- 1Ministry of Education (MOE) Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou, China
- 2Gansu Province Key Laboratory of Gene Editing for Breeding, School of Life Sciences, Lanzhou University, Lanzhou, China
- 3State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
- 4College of Life Science and Technology, Gansu Agricultural University, Lanzhou, China
- 5Basic Forestry and Proteomics Research Center, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
Auxin is one of the most important plant growth regulators of plant morphogenesis and response to environmental stimuli. Although the biosynthesis pathway of auxin has been elucidated, the mechanisms regulating auxin biosynthesis remain poorly understood. The transcription of auxin biosynthetic genes is precisely regulated by complex signaling pathways. When the genes are expressed, epigenetic modifications guide mRNA synthesis and therefore determine protein production. Recent studies have shown that different epigenetic factors affect the transcription of auxin biosynthetic genes. In this review, we focus our attention on the molecular mechanisms through which epigenetic modifications regulate auxin biosynthesis.
Introduction
As an important plant growth regulator, auxin plays a central role in regulating the growth and development of plants (Woodward and Bartel, 2005; Enders and Strader, 2015; Lavy and Estelle, 2016; Weijers et al., 2018; Semeradova et al., 2020). It is well-established that the key to auxin's control of growth and development is through its gradient distribution (Tanaka et al., 2006; Vieten et al., 2007). According to the classical view in the auxin field, auxin is produced in the shoot apical meristems, young leaves, and flower buds and is distributed over a gradient through Polar Auxin Transport (PAT) (Teale et al., 2006). More recently, local auxin biosynthesis has also been considered to play an important role in the formation of auxin gradients. Local auxin biosynthesis is regulated by diversified signaling pathways and guides plant growth and development and response to environmental stimuli (Brumos et al., 2018; Lv et al., 2019). The regulatory mechanisms underlying auxin biosynthesis include transcriptional regulation and posttranslational modifications (Casanova-Saez et al., 2021).
As one of the primary modes of transcriptional regulation, epigenetic modifications of the genome, which mainly include histone modifications in nucleosomes and DNA methylation modifications, change the structure of chromosomes or change the spatial structure of the modified DNA, resulting in gene silencing or overexpression. A number of reviews have summarized the functions of different epigenetic mechanisms in auxin signaling, transport, and metabolism (Yamamuro et al., 2016; Do et al., 2019; Mateo-Bonmati et al., 2019; Nguyen et al., 2020; Wojcikowska et al., 2020), two of which focused on epigenetic contributions to auxin biosynthesis. Mateo-Bonmati et al. (2019) comprehensively described the relationships between epigenetic modifications and auxin homeostasis. Do et al. (2019) highlighted the roles of epigenetic modifications in the YUC family genes, which are the most important genes in the auxin biosynthesis pathway. This review, based on emerging results, aims to provide an updated overview and some hypotheses about the effects of epigenetic modification on the regulation of auxin biosynthesis (Table 1).

Table 1. Epigenetically regulated auxin biosynthetic genes and the factors involved in the epigenetic modification.
Main pathway for IAA biosynthesis
Biochemical and genetic evidence has shown that the major natural auxin, indole-3-acetic acid (IAA) is synthesized through two major pathways: the Tryptophan-Independent (TI) pathway and the Tryptophan-Dependent (TD) pathway (Woodward and Bartel, 2005; Normanly, 2010; Ljung, 2013; Casanova-Saez and Voss, 2019; Casanova-Saez et al., 2021). While the TD pathway is fairly well-understood, little is known about the TI pathway (Gomes and Scortecci, 2021).
As shown in Figure 1, the Tryptophan (Trp) biosynthesis pathway includes six critical steps (Maeda and Dudareva, 2012). The rate-limiting step is catalyzed by anthranilate synthase, which contains two subunits, WEAK ETHYLENE INSENSITIVE 2 (WEI2)/ANTHRANILATE SYNTHASE ALPHA-SUBUNIT 1 (ASA1) and WEI7/ANTHRANILATE SYNTHASE BETA-SUBUNIT 1 (ASB1) (Stepanova et al., 2005, 2008). Some publications reported that both wei2 and wei7 mutants exhibit auxin-deficient phenotypes triggered by reduced endogenous auxin (Stepanova et al., 2005; Di et al., 2016a; Veloccia et al., 2016). In the TD pathway, there are several parallel pathways downstream of Trp for IAA biosyntheses, such as the indole-3-pyruvic acid (IPyA) pathway, the indole-3-acetamide (IAM) pathway, and the indole-3-acetaldoxime (IAOx) pathway (Ljung, 2013; Di et al., 2016c; Casanova-Saez and Voss, 2019; Casanova-Saez et al., 2021). The IPyA pathway has been established as the main pathway of auxin biosynthesis (Mashiguchi et al., 2011; Won et al., 2011; Zhao, 2014) and consists of a two-step reaction. The TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) family proteins have overlapping functions and catalyze the conversion of Trp to IPyA (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009; Zhou et al., 2011). This family of proteins include three homologous proteins TAA1/WEAK ETHYLENE INSENSITIVE 8 (WEI8)/TRANSPORT INHIBITOR RESPONSE 2 (TIR2)/SHADE AVOIDANCE 3 (SAV3)/CYTOKININ INDUCED ROOT CURLING 1 (CKRC1), TRYPTOPHAN AMINOTRANSFERASE RELATED 1 (TAR1), and TAR2 (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009; Zhou et al., 2011). The biosynthesis of IAA from IPyA is catalyzed by YUC (YUCCA) flavin monooxygenase-like proteins (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Di et al., 2016b), which is a large family of 11 members that are functionally redundant with each other (Zhao et al., 2001; Cheng et al., 2006, 2007; Kim et al., 2007; Stepanova et al., 2011; Chen et al., 2014; Liu et al., 2017).
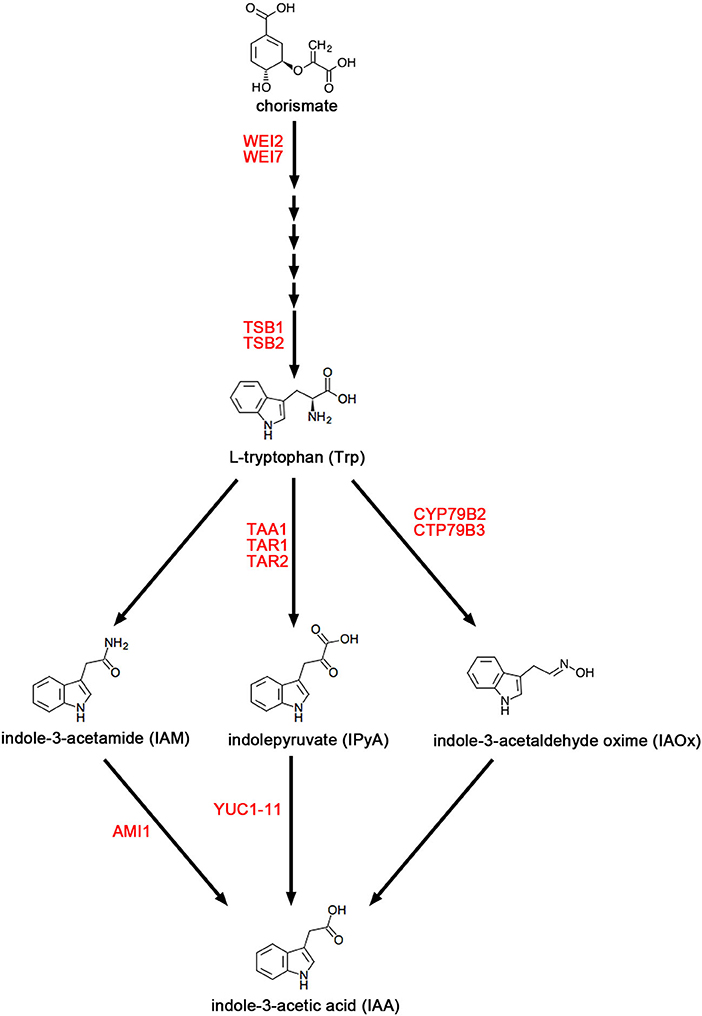
Figure 1. The main pathways of auxin biosynthesis. The figure shows three parallel Trp-dependent (TD) IAA biosynthesis pathways in Arabidopsis, namely the indole-3-acetamide (IAM) pathway, and the indole-3-pyruvic acid (IPyA) pathway, and the indole-3-acetaldoxime (IAOx) pathway. Enzymes mentioned in this review are in red font.
