
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 15 July 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.944721
This article is part of the Research Topic Marker-Assisted Selection (MAS) in Crop Plants View all 45 articles
Mungbean is a socioeconomically important legume crop in Asia that is currently in high demand by consumers and industries both as dried beans and in plant-based protein foods. Marker-assisted and genomics-assisted breeding are promising approaches to efficiently and rapidly develop new cultivars with improved yield, quality, and resistance to biotic and abiotic stresses. Although mungbean was at the forefront of research at the dawn of the plant genomics era 30 years ago, the crop is a “slow runner” in genome research due to limited genomic resources, especially DNA markers. Significant progress in mungbean genome research was achieved only within the last 10 years, notably after the release of the VC1973A draft reference genome constructed using next-generation sequencing technology, which enabled fast and efficient DNA marker development, gene mapping, and identification of candidate genes for complex traits. Resistance to biotic stresses has dominated mungbean genome research to date; however, research is on the rise. In this study, we provide an overview of the past progress and current status of mungbean genomics research. We also discuss and evaluate some research results to provide a better understanding of mungbean genomics.
Mungbean [V. radiata (L.) R. Wilczek var. radiata] is an ancient legume crop from Asia. The crop is believed to have been domesticated from its wild form, V. radiata var. sublobata (Roxb.) Vercourt, in India about 4,000–4,500 years ago (Fuller and Harvey, 2006). Mungbean has been cultivated and consumed as a common food in China and Thailand for more than 2,000 years (Castillo, 2013; Tang et al., 2014). It is well-known for its detoxification activity and is used to refresh the mind, alleviate heat stroke, and reduce swelling in summer (Tang et al., 2014). Mungbean adapts well to various cropping systems due to its rapid growth, early maturity (60–75 days), relative tolerance to drought, and ability to improve soil fertility through atmospheric nitrogen (N2) fixation in symbiosis with Rhizobium and Bradyrhizobium bacteria in the soil (Yimram et al., 2009). Hence, it plays an important role in crop diversification and sustainable agricultural intensification in Asia.
Mungbean seeds are a source of protein, amino acids, carbohydrates, vitamins, and minerals. The seeds contain about 20–30% protein and 60–70% carbohydrate. Mungbean is an important protein source for people in cereal-based societies, especially in South Asia. Mungbean seeds are cooked and consumed in a variety of ways, and mungbean flour is used to prepare several kinds of food. The seeds are also processed into sprouts, snacks, pastes, starches, noodles, protein isolates, and protein concentrates (Nair and Schreinemachers, 2020). There is a growing interest in mungbean consumption and use because of its high protein content, high-quality starch, and other nutritional contents. Mungbean seeds have become a major material used in the production of plant-based protein foods, such as egg and meat substitutes.
Although official statistical data on the global production area and seed yield of mungbean are not available, the production area is about 7.5–8.0 million ha, about 80–90% of which is in Asia (Nair and Schreinemachers, 2020; Anonymous, 2021). Mungbean production areas outside Asia, including in Australia, America, and Africa, are increasing. This is driven by increasing consumer demand for dry legumes and plant-based protein foods. The biggest producer and consumer of mungbean is India, with about 4.5 million ha cultivated and a total production of 2.5 million tons (Anonymous, 2021). Myanmar is the second-highest producer and largest exporter, with about 1.2 million ha cultivated and a total production of 1.5 million tons (MAOLI, 2019). Although mungbean production areas are increasing, the yield is low, at only about 115 kg/ha (Nair and Schreinemachers, 2020), and production is challenged by insect pests, diseases, and unsuitable environments (Pandey et al., 2018; Nair et al., 2019). Insects and diseases are mainly controlled by applying pesticides, which increases production costs and is hazardous to farmers, consumers, and the environment. There is a need to develop new mungbean cultivars that fulfill the needs of farmers, consumers, and processors.
Despite its high socioeconomic importance, mungbean is neglected and underfunded in breeding research at both the national and international level, particularly in the field of genomics, which holds great promise for modern plant breeding. Mungbean is an ideal crop for genomics study because it is self-pollinating and diploid (2n = 2x = 22) and has a small genome size of 493.6–579.0 megabase (Mb) pairs (Arumuganathan and Earle, 1991; Kang et al., 2014; Liu M. S. et al., 2016) and a short life cycle with early maturity (60–75 days). In fact, mungbean was among the first legumes subjected to genomics research, with the first genome mapping reported 30 years ago, but today, advances in mungbean genomics lag behind other legume crops with similar importance (Figure 1).
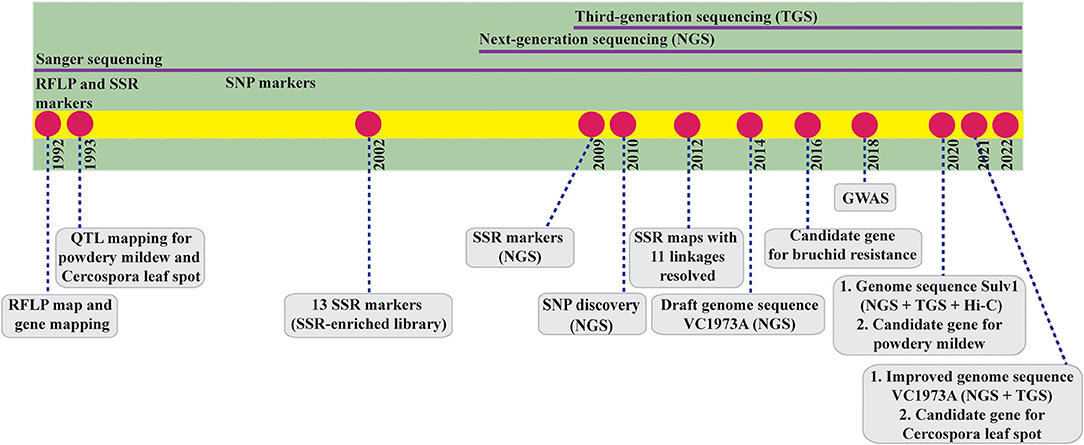
Figure 1. Timeline of mungbean genome research since the first publication of genome mapping in 1992. RFLP, restriction fragment length polymorphism; QTL, quantitative trait locus; SSR, simple sequence repeat; SNP, single nucleotide polymorphism; GWAS, genome-wide association study; Hi-C, high-throughput chromosome conformation capture.
The most recent review on mungbean genomics was published 7 years ago (Kim et al., 2015). In that review, the authors mainly described genomic and genetic resources for the genetic improvement of mungbean. In addition, rapid and extensive progress has been made in mungbean genome research. In this study, we review all aspects of mungbean genomics research, past and present, and discuss and evaluate some research results to provide in-depth, wide-ranging, and recent analysis of mungbean genome research.
Early mungbean genetics and breeding research occurred in India, the Philippines, and the USA. In India, germplasm collection and evaluation began in the 1920s, and pure lines were isolated (Bose, 1932). The first mungbean cultivar released for farmers was “Type 1” in 1936. The first cultivar developed through hybridization and selection and released to farmers was “Type 44,” and this was released in 1948. All cultivars used in this period were small-seeded, with non-uniform maturity, photoperiod sensitivity, and low yield. The average yield was only about 0.4–0.8 ton/ha. Since 1980s, many cultivars with improved yield, large seed, uniform maturity, non-sensitivity to photoperiod, and resistance to diseases have been developed.
In the Philippines, various landraces and introduced germplasms were tested before 1916 and the breeding programs began in 1933 (Catipon et al., 1988). Landraces were collected and pure lines were isolated, selected, and evaluated, resulting in six pure lines released to farmers and becoming very popular (Ballon et al., 1977). In 1956, a national breeding program was set up to develop cultivars with high yield that were semi-dwarf, with uniform maturity, large and shiny seed, non-shattering pods, and high seed protein and carbohydrate content by hybridization and selection, resulting in the release of cultivars “MG50-10A” and “MD15-2” in 1969 (Catipon et al., 1988). These cultivars yielded about 1.2 tons/ha, about double that of the pure-line cultivars. The improved cultivars revolutionized the yield, plant architecture, and phenology of mungbean, although they were still susceptible to diseases. In the mid-1970s, cultivars with high yield, good architecture, and disease resistance were successfully developed using resistance germplasm from India (Catipon et al., 1988).
In the USA, before 1914, the US Department of Agriculture (USDA) introduced and evaluated mungbean germplasm from several countries. Three cultivars, “Berken,” “Oklahoma 12,” and “Kiloga,” developed by selection from introduced germplasm, were released in 1963 (Matlock and Oswalt, 1963). In the early 1970s, the International Mungbean Nursery (IMN) was organized by the University of Missouri and later coordinated by the Asian Vegetable Research and Development Center in Taiwan [now the World Vegetable Center (WorldVeg)]. The IMN stimulated international cooperation in mungbean breeding.
Since WorldVeg was established in 1971, the center has played a key role in mungbean germplasm collection, characterization, and breeding. By exploiting the high yield and good agronomic traits of improved cultivars/lines from the Philippines and the disease resistance of Indian germplasm as parents, newly improved lines with better yield, agronomic traits, and disease resistance were bred (Fernandez and Shanmugasundaram, 1988). The improved lines yielded up to 1.8–2.2 tons/ha and boosted mungbean cultivation and production in Asia from the 1980s to 2000s. WorldVeg mungbean lines were also used as parents in breeding programs in several countries. The adoption and use of the WorldVeg breeding lines have transformed the mungbean from a marginal crop to a major crop in Asia. For example, the cultivars developed and released by the WorldVeg generated economic gains of USD 1.4 billion in Myanmar from 1980 to 2016 (Sequeros et al., 2020).
One of the goals of plant genomics research is to utilize tools, knowledge, and information generated from research to improve crops. Plant genomics research relies on the availability of polymorphic DNA markers and dense genetic linkages. The era of crop genomics began in the late 1980s with the development of restriction fragment length polymorphism (RFLP) markers and genetic linkage maps of several economic crops, including rice (McCouch et al., 1988), tomato (Bernatzky and Tanksley, 1986), and maize (Helentjaris, 1987). The publication of quantitative trait loci (QTL) mapping of tomato in 1988 (Paterson et al., 1988) stimulated gene mapping of crop plants. Genome research in legume crops began in the late 1980s to early 1990s. The first genome mapping of mungbean was published in 1992 (Fatokun et al., 1992; Young et al., 1992), and the first genetic linkage map of mungbean was officially published in 1993 (Menancio-Hautea et al., 1993). Genetic linkage maps of some important legume crops, such as soybean [Glycine max (L.) Merr.] (Keim et al., 1990), common bean (Phaseolus vulgaris L.) (Vallejos et al., 1992), and cowpea [Vigna unguiculata (L.) Walp.] (Fatokun et al., 1993), were developed at around the same time. The mungbean linkage map was based solely on RFLP markers and was used to identify QTLs controlling seed size (Fatokun et al., 1992), insect resistance (Young et al., 1992), and disease resistance (Young et al., 1993). Comparative linkage analyses between mungbean and cowpea, common bean, and soybean were also published (Fatokun et al., 1992; Boutin et al., 1995), revealing high conservation of synteny between mungbean and these legumes. The orthologous seed weight QTLs between mungbean and cowpea (Fatokun et al., 1992) was reported in the first comparative genome analysis in legumes. However, after this period, mungbean genome research progressed very slowly.
Prior to 2010, only seven genetic linkage maps of mungbean were constructed and all of them were principally based on RFLP markers, which did not resolve the 11 haploid chromosomes of the mungbean (Menancio-Hautea et al., 1993; Boutin et al., 1995; Lambrides et al., 2000; Chaitieng et al., 2002; Humphry et al., 2002; Zhao et al., 2010). The RFLP probes utilized in those studies were mainly from other legumes, including common bean, cowpea, soybean, and lablab [Lablab purpureus (L.) Sweet]. In addition, gene mapping of mungbean involved only six traits, including seed size, seed dormancy, bruchid, bean bug (Riptortus clavatus Thunb.), and powdery mildew resistance (Kaga and Ishimoto, 1998; Chaitieng et al., 2002; Humphry et al., 2003, 2005; Mei et al., 2009; Hong et al., 2015). The slow progress was mainly due to the lack of efficient genomic resources, especially genetic markers (the most important tool in genomics study), such as simple sequence repeat (SSR), which was the most efficient DNA marker system at that time, even though the first report on SSR variation in mungbean was in the late 1990s (Yu et al., 1999). Fewer than 150 polymorphic SSR markers were reported for mungbean during this period (Kumar et al., 2002a,b; Gwag et al., 2006; Somta P. et al., 2008; Seehalak et al., 2009; Somta et al., 2009; Tangphatsornruang et al., 2009).
Since 2010, relatively rapid progress in mungbean genome research has been made. Several SSR-based maps were constructed and utilized for QTL mapping of various traits, including agronomic and domestication-related traits, biotic and abiotic stress resistance, and seed chemical contents (Kasettranan et al., 2010; Zhao et al., 2010; Chankaew et al., 2011, 2013; Isemura et al., 2012; Kajonphol et al., 2012; Prathet et al., 2012; Sompong et al., 2012; Chen et al., 2013; Kitsanachandee et al., 2013; Alam et al., 2014a,b; Wang et al., 2016; Liu et al., 2017; Masari et al., 2017). However, all of those maps were low-resolution, with nearly all of them containing <100 polymorphic markers. Highly polymorphic SSR markers from azuki bean [Vigna angularis (Willd.) Ohwi and Ohashi] (Wang et al., 2004) comprised the core of these maps. The first linkage maps that resolved the 11 haploid chromosomes of mungbean were published by Isemura et al. (2012) and Kajonphol et al. (2012), which were constructed from SSR markers of various legumes. The map developed by Isemura et al. (2012) was the densest, with 430 SSR markers and an average marker density of 1.78 centimorgans.
Next-generation sequencing (NGS) is a technology that involves the sequencing of millions of small fragments of DNA in parallel and enables rapid, inexpensive, and comprehensive analysis of the genomes of individual organisms and complex populations. NGS has revolutionized genome research on all organisms and in all disciplines. There are several NGS platforms, including Illumina/Solexa, Roche 454, Ion Torrent, and SOLiD sequencing. These platforms became available in the late 2000s and early 2010s, and they differ in terms of template preparation, sequencing chemistry, read length, and outputs per run (Metzter, 2010). In the early stage of NGS, parts of nuclear genomes and transcriptomes of mungbean were sequenced, which led to the identification of a massive number of SSRs and single nucleotide polymorphisms (SNPs) and the development of new SSR markers (Tangphatsornruang et al., 2009; Moe et al., 2011; Van et al., 2013; Gupta et al., 2014). NGS is also a powerful tool in the discovery and genotyping of large numbers of SNPs at a drastically reduced expense, such as genotyping-by-sequencing (GBS) (Elshire et al., 2011), which allowed highly dense genetic linkage maps to be developed for mungbean (Kang et al., 2014; Hwang et al., 2017; Mathivathana et al., 2019; Ha et al., 2021). In addition, the chloroplast genome of mungbean was fully sequenced by NGS (Tangphatsornruang et al., 2010).
