- 1Biotechnology Division, CSIR-Institute of Himalayan Bioresource Technology, Palampur, India
- 2Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, Uttar Pradesh, India
- 3Agrotechnology Division, CSIR-Institute of Himalayan Bioresource Technology, Palampur, India
- 4ICAR-Central Potato Research Institute, Modipuram, Uttar Pradesh, India
- 5Division of Crop Improvement, ICAR-Central Potato Research Institute, Shimla, Himachal Pradesh, India
Potato is a temperate crop consumed globally as a staple food. High temperature negatively impacts the tuberization process, eventually affecting crop yield. DNA methylation plays an important role in various developmental and physiological processes in plants. It is a conserved epigenetic mark determined by the dynamic concurrent action of cytosine-5 DNA methyltransferases (C5-MTases) and demethylases (DeMets). However, C5-MTases and DeMets remain unidentified in potato, and their expression patterns are unknown under high temperatures. Here, we performed genome-wide analysis and identified 10 C5-MTases and 8 DeMets in potatoes. Analysis of their conserved motifs, gene structures, and phylogenetic analysis grouped C5-MTases into four subfamilies (StMET, StCMT3, StDRM, and StDNMT2) and DeMets into three subfamilies (StROS, StDML, and StDME). Promoter analysis showed the presence of multiple cis-regulatory elements involved in plant development, hormone, and stress response. Furthermore, expression dynamics of C5-MTases and DeMets were determined in the different tissues (leaf, flower, and stolon) of heat-sensitive (HS) and heat-tolerant (HT) genotypes under high temperature. qPCR results revealed that high temperature resulted in pronounced upregulation of CMT and DRM genes in the HT genotype. Likewise, demethylases showed strong upregulation in HT genotype as compared to HS genotype. Several positive (StSP6A and StBEL5) and negative (StSP5G, StSUT4, and StRAP1) regulators are involved in the potato tuberization. Expression analysis of these genes revealed that high temperature induces the expression of positive regulators in the leaf and stolon samples of HT genotype, possibly through active DNA demethylation and RNA-directed DNA methylation (RdDM) pathway components. Our findings lay a framework for understanding how epigenetic pathways synergistically or antagonistically regulate the tuberization process under high-temperature stress in potatoes. Uncovering such mechanisms will contribute to potato breeding for developing thermotolerant potato varieties.
Introduction
Global climate change is a serious threat to sustainable crop production. The global mean surface air temperature is expected to rise from 0.5°C to 4.5°C by the end of the twenty-first century (Change, 2013), which is predicted to result in a 17–24% loss in crop productivity (Zhou et al., 2019). The temperate crops are more vulnerable to global temperature rise, negatively affecting their development and final harvest (Lesk et al., 2016).
Potato is a temperate staple food crop with global production of 370.43 million tons (FAOSTAT, 2021). It is one of the richest sources of carbohydrates, fibers, vitamins, and antioxidants (Beals, 2019), feeding the global nutritional requirements (Birch et al., 2012). Potato is vegetatively propagated through tubers, which develop from underground stems known as stolons (Zierer et al., 2021). The stages of tuberization involve different phases, viz. stolon induction, elongation/branching, termination of stolon longitudinal growth, tuber induction by sub-apical stolon swelling, tuber growth, and bulking (Vreugdenhil and Struik, 1989).
Tuberization is a complex biological phenomenon affected by several environmental cues (Dutt et al., 2017). Short days, cool temperature, and low nitrogen supply favors tuberization, whereas long days, high temperature, and high nitrogen supply oppose the tuberization process (Gao et al., 2014). The optimal temperature requirement for tuberization is 15–20°C (Rykaczewska, 2015). High temperatures and long days impede the tuberization process (Dutt et al., 2017). The process of tuberization under high temperature involves altered assimilate partitioning and change in the source to sink balance (Hastilestari et al., 2018). Even a slight increase in temperature (e.g., 30°C Day/20°C night) can impact the process of tuberization, affecting crop yield severely (Hancock et al., 2014; Kim et al., 2017). Additionally, phytohormones are involved in the tuberization process. The high GA levels impair tuber development, whereas ABA, cytokinin, and sucrose promote tuberization (Chatterjee et al., 2007). The tuberization signaling network involves several key players, including promoters (StSP6A, StBEL5, StPOTH1, and StCDF) and inhibitors (StSUT4, StSP5G, and StRAP1) of tuberization (Dutt et al., 2017).
DNA methylation is a conserved epigenetic mark that regulates gene expression, transposable elements, and stress-mediated responses, eventually contributing to genomic stability (Zhang et al., 2018). DNA methylation in eukaryotes is determined by the concurrent activity of methyltransferases and demethylases (Zhang et al., 2018). In plants, DNA methylation occurs at cytosine residues in three sequence contexts: CpG, CpHpG, and CpHpH (H depicts A, C, T; Law and Jacobsen, 2010). DNA methylation is broadly categorized into de novo and maintenance methylation (Zhang et al., 2018). The basic function of any C5-MTase is to recognize specific nucleotide sequences and mark the transfer of methyl group from co-factor S-adenosyl-L methionine (SAM) to 5'-cytosine residue on the pyrimidine ring (Cheng et al., 1993). This function is attributed to the N-terminal regulatory and C-terminal catalytic domain (Pavlopoulou and Kossida, 2007). Plant cytosine DNA methylases (C5-MTase) are much more diverse and less explored than mammalian C5-MTase. Till now, four major types of C5-MTases have been identified including METHYLTRANSFERASE (MET), CHROMO METHYLTRANSFERASE (CMT), DOMAINS REARRANGED METHYLASE (DRM), and DNA METHYLASE HOMOLOG 2 (DNMT2; Cao et al., 2003). DNA demethylation occurs either passively by loss of maintenance methylation enzymes during consecutive DNA replication or actively by the action of a special set of DNA glycosylases (HhH-GPD); DNA glycosylases/lyases that excise 5-meC and operate through the base excision pathway (Roldán-Arjona et al., 2019). Also, two additional conserved domains named RNA recognition motif (RRM) and methylated CpG discriminating CXXC domain are found (Iyer et al., 2011). Thus, active DNA demethylation requires a set of DNA demethylases to initiate the process (Gallego-Bartolomé et al., 2018; Liu et al., 2021).
The concurrent action of methyltransferase and demethylases regulate several developmental features such as plant size, leaf size and shape, flowering time, and fruit ripening (Finnegan et al., 1996). Also, methyltransferases and demethylases are involved in regulating stress-responsive genes in plants (Zhu et al., 2020). For instance, the role of DNA methylation has been suggested in drought and salinity stress responses while studying the contrasting rice genotypes (Garg et al., 2015). Likewise, the role of DNA methylation in conferring plant immunity in A. thaliana has also been demonstrated (Dowen et al., 2012).
In the present study, DNA methyltransferase and demethylases were characterized in potato (Solanum tuberosum) for the first time. Also, their expression patterns were determined in several tissues such as the leaf, flower, and stolon in the heat-sensitive (HS) and heat-tolerant (HT) potato genotypes under high temperature. Furthermore, the expression of tuberization genes and heat shock factors (HSFs) were studied in these genotypes to delineate how epigenetic components are linked with tuberization under high temperature.
Materials and methods
Collection of candidate genes
Protein sequences were downloaded from the potato genome database (Spud DB; Hirsch et al., 2014) by using Pfam ID (PF00145, PF15628, PF00730, and PF15629) to identify the C5-MTases and demethylases. The resultant protein sequences were also verified by downloading the hidden Markov model (HMM) file from the HMMER search and identified through the ENSEMBL database (Potter et al., 2018). The identified potato C5-MTase and demethylase protein sequences were then classified based on the presence of conserved domain using available online software SMART (Schultz et al., 2000) and NCBI_CDD (Marchler-Bauer et al., 2015). Domain Graph 2.0 (DOG 2.0) was used to construct the figures of conserved domains of each class of proteins (Ren et al., 2009). The ExPASy server (http://www.expasy.org/) (Artimo et al., 2012) was used to predict the relative molecular weight (MW) and isoelectric point (PI) of potato C5-MTase and demethylase.
Analysis of conserved motif and gene structure
The conserved motifs of C5-MTase and demethylases were analyzed using MEME suite vs 5.05 (Bailey et al., 2009), and a total of 15 conserved motifs were identified which was further constructed using TBtool (Chen et al., 2018). Gene Structure Display Server (GSDS 2.0) was used to draw the gene structure of potato C5-MTase and demethylase (Hu et al., 2015).
Identification of promoter cis-regulatory elements
The sequence retrieval tab of the potato genome database (Spud DB) was used to extract 2 kb sequences that lie upstream of the Transcription Start Site (TSS) of genomic sequences of C5-MTases and demethylases. The online software PlantCare (Lescot et al., 2002) was used to list out the conserved elements of promoter sequences.
Analysis of multiple sequence alignment and phylogenetic tree
Full-length amino acid sequences of C5-MTase (Arabidopsis thaliana, Solanum lycopersicum, Solanum tuberosum, Glycine max, Zea mays, and Oryza sativa) and DNA demethylase (Arabidopsis thaliana, Solanum lycopersicum, Solanum tuberosum, Cucumis sativa, and Glycine max) were subjected to multiple sequence alignments by using muscle program of MEGA X (10.05) software (Kumar et al., 2018). The aligned protein sequences were used to construct a phylogenetic tree, using the Neighbor-Joining (NJ) program for 1,000 bootstrap replicates with Poisson correction (Yang, 2007).
