
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 08 September 2022
Sec. Sustainable and Intelligent Phytoprotection
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.957961
This article is part of the Research Topic Deep Learning in Crop Diseases and Insect Pests View all 17 articles
Early recognition of tomato plant leaf diseases is mandatory to improve the food yield and save agriculturalists from costly spray procedures. The correct and timely identification of several tomato plant leaf diseases is a complicated task as the healthy and affected areas of plant leaves are highly similar. Moreover, the incidence of light variation, color, and brightness changes, and the occurrence of blurring and noise on the images further increase the complexity of the detection process. In this article, we have presented a robust approach for tackling the existing issues of tomato plant leaf disease detection and classification by using deep learning. We have proposed a novel approach, namely the DenseNet-77-based CornerNet model, for the localization and classification of the tomato plant leaf abnormalities. Specifically, we have used the DenseNet-77 as the backbone network of the CornerNet. This assists in the computing of the more nominative set of image features from the suspected samples that are later categorized into 10 classes by the one-stage detector of the CornerNet model. We have evaluated the proposed solution on a standard dataset, named PlantVillage, which is challenging in nature as it contains samples with immense brightness alterations, color variations, and leaf images with different dimensions and shapes. We have attained an average accuracy of 99.98% over the employed dataset. We have conducted several experiments to assure the effectiveness of our approach for the timely recognition of the tomato plant leaf diseases that can assist the agriculturalist to replace the manual systems.
In accordance with a report issued by the Food and Agriculture Organization (FAO) of the United Nations, the population of humans will undergo a tremendous increase around the globe to 9.1 billion by 2050. Such an increase in the number of humans will also raise the demand for food (Bruinsma, 2009). Meanwhile, the decrease in agricultural land and the unavailability of clean water will limit the progress of nutriment amounts. Therefore, there is an urgent demand for improving food yields by consuming minimum cultivation space to fulfill the necessities of humans. The occurrence of several crop abnormalities results in a substantial decline in both the yield and quality of food. Hence, the timely recognition of such plant diseases is required as these diseases can affect the profit of farmers and can increase the purchase cost of food. Such implications can introduce economic instability in the markets. Moreover, the plant diseases at their adverse stage can destroy the crops which can create a starvation scenario within a region, specifically in low-income countries. Plant inspections are generally carried out with the help of human experts. However, this is a cumbersome and time-consuming activity that relies upon the presence of area experts. These plant examination procedures are not considered very reliable and it is practically impossible for humans to inspect every plant separately (Pantazi et al., 2019). To enhance the quantity and quality of food, there is a need to timeously and correctly recognize the various plant diseases which can also force the farmers into using the costly spray methods. To tackle the above-mentioned problems of manual processes, the research community is focusing on the development of automated plant disease detection and classification systems (Wolfenson, 2013).
The focus of this paper is the recognition of several tomato plant diseases as tomato has the largest consumption rate, of 15 kg per capita within a year when compared to other vegetables such as rice, potato, and cucumber. Moreover, the tomato crop counts for 15% of the entire vegetable ingestion globally (Chowdhury et al., 2021). Further, tomatoes have the highest cultivation rate with an annual growth rate of 170 tons worldwide (Valenzuela and Restović, 2019). The leading countries for its production are Egypt, India, the United States, and Turkey (Elnaggar et al., 2018). In a study conducted by the FAO (Sardogan et al., 2018), the occurrence of several tomato plant diseases caused a severe reduction in its quantity and most of the abnormalities originated from the leaves of tomato plants. It has been observed that such diseases reduce the tomato food quantity from 8 to 10% annually (Sardogan et al., 2018). Farmers or agriculturalists can guard against these huge monetary losses by adopting automated systems which can assist them in the timely detection of plant diseases and taking proactive measures. At first, technology experts utilized the methods used in the field of molecular biology and immunology for locating the presence of tomato plant leaf diseases (Sankaran et al., 2010; Dinh et al., 2020). However, these techniques were not fruitful due to their high processing requirements and dependence on the expertise of humans. Most agriculturists belong to poor or under-developed countries where the adaptability of such an expensive solution is not affordable (Patil and Chandavale, 2015; Ferentinos, 2018). The rapid progression in the area of machine learning (ML) has introduced low-cost solutions for the recognition of tomato plant diseases (Gebbers and Adamchuk, 2010). Many researchers have tested the conventional ML methods, such as hand-coded approaches, in the field of agriculture (Gebbers and Adamchuk, 2010). The availability of economical image-capturing gadgets has assisted researchers to take pictures in real-time and then give intelligent predictions via using ML-based approaches. Examples of such approaches include K-nearest neighbors (KNN), decision trees (DT) (Rokach and Maimon, 2005), and support vector machines (SVM) (Joachims, 1998), which are heavily evaluated by researchers for plant disease classification. Such techniques are simple in their architecture and can work well with a small amount of training data. However, they are unable to contend with image distortions such as intensity variations, color changes, and brightness alterations of suspected samples. Furthermore, the conventional approaches always impose a trade-off among the classification performance and processing time (Bello-Cerezo et al., 2019).
The empowerment of DL frameworks has assisted the researchers in dealing with the problems of conventional ML approaches (Agarwal et al., 2021d,2022). Several DL techniques such as CNN (Roska and Chua, 1993), recurrent neural networks (RNNs) (Zaremba et al., 2014), and long short-term memory (LSTM) (Salakhutdinov and Hinton, 2009) have been found to be reliable in recognizing plant leaf diseases. The DL approaches are inspired by the human brain and can learn to discriminate between a set of image features without relying on the intervention of domain experts. These frameworks recognize the objects in the same way as humans by visually examining several samples to accomplish a pattern recognition task. Because of such properties, the DL approaches are found to be more suitable in areas of agriculture, including plant disease classification (Gewali et al., 2018). Several well-known DL frameworks such as GoogLeNet (Szegedy et al., 2015), AlexNet (Yuan and Zhang, 2016), VGG (Vedaldi and Zisserman, 2016), and ResNet (Thenmozhi and Reddy, 2019) have been thoroughly tested for accomplishing several jobs in farming, i.e., estimating food yield, crop heads recognition, fruit totaling, plant leaf disease detection and categorization, among others. Such approaches show reliable performance by minimizing the processing complexity as well as by better analyzing the topological information of the input samples.
Numerous techniques have been evaluated to identify and classify tomato leaf diseases. However, the reliable and timely recognition of such abnormality is a complicated job because of the significant color resemblance between the healthy and diseased areas of plant leaves (Paul et al., 2020). Furthermore, the intense changes in the dimension of plant leaves, lightning conditions, the incidence of noise, and blurring in the input samples further problematize the disease recognition procedure. Hence, there is a need for a more reliable system to accurately perform the plant disease classification process with minimum time constraints. To deal with these issues, we have introduced a DL approach, namely the custom CornerNet model. We have utilized Dense-77 as the backbone of the CornerNet model for extracting the image features. These are later classified by the one-stage detection module of the CornerNet model. We have conducted extensive evaluation over a challenging dataset and confirm that our approach is proficient in classifying the numerous types of tomato plant leaf diseases. The major contributions of the proposed approach are listed as:
1. Modified an object detection approach named CornerNet for tomato plant leaf abnormality categorization which improves the classification performance with an accuracy value of 99.98%.
2. Exhibits robust performance for 10 classes of the tomato plant leaf diseases because of the empowerment of the custom CornerNet model to tackle the over-fitted model training data.
3. A cost-effective solution is presented for the classification of tomato plant leaf abnormalities which minimizes the test time to 0.22 s.
4. Efficient localization of diseased regions from the tomato plant samples due to the better keypoints calculation power of the Dense-77-based CornerNet model with the mean average precision (mAP) value of 0.984.
5. In contrast to several new methods, extensive experimentation has been carried out on a challenging database named the PlantVillage dataset to exhibit the robustness of the proposed work.
6. The presented work is capable of correctly identifying the abnormal area of the tomato plant leaves even from the distorted samples and under the influence of size, color, and light variations.
The article is structured as follows: existing studies are compared in section “Related work,” the details of the introduced approach are described in section “Materials and methods,” section “Results” contains the results, and the conclusion is drawn in section “Conclusion.”
