- 1Single Cell Research Center, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 2Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
Nitrate transporter (NRT) genes that participate in nitrate transport and distribution are indispensable for plant growth, development, and stress tolerance. Spirodela polyrhiza has the smallest genome among monocotyledon plants, and it has strong nitrate absorbance and phytoremediation abilities. However, the evolutionary history, expression patterns, and functions of the NRT gene family in S. polyrhiza are not well understood. Here, we identified 29 NRT members in the S. polyrhiza genome. Gene structure and phylogeny analyses showed that S. polyrhiza nitrate transporter (SpNRTs) genes were divided into eight clades without gene expansion compared with that in Arabidopsis. Transcriptomic analysis showed that SpNRT genes have spatiotemporal expression patterns and respond to abiotic stress. Functional analysis revealed that in S. polyrhiza, SpNRT1.1 expression was strongly induced by treatment with nitrate and ammonium. Overexpression of SpNRT1.1 significantly repressed primary root length, and the number and total length of lateral roots. This was more pronounced in high ammonium concentration medium. Overexpressed SpNRT1.1 in Arabidopsis significantly improved biomass and delayed flowering time, indicating that the nitrate transport ability of SpNRT1.1 differs from AtNRT1.1. In conclusion, our results provide valuable information about the evolution of the NRT family in higher plants and the function of SpNRT1.1.
Introduction
Nitrate, the most common form of nitrogen, is a limiting macronutrient for plant growth and development. Plants absorb nitrate mainly through their roots via nitrate transporter (NRT) proteins. Based on their nitrate affinity, NRT proteins in Arabidopsis can be clustered into three subfamilies. Most of the NRT1 family proteins are low-affinity nitrate transporters. AtNRT1.1 (CHL1) is the first nitrate transporter protein identified in Arabidopsis with high or low nitrate affinity levels switched by T101 phosphorylation states (Tsay et al., 1993; Liu et al., 1999; Liu and Tsay, 2003). NRT2 proteins are high-affinity nitrate transporters and NRT3 (also known as NAR2) proteins interact with NRT2 subfamily members to regulate nitrate uptake activity (Quesada et al., 1994; Orsel et al., 2006; Yan et al., 2011).
Plant NRT proteins are involved in numerous physiological processes and developmental stages, including tissue development (Li et al., 2017; Wang et al., 2018a), hormone (abscisic acid-ABA, gibberellins, or auxin) transport (Krouk et al., 2010; Kanno et al., 2012; Boursiac et al., 2013; Tal et al., 2016), and stress tolerance through nitrate absorbance, assimilation, and signaling pathways (Guo et al., 2003; Gojon and Gaymard, 2010; Li et al., 2010). AtNRT1.1 acts as a transceptor of nitrate, sensing environmental nitrate fluctuations and altering its nitrate transport activity (Krouk et al., 2006; Remans et al., 2006; Walch-Liu and Forde, 2008; Ho et al., 2009; Wang et al., 2009). Additionally, AtNRT1.1 is involved in nitrate-induced depolarization of guard cells; chl1 mutant showed reduced nitrate accumulation in guard cells and reduced stomatal opening that improved drought stress tolerance in chl1 mutant (Guo et al., 2003). AtNRT1.1 also displays auxin transport activity and mediates nitrate-modulated root development (Krouk et al., 2010; Bouguyon et al., 2015). NRT1.1 expression in Arabidopsis represses lateral root growth under low nitrate concentration, which is the sole nitrogen source, by promoting auxin transport out of roots (Krouk et al., 2010; Bouguyon et al., 2015), whereas AtNRT1.1 promotes lateral root formation under low nitrate condition and in the presence of ammonium (Guo et al., 2001). AtNRT1.2 has been characterized as a low-affinity nitrate transporter and an ABA transporter in Arabidopsis (Kanno et al., 2012; Boursiac et al., 2013). The stability and activities of NRT1.2 are regulated by the stress response, which could influence germination and vegetative growth in Arabidopsis (Li et al., 2020). In addition, NRTs display differences in spatiotemporal expression and nitrate transport routes in plants. NRT1.11, NRT1.12, and NRT1.7 function in the xylem-to-phloem transfer route for redistributing nitrate into development tissue (Fan et al., 2009; Hsu and Tsay, 2013); NPF2.3 is involved in nitrate translocation to shoots for acclimation to salt stress (Taochy et al., 2015). NRT1.6 expresses in the vascular tissues of the silique and funiculus for delivering nitrate into developing embryos (Almagro et al., 2008). These findings indicate that NRTs in different plant species have evolved more structural variation and functional diversity for acclimation to changing environments and stresses than expected.
Spirodela polyrhiza (duckweed), which belongs to the family Lemnaceae, is a fast-growing aquatic plant with a strong nutrient absorption capacity. Under optimal growth conditions, the biomass of duckweeds can double in 30 h (Wang et al., 2014). In addition, duckweed can tolerate high concentrations of ammonia and adapt to environments with a wide pH range. This makes them suitable for being widely used in phytoremediation. Genome sequence analysis showed that the size of the S. polyrhiza genome is 158 Mb with 19,623 protein-coding genes. Spirodela polyrhiza has the smallest genome among monocotyledons, being 50% smaller than that of rice (Wang et al., 2014). It has been reported that S. polyrhiza has a higher number of genes related to the nitrogen assimilation pathway and stronger nitrate assimilation ability than rice or Arabidopsis (Wang et al., 2014). To elucidate nitrate transport in S. polyrhiza, we analyzed NRT gene families in S. polyrhiza and characterized the spatiotemporal expression of SpNRTs in different tissues and under different stress conditions. We also confirmed the functions of SpNRT1.1 and analyzed its potential to improve plant nitrogen utilization.
Materials and methods
Plant materials and culture conditions
Arabidopsis ecotype Columbia (Col-0) and CHL1 mutant (chl1-5) seeds were obtained from the Nottingham Arabidopsis Stock Centre (NASC). All seeds were sterilized with 75% ethanol for 1 min and 3% sodium hypochlorite for 10 min and plated on 1/2 Murashige and Skoog (MS) medium for germination. To analyze the effects of nitrogen concentration changes on seedling growth, MS-modified basal medium without N (Phytotechnology Laboratories)1 was used or supplemented with different concentrations of KNO3 (final concentration: 1 mm, 3 mm, or 10 mm) as the only N source. The medium contained 1.5% sucrose and 0.6% Phytoblend agar. After stratification at 4°C for 2 days, the seedlings were grown at 22°C under long-day conditions (16 h light/8 h dark) and a light intensity of 120 μmol·m−2 s−1. Root length and physiological performance of seedlings were analyzed after 8 days of growth.
Spirodela polyrhiza plants were kindly provided by Dr. Wang’s laboratory (Wang et al., 2014). The aseptic seedlings were transferred to liquid MS medium without nitrogen and cultivated for 3 days. The plants were then transferred to liquid medium with different concentrations of nitrogen (final concentration: 0.5 mm, 1 mm, and 5 mm KNO3; 0.5 mm, 1 mm, and 5 mm NH4Cl). The seedlings were grown under the same conditions as those of the Arabidopsis seedlings at 22°C.
SpNRT1.1 gene cloning and phylogenetic analysis
To clone the SpNRT1.1 gene from S. polyrhiza, the amino acid sequences of OsNRT1.1 and AtNRT1.1 were used in the basic local alignment search tool BLASTX to search for homologous genes in the S. polyrhiza genome. The best-matched gene Spipo1G0085300 (Phytozome), probably encoding SpNRT1.1 protein in S. polyrhiza was selected as the candidate reference sequence. To clone the full-length SpNRT1.1 gene, the thermal asymmetric interlaced-polymerase chain reaction (Tail-PCR) method was performed using primers AD1-3 and R1-R3 (Supplementary Table 1; Liu et al., 1995; Liu and Chen, 2007). Phylogenetic analysis was performed to analyze the evolutionary relationship based on NRT1 proteins from the following nonvascular plants: Selaginella moellendorffii, Physcomitrella patens; Gymnospermae plants: Picea abies; and Angiospermae plants: Vitis vinifera, Populus trichocarpa, Brachypodium distachyon, Oryza sativa, Arabidopsis thaliana, and S. polyrhiza, using the maximum likelihood method by Mega X (Stecher et al., 2020).
Subcellular localization analysis of SpNRT1.1 protein
SpNRT1.1 CDS sequence was amplified and recombined into pEarlyGate 101 plasmid to generate the expression cassette CaMV35S:: SpNRT1.1-eYFP::NOS. The SpNRT1.1 expressing plasmid and pEarlyGate104 plasmid was transformed into the Agrobacterium strain GV3101. After PCR and DNA sequencing, positive clones were selected and cultured overnight. When the cultured Agrobacterium reached OD600 = 0.6–1.0, the bacterium was centrifuged at 4000 rpm for 10 min at room temperature. The collected pellets were dissolved in a transforming solution. One-month-old tobacco leaves were infiltrated with transformation solution according to a previously reported method (Grefen et al., 2010). The infiltrated tobacco plants were kept under dark conditions for 24 h, and then transferred to normal growth conditions. The fluorescent signals were imaged by Lecia TCS SPS-II 2–3 days after infiltration.
Generating transgenic SpNRT1.1 Arabidopsis plants
The open reading frame of SPNRT1.1 was inserted into the pHB binary vector to generate the expression cassette CaMV35S::SPNRT1.1-YFP::NOS. The plasmid harboring SPNRT1.1 gene expression cassette was then transformed into Agrobacterium GV3101 to generate transgenic Arabidopsis plants. The 3–4-week-old Col-0 or chl1-5 plants were used for flower dipping (Clough and Bent, 1998; Zhang et al., 2006). The positive plants were confirmed by hygromycin screening and PCR. Transgenic plants were self-crossed to generate T3 homozygous lines.
