Corrigendum: Development of plant systemic resistance by beneficial rhizobacteria: Recognition, initiation, elicitation and regulation
- 1Key Lab of Bio-Organic Fertilizer Creation, Ministry of Agriculture and Rural Affairs, Anhui Science and Technology University, Bengbu, China
- 2School of Life Sciences and Technology, Tongji University, Shanghai, China
- 3Jiangsu Provincial Key Lab of Solid Organic Waste Utilization, Jiangsu Collaborative Innovation Center of Solid Organic Wastes, Educational Ministry Engineering Center of Resource-Saving Fertilizers, Nanjing Agricultural University, Nanjing, China
A plant growing in nature is not an individual, but it holds an intricate community of plants and microbes with relatively stable partnerships. The microbial community has recently been demonstrated to be closely linked with plants since their earliest evolution, to help early land plants adapt to environmental threats. Mounting evidence has indicated that plants can release diverse kinds of signal molecules to attract beneficial bacteria for mediating the activities of their genetics and biochemistry. Several rhizobacterial strains can promote plant growth and enhance the ability of plants to withstand pathogenic attacks causing various diseases and loss in crop productivity. Beneficial rhizobacteria are generally called as plant growth-promoting rhizobacteria (PGPR) that induce systemic resistance (ISR) against pathogen infection. These ISR-eliciting microbes can mediate the morphological, physiological and molecular responses of plants. In the last decade, the mechanisms of microbial signals, plant receptors, and hormone signaling pathways involved in the process of PGPR-induced ISR in plants have been well investigated. In this review, plant recognition, microbial elicitors, and the related pathways during plant-microbe interactions are discussed, with highlights on the roles of root hair-specific syntaxins and small RNAs in the regulation of the PGPR-induced ISR in plants.
Introduction
Root exudates can shape highly specific micro-environments in plant rhizosphere, which are populated by a huge variety of soil-borne bacteria (Sasse et al., 2018; Pascale et al., 2020; Stassen et al., 2021). These root-associated bacteria are known as rhizobacteria, some of which can promote plant growth and are often referred to as plant growth-promoting rhizobacteria (PGPR) (Kloepper and Schroth, 1978). It has been well documented that PGPR can improve plant growth and stress adaptation by multiple strategies, such as secretion of hormone, reduction of host ethylene levels, and promotion of nitrogen fixation (Arshad et al., 2007; Reis and Teixeira, 2015; Backer et al., 2018). Besides their roles in plant growth promotion, PGPR can secrete antagonistic substances to suppress the growth of pathogens (Ajilogba and Babalola, 2013; Haskett et al., 2021).
Diverse biotic factors such as insects, bacterial and fungal pathogens, and viruses often lead to harmful effects, as reflected by biomass reduction, yield loss, and even plant mortality (Mertens et al., 2021). A vast number of PGPR strains such as Bacillus, Pseudomonas, Enterobacter, Klebsiella, Azosprillum, and Paenibacillus induce systemic resistance (ISR) of plants against biotic stress (Pérez-García et al., 2011; Beneduzi et al., 2012; Pieterse et al., 2014; Gkizi et al., 2016). PGPR can emit volatile organic compounds (VOCs) to stimulate the ISR responses in plants (Ryu et al., 2004). Different kinds of secondary metabolites such as bacterial quorum sensing (QS) molecules, siderophores and cyclic lipopeptides can also be released for provoking the ISR responses of plants by several defense-related signaling pathways (Aznar and Dellagi, 2015; Grady et al., 2016). Since the first report on the PGPR-induced ISR in plants (Van Peer et al., 1991), great progress has been made in understanding the mechanisms of recognition, initiation, elicitation, and regulation of plant ISR responses. In this review, we have provided an overview of the mechanisms and multiple processes associated with the PGPR-induced ISR in plants, and summarize recent advances about the roles of root hair-specific syntaxins and small RNAs in the process of the PGPR-mediated defense responses in plants.
Recognition of plant growth-promoting rhizobacteria by host plants
Most of soil-borne microbes have no direct impacts on plant growth and fitness, but there are a large number of beneficial or pathogenic microbes among the huge diversity of plant microbiomes (Trivedi et al., 2020). Beneficial associations involve diverse microbes colonized in the rhizosphere, such as root-associated bacteria and fungi that promote plant growth (Lugtenberg and Kamilova, 2009; Niu et al., 2020). Since beneficial microbes are considered as alien organisms, the active interference to plant defense systems is the basis for establishing a close and mutually beneficial relationship with the hosts. Like animals, plants have an innate immune system that is activated after identifying invading organisms. Recognition of non-self-signal molecules is a key step to achieve effective defense responses, which can be recognized by pattern recognition receptors (PRRs) in plants. Microbial associated molecular patterns (MAMPs), commonly called as pathogen associated molecular patterns (PAMPs), can be perceived by these PRRs (Dodds and Rathjen, 2010; Sanabria et al., 2010). PRRs recognize the MAMPs/PAMPs and further activate the PAMP-triggered immunity (PTI), conferring the first line of plant defense against pathogens. In plants, the most characteristic PRRs are the receptor like proteins (RLPs) or the receptor-like kinases (RLKs) (He and Wu, 2016; Tang et al., 2017). RLKs are the putative transmembrane proteins, harboring both extracellular ligand recognition and intracellular kinase domains that are responsible for signal transduction. The structures of RLPs are much similar, but lack kinase domains (Walker, 1994).
Plants can recognize common structures from different microbial species. Many common MAMPs recognized by plants have been documented, such as chitin, lipopolysaccharides (LPS), flagellin, and peptidoglycan (Mishra et al., 2012; Newman et al., 2013; D’Ambrosio et al., 2017; Ma et al., 2021). In nature, plants can not only interact with pathogens, but also form beneficial interactions with soil-borne microbes. Typical examples of symbiotic plant-microbe combinations are mycorrhizal fungi that form symbiotic relationships with many plant species and help absorb water and minerals, rhizobia that fix atmospheric nitrogen for plants, and PGPR that improve plant growth and inhibit disease occurrence (Lugtenberg and Kamilova, 2009; Suzaki et al., 2015; Lindström and Mousavi, 2020). Many of them exist outside the plant roots, while others are endophytic microbes that establish a closer relationship with the hosts. Due to many MAMPs that are widely existed and preserved in microbes, beneficial microbes display the similarity with pathogens (Hacquard et al., 2017). In order to achieve benefit services from these beneficial microbes, it is important for plants to identify the differences between pathogenic and beneficial microbes. Accumulative evidence has indicated that beneficial microbes were initially regarded as potential invaders, leading to activation of plant immunity (Pieterse et al., 2012). However, like pathogens, many PGPR strains can inhibit host defensive responses and thus establish successful relationships with their hosts. In addition, beneficial microbes seem to have similar strategies to avoid plant recognition systems (Park and Ryu, 2021).
PGPR are beneficial microbes, which establish symbiotic or non-symbiotic associations with their hosts and improve plant growth. PGPR can produce massive MAMPs (e.g., flagellin and LPS) to stimulate plant defense (Zamioudis and Pieterse, 2012). Different PGPR strains can be recognized by plant defense systems and trigger defense responses in the early stage in a way similar to PTI (Jacobs et al., 2011). LPS derived from cell wall of Pseudomonas fluorescens WCS417 is composed of lipid A/innercore/O-antigen side chains, which enhances plant defense against Fusarium pathogens (Leeman et al., 1995). However, unlike the PTI triggered by pathogens that usually lead to severe cell damage, the PGPR-mediated defense responses are transient and mild for establishing reciprocal relationships with the hosts. The flg22 peptide from beneficial Burkholderia species slightly induces oxidative burst and transiently activates the expression of defensive genes without repression of plant growth (Felix et al., 1999). Pseudomonas fluorescens WCS417r can inhibit the flagellin-triggered PTI reaction in Arabidopsis roots by secreting small molecular compounds (Millet et al., 2010). The colonization of PGPR on the roots requires local inhibition of PTI to protect PGPR from antibacterial compounds triggered by MAMPs, indicating that a coevolution results in the regulation of plant defense after perceiving specific signals of beneficial microbes.
Initiation of plant induce systemic resistance by plant growth-promoting rhizobacteria
In plants, PRRs can recognize common microbial signals, such as PAMPs and MAMPs (Lu and Tsuda, 2021). To achieve successful invasion, pathogens have evolved to weaken the activation of host immune systems. Moreover, it can deploy virulence effector proteins to inhibit the PTI signaling pathway or avoid recognition by the hosts (Zipfel, 2009; Hatsugai et al., 2017; Kud et al., 2019). Subsequently, a second line of defense has been acquired, in which specific effector molecules from pathogens are perceived by the nucleotide-binding leucine-rich repeat (NB-LRR) receptor proteins, leading to effector-triggered immunity (ETI). The gene-for-gene resistance in plants belongs to the ETI, usually accompanied by programmed cell death at infecting sites, thereby preventing the entry of biotrophic pathogens (Dodds and Rathjen, 2010). The occurrence of PTI and ETI often stimulates the ISR responses in plant tissues far from the pathogen-infecting sites and involves distant signals that propagate the enhanced defense in intact parts of plants. The classical mode of pathogen-induced resistance is often called as systemic acquired resistance (SAR), which confers the increased resistance of plants against diverse pathogens (Vlot et al., 2009). Like the pathogen recognition systems, herbivorous insects can be recognized by host plants, probably by similar signaling pathways (Pan et al., 2016; Aljbory and Chen, 2018).
Due to its broad-spectrum effectiveness, the pathogen-induced SAR is initially thought to be similar to the PGPR-induced ISR in the mechanistic way. Root colonization by PGPR induces a state of priming in host plants, in which plants can respond stronger and faster to pathogenic attacks, reflecting a common feature of ISR triggered by PGPR (Pieterse et al., 2014; Gkizi et al., 2016; Fan et al., 2018; Zehra et al., 2021). To date, diverse PGPR strains have been shown to provoke ISR in plants, which confers broad-spectrum disease resistance (Nishad et al., 2020; Samaras et al., 2021). Several PGPR strains can stimulate the SA-dependent ISR responses, which are similar to the SAR. Pseudomonas aeruginosa 7NSK2 that is not able to produce SA can increase the resistance of bean plants against pathogens, but not observed for the NahG-overexpressing plants (De Meyer et al., 1999; Audenaert et al., 2002). Also, P. fluorescens strain P3 overexpressing the biosynthetic gene of SA can trigger the SAR in plants (Maurhofer et al., 1998). In the cases that PGPR induces the SAR, the accumulation of reactive oxygen species (ROS) is essential for activating the SA-dependent SAR in plants (De Meyer et al., 1999; Niu et al., 2016a; Zehra et al., 2021). Bacillus cereus AR156 can stimulate the SAR responses by activating the SA signaling pathway in an NPR1-dependent manner (Niu et al., 2016a). Pseudomonas sp. 23S induces ISR in tomato plants, which is closely related to upregulation of PR1a transcripts (Takishita et al., 2018). More recently, several Bacillus strains have been reported to induce host ISR against the pepper bacterial spot disease by increasing the expression of PR genes such as CaPR1, CaPR4, and CaPR10 (Li et al., 2020). However, activation of the SA-independent ISR responses by PGPR also occurs in different plant species. P. fluorescens WCS417r triggers the ISR of radish plants against Fusarium oxysporum without increasing the expression of PR genes, which is a typical characteristic of SAR. Similarly, the WCS417r-induced ISR in Arabidopsis plants is not associated with the up-regulation of PR genes in leaves (Pieterse et al., 1996). The assays of NahG-overexpressing plants that are deficient in the accumulation of SA reveal that the WCS417r-induced ISR responses are independent on the SA signaling pathway (Pieterse et al., 1996, 2000). Similar phenomenon is also observed for Pseudomonas putida WCS358r-mediated activation of ISR responses in plants (Kloppholz et al., 2011). Besides the plant hormone SA, jasmonic acid (JA) and ethylene (ET) are essential for regulating plant defense responses. In Arabidopsis mutants deficient in the JA/ET signaling, the PGPR-induced ISR in plants is largely compromised (Pieterse et al., 1998). The WCS417r-induced ISR responses are defective in both the JA (e.g., coi1, jin1, and jar1) and ET signaling mutants (e.g., eir1, etr1, and ein3) (Pieterse et al., 1998; Knoester et al., 1999; Pozo et al., 2008). Mounting evidence has indicated that JA and ET are essential for activating the SA-independent defense responses induced by beneficial microbes. The PGPR-induced ISR responses are effective against pathogens and insect herbivores that are sensitive to the JA/ET-dependent defense (Van Wees et al., 2008; Pineda et al., 2010). Interestingly, the SA- and JA/ET-dependent signaling pathways are also involved in the regulation of the PGPR-induced ISR in plants. Both the SA and ET signaling are involved in the regulation of Bacillus velezensis CLA178-mediated ISR in Rosa multiflora (Chen et al., 2020). B. amyloliquefaciens CRN9 can trigger innate immunity and inhibit virus growth in plants via ISR activated by both the SA and JA/ET signaling pathways (Rajamanickam and Nakkeeran, 2020). Therefore, different PGPR strains can trigger host ISR against the attacks of pathogens and insects, involving activation of intricate signaling networks.
