
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 August 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.955590
This article is part of the Research TopicAdvances in Genetics and Molecular Breeding of Ornamental PlantsView all 12 articles
Paeonia rockii is well-known for its distinctive large dark-purple spot at the white petal base and has been considered to be the main genetic source of spotted tree peony cultivars. In this study, the petal base and petal background of Paeonia ostii (pure white petals without any spot), P. rockii, and other three tree peony cultivars were sampled at four blooming stages from the small bell-like bud stage to the initial blooming stage. There is a distinct difference between the pigmentation processes of spots and petal backgrounds; the spot pigmentation was about 10 days earlier than the petal background. Moreover, the cyanin and peonidin type anthocyanin accumulation at the petal base mainly contributed to the petal spot formation. Then, we identified a C1 subgroup R2R3-MYB transcription factor, PrMYB5, predominantly transcribing at the petal base. This is extremely consistent with PrDFR and PrANS expression, the contents of anthocyanins, and spot formation. Furthermore, PrMYB5 could bind to and activate the promoter of PrDFR in yeast one-hybrid and dual-luciferase assays, which was further verified in overexpression of PrMYB5 in tobacco and PrMYB5-silenced petals of P. rockii by comparing the color change, anthocyanin contents, and gene expression. In summary, these results shed light on the mechanism of petal spot formation in P. rockii and speed up the molecular breeding process of tree peony cultivars with novel spot pigmentation patterns.
Petal spot is a kind of regular variegated pattern of flower color, which exists in angiosperm, such as Nomocharis meleagrina, Torenia fournieri, Sparaxia elegans, Dendrobium nobile, and Paeonia suffruticosa (Gao et al., 2015; Zhang et al., 2015; Shi et al., 2017; Su et al., 2017; Gu et al., 2019). It not only imparts ornamental and commercial values but also affects the preferences of pollinators and promotes the success of pollination (Davies et al., 2012; Glover et al., 2013), so the molecular mechanism of petal spot formation has always been a research hotspot.
The formation of petal spots is determined by the specific spatiotemporal transcription of genes in the anthocyanin biosynthesis pathway (ABP), resulting in specific anthocyanins accumulation in defined petal regions (Suzuki et al., 2016). In ABP, chalcone synthase (CHS), chalcone isomerase (CHI), flavanoid 3'-hydroxylase (F3'H), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and flavonoid glucosyltransferase (UFGT) are mainly controlled by MYB transcription factor (TF), basic helix–loop–helix (bHLH), and WD40 protein in different plants (Cao et al., 2020; Yan et al., 2021). To date, the spatio-temporal regulation mechanisms involved in petal spot formation have been explored in many ornamental plants, including Antirrhinum spp., Petunia hybrid, Lilium spp., Mimulus spp., Clarkia gracilis, and Phalaenopsis spp. (Schwinn et al., 2006; Albert et al., 2011; Shang et al., 2011; Yamagishi, 2011, 2020; Yamagishi et al., 2010; Glover et al., 2013; Yuan et al., 2014; Hsu et al., 2015; Martins et al., 2017). In the same way, CgMYB1 activated CgDFR2 and CgANS to produce petal spots in C. gracilis (Martins et al., 2017). However, the specific regulation mechanism of petal spot is rarely studied in woody ornamental plants, especially wild tree peony species.
Tree peony is one of the most famous traditional flowers in China and is popular all over the world for its large, colorful, and distinct flowers. At present, there are more than 2,000 cultivars worldwide with various petal pigmentation patterns, such as spots, stripes, and more complex designs (Zhou et al., 2014). Paeonia rockii is the most important wild tree peony species in Northwest China and has a large dark-purple spot at the white petal base, which has been confirmed as a main genetic source of spot pigmentation in tree peony (Wang et al., 2000; Shi et al., 2017). Paeonia ostii has pure white petals without any spots, and other tree peony cultivars of P. suffruticosa, “Lanhudie” (LHD), “Mochi Jinhui” (MCJH), and {High Noon” (HN), possess different degrees of purple spots on differentially colored petal backgrounds, which can be considered as the control group for the spot pigmentation study of P. rockii. Therefore, it is urgent to explore the molecular mechanism of petal spot pigmentation of P. rockii to accelerate the molecular breeding process of a variegated tree peony in China. Previous research demonstrated that the spot formation at the petal base of tree peony was primarily attributed to the spatial biosynthesis of cyanidin (Cy) and peonidin (Pn) anthocyanins (Zhang et al., 2015; Shi et al., 2017). The anthocyanin structural genes and anthocyanin regulatory MYBs have been identified in tree peony cultivars, including PsMYB114L, PsMYB12L, PsMYB12, PsMYB111, PsMYB4, PsMYB57, and PsMYB58 (Gu et al., 2019; Zhang et al., 2019, 2020a,b; Luo et al., 2021). Among them, PsMYB12 directly activated the expression of PsCHS by forming an MBW regulatory complex with bHLH and WD40 proteins, which were specific to the petal blotch in tree peony cultivar “Qing Hai Hu Yin Bo” (Gu et al., 2019). However, whether and how other MYBs regulated the petal spot formation in P. rockii are almost unknown.
Previously comparative transcriptome analyses of P. rockii and P. ostii flowers suggested that CHS, DFR, ANS, glutathione S-transferase (GST), and two R2R3-MYB TFs might be related to petal spot formation in P. rockii (Shi et al., 2017). In the present study, we investigated one anthocyanin regulatory R2R3-MYB TF PrMYB5 in P. rockii and analyzed the expression patterns at different parts of petals during flower opening of P. rockii, P. ostii, and other three tree peony cultivars. Furthermore, we conducted gene overexpression in tobacco plants, virus-induced gene silencing (VIGS), and RNA in situ hybridization to verify that PrMYB5 can individually regulate the distinct petal pigmentation in P. rockii by activating PrDFR expression, resulting in anthocyanin accumulation. The results aim to provide valuable resources and theoretical supports for molecular breeding of variegation cultivars in tree peony and supply references for the studies on the spot formation mechanism in other woody plants.
The plants of P. rockii, P. ostii, and three tree peony cultivars P. suffruticosa, “Lanhudie,” “Mochi Jinhui,” and “High Noon,” were grown in the germplasm repository of the Northwest A&F University, Shaanxi, China. Petal backgrounds and petal bases at four different blooming stages were sampled separately from March to April, 2018 (Figure 1). The four blooming stages include: stage 1 (S1): a small bell-like bud stage when the petals are mainly yellow-green; stage 2 (S2): a large bell-like bud stage when the petal bases turn obvious color with the yellow-green background; stage 3 (S3): a bell-like bud extending stage when spots get bigger and darker; and stage 4 (S4): an initiating blooming stage when spots are completely formed (Figure 1). Tobacco plants (Nicotiana tabacum L. cv. K326) were grown in a conservatory under 16 h/8 h light photoperiod at 25°C. The color indices (L*, a*, b*, C, and h) were measured in the fresh petals at four stages using a colorimeter (CR-400, Konica Minolta, Osaka, Japan). The other materials were immediately frozen in liquid nitrogen and then stored at −80°C.
