
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 18 August 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.932594
This article is part of the Research TopicPlant Secondary Metabolic Regulation and EngineeringView all 27 articles
 Zhigeng Wu1,2
Zhigeng Wu1,2 Sanjay Kumar Singh3
Sanjay Kumar Singh3 Ruiqing Lyu3
Ruiqing Lyu3 Sitakanta Pattanaik3
Sitakanta Pattanaik3 Ying Wang1,2
Ying Wang1,2 Yongqing Li1,2
Yongqing Li1,2 Ling Yuan1,3*
Ling Yuan1,3* Yongliang Liu1,3*
Yongliang Liu1,3*Echinatin and licochalcone A (LCA) are valuable chalcones preferentially accumulated in roots and rhizomes of licorice (Glycyrrhiza inflata). The licorice chalcones (licochalcones) are valued for their anti-inflammatory, antimicrobial, and antioxidant properties and have been widely used in cosmetic, pharmaceutical, and food industries. However, echinatin and LCA are accumulated in low quantities, and the biosynthesis and regulation of licochalcones have not been fully elucidated. In this study, we explored the potential of a R2R3-MYB transcription factor (TF) AtMYB12, a known regulator of flavonoid biosynthesis in Arabidopsis, for metabolic engineering of the bioactive flavonoids in G. inflata hairy roots. Overexpression of AtMYB12 in the hairy roots greatly enhanced the production of total flavonoids (threefold), echinatin (twofold), and LCA (fivefold). RNA-seq analysis of AtMYB12-overexpressing hairy roots revealed that expression of phenylpropanoid/flavonoid pathway genes, such as phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), and flavanone 3’-hydroxylase (F3’H), is significantly induced compared to the control. Transient promoter activity assay indicated that AtMYB12 activates the GiCHS1 promoter in plant cells, and mutation to the MYB-binding motif in the GiCHS1 promoter abolished activation. In addition, transcriptomic analysis revealed that AtMYB12 overexpression reprograms carbohydrate metabolism likely to increase carbon flux into flavonoid biosynthesis. Further, AtMYB12 activated the biotic defense pathways possibly by activating the salicylic acid and jasmonic acid signaling, as well as by upregulating WRKY TFs. The transcriptome of AtMYB12-overexpressing hairy roots serves as a valuable source in the identification of potential candidate genes involved in LCA biosynthesis. Taken together, our findings suggest that AtMYB12 is an effective gene for metabolic engineering of valuable bioactive flavonoids in plants.
Glycyrrhiza species of the family Fabaceae, including Glycyrrhiza glabra L., Glycyrrhiza uralensis Fisch., and Glycyrrhiza inflata Bat., are valued greatly for their roots and rhizomes (licorice), which are widely used in cosmetics and herbal medicines (Zhang and Ye, 2009; Jiang et al., 2020). The bioactivity of licorice is mainly attributed to two groups of specialized metabolites:, namely, triterpene saponins and flavonoids (Wang D. et al., 2020; Wang Z.-F. et al., 2020). Glycyrrhizin is the most abundant saponin in licorice and has long been recognized as a potent sweetening agent (Pandey and Ayangla, 2018). Glycyrrhizin also has been explored for anti-coronavirus properties in the current COVID-19 pandemic (Luo et al., 2020; Chrzanowski et al., 2021). The other major group of bioactive components present in licorice are flavonoids (Zhu et al., 2016; Cheng et al., 2021). The licorice flavonoids are known to possess anti-inflammatory, antioxidant, and antimicrobial properties (Wang Z.-F. et al., 2020; Husain et al., 2021). Among the different flavonoids, echinatin and licochalcone A (LCA) are predominantly present in G. inflata (Lin et al., 2017; Rizzato et al., 2017; Song et al., 2017). A high LCA cosmetic formulation reduces UV-induced erythema formation in human healthy volunteers possibly by modulation of dendritic cell activity (Kolbe et al., 2006). Because of its anti-inflammatory and antimicrobial properties, LCA has been used for the treatment of facial skin diseases such as acne and rosacea (Schoelermann et al., 2016; Yang et al., 2018). Therefore, there is a great demand of LCA in cosmetic industries (Nguyen et al., 2020; Cerulli et al., 2022). However, LCA is naturally accumulated at low levels in wild G. inflata, even less in cultivated G. inflata plants. Metabolic engineering is thus viewed as a rational alternative to increase LCA production.
Chalcones are a subgroup of polyphenol compounds that are synthesized through the phenylpropanoid pathway (Figure 1A). The precursor phenylalanine (Phe) is derived from the primary metabolic pathways, including glycolysis, the shikimate pathway, and Phe biosynthetic pathway (Tzin and Galili, 2010). Chalcone synthase (CHS) is the first rate-limiting enzyme specific for flavonoid pathway (Saito et al., 2013). Regulation of the flavonoid biosynthesis has been extensively studied in numerous plant species including Arabidopsis thaliana (Saito et al., 2013). In Arabidopsis, three closely related MYB transcription factors (TFs), MYB11, MYB12, and MYB111, from subgroup 7 of the R2R3-MYB family, redundantly regulate the biosynthesis of flavonoids, especially flavonols (Mehrtens et al., 2005; Stracke et al., 2007). These MYBs bind to the promoters of key flavonoid biosynthetic pathway genes, such as CHS, to activate expression (Mehrtens et al., 2005; Stracke et al., 2007). The three MYBs exhibit distinct expression patterns, and AtMYB12 mainly controls flavonoid biosynthesis in Arabidopsis roots (Stracke et al., 2007). The regulatory role of AtMYB12 on flavonoid pathway has been further investigated through heterologous expression in tobacco leaves and tomato fruits (Luo et al., 2008; Pandey et al., 2015; Zhang et al., 2015). AtMYB12 induces the accumulation of flavonoids in tomato fruits by reprogramming the primary metabolism and directing the carbon flux toward flavonoid pathway (Zhang et al., 2015). Chlorogenic acid (CGA) is a subclass of polyphenols present in Solanaceous species (tomato and tobacco) and coffee, but not in Arabidopsis (Luo et al., 2008; Naveed et al., 2018). In addition to flavonoids, ectopic expression of AtMYB12 in tobacco significantly increases CGA biosynthesis (Luo et al., 2008; Zhang et al., 2015). AtMYB12 also activates CGA biosynthetic genes in tomato fruits (Luo et al., 2008; Zhang et al., 2015). AtMYB12 overexpression in kale increases total flavonoid and phenolics in leaves (Lännenpää, 2014). These findings suggest that AtMYB12 is a potential candidate for metabolic engineering to induce flavonoids and flavonoid-derived metabolites in heterologous plant species.