Emerging evidence has indicated that plants can use volatiles as the language to communicate with diverse microbes (e.g., bacteria, fungi and virus), insects and other neighboring plants (Ballhorn et al., 2013; Simpraga et al., 2016; Sharifi et al., 2018; de Almeida et al., 2021). Several kinds of volatiles (e.g., α-pinene and isothiocyanate) are often liberated by plants, which function as the cues for attracting insects (Elzen et al., 1983). Injured plants release different kinds of volatiles such as C6 fatty acid derivatives, isoprenoids (terpenes), methyl salicylate and indoles, which can be perceived by neighboring plants (Simpraga et al., 2016). Volatile cyanogen such as hydrogen cyanide (HCN) is also an important defense metabolite in plants (Figure 1). The production of cyanogenic glucosides by many plant species can be catalyzed by two key enzymes including β-glucosidases and α-hydroxynitrilase to release toxic HCN in response to insect herbivory and pathogens (Clausen et al., 2015; Hansen et al., 2018; Olsen et al., 2021). Although the release of HCN occurs only in response to cell injury, cyanogenesis has been considered as a constitutive plant defense (Pentzold et al., 2014; Bjarnholt et al., 2018). In addition, HCN can also act as a signaling molecule in plants, similar to other molecules such as nitric oxide (NO) and hydrogen sulfide (H2S) (Aroca et al., 2018; Feng et al., 2019; Wei et al., 2020). These signaling molecules can induce post-translationally modification of proteins including nitrosylation and persulfidation (Aroca et al., 2018; Gupta et al., 2020). HCN can promote the S-cyanylation of proteins by adding the SCN groups to cysteines, which leads to the alteration of protein functions (Gotor et al., 2019). Moreover, JA is also involved in the regulation of plant cyanogenesis and thus increases the resistance of lima bean to insect herbivory (Kautz et al., 2014). Interestingly, plants can emit similar kinds of volatiles after pathogenic attacks or inoculation with PGPR (Ballhorn et al., 2013). PGPR can regulate plant physiological processes and stress adaptation through different ways such as promotion of nutrient uptake and mediation of hormone signaling pathways (Sharifi and Ryu, 2017). Several PGPR strains have also been demonstrated to enhance plant defense against pathogens and insect pests by modification of host volatile profiles. Co-treatment with rhizobia and JA markedly reduces the emission of volatiles, but promotes the release of shikimic acid derivative indoles (Figure 2A). Indoles have been reported to mediate plant growth, disease resistance and bacterial pathogenesis (Lee et al., 2015; Sharifi and Ryu, 2017). It can also provoke the ISR responses of plants against herbivores by promoting the biosynthesis of terpenes and JA isoleucines in leaves. Veyrat et al. (2016) have shown that the indole-deficient plants exhibit the increased attractiveness to Spodoptera littoralis caterpillars. Before the attacks of Mamestra brassicae, pretreatment of Arabidopsis plants with Pseudomonas simiae WCS417r activates the ORA59-mediated JA/ET signaling pathways. Plants inoculated with WCS417r exhibit the increased attractivity to the parasitoid Microplitis mediator with less lilial, (E)-a-bergamotene and MeSA emissions compared with the control plants (Pangesti et al., 2015). Transcriptional changes of several genes associated with the biosynthesis of oxylipins are considerably induced by P. putida KT2440, which are closely related to the enhanced plant defense against pathogens and insects. The release of two defense-related volatiles including β-caryophyllene and indoles is remarkably promoted by the strain KT2440 (Planchamp et al., 2014).
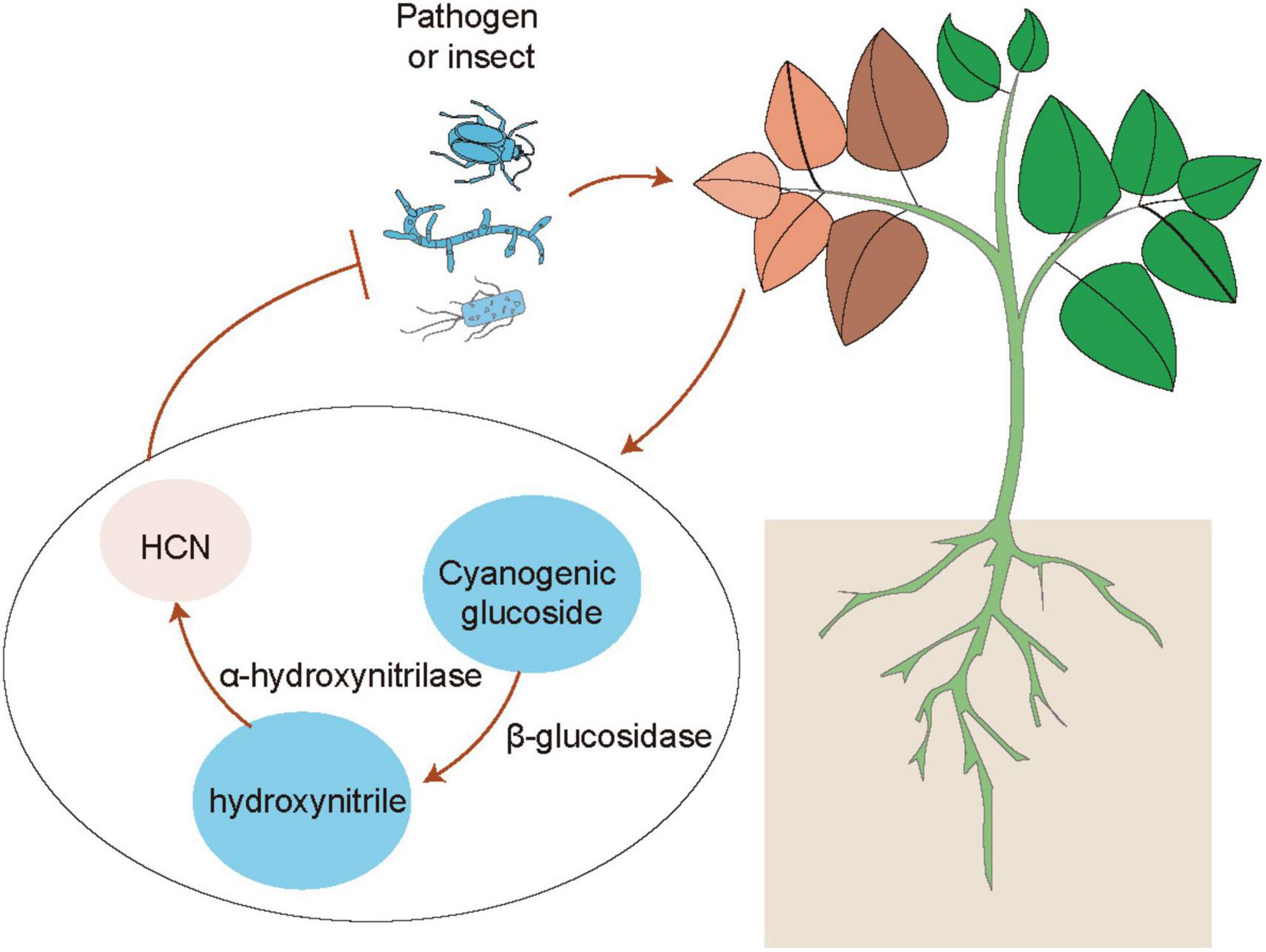
Figure 1. Mechanisms of hydrogen cyanide (HCN)-mediated plant defense. When plants are subjected to the attacks of herbivores and pathogens, the accumulation of cyanogenic glucosides in leaves is quickly catalyzed by two key enzymes including β-glucosidases and α-hydroxynitrilase into producing toxic HCN, which confers the enhanced plant defense against herbivores and pathogens.
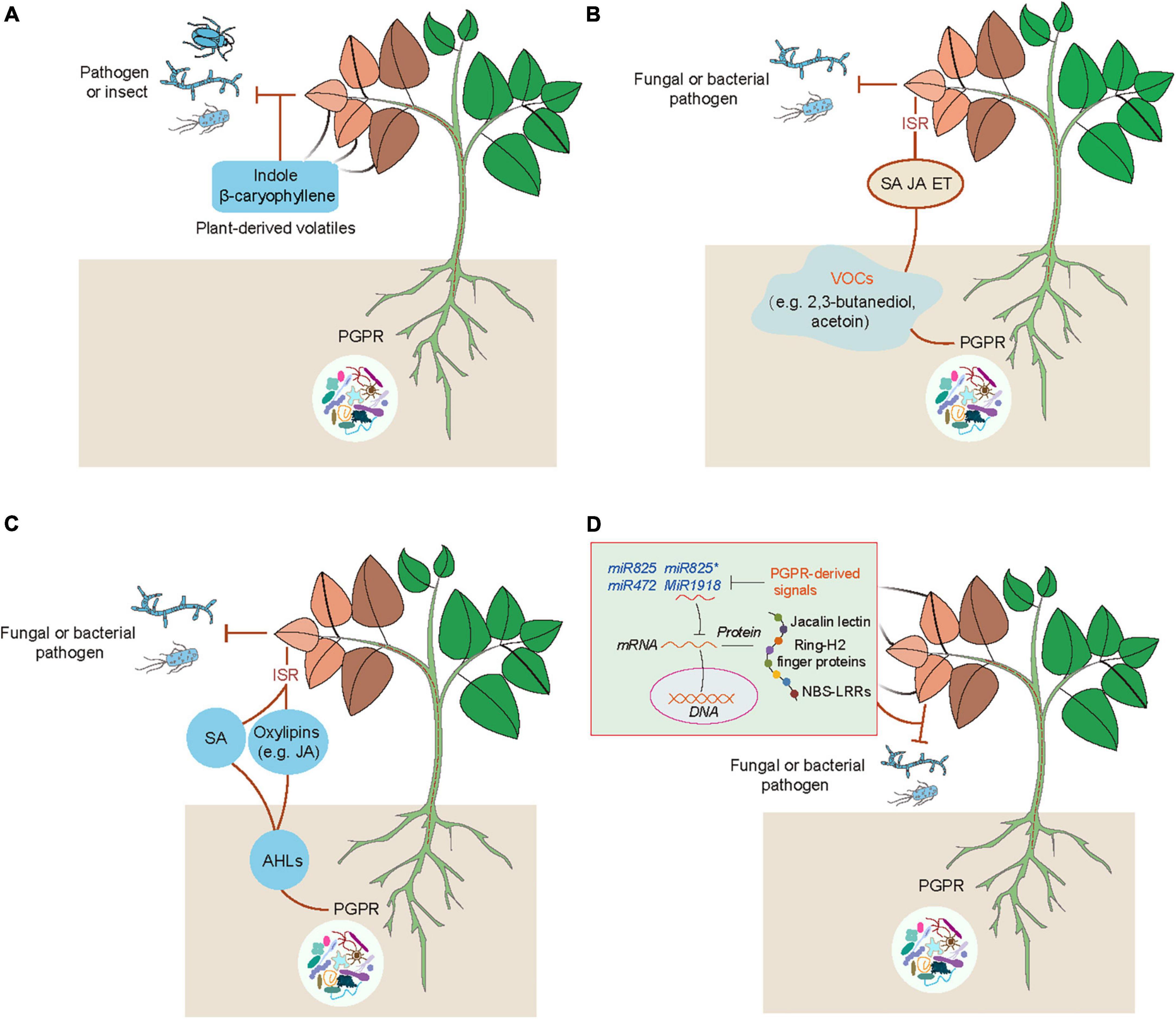
Figure 2. Multiple acting models of PGPR-induced ISR in plants. (A) PGPR-induced releases of plant volatiles act as defense-related substances against biotic stress. PGPR can induce the emission of plant volatiles such as indole and β-caryophyllene that enhance plant defense against the attacks of pathogens and insect pests. (B) PGPR can emit various kinds of VOCs such as 2,3-butanediol and acetoin, which lead to activation of hormone (SA, ET, and/or JA) signaling pathways that provoke plant ISR against pathogens. (C) PGPR can secrete the QS molecules (N-acyl-homoserine lactones, AHLs) to activate the SA- and JA-mediated pathways, MAPK cascades, and oxylipin-induced defense responses (e.g., promotion of stomatal closure, and the increased accumulation of callose, lignin, ROS and phenolic compounds). These effects lead to the enhanced plant defense against pathogens. (D) Suppression of plant miRNAs by PGPR enhances plant defense against pathogenic attacks. PGPR can release certain signals to repress negative regulators of plant defense systems such as miRNA825/miRNA825*, miR472 and miR1918, and thus enhance the expression of defense-related genes associated with jacalin lectin, Ring-H2 finger gene, and NBS-LRRs, which contribute to the increased resistance of plants against pathogens.
