- 1Department of Forestry, Shenyang Agricultural University, Shenyang, China
- 2Biotechnology and Analysis Test Center, Liaoning Academy of Forest Science, Shenyang, China
Auxin plays an essential role in flowering, embryonic development, seed dormancy, and germination. Auxin response factors (ARFs) are plant-specific key transcriptional factors in mediating the gene expression network of auxin signaling. Although ARFs in model plants such as Arabidopsis had been well characterized, their identities and potential roles in non-model plants are less studied. Here, we performed genome-wide identification of ARFs in Magnolia sieboldii K. Koch, a primitive species with high taxonomic importance and medicinal values. We found 25 ARF genes in M. sieboldii, which were widely distributed across multiple chromosomes. Based on sequence similarity, the encoded proteins could be either transcriptional repressors or activators. Gene expression analysis showed a dynamic pattern for many ARFs including MsARF5 during seed germination. In addition, overexpressing of MsARF5 showed that it restores many developmental defects in the Arabidopsis mutant. Moreover, two phenotypically distinct transgenic Arabidopsis lines were obtained, indicating a link between gene expression levels and developmental phenotypes. Taken together, we provided a systematic investigation of the ARF gene family in M. sieboldii and revealed an important role of MsARF5 in mediating auxin signaling.
1 Introduction
Auxin controls many critical events of plant life including lateral root formation, embryonic development, seed dormancy, and germination. In the model plant Arabidopsis, early embryonic patterning is largely dependent on the biosynthesis, transport, and signaling of auxin (Moller and Weijers, 2009). The role of auxin in seed germination has long been recognized due to its antagonistic effect on abscisic acid (ABA) (Rousselin et al., 1992). More recently, a role of indole-3-acetic acid (IAA) in directly regulating seed dormancy and germination has also been discovered (Belin et al., 2009; Wang et al., 2011; Boter et al., 2019).
Genetic studies had identified two families of key transcriptional factors (TFs) in mediating auxin signaling: auxin response factors (ARFs) and Aux/IAA proteins (Woodward and Bartel, 2005; Quint and Gray, 2006). ARFs are a group of plant-specific proteins typically consisting of an N-terminal DNA-binding domain (DBD), a non-conserved middle region (MR), and a carboxyl-terminal dimerization domain (CTD) in mediating protein-protein interaction. ARF proteins regulate the expression of auxin-responsive genes by binding to conserved promoter regions such as the auxin-response element (TGTCTC). They can also form dimers by interacting with Aux/IAA proteins (Tiwari et al., 2003). Interestingly, ARFs could act as either transcriptional activators or repressors depending on the amino acid compositions in the MR region. For instance, auxin contributes to seed dormancy by regulating ABSCISIC ACID INSENSITIVE 3 (ABI3) via ARF10 and ARF16, two transcriptional repressors (Liu et al., 2013). In addition, the transcriptional activity of ARFs is also controlled by IAA concentration and different expression levels of ARFs may lead to distinct phenotypes. Thus, the exact role of ARFs in specific physiological contexts deserves detailed investigation.
The first identification of plant ARFs was performed in Arabidopsis by yeast one-hybrid (Ulmasov et al., 1997). Following studies identified a total of 23 ARF genes in Arabidopsis (Okushima et al., 2005). These genes are distributed on all five chromosomes and may encode proteins of high sequence similarity (e.g., ARF1 and ARF2). With the availability of genome sequencing data, genome-wide identifications and functional characterization of ARFs had been performed in multiple species. For example, 31 ARFs were reported in maize (Zea mays) with dynamic expression patterns during embryo development, indicating a potential role of these ARFs in regulating pattern formation of the embryo (Xing et al., 2011). Importantly, 18 out of the 31 ARFs in maize were putative targets of small RNAs, which can regulate the expression of ARFs by degradation of the transcripts.
Among the 23 ARFs in Arabidopsis, ARF5/MONOPTEROS (MP) is essential in auxin signaling during embryo patterning (Hardtke and Berleth, 1998; Odat et al., 2014). ARF5/MP mediates the auxin-induced embryonic root initiation and promotes the transport of auxin from embryonic cells to extraembryonic suspensor cells (e.g., precursor cells of the quiescent center) during embryonic cell specification (Rademacher et al., 2012). In addition, AtARF5 also regulates plant growth and development such as the formation of cotyledons (Cole et al., 2009), apical meristem (Zhao et al., 2010), and vascular tissues in leaves (Donner et al., 2009). Mutation of ARF5/MP in Arabidopsis results in impaired body axis patterning during the early embryogenic stage (e.g., lack of embryonic root) and decreased complexity in vascular patterning. It has also been demonstrated that mutation of ARF5/MP leads to an abnormal expression of AMP1 (altered meristem program 1) and thus abnormal embryonic differentiation (Vidaurre et al., 2007). Analysis of ARF5/MP mutants featuring deletion of different parts of the protein also revealed distinct functions of each domain. For instance, deletion mutations toward the CTD tend to result in a stronger effect phenotypically, suggesting an important role of CTD (e.g., for dimerization) in protein functioning (Liscum and Reed, 2002).
Magnolia sieboldii K. Koch is a small deciduous tree of the Magnolia family (Magnoliaceae). As a relatively primitive angiosperm species with high taxonomic importance, it also has ornamental, aromatic, and medicinal values. Seed germination in M. sieboldii is difficult under both natural conditions and even after stratification (Li et al., 2006; Lu et al., 2014; Mei et al., 2017). Recently, a sequencing analysis revealed 13,877 genes showing differential expression patterns between germinated and non-germinated M. sieboldii seeds (Mei et al., 2021). Among them, 38 auxin response factors were up-regulated in the germinated seeds, confirming an important role of auxin in mediating seed germination in M. sieboldii. However, the molecular mechanism by which auxin and ARF genes regulate seed germination and other aspects of plant phenotypes remain unclear.
Here, we performed genome-wide identification of ARFs in M. sieboldii and characterized their chromosomal locations, gene structure, protein sequence similarity, and dynamic expression pattern during seed germination. Furthermore, we overexpressed MsARF5 in Arabidopsis and found that it could rescue multiple defects of the Arabidopsis ARF5/MP mutant. The impact of overexpressing MsARF5 on vegetive growth, flowering, and auxin response was also investigated.
2 Materials and methods
2.1 Sequence identification
ARF proteins in Arabidopsis and rice (Wang et al., 2007) were downloaded from TAIR (http://www.Arabidopsis.org) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), respectively. Sequences from other families were obtained according to previous studies that characterized ARFs in Medicago truncatula (Shen et al., 2015), apple (Wang et al., 2020), and Vitis vinifera (Wan et al., 2014). The obtained sequences were used to search the M. sieboldii protein database from NGDC with the BioProject No. of PRJCA009763. In addition, ARF domains (PF06507, PF02362, and PF02309) were used as queries to search the protein family database (PFAM). Top hits were checked by domain analysis using the NCBI CDD tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and Pfam (https://pfam.xfam.org).
2.2 Bioinformatics analysis
The chromosomal location of each MsARF was obtained from the M. sieboldii genome database. Motif analysis was performed using MEME Suite (version 4.12.0) with the following parameters: site distribution of 0 or 1 occurrence per sequence, number of motifs of 10, and motif size of 10 to 200 amino acid residues (http://meme-suite.org/tools/meme). Conserved domains were visualized using TBtools (v1.046, https://github.com/CJ-Chen/TBtools) (Chen et al., 2020). Multiple sequence alignment was performed using ClustalX (Larkin et al., 2007). A phylogenetic tree was constructed using MEGA-X (Kumar et al., 2018) with the neighbor-joining (NJ) algorithm and 1000 bootstrap replicates. Hidden Markov model (HMM 3.0) (Finn et al., 2011) software is used to obtain the starting position information of MsARFs gene on the chromosome from the genome annotation information of M. sieboldii. Homologous replication were evaluated via blastp of MsARF genes (Ai et al., 2022).
