
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 September 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.952732
This article is part of the Research Topic Abiotic Stress Signaling in Plants: Functional Genomic Intervention, Volume II View all 18 articles
 Guddimalli Rajasheker1
Guddimalli Rajasheker1 Marka Nagaraju2
Marka Nagaraju2 Rinku Polachirakkal Varghese3,4
Rinku Polachirakkal Varghese3,4 Naravula Jalaja3
Naravula Jalaja3 Anil Kumar Somanaboina3
Anil Kumar Somanaboina3 Prashant Singam1
Prashant Singam1 Chintala Ramakrishna5
Chintala Ramakrishna5 Suprasanna Penna6
Suprasanna Penna6 Nese Sreenivasulu7
Nese Sreenivasulu7 P. B. Kavi Kishor3*
P. B. Kavi Kishor3*Systematic genome-wide analysis of Sorghum bicolor revealed the identification of a total of 48 homologous genes comprising 21 proline-rich proteins (PRPs) and 27 hybrid proline-rich proteins (HyPRPs). Comprehensive scrutiny of these gene homologs was conducted for gene structure, phylogenetic investigations, chromosome mapping, and subcellular localization of proteins. Promoter analysis uncovered the regions rich with phosphorous- (BIHD), ammonium-, sulfur-responsive (SURE), and iron starvation-responsive (IRO2) along with biotic, abiotic, and development-specific cis-elements. Further, PRPs exhibit more methylation and acetylation sites in comparison with HyPRPs. miRNAs have been predicted which might play a role in cleavage and translation inhibition. Several of the SbPRP genes were stimulated in a tissue-specific manner under drought, salt, heat, and cold stresses. Additionally, exposure of plants to abscisic acid (ABA) and zinc (Zn) also triggered PRP genes in a tissue-dependent way. Among them, SbPRP17 has been found upregulated markedly in all tissues irrespective of the stress imposed. The expressions of SbHyPRPs, especially SbHyPRP2, SbHyPRP6, and SbHyPRP17 were activated under all stresses in all three tissues. On the other hand, SbHyPRP8 (root only) and SbHyPRP12 (all three tissues) were highly responsive to cold stress and ABA while SbHyPRP26 was induced by drought and Zn in the stem. Taken together, this study indicates the critical roles that SbPRPs and SbHyPRPs play during diverse abiotic stress conditions and notably the plausible roles that these genes play upon exposure to zinc, the crucial micronutrient in plants.
Plants are constantly exposed to both biotic and abiotic stresses because of which a lion’s share of the final productivity is lost globally. Plants can cope with them by deploying several complex physiological and molecular mechanisms depending upon the type, severity, and duration of stress. But unraveling the underlying mechanisms is crucial for us to design crops with superior tolerance to stresses. Extreme environmental conditions cause membrane damage, denaturation of proteins, and accumulation of excessive reactive oxygen species (ROS) that eventually dismantle the cellular fabric (Bokszczanin, 2013; Wang et al., 2019). To combat these stress-induced damages, plants accumulate osmotic agents, antioxidants, and also antioxidative enzymatic machinery (Bohnert et al., 2006; Bokszczanin, 2013). Plant cell wall proteins consist of both structural [arabinogalactan proteins, (AGPs) and extensins or hydroxyproline-rich O-glycoproteins (HRGPs)] and enzymatic proteins. They play key roles in cell wall formation, cell differentiation, development, and oxidative cross-linking mediated by extension peroxidases (McNeil et al., 1984; Jose-Estanyol and Puigdomenech, 1993; Showalter, 1993; Sommer-Knudsen et al., 1998). Proline rich-proteins (PRPs) and hybrid proline-rich proteins (HyPRPs) contain proline and hydroxyproline and are a part of the HRGPs super family proteins, widely distributed in plants and implicated in biotic and abiotic stress tolerance. PRPs contain at least two consecutive proline residues organized in the peptides and the domains contain 70% of proline residues (Zhang et al., 2021). Based on the presence or absence of N-terminal signal peptides and also differences in the domains, PRPs are divided into (1) PRPs that have two domains with signal peptides and highly rich repeat regions of proline at the N-terminus (Dvorakova et al., 2007), (2) PRPs that have three domains with N-terminal signal peptide, repeat regions of proline, aside cysteine-rich regions in the C-terminal end, and (3) PRPs that are devoid of any signal peptide, but with similar PPVYK repeat sequences at the C-terminus (Jose-Estanyol and Puigdomenech, 1994, 2000). The presence of signal peptides in these proteins infers that they are secreted-type of proteins in plants (Keller, 1993; Showalter, 1993; Cassab, 1998). The signal peptides permit their sorting into the rough endoplasmic reticulum and other cellular organelles. Newman and Cooper (2011) identified 31 distinct proline-rich tandem repeat protein classes targeted for secretion using a systems-level computational approach. Further, PRPs have been classified as an 8-cysteine motif or 8CM proteins, with 90–100 amino acids but the proteins are not integrated within the membrane. The 8-cysteine residues appear indispensable for the three-dimensional structure of these proteins. The 8-cysteine residues have a versatile structure in plant proteins domain and lipid transfer proteins (Jose-Estanyol et al., 2004; Dvorakova et al., 2012). In general, proline-rich repeats precede the 8CM, and such proteins are named as HyPRPs, but the N-terminus features like that of PRPs (Jose-Estanyol and Puigdomenech, 1994, 2000). HyPRPs differ from that of PRPs in the amino acid motif size and composition of proline repeats (Jose-Estanyol et al., 2004; Dvorakova et al., 2007). HyPRPs are subdivided into two groups based on cysteine residue positions in the peptidic sequence (Jose-Estanyol and Puigdomenech, 1994). Proline-rich domains of SbrHyPRP from Solanum brevidens contains repeated motifs PPHVKPPSTPK and PTPPIVSPP extended with TPKYP and TPKPPS motifs at N- or C-termini, respectively (Fischer et al., 2002). PRPs having tandem repeats of a hexapeptide PPPVHL were first characterized by corn endosperm (Esen et al., 1982). Function, localization, and differential expression of three PRPs from Glycine max (PRP1, PRP2, and PRP3) were studied (Hong et al., 1990; Wyatt et al., 1992). PRPs and PRP-like proteins have been found to play a crucial role during plant growth, germination of seeds (Bouton et al., 2005; Chen et al., 2014), and elongation of root hairs in Arabidopsis (Boron et al., 2014). Blocking OsPRP3 by RNAi leads to defects in floral organogenesis in Oryza sativa implying its role during plant development (Gothandam et al., 2010). Further, PRPs modulate both biotic and abiotic stresses such as chilling, drought, and salinity stresses (Zhan et al., 2012; Kavi Kishor et al., 2015). Pathogen-derived molecules usually trigger systemic acquired resistance (SAR) in plants, which can be abolished in a wide spectrum of pathogens. Many experiments have shown that HyPRPs are closely related to biotic stress resistance (Yu et al., 2013; Cecchini et al., 2015, 2021). HyPRPs with plastid pools modulates systemic immunity either positively or negatively against Pseudomonas syringae (Banday et al., 2022). It appears that HyPRPs are obligatory for both SAR and induced-systemic resistance (ISR), and SAR signal movement or action (Gao et al., 2015; Cecchini et al., 2019, 2021; Kachroo and Kachroo, 2020). Therefore, HyPRPs have distinct roles to play in plant immunity. Also, when useful bacteria attack the roots of plants, ISR occurs, which leads to the transport of mobile signals from the tissues or organs that are immunized (Cecchini et al., 2015, 2019; Carella, 2020; Kachroo and Kachroo, 2020). Signal molecules such as azelaic acid (AZA) work as mobile signals for SAR, and such signals alongside free radicals are imperative in imparting SAR (Jung et al., 2009; Wang et al., 2014; Kachroo and Kachroo, 2020). The experiments infer that HyPRPs may have a critical role to play in biotic stress tolerance. In Arabidopsis, FUSED OUTER CUTICULAR LEDGE1 (FOCL1) encodes a guard cell-expressed, secreted PRP protein. Interestingly, focl1 mutants display enhanced drought tolerance compared to wild-type plants (Hunt et al., 2017). HyPRPs are poorly glycosylated cell wall glycoproteins and also play critical roles during plant ontogeny and abiotic stress tolerance like PRPs (Holk et al., 2002; Blanco-Portales et al., 2004; Kavi Kishor et al., 2015; Gujjar et al., 2019).
Information on the number of PRPs in diverse taxa is scanty. Showalter et al. (2016) discovered 49 PtPRPs in Populus trichocarpa and Malus domestica, 9 MdPRP genes have been identified (Zhang et al., 2021). Surprisingly, no HyPRPs have been found in algae, mosses, and ferns, but Pinus taeda contains 21 PtHyPRPs (Dvorakova et al., 2007). While in dicots such as Arabidopsis thaliana 28 AtHyPRPs have been found, 16 StHyPRPs were recorded in Solanum tuberosum (Dvorakova et al., 2012), 19 complete or nearly complete from S. lycopersicon (SlHyPRPs) (Kapoor et al., 2019) and 14 from Medicago truncatula (MtHyPRPs) have been recorded from the publicly available sequence data (Dvorakova et al., 2012). In Glycine max, 35 GmHyPRP-encoding genes were identified by Neto et al. (2013). In monocots, like Oryza sativa genome, conflicting reports exist. Studies by Dvorakova et al. (2007); Boutrot et al. (2008), and Kapoor et al. (2019) showed 21, 31, and 45 OsHyPRP genes, respectively, in rice whereas, in Zea mays, 52 members of ZmHyPRPs have been detected (Dvorakova et al., 2007). Such a large variation could be because of different methods that researchers have employed for identification.
Sorghum [Sorghum bicolor (L.) Moench] is an important C4 cereal crop and the fifth largest grown in arid and semi-arid regions, globally utilized for both food and fodder (FAOSTAT1). In the semi-arid tropics of Asia and Africa, it serves as a staple food, animal feed, fodder, and notably a promising multipurpose feedstock crop for bioethanol production (Silva et al., 2022). It is moderately tolerant to both drought and heat and requires very low inputs. Traits that impart alkalinity, salt, water deficit, waterlogging, and high-temperature stresses make this crop attractive for research purposes in comparison to other cereal crops (Hadebe et al., 2017; Huang, 2018). Fujino et al. (2014) have detected 21 HyPRPs in Oryza sativa, 12 in Hordeum vulgare, 10 in Brachypodium, 20 in Zea mays, and 28 in S. bicolor using a BLAST search of qLTG3-1 as the query. Though 28 HyPRPs have been reported in sorghum, their subcellular localizations, cis-element characterization, and their tissue-specific expressions and implications under abiotic stress conditions have not been undertaken. So, the number of PRPs and HyPRPs, their tissue-specific expressions under cold, high temperature, salt, drought stress conditions, and their response to ABA have not yet been elucidated. Similarly, little is known if these genes play a molecular function in response to soil nutrients especially sulfur (S), zinc (Zn), and iron (Fe). In the present study, we have presented results on the genome-wide analysis of sorghum for both PRP and HyPRP homolog genes, their characterization, and tissue-specific expressions under abiotic stress conditions besides their response to ABA and Zn.
Seeds of S. bicolor variety BTx623 (an inbred line) were obtained from ICRISAT, Patancheru, Hyderabad. This line is moderately tolerant to water deficit conditions and its genome sequencing information is publicly available. Seeds were sown in the pots filled with 5 kg of garden soil, and seedlings were grown for 40-days under glasshouse conditions (28/20°C day/night temperatures). Seedlings were subjected to drought stress by imposing a 200 mM mannitol solution, and salt stress was induced by saturating the seedlings with 200 mM NaCl solution for 4 h. Seedlings were subjected to cold stress by keeping them at 4°C and high-temperature stress by exposing them to 40°C for 4 h. Seedlings were also treated with 100 μM ABA and 25 mM zinc chloride solution (based on literature search) for 4 h separately. After completion of treatment, roots, stems, and leaves were collected along with respective controls (without treatment) and frozen immediately in liquid nitrogen and stored at –80°C refrigerator for isolation of RNA and gene expression analysis by qRT-PCR. Biological triplicates and technical duplicates were maintained for gene expression analysis for each treatment.
Proline-rich proteins and HyPRP gene sequences of Oryza, and Arabidopsis were retrieved from Plant GDB2 and Gramene database3 (Tello-Ruiz et al., 2021). These gene sequences were blasted against S. bicolor genome in Gramene database to find out their homologs. Genscan4 (Burge and Karlin, 1998) was used to retrieve genes and their respective protein sequences. Based on the homology, the identified putative protein sequences were subjected to motif search5 to check the reliability and for identifying the conserved domains (Letunic et al., 2004).
To predict the putative cis-acting elements of PRP and HyPRP promoter regions, the 2,000 bp genomic sequences upstream of the start codon were extracted and PLANTCARE (Lescot et al., 2002) software was used to identify putative cis-acting elements responsible for plant development and biotic and abiotic stress tolerance.
The identified SbPRP and SbHyPRP genes were mapped to their respective chromosomes based on the information provided in the Gramene Genome Database. Gene Structure Display Server6 software was used for obtaining the gene structures of SbPRPs and SbHyPRPs – exons, introns, and untranslated sequence regions (UTRs) by aligning their gene and coding sequences (Guo et al., 2007).
TBtools (Chen et al., 2020) was used for identifying the synteny of PRP and HyPRP genes across S. bicolor (Sb), O. sativa (Os), and A. thaliana (At).
The Maximum Likelihood phylogenetic tree was constructed using MEGA 6.2 software (Tamura et al., 2013) by employing the Poisson correction, partial deletion, and bootstrap value (1,000 replicates) parameters to know the evolutionary relationships. The PAL2NAL7 (Suyama et al., 2006) software was used to calculate the substitution rates for non-synonymous and synonymous sites of each of the identified paralogs (sorghum) and orthologous gene pairs (between Sorghum/Oryza, and Sorghum/Arabidopsis) from phylogeny.
Molecular weight (MW), isoelectric point (pI), and GRAVY (grand average of hydropathy) of PRPs and HyPRPs were identified by using ProtParam Expasy tools8 (Gasteiger et al., 2005), while phosphorylation sites were predicted by employing NetPhos3.1 software of Expasy tools (Blom et al., 2004). The putative transmembrane helices within these genes were identified using TMHMM software (Moller et al., 2001). The sub-cellular localization of PRPs and HyPRPS was identified by the WOLFPSORT program9 (Horton et al., 2007). MEME software (Bailey and Gribskov, 1998) was employed to analyze new sequence patterns and their significance (Bailey et al., 2006). The software helped to identify the nature of motifs by setting different default parameters, number of motifs from 1 to 10 with a motif width of 5–50, and the number of motif sites from 5 to 10.
The 3D structures for the highly expressed 12 proteins encoded by SbPRP (SbPRP3, SbPRP6, SbPRP10, SbPRP12, SbPRP17, and SbPRP19) and SbHyPRP genes (SbHyPRP2, SbHyPRP5, SbHyPRP6, SbHyPRP13, SbHyPRP17, and SbHyPRP26) were predicted using SWISS-MODEL server (Biasini et al., 2014). The predicted 3D structures of proteins for the highly expressed genes were evaluated for stability using PROCHECK and protein structure verification server (PSVS)10. The stability of the proteins was analyzed by Ramachandran plots through the calculation of phi (F) and psi (ψ) torsion angles.
The acetylation of the internal lysines was predicted by using PAIL11 (Li et al., 2006), the PRmePRed12 (Kumar et al., 2017) was used to predict the methylation sites. The putative miRNAs in targeting the SbPRP and SbHyPRP genes were identified using the psRNATarget server (Dai and Zhao, 2011) by default parameters.
The simple sequence repeats (SSRs), the gene-specific molecular markers of PRPs and HyPRPs were identified within the transcripts by employing MISA-web13 (Beier et al., 2017).
