
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 August 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.933498
This article is part of the Research Topic Nutrient Dependent Signaling Pathways Controlling the Symbiotic Nitrogen Fixation Process, Volume II View all 6 articles
 Ahmed Abdelkhalek1*
Ahmed Abdelkhalek1* Hamada El-Gendi2
Hamada El-Gendi2 Abdulaziz A. Al-Askar3
Abdulaziz A. Al-Askar3 Viviana Maresca4
Viviana Maresca4 Hassan Moawad5
Hassan Moawad5 Mohsen M. Elsharkawy6
Mohsen M. Elsharkawy6 Hosny A. Younes7
Hosny A. Younes7 Said I. Behiry7
Said I. Behiry7Rhizobium spp. manifests strong nitrogen fixation ability in legumes. However, their significance as biocontrol agents and antivirals has rarely been investigated. Under greenhouse conditions, the molecularly identified nitrogen-fixing plant growth-promoting rhizobacteria (PGPR), Rhizobium leguminosarum bv. viciae strain 33504-Alex1, isolated from the root nodules of faba bean plants, was tested as a soil inoculum or a foliar application to trigger faba bean plants’ resistance against Bean yellow mosaic virus (BYMV) infection. Compared to the non-treated faba bean plants, the applications of 33504-Alex1 in either soil or foliar application significantly promoted growth and improved total chlorophyll content, resulting in a considerable reduction in disease incidence and severity and the inhibition index of BYMV in the treated faba bean plants. Furthermore, the protective activities of 33504-Alex1 were associated with significant reductions in non-enzymatic oxidative stress markers [hydrogen peroxide (H2O2) and malondialdehyde (MDA)] and remarkably increased DPPH free radical scavenging activity and total phenolic content compared to the BYMV treatment at 20 days post-inoculation. Additionally, an increase in reactive oxygen species scavenging enzymes [superoxide dismutase (SOD) and polyphenol oxidase (PPO)] and induced transcriptional levels of pathogenesis-related (PR) proteins (PR-1, PR-3, and PR-5) were observed. Of the 19 polyphenolic compounds detected in faba bean leaves by high-performance liquid chromatography (HPLC) analysis, gallic and vanillic acids were completely shut down in BYMV treatment. Interestingly, the 33504-Alex1 treatments were associated with the induction and accumulation of the most detected polyphenolic compounds. Gas chromatography-mass spectrometry (GC-MS) analysis showed hexadecanoic acid 2,3-dihydroxypropyl ester, tetraneurin-A-Diol, oleic acid, and isochiapin B are the major compounds in the ethyl acetate extract of 33504-Alex1 culture filtrate (CF), suggesting it acts as an elicitor for the induction of systemic acquired resistance (SAR) in faba bean plants. Consequently, the capacity of R. leguminosarum bv. viciae strain 33504-Alex1 to enhance plant growth and induce systemic resistance to BYMV infection will support the incorporation of 33504-Alex1 as a fertilizer and biocontrol agent and offer a new strategy for crop protection, sustainability, and environmental safety in agriculture production.
A sustainable increase in the human population and the global reduction in the agricultural area force the requirement for higher plant yields (per unit area) with enhanced quality (Shah et al., 2021). However, extensive agriculture for food supplementation and the toxic negative effect of industrial discharge are challenged by numerous milestones that restrict the continuous food supply and result in significant financial losses (Abd El-Rahim et al., 2016; Saeed et al., 2021). Plant viral infection is a dominant agriculture challenge that adversely affects yield and quality (Abdelkhalek and Hafez, 2020). Some plant viruses have a broad host range and multiple transmission mechanisms that complicate the control process and account for significant losses in several crops (Scholthof et al., 2011; Adhikari et al., 2021). Bean yellow mosaic virus (BYMV) is one of the most noteworthy plant viruses that affect more than 60 plant species in legumes and non-legumes (Chatzivassiliou, 2021). In addition, the various modes of viral transmission make BYMV a major economic threat to crop production. BYMV-infected beans are associated with mosaic, necrosis, and mottle symptoms. It can be spread by aphids, seeds, and mechanical inoculation (Ismail and Atef, 1998; El-Helaly et al., 2016).
Among the other Fabaceae members, the faba bean (Vicia faba L.) is an essential and economically important plant, severely affected by BYMV infection that directly affects the crop yield (Chatzivassiliou, 2021). Due to the highly beneficial and nutritional values of faba bean grains, they are directly utilized in the human diet in the Middle East and East Asia countries and are applied in animal feeding recipes worldwide (Duc et al., 2010; Adhikari et al., 2021). The protein content of faba bean grains ranged from 26 to 41%, representing an ideal protein source for plant-based diets (Fernández et al., 1996; Warsame et al., 2020). Despite the crop’s economic importance, its cultivation area was globally reduced to 2.6 million hectares in 2019, compared to 5.6 million hectares in 1961 (Adhikari et al., 2021). Crop losses due to faba bean sensitivity to biotic and abiotic stresses could partially explain this decline. It was found that BYMV infection caused a decrease in leaf area and flower production, as well as yellowing, mosaicing, mottling, and bending of plants, which could cause crop losses of up to 30% (Moury and Desbiez, 2020).
Developing new resistant plants and/or controlling the viral transmission vector as best as possible are more applicable approaches (Elbadry et al., 2006). Though developing new resistance cultivars through genetic engineering is an important step toward overcoming plant infection in the field, the infectious agent may manage to overcome this resistance and complicate the problem through resistance spreading (Melero-Vara et al., 2000). On the other hand, the widespread use of chemical pesticides exacerbates environmental issues by accumulating hazardous waste and negatively impacting crop quality and human health (Vejan et al., 2016). Increasing plant systemic immunity is an eco-friendly and more effective approach for plant viral control. Plant growth-promoting rhizobacteria (PGPR) have improved plant growth under various biotic and abiotic stresses (Al-Ani and Adhab, 2013; Karavidas et al., 2022; Rizvi et al., 2022). The PGPRs produce a wide range of bioactive chemicals that could induce the plant systemic resistance by activating many plant protection genes against viruses and/or enhancing the plant growth for better nutrient and water acquisition (Vejan et al., 2016; Abdelkhalek et al., 2020a,b).
Biological nitrogen fixation is a vital process in agriculture since it allows for nitrogen production through legume-rhizobia symbiosis, which contributes to rising nitrogen levels in the soil, resulting in enhanced plant growth (Moawad et al., 2005). The direct and indirect augmentation of plant growth and immunity by PGPR renders plants more resistant to surrounding stresses and infections (Saeed et al., 2021). Rhizobia, one of the PGPRs, has demonstrated a high level of nitrogen fixation in legumes and stimulating plant defense mechanisms. Al-Ani and Adhab (2013) and Elhelaly (2022) found that BYMV infection decreased root and shoot dry weights, while rhizobia inoculation increased these parameters significantly. Nonetheless, their potential as biocontrol agents, particularly antiviral agents, has received little attention. Therefore, this study aimed to evaluate the ability of Rhizobium leguminosarum bv. viciae strain 33504-Alex1 to promote faba bean growth and boost its resistance to BYMV infection either through soil inoculation or foliar application of culture filtrate (CF) to the plant leaves. Moreover, non-enzymatic oxidative stress markers [hydrogen peroxide (H2O2) and malondialdehyde (MDA)], DPPH free radical scavenging activity, total phenolic content, and reactive oxygen species scavenging enzymes [superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), and polyphenol oxidase (PPO)] were assessed. In addition, the expression of four faba bean pathogenesis-related (PR) proteins (PR-1, PR-2, PR-3, and PR-5) was also evaluated. Additionally, high-performance liquid chromatography (HPLC) and gas chromatography-mass spectrometry (GC-MS) analysis were used to figure out the metabolites in faba bean plant extract and the bioactive constituents of 33504-Alex1-CF.
Faba bean (Vicia faba L.) leaves showing BYMV-like symptoms were collected from an open field in the Alexandria governorate in 2021. The samples were checked for virus presence using a complete double antibody-sandwich enzyme-linked immunosorbent assay (DAS-ELISA) kit (DSMZ, RT-0717) as described previously (Clark and Adams, 1977). Infected faba bean leaves that reacted positively to DAS-ELISA were mechanically inoculated on Chenopodium amaranticolor leaves to induce chlorotic local lesion signs (Younes et al., 2021), which were later used as a source for BYMV. The Plant Virus RNA Kit PVR050 (Geneaid Biotech Ltd., New Taipei City, Taiwan) was used to extract viral RNA from faba bean plants. A 2 g of DNase I-treated RNA was reverse-transcribed into cDNA using Maxima Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, United States). As previously described, a PCR reaction was carried out with 2 μl of generated cDNA and BYMV-CP gene-specific primers (Table 1; Heflish et al., 2021). The PCR product was examined for specificity on a 1.5% agarose gel electrophoresis, purified with a PCR clean-up column kit, and sent for sequencing. NCBI-BLAST1 compared the annotated nucleotide sequences to sequences of previously reported BYMV isolates. The annotated sequences were then deposited in GenBank and given an accession number.
The Rhizobium isolates applied in the current work were locally isolated from faba bean root nodules collected from an open field in Alexandria governorate (Vincent, 1970). The isolates were tested for symbiotic effectiveness with faba bean plants cv. Giza 716 as described by Somasegaran and Hoben (2012). The best bacterial isolate was characterized and molecularly identified using the 16S rRNA sequencing methodology. The bacterial genomic DNA was extracted according to the manufacturer’s instructions of the Wizard Genomic DNA Purification Kit (Promega Corporation, WI, United States). The full-length 16S rRNA gene was subjected to PCR amplification using the universal 16S rRNA primers (Table 1). The purified 16S rRNA gene was subjected to identification using an automated DNA sequencer (ABI PRISM model 310, Sigma Scientific Services Company, United States). The resulting sequence was aligned through CLUSTALW (1.82) and compared with sequences in the GenBank database through BLASTN search tools at the National Center for Biotechnology Information site.2 The MEGA program generated the phylogenetic relationship (ver. 11) based upon the bootstrap neighbor-joining tree from the CLUSTALW alignment.
A pure colony of the Rhizobium isolate was inoculated in 100 ml of Yeast Extract Mannitol (YEM) broth media. The culture was grown at 28°C and shaken at 150 rpm for 48 h until it achieved 109 CFU/ml. On the other hand, the CF was obtained after spinning the bacterial culture at 10,000 rpm for 10 min. A pot experiment was conducted to evaluate the efficiency of the isolated Rhizobium isolate and its secondary metabolites for inducing systemic resistance of faba bean plants against BYMV. A virus-free seed of faba bean cultivar Giza 716 was obtained from the Agriculture Research Center, Egypt. After being surface-sterilized, the uniform seeds were grown in 40 cm plastic pots containing a sterilized equal mixture of clay, sand, and peat moss. Plants of similar sizes were chosen after 17 days of growth and distributed into four treatment groups. The first treatment (control group, G1) was faba bean plants inoculated with viral inoculation buffer and foliar sprayed with sterile non-inoculated broth medium. The second treatment (virus group, G2) was a group of plants mechanically inoculated with BYMV and foliar sprayed with a sterile non-inoculated broth medium. The third treatment (soil group, G3) includes soil inoculated plants 4 days before BYMV inoculation with the Rhizobium isolate. Each pot was inoculated with 2 ml of the culture having 109 CFU/ml. The fourth treatment (foliar group, G4) was plants treated by foliar spraying CF 24 h before inoculation with BYMV. The CF was sprayed on the whole plant shoots until the leaves looked like they were covered with it. Each treatment contained five biological replicates. Each of which consisted of a pool of 9 faba bean leaves collected from the three plants in each pot (3 leaves/plant). Each biological replicate was run in three technical replicates. The BYMV inoculation was done on the 21st day of the growth. Briefly, each plant’s two true upper leaves were carborundum-dusted (600 mesh) and mechanically inoculated with BYMV as described previously (Abdelkhalek et al., 2022). All plants were kept under insect-proof greenhouse conditions of 28/16°C (day/night) and 70% relative humidity and were observed daily for symptom development.
At 20 days post-viral inoculation (dpi), faba bean plants were harvested and evaluated in terms of plant shoot and root growth parameters and total chlorophyll content directly measured in the plant leaves through the Chlorophyll Meter SPAD-502Plus (Konica Minolta, Inc., Tokyo, Japan). Moreover, enzyme activity estimation and HPLC analysis were also estimated simultaneously. The DAS-ELISA complete kit (DSMZ, RT-0717) was used to detect BYMV and measure its concentrations within the inoculated tissues. ELISA was performed on both BYMV-challenged inoculated and non-inoculated leaves (to check systemic virus movement) of faba bean plants. The absorbance value of A405 of the sample was reported to be positive for the presence of viruses if it was double the threshold value of the healthy control samples. The viral inhibition index was measured according to El Gamal et al. (2022) as follows:
where “C” represents the mean average A405 value of untreated positive control plants (virus group, G2) and “T” represents the average A405 value of each treatment group (G3 and G4).
Disease incidence was calculated using the formula: Disease incidence (%) = Total number of infected plants/total number of plants × 100 (El Gamal et al., 2022). The disease severity (DS) was recorded using the rating scale: 0 indicates no symptoms, 1 indicates chlorotic local lesions and mild mosaic, 2 indicates severe mosaic, and 3 indicates malformation. Disease severity values were calculated using the following formula (Radwan et al., 2008).
