- College of Forestry, Co-Innovation Center for the Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
Protein S-acylation, also known as palmitoylation, is an important lipid post-translational modification of proteins in eukaryotes. S-acylation plays critical roles in a variety of protein functions involved in plant development and responses to abiotic and biotic stresses. The status of S-acylation on proteins is dynamic and reversible, which is catalyzed by protein S-acyltransferases (PATs) and reversed by acyl protein thioesterases. The cycle of S-acylation and de-S-acylation provides a molecular mechanism for membrane-associated proteins to undergo cycling and trafficking between different cell compartments and thus works as a switch to initiate or terminate particular signaling transductions on the membrane surface. In plants, thousands of proteins have been identified to be S-acylated through proteomics. Many S-acylated proteins and quite a few PAT-substrate pairs have been functionally characterized. A recently characterized acyl protein thioesterases family, ABAPT family proteins in Arabidopsis, has provided new insights into the de-S-acylation process. However, our understanding of the regulatory mechanisms controlling the S-acylation and de-S-acylation process is surprisingly incomplete. In this review, we discuss how protein S-acylation level is regulated with the focus on catalyzing enzymes in plants. We also propose the challenges and potential developments for the understanding of the regulatory mechanisms controlling protein S-acylation in plants.
Introduction
Protein S-acylation, referring to the addition of 16-carbon palmitate (predominant form) or 18-carbon stearate to specific cysteine residues via a thioester bond, has attracted much attention due to its exclusively reversible feature among lipid modifications and its vital roles in a variety of biological processes (Smotrys and Linder, 2004; Running, 2014; Li and Qi, 2017; Zheng et al., 2019; Hu and Ye, 2022). The status of S-acylation on proteins is dynamically changed, which is catalyzed by protein acyltransferases (PATs) and de-S-acylation enzymes, such as acyl-protein thioesterases (APTs) and recently reported α/α Hydrolase domain-containing Protein 17-like acyl protein thioesterases (ABAPTs; Liu et al., 2021). In plants, owing to the application of approaches for studying S-acylation and remarkable advances in high-resolution proteomics, enormous progress has been made in understanding S-acylation in the last two decades. Thousands of proteins have been identified as potential S-acylated proteins involved in vesicle trafficking, signal transduction, primary and secondary metabolism, and stress responses (Hemsley et al., 2013; Srivastava et al., 2016; Zhou et al., 2020; Kumar et al., 2022). S-acylated proteins and their functions have been well studied and nicely reviewed elsewhere (Hemsley et al., 2008; Li and Qi, 2017; Turnbull and Hemsley, 2017; Zheng et al., 2019). In this paper, we will review the current progress of S-acylation studies from the standpoint of the regulatory mechanisms of the protein S-acylation and de-S-acylation process with a focus on their catalyzing enzymes in plants.
Protein sequence requirements for S-acylation
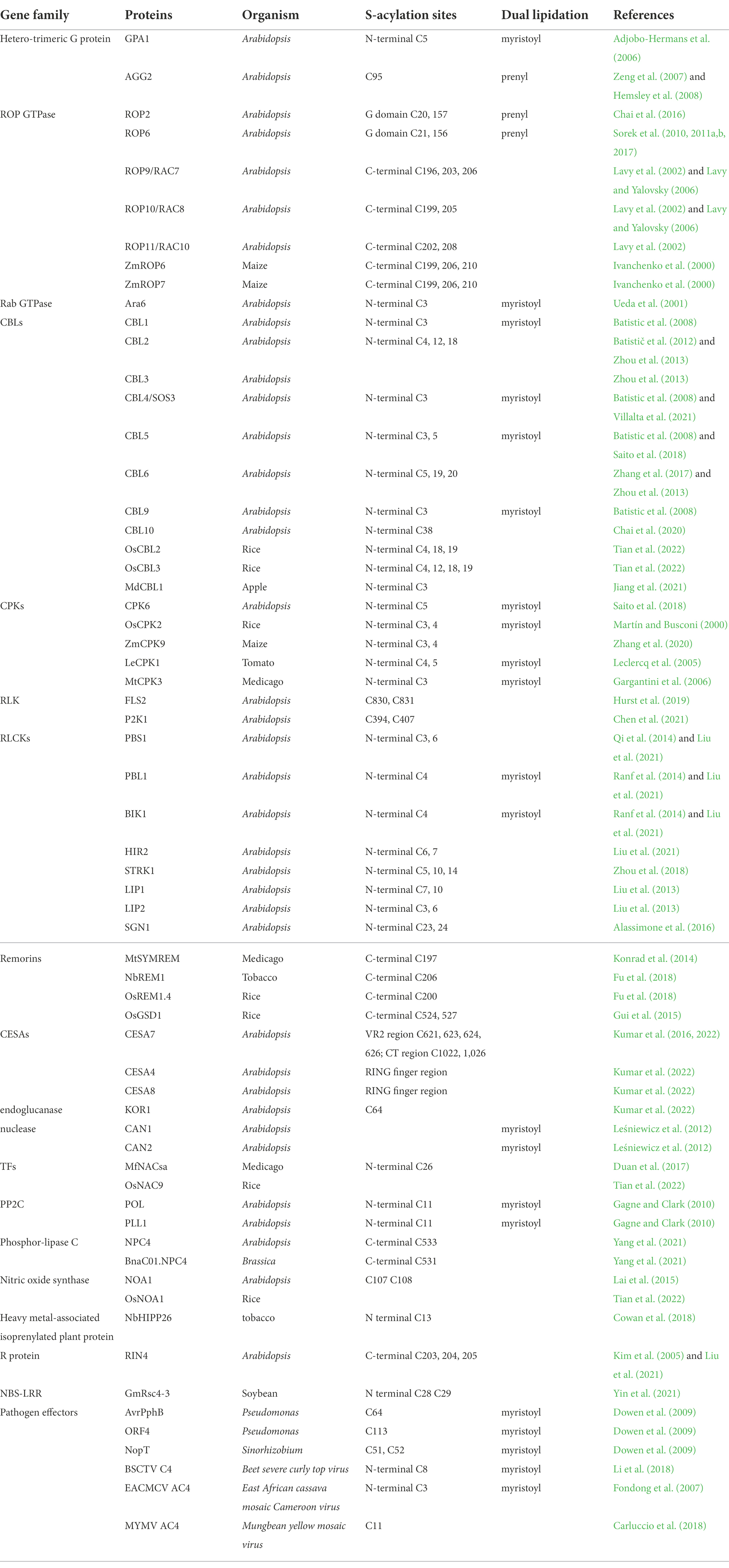
S-acylation is a widespread modification found in a variety of protein families. Functionally characterized S-acylated proteins are summarized in Table 1, including heterotrimeric G proteins, small G proteins Rho of Plants (ROPs), Calcineurin B-like proteins (CBLs), calcium-dependent protein kinases (CPKs), receptor-like kinases (RLKs), receptor-like cytoplasmic kinases (RLCKs), remorins, cellulose synthase subunits (CESAs), transcription factors (TFs), protein phosphatase type 2C proteins (PP2C), etc. Among these proteins, the S-acylation sites have been validated and functionally studied by point mutagenesis. In addition to these listed S-acylation sites, there may exist other uncharacterized modified sites in these proteins. Except for the requisite cysteines, no well-defined conserved motifs have been found for this modification so far. Nevertheless, certain motifs and some other post-translational modifications (PTMs) are required for the S-acylation of particular proteins.
Other lipid modification, either N-myristoylation or prenylation, frequently occurs together with S-acylation, and even acts as a prerequisite for S-acylation. In most reports, S-acylation itself or in combination with other lipidations are required for the correct trafficking and membrane targeting of the modified proteins; mutations at either the S-acylation site, the other lipidation site, or the dual lipidation sites show incorrect subcellular localization and compromised functions of the protein (Adjobo-Hermans et al., 2006; Batistic et al., 2008; Li and Qi, 2017; Hemsley, 2020).