2.3 Plant materials and treatments
M. sieboldii seeds were collected from the Botanical Garden of Shengyang. Seeds were air-dried and stored at 4°C. Treatment for seed germination was performed as described (Mei et al., 2017). After soaking in water for 72 h, the seed coat was removed. Surface disinfection was performed in 0.25% KMnO4 for 30 min. Seeds were then mixed with wet sand (v/v = 1:3) for stratification with the following regimen: low temperature (0 - 10°C) for 30 days, varying temperature (day of 8 - 10°C, night of -5 - 0°C) for 15 days, and high temperature (15°C) for 30 days. For imbibition experiments, seeds were sampled at various time points before (up to 12 h) and after the initiation of imbibition (every 6 h till 72 h). For experiments involving laminated samples, sampling was performed every 15 days for 90 days.
Arabidopsis plants including the wild type Col-0 and the T-DNA mutant of ARF5 (salk_001058) were obtained from The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/index.jsp). Seeds were surface-sterilized with 75% ethanol (30 s) and 1% NaClO (15 min) and then plated on half-strength MS plates. After stratification at 4°C for 3 days, they were maintained at 20-23°C with a 16/8 h of light/dark cycle. Seedlings with 2-4 true leaves were transferred to the soil for further growth.
2.4 Gene expression analysis
Real-time quantitative reverse transcription PCR (qRT-PCR) was used for gene expression analysis. Briefly, RNA was isolated using a RNAprep pure plant kit (Tiangen, China). cDNA was synthesized using a PrimeScript™ II 1st strand cDNA synthesis kit (TAKARA, Japan). qRT-PCR was carried out using FastFire qPCR PreMix(SYBR Green) (Tiangen, China) on an ABI Stepone plus system (CA, USA). Primers used were listed in Table S1. The 2-ΔΔCT method was used to calculate relative expression levels. Actin was used for normalization. For all experiments, three biological replicates with two technical replicates were performed.
2.5 Quantficaiotn of Auxin
Endogenous auxin was determined using an ELISA kit (Hailian Biological Company). Briefly, seed samples were homogenized with 4 mL of PBS (PH7.4) and then centrifuged at 2000 rpm. The supernatant was added to the wells and incubated with enzyme-linked antibodies at 37°C for 60 min. After washing and signal development, absorbance at 450 nm was quantified. A standard curve was built for concentration calculation.
2.6 Transgenic lines
Two vectors were created for reverse genetics studies: full-length MsARF5 driven by either the 35S promoter (35S::MsARF5) or its native promoter (MsARF5-Pro::MsARF5). For 35S::MsARF5, MsARF5 was amplified by PCR with the introduction of cutting sites of SalI and SacI. The PCR product was inserted into pRI101. For MsARF5-Pro::MsARF5, the promotor region of MsARF5 was fused to the coding sequence of MsARF5 and then inserted into pCAMBIA3301 as described previously (Lei, 2007). All constructs were confirmed by double digestion and sequencing. Both vectors were introduced to Agrobacterium tumefaciens GV3101 strain by the freeze-and-thaw method (Yu et al., 2003).
Transformation of Arabidopsis plants was performed as described (Clough and Bent, 1998). Three transformation experiments were conducted with the following constructs and plants (Table 1): OE1 (35S::MsARF5 in the Col-0 background), OE2 (35S::MsARF5 in the homozygous plant arf-/-), and OE3 (MsARF5-Pro::MsARF5 in the Col-0 background). Seeds of the transgenic lines were screened on half MS with 30 mg/L Kan and further validated by PCR.
2.7 Phenotypic analysis
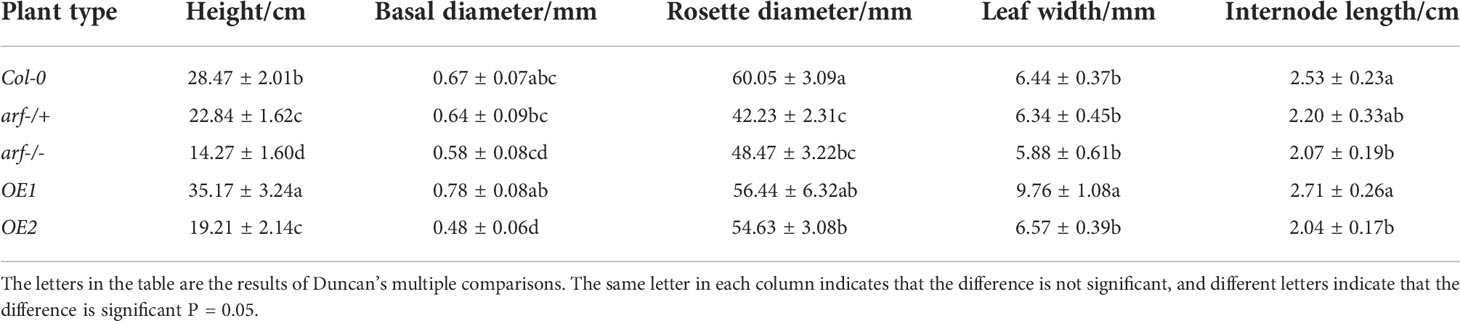
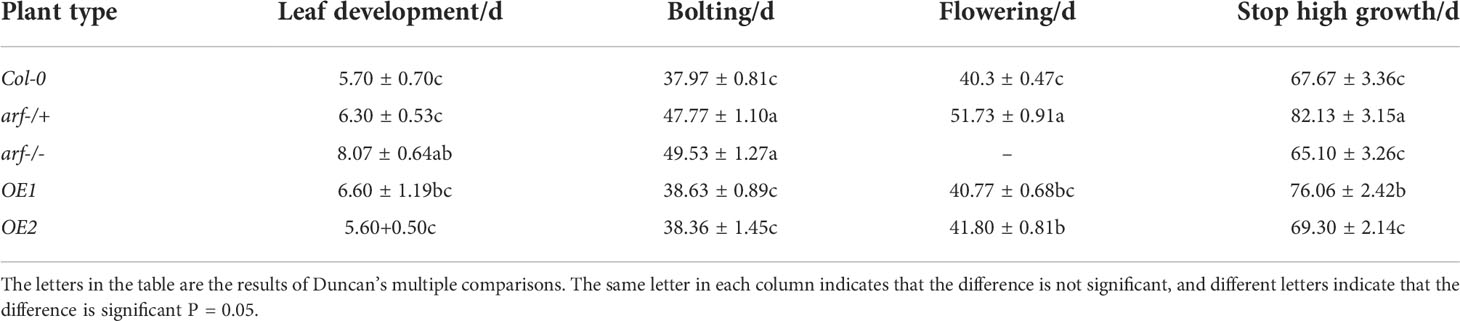
Seeds of the third generation of transgenic plants were used. Parameters on plant growth including plant height, ground diameter, rosette diameter, leaf length, leaf width, and internode were measured. In addition, seed germination, bolting, flowering, and senescence were also quantified. Thirty plants were used for each group, and three biological replicates were performed.
2.8 Auxin responses
Seeds of the third generation of transgenic plants were used. Half-strength MS medium was prepared either without hormone (for controls) or in the presence of 1 µM of IAA or IBA. Seeds were surface-sterilized and planted on the plate. Observations were made 5 days later under a microscope (Carl Zeiss AG). Forty seeds were used for each group, and three biological replicates were performed.
2.9 Statistical analysis
Statistical analysis was performed using the SPSS20.0. Gene expression analysis was performed using TBtools (v1.046). GraphPad Prism 5 was used for data visualization.