Microbial elicitation of plant induce systemic resistance
A large number of MAMPs from beneficial microbes such as flagellin, liposaccharides (LPS), peptidoglycans, and hairpins induce the MAMP-triggered plant immunity to inhibit pathogen infection (Newman et al., 2013; D’Ambrosio et al., 2017; Ma et al., 2021). The cascade of events closely related to plant ISR and SAR is activated by some regulatory genes such as MAPKs, WRKYs and MYCs (Kazan and Manners, 2013; Meng and Zhang, 2013; Jiang et al., 2016). In Arabidopsis, P. putida WCS358-derived flagellin can trigger ISR against Pseudomonas syringae. Similarly, LPS, tripartite amphipathic molecules with O-antigen side chains, from several PGPR strains can also enhance plant defense against pathogenic attacks (Meziane et al., 2005). Furthermore, diverse kinds of active molecules such as VOCs, QS molecules, siderophores, and cyclic lipopeptides can be released by PGPR and function as important ISR elicitors.
Bacterial volatile organic compounds
Plants are inevitably subjected to VOCs from diverse organisms including bacteria, fungi and neighboring plants (Bailly et al., 2014; Karamanoli et al., 2020; Vlot et al., 2021). More than 1000 VOCs (such as alcohols, alkenes and ketones) and non-organic compounds (such as HCN and NH3) can be liberated by a huge number of microbes (Audrain et al., 2015). Bacterial VOCs function as important regulators for plant growth and stress resistance (Sharifi and Ryu, 2016). Different PGPR strains can emit differential blends of VOCs, which are involved in the regulation of bacterial life cycles and interactions with their hosts. VOCs have also been reported to regulate the antibiotic sensitivity, motility and biofilm formation of bacteria, which function as the virulence-mediating factors for bacterial pathogens (Sharifi and Ryu, 2016). Indeed, single volatile compound can also benefit the emitters (Huang et al., 2012; Bailly et al., 2014). Bacteria-released indoles can promote the formation of bacterial biofilm, antibiotic resistance, and kill nematodes (Anyanful et al., 2005; Bailly et al., 2014; Audrain et al., 2015).
Bacterial VOCs are key inducers for stimulating the ISR responses of plants against pathogen infection (Figure 2B). Different PGPR strains can liberate diverse kinds of VOCs, which induce systemic defense of plants against pathogenic attacks in a strain-specific manner (Ryu et al., 2004; Song and Ryu, 2013; Sharifi and Ryu, 2016; Vlot et al., 2021). Bacillus subtilis GB03-emitted VOCs trigger the ISR responses of Arabidopsis plants to inhibit the attacks of Erwinia carotovora subsp. carotovora by activation of the ET signaling pathway rather than the JA and SA signaling pathways (Ryu et al., 2004). The release of VOCs by B. subtilis FB17 induces the ISR responses of Arabidopsis plants against the hemibiotrophic pathogen P. syringae pv. tomato (Pst) DC3000, which is attributable to the stimulation of both the SA and ET signaling pathways (Rudrappa et al., 2010). Bacterial VOCs can enhance the expression of PDF1.2 and PR1, and provoke both the SA- and JA-dependent pathways, which contribute to the increased host defense against pathogenic attacks (Sharifi and Ryu, 2016). It is increasingly evidenced that bacterial VOCs regulate multiple signaling pathways for enhancing plant defense. The release of 2,3-butanediol and acetoin by PGPR increases the resistance of plants against pathogen infection (Farag et al., 2006). Treatment with acetion greatly induces the expression of PR-4 and SA-related signaling pathways in Agrostis stolonifera (Cortes-Barco et al., 2010a). However, 2,3-butanediol induces systemic resistance of Nicotiana benthamiana against the fungal pathogen Colletotrichum orbiculare by activating the JA-dependent pathway, but not the SA-dependent pathway (Cortes-Barco et al., 2010b).
Recent genome sequencing have revealed that many bacterial species such as Deinococcus radiodurans, Bacillus halodurans, and B. subtilis possess the NOS-like proteins, which are essential for generating the gaseous molecule NO (Adak et al., 2002). NO is one of the most important bacterial VOCs that induce the plant ISR against microbial pathogens (Trapet et al., 2015). The mechanisms by which NO regulates plant defense signaling cascades have been well examined. The S-nitrosylation of proteins is an important regulatory event modulated by NO, in which NO can react with the cysteine-rich thiol groups in proteins to form the S-nitrosothiols (Yu et al., 2014). It has been well documented that several transcription factors can be S-nitrosylated in plants (Castillo et al., 2015; Kawabe et al., 2018). In Arabidopsis, NO can switch the translocation of the SA signaling component, NPR1, which takes part in the induction of PR genes, into the nucleus (Tada et al., 2008; Lindermayr et al., 2010). The S-nitrosylation of the zinc finger transcription factor SRG1 plays a critical role in regulating plant defense responses (Cui et al., 2018). Furthermore, the modified activity of the Arabidopsis NADPH oxidase, AtrbohD, is a typical example of the role of S-nitrosylation in plant defense (Yu et al., 2014; Sivakumaran et al., 2016). It has been clearly evidenced that SAR can be initiated by NO, which works together with ROS and SA signals (Wendehenne et al., 2014). In addition, NO has also been demonstrated to interact with both the JA and ET signaling pathways for regulating plant defense responses (Mur et al., 2013).
Bacterial quorum sensing molecules
QS is a wide-existed biological process, in which bacteria can synthesize and perceive QS molecules to mediate their cell density and collective behaviors (Papenfort and Bassler, 2016; Abisado et al., 2018; Mukherjee and Bassler, 2019). The secretion of N-acyl-homoserine lactones (AHLs) by Gram-negative bacteria can function as QS molecules to regulate the intra-population communications (Fu and Dong, 2013; Ortiz-Castro and López-Bucio, 2019). Bacteria can perceive the QS molecules for activating or inactivating the expression of several genes related to diverse processes such as biofilm formation and chemotaxis (Bellezza et al., 2014; Laganenka et al., 2016; Zhang et al., 2020). AHLs are one of the well-examined QS molecules that harbor an acyl side-chained homoserine lactone ring. The hydrogen at the C3 position from different length of the acyl chains can be substituted with a hydroxyl or a ketone group. The lactone ring is essential for the recognition of AHLs by its cognate receptors, and the specificity of cell-to-cell recognition and interaction is determined by both the fatty acid chain length and amide group (Whitehead et al., 2001; Churchill and Chen, 2011).
Bacterial QS molecules are also involved in the mediation of plant behaviors (Ortiz-Castro and López-Bucio, 2019). Although the mechanisms underlying plants perceive the QS molecules remain largely unclear, AHLs can regulate gene expression, protein profiles and root growth (Ortiz-Castro et al., 2008; Schenk et al., 2012). Proteomic analysis of the roots of Medicago truncatula has revealed that treatment with different AHLs changes the expression of 150 proteins involved in multiple processes such as flavonoid synthesis and oxidative stress (Mathesius et al., 2003). Similarly, treatment with oxo-C8-HSL remarkably induces the expression of proteins associated with carbon metabolism and plant defense in Arabidopsis seedlings (Ding et al., 2016). Shrestha et al. (2020) have shown that different forms of AHLs can trigger specific responses in plants, which depend on the length of the acyl moiety. Primary root growth can be promoted by the short acyl-chained AHLs, which is mainly attributable to activation of auxin signaling pathways (Von Rad et al., 2008; Schenk et al., 2012). The expression of several genes related to hormone signaling pathways is markedly increased in plants treated with the N-3-oxohexanoyl homoserine lactone (oxo-C6-HSL) (Von Rad et al., 2008). The increased auxin level is essential for promoting the formation of adventitious roots in Vigna radiate treated with the N-3-oxo-decanoyl-homoserine-lactone (3-O-C10-HL) (Bai et al., 2012).
Besides the roles of AHLs in the regulation of plant growth, AHLs can effectively ISR in plants (Figure 2C). The long acyl-chained AHLs have been reported to stimulate the ISR in different plant species (Schikora et al., 2011; Shrestha et al., 2019). The enhanced defense of AHL-treated plants is closely related to activation of multiple signal components. N-hexanoyl-homoserine lactone (HHL) promotes the biosynthesis of SA in plants, which contributes to enhancing the resistance of plants against Alternaria alternate (Schuhegger et al., 2006). N-decanoyl-homoserine lactone increases the resistance of tomato plants against Botrytis cinerea by activation of the JA signaling pathway (Hu et al., 2018). Exposure to N-3-oxo-tetradecanoyl-homoserine lactone (OTHL) provokes the mitogen-activated protein kinase (MAPK) cascades and thus enhances the transcription of defense-related transcription factors, thereby increasing the resistance of Arabidopsis and barley plants against obligate biotrophic fungi (Schikora et al., 2011). In addition, oxo-C14-HSL stimulates the production of oxylipins and further promotes the accumulation of callose and phenolic compounds, and stomatal closure, which result in the increased resistance of Arabidopsis plants against pathogen infection (Schenk and Schikora, 2014). The oxo-C14-HSL-induced disease resistance of cucumber plants has also been found to be associated with the enhanced deposition of lignin and callose, phenolics and ROS levels, and defense-related enzymatic activities (Pazarlar et al., 2020).
Siderophores
Iron (Fe) is an indispensable element for all living creature because of its redox catalyzing ability, while excess Fe often triggers overproduction of hydroxyl radicals that are harmful to cell metabolism and structures (Verbon et al., 2017). Dynamic regulation of Fe homeostasis is the most critical mechanism for mediating plant-pathogen interactions (López-Berges et al., 2013; Aznar et al., 2015; Liu et al., 2021). Plants can employ a Fe-withholding strategy to weaken pathogen virulence or elevate Fe levels for inducing oxidative burst (Cassat and Skaar, 2013). Many studies have confirmed that siderophores are secreted by plant pathogens to fight for Fe with host plants and attenuate the Fe-regulated immune responses. Beneficial soil bacteria can suppress plant disease occurrence by reducing the bioavailability of Fe in the rhizosphere (Loper and Buyer, 1991; Berendsen et al., 2015). They can also directly stimulate the ISR in plants by activating the Fe uptake-associated signaling pathways (Oide et al., 2006; Lemanceau et al., 2009). In the mammalian immune systems, the Fe-withholding strategy facilitates the hosts to prevent pathogen infection (Meziane et al., 2005; López-Berges et al., 2012). By contrast, the functions of Fe in plant defense responses are even more intricate, since it involves a tripartite interaction in the rhizosphere among beneficial bacteria, pathogens and plants (Harman et al., 2004; De Vleesschauwer et al., 2008; Verbon et al., 2017).
Plant pathogens can secrete siderophores to acquire Fe, which is required for their virulence and successful invasion (Martinez-Medina et al., 2016; Gu et al., 2020). Before pathogen infection, soil-borne pathogens need to fight for the scarcely available Fe in the rhizosphere with other microbes for growth. Microbial release of siderophores has been demonstrated to play vital roles during the warfare for rhizopsheric Fe (Expert et al., 1996; Lamont et al., 2002; Weller et al., 2002; Pozo et al., 2008; Taguchi et al., 2010; Franza and Expert, 2013; Aznar et al., 2015). The efficiency of siderophore-mediated acquisition for Fe is mainly attributable to the affinity of siderophores for Fe, their species-specificity and abundance. The siderophore-recognized receptors of microbes are highly specific, while different microbes can also recognize and assimilate heterologous siderophores (Loper and Buyer, 1991). Highly rhizosphere-competent microbes can synthesize and release specific siderophores and possess various receptors for recognizing heterologous siderophores (Berendsen et al., 2015). PGPR can secrete such high-affinity Fe uptake and species-specific siderophores to compete for Fe with soil-borne pathogens, thereby reducing plant disease occurrence (Lemanceau et al., 2009). During long-term evolution, soil-borne pathogens also develop adaptive strategies to confront antagonistic microbes. In plants, cellular Fe homeostasis can be mediated by plant pathogens and rhizosphere microbes (Oide et al., 2006; López-Berges et al., 2012). Many studies have unraveled a close connection between the Fe homeostasis and PGPR-induced ISR in plants. The release of siderophores by PGPR effectively elicits ISR in plants (Meziane et al., 2005; De Vleesschauwer et al., 2008; Grady et al., 2016; Fan et al., 2018; Li et al., 2020). Other beneficial fungi such as Piriformospora indica and Trichoderma species can also mediate the uptake of Fe and trigger the ISR in host plants (Harman et al., 2004; Peskan-Berghofer et al., 2004). However, the underlying mechanisms behind the associations between cellular Fe status and PGPR-induced ISR in plants remain obscure.
