- 1Department of Biology, Amherst College, Amherst, MA, United States
- 2Division of Natural and Applied Sciences, Duke Kunshan University, Kunshan, China
Plants utilize delicate mechanisms to effectively respond to changes in the availability of nutrients such as iron. The responses to iron status involve controlling gene expression at multiple levels. The regulation of iron deficiency response by a network of transcriptional regulators has been extensively studied and recent research has shed light on post-translational control of iron homeostasis. Although not as considerably investigated, an increasing number of studies suggest that histone modification and DNA methylation play critical roles during iron deficiency and contribute to fine-tuning iron homeostasis in plants. This review will focus on the current understanding of chromatin-based regulation on iron homeostasis in plants highlighting recent studies in Arabidopsis and rice. Understanding iron homeostasis in plants is vital, as it is not only relevant to fundamental biological questions, but also to agriculture, biofortification, and human health. A comprehensive overview of the effect and mechanism of chromatin-based regulation in response to iron status will ultimately provide critical insights in elucidating the complexities of iron homeostasis and contribute to improving iron nutrition in plants.
Introduction
Plants evolved complex regulatory mechanisms to cope with changes in the environment, including nutrient availability (Secco et al., 2017). At the molecular level, plants respond to nutritional status by modulating gene expression at multiple levels through a network of transcription factors and via post-translational regulation. Multiple studies have also revealed that changes in chromatin state by histone modification or DNA methylation play important roles in nutrient homeostasis in plants (Secco et al., 2017; Séré and Martin, 2020).
Post-translational modification of histone and DNA methylation lead to transcriptional regulation by altering chromatin packaging and chemical properties of the nucleosome surface, both of which influence association of DNA-binding transcriptional regulators (Berger, 2007). Emerging evidence reveals the importance of chromatin regulation in nutritional homeostasis in plants. For example, phosphate starvation-induced genes are regulated by the histone acetyltransferase GCN5 (Wang T. et al., 2019) and histone deacetylases HDA19 and HDC1 (Chen et al., 2015; Xu et al., 2020). Histone 3 lysine 4 trimethylation (H3K4me3) was also shown to regulate gene expression under phosphate deficiency (Chandrika et al., 2013a,b). In high nitrogen, increased H3K27me3 deposition, and decreased H3K4me3 and H3K36me3 contribute to the repression of the high affinity nitrate transporter gene, AtNRT2.1 (Widiez et al., 2011). H3K27me3 also modulates AtNRT2.1 by limiting its induction under low nitrogen (Bellegarde et al., 2018). Multiple genes involved in sulfate uptake and assimilation are direct targets of histone methylation and acetylation (Huang et al., 2019). In addition, global changes in DNA methylation were observed under phosphate starvation (Yong-Villalobos et al., 2015; Secco et al., 2017), sulfur deficiency (Huang et al., 2016), and zinc deficiency (Chen et al., 2018). Chromatin remodeling genes were differentially expressed upon zinc or iron treatment, implying chromatin-level responses to maintain mineral homeostasis (Darbani et al., 2015). Although chromatin remodeling has not been extensively studied in the context of metal homeostasis, reports increasingly suggest the involvement of histone modification and DNA methylation in regulating iron. This minireview will focus on the current knowledge of chromatin-based regulation of iron homeostasis in plants.
Iron is an essential micronutrient for plant growth and development. Iron is an indispensable cofactor in vital metabolic processes, but improperly regulated iron causes cytotoxicity by facilitating the generation of reactive oxygen species (ROS) (Halliwell and Gutteridge, 1992). Despite being abundant in the soil, iron is not readily accessible for plants, as it is highly insoluble in aerobic conditions at neutral or alkaline pH (Colombo et al., 2014). Iron’s importance as an essential micronutrient with low bioavailability and its potential for toxicity necessitates a tightly regulated system of iron acquisition and regulation in plants. Understanding iron homeostasis is important to answer fundamental biological questions, but also to improve agriculture and human health.
Iron deficiency response and iron uptake
In response to iron deficiency, plants induce iron uptake mechanisms that involve reducing or chelating iron (Connorton et al., 2017; Riaz and Guerinot, 2021). Dicots acquire iron via a reduction-based process known as Strategy I, which involves proton efflux to the rhizosphere by proton ATPases such as AHA2 to solubilize ferric chelates (Santi et al., 2005), coumarin secretion to facilitate iron mobilization (Clemens and Weber, 2016), reduction of ferric chelates to ferrous iron by FERRIC REDUCTASE OXIDASE 2 (FRO2) (Robinson et al., 1999), and ferrous iron import into root epidermal cells by IRON-REGULATED TRANSPORTER 1 (IRT1) (Eide et al., 1996). IRT1, FRO2, and AHA2 co-localize in interactomes, which likely optimize iron uptake (Martín-Barranco et al., 2020). Grasses use a chelation-based process or Strategy II for iron uptake. When iron is limited, phytosiderophores, mugineic acid (MA) and its derivatives, are synthesized (Mori and Nishizawa, 1987; Shojima et al., 1990) and secreted into the rhizosphere by Transporter of Mugineic acid family phytosiderophores 1 (TOM1) to chelate iron (Nozoye et al., 2011). Fe3+-phytosiderophore complexes are then transported into the root epidermal cells by the Yellow Stripe (YS) family transporters (Curie et al., 2001). Even though grasses are considered as Strategy II plants, Strategy I is used or its components exist in graminaceous plants (Bughio et al., 2002; Ishimaru et al., 2006; Cheng et al., 2007; Bashir et al., 2011; Li et al., 2016; Kaur et al., 2019; Wairich et al., 2019; Wang M. et al., 2019).
