- 1Henan Engineering Research Center of Crop Genome Editing, Henan International Joint Laboratory of Plant Genetic Improvement and Soil Remediation, College of Life Science and Technology, Henan Institute of Science and Technology, Xinxiang, China
- 2College of Landscape Architecture and Horticulture, Henan Institute of Science and Technology, Xinxiang, China
- 3College of Biological Engineering, Henan University of Technology, Zhengzhou, China
The ankyrin-transmembrane (ANKTM) subfamily is the most abundant subgroup of the ANK superfamily, with critical roles in pathogen defense. However, the function of ANKTM proteins in wheat immunity remains largely unexplored. Here, a total of 381 ANKTMs were identified from five Triticeae species and Arabidopsis, constituting five classes. Among them, class a only contains proteins from Triticeae species and the number of ANKTM in class a of wheat is significantly larger than expected, even after consideration of the ploidy level. Tandem duplication analysis of ANKTM indicates that Triticum urartu, Triticum dicoccoides and wheat all had experienced tandem duplication events which in wheat-produced ANKTM genes all clustered in class a. The above suggests that not only did the genome polyploidization result in the increase of ANKTM gene number, but that tandem duplication is also a mechanism for the expansion of this subfamily. Micro-collinearity analysis of Triticeae ANKTMs indicates that some ANKTM type genes evolved into other types of ANKs in the evolution process. Public RNA-seq data showed that most of the genes in class d and class e are expressed, and some of them show differential responses to biotic stresses. Furthermore, qRT-PCR results showed that some ANKTMs in class d and class e responded to powdery mildew. Silencing of TaANKTM2A-5 by barley stripe mosaic virus-induced gene silencing compromised powdery mildew resistance in common wheat Bainongaikang58. Findings in this study not only help to understand the evolutionary process of ANKTM genes, but also form the basis for exploring disease resistance genes in the ANKTM gene family.
Introduction
Wheat (Triticum aestivum L.) is a leading source of calories for the global human diet (Schilling et al., 2020). Changes in the climate and crop planting systems have not only caused environmental degradation, but also the prevalence of diseases (Guo et al., 2010; Cui et al., 2018). In the winter wheat area, the proportion of powdery mildew in the total sown area has increased yearly with the change in climate (Tang et al., 2017). The most environmentally friendly way to control plant fungal diseases is to mine the gene pool for resistance and the cultivation of disease-resistant varieties (Kuraparthy et al., 2007).
The ankyrin repeat (ANK) is a 33-residue, two alpha helix motifs in proteins and is one of the most common protein domains present in bacteria, virus, plants and humans (Sedgwick and Smerdon, 1999; Mosavi et al., 2004). In ANK-containing proteins, the number, spatial structure and primary sequence of the ANK domains can differ substantially, and ANK proteins have been found to play diverse biological functions, usually involving ANK domain-mediated protein–protein interactions (Shen et al., 2010; Fu et al., 2012; Vo et al., 2015). However, ANK repeat domain-containing proteins can also contain other domains, including zinc or ring fingers, calmodulin binding or transmembrane domains, thus forming the structurally and functionally diverse ANK protein superfamily (Becerra et al., 2004). Structurally, the most abundant protein subgroup contains ankyrin-transmembrane domains (ANKTM; Becerra et al., 2004) and functionally, ANKTM family genes have been described with roles in plant development, hormone signal transduction, but especially in the attenuation of abiotic and biotic stress (Vo et al., 2015).
Previous studies of the ANKTM proteins showed that these can interact with different ligands to participate in important physiological and developmental processes. For example, TIP1 from Arabidopsis, which contains ANK, TM and DHHC domains, the latter of which confers S-acyl transferase activity involved in the positive regulation of root hair formation (Hemsley et al., 2005). ANK1 from Nicotiana tabacum interacted with BZI-1 and BZI-2 in the nucleus, and is involved in both auxin signaling and the pathogen response (Kuhlmann et al., 2003). ANKTM proteins are also related to plant-microorganism interactions and abiotic stress, the ANKTM protein, IGN1 of Lotus japonicas, is involved in symbiotic nitrogen fixation in root nodules and the ign1 mutant plant grows abnormally due to nitrogen deficiency (Kumagai et al., 2007). ITN1, an Arabidopsis homolog of IGN1, is related to salt-stress tolerance through its effect on abscisic acid-induced production of reactive oxygen species under salt stress (Sakamoto et al., 2008).
In recent years, an increasing number of studies have shown that ANKTMs are involved in plant resistance to pathogens. The Arabidopsis ANKTM family gene BDA1 is related to plant disease resistance, and acts downstream of the receptor-like protein SNC2 and upstream of NPR1 and WRKY70 to regulate plant immunity (Yang et al., 2012). The ANKTM protein GBP, plays a role in the connection between defense response and carbohydrate metabolism and a loss-of-function mutant of gbp causes necrotic lesions (Wirdnam et al., 2004). ACCELERATED CELL DEATH6 (ACD6) is a widely studied ANKTM gene that play an important role in broad-spectrum resistance in Arabidopsis (Lu et al., 2005; Todesco et al., 2010). ACD6 acts in the plant immune response and is involved in salicylic acid signaling and salicylic acid-dependent cell death and defense (Rate et al., 1999; Lu et al., 2003). The Arabidopsis nucleotide-binding domain and leucine-rich-repeat-containing (NLR) resistance protein, SNC1, can modulate ACD6-dependent hyper immunity and link different arms of the plant immune system (Zhu et al., 2018). ACD6 can not only interact with pattern recognition receptors like BAK1 and CERK1 to form large complexes at the membrane (Tateda et al., 2014; Zhang et al., 2014b), but also regulates PAD4 and EDS1 to trigger the hypersensitive response (Feys et al., 2001). ZmACD6, the orthologous gene of ACD6 in Zea mays, confers resistance to Ustilago maydis (Zhang et al., 2019). Recently, the race-specific leaf rust resistance gene, Lr14a, from hexaploid wheat was shown to encode an ANKTM protein (Kolodziej et al., 2021). A further study showed that the wheat stripe rust resistance protein, YrU1, is an NLR protein with an integrated ANK domain, and the ANK domain of YrU1 is derived from ANKTM proteins (Wang et al., 2020; Kolodziej et al., 2021). The above studies suggest that several ANKTM-type genes are involved in basal resistance or effector-triggered immunity.
The gene expansion, evolution, expression pattern and function of ANKTM genes in Triticeae species remain largely unexplored. In this study, based on whole genome information of wheat and its related species, a new subfamily of the ANKTM-type proteins in Triticeae species was identified. The evolutionary relationship, gene distribution, tandem duplication of TaANKTM genes in Triticeae species, and expression pattern of these genes in response to different biotic stresses were systematically analyzed. The technology of barley stripe mosaic virus-induced gene silencing (BSMV-VIGS) was used to verify the functions of selected TaANKTM-type genes in wheat powdery mildew resistance, and the results showed that silencing of TaANKTM2A-5 compromised powdery mildew resistance in common wheat Bainongaikang58 (AK58). These findings not only help to understand the evolutionary process of ANKTM genes, but also provide gene resources for disease resistance breeding.
Materials and methods
Plant materials and Blumeria graminis f. sp. Tritici race preparation
Common wheat AK58 was used for the BSMV-VIGS and gene expression analysis assays, and was developed and maintained by the Henan Institute of Science and Technology (Xinxiang, China). For the BSMV-VIGS assay, AK58 was grown in a growth cabinet at 70% relative humidity with a 14 h light/10 h dark cycle at 16/12°C. For gene expression analysis, AK58 was grown in a growth cabinet at 70% relative humidity with a 14 h light/10 h dark cycle at 22/18°C. Mixed races of Blumeria graminis f. sp. Tritici (Bgt) were collected from an agricultural field in Xinxiang (China) and maintained on seedlings of the highly susceptibility wheat variety, Sumai 3, and cultured in the light incubator at 70% relative humidity with a 23°C/14 h light and 18°C/10 h dark cycle. The leaves of three individuals were collected at 0, 2, 6, 12, 24, 36, and 48 h post inoculation for RNA extraction.
Expression analysis of TaANKTMs by quantitative real-time reverse transcription-polymerase chain reaction
Total RNAs were extracted from collected samples using the RNA isolation kit, Total RNA Extraction Reagent (Vazyme, Nanjing, China), following the manufacturer’s protocol. The RNAs were reverse-transcribed using HiScript II 1st Strand cDNA Synthesis Kit (Vazyme). The qRT-PCR was performed using AceQ qPCR SYBR Green Master Mix (Vazyme) on a LC 480II platform (Roche, Germany). The procedure used was as follows: 95°C for 5 min, followed by 40 cycles at 95°C for 10 s, 60°C for 20 s. The comparative 2–ΔΔCT method was used to quantify relative gene expression. All the oligonucleotide primers used in this study (Supplementary Table 1) were synthesized by Gene create Corporation (Wuhan, China). The wheat TaTubulin gene was used as an internal control.
BSMV-VIGS
To silence the corresponding TaANKTM genes, fragments of the three selected TaANKTM2A-5/TaANKTM3A-2/ TaANKTM6A-1 genes with the length of 234 bp, 242 bp, and 276 bp were amplified with corresponding primer pairs (Supplementary Table 1). Each of the target fragment was inserted into the γ-strain of BSMV by the homogenous recombination method to produce BSMV:TaANKTM vectors. The second fully expanded leaves of AK58 were infected with the in vitro transcribed (mMESSAGEmMACHINE T7, Invitrogen, Waltham, MA, USA) viruses BSMV:TaANKTM2A-5, BSMV:TaANKTM3A-2 and BSMV:TaANKTM6A-1, while seedlings infected with BSMV:TaPDS and BSMV:γ served as controls. The infected plants were grown at 23°C, with a 14 h light/10 h dark cycle environment condition with 70% relative humidity. The fourth fully unfolded leaves with visible viral infection symptoms were detached and placed on 6BA-plate with mixed race of Bgt spores to evaluate disease resistance. The inoculated leaves were cultured in a light incubator with a cycle of 14 h light/22°C and 10 h dark at 18°C for 6 days. Target genes silencing efficiency were evaluated by qRT-PCR using the corresponding primer pair TaANKTM-Q (Supplementary Table 1).
