
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 14 October 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.955623
This article is part of the Research TopicFlowering Time Control in Agricultural and Horticultural CropsView all 10 articles
 Ben Trevaskis1*
Ben Trevaskis1* Felicity A. J. Harris2,3
Felicity A. J. Harris2,3 William D. Bovill1
William D. Bovill1 Allan R. Rattey4
Allan R. Rattey4 Kelvin H. P. Khoo5
Kelvin H. P. Khoo5 Scott A. Boden5
Scott A. Boden5 Jessica Hyles1
Jessica Hyles1Oat (Avena sativa) is an annual cereal grown for forage, fodder and grain. Seasonal flowering behaviour, or phenology, is a key contributor to the success of oat as a crop. As a species, oat is a vernalization-responsive long-day plant that flowers after winter as days lengthen in spring. Variation in both vernalization and daylength requirements broadens adaptation of oat and has been used to breed modern cultivars with seasonal flowering behaviours suited to different regions, sowing dates and farming practices. This review examines the importance of variation in oat phenology for crop adaptation. Strategies to advance understanding of the genetic basis of oat phenology are then outlined. These include the potential to transfer knowledge from related temperate cereals, particularly wheat (Triticum aestivum) and barley (Hordeum vulgare), to provide insights into the potential molecular basis of variation in oat phenology. Approaches that use emerging genomic resources to directly investigate the molecular basis of oat phenology are also described, including application of high-resolution genome-wide diversity surveys to map genes linked to variation in flowering behaviour. The need to resolve the contribution of individual phenology genes to crop performance by developing oat genetic resources, such as near-isogenic lines, is emphasised. Finally, ways that deeper knowledge of oat phenology can be applied to breed improved varieties and to inform on-farm decision-making are outlined.
Oat (Avena sativa) was potentially a wild food for Paleolithic hunter-gatherers (Lippi et al., 2015), before entering agriculture as a weed of wheat and barley during the Neolithic period (Murphy and Hoffman, 1992). As a weed, oat can dominate fields of wheat or barley and by 2000 years ago it was itself being grown as a crop (Harlan, 1982; Murphy and Hoffman, 1992). Factors that drove the acceptance of oat into early European farming systems included the ability to perform well in colder climates and marginal areas, with less inputs, together with end-use versatility that contributed to overall farm resilience (Moore-Colyer, 1995). The nutritional quality of oat grains, or groats, was also a key factor. Oat grains have high lipid content, contain lysine-rich protein and soluble fibre, particularly β-glucan (Cuddeford, 1995; Zwer, 2017). Oat grains are an ideal feed for horses, so there was strong demand for oats throughout the period when horses were a major contributor to transport and industry (Murphy and Hoffman, 1992). The same grain quality parameters are now driving interest in oats as a healthy food and as a non-animal protein source.
Oat is an important crop in Australia, Canada, China, Europe, North and South America. Variation in the seasonal timing of life cycle events (phenology), particularly the timing of flowering and grain production, enables adjustment of the oat life cycle to suit local constraints. This variation is critical for the cultivation of modern oats across such a broad geographical range. For example, cultivation of oat in Canada is typically limited to spring and summer, thereby avoiding harsh winters. This contrasts with regions such as the United Kingdom where crops can be sown in autumn, then over-winter before producing grain in spring and summer. Other climate factors that can define the timing and duration of the growing season for oat include seasonal rainfall patterns and extreme summer heat.
The history of the Australian grains industry provides an example of the importance of variation in phenology for geographical adaptation. Oat came to Australia with Europeans and was cultivated in the Sydney area as early as 1791 (Collins and King, 1798). It was then slow to expand as a crop due to a lack of varieties suited to Australian growing conditions. This was because the oat growing season in south-eastern Australia is constrained by the timing of autumn rains, which dictate sowing dates, combined with the need to flower in the “optimal flowering period” to avoid frost, heat and water-limitation disrupting flowering and grain development (Tashiro and Wardlaw, 1990; Dolferus et al., 2011; Flohr et al., 2017). The European oats first introduced to Australia were ill-suited to the local growing conditions because they were slow to mature and flower. This remained the case until the early 20th century when John Pridham, a cereal breeder trained by the pioneer wheat breeder William Farrer, began to breed locally adapted oat cultivars (Mengersen, 1960; Fitzsimmons, 1988). Pridham selected oats with earlier flowering, with his most successful variety being Belar (Fitzsimmons, 1988). Breeding of oats for Australian growing conditions continued with Pridham’s successors and this facilitated the expansion of the Australian oat industry (Mengersen, 1960). Now, on average, 1.4 million tons of milling oats are produced annually, with Western Australia and New South Wales being the major growing regions (ABARES, 2021). Additionally, around 0.7 million tons of oaten hay are exported annually, and more is used domestically as fodder (ABARES, 2021). Oat is also grown as a forage crop across large areas of New South Wales and Queensland.
Similar examples of how phenology adapts oat to local seasonal constraints can be found for other regions globally, though the specific of constraints differ to those in Australia (harsh winters in Canadian prairies, for example). In addition, as outlined in subsequent sections, phenology influences yield component traits, such as grain number, via determining pattens of plant growth and development, and so influences crop yield potential. The overall relationship between phenology and crop yield, through adaptation and yield potential, is not limited to Australian context. For example, phenology is a driver of yield in North American and European climates (Yan et al., 2007; Howarth et al., 2021).
From the perspective of overall farming systems, phenology also allows adaption to specific farm management practices. For example, selection for earlier flowering can allow double cropping in some farming systems (Islam et al., 2010). There is also evidence that in some regions breeders have selected for earlier maturity during the transition to mechanisation, which allowed sowing of large areas to occur within a narrower seasonal window (Grau Nersting et al., 2006). Similarly, phenology is an important component of optimisation of oat for different end uses. For example, the timing of maturation influences quality of milling oats (Howarth et al., 2021). Interactions between phenology and seasonal conditions also influence forage oat quality (Coblentz et al., 2012; Ki-Seung et al., 2014).
Phenology is likely to be of renewed global importance in an era of warming climates and increased climate variability. Re-optimisation of phenology might be required in some production zones. This could include adjustment of phenology to suit shifting sowing or harvest dates, and/or adjustment of phenology to compensate for more rapid growth and development driven by faster accumulation of growing degree days (Hunt et al., 2019; Peltonen-Sainto and Juahianen, 2020).
In summary, phenology enables adaption of oat to different climates, is a major determinant of crop yields, and can enable breeding of cultivars for different farming systems or end-uses. Subsequent sections review current understanding of oat phenology.
The time of year when oat flowers and produces grain is determined largely by genotype-dependent responses to seasonal temperature and daylength (photoperiod) cues. As a species, cultivated oat can be described as a “vernalization-responsive long-day plant”, which will flower after exposure to winter cold (i.e., vernalization) as days lengthen in spring (Figure 1) (Borodin, 1934; Wiggans and Frey, 1955). This archetypal combination of seasonal flowering-responses is found in “winter” oats, which are well-suited to situations where crops are sown autumn and then overwinter before flowering in spring (Sampson and Burrows, 1972; Sorrells and Simmons, 1992). There is variation in both vernalization and daylength requirements amongst oat accessions/cultivars (Sampson and Burrows, 1972: King and Bacon, 1992). This variation has been harnessed by plant breeders to selectively modify oat phenology to suit diverse environments and different management practices, as outlined above.