The H2O2 titers in all treated groups were evaluated by the method of Velikova et al. (2000), with slight modification. Initially, the H2O2 was extracted by homogenizing the dried plant cells in 0.1% trichloroacetic acid (TCA) in a ratio of 1:10 (w/v), where the clear supernatant was used as an H2O2 source. Plant leaf extract (0.5 ml) was added to 1 ml of potassium iodide (1 M), 0.5 ml of potassium phosphate buffer (10 mM, pH 7.0), and vortexed briefly. After 5 min at room temperature, the reaction absorbance was measured at 390 nm, where results were deduced using a standard curve of H2O2.
According to Heath and Packer (1968), lipid peroxidation was evaluated in all plant groups regarding MDA content. First, plant leaf samples were homogenized in TCA (0.1%) in a ratio of 1:5 (w/v) ratio, where the clear supernatant was used for MDA determination. Plant leaf extract (1 ml) was added to 4 ml of TCA- thiobarbituric acid solution (20% TCA supplemented with 0.5% thiobarbituric acid) and incubated for 30 min at 95°C. Subsequently, the reaction was terminated through direct cooling into ice. The developed color was measured at 600 nm. The MDA contents were deduced from the 155/mM/cm extinction coefficient.
The free radical scavenging activity in the different treatment groups (G1–G4) was evaluated using the 2,2-Diphenyl-1-picrylhydrazyl (DPPH) method as described in Shimada et al. (1992). The scavenging rate was determined by reducing absorbance at 517 nm for 30 min in a reaction mixture that included 100 μl of plant extract (in phosphate buffer pH 7.0) and 2 ml of DPPH (0.05 M in methanol). The radical scavenging capacity results were expressed as a percentage (%) using the following equation:
where A0 is the absorbance of the control and A30 is the absorbance of the treatment extract.
The effects of Rhizobium treatments upon total phenolic compounds in faba bean plants were evaluated through the Folin–Ciocalteu method according to Velioglu et al. (1998) with some modifications. First, dried plant samples (10 mg) were extracted for 24 h with 10 ml of methanol (80%) under shaking at 100 rpm. The resulting plant leaf extracts (1 mg/ml) were added (100 μl) separately to 75 μl of the Folin–Ciocalteu reagent. After 5 min, 750 μl of Na2CO3 was added and mixed. The reaction mixture was left at room temperature for 90 min, and then the UV-Vis spectrophotometer was used to measure it at 725 nm. A standard curve of gallic acid was used to figure out the TPC.
To evaluate the enzymes responsible for antioxidant activity in all treated groups (G1–G4), plant leave samples from each treatment group were collected, dried in liquid nitrogen, and powdered in a mortar. The enzyme contents were extracted from dried plant powder in 4 volumes of phosphate buffer (100 mM, pH 7.0), supplemented with Na-EDTA (100 mM) and polyvinylpyrrolidone (1% w/v). The clear supernatant was used as a source for enzyme activity.
Superoxide dismutase activity was assayed in all treatment groups by the photocatalytic reduction method of nitro blue tetrazolium (NBT) chloride according to Beauchamp and Fridovich (1971) with minor modification. The reaction mixture included 100 μl of plant extract, 20 μM riboflavin, 75 μM NBT, 0.05 mM EDTA, and 13 mM methionine. The final volume was adjusted to 1.5 ml by 50 mM potassium phosphate buffer (pH 7.8). The photochemical reaction was initiated by two fluorescent lamps (15-W) for 15 min at 25°C, followed by incubation for 15 min under dark conditions. The color reduction was measured at 560 nm, where a 50% reduction in the NBT represents one unit of SOD activity.
Catalase (CAT) activity was evaluated in various treated plant groups by the H2O2 decomposition method according to Cakmak and Marschner (1992) with minor modification. Plant extracts (50 μl) were added separately to 10 mM H2O2 in phosphate buffer (25 mM, pH 7.0) to a final volume of 1 ml. The H2O2 concentration was measured at 240 nm, where the decay of 1 μmol/min of H2O2 represented one unit of CAT activity under the assay conditions.
Ascorbate peroxidase activity was assayed according to the method of Nakano and Asada (1981) with slight modification as follows: 100 μl of plant enzyme extract was added to 0.25 mM ascorbic acid. The final reaction volume was adjusted to 1 ml with 25 mM phosphate buffer at pH 7.0, supplemented with 0.1 mM EDTA, and initiated by adding 10 μl of 10 mM H2O2. At 290 nm, the absorbance was measured. One unit of APX activity was equal to a 0.1 change in the reaction absorbance under the assay conditions.
Polyphenol oxidase activity was determined in plant groups through the quinone method described by Cho and Ahn (1999). The plant leaves (1 g) were extracted in phosphate buffer (100 mM, pH 7.0). Plant extract (500 μl) was incubated with 1 ml of quinone (50 mM in 100 mM Tris–HCl buffer pH 6.0). After setting the reaction mixture at 25°C for 10 min, the absorbance was measured at 420 nm. One unit of the enzyme activity (μM/g of fresh weight) is equivalent to increasing the absorbance by 0.001 at the assay conditions.
The relative expression levels of four PR (PR-1, PR-2, PR-3, and PR-5) genes of faba bean were evaluated in all treatment groups through quantitative real-time PCR (qRT-PCR). Plant total RNA was extracted from leaf samples of each treatment collected at 2, 4, and 5 dpi using the RNeasy plant mini kit (QIAGEN, Germany) according to the manufacturer’s instructions and quantified by the NanoDrop UV spectrophotometer (Labtech International Ltd., Sussex, United Kingdom). The total RNA was used as a template for cDNA synthesis. As described by AbdEl-Rahim et al. (2010), DNase I-treated RNA (2 μg) was reverse transcribed to cDNA using oligo (dT) and random hexamer primers with the reverse transcriptase enzyme of Super-Script II (Invitrogen, United States). The amplified cDNA was used as a template for qRT-PCR through sets of PR primers (Table 1) with β-actin gene (housekeeping gene) to normalize the transcription levels. The qPCR reaction was conducted according to the manufacturer’s instructions of the SYBR Green PCR Master Mix (Fermentas, United States) on Rotor-Gene 6000 (QIAGEN, ABI System, United States) (Hafez et al., 2013). The relative expression level of the target gene was accurately quantified and calculated according to the 2–ΔΔCt algorithm (Livak and Schmittgen, 2001). The results were expressed as relative gene expression compared to mocked-inoculated faba bean (a value of 1 is the base), where values >1 indicate gene accumulation (upregulation) and values <1 indicate the opposite (downregulation).
The titer of different phenolic and flavonoid compounds in all faba bean plants was evaluated in leaf samples collected from different treatment groups. Air-dried and milled leaf samples (2 g) were extracted for 5 h at 35°C with shaking in 15 ml of ethanol (96%). Afterward, the extract was filtrated and vacuum dried, where the phenolic and flavonoid contents were analyzed through HPLC (Agilent 1260 Infinity HPLC series, equipped with a Zorbax Eclipse Plus C18 column 100 mm × 4.6 mm). The analysis was conducted using a mixture of 0.2% phosphoric acid, acetonitrile, and methanol as mobile phase, an injection sample of 20 μl, and a VWD detector at 284 nm. The standard polyphenolic compounds used were benzoic acid, caffeic acid, catechin, catechol, chlorogenic acid, cinnamic acid, ellagic acid, ferulic acid, gallic acid, kaempferol, myricetin, naringenin, p-coumaric acid, p-hydroxybenzoic acid, quercetin, quinol, resvertol, rosemarinic, rutin, syringic acid, and vanillic acid.
The bioactive components in the cell-free supernatant were explored and identified through gas chromatography-mass spectroscopy (GC-MS). Cell-free supernatant from a 48 h Rhizobium culture was mixed with ethyl acetate in a final ratio of 1:1 under shaking for 20 min. The aqueous phase was separated and dried under a vacuum. The resulting residues were analyzed through GC-MS (TRACE 1300 Series, Thermo, United States), using a mass detector in split mode with a helium gas carrier at a 1 ml/min flow rate. The injector temperature was 250°C, whereas the oven temperature was between 60 and 250°C for 20 min with a scanning time of 0.2 s and a range of 50–650 amu. Running for 53 min at 70 eV was enough to get the CFS mass spectra, which were then matched to the data already in the GC-MS library.
All experiments were conducted in triplicates (or more), and the means (M) with standard deviations (SD) were represented as M ± SD. The significance of the represented data was analyzed through analysis of variance (ANOVA) in CoStat software using Tukey’s honest significant differences (H.S.D.) method at a probability value (P-Value) of ≤0.05. The significant difference between groups was shown in alphabetical order, from least important to most important (a > b > c), and the same letters showed no significant difference.
Systemically mild to severe mosaic, green vein banding, leaf twisting, and distortion of the leaves were common symptoms in the field-collected faba bean samples. In DAS-ELISA tests, almost 91% of these samples tested positive for BYMV. A single local lesion developed on Ch. amaranticolor leaves at 6–7 dpi was used as a source of pure BYMV isolation. Using BYMV-CP gene primers, the RT-PCR assay gave a positive result with a 230-bp amplicon in the infected tissues. After PCR product purification and sequencing, the annotated sequence was deposited in the GenBank database under BYMV isolate BY33504-Alx2 with accession number OM863965. The NCBI-BLAST alignment and phylogenetic tree analysis showed that the BY33504-Alx2 was closely related to other BYMV isolates in GenBank, especially the Spanish isolate (Acc # JN792520), with 99% similarity (Figure 1).
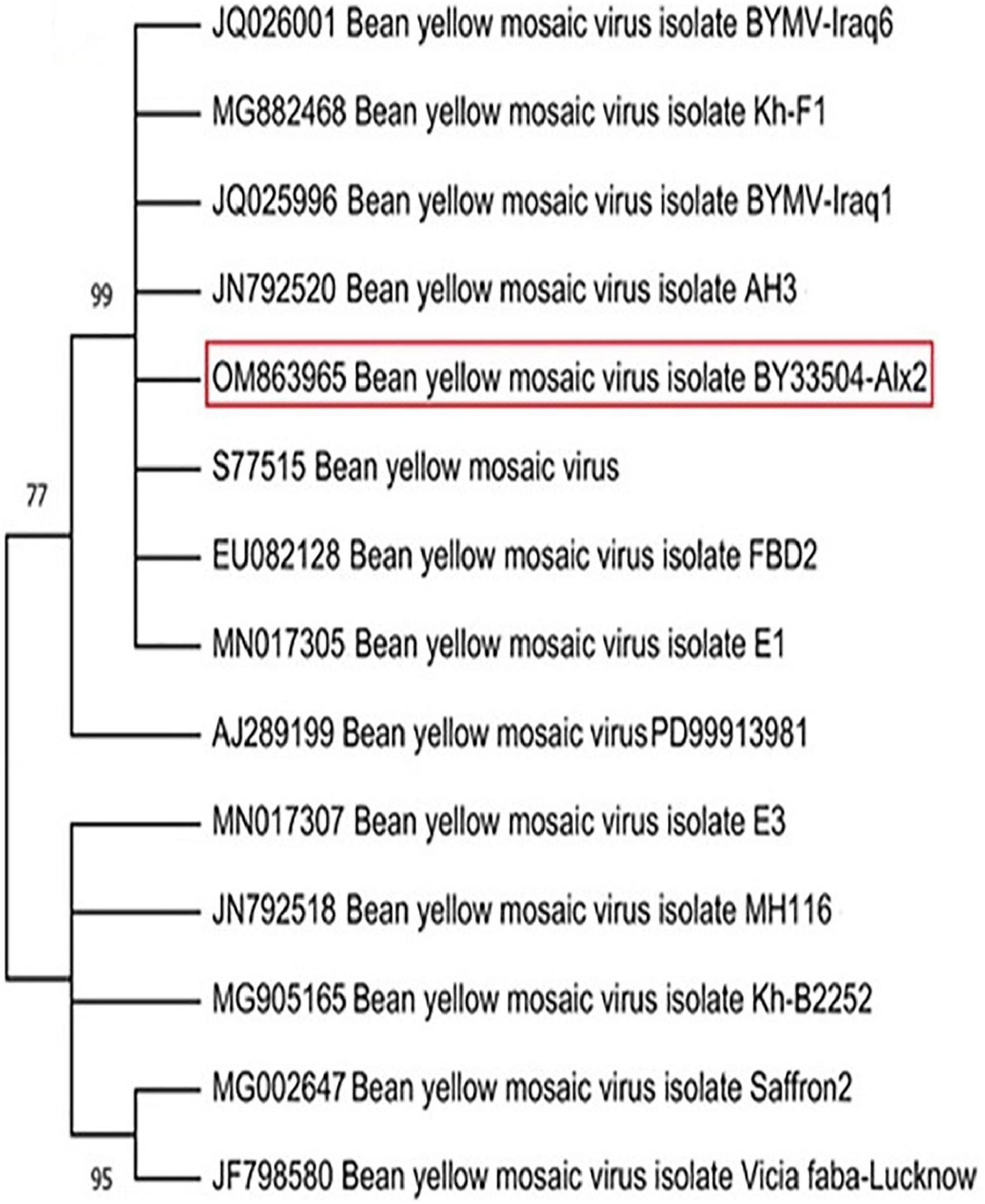
Figure 1. Phylogenetic tree illustrating the genetic relationship between the nucleotide sequence of coat protein gene of our Bean yellow mosaic virus (BYMV) isolate BY33504-Alx2 (OM863965) and other BYMV isolates obtained from GenBank. The phylogeny was created using the UPGMA statistical approach and tested using the bootstrap method with 2.000 replications.