Identification of the ANKTM genes in Triticeae species
The genomic data for T. aestivum (Chinese Spring) was obtained from IWGSC1 (IWGSC, 2018). The data for Triticum urartu (Tu 2.0) analysis was downloaded from the MBKBase website2 (Ling et al., 2018). Data for Triticum dicoccoides (WEWSeq_v.1.0), Hordeum vulgare (IBSC_v2), Aegilops tauschii (Aet_v4.0), and Arabidopsis thaliana (TAIR10) were downloaded from the Ensemble Plants website3 to construct a local database. The typical ANK domains (PF00023, PF12796, PF13606, PF13637, PF13857) used as the search models were downloaded from the Pfam database4 (El-Gebali et al., 2019). A new hidden Markov model (HMM) was built to ensure the search results were reliable. A high-quality protein set (E-value <1 × 10−20) with intact ANK and TM domains was obtained by the raw ANKTM HMM, and then used to construct a specific ANKTM HMM using hmmbuild from the HMMERv3 suite (Lozano et al., 2015; Xu et al., 2021). The specific ANKTM HMM was used to select the ANKTM protein, and as a result, the proteins with an E-value lower than 0.001 were retained. Both the conserved domains5 (Lu et al., 2020) and SMART6 (Simple Modular Architecture Research Tool; Letunic et al., 2021) were used to recheck the candidate ANKTM protein sequences. When a gene contained multiple transcripts, the longest transcript was retained for further analysis. The analyzed genes were renamed sequentially according to their species and chromosomal distributions on the chromosomes. The gene names and their corresponding gene IDs are listed in Supplementary Table 2.
Phylogenetic, chromosome localization, gene duplication and micro-collinearity analyses
All the full length ANKTM protein sequences were aligned by ClustalW with the default options in MEGA X and a phylogenetic tree was constructed using the Maximum Likelihood method with 1,000 bootstrap replicates (Kumar et al., 2018). The EvolView7 was used to visualize the phylogenetic tree (He et al., 2016). Multiple Collinearity Scan toolkit (MCScanX) was used to identify gene duplication (Wang et al., 2012). The shinyCircos software8 was used to express gene duplication events, the syntenic relationship of the gene pairs, and the chromosome localization of the analyzed genes (Yu et al., 2018). TGT (Triticeae-Gene Tribe9) was used to trace the evolutionary history of the target genes and for gene pair analyses (Chen et al., 2020b).
RNA-seq expression analysis
RNA-seq data of 145 wheat TaANKTM genes were downloaded from WheatOmics10 (Ma et al., 2021). Data for biotic stresses responses (powdery mildew and stripe rust) and after elicitation with PAMPs (chitin and flg22) were collected from N9134 (powdery mildew and stripe rust-resistant wheat) and Chinese Spring, respectively (Zhang et al., 2014a). The relative expressions of each TaANKTM gene in response to the different stresses were presented as a heat map constructed with TBtools (Chen et al., 2020a).
Results
Genome-wide identification and phylogenetic relationship analysis of the ANKTM genes in Triticeae species
In the present study, 145, 72, 36, 42, 42, and 44 ANKTM genes were identified from T. aestivum, T. dicoccoides, T. urartu, Ae. tauschii, H. vulgare and Arabidopsis, respectively. To study the evolutionary relationships of the ANKTM proteins, all the above 381 ANKTM protein sequences were used to construct a phylogenetic tree (Figure 1). ANKTM is divided into five classes (class a-e). Among them class a only contains proteins from Triticeae species, while the remaining classes include proteins from Triticeae species and Arabidopsis. Within classes b–e, the ANKTM members of Triticeae species are clustered together, in sub-classes distinct from those of Arabidopsis, indicating that the ANKTM genes of monocots and eudicots had experienced significant differentiation in the evolutionary process. The ANKTM protein sequences of the Triticeae species in all clades of the phylogenetic tree are highly similar, indicating that evolution of the ANKTM genes was relatively conservative after Triticeae species speciation.
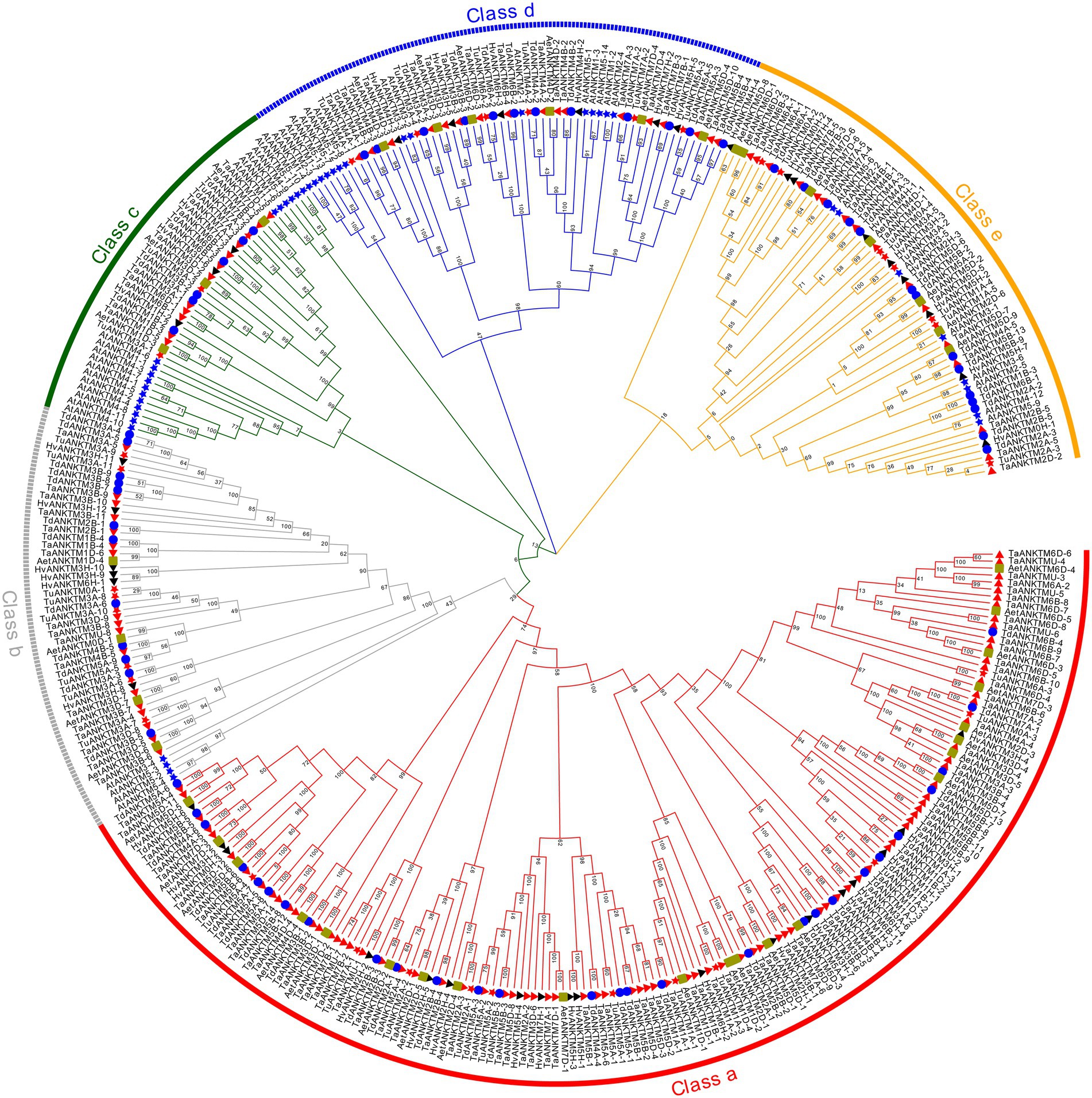
Figure 1. Phylogenetic relationship analysis of 381 ANKTM proteins from Triticum aestivum, Triticum dicoccoides, Aegilops tauschii, Triticum urartu, H. vulgare and Arabidopsis. The phylogenetic tree was built using the Maximum likelihood method with 1,000 bootstrap replicates by MEGA X. The diverse classes of ANKTM proteins were marked with different colors. The ANKTM proteins of T. aestivum, T. dicoccoides, A. tauschii, T. urartu, H. vulgare, and Arabidopsis were represented by red triangles, blue circles, yellow squares, red stars, black triangles and blue stars, respectively. Gene IDs of all the analyzed genes can be found in Supplementary Table 2.