Unlike S-acylation, N-myristoylation and prenylation are catalyzed by their enzymes on relatively conserved sequences. N-myristoylation refers to the addition of 14-carbon myristoyl groups to N-terminal glycine residues of proteins that harbor an MGXXXS/T motif. In contrast, prenylation modifies target proteins in the C-terminus by adding a 15-carbon farnesyl or a 20-carbon geranylgeranyl group to the cysteine of the C-terminal ‘CaaX’ box (Sorek et al., 2009; Running, 2014; Hemsley, 2015). As shown in Table 1, normally, S-acylation sites occur proximal to the N-myristoylation site when dual lipidation occurs, which is not applicable to the prenylation-S-acylation combination. Proteins from a gene family/subfamily are likely subjected to the same dual lipidation types due to the sequence and functional conservations (Table 2).
N-myristoylation-S-acylation combination has been found in the Gα subunit GPA1 in the heterotrimeric G proteins, Rab GTPase ARA6, CBLs, CPKs, RLCKs, staphylococcal-like nucleases, PP2Cs, and some of the pathogen effectors (Ueda et al., 2001; Adjobo-Hermans et al., 2006; Batistic et al., 2008; Dowen et al., 2009; Gagne and Clark, 2010; Leśniewicz et al., 2012; Liu et al., 2013; Saito et al., 2018). The S-acylation sites are always located adjacent to or nearby the N-myristoylated glycine (G2 site) at the cysteine residues from C3 to C11. This rule also applies to the bacterial type III effectors, including AvrPphB, ORF4, and NopT, after undergoing auto-proteolysis on specific sites (Dowen et al., 2009). N-myristoylation and S-acylation of calcium signaling participants, CBLs and CPKs, have been well studied. CBL1, 4, 5, and 9, which undergo both N-myristoylation and S-acylation, are localized on the plasma membrane (PM), whereas CBL2, 3, 6, and 10, containing only S-acylation modification at the N-terminal sites, are found on the tonoplast (Zhou et al., 2013; Saito et al., 2018; Chai et al., 2020; Villalta et al., 2021). Mutations at any of these sites significantly diminish the protein localization. It seems that S-acylation sites directly affect the membrane localization of CBLs, and myristoylation appears to direct their targeting to the PM or tonoplast. The case of CBLs is consistent with the prevailing model that the major function of myristoylation is to assist in the targeting of the modified protein to the membrane, and the addition of S-acylation can serve to stabilize the membrane association of a myristoylated protein (Farazi et al., 2001; Resh, 2013). However, many exceptions exist. PBS1 contains predicted N-terminal myristoylation and S-acylation signal. However, the myristoylation of PBS1 was not detected experimentally and mutation of the G2 site does not affect the protein localization on PM, indicating that the N-terminal S-acylation of itself is sufficient for PM localization of PBS1, which is independent of myristoylation (Dowen et al., 2009; Qi et al., 2014). Rab GTPase protein Ara6 is modified by both N-myristoylation and S-acylation. The Ara6G2A mutant was not S-acylated, indicating that myristoylation of Ara6 is a prerequisite for S-acylation (Ueda et al., 2001). Pathogens employ the host lipidation machinery to modify the effectors and thus anchor them on plant cellular PM for further avirulence activities. It has been demonstrated that the self-proteolysis process next to the GDK motif of AvrPphB, the VER motif of ORF4, and the DKM motif of NopT, exposes their respective embedded sites for fatty acylation. Therefore, these motifs and the auto-proteolysis are a prerequisite for dual lipidations and the acylation-dependent PM localization (Dowen et al., 2009).
Prenylation-S-acylation combination has been well characterized in Gγ subunit AGG2 in the heterotrimeric G proteins and type-I ROPs. S-acylation sites of ROPs are uniformly located at the protein C-terminus, but the two subfamilies undergo different mechanisms of membrane targeting. Type-I ROPs, including ROP1-ROP8, which terminate with a conserved CaaL box and a polybasic region, undergo prenylation and S-acylation on the G domain (Sorek et al., 2010, 2011a,b, 2017). PM localization of Type-II ROPs, including ROP9, ROP10, and ROP11, requires S-acylation on two or three C-terminal cysteines on the GC-CG box and is prenylation independent, even though ROP9 contains a CaaX box and could be prenylated in yeast (Lavy et al., 2002). Interestingly, heterotrimeric G-protein subunits undergo different lipidations. Gα subunit GPA1 is both myristoylated and S-acylated (Adjobo-Hermans et al., 2006); Gγ subunit AGG1 is prenylated while AGG2 is simultaneously prenylated and S-acylated. Moreover, polybasic residues also contribute to the efficient membrane-targeting of AGG2 (Zeng et al., 2007).
Dual lipidation or multiple adjacent S-acylations may enhance membrane attachment or trafficking of proteins by creating a more hydrophobic protein structure. Consistent with this assumption, consecutive or adjacent modification of S-acylation are commonly found in S-acylated proteins, especially for soluble proteins, e.g., double/triple/quad S-acylation sites in ROPs, CBLs, some RLCKs, CESAs, and triple S-acylation sites in the C-terminal domain of RIN4 (Lavy et al., 2002; Kim et al., 2005; Zhou et al., 2018; Alam et al., 2021; Tian et al., 2022).
Based on the functionally characterized sites, unlike myristoylation and prenylation, S-acylation modification is not restricted to the protein ends. The proteomics data strongly support it. A most recent acyl-RAC-based site-specific S-acylproteome analysis for Arabidopsis identified 1849 putative S-acylated cysteine sites from 1,094 proteins with high- and medium-confidence, representing around 6% of the detectable Arabidopsis proteome. These putative S-acylated proteins have diverse molecular functions and are from almost all cellular compartments including soluble and transmembrane proteins (Kumar et al., 2022). Cellulose synthase A proteins, including CESA4, CESA7, and CESA8, are S-acylated in both the centrally located variable region 2 (VR2) and the carboxy-terminal domain (Kumar et al., 2016, 2022). Several S-acylation sites have been found in both the kinase domain and non-kinase domains of BSK proteins, which belong to the RLCK family (Ren et al., 2019; Kumar et al., 2022). For RLKs, S-acylation occurs in the kinase domain, at the juxta-membrane sites in the intracellular domain, or in the extracellular domain (Kumar et al., 2022).
