
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 25 July 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.931275
This article is part of the Research Topic Omics Driven Research for the Improvement of Industrial Crops View all 5 articles
 Bindu Yadav1
Bindu Yadav1 Vikender Kaur1*
Vikender Kaur1* Om Prakash Narayan2
Om Prakash Narayan2 Shashank Kumar Yadav1
Shashank Kumar Yadav1 Ashok Kumar1
Ashok Kumar1 Dhammaprakash Pandhari Wankhede3*
Dhammaprakash Pandhari Wankhede3*Flax (Linum usitatissimum L.) or linseed is one of the important industrial crops grown all over the world for seed oil and fiber. Besides oil and fiber, flax offers a wide range of nutritional and therapeutic applications as a feed and food source owing to high amount of α-linolenic acid (omega-3 fatty acid), lignans, protein, minerals, and vitamins. Periodic losses caused by unpredictable environmental stresses such as drought, heat, salinity-alkalinity, and diseases pose a threat to meet the rising market demand. Furthermore, these abiotic and biotic stressors have a negative impact on biological diversity and quality of oil/fiber. Therefore, understanding the interaction of genetic and environmental factors in stress tolerance mechanism and identification of underlying genes for economically important traits is critical for flax improvement and sustainability. In recent technological era, numerous omics techniques such as genomics, transcriptomics, metabolomics, proteomics, phenomics, and ionomics have evolved. The advancements in sequencing technologies accelerated development of genomic resources which facilitated finer genetic mapping, quantitative trait loci (QTL) mapping, genome-wide association studies (GWAS), and genomic selection in major cereal and oilseed crops including flax. Extensive studies in the area of genomics and transcriptomics have been conducted post flax genome sequencing. Interestingly, research has been focused more for abiotic stresses tolerance compared to disease resistance in flax through transcriptomics, while the other areas of omics such as metabolomics, proteomics, ionomics, and phenomics are in the initial stages in flax and several key questions remain unanswered. Little has been explored in the integration of omic-scale data to explain complex genetic, physiological and biochemical basis of stress tolerance in flax. In this review, the current status of various omics approaches for elucidation of molecular pathways underlying abiotic and biotic stress tolerance in flax have been presented and the importance of integrated omics technologies in future research and breeding have been emphasized to ensure sustainable yield in challenging environments.
Flax (Linum usitatissimum L.) or linseed is one of the primeval crops domesticated for oil and fiber since beginning of civilization (Zohary and Hopf, 2000). It is believed to be originated in either the Middle East or Indian regions from where it spread to whole world (Vavilov, 1951; Green et al., 2008). Since ages, the oil from flax seed has been used in paints, varnishes, and polymer industries owing to its unique fatty acid composition (Przybylski, 2005; Shim et al., 2015) while the fiber extracted from flax stem has been used in textile industry to produce quality Linen fabrics. Nutritionally flaxseeds are very dense as they are packed with high amount of alpha linolenic acid (55–57%), proteins (upto 18.29%), fibers (27.3%), vitamin B1, and lignans particularly secoisolariciresinol diglucoside (SDG; 294–700 mg/100 g) making it among preeminent functional food (Singh et al., 2011; Goyal et al., 2014; Kajla et al., 2015). Flax seed consumption has proven beneficial effects on coronary heart disease, cancer, neurological/hormonal disorders, and atherosclerosis (Westcott and Muir, 2003; Hosseinian et al., 2006; Bassett et al., 2009). Presently, China occupies the paramount position in terms of flax consumption and is the largest importer valuing 31,108 M US$ in the past decade which accounts for 26.8% of total global flax import in the year 2020. Canada is the leading producer and exporter of flax worldwide over the past decade, while India ranks seventh in terms of production and eleventh in terms of export (FAOSTAT, 2022; Figure 1). Biotic and abiotic stress factors have been the major constraints in increasing flax production worldwide. The productivity of fiber flax is severely affected by devastating fungal diseases such as Fusarium wilt, Alternaria blight, powdery mildew, rust, and pasmo in European countries, whereas the oil type linseed mainly cultivated in Asian countries, particularly India suffers from drought, salinity, and heat in conjugation to varied diseases and insect-pests. In addition, the warmer climate of these tropical countries is not suitable for fiber flax which requires a prolonged cool season for effective yields and fiber quality. As a result, yields have been stagnated in these countries. The renewed interest in flax consumption as functional food has led to the increase in consumer demand for flax-based products such as multigrain breads, ready-to-eat breakfast cereals, breakfast drinks, salad dressings, biscuits, crackers, soups, and cakes (Coşkuner and Karababa, 2007; Ayelign and Alemu, 2016). Moreover, with the recent advances in material science, the flax fiber has new range of industrial applications, such as geotextiles, biopolymers, specialty papers, composites, and biofuels (Diederichsen and Ulrich, 2009; Cullis, 2011), and has gained new attention because of its quality, biodegradability, and recyclability. Thus, the burgeoning interest revolving around health promoting effects and natural fiber industry has fueled for enhanced demand worldwide. The increased demand is reflected by the up-scaling trend in global production of linseed from 2.5 million tonnes to more than 3.5 million tonnes as well as flax fiber from about 26,000 tonnes to approx. 1 million tonnes over past decade (FAOSTAT, 2022; Figure 1). However, environmental challenges, such as dwindling water resources, salinization or alkalinization of soil, extreme temperature fluctuations, fungal diseases, such as wilt, rust, and pasmo, have deleterious effects on plant growth resulting in huge yield loss in flax (Fofana et al., 2006; Saha et al., 2021; Zare et al., 2021).
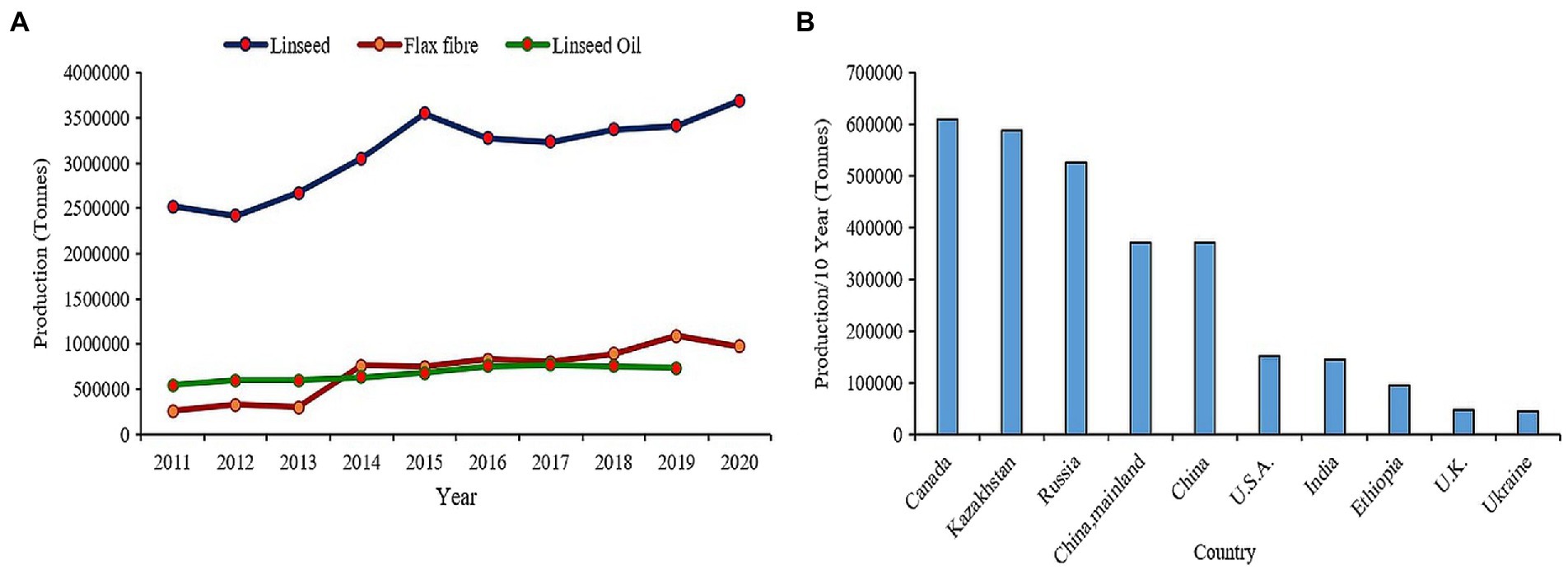
Figure 1. (A) Trends in global production of linseed and flax fiber in past decade. (B) Flaxseed production in top 10 countries in the world.
Among abiotic stresses, drought is one of the most prevalent and detrimental constraints to agricultural production, that negatively affects the overall crop growth, yield, and quality causing more than 50% average yield loss (Fahad et al., 2017; Kole, 2020). It is expected to wreak havoc on plant growth on more than half of arable land by 2050 (Jaggard et al., 2010). Western Canada, Russia, China, and India are important flax producing regions and during the last 100 years, annual precipitation has become less evenly distributed in these areas in addition to temperature change (Gitay et al., 2002). Scarcity of soil moisture can negatively impair the yield potential, oil content and fatty acid composition, and fiber quality traits in flax (Fofana et al., 2006; Abd El-Fatah, 2007; Heller and Byczyńska, 2015). Drought results in reduced leaf expansion, leaf senescence, abscission, oxidative damage, and increased membrane lipid peroxidation thereby disrupting normal metabolism (Hu and Xiong, 2014). Although flax tolerates drought better than many other oil and food crops due to its hardiness; however, at the same time, flax plants transpire very high amounts of water owing to high transpiration coefficient (the amount of water necessary to produce one unit of dry matter) value of 787–1,093 (Kozłowska, 2007; Heller and Byczyńska, 2015). Therefore, fiber flax requires annual precipitation of at least 600–650 mm for optimal yields, of which at least 110–150 mm of rain fall is essential in the vegetation period. Thus, water scarcity continues to be a significant impediment to flax production as it is a neglected crop in developing countries and is normally cultivated in rain-fed areas with poor management and low input conditions (Lisson and Mendham, 2000; Dash et al., 2014; Kaur et al., 2017). Drought is an erratic and highly unpredictable environmental phenomenon; therefore, selection should target drought tolerant genotypes having yield potential. Accordingly, long-term traditional breeding programs and later development of transgenic flax were initiated to combat these constraints and improve flax production (Tawfik et al., 2016). Since drought tolerance is a complex polygenic trait, understanding the adaptive mechanisms and identification of underlying genes/markers/QTLs could pave a way for genetic enhancement and productivity of flax in arid and semi-arid regions. Only a few studies have been reported identifying drought resilient genotypes in flax (Diederichsen et al., 2006; Qi et al., 2010; Sharma et al., 2012; Asgarinia et al., 2017) and genome-wide analysis of drought induced gene expression (Dash et al., 2014). The root system is shallow in flax compared to other oilseed crops such as rapeseed, sunflower, and safflower. Therefore, studying root system architecture is of pivotal importance for more efficient water acquisition in flax. The importance of root traits for efficient water and nutrient absorption under water scarce conditions have been realized recently in many crops, such as rice, wheat, and maize (Tuberosa et al., 2002; Manschadi et al., 2006; Gowda et al., 2011; Kaur et al., 2020); however, knowledge is still limited in flax (Soto-Cerda et al., 2019, 2020).
Soil salinity has risen exponentially in recent years due to a number of factors including excessive irrigation, low precipitation, high surface evaporation, rock weathering, ion exchange, and poor cultural practices (Bui, 2020; Dubey et al., 2020). Approximately 20% of total cultivated and 33% of irrigated land is currently affected by saline conditions, and more than 50% of arable land is predicted to be salinized by 2050 (Jamil et al., 2011; Shrivastava and Kumar, 2015). In flax, soil salinity-alkalinity leads to delayed germination and emergence, low seedling survival, irregular crop growth, and lower yield (Dubey et al., 2020). Few studies have reported screening of flax germplasm against salinity-alkalinity stresses (Kaya et al., 2012; Patil et al., 2015; Nasri et al., 2017; El-Afry et al., 2018; Kocak et al., 2022) and identified salinity tolerant lines based on germination, seedling characteristics, and biomass and K+/Na+ ratio. Genes conferring salt tolerance by increasing root length, improving membrane injury and ion distribution in flax were identified by Wu et al. (2019a). Since flax can tolerate the pH up to 9, thus can serve to utilize agricultural land where other crops cannot be successfully grown.
Heat stress adversely affects the growth, development, and physiological processes, and thus yield particularly in tropical and subtropical regions (Ramirez-Villegas et al., 2020). A sustained period of heat stress (40°C for 5–7 days) during flowering might have a significant impact on pollen production, pollen viability, flowering, boll development, seed set, oil quality, and quantity in flax (Cross, 2002; Cross et al., 2003; Saha et al., 2019, 2021). Fiber flax does not require high temperatures. The largest and highest quality fiber flax yields are obtained in humid, cloudy, and relatively cool (18°C–20°C) conditions. High temperature particularly terminal heat is limiting for flax growth, resulting in low adaptation of elite fiber flax genotypes to warmer climes. Although few studies have been conducted on the effects of higher temperatures on growth, physiological processes, and yields in flax, the molecular dissection is hitherto unknown (Cross et al., 2003; Pokhrel and Meyers, 2022).
Among biotic stresses, globally most widespread and devastating pathogen of flax is Fusarium oxysporum f. sp. lini which causes wilt disease and can result in an 80%–100% loss in yield (Rashid, 2003). The fungus infiltrates into the flax root cells and then advances intra-cellularly into vascular tissue. The fungal microconidia germinate and thus block the vascular vessels and prevent water and nutrient translocation resulting in epinasty followed by progressive wilting and death. Along with fusarium wilt, flax rust, caused by Melampsora lini is another important fungal disease limiting flax production worldwide. The gene-for-gene relationship was initially described for the flax rust interaction (Flor, 1956). Since then, it has served as a model pathosystem to study underlying genetics in host-pathogen interaction in plants. Extensive work has been done on flax-rust interaction at molecular (resistance gene R) and pathogen effectors (avirulence genes Avr) level (Ravensdale et al., 2011); however, whole genome responses involving signaling and defense remains largely unexplored. In addition to wilt and rust, other widespread disease of flax is pasmo caused by Septoria linicola, while anthracnose and powdery mildew (caused by Colletotrichum lagenarium and Oidium lini, respectively) are less common and endemic in nature.
Flax occupies an important position in global economy due to its wide industrial utility as well as regional and niche preferences. However, unprecedent climate changes may have detrimental impact on flax productivity, and therefore in depth understanding of various diseases and environmental stresses assumes importance for future planning from the perspective of growth, equity and sustainability. Recent technological advances in DNA sequencing and molecular biology have expedited genomics and transcriptomic research and thus paved way for accelerated development of other domains of omics such as proteomics, metabolomics, and phenomics. Amalgamation of omics assisted multidisciplinary approach is necessary for understanding and investigating complex stress tolerance mechanism to design climate resilient flax varieties. Despite multitudinous utility and being a model crop for research studies, there is scanty and scattered information regarding integration of omics approaches for flax improvement. Present review is intended to apprise the readers about the current status of omics interventions in flax in response to major biotic and abiotic stresses and underlying molecular pathways.
