- 1Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Resources and Environmental Sciences, China Agricultural University, Beijing, China
- 4College of Land and Environment, Shenyang Agricultural University, Shenyang, China
- 5Department of Horticulture and Natural Resources, Kansas State University, Manhattan, KS, United States
Calcium (Ca2+) is an essential plant nutrient, and Ca2+/H+ exchangers (CAXs) regulate Ca2+ partitioning between subcellular compartments. AtCAX1 activity is inhibited by its N-terminal regulatory region (NRR), which was initially defined as the sequence between the first two methionines. However, the accuracy of this NRR definition and the NRR regulatory mechanism remain unclear. Here, using tomato SlCAX1 as a model, we redefined the NRR of CAXs and demonstrated that our new definition is also applicable to Arabidopsis AtCAX1 and AtCAX3. The N-terminal-truncated SlCAX1 (SlCAX1Δ39) but not the full-length SlCAX1 was active in yeast, similar to Arabidopsis AtCAX1. Characterization of slcax1 mutants generated by CRISPR-Cas9 confirmed the calcium transport ability of SlCAX1. Sequence alignment between SlCAX1, AtCAX1, AtCAX3, and the Bacillus subtilis Ca2+/H+ antiporter protein YfkE revealed that SlCAX1 does not have the 2nd methionine and YfkE does not have any amino acid residues in front of the first transmembrane domain. Truncating the amino acid residues up to the first transmembrane of SlCAX1 (SlCAX1Δ66) further increased its activity. The same truncation had a similar effect on Arabidopsis AtCAX1 and AtCAX3. Expression of full-length SlCAX1 and SlCAX1Δ66 in tomato plants confirmed the results. Our results suggest that SlCAX1 is critical for Ca2+ homeostasis and all the amino acid residues in front of the first transmembrane domain inhibit the activity of CAXs. Our redefinition of the NRR will facilitate fine-tuning of Ca2+ partitioning to reduce the incidence of Ca2+-related physiological disorders in crops.
Introduction
Calcium (Ca2+) is an essential macronutrient for plants and functions as a second messenger that is generated in response to different stimuli. Precise regulation of its distribution between different subcellular compartments is critical for normal plant growth, development, and adaptation to the environment. Most Ca2+ is stored in the cell wall, apoplast, vacuole, and endoplasmic reticulum and is maintained at extremely low levels in the cytosol (Dodd et al., 2010). Ca2+ antiporters, such as Ca2+/H+ exchangers (CAXs), control the efflux of Ca2+ from the cytosol. This establishes a Ca2+ gradient between the cytosol and other compartments and maintains Ca2+ partitioning in plant cells (Emery et al., 2012).
Arabidopsis thaliana AtCAX1, the first identified plant CAX, was cloned based on its ability to complement the hypersensitivity of a Saccharomyces cerevisiae mutant with defects in vacuolar Ca2+ transport to high concentrations of Ca2+ (Hirschi et al., 1996). AtCAX1 is a high-affinity, high-capacity Ca2+/H+ antiporter. Five additional AtCAXs have been cloned from Arabidopsis (Shigaki et al., 2006). CAXs from other plant species have been intensively studied and the substrate range of these CAXs extends beyond Ca2+ (Kamiya et al., 2006; Pittman and Hirschi, 2016; Amagaya et al., 2020).
AtCAX1 activity is tightly regulated by its N-terminal regulatory region (NRR), which is 36 amino acids long (Pittman and Hirschi, 2001). The NRR was initially identified in the original AtCAX1 antiporter used in yeast complementation experiments. This antiporter is the product of a partial-length cDNA in which the N terminus is truncated, but there is a second in-frame start codon, allowing translation initiation from the methionine at the 37th amino acid position (referred to as AtCAX1Δ36 hereafter; Hirschi et al., 1996; Pittman and Hirschi, 2001). Heterologous expression of AtCAX1Δ36, but not full-length AtCAX1 (lCAX1), complements the hypersensitivity of the yeast mutant strain to calcium, suggesting that the first 36 amino acid residues inhibit AtCAX1 activity. The region deleted in AtCAX1Δ36 was defined as the NRR (Pittman and Hirschi, 2001). AtCAX3 has high amino acid sequence similarity with AtCAX1, including in the NRR. Neither full-length AtCAX3 nor the 36-amino-acid N-terminal truncation of AtCAX3 (AtCAX3Δ36) is as active as AtCAX1Δ36 in yeast expression assays. However, when longer truncations are performed, AtCAX3 is able to transport Ca2+ in both yeast and plants (Manohar et al., 2011). Further genetic evidence suggests that AtCAX3 functions redundantly with AtCAX1 because neither atcax1 nor atcax3 single mutants display an obvious phenotype, whereas the growth and development of atcax1 atcax3 double mutants are strongly impaired (Cheng et al., 2005; Hocking et al., 2017), suggesting that AtCAX3 is an active Ca2+ transporter though how it is activated remains unclear.
As CAXs are important for Ca2+ homeostasis, tremendous effort has been made to ectopically express these genes in different economically important species, such as tomatoes, potatoes, and carrots, with the aim of improving fruit quality, abiotic stress tolerance, or shelf life, or alleviating Ca2+-related physiological disorders (Park et al., 2004, 2005a,b, 2009; Kim et al., 2006; Han et al., 2009; Wu et al., 2011, 2012). However, ectopic expression of AtCAX1Δ36, which is constitutively active, in certain species including tobacco and tomato resulted in strong calcium disorder symptoms such as necrosis in leaf tips or blossom-end rot (Park et al., 2005a; Wu et al., 2012). In tomato, 100% of AtCAX1Δ36-expressing plants exhibited blossom-end rot due to perturbation of calcium partitioning in plant cells (Park et al., 2005a; De Freitas et al., 2011, 2012). Although bioinformatics analysis has been performed for tomato SlCAX genes (Amagaya et al., 2020), their functions in Ca2+ homeostasis and their roles in controlling calcium-related physiological disorders remain to be addressed.
Here, we characterized the tomato SlCAX1 gene and gained insight into its regulatory mechanism of Ca2+ antiporters. We redefined which region constitutes the NRR and demonstrated that our new definition could also apply to Arabidopsis AtCAX1 and AtCAX3. These results could facilitate efforts to mitigate physiological disorders in tomato by fine-tuning Ca2+ homeostasis.
Materials and Methods
Yeast Strains and Plant Materials
Yeast strain K667 (vcx1::hisG cnb1::LEU2 pmc1::TRP1 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1), which lacks the vacuolar Ca2+-ATPase (PMC1) and vacuolar Ca2+/H+ antiporter (VCX1), was provided by Dr. Kendal Hirschi (Cunningham and Fink, 1996). For metal ion treatments, tomato plants (Solanum lycopersicum “Rubion”) were grown on 1/2 Murashige and Skoog (MS) medium for 2 weeks and then transferred onto an equal volume of solution containing the different metal ions and incubated for 16 h. The tissues were rinsed with distilled water three times and dried on tissue paper before flash freezing with liquid nitrogen. The frozen tissues were kept at −80°C before analysis.
