- 1Department of Chemistry, Seoul National University, Seoul, South Korea
- 2Plant Genomics and Breeding Institute, Seoul National University, Seoul, South Korea
- 3Research Institute of Basic Sciences, Seoul National University, Seoul, South Korea
- 4Department of Biological Sciences, Sungkyunkwan University, Suwon, South Korea
Developmental plasticity contributes to plant adaptation and fitness in a given condition. Hypocotyl elongation is under the tight control of complex genetic networks encompassing light, circadian, and photoperiod signaling. In this study, we demonstrate that HISTONE DEACETYLASE 9 (HDA9) mediates day length-dependent hypocotyl cell elongation. HDA9 binds to the GIGANTEA (GI) locus involved in photoperiodic hypocotyl elongation. The short day (SD)-accumulated HDA9 protein promotes histone H3 deacetylation at the GI locus during the dark period, promoting hypocotyl elongation. Consistently, HDA9-deficient mutants display reduced hypocotyl length, along with an increase in GI gene expression, only under SD conditions. Taken together, our study reveals the genetic basis of day length-dependent cell elongation in plants.
Introduction
Hypocotyl elongation results from cell elongation, which is affected by various internal and environmental signals, such as phytohormone signaling, light quality and intensity, and ambient temperature (Gray et al., 1998; Niwa et al., 2009; Franklin et al., 2014; Gommers and Monte, 2018; Simon et al., 2018), and has been extensively investigated as a model for exploring the genetic networks controlling plant growth and development. PHYTOCHROME-INTERACTING FACTORS (PIFs), a group of basic helix-loop-helix (bHLH) transcription factors, are well-known as central players in hypocotyl elongation (Lorrain et al., 2008; Liu et al., 2011; Nusinow et al., 2011). Multilayered regulation of PIF4 and PIF5 at transcriptional and post-transcriptional levels underlies the elaborate control of hypocotyl growth (Kunihiro et al., 2010; Nusinow et al., 2011; Nieto et al., 2015; Hwang et al., 2019; Tavridou et al., 2020). Circadian clock components, such as PSEUDO-RESPONSE REGULATORS (PRRs) (Soy et al., 2016; Martin et al., 2018; Li et al., 2020), GIGANTEA (GI) (Li et al., 2016; Nohales et al., 2019), and the Evening Complex (Nusinow et al., 2011; Nieto et al., 2015), regulate PIF4 and PIF5 expression during the night (Nozue et al., 2007; Nusinow et al., 2011). Moreover, light-dependent degradation of PIFs by phytochromes further allows the accumulation of PIFs during the night (Nozue et al., 2007; Van Buskirk et al., 2014; Li et al., 2016; Pham et al., 2018; Hoang et al., 2021). The hormone- and temperature-dependent regulation of hypocotyl elongation also involves a PIF-dependent genetic pathway (Gray et al., 1998; Bernardo-Garcia et al., 2014; Li et al., 2016; Kim et al., 2020; Lee et al., 2021).
Notably, plants have evolved to coordinate their growth and development with seasonal variation in photoperiod (Wilczek et al., 2010). In Arabidopsis, diurnal hypocotyl elongation is accelerated to a greater extent in short days (SDs) than in long days (LDs), and requires both the circadian clock and light signaling to properly respond to the changing photoperiod (Nozue et al., 2007; Niwa et al., 2009). Several genetic factors are known to be involved in photoperiodic hypocotyl elongation. For example, the CYCLING DOF FACTOR (CDF) transcription factors, CDF1 and CDF5, which exhibit differential accumulation patterns under LD and SD conditions (Song et al., 2015), are crucial for hypocotyl elongation, especially in SDs (Martin et al., 2018, 2020). Consistently, genetic mutations of CDF1 or CDF5 reduce hypocotyl elongation in SDs but not in LDs (Martin et al., 2020), whereas constitutive overexpression of CDFs promotes hypocotyl growth only in SDs (Martin et al., 2020). In addition, GI, a key photoperiodic regulator, is also known to participate in hypocotyl elongation under various light conditions as shown by long hypocotyls under both red and blue light conditions (Huq et al., 2000; Tseng et al., 2004; Nohales and Kay, 2019; Park et al., 2020) and has been implicated in photoperiodic hypocotyl elongation (Kim et al., 2012; Li et al., 2016).
HISTONE DEACETYLASE 9 (HDA9) is involved in diverse aspects of plant growth and development, including seed dormancy (van Zanten et al., 2014; Baek et al., 2020), flowering time regulation (Kim et al., 2013, 2016; van Zanten et al., 2014; Kang et al., 2015; Mayer et al., 2019; Park et al., 2019; van der Woude et al., 2019; Zheng et al., 2020), leaf senescence (Chen et al., 2016; Mayer et al., 2019; Yang et al., 2020), cell differentiation (Lee et al., 2016), cell proliferation (Suzuki et al., 2018), stem cuticular wax accumulation (Wang et al., 2018), flower opening (Kang et al., 2015), and circadian oscillation (Lee et al., 2019). Moreover, HDA9 mediates responses to various environmental signals, such as salt and drought stresses (Zheng et al., 2016, 2020; Baek et al., 2020; Khan et al., 2020) and warm temperatures (Tasset et al., 2018; Shen et al., 2019; van der Woude et al., 2019). HDA9 negatively controls gene expression, in concert with interacting proteins, such as POWERDRESS (PWR), HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 15 (HOS15), WRKY53, ELONGATED HYPOCOTYL 5 (HY5), ABA INSENSITIVE 4 (ABI4), and EARLY FLOWERING 3 (ELF3) (Chen et al., 2016; Lee et al., 2019; Mayer et al., 2019; Park et al., 2019; Baek et al., 2020; Khan et al., 2020).
In this study, we found that the HDA9–GI module is important for photoperiodic hypocotyl elongation. The HDA9 protein accumulated particularly in SDs, whereas GI was expressed to higher levels in LDs than in SDs. Under SD conditions, HDA9 could bind to the GI promoter and suppress its expression by removing histone H3 acetylation. Consistently, hda9 and gi mutants displayed short and long hypocotyls, respectively, specifically under SD conditions. Overall, our results indicate that the negative regulation of GI by HDA9 is critical for hypocotyl elongation in SDs.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used in this study. Seeds were plated on half-strength Murashige and Skoog (1/2 MS) medium containing 1% sucrose, and then cold-stratified for 2 days at 4 °C in continuous darkness. The plates were incubated at 22–23°C under long day (LD; 16 h light/8 h dark) or short day (SD; 8 h light/16 h dark) photoperiods and cool white fluorescent light (120 mmol photons m–2 s–1), to promote seed germination and seedling growth. The hda9-1 (Suzuki et al., 2018; van der Woude et al., 2019), hda9-2 (Suzuki et al., 2018; van der Woude et al., 2019), gi-2 (Sawa and Kay, 2011), and pHDA9:HDA9-VENUS/hda9-1 (Suzuki et al., 2018) plants have been described previously. Seeds of the T-DNA insertion mutants hda9-1 (SALK_007123) and hda9-2 (CS370750) were also obtained from the Arabidopsis Biological Resource Center (ABRC), Columbus, OH, United States. To measure hypocotyl length, seedlings were grown horizontally on a plate for 7 days, and hypocotyls were measured using the ImageJ software1. This experiment was conducted in at least three independent biological replicates, with each replicate containing at least 30 seedlings per genotype.
Immunoblot Analysis
Total proteins were extracted from pHDA9:HDA9-VENUS/hda9-1 seedlings grown for 7 days under LD and SD conditions. Whole seedlings were harvested and ground in liquid nitrogen. For the nuclear protein enrichment, the ground tissue was resuspended in nuclei isolation buffer [50 mM Tris–Cl (pH 7.5), 1 mM EDTA, 75 mM NaCl, 0.1% Triton X-100, 5% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 × protease inhibitor cocktail]. After centrifugation at 15,000 × g for 10 min at 4 °C, the supernatant was discarded, and the pellet containing the nuclei was resuspended in 2 × sodium dodecyl sulfate (SDS) loading buffer. Nuclei suspension samples were transferred into clean tubes and 30 μg of each sample was separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE). The HDA9-VENUS and histone H3 (loading control) proteins were immunodetected using rabbit anti-GFP antibody (1:5000 dilution; ab290; Abcam, Cambridge) and rabbit anti-histone H3 antibody (ab1791; Abcam, Cambridge), respectively.
Quantitative Real-Time Reverse Transcription PCR
Total RNA was extracted using TransZol reagent (TransGen Biotech, Beijing, China) according to the manufacturer’s recommendations. Then, 2 μg of total RNA was reverse transcribed using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Dr. Protein, Seoul, South Korea) and dT18 oligos to synthesize first-strand cDNA. The cDNA samples were diluted to 100 μL with water, and 1 μL of diluted cDNA was used for PCR amplification.
Real-Time Reverse Transcription PCR reactions were performed in 96-well blocks on the Step-One Plus Real-Time PCR System (Applied Biosystems) using SYBR Master Mix (Enzynomics, Seoul, South Korea) and sequence-specific primers (Supplementary Table 5). The expression of genes was normalized relative to that of EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (eIF4A; At3g13920), and quantified using the ΔΔCt method. The specificity of the RT-qPCR primers was determined by melt curve analysis of the amplified products using the standard method installed in the system.
