- 1College of Grassland Agriculture, Northwest A&F University, Xianyang, China
- 2College of Agricultural, Ningxia University, Yinchuan, China
In the long-term growth process, alfalfa rhizosphere forms specific microbiome to provide nutrition for its growth and development. However, the effects of different perennial alfalfa cultivars on changes in the rhizosphere soil characteristics and microbiome are not well understood. In this study, 12 perennial alfalfa cultivars were grown continuously for eight years. Rhizosphere samples were tested using Illumina sequencing of the 16S rRNA gene coupled with co-occurrence network analysis to explore the relationship between alfalfa (biomass and crude protein content), soil properties, and the microbial composition and diversity. Redundancy analysis showed SOC and pH had the greatest impact on the composition of the rhizosphere microbial community. Moreover, microbial diversity also contributes to microbial composition. Soil properties (AP, EC, SOC and pH) exhibited a significant positive correlation with soil bacterial communities, which was attributed to the differences between plant cultivars. Partial least squares path modeling (PLS-PM) revealed that microbial biomass and community composition rather than diversity, are the dominant determinants in the rhizosphere soil nitrogen content of perennial alfalfa. Our findings demonstrate that the soil microbial biomass and composition of rhizosphere bacterial communities are strongly affected by cultivar, driving the changes in soil nitrogen content, and variances in the selective capacities of plants.
Introduction
Alfalfa (Medicago sativa L.) is a perennial forage legume with high yield and good nutritional quality that is widely considered important and cultivated (Annicchiarico, 2015). Owing to the high value in nutrients beneficial to livestock performance, alfalfa is known as “the queen of forage crops”(Hrbackova et al., 2020). The quality of alfalfa is mainly based on its protein content and fiber which affect animal feed digestibility and nutrient intake (Yang et al., 2021). Alfalfa being legume plants can fix nitrogen naturally through symbiotic nitrogen fixation (Fonouni-Farde et al., 2016). This characteristic can affect the content of crude protein, biomass of alfalfa plants, and soil N content. The crude protein content of alfalfa is relatively high (over 20% in its stems and leaves; Elfaki and Abdelatti, 2016), with significant differences generally found among different cultivars (Wang et al., 2022).
The rhizosphere is a critical hotspot for biogeochemical transformation and harbors one of the most complex, diverse, and active plant-associated microbial communities (Zhalnina et al., 2018), which is strongly influenced by both roots and the surrounding soil (Tkacz et al., 2020a). Rhizosphere effect describes the enrichment and activity of microbial cells near growing roots (Cúcio et al., 2016; Sasse et al., 2018). Previous studies have reported that some microbes are parasitic in plants and can interact with each other (Shelake et al., 2019). Rhizosphere microorganisms can also be recruited by the plant host to perform essential roles that contribute to soil nutrient cycling, healthy root growth, and plant community development in the soil ecosystem (Song et al., 2019). Therefore, plant growth and development are strongly affected by the responses of the rhizosphere microbial community, as it contributes directly in nutrient acquisition, defense, and abiotic stress responses (de Faria et al., 2021). Bulk soil is the main source of microbes colonizing the root rhizosphere, and plant genotype partially drives the selection of rhizosphere microorganisms (Mendes et al., 2018b). As a rich source of nutrients for soil microorganisms, plants affect the input and metabolism of microbial nutrition owing to differences in the distribution of core rhizosphere microorganisms of each plant variety (Zhang et al., 2021). Hence, understanding the influence of rhizosphere soil characteristics through differences in alfalfa cultivars is important for improving animal husbandry development and the soil N cycle (Samaddar et al., 2021).
The diversity and abundance of bacteria in plant rhizosphere soils are affected by several factors. Plants release root exudates, which significantly influence microbial responses, root architecture, and nutrient uptake (Wen et al., 2021). Cultivars can differ greatly in root exudate structure and release patterns, and therefore can strongly affect microbial activity and community structure. Previous reports have shown that changes in soil microbial diversity and structure are depend on plant species, soil physiochemical characteristics, and land use types (Mitchell et al., 2010; Brereton et al., 2020; Rossmann et al., 2020; Tkacz et al., 2020b; Xu et al., 2020). Soil bacterial communities are shaped by several soil factors (Llado et al., 2018), especially pH (Mod et al., 2021), along with other edaphic factors such as soil electrical conductivity (EC), soil organic carbon (SOC; Yashiro et al., 2016), total and available phosphorus (Dai et al., 2020). The diversity, composition, and biomass of soil microbial communities play key roles in nutrient acquisition, cycling, and movement patterns. Soil microbial biomass is a reserve of soil nutrients and has an excellent ability to decompose organic matter and convert complex N-rich organic matter into ammonia (Tao et al., 2019). Therefore, any changes in microbial responses can strongly affect crop growth.
A previous study reported the composition of naturally occurring rhizosphere bacterial populations and their metabolic activities differed between transgenic and unmodified alfalfa plants grown in the field (Tesfaye et al., 2003). While long-term planting of switchgrass did not significantly change the structure of the soil microbial community (He et al., 2017). To further explore these dynamics, the effects of plant genotype on the bacterial communities in maize (Aira et al., 2010), soybean (Sugiyama et al., 2014), rice (Shenton et al., 2016), common bean (Mendes et al., 2018a,b) and lima bean (Sousa et al., 2020) have been well studied. In addition, the mulberry and alfalfa intercropping system changed the soil physical and chemical properties and soil bacterial community (Zhang et al., 2018). However, little is known about the relationships between perennial alfalfa cultivars, rhizosphere soil properties, and soil bacteria. Plant–microbe–soil feedback can influence plant growth and the nutrient cycle, which play a fundamental role in plant community establishment and success (Baxendale et al., 2014). These interactions contribute to the nutrient acquisition of alfalfa and substantially increase the fitness of the host and of various soil properties.
Therefore, we determined the bacterial community composition as well as physiochemical properties and enzyme activities in rhizosphere soils of 12 perennial alfalfa cultivars. The goal of this experiment was to assess the effects of different cultivars on soil bacterial community diversity, composition, and networks. Finally, compared these effects to the relative influence of nitrogen content in long-term plant-soil histories. Differences in microbial enrichment in the alfalfa cultivars were observed based on the identification of the rhizosphere microbial community (Ma et al., 2020). In addition, the physiochemical properties of the rhizosphere soil were determined to explore the relationship between the microbial community composition and diversity. Our results illustrate how plant and soil microbes play a role in nitrogen content and which of these plant-mediated changes are most important in response to soil nitrogen content.
Materials and methods
Sample site and experimental design
The study was conducted at the experimental station of Northwest A&F University, located in Yangling Shaanxi (34°16′ N, 108°4′ E). The area has a warm temperate monsoon semi-humid climate with a mean annual temperature of 12.9°C and total annual precipitation of approximately 600 mm, and approximately 60% of the precipitation occurs between July and September. The soil type was Lou (Eum-Orthic Anthrosol; Zhong et al., 2020).
The planting area was 60 m × 18 m, dozens of alfalfa cultivars have been cultivated since 2012 under a green harvest management, between the different cultivars were separated by 1 m ridges. Before planting, the plots were plowed and homogenized to reduce soil variability. Clean cultivation, compound fertilizer (dissolved in water) and hand weeding are conventional practices for alfalfa cultivation in this region. Alfalfa sampled in this study was mowed three times per year and did not show evidence of pest attack, disease, or nutritional deficiency. After 20 days of cutting in July 2020, the alfalfa entered the budding stage (Fan et al., 2018). Twelve alfalfa cultivars samples (Table 1) were selected from dozens of alfalfa cultivars in the same growth stage, refers to budding stage.
In this study, the plots had 18 m2 of each alfalfa cultivar, being 6 m2 the usable area for soil sampling. Before soil sampling, we used the five-point sampling method, as previously described (Navarrete et al., 2015). Five samples were obtained at five cardinal points around the root of alfalfa, and collection was carried out by digging an area of 10 × 10 × 40 cm (length × width × depth) after removing the litter layer. A soil drill was used to obtain mixed samples of soil and roots, and fine roots were picked out. Soil that adhered to the roots was collected and mixed to form a composite sample for each plot. Five plants (biological replicate) were selected randomly from the plots of each cultivar of alfalfa, generating a total of 60 samples (12 alfalfa cultivars × five repeats). Before collecting the next soil sample, the residue on the shovel was wiped with sterile paper, and the sample was disinfected to avoid contamination between treatments. All samples were passed through a sterile 2 mm sieve before use. All soil samples were obtained and stored at –80°C for further analysis.
16S rRNA gene sample preparation, sequencing, and analysis
In total, 60 samples from the alfalfa rhizosphere were used for lumina-based 16S rRNA gene sequencing. Microbial DNA was extracted using the DNeasy 96 PowerSoil Pro QIAcube HT Kit (QIAGEN, Germany). The V3-V4 hypervariable region of the bacterial 16S rRNA gene was amplified using the primers 338F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT; Xu et al., 2016). PCR was performed as follows: 95°C for 3 min; 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s; and 72°C for 10 min. 10°C until it was halted by the user. The products were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States). The amplicons were pooled in equimolar concentrations and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, CA, United States) according to standard protocols at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
Growth and nutrition index of alfalfa
During harvesting, the total fresh weight of each plant (biomass of the aerial part of the plant), called aboveground biomass (BM), was collected to account for plant tissue nutrient characteristics. The crude protein content (CP) of alfalfa leaves was determined by H2SO4-H2O2 digestion using Kjeldahl nitrogen (Mulvaney et al., 1992).