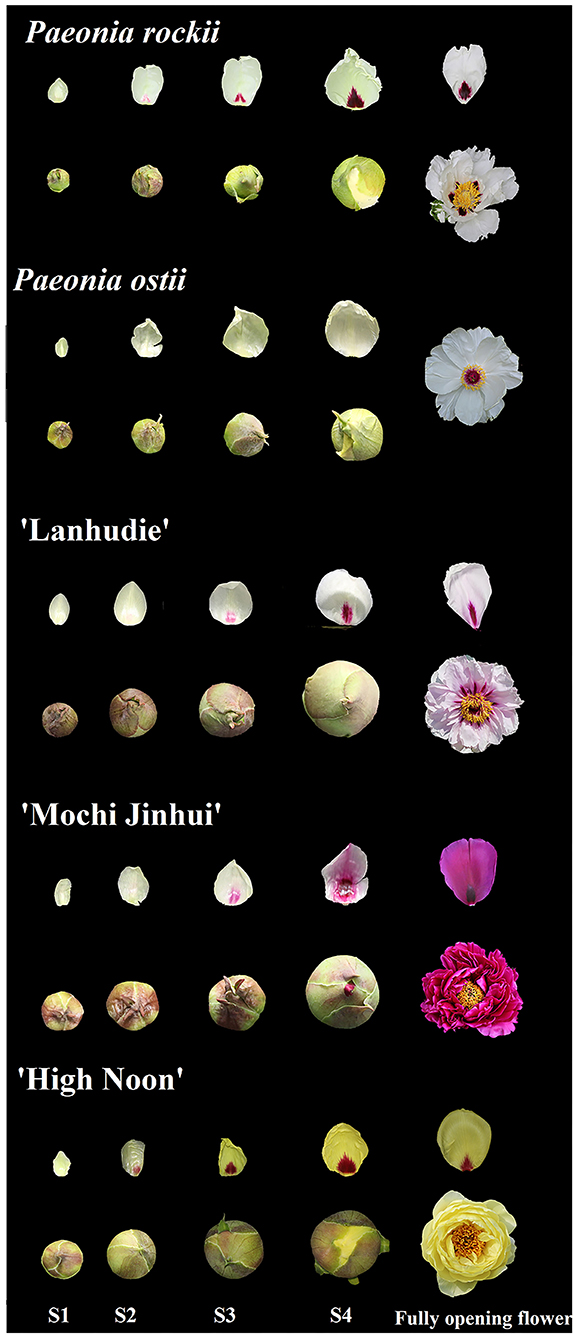
Figure 1. Flower phenotypes of Paeonia rockii, P. ostii, P. suffruticosa “Lanhudie,” “Mochi Jinhui,” and “High Noon” at four different opening stages. S1: stage 1, small bell-like bud stage when the petals were mainly yellow-green; S2: stage 2, large bell-like bud stage when the petal bases turn obvious color with the yellow-green background; S3: stage 3, bell-like bud extending stage when spots get bigger and darker; S4: stage 4, initiating blooming stage when spots are completely formed.
Flavonoids were detected in tree peony petals and tobacco petals using the high-performance liquid chromatography (HPLC) method, as previously reported (Luo et al., 2021). First, each sample was ground into powder in liquid nitrogen. Then, 1 mg of petal powder was extracted with 2 ml methanol-formic acid (99:1, V/V) at 4°C for 24 h in the dark. After ultrasonic-assisted extraction for 30 min and centrifugation at 10,000 rpm for 10 min, liquid supernatant was gathered and filtered using a 0.22 μm nylon microporous membrane. Flavonoid analysis was done as follows: eluent A: 0.04% formic acid; eluent B: acetonitrile. Elution procedure: 1–40 min: 5–40% B; 40–45 min, 40–100% B; 45–55 min, 100%B; 55–60 min, 100–5% B. Flow rate: 0.5 mL ∙min−1, column temperature: 40°C, injection volume: 10 μL. We used Cy3G, Cy3G5G, Pg3G, Pg3G5G, Pn3G, Pn3G5G, Ap, Ch, Is, Km, Lu, and Qu from the Shanghai Yuanye Bio-Technology Co., Ltd (Shanghai, China) as standards. All assays were conducted with three biological replicates.
Total RNA was extracted from 100 mg petal base of P. rockii at S4 using the Quick RNA Isolation Kit (Bioteke Corporation, Beijing, China). After RNA purity and integrity were checked, full-length cDNAs of PrMYB5 were cloned using the SMARTer RACE 5'/3' Kit (TAKARA Corporation, Beijing, China) according to the manufacturer's protocols. The primers are listed in Supplementary Table 1.
The multiple sequence alignment and phylogenetic tree of PrMYB5 and some anthocyanin biosynthesis and spot formation related R2R3-MYB proteins from tree peony and other plants were performed using the DNAMAN6.0.3.99 software and the MEGA 6.0 software using the neighbor-joining method, respectively.
The PrMYB5 ORF was cloned into the pC2300-GFP vector with the KpnIand SalI restriction sites using In-Fusion Cloning and then transformed into the onion epidermal cells through particle bombardment experiment. After onion tissues were cultured at 25°C for 24 h in the dark, we used a laser confocal microscope (Leica TCS SP2) to observe the fluorescence. All primers are listed in Supplementary Table 1.
RNA in situ hybridization was conducted according to the previously reported method (Li et al., 2018). The stage 4 petal background and petal base of P. rockii were fixed in 4% paraformaldehyde at 4°C for 24 h. After being dehydrated through a graded ethanol series and then embedded in paraffin, the petals were sectioned into 10 μm thick and followed by dewaxing with xylene, rehydrating through an ethanol series, pretreating with proteinase K (1μg mL−1) in 1×PBS at 37°C for 30 min, prehybriding and hybriding followed previous protocols: two times of 1× SSC at 45°C for 20 min and two times of 0.5× SSC at 42°C for 15 min (Hsu et al., 2015; Li et al., 2018). The antisense and sense probes of PrMYB5 were synthesized using gene-specific primers with T7 and SP6 RNA polymerase-binding sites, which are shown in Supplementary Table 1.
Total RNAs were extracted and reverse transcribed into first-strand cDNA for real-time quantitative PCR (qRT-PCR) using the PrimeScriptTM RT Master MIX reverse transcription kit (TaKaRa, Beijing, China). qRT-PCR was conducted using SYBR® Premix Ex TaqTM II (Perfect Real Time) Kit (TaKaRa, Beijing, China) on a Bio-Rad CFX96TM Real-Time system (Bio-Rad, Hercules, CA, USA) according to the reaction conditions mentioned previously (Luo et al., 2021). The transcription levels of genes were normalized to the PsUbiquitin expression level and calibrated to the expression level in P. ostii petal background at S1. Three biological replicates and technical replicates were performed for each reaction. All primers are listed in Supplementary Table 1.
Based on the transcripts of PrMYB5 and the structural genes and flavonoid components, a network was established using the R package (https:// www. r- project.org) with a coefficient of R ≥ 0.5 or R ≤ −0.5. The hub genes were determined to be co-expressed genes with strong interconnections. Finally, the network was visualized using Cytoscape (v.3.1.0; Shannon et al., 2013).