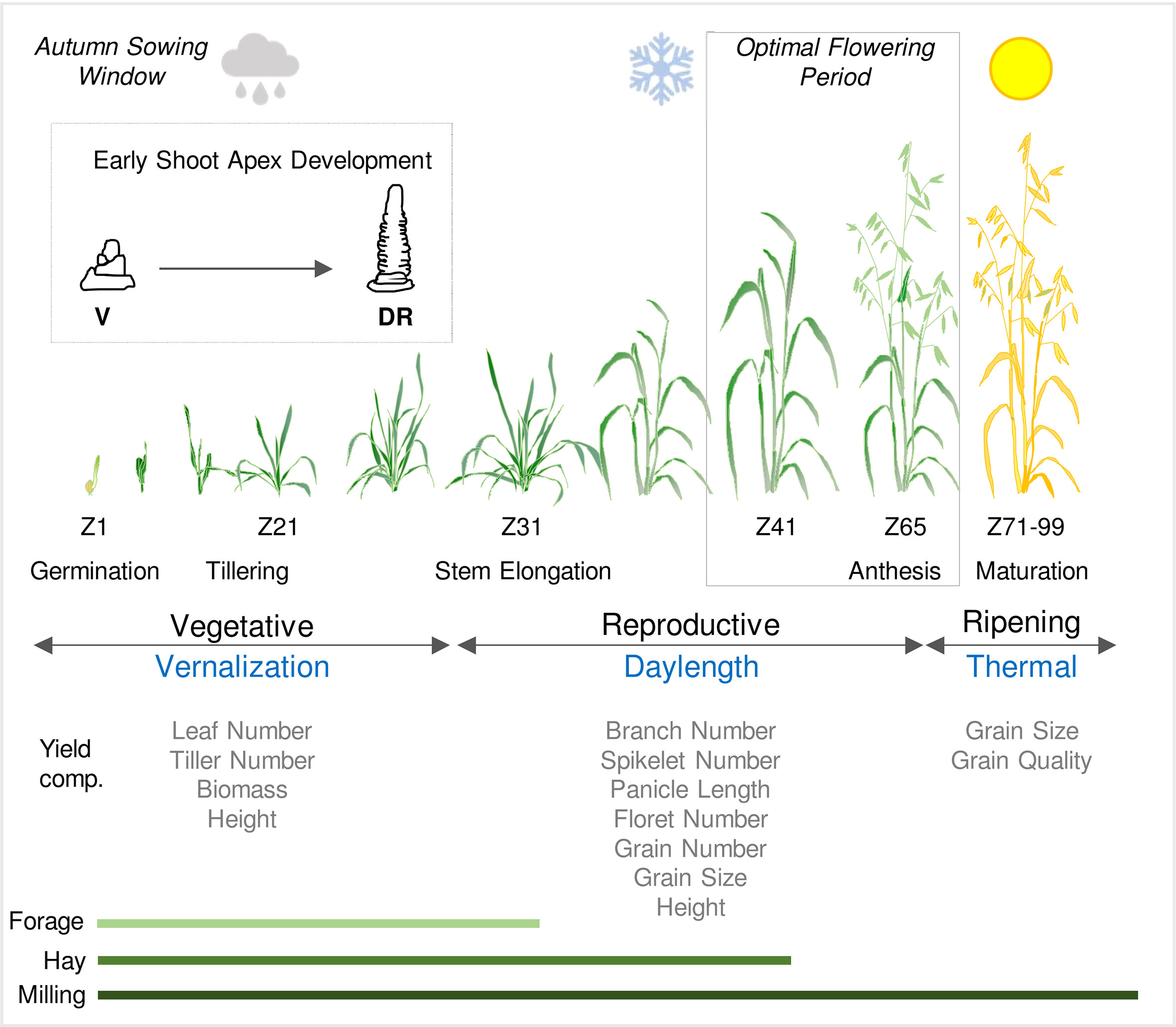
Figure 1 Schematic representation of the life cycle of oat in an Australian context. Sowing and establishment of crops is timed according to water availability in autumn (cloud symbol) and then plants experience cold temperatures through late autumn and winter (vernalization). Flowering and grain production occur during a optimal flowering period in spring, after the risk of frost subsides (snowflake symbol) and before the onset of heat and water limitation (sun symbol). Developmental stages are referenced by Zadoks scale (Zadoks et al., 1974). Inset shows early shoot apex development and the first visible sign of the transition from vegetative (V) to reproductive development; the double ridge stage (DR). The relationships between phases of development (vegetative and reproductive) and ripening are shown relative to the seasonal climate cues that influence the duration of each these phases. The impact of phase durations on physiological and yield related traits are indicated together with the relevance of growth phases to different end uses (forage, hay or milling).
Winter oats can require up to 50 days at low temperatures (below 10°C) to flower rapidly (Sorrells and Simmons, 1992). The response to cold is quantitative, such that increasing the duration of vernalization typically accelerates flowering to greater extents until the vernalization response is saturated (Sampson and Burrows, 1972). Vernalization accelerates the transition to flowering, the point when the shoot apex switches from vegetative to reproductive development (Bell, 1936; Bonnett, 1966; Aitken, 1974). A clear developmental indicator of the vernalization response, aside from earlier flowering, is reduction in the number of leaves produced on the main stem (final leaf number), since a vernalized plant spends less time in the vegetative phase, generating fewer phytomers and less leaves over the course of the plant life cycle (Aitken, 1974). A saturating vernalization treatment is one that generates the minimum final leaf number (Aitken, 1974). Whereas winter oats require vernalization to flower, spring oats generally show reduced or no requirement for vernalization (Borodin, 1934; Sampson and Burrows, 1972: King and Bacon, 1992). This allows spring oats to be sown when vernalization might not occur, after winter or in regions where winters are mild.
Daylengths exceeding 12 hours (long days) typically accelerate flowering of oat. The extent to which flowering is accelerated increases with longer daylengths, with the maximal response occurring when daylength exceeds 18 hours (Sampson and Burrows, 1972; Sorrells and Simmons, 1992). Daylength can influence the duration of the vegetative growth phase but, under typical field conditions, increasing daylengths during spring typically coincide with reproductive development and so have more impact on the duration of inflorescence development (Jenkins, 1973; Sorrells and Simmons, 1992) (Figure 1). While flowering of oat is usually delayed when plants are grown in daylengths shorter than 12 hours, some varieties flower rapidly irrespective of daylength (Burrows, 1984). There are examples of oats that flower rapidly irrespective of both vernalization and daylength (Aitken, 1974).
For the reasons outlined above, selecting optimal phenology is a key goal for oat breeding programs, so understanding the genetic basis for variation in oat phenology can contribute to future breeding strategies. The molecular basis of variation in oat phenology has not been resolved. There is, however, detailed knowledge of the gene sequences that underlie variation in the vernalization and daylength requirements of other temperate cereals, particularly wheat and barley (see Fjellheim et al., 2014; Hyles et al., 2020). The Avena genus is part of the same subfamily of grasses as wheat and barley (Pooideaea) and all are members of the “Core Pooid” clade. This close evolutionary relationship, combined with the similar flowering physiology, suggests that there are good prospects to transfer knowledge of the molecular basis of seasonal flowering from wheat and barley to oat.
The key gene controlling vernalization-induced flowering of wheat and barley is the MADS box transcription factor, VERNALIZATION1 (VRN1) (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). VRN1 promotes the transition to reproductive development but is transcribed at low levels in plants that have not been vernalized. Exposing plants to prolonged cold activates transcription of VRN1 and this subsequently accelerates flowering when plants are grown at warmer temperatures (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003; Chen and Dubcovsky, 2012). Activation of VRN1 by cold is quantitative such that VRN1 transcript levels increase more with longer cold treatments, accelerating flowering to greater extents (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003; von Zitzewitz et al., 2005; Sasani et al., 2009). Spring wheats that flower without vernalization typically carry alleles of VRN1 that are transcribed without cold and so bypass the requirement for vernalization (Pugsley, 1971; Yan et al., 2003; Yan et al., 2004a; Fu et al., 2005; Eagles et al., 2009; Eagles et al., 2011; Díaz et al., 2012; Oliver et al., 2013). These alleles have mutations in the promoter or first intron that are suggested to trigger elevated transcriptional activity (Yan et al., 2003; Yan et al., 2004a). VRN1 copy number variation and amino acid substitutions have also been linked to variation in vernalization requirement of wheat (Chen et al., 2009; Díaz et al., 2012; Dixon et al., 2019).
The mechanisms underlying the daylength flowering response were first resolved in Arabidopsis thaliana (Arabidopsis) where acceleration of flowering by long days is mediated by FLOWERING LOCUS T (FT) (Kardailsky et al., 1999; Kobayashi et al., 1999; Corbesier et al., 2007). FT encodes a small protein, often described as a florigen, that is expressed in leaves in long days and then transported to the shoot apex where it triggers flowering (Corbesier et al., 2007; Tamaki et al., 2007). Activation of FT expression in leaves is activated by a molecular network that includes phytochromes, which perceive external light cues, and the circadian oscillator, which gives rise to rhythmic day-night gene expression patterns that mediate daylength (or photoperiod) responses (Suarez-Lopez et al., 2001; Valverde et al., 2004). The photoperiod response pathway first elucidated in Arabidopsis seems to be widely conserved in plants, including wheat and barley, where transcription of a FT-like gene (FT1, also known as VERNALIZATION3, VRN3) is activated in the leaves by long days (Turner et al., 2005; Yan et al., 2006).
Mutations in genes that mediate circadian oscillator function and output have been linked to variation for daylength sensitivity. For example, the main gene that determines daylength sensitivity of wheat and barley is PHOTOPERIOD1 (PPD1) (Turner et al., 2005). PPD1 is related to PSEUDORESPONSE REGULATOR genes that contribute to circadian oscillator function (Turner et al., 2005; Beales et al., 2007). In photoperiod sensitive wheats, which require long days to flower, PPD1 is expressed with an oscillating expression pattern through day-night cycles and mediates activation of FT expression when days are long or increasing in length (i.e., spring and summer) (Beales et al., 2007). Mutations that alter PPD1 activity are a common basis for variation in the photoperiod sensitivity of wheat and barley. For example, a deletion in the promoter of the D genome of PPD1 is associated with high PPD1 expression throughout day-night cycles and, in turn, with elevated FT1 expression and earlier flowering in short days (Beales et al., 2007; Gauley and Boden, 2021). Conversely, coding sequence mutations in the barley PPD1 gene that likely inhibit function of the PPD1 protein are linked to delayed flowering under long days (Turner et al., 2005). Copy number variation for the PPD1 gene has also been linked to variation in photoperiod sensitivity of wheat (Díaz et al., 2012). Other genes that function in the circadian oscillator influence phenology of wheat and barley, including LUX/ARRYTHMO and EARLY FLOWERING 3 (Mizuno et al., 2012; Faure et al., 2012; Zakhrabekova et al., 2012; Campoli et al., 2013; Boden et al., 2014; Gawroński et al., 2014; Zikhali et al., 2016). Additionally, PHYTOCHROME C (PHYC) plays a major role in daylength perception in cereals and can influence the photoperiod flowering response (Chen et al., 2014). Similar to PPD1, all of these genes seem to influence flowering behaviour, at least in part, by influencing FT1 expression (Figure 2).
