTL for the number of VB in wheat stem with most studies being concentrated on the regions close to the neck of the spike (Sang et al., 2010). Here, we have found that the VB number in the third internode showed the greatest variation and several QTL were responsible for VB number. A major novel QTL affecting VB the third internode was identified on chromosome 5A [Qvb-5A (206.56–214.01 cM, 612.16–641.76 Mb); Table 3; Figure 5]. According to the annotation database,4 TraesCS5A01G445800 (625,908,587–625,908,859 bp) coding for an auxin efflux carrier family protein is supposed to participate in all aspects of vascular differentiation thus could be one of the candidate genes for Qvb-5A. One of the theories to explain how auxin regulates the formation of regular patterns of vascular tissue distribution in plants is the canalization of auxin flow (Biedroń and Banasiak, 2018). Narrow leaf1 (nal1) in rice, which is abundantly expressed in vascular tissues, affects polar auxin transport and vascular patterns in rice plants (Qi et al., 2008). Several auxin efflux carrier family proteins have been reported in Arabidopsis thaliana, which were involved in polar auxin transport and accumulation and the formation of vascular tissues (Forestan and Varotto, 2012).
Among all other minor QTL, the Qvb-2D on chromosome 2D was located at a similar position to the major wheat semi-dwarfing gene Rht8/RNHL-D1 [TraesCSU02G024900 (CS RefSeq v1.0, 24 Mb); Chai et al., 2022; Xiong et al., 2022]. The GA-sensitive Rht8 reduces plant height without scarifying coleoptile length (Chai et al., 2022), and the combinations of Rht8 and Rht4 (had moderate effects on plant height) could reduce plant height to a desirable level, and improve yield-related traits in the rainfed cultivation (Yingying et al., 2018). However, our results showed that the QTL for VB was independent on the QTL for plant height with no significant correlation between VB and PH. Hence, validation and/or fine mapping of those new QTL is necessary for finding reliable validated markers to be utilized in marker-assisted selection (MAS) or genomic selection (GS) for stem strength and high yield.
The combination of the positive VB alleles detected in our study significantly increased the number of vascular bundles from 30 to 31 (All−) to 35–38 (All+; Figure 6A). The reported QTL/gene on chromosome 2D only slightly improved the number of vascular bundles with no significant difference between the presence (2D+) and absence (2D−) of 2D allele. Therefore, the new discovered QTL for VB present great potential in improving stem strength by pyramiding of major QTL for large number of vascular bundles. The pattern of mechanical development of winter wheat can maximize its reproductive success (Crook et al., 1994). In this study, VB showed a significant correlation with RN which was confirmed by QTL analysis for RN using VB as a covariate (Table 4). The increasing allele represented by the closest marker 1120870 for VB 4B QTL also showed a significant increase in RN (Figure 6B). The major QTL on 5A (Qvb-5A) showed slight but positive impacts on RN.
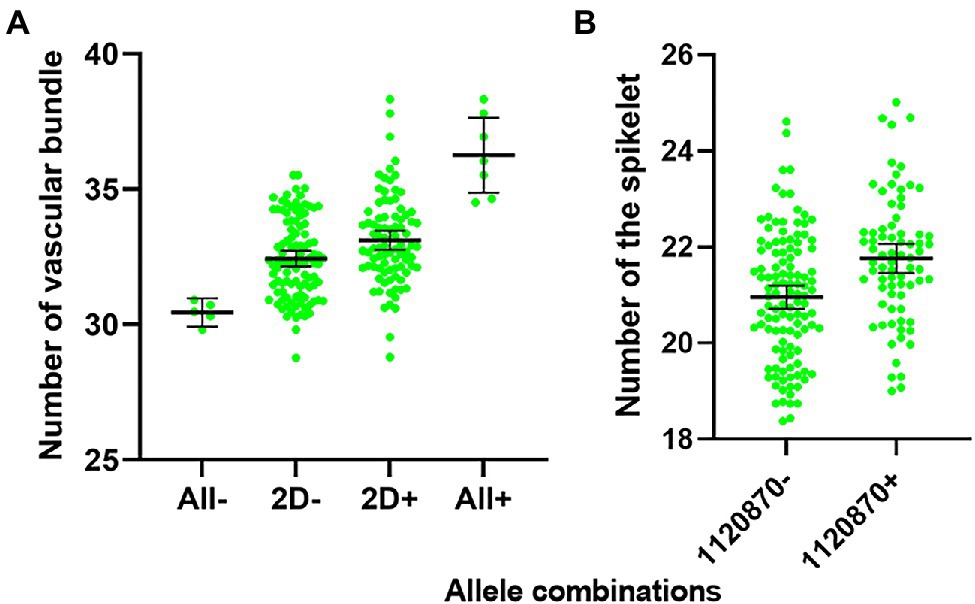
Figure 6. Allele effects of VB alleles on the number of big vascular bundle (A), and the number of the spikelet (B). ALL-stands for all alleles for VB were absent; 2D-stands for allele on Chromosome 2D (Qvb-2D) was absent; 2D+ stands for allele on Chromosome 2D (Qvb-2D) was present; ALL+ stands for all alleles for VB were present. 1120870 is the nearest marker for Qvb-4B, 1120870- means 1120870 was absent; 1120870+ means 1120870 was present. Error bars in the figures indicate 95% confidence interval.