In this section, we review existing studies that have attempted to classify tomato plant leaf diseases. Typically, the approaches for tomato plant leaf disease detection and classification are either conventional ML-based techniques or DL frameworks. Hand-coded features computation approaches with the ML-based classifiers were explored initially for the plant leaf disease classification. One such framework was presented in Le et al. (2020) where the suspected images were initially processed by applying the morphological opening and closing techniques to remove the undesired objects. Then, the filtered local binary pattern method, namely the k-FLBPCM, was used on the processed images to obtain the desired feature vector. Finally, the SVM classifier was trained on the computed features for classification. The technique in Le et al. (2020) improved classification results for the plant leaf diseases but was unable to show better results on the distorted samples. Another work, namely Directional Local Quinary Patterns (DLQP), was introduced in Ahmad et al. (2020) to extract the keypoints from the input images. The work also used the SVM classifier on the computed features for categorizing the several classes of plant leaf diseases. The solution introduced in Ahmad et al. (2020) was robust in classifying the affected areas of plant leaves into their respective groups but classification performance degraded for noisy images. Sun et al. (2019) proposed an automated solution to quickly locate the diseased portion of plant leaves. They used the Simple Linear Iterative Cluster (SLIC) algorithm for distributing the input images into numerous chunks. Then, for each block of the divided image, the GLCM approach was used to extract the features which were later combined and passed to the SVM classifier for classification. This approach (Sun et al., 2019) performed well in recognizing the several categories of plant diseases but suffered from extensive processing complexity. Another pattern recognition approach was used in Pantazi et al. (2019) where the input sample was initially segmented via applying the GrabCut method to locate the region of interest. Then, the LBP algorithm was applied for keypoints vector estimation. Finally, classification was carried out with the help of the SVM classifier. This technique (Pantazi et al., 2019) was proficient in locating the abnormal area of the plant leaves. However, detection performance degraded for the samples with intense noise attacks. Ramesh et al. (2018) proposed a computer-aided system for the automated detection and classification of several abnormalities of plant leaves. For feature estimation, the HOG filter was used on the input samples, and disease classification was performed using the Random Forest (RF) technique. This work, elaborated on in Ramesh et al. (2018), was found to be a lightweight solution for the recognition of plant leaf diseases but the classification accuracy required further improvements. Another technique was discussed in Kuricheti and Supriya (2019) where an ML-based approach was presented to classify the several abnormalities of the turmeric plant. In the first phase, the K-means clustering approach was used on the input sample to locate the area of interest. The GLCM algorithm was applied to this area to calculate the feature vector. Finally, the SVM classifier was adopted for classification using the computed keypoints. The work discussed in Kuricheti and Supriya (2019) showed better plant leaf disease classification results. However, detection performance degraded for images with large brightness changes. Another handcrafted feature estimation approach to recognize and categorize crop leaf diseases was found in Kaur and Education (2021). Several pattern-based approaches like the GLCM, LBP, and SIFT were used for feature vector estimation. Then, several well-known ML classifiers, named the SVM, RF, and KNN, were trained on the computed features to execute the classification task. The best results were reported for the RF classifier but the classification accuracy needed enhancement. A similar solution was elaborated on in Shrivastava and Pradhan (2021) where the fourteen color spaces approach was used to extract the keypoints from the test images with a length of 172. Then, the calculated keypoints were passed to the SVM algorithm to classify the samples into their respective classes based on the detected abnormal plant leaf areas. This solution (Shrivastava and Pradhan, 2021) provided superior plant leaf disease categorization results. However, this performance degraded for samples with significant color and light changes.
Due to the empowerment of DL frameworks and their ability to better deal with image transformations, researchers are now employing them for recognizing plant diseases.
The framework in Argüeso et al. (2020) used the DL technique named Few-Shot Learning (FSL) for recognizing the affected portions of crops and determining the related category. The InceptionV3 model was applied to capture the keypoints of the input image. The SVM classifier was used to classify the samples using the keypoints, according to the detected disease. The approach described in Argüeso et al. (2020) exhibited robust plant disease classification results but requires extensive data for the model training. Agarwal et al. (2020b) proposed a CNN framework containing 3 convolution layers as the feature extractor module before classification. The framework presented in Agarwal et al. (2020b) was a lightweight solution for the plant leaf disease classification but performance degraded for noisy samples. Another lightweight model was presented in Richey et al. (2020) to be used with cellphones. The ResNet50 approach was used as the end-to-end framework to compute the deep features and perform the classification task. The approach improved the processing complexity for plant disease classification. However, it was not supported by all mobile phones due to the memory requirements. Another framework was depicted in Batool et al. (2020) to classify the numerous types of tomato crop abnormalities. The AlexNet model was employed to extract the deep features of the plant images which were later passed as input to the KNN approach for the classification of the images into their respective category. This work was proficient in recognizing the various categories of tomato plant leaves. However, the KNN algorithm was a time-consuming approach. Similarly, an approach for categorizing the tomato plant leaf abnormalities was described in Karthik et al. (2020) that employed the residual method to compute the reliable feature set. A CNN-based classifier was introduced to categorize the samples based on the learned features of different classes. The approach (Karthik et al., 2020) classified the samples in the related categories better. However, it required a large number of samples for training, which further complicated the model. Dwivedi et al. (2021) applied the object detection approach named region-based CNN (RCNN) to automatically detect and localize the diseased area of grape plant leaves. The approach used the ResNet18 as the feature extractor unit which calculates the keypoints set from the plant images. In the next phase, the RCNN framework applied the region proposals approach to locate the affected portion and determine the associated class. The solution depicted in Dwivedi et al. (2021) worked well in recognizing the various diseases of the grape plant but was unable to generalize well from unseen training data. Another approach was discussed in Akshai and Anitha (2021) where several DL frameworks, namely VGG, DenseNet, and ResNet, were evaluated for the detection and classification of several types of plant leaf diseases. This approach (Akshai and Anitha, 2021) showed better results for the DenseNet model. Albattah et al. (2021) proposed an object detection approach, namely the CenterNet model, for the automated identification and classification of numerous types of plant leaf diseases. Initially, the dense model was used for the extraction of the keypoints set from the input images. These were then used to recognize the diseased portion of plant samples. This approach (Albattah et al., 2021) showed better plant leaf abnormality recognition ability. However, the model needed assessment on a more challenging dataset. Another DL approach was evaluated in Albattah et al. (2022) where the EfficientNetV2 model was tested for the classification of numerous types of plant diseases, that results in improving the classification performance. In Agarwal et al. (2021c), a DL approach, namely the VGG16 model, was used in the classification of tomato leaf diseases. The approach introduced the concept of model optimization, but the detection performance required extensive result improvements. Similarly, other works discussed the model optimization concept for the plant leaf diseases categorization (Agarwal et al., 2021a,b) but the recognition results needed improvement. Zhao et al. (2021) presented a model to recognize numerous tomato plant leaf abnormalities in which the CNN approach, merged with an attention mechanism, was utilized. The methodology attained classification results of 99.24%. Moreover, in Maeda-Gutiérrez et al. (2020), different DL networks, i.e., Inception V3, AlexNet, GoogleNet, ResNet-18, and SE-ResNet50 were tested for tomato plant disease classification. The GoogleNet approach worked well with classification results of 99.39%. Bhujel et al. (2022) also proposed a DL model, namely ResNet18, along with the CBAM for recognizing the tomato plant abnormalities and achieved an accuracy of 99.69%. The methods in Maeda-Gutiérrez et al. (2020), Zhao et al. (2021), and Bhujel et al. (2022) enhanced the tomato plant leaf diseases categorization results. However, these works accomplished classification at the image level and are incapable of identifying the precise diseased area.
A critical investigation of existing work is outlined in Table 1, which depicts that there is a performance gap that requires a more reliable model. This model must be proficient enough to recognize the numerous categories of tomato plant leaf disease and minimize the time complexity. In the presented work, we have tried to cover this gap by proposing a more accurate and robust approach for tomato plant leaf disease classification.
In this section, an in-depth discussion of the proposed technique for tomato plant leaf disease localization and classification is presented. The basic motivation of this framework is to present an accurate and computationally efficient approach that is empowered to automatically nominate a representative feature vector independent from executing any manual examination. Our work comprises two main steps to accomplish the automated recognition of plant leaf diseases. First, the images from the PlantVillage dataset are employed to develop the annotations to correctly identify the affected portions and their associated classes. Then, these annotations are used in training the DenseNet-77-based CornerNet approach. During the test phase, the images from the test set are used to validate the model’s performance. More precisely, we have customized the CornerNet model (Law and Deng, 2019) by introducing the DenseNet-77 network in its feature extraction unit. The DenseNet-77 approach as the base network computes the feature vector which is then passed to the one-stage detector of the CornerNet model to localize and classify the affected regions into 10 classes. Several standard evaluation measures are then used to quantitatively measure the performance of the introduced framework. The detailed model formulation of our framework is given in Algorithm 1, while the pictorial demonstrations showing the detailed steps of our approach are given in Figure 1.
Algorithm 1. Description of steps followed by the proposed work.
INPUT:
TS, AI
OUTPUT:
Bbx, CustomCoNet, C-score
TS - total no of samples used
for model training
AI - annotated images showing the
diseased area on the tomato plant
leaves
Bbx - rectangular box showing the
diseased region on the output image
CustomCoNet - CornerNet model with
the DenseNet-77 backbone
C-score - confidence score along
with predicted class
SampleSize ← [x y]
Bbx computation
β ← AnchorsCalculation (TS, AI)
CustomCustomCoNet-Model
CustomCoNet ← CornerNetWithDenseNet-77
(SampleSize, β)
[dr dt] ← Distribution of dataset into
train and test sets
The training module for tomato
plant leaf disease detection and
classfication
Foreach image m in → dr
Calculate DenseNet-77-based-
deepFeatures ← df
End For
Train CustomCoNet on df, and measure
network training time as t_d77
β _dense ← EstimateDiseasedPos(df)
V_dense ← Validate_Model
(DenseNet-77, β_dense)
Foreach images M in → dt
(i) Measure features with trained
model €→V_dense
(ii) [Bbx, C-score, class] ←
Predict (€)
(iii) Present output image with
Bbox, class
(iv) η ← [η bbox]
End For
Ap_€ ← Test framework € using η
Output_class ← CustomCoNet (Ap_€).