The molecular basis of initiation, signaling transduction and activation of ISR in plants has been well illustrated in the interactions between Arabidopsis plants and P. fluorescens WCS417 (Figure 3). In the plant-microbe system, the disease resistance of ISR-expressing plants is closely related to activation of both the JA and ET signaling pathways. However, the PGPR-induced ISR responses are not attributable to the promoted biosynthesis of these hormones or substantial expression of defense-related genes in plants. Conversely, a quicker and stronger stimulation of defense responses is observed in the ISR-expressing plants upon exposure to insect or pathogenic attacks (Pieterse et al., 2014). Although no marked alterations of transcriptome changes occur in the WCS417-colonized Arabidopsis leaves, bacterial colonization leads to massive changes in the roots (Verhagen et al., 2004). MYB72, plays a regulatory role in the metabolism of Fe-mobilizing phenolics under Fe deficiency, is among differentially expressed genes in the WCS417-treated roots (Zamioudis et al., 2015). Other beneficial microbes such as Trichoderma species also result in similarly changing patterns of MYB72 expression, but that is not found in the plants colonized by the non-ISR-inducing strain P. fluorescens WCS374 (Van der Ent et al., 2008; Berendsen et al., 2015). Treatment with WCS417 or Trichoderma species is not able to induce the establishment of ISR in the roots of the Arabidopsis myb72 mutant, indicating that MYB72 is essential for initiating ISR provoked by beneficial microbes (Segarra et al., 2009). MYB72 can directly regulate the expression of BGLU42 and PDR9 genes, which are required for root exudation of Fe-mobilizing phenolics under Fe deficiency (Zamioudis et al., 2014). Interestingly, phenolic compounds have been shown to be considerably secreted by the roots inoculated with PGPR (Van de Mortel et al., 2012; Zhou et al., 2016). The Arabidopsis bglu42 mutant cannot initiate the ISR responses upon exposure to WCS417, and overexpression of BGLU42 in Arabidopsis increases the resistance against broad-spectrum pathogens (Zamioudis et al., 2014). Therefore, the PGPR-induced ISR responses are closely related to the MYB72-regulated phenolic metabolisms and Fe uptake.
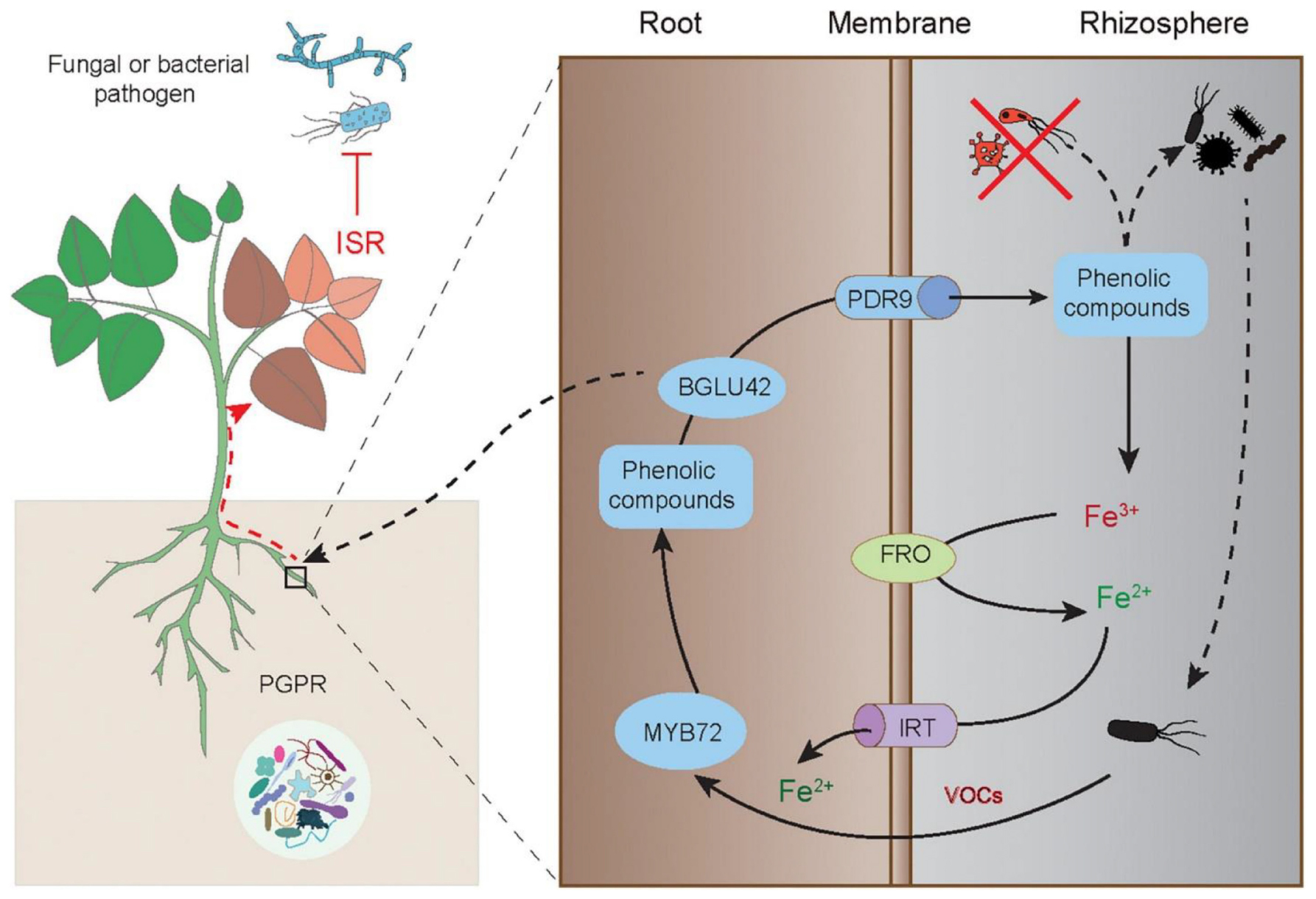
Figure 3. Elicitation of the MYB72-mediated ISR in plants by PGPR-released VOCs. The root-specific gene MYB72 initiates the ISR and Fe uptake in plants induced by PGPR. The ISR-eliciting PGPR can activate the expression of MYB72, which controls the biosynthesis of fluorescent phenolic compounds, and the expression of BGLU42 encoding the glucose hydrolase gene and PDR9 encoding the ABC transporter gene, thereby triggering root exudation of phenolics. The root-released phenolics further promote the mobilization of Fe3+ and make it available for reduction and uptake by plant roots. The phenolics can also shape specific rhizosphere microbiota. Moreover, the MYB72-dependent BGLU42 activity is essential for stimulating the ISR responses. This figure is acknowledged by Verbon et al. (2017).
Cylic lipopeptides
Cylic lipopeptides (CLPs) such as iturin, surfactin, and fengycin exhibit antibacterial activities, which can be generated by different Bacillus species (Raaijmakers et al., 2010). The surfactin family contains heptapeptide, which is linked to β -hydroxyl fatty acid chains with a length of 12–16 carbon atoms to form a ring lactone ring structure, and shows strong antimicrobial activity, but no obvious antifungal activity (Henry et al., 2011). Surfactin and fengycin (but not Iturin) trigger the plant ISR against fungal pathogens, although they show differential preferences for different plant cell types (Ongena and Jacques, 2008).
During extracellular matrix formation, the surface proteins secreted by B. subtilis can also function as signaling molecules to mediate ISR, root colonization, and biofilm formation (Shank and Kolter, 2011; Arnaouteli et al., 2021). B. amyloliquefaciens FZB42 is a natural isolate, which can stimulate plant growth and produce three families of lipopeptides including surface proteins (the surfactin family), bacilomycin D (the iturin family), and fonamycin (the fengycin family) (Koumoutsi et al., 2004; Idris et al., 2007). The secretion of surface proteins from B. amyloliquefaciens into perennial ryegrass is important for activating the plant ISR against rice blast infection. Surface protein molecules are mainly bound to cell membranes, and the perception of surface proteins by plant cells contributes to their ISR activity (Henry et al., 2011). This perception is required for sensitizing the activity of resistance in plants, which leads to the surveillance state being extremely sensitive to the penetration of fungal pathogens, thereby provoking rigorous stimulation of H2O2-mediated plant defense. However, the fungal pathogen Magnaporthe grisea induces changes in the metabolic profiles of host cells (Parker et al., 2009), and M. grisea can manipulate antioxidant systems to weaken the H2O2-mediated defense in plants (Chi et al., 2009). Thus, the activated H2O2-mediated defense in plants is essential for restricting pathogen proliferation. Samalova et al. (2014) have reported that the M. oryzae redox potential exceeds the H2O2-mediated oxidation potential in non-induced susceptible plants (Samalova et al., 2014). The surfactin-treated perennial ryegrass exhibits a rapid and powerful induction of H2O2-mediated defense responses, which contributes to the enhanced resistance against gray leaf spot disease.
In plants, oxylipins are a series of lipid metabolites generated from the oxidation of polyunsaturated fatty acids, which function as antimicrobial substances and signaling molecules that induce defense responses and regulate cell death (Griffiths, 2015). Interference with the oxylipin pathways such as the biosynthesis or perception of oxylipins affects plant defense against pathogens (Prost et al., 2005; Battilani et al., 2018; Deboever et al., 2020). Molecular oxygen can be introduced by lipoxygenase (LOX), a key enzyme involved in the oxylipin pathways, into unsaturated linolenic and linoleic acids for generating the 9- and 13-hydroperoxides, which can be further utilized as substrates by various enzymes to produce several secondary metabolites such as colneleic (CA) and colnelenic acids (CnA) (Göbel et al., 2001, 2002). These LOX-derived oxylipins display strong antimicrobial activities. The accumulation of CA and CnA is quickly increased at the pathogen-infecting sites, which confers the increased resistance of plants against pathogens (Prost et al., 2005). Moreover, application of CA to barley plants reduces disease occurrence imposed by the powdery mildew Blumeria graminis f. sp. hordei (Cowley and Walters, 2005). Activation of the oxylipin pathways in bean has been correlated with the induction of ISR by beneficial Pseudomonas putida BTP1 (Ongena et al., 2004). The activities of LOX involved in the metabolic route of plant oxylipins are also enhanced in tomato plants treated with the lipopeptide-overproducing Bacillus strains (Mariutto et al., 2011). Therefore, the metabolism of oxylipins can be mediated by PGPR, which is involved in the regulation of plant defense responses.
Regulation of plant induce systemic resistance by syntaxins and small RNAs
Root hair-specific syntaxins
Root hairs play pivotal functions during nutrient and water uptake, and microbial colonization. PGPR can modify root system architecture by repressing primary root growth, and promoting root hair formation (Zamioudis et al., 2013). PGPR can also prime plant defense systems against pathogen infection (Lugtenberg and Kamilova, 2009). It has recently been indicated that root hair-specific syntaxin genes (SYPs) can mediate the PGPR-induced ISR signaling pathways in plants (Rodriguez-Furlán et al., 2016). The structure of syntaxins consists of an N-terminal auto-regulatory region, a transmembrane domain, a linker and an N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) domain (Lipka et al., 2007). The Arabidopsis SYP1 family contains nine proteins that are specifically localized at plasma membrane, of which can regulate plant defense responses (Uemura et al., 2004). The SYP121 gene is involved in the regulation of exocytosis-mediated extracellular immune, which confers the enhanced plant defense against the powdery mildew Blumeria graminis f. sp. hordei, and mediates focal secretion at the pathogen-infected sites (Kwon et al., 2008). In tobacco, the SYP132 gene regulates plant resistance against bacterial pathogens by controlling the secretion of PR1. Plant resistance to pathogens is compromised in the syp123 mutants, which indicates that distinct plasma membrane-localized syntaxins are engaged by plants to confront pathogen infection (Kalde et al., 2007).
Cell wall in root hairs is one of the most important interacting sites with beneficial microbes. During the root hair-microbe interactions, the expression of genes encoding cell wall modifying-enzymes (e.g., pectin methyl esterase) is considerably increased for lowering cell wall rigidity (Bellincampi et al., 2014). The SYP132-deficient plants often display low methyl esterification in root hairs and the altered cell wall rigidity, which leads to the inhibition of plant-rhizobacteria interactions. Brisson et al. (1994) have shown that pathogenic attacks cause rapid insolubility of proline-rich proteins (PRPs) and thus strengthen the cell wall. The changes of PRP3 localization affect the activation of ISR responses in the syp123 plants. The prp3 mutants exhibit the increased susceptibility to the bacterial pathogen P. syringae. Since the expression of PRP3 is specifically expressed in root hairs, the increased susceptibility of the prp3 mutants confirms the idea that the proper localization of PRP3 at root cell wall is important for the PGPR-induced ISR in plants (Larson et al., 2014). Considering that plant defense actions are necessarily regulated by receptor molecules closely related to the plasma membrane, the SYP123 gene plays other roles in the mediation of PGPR-induced ISR in plants. Abnormal localization of these receptors in the syp123 mutants may lead to the impaired activation of ISR in plants. The induced resistance in the roots is manifested in the whole plants as the alleviation of disease severity upon the subsequent pathogen attacks. The PGPR-induced ISR partially overlaps with those of the pathogen-induced SAR in plants. Consequently, the syp123 mutants lack the ability to mount the SAR against the bacterial pathogen P. syringae. Many studies have indicated that plant ISR responses are associated with the activation of defense-related genes such as PR1, PDF1.2, and MYC2 (Ryu et al., 2003, 2004). Beneficial Pseudomonas species greatly increase the transcription of ISR priming marker genes as compared to the control plants, while a lower expression of these genes is observed in the syp123 mutants (Rodriguez-Furlán et al., 2016). These results strongly indicate that root hair-specific syntaxins are essential for regulating the PGPR-induced ISR responses in plants.