NaClO3 uptake analysis
The seeds of Arabidopsis ecotype Col-0, NRT1.1 mutant chl1-5, and transgenic plants (T3 homozygous lines) were sterilized with 70% ethanol for 1 min and 3% sodium hypochlorite for 10 min, and sown on 1/2 MS-modified basal medium with 1 mm NH4NO3. The seeds were stratified at 4°C for 3 days under dark conditions and then grown at 22°C under long-day conditions. 4 days later, Arabidopsis seedlings were transferred to 1/2 MS medium containing 2 mm NaClO3. The chlorophyll content and fresh weight of Arabidopsis seedlings were recorded after 4 days of treatment, and 6 to 10 plants were mixed as one sample. To determine the primary root length and lateral root length of Arabidopsis seedlings, they were photographed using a Canon camera and the measurements were calculated using ImageJ software.
RT-qPCR analysis of gene expression
Total RNA was extracted using a Total Plant RNAprep Plant Kit (TIANGEN, China). cDNA was synthesized using the HiFiScript gDNA Removal RT MasterMix (CWBIO, China). Quantitative RT-PCR was performed according to the manual from the MagicSYBR Mixture Kit (CWBIO, China) using a LightCycler 96 System (Roche, Germany). Gene expression was calculated using the ΔΔCT method, and SpACTIN6 (Spipo12G0023800) and AtACTIN2 (Wang et al., 2018b) were used as internal controls. RT-qPCR primers used are listed in Supplementary Table 1.
Data analysis
Student’s t-test were used to assess the statistical significance between the two groups. All analyses were repeated at least three times. Prism 8 software was used for the statistical analysis and to generate the diagrams.
Results
Genome-wide identification of NRT gene families in the Spirodela polyrhiza genome
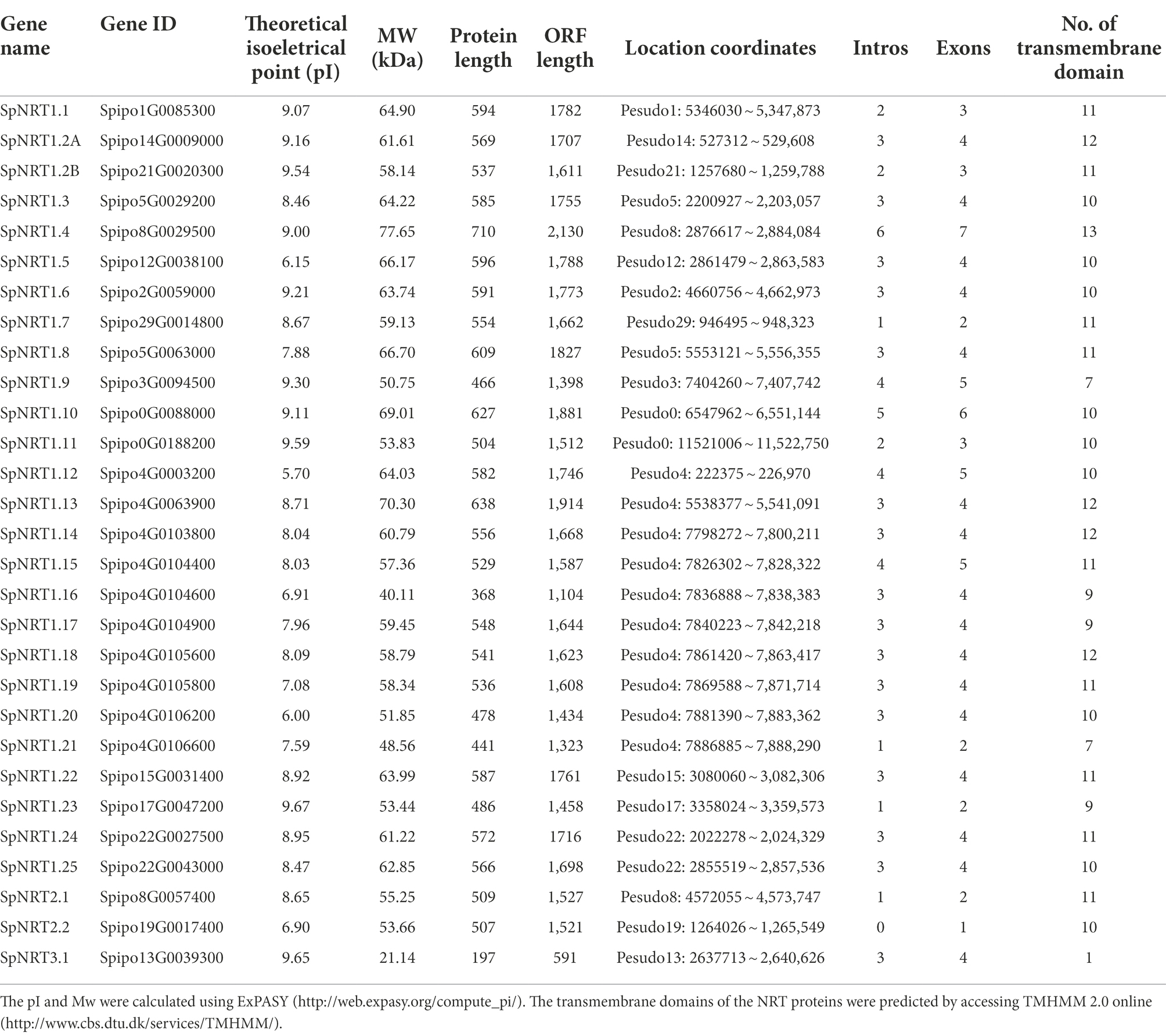
To identify NRT gene families in S. polyrhiza, we used 12 functionally identified NRT1, 7 NRT2, and 2 NRT3 proteins from Arabidopsis and 14 NRT1, 4 NRT2, and 2 NRT3 proteins from Oryza sativa. BLASTP and hidden Markov models (HMM) search methods were used to search NRT homologs in the S. polyhiza genome (GeneBank Accession No.: SRX5321175). We identified 26 NRT1 genes, 2 NRT2 genes, and 1 NRT3 gene from the S. polyhiza genome (Supplementary Table 2). Based on sequence coordinates and similarities with their homologs in Arabidopsis, we named all 29 SpNRT genes from SpNRT1.1 to SpNRT3.1 (Table 1). Ten SpNRT genes were distributed in pseudomolecules 4. Pseudomolecules 5, 8, and 22 contained two genes. Other pseudomolecules had only one gene. Two SpNRT genes were not assigned to any known pseudomolecules; therefore, they were labeled pseudomolecules 0. Exon/intron structure analysis showed greater structural divergence among SpNRT genes (Figure 1B). Most SpNRTs had four exons, whereas four SpNRT genes had two exons, three SpNRT genes had three exons, and three SpNRT genes had five exons. The difference between SpNRT genes in the exon/intron structure indicated that functional diversity probably existed in different NRT families.
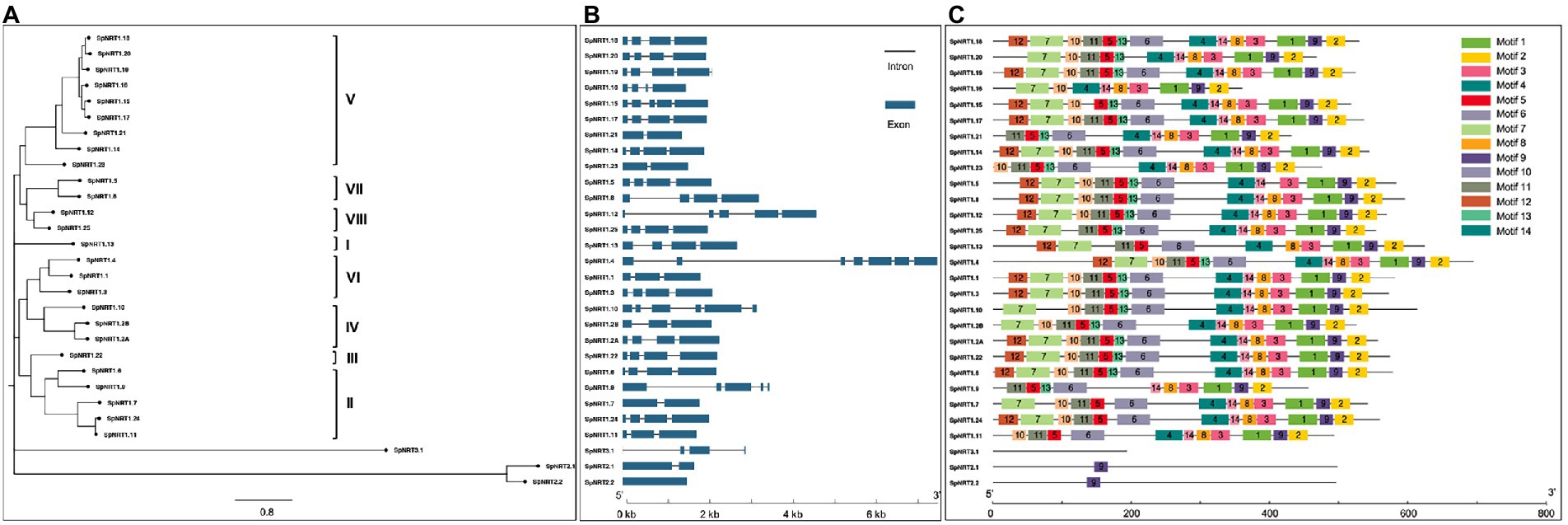
Figure 1. SpNRTs gene structure analysis and the conserved motifs in the SpNRT protein families. (A) Neighbor-joining method was used to construct a phylogenetic tree of SpNRT proteins by MEGA X software. All the parameters adopted the reported default values (Stecher et al., 2020). (B) Gene structure analysis of SpNRTs. The sequences of SpNRTs and gene structure information were obtained from the Ensemble plants database, and the GSDS program was used to display Gene structure (http://gsds.cbi.pku.edu.cn; Hu et al., 2015). (C) The predicted conserved motif of SpNRT proteins by the MEME program http://meme-suite.org/tools/meme (Bailey and Elkan, 1994; Bailey et al., 2015). TBtools was used to display the motif location (Chen et al., 2020).
