
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 September 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.946037
This article is part of the Research Topic Crop Population Genomics and Pan-omics for Gene Discovery and Breeding View all 6 articles
 Shuangxing Zhang1
Shuangxing Zhang1 Yang Yu1
Yang Yu1 Tianqi Song1
Tianqi Song1 Mingfei Zhang2
Mingfei Zhang2 Nan Li2
Nan Li2 Ming Yu1
Ming Yu1 Hongwei Zhou1
Hongwei Zhou1 Yanning Yang1
Yanning Yang1 Sihai Guo1
Sihai Guo1 Chunhong Xu1
Chunhong Xu1 Yongle Tu1
Yongle Tu1 Jishan Xiang2*
Jishan Xiang2* Xiaoke Zhang1*
Xiaoke Zhang1*Thioredoxins (TRXs) are small-molecule proteins with redox activity that play very important roles in the growth, development, and stress resistance of plants. Foxtail millet (Setaria italica) gradually became a model crop for stress resistance research because of its advantages such as its resistance to sterility and its small genome. To date, the thioredoxin (TRX) family has been identified in Arabidopsis thaliana, rice and wheat. However, studies of the TRX family in foxtail millet have not been reported, and the biological function of this family remains unclear. In this study, 35 SiTRX genes were identified in the whole genome of foxtail millet through bioinformatic analysis. According to phylogenetic analysis, 35 SiTRXs can be divided into 13 types. The chromosome distribution, gene structure, cis-elements and conserved protein motifs of 35 SiTRXs were characterized. Three nucleoredoxin (NRX) members were further identified by a structural analysis of TRX family members. The expression patterns of foxtail millet’s SiNRX members under abiotic stresses showed that they have different stress-response patterns. In addition, subcellular localization revealed that SiNRXs were localized to the nucleus, cytoplasm and membrane. Further studies demonstrated that the overexpression of SiNRX1 enhanced Arabidopsis’ tolerance to drought and salt stresses, resulting in a higher survival rate and better growth performance. Moreover, the expression levels of several known stress-related genes were generally higher in overexpressed lines than in the wild-type. Thus, this study provides a general picture of the TRX family in foxtail millet and lay a foundation for further research on the mechanism of the action of TRX proteins on abiotic stresses.
Drought, salt and temperature stresses are the main environmental factors that affect the geographical distribution of natural plants, limit the development of agriculture and endanger food security. In recent years, the frequent occurrence of extreme weather has intensified the adverse effects of abiotic stresses (Zhu, 2016). In the process of coping with external environmental pressure, plants produce various reactive oxygen species (ROS) as signaling molecules, but the excessive accumulation of ROS leads to the oxidative damage of key macromolecules in cells (Zandalinas et al., 2020). Dynamic redox balance is the basis of cell metabolism. Studies have shown that thioredoxin (TRX) can eliminate excess ROS by interacting with antioxidant enzymes to stabilize the redox equilibrium of the intracellular environment (Vieira Dos Santos and Rey, 2006; Rouhier et al., 2015).
Thioredoxins (TRXs) are the primary protein disulfide reductases found in a variety of biological cells, and they are essential for maintaining the dynamic protein redox equilibrium (Dal Piaz et al., 2010). The TRX protein was first discovered in Escherichia coli and has been quite conserved through evolution (Laurent et al., 1964). Its structural characteristics include a disulfide active center (CXXC). The first cysteine (Cys) can attack an oxidized mercaptan in the substrate target, leading to the formation of covalent mixed disulfide bonds between TRX and the substrate. Then, the second Cys can decompose the mixed disulfide and reduce the substrate, while the cysteine in the TRX’s active center forms a disulfide bond. Thereby, TRX is involved in many redox reactions through the reversible transformation of mercaptan–disulfide, thus enabling TRX to perform various biological functions in cells (Dal Piaz et al., 2010). Different from those in other species, plant TRXs are encoded by a large gene family, and the types of TRX are abundant in plants and located in different organelles, such as chloroplasts, mitochondria, peroxisomes, nuclei, plasma membranes and extracellular matrices, indicating that TRX family members have complex biological functions (Meyer et al., 2005; Rouhier et al., 2015). TRXs are divided into two types based on their active sites: typical TRXs (WCG/PPC active site) and atypical TRXs (XCXXC active site) (Chibani et al., 2021). The typical TRXs mainly include f, h, m, o, x, y, and z types, while the atypical TRXs mainly include CDSP32, NTRC, ACHT, APR, NRX (Nucleoredoxin), TRX-like, etc. (Michelet et al., 2013; Chibani et al., 2021). To date, the TRX family has been identified in Arabidopsis thaliana, rice (Oryza sativa) and wheat (Triticum aestivum L.) (Nuruzzaman et al., 2008; Nuruzzaman et al., 2012; Chibani et al., 2021; Bhurta et al., 2022). Numerous TRX types have been found to integrate gene expression and metabolic pathways, regulate various physiological processes (such as seed germination, seedling growth and flowering), and facilitate stress tolerance and defense responses to external stimuli in a wide number of studies (Apel and Hirt, 2004; Meyer et al., 2005; Richter et al., 2018; Nikkanen and Rintamäki, 2019; Jing et al., 2020; Nietzel et al., 2020; Sytykiewicz et al., 2020). These findings imply that TRXs play significant roles in plant growth and development as well as stress tolerance. However, the functions of chloroplasts and cytoplasmic TRXs in plants have been extensively studied, but the biochemical and biological functions of nuclear TRX proteins remain largely unknown.
Nucleoredoxins (NRX) are classified into the TRX family because they are densely localized in the nucleus, have two or three TRX-like domains and a C-terminal domain rich in cysteine, and have TRX-enzyme activity (Laughner et al., 1998; Marchal et al., 2014). Two NRX genes (AtNRX1 and AtNRX2) have been identified in Arabidopsis, which have dual localization in the nucleus and cytoplasm. Studies have demonstrated that both the AtNRX1 and AtNRX2 proteins can reduce disulfide bonds (Marchal et al., 2014). It was demonstrated that the redox activity of GbNRX1 (Gossypium barbadense) was higher than that of AtTRX h, and the redox activity of three distinct TRX domains was found to be different (Li et al., 2016). In Arabidopsis, AtNRX1 has been shown to regulate pollen tube growth and pollen fertility (Qin et al., 2009), and AtNRX2 plays a physiological role in jasmonic acid (JA)-mediated trichome formation in Arabidopsis (Lee et al., 2020). In poplar (Populus spp.), NRX1 participates in the salicylic acid (SA) signal transduction pathway (Xue et al., 2013). Studies have shown that the NRX1-mediated reduction of catalase (CAT) is an important mechanism for sustaining maximal CAT activity, and NRX1 may be involved in directly regulating the ability of cells to detoxify hydrogen peroxide (H2O2), thus protecting plant cells from oxidative stress induced by adverse environmental conditions (Kneeshaw et al., 2017). It was found that TaNRX1 was closely related to stress resistance in wheat (Zhang et al., 2014). The TaNRX1 promoter was then examined further, and it was discovered that a 36 bp fragment (-193~157 bp) was the critical sequence of the TaNRX1-D gene in the response to osmotic stress or ABA therapy (Cheng et al., 2021). Furthermore, it was discovered that the homologous overexpression of TaNRX1 in wheat positively regulated the drought resistance of wheat (Zhang et al., 2021). NRX has been proven to be an antioxidant protective mechanism that can regulate the status of reactive-oxygen-scavenging enzymes such as catalase and may also regulate the status of ascorbate peroxidase (APX), monodehydroascorbate reductase (MDHAR) and dehydroascorbate reductase (DHAR). These enzymes are potential targets for NRX proteins that regulate H2O2 metabolism through regulating target proteins to protect plant cells from oxidative stress (Kneeshaw et al., 2017; Calderón et al., 2018). NRX has significant impacts on plant growth, development and stress.
Foxtail millet (Setaria italica), a C4 crop of the Gramineae family, is widely cultivated in arid and semi-arid regions of Asia and Africa. It is known for its superior tolerance to drought, salinity and diseases, and has higher nutritional value than other major cereal crops due to its high protein and mineral content (Maharajan et al., 2021). Foxtail millet is better able to adapt to adverse agro-ecological conditions than rice, maize and wheat; requires minimal inputs; and is an indispensable plant genetic resource for agriculture and food security for poor farmers living on arid, non-cultivable and marginal lands (Neupane et al., 2022). The completion of the genome sequencing of foxtail millet in recent years has been useful for the study of the molecular biological stress resistance of foxtail millet. So far, plant TRXs have primarily focused on Arabidopsis, rice and wheat. A vast number of studies have indicated that TRX is vital for the maintenance of redox balance in plants, but few abiotic-stress-related studies of TRX in C4 foxtail millet crops have been reported. In this study, TRX family members in foxtail millet were retrieved, and their evolutionary relationships, physicochemical characteristics, subcellular localizations, gene structures, conserved motifs, promoter cis-elements and other parameters were predicted and analyzed. Following that, the abiotic expression patterns of SiNRXs under stress in the seedling stage were detected, and function analysis of SiNRX1 was performed. We conducted this study with the purpose of providing data to enable further research into the role of the TRX family in the signaling system for the abiotic stress response in foxtail millet and improving agricultural stress resistance through genetic engineering technologies.
The whole-genome sequence data for foxtail millet were downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/); then, the Hidden Markov Model (HMM) profile (ID: PF00085) with the TRX domain was downloaded from the Pfam database (http://pfam.xfam.org/). The HMMER software was used to search the genome data for foxtail millet for TRX proteins with an E-value cutoff of 1e-3. The candidate TRX protein sequences of foxtail millet were confirmed using the NCBI-CDD retrieval tool (https://www.ncbi.nlm.nih.gov/cdd) and SMART structure domain (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1#).
The chromosomal location information of SiTRXs were obtained from the Phytozome database, and the visual analysis of the SiTRX chromosomes’ location was performed using the online website of MapGene2Chrom web v2 (http://mg2c.iask.in/mg2c_v2.1/).
The amino acid number, molecular weight (MW) and isoelectric point (PI) of the SiTRX proteins were calculated using the online website ExPASy (https://web.expasy.org/protparam/). The subcellular locations of the TRX proteins were predicted using the CELLO online website (https://cello.life.nctu.edu.tw/).
