
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 September 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.949897
This article is part of the Research Topic XVII Spanish Portuguese Congress on Plant Biology (BP2021) - Plant Growth and Development View all 6 articles
 Shujing Chen1†
Shujing Chen1† Xinhuang Zeng1†
Xinhuang Zeng1† Yiqi Li1
Yiqi Li1 Shijun Qiu1
Shijun Qiu1 Xiaoqun Peng1
Xiaoqun Peng1 Xinjue Xie1
Xinjue Xie1 Yujie Liu1
Yujie Liu1 Chancan Liao1
Chancan Liao1 Xiaoyan Tang1,2,3*
Xiaoyan Tang1,2,3* Jianxin Wu1*
Jianxin Wu1*Plastid ribosomal proteins (PRPs) are necessary components for plastid ribosome biogenesis, playing essential roles in plastid development. The ribosomal protein L18 involved in the assemble of 5S rRNA and 23S rRNA, is vital for E. coli viability, but the functions of its homologs in plant plastid remain elusive. Here, we characterized the functions of the plant plastid ribosomal protein L18s (PRPL18s) in Arabidopsis and rice. AtPRPL18 was ubiquitously expressed in most of the plant tissues, but with higher expression levels in seedling shoots, leaves, and flowers. AtPRPL18 was localized in chloroplast. Genetic and cytological analyses revealed that a loss of function of AtPRPL18 resulted in embryo development arrest at globular stage. However, overexpression of AtPRPL18 did not show any visible phenotypical changes in Arabidopsis. The rice OsPRPL18 was localized in chloroplast. In contrast to AtPRPL18, knockout of OsPRPL18 did not affect embryo development, but led to an albino lethal phenotype at the seedling stage. Cytological analyses showed that chloroplast development was impaired in the osprpl18-1 mutant. Moreover, a loss-function of OsPRPL18 led to defects in plastid ribosome biogenesis and a serious reduction in the efficiency of plastid intron splicing. In all, these results suggested that PRPL18s play critical roles in plastid ribosome biogenesis, plastid intron splicing, and chloroplast development, and are essential for plant survival.
Plastid is a semiautonomous organelle that possesses its own genome and protein synthesis apparatus (Zoschke and Bock, 2018). The plastid ribosome is responsible for the synthesis of plastid genome-encoded proteins, playing a pivotal role in plastid biogenesis (Tiller and Bock, 2014). Similar to prokaryotic ribosomes, the plastid 70S ribosome consists of a 50S large subunit and a 30S small subunit. The 50S subunit contains 23S rRNA, 5S rRNA, 4.5S rRNA, and 33 ribosomal proteins, while the 30S subunit contains 16S rRNA, and 24 ribosomal proteins (Tiller and Bock, 2014). Interestingly, 36 out of the 57 ribosomal proteins are encoded by nuclear genes, and the others are encoded by plastid genes (Tiller and Bock, 2014), indicating that the biogenesis of plastid translational apparatus may be tightly regulated under the cooperation of plastid and nucleus.
In the past two decades, lots of researches have shed light on the functions of plastid ribosomal proteins (PRPs). In Arabidopsis, many PRPs were confirmed to be essential for plant viability, such as AtPRPL1, AtPRPL4, AtPRPL6, AtPRPL10, AtPRPL13, AtPRPL18, AtPRPL21, AtPRPL27, AtPRPL28, AtPRPL31, AtPRPL35, AtPRPS13, and AtPRPS20 (Tzafrir et al., 2004; Bryant et al., 2011; Romani et al., 2012; Yin et al., 2012). Indeed, AtPRPL1, AtPRPL4, AtPRPL21, AtPRPL27, AtPRPL35 and AtPRPS20 were vital for the transition from the globular to the heart stage of embryogenesis (Romani et al., 2012; Yin et al., 2012), and AtPRPL28 was essential for plant development during the embryo greening process (Romani et al., 2012). Some PRPs appeared to be dispensable for plant viability, but play critical roles in plant development, such as AtPRPL11, AtPRPL21, AtPRPL24, AtPRPS1, AtPRPS5, AtPSRP3, AtPSRP4, and AtPSRP5, which showed pale-green leaves and retarded plant growth (Pesaresi et al., 2001; Morita-Yamamuro et al., 2004; Romani et al., 2012; Tiller et al., 2012; Zhang et al., 2016). In contrast to Arabidopsis, a few of plastid ribosomal genes were characterized in rice. A loss function of either of the plastid ribosomal genes OsPRPS1/ASL4, OsPRPS20/ASL1, OsPRPL12/AL1, or OsPRPL21/ASL2 resulted in an albino seedling death phenotype (Gong et al., 2013; Lin et al., 2015; Zhao et al., 2016; Zhou et al., 2021). The mutant with a substitution from glycine to valine (G92V) of OsPRPS9/WGL2 showed an albino phenotype at the early seedling stage, and then gradually transitioned to green (Qiu et al., 2018). However, the knock-out mutant of OsPRPS9/WGL2 exhibited albino lethal phenotype at seedling stage (Qiu et al., 2018). Interestingly, a single amino acid residue mutation from Threonine to Isoleucine (T81I) of OsPRPL13/WLP1 led to an albino phenotype at 23°C, but no significant differences compared with wild-type plants at 30°C, while OsPRPL13 RNAi plants displayed albino lethal phenotype (Song et al., 2014). These results suggested that PRPs are crucial for plant development.
In prokaryotic organisms, the ribosomal protein L18 is essential for the assembly of 5S rRNA and 23S rRNA. The C-terminal of L18 contains a 5S rRNA binding site, and its N-terminal is involved in the interaction of 5S rRNA and 23S rRNA (Brosius et al., 1975; Newberry and Garrett, 1980). In Arabidopsis, eight homologs of L18 were identified, including μL18-L1, μL18m, μL18c/AtPRPL18 (Plastid Ribosomal Protein L18), μL18-L4, μL18-L5, μL18-L6, μL18-L7, and μL18-L8 (Wang et al., 2020). μL18 m was confirmed to be a component of mitochondria ribosome (Waltz et al., 2019), while μL18c/AtPRPL18 was proposed to be a plastid ribosomal protein (Wang et al., 2020). A screening of nuclear genes encoding chloroplast-localized proteins required for embryo development identified a mutant (emb3105) of AtPRPL18, which displayed an unconfirmed embryo-defective phenotype in Arabidopsis (Bryant et al., 2011). However, direct evidence of the role of AtPRPL18 in plant embryo development is still lacking, and the functions of PRPL18s in other plant species remains elusive.
In the present study, a comparison of the functions of PRPL18s in Arabidopsis and rice was performed. Both of AtPRPL18 and OsPRPL18 were expressed highly in green tissues, and their proteins were localized in chloroplast. However, a loss function of AtPRPL18 resulted in embryo development arrest at globular stage, while knockout of OsPRPL18 did not affect embryo development, but chloroplast development was aborted, leading to an albino lethal phenotype at seedling stage. Furthermore, OsPRPL18 is required for plastid ribosome biogenesis and plastid intron splicing. Taken together, these data highlight the essential roles of PRPL18s in plant development in Arabidopsis and rice.
Arabidopsis thaliana ecotype Columbia-0 (Col-0) and the japonica rice ecotype WuYunGeng (WYG) were used as the wild-types in this study. The atprpl18/+ T-DNA insertion mutant (SAIL_415_H08) was obtained from Arabidopsis Biological Resource Center (ABRC). Arabidopsis seeds were surface sterilized by 2.5% NaClO for 8 min followed by 5 times rinsing with sterile water and stratified at 4°C for 2 days. The Arabidopsis plants were cultivated in a greenhouse at 22 ± 2°C under a 16 h light and 8 h dark cycle (Wu et al., 2015). Rice seeds were surface sterilized and germinated on MS medium supplemented with 3% sucrose and 0.7% agar in a growth chamber under a 10 h light/14 h photoperiod with 12,000 lux light intensity at 28°C. The rice seedlings at three-leaf stage were then planted in rice paddy field under natural conditions with regular care.
BLASTP was performed in NCBI1 and Phytozome2 using the full-length sequence of AtPRPL18 as a query. Fifteen representative homologues of AtPRPL18 from different species were retrieved including Crahi.0013s0018.1 (Crambe hispanica), Ciclev10022561m (Citrus clementina), Potri.010G005000.3 (Populus trichocarpa), GlymaLee.09G068200.1. (Glycine max), Vigun09g043700.1 (Vigna unguiculata), BdiBd213.1G0042200.1 (Brachypodium distachyon), LOC_Os03g61260.1/OsPRPL18 (Oryza sativa), Pahal.9G024200.1 (Panicum hallii), ACF85829.1 (Zea mays), Joasc.10G094600.1 (Joinvillea ascendens), Ceric.13G022200.1 (Ceratopteris richardii), Pp3c11_5070V3.1 (Physcomitrium patens), Cre01.g052100.t1.2 (Chlamydomonas reinhardtii), GIL60489.1 (Volvox africanus), and WP_069790223.1 (Cyanobacterium). The protein sequences were aligned using Clustal Omega3 with default parameters, and the results were visualized by Jalview.4 The phylogenetic tree was constructed using the Neighbor-Joining algorithm (1,000 replicates) in MEGA11 (Tamura et al., 2021).
For gene expression analysis, quantitative RT-PCR (qRT-PCR) was carried out using SYBR Green fluorescence with LightCycler 96 System (Schmittgen and Livak, 2008). Various Arabidopsis tissues including 7-day-old seedlings, and roots, stems, mature leaves and inflorescence from 40-day-old plants, and siliques on the 5th and 15th day after fertilization (DAF) were collected. Rice vegetative tissues and panicles were collected from seedlings or mature plants at heading date stage. Total RNA was extracted and reverse-transcribed to cDNA. qRT-PCR was performed using specific primer pairs (Supplementary Table 1). The AtACT2 (AT3G18780) gene and OsUBI (LOC_Os03g13170) gene were used as internal controls in Arabidopsis and rice, respectively.