Digital expression profiling of SbPRP and SbHyPRP genes was analyzed using Genevestigator14. S. bicolor mRNA-seq data available for all the 21 SbPRP and 27 SbHyPRP genes for stress conditions like cold and drought, three tissues, and five developmental stages were collected and used for analysis. Gene expression profiles were developed using hierarchical clustering and heat maps were generated for anatomy, development, and perturbations separately.
The transcriptional profiling of PRPs and HyPRPs was further investigated using qRT-PCR. Total RNA from root, stem, and leaf tissues under various stresses was extracted by using a nucleo-spin plant RNA isolation kit (MACHEREY-NAGEL) according to the manufacturer’s instructions. A total of 3 μg of total RNA was transcribed into cDNA by using first-strand synthesis kit (Thermo Scientific, Waltham, MA, USA). The SYBR Green Master Mix (2×) (Takara, Kasatsu, Shiga, Japan) was used according to the manufacturer’s recommendations. The qRT-PCR analysis was conducted by using ABI 7500 real-time PCR system (Applied Biosystems, Waltham, MA, USA) and Takara SYBR Green Master Mix (2 ×) with the following thermal cycles: 1 cycle at 95oC for 10 min, followed by 40 cycles alternatively at 95°C for 15 s and 60°C for 1 min. The amplicon dissociation curves were recorded with a fluorescence lamp after the 40th cycle by heating from 58 to 95oC within 20 min. Three biological replicates and two technical replicates were taken for the study. The gene expression data were normalized with Acyl Carrier Protein 2 (SbACP2) and Elongation Factor P (SbEF-P) genes of sorghum (Reddy et al., 2016). The PRP and HyPRP gene-specific primers used for qRT-PCR and reference genes are listed in Supplementary Table 1. The relative gene expressions were calculated by employing REST software (Pfaffl et al., 2002).
The workflow of genome-wide screening and characterization of SbPRPs and SbHyPRPs is shown in Figure 1. Genome-wide analysis of sorghum resulted in the identification of a total of 48 homologous genes encompassing 21 SbPRPs and 27 SbHyPRPs. After checking their reliability by conserved domain search using the MOTIF search tool for convenience, the predicted PRPs, HyPRPs have been named as SbPRP1 to SbPRP21, and SbHyPRP1 to SbHyPRP27, respectively. Out of 21 SbPRP genes, 7 genes were localized on chromosome 1, followed by 3 and 6 (3 genes each). Chromosomes 4, 5, and 10 have 2 genes each, and chromosomes 7 and 8 have 1 gene each in S. bicolor (Table 1 and Figure 2A). Chromosome 1 also displays 9 HyPRP genes in S. bicolor out of 27 SbHyPRPs, followed by chromosomes 4 and 6 with 4 each, chromosome 3 with 3, chromosome 8 with 2, chromosome 5, 7, 10, 11, 12 with 1 gene each (Table 2 and Figure 2B). Chromosomes 2 and 9 totally lack the distribution of these genes. Thus, chromosome 1 has been found as the hot spot with 7 SbPRP and 8 SbHyPRP genes. The gene structure analysis provides possible mechanisms of structural evolution of SbPRP and SbHyPRP genes in sorghum and hence an attempt was made to compare the exon–intron structures of all the identified genes. Out of 21 SbPRP genes, 4 have no introns, 2 exons were found in 4 genes, 3 exons in 3 genes, 4 in 3, 6 in 1, 7 in 1, 8 in 1, 10 in 1, and 12 in 3 (Table 1 and Figure 3A). The SbPRP11, 13, and 18 have the highest number of exons, i.e., 12, while the least number of exons (one) was noticed in SbPRP8, 16, 19, and 20. Of the 27 SbHyPRPs, 16 genes have been found without any introns, but comprise only one exon, 2 exons in 7 genes, 3 in 3, and 4 exons in one gene. The majority of the SbHyPRPs were intron less in comparison with SbPRPs (Table 2 and Figure 3B).
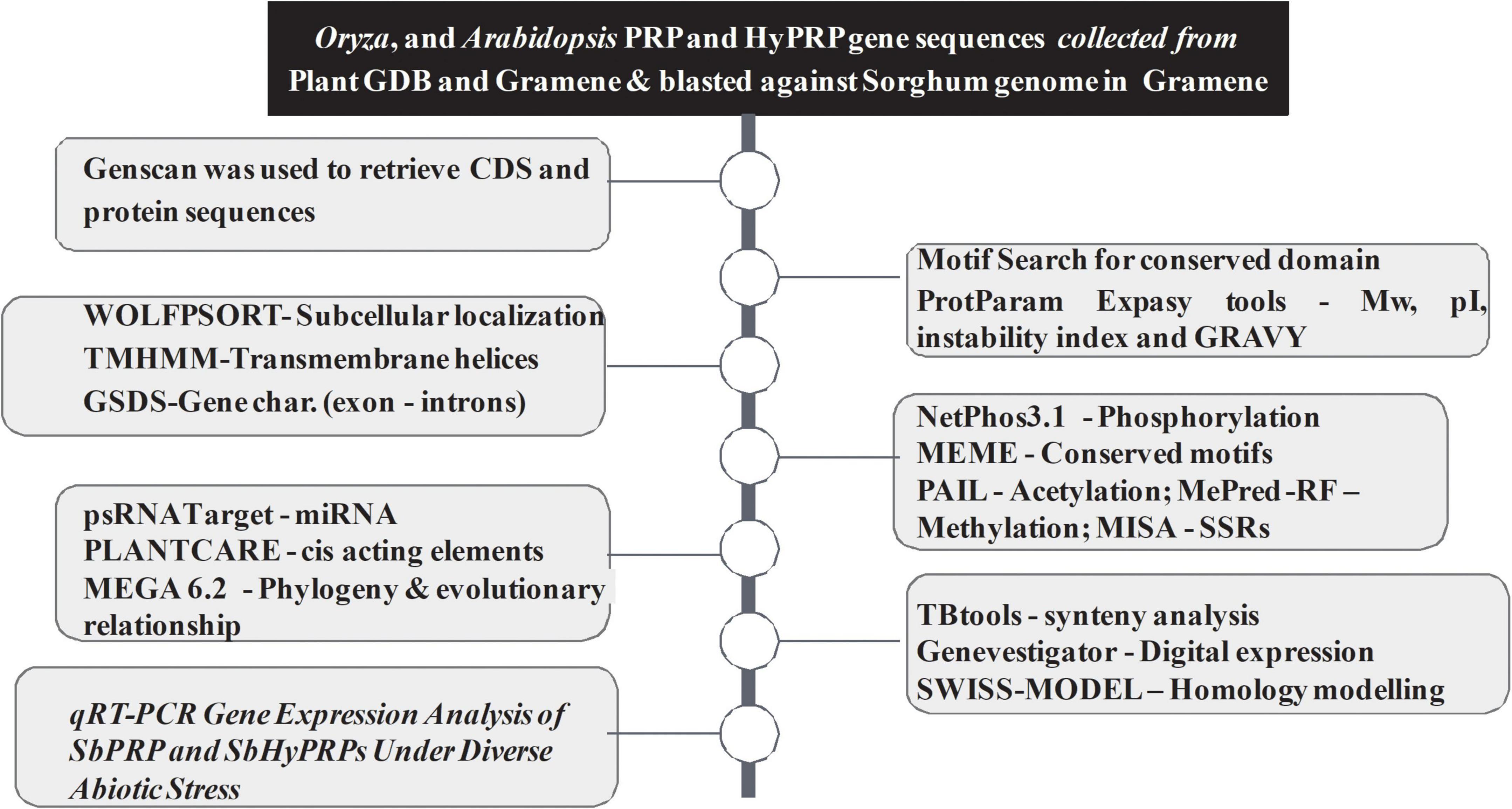
Figure 1. Methods, softwares, and pipelines used for genome-wide screening and characterization of SbPRPs and SbHyPRPs.
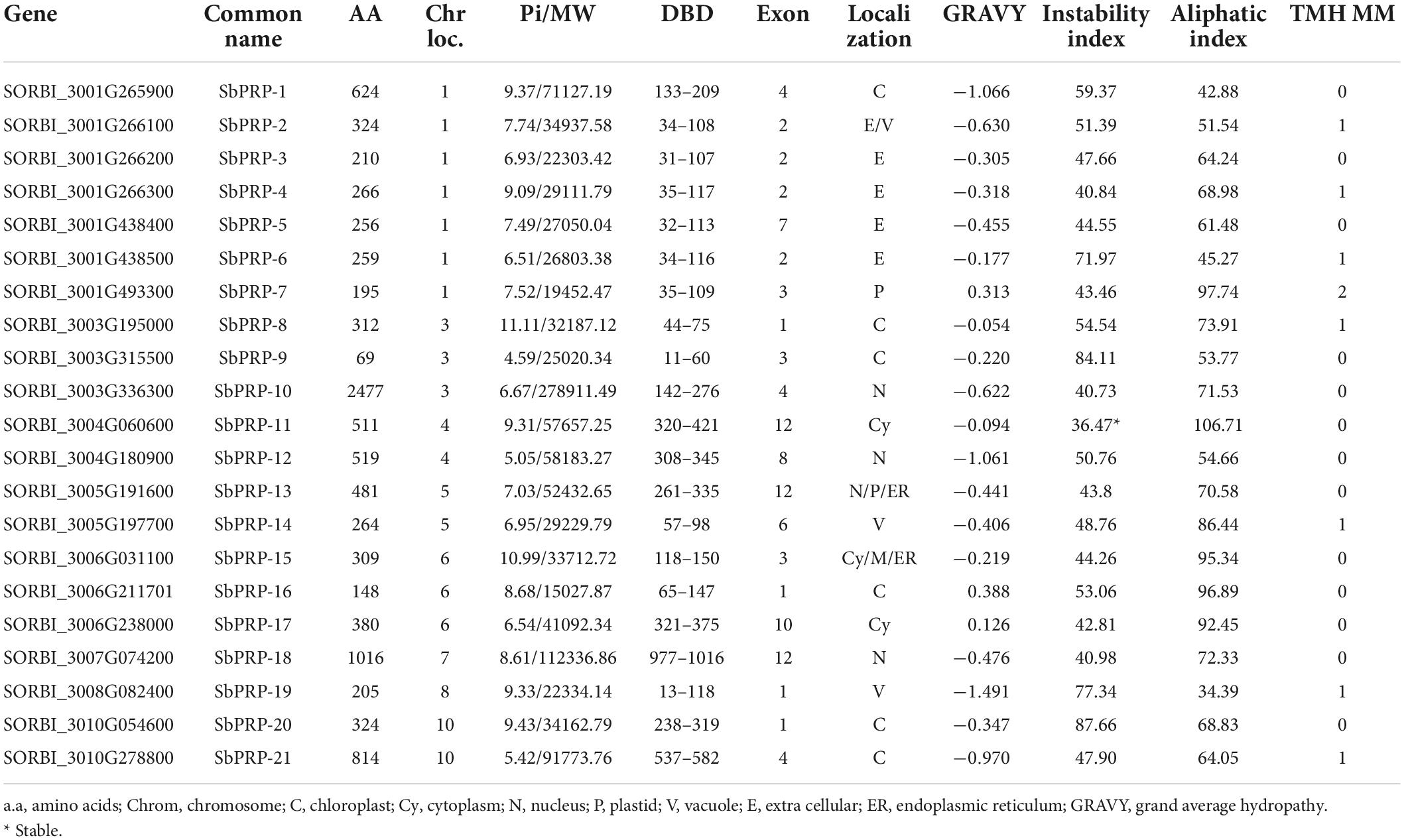
Table 1. List of identified PRPs exhibiting chromosomal location, sub group, length, DNA binding domains (DBD), molecular weight, (MW), iso-electric point (pI), GRAVY, no. of exons, localization, and instability and aliphatic indexes.
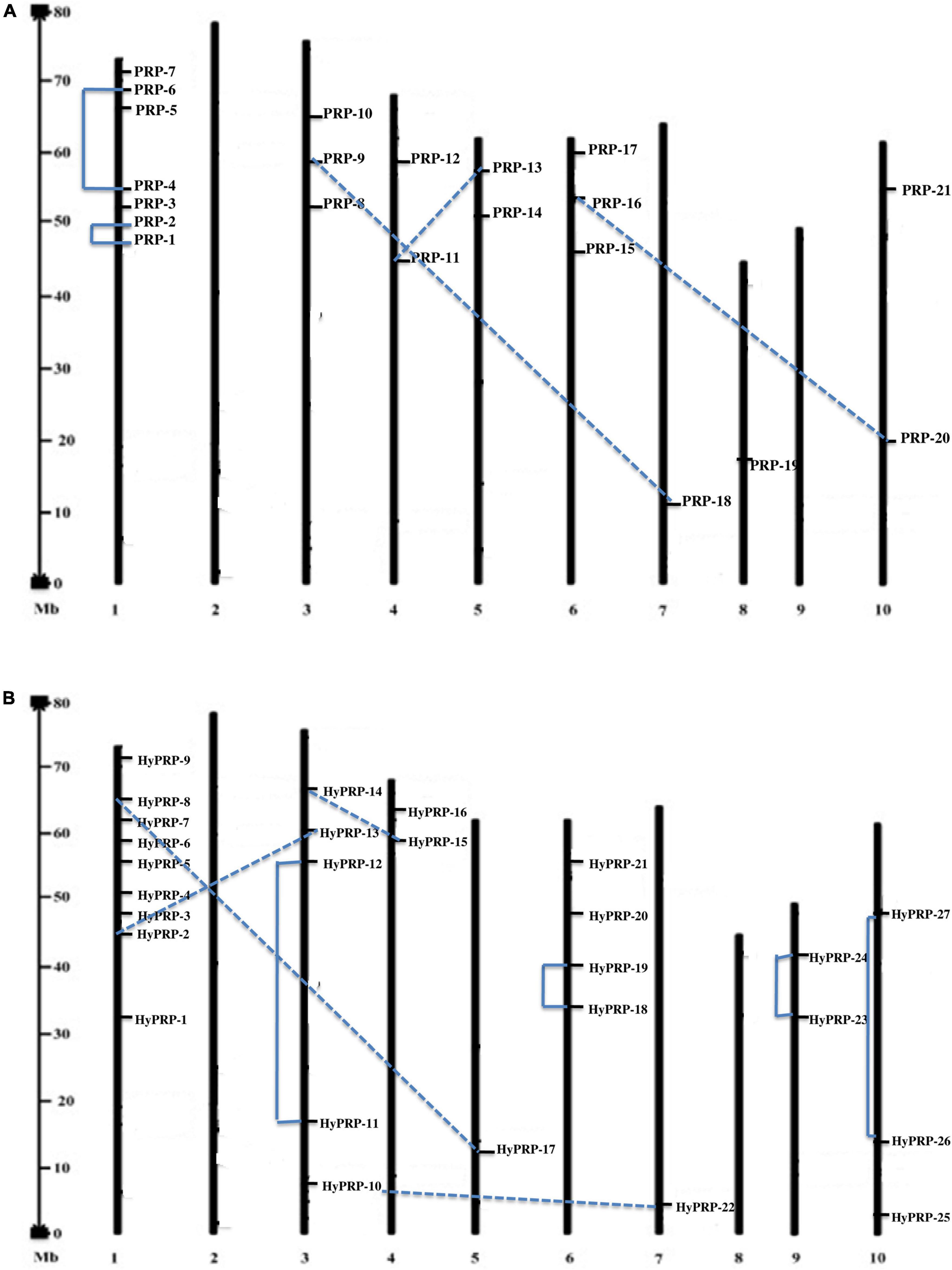
Figure 2. Location and duplications of SbPRP and SbHyPRP. (A) Location and duplication of SbPRPs on chromosomes. (B) Location and duplication of SbHyPRPs on chromosomes.