Protein S-acyltransferases and their substrates in plants
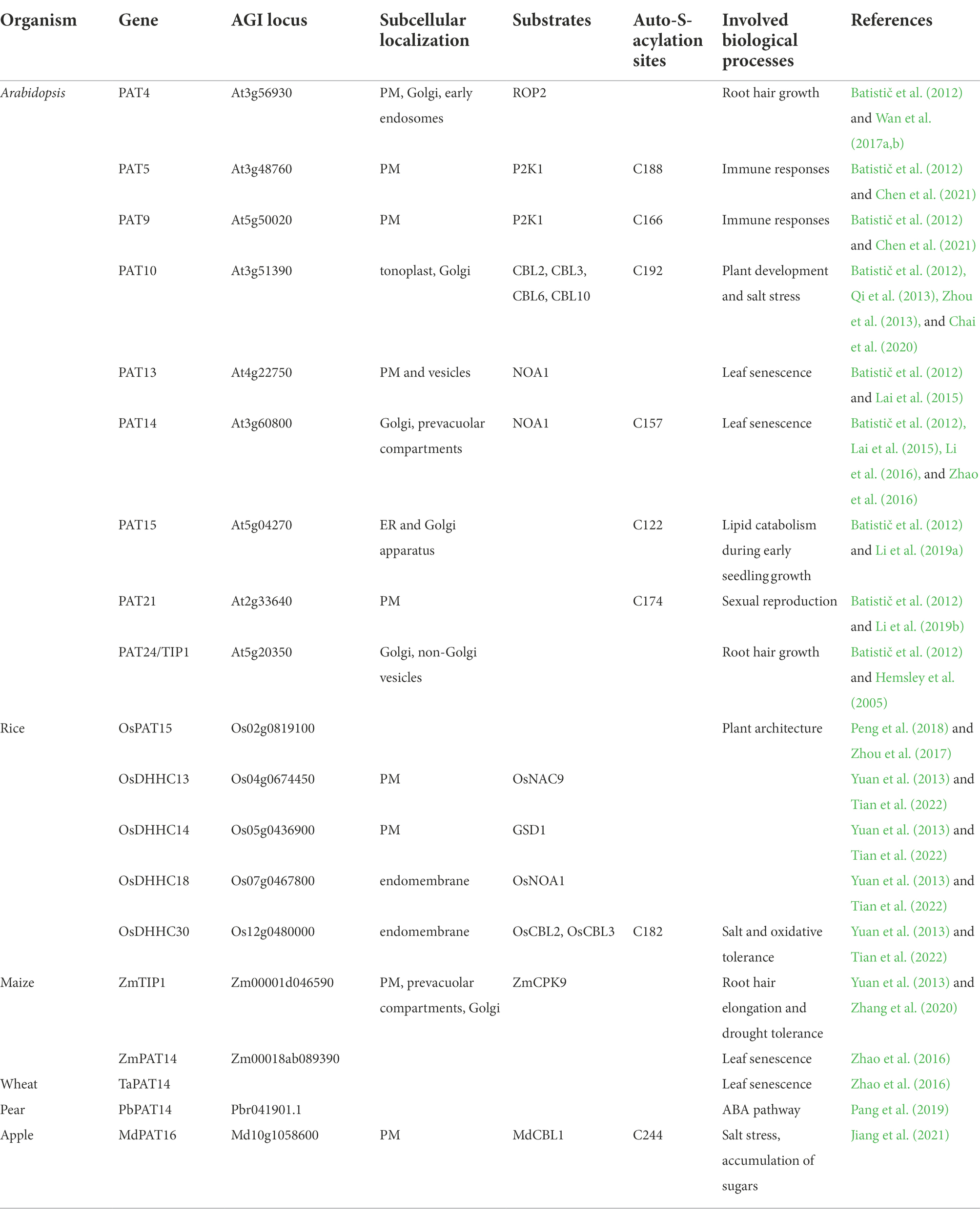
Protein S-acyltransferase (PAT) family proteins, featured by a highly conserved Asp-His-His-Cys cysteine-rich domain (DHHC-CRD), are responsible for protein S-acylation. Plant PATs are transmembrane proteins with 3–6 membrane-spanning domains and possess variable N-and C-terminal domains that might be essential for their substrate specificity (Smotrys and Linder, 2004; Batistič et al., 2012). Functions of PAT proteins from different organisms are highly conserved as evidenced by the complemented assay for the yeast akr mutant. Based on studies from the mammalian and yeast systems, the PATs catalyze target proteins through a two-step ping-pong mechanism: (1) the deprotonated and nucleophilic cysteine in the catalytic central DHHC tetrapeptide, undergoes auto-acylation and (2) transfer the acyl group to a cysteine residue on a target protein (Jennings and Linder, 2012; Zmuda and Chamberlain, 2020). Mutation analyses revealed that the cysteine residue of the DHHC domain is necessary for PAT auto-acylation and its enzyme activity (Peng et al., 2018; Li et al., 2019b; Chen et al., 2021; Jiang et al., 2021; Tian et al., 2022).
PATs in Arabidopsis
In Arabidopsis, the PAT family comprises 24 members (Batistic, 2012). Microarray data suggests that most of the Arabidopsis PATs display a broad and constant expression pattern at different developmental stages. Only a few PAT genes exhibited a specific transcription either in certain tissues and/or at a specific developmental stage. PATs are localized to a variety of intracellular membranes, including PM, ER, Golgi, tonoplast, and vesicles, indicating that proteins could possibly be S-acylated during the protein PTM and maturation processes on ER and Golgi, or during protein sorting and trafficking processes in Golgi and vesicles, or after arriving targeting compartments, such as PM and tonoplast (Batistič et al., 2012). Based on the subcellular localization, protein PTM time point, and the necessity for a functional protein, we propose that PATs localized on ER (PAT3, 15, 17, and 18), Golgi (PAT14, 15, 16, 23, and 24), and vesicles (PAT1, 2) may undertake most constant S-acylation modification that are necessary for protein maturation, sorting, and targeting; PATs localized on PM (PAT4, 5, 6, 7, 8, 9, 13, 19, 20, 21, and 22) and tonoplast (PAT10, 11, 12) may be responsible for local routine S-acylation processes and/or for protein S-acylation in response to external stimuli. Interestingly, about half of plant PATs reside on PM, indicating that PM might be one of the main locations for S-acylation occurrence and also suggesting the importance of S-acylation for the functions of PM proteins in plants. Since the number of S-acylated proteins far exceeds the number of PATs, PATs are likely to S-acylate multiple substrates (Hemsley, 2020). PATs may act redundantly due to their overlapping substrates.
Several PAT mutants from different plant species have been reported to show visible phenotypes and their substrates have been characterized. TIP1/PAT24 is the first reported PAT in plants, whose mutant tip1 shows growth defects including dwarf plants, smaller rosettes, and shorter root hair (Hemsley et al., 2005). TIP1 plays a key role in root hair growth polarity since tip1 mutants exhibited root hairs with multiple initiations and with branches (Hemsley et al., 2005; Zhang et al., 2015). Interestingly, TIP1 acts synergistically with PLURIPETALA (PLP), which involves in protein prenylation, during root hair growth (Chai et al., 2016). Besides TIP1, PAT4 also regulates root hair growth. PAT4 S-acylates ROP2 and facilitates the PM association of ROP2 at the root hair apex and thus regulates root hair growth (Wan et al., 2017a). In addition, PAT4 also functions in apex-associated re-positioning of the nucleus during root hair elongation (Wan et al., 2017b). PAT10 is critical for plant development and salt tolerance in Arabidopsis by regulating the tonoplast localization of several CBLs, including CBL2, CBL3, CBL6, and CBL10, whose membrane association depends on S-acylation (Qi et al., 2013; Zhou et al., 2013; Chai et al., 2020). PAT13 and PAT14 are involved in leaf senescence regulation by S-acylating chloroplast-localized Nitric Oxide Associated 1 (NOA1). pat13/pat14 double mutant displays a severely early leaf senescence phenotype (Lai et al., 2015). PAT13 is localized on PM and vesicles, PAT14 is localized on Golgi and vesicles, while NOA1 functions on chloroplast. It has been proposed that NOA1 is S-acylated by PAT13 and PAT14 during trafficking to chloroplasts by the endomembrane system (Lai et al., 2015). Interestingly, Li et al. (2016) and Zhao et al. (2016) discovered a different regulatory mechanism of PAT14 in leaf senescence regulation. They found that pat14 mutants were hypersensitive to salicylic acid (SA) and SA pathway-related genes were altered in pat14 mutants. Repressing SA biosynthesis or signaling by knock-out SA receptor NPR1 or SA hydroxylase NahG in pat14 can rescue its early senescence phenotype, demonstrating that AtPAT14 regulates senescence via SA pathways (Li et al., 2016; Zhao et al., 2016). In addition, PAT14 homologs from maize (ZmPAT14) and wheat (TaPAT14) fully rescued the precocious leaf senescence of Arabidopsis pat14, indicating the conservative role of PAT14 in regulating leaf senescence between dicots and monocots (Zhao et al., 2016). PAT5 and PAT9 are two of the few well-studied PATs in Arabidopsis (Chen et al., 2021). These two enzymes were shown to S-acylate the extracellular ATP receptor P2K1 at the PM to regulate the pathogen immune response in Arabidopsis. Interaction between PAT5, 9, and P2K1 is intricate and fine-tuned. PAT5 and PAT9 were activated by P2K1 phosphorylation, and activated PAT5, 9, in turn, impacted S-acylation level of P2K1. S-acylation of P2K1 plays an important role in mediating ATP-induced autophosphorylation and protein turnover, thus affecting the responses to extracellular ATP and pathogen infection (Chen et al., 2021). This study firstly reported the regulation of PAT activity by phosphorylation. Besides, functions of PAT15 and PAT21 have been studied, but their substrates have yet to be uncovered. PAT15 is involved in seed triacylglycerol catabolism during early seedling growth (Li et al., 2019a). AtPAT21 is essential for both male and female gametogenesis by participating in repairing SPO11-induced meiotic DNA double-stranded breaks (Li et al., 2019b).