Regulation of iron deficiency response
Responses to iron availability are controlled from transcriptional to post-translational levels (Vélez-Bermúdez and Schmidt, 2022). In particular, the complex network of basic helix-loop-helix (bHLH) family transcription factors involved in iron deficiency response has been extensively studied (Gao et al., 2020; Schwarz and Bauer, 2020). In Arabidopsis, FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT)/bHLH29 directly regulates IRT1, FRO2, FIT, and other genes involved in iron uptake under iron deficiency (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). FIT forms heterodimers with subgroup Ib bHLH transcription factors, bHLH038/39/100/101, to activate FIT-dependent gene expression (Yuan et al., 2008; Wang et al., 2013). FIT also interacts with subgroup IVa bHLHs, triggering the degradation of FIT via the 26S proteasome pathway (Cui et al., 2018). Alongside FIT, POPEYE (PYE)/bHLH47 is another major transcriptional regulator of iron deficiency response in Arabidopsis (Long et al., 2010). PYE is expressed under iron deficiency and negatively regulates its target genes, which include those involved in iron translocation, storage, and assimilation. ILR3/bHLH105 plays a dual role in iron homeostasis; depending on the heterodimer it forms, ILR3 activates PYE expression (Zhang J. et al., 2015) or represses PYE-target genes (Tissot et al., 2019). UPSTREAM REGULATOR OF IRT1 (URI)/bHLH121 directly or indirectly positively regulates multiple iron homeostasis genes of the bHLH network (Kim et al., 2019; Gao et al., 2020; Lei et al., 2020). URI controls nearly half of iron-regulated genes, including both FIT-dependent and independent genes (Kim et al., 2019; Gao et al., 2020). Although URI expression is not iron-regulated, phosphorylation of its protein stabilizes it to form heterodimers with subgroup IVc bHLH transcription factors and activate subgroup Ib bHLH genes under iron deficiency (Kim et al., 2019). Upon iron re-supply, phosphorylated URI is targeted by the E3 ligase BRUTUS (BTS) and subjected to proteasome-mediated degradation. The IRONMAN/FE-UPTAKE-INDUCING PEPTIDE (IMA/FEP) peptides also positively regulate iron deficiency response in Arabidopsis (Grillet et al., 2018; Hirayama et al., 2018) by sequestering BTS to prevent degradation of bHLH105/bHLH115 and activate iron uptake (Li et al., 2021).
Responses to iron deficiency in grasses also utilize several bHLH transcription factors (Gao and Dubos, 2021). OsFIT/OsbHLH156 positively regulates Strategy II-related genes such as those involved in MA biosynthesis and also regulates OsIRT1, a Strategy I-related gene (Liang et al., 2020; Wang et al., 2020). OsIRO2 interacts with OsFIT to promote its nuclear localization and positively regulate iron uptake by OsIRT1 (Ogo et al., 2006, 2007; Liang et al., 2020; Wang et al., 2020). OsIRO3/OsbHLH63 represses iron deficiency response possibly via antagonizing OsIRO2 to avoid iron overload by limiting iron uptake (Zheng et al., 2010; Gao and Dubos, 2021).
Iron homeostasis and histone modification
Each nucleosome consists of an octameric complex of histones subjected to a wide range of post-translational modifications. These modifications are reversible but are controlled by many histone modifying enzymes and play key roles in regulating chromatin structure and transcription (Bannister and Kouzarides, 2011; Zhang T. et al., 2015). Multiple iron homeostasis genes in Arabidopsis have been found to be controlled by histone modifications as discussed in this section.
H3K4me3
H3K4me3, the trimethylation of histone 3 lysine 4, generally leads to gene activation (Liu et al., 2010; Xiao et al., 2016). Using a forward genetics screen in Arabidopsis, Singh et al. (2021) identified a regulator of iron deficiency response, NON-RESPONSE TO Fe-DEFICIENCY2 (NRF2). In Arabidopsis, NRF2 is known as EARLY FLOWERING8 (ELF8), which regulates FLOWERING LOCUS C (FLC) expression via H3K4me3 (He, 2009). NRF2/ELF8 belongs to the trithorax group (TrxG) methyltransferases that modify histones to activate genes via relaxing chromatin structure and serve as antagonistic regulators of polycomb group proteins (Schuettengruber et al., 2011).
Under iron deficiency, AtNRF2/ELF8 is required for AtGRF11 expression as it modulates H3K4me3 levels at its transcription start site (Singh et al., 2021). While AtGRF11 does not directly interact with AtFIT, it acts downstream of NO to induce AtFIT expression in iron deficient roots (Singh et al., 2021). In the nrf2 mutant, AtGRF11-regulated iron uptake was repressed and iron transport and storage genes were downregulated. The mutant normally induced NO under iron deficiency, suggesting that the repression of AtGRF11 was solely responsible for the regulation of iron uptake genes (Singh et al., 2021).
H3K4me3 also likely regulates the expression of iron storage genes AtFERRITIN1 (FER1), AtFER3, and AtFER4 in iron sufficient seedlings (Tissot et al., 2019). At the promoter regions of these ferritin genes, activation marks such as H3K4me3 and histone 3 lysine 9 acetylation (H3K9ac) were detected in seedlings grown under iron sufficient conditions, whereas H3K27me3 was not present based on analysis of publicly available epigenome profiles (Tissot et al., 2019; Park et al., 2020).
H3K27me3
The trimethylation of histone 3 lysine 27 (H3K27me3) is typically associated with gene repression; it spreads along the chromatin, resulting in compaction and the silencing of targeted genes (Liu et al., 2010; Xiao et al., 2016). H3K27me3 is catalyzed by Polycomb Repressive Complex 2 (PRC2) (Margueron and Reinberg, 2011). CURLY LEAF (CLF) is a predominant methyltransferase of the core PRC2 complex (Chanvivattana et al., 2004; Schubert et al., 2006; Zhang et al., 2007). In Arabidopsis, H3K27me3 was found to modulate the expression of FIT-dependent genes by directly targeting their loci (Park et al., 2019). Under iron deficiency, the expression of FIT-dependent genes, such as AtFIT, AtIRT1, AtFRO2, and AtF6’H1, was significantly higher in clf than in wild type roots, and their transcript levels inversely correlated with H3K27me3 deposition on their loci (Park et al., 2019). However, expression of PYE-dependent genes was not significantly affected (Park et al., 2019). Transcriptomic analysis revealed that transcript levels of FIT-dependent genes were consistently higher in clf even under iron-sufficient conditions where FIT-dependent gene expression is extremely low, but the lack of the H3K27me3 mark in iron sufficient clf mutants was not sufficient to fully induce FIT-dependent genes when upstream iron-deficiency signals were not present. In iron-deficient conditions, the residual H3K27me3 on FIT-dependent genes may be attenuating the induction of iron acquisition genes to limit their maximum induction to prevent plants from iron-induced cytotoxicity.
