
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 July 2022
Sec. Plant Proteomics and Protein Structural Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.930644
This article is part of the Research Topic Plant Infection and Defense – A Proteomic Perspective View all 7 articles
Panax notoginseng (Burk) F.H. Chen is a rare and valuable Chinese herb, but root rot mainly caused by Fusarium solani severely affects the yield and quality of P. notoginseng herbal materials. In this study, we isolated 30 P. notoginseng WRKY transcription factors (TFs), which were divided into three groups (I, II, and III) on the basis of a phylogenetic analysis. The expression levels of 10 WRKY genes, including PnWRKY9, in P. notoginseng roots increased in response to a methyl jasmonate (MeJA) treatment and the following F. solani infection. Additionally, PnWRKY9 was functionally characterized. The PnWRKY9 protein was localized to the nucleus. The overexpression of PnWRKY9 in tobacco (Nicotiana tabacum) considerably increased the resistance to F. solani, whereas an RNAi-mediated decrease in the PnWRKY9 expression level in P. notoginseng leaves increased the susceptibility to F. solani. The RNA sequencing and hormone content analyses of PnWRKY9-overexpression tobacco revealed that PnWRKY9 and the jasmonic acid (JA) signaling pathway synergistically enhance disease resistance. The PnWRKY9 recombinant protein was observed to bind specifically to the W-box sequence in the promoter of a JA-responsive and F. solani resistance-related defensin gene (PnDEFL1). A yeast one-hybrid assay indicated that PnWRKY9 can activate the transcription of PnDEFL1. Furthermore, a co-expression assay in tobacco using β-glucuronidase (GUS) as a reporter further verified that PnWRKY9 positively regulates PnDEFL1 expression. Overall, in this study, we identified P. notoginseng WRKY TFs and demonstrated that PnWRKY9 positively affects plant defenses against the root rot pathogen. The data presented herein provide researchers with fundamental information regarding the regulatory mechanism mediating the coordinated activities of WRKY TFs and the JA signaling pathway in P. notoginseng responses to the root rot pathogen.
Panax notoginseng (Burk) F.H. Chen is a high-value medicinal herb in the family Araliaceae. The main active substances in P. notoginseng are triterpene saponins, which have many effects (e.g., hemostatic, anti-platelet, thrombolytic, anti-hypertensive, and anti-inflammatory effects) (Xie et al., 2018). Panax notoginseng plants must be cultivated under shade and humid conditions for at least 3 years before they can be used for medicine. Because of the specific environmental conditions required during its cultivation and its long growth cycle, P. notoginseng is susceptible to many diseases, especially root rot, which is mainly caused by Fusarium solani (Liu D. et al., 2019). Fusarium solani is a necrotrophic fungus widely distributed in soil. After conidia germination in soil, F. solani invades the plant host through the root tip or wound, then multiplies in roots and further spreads to the whole plant (Caroline and Olubukola, 2013). In addition, the chlamydospores of F. solani are formed when plants die, and these spores are released back into the soil, where they can remain viable for up to 30 years (Schollenberger et al., 2005). Root rot initially results in the decaying of P. notoginseng roots (i.e., main medicinal tissue), and it may eventually lead to the death of susceptible plants (Ma et al., 2019). Root rot seriously affects the yield and quality of P. notoginseng herbal materials. Additionally, the agricultural chemical treatments used to control root rot might make raw P. notoginseng herbal materials unsafe for human consumption (Hao et al., 2019; Khaliq et al., 2020). Therefore, the mechanism underlying P. notoginseng defenses against root rot must be clarified to enable the development of root rot-resistant cultivars.
Plant defense responses can be divided into the following two interrelated branches: pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) (Li et al., 2021b). The signaling associated with PTI and ETI leads to a large-scale transcriptional reprogramming, which is regulated by many transcription factors (TFs). WRKY TFs are specific in plants, and have an important role in resistance. Some WRKY TFs promote susceptibility to a few pathogens while others promote resistance (Jiang J. et al., 2017). A total of 62 WRKY genes were identified in the pepper (Capsicum annuum) genome, one of which, CaWRKY22, positively regulated pepper resistance to Ralstonia Solanacearum, but CaWRKY40b was a negative regulator of resistance (Hussain et al., 2018; Ifnan Khan et al., 2018). Fifty-six possible WRKY genes in melon (Cucumis melo) were detected after a protein database was screened for conserved WRKY domains (Jiao et al., 2018). CmWRKY6/31/47/48/55 positively responded to powdery mildew pathogen (Erysiphe cichoracearum) infection, while CmWRKY15 was depressed during defense response. More than 50 WRKY genes were identified in rose (Rosa chinensis), and there were 19 RcWRKY genes may regulate rose resistance against Botrytis cinerea infection (Liu X. et al., 2019).
The WRKY TFs have some conserved structures associated with their transcriptional regulatory functions, including nuclear localization signals, leucine zippers, and WRKY domains. The WRKY domain comprises a heptapeptide sequence (WRKYGQK) that is usually located in the N-terminal region, whereas the zinc finger domain is located at the C-terminal (Xu et al., 2020). The WRKY proteins are phylogenetically classified in three major groups (Groups I–III) according to the number of WRKY domains and the structure of the zinc finger motif. Both of these motifs are necessary for the interaction between WRKY TFs and the W-box (TGACT/C) cis-element present in the promoters of many defense-related genes (Hussain et al., 2018). In apple (Malus × domestica), WRKY31 binds specifically to W-box2 in the MdhIR4 promoter to inhibit gene expression and enhance the resistance to Botryosphaeria dothidea (Zhao et al., 2019a).
Plant stress hormones play an important role in response to abiotic and biotic stress (Afrin et al., 2015). The resistant soybean (Glycine max) had higher level of salicylic acid (SA) during F. solani infection than the susceptible (Bawa et al., 2019). The jasmonic acid (JA) content in peony (Paeonia lactiflora) leaves gradually increased, while the SA content significantly decreased after inoculation with Alternaria tenuissima (Wang X. et al., 2019). Our previous study demonstrated that the defense priming in P. notoginseng through exogenous application of methyl jasmonate (MeJA) evidently induced the resistance to root rot pathogen F. solani (Liu D. et al., 2019). Moreover, the JA-responsive defensin like protein (PnDEFL1) and snakin (PnSN1) act as antimicrobial peptide to enhance the P. notoginseng resistance to F. solani (Wang Q. et al., 2019; Qiu et al., 2020). These findings imply that JA contributes to resistance of P. notoginseng to root rot. Under stressful conditions, JA is induced to synthesize and binds to the F-box protein coronatine insensitive 1 receptor. Subsequently, the promoted degradation of JAZ transcriptional repressors by ubiquitination releases a variety of TFs, and then leads to the activation of their target genes, which finally cope with and adapt to multiple stress (Zhou and Memelink, 2016). In addition, JA often forms a crosstalk network with ethylene (ET) to respond the necrotrophic pathogens. JAZ can inhibit the activity of ethylene-stabilized transcription factors EIL2/EIN3 in the ET signaling pathway, and then activates the expression of ETHYLENE RESPONSE FACTOR 1, thereby resisting the necrotrophic pathogen Botrytis cinerea (Zhu et al., 2011). Overexpression of ETHYLENE RESPONSE FACTOR 96 in Arabidopsis thaliana up-regulated expression of JA/ET-responsive defense genes including PDF1.2a, PR-3, PR-4, and VSP2, and then enhanced resistance to B. cinerea (Catinot et al., 2015).
Previous research revealed that WRKY TFs usually function cooperatively with plant hormone signal transduction pathways during responses to biotic stresses, including pathways involving JA, ET, SA, and abscisic acid (ABA) (Jiang J. et al., 2017). The expression of lotus (Nelumbo nucifera) WRKY genes in roots is regulated by SA and JA treatments, with the NnWRKY40a and NnWRKY40b expression levels significantly induced by JA, in particular (Li et al., 2019). In addition, WRKY75 regulated the JA-mediated signaling pathway to modulate defense responses in Arabidopsis thaliana. Under normal conditions, WRKY75 interacted with JAZ to inhibit the JA signaling pathway. When A. thaliana plants were infected by Botrytis cinerea, JA biosynthesis was boosted, and JAZ was degraded. Subsequently, WRKY75 was released to activate the expression of its target gene ORA59 for enhancing disease resistance (Chen et al., 2021).
The WRKY genes have been widely studied in recent years, but the P. notoginseng WRKY gene family remains relatively uncharacterized. It remains unclear whether WRKY TFs help regulate the P. notoginseng defense response to root rot. Thus, in this study, the recently generated RNA sequencing (RNA-seq) data for P. notoginseng plants infected with F. solani (Liu D. et al., 2019) were used to isolate and identify WRKY gene transcripts. Moreover, exogenous MeJA treatment induced resistance of P. notogisneng to F. solani (Liu D. et al., 2019), the WRKY gene expression patterns resulting from the independent MeJA treatment and F. solani infection as well as the combined MeJA treatment and F. solani inoculation were examined on the basis of quantitative real-time PCR (qRT-PCR) analyses. To elucidate the transcriptional regulatory mechanism mediating P. notoginseng responses to root rot, a JA-responsive gene (PnWRKY9) was selected for an analysis of its function during an F. solani infection. Furthermore, the promoter of an F. solani resistance-related gene (PnDEFL1) was cloned and the activation of its transcription by PnWRKY9 was verified.