PATs in other higher plants
In addition to Arabidopsis, the PAT gene family has also been found in other higher plants (Yuan et al., 2013). There are 30 PATs in rice (Oryza sativa L.). A quick screening of PATs of proven S-acylated proteins in rice through constructing an OsDHHC cDNA library and the BiFC assay has identified several PAT-substrate pairs OsDHHC13-OsNAC9, OsDHHC14-OsGSD1, OsDHHC18-OsNOA1, and OsDHHC30-OsCBL2, 3 (Tian et al., 2022). Further functional characterization of OsDHHC30-OsCBL2, 3 pairs has proven that OsDHHC30 determines the endomembrane association of OsCBL2 and OsCBL3 by S-acylation and contributes to salt tolerance in rice (Tian et al., 2022). OsPAT15, an alternatively spliced model of Os02g0819100 in rice, is a homolog of AtPAT15/AtDHHC1 and contains a zinc finger DHHC domain. OsPAT15 regulates plant architecture by altering the rice tiller (Zhou et al., 2017). Heterologous overexpression of the OsPAT15 gene into the dicot Brassica napus L. exhibited increased primary branches and increased seed yield (Peng et al., 2018). In B. napus L., 24 proteins have been predicted as PATs based on protein sequences. Except for BnPAT3, 8, 18, 19, 20, 21, and 22, all the other BnPATs contain the DHHC-CRD domain. No function studies have been done for these genes (Yuan et al., 2013; Peng et al., 2018). ZmTIP1, a homolog of AtTIP1/PAT24, was reported to positively regulate root hair length and drought tolerance in maize (Zea mays L.) through S-acylating its substrate ZmCPK9, which facilitates the association of ZmCPK9 with the PM (Zhang et al., 2020). MdPAT16, a PAT member in apple (Malus domestica), was shown to regulate plant resistance to salt stress and the accumulation of soluble sugars by S-acylating MdCBL1 protein. S-acylation of MdCBL1 at the N-terminal site enhances the protein stability and the PM localization (Jiang et al., 2021). PbPAT14, an S-acyltransferase gene from pear (Pyrus bretschneideri), has been functionally characterized. Knockout transgenic pear lines of PbPAT14 using CRISPR/Cas9 system exhibited yellow dwarf phenotype. Endogenous hormone measurement and marker gene expression analysis indicated that PbPAT14 function was related to the ABA pathway (Pang et al., 2019).
de-S-acylation enzymes and their substrates in plants
Protein de-S-acylation is the other half indispensable process in the S-acylation/de-S-acylation cycle, which modulates protein membrane localization and protein properties by removing the thioester-linked fatty acids from protein substrates (Figure 1). In mammals, the G proteins, Ras-family small GTPases, and N-Ras have established a distinct mode of membrane protein directionality depending on an S-acylation and de-S-acylation cycle (Duncan and Gilman, 1998; Won et al., 2018). The Protein de-S-acylation seems to occur everywhere in the cell, and no specific conserved sequence or biunique substrate specificity has been described for this enzymatic reaction so far, which is also reflected by the fact that thousands of proteins could be S-acylated, but only a few acyl protein thioesterases have been identified.
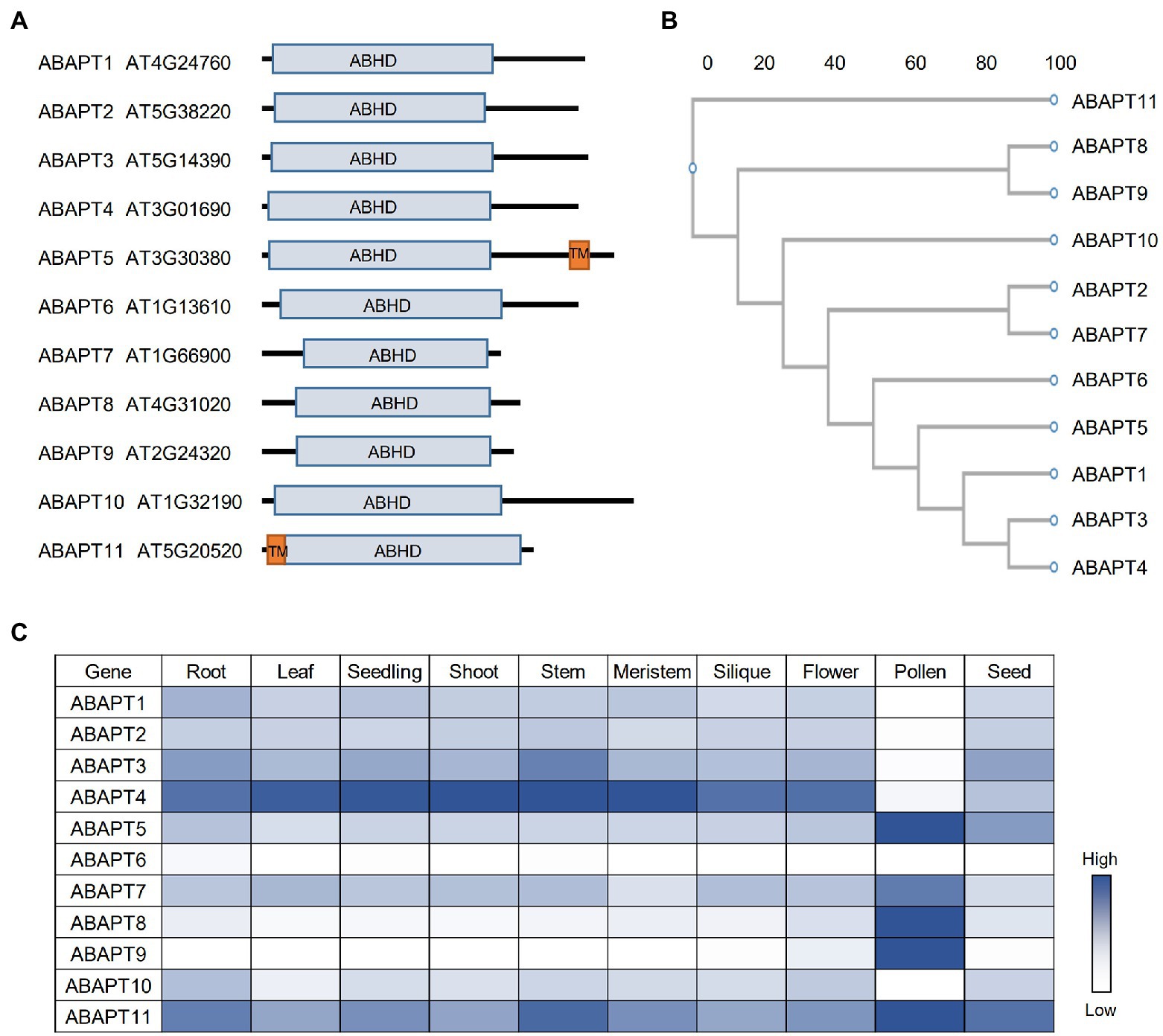
Figure 1. The ABAPT family in Arabidopsis. (A) Protein domain architecture of ABAPT family proteins. ABHD, alpha beta hydrolase domain; TM, transmembrane domain. (B) Phylogenetic tree of the Arabidopsis ABAPT family proteins. The phylogenic tree was conducted using protein sequences and the neighbor-joining method was used. Scale bar indicates genetic distance. (C) Expression profiles of all ABAPT genes in different tissues. The heat map shows the relative expression level of each gene. All the gene expression data were retrieved from Arabidopsis RNA-seq Database (http://ipf.sustech.edu.cn/pub/athrna/).