Chromatin Immunoprecipitation Assays
Seven-day-old pHDA9:HDA9-VENUS/hda9-1 and hda9-1 seedlings grown under SD conditions were used for ChIP assays. The harvested plant materials were fixed in 1% formaldehyde and ground in liquid nitrogen. Chromatin was resuspended in nuclei lysis buffer [50 mM Tris–HCl (pH 8.0), 10 mM EDTA, 1% SDS, 1 mM PMSF, and 1× protease inhibitor cocktail] and sonicated using Bioruptor Pico (Diagenode) to generate approximately 500 bp fragments. The immunoprecipitates were collected using anti-GFP antibody (ab290; Abcam, Cambridge), anti-H3ac antibody (06-599; Millipore, Billerica, MA, United States), and agarose A/G beads (SC-2003; Santa Cruz Biotechnology, Santa Cruz, CA, United States), and DNA fragments were purified using the DNA elution kit (CMA0112, LaboPass, South Korea). The amount of precipitated DNA was quantified by quantitative PCR (qPCR) using sequence-specific primers (Supplementary Table 6). Data were normalized relative to eIF4a, and values for control plants were set to 1.
Sequencing Data Analysis
The data of RNA-sequencing and ChIP-sequencing used in this study were obtained from previous studies. Up-regulated genes in hda9 were analyzed using the data source of SRA BioProject number PRJNA743930. RNA-seq reads were mapped using STAR(v.2.7.10a) with ‘–pOverlapNbasesMin 12 –peOverlapMMp 0.1 –twopassMode Basic’ options (Dobin et al., 2013). RSEM(v1.3.1) was used to quantify transcript abundance (Li and Dewey, 2011), and DEseq2(v1.34.0) was used to identify differentially expressed genes (DEGs) between two conditions (Love et al., 2014) with cut-off of log2-fold change > 1 and P-value < 0.05. HDA9-binding genes were analyzed using the data source of accession number GSE80056 (Chen et al., 2016). ChIP-seq reads were trimmed and mapped using Trim Galore(v0.6.7)2 and Bowtie2(v2.4.5) (Langmead and Salzberg, 2012) with default parameters. MACS2(v2.2.7.1) (Zhang et al., 2008) was employed to call peaks with default parameters. Genes containing peaks within upstream 2-kb and gene body regions were annotated as HDA9-binding genes using ChIPpeakAnno(v3.28.1) (Zhu et al., 2010). Additionally, the list of down-regulated genes in response to darkness was obtained from Yan et al. (2019).
Transient Expression Analysis Using Arabidopsis Protoplasts
For transient expression analysis using Arabidopsis protoplasts, coding sequences for HDA9 and GI were cloned into the vector containing the CaMV 35S promoter. Seven-day-old seedlings grown under the SD conditions were harvested in 20 mL 0.5 M mannitol solution (90Φ plate) and incubated for 1 h at room temperature (RT) for protoplast extraction. Then, the 0.5 M mannitol solution was replaced to a 20 mL enzyme solution (2% Viscozyme L, 1% Celluclast 1.5 L, 1% Pectinex Ultra SP-L in MMC, adjusted to pH 5.8 by NaOH and sterilized through a 0.2 μm syringe filtering) and incubated in the darkness for 16 h at 22–23°C. The protoplasts were collected by centrifugation at 100 g for 7 min and washed twice with the W5 solution containing 0.1% glucose, 0.08% KCl, 0.9% NaCl, 1.84% CaCl2, and 2 mM MES (pH 5.7). Arabidpsis protoplasts were transfected through PEG-mediated transfection. After 16 h incubation in the dark at 23 °C, transformed protoplasts were processed for total RNA extraction.
Results
hda9 Mutants Display Short Hypocotyls in Short Days
Accumulating evidence has shown that reversible histone acetylation and deacetylation underlie a variety of plant developmental processes. Here, we investigated hypocotyl elongation, which is regulated by environmental stimuli, such as ambient temperature, light intensity, and day length (Gray et al., 1998; Niwa et al., 2009; Franklin et al., 2014; Gommers and Monte, 2018). Notably, hypocotyl elongation is promoted under SD conditions (Nozue et al., 2007; Niwa et al., 2009), although the molecular basis of photoperiodic regulation of hypocotyl elongation remains to be fully elucidated.
Initial screening revealed that the HDA9 gene is particularly important for photoperiodic regulation of hypocotyl growth. We examined hypocotyl elongation in HDA9-deficient mutants, hda9-1 and hda9-2, under both SD and LD conditions. Hypocotyl growth was reduced in both hda9 mutants compared with the wild type in SDs; however, the hypocotyl length of both hda9 mutants was indistinguishable from that of the wild type in LDs (Figure 1). To confirm the results, we tested the effect of photoperiod on hypocotyl elongation in HDA9 complementation lines generated previously (Suzuki et al., 2018). The reduced hypocotyl length of hda9-1 mutant in SDs was recovered in pHDA9:HDA9-VENUS/hda9-1 seedlings (Figure 1). These results suggest that HDA9 promotes hypocotyl elongation under SD conditions.
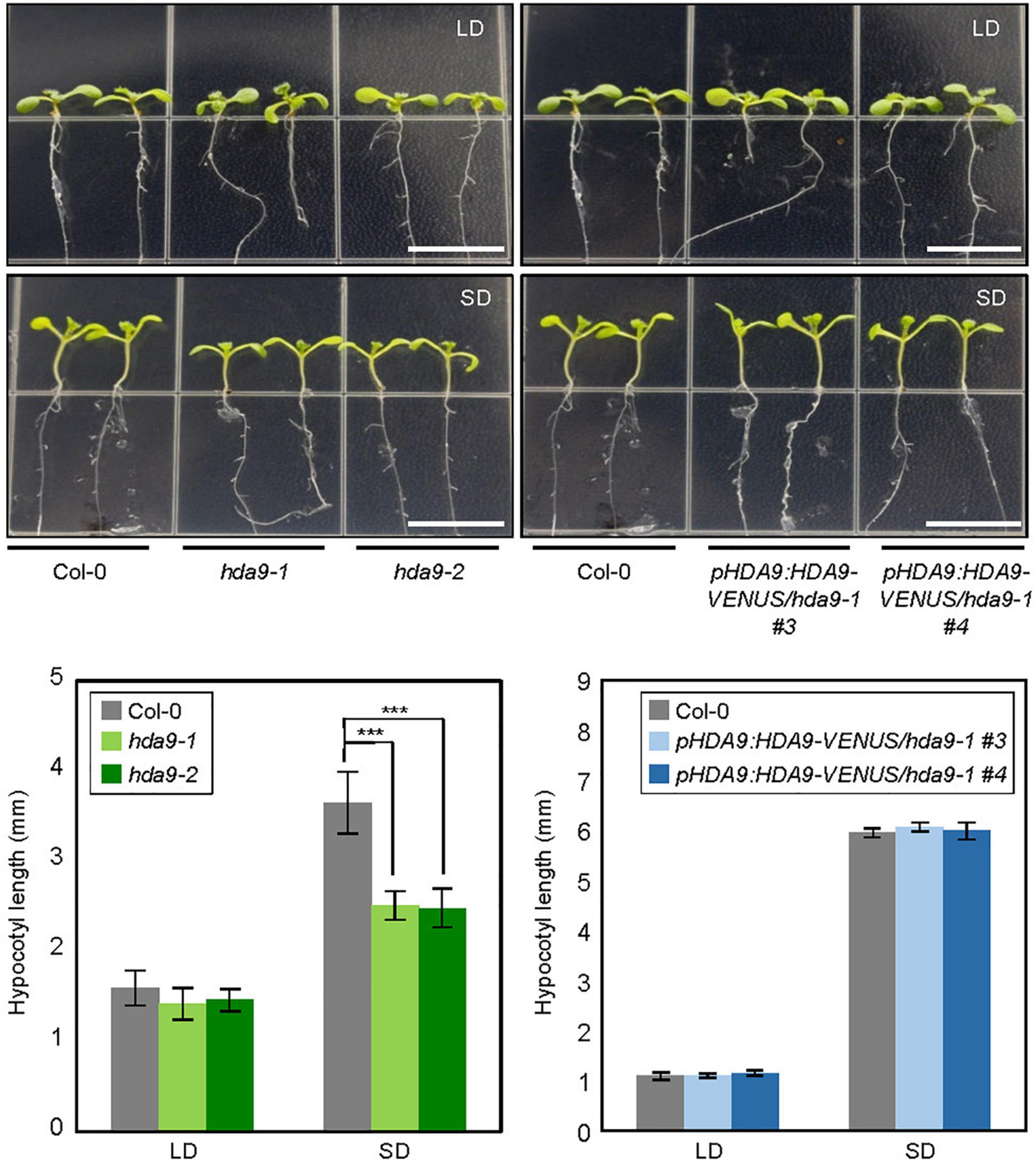
Figure 1. Hypocotyl elongation phenotypes of hda9 mutants under short day (SD) photoperiod. Seedlings were grown under long day (LD) and SD conditions at 22°C, and hypocotyl length was measured for 7 days. At least 30 seedlings of each genotype were used in a single replicate, and values of three biologically independent replicates were averaged. Error bars indicate the standard error of mean (SEM). Statistical significance of the measurements was analyzed by Student’s t-test (***P < 0.001). Scale bars = 1.0 cm.