3 Results
3.1 Genome-wide identification of ARF genes in M. sieboldii
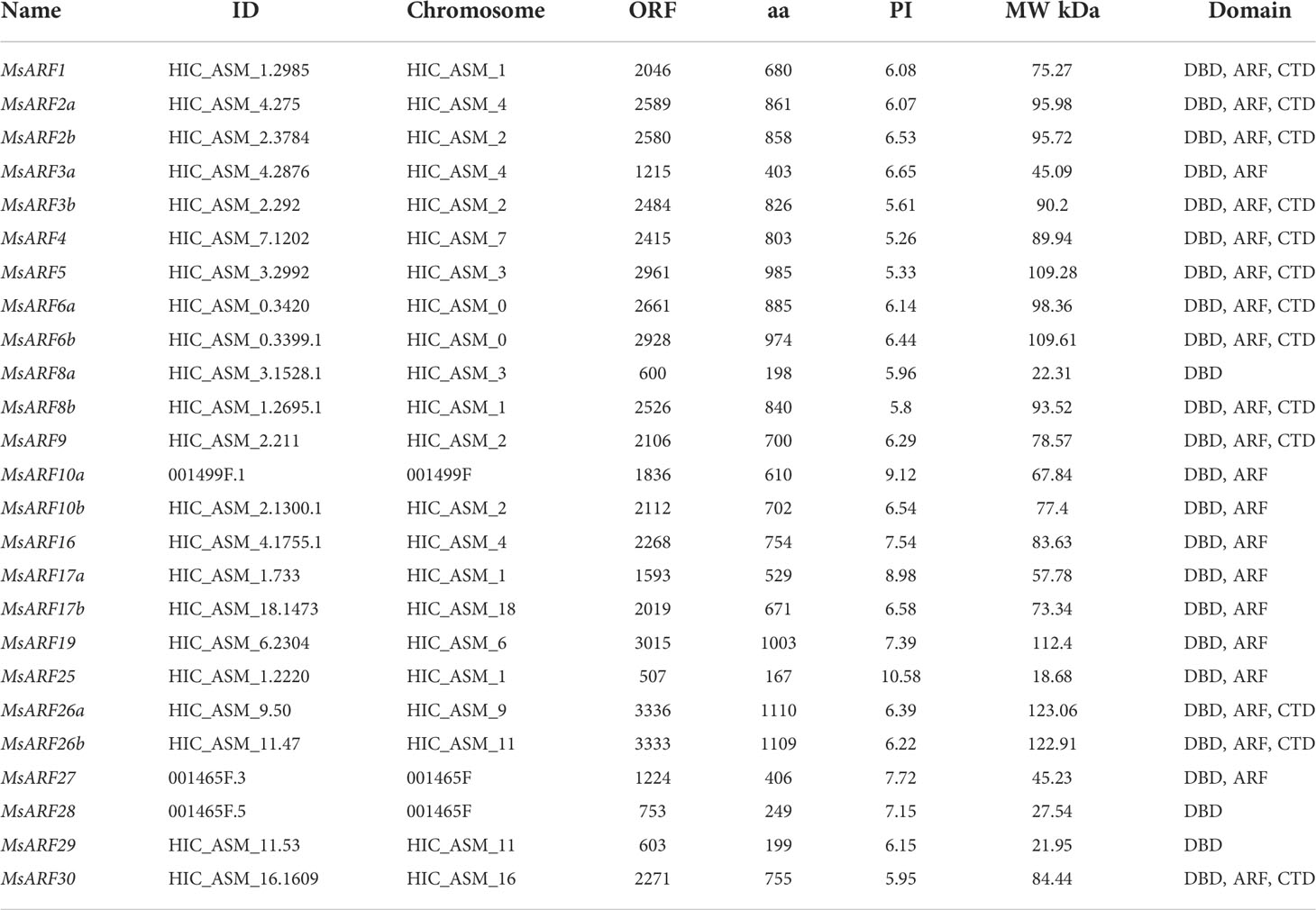
Homology search revealed that the genome of M. sieboldii encode 25 ARF genes, which were named based on similarity to that of Arabidopsis (Table 2). Amon the 25 genes, 23 were localized to 12 chromosomes including HIC_ASM_0 (n = 2), HIC_ASM_1 (n = 4), HIC_ASM_2 (n = 4), HIC_ASM_3 (n = 2), HIC_ASM_4 (n = 3), HIC_ASM_6 (n = 1), HIC_ASM_7 (n = 1), HIC_ASM_9 (n = 1), HIC_ASM_11 (n = 2), HIC_ASM_16 (n = 1), and HIC_ASM_18 (n = 1, Figure S1). The other two genes were localized to unassembled chromosomal fragments. In addition, these gene varied greatly in length, as the open reading frame (ORF) ranged from 507 to 3,336 bp. In line with the variation in gene length, there were different numbers of exons (2-14) in the putative MsARF genes (2-3 for 4 genes; 4-9 for 6 genes; 11-13 for 5 genes; and 14 for 10 genes).
3.2 Putative MsARF proteins and bioinformatic analysis
The deduced proteins of these genes ranged from 167 to 1,110 amino acid residues (18.68 to 123.06 kDa) with a wide range of pI values (5.26 to 10.58). Phylogenetic analysis classified 25 MsARFs into four classes (Figure 1). Class I consisted of MsARF10a/b, MsARF16, and MsARF17a/b (homologs of ARF10/16/17 in Arabidopsis). Class II consisted of MsARF5, MsARF6a/b, MsARF8a/b, MsARF19, MsARF25, MsARF26a/b, and MsARF27-29 (homologs of ARF5/8 in Arabidopsis). Class III consisted of MsARF3a/b, and MsARF4 (homologs of ARF3/4 in Arabidopsis). Lastly, class IV consisted of MsARF1, MsARF2a/b, MsARF9, and MsARF30 (homologs of ARF1/2 in Arabidopsis).
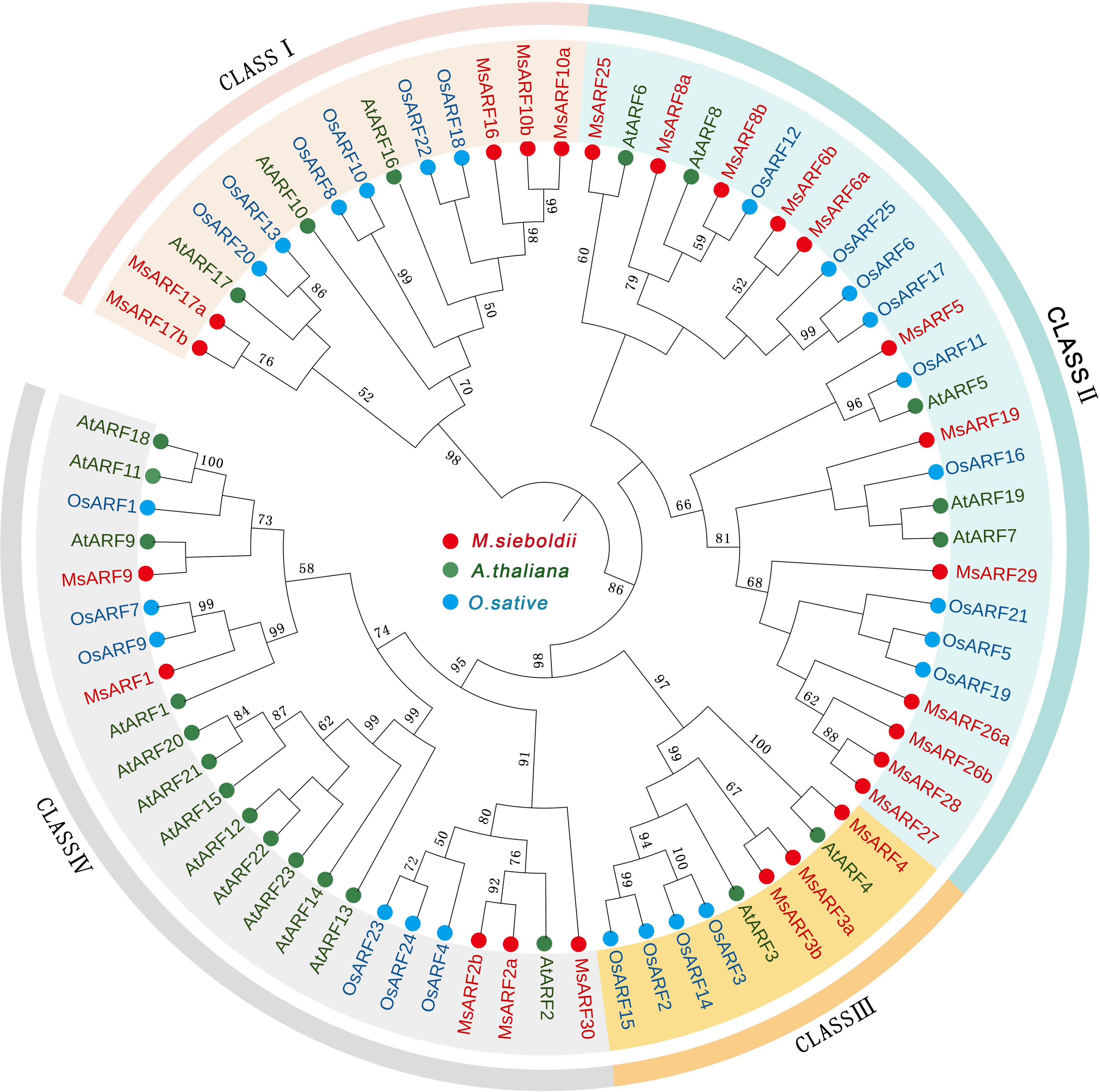
Figure 1 Identification of ARFs in M. sieboldii and phylogenetic analysis. The unrooted tree was constructed using MEGA-X with the neighbor-joining method (bootstrap of 1000).