Soil physiochemical properties
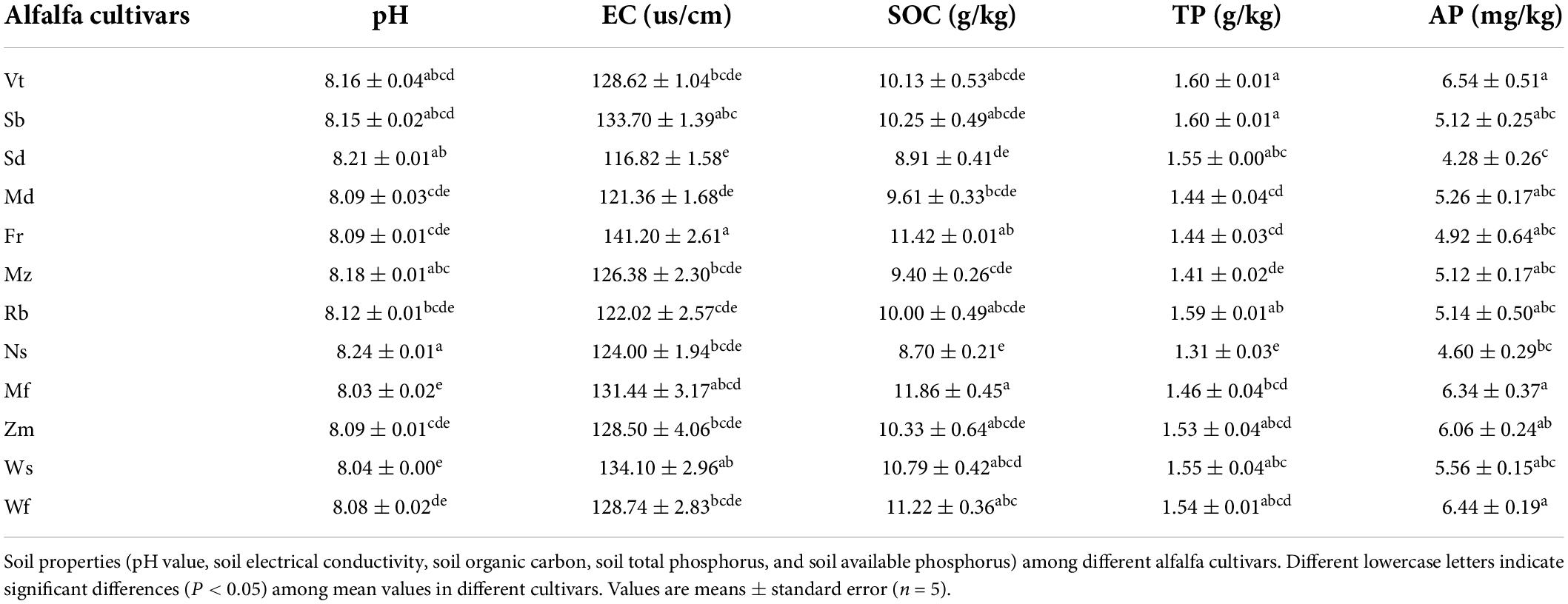
Soil characteristics such as pH, EC, and total phosphorus (TP), available phosphorus (AP), soil organic carbon (SOC), total nitrogen (TN), ammonium nitrogen (AN) and nitrate nitrogen (NT) were determined following standard methods (Jing et al., 2015). In brief, soil pH and EC were determined using a 5:1 water-to-soil ratio. TP was determined using HClO4-H2SO4 digestion molybdenum antimony anticolorimetry. AP was determined by Na2CO3 extraction using molybdenum–antimony inverse colorimetry. SOC was determined using the KCr2O7-H2SO4 external heating method. TN was determined using the semi-micro Kjeldahl method. AN and nitrate nitrogen (NT) were determined by 2 mol/L KCl extraction continuous flow analyzer (Wang et al., 2014).
Soil microbial biomass and extracellular enzyme activities
Soil microbial biomass carbon (MBC) and nitrogen (MBN) were determined using the chloroform fumigation extraction method followed by K2SO4 soil extraction (Vance et al., 1987), and a total organic carbon analyzer was used for analysis. Urease activity (URE) was determined by sodium phenolate sodium hypochlorite colorimetry using urea as the substrate. Leucine peptidase (LAP) and β-1,4-n-acetylglucosaminidase (NAG) activities were measured using a modified microplate fluorescence method (Saiya-Cork et al., 2002), and soil enzyme activity [nmol/(g⋅h)] was expressed as the matrix conversion rate of 1 g sample per unit time. Nitrogen fixation rates (NAT) were measured using acetylene conversion to ethylene (ARA), as previously described (Tao et al., 2019). The C2H4 concentration of each gas sample was measured using a gas chromatograph (Trace GC UL tra, Thermo Fisher, United States). ARA was determined using gas chromatography (SRI Instruments).
Data analyses
Data were tested for normality (Kolmogorov–Smirnov test) and homoscedasticity (Levene’s test). One-way analysis of variance (ANOVA) followed by Tukey’s HSD test (P < 0.05) was used to evaluate differences in plant crude protein content, aboveground biomass, soil properties, microbial diversity, and relative abundance of bacteria among the 12 cultivars. Tukey post hoc comparisons were used for all univariate analyses using SPSS v 25.0 (SPSS: IBM Corp). We performed redundancy analysis (RDA) to identify the effects of plants and soil on microbial community composition (Oksanen et al., 2013). SPSS 25.0 software was used to perform a Spearman correlation analysis of soil properties and phylum-level bacterial composition. The Spearman correlation coefficients of the top 50 most abundant bacterial genera, soil properties, and plant characteristics were calculated and displayed on the heat map. The relationship between microbial community composition, diversity attributes, and soil nitrogen was assessed using the corrplot package in R.
Network analysis was performed on the alfalfa rhizosphere microorganisms and soil properties based on strong and significant correlations (non-parametric Spearman’s correlation, P < 0.01, and absolute value of r > 0.6). Low-abundance genera were eliminated from the taxonomy table when they comprised less than 0.01% of the total relative abundance across all samples before network analysis. Statistical analysis of the network was carried out in the R environment, and correlation networks were visualized using the Gephi software (Bastian et al., 2009).
Structural equation modeling (SEM; Grace et al., 2012) was used to better understand the relationship between plant and soil variables and their impact on total nitrogen. Structural equation modeling was performed using the online software PLS (Ringle et al., 2015) based on the principle of the partial least squares path model. The priori SEM was originally constructed based on the bivariate correlation of plant and soil variables, in which direct and indirect pathways were considered. The normality of variables, linearity between variables, and non-multicollinearity between variables were verified before operating the model. To obtain the most concise model, insignificant and uninformative weak paths were removed from the final model. The maximum likelihood chi-square test was used (χ2, P > 0.05), Tucker-Lewis index (TLI ≥ 0.90), comparative fit index (CFI ≥ 0.90), the goodness of fit (GFI ≥ 0.90), and mean square error of root evaluation model fitting approximation (RMSEA < 0.10). The significance level was set at P < 0.05.
Results
Crude protein and biomass changes in alfalfa cultivars
Significant differences were found among alfalfa cultivars for crude protein and biomass (fresh weight per plant; Figure 1). The crude protein content ranged between 20.20 and 25.05 g kg–1 (Figure 1A), and the aboveground biomass ranged from 141.03 to 422.08 g plant–1 (Figure 1B). Crude protein of plants was highest in Fr, and comparatively lower in Zm and Mf compared to the other cultivars (P < 0.05). The cultivar producing largest biomass was Mf; and, Zm, Ns, Mz, and Sd were much lower than those of the other cultivars.
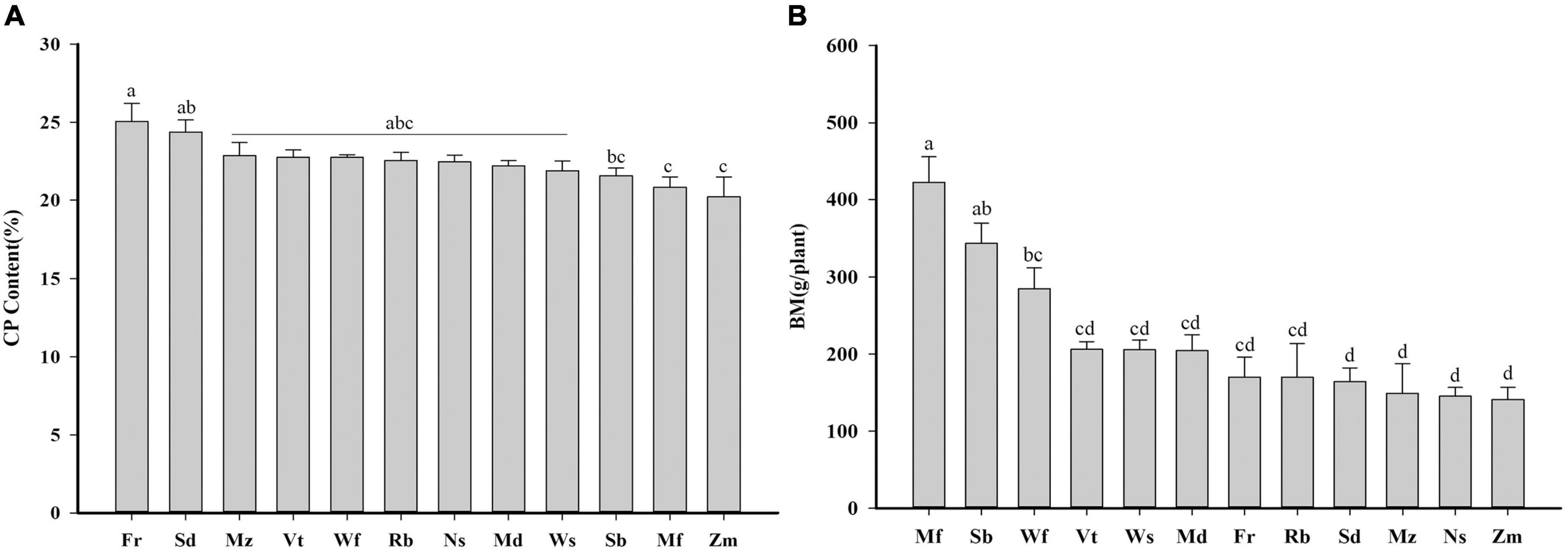
Figure 1. Average values of twelve alfalfa cultivars: (A) Crude protein content of alfalfa above ground (CP), (B) Aboveground biomass of alfalfa (BM, g plant– 1), fresh weight per plant. One-way analysis of variance, followed by Tukey’s honestly significant difference test was performed (P < 0.05). Different lowercase letters indicate significant differences among mean values in different cultivars. The alfalfa cultivars from high mean value to low mean value are shown from left to right.