H3K27me3 was also implicated to play a role in iron translocation from roots to shoots in Arabidopsis (Park et al., 2020). Iron-deficient clf mutants accumulated less iron in the roots than the shoots, but clf seedlings still had higher levels of iron compared to wild type. This phenotype and the higher expression of iron acquisition genes in clf roots (Park et al., 2019) suggest that clf mutants may still be acquiring more iron without retention in the roots due to greater translocation (Park et al., 2020). Indeed, the expression of AtYSL1, which encodes an iron-NA transporter involved in supplying iron to sink tissues (Waters et al., 2006), was significantly increased in clf compared to wild type and AtYSL1 was verified to be a direct target of H3K27me3 (Park et al., 2020). AtIMA1 was also revealed to be a direct target of H3K27me3, but under iron deficiency, H3K27me3 appears to play a limited role in regulating AtIMA1 expression (Park et al., 2020).
H4R3sme2
Shk1 binding protein 1 (SKB1) catalyzes the symmetric dimethylation of histone4 arginine3 (H4R3sme2) and regulates diverse biological processes including response to salt stress (Niu et al., 2007; Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008; Zhang et al., 2011). SKB1-mediated H4R3sme2 also affects iron homeostasis by negatively modulating the expression of Ib subgroup bHLH genes that encode FIT-interacting partners, such as AtbHLH38/39/100/101, in response to iron (Fan et al., 2014). While AtSKB1 expression is not regulated by iron, the level of SKB1 association and H4R3sme2 deposition on the Ib subgroup bHLH loci positively correlated with the iron status of plants. As a result, transcript levels of the Ib subgroup AtbHLH genes and its downstream genes including AtFRO2 and AtIRT1 that are not direct targets of SKB1 were higher in skb1 mutants than in wild type roots. Although SKB1 did not affect AtFIT expression, transcript levels of AtFRO2 and AtIRT1 were not significantly increased in the skb1 fit1 double mutant, indicating that the negative regulation of iron acquisition genes by SKB1 was dependent on FIT (Fan et al., 2014). The mechanism by which SKB1 perceives iron levels and other environmental signals to determine the degree of H4R3sme2 in specific genes remains to be understood.
Histone acetylation
Histone acetylation is generally associated with transcriptional activation, in contrast to the more complex effects of histone methylation on gene expression (Berger, 2007). The combined action of histone acetylation and deacetylation is crucial for regulating gene expression (Grunstein, 1997). GENERAL CONTROL NON-REPRESSED PROTEIN5 (GCN5) is responsible for the acetylation of H3K14 and facilitates the acetylation of H3K9 and H3K27, which are required for the expression of a large number of genes (Vlachonasios et al., 2003; Earley et al., 2007; Benhamed et al., 2008).
Xing et al. (2015) reported that AtGCN5 contributes to iron homeostasis by modulating the expression of Arabidopsis FERRIC REDUCTASE DETECTIVE3 (AtFRD3), which encodes a transporter that loads citrate into the xylem to aid translocation of iron-citrate complexes to the shoots (Durrett et al., 2007). AtGCN5 directly binds to the promoters of AtFRD3 and other iron responsive genes to control H3K9ac and/or H3K14ac levels. In the gcn5 mutant, iron-related phenotypes similar to those of frd3 were observed due to significantly decreased H3K9ac and/or H3K14ac deposition at the AtFRD3 locus and reduced expression of AtFRD3 (Xing et al., 2015). In the mutants of two histone deacetylases, hda7 and hda14, AtFRD3 transcript level was increased, providing an example of the coordination between histone acetylation and deacetylation to precisely regulate gene expression (Xing et al., 2015).
Iron homeostasis and DNA methylation
DNA methylation controls gene expression and contributes to silencing of transposons to maintain genome stability (Law and Jacobsen, 2010; Zhang et al., 2018). In plants, methylation of cytosine occurs in symmetric methylation at CG and CHG, where H represents A, T, or C, and asymmetric methylation at CHH (Matzke and Mosher, 2014; Matzke et al., 2015). While CG and CHG methylations are maintained during DNA replication, CHH methylations are established de novo after DNA replication via RNA-dependent mechanisms and are frequently found between condensed and relaxed chromatin near highly expressed genes (Gent et al., 2013; Martin et al., 2021).
A recent report suggested that CHH DNA methylation modulates iron deficiency response in rice via changing methylation status of genes encoding two major positive regulators of iron deficiency response, OsIRO2 and OsbHLH156 (Sun et al., 2021). In this study, widespread hypermethylation, mainly CHH methylation, was detected in rice roots and shoots grown in iron deficient conditions by mapping the DNA methylome at a single-base resolution. Although little correlation was found between CHH hypermethylation and expression of iron deficiency response genes, OsIRO2 and OsbHLH156 exhibited CHH hypermethylation and their expression increased under iron deficiency. Furthermore, treatment of 5-aza-2-deoxycytidine (Aza), a DNA methylation inhibitor, and the loss of OsDRM2, a key methyltransferase responsible for CHH methylation, resulted in lower expression of OsIRO2 and OsbHLH156, accumulation of less iron, and growth retardment under iron deficiency (Sun et al., 2021). It was speculated that small RNAs might play a critical role as rice acclimates to iron deficiency, as the levels of 24-nt siRNAs increased, whereas transcript levels of canonical RNA-dependent DNA methyltransferases involved in CHH methylation did not change under iron deficient conditions (Sun et al., 2021).
In barley, iron deficiency led to a general reduction of CG methylation, but the overall methylation and demethylation status was not recovered after iron resupply (Bocchini et al., 2015). Further studies are necessary to understand the extent to which DNA methylation or demethylation is maintained upon changes in iron conditions and mechanisms therein.