For protein subcellular localization analysis, the CDSs of AtPRPL18 and OsPRPL18 were cloned into the vector pSAT6-EYFP-N1 and pCAMBIA1300 driven by 35S promoter (Tzfira et al., 2005). The resultant pSAT6-AtPRPL18-EYFP and pSAT6-OsPRPL18-EYFP vectors were introduced into Arabidopsis or rice protoplasts by polyethylene glycol (PEG)–calcium-mediated transformation (Chen et al., 2006; Yoo et al., 2007). The transformed protoplasts were observed with a laser confocal scanning microscope (LSM-800; Carl Zeiss) after 12 h incubation. The resultant pCAMBIA1300-35S:AtPRPL18-GFP and pCAMBIA1300-35S:OsPRPL18-GFP vectors were introduced into Agrobacterium tumefaciens strain EHA105. For subcellular protein localization, Nicotiana benthamiana (N. benthamiana) leaves were infiltrated with the indicated Agrobacteria as previously described (Sparkes et al., 2006). The GFP-tagged proteins were examined 2 d after infiltration using a confocal laser scanning microscope (LSM810, Carl Zeiss).
For mutant complementation, a 4,650 bp genomic DNA including the gene body of AtPRPL18, a 2,363 bp upstream sequence and a 956 bp downstream sequence, was cloned into the vector pCAMBIA1300. A pCRISPR-AtPRPL18 plasmid for gene editing was constructed with the target sequence ACTAACCGCAGTGCGTTCTT according to a previously reported protocol (Yan et al., 2015). The overexpression construct was generated by introducing a gene expression cassette with 35S:AtPRPL18-NosR into pCAMBIA1300. The resulted constructs were introduced into Agrobacterium tumefaciens EHA105 and then transformed into heterozygous mutant atprpl18/+ or Columbia-0 with a floral dip method (Zhang et al., 2006).
The osprpl18 mutant was generated by employing a CRISPR-Cas9 gene editing tool with the target sequence AGTGATTGCCAAGTCTTGCT and AGGACTTGGAATACTCGGCAGG on OsPRPL18 (Ma et al., 2015), and an Agrobacterium-mediated rice transformation method (Toki et al., 2006).
For GUS staining analysis, the 2.4 kb upstream region of AtPRPL18 was amplified by PCR using the primers AtPRPL18-GUS-F and AtPRPL18-GUS-R and cloned into the vector pCAMBIA1301 with In-Fusion HD cloning kit. The resultant proAtPRPL18-GUS plasmid was transformed into Arabidopsis wild-type plants via the floral dip method (Zhang et al., 2006). The homozygous proAtPRPL18-GUS transgenic lines were identified and then used for GUS staining. The GUS staining solution includes 2 mM 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc), 0.1 % Triton X-100, 2 mM K4Fe (CN)6 and 2 mM K3Fe (CN)6 in 50 mM sodium phosphate buffer, pH 7.0 (Jefferson et al., 1987). The plant tissues were incubated in GUS staining solution at 37°C for 4 h then decolorized in 75% ethanol at 37°C overnight. The stained samples were photographed using a Leica DM6 microscope.
Seeds of wild-type and atprpl18/+ mutant were removed from siliques and totally cleared in Hoyer’s buffer [chloral hydrate: glycerol: water, 8:1:2 (w/v/v)] for 30 min to 8 h depending on the embryo developmental stage (Chen et al., 2015). Embryos were examined using a light microscope Carl Zeiss Axio Observer A1 with optimal differential interference contrast.
For transmission electron microscopy (TEM) analysis, leaves from the 3-leaf-stage seedlings of osprpl18-1 and wild-type were fixed and resin-embedded as previously described (Chang et al., 2016). Samples were sliced and stained with 0.5% toluidine blue for semithin sections and uranyl acetate and lead citrate for ultrathin sections. The stained semithin sections and stained ultrathin sections were photographed using a Zeiss Axio Imager A1 microscope and a transmission electron microscope (TEM) (JEOL JEM1400), respectively.
Total RNA was extracted from the seedling shoots of WYG and osprpl18-1 at the third-leaf stage using the RNA extraction kit (Omega Bio-tek, R6830). First strand cDNA was synthesized with random hexamers or RT primer mix using the PrimeScript II 1st strand cDNA Synthesis Kit (Takara, 6210A). For RT-PCR analyses, primers were designed situated on exons flanking with the concerned introns. For qRT-PCR, primers were designed positioned across intron/exon junctions of the indicated transcripts. Some primers used in this study were consistent with that in previous research (Lv et al., 2020).
In previously reports, AtPRPL18 was identified as a plastid ribosomal protein L18 in Arabidopsis (Wang et al., 2020). To identify plant PRPL18 proteins, BlastP was employed to search homologs of AtPRPL18, and 15 L18 protein sequences from representative species were selected. Plastid is proposed originating from the engulfment of a cyanobacteria ancestor into a unicellular heterotrophic protist (Jensen and Leister, 2014). Lots of plastid genes have undergone a transfer from plastid to nucleus (Jensen and Leister, 2014). Interestingly, all of the displayed PRPL18 proteins from various eukaryotic species were encoded by nuclear genes, indicating the transfer event of PRPL18 from plastid to nucleus occurred very long time ago. However, protein sequences alignment analysis revealed that PRPL18 proteins remained high identities, especially in the protein C terminals, such as AtPRPL18 and OsPRPL18, which shared 60 and 59% identities with the full length of cyanobacterium L18 protein, respectively (Figure 1A). Phylogenetic analysis showed that AtPRPL18, together with other PRPL18 homologs of dicotyledonous plants, belong to a clan that was separated from other clans formed by monocots, mosses, green algae and prokaryotes (Figure 1B), indicating PRPL18s may retain a conserved function from their ancestor and also evolve species-specific functions.
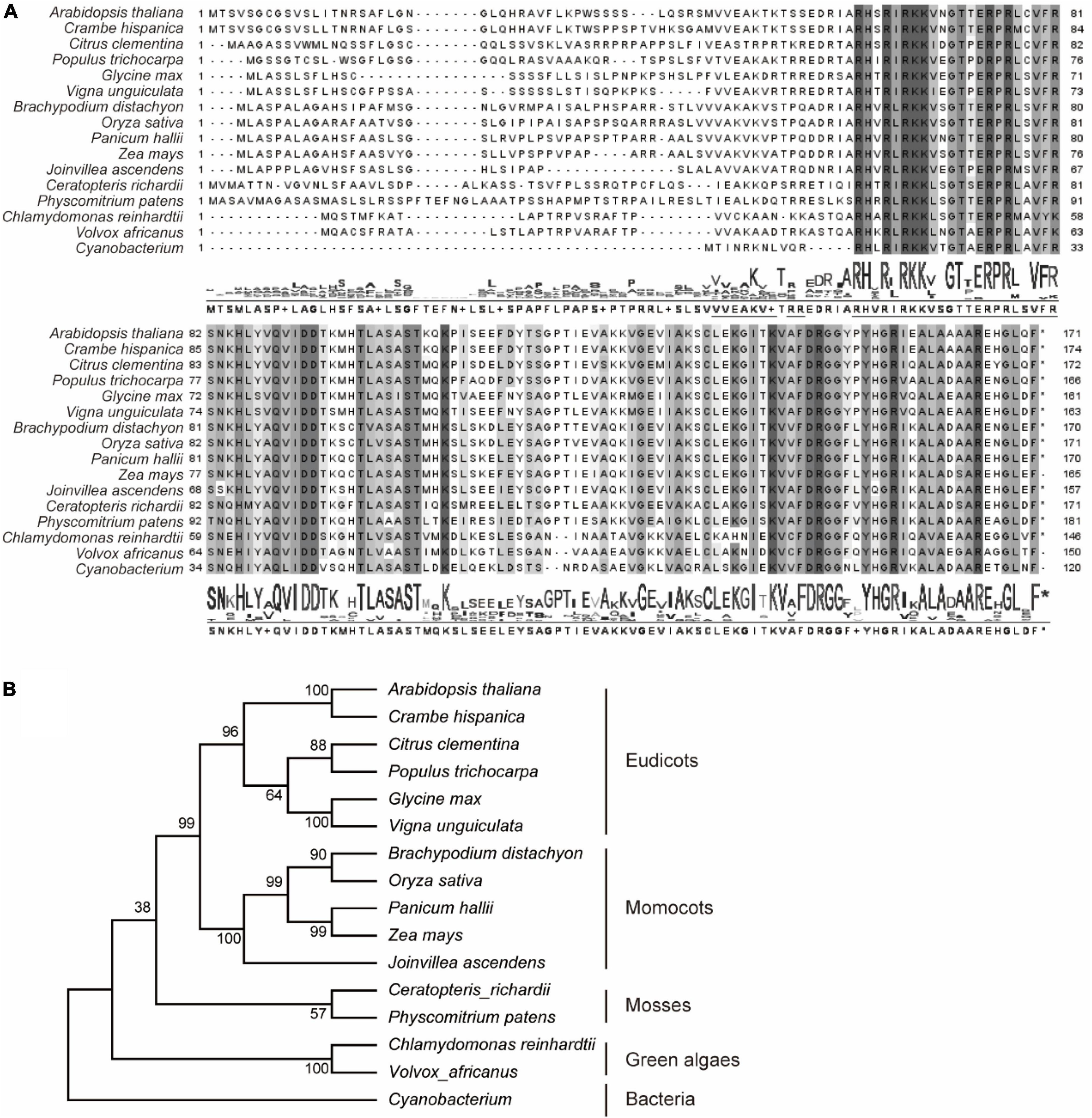
Figure 1. Phylogenetic analysis of RPL18s. Sixteen represented PRPL18 homologs were obtained from plants and cyanobacterium using BlastP of AtPRPL18. Multiple sequence alignment was performed using Clustal Omega (A), and phylogenetic tree was constructed by MEGA11 (B). The conserved amino acid residues were highlighted by gray backgrounds.
Gene function is closely related to its expression profiles and protein localization. qRT-PCR analysis showed that the transcripts of AtPRPL18 were more abundant in seedlings, stem, leaves, inflorescence and siliques than that in roots (Figure 2A). Moreover, GUS reporter assay was employed to further analyze the expression pattern of AtPRPL18. A 2,452 bp fragment upstream of the translational initiation site was fused to the GUS reporter gene and introduced into wild-type plants to generate pAtPRPL18-GUS transgenic plants. Consistent with the qRT-PCR results, strong GUS activities were detected in seedling shoots, leaves, flowers and young siliques, whereas in roots, only root tips displayed GUS activity (Figures 2B–F). Interestingly, a higher GUS activity was observed in embryos at heart stage (Figures 2G–J). These results suggested that AtPRPL18 may participate in the development of green tissues as well as reproductive tissues.
