
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 July 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.957566
 Xiaqing Wang†
Xiaqing Wang† Yining Chen†
Yining Chen† Xuan Sun†
Xuan Sun† Jinghuan Li
Jinghuan Li Ruyang Zhang
Ruyang Zhang Yanyan Jiao
Yanyan Jiao Ronghuan Wang
Ronghuan Wang Wei Song*
Wei Song* Jiuran Zhao*
Jiuran Zhao*Lodging is a major problem in maize production, which seriously affects yield and hinders mechanized harvesting. Improving stalk strength is an effective way to improve lodging. The maize inbred line Jing2416 (J2416) was an elite germplasm in maize breeding which had strong stalk mechanical strength. To explore the characteristics its stalk strength, we conducted physiological, metabolic and transcriptomic analyses of J2416 and its parents Jing24 (J24) and 5237. At the kernel dent stage, the stalk rind penetrometer strength of J2416 was significantly higher than those of its two parents in multiple environments. The rind thickness, sclerenchyma tissue thickness, and cellulose, hemicellulose, and lignin contents of J2416 were significantly higher than those of its parents. Based on the significant differences between J2416 and 5237, we detected metabolites and gene transcripts showing differences in abundance between these two materials. A total of 212 (68.60%) metabolites and 2287 (43.34%) genes were up-regulated in J2416 compared with 5237. The phenylpropanoid and glycan synthesis/metabolism pathways were enriched in metabolites and genes that were up-regulated in J2416. Twenty-eight of the up-regulated genes in J2416 were involved in lignin, cellulose, and hemicellulose synthesis pathways. These analyses have revealed important physiological characteristics and candidate genes that will be useful for research and breeding of inbred lines with excellent stalk strength.
Lodging, the breaking of the stalk, is a major problem in the production of maize (Zea mays L.). It causes maize plants to fall over, thereby affecting photosynthesis and development, and rendering the plants more vulnerable to damage from pests and diseases (Shah et al., 2017). Lodging can occur at different stages of development, but it causes the most serious problems when it occurs at the grain development stage. At this stage, lodging hinders the transport of photosynthates to the grain, thereby negatively affecting the maize yield (Wen et al., 2019; Zhao and Zhou, 2022). Stalk lodging also affects mechanized harvesting. It has been reported that when the lodging rate increases by 1%, the ear drop rate increases by 0.15% (Xue et al., 2018). With the development of mechanization for modern maize cultivation, the demand for maize varieties that retain upright stalks during harvesting is increasing, and consequently, there is a need for new varieties with strong stalk strength. Therefore, improving stalk strength is of great significance for maize lodging resistance, stabilizing maize production, and accelerating the modernization of maize production.
There are many indicators of stalk strength, such as rind penetrometer strength, stalk bending strength, and stalk bending angle. Rind penetrometer strength has been widely used by many researchers. Because it easy to measure, this trait can be measured multiple times in different positions on the same stalk to obtain more accurate data (Li et al., 2014; Wang et al., 2019). Stalk strength and the position of fracturing differ among developmental stages. At the early stage of tasseling, the stalk is easily broken at 1 cm above the first node above the ear (Wang et al., 2019). At the grain filling stage, the maize stalk is prone to breaking at the internode of the third node at the base. This may be because the height of the center of gravity moves upwards during growth and some of the photosynthates that stabilize the stalk are transported to the grain (Xu et al., 2017; Stein and Granot, 2019; Shah et al., 2021).
Stalk strength is affected by several physiological traits, including stalk shape, stalk anatomical characteristics, and cell wall components, i.e., the types and contents of various structural compounds and metabolites (Shah et al., 2017). Varieties with a short and thick internode at the base are more resistant to lodging (Kotake et al., 2011; Okuno et al., 2014). The internal structure of the stalk determines its external macroscopic morphology and mechanical properties. The anatomical features of the maize stalk include the epidermis, sclerenchyma cells, parenchyma cells, and vascular bundles. In recent years, X-ray microcomputed tomography technology has allowed for the observation of the total number of vascular bundles in a cross-section of the entire stalk. This technology has greatly facilitated the observation of the number and shape of vascular bundles (Du et al., 2016). Studies have shown that stalk strength is positively correlated with the thickness of the rind and sclerenchyma tissue, as well as the number and density of vascular bundles (Kong et al., 2013; Xue et al., 2016; Wang et al., 2019; Guo et al., 2021). The stalk consists of many chemical components, such as cellulose, hemicellulose, lignin, pectin, and starch. The most abundant components are cellulose, hemicellulose, and lignin. Cellulose is the largest polymer in plant cell walls and provides mechanical support for cells. Hemicellulose and cellulose together form a network; and lignin exists in a highly complexed state in thickened secondary cell walls, providing mechanical support (Qiu and Hu, 2013; Yoon et al., 2015; Deshavath et al., 2017). Cellulose, hemicellulose, and lignin contents are all positively related to stalk strength (Jiao et al., 2019; Wang et al., 2020; Zhang et al., 2020). With the development of phenomics technology, more novel phenotypes have been revealed. Metabolite analysis has been gradually applied in crop genetics research because of their diversity and extremely sensitive response to changes in gene expression (Wen et al., 2016). However, few studies have explored the roles of particular metabolites in stalk development and stalk strength.
The germplasm of the Huang-gai group is an extremely important heterotic group in maize breeding. Because of its excellent characteristics, this group is often used for gene mining and breeding of new maize varieties. The inbred lines J24 and 5237 are elite lines derived from the Huang-gai germplasm. Some popular maize hybrids such as Jingdan 28 and Yedan 22 have been bred from J24 and 5237, respectively (Chen et al., 2009). Recurrent selection breeding from the hybrids of these two inbred lines has resulted in some excellent inbred lines, among which J2416 ([(J24 × 5237) × J24]) is the most successful. To date, more than 21 commercial maize hybrids have been bred from J2416, and more than four hybrids are cultivated across a planting area of more than 67,000 ha (Zhao et al., 2020a). Eighteen varieties bred from J2416 have particularly strong stalk strength (unpublished). Therefore, J2416 has become a backbone inbred line for research and breeding.
To explore the mechanism of the excellent stalk strength of J2416, we analyzed the anatomical and biochemical traits of its stalk, and detected metabolites that differed significantly in abundance compared with its parents (J24 and 5237). Genes that were up-regulated in J2416 and closely related to stalk strength traits were identified. Our results reveal which characteristics and candidate genes are related to the strong stalk phenotype, and provide a reference for developing maize varieties with high-quality stalks.
The maizel inbred line J2416 and its parents J24 and 5237 were obtained from the Maize Research Institute, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China. All the three materials belong to the germplasm of the heterotic group of Huang-gai. J24 was bred from a hybrid of the early maturing line 302 (ZS302) and Huangyesi (HYS) (Figure 1). 5237 was bred from a hybrid of Huangzaosi (HZS) and Dan340. J2416 was bred from the backcross population of J24 and 5237 [(J24 × 5237) × J24] (Zhao et al., 2020a; Figure 1).
The three inbred lines J2416, J24, and 5237 were planted at Sanya, Hainan (HN, 108.56°E, 18.09°N) in winter 2020; Tongzhou, Beijing (TZ, 116.65°E, 39.92°N), and Changping Beijing (CP, 119.39°E, 40.17°N) in summer 2021. In all environments, each inbred line was planted in five rows, with 20 plants per row (plant spacing of 25 cm; row spacing of 60 cm). Irrigation and pest control were consistent with local agronomic practices throughout the growth and development of maize.