To identify the evolutionary relationship between SpNRTs and other homologous proteins, we constructed a multi-species phylogenetic tree using the maximum likelihood method based on 582 NRT proteins from Volvox carteri, Selaginella moellendorffii, Physcomitrella patens, Picea abies, Vitis vinifera, Populus trichocarpa, Brachypodium distachyon, Oryza sativa, Arabidopsis thaliana, and S. polyhiza. All 582 proteins were divided into three subfamilies, of which 516 proteins belonged to the NRT1 subfamily, 48 proteins belonged to the NRT2 subfamily, and 18 proteins belonged to the NRT3 subfamily (Supplementary Figure 1; Supplementary Table 3). SpNRT1 subfamilies were further divided into eight clades: Clade I (one protein), Clade II (five proteins), Clade III (one protein), Clade IV (three proteins), Clade V (nine proteins), Clade VI (three proteins), Clade VII (two proteins), and Clade VIII (two proteins; Figure 1A). Protein structure analysis indicated that each NRT subfamily had similar conserved motifs and transmembrane domains (TM), similar to homologous proteins from Arabidopsis. The number of TM in SpNRT1 proteins ranged from 7 to 13; SpNRT2 proteins had 10 or 11 TMs, whereas SpNRT3.1 had only 1 TM. The conserved TM domain structure in SpNRTs indicated that they might have nitrate transport ability similar to that of the orthologs in Arabidopsis.
Amino acid alignment of SpNRTs identified 14 motifs among SpNRT proteins, 13 of which were located in the TM areas of SpNRT1 proteins (Figure 1C; Supplementary Figure 2). Motif 1 is located in the TM10 and TM 10-TM 11 loops, in which the 34th amino acid is usually P, and its mutation can result in the loss of nitrate transporter function of AtNRT1.1 (Supplementary Figures 3, 4). Motif 4 was located in TM 7, in which the 24th amino acid is the nitrate-binding site affecting the nitrate uptake efficiency of NRT1.1 in maize (Sun et al., 2014; Wen et al., 2017). Motif 6 with interface residues was located in TM 5, TM 5-TM 6 loop, or TM6. Motif 10 was located in the TM 3 or TM 3-TM 4 loop, where the 21st amino C forms a disulfide linkage with the 5th C in motif 11. Motif 12, with the 20th-ExxERFxYY-28th motif, is functional in proton coupling (Supplementary Figures 3, 4). SpNRT3 had no motif similar to SpNRT1s and SpNRT2s. The SpNRT2 subfamily had a motif (motif 9) similar to SpNRT1s. Motif 9 is usually located from the 150th to 170th amino acid in SpNRT2.1 or from the 139th to 159th amino acid in SpNRT2.2, in which Phe511 could form a nitrate-binding pocket in AtNRT1.1 (Sun et al., 2014). These changes in the motif structure indicate that SpNRTs have their own characteristics in nitrate transport.
Expression patterns of SpNRTs in different tissues and under abiotic stresses
To analyze the expression patterns of SpNRTs in different tissues, we used RNA-seq data from leaves, stipules, and roots of S. polyrhiza to compare their transcriptional levels (Supplementary Tables 4, 5). The expression patterns of the 29 SpNRT genes were classified into three groups (Figure 2A). Group I was highly expressed in the leaves and contained seven genes, including SpNRT1.18, SpNRT1.13, SpNRT1.2A, SpNRT1.2B, SpNRT1.22, SpNRT1.19, and SpNRT1.25. Group II genes with higher expression in stipules included the following ten NRT genes: SpNRT1.23, SpNRT1.3, SpNRT1.6, SpNRT1.11, SpNRT1.21, SpNRT1.4, SpNRT1.20, SpNRT1.15, SpNRT1.1, and SpNRT1.12. Group III genes were highly expressed in roots, these were SpNRT1.14, SpNRT1.5, SpNRT1.7, SpNRT1.8, SpNRT1.9, SpNRT1.16, SpNRT1.17, SpNRT1.24, SpNRT2.1, SpNRT2.2, SpNRT3.1, SpNRT2.1, SpNRT2.2, and SpNRT3.1. This suggests that SpNRT3.1 may have the same function as AtNRT3.1 in Arabidopsis, in which SpNRT3.1 cooperates with SpNRT2s in nitrate transport. The difference in expression in different tissues indicated that SpNRTs function in different plant growth processes across tissues.
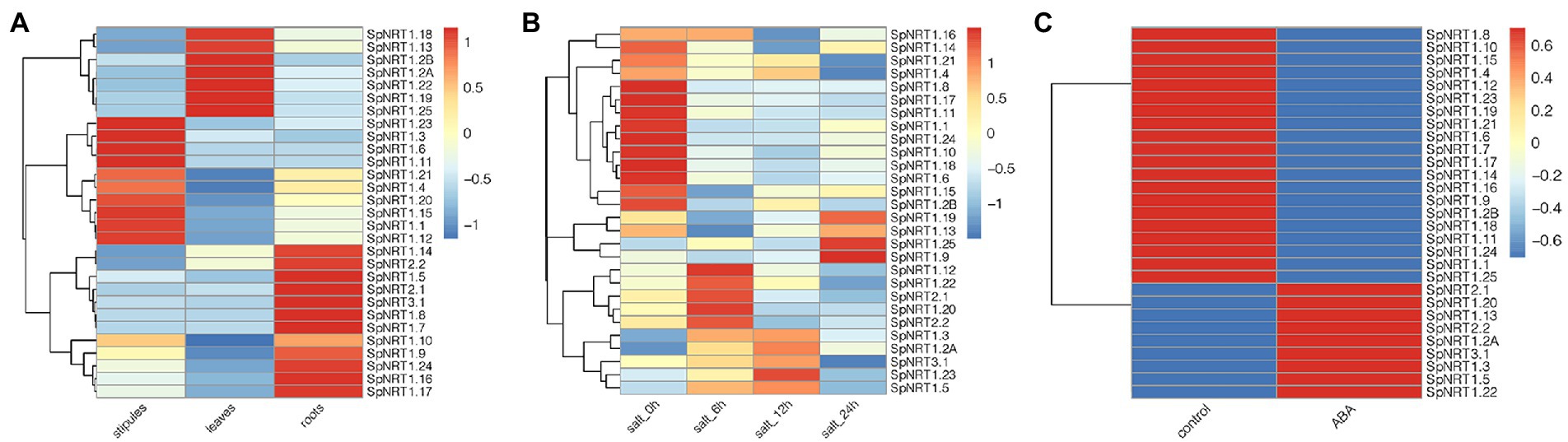
Figure 2. Expression patterns of SpNRT genes. (A) Relative expression level of SpNRT genes in various tissues determined by RNA-seq FPKM value. (B) Relative expression level of SpNRT genes response to salt stress determined by RNA-seq FPKM value. (C) Relative expression level of SpNRT gene response to ABA stress determined by RNA-seq FPKM value. Red means relative expression high and blue means lower. SpNRTs RNA-seq data downloaded from NCBI Sequence Read Archive (SRA) database were used to express profile analysis, all the data is listed in Supplementary Tables 4–7. FPKM (fragments per kilobase of transcript per million fragments mapped) value standardized by the Z-score method of every SpNRT gene was used to generate heatmaps by TopHat and Cuffilinks software.
Duckweeds are widely recognized in phytoremediation with a high tolerance to abiotic stresses. We further analyzed the expression patterns of SpNRTs in response to salt stress (Figure 2B; Supplementary Tables 4, 6). The expression of SpNRTs in Group I was strongly inhibited by salt stress treatment, which included 14 (48.3%) NRT genes and 6 from the NPF5 group (9 NPF5 genes in S. polyrhiza, Figure 1A). The expression of SpNRT genes in Group II was initially inhibited by salt stress after 6 h or after 12 h; thereafter, the expression increased significantly. Group II contained four SpNRTs from four different groups. In contrast to Group I and II, the expression of SpNRTs in Group III was clearly induced by salt stress at 6 h but significantly decreased after 24 h of treatment. Group III contained 10 genes, including all genes from the NRT2 and NRT3 subfamilies. Interestingly, the expression of SpNRTs showed two completely different responses to ABA treatment, either strongly increasing or decreasing after ABA treatment (Figure 2C; Supplementary Tables 4, 7), indicating that SpNRTs likely transport or redistribute nitrate under different abiotic stress conditions.