RNA Extraction and RT-qPCR
RNA was isolated using the RNeasy Plant Kit (Zymo Research), according to the manufacturer’s instructions. RT-qPCR was performed to detect SlCAX expression. First-strand cDNA was produced using the iScript™ cDNA Synthesis kit from BIO-RAD. The first-strand cDNA of 1 μl was used to amplify SlCAX-specific fragments and SlACTIN (acc. no. TC194780) was used as a control. Specific amplification primers are shown in Supplementary Table 1.
Plasmid Construction and Yeast Transformation
Different versions of the SlCAX (SlCAX1, SlCAX1Δ39, SlCAX1Δ58, SlCAX1Δ66, and SlCAX1Δ93) and AtCAX (AtCAX1Δ36, AtCAX1Δ63, AtCAX3Δ36, and AtCAX3Δ63) genes were amplified from the tomato and Arabidopsis cDNA libraries, respectively, and cloned into the yeast expression vector piHGpd (Pittman et al., 2002). The first 39 and 66 amino acids of SlCAX1 were randomly scrambled and codon optimized based on yeast codon preference and the DNA oligos were synthesized by Beijing Dahong Biotechnology. The scrambled-39-SlCAX1 and scrambled-66-SlCAX1 were fused with SlCAX1Δ39 and SlCAX1Δ66, respectively, and cloned into the yeast expression vector piHGpd. The scrambled DNA sequences were shown in Supplementary Table 2. All PCR amplifications were performed using Phusion® High-Fidelity DNA Polymerases from New England Biolabs; the restriction sites used were XbaI and SacI. Primer sequences used in this study are shown in Supplementary Table 3.
Yeast cells were transformed using the standard lithium acetate method and selected on a synthetic defined medium lacking His as described (Pittman et al., 2002). For the Ca2+ tolerance assays, the yeast cells were cultured on SD-His liquid medium to an OD value of 1. After series dilution, the yeast cells were cultured on solid YPD supplemented with different concentrations of CaCl2 and grown at 30°C for 3 days (Pittman and Hirschi, 2001; Manohar et al., 2011).
Preparation of Transgenic Plants
Agrobacterium-mediated transformation was used to generate transgenic tomato plants. Different versions of SlCAX1 were fused with 2× Sterp tag II- 3x Flag tag driven by the CaMV35S promoter and cloned into the PRI101 vector (Satoh et al., 2004; Sugio et al., 2008). Primer sequences used for cloning are shown in Supplementary Table 2. The plasmids were introduced into Agrobacterium strain LBA4404 following standard protocols and then transformed into tomato (S. lycopersicum “Micro-Tom”) following a previously described method (Cosson et al., 2006). The transgenic lines were confirmed by genomic PCR using the following primers: SlCAX1-PRI forward, 5′-CCA ACC ACG TCT TCA AAG CA-3′ and reverse, 5′-TCC TGT AGA GTG AAA GCT GTG A-3′.
Knock-Out of SlCAX1 Using CRISPR-Cas9
CRISPR-Cas9 was used to create slcax1 knockouts in tomato (Micro-Tom) through Agrobacterium-mediated transformation. We designed two guide RNAs (gRNAs) targeting the coding region of SlCAX1. The sequence of Guide1 was ‘TCG TAG CCA TGG ACG AAC GG’ and that of Guide2 was ‘CTT CTA AAT GCA ACA TGT GG’. The two guides were cloned into the CRISPR-Cas9 vector BGK012, which was provided by Jiangsu Baige Gene. The vector map is shown in Supplementary Figure 1. The CRISPR/Cas9 construct was transferred into the Agrobacterium strain LBA4404 and then into tomato, as described (Li et al., 2018). The genomic regions surrounding the target sites were PCR-amplified and Sanger sequenced. All sequence information is shown in Supplementary Table 4.
Measurement of Ca2+ Contents
Seeds (T2) of the SlCAX1, SlCAX1Δ66-expressing, SlCAX1 CRISPR knockout, and wild-type plants were germinated on MS medium containing MS inorganic salts and 0.8% agar (Sigma), and the pH was adjusted to 5.8–6.0 with KOH. After 4 weeks, plants of similar size from each of the lines were harvested and rinsed with deionized water two to three times before drying with filter paper (Wu et al., 2011). The plants were further dried at 80°C for 48 h. For each sample, a total of 0.5 g (dry weight) tissue was digested with 10 ml nitric acid and 0.5 ml perchloric acid at 120°C for 1 h. The amount of Ca2+ present in the digested tissue was determined by inductively coupled plasma optical emission spectroscopy (Tang et al., 2015).
Results
To identify SlCAX proteins from tomato, we conducted a BLAST analysis of the tomato protein database in Phytozome1 using the Arabidopsis AtCAX1 protein sequence as query. We identified six proteins with high similarity to Arabidopsis AtCAX1 (Solyc09g005260, Solyc12g055750, Solyc06g006110, Solyc03g123790, Solyc07g056110, and Solyc12g011070). Phylogenetic analysis using the Neighbour-Joining method identified two tomato proteins (Solyc09g005260 and Solyc06g006110) in the same clade as AtCAX1, AtCAX3, and AtCAX4, which we named SlCAX1 and SlCAX3, respectively (Figure 1A). This clade, named the I-A clade, contains specialized Ca2+ transporters (Shigaki et al., 2006). SlCAX2, SlCAX4, SlCAX5, and SlCAX6 were grouped with AtCAX2, AtCAX5, and AtCAX6 in the I-B clade; proteins in this group transport multiple cations, including Ca2+, Cd2+, and Mn2+ (Figure 1A; Hirschi et al., 2000). All the tomato SlCAX proteins, except SlCAX6, were predicted to have 11 transmembrane (TM) domains (Supplementary Figure 2). Gene expression data from the Tomato eFP browser showed that SlCAX1 and SlCAX3 are mainly expressed in leaves, and SlCAX2, SlCAX4, and SlCAX5 are expressed in fruits, whereas SlCAX6 is only expressed in immature flowers (Supplementary Figure 3; Sato et al., 2012). We treated the tomato seedlings with different types of ions. Only SlCAX1 and SlCAX3 were dramatically induced by Ca2+ treatment, indicating their roles in Ca2+ transport (Figure 1B).