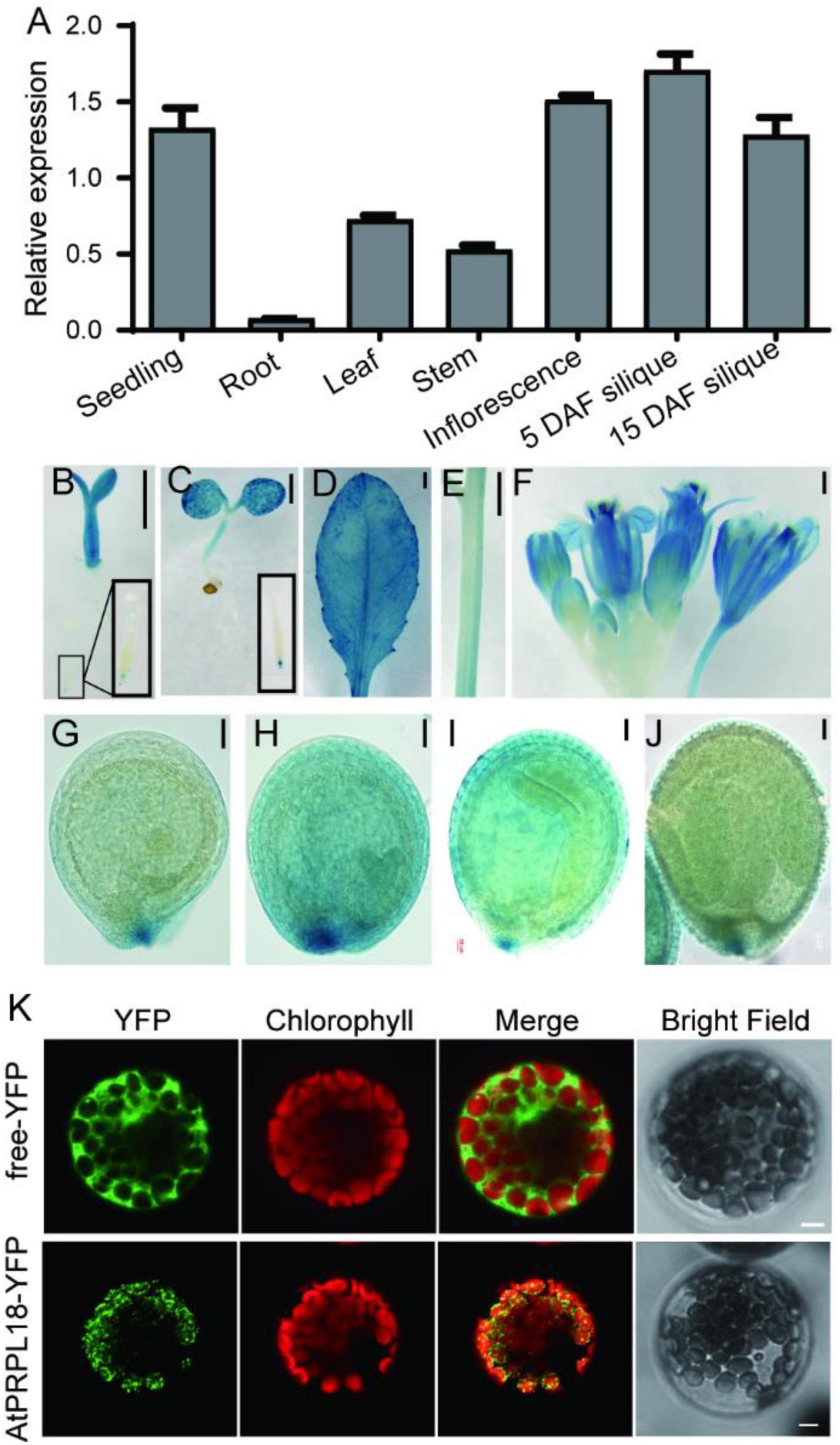
Figure 2. Gene expression patterns and protein subcellular localization of AtPRPL18. (A) The relative expression of AtPRPL18 in seedling, and stem, mature leaf, inflorescence and siliques from 40-old-day Arabidopsis plants. AtACT2 served as the internal control. Data represent means ± SD (n = 4). (B–J) GUS staining in the tissues of proAtPRPL18:GUS plants. (B) 3 day-old seedling, (C) 6 day-old seedling, (D) leaf, (E) stem, (F) inflorescence, (G) seeds at globular stage, (H) seeds at heart stage, (I) seeds at torpedo stage, (J) seeds with well-developed embryo. (K) Subcellular localization of AtPRPL18-YFP in Arabidopsis mesophyll protoplast. Free YFP was used as a control. The chlorophyll autofluorescence was used as the marker of chloroplast. Scale bars = 1 mm (B-F), 50 μm (G–J), and 5 μm (K).
The Arabidopsis PRPL18 protein was predicted to contain a typical signal peptide for chloroplast subcellular localization at its N-terminus using TargetP-2.0 software (Almagro Armenteros et al., 2019). To verify the prediction, a plasmid harboring AtPRPL18-YFP expression cassette was introduced into Arabidopsis protoplasts to examine the subcellular localization of AtPRPL18. In the transformed cells, the YFP fluorescence appeared in small dot-like structures and, was overlapped with the chlorophyll autofluorescence (Figure 2K). Transient expression of AtPRPL18-GFP in tobacco leaves showed that AtPRPL18-GFP was unevenly distributed in chloroplast, and also presented lots of dot-like structures (Supplementary Figure 1A). Though, these results were not well consistent with the previous report that μL18c-GFP/AtPRPL18-GFP was found to be localized diffusely throughout chloroplasts in the 35S: μL18c-GFP transgenic plants, it is obvious that AtPRPL18 is a chloroplast-localized protein in Arabidopsis. The difference in the distribution of the fluorescence signals in chloroplast may be resulted from the diverse protein expression levels in different protein localization assays.
In order to understand the biological roles of AtPRPL18 in Arabidopsis, a T-DNA insertion mutant (SAIL_415_H08) of AtPRPL18 was obtained from ABRC.5 The T-DNA was localized in the second exon of AtPRPL18 (Figure 3A), which was confirmed by PCR and sequencing (Figure 3B). However, no homozygous atprpl18 mutant was identified from the progenies of atprpl18 /+ heterozygotes (Figure 3C). The genotype ratio of wild-type and heterozygotes was 1 to 1.94 (85/167) in the progenies of the self-pollinated plants, consistent with the expected ratio 1:2 from a single mutational event (x2 = 0.9248,p < 0.05) (Figure 3C), suggesting that the homozygous mutant atprpl18 might be aborted during seed development. To address this hypothesis, siliques from wild-type and atprpl18/+ plants were collected and analyzed at the indicated days after self-pollination. The appearance of the mutant silique was similar to that of wild-type plants. However, a large number of albino seeds (24.6%) were found randomly distributed in the siliques of atprpl18/+ plants, whereas all seeds of the wild-type plants were green in color (Figure 3F). The albino seeds finally became shrunken in dark brown color and dried out and died in mature siliques. Reciprocal cross analysis of atprpl18/+ plant showed that the male transmission efficiency was 84% (86/102) and the female transmission efficiency was 96% (88/92), both of them had no significant differences from the expected value 100% (Table 1), indicating that AtPRPL18 did not affect gametophyte development. Together, these results suggested that the missing of the atprpl18 homozygous offspring from atprpl18/+ self-pollination was caused by impaired seed development.
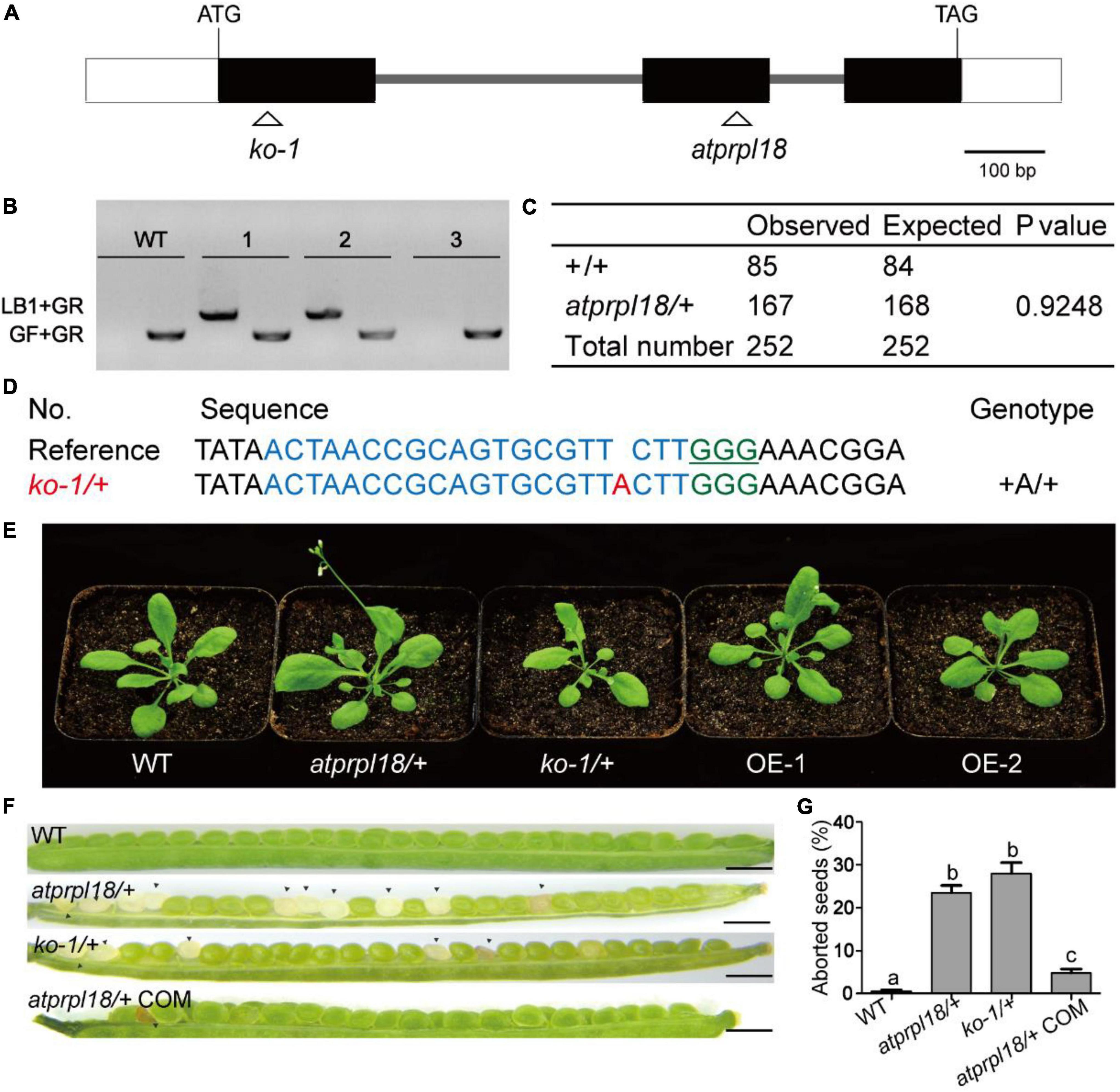
Figure 3. Characterization of the function of AtPRPL18 in plant development. (A) The gene structure of AtPRPL18. The gray line, white rectangle, and black rectangle represent intron, 5’/3’UTR, and exon, respectively. (B) PCR analysis of the genotype of the progenies of atprpl18/+ mutant. 1, 2, and 3 were the representative progenies from atprpl18/+ mutant. The T-DNA specific primer LB1 and genomic DNA primer GR were used to amplify T-DNA flanking sequences, GF and GR were used to amplify the genomic DNA sequence without T-DNA insertion. (C) Statistical analysis of the genotypes of atprpl18/+ progenies. (D) Generation of AtPRPL18 deficient mutants by CRISPR-Cas9. The guide RNA targeted DNA sequence and PAM motif were highlighted in blue and green, respectively. (E) 30-day-old plants of atprpl18/+, ko-1/+, and AtPRPL18 overexpression transgenic plants OE-1 and OE-2. (F,G) Comparison of the seed phenotypes of WT, atprpl18/+, ko-1/+, and atprpl18/+ COM plants. At least fourteen siliques of each genotype were examined. The rates of albino seeds in siliques were analyzed. Data represent means ± SD. Letters above bars indicate significant differences (P < 0.05, Student–Newman–Keuls test). This experiment was repeated twice with similar results. (a–c) Genetic analysis of the atprpl18/+ mutant.