MicroRNAs
In plants, microRNAs (miRNAs) are one of the most important non-coding RNA molecules that can mediate the expression of target mRNAs by translational repression or cleavage (Baulcombe, 2004; Ha and Kim, 2014; Song et al., 2019). MiRNAs can be converted into primary miRNA, and then processed to pre-miRNAs, which contain stem-loop hairpin structures. The resulting pre-miRNAs are cleaved for producing the duplexes of miRNA/miRNA. The mature miRNAs can be loaded into an RNA-induced silencing complex (RISC), in which they bind to target mRNAs for the control of their transcription (Martinez et al., 2002; Nakanishi, 2016; Michlewski and Cáceres, 2019). In plants, miRNAs have been demonstrated to regulate various processes, such as plant growth, disease resistance, stress adaptation, and cellular signal transduction (Zhang et al., 2021; Begum, 2022). Plant-derived miRNAs also function as molecular linkers that mediate plant growth and auxin signaling pathways under adverse conditions (Sunkar et al., 2007; Padmanabhan et al., 2009; Zhang, 2015; Begum, 2022).
Recently, miRNAs have been considered as key biomarkers of plants in response to biotic factors. The expression levels of miR393 and miR167 are down-regulated in the A. tumefaciens C58-induced plant tumors (Dunoyer et al., 2006). Navarro et al. (2006) have reported that bacterial PAMP flg22 significantly induces the expression of miR393 in Arabidopsis, which is involved in the regulation of PTI responses by silencing several auxin receptors such as TIR1, AFB2 and AFB3, and thereby weakens the auxin-mediated pathways (Navarro et al., 2006). Moreover, a non-pathogenic strain Pst (hrcC) enhances the expression of miR160 and miR167, which target several auxin-response factor (ARF) genes (Fahlgren et al., 2007). In addition, Pst hrcC infection induces the expression of miR825, which may silence several members of zinc finger homeobox gene family, remorin and frataxin-related genes. During fungal infection, the miRNA-mediated gene silencing can be employed by plants to defend against pathogenic attacks. This posttranscriptional gene silencing is essential for regulating plant defense responses against fungal pathogens (Katiyar-Agarwal and Jin, 2010). In Arabidopsis, several RNA silencing mutants such as rdr2, sgs2, and sgs3 display the increased susceptibility to Verticillium wilt (Ellendorff et al., 2009). During viral infection, the expression of miR158 and miR1885 is largely increased in Brassica rapa upon exposure to the Turnip mosaic virus (TuMV) infection. Plant miR1885 has been predicted to target a TIR-NBS-LRR gene, which positively regulates plant disease resistance (He et al., 2008).
It has been indicated that miRNAs are involved in the mediation of the process of plant-microbe interactions. The expression of miR172c is positively correlated with the efficiency of rhizobia infection and nodulation formation, indicating that miR172c acts as an important regulator for plant-rhizobium symbiosis (Nova-Franco et al., 2015; Wang et al., 2019). During the symbiosis process, miR2111 can translocate from shoots to roots, thereby regulating root symbiosis suppressors to control nodule symbiosis (Zhang et al., 2021). In Medicago truncatula, miR396 and miR171 can regulate plant-arbuscular mycorrhizal (AM) fungi symbiosis by silencing of the growth regulating factor and nodulation signaling pathway 2, respectively (De Luis et al., 2012; Bazin et al., 2013). Rhizo-colonization of Bacillus strains can benefit plants through diverse means, such as secretion of auxin and antibiotic substances, the increased bioavailability of nutrients and stimulation of ISR (Abriouel et al., 2011; Shao et al., 2015; Zhou et al., 2019). Several miRNAs have recently been reported to control the process of PGPR-induced ISR in plants (Figure 2D). In Arabidopsis, the inhibited transcription of miR846 by B. velezensis FZB42 leads to the increased expression of target jacalin lectin genes and the activation of ISR by the JA signaling pathway (Xie et al., 2018). The expression of miR825/miR825* is also remarkably suppressed by B. cereus AR156, which leads to the stimulation of ISR in plants (Niu et al., 2016b). In Arabidopsis, miR472 also takes part in mediating the B. cereus AR156-induced ISR of plants against Pst by the nucleotide-binding site and leucine-rich repeat type (NBS-LRR)-mediated basal immunity (Jiang et al., 2020). However, it remains unclear how PGPR can regulate the expression of miRNAs for inducing disease resistance in plants. More recently, inoculation of tomato plants with B. subtilis SL18r enhances the resistance against B. cinerea by activating the expression of long non-coding RNA, MSTRG18363, for the decoy of miR1918, which silences the defense-related gene SlATL20 encoding a putative RING-H2 finger gene (Zhou et al., 2021).
Future perspectives
Since the discovery that PGPR can ISR in plants (Van Peer et al., 1991; Wei et al., 1991), now about 30 years ago, accumulative knowledges have been illustrated for the mechanisms of the PGPR-induced plant ISR responses. The plant defense system can be activated for resisting various pathogenic attacks, and also be suppressed for allowing the colonization of beneficial microbes. Both aspects of plant defense mediation are operative in the phenomenon of plant ISR, and their interplay need to be further investigated. A large gap is how the recognition of PGPR drives whole plants to improve growth and enhance disease resistance. Massive efforts for probing into molecular dialogs between plants and ISR-inducing microbes have been made, but several puzzles need to be unlocked in future. For instance, do plant roots distinguish the signals from pathogens and beneficial microbes and make appropriate response? How are the PGPR-derived signal molecules perceived in plant roots and transformed into specific responses that prime plant defense against foliar pathogens?
Conclusion
Here, we have made a discussion about the mechanisms underlying plants recognize beneficial microbes. PGPR can be recognized as MAMPs by diverse plant PRRs and further trigger host defense responses. For establishing mutual benefits with the hosts, PGPR have developed strategies to weaken the activation of host defense systems. Moreover, the process of the PGPR-induced ISR in plants can be regulated by root hair-specific syntaxins and non-coding RNAs. However, it remains elusive how plants balance between microbial recognition and defense activation. Additionally, the transferring mechanisms of small RNAs from roots to shoots for provoking ISR need to be deeply explored.
Author contributions
CZ and LZ wrote the manuscript. JH and XL provided some suggestions for the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key R&D Program Projects of Anhui Province (202004a06020003), the Open Project of Key Laboratory of Bio-organic Fertilizer Creation of Ministry of Agriculture and Rural Affairs (BOFA202012), the Natural Science Foundation of the Anhui Science and Technology Committee (1908085QC110 and 2108085MC107), the Natural Science Foundation of Education Department of Anhui Province (KJ2021A0893), and the Outstanding Talent Cultivation Program in Colleges and Universities of Education Department of Anhui Province (gxypZD2020038).
Conflict of interest
The Editorial Office checked and made sure that Anhui Science and Technology University, Bengbu and Anhui University of Science and Technology, Huainan City were independent affiliations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., and Chandler, J. R. (2018). Bacterial quorum sensing and microbial community interactions. mBio 9:e02331–17.
Abriouel, H., Franz, C. M., Ben Omar, N., and Gálvez, A. (2011). Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 35, 201–232.
Adak, S., Aulak, K. S., and Stuehr, D. J. (2002). Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis. J. Biol. Chem. 277, 16167–16171.
Ajilogba, C. F., and Babalola, O. O. (2013). Integrated management strategies for tomato Fusarium wilt. Biocontrol. Sci. 18, 117–127. doi: 10.4265/bio.18.117
Aljbory, Z., and Chen, M. S. (2018). Indirect plant defense against insect herbivores: A review. Insect Sci. 25, 2–23.
Anyanful, A., Dolan Livengood, J. M., Lewis, T., Sheth, S., Dezalia, M. N., Sherman, M. A., et al. (2005). Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol. Microbiol. 57, 988–1007. doi: 10.1111/j.1365-2958.2005.04739.x
Arnaouteli, S., Bamford, N. C., Stanley-Wall, N. R., and Kovács, ÁT. (2021). Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 19, 600–614. doi: 10.1038/s41579-021-00540-9
Aroca, A., Gotor, C., and Romero, L. C. (2018). Hydrogen sulfide signaling in plants: Emerging roles of protein persulfidation. Front. Plant Sci. 9:1369. doi: 10.3389/fpls.2018.01369
Arshad, M., Saleem, M., and Hussain, S. (2007). Perspectives of bacterial ACC deaminase in phytoremediation. Trends Biotechnol. 25, 356–362. doi: 10.1016/j.tibtech.2007.05.005
Audenaert, K., Pattery, T., Cornelis, P., and Höfte, M. (2002). Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: Role of salicylic acid, pyochelin, and pyocyanin. Mol. Plant Microbe Interact. 15, 1147–1156. doi: 10.1094/MPMI.2002.15.11.1147
Audrain, B., Farag, M. A., Ryu, C. M., and Ghigo, J. M. (2015). Role of bacterial volatile compounds in bacterial biology. FEMS Microbiol. Rev. 39, 222–233.
Aznar, A., Chen, N. W. G., Thomine, S., and Dellagi, A. (2015). Immunity to plant pathogens and iron homeostasis. Plant Sci. 240, 90–97.
Aznar, A., and Dellagi, A. (2015). New insights into the role of siderophores as triggers of plant immunity: What can we learn from animals? J. Exp. Bot. 66, 3001–3010. doi: 10.1093/jxb/erv155
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Bai, M. Y., Fan, M., Oh, E., and Wang, Z. Y. (2012). A triple helix-loophelix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24, 4917–4929. doi: 10.1105/tpc.112.105163
Bailly, A., Groenhagen, U., Schulz, S., Geisler, M., Eberl, L., and Weisskopf, L. (2014). The inter-kingdom volatile signal indole promotes root development by interfering with auxin signalling. Plant J. 80, 758–771. doi: 10.1111/tpj.12666
Ballhorn, D. J., Kautz, S., and Schadler, M. (2013). Induced plant defense via volatile production is dependent on rhizobial symbiosis. Oecologia 172, 833–846. doi: 10.1007/s00442-012-2539-x
Battilani, P., Lanubile, A., Scala, V., Reverberi, M., Gregori, R., Falavigna, C., et al. (2018). Oxylipins from both pathogen and host antagonize jasmonic acid-mediated defence via the 9-lipoxygenase pathway in Fusarium verticillioides infection of maize. Mol. Plant Pathol. 19, 2162–2176. doi: 10.1111/mpp.12690
Bazin, J., Khan, G. A., Combier, J. P., Bustos-Sanmamed, P., Debernardi, J. M., Rodriguez, R., et al. (2013). miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 74, 920–934. doi: 10.1111/tpj.12178
Begum, Y. (2022). Regulatory role of microRNAs (miRNAs) in the recent development of abiotic stress tolerance of plants. Gene 821:146283.
Bellezza, I., Peirce, M. J., and Minelli, A. (2014). Cyclic dipeptides: From bugs to brain. Trends Mol. Med. 20, 551–558. doi: 10.1016/j.molmed.2014.08.003
Bellincampi, D., Cervone, F., and Lionetti, V. (2014). Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front. Plant Sci. 5:228. doi: 10.3389/fpls.2014.00228
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051.
Berendsen, R. L., Van Verk, M. C., Stringlis, I. A., Zamioudis, C., Tommassen, J., Tommassen, J., et al. (2015). Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genom. 16:539. doi: 10.1186/s12864-015-1632-z
Bjarnholt, N., Neilson, E. H. J., Crocoll, C., Jørgensen, K., Motawia, M. S., Olsen, C. E., et al. (2018). Glutathione transferases catalyze recycling of auto-toxic cyanogenic glucosides in sorghum. Plant J. 94, 1109–1125. doi: 10.1111/tpj.13923
Brisson, L. F., Tenhaken, R., and Lamb, C. (1994). Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 6, 1703–1712.
Cassat, J. E., and Skaar, E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13, 509–519.
Castillo, M. C., Lozano-Juste, J., González-Guzmán, M., Rodriguez, L., Rodriguez, P. L., and León, J. (2015). Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 8:ra89. doi: 10.1126/scisignal.aaa7981
Chen, L., Wang, X., Ma, Q., Bian, L., Liu, X., Xu, Y., et al. (2020). Bacillus velezensis CLA178-induced systemic resistance of Rosa multiflora against crown gall disease. Front. Microbiol. 11:587667. doi: 10.3389/fmicb.2020.587667
Chi, M. H., Park, S. Y., Kim, S., and Lee, Y. H. (2009). A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 5:e1000401. doi: 10.1371/journal.ppat.1000401
Churchill, M. E., and Chen, L. (2011). Structural basis of acyl-homoserine lactone-dependent signaling. Chem. Rev. 111, 68–85. doi: 10.1021/cr1000817
Clausen, M., Kannangara, R. M., Olsen, C. E., Blomstedt, C. K., Gleadow, R. M., Jørgensen, K., et al. (2015). The bifurcation of the cyanogenic glucoside and glucosinolate biosynthetic pathways. Plant J. 84, 558–573. doi: 10.1111/tpj.13023
Cortes-Barco, A., Goodwin, P., and Hsiang, T. (2010a). Comparison of induced resistance activated by benzothiadiazole, (2R,3R)-butanediol and an isoparaffin mixture against anthracnose of Nicotiana benthamiana. Plant Pathol. 59, 643–653.