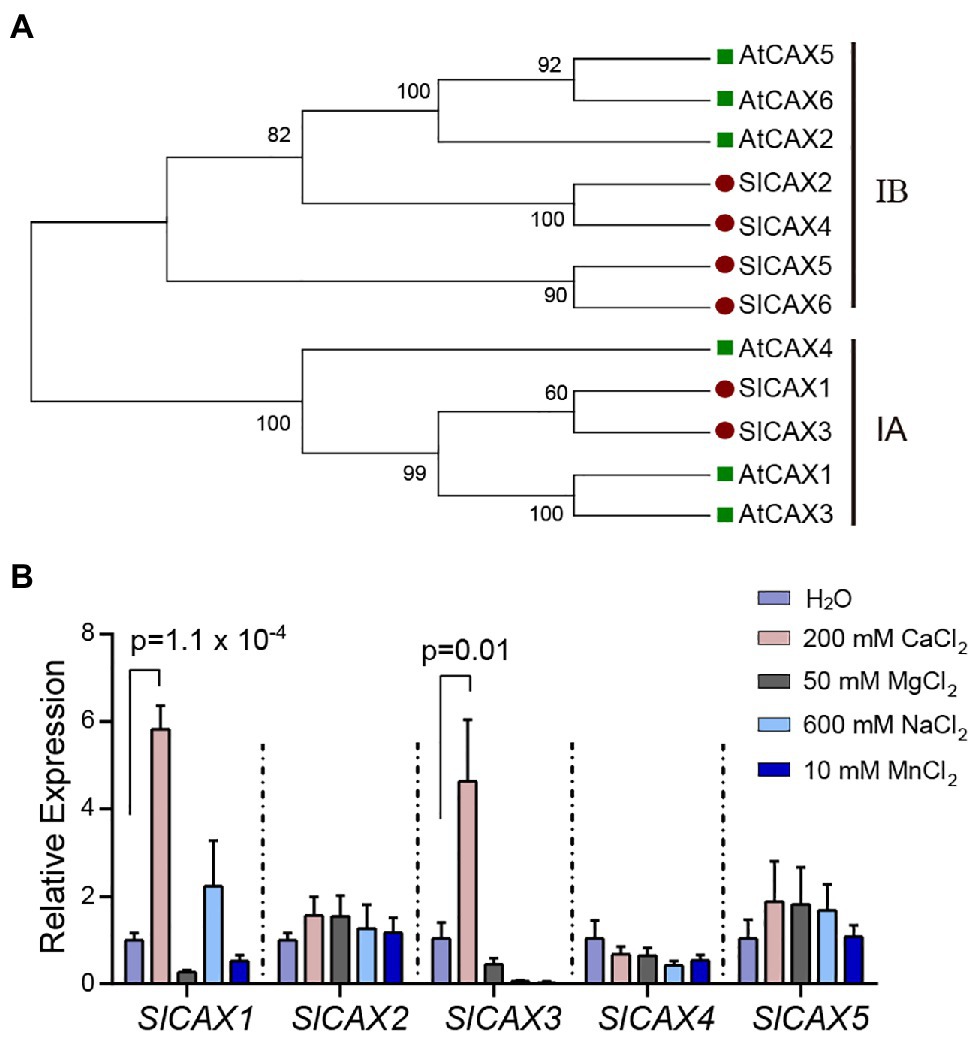
Figure 1. Phylogenetic and expression analysis of SlCAXs. (A) Neighbour-joining tree constructed with MEGA X software (Kumar et al., 2018). The percentages of supported bootstrap scores with 1,000 iterates are shown at the branch nodes. (B) Relative expression of the SlCAX family under different cation treatment conditions. The 14-day-old tomato seedlings were treated with different ion solutions for 16 h. Whole seedlings were collected for qPCR, and the expression of SlCAX genes was determined. The gene expression level under the H2O treatment was set as 1, and the data indicate the mean value ± SE from three biological replicates. Student’s t-test was used to analyse the data.
To further verify the Ca2+ transport ability of SlCAX1, we heterologously expressed the encoding gene in the yeast K667 strain (vcx1::hisG cnb1::LEU2 pmc1::TRP1 ade2-1 can1–100 his3-11,15 leu2-3,112 trp1-1 ura3-1), which has defects in vacuolar Ca2+ transport (Cunningham and Fink, 1996). Expression of full-length SlCAX1 failed to complement the hypersensitivity of the K667 strain to Ca2+, suggesting that, similar to AtCAX1, the full-length SlCAX1 is not active and an NRR may inhibit its activity (Pittman and Hirschi, 2001). Aligning the protein sequences of SlCAX1 and AtCAX1 revealed that the two proteins share 79% similarity and the N-terminal sequences of these two proteins are highly conserved. However, unlike AtCAX1, there is no second methionine in the N terminus of SlCAX1, and the analogous methionine at the 37th amino acid position of AtCAX1 corresponded to the valine residue at the 40th amino acid position in SlCAX1 (Figure 2A). We truncated the first 39 amino acid residues of SlCAX1 and changed valine to methionine to initiate translation. We named this N-terminal-truncated version SlCAX1Δ39 (Figure 2B). Complementation analysis in yeast showed that SlCAX1Δ39 was able to complement the hypersensitivity of the K667 strain to Ca2+, suggesting that SlCAX1Δ39 has Ca2+ transport ability (Figure 2C).
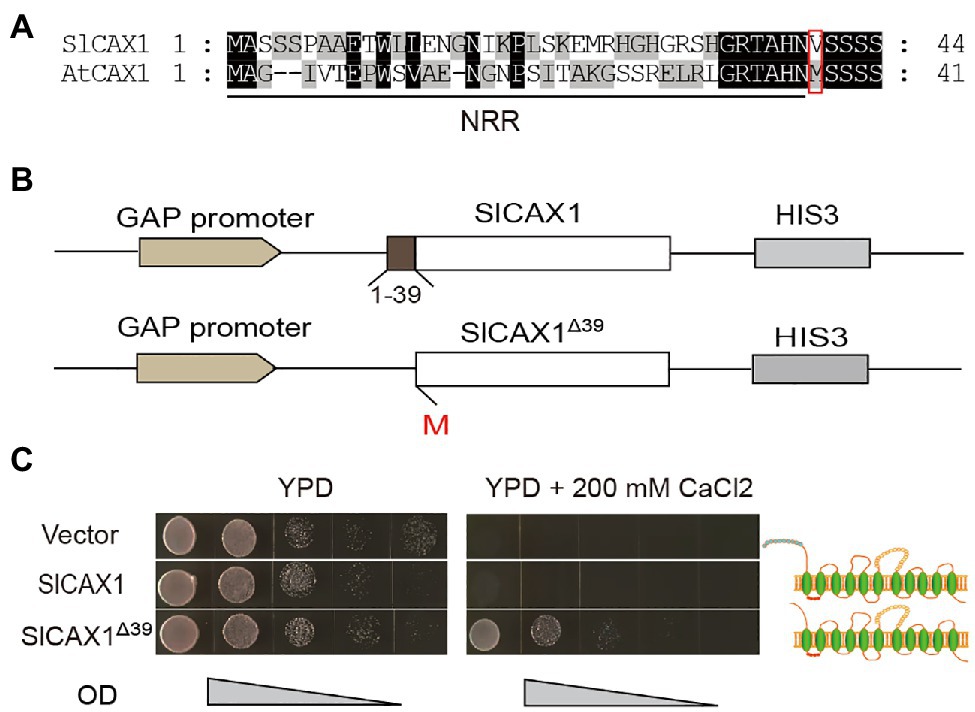
Figure 2. Truncating the first 39 amino acid residues of SlCAX1 increased the Ca2+ transport ability. (A) Partial amino acid sequence alignment of the N-terminal region of CAX1 from Arabidopsis and tomato. Consensus amino acid residues are boxed in black (identical) or grey (similar). Gaps introduced to maximize the alignments are denoted by hyphens. The red box indicates the 2nd methionine of AtCAX1 and the analogous valine of SlCAX1. (B) Schematic of SlCAX1 and SlCAX1Δ39 expression vectors in yeast. The first 39 amino acids of SlCAX1 were truncated, and the 40th amino acid, valine, was mutated to methionine, resulting in SlCAX1Δ39. HIS3 indicates that yeast containing this construct were able to grow on medium lacking histidine. (C) SlCAX1 and SlCAX1Δ39 were expressed in the yeast strain K667. The cultures were diluted 10, 100, 1,000, and 10,000 times from OD 1.0, respectively, and an equal amount was pipetted onto yeast YPD medium with or without 200 mM CaCl2. The yeast cells were cultured for 3 days at 30°C.