Increasing stem strength with a minimal investment in biomass can be achieved by increasing internode diameter and material strength rather than internode wall width (Berry et al., 2007; Piñera-Chavez et al., 2021). Among the six QTL for SD, Qsd-2D was at a similar position to Qvb-2D and the major QTL for PH. Qsd-3A and Qsd-6B have been reported earlier (Berry and Berry, 2015). Qsd-3A for SD (100.82–102.02 cM) is located at a similar position to Qvb-3A for VB (101.67–123.51 cM) with increasing alleles of both QTL being from the parent “Neixiang 5” (Table 3; Figure 5), indicating potential pleiotropic effects on overall stem strength or tightly linked genes in this QTL region. The other major QTL (Qsd-5A) was a novel one from this population (Table 3; Figure 5). Candidate genes, such as TraesCS5A01G239400 (Bri1 kinase inhibitor 1), and two cellulose synthase genes (TraesCS5A01G253100 and TraesCS5A01G253200) have great potential on regulating cell expansion and elongation (Hyles et al., 2017; Oh et al., 2020).
SWT showed no significant correlation with VB, but it was positively associated with SD (0.51, p < 0.001, Figure 4). To further investigate the relationship between SWT and SD, we performed the QTL analysis for SD using SWT as a covariate. Qsd-3B, Qsd-6B, and Qsd-7A became insignificant, and the R2 of the two major QTL (Qsd-3A and Qsd-5A) for SD reduced significantly from 12.6 to 5.9 and 11.5 to 7.6, respectively (Table 4). The major QTL for SWT (Qswt-3B) was located on the long arm of chromosome 3B with a QTL region of between 181.66 and 185.94 cM (771.56–775.09 Mb), which coincides with Qss.msub-3BL (761,188,585–773,049,079 bp) for stem solidness (Sherman et al., 2015; Oiestad et al., 2017). No candidate genes responsible for Qss.msub-3BL were identified. Qswt-3D has not been reported before. A gene encoding Rho of Plants (ROP) proteins (TraesCS3D01G057400, 24,090,998–24,095,444 bp), also known as RACs, is the most likely candidate gene for this QTL. ROPs are involved in the regulation of abscisic acid (ABA) and auxin signaling and transport (Wu et al., 2011; Liao et al., 2017), and also regulate cell polarization and secondary cell wall development in the xylem of plants (Bloch and Yalovsky, 2013; Oda and Fukuda, 2014; Feiguelman et al., 2018). The QTL interval of Qswt-7D (166.781–193.984 cM, 177.88–413.11 Mb) overlapped with Qph-7D (151.948–178.44 cM, 135.40–384.45 Mb) where a MADS-box gene TaSEP3-D1 (TraesCS7D02G261600, Refv1.0, chr7D:237.609–237.619 Mb) that regulates both heading date and plant height development (Zhang et al., 2021) is located.