The ORF of PrMYB5 was cloned into the pCAMBIA1300 vector, and the constructs were transformed into tobacco using the previously described method (Horsch and Klee, 1986). After T0 generation transgenic tobacco was obtained, we screened overexpressed-PrMYB5 transgenic lines (OE-PrMYB5) using PCR and collected seeds until the T2 generation OE-PrMYB5 plants were obtained. Finally, three T2 generation OE-PrMYB5 lines were selected to detect color indices, quantify flavonoid levels, and qRT-PCR verification as described above. All primers are listed in Supplementary Table 1.
According to the previous VIGS approach (Luo et al., 2022), about 200 bp fragment at the 3' end of PrMYB5 ORF was cloned into the TRV2 vector digested with KpnI and XhoI. The TRV2-PrMYB5 construct, TRV1, and TRV2 empty vectors were transferred into Agrobacterium strain GV3101. Then, the mixtures with Agrobacterium containing TRV1 and that containing TRV2, TRV2-PrMYB5 at a ratio of 1:1 were cultured for 4 h at 25°C in the dark, and then infected fresh petal discs with 1.2 cm diameter from the center of petal background and petal base of P. rockii at S4 through vacuum infiltration (Luo et al., 2022). After vacuuming for 2 days, petal discs were washed with deionized water and collected for further analysis. All primers are listed in Supplementary Table 1.
The promoters of PrDFR and PrANS were cloned from the S4 petal genomic DNA of P. rockii using the Genome Walking Kit (TAKARA, Beijing, China). Based on the mRNA sequences of PrDFR and PrANS, three reverse primers, SP1, SP2, and SP3, were used in three rounds of PCR for PrDFR and PrANS promoters cloning, respectively (Supplementary Table 1). The forward primer was AP4 in all PCR reactions, which was supplied in the kit. cis-elements analysis was detected using the online software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al., 2002).
First, the PrMYB5 ORF was inserted into the pGADT7 vector (SmaI and SacI restriction sites) to obtain pGADT7-PrMYB5. The PrDFRpro and PrANSpro were cloned into the pHIS2 vector with EcoR I and Sac I restriction sites. Then, all constructs co-transformed pairwise into yeast strain AfHY187 according to the lithium acetate method (Zhang et al., 2018). All primers used in Y1H are shown in Supplementary Table 1.
The PrDFRpro and PrANSpro were cloned into the pGreenII 0800-LUC vector between Sal I and Hind III restriction sites to obtain PrANSpro-pGreenII 0800-LUC and PrDFRpro-pGreenII 0800-LUC constructs, respectively. In the meantime, the ORF of PrMYB5 was inserted into the pGreenII 62-SK vector using BamH I and EcoR I restriction sites to obtain the PrMYB5-pGreenII 62-SK construct. All primers are listed in Supplementary Table 1. After transforming into Agrobacterium strain GV3101, the mixtures of cultures (PrMYB5-SK+PrANS-0800, PrMYB5-SK+PrDFR-0800) were infiltrated into N. benthamiana leaves with needleless syringes (Zhang et al., 2018). After 72 h infiltration, firefly luciferase (LUC) and renilla luciferase (REN) were assayed. The LUC to REN ratio was calculated to measure the binding activity of PrMYB5 to the promoters. Each combination was conducted in three biological replicates and three technical replicates.
Petal basal spots began to emerge at the small bell-like bud stage (S1) when petals were not colored and fully formed until the blooming stage (S4). However, petals began to color from the bell-like bud extending stage (S3) after 10 days (Figure 1). The same phenomenon also appeared in other tree peony cultivars with different petal spot patterns (Figure 1).
Then, the color indices of petal tissues of P. rockii (PR), P. ostii (PO), “Lanhudie” (LHD), “Mochi Jinhui” (MCJH), and “High Noon” (HN) were measured at S1–S4 (Supplementary Figure 1). According to the Royal Horticultural Society Color Chart (RHSCC), the background color of PR at S1–S4 was the same as that of PO petals. The petal base colors of PR and PO were green at S1, and then the petal base color of PR varied greatly from green to dark purple at S2–S4 (Supplementary Table 2). At the petal bases of PR, a* (redness) value raised gradually from S1 to S4, while b* (yellowness) value decreased; the L* (lightness) values peaked at S2, and then subsequently declined from S3 to S4 (Supplementary Figure 1A); h (hue angle) and C* (chroma) decreased from S1 to S3, and then increased at S4; h-value at S4 was ~ 0° (360°), consistent with the dark-purple color (Supplementary Figure 1A). However, color indices of petal bases in PO changed slightly during flower opening, which were similar to those of petal background in PO and PR (Supplementary Figure 1B). Furthermore, the color of petal spots at the S4 stage in LHD, MCJH, and HN were moderate purplish-red, dark purple, and strong red, respectively (Supplementary Table 2). Similarly, L* and b* values declined, and conversely, a* value increased from S1 to S4 at the petal base of LHD, MCJH, and HN (Supplementary Figures 1C–E). The values of h and C* were all consistent with their petal base colors (Supplementary Figures 1C–E).
Significant differences in flavonoid compositions and contents were measured in petal backgrounds and bases of PR, PO, LHD, MCJH, and HN during petal spot formation (Figure 2). Generally, flavones and flavonols were predominantly accumulated at the petal background of white and yellow flowers, including kaempferol (Km), apigenin (Ap), isorhamnetin (Is), luteolin (Lu), and chrysoeriol (Ch), which also occurred at petal bases at S1. Anthocyanins were abundantly accumulated from S2 and reached the highest peak at S4. For pink and purple flowers, the anthocyanin accumulation in petal spots was relatively higher than that of petal backgrounds. During dark-purple petal spot formation in PR and MCJH, peonidin 3, 5-O-glucoside (Pn3G5G), cyanidin 3, 5-O-glucoside (Cy3G5G), and cyanidin 3-O-glucoside (Cy3G) were the dominated anthocyanins and most abundant at S4. In addition, Pn3G5G and Cy3G were considered as the major anthocyanins and accumulated more and more abundantly during petal spots formation of LHD, whereas peonidin 3-O-glucoside (Pn3G) and Pn3G5G in HN. In contrast, flavonoids were accumulated similarly in petal bases with petal background in PO and PR (Figure 2).
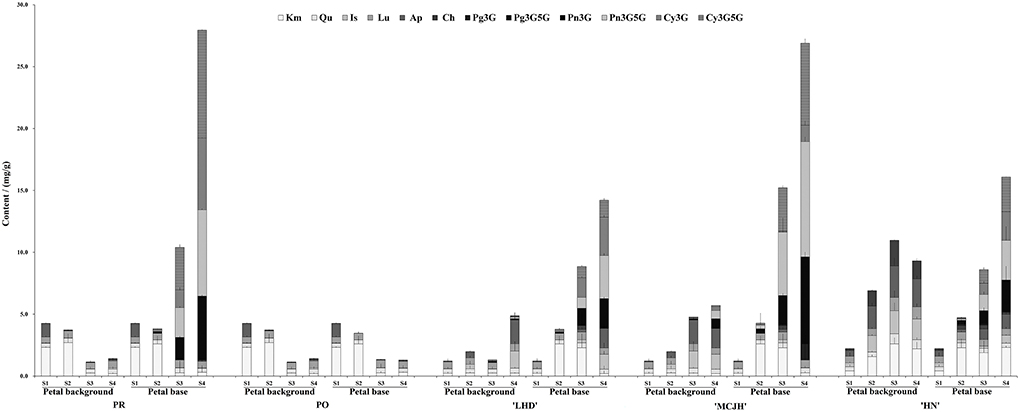
Figure 2. The contents of flavonoids in petal background and base of P. rockii (PR), P. ostii (PO), and three tree peony cultivars “Lanhudie” (LHD), “Mochi Jinhui” (MCJH), and “High Noon” (HN) during petal spot formation. S1–S4 represent four different blooming stages. Cy3G, cyanidin 3-O-glucoside; Cy3G5G, cyanidin 3, 5-O-glucoside; Pg3G, pelargonidin 3-O-glucoside; Pg3G5G, pelargonidin 3, 5-O-glucoside; Pn3G, peonidin 3-O-glucoside; Pn3G5G, peonidin 3, 5-O-glucoside; Km, kaempferol; Qu, quercetin; Lu, luteolin; Is, isorhamnetin; Ap, apigenin; Ch, chrysoeriol.