The greatest contribution of NGS to mungbean genomics is whole-genome sequencing (WGS). A draft reference genome of the elite mungbean breeding line “VC1973” from WorldVeg was constructed on the chromosome level using Illumina/Solexa and Roche 454 sequencing (Kang et al., 2014). A 431-megabase (Mb) part (80%) of the total estimated mungbean genome (579 Mb) was sequenced and generated into 2,748 scaffolds with an N50 length of 1.62 Mb. Based on a linkage map of 1,321 SNP markers, scaffolds were assembled into 11 pseudochromosomes covering 314 Mb, equivalent to 73% of the assembled sequence. In total, only 22,427 protein-coding genes were identified, of which 18,378 genes were located on the pseudochromosomes. In addition to “VC1973A,” the whole genome of the wild mungbean accession “TC1966” was also sequenced (423 Mb, corresponding to about 84% of the estimated genome size). By comparing the genome sequences of mungbean “VC1973A,” “V2984,” and “TC1966,” millions of SNPs and hundreds of thousands of indels were discovered (Kang et al., 2014). Draft genome sequences, SNPs, and indels are highly useful resources for gene mapping and mining. A few years after the release of the draft genome sequence of “VC1973A,” the WGS of the breeding line “RIL59” was reported (Liu M. S. et al., 2016). A 455.2-Mb portion (88%) of the genome of RIL59 (517.1 Mb) was sequenced using Illumina sequencing and assembled into 2,509 scaffolds with an N50 of 676.7 kb and an average sequence length 181.4 Mb. A total of 36,939 protein-coding genes were predicted.
A complete and high-quality reference genome sequence serves as the basis for advanced genomic analysis, providing an opportunity to systemically identify and characterize the functions of genes. Third-generation sequencing, such as single-molecule real-time (SMRT) sequencing and nanopore sequencing, generates sequence reads with unprecedented lengths in less time and with more accuracy, but at a lower cost. Such technology is highly useful for the construction of contiguous and chromosome-scale genome sequences (Amarasinghe et al., 2020). Recently, a high-quality chromosome-scale genome assembly of the mungbean cultivar “Sulv1” was constructed using a combination of second-generation sequencing, third-generation sequencing, and Hi-C analysis (Yan et al., 2020). In the Sulv1 genome assembly, 473.7 Mb, covering 87.8% of the estimated genome size, was assembled and 99.32% of sequences were assigned to 11 pseudochromosomes, with scaffold N50 of 42.4 Mb and 33,924 predicted protein-coding genes. More recently, the genome sequence of VC1973 was improved using SMRT sequencing (Ha et al., 2021). The new genome assembly covered the total sequence of 476 Mb from 557 scaffolds with an N50 length of 5.2 Mb and only a 0.4% gap. The sequence corresponded to 87.5% of the VC1973A genome, oriented into 11 pseudochromosomes, and contained 30,958 genes. The new assembly was compared with the previous ones by mapping common SNP markers to both genome assemblies, which revealed many disagreements between the two assemblies. Nonetheless, comparative genome analysis of the new genome assembly and genomes of legumes closely related to mungbean demonstrated that the new mungbean assembly was more reliable that the previous one. The large difference in the number of identified genes in the genomes of VC1973A, RIL59, and Sulv1 suggests a substantial genome variation in mungbean.
Having more than one reference genome for mungbean is an advantage when analyzing gene discovery and genome structural variations (e.g., presence–absence variation, copy number variation, and chromosomal rearrangements) because different genomes of a species vary in both gene content and repetitive portions of the genome (Della Coletta et al., 2021). To date, the improved genome sequence of VC1973A and the genome sequences of Sulv1 and wild mungbean TC1966 have not been released to the public. Moreover, a large portion (12%) of VC1973A, RIL59, and Sulv1 genome sequences is still not covered by WGS. Additional genome sequencing is necessary to increase genome coverage as much as possible. The puzzle of mungbean genome variation may be resolved through pangenome construction. Additional reference genome sequences of mungbean are being constructed and a mungbean pangenome project has been initiated (Nair et al., 2022).
The NGS technology also provides access to genome-wide transcript variation for understanding the functional elements of the genome, molecular constituents of cells and tissues, and phenotypic variations via transcriptome sequencing (Wang et al., 2009). Before 2000, there was only one transcriptomics study on mungbean (Chen et al., 2008). When the cost of NGS became affordable around the mid-2010s, the number of transcriptomics sequencing studies on mungbean increased sharply (Moe et al., 2011; Chen et al., 2015; Li et al., 2015; Lin et al., 2016; Liu C. et al., 2016; Liu M. S. et al., 2016; Tian et al., 2016; Kumar et al., 2020; Chang et al., 2021; Dasgupta et al., 2021; Hu et al., 2022; Sreeratree et al., 2022; Sudha et al., 2022; Zhao et al., 2022). The majority of these transcriptome analyses were related to responses to biotic and abiotic stresses, for example, drought, chilling temperature, waterlogging, heat, bruchids (C. chinensis), and Fusarium wilt (Fusarium oxysporum).
Quantitative trait loci mapping and gene cloning play pivotal roles in understanding the genetic basis of variation in quantitative and complex traits that are important in molecular breeding. QTLs can be statistically localized onto genetic linkage maps by exploiting linkage relationships between markers and phenotypes in experimental populations. Several traits in mungbean have been dissected by QTL mapping (Table 1). In this review, we focus on some important traits, as follows.
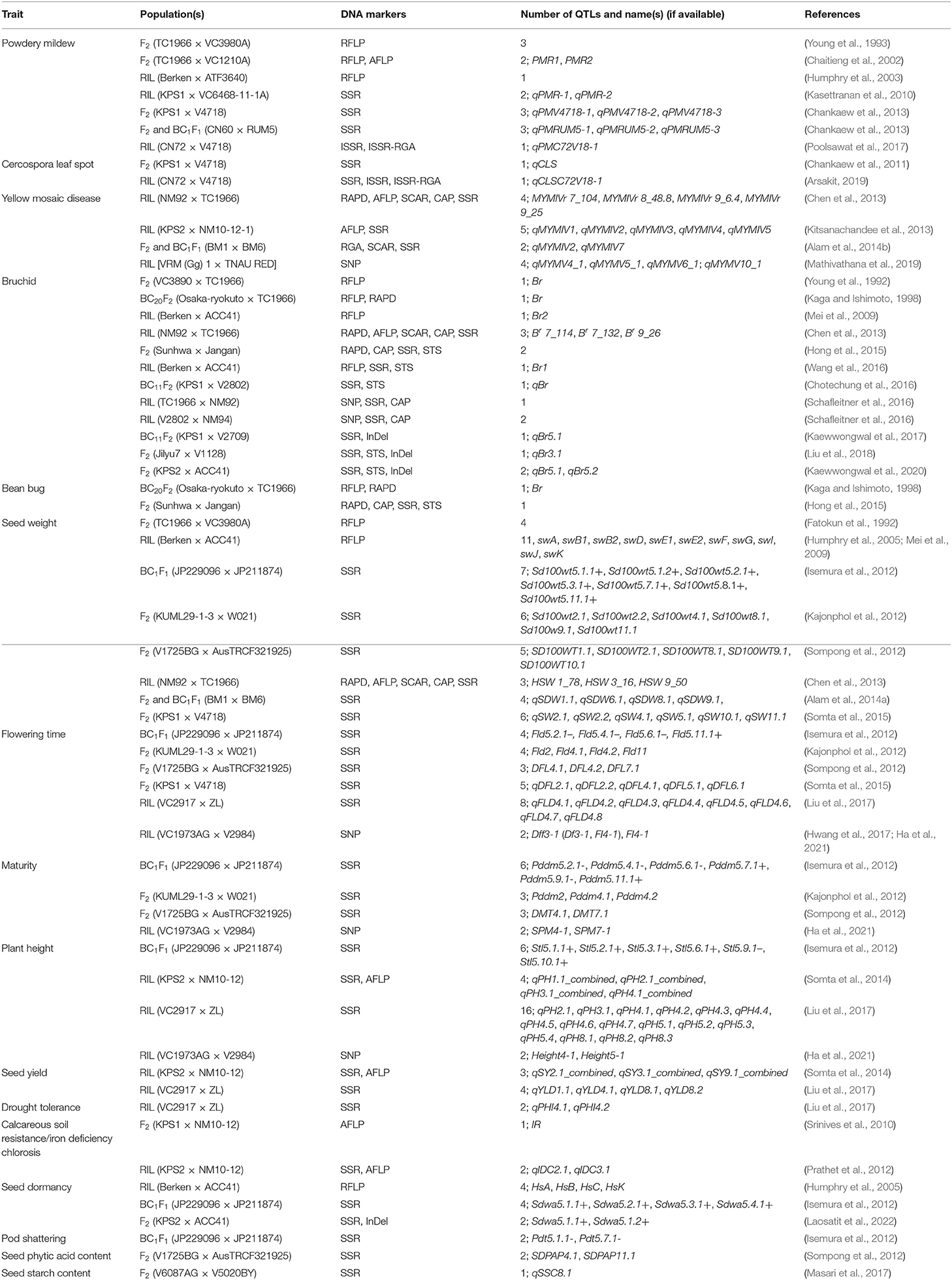
Table 1. Quantitative trait loci identified for various traits in mungbean by gene mapping using experimental populations.
Bruchids, or seed weevils, are stored-product insects that infest the starchy seeds of various crops. Azuki bean weevil (C. chinensis L.) and cowpea weevil (Callosobruchus maculatus Fab.) are the most important bruchids in the storage of mungbean and several other legume crops (Srinives et al., 2007). Seed resistance to bruchids in mungbean is due to antibiosis in cotyledons (Somta C. et al., 2008) and is controlled by a single dominant locus, Br, with modifiers (Kitamura et al., 1988; Somta et al., 2007). Br was the first gene to be molecularly located on the mungbean genetic map (Young et al., 1992). The first fine-mapping study in mungbean attempted to narrow down the Br genomic region in wild mungbean “TC1966” (Kaga and Ishimoto, 1998) using RFLPs and RAPD markers. Several years later, map-based cloning of the Br locus in mungbean cultivar “Jangannogdu,” deriving resistance from landrace cultivar “V2709,” identified VrBURP1, encoding a protein containing the BURP (BNM2, USP, RD22, and PG1b) domain, as the gene responsible for resistance (Jeong et al., 2015). By combining transcriptomic, proteomic, and gene expression analyses of the near-isogenic line VC6089A carrying the Br locus from TC1966 and its recurrent parent VC1973A, gene g5551, encoding aspartic proteinase, and g34458 and g39185, each encoding a protein containing a BURP domain, were identified as candidate genes for resistance (Lin et al., 2016). g34458 and g39185 are located on mungbean chromosome 5 near the Br locus (Lin et al., 2016; Liu M. S. et al., 2016; Schafleitner et al., 2016). The protein sequences encoded by VrBURP1, g34458, and g39185 are similar, and they all match polygalacturonase (Laosatit et al., 2020).
Chotechung et al. (2016) exploited information from fine mapping of the Br locus in TC1966 (Kaga and Ishimoto, 1998) and the draft genome sequence of mungbean to find candidate genes for resistance in the mungbean landrace “V2802,” and also found that the Br locus is localized on chromosome 5 between markers VrBr-SSR013 and DMB-SSR158. DMB-SSR158 co-segregated perfectly with resistance. VrBr-SSR013 and DMB-SSR158 corresponded to Vradi05g03940 (VrPGIP1) and Vradi05g03950 (VrPGIP2), respectively. No other genes were found between these two genes. VrPGIP1 and VrPGIP2 each encode a polygalacturonase inhibitor (polygalacturonase-inhibiting protein; PGIP). Sequence alignment of VrPGIP1 and VrPGIP2 between the mapped mungbean parents revealed SNPs in only the coding sequence of VrPGIP2. The SNPs cause amino acid changes in the VrPGIP2 protein in V2802. The same resistance allele is also found in other bruchid-resistant mungbean, including TC1966, “V1128,” “V2066,” and “V2817.” Later, new resistance alleles of VrPGIP2 and/or VrPGIP1 were identified in the cultivated mungbean accession “V2709” and wild mungbean accession “ACC41” (Kaewwongwal et al., 2017, 2020). Polygalacturonase is an important digestive enzyme secreted in the gut of C. maculatus (Nogueira et al., 2012). Recently, VrPGIP1 and VrPGIP2 proteins isolated from V2802 were shown to enzymatically inhibit polygalacturonases (PGs) isolated from larvae and adults of C. maculatus (Zhang et al., 2021). A feeding test also confirmed the antibiosis effect of the proteins VrPGIP1 and VrPGIP2 against bruchids (Zhang et al., 2021). These results strongly indicate that VrPGIP1 and VrPGIP2 are the genes at the Br locus that confer resistance to bruchids in seeds by inhibiting C. maculatus PGs to degrade pectin and the cell wall of seeds, thus limiting energy and other digestive enzymes to access their substrates in the ingested seeds. VrPGIP1 and VrPGIP2 represent a new class of insect resistance genes in plants.
Erysiphales fungi are obligate biotrophic plant pathogens that produce white mycelia on the leaves, stems, and fruits of angiosperm plants. Powdery mildew (PM) disease caused by obligate fungus E. polygoni is a common leaf disease of mungbean crops grown in different regions. The disease is also caused by Sphaerotheca phaseoli (Z. Y. Zhao) U. Braun (Lee et al., 2002), Podosphaera xanthii (Castagne) U. Braun and Shishkoff (Sheu et al., 2021), and Erysiphe vignae sp. nov. (Kelly et al., 2021). The disease is prevalent in cool and dry environments and is favored by high-density plant populations and cloudy weather (Laosatit et al., 2020). It can cause reduced seed yield of susceptible mungbean cultivars by up to 40% if there is no protection.
Young et al. (1993) was the first to identify three QTLs for partial resistance in “VC3890A” using RFLP markers. Nearly 10 years later, Chaitieng et al. (2002) used RFLP and amplified fragment length polymorphism (AFLP) markers and identified one major and one minor QTL, PMR1, and PMR2, controlling resistance to PM in “VC1210A.” PMR1 explained up to 65% of the disease resistance variation. A major QTL governing resistance in “ATF3640” was mapped using RFLP markers (Humphry et al., 2003). This QTL accounted for up to 86% of the variation in disease reaction. SSR and sequence-tagged site (STS) markers were added to the QTL region identified in ATF3640 (Zhang et al., 2008). Using SSR markers, two major QTLs, qPMR1 and qPMR2, were detected for PM resistance in “VC6468-11-1A” (Kasettranan et al., 2010). These two QTLs accounted for 20.10 and 57.81% of the variation in disease reaction, respectively. Chankaew et al. (2013) located QTLs controlling resistance in highly resistant accessions “RUM5” and “V4718.” Two major (qPMRUM5-2 and qPMRUM5-3) and one minor (qPMRUM5-1) QTL were detected in RUM5, while one major (qPMV4718-2) and two minor (qPMV4718-1 and qPMV4718-3) QTLs were identified in V4718. Comparative linkage analysis of the QTLs for PM resistance in different mungbean accessions revealed that qPMRUM5-3 in RUM5, qPMV4718-3 in V4718, qPMR2 in VC6468-11-1A, and the resistance QTL in ATF3640 are in the same linkage group and possibly the same locus, whereas qPMRUM5-2 in RUM5 and qPMR1 in VC6468-11-1A are on the same linkage group and very likely the same locus (Chankaew et al., 2013). In contrast to the above studies involving E. polygoni, Arsakit (2019) showed that the resistance to PM caused by S. phaseoli in V4718 is controlled by a single major QTL, qPMC72V18, localized on linkage group 3 between SSR markers VR108 and VR393. qPMC72V18 was on a different linkage group from other QTLs for PM resistance.