To study the stalk strength of these materials at different stages after silking, stalk strength was measured for 5237, J2416, and J24 at the kernel dent stage, corresponding to about 31 to 33 days after the R1 stage (silks extending outside the husk leaves) of maize, respectively (Ciampitti et al., 2011). During this period, kernels are “dented” at the tip because of declining moisture content and increasing starch content, and husk leaves are fading to a pale green with browning on the edges (Ciampitti et al., 2011). At this stage, the stalks of the three materials were still standing upright and were not dry. Rind penetrometer strength was measured at the third internode with 9–60 uniform plants using a YYD-1 instrument (Zhejiang Top Cloud-Agri Technology Co., Ltd., Zhejiang, China) (Wang et al., 2019).
To investigate anatomic traits, twenty 200-μm hand-cut sections were prepared from the third stalk at the base of the stalk of the three materials. The materials were collected at 30DAS_21TZ (30 days after silking in Tongzhou in summer 2021). The sections were stained with 2% phloroglucinol dye for 30–60 s (Sindhu et al., 2007). The anatomical characteristics were observed under an Olympus DP80 compound microscope (Olympus Corp., Shinjuku, Japan). Images were analyzed with Image-Pro Plus 6.0 to measure the thickness of the stalk rind and sclerenchyma. The results were statistically analyzed using the R software package.
After measuring the stalk strength of each material, stalks were collected with three to five replicates per material, with five plants per replicate on 30DAS_21TZ. The material was naturally air-dried, and then dried at 40°C before grinding and sieving. Samples were passed through 40 to 80 mesh sieves (0.425 mm–0.180 mm fineness) before extracting structural carbohydrates and lignin using a two-step sulfuric acid hydrolysis process. The contents of cellulose, hemicellulose and lignin were determined using a high-performance liquid chromatography system (HPLC, 1260 series, Agilent Technologies, Santa Clara, CA, United States) (Sluiter et al., 2010).
For each material, stalk samples were taken at 1 cm above the third stalk node, and were stored at −80°C. Three replicates of each material, with each replicate consisting of three mixed samples, were collected on 15DAS_21TZ (15 days after silking in 2021 at Tongzhou base). The extraction, detection and analysis methods of metabolites were as follows.
Extraction process of metabolite. 50 mg of the sample powder was added to 1000 μL of the extraction solution containing the internal standard (volume ratio of methanol to acetonitrile to water as 2:2:1, internal standard concentration as 2 mg/L), and vortexed for 30 s. The samples were grinded for 10 min, and then used for ultrasonic extraction for 10 min at low temperature. Next, the samples were kept in −20°C for 1 h. The supernatant was dried in a vacuum concentrator after centrifugation at 4°C for 15 min (12000 rpm). The extraction solution (volume ratio of acetonitrile to water as 1:1) was added to the dried substance to reconstitute. The solution was vortexed again for 30 s, and then used for ultrasonic extraction for 10 min at low temperature. The mixture was centrifuged at 12000 rpm for 15 min at low temperature, and the supernatant liquid was transferred into the injection vial.
Detection of metabolites. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) system used for metabolites detection consisted of a Waters Acquity I-Class PLUS UHPLC coupled with a Waters Xevo G2-XS QTof high-resolution mass spectrometer (Song et al., 2016). The column used in this system is an Acquity UPLC HSS T3 column (100mm × 2.1 mm, 1.8 mm, Waters, Milford, MA, United States), with the injection volume of 1 μL. The mobile phases A consisted of 0.1% formic acid aqueous solution. The mobile phase B consisted of 0.1% formic acid acetonitrile. The gradient parameters were based on Song et al., 2016. The Waters Xevo G2-XS QTof high-resolution mass spectrometer can perform primary and secondary mass spectrometry data acquisition in MSE mode under the control of the acquisition software (MassLynx V4.2, Waters). Dual-channel data acquisition can be performed simultaneously for both low collision energy and high collision energy in each data acquisition cycle. The low collision energy was 2 V, and the high collision energy was 10∼40 V. The scanning frequency was 0.2 s for a mass spectrum. ESI ion source parameters were set as follows: capillary voltage as 2000 V (positive ion mode) or −1500 V (negative ion mode); cone voltage as 30 V; ion source temperature as 150°C; desolvation gas temperature as 500°C; backflush gas flow rate as 50 L/h; flow rate of desolventizing gas as 800 L/h.
Analysis of metabolites. The raw data collected by MassLynx V4.2 was processed by Progenesis QI (Waters, Milford, MA, United States) software for peak extraction, peak alignment and other data processing operations. Metabolites were identified based on METLIN database (Smith et al., 2005). The differentially accumulated metabolites were selected on the basis of the P-value of the Student’s t-test and the variable importance value (VIP) from the Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) model. Metabolites with a P-value < 0.05 and VIP > 1 were considered to be differentially accumulated between the two compared materials (Wiklund et al., 2008). The function of the differentially accumulated metabolites was determined by searches at the KEGG database (Kanehisa et al., 2019). Enrichment and hierarchical clustering analyses were conducted using KOBAS2.0 software1 (Altschul et al., 1997; Xie et al., 2011).
The samples used for RNA sequencing and analysis were the same as those used for metabolite detection. Total RNA was extracted using TRIZOL reagent (Invitrogen, Carlsbad, CA, United States). Sequencing libraries were generated using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, United States), and sequenced using an Illumina HiSeq™ 2000 system. The clean reads were mapped to the maize reference genome (B73 AGPv4) using Hisat2 software (Kim et al., 2019). Gene expression levels were calculated as fragments per kilobase of transcript per million fragments mapped (FPKM) values (Florea et al., 2013). Genes with an abs (log2(fold change)) > 1 and false discovery rate (FDR) < 0.01 were considered as differentially expressed genes (DEGs) (Anders and Huber, 2010). The function of DEGs was determined according to annotations at the KEGG database (Kanehisa et al., 2019).
The RNA samples used for RT-qPCR were the same as those used for transcriptome sequencing. The cDNA synthesis was performed using cDNA synthesis kit (HiScriptIII 1st Strand cDNA Synthesis Kit, Vazyme Biotech Company, China), according to the manufacturer’s protocol. The primers used for RT-qPCR were synthesized at the Tanyibiotech Company. The PCR system consisted of cDNA (2.5 uL), 2x Taq Pro Universal SYBR qPCR Master Mix (Q172, vazyme, 10 μL), 10 μM F and R primers (0.4 μL each), and water (6.7 μL). The PCR program was set as 95°C, 30 s; 40 cycles (95°C, 10 s; and 60°C, 30 s). Melting curves were established for the PCR products at the end of the amplification reaction (95°C, 15 s; 60°C, 60 s; 95°C, 15 s) with slow heating from 60°C to 99°C (automatically ramped the temperature at a rate of 0.05°C/s). The procedure was carried out on Quant Studio 6 Flex Real-Time PCR System (Thermo Fisher Scientific ABI, United States). ZmActin1 was used as the internal control (Zhang et al., 2018). The expression level of target genes was calculated using the relative 2–ΔΔCt method (Livak and Schmittgen, 2001). All primers used for RT–qPCR was listed in Supplementary Table 1.
To compare stalk strength among J2416, J24 and 5237, we measured their rind penetrometer strength at the kernel dent stage in two different environments (Figure 2A). Although they were all 30 days after silking, the rind penetrometer strength of the same material in different environments was different. However, we detected significant differences in rind penetrometer strength among the three inbred lines across 22 degrees north latitude and 11 degrees east longitude, regardless of whether planting was in summer or winter. We found the rind penetrometer strength of J2416 was slightly higher than that of J24, but significantly higher than that of 5237 (Figure 2B).