To analyze the phylogenetic relationship of the TRXs, a phylogenetic tree was constructed using the TRX protein sequences for foxtail millet, rice (Nuruzzaman et al., 2008) and Arabidopsis (Meyer et al., 2005). The protein sequence information of the family members in rice and Arabidopsis was downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/). Multiple alignments of the TRX family from foxtail millet, rice and Arabidopsis were performed by using ClustalW with the default settings. A phylogenetic tree was constructed by using the method of maximum likelihood (ML) with 1000 bootstrap repetitions in the MEGA 7.0 software (https://www.megasoftware.net).
SiTRX gene sequence files (coding sequence and genome sequence) were retrieved from the Phytozome database. The gene structure was analyzed through the Gene Structure Display Server (GSDS 2.0, http://gsds.gao-lab.org/). MEME (https://meme-suite.org/meme/tools/meme) was used to analyze conserved motifs, and the maximum number parameter for a conserved base sequence was set to 10.
The genomic sequence (2000 bp upstream of the ATG initiation codon) of the each SiTRX member was obtained from the Phytozome database, and cis-regulatory elements were identified using the Plant CARE online website tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
The seeds of the foxtail millet variety Yugu 1 were provided by Research Fellow Jishan Xiang of Chifeng University, China, and were sown in pots and grown in a controlled incubator under controlled conditions (30°C/18°C temperature, 14 h light/10 h dark photoperiod, 60-70% humidity, 15000 lux light intensity). After reaching the three-leaf stage, the seedlings were transferred and precultured in half-strength Hoagland’s solution. When the growing seedlings reached the five-leaf stage, stress treatments were applied by transferring the seedlings to the same solution containing 20% PEG6000 and 100 mM NaCl. For cold treatment, the seedlings were transferred to a growth chamber set at 4°C. As a control, cultures were unvaryingly grown in half-strength Hoagland’s solution. Whole plants under a time series treatment (0, 1, 3, 6, 12 and 24 h; three biological replicates) were harvested, immediately frozen in liquid nitrogen and stored at -80°C for further RNA extraction.
Wild-type (WT) Arabidopsis is a Columbian ecotype, which was preserved by our laboratory for the genetic transformation and functional analysis of SiNRX1. Arabidopsis plants were grown both in pots with a soil mixture (vegetative soil/vermiculite = 3:1, v/v) and in a controlled incubator (25°C/20°C temperature, 16 h light/8 h dark photoperiod, 60-70% humidity, 15000 lux light intensity). All seeds were sterilized with 3% NaClO and 70% ethanol before sowing.
The total RNA was extracted from the leaves of the foxtail millet variety Yugu 1 using the TRIzol method, and hydroponic seedlings were tested under normal condition and different stresses (20% PEG6000, 100 mM NaCl and 4°C) for 0, 1, 3, 6, 12 and 24 h. The first-strand cDNA was synthesized by reverse transcription using the EasyScript® One-step gDNA Removal and cDNA Synthesis SuperMix according to the manufacturer’s instructions. qRT-PCR was carried out using a PerfectStart® Green qPCR SuperMix on a QuantStudio 7 Flex Real-Time PCR System following the manufacturer’s instructions. The PCR thermal profile comprised an initial denaturation at 94°C for 30 s, followed by 43 cycles of 94°C for 5 s and 60°C for 30 s. The relative expression level of each NRX gene (SiTRX33 (SiNRX1), SiTRX20 (SiNRX2) and SiTRX25 (SiNRX3)) was then calculated based on the Eq. 2−ΔΔCT. All the samples were processed with at least three biological and three technical replicates. SiActin (ID: Seita.5G393000) was used as an internal control. The specific primers used above are shown in Table S2.
The cDNA was obtained as described above. Specific primers were designed using the Oligo 7 software and were used to amplify each gene (SiNRX1, SiNRX2 and SiNRX3) fragment (Table S2). The PCR system used 1.0 μL of 100 ng μL-1 cDNA, 10 μmol L-1 of forward and reverse primers at 2 μL each, 2 × Phanta Max Buffer at 25.0 μL, dNTP Mix 1 μL, Phanta Max Super-Fidelity DNA Polymerase 1 μL (Vazyme, Nanjing, China), ddH2O 18 μL, at a total of 50.0 μL. The PCR procedure was as follows: pre degeneration at 95°C for 5 min; degeneration at 95°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 1 min 30 s, over 35 cycles; 72°C for 5 min. At a voltage of 130 V and a current of 250 mA, gel electrophoresis was used to detect whether the PCR product contained the target band. After gel recovery and purification, the PCR products were introduced into the pEASY-T1 cloning vector (TransGen, Beijing, China), which was used to transform DH5α competent cells (Vazyme, Nanjing, China) via the heat shock method. Then, positive clones were identified by PCR and sequencing.
The subcellular localizations of green fluorescent protein (GFP) tags were determined for protein localization analysis. The SiNRX1, SiNRX2 and SiNRX3 coding sequences were amplified without a termination codon and fused to the Pcambia1302-GFP vector under the control of the CaMV35S promoter (for the primers, see Table S2). The GFP fusion vector was transformed into tobacco epidermal cells, and the GFP fluorescence was observed using a confocal laser scanning microscope (Olympus, Japan) 48 hours later. The empty vector pGBKT7 and recombinant vector pGBKT7-SiNRX were transferred into a yeast two-hybrid (Y2H) gold strain (for the primers, see Table S2), and cultured upside down in SD/-Trp medium at 30°C. The colony growth was observed 2-3 days later. Then, monoclonal colonies were cultured in SD/-Trp medium, and 1 μL was extracted and inoculated on SD/-Trp and SD/-Trp/-His/-Ade media, respectively. At a temperature of 30°C, the colony growth was observed and recorded after the inverted culture for 3-5 days.
For Arabidopsis transformation, the ORF of SiNRX1 was cloned into the pBI121 vector at the BamHI and SacI sites in a manner that created a pBI121–SiNRX1 construct under the control of the CaMV35S promoter (for the primers, see Table S2). The transformation of Arabidopsis was performed using the Agrobacterium-mediated transformation method (Clough and Bent, 1998). Agrobacterium tumefaciens inflorescence of Arabidopsis was immersed in a transformation medium containing Agrobacterium for 1 min. With the help of transformation media, Agrobacterium tumefaciens can penetrate the plant cell wall and plasma membrane, infect the female gametophyte, or embryo sac, and integrate the T-DNA fragments with the target genes and components into the genome of the female gametophyte (Zhang et al., 2006). After infection with WT Arabidopsis (T0), seeds collected from the T0 generation were considered T1 generation seeds (mixed collection). The T1 generation seeds were screened on Murashige and Skoog (MS) medium containing 50 mg L-1 kanamycin, and the transgenic T1-generation positive seedlings were directly screened out and the T2 generation seeds were harvested (single plant). T2 generation seeds were screened on MS medium containing 50 mg L-1 kanamycin. If the ratio of positive to negative seedlings met the separation ratio of 3:1, positive seedlings were transplanted and T3 seeds were collected (single plant). T3 seeds were screened on MS medium containing 50 mg L-1 kanamycin, transplanting and culture were carried out, and transgenic homozygous lines were finally obtained. Three independent homozygous T3 seedling lines were chosen for subsequent experiments.
For the growth assay, 7-day-old WT and transgenic Arabidopsis seedlings were grown on MS medium and then transferred to pots filled with soil and vermiculite (3:1, v/v) for an additional 2 weeks. By counting the days since the last watering, the drought-stress treatment was performed for 20 days, and rehydration was performed for 3 days. Ultimately, the number of surviving plants was recorded, and the survival rates were determined. Each experiment (including approximately 36 plants) was repeated three times. For the salt-stress treatment, three-week-old Arabidopsis were irrigated with 150 mM NaCl every 5 days and irrigated 3 times for 15 days (3 irrigations in total). Then, the number of surviving plants was recorded, and the survival rates were counted. Each experiment (including approximately 36 plants) was repeated three times. For the water-loss-rate assay, when the seedlings had grown to three-week-old, the in vitro water loss rate (WLR) of the WT and the overexpression (OE) plants of the same size and growth rate were measured. The weights (W2) of the plants at 0, 0.5, 1, 2, 3 and 5 h were measured under constant conditions, where 0 h was determined as the fresh weight (W1), and the calculation formula was WLR= (W2-W1)/W1×100% (Mao et al., 2021).
To analyze physiological and biochemical indexes under drought stress, the three-week-old Arabidopsis seedlings were treated with normal and water withholding for 15 days. To analyze physiological and biochemical indexes under salt stress, the three-week-old Arabidopsis seedlings were kept in normal conditions and treated with 150 mM NaCl for 10 days. The leaves were harvested to determine the MDA content, as well as the proline content (Li et al., 2020). The antioxidant enzymes were extracted, and the activities of cellular CAT, POD and SOD were determined according to the instructions of the antioxidant enzyme assay kits (Solarbio, cat nos. BC0200, BC0090 and BC0170). qRT-PCR was used to analyze the expression levels of stress-related and antioxidant-related genes under drought and salt stresses, including AtP5CS1, AtRD29A, AtDREB2A, AtCAT1, AtPOD1 and AtSOD (Cu/Zn). In addition, qRT-PCR was used to analyze whether the overexpression of SiNRX1 in Arabidopsis affected the expression level of AtNRXs in Arabidopsis. Tubulin beta-2 (ID: AT5G62690) was used as an internal control (for the primers, see Table S2).
The statistical analysis was performed using Excel (version 2016) and SPSS (version 2020). The mean values were statistically compared using Student’s t-test (* indicates p < 0.05, and ** indicates p < 0.01).
The NCBI-CDD retrieval tool and a SMART domain search were used to select protein sequences with complete TRX or TRX-like domains. Thereby, 35 candidate SiTRX genes were identified in the foxtail millet genome. The SiTRX genes were mapped to all nine foxtail millet chromosomes and were named according to the distribution of chromosomes (Figure S1). Among all the chromosomes, the SiTRX genes were unevenly distributed across the genome. Seven SiTRX genes were found on chromosome 9, while five SiTRX genes were found on each of chromosomes 2, 3 and 7, and at least one gene was found on each of chromosomes 6 and 8.