Differing rhizosphere soil properties of alfalfa cultivars
Soil physiochemical characteristics were also significantly affected by cultivars. The pH values were moderately alkaline and generally remained consistent among cultivars; with the highest and lowest pH values found in Ns (8.24) and Mf (8.03), respectively (Table 2). The SOC contents were highest in Mf (11.86 g kg–1) and lowest in Ns (8.70 g kg–1), and were significantly different (P < 0.05; Table 2). The AP and TP contents showed similar trends, except for Sb and Sd, which had higher TP and lower in AP levels compared to other cultivars for all cultivars, except Sb and Sd, which had higher TP and lower AP levels compared to other cultivars (Table 2). In addition, significant changes in EC were found in some cultivars (Table 2).
Nitrogen content and microbial biomass in plant rhizosphere soil
The total nitrogen content of all soil samples ranged between 0.94 and 1.25 g kg–1 (Figure 2A). TN contents of Mf were significantly higher than Mz, Sd and Ns (P < 0.05). The soil NO3–-N (NT) content varied widely among different cultivars, ranged between 11.28 and 23.53 mg kg–1 (Figure 2B). NH4+-N (AN) content ranged between 3.29 and 4.22 mg kg–1 (Figure 2C). Md had the highest AN content and lowest NT content. Mf had the highest MBN content (21.98 mg kg–1; Figure 2D). Mz showed the lowest content of MBN and MBC compared to the other cultivars (Figure 2D and Supplementary Figure 1D).
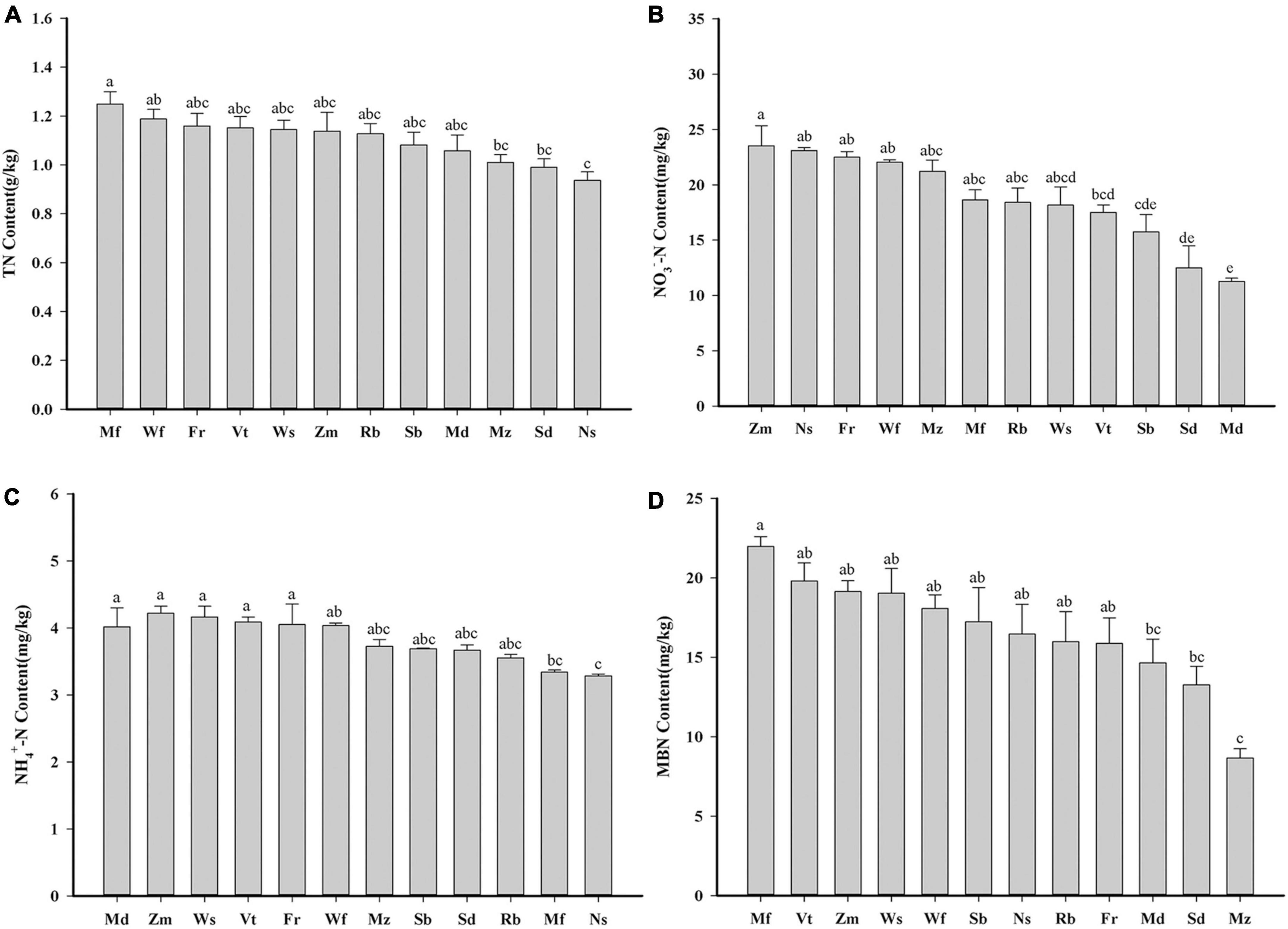
Figure 2. Average values of nitrogen in soil: (A) total nitrogen (TN, g kg–1), (B) nitrate nitrogen (NT, mg kg–1), (C) ammonium nitrogen (AN, mg kg–1) and (D) microbial biomass nitrogen (MBN, mg kg–1) of different alfalfa cultivars. Data were obtained from five biological replicates. One-way analysis of variance, followed by Tukey’s honestly significant difference test was performed (P < 0.05). Statistical classes sharing a lowercase letter are not significantly different. The alfalfa cultivars from high mean value to low mean value are shown from left to right.
Taxonomic classification, microbial rhizosphere community diversity, and community composition
The number of bacterial sequences varied from 38,220 to 73,626 per sample (mean: 55,209), and the average read length of the bacteria was 418 bp. For downstream analysis of the bacterial sequences, the datasets were rarefied to 3,312,565 valid sequences. Among the valid sequences obtained from alfalfa soil samples, an average of 7592 OTUs were obtained. The Sobs index showed that the total number of OTUs in addition to Wf was significantly different from that of other alfalfa cultivars (Figure 3A); among them, the total number of OTUs in Md was the highest and that in Zm was the lowest (Supplementary Figure 3A). There were significant differences between the two cultivars Mf and Sd. In terms of Simpson index (Figure 3B), only Ns and Fr were significantly higher than Sd. The Chao1 index was significantly higher in Sb, Fr, and Sd than that in Ns and Zm. According to the Shannon index, only Ns and Sd differed significantly (Figure 3C). No significant difference was found in the ACE index among the 12 alfalfa cultivars in this study (Figure 3D).
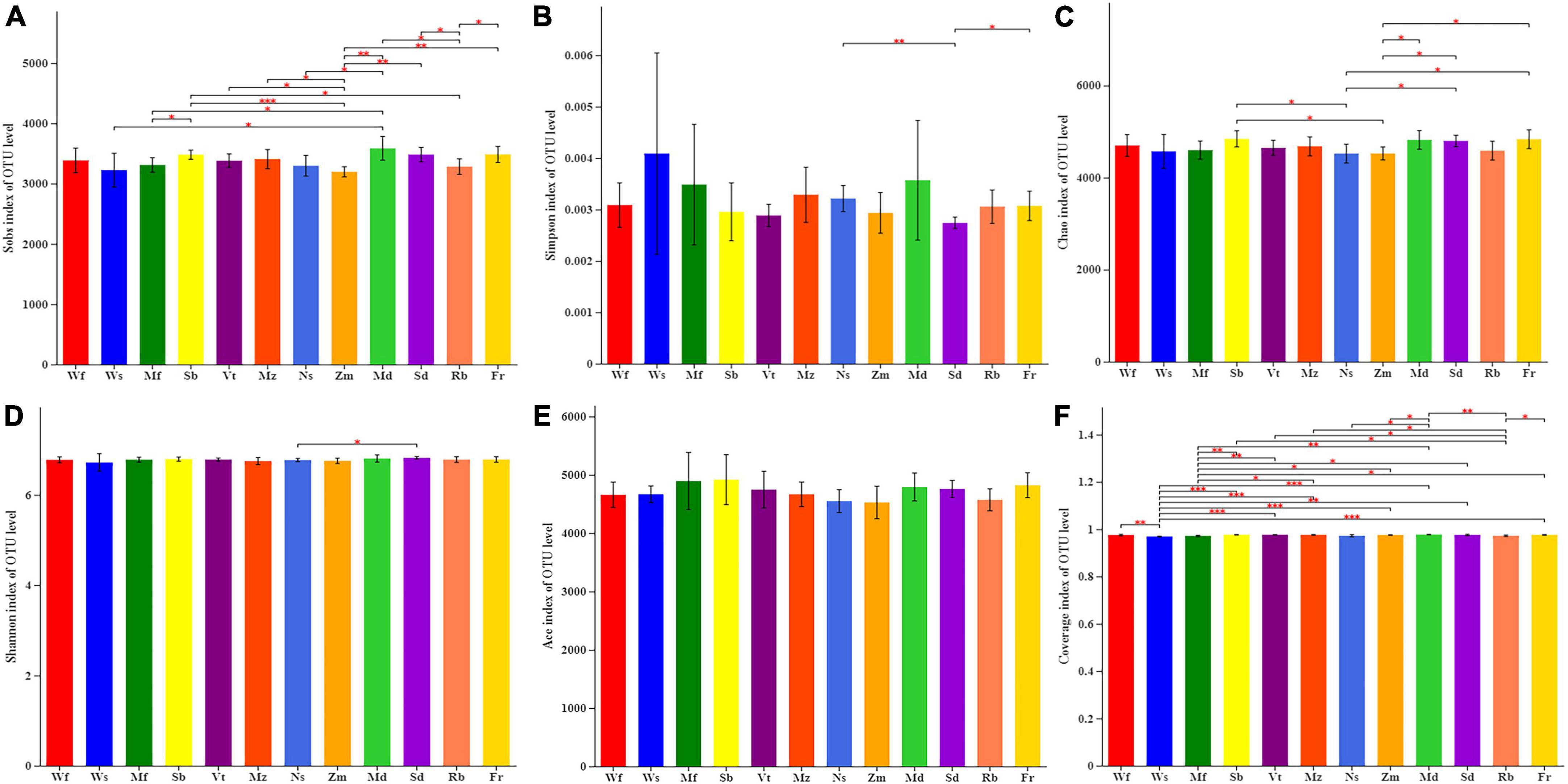
Figure 3. Average values of twelve alfalfa cultivars on bacterial alpha-diversity of OTU: (A) Sobs index, (B) Simpson diversity, (C) Chao richness, (D) Shannon diversity, (E) Ace richness, (F) Coverage index. *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001.