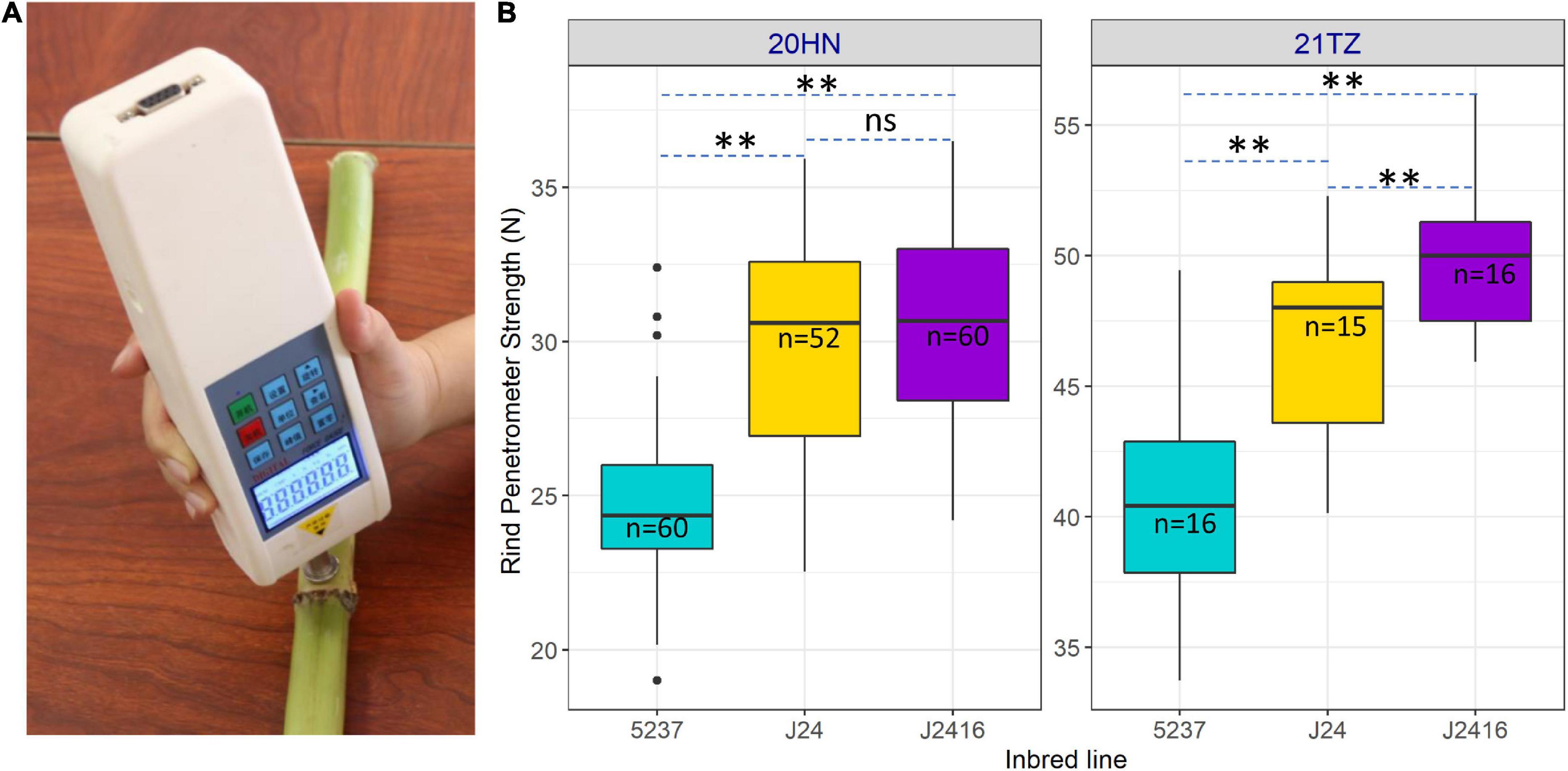
Figure 2. Rind penetrometer strength of J2416 and its two parents. (A) Measurement of rind penetrometer strength. (B) Rind penetrometer strength of three inbred lines in different locations. **Means the significant difference at 0.01 level. ns means no significant difference.
To understand the internal structure of the superior stalk-strength materials, we carried out anatomical observations of stalk cross-sections of the three materials collected at the kernel dent stage. Stalk sections of each material were stained with phloroglucinol, and then the internal structural characteristics of the stalk were observed under a microscope (Figure 3A). All the three materials had normal and similar organizational structure, with well-developed secondary cell walls (Figure 3A). The rind thickness of J2416 was significantly greater than that of 5237, but no significant difference with J24 (Figure 3B). The sclerenchyma thickness varied substantially among the three materials. The sclerenchyma thickness of J2416 was significantly greater than those of J24 and 5237. The sclerenchyma thickness of J24 was also significantly greater than that of 5237 (Figure 3B).
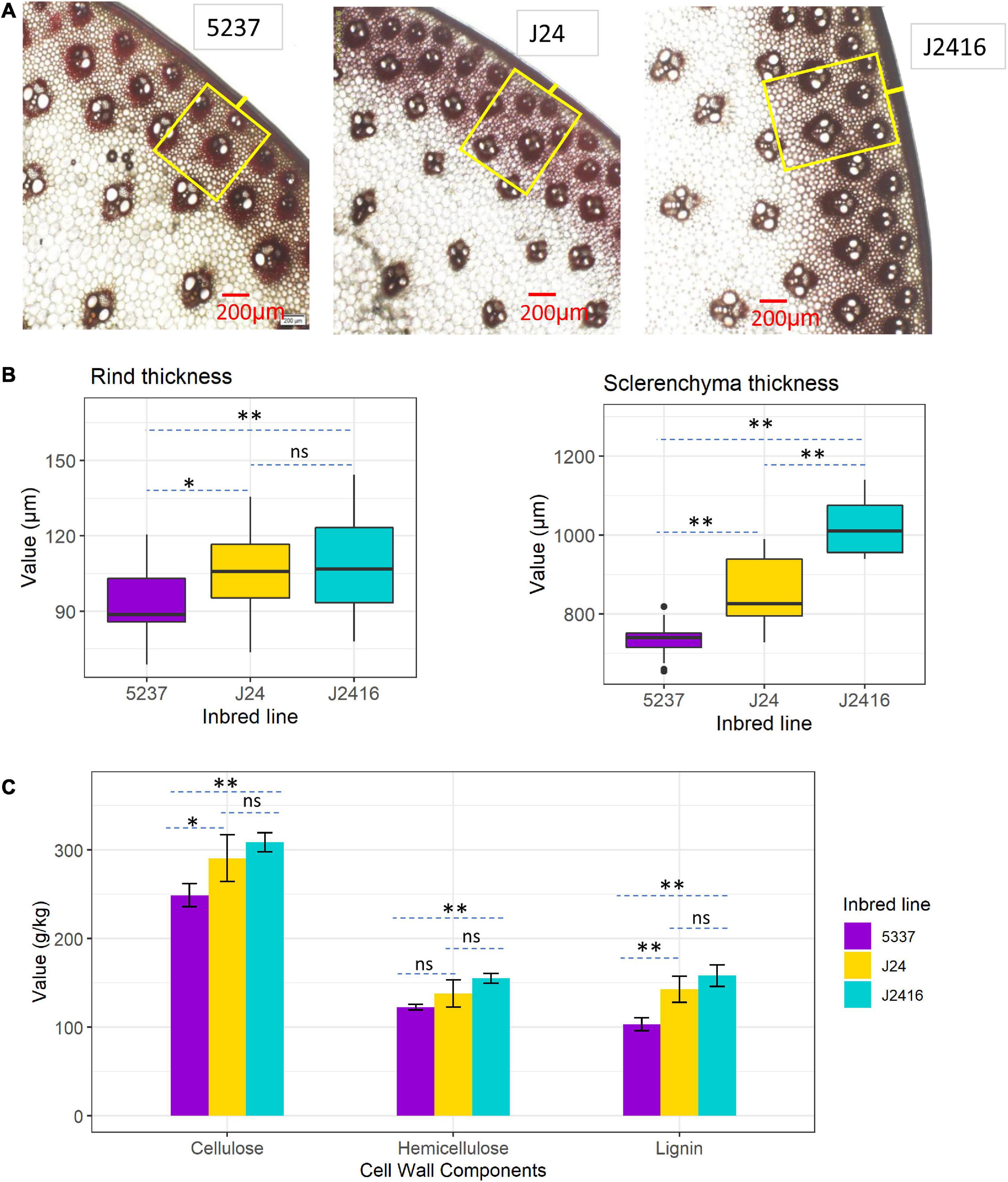
Figure 3. Stalk anatomical characteristics and cell wall components in J2416 and its two parents. (A) Anatomical characteristics observed in stalk cross sections (200 μm field) under a microscope. Yellow box represents sclerenchyma tissue; short yellow line in the middle of the box represents rind cells. (B) Rind thickness and sclerenchyma thickness in stalks of the three materials. (C) Contents of cell wall components in stalks of J2416 and its two parents. *, **Means the significant difference at 0.05 and 0.01 level, respectively. ns means no significant difference.