Major components of omics include genomics (generation of genetic and genomic resources, gene mapping, functional genomics, and genomic selection), transcriptomics (gene regulation and expression profiling), proteomics (protein identification and effects), metabolomics (metabolite profiling, regulation, pathway and intermediates), phenomics (automated study and analysis of phenotypic and physiological effects), and ionomics (elemental identification, composition, effects, and interactions). Different omics mechanism and their integration has pivotal role in understanding plant systems biology as elaborated in extensive reviews (Fukushima et al., 2009; Weckwerth et al., 2020; Pazhamala et al., 2021). Omics assisted technologies have been advocated and utilized for engineering stress tolerance in reviews on rice (Kumari et al., 2022), wheat (Shah et al., 2018), soybean (Chaudhary et al., 2015), tomato (Chaudhary et al., 2019), and flax (Shivaraj et al., 2019). However, relatively less efforts have been made to utilize the available genetic and genomic resources for flax improvement compared to other crops. The advanced tools like genome-wide association studies (GWAS) and genomic selection in conjugation with other omic technologies provide an opportunity to increase the precision of plant selection for flax improvement as suggested by Shivaraj et al. (2019) and Akhmetshina et al. (2020) while reviewing the utilization of high-throughput sequencing technologies and omics-assisted breeding for development of climate-smart flax. Therefore, a holistic approach involving diverse technologies can greatly facilitate the introduction of climate-resilient traits into flax genotypes for sustainable productivity. A schematic view of integration of key omics approaches that can be utilized for the improvement of flax under various biotic and abiotic stresses is presented in Figure 2. In further sections of the review, we have elaborated the advancement made in various omic technologies and the amalgamation of omics data in future flax breeding for economic and sustainable yield.
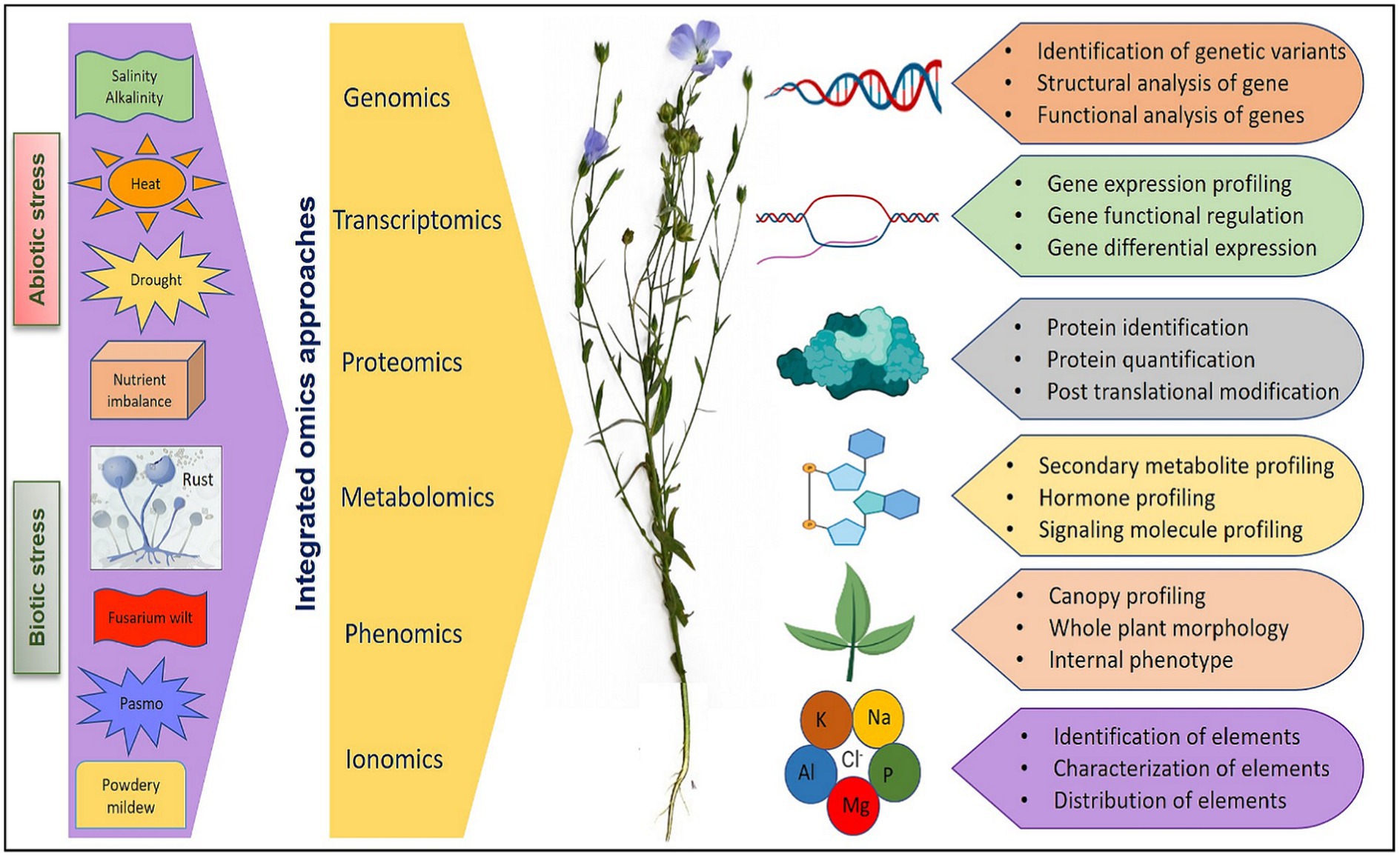
Figure 2. An overview of integration of different omics approaches for flax improvement under various abiotic and biotic stresses.
In the initial years of the century, many molecular markers such as rapid amplification of polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), inter-simple sequence repeat (ISSR), and expressed sequence tags-simple sequence repeats (EST-SSR) were used to assess the genetic diversity in flax (Oh et al., 2000; Fu et al., 2002a,b, 2003; Green et al., 2008; Cloutier et al., 2009, 2011, 2012; Rajwade et al., 2010; Uysal et al., 2010; Kaur et al., 2018; Saroha et al., 2022a). The substantial lead in the generation of genomic resources was made with the availability of whole genome sequence of flax (Wang et al., 2012). Subsequently, the whole genome resequencing and reduced representation sequencing information has been effectively utilized to understand crop diversity, marker identification, linkage map construction and QTL identification in flax. Genome wide SNP discovery through genotyping-by-sequencing (GBS) approach has been used to identify 258,873 SNPs distributed on all 15 flax chromosomes (Kumar et al., 2012). SNPs linked to major agro-morphological traits (Deng, 2013; Soto-Cerda et al., 2013, 2014; Xie et al., 2018; Saroha et al., 2022b), oil quality attributes (Soto-Cerda et al., 2014, 2018; You et al., 2018b), fiber length and plant height (Xie et al., 2018), mucilage and hull content (Soto-Cerda et al., 2018), and disease resistance (He et al., 2019a,b) have been identified in flax through GWAS. For improving abiotic stress tolerance, GWAS has been reported in many recent studies to identify potential SNPs for different traits such as oil content, yield, or improved stress tolerance indices in sunflower (Mangin et al., 2017), maize (Millet et al., 2016; Shikha et al., 2017), sorghum (Lasky et al., 2015; Badigannavar et al., 2018; Spindel et al., 2018), rice (Guo et al., 2018), and sesamum (Dossa et al., 2019). Although, a number of genes were discovered and functionally characterized for their role in abiotic stress tolerance in flax, for instance, NAC-domain transcription factor genes (LuNACs) associated with drought, salinity, cold and heat (Saha et al., 2021), putative heat shock factor (HSF) candidate genes for high temperature tolerance (Saha et al., 2019), transporter gene family detoxification efflux carriers (DTX)/multidrug and toxic compound extrusion (MATE) to mediate the response to abiotic stresses (Ali et al., 2020), and aquaporin (AQP) gene family in improving drought tolerance (Shivaraj et al., 2017), however, the progress is relatively slow in flax compared to other crops. Regarding biotic stress, Asgarinia et al. (2013) conducted QTL-analysis for powdery mildew resistance and detected loci by homology search in the whole-genome sequencing database using information about nucleotide sequences of ESTs and ВАС-clones. The de novo genome of flax rust pathogen Melampsora lini was sequenced and assembled and 16,271 putative protein coding genes were identified (Nemri et al., 2014). This could help to understand the previously unknown facts about number of virulence effectors, their function and degree of conservation. He et al. (2019a) conducted GWAS to identify genetic regions associated with pasmo resistance in 370 flax accessions of Canadian core collection and detected 258,873 SNPs using GBS. They identified 500 putative QTL, 45 of which spanned 85 resistance genes. Further, based on orthology with genes of Arabidopsis thaliana, two candidate genes, Lus10031043 and Lus10020016 for flax resistance to this pathogen were detected. Recently, You et al. (2022) performed both GWAS and GS analyses in 447 flax accessions comprising 372 core collection accessions and 75 breeding lines which were evaluated for powdery mildew resistance for 5–8 years across three locations. They identified a total of 349 QTNs (of which 44 were highly stable large-effect QTNs) and 445 candidate resistant gene analogs (RGAs) associated with powdery mildew resistance in flax. Interestingly, 45 of the identified QTNs were in RGAs of which 14 QTNs were with large effect (R2 = 10%–30%). Table 1 enlists various QTN/QTLs linked to major abiotic and biotic stresses in flax. However, much work has been done on agronomic and quality evaluation work while little attention has been paid to high throughput sequencing and GWAS for response to climatic threats and pathogen attack in flax. Therefore, comprehensive physiological, biochemical and molecular evaluation under different stress regimes followed by structural and functional genomics strategies as outlined in Figure 3 is required for improving biotic and abiotic stress tolerance in flax.
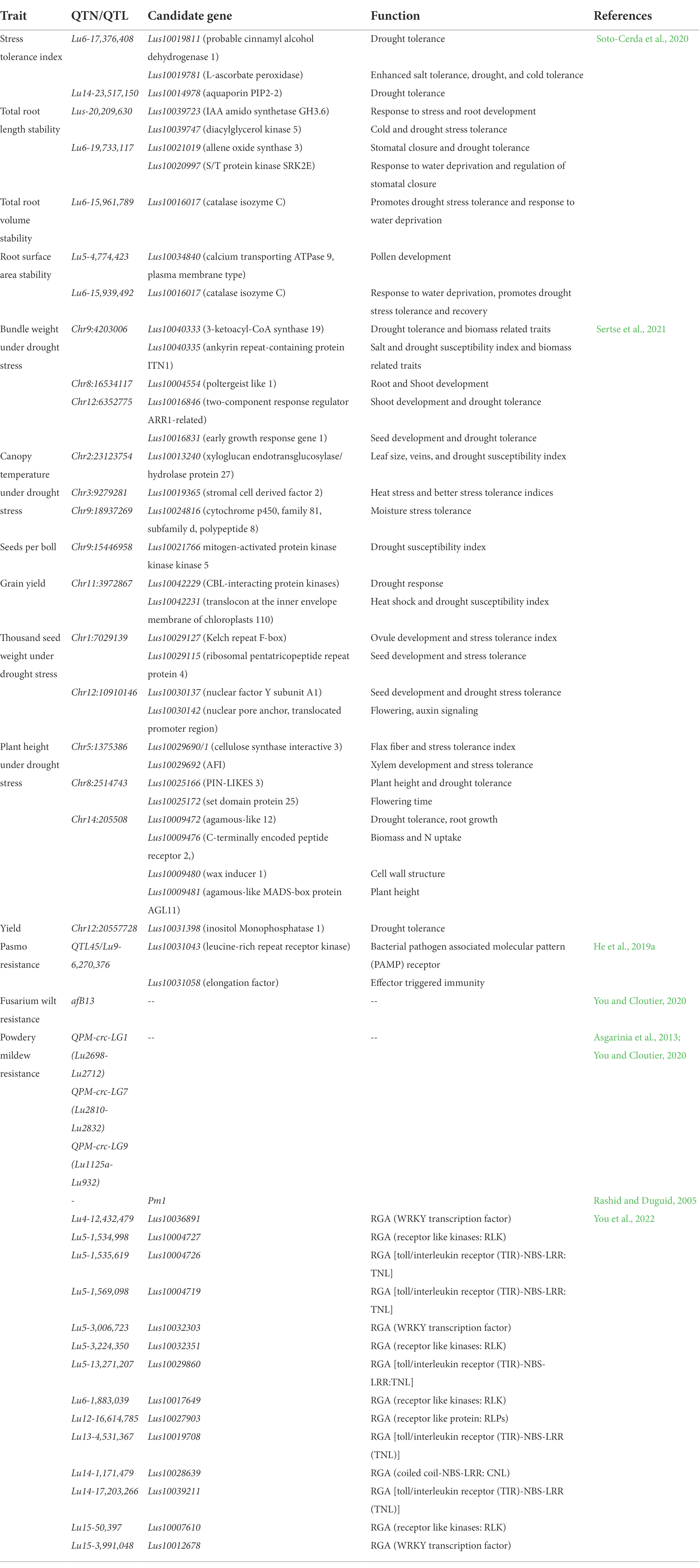
Table 1. Quantitative trait nucleotides/loci identified by Genome wide association studies for major abiotic and biotic stresses in flax.
Figure 3. Genomics enabled strategies for flax improvement in response to adverse climatic conditions and pathogenic invasion.
Genomic selection (GS) is a breeding approach that determines the genetic potential instead of identifying specific QTL and thus it overcomes restrictions involved in marker assisted selection (MAS) for speed breeding. GS has the potential to fix all the genetic variation of complex traits contrary to classical plant breeding approach which is slow in targeting the complex and low heritable quantitative traits. That is why it is emerging as promising technique exploiting molecular genetic markers to develop novel markers-based models for genetic evaluation. It involves the precise phenotyping of a selected varied group of genotypes (training population) in multiple environments and genotyping to develop statistical model (GS model) which is employed for the estimation of genomic estimated breeding values (GEBVs) in the breeding population. GS method has many advantages over conventional as well as marker assisted breeding as it deals with minor effect of QTL (Crossa et al., 2017). As a result, GS has been advocated as the most effective method for predicting genetic values for selection by combining all available molecular markers with phenotypic data (Deshmukh et al., 2014; Chaudhary et al., 2015; Abed et al., 2018). GS studies conducted in flax resulted in increased genetic gain per unit time during the breeding cycle (You et al., 2016). They used three bi-parental populations developed by crossing high-yielding, high and low alpha linolenic acid content flax lines for QTL mapping to optimize GS model. He et al. (2019b) developed a high-throughput prediction model of genetic resistance of flax to Septoria linicola, which is one of the most accurate genomic prediction model for disease resistance in plants. The latest prediction model by You et al. (2022) has been constructed using 447 flax accessions as a training population and the powdery mildew ratings over 5 years at three locations. All the 349 QTNs identified through GWAS explained 96% of powdery mildew variation showing high predictive ability and the potential of this model in applied in genomic prediction. With the increased genetic and genomic resources in flax, more extensive GS research is expected in the near future which may contribute in releasing new cultivars tailored to specific needs. Presently the more extensive use of GS remains a challenge owing to higher expenses than MAS. However, the availability of low cost, flexible and high-density marker system, cheaper NGS technologies are expected to make the whole genome re-sequencing feasible and cost effective for the GS in near future (Bhat et al., 2016). The current status of GS studies in crop plants, and perspectives for its successful implementation in the development of climate-resilient crops has been reviewed by Budhlakoti et al. (2022) who emphasized that the studies on genetic architecture under drought and heat stress can significantly accelerate the development of stress-resilient crop varieties through GS.
Transcriptome profiling provides a comprehensive overview of gene expression, regulation and helps in identification of key genes involved in stress tolerance mechanism. Various approaches are used to study transcriptome such as expressed sequence tags (ESTs), spotted micro arrays, sequencing along with suppression subtractive hybridization, Affymetrix GeneChips and RNA-sequencing depending upon the availability of genomic resources generated and plant type. With the rapid advancement in next-generation sequencing technologies, RNA-sequencing has become the most efficient, cost-effective and high-throughput transcriptomic method. So far, ample of transcriptomics studies has been carried out in oilseed crops such as flax (Wu et al., 2018), sesame (Dossa et al., 2019), soybean (Leisner et al., 2017), Jatropha (Cartagena and Marquez, 2021), and sunflower (Moschen et al., 2017) to ascertain the effect of drought and salinity.