MsARFs can also be classified based on the presence of a C-terminal dimerization domain (CTD) and the nature of the middle region (MR), which could function as either transcriptional repression or activation domain (Table 2). Thus, while seven MsARFs (MsARF5, MsARF6a/b, MsARF8b, MsARF19, and MsARF26a/b) were identified as putative activator due to the presence of a Q/S/L-rich MR domain, seven MsARFs (MsARF1, MsARF2a/b, MsARF3b, MsARF4, MsARF9, and MsARF30) were identified as putative repressors since they harbor a P/G-rich MR domain. In addition, three MsARFs had only the conserved N-terminal DNA binding domain (DBD) but no CTD or MR. Transcriptional activities for these MsARFs are unclear. Furthermore, there were eight MsARFs showing no CTD domain that could be either activators (MsARF3a, MsARF10a/b, MsARF16, and MsARF17a/b) or repressors (MsARF25 and MsARF27).
In addition, 10 conserved motifs ranging from 16 to 50 amino acid residues were identified. Among these motifs, motifs 1-5 belong to DBD (B3 domains), motifs 6-8 belong to ARF (Auxin responsive domains), and motifs 9/10 belong to CTD (AUX/IAA domains). Notably, motifs 1-5 were found in all MsARFs expect MsARF10a, MsARF17a, and MsARF25. While motifs 6-8 resided in the MR region, motifs 9 and 10 were within the CTD domain. Although the arrangement of these motifs was consistent in most MsARFs, duplication of some motifs was also found including an extra motif 1 in the MR region of MsARF5 and an extra motif 7 in MsARF3b. The presence of these duplicated motifs may indicate their key roles in mediating specific functions.
3.3 MsARF5 is significantly upregulated during seed germination
Given the important role of ARF during seed development, we examined the gene expression pattern of MsARF genes at various seed developmental stages. As shown in Figure 2A, multiple genes showed a dynamic expression pattern after various stratification times. The expression pattern of MsARF10/16, which are related to root crown development and root tropism, was the same with an up-regulation at 45 d and 90 d. A similar expression pattern was seen for MsARF17a/b, indicating a similar structure and function of MsARF17a/b compared to that of MsARF10/16. It is noteworthy that a negative correlation in expression between ARF10/16 and ARF5, which promote seed dormancy and germination, respectively, was also found. Because of the dominant role of ARF5 in seed germination (Galstyan and Nemhauser, 2019), we also determined the gene expression pattern of MsARF5 in various tissues and seed germination stages. We found a relatively low expression in dry seed, a stable expression in shoot and leaf, and a significantly high level in germinating seeds (Figure 2B).
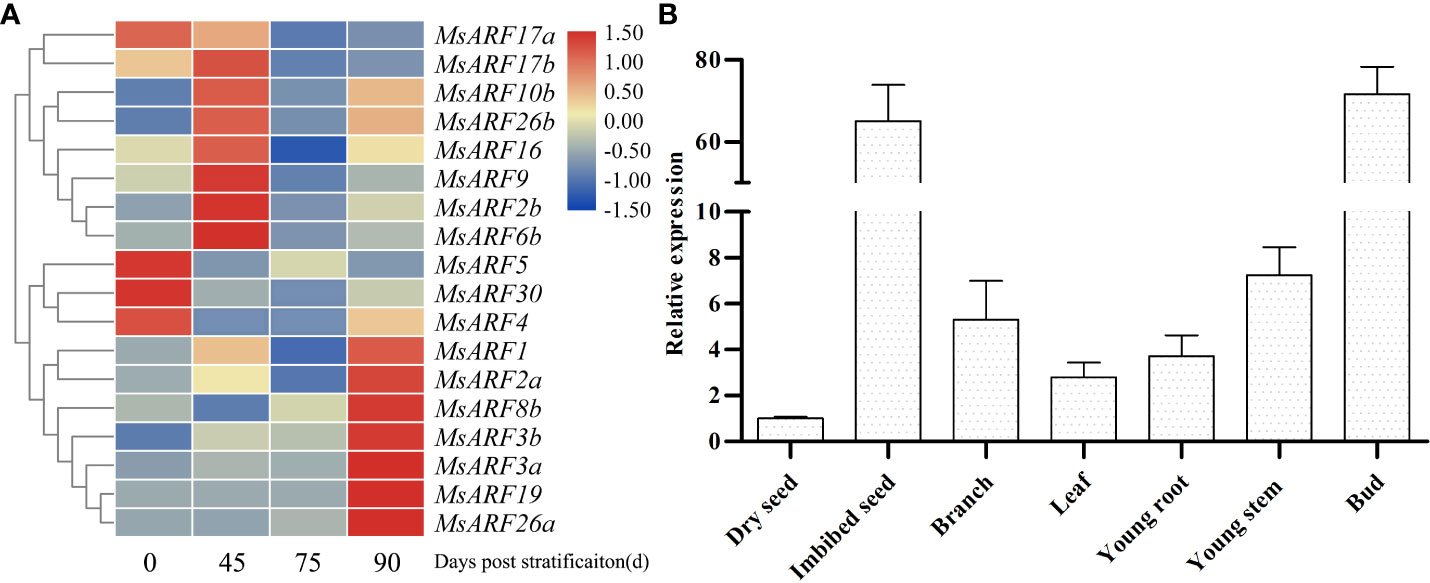
Figure 2 Gene expression analysis of MsARFs. (A) Expression of 25 MsARFs during seed germination. (B) Expression of MsARF5 in various tissues.
To further characterize the potential role of MsARF5 during seed germination, we performed a time-course gene expression analysis after imbibition. The expression of MsARF5 showed an initial decrease at 1 h after imbibition, followed by a gradual increase, and a sharp increase at 6-12 h after imbibition. Such a high-level expression was maintained up to 72 h after imbibition (Figure 3A). Similarly, we profiled the impact of stratification time on MsARF5 expression and found a general decrease trend (Figure 3B). Significant, we also quantified auxin and found a similar trend between the level of auxin and MsARF5 expression in both events (seed imbibition and stratification in Figures 3C, D, respectively). This indicated that MsARF5 is positively regulated by endogenous auxin in seeds. Given the morphological changes in the embryo during stratification, it was speculated that MsARF5 is involved in embryonic development after maturation (0-30 days after stratification), cell elongation, and rapid growth of seed embryos (60-75 days).