SpNRT1.1 gene cloning and phylogenetic analysis
NRT1.1 is an important nitrate sensor and nitrate transporting protein in Arabidopsis; therefore, we cloned the full-length cDNA of SpNRT1.1 gene by Tail-PCR and analyzed its function. SpNRT1.1 gene encodes a protein with 594 amino acids and a theoretical isoelectric point of 9.07. SpNPF6 subfamily proteins aligned with NPF6 proteins from gymnosperms and angiosperms, indicating that SpNRT1.1 was close to OsNRT1.1A with 70% similarity. The phylogenetic tree showed that the NPF6s from nonvascular plants, including Selaginella tamariscina and Physcomitrella patens, were clustered into one independent clade with the greatest genetic distance from other NPF6s (Supplementary Figure 5). The subcategory number of NPF6 proteins tended to show increased levels in angiosperms relative to gymnosperms. No NRT1.1 homolog was identified in gymnosperms, but NRT1.1, NRT1.3, and NRT1.4 subcategories were found in angiosperms. The conserved domain showed that SpNRT1.1 has 12 TMs, the conserved phosphorylation site is T102 on TM3, and the T101 site corresponds to a functional switch from high-affinity to low-affinity nitrate transport in Arabidopsis (Supplementary Figure 3). The NO3− binding site on TM7 (Y353) in SpNRT1.1 was similar to that in OsNRT1.1A and OsNRT1.1C but different from those of AtNRT1.1 and OsNRT1.1B (Supplementary Figure 4). The structure of SpNRT1.1 indicated that the nitrate transporting activity of SpNRT1.1 may be different from that of AtNRT1.1.
SpNRT1.1 protein is localized on the plasma membrane
To analyze the subcellular localization of the SpNRT1.1 protein, SpNRT1.1-eYFP fusion protein was transiently expressed in the epidermal cells of tobacco leaves. The fluorescence signals of SpNRT1.1-eYFP protein were found on the plasma membrane, compared with eYFP signals appearing in the nucleus, cytoplasm, and cell membrane (Figure 3), indicating that SpNRT1.1 is a transmembrane protein.
Figure 3. Localization of SpNRT1.1 protein in the plasma membrane of tobacco epidermal leaves. The CDS sequence of SpNRT1.1 gene fused with the eYFP sequence under the control 35S promoter was transiently expressed in tobacco leaves (A–C), using the eYFP protein as the control (D–F). (A,D) eYFP fluorescence; (B,E) bright field images; (C,F) merged eYFP fluorescence with bright field. Scale bar = 25 μm.
SpNRT1.1 gene expression induced by nitrate and ammonium in Spirodela polyrhiza
To analyze the expression patterns of SpNRT1.1 gene in S. polyrhiza, we transferred cultured duckweed plants grown in a nitrogen-free medium into the liquid 1/2 MS medium with different concentrations of nitrogen (0.5 mm, 1 mm, and 5 mm NO3−; or 0.5 mm, 1 mm, and 5 mm NH4+). RT-qPCR analysis showed that SpNRT1.1 expression was induced by both nitrate and ammonium, with transcription levels reaching the peak 30 min after treatment under both low and high concentrations (Supplementary Figure 6). Compared with the AtNRT1.1 expression in Col-0 plants, the SpNRT1.1 transcripts were 100 times higher than that of AtNRT1.1 in Arabidopsis (Supplementary Figure 6C), indicating that SpNRT1.1 is functional in both ammonium and nitrate in S. polyrhiza, which is completely different from AtNRT1.1 in Arabidopsis.
SpNRT1.1 complements the phenotype of chl1-5 mutant in arabidopsis under chlorate-supplied conditions
ClO3− can be reduced by nitrate reductase to chlorite, which is toxic to plants, resulting in etiolated leaves of chl1-5 mutant plants. Therefore, it can be used for nitrate uptake analysis (Tsay et al., 1993; Huang et al., 1999). To check whether SpNRT1.1 functions in the uptake of ClO3−, 2 mm ClO3− was added to 1/2 MS medium to treat chl1-5 mutant plants which had been complemented by SpNRT1.1. Chlorophyll content and fresh weight of shoots were significantly decreased in both Col-0 and SpNRT1.1 complemented plants and SpNRT1.1 overexpressed plants compared with chl1-5 mutant plants (Figure 4). These results suggest that SpNRT1.1, which functions in nitrate absorption, is sensitive to chlorate.
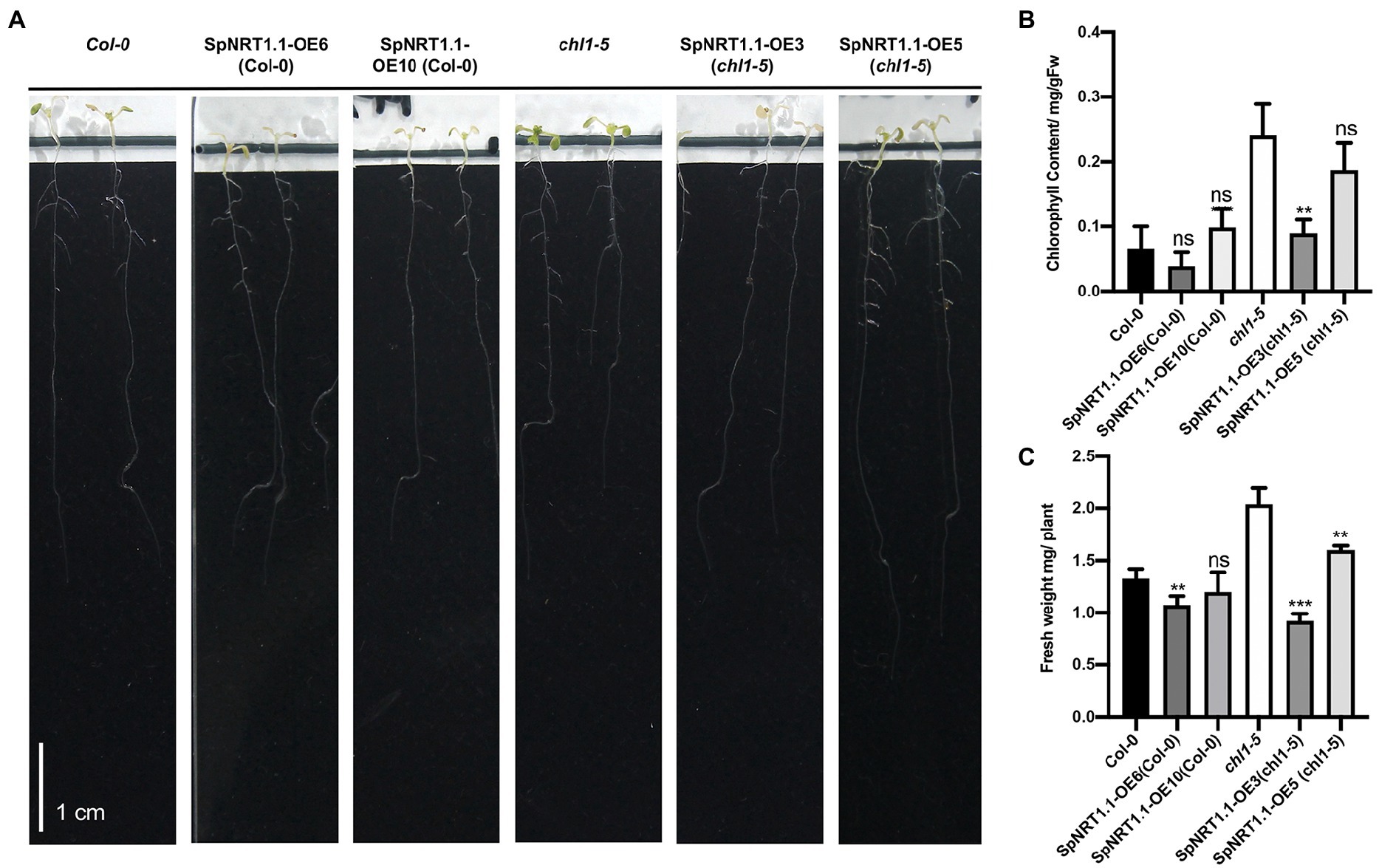
Figure 4. Overexpression of SpNRT1.1 in Col-0 and chl1-5 in Arabidopsis leads to etiolated phenotype under chlorate-supplied conditions. (A) Phenotype of SpNRT1.1 overexpressed lines and complemented lines under chlorate-supplied conditions. (B) Chlorophyll content of Col-0 and transgenic lines compared with chl1-5 plant. (C) Shoot fresh weight of Col-0 and transgenic lines compared with chl1-5 plant. Student’s t-test was used for significant difference analysis. Asterisks indicate significance levels compared with chl1-5 mutant plants, n ≥ 3, **p < 0.01, ***p < 0.001. Scale bar = 1 cm.
Overexpressed SpNRT1.1 gene in Arabidopsis represses root development
To analyze the function of SpNRT1.1 in Arabidopsis, we analyzed the root phenotypes of Col-0 and chl1-5 with ectopically expressed SpNRT1.1 in plants growing in 1/2 MS medium under different concentrations of nitrate (0.3 mm, 3 mm, and 10 mm KNO3; Figure 5A). Primary root length of SpNRT1.1 overexpressing plants slightly decreased (Figures 5B,E,H); the total lateral root length (Figures 5C,F,I) and lateral root number (Figures 5D,G,J) of SpNRT1.1 overexpressing plants were significantly repressed compared to those of Col-0 and chl1-5 plants after 8 d of growth. These results indicate that overexpression of SpNRT1.1 in Arabidopsis has negative effects on root growth.