Twenty-three rhizobia isolates were isolated from faba bean root nodules. All the isolates were Gram-negative, rod-shaped cells, and absorbed a small amount of Congo red, resulting in pale pink to white colonies. The rhizobia isolate exhibiting the most potent symbiotic effectiveness with faba bean plants was selected and subjected to characteristics. On the other hand, the nucleotide sequencing analysis of the amplified 16S rRNA gene revealed that the chosen bacterial isolate was identified as R. leguminosarum bv. viciae. The annotated sequence was deposited in the GenBank database under R. leguminosarum bv. viciae strain 33504-Alex1 with accession number OM744206. Compared to related sequences in the GenBank, the NCBI-BLAST and phylogenic tree relatedness showed that the 33504-Alex1 strain was closely related to other R. leguminosarum bv. viciae strains with 100% identity (Figure 2).
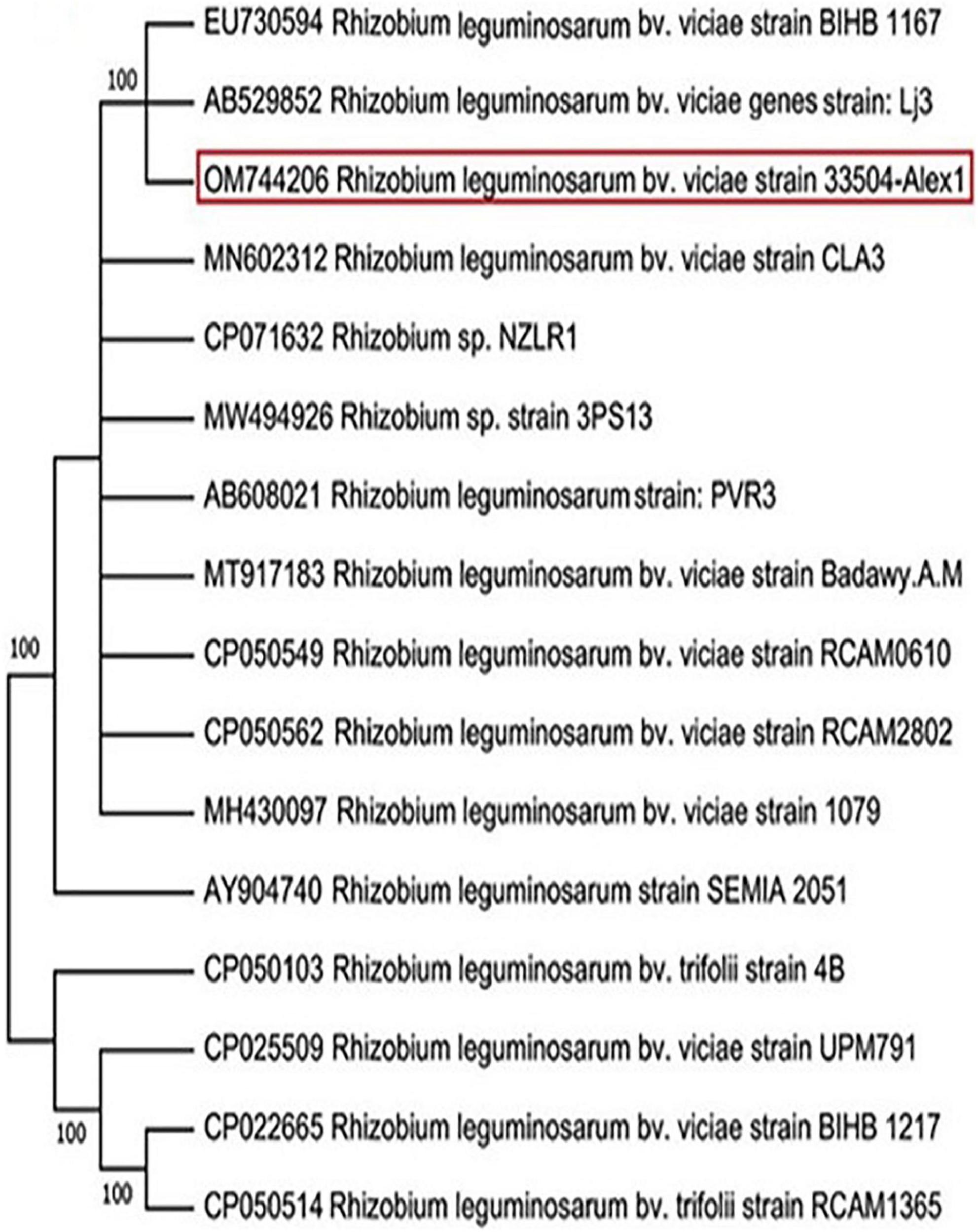
Figure 2. Phylogenetic tree illustrating the genetic link between 16s rRNA nucleotide sequence of our Rhizobium leguminosarum bv. viciae strain 33504-Alex1 (OM744206) and other Rhizobium isolates were obtained from GenBank. The phylogeny was created using the UPGMA statistical approach and tested using the bootstrap method with 2.000 replications.
The effects of Rhizobium treatment on the growth of faba bean plants were evaluated in terms of shoot and root properties. As indicated in Table 2, the plant height was remarkably reduced upon viral infection, as shown by the shoot and root length reduction in the G2 treatment compared to the control (G1), asserting the adverse impact of BYMV on faba bean growth. The soil treatment with 33504-Alex1 (G3) significantly enhanced both shoot and root length (40.31 and 25.31, respectively), representing a 1.34- and 1.65-fold increase in shoot and root length compared to the G2 treatment (30.03 and 15.31 cm, respectively). Foliar spraying of 33504-Alex1 (G4) comes next to G3 treatment in alleviating the impact of viral infection on plant growth (33.72 and 20.71 for the shoot and root length, respectively) compared to G2 treatment. The plant weight results were consistent with plant length results, where G2 treatment showed lower weights in the shoot (6.89 and 2.04 g for fresh and dry weight, respectively) and root system (5.22 and 1.27 g for fresh and dry weight, respectively) compared to the G1 group (Table 2). Compared to the G2 treatment, the G3 and G4 treatments significantly increased the fresh and dry weights of shoot and root (Table 2). On the other hand, the G4 treatment slightly boosted the growth of faba bean plants, with fresh and dry weights of shoot of 7.84 and 2.69 g and root of 6.11 and 1.62 g, respectively, though the weight gain was slightly less than the G3 treatment.
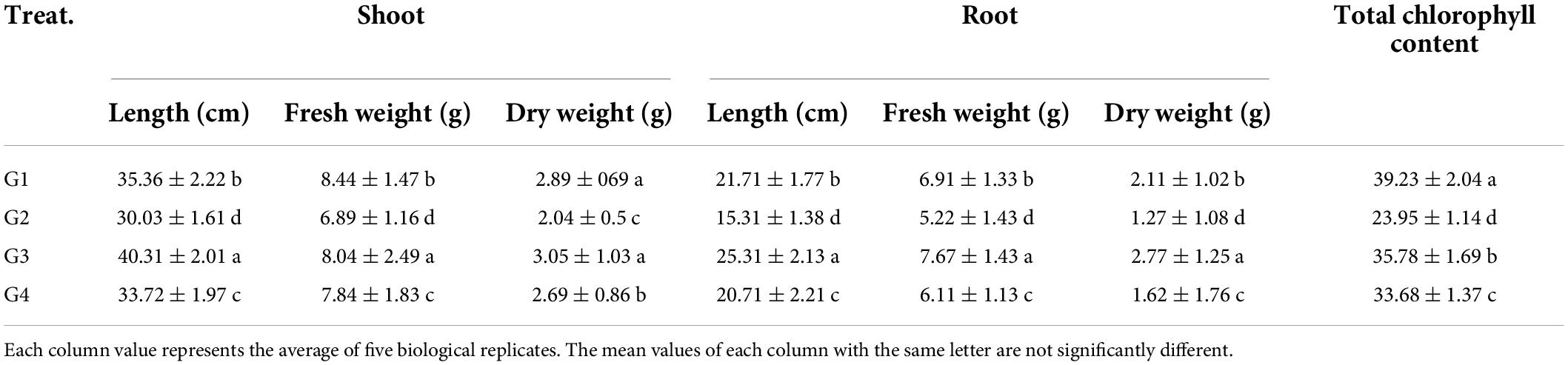
Table 2. The effect of Rhizobium leguminosarum bv. viciae strain 33504-Alex1 on faba bean growth upon infection with Bean yellow mosaic virus and total chlorophyll content.
Furthermore, the total chlorophyll content in all treatments was analyzed directly in the fresh leaf samples (Table 2). The results indicate a significant reduction in the chlorophyll content due to viral infection as indicated in the G2 treatment (23.95 SPAD unit) with a 1.61-fold decrease (about 39%) when compared to the G1 treatment (39.23 SPAD unit). On the other hand, G3 and G4 treatments enhanced the chlorophyll content with a maximum level in the G3 group of 35.78 SPAD units, followed by the G4 group of 33.68 SPAD units (Table 2). The maximum amount of chlorophyll in the treatment groups was 35.78 SPAD units, slightly less than in the G1 treatment but 1.5 times more than in the G2 treatment.
Under greenhouse conditions, the G2 treatment developed mosaic signs at 10 dpi. At 12 dpi, severe mosaic and green vein banding are clearly visible (Figure 3). Interestingly, the application of 33504-Alex1 in either soil (G3 treatment) or foliar application (G4 treatment) was associated with a delayed symptom appearance of approximately 3 days with mild symptoms at 15 dpi, compared to G2 treatment (Figure 3). No symptoms were observed during the G1 treatment. The G2 treatment reported 100% of disease incidence with a DS of 91.34% (Table 3). However, G3 treatment plants significantly reduced disease incidence and severity to 40% (6 infected plants/15 plants) and 15.46%, respectively. Moreover, G4 treatment exhibited 53.33% (8 infected plants/15 plant) disease incidence and 21.38% DS (Table 3). Furthermore, treatment with 33504-Alex1 in soil (G3) or foliar (G4) considerably increases the inhibition index of BYMV in the treatment of faba bean plants. As presented in Table 3, the highest ELISA value (1.85) was recorded in the G2 treatment, followed by the G4 and G3 treatments, with ELISA values of 0.35 and 0.49, respectively.

Figure 3. Morphological variation in faba bean leaves under Bean yellow mosaic virus (BYMV) challenge in different treatment groups, including G1: control plants, G2: BYMV-infected plants, G3: plants of soil treated with a Rhizobium isolate 4 days before BYMV inoculation, G4: plants treated by foliar spraying of culture filtrate, 24 h before inoculation with BYMV.
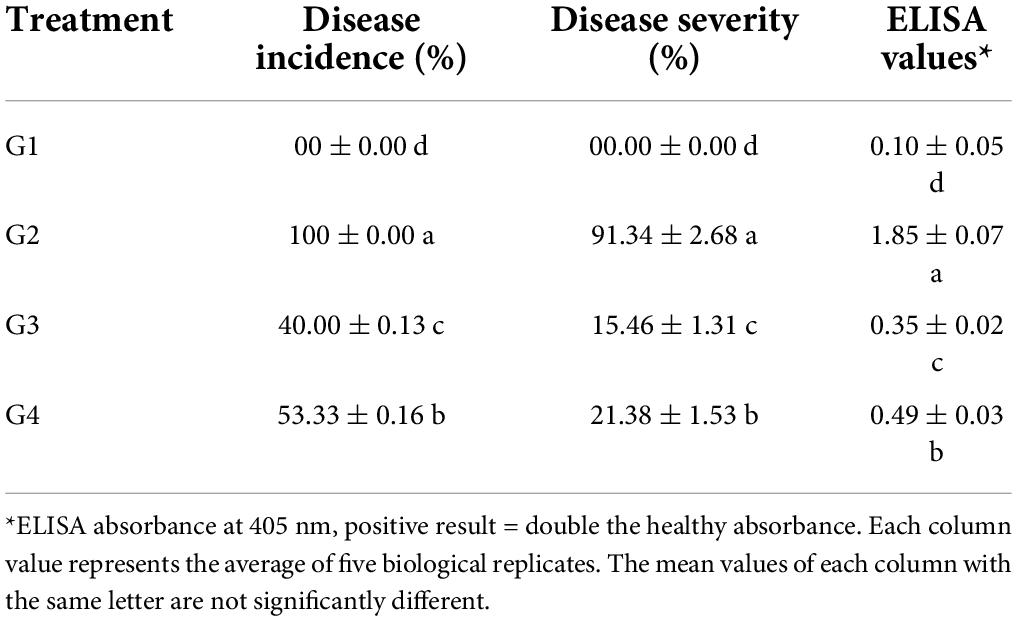
Table 3. Effect of Rhizobium treatments and Bean yellow mosaic virus (BYMV) on faba bean disease incidence, disease severity, and virus accumulation level at 21 dpi.