The repressive histone mark H3K27me3 controls the expression of auxin biosynthetic genes
The DNA sequences in eukaryotes are assembled with proteins into nucleosomes, which are 146-bp of DNA wrapped 1.7 times around histone core protein complexes (H2A, H2B, H3, and H4). Surrounding the nucleosome is the junction histone H1, which compresses the nucleosome into higher-order structures to form chromatin fibers (Carlberg and Molnár, 2020b). In Arabidopsis, gene transcription is regulated by the chromatin state, which is determined by histone H3 methylation and acetylation and histone H2B monoubiquitination (H2Bub1) (Roudier et al., 2011). Compared with acetylation and monoubiquitination, methylation also occurs on lysine residues of histones but varies in the degree of modification, i.e., mono-, di-, or tri-methylation (Carlberg and Molnár, 2020a). Different methylation sites have different regulatory functions on gene transcription. For instance, H3K4me3 (H3 lysine 4 trimethylation) is detected specifically at active promoters, while H3K27me3 is correlated with gene repression over larger genomic regions (Carlberg and Molnár, 2020a).
About a decade ago, whole-genome occupancy studies showed that H3K27me3 was associated with the transcriptional repression of auxin-related genes. In comparisons of dividing and differentiated cells, differential H3K27me3 modifications were observed at the loci of YUCs, Cytochrome P450s (CYPs), and TAA1/TARs (Lafos et al., 2011; He et al., 2012). In Arabidopsis, H3K27me3 is catalyzed by Polycomb Repressive Complex 2 (PRC2), which is an important epigenetic regulator of developmental processes (Schuettengruber et al., 2007; Muller and Verrijzer, 2009). In Drosophila, the PRC2 complex consists of four core subunits: the histone methyltransferase Enhancer of zeste [E(z)], Suppressor of zeste 12 [Su(z)12], Extra sex combs (Esc), and the histone-binding Nurf55 (nucleosome remodeling factor 55 kDa) (Schuettengruber and Cavalli, 2009; Simon and Kingston, 2013; Mozgova and Hennig, 2015). There are three PRC2 complexes in Arabidopsis, one of which is called FIS-PRC2 [including FERTILIZATION INDEPENDENT SEED2 (FIS2), Homolog of Su(z)12, CURLY LEAF (CLF), Homolog of E(z), FERTILIZATION INDEPENDENT ENDOSPERM (FIE), Homolog of Esc, and MULTIPLE SUPPRESSOR OF IRA 1 (MSI1), Homolog of Nurf55] (Mozgova and Hennig, 2015). During angiosperm fertilization, the auxin biosynthetic gene YUC10 is constitutively repressed in maternal-derived tissues by the action of the FIS-PRC2 complex (Figueiredo et al., 2015). When FIS2, MEA/FIS, and MSI1 functions are deficient, the absence of H3K27me3 results in heterochronic expression of auxin biosynthetic genes, resulting in fertilization-independent development of empty seeds (Figueiredo et al., 2015).
A model for the regulation of auxin biosynthetic genes by H3K27me3 was established through studies of LHP1/TFL2 (LIKE HETEROCHROMATIN PROTEIN 1/TERMINAL FLOWER 2), which is a unique homolog of Drosophila HETEROCHROMATIN PROTEIN 1 (HP1) in the Arabidopsis genome (James and Elgin, 1986; Gaudin et al., 2001; Kotake et al., 2003). It was initially thought that LHP1 binds to methylated H3K27, interact with a plant Ring-Finger protein, and catalyzes H2A ubiquitination as part of the plant PRC1-like complex (Xu and Shen, 2008; Bratzel et al., 2010). However, Derkacheva et al. (2013) found that the MSI1 subunit links PRC2 to LHP1 for H3K27me3. There is evidence of this LHP1-PRC2 association in other reports, such as LHP1 is proved to be co-purified with PRC2 and impacts H3K27me3 levels at thousands of loci (Wang et al., 2016), and the involvement of LHP1-PRC2 complex in maintaining H3K27me3 levels in dividing cells in Arabidopsis (Zhou et al., 2017). Early studies showed that TFL2/LHP1 is involved in auxin biosynthesis by activating the expression of YUCs (Rizzardi et al., 2011). Chromatin immunoprecipitation (ChIP) analysis showed TFL2/LHP1 enrichment over the transcribed regions of YUC1, YUC2, YUC4, YUC5, YUC6, YUC8, YUC9, and YUC10 (Rizzardi et al., 2011). At the late stage of floral organ development, the C2H2-type zinc-finger transcription factor SUPERMAN (SUP) was expressed at the floral meristem-organ boundaries to define flower patterning. The loss function of SUP leads to higher levels of auxin in these boundaries, resulting in flowers with supernumerary stamens. As shown in Figure 2A, while binding to the YUC1 and YUC4 promoters through the C2H2 zinc-finger domain, SUP recruits the PCR2 complex by interacting with the CLF subunit and LHP1 to form the SUP-LHP1-PRC2 complex, which represses YUC1 and YUC4 expression by trimethylation modification of histone H3 at K27 (Xu et al., 2018). Interestingly, LHP1 functions by linking with PRC2 and participating in maintaining H3K27me3, which contradicts earlier studies that showed that LHP1 positively regulates auxin biosynthetic genes (Rizzardi et al., 2011), suggesting that the function of LHP1 seemed to be controlled by complex mechanisms. Perhaps LHP1 plays various biological functions with different epigenetic regulators in different tissues, at different developmental stages, or under different environmental stimuli. It remains necessary to uncover the precise mechanism by which LHP1 regulates auxin biosynthesis at the YUC loci in the future.
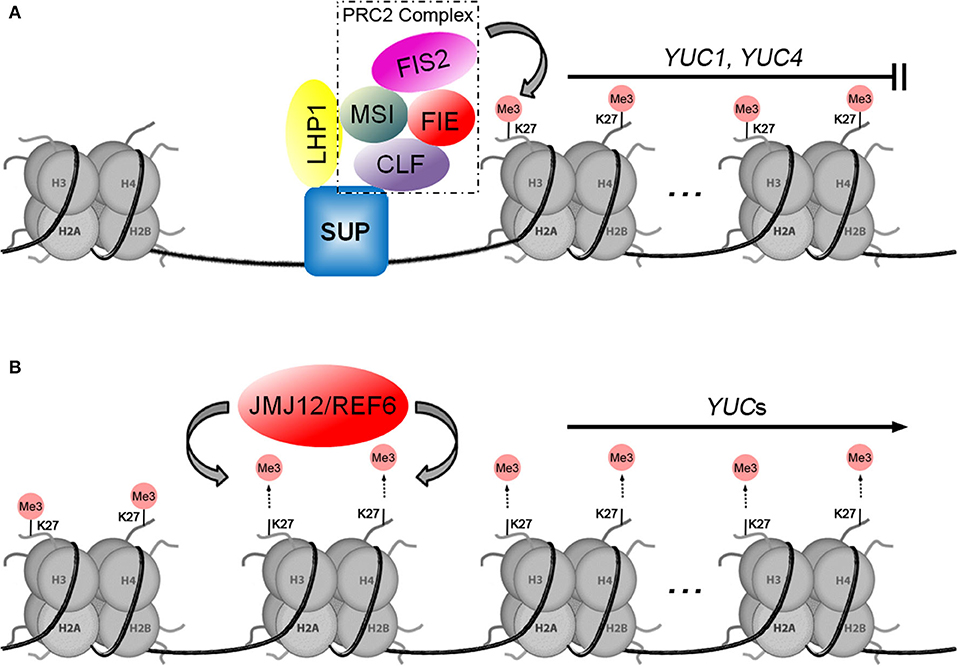
Figure 2. Two models for histone methylation are involved in the transcriptional regulation of YUCs. (A) Model of how H3K27me3 by the PRC2 complex inhibits the expression of auxin biosynthetic genes. (B) The JMJ12/REF6 histone demethylase positively regulates YUCs by demethylation of histone H3K27.
Gene repression by H3K27me3 is also orchestrated by other proteins that act antagonistically to PRC2. The JUMONJI DOMAIN-CONTAINING (JMJ) proteins have been identified to function as Histone DeMethylases (HDMs) in mammalian and plant cells (Agger et al., 2008; Lu et al., 2011; Cheng et al., 2018). As shown in Figure 2B, a number of studies have documented that JMJ12/REF6 (RELATIVE OF EARLY FLOWERING 6) was found to regulate YUC1, YUC3, YUC7, YUC8, YUC9, and YUC11 via specifically demethylating H3K27me3 and H3K27me2 in Arabidopsis (Cui et al., 2016; Li et al., 2016; Yan et al., 2018). In leaf explants upon wounding, upregulation of YUC1 and YUC4 is accompanied by the removal of H3K27me3 (Chen et al., 2016). Do et al. (2019) speculated in their review that the wound-jasmonate (JA) signal or even other environmental signals may trigger the recruitment of JMJ12/REF6. A recent study showed that JMJ14, JMJ15, and JMJ18 play key roles in high ambient temperature-induced YUC8 expression. However, unlike JMJ12/REF6, the JMJ14, JMJ15, and JMJ18 mediated removal of H3K4me3 (activating) marks activates YUC8 transcription by inhibiting the expression of some negative regulators (Cui et al., 2021).