Prediction of subcellular localization
The subcellular localization of candidate genes was determined by using the protein sequences in the web tool CELLO v.2.5 (Yu et al., 2014). The localization of genes was further used to map them.
Experimental set-up
Heat-sensitive (Kufri Chandramukhi, CP2141/A-2708) and -tolerant (Kufri Kiran, CP4803/A-2708) potato tubers were obtained from the Central Potato Research Institute (CPRI), Shimla, India. Well-sprouted tubers were grown in 0.2 L and 12 cm pots containing soil mixture (soil: vermicompost:sand, 2:1:1). Six replicates per genotype were raised in a Percival growth chamber under 16 h light and 8 h darkness conditions with day and night temperatures of 22 and 18°C, respectively. The light intensity of the growth chamber was 300 μmol/s/m2. The plants were watered and rotated daily to avoid water deficit and minimize positional effects. After 50 days, half of the plants were subjected to 14 days high-temperature stress (32°C day and 27°C night). The leaves, flowers, and stolon tissues were harvested from both the control and heat treatments around 10 AM (4 h of light onset). The harvested tissues, viz. leaf, flower, and stolon of similar size/age, were collected in aluminum zippers, frozen immediately in liquid nitrogen, and stored at −80°C.
RNA isolation, cDNA synthesis, and quantitative real-time PCR (qPCR)
About 100 mg of the frozen tissue was finely ground with a mortar and pestle (Yu et al., 2007). RNA was isolated with Spectrum Plant Total RNA Kit (Sigma Aldrich, USA) by following the manufacturer's protocol. The isolated RNA was treated with 1 U/μl of DNase I (ThermoFisher Scientific Baltics, UAB, Lithuania). Further, 1 μg of RNA was used to synthesize cDNA using the Verso cDNA synthesis kit (ThermoFisher Scientific Baltics, UAB, Lithuania). qPCR primers were designed from the CDS sequence using the ApE software (Davis and Jorgensen, 2022) (Supplementary Table S1). The quantitative real-time PCR was carried out using the Dynamo Colorflash SYBR Green (ThermoFisher Scientific Baltics, UAB, Lithuania) in an Applied Biosystem step-plus real-time PCR system. Three biological and three technical replicates were used to perform qPCR analysis. The expression of the housekeeping gene tubulin was found to be stable across tissues, treatments, and genotypes. Hence, tubulin was used as an internal control to measure target gene expression using the 2–ΔΔCt method (Schmittgen and Livak, 2008).
Statistical analysis
The gene expression data obtained by qPCR were expressed as mean ± standard error (SE) for three biological and technical replicates. Statistical analysis was carried out by using the SPSS 16.0 software (SPSS Science, UK). The significant difference between control and heat-treated plants was analyzed based on two-tailed Student's t-test. The significant difference between control and heat-treated samples at p < 0.05, p < 0.01, and p < 0.001 are denoted by one, two, and three asterisks.
Results
Gene identification, structure analysis, and chromosome localization of C5-MTase and demethylase genes
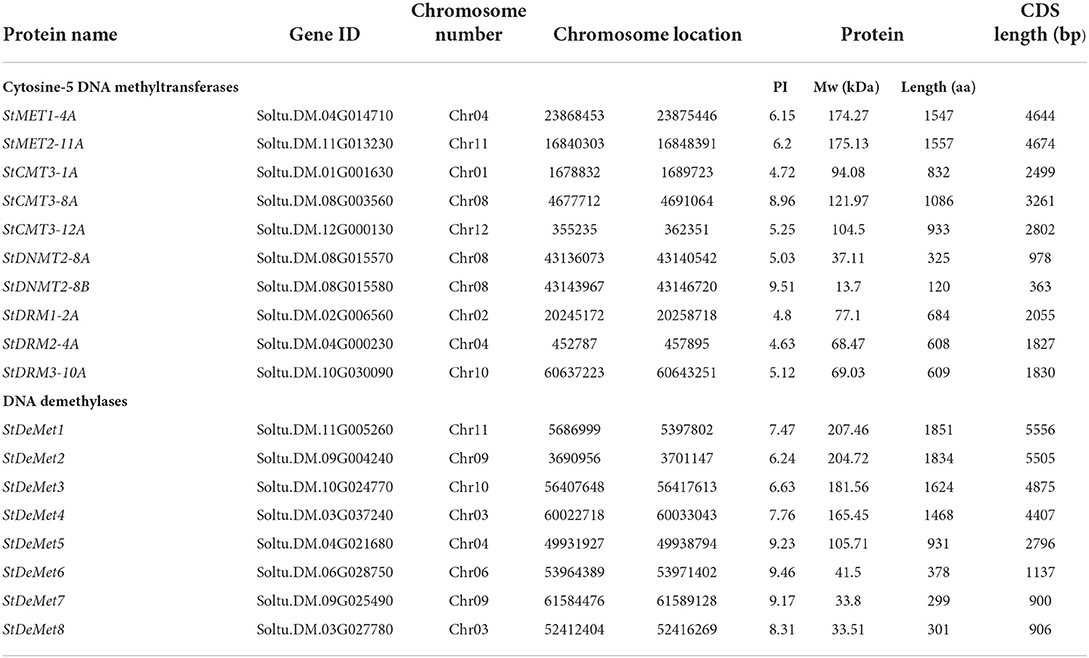
In our present study, we identified 10 C5-MTase (StMET1-4A, StMET2-11A, StCMT3-1A, StCMT3-8A, StCMT3-12A, StDNMT2-8A, StDNMT2-8B, StDRM1-2A, StDRM2-4A, and StDRM3-10A) and 8 DNA demethylase genes (StDeMet1, StDeMet2, StDeMet3, StDeMet4, StDeMet5, StDeMet6, StDeMet7, and StDeMet8). The CDS and protein length of methyltransferase genes varied from 363 bp (StDNMT2-8B) to 4,674 bp (StMET2-11A) and 120 amino acids (aa) (StDNMT2-8B) to 1,557 aa (StMET2-11A), respectively. Similarly, gene transcript length and polypeptide length of demethylase genes ranges from 906 bp (StDeMet8) to 5,556 bp (StDeMet1) and 301aa (StDeMet8) to 1,851 aa (StDeMet1), respectively (Figure 1). The predicted molecular weight and pI value range from 174.27 to 13.7 kDa and 4.63 to 9.51 in C5-MTase; and 207.46 to 33.51 kDa and 6.24 to 9.46 in DeMets (Table 1). The chromosome localization of the candidate proteins has also been incorporated (Table 1). Interestingly, the classification of C5-MTase is based on the linear arrangement of distinct and conserved domains across members of a different enzyme family (Pavlopoulou and Kossida, 2007). Pfam analysis revealed that all C5-MTases (PF00145) have conserved DNA_Methylase (DNA_M) domain. However, different C5-MTase are characterized due to the presence of distinctive domains at their N-terminal: (i) CMT family possesses CHROMO (Chromatin organization modifier) and one BAH (Bromo Adjacent Homology) in contrast to (ii) MET which has two BAH and an RFD domains (iii) DRM family has unique UBA domain (Ubiquitin-associated), whereas (iv) DNMT has shorter protein length as compared to other C5-MTase owing to lack of N-terminal regulatory domain (Figure 2). Additionally, DeMets have conserved HhH-GPD (PF00730), Perm-CXXC (PF15629), and RRM_DME (PF15628). The distribution of conserved motifs in the StC5-MTase and StDeMets proteins were predicted using the MEME online suite. A total of 15 conserved motifs were explored in StC5-MTase and StDeMets proteins. Motifs 9, 11, and 12 were highly conserved in the MET subfamily. Motifs 5, 13, and 15 were major motifs in DRM, while Motif 8 was unique to CMT3 subfamilies. The motif distribution in StDeMets proteins showed that Motifs 1, 3, 4, 5, 6, 7, 8, 9, 11, and 13 were highly conserved in StDeMet 1- 5 proteins, while DeMet 2 and 3 have additional unique Motif 14 (Figure 2). The structure of StC5-MTase and StDeMets of potato is identical to Arabidopsis thaliana and is presumed to catalyze similar functions as that of Arabidopsis thaliana in potato.