The two oxidative stress markers, H2O2 and MDA, were evaluated in the four faba bean treatment groups in terms of content (Figure 4). The H2O2 level was considerably elevated in the G2 treatment plants, up to 10.29 μM/g f.wt., representing about a 2.3-fold increase compared to the G1 treatment (4.48 μM/g f.wt.), as indicated in Figure 4A. Soil application of 33504-Alex1 significantly diminished the H2O2 in the G3 group (6.99 μM/g f.wt.). The foliar application of 33504-Alex1 showed the maximum H2O2 reduction with about a 59% reduction from the H2O2 level reported due to viral infection in the G2 group (Figure 4A). Similarly, the results of MDA indicate upregulated high lipid peroxidation in the G2 group (132.14 μM/g f.wt.) compared to the G1 treatment (99.98 μM/g f.wt.), which is about 32% higher (Figure 4B). On the other hand, the G3 and G4 treatments showed significant reductions in MDA levels, indicating lower lipid peroxidation (80.59 and 89.13 μM/g f.wt., respectively) compared to the G2 group. Figure 4B shows that the MDA level was lower in G3 and G4 than in G1. For G3, the reduction was 20%, and for G4, it was 11%.

Figure 4. The histogram shows the evaluation of phenolic compounds and various oxidative stress markers in faba bean plants under the Bean yellow mosaic virus (BYMV) challenge, including (A) total phenolic compounds, (B) free radical quenching activity (DPPH), (C) hydrogen peroxides (H2O2), and (D) malondialdehyde (MDA). All compounds were evaluated in the treatment groups: G1: control plants, G2: BYMV-infected plants, G3: plants of soil treated with a Rhizobium isolate 4 days before BYMV inoculation, G4: plants treated by foliar spraying of culture filtrate, 24 h before inoculation with BYMV. The columns reflect the mean of five biological replicates, while the bars represent the standard deviation (±SD). Columns with the same letter meaning do not differ significantly.
The free radical quenching activity in the different treatment groups (G1–G4) was evaluated based on the DPPH approach. As shown in Figure 4C, the results showed that the free radical scavenging was slightly more active in the G2 group (86.28%) than in the G1 group (83.05%). The G3 and G4 treatments significantly increased free radical scavenging activity, with the G3 treatment reporting the highest level at about 94.67%, representing a 10 and 14% increase over the G2 and G1 treatment groups, respectively (Figure 4C). Regarding the phenolic compounds, the results revealed a slight increase in the total phenolic content in the G2 treatment, which could be attributed to the systemic defense of faba bean plants against viral infection (Figure 4D). Figure 4D shows that treatment with 33504-Alex1 significantly increased the total phenolic content in the G3 (12.76 mg/g d.wt.) and G4 (12.71 mg/g d.wt.) groups by about 25% compared to the G1 (10.17 mg/g d.wt.) group.
Four important antioxidant enzymes, such as SOD, CAT, APX, and PPO, were measured to find out how much antioxidant activity each treatment group had. For SOD, the G3 and G4 treatments revealed significantly superior SOD production (0.807 and 0.586 μM/g f.wt., respectively) compared to the SOD level in the G1 and G2 treatments (0.493 and 0.537 μM/g f.wt.) (Figure 5A). The SOD level of the G3 treatment represents about a 1.5-fold increase compared to the control (Figure 5A). Regarding CAT, the statistical analysis revealed that no significant changes were reported between different treatments (Figure 5B). Concerning the APX activity, the data represented a significant increase in the enzyme level in the G2, G3, and G4 treatments (0.059, 0.077, 0.061 μM/g f.wt., respectively), compared to G1 (0.026 μM/g f.wt.), asserting the vital role of APX in the protection against BYMV infection (Figure 5C). The PPO activity showed that the G3 treatment exhibited maximum activity (1.54 μM/g f.wt.), representing 2.5- and 2.3-fold significant increases compared to G1 and G2, respectively (Figure 5D).
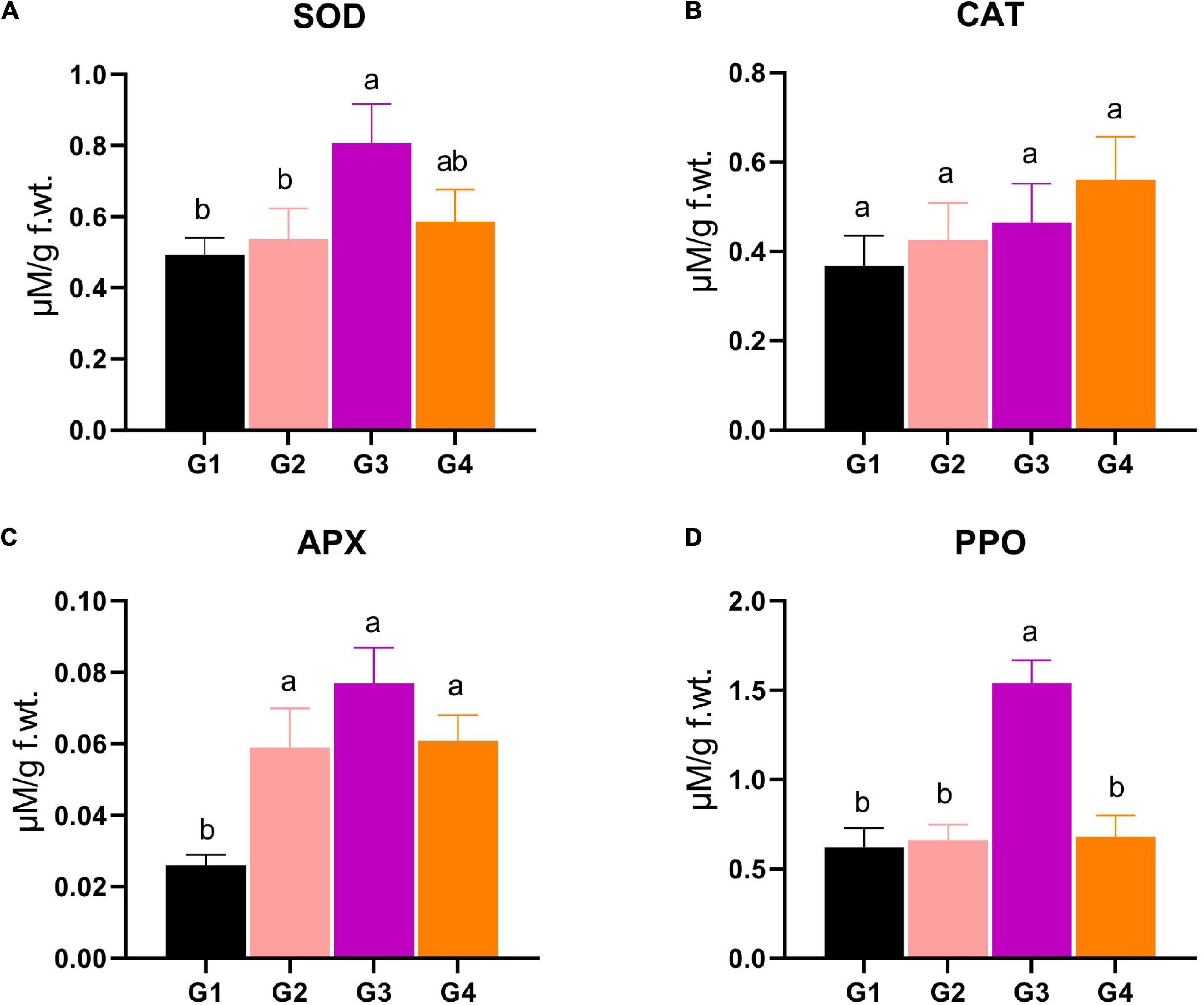
Figure 5. Evaluation of various antioxidant enzymes production, including (A) superoxide dismutase (SOD), (B) catalase (CAT), (C) ascorbate peroxidase (APX), and (D) polyphenol oxidase (PPO) in Bean yellow mosaic virus (BYMV)-infected fab bean plants: G1: control plants, G2: BYMV-infected plants, G3: plants of soil treated with a Rhizobium isolate 4 days before BYMV inoculation, G4: plants treated by foliar spraying of culture filtrate, 24 h before inoculation with BYMV. The columns reflect the mean of five biological replicates, while the bars represent the standard deviation (±SD). Columns with the same letter meaning do not differ significantly.
The expression of four faba bean PRs (PR-1, PR-2, PR-3, and PR-5) was evaluated in all treatment groups after 2, 4, and 5 dpi (Figure 6). For PR-1 expression level, the G2 treatment significantly reduced its transcriptional level at 2 and 4 dpi, with a maximum reduction level at 4 dpi, reporting a relative transcriptional level of 0.36-fold change lower than the G1 treatment. Interestingly, soil treatment with 33504-Alex1 (G3) significantly enhanced the PR-1 overexpression from 2 dpi, with a maximum level detected at 5 dpi, representing a 2.41-fold increase compared to the G1 treatment (Figure 6A). On the other hand, the foliar spraying of 33504-Alex1-CF was the most potent upregulator for the PR-1 gene, with relative expression levels of 1.44, 2.48, and 2.54-fold change higher than control at 2, 4, and 5 dpi, respectively (Figure 6A). The results indicated nearly equal PR-1 induction potential for G3 and G4 at 2 and 5 dpi. However, the 4 dpi results showed that foliar application of 33504-Alex1-CF increased the PR-1 expression level. This means that spraying 33504-Alex1-CF has a faster effect on increasing plant resistance by over-activating the PR-1 gene.
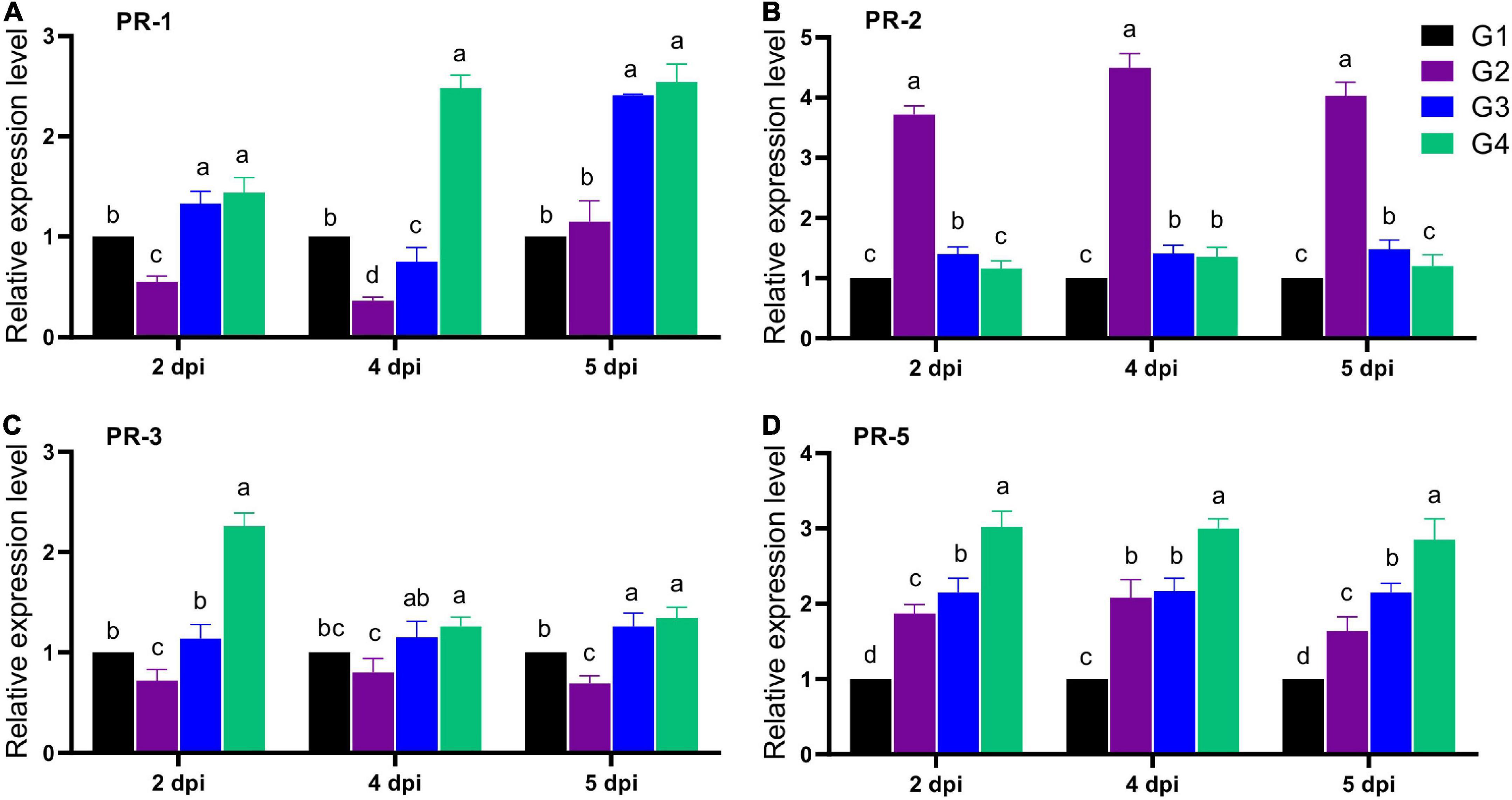
Figure 6. The effect of Rhizobium cells and supernatant treatment upon the relative expression levels of four pathogen-related genes after 2, 4, and 5 dpi of Bean yellow mosaic virus (BYMV) infection for faba bean plants, including (A) PR-I, (B) PR-2, (C) PR-2, and (D) PR-5 genes. G1: control plants, G2: BYMV-infected plants, G3: plants of soil treated with a Rhizobium isolate 4 days before BYMV inoculation, G4: plants treated by foliar spraying of culture filtrate, 24 h before inoculation with BYMV. The columns reflect the mean of five biological replicates, while the bars represent the standard deviation (±SD). Columns with the same letter meaning do not differ significantly.