DNA methylation status was also proposed to be involved in feedback mechanisms between iron status and tolerance to cadmium stress (Fan et al., 2020). Arabidopsis plants exposed to cadmium stress expressed lower levels of the three DNA demethylase genes AtROS1/DML2/DML3 (RDD) and exhibited increased global DNA methylation that resembled the methylation profile of rdd triple mutants (Fan et al., 2020). The rdd mutants were more tolerant against cadmium stress and accumulated more iron in the shoots by expressing higher levels of iron deficiency response genes than wild type. However, inadequate iron supply abolished cadmium tolerance in rdd mutants (Fan et al., 2020).
Conclusion and perspectives
Increasing evidence has shown that iron homeostasis gene expression is affected by histone modification (Figure 1) and DNA methylation (Figure 2). Such chromatin-based regulation is critical during iron deficiency and allows to fine-tune iron homeostasis in plants. Given that chromatin-based regulation is a dynamic process, it will be important to understand the mechanistic details regarding changes in histone modification or DNA methylation in response to changes in iron status. Research to date has mainly focused on iron deficiency and little is known about the effect of iron overload on chromatin remodeling via histone modification or DNA methylation. Global changes in H3K9me2 and H3K4me3 levels under high iron stress conditions were detected in the proximal root meristem in rice (Polosoro et al., 2019), but further studies are needed to understand the underlying mechanisms and the biological implications. Furthermore, it will be necessary to integrate large scale datasets of various histone modifications, DNA methylation, and the combinatorial effect of different modifications, as well as comparative analyses of transcriptomics and epigenetics of specific cell-types or at a single cell level. Although chromatin-based regulation is an integral part of epigenetics, some chromatin modifications are not heritable or considered epigenetic (Eichten et al., 2014). Thus, transgenerational studies to determine the heritability of chromatin modifications in response to iron will lead to insightful information. Considering the growing evidence that reveal the significance of dynamic adjustment in chromatin structure and subsequent transcriptional changes in response to nutritional status, a clear understanding of chromatin-based iron homeostasis is necessary for a comprehensive understanding of iron homeostasis. Such efforts will contribute insights toward developing crops with improved nutritional profiles and enhanced tolerance to undesirable conditions in the long run.
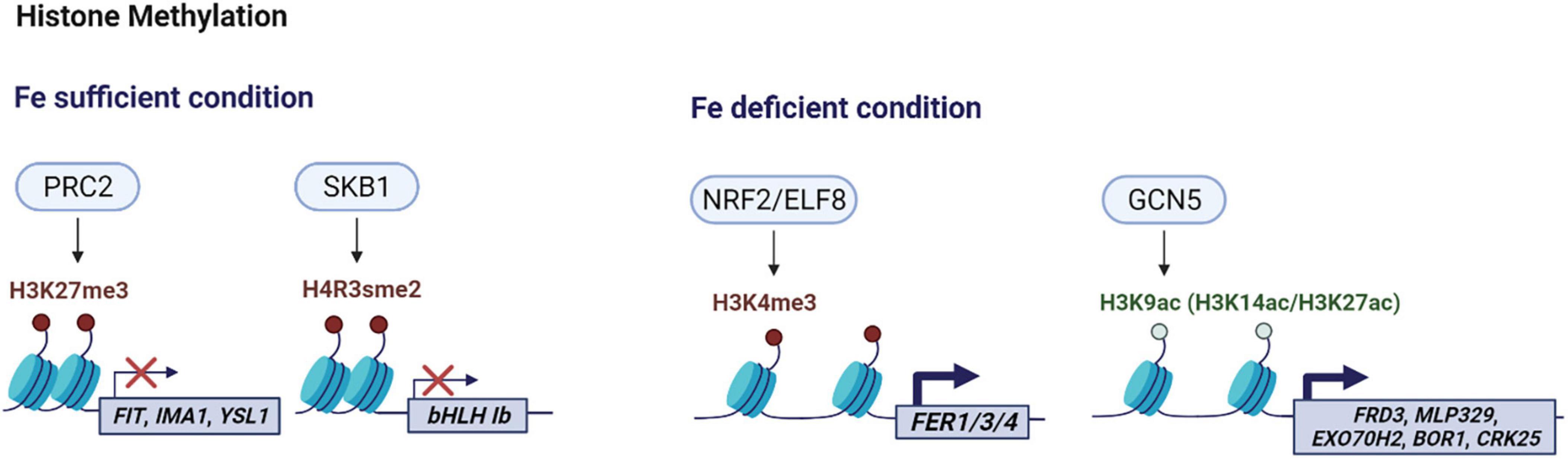
Figure 1. Schematic overview of histone modifications involved in iron homeostasis in Arabidopsis. Under iron sufficiency, PRC2-mediated H3K27me3 induces chromatin condensation in Arabidopsis, resulting in gene silencing of the target genes such as AtFIT, AtIMA1 and AtYSL1. AtSKB1-induced H4R3sme2 also suppresses AtbHLH1b transcripts when iron is sufficient. Under iron deficiency, AtNRF2/ELF8 catalyzes the trimethylation of H3K4 generating H3K4me3, and AtGCN5 acetylates H3K9 producing H3K9ac and facilitates the generation of H3K14ac and H3K27ac to activate corresponding target genes. The color scheme denotes methylation (red), acetylation (light green), histone (light blue). This figure was created with BioRender.com.