HDA9 Accumulation Is Enhanced in Short Days
Since HDA9 promotes hypocotyl elongation specifically in SDs, we asked whether the HDA9 gene is activated only under the SD photoperiod. Total RNA was isolated from 7-day-old seedlings grown in LDs and SDs, and subjected to RT-qPCR. The results showed that the diurnal expression pattern of HDA9 was indistinguishable between seedlings grown under LD and SD conditions (Figure 2A and Supplementary Figure 1).
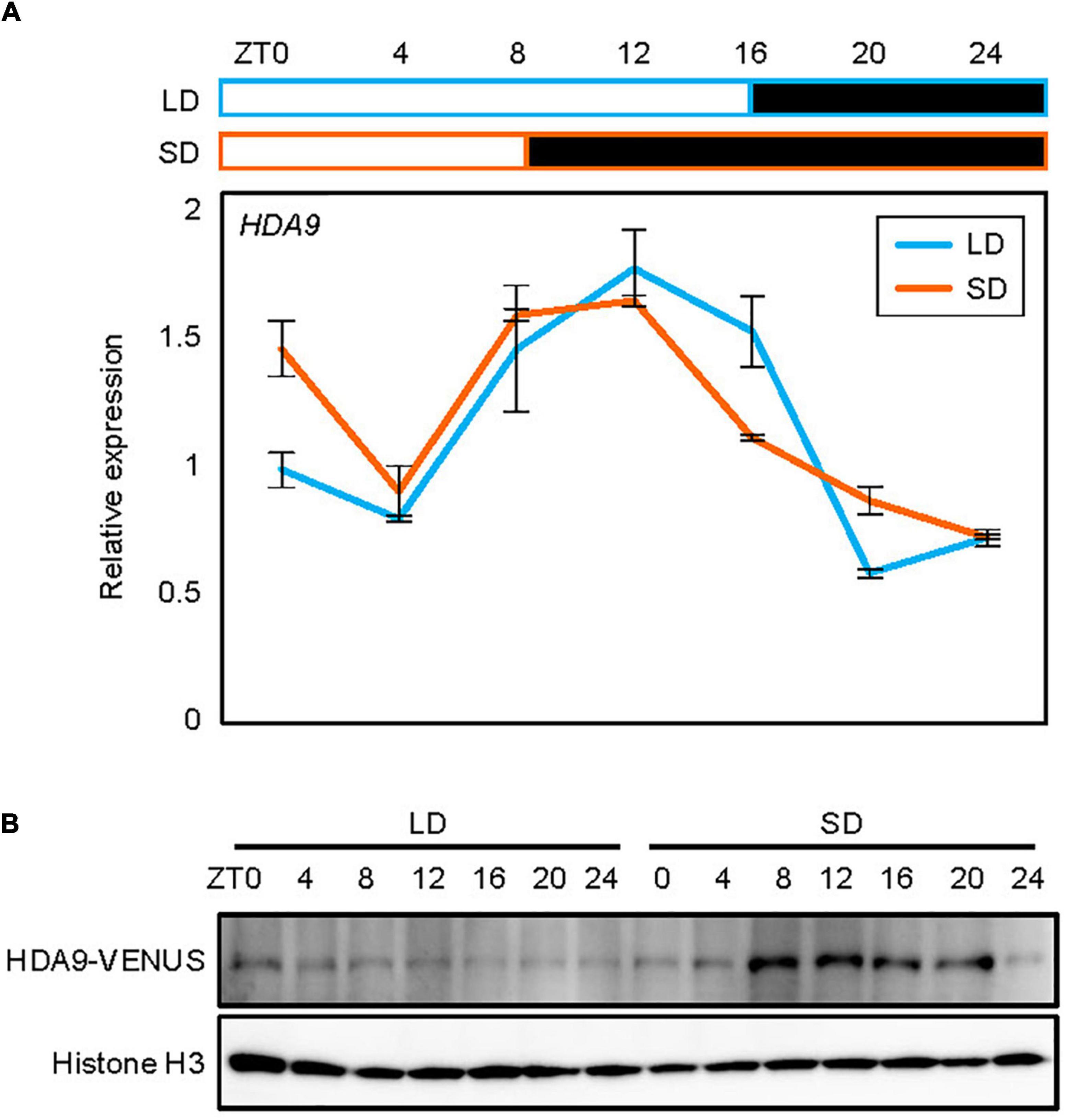
Figure 2. Accumulation of HDA9 protein in short days (SDs) relative to that in long days (LDs). Seedlings grown under LD and SD conditions for 7 days were harvested at 4 h intervals within a 24 h period. (A) Expression analysis of HDA9. Transcript levels of HDA9 were analyzed by quantitative real-time reverse transcription PCR (RT-qPCR) and normalized relative to that of eIF4A. Error bars indicate the standard error of mean (SEM). White and black boxes indicate subjective day and night, respectively. (B) Analysis of nuclear HDA9 protein level. The HDA9-VENUS fusion protein in pHDA9:HDA9-VENUS/hda9-1 plants were immunologically detected by Western blot analysis. Histone H3 accumulation was used as a loading control.
To further examine whether HDA9 accumulation is regulated at the translational and/or post-translational levels, we obtained transgenic plants expressing the HDA9-VENUS fusion under the control of the native HDA9 promoter, and analyzed the diurnal pattern of HDA9 accumulation in LDs and SDs. Notably, while total HDA9 levels were unchanged between LDs and SDs (Supplementary Figure 2), the nuclear HDA9 protein accumulated to higher levels in SDs than in LDs, suggesting that day length-dependent nuclear localization and/or enrichment might be involved in photoperiodic hypocotyl elongation. The higher protein accumulation in SDs was particularly observed during the subjective night (ZT8–20) (Figure 2B). Although HDA9 accumulation increased during the dark period in SDs, it is unlikely that HDA9 accumulation was simply responsive to the dark, as the level of HDA9 protein remained unchanged during the dark period under LD conditions (Figure 2B). These observations suggest that the HDA9 protein level is mainly regulated by photoperiod rather than by dark exposure, and that SD-driven HDA9 accumulation is responsible for hypocotyl elongation.
HDA9 Represses GI Expression in Short Days
Next, we investigated a molecular factor regulated by HDA9 to control photoperiodic hypocotyl elongation. We first obtained a list of HDA9 target genes from the HDA9-Flag ChIP-seq data published previously (Chen et al., 2016; Supplementary Table 1). To further narrow down the gene candidates responsible for HDA9-mediated regulation of photoperiodic hypocotyl elongation, we also collected genes repressed by HDA9 from Sequence Read Archive (SRA) data PRJNA743930 (Supplementary Table 2) as well as by darkness (Yan et al., 2019; Supplementary Table 3). Venn diagram analysis3 revealed 17 common genes (Figure 3A and Supplementary Table 4), including GI, CDF3, INDOLE-3-ACETIC ACID INDUCIBLE 19 (IAA19), and SMALL AUXIN UPREGULATED RNA 6 (SAUR6) (Supplementary Table 4).