In our previous flower transcriptome database of P. rockii (Shi et al., 2017), an R2R3-MYB TF related to anthocyanin regulation named PrMYB5 (Genbank accession number: c117848) was obtained. PrMYB5 contained an 828 bp open reading frame (ORF), encoding a protein of 275 amino acids (Supplementary Figure 2A). PrMYB5 contained highly conserved R2 and R3 domains at the N-terminus, indicating that it belonged to the typical R2R3-MYB family. Meanwhile, phylogenetic analysis showed PrMYB5 in the same clade as petal spot formation regulated MYBs, including OgMYB1 and PeMYB11, full-red pigmentation regulatory PeMYB2, venation pattern regulatory PeMYB12, and anthocyanin-promoting MYBs, such as Zea mays C1 (ZmC1), ZmPL, PsMYB12L, and PsMYB114L (Supplementary Figure 2B). Furthermore, we detected the fluorescence PrMYB5-GFP (green fluorescent protein) fusion protein in the nucleus of Allium cepa cell protoplasts, suggesting that PrMYB5 is a nuclear-localized TF in regulating petal spot formation (Supplementary Figure 2C).
RNA in situ hybridization in the petal background and petal base of P. rockii at S4 showed that PrMYB5 expressed more highly on the adaxial surface than on the abaxial surface of the petal base (Figure 3A). On the contrary, no expression of PrMYB5 was detected in the epidermal cells of the petal background (Figure 3D). These results were consistent with the transverse section phenotypes (Figures 3C,F). As a negative control, hybridization with sense probes was conducted (Figures 3B,E). Therefore, PrMYB5 showed distinct expression patterns in the petal background and petal base of P. rockii.
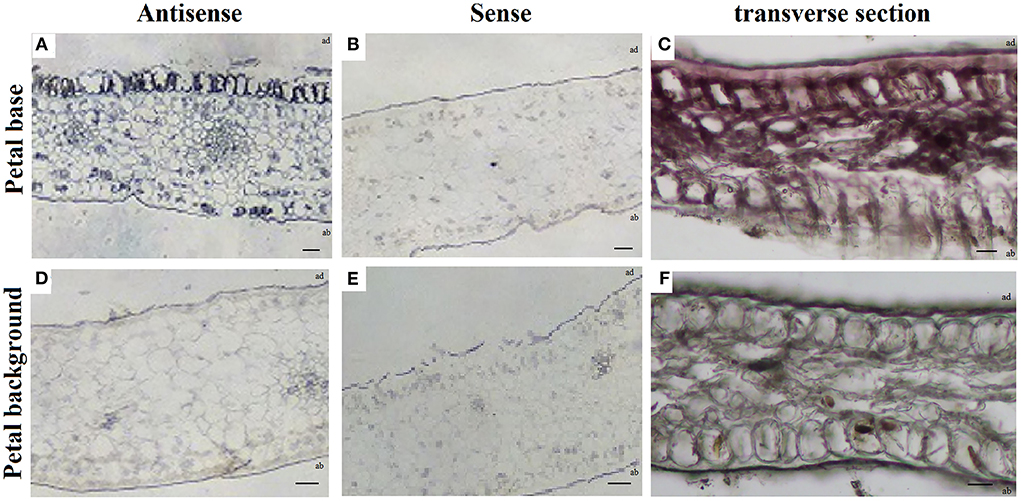
Figure 3. RNA in situ hybridization of PrMYB5 transcripts in the stage 4 petals of P. rockii. (A,B) Transverse sections of the petal base of P. rockii were hybridized with antisense or sense RNA probes of PrMYB5. (C) Transverse section of the petal base of P. rockii. (D,E) Transverse sections of the petal background of P. rockii were hybridized with antisense or sense RNA probes of PrMYB5. (F) Transverse sections of the petal background of P. rockii. ab indicates Abaxial surface; ad indicates Adaxial surfaces. Bars = 100 mm.
To further identify the function of PrMYB5 associated with the petal spot formation, we analyzed the expression patterns of PrMYB5 and four key structural genes (PrCHS, PrF3′H, PrDFR, and PrANS) that have been screened in our previous study (Shi et al., 2017). The expression levels of PrMYB5 at the petal spot increased first and then decreased and peaked at S3 in PR, which were significantly higher than those at the petal background of P. rockii all the time. Compared to PR, PrMYB5 is hardly expressed in all petals of PO (Figure 4A). In addition, the expression levels of PrMYB5 in LHD, MCJH, and HN increased continuously from S1 to S4 (Figure 4A), and the transcript of PrMYB5 at the petal base exhibited higher abundance than that at the petal background. In general, the expression level of PrMYB5 increased dramatically with the spot formation in spotted species and cultivars. Especially, it is expressed in the highest abundance in the petal base of MCJH with darker petal spot (Figure 4A).
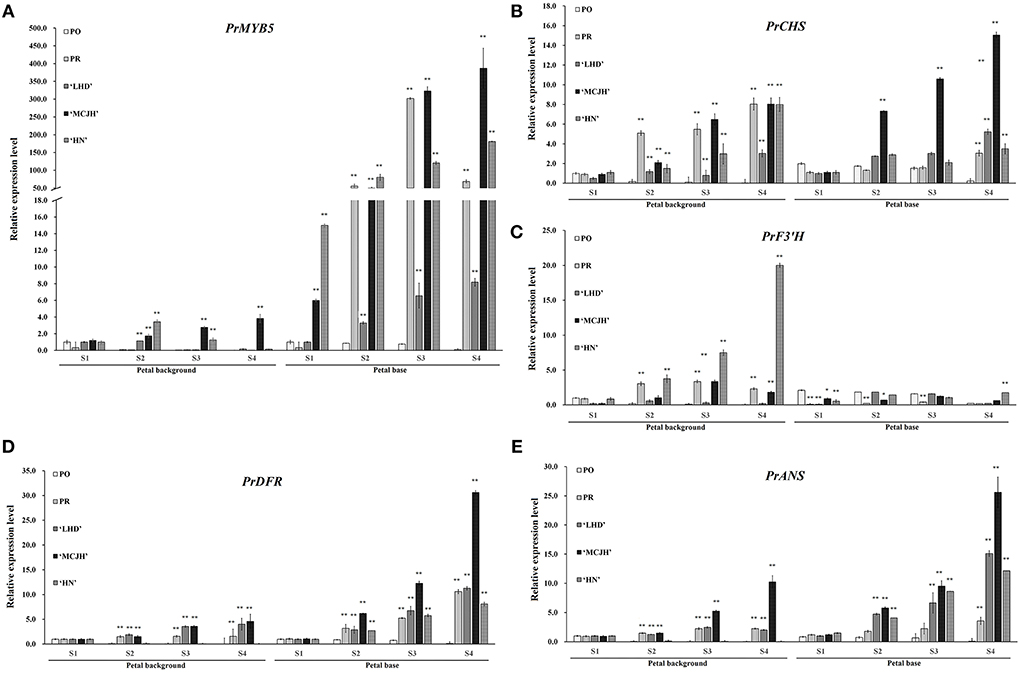
Figure 4. The expression profiles of PrMYB5 and four related structural genes in P. rockii (PR), P. ostii (PO), and three tree peony cultivars “Lanhudie” (LHD), “Mochi Jinhui” (MCJH) and “High Noon” (HN) using qRT-PCR. The expression profiles of PrMYB5 (A), PrCHS (B), PrF3'H (C), PrDFR (D), and PrANS (E) in all samples. S1–S4 represent four different blooming stages.