From all alfalfa cultivars in this study, we identified ten bacterial phyla: Actinobacteria (25.91%), Proteobacteria (22.79%), Acidobacteria (17.03%), Chloroflexi (10.10%), Firmicutes (5.46%), Myxococcota (3.77%), Bacteroidetes (2.91%), Gemmatimonadota (2.58%), Methylomirabilota (2.30%), Planctomycetota (1.78%) and others (5.35%; Supplementary Figure 2). For each alfalfa cultivar, the percentage of the total reads corresponding to each phylum (Figure 4A). Significant differences were observed in Actinobacteriota, Gemmatimonadota and Entotheonellaeota in bacterial phyla of different alfalfa cultivars (Supplementary Figure 3). At a genus level (Figure 4B), the most abundant genus in the rhizosphere soil of all alfalfa cultivars was an unknown genus, followed by Vicinamibacterales and Vicinamibacteraceae which belong to the Acidobacteriota phylum.
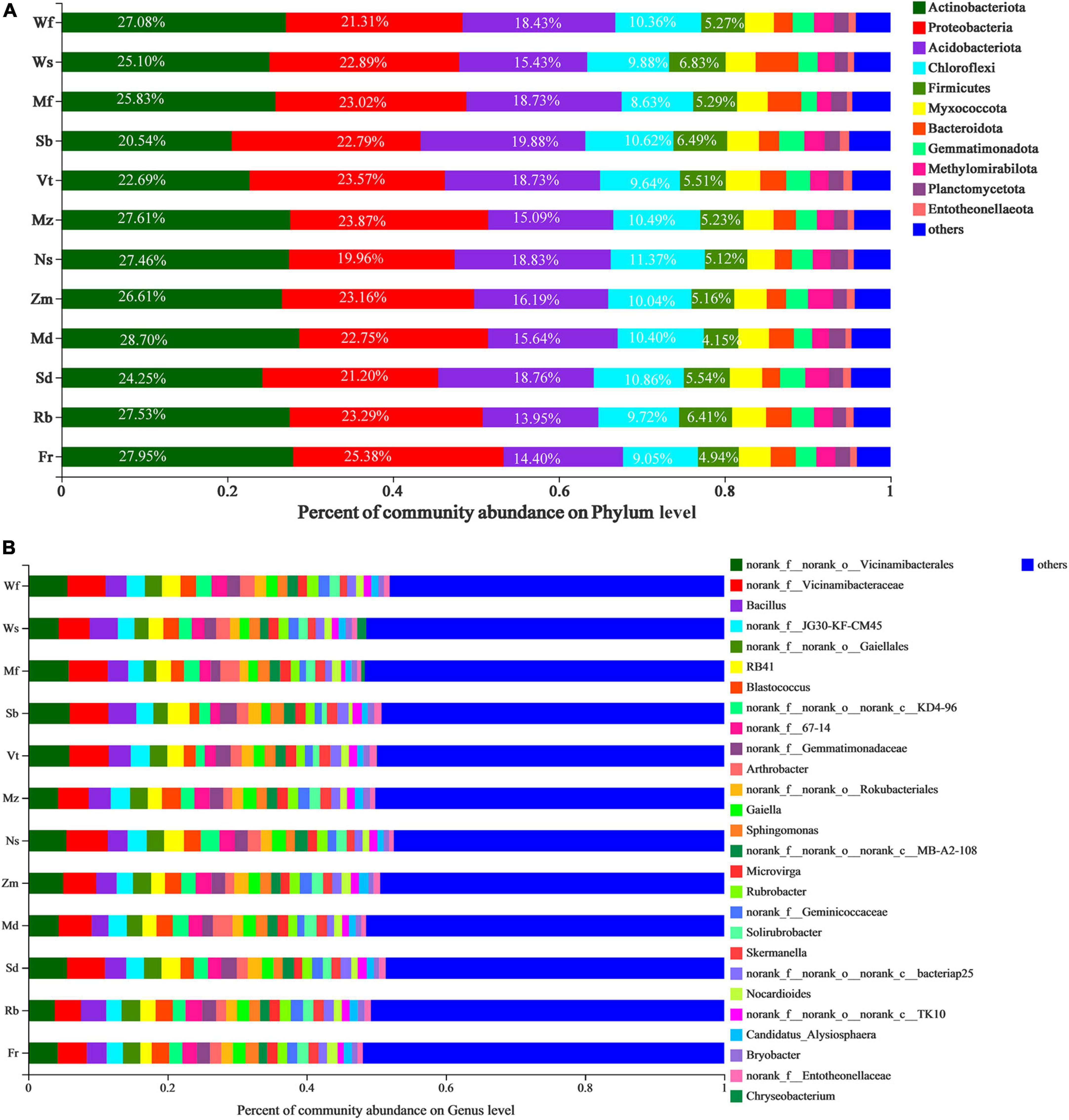
Figure 4. Taxonomic profiles of twelve alfalfa cultivars at the phylum (A) and genus (B) level, with higher relative richness are shown in the form of percentage in the figure, the remainder were combined and lumped in a category designated as “Other.” Different colors represent different species, and the length of each bar represents the proportion contributed by each species.
Correlations between physiochemical properties, microbial biomass, bacterial community composition and diversity
RDA (Figure 5A) was used to quantify the variations in plant and soil properties and its relationships with soil microbial community composition and structure. It was revealed that soil properties (pH, SOC, and TP) and microbial diversity were more influential on soil bacterial communities than plant crude protein and biomass. RDA supported the driving forces of soil properties on bacterial communities at the phylum level (Figure 5A) and showed a correlation between the two variables. The pH value was positively correlated with Acidobacteria, Chloroflexi, and Gemmatimonadota but negatively correlated with Bacteroidetes. TP content was positively correlated with Firmicutes and Gemmatimonadota but negatively correlated with Actinobacteria; however, available P content was negatively correlated with Gemmatimonadota and positively correlated with Firmicutes. SOC was negatively correlated with Acidobacteria, Chloroflexi, and Gemmatimonadota but positively correlated with Bacteroidota (Supplementary Table 3).
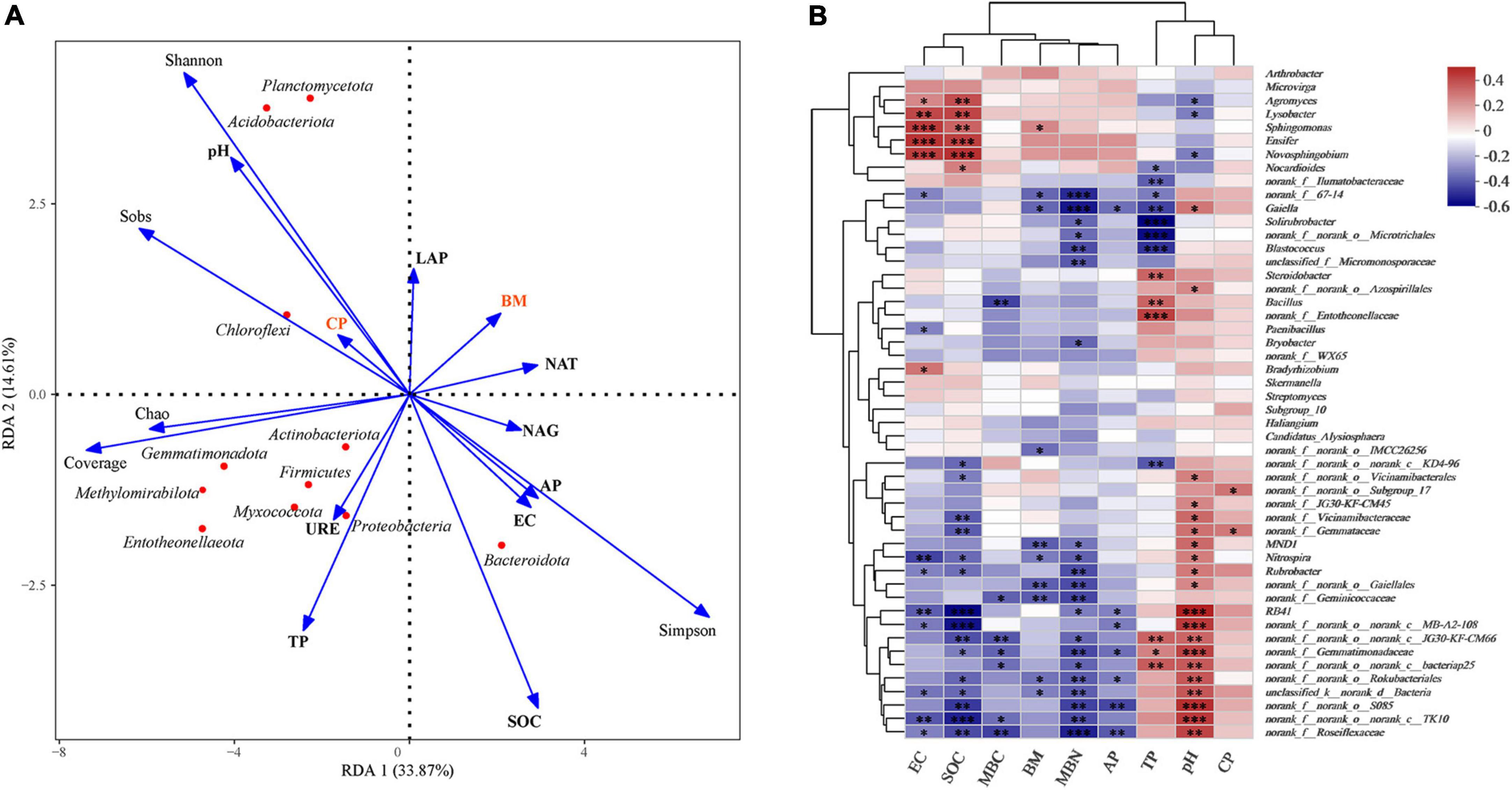
Figure 5. (A) Redundancy analysis (RDA) shows relationships between plant (BM and CP), soil physiochemical characteristics (pH, SOC, EC, AP, TP), microbial communities. The values of axes 1 and 2 are the percentages explained by the corresponding axis. Analysis of the level of contribution of soil, plant characteristics and bacterial diversity to changes in bacterial communities. (B) Correlation heat map of the top fifty genera and soil properties, the R value is shown in different colors in the figure. *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001.