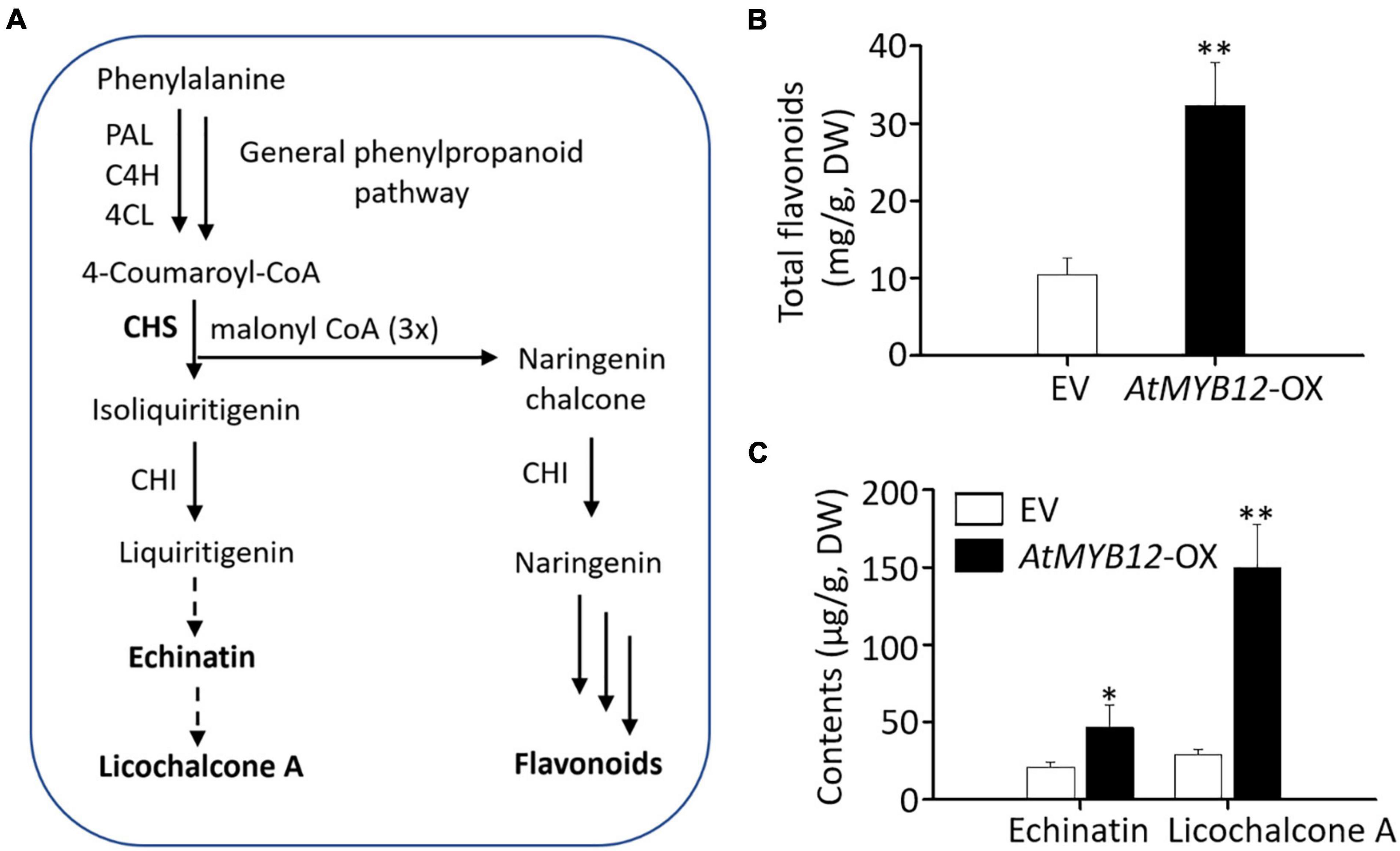
Figure 1. Proposed biosynthetic pathway of licorice chalcones and measurement of metabolites in empty vector (EV) control and AtMYB12-OX hairy roots of G. inflata. (A) A simplified, schematic diagram of the proposed licorice chalcone and general flavonoid biosynthetic pathway. PAL, phe ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate; CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase. Solid arrows indicate known enzymes; dotted arrows indicate pathway enzymes are not known. (B,C) Echinatin and licochalcone A contents on dry weight basis. Data are presented as the mean ± SD (n = 3). Asterisks indicate statistically significant differences compared with EV lines (*p < 0.05, **p < 0.01, Student’s t-test). Total flavonoids based on dry weight (DW).
As the biosynthetic pathway and gene regulation of licorice chalcones are not well elucidated, we aimed to explore the potential of AtMYB12 for metabolic engineering of licorice chalcones in G. inflata. We hypothesized that ectopic expression of AtMYB12 in G. inflata will lead to higher accumulation of licorice chalcones and identification of potential chalcone pathway genes. As protocol for the generation of stable transgenic lines is not established in G. inflata, we thus generated hairy roots overexpressing AtMYB12. Molecular and biochemical analyses of AtMYB12-overexpressing hairy roots showed higher expression of phenylpropanoid/flavonoid pathway genes, including GiCHS, and increased accumulation of total flavonoids and licorice-specific flavonoids, such as echinatin and LCA, confirming the regulatory roles of AtMYB12 on early flavonoid pathway genes in a heterogeneous plant species. In addition, RNA-seq data showed that the carbon flux was reprogrammed toward the flavonoid pathway. Our findings suggest that AtMYB12 is an effective regulator for engineering the production of licorice chalcones in G. inflata.
Arabidopsis thaliana Col-0 accession was used for RNA isolation and AtMYB12 cloning. G. inflata seeds were provided by Gansu Jin You Kang Pharmaceutical Technology Co., Ltd., Lanzhou, China. G. inflata seeds were soaked in H2SO4 for 30 min, washed with water five times, then treated with 1% NaClO for 10 min, and washed with sterilized distilled water five times. Surface-sterilized seeds were germinated on Murashige and Skoog (MS) medium and kept in dark for 2 days before being transferred to light condition. Then, 8-day-old G. inflata seedlings were used for DNA, RNA isolation, transient gene expression, and generation of transgenic hairy roots.
AtMYB12 was amplified from Arabidopsis cDNA and cloned into the pCAMBIA2301 vector containing CaMV35S promoter and rbcS terminator to generate pCAMBIA2301-AtMYB12. Primers used for cloning of AtMYB12 and other genes in this study are all listed in Supplementary Table 1. The empty vector (EV; pCAMBIA2301) and pCAMBIA2301-AtMYB12 plasmids were separately transformed into Agrobacterium rhizogenes R1000 by freeze–thaw method. The hypocotyl segments from 8-day-old G. inflata seedlings were submerged in the A. rhizogenes R1000 suspension for 30 min, blot-dried on sterile filter paper, and then placed on MS medium at 22°C in darkness. After co-cultivation for 2 days, the hypocotyl segments were transferred to MS medium supplemented with 400 mg/l cefotaxime. After 2–3 weeks of culture, hairy roots developed from hypocotyls and the rapidly growing hairy roots were excised and cultivated individually on solid MS medium supplemented with 400 mg/l cefotaxime and 100 mg/l kanamycin for 2 weeks at 25°C in dark. Rapidly growing root lines that showed kanamycin resistance were selected for further analysis. These hairy root lines were cultured in 125 ml flasks each containing 10 ml MS liquid medium on an orbital shaker at 100 rpm at 25°C. The hairy roots clones were routinely subcultured every 2 weeks and harvested after 2 months for RNA isolation and metabolite extraction.
Total RNA was isolated from EV control and AtMYB12-overexpressing seedlings and hairy roots using the RNeasy Plant Mini Kit following the instructions of the manufacturer (QIAGEN, United States). Approximately 2 μg of total RNA was used for DNase I digestion. Synthesis of first-strand cDNA was performed using Superscript III reverse transcriptase (Invitrogen) in a total volume of 20 μl. To verify the transgenic status of AtMYB12-OX and EV control hairy root lines, gene-specific primers were used to PCR-amplify the rol B, rol C, vir C, and kanamycin-resistant (nptII) genes. PCR products were analyzed on a 1% ethidium bromide-stained agarose gel.
Total flavonoid contents were determined by sodium nitrite–aluminum nitrate colorimetric method using rutin as standard (Hao et al., 2018). The standards rutin, echinatin, and LCA were purchased from Biosynth Carbosynth, United States. The contents of echinatin and LCA were determined by LC-MS/MS.
Three independent lines of both EV and AtMYB12-OX hairy roots were used for RNA-seq. Total RNA was isolated from hairy roots using the RNeasy Plant Mini Kit (QIAGEN, United States) following the instructions of the manufacturer. The RNA samples with RNA integrity number (RIN) 8 or above were used for library preparation and sequencing. The TruSeq RNA Sample Prep Kit (Illumina, United States) was used for making libraries according to the protocol of the manufacturer. Individually indexed libraries were combined at equal proportions and loaded onto a single lane of a flow cell. A 50-cycle single-end sequencing run was performed on the Illumina HiSeq2500 at the Duke Center for Genomic and Computational Biology.