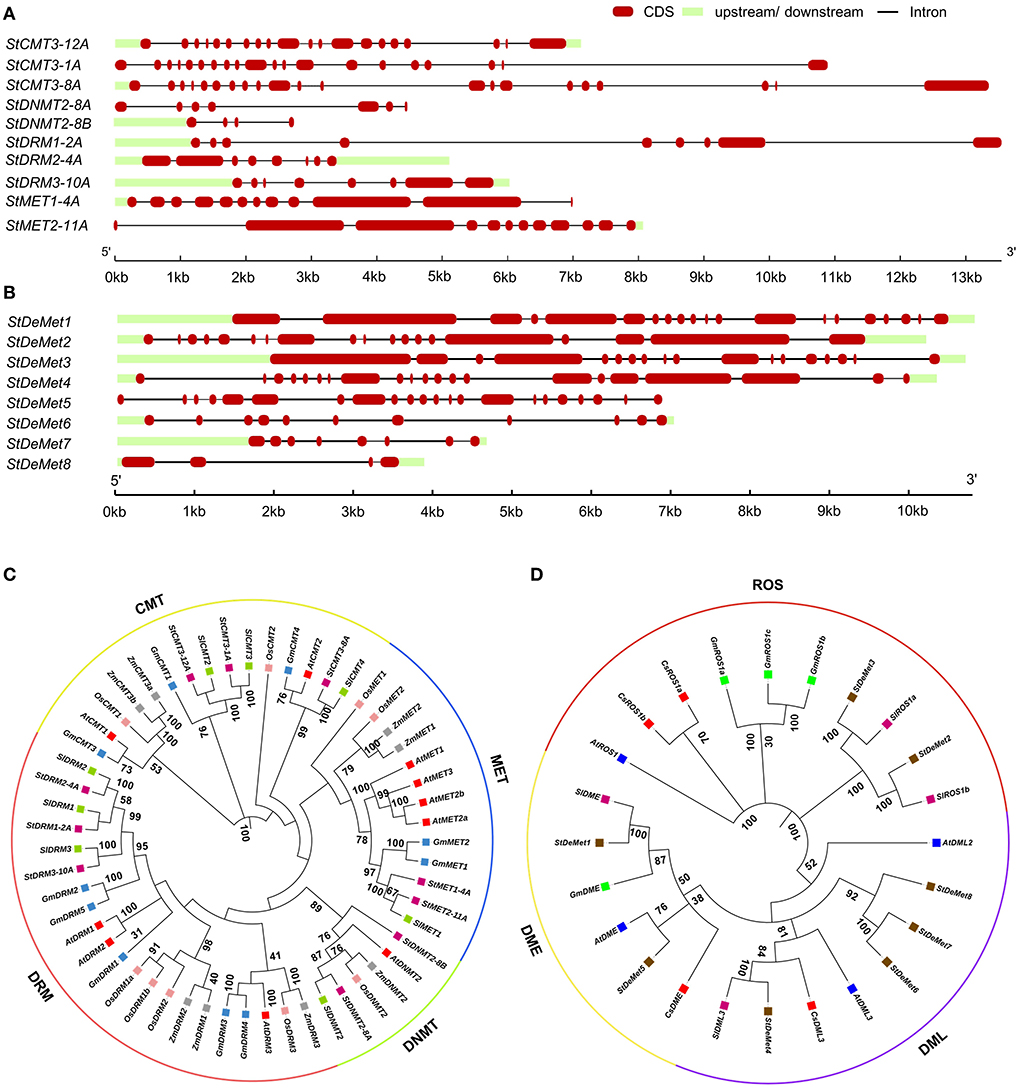
Figure 1. Gene structures of StC5-MTase (A) and StDeMets (B) in S. tuberosum. Phylogenetic tree analysis of C5-MTase (C) and DeMet proteins (D) with 1,000 bootstrap replicates. The lines in gene structure represent introns and filled red and light green boxes represent CDS and UTR, respectively. The lengths of CDS and introns can be determined using the scale bar on the bottom.

Figure 2. Schematic structures of conserved domains present in StC5-MTase (A) and StDeMet (B) proteins. The conserved motifs present in C5-MTase (C) and DNA demethylase (D) genes in Solanum tuberosum. (i) The conserved domains in the CMT family possess CHROMO (Chromatin organization modifier) and one BAH (Bromo Adjacent Homology); (ii) MET has two BAH and an RFD domains; (iii) DRM family has a unique UBA domain (Ubiquitin-associated); whereas (iv) DNMT has shorter protein length due to lack of N-terminal regulatory domain. StDeMet family possesses DNA glycosylase (ENDO_3C), RNA Recognition Motif (RRM), and methylated CpG discriminating (CXXC). The color boxes in the motif represent the position of different motifs and box sizes show the length (aa) of motifs.
Phylogenetic analysis
To exemplify the evolutionary relationship among C5-MTase subfamilies in plants, we used 55 protein sequences from four dicots (Arabidopsis thaliana, Solanum lycopersicum, Solanum tuberosum, and Glycine max) and two monocots (Zea mays and Oryza sativa) to construct a phylogenetic tree (Supplementary Table S2). C5-MTase is grouped into four subfamilies: DRM, CMT, MET, and DNMT having 21, 15, 13, and six members, respectively (Figure 1). Subsequently, the largest and smallest group in the tree were DRM and DNMT subfamily. Further, the subfamilies could also be divided into monocot and dicot distinct subgroups, based upon the significant differences in protein sequences. Interestingly, in almost all clades, tomato C5-MTase was closer to potato C5-MTase protein, suggesting that they are the members of the same taxonomic family, Solanaceae, and predicted to share functional similarity.
As for DNA Demethylases, 24 protein sequences from four dicots (Arabidopsis thaliana, Solanum lycopersicum, Solanum tuberosum and Cucumis sativa, and Glycine max) were extracted to construct a phylogenetic tree (Supplementary Table S3) arranged in ascending order in accordance to the number of members: DME, DML, and ROS. However, we analyzed DeMets from dicots as DME evolved monophyletically in dicots, such results are consistent with the previous report (Yu et al., 2021). We also observed DML group constituting four out of eight StDeMets genes as compared to ROS and DME.
Analysis of regulatory elements in the promoter region
Cis-regulatory elements are the core components governing biological processes. A sum of 30 major elements was represented that were predicted in the 2 kb upstream of the TSS in the promoter region. They were further categorized into light, development, hormone, and stress-responsive cis elements (Figure 3). C5-MTase genes showed several stress-responsive cis elements, such as CAAT and TATA box, which were predominant across the subfamilies/members (MET, CMT3, DRM, and DNMT2). Among all the subfamilies, CMT3 had the largest number of stress-involved elements. We found nine hormone-responsive elements such as ABRE and ERE that were abundant in all members. However, AuxRR-core is present exclusively in StDNMT2-8A. Elements involved in plant development were also screened, and five major elements including A box, CCGTCC, CAT box, ACA motif, and Circadian were identified. A box and CCGTCC were present only in StCMT3-12A, while circadian control element and ACA motif were found exclusively in DNMT2 and MET subfamilies. Further, eight CRE elements of the light-responsive module were also identified, out of which G box and Box 4 were found in all the naturally grouped subfamilies.
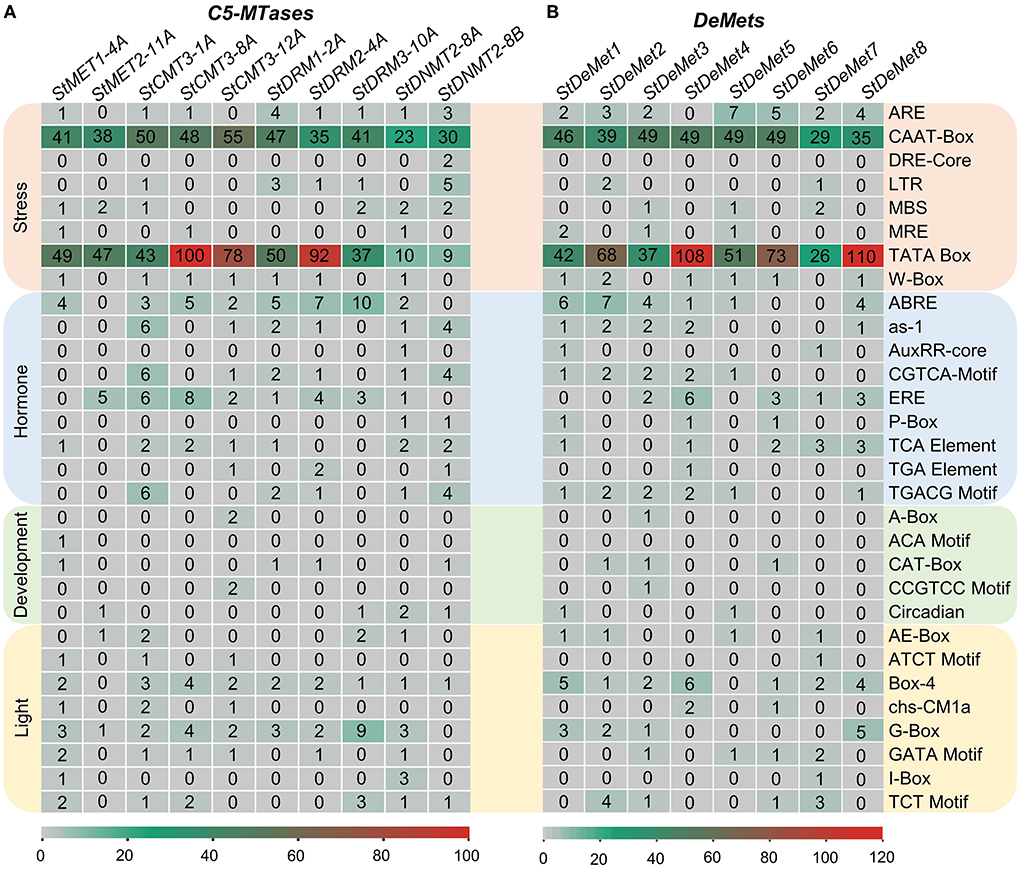
Figure 3. Cis-acting regulatory elements in the promoters of (A) StC5-MTase and (B) StDeMets. Cis-regulatory elements are classified into four categories (light, development, hormones and stress responses). The numbers in the heat map depict the abundance of cis-regulatory elements in the promoter region.
Similarly, cis-regulatory elements were also analyzed in the potato DNA methyltransferase proteins. ARE, CAAT, and TATA control elements were present in all the eight DeMet genes. Surprisingly, the abscisic hormone element ABRE was not found in StDeMet6 and StDeMet7, while TCA elements involved in salicylic acid response were present in StDeMet6 and StDeMet7. Furthermore, light module Box4 was present in all members. Altogether, our results suggest that C5-MTase and demethylase genes are involved in plant growth, development, and various abiotic stress responses.