To further confirm that the seed abortion phenotype was caused by AtPRPL18 mutation, CRISPR/Cas9 gene editing experiment was performed to generate AtPRPL18 mutants. A series of heterozygous knockout mutants were obtained from the T1 transgenic plants (Figure 3D). The T2 plants of ko1/+(knockout 1) without T-DNA were identified for further study (Supplementary Figure 2). Consistent with the findings in atprpl18/+ mutant, albino seeds were observed in the 9-day siliques of ko1/+ plants, and homozygous mutant could not be detected from the offspring of ko1/+ plants (Figures 3F,G). Additionally, a construct containing the genomic DNA fragment encompassing the AtPRPL18 gene body was generated and transformed into atprpl18/+ mutant. Heterozygous mutant plants complemented by the transgene, atprpl18/+ COM, were identified by PCR and sequencing analysis. atprpl18/+ COM plants exhibited a similar vegetative appearance to wild-type plants, and had only 5% (n = 1098) albino seeds compared the expected value 5.75% (Figures 3F,G). These results indicated that AtPRPL18 is essential for seed development. However, plants overexpressing AtPRPL18 did not show any significant difference from wild-type plants (Figure 3E and Supplementary Figure 3).
To investigate the function of AtPRPL18 in seed development, successive stages of embryo development were examined in wild-type and the atprpl18/+ mutant plants. In wild-type plants, the embryos underwent a series of development stages to become mature embryos (Figures 4A–E). There was no detectable difference in embryo development between WT and the atprpl18/+ mutant in the first 2 days after fertilization (before heart stage) (Figures 4A,B,F,G). However, on the third day after fertilization(3 DAF), some seeds appeared in white color, and the others were in green color. The green seeds from wild-type and atprpl18/+ mutant developed into late globular and early heart stages, while all of the embryos in albino seeds remained at globular stage (Figures 4C,H,K). When embryos in green seeds developed into torpedo stage, even finished embryogenesis (Figures 4D,E,I,J), the embryos in albino seeds were still at globular stage but with enlarged irregular structure (Figures 4L–M). On the 9th DAF, almost all of the embryos from wild-type plants were established, whereas, about 22.01% seeds of atprpl18/+ mutant were arrested at globular stage (Figure 4N). These data revealed that AtPRPL18 is indispensable for embryo development in the transition from globular stage to heart stage in Arabidopsis.
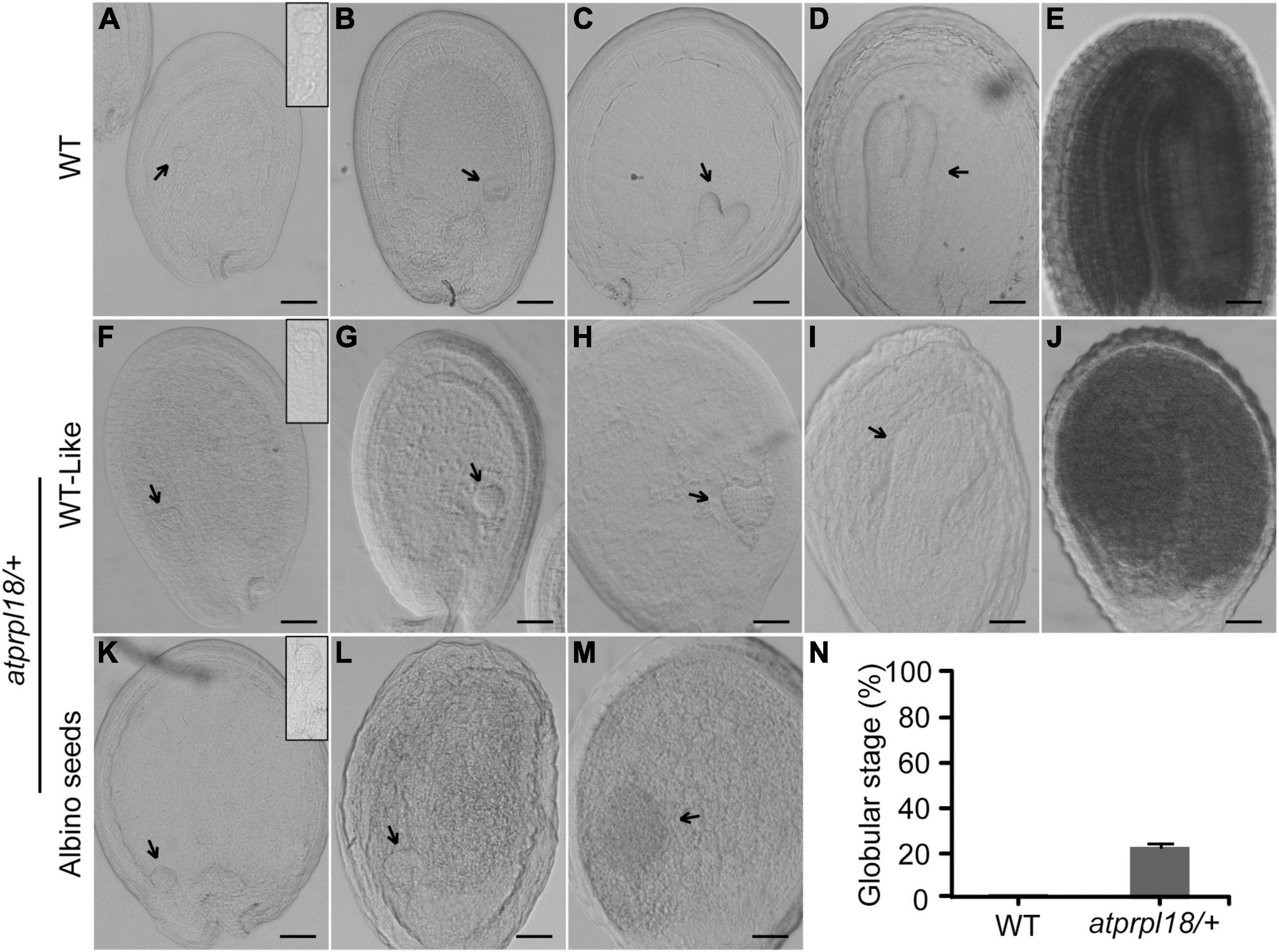
Figure 4. Comparison of the embryo development in WT and atprpl18/+ mutant. (A–E) Various developmental stages of WT embryos. (A) 4-cell, (B) globular, (C) heart, (D) torpedo, (E) mature. (F–J) WT-like embryos from the developing siliques of atprpl18/+ mutant. (K–M) Abnormal embryos from the albino seeds of atprpl18/+ mutant. The magnified views of embryos from (A,F,K) were shown on the top right of the graphs. (N) The rate of arrested embryos in the 9 DAF siliques of WT and atprpl18/+ mutant plants. Eight siliques for each genotype plants were examined. The rate of embryos arrested at globular stage per silique was calculated. Data represent means ± SD. This experiment was repeated twice with similar results. Scale bars = 50 μm (A–M).
Dicotyledonous plants and monocotyledonous plants are very distinct in embryo patterning and seed structure (Radoeva et al., 2019). To compare the biological functions of PRPL18 proteins in dicotyledonous plants and monocotyledonous plants, we further characterized the function of OsPRPL18 in the model monocotyledonous plant rice. The real-time RT-PCR analysis revealed that OsPRPL18 was expressed highly in seedling shoot and leaf sheath, lower in leaf blade, stem and panicle, and very low in root (Figure 5A), suggesting that OsPRPL18 may be critical for green tissues. Protein subcellular localization analysis showed that the fusion proteins OsPRPL18-YFP and OsPRPL18-eGFP were colocalized with the autofluorescence of chlorophyll in rice protoplast and tobacco leaf cells, respectively, indicating that OsPRPL18 is a chloroplast-localized protein (Figure 5B and Supplementary Figure 1B).