Raw Illumina sequence reads were processed as described previously (Singh et al., 2015). Read mapping was performed by Bowtie2 (Langmead and Salzberg, 2012) using an in-house-generated G. inflata transcriptome (unpublished data). Differential gene expression analysis was carried out using the DESeq2 Bioconductor package in R (Love et al., 2014). The differentially expressed genes (DEGs) were identified following two criteria: (i) fold change ≥ 2 and (ii) false discovery rate p-value correction of ≤ 0.05. Heatmaps were constructed using the Complex Heatmap (Gu et al., 2016) function in R through the Bioconductor package (R Core Team, 2022). Functional annotation of DEGs was performed with eggNOG 4.5 (Huerta-Cepas et al., 2016) database. Gene Ontology (GO) analysis of the enriched functional categories was performed using BiNGO (version 2.44) (Maere et al., 2005).
Reverse transcription quantitative PCR (RT-qPCR) was used to measure transcripts levels of GiCHS genes. The GiActin gene was used as an internal control. Relative gene expression was measured as previously described (Liu et al., 2019). All qRT-PCRs were performed in triplicate and repeated twice. Primers used in qRT-PCR are listed in Supplementary Table 1.
The EV and pCAMBIA2301-AtMYB12 were transformed into Agrobacterium tumefaciens GV3101 by freeze–thaw method and was plated on Luria–Bertani (LB) medium containing 100 μg ml–1 kanamycin, 50 μg ml–1 gentamicin, and 30 μg ml–1 rifampicin. A single colony was transferred to 1 ml liquid LB medium containing the same antibiotics and incubated at 250 rpm and 28°C overnight. The overnight culture was diluted in 25 ml liquid LB medium and grown for 16 h at 250 rpm and 28°C. The cells were then centrifuged, and the pellet was resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, 100 μM acetosyringone) to an OD600 of 1.0, and incubated at 28°C for at least 3 h. Then, 8-day-old G. inflata seedlings were immersed in the infiltration solution under vacuum pressure for 1 h. After vacuum infiltration, seedlings were washed five times with sterile distilled water and laid on sterile wet filter papers in Petri dishes. After 5 days of incubation at room temperature, the transfected seedlings were collected for RNA isolation.
Genomic DNA was extracted from G. inflata seedlings for promoter cloning. A forward primer (CHS1-pro-F) was designed based on the genomic sequence upstream of the coding region of G. uralensis homolog of GiCHS1. A reverse primer (GiCHS1-cds-R) was designed within the coding sequence of GiCHS1. PCR product of GuCHS1-pro-F and GiCHS1-cds-R was sequenced. CHS1 promoter sequences of G. inflata and G. uralensis were aligned using ClustalW software (Thompson et al., 2003). Based on the GiCHS1 promoter sequence, another pair of primers (GiCHS1-pro-F2 and GiCHS1-pro-R2) was designed for vector construction.
Tobacco cell line described before (Pattanaik et al., 2010) was used for protoplast isolation and promoter activity assay. The effector plasmid was constructed by cloning AtMYB12 into a modified pBS vector under the control of the CaMV35S promoter and rbcS terminator. The reporter plasmid was generated by cloning GiCHS1 promoter in a modified pUC vector containing the firefly luciferase (LUC) reporter and rbcS terminator. The MYB binding motif in GiCHS1 promoter was mutated by site-directed mutagenesis to generate mutant promoter. The GUS reporter driven by CaMV35S promoter and rbcS terminator was used as an internal control in the protoplast assay. The reporter, effector, and internal control plasmids were electroporated into tobacco protoplasts in different combinations; luciferase and GUS activities in transfected protoplasts were measured as described previously (Pattanaik et al., 2010).
We generated transgenic hairy roots overexpressing AtMYB12 (AtMYB12-OX) aiming to increase flavonoid production. EV hairy root lines (EV-1, EV-2, and EV-3) served as control. Three independent AtMYB12-OX hairy root lines (AtMYB12-OX-1, AtMYB12-OX-2, and AtMYB12-OX-3) were selected for further analysis. The transgenic status of the independent EV and AtMYB12-OX hairy root lines was verified by PCR (Supplementary Figure 1). Total flavonoid contents of the three AtMYB12-OX lines were significantly higher (threefold) than those of the EV lines (Figures 1A,B). While echinatin and LCA in EV hairy root lines were approximately 18–24 and 25–31 ng mg–1, respectively (Figures 1A,C); AtMYB12-OX lines showed a significant increase in the accumulation of echinatin (30–59 ng mg–1; ∼2.2-fold increase) and LCA (119–174 ng mg–1; ∼5.2-fold increase) (Figure 1C). The metabolic outcomes of AtMYB12 overexpression suggest that AtMYB12 is an effective gene for metabolic engineering of the licorice flavonoid pathway.
The metabolic outcomes of AtMYB12-OX hairy roots prompted us to generate and analyze the transcriptome data of EV and AtMYB12-OX lines. Sequencing of RNA libraries of EV and AtMYB12-OX lines generated a total of 1,110 million (M) clean reads. Each biological replicate was represented by an average of more than 170 M reads. On average, more than 70% of the total reads from EV and overexpression line libraries were successfully mapped to the G. inflata transcriptome (Supplementary Figure 2). Compared to the EV lines, 3,236 genes were differentially expressed in AtMYB12-OX lines, in which 1,722 genes were upregulated and 1,514 genes were downregulated (Supplementary Table 2). We particularly examined genes in the phenylpropanoid/flavonoid pathway. CHS is a key rate-limiting enzyme in flavonoid biosynthetic pathway (Zhang et al., 2017). Noticeably, 13 GiCHSs were identified among the DEGs, and 12 of them were upregulated in AtMYB12-OX hairy roots (Figure 2A and Table 1). In addition, we identified three G. inflata phenylalanine ammonia-lyase (PAL) and 9 flavanone 3’-hydroxylase (F3’H) genes among the DEGs, and all of them were induced in AtMYB12-OX hairy roots (Table 1). To verify the expression of selected DEGs in RNA-seq, we conducted RT-qPCR to measure the expression of two CHS genes, Gin33862 (hereafter designed as GiCHS1) and Gin35437, using independently isolated RNAs from the AtMYB12-OX hairy roots. The results confirmed the induction of both CHS genes in AtMYB12-OX hairy root lines (Figure 2B). These results suggest that the upregulation of GiPALs, GiCHSs, and GiF3’Hs likely leads to the enrichment of flavonoids in G. inflata hairy roots.