Mildew locus O (MLO) genes are known to be involved in susceptibility/resistance to PM diseases in plants. In fact, MLOs are susceptibility genes (S-genes), genes that facilitate infection and support compatibility for a pathogen (van Schie and Takken, 2014). Loss of MLO function is associated with resistance to the disease and is known as mlo-based resistance (Kusch and Panstruga, 2017). In barley, mlo-based resistance has been durable under field conditions and has been utilized for over 40 years with no reported breakdown of resistance (Kusch and Panstruga, 2017). The binding of Ca+-dependent calmodulin (CaM) to the CaMBD domain of the MLO protein promotes susceptibility to PM during infection (Consonni et al., 2006). Mutation in the CaMBD domain, which reduces or abolishes CMD binding, results in resistance to disease. Eighteen MLO genes have been identified in the mungbean reference genome (Rispail and Rubiales, 2016). Bioinformatic analysis and fine mapping of qPMRUM5-3, which confers PM resistance in RUM5, demonstrated that VrMLO12 is the only candidate gene at the qPMRUM5-3 (Yundaeng et al., 2020). Some SNPs in the VrMLO12 of RUM5 cause amino acid changes in functional domains, transmembrane 6 (TM6) domain, and calmodulin-binding domain (CaMBD) of the VrMLO12 protein. Similar mutations have also been found in V4718. Therefore, amino acid change in the CaMBD domain of VrMLO12 is likely the cause of PM resistance in RUM5 and V4718, although the amino acid change in TM6 may also contribute to the resistance. The mlo-based resistance appears to be a major PM resistance mechanism in mungbean.
Mildew locus O proteins from plants and algae are classified into seven clades, I to VII (Pessina et al., 2016). Interestingly, in contrast to the finding in other plant species that all MLO proteins involved in susceptibility/resistance to PM disease are restricted to clade IV (for monocots) and clade V (for dicots), VrMLO12 belongs to MLO clade II (Rispail and Rubiales, 2016; Yundaeng et al., 2020). It has been proposed that MLO proteins negatively regulate vesicle-associated and actin-dependent defense pathways at the site of powdery mildew fungi penetration (Panstruga, 2005). The secretory vesicle traffic modulates pathogen penetration, leading to the formation of papillae or cell wall appositions (Feechan et al., 2011), which are associated with mlo-based resistance (Consonni et al., 2006). It is not yet known how VrMLO12 modulates susceptibility to E. polygoni. Molecular characterization of the function of VrMLO12 could allow a better understanding of the interaction between MLO proteins and powdery mildew fungi, and thus of mlo-based resistance.
Resistance to PM appears to be controlled by several loci. To date, however, only one gene, VrMLO12, has been identified as a candidate gene for resistance. Additional attempts should be made to identify candidate genes for other loci for resistance, for example, the genes for qPMRUM5-2 that express stable resistance. Since the loss-of-function mutations in clade V MLO genes lead to broad-spectrum resistance to PM in several plant species, the clade V MLO genes in mungbean, namely VrMLO1, VrMLO2, VrMLO5, VrMLO6, VrMLO11, and VrMLO17 (Rispail and Rubiales, 2016), can be suitably targeted as selected candidate genes for association analysis of resistance to PM disease in mungbean. They could also be candidates for the development of mlo-based resistance in mungbean through genetic engineering or gene editing.
Cercospora leaf spot (CLS) disease is an important foliar disease in mungbean crops grown in tropical and subtropical regions. The disease is caused by the obligate biotrophic fungus Cercospora canescens Ellis and Martin and occurs widely in wet seasons (Laosatit et al., 2020). This fungus causes defoliation and up to 50% seed yield reduction in mungbean. Despite its importance, there have been only a few studies reporting the genetics and genomics of resistance to CLS disease in mungbean. All of those studies used the same resistant germplasm, V4718, which is also resistant to PM. CLS resistance in V4718 is controlled by a single resistance gene (Chankaew et al., 2011). A single and stable major QTL, qCLS, controlling resistance was mapped onto linkage group 3 between SSR markers CEDG117 and VR393 (Chankaew et al., 2011). The location of qCLS was validated in an independent study (Arsakit, 2019). Fine mapping targeting the CEDG117-VR393 region in an F2 population and an advanced back-cross population was carried out, and the qCLS was consistently narrowed down to a genome region of 13.0 kb on chromosome 6 containing only a single gene, LOC106765332 (VrTAF5), which encodes general transcription initiation factor IID (TFIID) subunit 5 [TATA-box-binding protein (TBP)-associated factor 5 (TAF5)] (Yundaeng et al., 2021). A VrTAF5 sequence comparison between V4178 and KPS1 revealed an SNP in the coding sequence. This SNP causes an amino acid change in VrTAF5. These results indicate that VrTAF5 is the gene at the qCLS locus and is responsible for resistance to C. canescens. This is the first line of evidence showing an association between TAF5 and stress resistance in plants. Since TAF5 is a key protein in the formation of the TFIID and Spt-Ada-Gcn5 acetyltransferase (SAGA) complexes, which regulate gene expression (Soffers and Workman, 2020), VrTAF5 possibly modulates the expression of resistance genes to C. canescens. It is worth noting that qCLS is co-localized with qPMC72V18, conferring PM resistance caused by S. phaseoli in V4718 (Arsakit, 2019). It is tempting to speculate that VrTAF5 controls resistance to both C. canescens and S. phaseoli.
Yellow mosaic disease (YMD) is the most important and devastating disease in mungbean production in South Asia and is becoming a major threat in Myanmar and Thailand. YMD is caused by three species of begomovirus: mungbean yellow mosaic virus (MYMV), mungbean yellow mosaic India virus (MYMIV), and horse gram yellow mosaic virus (HgYMV). These viruses are transmitted and spread via whitefly (Bemisia tabaci Gennadius). The symptoms of YMD include leaf yellowing and/or chlorosis followed by leaf necrosis, fewer pods, small and abnormal pods and seeds, and stunted plants. YMD causes up to 100% yield loss. Resistance to YMD in mungbean has been reported to be controlled by a single dominant gene or a single recessive gene, or two recessive genes or complementary recessive genes (reviewed in Laosatit et al., 2020).
Quantitative trait loci mapping in an RIL population in India revealed four QTLs for resistance, with QTL MYMIVr9_25 showing the largest effect, and only QTL MYMIVr8_48.8 was consistently identified across different years (Chen et al., 2013). Evaluating MYMIV resistance of an RIL population in India and Pakistan, Kitsanachandee et al. (2013) detected three QTLs, qMYIMV1, qMYIMV2, and qMYIMV3, for resistance in India and two QTLs, qMYIMV4 and qMYIMV5, in Pakistan. All of the QTLs except qMYMIV4 accounted for <20% of disease variation. Two major QTLs, qMYMIV2.1 and qMYMIV7.1, for MYMIV resistance in two locations in Bangladesh were consistently identified using F2 and BC1F1 populations (Alam et al., 2014a). Each QTL accounted for more than 40% of disease resistance variation.
When the reference mungbean genome sequence became available, Mathivathana et al. (2019) used an RIL population of an interspecific cross between mungbean (VRM (Gg) 1, susceptible) and rice bean (TNAU RED, resistant) to map the QTLs for MYMV resistance using SNP markers. Two and three QTLs on chromosomes 4, 5, 6, and 10 were identified for the years 2015 and 2016, respectively. The QTL qMYMV4_1 on chromosome 4 was identified in both years and showed phenotypic variance of about 20%. The physical distance of markers VigSNP_04_32 and VigSNP_04_36 flanking qMYMV4_1 was about 1.28 Mb and contained 83 annotated genes. Laosatit et al. (2020) explored the linkages of QTLs for YMD resistance and found that the QTLs MYMIVr9_25, qMYMIV4 (qMYMIV1), and qMYMIV2.1 were mapped to the same linkage group and were possibly the same QTL because the resistance sources used in all studies derived their resistance from the same germplasm. If this is the case, this QTL will be useful for breeding YMD-resistant mungbean cultivars, since it is stable across environments. One of the QTLs derived from rice bean, qMYMV5_1, was located in the qMYMIV2.1 region (Mathivathana et al., 2019). This suggests a common mechanism of resistance to YMD between mungbean and rice bean.
A transcriptome analysis was conducted to identify genes involved in MYMV resistance in the resistant mungbean “VGGRU-1,” which is a derivative of the cross VRM (Gg) 1 × TNAU RED (Sudha et al., 2022), by comparing the differentially expressed genes (DEGs) between VGGRU-1 and VRM (Gg) 1. We checked those DEGs and found that Vradi04g06840 is of interest, as it encodes a protein similar to suppressors of TYPE ONE PROTEIN PHOSPHATASE 4 MUTATION (TOPP4-1) (SUT). SUT1 is a nucleotide-binding, leucine-rich repeat protein of the C-NB-LRR type and is involved in TOPP4-mediated immune response in Arabidopsis (Yan et al., 2019). In addition, Vradi04g06840 is within the qMYMV4_1 region, and therefore can be considered a lead candidate for further molecular investigation of resistance.
Seed size directly contributes to seed yield and is important in the mungbean industry. Farmers, processors, and buyers prefer large-seeded mungbean. The trait has certainly been a major target for selection by farmers and breeders. Seed weight is a polygenic but highly heritable trait with an H2 value of more than 80% (Humphry et al., 2005; Yimram et al., 2009; Isemura et al., 2012). Together with PM and CLS, seed weight was one of the first traits molecularly mapped in mungbean (Fatokun et al., 1992). There have been some studies on QTLs controlling seed weight, most of them using populations derived from hybridization between wild and cultivated lines, and 4–11 QTLs were identified for the trait (Fatokun et al., 1992; Humphry et al., 2005; Mei et al., 2009; Isemura et al., 2012; Kajonphol et al., 2012; Sompong et al., 2012; Alam et al., 2014b; Somta et al., 2015).
In the study of Alam et al. (2014b), four QTLs, qSDWT1.1, qSDWT6.1, qSDWT8.1, and qSDWT9.1, were consistently identified for seed weight in an F2:3 population grown in three environments. They noted that (i) qSDWT1.1 and qSDWT8.1 appeared to be the same as QTLs Sd100w5.1.1 and Sd100w8.1.1 reported by Isemura et al. (2012) and the same as the QTLs on LG VI and LG II, identified by Fatokun et al. (1992); (ii) qSDWT8.1 and qSDWT9.1 corresponded to the QTLs Sd100wt8 and Sd100wt9, identified by Kajonphol et al. (2012); and (iii) qSDWT1.1, qSDWT8.1, and qSDWT9.1 corresponded to the QTLs SD100WT1.1, SD100WT8.1, and qSDWT9.1 reported by Sompong et al. (2012). Some mungbean seed weight QTLs are conserved in other related legume species, such as azuki bean, rice bean, and cowpea (Fatokun et al., 1992; Isemura et al., 2012; Alam et al., 2014b; Somta et al., 2015). For instance, qSDWT1.1 was also found in all of these legume crops, qSDWT6.1 was identified in azuki bean and cowpea, qSDWT8.1 was found in cowpea, and qSDWT9.1 was detected in azuki bean (Alam et al., 2014b). Since qSDWT8.1 expressed the largest or second-largest effect (Isemura et al., 2012; Sompong et al., 2012; Alam et al., 2014b), this QTL should be further studied to identify genes underlying seed size.
Stem determinacy (determinate growth) is an agronomically important trait that is selected during mungbean domestication and breeding, as it has been useful for farmers since ancient times when harvesting mungbean pods both by hand and using machines (Isemura et al., 2012). Stem determinacy, flowering time, and photoperiod insensitivity have been genetically improved by breeders to develop modern mungbean cultivars with a short flowering period and early maturity for the ease of harvest (Fernandez and Shanmugasundaram, 1988). In general, improved mungbean cultivars have a determinate habit, whereas all wild mungbean germplasms have an indeterminate habit. The determinate habit is recessive and is controlled by a single gene, originally named stdet5.9.1, and mapped onto linkage group 9 between SSR markers GATS11 and CEDG172 (Isemura et al., 2012). Later, stdet5.9.1 was renamed VrDet1 (Li et al., 2018). In soybean, stem determinacy is controlled by Dt1, which is orthologous to Arabidopsis Terminal Flowering 1 (TFL1), encoding a shoot apical meristem signaling protein (Liu et al., 2010). Vradi03g04510 (VrDet1) in mungbean is a TFL1 ortholog (Li et al., 2018). Sequencing of the VrDet1 gene and its upstream region between cultivated mungbean “Dahyeon” and wild mungbean “W164” revealed no polymorphism in exons, but 15 SNPs, 12 of which are in the promoter region. Association analysis revealed perfect co-segregation between VrDet1 and the stem determinacy trait. Compared to wild (indeterminate growth) mungbean, VrDet1 is much less expressed in cultivated (determinate growth) mungbean. Complementation tests showed that the VrDet1 allele from cultivated mungbean complements TFL1 function in Arabidopsis, whereas the VrDet1 allele from wild mungbean does not, and that mutations in the promoter region of VrDet1 causes the change from indeterminacy in wild mungbean to determinacy in cultivated mungbean. A VrDet1 and upstream sequence comparison between 13 wild and 14 cultivated mungbean accessions revealed three unique SNPs in the promoter region between the two groups. Further investigation elucidated that only an SNP belonging to a cis-regulatory element of MARTBOX and one belonging to a NODCON2GM/OSE2ROOTNODULE cis-regulatory element were essential for VrDet1 function and indeterminacy. These results indicate that the transition from indeterminacy to determinacy in cultivated mungbean is due to the loss-of-function mutations in the two cis-regulatory elements in the promoter region of VrDet1. It is believed that the SNPs in the cis-regulatory elements in cultivated mungbean reduce or cancel the expression of VrDet1, thereby suppressing the expression of floral identity genes in the apical meristems of stem tips in determinate mungbean to produce terminal flowers that develop into terminal pods (Li et al., 2018).
Photoperiod and temperature are key environmental factors that affect the phenological development of mungbean in all growth stages, and thus determine yield and adaptability. Mungbean is a quantitative short-day (SD) and warm-season plant that can be cropped in a wide range of latitudes between 51°N and 40°S. Many landraces and most wild mungbean are sensitive to photoperiod, whereas modern cultivars are less sensitive to photoperiod and are early flowering. Flowering time is a quantitative trait and is highly correlated with maturity time. Understanding the genetic mechanism underlying the photoperiod response in mungbean is crucial for expanding mungbean production to high-latitude regions.
Four QTLs on LGs 2, 4, and 11 were identified for days to first flowering (DFL) in an F2 population of a cross between cultivated mungbean showing early flowering (DFL = 31 days) and wild mungbean showing late flowering (DFL = 66 days) grown during SD and long-day (LD) conditions in Thailand (Kajonphol et al., 2012). Fld2 on LG2 and Fld4.2 on LG4 were the major QTLs, contributing 15.9 and 28.6% of the flowering variation. Similarly, four QTLs each on LGs 2, 4, 6, and 11 were detected for DFL in an F2 population of a cross of wild and late-flowering mungbean (DFL = 81 days) × cultivated and early-flowering mungbean (DFL = 35 days) grown from July (day length about 14 h) to December (day length about 10 h) 2007 (Isemura et al., 2012). The Fld5.2.1- on LG2 had the largest effect (PVE = 32.9%), followed by that on LG4 (PVE = 24.0%). A study of an F2 population of a cross between cultivated mungbean (DFL = 34 days) and wild mungbean (DFL = 44 days) grown in summer (LD condition) in Thailand revealed three QTLs on LGs 4 and 7 governing DFL (Sompong et al., 2012). DFL4.1 and DFL4.2 on LG4 were the major loci, accounting for 33.4 and 25.7% of the flowering variation, respectively. In another study, using an F2 population of a cross between early- and late-flowering cultivated mungbean evaluated in three seasons (one LD and two SD conditions) in Thailand, five QTLs on LGs 2, 4, 5, and 6 were identified for rainy seasons and two QTLs on LG2 were detected for dry seasons (Somta et al., 2015). These QTLs accounted for between 12.6% (qDFL2.2) and 32.0% (qDFL5.1) of the flowering variation. It should be noted that there was a 11-day difference between the parents in DFL under LD conditions, but they flowered on the exact same or nearly the same day under SD conditions. The QTLs on LGs 2, 4, 6, and 11 were always detected, especially those on LGs 2 and 4. Two QTLs on LG2 were always detected in both SD and LD conditions, indicating their importance in regulating flowering in mungbean, while the QTLs on LGs 4, 6, and 11 were responsive to LD.