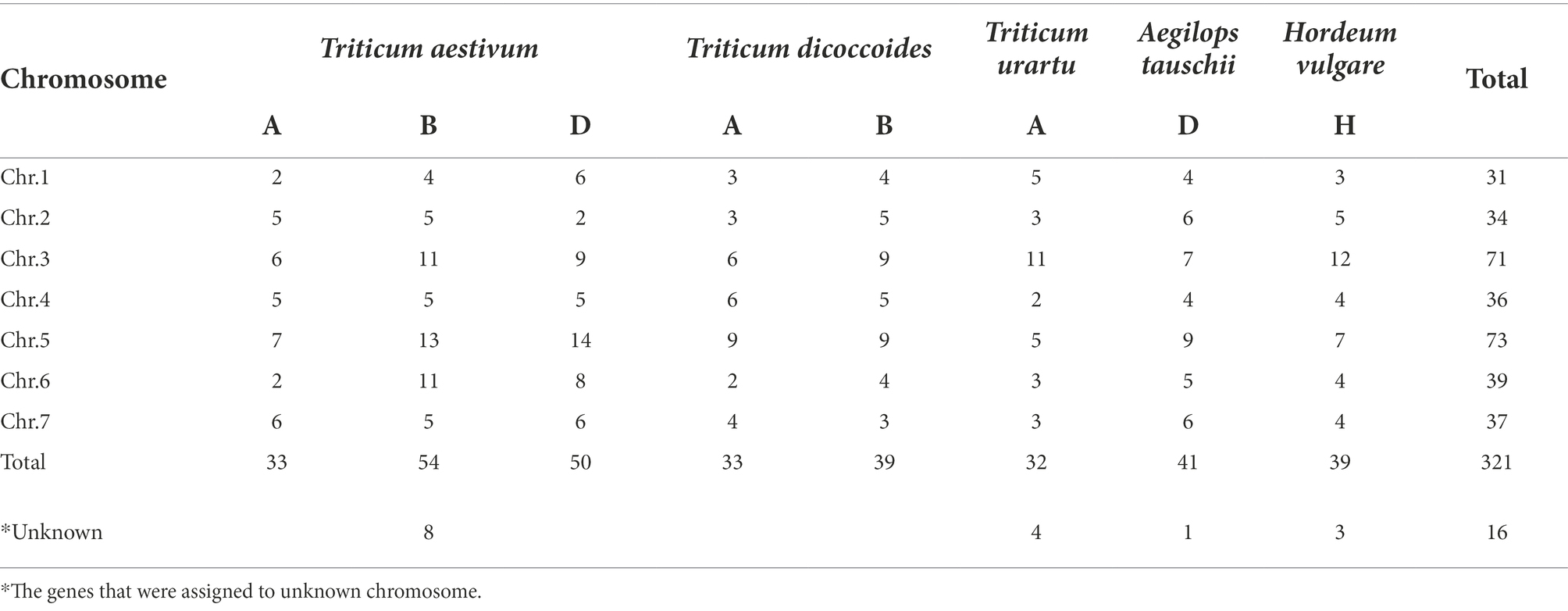
The numbers of ANKTM in each class of all the analyzed species are shown in Table 1. As a heterohexaploid species, wheat resulted from two rounds of hybridization (Shewry, 2009; Zhou et al., 2020). Generally, the number of gene family members in T. aestivum (AABBDD) and T. dicoccoides (AABB) is about 3 and 2 times that of other diploid Triticeae species, respectively. In this study, the number of ANKTM genes in T. dicoccoides was 2, 1.71, and 1.71 times that in T. urartu, Ae. tauschii and H. vulgare, respectively, whereas the quantity of ANKTM in wheat was 4.03, 3.45, 3.45, and 2.01 times greater that in T. urartu, Ae. tauschii, H. vulgare and T. dicoccoides, respectively. This is partly due to the polyploidization of common wheat. However, even after the ploidy level is considered, the number of ANKTM genes in common wheat was significantly higher than that in diploid and tetraploid species. On the contrary, the number of ANKTM in T. dicoccoides was slightly less.
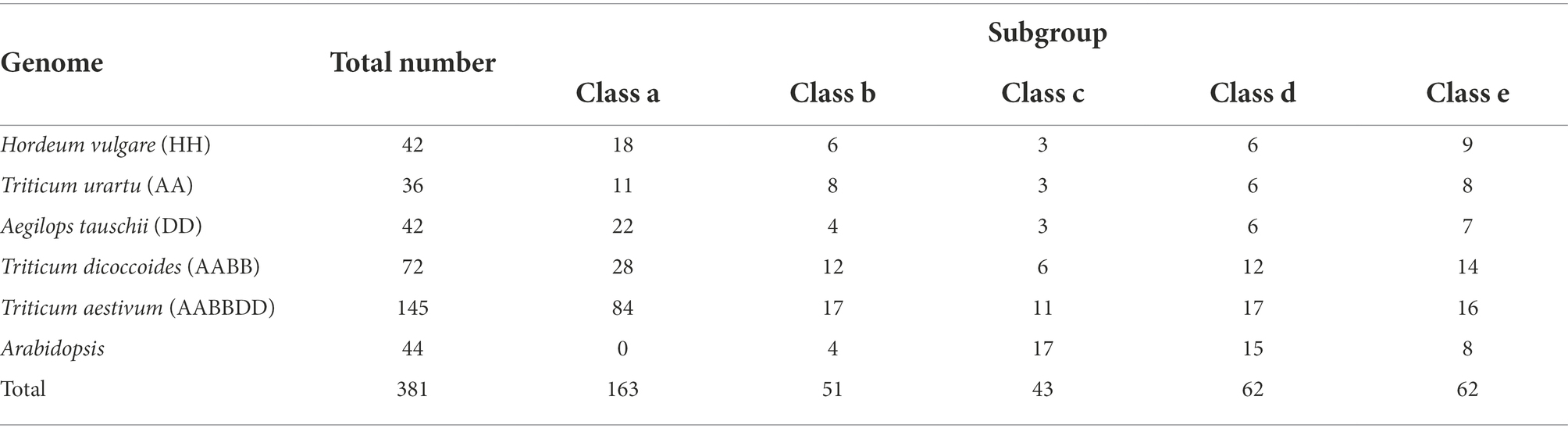
Table 1. Numbers of ANKTMs in the five analyzed Triticeae species and Arabidopsis genomes in total and each class.
For the dicotyledonous plant Arabidopsis, most of the AtANKTMs were mainly distributed in class c and class d, accounting for 72.73% of the total AtANKTM genes, while the proportion of Triticeae ANKTM genes in these two classes was only 21.66%. Overall, 48.73% of Triticeae ANKTM genes were concentrated in class a (Table 1). Class a and class b had significant variation in the proportion of ANKTM among the different Triticeae species (Table 1). The numbers of ANKTM genes of wheat in class d and class e were below the expected 3:2 or 3:1 ratio compared with the analyzed tetraploid and diploid species, respectively. Conversely, the numbers of wheat ANKTM in classes a and class c were significantly larger than expected. The number of TaANKTM in class a of T. aestivum was about 3, 3.82, 7.64 times that in T. dicoccoides, Ae. tauschii and T. urartu, respectively (Table 1); and in class c the number of TaANKTM from T. aestivum was about 1.83, 3.66, 3.66 times that in T. dicoccoides, Ae. tauschii and T. urartu, respectively (Table 1); suggesting that wheat ANKTM in class a and class c underwent gene expansion.
Chromosome distribution and gene duplication
The chromosome and subgenome distribution of ANKTMs from the five Triticeae species are shown in Table 2. ANKTM genes were generally equally distributed among the chromosomes, except the homologous groups 3 and 5, which contained significantly more genes relative to the other homologous groups. The ANKTM gene number in the B and D subgenomes were higher than that of the A subgenome (Table 2). From diploid to tetraploid and even hexaploid, the ANKTMs number on chromosomes 1A and 3A were decreased whereas the ANKTMs number on chromosomes 4A and 7A were increased (Table 2). Compared with T. urartu, Ae. tauschii, T. dicoccoides and H. vulgare, which contained 36, 42, 72 and 42 ANKTMs genes, respectively, the common wheat family of ANKTMs was remarkably large, with 145 members. The data suggest that the evolution of ANKTM genes in Triticeae was a complex evolutionary process and that the expansion of ANKTMs in common wheat possibly involved more than can be expected from two rounds of polyploidization.
During the evolution of plant genome and genetic systems, gene duplications have been one of the leading causes for the expansion of gene families (Cannon et al., 2004; Wang et al., 2021; Lin et al., 2022). To further understand the evolution of ANKTM genes in wheat and T. dicoccoides, the tandem duplications of the common wheat, T. dicoccoides and T. urartu of the ANKTM family were investigated. In this study, the MCScanX analysis was used to investigate the tandem duplication of the ANKTM gene family, and shinyCircos software was used to show the tandem duplication genes, the syntenic relationship of the gene pairs and their respective loci in the wheat and T. dicoccoides genomes (Figure 2). Tandem duplication analysis of ANKTM indicated that wheat, T. dicoccoides and T. urartu had experienced three, three and two tandem duplication events, respectively (Figure 2; Supplementary Table 2). The above indicates that ANKTM is active in the process of evolution and has experienced multiply tandem duplications at different evolutionary stages. The tandem duplication of ANKTM in wheat are located on chromosome 5D (TaANKTM5D-2, TaANKTM5D-3 and TaANKTM5D-4), 5B (TaANKTM5B-7, TaANKTM5B-8, TaANKTM5B-9 and TaANKTM5B-10) and 7B (TaANKTM7B-1 and TaANKTM7B-2; Figure 2A). In T. dicoccoides are located on chromosome 3A (TdANKTM3A-4 and TdANKTM3A-5), 3B (TdANKTM3B-7, TdANKTM3B-8 and TdANKTM3B-9) and 4B (TdANKTM4B-2 and TdANKTM4B-3; Figure 2B). The tandem duplications of ANKTM in T. urartu are located on chromosome 1A (TuANKTM1A-4 and TuANKTM1A-5) and 3A (TuANKTM3A-9 and TuANKTM3A-10; Supplementary Table 2). The above results may partly explain why the ANKTM genes in Triticeae are mainly concentrated in homologous groups 3 and 5, and the number in the B and D subgenomes are higher than that of the A subgenome. Interestingly, the tandem ANKTM duplicates in wheat all clustered in class a, whereas those in T. dicoccoides all clustered in class b and in T. urartu were clustered in classes b and e (Figure 1). This may also be part of the reason that the number ratio of ANKTM between wheat and other diploid Triticeae species in class a is higher than 3:1, and in class b and class e are lower than 3:1 (except class b of Ae. tauschii). The above indicates that besides the genome polyploidization result in the increase of ANKTM gene number, tandem duplication is also a mechanism for the subfamily expansion. A few ANKTMs from T. dicoccoides and wheat produced gene pairs across homologous groups. For example, TdANKTM4A-5 of class e was located on the fourth homologous group. However, the gene pairs of TdANKTM4A-5 from B (TdANKTM5B-9) subgenomes of T. dicoccoides was located on the fifth homologous group (Figure 2B). This may be due to structural rearrangements of chromosomes 4A–5A–7B in the formation of the Triticum-Aegilops joint genus in two major translocation events (Chen et al., 2020b).