Other studies have also suggested that H3K36me3 and H3K4me2 are involved in auxin biosynthesis. The H3K36me3 histone mark readers EMSY-Like protein 1 (EML1) and EML3 in the Tudor/Agent family repress YUC10 expression during seed coat and endosperm development (Milutinovic et al., 2019). This is contrary to the common belief that H3K36me3 positively regulates gene transcription (Liu et al., 2010; Milutinovic et al., 2019), although the molecular mechanism remains unknown. The RNA binding protein FLOWERING CONTROL LOCUS A (FCA) is involved in chromatin silencing by promoting histone demethylation (Tian et al., 2019). A large number of studies have shown that PIF4 promotes growth by inducing the expression of YUC8 at high ambient temperatures (Sun et al., 2012; van der Woude et al., 2019; Xue et al., 2021), but there is also a mechanism that decreases transcriptional activation of PIF4 to avoid heat stress damage. PIF4 recruits FCA to trigger histone H3K4me2 (activating mark) demethylation at the YUC8 promoter region, which triggers PIF4 dissociation from the DNA to inhibit its expression (Lee et al., 2014).
Histone acetylation and chromatin remodeling in activating or repressing auxin biosynthetic genes
Since histone acetylation was first reported (Allfrey et al., 1964), it has been extensively studied in microorganisms, animals, and plants (Anonymous, 2021). Histone acetylation neutralizes the positive charge of lysine at the histone tail, decreasing the interaction between the histone protein and the negatively charged DNA to promote an “open” and transcriptionally permissive chromatin structure (Lee and Workman, 2007; Lawrence et al., 2016). Increased levels of histone acetylation are associated with the activation of many genes involved in different plant biological processes, such as root morphogenesis (Nguyen et al., 2020), photosynthesis (Zhou et al., 2022), shade avoidance response (Peng et al., 2018), wound-induced cellular reprogramming (Rymen et al., 2019), and salt stress response (Li et al., 2014). The dynamic changes in histone acetylation are often controlled by two enzymes with opposing functions, HISTONE ACETYLTRANSFERASEs (HATs), and HISTONE DEACETYLASEs (HDAs) (McGinty and Tan, 2014). Current studies have elucidated that homeostasis of histone acetylation is not only critical for AUXIN/INDOLE-3-ACETIC ACID-AUXIN RESPONSE FACTOR (AUX/IAA-ARF) mediated auxin signaling (Nguyen et al., 2020), but also for regulation of auxin biosynthesis.
Local biosynthesis of auxin plays a key role in the shade avoidance response of plants. Disruption of auxin biosynthesis by mutation of the TAA1 gene reduced hypocotyl elongation responses (Tao et al., 2008; Won et al., 2011). Shade can activate the expression of YUC2, 5, 8, and 9 (Tao et al., 2008; Hornitschek et al., 2012; Muller-Moule et al., 2016; Peng et al., 2018), which depend on different PIF transcription factors such as PIF4, PIF5, and PIF7 (Hornitschek et al., 2012; Peng et al., 2018). As shown in Figure 3A, Peng et al. (2018) have revealed the molecular mechanism of PIF7 activating YUC8 transcription. When plants are exposed to shade, the PIF7 is dephosphorylated and subsequently binds to the G-box (CACGTG) cis-element of the YUC8 promoter (Li et al., 2012; Peng et al., 2018). Secondly, PIF7 interacts with MRG2 (MORF-RELATED GENE 2), which binds to H3K4me3/H3K36me3 at the 5′-end of YUC8 (Bu et al., 2014; Xu et al., 2014; Peng et al., 2018). Finally, the PIF7-MRG2 complex recruits HATs to promote histone acetylation at H4K5, H3K9, and H3K27 at the coding region of YUC8, resulting in the induction of its expression (Peng et al., 2018). A similar mechanism for transcriptional regulation of GH3.3, which converts IAA to IAA amino acid conjugates (Chen et al., 2010), is driven by the BASIC LEUCINE ZIPPER 11—TRANSCRIPTIONAL ADAPTOR 2b (bZIP11-ADA2b) complex, which is able to recruit GENERAL CONTROL NON-REPRESSIBLE 5/HISTONE ACETYLTRANSFERASE OF THE GNAT FAMILY 1 (GCN5/HAG1) to the GH3.3 promoter, inducing the activation of GH3.3 expression (Weiste and Droge-Laser, 2014).
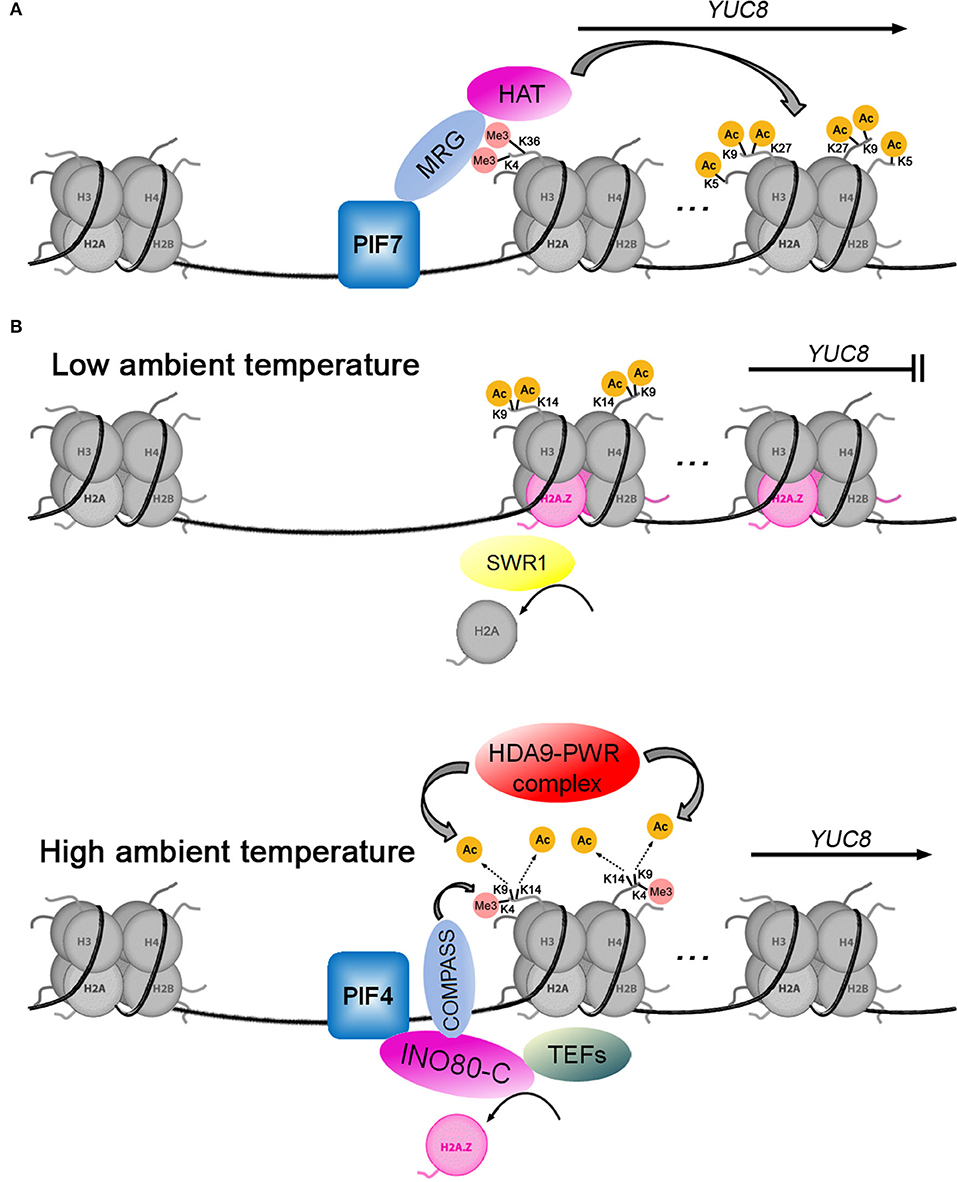
Figure 3. Two models for histone acetylation involvement in transcriptional regulation of YUC8. (A) The transcription factor PIF7 positively regulates YUC8 by recruiting the MRG-HAT complex. (B) When the ambient temperature rises, the HDA9-PWR complex mediates the deacetylation of H3K9 and H3K14, which opens YUC8 chromatin to allow PIF4 binding to activate YUC8 transcription. PIF4 recruits the INO80-C-COMPASS-TEFs complex to promote H2A.Z eviction, H3K4me3, and transcription elongation at the YUC8 loci.