The second evaluated gene was the PR-2 gene. The G2 treatment showed significant up-regulation in the PR-2 expression level with a 3.72- and 4.03-fold increase in the gene expression at 2 and 5 dpi. The maximum level was at 4 dpi, with a relative expression level of 4.49-fold higher than the control (Figure 6B). Compared to the G2 treatment, the G3 and G4 treatment groups showed a significant reduction in PR-2 expression levels with values nearly the control (Figure 6B). Treatment with 33504-Alex1 CF (G4) was slightly better at reducing PR-2 than the G3 treatment. The G3 treatment showed a slight increase in PR-2 expression levels, about 1.40-fold higher than the control at all dpi (Figure 6B). Regarding PR-3, the qPCR results indicated a significant reduction in PR-3 gene expression in the G2 treatment by about 26% at all dpi compared to the G1 treatment (Figure 6C). At 2 dpi, the foliar application of 33504-Alex1-CF significantly increased the PR-3 expression level by 2.26-fold compared to the G1 treatment (Figure 6C). At 5 days post-infection (dpi), the PR-3 transcript levels were about 1.3 times higher in the G3 and G4 groups than in the control group (Figure 6C). Like the PR-3 transcript profile, the G4 treatment exhibited the highest transcriptional levels of PR-5 with relative expression levels of 3. 02-, 3. 01-, and 2.85-fold change higher than control at 2, 4, and 5 dpi, respectively (Figure 6D). Also, the G2 (1. 87-, 2. 08-, and 1.64-fold) and G3 (2. 15-, 2. 17-, and 2.15-fold) treatments significantly increased PR-5 transcriptional levels at 2, 4, and 5 dpi, respectively (Figure 6D).
The variations in the phytochemical constituents within the different faba bean treatment groups (G1-G4) were evaluated in the leaf samples through HPLC after 20 dpi. The results showed that the other treatment groups had different amounts of polyphenolic compounds. The total amount of the 19 polyphenolic compounds found was 115,405 mg/kg for G1, 65,621.8 mg/kg for G2, 83,499.3 mg/kg for G3, and 60,998.9 mg/kg for G4. The major detected compounds were benzoic acid, rutin, ellagic, rosemarinic, and quercetin, with more than 1,000 mg/kg (Figure 7). The HPLC analysis revealed that the four flavonoid compounds, pyrogallol, neringein, myricetin, and kaempferol, were induced after BYMV inoculation (Figure 7). The fact that this compound was found in all virus-infected groups (G2, G3, and G4) but not in the control group showed that it was part of a plant’s defense against virus infection. On the other hand, the suppressor activity of BYMV inhibited the accumulation of gallic and vanillic acids in the G2 treatment. At the same time, they were detected in the G1 control group (130.42 and 265.67 mg/kg, respectively) and the treatment groups G3 and G4 (150.49 and 205.17 for gallic acid and 178.68 and 94.73 for vanillic acid in both groups, respectively). Interestingly, the application of 33504-Alex1 led to the production of cinnamic acid, which was different from the G1 and G2 treatments (Figure 7). Moreover, syringic acid was exclusively detected in the G2 treatment.
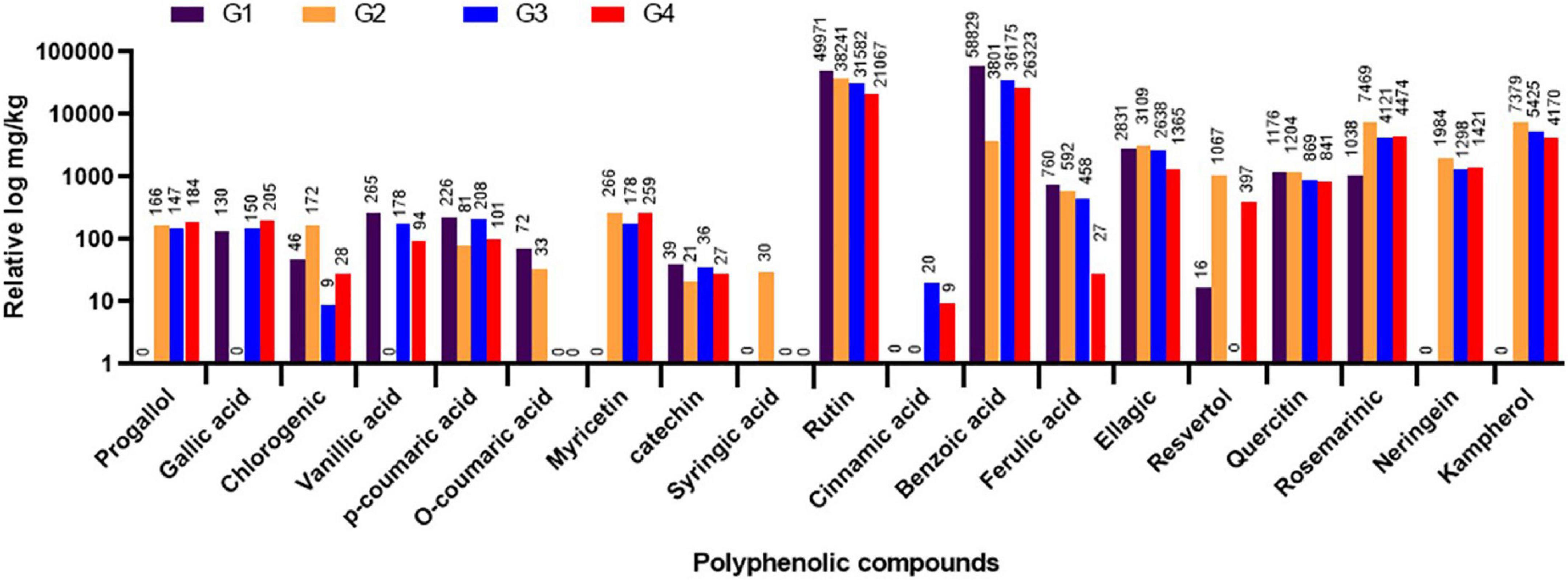
Figure 7. The high-performance liquid chromatography (HPLC) analysis of different polyphenolic and flavonoids in leaf samples of faba bean as detected at 20 dpi. G1: control plants, G2: Bean yellow mosaic virus (BYMV)-infected plants, G3: plants of soil treated with a Rhizobium isolate 4 days before BYMV inoculation, G4: plants treated by foliar spraying of culture filtrate, 24 h before inoculation with BYMV. The results were represented with the relative log, where compound values were represented above each column.
The bioactive components of 33504-Alex1-CF were elucidated in the ethyl acetate extracted supernatant through GC-MS analysis. Figure 8 shows that the result showed that there were several compounds because there were many peaks at different retention times (RT). In Table 4, 17 detected components are listed. These include 3,6-octadecadiynoic acid, methyl ester; octanoic acid, ethyl ester; 4-trifluoroacetoxytetradecane; 5-heptenoic acid, 6-methyl-4-[(4-ethylphenyl) sulfonyl]; methyltricyclo[6.5.2(13,14)0.0(7,15)]pentadeca-1,3,5,7,9,11,13-heptene; 1-hexadecanol, 2-methyl-; 1,3,5-triazine-2,4-diamine, 6-chloro-N-ethyl-; (3-methyl-1,4-diphenylbicyclo[2.2.0]hex-2- yl)methanol; isochiapin B; tetraneurin-A-Diol; hexadecanoic acid 2,3-dihydroxypropyl ester; oleic acid; dotriacontane; 2,2,3,3,4,4 hexadeutero octadecanal; 9,12,15-octadecatrienoic acid; 4,7-octadecadiynoic acid, methyl ester; diisooctyl phthalate were detected at different RT. The GC-MS analysis also revealed the presence of retinal (aldehyde form of vitamin A) at RT of 21.26 min and several bioactive fatty acids, including oleic acid at RT of 26.95, cis-11-eicosenoic acid at RT of 26.28 min, and hexadecanoic acid 2,3-dihydroxypropyl ester at RT of 26.71 min.
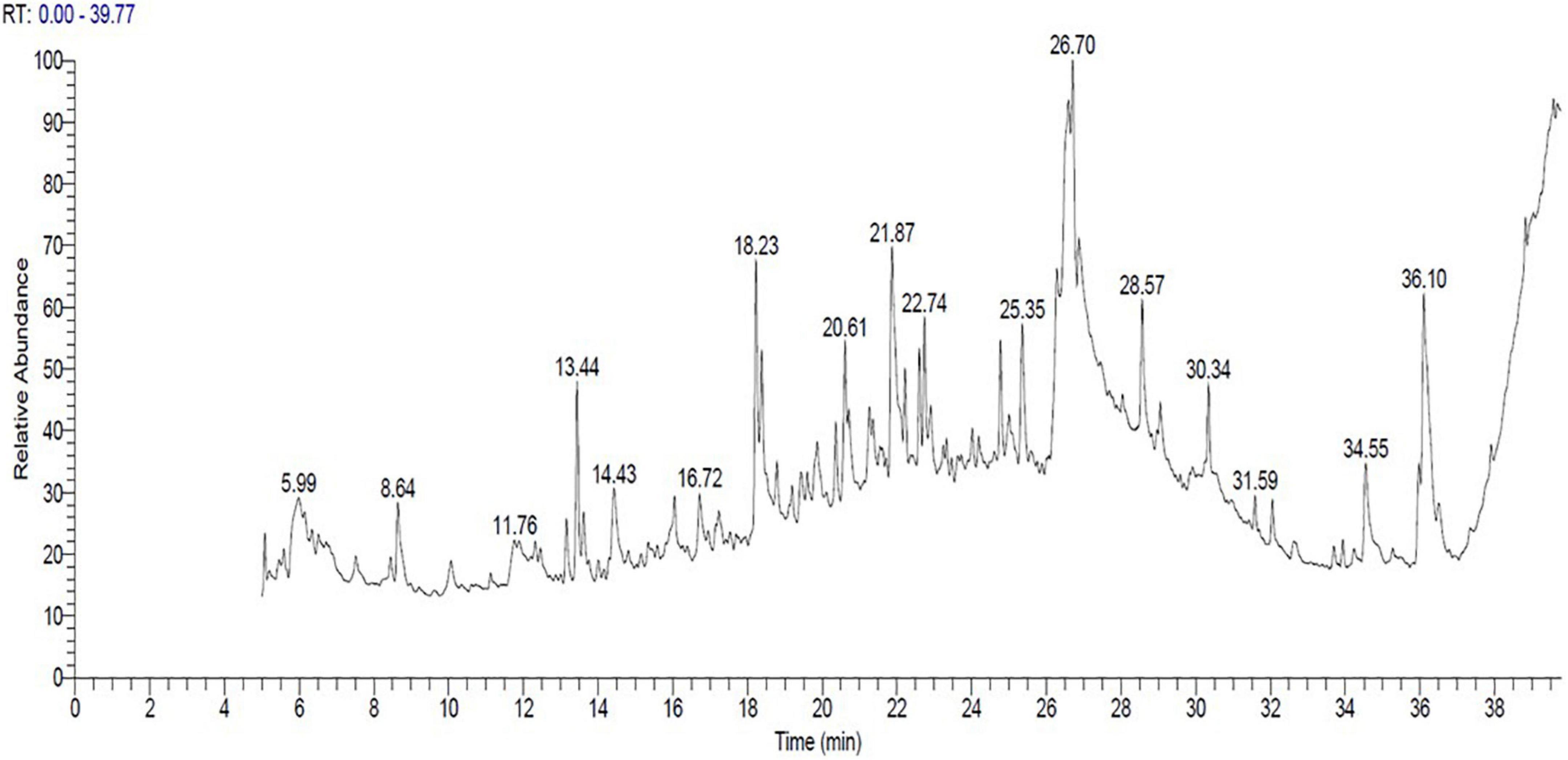
Figure 8. The cell-free supernatant chromatogram for Rhizobium leguminosarum bv. viciae strain 33504-Alex1 ethyl acetate extract constituents as elucidated with gas chromatography-mass spectrometry (GC-MS).