To confirm the Ca2+ transport activity of SlCAX1, we used CRISPR-Cas9 to knock out this gene. We obtained different deletions, including 37-bp and 4-bp deletions with premature stop codons predicted to result in null alleles (Figures 3A,B). The slcax1 mutants were smaller than the wild-type plants but were able to complete the lifecycle (Figures 3C,D), and the plant height and number of flowers of slcax1 mutants were significantly lower than those of wild-type (Figure 3E). Quantification of SlCAX3 expression revealed that SlCAX3 was upregulated in the slcax1 mutants, supporting the idea of genetic redundancy between SlCAX1 and SlCAX3 (Figure 3F). Measuring the Ca2+ concentration of the wild type and mutants showed that mutants had significantly lower Ca2+ content (Figure 3G), confirming the Ca2+-transporting ability of SlCAX1. Together, the results demonstrate that SlCAX1 has a Ca2+ transport ability in both yeast and tomato.
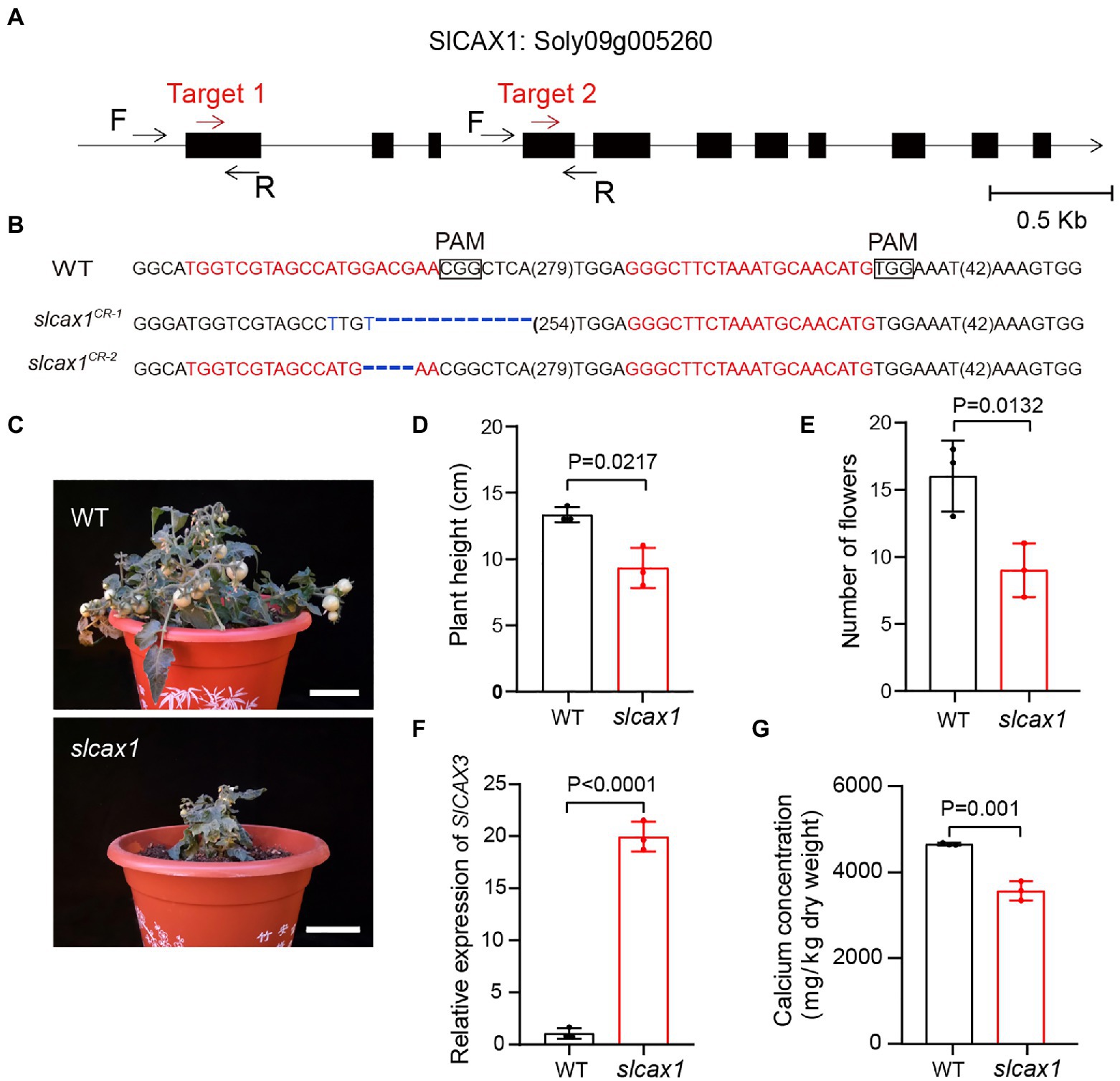
Figure 3. CRISPR/Cas9-engineered mutations of SlCAX1 resulted in low Ca2+ concentrations in tomatoes. (A) The first exon of SlCAX1 was targeted by CRISPR-Cas9 using two single guide RNAs (sgRNA; target 1 and target 2; red arrows). Black arrows indicate forward (F) and reverse (R) primers used for PCR genotyping and sequencing. (B) Sequences of slcax1 alleles were identified from two independent T1 transgenic plants. sgRNA targets and protospacer adjacent motif sequences are in red and bold font, respectively; deletions are indicated by blue dashes; and base mutations are shown in blue. The sequence gap length is shown in parentheses. (C) Phenotypes of the wild type and slcax1 mutants. (D) Plant height of wild type and slcax1 mutants grown for 2 months. (E) The number of flowers of wild type and slcax1 mutants. The pictures, plant height, and flower number data were collected from 2-month-old plants. (F) Analysis of SlCAX3 expression by RT-qPCR. Wild-type and slcax1 mutant leaves were harvested 2 weeks after germination for RNA extraction from three biological replicates. (G) Shoot calcium content in the wild type and slcax1 mutants. The wild type and mutants were grown on MS medium for 1 month, and then the shoots were harvested for calcium content measurement (n = 3). The Student’s t-test was used to analyze the data for (D-G). Error bars represent standard error of the mean.