The contents of cellulose, hemicellulose and lignin in the stalk were determined by HPLC. All the contents of the three substances were significantly different between J2416 and 5237, but no significant difference between J2416 and J24 (Figure 3C). In addition, cellulose and lignin were also significantly different between J24 and 5237. The results of cell wall components together with stalk anatomic feature indicated that there were minor differences between J2416 and J24.
To explore the mechanisms of the differences in physiological characteristics of the stalks among the three inbred lines, we detected metabolites using LC-MS/MS technology. We detected 714 and 1082 metabolites in the positive and negative ion modes, respectively. Next, we divided the three materials into two comparison groups (group 1, J2416 vs. 5237; group 2, J2416 vs. J24) to detect differentially accumulated metabolites.
We detected 309 differentially accumulated metabolites between J2416 and 5237, consisting of 179 and 130 detected in the positive and negative ion modes, respectively (Figure 4A). In total, 212 metabolites (68.60%) were up-regulated in J2416. In order to obtain the specific biological functions and the metabolic networks of 212 metabolites, we used KOBAS2.0 software to annotate these metabolites into the KEGG database (Kanehisa et al., 2019). However, as the limitation of this database, only 30 metabolites were annotated into 16 KEGG pathways (Figure 4B). The number of up-regulated metabolites in each of these KEGG pathways varied from one to four. The phenylpropanoid pathway contained the largest number of differentially accumulated metabolites (four), followed by the glycan synthesis and metabolism pathways, flavonoid biosynthesis, purine metabolism, and stilbenoid diarylheptanoid biosynthesis pathways (three metabolites each). The other pathways contained one or two differentially accumulated metabolites.
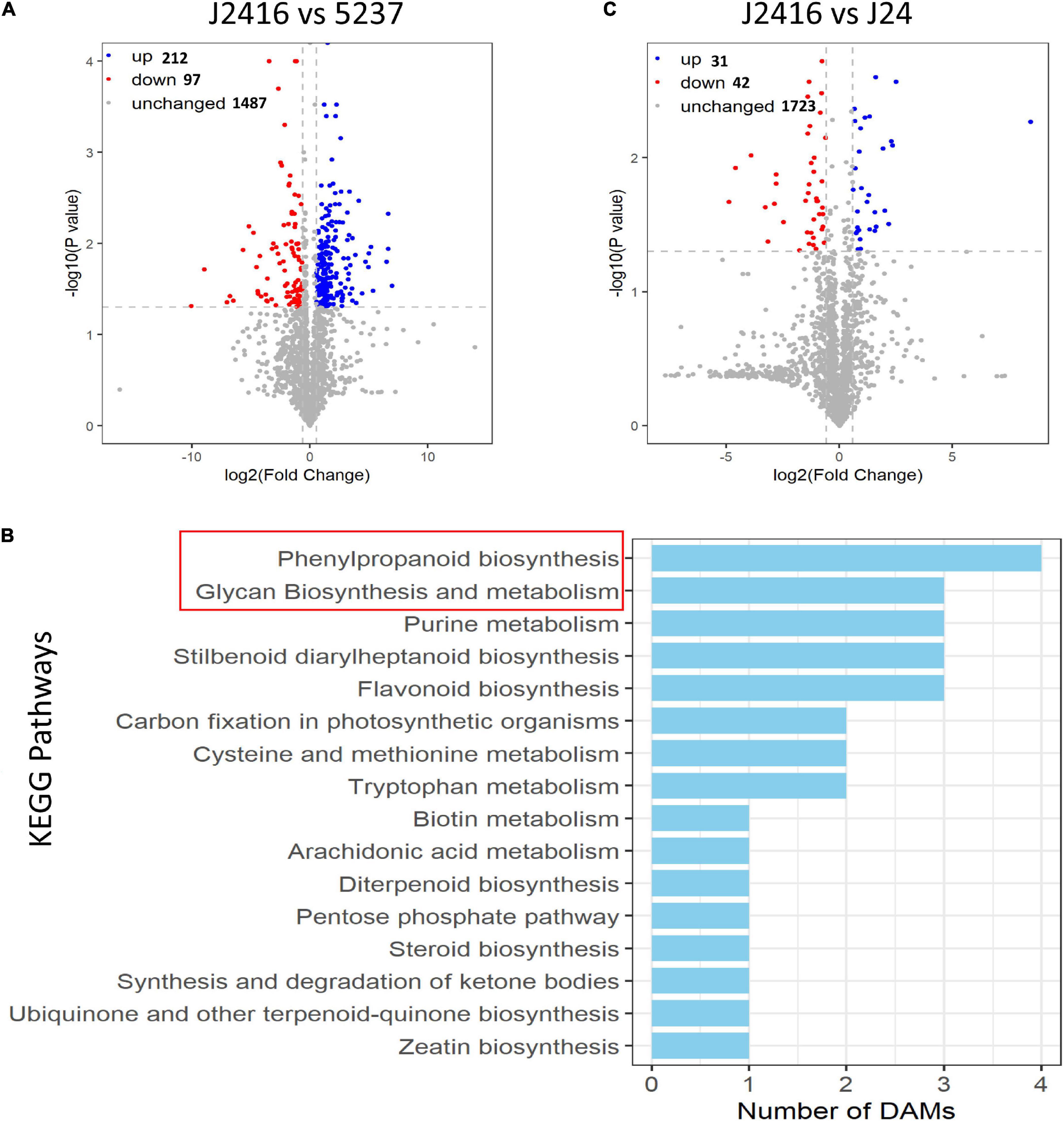
Figure 4. Differentially accumulated metabolites (DAMs). (A) Volcano plot of differentially accumulated metabolites between J2416 and 5237. Blue and red dots represent significantly up- and down-regulated metabolites, respectively, in J2416 compared with 5237. (B) KEGG pathways enriched with up-regulated metabolites in J2416. (C) (A) Volcano plot of differentially accumulated metabolites between J2416 and J24.
Only 73 differentially accumulated metabolites were detected between J2416 and J24, consisting of 51 and 22 detected in the positive and negative ion modes, respectively. Of these, 31 were up-regulated in J2416, and six of those were in the polysaccharide biosynthesis and metabolism pathways (Figure 4C).
According to the above phenotypes and metabolome analysis, we found there were few differences between J2416 and J24. Therefore, we only compared J2416 and 5237 to detect differentially expressed genes (DEGs).