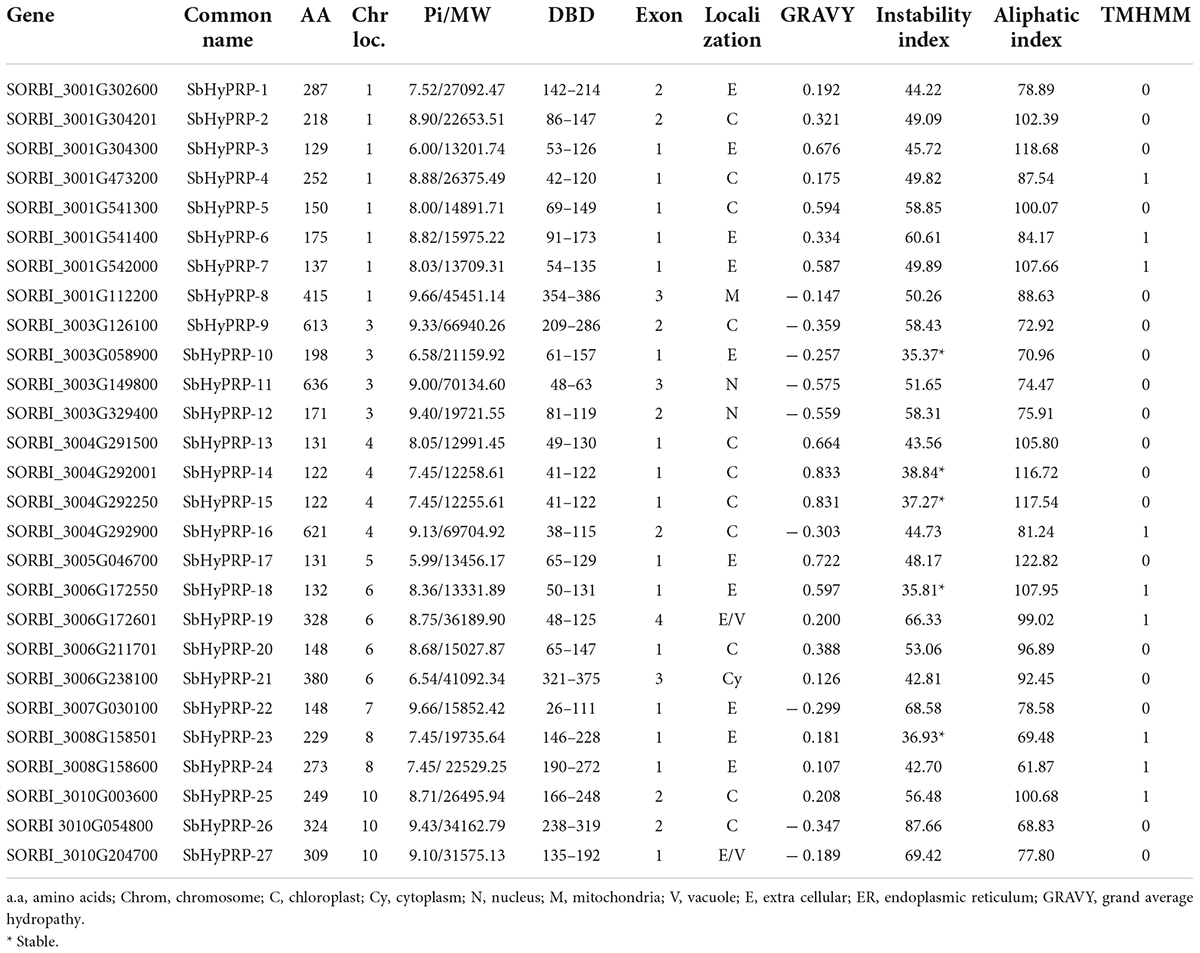
Table 2. List of identified HyPRPs exhibiting chromosomal location, length of aa, DNA binding domains (DBD), molecular weight (MW), iso-electric point (pI), GRAVY, no. of exons, localization, and instability and aliphatic indexes.
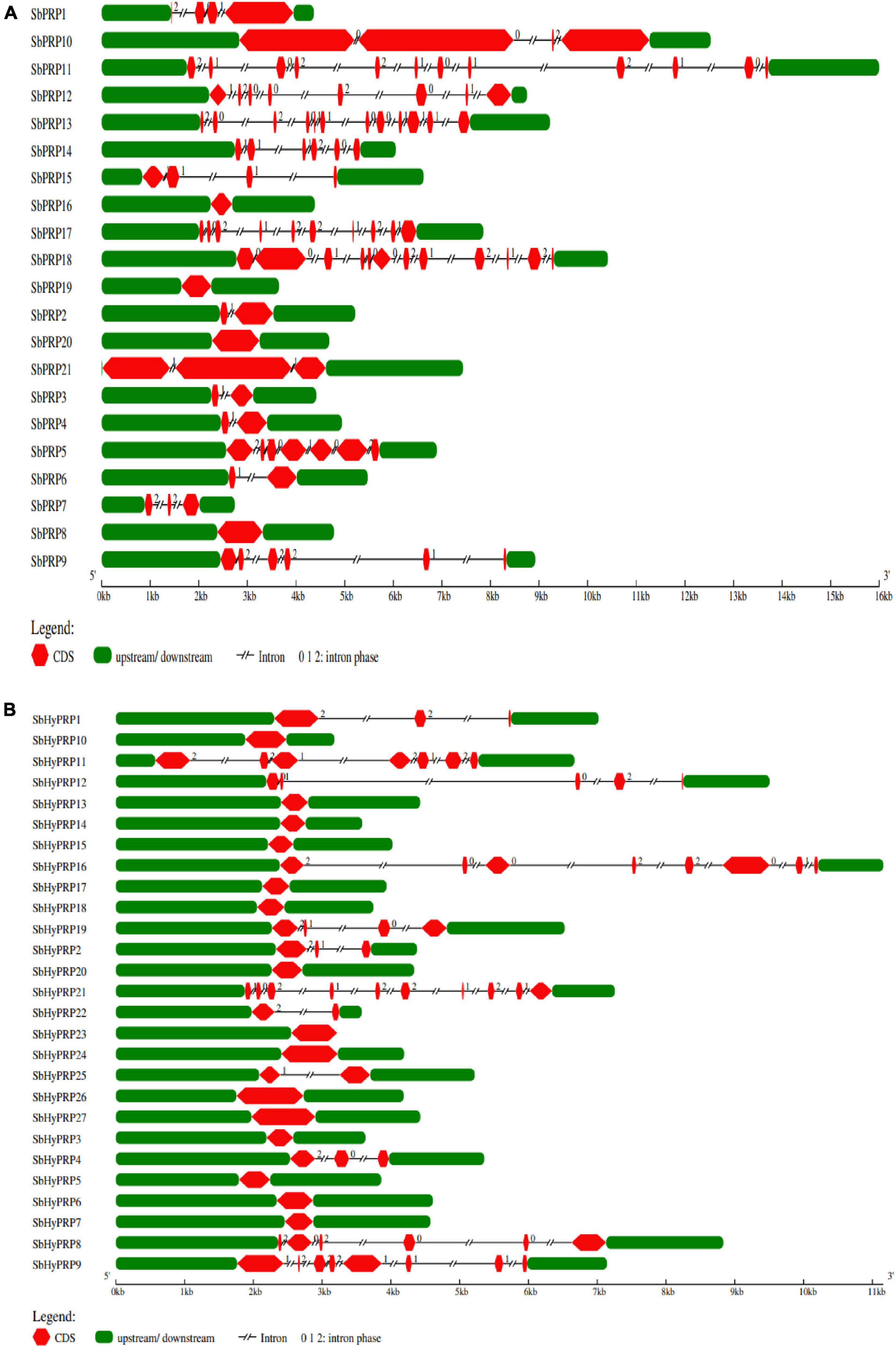
Figure 3. Gene characterization of SbPRP and SbHyPRP genes. (A) Distribution of exons, introns, upstream, and downstream regions in SbPRPs, (B) distribution of exons, introns, upstream, and downstream regions in SbHyPRPs.
Promoter analysis predicted that SbPRP and SbHyPRP genes have potential cis-regulatory elements such as Dc3 Promoter Binding Factor (DPBF, participate in seed specific and/or ABA-inducible expression), MYC, MYB, etiolation, G-box, I-box, ANAERO, associated with abiotic stress, hormone-specific (ABRE, and ERE), T/GBOX for jasmonic acid, biotic stress-responsive WBox, development-specific (pollen, endosperm, and meristem specific) and guard cell-specific elements. Phosphorous (BIHD)-, ammonium-, sulfur-responsive (SURE), and iron (IRO2) starvation-responsive elements have also been predicted. They were rich with EECCRCAH1 elements, the binding site of MYB transcription factor; rbcS general consensus sequence; XYLAT, the core xylem gene set; Telo-box elements for activation of expression in root primordia (Supplementary Tables 2, 3).
The phylogenetic analysis of SbPRPs has been clustered into two clades, based on homology and conserved motifs. Clade I is subdivided into 7, whereas clade II is grouped into 4. A total of five paralogous pairs were observed in the SbPRP family, of which two tandem (SbPRP1/2 and SbPRP4/6 on chromosome 1) and the remaining segmental duplications (Figures 2A, 4A). A phylogenetic tree was constructed to see the evolutionary relationship of SbPRPs with Oryza and Arabidopsis ancestors for monocot and dicots. All of them exhibited lesser evolutionary relationships, but species-specific clusters were noticed. SbPRPs showed five orthologous relationships, of which four are with Oryza and only one with Arabidopsis (Sorbi_3001G266100/AT5G9170) (Supplementary Figure 1). The phylogenetic tree of SbHyPRPs clustered into two clades, based on their conserved motifs and subcellular localizations. Clade I is divided into two groups, whereas clade II is clustered into eight groups. A total of nine paralogs were predicted, of which four tandem (SbHyPRP11/12 on chromosome 3; SbHyPRP18/19 on 6; SbHyPRP23/24 on chromosome 9; SbHyPRP26/27 on chromosome 10) and five segmental duplications (Figures 2B, 4B). The SbHyPRPS have been predicted to have 10 orthologous relationships with Oryza, and only one with Arabidopsis (Sorbi_3010G204700/AT2G10940) (Supplementary Figure 2). Comprehensive phylogenetic trees were constructed by including Oryza, Arabidopsis, Zea, Medicago, Lycopersicum, Solanum, Medicago, Hordeum, Beta vulgaris, and Setaria for PRP and HyPRPs. They display species and family specificities. While SbPRPs showed 12 orthologous relationships with Zea, SbHyPRPS depicted 3 with Zea (Supplementary Figures 3, 4). Darwinian selection in paralogous and orthologous duplications was uncovered by calculating the non-synonymous substitution (dN) to synonymous (dS) ratios. Among the five paralogs of SbPRPs, only one event (SbPRP9/SbPRP18) showed the dN/dS ratio below 1, implying purifying selection, while the remaining four followed a positive/Darwinian selection (Supplementary Table 4). Out of the 5 PRP orthologous events, three events (SORBI_3006G211701/Os06G0104800; SORBI_3010G054600/Os06G0168700; SORBI_3001G266100/At5G59170) exhibited the dN/dS ratio less than 1, while the remaining two showed Darwinian/positive selection (Supplementary Table 5). Of the nine paralogous events of SbHyPRPs, only two (SbHyPRP14/SbHyPRP15, SbHyPRP18/SbHyPRP19) exhibited purifying selection (Supplementary Table 6). Out of six orthologs of SbHyPRPs, only one event (SORBI_3001G304201/Os10G40420) followed purifying selection and the remaining positive selection (Supplementary Table 7). Syntenic genes for sorghum PRP and HyPRPs were mapped to Oryza and Arabidopsis. For SbPRP genes, chromosome 1 displays seven genes, followed by chromosomes 3 and 6 with three genes, chromosomes 4, 5, and 10 with two genes, and chromosomes 7 and 8 with one gene each, respectively (Figure 5A). For SbHyPRP, S. bicolor chromosome 1 displays nine genes, followed by chromosomes 4 and 6 with 4, chromosome 3 with 3, chromosome 8 with 2, and chromosomes 5, 7, 10, 11, and 12 with one gene each, respectively (Figure 5B).
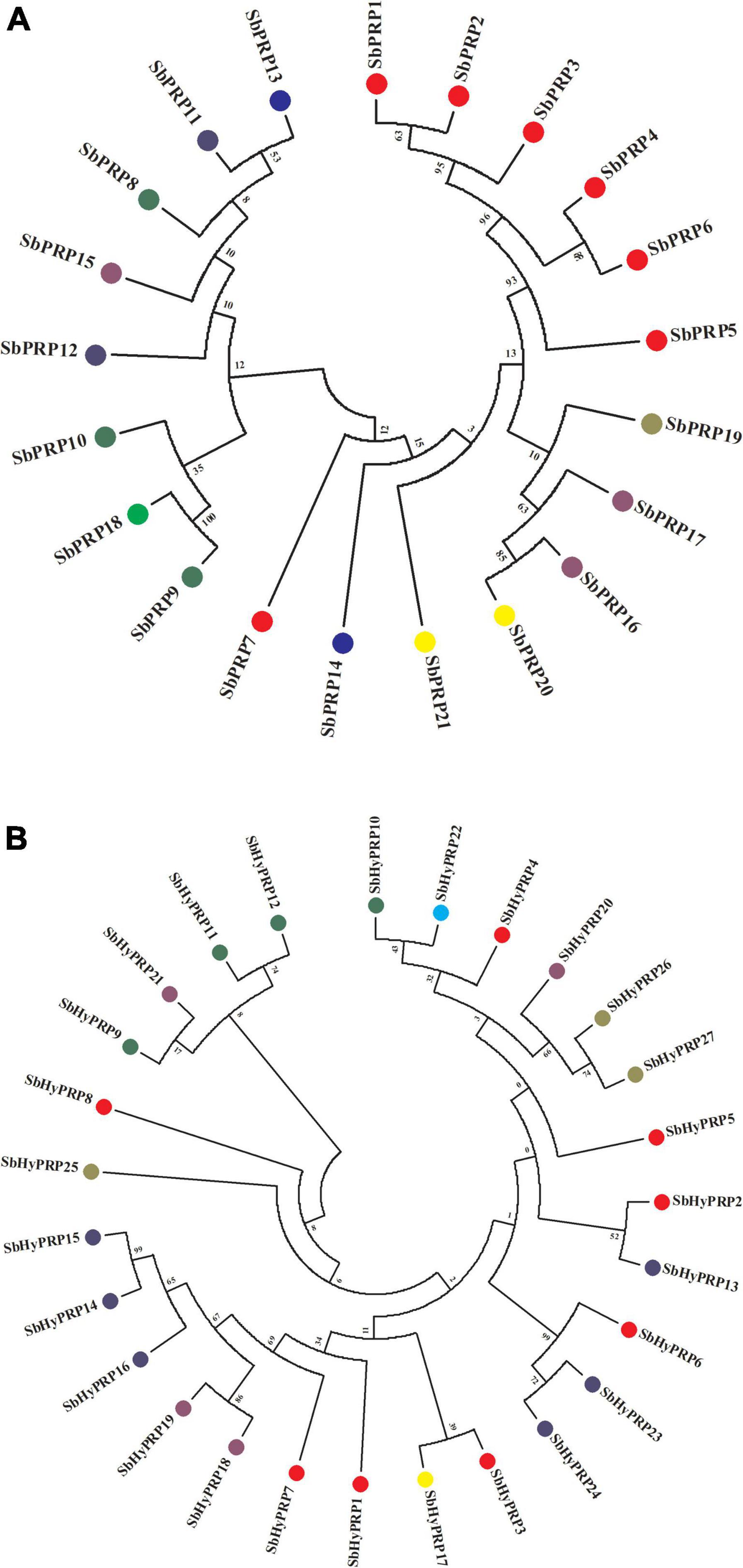
Figure 4. Phylogenetic analysis of SbPRP and SbHyPRP genes. (A) Phylogenetic tree of PRPs in Sorghum.Sub-groups are distinguished by different colors based on their chromosome locations, (B) phylogenetic tree of HyPRPs in Sorghum. Sub-groups are distinguished by different colors based on their chromosome locations.