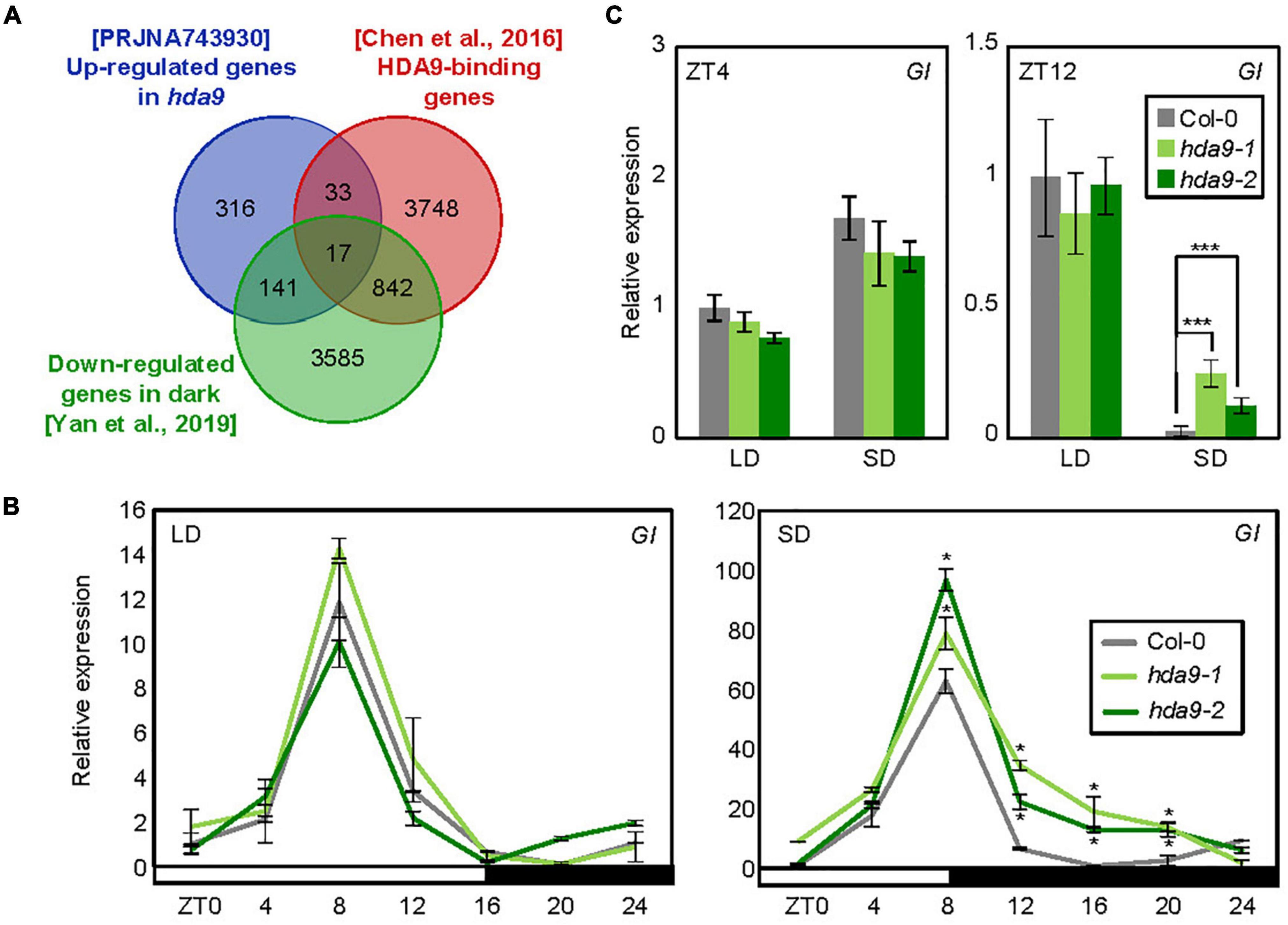
Figure 3. HDA9 represses GIGANTEA (GI) expression during the dark period in short days (SDs). (A) Venn diagram analysis. The intersection of genes targeted by HDA9, up-regulated in hda9 mutants and down-regulated by darkness, was determined for downstream analysis. (B) GI expression in hda9-1 and hda9-2 mutants. Seven-day-old seedlings grown under long day (LD) and short day (SD) conditions (n > 15) were harvested at different time points within a 24 h period (from ZT0 to ZT24). Gene expression levels were normalized relative to eIF4A expression. Error bars indicate standard error of mean (SEM). (C) Transcript level of GI in hda9 mutants in long days (LDs) and SDs. Whole seedlings were harvested at ZT4 and ZT12. Biological triplicates were averaged. Error bars indicate the SEM. Statistical significance of the measurements was analyzed by Student’s t-test (*P < 0.05).
We were particularly interested in investigating the GI gene, which is known as a key regulator of photoperiodic response (Kim et al., 2012; Song et al., 2014; Lee et al., 2019; Park et al., 2020) and has been shown to regulate hypocotyl elongation under various light conditions (Huq et al., 2000; Crepy et al., 2007; Li et al., 2016). We thus examined whether HDA9 regulates GI expression, and whether the effect of HDA9 on GI expression varies with day length. In wild-type seedlings, the expression of GI was diurnally regulated, with peak transcript levels at ZT8 in both LDs and SDs (Figure 3B). Notably, while the relative expression of GI was unaffected by absence of HDA9 in LDs, its transcript level was increased in hda9 mutants specifically under SD conditions, especially during the dark period (ZT8–20) (Figures 3B,C), when HDA9 accumulation was relatively high (Figure 2B). These results indicate that SD-induced accumulation of HDA9 represses GI expression, especially in dark period.
HDA9 Directly Binds to the GI Promoter to Catalyze H3 Deacetylation in Short Days
To investigate whether HDA9 directly binds to the GI locus, we performed ChIP assays using pHDA9:HDA9-VENUS/hda9-1 transgenic plants grown under LD and SD conditions. ChIP-qPCR analysis of the plants harvested at ZT4 and ZT12 revealed that although HDA9 did not bind to the GI locus in LDs, its binding to the GI promoter was observed in SDs, specifically at ZT12 (Figures 4A,B). This is in agreement with the accumulation pattern of HDA9 (Figure 2B). Furthermore, the HDA9 protein showed strong association with the proximal region of the GI promoter, while other chromatin regions were not targeted by HDA9 (Figures 4A,B).
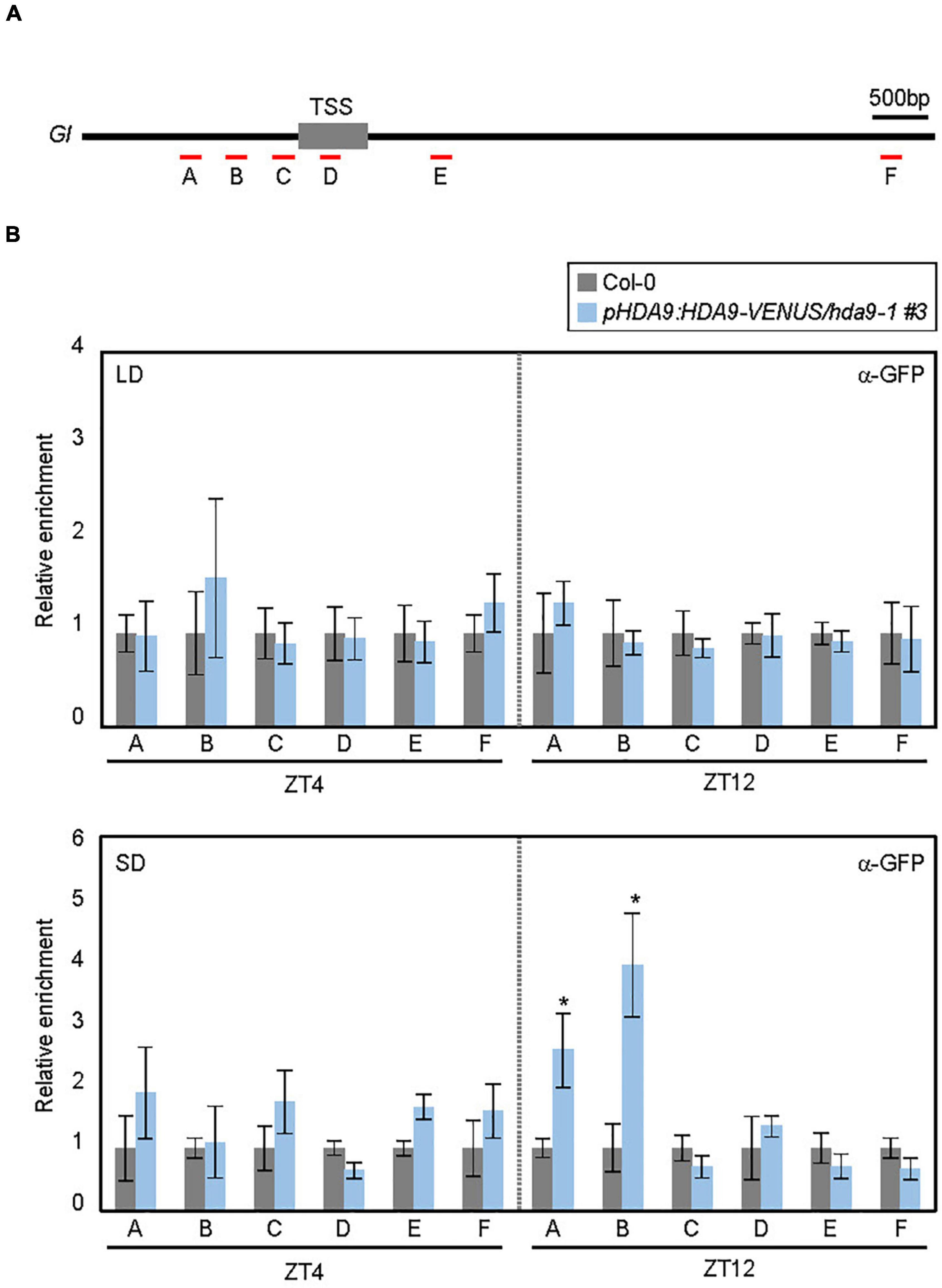
Figure 4. HDA9 binds to the GIGANTEA (GI) promoter. (A) Genomic structure of GI. Underbars indicate the regions amplified by PCR following chromatin immunoprecipitation (ChIP). (B) HDA9 binds to the GI locus in short days (SDs). Seven-day-old seedlings grown under long day (LD) and short day (SD) conditions were harvested at ZT4 and ZT12, and subjected to the ChIP assay using anti-GFP antibody. Three biological replicates were averaged. Error bars indicate the SEM. Statistical significance of the differences between Col-0 (wild type) and pHDA9:HDA9-VENUS/hda9-1 transgenic plants were analyzed by Student’s t-test (*P < 0.05).
The SD-specific recruitment of HDA9 to the GI locus was likely to facilitate photoperiodic histone deacetylation. Since HDA9 mainly determines histone H3 acetylation (H3ac) levels (Chen et al., 2016; Kim et al., 2016; Lee et al., 2019; Mayer et al., 2019; Baek et al., 2020), we examined H3ac accumulation at the GI promoter (Figures 5A,B). In wild-type seedlings, the H3ac level at the GI locus was higher in LDs, especially at ZT12, than in SDs (‘C’ and ‘D’ regions; Figure 5B), which is consistent with the higher expression level of GI in LDs (Figure 3C). The reduction of H3ac levels at the GI locus in SDs was likely caused by HDA9. Low H3ac deposition at ZT12 in SDs was at least partially impaired in the hda9-1 mutant (Figure 5B). Consistent with the enhanced accumulation of HDA9 during the dark period in SDs (Figure 2B), the level of H3ac at the GI locus was unaffected in the hda9-1 mutant at ZT4 under SD conditions (Figure 5B). Overall, our results indicate that HDA9 binds to and catalyzes histone deacetylation at the GI promoter to repress gene expression in SDs.