The NRRs of Arabidopsis AtCAXs were initially defined by the sequence between the first and second methionine of AtCAX1 and AtCAX3; however, we found that this methionine residue is not conserved in tomato and other species (Figure 2A). Indeed, the Bacillus subtilis Ca2+/H+ antiporter protein YfkE, which shares high sequence similarity with Arabidopsis AtCAX1, does not even have any peptides in upstream of the first TM domain (Wu et al., 2013; Supplementary Figure 4). Therefore, we hypothesized that all the amino acid residues in upstream of the first TM domain of AtCAX1 negatively regulate its activity. To test this hypothesis, instead of truncating the first 39 amino acids, we truncated all 66 upstream amino acids of the first TM domain and named it SlCAX1Δ66 (Figure 4A). As expected, SlCAX1Δ66 showed higher activity than SlCAX1Δ39 in a yeast assay (Figure 4B), suggesting that all the amino acids in upstream of the first TM domain of SlCAX1 negatively regulate its Ca2+ transporting ability. To further validate our hypothesis, we made two extra truncations, SlCAX1Δ58 and SlCAX1Δ93 with the first TM domain deleted (Supplementary Figure 4). The results showed that SlCAX1Δ66 still displayed the highest activity in the yeast assay and contrasting to AtCAX1Δ90, SlCAX1Δ93 was not active at all, suggesting that the first TM of SlCAX1 is critical for its activity (Supplementary Figure 5). Together, our results indicate that all the amino acids in upstream of the first TM domain of SlCAX1 negatively regulate its Ca2+ transporting ability.
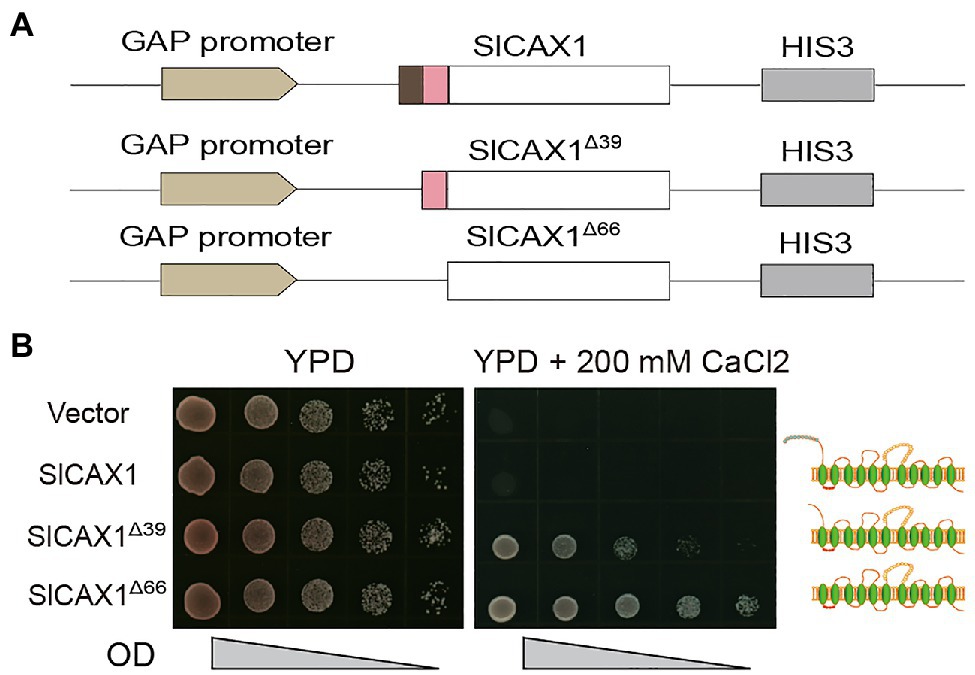
Figure 4. Amino acid residues before the first TM domain of SlCAX1 negatively regulate the Ca2+ transporting ability of the antiporter. (A) Schematics of the vectors used to express different truncated versions of SlCAX1. (B) SlCAX1, SlCAX1Δ39 and SlCAX1Δ66 were expressed in yeast strain K667. The cultures were diluted 10, 100, 1,000, and 10,000 times from OD 1.0, respectively, and an equal amount of sample was pipetted onto YPD medium with or without 200 mM CaCl2. The yeast cells were cultured at 30°C for 3 days.
To determine if this is also applicable to other CAXs, such as Arabidopsis AtCAX1 and AtCAX3, we tested the Ca2+ transport activity of variants with different truncations in the K667 strain. Alignment of the protein sequences of SlCAX1, SlCAX3, AtCAX1, and AtCAX3 revealed that the sequences in upstream of the first TM domain were conserved between AtCAX1, AtCAX3 and SlCAX1, but not SlCAX3 (Figure 5A). Therefore, we truncated the first 63 amino acid residues before the first TM domains of AtCAX1 and AtCAX3 to create AtCAX1Δ63 and AtCAX3Δ63, respectively, and evaluated whether this truncation increased the activity of these proteins. Similar to SlCAX1Δ66, both AtCAX1Δ63 and AtCAX3Δ63 showed higher activity than AtCAX1Δ36 and AtCAX3Δ36, respectively (Figures 5B,C). The difference was particularly significant for AtCAX3 because, as indicated above, truncating the initial NRR domain of AtCAX3 does not activate this antiporter (Shigaki and Hirschi, 2000). Together, our results suggest that CAXs can be completely activated after truncating all of the amino acid residues before the first TM domain, thus our results redefine the NRR of CAXs.