The flax genome sequencing and availability of genetic maps (Wang et al., 2012; You et al., 2018a; Cullis, 2019; Akhmetshina et al., 2020) laid the foundation for significant number of transcriptomic studies and identification of genes underlying traits of agronomic and economic importance. High-throughput sequencing had been carried out for studying flax response to drought (Dash et al., 2017), alkalinity and salt (Yu et al., 2014, 2016; Dmitriev et al., 2019), metal stress (Dmitriev et al., 2016a; Zyablitsin et al., 2018), and nutrient stress (Melnikova et al., 2015, 2016). Transcriptome study from a moderately drought tolerant flax cultivar (T-397) of Indian origin was conducted by Dash et al. (2017) and expression profiling helped to identify loci/markers for selection of drought resilient varieties. Using transcriptome analysis data, Shivaraj et al. (2017) demonstrated high expression of integral membrane proteins, mostly aquaporins and low expression of integral nodulin-26-like proteins leading to better understanding of their physiological functioning. Another study reported overexpression of drought responsive element binding protein 2A (DREB2A) gene imparting drought tolerance in transgenic line of flax cv. Blanka (Tawfik et al., 2016). Similarly, for high temperature stress, few genes have been discovered and functionally characterized in flax. Saha et al. (2019) reported the genome-wide identification of 34 putative HSF genes from the flax genome. Heat shock factors and NAC domain transcription factors bestow distinct expression patterns under heat stress. Wu et al. (2019a) identified two salt-tolerant genes homologous with Arabidopsis Senescence-Associated Gene 29 (SAG29) having putative role in enhancing salt tolerance by increasing root length, improving membrane injury and ion distribution. Transcriptome of response of flax to unfavorable soil pH led to revelation of genes with altered expression profiles (Yu et al., 2014; Dmitriev et al., 2016b, 2020; Wu et al., 2019b). Flax response to non-optimal soil acidity (increased pH) and zinc deficiency revealed genes involved in ion transport, cell wall biogenesis and photosynthesis through transcriptomics (Dmitriev et al., 2019). The induction of several pathogen related dominant genes in high pH tolerant flax cultivars were associated to overcome unfavorable effects of reduced Zn content. Melnikova et al. (2016) identified 96 conservative homologs of microRNA belonging to 21 families, and reported the role of seven microRNAs (miR168, miR169, miR395, miR398, miR399, miR408, and lus-miR-N1) in the regulation of gene expression and metabolism in plants under nutrient stress. Changes in the expressions of miR319, miR390, and miR393 associated with significant increase of gene expression in glutathione-S-transferase and UDP-glycosyl-transferase provided insight into putative role of these genes in providing protection against aluminum stress via scavenging of reactive oxygen forms and modification of the cell wall (Dmitriev et al., 2017). Similarly, altered expression profiles of lus-miR-N1 and miR399 under phosphate deficiency (Melnikova et al., 2015) were detected. Yu et al. (2016) reported differentially expressed genes (DEGs) and saline-alkaline tolerant miRNAs in flax (Lus-miRNAs) for the first time and selected 17 known lus-miRNA and 36 new lus-miRNA after assessment of the DEG profiles to predict the target genes. It was suggested that the miR398 and miR530, coding for superoxide dismutase and transcription factors of the WRK family could play significant roles in flax stress resistance. Genome-wide annotation of miRNAs and phasiRNAs encoding genes along with sRNA transcriptomics (reproductive stage) showed downregulation of phasiRNAs in flax reproductive organs under heat stress (Pokhrel and Meyers, 2022).
Pathogen attack also triggers alterations in the transcriptional and translational profile of plants leading to activation of a number of genes and metabolic pathways as defense mechanism. Kostyn et al. (2012) evaluated the gene response in early stages of infection by Fusarium and identified 47 genes including genes responsible for phenylpropanoid pathway enzymes and antioxidant biosynthesis in flax. Transcriptome of dominant Canadian cv. CDC Bethune, an oil type flax resistant to Fusarium wilt and sensitive variety, Lutea identified 100 genes that were differentially expressed in response to early pathogenesis (Galindo-Gonzalez and Deyholos, 2016). Among these, several key genes that are involved in activation of pathogenesis-related (PR) interactions, secondary metabolism and lignin formation had increased transcript abundance in congruence with other pathogenesis related studies done earlier. Similarly, in another study, transcriptome of four fibrous flax cultivars (two resistant and two susceptible) as well as two resistant BC2F5 populations with respect to Fusarium wilt, showed predominant overexpression of numerous genes involved in defense response such as PR protein encoding genes, ROS production, and related to cell wall biogenesis (Dmitriev et al., 2017). Recently, Boba et al. (2020) reported that upregulation of the terpenoid pathway leading to increased ABA content upon Fusarium infection in flax activates the early plant’s response and PR genes especially chitinase and β-1,3-glucanase play an essential role for resistance. Earlier study reported that transgenic flax plants overexpressing the β-1,3-glucanase gene showed lower susceptibility to this pathogen (Wróbel-Kwiatkowska et al., 2004). The transcriptomal response of the resistant flax cultivar was found to be quicker and more effective allowing translation to a higher number of activated and repressed genes in response to infection by F. oxysporum lini (Boba et al., 2021). The numbers of the differentially expressed PR genes in resistant variety were higher initially (24 hpi) but similar later (48 hpi) in comparison to susceptible variety further established that the degree of the response plays deciding role in the differential resistance reaction, even though the similar qualitative response. RNA-Seq analysis of M. lini transcriptome was performed during early establishment of disease in flax and the expression profiles of Avrs and effector genes revealed 58 previously uncharacterized genes encoding secreted proteins (Wu et al., 2019b).
Major transcriptomic studies revealing genes that were upregulated/downregulated in response to different abiotic and biotic stresses in flax are listed in Tables 2 and 3. Flax transcriptome sequences and gene expression information are available in NCBI Sequence Read Archive and NCBI Gene Expression Omnibus databases.1 Flax microRNA data are deposited in miRbase database, wherein sequences of 124 microRNA of L. usitatissimum are presented along with primary and secondary structures and localization in flax genome.2 Importantly, there are more research publications regarding tolerance to abiotic stresses in comparison to resistance to the biotic stress in the area of flax transcriptomics, which may be due to targeted traits under breeding programs for specific regions. Most of the transcriptomic studies were limited to only one or two cultivars, however more number of diverse genotypes should be investigated for the comparative analysis and gene function annotation. Study of microRNAs and their role is in the initial stages in flax and several key questions remain unanswered. Further knowledge in this domain will assist scientists to develop artificial microRNA as effective tools to regulate gene expression.
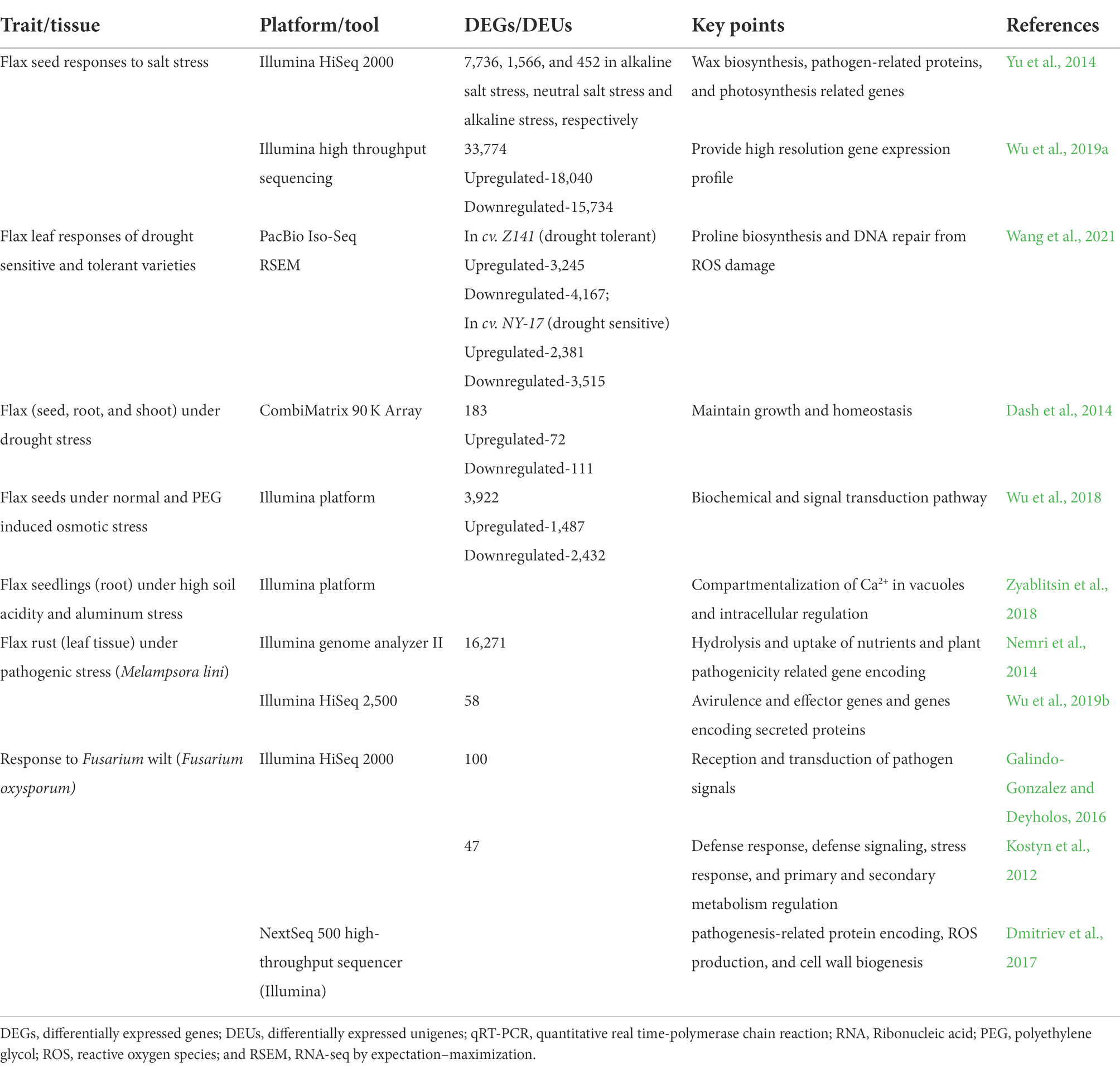
Table 2. Global transcriptomic analysis revealing gene expression profiles in response to major abiotic and biotic stresses in flax.
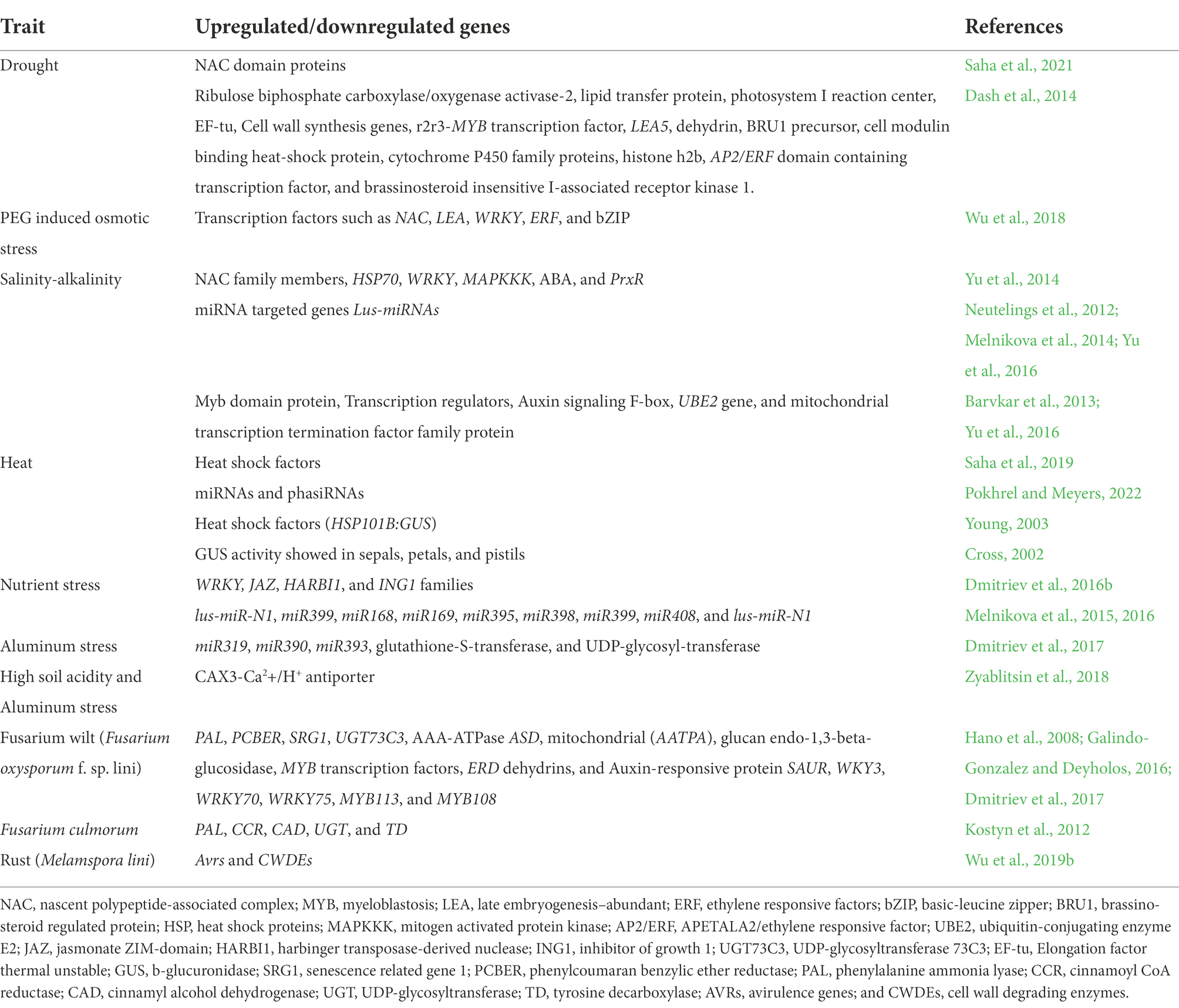
Table 3. Important genes which are upregulated and downregulated in response to various biotic and abiotic stresses in flax.