Cortes-Barco, A., Hsiang, T., and Goodwin, P. (2010b). Induced systemic resistance against three foliar diseases of Agrostis stolonifera by (2R,3R)-butanediol or an isoparaffin mixture. Ann. Appl. Biol. 157, 179–189.
Cowley, T., and Walters, D. (2005). Local and systemic effects of oxylipins on powdery mildew infection in barley. Pest Manag. Sci. 61, 572–576. doi: 10.1002/ps.1026
Cui, B., Pan, Q., Clarke, D., Villarreal, M. O., Umbreen, S., Yuan, B., et al. (2018). S-nitrosylation of the zinc finger protein SRG1 regulates plant immunity. Nat. Commun. 9:4226.
D’Ambrosio, J. M., Couto, D., Fabro, G., Scuffi, D., Lamattina, L., Munnik, T., et al. (2017). Phospholipase C2 affects MAMP-triggered immunity by modulating ROS production. Plant Physiol. 175, 970–981. doi: 10.1104/pp.17.00173
de Almeida, J. R., Bonatelli, M. L., Batista, B. D., Teixeira-Silva, N. S., Mondin, M., Dos Santos, R. C., et al. (2021). Bacillus thuringiensis RZ2MS9, a tropical plant growth-promoting rhizobacterium, colonizes maize endophytically and alters the plant’s production of volatile organic compounds during co-inoculation with Azospirillum brasilense Ab-V5. Environ. Microbiol. Rep. 13, 812–821. doi: 10.1111/1758-2229.13004
De Luis, A., Markmann, K., Cognat, V., Holt, D. B., Charpentier, M., Parniske, M., et al. (2012). Two microRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol. 160, 2137–2154. doi: 10.1104/pp.112.204883
De Meyer, G., Capieau, K., Audenaert, K., Buchala, A. M., ’etraux, J. P., and Höfte, M. (1999). Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol. Plant Microbe Interact. 12, 450–458. doi: 10.1094/MPMI.1999.12.5.450
De Vleesschauwer, D., Djavaheri, M., Bakker, P. A., and Höfte, M. (2008). Pseudomonas fluorescens WCS374r–induced systemic resistance in rice against Magnaporthe oryzae is based on pseudobactin-mediated priming for a salicylic acid–repressible multifaceted defense response. Plant Physiol. 148, 1996–2012. doi: 10.1104/pp.108.127878
Deboever, E., Deleu, M., Mongrand, S., Lins, L., and Fauconnier, M. L. (2020). Plant-pathogen interactions: Underestimated roles of phyto-oxylipins. Trends Plant Sci. 25, 22–34. doi: 10.1016/j.tplants.2019.09.009
Ding, L., Cao, J., Duan, Y., Li, J., Yang, Y., Yang, G., et al. (2016). Proteomic and physiological responses of Arabidopsis thaliana exposed to salinity stress and N-acyl-homoserine lactone. Physiol. Plant 158, 414–434. doi: 10.1111/ppl.12476
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dunoyer, P., Himber, C., and Voinnet, O. (2006). Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 38, 258–263.
Ellendorff, U., Fradin, E. F., de Jonge, R., and Thomma, B. P. (2009). RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602. doi: 10.1093/jxb/ern306
Elzen, G. W., Williams, H. J., and Vinson, S. B. (1983). Response by the parasitoid Campoletis sonorensis (hymenoptera: Ichneumonidae) to chemicals (synomones) in plants: Implications for host habitat location. Environ. Entomol. 12, 1872–1876.
Expert, D., Enard, C., and Masclaux, C. (1996). The role of iron in plant host-pathogen interactions. Trends Microbiol. 4, 232–237.
Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS One 2, e219. doi: 10.1371/journal.pone.0000219
Fan, B., Wang, C., Song, X., Ding, X., Wu, L., Wu, H., et al. (2018). Bacillus velezensis FZB42 in 2018: The Gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 9:2491. doi: 10.3389/fmicb.2018.02491
Farag, M. A., Ryu, C. M., Sumner, L. W., and Paré, P. W. (2006). GC–MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry 67, 2262–2268. doi: 10.1016/j.phytochem.2006.07.021
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313x.1999.00265.x
Feng, J., Chen, L., and Zuo, J. (2019). Protein S-Nitrosylation in plants: Current progresses and challenges. J. Integr. Plant Biol. 61, 1206–1223. doi: 10.1111/jipb.12780
Franza, T., and Expert, D. (2013). Role of iron homeostasis in the virulence of phytopathogenic bacteria: An ‘à la carte’ menu. Mol. Plant Pathol. 14, 429–438. doi: 10.1111/mpp.12007
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gkizi, D., Lehmann, S., L’Haridon, F., Serrano, M., Paplomatas, E. J., Métraux, J. P., et al. (2016). The innate immune signaling system as a regulator of disease resistance and induced systemic resistance activity against Verticillium dahlia. Mol. Plant Microbe Interact. 29, 313–323. doi: 10.1094/MPMI-11-15-0261-R
Göbel, C., Feussner, I., Hamberg, M., and Rosahl, S. (2002). Oxylipin profiling in pathogen-infected potato leaves. Biochem. Biophys. Acta 1584, 55–64. doi: 10.1016/s1388-1981(02)00268-8
Göbel, C., Feussner, I., Schmidt, A., Scheel, D., Sanchez-Serrano, J., Hamberg, M., et al. (2001). Oxylipin profiling reveals the preferential stimulation of the 9-lipoxygenase pathway in elicitor treated potato cells. J. Biol. Chem. 276, 6267–6273. doi: 10.1074/jbc.M008606200
Gotor, C., García, I., Aroca, Á, Laureano-Marín, A. M., Arenas-Alfonseca, L., Jurado-Flores, A., et al. (2019). Signaling by hydrogen sulfide and cyanide through post-translational modification. J. Exp. Bot. 70, 4251–4265. doi: 10.1093/jxb/erz225
Grady, E. N., MacDonald, J., Liu, L., Richman, A., and Yuan, Z. C. (2016). Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 15:203.
Gu, S., Wei, Z., Shao, Z., Friman, V. P., Cao, K., Yang, T., et al. (2020). Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 5, 1002–1010. doi: 10.1038/s41564-020-0719-8
Gupta, K. J., Kolbert, Z., Durner, J., Lindermayr, C., Corpas, F. J., Brouquisse, R., et al. (2020). Regulating the regulator: Nitric oxide control of post-translational modifications. New Phytol. 227, 1319–1325.
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524.
Hacquard, S., Spaepen, S., Garrido-Oter, R., and Schulze-Lefert, P. (2017). Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 55, 565–589.
Hansen, C. C., Sørensen, M., Veiga, T. A. M., Zibrandtsen, J. F. S., Heskes, A. M., Olsen, C. E., et al. (2018). Reconfigured cyanogenic glucoside biosynthesis in Eucalyptus cladocalyx involves a cytochrome P450 CYP706C55. Plant Physiol. 178, 1081–1095. doi: 10.1104/pp.18.00998
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Haskett, T. L., Tkacz, A., and Poole, P. S. (2021). Engineering rhizobacteria for sustainable agriculture. ISME J. 15, 949–964.
Hatsugai, N., Igarashi, D., Mase, K., Lu, Y., Tsuda, Y., Chakravarthy, S., et al. (2017). A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J. 36, 2758–2769.
He, K., and Wu, Y. (2016). Receptor-like kinases and regulation of plant innate immunity. Enzymes 40, 105–142.
He, X. F., Fang, Y. Y., Feng, L., and Guo, H. S. (2008). Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 582, 2445–2452. doi: 10.1016/j.febslet.2008.06.011
Henry, G., Deleu, M., Jourdan, E., Thonart, P., and Ongena, M. (2011). The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cell. Microbiol. 13, 1824–1837. doi: 10.1111/j.1462-5822.2011.01664.x
Hu, Z., Shao, S., Zheng, C., Sun, Z., Shi, J., Yu, J., et al. (2018). Induction of systemic resistance in tomato against Botrytis cinerea by N-decanoyl-homoserine lactone via jasmonic acid signaling. Planta 247, 1217–1227. doi: 10.1007/s00425-018-2860-7
Huang, M., Sanchez-Moreiras, A. M., Abel, C., Sohrabi, R., Lee, S., Gershenzon, J., et al. (2012). The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-beta-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 193, 997–1008. doi: 10.1111/j.1469-8137.2011.04001.x
Idris, E. E., Iglesias, D. J., Talon, M., and Borriss, R. (2007). Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant Microbe Interact. 20, 619–626. doi: 10.1094/MPMI-20-6-0619
Jacobs, S., Zechmann, B., Molitor, A., Trujillo, M., Petutschnig, E., Lipka, V., et al. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 156, 726–740. doi: 10.1104/pp.111.176446
Jiang, C., Fan, Z., Li, Z., Niu, D., Li, Y., Zheng, M., et al. (2020). Bacillus cereus AR156 triggers induced systemic resistance against Pseudomonas syringae pv. tomato DC3000 by suppressing miR472 and activating CNLs-mediated basal immunity in Arabidopsis. Mol. Plant Pathol. 21, 854–870. doi: 10.1111/mpp.12935
Jiang, C. H., Huang, Z. Y., Xie, P., Gu, C., Li, K., Wang, D. C., et al. (2016). Transcription factors WRKY70 and WRKY11 served as regulators in rhizobacterium Bacillus cereus AR156-induced systemic resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. J. Exp. Bot. 67, 157–174. doi: 10.1093/jxb/erv445
Kalde, M., Nühse, T. S., Findlay, K., and Peck, S. C. (2007). The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. U.S.A. 104, 11850–11855. doi: 10.1073/pnas.0701083104
Karamanoli, K., Kokalas, V., Koveos, D. S., Junker, R. R., and Farré-Armengol, G. (2020). Bacteria affect plant-mite interactions via altered scent emissions. J. Chem. Ecol. 46, 782–792. doi: 10.1007/s10886-020-01147-9
Katiyar-Agarwal, S., and Jin, H. (2010). Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48, 225–246.
Kautz, S., Trisel, J. A., and Ballhorn, D. J. (2014). Jasmonic acid enhances plant cyanogenesis and resistance to herbivory in lima bean. J. Chem. Ecol. 40, 1186–1196. doi: 10.1007/s10886-014-0524-z
Kawabe, H., Ohtani, M., Kurata, T., Sakamoto, T., and Demura, T. (2018). Protein S-nitrosylation regulates xylem vessel cell differentiation in Arabidopsis. Plant Cell Physiol. 59, 17–29. doi: 10.1093/pcp/pcx151
Kloepper, J. W., and Schroth, M. N. (1978). “Plant growth promoting rhizobacteria on radishes,” in Proceedings of the fourth International Conference on Plant Pathogenic Bacteria (Angers: INRA).
Kloppholz, S., Kuhn, H., and Requena, N. (2011). A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 21, 1204–1209. doi: 10.1016/j.cub.2011.06.044
Knoester, M., Pieterse, C. M. J., Bol, J. F., and Van Loon, L. C. (1999). Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol. Plant Microbe Interact. 12, 720–727. doi: 10.1094/MPMI.1999.12.8.720
Koumoutsi, A., Chen, X. H., Henne, A., Liesegang, H., Hitzeroth, G., Franke, P., et al. (2004). Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186, 1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004
Kud, J., Wang, W., Gross, R., Fan, Y., Huang, L., Yuan, Y., et al. (2019). The potato cyst nematode effector RHA1B is a ubiquitin ligase and uses two distinct mechanisms to suppress plant immune signaling. PLoS Pathog. 15:e1007720. doi: 10.1371/journal.ppat.1007720
Kwon, C., Neu, C., Pajonk, S., Yun, H. S., Lipka, U., Humphry, M., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840. doi: 10.1038/nature06545
Laganenka, L., Colin, R., and Sourjik, V. (2016). Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat. Commun. 7:12984.
Lamont, I. L., Beare, P. A., Ochsner, U., Vasil, A. I., and Vasil, M. L. (2002). Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 99, 7072–7077.
Larson, E. R., Tierney, M. L., Tinaz, B., and Domozych, D. S. (2014). Using monoclonal antibodies to label living root hairs: A novel tool for studying cell wall microarchitecture and dynamics in Arabidopsis. Plant Methods 10:30. doi: 10.1186/1746-4811-10-30
Lee, J. H., Wood, T. K., and Lee, J. (2015). Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 23, 707–718.
Leeman, M., Van Pelt, J. A., Den Ouden, F. M., Heinsbroek, M., Bakker, P. A. H. M., and Schippers, B. (1995). Induction of systemic resistance against Fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 85, 1021–1027.
Lemanceau, P., Expert, D., Gaymard, F., Bakker, P., and Briat, J. F. (2009). Role of iron in plant-microbe interactions. Adv. Bot. Res. 51, 491–549.
Li, W., Lee, S. Y., Cho, Y. J., Ghim, S. Y., and Jung, H. Y. (2020). Mediation of induced systemic resistance by the plant growth-promoting rhizobacteria Bacillus pumilus S2-3-2. Mol. Biol. Rep. 47, 8429–8438. doi: 10.1007/s11033-020-05883-9
Lindermayr, C., Sell, S., Müller, B., Leister, D., and Durner, J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. doi: 10.1105/tpc.109.066464
Lindström, K., and Mousavi, S. A. (2020). Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 13, 1314–1335.