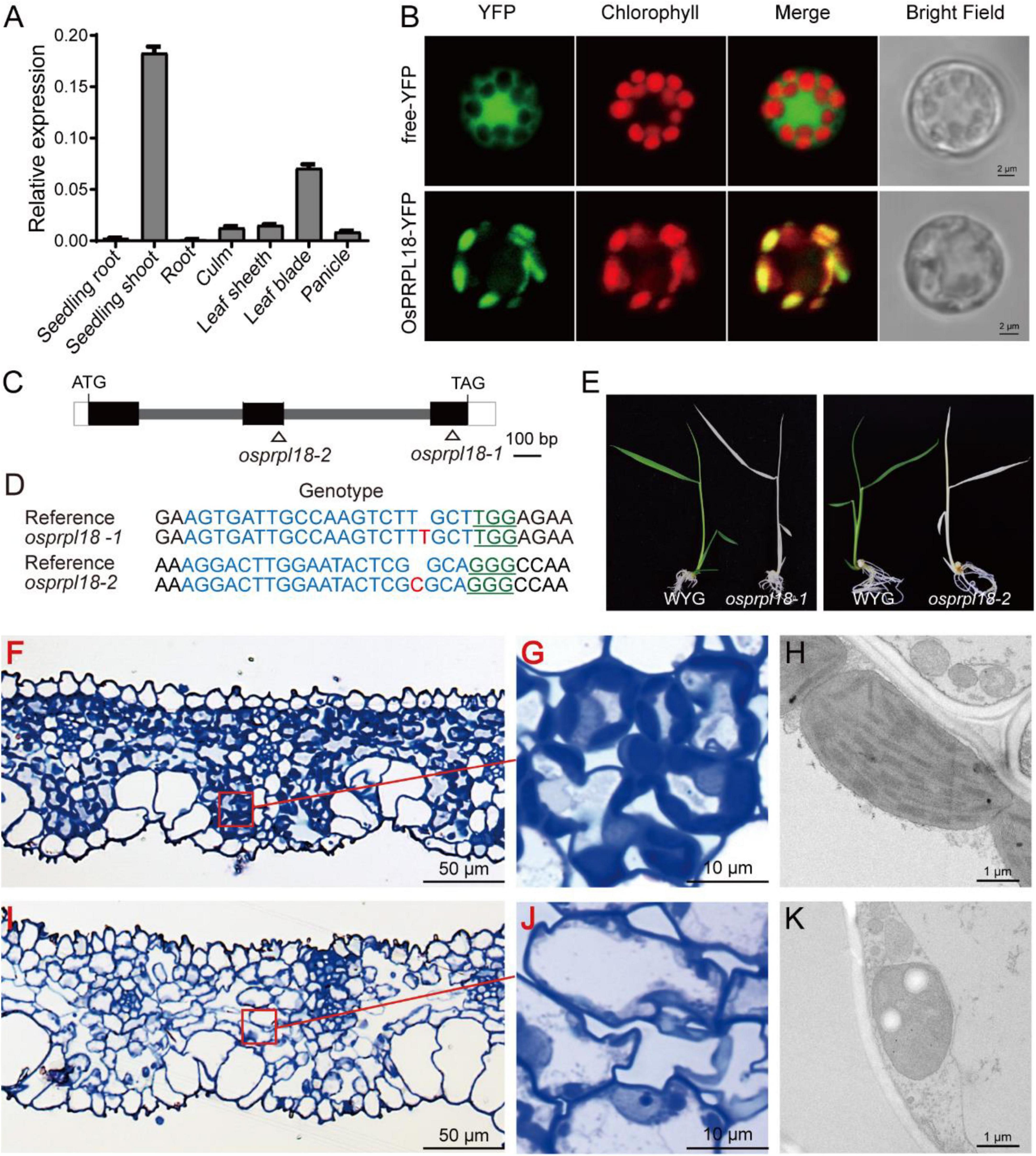
Figure 5. Characterization of the function of OsPRPL18 in rice. (A) The gene expression of OsPRPL18 in various rice tissues. UBQ served as the internal control. Data represent means ± SD from three biological replicates. (B) Subcellular location of OsPRPL18-YFP in rice protoplast. Free YFP was used as a control. The chlorophyll autofluorescence indicates chloroplast. Scale bars = 5 μm (C) The gene structure of OsPRPL18. (D) The genotypes of osprpl18 mutants. The guide RNA targeted DNA sequence and PAM motif were highlighted in blue and green, respectively. The targeted site was marked as triangle in (C). (E) 11-day-old seedlings of wild-type rice (WYG) and osprpl18-1 and osprpl18-2 mutants. (F–K) The structure of leaf blade and chloroplast of WYG (F–G) and the osprpl18-1 mutant (I–K).
To test the function of OsPRPL18 in rice, CRISPR/Cas9-based gene editing constructs targeting the second or the third exon of OsPRPL18 were introduced into wild-type rice (WYG) by Agrobacteria-mediated rice transformation. Two independent homozygous mutants were identified, in which the OsPRPL18 gene exhibited frame shift mutation (Figures 5C,D). However, these homozygous mutants displayed albino seedling phenotypes and died at the three-leaf stage (Figure 5E).
The albino leaf phenotype was usually caused by an impairment of chloroplast (Puthur et al., 2013). To examine the chloroplast development in osprpl18-1 plants, the albino leaves were cross-sectioned and analyzed using light microscope and TEM, respectively. In the semi-thin sections of wild-type leaves, chloroplasts were stained in dark blue by toluidine blue (Figures 5F,G), whereas no typical chloroplast-like structure was observed in osprpl18-1 plants under light microscope (Figures 5I,J). Interestingly, chloroplast-like structures were clearly visible in the osprpl18-1 mutant under TEM (Figure 5K). However, these chloroplast-like structures did not contain any thylakoids, compared with the well-developed chloroplasts with stacked grana and thylakoid membranes in wide-type plants (Figures 5H,K). These results suggested that OsPRPL18 is required for chloroplast development in rice.
Ribosomal proteins are essential for maintaining rRNA stability and ribosome assembly (Saez-Vasquez and Delseny, 2019). The ribosomal protein L18 directly interacted with 5S rRNA and 23S rRNA, thus incorporating the 5S rRNA into the 50S ribosomal subunit (Newberry and Garrett, 1980). There are four kinds of rRNA in plastid, including 4.5S, 5S, 16S, and 23S rRNA (Stern et al., 2010). To assess the effects of OsPRPL18 in plastid ribosome biogenesis, rRNA was examined by an Agilent 2,100 bioanalyzer. It was clearly to identify the peak of cytosolic 18S rRNA and 25S rRNA, and the peak of plastid 16S rRNA in the total RNA sample of wild-type (WYG) (Figure 6A), whereas the amount of plastid 16S rRNA was barely detectable in the osprpl18-1 mutant (Figure 6B). qRT-PCR analysis showed that the transcripts of plastid rRNAs were dramatically reduced in the osprpl18-1 mutant (Figure 6C). rbcL (large subunit of the ribulose-bisphosphate carboxylase), the most abundant plant protein, is encoded by a plastid gene and synthesized by chloroplast translation apparatus (Vitlin Gruber and Feiz, 2018). The plastid-encoded protein psaA comprises the reaction center for photosystem I along with psaB (Kapoor et al., 1994). Western blotting analyses revealed that the synthesis of rbcL and psaA were almost aborted in the osprpl18-1 mutant (Figure 6D). Collectively, these data suggested that OsPRPL18 is indispensable for plastid ribosome biogenesis.
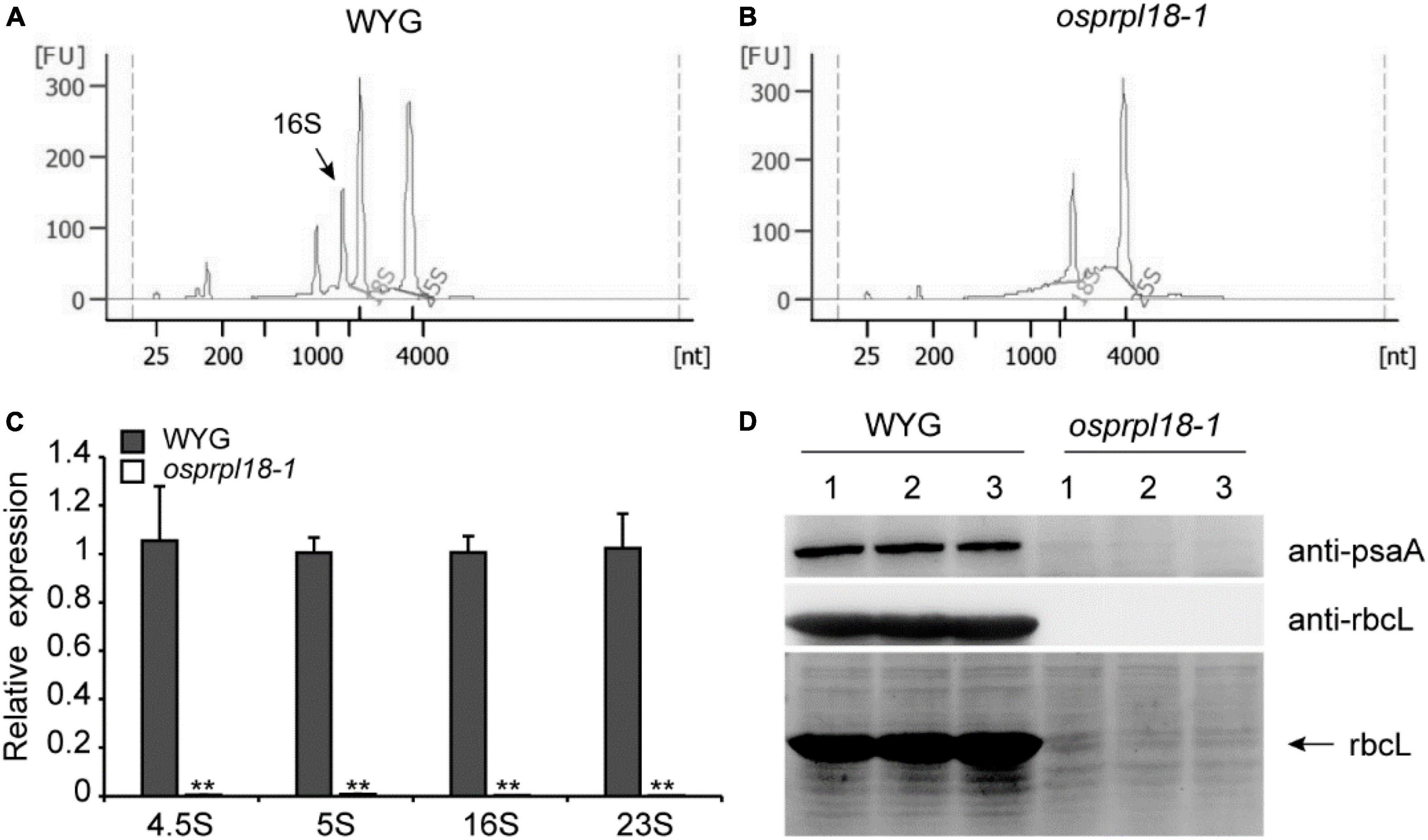
Figure 6. Comparison of the expression level of plastid rRNA and plastid-encoded protein rbcL in the seedling shoots of WYG and osprpl18-1 mutant. rRNA analysis of WYG (A) and osprpl18-1 mutant (B) with an Agilent 2,100 bioanalyzer. (C) qRT-PCR analysis of the transcripts level of plastid rRNA. UBQ served as the internal control. Gene expression values are presented relative to WYG levels (set as 1). Data represent means ± SD from three biological replicates (**P < 0.01; Student’s t-test). (D) Western blot analyses of the expression of rbcL and psaA. Immunoblotting was performed with specific antibodies, anti-rbcL and anti-psaA (Beijing Protein Innovation Co., Ltd, AbP80037-A-SE, AbP80033-A-SE), respectively (upper panels). The coomassie blue stained gel was used as a loading control (lower panel). For each genotype, three independent biological samples were analyzed.