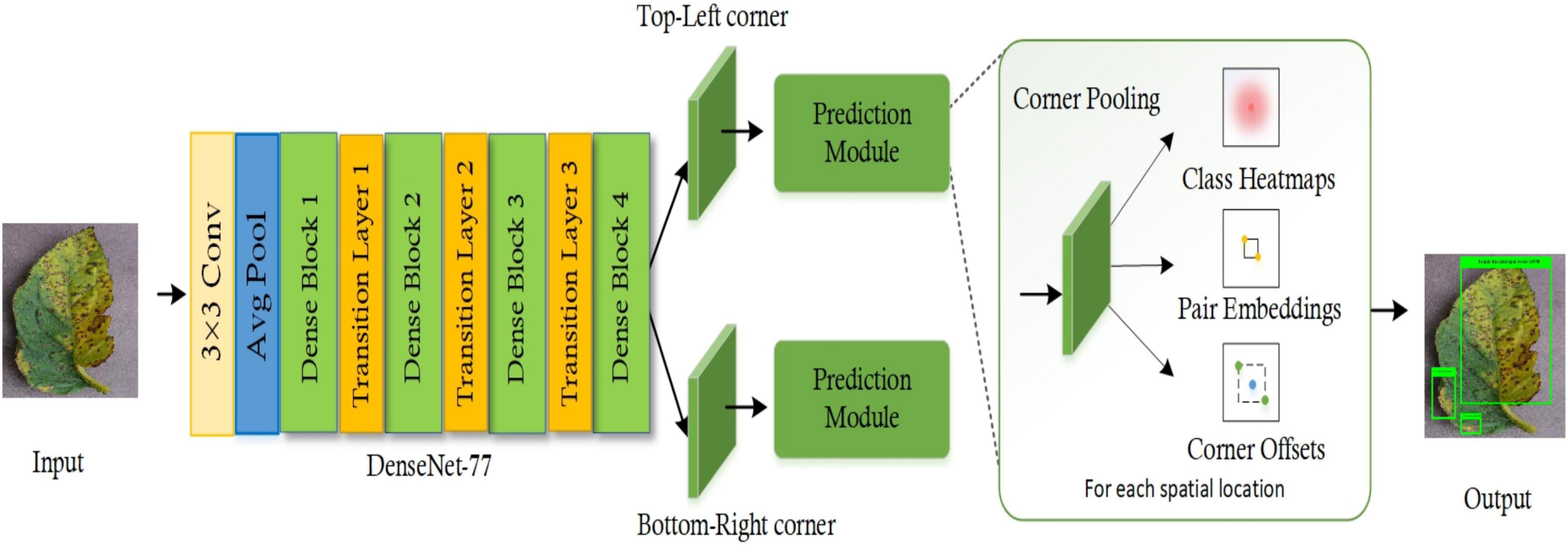
Figure 1. Pictorial depiction of the DenseNet-77-based CornerNet model for the tomato plant leaf diseases classification.
The training of the object detection model was based on annotations development. This was focused on clearly localizing the affected region from the training samples and their associated category. Therefore, in the first step, we have used the images from the training set of the plant samples from the PlantVillage dataset and used the LabelImg software (Lin, 2020) for relevant annotation generation. These annotations assist in exactly outlining the diseased areas of leaves by developing the bounding box (bbx) around them. The dimensions of the annotations are saved as an XML file which is later employed for model training. A few examples of annotated samples are given in Figure 2.
The CornerNet (Law and Deng, 2019) is a well-known one-stage object detection model that recognizes the region of interest (ROI) (the diseased region of the tomato plants in this case) from the input samples through keypoint calculation. The CornerNet model estimates the Top-Left (TL) and Bottom-Right (BR) corners to draw the bbx with more accuracy when compared to other object detection models (Girshick, 2015; Ren et al., 2016). The CornerNet framework is comprised of two basic units: the feature computation backbone and the prediction module (Figure 1). At the start, a keypoints extractor unit is used which extracts the reliable feature vector that is employed to estimate the heatmaps (Hms), embeddings, offset, and class (C). The Hms give an approximation of a location in a sample where a TL/BR corner is associated with a particular category (Nawaz et al., 2021). The embeddings are used to discriminate the detected pairs of corners and offsets to fine-tune the bbx position. The corners with high scored TL and BR coordinates are used to determine the exact position of the bbx, whereas the associated category for each detected diseased region is specified by using the embedding distances on the computed feature vector.
The CornerNet framework shows robust performance in detecting and classifying several types of objects (Girshick, 2015; Raj et al., 2015; Redmon et al., 2016; Zhao et al., 2016). However, the abnormalities of tomato plant leaves have some distinct characteristics. These include leaves of different shapes and sizes and high color resemblance in the affected and healthy regions of plant leaves which complicates the classification procedure. Moreover, the existence of several image distortions such as differences in the light, color, and brightness of the samples and the incidence of noise and blurring effect further increase the complexity of the tomato plant leaf disease classification process. Therefore, to better tackle the complexities of samples, we have customized the CornerNet model by introducing a more effective feature extractor, namely the DenseNet-77, as its base network. The introduced base network is capable of locating and extracting the more relevant sample attributes which assist the CornerNet approach and enhance its recall ability in comparison to the conventional model.
The reason for selecting the CornerNet approach for classifying the diseases of tomato plants in this study is its capability for effectively detecting objects by utilizing keypoint approximation in comparison to earlier approaches (Girshick, 2015; Girshick et al., 2015; Liu et al., 2016; Ren et al., 2016; Redmon and Farhadi, 2018). The framework utilizes detailed keypoints and identifies the object by employing a one-stage detector. This eliminates the need to use large anchor boxes for diverse target dimensions as used in other one-stage object recognition models, i.e., single-shot detector (SSD) (Liu et al., 2016), and You Only Look Once (YOLO) (v2, v3) (Redmon and Farhadi, 2018). Moreover, the CornerNet model is more computationally robust than the other anchor-based two-stage approaches, i.e., RCNN (Girshick et al., 2015), Fast-RCNN (Girshick, 2015; Nazir et al., 2020), and Faster-RCNN (Ren et al., 2016; Albahli et al., 2021), as these techniques employ two phases to accomplish the object localization and categorization. Consequently, the DenseNet-77-based CornerNet framework efficiently deals with the issues of existing models by presenting a more proficient network that extracts more nominative sample features and reduces the computational cost.
The base of a model is responsible for identifying and computing the reliable feature vector that gives the semantic information and reliable location of a target in an image. The affected regions of tomato plant leaves are small, therefore a robust and representative set of keypoints is mandatory to recognize the diseased portion from complex backgrounds such as changing acquisition positions, lightning conditions, noise, and blurring. The conventional CornerNet approach was introduced along with the Hourglass104 as the base network (Law and Deng, 2019). The major drawback of the Hourglass104 network is its huge structural complexity. The larger number of framework parameters increases the computational burden on the CornerNet model and slows down the target identification procedure. Further, the Hourglass104 approach is inefficient when computing reliable keypoints for all types of image distortions, e.g., extensive changes in the size, color, and orientation of the affected areas (Zhao et al., 2019). Therefore, we have changed the feature extractor layer of the CornerNet model to enhance the identification and categorization performance for tomato plant leaf diseases. To this end, we have utilized the DenseNet-77 (Huang et al., 2017) as the base network of the CornerNet model in our proposed approach.
The DenseNet-77 network is a lightweight model from the DenseNet family and has two major benefits over the conventional DenseNet approach: first, the number of model parameters is smaller than the original DenseNet model (Masood et al., 2021); secondly, the layers within each dense block (Db) are also reduced to further simplify its structure. The employed DenseNet-77 model is a shallower framework compared to the Hourglass104 approach and comprises four Dbs in total. A detailed demonstration of the architectural representation of the DenseNet-77 is given in Figure 3. The DenseNet-77 approach comprises a smaller number of model parameters (6.2M) in comparison to the Hourglass104 base network (187M). Such architectural settings give it a computational advantage over the original base network. In all Dbs, the convolution layers are directly linked and the computed feature maps from starting layers are communicated to the subsequent layers. The DenseNet model encourages the reemployment of the computed features and facilitates the communication of the computed data in the entire network structure. This empowers it to deal with the image distortions effectively (Huang et al., 2017). Table 2 shows the network depiction of the DenseNet-77 model.
The network consists of numerous Convolutional Layers (CnL), Dbs, and Transition Layers (TnL). A pictorial depiction of the Db is given in Figure 3 and is the fundamental part of the DenseNet framework. In Figure 3, i0 represents the input layer and k0 depicts the feature maps. Furthermore, Cn(.) is a compound function containing 3 consecutive actions: a 3 × 3 CnL filter, Batch Normalization (BtN), and ReLU. Each CN(.) operation produces keypoint maps (k), that are used as input iN succeeding layers. The employment of all earlier computed features to the next layers introduces the k × (t−1)+k0 feature maps at the t-th layer of Db, which increases the feature space immensely. Hence, the TnL is used between the Db to lessen the computed features. The TnL is calculated as BtN and 1 × 1 CnL and the average pooling layer is represented as ApL, as depicted in Figure 3.
The feature computation framework consists of two separate output units that denote the TL and the BR corners estimation branches, respectively. Each branch unit comprises a corner pooling layer (CPL) positioned on the top of the backbone to pool keypoints and produces three results: Hms, embeddings, and offsets. The prediction module is an improved residual block (RB) containing two 3 × 3 CnL and one 1 × 1 residual network, followed by a CPL. The CPL assists the framework to identify the potential corners. The reduced keypoints are used as the input into a 3 × 3 CnL-BtN layer and then the reverse projection is performed. This improved RB is followed by a 3 × 3 CnL which produces Hms, embeddings, and offsets. The Hms give the approximation of a location in a sample, as a TL/BR corner, that is associated with a particular category. The embeddings are used to discriminate between the detected pairs of corners and offsets to fine-tune the bbx position. A suspected image can contain more than one affected region, therefore, embeddings assist the model to determine if the predicted corner points belong to a single disease class or different disease classes.
The CornerNet model is a deep learning framework that is independent of the selective search and proposal generation techniques. The test image and the associated annotated sample are given as input to the trained model. The improved CornerNet model extracts the corner points for the diseased area of the tomato plants and computes the associated offsets to the x and y coordinates, the measurements of bbx, and the associated class.
The employed framework for the detection and classification of tomato leaf disease is an end-to-end learning method that practices multi-task loss during the training to increase its recognition ability and precisely locate affected leaf regions. The total training loss, designated by Lt, is the combination of four different losses, given as:
Here, the Ld signifies detection loss accountable for corner identification, while Lpl denotes the group loss of group corners of the same bbx. Moreover, Lps is the corner separation loss used to separate the corners of different bbx, and Loff is the smooth L1 loss designated for offset adjustment. The symbols α, β, and γ are the constants for our approach, with the values of 0.1, 0.1, and 1, respectively. The mathematical description of the Ld is given in Eq. 2.
In this equation, R is the total number of detected diseased areas in a given image. For a given image, c, h, and w designate its total channels, width, and height. Moreover, tjuv indicates the estimated score at a given position (u, v) for the diseased area of class (j) in the suspected sample, and gjuv is the related ground-truth value. The ∅ and ω indicates the model hyperparameters that govern the influence of every selected point and have the values of 2 and 4 for our framework, respectively.