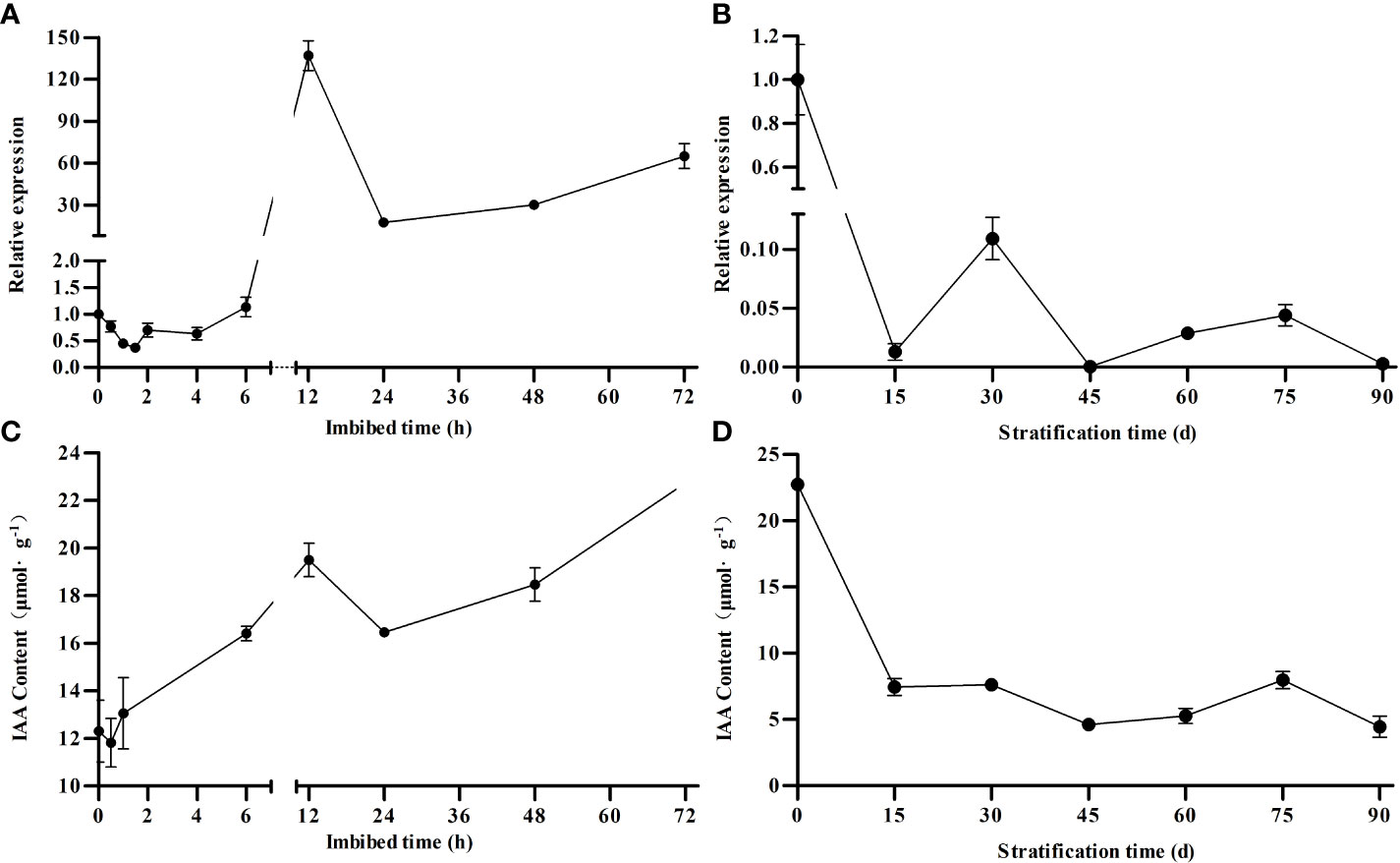
Figure 3 Correlation between MsARF5 expression and auxin concentration following seed stratification and imbibition. (A, B): relative expression of MsARF5 following stratification and imbibition. (C, D): auxin concentration following stratification and imbibition.
3.4 Overexpressing MsARF5 in Arabidopsis rescues the Arabidopsis arf5 mutant in vegetative growth
For further functional investigation, we created transgenic plants overexpressing (OE) MsARF5 in the Col-0 A. thaliana. And the MsARF5 expression results of the transgenic lines were obtained (Figure 4E).The expression of OE1 was significantly higher than that of OE2, and MsARF5 was not detected in Col-0, arf-/- or arf+/-. It is speculated that the expression of MsARF5 may be affected by AtARF5. The phenotypic difference of OE1/OE2 may be related to the expression of MsARF5.
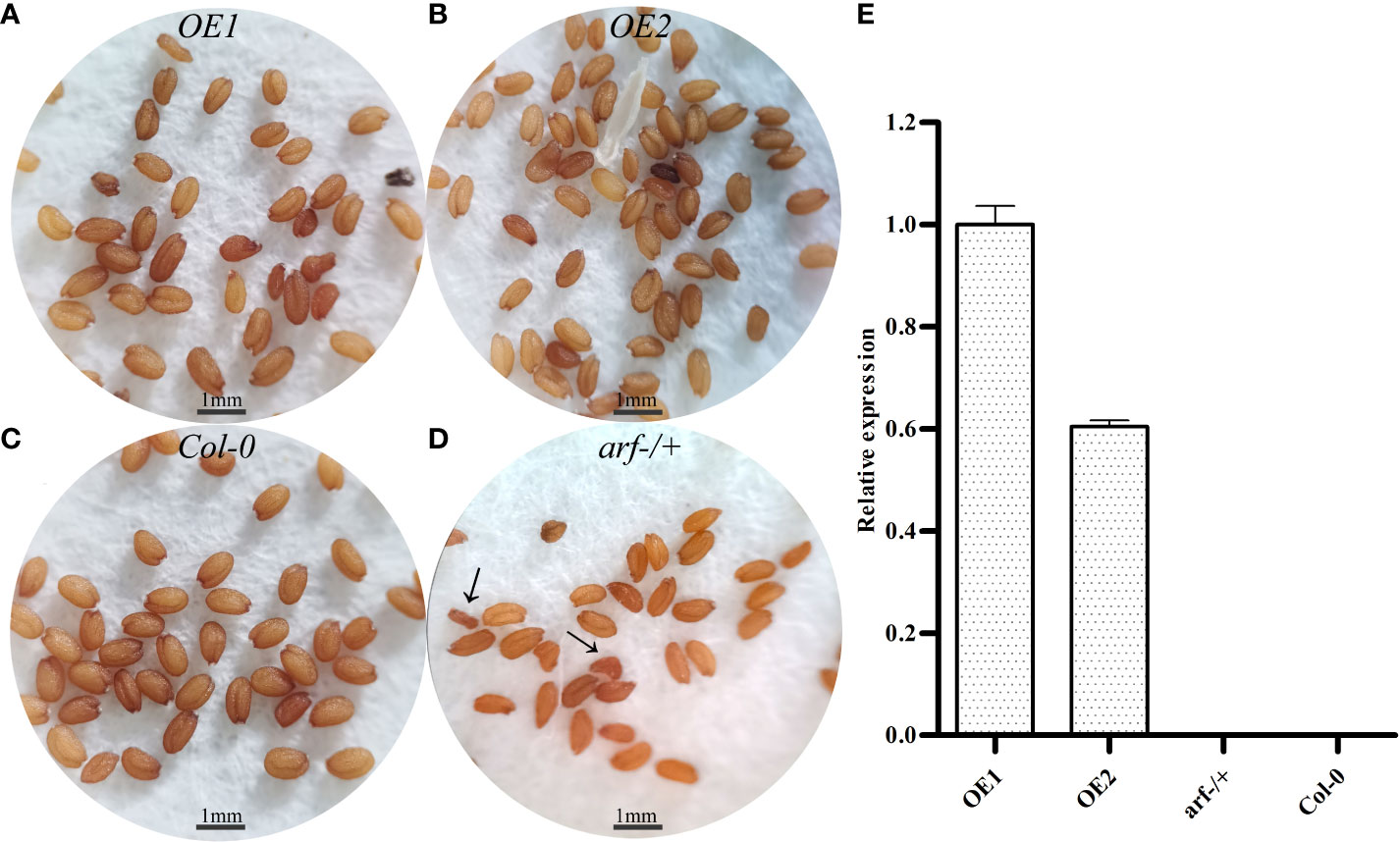
Figure 4 Phenotypes of transgenic A. thaliana overexpressing MsARF5. (A-D): Seed morphology of OE1, OE2, WT (Col-0) and arf-/+ (heterozygous mutant of AtARF5) (E): Results on RT-qPCR of transgenic plants overexpressing.
The phenotypes of OE1 and OE2 plants were compared to that of wild type (Col-0), heterozygous/homozygous Arabidopsis mutants of AtARF5 (arf-/+ and arf-/-, respectively). Our results (summarized in Table 3) showed that the OE1 plants were significantly taller (35.17 ± 3.24 cm in height) with broader leaves (9.76 ± 1.08 mm) compared to the Col-0 plants. Such an enhancement in the growth of the OE1 plants was accompanied by well-developed vascular tissues. Thus, these results showed that overexpressing MsARF5 promotes plant growth. We also overexpressed MsARF5 in the homozygous arf-/- mutant (designated as OE2) and found that the transgenic plants resembled the heterozygous mutant phyenotypically. In addition, OE2 outperformed the arf-/- mutant in multiple plant growth parameters such as plant height, rosette leaf diameter, and leaf width. This suggested that MsARF5 was able to rescue the Arabidopsis arf 5 mutant in vegetative growth.