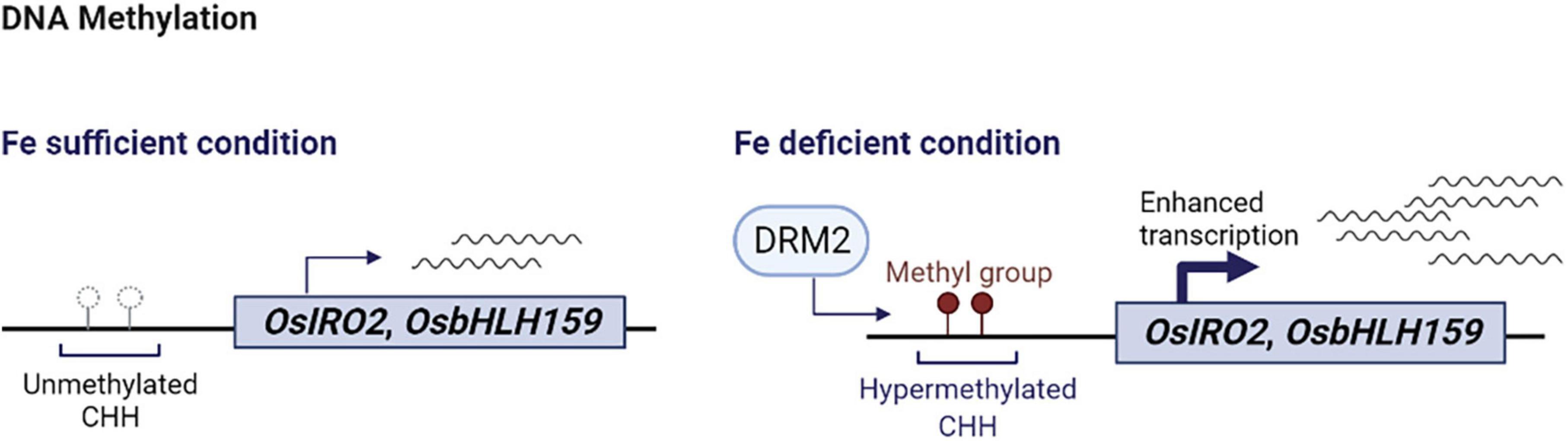
Figure 2. Schematic summary of DNA hypermethylation and iron deficiency response in rice. Under iron sufficient conditions, the CHH sequences of OsIRO2 and OsbHLH159 promoters remain unmethylated and basal levels of OsIRO2 and OsbHLH159 are expressed. Upon iron deficiency, hypermethylation of CHH nucleotides on the promoters of OsIRO2 and OsbHLH159 by DRM2 leads to activation of the expression of the corresponding downstream genes in response to iron deficiency. This figure was created with BioRender.com.
Author contributions
JS primarily wrote the initial draft of the manuscript. ZY and YW contributed to the manuscript writing and generated the figures. JL and JJ conceived the idea and made final edits. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Gregory Call Student Research Fund to JS, the Interdisciplinary Seed Grant, Synear and Wang-Cai Seed Grant, and Wang-Cai Biochemistry Lab Grant to JL, and the National Science Foundation grant (IOS#1754969) and the Alex Schupf ’57 Fund for Intellectual Life to JJ.
Acknowledgments
We regret that we were unable to cite all relevant literature due to restrictions in the length of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi: 10.1038/cr.2011.22
Bashir, K., Ishimaru, Y., Shimo, H., Kakei, Y., Senoura, T., Takahashi, R., et al. (2011). Rice phenolics efflux transporter 2 (PEZ2) plays an important role in solubilizing apoplasmic iron. Soil Sci. Plant Nutr. 57, 803–812. doi: 10.1080/00380768.2011.637305
Bellegarde, F., Herbert, L., Séré, D., Caillieux, E., Boucherez, J., Fizames, C., et al. (2018). Polycomb repressive complex 2 attenuates the very high expression of the Arabidopsis gene NRT2.1. Sci. Rep. 8:7905. doi: 10.1038/s41598-018-26349-w
Benhamed, M., Martin-Magniette, M.-L., Taconnat, L., Bitton, F., Servet, C., De Clercq, R., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56, 493–504. doi: 10.1111/j.1365-313X.2008.03606.x
Berger, S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447, 407–412. doi: 10.1038/nature05915
Bocchini, M., Bartucca, M. L., Ciancaleoni, S., Mimmo, T., Cesco, S., Pii, Y., et al. (2015). Iron deficiency in barley plants: Phytosiderophore release, iron translocation, and DNA methylation. Front. Plant Sci. 6:514. doi: 10.3389/fpls.2015.00514
Bughio, N., Yamaguchi, H., Nishizawa, N. K., Nakanishi, H., and Mori, S. (2002). Cloning an iron-regulated metal transporter from rice. J. Exp. Bot. 53, 1677–1682. doi: 10.1093/jxb/erf004
Chandrika, N. N. P., Sundaravelpandian, K., and Schmidt, W. (2013a). A PHD in histone language: On the role of histone methylation in plant responses to phosphate deficiency. Plant Signal. Behav. 8:e24381. doi: 10.4161/psb.24381
Chandrika, N. N. P., Sundaravelpandian, K., Yu, S.-M., and Schmidt, W. (2013b). ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. N. Phytol. 198, 709–720. doi: 10.1111/nph.12194
Chanvivattana, Y., Bishopp, A., Schubert, D., Stock, C., Moon, Y.-H., Sung, Z. R., et al. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276. doi: 10.1242/dev.01400
Chen, C.-Y., Wu, K., and Schmidt, W. (2015). The histone deacetylase HDA19 controls root cell elongation and modulates a subset of phosphate starvation responses in Arabidopsis. Sci. Rep. 5:15708. doi: 10.1038/srep15708
Chen, X., Schï Nberger, B., Menz, J., and Ludewig, U. (2018). Plasticity of DNA methylation and gene expression under zinc deficiency in Arabidopsis roots. Plant Cell Physiol. 59, 1790–1802. doi: 10.1093/pcp/pcy100
Cheng, L., Wang, F., Shou, H., Huang, F., Zheng, L., He, F., et al. (2007). Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol. 145, 1647–1657. doi: 10.1104/pp.107.107912
Clemens, S., and Weber, M. (2016). The essential role of coumarin secretion for Fe acquisition from alkaline soil. Plant Signal. Behav. 11:e1114197. doi: 10.1080/15592324.2015.1114197
Colangelo, E. P., and Guerinot, M. L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16, 3400–3412. doi: 10.1105/tpc.104.024315
Colombo, C., Palumbo, G., He, J.-Z., Pinton, R., and Cesco, S. (2014). Review on iron availability in soil: Interaction of Fe minerals, plants, and microbes. J. Soils Sediments 14, 538–548. doi: 10.1007/s11368-013-0814-z
Connorton, J. M., Balk, J., and Rodríguez-Celma, J. (2017). Iron homeostasis in plants - a brief overview. Metallomics 9, 813–823. doi: 10.1039/c7mt00136c
Cui, Y., Chen, C.-L., Cui, M., Zhou, W.-J., Wu, H.-L., and Ling, H.-Q. (2018). Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol. Plant 11, 1166–1183. doi: 10.1016/j.molp.2018.06.005
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., and Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
Darbani, B., Noeparvar, S., and Borg, S. (2015). Deciphering mineral homeostasis in barley seed transfer cells at transcriptional level. PLoS One 10:e0141398. doi: 10.1371/journal.pone.0141398