Compared to a significant number of studies in mammals, the de-S-acylated proteins and related enzymes have been rarely reported in plant cells. Unlike the plant PAT family genes, which are highly homologous to PATs from mammals and yeast, there are no homologs in plants with high similarity to mammalian APT1/2 (Hemsley, 2020). Nevertheless, MtAPT1, a putative homolog of human APT1 in Medicago falcata, is the first reported candidate acyl protein thioesterases in plant. MtAPT1 mediates the nuclear translocation of a NAC transcription factor MfNACsa in response to drought stress, which is suggested to be realized by its de-S-acylation (Duan et al., 2017). However, unlike mammalian APTs, MtAPT1 belongs to an acyl-ACP thioesterase from the Hotdog fold superfamily rather than the serine hydrolase superfamily. Further biochemical evidence is needed to support the role of MtAPT1 in de-S-acylation (Duan et al., 2017; Hemsley, 2020). Bürger et al. suggested ZmB6T1C9 be a bona fide APT member based on the phylogenetic and structural analysis of plant hydrolases. Due to the structural similarity to human APT2, ZmB6T1C9 was also assigned as ZmAPT2 (Bürger et al., 2017). However, the de-S-acylation activity, substrates, and biological functions of this enzyme have not been studied.
Recently, ABAPT family proteins (ABHD17-like acyl protein thioesterases, ABAPTs), which share a conserved ABHD region with mammalian ABHD17 proteins, were identified as putative protein acyl thioesterases in Arabidopsis (Liu et al., 2021). There are 11 members in the ABAPT family (Figure 1A), and all of them share a conserved ABHD protein domain with a serine, an aspartate, and a histidine residue, which are proved to be essential for ABDH17 de-S-acylation activity (Lin and Conibear, 2015). The de-S-acylation activity of ABAPTs was tested and the ABAPT-substrate pairs were identified using a robust screen system, in which five RFP-tagged plant immunity-related protein substrate candidates (RIN4, PBS1, PBL1, HIR2, and BIK1) were co-expressed with ABAPTs in protoplasts and the effects of ABAPTs on the subcellular localization and S-acylation level of these candidate substrates were examined. By this method, these S-acylated proteins were proved to undergo a de-S-acylation process mediated by ABAPTs (Liu et al., 2021). In addition, ABAPT8-RIN4/PBL1/BIK1, ABAPT7-HIR2, and ABAPT11-PBS1 catalytic pairs have been identified. A further functional study demonstrated that de-S-acylation of RIN4 mediated by ABAPT8 stimulated cell death, implying that ABAPT8 may function in the regulation of plant immunity responses (Liu et al., 2021). Although the ABHD region of ABAPTs is conserved with mammalian ABHD17 proteins, the other regions show very low levels of identity. The striking differences between ABAPTs and mammalian ABHD17 proteins suggest distinct mechanisms of the thioesterase activity regulation or/and substrate selectivity.
Among the ABAPT family, ABAPT5 and ABAPT11 harbor a transmembrane domain, while other ABAPTs do not (Figure 1A). Interestingly, all 11 ABAPTs could localize on the plasma membrane, which is probably implemented by their own S-acylation modification similar to their mammalian homologs. However, future biochemical evidence is needed to prove their S-acylation. Other than the membrane localization, the ABAPTs are also localized in the cytosol (Liu et al., 2021), reflecting the diverse subcellular localization of the detected S-acylated proteins (Hemsley et al., 2013; Kumar et al., 2022). In mammals, APT1 is even an active mitochondrial acyl thioesterases (Kathayat et al., 2018). It would be interesting to investigate the roles of ABAPTs in distinct cell organelles other than plasma membrane.
The spatial and temporal expression patterns of genes are very important for their cellular functions. In Arabidopsis, similar to PATs, the ABAPT expression could also be detected in different Arabidopsis tissues and developmental stages (Figure 1C). ABAPTs can be divided into three groups based on their expression patterns: (1) ABAPT5/7/8/9/11 are predominantly expressed in pollen; (2) ABAPT1/2/3/4/10 are constantly expressed in other tissues except pollen; and (3) ABAPT6 shows fairly low or undetectable expression in all tissues shown here. ABAPT11 was previously identified as Wavy Growth 2 (WAV2), which functions in root bending in response to environmental stimuli (Mochizuki et al., 2005). However, the function of WAV2 in pollen has not been studied. Interestingly, ABAPT5 and ABAPT11, the two putative transmembrane domain-containing proteins, are highly expressed in pollen. ABAPT1, ABAPT3, and ABAPT4 are clustered into one sub-branch in the phylogenetic tree (Figure 1B), and they display similar expression patterns, suggesting that they may act redundantly in plants. ABAPT8 and ABAPT89 are located in a sub-branch and also share similar expression patterns, which may suggest functional redundancy. However, they could not share protein substrates, at least for RIN4, PBL1, and BIK1 (Liu et al., 2021).
Conclusion and perspective
The reversible nature represents one of the most important aspects of protein S-acylation. Since the discovery of the first S-acyltransferase, Akr1 from yeast in 2002, research on protein S-acylation has been accelerated at a remarkable speed in both mammals and plants (Roth et al., 2002; Li and Qi, 2017; Zheng et al., 2019; Hemsley, 2020). However, the advancement of S-acylation studies in plants still lags far behind compared to that in mammals in almost every aspect, including S-acylation proteomics, functional and regulation study of S-acylation-related enzymes, as well as their corresponding substrates. The knowledge from other organisms sheds light on the plant S-acylation research, although there are still many unanswered questions.
The cycle of S-acylation and de-S-acylation provides a molecular mechanism for membrane-associated proteins to undergo cycling and trafficking between different cell compartments. In Medicago, MfNACsa translocation is possibly mediated by de-S-acylation through MtAPT1 (Duan et al., 2017). In Arabidopsis, RIN4, PBS1, PBL1, HIR2, and BIK1 also undergo both S-acylation and de-S-acylation processes. The sensing factors in response to stimuli, regulatory mechanisms, and functions remain to be in-depth investigated for the dynamic change.
Interestingly, ABHD17s themselves are S-acylated in mammals, which is possibly catalyzed by PATs (Yokoi et al., 2016). Similarly, ABAPT proteins could be possibly S-acylated by plant PATs. Membrane association of ABHD17s facilitated by S-acylation enables ABHD17s to de-S-acylate other membrane proteins. In turn, the increased de-S-acylating activity of the membrane-anchored ABHD17s may reduce the auto-S-acylation of PAT proteins in the DHHC domain, which is essential for their PAT activities. It is possible that a feedback inhibition loop between PATs and ABAPTs may regulate the proteome-level S-acylation.
Compared to humans and yeast, research progress about plant PATs has been very limited. Other forms of PTMs, including phosphorylation, S-acylation, ubiquitination, acetylation, and methylation, could regulate the stability, localization, and function of several PAT proteins in mammals (Zmuda and Chamberlain, 2020). However, in plants, most of the PAT studies to date are limited to phenotypic observations and substrate validations. Precise regulatory mechanisms for PAT activities, substrate specificity, and responses to external stimuli are poorly understood, which are interesting and important questions needed to be answered in the future.
ABAPT protein family has 11 members, and their diverse subcellular localizations allow them to catalyze many substrates in different cell compartments. However, considering the large number of S-acylated proteins in the plant (Hemsley et al., 2013; Srivastava et al., 2016; Zhou et al., 2020; Kumar et al., 2022), it remains obscure how many proteins actually undergo palmitate cycling in plant cells and how their S-acylation status respond to stimuli. Based on the above knowledge, we could put forward the following hypotheses: (1) ABAPTs are not substrate-specific enzymes, each of which can de-S-acylate a wide range of substrates; (2) other acyl thioesterases, homologs to APTs, or a completely new acyl thioesterases family may exist in plant, which is waiting to be discovered; and (3) not all S-acylated proteins undergo de-S-acylation. Clearly, future efforts are needed to explore the substrate specificity, redundancy between ABAPT isoforms, and physiological roles of each ABAPT enzyme. The chemical tools, S-acylation process inhibitor 2-bromopalmitate and APT inhibitors Palmostatin B/M for instance, are able to overcome the redundancy of proteins and advance our understanding of the S-acylation function without gene mutations (Davda and Martin, 2014; Remsberg et al., 2021). At the same time, the specificity and selectivity of inhibitors against PATs, APTs, and ABAPTs need to be investigated.