The transcripts of PrCHS and PrF3'H in petal background were higher than those at petal spot except for PrCHS in LHD and MCJH, whereas they displayed higher expression levels at petal base than petal background in PO (Figures 4B,C). However, PrDFR showed a gradually increasing expression trend at the petal bases of PR, LHD, MCJH, and HN from S1 to S4, which is similar with those of PrANS (Figures 4D,E). Especially, the expression levels of PrDFR were significantly more abundant at petal bases than those of PrANS from S1 to S4 in PR and MCJH. The interaction network based on flavonoid accumulation and gene expression profiles revealed that PrMYB5 was positively correlated with PrDFR and PrANS, directly associating with Pn3G5G and Cy3G5G (Supplementary Figure 3).
To characterize PrMYB5 function in anthocyanin biosynthesis, we overexpressed PrMYB5 in tobacco plants and obtained three PrMYB5 transgenic lines (OE-PrMYB5 1–3) (Figure 5). The petals of OE-PrMYB5 displayed deeper red color compared with wild-type plants, while the sepals, stamens, pistils, and leaves of transgenic lines had no obvious difference (Figure 5A). All OE lines displayed higher transcripts than WT (Figure 5A). Consistently, L* and b* values of corollas in all OE-PrMYB5 were observably lower than those in wild-type, whereas a* value was higher in all OE-PrMYB5 (Figure 5B). Furthermore, the corollas of three OE-PrMYB5 lines accumulated more Pg3G, Pn3G5G, Pn3G, Cy3G5G, and Cy3G than those of wild-type (Figure 5C), but not obviously different from other flavonoid components (Figure 5C). The above results indicate that PrMYB5 could significantly promote anthocyanin biosynthesis in tobacco.
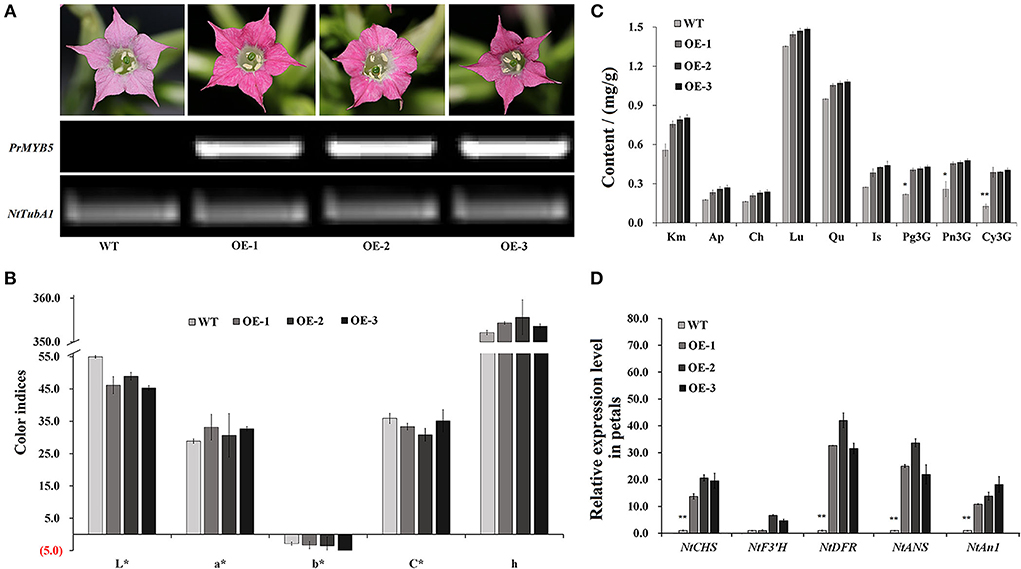
Figure 5. Overexpression of PrMYB5 promotes the accumulation of anthocyanins in tobacco. (A) Flower phenotypes of OE-lines (OE-1, 2, and 3) and wild-type (WT) petals, and their corresponding PrMYB5 transcription levels. NtTubA1 was used as an endogenous control. (B) The color indices in OE-lines and wild-type petals. L* represents the lightness. a* represents the redness. b* represents the yellowness. C* represents chroma. h represents the hue angle. (C) The contents of pigments in OE-lines and wild-type petals. (D) Expression profiles of PrMYB5 and structural genes in OE-lines petals by qRT-PCR. *Indicates significant differences at P < 0.05; **indicates relatively significant differences at P < 0.01.
The transcript of PrMYB5 was 70-fold higher in petals of OE-PrMYB5 than that in wild type, and those in leaves were also 26-fold more abundant (Figure 5D). Correspondingly, four structural genes were also upregulated in the petals of three OE-PrMYB5 lines. Particularly, NtDFR and NtANS exhibited the most significant high transcripts in petals of three transgenic lines. The bHLH TF NtAn1, stimulating the anthocyanin biosynthesis in tobacco, upregulated in the petals of all three transgenic lines. In short, PrMYB5 acted as a positive TF to activate anthocyanin accumulation in the petals.
After VIGS were conducted in petal discs of P. rockii, the colors of all PrMYB5-silenced petal base discs were visibly lighter than those of the blank-control and empty vector control (Figures 6A,B). The changes of flavonoid contents were mainly concentrated in down-accumulation of Km and Lu in PrMYB5-silenced petal background discs (Figure 6C), and the contents of Cy3G, Cy3G5G, Pn3G, and Pn3G5G observably decreased in PrMYB5-silenced petal base discs (Figure 6D). Correspondingly, PrMYB5 is down-regulated in PrMYB5-silenced petal background discs and base discs, accompanied by decreased expression levels of PrCHS, PrDFR, and PrANS (Figures 6E,F). It is noteworthy that PrDFR was significantly downregulated in PrMYB5-silenced petal base discs, indicating the direct positive regulatory relationship between PrMYB5 and PrDFR contributing to petal spot formation.

Figure 6. Transient silencing of PrMYB5 in P. rockii petals using virus gene silencing (VIGS). Phenotypes of petal background disc (A,B). The flavonoids accumulation in the petal background disc (C) and petal base disc (D). The expression levels of PrMYB5 and structural genes in petal background disc (E) and petal base disc (F). *Indicates significant differences at P < 0.05; **indicates relatively significant differences at P < 0.01).