Spearman correlation of plant characteristics, soil properties, and microbial biomass in genus taxa, and the results are displayed as heat maps (Figure 5B). Agromyces, Lysobacter, Sphingomonas, Ensifer and Novosphingobium showed an extremely positive correlation with EC and SOC, in which Ensifer belongs to Rhizobiales. Nocardioides showed a significant positive correlation with SOC and Bradyrhizobium belongs to Rhizobiales. In our study, we found that most bacteria, such as Gemmatimonadaceae, RB41, Rokubacteriales, Gaiella, Vicinamibacteraceae, Rubrobacter, and JG30-KF-CM45, demonstrated an extremely significant positive correlation with pH, whereas Vicinamibacteraceae, RB41, Gemmatimonadaceae, and Rubrobacter were significantly negative correlated with SOC. MBN was significantly negatively correlated with Roseiflexaceae, Gemmatimonadaceae, Gaiella, Geminicoccaceae, and Rokubacteriales (Figure 5B).
Correlations between soil nitrogen content, bacterial community composition, and diversity
TN and NT were significantly negatively correlated with Acidobacteria, Chloroflexi, and Gemmatimonadota. NT was significantly negatively correlated with Firmicutes, Myxococcota, and Entotheonellaeota. In contrast, AN was significantly positively correlated with Proteobacteria and Bacteroidetes. The bacterial alpha diversity indices indicated no significant effects on TN and AN content (Figure 6). The exception was NT, which showed the same results as the cultivars in the relationship with bacterial diversity. The major factor affecting nitrogen content is the dominant bacterial community in terms of bacterial diversity, with differences in total and inorganic nitrogen.
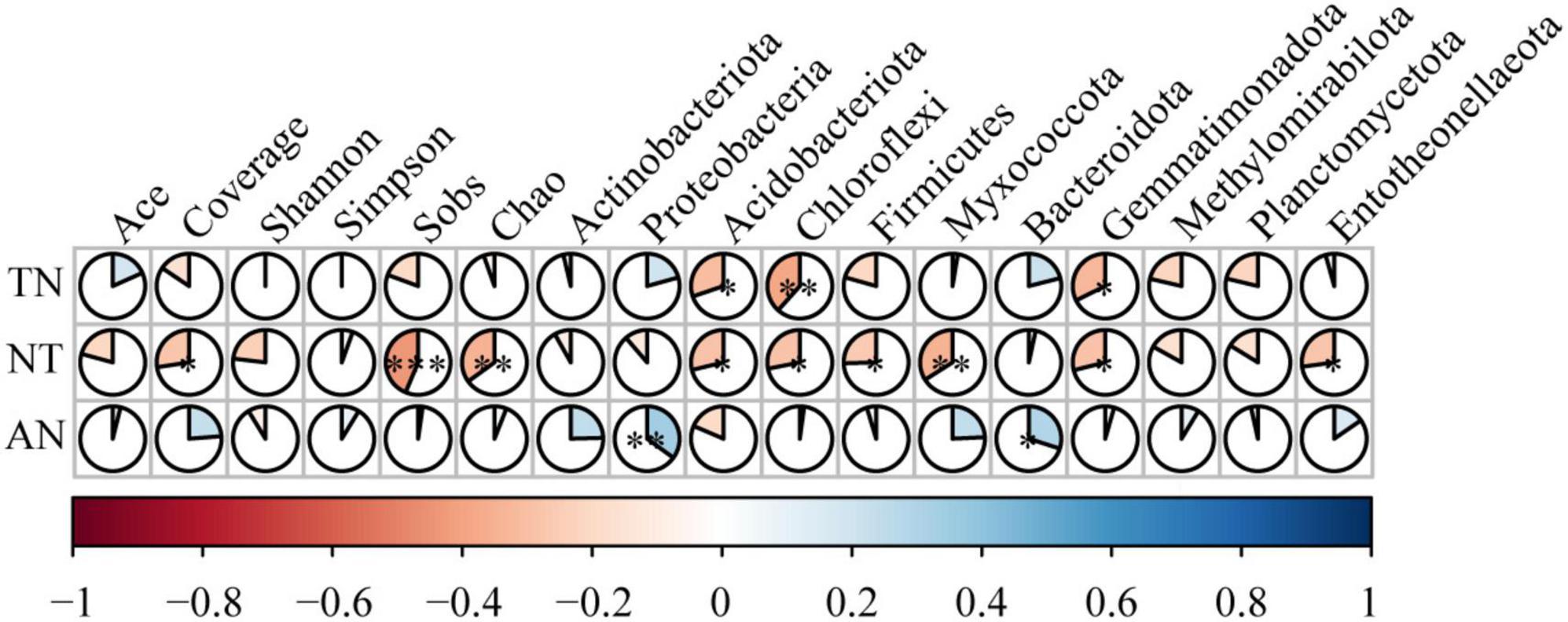
Figure 6. Spearman correlation coefficients between the soil nitrogen content, soil microbial diversity and soil microbial communities. *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001.
Soil bacterial community responses to nitrogen content in alfalfa
Based on partial least squares path models, we established that plants, soil, and microbiota were significantly associated with both direct and indirect effects on the response to soil nitrogen through our hypothesized pathways, involving the plant index, soil properties, microbial biomass, microbial composition, and diversity. The results revealed that the rhizosphere soil properties were positively associated with the plant index (Figure 7). Moreover, most soil properties had a significantly positive effect on microbial biomass and composition, including SOC, EC, and AP, except for the pH value. Soil microbial biomass had a direct positive correlation with the soil total nitrogen content. Interestingly, most microbial communities were shown to have a direct negative effect on soil nitrogen content, including Acidobacteria, Chloroflexi, Gemmatimonadota, and Methylomirabilota, whereas Proteobacteria and Bacteroidetes had direct positive effects on soil nitrogen content. Notably, soil microbial diversity appeared to have weaker correlations with plant and soil nitrogen content than with soil microbial biomass and composition.