In downsampling, the dimension of the output sample is reduced than the actual sample size. The position (u, v) of the diseased portion in the test sample is plotted to the position in the Hms, where N indicates the downsampling factor. The remapping of Hms to the actual sample size introduces precision loss that eventually degrades the IoU performance for small bbx. To tackle this problem, the offsets for all locations are computed to fine-tune the corner dimensions as described in Eq. 3.
Here, Oi shows calculated offset, while for corner i, ui, and vi represents the coordinators of u and v. Furthermore, the Loff, employs the smooth L1 method for adjusting the corner positions and is represented as:
There could be several affected regions on a single image. Therefore, several BR and TL corners are nominated. For all corners, the model estimates an embedding vector to decide whether a group of BR and TL corners is associated with the same disease class or different disease classes. For this purpose, the CornerNet model uses the “pull and push” losses for framework training and are given as:
Here, elx shows the TL while the e_rx denotes the BR corners for a diseased region x and ex is the average value of erx and erx. The distance value to declare two detected corners belonging to different categories is set as 1, while the value of Δ is also 1 for all experiments.
In this section, we will outline detailed information about the dataset employed for the detection and classification of tomato plant leaf diseases. Moreover, the mathematical description of the used performance measures is also given. Finally, the results of the extensive experiments that have been conducted to show the efficacy of the proposed approach for tomato plant leaf disease recognition will be discussed.
We have used the PlantVillage database (Hughes and Salathé, 2015), a large repository accessible online, to evaluate the effectiveness of the model in detecting and classifying tomato leaf diseases. This dataset is comprised of a total of 54,306 images for 14 crop types. As this study is focused on the diseases of the tomato plant, we have utilized the tomato plant samples belonging to 10 different diseases. The main reason to employ the PlantVillage dataset for our work is that its images contain severe alterations in the size, chrominance, and position of the affected leaf regions. Furthermore, the images contain noise, brightness changes, blurring, and color alterations. An in-depth demonstration of the employed dataset is elaborated in Figure 4 while a few samples are shown in Figure 5.
For measuring the performance of the custom CornerNet model in detecting and classifying tomato plant leaf diseases, we have selected several standard metrics such as accuracy, mAP, intersection over union (IOU), precision, and recall. The mathematical description of accuracy and the mAP measure are given in Eqs 7, 8, respectively, while a graphical demonstration of precision, recall, and IOU is given in Figure 6.
The distinguishing attribute of a robust plant leaf disease classification framework is its ability to differentiate among the different classes of disease. To measure this, we designed an experiment. To visually elaborate on the detection performance of the custom CornerNet model, we have depicted the localized samples from the used dataset in Figure 7. The samples in Figure 7 clearly show that our technique is quite efficient in detecting the affected portion of the plant leaves and recognizing the associated classes even under the incidence of color, size, light, chrominance, and brightness changes.
The high recall power of the custom CornerNet model allows it to appropriately identify and categorize the several classes of tomato plant abnormalities. To numerically show the robustness of the proposed solution for tomato plant leaf disease classification, we have used two measures, namely the mAP and IOU score. These are the standard and most heavily employed metrics by the research community for object detection models. The proposed CornerNet model has localized the diseased portion from the plant samples with mAP and IOU scores of 0.984, and 0.979, respectively, which shows the effectiveness of our approach.
An efficient plant leaf disease recognition system must be powerful enough to accurately discriminate among the different types of diseases. We tested the class-wise performance of the presented model with the help of several standard metrics such as precision, recall, accuracy, and F1-score. Initially, we computed the precision and recall values for the custom CornerNet model in locating and classifying the 10 categories of plant leaf abnormalities. We have used the boxplot to show the obtained results as these plots provide a better understanding of the results by showing the minimum, maximum, and average values for the employed metrics (Figures 8, 9). The results reported in Figures 8, 9 show that the introduced approach is capable of correctly classifying the 10 classes of tomato plant leaf diseases.
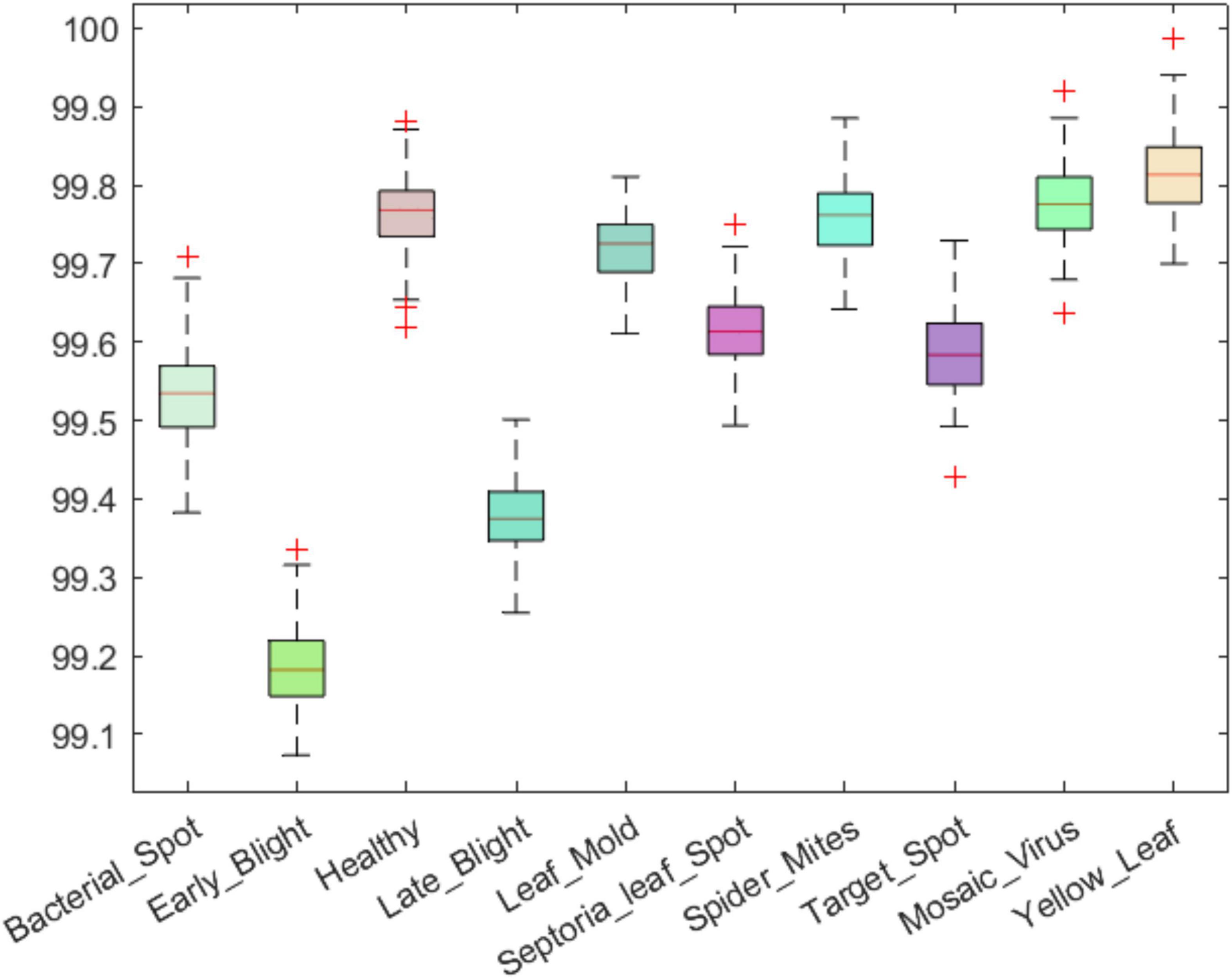
Figure 8. A pictorial depiction of the class-wise precision values obtained for the DenseNet-77-based CornerNet model.