A BLASTN search for chromosomal locations of SSR markers surrounding the QTLs on LG2 against the genome sequence of wild mungbean “NI1135” (https://viggs.dna.affrc.go.jp) revealed a physical association between the markers on LG2 and a Phytochrome gene, and between the markers on LG4 and Flowering Locus T-like (FT-like), Flowering Locus K homology domain (FLK), APETALA2/Ethylene-Responsive Factor (AP2/ERF), and CONSTANS genes (Somta, unpublished data). FT encodes florigen, which serves as an integrator of flowering time control in several plant species. In soybean, FT-like genes GmFT2a and GmFT5a promote flowering, whereas GmFT1a and GmFT4 inhibit flowering and maturity (reviewed in Lin et al., 2021). In Arabidopsis, FLK positively regulates flowering by repressing the expression of a flowering-time gene (Mockler et al., 2004). AP2/ERF proteins are key regulators of various growth and development processes, including reproductive and vegetative organ development (Xie et al., 2019), whereas CONSTANS plays a central role in integrating various external and internal signals into a photoperiodic flowering pathway (Shim et al., 2017). Correlative relationships between these genes and flowering time in mungbean should be molecularly confirmed.
In addition to the low-resolution mapping of QTLs for DFL mentioned above, high-resolution QTL mapping for DFL was carried out using 1,321 SNPs in an RIL population derived from a cross between the elite breeding line VC1973A (DFL = 38 days) and the landrace mungbean V2984 (DFL = 44 days) grown under LD conditions in Korea (Hwang et al., 2017). Only a single QTL, Dff 3-1, was mapped on chromosome 3. Dff 3-1 accounted for 33.4% of the FLD variation detected, and Vradi03g07170 (VrPHYA), encoding phytochrome A (PHYA), was identified as a candidate gene for this locus (Hwang et al., 2017). VrPHYA is homologous to soybean E3 (GmPHYA3) and E4 (GmPHYA2) genes, known to be involved in the control of flowering under LD conditions (reviewed in Lin et al., 2021). In a recent study, high-resolution mapping of QTLs for plant height (PLH), flower initiation (FLI), number of branches (BRN), number of nodes (NDN), and synchronous pod maturity (SPDM) was carried out using 8,966 SNPs in an RIL population grown in Korea (Ha et al., 2021). This RIL population was possibly the same one used by Hwang et al. (2017). QTL analysis detected one major QTL on chromosome 4 and one minor QTL on another chromosome for all the traits except BRN, for which only one major QTL on chromosome 3 was detected. The QTLs on chromosome 4 detected for PLH, FLI, NDN, and SPDM were in the same or nearly the same region, suggesting a pleiotropic effect on these traits. Based on the newly assembled genome of VC1973A, candidate genes for the major QTLs of these traits were identified. Vradi04g00002773 (VrPHYA in Hwang et al., 2017) was identified as the candidate gene for PLH, BRN, and SPDM, while Vradi04g00002764 (VrBONSAI), encoding a protein with similarity to the APC13 component of the anaphase-promoting complex, and Vradi03g00002294 (VrERF4), producing ERF4, were identified as candidate genes for FLI and BRN, respectively.
In Arabidopsis, as compared to wild type, bonsai plants are shorter and with more compact inflorescence, resulting in reduced PLH (Saze and Kakutani, 2007). In Arabidopsis, ERF4 is a negative regulator that can modulate ethylene and abscisic acid responses. The ERF4 gene appears to affect plant senescence. Transgenic Arabidopsis lines overexpressing ERF4-R showed slightly delayed flowering and silique emergence (Riester et al., 2019). Candidate genes Vradi09g00002773 (VrPHYA homolog) and Vradi11g00000623 (VrELF3), Vradi05g00000394 (VrPKpl), and Vradi07g00000916 (VrHAK1) were identified for minor QTLs controlling for PLH, NDN, and SPDM, PLH, and SPDM (Ha et al., 2021). All of these candidate genes are likely to have experienced breeding selection in the process of mungbean breeding that transformed landraces into modern cultivars with early flowering and maturity, compact plant habit, and synchronous pod maturity. Nonetheless, although VrPHYA appears to be useful in increasing early flowering (FLI and DFL) and SPDM, it decreases PLH and NDN. The pleiotropy of VrPHYA must be carefully considered to allow its exploitation in mungbean breeding. The genomes of mungbean and soybean are highly conserved (Kang et al., 2014; Yan et al., 2020). Soybean is molecularly well-studied for the photoperiodic regulation of flowering (Lin et al., 2021). The molecular mechanisms controlling photoperiod response and flowering in soybean can be used as a model in mungbean through comparative genomics. Mungbean genes corresponding to some key genes for the photoperiod response and flowering in soybean have been identified (Ha et al., 2021).
Quantitative trait loci mapping and candidate genes have also been reported for other traits related to plant architecture and phenology, including the morphology of leaves (Jiao et al., 2016; Wang et al., 2020), inflorescence (Lee et al., 2021), and flowers (Chen et al., 2016; Lin et al., 2020). Interestingly, candidate genes Vradi04g00002481 for compound raceme, Vradi04g00002442 (Vradi03g04510 in Li et al., 2018) for stem determinacy, Vradi04g00002773 (Vradi03g07170 in Hwang et al., 2017) for DFL, PLH, BRN, and SPDM, and Vradi04g00002764 for FLI are clustered on chromosome four. It would be difficult to break the close/tight linkage of these genes by gene recombination.
Non-dormancy of seeds and non-shattering of pods are the first two traits that are selected for the process of legume domestication. Nondormant seeds provide uniform germination and timely cultivation. In contrast, seed dormancy prevents germination of wild species in unsuitable environments. In legume and cereal crops grown in tropical and subtropical regions, the non-dormancy of seeds can result in preharvest sprouting, leading to yield and quality losses. Like other wild legume species, wild mungbean shows strong seed dormancy, which can provide protection against preharvest sprouting (Imrie et al., 1988; Lawn et al., 1988). Classical genetic studies suggested that seed dormancy in wild mungbean is controlled by a single dominant gene with or without modifying genes (Singh et al., 1983; Lawn et al., 1988). QTL analysis revealed that dormancy in wild mungbean ACC41 is controlled by one major and three minor QTLs, and only the major QTL, HsA, explaining 23.2% of the dormancy variation, could be confirmed for the trait (Humphry et al., 2005). HsA was located between RFLP markers cgO103 and VrCS364. Isemura et al. (2012) identified QTLs for domestication-related traits and found that seed dormancy in wild mungbean accession “JP211874” was conditioned by two major and two minor QTLs. Sdwa5.1.1+ showed the largest effect, explaining 33.7% of the seed dormancy variation. It was located between SSR markers cp05137 and CEDG074b. Laosatit et al. (2022) speculated that HsA and Sdwa5.1.1+ are the same loci and finely mapped Sdwa5.1.1+ using ACC41 as the source of seed dormancy; they found that the cp05137–CEDG074b interval contained two QTLs for dormancy, Sdwa5.1.1+ and Sdwa5.1.2+, with more or less equal effects. Sdwa5.1.1+ was located between markers VrSdp-SSR5 and VrKNAT7-SSR4, where there is only one gene, Vradi07g13210 (VrKNAT7-1), in this genomic interval, while Sdwa5.1.2+ was localized between markers VrSdp-SSR102 and VrSdp-SSR104, which encompasses five genes: Vradi07g10460, Vradi07g10470, Vradi07g10480, Vradi07g10490, and Vradi07g10500.
Sequence alignment of VrKNAT7-1 between ACC41 and KPS2 (cultivated mapping parent) revealed SNP and indel polymorphisms in CDS, 5′UTR and 3′UTR, and upstream sequences, although the SNPs in CDS cause synonymous mutation. Expression analysis of seed coat demonstrated higher expression of VrKNAT7-1 and Vradi06g06960 (VrCYP86A, a downstream gene of VrKNAT7-1) in ACC41 than in KPS2. Vradi07g13210 encodes KNOTTED ARABIDOPSIS THALIANA 7 (KNAT7), a homolog of MtKNOX4 protein that controls physical seed dormancy by regulating the expression of MtCYP86A, which is involved in the cuticle biosynthesis pathway, and MtKCS12, which controls the production of very-long-chain lipids in the seed coat (Chai et al., 2016, 2021). Scarified seeds of ACC41 germinated rapidly under wet conditions, while the intact seeds of ACC41 remained dormant for several days. Microscopic observation revealed that ACC41 seeds possess a cuticle layer, while KPS2 seeds lack such a layer. These results indicate that VrKNAT7-1 controls physical dormancy in wild mungbean.
At the Sdwa5.1.2+ region, containing five genes, Vradi07g10480, encoding calmodulin-like (CML) protein 1, is likely the gene controlling seed dormancy (Laosatit et al., 2022). Calmodulin (CaM) and CML proteins have been shown to be involved in the ABA-induced inhibition of seed germination and seedling growth. Sdwa5.3.1+, with PVE of about 14.2%, is another major QTL controlling seed dormancy in wild mungbean JP211874 (Isemura et al., 2012). This QTL is located between SSR markers GMES6583 and GMES0294a on LG3. We inspected the physical locations of these markers by blasting their primer sequences against the VC1973A genome and reanalyzed LG3 and Sdwa5.3.1+ by removing a few markers that seemed to be problematic and found that Sdwa5.3.1+ was mapped to nearly the same position as marker GMES0591 (Supplementary Figure 1). GMES0591 corresponds with the LOC106764165 gene, encoding gibberellin 2-beta-dioxygenase 1 (GA2OX1), which controls gibberellin catabolism. The balance between GA and ABA biosynthesis and catabolism determines seed dormancy and germination (Finch-Savage and Leubner-Metzger, 2006). Therefore, both physical and physiological dormancies appear to control seed dormancy in wild mungbean.
Quantitative trait loci for bean bug (Riptortus clavatus Thunberg) resistance (Kaga and Ishimoto, 1998; Hong et al., 2015), seed phytic content (Sompong et al., 2012), iron-deficiency chlorosis (calcareous soil resistance) (Srinives et al., 2010; Prathet et al., 2012), drought tolerance (Liu et al., 2017), and domestication-related traits (Isemura et al., 2012; Kajonphol et al., 2012) have also been identified for mungbean. However, most of these QTLs underwent low-density mapping and have not been confirmed/validated.
This study, also known as linkage disequilibrium (LD) mapping, is a form of gene mapping used to unravel the genetic basis of complex traits by identifying DNA markers, generally SNPs, associated with a particular trait of interest. GWAS takes advantage of past recombination events occurring in a set of germplasms, particularly landraces, and is performed by scanning genotype–phenotype associations along the chromosomes of the given germplasms. GWAS requires a large number of germplasms (several hundred to a few thousand) with high diversity and a large number of SNPs (several hundreds to tens of thousands). Compared to biparental mapping, GWAS provides higher QTL resolution, often to the gene level. Therefore, causative genes for a given trait can be identified by GWAS. In the past, GWAS could only be used for models and major crops where large numbers of SNP markers were available. With the advanced development of sequencing technologies, large numbers of SNPs in all crop species can now be discovered and genotyped by NGS, thus allowing the possibility of using GWAS in orphan crops like mungbean. The power of GWAS depends on the strength of correlation (the degree of LD) between the genotypes of markers and those of causative genes, which is a function of the distance between them, and the resolution of a QTL mapped by GWAS depends on how rapidly the LD decays over that distance (Myles et al., 2009). The LD extent is about 72–290 kb in cultivated mungbean (Noble et al., 2018; Ha et al., 2021; Sandhu and Singh, 2021) and 3–60 kb in wild mungbean (Noble et al., 2018).
GWAS for agronomic traits (PLH and DFL) and seed size (100 SDW) in a USDA mungbean collection of 482 accessions was carried out using 264,550 SNPs (Sandhu and Singh, 2021). Three SNP loci on different chromosomes were detected for each trait. For DFL, SNP 1_11367629 on chromosome 1 and 5_4604047 on chromosome 5 showed R2-values higher than 25%, while the other SNPs showed R2-values of about only 1%. Among the three loci for PLH, only SNPs 1_11367629, with R2 of about 30%, appeared to be correctly identified, while the others, with R2-values of 0%, were likely false positives. The major loci for 100 SDW were on chromosomes 1 and 7, and each QTL contributed 10–13% of the seed weight variation. Notably, SNP 1_11367629, located in LOC106774729 and producing receptor-like protein kinase FERONIA (FER), was associated with all three traits. FER has diverse functions in plant growth and development, including hypocotyl and root elongation, root hair development, and flowering time (Deslauriers and Larsen, 2010; Duan et al., 2010; Wang et al., 2020; Zhu et al., 2020). We looked into an 83 kb region (Vr01:11309527..11393240) covering LOC106774729 and found five other FER genes located next to LOC106774729. It will therefore be difficult to determine the causative FER genes for these traits. We also surveyed the region around SNP 5_4604047 and found that this marker is about 25 kb away from a Phytochrome gene. However, none of the DFL QTLs detected in this study were in the same region as VrPHYA, the candidate gene for DFL reported by Hwang et al. (2017). Nonetheless, the relationship between the seed weight QTLs identified by this GWAS and those detected by biparental mappings is not known.
The seed coat color of mungbean can be green, black, yellow, or brown. The color affects the consumer and industry preferences. Although seed coat color is a qualitative trait, several genetic models with up to five genes have been proposed for the inheritance of this trait in mungbean (reviewed in Dikshit et al., 2020). A GWAS of 209 cultivated and 9 wild mungbean accessions from 23 countries using 18,171 SNP markers detected a single SNP located on chromosome 4 in Vradi04g08950 (positions 1,7682,006 to 17,685,676) encoding a pectinesterase associated with seed coat color variation determined by red, green, and blue (R, G, B) color components (Daovongdeuan, 2017). A GWAS for seed coat color variation in 466 cultivated accessions from various regions and 16 wild mungbean accessions using 22,230 SNPs revealed 9 SNPs located on four chromosomes associated with color variation (Noble et al., 2018). Five SNPs clustering and spanning a 22-kb region (positions 17,668,384 to 17,690,573) on chromosome 4 harbored a gene encoding a transcription factor, MYB113, known to be involved in anthocyanin biosynthesis, which is the candidate gene for black seed coat coloring, and an SNP on chromosome 5 was associated with a gene encoding flavonoid 3'-hydroxylase, which controls yellow seed color in soybean (Noble et al., 2018). The 22 kb region covered the SNP for seed color reported by Daovongdeuan (2017). In another study, GWAS was used to identify SNPs at positions 4,639,979 and 17,694,108 on chromosomes 2 and 4, respectively, associated with seed coat color (Sandhu and Singh, 2021). Anthocyanins are present in the seed coats of black mungbean seeds but absent in the seed coats of green, yellow, and brown seeds (Pandey et al., 1989). However, our exploration into the 22 kb region reported by Noble et al. (2018) found no MYB gene, but we did find Vradi04g08920 (VrMYB16) encoding MYB16 and LOC106758748 (VrMYB90) encoding MYB90, located about 38 and 10 kb from the region, respectively.