The unigenes with an open reading frame (ORF) encoding a WRKY protein were obtained by analyzing the P. notoginseng transcriptome sequencing data from our previous study (Liu D. et al., 2019). The susceptible 1-year-old P. notoginseng plants grown in a greenhouse under shading nets were used for the cloning of WRKY transcripts ORF and the subsequent gene expression analysis. The F. solani isolate used in this study, which was obtained from P. notoginseng plants exhibiting typical root rot symptoms, was stored at 4°C in our laboratory. After being activated on potato dextrose agar medium, it was used in the following experiments. The susceptible wild-type (WT) tobacco (Nicotiana tabacum) seeds used for genetic transformations were surface-sterilized in 75% alcohol and a 0.1% HgCl2 solution prior to sowing on 1/4 Murashige and Skoog culture medium.
Total RNA of P. notoginseng roots under normal conditions was extracted using the TRIGene kit (GeneStar, China), and then cDNA was synthesized via reverse transcription using the Eastep® RT Master Mix kit (Promega, USA). Specific primers were designed for the RT-PCR amplification of P. notoginseng WRKY transcripts from cDNA. The PCR products were sequenced after T-A cloning to verify the accuracy of WRKY unigene sequences. Moreover, a phylogenetic tree was constructed using the MEGA 6.0 software. The PnWRKY9 (OM273051) protein served as the query for a BLAST search of the NCBI database (https://www.ncbi.nlm.nih.gov/). Several WRKY protein sequences highly homologous to PnWRKY9 were included in a multiple sequence alignment using the DNAMAN software to identify their conserved domains. Photoshop was used to visualize the structure of PnWRKY9. The subcellular localization of PnWRKY9 was predicted using the PredictProtein online program (http://www.predictprotein.org/).
To analyse whether these WRKYs responded to JA signaling and F. solani, the roots of P. notoginseng plants were wounded with sterile scissors and then immersed in 100 μM MeJA solution (Solarbio, China) and F. solani spore solution (106 spores/mL) for 30 min, respectively. The roots were collected at 24, 48, and 72 h post treatment or inoculation, and the roots treated with sterile water for 30 min were used as control. In addition, to investigate whether P. notoginseng WRKYs cooperated with JA signaling in response to F. solani infection, the P. notoginseng roots were wounded with a small pair of scissors and then immersed in 100 μM MeJA solution for 30 min. At 24 h post treatment with MeJA, these P. notoginseng roots were further inoculated with F. solani spore solution for 30 min, and then were collected at 12, 48, and 72 h post-inoculation (hpi). Meanwhile, the roots only treated with sterile water were collected as the control.
To eliminate differences among individual plants, the roots of three similarly treated or inoculated P. notoginseng plants were collected and combined for the qRT-PCR analysis. Total RNA was extracted from the root samples using the TRIGene kit (GeneStar, China) and then cDNA was synthesized via reverse transcription using the Eastep® RT Master Mix kit (Promega, USA). Primers designed to anneal to specific P. notoginseng WRKY transcripts were used for the qRT-PCR analysis (Supplementary Table 1). The P. notoginseng actin gene (PnACT2, KF815706.1) was selected as the internal reference gene. The qRT-PCR analysis was completed using the Eastep® qRT-PCR Master Mix kit (Promega, USA). The PnWRKY expression levels were calculated according to the 2−ΔΔCt method.
Gene-specific primers (Supplementary Table 2) were designed to clone the PnWRKY9 ORF (without the stop codon), which was then inserted into the pGEM-T® Easy vector (Promega, USA). The pGEM-T Easy-PnWRKY9 recombinant plasmid was digested with BamHI and XbaI and the PnWRKY9 sequence was inserted into the pBIN m-gfp5-ER vector for the subsequent expression of the PnWRKY9-green fluorescent protein (GFP). The pBIN m-gfp5-ER-PnWRKY9 recombinant plasmid was inserted into Agrobacterium tumefaciens EHA105 cells according to a CaCl2 freeze–thaw method (Holsters et al., 1978). The A. tumefaciens positive clones, which were identified on the basis of a PCR amplification, were used to infect onion (Allium cepa) inner epidermal cells. The green fluorescence of the PnWRKY9-GFP fusion protein was detected using a confocal laser scanning microscope (Nikon, Japan). The cells were then stained with propidium iodide (PI), after which the subcellular localization of PnWRKY9 was determined on the basis of the observed fluorescence.
Gene-specific primers (Supplementary Table 2) were designed to clone PnWRKY9 from pGEM-T-PnWRKY9. Next, the pCAMBIA2300s-PnWRKY9 recombinant plasmid was constructed via a double digestion with EcoRI and BamHI and a ligation mediated by T4 DNA ligase. The pCAMBIA2300s-PnWRKY9 recombinant plasmid was inserted into A. tumefaciens LBA4404 cells according to the CaCl2 freeze–thaw method. The A. tumefaciens positive clones were used for the transformation of WT tobacco plants (Horsch et al., 1985). Genomic DNA was extracted from transgenic tobacco plants using cetyltrimethylammonium bromide as previously described (Allen et al., 2006). The positive transgenic tobacco plants were identified on the basis of a PCR amplification. The T0 generation tobacco plants were grown in a greenhouse to obtain the T2 generation lines for further analyses.
To analyse PnWRKY9 transcripts levels, total RNA was extracted from T2 transgenic tobacco lines and used to synthesize cDNA as described above. The gene-specific primers were listed in Supplementary Table 1. The resistance of T2 PnWRKY9-OE transgenic tobacco plants to F. solani was evaluated after inoculating the roots. Specifically, the roots of WT and transgenic tobacco plants were injured and then soaked in an F. solani spore solution (2 × 106 spores/mL) for 30 min. The plants were subsequently maintained in an incubator at 25°C with a 16-h photoperiod for 7 days. The disease symptoms at 7 days post-inoculation were recorded using a digital camera (Nikon, Japan).
Gene-specific primers (Supplementary Table 2) with attB linkers (F: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′; R: 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′) were used for the PCR amplification performed to obtain a PnWRKY9 RNAi fragment. The pHellsgate2-PnWRKY9 recombinant plasmid was constructed using the Gateway BP Clonase™ II Enzyme Mix kit (Invitrogen, USA). The pHellsgate2-PnWRKY9 recombinant plasmid and the empty pHellsgate2 vector were inserted into separate A. tumefaciens EHA105 cells. Fresh P. notoginseng leaves with similarly sized wounds were inoculated with 100 μL solutions of A. tumefaciens cells containing the pHellsgate2-PnWRKY9 recombinant vector or the empty pHellsgate2 vector (control). The leaves were placed in a foam box containing moistened filter paper and kept in an incubator for 24 h before being inoculated with F. solani (2 × 106 spores/mL). The disease symptoms after a 72-h incubation were recorded using a digital camera. The diseased area on leaves was calculated using the Photoshop software. Additionally, total RNA was extracted from the P. notoginseng leaves for an examination of PnWRKY9 expression in a qRT-PCR assay.
High-quality total RNA was extracted from the PnWRKY9-OE tobacco leaves (line 9-11) and WT tobacco leaves using the TRIzol® reagent according to the manufacturer's instructions (Invitrogen, USA). Genomic DNA was removed using DNase I (Takara, Japan) prior to the RNA-seq analysis, which was completed using the Illumina platform (Majorbio, China). The following data processing and analysis steps were performed using the online platform of Majorbio Cloud Platform (www.majorbio.com): generation of clean reads, alignment with the reference genome, expression analysis, identification of differentially expressed genes (DEGs), gene ontology (GO) functional annotation, and kyoto encyclopedia of genes and genomes (KEGG) pathway analysis. The RNA-seq analysis involved three biological replicates.
The JA and SA contents in the PnWRKY9-OE tobacco and WT tobacco plants were determined using an ultra-high performance liquid chromatography–mass spectrometry (UPLC-MS) system. Briefly, tobacco leaves (50 mg) were collected from two PnWRKY9-OE tobacco lines 9-3/11 and WT tobacco, respectively. Three biological replicates were included in this experiment. Samples were processed and their hormone contents were determined as described by Qiu et al. (2020).