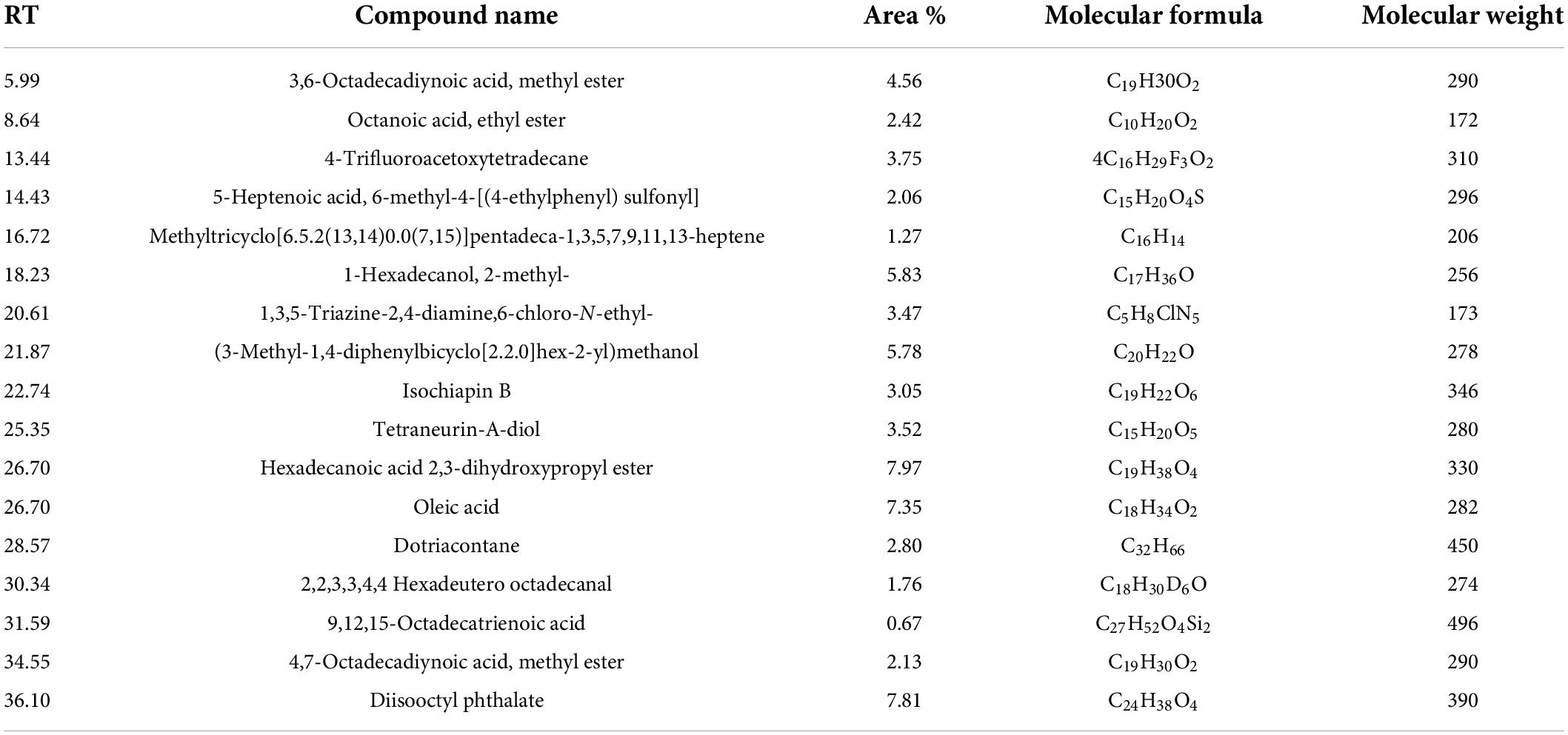
Table 4. The constituents of Rhizobium leguminosarum cell-free supernatant as elucidated through gas chromatography-mass spectrometry (GC-MS) analysis.
Legume crops are an essential source of food, nutrition, and industrial materials for humans, animals, and the environment (Moawad et al., 2004). Many phytohormones, such as brassinosteroids, cytokinins, enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase, ethylene, gibberellins, IAA, jasmonates, lipo-chito-oligossacharide Nod factors, lumichrome, rhizobitoxine, and riboflavin, were synthesized and released during the symbiotic interaction process between rhizobia and legumes (Jaiswal et al., 2021). Such compounds can directly or indirectly stimulate plant growth and may promote plant adaptation to various stresses (Purwaningsih et al., 2021). Rhizobium is a natural PGPR in the rhizosphere used in farming to enhance legume crops’ growth through mutually beneficial relationships (Allito et al., 2021; Boivin et al., 2021). This way, finding new biocontrol agents that are safe for the environment, control plant viruses, and fix nitrogen is a top priority for food security.
Under greenhouse conditions, R. leguminosarum bv. viciae strain 33504-Alex1 was applied to alleviate the BYMV infection in faba beans. The results showed that the growth parameters of faba beans significantly improved when 33504-Alex1 was applied to the soil (G3 treatment) or the leaves (G4 treatment). The soil treatment had a more considerable effect than the foliar treatment. The importance of 33504-Alex1 in plant growth enhancement can be attributed to PGPR’s direct effect on increasing nutrient availability, production of plant growth-promoting phytohormones, and water uptake (Elhelaly, 2022; Rizvi et al., 2022). The total chlorophyll content was reduced in the BYMV-challenged non-treated plants (G2 treatment), which is in accordance with Sofy et al. (2020), who reported a decrease in the chlorophyll by about 43% in faba bean plants under the BYMV challenge compared to control. The chlorophyll reduction could be attributed to the direct adverse effect of BYMV on chloroplast functions (Radwan et al., 2008). Interestingly, the application of 33504-Alex1 significantly increased chlorophyll content by 49.39 and 40.62% in G3 and G4, respectively, compared to the G2 treatment. On the other hand, the results showed that 33504-Alex1 could protect faba bean plants from BYMV by lowering disease incidence (up to 60%), decreasing its severity (up to 83%), and increasing inhibition index (up to 81%) in treated plant tissues. These results were in harmony with those obtained by Elhelaly (2022), who showed that the treatment of faba bean seeds with R. leguminosarum significantly decreased the disease incidence of BYMV infection compared to untreated seeds. The significant increase in the inhibition index of BYMV confirmed the protective efficacy of 33504-Alex1 against BYMV infestation. Thus, using 33504-Alex1 could boost the host’s innate immune system or cause SAR, preventing BYMV from spreading or inhibiting its replication.
The elevation of the two oxidative stress markers, H2O2 and MDA, was consistent with the common BYMV-plant infection feature combined with a high titer of reactive oxygen species (Radwan et al., 2010; Sofy et al., 2020). Lipid peroxidation (MDA) directly indicates membrane disruption and deterioration of the plant cells due to intolerable oxidative stress (Loreto and Velikova, 2001; Alché, 2019). Both markers have increased in the G2 group, whereas they have been significantly reduced in the G3 and G4 groups. The results were similar to Hamzah et al. (2021), who reported the induction of MDA in faba bean plants upon inoculation with BYMV. The positive effects of rhizobia are mediated by creating various compounds and enzymes directly or indirectly induced by rhizobia and plants during nodule formation (Jaiswal et al., 2021). As a result, the 33504-Alex1 application may increase phenolic compounds and free radicals while activating the non-enzymatic pathway to reduce the risk of oxidative stress. The results were similar to those reported by Hamzah et al. (2021), who reported that the inoculation of faba bean plants with rhizobia was associated with an increase in total phenolic content compared to non-treated plants. In the same regard, the antioxidant enzyme results revealed a slight increase in all enzymes under the current study in plants under BYMV challenge (G2), which could be related to the activation of the faba bean defense system as an initial response to BYMV infection. However, the antioxidant enzyme levels in G2 were insufficient to alleviate the adverse effects of the reactive oxygen species on the plant, as indicated by the lower growth parameters and higher oxidative stress markers in the G2 treatment results. The generation of reactive oxygen species is well documented under several viral infections due to the impairment of the electron transport chain (Chandra et al., 2015; Sofy et al., 2020); hence, the overproduction of the antioxidant enzyme is crucial for mitigating oxidative stress. When 33504-Alex1 was used, the antioxidant enzymes’ production increased drastically, and soil treatment was much better than foliar treatment in all enzyme assays.
Pathogenesis-related genes encode essential plant defense proteins in response to several biotic and abiotic stressors. The expression levels of four PR genes were analyzed to investigate the molecular processes underlying faba bean resistance to BYMV infection following treatment with 33504-Alex1. The PR-1 expression level was significantly reduced due to viral infection, with up to a 64% decrease at 4 dpi compared to the G1 treatment. Notably, the G3 or G4 treatments significantly increased PR-1 overexpression, with a maximum level detected at 5 dpi. The results are consistent with the reported role of the PR-1 gene in mitigating BYMV and other mosaic viruses’ infection (Sofy et al., 2020; Abdelkhalek et al., 2021). On the contrary, the PR-2 gene was significantly elevated in the G2 treatment, with a maximum relative expression level of 4.49-fold increase at 4 dpi compared to the G1 treatment. The PR-2 gene encodes a β-1,3-glucanase protein, an essential enzyme in cell-to-cell translocation and signaling. This enzyme hydrolyzes the callose deposits between plant cells, so its overexpression is necessary for most viruses to facilitate cell-cell viral translocation (Kawakami et al., 2004; Oide et al., 2013; Otulak-Kozieł et al., 2018). Interestingly, G3 and G4 treatments revealed a significant reduction in PR-2 expression levels, nearly to the G1 level. The foliar application of 33504-Alex1-CF on the leaves was the most effective way to reduce PR-2 expression, with a 3.2-fold drop compared to the G2 treatment.
Furthermore, the PR-5 (Thaumatin-like proteins) was upregulated upon treatment with 33504-Alex1. The results are consistent with the reported role of PR-5 in improving the plant’s resistance to numerous infections (Zhang et al., 2017). In the same line with PR-1 and PR-3 gene results, the 33504-Alex1-CF foliar spraying was most effective in inducing PR-5 accumulation, which is composed of the accumulation of PR proteins in the infected and surrounding plant tissue. Hence, foliar treatment is more logical for infected plants (Chandrashekar et al., 2018). The PR-1 and PR-5 gene expression are regulated by salicylic acid (SA), which is the primary mediator of the systemic acquired resistance (SAR) mechanism (Breen et al., 2017; Wang et al., 2021). Thus, the results showed that 33504-Alex1 could boost SA-mediated SAR in faba bean when infected with BYMV. Ali et al. (2018) found that jasmonic acid (JA) and ethylene are the main mediators of induced systemic resistance. JA and ethylene control the PR-3 gene that codes for chitinase activity. In the current study, the PR-3 was significantly activated in G4 treatment at 2 dpi. According to the TMV infection study (Zhu et al., 2014), the slight induction of JA at the beginning is important for the later buildup of SA. Despite its role in SA accumulation, evidence for JA’s direct participation in plant resistance to viral challenges is inconclusive (Oka et al., 2013; Zhao and Li, 2021). The PR-3 gene upregulation is widely reported as a part of plant resistance under fungal infection (Eslahi et al., 2021; Heflish et al., 2021), with a limited role in plant viral infection (Abo-Zaid et al., 2020). This could explain why its level dropped in the BYMV-challenged plants at 4 and 5 dpi.
The HPLC analysis demonstrated that the four flavonoid compounds, namely, pyrogallol, neringein, myricetin, and kaempferol, were induced following BYMV inoculation. Kaempferol, neringein, and myricetin are powerful antioxidants, so they could be made more during viral infection to help with the higher oxidative stress (Marín et al., 2018). It was reported that myricetin could directly interact with viral glycoprotein D to prevent its adsorption and membrane fusion to host cells, thereby inhibiting viral infection and replication (Li et al., 2020). Myricetin reportedly inhibited Zika virus replication and African swine fever virus protease (Jo et al., 2020; Zou et al., 2020). Kaempferol is one of the important members of the flavonol subgroup of flavonoids. It is a critical component of the auxin-dependent defense response that prevents plant viruses from spreading systemically. This defense response is engaged before the well-known SA-dependent defense response (Likic̀ et al., 2014). These compounds were detected only in BYMV-inoculated plants, not control plants. This suggests that they play a big role in plant defense against viruses. Interestingly, gallic acid and vanillic acid were suppressed in the G2 treatment, which could be attributed to their role as natural polyphenolics with reported antioxidant, antiviral, and antibacterial activities (Sethupathy et al., 2017; Bai et al., 2021). Gallic acid revealed excellent antibacterial and antiviral activity against Salmonella and herpes viruses (Shahidi and Yeo, 2018). Based on the findings, we hypothesized that these two compounds, gallic and vanillic acids, may have antiviral activity against BYMV infection. However, more research is needed to confirm this hypothesis. Benzoic acid is an important building block for plants’ phytohormones, enzymes, and cofactors (Widhalm and Dudareva, 2015). Benzoic acid and rutin were the most detected polyphenolic compounds in all treatments. Interestingly, the application of 33504-Alex1 resulted in the synthesis of cinnamic acid, which was distinct from the G1 and G2 treatments. Moreover, syringic acid was exclusively found in the G2 treatment.