The TRX proteins of foxtail millet range in size between 100 and 580 amino acids. The relative molecular weights (MWs) vary from 11,086.94 to 64,177.87 Da, and about half (51.43%) have low isoelectric points (PIs) (PI < 7.0) (Table S1). Through the prediction of the subcellular localization of the TRX family, TRX members may be distributed in different organelles, such as chloroplasts, cytoplasm, nuclei, mitochondria or plasma membranes. These details indicate that TRX members may play roles in different parts of cells. Therefore, TRX members presumably play roles in different aspects of plant growth and development.
According to the evolutionary relationships and BLAST search against the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast), the TRX proteins were divided into h, o, y, z, x, m, f, ACHT, TRX-like, APR, NRX, NTRC, PDIL and CDSP32 types (Figure 1). Type h has the most members, including ten SiTRXs, eleven AtTRXs and ten OsTRXs. This is followed by type m, which includes five SiTRX members, four AtTRX members and three OsTRX members. Type f includes one SiTRX member, two AtTRX members and one OsTRX member. Type z includes one SiTRX member and one OsTRX member. Type o includes one SiTRX member, two AtTRX members and one OsTRX member. Type y includes one SiTRX member and two AtTRX members. Type x includes one SiTRX member and one OsTRX member. ACHT includes four SiTRXs, five AtTRXs and one OsTRX. TRX-like includes four SiTRXs, two AtTRXs and one OsTRX. APR includes three AtTRXs. NRX includes four SiTRXs and two AtTRXs. NTRC includes one SiTRX and one OsTRX. PDIL includes one SiTRX and eight OsTRXs. However, none of the SiTRXs in foxtail millet are classified as the APR type. Type h is divided into three branches, indicating different evolutionary origins. Overall, the results show that the relationship between TRX family proteins in foxtail millet and rice was closer than that in Arabidopsis, which is in line with the evolutionary relationships of the plants.

Figure 1 Phylogenetic relationships of TRX family proteins in foxtail millet, Arabidopsis and rice. The phylogenetic tree was constructed using the maximum likelihood method in the MEGA 7.0 software. The red star represents SiTRX. The species’ acronyms are shown before each TRX-protein name: Si, Setaria italica; At, Arabidopsis thaliana; Os, Oryza sativa.
On the basis of the phylogeny and gene structure, the 35 SiTRX members were divided into multiple branches, including h, o, y, z, x, m, f, ACHT, TRX-like, NRX, NTRC, PDIL and CDSP32 types, which were consistent with the phylogenetic relationship (Figure 2A). Exon/intron organization analysis further revealed that most of the SiTRXs contained two to five exons, with similar gene structures for each type (Figure 2B). The gene lengths greatly differed among the members of the TRXs, indicating that great variation in the exon/intron lengths of this family of genes occurred during evolution, which may have led to diversity in function.
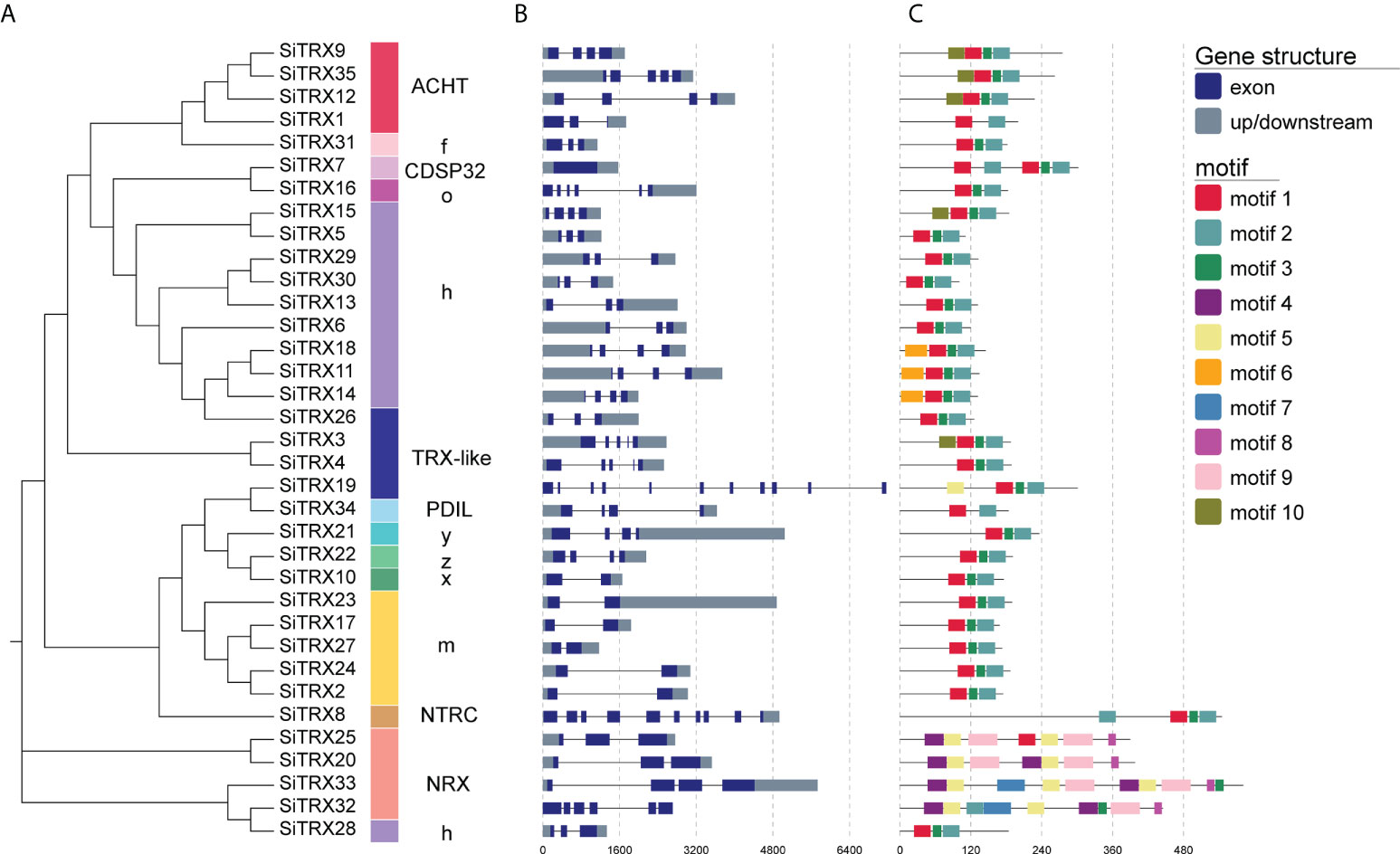
Figure 2 Evolutionary relationship, gene structure and conserved motif analysis of SiTRX members. (A) The evolutionary relationship of TRX in foxtail millet. (B) Gene structures of SiTRXs. The intron–exon structures were obtained by comparing the coding and genomic sequences using the GSDS website. (C) Motif composition, with the ten conserved motifs indicated with boxes of different colors.
In addition, whether SiTRX proteins had a conserved domain was analyzed (Figure S2). SiTRXs encode proteins with conserved TRX or TRX-like domains. In addition to the TRX-like domain, SiTRX20, SiTRX25 and SiTRX33 have a C-terminal domain rich in cysteine, and SiTRX8 contains another redox domain. To better understand the structural characteristics of SiTRX proteins and provide clues regarding their functions, the conserved motifs of SiTRX proteins were analyzed (Figure 2C). All of motif 1 and motif 2 are present in typical TRXs. The motif found in atypical TRXs differs from the conserved motifs observed in typical TRXs. Interestingly, conserved motifs of NRX are included in motif 4, motif 5, motif 8 and motif 9, which are different from the other types of conserved motifs in TRX. The functions of most of these conserved motifs remain to be elucidated.
A further analysis using Plant CARE revealed cis-regulatory elements in the promoters of the SiTRX genes, and these elements were related to different functions (Figure S3). In addition to the basic elements (CAAT-box and TATA-box), a large number of light-responsive, environmental-stress-related, hormone-responsive, development-related, site-binding-related and other cis-regulatory elements were identified in 35 SiTRX genes in the promoter region. Most of the SiTRX genes had light-response elements (G-box, ACE, GT1-motif, GATA-motif, Box 4 and TCT-motif). About 54% of the SiTRX genes contained low-temperature-response elements (LTR), 57% of the SiTRX genes contained drought-induction-response elements (MBS), 82% of the SiTRX genes contained antioxidant-stress-response elements (ARE), and 48% of the SiTRX genes contained hypoxia-specific-induction elements (GC-motif). About 88% of the SiTRX genes contained hormone-response elements and methyl-jasmonate response elements (CGTCA-motif and TGACG-motif), about 82% of the SiTRX genes contained abscisic-acid-response elements (ABRE, ABRE4, and ABRE 3A), and about 45% of the SiTRX genes contained gibberellin-response elements (P-box and GARE-motif). In addition, some genes contained elements related to growth and development, such as meristem expression (CAT-box), the material metabolism regulatory element (O2-site), MYB (MER), 45% of the MYBHv1 binding site (CCAAT-box), etc. These results indicate that SiTRX genes may be regulated by multiple cis-acting elements and play important roles in plant photosynthesis, growth and development, hormone regulation and the stress response.
NRX is the first TRX found to be located in the nucleus, and its specific biological function remains to be further explored. In order to verify the results of the cis-element analysis and further reveal the responses of SiNRX genes (SiTRX33 (renamed SiNRX1), SiTRX20 (renamed SiNRX2) and SiTRX25 (renamed SiNRX3)) to different stresses, qRT-PCR was used to detect the expression of SiNRX genes in foxtail millet leaves treated with normal condition, 20% PEG6000, 100 mM NaCl and 4°C (Figure 3). Under drought treatment, compared with the normal condition, the expression level of SiNRX1 decreased rapidly at 1 h, then increased and reached the highest level at 3 h; the expression of SiNRX2 decreased at 6 h, then increased and reached the highest level at 12 h, and the expression level began to decline and returned to normal at 24 h; the expression of SiNRX3 decreased dramatically at 24 h. Compared with that of the other two genes, the expression of SiNRX1 increased the most under drought stress. During salt treatment, relative to the normal condition, the expression level of SiNRX1 gradually increased from 1 h to 12 h and reached the highest level; SiNRX2 and SiNRX3 reached their highest expression level at 6 h. On the whole, the mRNAs of SiNRX1, SiNRX2 and SiNRX3 showed a trend of first reaching their highest levels and then declining. Under a low-temperature treatment at 4°C, compared with the normal condition, the level of SiNRX1 showed a trend of first increasing and then decreasing; SiNRX2 reached its highest level at 6 h, SiNRX3;s mRNA level increased gradually over 24 h. Our results clearly show that different SiNRX genes respond differently to different abiotic stresses in foxtail millet leaves.