3.5 Impact of MsARF5 overexpression on flowering and seed development
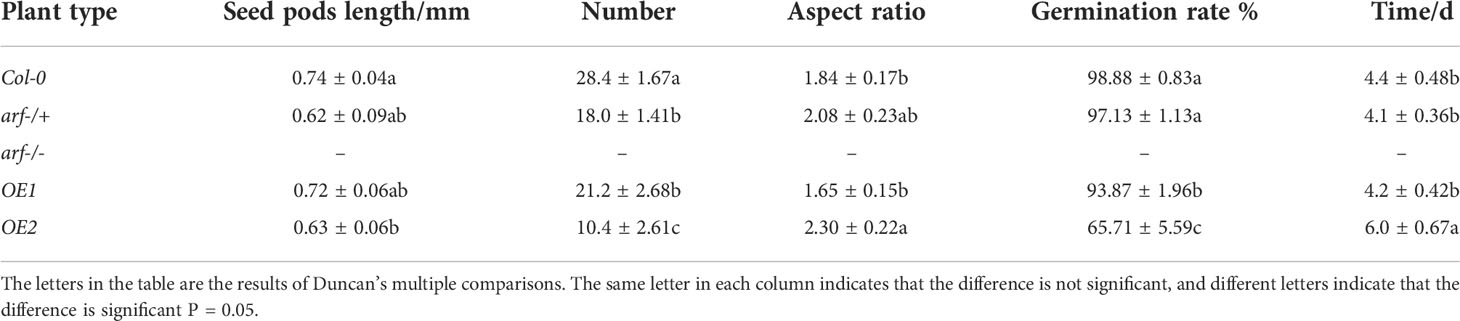
We next investigated the impact of MsARF5 overexpression on flowering and seed development. Compared to Col-0, the homozygous mutant arf-/- plants showed a delay in leaf development and bolting (Table 4). No flowering was observed in the homozygous mutant arf-/- plants. By contrast, the OE1 and the OE2 plants show no difference compared to the Col-0 except that OE1 had a longer life cycle. Seed phenotypes in various Arabidopsis lines were summarized in Table 5. In line with the notion that insufficient AtARF5 expression could lead to defects in seed development, we found that ~12.5% of the arf-/+ plants showed abnormal development (Figure 4D). Although overexpressing MsARF5 in Arabidopsis allowed the flower formation in the arf-/- mutant, significant differences in seedpod length, seed number, and the aspect ratio were found between the arf-/+ and Col-0 plants. Except for OE2, the seed number of OE1, WT and arf-/+ was inversely proportional to the growth cycle. It can be speculated that the overexpression of MsARF5 can preferentially promote the vegetative growth of plants. However, arf-/- seeds could not develop normally.
3.6 Response to exogenous auxin in transgenic plants
Next, we determined the response to IAA and indole-3-butyric acid (IBA) in Arabidopsis plants of different genetic backgrounds (Figure 5). For this experiment, we also created a transgenic line in which the expression of MsARF5 is driven by the native promoter (designated as OE3). First, we observed that both IAA and IBA promoted the primary root elongation in the Col-0, OE1, and OE3 plants (Figures 5A-C). Notably, the impact of IBA was far more significant than that of IAA, especially in the OE3 plants. Moreover, a significant increase in root elongation was observed in the OE2 plants following exposure to IBA, but not IAA (Figure 5D). This indicates that MsARF5 is sensitive to exogenous auxin, and the response to 1 μM IBA is higher than that of 1 μM IAA. These results also verified that ARF5 is an essential gene for seed embryo morphogenesis under auxin signaling.
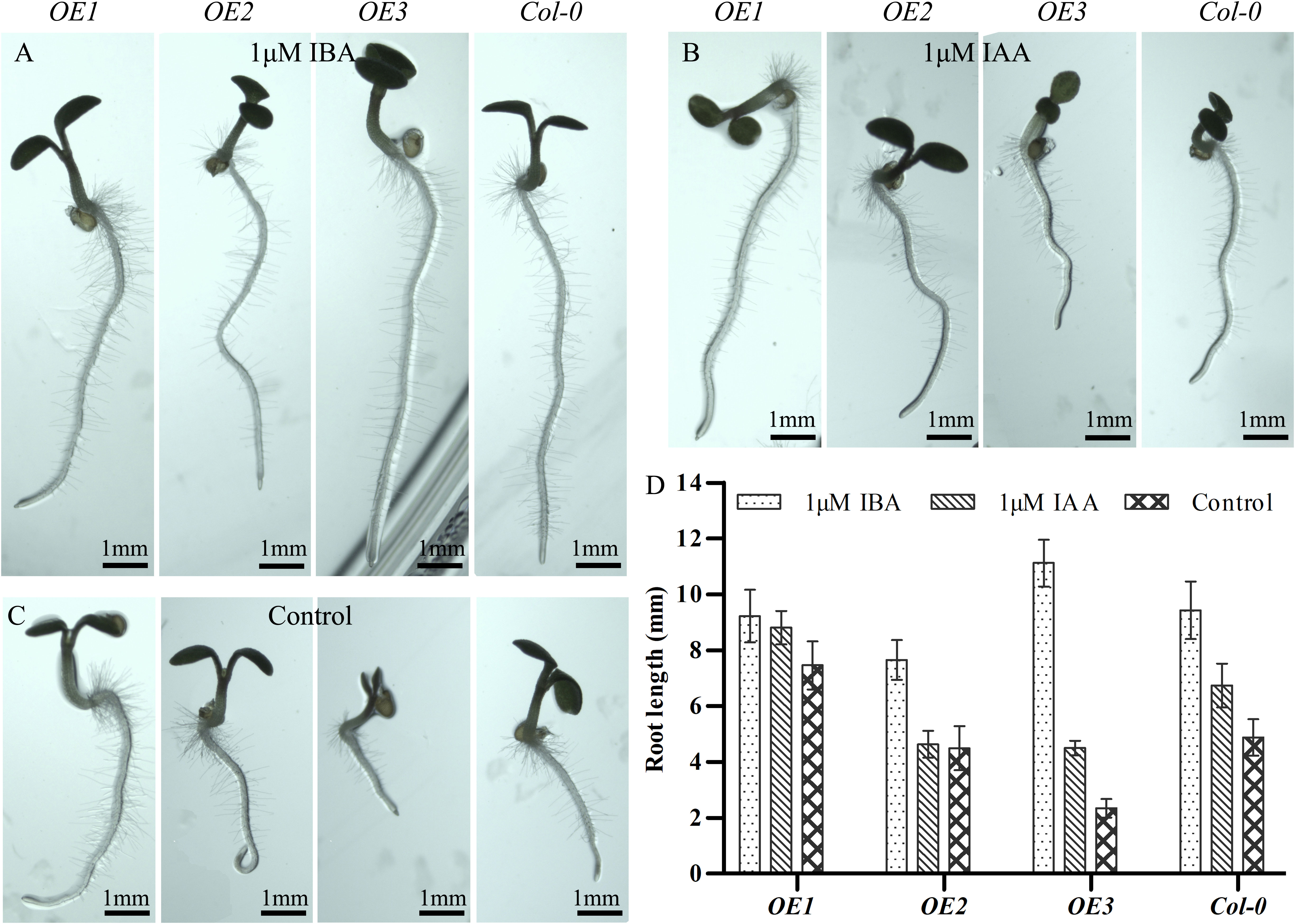
Figure 5 Response to exogenous auxin in transgenic Arabidopsis overexpressing MsARF5.(A–C). Impact of exogenous IBA (A) and IAA (B) on transgenic seedlings overexpressing MsARFs compared to controls (C). (D) Quantitative analysis on root length following exposure to IBA/IAA. The letters in the figure are the results of Duncan’s multiple comparisons. The same letter in each column indicates that the difference is not significant, and different letters indicate that the difference is significant P = 0.05.
4 Discussion
Here we found 4 groups of ARFs in M. sieboldii, Class I–IV. Each class has a counterpart in M Sieboldii with Class I–III in A. thaliana corresponding to Class V, I and II, respectively. Similar analysis also identified pairing class of ARFs between M. sieboldii and M. truncatula. Interestingly, M. truncatula showed 8 groups of ARFs and class VIII may be a result of hybridization during evolution (Shen et al., 2015). We noted that no MsARF protein clustered with AtARF12-15,21-23. These genes form a gene cluster on Arabidopsis chromosome I and are mainly expressed during embryogenesis and seed development (Okushima et al., 2005). While there is no similar gene form AtARF12-15,21-23 in M. Sieboldii and O. sativa. In addition, MsARFs proteins often cluster independently, or at the periphery of monocotyledons and dicotyledons, indicating that in terms of evolutionary relationship, M. Sieboldii may be more primitive. The update of APG IV shows that Magnoliaceae belongs to the base of angiosperms, and its evolutionary relationship is earlier than that of monocotyledons and dicotyledons (Byng et al., 2016). This also supports the results of phylogenetic analysis in this paper.