Plant plastid genome harbors two main types of introns, group I and group II, defined by their different splicing mechanisms and conserved structural elements (Stern et al., 2010). In land plants, there are 18 introns distributed in 10 protein-encoding genes and 6 tRNA genes in plastid genome (Hess et al., 1994), and only rps12 contains a trans-splicing intron (Koller et al., 1987). Recent study revealed that two homologs of AtPRPL18, μL18-L1 and μL18-L8, were necessary for the splicing of certain mitochondrial and plastid group II introns, respectively (Wang et al., 2020). μL18-L8 was required for the trans splicing of the first intron of rps12 in Arabidopsis plastid. To test whether OsPRPL18 is involved in the intron splicing in plastid, intron splicing efficiency of the representative genes were examined by RT-PCR and qRT-PCR. Surprisingly, the trans-splicing efficiency of the first intron of rps12 was also remarkably reduced in the osprpl18-1 mutant (Figures 7A,B). Furthermore, all of the other tested genes also showed obvious reductions of intron splicing efficiency in the osprpl18-1 mutant compared with that in wild-type (WYG) (Figure 7). These results indicated that OsPRPL18 is required for intron splicing in rice plastid.
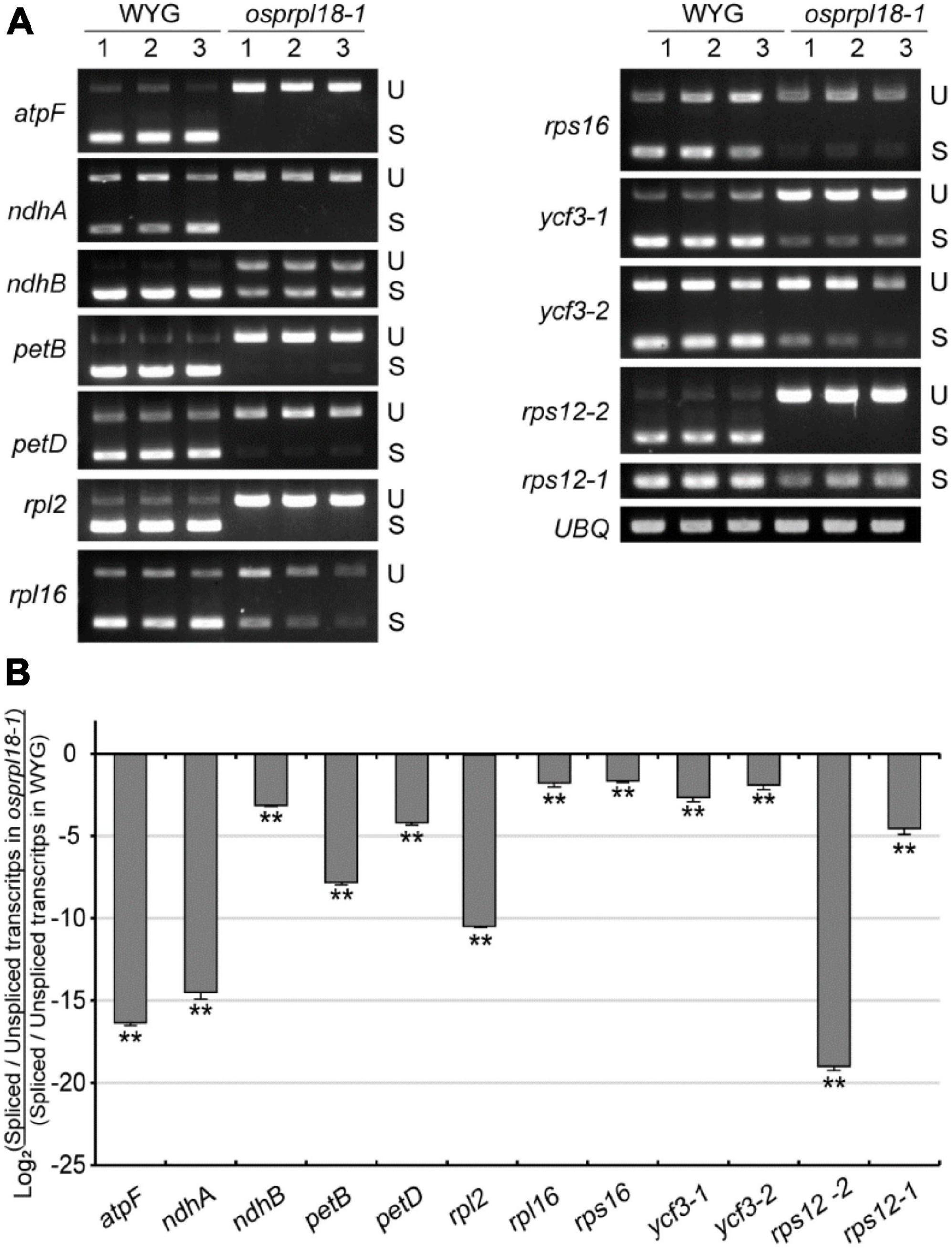
Figure 7. Comparison of the efficiency of intron splicing in the plastids of WYG and osprpl18-1 mutant. (A) Spliced (S) and unspliced (U) transcripts examined by RT-PCR. (B) Relative quantitative analyses of plastid intron splicing efficiency of WYG and osprpl18-1 mutant by qRT-PCR. Data represent the mean log2 ratios of splicing efficiencies in osprpl18-1 mutant to the wild type (WYG). Three independent biological replicates were used for each genotype. The asterisks indicate significant difference between WYG and osprpl18-1 mutant (**P < 0.01; Student’s t-test).
Plastid is a semiautonomous organelle. Lots of essential plastid proteins are translated by plastid ribosomes. PRPs are fundamental components of plastid ribosome, which play diverse roles in plant viability and development (Tiller and Bock, 2014). In this study, we report the different functions of PRPL18s in Arabidopsis and rice. Both of AtPRPL18 and OsPRPL18 were essential for plant development. However, knockout of AtPRPL18 retarded the transition from globular stage to heart stage in Arabidopsis, while knockout of OsPRPL18 did not affect embryo development, but resulted in an albino lethal phenotype at seedling stage in rice.
In bacteria, ribosome is consisted of a 50S large subunit and a 30S small subunit. The 50S subunit contains 23S rRNA, 5S rRNA and 34 ribosomal proteins. The 5S rRNA collaborates with ribosomal large subunit protein L5, L18 and L25 to form an essential autonomous structural domain of 50S subunit. L5 and L18 were very important for the incorporation of 5S rRNA into 50S ribosomal subunit (Newberry and Garrett, 1980). The C-terminal part of L18 functions in binding with 5S rRNA, and its N-terminal region is involved in the interaction of 5S rRNA with 23S rRNA (Tiller and Bock, 2014). Disruption of the coding genes of L5, L18 and L25 revealed that L5 and L18 but not L25 were essential for E. coli viability (Korepanov et al., 2007). Plants have two semiautonomous organelles, chloroplast and mitochondria, which are proposed to be evolved from their prokaryotic ancestors. The structure and constitution of ribosomes of chloroplast and mitochondria are similar to that in bacteria (Zoschke and Bock, 2018). Protein sequence-based search for L18 homologues in Arabidopsis found two close homologs, AtPRPL18/μL18c and μL18m (Wang et al., 2020). μL18m was localized in mitochondria and appeared to be a mitochondrial ribosomal protein. AtPRPL18 was localized in chloroplast and was proposed to be a plastid ribosomal protein (Wang et al., 2020). Phylogenetic analysis showed that the plastid ribosomal protein L18s were very conserved in plant kingdom (Figure 1), indicating they may have a similar molecular function. Gene expression analyses showed both of OsPRPL18 and AtPRPL18 were highly expressed in green tissues, and their encoding proteins were localized in chloroplast. In the heterozygous mutants atprpl18/+ and ko-1/+ of AtPRPL18, about a quarter of albino developing seeds were observed in silique (Figures 3F,G). Cytological analysis revealed the embryos were arrested at globular stage in these albino seeds (Figure 4). The albino seeds were then shrunken and aborted. Genetic and complementation analyses showed the albino seed phenotype was caused by the mutation of AtPRPL18, suggesting AtPRPL18 was essential for Arabidopsis seed development. Moreover, knockout of OsPRPL18 led to an albino lethality of rice seedlings (Figure 5E). Cytological analysis found that the chloroplast development was impaired in the leaves of osprpl18-1 mutants (Figure 5K). Further studies uncovered the indispensable role of OsPRPL18 in plastid ribosome biogenesis (Figure 6). Overall, consistent with the function of L18 in E. coli, PRPL18s are essential for plant viability.