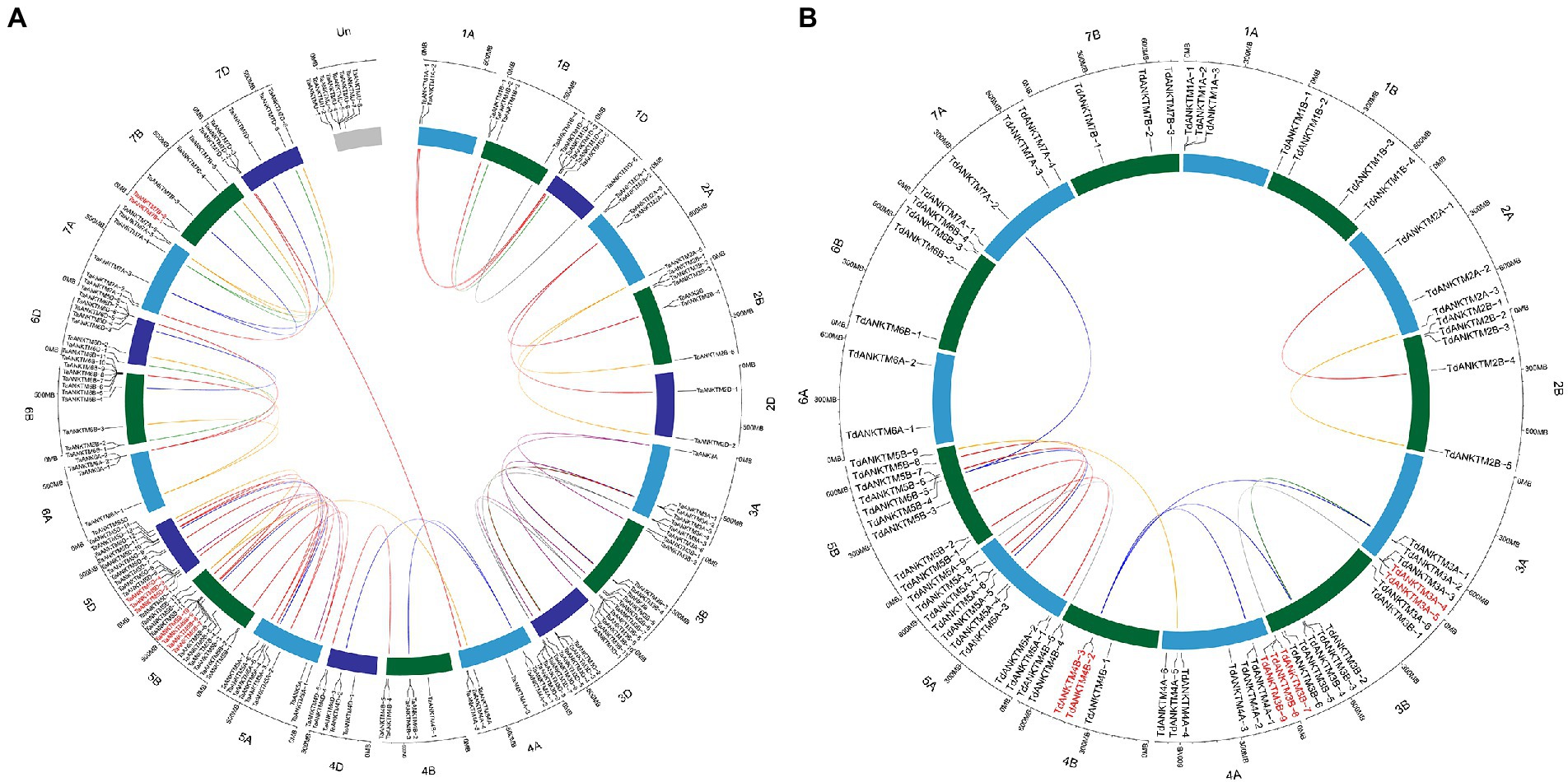
Figure 2. Homologous gene pairs, tandem duplication and location of ANKTM genes in wheat and T. dicoccoides genomes. All ANKTM genes were mapped to their respective loci in the wheat (A) and T. dicoccoides (B) genome in a circular diagram using shinyCircos. Homologous genes were inferred by TGT and linked with specific colors. Tandem duplication was analyzed by MCScanX, and the genes of red font indicated these genes were produced by tandem duplications. Subgenomes A and B are indicated by light blue and dark green, respectively; subgenomes D of wheat is indicated by dark blue.
Micro-collinearity analysis of Triticeae ANKTMs
The tandem duplications of ANKTM in T. urartu are located on chromosomes 1A and 3A, and the ANKTMs on chromosomes 1A and 3A of T. urartu are more than that of T. dicoccoides and wheat (Table 2; Supplementary Table 2). To trace the evolutionary history of the target genes, a micro-collinearity analysis was performed to help understand their evolution in a local region (Chen et al., 2020b). When the tandem duplication produced genes of TuANKTM1A-4 and TuANKTM1A-5 were used as query genes, the results showed that its neighboring genes were relatively conserved across the investigated genomes (Figure 3A), and homologs of TuANKTM1A-4 and TuANKTM1A-5 were found in the collinearity regions of the A subgenome of T. dicoccoides and wheat was TRIDC1AG012570 and TraesCS1A02G088100, respectively (Figure 3A). The prediction of protein conserved domains indicated TRIDC1AG012570 and TraesCS1A02G088100 as ANK proteins with a RING finger domain, but no TM domain (Figure 3B), belonging to the ANKRF type protein subgroup (Vo et al., 2015). Using the tandem repeats of TuANKTM3A-9 and TuANKTM3A-10 as query genes, the micro-collinearity relationship showed that both TuANKTM3A-9 and TuANKTM3A-10 from T. urartu had a “1-to-many” pairwise homology with TdANKTM3A-4, TdANKTM3A-5 and TdANKTM3A-6 in the A-subgenome of T. dicoccoides (Figure 3C). However, in the collinearity regions of common wheat, only TaANKTM3A-5 was found to show homology (Figure 3C). Among the subgenomes of the three analyzed species, the number of genes in the micro-collinearity region varies greatly. Nevertheless, the number of collinear genes found in this region of wheat is much less than that of T. dicoccoides and T. urartu (Figure 3C). The above suggests that ANKTM type genes evolved into other types of ANK genes in the process of Triticeae evolution and that the evolution of ANKTM type genes is complex. However, we also cannot exclude the possibility that some of the diploid and tetraploid species experienced gene loss or generated genes by duplication after T. aestivum speciation.