We detected 5276 DEGs between J2416 and 5237, of which 2287 (43.34%) were up-regulated in J2416 (Figure 5A). Among the up-regulated genes in J2416, 375 were annotated in 72 KEGG networks, 11 of which matched with the pathways of differentially accumulated metabolites (Figure 5B). The number of up-regulated genes in each of these KEGG pathways varied from 1 to 16. Among them, the phenylpropanoid pathway was related to lignin metabolism, and the glycan pathway was related to cellulose and hemicellulose metabolism. As we found that cellulose, hemicellulose and lignin were significantly enriched in J2416, and these substances were reported to be related to stalk strength, we focused on the genes related to these three substances. By searching the MAIZEWALL database and other reports (Penning et al., 2009; Wang et al., 2021), we listed 28 candidate genes which were related to cellulose, hemicellulose and were up regulated in J2416 and (Table 1).
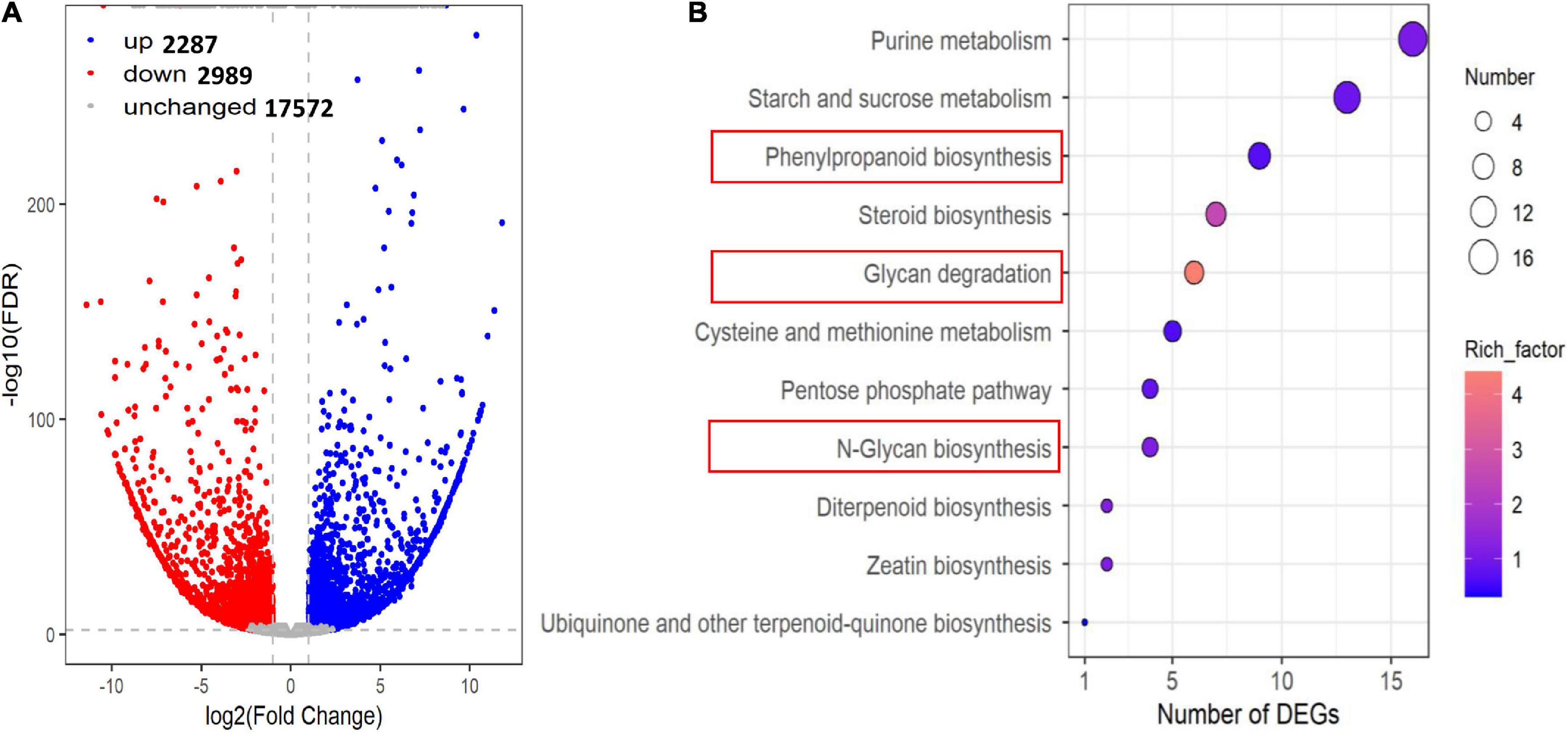
Figure 5. Differentially expressed genes (DEGs) between J2416 and 5237. (A) Volcano plot of DEGs. Blue and red dots represent significantly up- and down-regulated genes, respectively, in J2416 compared with 5237. (B) KEGG pathways enriched with up-regulated genes in J2416.
Three of the up-regulated genes in J2416 were related to cellulose content, and their -fold change values in J2416 ranged from 1.86 to 5.42. The genes were found to encode cellulose synthase, chitinase, and COBRA-like protein 4 (bk2), which was first identified from a brittle stalk mutant (Sindhu et al., 2007). The functions of other genes were related to cellulose synthesis and stalk lodging resistance (Table 1).
Sixteen of the up-regulated genes in J2416 were related to hemicellulose, and their fold change values ranged from 1 to 8.2. These genes encoded glycosyltransferase family members, UDP-glycosyltransferase, and xyloglucan endotransglucosylase. Zm00001d021755 encoding UDP-glycosyltransferase 88A1 was previously identified from a comparison of lodging-resistant and lodging-sensitive accessions (Guo et al., 2021). The other genes encoded products with roles in hemicellulose synthesis or stalk lodging resistance (Table 1).
The phenylpropanoid pathway which involved in the biosynthesis and degradation of lignin contained nine up-regulated genes with their fold change values in J2416 ranging from 1.19 to 4.25 (Table 1). These genes encoded multiple key enzymes in lignin metabolism, such as phenylalanine ammonia lyase2 (PAL2), cinnamyl alcohol dehydrogenase 6 (CAD6), peroxidase 53, and hydroxy cinnamoyl transferase (Table 1).
To validate the expression profiles of the genes in Table 1, five DEGs were randomly selected for RT-qPCR (Figure 6). From the results of RT-qPCR of these genes, we can see the expression level of J2416 were significantly higher than that of 5237. The concordance between RT-qPCR and RNA-Seq results confirmed that the findings from RNA-Seq were credible.