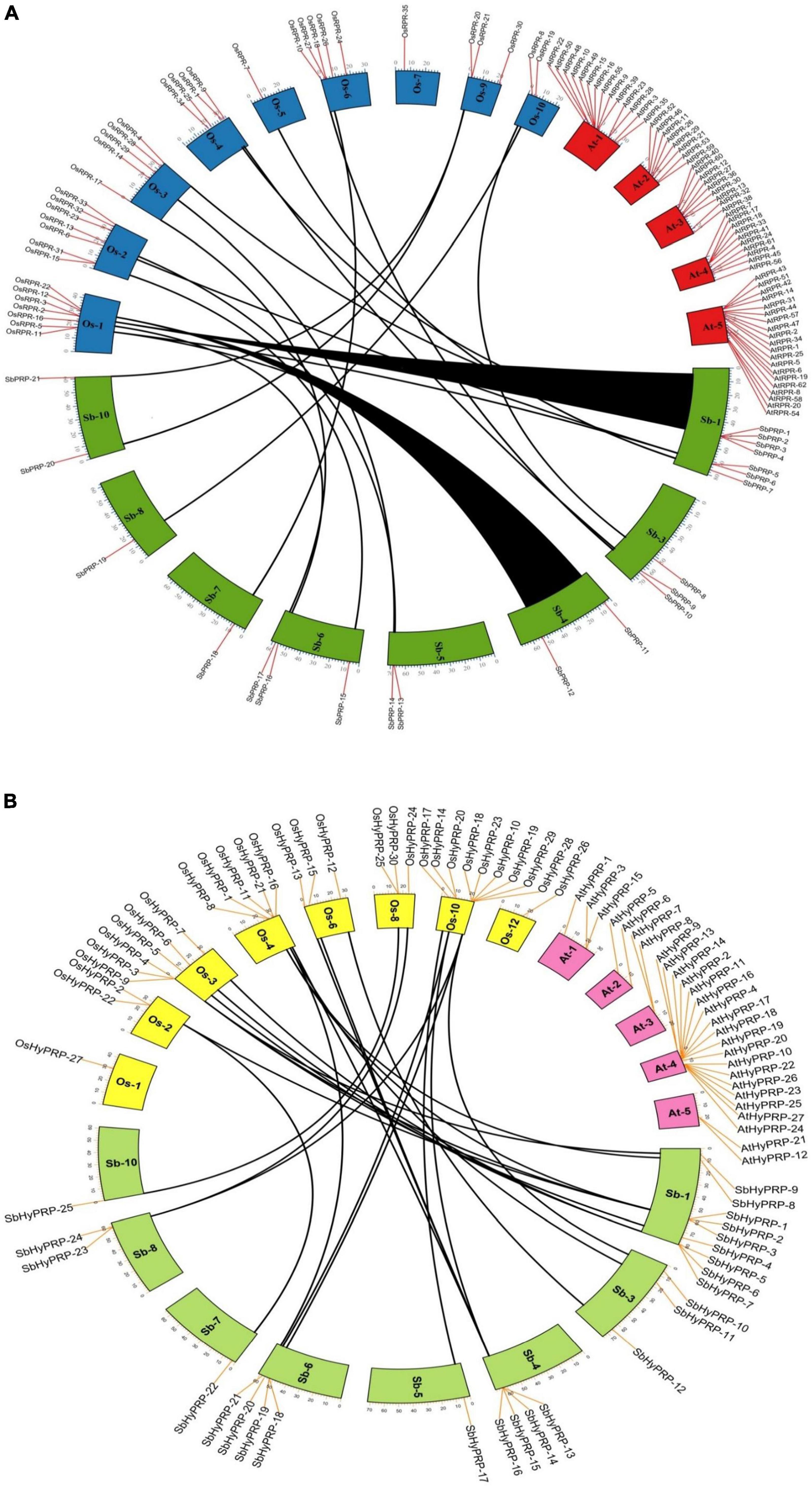
Figure 5. Synteny analysis of SbPRP and SbHyPRP genes. (A) Synteny analysis of PRP in Sorghum bicolor, Oryza sativa,and Arabidopsis thaliana. (B) Synteny analysis of HyPRP in Sorghum bicolor, Oryza sativa,and Arabidopsis thaliana. The map was built with TB tools software. Sb, Sorghum bicolor; Os, Oryza sativa; PRP, Proline-rich proteins; HyPRP, Hybrid proline-rich proteins.
It has been predicted that SbPRP genes encoding peptides span from 69 (SbPRP9) to 2477 (SbPRP10) amino acids with the pI value varying from 4.59 (SbPRP9) to 11.11 (SbPRP8). The molecular weights range from 15027.87 (SbPRP-16) to 278911.49 (SbPRP10) Da. The pI values ranges between 5.99 (SbHyPRP17) and 9.66 (SbHyPRP8 and 22), and the molecular weights extend from 12255.61 (SbHyPRP15) to 70134.60 Da (SbHyPRP11). The analyses reveal that majority of them are basic in nature with few exceptions. All the predicted SbPRPs are unstable, (if instability index > 40, considered stable in nature) and the instability index ranges between 40.73 (SbPRP10) and 87.66 (SbPRP20). In the case of SbHyPRPs, 81.48% of proteins were unstable and the remaining were stable (Table 1). The instability index varies from 35.37 (SbHyPRP1) to –87.66 (SbHyPRP26) (Table 2). SbPRPs are hydrophilic proteins (86%) with lower aliphatic indices. When compared, the majority of the SbHyPRPs (66.66%) are found to be hydrophobic in nature, with a high aliphatic index. Out of 21 SbPRPs, only 38.09% contain transmembrane helices, while 33.33% of SbHyPRPs consist of the same (Tables 1, 2). Six (28.57%) of the SbPRPs target chloroplasts, four extracellular, one plastid, two the cytoplasm, three the nucleus, two the vacuole, and the remaining three target different localizations (Table 1). Out of 27 SbHyPRPs, 11 have been distributed in the chloroplast, 10 extracellular, one in mitochondria, two in the nucleus, one in the cytoplasm, and two extracellular or vacuolar (Table 2). Based on the analysis, it has been predicted that chloroplast and extracellular are the major target sites for their localization. Most of the SbPRPs have been predicted in the cytoplasm, indicating that they are secretary proteins. SbPRPs have been predicted to have a signal peptide at the N-terminus and a hydrophobic cysteine-rich region at the C-terminus. These results indicate that PRP proteins are secreted to the extracellular matrix through the signal peptide. The majority of the SbPRP proteins phosphorylate at serine and threonine sites and very few of them at tyrosine residues, threonine being the predominant one. Protein kinase C (PKC) and unspecified (unsp) are the most dominant types present in higher amounts in all the SbPRP proteins. Next to CKII, PKA P38MAPK, CDK5, PKG, and GSK3 are the most common kinases associated with phosphorylation (Supplementary Table 8). Whereas, in the case of SbHyPRPs, serine is the governing phosphorylation site followed by threonine and tyrosine. TPKC, unsp, PKA, CKII, cdc2, P38MAPK, CDK5, PKG, and GSK3 are the common kinases for SbHyPRP protein phosphorylations (Supplementary Table 9).
The MEME web server was used to analyze the motif distribution of SbPRPs and SbHyPRPs. In SbPRPs, the predicted motif 1 appears as the biggest and motif 10, the smallest. The SbPRP family does not exhibit a common motif distribution pattern. Motif 1 is the common motif found at N-terminus in fewer proteins. The motifs 2, 3, 4, and 5 are the structural conserved motifs, present in the middle of the proteins (Supplementary Figures 5, 6). In SbHyPRPs, motif 6 is the largest with 43 amino acids in length, and motifs 7 and 8 with 11 amino acids, are the smallest. Motif 4 is the common conserved motif present at N-terminus, while motif 3 is at C-terminus. Motif 8 exhibited repeated distribution in SbHyPRP 11, 26, and 27 (Supplementary Figures 7, 8). These conserved motifs reflect features for PRP and HyPRP proteins as shown by Wang et al. (2006) and hence provide confirmatory identification of proteins from the sorghum genome.
Protein models help to understand structure-function relationships (Rasheed et al., 2020). So, the 3D structures of SbPRP and SbHyPRP proteins were predicted with the best PDB templates (Supplementary Figure 9). The template PDB id, template description, chain, model of the oligomer, and their structure validations are represented in Supplementary Table 10. 3D structures of one SbPRP (SbPRP12) and six SbHyPRP (SbHyPRP2, SbHyPRP5, SbHyPRP6, SbHyPRP13, SbHYPRP17, and SbHYPRP26) proteins displayed significant sequence similarity percent ranging from 31.1% (SbPRP12) to 55.26% (SbHyPRP6). 3D structures of five SbPRP proteins (SbPRP3, SbPRP6, SbPRP10, SbPRP17, and SbPRP19) did not show any significant (<30%) sequence similarity (Supplementary Table 10). All the generated Ramachandran plots for structure validation are represented in Supplementary Figure 10.
Plant regulatory small RNAs (sRNAs) include microRNAs (miRNAs) and small interfering RNAs (siRNAs) like phasiRNAs which regulate gene expressions, thus, modulating plant growth as well as stress tolerance (Haak et al., 2017). The psRNA Target Server predicts the miRNAs and their target sites in SbPRP and SbHyPRP genes. The 21 SbPRPS predicted in the present study appear to be the targets for 34 different miRNAs. The sbi-miR437 and sbi-miR5568 have been predicted to target the SbPRP family. The miRNAs participate in cleavage and translation inhibition events. The SbPRP13, 18, and 20 appear as the major targets for different miRNAs (Supplementary Table 11). Interestingly, 29 different miRNAs target 25 SbHyPRPs and moderate the activity. The SbHyPRP2, 3, 14, 16, 18, and 24 are the major gene targets for the majority of the miRNAs (Supplementary Table 12). SbPRP9 and SbPRP21 showed a high number of acetylations (Supplementary Table 13). When compared with SbPRPs, SbHyPRPs displayed a diminished number of acetylation and methylation sites. SbHyPRP 11 and 16 have been predicted to have myriad acetylation, and methylation sites (Supplementary Table 14). Prediction of miRNAs, methylation and acetylation sites indicate that SbPRP and SbHyPRP genes might be regulated epigenetically.
Out of a total of 36 SSRs, 18 were predicted in PRPs and HyPRPS. They were present on chromosomes 1, 3, 4, 5, 6, 7, and 10. Chromosome 1 has been found as the hot spot with three PRPs and five HyPRPs. While the majority of them carried single SSRs, PRP8, 10, HyPRP1, 4, and 6, have 2, and PRP6 and 8 have 3 SSRs. Tri-nucleotide repeats have been predicted as the major repeats (25), followed by hexamer (7) and tetra-nucleotide repeats (4) (Supplementary Table 15).
Using digital data, the expression of 21 SbPRP genes was analyzed in the root, shoot, and leaf tissues exposed to cold and drought stress conditions. Overall, high expression levels of SbPRPs were noticed in the leaf compared to root and shoot tissues. Out of 21 SbPRPs, SbPRP16, SbPRP17, SbPRP19, SbPRP1, and SbPRP3 displayed high expressions in shoot tissues (Figure 6A). Among diverse developmental stages such as maturity, dough, milking, flowering, and seedling, the seedling stage showed superior expressions followed by the flowering stage. SbPRP19, SbPRP16, SbPRP17, SbPRP1, SbPRP13, SbPRP13, and SbPRP20 showed high expression levels in seedlings treated with cold and drought stresses in comparison with other developmental stages (Figure 6B). SbPRP18 displayed higher expression levels, followed by 16, and 19 under both the stresses (Figure 6C). SbHyPRP5, SbHyPRP20, SbHyPRP13, and SbHyPRP21 showed elevated expressions in the leaf. Expressions of SbHyPRP12, SbHyPRP17, and SbHyPRP11 were strikingly high in roots (Figure 7A). SbHyPRP12 and SbHyPRP26 were markedly upregulated during maturity, dough, milking, and flowering stages in comparison with other genes. The majority of the SbHyPRP genes are also expressed during the seedling stage (Figure 7B). The heat map of SbHyPRPs showed both up- and down-regulation up to 2.5-folds under cold and drought stresses (Figure 7C). SbHyPRP17, SbHyPRP2, SbHyPRP18, and SbHyPRP20 showed upregulation in comparison with other genes under stress conditions. SbHyPRP2, SbHyPRP8, and SbHyPRP17 displayed high expressions under drought compared to cold stress conditions (Figure 7C).
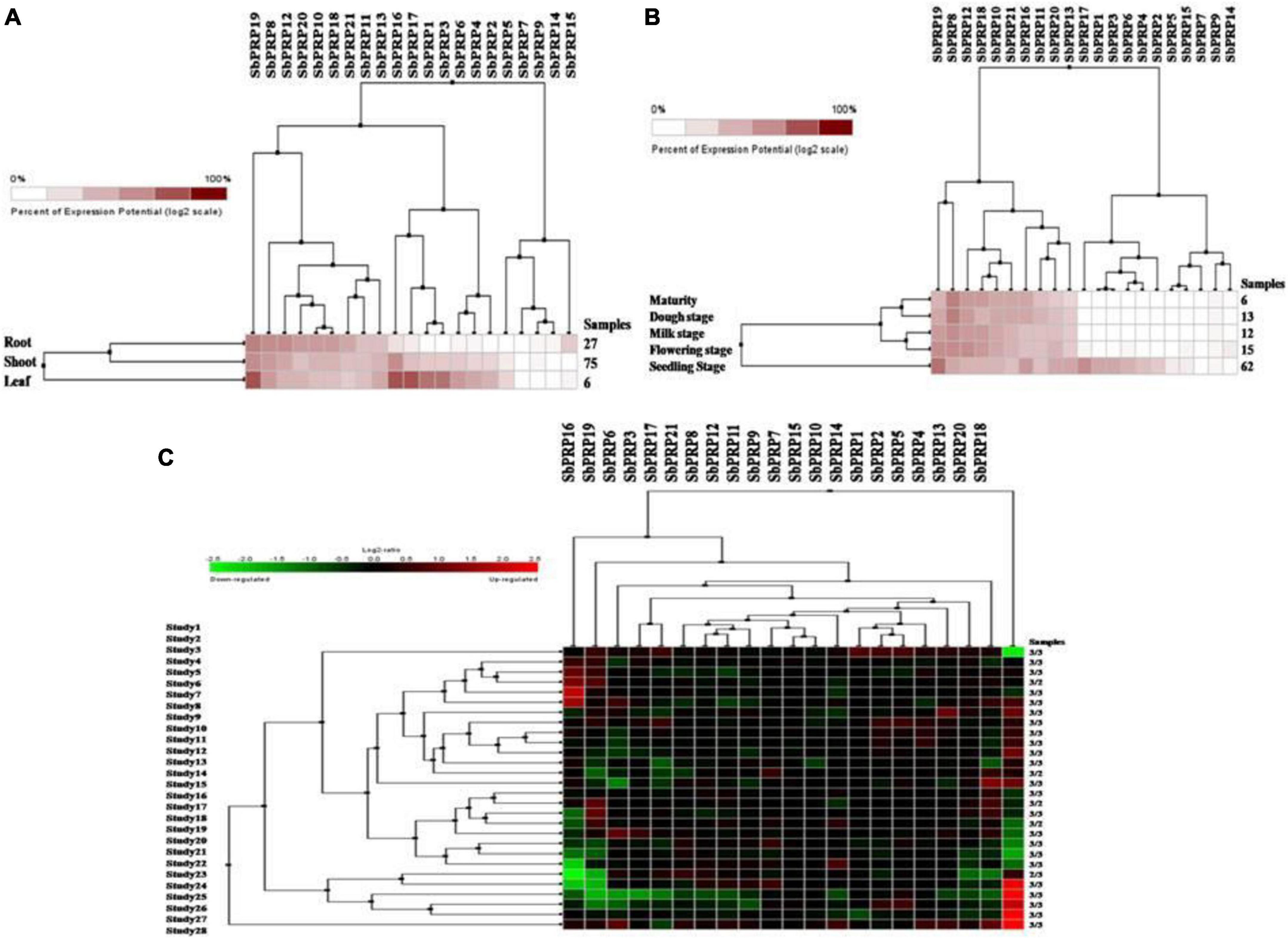
Figure 6. Digital expression profile of SbPRP genes in different organs, developmental stages under cold and drought stresses. (A) SbPRP in anatomical tissues, (B) SbPRP genes in developmental stages, (C) SbPRP gene expressions under cold and drought stress conditions.