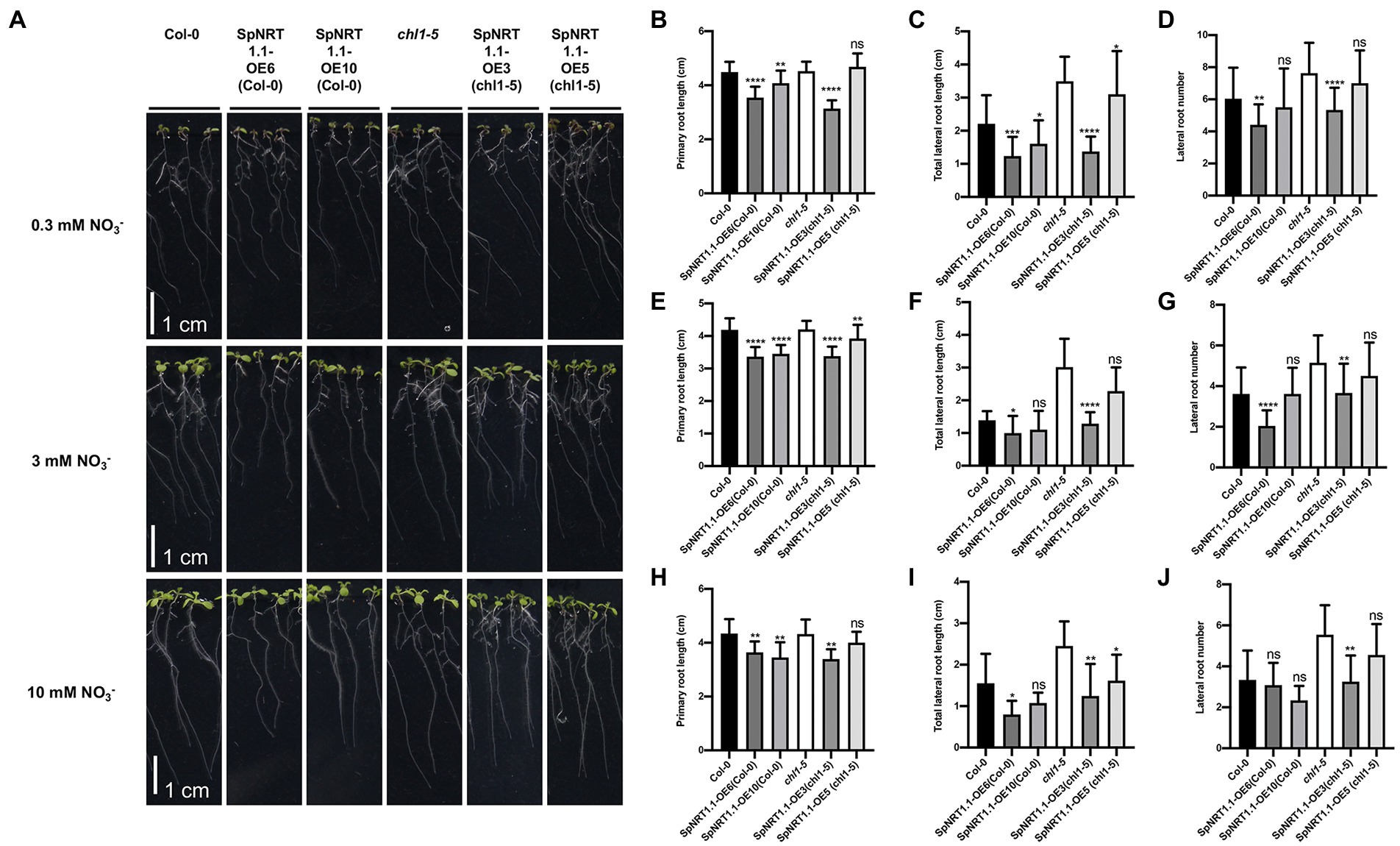
Figure 5. Overexpression of SpNRT1.1 in Col-0 and chl1-5 plants suppress primary and lateral root growth under different nitrate concentrations. (A) Phenotype of SpNRT1.1 overexpression lines and complemented lines under different nitrate-supplied concentrations. (B–J) Primary root length, lateral root number, and total lateral root lengths of SpNRT1.1 overexpression lines and complemented plants under 0.3 mm NO3−-supplied condition (B–D), 3 mm (E,F), 10 mm (H–J). Student’s t-test was used for significant difference analysis. Asterisks indicate significance levels compared with chl1-5 mutant plants, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n ≥ 10, Scale bar = 1 cm.
AtNRT1.1 can regulate ammonium absorbance, leading to pH and auxin changes in roots, and AtNRT1.1 mutant plants are tolerant to ammonium/low pH in the growing medium (Hachiya and Noguchi, 2011; Meier et al., 2020). To analyze the effects of SpNRT1.1 on root growth, Col-0, chl1-5, and SpNRT1.1 overexpressing plants were grown in 1/2 MS medium with different concentrations of ammonium. Under conditions of high concentrations of ammonium (3 mm and 10 mm ammonium), primary root length and the number of lateral roots in the SpNRT1.1 overexpressing lines were significantly decreased (Figure 6), indicating that SpNRT1.1 also functions in ammonium absorbance.
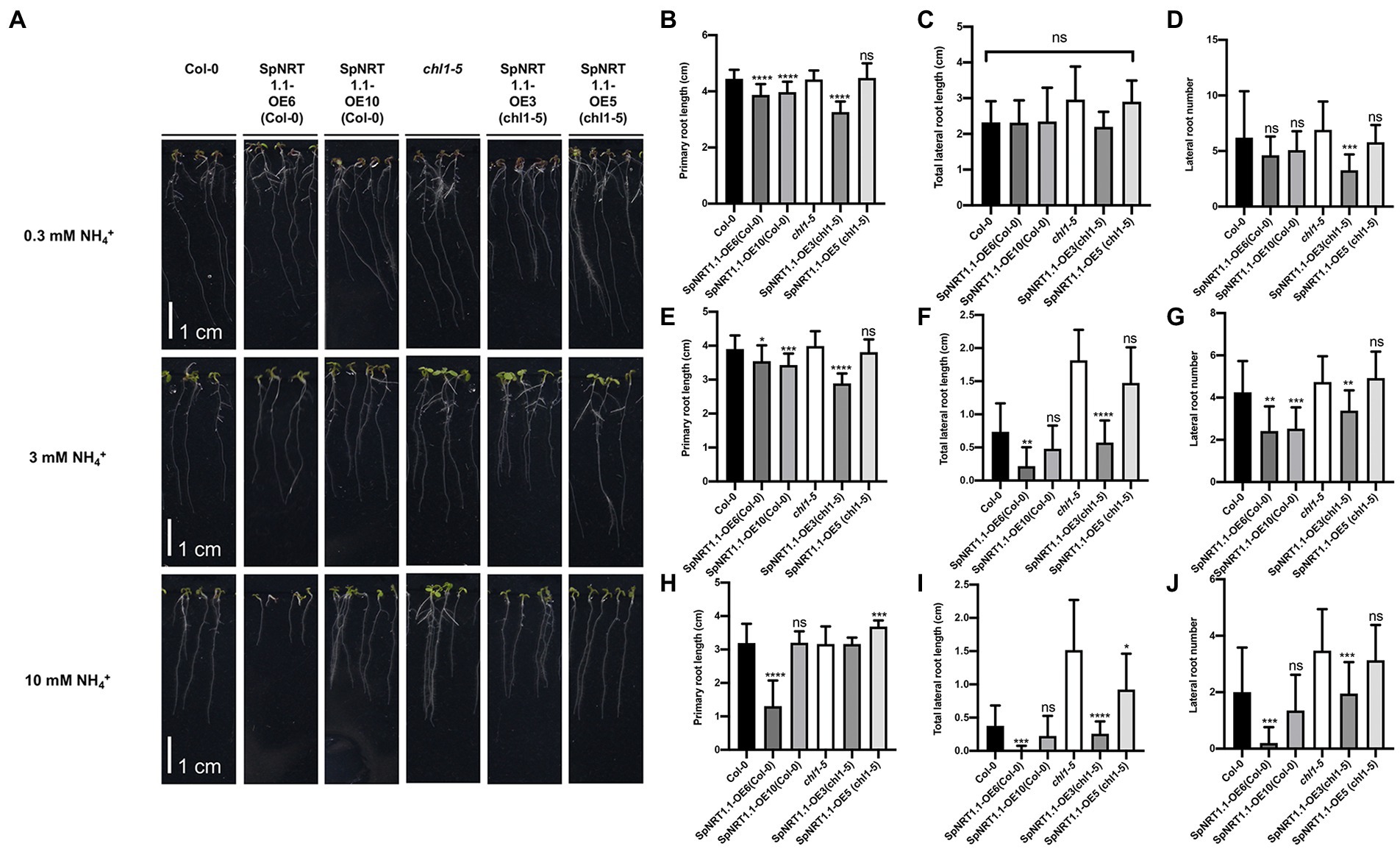
Figure 6. Overexpression of SpNRT1.1 in Col-0 and chl1-5 in Arabidopsis suppresses primary and lateral root growth under different ammonium concentrations. (A) Phenotype of SpNRT1.1 overexpressed lines and complementary lines under different ammonium-supplied concentrations. (B–J) Primary root length, lateral root number, and total lateral root length of SpNRT1.1 overexpressed and complementary plants under 0.3 mm NH4+-supplied condition (B–D), 3 mm (E,F), 10 mm (H–J). Student’s t-test was used for significant difference analysis. Asterisks indicate significance levels compared with chl1-5 mutant plants, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n ≥ 10, Scale bar = 1 cm.