As an evolutionarily conserved variant of the canonical histone H2A, H2A.Z plays a critical role in multiple cellular processes through its influence on chromatin structure and dynamics. In plants, H2A.Z is involved in regulating growth and development, phase transitions, and response to environmental stimuli (Kumar, 2018). One well-studied model of H2A.Z is the regulation of thermomorphogenesis through temperature-responsive nucleosome dynamics (Kumar and Wigge, 2010; Casal and Balasubramanian, 2019). The histone variant H2A.Z is enriched at gene loci with high thermo-responsiveness, and its presence is often associated with transcriptional repression (Coleman-Derr and Zilberman, 2012; Dai et al., 2017; Sura et al., 2017). Numerous auxin-related genes, such as YUCs, AUX/IAAs, and SMALL AUXIN UP RNA (SAURs), have been shown to mediate high-temperature-associated plant growth (Koini et al., 2009; Franklin et al., 2011; Sun et al., 2012; Lee et al., 2014; Perrella et al., 2022). As shown in Figure 3B, at low ambient temperature, the transcription of YUC8 is repressed by the SWR1 chromatin remodeling complex (Tasset et al., 2018; van der Woude et al., 2019), which is necessary for inserting the alternative histone H2A.Z into nucleosomes in place of H2A (Krogan et al., 2003; Kobor et al., 2004; Mizuguchi et al., 2004). When the ambient temperature increases, the HDA9-PWR (POWERDRESS) complex mediates the deacetylation of H3K9 and H3K14 in the first nucleosome at the 5'-end of the YUC8 gene, which reduces H2A.Z occupancy in nucleosomes (Tasset et al., 2018; van der Woude et al., 2019). The transcription factor PIF4 directly interacts with the INO80 chromatin remodeling complex (INO80-C), which mediates H2A.Z eviction at the YUC8 loci (Xue et al., 2021). At the same time, INO80-C can interact with COMPASS (methyltransferase for H3K4 trimethylation) and TEFs (transcription elongation factors) to enhance H3K4me3 and transcription elongation (Xue et al., 2021). These dynamic changes in the chromatin promote YUC8 expression, leading to plant morphological changes in response to elevated temperatures (Tasset et al., 2018; van der Woude et al., 2019; Xue et al., 2021).
There are two other reports which suggested that the chromosome remodeling resulting from histone deacetylation may also be involved in the transcriptional regulation of other YUCs. In the Arabidopsis gynoecium, the apical-basal and mediolateral polarity of auxin involve regulation at the YUC4 loci through histone acetylation at H3K9 and H3K14 by GCN5/HAG1 (at −174 to −84 in the YUC4 promoter, also the first nucleosome at the 5′-end of YUC4), which inhibits the transcription of YUC4 (Poulios and Vlachonasios, 2018). The SWR1 complex consists of several subunits, such as PHOTOPERIOD INDEPENDENT EARLY FLOWERING 1 and ARP6 (ACTIN-RELATED PROTEIN 6) (Aslam et al., 2019). As a homologous protein of ARP6, ARP4 represses the transcription of YUC9 through exchanging H2A with the histone variant H2A.Z (Lee and Seo, 2017), but it remains unknown if the release of YUC9 transcriptional inhibition is related to HDA-PWR complex-mediated histone deacetylation. Moreover, in Arabidopsis leaf development, the expression of the auxin metabolism gene IAMT1 (encoding an enzyme that converts IAA into its methyl ester) and biosynthetic gene YUC6 were regulated by the subunit SWI3B of the chromatin remodeling complex SWI/SNF (Han et al., 2018; Lin et al., 2021).
Interestingly, the effects of histone acetylation on the transcription of YUC8 were opposite in the two models (Figures 3A,B), the acetylation of histone activated YUC8 transcription (Peng et al., 2018) during shade avoidance but inhibited YUC8 transcription during thermomorphogenesis (van der Woude et al., 2019). We noticed that the nucleosomes containing acetylated histone were located in the coding region of YUC8 in the study on shade avoidance, but were located at the 5′-end of YUC8 in the study on thermomorphogenesis (Peng et al., 2018; van der Woude et al., 2019). In addition, the acetylated lysine sites were different, at H4K5, H3K9, and H3K27 during shade avoidance (van der Woude et al., 2019) and at H3K9 and H3K14 during high ambient temperature (Peng et al., 2018). This means that nucleosomes may have different functions based on the position of the acetylated histone in the chromatin and on which lysine residues in the histone are modified by acetylation. In addition, the histone acetyltransferase GCN5/HAG1 not only acetylates H3K27 in the GH3.3 promoter to activate GH3.3 expression (Weiste and Droge-Laser, 2014), but also acetylates H3K9 and H3K14 in the YUC4 promoter to inhibit YUC4 expression (Poulios and Vlachonasios, 2018). This further shows that compared with the enzyme that mediates histone acetylation modification, the position of acetylated lysine sites in histone H3 may determine whether transcription is activated or inhibited.
H2Bub1 is involved in the regulation of auxin biosynthesis
Like histone acetylation, monoubiquitination of histone H2B (H2Bub1) is another important epigenetic modification related to gene transcriptional activation (Roudier et al., 2011). In nucleosomes embedded in the coding regions of genes with strong transcriptional activity, there are often increased levels of histone H2Bub1, which changes the chromatin state and is specifically linked with transcript elongation (Pavri et al., 2006; Bourbousse et al., 2012; Himanen et al., 2012; Feng and Shen, 2014; Van Lijsebettens and Grasser, 2014; Woloszynska et al., 2019). H2Bub1 participates in a variety of physiological processes by activating the transcription of related genes, such as seed germination and dormancy (Wang et al., 2022), flowering time (Cao et al., 2008; Woloszynska et al., 2019), defense (Zhao et al., 2020; Ma et al., 2021), and stress responses (Chen et al., 2019; Ma et al., 2019; Sun et al., 2020).
The monoubiquitination of histone H2B is catalyzed by the ubiquitin ligase E3 heterotetrameric complex, consisting of two Histone Monoubiquitination 1 proteins (HUB1s) and two homologous HUB2s (Cao et al., 2008). The function of HUB1 was tied to auxin biosynthesis through the Cytokinin-Induced Root Curling (CKRC) system for screening auxin deficient mutants. There was a reduced level of IAA in the ckrw2 mutant (cytokinin-induced root waving 2) that was linked to the loss of function of the gene HUB1 (Wu et al., 2015). Recent ChIP analysis showed that the ckrw1/hub1 mutant had corresponding defects in H2Bub1 in the coding regions of the auxin biosynthetic genes TSB1, WEI7/ASB1, YUC7, and AMI1 (amidase 1), indicating that H2Bub1 is required for the transcriptional regulation of these genes (Figure 4A; Zhang et al., 2021).
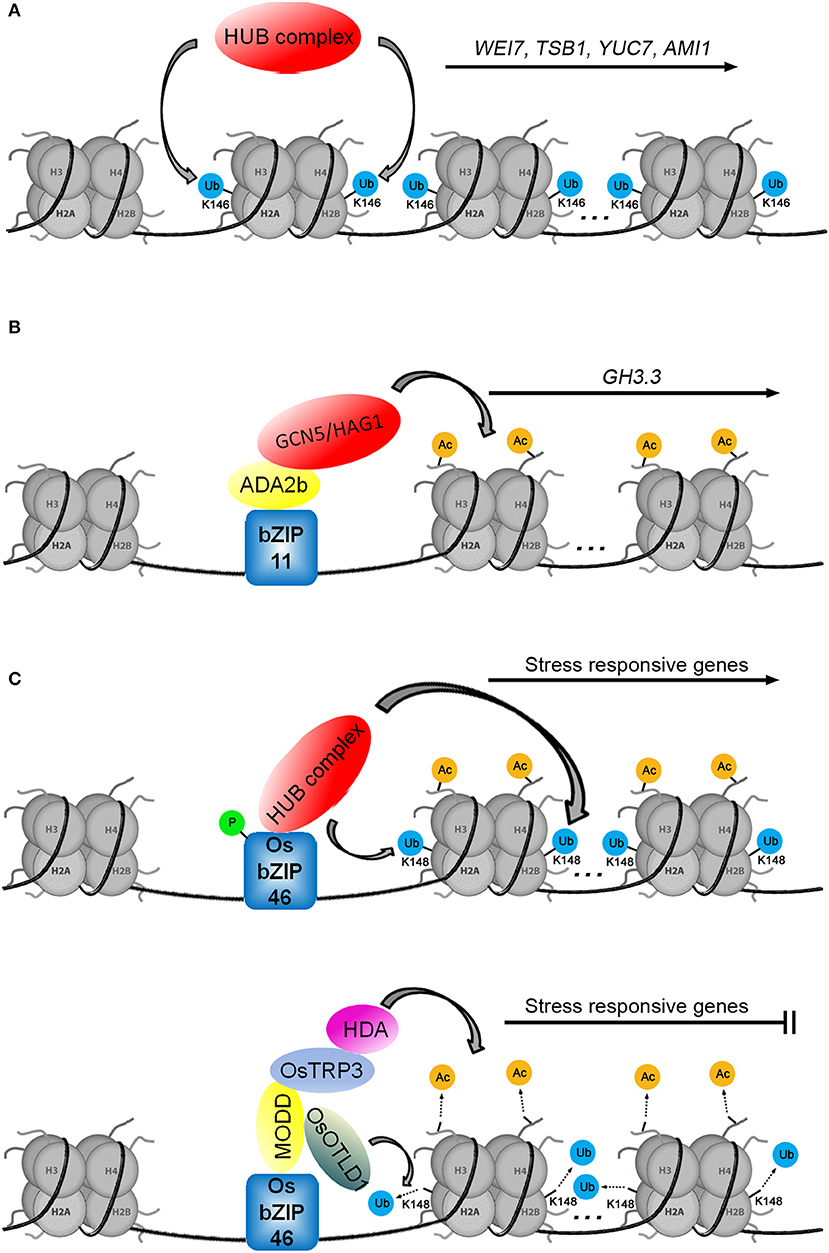
Figure 4. Three models for histone ubiquitination and/or acetylation are involved in transcriptional regulation. (A) The auxin biosynthetic genes WEI7, TSB1, YUC7, and AMI1 are regulated by H2Bub1. (B) The transcription factor bZIP11 positively regulates GH3.3 by recruiting the ADA2b-GCN5/HAG1 complex. (C) H2Bub1 (according to the study of Cao et al., 2008, we speculate that the monoubiquitination modification of H2B occurs at the K148 of H2B in Oryza sativa L.) and H3ac are involved in the regulation of stress-responsive genes (in Tang et al., 2016, the sites of histone lysine residues modified by acetylation were not mentioned).