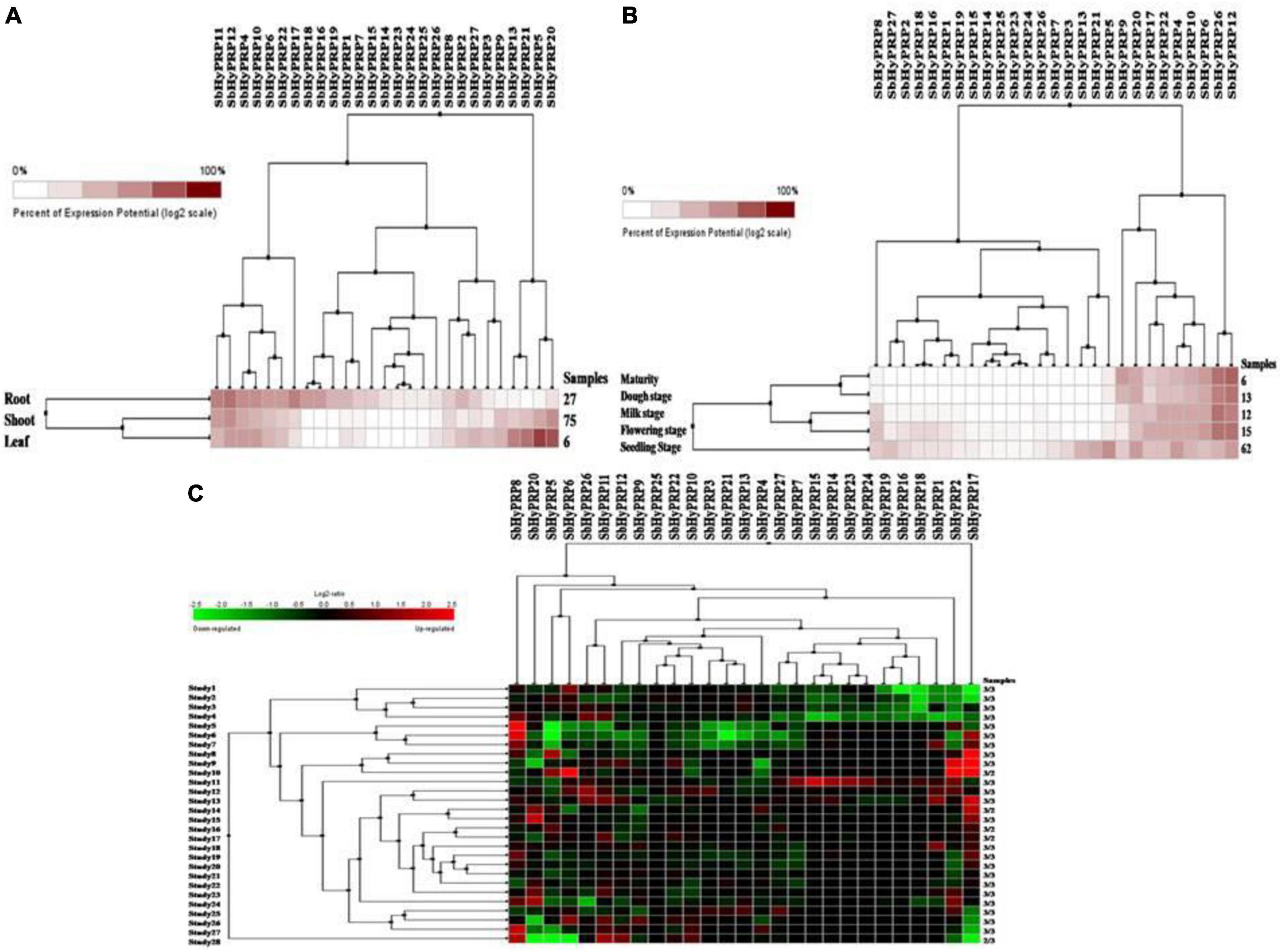
Figure 7. Digital expression profile of SbHyPRP genes in different organs, developmental stages under cold and drought stresses. (A) SbHyPRPin anatomical tissues, (B) SbHyPRP in developmental stages, (C) SbHyPRP gene expressions under cold and drought stresses.
Expression analysis of SbPRP and SbHyPRP genes were carried out in root, stem, and leaf to address their role in diverse organs. There is no single gene that has exhibited uniform levels of expression in all the three tissues. Under control conditions (without any stress), only SbPRP2 (1.303-folds) and SbPRP4 (0.824-folds) were constitutively expressed in roots, while SbPRP11 (1.820-folds) and SbPRP18 (1.473-folds) in the stem, and SbPRP3 (0.985) in leaf tissues (Figure 8A and Supplementary Table 16). Drought has considerably stimulated the expression of SbPRP17 in leaf (4.73-folds), stem (4.08-folds), and root (3.94-folds), but only in the roots of SbPRP12 (3.61-folds). Salt stress induced the activity of SbPRP17 by 4.25-, 3.53-, and 4.44-folds in leaf, stem, and root tissues, respectively, and also only in the roots of SbPRP19 (4.34-folds). Heat stress has upregulated SbPRP3 (3.31-folds), SbPRP6 (3.34-folds) and SbPRP17 (4.94-folds), and PRP19 (4.00-folds) in the leaf, SbPRP8 (4.58-folds) and SbPRP17 (3.744-folds) in the stem, SbPRP6, SbPRP12 and SbPRP17 by 4.05-, 3.84-, and 3.65-folds respectively in the root (Figure 8B and Supplementary Table 16). Cold stress has stimulated SbPRP4, SbPRP5, SbPRP6, SbPRP7, SbPRP9, SbPRP10, SbPRP13, SbPRP15, SbPRP17, SbPRP20, and SbPRP21 in the leaf between 3.08 to 4.99-folds. The fold-wise increase in the stem of SbPRP3, SbPRP6, SbPRP12, SbPRP13, SbPRP17, and SbPRP19 ranged from 3.01 to 5.20-folds. In the case of roots, the elevated transcript levels were 4.53, 5.38, 4.82, 3.03, 4.82, 3.24, and 4.59-folds in SbPRP3, SbPRP6, SbPRP12, SbPRP13, SbPRP15, SbPRP17, and SbPRP19, respectively. While ABA stimulated SbPRP6 and SbPRP17 by 3.85-, 3.65- (SbPRP6), 4.94-, and 4.20-folds (SbPRP17) in the leaf and root, it activated SbPRP3, SbPRP12, and SbPRP17 in the stems by 4.00, 3.04, and 4.00-folds, respectively (Figure 8B and Supplementary Table 16). Zn has jacked up only SbPRP17 by 3.58-folds in the leaf, SbPRP3 and SbPRP17 in the stems by 3.51- and 3.37-, and in the roots by 4.81- and 3.52-folds respectively. From the results obtained, it is inferred that SbPRP17 is critical for stress tolerance since it is expressed under various abiotic stress conditions in all three tissues, followed by SbPRP3, SbPRP6, and SbPRP12 (Figure 8B, Supplementary Figure 11, and Supplementary Table 16).
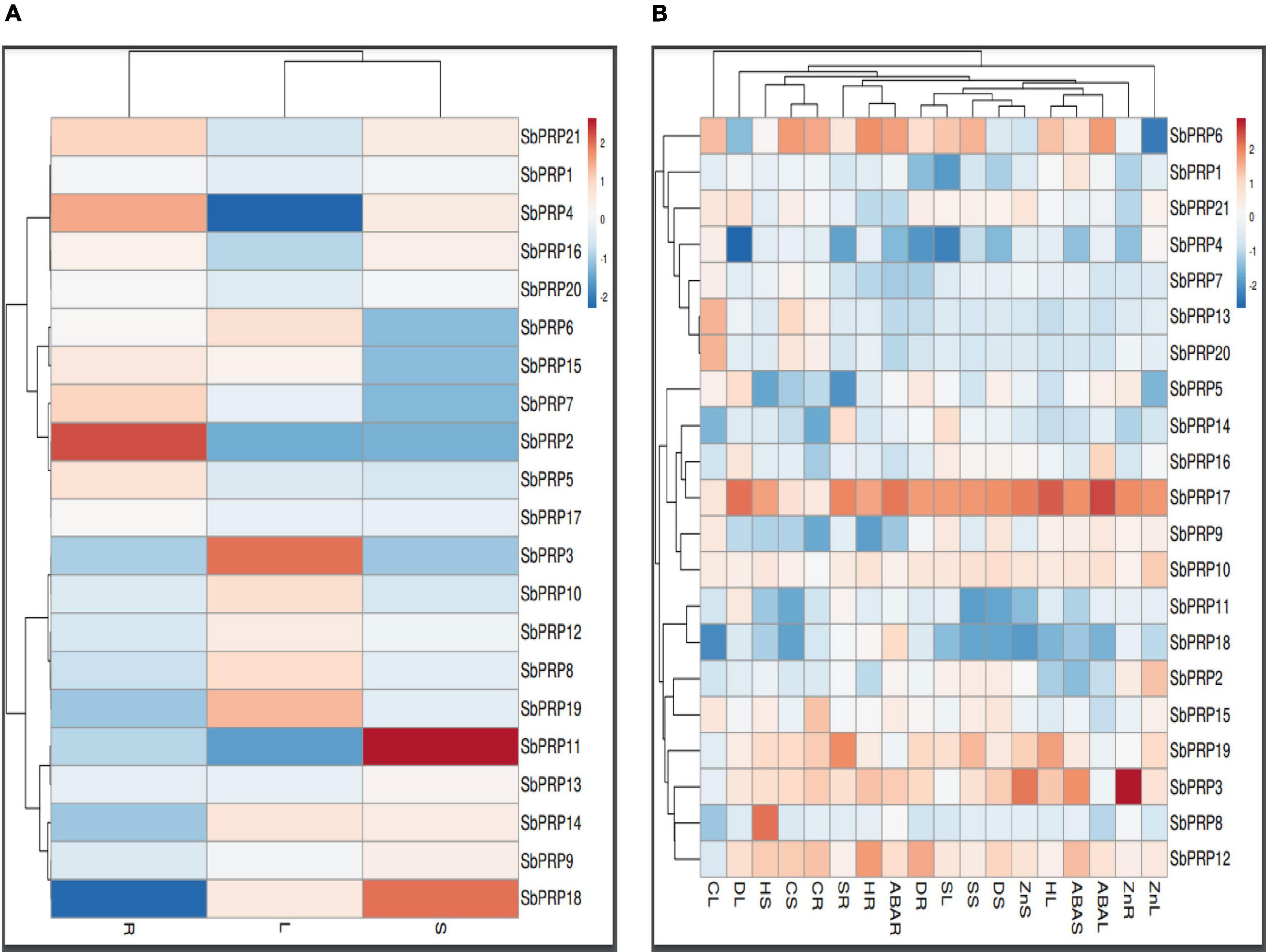
Figure 8. qRT-PCR expression profiling of SbPRPs. (A) Native expression analysis in root, stem, and leaf, (B) relative expression analysis in root, stem, and leaf under drought, salt, heat, cold, ABA, and Zn stresses. R, root; S, stem; L, leaf.
Unlike SbPRPs, SbHyPRPs displayed uniform constitutive expressions in all the tissues. In roots, SbHyPRP12 was enhanced by 1.75-, SbHyPRP10 by 1.60-, and SbHyPRP21 by 1.54-folds when compared to the expression levels of the housekeeping gene actin. In the case of the stem, SbHyPRP3 was upregulated by 2.77-, SbHyPRP20 by 2.03-, SbHyPRP16 by 1.95, and SbHyPRP13 by 1.91-folds. While in leaf tissues, gene expressions increased by 1.77- in SbHyPRP14, 1.35- in SbHyPRP11, and 1.32-folds in SbHyPRP18 devoid of any stress (Figure 9A and Supplementary Table 17). Significant upregulation of the SbHyPRP2 and SbHyPRP17 genes was noticed in comparison with control tissues regardless of the tissue and type of stress imposed. Drought stress has increased the expressions of SbHyPRP2 by 5.16-, 9.67-, and 8.17-folds in the leaf, stem, and root, respectively. SbHyPRP17 registered an upregulation of 26.66-, 17.01-, and 15.35-folds in leaf, stem, and root, respectively, under drought. The highest expression of 44.15-folds was recorded in SbHyPRP20 in the stem tissues under water-deficit conditions. Expressions varied between 3.45-, and 24.00-folds in SbHyPRP5, SbHyPRP6, SbHyPRP10, SbHyPRP13, SbHyPRP16, SbHyPRP27 genes under drought stress (Figure 9B and Supplementary Table 17). Under salt stress, the highest expression of 21.79-folds was noticed in the root tissues of SbHyPRP17, followed by leaf tissues (19.10-folds). SbHyPRP2 was upregulated by 15.60-folds in the leaf under saline conditions. Activities were also high under salt stress in SbHyPRP6, SbHyPRP7, SbHyPRP10, SbHyPRP14, and SbHyPRP26. SbHyPRP17 displayed a dramatic increase in the activity (30.82-folds) under heat stress in the leaves, followed by SbHyPRP2 in the leaf (23.44-folds). SbHyPRP2 and SbHyPRP17 also recorded markedly high activities in the stem and root tissues (15.3-, 12.58-, 8.54-, 13.40-folds respectively). Cold temperature induced the transcript levels of SbHyPRP2, SbHyPRP6, SbHyPRP9, SbHyPRP12, SbHyPRP17, SbHyPRP26, and SbHyPRP27 in the leaves in the range of 3.34- (SbHyPRP26) to 19.96-folds (SbHyPRP12). Again in the stem, the expression level was high in SbHyPRP12 (11.35-folds). SbHyPRP2 and SbHyPRP13 exhibited 10.13- and 17.83-folds enhanced transcript levels in the roots under cold stress (Figure 9B and Supplementary Table 17). Significant upregulation in the activity of SbHyPRP17 was noticed in the leaf, stem, and root tissues (30.87-, 16.10-, and 18.46-folds), and the activities ranged between 3.13- and 9.57-folds in other genes when exposed to the phytohormone ABA. Zn treatment boosted the activities of SbHyPRP26 (25.39-folds) and SbHyPRP17 (11.96-folds) in the leaves. While SbHyPRP2 displayed the maximum activity (16.83-folds) in the stem, the roots of SbHyPRP17 showed 11.47-fold higher activities compared to the controls (Figure 9B, Supplementary Figure 12, and Supplementary Table 17).

Figure 9. qRT-PCR expression profiling of SbHyPRPs. (A) Native expression analysis in root, stem, and leaf, (B) relative expression analysis in root, stem, and leaf under drought, salt, heat, cold, ABA, and Zn stresses. R, root; S, stem; L, leaf.