Sub-cellular localization of potato C5-MTase and DeMets
The in silico subcellular localization revealed that all DeMets were localized in the nucleus; however, StDeMet7 was additionally found in mitochondria. The C5-MTases were localized in the nucleus (StMET1-4A, StMET2-11A, StCMT3-8A, StCMT3-12A, StDRM1-2A, StDRM2-4A, and StDRM3-10A) and cytoplasm (StCMT3-1A, StDNMT2-8A, and StDNMT2-8B). Apart from being in the nucleus, a couple of DRM members (StDRM1-2A and StDRM2-4A) were also localized in the cytoplasm (Figure 4).
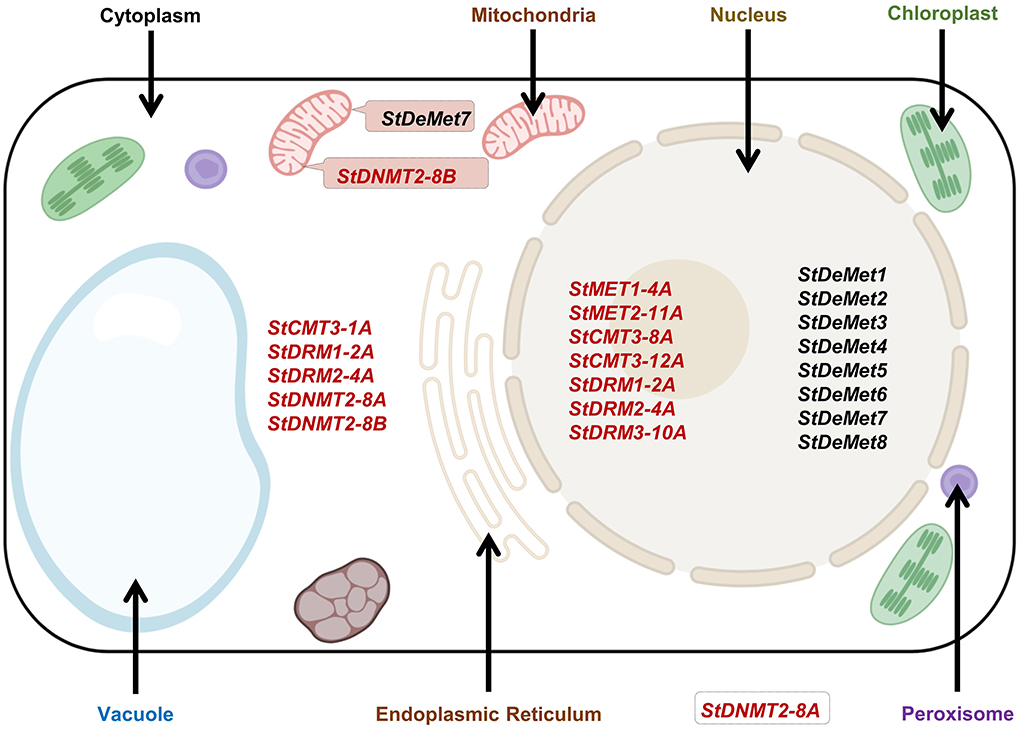
Figure 4. Subcellular localization of StC5-MTase and StDeMet proteins in potato predicted using the CELLO program. Partially adapted from Gahlaut et al. (2020).
Expression patterns of C5-MTase and DeMet genes
The expression dynamics of C5-MTase and DeMet genes were studied in different tissues, viz. leaf, flower, and stolon in the heat-sensitive (HS) and heat-tolerant (HT) potato genotypes under high temperature (Figures 5, 6). The expression of StMET1-4A and StMET2-11A showed a significant decline both in leaf and floral tissues in heat-sensitive genotype as compared to heat tolerant. However, stolon tissue shows upregulation and downregulation of StMET1-4A and StMET2-11A, respectively, in heat-sensitive cultivar. StDNMT-8A and StDNMT-8B expression level is predominantly detected in stolon tissues of HS. Moreover, heat induced the expression of StDNMT-8B in the flower of HT. The expression profile of the CMT subfamily is significantly higher in HT as compared to HS across three tissues (leaf, flower, and stolon) except StCMT3-1A. StDRM1-2A is downregulated in HT and HS in all samples. Interestingly, the expression level of StDRM2-4A increased in leaves, flowers, and stolon in heat-tolerant genotype. Overall, C5-MTase genes respond differently in different tissues of two genotypes, wherein CMT and DRM subfamilies showed pronounced heat-induced upregulation in the tolerant genotype.
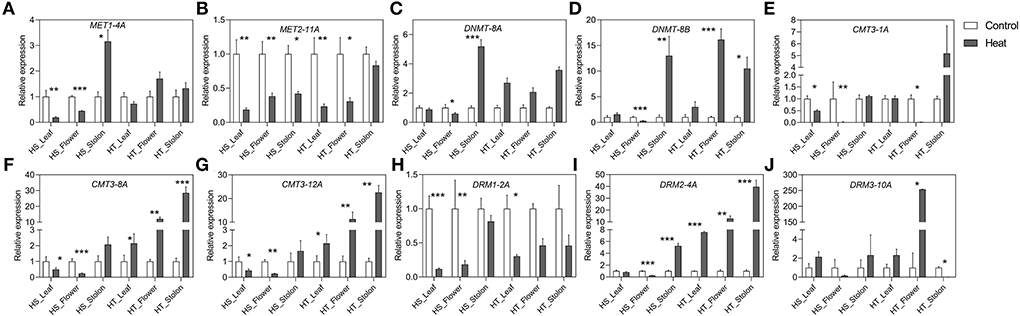
Figure 5. (A–J) Relative expression levels of potato C5-MTase genes in different tissues in heat sensitive (HS) and tolerant (HT) genotypes under high-temperature stress. Three biological samples were measured with three technical replicates. The fold change in RNA levels was calculated as the 2–ΔΔCt value relative to the mean values obtained in the leaf, flower and stolon samples (set at a value of 1.0). Standard errors of the means are shown with one, two and three asterisks indicating significant differences (p < 0.05, p < 0.01, p < 0.001, respectively) using a student's t-test.
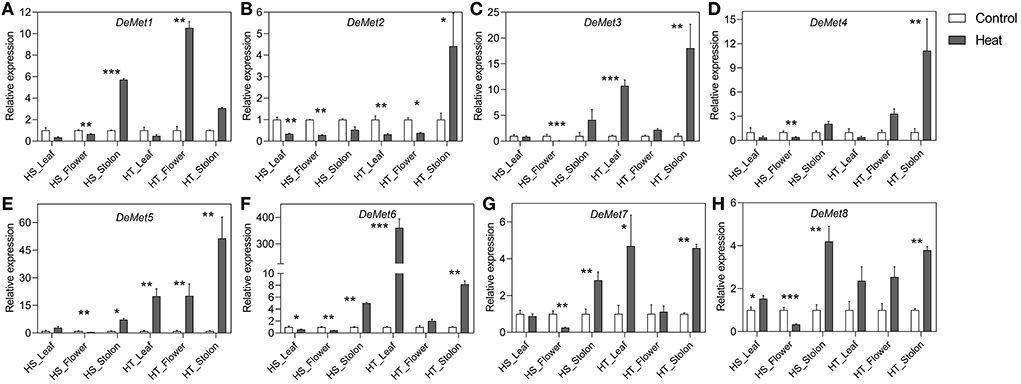
Figure 6. (A–H) Relative expression levels of potato DeMet genes in different tissues in heat sensitive (HS) and tolerant (HT) genotypes under high temperature stress. Three biological samples were measured with three technical replicates. The fold change in RNA levels was calculated as the 2–ΔΔCt value relative to the mean values obtained in the leaf, flower and stolon samples (set at a value of 1.0). Standard errors of the means are shown with one, two and three asterisks indicating significant differences (p < 0.05, p < 0.01, p < 0.001, respectively) using a student's t-test.
DeMets are involved in DNA demethylation, and we observed a varied expression pattern across different tissues and genotypes. Heat resulted in the downregulation of StDeMet1 in flower; however, the stolon tissue showed upregulation in the HS genotype. In contrast, StDeMet1 showed upregulation in flower. In general, StDeMet2 expression was downregulated in all the tissues of both the genotypes under heat stress, except the stolon of HT where it showed upregulation. Importantly, the expression of StDeMet3, StDeMet5, StDeMet6, and StDeMet7 was induced under heat in the leaf and stolon tissues of the HT genotype. The expression of StDeMet4, StDeMet5, and StDeMet6 were downregulated in the flower of HS. In the case of StDeMet8, HS leaf showed upregulation, while flower showed downregulation; however, the stolon tissue showed upregulation in both the genotypes.
Expression patterns of tuberization and HSFs genes
Several genes play an important role in tuber formation in potato, wherein the concurrent action of positive (e.g., StSP6A and StBEL5) and negative regulators (e.g., StSUT4, StSP5G, and StRAP1) determine the tuber yield. Also, Heat Shock Factors (HSFs) are well-known to impart thermotolerance in plants. Thus, we analyzed the expression of tuberization and HSF genes in the leaf and stolon tissues (Figure 7).