MYB16 and MYB90 are R2R3–MYB transcription factors that regulate various biological processes in land plants, including anthocyanin biosynthesis. Anthocyanins are pigments that give rise to red, purple, blue, and black colors in plants. MYB16 has been demonstrated to inhibit anthocyanin synthesis in apples (Xu et al., 2017), while MYB90 has been shown to regulate anthocyanin production in the tissues of several plant species (Qi et al., 2020), including mungbean (Lin et al., 2022). Map-based cloning, gene overexpression analysis, and haplotype analysis revealed that VrMYB90 is responsible for anthocyanin synthesis in the mungbean hypocotyl, possibly through the upregulation of VrDFR1, encoding dihydroflavonol 4-reductase, at the last step of the synthesis, but the gene is not responsible for the black color of the seed coat (Lin et al., 2022). Thus, at this stage, it is reasonable to hypothesize that VrMYB16 may control black seed coloring in mungbean. It is noteworthy that the germplasm used in the GWAS of Daovongdeuan (2017) and Noble et al. (2018) contained a small portion of black mungbean seeds (<5%). Most of the black mungbean in these studies were wild varieties. A strong population structure and genetic relatedness can prevent GWAS from correctly identifying causal variants and cause the identification of spurious associations. Moreover, in wild mungbean, the seeds are generally dull and brownish-black or black, and when the seed texture is removed, the green or mottled green-black color of the shiny seed is revealed (Watt et al., 1977; Sarikarin et al., 1999). Therefore, phenotyping seed coat colors must be performed with caution. Additional genomics studies should be conducted to find out the relationship between VrMYB16 and seed coat color.
The GWAS of a small set of germplasms of 95 cultivated mungbean accessions (mostly from India and Pakistan) with 6,486 SNPs detected 43 SNP markers from 35 chromosome regions associated with calcium, iron, potassium, manganese, phosphorus, sulfur, and zinc concentrations in seeds (Wu et al., 2020). Some of the SNPs/regions were associated with more than one mineral, suggesting pleiotropic effects. In most cases, the significant SNP markers explained about 10–34% of the mineral variations. Twenty-six candidate genes related to various biological functions were identified for the seed mineral content, generally related to metal binding, metal translocation, and mineral uptake.
The GWAS for mungbean tolerance to biotic and abiotic stresses has also been carried out. GWAS for resistance to Fusarium wilt disease, caused by Fusarium oxysporum, was conducted in a USDA mungbean collection of 482 accessions, with 264,550 SNPs identified 3 loci associated with resistance (Sandhu and Singh, 2021). However, the highest R2 among these loci was only 6.8%. GWAS of phosphorus use efficiency in 144 mungbean accessions (mostly from India, along with Thailand, Taiwan, and Bhutan) with 55,634 SNPs identified 136 SNPs of 77 annotated genes of all 11 mungbean chromosomes associated with response to low-P and optimum-P concentrations under hydroponic conditions; 84 and 29 SNPs were related to root architecture and leaf area, respectively (Reddy et al., 2020). Based on the functions of candidate genes, 13 genes were chosen as candidate genes for the response and genes Vradi01g05520, Vradi04g10750, and Vradi11g08340 were found to possess SNPs causing amino acid changes (Reddy et al., 2020). GWAS of salt stress tolerance (seed germination at 50 mM NaCl) of the minicore collection of WorldVeg mungbean with 5,288 SNPs (see details below) identified two genome regions on chromosomes 7 and 9 associated with tolerance (Breria et al., 2020). Although the QTL regions were very large (about 1 Mb), Vradi07g01630, encoding the ammonium transport protein, was selected as the candidate gene for QTL on chromosome 7, while Vradi09g09510 and Vradi09g09600, encoding OsGrx_S16-glutaredoxin subgroup II and dnaJ domain proteins, respectively, were considered as candidate genes for QTL on chromosome 9 (Breria et al., 2020). Since salt stress tolerance is a complex trait and affects plants at all stages of growth and development, additional genomics studies should be conducted to elucidate the salt stress tolerance mechanism in mungbean.
In addition to the traits mentioned above, GWAS has been carried out for seed coat luster, hypocotyl color, pod color, and leaf drop at maturity (Sandhu and Singh, 2021).
Genetic resources are essential for breeding new crop cultivars and useful for gene mining via genomics research. Considerable genetic resources are available for mungbean. The largest collection of mungbean germplasm is maintained by the World Vegetable Center in Taiwan (10,438 accessions; https://genebank.worldveg.org). Mungbean germplasm collections with large numbers of accessions are also conserved in the national genebanks of India (11,000 accession) (Gayacharan et al., 2020), USA (3,931 accessions; www.genesys-pgr.org), the Philippines (1,623 accession), Australia (1,385 accessions; www.genesys-pgr.org), Japan (1,455 accessions; www.gene.affrc.go.jp), Korea (1,091 accessions; http://genebank.rda.go.kr), Russia (877 accessions; www.genesys-pgr.org), and the United Arab Emirates (408 accessions; www.genesys-pgr.org). Several thousand mungbean accessions are also conserved in the national genebank of China, although the exact number is not known. The mungbean collection in Russia is the oldest, with most of the accessions collected between 1910 and 1927 (Gayacharan et al., 2020), while the collection maintained in Japan has the largest number of wild mungbean, 211 accessions (www.gene.affrc.go.jp). The availability of these large collections of mungbean germplasm provides opportunities to discover new, beneficial genes and study their functions for breeding through genomics analysis.
There are only a few reports on the molecular genetic diversity of large collections of mungbean. A collection of 615 mungbean accessions (415 cultivated, 189 wild, and 11 weedy) covering all distribution areas were analyzed with 19 azuki bean SSR markers (Sangiri et al., 2007). The number of alleles found in wild mungbean (257) was about double that found in cultivated mungbean (138), while the gene diversity (HE) in cultivated mungbean was 65.01% of that in wild mungbean. For cultivated mungbean, the greatest diversity was found in India and Pakistan. For wild mungbean, the highest diversity was found in South Asia (HE = 0.68), but the level of diversity was not much different from that in Australia (HE = 0.59). These results support the hypotheses of Tomooka et al. (1992). However, the Australian wild mungbean is distinctly different from Asian and African wild mungbean at both the genetic and phenotypic levels (Lawn and Rebetzke, 2006; Sangiri et al., 2007; Ha et al., 2021); thus, the wild mungbean can be used to enrich the gene pool of cultivated mungbean in breeding.
A core collection comprising 52 cultivated and 49 wild accessions was developed based on SSR profiles and phenotypic data (Sangiri et al., 2007). In another study, 705 cultivated mungbean accessions from 26 countries (about 60% from Korea) were assessed for diversity using 695 AFLP and 15 SSR markers (Moe et al., 2012). The gene diversity of the germplasm estimated by AFLP and SSR markers was 0.69 and 0.36, respectively. A core collection of 222 accessions was constructed based on AFLP markers (Moe et al., 2012). A core collection of 1,481 mungbean accessions was developed from 5,234 accessions of cultivated mungbean maintained at WorldVeg based on geographical information and eight agronomic traits (Schafleitner et al., 2015). In addition, this core collection was molecularly assessed using 20 SSR markers and, as a result, a minicore collection of 297 accessions was developed. Genetic diversity in the core and minicore collections was moderate, with 122 and 166 alleles and HE of 0.49 and 0.57, respectively. This minicore collection was further characterized using 5,288 SNP markers generated by genotyping-by-sequencing (GBS), revealing that the collection is composed of four genetic clusters that are generally related to the geographical origins of the mungbean. In general, mungbean from South Asia (India and Pakistan) were clustered with accessions from West Asia, but separated into different subclusters, while those from other regions were subdivided and clustered together.
The core/minicore collections developed by Sangiri et al. (2007) and Schafleitner et al. (2015) are useful resources for QTL and gene identification by GWAS and are available to the research community. Combining the core collection developed by Sangiri et al. (2007) and the minicore collection reported by Schafleitner et al. (2015) can enhance genome diversity, utilization of genetic resources in mungbean genomics research, and genetic improvement. Recently, a set of mungbean germplasms with 276 accessions (233 cultivated and 42 wild) from 23 countries was assessed for diversity using genome-wide SNP variations analyzed by GBS (Ha et al., 2021). The assessment showed that the average nucleotide diversity in cultivated mungbean was more than 3-fold lower than that in wild mungbean, and the cultivated mungbean from India showed the closest relationship with the wild mungbean. Hence, it is clear that cultivated mungbean have lost much of their genetic diversity through domestication and breeding. Wild mungbean germplasm is an important source of resistance to biotic and abiotic stresses (Fujii and Miyazaki, 1987; Lawn et al., 1988; Lawn and Rebetzke, 2006), which have become more prominent and serious owing to climate change. The genomes of wild mungbean should be explored by WGS and whole-genome resequencing to harness their natural allelic variation for breeding climate-resilient mungbean.
Conventional and molecular breeding are the two approaches used to develop crop cultivars. Genomics research provides tools (such as DNA markers), informatics, and knowledge that can enhance the efficiency and precision of conventional plant breeding. The application of DNA markers for decision-making in plant selection processes is called marker-assisted breeding (MAB), which is a kind of molecular breeding (Collard and Mackill, 2008). MAB is composed of marker-assisted selection (MAS) and genomic selection (GS). MAS utilizes markers that are closely or tightly linked with traits of interest to select plants with desirable allele-affecting target traits (Collard and Mackill, 2008), while GS employs genome-wide markers to predict the effects of all loci and thereby calculate genomic estimated breeding values for all individuals of a breeding population (Goddard, 2009).
Simple sequence repeat markers are widely used in MAS because they are readily available and comparatively cheaper than others, and they require a simple technique with a higher polymorphism rate. MAS has been successfully applied in several major crops, but rarely in minor and orphan crops, including mungbean. As a result of limited progress in mungbean genomics research, only a few simple and efficient markers closely linked to phenotypic traits have been reported and validated recently, as mentioned above; thus, few MASs have been reported in mungbean. Marker-assisted backcrossing (MABC) was practiced to introduce VrPGIP2 from V2802 and VrMLO12 from V4718 to mungbean cultivar “Chai Nat 84-1” to improve resistance to bruchids and powdery mildew, respectively (Jittawimon, 2021). Papan et al. (2021) enhanced the CLS and PM resistance of mungbean cultivar “KING” via MABC using qCLSC72V18-1 (VrTAF5?) from V4718 CLS, and qPMC72V18-1 from V4718 and qPMC72V85 from V4785 for PM. Wu et al. (2022) improved bruchid resistance in the mungbean cultivar “Kamphaeng Saen 1” using VrPGIP2 from V2802 by MABC.
Since the publication of the first genomic mapping of mungbean in 1992, mungbean has entered the genomics era. Unfortunately, like other minor or orphan crops, mungbean has received little attention in the research community over the past 30 years; thus, it has not been established as a model crop species for genomics research. Despite the slow progress in genome research due to the limited genomic resources, the last 10 years have witnessed an increase in genomic studies in mungbean, resulting in significant findings and knowledge regarding some complex and important traits, especially insect and disease resistance, accumulated, thanks to advanced sequencing technologies and aiding in the construction of reference genome sequences of mungbean and genotyping of SNP markers.
The majority of mungbean genomics research is concentrated on resistance to a few diseases (PM, CLS, and YMD) and insect pests (bruchids and bean bug), while research on other traits, such as resistance to root diseases, tolerance to environmental stresses (e.g., scarce moisture, excessive moisture (waterlogging and preharvest sprouting), and high temperature), and the nutrient and chemical composition of seeds, has not been done or is very rare. At present, several omics areas, such as transcriptomics, proteomics, and phenomics technologies, have advanced, but only a few have been integrated into mungbean genomics research. Applying omics methods could accelerate and strengthen genomics research to solve the complexity of these traits in mungbean.
QTL mapping and information are the basis for MAB and GAB. Only biparental mapping populations have been used in mungbean. Generally, biparental populations provide low-resolution and imprecise QTLs of complex traits. Populations derived from intercrossing multiple parents, including nested association mapping (NAM) (McMullen et al., 2009) and multiparent advanced generation intercross (MAGIC) (Cavanagh et al., 2008) populations, have been proposed for use in high-resolution, precise QTL mapping. To date, two NAM mungbean populations (Nair et al., 2022) and one MAGIC mungbean population (Somta, unpublished data) have been developed, which will improve efficiency and precision in the genetic dissection of complex traits in mungbean.
In recent years, mungbean has been gaining interest from consumers and food industries and growing in popularity globally due to its high nutritional density, leading to increased attention being paid to the development of new mungbean cultivars that meet the demands of farmers, consumers, and industries in different countries by exploiting natural genetic variation through genomics. This is reflected by a considerable number of publications on the use of genomics and other omics sciences in mungbean from several laboratories within the last 5 years. Although advanced sequencing technology has revolutionized mungbean genome research, the genomics resources (such as high-quality genome sequences) still lag behind other crops, and genome information (such as QTLs) is still limited to a few traits and is insufficient for MAB and GAB. In the future, therefore, the main tasks in mungbean genome research will be to (i) construct complete, high-quality chromosome-scale reference genomes; (ii) identify the QTLs and genes controlling agronomically important traits, yield, quality, and resistance to biotic and abiotic stresses, as well as the molecular mechanisms underlying these traits; (iii) unravel genome variation in diverse mungbean germplasms (pan-genome analysis and assembly); (iv) create comprehensive databases of mungbean genomes and resources with the ability to analyze the genomic data; and (v) develop a highly efficient regeneration protocol and transformation system to support functional genomics. In addition, besides GSB, other SNP genotyping platforms, such as kompetitive allele-specific PCR (KASP) assay and SNP array, should be developed for mungbean to facilitate MAB and GAB.
To date, genomes of several other cultivated and wild Vigna species have been sequenced and are publicly available, and genomes of these species are highly syntenic with mungbean at both the micro and macro scale (Sakai et al., 2015; Lonardi et al., 2019; Pootakham et al., 2021; Guan et al. submitted) https://viggs.dna.affrc.go.jp). These Vigna genome sequences are valuable resources for gene identification and functional gene analysis of mungbean via comparative genome analysis. Since mungbean is a minor crop with a small research community and limited research funds, we propose that an international consortium for mungbean genomics research, making a collaborative effort to produce a body of genetic and genomic resources, should be established to accelerate mungbean genome research. Recently, the International Mungbean Improvement Network (IMIN), with the aim of strengthening local mungbean research, pre-breeding, and variety development capacity to generate improved, farmer-accepted varieties, was established (Nair et al., 2022). The IMIN is led by the WorldVeg with partners from Australia, Bangladesh, India, Myanmar, Kenya, Tanzania, and Uganda, and has generated resources and knowledge on mungbean genomics.
PS conceived the idea and designed the structure of this manuscript. PS and KL revised the manuscript. All authors collected literature, wrote the manuscript, contributed to the article, and approved the submitted version.
This work was supported by the National Research Council of Thailand (NRCT) (Grant No. N42A650274), Kasetsart University Research and Development Institute, the National Key R&D Program of China (2019YFE0109100), the China Agriculture Research System of MOF and MARA-Food Legumes (CARS-08-G15), and the Jiangsu Seed Industry Revitalization Project [JBGS(2021)004].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.944721/full#supplementary-material
Alam, A. M., Somta, P., Muktadir, M. A., and Srinives, P. (2014a). Quantitative trait loci associated with seed weight in mungbean [Vigna radiata (L.) Wilczek]. Kasetsart J. Nat. Sci. 48, 197–204.
Alam, A. M., Somta, P., and Srinives, P. (2014b). Identification and confirmation of quantitative trait loci controlling resistance to mungbean yellow mosaic disease in mungbean [Vigna radiata (L.) Wilczek]. Mol. Breed. 34, 1497–1506. doi: 10.1007/s11032-014-0133-0
Amarasinghe, S. L., Su, S., Dong, X., Zappia, L., Ritchie, M. E., and Gouil, Q. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 30. doi: 10.1186/s13059-020-1935-5
Anonymous (2021). Greengram Outlook Report. Available online at: https://angrau.ac.in/downloads/AMIC/GREENGRAM%20OUTLOOK%20REPORT%20-January%20to%20May%202021.pdf (accessed April 7, 2022).