We identified 25 ARF genes in M. sieboldii via homology searching. The encoded proteins could act as either transcriptional activators or repressors, depending on the amino acid compositions in the MR region. Previous studies using transfected protoplasts showed transcriptional activation activity of Q-rich ARFs in Arabidopsis (Tiwari et al., 2003). Later, ARFs proteins with different transcriptional activities had also been identified in non-model plants. For instance, ARFs with MRs that are rich in serine (S), proline (P), and glutamine (Q) exhibit transcriptional repressing activities in Dimocarpus longan (Lin et al., 2013). Similarly, ARFs with MRs rich in Q/S/L(leucine) function as activators in multiple species including Prumus persica (Shi et al., 2012)、Vitis vinifera (Yuan et al., 2015), Lycopersicon esculentum (Wu, 2011), and Malus pumila (Li et al., 2015). In our study, MsARF5 was predicted as an activator due to the same amino acid composition as found in transcriptional activators in M. pumila. In addition, we also found that all MsARFs with Q-rich MRs belong to Class II.
The involvement of auxin in vascular differentiation had been demonstrated decades ago (Aloni, 1995; Mattsson et al., 2003). Active transportation of auxin and an elevated concentration had been found in regions where vascular differentiation occurs. In addition, impaired patterning in vascular differentiation had been found in various mutants that are defective in the biosynthesis, transportation, or signaling of auxin. At the molecular level, MP/AtARF5 had been identified as a central regulator in leaf vascularization (Krogan and Berleth, 2012; Krogan et al., 2012) and root growth (Krogan et al., 2016). In M. sieboldii, vascular tissue differentiation was preliminarily observed after stratification for 30 days. Notably, the expression of MsARF5 also showed a significant increase at this time, indicating a role of MsARF5 in regulating vascular tissue differentiation in the seeds.
In addition to vascular differentiation, ARF5/MP also plays an essential role in embryo development as the Arabidopsis arf/mp mutant is impaired in flower organ formation and seed development (Przemeck et al., 1996; Hardtke and Berleth, 1998). In this study, we not only confirmed the infertility of the arf/mp mutant but also showed that the MsARF5-overexpressing line is also infertile. While no flower organ formation was observed in the former, the latter exhibited normal development of floral organs and pod but no seeds. That fact either complete loss-of-function or overexpression leads to abnormal embryo development suggests that the expression level of ARF5 controls the final phenotype. These observations also indicated a similar function of MsARF5 and its homolog in Arabidopsis (Schlereth et al., 2010; Krogan et al., 2016).
Our finding on the two different phenotypes in the MsARF5-overexpressing Arabidopsis plants indicated the expression level of ARF5 is behind the final plant phenotype. Specifically, OE1 and OE2 showed significant altered vegetative growth including height, basal diameter, and leaf width compared to WT plants. These observations are in line with previous findings that ARF5/MP in Arabidopsis is a quantitative gene and that it plays an essential role in mediating auxin signaling and its polar transport (Krogan et al., 2016). The patterning events in embryos were under the direct control of ARF5/MP-mediated auxin signaling and transportation. Accordingly, ARF5/MP acts as a switch to regulate the expression of PIN (pin formed) auxin efflux facilitators during pattern formation of root and shoot (Wenzel et al., 2007). This is further supported by previous studies showing the binding of ARF5/MP on the promoters of multiple PIN genes (Okushima et al., 2005). Together, our data indicate a role of MsARF5 in mediating auxin signaling and transportation to control the patterning of multiple plant organs.
5 Conclusions
A total of 25 ARF genes were identified in M sieboldii. Diversity among these genes was found due to the presence of different functional domains, suggesting diversity in function. Gene expression analysis of MsARF5 showed a similar pattern to that of auxin during seed germination. The function of MsARF5 was further investigated by overexpressing in Arabidopsis, which showed an increase in the growth of vascular tissues and promotion of seed germination. In addition, MsARF5 can rescue the deficiency in flowering of the arf mutant.
Data availability statement
The data presented in the study are deposited in the National Genomics Data Center (NGDC) repository, accession number PRJCA009763.
Ethics statement
M. sieboldii seeds were used in this study. M. sieboldii is a National level 3 protection plants of China and the identification information is stored in the Plant Photo Bank of China (PPBC ID: 2657292). The seed sample was kindly provided by Benxi Botanical Garden (Benxi, Liaoning, China). We complied with all the relevant institutional, national and international guidelines.
Author contributions
XL conceived the experiment. MM and LL collected resources. MM conducted the experiment. MM and WA analyzed the results. MM wrote the manuscript. XL and XX revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research and the APC was funded by National Natural Science Foundation of China, grant number 31570621 and 32101508.
Acknowledgments
We thank Prof. Yongxiu Liu (Institute of Botany, Chinese Academy of Sciences) for providing the T-DNA mutant of ARF5 (salk_001058) seeds. All sequencing was outsourced to Novogene Bioinformatics Institute, Beijing, China. We thank Novogene Co., Ltd. for the technology support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.958816/full#supplementary-material
Supplementary Figure 1 | Chromosomal distribution of MsARFs.
Supplementary Figure 2 | Gene structure analysis and motif characterization of MsARFs.
Supplementary Figure 3 | Identification of ARFs in M. sieboldii, Medicago truncatula, Vitis vinifera and apple. The unrooted tree was constructed using MEGA-X with the neighbor-joining method (bootstrap of 1000).
Supplementary Table 1 | PCR primer names and sequences.
References
Ai, W., Liu, Y., Mei, M., Zhang, X., Tan, E., Liu, H., et al. (2022). A chromosome-scale genome assembly of the Mongolian oak (Quercus mongolica). Mol. Ecol. Resour. 22 2396–2410. doi: 10.1111/1755-0998.13616
Aloni, R. (1995). The induction of vascular tissues by auxin and cytokinin Plant Hormones (Kluwer Academic) 531–546. doi: 10.1007/978-94-011-0473-9_25
Belin, C., Megies, C., Hauserova, E., Lopez-Molina, L. (2009). Abscisic acid represses growth of the arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21, 2253–2268. doi: 10.1105/tpc.109.067702
Boter, M., Calleja-Cabrera, J., Carrera-Castao, G., Wagner, G., Oate-Sánchez, L. (2019). An integrative approach to analyze seed germination in brassica napus. Front. Plant Sci. 10, 1342. doi: 10.3389/fpls.2019.01342
Byng, J. W., Chase, M. W., Christenhusz, M. J. M., Fay, M. F., Judd, W. S. (2016). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Botanical J. Linn. Soc. 181, 1–20. doi: 10.1111/boj.12385
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cole, M., Chandler, J., Weijers, D., Jacobs, B., Comelli, P., Werr, W. (2009). DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the arabidopsis embryo. Development 136, 1643–1651. doi: 10.1242/dev.032177
Donner, T. J., Sherr, I., Scarpella, E. (2009). Regulation of preprocambial cell state acquisition by auxin signaling in arabidopsis leaves. Development 136, 3235–3246. doi: 10.1242/dev.037028
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, 29–37. doi: 10.1093/nar/gkr367
Galstyan, A., Nemhauser, J. L. (2019). Auxin promotion of seedling growth via ARF5 is dependent on the brassinosteroid-regulated transcription factors BES1 and BEH4. Plant Direct 3, e00166. doi: 10.1002/pld3.166
Hardtke, C. S., Berleth, T. (1998). The arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. doi: 10.1093/emboj/17.5.1405
Krogan, N. T., Berleth, T. (2012). A dominant mutation reveals asymmetry in MP/ARF5 function along the adaxial-abaxial axis of shoot lateral organs. Plant Signaling Behav. 7, 940–943. doi: 10.4161/psb.20790
Krogan, N. T., Ckurshumova, W., Marcos, D., Caragea, A. E., Berleth, T. (2012). Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in arabidopsis leaf vascular patterning. New Phytol. 194:391–401. doi: 10.1111/j.1469-8137.2012.04064.x
Krogan, N. T., Marcos, D., Weiner, A. I., Berleth, T. (2016). The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytol. 212, 45–50. doi: 10.1111/nph.14107
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lei, Y. (2007). Cloning and expression of the resveratrol synothase gene from polygonum cuspidatum (Beijing:Chinese Academy of Agricultural Sciences).