In conclusion, QTL were identified for the stem traits of the third internode, which showed the greatest variation and is probably the most important part linked to lodging resistance. The number of VB in the third internode showed significant correlation with the number spikelets. The combination of positive alleles of the QTL for VB can increase VB by more than 15%. QTL were also identified from stem wall thickness and diameter. Most of the QTL for these traits showed no significant correlation with plant height. The results offer great opportunities for improving stem lodging resistance and improving yield components with less effect on PH.
The original contributions presented in the study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding author.
MZ acquired the funding and participated in supervision. YN and TC conducted the field trials, data collection, and data analysis and carried out data visualization. CG collected the data. YN wrote the draft. MZ and YN wrote, reviewed, and edited the draft. CZ participated in supervision. All authors contributed to the article and approved the submitted version.
This project is funded by the Grains Research and Development Corporation (GRDC) of Australia. The funder had the following involvement with the study: Identification of QTL for stem traits in wheat (T. aestivum L.).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We warmly thank Peter Johnson for providing us professional field management suggestions. We would also like to thank the reviewers for their insightful comments that helped us to improve our manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.962253/full#supplementary-material
Supplementary Figure S1 | Collinearity of marker orders between the genetic and reported consensus maps.
Supplementary Figure S2 | Phenotypic distributions of two yield-related traits and PH in different environments. RN: the number of the spikelet; PW: panicle weight; PH: plant height.
Supplementary Figure S3 | Correlations between lodging-related traits and yield-related traits across different environments. The number in the middle of the cell is the correlation coefficient; “*,” “**,” and “***” refer to significant correlations (p < 0.05, p < 0.01, and p < 0.001). VB: the number of big vascular bundle; SD: stem diameter; SWT: stem wall thickness; RN: the number of the spikelet; PW: panicle weight; PH: plant height.
Supplementary Table S1 | Phenotypic data of wheat DH population from Baiqimai and Neixiang 5.
Supplementary Table S2 | Genotypic data of wheat DH population from Baiqimai and Neixiang 5.
Supplementary Table S3 | Map positions of different SNP and DArT markers.
BLUE, Best linear unbiased estimation; DArT, Diversity array technology; DH, Doubled haploid; GS, Genomic selection; MAS, Marker-assisted selection; PH, Plant height; PW, Total grain weight; QTL, Quantitative trait loci; RN, Number of the spikelet; ROP, Rho of Plants; SD, Stem diameter; SNP, Single nucleotide polymorphism; SWT, Stem wall thickness; VB, Number of big vascular bundle; WAPO1, WHEAT ORTHOLOG OF APO1.
2. ^https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Assemblies/v2.0/
Aksoy, M. A., and Beghin, J. C. (2004). Global Agricultural Trade and Developing Countries. Washington, DC: World Bank Publications.
Alaux, M., Rogers, J., Letellier, T., Flores, R., Alfama, F., Pommier, C., et al. (2018). Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 19:111. doi: 10.1186/s13059-018-1491-4
Al-Qaudhy, W., Morris, R., Mumm, R. F., and Hanna, M. A. (1988). Chromosomal locations of genes for traits associated with lodging in winter wheat. Crop Sci. 28, 631–635. doi: 10.2135/cropsci1988.0011183X002800040012x
Berry, P. M., and Berry, S. T. (2015). Understanding the genetic control of lodging-associated plant characters in winter wheat (Triticum aestivum L.). Euphytica 205, 671–689. doi: 10.1007/s10681-015-1387-2
Berry, P., Sterling, M., Spink, J., Baker, C. J., Sylvester-Bradley, R., Mooney, S., et al. (2004). Understanding and reducing lodging in cereals. Adv. Agron. 84, 217–271. doi: 10.1016/S0065-2113(04)84005-7
Berry, P. M., Sylvester-Bradley, R., and Berry, S. (2007). Ideotype design for lodging-resistant wheat. Euphytica 154, 165–179. doi: 10.1007/s10681-006-9284-3
Biedroń, M., and Banasiak, A. (2018). Auxin-mediated regulation of vascular patterning in Arabidopsis thaliana leaves. Plant Cell Rep. 37, 1215–1229. doi: 10.1007/s00299-018-2319-0
Bloch, D., and Yalovsky, S. (2013). Cell polarity signaling. Curr. Opin. Plant Biol. 16, 734–742. doi: 10.1016/j.pbi.2013.10.009
Chai, L., Xin, M., Dong, C., Chen, Z., Zhai, H., Zhuang, J., et al. (2022). A natural variation in Ribonuclease H-like gene underlies Rht8 to confer “green revolution” trait in wheat. Mol. Plant 15, 377–380. doi: 10.1016/j.molp.2022.01.013
Chen, T., Liu, Y.-X., and Huang, L. (2022). ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 1:e5. doi: 10.1002/imt2.5
Crook, M. J., Ennos, A. R., and Sellers, E. K. (1994). Structural development of the shoot and root systems of two winter wheat cultivars. Triticum aestivum L. J. Exp. Bot. 45, 857–863. doi: 10.1093/jxb/45.6.857
Cui, D., Wei, J., Nie, L., and Chen, F. (2002). Study on the genetic model stem characters in wheat. J. Henan Agric. Sci 9, 4–7.