Metabolic profiling gives the precise depiction of biological and physiological state of an organism as metabolites are the end products of gene expression and integration of metabolomics has pivotal role in understanding plant systems biology (Weckwerth, 2003; Ghatak et al., 2018; Pontarin et al., 2020). However, the actual size of the plant metabolome being unknown, and owing to the greater diversity of metabolites in plants than other organisms, metabolomic analysis faces some challenges as reviewed by Hall (2006), Schauer and Fernie (2006), and Harrigan et al. (2007). Several analytical platforms have been used to identify and quantify the wide range of primary and secondary metabolites in response to stress, these include a group of well-established analytical techniques, namely, nuclear magnetic resonance (NMR) and mass spectrometry (MS)-based techniques such as GC–MS (Gas Chromatography–Mass Spectrometry), CE-MS (Capillary electrophoresis-Mass spectrometry), LC–MS (Liquid Chromatography – Mass Spectrometry), and FTIR (Fourier transform infrared; Schripsema, 2010; Kaspar et al., 2011; Putri et al., 2013; Simó et al., 2014). NMR requires limited sample preparation and medium to high abundance metabolites are usually detected using this technique. Further, the recent advancements in field strength in NMR superconducting magnets have resulted in improved spectral resolution and detection sensitivity. Contrary to NMR, high and ultra-high resolution mass spectrometers used in current MS-based approaches yield higher sensitivity when analyzing complex plant metabolite mixtures. Ibáñez et al. (2013) presented an overview of recent novel direct ionization or desorption/ionization techniques developed and combined for applications in food metabolomics in their review article. The beneficial effect of metabolites such as lignans, polyunsaturated fatty acids (PUFA), specifically ω-3 fatty acids have been well documented for nutritional enhancement and prevention of certain ailments (McCann et al., 2005; Kouba and Mourot, 2011). Flaxseed being an important source of bioactive compounds of interest in human health (lignans and ω-3 fatty acids) and have multitudinous applications in food industry, Ramsay et al. (2014) developed an NMR metabolomics-based tool for selection of flaxseed varieties with better nutrient profile. In addition to metabolomics of nutritional compounds, the detection of the accumulation of many secondary metabolites such as proline, glycine betaine, sugars, and inorganic ions has been reported in oilseeds to help adaption of plant to abiotic stress (El Sabagh et al., 2019). Metabolites, such as β-Aminobutyric acid (BABA), have proven role in inducing drought tolerance in Arabidopsis (Jakab et al., 2005), spring wheat (Du et al., 2012), apple (Macarisin et al., 2009), rice (Garg et al., 2002), tomato (Cortina and Culianez-Macia, 2005), and potato (Bengtsson et al., 2014). The overexpression of BABA resulted in enhanced accumulation of osmoprotectants namely anthocyanins and proline, overexpression of the pathogenesis related genes PR1, PR2, and PR5 in Arabidopsis (Jakab et al., 2001; Singh et al., 2010; Wu et al., 2010), trehalose biosynthesis induced drought tolerance in tobacco (Romero et al., 1997). In flax, BABA causes accumulation of proline and non-structural carbohydrates and reduction in aspartate content and inorganic solutes in response to water stress (Quéro et al., 2015). Proline and glycine-betaine contents were found to be relatively high under salinity stress in flax (Qayyum et al., 2019) and rice (Cha-um et al., 2006). Total soluble sugars, total protein content and compatible solutes, such as proline, betaine were found to increase with increasing salinity in flax genotypes, suggesting that they may play a role in adjusting osmotic stress under PEG induced water stress and saline-alkaline environments (Guo et al., 2012, 2014; Naz et al., 2016). Differential level of lipid peroxidation and metabolic profile of MDA in wild-type and PLR-RNAi transgenic flax has been reported under salinity and or osmotic stress (Qayyum et al., 2019; Hamade et al., 2021).
Pathogen attack also triggers alterations in the translational profile of plant resulting in synthesis of many secondary metabolites such as flavonoids, catecholamines, polyamines, lignins, terpenoids, tannins, phenolic, and phenylpropanoic acids as defense mechanism. Metabolomics studies have been carried out extensively in rice to find key metabolic products and pathways in response to various biotic stress (Vo et al., 2021). These studies were aimed to understand the induction of defense mechanism involving Pathogen associated molecular pattern (PAMP)-triggered immunity (PTI) and effector triggered immunity (ETI) in model crop rice. The first report to describe metabolites of early flax to Fusarium infection was by Kostyn et al. (2012) who determined the level of metabolites produced in phenylpropanoid pathway (flavonoids and phenolic acids) by GC–MS. Wojtasik et al. (2015) identified for the first-time genes involved in polyamine synthesis pathway and reported increase in content of polyamines putrescine, spermidine, and spermine during Fusarium infection in flax. The main polyamine identified was putrescine. Furthermore, differential content of polyamine was measured in response to infection by pathogenic and non-pathogenic Fusarium strains in flax which indicate different defense mechanisms. Thus, stress induces drastic changes in the metabolic profile of a plant and therefore complete metabolite profiling may provide valuable insights into stress tolerance mechanisms (Supplementary Figure 1). The prior knowledge of metabolomics in conjugation with other allied omics technologies such as genomics, transcriptomics and proteomics is essential to understand the complete overview of biochemical and molecular mechanisms in response to various biotic and abiotic stress elicitors. However, this is a new research area and no metabolomic databases with reference to environmental stress are available until now.
Proteomics is the study of the structural and functional characteristics of all proteins in a living organism in real-time. It includes two-dimensional (2-D) gel electrophoresis, mass spectrometry (MS), ELISA, Western Blotting, and matrix-assisted laser desorption ionization-time of flight (MALDI TOF) along with various bioinformatic tools (Baggerman et al., 2005; Gevaert and Vandekerckhove, 2011; Chaudhary et al., 2019). Recent achievement in proteomics has reduced the errors in protein assessment and provided new possibilities for high-throughput proteome analyses. Mostly proteomic investigations have been focused on rice, wheat, barley, maize, potato, and soybean, all of which have whole genome sequences available in public domain. In oilseeds, proteomic studies on Indian mustard (Alvarez et al., 2009), flax (Hradilová et al., 2010; Klubicová et al., 2011), and sunflower (Balbuena et al., 2011) have been reported recently. The proteome analyses revealed that continuous higher level of stress responsive proteins (that includes transcriptional regulators such as SWIB/MDM2 protein, Myb protein, B-Peru-like protein involved in anthocyanin biosynthesis) in tolerant plants help them to cope up with adverse effects of stress compared to sensitive counterpart (Pang et al., 2010; Wendelboe-Nelson and Morris, 2012). Enhanced level of specific proteins, lipoxygenase (LOX), several chaperons (HSP70, HSP90, CPN60-α, β, and cyclophilin A), and glutathione-S-transferase (GST) were found in drought tolerant barley and wheat varieties with respect to sensitive counterpart (Kosová et al., 2014). Another study reported reduction in RubisCo (smaller and larger subunits) as well as calcium cycle enzymes such as phosphoribulokinase (PRK), phosphoglycerokinase (PGK) and transketolase in wheat under salt (Caruso et al., 2008), drought (Caruso et al., 2009) and low temperature (Rinalducci et al., 2011). Similarly, changes in OEE1 and OEE2 proteins were frequently found in barley under salt stress (Rasoulnia et al., 2011; Fatehi et al., 2012) and drought stress (Ghabooli et al., 2013). Also, in developing wheat grains subjected to a heat phase, a rise in many minor HSP proteins, as well as HSP82 from the HSP90 family was detected in the endosperm (Skylas et al., 2002; Majoul et al., 2004). Similarly, proteome analysis conjugated with physiological response in two maize varieties resistant to drought stress reported the role of HSP to be important in protecting plants from drought stress (Li et al., 2021b). Lately, Halder et al. (2022) reviewed the role of proteomics for abiotic stress tolerance in wheat and presented a summary of proteomic studies on salinity, drought stress tolerance, and root system architecture conducted in the last decade.
Proteomic analysis of biotic stress has been advantageous to describe the proteome of plants and pathogens infected tissues. The global proteomics studies investigating biotic stress responses in rice have been extensively reviewed (Vo et al., 2021) and many potent metabolites responsible for resistance have been enlisted. The changed proteome response in response to biotic stress has been elucidated in many crops such as grapevine resistance to downy mildew (Milli et al., 2012; Palmieri et al., 2012), tomato infected with Botrytis cinerea (Shah et al., 2012), avocado resistance to root rot (Acosta-Muniz et al., 2012), and resistance related proteins mainly involved in pathogenesis response were identified. Proteomic analysis has also been used to explore plant-virus interaction to unravel proteins corresponding to enzymes involved in photosynthesis, primary metabolism, and defense (Di Carli et al., 2010). On a similar note, proteomics and phosphoproteomics analyses may assist in identification of candidate protein under various stress conditions in flax (Figure 4). Presently, this domain has been explored to a very limited extent in flax (Hradilová et al., 2010; Klubicová et al., 2011).
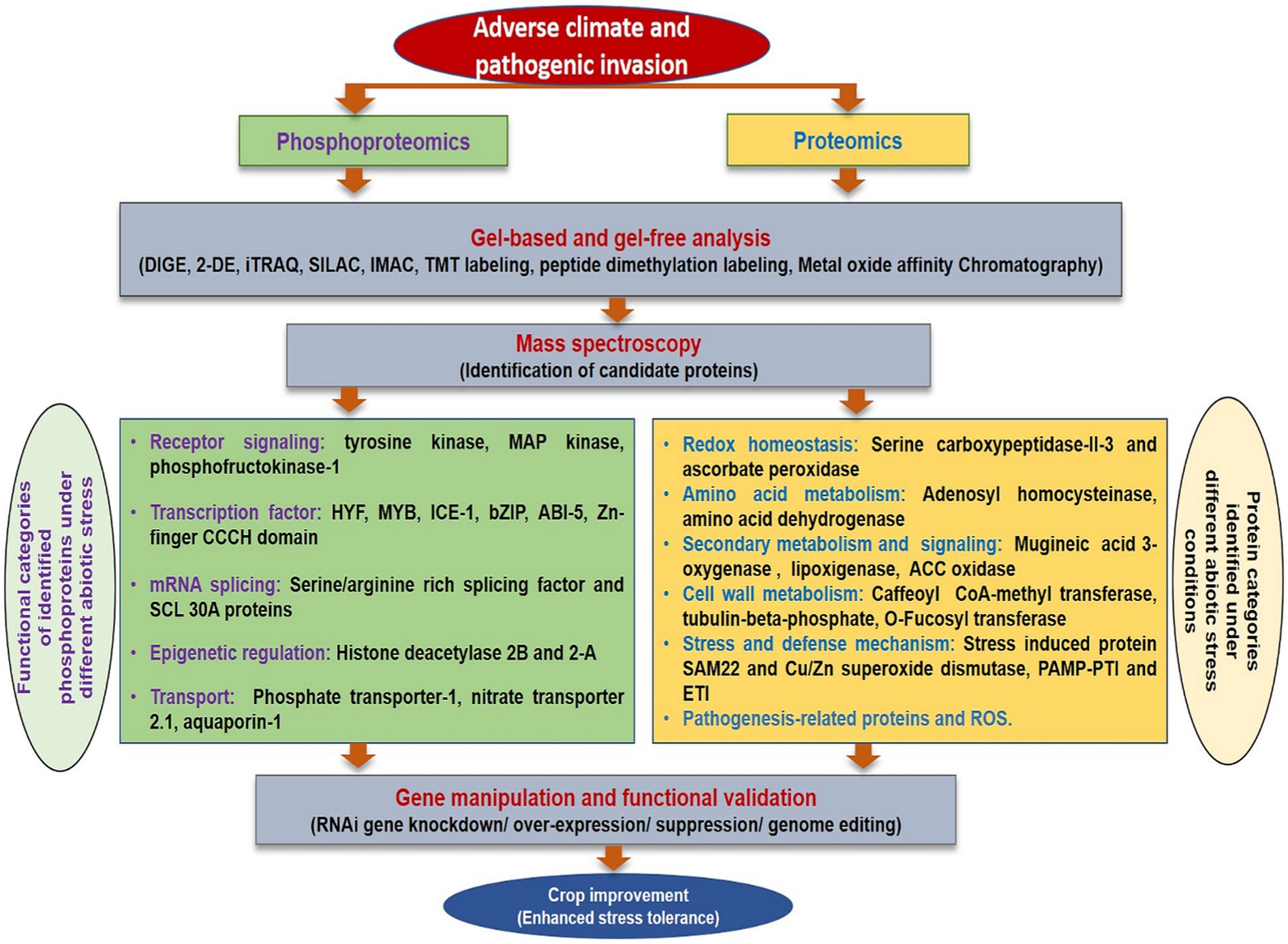
Figure 4. A comprehensive overview of proteomics and phosphoproteomics analysis under different stresses.
Ionomics is the study of an elemental composition of metal, metalloids and non-metal of the various types of plant species, with a focus on high-throughput detection and measurement (Supplementary Figure 2). Ionomic profile of plant species can be detected using high-throughput technologies such as Inductively Coupled Plasma-Mass Spectrometry (ICP-MS) and Inductively Coupled Plasma-Atomic Emission Spectrometry (ICP-AES; Salt et al., 2008). It provides the important role in understanding the different composition of elements along with their involvement in plant biochemistry, physiology, and nutrition. Plants have evolved with a variety of element uptake abilities owing to numerous soil types and other edaphic factors associated with growth and development (Fujita et al., 2013). Additionally, ionomic profile of a crop is affected by element availability, uptake ability of roots, transport, and environmental stress. A wide range of investigations have been carried out in the realm of ionomics. The ionome of wild and cultivated barley subjected to different salt tolerance levels revealed a substantial negative relationship between the amount of accumulated Na+ and metabolites involved in glycolysis and tricarboxylic acid (TCA) cycle (Wu et al., 2013). Studies performed in wheat (Guo et al., 2009) and other grasses, such as Aneurolepidium chinense (Shi and Wang, 2005), and Setaria viridis (Guo et al., 2011) and flax (Guo et al., 2014) showed that concentration of Na+ increases with increasing alkalinity stress as plants accumulate significant levels of Na+ in their vacuoles to reduce cell water potential. Under PEG induced water stress in flax, the main inorganic ions involved in osmotic adjustment were K+, Na+, Ca2+, and Cl− thereby increasing drought resistance (Guo et al., 2012). However, no significant differences were observed between the effects of salinity and alkalinity stress on the concentration of Na+ and K+ in shoots in case of flax (Guo et al., 2014). This suggests that the adaptive mechanism of flax shoots to the alkaline stress may differ from that of other plants such as barley (Guo et al., 2009) and Chloris virgata (Yang et al., 2010) where K+ concentration of shoots was found to be lower under alkaline stress. Another study in flax showed significant increase in the uptake of Cl−, H2PO4−, and SO42, whereas the levels of NO3− decreased in flax shoots under salt stress. This depicts that these anions build up in the vacuoles to counteract the input of Na+ and together they keep the cell hydrated (Parida and Das, 2005). Also, the concentrations of inorganic anions were much lower under alkali stress than under salt stress with the same osmotic potential, implying that the high pH of alkali stress may block anions such as NO3−, H2PO4−, and SO42− absorption in flax (Guo et al., 2013, 2014). Application of many inorganic elements can result in enhanced tolerance to abiotic stress, for example, Silicon has proven beneficial against drought, salinity, heat, heavy metals and UV-b (Liang et al., 2007; Pilon-Smits et al., 2009; Deshmukh et al., 2014). Hyperaccumulation of manganese (Mn) in the leaves of grapevine has been reported to delay pathogen spread and thus induction of powdery mildew resistance (Yao et al., 2012). Therefore, studying the elemental profile can aid to better understanding of stress tolerance mechanism. However, ionomics studies are yet to get more attention in flax.