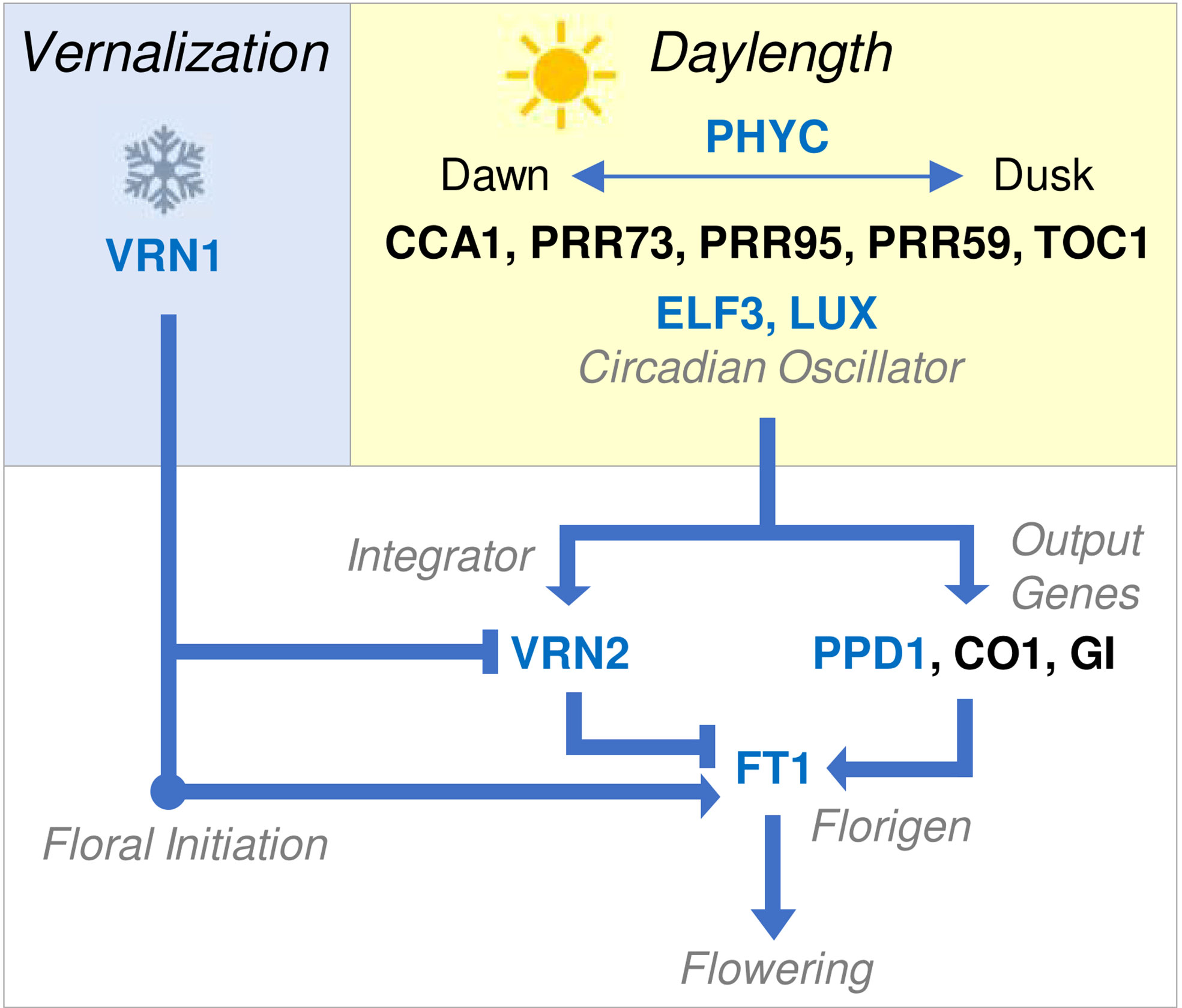
Figure 2 Overview of molecular genetic network controlling the seasonal flowering behaviour of temperate cereals. Vernalization (blue box) activates VRN1. This is sufficient to trigger the transition to reproductive development or floral initiation, as indicated by the double ridge stage of shoot apex development. Additionally, VRN1 represses VRN2 and activates FT1, allowing the daylength flowering response to occur after vernalization. Daylength perception (yellow box) is mediated by the circadian oscillator, which generates internal biological rhythms that allow perception of external photoperiod. The circadian oscillator regulates output genes (white box) that activate FT1, the key trigger for daylength-induced flowering also known as florigen. VRN2 is also daylength responsive and blocks long-day induction of FT1, until VRN2 is itself repressed by VRN1. FT1 accelerates reproductive development and stem elongation, leading to flowering. Arrows indicate activation of a target gene, lines ending in bars indicate repression. Blue text indicates that a gene has been linked to (or associated with) natural variation for seasonal flowering behaviour. Gene names are shown without italics for clarity and blue text indicates genes known to mediate natural variation in phenology of other cereals.
Vernalization is normally a pre-requisite for long-day induction of FT1 (Hemming et al., 2008). This is mediated by the VERNALIZATION2 (VRN2) gene, which limits expression of FT1 in long days to repress flowering prior to vernalization (Yan et al., 2004b; Trevaskis et al., 2007; Hemming et al., 2008). Loss of VRN2 function (e.g., through gene deletion) allows long-day induction of FT1 expression without plants experiencing prolonged cold and thereby bypasses the vernalization requirement (Yan et al., 2004b; Hemming et al., 2008). This occurs mainly in barley and diploid einkorn wheat but is unlikely to be common in tetraploid durum or hexaploid bread wheats, where gene redundancy reduces the likelihood of complete loss of VRN2 function. Alleles of FT1 that are expressed at high basal levels can bypass VRN2 and trigger rapid flowering irrespective of vernalization status or daylength (Yan et al., 2006). The precise genetic basis for these alleles has not been resolved; copy number, local rearrangements and insertions/deletions have all been associated with ‘early’ alleles of FT1 (Yan et al., 2006; Loscos et al., 2014; Nitcher et al., 2014).
Phenology influences other aspects of oat biology in addition to determining when flowering and grain production occur. Vernalization can alter tiller number and there is a strong relationship between the duration of vegetative growth phase and the capacity to survive freezing winter temperatures, as is the case for other temperate cereals (Sorrells and Simmons, 1992; Wooten et al., 2009). By influencing the duration of inflorescence development, the daylength flowering response influences inflorescence structure, such that longer daylengths reduce the duration of inflorescence development and decrease the number of spikelets produced by the oat panicle (Finkner et al., 1973; Peltonen-Sainto, 1994). Genes that determine the duration of vegetative or reproductive development can influence inflorescence node and spikelet number of wheat, barley and rice (Oryza sativa). Examples include the PPD1 and FT1 genes of wheat and also the HEADING DATE1 and EARLY HEADING DATE 1 genes of rice (Endo-Higashi and Izawa, 2011; Boden et al., 2015; Dixon et al., 2018; Finnegan et al., 2018; Brassac et al., 2021). The common theme from these examples is that genes that reduce the duration of reproductive development can decrease the number of inflorescence nodes produced and thus the number of spikelets and florets produced by each plant. This can in turn impact the total grain number per unit area and thereby influence yield. This parallels observations that vernalization and daylength influence tiller or spikelet number of oat (see above). So, in addition to playing a central role in adaptation, oat phenology genes are likely to influence other traits that underpin grain yield, as is the case for other temperate cereals and rice (see Trevaskis, 2018) (Figure 1).
A role for VRN1-like genes in mediating vernalization-induced flowering appears to be broadly conserved in Pooid grasses (Ream et al., 2014; McKeown et al., 2016; Feng et al., 2017; Woods et al., 2017). This suggests that VRN1-like genes are likely to play a role in vernalization-induced flowering of oat, a member of the core Pooid grasses. Direct support for this idea comes from observations that an oat VRN1 orthologue is induced by vernalization in winter cultivars but is expressed at high levels irrespective of vernalization status in spring cultivars (Preston and Kellogg, 2008). Additionally, analysis of FT1 expression patterns across the Pooid clade suggests that long-day induction of FT1 was also a feature of the common ancestor of this group of grasses (Ream et al., 2014; McKeown et al., 2016). If the molecular networks controlling the seasonal flowering behaviour of oat are similar to those of wheat and barley, a key question then becomes; does variation for vernalization requirement or photoperiod sensitivity map to oat homologues of VRN1 or FT?