The accessibility of the whole sorghum genome sequence (Paterson et al., 2009) has provided an excellent opportunity for genome annotation, comparative genomic analysis, and understanding of the structural and functional variations among the diverse gene families. Publicly available genome sequence data have helped us in identifying different gene families in sorghum (Kumari et al., 2018; Maheshwari et al., 2019; Nagaraju et al., 2020, 2021). PRPs and HyPRPs play crucial roles in abiotic stress and growth functions as well (Zhang and Schläppi, 2007; Priyanka et al., 2010; Huang et al., 2011; Berendsen et al., 2012; Li et al., 2016; Efthimiadou et al., 2020). Recently, OsHyPRP06/R3L1 has been found to modulate root system development besides salt tolerance via apoplastic ROS homeostasis in rice (Zhao et al., 2021). Great variability exists in the number of PRPs and HyPRPs in different taxa (Dvorakova et al., 2007; Neto et al., 2013; Fujino et al., 2014; Showalter et al., 2016; Kapoor et al., 2019; Zhang et al., 2021), which could be due to differences in the genera or the method employed. Genome-wide analyses of sorghum resulted in the identification of 21 SbPRPs and 27 SbHyPRPs in the present study. Detection of both SbPRPs and SbHyPRPs in the present study in S. bicolor indicates their conservation in evolution.
The exon-intron structure in the genomic sequences furnishes an insight into evolutionary relationships among the genes or organisms and perhaps needs intensified investigations (Koralewski and Krutovsky, 2011). Our predicted results on intron-exon structure organization both in SbPRPs and SbHyPRPs conform to the results obtained from other species (Fujino et al., 2014; Kapoor et al., 2019; Zhang et al., 2021). Several SbPRP and SbHyPRP genes have been found with no or few introns. It has been shown in yeast, A. thaliana, and mice that genes with rapidly changing expression levels in response to environmental stresses contain lower intron densities (Jeffares et al., 2008). Introns delay the regulatory responses, hence genes that need rapid adjustment for survival against the changing environment are intron poor (Jeffares et al., 2008; Lan et al., 2013). It appears, therefore, that there is an intron loss during the evolution of a few eukaryotic lineages. Besides structural diversity, the complexity of the eukaryotic genes might contribute to the evolution of gene families. Our results show that the SbPRP and SbHyPRP genes have been distributed randomly on different chromosomes.
In the present study, cis-elements such as DPBF, MYC, MYB, G-box, ABRE, and ERE have been identified in sorghum besides phosphorous, ammonium, sulfur, and iron starvation-responsive elements. SbPRPs have been found rich in phosphorous, iron, ammonium, and sulfur-responsive elements, while SbHyPRPs in sulfur-responsive elements, indicating the diverse roles they play in nutrient homeostasis. These elements might target transcription factors like SPB, zinc finger, WRKY, WD-40, NAC, MYB, HSFs, GRAS, ARFs, and bHLH families and play a vital role in abiotic stress tolerance (Katiyar et al., 2015). MYB and ABRE elements might play a role in the ABA-dependent signaling pathway in response to drought, salt, and cold stresses. Promoter analysis also revealed the presence of elements that participate in sugar signaling, nuclear factor binding, and biotic stress-responsive cis-elements such as WBOXNTERF3, WBOXATNPR1, and CGTCA that respond to wounds, pathogens, and salicylic acid (Hwang et al., 1998; Schumann et al., 2017). In cotton, the GhHyPRP4 promoter is cold-inducible (Huang et al., 2011). Kapoor et al. (2019) reported a good correlation between inducible expression in OsHyPRP16 in response to the pathogen Magnaporthe oryzae, hormones ABA, methyl jasmonate, salicylic acid, and abiotic stresses like cold, and salt in the promoter sequences of rice.
Phylogenetic trees infer that PRPs are highly conserved in different taxa. Analysis of the phylogenetic trees reveals that homologous gene pairs have high sequence identities with other taxa compared, thus displaying their evolutionary relationships. The clustering of the PRP and HyPRPs of sorghum, rice, maize, and disconnection from Arabidopsis indicates their evolutionary divergence during monocotyledon lineage-specific evolution. Expansion of gene families occurs through segmental, and tandem duplications besides transposition events (Kong et al., 2007). In the present study, a total of eight paralogs were observed including two regional and six segmental duplication events in SbPRPs, while three regional and five segmental duplications in SbHyPRPs, indicating that segmental and tandem duplications are responsible for gene family expansion. Gene order conservation of PRP and HyPRP has been identified by circos maps. High conservation of PRP and HyPRP genes has been noticed between Sorghum and Oryza when compared to Sorghum and Arabidopsis, indicating that rice and sorghum are closely related.
Predictions related to the subcellular localization of the proteins in the extracellular matrix, identification of signal peptides, and also cysteine-rich regions indicate that all these proteins might not be secretory type as has also been pointed out by Banday et al. (2022). PRPs and HyPRPs are located on the cell wall and plasma membrane and regulate the synthesis of cell walls aside from responding to abiotic stresses (Jose-Estanyol et al., 2004; Kavi Kishor et al., 2015; Mellacheruvu et al., 2016; Gujjar et al., 2018; Li et al., 2019; Zhang et al., 2021). In citrus, CsPRP4 protein was localized in the plasma membrane (Ma et al., 2013), and in Juglans sigillata, JsPRP1 in the epidermal cells (Liu et al., 2018). While HyPRP protein from Arabidopsis has been identified on the cell wall (Zhang and Schläppi, 2007), GhHyPRP3 from cotton has been noticed on the plasma membrane (Qin et al., 2013). In apple, PRP proteins have been detected in the cell membrane, nucleus, and chloroplast (Zhang et al., 2021). AZELAIC ACID INDUCED 1 (AZI1) is a HyPRP, a member of the HyPRP superfamily (Dvorakova et al., 2007), and a signal-anchored protein that has a proline-rich region (PRR) for targeting the protein to plastids (Cecchini et al., 2015, 2021). HyPRPs possess transmembrane domains besides a PRR and a lipid transfer domain. A feedback regulatory loop exists between glycerol-3-phosphate (G3P) and lipid transfer protein Defective in Induced Resistance (DIR1) in plants and AZI1 mediates azelaic-acid-induced systemic immunity (Yu et al., 2013). But, the subcellular locations of HyPRPs and their precise functions during development and biotic and abiotic stress tolerance have mostly not yet been identified. HyPRPs may be localized to the plastid outer membranes, thylakoid membranes, membranes of the endoplasmic reticulum, plasma membrane, or plasmodesmata (Banday et al., 2022). Few of the HyPRPs have a set of proteins that target the outer membrane of the plastid with the aid of PRRs. HyPRPs modulate systemic immunity against Pseudomonas syringae and also help in the colonization of P. simiae WCS417 in the roots. Taken together, it appears that HyPRPs may facilitate signal molecule transport and help in plant immunity, growth, and development besides abiotic stress tolerance. Similar to the studies carried out by He et al. (2002); Fujino et al. (2014), and Kapoor et al. (2019), our findings suggest that SbPRP and SbHyPRP proteins are localized in different subcellular locations.
The highly expressed SbPRP and SbHyPRP protein structures were predicted and structurally validated for accuracy using Ramachandran plots. Similar studies were reported previously by Kumar et al. (2018). Understanding the prediction of protein structures can facilitate the understanding of their function (Jumper et al., 2021).
SbPRPs and SbHyPRPs showed methylation as well as acetylation sites. Elements of epigenetic regulation such as histone variants, chromatin remodeling, non-coding RNAs, and DNA methylations are highly conserved among plant systems and represent an important component of the omnifarious biological process (Goldberg et al., 2007; Allis and Jenuwein, 2016; Ueda and Seki, 2020). It is known that epigenetic regulation including acetylation and methylation coordinates abiotic stress responses (Kim et al., 2015; Asensi-Fabado et al., 2017; Luo et al., 2017; Ueda and Seki, 2020). Methylations and acetylations found in sorghum might regulate the genes post-transcriptionally which are implicated in abiotic stresses.
Micro RNAs (miRNAs) are small, non-coding, regulatory elements, which play overriding roles in gene regulation by disturbing the transcripts of genes and mediating the plant adaptation to the changing environmental conditions (Bartel, 2004). In the present study, miRNAs that target SbPRPs and SbHyPRPs have been predicted. We assume that these miRNAs regulate these genes. miRNAs are critical not only for stress responses but also for plant development (Sunkar et al., 2007; Rubio-Somoza et al., 2009). In the present investigation, miR437 and miR5568 have been identified as dominant miRNAs, specific for monocots and absent from eudicots (Sunkar et al., 2007). The conservation and divergence of miRNAs may help to understand miRNA evolution and identify the species-specific miRNAs and the roles that they play in gene regulation. SICKLE (SIC), a proline-rich protein has been found vital in A. thaliana for development and abiotic stress tolerance. It is also involved in microRNA biogenesis (Zhan et al., 2012). Loss-of-function sic-1 mutant Arabidopsis displayed higher levels of introns, sickle-like margin, delayed flowering, hypersensitivity to chilling, and salt stresses. The SIC protein has been observed to colocalize with the miRNA biogenesis component hyponastic leaves 1 (HYL1). These observations have led Zhan et al. (2012) to conclude that SIC is necessary for the biogenesis of some miRNAs and spliced introns. DNA and chromatin modifications modulate the expression of stress-responsive genes. Such modifications are also heritable inferring their role in the evolutionary mechanisms associated with abiotic stress tolerance (Singroha et al., 2022).
Simple sequence repeats are used generally to study genetic relatedness or variation among the species. SSRs get mutated quickly; hence, differentiation of closely related species is possible. So far, several QTLs associated with diverse traits were identified in many crops. The predicted SSRs associated with PRPs and HyPRPs could be informative and used to evaluate the genetic diversity and relationships among diverse sorghum lines. Such markers may also be useful to the breeders who are aiming to generate abiotic stress tolerant sorghum lines.
Digital expression data indicated that the expressions of PRP and HyPRP genes were higher in leaf tissues during the seedling stage of plant development. The results indicate the importance of SbPRP19, SbPRP16, SbPRP17, SbPRP1, SbPRP13, SbPRP13, and SbPRP20 genes during seedling when compared to other stages. Such genes need to be functionally validated to find out the seedling specific roles they play especially under water-deficit conditions. Increased expressions of both SbHyPRP12 and SbHyPRP26 during maturity, dough, milking, and flowering stages infer the crucial role they might play during these stages. High expressions of SbHyPRP2, SbHyPRP8, and SbHyPRP17 under water-deficit conditions compared to cold stress surmise that functional validation of these genes needs to be carried out.
Insolubilization of PRPs leads to the formation of protein–protein or protein–carbohydrate linkages within cell walls and assists in the stability of extracellular matrix in plants (Bernhardt and Tierney, 2000; Akiyama and Pillai, 2003). PRPs respond to the changing environments and thus abet in coping with stress situations (Kavi Kishor et al., 2015; Zhang et al., 2021). Literature on the transcript expressions of PRPs is scanty. Munoz et al. (1998) in Cicer arietinum and Xu et al. (2007) in Gossypium hirsutum have shown tissue-specific expression of PRP genes. GmPRP1 mRNAs have been found in elongating and mature regions of the seedling hypocotyl cells in Glycine max, GmPRP2 in phloem cells, and GmPRP3 in the endodermoid layer of cells in the hypocotyl elongating region (Wyatt et al., 1992). In the case of Arabidopsis, AtPRP1 and AtPRP3 gene expressions were noticed in roots, and AtPRP2 and AtPRP4 in aerial organs (Fowler et al., 1999; Bernhardt and Tierney, 2000). SbPRP genes were activated under different abiotic stress conditions aside from ABA and Zn treatments in a tissue-specific manner in sorghum. This reveals that these genes are spatially regulated during plant development in a specific tissue or cell type as has been shown by others (Munoz et al., 1998; Xu et al., 2007; Zhang et al., 2021). Out of 21 SbPRPs, SbPRP17 appeared to have ubiquitous expressions in all tissues though the magnitude differed, suggesting that SbPRPs function in many physiological processes during stress. It is known that PRP genes are up- and down-regulated, involved in plant growth and environmental challenges. In support of this statement, Gothandam et al. (2010) overexpressed rice OsPRP3 and showed that transgenics acquire cold tolerance, but knockdown mutants display defects in flower morphogenesis. In Arabidopsis, PRP is critical for development, and abiotic stress tolerance is associated with microRNA biogenesis (Zhan et al., 2012). When Gossypium hirsutum GhPRP5 gene was overexpressed in Arabidopsis, transgenics exhibited diminished growth in comparison with wild-type plants. On the other hand, GhPRP5 knock-down mutants in cotton improved fiber development with longer fiber length (Xu et al., 2013). Thus, PRPs have been proved to be associated with positive or negative regulation of plant growth. In the present study, it has been noticed that SbPRPs respond to drought, salt, heat, cold, ABA, and Zn inferring that SbPRPs play critical roles, but the responses differ among various environmental stimuli and Zn treatment. Such a phenomenon was noticed in Brassica napus, and BnPRP responds to low and high temperatures, and drought, but not injury (Goodwin et al., 1996). In Glycine max, GmPRP was triggered by salicylic acid, an endogenous circadian rhythm and diverse abiotic stress conditions (He et al., 2002). Contrary to the above, OsPRP was down-regulated during submergence in Oryza sativa (Akiyama and Pillai, 2003). In the case of Arabidopsis, out of 18 PRP genes, nematode infection upregulated the expression of PRP4, PRP11, and PRP16 and P. syringae infection induced PRP9 and PRP10 (Showalter et al., 2010). Overexpression of Juglans sigillata JsPRP1 resulted in drought tolerance, CdCl2 stress, Colletotrichum gloeosporiodes infection (Liu et al., 2018). In apple also, many MdPRP genes were expressed in all tissues except MdPRP4, MdPRP5, MdPRP7, and MdPRP8 (Zhang et al., 2021). Interestingly, Zn also stimulated SbPRP3 and SbPRP17 genes in all three tissues, but their implication in zinc nutrition or starvation await functional assays to demonstrate the precise role of ABA and Zn in PRP functioning. Overall, these findings indicate that PRPs are differentially regulated under diverse abiotic stress conditions and might play decisive roles in modulating growth in addition to stress responses.
HyPRPs are a large family of cell wall protein with a variable N-terminal domain and a conserved C-terminal domain which is related to non-specific lipid transfer proteins. Initially, they have been regarded as only cell wall proteins, but slowly their multifunctional nature is being unraveled. HyPRPs take an active part in abscission, cell elongation, cell wall synthesis, growth, development, and biotic and abiotic stress tolerance (Wu et al., 1993; Zhang and Schläppi, 2007; Dvorakova et al., 2012; Sundaresan et al., 2018). Compared to SbPRP genes, multiple SbHyPRP genes have been found upregulated upon exposure to different abiotic stress conditions. Several HyPRP genes have been cloned and overexpressed and the transgenics displayed freezing or low-temperature stress tolerance inferring their positive regulatory role in stress response (Zhang and Schläppi, 2007; Huang et al., 2011; Peng et al., 2015). Overexpression of other HyPRP genes such as CcHyPRP (Cajanus cajan), GhHyPRP3 (Gossypium hirsutum), and MfHyPRP (Medicago falcata) increased the levels of tolerance to multiple abiotic stresses like salt, and cold temperature (Priyanka et al., 2010; Qin et al., 2013; Tan et al., 2013). Pitzschke et al. (2014) identified and characterized AZI1, a lipid transfer protein (LTP)-related HyPRP, a target of mitogen-activated protein kinase 3 (MPK3). While null mutants of azi1 are salt susceptible, overexpressing lines exhibit tolerance. Thus, MPK3 appears as a positive regulator of AZI1 and a key interactant. HyPRP from Arabidopsis has been found to play a key role in SAR when infected with Pseudomonas syringae (Jung et al., 2009). Neto et al. (2013) identified soybean HyPRP family members and their responses to Asian soybean rust disease. EARLI1 is an Arabidopsis HyPRP, protects plants against cold stress, and improves germination under low temperatures and salt stress conditions (Zhang and Schläppi, 2007). Experiments performed by Li et al. (2012) using recombinant EARLI1 protein indicate that its application to Botrytis cinerea, Fusarium oxysporum inhibits the growth and cell viability. These results point out that HyPRPs are positively associated with biotic stress tolerance. Conversely, Yeom et al. (2012) identified the HyPRP1 gene in Capsicum annuum and Nicotiana benthamiana which is constitutively expressed in different organs. But the gene was down-regulated when inoculated with pathogens. Overexpression of the gene resulted in accelerated death along with the down-regulation of ROS-scavenging genes. Gene silencing suppressed pathogen-induced cell death but increased disease resistance. It appears therefore that HyPRP acts as a positive regulator of cell death and a negative regulator of basal defense against pathogens. A DOUBLE HYBRID PROLINE-RICH PROTEIN 1 (AtDHyPRP1) detected in Arabidopsis contains two tandem proline-rich domain-eight cysteine motifs (PRD-8CMs) and is a novel HyPRP and induced by salicylic acid, methyl jasmonate, virulent and non-virulent strains of the pathogen P. synringae. Overexpression of this gene resulted in increased pathogen resistance, but RNAi lines of AtDHyPRP1 exhibited susceptibility, indicating its involvement in defense response (Li et al., 2014). Li et al. (2016) noticed that the SpHyPRP1 gene isolated from Solanum pennellii play a negative role in abiotic stress tolerance in tomato. Similarly, GhHyPRP1 negatively regulates the resistance to Verticillium dahliae in cotton via the thickening of cell walls and ROS accumulation (Yang et al., 2018). These studies underpin the significant role that HyPRPs play during biotic and abiotic stress tolerance.