To further analyze whether SpNRT1.1 has a similar function in ammonium tolerance (Hachiya and Noguchi, 2011), we measured the chlorophyll content and fresh weight of SpNRT1.1 complemented plants and Col-0 plants in the medium containing 3 and 10 mm ammonium, respectively. The chlorophyll content and fresh weight of SpNRT1.1 complemented and Col-0 plants were decreased compared with those of chl1-5 mutant plants (Figure 7); they were found to decrease with the increase in ammonium concentration in the medium. The chlorophyll content and fresh weight of SpNRT1.1 complemented plants decreased slightly compared with Col-0 plants. These results suggest that SpNRT1.1 has a similar but weaker sensitivity to ammonium compared with AtNRT1.1 protein.
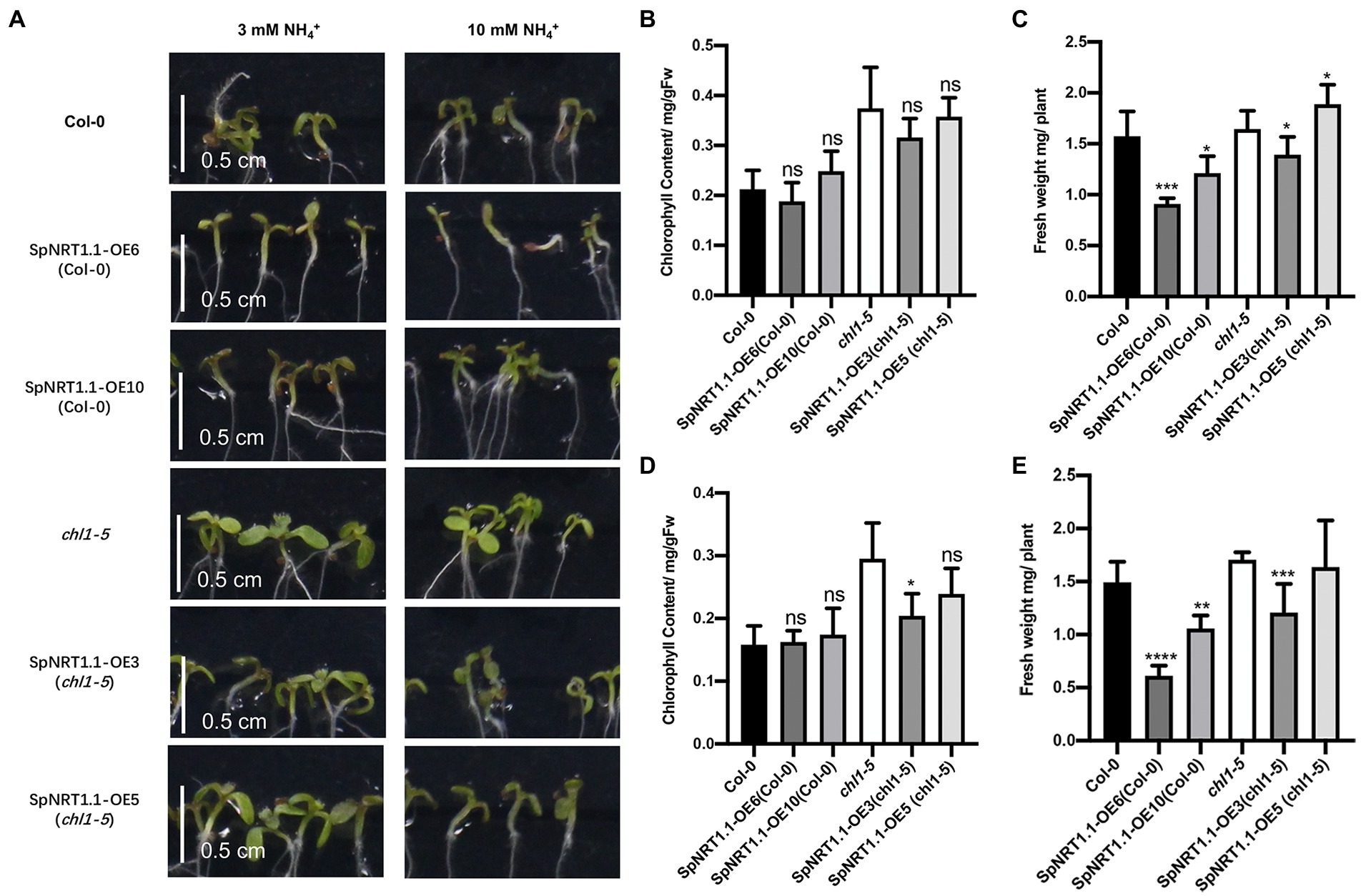
Figure 7. Overexpression of SpNRT1.1 in Col-0 and chl1-5 plants leads to sensitivity to ammonium. (A) Phenotype of SpNRT1.1 overexpressed and complemented plants under different ammonium concentrations. Chlorophyll content and fresh weight under 3 mm NH4+ (B,C) and 10 mm NH4+ (D,E). Student’s t-test was used for significant difference analysis. Asterisks indicate significance levels compared with chl1-5 mutant plants, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, n ≥ 5, Scale bar = 0.5 cm.
Overexpression of SpNRT1.1 in Arabidopsis delayed flowering and increased biomass production
To analyze the effect of SpNRT1.1 overexpression on plant growth, Col-0 and SpNRT1.1 overexpressing plants were grown in vermiculite soil with 10 mm nitrate. After 1 month of growth, the flowering time of SpNRT1.1 overexpressing lines was delayed by approximately 5 days, gene expression analysis that SpNRT1.1 overexpression repressed the transcription levels of SOC1 gene, which directly activates floral identity gene LEAFY (Lee and Lee, 2010). Reproduction transition gene FT and FD gene expression were repressed to some extent by SpNRT1.1 overexpression (Supplementary Figure 7). And their biomass increased by approximately 60% compared with Col-0 plants (Figure 8), indicating that SpNRT1.1 expression influences nutrition absorbance and maturation in Arabidopsis.
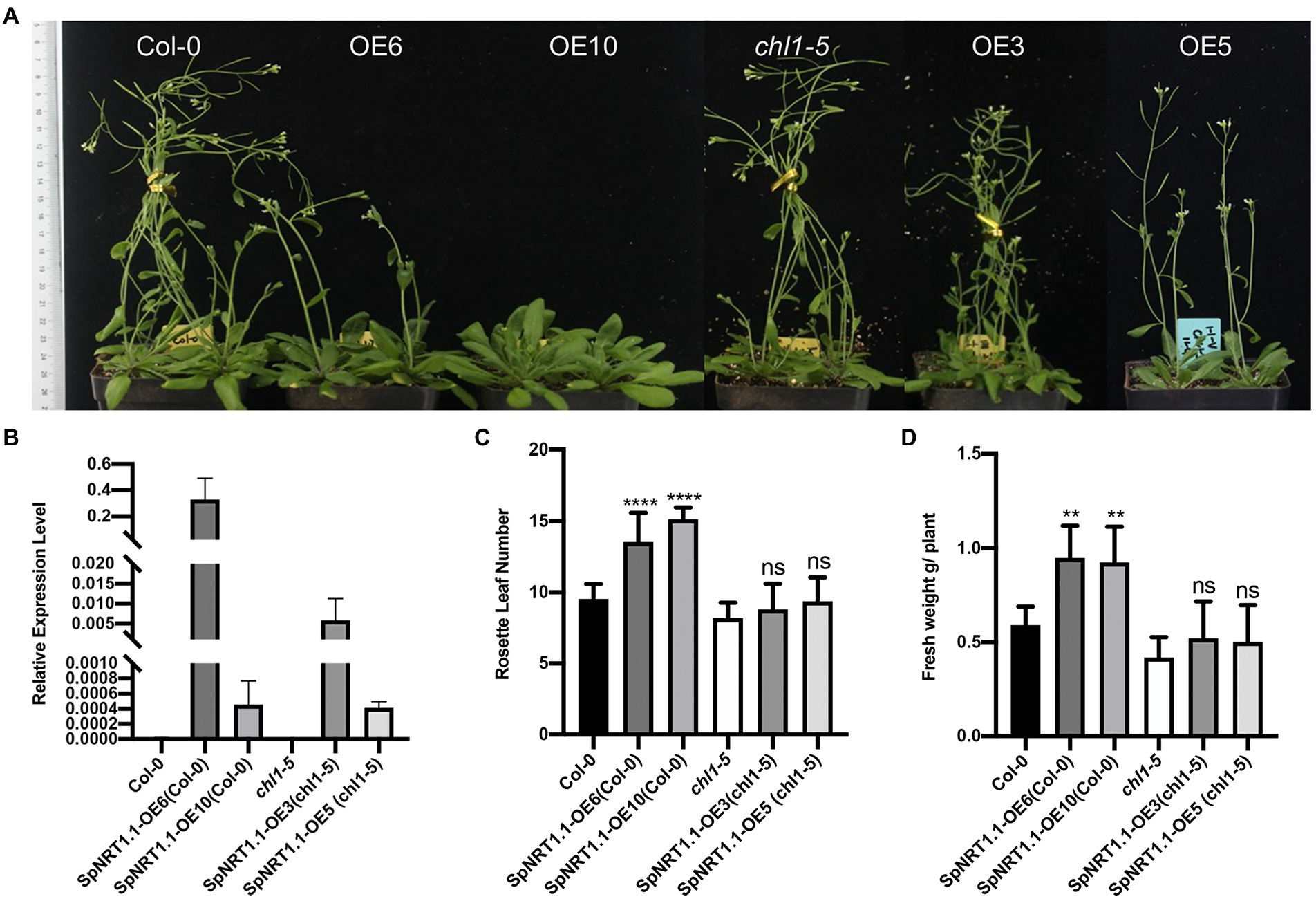
Figure 8. Overexpression of SpNRT1.1 in Col-0 and chl1-5 plants leads to late flowering time and improved biomass. Ordinary one-way ANOVA was used for statistical significance analysis, n ≥ 5, p < 0.05. (A) Phenotype of SpNRT1.1 overexpressed and complemented plants. (B) Relative expression level of SpNRT1.1 gene. Expression data display in Supplementary Table 8. (C) Rosette leaf number and (D) Fresh weight of SpNRT1.1 overexpressed and complemented plants. Student’s t-test was used for significant difference analysis. Asterisks indicate significance levels compared with chl1-5 mutant plants, **p < 0.01, ****p < 0.0001, n ≥ 5.