Several studies describe genetic mapping of oat loci that contribute to variation in the timing of flowering (Wight et al., 1994; Holland et al., 1997; Holland et al., 2002; Portyanko et al., 2005; Locatelli et al., 2006; Wooten et al., 2009; Nava et al., 2012; Herrmann et al., 2014; Esvelt Klos et al., 2016; Sunstrum et al., 2019). These studies used a range of mapping populations, including recombinant inbred lines and diversity panels, which were genotyped using different marker systems. Phenotyping was conducted in diverse conditions including controlled environments, with different vernalization or daylength treatments, and at a range of field locations with different sowing dates across different years. Many of these studies linked phenological variation to chromosomal locations where VRN1 or FT1 are likely to be located according to cross-species comparisons of genetic maps. For example, Holland et al. (2002) suggested the marker BCD808b, which is linked to variation in vernalization responsiveness, is located near a copy of the VRN1 gene. Another study amplified VRN1 and FT1 gene sequences directly from oat, identified nucleotide polymorphisms in these sequences, and then used these to assign potential locations for these genes on genetic maps (Nava et al., 2012). This study also concluded that VRN1 is linked to variation in oat phenology.
The assembly of hexaploid (AACCDD) oat reference genome sequences (Maughan et al., 2019; Kamal et al., 2022; Peng et al., 2022) now allows direct comparisons between the physical position of candidate phenology genes and the locations of genetic markers linked to variation in flowering behaviour. These comparisons support the idea that VRN1 and FT1 are likely to underlie variation in oat phenology, with VRN1 homeoalleles on chromosomes 4D, 7A and 7D, and FT1 homeoallele on 7A all linked to phenological variation (Tinker et al., 2022). FT1 is also potentially linked to the DAYLENGTH INSENSITIVITY1 (Di1) gene, on chromosome 7D, which reduces daylength sensitivity by accelerating flowering in short days and has been used to breed rapid cycling oats for Canada and Brazil (Burrows, 1984; Locatelli et al., 2006; Tinker et al., 2022).
It should be noted that there are other candidate genes within the chromosomal regions of interest outlined above. For example, PHYC and an oat homologue of HEADING DATE 6 (HD6) (Takahashi et al., 2001) are co-located with VRN1. This is consistent with the suggestion of Sunstrum et al. (2019) that the genetic interval where VRN1 is located might combine effects from multiple genes, including genes that influence flowering in a vernalization-independent manner. Alternative candidate genes have also been suggested for the region linked to Di1, including a CONSTANS-like gene. (Chaffin et al., 2016; Sunstrum et al., 2019; Tinker et al., 2022). This region is difficult to resolve because marker-trait relationships are inconsistent across populations and there is also evidence for reduced recombination rates (Chaffin et al., 2016; Sunstrum et al., 2019; Tinker et al., 2022). Nevertheless, VRN1 and FT1 genes are promising candidates for more detailed analyses, while PHYC and HD6 also warrant further characterisation.
In summary, variation in oat phenology has been mapped to chromosomal regions containing genes related to those that control vernalization and daylength sensitivity in other cereals. Detailed understanding into the nature of the variation in these genes, and how this affects gene function and overall flowering behaviour, is not yet available.
A deeper understanding of the role that oat phenology genes play in adaptation can contribute to the future success of Australian oats through breeding and by contributing to on farm management decision-making. Additionally, there is potential to use phenology genes to optimise plant architecture and thereby increase crop yield.
A first step towards developing an increased understanding of the genetic basis of oat phenology is to identify “major genes” that control vernalization requirement and photoperiod sensitivity. Current knowledge allows the genetic intervals containing these genes to be identified (see above). Future studies will focus on validating and further characterising “causal genes”, as has been achieved in wheat and barley. A number of high-resolution mapping approaches can be used to achieve this aim, but ultimately a test of gene function, such as mutagenesis or gene editing, will be required to verify candidate gene functions (e.g., Boden et al., 2015). Additionally, as has been the case in wheat and barley, detailed analysis of gene expression patterns in diverse oat varieties, in response to different vernalization or photoperiod treatments, can provide further insights into the potential roles of candidate phenology genes.
Already there is scope for surveys of genetic diversity using high-throughput sequencing, either of whole genomes or by targeting candidate genes in the regions already identified by genetic mapping and cross-species comparisons. Sequencing of oat pangenomes (https://wheat.pw.usda.gov/GG3/PanOat) will further accelerate this approach. The collected pedigrees of oat varieties, which have been recorded and curated by breeders over many decades (Tinker and Deyl, 2005), are a valuable resource for surveying genetic diversity of major phenology genes. Access to pedigrees allows diversity surveys to be targeted to accessions and cultivars that represent key lines from the history of oat breeding, including key founders and breeding parents spanning the entire period from development of early modern varieties to current elite cultivars (see Eagles et al., 2009). Understanding the genetic basis of phenology and adaptation of oats that were bred for different regions is a logical approach to understand adaptation to local climate and farming practices.
An example of an oat pedigree is presented for Mitika, a South Australian milling oat that was registered in 2003 (Figure 3). This example highlights how pedigrees can be used to identify the donors of key traits into a local breeding program, including breeding line OT207 for the DWARF6 (DW6) reduced height gene, the Canadian cultivars Terra for the NAKED1 gene for hull-less grain and Dumont for crown rust (Puccinia coronata) resistance genes Pc38 and Pc39 (Brown et al., 1980; McKenzie et al., 1984; Ubert et al., 2017). The pedigree of Mitika also shows extensive mixing of Australian and North American germplasm. Also noteworthy is that all ancestors shown are described as spring types. This suggests that there is potential to introduce more variation for vernalization requirement into Australian milling oats to suit specific environments or farming systems opportunities, such as earlier sowing and dual-purpose crops (graze and grain).
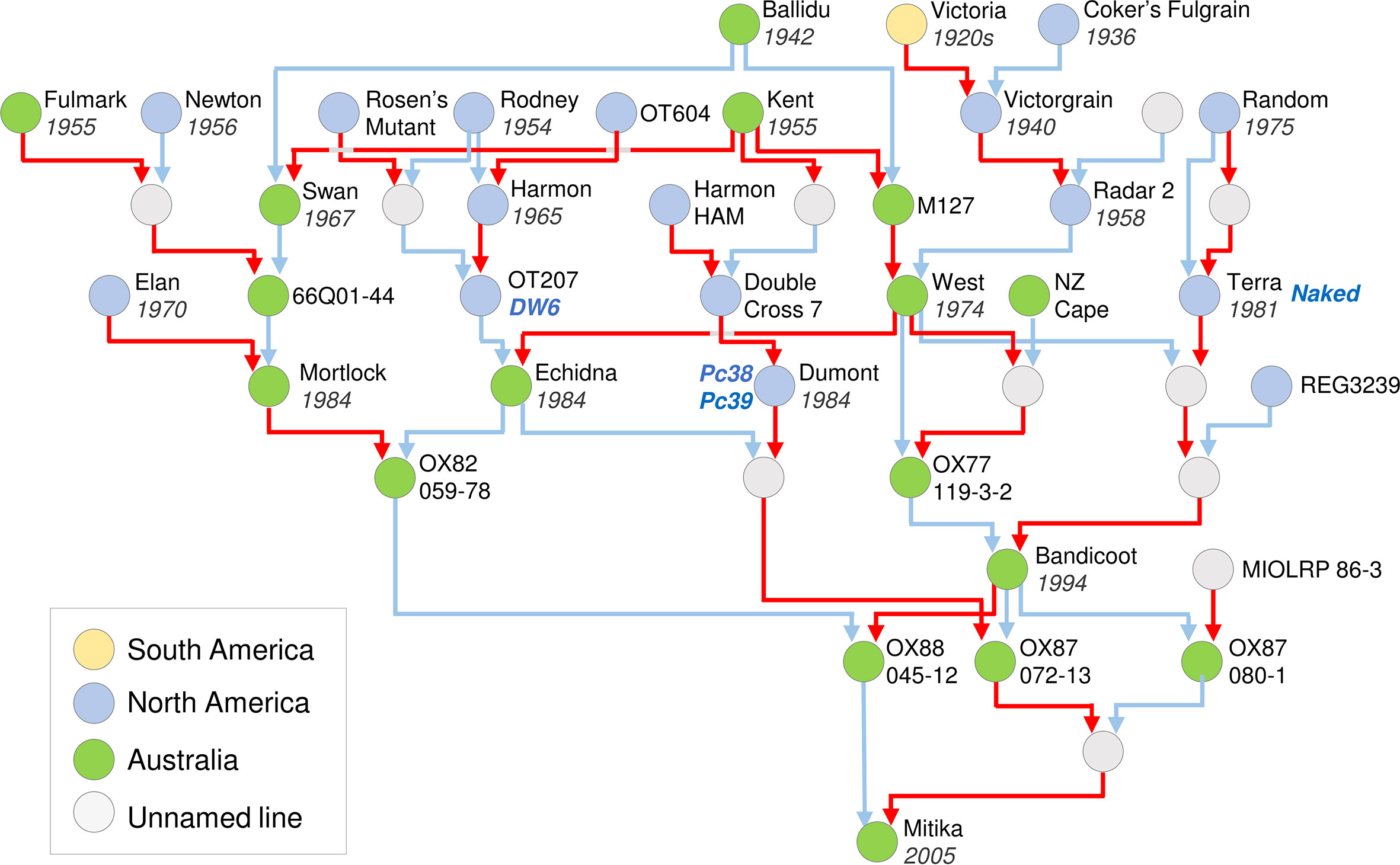
Figure 3 Pedigree of an elite Australian spring oat cultivar, cv. Mitika. A simplified pedigree of cultivar Mitika, modified from a Helium output (Shaw et al., 2014) generated from the pedigrees of over 1000 international accessions spanning diversity relevant to the history of Australian oat breeding. Pedigrees are shown as far back as early 20th century oats from Australian and North America. For ease of representation, multiple rounds of backcrossing are not shown. Pedigrees were obtained from the “Pedigrees of Oat Lines” POOL database (Tinker and Deyl, 2005), from Fitzsimmons et al. (1983) and directly from oat breeders (Dr Pamela Zwer and Dr Bruce Winter, personal communication). The year of release is indicated for cultivars. Red line indicates maternal parent connection and blue indicates paternal parent (pollen donor). Colours of circles indicate region of origin for released lines. Grey circles indicate unnamed intermediate lines used in crossing.