Proline-rich proteins and HyPRPs are dominant constituents of cell wall proteins and help in the stability of the extracellular matrix. Their role in plant growth, development, and biotic and abiotic stresses are being rolled in recent times. The present study predicts the identification of 21 PRPs and 27 HyPRPs in sorghum and also the presence of cis-regulating elements implicated in abiotic stress tolerance, and nutritional starvation. Further, qRT-PCR analysis indicates that SbPRP and SbHyPRP genes are stimulated under cold, drought, high temperature, and salt stress conditions in sorghum. However, it is necessary to understand the molecular regulation of PRP and HyPRP proteins and also the functional validation of these genes in a wide array of biotic and abiotic stress conditions.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
PK and NS designed the experiments. MN, RV, and GR carried out the bioinformatics analysis. GR performed the qRT-PCR experiments. PK, NS, GR, MN, RV, NJ, AKS, PS, and SP prepared the manuscript and refined it. PK, MN, GR, NS, and AKS revised the manuscript. All authors have read and approved the final version of the manuscript.
This work was financially supported from the German Federal Ministry for Economic Cooperation and Development (BMZ) commissioned and administered through the Deutsche Gesellschaft für Internationale Zusammenarbeit (GIZ) Fund for International Agricultural Research (FIA), grant number: 81206686 and the Bill & Melinda Gates Foundation AGGRi Alliance (grant No. OPP1194925), and Stress Tolerant Rice for Africa and South Asia (STRASA) Phase III (grant No. OPP1088843).
PK acknowledges VFSTR, Guntur for providing emeritus fellowship. GR is grateful to ICMR for providing fellowship. RV is a graduate student worked in the laboratory of PK and is grateful to VFSTR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.952732/full#supplementary-material
Akiyama, T., and Pillai, M. A. (2003). Isolation and characterization of a gene for a repetitive proline-rich protein (OsPRP) down-regulated during submergence in rice (Oryza sativa). Physiol. Plant. 118, 507–513.
Allis, C. D., and Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500. doi: 10.1038/nrg.2016.59
Asensi-Fabado, M. A., Amtmann, A., and Perrella, G. (2017). Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 106–122. doi: 10.1016/j.bbagrm.2016.07.015
Bailey, T. L., and Gribskov, M. (1998). Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 14, 48–54. doi: 10.1093/bioinformatics/14.1.48
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Banday, Z. Z., Cecchini, N. M., Speed, D. J., Scott, A. T., Parent, C., Hu, C. T., et al. (2022). Friend or foe: Hybrid proline-rich proteins determine how plants respond to beneficial and pathogenic microbes. Plant Physiol. 1:kiac263. doi: 10.1093/plphys/kiac263
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-web: A web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bernhardt, C., and Tierney, M. L. (2000). Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol. 122, 705–714. doi: 10.1104/pp.122.3.705
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL:modeling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Blanco-Portales, R., Lopez-Raez, J. A., Bellido, M. L., Moyano, E., Dorado, G., Gonzalez-Reyes, A., et al. (2004). A strawberry fruit-specific and ripening-related gene codes for a HyPRP protein involved in polyphenol anchoring. Plant Mol. Biol. 55, 763–780. doi: 10.1007/s11103-004-1966-4
Blom, N., Sicheritz-Ponten, T., Gupta, R., Gammeltoft, S., and Brunak, S. (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649. doi: 10.1002/pmic.200300771
Bohnert, H. J., Gong, Q., Li, P., and Ma, S. (2006). Unraveling abiotic stress tolerance mechanisms-getting genomics going. Curr. Opin. Plant Biol. 9, 180–188. doi: 10.1016/j.pbi.2006.01.003
Bokszczanin, K. (2013). Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 4:315. doi: 10.3389/fpls.2013.00315
Boron, A. K., Orden, J. V., Markakis, M. N., Mouille, G., Adriaensen, D., Verbelen, J., et al. (2014). Proline-rich protein-like PRPL1 controls elongation of root hairs in Arabidopsis thaliana. J. Exp. Bot. 65, 5485–5495. doi: 10.1093/jxb/eru308
Bouton, S., Viau, L., Lelievre, E., and Limami, A. M. (2005). A gene encoding a protein with a proline-rich domain (MtPPRD1), revealed by suppressive subtractive hybridization (SSH), is specifically expressed in the Medicago truncatula embryo axis during germination. J. Exp. Bot. 56, 825–832. doi: 10.1093/jxb/eri077
Boutrot, F., Chantret, N., and Gautier, M. F. (2008). Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genom. 9:86. doi: 10.1186/1471-2164-9-86
Burge, C. B., and Karlin, S. (1998). Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 8, 346–354. doi: 10.1016/s0959-440x(98)80069-9
Carella, P. (2020). Xylem-mobile oxylipins are critical regulators of induced systemic resistance in maize. Plant Cell 32, 13–14. doi: 10.1105/tpc.19.00924
Cassab, G. I. (1998). Plant cell wall proteins. Annu. Rev. Plant Biol. Plant Mol. Biol. 49, 281–309. doi: 10.1146/annurev.arplant.49.1.281
Cecchini, N. M., Roychoudhry, S., Speed, D. J., Steffes, K., Tambe, A., Zodrow, K., et al. (2019). Underground azelaic acid-conferred resistance to Pseudomonas syringae in Arabidopsis. Mol. Plant Microbe Interact. 32, 86–94. doi: 10.1094/MPMI-07-18-0185-R
Cecchini, N. M., Speed, D. J., Roychoudhry, S., and Greenberg, J. T. (2021). Kinases and protein motifs required for AZI1 plastid localization and trafficking during plant defense induction. Plant J. 105, 1615–1629. doi: 10.1111/tpj.15137
Cecchini, N. M., Steffes, K., Schlappi, M. R., Gifford, A. N., and Greenberg, J. T. (2015). Arabidopsis AZI1 family proteins mediate signal mobilization for systemic defence priming. Nat. Commun. 6:7658. doi: 10.1038/ncomms8658
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J., Zhao, J., Ning, J., Liu, Y., Xu, J., Tian, S., et al. (2014). NtProRP1, a novel proline-rich protein, is an osmotic stress-responsive factor and specifically functions in pollen tube growth and early embryogenesis in Nicotiana tabacum. Plant Cell Environ. 37, 499–511. doi: 10.1111/pce.12174
Dai, X., and Zhao, P. X. (2011). psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
Dvorakova, L., Cvrckova, F., and Fischer, L. (2007). Analysis of the hybrid proline-rich protein families from seven plant species suggests rapid diversification of their sequences and expression patterns. BMC Genom. 8:412. doi: 10.1186/1471-2164-8-412
Dvorakova, L., Srba, M., Opatrny, Z., and Fischer, L. (2012). Hybrid proline-rich proteins: Novel players in plant cell elongation? Ann. Bot. 109, 453–462. doi: 10.1093/aob/mcr278
Efthimiadou, A., Katsenios, N., Chanioti, S., Giannoglou, M., Djordjevic, N., and Katsaros, G. (2020). Effect of foliar and soil application of plant growth promoting bacteria on growth, physiology, yield and seed quality of maize under Mediterranean conditions. Sci. Rep. 10:21060. doi: 10.1038/s41598-020-78034-6
Esen, A., Bietz, J. A., Paulis, J. W., and Wall, J. S. (1982). Tandem repeats in the N-terminal sequence of a proline-rich protein from corn endosperm. Nature 296, 678–679. doi: 10.1038/296678a0
Fischer, L., Lovas, A., Opatrny, Z., and Banfalvi, Z. (2002). Structure and expression of a hybrid proline-rich protein gene in the Solanaceous species, Solanum brevidens, Solanum tuberosum and Lycopersicum esculentum. J. Plant Physiol. 159, 1271–1275. doi: 10.1078/0176-1617-00744
Fowler, T. J., Bernhardt, C., and Tierney, M. L. (1999). Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol. 121, 1081–1091. doi: 10.1104/pp.121.4.1081
Fujino, K., Obara, M., and Sato, K. (2014). Diversification of the plant-specific hybrid glycine-rich protein (HyGRP) genes in cereals. Front. Plant Sci. 5:489. doi: 10.3389/fpls.2014.00489
Gao, Q. M., Zhu, S., Kachroo, P., and Kachroo, A. (2015). Signal regulators of systemic acquired resistance. Front. Plant Sci. 6:228. doi: 10.3389/fpls.2015.00228
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., and Bairoch, A. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook. Springer Protocols Handbooks, ed. J. M. Walker (Totowa, NJ: Humana Press), 571–607. doi: 10.1385/1-59259-890-0:571
Goldberg, A. D., Allis, C. D., and Bernstein, E. (2007). Epigenetics: A landscape takes shape. Cell 128, 635–638. doi: 10.1016/j.cell.2007.02.006
Goodwin, W., Pallas, J. A., and Jenkins, G. I. (1996). Transcripts of a gene encoding a putative cell wall-plasma membrane linker protein are specifically cold-induced in Brassica napus. Plant Mol. Biol. 31, 771–781. doi: 10.1007/BF00019465
Gothandam, K. M., Nalini, E., Karthikeyan, S., and Shin, J. S. (2010). OsPRP3, a flower specific proline-rich protein of rice, determines extracellular matrix structure of floral organs and its overexpression confers cold-tolerance. Plant Mol. Biol. 72, 125–135. doi: 10.1007/s11103-009-9557-z
Gujjar, R. S., Karkute, S. G., Rai, A., Singh, M., and Singh, B. (2018). Proline-rich proteins may regulate free cellular proline levels during drought stress in tomato. Curr. Sci. 114, 915–920. doi: 10.18520/cs/v114/i04/909-915
Gujjar, R. S., Pathak, A. D., Karkute, S. G., and Watana, K. S. (2019). Multifunctional proline rich proteins and their role in regulating cellular proline content in plants under stress. Biol. Plant 63, 448–454. doi: 10.32615/bp.2019.078
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). GSDS: A gene structure display server. Yi Chuan Hered. 29, 1023–1026.
Haak, D. C., Fukao, T., Grene, R., Hua, Z., Ivanov, R., Perrella, G., et al. (2017). Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 8:1564. doi: 10.3389/fpls.2017.01564
Hadebe, S. T., Modi, A. T., and Mahabudhi, T. (2017). Drought tolerance and water use of cereal crops: A focus on sorghum as a food security crop in sub-saharan Africa. J. Agron. Crop Sci. 203, 177–191. doi: 10.1111/jac.12191
He, C. Y., Zhang, J. S., and Chen, S. Y. (2002). A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor. Appl. Genet. 104, 1125–1131. doi: 10.1007/s00122-001-0853-5
Holk, A., Klumpp, L., and Scherer, G. F. E. (2002). A cell wall protein down-regulated by auxin suppressed cell expansion in Daucus carota (L.). Plant Mol. Biol. 50, 295–305. doi: 10.1023/a:1016052613196
Hong, J. C., Nagao, R. T., and Key, J. L. (1990). Characterization of a proline-rich cell wall protein gene family of soybean. A comparative analysis. J. Biol. Chem. 265, 2470–2475. doi: 10.1016/S0021-9258(19)39823-0
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Huang, G., Gong, S., Xu, W., Li, P., Zhang, D., Qin, L., et al. (2011). GhHyPRP4, a cotton gene encoding putative hybrid proline-rich protein, is preferentially expressed in leaves and involved in plant response to cold stress. Acta Biochim. Biophys. Sin. 43, 519–527. doi: 10.1093/abbs/gmr040
Huang, R. (2018). Research progress on plant tolerance to soil salinity and alkalinity in Sorghum. J. Integr. Agric. 17, 739–746. doi: 10.1016/S2095-3119(17)61728-3
Hunt, L., Amsbury, S., Baillie, A., Movahedi, M., Mitchell, A., Afsharinafar, M., et al. (2017). Formation of the stomatal outer cuticular ledge requires a guard cell wall proline-rich protein. Plant Physiol. 174, 689–699. doi: 10.1104/pp.16.01715
Hwang, Y. S., Karrer, E. E., Thomas, B. R., Chen, L., and Rodriguez, R. L. (1998). Three cis-elements required for rice α-amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 36, 331–341. doi: 10.1023/A:1005956104636
Jeffares, D. C., Penkett, C. J., and Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Jose-Estanyol, M., Gomis-Ruth, F. X., and Puigdomènech, P. (2004). The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 42, 355–365. doi: 10.1016/j.plaphy.2004.03.009
Jose-Estanyol, M., and Puigdomenech, P. (1993). Structure and expression of genes coding for structural proteins of the plant cell wall. New Phytol. 125, 259–282. doi: 10.1111/j.1469-8137.1993.tb03881.x
Jose-Estanyol, M., and Puigdomenech, P. (1994). Hybrid-proline rich and related proteins. Curr. Top. Mol. Genet. 2, 109–126.