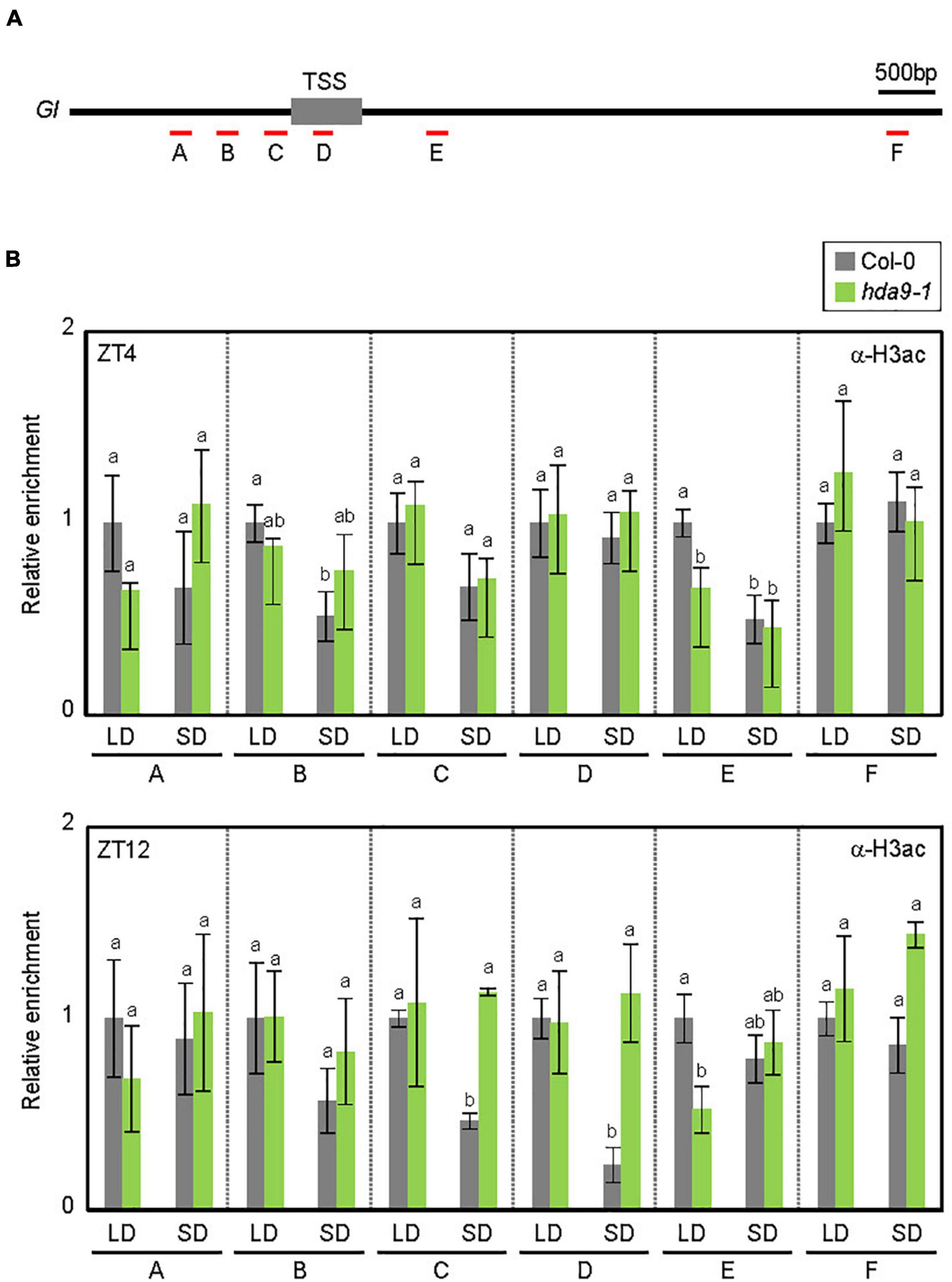
Figure 5. HDA9 regulates histone H3 acetylation levels at the GIGANTEA (GI) locus. (A) Genomic structure of GI. Underbars indicate the regions amplified by PCR following chromatin immunoprecipitation (ChIP). (B) HDA9 mediates the histone deacetylation at the GI locus in SDs. Seven-day-old seedlings grown under long day (LD) and short day (SD)conditions were harvested at ZT4 and ZT12, and subjected to the ChIP using anti-H3ac antibody. Three biological replicates were averaged. Error bars indicate the standard error of mean (SEM). Different letters represent a significant difference at P < 0.05 (one-way ANOVA with Fisher’s post hoc test).
HDA9–GI Module Ensures Photoperiodic Regulation of Hypocotyl Elongation
Our results showed that HDA9 directly binds to the GI promoter and represses its expression, particularly under SD conditions. Although the role of GI in hypocotyl elongation has been demonstrated under various light conditions (Tseng et al., 2004; Kim et al., 2012; Li et al., 2016), its role in photoperiodic hypocotyl elongation needed to be verified under our conditions. Therefore, we measured the hypocotyl length of the GI-deficient mutant (gi-2) plants in LDs and SDs. Under the LD photoperiod, the gi-2 mutant displayed marginally increased hypocotyl length compared with wild-type seedlings (Figure 6A). However, in SDs, the gi-2 mutant exhibited much longer hypocotyls than the wild type (Figure 6A), whereas the hda9-2 mutant displayed shorter hypocotyl length (Figure 6A), indicating that the HDA9–GI module plays a critical role in photoperiodic regulation of hypocotyl elongation.
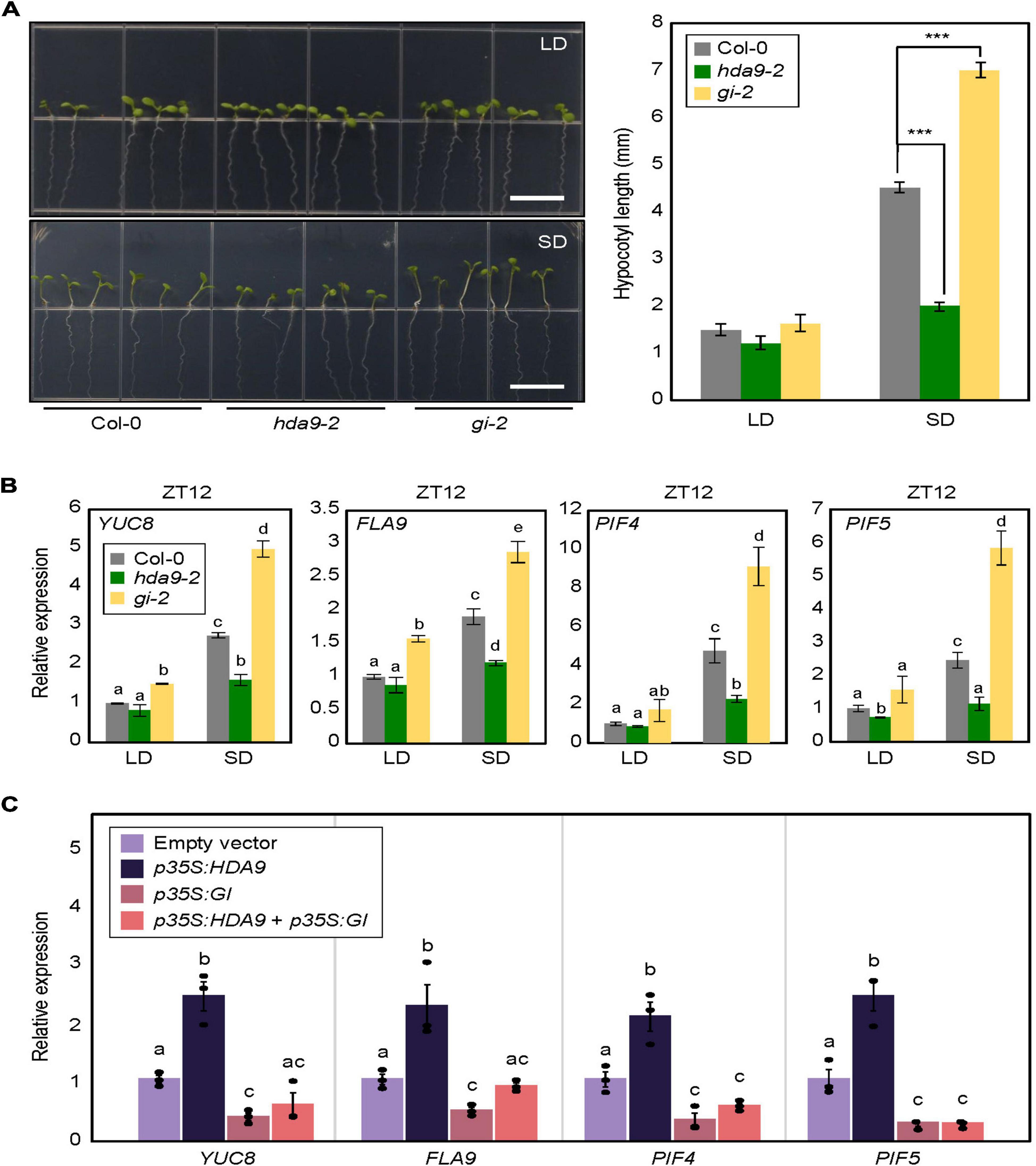
Figure 6. Photoperiodic regulation of hypocotyl elongation in hda9-2 and gi-2 mutants. (A) Analysis of hypocotyl growth in hda9-2 and gi-2 mutants in short days (SDs). Seven-day-old seedlings grown under long day (LD) and short day (SD) conditions were photographed (left panel). Hypocotyl length was measured using the ImageJ software (right panel). At least 30 seedlings of each genotype were used to measure the hypocotyl length. Three biological replicates were averaged. Error bars indicate the standard error of mean (SEM). Statistical significance of the measurements was analyzed by Student’s t-test (***P < 0.001). Scale bars = 1.0 cm. (B) Expression analysis of YUC8, FLA9, PIF4, and PIF5 at ZT12. Transcript accumulation was measured by RT-qPCR. Three biological replicates were averaged. Error bars indicate the SEM. Different letters represent a significant difference at P < 0.05 (one-way ANOVA with Fisher’s post hoc test). (C) Expression of YUC8, FLA9, PIF4, and PIF5 in protoplasts transiently overexpressing HDA9 and GIGANTEA (GI). Seven-day-old seedlings grown under SD conditions were used for protoplast isolation. The recombinant constructs were coexpressed transiently in Arabidopsis protoplasts, and then protoplast samples were harvested at ZT12 24 h after transfection to extract total RNAs. Transcript accumulation was determined by RT-qPCR analysis. Biological triplicates were averaged, and statistical significance of the measurements was analyzed by one-way analyzed by one-way analysis of variance (ANOVA) with Fisher’s post hoc test (P < 0.05). Bars indicate the standard error of the mean.