Discussion
Spirodela polyrhiza, a duckweed, is the smallest, fast-growing, and morphologically simplest flowering plant. The rapid growth rate of S. polyrhiza requires sufficient nutrients, including nitrogen and phosphorus, to maintain its development and normal life cycle. Many S. polyrhiza protein families have been identified such as the lipoxygenase gene family (Upadhyay et al., 2020), WRKY gene family (Zhao et al., 2021), and polyamine biosynthesis pathway (Upadhyay et al., 2021). NRT proteins act as transporters that acquire nitrogen from the environment. However, there is limited knowledge regarding the evolution, protein structure, and function of NRT families in S. polyhiza. The SpNRTs gene family was identified in this study using BLASTP and HMM search methods. A total of 29 NRT genes were identified from the S. polyrhiza genome, and they were divided into three NRT subfamilies: 26 NPF (NRT1/PTR, NPF), 2 NRT2, and 1 NRT3. Compared with NRT proteins in the rice genome, S. polyrhiza has fewer NRT1 family members. The stronger nitrate absorbance ability of S. polyrhiza does not result from the expansion of NRT proteins during evolution compared with other monocotyledons. For example, S. polyrhiza has only one NRT1.1 gene, whereas rice has four NRT1.1 members (NRT1.1A, NRT1.1B, NRT1.1C, and NRT1.1D), and maize has four NRT1.1 genes. SpNRT1.1 s possess low identity with their counterparts from Arabidopsis (61%) and rice (70%). This indicates that the SpNRT1 protein may have a different protein structure and nitrate transport ability from NRT1 in Arabidopsis or rice.
Conserved domain analysis showed that SpNRTs contain all the conserved TM domains and motifs in the NRTs of Arabidopsis, and motif 9 specifically exists in SpNRT1 and SpNRT2 proteins. The 10th amino acid of motif 9 is the hydrophobic residue Phe511, which, in association with Leu49, Val53, and Leu78 can form a nitrate-binding pocket similar to AtNRT1.1. This result indicated that motif 9 in SpNRT1s and SpNRT2s is probably involved in nitrate uptake activity. In addition, SpNRT1 proteins also have conserved amino acid residues responsible for nitrate transport in AtNRT1.1 (the 24th amino acid in Motif4, and the 34th amino acid in motif 1). Taken together, these results indicate that SpNRT1.1 has nitrate transport ability, and its activities are slightly different from those of AtNRT1.1.
NRT1.1 is the most important nitrate transporter and receptor. It is identified and characterized in various species. NRT1.1 members in different plant species have different nitrate absorbance and nitrate signal sensing characteristics. Rice OsNRT1.1A is a low-affinity nitrate transporter, and overexpression OsNRT1.1A in rice improves rice yield and significantly promotes early maturation (Wang et al., 2018b). OsNRT1.1B is a constitutive nitrate transporter, and overexpression of OsNRT1.1B increased nitrogen accumulation under both low and high nitrate conditions, but OsNRT1.1A increased nitrogen accumulation in rice only under high nitrate conditions (Fan et al., 2016). Maize has 4 NRT1.1 genes; ZmNRT1.1A-D, of which ZmNRT1.1A (ZmNPF6.4) is a low-affinity nitrate transporter, and ZmNRT1.1B (ZmNPF6.6) is a pH-dependent non-biphasic high-affinity nitrate transporter (Plett et al., 2010; Wen et al., 2017; Wang et al., 2020). These diverse characteristics prompted us to investigate the functions of SpNRT1.1.
SpNRT1.1 expression was strongly induced by nitrate and ammonium. This inducible expression pattern is similar to that of OsNRT1.1A, indicating that SpNRT1.1 is probably involved in the rapid ammonium utilization in aquatic plants (Wang et al., 2018b). The chl1-5 plants complemented by SpNRT1.1 displayed decreased ammonium toxicity compared with Col-0 and SpNRT1.1 overexpression lines. We deduced that SpNRT1.1 has a greater ammonium tolerance than AtNRT1.1 (Hachiya and Noguchi, 2011), which benefits the growth of S. polyrhiza in water condition. The biomass and flowering time of SpNRT1.1 overexpressing plants were drastically changed compared with those of Col-0. However, the phenotypes of SpNRT1.1 complemented lines were slightly changed. One probable reason for the inconsistency between the phenotype of SpNRT1.1 overexpression lines and SpNRT1.1 complemented lines was that SpNRT1.1 and AtNRT1.1 had the different sensitivity to ammonium or pH and different genetic backgrounds. Additionally, SpNRT1.1 and AtNRT1.1 proteins form heterodimers, and the function of SpNRT1.1-AtNRT1.1 heterodimers differs from that of AtNRT1.1-AtNRT1.1 homodimers. In Arabidopsis, nitrate treatment can change the phosphorylation profile of NRT1.1 mutant plants, including the auxin transporter PIN2, (Vega et al., 2021). Auxin accumulation depressed root growth (Li et al., 2015), and many auxin transporter induced by nitrate, such as PIN1, PIN2, PIN3, PIN4, PIN7, AUX1, LAX3 and so on (Maghiaoui et al., 2020). Auxin transporter AUX1, LAX3 and auxin signaling F-box 3 (AFB3) gene expression was induced by SpNRT1.1 (Supplementary Figure 8), mutation of AUX1 leads to increased root length under high nitrogen condition (Li et al., 2015), LAX3 or AFB3 probably play the similar role with AUX1, although they positively regulated root growth under low nitrogen condition. CLV1 repressed lateral root emergence (Liu et al., 2020), which expression induced in our experiment. These indicted SpNRT1.1 negative regulated root growth under high nitrogen cultured condition. Local ammonium promotes lateral root emergence to develop a highly branched root system, as ammonium acidifies the root apoplast and increases auxin content in the cortex and epidermis (Meier et al., 2020). This suggests that ectopic expression SpNRT1.1 in Arabidopsis improved nitrogen absorbance and prolonged vegetative growth by affecting auxin transport. How SpNRT1.1 regulates root structure and ammonium tolerance through auxin transduction needs to be determined in future studies.
To conclude, we identified all NRT proteins in S. polyrhiza at the genomic level and analyzed their conserved domains and motifs. Transcriptomic analysis showed that SpNRTs had spatiotemporal expression patterns and quickly responded to various abiotic stresses, indicating their complex and diverse functions in nitrate uptake and stress tolerance. Functional analysis of SpNRT1.1 in Arabidopsis presented the potential use of SpNRT1.1 in increasing crop biomass and should be extensively studied in future. Future studies should determine why SpNRT1.1 had a different function in case of sensitivity towards ammonium and whether the conserved T102 is an important phosphorylation site similar to T101 in AtNRT1.1.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
KZ and ML designed and conducted the experiments. ML and TD performed the experiments. ML analyzed the data. ML, JW, and KZ wrote and revised the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Key R&D Program of China (No. 2018YFA0901000 and 2018YFA0901003). We also appreciate the support of BIO-Agri project of SJTU and the Agricultural Science and Technology Innovation Program of CAAS.
Acknowledgments
We thank Wang’s laboratory for kindly providing the S. polyrhiza plant. We thank NASC for providing the Arabidopsis seeds described in this article. We would like to thank Editage (www.editage.cn) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.945470/full#supplementary-material
Abbreviations
ANOVA, Analysis of variance; BLAST, Basic local alignment search tool; HMM, Hidden Markov models; NRT, Nitrate transporter; MS medium, Murashige and Skoog medium; PCR, Polymerase chain reaction; SpNRTs, S. polyrhiza nitrate transporter; TM, Transmembrane domains.