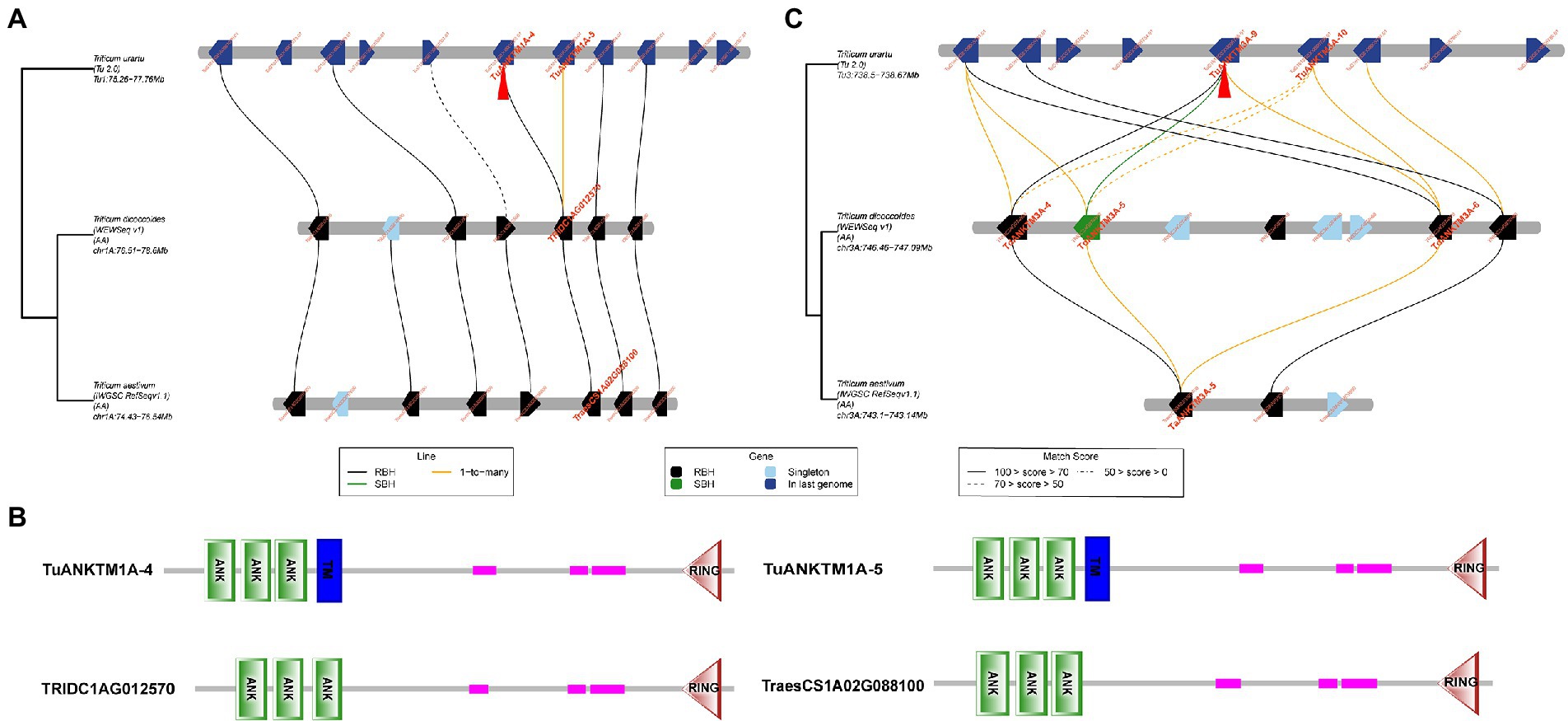
Figure 3. Micro-collinearity analysis by TGT to track the evolutionary history of the tandem duplicated TuANKTM homologs. (A) The tandem repeats TuANKTM1A-4 and TuANKTM1A-5 were used as query genes. The micro-collinearity relationship showed that homologs of the analyzed TuANKTMs were found in all investigated genomes, and the homologous of TuANKTM1A-4 and TuANKTM1A-5 was TRIDC1AG012570 in T. dicoccoides and TraesCS1A02G088100 in wheat, respectively. (B) Conservative domain prediction of TRIDC1AG012570 and TraesCS1A02G088100 proteins. The conserved domain prediction indicates that TRIDC1AG012570 and TraesCS1A02G088100 are ANK proteins with a RING finger domain. Abbreviations: ANK, ankyrin repeat; TM, transmembrane domain; RING, RING finger domain. (C) The tandem repeats TuANKTM3A-9 and TuANKTM3A-10 were used as query genes. TuANKTM3A-9 and TuANKTM3A-10 from T. urartu had a “1-to-many” pairwise homology with TdANKTM3A-4, TdANKTM3A-5 and TdANKTM3A-6 in the A-subgenome of T. dicoccoides; meanwhile the homologous of the above genes in the collinearity regions of common wheat was just TaANKTM3A-5. Blackline, 1-to-1-mutual-best. Greenline, 1-to-its-best. Yellowline, 1-to-many. RBH, reciprocal best hits; SBH, single-side best hits.
Expression patterns analysis of TaANKTMs
The expression patterns of genes are helpful in predicting their potential biological functions (Hu et al., 2022). To elucidate the potential role of TaANKTMs in biotic stress, their expression patterns were studied by in silico expression profiling. Expression patterns of TaANKTMs under two biotic stresses (powdery mildew pathogen and stripe rust) and two PAMPs (flag 22 and chitin) were analyzed using the wheat RNA-seq data from public databases (Ma et al., 2021). The data are displayed in a heat map (Figure 4). Interestingly, the expression of almost all TaANKTM genes in classes a-c could not be detected under the different treatments. In contrast, most of the genes in class d and class e are expressed, and some of them show differential responses to infections by Bgt, Puccinia striiformis f. sp. tritici (Pst) or elicitation with chitin or flg22 (Figure 4). For example, the gene pairs of TaANKTM3A-2 (TaANKTM3B-3 and TaANKTM3D-3) and TaANKTM6A-1(TaANKTM6B-3 and TaANKTM6D-2) were up-regulated upon Bgt or Pst infection at 24 h and were also up-regulated upon flg22 and chitin treatment at 0.5 h. The gene pairs, TaANKTM7B-5 (TaANKTM7A-5 and TaANKTM7D-6), were up-regulated after Bgt infection, down-regulated after the infection with Pst, but displayed only insignificant alterations in expression after chitin and flg22 treatments. The expression of the gene pair, TaANKTM5A-3 (TaANKTM5B-4 and TaANKTM5D-10), was obviously up-regulated upon the treatments of chitin and flg22. The gene pair of TaANKTM7A-3 (TaANKTM7B-3 and TaANKTM7D-4), was obviously down-regulated upon flg22 and chitin treatment at 0.5 h; specially, the expression of TaANKTM7D-4 was up-regulated upon Bgt infection at 48 h and Pst infection at 24 h. The expression of TaANKTM6B-4 and TaANKTM6D-3 were slightly up-regulated after Bgt infection at 48 h and Pst infection at 24 h, however, were down-regulated upon flg22 and chitin treatment at 0.5 h. The gene pair of TaANKTM4A-1(TaANKTM4B-3 and TaANKTM4D-3) was slightly down-regulated and up-regulated upon Bgt and Pst infection at 24 h, respectively; and was down-regulated upon flg22 and chitin treatment at 0.5 h. In addition, the expression levels of the gene pairs TaANKTM2A-5 (TaANKTM2D-2 and TaANKTM2B-5) were up-regulated after infection by Bgt and Pst and transiently down-regulated after treatments with chitin and flg22 at 0.5 h and returned to the pretreatment levels at 3 h. The RNA-Seq data suggests the above genes can response to rust, powdery mildew infections or PAMP elicitors; however, the absolute times of relative expression change of some genes after Bgt infection were small, and change multiples were less than two (Figure 4). Therefore, the genes TaANKTM2A-5 (TaANKTM2D-2 and TaANKTM2B-5) and TaANKTM6A-1 (TaANKTM6B-3 and TaANKTM6D-2; class e), and TaANKTM3A-2 (TaANKTM3B-3 and TaANKTM3D-3; class d) induced by Bgt obviously from the RNA-Seq data were selected for further expression analysis by qRT-PCR following powdery mildew stress.
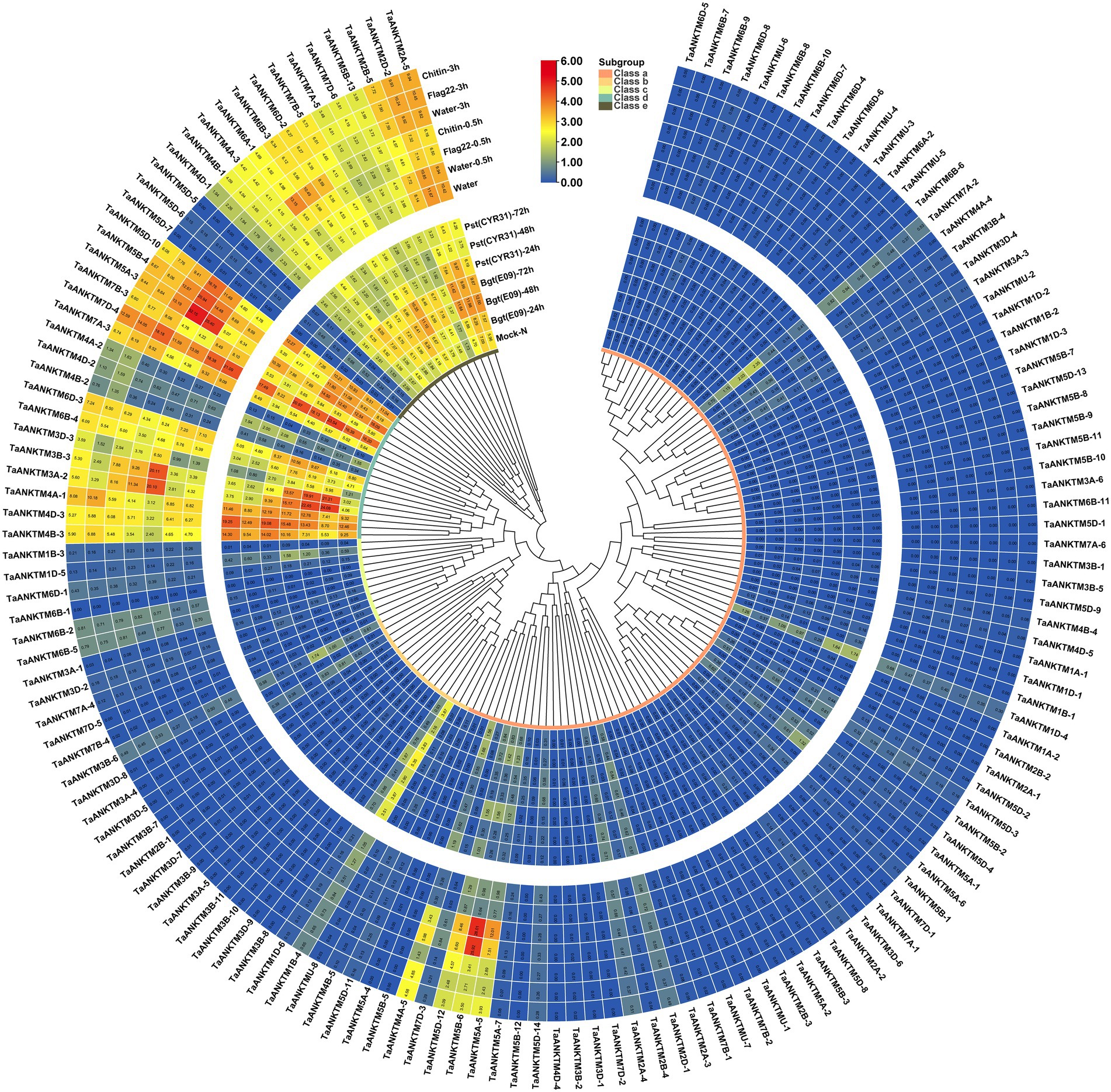
Figure 4. Heat map of the expression profiling of wheat ANKTM genes under different stress. The color scale bar represents the expression values (in log2-based tags per million values) of the genes, and the values in square frames represent the tags per million values. The phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates by MEGA X. Bgt, Blumeria graminis f. sp. tritici; Pst, Puccinia striiformis f. sp. tritici; Mock-N, Disease-resistant wheat varieties N9134 without the infection of Bgt and Pst.