In P. notoginseng, PnDEFL1 is a JA-responsive defensin gene that mediates the resistance to F. solani (Wang Q. et al., 2019). The PnDEFL1 promoter (PPnDEFL1) was amplified via genome walking for an analysis of the transcriptional regulation of PnDEFL1 by PnWRKY9. According to the PnDEFL1 (MK238492.1) sequence, two gene-specific primers (GSP1 and GSP2; Supplementary Table 2) were designed. The PPnDEFL1 sequence was cloned according to the Universal GenomeWalker 2.0 user manual (Takara, Japan). The putative cis-elements in PPnDEFL1 were predicted using the PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
A pET32a plasmid expressing recombinant PnWRKY9 with N-terminal His-tag was constructed using the ClonExpress II One Step Cloning Kit (Vazyme, China) to produce the PnWRKY9 recombinant protein for EMSA. Gene-specific primers were designed and used to amplify the PnWRKY9 ORF (without the stop codon) (Supplementary Table 2). The pET32a vector was linearized via a digestion with BamHI and EcoRV before incorporating the PnWRKY9 ORF. The pET32a-PnWRKY9 recombinant plasmid was inserted into Escherichia coli BL21 (DE3) cells (Tsingke, China). The production of the PnWRKY9 recombinant protein was induced by adding 0.5 mM isopropyl-β-D-1-thiogalactopyranoside to the bacterial solution, which were then cultured for 6 h at 28°C. The proteins in inclusion bodies were denatured and renatured as described by Li et al. (2021b). The denatured recombinant protein was purified and concentrated using the Ni-NTA Sepharose column (Sangon Biotech, China) and the Amicon® Ultra-15 centrifugal filter device (Merck Millipore, USA), respectively.
The PPnDEFL1 sequence was used to design probes (Supplementary Table 2) containing a W-box or a mutated W-box. The probes were synthesized and labeled with biotin (Sangon Biotech, China). The free probes used for the EMSA were the DNA fragments containing a W-box labeled with biotin, whereas the competitor probes lacked biotin and were used at 50-fold higher concentrations than the free probes. Additionally, the mutant probes contained a mutated W-box and were labeled with biotin. The EMSA was performed according to the standard procedure of the LightShift™ Chemiluminescence EMSA kit (Pierce, USA).
Genomic DNA extracted from P. notoginseng plants served as the template for cloning the PPnDEFL1 sequence using a pair of primers (Supplementary Table 2). The PPnDEFL1 sequence was inserted into the pAbAi yeast bait vector (Clontech, USA) that had been digested with BamHI and HindIII. The PPnDEFL1-pAbAi recombinant plasmid was inserted into Y1Hgold yeast cells, which were then grown on SD/–Ura medium. The PnWRKY9 ORF was amplified by PCR using specific primers (Supplementary Table 2) and then inserted into the pGADT7 yeast prey vector (Clontech, USA). The resulting pGADT7-PnWRKY9 recombinant plasmid was inserted into yeast cells containing PPnDEFL1-pAbAi. The p53-pAbAi yeast bait vector and the pGADT7-P53 yeast prey vector were from the Matchmaker® Gold Yeast One-Hybrid Library Screening System kit (Clontech, USA). The positive transformants on SD/–Leu medium were identified on the basis of a PCR amplification and then transferred to SD/–Leu medium containing 200 ng/mL aureobasidin A (AbA) to analyze the potential interaction between PnDEFL1 promoter and PnWRKY9 protein.
The PPnDEFL1 sequence was amplified from P. notoginseng genomic DNA by PCR using specific primers (Supplementary Table 2) and then inserted into the pBI121 vector to drive the expression of reporter gene (GUS). Moreover, the pBI121-GUS recombinant plasmid was inserted into A. tumefaciens LBA4404 cells, which were then used to infect WT tobacco and PnWRKY9-OE tobacco according to the leaf disk transformation method as described by Horsch et al. (1985). Finally, the infected leaf discs were kept in an incubator (25°C, 16-h photoperiod) to regenerate tobacco seedlings. The PPnDEFL1-GUS/PnWRKY9-OE tobacco plants and the PPnDEFL1-GUS transgenic tobacco plants were identified on the basis of a PCR amplification. Total RNA was extracted from both types of transgenic tobacco and then cDNA was synthesized using the Eastep® RT Master Mix kit (Promega, USA). Primers specific for the GUS (Supplementary Table 1) were used for the qRT-PCR analysis of GUS expression levels, with the tobacco actin gene (NtACT, AB158612.1) selected as an internal reference gene. The GUS relative expression level was calculated according to the 2−ΔΔCt method. The significance of any differences in expression was determined on the basis of Student's t-test, which was conducted using the SPSS software. Furthermore, GUS activities in the PPnDEFL1-GUS/PnWRKY9-OE and PPnDEFL1-GUS transgenic tobacco plants were determined using a fluorescence spectrophotometer as described by Chen R. et al. (2017).
Thirty unigenes encoding putative WRKYs were identified following an analysis of published P. notoginseng transcriptome sequencing data (Liu D. et al., 2019). Additionally, the full-length cDNA sequences were amplified, after which a sequencing analysis confirmed the accuracy of these WRKY genes, which were named PnWRKY1–PnWRKY30 (KM925138, OM273030-OM273058). The phylogenetic tree constructed for the P. notoginseng WRKY gene family using MEGA 6.0 (Figure 1) revealed the WRKY TFs could be divided into three groups (I, II, and III). The Group I WRKY TFs, which contained two WRKY motifs and one C2-H2(C-X4−5-CX22−23-H-X1-H) zinc finger motif, included five PnWRKYs (PnWRKY13, 15, 18, 22, and 23). The Group II members had the same zinc finger motif as the Group I WRKY TFs, but they contained only one WRKY motif. Most of the 30 PnWRKYs belonged to Group II, which was further divided into five subgroups (IIa, IIb, IIc, IId, and IIe). The Group III WRKY TFs, which had a WRKY motif and a zinc finger motif [C2-HC(C-X7-C-X23-H-X-C)] that differed from the one in the WRKY TFs in Groups I and II, included four PnWRKYs (PnWRKY16, 17, 26, and 29).
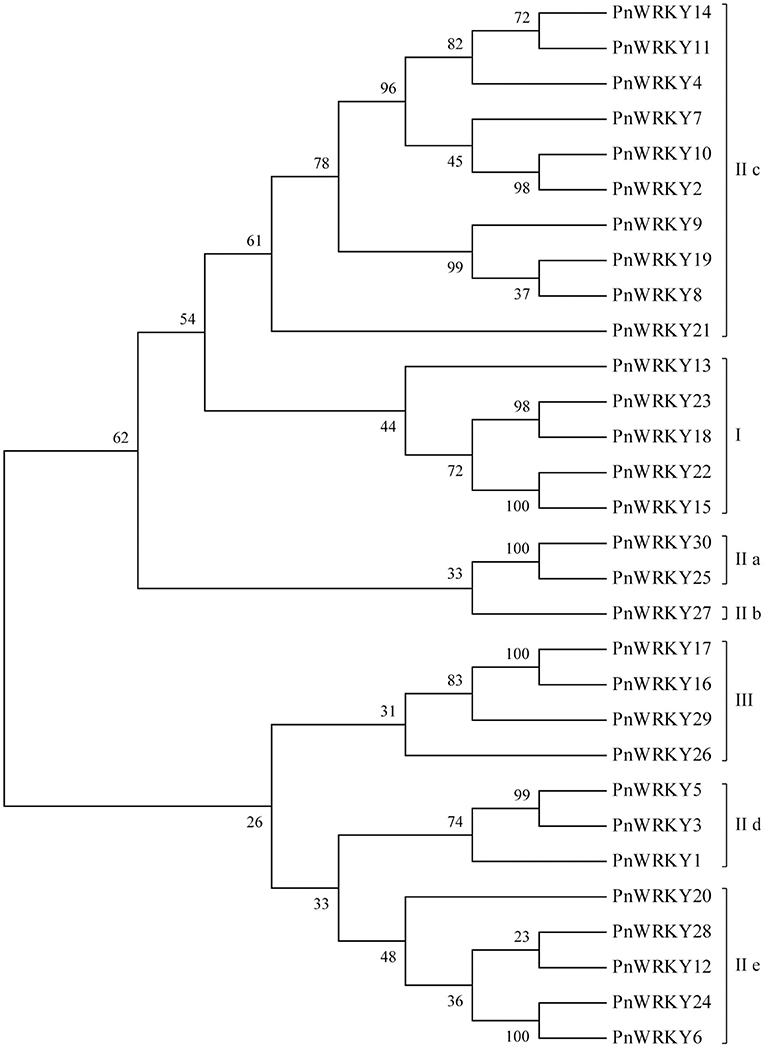
Figure 1. Phylogenetic analysis of the Panax notoginseng WRKY family. The phylogenetic tree of P. notoginseng WRKY family was completed with MEGA6 software. The PnWRKYs were divided into groups I, II and III, in which the group II was further divided into five subgroups (IIa, IIb, IIc, IId, and IIe).
The expression patterns of some PnWRKY genes following the MeJA treatment and the F. solani infection were analyzed by qRT-PCR. The results revealed that the expression levels of several P. notoginseng WRKY genes increased after the MeJA treatment (Figure 2). Among these induced genes, PnWRKY5/6/9/21/25/28/30 expression levels were maximum at 24 h post-treatment with MeJA. Moreover, the transcription of these WRKY genes was also enhanced by the F. solani infection. Following the inoculation with F. solani, the relative expression values of 5 WRKY genes (PnWRKY5/11/15/25/28) were maximum at 48 hpi (Figure 2). Notably, the PnWRKY6/21/22 expression levels were highest during the early stage of F. solani infection. The expression levels of PnWRKY9 and PnWRKY30 were maximum at 72 hpi. These WRKY genes were clearly responsive to JA signaling and F. solani in P. notoginseng roots.