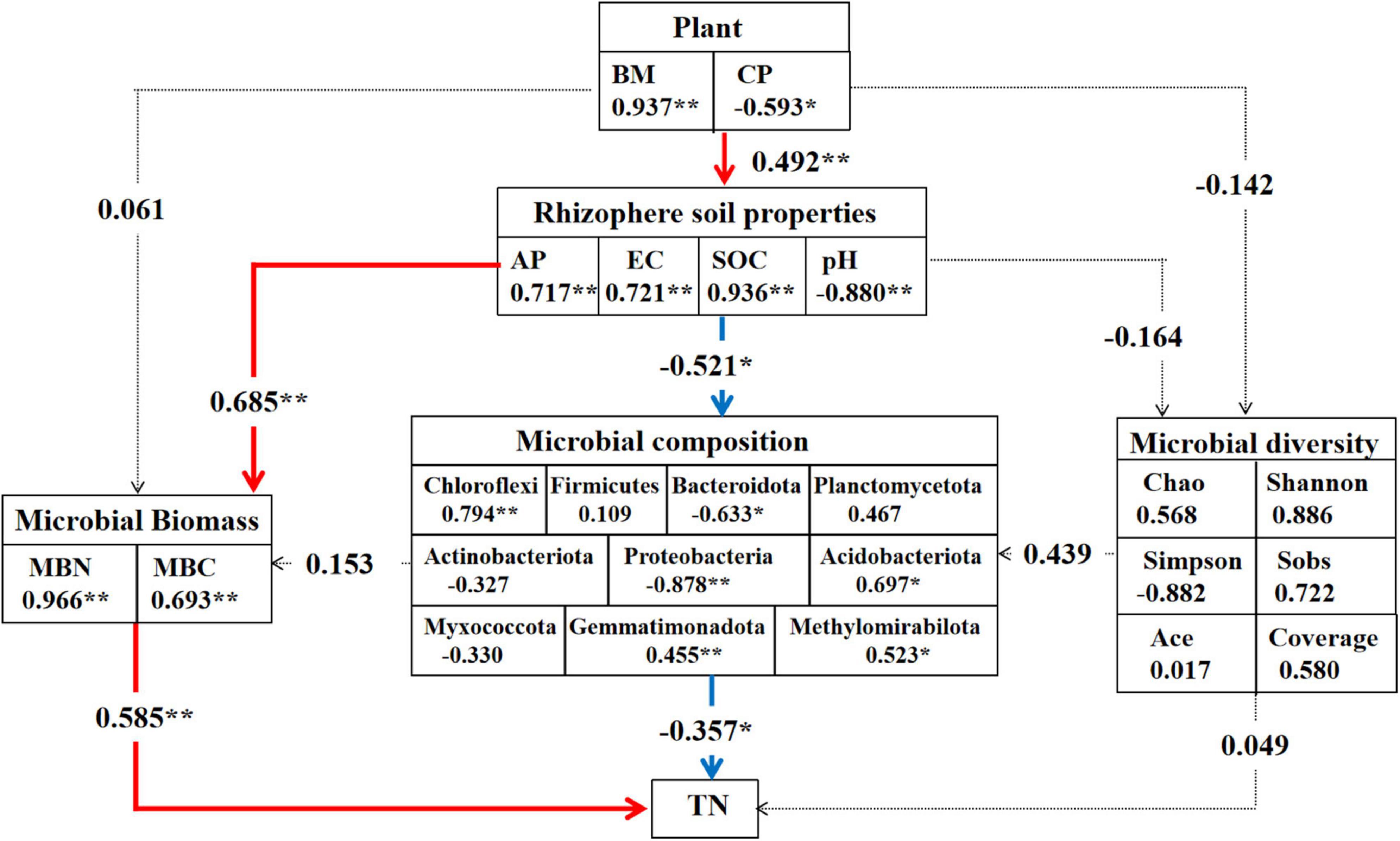
Figure 7. Partial least squares path models (PLS-PM) of the drivers of soil nitrogen content. Path analysis results for direct and indirect effects of the plant, rhizosphere soil properties, microbial biomass and composition on soil nitrogen content. The asterisks denote statistically significant differences among different alfalfa cultivars (*P < 0.05, **P < 0.01). Numbers on arrows are path coefficients indicating a positive (positive number) in red or negative effect in green (negative number).
Discussion
Effects of plant cultivars on soil properties
In natural ecosystems, plants grow in their native soil, and the characteristics of the soil near plant roots are modified by a range of plant growth processes, which in turn affect the rhizosphere microbiota (Philippot et al., 2013). Plant genotypes have a significant effect on the phylogenetic structure of root-associated microbiota (Hamonts et al., 2018). Under the same environmental conditions and soil, plant genotype is the main factor affecting the structure and function of rhizosphere microbiome (Zhang et al., 2017). By changing the properties of the surrounding soil, plants can modify their growth environments (Hu et al., 2018). Based on previous research, we hypothesized that differences in alfalfa cultivars induce changes in soil properties, which can significantly affect the soil nitrogen content and structure of rhizosphere microbial communities. We examined 12 different alfalfa cultivars from the same area with the same growth period, cutting method, cutting time, and similar initial soil. Some differences were observed in the crude protein and biomass per plant among the different alfalfa cultivars (Figure 1), which might be related to the productivity of plants, since it depends on the ability to acquisition of water and nutrients from the soil. In other words, the chemical characteristics of a plant species affect the properties of the surrounding soil (Song et al., 2019). In addition, Plants can directly alter the soil microbiome by secreting and releasing bioactive molecules and metabolites (Pang et al., 2021). Root exudates may differ between plant species, different growth stages, and even between the same plant species (Micallef et al., 2009; Li et al., 2013; Dietz et al., 2019), therefore, root exudates from the twelve alfalfa cultivars might have led to changes in soil physiochemical properties (Zhang et al., 2020), and most likely contributed to the effects on soil microbial activity and biomass (Gomez-Sagasti et al., 2021).
Effects of rhizosphere soil properties on soil microorganisms
Plant–soil feedback is associated with maintaining or changing the soil community, and soil rhizosphere bacteria can be influenced by both soil physiochemical properties and enzyme activities (Bani et al., 2018). We illustrated that, among specific perennial alfalfa cultivars, there were differences in changing soil properties and the recruitment of microorganisms. Hence, this is a fact that plants indirectly affect nutrient cycling by affecting the activities of soil decomposers and the decomposition of organic matter (Henneron et al., 2020). The critical roles of pH and SOC in shaping the bacterial community structure are characterized well (Figure 5A; Ling et al., 2016; Qu et al., 2020). The pH value can be increased or decreased by plant roots owing to the release or absorption of ions (Hinsinger et al., 2009), which gives rise to differences in soil pH values between species and even cultivars in the plant rhizosphere (Table 2). All these changes indicates that although the soil was uniform before sowing, but after years of growth of different cultivars has caused significant alterations in soil properties. Soil pH has significant effects on alfalfa rhizome microecosystems (Xiao et al., 2017), which mainly causes different microorganisms to favor different optimal pH values. The strong correlation between soil pH and microbial distribution could be due to the relatively narrow growth tolerance exhibited by most bacterial taxa (Xue et al., 2017). Soil organic carbon act as the soil carbon pool, provide rich carbon sources for the activities of microorganisms (Gan et al., 2013). This may coincide with the results that the rhizosphere soil properties directly affect the soil microbial biomass (Figure 7). RDA showed that soil properties (pH, SOC, and EC) and enzyme activity (NAT and URE) were the primary factors influencing bacterial community composition (Figure 5A). Meanwhile, soil microbial biomass is the source and sink of plant nutrients and actively participates in the nutrient cycle, representing the active part of the soil nutrients (Jia et al., 2017; Henneron et al., 2020). Taken together, these results indicate that soil properties, especially pH and SOC play a vital role in determining the bacterial community composition in perennial alfalfa.
Microbial biomass and community can directly regulate soil nitrogen content
The composition of the soil microbial community is considered an important component of the plant–soil feedback process (Bever et al., 2012). Soil microbial biomass nitrogen content is a reflection of soil microorganisms on nitrogen mineralization and fixation (Li et al., 2018) and further affects the content of total nitrogen and inorganic nitrogen in the soil. Concurrent with changes in MBN and MBC, the SEM model revealed a positive correlation with TN content (Figure 7). Our research indicates that soil bacterial diversity has a minimal effect on soil nitrogen (Figures 6, 7), possibly because all cultivars are closely related and belong to the legume family. Microbial species interactions are crucial to the structure and dynamics of soil bacterial communities (Chai et al., 2019). Thus, we further used co-occurrence network analysis to explore the complexity of the connections within the rhizosphere microbiomes of the different perennial alfalfa cultivars. First, the network complexity of the root system has a profound impact on nitrogen content. The higher content of TN in Mf indicates that the overall impact of the root network on the crude protein was much greater than the soil properties (Supplementary Figure 4). In Ws, the dominant bacteria belonging to Myxococcota, Chloroflexi, Entotheoellaeota, Bacteroides, and Acidobacterita had lower network complexity and accounted for a lower proportion of the composition of the total bacterial community. In Ns and Fr, the dominant bacteria belonged to Proteobacteria, Actinobacteria, Chloroflexi, and Myxococcota, which play an important role in the composition of the whole bacterial community. Nevertheless, the TN content of Md was the lowest, which demonstrated stronger correlations in this ecological network, possibly because the number of negative correlations was the highest among the edges compared with the other cultivars, implying that inter-competitive exclusion occurred within the bacterial community (Feng et al., 2017). The results showed that the dominant microbial population plays a dominant role in the rhizosphere. The most abundant phyla studied in the co-occurrence network were Acidobacteria and Proteobacteria, indicating that these generalists are adapted to a variety of environments (Xue et al., 2017). Proteobacteria are capable of inducing nitrogen fixation in symbiosis with plants (Emmert and Handelsman, 1999). The high abundance of Proteobacteria confirms that this phylum seems to benefit from the characteristics of leguminous plants (Sousa et al., 2020) and exhibited the variation by cultivar in our study (Figure 6). Plants can alter their rhizosphere microbiomes by influencing nutrient availability (Bell et al., 2015). These results indicate that rhizosphere microbial community composition, rather than diversity, plays a dominant role in affecting soil nitrogen. Similar to our findings (Figure 6), different biological and chemical drivers of each lima bean genotype influence the bacterial structure (Sousa et al., 2020). In this study, a significant relationship between bacterial community structure and soil total nitrogen content was also observed, indicating that variables associated with bacterial community composition may be crucial determinants of total nitrogen content (Figure 7).
Our results suggest that the differences in perennial alfalfa cultivars influenced soil physiochemical properties and further influenced soil microbial composition and biomass (Figure 7). The ability of plants to establish beneficial rhizosphere bacterial communities may be an important aspect of plant adaptability and should be considered an intrinsic plant trait that may be selected (Bell et al., 2015). The community structure of a certain cultivar in the rhizosphere showed a relatively low abundance, which may be due to its low ability to change the community structure of rhizosphere bacteria. The dynamics of soil microbial communities are likely determined by plants and the mediation of soil nutrient availability (Cabugao et al., 2017). We postulated that the difference in alfalfa may have unintentionally co-selected, affecting the recruitment of beneficial cultivar-specific microbiota, and finally affecting the soil nitrogen content. Further investigations are required to reveal the mechanisms underlying this specific microbial recruitment process.
Conclusion
This study reported differences in the rhizosphere soil microbial community and soil nitrogen content among different perennial alfalfa cultivars. According to the PLS-PM analysis, changing soil properties indirectly affected microbial biomass and community composition. Soil pH and SOC had a strong correlation with bacterial composition and diversity. Microbial biomass and soil total nitrogen are positively correlated, and some key microbial species play a major role. This study provides insight into the distributional patterns and drivers of soil total nitrogen in rhizosphere soil of perennial alfalfa and improves our understanding of the relationship between alfalfa, physiochemical properties and microbial.