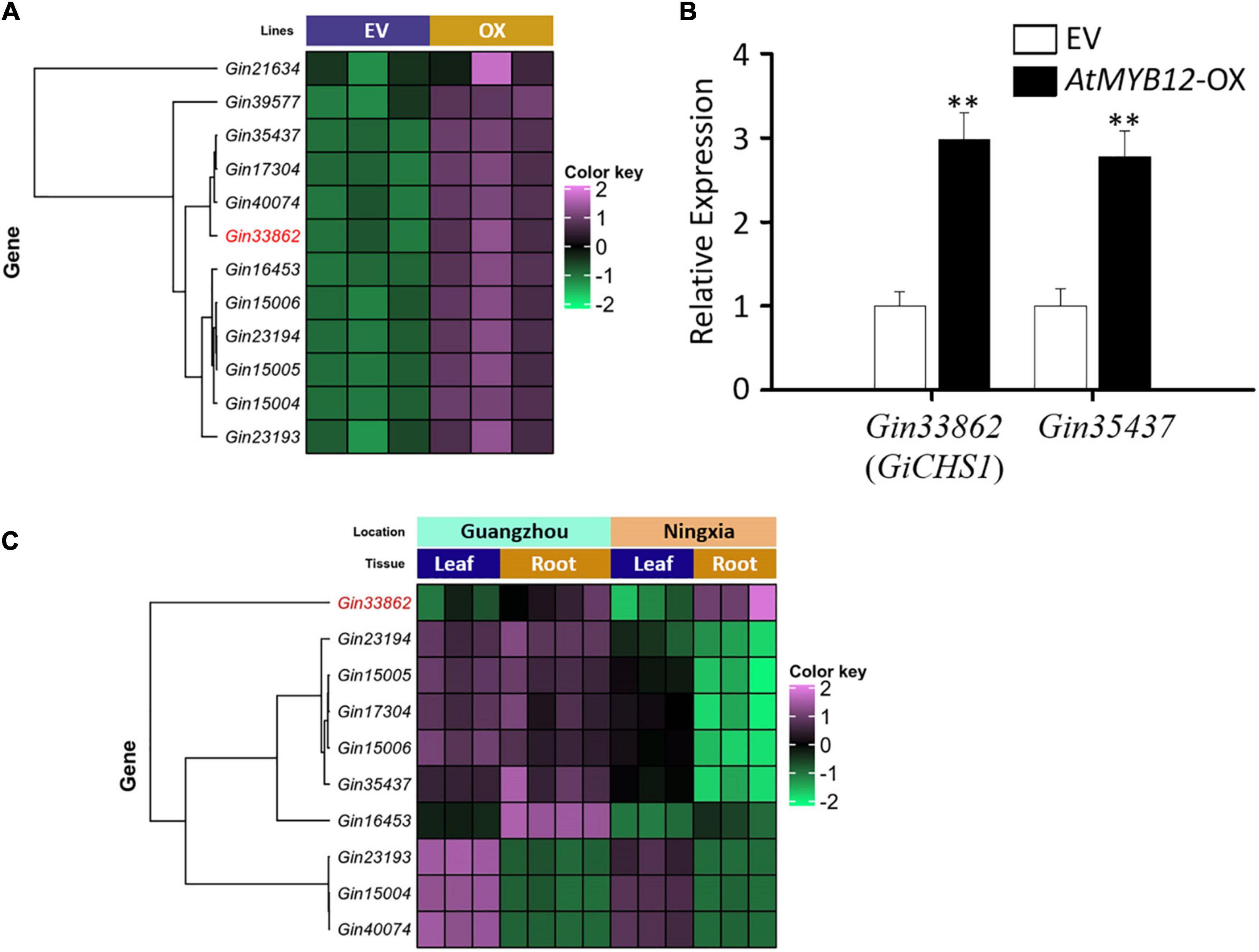
Figure 2. Expression profiles of the G. inflata CHS genes induced by AtMYB12. (A) Heatmap shows the expression patterns of 12 GiCHS genes in EV and AtMYB12-OX (OX) hairy root lines. Gin33862 (GiCHS1) is highlighted in red. (B) Relative expression of two selected GiCHS genes in EV and AtMYB12-OX hairy root lines measured by qRT-PCR. Values were normalized to the expression level of an internal control, the actin gene (GiActin). Data presented are the means of three biological replicates ± SD (n = 3). Asterisks indicate statistically significant differences compared with EV lines (**p < 0.01, Student’s t-test). (C) Heatmap shows the expression patterns of 10 GiCHS genes in two tissues (root and leaf) collected from two locations (Guangzhou and Ningxia) in China.
LCA and echinatin are preferentially accumulated in G. inflata roots and rhizomes. We therefore analyzed the transcriptomes (SRA accession: PRJNA574093) of G. inflata leaves and roots, collected from two geographical locations in China (Guangzhou and Ningxia), to determine the tissue-specific expression of CHS. Among the 12 GiCHSs upregulated in AtMYB12-OX roots, expression of two GiCHSs was not detected in leaf and root transcriptomes. Among the other 10 GiCHSs, Gin33862 (GiCHS1) is preferentially expressed in G. inflata roots from both locations (Figure 2C). Two other GiCHSs, Gin35437 and Gin16453, showed increased expression only in the roots collected from Guangzhou (Figure 2C).
To further verify the effect of AtMYB12 on LCA biosynthesis in G. inflata, we developed an Agrobacteria-mediated transient gene expression assay in G. inflata seedlings. Similar to that in the hairy roots, expression of GiCHS1 and Gin35437 was significantly induced by ectopic expression of AtMYB12 in G. inflata seedlings (Figures 3A,B). These results indicate that the heterologous AtMYB12 positively regulates flavonoid biosynthesis in G. inflata plants.
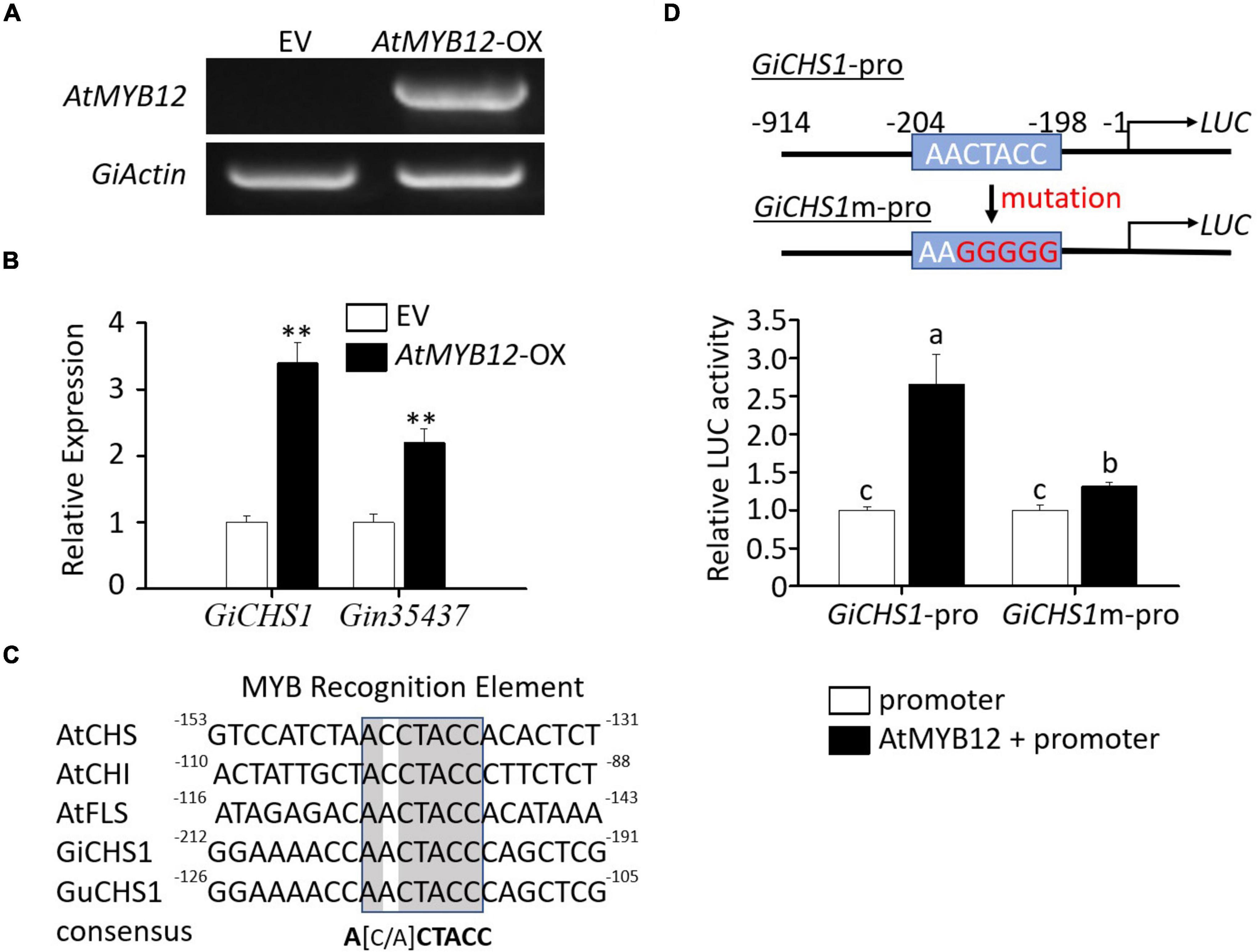
Figure 3. Molecular analysis of G. inflata seedlings transiently overexpressing AtMYB12 and transactivation assay of the GiCHS1 promoter. (A) RT-PCR analysis showed AtMYB12 expression in G. inflata AtMYB12-OX seedlings but not in EV seedlings. The G. inflata actin gene (GiActin) served as internal control. (B) Relative expression of the two GiCHS genes in AtMYB12-OX seedlings was measured using qRT-PCR. GiActin was used as an internal control for normalization. Data are presented as the mean of three biological replicates ± SD. Asterisks indicate statistically significant differences compared with EV lines (**p < 0.01, Student’s t-test). (C) Similar to the promoters of Arabidopsis CHS, CHI, and FLS, GiCHS1 and GuCHS1 promoters also contain the MYB recognition elements (MRE). Numbers by the ends of the DNA sequences, such as -130 at the right end of AtCHS promoter sequence, represent positions relative to translation start site. (D) The diagram in the right shows the MRE in the GiCHS1 promoter (GiCHS1-pro) and the mutated MRE sequence in the mutant promoter (GiCHS1m-pro). LUC, the reporter luciferase gene. Left panel shows transactivation of GiCHS1-pro and GiCHS1m-pro after infiltration of the promoter vector alone or in combination with the AtMYB12-expression vector into tobacco cells. Data presented as the mean of biological replicates ± SD (n = 3). Asterisks indicate statistically significant differences compared with EV lines (**p < 0.01, Student’s t-test).