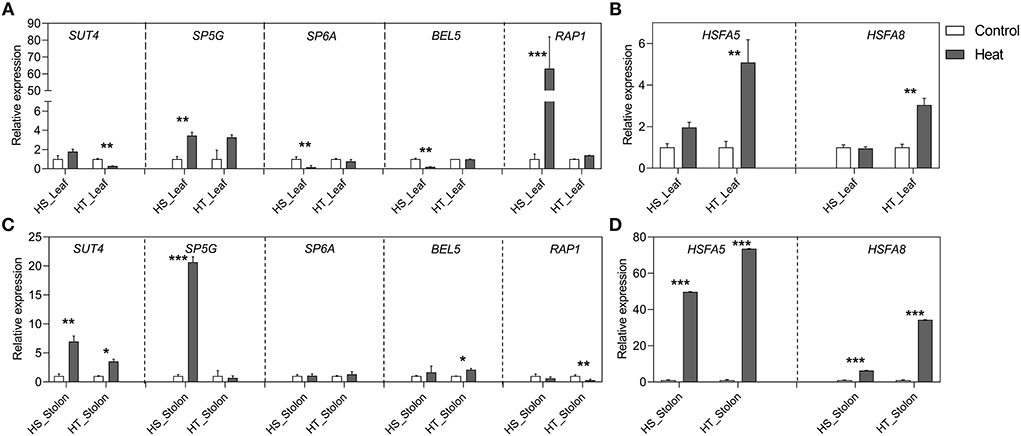
Figure 7. Relative expression levels of potato tuberization genes and HSFs in leaf (A,B) and stolon (C,D) tissues in heat-sensitive (HS) and -tolerant (HT) genotypes under high-temperature stress. Three biological samples were measured with three technical replicates. The fold change in RNA levels was calculated as the 2–ΔΔCt value relative to the mean values obtained in the leaf and stolon samples (set at a value of 1.0). Standard errors of the means are shown with one, two, and three asterisks indicating significant differences (p < 0.05, p < 0.01, p < 0.001, respectively) using a Student's t-test.
Heat resulted in the downregulation of StSUT4 expression in the leaf tissues of the HT genotype, whereas StSP5G showed significant upregulation in stolon tissues of the HS genotype. StSP6A is a master regulator of tuberization that is affected by high-temperature stress. Heat resulted in the significant suppression of StSP6A in leaves of HS genotype compared to HT genotype (Figure 7). However, unexpectedly intrinsic basal transcript level was maintained in stolon tissues of HS and HT genotypes. StBEL5 expression was downregulated by high temperature in the leaf tissue of the HS genotype, while it was stable across leaf and stolon tissues of the HT genotype. StRAP1 showed enhanced expression in leaves while it remained unaltered in stolons of HS genotype. Furthermore, the expression of HSFs was quantified. We found that StHSFA5 and StHSFA8 were strongly upregulated in the HT genotype both in leaves and stolons tissues under heat stress, suggesting its role in thermotolerance.
Discussion
Potato is a temperate crop that is challenged by several environmental cues such as high temperature. High-temperature exposure alters the partitioning of assimilates and accumulation of biomass in plant organs, which results in stem elongation, an increase in leaf number, and a decline in tuber yield (Wolf et al., 1991; Lafta and Lorenzen, 1995). A complex gene regulatory network comprising FT orthologs SELF PRUNING 6A (StSP6A) and SELF PRUNING 5G (StSP5G), CONSTANS-LIKE 1 (StCOL1), BEL1-like homeodomain (StBEL5), and an AP2-like gene (StRAP1) plays an important role in potato tuberization (Navarro et al., 2011; Hancock et al., 2014). For example, SP6A is a master regulator that interacts with its FD partner (StFDL) and scaffold protein St14-3-3 to form Tuber Activating Complex (TAC) to induce stolon–tuber induction (Hannapel and Banerjee, 2017). Recent developments have established that the StSP6A transcript is suppressed under elevated temperature, possibly through another FT homolog, StSP5G, which functions to repress tuberization through the accumulation of StCO. Similarly the antagonistic role of FT homologs in controlling flowering time in sugar beet has been reported (Pin et al., 2010). Furthermore, high temperature causes SP6A repression by a micro-RNA named SUPPRESSING OF SP6A (SES; Lehretz et al., 2019). Also, recently it has been shown that SP6A is suppressed transcriptionally and post-transcriptionally under high-temperature stress (Park et al., 2022). Epigenetic components are also involved in potato tuberization, wherein Polycomb Repressive Complex (PRC)-mediated histone modifications repress several target genes by H3K27 trimethylation (Kumar et al., 2021). The role of DNA methylation in photoperiod-mediated tuberization has also been recently reported (Ai et al., 2021). However, DNA methylation and demethylation enzymes remain uncharacterized in potatoes and their role in potato tuberization under high temperature is unexplored. The concurrent action of DNA methyltransferase and demethylase carries out DNA methylation and demethylation respectively (Lang et al., 2017).
In the present study, we identified several C5-MTases and DeMets in potatoes and studied their expression dynamics in the different tissues of the two contrasting genotypes, viz. Kufri Chandramukhi (heat sensitive) and Kufri Kiran (heat tolerant) under high temperature stress. We found 10 C5-MTase and 8 DeMets in potato, and several C5-MTase and DeMets were also reported in other allotetraploid soybean and peanut (Song et al., 2013; Wang et al., 2016), which is comparatively higher than the diploid species (Kong et al., 2020). This suggests that polyploidization events potentially contribute to the enhancement of plant epigenetic factors. Furthermore, based on their domain structure and phylogenetic analysis, C5-MTases were divided into four different subfamilies, viz. METs, CMTs, DRMs, and DNMTs. The members within the same subfamily shared similar domain architecture and exon-intron boundary, but it differed between sub-families. Our results are consistent with the previous reports on Solanum lycopersicum (Cao et al., 2014). The principal maintenance of symmetric CG methylation is operated by METs, an ortholog of mammalian DNMT1 (Zhang et al., 2018). Contrastingly, METs in plants cannot distinguish hemimethylated CG dinucleotides from non-methylated CG, unlike DNMT1, as it lacks a cysteine-rich CXXC domain (Song et al., 2011). BAH domain of METs facilitates protein-protein interactions, which result in inhibition of gene expression and establish an interconnection among cytosine methylation, replication, and transcription (Callebaut et al., 1999). CMTs are involved in maintaining CHG methylation in centromeric and transposable regions (Gehring and Henikoff, 2008). The chromodomain of CMTs is responsible for protein association with heterochromatin (Gehring and Henikoff, 2008). The protein structure of maize revealed the association of chromo and BAH domain with H3K9me2 and shed light on the functional activity of CMTs (Stroud et al., 2014). The maintenance of “non-symmetrical” CHH methylation is catalyzed by DRMs that operates through the RNA-directed DNA methylation pathway (RdDM). The N-terminal UBA domain provides a connecting link between DNA methylation and ubiquitin-mediated protein degradation. This domain also promotes the degradation of DRMs at specific points in the cell cycle (Cao et al., 2000).
DeMets in potato also has conserved domains that are responsible for functional identity and activity (Zhang et al., 2018). The functional demethylases contain HhH-GPD (Endo_3c) domain that is responsible for cleaving glycosidic bonds and generating AP sites (Gehring and Henikoff, 2008). Sometimes, two additional conserved domains named RNA recognition motif (RRM) and methylated CpG discriminating CXXC domain are also found (Iyer et al., 2011). The evolutionary studies revealed that DeMets expression varied across species of different genotypes (Yang et al., 2019).
The sub-cellular localization of any protein is important for determining its function in plants. Therefore, we performed in-silico subcellular prediction to identify the location of C5-MTases and DeMets. C5-MTases were localized either in the nucleus or in the cytoplasm. These results are consistent with the previous studies on Solanum lycopersicum, which reported the localization of C5-MTase in the nucleus and cytoplasm (Cao et al., 2014). The presence of C5-MTase in both these organelles indicates its possible involvement in stress management machinery (Gahlaut et al., 2020). The exclusive nuclear localization of DeMets suggests its potential role in replication and transcriptional regulation (Wang et al., 2016).
Cis-regulatory elements play an important role in the regulation of biological processes and molecular function under developmental transitions and stress episodes (Yamaguchi-Shinozaki and Shinozaki, 2005). The promoter analysis of potato C5-MTase and DeMets revealed that cis-regulatory elements are classified into four categories such as light response, development, hormones, and stress responses. The presence of CAAT box in the promoter region plays an essential role in leaf development by modulating jasmonate pathways in proliferating tissues as reported in Nicotiana benthamiana (NbCMT3–2; Lin et al., 2015). The predominance of TATA box elements in the promoter region confers stress responsiveness, whereas TATA-less genes are related to cell growth or housekeeping activity (Bae et al., 2015). The abundance of ABRE and ERE in all the members demonstrates the active role of C5-MTase and DeMets in abscisic acid and ethylene responsiveness (Kaur and Asthir, 2017). Also, stress-specific regulatory elements such as ARE and MBS were found in the C5-MTase of the tea plant (Zhu et al., 2020). Overall, these findings highlight the role of potato's C5-MTase and DeMets in plant development and stress responses (Gahlaut et al., 2020).
The relationship between DNA methylation and heat stress is less explored. DNA methylation level in different tissues regulates plant growth and development via controlling the transcript levels of developmentally important genes (Zhang et al., 2018). We investigated the expression pattern of C5-MTase and DeMets in two contrasting genotypes across different tissues and observed differential DNA methylation. Our results are similar to studies on rice and rapeseed (Karan et al., 2012; Gao et al., 2014). The decline in expression of StMET1-4A and StMET2-11A in leaf and floral tissues in heat-sensitive genotype possibly led to reduced CG methylation.