The substrate specificity of both PATs and ABAPTs deserves more attention. Future work on S-acylation in plants should focus on determining how S-acylation and de-S-acylation impact substrate protein function. Revealing the determination and regulation mechanisms of the dynamics of S-acylation and the biological functions it impacted will help us better understand the significance of this lipid modification in plants.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This paper and related research are funded by the Shuishan Scholars Program of Nanjing Forestry University (grant no. 163010299).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adjobo-Hermans, M. J., Goedhart, J., and Gadella, T. W. Jr. (2006). Plant G protein heterotrimers require dual lipidation motifs of Galpha and Ggamma and do not dissociate upon activation. J. Cell Sci. 119, 5087–5097. doi: 10.1242/jcs.03284
Alam, M., Tahir, J., Siddiqui, A., Magzoub, M., Shahzad-Ul-Hussan, S., Mackey, D., et al. (2021). RIN4 homologs from important crop species differentially regulate the Arabidopsis NB-LRR immune receptor, RPS2. Plant Cell Rep. 40, 2341–2356. doi: 10.1007/s00299-021-02771-9
Alassimone, J., Fujita, S., Doblas, V. G., van Dop, M., Barberon, M., Kalmbach, L., et al. (2016). Polarly localized kinase SGN1 is required for Casparian strip integrity and positioning. Nat. Plants 2:16113. doi: 10.1038/nplants.2016.113
Batistic, O. (2012). Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiol. 160, 1597–1612. doi: 10.1104/pp.112.203968
Batistič, O., Rehers, M., Akerman, A., Schlücking, K., Steinhorst, L., Yalovsky, S., et al. (2012). S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res. 22, 1155–1168. doi: 10.1038/cr.2012.71
Batistic, O., Sorek, N., Schültke, S., Yalovsky, S., and Kudla, J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20, 1346–1362. doi: 10.1105/tpc.108.058123
Bürger, M., Willige, B. C., and Chory, J. (2017). A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors. Nat. Commun. 8:2201. doi: 10.1038/s41467-017-02347-w
Carluccio, A. V., Prigigallo, M. I., Rosas-Diaz, T., Lozano-Duran, R., and Stavolone, L. (2018). S-acylation mediates Mungbean yellow mosaic virus AC4 localization to the plasma membrane and in turns gene silencing suppression. PLoS Pathog. 14:e1007207. doi: 10.1371/journal.ppat.1007207
Chai, S., Ge, F. R., Feng, Q. N., Li, S., and Zhang, Y. (2016). PLURIPETALA mediates ROP2 localization and stability in parallel to SCN1 but synergistically with TIP1 in root hairs. Plant J 86, 413–425. doi: 10.1111/tpj.13179
Chai, S., Ge, F. R., Zhang, Y., and Li, S. (2020). S-acylation of CBL10/SCaBP8 by PAT10 is crucial for its tonoplast association and function in salt tolerance. J. Integr. Plant Biol. 62, 718–722. doi: 10.1111/jipb.12864
Chen, D., Hao, F., Mu, H., Ahsan, N., Thelen, J. J., and Stacey, G. (2021). S-acylation of P2K1 mediates extracellular ATP-induced immune signaling in Arabidopsis. Nat. Commun. 12:2750. doi: 10.1038/s41467-021-22854-1
Cowan, G. H., Roberts, A. G., Jones, S., Kumar, P., Kalyandurg, P. B., Gil, J. F., et al. (2018). Potato mop-top virus co-opts the stress sensor HIPP26 for Long-distance movement. Plant Physiol. 176, 2052–2070. doi: 10.1104/pp.17.01698
Davda, D., and Martin, B. R. (2014). Acyl protein thioesterase inhibitors as probes of dynamic S-palmitoylation. MedChemComm 5, 268–276. doi: 10.1039/C3MD00333G
Dowen, R. H., Engel, J. L., Shao, F., Ecker, J. R., and Dixon, J. E. (2009). A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and-independent strategies to localize to plasma membranes. J. Biol. Chem. 284, 15867–15879. doi: 10.1074/jbc.M900519200
Duan, M., Zhang, R., Zhu, F., Zhang, Z., Gou, L., Wen, J., et al. (2017). A lipid-anchored NAC transcription factor is Translocated into the nucleus and activates Glyoxalase I expression during drought stress. Plant Cell 29, 1748–1772. doi: 10.1105/tpc.17.00044
Duncan, J. A., and Gilman, A. G. (1998). A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J. Biol. Chem. 273, 15830–15837. doi: 10.1074/jbc.273.25.15830
Farazi, T. A., Waksman, G., and Gordon, J. I. (2001). The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276, 39501–39504. doi: 10.1074/jbc.R100042200
Fondong, V. N., Reddy, R. V., Lu, C., Hankoua, B., Felton, C., Czymmek, K., et al. (2007). The consensus N-myristoylation motif of a geminivirus AC4 protein is required for membrane binding and pathogenicity. Mol. Plant Microbe Interact. 20, 380–391. doi: 10.1094/MPMI-20-4-0380
Fu, S., Xu, Y., Li, C., Li, Y., Wu, J., and Zhou, X. (2018). Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 11, 269–287. doi: 10.1016/j.molp.2017.11.011
Gagne, J. M., and Clark, S. E. (2010). The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell 22, 729–743. doi: 10.1105/tpc.109.068734
Gargantini, P. R., Gonzalez-Rizzo, S., Chinchilla, D., Raices, M., Giammaria, V., Ulloa, R. M., et al. (2006). A CDPK isoform participates in the regulation of nodule number in Medicago truncatula. Plant J. 48, 843–856. doi: 10.1111/j.1365-313X.2006.02910.x
Gui, J., Zheng, S., Shen, J., and Li, L. (2015). Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front. Plant Sci. 6:804. doi: 10.3389/fpls.2015.00804
Hemsley, P. A. (2015). The importance of lipid modified proteins in plants. New Phytol. 205, 476–489. doi: 10.1111/nph.13085
Hemsley, P. A. (2020). S-acylation in plants: an expanding field. Biochem. Soc. Trans. 48, 529–536. doi: 10.1042/BST20190703
Hemsley, P. A., Kemp, A. C., and Grierson, C. S. (2005). The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 17, 2554–2563. doi: 10.1105/tpc.105.031237
Hemsley, P. A., Taylor, L., and Grierson, C. S. (2008). Assaying protein palmitoylation in plants. Plant Methods 4:2. doi: 10.1186/1746-4811-4-2
Hemsley, P. A., Weimar, T., Lilley, K. S., Dupree, P., and Grierson, C. S. (2013). A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol. 197, 805–814. doi: 10.1111/nph.12077
Hu, X., and Ye, Y. (2022). Improving infant formula using algae from the sea. Plant Physiol. 189, 450–451. doi: 10.1093/plphys/kiac108
Hurst, C. H., Wright, K. M., Turnbull, D., Leslie, K., Jones, S., and Hemsley, P. A. (2019). Juxta-membrane S-acylation of plant receptor-like kinases is likely fortuitous and does not necessarily impact upon function. Sci. Rep. 9:12818. doi: 10.1038/s41598-019-49302-x
Ivanchenko, M., Vejlupkova, Z., Quatrano, R. S., and Fowler, J. E. (2000). Maize ROP7 GTPase contains a unique, CaaX box-independent plasma membrane targeting signal. Plant J. 24, 79–90. doi: 10.1046/j.1365-313x.2000.00855.x
Jennings, B. C., and Linder, M. E. (2012). DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J. Biol. Chem. 287, 7236–7245. doi: 10.1074/jbc.M111.337246