Durrett, T. P., Gassmann, W., and Rogers, E. E. (2007). The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144, 197–205. doi: 10.1104/pp.107.097162
Earley, K. W., Shook, M. S., Brower-Toland, B., Hicks, L., and Pikaard, C. S. (2007). In vitro specificities of Arabidopsis co-activator histone acetyltransferases: Implications for histone hyperacetylation in gene activation. Plant J. 52, 615–626. doi: 10.1111/j.1365-313X.2007.03264.x
Eichten, S. R., Schmitz, R. J., and Springer, N. M. (2014). Epigenetics: Beyond chromatin modifications and complex genetic regulation. Plant Physiol. 165, 933–947. doi: 10.1104/pp.113.234211
Eide, D., Broderius, M., Fett, J., and Guerinot, M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 5624–5628. doi: 10.1073/pnas.93.11.5624
Fan, H., Zhang, Z., Wang, N., Cui, Y., Sun, H., Liu, Y., et al. (2014). SKB1/PRMT5-mediated histone H4R3 dimethylation of Ib subgroup bHLH genes negatively regulates iron homeostasis in Arabidopsis thaliana. Plant J. 77, 209–221. doi: 10.1111/tpj.12380
Fan, S. K., Ye, J. Y., Zhang, L. L., Chen, H. S., Zhang, H. H., Zhu, Y. X., et al. (2020). Inhibition of DNA demethylation enhances plant tolerance to cadmium toxicity by improving iron nutrition. Plant Cell Environ. 43, 275–291. doi: 10.1111/pce.13670
Gao, F., and Dubos, C. (2021). Transcriptional integration of plant responses to iron availability. J. Exp. Bot. 72, 2056–2070. doi: 10.1093/jxb/eraa556
Gao, F., Robe, K., Bettembourg, M., Navarro, N., Rofidal, V., Santoni, V., et al. (2020). The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. Plant Cell 32, 508–524. doi: 10.1105/tpc.19.00541
Gent, J. I., Ellis, N. A., Guo, L., Harkess, A. E., Yao, Y., Zhang, X., et al. (2013). CHH islands: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23, 628–637. doi: 10.1101/gr.146985.112
Grillet, L., Lan, P., Li, W., Mokkapati, G., and Schmidt, W. (2018). IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 4, 953–963. doi: 10.1038/s41477-018-0266-y
Grunstein, M. (1997). Histone acetylation in chromatin structure and transcription. Nature 389, 349–352. doi: 10.1038/38664
Halliwell, B., and Gutteridge, J. M. C. (1992). Biologically relevant metal ion-dependent hydroxyl radical generation. FEBS Lett. 307, 108–12. doi: 10.1016/0014-5793(92)80911-y
He, Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2, 554–564. doi: 10.1093/mp/ssp005
Hirayama, T., Lei, G. J., Yamaji, N., Nakagawa, N., and Ma, J. F. (2018). The putative peptide gene FEP1 regulates iron deficiency response in arabidopsis. Plant Cell Physiol. 59, 1739–1752. doi: 10.1093/pcp/pcy145
Huang, X.-Y., Chao, D.-Y., Koprivova, A., Danku, J., Wirtz, M., Müller, S., et al. (2016). Nuclear Localised MORE SULPHUR ACCUMULATION1 epigenetically regulates sulphur homeostasis in Arabidopsis thaliana. PLoS Genet. 12:e1006298. doi: 10.1371/journal.pgen.1006298
Huang, X.-Y., Li, M., Luo, R., Zhao, F.-J., and Salt, D. E. (2019). Epigenetic regulation of sulfur homeostasis in plants. J. Exp. Bot. 70, 4171–4182. doi: 10.1093/jxb/erz218
Ishimaru, Y., Suzuki, M., Tsukamoto, T., Suzuki, K., Nakazono, M., Kobayashi, T., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45, 335–346. doi: 10.1111/j.1365-313X.2005.02624.x
Jakoby, M., Wang, H.-Y., Reidt, W., Weisshaar, B., and Bauer, P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577, 528–534. doi: 10.1016/j.febslet.2004.10.062
Kaur, G., Shukla, V., Kumar, A., Kaur, M., Goel, P., Singh, P., et al. (2019). Integrative analysis of hexaploid wheat roots identifies signature components during iron starvation. J. Exp. Bot. 70, 6141–6161. doi: 10.1093/jxb/erz358
Kim, S. A., LaCroix, I. S., Gerber, S. A., and Guerinot, M. L. (2019). The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. U.S.A. 116, 24933–24942. doi: 10.1073/pnas.1916892116
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Lei, R., Li, Y., Cai, Y., Li, C., Pu, M., Lu, C., et al. (2020). bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining fe homeostasis in Arabidopsis. Mol. Plant 13, 634–649. doi: 10.1016/j.molp.2020.01.006
Li, S., Zhou, X., Chen, J., and Chen, R. (2016). Is there a strategy I iron uptake mechanism in maize? Plant Signal. Behav. 13:e1161877. doi: 10.1080/15592324.2016.1161877
Li, Y., Lu, C. K., Li, C. Y., Lei, R. H., Pu, M. N., Zhao, J. H., et al. (2021). IRON MAN interacts with BRUTUS to maintain iron homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118:e2109063118. doi: 10.1073/pnas.2109063118
Liang, G., Zhang, H., Li, Y., Pu, M., Yang, Y., Li, C., et al. (2020). Oryza sativa fer-like fe deficiency-induced transcription factor (OsFIT/OsbHLH156) interacts with OsIRO2 to regulate iron homeostasis. J. Integr. Plant Biol. 62, 668–689. doi: 10.1111/jipb.12933
Liu, C., Lu, F., Cui, X., and Cao, X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., and Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236. doi: 10.1105/tpc.110.074096