Lipka, V., Kwon, C., and Panstruga, R. (2007). SNARE-ware: The role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23, 147–174. doi: 10.1146/annurev.cellbio.23.090506.123529
Liu, Y., Kong, D., Wu, H. L., and Ling, H. Q. (2021). Iron in plant-pathogen interactions. J. Exp. Bot. 72, 2114–2124.
Loper, J. E., and Buyer, J. S. (1991). Siderophore in microbial interaction on plant surface. Mol. Plant Microbe Interact. 4, 5–13.
López-Berges, M. S., Capilla, J., Turrà, D., Schafferer, L., Matthijs, S., Jöchl, C., et al. (2012). HapX-mediated iron homeostasis is essential for rhizosphere competence and virulence of the soilborne pathogen Fusarium oxysporum. Plant Cell 24, 3805–3822. doi: 10.1105/tpc.112.098624
López-Berges, M. S., Turrà, D., Capilla, J., Schafferer, L., Matthijs, S., Jöchl, C., et al. (2013). Iron competition in fungus-plant interactions: The battle takes place in the rhizosphere. Plant Signal. Behav. 8:e23012. doi: 10.4161/psb.23012
Lu, Y., and Tsuda, K. (2021). Intimate association of PRR- and NLR-mediated signaling in plant immunity. Mol. Plant Microbe Interact. 34, 3–14. doi: 10.1094/MPMI-08-20-0239-IA
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556.
Ma, K. W., Niu, Y., Jia, Y., Ordon, J., Copeland, C., Emonet, A., et al. (2021). Coordination of microbe-host homeostasis by crosstalk with plant innate immunity. Nat. Plants 7, 814–825. doi: 10.1038/s41477-021-00920-2
Mariutto, M., Duby, F., Adam, A., Bureau, C., Fauconnier, M. L., Ongena, M., et al. (2011). The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Biol. 11:29. doi: 10.1186/1471-2229-11-29
Martinez, J., Patkaniowska, A., Urlaub, H., Lührmann, R., and Tuschl, T. (2002). Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574. doi: 10.1016/s0092-8674(02)00908-x
Martinez-Medina, A., Flors, V., Heil, M., Mauch-Mani, B., Pieterse, C. M. J., Pozo, M. J., et al. (2016). Recognizing plant defense priming. Trends Plant Sci. 21, 818–822.
Mathesius, U., Mulders, S., Gao, M., Teplitski, M., Caetano-Anolle’s, G., Rolfe, B. G., et al. (2003). Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. U.S.A. 100, 1444–1449.
Maurhofer, M., Reimmann, C., Schmidli-Sacherer, P., Heeb, S. D., and Défago, G. (1998). Salicylic acid biosynthesis genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88, 678–684. doi: 10.1094/PHYTO.1998.88.7.678
Meng, X. Z., and Zhang, S. Q. (2013). MAPK Cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266.
Mertens, D., Boege, K., Kessler, A., Koricheva, J., Thaler, J. S., Whiteman, N. K., et al. (2021). Predictability of biotic stress structures plant defence evolution. Trends Ecol. Evol. 36, 444–456.
Meziane, H., Van, D. E. R., Sluis, I., Van Loon, L. C., Höfte, M., and Bakker, P. A. (2005). Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol. Plant Pathol. 6, 177–185. doi: 10.1111/j.1364-3703.2005.00276.x
Michlewski, G., and Cáceres, J. F. (2019). Post-transcriptional control of miRNA biogenesis. RNA 25, 1–16.
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis Roots by microbe-associated molecular patterns. Plant Cell 22, 973–990. doi: 10.1105/tpc.109.069658
Mishra, A. K., Sharma, K., and Misra, R. S. (2012). Elicitor recognition, signal transduction and induced resistance in plants. J. Plant Interact. 7, 95–120.
Mukherjee, S., and Bassler, B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382.
Mur, L. A., Prats, E., Pierre, S., Hall, M. A., and Hebelstrup, K. H. (2013). Integrating nitric oxide into salicylic acid and jasmonic acid/ethylene plant defense pathways. Front. Plant Sci. 4:215. doi: 10.3389/fpls.2013.00215
Nakanishi, K. (2016). Anatomy of RISC: How do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip. Rev. RNA 7, 637–660.
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439.
Newman, M. A., Sundelin, T., Nielsen, J. T., and Erbs, G. (2013). MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4:139. doi: 10.3389/fpls.2013.00139
Nishad, R., Ahmed, T., Rahman, V. J., and Kareem, A. (2020). Modulation of plant defense system in response to microbial interactions. Front. Microbiol. 11:1298. doi: 10.3389/fmicb.2020.01298
Niu, B., Wang, W., Yuan, Z., Sederoff, R. R., Sederoff, H., Chiang, V. L., et al. (2020). Microbial interactions within multiple-strain biological control agents impact soil-borne plant disease. Front. Microbiol. 11:585404. doi: 10.3389/fmicb.2020.585404
Niu, D., Wang, X., Wang, Y., Song, X., Wang, J., Guo, J., et al. (2016a). Bacillus cereus AR156 activates PAMP-triggered immunity and induces a systemic acquired resistance through a NPR1-and SA-dependent signaling pathway. Biochem. Biophys. Res. Commun. 469, 120–125. doi: 10.1016/j.bbrc.2015.11.081
Niu, D., Xia, J., Jiang, C., Qi, B., Ling, X., Lin, S., et al. (2016b). Bacillus cereus AR156 primes induced systemic resistance by suppressing miR825/825 and activating defense-related genes in Arabidopsis. J. Integr. Plant Biol. 58, 426–439. doi: 10.1111/jipb.12446
Nova-Franco, B., Íñiguez, L. P., Valdés-López, O., Alvarado-Affantranger, X., Leija, A., Fuentes, S. I., et al. (2015). The micro-RNA72c-APETALA2-1 node as a key regulator of the common bean-Rhizobium etli nitrogen fixation symbiosis. Plant Physiol. 168, 273–291. doi: 10.1104/pp.114.255547
Oide, S., Moeder, W., Krasnoff, S., Gibson, D., Haas, H., Yoshioka, K., et al. (2006). NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18, 2836–2853. doi: 10.1105/tpc.106.045633
Olsen, K. M., Goad, D. M., Wright, S. J., Dutta, M. L., Myers, S. R., Small, L. L., et al. (2021). Dual-species origin of an adaptive chemical defense polymorphism. New Phytol. 232, 1477–1487. doi: 10.1111/nph.17654
Ongena, M., Duby, F., Rossignol, F., Fauconnier, M. L., Dommes, J., and Thonart, P. (2004). Stimulation of the lipoxygenase pathway is associated with systemic resistance induced in bean by a nonpathogenic Pseudomonas strain. Mol. Plant Microbe Interact. 17, 1009–1018. doi: 10.1094/MPMI.2004.17.9.1009
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ortiz-Castro, R., and López-Bucio, J. (2019). Review: Phytostimulation and root architectural responses to quorum-sensing signals and related molecules from rhizobacteria. Plant Sci. 284, 135–142. doi: 10.1016/j.plantsci.2019.04.010
Ortiz-Castro, R., Valencia-Cantero, E., and Lopez-Bucio, J. (2008). Plant growth promotion by Bacillus megaterium involves cytokinin signaling. Plant Signal. Behav. 3, 263–265. doi: 10.4161/psb.3.4.5204
Padmanabhan, C., Zhang, X., and Jin, H. (2009). Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 12, 465–472. doi: 10.1016/j.pbi.2009.06.005
Pan, H., Preisser, E. L., Su, Q., Jiao, X., Xie, W., Wang, S., et al. (2016). Natal host plants can alter herbivore competition. PLoS One 11:e0169142. doi: 10.1371/journal.pone.0169142
Pangesti, N., Weldegergis, B. T., Langendorf, B., van Loon, J. J., Dicke, M., and Pineda, A. (2015). Rhizobacterial colonization of roots modulates plant volatile emission and enhances the attraction of a parasitoid wasp to host-infested plants. Oecologia 178, 1169–1180. doi: 10.1007/s00442-015-3277-7
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588.
Park, Y. S., and Ryu, C. M. (2021). Understanding plant social networking system: Avoiding deleterious microbiota but calling beneficials. Int. J. Mol. Sci. 22:3319. doi: 10.3390/ijms22073319
Parker, D., Beckmann, M., Zubair, H., Enot, D. P., Caracuel-Rios, Z., Overy, D. P., et al. (2009). Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 59, 723–737. doi: 10.1111/j.1365-313X.2009.03912.x
Pascale, A., Proietti, S., Pantelides, I. S., and Stringlis, I. A. (2020). Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10:1741. doi: 10.3389/fpls.2019.01741
Pazarlar, S., Cetinkaya, N., Bor, M., and Kara, R. S. (2020). N-acyl homoserine lactone-mediated modulation of plant growth and defense against Pseudoperonospora cubensis in cucumber. J. Exp. Bot. 71, 6638–6654. doi: 10.1093/jxb/eraa384
Pentzold, S., Zagrobelny, M., Roelsgaard, P. S., Møller, B. L., and Bak, S. (2014). The multiple strategies of an insect herbivore to overcome plant cyanogenic glucoside defence. PLoS One. 9:e91337. doi: 10.1371/journal.pone.0091337
Pérez-García, A., Romero, D., and de Vicente, A. (2011). Plant protection and growth stimulation by microorganisms: Biotechnological applications of Bacilli in agriculture. Curr. Opin. Biotechnol. 22, 187–193. doi: 10.1016/j.copbio.2010.12.003
Peskan-Berghofer, T., Shahollari, B., Giong, P. H., Hehl, S., Markert, C., Blanke, V., et al. (2004). Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol. Plant. 122, 465–477.
Pieterse, C. M., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521.
Pieterse, C. M., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C., and Bakker, P. A. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375.
Pieterse, C. M. J., Van Pelt, J. A., Ton, J., Parchmann, S., Mueller, M. J., Buchala, A. J., et al. (2000). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant Pathol. 57, 123–134.
Pieterse, C. M. J., Van Wees, S. C. M., Hoffland, E., Van Pelt, J. A., and Van Loon, L. C. (1996). Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis related gene expression. Plant Cell 8, 1225–1237. doi: 10.1105/tpc.8.8.1225
Pieterse, C. M. J., Van Wees, S. C. M., Van Pelt, J. A., Knoester, M., Laan, R., Gerrits, H., et al. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. doi: 10.1105/tpc.10.9.1571
Pineda, A., Zheng, S. J., Van Loon, J. J. A., Pieterse, C. M. J., and Dicke, M. (2010). Helping plants to deal with insects: The role of beneficial soil-borne microbes. Trends Plant Sci. 15, 507–514. doi: 10.1016/j.tplants.2010.05.007
Planchamp, C., Glauser, G., and Mauch-Mani, B. (2014). Root inoculation with Pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front. Plant Sci. 5, 719. doi: 10.3389/fpls.2014.00719
Pozo, M. J., Van der Ent, S., Van Loon, L., and Pieterse, C. M. J. (2008). Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 180, 511–523. doi: 10.1111/j.1469-8137.2008.02578.x
Prost, I., Dhondt, S., Rothe, G., Vicente, J., Rodriguez, M. J., Kift, N., et al. (2005). Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 139, 1902–1913.
Raaijmakers, J. M., De Bruijn, I., Nybroe, O., and Ongena, M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x
Rajamanickam, S., and Nakkeeran, S. (2020). Flagellin of Bacillus amyloliquefaciens works as a resistance inducer against groundnut bud necrosis virus in chilli (Capsicum annuum L.). Arch. Virol. 165, 1585–1597. doi: 10.1007/s00705-020-04645-z
Reis, V. M., and Teixeira, K. R. (2015). Nitrogen fixing bacteria in the family Acetobacteraceae and their role in agriculture. J. Basic Microbiol. 55, 931–949.