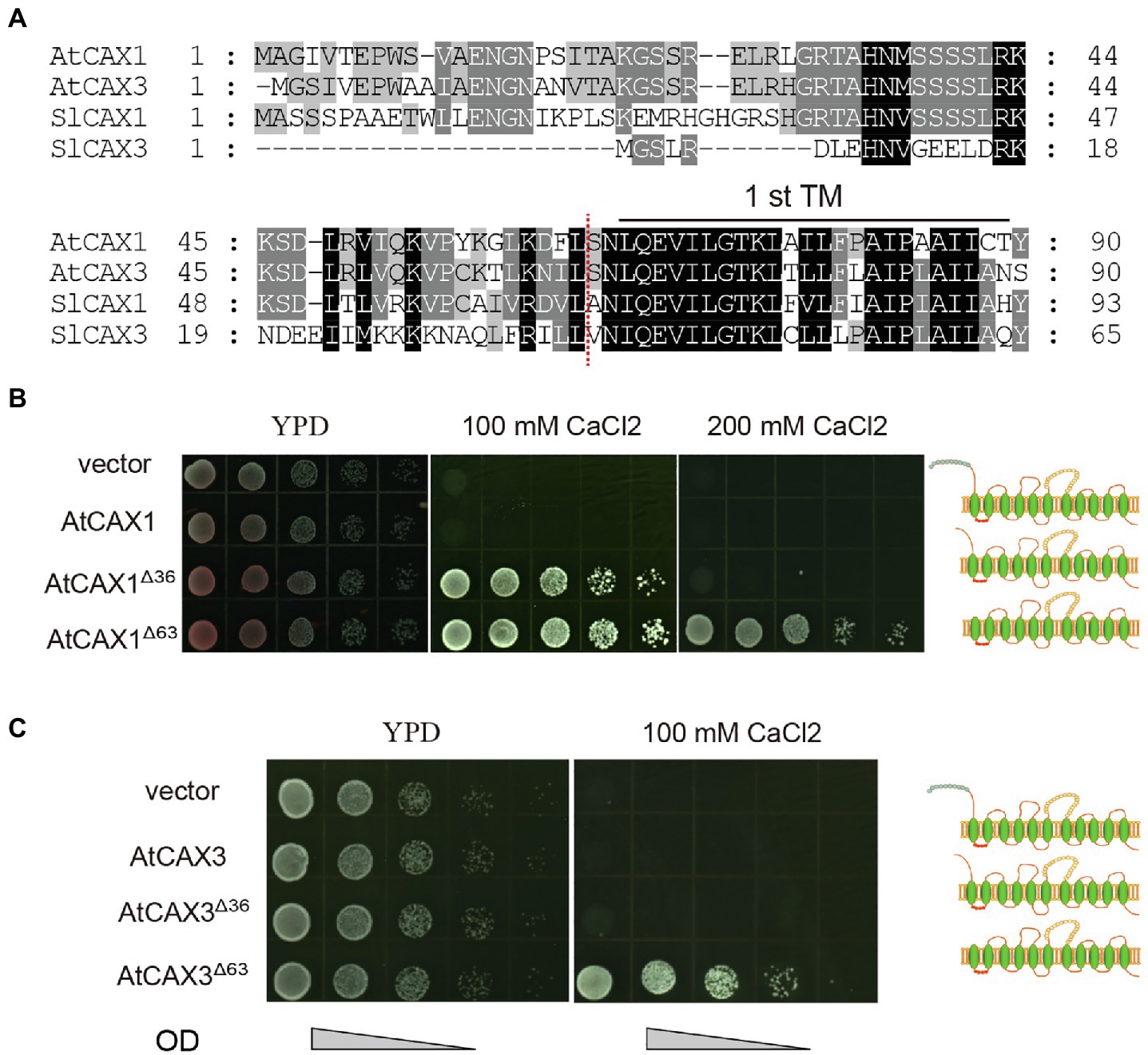
Figure 5. Truncating the amino acids before the first TM further increases the activity of CAX1 and CAX3. (A) Alignment of deduced amino acid sequences of polypeptides encoded by Arabidopsis CAX1 and CAX3 and tomato SlCAX1 and SlCAX3 (partial). Consensus amino acid residues are boxed in black (identical) or grey (similar). Gaps introduced to maximize the alignments are denoted by hyphens. The amino acids residues before the red dotted lines are deleted to form AtCAX1Δ63 and AtCAX3Δ63. (B,C) Truncated AtCAX1 and AtCAX3 were expressed in yeast mutant strain K667, respectively. The cultures were diluted 10, 100, 1,000, and 10,000 times from OD 1.0, respectively, and an equal amount of sample was pipetted onto yeast YPD medium with or without CaCl2. The yeast cells were cultured at 30°C for 3 days.
To confirm that SlCAX1Δ66 has enhanced Ca2+ transport ability in planta, we expressed SlCAX1 and SlCAX1Δ66 driven by the CaMV35S promoter in tomato plants and quantified the Ca2+ content (Figure 6A). We confirmed the expression using PCR (Supplementary Figure 6). Overexpression of full-length SlCAX1 did not enhance plant Ca2+ content, consistent with its non-active nature. By contrast, overexpression of SlCAX1Δ66 significantly increased plant Ca2+ content, indicating that truncating the amino acid residues upstream of the first TM domain can activate SlCAX1 in vivo (Figure 6B). Together, the results from the yeast assays and the in planta data demonstrate that the region constituting the NRR of CAXs should be re-defined.
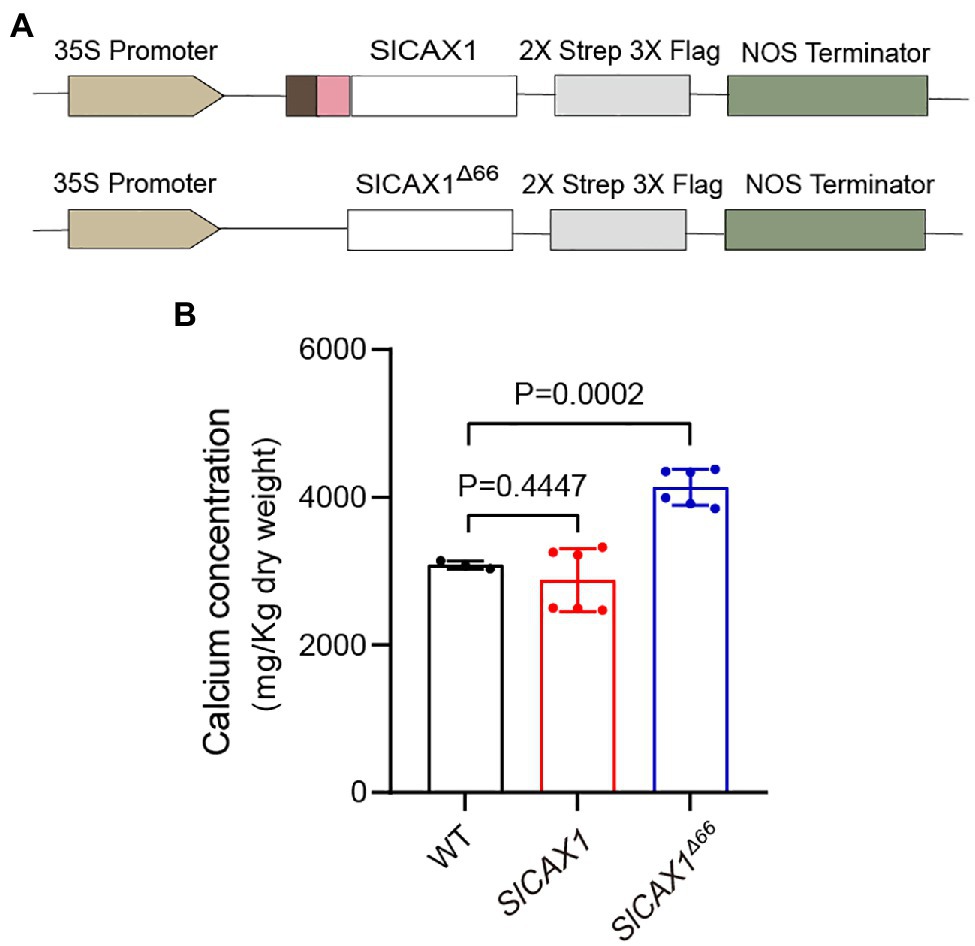
Figure 6. Overexpressing SlCAX1Δ66 in tomato increased the calcium content. (A) Schematics of vectors used to express different truncated versions of SlCAX1 in tomato. (B) Calcium content of different lines. A Student’s t-test was used to analyze the data. Error bars represent standard error of the mean of six biological replicates.