Because of the high sequence similarity of the three copy genes in different subgenomes of wheat, qRT-PCR primers could not effectively distinguish the three copy genes; therefore, the gene of subgenomes A was used to represent the relative expression of the three copy genes. After Bgt inoculation, the expression patterns of TaANKTM2A-5, TaANKTM3A-2, and TaANKTM6A-1 were similar, the relative expression levels of the three genes were rapidly up-regulated and reached to the expression peak at 2 h (Figure 5). The relative expression levels of TaANKTM2A-5 returned to the original level at 6, 12 and 24 h, and significantly up-regulated at 36 h and 48 h (Figure 5). For TaANKTM3A-2, the relative expression reached a new peak at 36 h and then returned to the original expression level at 48 h (Figure 5). For TaANKTM6A-1, the relative expression reached to the expression peak at 2 h and then returned to the original level at 6 h, and there was no significant change at each subsequent time point (Figure 5). Since all the three genes can respond to the induction of Bgt, they were therefore selected for further analysis of their potential biological functions.
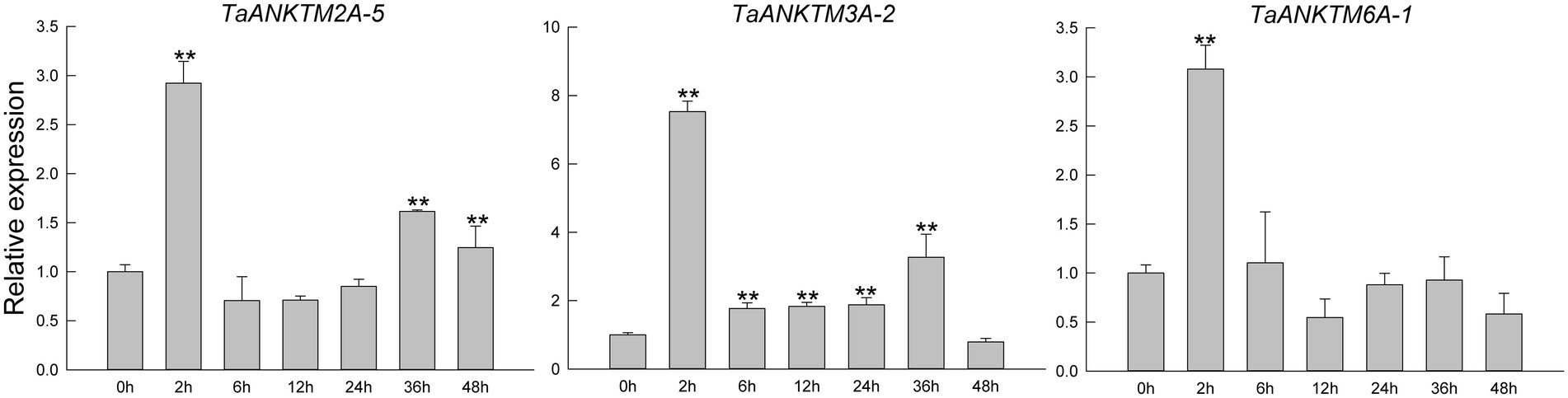
Figure 5. Expression profiling of three TaANKTMs in common wheat AK58 after Bgt inoculation by qRT-PCR. Relative expression of three TaANKTMs in response to Bgt. Data were normalized to the TaTubulin gene. The values are the means of three technical replicates of one biological experiment. Asterisks indicate significant differences (assessed using Duncan’s honestly significant difference test), **p < 0.01. All the raw data for qRT-PCR are listed in Supplementary Table 3.
Silencing of TaANKTM2A-5 compromises powdery mildew resistance in common wheat AK58
In order to further explore the potential roles of TaANKTM2A-5, TaANKTM3A-2, and TaANKTM6A-1 in the resistance to powdery mildew disease in the common wheat variety, AK58, constructs for their virus (BSMV) induced silencing were produced. Six days after Bgt infection, TaANKTM2A-5-silenced leaves were seen to be more susceptible to Bgt than those of the control (BSMV:γ-innoculated plants; Figure 6A). TaANKTM3A-2- and TaANKTM6A-1-silenced leaves showed no obvious difference to Bgt infection than those from BSMV:γ-innoculated individuals (Supplementary Figures 1A1,A2). The expression levels of TaANKTM2A-5, TaANKTM3A-2 and TaANKTM6A-1 were assessed by qRT-PCR and were shown to be significantly decreased by 2–5-fold (Figure 6B; Supplementary Figures 1B1,B2). Therefore, silencing the TaANKTM2A-5 gene could compromise the resistance of AK58 to powdery mildew.
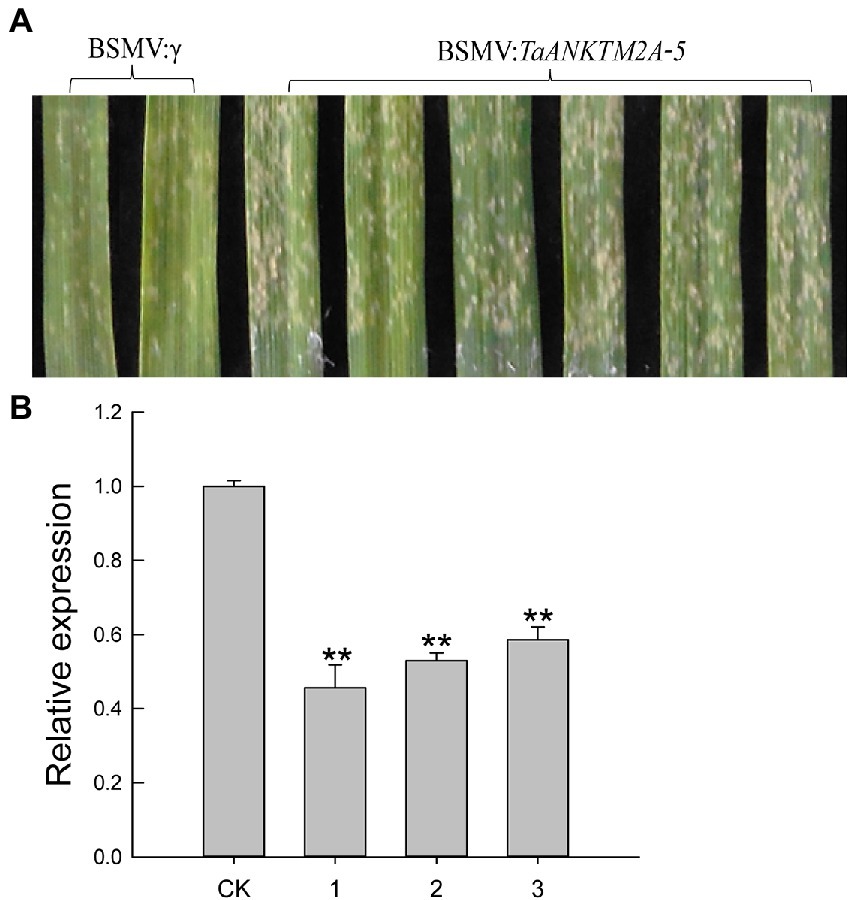
Figure 6. Functional analysis of TaANKTM2A-5 by Barley stripe mosaic virus-based virus-induced gene silencing (BSMV-VIGS) in AK58. (A) BSMV: TaANK2A-5 infected plants were inoculated with Blumeria graminis f. sp. Tritici, and their leaves were photographed 6 days post-inoculation. BSMV:γ infected plants were performed as controls. The experiment was repeated independently three times, and the same results were obtained. (B) Expression of TaANKTM2A-5 in BSMV: TaANKTM2A-5-infected leaves was compared with that in BSMV:γ-infected controls of AK58. CK represents plants inoculated with BSMV:γ, and 1–3 represents plants inoculated with BSMV: TaANK2A-5. Significant differences assessed using Duncan’s honestly significant difference test, **p < 0.01.
Discussion
Tandem duplication is one of the major mechanisms for ANKTM gene subfamily expansion
In the process of evolution, duplicated genes can experience functional divergence, which is essential for speciation and environmental adaptability (Prince and Pickett, 2002; Hittinger and Carroll, 2007). As a heterologous hexaploid species, wheat resulted from two rounds of hybridizations and has experienced complex evolutionary mechanisms (Shewry, 2009; Zhou et al., 2020), which makes it more challenging to explore its evolutionary relationships and functional genomics. The recent rapid development of interdisciplinary bioinformatics and high-quality genome assembly of Triticeae species (Avni et al., 2017; Luo et al., 2017; IWGSC, 2018; Ling et al., 2018) have provided the opportunity for a more detailed study of the phylogenetics of ANKTM genes and inferring how gene members of this subfamily replicated and expanded.