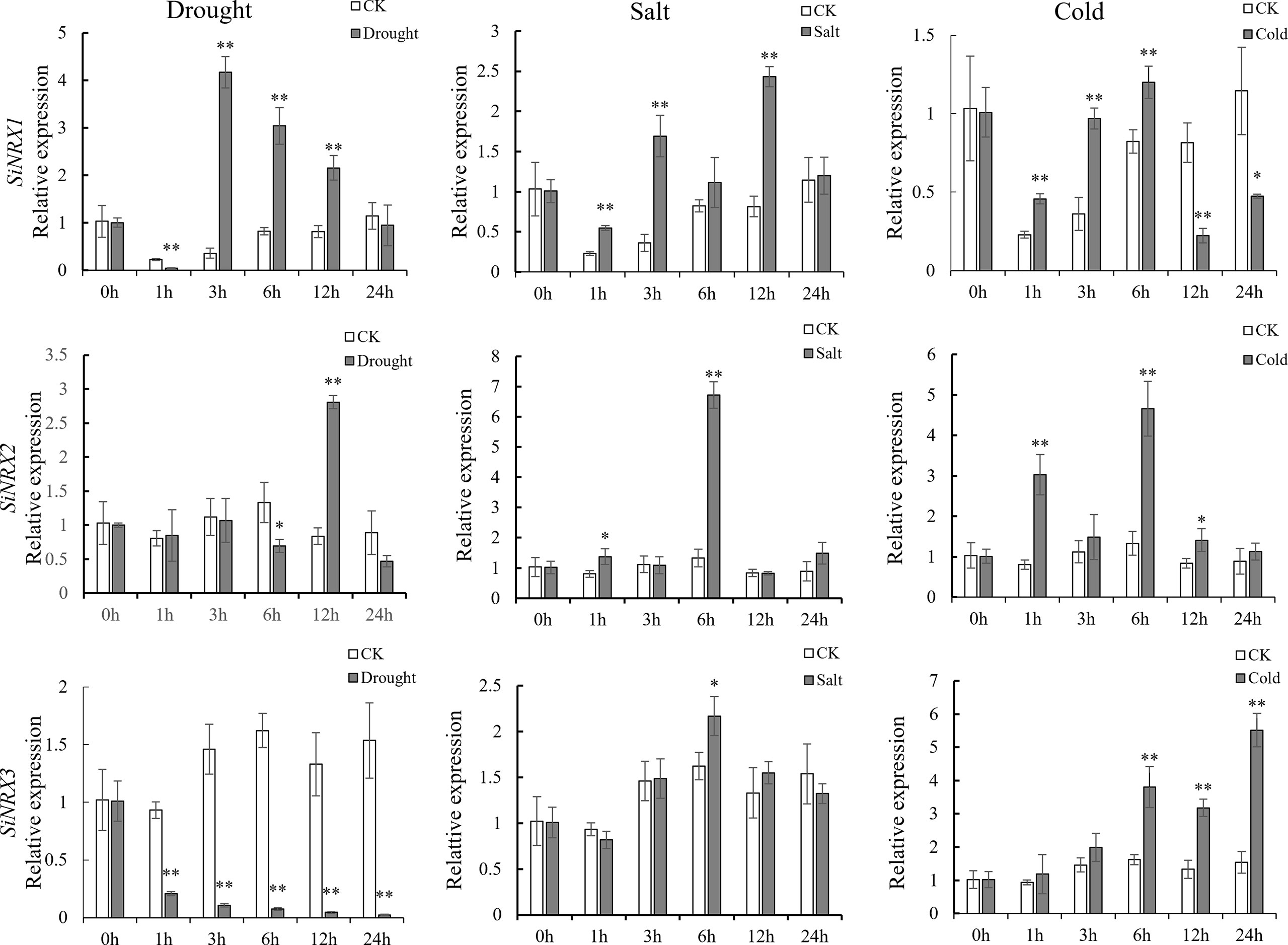
Figure 3 Expression patterns of SiNRX1, SiNRX2, SiNRX3 under abiotic stresses. qRT-PCR analysis of SiNRX1, SiNRX2, and SiNRX3 in foxtail millet leaves in response to diverse treatments (drought [20% PEG6000), salt (100 mM NaCl), and cold (4°C)]. CK as control (without stress treatment). Standard deviations are indicated by error bars (mean ± SD and n = 3). Each time point was compared to the control group, and student’s t-test was used to calculate significance: *indicates p < 0.05, and **indicates p < 0.01.
NRX, named nucleoredoxin because it was originally found in the nucleus, was predicted to be localized in the cytoplasm, so subcellular localization was used to verify this prediction. As a control, the fluorescence signal from a GFP–null fusion protein was measured in the nucleus, cytoplasm and membrane, and the fluorescence signal from a GFP–SiNRX fusion protein was measured in the same manner as that for the control. The fusion proteins SiNRX1–GFP, SiNRX2–GFP and SiNRX3–GFP showed green fluorescence signals on the nucleus, cytoplasm and membrane (Figure 4A). The subcellular localization analysis indicated that SiNRX1, SiNRX2 and SiNRX3 were all localized in the nucleus, cytoplasm and membrane. The experimental results are reasonably consistent with the predictions. Yeast transfected with pGBKT7 and the recombinant vector pGBKT7–SiNRX could grow in SD/-Trp medium with no significant difference in growth rate. The yeast could not grow on SD/-Trp/-His/-Ade medium, indicating that SiNRX had no self-activation effect (Figure 4B).
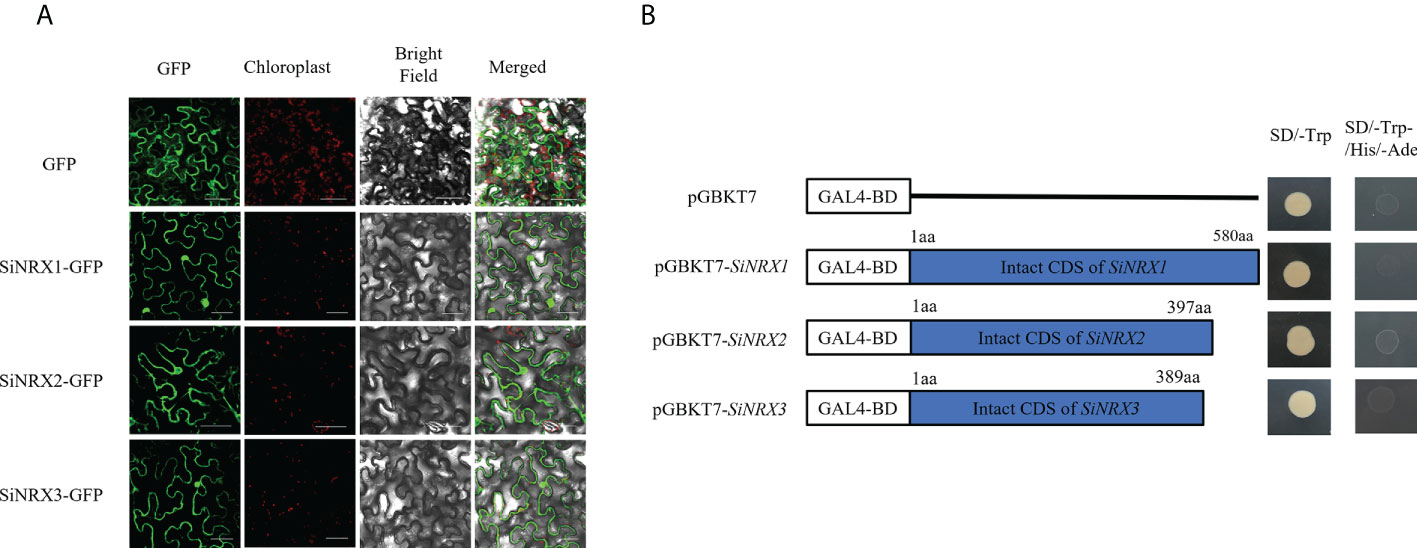
Figure 4 Subcellular localization and transcriptional activation of SiNRX1, SiNRX2 and SiNRX3. (A) Recombinant plasmid and control plasmid were transiently expressed in tobacco cells. The scale bar represents 50 μm. (B) The ORF fragments of SiNRX genes were ligated with pGBKT7 to construct a fusion vector. The yeast cells harboring SiNRX fusion vectors were cultured on the SD/-Trp and SD/-Trp/-His/-Ade.
From the perspective of the structural domains of different NRX members, compared with the other two types, NRX1 has three TRX-like structural domains, so it was presumed that its redox activity would be higher than that of the other two types. According to the qRT-PCR, SiNRX1 was more highly expressed under different stresses. Consequently, SiNRX1 was taken as the research object for further functional analysis. To analyze the function of SiNRX1 in drought resistance and salt tolerance, SiNRX1-transgenic Arabidopsis plants were generated under the control of the CaMV35S promoter. After RT-PCR, no bands were detected in wild-type (WT) lines, while bands were detected in overexpression (OE) lines, indicating the successful transformation of SiNRX1 in Arabidopsis (Figure 5A; Table S2). Then, three independent T3 lines with homozygous SiNRX1 overexpression (OE1, OE2 and OE3) were selected for further functional analyses. WT and transgenic seedlings were grown for 3 weeks under normal conditions. After 20 days of drought treatment, WT plants were severely withered or even bleached, while OE lines suffered less damage than WT. After 3 days of rehydration treatment, it was found that the new leaves of OE lines recovered better than those of the WT (Figure 5B). The survival rates of the three OE lines were 86.11%, 91.66% and 88.88%, respectively, while the survival rate of the WT lines was 44%. Moreover, the survival rates of the OE lines were about 42% higher than those of the WT (Figure 5C). WT and transgenic seedlings at three weeks old were each treated with 150 mM NaCl for 15 days. OE lines had deeper leaves, while WT lines had wilted or even white leaves (Figure 5E). The survival rates of the OE lines (96.29%, 98.14%, 97.22%) were higher than those of the WT (85.19%) (Figure 5F). In general, SiNRX1 overexpression improves the survival rate of Arabidopsis plants under drought and salt stresses. The analysis of the water-loss rate at 5 h showed that the in vitro water-loss rate of the leaves of the WT was 80.33%, and that of the OE lines was 55.06%, on average. The water-loss rate of the WT lines was about 25% higher than that of the OE lines (Figure 5D).