Margueron, R., and Reinberg, D. (2011). The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349. doi: 10.1038/nature09784
Martin, G. T., Seymour, D. K., and Gaut, B. S. (2021). CHH Methylation Islands: A nonconserved feature of grass genomes that is positively associated with transposable elements but negatively associated with gene-body methylation. Genome Biol. Evol. 13:evab144. doi: 10.1093/gbe/evab144
Martín-Barranco, A., Spielmann, J., Dubeaux, G., Vert, G., and Zelazny, E. (2020). Dynamic control of the high-affinity iron uptake complex in root epidermal cells. Plant Physiol. 184, 1236–1250. doi: 10.1104/pp.20.00234
Matzke, M. A., and Mosher, R. A. (2014). RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. doi: 10.1038/nrg3683
Matzke, M. A., Kanno, T., and Matzke, A. J. M. (2015). RNA-directed DNA Methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66, 243–267. doi: 10.1146/annurev-arplant-043014-114633
Mori, S., and Nishizawa, N. (1987). Methionine as a dominant precursor of phytosiderophores in graminaceae plants. Plant Cell Physiol. 28, 1081–1092. doi: 10.1093/oxfordjournals.pcp.a077388
Niu, L., Lu, F., Pei, Y., Liu, C., and Cao, X. (2007). Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 8, 1190–1195. doi: 10.1038/sj.embor.7401111
Nozoye, T., Nagasaka, S., Kobayashi, T., Takahashi, M., Sato, Y., Sato, Y., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454. doi: 10.1074/jbc.M110.180026
Ogo, Y., Itai, R. N., Nakanishi, H., Inoue, H., Kobayashi, T., Suzuki, M., et al. (2006). Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 57, 2867–2878. doi: 10.1093/jxb/erl054
Ogo, Y., Itai, R. N., Nakanishi, H., Kobayashi, T., Takahashi, M., Mori, S., et al. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51, 366–377. doi: 10.1111/j.1365-313X.2007.03149.x
Park, E. Y., Tsuyuki, K. M., Hu, F., Lee, J., and Jeong, J. (2019). PRC2-Mediated H3K27me3 contributes to transcriptional regulation of FIT-dependent iron deficiency response. Front. Plant Sci. 10:627. doi: 10.3389/fpls.2019.00627
Park, E. Y., Tsuyuki, K. M., Parsons, E. M., and Jeong, J. (2020). PRC2-mediated H3K27me3 modulates shoot iron homeostasis in Arabidopsis thaliana. Plant Signal. Behav. 15:1784549. doi: 10.1080/15592324.2020.1784549.
Pei, Y., Niu, L., Lu, F., Liu, C., Zhai, J., Kong, X., et al. (2007). Mutations in the type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 144, 1913–1923. doi: 10.1104/pp.107.099531
Polosoro, A., Enggarini, W., and Ohmido, N. (2019). Global epigenetic changes of histone modification under environmental stresses in rice root. Chromosome Res. 27, 287–298. doi: 10.1007/s10577-019-09611-3
Riaz, N., and Guerinot, M. L. (2021). All together now: Regulation of the iron deficiency response. J. Exp. Bot. 72, 2045–2055. doi: 10.1093/jxb/erab003
Robinson, N. J., Procter, C. M., Connolly, E. L., and Guerinot, M. L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. doi: 10.1038/17800
Santi, S., Cesco, S., Varanini, Z., and Pinton, R. (2005). Two plasma membrane H(+)-ATPase genes are differentially expressed in iron-deficient cucumber plants. Plant Physiol. Biochem. 43, 287–292. doi: 10.1016/j.plaphy.2005.02.007
Schmitz, R. J., Sung, S., and Amasino, R. M. (2008). Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 411–416. doi: 10.1073/pnas.0710423104
Schubert, D., Primavesi, L., Bishopp, A., Roberts, G., Doonan, J., Jenuwein, T., et al. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25, 4638–4649. doi: 10.1038/sj.emboj.7601311
Schuettengruber, B., Martinez, A.-M., Iovino, N., and Cavalli, G. (2011). Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814. doi: 10.1038/nrm3230
Schwarz, B., and Bauer, P. (2020). FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. J. Exp. Bot. 71, 1694–1705. doi: 10.1093/jxb/eraa012
Secco, D., Whelan, J., Rouached, H., and Lister, R. (2017). Nutrient stress-induced chromatin changes in plants. Curr. Opin. Plant Biol. 39, 1–7. doi: 10.1016/j.pbi.2017.04.001
Séré, D., and Martin, A. (2020). Epigenetic regulation: Another layer in plant nutrition. Plant Signal. Behav. 15:1686236. doi: 10.1080/15592324.2019.1686236
Shojima, S., Nishizawa, N. K., Fushiya, S., Nozoe, S., Irifune, T., and Mori, S. (1990). Biosynthesis of phytosiderophores : In vitro biosynthesis of 2’-deoxymugineic acid from l-methionine and nicotianamine. Plant Physiol. 93, 1497–1503. doi: 10.1104/pp.93.4.1497
Singh, S., Kailasam, S., Lo, J.-C., and Yeh, K.-C. (2021). Histone H3 lysine4 trimethylation-regulated GRF11 expression is essential for the iron-deficiency response in Arabidopsis thaliana. N. Phytol. 230, 244–258. doi: 10.1111/nph.17130
Sun, S., Zhu, J., Guo, R., Whelan, J., and Shou, H. (2021). DNA methylation is involved in acclimation to iron-deficiency in rice (Oryza sativa). Plant J. 107, 727–739. doi: 10.1111/tpj.15318
Tissot, N., Robe, K., Gao, F., Grant-Grant, S., Boucherez, J., Bellegarde, F., et al. (2019). Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. N. Phytol. 223, 1433–1446. doi: 10.1111/nph.15753