Phenomics is the study of set of all phenotypes involving genotype, phenotype and environment (GxPxE) interactions in specific environmental conditions using high-throughput analysis (Ichihashi and Sinha, 2014; Tardieu et al., 2017; Zhao et al., 2019; Weckwerth et al., 2020; Ninomiya, 2022). Therefore, phenotype provides ultimate association between environment and plant genotype. In the last decade, advances in sequencing technologies have increased genotyping efficiency, but phenotypic characterization has proceeded more slowly, limiting the identification of quantitative features, particularly those related to stress tolerance (White et al., 2012). Due to complicated biosynthetic processes that address response of plants to external stimuli, phenotyping in response to abiotic stress remains a big challenge (Pratap et al., 2019). In the postgenomic era, the importance of precise phenotyping has become more important owing to dependence of genomic approaches such as GWAS, GS and QTL on the high-throughput phenotyping for the improvement of targeted traits (Walter et al., 2015). Phenomics combined with other omic techniques has the greatest potential for plant breeding. Therefore, non-invasive technologies such as color imaging of biomass, far infrared imaging of the canopy, lidar (includes RBG digital imaging) to assess growth parameters and magnetic resonance imaging (MRI) have been explored to estimate above ground canopy and hidden half (root system) of the plants (Yang et al., 2021), few examples include PHENOPSIS (an automated platform to examine water stress in Arabidopsis (Granier et al., 2006) and soil-filled rhizoboxes for study of root system architecture using RGB imaging in wheat (Bodner et al., 2017), RGB digital imaging for phenotyping of plant shoots (Humplík et al., 2015), infrared thermography to validate role of stomatal conductance in barley and wheat seedlings under salinity stress (Sirault et al., 2009) and chlorophyll fluorescence imaging to screen abiotic stress response in tobacco, canola and cotton (Saranga et al., 2004; Baker, 2008) have been explored. X-ray, computed tomography (CT) and nuclear magnetic resonance (NMR) has been used for 3D visualization of root architecture in situ. To automate the analysis of root traits, there has been a proliferation of semi-automated such as SmartRoot, GROWSCREEN_ROOT, EZ-Rhizo, and automated softwares WinRhizo, Root Reader 3D and GiaRoots in recent years. Advanced phenomics platforms for a larger range of crop plants such as state of the art “The Australian Plant Phenomics Facility” (APPF),3 multispectral and fluorescence imaging for physiological phenotyping4 and many others covering ground-based proximal phenotyping to aerial large-scale remote sensing have been developed. Li et al. (2021a) have elaborated the current developments, configurations, novelties, as well as strengths and weaknesses of diverse high-throughput plant phenotyping platforms in a recent review. Few online databases, such as http://www.plant-image-analysis.org are available to assist users in image processing. Thus, high-resolution IR/NIR cameras, fluorescence imaging systems, laser scanners, hyperspectral imaging systems and high throughput advance plant phenotyping platforms are modern tools to get real time phenome in response to external environment, nutrients and disease. However, deep learning tools are needed to extract phenome information through advanced algorithms from huge datasets generated while phenotyping. In addition, comprehensive management of platforms and softwares are considerable challenges limiting this application to few major crops such as rice, maize and wheat.
Globally, enormous data are being rapidly generated and annotated to better understand the complicated biological pathways involved in stress tolerance of plants. The availability of diverse genomic resources, such as whole genome sequences, transcriptomes, molecular markers, and linkage maps, has increased significantly in many crops including flax over the last decade. These resources can be efficiently utilized for wider climatic adaptability and biotic stress tolerance in flax through varietal improvement program. Flax being a high value economic crop, finds wide range of uses in the culinary, bioenergy, nutritional, nutraceutical industries. Different omic tools and integrated approaches discussed in the present review provide glimpses of current scenarios and future perspectives for the effective management of abiotic stress and disease resistance in flax. Under integrated approach of omics utilization, the techniques of genomics, transcriptomics, and metabolomics have been employed in flax, but other significant areas such as proteomics, phenomics, and ionomics are yet to be explored. Deeper insight into genetic architecture, signaling pathways, and adaptation under stress through the lenses of different omics technologies are critical to understand the stress response and the underlying regulatory mechanism. Integration of these omics technologies on diverse flax genotypes with substantial trait variation are expected to unravel hitherto unknown factors in flaxseed which would pave way for the breeding of stress tolerant varieties for the larger good.
BY prepared the initial draft. BY, ON, VK, DW, and SY wrote, edited, and reviewed the original draft. ON and BY helped in the preparation of figures and tables. VK and AK conceptualized the theme. VK, DW, and AK did supervision, reviewing, and editing of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by funding for the project (No. BT/Ag/Network/Linseed/2019-20) from Department of Biotechnology (DBT), Government of India.
Authors acknowledge the funding support for the project (No. BT/Ag/Network/Linseed/2019-20) from Department of Biotechnology (DBT), Government of India. Authors thank former and present Director, ICAR-National Bureau of Plant Genetic Resources (NBPGR), New Delhi and Heads, DGE, ICAR-NBPGR for facilitation, guidance and inspiration.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls. 2022.931275/full#supplementary-material
Supplementary Figure 1 | Schematic diagram showing role of metabolomics in improving abiotic and biotic stress tolerance.
Supplementary Figure 2 | Information flow for ionomics and their role in stress response.
1. ^https://www.ncbi.nlm.nih.gov/geo/
2. ^https://www.mirbase.org/textsearch.shtml?q=Linum
Abd El-Fatah, A. A. E. (2007). Comparative study on some flax cultivars. J. Agric. Sci. Mansoura Univ. 32, 7111–7119. doi: 10.21608/jpp.2007.220613
Abed, A., Pérez-Rodríguez, P., Crossa, J., and Belzile, F. (2018). When less can be better: how can we make genomic selection more cost-effective and accurate in barley? Theor. Appl. Genet. 131, 1873–1890. doi: 10.1007/S00122-018-3120-8
Acosta-Muniz, C. H., Escobar-Tovar, L., and Valdes-Rodriguez, S. (2012). Identification of avocado (Persea americana) root proteins induced by infection with the oomycete Phytophthora cinnamomi using a proteomic approach. Physiol. Plant. 144, 59–72. doi: 10.1111/j.1399-3054.2011.01522.x
Akhmetshina, A. O., Strygina, K. V., Khlestkina, E. K., Porokhovinova, E. A., and Brutch, N. B. (2020). High-throughput sequencing techniques to flax genetics and breeding. Ecol. Genet. 18, 103–124. doi: 10.17816/ecogen16126
Ali, E., Saand, M. A., Khan, A. R., Shah, J. M., Feng, S., Ming, C., et al. (2020). Genome-wide identification and expression analysis of detoxification efflux carriers (DTX) genes family under abiotic stresses in flax. Physiol. Plant. 171, 483–501. doi: 10.1111/ppl.13105
Alvarez, S., Berla, B. M., Sheffield, J., Cahoon, R. E., Jez, J. M., and Hicks, L. M. (2009). Comprehensive analysis of the Brassica juncea root proteome in response to cadmium exposure by complementary proteomic approaches. Proteomics 9, 2419–2431. doi: 10.1002/pmic.200800478
Asgarinia, P., Cloutier, S., Duguid, S. R., Rashid, K., Mirlohi, A., Banik, M., et al. (2013). Mapping quantitative trait loci for powdery mildew resistance in flax. Crop. Sci. 53, 2462–2472. doi: 10.2135/cropsci2013.05.0298
Asgarinia, P., Mirlohi, A., Saeidi, G., Mohamadi-Mirik, A. A., Gheysari, M., Razavi, V., et al. (2017). Selection criteria for assessing drought tolerance in a segregating population of flax (Linum usitatissimum L.). Can. J. Plant Sci. 97, 424–437. doi: 10.1139/cjps-2016-0140
Ayelign, A., and Alemu, T. (2016). The functional nutrients of flaxseed and their effect on human health: a review. European J. Nutr. Food Saf. 6, 83–92. doi: 10.9734/ejnfs/2016/16318
Badigannavar, A., Teme, N., de Oliveira, A. C., Li, G., Vaksmann, M., Viana, V. E., et al. (2018). Physiological, genetic and molecular basis of drought resilience in sorghum (Sorghum bicolor (L.)). Indian J. Plant Physiol. 23, 670–688. doi: 10.1007/s40502-018-0416-2
Baggerman, G., Vierstraete, E., De Loof, A., and Schoofs, L. (2005). Gel-based versus gel-free proteomics: A review. Comb. Chem. High Throughput Screen. 8, 669–677. doi: 10.2174/138620705774962490
Baker, N. R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Balbuena, T. S., Salas, J. J., Martinez-Force, E., Garces, R., and Thelen, J. J. (2011). Proteome analysis of cold acclimation in sunflower. J. Proteome Res. 10, 2330–2346. doi: 10.1021/pr101137q
Barvkar, V. T., Pardeshi, V. C., Kale, S. M., Qiu, S. Q., Rollins, M., Datla, R., et al. (2013). Genome-wide identification and characterization of microRNA genes and their targets in flax (Linum usitatissimum). Planta 237, 1149–1161. doi: 10.1007/s00425-012-1833-5
Bassett, C. M., Rodriguez-Leyva, D., and Pierce, G. N. (2009). Experimental and clinical research findings on the cardiovascular benefits of consuming flaxseed. Appl. Physiol. Nutr. Metab. 34, 965–974. doi: 10.1139/H09-087
Bengtsson, T., Weighill, D., Proux-Wéra, E., Levander, F., Resjö, S., Burra, D. D., et al. (2014). Proteomics and transcriptomics of the BABA-induced resistance response in potato using a novel functional annotation approach. BMC Genomics 15:315. doi: 10.1186/1471-2164-15-315
Bhat, J. A., Ali, S., Salgotra, R. K., Mir, Z. A., Dutta, S., Jadon, V., et al. (2016). Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 7:221. doi: 10.3389/fgene.2016.00221
Boba, A., Kostyn, K., Kozak, B., Wojtasik, W., Preisner, M., Prescha, A., et al. (2020). Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 251:50. doi: 10.1007/s00425-020-03339-9
Boba, A., Kostyn, K., Kozak, B., Zalewski, I., Szopa, J., and Kulma, A. (2021). Transcriptomic profiling of susceptible and resistant flax seedlings after Fusarium oxysporum lini infection. PLoS One 16:e0246052. doi: 10.1371/journal.pone.0246052
Bodner, G., Alsalem, M., Nakhforoosh, A., Arnold, T., and Leitner, D. (2017). RGB and spectral root imaging for plant phenotyping and physiological research: experimental setup and imaging protocols. J. Vis. Exp. 126:56251. doi: 10.3791/56251
Budhlakoti, N., Kushwaha, A. K., Rai, A., Chaturvedi, K. K., Kumar, A., Pradhan, A. K., et al. (2022). Genomic selection: A tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops. Front. Genet. 13:832153. doi: 10.3389/fgene.2022.832153
Bui, E. N. (2020). “Causes of soil salinization, sodification, and alkalinization.” in The Oxford Encyclopedia of Agriculture and the Environment. ed. R. W. Hazlett (Oxford University Press).
Cartagena, J. A., and Marquez, G. P. B. (2021). “Transcriptome analysis in Jatropha during abiotic stress response,” in Oil Crop Genomics, ed. H. Tombuloglu, T. Unver, G. Tombuloglu, and K. R. Hakeem (Cham: Springer), 317–337.
Caruso, G., Cavaliere, C., Foglia, P., Gubbiotti, R., Samperi, R., and Laganà, A. (2009). Analysis of drought responsive proteins in wheat (Triticum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant Sci. 177, 570–576. doi: 10.1016/j.plantsci.2009.08.007
Caruso, G., Cavaliere, C., Guarino, C., Gubbiotti, R., Foglia, P., Laganà, A., et al. (2008). Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal. Bioanal. Chem. 391, 381–390. doi: 10.1007/s00216-008-2008-x
Chaudhary, J., Khatri, P., Singla, P., Kumawat, S., Kumari, A., Vikram, A., et al. (2019). Advances in omics approaches for abiotic stress tolerance in tomato. Biology 8:90. doi: 10.3390/biology8040090
Chaudhary, J., Patil, G. B., Sonah, H., Deshmukh, R. K., Vuong, T. D., Valliyodan, B., et al. (2015). Expanding omics resources for improvement of soybean seed composition traits. Front. Plant Sci. 6:1021. doi: 10.3389/fpls.2015.01021
Cha-um, S., Supaibulwatana, K., and Kirdmanee, C. (2006). Water relation, photosynthetic ability and growth of Thai jasmine rice (Oryza sativa L. ssp. Indica cv. KDML105) to salt stress by application of exogenous glycine betaine and choline. J. Agron. Crop Sci. 192, 25–36. doi: 10.1111/j.1439-037x.2006.00186.x
Cloutier, S., Miranda, E., Ragupathy, R., Radovanovic, N., Reimer, E., and Walichnowski, A. (2012). Integrated consensus genetic and physical maps of flax (Linum usitatissimum L.). Theor. Appl. Genet. 125, 1783–1795. doi: 10.1007/s00122-012-1953-0
Cloutier, S., Niu, Z., Datla, R., and Duguid, S. (2009). Development and analysis of EST-SSRs for flax (Linum usitatissimum L.). Theor. Appl. Genet. 119, 53–63. doi: 10.1007/s00122-009-1016-3
Cloutier, S., Ragupathy, R., Niu, Z., and Duguid, S. (2011). SSR-based link-age map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol. Breed. 28, 437–451. doi: 10.1007/s11032-010-9494-1
Cortina, C., and Culianez-Macia, F. A. (2005). Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci. 169, 75–82. doi: 10.1016/j.plantsci.2005.02.026
Coşkuner, Y., and Karababa, E. (2007). Some physical properties of flaxseed (Linum usitatissimum L.). J. Food Eng. 78, 1067–1073. doi: 10.1016/j.jfoodeng.2005.12.017
Cross, R. H. (2002). Heat stress effects on flowering and reproduction in Linum usitatissimum (flax). MSc, University of Saskatchewan, Saskatoon.
Cross, R. H., McKay, S. A. B., McHughen, A. G., and Bonham-Smith, P. C. (2003). Heat stress effects on reproduction and seed set in Linum usitatissimum L. (flax). Plant Cell Environ. 26, 1013–1020. doi: 10.1046/j.1365-3040.2003.01006.x
Crossa, J., Pérez-Rodríguez, P., Cuevas, J., Montesinos-López, O., Jarquín, D., Campos, G. D. L., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. doi: 10.1016/j.tplants.2017.08.011
Cullis, C. (2011). “Linum,” in Wild Crop Relatives: Genomic and Breeding Resources. ed. C. Kole (Berlin, Heidelberg: Springer), 177–189.
Cullis, C. A. (2019). Genetics and Genomics of Linum. Switzerland: Springer International Publishing.
Dash, P. K., Cao, Y., Jailani, A. K., Gupta, P., Venglat, P., Xiang, D., et al. (2014). Genome-wide analysis of drought induced gene expression changes in flax (Linum usitatissimum). GM Crops Food. 5, 106–119. doi: 10.4161/gmcr.29742
Dash, P. K., Rai, R., Mahato, A. K., Gaikwad, K., and Singh, N. K. (2017). Transcriptome landscape at different developmental stages of a drought tolerant cultivar of flax (Linum usitatissimum). Front. Chem. 5:82. doi: 10.3389/fchem.2017.00082
Deng, X. (2013). Development of molecular markers and association analysis of yield related traits in flax (Linum usitatissimum L). Ph.D thesis. Chin. Acad. Agric. Sci.
Deshmukh, R., Sonah, H., Patil, G., Chen, W., Prince, S., Mutava, R., et al. (2014). Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 5:244. doi: 10.3389/fpls.2014.00244
Di Carli, M., Villani, M. E., and Bianco, L. (2010). Proteomic analysis of the plant-virus interaction in cucumber mosaic virus (CMV) resistant transgenic tomato. J. Proteome Res. 9, 5684–5697. doi: 10.1021/pr100487x
Diederichsen, A., Rozhmina, T. A., Zhuchenko, A. A., and Richards, K. W. (2006). Screening for broad adaptation in 96 flax (Linum usitatissimum L.) accessions under dry and warm conditions in Canada and Russia. Plant Genet. Resour. Newsl. 146, 9–16.