Footnotes
References
Almagro, A., Lin, S. H., and Tsay, Y. F. (2008). Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20, 3289–3299. doi: 10.1105/tpc.107.056788
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bouguyon, E., Brun, F., Meynard, D., Kubeš, M., Pervent, M., Leran, S., et al. (2015). Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants. 1:15015. doi: 10.1038/nplants.2015.15
Boursiac, Y., Léran, S., Corratgé-Faillie, C., Gojon, A., Krouk, G., and Lacombe, B. (2013). [Epub 2013 march 1]. ABA transport and transporters. Trends Plant Sci. 18, 325–333. doi: 10.1016/j.tplants.2013.01.007
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Fan, X., Feng, H., Tan, Y., Xu, Y., Miao, Q., and Xu, G. (2016). A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. J. Integr. Plant Biol. 58, 590–599. doi: 10.1111/jipb.12382
Fan, S. C., Lin, C. S., Hsu, P. K., Lin, S. H., and Tsay, Y. F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761. doi: 10.1105/tpc.109.067603
Gojon, A., and Gaymard, F. (2010). Keeping nitrate in the roots: an unexpected requirement for cadmium tolerance in plants. J. Mol. Cell Biol. 2, 299–301. doi: 10.1093/jmcb/mjq019
Grefen, C., Donald, N., Hashimoto, K., Kudla, J., Schumacher, K., and Blatt, M. R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64, 355–365. doi: 10.1111/j.1365-313X.2010.04322.x
Guo, F. Q., Wang, R., Chen, M., and Crawford, N. M. (2001). The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13, 1761–1777. doi: 10.11054/tpc.010126
Guo, F. Q., Young, J., and Crawford, N. M. (2003). The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell 15, 107–117. doi: 10.1105/tpc.006312
Hachiya, T., and Noguchi, K. (2011). Mutation of NRT1.1 enhances ammonium/low pH-tolerance in Arabiopsis thaliana. Plant Signal. Behav. 6, 706–708. doi: 10.4161/psb.6.5.15068
Ho, C. H., Lin, S. H., Hu, H. C., and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hsu, P. K., and Tsay, Y. F. (2013). Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol. 163, 844–856. doi: 10.1104/pp.113.226563
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, N. C., Liu, K. H., Lo, H. J., and Tsay, Y. F. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392. doi: 10.1105/tpc.11.8.1381
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U. S. A. 109, 9653–9658. doi: 10.1073/pnas.1203567109
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Krouk, G., Tillard, P., and Gojon, A. (2006). Regulation of the high-affinity NO3− uptake system by NRT1.1-mediated NO3− demand signaling in Arabidopsis. Plant Physiol. 142, 1075–1086. doi: 10.1104/pp.106.087510
Lee, J., and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Li, J. Y., Fu, Y. L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Li, H., Hu, B., and Chu, C. (2017). Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J. Exp. Bot. 68, 2477–2488. doi: 10.1093/jxb/erx101
Li, J., Xu, H. H., Liu, W. C., Zhang, X. W., and Lu, Y. T. (2015). Ethylene inhibits root elongation during alkaline stress through AUXIN1 and associated changes in AUXIN accumulation. Plant Physiol. 168, 1777–1791. doi: 10.1104/pp.15.00523
Li, J., Zhao, C., Hu, S., Song, X., Lv, M., Yao, D., et al. (2020). Arabidopsis NRT1.2 interacts with the phospholipase Dα1 (PLDα1) to positively regulate seed germination and seedling development in response to ABA treatment. Biochem. Biophys. Res. Commun. 533, 104–109. doi: 10.1016/j.bbrc.2020.08.025
Liu, Y. G., and Chen, Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTechniques 43, 649–656. doi: 10.2144/000112601
Liu, K. H., Huang, C. Y., and Tsay, Y. F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874. doi: 10.1105/tpc.11.5.865
Liu, Y. G., Mitsukawa, N., Oosumi, T., and Whittier, R. F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. doi: 10.1046/j.1365-313x.1995.08030457.x
Liu, K. H., and Tsay, Y. F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013. doi: 10.1093/emboj/cdg118
Liu, B., Wu, J., Yang, S., Schiefelbein, J., and Gan, Y. (2020). Nitrate regulation of lateral root and root hair development in plants. J. Exp. Bot. 71, 4405–4414. doi: 10.1093/jxb/erz536
Maghiaoui, A., Bouguyon, E., Cuesta, C., Perrine-Walker, F., Alcon, C., Krouk, G., et al. (2020). The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J. Exp. Bot. 71, 4480–4494. doi: 10.1093/jxb/eraa242
Meier, M., Liu, Y., Lay-Pruitt, K. S., Takahashi, H., and von Wirén, N. (2020). Auxin-mediated root branching is determined by the form of available nitrogen. Nat. Plants. 6, 1136–1145. doi: 10.1038/s41477-020-00756-2
Orsel, M., Chopin, F., Leleu, O., Smith, S. J., Krapp, A., Daniel-Vedele, F., et al. (2006). Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 142, 1304–1317. doi: 10.1104/pp.106.085209
Plett, D., Toubia, J., Garnett, T., Tester, M., Kaiser, B. N., and Baumann, U. (2010). Dichotomy in the NRT gene families of dicots and grass species. PLoS One 5:e15289. doi: 10.1371/journal.pone.0015289
Quesada, A., Galván, A., and Fernández, E. (1994). Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 5, 407–419. doi: 10.1111/j.1365-313x.1994.00407.x
Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E., Mounier, E., et al. (2006). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. U. S. A. 103, 19206–19211. doi: 10.1073/pnas.0605275103
Stecher, G., Tamura, K., and Kumar, S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239. doi: 10.1093/molbev/msz312
Sun, J., Bankston, J. R., Payandeh, J., Hinds, T. R., Zagotta, W. N., and Zheng, N. (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77. doi: 10.1038/nature13074
Tal, I., Zhang, Y., Jørgensen, M. E., Pisanty, O., Barbosa, I. C., Zourelidou, M., et al. (2016). The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 7, 11486. doi: 10.1038/ncomms11486
Taochy, C., Gaillard, I., Ipotesi, E., Oomen, R., Leonhardt, N., Zimmermann, S., et al. (2015). The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J. 83, 466–479. doi: 10.1111/tpj.12901
Tsay, Y. F., Schroeder, J. I., Feldmann, K. A., and Crawford, N. M. (1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713. doi: 10.1016/0092-8674(93)90399-b
Upadhyay, R. K., Edelman, M., and Mattoo, A. K. (2020). Identification, phylogeny, and comparative expression of the lipoxygenase gene family of the aquatic duckweed, Spirodela polyrhiza, during growth and in response to methyl jasmonate and salt. Int. J. Mol. Sci. 21:9527. doi: 10.3390/ijms21249527
Upadhyay, R. K., Shao, J., and Mattoo, A. K. (2021). Genomic analysis of the polyamine biosynthesis pathway in duckweed Spirodela polyrhiza L.: presence of the arginine decarboxylase pathway, absence of the ornithine decarboxylase pathway, and response to abiotic stresses. Planta 254:108. doi: 10.1007/s00425-021-03755-5
Vega, A., Fredes, I., O’Brien, J., Shen, Z., Ötvös, K., Abualia, R., et al. (2021). Nitrate triggered phosphoproteome changes and a PIN2 phosphosite modulating root system architecture. EMBO Rep. 22:e51813. doi: 10.15252/embr.202051813
Walch-Liu, P., and Forde, B. G. (2008). Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 54, 820–828. doi: 10.1111/j.1365-313X.2008.03443.x
Wang, Y. Y., Cheng, Y. H., Chen, K. E., and Tsay, Y. F. (2018a). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Wang, W., Haberer, G., Gundlach, H., Gläßer, C., Nussbaumer, T., Luo, M. C., et al. (2014). The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 5, 3311. doi: 10.1038/ncomms4311
Wang, W., Hu, B., Li, A., and Chu, C. (2020). NRT1.1s in plants: functions beyond nitrate transport. J. Exp. Bot. 71, 4373–4379. doi: 10.1093/jxb/erz554
Wang, W., Hu, B., Yuan, D., Liu, Y., Che, R., Hu, Y., et al. (2018b). Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30, 638–651. doi: 10.1105/tpc.17.00809
Wang, R., Xing, X., Wang, Y., Tran, A., and Crawford, N. M. (2009). A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 151, 472–478. doi: 10.1104/pp.109.140434
Wen, Z., Tyerman, S. D., Dechorgnat, J., Ovchinnikova, E., Dhugga, K. S., and Kaiser, B. N. (2017). Maize NPF6 proteins are homologs of Arabidopsis CHL1 that are selective for both nitrate and chloride. Plant Cell 29, 2581–2596. doi: 10.1105/tpc.16.00724
Yan, M., Fan, X., Feng, H., Miller, A. J., Shen, Q., and Xu, G. (2011). Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 34, 1360–1372. doi: 10.1111/j.1365-3040.2011.02335.x
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhao, X., Yang, J., Li, G., Sun, Z., Hu, S., Chen, Y., et al. (2021). Genome-wide identification and comparative analysis of the WRKY gene family in aquatic plants and their response to abiotic stresses in giant duckweed (Spirodela polyrhiza). Genomics 113, 1761–1777. doi: 10.1016/j.ygeno.2021.03.035
Keywords: nitrate transporter genes, root development, ammonium, flowering time, biomass production
Citation: Lv M, Dong T, Wang J and Zuo K (2022) Genome-wide identification of nitrate transporter genes from Spirodela polyrhiza and characterization of SpNRT1.1 function in plant development. Front. Plant Sci. 13:945470. doi: 10.3389/fpls.2022.945470
Edited by:
Sajid Fiaz, The University of Haripur, PakistanReviewed by:
Antt Htet Wai, Yangon University of Education (YUOE), MyanmarRakesh K. Upadhyay, Agricultural Research Service (USDA), United States
Copyright © 2022 Lv, Dong, Wang and Zuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Wang, d2FuZ2ppbkBjYWFzLmNu; Kaijing Zuo, a2p6dW9Ac2p0dS5lZHUuY24=
 Mengli Lv
Mengli Lv Tiantian Dong1
Tiantian Dong1 Kaijing Zuo
Kaijing Zuo