Jose-Estanyol, M., and Puigdomenech, P. (2000). Plant cell wall glycoproteins and their genes. Plant Physiol. Biochem. 38, 97–108. doi: 10.1016/S0981-9428(00)00165-0
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Jung, H. W., Tschaplinski, T. J., Wang, L., Glazebrook, J., and Greenberg, J. T. (2009). Priming in systemic plant immunity. Science 324, 89–91. doi: 10.1126/science.1170025
Kachroo, A., and Kachroo, P. (2020). Mobile signals in systemic acquired resistance. Curr. Opin. Plant Biol. 58, 41–47. doi: 10.1016/j.pbi.2020.10.004
Kapoor, R., Kumar, G., Arya, P., Jaswal, R., Jain, P., Singh, K., et al. (2019). Genome-wide analysis and expression profiling of rice hybrid proline-rich proteins in response to biotic and abiotic stresses, and hormone treatment. Plants 8:343. doi: 10.3390/plants8090343
Katiyar, A., Smita, S., Muthusamy, S. K., Chinnusamy, V., Pandey, D. M., and Bansal, K. C. (2015). Identification of novel drought-responsive microRNAs and trans-acting siRNAs from Sorghum bicolor (L.) Moench by high-throughput sequencing analysis. Front. Plant Sci. 6:506. doi: 10.3389/fpls.2015.00506
Kavi Kishor, P. B. K., Hima Kumari, P., Sunita, M. S. L., and Sreenivasulu, N. (2015). Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front. Plant Sci. 6:544. doi: 10.3389/fpls.2015.00544
Keller, B. (1993). Structural cell wall proteins. Plant Physiol. 101, 1127–1130. doi: 10.1104/pp.101.4.1127
Kim, J. M., Sasaki, T., Ueda, M., Sako, K., and Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6:114. doi: 10.3389/fpls.2015.00114
Kong, H., Landherr, L., Frohlich, M., Leebens-Mack, J., Ma, H., and dePamphilis, C. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Koralewski, T. E., and Krutovsky, K. V. (2011). Evolution of exon-intron structure and alternative splicing. PLoS One 6:e18055. doi: 10.1371/journal.pone.0018055
Kumar, A., Batra, R., Gahlaut, V., Gautam, T., Kumar, S., Sharma, M., et al. (2018). Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.). PLoS One 13:e0208409. doi: 10.1371/journal.pone.0208409
Kumar, P., Joy, J., Pandey, A., and Gupta, D. (2017). PRmePRed: A protein arginine methylation prediction tool. PLoS One 12:e0183318. doi: 10.1371/journal.pone.0183318
Kumari, P. H., Anil Kumar, S., Katam, R., Reddy, P. S., Nagaraju, M., Bhanu Prakash, A., et al. (2018). Genome-wide identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHE) homologs in Sorghum bicolor. Genes 9:236. doi: 10.3390/genes9050236
Lan, P., Li, W., Lin, W. D., Santi, S., and Schmidt, W. (2013). Mapping gene activity of Arabidopsis root hairs. Genome Biol. 14:R67. doi: 10.1186/gb-2013-14-6-r67
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., et al. (2004). SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32, D142–D144. doi: 10.1093/nar/gkh088
Li, A., Xue, Y., Jin, C., Wang, M., and Yao, X. (2006). Prediction of Nepsilon-acetylation on internal lysines implemented in Bayesian discriminant method. Biochem. Biophys. Res. Commun. 350, 818–824. doi: 10.1016/j.bbrc.2006.08.199
Li, B. C., Zhang, C., Chai, Q. X., Han, Y. Y., Wang, X. Y., Liu, M. X., et al. (2014). Plasmalemma localisation of double hybrid proline-rich protein 1 and its function in systemic acquired resistance of Arabidopsis thaliana. Funct. Plant Biol. 41, 768–779. doi: 10.1071/FP13314
Li, J., Ouyang, B., Wang, T., Luo, Z., Yang, C., Li, H., et al. (2016). HyPRP1 gene suppressed by multiple stresses play a negative role in abiotic stress tolerance in tomato. Front. Plant Sci. 7:967. doi: 10.3389/fpls.2016.00967
Li, L., Zhang, C., Xu, D., Schläppi, M., and Xu, Z. Q. (2012). Expression of recombinant EARLI1, a hybrid proline-rich protein of Arabidopsis, in Escherichia coli and its inhibition effect to the growth of fungal pathogens and Saccharomyces cerevisiae. Gene 506, 50–61. doi: 10.1016/j.gene.2012.06.070
Li, S., Zhang, Y., Ding, C., Gao, X., Wang, R., Mo, W., et al. (2019). Proline-rich protein gene PdPRP regulates secondary wall formation in poplar. J. Plant Physiol. 233, 58–72. doi: 10.1016/j.jplph.2018.12.007
Liu, D. Q., Han, Q., Shah, T., Chen, C. Y., Wang, Q., Tang, B. F., et al. (2018). A hybrid proline-rich cell-wall protein gene JsPRP1 from Juglans sigillata Dode confers both biotic and abiotic stresses in transgenic tobacco plants. Trees 32, 1199–1209. doi: 10.1007/s00468-018-1702-3
Luo, M., Cheng, K., Xu, Y., Yang, S., and Wu, K. (2017). Plant responses to abiotic stress regulated by histone deacetylases. Front. Plant Sci. 8:2147. doi: 10.3389/fpls.2017.02147
Ma, Y. Y., Wu, T. L., Zhong, G. Y., and Zheng, Y. Q. (2013). Overexpression of a new proline-rich protein encoding gene CsPRP4 increases starch accumulation in Citrus. Sci. Hortic. 260:108744. doi: 10.1016/j.scienta.2019.108744
Maheshwari, P., Divya, K., Reddy, P. S., Tejaswi, U. N., Nagaraju, M., Rajasheker, G., et al. (2019). Genome-wide identification and expression profile analysis of NUCLEAR FACTOR Y family genes in Sorghum bicolor L. (Moench). PLoS One 14:e0222203. doi: 10.1371/journal.pone.0222203
McNeil, M., Darvill, A. G., Fry, S. C., and Albersheim, P. (1984). Structure and function of the primary cell walls of plants. Annu. Rev. Biochem. 53, 625–663. doi: 10.1146/annurev.bi.53.070184.003205
Mellacheruvu, S., Tamirisa, S., Vudem, D. R., and Khareedu, V. R. (2016). Pigeonpea hybrid-proline-rich protein (CcHyPRP) confers biotic and abiotic stress tolerance in transgenic rice. Front. Plant Sci. 6:1167. doi: 10.3389/fpls.2015.01167
Moller, S., Croning, M. D., and Apweiler, R. (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17, 646–653. doi: 10.1093/bioinformatics/17.7.646
Munoz, F. J., Dopico, B., and Labrador, E. (1998). A cDNA encoding a proline-rich protein from Cicer arietinum. changes in expression during development and abiotic stresses. Physiol. Plant. 102, 582–590. doi: 10.1111/j.1467-7652.2009.00467.x
Nagaraju, M., Anuj Kumar, S., Jalaja, N., Rao, D. M., and Kavi Kishor, P. B. (2021). Functional exploration of chaperonin (HSP60/10) family genes and their abiotic stress-induced expression patterns in Sorghum bicolor. Curr. Genom. 22, 137–155. doi: 10.2174/1389202922666210324154336
Nagaraju, M., Reddy, P. S., Anil Kumar, S., Kumar, A., Rajasheker, G., Rao, D. M., et al. (2020). Genome-wide identification and transcriptional profiling of small heat shock protein gene family under diverse abiotic stress conditions in Sorghum bicolor (L.). Int. J. Biol. Macromol. 142, 822–834. doi: 10.1016/j.ijbiomac.2019.10.023
Neto, L. B., de Oliveira, R. R., Wiebke-Strohm, B., Bencke, M., Weber, R. L. M., Cabreira, C., et al. (2013). Identification of the soybean HyPRP family and specific gene response to Asian soybean rust disease. Genet. Mol. Biol. 36, 214–224. doi: 10.1590/S1415-47572013005000017
Newman, A. M., and Cooper, J. B. (2011). Global analysis of proline-rich tandem repeat proteins reveals broad phylogenetic diversity in plant secretomes. PLoS One 6:e23167. doi: 10.1371/journal.pone.0023167
Paterson, A. H., Bowers, J. E., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. doi: 10.1038/nature07723
Peng, L., Jia, M. M., and Liu, J. H. (2015). RNAi-based functional elucidation of PtrPRP, a gene encoding a hybrid proline rich protein, in cold tolerance of Poncirus trifoliata. Front. Plant Sci. 6:808. doi: 10.3389/fpls.2015.00808
Pfaffl, M. W., Horgan, G. W., and Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. doi: 10.1093/nar/30.9.e36
Pitzschke, A., Datta, S., and Persak, H. (2014). Salt stress in Arabidopsis: Lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol. Plant 7, 722–738. doi: 10.1093/mp/sst157
Priyanka, B., Sekhar, K., Reddy, V. D., and Rao, K. V. (2010). Expression of pigeonpea hybrid-proline-rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance. Plant Biotechnol. J. 8, 76–87. doi: 10.1111/j.1467-7652.2009.00467.x
Qin, L. X., Zhang, D. J., Huang, G. Q., Li, L., Li, J., Gong, S. Y., et al. (2013). Cotton GhHyPRP3 encoding a hybrid proline-rich protein is stress inducible and its overexpression in Arabidopsis enhances germination under cold temperature and high salinity stress conditions. Acta Physiol. Plant. 35, 1531–1542. doi: 10.1007/s11738-012-1194-5
Rasheed, F., Markgren, J., Hedenqvist, M., and Johansson, E. (2020). Modeling to understand plant protein structure-function relationships-implications for seed storage proteins. Molecules 25:873. doi: 10.3390/molecules25040873
Reddy, P. S., Reddy, D. S., Sivasakthi, K., Bhatnagar-Mathur, P., Vadez, V., and Sharma, K. K. (2016). Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front. Plant Sci. 7:529. doi: 10.3389/fpls.2016.00529
Rubio-Somoza, I., Cuperus, J. T., Weigel, D., and Carrington, J. C. (2009). Regulation and functional specialization of small RNA-target nodes during plant development. Curr. Opin. Plant Biol. 12, 622–627. doi: 10.1016/j.pbi
Schumann, U., Lee, J., Kazan, K., Ayliffe, M., and Wang, M. B. (2017). DNA-demethylase regulated genes show methylation-independent spatiotemporal expression patterns. Front. Plant Sci. 8:1449. doi: 10.3389/fpls.2017.01449
Showalter, A. M. (1993). Structure and function of plant cell wall proteins. Plant Cell 5, 9–23. doi: 10.1105/tpc.5.1.9
Showalter, A. M., Keppler, B., Lichtenberg, J., Gu, D. Z., and Welch, L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513. doi: 10.1104/pp.110.156554
Showalter, A. M., Keppler, B. D., Liu, X., Lichtenberg, J., and Welch, L. R. (2016). Bioinformatic identification and analysis of hydroxyproline-rich glycoproteins in Populus trichocarpa. BMC Plant Biol. 16:229. doi: 10.1186/s12870-016-0912-3
Silva, T. N., Thomas, J. B., Dahlberg, J., Rhee, S. Y., and Mortimer, J. C. (2022). Progress and challenges in sorghum biotechnology, a multipurpose feedstock for the bioeconomy. J. Exp. Bot. 73, 646–664. doi: 10.1093/jxb/erab450
Singroha, G., Kumar, S., Gupta, O. P., Singh, G. P., and Sharma, P. (2022). Uncovering the epigenetic marks involved in mediating salt stress tolerance in plants. Front. Genet. 13:811732. doi: 10.3389/fgene.2022.811732
Sommer-Knudsen, J., Bacic, A., and Clarke, A. E. (1998). Hydroxyproline-rich plant glycoproteins. Phytochem 47, 483–497. doi: 10.1016/S0031-9422(97)00724-3
Sundaresan, S., Philosoph-Hadas, S., Ma, C., Jiang, C. Z., Riov, J., Mugasimangalam, R., et al. (2018). The tomato hybrid proline-rich protein regulates the abscission zone competence to respond to ethylene signals. Hortic. Res. 5:28. doi: 10.1038/s41438-018-0033-2
Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309. doi: 10.1016/j.tplants.2007.05
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi: 10.1093/nar/gkl315
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, J., Zhuo, C., and Guo, Z. (2013). Nitric oxide mediates cold-and dehydration-induced expression of a novel MfHyPRP that confers tolerance to abiotic stress. Physiol. Plant. 149, 310–320. doi: 10.1111/ppl.12032
Tello-Ruiz, M. K., Naithani, S., Gupta, P., Olson, A., Wei, S., Preece, J., et al. (2021). Gramene 2021: Harnessing the power of comparative genomics and pathways for plant research. Nucleic Acids Res. 49, D1452–D1463. doi: 10.1093/nar/gkaa979
Ueda, M., and Seki, M. (2020). Histone modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiol. 182, 15–26. doi: 10.1104/pp.19.00988
Wang, C., El-Shetehy, M., Shine, M. B., Yu, K., Navarre, D., Wendehenne, D., et al. (2014). Free radicals mediate systemic acquired resistance. Cell Rep. 7, 348–355. doi: 10.1016/j.celrep.2014.03.032
Wang, R., Chong, K., and Wang, T. (2006). Divergence in spatial expression patterns and in response to stimuli of tandem-repeat paralogues encoding a novel class of proline-rich proteins in Oryza sativa. J. Exp. Bot. 57, 2887–2897. doi: 10.1093/jxb/erl057
Wang, X., Liu, H., Yu, F., Hu, B., Jia, Y., Sha, H., et al. (2019). Differential activity of the antioxidant defence system and alterations in the accumulation of osmolyte and reactive oxygen species under drought stress and recovery in rice (Oryza sativa L.) tillering. Sci. Rep. 9:8543. doi: 10.1038/s41598-019-44958-x
Wu, H., Zou, J., May, B., Gu, Q., and Cheung, A. Y. (1993). A tobacco gene family for flower cell wall proteins with a proline-rich domain and a cysteine-rich domain. Proc. Natl. Acad. Sci. U. S. A. 90, 6829–6833. doi: 10.1073/pnas.90.14.6829
Wyatt, R. E., Nagao, R. T., and Key, J. L. (1992). Patterns of soybean proline-rich protein gene expression. Plant Cell 4, 99–110. doi: 10.1105/tpc.4.1.99
Xu, W. L., Huang, G. Q., Wang, X. L., Wang, H., and Li, X. B. (2007). Molecular characterization and expression analysis of five novel genes encoding proline-rich proteins in cotton (Gossypium hirsutum). Prog. Biochem. Biophys. 34, 509–517. doi: 10.1104/pp.005538
Xu, W. L., Zhang, D. J., Wu, Y. F., Qin, L. X., Huang, G. Q., Li, J., et al. (2013). Cotton PRP5 gene encoding a proline-rich protein is involved in fiber development. Plant Mol. Biol. 82, 353–365. doi: 10.1007/s11103-013-0066-8
Yang, J., Zhang, Y., Wang, X., Wang, W., Li, Z., Wu, J., et al. (2018). HyPRP1 performs a role in negatively regulating cotton resistance to Verticillum dahliae via the thickening of cell walls and ROS accumulation. BMC Plant Biol. 18:339. doi: 10.1186/s12870-018-1565-1
Yeom, S., Seo, E., Oh, S., Kim, K. W., and Choi, D. (2012). A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J. 69, 755–768. doi: 10.1111/j.1365-313X.2011.04828.x
Yu, K., Soares, J. M., Mandal, M. K., Wang, C., Chanda, B., Gifford, A. N., et al. (2013). A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 3, 1266–1278. doi: 10.1016/j.celrep.2013.03.030
Zhan, X., Wang, B., Li, H., Liu, R., Kalia, R., Zhu, J. K., et al. (2012). Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc. Nat. Acad. Sci. U. S. A. 109, 18198–18203. doi: 10.1073/pnas.1216199109
Zhang, X., Gong, X., Li, D., Yue, H., Qin, Y., Liu, Z., et al. (2021). Genome-wide identification of PRP genes in apple genome and the role of MdPRP6 in response to heat stress. Int. J. Mol. Sci. 22:5942. doi: 10.3390/ijms22115942
Zhang, Y., and Schläppi, M. (2007). Cold responsive EARLI1 type HyPRPs improve freezing survival of yeast cells and form higher order complexes in plants. Planta 227, 233–243. doi: 10.1007/s00425-007-0611-2
Keywords: proline-rich proteins, hybrid proline-rich proteins, gene expressions, abiotic stresses, Sorghum bicolor
Citation: Rajasheker G, Nagaraju M, Varghese RP, Jalaja N, Somanaboina AK, Singam P, Ramakrishna C, Penna S, Sreenivasulu N and Kishor PBK (2022) Identification and analysis of proline-rich proteins and hybrid proline-rich proteins super family genes from Sorghum bicolor and their expression patterns to abiotic stress and zinc stimuli. Front. Plant Sci. 13:952732. doi: 10.3389/fpls.2022.952732
Received: 25 May 2022; Accepted: 16 August 2022;
Published: 26 September 2022.
Edited by:
Ashish Kumar Srivastava, Bhabha Atomic Research Centre (BARC), IndiaReviewed by:
Anuj Kumar, Dalhousie University, CanadaCopyright © 2022 Rajasheker, Nagaraju, Varghese, Jalaja, Somanaboina, Singam, Ramakrishna, Penna, Sreenivasulu and Kishor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: P. B. Kavi Kishor, cGJrYXZpQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.