Deng, Q., Kong, Z., Wu, X., Ma, S., Yuan, Y., Jia, H., et al. (2019). Cloning of a COBL gene determining brittleness in diploid wheat using a MapRseq approach. Plant Sci. 285, 141–150. doi: 10.1016/j.plantsci.2019.05.011
Du, Q., and Wang, H. (2015). The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal. Behav. 10:e1078955. doi: 10.1080/15592324.2015.1078955
Duan, C., Wang, B., Wang, P., Wang, D., and Cai, S. (2004). Relationship between the minute structure and the lodging resistance of rice stems. Colloids Surf. B Biointerfaces 35, 155–158. doi: 10.1016/j.colsurfb.2004.03.005
Evans, L., Dunstone, R., Rawson, H., and Williams, R. (1970). The phloem of the wheat stem in relation to requirements for assimilate by the ear. Aust. J. Biol. Sci. 23, 743–752. doi: 10.1071/BI9700743
Fan, Y., Shabala, S., Ma, Y., Xu, R., and Zhou, M. (2015). Using QTL mapping to investigate the relationships between abiotic stress tolerance (drought and salinity) and agronomic and physiological traits. BMC Genomics 16:43. doi: 10.1186/s12864-015-1243-8
Fei, C., Geng, X., Xu, Z., and Xu, Q. (2019). Multiple areas investigation reveals the genes related to vascular bundles in rice. Rice 12:17. doi: 10.1186/s12284-019-0278-x
Feiguelman, G., Fu, Y., and Yalovsky, S. (2018). ROP GTPases structure-function and signaling pathways. Plant Physiol. 176, 57–79. doi: 10.1104/pp.17.01415
Ferreira, A., Silva, M. F. D., Silva, L. D. C. E., and Cruz, C. D. (2006). Estimating the effects of population size and type on the accuracy of genetic maps. Genet. Mol. Biol. 29, 187–192. doi: 10.1590/s1415-47572006000100033
Fischer, R. A., and Stapper, M. (1987). Lodging effects on high-yielding crops of irrigated semidwarf wheat. Field Crops Res. 17, 245–258. doi: 10.1016/0378-4290(87)90038-4
Flintham, J. E., Borner, A., Worland, A. J., and Gale, M. D. (1997). Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 128, 11–25. doi: 10.1017/s0021859696003942
Forestan, C., and Varotto, S. (2012). The role of PIN Auxin efflux carriers in polar Auxin transport and accumulation and their effect on shaping maize development. Mol. Plant 5, 787–798. doi: 10.1093/mp/ssr103
Fujita, D., Trijatmiko, K. R., Tagle, A. G., Sapasap, M. V., Koide, Y., Sasaki, K., et al. (2013). NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc. Natl. Acad. Sci. U. S. A. 110, 20431–20436. doi: 10.1073/pnas.1310790110
Gill, M. B., Zeng, F., Shabala, L., Zhang, G., Fan, Y., Shabala, S., et al. (2017). Cell-based phenotyping reveals QTL for membrane potential maintenance associated with hypoxia and salinity stress tolerance in barley. Front. Plant Sci. 8:1941. doi: 10.3389/fpls.2017.01941
Hai, L., Guo, H., Xiao, S., Jiang, G., Zhang, X., Yan, C., et al. (2005). Quantitative trait loci (QTL) of stem strength and related traits in a doubled-haploid population of wheat (Triticum aestivum L.). Euphytica 141, 1–9. doi: 10.1007/s10681-005-4713-2
Hardtke, C. S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. doi: 10.1093/emboj/17.5.1405
Housley, T. L., and Peterson, D. M. (1982). Oat stem vascular size in relation to kernel number and weight. I. Controlled Environment1. Crop Sci. 22, 259–263. doi: 10.2135/cropsci1982.0011183X002200020014x
Hyles, J., Vautrin, S., Pettolino, F., MacMillan, C., Stachurski, Z., Breen, J., et al. (2017). Repeat-length variation in a wheat cellulose synthase-like gene is associated with altered tiller number and stem cell wall composition. J. Exp. Bot. 68, 1519–1529. doi: 10.1093/jxb/erx051
Kelbert, A. J., Spaner, D., Briggs, K. G., and King, J. R. (2004). The association of culm anatomy with lodging susceptibility in modern spring wheat genotypes. Euphytica 136, 211–221. doi: 10.1023/B:EUPH.0000030668.62653.0d