The current challenge in understanding HUB function is the identification of mechanisms that recruit the HUB complex to the auxin biosynthetic genes. Research on the rice transcription factor OsbZIP46 provides clues. Similar to how the transcription factor bZIP11 recruits the ADA2b-GCN5/HAG1 complex (Figure 4B; Weiste and Droge-Laser, 2014), phosphorylated OsbZIP46 recruits the HUB complex to activate the expression of stress-responsive genes by histone H2B monoubiquitination (Figure 4C; Ma et al., 2019). Working in opposition, the MEDIATOR OF OSBZIP46 DEACTIVATION AND DEGRADATION (OsbZIP46-MODD) complex, consisting of dephosphorylated OsbZIP46 and MODD, simultaneously recruits OTUBAIN-LIKE DEUBIQUITINASE 1 (OsOTLD1) and the TPL/TPR COREPRESSOR (OsTRP3)-HDA complex to shut down the transcription of stress-responsive genes via deubiquitination and deacetylation, respectively (Tang et al., 2016; Ma et al., 2019). The transcriptional activity of OsbZIP46 is blocked by the negative regulatory domain D. Phosphorylation promotes the interaction between OsbZIP46 and the HUB complex to activate the transcriptional activity of OsbZIP46 while also suppressing the negative regulatory function of domain D and preventing MODD interaction with domain D (Tang et al., 2016; Ma et al., 2019). The function of the OsbZIP46 transcription factor is involved not only in histone ubiquitination and deubiquitination but also in histone acetylation and deacetylation, suggesting that ubiquitination and acetylation may play a synergistic role in the regulation of gene transcription (Figure 4C). We speculate that some unknown factors may activate the transcription of auxin biosynthetic genes by recruiting the HUB complex to promote H2Bub1 on specific loci. Of course, there may be other epigenetic factors involved in this process.
DNA methylation and small RNAs' role in auxin biosynthesis
Another epigenetic modification is DNA methylation which is usually associated with the transcriptional silencing of genes. DNA methylation is relatively stable but also reversible, and usually occurs on a cytosine base of DNA, to form 5-methylcytosine, in eukaryotes. Unlike mammalian methylation which occurs only in the CG sequence context, plant DNA can be methylated in CG, CHH, and CHG sequence contexts (with H representing A, T, or C) (Zhang et al., 2018; Gallego-Bartolome, 2020). There are two types of DNA methylation, one is de novo methylation, the methylation of DNA with two unmethylated chains; the other is maintenance methylation of a newly synthesized DNA strand after semi-conservative replication of methylated DNA. DNA methylation is mainly established by the de novo DNA methyltransferases DOMAINS REARRANGED METHYLTRANSFERASE 1/2 (DRM1/2), which are directed by 24-nt small interfering RNAs (siRNAs), and methylates all three sequence contexts via the RNA-directed DNA methylation (RdDM) pathway (Zhang et al., 2018). During DNA replication, methylation at the CG, CHH, and CHG contexts is maintained by METHYLTRANSFERASE 1 (MET1), CHROMOMETHYLASE 2 (CMT2), and CMT3, respectively (Stroud et al., 2014; Zhang et al., 2018).
In the previous section, we reviewed the regulation of auxin biosynthetic genes by PIFs and histone acetylation during thermomorphogenesis. Another study on thermomorphogenesis reported the temperature-related regulation of endogenous auxin biosynthesis by a PIF4-independent epigenetic pathway (Figure 5; Gyula et al., 2018). The expression of the auxin biosynthetic gene YUC2 is negatively regulated by miRNA169 which prevents the binding of transcription factors NF-YA2 and NF-YA10 to the YUC2 promoter (Zhang et al., 2017). A 24-nt siRNA, Locus_77297, directs DNA methylation in the promoter of YUC2, which blocks the binding of NF-YA2 to inhibit the expression of YUC2 (Gyula et al., 2018). At low ambient temperature, the high levels of miRNA169 and Locus_77297 result in a low concentration of active NF-YA2 and a methylated YUC2 promoter. In contrast, at high ambient temperature, miRNA169 levels decrease, as do Locus_77297 levels and the methylation level at the YUC2 promoter (Gyula et al., 2018).

Figure 5. Model for the involvement of DNA methylation and small RNAs in the transcriptional regulation of the auxin biosynthetic gene YUC2.
It has also been reported that the auxin biosynthetic genes YUC2 and TAA1 are specifically up-regulated in leaves of the drm1 drm2 cmt3 triple mutant, which has a low level of DNA methylation (Forgione et al., 2019). Surprisingly, the transcription of YUC2 and TAA1 showed almost no differences in the roots of the triple mutant, thus suggesting that DNA methylation is involved in tissue-specific patterns of gene expression (Forgione et al., 2019) and may be a potential mechanism to regulate local auxin biosynthesis. Recently, Markulin et al. (2021) found several gene loci targeted by RdDM in a whole-genome analysis. Most of the auxin biosynthesis-related genes (TAA1, TAR1, TAR2, YUC1, YUC2, YUC5, YUC10, LEC2) were targeted by RdDM. This means that DNA methylation is a very important yet poorly understood mechanism regulating auxin biosynthesis, marking methylation as an interesting subject that requires further study in plants.
Concluding remarks
Auxin is an essential plant growth regulator that governs growth and development in concert with other signaling pathways. Therefore, understanding the auxin biosynthesis pathway and its regulation is crucial for plant science and agriculture. The biosynthesis pathway of auxin has two remarkable characteristics: the biosynthesis pathway includes multi-step reactions and has several parallel pathways (Figure 1). The complexity of auxin biosynthesis determines the complexity of its regulation. At present, there are only a few genes known to participate in the epigenetic regulation of auxin biosynthesis (Table 1), a simplicity that does not match the complexity of the pathway and suggests a lack of understanding of this topic. For example, little is known about the epigenetic regulation of the TAA gene family, which participates in the main auxin biosynthesis pathway. Moreover, multiple histone modifications act in a combinatorial fashion to specify distinct chromatin states (Carlberg and Molnár, 2020a). Therefore, future research should not only study the effect of certain histone modifications on the transcription of auxin biosynthetic genes but also study how various histone modifications determine chromatin structure and ultimately determine the specific expression level of genes. At the same time, the relationships between these histone modifications and DNA methylation, transcription factor binding, chromatin remodeling as well as RNA expression need to be further studied.
The three of the most critical questions for local auxin biosynthesis in plant growth and development are: in which tissues and at what time are the expression of auxin biosynthetic genes controlled by which internal and external signals and how these signaling pathways regulate chromosomal states. Although epigenetic regulation of gene expression during development has been known for decades, the specific relationship between auxin biosynthesis and epigenetic modifications in plants is only just being elucidated. The epigenetic regulation of auxin biosynthesis in plants is a fascinating subject requiring further study.
Author contributions
LW drafted, wrote, and edited this review. J-LW helped LW to organize and collect information and participated in the writing of sections introduction, main pathway for IAA biosynthesis, and the drawing of figures. G-QG reviewed the manuscript before submitting it and gave many constructive comments. J-LW, D-WD, PL, LZ, X-FL, and LW participated in the discussion of this review. All authors have read and agreed to the published version of this manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31970713), the Fundamental Research Funds for the Central Universities (lzujbky-2020-33 and lzujbky-2021-kb05), and the Natural Science Foundation of Gansu Province (20JR5RA270).