Arsakit, K. (2019). ISSR and SSR Markers Linked to Powdery Mildew and Cercospora Leaf Spot Resistance in Mungbean. [Master's thesis], Mueang Nakhon Ratchasima: Suranaree University of Technology (Thailand).
Arumuganathan, K., and Earle, E. D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. doi: 10.1007/BF02672069
Ballon, F. B., Legaspi, B. M., and Catipon, E. M. (1977). “Mungbean varietal improvement program of the Bureau of plant industry” in The First International Mungbean Symposium, ed R. Cowell (Shanhua: AVRDC), 217–219.
Bernatzky, R., and Tanksley, S. D. (1986). Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112, 887–898. doi: 10.1093/genetics/112.4.887
Bose, R. D. (1932). Studies in Indian pulses. IV mung or green gram (Phaseolus aureus Roxb). Indian J. Agric. Sci. 2, 607–624.
Boutin, S. R., Young, N. D., Olson, T. C., Yu, Z. H., Vallejos, C. E., and Shoemaker, R. C. (1995). Genome conservation among three legume genera detected with DNA markers. Genome 38, 928–937. doi: 10.1139/g95-122
Breria, C. M., Hsieh, C. H., Yen, J. Y., Nair, R., Lin, C. Y., Huang, S. M., et al. (2020). Population structure of the world vegetable center mungbean mini core collection and genome-wide association mapping of loci associated with variation of seed coat luster. Trop. Plant Biol. 13, 1–12. doi: 10.1007/s12042-019-09236-0
Castillo, C. (2013). The Archaeobotany of Khao Sam Kaeo and Phu Khao Thong: The Agriculture of Late Prehistoric Southern Thailand. [Unpublished Ph.D. thesis], London: University College London.
Catipon, E. M., Legaspi, B. M., and Jarilla, F. A. (1988). “Development of mungbean varieties from AVRDC lines for the Philippines,” in Mungbean: Proceeding of the Second International Symposium, eds S. Shanmugasundaram, and B. T. McLean (Shanhua: AVRDC), 88–97.
Cavanagh, C., Morell, M., Mackay, I., and Powell, W. (2008). From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11, 215–221 doi: 10.1016/j.pbi.2008.01.002
Chai, M., Castillo, I. Q., Sonntag, A., Wang, S., Zhao, S., Liu, W., et al. (2021). A seed coat-specific β-ketoacyl-CoA synthase, KCS12, is critical for preserving seed physical dormancy. Plant Physiol. 186, 1606–1615. doi: 10.1093/plphys/kiab152
Chai, M., Zhou, C., Molina, I., Fu, C., Nakashima, J., Li, G., et al. (2016). A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. U.S.A. 113, 6997–7002. doi: 10.1073/pnas.1601256113
Chaitieng, B., Kaga, A., Han, O. K., Wang, X. W., Wongkaew, S., Laosuwan, P., et al. (2002). Mapping a new source of resistance to powdery mildew in mungbean. Plant Breed. 121, 521–525. doi: 10.1046/j.1439-0523.2002.00751.x
Chang, Y., Sun, F., Sun, S., Wang, L., Wu, J., and Zhu, Z. (2021). Transcriptome analysis of resistance to Fusarium wilt in mung bean (Vigna radiata L.). Front. Plant Sci. 12, 679629. doi: 10.3389/fpls.2021.679629
Chankaew, S., Somta, P., Isemura, T., Tomooka, N., Kaga, A., Vaughan, D. A., et al. (2013). Quantitative trait locus mapping reveals conservation of major and minor loci for powdery mildew resistance in four sources of resistance in mungbean [Vigna radiata (L.) Wilczek]. Mol. Breed. 32, 121–130. doi: 10.1007/s11032-013-9856-6
Chankaew, S., Somta, P., Sorajjapinun, W., and Srinives, P. (2011). Quantitative trait loci mapping of cercospora leaf spot resistance in mungbean, Vigna radiata (L.) Wilczek. Mol. Breed. 28, 255–264. doi: 10.1007/s11032-010-9478-1
Chen, H., Wang, L., Wang, S., Liu, C., Blair, M. W., and Cheng, X. (2015). Transcriptome sequencing of mung bean (Vigna radiata L.) genes and the identification of EST-SSR markers. PLoS ONE 10, e0120273. doi: 10.1371/journal.pone.0120273
Chen, H. M., Ku, H. S., Schafleitner, R., Bains, T. S., Kuo, C. G., Liu, C. A., et al. (2013). The major quantitative trait locus for mungbean yellow mosaic Indian virus resistance is tightly linked in repulsion phase to the major bruchid resistance locus in a cross between mungbean [Vigna radiata (L.) Wilczek] and its wild relative Vigna radiata ssp. sublobata. Euphytica 192, 205–216. doi: 10.1007/s10681-012-0831-9
Chen, J. B., Somta, P., Chen, X., Cui, X., Yuan, X., and Srinives, P. (2016). Gene mapping of a mutant mungbean (Vigna radiata L.) using new molecular markers suggests a gene encoding a YUC4-like protein regulates the chasmogamous flower trait. Front. Plant Sci. 7, 830. doi: 10.3389/fpls.2016.00830
Chen, L. R., Markhart, A. H., Shanmugasundaram, S., and Lin, T. Y. (2008). Early developmental and stress responsive ESTs from mungbean, Vigna radiata (L.) Wilczek, seedlings. Plant Cell Rep. 27, 535–552. doi: 10.1007/s00299-007-0488-3
Chotechung, S., Somta, P., Chen, J., Yimram, T., Chen, X., and Srinives, P. (2016). A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 129, 1673–1683. doi: 10.1007/s00122-016-2731-1
Collard, B. C., and Mackill, D. J. (2008). Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 363, 557–572. doi: 10.1098/rstb.2007.2170
Consonni, C., Humphry, M. E., Hartmann, H. A., Livaja, M., Durner, J., Wesphal, L., et al. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. doi: 10.1038/ng1806
Daovongdeuan, S. (2017). Genome-Wide Association Study for Seed Size and Color in Mungbean [Vigna radiata (L.) Wilczek]. [Master's thesis]. Seoul: Seoul National University.
Dasgupta, U., Mishra, G. P., Dikshit, H. K., Mishra, D. C., Bosamia, T., Roy, A., et al. (2021). Comparative RNA-Seq analysis unfolds a complex regulatory network imparting yellow mosaic disease resistance in mungbean [Vigna radiata (L.) R. Wilczek]. PLoS ONE 16, e0244593. doi: 10.1371/journal.pone.0244593
Della Coletta, R., Qiu, Y., Ou, S., Hufford, M. B., and Hirsch, C. N. (2021). How the pan-genome is changing crop genomics and improvement. Genome Biol. 22, 3. doi: 10.1186/s13059-020-02224-8
Deslauriers, S. D., and Larsen, P. B. (2010). FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol. Plant 3, 626–640. doi: 10.1093/mp/ssq015
Dikshit, H. K., Mishra, G. P., Somta, P., Shwe, T., Alam, A. K. M. M., Bains, T. S., et al. (2020). “Classical genetics and traditional breeding in mungbean,” in The Mungbean Genome, eds R.M. Nair, R. Schafleitner, and S. H. Lee (Cham: Springer Nature), 43–54. doi: 10.1007/978-3-030-20008-4_4
Duan, Q., Kita, D., Li, C., Cheung, A. Y., and Wu, H. M. (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U.S.A. 107, 17821–17826. doi: 10.1073/pnas.1005366107
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6, e19379. doi: 10.1371/journal.pone.0019379
Fatokun, C. A., Danesh, D., Stewart, E. L., and Young, N. D. (1993). “A linkage map for cowpea [Vigna unguiculata (L.) Walp.] based on DNA markers,” in Genetic Maps. Locus Maps of Complex Genomes, 6th Edn, ed S. J. O'Brien (New York, NY: Cold Spring Harbour Laboratory Press), 256–258.
Fatokun, C. A., Menancio-Hautea, D. I., Danesh, D., and Young, N. D. (1992). Evidence for orthologous seed weight genes in cowpea and mung bean based on RFLP mapping. Genetics 132, 841–846. doi: 10.1093/genetics/132.3.841
Feechan, A., Kabbara, S., and Dry, I. B. (2011). Mechanisms of powdery mildew resistance in the vitaceae family. Mol. Plant Pathol. 12, 263–274. doi: 10.1111/j.1364-3703.2010.00668.x
Fernandez, G. C. J., and Shanmugasundaram, S. (1988). “The AVRDC mungbean improvement programme: the past, present and future,” in Proceedings of the 2nd International Mungbean Sypmposium (Tainan: Asian Vegetable Research and Development Center), 58–70.
Finch-Savage, W. E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Fujii, K., and Miyazaki, S. (1987). Infestation resistance of wild legumes (Vigna sublobata) to azuki bean weevil, Callosobruchus chinensis (L.) (Coleoptera: Bruchidae) and its relationship with cytogenic classification. Appl. Entomol. Zool. 22, 229–230. doi: 10.1303/aez.22.229
Fuller, D. Q., and Harvey, E. (2006). The archaeobotany of Indian pulses: identification, processing and evidence for cultivation. Environ. Archaeol. 11, 219–246. doi: 10.1179/174963106x123232
Gayacharan, A. S., Archak, S., Gupta, K., Gupta, V., Tyagi, V., and Singh, K. (2020). “Mungbean genetic resources and utilization,” in The Mungbean Genome, eds R.M. Nair, R. Schafleitner, and S. H. Lee (Cham: Springer Nature), 9–26. doi: 10.1007/978-3-030-20008-4_2
Goddard, M. (2009). Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136, 245–257. doi: 10.1007/s10709-008-9308-0
Gupta, S. K., Bansal, R., and Gopalakrishna, T. (2014). Development and characterization of genic SSR markers for mungbean [Vigna radiata (L.) Wilczek]. Euphytica 195, 245–258. doi: 10.1007/s10681-013-0993-0
Gwag, J. G., Chung, J. W., Chung, H. K., Lee, G. H., Ma, K. H., Dixit, A., et al. (2006). Characterization of new microsatellite markers in mung bean, Vigna radiata (L.). Mol. Ecol. Notes 6, 1132–1134. doi: 10.1111/j.1471-8286.2006.01461.x
Ha, J., Satyawan, D., Jeong, H., Lee, E., Cho, K. H., Kim, M. Y., et al. (2021). A near-complete genome sequence of mungbean (Vigna radiata L.) provides key insights into the modern breeding program. Plant Genome 14, e20121. doi: 10.1002/tpg2.20121
Helentjaris, T. (1987). A genetic linkage map for maize based on RFLPs. Trends Genet. 3, 217–221. doi: 10.1016/0168-9525(87)90239-3
Hong, M. G., Kim, K. H., Ku, J. H., Jeong, J. K., Seo, M. J., Park, C. H., et al. (2015). Inheritance and quantitative trait loci analysis of resistance genes to bruchid and bean bug in mungbean (Vigna radiata L. Wilczek). Plant Breed. Biotechnol. 3, 39–46. doi: 10.9787/PBB.2015.3.1.039
Hu, H., Feng, N., Shen, X., Zhao, L., and Zheng, D. (2022). Transcriptomic analysis of Vigna radiata in response to chilling stress and uniconazole application. BMC Genomics 23, 205. doi: 10.1186/s12864-022-08443-6
Humphry, M., Konduri, V., Lambrides, C., Magner, T., McIntyre, C., Aitken, E., et al. (2002). Development of a mungbean (Vigna radiata) RFLP linkage map and its comparison with lablab (Lablab purpureus) reveals a high level of colinearity between the two genomes. Theor. Appl. Genet. 105, 160–166. doi: 10.1007/s00122-002-0909-1
Humphry, M. E., Lambrides, C. J., Chapman, S. C., Aitken, E. A. B., Imrie, B. C., Lawn, R. J., et al. (2005). Relationships between hard-seededness and seed weight in mungbean (Vigna radiata) assessed by QTL analysis. Plant Breed. 124, 292–298. doi: 10.1111/j.1439-0523.2005.01084.x
Humphry, M. E., Magner, T., McIntyre, C. L., Aitken, E. A. B., and Liu, C. J. (2003). Identification of major locus conferring resistance to powdery mildew (Erysiphe polygoni D.C.) in mungbean (Vigna radiata L. Wiczek) by QTL analysis. Genome 46, 738–744. doi: 10.1139/g03-057
Hwang, W. J., Ha, J., Lee, T., Jeong, H., Kum, M. Y., Kim, S. K., et al. (2017). A candidate flowering gene in mungbean is homologous to a soybean Phytochrome A gene. Euphytica 213, 79. doi: 10.1007/s10681-017-1866-8
Imrie, B. C., Williams, R. W., and Lawn, R. J. (1988). “Breeding for resistance to weather damage in mungbean,” in Mungbean: Proceedings of the Second International Symposium, eds S. Shanmugasundaram, and B. T. McLean (Shanhua: Asian Vegetable Research and Development Center), 130–135.
Isemura, T., Kaga, A., Tabata, S., Somta, P., Srinives, P., Shimizu, T., et al. (2012). Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS ONE 7, e41304. doi: 10.1371/journal.pone.0041304
Jeong, J. K., Kim, Y. H., Hwang, T. Y., Lee, I. J., Seo, B. Y., Moon, C. K., et al. (2015). Identification of Genes and Active Compounds Related to Insect-Resistance From Mungbean, and Analysis of Their Function. Jeonju: Final report, National Institute of Food Science and Technology, Rural Development Administration, Republic of Korea.
Jiao, K., Li, X., Guo, W., Yuan, X., Ciu, X., and Chen, X. (2016). Genome re-sequencing of two accessions and fine mapping the locus of lobed leaflet margins in mungbean. Mol. Breed. 36, 128. doi: 10.1007/s11032-016-0552-1
Jittawimon, P. (2021). Improving Resistance to Bruchids and Powdery Mildew Disease in Mungbean Cultivar ‘Chai Nat 84-1’ by Marker-Assisted Backcross Selection. [Master's thesis], Bangko: Kasetsart University.
Kaewwongwal, A., Chen, J., Somta, P., Kongjaimun, A., Yimram, T., Chen, X., et al. (2017). Novel alleles of two tightly linked genes encoding polygalacturonase inhibiting proteins (VrPGIP1 and VrPGIP2) associated with the Br locus that confer bruchid (Callosobruchus spp.) resistance to mungbean (Vigna radiata) accession V2709. Front. Plant Sci. 8, 1692. doi: 10.3389/fpls.2017.01692
Kaewwongwal, A., Liu, C., Somta, P., Chen, J., Tian, J., Yuan, X., et al. (2020). A second VrPGIP1 allele is associated with bruchid resistance (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) accession ACC41. Mol. Genet. Genomics 295, 275–286. doi: 10.1007/s00438-019-01619-y
Kaga, A., and Ishimoto, M. (1998). Genetic localization of a bruchid resistance gene and its relationship to insecticidal cyclopeptide alkaloids, the vignatic acids, in mungbean (Vigna radiata L. Wilczek). Mol. Gen. Genet. 258, 378–384. doi: 10.1007/s004380050744
Kajonphol, T., Sangsiri, C., Somta, T. T., and Srinives, P. (2012). SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean [Vigna radiata (L.) Wilczek]. SABRAO J. Breed. Genet. 44, 71–86.