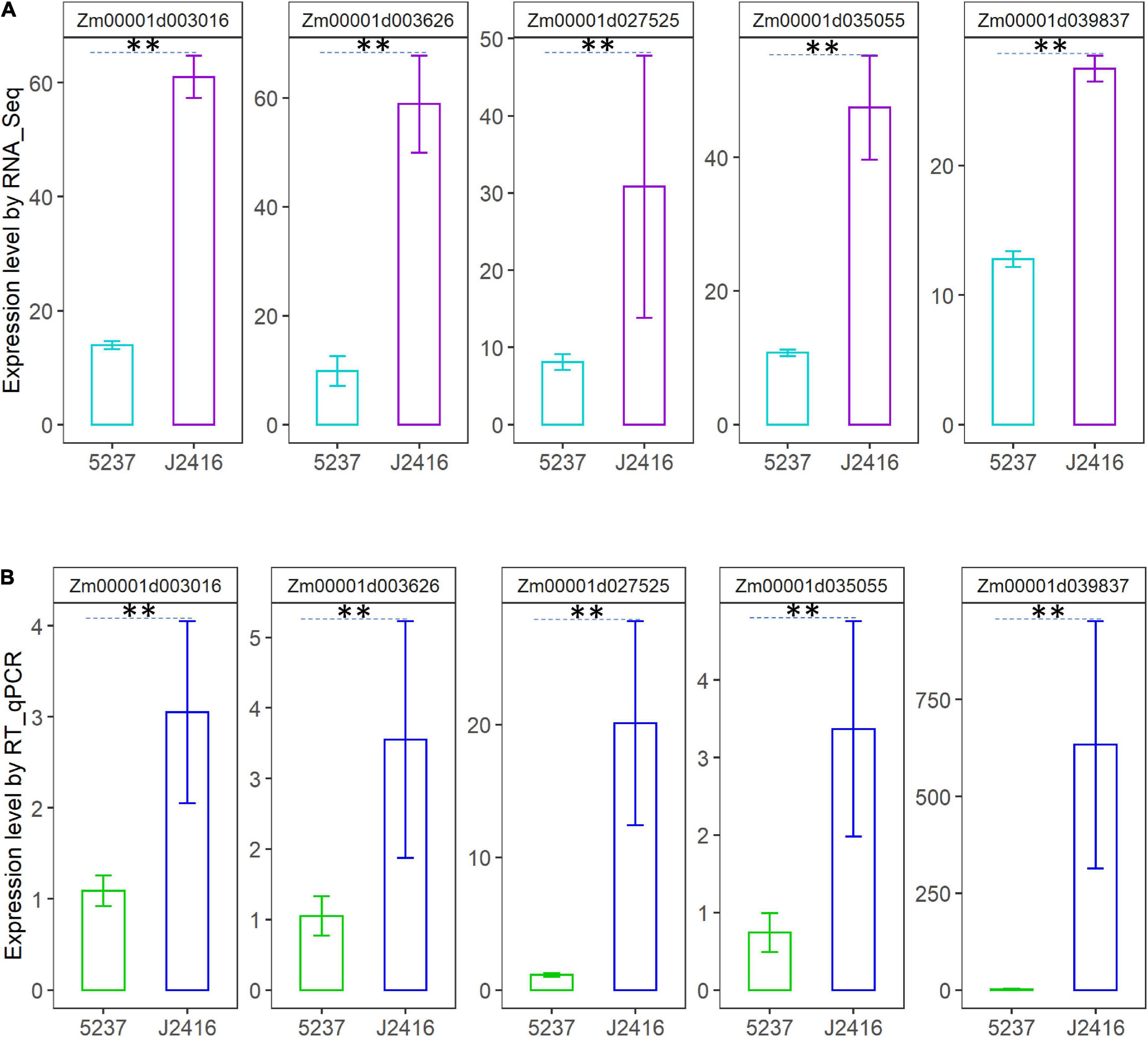
Figure 6. Expression levels of five randomly selected genes related to cellulose, hemicellulose and lignin in J2416 and 5237. (A) Is the expression level detected by RNA_Seq. (B) Is the expression level detected by RT-qPCR. **Means the significant difference at 0.01 level.
The field practice showed that J2416 and many cultivars bred of J2416 have good lodging resistance. In order to reveal the reason of the excellent stalk strength of J2416, we analyzed the stalk strength and related traits of J2416 and its parents (J24 and 5237). The results showed that the stalk strength of J2416 was significantly greater than that of 5237 in multiple environments, but just slightly higher than J24. Differential accumulated metabolites were also very few between J2416 and J24. The main reason was the bred process and the genetic background. From the pedigree, J2416 was bred from the cross of J24 and 5237 and the F1 backcrossed with J24 (Figure 1). The proportions of J2416 retained nearly 80.96% and 19.04% of identity by descend (IBD) segments from J24 and 5237, respectively, suggested the J24 had a greater genetic contribution to J2416 (Zhao et al., 2020b). Therefore, we hold the view that the stalk strength characteristics of J2416 inherited and better than the characteristics of J24.
Stalk strength is a mechanical characteristic that results from the contributions of multiple factors, including the internal structure of stalk and its chemical composition. In this study, we analyzed the inbred line J2416 with excellent stalk strength and its parents to determine which phenotypic, biochemical, and genetic factors are related to stalk strength.
We found that the stalk strength of J2416 was positively correlated with the rind thickness and sclerenchyma cell thickness. The main components of the cell wall are cellulose, hemicellulose, and lignin, so we quantified each of these components in the stalks of the three inbred lines. The contents of cellulose, hemicellulose, and lignin in J2416 were slightly higher than those in J24 and significantly higher than those in 5237. This result was consistent with the analyses of stalk mechanical strength and stalk anatomic structure, indicating that these three substances play vital roles in the stalk strength of J2416. Many studies have detected strong correlations between the mechanical strength of the stalk and the contents of cellulose, hemicellulose, and lignin (Vignols et al., 1995; Halpin et al., 1998; Sindhu et al., 2007; Tang et al., 2014; Li et al., 2015; Xiong et al., 2019). For example, the weaker stalk strength of the maize brittle stalk mutant bk2 was found to be due to lower cellulose and hemicellulose contents resulting from a mutation in a cellulose-related gene (Sindhu et al., 2007). The brown midrib mutants bm1–bm5 were found to have mutations in genes encoding lignin biosynthetic enzymes, leading to decreased contents of total lignin or lignin monomers, or a change in the ratio of monomers, resulting in reduced stalk strength (Vignols et al., 1995; Halpin et al., 1998; Tang et al., 2014; Li et al., 2015; Xiong et al., 2019).
Previous studies have shown that genes and metabolites related to the phenylpropanoid pathway were expressed at significantly lower levels at the fractured position of the stalk than in other parts of the stalk (Wang et al., 2019). The phenylpropanoid pathway is responsible for lignin synthesis. This pathway begins with phenylalanine, which serves as the substrate for the synthesis of lignin monomers via the actions of phenylalanine ammonia lyase (PAL), cinnamic acid 4-hydroxylase (C4H), and 4-coumarate-coenzyme A ligase (4CL) (Weng and Chapple, 2010). In this study, we found that the phenylpropanoid pathway, followed by the glycan synthesis and metabolism pathways, were most enriched with up-regulated metabolites in J2416.
To identify genes related to the control of phenylpropanoid metabolism, we detected DEGs between J2416 and 5237 by analyzing transcriptomic data. Nine genes in the phenylpropanoid pathway were up-regulated in J2416, encoding enzymes including PAL2, CAD6, and peroxidase. The first step in the phenylpropanoid pathway is catalyzed by PAL (Yoon et al., 2015). Another study found that the lignin content in the pal1 pal2 double mutant was reduced in parallel with an increase in the ratio of the S unit to G unit of lignin monolignol (Rohde et al., 2004). The last step of monolignol biosynthesis is catalyzed by CAD, which converts hydroxyl-cinnamaldehydes into their corresponding alcohols (Hirano et al., 2012). Analyses of the midrib mutant bm1 revealed a mutation in the CAD gene that resulted in severely reduced CAD activity, which affected both the total amount of lignin and the structure of lignin monomers (Halpin et al., 1998). The main function of peroxidase is to oxidize or dehydrogenate free lignin monomers and couple them to form lignin polymers (Marjamaa et al., 2009).
Three genes related to cellulose and 16 related to hemicellulose were among the genes that were up-regulated in J2416 compared with 5237. Bk2 was first identified in a maize brittle stalk mutant, in which the mutation resulted in a 40% reduction in total cellulose (Sindhu et al., 2007). Cellulose synthase catalyzes the polymerization of UDP-glucose to form glucan chains. Mutations in this type of gene in Arabidopsis led to defects in cellulose synthesis (Zhang et al., 2011). Chitin is a high molecular weight polysaccharide polymer. Another maize brittle stalk mutant, bk4, was found to be due to the mutation of a gene encoding a chitinase-like protein, and resulted in significantly decreased p-coumaric acid, glucose, mannose, and cellulose contents in the stalk (Jiao et al., 2019). The 16 hemicellulose-like genes family can be found in MAIZEWALL database (Penning et al., 2009). Especially, the gene Zm00001d021755 encoding UDP-glycosyltransferase 88A1was also identified in another research with high lodging resistance (Guo et al., 2021). The genes identified in this study can be as the candidate genes related to stalk strength.