In the present study, bioinformatics methods were used to analyze the ANKTM gene family in Triticeae species and investigate the expansion mode of ANKTM. The number of ANKTM genes identified from the whole genome of wheat (BBAADD), T. dicoccoides (BBAA), T. urartu (AA), Ae. tauschii and H.vulgare (HH) were 145, 72, 36, 42 and 42, respectively (Table 1). The increase in ANKTM numbers in wheat was proportionally larger than what could be expected from increase in its ploidy. Furthermore, the numbers of ANKTM in class a and class c of wheat were significantly larger than expected (Figure 1; Table 1). Several tandem duplication events occurred in wheat, T. dicoccoides and T. urartu ANKTM genes (Figure 2; Supplementary Table 2), and most of the tandem duplication produced genes distributed in the distal chromosome segments. This finding is consistent with that many fast-evolving genes are distributed in these segments and possibly to facilitate the adaption to different conditions (Glover et al., 2015; Chen et al., 2018; Schilling et al., 2020). The tandem duplication produced ANKTM genes of wheat all clustered in class a, thus contributing to the unexpectedly high number of wheat ANKTMs in class a. Previous studies indicate that the seven ANKTM-type genes At4g03440, At4g03450, At4g03460, At4g03470, At4g03480, At4g03490 and At4g03500 on chromosome 4, are tandem duplicated genes, which are linked and clustered tightly (Du et al., 2007). The seven genes in this study are AtANKTM4-1, AtANKTM4-2, AtANKTM4-3, AtANKTM4-4, AtANKTM4-5, AtANKTM4-6 and AtANKTM4-7, respectively (Supplementary Table 2), this consistent with the previous research. The ANKTM protein Lr14a confers leaf rust disease resistance in wheat, and its gene resides in a locus with tandem repeats (Kolodziej et al., 2021). TaANKTM5B-9 (TraesCS5B02G352300) is a homolog of Lr14a (Kolodziej et al., 2021), and in this study, we find that TaANKTM5B-7, TaANKTM5B-8, TaANKTM5B-9 and TaANKTM5B-10 are tandem repeats (Figure 2). These results therefore agree with previous studies indicating that the increase of ANKTM gene family members in wheat is not only due to the increased ploidy level, but that self-replication is also a major mechanism driving the gene family expansion (Cannon et al., 2004; Du et al., 2007; Kolodziej et al., 2021).
TaANKTM2A-5 positively regulates powdery mildew resistance in wheat
Wheat powdery mildew, caused by Bgt, is one of the most destructive diseases of wheat (Hu et al., 2018). Cultivating broad-spectrum and durable disease-resistant varieties is an effective and environment-friendly strategy to improve plant diseases (Li et al., 2020). Resistance genes play important roles in disease resistance breeding and are frequently pathogen-specific (Dangl et al., 2013; Saintenac et al., 2018; Xing et al., 2018). However, the natural evolution of new races of the pathogen can overcome the resistance (Dodds and Rathjen, 2010), so that new types of resistance-related genes need to be explored continuously.
Several ANKTMs have been identified to be involved in plant immune responses. For example, the Arabidopsis ANKTM family gene, BDA1, is an important regulator of plant immunity acting downstream of the receptor-like protein SNC2 and upstream of NPR1 and WRKY70 (Yang et al., 2012). The widely studied Arabidopsis ANKTM, ACD6, which plays a key role in growth and pathogen defense (Todesco et al., 2010), can form large complexes with the membrane bound PAMP receptors, BAK1 and CERK1 (Tateda et al., 2014; Zhang et al., 2014b). Lr14a encodes an ANKTM-like type protein that confers race-specific leaf rust resistance in wheat (Kolodziej et al., 2021). The NLR protein, YrU1, confers stripe rust resistance in wheat also contains an ANK domain, and the ANK domain of YrU1 may be derived from ANKTM proteins (Wang et al., 2020; Kolodziej et al., 2021). The above studies indicated that some of the ANKTM-type genes are involved in basal resistance and effector-triggered immunity. In this study, the expression of some TaANKTMs were found to be differentially responsive to infection by Pst or Bgt, or challenge by PAMPs (chitin, flg22; Figure 4), which is agreement with the earlier studies. In particular, the expression levels of TaANKTM genes TaANKTM2A-5, TaANKTM3A-2 and TaANKTM6A-1 displayed interesting responses to infection by Bgt and Pst, and elicitation by chitin and flg22 (Figure 4), and the relative expression of the three genes were further analyzed by qRT-PCR. The results showed that the expression of TaANKTM2A-5, TaANKTM3A-2, and TaANKTM6A-1 in powdery mildew susceptible cultivar AK58 were rapidly up-regulated and reached to the expression peak at 2 h after Bgt inoculation (Figure 5). The silencing of the TaANKTM genes TaANKTM3A-2 and TaANKTM6A-1 produced inconsistent phenotypes and no conclusions could be drawn concerning their effects on the resistance of AK58 to Bgt (Supplementary Figure 1). The silencing of TaANKTM2A-5 did however produce a consistent phenotype and plants were clearly compromised in their resistance to Bgt (Figure 6).
Based on the above research, we speculate that the ANKTM type protein may represent a new family of disease resistance related genes in plants. In this study, the function of three ANKTM genes on powdery mildew resistance was verified through the BSMV-VIGS assay. In future studies, the function of the TaANKTM2A-5 gene on wheat powdery mildew needs to be further verified in stable genetically transformed plants, and the three-dimensional structural characteristics of proteins with similar sequences and expression patterns but display different functions will be further analyzed. Findings in this study not only help in understand their evolutionary process, but also provide gene resources for disease resistance breeding.
Conclusion
In summary, a total of 381 ANKTM genes were identified from five Triticeae species and Arabidopsis, which could be divided into five classes. Among them class a only contains proteins from Triticeae species, and the numbers of ANKTM in class a of wheat are significantly larger than expected even after consideration of the ploidy level. T. urartu, T. dicoccoides and wheat all experienced tandem duplication events in the evolution process. Furthermore, the tandem duplication produced ANKTM genes of wheat all clustered in class a. Tandem duplication is one of the major mechanisms for ANKTM gene subfamily expansion. The expression pattern showed that almost all TaANKTM genes in classes a-c could not be detected under different treatments. In contrast, most of the genes in class d and class e are expressed, and some of them are responsive to biotic stress. Furthermore, silencing TaANKTM2A-5 belonging to class e compromised powdery mildew resistance in common wheat AK58. The findings in this study not only help our understanding of the evolutionary process of ANKTM genes, but also provide a valuable reference for mining disease resistance genes in the ANKTM gene family.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PH and JX designed the experiments and wrote the manuscript. PH, YR, JX, QW, PS, YG, YZ, and HG contributed to the experiments and performed the data analysis. HH and CL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (nos. 31901538, 31872129, and 31900240) and the Key Scientific and Technological Research Projects in Henan Province (no. 212102110052).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.943217/full#supplementary-material
Footnotes
1. ^http://www.wheatgenome.org/
2. ^http://www.mbkbase.org/Tu/
3. ^http://plants.ensembl.org/index.html
5. ^https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
6. ^http://smart.embl-heidelberg.de/
7. ^https://www.evolgenius.info/
8. ^http://shinycircos.ncpgr.cn
References
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. doi: 10.1126/science.aan0032
Becerra, C., Jahrmann, T., Puigdomenech, P., and Vicient, C. M. (2004). Ankyrin repeat-containing proteins in Arabidopsis: characterization of a novel and abundant group of genes coding ankyrin-transmembrane proteins. Gene 340, 111–121. doi: 10.1016/j.gene.2004.06.006
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Song, W., Xie, X., Wang, Z., Guan, P., Peng, H., et al. (2020b). A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the Triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant 13, 1694–1708. doi: 10.1016/j.molp.2020.09.019
Chen, N. W. G., Thareau, V., Ribeiro, T., Magdelenat, G., Ashfield, T., Innes, R. W., et al. (2018). Common bean subtelomeres are hot spots of recombination and favor resistance gene evolution. Front. Plant Sci. 9:1185. doi: 10.3389/fpls.2018.01185
Cui, Z., Zhang, H., Chen, X., Zhang, C., Ma, W., Huang, C., et al. (2018). Pursuing sustainable productivity with millions of smallholder farmers. Nature 555, 363–366. doi: 10.1038/nature25785
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Du, J., Wang, X., Zhang, M., Tian, D., and Yang, Y. H. (2007). Unique nucleotide polymorphism of ankyrin gene cluster in Arabidopsis. J. Genet. 86, 27–35. doi: 10.1007/s12041-007-0004-0
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Feys, B. J., Moisan, L. J., Newman, M. A., and Parker, J. E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. doi: 10.1093/emboj/20.19.5400
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Glover, N. M., Daron, J., Pingault, L., Vandepoele, K., Paux, E., Feuillet, C., et al. (2015). Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B. Genome Biol. 16:188. doi: 10.1186/s13059-015-0754-6
Guo, J. H., Liu, X. J., Zhang, Y., Shen, J. L., Han, W. X., Zhang, W. F., et al. (2010). Significant acidification in major Chinese croplands. Science 327, 1008–1010. doi: 10.1126/science.1182570