Figure 5 The phenotype analysis of OE lines and WT under drought and salt stresses. (A) Gene verification of SiNRX1 in OE lines by PCR. (B) The phenotype of OE lines after drought treatment for 20 days. (C) The survival rate of OE lines and WT after water withholding for 20 days. (D) The water loss rate of leaves in vitro measured at 25°C. (E) The phenotype of OE lines and WT after salt treatment (150 mM NaCl) for 15 days. (F) The survival rate of OE lines and WT after salt treatment for 15 days. OE: SiNRX1 transgenic Arabidopsis lines; WT: wild-type Arabidopsis lines. Standard deviations are indicated by error bars (mean ± SD and n = 3). Student’s t-test was used to calculate significance: ** indicates p > 0.01.
To investigate the physiological changes in the OE and WT lines, the malondialdehyde (MDA) content; proline content; and catalase (CAT), peroxidase (POD) and superoxide dismutase (SOD) activity were each measured under normal growth conditions, as well as drought and salt stresses. Under normal conditions, there were no significant differences in the MDA content; proline content; and CAT, POD and SOD activity between the WT and OE lines. Under drought stress, the MDA contents of the OE lines were 15.00 nmol g-1 FW, 14.01 nmol g-1 FW, and 13.31 nmol g-1 FW, respectively, while the MDA contents of the WT were 23.03 nmol g-1 FW (Figure 6A). Finally, the MDA content in the OE lines and WT was significantly upregulated under drought stress, but the increased MDA content in the OE lines was significantly lower than that in the WT. However, the trend of the proline content was opposite to that for the MDA content under drought stress (Figure 6B). For the antioxidant enzyme system (CAT, POD and SOD), the CAT activity of the OE lines was 2.48 U mg-1 protein, 2.34 U mg-1 protein, and 2.31 U mg-1 protein, respectively. On the other hand, the CAT activity of the WT was 1.55 U mg-1 protein under drought stress (Figure 6C). The trend of the POD and SOD activity was consistent with that for the CAT activity under drought stress (Figures 6D, E). Under salt stress, the MDA contents of the OE lines (9.77 nmol g-1 FW, 9.90 nmol g-1 FW, 9.48 nmol g-1 FW) were lower than those of the WT plants (15.16 nmol g-1 FW) (Figure 7A). However, The proline content and antioxidant enzyme (CAT, POD, SOD) activity of OE lines were higher than WT under salt stress (Figures 7B–E). In a word, under drought and salt stresses, the activities of CAT, POD and SOD in the OE lines were higher than those of the WT. These results indicate that the antioxidant capacity and ROS-scavenging capacity of the OE lines increased compared to those of the WT under drought and salt stresses, which resulted in the observed enhanced drought resistance and salt tolerance of the transgenic lines.
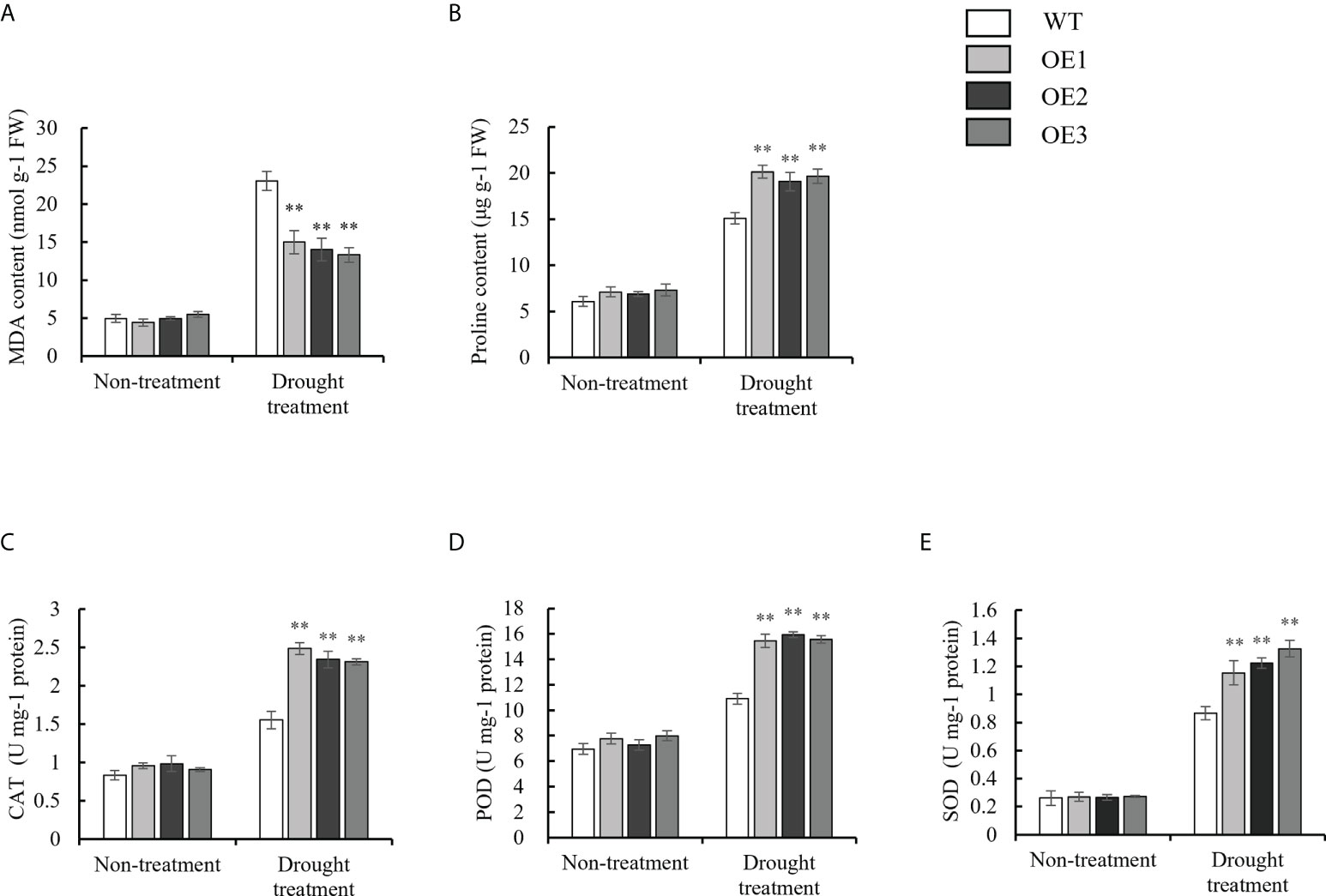
Figure 6 Evaluation of physiological indices of OE lines and WT responsive to drought stress. (A) MDA content. (B) Proline content. (C-E) Alterations of Activities of CAT, POD, and SOD. Standard deviations are indicated by error bars (mean ± SD and n = 3). Student’s t-test was used to calculate significance: ** indicates p > 0.01.
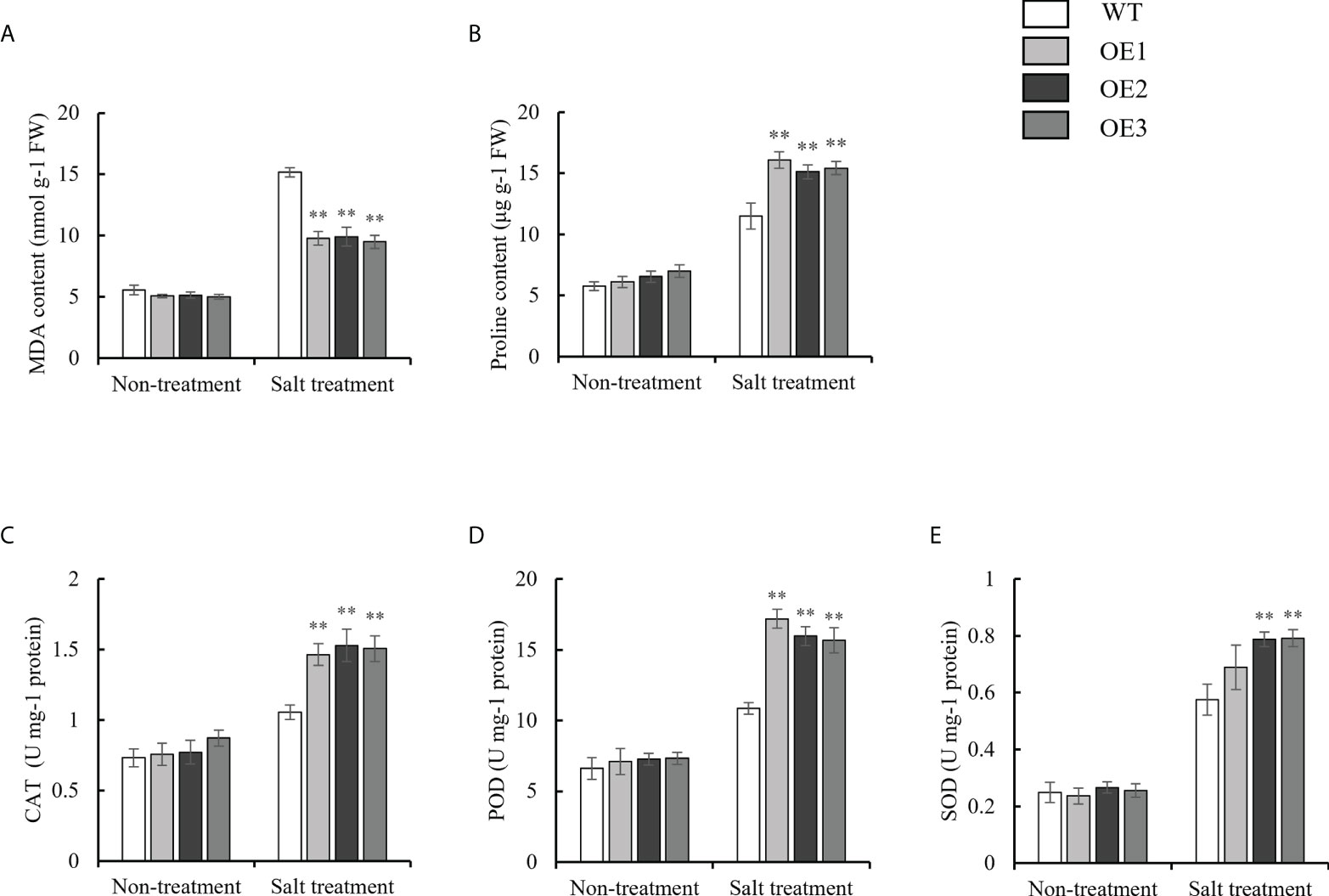
Figure 7 Evaluation of physiological indices of OE lines and WT responsive to salt stress. (A) MDA content. (B) Proline content. (C-E) Alterations of Activities of CAT, POD, and SOD. Standard deviations are indicated by error bars (mean ± SD and n = 3). Student’s t-test was used to calculate significance: ** indicates p > 0.01.