Vélez-Bermúdez, I. C., and Schmidt, W. (2022). How plants recalibrate cellular iron homeostasis. Plant Cell Physiol. 63, 154–162. doi: 10.1093/pcp/pcab166
Vlachonasios, K. E., Thomashow, M. F., and Triezenberg, S. J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15, 626–638. doi: 10.1105/tpc.007922
Wairich, A., de Oliveira, B. H. N., Arend, E. B., Duarte, G. L., Ponte, L. R., Sperotto, R. A., et al. (2019). The combined strategy for iron uptake is not exclusive to domesticated rice (Oryza sativa). Sci. Rep. 9:16144. doi: 10.1038/s41598-019-52502-0
Wang, M., Kawakami, Y., and Bhullar, N. K. (2019). Molecular analysis of iron deficiency response in hexaploid wheat. Front. Sustain. Food Syst. 3:67. doi: 10.3389/fsufs.2019.00067
Wang, N., Cui, Y., Liu, Y., Fan, H., Du, J., Huang, Z., et al. (2013). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6, 503–513. doi: 10.1093/mp/sss089
Wang, S., Li, L., Ying, Y., Wang, J., Shao, J. F., Yamaji, N., et al. (2020). A transcription factor OsbHLH156 regulates strategy II iron acquisition through localising IRO2 to the nucleus in rice. N. Phytol. 225, 1247–1260. doi: 10.1111/nph.16232
Wang, T., Xing, J., Liu, Z., Zheng, M., Yao, Y., Hu, Z., et al. (2019). Histone acetyltransferase GCN5-mediated regulation of long non-coding RNA At4 contributes to phosphate starvation response in Arabidopsis. J. Exp. Bot. 70, 6337–6348. doi: 10.1093/jxb/erz359
Wang, X., Zhang, Y., Ma, Q., Zhang, Z., Xue, Y., Bao, S., et al. (2007). SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26, 1934–1941. doi: 10.1038/sj.emboj.7601647
Waters, B. M., Chu, H.-H., Didonato, R. J., Roberts, L. A., Eisley, R. B., Lahner, B., et al. (2006). Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141, 1446–1458. doi: 10.1104/pp.106.082586
Widiez, T., El Kafafi, E. S., Girin, T., Berr, A., Ruffel, S., Krouk, G., et al. (2011). High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3- uptake is associated with changes in histone methylation. Proc. Natl. Acad. Sci. U.S.A. 108, 13329–13334. doi: 10.1073/pnas.1017863108
Xiao, J., Lee, U.-S., and Wagner, D. (2016). Tug of war: Adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi: 10.1016/j.pbi.2016.08.002
Xing, J., Wang, T., Liu, Z., Xu, J., Yao, Y., Hu, Z., et al. (2015). General control nonrepressed protein5-mediated histone acetylation of ferric reductase defective3 contributes to iron homeostasis in Arabidopsis. Plant Physiol. 168, 1309–1320. doi: 10.1104/pp.15.00397
Xu, J. M., Wang, Z. Q., Wang, J. Y., Li, P. F., Jin, J. F., Chen, W. W., et al. (2020). Low phosphate represses histone deacetylase complex1 to regulate root system architecture remodeling in Arabidopsis. N. Phytol. 225, 1732–1745. doi: 10.1111/nph.16264
Yong-Villalobos, L., González-Morales, S. I., Wrobel, K., Gutiérrez-Alanis, D., Cervantes-Peréz, S. A., Hayano-Kanashiro, C., et al. (2015). Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc. Natl. Acad. Sci. U.S.A. 112, E7293–E7302. doi: 10.1073/pnas.1522301112
Yuan, Y. X., Zhang, J., Wang, D. W., and Ling, H. Q. (2005). AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 15, 613–621. doi: 10.1038/sj.cr.7290331
Yuan, Y., Wu, H., Wang, N., Li, J., Zhao, W., Du, J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18, 385–397. doi: 10.1038/cr.2008.26
Zhang, H., Lang, Z., and Zhu, J.-K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhang, J., Liu, B., Li, M., Feng, D., Jin, H., Wang, P., et al. (2015). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27, 787–805. doi: 10.1105/tpc.114.132704
Zhang, T., Cooper, S., and Brockdorff, N. (2015). The interplay of histone modifications - writers that read. EMBO Rep. 16, 1467–1481. doi: 10.15252/embr.201540945
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Zhang, Z., Zhang, S., Zhang, Y., Wang, X., Li, D., Li, Q., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23, 396–411. doi: 10.1105/tpc.110.081356
Keywords: iron, chromatin, histone modification, DNA methylation, nutrition, epigenetics
Citation: Su J, Yao Z, Wu Y, Lee J and Jeong J (2022) Minireview: Chromatin-based regulation of iron homeostasis in plants. Front. Plant Sci. 13:959840. doi: 10.3389/fpls.2022.959840
Received: 02 June 2022; Accepted: 31 August 2022;
Published: 16 September 2022.
Edited by:
Anja Schneider, Ludwig Maximilian University of Munich, GermanyReviewed by:
Ajay Kumar Pandey, National Agri-Food Biotechnology Institute, IndiaAnamika Pandey, Selçuk University, Turkey
Jon Lucas Boatwright, Clemson University, United States
Behrooz Darbani, Aarhus University, Denmark
Copyright © 2022 Su, Yao, Wu, Lee and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joohyun Lee, am9vaHl1bi5sZWVAZHVrZS5lZHU=; Jeeyon Jeong, amplb25nQGFtaGVyc3QuZWR1
†These authors share senior authorship
 Justin Su
Justin Su Zhujun Yao
Zhujun Yao Yixuan Wu
Yixuan Wu Joohyun Lee
Joohyun Lee Jeeyon Jeong
Jeeyon Jeong