We next asked whether AtMYB12 directly activates the flavonoid pathway gene promoters in G. inflata. As GiCHS1 expression is upregulated by AtMYB12 and highly expressed in roots, we cloned the GiCHS1 promoter for activity assay. Due to the lack of genomic sequences for G. inflata, the GiCHS1 promoter was cloned based on the G. uralensis genome sequences (Mochida et al., 2017) as G. inflata and G. uralensis are two closely related species. The amino acid sequence identity between GiCHS1 and its G. uralensis homolog is 99%. The promoter of GiCHS1 also shares high sequence identity (96%) with that of G. uralensis CHS1 (Supplementary Figure 3). As shown in Figure 3D, transcriptional activity of the GiCHS1 promoter (GiCHS1-pro) was significantly induced by AtMYB12, suggesting that AtMYB12 directly activates the GiCHS1 promoter in plant cells. To further confirm the activation of the GiCHS1 promoter by AtMYB12, we surveyed the promoter sequence for MYB recognition element (MRE) (A[A/C]CTACC) and identified a putative MRE (AACTACC) at -204 to -198 relative to ATG. This MRE is conserved among the Arabidopsis CHS, CHI, and FLS promoters and also present in the CHS promoters from other plants (Figure 3C). It is predicted to be targeted by R2R3 MYBs, including MYB11, MYB111, and MYB12 (Stracke et al., 2007). We speculated that this MRE (AACTACC) in the GiCHS1 promoter is targeted by AtMYB12. We mutated this motif (to AAGGGGG) to generate the mutant GiCHS1 promoter (GiCHS1m-pro) (Figure 3D). Results of promoter activity assay showed that AtMYB12 is unable to activate GiCHS1m-pro (Figure 3D), suggesting that AtMYB12 directly binds to the MYB binding site in the GiCHS1 promoter.
Carbon resources of phenylpropanoids are derived from monosaccharides, such as glucose. The monosaccharides are directed to phenylpropanoid pathway through several primary pathways, including pentose phosphate pathway, glycolysis, and the shikimate pathway (Zhang et al., 2015). We observed that three genes related to glycolysis and shikimate pathways were induced by AtMYB12 in G. inflata hairy roots (Supplementary Table 3). Glucose also serves as a precursor of the polysaccharide cellulose, the major component of plant cell wall (Taylor, 2008; Yang et al., 2018). Further, GO enrichment analysis (Figure 4 and Supplementary Table 4) showed that several pathway genes related to cellulose synthesis and cell wall synthesis, including “cell wall biogenesis” and “cellulose metabolic process,” are downregulated in AtMYB12-OX lines. Cellulose production during cell wall biosynthesis has been shown to be dependent on cellulose synthase A (CESA). We identified seven G. inflata CESA genes in the DEGs that are downregulated in AtMYB12-OX lines (Table 2). These results indicate an increased carbon flux toward the phenylpropanoid pathway at the cost of cellulose synthesis in AtMYB12-OX hairy roots.
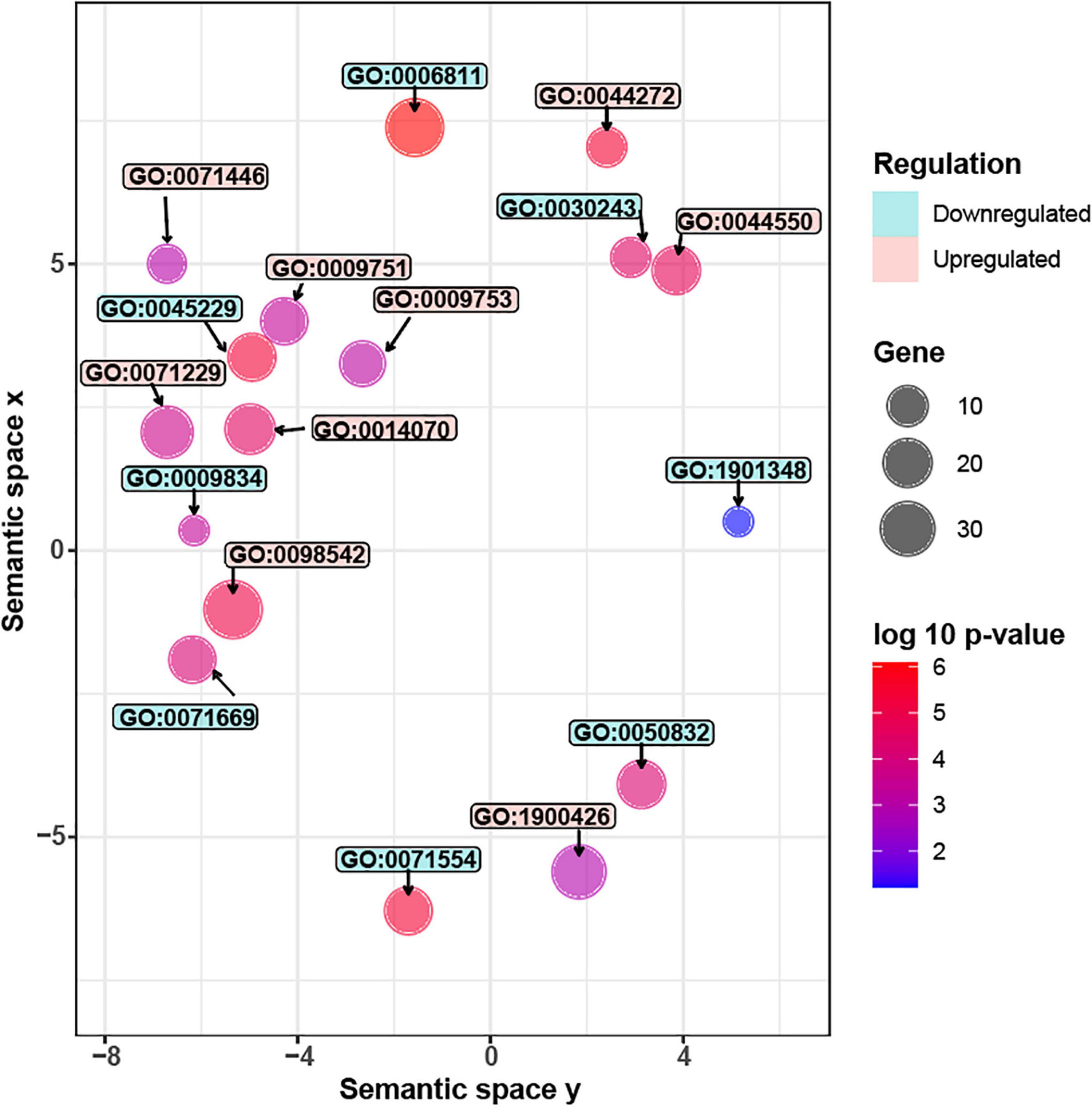
Figure 4. Significantly enriched GO terms in G. inflata AtMYB12-OX hairy roots. Gene Ontology (GO) analyses of differentially expressed genes (DEGs). Upregulated GO terms are colored in red while downregulated terms are in blue. Each circle represents one GO term. The circle size represents the number of genes in each GO category while the color represents the significance level. Description of the upregulated GO terms (from top to bottom): GO:0042742, defense response to bacterium; GO:0071446, cellular response to salicylic acid stimulus; GO:0044550, secondary metabolite biosynthetic process; GO:0009751, response to salicylic acid; GO:0009753, response to jasmonic acid; GO:0071229, cellular response to acid chemical; GO:0014070, response to organic cyclic compound; GO:0098542, defense response to other organism; GO:1900426, positive regulation of defense response to bacterium. Description of the downregulated GO terms (from top to bottom): GO:0006811, ion transport; GO:0030243, cellulose metabolic process; GO:0045229, external encapsulating structure organization; GO:0009834, plant-type secondary cell wall biogenesis; GO:1901348, positive regulation of secondary cell wall biogenesis; GO:0071669, plant-type cell wall organization or biogenesis; GO:0050832, defense response to fungus; GO:0071554, cell wall organization or biogenesis.