In order to further analyze the mechanism of the response of SiNRX1 genes to drought and salt stresses, several stress-related genes and antioxidant-related genes were chosen for observation at the transcriptional level. Three stress-related genes (AtP5CS1, AtRD29A and AtDREB2A) and three antioxidant-related genes (AtCAT1, AtPOD1 and AtSOD (Cu/Zn)) were selected for the gene expression analysis of the WT and OE lines after drought and salt stresses (Figure 8). Under normal conditions, there was no significant difference in the expression levels of the six genes (AtP5CS1, AtRD29A, AtDREB2A, AtCAT1, AtPOD1 and AtSOD (Cu/Zn)). Under drought stress, the expression level of AtP5CS1 in the OE3 line was 5.8-fold higher than WT; the expression level of AtRD29A in the OE3 line was 4.1-fold higher than WT; the expression level of AtDREB2A in the OE3 line was 5-fold higher than WT; the expression level of AtCAT1 in the OE3 line was 2.5-fold higher than WT; the expression level of AtPOD1 in the OE3 the line was 2.6-fold higher than WT; the expression level of AtSOD (Cu/Zn) in the OE3 the line was 5.3-fold higher than WT. Under salt stress, the expression level of AtP5CS1 in the OE3 line was 5.1-fold higher than WT; the expression level of AtRD29A in OE3 the line was 7.2-fold higher than WT; the expression level of AtDREB2A in the OE3 line was 4.1-fold higher than WT; the expression level of AtCAT1 in the OE3 line was 4.8-fold higher than WT; the expression level of AtPOD1 in the OE3 line was 3.6-fold higher than WT; and the expression level of AtSOD (Cu/Zn) in the OE3 line was 3-fold higher than WT. In general, the expression levels of six genes in the OE3 line were significantly upregulated under drought and salt stresses, although the levels of the upregulation of each gene were different under different stresses. The results indicate that the heterologous expression of SiNRX1 may enhance the drought and salt resistance/tolerance of Arabidopsis by enhancing the expression of stress-related genes. In addition, to analyze whether the overexpression of SiNRX1 in Arabidopsis would affect the expression of AtNRXs, AtNRX1 and AtNRX2 were selected to observe the changes at the transcription level (Figure S4). Under normal conditions, there was no significant difference in the expression levels of AtNRX1 and AtNRX2 in WT and the OE3 line, suggesting that overexpression of SiNRX1 may have no affect the expression level of AtNRXs in Arabidopsis. However, under drought and salt stresses, the expression level of AtNRX1 in OE3 line was significantly higher than that in WT. The expression level of AtNRX2 in the OE3 line was lower than WT under drought stress, but higher than WT under salt stress.
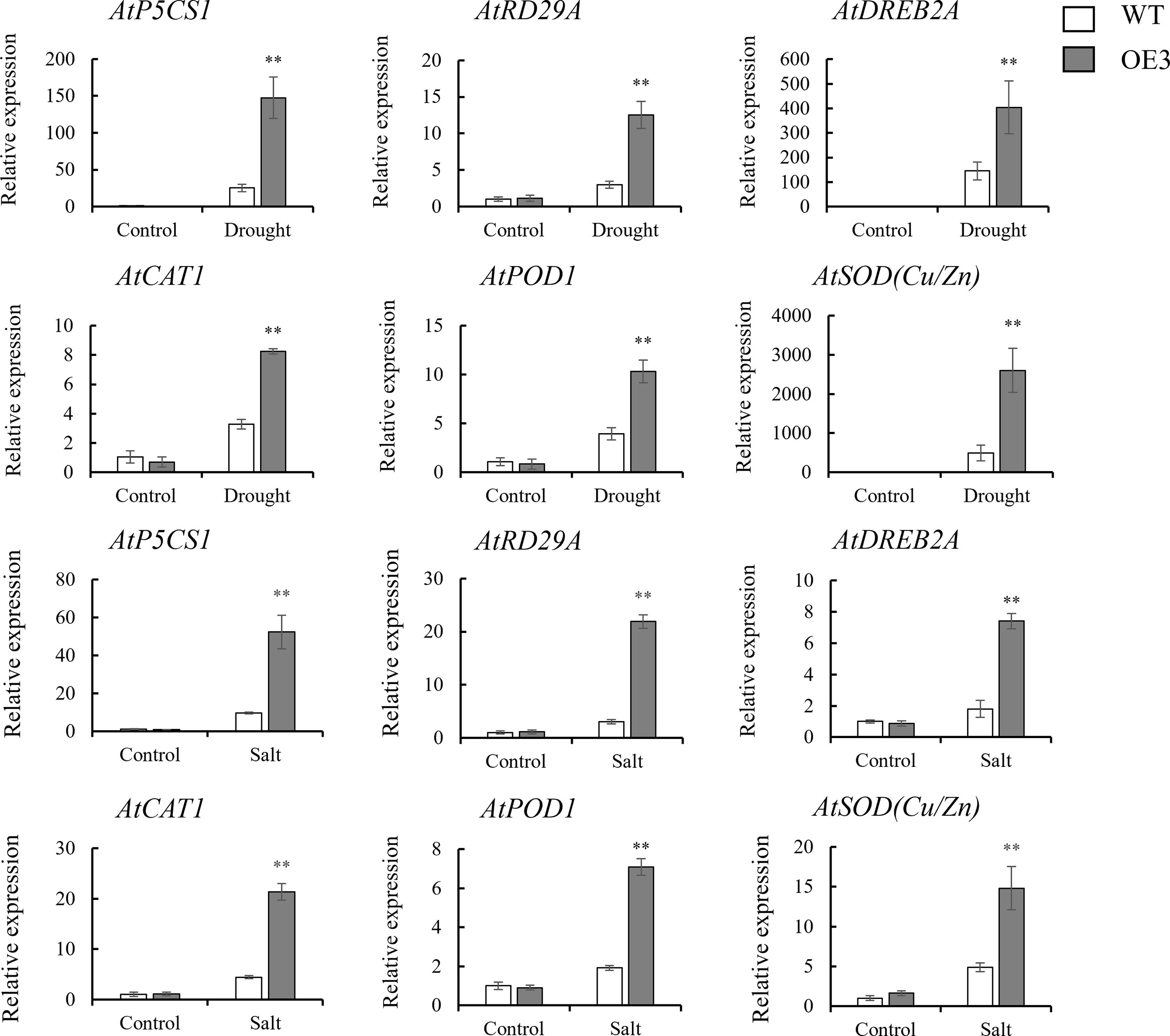
Figure 8 The expression levels of stress-related genes in the OE3 line and WT under stresses. stress-related genes—AtP5CS1, AtRD29A and AtDREB2A; antioxidant-related genes —AtCAT1, AtPOD1 and AtSOD (Cu/Zn). Stresses include drought stress and salt stress. Standard deviations are indicated by error bars (mean ± SD and n = 3). Student’s t-test was used to calculate significance: ** indicates p > 0.01.
Foxtail millet has become a research hotspot in recent years due to its small genome, strong stress resistance and high nutritional value. TRX family genes in rice, Arabidopsis and wheat have been confirmed to participate in the regulation of plant growth and development, environmental stresses and other biological processes, with very important biological significance (Nuruzzaman et al., 2012; Chibani et al., 2021; Bhurta et al., 2022). It is worth mentioning that, due to its unique redox regulatory activity, TRX has received extensive attention in the study of plant resistance. There are still few reports about foxtail millet TRX family members. The systematic identification of the SiTRX family in the full genome will assist us in excavating the gene resources for stress resistance in foxtail millet, thereby producing available gene resources for the breeding and genetic engineering of crops. In this study, 35 candidate SiTRX members were identified from the foxtail millet genome. Gene structure analysis showed that exon–introns located in the same branch of the TRX family were similar in distribution (Figure 2B). Plants with fewer introns have been shown to be better able to handle stress. TaTRX genes with fewer introns have been identified in wheat and can respond quickly to leaf rust pathogens (Bhurta et al., 2022). However, the function of introns in gene expression requires further verification. It is speculated that the difference between NRXs and other types may be the result of long-term evolution and adaptation to environmental changes through analysis of conserved motif. Promoters, as important regulatory regions upstream of genes, contain many cis-acting elements, which play very important roles in regulating gene responses to biotic stress, abiotic stresses and specific expression in plant tissues (Sheshadri et al., 2016). A large number of abscisic acid response elements (ABRE), stress response elements (MBS) and antioxidant stress elements (AREs) were found in the promoter regions of SiTRX family genes (Figure S3). These results suggest that SiTRXs may play important roles in plant stress responses and stress resistance.