Similarly, Arabidopsis met1 mutant with reduced MET level showed delayed flowering and juvenile-to-adult transition (Kankel et al., 2003). Such delayed flowering was due to hypomethylation of transposable element (TE) upstream of the FLOWERING WAGENINGEN (FWA) gene (Kankel et al., 2003). This shows how MET regulates the morphological characteristics and flowering time of plants (Finnegan and Kovac, 2000). We also found that CMT and DRM subfamilies showed pronounced heat-induced upregulation in the tolerant genotype. These findings indicate the role of CMT and DRM in the suppression of TEs (Ramakrishnan et al., 2021). High-temperature stress causes genomic instability and impairs plant health by activating TEs such as ONSEN (Chang et al., 2020). Several plant developmental abnormalities have been observed in the DNA methylation-free C5-MTase mutants of Arabidopsis, suggesting their role in controlling gene expression and developmental processes in plants (He et al., 2022). However, the reduced expression of CMT and DRM subfamilies in heat-sensitive genotypes across different tissues might be an alternative strategy to recover from heat stress by causing activation of SUPPRESSOR OF DRM1 DRM2 CMT3 (SDC), thus leading to loss of non-CG methylation (Popova et al., 2013). Differential methylation landscapes between genotypes can potentially contribute to stress adaptation (Eriksson et al., 2020).
Heat-induced DNA demethylase gene expression was observed in both genotypes. The expression of the demethylase gene family was more pronounced in HT genotype as compared to the HS genotype. Our results showed that DeMets-like StDeMet2,StDeMet3, StDeMet4, StDeMet5, StDeMet6, and StDeMet7, were significantly expressed in stolons of HT genotype. Such results are consistent with the increased expression of ROS and DML like demethylase gene family in Arabidopsis (Hsieh et al., 2009), suggesting its involvement in combating abiotic stress events. Consistently, active DNA demethylation is also associated with the upregulation of stress-responsive genes (Verhoeven et al., 2010).
DNA methylation and demethylation events are correlated with the gene expression patterns, thus regulating plant development and stress responses (Choi and Sano, 2007). Also, the genetic control of potato tuberization is well-studied. We observed that the expression of positive regulators of tuberization (StSP6A and StBEL5) was found to be stable in the leaves and stolon tissues in the HT genotype as compared to the HS genotype under high-temperature stress (Figure 8). The stability of the StSP6A transcript under high temperature in HT genotype might be due to the reduced expression of upstream negative regulators (StSUT4, StSP5G, and StRAP1). Heat induced the expression of StSUT4 in the leaves and stolons of the HS genotype, which might have led to the activation of the StCOL1-StSP5G regulatory module. Such studies are consistent with the recent finding that elevated temperature caused higher StCOL1 accumulation and suppressed potato tuberization (Park et al., 2022). Our expression analysis revealed the higher expression of StSP5G transcript under high temperature in the HS genotype. However, such enhanced expression of StSP5G did not alter the basal transcription of StSP6A in stolons of HS genotype. The expression levels of StSP5G and StSP6A are usually difficult to interpret in vegetative tissues like stolons and tubers (Hancock et al., 2014). In addition, different regulatory pathways operate for photoperiod and temperature-dependent tuberization, suggesting that a complex regulatory network is involved in the regulation of potato tuberization (Park et al., 2022).
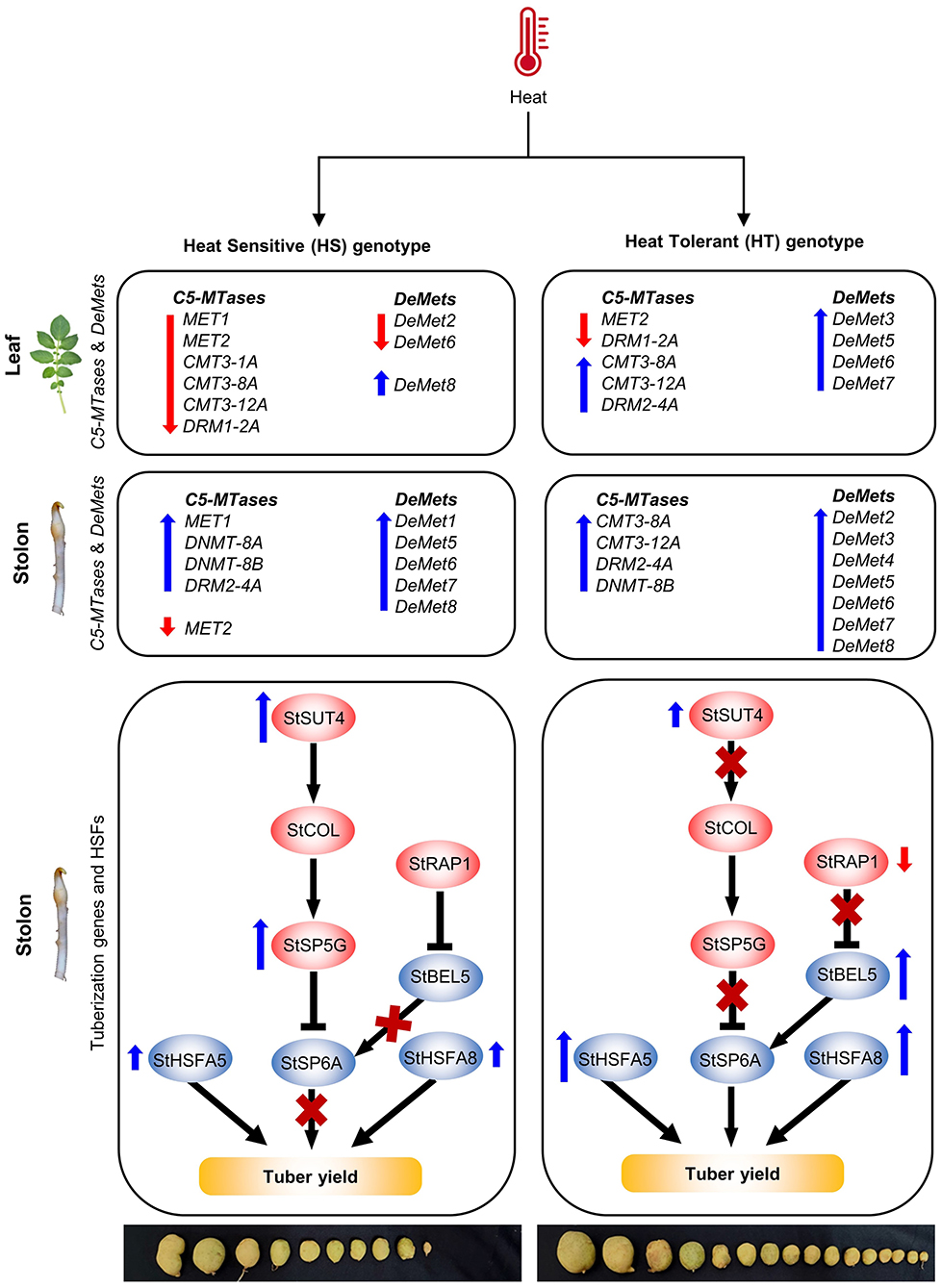
Figure 8. The schematic model depicting the role of C5-MTase and DeMets gene with potato tuberization genes under high temperature in heat-tolerant (HT) and heat-sensitive (HS) genotype: Tuberization is regulated by positive (StSP6A and StBEL5) and negative regulators (StSUT4, StSP5G, and StRAP1). Under high temperature, the positive regulators were relatively stable in the HT genotype as compared to the HS genotype. The stability might be attributed by reduced expression of upstream negative regulators, increased DNA demethylation (StDeMet2, StDeMet3, StDeMet4, StDeMet5, StDeMet6, StDeMet7, and StDeMet8) and active RdDM (DRM and CMT subfamilies) pathway in leaves and stolon samples of HT genotype. On the contrary, the HS genotype has heat-induced upregulation of negative regulators, thereby affecting the process of tuberization. Stronger upregulation of HSFs (StHSFA5 and StHSFA8) was observed in leaves and stolon tissues of HT genotype as compared to HS genotype maintaining the health status of the tolerant cultivar.
Additionally, we observed a spike in StRAP1 expression (<50-fold) in leaves of HS genotype, which acts to suppress a phloem mobile transcript (StBEL5), thereby delaying tuber induction (Banerjee et al., 2006). Other than genetic factors, the role of DNA methylation machinery components was sought in potato tuberization under high temperature. The increased expression of positive regulators of tuberization in HT genotype might be due to increased DNA demethylation and the active RdDM pathway in the stolons. Furthermore, we observed stronger upregulation of StHSFA5 and StHSFA8 (<3 and <5-fold, respectively) in leaves and (<50- and <30-fold) in stolons of the tolerant genotype as compared to the HS genotype. This may contribute to the better heat stress resilience of the HT genotype (Tang et al., 2016; Ramakrishnan et al., 2022). Altogether, a complex interplay of genetic and epigenetic factors controls potato tuberization under high-temperature conditions.