Data availability statement
The original contributions presented in this study are publicly available. This data can be found here: NCBI; PRJNA812793.
Author contributions
YA and PY contributed for experimental design, sampling, lab analysis, interpretation and analysis of data, and writing the manuscript. HS, WZ, and ZY contributed in experimental design, interpretation of data, and manuscript revisions. SL and YS contributed in interpretation of data. RY contributed in lab analysis. TH corrected the draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by China Agriculture Research System of MOF and MARA, and the Project of Innovation-Driven Plan of Shaanxi Academy of Forestry Sciences (No. SXLK2020-0204).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.938865/full#supplementary-material
References
Aira, M., Gomez-Brandon, M., Lazcano, C., Baath, E., and Dominguez, J. (2010). Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 42, 2276–2281. doi: 10.1016/j.soilbio.2010.08.029
Annicchiarico, P. (2015). Alfalfa forage yield and leaf/stem ratio: narrow-sense heritability, genetic correlation, and parent selection procedures. Euphytica 205, 409–420. doi: 10.1007/s10681-015-1399-y
Bani, A., Pioli, S., Ventura, M., Panzacchi, P., Borruso, L., Tognetti, R., et al. (2018). The role of microbial community in the decomposition of leaf litter and deadwood. Appl. Soil Ecol. 126, 75–84. doi: 10.1016/j.apsoil.2018.02.017
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: an open source software for exploring and manipulating networks,” in Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA.
Baxendale, C., Orwin, K. H., Poly, F., Pommier, T., and Bardgett, R. D. (2014). Are plant-soil feedback responses explained by plant traits? New Phytol. 204, 408–423. doi: 10.1111/nph.12915
Bell, C. W., Asao, S., Calderon, F., Wolk, B., and Wallenstein, M. D. (2015). Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biol. Biochem. 85, 170–182. doi: 10.1016/j.soilbio.2015.03.006
Bever, J. D., Platt, T. G., and Morton, E. R. (2012). Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu. Rev. Microbiol. 66, 265–283.
Brereton, N. J. B., Gonzalez, E., Desjardins, D., Labrecque, M., and Pitre, F. E. (2020). Co-cropping with three phytoremediation crops influences rhizosphere microbiome community in contaminated soil. Sci. Total Environ. 711:135067. doi: 10.1016/j.scitotenv.2019.135067
Cabugao, K. G., Timm, C. M., Carrell, A. A., Childs, J., Lu, T.-Y. S., Pelletier, D. A., et al. (2017). Root and rhizosphere bacterial phosphatase activity varies with tree species and soil phosphorus availability in Puerto Rico tropical forest. Front. Plant Sci. 8:1834. doi: 10.3389/fpls.2017.01834
Chai, Y. F., Cao, Y., Yue, M., Tian, T. T., Yin, Q. L., Dang, H., et al. (2019). Soil abiotic properties and plant functional traits mediate associations between soil microbial and plant communities during a secondary forest succession on the loess plateau. Front. Microbiol. 10:895. doi: 10.3389/fmicb.2019.00895
Cúcio, C., Engelen, A. H., Costa, R., and Muyzer, G. (2016). Rhizosphere microbiomes of European seagrasses are selected by the plant, but are not species specific. Front. Microbiol. 7:440. doi: 10.3389/fmicb.2016.00440
Dai, Z., Liu, G., Chen, H., Chen, C., Wang, J., Ai, S., et al. (2020). Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J. 14, 757–770. doi: 10.1038/s41396-019-0567-9
de Faria, M. R., Costa, L. S. A. S., Chiaramonte, J. B., Bettiol, W., and Mendes, R. (2021). The rhizosphere microbiome: functions, dynamics, and role in plant protection. Trop. Plant Pathol. 46, 13–25.
Dietz, S., Herz, K., Doll, S., Haider, S., Jandt, U., Bruelheide, H., et al. (2019). Semi-polar root exudates in natural grassland communities. Ecol. Evol. 9, 5526–5541. doi: 10.1002/ece3.5043
Elfaki, M. O., and Abdelatti, K. A. (2016). Rumen content as animal feed: a review. J. Vet. Med. Anim. Prod. 7, 80–88.
Emmert, E. A. B., and Handelsman, J. (1999). Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol. Lett. 171, 1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x
Fan, W. Q., Ge, G. T., Liu, Y. H., Wang, W., Liu, L. Y., and Jia, Y. S. (2018). Proteomics integrated with metabolomics: analysis of the internal causes of nutrient changes in alfalfa at different growth stages. BMC Plant Biol. 18:78. doi: 10.1186/s12870-018-1291-8
Feng, K., Zhang, Z. J., Cai, W. W., Liu, W. Z., Xu, M. Y., Yin, H. Q., et al. (2017). Biodiversity and species competition regulate the resilience of microbial biofilm community. Mol. Ecol. 26, 6170–6182. doi: 10.1111/mec.14356
Fonouni-Farde, C., Tan, S., Baudin, M., Brault, M., Wen, J. Q., Mysore, K. S., et al. (2016). DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 7:12636. doi: 10.1038/ncomms12636
Gan, Y., Siddique, K. H., Turner, N. C., Li, X.-G., Niu, J.-Y., Yang, C., et al. (2013). Ridge-furrow mulching systems—an innovative technique for boosting crop productivity in semiarid rain-fed environments. Adv. Agron. 118, 429–476.
Gomez-Sagasti, M. T., Garbisu, C., Urra, J., Miguez, F., Artetxe, U., Hernandez, A., et al. (2021). Mycorrhizal-assisted phytoremediation and intercropping strategies improved the health of contaminated soil in a peri-urban area. Front. Plant Sci. 12:693044. doi: 10.3389/fpls.2021.693044
Grace, J. B., Schoolmaster, D. R., Guntenspergen, G. R., Little, A. M., Mitchell, B. R., Miller, K. M., et al. (2012). Guidelines for a graph-theoretic implementation of structural equation modeling. Ecosphere 3, 1–44. doi: 10.1890/Es12-00048.1
Hamonts, K., Trivedi, P., Grinyer, J., Holford, P., Drigo, B., Anderson, I. A., et al. (2018). Yellow canopy syndrome in sugarcane is associated with shifts in the rhizosphere soil metagenome but not with overall soil microbial function. Soil Biol. Biochem. 125, 275–285. doi: 10.1016/j.soilbio.2018.07.019
He, S. B., Guo, L. X., Niu, M. Y., Miao, F. H., Jiao, S., Hu, T. M., et al. (2017). Ecological diversity and co-occurrence patterns of bacterial community through soil profile in response to long-term switchgrass cultivation. Sci. Rep. 7:3608. doi: 10.1038/s41598-017-03778-7
Henneron, L., Kardol, P., Wardle, D. A., Cros, C., and Fontaine, S. (2020). Rhizosphere control of soil nitrogen cycling: a key component of plant economic strategies. New Phytol. 228, 1269–1282. doi: 10.1111/nph.16760
Hinsinger, P., Bengough, A. G., Vetterlein, D., and Young, I. M. (2009). Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152.
Hrbackova, M., Dvorak, P., Takac, T., Ticha, M., Luptovciak, I., Samajova, O., et al. (2020). Biotechnological perspectives of omics and genetic engineering methods in Alfalfa. Front. Plant Sci. 11:592. doi: 10.3389/fpls.2020.00592
Hu, L., Robert, C. A., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9:2738. doi: 10.1038/s41467-018-05122-7
Jia, X. H., Li, Y. S., Wu, B., Zhou, Y. Y., and Li, X. R. (2017). Effects of plant restoration on soil microbial biomass in an arid desert in northern China. J. Arid Environ. 144, 192–200. doi: 10.1016/j.jaridenv.2017.04.014
Jing, X., Sanders, N. J., Shi, Y., Chu, H. Y., Classen, A. T., Zhao, K., et al. (2015). The links between ecosystem multifunctionality and above- and belowground biodiversity are mediated by climate. Nat. Commun. 6:8159. doi: 10.1038/ncomms9159
Li, L., Xu, M., Eyakub Ali, M., Zhang, W., Duan, Y., and Li, D. (2018). Factors affecting soil microbial biomass and functional diversity with the application of organic amendments in three contrasting cropland soils during a field experiment. PLoS One 13:e0203812. doi: 10.1371/journal.pone.0203812
Li, X. G., Zhang, T. L., Wang, X. X., Hua, K., Zhao, L., and Han, Z. M. (2013). The composition of root exudates from two different resistant peanut cultivars and their effects on the growth of soil-borne pathogen. Int. J. Biol. Sci. 9, 164–173. doi: 10.7150/ijbs.5579
Ling, N., Zhu, C., Xue, C., Chen, H., Duan, Y. H., Peng, C., et al. (2016). Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol. Biochem. 99, 137–149. doi: 10.1016/j.soilbio.2016.05.005
Llado, S., Lopez-Mondejar, R., and Baldrian, P. (2018). Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 102, 4331–4338. doi: 10.1007/s00253-018-8950-4
Ma, C., Qi, J., Yu, X., Su, L., He, T., Wang, X., et al. (2020). Comparison of Rhizosphere Soil Microbial Diversity of Different Introduced Alfalfa Varieties.