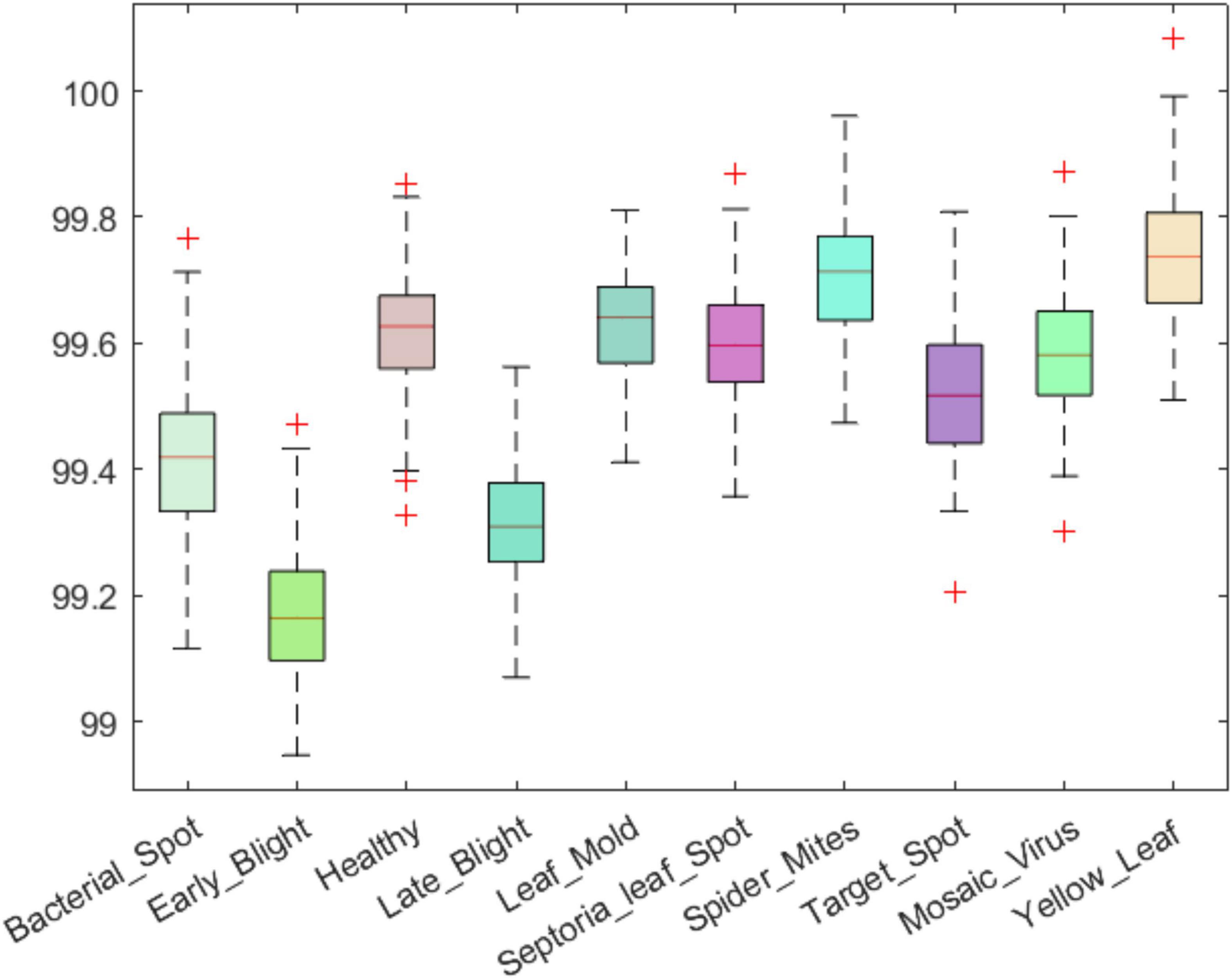
Figure 9. A pictorial depiction of the class-wise recall values obtained for the DenseNet-77-based CornerNet model.
Secondly, we show the calculated F1-score together with the error rate over the employed dataset and acquired values in Figure 10. The custom CornerNet model attains the average F1-score of 99.57% with the maximum and minimum error rates of 0.23 and 0.82%, respectively. The reported values demonstrate the robustness of the custom CornerNet model in locating and classifying all classes of tomato leaf disease efficiently.
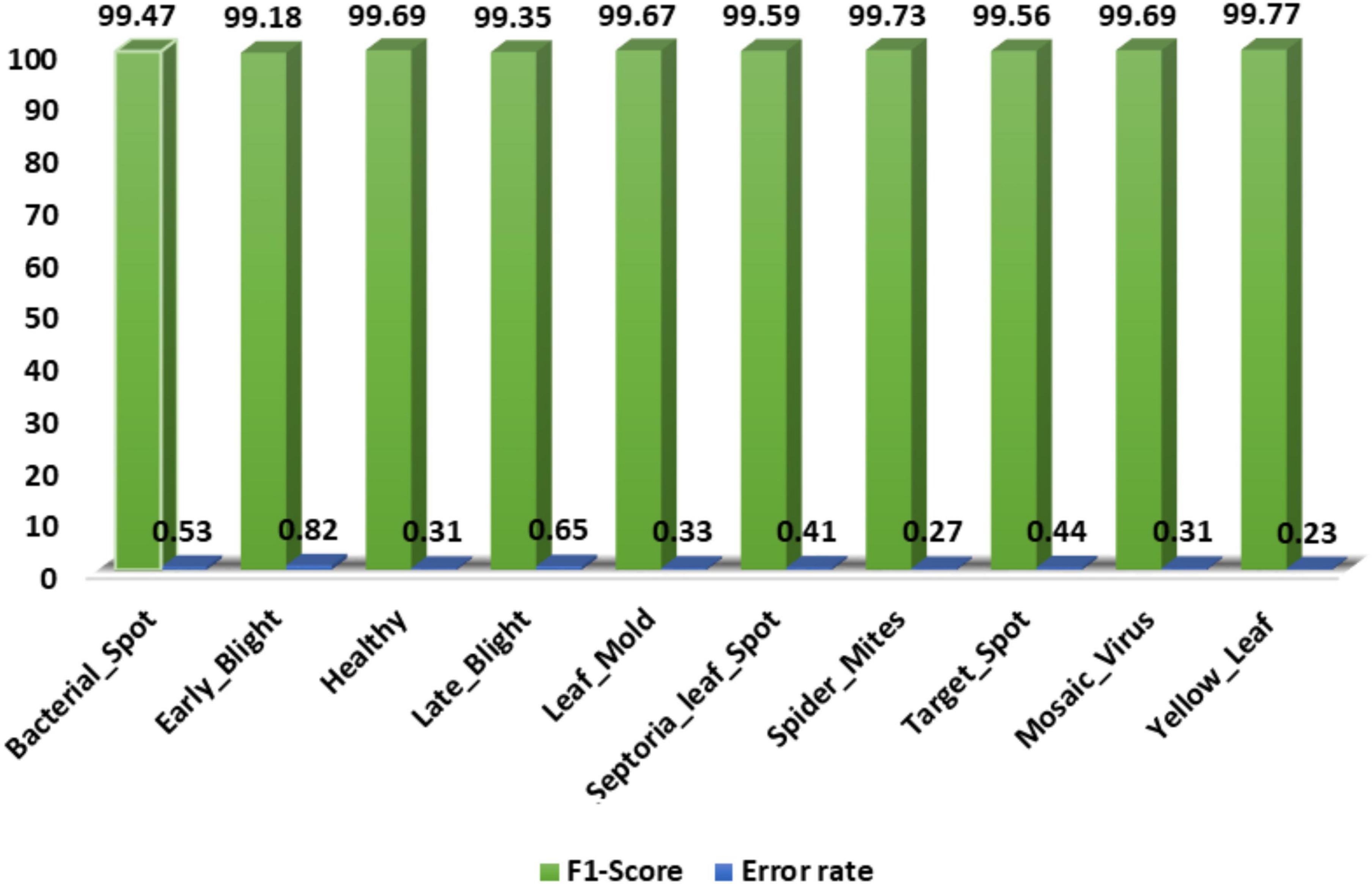
Figure 10. A pictorial depiction of the class-wise F1-score values obtained for the DenseNet-77-based CornerNet model.
Additionally, we have measured the class-wise accuracy values of the proposed technique and the acquired results are demonstrated in Figure 11. The introduced DenseNet-77-based CornerNet model attains the accuracy values of % for the 10 disease categories of the tomato plant and confirms the effectiveness of our approach.
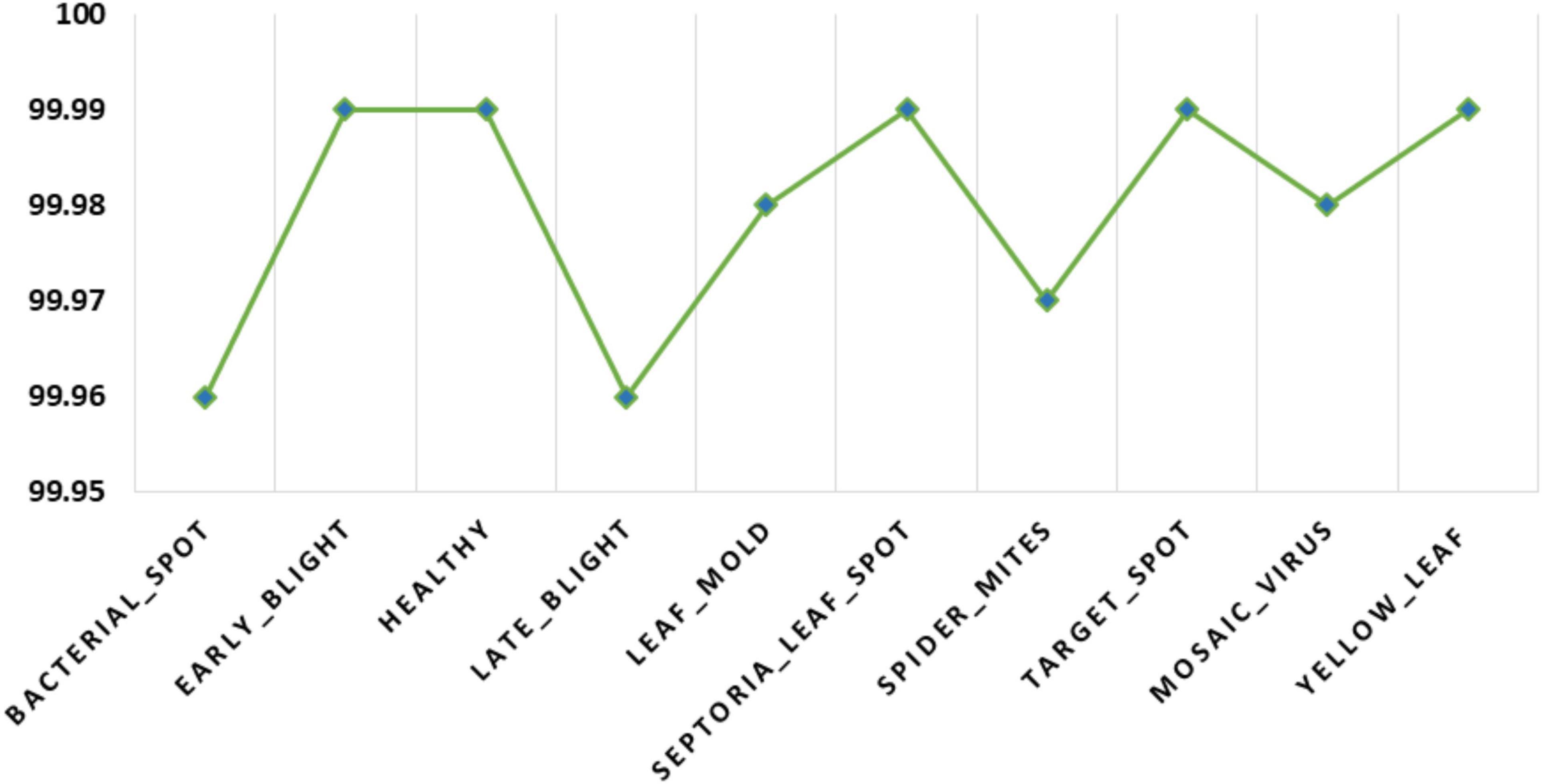
Figure 11. A pictorial depiction of the class-wise accuracy values obtained for the DenseNet-77-based CornerNet model.