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5, 5443. doi: 10.1038/ncomms6443
Kasettranan, W., Somta, P., and Srinives, P. (2010). Mapping of quantitative trait loci controlling powdery mildew resistance in mungbean [Vigna radiata (L.) Wilczek]. J. Crop Sci. Biotechnol. 13, 155–161. doi: 10.1007/s12892-010-0052-z
Keim, P., Diers, B. W., Olson, T. C., and Shoemaker, R. G. (1990). RFLP mapping in the soybean: association between marker loci and variation in quantitative traits. Genetics 126, 735–742. doi: 10.1093/genetics/126.3.735
Kelly, L. A., Vaghefi, N., Bransgrove, K., Fechner, N., Stuart, K., Pandey, A. K., et al. (2021). One crop disease, how many pathogens? Podosphaera xanthii and Erysiphe vignae sp. nov. identified as the two species that cause powdery mildew of mungbean (Vigna radiata) and black gram (V. mungo) in Australia. Phytopathology 111, 1193–1206. doi: 10.1094/PHYTO-12-20-0554-R
Kim, S. K., Nair, R. M., Lee, J., and Lee, S. H. (2015). Genomic resources in mungbean for future breeding programs. Front. Plant Sci. 6, 626. doi: 10.3389/fpls.2015.00626
Kitamura, K., Ishimoto, M., and Sawa, M. (1988). Inheritance of resistance to infestation with azuki bean weevil in Vigna sublobata and successful incorporation to V. radiata. Jap. J. Breed. 38, 459–464. doi: 10.1270/jsbbs1951.38.459
Kitsanachandee, R., Somta, P., Chatchawankanphanich, O., Akhtar, K. P., Shah, T. M., Nair, R. M., et al. (2013). Detection of quantitative trait loci for mungbean yellow mosaic India virus (MYMIV) resistance in mungbean [Vigna radiata (L.) Wilczek] in India and Pakistan. Breed. Sci. 63, 367–373. doi: 10.1270/jsbbs.63.367
Kumar, S., Ayachit, G., and Sahoo, L. (2020). Screening of mungbean for drought tolerance and transcriptome profiling between drought-tolerant and susceptible genotype in response to drought stress. Plant Physiol. Biochem. 157, 229–238. doi: 10.1016/j.plaphy.2020.10.021
Kumar, S. V., Tan, G., Quah, S. C., and Yusoff, K. (2002b). Isolation of microsatellite markers in mungbean, Vigna radiata. Mol. Ecol. Notes 2, 96–98. doi: 10.1046/j.1471-8286.2002.00158.x
Kumar, S. V., Tan, S. G., Quah, S. C., and Yusoff, K. (2002a). Isolation and characterization of seven tetranucleotide microsatellite loci in mungbean, Vigna radiata. Mol. Ecol. Notes 2, 293–295. doi: 10.1046/j.1471-8286.2002.00239.x
Kusch, S., and Panstruga, R. (2017). mlo-based resistance: an apparently universal “weapon” to defeat powdery mildew disease. MPMI 30, 179–189. doi: 10.1094/MPMI-12-16-0255-CR
Lambrides, C. J., Lawn, R. J., Godwin, I. D., Manners, J., and Imrie, B. C. (2000). Two genetic linkage maps of mungbean using RFLP and RAPD markers. Aust. J. Agric. Res. 51, 415–425. doi: 10.1071/AR99052
Laosatit, K., Amkul, K., Yimram, T., Chen, J., Lin, Y., Yuan, X., et al. (2022). A class II KNOX gene, KNAT7-1, regulates physical seed dormancy in mungbean [Vigna radiata (L.) Wilczek]. Front. Plant Sci. 13, 852373. doi: 10.3389/fpls.2022.852373
Laosatit, K., Somta, P., Chen, X., and Srinives, P. (2020). “Genomic approaches to biotic stresses,” in The Mungbean Genome, eds R. M. Nair, R. Schafleitner, and S. H. Lee (Cham: Springer Nature). 133–168. doi: 10.1007/978-3-030-20008-4_10
Lawn, R. J., and Rebetzke, G. J. (2006). Variation among Australian accessions of the wild mungbean (Vigna radiata ssp. sublobata) for traits of agronomic, adaptive, or taxonomic interest. Aust. J. Agric. Res. 57, 119–132. doi: 10.1071/AR05215
Lawn, R. J., Williams, R. W., and Imrie, B. C. (1988). “Potential of wild germplasm as a source of tolerance to environmental stresses in mungbean,” in Mungbean: Proceedings of the Second International Symposium, eds S. Shanmugasundaram, and B. T. McLean (Shanhua: Asian Vegetable Research and Development Centre), 136–145.
Lee, E., Yang, X., Ha, J., Kim, M. Y., Park, K. Y., and Lee, S. H. (2021). Identification of a locus controlling compound raceme inflorescence in mungbean [Vigna radiata (L.) R. Wilczek]. Front. Genet. 12, 642518. doi: 10.3389/fgene.2021.642518
Lee, S. Y., Hwang, S. J., and Lee, S. B. (2002). Occurrence of powdery mildew on mung bean (Vigna radiatus L.) caused by Sphaerotheca phaseoli. Res. Plant Dis. 8, 166–170. doi: 10.5423/RPD.2002.8.3.166
Li, S., Ding, Y., Zhang, D., Wang, X., Tang, X., Dai, D., et al. (2018). Parallel domestication with a broad mutational spectrum of determinate stem growth habit in leguminous crops. Plant J. 96, 761–771. doi: 10.1111/tpj.14066
Li, S. W., Shi, R. F., and Leng, Y. (2015). De novo characterization of the mung bean transcriptome and transcriptomic analysis of adventitious rooting in seedlings using RNA-seq. PLoS ONE 10, e0132969. doi: 10.1371/journal.pone.0132969
Lin, W. J., Ko, C. Y., Liu, M. S., Kuo, C. Y., Wu, D. C., Chen, C. Y., et al. (2016). Transcriptomic and proteomic research to explore bruchid-resistant genes in mungbean isogenic lines. J. Agric. Food Chem. 64, 6648–6658. doi: 10.1021/acs.jafc.6b03015
Lin, X., Liu, B., Weller, J. L., Abe, J., and Kong, F. (2021). Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 63, 981–994. doi: 10.1111/jipb.13021
Lin, Y., Laosatit, K., Chen, J., Yuan, X., Wu, R., Amkul, K., et al. (2020). Mapping and functional characterization of stigma exposed 1, a DUF1005 gene controlling petal and stigma cells in mungbean (Vigna radiata). Front. Plant Sci. 11, 575922. doi: 10.3389/fpls.2020.575922
Lin, Y., Laosatit, K., Liu, J., Chen, J., Yuan, X., Somta, P., et al. (2022). The mungbean VrP locus encoding MYB90, an R2R3-type MYB protein, regulates anthocyanin biosynthesis. Front. Plant Sci (in press).
Liu, B., Watanabe, S., Uchiyama, T., Kong, F., Kanazawa, A., Xia, Z., et al. (2010). The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 153, 198–210. doi: 10.1104/pp.109.150607
Liu, C., Fan, B., Cao, Z., Su, Q., Wang, Y., Zhang, Z., et al. (2016). A deep sequencing analysis of transcriptomes and the development of EST-SSR markers in mungbean (Vigna radiata). J. Genet. 95, 527–535. doi: 10.1007/s12041-016-0663-9
Liu, C., Wu, J., Wang, L., Fan, B., Cao, Z., Su, Q., et al. (2017). Quantitative trait locus mapping under irrigated and drought treatments based on a novel genetic linkage map in mungbean (Vigna radiata L.). Theor. Appl. Genet. 130, 2375–2393. doi: 10.1007/s00122-017-2965-6
Liu, C. Y., Su, Q. Z., Fan, B. J., Cao, Z. M., Zhang, Z. X., Wu, J., et al. (2018). Genetic mapping of bruchid resistance gene in mungbean V1128. Acta. Agron Sin. 44, 1875–1881. doi: 10.3724/SP.J.1006.2018.01875
Liu, M. S., Kuo, T. C. Y., Ko, C. Y., Wu, D. C., Li, K. Y., Lin, W. J., et al. (2016). Genomic and transcriptomic comparison of nucleotide variations for insights into bruchid resistance of mungbean [Vigna radiata (L.) R. Wilczek]. BMC Plant Biol. 16, 46. doi: 10.1186/s12870-016-0736-1
Lonardi, S., Muñoz-Amatriaín, M., Liang, Q., Shu, S., Wanamaker, S. I., Lo, S., et al. (2019). The genome of cowpea [Vigna unguiculata (L.) Walp.]. Plant J. 98, 767–782. doi: 10.1111/tpj.14349
MAOLI (2019). Myanmar Agriculture in Brief, 2018 and 2019. Naypyidaw: The Ministry of Agriculture, Livestock and Irrigation (MAOLI).
Masari, A., Kaewwongwal, A., Somta, P., and Srinives, P. (2017). Inheritance and a major quantitative trait locus of seed starch content in mungbean [Vigna radiata (L.) Wilczek]. Euphytica 213, 166. doi: 10.1007/s10681-017-1960-y
Mathivathana, M. K., Murukarthick, J., Karthikeyan, A., Jang, W., Dhasarathan, M., Jagadeeshselvam, N., et al. (2019). Detection of QTLs associated with mungbean yellow mosaic virus (MYMV) resistance using the interspecific cross of Vigna radiata × Vigna umbellata. J. Appl. Genet. 60, 255–268. doi: 10.1007/s13353-019-00506-x
Matlock, R. S., and Oswalt, R. M. (1963). Mungbean Varieties for Oklahoma. Stillwater: Bulletin B-612, Oklahoma Agricultural Experiment Station.
McCouch, S. R., Kochert, G., Yu, Z. H., Wang, Z. Y., Khush, G. S., Coffman, W. R., et al. (1988). Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76, 815–829. doi: 10.1007/BF00273666
McMullen, M. D., Kresovich, S., Villeda, H. S., Bradbury, P., Li, H., Sun, Q., et al. (2009). Genetic properties of the maize nested association mapping population. Science 325, 737–740. doi: 10.1126/science.1174320
Mei, L., Cheng, X. Z., Wang, S. H., Liu, C. Y., Sun, L., Xu, N., et al. (2009). Relationship between bruchid resistance and seed mass in mungbean based on QTL analysis. Genome 52, 589–596. doi: 10.1139/G09-031
Menancio-Hautea, D., Kumar, L., Danesh, D., and Young, N. D. (1993). “A genome map for mungbean [Vigna radiata (L.) Wilczek] based on DNA genetic markers (2n = 2x = 22),” in Genetic Maps 1992. A Compilation of Linkage and Restriction Maps of Genetically Studied Organisms, ed J. S. O'Brien (New York, NY: Cold Spring Harbor Laboratory Press).
Metzter, M. L. (2010). Sequencing technologies-the next generation. Nat. Rev. Genet. 11, 31–46. doi: 10.1038/nrg2626
Mockler, T. C., Yu, X., Shalitin, D., Parikh, D., Michael, T. P., Liou, J., et al. (2004). Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12759–12764. doi: 10.1073/pnas.0404552101
Moe, K. T., Chung, J. W., Cho, Y. I., Moon, J. K., Ku, J. H., Jung, J. K., et al. (2011). Sequence information on simple sequence repeats and single nucleotide polymorphisms through transcriptome analysis of mungbean. J. Integr Plant Biol. 53, 63–73. doi: 10.1111/j.1744-7909.2010.01012.x
Moe, K. T., Gwag, J. G., and Park, Y. J. (2012). Efficiency of PowerCore in core set development using amplified fragment length polymorphic markers in mungbean. Plant Breed. 131, 110–117. doi: 10.1111/j.1439-0523.2011.01896.x
Myles, S., Peiffer, J., Brown, P. J., Ersoz, E. S., Zhang, Z., Costich, D. E., et al. (2009). Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 21, 2194–2202. doi: 10.1105/tpc.109.068437
Nair, R., and Schreinemachers, P. (2020). “Global status and economic importance of mungbean”, in The Mungbean Genome, ed R. M. Nair, R. Schafleitner, and S. H. Lee (Cham: Springer International Publishing), 1–8. doi: 10.1007/978-3-030-20008-4_1
Nair, R. M., Alam, A. K. M. M., Douglas, C., Gowda, A., Pratap, A., Win, M. M., et al. (2022). Establishing the International Mungbean Improvement Network: Final report. Canberra, ACT: Australian Centre for International Agricultural Research.