Knowledge of diversity in the major phenology genes can be used in breeding to facilitate choice of parents with compatible phenology for crossing and/or to select progeny of crosses, using molecular markers. Based on knowledge of wheat and barley there is potential for multiple functional alleles of major phenology genes (i.e., many variants of each gene) that have different impacts on phenology and/or yield (Eagles et al., 2009; Hemming et al., 2009; Eagles et al., 2010; Eagles et al., 2011; Díaz et al., 2012). Molecular markers should ideally have the capacity to resolve different functional haplotypes or alleles (Eagles et al., 2010; Eagles et al., 2011). Design of such markers requires high-resolution assays of genetic variation at key loci, using gene resequencing or high-density genome-wide Single Nucleotide Polymorphism (SNP) assays, for example. There then needs to be functional understanding of how different haplotypes influence phenology. Development of near-isogenic lines, by recurrent backcrossing, is a reliable way to contrast the effects of different alleles on phenology; different haplotypes identified by diversity surveys can be introgressed into a common genetic background for phenotyping in different controlled and field conditions (e.g., Hunt et al., 2019). Ideally a successful modern oat that performs well in target environments would be chosen as the recurrent parent that provides the genetic background for near-isogenic lines (e.g., Mitika or Bannister for Australia). Since oat is transformable there is also potential to develop and test novel variation in phenology using gene editing (Gasparis et al., 2008; Matres et al., 2021). Ideally this will target modern elite cultivars that are most relevant to the grains industry.
If the impact of different alleles of the major phenology genes can be resolved then a key question becomes; how can detailed knowledge of individual genes derived from reductionist approaches be re-integrated to understand, predict or optimise overall crop performance? This is a complex challenge that needs to consider both gene-gene and gene-environment interactions. There is also a “sparse data” challenge because detailed phenological observations will only ever be available for specific genotypes grown at particular field sites or sowing dates, representing only a subset of all the scenarios that potentially exist. One effective approach to address these challenges is to integrate genetic knowledge into physiology-based predictive models, such as Agricultural Production Systems Simulation (APSIM, Keating et al., 2003; Zheng et al., 2013). Predictive tools like APSIM can use genetic knowledge to help inform on-farm decision-making. For example, genotypes for vernalization and photoperiod genes can be incorporated into simulation models that predict the date of flowering for specific cultivars, from different sowing dates. Combined with climate data, these predictions can be used by farmers to select sowing dates for cultivars that minimise frost or heat risk. (Zheng et al., 2013).
The research strategies outlined above were tried and tested in wheat and barley research over a period of more than two decades. Advances in genomics, phenomics and biological data-science allow an alternative “genome-to-phenome” approach to be applied to understand phenology and adaptation of oats (Figure 4). Core to this approach is to survey genetic diversity at high-resolution and genome-wide scale in a representative population that captures the flow of alleles through the history of oat breeding, spanning founders, international imports, key parents and modern elite cultivars. The advent of high-resolution oat genotyping-by-sequencing assays can facilitate this (Bekele et al., 2018; Bekele et al., 2020). Another approach is to generate population-scale transcriptomes from key organs or timepoints in the plant lifecycle. Transcriptome data can be used to detect SNP variation in the coding regions of genes that are likely to function in the relevant tissues/timepoints of interest and can also assay variation in gene expression states across the genome, in different accessions (Xu et al., 2015). By phenotyping the same panel under controlled conditions (long versus short days, with or without vernalization), together with industry relevant field trials, it is possible to link variation in phenology to chromosomal regions. This generates data at scale that can be used to resolve gene-by-environment interactions through the application of emerging analytical tools, such as machine learning. Access to marker-trait associations for phenology at genome-wide scale in controlled environments that resolve variation for discrete aspects of phenology (i.e., vernalization and daylength requirements) also allows genomic parameterisation of simulation models or other data-driven prediction methods. Specifically, marker-trait association data can be used to set vernalization and daylength response parameters in APSIM to predict the flowering behaviour of individual oat cultivars. The same marker-trait association data can then be utilised in crop breeding through genomic selection/prediction. A “genome-to-phenome” strategy could be used alongside a more traditional approach and has the advantage that the genomic resources developed can be used to explore other aspects of oat biology.
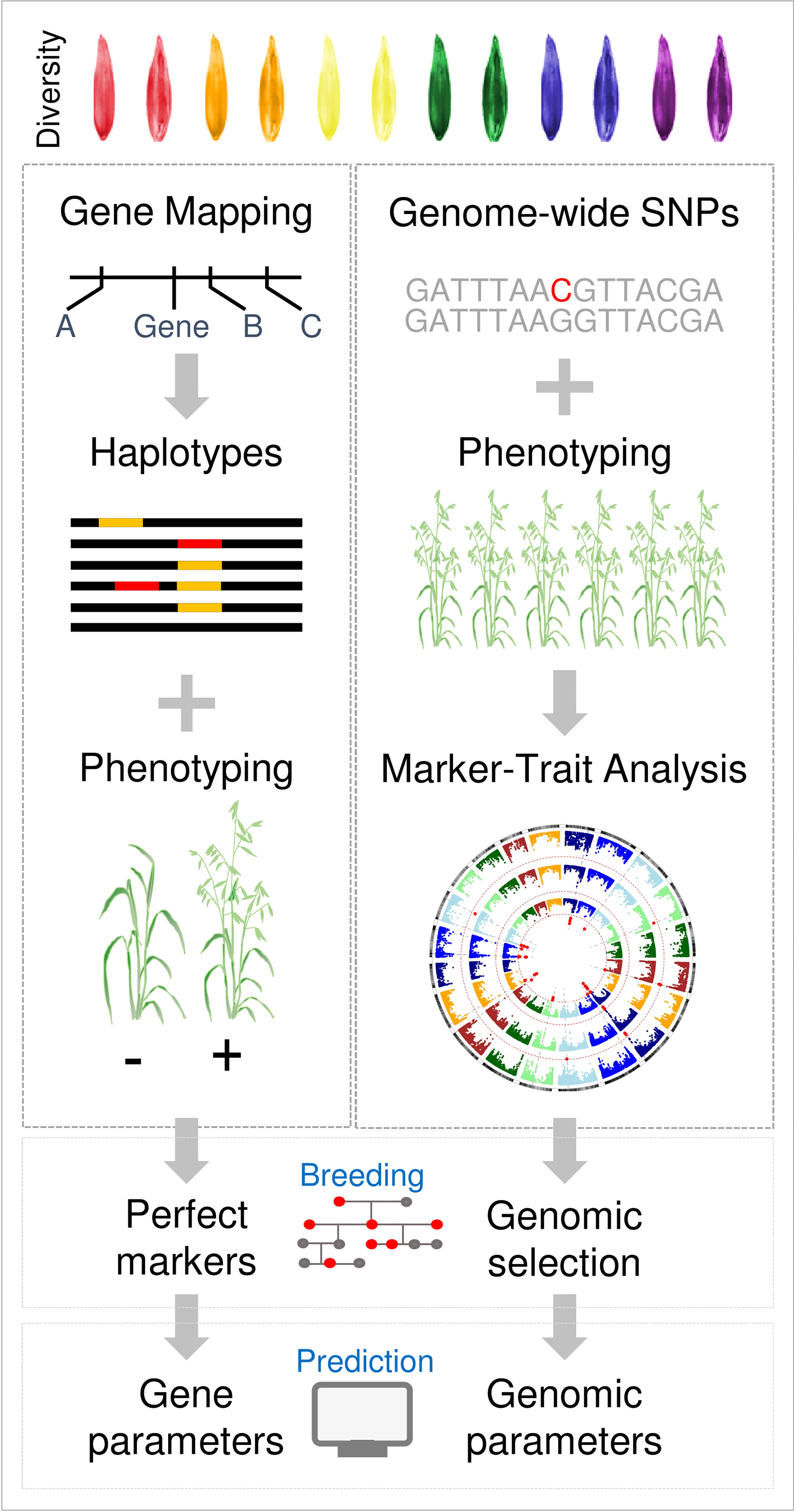
Figure 4 Overview of future oat phenology research strategies. A traditional strategy, based on approaches applied to wheat and barley, is compared with a “genome-to-phenome” strategy. In the traditional approach individual phenology traits are mapped to genes, using bi-parental mapping populations, for example. Knowledge of variation in these genes is then assembled by gene resequencing and used to design molecular markers for genotyping. In parallel, bespoke genetic resources such as near-isogenic lines or induced mutants are used to test gene function. In the genome-to-phenome strategy diversity panels are used to link genome-wide genetic diversity to broad phenotypic variation (phenology in this case). Marker-trait associations are then used for genomic prediction or to inform crop simulation models that predict phenology.