Three maize inbred lines with pedigree relationships but significant differences in stalk strength were analyzed to identify traits related to superior stalk strength at the cellular, physiological, metabolic and transcript levels. Our analyses revealed nine genes related to lignin content, three related to cellulose content, and sixteen related to hemicellulose content that were up-regulated in the line with the strongest stalks. High transcript levels of these genes in the line with excellent stalk strength resulted in increased total amounts of related metabolites and macromolecular polymers and thicker cell walls, resulting in a stiff stalk.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA843174.
XW and JZ conceived and designed the experiments. YC, XS, JL, and RZ carried out the experiments. XW, YC, WS, and RW analyzed the data. XW wrote the manuscript. YJ revised the manuscript. All authors have read and agreed to the published version of the manuscript.
This research was supported by the Youth Fund of Beijing Academy of Agriculture and Forestry Sciences (QNJJ202231 and QNJJ202208) and Beijing Scholars Program (BSP041).
We thank BMKCloud (www.biocloud.net) for the help in transcriptome data analysis. We also thank Jennifer Smith, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.957566/full#supplementary-material
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Chen, G., Fan, H. W., Mao, Z. W., and Liu, C. G. (2009). Breeding and application of new maize variety Jingdan 28. Bull. Agric. Sci. Technol. 6, 125–127.
Chiniquy, D., Sharma, V., Schultink, A., Baidoo, E. E., Rautengarten, C., Cheng, K., et al. (2012). XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc. Natl. Acad. Sci. U S A. 109, 17117–17122. doi: 10.1073/pnas.1202079109
Deshavath, N. N., Mohan, M., Veeranki, V. D., Goud, V. V., Pinnamaneni, S. R., and Benarjee, T. (2017). Dilute acid pretreatment of sorghum biomass to maximize the hemicellulose hydrolysis with minimized levels of fermentative inhibitors for bioethanol production. 3 Biotech. 7:139. doi: 10.1007/s13205-017-0752-3
Du, J., Zhang, Y., Guo, X., Ma, L., Shao, M., Pan, X., et al. (2016). Micron-scale phenotyping quantification and three-dimensional microstructure reconstruction of vascular bundles within maize stalks based on micro-CT scanning. Funct. Plant Biol. 44, 10–22. doi: 10.1071/FP16117
Florea, L., Song, L., and Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res 2:188. doi: 10.12688/f1000research.2-188.v1
Furukawa, K., Ichikawa, S., Nigorikawa, M., Sonoki, T., and Ito, Y. (2014). Enhanced production of reducing sugars from transgenic rice expressing exo-glucanase under the control of a senescence-inducible promoter. Transgenic Res. 23, 531–537. doi: 10.1007/s11248-014-9786-z
Gao, D., Sun, W., Wang, D., Dong, H., Zhang, R., and Yu, S. (2020). A xylan glucuronosyltransferase gene exhibits pleiotropic effects on cellular composition and leaf development in rice. Sci. Rep. 10:3726. doi: 10.1038/s41598-020-60593-3
Guo, Y., Hu, Y., Chen, H., Yan, P., Du, Q., Wang, Y., et al. (2021). Identification of traits and genes associated with lodging resistance in maize. Crop J. 9, 1408–1417. doi: 10.1016/j.cj.2021.01.002
Halpin, C., Holt, K., Chojecki, J., Oliver, D., Chabbert, B., Monties, B., et al. (1998). Brown-midrib maize (bm1)—a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 14, 545–553. doi: 10.1046/j.1365-313X.1998.00153.x
Hirano, K., Aya, K., Kondo, M., Okuno, A., Morinaka, Y., and Matsuoka, M. (2012). OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep. 31, 91–101. doi: 10.1007/s00299-011-1142-7
Jiao, S., Hazebroek, J. P., Chamberlin, M. A., Perkins, M., Sandhu, A. S., Gupta, R., et al. (2019). Chitinase-like1 plays a role in stalk tensile strength in maize. Plant Physiol. 181, 1127–1147. doi: 10.1104/pp.19.00615
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K., and Tanabe, M. (2019). New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, 590–595. doi: 10.1093/nar/gky962
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kong, E., Liu, D., Guo, X., Yang, W., Sun, J., Li, X., et al. (2013). Anatomical and chemical characteristics associated with lodging resistance in wheat. Crop J. 1, 43–49. doi: 10.1016/j.cj.2013.07.012
Kotake, T., Aohara, T., Hirano, K., Sato, A., Kaneko, Y., Tsumuraya, Y., et al. (2011). Rice brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J. Exp. Bot. 62, 2053–2062. doi: 10.1093/jxb/erq395
Kwon, Y., Kim, S. H., Jung, M. S., Kim, M. S., Oh, J. E., Ju, H. W., et al. (2007). Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat, salt and drought stresses. Plant J. 49, 184–193. doi: 10.1111/j.1365-313X.2006.02950.x
Li, F., Xie, G., Huang, J., Zhang, R., Li, Y., Zhang, M., et al. (2017). OsCESA9 conserved-site mutation leads to largely enhanced plant lodging resistance and biomass enzymatic saccharification by reducing cellulose DP and crystallinity in rice. Plant Biotechnol. J. 15, 1093–1104. doi: 10.1111/pbi.12700
Li, K., Yan, J., Li, J., and Yang, X. (2014). Genetic architecture of rind penetrometer resistance in two maize recombinant inbred line populations. BMC Plant Biol. 14:152. doi: 10.1186/1471-2229-14-152
Li, L., Hill-Skinner, S., Liu, S., Beuchle, D., Tang, H. M., Yeh, C. T., et al. (2015). The maize brown midrib4 (bm4) gene encodes a functional folylpolyglutamate synthase. Plant J. 81, 493–504. doi: 10.1111/tpj.12745
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marjamaa, K., Kukkola, E. M., and Fagerstedt, K. V. (2009). The role of xylem class III peroxidases in lignification. J. Exp. Bot. 60, 367–376. doi: 10.1093/jxb/ern278
Okuno, A., Hirano, K., Asano, K., Takase, W., Masuda, R., Morinaka, Y., et al. (2014). New approach to increasing rice lodging resistance and biomass yield through the use of high gibberellin producing varieties. PLoS One 9:e86870. doi: 10.1371/journal.pone.0086870
Penning, B. W., Hunter, C. T., Tayengwa, R., Eveland, A. L., Dugard, C. K., Olek, A. T., et al. (2009). Genetic resources for maize cell wall biology. Plant Physiol. 151, 1703–1728. doi: 10.1104/pp.109.136804
Qiu, X., and Hu, S. (2013). “Smart” materials based on cellulose: a review of the preparations, properties, and applications. Materials 6, 738–781. doi: 10.3390/ma6030738
Rohde, A., Morreel, K., Ralph, J., Goeminne, G., Hostyn, V., De Rycke, R., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16, 2749–2771. doi: 10.1105/tpc.104.023705
Schomburg, D., and Salzmann, M. (1990). “Phospho-2-dehydro-3-deoxyheptonate aldolase,” in Enzyme Handbook 1, eds D. Schomburg and M. Salzmann (Berlin: Springer). doi: 10.1007/978-3-642-86605-0_74
Serrani-Yarce, J. C., Escamilla-Trevino, L., Barros, J., Gallego-Giraldo, L., Pu, Y., Ragauskas, A., et al. (2021). Targeting hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase for lignin modification in Brachypodium distachyon. Biotechnol. Biofuels 14:50. doi: 10.1186/s13068-021-01905-1
Shah, A. N., Tanveer, M., Abbas, A., Yildirim, M., Shah, A. A., Ahmad, M. I., et al. (2021). Combating dual challenges in maize under high planting density: stem lodging and kernel abortion. Front. Plant Sci. 12:699085. doi: 10.3389/fpls.2021.699085
Shah, A. N., Tanveer, M., Rehman, A. U., Anjum, S. A., Iqbal, J., and Ahmad, R. (2017). Lodging stress in cereal-effects and management: an overview. Environ. Sci. Pollut. Res. Int. 24, 5222–5237. doi: 10.1007/s11356-016-8237-1
Sindhu, A., Langewisch, T., Olek, A., Multani, D. S., McCann, M. C., Vermerris, W., et al. (2007). Maize brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol. 145, 1444–1459. doi: 10.1104/pp.107.102582
Sluiter, A., Hames, B., Ruiz, R., Scarlata, C., Sluiter, J., Templeton, D., et al. (2010). Determination of structural carbohydrates and lignin in biomass. Lab. Anal. Procedure 1617, 1–16.