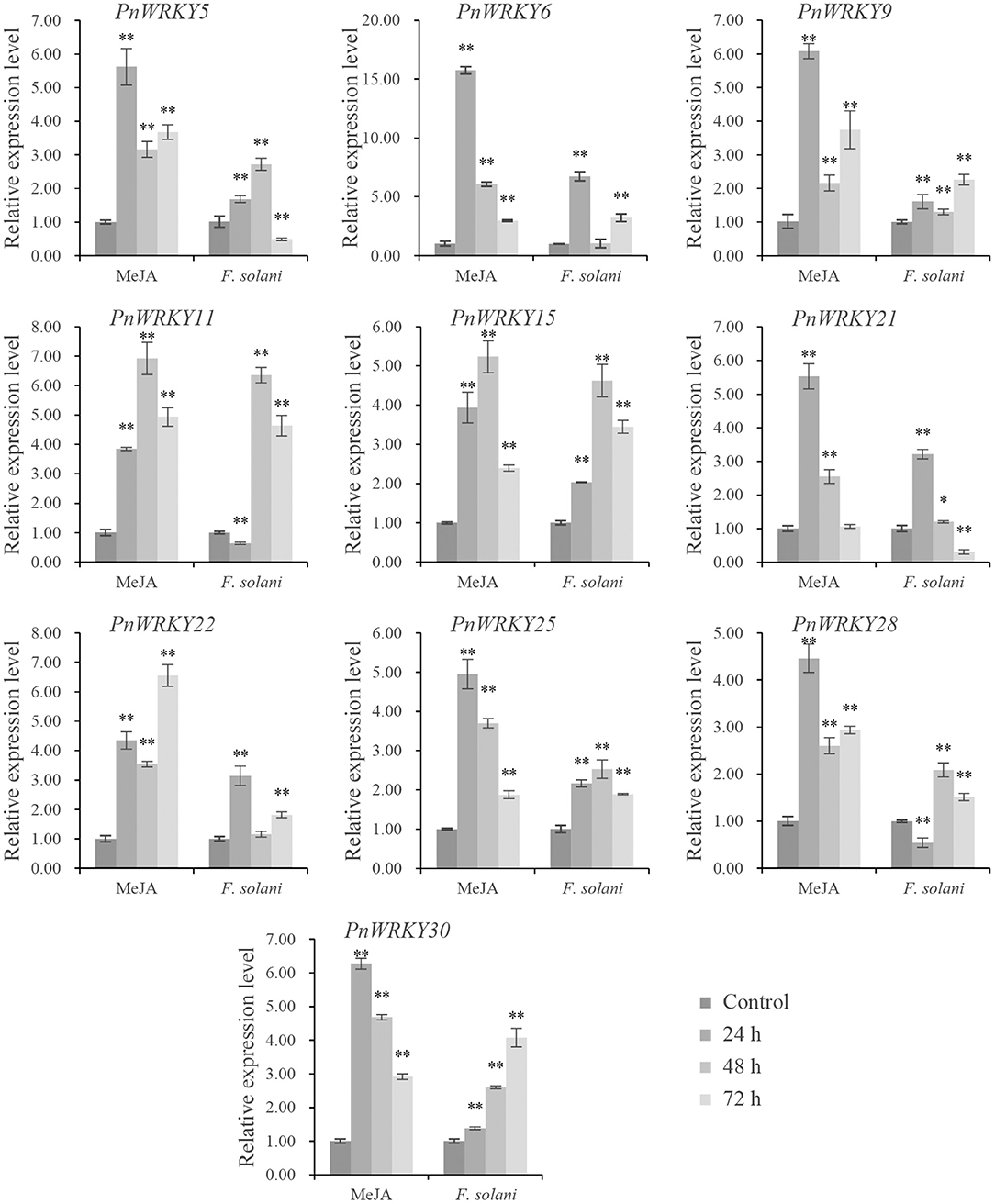
Figure 2. The expression levels of PnWRKYs during independent MeJA treatment or Fusarium solani infection. P. notoginseng roots were treated with MeJA and inoculated with F. solani, respectively. The roots were collected after treatment or inoculation for 24, 48, and 72 h. P. notoginseng roots treated with sterile water for 30 min were harvested as control. The 2−ΔΔCt method and t test (*p < 0.05, **p < 0.01) were used to calculate the relative gene expression levels and statistical difference.
Furthermore, the P. notoginseng roots were pre-treated with MeJA and then inoculated with F. solani in order to analyze the effect of JA signal on PnWRKYs expression during F. solani infection. The result confirmed that these WRKY genes responded to MeJA pre-treatment, in addition, their transcription levels were further enhanced during F. solani infection (Figure 3). Following the inoculation with F. solani, the expression levels of eight WRKY genes (PnWRKY5/6/9/15/22/25/28/30) were maximum at 48 hpi (Figure 3). Notably, the PnWRKY11 and PnWRKY21 expression levels were highest at 12 hpi. Among the examined PnWRKY genes, the expression of PnWRKY5 and PnWRKY6 was induced the most by F. solani infection. The above data showed that P. notoginseng WRKY genes may cooperate with JA signal in response to F. solani infection.
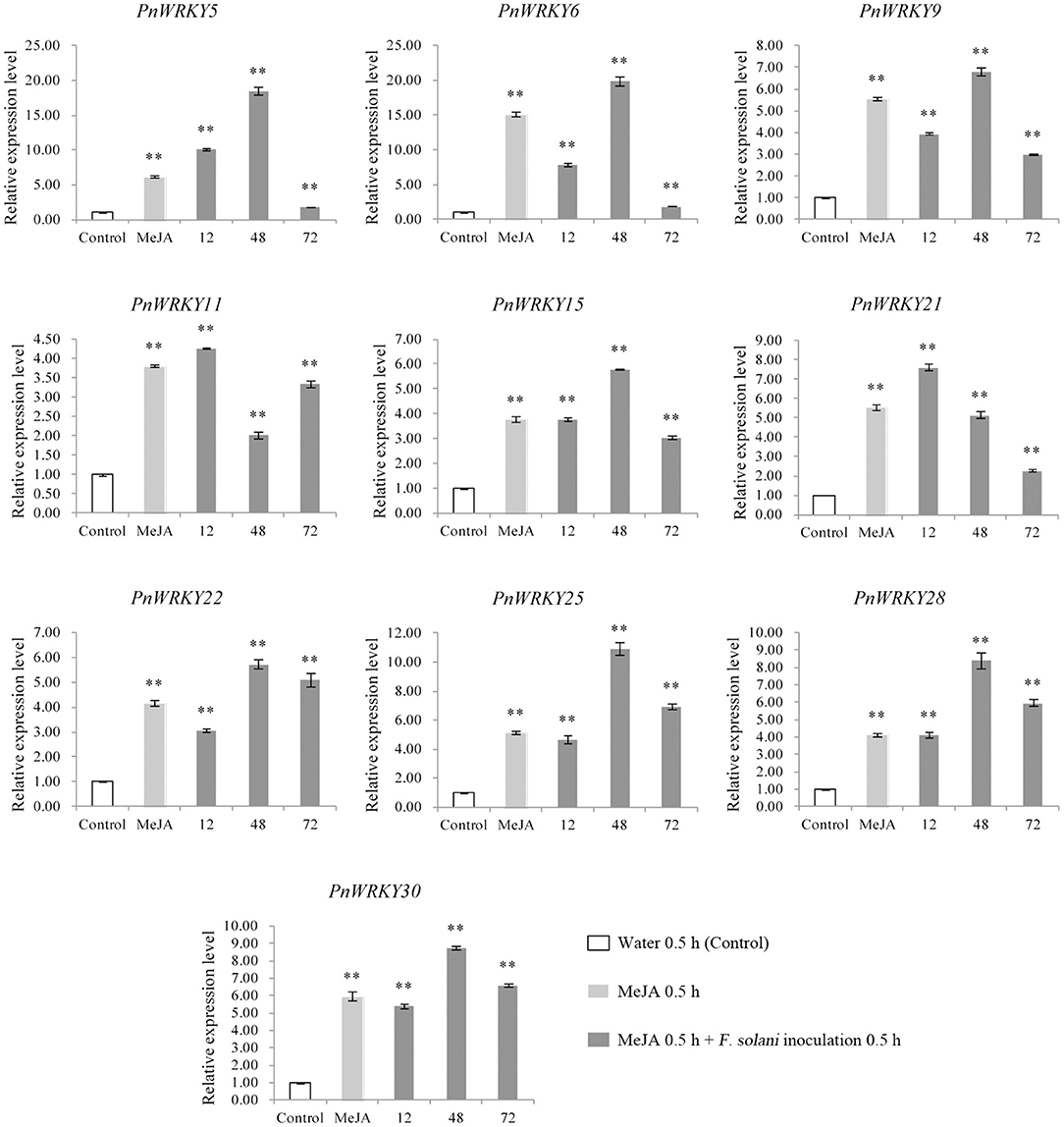
Figure 3. The expression levels of PnWRKYs were estimated in P. notoginseng roots pretreated with MeJA followed by F. solani infection. Panax notoginseng roots were pre-treated with MeJA, and after treatment for 24 h, the treated-roots were further inoculated with F. solani. The MeJA-treated roots were collected after treatment for 24 h, and the inoculated roots were collected at 12, 48, and 72 h post F. solani inoculation. P. notoginseng roots treated with sterile water for 30 min were used as control. The 2−ΔΔCt method and t test (**p < 0.01) were used to calculate the relative gene expression levels and statistical difference.