Most studies have focused on the characterization of typical TRX functions, while less attention has been paid to atypical TRXs. With the deepening of research, the biological functions of atypical TRXs have gradually attracted the attention of researchers. The NRXs, as a class of atypical TRX members, contain multiple redox-active sites and are located in the nucleus, which may indicate that the NRXs have a stronger biological activity. In this study, three SiNRXs, SiTRX33 (Type I), SiTRX20 (Type II) and SiTRX25 (Type III), were identified according to the conserved domain, active site and subcellular localization. NRX was originally found in the nucleus of the mouse, so it was named “nucleoredoxin”. Studies have shown that NRX may be a redox regulator of the transcription factors (Kurooka et al., 1997). However, Funato et al. (2006) found that NRX was mainly located in the cytoplasm, and NRX appeared to be a specific suppressor of Wnt-β-catenin signaling. Marchal et al. (2014) describe a new TRX system, NTR/NRX, which is located in the nucleus and cytoplasmic compartment of Arabidopsis. It was found that the AtNRX1 of Arabidopsis has a dual localization—nucleus and cytoplasm. Li et al. (2016) found the GbNRX1 of island cotton is located in the exoplasm under stress. In this study, the localization analysis of the NRXs shows that their members are located in the nucleus, cytoplasm and membrane. The subcellular localization of NRX in different species may be different and may play different functions in different locations. Biotic and abiotic stresses have been reported to lead to cellular translocations of TRX enzymes or the entire TRX/NTR system, suggesting that these enzymes are recruited to specific sites for their signaling or protective functions (Mata-Pérez and Spoel, 2019), so it is speculated that the subcellular localization of NRX in different plants may be different or displaced. In addition, SiNRX1, SiNRX2 and SiNRX3 showed no self-activation activity in yeast, suggesting that NRX may not function as a transcription factor in plants, but as a functional protein regulating the redox state of biological targets in vivo. The analysis of cis-acting elements showed that the promoter regions of SiNRX1, SiNRX2 and SiNRX3 all contained abscisic acid response elements (ABRE), a low-temperature-response element (LTR), a hypoxia-specific-induction element (GC-motif) and other plant-hormone-response elements and stress-response elements. These results indicate that NRX might be involved in the anti-retroviral response, which was also confirmed by qRT-PCR (Figure 3). Therefore, we speculate that SiNRX1 also may play a critical role in stress tolerance in plants. This hypothesis was verified by further functional identification of SiNRX1 transgenic Arabidopsis. SiNRX1 overexpression Arabidopsis significantly increased the growth potential and survival rate under drought or salt stress (Figure 5).
Plant responses to water and salt stresses have much similarity in most metabolic processes (Datta et al., 2012). Osmotic stress is a common adverse outcome of the response to drought and salt stresses. Proline accumulation is the first response of plants under osmotic stress (Grzesiak et al., 2013). It has been reported that proline, as a non-enzymatic antioxidant, may be involved in ROS signal transduction, and proline metabolism is related to the tolerance of plants to environmental stress. It has been proven that, the higher the content of proline, the stronger the stress resistance of plants (Szabados and Savouré, 2010; Liang et al., 2013). P5CS1 is a key gene for proline accumulation, and its increased expression is induced by drought and salt stresses (Ben Rejeb et al., 2014). In this study, the proline content of overexpressed plants was highly up-regulated compared with WT under stresses (Figures 6B, 7B). The AtP5CS1 transcript level was strongly induced in the transgenic lines under drought and salt stresses (Figure 8). MDA is considered an indicator of oxidative damage at the cellular level (Sevanian and Ursini, 2000). The MDA content in the OE lines was much lower than that in the WT (Figures 6A, 7A). Previous studies have demonstrated that the lower the MDA content, the higher the tolerance to abiotic stress (Zhu et al., 2021).
Drought and salt stresses also can lead to the excessive accumulation of ROS, especially O2·- and H2O2, leading to severe oxidative damage in plants (Skopelitis et al., 2006; Wei et al., 2017). The plant’s antioxidant enzyme system (POD, SOD and CAT) protects cells from stress by maintaining ROS at normal levels (Yadav et al., 2019; Zhong et al., 2020; Ampofo and Ngadi, 2021). To determine whether SiNRX1 modulates ROS accumulation, we tested the activities of several antioxidant enzymes (CAT, POD, SOD) and analyzed the expression levels of stress-related and antioxidant-related genes by qRT-PCR. They have previously been reported to be associated with the stress response (Sakuma et al., 2002; Narusaka et al., 2003; Li et al., 2020). The results showed that the activities of CAT, POD and SOD in SiNRX1 overexpressed Arabidopsis were higher than those in WT under osmotic stress (Figures 6, 7). Accordingly, several genes related to ROS-scavenging were strongly induced in overexpression lines (Figure 8). Li et al. (2016) proved that GbNRX1 is involved in regulating ROS homeostasis in response to pathogen invasion. Studies in Arabidopsis suggest that AtNRX1 plays an important role in plant immune signal transduction by maintaining CAT in a reduced state, thereby protecting its H2O2-detoxifying activity (Kneeshaw et al., 2017). TaNRX1-overexpressed wheat also displayed increased antioxidant enzyme activity under drought stress, which had a positive regulatory effect on transgenic wheat (Zhang et al., 2021). This was consistent with our experimental results. Moreover, AtNRXs was not affected under normal conditions, but AtNRX1 of the OE line showed higher expression levels than WT after stress (Figure S4). It is speculated that SiNRX1 could induce an increase in AtNRX1 expression levels under abiotic stress. AtNRX1, as a redox protein, directly or indirectly regulates the antioxidant enzyme system, eliminates excessive reactive oxygen species and maintains the stable state of reactive oxygen species. As a result, SiNRX1-overexpressed Arabidopsis reduces the oxidative damage of plants and shows a better growth phenotype. However, further research is needed to elucidate the molecular mechanism of SiNRX1. In addition, the above studies indicated that SiNRX1 is located in the nucleus, cytoplasm and membrane, supposing that SiNRX1 also plays an important role in the nucleus. It may improve the tolerance of plants by regulating transcription factors related to stress tolerance. What NRX does in the nucleus may be the key to further research.
In summary, through the genome-wide analysis of foxtail millet, a total of 35 candidate TRX members were identified, which were divided into 13 groups according to their evolutionary relationships. In this study, the phylogenetic relationships, chromosome distributions, gene structures, conserved motifs and cis-elements of 35 SiTRX members were characterized. In addition, the expression pattern of SiNRXs under abiotic stresses was analyzed. Due to the special structure of SiNRX1 and its strong induction under stress, the stress resistance function of SiNRX1 was identified through the overexpression of SiNRX1 in the model plant Arabidopsis. These results show that SiNRX1 was involved in the responses to drought and salt stresses, and results provide evidence for the relationship between TRX family genes and abiotic stresses in foxtail millet.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XZ and JX designed the study. SZ and YY performed the bioinformatics analysis. SZ, TS, and HZ collected samples and performed data analyses. MZ and NL conducted qRT-PCR experiment. TS and MY performed subcellular localization analysis. SZ wrote the manuscript. YNY, SG, CX, and YT revised the manuscript. All authors read and approved the final manuscript.
This study was funded by the National Natural Science Foundation of China (32171992, 31671693), Research Program of Science and Technology at Universities of Inner Mongolia Autonomous Region (NJZZ22132), Central Guidance on Local Science and Technology Development Fund of Mongolia Autonomous Region (ZY20200089), Program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (NJYT-19-A25), Key Research and Development Project of Shaanxi Province (2019ZDLNY04-05), Innovation and Entrepreneurship Training program for college students of Northwest A&F University (S202110712347).
We would like to thank Yi Ding and Jiaxin Ma for their assistance in sample collections and RNA extraction.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.946037/full#supplementary-material
Ampofo, J. O., Ngadi, M. (2021). Stimulation of the phenylpropanoid pathway and antioxidant capacities by biotic and abiotic elicitation strategies in common bean (Phaseolus vulgaris) sprouts. Process Biochem. 100, 98–106. doi: 10.1016/j.procbio.2020.09.027
Apel, K., Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Ben Rejeb, K., Abdelly, C., Savouré, A. (2014). How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 80, 278–284. doi: 10.1016/j.plaphy.2014.04.007
Bhurta, R., Hurali, D. T., Tyagi, S., Sathee, L., Adavi B, S., Singh, D., et al. (2022). Genome-wide identification and expression analysis of the thioredoxin (trx) gene family reveals its role in leaf rust resistance in wheat (Triticum aestivum l.). Front. Genet. 13. doi: 10.3389/fgene.2022.836030
Calderón, A., Sevilla, F., Jiménez, A. (2018). “Redox protein thioredoxins: Function under salinity, drought and extreme temperature conditions," in Antioxidants and antioxidant enzymes in higher plants, (Cham: Springer) 123–162. doi: 10.1007/978-3-319-75088-0_7
Cheng, J., Wei, F., Zhang, M., Li, N., Song, T., Wang, Y., et al. (2021). Identification of a 193 bp promoter region of TaNRX1-d gene from common wheat that contributes to osmotic or ABA stress inducibility in transgenic Arabidopsis. Genes Genomics 43, 1035–1048. doi: 10.1007/s13258-021-01115-x