GO enrichment analysis showed that, in AtMYB12-OX lines, most of the upregulated pathways are related to pathogen defense responses, including “defense response to bacterium,” “defense response to oomycetes,” “response to fungus,” and “innate immune response” (Figure 4). Salicylic acid (SA) and jasmonic acid (JA) are two important phytohormones that are particularly involved in pathogen defense (Yang et al., 2019; Chen et al., 2020). We observed that the SA and JA signaling pathways, including “response to salicylic acid,” “cellular response to salicylic acid stimulus,” and “response to jasmonic acid,” were activated (Figure 4 and Supplementary Table 4). In addition, a number of TF families were identified among the DEGs. In particular, members of the WRKY TFs are enriched in the DEGs (Figure 5). A growing body of research suggests that WRKY TFs are involved in pathogen resistance (Wani et al., 2021). Therefore, it is possible that AtMYB12-mediated defense responses are activated through SA and JA signaling, as well as the activation of WRKY TFs.
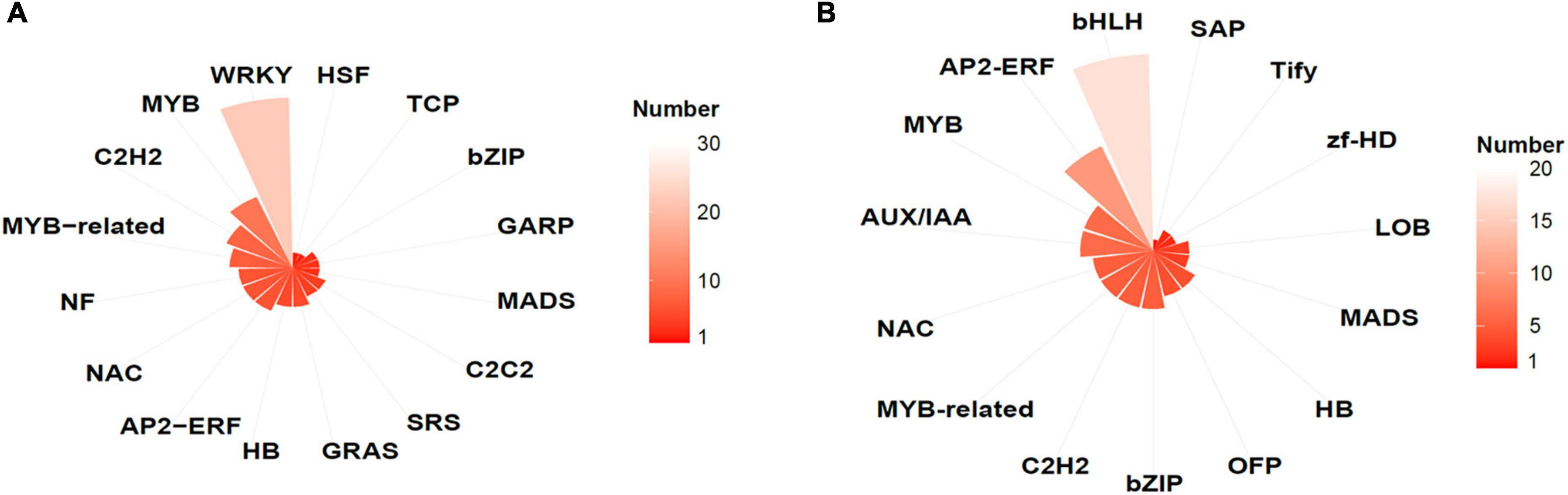
Figure 5. Transcription factor (TF) families identified in DEGs in EV and AtMYB12 transcriptomes. (A) Upregulated TFs represented by gene numbers (the lighter the color, the higher the number); e.g., the most upregulated TFs belong to WRKY family. (B) Downregulated TFs represented by gene numbers (the lighter the color, the higher the number); e.g., the bHLH family is most affected.
TFs are ideal candidates for metabolic engineering because of their broad regulatory roles in metabolic pathways (Broun, 2004; Lu et al., 2016). Increasing evidence suggests that many TF functions and regulatory mechanisms are conserved across the species (Feller et al., 2011; Schluttenhofer and Yuan, 2015). For instance, ectopic expression of the maize bHLH TF Lc induces anthocyanin accumulation in tobacco and Arabidopsis (Lloyd et al., 1992). Similarly, expression of snapdragon R2R3 MYB Rosea1 and bHLH TF Delila, both driven by a fruit-specific promoter, induces anthocyanin accumulation in tomato fruit (Butelli et al., 2008). Ectopic expression of the Arabidopsis R2R3 MYB PAP1 induces anthocyanin accumulation in tobacco (Borevitz et al., 2000). These findings support the conserved regulatory roles of TFs in metabolic pathways in plants. Licorice chalcones, including echinatin and LCA, are a characteristic group of flavonoids that are exclusively produced in G. inflata roots and rhizomes (Lin et al., 2017; Rizzato et al., 2017; Song et al., 2017). Although echinatin and LCA possess several important bioactive properties (Wang D. et al., 2020; Wang Z.-F. et al., 2020), key enzymes involved in the biosynthesis and molecular mechanism of regulation have not been fully elucidated. AtMYB12, along with AtMYB11 and AtMYB111, regulates flavonoid biosynthesis in Arabidopsis (Stracke et al., 2007). Heterologous expression of AtMYB12 in tomato not only activates flavonoid pathway genes but also induces genes in the upstream primary metabolic pathways (Luo et al., 2008; Zhang et al., 2015). Here, we demonstrated that ectopic expression of AtMYB12 upregulates the expression of GiCHSs and enhances echinatin and LCA accumulation in G. inflata roots (Figures 1, 3).
A transformation protocol to generate stable transgenic plants is not available for G. inflata. As licorice-specific flavonoids are preferentially accumulated in rhizomes and roots of G. inflata (Lin et al., 2017; Rizzato et al., 2017; Song et al., 2017), we used transgenic hairy roots to explore the potential of AtMYB12 to enhance the accumulation of echinatin and LCA. RNA-seq analysis of three independent hairy root lines showed upregulation of flavonoid pathway genes, including PAL, CHS, and F3’H (Table 1). CHS belongs to the plant polyketide synthase superfamily and is a key enzyme in the flavonoid pathway (Dao et al., 2011). CHS catalyzes the condensation of one molecule cinnamic acid or its derivatives and three molecules of malonyl Co-A to produce the narigenin chalcone, which serves as a precursor for diverse sets of flavonoids (Dao et al., 2011). Our RNA-seq data showed that expression of 12 G. inflata CHSs is upregulated by AtMYB12 (Figure 2A and Table 1).