Rodriguez-Furlán, C., Salinas-Grenet, H., Sandoval, O., Recabarren, C., Arraño-Salinas, P., Soto-Alvear, S., et al. (2016). The root hair specific SYP123 regulates the localization of cell wall components and contributes to rizhobacterial priming of induced systemic resistance. Front. Plant Sci. 7:1081. doi: 10.3389/fpls.2016.01081
Rudrappa, T., Biedrzycki, M. L., Kunjeti, S. G., Donofrio, N. M., Czymmek, K. J., Paré, P. W., et al. (2010). The rhizobacterial elicitor acetoin induces systemic resistance in Arabidopsis thaliana. Commun. Integr. Biol. 3, 130–138. doi: 10.4161/cib.3.2.10584
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Wei, H. X., Kloepper, J. W., et al. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. doi: 10.1104/pp.103.026583
Ryu, C. M., Hu, C. H., Reddy, M., and Kloepper, J. W. (2003). Different signaling pathways of induced resistance by rhizobacteria in Arabidopsis thaliana against two pathovars of Pseudomonas syringae. New Phytol. 160, 413–420. doi: 10.1046/j.1469-8137.2003.00883.x
Samalova, M., Meyer, A. J., Gurr, S. J., and Fricker, M. D. (2014). Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol. 201, 556–573. doi: 10.1111/nph.12530
Samaras, A., Roumeliotis, E., Ntasiou, P., and Karaoglanidis, G. (2021). Bacillus subtilis MBI600 promotes growth of tomato plants and induces systemic resistance contributing to the control of soil borne pathogens. Plants 10:1113. doi: 10.3390/plants10061113
Sanabria, N. M., Huang, J. C., and Dubery, I. A. (2010). Self/non-self perception in plants in innate immunity and defense. Self Nonself 1, 40–54. doi: 10.4161/self.1.1.10442
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Schenk, S. T., and Schikora, A. (2014). AHL-priming functions via oxylipin and salicylic acid. Front. Plant Sci. 5:784. doi: 10.3389/fpls.2014.00784
Schenk, S. T., Stein, E., Kogel, K. H., and Schikora, A. (2012). Arabidopsis growth and defense are modulated by bacterial quorum sensing molecules. Plant Signal. Behav. 7, 178–181. doi: 10.4161/psb.18789
Schikora, A., Schenk, S. T., Stein, E., Molitor, A., Zuccaro, A., and Kogel, K. H. (2011). N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol. 157, 1407–1418. doi: 10.1104/pp.111.180604
Schuhegger, R., Ihring, A., Gantner, S., Bahnweg, G., Knappe, C., Vogg, G., et al. (2006). Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918. doi: 10.1111/j.1365-3040.2005.01471.x
Segarra, G., Van der Ent, S., Trillas, I., and Pieterse, C. M. J. (2009). MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 11, 90–96. doi: 10.1111/j.1438-8677.2008.00162.x
Shank, E. A., and Kolter, R. (2011). Extracellular signaling and multicellularity in Bacillus subtilis. Curr. Opin. Microbiol. 14, 741–747. doi: 10.1016/j.mib.2011.09.016
Shao, J., Li, S., Zhang, N., Cui, X., Zhou, X., Zhang, G., et al. (2015). Analysis and cloning of the synthetic pathway of the phytohormone indole-3-acetic acid in the plant-beneficial Bacillus amyloliquefaciens SQR9. Microb. Cell Fact. 14:130. doi: 10.1186/s12934-015-0323-4
Sharifi, R., Lee, S. M., and Ryu, C. M. (2018). Microbe-induced plant volatiles. New Phytol. 220, 684–691.
Sharifi, R., and Ryu, C. M. (2016). Are bacterial volatile compounds poisonous odors to a fungal pathogen Botrytis cinerea, alarm signals to Arabidopsis seedlings for eliciting induced resistance, or both? Front. Microbol. 7:196. doi: 10.3389/fmicb.2016.00196
Sharifi, R., and Ryu, C. M. (2017). Chatting with a tiny belowground member of the holobiome: Communication between plants and growth-promoting rhizobacteria. Adv. Bot. Res. 82, 135–160.
Shrestha, A., Elhady, A., Adss, S., Wehner, G., Böttcher, C., Heuer, H., et al. (2019). Genetic differences in barley govern the responsiveness to N-Acyl homoserine lactone. Phytob. J. 3, 191–202.
Shrestha, A., Grimm, M., Ojiro, I., Krumwiede, J., and Schikora, A. (2020). Impact of quorum sensing molecules on plant growth and immune system. Front. Microbiol. 11:1545. doi: 10.3389/fmicb.2020.01545
Simpraga, M., Takabayashi, J., and Holopainen, J. K. (2016). Language of plants: Where is the word? J. Integr. Plant Biol. 58, 343–349.
Sivakumaran, A., Akinyemi, A., Mandon, J., Cristescu, S. M., Hall, M. A., Harren, F. J., et al. (2016). ABA suppresses Botrytis cinerea elicited NO production in tomato to influence H2O2 generation and increase host susceptibility. Front. Plant Sci. 7:709. doi: 10.3389/fpls.2016.00709
Song, G. C., and Ryu, C. M. (2013). Two volatile organic compounds trigger plant self-defense against a bacterial pathogen and a sucking insect in cucumber under open field conditions. Int. J. Mol. Sci. 14, 9803–9819. doi: 10.3390/ijms14059803
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 70, 489–525.
Stassen, M. J. J., Hsu, S. H., Pieterse, C. M. J., and Stringlis, I. A. (2021). Coumarin communication along the microbiome-root-shoot axis. Trends Plant Sci. 26, 169–183. doi: 10.1016/j.tplants.2020.09.008
Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309. doi: 10.1016/j.tplants.2007.05.001
Suzaki, T., Yoro, E., and Kawaguchi, M. (2015). Leguminous plants: Inventors of root nodules to accommodate symbiotic bacteria. Int. Rev. Cell Mol. Biol. 316, 111–158. doi: 10.1016/bs.ircmb.2015.01.004
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Taguchi, F., Suzuki, T., Inagaki, Y., Toyoda, K., Shiraishi, T., and Ichinose, Y. (2010). The siderophore pyoverdine of Pseudomonas syringae pv. tabaci 6605 is an intrinsic virulence factor in host tobacco infection. J. Bacteriol. 192, 117–126. doi: 10.1128/JB.00689-09
Takishita, Y., Charron, J. B., and Smith, D. L. (2018). Biocontrol rhizobacterium Pseudomonas sp. 23S induces systemic resistance in tomato (Solanum lycopersicum L.) against bacterial canker Clavibacter michiganensis subsp. michiganensis. Front. Microbiol. 9:2119. doi: 10.3389/fmicb.2018.02119
Tang, D., Wang, G., and Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 29, 618–637.
Trapet, P., Kulik, A., Lamotte, O., Jeandroz, S., Bourque, S., Nicolas-Francès, V., et al. (2015). NO signaling in plant immunity: A tale of messengers. Phytochemistry 112, 72–79. doi: 10.1016/j.phytochem.2014.03.015
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol 18, 607–621.
Uemura, T., Ueda, T., Ohniwa, R. L., Nakano, A., Takeyasu, K., and Sato, M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29, 49–65.
Van de Mortel, J. E., De Vos, R. C. H., Dekkers, E., Pineda, A., Guillod, L., Bouwmeester, K., et al. (2012). Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 160, 2173–2188. doi: 10.1104/pp.112.207324
Van der Ent, S., Verhagen, B. W., Van Doorn, R., Bakker, D., Verlaan, M. G., Pel, M. J., et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 146, 1293–1304. doi: 10.1104/pp.107.113829
Van Peer, R., Niemann, G. J., and Schippers, B. (1991). Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS417r. Phytopathology 81, 728–734.
Van Wees, S. C. M., Van der Ent, S., and Pieterse, C. M. J. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448.
Verbon, E. H., Trapet, P. L., Stringlis, I. A., Kruijs, S., Bakker, P. A. H. M., and Pieterse, C. M. J. (2017). Iron and immunity. Annu. Rev. Phytopathol. 55, 355–375. doi: 10.1146/annurev-phyto-080516-035537
Verhagen, B. W., Glazebrook, J., Zhu, T., Chang, H. S., van Loon, L. C., and Pieterse, C. M. (2004). The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol. Plant Microbe Interact. 17, 895–908. doi: 10.1094/MPMI.2004.17.8.895
Veyrat, N., Robert, C. A. M., Turlings, T. C. J., Erb, M., and De Deyn, G. (2016). Herbivore intoxication as a potential primary function of an inducible volatile plant signal. J. Ecol. 104, 591–600.
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206.
Vlot, A. C., Sales, J. H., Lenk, M., Bauer, K., Brambilla, A., Sommer, A., et al. (2021). Systemic propagation of immunity in plants. New Phytol. 229, 1234–1250.
Von Rad, U., Klein, I., Dobrev, P. I., Kottova, J., Zazimalova, E., Fekete, A., et al. (2008). Response of Arabidopsis thaliana to N-hexanoyl-DL-homoserinelactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 229, 73–85. doi: 10.1007/s00425-008-0811-4
Walker, J. C. (1994). Structure and function of the receptor-like protein kinases of higher plants. Plant Mol. Biol. 26, 1599–1609.
Wang, L., Sun, Z., Su, C., Wang, Y., Yan, Q., Chen, J., et al. (2019). A GmNINa-miR172c-NNC1 regulatory network coordinates the nodulation and autoregulation of nodulation pathways in soybean. Mol. Plant 12, 1211–1226. doi: 10.1016/j.molp.2019.06.002
Wei, G., Kloepper, J. W., and Tuzun, S. (1991). Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathology 81, 1508–1512.
Wei, L., Zhang, M., Wei, S., Zhang, J., Wang, C., and Liao, W. (2020). Roles of nitric oxide in heavy metal stress in plants: Cross-talk with phytohormones and protein S-nitrosylation. Environ. Pollut. 259:113943. doi: 10.1016/j.envpol.2020.113943
Weller, D., Raaijmakers, J. M., McSpadden Gardener, B., and Thomashow, L. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Wendehenne, D., Gao, Q. M., Kachroo, A., and Kachroo, P. (2014). Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20, 127–134. doi: 10.1016/j.pbi.2014.05.012
Whitehead, N. A., Barnard, A. M. L., Slater, H., Simpson, N. J. L., and Salmond, G. P. C. (2001). Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25, 365–404.
Xie, S., Jiang, H., Ding, T., Xu, Q., Chai, W., and Cheng, B. (2018). Bacillus amyloliquefaciens FZB42 represses plant miR846 to induce systemic resistance via a jasmonic acid-dependent signalling pathway. Mol. Plant Pathol. 19, 1612–1623. doi: 10.1111/mpp.12634
Yu, M., Lamattina, L., Spoel, S. H., and Loake, G. J. (2014). Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 202, 1142–1156. doi: 10.1111/nph.12739
Zamioudis, C., Hanson, J., and Pieterse, C. M. J. (2014). β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204, 368–379. doi: 10.1111/nph.12980
Zamioudis, C., Korteland, J., Van Pelt, J. A., Van Hamersveld, M., Dombrowski, N., Bai, Y., et al. (2015). Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses. Plant J. 84, 309–322. doi: 10.1111/tpj.12995
Zamioudis, C., Mastranesti, P., Dhonukshe, P., Blilou, I., and Pieterse, C. M. (2013). Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 162, 304–318. doi: 10.1104/pp.112.212597
Zamioudis, C., and Pieterse, C. M. J. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25, 139–150.
Zehra, A., Raytekar, N. A., Meena, M., and Swapnil, P. (2021). Efficiency of microbial bio-agents as elicitors in plant defense mechanism under biotic stress: A review. Curr. Res. Microb. Sci. 2:100054. doi: 10.1016/j.crmicr.2021.100054
Zhang, B. (2015). MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761.
Zhang, L., Li, S., Liu, X., Wang, Z., Jiang, M., Wang, R., et al. (2020). Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat. Commun. 11:5371.
Zhang, M., Su, H., Gresshoff, P. M., and Ferguson, B. J. (2021). Shoot-derived miR2111 controls legume root and nodule development. Plant Cell Environ. 44, 1627–1641. doi: 10.1111/pce.13992
Zhou, C., Ge, N., Guo, J., Zhu, L., Ma, Z., Cheng, S., et al. (2019). Enterobacter asburiae reduces cadmium toxicity in maize plants by repressing iron uptake-associated pathways. J. Agric. Food Chem. 67, 10126–10136. doi: 10.1021/acs.jafc.9b03293
Zhou, C., Guo, J., Zhu, L., Xiao, X., Xie, Y., Zhu, J., et al. (2016). Paenibacillus polymyxa BFKC01 enhances plant iron absorption via improved root systems and activated iron acquisition mechanisms. Plant Physiol. Biochem. 105, 162–173. doi: 10.1016/j.plaphy.2016.04.025
Zhou, C., Zhu, J., Qian, N., Guo, J., and Yan, C. (2021). Bacillus subtilis SL18r induces tomato resistance against Botrytis cinerea, involving activation of long non-coding RNA, MSTRG18363, to decoy miR1918. Front. Plant Sci. 11:634819. doi: 10.3389/fpls.2020.634819
Keywords: induction of systemic resistance, microRNAs, beneficial rhizobacteria, syntaxins, volatile organic compounds
Citation: Zhu L, Huang J, Lu X and Zhou C (2022) Development of plant systemic resistance by beneficial rhizobacteria: Recognition, initiation, elicitation and regulation. Front. Plant Sci. 13:952397. doi: 10.3389/fpls.2022.952397
Received: 25 May 2022; Accepted: 21 July 2022;
Published: 09 August 2022.
Edited by:
Jianfei Wang, Anhui University of Science and Technology, ChinaReviewed by:
Anelise Beneduzi, State Secretariat of Agriculture, Livestock and Irrigation, BrazilFiroz Ahmad Ansari, Aligarh Muslim University, India
Shrivardhan Dheeman, Sardar Bhagwan Singh Post Graduate Institute of Biomedical Sciences and Research, India
Copyright © 2022 Zhu, Huang, Lu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Zhou, emhvdWNoZW5nQG5qYXUuZWR1LmNu
 Lin Zhu
Lin Zhu Jiameng Huang1
Jiameng Huang1 Cheng Zhou
Cheng Zhou