Chibani, K., Pucker, B., Dietz, K. J., Cavanagh, A. (2021). Genome-wide analysis and transcriptional regulation of the typical and atypical thioredoxins in Arabidopsis thaliana. FEBS Lett. 595, 2715–2730. doi: 10.1002/1873-3468.14197
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dal Piaz, F., Braca, A., Belisario, M. A., De Tommasi, N. (2010). Thioredoxin system modulation by plant and fungal secondary metabolites. Curr. Med. Chem. 17, 479–494. doi: 10.2174/092986710790226165
Datta, K., Baisakh, N., Ganguly, M., Krishnan, S., Yamaguchi Shinozaki, K., Datta, S. K. (2012). Overexpression of Arabidopsis and rice stress genes' inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol. J. 10, 579–586. doi: 10.1111/j.1467-7652.2012.00688.x
Funato, Y., Michiue, T., Asashima, M., Miki, H. (2006). The thioredoxin-related redox-regulating protein nucleoredoxin inhibits wnt-beta-catenin signalling through dishevelled. Nat. Cell Biol. 8 (5), 501–508. doi: 10.1038/ncb1405
Grzesiak, M., Filek, M., Barbasz, A., Kreczmer, B., Hartikainen, H. (2013). Relationships between polyamines, ethylene, osmoprotectants and antioxidant enzymes activities in wheat seedlings after short-term PEG-and NaCl-induced stresses. Plant Growth Regul. 69 (2), 177–189. doi: 10.1007/s10725-012-9760-9
Jing, X., Yao, J., Ma, X., Zhang, Y., Sun, Y., Xiang, M., et al. (2020). Kandelia candel thioredoxin f confers osmotic stress tolerance in transgenic tobacco. Int. J. Mol. Sci. 21, 3335. doi: 10.3390/ijms21093335
Kneeshaw, S., Keyani, R., Delorme-Hinoux, V., Imrie, L., Loake, G. J., Le Bihan, T., et al. (2017). Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. Proc. Natl. Acad. Sci. U.S.A. 114, 8414–8419. doi: 10.1073/pnas.1703344114
Kurooka, H., Kato, K., Minoguchi, S., Takahashi, Y., Ikeda, J., Habu, S., et al. (1997). Cloning and characterization of the nucleoredoxin gene that encodes a novel nuclear protein related to thioredoxin. Genomics 39 (3), 331–339. doi: 10.1006/geno.1996.4493
Laughner, B. J., Sehnke, P. C., Ferl, R. J. (1998). A novel nuclear member of the thioredoxin superfamily. Plant Physiol. 118, 987–996. doi: 10.1104/pp.118.3.987
Laurent, T. C., Moore, E. C., Reichard, P. (1964). Enzymatic synthesis of deoxyribonucleotides: IV. isolation and characterization of thioredoxin, the hydrogen donor from escherichia coli b. J. Biol. Chem. 239, 3436–3444. doi: 10.1016/S0021-9258(18)97742-2
Lee, E. S., Park, J. H., Hwang, G. Y., Chi, Y. H., Kang, C. H., Chae, H. B., et al. (2020). Nucleoredoxin 2 (NRX2) promotes jasmonate-mediated trichome formation in Arabidopsis. J. Plant Biol. 63, 495–503. doi: 10.1007/s12374-020-09277-z
Liang, X., Zhang, L., Natarajan, S. K., Becker, D. F. (2013). Proline mechanisms of stress survival. Antioxid Redox Signal 19, 998–1011. doi: 10.1089/ars.2012.5074
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Li, X., Tang, Y., Li, H., Luo, W., Zhou, C., Zhang, L., et al. (2020). A wheat R2R3 MYB gene TaMpc1-D4 negatively regulates drought tolerance in transgenic Arabidopsis and wheat. Plant Sci. 299, 110613. doi: 10.1016/j.plantsci.2020.110613
Maharajan, T., Antony Ceasar, S., Ajeesh Krishna, T. P., Ignacimuthu, S. (2021). Finger millet [Eleusine coracana (L.) gaertn]: an orphan crop with a potential to alleviate the calcium deficiency in the semi-arid tropics of Asia and Africa. Front Sustain. Food Syst. 5. doi: 10.3389/fsufs.2021.684447
Mao, Y., Xu, J., Wang, Q., Li, G., Tang, X., Liu, T., et al. (2021). A natural antisense transcript acts as a negative regulator for the maize drought stress response gene ZmNAC48. J. Exp. Bot. 72, 2790–2806. doi: 10.1093/jxb/erab023
Marchal, C., Delorme-Hinoux, V., Bariat, L., Siala, W., Belin, C., Saez-Vasquez, J., et al. (2014). NTR/NRX define a new thioredoxin system in the nucleus of Arabidopsis thaliana cells. Mol. Plant 7, 30–44. doi: 10.1093/mp/sst162
Mata-Pérez, C., Spoel, S. H. (2019). Thioredoxin-mediated redox signalling in plant immunity. Plant Sci. 279, 27–33. doi: 10.1016/j.plantsci.2018.05.001
Meyer, Y., Reichheld, J. P., Vignols, F. (2005). Thioredoxins in Arabidopsis and other plants. Photosynth Res. 86, 419–433. doi: 10.1007/s11120-005-5220-y
Michelet, L., Zaffagnini, M., Morisse, S., Sparla, F., Pérez-Pérez, M. E., Francia, F., et al. (2013). Redox regulation of the calvin-benson cycle: something old, something new. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00470
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-regulatory elements, ABRE and DRE, in ABA-dependent expression of arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313x.2003.01708.x
Neupane, D., Adhikari, P., Bhattarai, D., Rana, B., Ahmed, Z., Sharma, U., et al. (2022). Does climate change affect the yield of the top three cereals and food security in the world? Earth 3, 45–71. doi: 10.3390/earth3010004
Nietzel, T., Mostertz, J., Ruberti, C., Née, G., Fuchs, P., Wagner, S., et al. (2020). Redox-mediated kick-start of mitochondrial energy metabolism drives resource-efficient seed germination. Proc. Natl. Acad. Sci. U.S.A. 117, 741–751. doi: 10.1073/pnas.1910501117
Nikkanen, L., Rintamäki, E. (2019). Chloroplast thioredoxin systems dynamically regulate photosynthesis in plants. Biochem. J. 476, 1159–1172. doi: 10.1042/BCJ20180707
Nuruzzaman, M., Gupta, M., Zhang, C., Wang, L., Xie, W., Xiong, L., et al. (2008). Sequence and expression analysis of the thioredoxin protein gene family in rice. Mol. Genet. Genomics 280, 139–151. doi: 10.1007/s00438-008-0351-4
Nuruzzaman, M., Sharoni, A. M., Satoh, K., Al-Shammari, T., Shimizu, T., Sasaya, T., et al. (2012). The thioredoxin gene family in rice: genome-wide identification and expression profiling under different biotic and abiotic treatments. Biochem. Biophys. Res. Commun. 423, 417–423. doi: 10.1016/j.bbrc.2012.05.142
Qin, Y., Leydon, A. R., Manziello, A., Pandey, R., Mount, D., Denic, S., et al. (2009). Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PloS Genet. 5, e1000621. doi: 10.1371/journal.pgen.1000621
Richter, A. S., Pérez-Ruiz, J. M., Cejudo, F. J., Grimm, B. (2018). Redox-control of chlorophyll biosynthesis mainly depends on thioredoxins. FEBS Lett. 592, 3111–3115. doi: 10.1002/1873-3468.13216
Rouhier, N., Cerveau, D., Couturier, J., Reichheld, J. P., Rey, P. (2015). Involvement of thiol-based mechanisms in plant development. Biochim. Biophys. Acta 1850, 1479–1496. doi: 10.1016/j.bbagen.2015.01.023
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., Yamaguchi-Shinozaki, K. (2002). DNA-Binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sevanian, A., Ursini, F. (2000). Lipid peroxidation in membranes and low-density lipoproteins: similarities and differences. Free Radic. Biol. Med. 29, 306–311. doi: 10.1016/s0891-5849(00)00342-7
Sheshadri, S. A., Nishanth, M. J., Simon, B. (2016). Stress-mediated cis-element transcription factor interactions interconnecting primary and specialized metabolism in planta. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01725
Skopelitis, D. S., Paranychianakis, N. V., Paschalidis, K. A., Pliakonis, E. D., Delis, I. D., Yakoumakis, D. I., et al. (2006). Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18, 2767–2781. doi: 10.1105/tpc.105.038323
Sytykiewicz, H., Łukasik, I., Goławska, S., Sprawka, I., Goławski, A., Sławianowska, J., et al. (2020). Expression of thioredoxin/thioredoxin reductase system genes in aphid-challenged maize seedlings. Int. J. Mol. Sci. 21, 6296. doi: 10.3390/ijms21176296
Szabados, L., Savouré, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Vieira Dos Santos, C., Rey, P. (2006). Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 11, 329–334. doi: 10.1016/j.tplants.2006.05.005
Wei, Q., Luo, Q., Wang, R., Zhang, F., He, Y., Zhang, Y., et al. (2017). A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants. Front. Plant Sci. 81374. doi: 10.3389/fpls.2017.01374
Xue, L. J., Guo, W., Yuan, Y., Anino, E. O., Nyamdari, B., Wilson, M. C., et al. (2013). Constitutively elevated salicylic acid levels alter photosynthesis and oxidative state but not growth in transgenic populus. Plant Cell 25, 2714–2730. doi: 10.1105/tpc.113.112839
Yadav, S., Gill, S. S., Passricha, N., Gill, R., Badhwar, P., Anjum, N. A., et al. (2019). Genome-wide analysis and transcriptional expression pattern-assessment of superoxide dismutase (SOD) in rice and Arabidopsis under abiotic stresses. Plant Gene 17, 100165. doi: 10.1016/j.plgene.2018.10.001
Zandalinas, S. I., Fichman, Y., Devireddy, A. R., Sengupta, S., Azad, R. K., Mittler, R. (2020). Systemic signaling during abiotic stress combination in plants. Proc. Natl. Acad. Sci. U.S.A. 117, 13810–13820. doi: 10.1073/pnas.2005077117
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhang, F., Jiang, L., Ju, L. P., Jin, X. F., Wang, X., Zhang, X. K., et al. (2014). Cloning a novel gene of trx superfamily and developing its molecular markers related to drought resistance in common wheat. Acta Agronomica Sin. 40, 29. doi: 10.3724/SP.J.1006.2014.00029
Zhang, Y., Zhou, J., Wei, F., Song, T., Yu, Y., Yu, M., et al. (2021). Nucleoredoxin gene TaNRX1 positively regulates drought tolerance in transgenic wheat (Triticum aestivum l.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.756338
Zhong, M., Song, R., Wang, Y., Shu, S., Sun, J., Guo, S. (2020). TGase regulates salt stress tolerance through enhancing bound polyamines-mediated antioxidant enzymes activity in tomato. Environ. Exp. Bot. 179, 104191. doi: 10.1016/j.envexpbot.2020.104191
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: foxtail millet, thioredoxin, SiNRX1, drought, salt
Citation: Zhang S, Yu Y, Song T, Zhang M, Li N, Yu M, Zhou H, Yang Y, Guo S, Xu C, Tu Y, Xiang J and Zhang X (2022) Genome-wide identification of foxtail millet’s TRX family and a functional analysis of SiNRX1 in response to drought and salt stresses in transgenic Arabidopsis. Front. Plant Sci. 13:946037. doi: 10.3389/fpls.2022.946037
Received: 17 May 2022; Accepted: 02 September 2022;
Published: 26 September 2022.
Edited by:
Bo Song, Agricultural Genomics Institute at Shenzhen (CAAS), ChinaReviewed by:
Ben Zhang, Shanxi University, ChinaCopyright © 2022 Zhang, Yu, Song, Zhang, Li, Yu, Zhou, Yang, Guo, Xu, Tu, Xiang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jishan Xiang, eGlhbmdqc2hAMTI2LmNvbQ==; Xiaoke Zhang, emhhbmd4aWFva2U2NkAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.