To further support these observations, we examined the expression of hypocotyl elongation-related genes, including YUCCA 8 (YUC8), FASCICLIN-LIKE ARABINOOGALACTAN 9 (FLA9), PIF4, and PIF5, in gi-2 and hda9-2 mutants. In LDs, all of these genes showed comparable expression levels in wild-type and mutant seedlings. However, under SD conditions, these genes were significantly downregulated in hda9-2 seedlings but upregulated in gi-2 seedlings (Figure 6B), in agreement with their hypocotyl phenotypes (Figure 6A).
We wanted to confirm genetic hierarchy between HDA9 and GI. To this end, 35S:HDA9 and 35S:GI constructs were generated and used for transient expression analysis using protoplasts obtained from SD-grown seedlings. Overexpression of HDA9 led to the increase in YUC8, FLA9, PIF4, and PIF5 expression, whereas GI overexpression lowered their expression in transfected protoplasts (Figure 6C). Notably, overexpression of both HDA9 and GI reduced the expression of all genes comparable to GI overexpression (Figure 6C), indicating that GI is epistatic to HDA9 in the control of photoperiodic hypocotyl elongation.
Taken together, our results show that HDA9 accumulates by SD conditions and directly binds to the GI locus to repress its expression. When GI reaches peak expression in SDs, HDA9 promotes H3 deacetylation at the GI promoter in the dark to reduce its expression during the night, thereby promoting hypocotyl elongation (Figure 7). Given the SD-specific impact of mutations in the GI gene on hypocotyl elongation, we speculate that GI and/or its downstream target are involved in crosstalk with SD-activated factors (see Discussion).
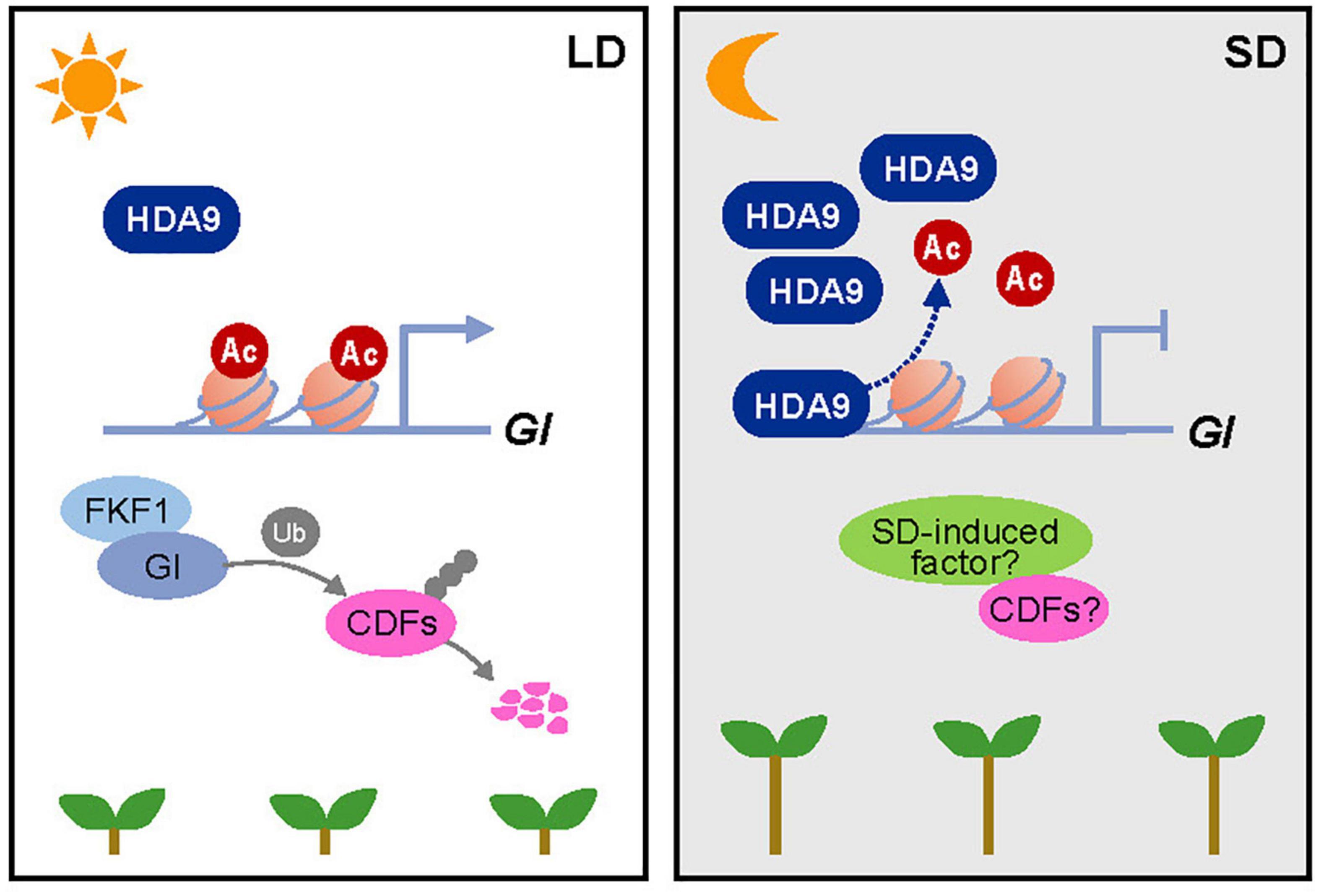
Figure 7. Proposed working model of the HDA9–GI module demonstrating its role in photoperiodic hypocotyl elongation. In short days (SDs), the HDA9 protein accumulates at dusk and then binds to the GIGANTEA (GI) promoter. This binding of HDA9 to the GI promoter facilitates the removal of H3ac from the region surrounding the transcription start site (TSS) of GI, thus repressing its expression in SDs. The HDA9–GI module regulates hypocotyl elongation in a photoperiod-dependent manner, possibly by acting together with as-yet-unidentified SD-activated molecular factors.
Discussion
Plants have evolved sophisticated molecular mechanisms for sensing changes in day length and inducing appropriate physiological responses. The representative signaling axis has been demonstrated in the control of photoperiodic flowering. Under LD conditions, GI interacts with the F-box protein FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) in a blue-light-dependent manner, inducing the 26S proteasome-dependent degradation of CYCLING DOF FACTOR 1 (CDF1), which acts as a floral repressor by directly repressing the expression of CONSTANS (CO) (Imaizumi et al., 2005; Sawa et al., 2007). The FKF1–GI complex-induced CO transcription promotes expression of the FT gene (Imaizumi et al., 2003, 2005; Sawa et al., 2007; Song et al., 2012). By contrast, under the SD photoperiod, FKF1 and GI do not interact, and their inability to form the degradation complex leads to low CO transcript abundance (Sawa et al., 2007; Song et al., 2014). In parallel, GI transcriptionally activates the transcription of miR172, which inhibits expression of TARGET OF EAT1 (TOE1), a negative regulator of FT in LDs (Jung et al., 2007). Additionally, GI binds directly to the FT promoter, possibly to activate its expression in LDs (Sawa and Kay, 2011). Furthermore, GI also binds to the transcriptional repressors of FT, such as SHORT VEGETATIVE PHASE (SVP), TEMPRANILLO 1 (TEM1), and TEMPRANILLO 2 (TEM2) (Sawa and Kay, 2011). The degradation of FT transcriptional repressors or unavailability of corresponding binding sites in the FT promoter because of the presence of GI relieves FT from transcriptional repression (Sawa and Kay, 2011; Jang et al., 2015). Consistently, gi mutants are non-responsive to day length, and display constitutive late flowering under both LD and SD conditions (Redei, 1962; Mizoguchi et al., 2005).