Diederichsen, A., and Ulrich, A. (2009). Variability in stem fibre content and its association with other characteristics in 1177 flax (Linum usitatissimum L) genebank accessions. Ind. Crop Prod. 30, 33–39. doi: 10.1016/j.indcrop.2009.01.002
Dmitriev, A. A., Krasnov, G. S., and Rozhmina, T. A. (2019). Flax (Linum usitatissimum L.) response to non-optimal soil acidity and zinc deficiency. BMC Plant Biol. 19:54. doi: 10.1186/s12870-019-1641-1
Dmitriev, A. A., Krasnov, G. S., Rozhmina, T. A., Kishlyan, N. V., Zyablitsin, A. V., Sadritdinova, A. F., et al. (2016a). Glutathione S-transferases and UDP-glycosyltransferases are involved in response to aluminum stress in flax. Front. Plant Sci. 7:1920. doi: 10.3389/fpls.2016.01920
Dmitriev, A. A., Krasnov, G. S., Rozhmina, T. A., Novakovskiy, R. O., Snezhkina, A. V., Fedorova, M. S., et al. (2017). Differential gene expression in response to Fusarium oxysporum infection in resistant and susceptible genotypes of flax (Linum usitatissimum L.). BMC Plant Biol. 17:253. doi: 10.1186/s12870-017-1192-2
Dmitriev, A. A., Kudryavtseva, A. V., Krasnov, G. S., Koroban, N. V., Speranskaya, A. S., Krinitsina, A. A., et al. (2016b). Gene expression profiling of flax (Linum usitatissimum L.) under edaphic stress. BMC Plant Biol. 16:237. doi: 10.1186/s12870-016-0927-9
Dmitriev, A. A., Novakovskiy, R. O., Pushkova, E. N., Rozhmina, T. A., Zhuchenko, A. A., Bolsheva, N. L., et al. (2020). Transcriptomes of different tissues of flax (Linum usitatissimum L.) cultivars with diverse characteristics. Front. Genet. 11:565146. doi: 10.3389/fgene.2020.565146
Dossa, K., Li, D., Zhou, R., Yu, J., Wang, L., Zhang, Y., et al. (2019). The genetic basis of drought tolerance in the high oil crop Sesamum indicum. J. Plant Biotechnol. 17, 1788–1803. doi: 10.1111/pbi.13100
Du, Y. L., Wang, Z. Y., Fan, J. W., Turner, C., Wang, T., Li, F. M., et al. (2012). B-aminobutyric acid increases abscisic acid accumulation and desiccation tolerance and decreases water use but fails to improve grain yield in two spring wheat cultivars under soil drying. J. Exp. Bot. 63, 4849–4860. doi: 10.1093/jxb/ers164
Dubey, S., Bhargava, A., Fuentes, F., Shukla, S., and Srivastava, S. (2020). Effect of salinity stress on yield and quality parameters in flax (Linum usitatissimum L.). Not. Bot. Horti. Agrobot Cluj-Napoca. 48, 954–966. doi: 10.15835/nbha48211861
El Sabagh, A., Hossain, A., Barutcular, C., Gormus, O., Ahmad, Z., Hussain, S., et al. (2019). Effects of drought stress on the quality of major oilseed crops: implications and possible mitigation strategies—a review. Appl. Ecol. Environ. Res. 17, 4019–4043. doi: 10.15666/aeer/1702_40194043
El-Afry, M. M., El-Okkiah, S. A., El-Kady, E. S. A., and El-Yamanee, G. S. A. (2018). Exogenous application of ascorbic acid for alleviation the adverse effects of salinity stress in flax (Linum usitatissimum L.). Middle East J. 7, 716–739.
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop production under drought and heat stress: plant responses and management options. Front. Plant Sci. 8:1147. doi: 10.3389/fpls.2017.01147
FAOSTAT (2022). Available at: https://www.fao.org/faostat/en/#data/QCL (Accessed June 5, 2022).
Fatehi, F., Hosseinzadeh, A., Alizadeh, H., Brimavandi, T., and Struik, P. C. (2012). The proteome response of salt-resistant and salt-sensitive barley genotypes to long-term salinity stress. Mol. Biol. Rep. 39, 6387–6397. doi: 10.1007/s11033-012-1460-z
Flor, H. H. (1956). The complementary genic systems in flax and flax rust. Adv. Genet. 8, 29–54. doi: 10.1016/s0065-2660(08)60498-8
Fofana, B., Cloutier, S., Duguid, S., Ching, J., and Rampitsch, C. (2006). Gene expression of stearoyl-ACP desaturase and D12 fatty acid desaturase 2 is modulated during seed development of flax (Linum usitatissimum). Lipids 41, 705–720. doi: 10.1007/s11745-006-5021-x
Fu, Y.-B., Diederichsen, A., Richards, K., and Peterson, G. (2002a). Genetic diversity within a range of cultivars and landraces of flax (Linum usitatissimum L.) as revealed by RAPDs. Genet. Resour. Crop. Evol. 49, 167–174. doi: 10.1023/A:1014716031095
Fu, Y.-B., Peterson, G., Diederichsen, A., and Richards, K. (2002b). RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet. Resour. Crop. Evol. 49, 253–259. doi: 10.1023/A:1015571700673
Fu, Y.-B., Rowland, G. G., Duguid, S. D., and Richards, K. W. (2003). RAPD analysis of 54 north American flax cultivars. Crop. Sci. 43, 1510–1515. doi: 10.2135/cropsci2003.1510
Fujita, Y., Venterink, H. O., Van Bodegom, P. M., Douma, J. C., Heil, G. W., Hölzel, N., et al. (2013). Low investment in sexual reproduction threatens plants adapted to phosphorus limitation. Nature 505, 82–86. doi: 10.1038/nature12733
Fukushima, A., Kusano, M., Redestig, H., Arita, M., and Saito, K. (2009). Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 13, 532–538. doi: 10.1016/j.cbpa.2009.09.022
Galindo-Gonzalez, L., and Deyholos, M. K. (2016). RNA-seq transcriptome response of flax (Linum usitatissimum L.) to the pathogenic fungus Fusarium oxysporum f. sp. lini. Front. Plant Sci. 7:1766. doi: 10.3389/fpls.2016.01766
Garg, A. K., Kim, J. K., Owens, T. G., Ranwala, A. P., Do Choi, Y., Kochian, L. V., et al. (2002). Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. 99, 15898–15903. doi: 10.1073/pnas.252637799
Gevaert, K., and Vandekerckhove, J. (2011). Gel-free proteomics: Methods and protocols. (Berlin: Springer).
Ghabooli, M., Khatabi, B., Ahmadi, F. S., Sepehri, M., Mirzaei, M., Amirkhani, A., et al. (2013). Proteomics study reveals the molecular mechanisms underlying water stress tolerance induced by Piriformospora indica in barley. J. Proteomics 94, 289–301. doi: 10.1016/j.jprot.2013.09.017
Ghatak, A., Chaturvedi, P., and Weckwerth, W. (2018). Metabolomics in plant stress physiology. Adv. Biochem. Engin./Biotechnol. 164, 187–236. doi: 10.1007/10_2017_55
Gitay, H., Suarez, A., and Watson, R.T. (2002). Climate change and biodiversity: IPCC Technical Paper V. Intergovernmental Panel on Climate Change. Geneva, 77.
Gowda, V. R. P., Henry, A., Yamauchi, A., Shashidhar, H. E., and Serraj, R. (2011). Root biology and genetic improvement for drought avoidance in rice. Field Crop Res 122, 1–13. doi: 10.1016/j.fcr.2011.03.001
Goyal, A., Sharma, V., Upadhyay, N., Gill, S., and Sihag, M. (2014). Flax and flaxseed oil: an ancient medicine & modern functional food. J. Food Sci. Technol. 51, 1633–1653. doi: 10.1007/s13197-013-1247-9
Granier, C., Aguirrezabal, L., Chenu, K., Cookson, S. J., Dauzat, M., Hamard, P., et al. (2006). PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 169, 623–635. doi: 10.1111/j.1469-8137.2005.01609.x
Green, A. G., Chen, Y., Singh, S. P., and Dribnenki, J. C. P. (2008). “Flax,” in Compendium of transgenic crop plants. eds. C. Kole and T. C. Hall (Oxford: Blackwell Publishing Ltd.), 199–226.
Guo, P., Baum, M., Grando, S., Ceccarelli, S., Bai, G., Li, R. V. K., et al. (2009). Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J. Exp. Bot. 60, 3531–3544. doi: 10.1093/jxb/erp194
Guo, B., Fedorova, N. D., Chen, X., Wan, C. H., Wang, W., Nierman, W. C., et al. (2011). Gene expression profiling and identification of resistance genes to aspergillus flavus infection in peanut through EST and microarray strategies. Toxins 3, 737–753. doi: 10.3390/toxins3070737
Guo, R., Hao, W., and Gong, D. Z. (2012). Effects of water stress on germination and growth of linseed seedlings (Linum usitatissimum L.), photosynthetic efficiency and accumulation of metabolites. J. Agric. Sci. 4, 253–265. doi: 10.5539/jas.v4n10p253
Guo, Z., Yang, W., Chang, Y., Ma, X., Tu, H., Xiong, F., et al. (2018). Genome-wide association studies of image traits reveal genetic architecture of drought resistance in rice. Mol. Plant 11, 789–805. doi: 10.1016/j.molp.2018.03.018
Guo, R., Zhou, J., Hao, W., Gu, F., Liu, Q., Li, H., et al. (2014). Germination, growth, chlorophyll fluorescence and ionic balance in linseed seedlings subjected to saline and alkaline stresses. Plant Prod. Sci. 17, 20–31. doi: 10.1626/pps.17.20
Guo, R., Zhou, J., Ren, G., and Hao, W. (2013). Physiological responses of linseed seedlings to iso osmotic polyethylene glycol, salt, and alkali stresses. Agron. J. 105, 764–772. doi: 10.2134/agronj2012.0442
Halder, T., Choudhary, M., Liu, H., Chen, Y., Yan, G., and Siddique, K. (2022). Wheat proteomics for abiotic stress tolerance and root system architecture: current status and future prospects. Proteomes 10:17. doi: 10.3390/proteomes10020017
Hall, R. D. (2006). Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol. 169, 453–468. doi: 10.1111/j.1469-8137.2005.01632.x
Hamade, K., Fliniaux, O., Fontaine, J. X., Molinié, R., Otogo Nnang, E., Bassard, S., et al. (2021). NMR and LC-MS-based metabolomics to study osmotic stress in lignan-deficient flax. Molecules 26:767. doi: 10.3390/molecules26030767
Hano, C., Addi, M., Fliniaux, O., Bensaddek, L., Duverger, E., Mesnard, F., et al. (2008). Molecular characterization of cell death induced by a compatible interaction between Fusarium oxysporum f. sp. linii and flax (Linum usitatissimum) cells. Plant Physiol. Biochem. 46, 590–600. doi: 10.1016/j.plaphy.2008.02.004
Harrigan, G. G., Martino-Catt, S., and Glenn, K. C. (2007). Metabolomics, metabolic diversity and genetic variation in crops. Metabolomics 3, 259–272. doi: 10.1007/s11306-007-0076-0
He, L., Xiao, J., Rashid, K. Y., Jia, G., Li, P., Yao, Z., et al. (2019b). Evaluation of genomic prediction for pasmo resistance in flax. Int. J. Mol. Sci. 20:359. doi: 10.3390/ijms20020359
He, L., Xiao, J., Rashid, K. Y., Yao, Z., Li, P., Jia, G., et al. (2019a). Genome-wide association studies for pasmo resistance in flax (Linum usitatissimum L.). Front. Plant Sci. 9:1982. doi: 10.3389/fpls.2018.01982
Heller, K., and Byczyńska, M. (2015). The impact of environmental factors and applied agronomy on quantitative and qualitative traits of flax fiber. J. Nat. Fibers 12, 26–38. doi: 10.1080/15440478.2013.879088
Hosseinian, F. S., Muir, A. D., Westcott, N. D., and Krol, E. S. (2006). Antioxidant capacity of flaxseed lignans in two model systems. J. Am. Oil Chem. Soc. 83:835. doi: 10.1007/s11746-006-5034-x
Hradilová, J., Řehulka, P., Řehulková, H., Vrbová, M., Griga, M., and Brzobohatý, B. (2010). Comparative analysis of proteomic changes in contrasting flax cultivars upon cadmium exposure. Electrophoresis 31, 421–431. doi: 10.1002/elps.200900477
Hu, H., and Xiong, L. (2014). Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 65, 715–741. doi: 10.1146/annurev-arplant-050213-040000
Humplík, J. F., Lazár, D., Husičková, A., and Spíchal, L. (2015). Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses—a review. Plant Methods 11:29. doi: 10.1186/s13007-015-0072-8
Ibáñez, C., García-Cañas, V., Valdés, A., and Simó, C. (2013). Novel MS-based approaches and applications in food metabolomics. Trends Anal. Chem. 52, 100–111. doi: 10.1016/j.trac.2013.06.015
Ichihashi, Y., and Sinha, N. R. (2014). From genome to phenome and back in tomato. Curr. Opin. Plant Biol. 18, 9–15. doi: 10.1016/j.pbi.2013.12.004
Jaggard, K. W., Qi, A., and Ober, E. S. (2010). Possible changes to arable crop yields by 2050. Philos. Trans. R. Soc. B Biol. Sci. 365, 2835–2851. doi: 10.1098/rstb.2010.0153
Jakab, G., Cottier, V., Toquin, V., Rigoli, G., Zimmerli, L., Metraux, J. P., et al. (2001). β-Aminobutyric acid-induced resistance in plants. Eur. J. Plant Pathol. 107, 29–37. doi: 10.1023/A:1008730721037
Jakab, G., Ton, J., Flors, V., Zimmerli, L., Metraux, J. P., Mauch-Mani, B., et al. (2005). Enhancing Arabidopsis salt and drought stress synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta 201, 293–297. doi: 10.1104/pp.105.065698
Jamil, A., Riaz, S., Ashraf, M., and Foolad, M. R. (2011). Gene expression profiling of plants under salt stress. Crit. Rev. Plant Sci. 30, 435–458. doi: 10.1080/07352689.2011.605739
Kajla, P., Sharma, A., and Sood, D. R. (2015). Flaxseed—a potential functional food source. J. Food Sci. Technol. 52, 1857–1871. doi: 10.1007/s13197-014-1293-y
Kaspar, S., Peukert, M., Mock, H. P., Svatos, A., Matros, A., and Mock, H. (2011). MALDI-imaging mass spectrometry—An emerging technique in plant biology. Proteomics 11, 1840–1850. doi: 10.1002/pmic.201000756
Kaur, V., Kumar, S., Yadav, R., Wankhede, D. P., Aravind, J., Radhamani, J., et al. (2018). Analysis of genetic diversity in Indian and exotic linseed germplasm and identification of trait specific superior accessions. J. Environ. Biol. 39, 702–709. doi: 10.25174/2249-4065/2018/83620
Kaur, V., Yadav, R., and Wankhede, D. P. (2017). Linseed (Linum usitatissimum L.) genetic resources for climate change intervention and its future breeding. J. App. Nat. Sci. 9, 1112–1118. doi: 10.31018/jans.v9i2.1331
Kaur, V., Yadav, S. K., Wankhede, D. P., Pulivendula, P., Kumar, A., and Chinnusamy, V. (2020). Cloning and characterization of a gene encoding MIZ1, a domain of unknown function protein and its role in salt and drought stress in rice. Protoplasma 257, 475–487. doi: 10.1007/s00709-019-01452-5
Kaya, M. D., Day, S., Cikili, Y., and Arslan, N. (2012). Classification of some linseed (Linum usitatissimum L.) genotypes for salinity tolerance using germination, seedling growth, and ion content. Chil. J. Agric Res. 72, 27–32. doi: 10.4067/s0718-58392012000100005
Klubicová, K., Danchenko, M., Skultéty, L., Berezhna, V. V., Hricová, A., Rashydav, N. M., et al. (2011). Agricultural recovery of a formerly radioactive area: II. Systematic proteomic characterization of flax seed development in the remediated Chernobyl area. J. Proteomics 74, 1378–1384. doi: 10.1016/j.jprot.2011.02.029
Kocak, M. Z., Göre, M., and Kurt, O. (2022). The effect of different salinity levels on germination development of some flax (Linum usitatissimum L.) varieties. J. Food Sci. Technol. 10, 657–662. doi: 10.24925/turjaf.v10i4.657-662.4758
Kosová, K., Vítámvás, P., and Prášil, I. T. (2014). Proteomics of stress responses in wheat and barley—search for potential protein markers of stress tolerance. Front. Plant Sci. 5:711. doi: 10.3389/fpls.2014.00711