Agrobacterium-mediated transient transformation of whole seedlings has been used to study the regulation of metabolic pathways in different plant species, such as Catharanthus roseus (Liu et al., 2019; Mortensen et al., 2019). We transiently transformed G. inflata seedlings with Agrobacterium harboring AtMYB12 and measured the expression of selected CHSs. Similar to the hairy roots, expression of two selected GiCHSs was significantly induced in seedlings transformed with AtMYB12 (Figure 3B). We demonstrated that AtMYB12 activates the GiCHS1 promoter in plant cells by binding to a MYB-recognition element (MRE), and mutation of the MRE abolished the activation (Figure 3D). In Arabidopsis (Mehrtens et al., 2005) and tomato (Zhang et al., 2015), AtMYB12 is shown to bind and activate the CHS promoter, further reinforcing the functional conservation of AtMYB12.
In tomato fruits, in addition to flavonoids, ectopic expression of AtMYB12 upregulates CGA biosynthetic pathway genes and increases CGA accumulation (Luo et al., 2008; Zhang et al., 2015). Given the fact that echinatin and LCA contents were higher in AtMYB12-OX hairy roots (Figure 1), we speculated that licorice chalcone biosynthetic genes are regulated by AtMYB12. Although genes encoding enzymes in licorice chalcone pathway are not fully characterized, structural differences between echinatin and LCA suggest two modification steps: O-methylation and prenylation likely occur in the biosynthetic steps from echinatin to LCA. From our RNA-seq data set, we identified four genes that are annotated as O-methyltransferases and two as prenyltransferases, all differentially expressed in AtMYB12-OX hairy roots (Supplementary Table 5). These genes are thus putative candidates of future investigation of LCA biosynthesis.
In tomato fruits, other than increased accumulation of flavonoids, ectopic expression of AtMYB12 decreases the contents of carbon resources, including glucose and fructose (Zhang et al., 2015). Genes in the primary metabolic pathways were mostly induced by AtMYB12 (Zhang et al., 2015). Our results confirmed the induction of the genes in the primary pathways upstream of flavonoids (Supplementary Table 3). We also found that the genes related to cellulose synthesis were downregulated (Figure 4 and Table 2), suggesting that, in both tomato fruits (Zhang et al., 2015) and G. inflata hairy roots, AtMYB12 redirects carbon flux from other carbohydrate resources toward the flavonoid pathway. Therefore, we suggest that the functionally conserved nature of AtMYB12 makes it a promising candidate for metabolic engineering in other plant species.
AtMYB12 overexpression improves pathogen resistance in transgenic tobacco plants (Ding et al., 2021). In transgenic tobacco, AtMYB12 induces the production of flavonoid compounds, such as rutin, as well as reactive oxygen species, H2O2, and NO (Pandey et al., 2015; Ding et al., 2021). Similar to tobacco, our results revealed that the genes involved in pathogen resistance pathways (Figure 4), defense-associated plant hormone signaling (Figure 4), and WRKY TFs (Figure 5) were significantly enriched in AtMYB12-OX hairy roots. SA and JA play essential roles in plant defense against different pathogens (Yang et al., 2019; Chen et al., 2020). WRKY TFs are among the largest families of transcriptional regulators and contribute to various plant processes, including disease defense (Wani et al., 2021). Some WRKYs like AtWRKY33 could regulate SA/JA biosynthesis while others are regulated by SA/JA signaling (Birkenbihl et al., 2012; Wani et al., 2021). We speculate that AtMYB12 activated the SA/JA-WRKY network that contributes to the pathogen defense responses in G. inflata hairy roots.
Metabolic engineering offers an excellent approach for producing various bioactive, health-promoting phytochemicals in plants. This study underscores the importance of metabolic engineering for enhancing accumulation of valuable metabolites, such as licorice chalcones in G. inflata, through heterologous expression of a known flavonoid regulator. Metabolic pathways of non-model plants are relatively less studied. Transcriptomic and genomic resources will help unravel the biosynthetic pathways in non-model plants, such as G. inflata, and aid the bioengineering of bioactive compounds.
The data presented in the study are deposited in the NCBI repository, accession number: PRJNA842240.
ZW and YLL performed the experiments, analyzed the data, and wrote the article. SS analyzed the RNA-seq data. RL, SP, YW, YQL, and LY evaluated the experiments and revised the article. YW, YQL, and LY initiated and supervised the project. All authors contributed to the article and approved the submitted version.
This study was partially supported by the National Key R&D Program of China (2019YFC1711100), by the grant nos. 2018530000241001 and 2019530000241005 from the Yunnan Tobacco Company, Harold R. Burton Endowed Professorship to LY and the Kentucky Tobacco Research and Development Center.
We thank Megan Combs (Department of Civil Engineering and Environmental Research Training Laboratories, University of Kentucky) for assistance on LC-MS/MS analyses.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.932594/full#supplementary-material
Birkenbihl, R. P., Diezel, C., and Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285. doi: 10.1104/pp.111.192641
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2393. doi: 10.1105/tpc.12.12.2383
Broun, P. (2004). Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 7, 202–209. doi: 10.1016/j.pbi.2004.01.013
Butelli, E., Titta, L., Giorgio, M., Mock, H.-P., Matros, A., Peterek, S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308.
Cerulli, A., Masullo, M., Montoro, P., and Piacente, S. (2022). Licorice (Glycyrrhiza glabra, G. uralensis, and G. inflata) and their constituents as active cosmeceutical ingredients. Cosmetics 9:7.
Chen, J., Clinton, M., Qi, G., Wang, D., Liu, F., and Fu, Z. Q. (2020). Reprogramming and remodeling: Transcriptional and epigenetic regulation of salicylic acid-mediated plant defense. J. Exp. Bot. 71, 5256–5268. doi: 10.1093/jxb/eraa072
Cheng, M., Zhang, J., Yang, L., Shen, S., Li, P., Yao, S., et al. (2021). Recent advances in chemical analysis of licorice (Gan-Cao). Fitoterapia 149:104803. doi: 10.1016/j.fitote.2020.104803
Chrzanowski, J., Chrzanowska, A., and Graboń, W. (2021). Glycyrrhizin: An old weapon against a novel coronavirus. Phytother. Res. 35, 629–636. doi: 10.1002/ptr.6852
Dao, T., Linthorst, H., and Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412.