Acknowledgments
We are grateful to Yuan Song (Lanzhou University) for the critical reading of the manuscript. We feel sorry for not being able to cite many other excellent papers due to space limitations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agger, K., Christensen, J., Cloos, P. A., and Helin, K. (2008). The emerging functions of histone demethylases. Curr. Opin. Genet. Dev. 18, 159–168. doi: 10.1016/j.gde.2007.12.003
Allfrey, V. G., Faulkner, R., and Mirsky, A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794. doi: 10.1073/pnas.51.5.786
Anonymous (2021). “Histone acetylation,” in Encyclopedia of Molecular Pharmacology, eds S. Offermanns, W. and Rosenthal (Cham: Springer International Publishing), 810. doi: 10.1007/978-3-030-57401-7_300265
Aslam, M., Fakher, B., Jakada, B. H., Cao, S., and Qin, Y. (2019). SWR1 chromatin remodeling complex: a key transcriptional regulator in plants. Cells 8, 1621. doi: 10.3390/cells8121621
Bourbousse, C., Ahmed, I., Roudier, F., Zabulon, G., Blondet, E., Balzergue, S., et al. (2012). Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 8, e1002825. doi: 10.1371/journal.pgen.1002825
Bratzel, F., Lopez-Torrejon, G., Koch, M., Del Pozo, J. C., and Calonje, M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859. doi: 10.1016/j.cub.2010.09.046
Brumos, J., Robles, L. M., Yun, J., Vu, T. C., Jackson, S., Alonso, J. M., et al. (2018). Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 47, 306–318.e305. doi: 10.1016/j.devcel.2018.09.022
Bu, Z., Yu, Y., Li, Z., Liu, Y., Jiang, W., Huang, Y., et al. (2014). Regulation of arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet. 10, e1004617. doi: 10.1371/journal.pgen.1004617
Cao, Y., Dai, Y., Cui, S., and Ma, L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20, 2586–2602. doi: 10.1105/tpc.108.062760
Carlberg, C., and Molnár, F. (2020a). “Chromatin modifiers,” in Mechanisms of Gene Regulation: How Science Works, eds C. Carlberg, and F. Molnár (Cham: Springer International Publishing), 83–98. doi: 10.1007/978-3-030-52321-3_6
Carlberg, C., and Molnár, F. (2020b). “Genes and chromatin,” in Mechanisms of Gene Regulation: How Science Works, eds C. Carlberg, and F. Molnár (Cham: Springer International Publishing), 1–17. doi: 10.1007/978-3-030-52321-3_1
Casal, J. J., and Balasubramanian, S. (2019). Thermomorphogenesis. Annu. Rev. Plant Biol. 70, 321–346. doi: 10.1146/annurev-arplant-050718-095919
Casanova-Saez, R., Mateo-Bonmati, E., and Ljung, K. (2021). Auxin metabolism in plants. Cold Spring Harb. Perspect. Biol. 13, a039867. doi: 10.1101/cshperspect.a039867
Casanova-Saez, R., and Voss, U. (2019). Auxin metabolism controls developmental decisions in land plants. Trends Plant Sci. 24, 741–754. doi: 10.1016/j.tplants.2019.05.006
Chen, H., Feng, H., Zhang, X., Zhang, C., Wang, T., and Dong, J. (2019). An Arabidopsis E3 ligase HUB2 increases histone H2B monoubiquitination and enhances drought tolerance in transgenic cotton. Plant Biotechnol. J. 17, 556–568. doi: 10.1111/pbi.12998
Chen, L., Tong, J., Xiao, L., Ruan, Y., Liu, J., Zeng, M., et al. (2016). YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 67, 4273–4284. doi: 10.1093/jxb/erw213
Chen, Q., Dai, X., De-Paoli, H., Cheng, Y., Takebayashi, Y., Kasahara, H., et al. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079. doi: 10.1093/pcp/pcu039
Chen, Q., Westfall, C. S., Hicks, L. M., Wang, S., and Jez, J. M. (2010). Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 285, 29780–29786. doi: 10.1074/jbc.M110.146431
Cheng, S., Tan, F., Lu, Y., Liu, X., Li, T., Yuan, W., et al. (2018). WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res. 46, 2356–2369. doi: 10.1093/nar/gky017
Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Cheng, Y., Dai, X., and Zhao, Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439. doi: 10.1105/tpc.107.053009
Coleman-Derr, D., and Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8, e1002988. doi: 10.1371/journal.pgen.1002988
Cui, X., Lu, F., Qiu, Q., Zhou, B., Gu, L., Zhang, S., et al. (2016). REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 48, 694–699. doi: 10.1038/ng.3556
Cui, X., Zheng, Y., Lu, Y., Issakidis-Bourguet, E., and Zhou, D. X. (2021). Metabolic control of histone demethylase activity involved in plant response to high temperature. Plant Physiol. 185, 1813–1828. doi: 10.1093/plphys/kiab020
Dai, X., Bai, Y., Zhao, L., Dou, X., Liu, Y., Wang, L., et al. (2017). H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in Arabidopsis. Mol. Plant 10, 1274–1292. doi: 10.1016/j.molp.2017.09.007
Derkacheva, M., Steinbach, Y., Wildhaber, T., Mozgova, I., Mahrez, W., Nanni, P., et al. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32, 2073–2085. doi: 10.1038/emboj.2013.145
Di, D. W., Wu, L., Luo, P., Zhang, L., Zhang, T. Z., Sun, X., et al. (2016a). Analysis the role of Arabidopsis CKRC6/ASA1 in auxin and cytokinin biosynthesis. J. Plant Biol. 59, 162–171. doi: 10.1007/s12374-016-0396-6
Di, D. W., Wu, L., Zhang, L., An, C. W., Zhang, T. Z., Luo, P., et al. (2016b). Functional roles of Arabidopsis CKRC2/YUCCA8 gene and the involvement of PIF4 in the regulation of auxin biosynthesis by cytokinin. Sci. Rep. 6, 36866. doi: 10.1038/srep36866
Di, D. W., Zhang, C. G., Luo, P., An, C. W., and Guo, G. Q. (2016c). The biosynthesis of auxin: how many paths truly lead to IAA? Plant Growth Regul. 78, 275–285. doi: 10.1007/s10725-015-0103-5
Do, B. H., Phuong, V. T. B., Tran, G.-B., and Nguyen, N. H. (2019). Emerging functions of chromatin modifications in auxin biosynthesis in response to environmental alterations. Plant Growth Regul. 87, 165–174. doi: 10.1007/s10725-018-0453-x
Enders, T. A., and Strader, L. C. (2015). Auxin activity: past, present, and future. Am. J. Bot. 102, 180–196. doi: 10.3732/ajb.1400285
Feng, J., and Shen, W. H. (2014). Dynamic regulation and function of histone monoubiquitination in plants. Front. Plant Sci. 5, 83. doi: 10.3389/fpls.2014.00083
Figueiredo, D. D., Batista, R. A., Roszak, P. J., and Kohler, C. (2015). Auxin production couples endosperm development to fertilization. Nat Plants 1, 15184. doi: 10.1038/nplants.2015.184
Forgione, I., Woloszynska, M., Pacenza, M., Chiappetta, A., Greco, M., Araniti, F., et al. (2019). Hypomethylated drm1 drm2 cmt3 mutant phenotype of Arabidopsis thaliana is related to auxin pathway impairment. Plant Sci. 280, 383–396. doi: 10.1016/j.plantsci.2018.12.029
Franklin, K. A., Lee, S. H., Patel, D., Kumar, S. V., Spartz, A. K., Gu, C., et al. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108, 20231–20235. doi: 10.1073/pnas.1110682108
Gallego-Bartolome, J.. (2020). DNA methylation in plants: mechanisms and tools for targeted manipulation. New Phytol. 227, 38–44. doi: 10.1111/nph.16529
Gaudin, V., Libault, M., Pouteau, S., Juul, T., Zhao, G., Lefebvre, D., et al. (2001). Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128, 4847–4858. doi: 10.1242/dev.128.23.4847
Gomes, G. L. B., and Scortecci, K. C. (2021). Auxin and its role in plant development: structure, signalling, regulation and response mechanisms. Plant Biol. 23, 894–904. doi: 10.1111/plb.13303
Gyula, P., Baksa, I., Toth, T., Mohorianu, I., Dalmay, T., and Szittya, G. (2018). Ambient temperature regulates the expression of a small set of sRNAs influencing plant development through NF-YA2 and YUC2. Plant Cell Environ. 41, 2404–2417. doi: 10.1111/pce.13355
Han, W., Han, D., He, Z., Hu, H., Wu, Q., Zhang, J., et al. (2018). The SWI/SNF subunit SWI3B regulates IAMT1 expression via chromatin remodeling in Arabidopsis leaf development. Plant Sci. 271, 127–132. doi: 10.1016/j.plantsci.2018.03.021
He, C., Chen, X., Huang, H., and Xu, L. (2012). Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 8, e1002911. doi: 10.1371/journal.pgen.1002911
Himanen, K., Woloszynska, M., Boccardi, T. M., De Groeve, S., Nelissen, H., Bruno, L., et al. (2012). Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J. 72, 249–260. doi: 10.1111/j.1365-313X.2012.05071.x
Hornitschek, P., Kohnen, M. V., Lorrain, S., Rougemont, J., Ljung, K., Lopez-Vidriero, I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
James, T. C., and Elgin, S. C. (1986). Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6, 3862–3872. doi: 10.1128/mcb.6.11.3862-3872.1986