Variation in phenology underlies adaptation of oat to climates, farming systems and end uses. Rapid progress in the development of genomic resourcess and the assembly of an oat reference genome sequence is accelerating progress towards understanding genes that underlie variation in phenology. Comparison of the physical locations of genetic markers linked to variation phenology with the likely locations of candidate phenology genes in the oat genome suggests that VRN1 and FT1 contribute to natural variation in oat phenology. There is now a need for detailed studies of the expression of these genes in diverse oat varieties, in response to different vernalization or photoperiod treatments, together with functional validation by mutagenesis or gene editing. Other future priorities will be to determine how haplotypes of phenology genes contribute to variation in oat phenology and to overall crop performance, by generating near-isogenic lines, for example. This knowledge can then be incorporated into genomic selection strategies, which can be used in crop breeding, and into phenology prediction tools that can be used to inform on-farm decision-making.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was jointly funded by the Grains Research and Development Corporation and CSIRO, project code CSP2007.
The authors wish to thank Dr Howard Eagles for his guidance and mentorship over several years. We also thank Dr Pamela Zwer and Dr Bruce Winter for providing pedigree information.
The authors declare that this study received funding from the Grains Research and Development Corporation (GRDC). The funder was not involved in the study design, collection, analysis interpretation of data, the writing of this article or the decision to submit it for publication.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ABARES (2021). Australian Crop report, Australian bureau of agricultural and resource economics and sciences. (Canberra) doi: 10.25814/xqy3-sx57
Aitken, Y. (1974). Flowering time, climate and genotype: the adaptation of agricultural species to climate through flowering responses (Melbourne: Melbourne University Press).
Beales, J., Turner, A., Griffiths, S., Snape, J. W., Laurie, D. A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum l.). Theor. Appl. Genet. 115, 721–733. doi: 10.1007/s00122-007-0603-4
Bekele, W. A., Itaya, A., Boyle, B., Yan, W., Fetch, J. M., Tinker, N. A. (2020). A targeted genotyping-by-sequencing tool (RAPTURE) for genomics-assisted breeding in oat. Theor. Appl. Genet. 133, 653–664. doi: 10.1007/s00122-019-03496-w
Bekele, W. A., Wight, C. P., Chao, S., Howarth, C. J., Tinker, N. A. (2018). Haplotype based genotyping-by-sequencing in oat genome research. Plant Biotechnol. J. 16, 1452–1463. doi: 10.1111/pbi.12888
Bell, G. H. D. (1936). Experiments on vernalization. J. Agric. Sci. 26:155–171. doi: 10.1017/S0021859600021869
Boden, S., Cavanagh, C., Cullis, B., Ramm, K., Greenwood, J., Finnegan, E. J., et al. (2015). Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat. Plant 1 (2), 14016. doi: 10.1038/nplants.2014.16
Boden, S. A., Weiss, D., Ross, J. J., Davies, N. W., Trevaskis, B., Chandler, P. M., et al. (2014). EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS t expression. Plant Cell 26, 1557–1569. doi: 10.1105/tpc.114.123794
Bonnett, O. T. (1966). Development of the oat panicle, in inflorescences of maize, wheat, rye, barley and oats: their initiation and development. Bulletin Illinois Agric. Exp. Station 721, 92–102.
Brassac, J., Muqaddsi, Q. H., Plieske, J., Ganal, M. W., Röder, M. S. (2021). Linkage mapping identifies a non-synonymous mutation in FLOWERING LOCUS T (FT-B1) increasing spikelet number per spike. Sci. Rep. 11, 1585. doi: 10.1038/s41598-020-80473-0
Brown, P. D., McKenzie, R. I. H., Mikaelsen, K. (1980). Agronomic, genetic, and cytologic evaluation of a vigorous new semidwarf oat. Crop Sci. 20, 303–306. doi: 10.2135/cropsci1980.0011183X002000030003x
Campoli, C., Pankin, A., Drosse, B., Casoa, C. M., Davis, S. J., von Korff, M. (2013). HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 199, 1045–1059. doi: 10.1111/nph.12346
Chaffin, A. S., Huang, Y. F., Smith, S., Bekele, W. A., Babiker, E., Gnanesh, B. N., et al. (2016). A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial subgenome rearrangement. Plant Genome 9 (2). doi: 10.3835/plantgenome2015.10.0102
Chen, Y., Carver, B. F., Wang, S., Zhang, F., Yan, L. (2009). Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor. Appl. Genet. 118, 881–889. doi: 10.1007/s00122-008-0946-5
Chen, A., Dubcovsky, J. (2012). Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PloS Genet. 8, e1003134. doi: 10.1371/journal.pgen.1003134
Chen, A., Li, C., Hu, W., Lau, M. L., Lin, H., Rockwell, N. C., et al. (2014). PHYTOCHROME c plays a major role in the acceleration of wheat flowering under long day photoperiod. Proc. Natl. Acad. Sci. U.S.A. 111, 10037–10044. doi: 10.1073/pnas.1409795111
Coblentz, W. K., Bertram, M. G., Martin, N. P., Berzaghi, P. (2012). Planting date effects on the nutritive value of fall-grown oat cultivars. Agron. J. 104, 312–323. doi: 10.2134/agronj2011.0273
Collins, D., King, G. D. (1798). “An account of the English colony in new south wales. London,” in: (London: Cadell and Davies in the Strand) Available at: http://gutenberg.net.au/ebooks/e00010.html.
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Cuddeford, D. (1995). ““Oats for animal feed”,” in The oat crop: production and utilization. Ed. Welch, R. W. (Dordrecht: Springer), 321–368. doi: 10.1007/978-94-011-0015-1_11
Danyluk, J., Kane, N. A., Breton, G., Limin, A. E., Fowler, D. B., Sarhan, F. (2003). TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 132, 1849–1860. doi: 10.1104/pp.103.023523
Díaz, A., Zikhali, M., Turner, A. S., Isaac, P., Laurie, D. A. (2012). Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PloS One 7 (3), e33234. doi: 10.1371/journal.pone.0033234
Dixon, L. E., Farré, A., Finnegan, E. J., Orford, S., Griffiths, S., Boden, S. A. (2018). Developmental responses of bread wheat to changes in ambient temperature following deletion of a locus that includes FLOWERING LOCUS T1. Plant Cell Environ. 41, 1715–1725. doi: 10.1111/pce.13130
Dixon, L. E., Karsai, I., Kiss, T., Adamski, N. M., Liu, Z., Ding, Y., et al. (2019). VERNALIZATION1 controls developmental responses of winter wheat under high ambient temperatures. Development 146, dev172684. doi: 10.1242/dev.172684
Dolferus, R., Ji, X., Richards, R. A. (2011). Abiotic stress and control of grain number in cereals. Plant Sci. 181, 331–341. doi: 10.1016/j.plantsci.2011.05.015
Eagles, H. A., Cane, K., Kuchel, H., Hollamby, G. J., Vallance, N., Eastwood, R. F., et al. (2010). Photoperiod and vernalization gene effects in southern Australian wheat. Crop Past. Sci. 61, 721–730. doi: 10.1071/CP10121
Eagles, H. A., Cane, K., Neil, V. (2009). The flow of alleles of important photoperiod and vernalisation genes through Australian wheat. Crop Past. Sci. 60, 646–657. doi: 10.1071/CP09014
Eagles, H. A., Cane, K., Trevaskis, B. (2011). Veery wheats carry an allele of Vrn-A1 that has implications for freezing tolerance in winter wheats. Plant Breed. 130, 413–418. doi: 10.1071/CP09014
Endo-Higashi, N., Izawa, T. (2011). Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol. 52, 1083–1094. doi: 10.1093/pcp/pcr059
Esvelt Klos, K., Huang, Y. F., Bekele, W. A., Obert, D. E., Babiker, E., Beattie, A. D., et al. (2016). Population genomics related to adaptation in elite oat germplasm. Plant Genome 9 (2). doi: 10.3835/plantgenome2015.10.0103
Faure, S., Turner, A. S., Gruszka, D., Christodoulou, V., Davis, S. J., von Korff, M., et al. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. U.S.A. 109, 8328–8333. doi: 10.1073/pnas.1120496109
Feng, Y., Yin, Y., Fei, S. (2017). BdVRN1 expression confers flowering competency and is negatively correlated with freezing tolerance in Brachypodium distachyon. front. Plant Sci. 8. doi: 10.3389/fpls.2017.01107
Finkner, V. C., Ponoleit, C. G., Davis, D. L. (1973). Heritability of rachis node number of Avena sativa l. Crop Sci. 13, 84–85. doi: 10.2135/cropsci1973.0011183X001300010026x
Finnegan, E. F., Ford, B., Wallace, X., Pettolino, F., Griffith, P., Schmitz, R. J., et al. (2018). Zebularine treatment is associated with deletion of FT-B1 leading to an increase in spikelet number in bread wheat. Plant Cell Environ. 41, 1346–1360. doi: 10.1111/pce.13164
Fitzsimmons, R. W. (1988). Pridham, John Theodore (1879-1954) Australian Dictionary of Biography, Volume 11, 329 (Melbourne University Press).