Conclusion
Over the last few decades, the mechanisms underlying plant responses to abiotic stresses have been studied in most of the model plants such as Arabidopsis, rice, and tomato. However, only a few studies are available on other important crops such as potato. More detailed studies are needed to shed light on the signaling networks and downstream target genes controlling potato response to heat stress. Importantly, understanding the interplay of genetic and epigenetic factors in plant stress response will provide new avenues for crop improvement. In this work, we report the characterization of DNA methyltransferase and demethylase gene families in the potato. The expression of C5-MTases and DeMets were studied in the different tissues of heat-tolerant and heat-sensitive genotypes followed by the expression analysis of tuberization genes. We found enhanced expression of DeMets and RdDM genes in HT genotype as compared to HS genotype. These results suggest that increased activity of DeMets might have contributed to transcriptional activation of tuberization genes wherein RdDM stabilized genomic integrity of HT genotype. Overall, this work will open new research directions on the role of DNA methylation machinery in potato tuberization under high temperature, which is important in the context of global temperature rise.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
MD performed in silico analysis, experiments, analyzed data, and wrote the original draft with figures. VR performed in silico analysis and experiments. VG performed in silico analysis and writing. AK, PS, and VV performed experiments. VKG and SS provided material and discussed data. GZ conceived the idea, edited, and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
MD and VR acknowledge UGC for Junior Research Fellowship. SS and VKG acknowledge the support of institutional project from ICAR-CPRI, Shimla. GZ acknowledges the financial support obtained from the projects viz. MLP-201, CSIR-FIRST (MLP-178), and DST-SERB-SRG (GAP-294). This manuscript represents CSIR-IHBT publication number 5102.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.933740/full#supplementary-material
References
Ai, Y., Jing, S., Cheng, Z., Song, B., Xie, C., Liu, J., et al. (2021). DNA methylation affects photoperiodic tuberization in potato (Solanum tuberosum L.) by mediating the expression of genes related to the photoperiod and GA pathways. Hortic. Res. 8, 181. doi: 10.1038/s41438-021-00619-7
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucl. Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Bae, S.-H., Han, H. W., and Moon, J. (2015). Functional analysis of the molecular interactions of TATA box-containing genes and essential genes. PLoS ONE 10, e0120848. doi: 10.1371/journal.pone.0120848
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acid. Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Banerjee, A. K., Chatterjee, M., Yu, Y., Suh, S.-G., Miller, W. A., and Hannapel, D. J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18, 3443–3457. doi: 10.1105/tpc.106.042473
Beals, K. A. (2019). Potatoes, nutrition, and health. Am. J. Potato Res. 96, 102–110. doi: 10.1007/s12230-018-09705-4
Birch, P. R., Bryan, G., Fenton, B., Gilroy, E. M., Hein, I., Jones, J. T., et al. (2012). Crops that feed the world 8: potato: are the trends of increased global production sustainable? Food Secur. 4, 477–508. doi: 10.1007/s12571-012-0220-1
Callebaut, I., Courvalin, J.-C., and Mornon, J.-P. (1999). The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446, 189–193. doi: 10.1016/S0014-5793(99)00132-5
Cao, D., Ju, Z., Gao, C., Mei, X., Fu, D., Zhu, H., et al. (2014). Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene 550, 230–237. doi: 10.1016/j.gene.2014.08.034
Cao, X., Aufsatz, W., Zilberman, D., Mette, M. F., Huang, M. S., Matzke, M., et al. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13, 2212–2217. doi: 10.1016/j.cub.2003.11.052
Cao, X., Springer, N. M., Muszynski, M. G., Phillips, R. L., Kaeppler, S., and Jacobsen, S. E. (2000). Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. National Acad. Sci. U. S. A. 97, 4979–4984. doi: 10.1073/pnas.97.9.4979
Chang, Y. N., Zhu, C., Jiang, J., Zhang, H., Zhu, J. K., and Duan, C. G. (2020). Epigenetic regulation in plant abiotic stress responses. J. Intergrat. Plant Biol. 62, 563–580. doi: 10.1111/jipb.12901
Chatterjee, M., Banerjee, A. K., and Hannapel, D. J. (2007). A BELL1-like gene of potato is light activated and wound inducible. Plant Physiol. 145, 1435–1443. doi: 10.1104/pp.107.105924
Chen, C., Chen, H., He, Y., and Xia, R. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660. doi: 10.1101/289660
Cheng, X., Kumar, S., Posfai, J., Pflugrath, J. W., and Roberts, R. J. (1993). Crystal structure of the Hhal DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell 74, 299–307. doi: 10.1016/0092-8674(93)90421-L
Choi, C.-S., and Sano, H. (2007). Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genom. 277, 589–600. doi: 10.1007/s00438-007-0209-1
Davis, M. W., and Jorgensen, E. M. (2022). ApE, A plasmid Editor: a freely available DNA manipulation and visualization program. Front. Bioinf. 2, 818619. doi: 10.3389/fbinf.2022.818619
Dowen, R. H., Pelizzola, M., Schmitz, R. J., Lister, R., Dowen, J. M., Nery, J. R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. National Acad. Sci. U. S. A. 109, E2183–E2191. doi: 10.1073/pnas.1209329109
Dutt, S., Manjul, A. S., Raigond, P., Singh, B., Siddappa, S., Bhardwaj, V., et al. (2017). Key players associated with tuberization in potato: potential candidates for genetic engineering. Crit. Rev. Biotechnol. 37, 942–957. doi: 10.1080/07388551.2016.1274876
Eriksson, M. C., Szukala, A., Tian, B., and Paun, O. (2020). Current research frontiers in plant epigenetics: an introduction to a Virtual Issue. New Phytol. 226, 285. doi: 10.1111/nph.16493
FAOSTAT (2021). Food and Agriculture Data. Available online at: http://faostat.fao.org (accessed April 15, 2022).
Finnegan, E., and Kovac, K. (2000). Plant DNA methyltransferases. Plant Mol. Biol. 43, 189–201. doi: 10.1023/a:1006427226972
Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. U. S. A. 93, 8449–8454. doi: 10.1073/pnas.93.16.8449
Gahlaut, V., Samtani, H., and Khurana, P. (2020). Genome-wide identification and expression profiling of cytosine-5 DNA methyltransferases during drought and heat stress in wheat (Triticum aestivum). Genomics 112, 4796–4807. doi: 10.1016/j.ygeno.2020.08.031
Gallego-Bartolomé, J., Gardiner, J., Liu, W., Papikian, A., Ghoshal, B., Kuo, H. Y., et al. (2018). Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. National Acad. Sci. U. S. A. 115, E2125–E2134. doi: 10.1073/pnas.1716945115
Gao, Y., Jia, L., Hu, B., Alva, A., and Fan, M. (2014). Potato stolon and tuber growth influenced by nitrogen form. Plant Prod. Sci. 17, 138–143. doi: 10.1626/pps.17.138
Garg, R., Narayana Chevala, V., Shankar, R., and Jain, M. (2015). Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 5, 1–16. doi: 10.1038/srep14922
Gehring, M., and Henikoff, S. (2008). DNA methylation and demethylation in Arabidopsis. Arabidopsis Book 6, e0102. doi: 10.1199/tab.0102
Hancock, R. D., Morris, W. L., Ducreux, L. J., Morris, J. A., Usman, M., Verrall, S. R., et al. (2014). Physiological, biochemical and molecular responses of the potato (Solanum tuberosum L.) plant to moderately elevated temperature. Plant Cell Environ. 37, 439–450. doi: 10.1111/pce.12168
Hannapel, D. J., and Banerjee, A. K. (2017). Multiple mobile mRNA signals regulate tuber development in potato. Plants 6, 8. doi: 10.3390/plants6010008
Hastilestari, B. R., Lorenz, J., Reid, S., Hofmann, J., Pscheidt, D., Sonnewald, U., et al. (2018). Deciphering source and sink responses of potato plants (Solanum tuberosum L.) to elevated temperatures. Plant Cell Environ. 41, 2600–2616. doi: 10.1111/pce.13366
He, L., Huang, H., Bradai, M., Zhao, C., You, Y., Ma, J., et al. (2022). DNA methylation-free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nat. Commun. 13, 1–16. doi: 10.1038/s41467-022-28940-2
Hirsch, C. D., Hamilton, J. P., Childs, K. L., Cepela, J., Crisovan, E., Vaillancourt, B., et al. (2014). Spud DB: A resource for mining sequences, genotypes, and phenotypes to accelerate potato breeding. Plant Genome 7, plantgenome2013.12.0042. doi: 10.3835/plantgenome2013.12.0042
Hsieh, T.-F., Ibarra, C. A., Silva, P., Zemach, A., Eshed-Williams, L., Fischer, R. L., et al. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324, 1451–1454. doi: 10.1126/science.1172417
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Iyer, L. M., Abhiman, S., and Aravind, L. (2011). Natural history of eukaryotic DNA methylation systems. Progr. Mol. Biol. Trans. Sci. 101, 25–104. doi: 10.1016/B978-0-12-387685-0.00002-0
Kankel, M. W., Ramsey, D. E., Stokes, T. L., Flowers, S. K., Haag, J. R., Jeddeloh, J. A., et al. (2003). Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163, 1109–1122. doi: 10.1093/genetics/163.3.1109
Karan, R., Deleon, T., Biradar, H., and Subudhi, P. K. (2012). Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE 7, e40203. doi: 10.1371/journal.pone.0040203