Kim, J. I., Sharkhuu, A., Jin, J. B., Li, P., Jeong, J. C., Baek, D., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145, 722–735. doi: 10.1104/pp.107.104935
Kobor, M. S., Venkatasubrahmanyam, S., Meneghini, M. D., Gin, J. W., Jennings, J. L., Link, A. J., et al. (2004). A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, E131. doi: 10.1371/journal.pbio.0020131
Koini, M. A., Alvey, L., Allen, T., Tilley, C. A., Harberd, N. P., Whitelam, G. C., et al. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19, 408–413. doi: 10.1016/j.cub.2009.01.046
Kotake, T., Takada, S., Nakahigashi, K., Ohto, M., and Goto, K. (2003). Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 44, 555–564. doi: 10.1093/pcp/pcg091
Krogan, N. J., Keogh, M. C., Datta, N., Sawa, C., Ryan, O. W., Ding, H., et al. (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576. doi: 10.1016/S1097-2765(03)00497-0
Kumar, S. V.. (2018). H2A.Z at the core of transcriptional regulation in plants. Mol. Plant 11, 1112–1114. doi: 10.1016/j.molp.2018.07.002
Kumar, S. V., and Wigge, P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147. doi: 10.1016/j.cell.2009.11.006
Lafos, M., Kroll, P., Hohenstatt, M. L., Thorpe, F. L., Clarenz, O., and Schubert, D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7, e1002040. doi: 10.1371/journal.pgen.1002040
Lavy, M., and Estelle, M. (2016). Mechanisms of auxin signaling. Development 143, 3226–3229. doi: 10.1242/dev.131870
Lawrence, M., Daujat, S., and Schneider, R. (2016). Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 32, 42–56. doi: 10.1016/j.tig.2015.10.007
Lee, H. J., Jung, J. H., Cortes Llorca, L., Kim, S. G., Lee, S., Baldwin, I. T., et al. (2014). FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat. Commun. 5, 5473. doi: 10.1038/ncomms6473
Lee, K., and Seo, P. J. (2017). Coordination of matrix attachment and ATP-dependent chromatin remodeling regulate auxin biosynthesis and Arabidopsis hypocotyl elongation. PLoS ONE 12, e0181804. doi: 10.1371/journal.pone.0181804
Lee, K. K., and Workman, J. L. (2007). Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295. doi: 10.1038/nrm2145
Li, C., Gu, L., Gao, L., Chen, C., Wei, C. Q., Qiu, Q., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48, 687–693. doi: 10.1038/ng.3555
Li, H., Yan, S., Zhao, L., Tan, J., Zhang, Q., Gao, F., et al. (2014). Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 14, 105. doi: 10.1186/1471-2229-14-105
Li, L., Ljung, K., Breton, G., Schmitz, R. J., Pruneda-Paz, J., Cowing-Zitron, C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. doi: 10.1101/gad.187849.112
Lin, X., Yuan, C., Zhu, B., Yuan, T., Li, X., Yuan, S., et al. (2021). LFR physically and genetically interacts with SWI/SNF component SWI3B to regulate leaf blade development in Arabidopsis. Front. Plant Sci. 12, 717649. doi: 10.3389/fpls.2021.717649
Liu, C., Lu, F., Cui, X., and Cao, X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Liu, Y., Xu, M., Liang, N., Zheng, Y., Yu, Q., and Wu, S. (2017). Symplastic communication spatially directs local auxin biosynthesis to maintain root stem cell niche in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, 4005–4010. doi: 10.1073/pnas.1616387114
Ljung, K.. (2013). Auxin metabolism and homeostasis during plant development. Development 140, 943–950. doi: 10.1242/dev.086363
Lu, F., Cui, X., Zhang, S., Jenuwein, T., and Cao, X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Lv, B., Yan, Z., Tian, H., Zhang, X., and Ding, Z. (2019). Local auxin biosynthesis mediates plant growth and development. Trends Plant Sci. 24, 6–9. doi: 10.1016/j.tplants.2018.10.014
Ma, S., Tang, N., Li, X., Xie, Y., Xiang, D., Fu, J., et al. (2019). Reversible histone H2B monoubiquitination fine-tunes abscisic acid signaling and drought response in rice. Mol. Plant 12, 263–277. doi: 10.1016/j.molp.2018.12.005
Ma, T., Zhang, L., Wang, M., Li, Y., Jian, Y., Wu, L., et al. (2021). Plant defense compound triggers mycotoxin synthesis by regulating H2B ub1 and H3K4 me2/3 deposition. New Phytol. 232, 2106–2123. doi: 10.1111/nph.17718
Maeda, H., and Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Markulin, L., Skiljaica, A., Tokic, M., Jagic, M., Vuk, T., Bauer, N., et al. (2021). Taking the wheel—de novo DNA methylation as a driving force of plant embryonic development. Front. Plant Sci. 12, 764999. doi: 10.3389/fpls.2021.764999
Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H., Natsume, M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517. doi: 10.1073/pnas.1108434108
Mateo-Bonmati, E., Casanova-Saez, R., and Ljung, K. (2019). epigenetic regulation of auxin homeostasis. Biomolecules 9, 623. doi: 10.3390/biom9100623
McGinty, R. K., and Tan, S. (2014). “Histone, nucleosome, and chromatin structure,” in Fundamentals of Chromatin, eds J. L. Workman and S. M. Abmayr (New York, NY: Springer), 1–28. doi: 10.1007/978-1-4614-8624-4_1
Milutinovic, M., Lindsey, B. E. 3rd, Wijeratne, A., Hernandez, J. M., Grotewold, N., Fernandez, V., Grotewold, E., et al. (2019). Arabidopsis EMSY-like (EML) histone readers are necessary for post-fertilization seed development, but prevent fertilization-independent seed formation. Plant Sci. 285, 99–109. doi: 10.1016/j.plantsci.2019.04.007
Mizuguchi, G., Shen, X., Landry, J., Wu, W. H., Sen, S., and Wu, C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. doi: 10.1126/science.1090701
Mozgova, I., and Hennig, L. (2015). The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296. doi: 10.1146/annurev-arplant-043014-115627
Muller, J., and Verrijzer, P. (2009). Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19, 150–158. doi: 10.1016/j.gde.2009.03.001
Muller-Moule, P., Nozue, K., Pytlak, M. L., Palmer, C. M., Covington, M. F., Wallace, A. D., et al. (2016). YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 4, e2574. doi: 10.7717/peerj.2574
Nguyen, C. T., Tran, G. B., and Nguyen, N. H. (2020). Homeostasis of histone acetylation is critical for auxin signaling and root morphogenesis. Plant Mol. Biol. 103, 1–7. doi: 10.1007/s11103-020-00985-1
Normanly, J.. (2010). Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2, a001594. doi: 10.1101/cshperspect.a001594
Pavri, R., Zhu, B., Li, G., Trojer, P., Mandal, S., Shilatifard, A., et al. (2006). Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717. doi: 10.1016/j.cell.2006.04.029
Peng, M., Li, Z., Zhou, N., Ma, M., Jiang, Y., Dong, A., et al. (2018). Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiol. 176, 1341–1351. doi: 10.1104/pp.17.01189
Perrella, G., Baurle, I., and van Zanten, M. (2022). Epigenetic regulation of thermomorphogenesis and heat stress tolerance. New Phytol. 234, 1144–1160. doi: 10.1111/nph.17970
Poulios, S., and Vlachonasios, K. E. (2018). Synergistic action of GCN5 and CLAVATA1 in the regulation of gynoecium development in Arabidopsis thaliana. New Phytol. 220, 593–608. doi: 10.1111/nph.15303
Rizzardi, K., Landberg, K., Nilsson, L., Ljung, K., and Sundas-Larsson, A. (2011). TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J. 65, 897–906. doi: 10.1111/j.1365-313X.2010.04470.x
Roudier, F., Ahmed, I., Berard, C., Sarazin, A., Mary-Huard, T., Cortijo, S., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30, 1928–1938. doi: 10.1038/emboj.2011.103
Rymen, B., Kawamura, A., Lambolez, A., Inagaki, S., Takebayashi, A., Iwase, A., et al. (2019). Histone acetylation orchestrates wound-induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun. Biol. 2, 404. doi: 10.1038/s42003-019-0646-5
Schuettengruber, B., and Cavalli, G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542. doi: 10.1242/dev.033902
Schuettengruber, B., Chourrout, D., Vervoort, M., Leblanc, B., and Cavalli, G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745. doi: 10.1016/j.cell.2007.02.009
Semeradova, H., Montesinos, J. C., and Benkova, E. (2020). All roads lead to auxin: post-translational regulation of auxin transport by multiple hormonal pathways. Plant Commun. 1, 100048. doi: 10.1016/j.xplc.2020.100048
Simon, J. A., and Kingston, R. E. (2013). Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824. doi: 10.1016/j.molcel.2013.02.013
Stepanova, A. N., Hoyt, J. M., Hamilton, A. A., and Alonso, J. M. (2005). A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242. doi: 10.1105/tpc.105.033365