Jiang, H., Ma, Q. J., Zhong, M. S., Gao, H. N., Li, Y. Y., and Hao, Y. J. (2021). The apple palmitoyltransferase MdPAT16 influences sugar content and salt tolerance via an MdCBL1-MdCIPK13-MdSUT2.2 pathway. Plant J. 106, 689–705. doi: 10.1111/tpj.15191
Kathayat, R. S., Cao, Y., Elvira, P. D., Sandoz, P. A., Zaballa, M. E., Springer, M. Z., et al. (2018). Active and dynamic mitochondrial S-depalmitoylation revealed by targeted fluorescent probes. Nat. Commun. 9:334. doi: 10.1038/s41467-017-02655-1
Kim, H. S., Desveaux, D., Singer, A. U., Patel, P., Sondek, J., and Dangl, J. L. (2005). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. U. S. A. 102, 6496–6501. doi: 10.1073/pnas.0500792102
Konrad, S. S., Popp, C., Stratil, T. F., Jarsch, I. K., Thallmair, V., Folgmann, J., et al. (2014). S-acylation anchors remorin proteins to the plasma membrane but does not primarily determine their localization in membrane microdomains. New Phytol. 203, 758–769. doi: 10.1111/nph.12867
Kumar, M., Carr, P., and Turner, S. R. (2022). An atlas of Arabidopsis protein S-acylation reveals its widespread role in plant cell organization and function. Nat. Plants 8, 670–681. doi: 10.1038/s41477-022-01164-4
Kumar, M., Wightman, R., Atanassov, I., Gupta, A., Hurst, C. H., Hemsley, P. A., et al. (2016). S-acylation of the cellulose synthase complex is essential for its plasma membrane localization. Science 353, 166–169. doi: 10.1126/science.aaf4009
Lai, J., Yu, B., Cao, Z., Chen, Y., Wu, Q., Huang, J., et al. (2015). Two homologous protein S-acyltransferases, PAT13 and PAT14, cooperatively regulate leaf senescence in Arabidopsis. J. Exp. Bot. 66, 6345–6353. doi: 10.1093/jxb/erv347
Lavy, M., Bracha-Drori, K., Sternberg, H., and Yalovsky, S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14, 2431–2450. doi: 10.1105/tpc.005561
Lavy, M., and Yalovsky, S. (2006). Association of Arabidopsis type-II ROPs with the plasma membrane requires a conserved C-terminal sequence motif and a proximal polybasic domain. Plant J. 46, 934–947. doi: 10.1111/j.1365-313X.2006.02749.x
Leclercq, J., Ranty, B., Sanchez-Ballesta, M. T., Li, Z., Jones, B., Jauneau, A., et al. (2005). Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J. Exp. Bot. 56, 25–35. doi: 10.1093/jxb/eri003
Leśniewicz, K., Poręba, E., Smolarkiewicz, M., Wolff, N., Stanisławski, S., and Wojtaszek, P. (2012). Plant plasma membrane-bound staphylococcal-like DNases as a novel class of eukaryotic nucleases. BMC Plant Biol. 12:195. doi: 10.1186/1471-2229-12-195
Li, Y., Li, H. J., Morgan, C., Bomblies, K., Yang, W., and Qi, B. (2019b). Both male and female gametogenesis require a fully functional protein S-acyl transferase 21 in Arabidopsis thaliana. Plant J. 100, 754–767. doi: 10.1111/tpj.14475
Li, Y., and Qi, B. (2017). Progress toward understanding protein S-acylation: prospective in plants. Front. Plant Sci. 8:346. doi: 10.3389/fpls.2017.00346
Li, Y., Scott, R., Doughty, J., Grant, M., and Qi, B. (2016). Protein S-acyltransferase 14: a specific role for palmitoylation in leaf senescence in Arabidopsis. Plant Physiol. 170, 415–428. doi: 10.1104/pp.15.00448
Li, Y., Xu, J., Li, G., Wan, S., Batistič, O., Sun, M., et al. (2019a). Protein S-acyl transferase 15 is involved in seed triacylglycerol catabolism during early seedling growth in Arabidopsis. J. Exp. Bot. 70, 5205–5216. doi: 10.1093/jxb/erz282
Li, H., Zeng, R., Chen, Z., Liu, X., Cao, Z., Xie, Q., et al. (2018). S-acylation of a geminivirus C4 protein is essential for regulating the CLAVATA pathway in symptom determination. J. Exp. Bot. 69, 4459–4468. doi: 10.1093/jxb/ery228
Lin, D. T., and Conibear, E. (2015). ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. elife 4:e11306. doi: 10.7554/eLife.11306
Liu, X., Li, M., Li, Y., Chen, Z., Zhuge, C., Ouyang, Y., et al. (2021). An ABHD17-like hydrolase screening system to identify de-S-acylation enzymes of protein substrates in plant cells. Plant Cell 33, 3235–3249. doi: 10.1093/plcell/koab199
Liu, J., Zhong, S., Guo, X., Hao, L., Wei, X., Huang, Q., et al. (2013). Membrane-bound RLCKs LIP1 and LIP2 are essential male factors controlling male-female attraction in Arabidopsis. Curr. Biol. 23, 993–998. doi: 10.1016/j.cub.2013.04.043
Martín, M. L., and Busconi, L. (2000). Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 24, 429–435. doi: 10.1046/j.1365-313x.2000.00889.x
Mochizuki, S., Harada, A., Inada, S., Sugimoto-Shirasu, K., Stacey, N., Wada, T., et al. (2005). The Arabidopsis WAVY GROWTH 2 protein modulates root bending in response to environmental stimuli. Plant Cell 17, 537–547. doi: 10.1105/tpc.104.028530
Pang, H., Yan, Q., Zhao, S., He, F., Xu, J., Qi, B., et al. (2019). Knockout of the S-acyltransferase gene, PbPAT14, confers the dwarf yellowing phenotype in first generation pear by ABA accumulation. Int. J. Mol. Sci. 20, 6347. doi: 10.3390/ijms20246347
Peng, D., Tan, X., Zhang, L., Yuan, D., Lin, J., Liu, X., et al. (2018). Increasing branch and seed yield through heterologous expression of the novel rice S-acyl transferase gene OsPAT15 in Brassica napus L. Breed. Sci. 68, 326–335. doi: 10.1270/jsbbs.17126
Qi, B., Doughty, J., and Hooley, R. (2013). A Golgi and tonoplast localized S-acyl transferase is involved in cell expansion, cell division, vascular patterning and fertility in Arabidopsis. New Phytol. 200, 444–456. doi: 10.1111/nph.12385
Qi, D., Dubiella, U., Kim, S. H., Sloss, D. I., Dowen, R. H., Dixon, J. E., et al. (2014). Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease RESISTANCE protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on s-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol. 164, 340–351. doi: 10.1104/pp.113.227686
Ranf, S., Eschen-Lippold, L., Fröhlich, K., Westphal, L., Scheel, D., and Lee, J. (2014). Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 14:374. doi: 10.1186/s12870-014-0374-4
Remsberg, J. R., Suciu, R. M., Zambetti, N. A., Hanigan, T. W., Firestone, A. J., Inguva, A., et al. (2021). ABHD17 regulation of plasma membrane palmitoylation and N-Ras-dependent cancer growth. Nat. Chem. Biol. 17, 856–864. doi: 10.1038/s41589-021-00785-8
Ren, H., Willige, B. C., Jaillais, Y., Geng, S., Park, M. Y., Gray, W. M., et al. (2019). BRASSINOSTEROID-SIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 15:e1007904. doi: 10.1371/journal.pgen.1007904
Resh, M. D. (2013). Covalent lipid modifications of proteins. Curr. Biol. 23, R431–R435. doi: 10.1016/j.cub.2013.04.024
Roth, A. F., Feng, Y., Chen, L., and Davis, N. G. (2002). The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28. doi: 10.1083/jcb.200206120
Running, M. P. (2014). The role of lipid post-translational modification in plant developmental processes. Front. Plant Sci. 5:50. doi: 10.3389/fpls.2014.00050
Saito, S., Hamamoto, S., Moriya, K., Matsuura, A., Sato, Y., Muto, J., et al. (2018). N-myristoylation and S-acylation are common modifications of Ca2+ −regulated Arabidopsis kinases and are required for activation of the SLAC1 anion channel. New Phytol. 218, 1504–1521. doi: 10.1111/nph.15053