Keller, M., Karutz, C., Schmid, J. E., Stamp, P., Winzeler, M., Keller, B., et al. (1999). Quantitative trait loci for lodging resistance in a segregating wheat×spelt population. Theor. Appl. Genet. 98, 1171–1182. doi: 10.1007/s001220051182
Khanna, V. (1991). Relationship of lodging resistance yield to anatomical characters of stem in wheat [Triticum spp.], triticale and rye. Wheat Information Service (Japan). 73, 19–24.
Kokubo, A., Kuraishi, S., and Sakurai, N. (1989). Culm strength of barley 1: correlation among maximum bending stress, cell wall dimensions, and cellulose content. Plant Physiol. 91, 876–882. doi: 10.1104/pp.91.3.876
Kong, E., Liu, D., Guo, X., Yang, W., Sun, J., Li, X., et al. (2013). Anatomical and chemical characteristics associated with lodging resistance in wheat. Crop J. 1, 43–49. doi: 10.1016/j.cj.2013.07.012
Kuzay, S., Xu, Y., Zhang, J., Katz, A., Pearce, S., Su, Z., et al. (2019). Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Theor. Appl. Genet. 132, 2689–2705. doi: 10.1007/s00122-019-03382-5
Liao, H., Tang, R., Zhang, X., Luan, S., and Yu, F. (2017). FERONIA receptor kinase at the crossroads of hormone signaling and stress responses. Plant Cell Physiol. 58, 1143–1150. doi: 10.1093/pcp/pcx048
Liu, Y., Salsman, E., Wang, R., Galagedara, N., Zhang, Q., Fiedler, J. D., et al. (2020). Meta-QTL analysis of tan spot resistance in wheat. Theor. Appl. Genet. 133, 2363–2375. doi: 10.1007/s00122-020-03604-1
Lucas, W. J., Groover, A., Lichtenberger, R., Furuta, K., Yadav, S.-R., Helariutta, Y., et al. (2013). The plant vascular system: evolution, development and functions F. J. Integr. Plant Biol. 55, 294–388. doi: 10.1111/jipb.12041
Ma, Q.-H. (2009). The expression of caffeic acid 3-O-methyltransferase in two wheat genotypes differing in lodging resistance. J. Exp. Bot. 60, 2763–2771. doi: 10.1093/jxb/erp132
Maccaferri, M., Ricci, A., Salvi, S., Milner, S. G., Noli, E., Martelli, P. L., et al. (2015). A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 13, 648–663. doi: 10.1111/pbi.12288
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
Milach, S. C. K., and Federizzi, L. C. (2001). Dwarfing genes in plant improvement. Adv. Agron. 73, 35–63. doi: 10.1016/s0065-2113(01)73004-0
Mohammad, A., Muhammad, A., and Muhammad, S. (2021). Genetic improvement in physiological traits of Rice yield. Genetic Improvement of Field Crops 73, 413–455. doi: 10.1201/9781003210238-9
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Niu, Y., Chen, T., Wang, C., Chen, K., Shen, C., Chen, H., et al. (2021). Identification and allele mining of new candidate genes underlying rice grain weight and grain shape by genome-wide association study. BMC Genomics 22:602. doi: 10.1186/s12864-021-07901-x
Oda, Y., and Fukuda, H. (2014). Emerging roles of small GTPases in secondary cell wall development. Front. in Plant Sci. 5:428. doi: 10.3389/fpls.2014.00428
Oh, M.-H., Honey, S. H., and Tax, F. E. (2020). The control of cell expansion, cell division, and vascular development by Brassinosteroids: A historical perspective. Int. J. Mol. Sci. 21:1743. doi: 10.3390/ijms21051743
Oiestad, A. J., Martin, J. M., Cook, J., Varella, A. C., and Giroux, M. J. (2017). Identification of candidate genes responsible for stem pith production using expression analysis in solid-stemmed wheat. Plant Genome 10:0008. doi: 10.3835/plantgenome2017.02.0008
Ooijen, J. W. V., Verlaat, J. V. T., Tol, J., Dalh, N. J., et al. (2006). JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen, Netherlands: Plant Research International BV and Kayazma BV.