Nair, R. M., Pandey, A. K., War, A. R., Hanumantharao, B., Shwe, T., Alam, A. K. M. M., et al. (2019). Biotic and abiotic constraints in mungbean production-progress in genetic improvement. Front. Plant Sci. 10, 1340. doi: 10.3389/fpls.2019.01340
Noble, T. J., Tao, Y., Mace, E. S., Williams, B., Jordan, D. R., Douglas, C. A., et al. (2018). Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Front. Plant Sci. 8, 2102. doi: 10.3389/fpls.2017.02102
Nogueira, F. C., Silva, C. P., Alexandre, D., Samuels, R. I., Soares, E. L., Aragão, F. J., et al. (2012). Global proteome changes in larvae of Callosobruchus maculatus (Coleoptera: Chrysomelidae: Bruchinae) following ingestion of a cysteine proteinase inhibitor. Proteomics 12, 2704–2715. doi: 10.1002/pmic.201200039
Pandey, A. K., Burlakoti, R. R., Kenyon, L., and Nair, R. M. (2018). Perspectives and challenges for sustainable management of fungal diseases of mungbean [Vigna radiata (L.) R. Wilczek var. radiata]: a review. Front. Environ. Sci. 6, 53. doi: 10.3389/fenvs.2018.00053
Pandey, R. N., Pawar, S. E., Chintalwar, G. J., and Bhatia, C. R. (1989). Seed coat and hypocotyl pigments in greengram and blackgram. Proc. Indian Acad. Sci. 99, 301. doi: 10.1007/BF03053572
Panstruga, R. (2005). Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 33, 389–392. doi: 10.1042/BST0330389
Papan, P., Chueakhunthod, W., Jinagool, W., Masari, A., Kaewkasi, C., Ngampongsai, S., et al. (2021). Improvement of cercospora leaf spot and powdery mildew resistance of mungbean variety KING through marker-assisted selection. J. Agric. Sci. 159, 676–687. doi: 10.1017/S0021859621000976
Paterson, A. H., Lander, E. S., Hewitt, J. D., Peterson, S., Lincoln, S. E., and Tanksley, S. D. (1988). Resolution of quantitative traits into mendelian factors, using a complete linkage map of restriction fragment length polymorphism. Nature 335, 721–726. doi: 10.1038/335721a0
Pessina, S., Lenzi, L., Perazzolli, M., Campa, M., Costa, L. A., Urso, S., et al. (2016). Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 3, 16016. doi: 10.1038/hortres.2016.16
Poolsawat, O., Kativat, C., Arsakit, K., and Tantasawat, P. A. (2017). Identification of quantitative trait loci associated with powdery mildew resistance in mungbean using ISSR and ISSR-RGA markers. Mol. Breed. 37, 150. doi: 10.1007/s11032-017-0753-2
Pootakham, W., Nawae, W., Naktang, C., Sonthirod, C., Yoocha, T., Kongkachana, W., et al. (2021). A chromosome-scale assembly of the black gram (Vigna mungo) genome. Mol. Ecol. Resour. 21, 238–250. doi: 10.1111/1755-0998.13243
Prathet, P., Somta, P., and Srinives, P. (2012). Mapping QTL conferring resistance to iron deficiency chlorosis in mungbean [Vigna radiata (L.) Wilczek]. Field Crops Res. 137, 230–236. doi: 10.1016/j.fcr.2012.08.002
Qi, Y., Gu, C., Wang, X., Gao, S., Li, C., Zhao, C., et al. (2020). Identification of the Eutrema salsugineum EsMYB90 gene important for anthocyanin biosynthesis. BMC Plant Biol. 20, 186. doi: 10.1186/s12870-020-02391-7
Reddy, V. R. P., Das, S., Dikshit, H. K., Mishra, G. P., Aski, M., Meena, S. K., et al. (2020). Genome-wide association analysis for phosphorus use efficiency traits in mungbean (Vigna radiata L. Wilczek) using genotyping by sequencing approach. Front. Plant Sci. 11, 537766. doi: 10.3389/fpls.2020.537766
Riester, L., Köster-Hofmann, S., Doll, B. K. W., and Zentgraf, U. (2019). Impact of alternatively polyadenylated isoforms of ETHYLENE RESPONSE FACTOR4 with activator and repressor function on senescence in Arabidopsis thaliana L. Genes 10, 91. doi: 10.3390/genes10020091
Rispail, N., and Rubiales, D. (2016). Genome-wide identification and comparison of legume MLO gene family. Sci. Rep. 6, 32673. doi: 10.1038/srep32673
Sakai, H., Naito, K., Ogiso-Tanaka, E., Takahashi, Y., Iseki, K., Muto, C., et al. (2015). The power of single molecule real-time sequencing technology in the de novo assembly of a eukaryotic genome. Sci. Rep. 5, 16780. doi: 10.1038/srep16780
Sandhu, K., and Singh, A. (2021). Strategies for the utilization of the USDA mung bean germplasm collection for breeding outcomes. Crop Sci. 61, 422–442. doi: 10.1002/csc2.20322
Sangiri, C., Kaga, A., Tomooka, N., Vaughan, D. A., and Srinives, P. (2007). Genetic diversity of the mungbean (Vigna radiata, Leguminosae) genepool based on microsatellite analysis. Aust. J. Bot. 55, 837–847. doi: 10.1071/BT07105
Sarikarin, N., Srinives, P., Kaveeta, R., and Saksoong, P. (1999). Effect of seed texture layer on bruchid infestation in mungbean Vigna radiata (L.) Wilczek. Sci. Asia 25, 203–206. doi: 10.2306/scienceasia1513-1874.1999.25.203
Saze, H., and Kakutani, T. (2007). Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 26, 3641–3652. doi: 10.1038/sj.emboj.7601788
Schafleitner, R., Huang, S., Chu, S., Yen, J., Lin, C., Yan, M., et al. (2016). Identification of single nucleotide polymorphism markers associated with resistance to bruchids (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) and cultivated V. radiata through genotyping by sequencing and quantitative trait locus analysis. BMC Plant Biol 16, 159. doi: 10.1186/s12870-016-0847-8
Schafleitner, R., Nair, R., Rathore, A., Wang, Y. W., Lin, C. Y., Chu, S. H., et al. (2015). The AVRDC – the world vegetable center mungbean (Vigna radiata) core and mini core collections. BMC Genomics 16, 344. doi: 10.1186/s12864-015-1556-7
Seehalak, W., Somta, P., Sommanas, W., and Srinives, P. (2009). Microsatellite markers for mungbean developed from sequence database. Mol. Ecol. Resour. 9, 862–864. doi: 10.1111/j.1755-0998.2009.02655.x
Sequeros, T., Schreinemachers, P., Depenbusch, L., Shwe, T., and Nair,. R. M. (2020). Impact and returns on investment of mungbean research and development in Myanmar. Agric Food Secur. 9, 5. doi: 10.1186/s40066-020-00260-y
Sheu, Z. M., Chiu, M. H., and Kenyon, L. (2021). First report of Podosphaera xanthii causing powdery mildew on mungbean (Vigna radiata) in Taiwan. Plant Dis. 105, 1856. doi: 10.1094/PDIS-09-20-2092-PDN
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Singh, D. P., Sharma, B. L., and Dwivedi, S. (1983). Inheritance of hard seeds in interspecific crosses of mungbean. Indian J. Genet. 43, 378–379.
Soffers, J. H. M., and Workman, J. L. (2020). The SAGA chromatin-modifying complex: the sum of its parts is greater than the whole. Genes Dev. 34, 1287–1303. doi: 10.1101/gad.341156.120
Sompong, U., Somta, P., Raboy, V., and Srinives, P. (2012). Mapping quantitative trait loci for phytic acid and phosphorus content in seed and seedlings of mungbean [Vigna radiata (L.) Wilczek]. Breed. Sci. 62, 87–92. doi: 10.1270/jsbbs.62.87
Somta, C., Somta, P., Tomooka, N., Ooi, P. A. C., Vaughan, D. A., and Srinives, P. (2008). Characterization of new sources of mungbean [Vigna radiata (L) Wilczek] resistance to bruchids, Callosobruchus spp. (Coleoptera: Bruchidae). J. Stored Prod. Res. 44, 316–321. doi: 10.1016/j.jspr.2008.04.002
Somta, P., Ammaranan, C., Ooi, P. A. C., and Srinives, P. (2007). Inheritance of seed resistance to bruchids in cultivated mungbean (Vigna radiata, L. Wilczek). Euphytica 155, 47–55. doi: 10.1007/s10681-006-9299-9
Somta, P., Chankaew, S., Kongjaimun, A., and Srinives, P. (2015). QTLs controlling seed weight and days to flowering in mungbean [Vigna radiata (L.) Wilczek], their conservation in azuki bean [V. angularis (Ohwi) Ohwi and Ohashi] and rice bean [V. umbellata (Thunb.) Ohwi and Ohashi]. Agrivita 37, 159–168. doi: 10.17503/Agrivita-2015-37-2-p159-168
Somta, P., Musch, W., Kongsamai, B., Chanprame, S., Nakasathien, S., Toojinda, T., et al. (2008). New microsatellite markers isolated from mungbean [Vigna radiata (L.) Wilczek]. Mol. Ecol. Resour. 8, 1155–1157. doi: 10.1111/j.1755-0998.2008.02219.x
Somta, P., Prathet, P., Kongjaimun, A., and Srinives, P. (2014). Dissecting quantitative trait loci for agronomic traits responding to iron deficeincy in mungbean [Vigna radiata (L.) Wilczek]. Agrivita 36, 101–111. doi: 10.17503/Agrivita-2014-36-2-p101-111
Somta, P., Seehalak, W., and Srinives, P. (2009). Development, characterization and cross-species amplification of mungbean (Vigna radiata) genic microsatellite markers. Conserv. Genet. 10, 1939. doi: 10.1007/s10592-009-9860-x
Sreeratree, J., Butsayawarapat, P., Chaisan, T., Somta, P., and Juntawong, P. (2022). RNA-Seq reveals waterlogging-triggered root plasticity in mungbean associated with ethylene and jasmonic acid signal integrators for root regeneration. Plants. 11, 930. doi: 10.3390/plants11070930
Srinives, P., Kitsanachandee, R., Thitaporn, C., Sommanas, W., and Chanprame, P. (2010). Inheritance of resistance to iron deficiency and identification of AFLP markers associated with the resistance in mungbean [Vigna radiata (L.) Wilczek]. Plant Soil. 335, 423–437. doi: 10.1007/s11104-010-0431-1
Srinives, P., Somta, P., and Somta, C. (2007). Genetics and breeding of resistance to bruchids (Callosobruchus spp.) in Vigna crops: a review. NU Int. J. Sci. 4, 1–17.
Sudha, M., Karthikeyan, A., Madhumitha, B., Ranjani, R. V., Mathivathana, M. K., Dhasarathan, M., et al. (2022). Dynamic transcriptome profiling of mungbean genotypes unveil the genes respond to the infection of mungbean yellow mosaic virus. Pathogens 11, 190. doi: 10.3390/pathogens11020190
Tang, D., Dong, Y., Ren, H., Li, L., and He, C. (2014). A review of phytochemistry, metabolite changes, and medicinal uses of the common food mung bean and its sprouts (Vigna radiata). Chem. Cent. J. 8, 4. doi: 10.1186/1752-153X-8-4
Tangphatsornruang, S., Sangsrakru, D., Chanprasert, J., Uthaipaisanwong, P., Yoocha, T., Jomchai, N., et al. (2010). The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 17, 11–22. doi: 10.1093/dnares/dsp025
Tangphatsornruang, S., Somta, P., Uthaipaisanwong, P., Chanprasert, J., Sangsrakru, D., Seehalak, W., et al. (2009). Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean [Vigna radiata (L.) Wilczek]. BMC Plant Biol. 9, 137. doi: 10.1186/1471-2229-9-137
Tian, X., Li, S., Liu, Y., and Liu, X. (2016). Transcriptomic profiling reveals metabolic and regulatory pathways in the desiccation tolerance of mungbean [Vigna radiata (L.) R. Wilczek]. Front. Plant Sci. 7, 1921. doi: 10.3389/fpls.2016.01921
Tomooka, N., Lairungreang, C., Nakeeraks, P., Egawa, Y., and Thavarasook, C. (1992). Center of genetic diversity and dissemination pathways in mung bean deduced from seed protein electrophoresis. Theor Appl Genet. 83, 289–293 doi: 10.1007/BF00224273
Vallejos, E. C., Sakiyama, N. S., and Chase, C. D. (1992). A molecular marker based linkage map of Phaseolus vulgaris L. Genetics 131, 733–740. doi: 10.1093/genetics/131.3.733
van Schie, C. C., and Takken, F. L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Van, K., Kang, Y. J., Han, K. S., Lee, Y. H., Gwag, J. G., Moon, J. K., et al. (2013). Genome-wide SNP discovery in mungbean by illumina HiSeq. Theor. Appl. Genet. 126, 2017–2027. doi: 10.1007/s00122-013-2114-9
Wang, L., Wu, C., Zhong, M., Zhao, D., Mei, L., Chen, H., et al. (2016). Construction of an integrated map and location of a bruchid resistance gene in mung bean. Crop J. 4, 360–366. doi: 10.1016/j.cj.2016.06.010
Wang, L., Yang, T., Lin, Q., Li, X., Luan, S., and Yu, F. (2020). Receptor kinase FERONIA regulates flowering time in Arabidopsis. BMC Plant Biol. 20, 26. doi: 10.1186/s12870-019-2223-y
Wang, X., Kaga, A., Tomooka, N., and Vaughan, D. A. (2004). The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean [Vigna angularis (Willd.) Ohwi and Ohashi]. Theor. Appl. Genet. 109, 352–360. doi: 10.1007/s00122-004-1634-8
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10:57–63. doi: 10.1038/nrg2484
Watt, E. E., Poehlman, J. M., and Cumbie, B. G. (1977). Origin and composition of texture layer on seed of mungbean. Crop Sci. 17, 121–125. doi: 10.2135/cropsci1977.0011183X001700010033x
Wu, R., Zhang, Q., Lin, Y., Chen, J., Somta, P., Yan, Q., et al. (2022). Marker-assisted backcross breeding for improving bruchid (Callosobruchus spp.) resistance in mung bean (Vigna radiata L.). Agronomy. 12:1271. doi: 10.3390/agronomy12061271
Wu, X., Islam, A. S. M. F., Limpot, N., Mackasmiel, L., Mierzwa, J., Cortés, A. J., et al. (2020). Genome-wide SNP identification and association mapping for seed mineral concentration in mung bean (Vigna radiata L.). Front. Genet. 11, 656. doi: 10.3389/fgene.2020.00656
Xie, Z., Nolan, T. M., Jiang, H., and Yin, Y. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10, 228. doi: 10.3389/fpls.2019.00228
Xu, H., Wang, N., Liu, J., Qu, C., Wang, Y., Jiang, S., et al. (2017). The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 94, 149–165. doi: 10.1007/s11103-017-0601-0
Yan, J., Liu, Y., Huang, X., Li, L., Hu, Z., Zhang, J., et al. (2019). An unreported NB-LRR protein SUT1 is required for the autoimmune response mediated by type one protein phosphatase 4 mutation (topp4-1) in Arabidopsis. Plant J. 100, 357–373. doi: 10.1111/tpj.14447
Yan, Q., Wang, Q., Xuzhen, C., Wang, L., Somta, P., Xue, C., et al. (2020). High-quality genome assembly, annotation and evolutionary analysis of the mungbean (Vigna radiata) genome. Authorea 2020, 20. doi: 10.22541/au.160587196.63922177/v1
Yimram, T., Somta, P., and Srinives, P. (2009). Genetic variation in cultivated mungbean germplasm and its implication in breeding for high yield. Field Crops Res. 112, 260–266. doi: 10.1016/j.fcr.2009.03.013
Young, N. D., Danesh, D., Menancio-Hautea, and Kumar, L. (1993). Mapping oligogenic resistance to powdery in mungbean with RFLPs. Theor. Appl. Genet. 87, 243–249. doi: 10.1007/BF00223772
Young, N. D., Kumar, L., Menancio-Hautea, D., Danesh, D., Talekar, N. S., Shanmugasundaram, S., et al. (1992). RFLP mapping of a major bruchid resistance gene in mungbean (Vigna radiata, L. Wilczek). Theor. Appl. Genet. 84, 839–844. doi: 10.1007/BF00227394
Yu, K., Park, S. J., and Poysa, V. (1999). Abundance and variation of microsatellite DNA sequences in beans (Phaseolus and Vigna). Genome 42, 27–34. doi: 10.1139/g98-100
Yundaeng, C., Somta, P., Chen, J., Yuan, X., Chankaew, S., and Chen, X. (2021). Fine mapping of QTL conferring cercospora leaf spot disease resistance in mungbean revealed TAF5 as candidate gene for the resistance. Theor. Appl. Genet. 134, 701–714. doi: 10.1007/s00122-020-03724-8
Yundaeng, C., Somta, P., Chen, J., Yuan, X., Chankaew, S., Srinives, P., et al. (2020). Candidate gene mapping reveals VrMLO12 (MLO Clade II) is associated with powdery mildew resistance in mungbean [Vigna radiata (L.) Wilczek]. Plant Sci. 298, 110594. doi: 10.1016/j.plantsci.2020.110594
Zhang, M. C., Wang, D. M., Zheng, Z., Humphry, M., and Liu, C. J. (2008). Development of PCR-based markers for a major locus conferring powdery mildew resistance in mungbean (Vigna radiata). Plant Breed. 127, 429–432. doi: 10.1111/j.1439-0523.2008.01521.x
Zhang, Q., Yan, Q., Yuan, X., Lin, Y., Chen, J., Wu, R., et al. (2021). Two polygalacturonase-inhibiting proteins (VrPGIP) of Vigna radiata confer resistance to bruchids (Callosobruchus spp.). J. Plant Physiol. 258–259, 153376. doi: 10.1016/j.jplph.2021.153376
Zhao, D., Cheng, X. Z., Wang, L. X., Wang, S. H., and Ling, M. A. Y. (2010). Integration of mungbean (Vigna radiata) genetic linkage map. Acta. Agron. Sin. 36, 932–939. doi: 10.3724/SP.J.1006.2010.00932
Zhao, P., Ma, B., Cai, C., and Xu, J. (2022). Transcriptome and methylome changes in two contrasting mungbean genotypes in response to drought stress. BMC Genomics 23, 80. doi: 10.1186/s12864-022-08315-z
Keywords: mungbean, gene mapping, genome research, genomics, QTL, marker-assisted breeding
Citation: Somta P, Laosatit K, Yuan X and Chen X (2022) Thirty Years of Mungbean Genome Research: Where Do We Stand and What Have We Learned? Front. Plant Sci. 13:944721. doi: 10.3389/fpls.2022.944721
Received: 15 May 2022; Accepted: 17 June 2022;
Published: 15 July 2022.
Edited by:
Baohua Wang, Nantong University, ChinaReviewed by:
Xin Li, Nanjing Agricultural University, ChinaCopyright © 2022 Somta, Laosatit, Yuan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prakit Somta, YWdycGtzQGt1LmFjLnRo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.