Considering the variability in the P. notoginseng WRKY gene expression patterns following the MeJA treatment and root rot pathogen infection, PnWRKY9 was randomly selected for a functional analysis. An examination of protein homology indicated that PnWRKY9 was 74.59, 68, and 67.35% homologous to WRKY75 in carrot (Daucus carota, XP_017248451.1), pistachio (Pistacia vera, XP_031272738.1), and apple (Malus baccata, AhJ78583.1), respectively. Additionally, the alignment of PnWRKY9 with DcWRKY75, PvWRKY75, and MbWRKY75 revealed WRKYGQK and the CTYQGCX23HTH zinc finger motif were highly conserved (Figures 4A,B).
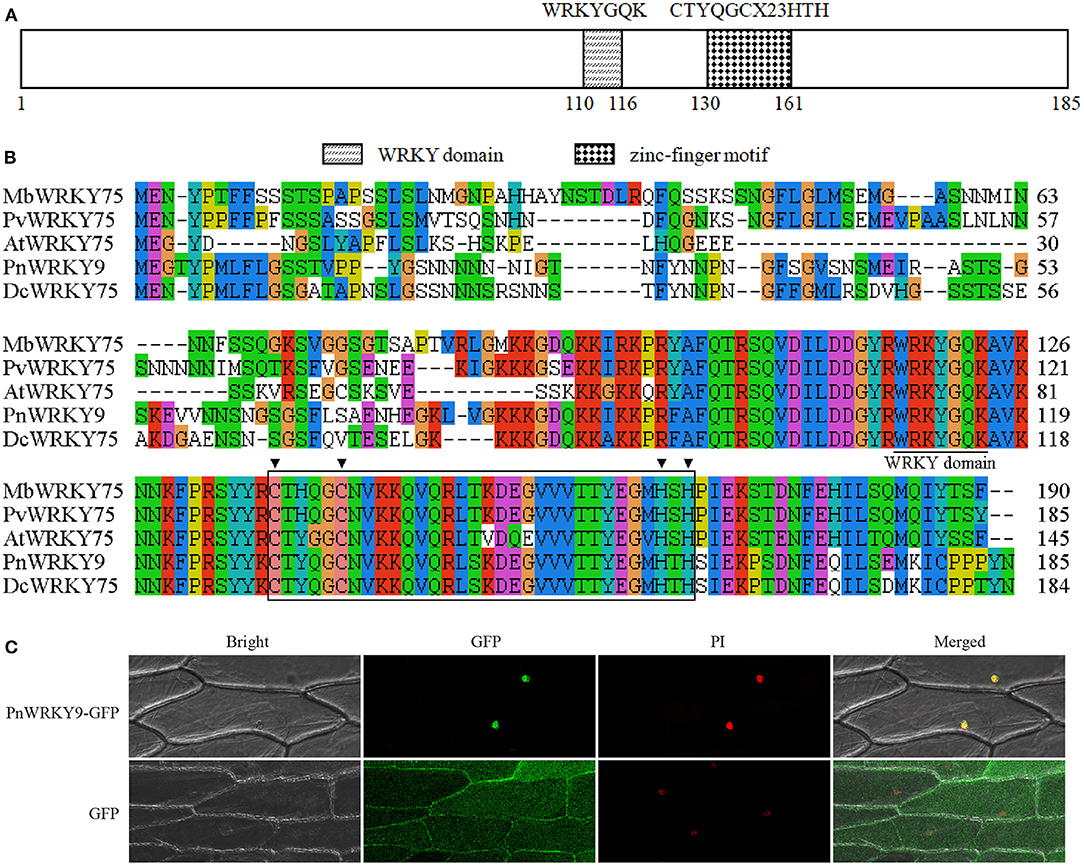
Figure 4. Sequence and subcellular localization analyses of PnWRKY9. (A) The structure schematic of PnWRKY9 protein. (B) Multiple sequence alignment of PnWRKY9 and homologous WRKY proteins using clustalx1.83 software. (C) Subcellular localization analysis of PnWRKY9-GFP fusion protein. Bright, white light field; GFP, green fluorescent field; PI, propidium iodide, which was used to indicate the nucleus; Merged, mixed field.
The PnWRKY9-GFP fusion protein was transiently expressed in onion epidermal cells. The green fluorescence of PnWRKY9-GFP was detected exclusively in the nucleus, whereas the green fluorescence of GFP alone was observed throughout the onion epidermal cells (Figure 4C). Moreover, the red fluorescence from the nuclear dye PI confirmed that PnWRKY9 is a nuclear protein.
The qRT-PCR analysis revealed PnWRKY9 expression levels varied among 13 T2 generation transgenic tobacco lines. The expression level was highest in lines 9-3 and 9-11. It was also relatively high in lines 9-1, 9-2, 9-4, 9-7, and 9-8 (Figure 5A). The data confirmed that PnWRKY9 was stably expressed in the T2 transgenic tobacco lines. The tobacco roots were inoculated with F. solani and examined 7 days later. The PnWRKY9-OE tobacco lines had no obvious disease symptoms (i.e., plants were green and healthy). In contrast, the WT tobacco plants had withered leaves and roots that were black and rotted (Figure 5B). Thus, the overexpression of PnWRKY9 clearly increased the resistance of transgenic tobacco plants to F. solani.
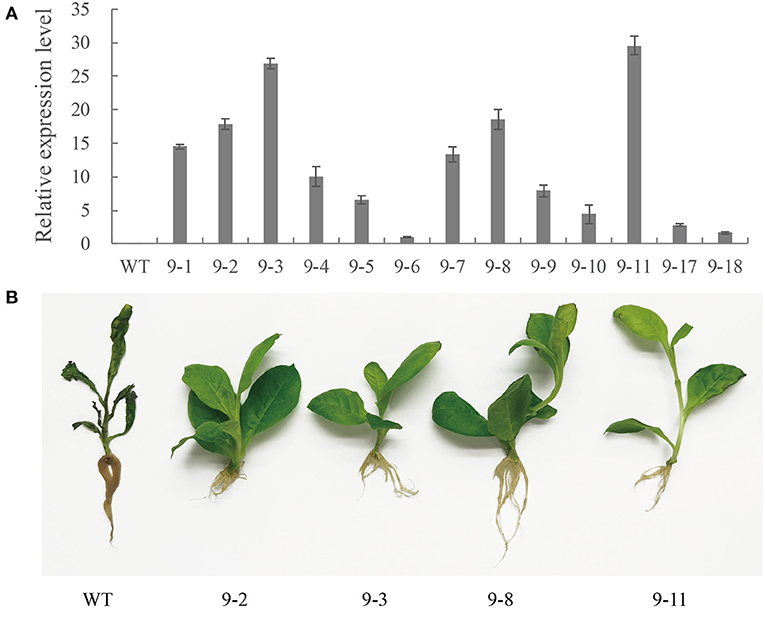
Figure 5. Analysis of PnWRKY9-OE tobacco. (A) PnWRKY9 expression levels in the overexpression tobacco. The PnWRKY9 was stably expressed in overexpression tobacco lines 9-1/2/3/4/5/6/7/8/9/10/11/17/18. (B) Resistance assay of PnWRKY9-OE tobacco against F. solani. The PnWRKY9-OE tobacco lines (9-2/3/8/11) and wild type (WT) tobacco roots were inoculated with F. solani. and the PnWRKY9-OE tobacco showed higher level of resistance to F. solani than the WT.
The PnWRKY9 RNAi fragments were transiently expressed in young P. notoginseng leaves. The leaves transformed with the RNAi empty vector served as the control samples. Although the extent of the damage caused by the F. solani infection varied, the lesions were almost 4-times larger on the leaves with the PnWRKY9 RNAi fragments than on the control leaves (Figures 6A,B). Additionally, the qRT-PCR data indicated the PnWRKY9 expression level in the leaves with the PnWRKY9 RNAi fragments decreased to about half of that in the control leaves (Figure 6C). Accordingly, the decrease in PnWRKY9 expression increased the susceptibility of P. notoginseng to F. solani.