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L. M., Xie, D. Y., Dolezal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. doi: 10.1016/j.cell.2008.01.047
Stepanova, A. N., Yun, J., Robles, L. M., Novak, O., He, W., Guo, H., et al. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973. doi: 10.1105/tpc.111.088047
Stroud, H., Do, T., Du, J., Zhong, X., Feng, S., Johnson, L., et al. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21, 64–72. doi: 10.1038/nsmb.2735
Sun, J., Qi, L., Li, Y., Chu, J., and Li, C. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 8, e1002594. doi: 10.1371/journal.pgen.1002594
Sun, Y., Zhao, J., Li, X., and Li, Y. (2020). E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in Arabidopsis. New Phytol. 227, 455–472. doi: 10.1111/nph.16538
Sura, W., Kabza, M., Karlowski, W. M., Bieluszewski, T., Kus-Slowinska, M., Paweloszek, L., et al. (2017). Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 29, 791–807. doi: 10.1105/tpc.16.00573
Tanaka, H., Dhonukshe, P., Brewer, P. B., and Friml, J. (2006). Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell. Mol. Life Sci. 63, 2738–2754. doi: 10.1007/s00018-006-6116-5
Tang, N., Ma, S., Zong, W., Yang, N., Lv, Y., Yan, C., et al. (2016). MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28, 2161–2177. doi: 10.1105/tpc.16.00171
Tao, Y., Ferrer, J. L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Tasset, C., Singh Yadav, A., Sureshkumar, S., Singh, R., van der Woude, L., Nekrasov, M., et al. (2018). POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 14, e1007280. doi: 10.1371/journal.pgen.1007280
Teale, W. D., Paponov, I. A., and Palme, K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7, 847–859. doi: 10.1038/nrm2020
Tian, Y., Zheng, H., Zhang, F., Wang, S., Ji, X., Xu, C., et al. (2019). PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 5, eaau7246. doi: 10.1126/sciadv.aau7246
van der Woude, L. C., Perrella, G., Snoek, B. L., van Hoogdalem, M., Novak, O., van Verk, M. C., et al. (2019). HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc. Natl. Acad. Sci. U.S.A. 116, 25343–25354. doi: 10.1073/pnas.1911694116
Van Lijsebettens, M., and Grasser, K. D. (2014). Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci. 19, 717–726. doi: 10.1016/j.tplants.2014.07.002
Veloccia, A., Fattorini, L., Della Rovere, F., Sofo, A., D'Angeli, S., Betti, C., et al. (2016). Ethylene and auxin interaction in the control of adventitious rooting in Arabidopsis thaliana. J. Exp. Bot. 67, 6445–6458. doi: 10.1093/jxb/erw415
Vieten, A., Sauer, M., Brewer, P. B., and Friml, J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 12, 160–168. doi: 10.1016/j.tplants.2007.03.006
Wang, H., Liu, C., Cheng, J., Liu, J., Zhang, L., He, C., et al. (2016). Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of polycomb group proteins in association with distinct sets of cis-regulatory elements. PLoS Genet. 12, e1005771. doi: 10.1371/journal.pgen.1005771
Wang, Z., Cao, H., Zhang, C., Chen, F., and Liu, Y. (2022). The SNF5-type protein BUSHY regulates seed germination via the gibberellin pathway and is dependent on HUB1 in Arabidopsis. Planta 255, 34. doi: 10.1007/s00425-021-03767-1
Weijers, D., Nemhauser, J., and Yang, Z. (2018). Auxin: small molecule, big impact. J. Exp. Bot. 69, 133–136. doi: 10.1093/jxb/erx463
Weiste, C., and Droge-Laser, W. (2014). The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 5, 3883. doi: 10.1038/ncomms4883
Wojcikowska, B., Wojcik, A. M., and Gaj, M. D. (2020). Epigenetic regulation of auxin-induced somatic embryogenesis in plants. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21072307
Woloszynska, M., Le Gall, S., Neyt, P., Boccardi, T. M., Grasser, M., Langst, G., et al. (2019). Histone 2B monoubiquitination complex integrates transcript elongation with RNA processing at circadian clock and flowering regulators. Proc. Natl. Acad. Sci. U.S.A. 116, 8060–8069. doi: 10.1073/pnas.1806541116
Won, C., Shen, X., Mashiguchi, K., Zheng, Z., Dai, X., Cheng, Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523. doi: 10.1073/pnas.1108436108
Woodward, A. W., and Bartel, B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735. doi: 10.1093/aob/mci083
Wu, L., Luo, P., Di, D. W., Wang, L., Wang, M., Lu, C. K., et al. (2015). Forward genetic screen for auxin-deficient mutants by cytokinin. Sci. Rep. 5, 11923. doi: 10.1038/srep11923
Xu, L., and Shen, W. H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18, 1966–1971. doi: 10.1016/j.cub.2008.11.019
Xu, Y., Gan, E. S., Zhou, J., Wee, W. Y., Zhang, X., and Ito, T. (2014). Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res. 42, 10960–10974. doi: 10.1093/nar/gku781
Xu, Y., Prunet, N., Gan, E. S., Wang, Y., Stewart, D., Wellmer, F., et al. (2018). SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 37. doi: 10.15252/embj.201797499
Xue, M., Zhang, H., Zhao, F., Zhao, T., Li, H., and Jiang, D. (2021). The INO80 chromatin remodeling complex promotes thermomorphogenesis by connecting H2A.Z eviction and active transcription in Arabidopsis. Mol. Plant 14, 1799–1813. doi: 10.1016/j.molp.2021.07.001
Yamada, M., Greenham, K., Prigge, M. J., Jensen, P. J., and Estelle, M. (2009). The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 151, 168–179. doi: 10.1104/pp.109.138859
Yamamuro, C., Zhu, J. K., and Yang, Z. (2016). Epigenetic modifications and plant hormone action. Mol. Plant 9, 57–70. doi: 10.1016/j.molp.2015.10.008
Yan, W., Chen, D., Smaczniak, C., Engelhorn, J., Liu, H., Yang, W., et al. (2018). Dynamic and spatial restriction of polycomb activity by plant histone demethylases. Nat. Plants 4, 681–689. doi: 10.1038/s41477-018-0219-5
Zhang, H., Lang, Z., and Zhu, J. K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhang, L., Luo, P., Bai, J., Wu, L., Di, D. W., Liu, H. Q., et al. (2021). Function of histone H2B monoubiquitination in transcriptional regulation of auxin biosynthesis in Arabidopsis. Commun. Biol. 4, 206. doi: 10.1038/s42003-021-01733-x
Zhang, M., Hu, X., Zhu, M., Xu, M., and Wang, L. (2017). Transcription factors NF-YA2 and NF-YA10 regulate leaf growth via auxin signaling in Arabidopsis. Sci. Rep. 7, 1395. doi: 10.1038/s41598-017-01475-z
Zhao, J., Chen, Q., Zhou, S., Sun, Y., Li, X., and Li, Y. (2020). H2Bub1 regulates RbohD-dependent hydrogen peroxide signal pathway in the defense responses to verticillium dahliae toxins. Plant Physiol. 182, 640–657. doi: 10.1104/pp.19.00913
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., et al. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. doi: 10.1126/science.291.5502.306
Zhou, J. X., Su, X. M., Zheng, S. Y., Wu, C. J., Su, Y. N., Jiang, Z., et al. (2022). The Arabidopsis NuA4 histone acetyltransferase complex is required for chlorophyll biosynthesis and photosynthesis. J. Integr. Plant Biol. 64, 901–914. doi: 10.1111/jipb.13227
Zhou, Y., Tergemina, E., Cui, H., Forderer, A., Hartwig, B., Velikkakam James, G., et al. (2017). Ctf4-related protein recruits LHP1-PRC2 to maintain H3K27me3 levels in dividing cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 114, 4833–4838. doi: 10.1073/pnas.1620955114
Keywords: auxin biosynthesis, epigenetic modifications, histone H3 methylation, histone acetylation, chromatin remodeling, histone H2B monoubiquitination, DNA methylation
Citation: Wang J-L, Di D-W, Luo P, Zhang L, Li X-F, Guo G-Q and Wu L (2022) The roles of epigenetic modifications in the regulation of auxin biosynthesis. Front. Plant Sci. 13:959053. doi: 10.3389/fpls.2022.959053
Received: 01 June 2022; Accepted: 15 July 2022;
Published: 09 August 2022.
Edited by:
Jungnam Cho, Center for Excellence in Molecular Plant Sciences (CAS), ChinaReviewed by:
Eun Yu Kim, Duke Kunshan University, ChinaSoon-Ki Han, Daegu Gyeongbuk Institute of Science and Technology (DGIST), South Korea
Ying Hua Su, Shandong Agricultural University, China
Copyright © 2022 Wang, Di, Luo, Zhang, Li, Guo and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Wu, bGVpd3VAbHp1LmVkdS5jbg==
 Jun-Li Wang
Jun-Li Wang Dong-Wei Di
Dong-Wei Di Pan Luo
Pan Luo Li Zhang
Li Zhang Xiao-Feng Li
Xiao-Feng Li Guang-Qin Guo
Guang-Qin Guo Lei Wu
Lei Wu