Fitzsimmons, R. W., Roberts, G. L., Wrigley, C. W. (1983). Australian Oat varieties (Melbourne: CSIRO Publishing). doi: 10.1071/9780643105447
Fjellheim, S., Boden, S., Trevaskis, B. (2014). The role of seasonal flowering responses in adaptation of grasses to temperate climates. Front. Plant Sci. 14. doi: 10.3389/fpls.2014.00431
Flohr, B. M., Hunt, J. R., Kirkegaard, J. A., Evans, J. R. (2017). Water and temperature stress define the optimal flowering period for wheat in south-eastern Australia. Field Crops Res. 209, 108–119. doi: 10.1016/j.fcr.2017.04.012
Fu, D. L., Szucs, P., Yan, L. L., Helguera, M., Skinner, J. S., von Zitzewitz, J., et al. (2005). Large Deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Gen. Genom. 273, 54–65. doi: 10.1007/s00438-004-1095-4
Gasparis, S., Bregier, C., Orczyk, W., Nadolska-Orczyk., A. (2008). Agrobacterium-mediated transformation of oat (Avena sativa l.) cultivars via immature embryo and leaf explants. Plant Cell Rep. 27, 1721–1729. doi: 10.1007/s00299-008-0593-y
Gauley, A., Boden, S. A. (2021). Stepwise increases in FT1 expression regulate seasonal progression of flowering in wheat (Triticum aestivum). New Phytol. 229, 1163–1176. doi: 10.1111/nph.16910
Gawroński, P., Ariyadasa, R., Himmelbach, A., Poursarebani, N., Kilian, B., Stein, N., et al. (2014). A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the eps-3Am mutant of einkorn wheat. Genetics 196, 1253–1261. doi: 10.1534/genetics.113.158444
Grau Nersting, L., Bode Andersen, S., von Bothmer, R., Jorgensen, R. B. (2006). Morphological and molecular diversity of Nordic oat through one hundred years of breeding. Euphytica 150, 327–337. doi: 10.1007/s10681-006-9116-5
Harlan, J. R. (1982). ““Relationships between weeds and crops”,” in Biology and ecology of weeds. Eds. Holzner, M., Numata, M. (W. Junk Publishers: The Hague), 91–96.
Hemming, M. N., Fieg, S., Peacock, W. J., Dennis, E. S., Trevaskis, B. (2009). Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol. Gen. Genom. 282, 107–117. doi: 10.1007/s00438-009-0449-3
Hemming, M. N., Peacock, W. J., Dennis, E. S., Trevaskis, B. (2008). Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS t in barley. Plant Physiol. 147, 355–366. doi: 10.1104/pp.108.116418
Herrmann, M. H., Yu, J., Beuch, S., Weber, W. E. (2014). Quantitative trait loci for quality and agronomic traits in two advanced backcross populations in oat (Avena sativa l.). Plant Breed. 133, 588–601. doi: 10.1111/pbr.12188
Holland, J., Moser, H., O'Donoughue, L., Lee, M. (1997). QTLs and epistasis associated with vernalization responses in oat. Crop Sci. 37, 1306–1316. doi: 10.2135/cropsci1997.0011183X003700040047x
Holland, J., Portyanko, A., Hoffman, L., Lee, M. (2002). Genomic regions controlling vernalization and photoperiod responses in oat. Theor. Appl. Genet. 105, 113–126. doi: 10.1007/s00122-001-0845-5
Howarth, C. J., Martinez, P. M. J., Cowan, A. A., Griffiths, I. A., Lister, S. J., Langdon, T., et al. (2021). Genotype and environment affect the grain quality and yield of winter oats (Avena sativa l.). Foods 10, 2356. doi: 10.3390/foods10102356
Hunt, J., Lilley, J., Trevaskis, B., Flohr, B., Peake, A., Fletcher, A., et al. (2019). Early sowing systems can boost Australian wheat yields despite recent climate change. Nat. Clim. Change 9 (3), 244–247. doi: 10.1038/s41558-019-0417-9
Hyles, J., Bloomfield, M. T., Hunt, J. R., Trethowan, R. M., Trevaskis, B. (2020). Phenology and related traits for wheat adaptation. Heredity 125, 417–430. doi: 10.1038/s41437-020-0320-1
Islam, M. R., Egrinya Eneji, A., Changzhomg, R., Yuegao, H., Gong, C., Xuzhang, X. (2010). Oat-based cropping system for sustainable agricultural development in arid regions of northern China. J. Agriculture Biotechnol. Ecol. 3, 1–8.
Jenkins, G. (1973). The effect of sowing date and photoperiod on panicle morphology in naked oats (Avena nuda). Ann. Appl. Biol. 73, 85–94. doi: 10.1111/j.1744-7348.1973.tb01312.x
Kamal, N., Tsardakas Renhuldt, N., Bentzer, J., Gundlach, H., Haberer, G., Juhász, A., et al. (2022). The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 606, 113–119. doi: 10.1038/s41586-022-04732-y
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
Keating, B. A., Carberry, P. S., Hammer, G. L., Probert, M. E., Robertson, M. J., Holzworth, D., et al. (2003). An overview of APSIM, a model designed for farming systems simulation. Eur. J. Agron. 18, 267–288. doi: 10.1016/S1161-0301(02)00108-9
King, S. R., Bacon, R. K. (1992). Vernalization requirement of winter and spring oat genotypes. Crop Sci. 32, 677–680. doi: 10.2135/cropsci1992.0011183X003200030019x
Ki-Seung, K., Tinker, N., Newell, M. (2014). Improvement of oat as a winter forage crop in the southern united states. Crop Sci. 54, 1336–1346. doi: 10.2135/cropsci2013.07.0505
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Lippi, M. M., Foggi, B., Aranguren, B., Ronchitelli, A., Revedin, A. (2015). Multistep food plant processing at grotta paglicci (Southern Italy) around 32,600 cal B.P. Proc. Natl. Acad. Sci. U.S.A. 112, 12075–12080. doi: 10.1073/pnas.1505213112
Locatelli, A. B., Federizzi, L. C., Milach, S. C. K., Wight, C. P., Molnar, S. J., Chapados, J. T., et al. (2006). Loci affecting flowering time in oat under short-day conditions. Genome 49, 1528–1538. doi: 10.1139/g06-108
Loscos, J., Igartua, E., Contreras-Moreira, B., Gracia, M., Casas, A. (2014). HvFT1 polymorphism and effect–survey of barley germplasm and expression analysis. Front. Plant Sci. 5. doi: 10.3389/fpls.2
Matres, J. M., Hilscher, J., Datta, A., Armario-Nájera, V., Baysal, C., He, W., et al. (2021). Genome editing in cereal crops: an overview. Transgenic Res. 30, 461–498. doi: 10.1007/s11248-021-00259-6
Maughan, P. J., Lee, R., Walstead, R., Vickerstaff, R. J., Fogarty, M. C., Brouwer, C. R., et al. (2019). Genomic insights from the first chromosome-scale assemblies of oat (Avena spp.) diploid species. BMC Biol. 22, 92. doi: 10.1186/s12915-019-0712-y.014.00251
McKenzie, R. I. H., Brown, P. D., Martens, J. W., Harder, D. E., Nielsen, J., Gill, C. C., et al. (1984). Registration of Dumont oats. Crop Sci. 24, 207. doi: 10.2135/cropsci1984.0011183X002400010051x
McKeown, M., Schubert, M., Marcussen, T., Fjellheim, S., Preston, J. C. (2016). Evidence for an early origin of vernalization responsiveness in temperate pooideae grasses. Plant Physiol. 172, 416–426. doi: 10.1104/pp.16.01023
Mengersen, F. (1960). Oat improvement in N.S.W. i. early history and development. II. development of new varieties in southern New South Wales. Agric. Gazette NSW 71, 449–461.