To further validate the class-wise accurateness of the introduced approach for distinguishing the numerous categories of plant leaf disease, we have created a confusion matrix (Figure 12). This plot can show the actual and estimated classes recognized by a model. The values shown in figure demonstrate that the custom CornerNet model is proficient at recognizing all classes of tomato plant leaf diseases due to its higher recall rate which empowered it to differentiate all categories reliably.
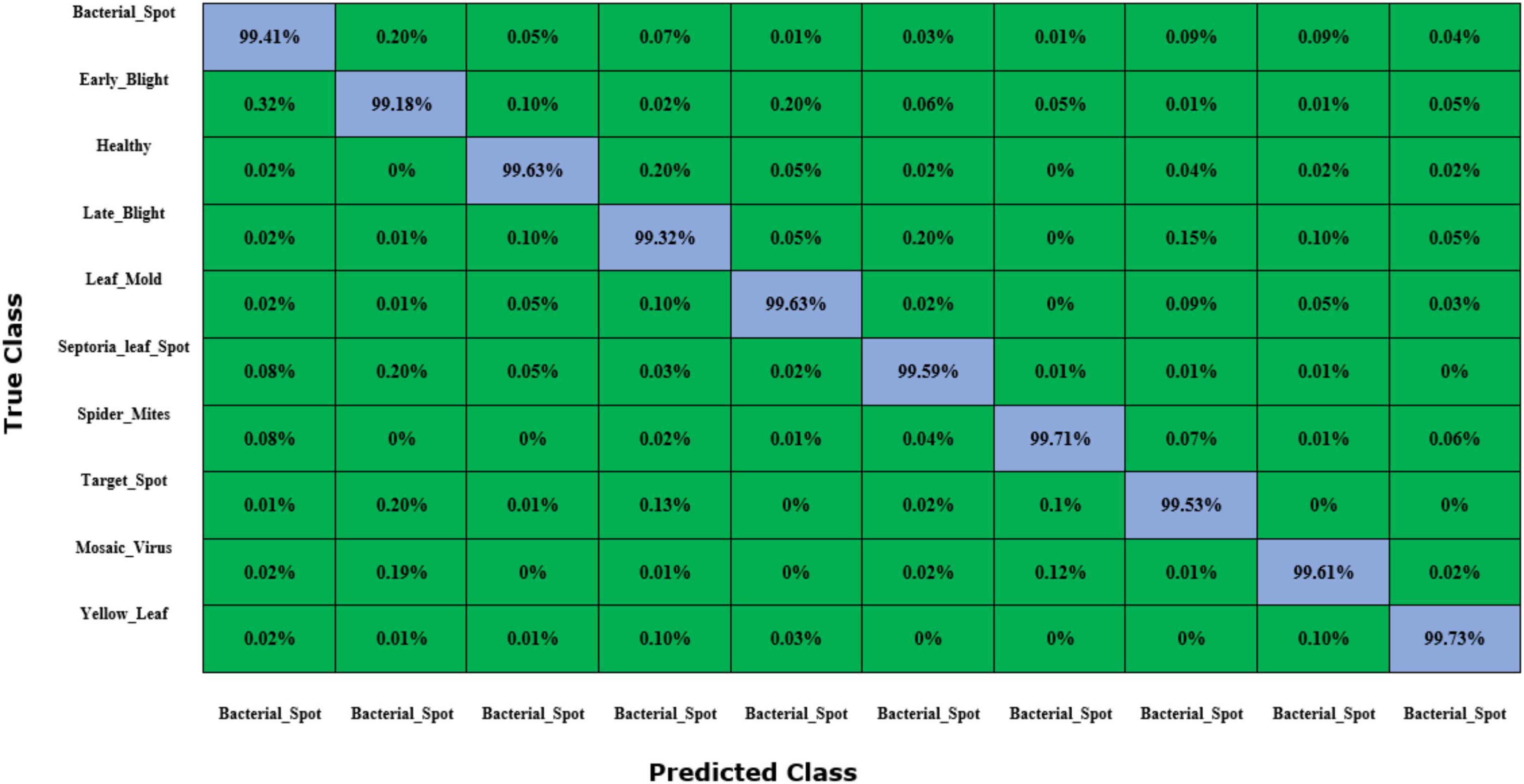
Figure 12. Confusion matrix results for tomato plant leaf diseases classification obtained using the DenseNet-77-based CornerNet model.
In this section, we outline an experiment to compare the tomato plant leaf disease recognition capability of the improved CornerNet model against the base networks. We chose several well-known DL frameworks, i.e., GoogleNet, ResNet-101, Xception, VGG-19, and SE-ResNet50. The comparison is depicted in Table 3. The performance analysis shown in Table 3 illustrates that our technique is more accurate than the peer approaches. The DenseNet-77-based CornerNet model attains the highest results for the precision, recall, F1-score, and accuracy measures with the numeric count of 0.9962, 0.9953, 0.9957, and 99.98%, respectively. The second-highest results are reported by the SE-ResNet50 model with 0.9677, 0.9681, 0.9679, and 96.81% for the precision, recall, F1-score, and accuracy metrics, respectively. Moreover, the GoogleNet model attains the lowest results in classifying the leaf diseases of the tomato plant and attains the scores for precision, recall, F1-score, and accuracy measures of 0.8716, 0.8709, 0.8712, and 87.27%, respectively. The second-lowest values are attained by the Xception model with the numeric stats of 0.8825, 0.8814, 0.8819, and 88.16%. The comparison illustrates the effectiveness of our approach. Specifically, for the precision measurement, the selected methods have an average value of 0.9050, while the DenseNet-77-based CornerNet model acquires the value of 0.9962 and shows a performance gain of 9.12%. For the recall and F1-score, the selected models have attained the average numeric score of 0.9053 and 0.9091, while in comparative analysis the presented solution has shown the average recall and F1-score of 0.9953 and 0.9957, respectively. Therefore, we can demonstrate average performance gains for the recall and F1-score of 9 and 8.66%, respectively. Moreover, in terms of accuracy, the base models attain an average value of 90.56%. The proposed model attains 99.98% accuracy, representing a performance gain of 9.42%. Furthermore, we outline the time taken for each model. It should be noted that the proposed approach shows the minimum test time. The values show the efficacy of our work to better recognize the several classes of tomato plant leaf abnormalities. The basic cause of this better classification performance of the proposed improved CornerNet model is the employment of the DenseNet-77 model as the keypoints extractor. This uplifts the model to better select the image information to identify the affected areas of the plant leaves and better recognize the associated class.
We have employed an object detection-based model for the localization and classification of the tomato plant leaf diseases and compared the performance of the proposed approach with other object detection techniques. The major reason for performing this simulation was to verify the reliability of the proposed DenseNet-77-based CornerNet model against other competitor techniques while locating the diseased areas from the tomato plant leaves under the occurrence of noise, light alteration, color changes, size variations, etc.
To execute this analysis, we have chosen numerous well-known object detection approaches, namely the Fast-RCNN (Girshick, 2015), Faster-RCNN (Ren et al., 2016) YOLO (Redmon and Farhadi, 2018), the SSD (Liu et al., 2016), and CornerNet (Law and Deng, 2019) models. To measure the performance of the model, the mAP metric is used as it is the standard evaluation measure used by the researchers to assess the classification performance of the object detection techniques. Furthermore, we have compared the test time of models as well to evaluate the time complexities of the comparative approaches as well. The comparison shows the efficiency and effectiveness of our approach and is illustrated in Table 4. The results in Table 4 show that the proposed approach has the highest mAP score and lowest test time with a numeric score of 0.984 and 0.22 s, respectively. The second highest mAP score is the Faster-RCNN model with a numeric count of 0.884. However, it is computationally inefficient and shows a time complexity of 0.28 s due to its two-stage classification network architecture. The SSD model has the lowest mAP score of 0.883 and a test time of 0.27 s. Furthermore, this approach does not perform well for very small plant leaf sizes. The conventional CornerNet model also has less promising results with a mAP score of 0.883 and a test time of 0.25 s. Whereas, the DenseNet-77-based CornerNet approach better tackles the issues of existing object detection approaches for identifying and classifying the numerous categories of the tomato plant leaves and shows the highest results. The comparison object detection approaches have an average mAP value of 0.859, compared to 0.984 for the proposed algorithm. Therefore, we have attained an average performance gain of 12.42% for the mAP metric. The one-stage detection ability of the proposed approach reduces the network structure complexity which, in turn, gives it a computational advantage.