Ding, X., Zhang, H., Li, M., Yin, Z., Chu, Z., Zhao, X., et al. (2021). AtMYB12-expressing transgenic tobacco increases resistance to several phytopathogens and aphids. Front Agron. 3:694333. doi: 10.3389/fagro.2021.694333
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Hao, P.-Y., Feng, Y.-L., Zhou, Y.-S., Song, X.-M., Li, H.-L., Ma, Y., et al. (2018). Schaftoside interacts with NlCDK1 protein: A mechanism of rice resistance to brown planthopper, Nilaparvata lugens. Front. Plant Sci. 9:710. doi: 10.3389/fpls.2018.00710
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Husain, I., Bala, K., Khan, I. A., and Khan, S. I. (2021). A review on phytochemicals, pharmacological activities, drug interactions, and associated toxicities of licorice (Glycyrrhiza sp.). Food Front. 2, 449–485. doi: 10.1002/fft2.110
Jiang, M., Zhao, S., Yang, S., Lin, X., He, X., Wei, X., et al. (2020). An “essential herbal medicine”—licorice: A review of phytochemicals and its effects in combination preparations. J. Ethnopharmacol. 249:112439. doi: 10.1016/j.jep.2019.112439
Kolbe, L., Immeyer, J., Batzer, J., Wensorra, U., Mundt, C., Wolber, R., et al. (2006). Anti-inflammatory efficacy of Licochalcone A: Correlation of clinical potency and in vitro effects. Arch. Dermatol. Res. 298, 23–30. doi: 10.1007/s00403-006-0654-4
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lännenpää, M. (2014). Heterologous expression of AtMYB12 in kale (Brassica oleracea var. acephala) leads to high flavonol accumulation. Plant Cell Rep. 33, 1377–1388. doi: 10.1007/s00299-014-1623-6
Lin, Y., Kuang, Y., Li, K., Wang, S., Song, W., Qiao, X., et al. (2017). Screening for bioactive natural products from a 67-compound library of Glycyrrhiza inflata. Bioorg. Med. Chem. 25, 3706–3713. doi: 10.1016/j.bmc.2017.05.009
Liu, Y., Patra, B., Pattanaik, S., Wang, Y., and Yuan, L. (2019). GATA and phytochrome interacting factor transcription factors regulate light-induced vindoline biosynthesis in Catharanthus roseus. Plant Physiol. 180, 1336–1350. doi: 10.1104/pp.19.00489
Lloyd, A. M., Walbot, V., and Davis, R. W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. doi: 10.1126/science.1465611
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, X., Tang, K., and Li, P. (2016). Plant metabolic engineering strategies for the production of pharmaceutical terpenoids. Front. Plant Sci. 7:1647. doi: 10.3389/fpls.2016.01647
Luo, J., Butelli, E., Hill, L., Parr, A., Niggeweg, R., Bailey, P., et al. (2008). AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J. 56, 316–326. doi: 10.1111/j.1365-313X.2008.03597.x
Luo, P., Liu, D., and Li, J. (2020). Pharmacological perspective: Glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int. J. Antimicrob. Agents 55:105995. doi: 10.1016/j.ijantimicag.2020.105995
Maere, S., Heymans, K., and Kuiper, M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. doi: 10.1093/bioinformatics/bti551
Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. doi: 10.1104/pp.104.058032
Mochida, K., Sakurai, T., Seki, H., Yoshida, T., Takahagi, K., Sawai, S., et al. (2017). Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 89, 181–194. doi: 10.1111/tpj.13385
Mortensen, S., Bernal-Franco, D., Cole, L. F., Sathitloetsakun, S., Cram, E. J., and Lee-Parsons, C. (2019). EASI transformation: An efficient transient expression method for analyzing gene function in Catharanthus roseus seedlings. Front. Plant Sci. 10:755. doi: 10.3389/fpls.2019.00755
Naveed, M., Hejazi, V., Abbas, M., Kamboh, A. A., Khan, G. J., Shumzaid, M., et al. (2018). Chlorogenic acid (CGA): A pharmacological review and call for further research. Biomed. Pharmacother. 97, 67–74. doi: 10.1016/j.biopha.2017.10.064
Nguyen, J. K., Masub, N., and Jagdeo, J. (2020). Bioactive ingredients in Korean cosmeceuticals: Trends and research evidence. J. Cosmet. Dermatol. 19, 1555–1569. doi: 10.1111/jocd.13344
Pandey, A., Misra, P., Choudhary, D., Yadav, R., Goel, R., Bhambhani, S., et al. (2015). AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues. Sci. Rep. 5:12412. doi: 10.1038/srep12412
Pandey, D. K., and Ayangla, N. J. P. R. (2018). Biotechnological aspects of the production of natural sweetener glycyrrhizin from Glycyrrhiza sp. Phytochem. Rev. 17, 397–430.
Pattanaik, S., Werkman, J. R., Kong, Q., and Yuan, L. (2010). Site-directed mutagenesis and saturation mutagenesis for the functional study of transcription factors involved in plant secondary metabolite biosynthesis. Methods Mol. Biol. 643, 47–57. doi: 10.1007/978-1-60761-723-5_4
R Core Team (2022). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Rizzato, G., Scalabrin, E., Radaelli, M., Capodaglio, G., and Piccolo, O. (2017). A new exploration of licorice metabolome. Food Chem. 221, 959–968.
Saito, K., Yonekura-Sakakibara, K., Nakabayashi, R., Higashi, Y., Yamazaki, M., Tohge, T., et al. (2013). The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 72, 21–34. doi: 10.1016/j.plaphy.2013.02.001
Schluttenhofer, C., and Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Schoelermann, A., Weber, T., Arrowitz, C., Rizer, R., Qian, K., and Babcock, M. (2016). Skin compatibility and efficacy of a cosmetic skin care regimen with licochalcone A and 4-t-butylcyclohexanol in patients with rosacea subtype I. J. Eur. Acad. Dermatol. Venereol. 30, 21–27. doi: 10.1111/jdv.13531
Singh, S. K., Wu, Y., Ghosh, J. S., Pattanaik, S., Fisher, C., Wang, Y., et al. (2015). RNA-sequencing reveals global transcriptomic changes in Nicotiana tabacum responding to topping and treatment of axillary-shoot control chemicals. Sci. Rep. 5:18148. doi: 10.1038/srep18148
Song, W., Qiao, X., Chen, K., Wang, Y., Ji, S., Feng, J., et al. (2017). Biosynthesis-based quantitative analysis of 151 secondary metabolites of licorice to differentiate medicinal Glycyrrhiza species and their hybrids. Anal. Chem. 89, 3146–3153. doi: 10.1021/acs.analchem.6b04919
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Taylor, N. G. (2008). Cellulose biosynthesis and deposition in higher plants. New Phytol. 178, 239–252.
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2003). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2:2.3.
Tzin, V., and Galili, G. (2010). The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. Arabidopsis Book 8:e0132. doi: 10.1199/tab.0132
Wang, D., Liang, J., Zhang, J., Wang, Y., and Chai, X. (2020). Natural chalcones in Chinese materia medica: Licorice. Evid. Based Complementary Altern. Med. 2020, 1–14.
Wang, Z.-F., Liu, J., Yang, Y.-A., and Zhu, H.-L. (2020). A review: The anti-inflammatory, anticancer and antibacterial properties of four kinds of licorice flavonoids isolated from licorice. Curr. Med. Chem. 27, 1997–2011. doi: 10.2174/0929867325666181001104550
Wani, S. H., Anand, S., Singh, B., Bohra, A., and Joshi, R. (2021). WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Yang, G., Lee, H. E., Yeon, S. H., Kang, H. C., Cho, Y. Y., Lee, H. S., et al. (2018). Licochalcone A attenuates acne symptoms mediated by suppression of NLRP3 inflammasome. Phytother. Res. 32, 2551–2559. doi: 10.1002/ptr.6195
Yang, J., Duan, G., Li, C., Liu, L., Han, G., Zhang, Y., et al. (2019). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10:1349. doi: 10.3389/fpls.2019.01349
Zhang, Q., and Ye, M. (2009). Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice). J. Chromatogr A 1216, 1954–1969.
Zhang, X., Abrahan, C., Colquhoun, T. A., and Liu, C.-J. (2017). A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. Plant Cell 29, 1157–1174. doi: 10.1105/tpc.16.00855
Zhang, Y., Butelli, E., Alseekh, S., Tohge, T., Rallapalli, G., Luo, J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6:8635. doi: 10.1038/ncomms9635
Keywords: Glycyrrhiza inflata, flavonoids, echinatin, licochalcone A, AtMYB12 gene, metabolic engineering
Citation: Wu Z, Singh SK, Lyu R, Pattanaik S, Wang Y, Li Y, Yuan L and Liu Y (2022) Metabolic engineering to enhance the accumulation of bioactive flavonoids licochalcone A and echinatin in Glycyrrhiza inflata (Licorice) hairy roots. Front. Plant Sci. 13:932594. doi: 10.3389/fpls.2022.932594
Received: 30 April 2022; Accepted: 18 July 2022;
Published: 18 August 2022.
Edited by:
Supaart Sirikantaramas, Chulalongkorn University, ThailandReviewed by:
Wei Zhou, Zhejiang Chinese Medical University, ChinaCopyright © 2022 Wu, Singh, Lyu, Pattanaik, Wang, Li, Yuan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Yuan, bHl1YW4zQHVreS5lZHU=; Yongliang Liu, eW9uZ2xpYW5nbGl1QHVreS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.