The GC-MS analysis indicated several bioactive molecules, including 17 compounds of polyphenolic, flavonoids, and fatty acids. Isochiapin-B was among the most detected compounds in the 33504-Alex1-CF and is a volatile terpenoid compound with several biological activities. Isochiapin B was detected in the Achillea fragmmentissma and Citrus aurantium plant extracts with potent anti-inflammatory, antimicrobial, and antioxidant activities (Elsharkawy, 2016; Değirmenci and Erkurt, 2020). Hexadecanoic acid 2,3-dihydroxypropyl ester (known as palmitate) is a biologically active fatty acid with reported antitumor activity against human leukemia cells, with no side effects or cytotoxicity to normal human dermal fibroblast (Al-Wahaibi et al., 2020). It was reported that the broad-spectrum antimicrobial activity of Conocarpus lancifolius leaf extract and its antitumor activity against breast cancer might be attributed to the presence of hexadecanoic acid, 2,3-dihydroxypropyl ester (Moni et al., 2021). Furthermore, oleic acids are biologically active monounsaturated fatty acids with reported antibacterial, antioxidant, and antiviral activities (Dilika et al., 2000; Wei et al., 2016; Zhao et al., 2017). Dotriacontane and tetraneurin-A-diol were also found in the GC-MS profile. Dotriacontane has high antimicrobial and antioxidant activities (Asong et al., 2019), while tetraneurin-A-diol has pesticide activity and has been found in many plants and microbial extracts (Abdelhamid et al., 2015). As a result, the GC–MS analysis indicated the existence of many active compounds with different biological activities in the 33504-Alex1-CF. The current study indicated that 33504-Alex1-CF has considerable antiviral activity against BYMV and growth-stimulating effects on faba bean plants. Consequently, 33504-Alex1-CF may be effective as a preventative biocontrol agent against plant viral infection. However, additional research is required to confirm potential field applications.
Under experimental greenhouse conditions, the R. leguminosarum bv. viciae strain 33504-Alex1 isolated from faba bean root nodules seems to be a promising inducer for systemic resistance in faba bean against challenge BYMV infection. Compared to non-treated faba bean plants, the application of 33504-Alex1 in either soil or foliar application was associated with lowering disease incidence (up to 60%), decreasing DS (up to 83%), and increasing inhibition index (up to 81%), as well as promoting growth and increasing the total chlorophyll content. It also reduced H2O2 and MDA, boosted DPPH and total phenolic content, and antioxidant enzymes and PR transcription levels. Moreover, the most common phenolic compounds were found to be activated and build up, except for gallic and vanillic acids, which were completely turned off in non-treated plants. These results could be very important since they may offer a simple, environmentally safe, and economically accepted means to protect faba bean plants from BYMV infection. However, additional studies are needed to confirm these results under field conditions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
AA, HE-G, and SB designed the research, wrote the manuscript, performed the experiments, and analyzed the data. AA-A, VM, HM, ME, and HY helped with editing and provided suggestions for the experiments. All authors contributed to the article and approved the submitted version.
This work was partially funded by the Science and Technology Development Fund (STDF), Egypt (Grant No. 33504). This research was financially supported by the Researchers Supporting Project number (RSP2022R505), King Saud University, Riyadh, Saudi Arabia.
This paper was based upon work supported by Science, Technology & Innovation Funding Authority (STDF) under the grant (No. 33504). The authors would like to extend their appreciation to the Researchers Supporting Project number (RSP2022R505), King Saud University, Riyadh, Saudi Arabia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abd El-Rahim, W. M., Mostafa, E. M., and Moawad, H. (2016). High cell density cultivation of six fungal strains efficient in azo dye bioremediation. Biotechnol. Rep. 12, 1–5. doi: 10.1016/j.btre.2016.08.001
Abdelhamid, M. S., Kondratenko, E. I., and Lomteva, N. A. (2015). GC-MS analysis of phytocomponents in the ethanolic extract of Nelumbo nucifera seeds from Russia. J. Appl. Pharm. Sci. 5, 115–118. doi: 10.7324/JAPS.2015.50419
Abdelkhalek, A., Al-Askar, A. A., Alsubaie, M. M., and Behiry, S. I. (2021). First report of protective activity of paronychia argentea extract against tobacco mosaic virus infection. Plants 10:2435. doi: 10.3390/plants10112435
Abdelkhalek, A., Al-Askar, A. A., Arishi, A. A., and Behiry, S. I. (2022). Trichoderma hamatum Strain Th23 Promotes Tomato Growth and Induces Systemic Resistance against Tobacco Mosaic Virus. J. Fungi 8:228. doi: 10.3390/jof8030228
Abdelkhalek, A., Al-Askar, A. A., and Behiry, S. I. (2020a). Bacillus licheniformis strain POT1 mediated polyphenol biosynthetic pathways genes activation and systemic resistance in potato plants against Alfalfa mosaic virus. Sci. Rep. 10:16120. doi: 10.1038/s41598-020-72676-2
Abdelkhalek, A., Behiry, S. I., and Al-Askar, A. A. (2020b). Bacillus velezensis pea1 inhibits fusarium oxysporum growth and induces systemic resistance to cucumber mosaic virus. Agronomy 10:1312. doi: 10.3390/agronomy10091312
Abdelkhalek, A., and Hafez, E. (2020). “Plant Viral Diseases in Egypt and Their Control,” in Cottage Industry of Biocontrol Agents and Their Applications, eds N. El-Wakeil, M. Saleh, and M. Abu-hashim (Cham: Springer), 403–421. doi: 10.1007/978-3-030-33161-0_13
AbdEl-Rahim, W. M., Khalil, W. K. B., and Eshak, M. G. (2010). Evaluation of the gene expression changes in Nile tilapia (Oreochromis niloticus) as affected by the bio-removal of toxic textile dyes from aqueous solution in small-scale bioreactor. Environmentalist 30, 242–253. doi: 10.1007/s10669-010-9268-7
Abo-Zaid, G. A., Matar, S. M., and Abdelkhalek, A. (2020). Induction of plant resistance against tobacco mosaic virus using the biocontrol agent streptomyces cellulosae isolate actino 48. Agronomy 10:1620. doi: 10.3390/agronomy10111620
Adhikari, K. N., Khazaei, H., Ghaouti, L., Maalouf, F., Vandenberg, A., Link, W., et al. (2021). Conventional and molecular breeding tools for accelerating genetic gain in faba bean (Vicia Faba L.). Front. Plant Sci. 12:744259. doi: 10.3389/fpls.2021.744259
Al-Ani, R. A., and Adhab, M. A. (2013). Bean Yellow Mosaic Virus (BYMV) on broadbean: Characterization and resistance induced by Rhizobium leguminosarum. J. Pure Appl. Microbiol. 7, 135–142.
Alché, J. D. (2019). A concise appraisal of lipid oxidation and lipoxidation in higher plants. Redox Biol. 23:101136. doi: 10.1016/j.redox.2019.101136
Ali, S., Ganai, B. A., Kamili, A. N., Bhat, A. A., Mir, Z. A., Bhat, J. A., et al. (2018). Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 212, 29–37. doi: 10.1016/j.micres.2018.04.008
Allito, B. B., Ewusi-Mensah, N., Logah, V., and Hunegnaw, D. K. (2021). Legume-Rhizobium specificity effect on nodulation, biomass production and partitioning of faba bean (Vicia faba L.). Sci. Rep. 11:3678. doi: 10.1038/s41598-021-83235-8
Al-Wahaibi, L. H. N., Mahmood, A., Khan, M., and Alkhathlan, H. Z. (2020). Comparative study on the essential oils of Artemisia judaica and A. herba-alba from Saudi Arabia. Arab. J. Chem. 13, 2053–2065. doi: 10.1016/j.arabjc.2018.03.004
Asong, J. A., Amoo, S. O., McGaw, L. J., Nkadimeng, S. M., Aremu, A. O., and Otang-Mbeng, W. (2019). Antimicrobial activity, antioxidant potential, cytotoxicity and phytochemical profiling of four plants locally used against skin diseases. Plants 8:350. doi: 10.3390/plants8090350
Bai, J., Zhang, Y., Tang, C., Hou, Y., Ai, X., Chen, X., et al. (2021). Gallic acid: pharmacological activities and molecular mechanisms involved in inflammation-related diseases. Biomed. Pharmacother. 133:110985. doi: 10.1016/j.biopha.2020.110985
Beauchamp, C., and Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. doi: 10.1016/0003-2697(71)90370-8
Boivin, S., Mahé, F., Debellé, F., Pervent, M., Tancelin, M., Tauzin, M., et al. (2021). Genetic variation in host-specific competitiveness of the symbiont rhizobium leguminosarum symbiovar viciae. Front. Plant Sci. 12:719987. doi: 10.3389/fpls.2021.719987
Breen, S., Williams, S. J., Outram, M., Kobe, B., and Solomon, P. S. (2017). Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 22, 871–879. doi: 10.1016/j.tplants.2017.06.013
Cakmak, I., and Marschner, H. (1992). Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 98, 1222–1227. doi: 10.1104/pp.98.4.1222
Chandra, S., Chakraborty, N., Dasgupta, A., Sarkar, J., Panda, K., and Acharya, K. (2015). Chitosan nanoparticles: a positive modulator of innate immune responses in plants. Sci. Rep. 5:15195. doi: 10.1038/srep15195
Chandrashekar, N., Ali, S., and Grover, A. (2018). Exploring expression patterns of PR-1, PR-2, PR-3, and PR-12 like genes in Arabidopsis thaliana upon Alternaria brassicae inoculation. 3 Biotech 8:230. doi: 10.1007/s13205-018-1259-2
Chatzivassiliou, E. K. (2021). An annotated list of legume-infecting viruses in the light of metagenomics. Plants 10:1413. doi: 10.3390/plants10071413
Cho, Y. K., and Ahn, H. K. (1999). Purification and characterization of polyphenol oxidase from potato: II. Inhibition and catalytic mechanism. J. Food Biochem. 23, 593–605. doi: 10.1111/j.1745-4514.1999.tb00588.x
Clark, M. F., and Adams, A. N. (1977). Characteristics of the microplate method of enzyme linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 34, 475–483. doi: 10.1099/0022-1317-34-3-475
Değirmenci, H., and Erkurt, H. (2020). Relationship between volatile components, antimicrobial and antioxidant properties of the essential oil, hydrosol and extracts of Citrus aurantium L. flowers. J. Infect. Public Health 13, 58–67. doi: 10.1016/j.jiph.2019.06.017
Dilika, F., Bremner, P. D., and Meyer, J. J. M. (2000). Antibacterial activity of linoleic and oleic acids isolated from Helichrysum pedunculatum: a plant used during circumcision rites. Fitoterapia 71, 450–452. doi: 10.1016/S0367-326X(00)00150-7
Duc, G., Bao, S., Baum, M., Redden, B., Sadiki, M., Suso, M. J., et al. (2010). Diversity maintenance and use of Vicia faba L. genetic resources. F. Crop. Res. 115, 270–278. doi: 10.1016/j.fcr.2008.10.003
El Gamal, A. Y., Tohamy, M. R., Abou-Zaid, M. I., Atia, M. M., El Sayed, T., and Farroh, K. Y. (2022). Silver nanoparticles as a viricidal agent to inhibit plant-infecting viruses and disrupt their acquisition and transmission by their aphid vector. Arch. Virol. 167, 85–97. doi: 10.1007/s00705-021-05280-y
Elbadry, M., Taha, R. M., Eldougdoug, K. A., and Gamal-Eldin, H. (2006). Induction of systemic resistance in faba bean (Vicia faba L.) to bean yellow mosaic potyvirus (BYMV) via seed bacterization with plant growth promoting rhizobacteria. J. Plant Dis. Prot. 113, 247–251. doi: 10.1007/BF03356189
El-Helaly, S., Awad, H., Ahmed, A., and Al-Attar, A. (2016). Studies On Bean Yellow Mosaic Virus Infecting Some Leguminous Crops In Egypt. Egypt. J. Crop Prot. 11, 1–9. doi: 10.21608/ejcp.2016.229440
Elhelaly, S. H. (2022). The effectiveness of the bacteria Rhizobium leguminosarum against bean yellow mosaic (BYMV) potyvirus infecting faba bean (Vicia faba L.) Plants. Ann. Agric. Sci. Moshtohor. 60, 181–190. doi: 10.21608/assjm.2022.227530
Elsharkawy, E. (2016). Anti-inflammatory activity and chemical compositions of essential oil of Achillea fragmmentissma. Natl. J. Physiol. Pharm. Pharmacol. 6, 258–262. doi: 10.1016/j.jep.2017.02.010
Eslahi, N., Kowsari, M., Zamani, M. R., and Motallebi, M. (2021). The profile change of defense pathways in phaseouls vulgaris L. by biochemical and molecular interactions of Trichoderma harzianum transformants overexpressing a chimeric chitinase. Biol. Control 152:104304. doi: 10.1016/j.biocontrol.2020.104304
Fernández, M., López-Jurado, M., Aranda, P., and Urbano, G. (1996). Nutritional assessment of raw and processed faba bean (Vicia faba L.) cultivar major in growing rats. J. Agric. Food Chem. 44, 2766–2772. doi: 10.1021/jf9505483
Hafez, E. E., Abdelkhalek, A. A., Abd El-Wahab, A. S. E.-D., and Galal, F. H. (2013). Altered gene expression: Induction/suppression in leek elicited by Iris Yellow Spot Virus infection (IYSV) Egyptian isolate. Biotechnol. Biotechnol. Equip. 27, 4061–4068. doi: 10.5504/BBEQ.2013.0068
Hamzah, K. A., Younes, H. A., Behiry, S. I., and Abdelkhalek, A. (2021). Act of malondialdehyde and total phenolic content under bean yellow mosaic virus infection and biostimulants application. Egypt. Acad. J. Biol. Sci. H. Bot. 12, 39–42. doi: 10.21608/eajbsh.2021.148885
Heath, R. L., and Packer, L. (1968). Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Heflish, A. A., Abdelkhalek, A., Al-Askar, A. A., and Behiry, S. I. (2021). Protective and Curative Effects of Trichoderma asperelloides Ta41 on Tomato Root Rot Caused by Rhizoctonia solani Rs33. Agronomy 11:1162. doi: 10.3390/agronomy11061162
Ismail, M. H., and Atef, N. M. (1998). Impact of bean yellow mosaic virus (BYMV) on Rhizobium nodulation in faba bean. Phytopathol. Mediterr. 37, 58–68.