To verify whether PrMYB5 is regulating PrDFR and PrANS, we cloned the promoter sequences of PrDFR (PrDFRpro) and PrANS (PrANSpro), with the length of 1,067 and 1,021 bp, respectively (Supplementary Figure 5). PrDFRpro included one MYB-binding site (5'-CAAC(A/G) G-3'), four MYB sites (5'-(T/C) AACC (A/G)-3'), two MYC sites (CA(A/T) (T/G) TG), and four G-box (5'-CACGT(T/G)-3'), while PrANSpro had two MYB sites, two MYC sites, and two G-box (5'-CACGT (T/G)-3') (Figure 7A), suggesting a potential regulatory relationship among PrMYB5, PrDFR, and PrANS.
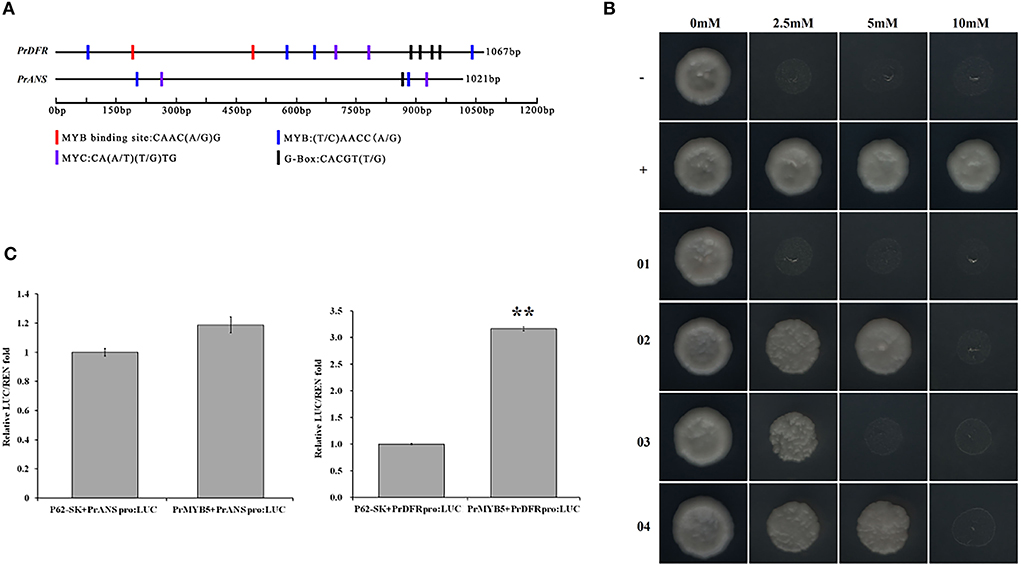
Figure 7. Transcriptional activity analysis of PrMYB5 against the promoters of PrDFR and PrANS. (A) Schematic overview of PrDFR and PrANS promoters. The lengths, MYB binding sites, MYB sites, MYC sites, and G-box sites are marked with black solid lines and colored rectangles, respectively. (B) Recombinant yeast on the SD medium without His, Leu, and Trp with 0 mM, 2.5 mM, 5 mM, and 10 mM indicate the SD medium without His, Leu, and Trp with 3-AT at concentrations of 0, 2.5, 5, and 10 mM, respectively. -, a negative control (pGADT7+p53::pHis2); +, a positive control (pGADT7::53+p53::pHis2); 01, pGADT7+PrDFR::pHis2; 02, PrMYB5::pGADT7+PrDFR::pHis2; 03, pGADT7+PrANS::pHis2; 04, PrMYB5::pGADT7+PrANS::pHis2. (C) Dual-luciferase assay in tobacco leaves. **Indicates relatively significant differences at P < 0.01.
To further verify whether PrMYB5 activated PrDFR or PrANS, we first performed yeast one-hybrid (Y1H) assays (Figure 7B). Compared with the corresponding control groups, the yeast cells co-transformed with PrMYB5::pGADT7+PrDFRpro::pHIS2 and PrMYB5::pGADT7+PrANSpro::pHIS2 could grow normally on SD/-His/-Leu/-Trp selective medium with 2.5 mM 3-AT and 10 mM 3-AT, respectively (Figure 7B). Then, a dual-luciferase assay was conducted. The relative luciferase level of PrMYB5::SK+PrDFR::LUC was more than 3-folds than the control (SK+PrDFR::LUC), while no obvious enhancement was found in PrMYB5::SK+PrANS::LUC. Taken together, PrMYB5 preferred directly activating the PrDFR promoter, promoting Cy- and Pn- anthocyanin at the petal base of P. rockii (Figure 7C).
The spot at the petal base of P. rockii mainly contributes to various spot patterns of tree peony (Wang et al., 2000; Shi et al., 2017). Pigmentation at the petal base begins much earlier than petal background pigmentation in tree peony (Figure 1, Supplementary Table 2), which is similar to those in snapdragons and lilies (Shang et al., 2011; Yamagishi et al., 2014). That is, petal spot pigmentation and petal background pigmentation may be independent.
Cy-based anthocyanins were the major cause of dark-purple spot, while Pn-based anthocyanins were for a vivid purple-red spot in tree peony (Shi et al., 2017; Gu et al., 2019). As expected, Cy3G, Cy3G5G, and Pn3G5G were the dominant anthocyanins in the dark-purple petal spot of PR and MCJH, whereas Pn3G5G and Cy3G in purplish red petal spots of LHD, and Pn3G5G and Pn3G in vivid red petal spots of HN (Figure 2). Whether the flower color is white, yellow, pink, or purplish red, the contents of anthocyanins were markedly higher in the petal spots, while flavones and flavonols accumulated at little different levels. Similar results have been found for petal spot formation in Viola × wittrockiana Gams. and C. gracilis (Martins et al., 2013; Li et al., 2014).
The biosynthesis of distinct Cy and Pn types of anthocyanins is regulated by structural genes and TFs (Suzuki et al., 2016). Among them, DFR and ANS catalyze the dihydroflavonols into the corresponding anthocyanins. Although they were expressed both in the red and purple colored petal areas, the expression levels of PrDFR in petal spot were observably different from those in the colored petal background, positively correlating with the accumulation of Pn3G5G and Cy3G5G (Figure 4; Supplementary Figure 3). Similar results in C. gracilis, Oncidium Gower Ramsey, and Viola × wittrockiana. Gams. showed that the different expression of DFR genes in petal spot activated anthocyanin production resulting in spot pattern (Chiou and Yeh, 2008; Martins et al., 2013; Li et al., 2014). Thus, the differences in anthocyanin accumulation between petal background and spot may be explained by the special spatial and temporal expression of PrDFR.