Discussion
In this study, we confirmed the Ca2+ transporting ability of SlCAX1 using knockout mutants generated by CRISPR-Cas9 and redefined the region constituting the NRR of CAX proteins. Initially, the NRR was defined as the region between the first and second methionine of the antiporter, because both Arabidopsis AtCAX1 and AtCAX3 have a second methionine at the 36th amino acid position, and truncating this region increased the activity of AtCAX1 (Pittman and Hirschi, 2001). However, amino acid sequence alignment showed that the SlCAX from tomato do not have a second in-frame start codon, and some CAXs do not even have any amino acid residues in front of the first TM domain, promoting us to re-evaluate the NRR region. By testing different variants of CAXs, we established that the entire region upstream of the first TM domain inhibits CAX activity. The work by Manohar et al. also revealed that AtCAX3 with N-terminal truncation of 57 amino acids and more has the higher activity than the full length and 36-amino-acid truncation of AtCAX3 (AtCAX3Δ36; Manohar et al., 2011). However, how this region inhibits CAX activity remains elusive. Previous studies showed that AtCAX1 and AtCAX3 interact with each other (Cheng et al., 2005; Hocking et al., 2017), and structural analyses of the Ca2+ antiporter YfkE and ScVCX1 suggest that CAXs form oligomer (Waight et al., 2013; Wu et al., 2013). Therefore, it is possible that plant CAXs also form oligomeric complex. Interestingly, NRR has a high probability to consist of an intrinsically disordered region (IDR), as predicted by IUPRED3,2 and scrambling of the first 39 or 66 amino acids of SlCAX1 did not fully unleash its activity (Supplementary Figure 7), suggesting that the sequence of NRR does not need stringently specific to be functional. We speculate that the NRR of CAX consisting of IDR may fold back and loosely binds to its own or other CAXs’ Ca2+ binding sites in the complex to inhibit the Ca2+ transport ability. The NRR of AtCAX1 can also be phosphorylated by CBL-INTERACTING PROTEIN KINASE24 (CIPK24), and AtCAX1 is activated upon the phosphorylation (Cheng et al., 2004), indicating that activation of the CAX complex relies on other proteins. The low complexity region in the NRR consists of four serine amino acids. It is possible that phosphorylation of NRR at these amino acids could affect the interaction of NRR with the Ca2+ binding sites of CAX proteins and also affects the CAX activity by increasing the accessibility to Ca2+. However, a detailed mechanism will not be elucidated until the structure of the CAX complex is determined. Given recent advances in the cryo-EM technique, solving the structure of this membrane complex is feasible. A complete understanding of the regulatory mechanism of plant CAX proteins will benefit efforts to improve Ca2+ use efficiency by crop plants.
slcax1 mutants can still complete the lifecycle, and this may be because SlCAX1 and SlCAX3 are genetically redundant. Similarly, neither the Arabidopsis atcax1 nor atcax3 single mutants showed strong growth defects, but the growth of atcax1 atcax3 double mutants was severely hindered (Hocking et al., 2017). Our RT-qPCR results revealed that upregulation of SlCAX3 in slcax1 mutants allowed for functional compensation, indicating the genetic robustness of the Ca2+ homeostasis pathway. Genetic robustness is also common for other biological processes, such as regulation of shoot meristem development. For instance, disruption of tomato SlCLAVATA3 resulted in upregulation of its paralogous gene CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (SlCLE9), and double mutants show synergistic phenotypes (Rodriguez-Leal et al., 2019). Further characterization of slcax1 slcax3 double mutants would help dissect the functions of SlCAX1 and SlCAX3.
Here, we used tomato, an economically important crop, as a model and determined the regulatory mechanism of CAX proteins. This study will facilitate the manipulation of Ca2+ antiporters in efforts to fine-tune Ca2+ homeostasis and thereby reduce the incidence of Ca2+-related physiological disorders and improve the fruit quality of crops.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
BH, YT, SL, JS, XW, and TK performed the experiments. BH, YT, and QW designed research. JW, XC, SP, and QW wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFF1000400), National Natural Science Foundation of China (32171925 and U21A20210), and Innovation Program of Chinese Academy of Agricultural Sciences (GJ2022-14-5 and CAAS-ZDRW202004). The open access publication fees will be from Innovation Program of Chinese Academy of Agricultural Sciences (GJ2022-14-5).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Kendal Hirschi for providing the yeast strain.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.938839/full#supplementary-material
Footnotes
References
Amagaya, K., Shibuya, T., Nishiyama, M., Kato, K., and Kanayama, Y. (2020). Characterization and expression analysis of the Ca2+/Cation Antiporter gene family in tomatoes. Plants 9:25. doi: 10.3390/plants9010025
Cheng, N. H., Pittman, J. K., Shigaki, T., Lachmansingh, J., Leclere, S., Lahner, B., et al. (2005). Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 138, 2048–2060. doi: 10.1104/pp.105.061218
Cheng, N. H., Pittman, J. K., Zhu, J. K., and Hirschi, K. D. (2004). The protein kinase SOS2 activates the Arabidopsis H(+)/Ca(2+) antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279, 2922–2926. doi: 10.1074/jbc.M309084200
Cosson, V., Durand, P., D’Erfurth, I., Kondorosi, A., and Ratet, P. (2006). Medicago truncatula transformation using leaf explants. Mol. Biol. 343, 115–127. doi: 10.1385/1-59745-130-4:115
Cunningham, K. W., and Fink, G. R. (1996). Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2226–2237. doi: 10.1128/MCB.16.5.2226
De Freitas, S. T., Handa, A. K., Wu, Q. Y., Park, S., and Mitcham, E. J. (2012). Role of pectin methylesterases in cellular calcium distribution and blossom-end rot development in tomato fruit. Plant J. 71, 824–835. doi: 10.1111/j.1365-313X.2012.05034.x
De Freitas, S. T., Padda, M., Wu, Q. Y., Park, S., and Mitcham, E. J. (2011). Dynamic alternations in cellular and molecular components during blossom-end rot development in tomatoes expressing sCAX1, a constitutively active Ca2+/H+ Antiporter from Arabidopsis. Plant Physiol. 156, 844–855. doi: 10.1104/pp.111.175208