Figure 6. The transiently expression analysis of PnWRKY9 RNAi vector in the leaves of P. notoginseng. (A) The symptoms of P. notoginseng leaves inoculated with F. solani, that were first infected with A. tumefaciens containing PnWRKY9 RNAi vector. The P. notoginseng leaves with expressed empty RNAi vector and inoculated F. solani was as control. (B) The lesion size in P. notoginseng leaves expressing the PnWRKY9 RNAi vector and empty RNAi vector, respectively (**p < 0.01). (C) The PnWRKY9 expression levels in P. notoginseng leaves expressing PnWRKY9 RNAi vector and empty RNAi vector, respectively. The results were calculated by the 2−ΔΔCt method and analyzed by the t test (**p < 0.01).
The RNA-seq analysis of PnWRKY9-OE tobacco and WT tobacco detected 4,286 DEGs, including 976 up-regulated genes and 3,310 down-regulated genes. The KEGG analysis of the DEGs revealed that the three most significantly enriched pathways were plant hormone signal transduction, plant–pathogen interaction, and phenylpropanoid biosynthesis (Figure 7A). There were 76 DEGs associated with the plant hormone signal transduction pathway (Figure 7B). Some related genes of JA biosynthesis and signal transduction pathway were significantly up-regulated (Supplementary Table 5), such as allene oxide synthase and bZIP transcription factor, moreover, the expression of some JA signal repressor genes was down-regulated including JAZs and MYC2. However, the expression of SA biosynthesis related genes including pyridoxal phosphate-dependent transferases, amidase, aspartate aminotransferase 3, and tyrosine transaminase were down-regulated. The 61 DEGs involved in the plant–pathogen interaction pathway (Figure 7C) included up-regulated genes encoding heat shock protein 90 (HSP90), which is associated with plant hypersensitive responses (HRs), many disease resistance-related proteins, and cytochrome P450. In contrast, the expression of WRKY33 encoding an HR-suppressor was down-regulated. Moreover, 52 DEGs were associated with the phenylpropanoid biosynthesis pathway, including some up-regulated genes encoding the peroxidase superfamily protein, UDP-glucosyl transferases, and cinnamoyl CoA reductase (Figure 7D).
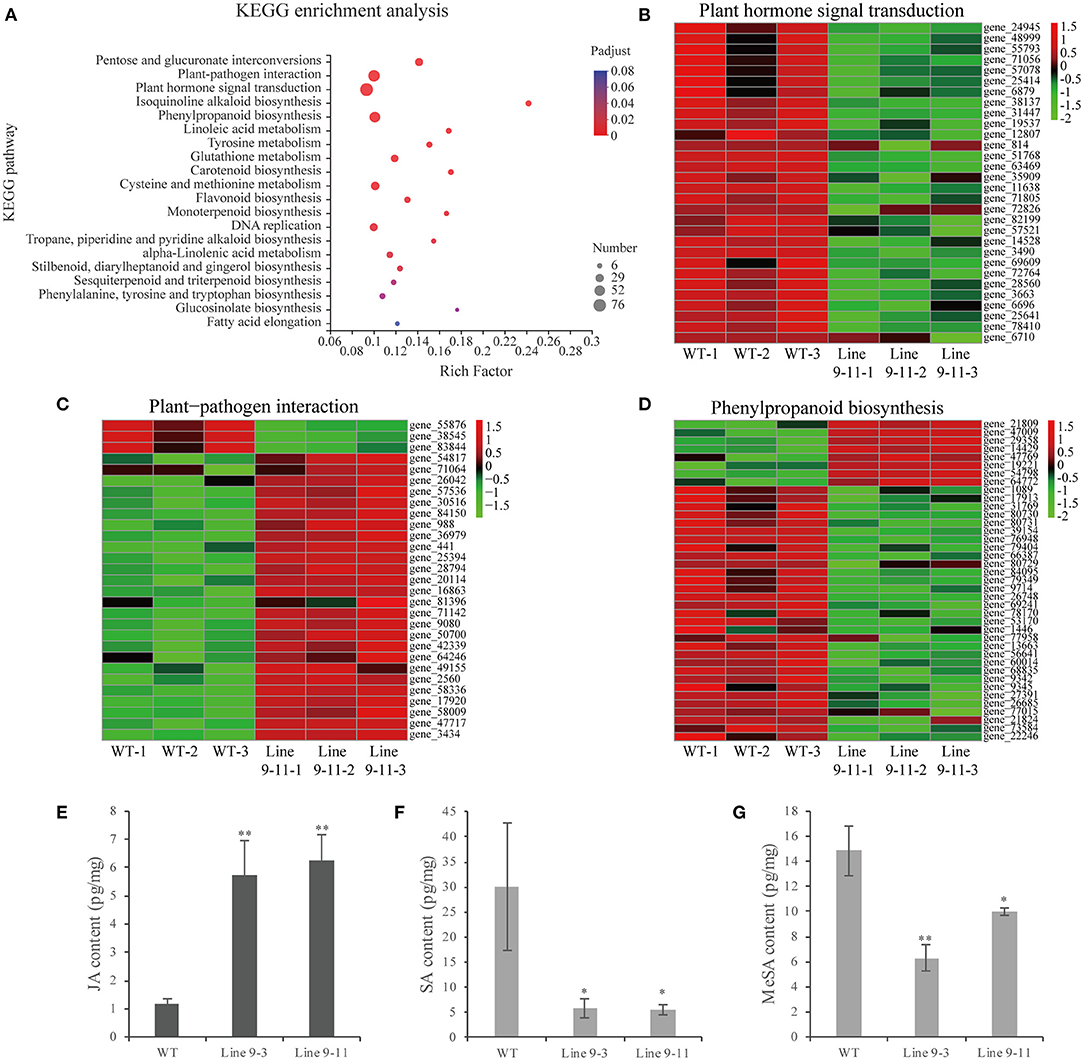
Figure 7. RNA sequencing analysis and JA/SA content in PnWRKY9-OE tobacco. (A) KEGG enrichment analysis of DEGs in PnWRKY9-OE tobacco line 9-11 compared with WT tobacco. The color showed the significance level, and the bubble size indicated the numbers of the enriched genes. (B–D) DEGs heat map in plant hormone signal, plant-pathogen interactions, and phenylpropanoid biosynthesis pathways. The red and green indicated the up-regulated and down-regulated gene expression, respectively. (E–G) JA, SA, and MeSA contents in PnWRKY9-OE tobacco line 9-3/11 and the WT tobacco (*p < 0.05; **p < 0.01).
The RNA-seq analysis indicated that the JA signaling pathway was activated, but the SA signaling pathway was suppressed, in PnWRKY9-OE tobacco. Therefore, the accumulation of endogenous JA and SA in PnWRKY9-OE tobacco and WT tobacco was analyzed to validate the RNA-seq data. The JA content was higher in PnWRKY9-OE tobacco than in WT tobacco (Figure 7E), whereas the opposite trend was observed for the SA and methyl salicylate contents (Figures 7F,G). These observations were consistent with the RNA-seq data. Accordingly, PnWRKY9 and the JA signaling pathway may cooperatively enhance disease resistance.
The promoter sequence of a JA-responsive and F. solani resistance-related gene (PnDEFL1) was cloned for an analysis of the interaction between PnWRKY9 and a P. notoginseng disease resistance-related gene. The PPnDEFL1 sequence (475 bp) was obtained by genome walking. The promoter sequence was predicted to contain many cis-elements (Supplementary Table 3), including G-box, MYB, and W-box. Moreover, the PnWRKY9 recombinant protein was expressed in E. coli cells and purified to verify the specific binding between PnWRKY9 and PPnDEFL1. The EMSA results revealed a lack of bands shifts in lanes 1 and 4 (Figure 8A), which, respectively, corresponded to the free probes (W-box sequence of PPnDEFL1) without PnWRKY9 and the mutant probes (mutated W-box sequence) with PnWRKY9. Therefore, PnWRKY9 did not bind to the mutant W-box sequence. Specifically, bands shifts were detected in lanes 2 and 3 (Figure 8A). Moreover, the band shift in lane 2 was more obvious than in lane 3, likely because the abundance of competitor probes without biotin was 50-times greater than the abundance of free probes. These results indicated that PnWRKY9 can bind specifically to the W-box sequence of PPnDEFL1.