Among the TFs involved in anthocyanin biosynthesis, R2R3-MYB TFs play major roles in determining spot formation (Hsu et al., 2015). In this study, the R2R3-MYB anthocyanin regulator, PrMYB5 was grouped in the same clade with OgMYB1, PeMYB11, ZmC1, ZmPL, and PsMYB12L (Supplementary Figure 2B), of which ZmC1, ZmPL, OgMYB1, and PeMYB11 belong to C1 subgroup of R2R3-MYBs, which is involved in upregulating anthocyanin biosynthesis (Jiang et al., 2004; Kranz et al., 2010; Stracke et al., 2010). In addition, OgMYB1 was critical for the mosaic red pigmentation in the lip crest of Oncidium spp. (Chiou and Yeh, 2008), and PeMYB11 was responsive to the red spots in the callus of the lip in Phalaenopsis spp. (Hsu et al., 2015). PrMYB5 participating in the petal spot formation was further verified using in situ hybridization (Figure 3). In particular, PrMYB5 showed a more abundant transcript at petal spots and upregulated gradually with the formation of spots at the petal base (Figure 4). In other plants, R2R3-MYBs in the C1 subgroup can regulate all structural genes in the flavonoid biosynthesis pathway, including CHS, CHI, F3H, DFR, and ANS (Chiu et al., 2010; Petroni and Tonelli, 2011; Liu et al., 2016; Schwinn et al., 2016), through binding MYB-binding sites or MYB sites' cis-elements with their promoters (Allan et al., 2008; Liu et al., 2016). Notably, a highly clear association existed among PrMYB5, PrDFR, and PrANS transcriptions during petal coloration and spot formation (Figure 4). In addition, Y1H and dual-luciferase assays revealed that PrMYB5 had a significant activation effect on the PrDFR promoter. It has been reported that McMYB10 can promote McDFR1 expression and increase anthocyanins accumulation in Malus crabapple (Tian et al., 2017). MdMYB10 and MdMYB1 can promote the accumulation of anthocyanins in apple peels through binding to the DFR promoter (Fornale et al., 2014). Overexpression of PrMYB5 in tobacco made petals appear deeper red, consisting with the increased Pg3G, Pn3G5G, Pn3G, Cy3G5G, and Cy3G levels and significantly upregulated NtDFR expression in petals of transgenic tobacco lines (Figures 5B–D). On the other hand, the reduction of Cy3G, Cy3G5G, Pn3G, and Pn3G5G caused by PrMYB5 silencing in petal spots of P. rockii was consistent with the downregulation of PrCHS, PrDFR, and PrANS, particularly PrDFR (Figure 6), indicating that PrMYB5 probably played a key role in petal spot formation of P. rockii through promoting PrDFR abundant expression at the petal base. Likewise, LhMYB18 activated the promoter of the DFR gene in tobacco plants (Yamagishi, 2018), and CgMYB1 activates CgDFR2 to produce petal spots in C. gracilis (Martins et al., 2017).
In summary, we compared the distinct pigmentation processes between petal bases and petal backgrounds of P. rockii and other tree peony cultivars, and further demonstrated that the petal spot formation was mainly attributed to the spatiotemporal transcription of PrMYB5 activating PrDFR promoters, which was associated with the Cy and Pn type anthocyanin accumulation at the petal base. However, the complex and exact regulation mechanism of PrMYB5 on petal spot formation requires to further research. These results not only provide a reference for further exploring the molecular mechanism of novel spot pigmentation patterns in P. rockii, but also lay the theoretical foundation for molecular breeding of novel spotted tree peony cultivars.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
QS and LL designed this research, performed data analysis, and wrote the manuscript. MY and SW performed the experiments and data analysis. XLu assisted in bioinformatics analyses. SL, YF, and XLi helped in the preparation of plant material. YZ provided some supervision of the study. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant No. 31800599), the Natural Science Foundation of Shannxi Province, China (Grant No. 2017JQ3024), and the National Key R&D Program of China (Grant No. 2019YFD1001505).
We thank YZ for field and laboratory support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.955590/full#supplementary-material
Albert, N. W., Lewis, D. H., Zhang, H., Schwinn, K., Jameson, P., and Davies, K. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65, 771–784. doi: 10.1111/j.1365-313X.2010.04465.x
Allan, A. C., Hellens, R. P., and Laing, W. A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. doi: 10.1016/j.tplants.2007.11.012
Cao, Y., Li, K., Li, Y., Zhao, X., and Wang, L. (2020). MYB transcription factors as regulators of secondary metabolism in plants. Biology 9, 61. doi: 10.3390/biology9030061
Chiou, C. Y., and Yeh, K. W. (2008). Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey. Plant Mol. Biol. 66, 379–388. doi: 10.1007/s11103-007-9275-3
Chiu, L., Zhou, X., Burke, S., Wu, X., Prior, R. L., and Li, L. (2010). The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 154, 1470–1480. doi: 10.1104/pp.110.164160
Davies, K. M., Albert, N. W., and Schwinn, K. E. (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39, 619–638. doi: 10.1071/FP12195
Fornale, S., Lopez, E., Salazarhenao, J. E., Fernanadeznohales, P., Rigau, J., and Caparrosruiz, D. (2014). AtMYB7, a new player in the regulation of UV-Sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 55, 507–516. doi: 10.1093/pcp/pct187
Gao, Y., Harris, A., and He, X. (2015). Morphological and ecological divergence of Lilium and Nomocharis within the hengduan Mountains and Qinghai-Tibetan Plateau may result from habitat specialization and hybridization. BMC Evol. Biol. 15, 147–168. doi: 10.1186/s12862-015-0405-2
Glover, B. J., Walker, R. H, Moyroud, E., and Brockington, S. F. (2013). How to spot a flower. New Phytol. 197, 687–689. doi: 10.1111/nph.12112
Gu, Z., Zhu, J., Hao, Q., Yuan, Y., Duan, Y., Men, S., et al. (2019). A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 60, 599–611. doi: 10.1093/pcp/pcy232
Horsch, R., and Klee, H. (1986). Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: role of T-DNA borders in the transfer process. Proc. Natl. Acad. Sci. U. S. A. 83, 4428–4432. doi: 10.1073/pnas.83.12.4428
Hsu, C. C., Chen, Y. Y., Tsai, W. C., Chen, W. H., and Chen, H. H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168, 175. doi: 10.1104/pp.114.254599
Jiang, C., Gu, X., and Peterson, T. (2004). Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 5, R46. doi: 10.1186/gb-2004-5-7-r46
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Messner, R. C., et al. (2010). Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van, D. P., et al. (2002). Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl. Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, L., Cheng, Z., Ma, Y., Bai, Q., Li, X., and Gao, J. (2018). The association of hormone signaling genes, transcription, and changes in shoot anatomy during moso bamboo growth. Plant Biotechnol. J. 16, 72–85. doi: 10.1111/pbi.12750