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Emery, L., Whelan, S., Hirschi, K. D., and Pittman, J. K. (2012). Protein phylogenetic analysis of Ca(2+)/cation Antiporters and insights into their evolution in plants. Front. Plant Sci. 3:1. doi: 10.3389/fpls.2012.00001
Han, J. S., Park, S., Shigaki, T., Hirschi, K. D., and Kim, C. K. (2009). Improved watermelon quality using bottle gourd rootstock expressing a Ca2+/H+ antiporter. Mol. Breed. 24, 201–211. doi: 10.1007/s11032-009-9284-9
Hirschi, K. D., Korenkov, V. D., Wilganowski, N. L., and Wagner, G. J. (2000). Expression of arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 124, 125–134. doi: 10.1104/pp.124.1.125
Hirschi, K. D., Zhen, R. G., Cunningham, K. W., Rea, P. A., and Fink, G. R. (1996). CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 93, 8782–8786. doi: 10.1073/pnas.93.16.8782
Hocking, B., Conn, S. J., Manohar, M., Xu, B., Athman, A., Stancombe, M. A., et al. (2017). Heterodimerization of Arabidopsis calcium/proton exchangers contributes to regulation of guard cell dynamics and plant defense responses. J. Exp. Bot. 68, 4171–4183. doi: 10.1093/jxb/erx209
Kamiya, T., Akahori, T., Ashikari, M., and Maeshima, M. (2006). Expression of the vacuolar Ca2+/H+ exchanger, OsCAX1a, in rice: cell and age specificity of expression, and enhancement by Ca2+. Plant Cell Physiol. 47, 96–106. doi: 10.1093/pcp/pci227
Kim, C. K., Han, J. S., Lee, H. S., Oh, J. Y., Shigaki, T., Park, S. H., et al. (2006). Expression of an Arabidopsis CAX2 variant in potato tubers increases calcium levels with no accumulation of manganese. Plant Cell Rep. 25, 1226–1232. doi: 10.1007/s00299-006-0214-6
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Benzhong, Z., et al. (2018). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Frontiers in Plant Sci. 9:559. doi: 10.3389/fpls.2018.00559
Manohar, M., Shigaki, T., Mei, H., Park, S., Marshall, J., Aguilar, J., et al. (2011). Characterization of Arabidopsis Ca2+/H+ exchanger CAX3. Biochemistry 50, 6189–6195. doi: 10.1021/bi2003839
Park, S., Cheng, N. H., Pittman, J. K., Yoo, K. S., Park, J., Smith, R. H., et al. (2005a). Increased calcium levels and prolonged shelf life in tomatoes expressing Arabidopsis H+/Ca-2 transporters. Plant Physiol. 139, 1194–1206. doi: 10.1104/pp.105.066266
Park, S., Elless, M. P., Park, J., Jenkins, A., Lim, W., Chambers, E., et al. (2009). Sensory analysis of calcium-biofortified lettuce. Plant Biotechnol. J. 7, 106–117. doi: 10.1111/j.1467-7652.2008.00379.x
Park, S., Kang, T. S., Kim, C. K., Han, J. S., Kim, S., Smith, R. H., et al. (2005b). Genetic manipulation for enhancing calcium content in potato tuber. J. Agric. Food Chem. 53, 5598–5603. doi: 10.1021/jf050531c
Park, S., Kim, C. K., Pike, L. M., Smith, R. H., and Hirschi, K. D. (2004). Increased calcium in carrots by expression of an Arabidopsis H+/Ca2+ transporter. Mol. Breed. 14, 275–282. doi: 10.1023/B:MOLB.0000047773.20175.ae
Pittman, J. K., and Hirschi, K. D. (2001). Regulation of CAX1, an arabidopsis Ca2+/H+ antiporter. Identification of an N-terminal autoinhibitory domain. Plant Physiol. 127, 1020–1029. doi: 10.1104/pp.010409
Pittman, J. K., and Hirschi, K. D. (2016). Phylogenetic analysis and protein structure modelling identifies distinct Ca(2+)/Cation antiporters and conservation of gene family structure within Arabidopsis and rice species. Rice (NY) 9:3. doi: 10.1186/s12284-016-0075-8
Pittman, J. K., Shigaki, T., Cheng, N. H., and Hirschi, K. D. (2002). Mechanism of N-terminal autoinhibition in the arabidopsis Ca2+/H+ antiporter CAX1. J. Biol. Chem. 277, 26452–26459. doi: 10.1074/jbc.M202563200
Rodriguez-Leal, D., Xu, C., Kwon, C. T., Soyars, C., Demesa-Arevalo, E., Man, J., et al. (2019). Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 51, 786–792. doi: 10.1038/s41588-019-0389-8
Satoh, J., Kato, K., and Shinmyo, A. (2004). The 5′-untranslated region of the tobacco alcohol dehydrogenase gene functions as an effective translational enhancer in plant. J. Biosci. Bioeng. 98, 1–8.
Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Shigaki, T., and Hirschi, K. (2000). Characterization of CAX-like genes in plants: implications for functional diversity. Gene 257, 291–298. doi: 10.1016/S0378-1119(00)00390-5
Shigaki, T., Rees, I., Nakhleh, L., and Hirschi, K. D. (2006). Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. J. Mol. Evol. 63, 815–825. doi: 10.1007/s00239-006-0048-4
Sugio, T., Satoh, J., Matsuura, H., Shinmyo, A., and Kato, K. (2008). The 5′-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J. Biosci. Bioengineering. 105, 300–302.
Tang, R. J., Zhao, F. G., Garcia, V. J., Kleist, T. J., Yang, L., Zhang, H. X., et al. (2015). Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 112, 3134–3139. doi: 10.1073/pnas.1420944112
Waight, A. B., Pedersen, B. P., Schlessinger, A., Bonomi, M., Chau, B. H., Roe-Zurz, Z., et al. (2013). Structural basis for alternating access of a eukaryotic calcium/proton exchanger. Nature 499, 107–110. doi: 10.1038/nature12233
Wu, Q. Y., Shigaki, T., Han, J. S., Kim, C. K., Hirschi, K. D., and Park, S. (2012). Ectopic expression of a maize calreticulin mitigates calcium deficiency-like disorders in sCAX1-expressing tobacco and tomato. Plant Mol. Biol. 80, 609–619. doi: 10.1007/s11103-012-9970-6
Wu, Q., Shigaki, T., Williams, K. A., Han, J.-S., Kim, C. K., Hirschi, K. D., et al. (2011). Expression of an Arabidopsis Ca2+/H+ antiporter CAX1 variant in petunia enhances cadmium tolerance and accumulation. J. Plant Physiol. 168, 167–173. doi: 10.1016/j.jplph.2010.06.005
Keywords: calcium, CAX, NRR, tomato, CRISPR
Citation: Han B, Tai Y, Li S, Shi J, Wu X, Kakeshpour T, Weng J, Cheng X, Park S and Wu Q (2022) Redefining the N-Terminal Regulatory Region of the Ca2+/H+ Antiporter CAX1 in Tomato. Front. Plant Sci. 13:938839. doi: 10.3389/fpls.2022.938839
Edited by:
Guillaume Pilot, Virginia Tech, United StatesReviewed by:
Jon Pittman, The University of Manchester, United KingdomDebajyoti Dutta, Thapar Institute of Engineering and Technology, India
Copyright © 2022 Han, Tai, Li, Shi, Wu, Kakeshpour, Weng, Cheng, Park and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingyu Wu, d3VxaW5neXVAY2Fhcy5jbg==
†These authors have contributed equally to this work
 Beibei Han
Beibei Han Yuxin Tai2†
Yuxin Tai2† Jianfeng Weng
Jianfeng Weng Xianguo Cheng
Xianguo Cheng Sunghun Park
Sunghun Park Qingyu Wu
Qingyu Wu