Ouellette, L. A., Reid, R. W., Blanchard, S. G., and Brouwer, C. R. (2017). LinkageMapView—rendering high-resolution linkage and QTL maps. Bioinformatics 34, 306–307. doi: 10.1093/bioinformatics/btx576
Packa, D., Wiwart, M., Suchowilska, E., and Bienkowska, T. (2015). Morpho-anatomical traits of two lowest internodes related to lodging resistance in selected genotypes of Triticum. Int. Agrophysics 29, 475–483. doi: 10.1515/intag-2015-0053
Piñera-Chavez, F. J., Berry, P. M., Foulkes, M. J., Sukumaran, S., and Reynolds, M. P. (2021). Identifying quantitative trait loci for lodging-associated traits in the wheat doubled-haploid population Avalon × cadenza. Crop Sci. 61, 2371–2386. doi: 10.1002/csc2.20485
Qi, J., Qian, Q., Bu, Q., Li, S., Chen, Q., Sun, J., et al. (2008). Mutation of the rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959. doi: 10.1104/pp.108.118778
Qu, P., Wang, J., Wen, W., Gao, F., Liu, J., Xia, X., et al. (2021). Construction of consensus genetic map with applications in gene mapping of wheat (Triticum aestivum L.) using 90K SNP array. Front. in Plant Sci. 12:727077. doi: 10.3389/fpls.2021.727077
Sang, Y., Deng, Z. Y., Zhao, L., Zhang, K. P., Tian, J. C., and Ye, B. X. (2010). QTLs for the vascular bundle system of the uppermost internode using a doubled haploid population of two elite Chinese wheat cultivars. Plant Breed. 129, 605–610. doi: 10.1111/j.1439-0523.2009.01727.x
Shah, L., Yahya, M., Shah, S. M. A., Nadeem, M., Ali, A., Ali, A., et al. (2019). Improving lodging resistance: using wheat and rice as classical examples. Int. J. Mol. Sci. 20:4211. doi: 10.3390/ijms20174211
Sherman, J., Blake, N. K., Martin, J. M., Kephart, K. D., Smith, J., Clark, D. R., et al. (2015). Agronomic impact of a stem solidness gene in near-isogenic lines of wheat. Crop Sci. 55, 514–520. doi: 10.2135/cropsci2014.05.0403
Sourdille, P., Singh, S., Cadalen, T., Brown-Guedira, G. L., Gay, G., Qi, L., et al. (2004). Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genomics 4, 12–25. doi: 10.1007/s10142-004-0106-1
Terao, T., Nagata, K., Morino, K., and Hirose, T. (2010). A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor. Appl. Genet. 120, 875–893. doi: 10.1007/s00122-009-1218-8
Van Ooijen, J. (2009). MapQTL® 6, Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species. Kyazma BV: Wageningen, Netherlands, 64.