Kaur, G., and Asthir, B. (2017). Molecular responses to drought stress in plants. Biol. Plant. 61, 201–209. doi: 10.1007/s10535-016-0700-9
Kim, Y.-U., Seo, B.-S., Choi, D.-H., Ban, H.-Y., and Lee, B.-W. (2017). Impact of high temperatures on the marketable tuber yield and related traits of potato. Eur. Agron. 89, 46–52. doi: 10.1016/j.eja.2017.06.005
Kong, W., Xia, X., Wang, Q., Liu, L.-W., Zhang, S., Ding, L., et al. (2020). Impact of DNA demethylases on the DNA methylation and transcription of Arabidopsis NLR Genes. Front. Genet. 11, 460. doi: 10.3389/fgene.2020.00460
Kumar, A., Kondhare, K. R., Malankar, N. N., and Banerjee, A. K. (2021). The Polycomb group methyltransferase StE (z) 2 and deposition of H3K27me3 and H3K4me3 regulate the expression of tuberization genes in potato. J. Exp. Bot. 72, 426–444. doi: 10.1093/jxb/eraa468
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/molbev/msy096
Lafta, A. M., and Lorenzen, J. H. (1995). Effect of high temperature on plant growth and carbohydrate metabolism in potato. Plant Physiol. 109, 637–643. doi: 10.1104/pp.109.2.637
Lang, Z., Wang, Y., Tang, K., Tang, D., Datsenka, T., Cheng, J., et al. (2017). Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. U. S. A. 114, E4511–E4519. doi: 10.1073/pnas.1705233114
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Lehretz, G. G., Sonnewald, S., Hornyik, C., Corral, J. M., and Sonnewald, U. (2019). Post-transcriptional regulation of FLOWERING LOCUS T modulates heat-dependent source-sink development in potato. Curr. Biol. 29 1614–1624. e1613. doi: 10.1016/j.cub.2019.04.027
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Lesk, C., Rowhani, P., and Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Lin, Y.-T., Wei, H.-M., Lu, H.-Y., Lee, Y.-I., and Fu, S.-F. (2015). Developmental-and tissue-specific expression of NbCMT3-2 encoding a chromomethylase in Nicotiana benthamiana. Plant Cell Physiol. 56, 1124–1143. doi: 10.1093/pcp/pcv036
Liu, P., Nie, W. F., Xiong, X., Wang, Y., Jiang, Y., Huang, P., et al. (2021). A novel protein complex that regulates active DNA demethylation in Arabidopsis. J. Int. Plant Biol. 63, 772–786. doi: 10.1111/jipb.13045
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Navarro, C., Abelenda, J. A., Cruz-Oró, E., Cuéllar, C. A., Tamaki, S., Silva, J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. doi: 10.1038/nature10431
Park, J.-S., Park, S.-J., Kwon, S.-Y., Shin, A.-Y., Moon, K.-B., Park, J. M., et al. (2022). Temporally distinct regulatory pathways coordinate thermo-responsive storage organ formation in potato. Cell Rep. 38, 110579. doi: 10.1016/j.celrep.2022.110579
Pavlopoulou, A., and Kossida, S. (2007). Plant cytosine-5 DNA methyltransferases: structure, function, and molecular evolution. Genomics 90, 530–541. doi: 10.1016/j.ygeno.2007.06.011
Pin, P. A., Benlloch, R., Bonnet, D., Wremerth-Weich, E., Kraft, T., Gielen, J. J., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400. doi: 10.1126/science.1197004
Popova, O. V., Dinh, H. Q., Aufsatz, W., and Jonak, C. (2013). The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 6, 396–410. doi: 10.1093/mp/sst023
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Ramakrishnan, M., Papolu, P. K., Satish, L., Vinod, K. K., Wei, Q., Sharma, A., et al. (2022). Redox status of the plant cell determines epigenetic modifications under abiotic stress conditions and during developmental processes. J. Adv. Res. 4, 7. doi: 10.1016/j.jare.2022.04.007
Ramakrishnan, M., Satish, L., Kalendar, R., Narayanan, M., Kandasamy, S., Sharma, A., et al. (2021). The dynamism of transposon methylation for plant development and stress adaptation. Int. J. Mol. Sci. 22, 11387. doi: 10.3390/ijms222111387
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., and Yao, X. (2009). DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273. doi: 10.1038/cr.2009.6
Roldán-Arjona, T., Ariza, R. R., and Córdoba-Cañero, D. (2019). DNA base excision repair in plants: an unfolding story with familiar and novel characters. Front. Plant Sci. 10, 1055. doi: 10.3389/fpls.2019.01055
Rykaczewska, K . (2015). The effect of high temperature occurring in subsequent stages of plant development on potato yield and tuber physiological defects. Am. J. Potato Res. 92, 339–349. doi: 10.1007/s12230-015-9436-x
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protocols 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schultz, J., Copley, R. R., Doerks, T., Ponting, C.P., and Bork, P. (2000). SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acid. Res. 28, 231–234. doi: 10.1093/nar/28.1.231
Song, J., Rechkoblit, O., Bestor, T. H., and Patel, D. J. (2011). Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331, 1036–1040. doi: 10.1126/science.1195380
Song, Q.-X., Lu, X., Li, Q.-T., Chen, H., Hu, X.-Y., Ma, B., et al. (2013). Genome-wide analysis of DNA methylation in soybean. Mol. Plant 6, 1961–1974. doi: 10.1093/mp/sst123
Stroud, H., Do, T., Du, J., Zhong, X., Feng, S., Johnson, L., et al. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21, 64–72. doi: 10.1038/nsmb.2735
Tang, R., Zhu, W., Song, X., Lin, X., Cai, J., Wang, M., et al. (2016). Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Sci. 7, 490. doi: 10.3389/fpls.2016.00490
Verhoeven, K. J., Jansen, J. J., Van Dijk, P. J., and Biere, A. (2010). Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 185, 1108–1118. doi: 10.1111/j.1469-8137.2009.03121.x
Vreugdenhil, D., and Struik, P. C. (1989). An integrated view of the hormonal regulation of tuber formation in potato (Solanum tuberosum). Physiol. Plant. 75, 525–531. doi: 10.1111/j.1399-3054.1989.tb05619.x
Wang, P., Gao, C., Bian, X., Zhao, S., Zhao, C., Xia, H., et al. (2016). Genome-wide identification and comparative analysis of cytosine-5 DNA methyltransferase and demethylase families in wild and cultivated peanut. Front. Plant Sci. 7, 7. doi: 10.3389/fpls.2016.00007
Wolf, S., Marani, A., and Rudich, J. (1991). Effect of temperature on carbohydrate metabolism in potato plants. J. Exp. Bot. 42, 619–625. doi: 10.1093/jxb/42.5.619
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yang, X., Lu, X., Chen, X., Wang, D., Wang, J., Wang, S., et al. (2019). Genome-wide identification and expression analysis of DNA demethylase family in cotton. J. Cotton Res. 2, 1–9. doi: 10.1186/s42397-019-0033-2
Yang, Z . (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yu, C.-S., Cheng, C.-W., Su, W.-C., Chang, K.-C., Huang, S.-W., Hwang, J.-K., et al. (2014). CELLO2GO: a web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS ONE 9, e99368. doi: 10.1371/journal.pone.0099368
Yu, Y., Lashbrook, C., and Hannapel, D. J. (2007). Tissue integrity and RNA quality of laser microdissected phloem of potato. Planta 226, 797–803. doi: 10.1007/s00425-007-0509-z
Yu, Z., Zhang, G., Teixeira Da Silva, J. A., Li, M., Zhao, C., He, C., et al. (2021). Genome-wide identification and analysis of DNA methyltransferase and demethylase gene families in Dendrobium officinale reveal their potential functions in polysaccharide accumulation. BMC Plant Biol. 21, 1–17. doi: 10.1186/s12870-020-02811-8
Zhang, H., Lang, Z., and Zhu, J.-K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mo. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhou, R., Kong, L., Yu, X., Ottosen, C.-O., Zhao, T., Jiang, F., et al. (2019). Oxidative damage and antioxidant mechanism in tomatoes responding to drought and heat stress. Acta. Physiol. Plant. 41, 1–11. doi: 10.1007/s11738-019-2805-1
Zhu, C., Zhang, S., Zhou, C., Chen, L., Fu, H., Li, X., et al. (2020). Genome-wide investigation and transcriptional analysis of cytosine-5 DNA methyltransferase and DNA demethylase gene families in tea plant (Camellia sinensis) under abiotic stress and withering processing. PeerJ. 8, e8432. doi: 10.7717/peerj.8432
Keywords: DNA methylation, epigenetics, climate change, tuberization, heat stress, thermotolerance
Citation: Dutta M, Raturi V, Gahlaut V, Kumar A, Sharma P, Verma V, Gupta VK, Sood S and Zinta G (2022) The interplay of DNA methyltransferases and demethylases with tuberization genes in potato (Solanum tuberosum L.) genotypes under high temperature. Front. Plant Sci. 13:933740. doi: 10.3389/fpls.2022.933740
Received: 01 May 2022; Accepted: 08 July 2022;
Published: 16 August 2022.
Edited by:
Girdhar Kumar Pandey, University of Delhi, IndiaReviewed by:
Jonathan Fresnedo Ramirez, The Ohio State University, United StatesMuthusamy Ramakrishnan, Nanjing Forestry University, China
Wen-Feng Nie, Yangzhou University, China
Copyright © 2022 Dutta, Raturi, Gahlaut, Kumar, Sharma, Verma, Gupta, Sood and Zinta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaurav Zinta, Z3ppbnRhQGdtYWlsLmNvbQ==; Z2F1cmF2emludGFAaWhidC5yZXMuaW4=
†These authors have contributed equally to this work and share first authorship
 Madhushree Dutta
Madhushree Dutta Vidhi Raturi
Vidhi Raturi Vijay Gahlaut
Vijay Gahlaut Akhil Kumar
Akhil Kumar Paras Sharma
Paras Sharma Vipasha Verma
Vipasha Verma Vijai Kishor Gupta4
Vijai Kishor Gupta4 Salej Sood
Salej Sood Gaurav Zinta
Gaurav Zinta