Because of the conserved evolutionary relationship of the ribosomes in bacteria and plastid, lots of bacterial ribosomal proteins and its plant homologs were proved to be essential for bacteria and plant viability respectively, such as S2, S3, S4, S5, S9, S13, S14, S16, S18, L3, L4, L6, L10, L13, L18, L20, L22, L23, and L35 and their plant homologs (Ahlert et al., 2003; Rogalski et al., 2006; Baba et al., 2008; Bryant et al., 2011; Fleischmann et al., 2011; Shoji et al., 2011; Romani et al., 2012). However, some other ribosomal proteins are in a different case. For example, S9, S13, S20, L1, L21, L27, L28, and L31 were shown to be dispensable in bacteria, but their homologs were essential for plant viability (Dabbs, 1991; Bubunenko et al., 2007; Baba et al., 2008; Bryant et al., 2011; Shoji et al., 2011; Romani et al., 2012; Yin et al., 2012). On the contrary, S1 was proved to be essential in bacteria, but its homolog was reported to be non-essential for plant viability (Baba et al., 2008; Bryant et al., 2011; Romani et al., 2012). Interestingly, even in plants, some PRPs and their homologs also appeared to have diverse functions. In the siliques of the Arabidopsis heterozygous mutant atprps20/+, the albino/aborted seeds accounted for about one quarter of the total seeds, of which the embryos were arrested at globular stage (Romani et al., 2012). However, the null mutant of OsPRPS20 was defective in chloroplast development and exhibited an albino lethal phenotype at seedling stage (Gong et al., 2013). Similar to the case of PRPS20, albino/aborted seeds were also observed in the atprps9/+, atprpl13/+, and atprpl21/+ plants in Arabidopsis (Bryant et al., 2011; Yin et al., 2012), while an albino lethal phenotype was observed in the seedlings of null mutants osprps9, osprpl13 and osprpl21 in rice (Song et al., 2014; Lin et al., 2015; Qiu et al., 2018). In this study, we revealed that knockouts of PRPL18s in Arabidopsis and rice resulted in different phenotypical consequences. AtPRPL18 was essential for the transition of global stage to heart stage during embryo development, while OsPRPL18 did not affect embryogenesis. However, the osprp18 mutant was impaired in chloroplast developing, and displayed an albino lethal phenotype at seedling stage. In Arabidopsis, plastids are few in zygote, but gradually increase in number during embryo development. At the globular stage, plastids differentiate into chloroplasts with chlorophyll accumulation, and thereby embryos start greening (Tejos et al., 2010). It was proposed that chloroplasts are vital in supplying nutrition to the developing embryo, therefore deficiency of chloroplast may disrupt embryogenesis in Arabidopsis (Tiller and Bock, 2014). In contrast, embryo development is largely dependent on the nutrients supplied from endosperm in rice (An et al., 2020). Thus, Arabidopsis may be more susceptible to the loss of PRPL18 than rice during embryogenesis. Intriguingly, although OsPRPS9 and its maize homolog ZmPRPS9 share 82% identity, OsPRPS9 was proved to be a plastid protein, while ZmPRPS9 was shown to be localized on both chloroplast and nucleus (Ma and Dooner, 2004; Qiu et al., 2018). Moreover, OsPRPS9 was dispensable for rice embryogenesis, while the zmprps9 mutant exhibited an early embryo lethal phenotype (Ma and Dooner, 2004; Qiu et al., 2018). So, another possibility is that PRPL18 might have evolved species-specific functions. To address these hypothesis, future works on the molecular functions of PRPs and heterological complementary works are required.
The intron splicing efficiency in plastid is largely dependent on the plastid-encoded components. The intron splicing of rps2 and rps12 were impaired in the barley mutants with ribosome-deficient plastids (Hess et al., 1994; Hubschmann et al., 1996). In land plants, the intron in the trnK gene encodes a conserved maturase called MatK (Maturase K) (Zoschke et al., 2010). MatK directly interacts with the introns of trnA, trnI, trnV, trnK, atpF, rpl2 and the second intron of rps12 (Zoschke et al., 2010). Application of heterologously expressed MatK protein increased the intron self-splicing efficiency of the second intron of rps12 in vitro (Barthet et al., 2020). Thus, MatK was proposed to be a plastid-encoded splicing factor. However, addition of MatK protein did not increased the intron self-splicing efficiency of rpl2 (Barthet et al., 2020), and a functioning translational apparatus was not prerequisite for the intron splicing of rps16, rpl16, ndhB, petD, and trnL (Hess et al., 1994), suggesting the presence of other factors in the regulation of plastid intron splicing. Ribosomal proteins maybe participate in intron splicing. Human ribosomal protein S13 was reported to inhibit excision of RPS13 intron 1 in vitro (Malygin et al., 2007). Ribosomal protein L10a regulated its own alternative pre-mRNA splicing by directly and specifically binds to an evolutionarily conserved stretch between the two alternative 5’ splice sites in pre-mRNA to switch the splice site choice (Takei et al., 2016). A recent study reported that two homologs of PRPL18, μL18-L1, and μL18-L8, play essential roles in the splicing of the fourth intron of nad5 pre-mRNAs in mitochondria, and the removal of rps12 intron 1 in Arabidopsis plastid, respectively (Wang et al., 2020). μL18-L1 and μL18-L8 specifically associate with nad5 intron 4 and rps12 intron 1, respectively, indicating direct roles in the splicing of their targeted introns (Wang et al., 2020). Nevertheless, the function of PRPL18s in intron splicing remains elusive. In this report, we found that loss-function of OsPRPL18 resulted in a serious reduction of the intron splicing efficiency of all of the tested plastid introns, As the biogenesis of chloroplast ribosome was defective in osprpl18-1, OsPRPL18 may affect intron splicing by maintaining biosynthesis of the regulatory proteins such as MatK and so on. Alternatively, OsPRPL18 may participate in the process of plastid intron splicing directly. Employing the in vitro activity assay to test the role of OsPRPL18 in plastid intron excision would be helpful to uncover the direct function of OsPRPL18 in plastid intron splicing (Barthet et al., 2020).
In summary, our results provide the evidence that PRPL18s are essential for plant viability. However, AtPRPL18 is required for embryogenesis in Arabidopsis, while OsPRPL18 is dispensable for embryogenesis but required for seedling development in rice, although the underlying mechanisms need to be further clarified. Moreover, the surprising finding of the prerequisite role of OsPRPL18 in plastid intron splicing highlights the functions of ribosomal proteins in other biological process.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
JW, XT, and SC designed the experiments and wrote the manuscript. SC, XZ, JW, YQL, SQ, XP, XX, YJL, and CL performed the experiments. SC and JW analyzed the data. All authors read and approved the manuscript for publication.
This work was supported by the grants from the Laboratory of Lingnan Modern Agriculture Project (NZ2021003), the Natural Science Foundation of Guangdong Province (2021A1515010507, 2022A1515012353, and 2019A1515110671), and the National Natural Science Foundation of China (U1901203).
We thank ABRC (http://abrc.osu.edu), Qi Xie (University of Chinese Academy of Sciences), and Yaoguang Liu (South China Agricultural University) for providing the mutant SAIL_415_H08, YAO-based CRISPR-Cas9 vector, and rice CRISPR-Cas9 vector, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.949897/full#supplementary-material
Supplementary Figure 1. Subcellular localizations of AtPRPL18-GFP and OsPRPL18-GFP in N. benthamiana leaf cells.
Supplementary Figure 2. Identification of the T-DNA free ko-1/+ mutant plants.
Supplementary Figure 3. Analyses of the transcript levels of AtPRPL18 in OE-1 and OE-2.