Jaiswal, S. K., Mohammed, M., Ibny, F. Y. I., and Dakora, F. D. (2021). Rhizobia as a source of plant growth-promoting molecules: potential applications and possible operational mechanisms. Front. Sustain. Food Syst. 4:619676. doi: 10.3389/fsufs.2020.619676
Jo, S., Kim, S., Shin, D. H., and Kim, M.-S. (2020). Inhibition of African swine fever virus protease by myricetin and myricitrin. J. Enzyme Inhib. Med. Chem. 35, 1045–1049. doi: 10.1080/14756366.2020.1754813
Karavidas, I., Ntatsi, G., Vougeleka, V., Karkanis, A., Ntanasi, T., Saitanis, C., et al. (2022). Agronomic practices to increase the yield and quality of common bean (Phaseolus vulgaris L.): a systematic review. Agronomy 12:271. doi: 10.3390/agronomy12020271
Kawakami, S., Watanabe, Y., and Beachy, R. N. (2004). Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. 101, 6291–6296. doi: 10.1073/pnas.0401221101
Li, W., Xu, C., Hao, C., Zhang, Y., Wang, Z., Wang, S., et al. (2020). Inhibition of herpes simplex virus by myricetin through targeting viral gD protein and cellular EGFR/PI3K/Akt pathway. Antiviral Res. 177:104714. doi: 10.1016/j.antiviral.2020.104714
Likic̀, S., Šola, I., Ludwig-Müller, J., and Rusak, G. (2014). Involvement of kaempferol in the defence response of virus infected Arabidopsis thaliana. Eur. J. Plant Pathol. 138, 257–271. doi: 10.1007/s10658-013-0326-0
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real- time quantitative PCR and the 2 ? ?? C T method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loreto, F., and Velikova, V. (2001). Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 127, 1781–1787. doi: 10.1104/pp.010497
Marín, L., Gutiérrez-del-Río, I., Entrialgo-Cadierno, R., Villar, C. J., and Lombó, F. (2018). De novo biosynthesis of myricetin, kaempferol and quercetin in Streptomyces albus and Streptomyces coelicolor. PLoS One 13:e0207278. doi: 10.1371/journal.pone.0207278
Melero-Vara, J. M., Dominguez, J., and Fernández-Martinez, J. M. (2000). Update on sunflower broomrapesituation in spain: racial status and sunflower breeding for resistance/actualización de la situacion del jopo de girasol en españa: status racial y mejora del girasol para resistencia/situation de l’orobanche et du tournesol. Helia 23, 45–56. doi: 10.1515/helia.2000.23.33.45
Moawad, H., Abd El-Rahim, W. M., and Abd El-Haleem, D. (2004). Performance of phaseolus bean rhizobia in soils from the major production sites in the Nile Delta. C. R. Biol. 327, 445–453. doi: 10.1016/j.crvi.2004.03.005
Moawad, H., Abd el-Rahim, W. M., Abd el-Aleem, D., and Abo Sedera, S. A. (2005). Persistence of two Rhizobium etli inoculant strains in clay and silty loam soils. J. Basic Microbiol. 45, 438–446. doi: 10.1002/jobm.200510590
Moni, S. S., Alam, M. F., Sultan, M. H., Makeen, H. A., Alhazmi, H. A., Mohan, S., et al. (2021). Spectral analysis, in vitro cytotoxicity and antibacterial studies of bioactive principles from the leaves of Conocarpus lancifolius, a common tree of Jazan, Saudi Arabia. Brazilian J. Biol. 83:e244479. doi: 10.1590/1519-6984.244479
Moury, B., and Desbiez, C. (2020). Host range evolution of potyviruses: a global phylogenetic analysis. Viruses 12:111. doi: 10.3390/v12010111
Nakano, Y., and Asada, K. (1981). Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22, 867–880.
Oide, S., Bejai, S., Staal, J., Guan, N., Kaliff, M., and Dixelius, C. (2013). A novel role of PR 2 in abscisic acid (ABA) mediated, pathogen-induced callose deposition in Arabidopsis thaliana. New Phytol. 200, 1187–1199. doi: 10.1111/nph.12436
Oka, K., Kobayashi, M., Mitsuhara, I., and Seo, S. (2013). Jasmonic acid negatively regulates resistance to Tobacco mosaic virus in tobacco. Plant Cell Physiol. 54, 1999–2010. doi: 10.1093/pcp/pct137
Otulak-Kozieł, K., Kozieł, E., and Lockhart, B. (2018). Plant cell wall dynamics in compatible and incompatible potato response to infection caused by Potato virus Y (PVYNTN). Int. J. Mol. Sci. 19:862. doi: 10.3390/ijms19030862
Purwaningsih, S., Agustiyani, D., and Antonius, S. (2021). Diversity, activity, and effectiveness of Rhizobium bacteria as plant growth promoting rhizobacteria (PGPR) isolated from Dieng, central Java. Iran. J. Microbiol. 13, 130–136. doi: 10.18502/ijm.v13i1.5504
Radwan, D. E. M., Fayez, K. A., Mahmoud, S. Y., and Lu, G. (2010). Modifications of antioxidant activity and protein composition of bean leaf due to Bean yellow mosaic virus infection and salicylic acid treatments. Acta Physiol. Plant 32, 891–904.
Radwan, D. E. M., Lu, G., Fayez, K. A., and Mahmoud, S. Y. (2008). Protective action of salicylic acid against bean yellow mosaic virus infection in Vicia faba leaves. J. Plant Physiol. 165, 845–857. doi: 10.1016/j.jplph.2007.07.012
Rizvi, A., Ahmed, B., Khan, M. S., El-Beltagi, H. S., Umar, S., and Lee, J. (2022). bioprospecting plant growth promoting rhizobacteria for enhancing the biological properties and phytochemical composition of medicinally important crops. Molecules 27:1407. doi: 10.3390/molecules27041407
Saeed, Q., Xiukang, W., Haider, F. U., Kuèerik, J., Mumtaz, M. Z., Holatko, J., et al. (2021). Rhizosphere bacteria in plant growth promotion, biocontrol, and bioremediation of contaminated sites: A comprehensive review of effects and mechanisms. Int. J. Mol. Sci. 22:10529. doi: 10.3390/ijms221910529
Scholthof, K. G., Adkins, S., Czosnek, H., Palukaitis, P., Jacquot, E., Hohn, T., et al. (2011). Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. doi: 10.1111/j.1364-3703.2011.00752.x
Sethupathy, S., Ananthi, S., Selvaraj, A., Shanmuganathan, B., Vigneshwari, L., Balamurugan, K., et al. (2017). Vanillic acid from Actinidia deliciosa impedes virulence in Serratia marcescens by affecting S-layer, flagellin and fatty acid biosynthesis proteins. Sci. Rep. 7:16328. doi: 10.1038/s41598-017-16507-x
Shah, A., Nazari, M., Antar, M., Msimbira, L. A., Naamala, J., Lyu, D., et al. (2021). PGPR in agriculture: a sustainable approach to increasing climate change resilience. Front. Sustain. Food Syst. 5:667546. doi: 10.3389/fsufs.2021.667546
Shahidi, F., and Yeo, J. (2018). Bioactivities of phenolics by focusing on suppression of chronic diseases: a review. Int. J. Mol. Sci. 19:1573. doi: 10.3390/ijms19061573
Shimada, K., Fujikawa, K., Yahara, K., and Nakamura, T. (1992). Antioxidative properties of xanthan on the autoxidation of soybean oil in cyclodextrin emulsion. J. Agric. Food Chem. 40, 945–948. doi: 10.1021/jf00018a005
Sofy, A. R., Hmed, A. A., Aleem, M. A., Dawoud, R. A., Elshaarawy, R. F. M., and Sofy, M. R. (2020). Mitigating effects of Bean yellow mosaic virus infection in faba bean using new carboxymethyl chitosan-titania nanobiocomposites. Int. J. Biol. Macromol. 163, 1261–1275. doi: 10.1016/j.ijbiomac.2020.07.066
Somasegaran, P., and Hoben, H. J. (2012). Handbook for Rhizobia: Methods in Legume-Rhizobium Technology. Berlin: Springer Science & Business Media.
Vejan, P., Abdullah, R., Khadiran, T., Ismail, S., and Nasrulhaq Boyce, A. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability-A review. Molecules 21:573. doi: 10.3390/molecules21050573
Velikova, V., Yordanov, I., and Edreva, A. (2000). Oxidative stress and some antioxidant systems in acid rain-treated bean plants: protective role of exogenous polyamines. Plant Sci. 151, 59–66. doi: 10.1016/S0168-9452(99)00197-1
Velioglu, Y., Mazza, G., Gao, L., and Oomah, B. D. (1998). Antioxidant activity and total phenolics in selected fruits, vegetables, and grain products. J. Agric. Food Chem. 46, 4113–4117. doi: 10.1021/jf9801973
Vincent, J. M. (1970). A Manual for the Practical Study of Root-Nodule Bacteria. Oxford: Black Well scientific publications.
Wang, H., Liu, R., You, M. P., Barbetti, M. J., and Chen, Y. (2021). Pathogen biocontrol using plant growth-promoting bacteria (PGPR): Role of bacterial diversity. Microorganisms 9:1988. doi: 10.3390/microorganisms9091988
Warsame, A. O., Michael, N., O’Sullivan, D. M., and Tosi, P. (2020). Identification and quantification of major faba bean seed proteins. J. Agric. Food Chem. 68, 8535–8544. doi: 10.1021/acs.jafc.0c02927
Wei, C.-C., Yen, P.-L., Chang, S.-T., Cheng, P.-L., Lo, Y.-C., and Liao, V. H.-C. (2016). Antioxidative activities of both oleic acid and Camellia tenuifolia seed oil are regulated by the transcription factor DAF-16/FOXO in Caenorhabditis elegans. PLoS One 11:e0157195. doi: 10.1371/journal.pone.0157195
Widhalm, J. R., and Dudareva, N. (2015). A Familiar Ring to It: Biosynthesis of Plant Benzoic Acids. Mol. Plant 8, 83–97. doi: 10.1016/j.molp.2014.12.001
Younes, H. A., Abd El-Aziz, M. H., and Zeid, A. H. (2021). Detection of Bean yellow mosaic potyvirus in infected faba bean plants (Vicia faba L.) from Northern Egypt. Arch. Phytopathol. Plant Prot. 54, 1633–1648.
Zhang, Y., Yan, H., Wei, X., Zhang, J., Wang, H., and Liu, D. (2017). Expression analysis and functional characterization of a pathogen-induced thaumatin-like gene in wheat conferring enhanced resistance to Puccinia triticina. J. Plant Interact. 12, 332–339.
Zhao, L., Chen, Y., Wu, K., Yan, H., Hao, X., and Wu, Y. (2017). Application of fatty acids as antiviral agents against tobacco mosaic virus. Pestic. Biochem. Physiol. 139, 87–91. doi: 10.1016/j.pestbp.2017.05.005
Zhao, S., and Li, Y. (2021). Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog. 17:e1009242. doi: 10.1371/journal.ppat.1009242
Zhu, F., Xi, D.-H., Yuan, S., Xu, F., Zhang, D.-W., and Lin, H.-H. (2014). Salicylic acid and jasmonic acid are essential for systemic resistance against tobacco mosaic virus in Nicotiana benthamiana. Mol. Plant-Microbe Interact. 27, 567–577. doi: 10.1094/MPMI-11-13-0349-R
Keywords: Rhizobium leguminosarum bv. viciae, Bean yellow mosaic virus, faba bean, induced systemic resistance, oxidative stress, pathogenesis-related proteins, HPLC, GC-MS
Citation: Abdelkhalek A, El-Gendi H, Al-Askar AA, Maresca V, Moawad H, Elsharkawy MM, Younes HA and Behiry SI (2022) Enhancing systemic resistance in faba bean (Vicia faba L.) to Bean yellow mosaic virus via soil application and foliar spray of nitrogen-fixing Rhizobium leguminosarum bv. viciae strain 33504-Alex1. Front. Plant Sci. 13:933498. doi: 10.3389/fpls.2022.933498
Received: 01 May 2022; Accepted: 07 July 2022;
Published: 02 August 2022.
Edited by:
Katharina Pawlowski, Stockholm University, SwedenReviewed by:
Maria Luisa Izaguirre-Mayoral, Technical University of Manabi, EcuadorCopyright © 2022 Abdelkhalek, El-Gendi, Al-Askar, Maresca, Moawad, Elsharkawy, Younes and Behiry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed Abdelkhalek, YWFiZGVsa2hhbGVrQHNydGFjaXR5LnNjaS5lZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.