Kostyn, K., Czemplik, M., Kulma, A., Bortniczuk, M., Skała, J., and Szopa, J. (2012). Genes of phenylpropanoid pathway are activated in early response to fusarium attack in flax plants. Plant Sci. 190, 103–115. doi: 10.1016/j.plantsci.2012.03.011
Kouba, M., and Mourot, J. (2011). A review of nutritional effects on fat composition of animal products with special emphasis on n-3 polyunsaturated fatty acids. Biochimie 93, 13–17. doi: 10.1016/j.biochi.2010.02.027
Kumar, S., You, F. M., and Cloutier, S. (2012). Genome wide SNP discovery in flax through next generation sequencing of reduced representation libraries. BMC Genomics 13:684. doi: 10.1186/1471-2164-13-684
Kumari, J., Mahatman, K. K., Sharma, S., Singh, A. K., Adhikari, S., Bansal, R., et al. (2022). Recent advances in different omics mechanism for drought stress tolerance in rice. Russ. J. Plant Physiol. 69:18. doi: 10.1134/S1021443722010095
Lasky, J. R., Upadhyaya, H. D., Ramu, P., Deshpande, S., Hash, C. T., Bonnette, J., et al. (2015). Genome-environment associations in sorghum landraces predict adaptive traits. Sci. Adv. 1:e1400218. doi: 10.1126/sciadv.1400218
Leisner, C. P., Yendrek, C. R., and Ainsworth, E. A. (2017). Physiological and transcriptomic responses in the seed coat of field-grown soybean (Glycine max L. Merr.) to abiotic stress. BMC Plant Biol. 17, 1–11. doi: 10.1186/s12870-017-1188-y
Li, D., Quan, C., Song, Z., Li, X., Yu, G., Li, C., et al. (2021a). High-throughput plant phenotyping platform (HT3P) as a novel tool for estimating agronomic traits from the lab to the field. Front. Bioeng. Biotechnol. 8:623705. doi: 10.3389/fbioe.2020.623705
Li, H., Yang, M., Zhao, C., Wang, Y., and Zhang, R. (2021b). Physiological and proteomic analyses revealed the response mechanisms of two different drought-resistant maize varieties. BMC Plant Biol. 21:513. doi: 10.1186/s12870-021-03295-w
Liang, Y. C., Sun, W. C., Zhu, Y. G., and Christie, P. (2007). Mechanisms of silicon-mediated alleviation of abiotic stresses in higher plants: a review. Environ. Pollut. 147, 422–428. doi: 10.1016/j.envpol.2006.06.008
Lisson, S. N., and Mendham, N. J. (2000). Agronomic studies of flax (Linum usitatissimum L.) in South-Eastern Australia. Aust. J. Exp. Agric. 40, 1101–1112. doi: 10.1071/ea00059
Macarisin, D., Wisniewski, M. E., Bassett, C., and Thannhauser, T. W. (2009). Proteomic analysis of β-aminobutyric acid priming and abscisic acid—induction of drought resistance in crabapple (Malus pumila): effect on general metabolism, the phenyl-propanoid pathway and cell wall enzymes. Plant Cell Environ. 32, 1612–1631. doi: 10.1111/j.1365-3040.2009.02025.x
Majoul, T., Bancel, E., Triboi, E., Ben Hamida, J., and Branlard, G. (2004). Proteomic analysis of the effect of heat stress on hexaploid wheat grain: characterization of heat-responsive proteins from non-prolamins fraction. Proteomics 4, 505–513. doi: 10.1002/pmic.200300570
Mangin, B., Bonnafous, F., Blanchet, N., Boniface, M. C., Bret-Mestries, E., Carrère, S., et al. (2017). Genomic prediction of sunflower hybrids oil content. Front. Plant Sci. 8:1633. doi: 10.3389/fpls.2017.01633
Manschadi, A. M., Christopher, J., deVoil, P., and Hammer, G. L. (2006). The role of root architectural traits in adaptation of wheat to water-limited environments. Funct. Plant Biol. 33, 823–837. doi: 10.1071/fp06055
McCann, M. J., Gill, C. I. R., McGlynn, H., and Rowland, I. R. (2005). Role of mammalian lignans in the prevention and treatment of prostate cancer. Nutr. Cancer 52, 1–14. doi: 10.1207/s15327914nc5201_1
Melnikova, N. V., Belenikin, M. S., Bolsheva, N. L., Dmitriev, A. A., Speranskaya, A. S., Krinitsina, A. A., et al. (2014). Flax inorganic phosphate deficiency responsive miRNAs. J. Agric. Sci. 6, 156–160. doi: 10.5539/jas.v6n1p156
Melnikova, N. V., Dmitriev, A. A., Belenikin, M. S., Koroban, N. V., Speranskaya, A. S., Krinitsina, A. A., et al. (2016). Identification, expression analysis, and target prediction of flax genotroph MicroRNAs under normal and nutrient stress conditions. Front. Plant Sci. 7:399. doi: 10.3389/fpls.2016.00399
Melnikova, N. V., Dmitriev, A. A., Belenikin, M. S., Speranskaya, A. S., Krinitsina, A. A., and Rachinskaia, O. A. (2015). Excess fertilizer responsive miRNAs revealed in Linum usitatissimum L. Biochimie 109, 36–41. doi: 10.1016/j.biochi.2014.11.017
Millet, E. J., Welcker, C., Kruijer, W., Negro, S., Coupel-Ledru, A., Nicolas, S. D., et al. (2016). Genome-wide analysis of yield in Europe: allelic effects vary with drought and heat scenarios. Plant Physiol. 172, 749–764. doi: 10.1104/pp.16.00621
Milli, A., Cecconi, D., Bortesi, L., Persi, A., Rinalducci, S., and Zamboni, A. (2012). Proteomic analysis of the compatible interaction between Vitis vinifera and Plasmopara viticola. J. Proteomics 75, 1284–1302. doi: 10.1016/j.jprot.2011.11.006
Moschen, S., Di Rienzo, J. A., Higgins, J., Tohge, T., Watanabe, M., González, S., et al. (2017). Integration of transcriptomic and metabolic data reveals hub transcription factors involved in drought stress response in sunflower (Helianthus annuus L.). Plant Mol. Biol. 94, 549–564. doi: 10.1007/s11103-017-0625-5
Nasri, N., Maatallah, S., Saidi, I., and Lachaal, M. (2017). Influence of salinity on germination, seedling growth, ion content and acid phosphatase activities of Linum usitatissimum L. J. Anim. Plant Sci. 27, 517–521.
Naz, T., Akhtar, J., Iqbal, M. M., Haq, M. A. U., and Saqib, M. (2016). “Boron toxicity in salt-affected soils and effects on plants,” in Soil Science: Agricultural and Environmental Prospectives. eds. K. R. Hakeem, J. Akhtar, and M. Sabir (Cham: Springer), 259–286.
Nemri, A., Saunders, D. G., Anderson, C., Upadhyaya, N. M., Win, J., Lawrence, G. J., et al. (2014). The genome sequence and effector complement of the flax rust pathogen Melampsora lini. Front. Plant Sci. 5:98. doi: 10.3389/fpls.2014.00098
Neutelings, G., Fénart, S., Lucau-Danilaa, A., and Hawkins, S. (2012). Identification and characterization of miRNAs and their potential targets in flax. J. Plant Physiol. 169, 1754–1766. doi: 10.1016/j.jplph.2012.06.011
Ninomiya, S. (2022). High-throughput field crop phenotyping: current status and challenges. Breed. Sci. 72, 3–18. doi: 10.1270/jsbbs.21069
Oh, T. J., Gorman, M., and Cullis, C. A. (2000). RFLP and RAPD mapping in flax (Linum usitatisimum). Theor. Appl. Genet. 101, 590–593. doi: 10.1007/s001220051520
Palmieri, M. C., Perazzolli, M., Matafora, V., Moretto, M., Bachi, A., and Pertot, I. (2012). Proteomic analysis of grapevine resistance induced by Trichoderma harzianum T39 reveals specific defence pathways activated against downy mildew. J. Exp. Bot. 63, 6237–6251. doi: 10.1093/jxb/ers279
Pang, Q., Chen, S., Dai, S., Chen, Y., Wang, Y., and Yan, X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. doi: 10.1021/pr100034f
Parida, A. K., and Das, A. B. (2005). Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 60, 324–349. doi: 10.1016/j.ecoenv.2004.06.010
Patil, N. M., Datir, S. S., and Shah, P. V. (2015). Salt-induced physiological and biochemical changes in two varieties of Linum usitatissimum L. Int. J. Curr. Microbiol. App. Sci. 4, 296–304.
Pazhamala, L. T., Kudapa, H., Weckwerth, W., Millar, A. H., and Varshney, R. K. (2021). Systems biology for crop improvement. Plant Genome 14:e20098. doi: 10.1002/tpg2.20098
Pilon-Smits, E. A., Quinn, C. F., Tapken, W., Malagoli, M., and Schiavon, M. (2009). Physiological functions of beneficial elements. Curr. Opin. Plant Biol. 12, 267–274. doi: 10.1016/j.pbi.2009.04.009
Pokhrel, S., and Meyers, B. C. (2022). Heat-responsive microRNAs and phased small interfering RNAs in reproductive development of flax. Plant Direct. 6:385. doi: 10.1002/pld3.385
Pontarin, N., Molinié, R., Mathiron, D., Tchoumtchoua, J., Bassard, S., Gagneul, D., et al. (2020). Age-dependent metabolic profiles unravel the metabolic relationships within and between flax leaves (Linum usitatissimum). Metabolites 10:218. doi: 10.3390/metabo10060218
Pratap, A., Gupta, S., Nair, R. M., Gupta, S., Schafleitner, R., Basu, P., et al. (2019). Using plant phenomics to exploit the gains of genomics. Agronomy 9:126. doi: 10.3390/agronomy9030126
Przybylski, R. (2005). “Flax oil and high linolenic oils,” in Bailey’s Industrial Oil and Fat Products. eds. S. F. Wiley and N. J. Hoboken (John Wiley & Sons), 281–302.
Putri, S. P., Yamamoto, S., Tsugawa, H., and Fukusaki, E. (2013). Current metabolomics: technological advances. J. Biosci. Bioeng. 116, 9–16. doi: 10.1016/j.jbiosc.2013.01.004
Qayyum, M. A., Bashir, F., Maqbool, M. M., Ali, A., Bashir, S., and Abbas, Q. (2019). Implications of saline water irrigation for linseed on seed germination, seedling survival and growth potential. Sarhad J. Agric. 35, 1289–1297. doi: 10.17582/journal.sja/2019/35.4.1289.1297
Qi, X. S., Wang, X. R., Xu, J., Zhang, J. P., and Mi, J. (2010). Drought-resistance evaluation of flax germplasm at adult plant stage. Sci. Agric. Sin. 43, 3076–3087.
Quéro, A., Fliniaux, O., Elboutachfaiti, R., Petit, E., Guillot, X., Hawkins, S., et al. (2015). Β-aminobutyric acid increases drought tolerance and reorganizes solute content and water homeostasis in flax (Linum usitatissimum). Metabolomics 11, 1363–1375. doi: 10.1007/s11306-015-0792-9
Rajwade, A. V., Arora, R. S., Kadoo, N. Y., Harsulkar, A. M., Ghorpade, P. B., Gupta, V. S., et al. (2010). Relatedness of Indian flax genotypes (Linum usitatissimum L.): An inter-simple sequence repeat (ISSR) primer assay. Mol. Biotechnol. 45, 161–170. doi: 10.1007/s12033-010-9256-7
Ramirez-Villegas, J., Molero Milan, A., Alexandrov, N., Asseng, S., Challinor, A. J., Crossa, J., et al. (2020). CGIAR modeling approaches for resource-constrained scenarios: I. accelerating crop breeding for a changing climate. Crop. Sci. 60, 547–567. doi: 10.1002/csc2.20048
Ramsay, A., Fliniaux, O., Fang, J., Molinie, R., Roscher, A., Grand, E., et al. (2014). Development of an NMR metabolomics-based tool for selection of flaxseed varieties. Metabolomics 10, 1258–1267. doi: 10.1007/s11306-014-0664-8
Rashid, K., and Duguid, S. (2005). Inheritance of resistance to powdery mildew in flax. Can. J. Plant Pathol. 27, 404–409. doi: 10.1080/07060660509507239
Rasoulnia, A., Bihamta, M. R., Peyghambari, S. A., Alizadeh, H., and Rahnama, A. (2011). Proteomic response of barley leaves to salinity. Mol. Biol. Rep. 38, 5055–5063. doi: 10.1007/s11033-010-0651-8
Ravensdale, M., Nemri, A., Thrall, P. H., Ellis, J. G., and Dodds, P. N. (2011). Co-evolutionary interactions between host resistance and pathogen effector genes in flax rust disease. Mol. Plant Pathol. 12, 93–102. doi: 10.1111/j.1364-3703.2010.00657.x
Rinalducci, S., Egidi, M. G., Karimzadeh, G., Jazii, F. R., and Zolla, L. (2011). Proteomic analysis of a spring wheat cultivar in response to prolonged cold stress. Electrophoresis 32, 1807–1818. doi: 10.1002/elps.201000663
Romero, C., Bellés, J. M., Vayá, J. L., Serrano, R., and Culiáñez-Macià, F. A. (1997). Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta 201, 293–297. doi: 10.1007/s004250050069
Saha, D., Mukherjee, P., Dutta, S., Meena, K., Sarkar, S. K., Mandal, A. B., et al. (2019). Genomic insights into HSFs as candidate genes for high-temperature stress adaptation and gene editing with minimal off-target effects in flax. Sci. Rep. 9, 1–18. doi: 10.1038/s41598-019-41936-1
Saha, D., Shaw, A. K., Datta, S., and Mitra, J. (2021). Evolution and functional diversity of abiotic stress-responsive NAC transcription factor genes in Linum usitatissimum L. Environ. Exp. Bot. 188:104512. doi: 10.1016/j.envexpbot.2021.104512
Salt, D. E., Baxter, I., and Lahner, B. (2008). Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 59, 709–733. doi: 10.1146/annurev.arplant.59.032607.092942
Saranga, Y., Jiang, C. X., Wright, R. J., Yakir, D., and Paterson, A. H. (2004). Genetic dissection of cotton physiological responses to arid conditions and their inter-relationships with productivity. Plant Cell Environ. 27, 263–277. doi: 10.1111/j.1365-3040.2003.01134.x
Saroha, A., Pal, D., Gomashe, S. S., Akash,, Kaur, V., Ujjainwal, S., et al. (2022b). Identification of QTNs associated with flowering time, maturity, and plant height traits in Linum usitatissimum L. using genome-wide association study. Front. Genet. 13:811924. doi: 10.3389/fgene.2022.811924
Saroha, A., Pal, D., Kaur, V., Kumar, S., Bartwal, A., Aravind, J., et al. (2022a). Agro-morphological variability and genetic diversity in linseed (Linum usitatissimum L.) germplasm accessions with emphasis on flowering and maturity time. Genet. Resour. Crop. Evol. 69, 315–333. doi: 10.1007/s10722-021-01231-3
Schauer, N., and Fernie, A. R. (2006). Plant metabolomics: towards biological function and mechanism. Trends Plant Sci. 11, 508–516. doi: 10.1016/j.tplants.2006.08.007
Schripsema, J. (2010). Application of NMR in plant metabolomics: techniques, problems and prospects. Phytochem. Anal. 21, 14–21. doi: 10.1002/pca.1185
Sertse, D., You, F. M., Ravichandran, S., Soto-Cerda, B. J., Duguid, S., and Cloutier, S. (2021). Loci harboring genes with important role in drought and related abiotic stress responses in flax revealed by multiple GWAS models. Theoret. Appl. Genet. 134, 191–212. doi: 10.1007/s00122-020-03691-0
Shah, P., Powell, A. L. T., Orlando, R., Bergmann, C., and Gutierrez-Sanchez, G. (2012). Proteomic analysis of ripening tomato fruit infected by Botrytis cinerea. J. Proteome Res. 11, 2178–2192. doi: 10.1021/pr200965c
Shah, T., Xu, J., Zou, X., Cheng, Y., Nasir, M., and Zhang, X. (2018). Omics approaches for engineering wheat production under abiotic stresses. Int. J. Mol. Sci. 19:2390. doi: 10.3390/ijms19082390
Sharma, J. C., Tomar, S. S., Shivran, R. K., and Prakash, C. (2012). Water requirement water use efficiency consumptive use yield and quality parameters of linseed (Linum usitatissimum L.) varieties as influenced by fertility levels irrigation scheduling. Adv. Life Sci. 1, 180–182.