Li, P., Lu, X., Yao, F., Guo, R. (2006). Preliminary study on reasons of seed dormancy of magnolia sieboldii K.Koch. Seed 25:36–39. doi: 10.16590/j.cnki.1001-4705.2006.02.045
Lin, L. X., Lin, Y. L., Lai, Z. X. (2013). Analyses of DIARF5-1 expression levels by qPCR under the treatment of different hormones in longan. Horticult. Seed 10 17–20. doi: 10.3969/j.issn.2095-0896.2013.10.005
Li, H. F., Ran, K., He, P., Wang, H. B., Chang, Y. S., Sun, Q. R., et al. (2015). Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in apple. Plant Physiol. J 51:1045–1054. doi: 10.13592/j.cnki.ppj.2015.0173
Liscum, E., Reed, J. W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. doi: 10.1023/A:1015255030047
Liu, X., Zhang, H., Zhao, Y., Feng, Z., Li, Q., Yang, H. Q., et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in arabidopsis. Proc. Natl. Acad. Sci. United States America 110, 15485–15490. doi: 10.1073/pnas.1304651110
Lu, X., Mei, M., Liu, Y., Zhang, X., Ma, B. (2014). Effect of treatment with GA_3 and variable temperature stratification on germination and endogenous hormones of magnolia sieboldii seeds. Acta Botanica Boreali-Occidentalia Sin 34:1828–1835. doi: 10.7606/j.issn.1000-4025.2014.09.1828
Mattsson, J., Ckurshumova, W., Berleth, T. (2003). Auxin signaling in arabidopsis leaf vascular development. Plant Physiol. 131, 1327–1339. doi: 10.1104/pp.013623
Mei, M., Lu, X. J., Zhang, X. L., Liu, G. L., Sun, X. M. (2017). Variation in carbohydrates and screening of related differential proteins during the seed germination of magnolia sieboldii k. Koch. Trees 31, 63–75. doi: 10.1007/s00468-016-1456-8
Mei, M., Wei, J., Ai, W., Zhang, L., Lu, X. J. (2021). Integrated RNA and miRNA sequencing analysis reveals a complex regulatory network of magnolia sieboldii seed germination. Sci. Rep. 11, 10842. doi: 10.1038/s41598-021-90270-y
Moller, B., Weijers, D. (2009). Auxin control of embryo patterning. Cold Spring Harbor Perspect. Biol. 1, a001545. doi: 10.1101/cshperspect.a001545
Odat, O., Gardiner, J., Sawchuk, M. G., Verna, C., Donner, T. J., Scarpella, E. (2014). Characterization of an allelic series in the MONOPTEROS gene of arabidopsis. Genesis 52, 127–133. doi: 10.1002/dvg.22729
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. doi: 10.1105/tpc.104.028316
Przemeck, G. K., Mattsson, J., Hardtke, C. S., Sung, Z. R., Berleth, T. (1996). Studies on the role of the arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. doi: 10.1007/BF00208313
Quint, M., Gray, W. M. (2006). Auxin signaling. Curr. Opin. Plant Biol. 9, 448–453. doi: 10.1016/j.pbi.2006.07.006
Rademacher, E. H., Lokerse, A. S., Schlereth, A., Llavata-Peris, C. I., Bayer, M., Kientz, M., et al. (2012). Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 22, 211–222. doi: 10.1016/j.devcel.2011.10.026
Rousselin, P., Kraepiel, Y., Maldiney, R., Miginiac and, E. (1992). Characterization of three hormone mutants of nicotiana plumbaginifolia: evidence for a common ABA deficiency. Theor. Appl. Genet. 85, 213–221. doi: 10.1007/BF00222862
Schlereth, A., Möller, B., Liu, W., Kientz, M., Flipse, J., Rademacher, E. H., et al. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916. doi: 10.1038/nature08836
Shen, C., Yue, R., Sun, T., Zhang, L., Xu, L., Tie, S., et al. (2015). Genome-wide identification and expression analysis of auxin response factor gene family in medicago truncatula. Front. Plant Sci. 6, 73. doi: 10.3389/fpls.2015.00073
Shi, M., Li, Y., Zhang, W., Liu, Y. (2012). Progress in mechanism of auxin response factors. Biotechnol. Bull. 8 24–28. doi: 10.13560/j.cnki.biotech.bull.1985.2012.08.028
Tiwari, S. B., Hagen, G., Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. doi: 10.1105/tpc.008417
Ulmasov, T., Hagen, G., Guilfoyle, T. J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. doi: 10.1126/science.276.5320.1865
Vidaurre, D. P., Ploense, S., Krogan, N. T., Berleth, T. (2007). AMP1 and MP antagonistically regulate embryo and meristem development in arabidopsis. Development 134, 2561–2567. doi: 10.1242/dev.006759
Wang, C. K., Han, P. L., Zhao, Y. W., Yu, J. Q., You, C. X., Hu, D. G., et al. (2020). Genome-wide analysis of auxin response factor (ARF) genes and functional identification of MdARF2 reveals the involvement in the regulation of anthocyanin accumulation in apple New Zealand Journal of Crop and Horticultural Science 49, 78–91. doi: 10.1080/01140671.2020.1779756
Wang, L., Hua, D., He, J., Duan, Y., Chen, Z., Hong, X., et al. (2011). Auxin response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in arabidopsis. PloS Genet. 7, e1002172. doi: 10.1371/journal.pgen.1002172
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wan, S., Li, W., Zhu, Y., Liu, Z., Zhan, J. (2014). Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in vitis vinifera. Plant Cell Rep. 33, 1365–1375. doi: 10.1007/s00299-014-1622-7
Wenzel, C. L., Schuetz, M., Yu, Q., Mattsson, J. (2007). Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in arabidopsis thaliana. Plant J. 49, 387–398. doi: 10.1111/j.1365-313X.2006.02977.x
Woodward, A. W., Bartel, B. (2005). A receptor for auxin. Plant Cell 17, 2425–2429. doi: 10.1105/tpc.105.036236
Wu, J. (2011). Identification and expression profile analysis of auxin response genes and preliminary functional characterization of SlARF5 in tomato (Solanum lycopersicum) (Zhejiang University, Hangzhou: College of Agriculture and Biotechnology).
Xing, H., Pudake, R. N., Guo, G., Xing, G., Hu, Z., Zhang, Y., et al. (2011). Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12, 178. doi: 10.1186/1471-2164-12-178
Yuan, H., Zhao, M., Wu, W., Yu, H., Qian, Y., Wang, Z., et al. (2015). Genome-wide identification and expression analysis of auxin-related gene families in grape. Yi Chuan 37, 720–730. doi: 10.16288/j.yczz.14-390
Yu, Y. Z., Du, J., Wang, G., Ji, J. (2003). Studies on the freeze-thaw method of transforming recombinant plasmid DNA into agrobacterium tumefaciens. J. Jilin Agric. Univ. 25, 257–259, 262. doi: 10.3969/j.issn.1000-5684.2003.03.006
Keywords: auxin response factor, Magnolia sieboldii, auxin, embryo, seeds
Citation: Mei M, Ai W, Liu L, Xu X and Lu X (2022) Genome-wide identification of the auxin response factor (ARF) gene family in Magnolia sieboldii and functional analysis of MsARF5. Front. Plant Sci. 13:958816. doi: 10.3389/fpls.2022.958816
Received: 01 June 2022; Accepted: 20 September 2022;
Published: 05 October 2022.
Edited by:
Tao Zhang, Yangzhou University, ChinaReviewed by:
Li Zhengguo, Chongqing University, ChinaDavid Burks, University of North Texas, United States
Copyright © 2022 Mei, Ai, Liu, Xu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiujun Lu, bHhqc3lhdUBzeWF1LmVkdS5jbg==
 Mei Mei1,2
Mei Mei1,2 Xiujun Lu
Xiujun Lu