Hypocotyl elongation is promoted particularly in SDs (Niwa et al., 2009). The photoperiodic regulation of hypocotyl elongation most likely involves GI-regulated signaling components. The SD-repressed GI accumulation allows photoperiod-dependent hypocotyl growth, probably via multiple routes. GI promotes the degradation of CDFs in LDs (Fornara et al., 2009); however, post-translational regulation of CDFs by GI may be impaired in SDs. CDF proteins accumulated under SD conditions act as positive regulators of hypocotyl elongation (Martin et al., 2018, 2020). Consistently, the cdf5 mutant shows shorter hypocotyls only in SDs, whereas constitutive overexpression of CDF5 promotes hypocotyl growth under SD conditions (Martin et al., 2020). The gi mutant and CDF5-overexpressing seedlings showed no difference in hypocotyl length, compared with wild type, under LD conditions (Martin et al., 2020; Park et al., 2020), suggesting that GI- and CDF-regulated hypocotyl elongation requires additional SD-activated components. For instance, CDFs may interact with SD-induced factors to robustly promote hypocotyl elongation in SDs (Figure 7).
GI may also affect PIF4 expression. It has been demonstrated that while PIF4 expression levels are unaffected in gi mutants under LD conditions (Kim et al., 2012), its expression is promoted in gi mutants under SD conditions (Kim et al., 2012). PIF activity is gated during the dark phase in SDs and induces the expression of growth-promoting genes, including auxin signaling and other hormone-related genes (Michael et al., 2008; Nozue et al., 2011; Li et al., 2016; Soy et al., 2016; Martin et al., 2018). CDF5 is also regulated by PIFs. While CDF5 expression is maintained at low levels from the subjective day to early night by sequential waves of PRRs, PIFs activate CDF5 expression during the subjective night, inducing cell elongation (Martin et al., 2018, 2020). Moreover, it is also possible that GI acts as a positive regulator of TOC1 or in parallel with TOC1 to regulate hypocotyl elongation (Martin-Tryon et al., 2007).
Although GI is known as a pivotal regulator of photoperiodic responses in plants, how the activity of GI is shaped by the photoperiod remains elusive. In this study, we found that SD-activated HDA9 represses GI expression, and the HDA9–GI module regulates hypocotyl elongation only in SDs. HDA9 binds to the GI locus and catalyzes H3 deacetylation in SDs. Importantly, the regulation of GI by HDA9 was relevant particularly in SDs. The activity of HDA9 is also likely associated with skotomorphogenesis, since the hda9 mutants displayed reduced hypocotyl elongation in darkness (Supplementary Figure 3). Consistently, the accumulation of HDA9 protein was also increased in response to the continuous dark treatment (Supplementary Figure 4). However, the role of HDA9 in skotomorphogenesis is likely independent of GI because the gi-2 mutant showed no defect in skotomorphogenesis (Supplementary Figure 3; Huq et al., 2000; Tseng et al., 2004). Overall, our results demonstrate that HDA9 regulates GI expression in a photoperiod-dependent manner, ultimately allowing photoperiodic plant development.
Data Availability Statement
Publicly available datasets were analyzed in this study. The original data were deposited in the NCBI repository, accession numbers: PRJNA743930 and GSE80056.
Author Contributions
PJS and HGL participated in the design of the study and wrote the manuscript. HGL and YYJ executed the molecular experiments. HL performed the bioinformatic analysis of sequencing data. All authors approved the submitted version.
Funding
This work was supported by the Basic Science Research (NRF-2022R1A2B5B02001266) and Basic Research Laboratory (NRF-2022R1A4A3024451) programs funded by the National Research Foundation of Korea, and by Creative-Pioneering Researchers Program through Seoul National University (0409–20210288).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Gorou Horiguchi (Rikkyo University) for kindly providing pHDA9:HDA9-VENUS/hda9-1 seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.950378/full#supplementary-material
Footnotes
- ^ http://rsb.info.nih.gov/ij
- ^ https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- ^ http://bioinformatics.psb.ugent.be/webtools/Venn/
References
Baek, D., Shin, G., Kim, M. C., Shen, M., Lee, S. Y., and Yun, D. J. (2020). Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front. Plant Sci. 11:143. doi: 10.3389/fpls.2020.00143
Bernardo-Garcia, S., de Lucas, M., Martinez, C., Espinosa-Ruiz, A., Daviere, J. M., and Prat, S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28, 1681–1694. doi: 10.1101/gad.243675.114
Chen, X., Lu, L., Mayer, K. S., Scalf, M., Qian, S., Lomax, A., et al. (2016). POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. Elife 5:e17214. doi: 10.7554/eLife.17214
Crepy, M., Yanovsky, M. J., and Casal, J. J. (2007). Blue Rhythms between GIGANTEA and phytochromes. Plant Signal. Behav. 2, 530–532. doi: 10.4161/psb.2.6.4744
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Fornara, F., Panigrahi, K. C., Gissot, L., Sauerbrunn, N., Ruhl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Franklin, K. A., Toledo-Ortiz, G., Pyott, D. E., and Halliday, K. J. (2014). Interaction of light and temperature signalling. J. Exp. Bot. 65, 2859–2871.
Gommers, C. M. M., and Monte, E. (2018). Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiol. 176, 1061–1074. doi: 10.1104/pp.17.01460
Gray, W. M., Ostin, A., Sandberg, G., Romano, C. P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 95, 7197–7202. doi: 10.1073/pnas.95.12.7197
Hoang, Q. T. N., Cho, J. Y., Choi, D. M., Shin, A. Y., Kim, J. A., Han, Y. J., et al. (2021). Protein kinase activity of phytochrome A positively correlates with photoresponses in Arabidopsis. Front. Plant Sci. 12:706316. doi: 10.3389/fpls.2021.706316
Huq, E., Tepperman, J. M., and Quail, P. H. (2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 97, 9789–9794.
Hwang, G., Kim, S., Cho, J. Y., Paik, I., Kim, J. I., and Oh, E. (2019). Trehalose-6-phosphate signaling regulates thermoresponsive hypocotyl growth in Arabidopsis thaliana. EMBO Rep. 20:e47828. doi: 10.15252/embr.201947828
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Imaizumi, T., Tran, H. G., Swartz, T. E., Briggs, W. R., and Kay, S. A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306. doi: 10.1038/nature02090
Jang, K., Lee, H. G., Jung, S. J., Paek, N. C., and Seo, P. J. (2015). The E3 Ubiquitin ligase COP1 regulates thermosensory flowering by triggering GI degradation in Arabidopsis. Sci. Rep. 5:12071. doi: 10.1038/srep12071
Jung, J. H., Seo, Y. H., Seo, P. J., Reyes, J. L., Yun, J., Chua, N. H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748. doi: 10.1105/tpc.107.054528
Kang, M. J., Jin, H. S., Noh, Y. S., and Noh, B. (2015). Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol. 206, 281–294. doi: 10.1111/nph.13161
Khan, I. U., Ali, A., Khan, H. A., Baek, D., Park, J., Lim, C. J., et al. (2020). PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in Arabidopsis. Front. Plant Sci. 11:623. doi: 10.3389/fpls.2020.00623
Kim, S., Hwang, G., Kim, S., Thi, T. N., Kim, H., Jeong, J., et al. (2020). The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat. Commun. 11:1053. doi: 10.1038/s41467-020-14905-w
Kim, W., Latrasse, D., Servet, C., and Zhou, D. X. (2013). Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 432, 394–398. doi: 10.1016/j.bbrc.2012.11.102
Kim, Y., Yeom, M., Kim, H., Lim, J., Koo, H. J., Hwang, D., et al. (2012). GIGANTEA and EARLY FLOWERING 4 in Arabidopsis exhibit differential phase-specific genetic influences over a diurnal cycle. Mol. Plant 5, 678–687. doi: 10.1093/mp/sss005
Kim, Y. J., Wang, R., Gao, L., Li, D., Xu, C., Mang, H., et al. (2016). POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 113, 14858–14863. doi: 10.1073/pnas.1618618114
Kunihiro, A., Yamashino, T., and Mizuno, T. (2010). PHYTOCHROME-INTERACTING FACTORS PIF4 and PIF5 are implicated in the regulation of hypocotyl elongation in response to blue light in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 74, 2538–2541. doi: 10.1271/bbb.100586
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, K., Mas, P., and Seo, P. J. (2019). The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis. Commun. Biol. 2:143. doi: 10.1038/s42003-019-0377-7
Lee, K., Park, O. S., Jung, S. J., and Seo, P. J. (2016). Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis. J. Plant Physiol. 191, 95–100. doi: 10.1016/j.jplph.2015.12.006
Lee, S., Wang, W., and Huq, E. (2021). Spatial regulation of thermomorphogenesis by HY5 and PIF4 in Arabidopsis. Nat. Commun. 12:3656. doi: 10.1038/s41467-021-24018-7