In this section, we have selected several new approaches (Tm et al., 2018; Kaur and Bhatia, 2019; Agarwal et al., 2020a) that worked for tomato plant leaf disease classification and have used analysis to compare the performance of the improved CornerNet model with them. For this purpose, we have utilized three standard measures: precision, recall, and accuracy. Agarwal et al. (2020a) proposed the EfficientNet model for the automated detection and classification of tomato plant leaf diseases and attained an average accuracy value of 91.20%. Tm et al. (2018) proposed a CNN framework for categorizing the affected area of plant leaves and demonstrated an accuracy value of 94%. Similarly, Kaur and Bhatia (2019) employed a deep learning framework for recognizing the 10 types of plant leaf diseases with an accuracy rate of 98.80%. Hence, the comparative analysis is depicted in Table 5 and illustrates that our work has attained the highest results for all selected performance measures. From Table 5, it can be viewed that the techniques in Tm et al. (2018), Kaur and Bhatia (2019), and Agarwal et al. (2020a) achieve the precision of 0.90, 0.9481, and 0.9880, respectively, whereas the introduced improved CornerNet model obtains the precision of 0.9962. This is the highest of all the reported numeric scores for the selected works. The improved CornerNet model gains the largest value of 0.9953 for the recall performance measure, while the approaches in Tm et al. (2018), Kaur and Bhatia (2019), and Agarwal et al. (2020a) have recall scores of 0.92, 0.9478, and 0.9880, respectively. Moreover, with regards to accuracy, the proposed approach gains the numeric score of 99.98% while the approaches in Tm et al. (2018), Kaur and Bhatia (2019), and Agarwal et al. (2020a) have accuracy values of 91.20, 94, and 98.80%, respectively. The peer works (Tm et al., 2018; Kaur and Bhatia, 2019; Agarwal et al., 2020a) have the average precision, recall, and accuracy values of 0.9453, 0.9519, and 94.67%, respectively, as opposed to 0.9962, 0.9953, and 99.97%, respectively, for the presented work. Therefore, the DenseNet-77-based CornerNet model provides performance gains of 5.08, 4.34, and 5.30% for the precision, recall, and accuracy evaluation measures.
The reason for the competent classification results of the improved CornerNet model is that the techniques in Tm et al. (2018), Kaur and Bhatia (2019), and Agarwal et al. (2020a) are quite complex in network structure. This creates a framework over-fitting problem. The proposed solution is quite simple in structure and the employment of DenseNet-77 as the base network further empowered the CornerNet model to nominate a more reliable set of the sample feature vector. Such a model setting enhances its recognition ability by eliminating redundant information and reducing the model complexity. Further, the one-stage detection and classification ability of the CornerNet model prevents the framework from over-fitting issues and enables it to robustly deal with several image distortions like color, size, brightness, light variation, etc.
The manual screening of tomato plant leaf diseases relies highly on domain experts to detect the detailed information from the samples under observation. AI-based solutions are trying to fill this gap by automating the manual screening system. However, excessive changes in the mass, color, and size of plant leaves, and the incidence of noise, blurring, and brightness variations in the images complicate the classification task. In this work, we have attempted to overcome the existing issues by proposing a deep learning-based approach namely the DenseNet-77-based CornerNet model. We have carried out extensive experimentations on a standard dataset, namely the PlantVillage, and have confirmed through both the visual and numeric computations that the proposed approach is both efficient and effective in recognizing tomato plant leaf disease. Furthermore, the proposed approach is capable of efficiently detecting the diseased area of the plant leaves from the distorted samples containing several image transformations. However, the approach shows small detection degradation for images with huge angular variations which will be a major focus of our future work. Moreover, we plan to test the proposed model on other plant diseases and evaluate other DL-based frameworks.
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
SA: conceptualization, methodology, validation, software, supervision, and writing—reviewing and editing. MN: data curation, coding, validation, and writing—original draft preparation. Both authors contributed to the article and approved the submitted version.
We would like to thank the Deanship of Scientific Research, Qassim University for funding the publication of this project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agarwal, M., Gupta, S. K., and Biswas, K. (2020a). Development of Efficient CNN model for Tomato crop disease identification. Sustain. Comput. Inform. Syst. 28:100407. doi: 10.1016/j.suscom.2020.100407
Agarwal, M., Gupta, S. K., and Biswas, K. (2021b). “A compressed and accelerated SegNet for plant leaf disease segmentation: a differential evolution based approach,” in Pacific-Asia Conference On Knowledge Discovery And Data Mining, (Berlin: Springer). doi: 10.1007/978-3-030-75768-7_22
Agarwal, M., Gupta, S. K., and Biswas, K. (2021a). “Plant leaf disease segmentation using compressed UNet architecture,” in Pacific-Asia Conference on Knowledge Discovery and Data Mining, (Berlin: Springer). doi: 10.1007/978-3-030-75015-2_2
Agarwal, M., Gupta, S. K., Biswas, M., and Garg, D. (2022). Compression and acceleration of convolution neural network: A Genetic Algorithm based approach. J. Ambient Intell. Humaniz. Comput. 167, 1–11. doi: 10.1007/s12652-022-03793-1
Agarwal, M., Gupta, S. K., Garg, D., and Khan, M. M. (2021c). “A Partcle Swarm Optimization Based Approach for Filter Pruning in Convolution Neural Network for Tomato Leaf Disease Classification,” in International Advanced Computing Conference, (Berlin: Springer). doi: 10.1007/978-3-030-95502-1_49
Agarwal, M., Gupta, S. K., Garg, D., and Singh, D. (2021d). “A Novel Compressed and Accelerated Convolution Neural Network for COVID-19 Disease Classification: A Genetic Algorithm Based Approach,” in International Advanced Computing Conference, (Berlin: Springer). doi: 10.1007/978-3-030-95502-1_8
Agarwal, M., Singh, A., Arjaria, S., Sinha, A., and Gupta, S. (2020b). ToLeD: Tomato leaf disease detection using convolution neural network. Procedia Comput. Sci. 167, 293–301. doi: 10.1016/j.procs.2020.03.225
Ahmad, W., Shah, S., and Irtaza, A. (2020). Plants disease phenotyping using quinary patterns as texture descriptor. KSII Trans. Internet Inform. Syst. 14, 3312–3327. doi: 10.3837/tiis.2020.08.009
Akshai, K., and Anitha, J. (2021). “Plant disease classification using deep learning,” in 2021 3rd International Conference on Signal Processing and Communication (ICPSC), (Manhattan, NY: IEEE).
Albahli, S., Nawaz, M., Javed, A., and Irtaza, A. (2021). An improved faster-RCNN model for handwritten character recognition. Arab. J. Sci. Eng. 46, 8509–8523. doi: 10.1007/s13369-021-05471-4
Albattah, W., Javed, A., Nawaz, M., Masood, M., and Albahli, S. (2022). Artificial intelligence-based drone system for multiclass plant disease detection using an improved efficient convolutional neural network. Front. Plant Sci. 13.
Albattah, W., Nawaz, M., Javed, A., Masood, M., and Albahli, S. (2021). A novel deep learning method for detection and classification of plant diseases. Complex Intell. Syst. 8, 507–524. doi: 10.1007/s40747-021-00536-1
Argüeso, D., Picon, A., Irusta, U., Medela, A., San-Emeterio, M. G., Bereciartua, A., et al. (2020). Few-Shot Learning approach for plant disease classification using images taken in the field. Comput. Electron. Agric. 175:105542. doi: 10.1016/j.compag.2020.105542
Batool, A., Hyder, S. B., Rahim, A., Waheed, N., and Asghar, M. A. (2020). “Classification and Identification of Tomato Leaf Disease Using Deep Neural Network,” in 2020 International Conference on Engineering and Emerging Technologies (ICEET), (Bellingham, WA: IEEE). doi: 10.1109/ICEET48479.2020.9048207
Bello-Cerezo, R., Bianconi, F., Maria, F. Di, Napoletano, P., and Smeraldi, F. (2019). Comparative evaluation of hand-crafted image descriptors vs. off-the-shelf CNN-based features for colour texture classification under ideal and realistic conditions. Appl. Sci. 9:738. doi: 10.3390/app9040738
Bhujel, A., Kim, N.-E., Arulmozhi, E., Basak, J. K., and Kim, H.-T. (2022). A lightweight Attention-based convolutional neural networks for tomato leaf disease classification. Agriculture 12:228. doi: 10.3390/agriculture12020228
Bruinsma, J. (2009). The resource outlook to 2050: By how much do land, water and crop yields need to increase by 2050: Expert meeting on how to feed the world in 2009. Rome: Food and Agriculture Organization of the United Nations.
Chowdhury, M. E., Rahman, T., Khandakar, A., Ayari, M. A., Khan, A. U., Khan, M. S., et al. (2021). Automatic and reliable leaf disease detection using deep learning techniques. AgriEngineering 3, 294–312. doi: 10.3390/agriengineering3020020
Dinh, H. X., Singh, D., Periyannan, S., Park, R. F., and Pourkheirandish, M. (2020). Molecular genetics of leaf rust resistance in wheat and barley. Theor. Appl. Genet. 133, 2035–2050. doi: 10.1007/s00122-020-03570-8
Dwivedi, R., Dey, S., Chakraborty, C., and Tiwari, S. (2021). Grape disease detection network based on multi-task learning and attention features. IEEE Sen. J. 21, 17573–17580. doi: 10.1109/JSEN.2021.3064060
Elnaggar, S., Mohamed, A. M., Bakeer, A., and Osman, T. A. (2018). Current status of bacterial wilt (Ralstonia solanacearum) disease in major tomato (Solanum lycopersicum L.) growing areas in Egypt. Arch. Agric. Environ. Sci. 3, 399–406. doi: 10.26832/24566632.2018.0304012
Ferentinos, K. P. (2018). Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 145, 311–318. doi: 10.1016/j.compag.2018.01.009
Gebbers, R., and Adamchuk, V. I. (2010). Precision agriculture and food security. Science 327, 828–831. doi: 10.1126/science.1183899
Gewali, U. B., Monteiro, S. T., and Saber, E. (2018). Machine learning based hyperspectral image analysis: A survey. arXiv [Preprint].
Girshick, R. (2015). “Fast R-CNN,” in Proceedings Of The Ieee International Conference On Computer Vision, (Santiago: IEEE). doi: 10.1109/ICCV.2015.169
Girshick, R., Donahue, J., Darrell, T., and Malik, J. (2015). Region-based convolutional networks for accurate object detection and segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 38, 142–158. doi: 10.1109/TPAMI.2015.2437384
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger, K. Q. (2017). “Densely connected convolutional networks,” in Proceedings Of The Ieee Conference On Computer Vision And Pattern Recognition, (Honolulu, HI: IEEE) doi: 10.1109/CVPR.2017.243
Hughes, D., and Salathé, M. (2015). An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv [Preprint].