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Hemsley, P. A., Kemp, A. C., and Grierson, C. S. (2005). The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 17, 2554–2563. doi: 10.1105/tpc.105.031237
Hittinger, C. T., and Carroll, S. B. (2007). Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681. doi: 10.1038/nature06151
Hu, P., Liu, J., Xu, J., Zhou, C., Cao, S., Zhou, W., et al. (2018). A malectin-like/leucine-rich repeat receptor protein kinase gene, RLK-V, regulates powdery mildew resistance in wheat. Mol. Plant Pathol. 19, 2561–2574. doi: 10.1111/mpp.12729
Hu, P., Song, P., Xu, J., Wei, Q., Tao, Y., Ren, Y., et al. (2022). Genome-wide analysis of serine hydroxymethyltransferase genes in Triticeae species reveals that TaSHMT3A-1 regulates Fusarium Head Blight resistance in wheat. Front. Plant Sci. 13:847087. doi: 10.3389/fpls.2022.847087
IWGSC (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Kolodziej, M. C., Singla, J., Sanchez-Martin, J., Zbinden, H., Simkova, H., Karafiatova, M., et al. (2021). A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat. Nat. Commun. 12:956. doi: 10.1038/s41467-020-20777-x
Kuhlmann, M., Horvay, K., Strathmann, A., Heinekamp, T., Fischer, U., Bottner, S., et al. (2003). The alpha-helical D1 domain of the tobacco bZIP transcription factor BZI-1 interacts with the ankyrin-repeat protein ANK1 and is important for BZI-1 function, both in auxin signaling and pathogen response. J. Biol. Chem. 278, 8786–8794. doi: 10.1074/jbc.M210292200
Kumagai, H., Hakoyama, T., Umehara, Y., Sato, S., Kaneko, T., Tabata, S., et al. (2007). A novel ankyrin-repeat membrane protein, IGN1, is required for persistence of nitrogen-fixing symbiosis in root nodules of Lotus japonicus. Plant Physiol. 143, 1293–1305. doi: 10.1104/pp.106.095356
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuraparthy, V., Chhuneja, P., Dhaliwal, H. S., Kaur, S., Bowden, R. L., and Gill, B. S. (2007). Characterization and mapping of cryptic alien introgression from Aegilops geniculata with new leaf rust and stripe rust resistance genes Lr57 and Yr40 in wheat. Theor. Appl. Genet. 114, 1379–1389. doi: 10.1007/s00122-007-0524-2
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, W., Deng, Y., Ning, Y., He, Z., and Wang, G. L. (2020). Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603. doi: 10.1146/annurev-arplant-010720-022215
Lin, H., Han, X., Feng, X., Chen, X., Lu, X., Yuan, Z., et al. (2022). Molecular traits and functional analysis of Rapid Alkalinization factors (RALFs) in four Gossypium species. Int. J. Biol. Macromol. 194, 84–99. doi: 10.1016/j.ijbiomac.2021.11.127
Ling, H. Q., Ma, B., Shi, X., Liu, H., Dong, L., Sun, H., et al. (2018). Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557, 424–428. doi: 10.1038/s41586-018-0108-0
Lozano, R., Hamblin, M. T., Prochnik, S., and Jannink, J. L. (2015). Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics 16:360. doi: 10.1186/s12864-015-1554-9
Lu, H., Liu, Y., and Greenberg, J. T. (2005). Structure-function analysis of the plasma membrane-localized Arabidopsis defense component ACD6. Plant J. 44, 798–809. doi: 10.1111/j.1365-313X.2005.02567.x
Lu, H., Rate, D. N., Song, J. T., and Greenberg, J. T. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15, 2408–2420. doi: 10.1105/tpc.015412
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Luo, M. C., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502. doi: 10.1038/nature24486
Ma, S., Wang, M., Wu, J., Guo, W., Chen, Y., Li, G., et al. (2021). WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14, 1965–1968. doi: 10.1016/j.molp.2021.10.006
Mosavi, L. K., Cammett, T. J., Desrosiers, D. C., and Peng, Z. Y. (2004). The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448. doi: 10.1110/ps.03554604
Prince, V. E., and Pickett, F. B. (2002). Splitting pairs: the diverging fates of duplicated genes. Nat. Rev. Genet. 3, 827–837. doi: 10.1038/nrg928
Rate, D. N., Cuenca, J. V., Bowman, G. R., Guttman, D. S., and Greenberg, J. T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. doi: 10.1105/tpc.11.9.1695
Saintenac, C., Lee, W. S., Cambon, F., Rudd, J. J., King, R. C., Marande, W., et al. (2018). Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 50, 368–374. doi: 10.1038/s41588-018-0051-x
Sakamoto, H., Matsuda, O., and Iba, K. (2008). ITN1, a novel gene encoding an ankyrin-repeat protein that affects the ABA-mediated production of reactive oxygen species and is involved in salt-stress tolerance in Arabidopsis thaliana. Plant J. 56, 411–422. doi: 10.1111/j.1365-313X.2008.03614.x
Schilling, S., Kennedy, A., Pan, S., Jermiin, L. S., and Melzer, R. (2020). Genome-wide analysis of MIKC-type MADS-box genes in wheat: pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 225, 511–529. doi: 10.1111/nph.16122
Sedgwick, S. G., and Smerdon, S. J. (1999). The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316. doi: 10.1016/S0968-0004(99)01426-7
Shen, G., Kuppu, S., Venkataramani, S., Wang, J., Yan, J., Qiu, X., et al. (2010). ANKYRIN REPEAT-CONTAINING PROTEIN 2A is an essential molecular chaperone for peroxisomal membrane-bound ASCORBATE PEROXIDASE3 in Arabidopsis. Plant Cell 22, 811–831. doi: 10.1105/tpc.109.065979
Tang, X., Cao, X., Xu, X., Jiang, Y., Luo, Y., Ma, Z., et al. (2017). Effects of climate change on epidemics of powdery mildew in winter wheat in China. Plant Dis. 101, 1753–1760. doi: 10.1094/PDIS-02-17-0168-RE
Tateda, C., Zhang, Z., Shrestha, J., Jelenska, J., Chinchilla, D., and Greenberg, J. T. (2014). Salicylic acid regulates Arabidopsis microbial pattern receptor kinase levels and signaling. Plant Cell 26, 4171–4187. doi: 10.1105/tpc.114.131938
Todesco, M., Balasubramanian, S., Hu, T. T., Traw, M. B., Horton, M., Epple, P., et al. (2010). Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465, 632–636. doi: 10.1038/nature09083
Vo, K. T. X., Kim, C. Y., Chandran, A. K. N., Jung, K. H., An, G., and Jeon, J. S. (2015). Molecular insights into the function of ankyrin proteins in plants. J. Plant Biol. 58, 271–284. doi: 10.1007/s12374-015-0228-0
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, X., Zheng, Y., Chen, B., Zhi, C., Qiao, L., Liu, C., et al. (2021). Genome-wide identification of small heat shock protein (HSP20) homologs in three cucurbit species and the expression profiles of CsHSP20s under several abiotic stresses. Int. J. Biol. Macromol. 190, 827–836. doi: 10.1016/j.ijbiomac.2021.08.222
Wang, H., Zou, S., Li, Y., Lin, F., and Tang, D. (2020). An ankyrin-repeat and WRKY-domain-containing immune receptor confers stripe rust resistance in wheat. Nat. Commun. 11:1353. doi: 10.1038/s41467-020-15139-6
Wirdnam, C., Motoyama, A., Arn-Bouldoires, E., Van Eeden, S., Iglesias, A., and Meins, F. Jr. (2004). Altered expression of an ankyrin-repeat protein results in leaf abnormalities, necrotic lesions, and the elaboration of a systemic signal. Plant Mol. Biol. 56, 717–730. doi: 10.1007/s11103-004-4679-9
Xing, L., Hu, P., Liu, J., Witek, K., Zhou, S., Xu, J., et al. (2018). Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 11, 874–878. doi: 10.1016/j.molp.2018.02.013
Xu, J., Hu, P., Tao, Y., Song, P., Gao, H., and Guan, Y. (2021). Genome-wide identification and characterization of the Lateral Organ Boundaries Domain (LBD) gene family in polyploid wheat and related species. PeerJ 9:e11811. doi: 10.7717/peerj.11811
Yang, Y., Zhang, Y., Ding, P., Johnson, K., Li, X., and Zhang, Y. (2012). The ankyrin-repeat transmembrane protein BDA1 functions downstream of the receptor-like protein SNC2 to regulate plant immunity. Plant Physiol. 159, 1857–1865. doi: 10.1104/pp.112.197152
Yu, Y., Ouyang, Y., and Yao, W. (2018). shinyCircos: an R/shiny application for interactive creation of Circos plot. Bioinformatics 34, 1229–1231. doi: 10.1093/bioinformatics/btx763
Zhang, Z., Guo, J., Zhao, Y., and Chen, J. (2019). Identification and characterization of maize ACD6-like gene reveal ZmACD6 as the maize orthologue conferring resistance to Ustilago maydis. Plant Signal. Behav. 14:e1651604. doi: 10.1080/15592324.2019.1651604
Zhang, Z., Shrestha, J., Tateda, C., and Greenberg, J. T. (2014b). Salicylic acid signaling controls the maturation and localization of the arabidopsis defense protein ACCELERATED CELL DEATH6. Mol. Plant 7, 1365–1383. doi: 10.1093/mp/ssu072
Zhang, H., Yang, Y., Wang, C., Liu, M., Li, H., Fu, Y., et al. (2014a). Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics 15:898. doi: 10.1186/1471-2164-15-898
Zhou, Y., Zhao, X., Li, Y., Xu, J., Bi, A., Kang, L., et al. (2020). Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 52, 1412–1422. doi: 10.1038/s41588-020-00722-w
Keywords: ankyrin-transmembrane protein, evolutionary progress, powdery mildew, virus-induced gene silencing, wheat
Citation: Hu P, Ren Y, Xu J, Wei Q, Song P, Guan Y, Gao H, Zhang Y, Hu H and Li C (2022) Identification of ankyrin-transmembrane-type subfamily genes in Triticeae species reveals TaANKTM2A-5 regulates powdery mildew resistance in wheat. Front. Plant Sci. 13:943217. doi: 10.3389/fpls.2022.943217
Edited by:
Genlou Sun, Saint Mary's University, CanadaReviewed by:
Lingli Dong, Institute of Genetics and Developmental Biology (CAS), ChinaHongqi Si, Anhui Agricultural University, China
Longqing Sun, Huazhong Agricultural University, China
Copyright © 2022 Hu, Ren, Xu, Wei, Song, Guan, Gao, Zhang, Hu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyan Hu, aGFpeWFuaHVoaHlAMTI2LmNvbQ==; Chengwei Li, bGN3QGhhdXQuZWR1LmNu
 Ping Hu
Ping Hu Yueming Ren
Yueming Ren Jun Xu
Jun Xu Qichao Wei1
Qichao Wei1