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wang, J., Zhu, J., Lin, Q., Li, X., Teng, N., Li, Z., et al. (2006). Effects of stem structure and cell wall components on bending strength in wheat. Chin. Sci. Bull. 51, 815–823. doi: 10.1007/s11434-006-0815-z
Wen, W., He, Z., Gao, F., Liu, J., Jin, H., Zhai, S., et al. (2017). A high-density consensus map of common wheat integrating four mapping populations scanned by the 90K SNP array. Front. in Plant Sci. 8:1389. doi: 10.3389/fpls.2017.01389
Wingen, L. U., West, C., Leverington-Waite, M., Collier, S., Orford, S., Goram, R., et al. (2017). Wheat landrace genome diversity. Genetics 205, 1657–1676. doi: 10.1534/genetics.116.194688
Wolde, G. M., and Schnurbusch, T. (2019). Inferring vascular architecture of the wheat spikelet based on resource allocation in the branched headt (bht-A1) near isogenic lines. Funct. Integr. Genomics 46:1023, –1035. doi: 10.1071/fp19041
Wu, H.-M., Hazak, O., Cheung, A. Y., and Yalovsky, S. (2011). RAC/ROP GTPases and auxin signaling. Plant Cell 23, 1208–1218. doi: 10.1105/tpc.111.083907
Xiong, H., Zhou, C., Fu, M., Guo, H., Xie, Y., Zhao, L., et al. (2022). Cloning and functional characterization of Rht8, a “green revolution” replacement gene in wheat. Mol. Plant 15, 373–376. doi: 10.1016/j.molp.2022.01.014
Xu, B.-J., Chen, Q., Zheng, T., Jiang, Y.-F., Qiao, Y.-Y., Guo, Z.-R., et al. (2018). An overexpressed Q allele leads to increased spike density and improved processing quality in common wheat (Triticum aestivum). G3 Genes|Genomes|Genetics 8, 771–778. doi: 10.1534/g3.117.300562
Yingying, D., Chen, L., Wang, Y., Yang, Z., Saeed, I., Daoura Goudia, B., et al. (2018). The combination of dwarfing genes Rht4 and Rht8 reduced plant height, improved yield traits of rainfed bread wheat (Triticum aestivum L.). Field Crops Res. 215, 149–155. doi: 10.1016/j.fcr.2017.10.015
Yong, W., Sishen, L., Zengjun, Q., Anfei, L., Honggang, W., and Qingqi, L. (1998). Gene effects and heterosis of lodging resistance traits in wheat. Acta Bot. Sin. 18, 514–520.
Zhai, L., Zheng, T., Wang, X., Wang, Y., Chen, K., Wang, S., et al. (2018). QTL mapping and candidate gene analysis of peduncle vascular bundle related traits in rice by genome-wide association study. Rice 11:13. doi: 10.1186/s12284-018-0204-7
Zhang, W. J., Wu, L. M., Ding, Y. F., Weng, F., Wu, X. R., Li, G. H., et al. (2016a). Top-dressing nitrogen fertilizer rate contributes to decrease culm physical strength by reducing structural carbohydrate content in japonica rice. J. Integr. Agric. 15, 992–1004. doi: 10.1016/S2095-3119(15)61166-2
Zhang, Y., Xu, W., Wang, H., Fang, Y., Dong, H., and Qi, X. (2016b). Progress in improving stem lodging resistance of Chinese wheat cultivars. Euphytica 212, 275–286. doi: 10.1007/s10681-016-1768-1
Zhang, L., Zhang, H., Qiao, L., Miao, L., Yan, D., Liu, P., et al. (2021). Wheat MADS-box gene TaSEP3-D1 negatively regulates heading date. Crop J. 9, 1115–1123. doi: 10.1016/j.cj.2020.12.007
Zhong, R., and Ye, Z.-H. (2004). Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45, 369–385. doi: 10.1093/pcp/pch051
Keywords: lodging, genetic map, vascular bundle characters, plant height, yield contributing traits
Citation: Niu Y, Chen T, Zhao C, Guo C and Zhou M (2022) Identification of QTL for Stem Traits in Wheat (Triticum aestivum L.). Front. Plant Sci. 13:962253. doi: 10.3389/fpls.2022.962253
Received: 06 June 2022; Accepted: 21 June 2022;
Published: 14 July 2022.
Edited by:
Yuanhu Xuan, Shenyang Agricultural University, ChinaReviewed by:
Jianmin Bian, Jiangxi Agricultural University, ChinaCopyright © 2022 Niu, Chen, Zhao, Guo and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meixue Zhou, bWVpeHVlLnpob3VAdXRhcy5lZHUuYXU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.