Smith, C. A., O’Maille, G., Want, E. J., Qin, C., Trauger, S. A., Brandon, T. R., et al. (2005). METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751. doi: 10.1097/01.ftd.0000179845.53213.39
Song, G., Jin, M., Du, Y., Cao, L., and Xu, H. (2016). UPLC-QTOF-MS/MS based screening and identification of the metabolites in rat bile after oral administration of imperatorin. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1022, 21–29. doi: 10.1016/j.jchromb.2016.04.007
Stein, O., and Granot, D. (2019). An overview of sucrose synthases in plants. Front. Plant Sci. 10:95. doi: 10.3389/fpls.2019.00095
Tang, H. M., Liu, S., Hill-Skinner, S., Wu, W., Reed, D., Yeh, C. T., et al. (2014). The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. 77, 380–392. doi: 10.1111/tpj.12394
Vignols, F., Rigau, J., Torres, M. A., Capellades, M., and Puigdomènech, P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7, 407–416. doi: 10.1105/tpc.7.4.407
Wang, X., Shi, Z., Zhang, R., Sun, X., Wang, J., Wang, S., et al. (2020). Stalk architecture, cell wall composition, and QTL underlying high stalk flexibility for improved lodging resistance in maize. BMC Plant Biol. 20:515. doi: 10.1186/s12870-020-02728-2
Wang, X., Zhang, R., Shi, Z., Zhang, Y., Sun, X., Ji, Y., et al. (2019). Multi-omics analysis of the development and fracture resistance for maize internode. Sci. Rep. 9:8183. doi: 10.1038/s41598-019-44690-6
Wang, X. Q., Song, W., Zhang, R. Y., Chen, Y. N., Sun, X., and Zhang, J. R. (2021). Genetic research advances on maize stalk lodging resistance. Sci. Agric. Sinic. 54, 2261–2272.
Wen, W., Gu, S., Xiao, B., Wang, C., Wang, J., Ma, L., et al. (2019). In situ evaluation of stalk lodging resistance for different maize (Zea mays L.) cultivars using a mobile wind machine. Plant Methods 15:96. doi: 10.1186/s13007-019-0481-1
Wen, W., Liu, H., Zhou, Y., Jin, M., Yang, N., Li, D., et al. (2016). Combining quantitative genetics approaches with regulatory network analysis to dissect the complex metabolism of the maize kernel. Plant Physiol. 170, 136–146. doi: 10.1104/pp.15.01444
Weng, J. K., and Chapple, C. (2010). The origin and evolution of lignin biosynthesis. New Phytol. 187, 273–285. doi: 10.1111/j.1469-8137.2010.03327.x
Wiklund, S., Johansson, E., Sjöström, L., Mellerowicz, E. J., Edlund, U., Shockcor, J. P., et al. (2008). Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 80, 115–122. doi: 10.1021/ac0713510
Wu, A. M., Hörnblad, E., Voxeur, A., Gerber, L., Rihouey, C., Lerouge, P., et al. (2010). Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 153, 542–554. doi: 10.1104/pp.110.154971
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, 316–322. doi: 10.1093/nar/gkr483
Xie, L., Wen, D., Wu, C., and Zhang, C. (2022). Transcriptome analysis reveals the mechanism of internode development affecting maize stalk strength. BMC Plant Biol. 22:49. doi: 10.1186/s12870-022-03435-w
Xiong, W., Wu, Z., Liu, Y., Li, Y., Su, K., Bai, Z., et al. (2019). Mutation of 4-coumarate: coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol. Biofuels 12:82. doi: 10.1186/s13068-019-1421-z
Xu, C., Gao, Y., Tian, B., Ren, J., Meng, Q., and Wang, P. (2017). Effects of EDAH, a novel plant growth regulator, on mechanical strength, stalk vascular bundles and grain yield of summer maize at high densities. Field Crops Res. 200, 71–79. doi: 10.1016/j.fcr.2016.10.011
Xue, J., Gou, L., Zhao, Y., Yao, M., Yao, H., Tian, J., et al. (2016). Effects of light intensity within the canopy on maize lodging. Field Crops Res. 188, 133–141. doi: 10.1016/j.fcr.2016.01.003
Xue, J., Li, L., Xie, R. Z., Wang, K. R., Hou, P., Ming, B., et al. (2018). Effect of lodging on maize grain losing and harvest efficiency in mechanical grain harvest. Acta Agron Sinic. 44, 1774–1781. doi: 10.3724/SP.J.1006.2018.01774
Yoon, J., Choi, H., and An, G. (2015). Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant Biol. 57, 902–912. doi: 10.1111/jipb.12422
Zhang, B., Liu, X., Qian, Q., Liu, L., Dong, G., Xiong, G., et al. (2011). Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc. Natl. Acad. Sci. U S A. 108, 5110–5115. doi: 10.1073/pnas.1016144108
Zhang, D., Wu, S., An, X., Xie, K., Dong, Z., Zhou, Y., et al. (2018). Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 16, 459–471. doi: 10.1111/pbi.12786
Zhang, Z., Zhang, X., Lin, Z., Wang, J., Liu, H., Zhou, L., et al. (2020). A large transposon insertion in the stiff1 promoter increases stalk strength in maize. Plant Cell 32, 152–165. doi: 10.1105/tpc.19.00486
Zhao, J. R., Li, C. H., Song, W., Liu, X. X., Wang, Y. D., Zhang, R. Y., et al. (2020a). Heterosis and genetic recombination dissection of maize key inbred line Jing2416. Sci. Agric. Sinic. 53, 4527–4536.
Zhao, J. R., Wang, Y. D., Song, W., Zhang, R. Y., Li, C. H., and Liu, X. X. (2020b). Breeding and application of maize founder inbred line Jing2416. J. Plant Genet. Res. 21, 1051–1057.
Zhao, X., and Zhou, S. L. (2022). Research progress on traits and assessment methods of stalk lodging resistance in maize. Acta Agron Sinic. 48, 15–26.
Keywords: maize, stalk strength, cell wall, phenylpropanoid, cellulose, hemicellulose, differentially expressed genes
Citation: Wang X, Chen Y, Sun X, Li J, Zhang R, Jiao Y, Wang R, Song W and Zhao J (2022) Characteristics and candidate genes associated with excellent stalk strength in maize (Zea mays L.). Front. Plant Sci. 13:957566. doi: 10.3389/fpls.2022.957566
Received: 01 June 2022; Accepted: 11 July 2022;
Published: 28 July 2022.
Edited by:
Peng Hou, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Xuehai Zhang, Henan Agricultural University, ChinaCopyright © 2022 Wang, Chen, Sun, Li, Zhang, Jiao, Wang, Song and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Song, c29uZ3dlaTEwMDdAMTI2LmNvbQ==; Jiuran Zhao, bWFpemV6aGFvQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.