Li, Q., Wang, J., Sun, H. Y., and Shang, X. F. (2014). Lower color patterning in pansy (Violawittrockiana Gams.) is caused by the differential expression of three genes from the anthocyanin pathway in acyanic and cyanic flower areas. Plant Physiol. Bioch. 84, 134–141. doi: 10.1016/j.plaphy.2014.09.012
Liu, C., Long, J., Zhu, K., Liu, L., Yang, W., Zhang, H., et al. (2016). Characterization of a citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci. Rep. 6, 25352. doi: 10.1038/srep25352
Luo, X. N., Luo, S., Fu, Y. Q., Kong, C., Wang, K., Sun, D. Y., et al. (2022). Genome-wide identification and comparative profiling of microRNAs reveal flavonoid biosynthesis in two contrasting flower color cultivars of tree peony. Front. Plant Sci. 2021:797799. doi: 10.3389/fpls.2021.797799
Luo, X. N., Sun, D. Y., Wang, S., Luo, S., Fu, Y. Q., Niu, L. X., et al. (2021). Integrating full-length transcriptomics and metabolomics reveals the regulatory mechanisms underlying yellow pigmentation in tree peony (Paeonia suffruticosa Andr.) flower. Hortic. Res. 8, 235. doi: 10.1038/s41438-021-00666-0
Martins, T. R., Berg, J. J., Blinka, S., Rausher, M. D., and Baum, D. A. (2013). Precise spatial-temporal regulation of the anthocyanin biosynthetic pathway leads to petal spot formation in Clarkia gracilis (Onagraceae). New Phytol. 197, 958–969. doi: 10.1111/nph.12062
Martins, T. R., Jiang, P., and Rausher, M. D. (2017). How petals change their spots: cis-regulatory re-wiring in Clarkia (Onagraceae). New Phytol. 216, 510–518. doi: 10.1111/nph.14163
Petroni, K., and Tonelli, C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181, 219–229. doi: 10.1016/j.plantsci.2011.05.009
Schwinn, K., Ngo, H., Kenel, F., Brummell, D. A., Albert, N. W., Mccallum, J. A., et al. (2016). The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front. Plant Sci. 7, 1865. doi: 10.3389/fpls.2016.01865
Schwinn, K., Venail, J., Shang, Y., Mackay, S., Alm, V., Betelli, E., et al. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18, 831–851. doi: 10.1105/tpc.105.039255
Shang, Y. J., Venail, J., Mackay, S., Bailcy, P., Schwinn, K., Jamcson, P., et al. (2011). The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 189, 602–615. doi: 10.1111/j.1469-8137.2010.03498.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2013). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shi, Q., Li, L., Zhang, X., Luo, J. R., Li, X., Zhai, L. J., et al. (2017). Biochemical and comparative transcriptomic analyses identify candidate genes related to variegation formation in Paeonia rockii. Molecules 22, 1364. doi: 10.3390/molecules22081364
Stracke, R., ishihara, H. G., Barsch, A., Mehrten, F., Niehaus, K., and Weisshaar, B. (2010). Different regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Su, S., Xiao, W., Guo, W., Yao, X., Xiao, J., Ye, Z., et al. (2017). The CYCLOIDEA-RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol. 215, 1582–1593. doi: 10.1111/nph.14673
Suzuki, K., Suzuki, T., Nakatsuka, T., Dohra, H., Yamagishi, M., Matsuyama, K., et al. (2016). RNA-seq-based evaluation of bicolor tepal pigmentation in Asiatic hybrid lilies (Lilium spp.). BMC Genom. 17, 61. doi: 10.1186/s12864-016-2995-5
Tian, J., Chen, M. C., Zhang, J., Li, K. T., Song, T. T., Zhang, X., et al. (2017). Characteristics of dihydroflavonol 4-reductase gene promoters from different leaf colored Malus crabapple cultivars. Hortic. Res. 4, 17070. doi: 10.1038/hortres.2017.70
Wang, L., Hashimoto, F., Shiraishi, A., Shimizu, K., Aoki, N., and Sakata, Y. (2000). Petal coloration and pigmentation of tree peony cultivars of Xibei (the Northwest of China). J. Japanese Soc. Hortic. Sci. 69, 233.
Yamagishi, M. (2011). Oriental hybrid lily Sorbonne homologue of LhMYB12 regulates anthocyanin biosynthesis in flower tepals and tepal spots. Mol. Breeding 28, 381–389. doi: 10.1007/s11032-010-9490-5
Yamagishi, M. (2018). Involvement of a LhMYB18 transcription factor in large anthocyanin spot formation on the flower tepals of the Asiatic hybrid lily (Lilium spp.) cultivar “Grand Cru”. Mol. Breeding 38, 60. doi: 10.1007/s11032-018-0806-1
Yamagishi, M. (2020). MYB19LONG is involved in brushmark pattern development in Asiatic hybrid lily (Lilium spp.) flowers. Sci. Hortic. 272, 109570. doi: 10.1016/j.scienta.2020.109570
Yamagishi, M., Shimoyamada, Y., Nakatsuka, T., and Masuda, K. (2010). Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosynthesis in flower Tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol 51:463–474. doi: 10.1093/pcp/pcq011
Yamagishi, M., Toda, S., and Tasaki, K. (2014). The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol. 201, 1009–1020. doi: 10.1111/nph.12572
Yan, H., Pei, X., Zhang, H., Li, X., Zhang, X., Zhao, M., et al. (2021). MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 22, 3103. doi: 10.3390/ijms22063103
Yuan, Y. W., Sagawa, J. M., Frost, L., Vela, J. P., and Bradshaw, H. D. (2014). Transcriptional control of floral anthocyanin pigmentation in monkeyflower (Mimulus). New Phytol. 204, 1013–1027. doi: 10.1111/nph.12968
Zhang, X. P., Xu, Z. D., Yu, X. Y., Zhao, L. Y., Zhao, M. Y., Han, X., et al. (2019). Identification of two novel R2R3-MYB transcription factors, PsMYB114L and PsMYb12L, related to anthocyanin biosynthesis in Paeonia suffruticosa. Int. J. Mol. Sci. 20, 1055. doi: 10.3390/ijms20051055
Zhang, Y., Cheng, Y., Xu, S., Ma, H., Han, J., and Zhang, Y. (2020a). Tree peony variegated flowers show a small insertion in the F3'H gene of the acyamic flower parts. BMC Plant Biol. 20, 211. doi: 10.1186/s12870-020-02428-x
Zhang, Y., Cheng, Y., Ya, H., Xu, S., and Han, J. (2015). Transcriptome sequencing of purple petal spot region in tree peony reveals differentially expressed anthocyanin structural genes. Trends Plant Sci. 6, 964. doi: 10.3389/fpls.2015.00964
Zhang, Y., Xu, S., Cheng, Y., Wang, J., Wang, X., Liu, R., et al. (2020b). Functional identification of PsMYB57 involved in anthocyanin regulation of tree peony. BMC Genet. 21, 124. doi: 10.1186/s12863-020-00930-7
Zhang, Y., Yin, X., Xiao, Y., Zhang, Z., Li, S., Liu, X., et al. (2018). An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiol. 178, 189. doi: 10.1104/pp.18.00598
Keywords: Paeonia rockii, PrMYB5, PrDFR, anthocyanin biosynthesis, petal spot
Citation: Shi Q, Yuan M, Wang S, Luo X, Luo S, Fu Y, Li X, Zhang Y and Li L (2022) PrMYB5 activates anthocyanin biosynthetic PrDFR to promote the distinct pigmentation pattern in the petal of Paeonia rockii. Front. Plant Sci. 13:955590. doi: 10.3389/fpls.2022.955590
Received: 28 May 2022; Accepted: 08 July 2022;
Published: 03 August 2022.
Edited by:
Daqiu Zhao, Yangzhou University, ChinaReviewed by:
Antje Feller, University of Tübingen, GermanyCopyright © 2022 Shi, Yuan, Wang, Luo, Luo, Fu, Li, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long Li, bGlsb25nMTk0OUAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.