Mendes, L. W., Raaijmakers, J. M., de Hollander, M., Mendes, R., and Tsai, S. M. (2018b). Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 12, 212–224. doi: 10.1038/ismej.2017.158
Mendes, L. W., Mendes, R., Raaijmakers, J. M., and Tsai, S. M. (2018a). Breeding for soil-borne pathogen resistance impacts active rhizosphere microbiome of common bean. ISME J. 12, 3038–3042. doi: 10.1038/s41396-018-0234-6
Micallef, S. A., Shiaris, M. P., and Colon-Carmona, A. (2009). Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J. Exp. Bot. 60, 1729–1742. doi: 10.1093/jxb/erp053
Mitchell, R. J., Campbell, C. D., Chapman, S. J., and Cameron, C. M. (2010). The ecological engineering impact of a single tree species on the soil microbial community. J. Ecol. 98, 50–61. doi: 10.1111/j.1365-2745.2009.01601.x
Mod, H. K., Buri, A., Yashiro, E., Guex, N., Malard, L., Pinto-Figueroa, E., et al. (2021). Predicting spatial patterns of soil bacteria under current and future environmental conditions. ISME J. 15, 2547–2560. doi: 10.1038/s41396-021-00947-5
Mulvaney, R. L., Azam, F., Stein, S. N., and Simmons, F. W. (1992). Chloride interference in total nitrogen analysis by the Kjeldahl method. Commun. Soil Sci. Plant Anal. 23, 1805–1813. doi: 10.1080/00103629209368705
Navarrete, A. A., Tsai, S. M., Mendes, L. W., Faust, K., de Hollander, M., Cassman, N. A., et al. (2015). Soil microbiome responses to the short-term effects of Amazonian deforestation. Mol. Ecol. 24, 2433–2448. doi: 10.1111/mec.13172
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R., et al. (2013). Vegan: Community Ecology Package Version 2.0-2. R Package.
Pang, Z. Q., Chen, J., Wang, T. H., Gao, C. S., Li, Z. M., Guo, L. T., et al. (2021). Linking plant secondary metabolites and plant microbiomes: a review. Front. Plant Sci. 12:621276. doi: 10.3389/fpls.2021.621276
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Qu, Z. L., Liu, B., Ma, Y., Xu, J., and Sun, H. (2020). The response of the soil bacterial community and function to forest succession caused by forest disease. Funct. Ecol. 34, 2548–2559. doi: 10.1111/1365-2435.13665
Ringle, C. M., Wende, S., and Becker, J.-M. (2015). SmartPLS 3. Boenningstedt. Germany: SmartPLS GmbH.
Rossmann, M., Perez-Jaramillo, J. E., Kavamura, V. N., Chiaramonte, J. B., Dumack, K., Fiore-Donno, A. M., et al. (2020). Multitrophic interactions in the rhizosphere microbiome of wheat: from bacteria and fungi to protists. FEMS Microbiol. Ecol. 96:fiaa032. doi: 10.1093/femsec/fiaa032
Saiya-Cork, K. R., Sinsabaugh, R. L., and Zak, D. R. (2002). The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol. Biochem. 34, 1309–1315. doi: 10.1016/S0038-0717(02)00074-3
Samaddar, S., Schmidt, R., Tautges, N. E., and Scow, K. (2021). Adding alfalfa to an annual crop rotation shifts the composition and functional responses of tomato rhizosphere microbial communities. Appl. Soil Ecol. 167:104102. doi: 10.1016/j.apsoil.2021.104102
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Shelake, R. M., Pramanik, D., and Kim, J. Y. (2019). Exploration of plant-microbe interactions for sustainable agriculture in CRISPR Era. Microorganisms 7:269. doi: 10.3390/microorganisms7080269
Shenton, M., Iwamoto, C., Kurata, N., and Ikeo, K. (2016). Effect of wild and cultivated rice genotypes on rhizosphere bacterial community composition. Rice 9:42. doi: 10.1186/s12284-016-0111-8
Song, Z. L., Liu, G. B., and Zhang, C. (2019). Response of rhizosphere microbial communities to plant succession along a grassland chronosequence in a semiarid area. J. Soils Sediments 19, 2496–2508. doi: 10.1007/s11368-019-02241-6
Sousa, R. M. S., Mendes, L. W., Antunes, J. E. L., Oliveira, L. M. D., Sousa, A. M. D. B., Gomes, R. L. F., et al. (2020). Diversity and structure of bacterial community in rhizosphere of lima bean. Appl. Soil Ecol. 150:103490. doi: 10.1016/j.apsoil.2019.103490
Sugiyama, A., Ueda, Y., Zushi, T., Takase, H., and Yazaki, K. (2014). Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS One 9:e100709. doi: 10.1371/journal.pone.0100709
Tao, K., Kelly, S., and Radutoiu, S. (2019). Microbial associations enabling nitrogen acquisition in plants. Curr. Opin. Microbiol. 49, 83–89. doi: 10.1016/j.mib.2019.10.005
Tesfaye, M., Dufault, N. S., Dornbusch, M. R., Allan, D. L., Vance, C. P., and Samac, D. A. (2003). Influence of enhanced malate dehydrogenase expression by alfalfa on diversity of rhizobacteria and soil nutrient availability. Soil Biol. Biochem. 35, 1103–1113. doi: 10.1016/S0038-0717(03)00162-7
Tkacz, A., Bestion, E., Bo, Z., Hortala, M., and Poole, P. S. (2020a). Influence of plant fraction, soil, and plant species on microbiota: a multikingdom comparison. MBio 11, e2785–19. doi: 10.1128/mBio.02785-19
Tkacz, A., Pini, F., Turner, T. R., Bestion, E., Simmonds, J., Howell, P., et al. (2020b). Agricultural selection of wheat has been shaped by plant-microbe interactions. Front. Microbiol. 11:132. doi: 10.3389/fmicb.2020.00132
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707.
Wang, S. C., Guo, K., Ameen, A., Fang, D., Li, X. L., Liu, X. J., et al. (2022). Evaluation of different shallow groundwater tables and alfalfa cultivars for forage yield and nutritional value in coastal saline soil of North China. Life (Basel) 12:217. doi: 10.3390/life12020217
Wang, Z. N., Lu, J. Y., Yang, H. M., Zhang, X., Luo, C. L., and Zhao, Y. X. (2014). Resorption of nitrogen, phosphorus and potassium from leaves of lucerne stands of different ages. Plant Soil 383, 301–312. doi: 10.1007/s11104-014-2166-x
Wen, T., Zhao, M., Yuan, J., Kowalchuk, G. A., and Shen, Q. (2021). Root exudates mediate plant defense against foliar pathogens by recruiting beneficial microbes. Soil Ecol. Lett. 3, 42–51.
Xiao, X., Fan, M. C., Wang, E. T., Chen, W. M., and Wei, G. H. (2017). Interactions of plant growth-promoting rhizobacteria and soil factors in two leguminous plants. Appl. Microbiol. Biotechnol. 101, 8485–8497. doi: 10.1007/s00253-017-8550-8
Xu, N., Tan, G. C., Wang, H. Y., and Gai, X. P. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi: 10.13227/j.hjkx.202003113
Xu, Y., Ge, Y., Song, J. X., and Rensing, C. (2020). Assembly of root-associated microbial community of typical rice cultivars in different soil types. Biol. Fertil. Soils 56, 249–260. doi: 10.1007/s00374-019-01406-2
Xue, L., Ren, H. D., Li, S., Leng, X. H., and Yao, X. H. (2017). Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Front. Microbiol. 8:2377. doi: 10.3389/fmicb.2017.02377
Yang, C., Zhang, F., Jiang, X., Yang, X., He, F., Wang, Z., et al. (2021). Identification of genetic loci associated with crude protein content and fiber composition in alfalfa (Medicago sativa L.) using QTL mapping. Front. Plant Sci. 12:608940. doi: 10.3389/fpls.2021.608940
Yashiro, E., Pinto-Figueroa, E., Buri, A., Spangenberg, J. E., Adatte, T., Niculita-Hirzel, H., et al. (2016). Local environmental factors drive divergent grassland soil bacterial communities in the western Swiss Alps. Appl. Environ. Microbiol. 82, 6303–6316. doi: 10.1128/AEM.01170-16
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S. J., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, J. G., Fan, S. H., Qin, J., Dai, J. C., Zhao, F. J., Gao, L. Q., et al. (2020). Changes in the microbiome in the soil of an American ginseng continuous plantation. Front. Plant Sci. 11:572199. doi: 10.3389/fpls.2020.572199
Zhang, L., Zhang, J., Wei, Y., Hu, W., Liu, G., Zeng, H., et al. (2021). Microbiome-wide association studies reveal correlations between the structure and metabolism of the rhizosphere microbiome and disease resistance in cassava. Plant Biotechnol. J. 19, 689–701. doi: 10.1111/pbi.13495
Zhang, M. M., Wang, N., Hu, Y. B., and Sun, G. Y. (2018). Changes in soil physicochemical properties and soil bacterial community in mulberry (Morus alba L.)/alfalfa (Medicago sativa L.) intercropping system. Microbiologyopen 7:e00555. doi: 10.1002/mbo3.555
Zhang, R. F., Vivanco, J. M., and Shen, Q. R. (2017). The unseen rhizosphere root-soil-microbe interactions for crop production. Curr. Opin. Microbiol. 37, 8–14. doi: 10.1016/j.mib.2017.03.008
Keywords: alfalfa cultivars, soil properties, rhizosphere soil microbial community, microbial diversity, soil nitrogen
Citation: An Y, Sun H, Zhang W, Sun Y, Li S, Yu Z, Yang R, Hu T and Yang P (2022) Distinct rhizosphere soil responses to nitrogen in relation to microbial biomass and community composition at initial flowering stages of alfalfa cultivars. Front. Plant Sci. 13:938865. doi: 10.3389/fpls.2022.938865
Received: 08 May 2022; Accepted: 04 August 2022;
Published: 24 August 2022.
Edited by:
Linkun Wu, Fujian Agriculture and Forestry University, ChinaReviewed by:
Babak Motesharezadeh, University of Tehran, IranSajjad Raza, Nanjing University of Information Science and Technology, China
Shusheng Zhu, Yunnan Agricultural University, China
Copyright © 2022 An, Sun, Zhang, Sun, Li, Yu, Yang, Hu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianming Hu, aHV0aWFubWluZ0AxMjYuY29t; Peizhi Yang, eWFuZ3BlaXpoaUAxMjYuY29t
 Yunru An1
Yunru An1 Haoyang Sun
Haoyang Sun Shuxia Li
Shuxia Li Tianming Hu
Tianming Hu Peizhi Yang
Peizhi Yang