Ahlert, D., Ruf, S., and Bock, R. (2003). Plastid protein synthesis is required for plant development in tobacco. Proc. Natl. Acad. Sci. U.S.A. 100, 15730–15735. doi: 10.1073/pnas.2533668100
Almagro Armenteros, J. J., Salvatore, M., Emanuelsson, O., Winther, O., Von Heijne, G., Elofsson, A., et al. (2019). Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2:e201900429. doi: 10.26508/lsa.201900429
An, L., Tao, Y., Chen, H., He, M., Xiao, F., Li, G., et al. (2020). Embryo-Endosperm interaction and its agronomic relevance to rice quality. Front. Plant Sci. 11:587641. doi: 10.3389/fpls.2020.587641
Baba, T., Huan, H. C., Datsenko, K., Wanner, B. L., and Mori, H. (2008). The applications of systematic in-frame, single-gene knockout mutant collection of Escherichia coli K-12. Methods Mol. Biol. 416, 183–194. doi: 10.1007/978-1-59745-321-9_12
Barthet, M. M., Pierpont, C. L., and Tavernier, E. K. (2020). Unraveling the role of the enigmatic MatK maturase in chloroplast group IIA intron excision. Plant Direct 4:e00208. doi: 10.1002/pld3.208
Brosius, J., Schiltz, E., and Chen, R. (1975). The primary structure of the 5S RNA binding protein L18 from Escherichia coli ribosomes. FEBS Lett. 56, 359–361. doi: 10.1016/0014-5793(75)81127-6
Bryant, N., Lloyd, J., Sweeney, C., Myouga, F., and Meinke, D. (2011). Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 155, 1678–1689. doi: 10.1104/pp.110.168120
Bubunenko, M., Baker, T., and Court, D. L. (2007). Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 189, 2844–2853. doi: 10.1128/JB.01713-06
Chang, Z., Chen, Z., Wang, N., Xie, G., Lu, J., Yan, W., et al. (2016). Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. U.S.A. 113, 14145–14150. doi: 10.1073/pnas.1613792113
Chen, H., Zou, W., and Zhao, J. (2015). Ribonuclease J is required for chloroplast and embryo development in Arabidopsis. J. Exp. Bot. 66, 2079–2091. doi: 10.1093/jxb/erv010
Chen, S., Tao, L., Zeng, L., Vega-Sanchez, M. E., Umemura, K., and Wang, G. L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7, 417–427. doi: 10.1111/j.1364-3703.2006.00346.x
Dabbs, E. R. (1991). Mutants lacking individual ribosomal proteins as a tool to investigate ribosomal properties. Biochimie 73, 639–645. doi: 10.1016/0300-9084(91)90043-z
Fleischmann, T. T., Scharff, L. B., Alkatib, S., Hasdorf, S., Schottler, M. A., and Bock, R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23, 3137–3155. doi: 10.1105/tpc.111.088906
Gong, X., Jiang, Q., Xu, J., Zhang, J., Teng, S., Lin, D., et al. (2013). Disruption of the rice plastid ribosomal protein s20 leads to chloroplast developmental defects and seedling lethality. G3 3, 1769–1777. doi: 10.1534/g3.113.007856
Hess, W. R., Hoch, B., Zeltz, P., Hubschmann, T., Kossel, H., and Borner, T. (1994). Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell 6, 1455–1465. doi: 10.1105/tpc.6.10.1455
Hubschmann, T., Hess, W. R., and Borner, T. (1996). Impaired splicing of the rps12 transcript in ribosome-deficient plastids. Plant Mol. Biol. 30, 109–123. doi: 10.1007/BF00017806
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jensen, P. E., and Leister, D. (2014). Chloroplast evolution, structure and functions. F1000Prime Rep. 6:40. doi: 10.12703/P6-40
Kapoor, S., Maheshwari, S. C., and Tyagi, A. K. (1994). Developmental and light-dependent cues interact to establish steady-state levels of transcripts for photosynthesis-related genes (psbA, psbD, psaA and rbcL) in rice (Oryza sativa L.). Curr. Genet. 25, 362–366. doi: 10.1007/BF00351491
Koller, B., Fromm, H., Galun, E., and Edelman, M. (1987). Evidence for in vivo trans splicing of pre-mRNAs in tobacco chloroplasts. Cell 48, 111–119. doi: 10.1016/0092-8674(87)90361-8
Korepanov, A. P., Gongadze, G. M., Garber, M. B., Court, D. L., and Bubunenko, M. G. (2007). Importance of the 5 S rRNA-binding ribosomal proteins for cell viability and translation in Escherichia coli. J. Mol. Biol. 366, 1199–1208. doi: 10.1016/j.jmb.2006.11.097
Lin, D., Jiang, Q., Zheng, K., Chen, S., Zhou, H., Gong, X., et al. (2015). Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol. 17, 599–607. doi: 10.1111/plb.12271
Lv, J., Shang, L., Chen, Y., Han, Y., Yang, X., Xie, S., et al. (2020). OsSLC1 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling survival. Rice 13:25. doi: 10.1186/s12284-020-00385-5
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Ma, Z., and Dooner, H. K. (2004). A mutation in the nuclear-encoded plastid ribosomal protein S9 leads to early embryo lethality in maize. Plant J. 37, 92–103. doi: 10.1046/j.1365-313x.2003.01942.x
Malygin, A. A., Parakhnevitch, N. M., Ivanov, A. V., Eperon, I. C., and Karpova, G. G. (2007). Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 35, 6414–6423. doi: 10.1093/nar/gkm701
Morita-Yamamuro, C., Tsutsui, T., Tanaka, A., and Yamaguchi, J. (2004). Knock-out of the plastid ribosomal protein S21 causes impaired photosynthesis and sugar-response during germination and seedling development in Arabidopsis thaliana. Plant Cell Physiol. 45, 781–788. doi: 10.1093/pcp/pch093
Newberry, V., and Garrett, R. A. (1980). The role of the basic N-terminal region of protein L18 in 5S RNA-23S RNA complex formation. Nucleic Acids Res. 8, 4131–4142. doi: 10.1093/nar/8.18.4131
Pesaresi, P., Varotto, C., Meurer, J., Jahns, P., Salamini, F., and Leister, D. (2001). Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J. 27, 179–189. doi: 10.1046/j.1365-313x.2001.01076.x
Puthur, J. T., Shackira, A. M., Saradhi, P. P., and Bartels, D. (2013). Chloroembryos: a unique photosynthesis system. J. Plant Physiol. 170, 1131–1138. doi: 10.1016/j.jplph.2013.04.011
Qiu, Z., Chen, D., He, L., Zhang, S., Yang, Z., Zhang, Y., et al. (2018). The rice white green leaf 2 gene causes defects in chloroplast development and affects the plastid ribosomal protein S9. Rice 11:39. doi: 10.1186/s12284-018-0233-2
Radoeva, T., Vaddepalli, P., Zhang, Z., and Weijers, D. (2019). Evolution, initiation, and diversity in early plant embryogenesis. Dev. Cell 50, 533–543. doi: 10.1016/j.devcel.2019.07.011
Rogalski, M., Ruf, S., and Bock, R. (2006). Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34, 4537–4545. doi: 10.1093/nar/gkl634
Romani, I., Tadini, L., Rossi, F., Masiero, S., Pribil, M., Jahns, P., et al. (2012). Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J. 72, 922–934. doi: 10.1111/tpj.12000
Saez-Vasquez, J., and Delseny, M. (2019). Ribosome biogenesis in plants: from functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell 31, 1945–1967. doi: 10.1105/tpc.18.00874
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shoji, S., Dambacher, C. M., Shajani, Z., Williamson, J. R., and Schultz, P. G. (2011). Systematic chromosomal deletion of bacterial ribosomal protein genes. J. Mol. Biol. 413, 751–761. doi: 10.1016/j.jmb.2011.09.004
Song, J., Wei, X., Shao, G., Sheng, Z., Chen, D., Liu, C., et al. (2014). The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol. Biol. 84, 301–314. doi: 10.1007/s11103-013-0134-0
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Stern, D. B., Goldschmidt-Clermont, M., and Hanson, M. R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61, 125–155. doi: 10.1146/annurev-arplant-042809-112242
Takei, S., Togo-Ohno, M., Suzuki, Y., and Kuroyanagi, H. (2016). Evolutionarily conserved autoregulation of alternative pre-mRNA splicing by ribosomal protein L10a. Nucleic Acids Res. 44, 5585–5596. doi: 10.1093/nar/gkw152
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tejos, R. I., Mercado, A. V., and Meisel, L. A. (2010). Analysis of chlorophyll fluorescence reveals stage specific patterns of chloroplast-containing cells during Arabidopsis embryogenesis. Biol. Res. 43, 99–111.
Tiller, N., and Bock, R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7, 1105–1120. doi: 10.1093/mp/ssu022
Tiller, N., Weingartner, M., Thiele, W., Maximova, E., Schöttler, M. A., and Bock, R. (2012). The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J. 69, 302–316. doi: 10.1111/j.1365-313X.2011.04791.x
Toki, S., Hara, N., Ono, K., Onodera, H., Tagiri, A., Oka, S., et al. (2006). Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47, 969–976. doi: 10.1111/j.1365-313X.2006.02836.x
Tzafrir, I., Pena-Muralla, R., Dickerman, A., Berg, M., Rogers, R., Hutchens, S., et al. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220. doi: 10.1104/pp.104.045179
Tzfira, T., Tian, G.-W., Lacroix, B. T., Vyas, S., Li, J., Leitner-Dagan, Y., et al. (2005). PSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57:503. doi: 10.1007/s11103-005-0340-5
Vitlin Gruber, A., and Feiz, L. (2018). Rubisco assembly in the chloroplast. Front. Mol. Biosci. 5:24. doi: 10.3389/fmolb.2018.00024
Waltz, F., Nguyen, T. T., Arrive, M., Bochler, A., Chicher, J., Hammann, P., et al. (2019). Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 5, 106–117. doi: 10.1038/s41477-018-0339-y
Wang, C., Fourdin, R., Quadrado, M., Dargel-Graffin, C., Tolleter, D., Macherel, D., et al. (2020). Rerouting of ribosomal proteins into splicing in plant organelles. Proc. Natl. Acad. Sci. U.S.A. 117, 29979–29987. doi: 10.1073/pnas.2004075117
Wu, J. X., Wu, J. L., Yin, J., Zheng, P., and Yao, N. (2015). Ethylene modulates sphingolipid synthesis in Arabidopsis. Front. Plant Sci. 6:1122. doi: 10.3389/fpls.2015.01122
Yan, L., Wei, S., Wu, Y., Hu, R., Li, H., Yang, W., et al. (2015). High-efficiency genome editing in Arabidopsis using YAO Promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823. doi: 10.1016/j.molp.2015.10.004
Yin, T., Pan, G., Liu, H., Wu, J., Li, Y., Zhao, Z., et al. (2012). The chloroplast ribosomal protein L21 gene is essential for plastid development and embryogenesis in Arabidopsis. Planta 235, 907–921. doi: 10.1007/s00425-011-1547-0
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, J., Yuan, H., Yang, Y., Fish, T., Lyi, S. M., Thannhauser, T. W., et al. (2016). Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 67, 2731–2744. doi: 10.1093/jxb/erw106
Zhang, X. R., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhao, D. S., Zhang, C. Q., Li, Q. F., Yang, Q. Q., Gu, M. H., and Liu, Q. Q. (2016). A residue substitution in the plastid ribosomal protein L12/AL1 produces defective plastid ribosome and causes early seedling lethality in rice. Plant Mol. Biol. 91, 161–177. doi: 10.1007/s11103-016-0453-z
Zhou, K., Zhang, C., Xia, J., Yun, P., Wang, Y., Ma, T., et al. (2021). Albino seedling lethality 4; Chloroplast 30S ribosomal protein S1 is required for chloroplast ribosome biogenesis and early chloroplast development in rice. Rice 14:47. doi: 10.1186/s12284-021-00491-y
Zoschke, R., and Bock, R. (2018). Chloroplast translation: structural and functional organization, operational control, and regulation. Plant Cell 30, 745–770. doi: 10.1105/tpc.18.00016
Keywords: plastid ribosomal protein, chloroplast, albino seedling, Arabidopsis, rice, intron splicing, embryo development
Citation: Chen S, Zeng X, Li Y, Qiu S, Peng X, Xie X, Liu Y, Liao C, Tang X and Wu J (2022) The nuclear-encoded plastid ribosomal protein L18s are essential for plant development. Front. Plant Sci. 13:949897. doi: 10.3389/fpls.2022.949897
Received: 21 May 2022; Accepted: 29 August 2022;
Published: 23 September 2022.
Edited by:
Munetaka Sugiyama, The University of Tokyo, JapanReviewed by:
Miyuki T. Nakata, Nara Institute of Science and Technology (NAIST), JapanCopyright © 2022 Chen, Zeng, Li, Qiu, Peng, Xie, Liu, Liao, Tang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyan Tang, dHh5QGZyb250aWVyLWFnLmNvbQ==; Jianxin Wu, d2p4aW5AbS5zY251LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.