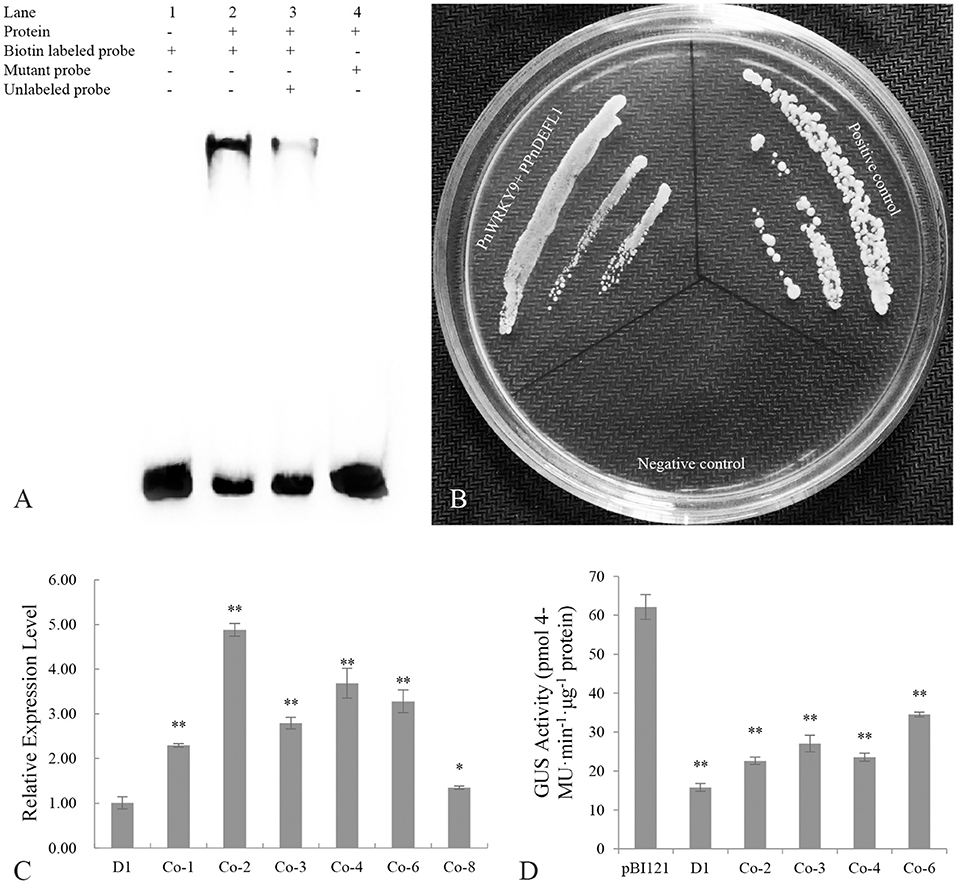
Figure 8. Transcriptional regulation of PnWRKY9 on PPnDEFL1. (A) EMSA assay verified the specific binding of PnWRKY9 with PPnDEFL1. Lane 1, the biotin labeled free probes containing the W-box sequence of PPnDEFL1; Lane 2, PnWRKY9 can interact with biotin labeled probes containing W-box sequence of PPnDEFL1; Lane 3, PnWRKY9 showed the interaction with the unlabeled probes and biotin labeled probes containing W-box sequence of PPnDEFL1; Lane 4, PnWRKY9 cannot interact with the mutant probes labeled by biotin. (B) Y1H assay verified the transcriptional activation of PnWRKY9 on PPnDEFL1. PnWRKY9 + PPnDEFL1, pGADT7-PnWRKY9 and PPnDEFL1-pAbAi co-transformed yeast cells; Positive control, p53-pAbAi and pGADT7-p53 co-transformed yeast cells; Negative control, PPnDEFL1-pAbAi and pGADT7-P53 co-transformed yeast cells. (C) Analysis of GUS gene expression level in PPnDEFL1-GUS/PnWRKY9 co-expression tobacco (Co-1/2/3/4/6/8). The transgenic tobacco (D1) just expressing the PPnDEFL1-GUS was control. (D) Determination of GUS activity in PPnDEFL1-GUS/PnWRKY9 co-expression tobacco (Co-2/3/4/6). The transgenic tobacco just expressing the PPnDEFL1-GUS (D1) and pBI121-GUS were control (*p < 0.05; **p < 0.01).
The ability of PnWRKY9 to activate the transcription of PnDEFL1 was assessed in a Y1H assay (Figure 8B). The Y1Hgold yeast cells with p53-pAbAi and pGADT7-P53 were used as the positive control, whereas the Y1Hgold yeast cells with PPnDEFL1-pAbAi and pGADT7-P53 served as the negative control. The Y1Hgold yeast cells with PPnDEFL1-pAbAi/pGADT7-PnWRKY9 grew normally on the SD/–Leu medium supplemented with 200 ng/mL AbA (i.e., similar to the control). In contrast, the negative control cells were unable to survive on the selective medium. The fusion protein GAL4-PnWRKY9 in the yeast cells transformed with the prey vector pGADT7-PnWRKY9 was expected to bind to the cis-element W-box in PPnDEFL1 and activate the expression of AUR1-C in the pAbAi recombinant plasmid, enabling cells to grow normally on the SD/–Leu/AbA medium. The Y1H results confirmed PnWRKY9 can activate PnDEFL1 transcription.
The GUS under the control of PPnDEFL1 was inserted into WT tobacco and PnWRKY9-OE tobacco. The subsequent qRT-PCR analysis revealed that GUS expression differed between the PPnDEFL1-GUS/PnWRKY9-OE and PPnDEFL1-GUS transgenic tobacco plants. The highest expression level, which was detected in line Co-2, was 3.88-times higher than that in the control (i.e., transgenic tobacco with only PPnDEFL1-GUS, D1). The second highest expression level was detected in line Co-4. The expression levels in Co-1, Co-3, Co-6, and Co-8 were, respectively, 1.3-, 1.8-, 2.3-, and 0.4-times higher than the control expression level (Figure 8C). Hence, lines Co-2, Co-3, Co-4, and Co-6 (PPnDEFL1-GUS/PnWRKY9-OE tobacco) were selected for an examination of GUS activity. Compared with the D1 control, the GUS activity was significantly higher in lines Co-2, Co-3, Co-4, and Co-6 (Figure 8D). Specifically, the GUS activity of the PPnDEFL1-GUS/PnWRKY9-OE tobacco exceeded 20 pmol 4-MU min−1 μg−1 protein, whereas the GUS activity of the control was 15 pmol 4-MU min−1 μg−1 protein. These observations suggested that PPnDEFL1 was activated by PnWRKY9.
The recent development of high-throughput sequencing technology has resulted in the identification of an increasing number of WRKY genes in various plant species. For example, an RNA-seq analysis identified 58 WRKY genes in eggplant (Solanum melongena), among which SmWRKY26 and SmWRKY32 were revealed to negatively regulate the cold stress tolerance of eggplant on the basis of virus-induced gene silencing (VIGS) (Yang et al., 2020). The Arabidopsis thaliana WRKY gene family comprises 74 genes, including AtWRKY38 and AtWRKY62, which are involved in defense responses to Pseudomonas aeruginosa, and AtWRKY57, which helps regulate senescence (Birkenbihl et al., 2018). The WRKY domain and the zinc finger motif are conserved structures that are used to divide WRKY TFs into different groups (Rushton et al., 2010). In this study, 30 P. notoginseng WRKY genes were identified by analyzing transcriptome sequencing data (Liu D. et al., 2019). The conserved domains and phylogenetic relationships among P. notoginseng WRKY TFs were examined. Additionally, the 30 WRKY TFs in P. notoginseng were divided into three groups, which is in accordance with the classification of WRKYs in other plants including pepper (C. annuum) and eggplant (Zheng et al., 2019; Yang et al., 2020). As regards the WRKY gene evolution, the ancient group IIc WRKY genes were considered to be ancestor of all WRKY genes, therefore, group IIc WRKYs in plant were the most diverse members (Luise et al., 2013). In the phylogenetic tree of PnWRKYs, some coefficients located in left branch of group IIc were big, but those in the right branch were small, which was similar to the phylogenetic tree of wild rice (Oryza nivara) group IIc WRKYs (Xu et al., 2016). These results hinted that the group IIc members of WRKY super gene family had rich diversity. Moreover, because the P. notoginseng genome has been sequenced (Chen W. et al., 2017), the WRKY genes isolated in this study provide the basis for future investigations conducted to identify all members of the P. notoginseng WRKY superfamily.
Earlier research demonstrated that TFs involved in various plant physiological processes are usually responsive to treatment with multiple plant hormones (Jiang J. et al., 2017). The expression of TaWRKY40-D, which encodes a positive regulator of leaf senescence in wheat (Triticum aestivum), is reportedly up-regulated after JA and ABA treatments (Zhao L. et al., 2020). In lotus (N. nucifera) roots, NnWRKY40a and NnWRKY40b expression is significantly induced following a JA treatment (Li et al., 2019). Similarly, the expression of GbWRKY20 in Ginkgo biloba is up-regulated at 12 h after the application of exogenous MeJA (Zhou et al., 2021). In another study, SA and JA treatments enhanced the expression of PacMYBA in sweet cherry (Prunus avium L.) leaves; the heterologous expression of this gene in A. thaliana increases the resistance to Pseudomonas syringae pv. tomato DC3000 (Shen et al., 2017). In the current study, the expression levels of several WRKY genes in P. notoginseng roots were up-regulated to varying degrees by the MeJA treatment. Moreover, the expression levels of 10 PnWRKY genes increased further in response to an F. solani infection. Thus, these P. notoginseng WRKY TFs and the JA signaling pathway may be involved in the early defense response to F. solani.