Mizuno, N., Nitta, M., Sato, K., Nasuda, S. (2012). A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotype to einkorn wheat. Genes Genet. Syst. 87, 357–367. doi: 10.1266/ggs.87.357
Moore-Colyer, R. J. (1995). “Oats and oat production in history and pre-history”,” in The oat crop: production and utilization. Ed. Welch, R. W. (Dordrecht: Springer), 1–33. doi: 10.1007/978-94-011-0015-1_1
Murphy, J. P., Hoffman, L. A. (1992). ““The origin, history, and production of oat”,” in Oat science and technology. Eds. Marshall, H. G., Sorrells, M. E. (Madison: Wiley), 1–28. doi: 10.2134/agronmonogr33.c1
Nava, I. C., Wight, C. P., Pacheco, M. T., Federizzi, L. C., Tinker, N. A. (2012). Tagging and mapping candidate loci for vernalization and flower initiation in hexaploid oat. Mol. Breed. 30, 1295–1312. doi: 10.1007/s11032-012-9715-x
Nitcher, R., Pearce, S., Tranquilli, G., Zhang, X., Dubcovsky, J. (2014). Effect of the hope FT-B1 allele on wheat heading time and yield components. J. Heredity 105, 666–675. doi: 10.1093/jhered/esu042
Oliver, S. N., Deng, W., Casao, M. C., Trevaskis, B. (2013). Low temperatures induce rapid changes in chromatin state and transcript levels of the cereal VERNALIZATION1 gene. J. Exp. Bot. 64, 2413–2422. doi: 10.1093/jxb/ert095
Peltonen-Sainto, P. (1994). Response to daylength in oats: pre-anthesis development and set of spikelets and florets. J. Agron. Crop Sci. 172, 104–112. doi: 10.1111/j.1439-037X.1994.tb0053x
Peltonen-Sainto, P., Juahianen, L. (2020). Large Zonal and temporal shifts in crops and cultivars coincide with warmer growing seasons in Finland. Regional Environ. Change 20, 89. doi: 10.1007/s10113-020-01682-x
Peng, Y., Yan, H., Guo, L., Deng, C., Wang, C., Wang., Y., et al. (2022). Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 54, 1248–1258. doi: 10.1038/s41588-022-01127-7
Portyanko, V. A., Chen, G., Rines, H. W., Phillips, R. L., Leonard, K. J., Ochocki, G. E., et al. (2005). Quantitative trait loci for partial resistance to crown rust, puccinia coronata, in cultivated oat, Avena sativa l. Theor. Appl. Genet. 111, 313–324. doi: 10.1007/s00122-005-2024-6
Preston, J. C., Kellogg, E. A. (2008). Discrete developmental roles for temperate cereal grass VERNALIZATION1/FRUITFULL-like genes in flowering competency and the transition to flowering. Plant Physiol. 146, 265–276. doi: 10.1104/pp.107.109561
Pugsley, A. T. (1971). A genetic analysis of the spring-winter habit of growth in wheat. Aust. J. Agric. Res. 22, 21–31. doi: 10.1071/AR9710021
Ream, T. S., Woods, D. P., Schwartz, C. J., Sanabria, C. P., Mahoy, J. A., Walters, E. M., et al. (2014). Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol. 164, 694–709. doi: 10.1104/pp.113.232678
Sampson, D. R., Burrows, V. D. (1972). Influence of photoperiod, short-day vernalization, and cold vernalization on days to heading in Avena species and cultivars. Can. J. Plant Sci. 52, 471–482. doi: 10.4141/cjps72-077
Sasani, S., Hemming, M. N., Oliver, S., Greenup, A., Tavakkol-Afshari, R., Mahfoozi, S., et al. (2009). The influence of vernalization and daylength cues on the expression of flowering-time genes in the leaves and shoot apex of barley (Hordeum vulgare). J. Exp. Bot. 60, 2169–2178. doi: 10.1093/jxb/erp098
Shaw, P. D., Graham, M., Kennedy, J., Milne, I., Marshall, DF (2014). Helium: visualization of large scale plant pedigrees. BMC Bioinf. 15, 259. doi: 10.1186/1471-2105-15-259
Sorrells, M. E., Simmons, S. R. (1992). “Influence of environment on the development and adaptation of oat”, in Oat science and technology. Eds. Marshall, H. G., Sorrells, M. E. (Madison: Wiley), 115–163. doi: 10.2134/agronmonogr33.c1
Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Sunstrum, F. G., Bekele, W. A., Wight, C. P., Yan, W., Chen, Y., Tinker, N.A (2019). A genetic linkage map in southern-by-spring oat identifies multiple quantitative trait loci for adaptation and rust resistance. Plant Breed. 138, 82–94. doi: 10.1111/pbr.12666
Takahashi, Y., Shomura, A., Sasaki, T., Yano, M. (2001). Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc. Natl. Acad. Sci. U.S.A. 98, 7922–7927. doi: 10.1073/pnas.111136798
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Tashiro, T., Wardlaw, I. F. (1990). The response to high temperature shock and humidity changes prior to and during the early stages of grain development in wheat. Aust. J. Plant Physiol. 17, 551–561. doi: 10.1071/PP9900551
Tinker, N. A., Deyl, J. K. (2005). A curated internet database of oat pedigrees. Crop Sci. 45, 2269–2272. doi: 10.2135/cropsci2004.0687
Tinker, N. A., Wight, C. P., Bekele, W. A., Yan, W., Jellen, E. N., Tsardakus Renhuldt, N., et al. (2022). Genome analysis in Avena sativa reveals hidden breeding barriers and opportunities for oat improvement. Commun. Biol. 5, 474. doi: 10.1038/s42003-022-03256-5
Trevaskis, B. (2018). Developmental pathways are blueprints for designing successful crops. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00745
Trevaskis, B., Bagnall, D. J., Ellis, M. H., Peacock, W. J., Dennis, E. S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. U.S.A. 100, 13099–13104. doi: 10.1073/pnas.1635053100
Trevaskis, B., Hemming, M. N., Dennis, E. S., Peacock, W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357. doi: 10.1016/j.tplants.2007.06.010
Turner, A., Beales, J., Faure, S., Dunford, R. P., Laurie, D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
Ubert, I. P., Zimmer, C. M., Pellizzaro, K., Federizzi, L. C., Nava, I. C. (2017). Genetics and molecular mapping of the naked grains in hexaploid oat. Euphytica 213:41. doi: 10.1007/s10681-017-1836-1
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
von Zitzewitz, J., Szucs, P., Dubcovsky, J., Yan, L. L., Francia, E., Pecchioni, N., et al. (2005). Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 59, 449–467. doi: 10.1007/s11103-005-0351-2
Wiggans, S. C., Frey, K. J. (1955). Photoperiodism in oats. Proc. Iowa Acad. Sci. 62, 125–130. Available at: https://scholarworks.uni.edu/pias/vol62/iss1/13
Wight, C. P., Penner, G. A., O’Donoughue, L. S., Burrows, V. D., Molnar, S. J., Fedak, G. (1994). The identification of random amplified polymorphic DNA (RAPD) markers for daylength insensitivity in oat. Genome 37, 910–914. doi: 10.1139/g94-130
Woods, D. P., Ream, T. S., Bouché, F., Lee, J., Thrower., N., Wilkerson, C., et al. (2017). Establishment of a vernalization requirement in Brachypodium distachyon requires REPRESSOR OF VERNALIZATION1. Proc. Natl. Acad. Sci. U.S.A. 114, 6623–6628. doi: 10.1073/pnas.1700536114
Wooten, D. R., Livingston, D. P., Lyerly, H. J., Holland, J. B., Jellen, E. N., Marshall, D. S., et al. (2009). Quantitative trait loci and epistasis for oat winter-hardiness component traits. Crop Sci. 49, 1989–1998. doi: 10.2135/cropsci2008.10.0612
Xu, Q., Xing, S., Zhu, C., Liu, W., Fan, Y., Wang, Q., et al. (2015). Population transcriptomics reveals a potentially positive role of expression diversity in adaptation. J. Int. Plant Biol. 57, 284–299. doi: 10.1111/jipb.12287
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103, 19581–19586. doi: 10.1073/pnas.0607142103
Yan, L., Helguera, M., Kato, K., Fukuyama, S., Sherman., J., Dubcovsky, J. (2004a). Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor. Appl. Genet. 109, 1677–1686. doi: 10.1007/s00122-004-1796-4
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004b). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. doi: 10.1126/science.1094305
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100, 6263–6268. doi: 10.1073/pnas.0937399100
Yan, W., Molnar, S. J., Fregeau-Reid, J., McElroy, A., Tinker, N. A. (2007). Associations among oat traits and their responses to the environment. J. Crop Improvement 20, 1–29. doi: 10.1300/J411v20n01.01
Zadoks, J. C., Chang, T. T., Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x
Zakhrabekova, S., Gough, S. P., Braumann, I., Müller, A. H., Lundqvist, J., Ahmann, K., et al. (2012). Induced mutations in circadian clock regulator mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. U.S.A. 109, 4326–4331. doi: 10.1073/pnas.1113009109
Zheng, B., Biddulph, B., Li, D., Kuchel, H., Chapman, S. (2013). Quantification of the effects of VRN1 and Ppd-D1 to predict spring wheat (Triticum aestivum) heading time across diverse environments. J. Exp. Bot. 64, 3747–3761. doi: 10.1093/jxb/ert209
Zikhali, M., Wingen, L. U., Griffiths, S. (2016). Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum). J. Exp. Bot. 67, 287–929. doi: 10.1093/jxb/erv458
Keywords: oat, flowering, seasons, vernalization, photoperiod
Citation: Trevaskis B, Harris FAJ, Bovill WD, Rattey AR, Khoo KHP, Boden SA and Hyles J (2022) Advancing understanding of oat phenology for crop adaptation. Front. Plant Sci. 13:955623. doi: 10.3389/fpls.2022.955623
Received: 29 May 2022; Accepted: 20 September 2022;
Published: 14 October 2022.
Edited by:
Liang Wu, Zhejiang University, ChinaReviewed by:
Koen Geuten, KU Leuven, BelgiumCopyright © 2022 Trevaskis, Harris, Bovill, Rattey, Khoo, Boden and Hyles. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ben Trevaskis, YmVuLnRyZXZhc2tpc0Bjc2lyby5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.