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, K., Yu, R., Fan, L. M., Wei, N., Chen, H., and Deng, X. W. (2016). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7:11868. doi: 10.1038/ncomms11868
Li, N., Zhang, Y., He, Y., Wang, Y., and Wang, L. (2020). Pseudo response regulators regulate photoperiodic hypocotyl growth by repressing PIF4/5 transcription. Plant Physiol. 183, 686–699. doi: 10.1104/pp.19.01599
Liu, Z., Zhang, Y., Liu, R., Hao, H., Wang, Z., and Bi, Y. (2011). Phytochrome interacting factors (PIFs) are essential regulators for sucrose-induced hypocotyl elongation in Arabidopsis. J. Plant Physiol. 168, 1771–1779. doi: 10.1016/j.jplph.2011.04.009
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. doi: 10.1111/j.1365-313X.2007.03341.x
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Martin, G., Rovira, A., Veciana, N., Soy, J., Toledo-Ortiz, G., Gommers, C. M. M., et al. (2018). Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr. Biol. 28, 311–318.e5. doi: 10.1016/j.cub.2017.12.021
Martin, G., Veciana, N., Boix, M., Rovira, A., Henriques, R., and Monte, E. (2020). The photoperiodic response of hypocotyl elongation involves regulation of CDF1 and CDF5 activity. Physiol. Plant 169, 480–490. doi: 10.1111/ppl.13119
Martin-Tryon, E. L., Kreps, J. A., and Harmer, S. L. (2007). GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143, 473–486. doi: 10.1104/pp.106.088757
Mayer, K. S., Chen, X., Sanders, D., Chen, J., Jiang, J., Nguyen, P., et al. (2019). HDA9-PWR-HOS15 is a core histone deacetylase complex regulating transcription and development. Plant Physiol. 180, 342–355. doi: 10.1104/pp.18.01156
Michael, T. P., Breton, G., Hazen, S. P., Priest, H., Mockler, T. C., Kay, S. A., et al. (2008). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6:e225. doi: 10.1371/journal.pbio.0060225
Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., et al. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270. doi: 10.1105/tpc.105.033464
Nieto, C., Lopez-Salmeron, V., Daviere, J. M., and Prat, S. (2015). ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr. Biol. 25, 187–193. doi: 10.1016/j.cub.2014.10.070
Niwa, Y., Yamashino, T., and Mizuno, T. (2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50, 838–854. doi: 10.1093/pcp/pcp028
Nohales, M. A., and Kay, S. A. (2019). GIGANTEA gates gibberellin signaling through stabilization of the DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 116, 21893–21899. doi: 10.1073/pnas.1913532116
Nohales, M. A., Liu, W., Duffy, T., Nozue, K., Sawa, M., Pruneda-Paz, J. L., et al. (2019). Multi-level modulation of light signaling by GIGANTEA regulates both the output and pace of the circadian clock. Dev. Cell 49, 840–851.e8. doi: 10.1016/j.devcel.2019.04.030
Nozue, K., Covington, M. F., Duek, P. D., Lorrain, S., Fankhauser, C., Harmer, S. L., et al. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361.
Nozue, K., Harmer, S. L., and Maloof, J. N. (2011). Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 156, 357–372. doi: 10.1104/pp.111.172684
Nusinow, D. A., Helfer, A., Hamilton, E. E., King, J. J., Imaizumi, T., Schultz, T. F., et al. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402. doi: 10.1038/nature10182
Park, H. J., Baek, D., Cha, J. Y., Liao, X., Kang, S. H., McClung, C. R., et al. (2019). HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 31, 37–51. doi: 10.1105/tpc.18.00721
Park, Y. J., Kim, J. Y., Lee, J. H., Lee, B. D., Paek, N. C., and Park, C. M. (2020). GIGANTEA shapes the photoperiodic rhythms of thermomorphogenic growth in Arabidopsis. Mol. Plant 13, 459–470. doi: 10.1016/j.molp.2020.01.003
Pham, V. N., Kathare, P. K., and Huq, E. (2018). Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038.
Sawa, M., and Kay, S. A. (2011). GIGANTEA directly activates flowering locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 108, 11698–11703. doi: 10.1073/pnas.1106771108
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Shen, Y., Lei, T., Cui, X., Liu, X., Zhou, S., Zheng, Y., et al. (2019). Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J. 100, 991–1006.
Simon, N. M. L., Kusakina, J., Fernandez-Lopez, A., Chembath, A., Belbin, F. E., and Dodd, A. N. (2018). The energy-signaling hub SnRK1 is important for sucrose-induced hypocotyl elongation. Plant Physiol. 176, 1299–1310. doi: 10.1104/pp.17.01395
Song, Y. H., Estrada, D. A., Johnson, R. S., Kim, S. K., Lee, S. Y., MacCoss, M. J., et al. (2014). Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. U. S. A. 111, 17672–17677. doi: 10.1073/pnas.1415375111
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464.
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., and Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. doi: 10.1126/science.1219644
Soy, J., Leivar, P., Gonzalez-Schain, N., Martin, G., Diaz, C., Sentandreu, M., et al. (2016). Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U. S. A. 113, 4870–4875. doi: 10.1073/pnas.1603745113
Suzuki, M., Shinozuka, N., Hirakata, T., Nakata, M. T., Demura, T., Tsukaya, H., et al. (2018). OLIGOCELLULA1/HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES15 promotes cell proliferation with HISTONE DEACETYLASE9 and POWERDRESS during leaf development in Arabidopsis thaliana. Front. Plant Sci. 9:580. doi: 10.3389/fpls.2018.00580
Tasset, C., Singh Yadav, A., Sureshkumar, S., Singh, R., van der Woude, L., Nekrasov, M., et al. (2018). POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 14:e1007280. doi: 10.1371/journal.pgen.1007280
Tavridou, E., Pireyre, M., and Ulm, R. (2020). Degradation of the transcription factors PIF4 and PIF5 under UV-B promotes UVR8-mediated inhibition of hypocotyl growth in Arabidopsis. Plant J. 101, 507–517. doi: 10.1111/tpj.14556
Tseng, T. S., Salome, P. A., McClung, C. R., and Olszewski, N. E. (2004). SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16, 1550–1563. doi: 10.1105/tpc.019224
Van Buskirk, E. K., Reddy, A. K., Nagatani, A., and Chen, M. (2014). Photobody localization of phytochrome B is tightly correlated with prolonged and light-dependent inhibition of hypocotyl elongation in the dark. Plant Physiol. 165, 595–607. doi: 10.1104/pp.114.236661
van der Woude, L. C., Perrella, G., Snoek, B. L., van Hoogdalem, M., Novak, O., van Verk, M. C., et al. (2019). HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc. Natl. Acad. Sci. U. S. A. 116, 25343–25354. doi: 10.1073/pnas.1911694116
van Zanten, M., Zoll, C., Wang, Z., Philipp, C., Carles, A., Li, Y., et al. (2014). HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. Plant J. 80, 475–488. doi: 10.1111/tpj.12646
Wang, T., Xing, J., Liu, X., Yao, Y., Hu, Z., Peng, H., et al. (2018). GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exp. Bot. 69, 2911–2922. doi: 10.1093/jxb/ery077
Wilczek, A. M., Burghardt, L. T., Cobb, A. R., Cooper, M. D., Welch, S. M., and Schmitt, J. (2010). Genetic and physiological bases for phenological responses to current and predicted climates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 3129–3147. doi: 10.1098/rstb.2010.0128
Yan, H., Liu, Y., Zhang, K., Song, J., Xu, W., and Su, Z. (2019). Chromatin state-based analysis of epigenetic H3K4me3 marks of Arabidopsis in response to dark stress. Front. Genet. 10:306. doi: 10.3389/fgene.2019.00306
Yang, C., Shen, W., Yang, L., Sun, Y., Li, X., Lai, M., et al. (2020). HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol. Plant 13, 515–531. doi: 10.1016/j.molp.2020.02.011
Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137.
Zheng, Y., Ding, Y., Sun, X., Xie, S., Wang, D., Liu, X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713. doi: 10.1093/jxb/erv562
Zheng, Y., Ge, J., Bao, C., Chang, W., Liu, J., Shao, J., et al. (2020). Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant 13, 598–611. doi: 10.1016/j.molp.2019.12.011
Keywords: Arabidopsis, hypocotyl elongation, photoperiod, HDA9, GI
Citation: Lee HG, Jeong YY, Lee H and Seo PJ (2022) Arabidopsis HISTONE DEACETYLASE 9 Stimulates Hypocotyl Cell Elongation by Repressing GIGANTEA Expression Under Short Day Photoperiod. Front. Plant Sci. 13:950378. doi: 10.3389/fpls.2022.950378
Received: 22 May 2022; Accepted: 20 June 2022;
Published: 18 July 2022.
Edited by:
Ming Luo, South China Botanical Garden (CAS), ChinaReviewed by:
Xiangsong Chen, Wuhan University, ChinaWannian Yang, Central China Normal University, China
Copyright © 2022 Lee, Jeong, Lee and Seo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pil Joon Seo, cGpzZW8xQHNudS5hYy5rcg==
 Hong Gil Lee1,2
Hong Gil Lee1,2 Pil Joon Seo
Pil Joon Seo