Smotrys, J. E., and Linder, M. E. (2004). Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73, 559–587. doi: 10.1146/annurev.biochem.73.011303.073954
Sorek, N., Bloch, D., and Yalovsky, S. (2009). Protein lipid modifications in signaling and subcellular targeting. Curr. Opin. Plant Biol. 12, 714–720. doi: 10.1016/j.pbi.2009.09.003
Sorek, N., Gutman, O., Bar, E., Abu-Abied, M., Feng, X., Running, M. P., et al. (2011a). Differential effects of prenylation and s-acylation on type I and II ROPS membrane interaction and function. Plant Physiol. 155, 706–720. doi: 10.1104/pp.110.166850
Sorek, N., Henis, Y. I., and Yalovsky, S. (2011b). How prenylation and S-acylation regulate subcellular targeting and function of ROP GTPases. Plant Signal. Behav. 6, 1026–1029. doi: 10.4161/psb.6.7.15578
Sorek, N., Poraty, L., Sternberg, H., Buriakovsky, E., Bar, E., Lewinsohn, E., et al. (2017). Corrected and republished from: activation status-coupled transient S-acylation determines membrane partitioning of a plant rho-related GTPase. Mol. Cell. Biol. 37, e00333–e00317. doi: 10.1128/MCB.00333-17
Sorek, N., Segev, O., Gutman, O., Bar, E., Richter, S., Poraty, L., et al. (2010). An S-acylation switch of conserved G domain cysteines is required for polarity signaling by ROP GTPases. Current Biol.: CB 20, 914–920. doi: 10.1016/j.cub.2010.03.057
Srivastava, V., Weber, J. R., Malm, E., Fouke, B. W., and Bulone, V. (2016). Proteomic analysis of a poplar cell suspension culture suggests a major role of protein S-acylation in diverse cellular processes. Front. Plant Sci. 7:477. doi: 10.3389/fpls.2016.00477
Tian, Y., Zeng, H., Wu, J., Huang, J., Gao, Q., Tang, D., et al. (2022). Screening DHHCs of S-acylated proteins using an OsDHHC cDNA library and bimolecular fluorescence complementation in rice. Plant J. 110, 1763–1780. doi: 10.1111/tpj.15769
Turnbull, D., and Hemsley, P. A. (2017). Fats and function: protein lipid modifications in plant cell signalling. Curr. Opin. Plant Biol. 40, 63–70. doi: 10.1016/j.pbi.2017.07.007
Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741. doi: 10.1093/emboj/20.17.4730
Villalta, I., García, E., Hornero-Mendez, D., Carranco, R., Tello, C., Mendoza, I., et al. (2021). Distinct roles of N-terminal fatty acid acylation of the salinity-sensor protein SOS3. Front. Plant Sci. 12:691124. doi: 10.3389/fpls.2021.691124
Wan, Z. Y., Chai, S., Ge, F. R., Feng, Q. N., Zhang, Y., and Li, S. (2017a). Arabidopsis PROTEIN S-ACYL TRANSFERASE4 mediates root hair growth. Plant J. 90, 249–260. doi: 10.1111/tpj.13484
Wan, Z. Y., Zhang, Y., and Li, S. (2017b). Protein S-acyl transferase 4 controls nucleus position during root hair tip growth. Plant Signal. Behav. 12:e1311438. doi: 10.1080/15592324.2017.1311438
Won, S. J., Cheung See Kit, M., and Martin, B. R. (2018). Protein depalmitoylases. Crit. Rev. Biochem. Mol. Biol. 53, 83–98. doi: 10.1080/10409238.2017.1409191
Yang, B., Zhang, K., Jin, X., Yan, J., Lu, S., Shen, Q., et al. (2021). Acylation of non-specific phospholipase C4 determines its function in plant response to phosphate deficiency. Plant J. 106, 1647–1659. doi: 10.1111/tpj.15260
Yin, J., Wang, L., Jin, T., Nie, Y., Liu, H., Qiu, Y., et al. (2021). A cell wall-localized NLR confers resistance to soybean mosaic virus by recognizing viral-encoded cylindrical inclusion protein. Mol. Plant 14, 1881–1900. doi: 10.1016/j.molp.2021.07.013
Yokoi, N., Fukata, Y., Sekiya, A., Murakami, T., Kobayashi, K., and Fukata, M. (2016). Identification of PSD-95 Depalmitoylating enzymes. J. Neurosci. Off. J. Soc. Neurosci. 36, 6431–6444. doi: 10.1523/JNEUROSCI.0419-16.2016
Yuan, X., Zhang, S., Sun, M., Liu, S., Qi, B., and Li, X. (2013). Putative DHHC-cysteine-rich domain S-acyltransferase in plants. PLoS One 8:e75985. doi: 10.1371/journal.pone.0075985
Zeng, Q., Wang, X., and Running, M. P. (2007). Dual lipid modification of Arabidopsis Ggamma-subunits is required for efficient plasma membrane targeting. Plant Physiol. 143, 1119–1131. doi: 10.1104/pp.106.093583
Zhang, C., Beckmann, L., Kudla, J., and Batistič, O. (2017). N-terminal S-acylation facilitates tonoplast targeting of the calcium sensor CBL6. FEBS Lett. 591, 3745–3756. doi: 10.1002/1873-3468.12880
Zhang, X., Mi, Y., Mao, H., Liu, S., Chen, L., and Qin, F. (2020). Genetic variation in ZmTIP1 contributes to root hair elongation and drought tolerance in maize. Plant Biotechnol. J. 18, 1271–1283. doi: 10.1111/pbi.13290
Zhang, Y. L., Li, E., Feng, Q. N., Zhao, X. Y., Ge, F. R., Zhang, Y., et al. (2015). Protein palmitoylation is critical for the polar growth of root hairs in Arabidopsis. BMC Plant Biol. 15:50. doi: 10.1186/s12870-015-0441-5
Zhao, X. Y., Wang, J. G., Song, S. J., Wang, Q., Kang, H., Zhang, Y., et al. (2016). Precocious leaf senescence by functional loss of PROTEIN S-ACYL TRANSFERASE14 involves the NPR1-dependent salicylic acid signaling. Sci. Rep. 6:20309. doi: 10.1038/srep20309
Zheng, L., Liu, P., Liu, Q., Wang, T., and Dong, J. (2019). Dynamic protein S-acylation in plants. Int. J. Mol. Sci. 20:560. doi: 10.3390/ijms20030560
Zhou, L. Z., Li, S., Feng, Q. N., Zhang, Y. L., Zhao, X., Zeng, Y. L., et al. (2013). Protein S-ACYL Transferase10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 25, 1093–1107. doi: 10.1105/tpc.112.108829
Zhou, B., Lin, J. Z., Peng, D., Yang, Y. Z., Guo, M., Tang, D. Y., et al. (2017). Plant architecture and grain yield are regulated by the novel DHHC-type zinc finger protein genes in rice (Oryza sativa L.). Plant Sci. 254, 12–21. doi: 10.1016/j.plantsci.2016.08.015
Zhou, Y. B., Liu, C., Tang, D. Y., Yan, L., Wang, D., Yang, Y. Z., et al. (2018). The receptor-Like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in Rice. Plant Cell 30, 1100–1118. doi: 10.1105/tpc.17.01000
Zhou, L., Zhou, M., Gritsenko, M. A., and Stacey, G. (2020). Selective enrichment coupled with proteomics to identify S-Acylated plasma membrane proteins in Arabidopsis. Curr. Protoc. Plant Biol. 5:e20119. doi: 10.1002/cppb.20119
Keywords: protein S-acyltransferases, acyl protein thioesterases, protein S-acylation, plant, signal transduction, post-translational modification
Citation: Li J, Zhang M and Zhou L (2022) Protein S-acyltransferases and acyl protein thioesterases, regulation executors of protein S-acylation in plants. Front. Plant Sci. 13:956231. doi: 10.3389/fpls.2022.956231
Edited by:
Agnieszka Sirko, Institute of Biochemistry and Biophysics (PAN), PolandReviewed by:
Mark Paul Running, University of Louisville, United StatesManoj Kumar, The University of Manchester, United Kingdom
Copyright © 2022 Li, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Zhou, bGp6aG91QG5qZnUuZWR1LmNu
 Jincheng Li
Jincheng Li Manqi Zhang
Manqi Zhang Lijuan Zhou
Lijuan Zhou