Joachims, T. (1998). Making Large-Scale SVM Learning Practical Technical Report. Dortmund: Technical University Dortmund.
Karthik, R., Hariharan, M., Anand, S., Mathikshara, P., Johnson, A., and Menaka, R. (2020). Attention embedded residual CNN for disease detection in tomato leaves. Appl. Soft Comput. 86:105933. doi: 10.1016/j.asoc.2019.105933
Kaur, M., and Bhatia, R. (2019). “Development of an improved tomato leaf disease detection and classification method,” in 2019 IEEE Conference on Information and Communication Technology, (Manhattan, NY: IEEE). doi: 10.1109/CICT48419.2019.9066230
Kaur, N. J. T. J. O. C., and Education, M. (2021). Plant leaf disease detection using ensemble classification and feature extraction. Turkish J. Comput. Math. Educ. 12, 2339–2352.
Kuricheti, G., and Supriya, P. (2019). “Computer Vision Based Turmeric Leaf Disease Detection and Classification: A Step to Smart Agriculture,” in 2019 3rd International Conference on Trends in Electronics and Informatics (ICOEI), (Manhattan, NY: IEEE). doi: 10.1109/ICOEI.2019.8862706
Law, H., and Deng, J. (2019). CornerNet: Detecting objects as paired keypoints. Int. J. Comput. Vis. 128, 642–656. doi: 10.1007/s11263-019-01204-1
Le, V. N. T., Ahderom, S., Apopei, B., and Alameh, K. (2020). A novel method for detecting morphologically similar crops and weeds based on the combination of contour masks and filtered Local Binary Pattern operators. GigaScience 9:giaa017. doi: 10.1093/gigascience/giaa017
Lin, T. (2020). Labelimg. Available online at: https://github.com/tzutalin/ImageNet_Utils.
Liu, W., Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu, C.-Y., et al. (2016). “Ssd: Single shot multibox detector,” in European Conference On Computer Vision, (Berlin: Springer). doi: 10.1007/978-3-319-46448-0_2
Maeda-Gutiérrez, V., Galvan-Tejada, C. E., Zanella-Calzada, L. A., Celaya-Padilla, J. M., Galván-Tejada, J. I., Gamboa-Rosales, H., et al. (2020). Comparison of convolutional neural network architectures for classification of tomato plant diseases. Appl. Sci. 10:1245. doi: 10.3390/app10041245
Masood, M., Nazir, T., Nawaz, M., Mehmood, A., Rashid, J., Kwon, H.-Y., et al. (2021). A novel deep learning method for recognition and classification of brain tumors from MRI images. Diagnostics 11:744. doi: 10.3390/diagnostics11050744
Nawaz, M., Nazir, T., Masood, M., Mehmood, A., Mahum, R., Khan, M. A., et al. (2021). Analysis of brain MRI images using improved cornernet approach. Diagnostics 11:1856. doi: 10.3390/diagnostics11101856
Nazir, T., Irtaza, A., Javed, A., Malik, H., Hussain, D., and Naqvi, R. A. (2020). Retinal image analysis for diabetes-based eye disease detection using deep learning. Appl. Sci. 10:6185. doi: 10.3390/app10186185
Pantazi, X. E., Moshou, D., and Tamouridou, A. A. (2019). Automated leaf disease detection in different crop species through image features analysis and One Class Classifiers. Comput. Electron. Agric. 156, 96–104. doi: 10.1016/j.compag.2018.11.005
Patil, S., and Chandavale, A. (2015). A survey on methods of plant disease detection. Int. J. Sci. Res. 4, 1392–1396.
Paul, A., Ghosh, S., Das, A. K., Goswami, S., Choudhury, S. D., and Sen, S. (2020). “A review on agricultural advancement based on computer vision and machine learning,” in Emerging Technology In Modelling And Graphics, eds J. Mandal and D. Bhattacharya (Berlin: Springer), 567–581. doi: 10.1007/978-981-13-7403-6_50
Raj, A., Namboodiri, V. P., and Tuytelaars, T. (2015). Subspace alignment based domain adaptation for rcnn detector. arXiv [Preprint]. doi: 10.5244/C.29.166
Ramesh, S., Hebbar, R., Niveditha, M., Pooja, R., Shashank, N., and Vinod, P. (2018). “Plant disease detection using machine learning,” in 2018 International Conference On Design Innovations For 3cs Compute Communicate Control (ICDI3C), (Manhattan, NY: IEEE). doi: 10.1109/ICDI3C.2018.00017
Redmon, J., Divvala, S., Girshick, R., and Farhadi, A. (2016). “You only look once: Unified, real-time object detection,” in Proceedings Of The IEEE Conference On Computer Vision And Pattern Recognition, (Las Vegas, NV: IEEE) doi: 10.1109/CVPR.2016.91
Ren, S., He, K., Girshick, R., and Sun, J. (2016). Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 39, 1137–1149. doi: 10.1109/TPAMI.2016.2577031
Richey, B., Majumder, S., Shirvaikar, M., and Kehtarnavaz, N. (2020). “Real-time detection of maize crop disease via a deep learning-based smartphone app,” in Proceedings of the Real-Time Image Processing and Deep Learning 2020, (Bellingham, WA: International Society for Optics and Photonics). doi: 10.1117/12.2557317
Rokach, L., and Maimon, O. (2005). “Decision trees,” in Data Mining And Knowledge Discovery Handbook, eds O. Maimon and L. Rokach (Berlin: Springer), 165–192. doi: 10.1007/0-387-25465-X_9
Roska, T., and Chua, L. O. (1993). The CNN universal machine: An analogic array computer. IEEE Trans. Circuits Syst. II Analog Digit. Signal Process. 40, 163–173. doi: 10.1109/82.222815
Salakhutdinov, R., and Hinton, G. (2009). “Deep Boltzmann machines,” in Proceedings of the artificial intelligence and statistics (PMLR), Birmingham.
Sankaran, S., Mishra, A., Ehsani, R., and Davis, C. (2010). A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 72, 1–13. doi: 10.1016/j.compag.2010.02.007
Sardogan, M., Tuncer, A., and Ozen, Y. (2018). “Plant leaf disease detection and classification based on CNN with LVQ algorithm,” in 2018 3rd International Conference on Computer Science and Engineering (UBMK), (Manhattan, NY: IEEE). doi: 10.1109/UBMK.2018.8566635
Shrivastava, V. K., and Pradhan, M. K. (2021). Rice plant disease classification using color features: A machine learning paradigm. J. Plant Pathol. 103, 17–26. doi: 10.1007/s42161-020-00683-3
Sun, Y., Jiang, Z., Zhang, L., Dong, W., and Rao, Y. (2019). SLIC_SVM based leaf diseases saliency map extraction of tea plant. Comput. Electron. Agric. 157, 102–109. doi: 10.1016/j.compag.2018.12.042
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., et al. (2015). “Going deeper with convolutions,” in Proceedings Of The IEEE Conference On Computer Vision And Pattern Recognition, (Berlin: IEEE). doi: 10.1109/CVPR.2015.7298594
Thenmozhi, K., and Reddy, U. S. (2019). Crop pest classification based on deep convolutional neural network and transfer learning. Comput. Electron. Agric. 164:104906. doi: 10.1016/j.compag.2019.104906
Tm, P., Pranathi, A., SaiAshritha, K., Chittaragi, N. B., and Koolagudi, S. G. (2018). “Tomato leaf disease detection using convolutional neural networks,” in 2018 Eleventh International Conference On Contemporary Computing (IC3), (Manhattan, NY: IEEE). doi: 10.1109/IC3.2018.8530532
Valenzuela, M. E. M., and Restović, F. (2019). “Valorization of Tomato Waste for Energy Production,” in Tomato Chemistry, Industrial Processing and Product Development, ed. S. Porretta (London: Royal Society of Chemistry), 245–258. doi: 10.1039/9781788016247-00245
Vedaldi, A., and Zisserman, A. (2016). Vgg convolutional neural networks practical. Dep. Eng. Sci. Univ. Oxford 2016:66.
Wolfenson, K. D. M. (2013). Coping With The Food And Agriculture Challenge: Smallholders’ Agenda. Rome: Food Agriculture Organisation of the United Nations.
Yuan, Z.-W., and Zhang, J. (2016). “Feature extraction and image retrieval based on AlexNet,” in Eighth International Conference on Digital Image Processing (ICDIP 2016), (Bellingham, WA: International Society for Optics and Photonics). doi: 10.1117/12.2243849
Zaremba, W., Sutskever, I., and Vinyals, O. (2014). Recurrent neural network regularization. arXiv [Preprint].
Zhao, S., Peng, Y., Liu, J., and Wu, S. (2021). Tomato leaf disease diagnosis based on improved convolution neural network by attention module. Agriculture 11:651.
Zhao, X., Li, W., Zhang, Y., Gulliver, T. A., Chang, S., and Feng, Z. (2016). “A faster RCNN-based pedestrian detection system,” in 2016 IEEE 84th Vehicular Technology Conference (VTC-Fall), (Manhattan, NY: IEEE). doi: 10.1109/VTCFall.2016.7880852
Keywords: CornerNet, classification, DenseNet, tomato plant diseases, localization
Citation: Albahli S and Nawaz M (2022) DCNet: DenseNet-77-based CornerNet model for the tomato plant leaf disease detection and classification. Front. Plant Sci. 13:957961. doi: 10.3389/fpls.2022.957961
Received: 31 May 2022; Accepted: 12 August 2022;
Published: 08 September 2022.
Edited by:
Peng Chen, Anhui University, ChinaCopyright © 2022 Albahli and Nawaz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saleh Albahli, c2FsYmFobGlAcXUuZWR1LnNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.