Shi, D. C., and Wang, D. (2005). Effects of various salt-alkaline mixed stresses on Aneurolepidium chinense (Trin.) Kitag. Plant and Soil 271, 15–26. doi: 10.1007/s11104-004-1307-z
Shikha, M., Kanika, A., Rao, A. R., Mallikarjuna, M. G., Gupta, H. S., and Nepolean, T. (2017). Genomic selection for drought tolerance using genome-wide SNPs in maize. Front. Plant Sci. 8:63. doi: 10.3389/fpls.2017.00550
Shim, Y. Y., Gui, B., Wang, Y., and Reaney, M. J. T. (2015). Flaxseed (Linum usitatissimum L.) oil processing and selected products. Trends Food Sci. Technol. 43, 162–177. doi: 10.1016/j.tifs.2015.03.001
Shivaraj, S. M., Deshmukh, R. K., Rai, R., Bélanger, R., Agrawal, P. K., Dash, P. K., et al. (2017). Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci. Rep. 7, 1–17. doi: 10.1038/srep46137
Shivaraj, S. M., Dhakate, P., Sonah, H., Vuong, T., Nguyen, H. T., and Deshmukh, R. (2019). “Progress toward development of climate-smart flax: a perspective on omics-assisted breeding,” in Genomic Designing Climate-Smart Oilseed Crops (Cham: Springer), 239–274.
Shrivastava, P., and Kumar, R. (2015). Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Aust. J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Simó, C., Ibáez, C., Valdés, A., Cifuentes, A., and García-Cañas, V. (2014). Metabolomics of genetically modified crops. Int. J. Mol. Sci. 15, 18941–18966. doi: 10.3390/ijms151018941
Singh, K. K., Mridula, D., Rehal, J., and Barnwal, P. (2011). Flaxseed: a potential source of food, feed and fiber. Crit. Rev. Food Sci. Nutr. 51, 210–222. doi: 10.1080/10408390903537241
Singh, P., Wu, C. C., and Zimmerli, L. (2010). β-Aminobutyric acid priming by stress imprinting. Plant Signal. Behav. 5, 878–880. doi: 10.4161/psb.5.7.11903
Sirault, X. R. R., James, R. A., and Furbank, R. T. (2009). A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Funct. Plant Biol. 36, 970–977. doi: 10.1071/FP09182
Skylas, D. J., Cordwell, S. J., Hains, P. G., Larsen, M. R., Basseal, D. J., Walsh, B. J., et al. (2002). Heat shock of wheat during grain filling: proteins associated with heat-tolerance. J. Cereal Sci. 35, 175–188. doi: 10.1006/jcrs.2001.0410
Soto-Cerda, B. J., Cloutier, S., Gajardo, H. A., Aravena, G., and Quian, R. (2019). Identifying drought-resilient flax genotypes and related candidate genes based on stress indices, root traits and selective sweep. Euphytica 215:4. doi: 10.1007/s10681-019-2362-0
Soto-Cerda, B. J., Cloutier, S., Gajardo, H. A., Aravena, G., Quian, R., and You, F. M. (2020). Drought response of flax accessions and identification of quantitative trait nucleotides (QTNs) governing agronomic and root traits by genome-wide association analysis. Mol. Breed. 40, 1–24. doi: 10.1007/s11032-019-1096-y
Soto-Cerda, B. J., Cloutier, S., Quian, R., Gajardo, H. A., Olivos, M., and You, F. M. (2018). Genome-wide association analysis of mucilage and hull content in flax (Linum usitatissimum L.) seeds. Int. J. Mol. Sci. 19:2870. doi: 10.3390/ijms19102870
Soto-Cerda, B. J., Diederichsen, A., Ragupathy, R., and Cloutier, S. (2013). Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol. 13:78. doi: 10.1186/1471-2229-13-78
Soto-Cerda, B. J., Westermeyer, F., Iñiguez-Luy, F., Muñoz, G., Montenegro, A., and Cloutier, S. (2014). Assessing the agronomic potential of linseed genotypes by multivariate analyses and association mapping of agronomic traits. Euphytica 196, 35–49. doi: 10.1007/s10681-013-1012-1
Spindel, J. E., Dahlberg, J., Colgan, M., Hollingsworth, J., Sievert, J., Staggenborg, S. H., et al. (2018). Association mapping by aerial drone reveals 213 genetic associations for Sorghum bicolor biomass traits under drought. BMC Genomics 19:679. doi: 10.1186/s12864-018-5055-5
Tardieu, F., Cabrera-Bosquet, L., Pridmore, T., and Bennett, M. (2017). Plant phenomics, from sensors to knowledge. Curr. Biol. 27, R770–R783. doi: 10.1016/j.cub.2017.05.055
Tawfik, R. S., Badr, A., Sammour, R., Ibrahim, U., Matter, M., and Sakr, M. (2016). Improvement of flax drought tolerance using gene transfer. Plant Tissue Cult. Biotechnol. 26, 197–207. doi: 10.3329/ptcb.v26i2.30570
Tuberosa, R., Sanguineti, M. C., Landi, P., Giuliani, M. M., Salvi, S., and Conti, S. (2002). Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol. Biol. 48, 697–712. doi: 10.1023/a:1014897607670
Uysal, H., Fu, Y.-B., Kurt, O., Peterson, G. W., Diederichsen, A., and Kusters, P. (2010). Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne mill.) as revealed by ISSR markers. Genet. Resour. Crop. Evol. 57, 1109–1119. doi: 10.1007/s10722-010-9551-y
Vavilov, N. I. (1951). The origin, variation, immunity and breeding of cultivated plants. Chron. Bot. 13, 1–366. doi: 10.2134/agronj1952.00021962004400020016x
Vo, K. T. X., Rahman, M. M., Rahman, M. M., Trinh, K. T. T., Kim, S. T., and Jeon, J. S. (2021). Proteomics and metabolomics studies on the biotic stress responses of rice: an update. Rice 14, 1–16. doi: 10.1186/s12284-021-00461-4
Walter, A., Liebisch, F., and Hund, A. (2015). Plant phenotyping: from bean weighing to image analysis. Plant Methods 11, 1–11. doi: 10.1186/s13007-015-0056-8
Wang, Z., Hobson, N., Galindo, L., Zhu, S., Shi, D., McDill, J., et al. (2012). The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 72, 461–473. doi: 10.1111/j.1365-313x.2012.05093.x
Wang, W., Wang, L., Wang, L., Tan, M., Ogutu, C. O., Yin, Z., et al. (2021). Transcriptome analysis and molecular mechanism of linseed (Linum usitatissimum L.) drought tolerance under repeated drought using single-molecule long-read sequencing. BMC Genomics 22, 1–23. doi: 10.1186/s12864-021-07416-5
Weckwerth, W. (2003). Metabolomics in systems biology. Annu. Rev. Plant Biol. 54, 669–689. doi: 10.1146/annurev.arplant.54.031902.135014
Weckwerth, W., Ghatak, A., Bellaire, A., Chaturvedi, P., and Varshney, R. K. (2020). PANOMICS meets germplasm. Plant Biotechnol. J. 18, 1507–1525. doi: 10.1111/pbi.13372
Wendelboe-Nelson, C., and Morris, P. C. (2012). Proteins linked to drought tolerance revealed by DIGE analysis of drought resistant and susceptible barley varieties. Proteomics 12, 3374–3385. doi: 10.1002/pmic.201200154
Westcott, N. A., and Muir, A. D. (2003). Flax seed lignan in disease prevention and health promotion. Phytochem. Rev. 2, 401–417. doi: 10.1023/B:PHYT.0000046174.97809.b6
White, J. W., Andrade-Sanchez, P., Gore, M. A., Bronson, K. F., Coffelt, T. A., Conley, M. M., et al. (2012). Field-based phenomics for plant genetics research. Field Crop Res 133, 101–112. doi: 10.1016/j.fcr.2012.04.003
Wojtasik, W., Kulma, A., Namysł, K., Preisner, M., and Szopa, J. (2015). Polyamine metabolism in flax in response to treatment with pathogenic and non-pathogenic fusarium strains. Front. Plant Sci. 6:291. doi: 10.3389/fpls.2015.00291
Wróbel-Kwiatkowska, M., Lorenc-Kukula, K., Starzycki, M., Oszmiański, J., Kepczyńska, E., and Szopa, J. (2004). Expression of β-1, 3-glucanase in flax causes increased resistance to fungi. Physiol. Mol. Plant Pathol. 65, 245–256. doi: 10.1016/j.pmpp.2005.02.008
Wu, W., Nemri, A., Blackman, L. M., Catanzariti, A.-M., Sperschneider, J., Lawrence, G. J., et al. (2019b). Flax rust infection transcriptomics reveals a transcriptional profile that may be indicative for rust Avr genes. PLoS One 14:e0226106. doi: 10.1371/journal.pone.0226106
Wu, J. W., Shi, Y., Zhu, Y. X., Wang, Y. C., and Gong, H. J. (2013). Mechanisms of enhanced heavy metal tolerance in plants by silicon, a review. Pedosphere 23, 815–825. doi: 10.1016/s1002-0160(13)60073-9
Wu, C. C., Singh, P., Chen, M. C., and Zimmerli, L. (2010). L-glutamine inhibits beta-aminobutyric acid-induced stress resistance and priming in Arabidopsis. J. Exp. Bot. 61, 995–1002. doi: 10.1093/jxb/erp363
Wu, J., Zhao, Q., Sun, D., Wu, G., Zhang, L., Yuan, H., et al. (2018). Transcriptome analysis of flax (Linum usitatissimum L.) undergoing osmotic stress. Ind. Crop Prod. 116, 215–223. doi: 10.1016/j.indcrop.2018.02.035
Wu, J., Zhao, Q., Wu, G., Yuan, H., Ma, Y., Lin, H., et al. (2019a). Comprehensive analysis of differentially expressed unigenes under NaCl stress in flax (Linum usitatissimum L.) using RNA-Seq. Int. J. Mol. Sci. 20, 369. doi: 10.3390/ijms20020369
Xie, D., Dai, Z., Yang, Z., Sun, J., Zhao, D., Yang, X., et al. (2018). Genome-wide association study identifying candidate genes influencing important agronomic traits of flax (Linum usitatissimum L.) using SLAF-seq. Front. Plant Sci. 8:2232. doi: 10.3389/fpls.2017.02232
Yang, C., Guo, W., and Shi, D. (2010). Physiological roles of organic acids in alkali-tolerance of alkali-tolerant halophyte Chloris virgata. Agron. J. 102, 1081–1089. doi: 10.2134/agronj2009.0471
Yang, Y., Saand, M. A., Huang, L., Abdelaal, W. B., Zhang, J., Wu, Y., et al. (2021). Applications of multi-omics technologies for crop improvement. Front. Plant Sci. 12:563953. doi: 10.3389/fpls.2021.563953
Yao, Y. A., Wang, J. R., Ma, X. M., Lutts, S., Sun, C., Ma, J., et al. (2012). Proteomic analysis of Mn-induced resistance to powdery mildew in grapevine. J. Exp. Bot. 63, 5155–5170. doi: 10.1093/jxb/ers175
You, F. M., Booker, H. M., Duguid, S. D., Jia, G., and Cloutier, S. (2016). Accuracy of genomic selection in biparental populations of flax (Linum usitatissimum L.). Crop J. 4, 290–303. doi: 10.1016/j.cj.2016.03.001
You, F. M., and Cloutier, S. (2020). Mapping quantitative trait loci onto chromosome-scale pseudomolecules in flax. Methods protoc. 3:28. doi: 10.3390/mps3020028
You, F. M., Rashid, K. Y., Zheng, C., Khan, N., Li, P., Xiao, J., et al. (2022). Insights into the genetic architecture and genomic prediction of powdery mildew resistance in flax (Linum usitatissimum L.). Int. J. Mol. Sci. 23:4960. doi: 10.3390/ijms23094960
You, F. M., Xiao, J., Li, P., Yao, Z., Jia, G., He, L., et al. (2018a). Chromosome-scale pseudomolecules refined by optical, physical and genetic maps in flax. Plant J. 95, 371–384. doi: 10.1111/tpj.13944
You, F., Xiao, J., Li, P., Yao, Z., Jia, G., He, L., et al. (2018b). Genome-wide association study and selection signatures detect genomic regions associated with seed yield and oil quality in flax. Int. J. Mol. Sci. 19:2303. doi: 10.3390/ijms19082303
Young, L.W. (2003). High temperature stress and flowering in Brassica napus L (Doctoral dissertation, University of Saskatchewan).
Yu, Y., Huang, W. G., Chen, H. Y., Wu, G. W., Yuan, H. M., Song, X. X., et al. (2014). Identification of differentially ex-pressed genes in flax (Linum usitatissimum L.) under saline-alkaline stress by digital gene expression. Gene 549, 113–122. doi: 10.1016/j.gene.2014.07.053
Yu, Y., Wu, G., Yuan, H., Cheng, L., Zhao, D., Huang, W., et al. (2016). Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biol. 16:124. doi: 10.1186/s12870-016-0808-2
Zare, S., Mirlohi, A., Saeidi, G., Sabzalian, M. R., and Ataii, E. (2021). Water stress intensified the relation of seed color with lignan content and seed yield components in flax (Linum usitatissimum L.). Sci. Rep. 11:23958. doi: 10.1038/s41598-021-02604-5
Zhao, C., Zhang, Y., Du, J., Guo, X., Wen, W., Gu, S., et al. (2019). Crop phenomics: current status and perspectives. Front. Plant Sci. 10:714. doi: 10.3389/fpls.2019.00714
Zohary, D., and Hopf, M., (2000). Domestication of plants in the Old World: The origin and spread of cultivated plants in West Asia. Europe and the Nile Valley: Oxford University Press.
Keywords: abiotic and biotic stress, climate change, flax, fungal diseases, omics, linseed
Citation: Yadav B, Kaur V, Narayan OP, Yadav SK, Kumar A and Wankhede DP (2022) Integrated omics approaches for flax improvement under abiotic and biotic stress: Current status and future prospects. Front. Plant Sci. 13:931275. doi: 10.3389/fpls.2022.931275
Received: 28 April 2022; Accepted: 27 June 2022;
Published: 25 July 2022.
Edited by:
Dinesh Yadav, Deen Dayal Upadhyay Gorakhpur University, IndiaReviewed by:
Yuri Shavrukov, Flinders University, AustraliaCopyright © 2022 Yadav, Kaur, Narayan, Yadav, Kumar and Wankhede. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vikender Kaur, dmlrZW5kZXIua2F1ckBpY2FyLmdvdi5pbg==; Dhammaprakash Pandhari Wankhede, ZC53YW5raGVkZUBpY2FyLmdvdi5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.