Proteins in plant cells are generally functional only in specific subcellular locations (Yousefi et al., 2021). Hence, analyzing the subcellular localization of proteins may provide important clues regarding protein functions. Most WRKY TFs are nuclear proteins (Zhao Y. et al., 2019). For example, in sugarcane (Saccharum spp.), WRKY3 is localized in the nucleus (Wang et al., 2018). A laser confocal microscopy-based examination of Populus tremula confirmed WRKY18 and WRKY35 are localized in the nucleus (Jiang Y. et al., 2017). Similarly, P. notoginseng WRKY9 was identified as a nuclear protein in the present study. Furthermore, the overexpression of PnWRKY9 in tobacco was observed to enhance the resistance to F. solani. In accordance with this finding, the transient expression of the PnWRKY9 RNAi fragment in P. notoginseng leaves increased the susceptibility of the plants to an infection by F. solani. The transcriptional regulation of TF genes occurs in the nucleus. Therefore, the identification of PnWRKY9 as a nuclear protein and the results of the reverse genetics analysis in this study confirmed that PnWRKY9 is a positive regulator of the root rot defense response in P. notoginseng.
Regulatory genes are crucial for the transcriptome reprogramming during plant responses to pathogens, and their functions are further amplified by the crosstalk of some signal transduction pathways (Shen et al., 2017). The expression levels of CsWRKY50 and some hormone signaling pathway-related genes, including JA-responsive genes and genes involved in SA biosynthesis, are up-regulated in cucumber (Cucumis sativus) during a Pseudoperonospora cubensis infection (Luan et al., 2019). The overexpression of cotton (Gossypium barbadense) WRKY1 enhances the resistance to Verticillium dahliae, but the silencing of this gene by VIGS has suppressive effects on JA- and ET-induced defense activities (Zhang et al., 2019). The cross-talk of hormone signal transduction pathways is complex, for example, the SA and JA signal pathways showed synergistic or antagonistic effects under different conditions. Arabidopsis WRKY70 is a positive regulator of SA-signaling pathway in response to P. syringae, but it functions as a negative regulator of JA-inducible genes (Li et al., 2004). In addition, the decreased expression of cotton (G. hirsutum) WRKY70 enhanced the resistance to V. dahliae, which resulted in up-regulated expression of JA responding genes and down-regulated expression of SA signaling pathway genes (Xiong et al., 2020). In the present study, the comparison between PnWRKY9-overexpressing tobacco and the control plants revealed plant hormone signal transduction and plant–pathogen interaction were the most enriched KEGG pathways among the DEGs. Some JA signaling pathway genes were up-regulated, nevertheless some SA signaling pathway genes were down-regulated. Additionally, the JA content was higher in PnWRKY9-OE tobacco than in WT tobacco, whereas the opposite trend was detected for the SA level. These findings imply that PnWRKY9 may synergistically JA signal and inhibit SA accumulation to regulate P. notoginseng defenses against the root rot pathogen.
The WRKY TFs bind to the W-box sequence in the promoters of their target genes, including disease resistance-related genes, through the WRKY domain, and then activate or repress the expression of the target genes (Li et al., 2021a). The interactions between WRKY TFs and their target genes have been elucidated in a variety of plant species on the basis of EMSA and Y1H analyses. In apple (Malus × domestica) WRKY46 binds to the MdPBS3.1 promoter, which includes a W-box sequence, and the resulting activation of MdPBS3.1 expression enhances the resistance to powdery mildew (Zhao et al., 2019b). In Citrus sinensis, WRKY65 regulates the expression of some Penicillium digitatum resistance-related genes (e.g., CsRbohB, CsRbohD, CsCDPK33, and CsPR10) by binding to the W-box in their promoters (Wang et al., 2021). In this study, EMSA and Y1H results indicated that PnWRKY9 directly regulates the expression of the F. solani resistance-related defensin gene PnDEFL1. This was confirmed by the significantly increased GUS expression level and GUS activity in the PPnDEFL1-GUS/PnWRKY9-OE tobacco. Thus, the regulation of PnDEFL1 expression may partially explain the synergism between JA and PnWRKY9 during the molecular interaction between P. notoginseng and the root rot pathogen.
In summary, we proposed a model of positive regulation of PnWRKY9 in P. notoginseng during response to F. solani infection (Figure 9). When P. notoginseng plants were threatened by root rot pathogen F. solani, PnWRKY9 was greatly induced to be expressed, and then PnWRKY9 specifically bound to the W-box sequence in promoter of P. notoginseng disease resistance gene PnDEFL1, which activated the expression of PnDEFL1 and enhanced the resistance of P. notoginseng to F. solani. Meanwhile, the accumulation of JA in P. notoginseng was also induced by F. solani, and the expression of JA-responsive resistance genes in P. notoginseng, such as PnSN1 and PnDEFL were further induced by the JA signaling to enhance resistance to root rot.
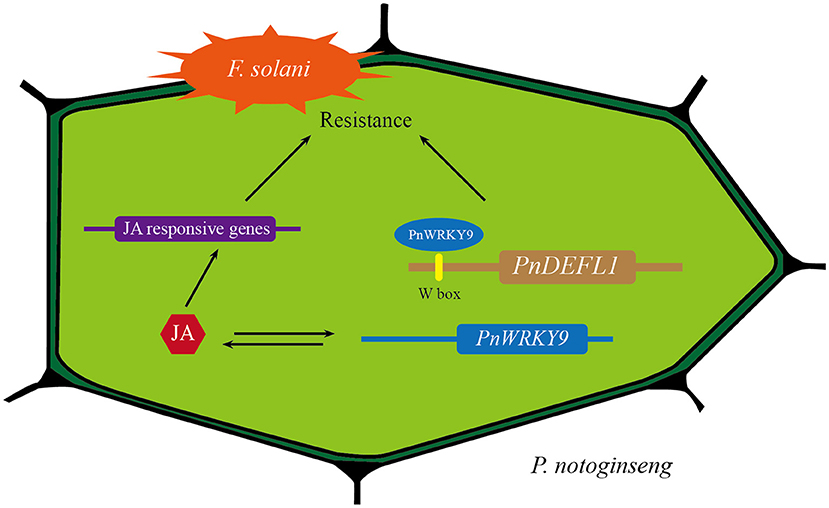
Figure 9. A model of positive regulation of PnWRKY9 in P. notoginseng in response to F. solani infection. After P. notoginseng was infected by root rot pathogen F. solani, PnWRKY9 activated the expression of JA-responsive resistance gene PnDEFL1 by specifically binding W-box in PnDEFL1 promoter, which enhanced resistance to F. solani. Moreover, the activated JA signaling induced the expression of JA-responsive genes to boost defense response against F. solani.
In this study, we identified P. notoginseng WRKY genes and determined that the expression of 10 PnWRKY genes was inducible by the MeJA treatment and F. solani infection. Moreover, PnWRKY9 was revealed to encode a nuclear protein that positively regulates the resistance of P. notoginseng to F. solani. Additionally, PnWRKY9 and the JA signaling pathway synergistically affect the defense response of P. notoginseng to root rot through the regulated expression of the defensin gene PnDEFL1.
The original contributions presented in the study are publicly available. The data can be found in the NCBI: PnWRKY1-PnWRKY30 (accession numbers KM925138, OM273030-OM273058); RNA sequencing data of PnWRKY9 transgenic tobacco accession number PRJNA827632. The sequence of PnDEFL1 promoter was showed in the Supplementary Materials.
LZ: methodology, data curation, experiment execution, and writing—original draft preparation. BQ: methodology, material collection, and experiment execution. LS: validation. HW: software and conceptualization. XC and FG: supervision. DL: methodology, supervision, and writing—reviewing and editing. All authors contributed to the article and approved the submitted version.
This work was financially supported by one grant received from the National Natural Sciences Foundation of China (Grant No. 82060693).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.930644/full#supplementary-material
Afrin, S., Huang, J. J., and Luo, Z. Y. (2015). JA-mediated transcriptional regulation of secondary metabolism in medicinal plants. Sci. Bull. 60, 1062–1072. doi: 10.1007/s11434-015-0813-0
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Bawa, G., Feng, L., Yan, L., Du, Y., Shang, J., Sun, X., et al. (2019). Pre-treatment of salicylic acid enhances resistance of soybean seedlings to Fusarium solani. Plant Mol. Biol. 101, 315–323. doi: 10.1007/s11103-019-00906-x
Birkenbihl, R. P., Kracher, B., Ross, A., Kramer, K., Finkemeier, I., and Somssich, I. E. (2018). Principles and characteristics of the Arabidopsis WRKY regulatory network during early MAMP-triggered immunity. Plant J. 96, 487–502. doi: 10.1111/tpj.14043
Caroline, F. A., and Olubukola, O. (2013). Integrated management strategies for tomato Fusarium wilt. Biocontrol Sci. 18, 117–127. doi: 10.4265/bio.18.117
Catinot, J., Huang, J. B., Huang, P. Y., Tseng, M. Y., Chen, Y. L., Gu, S. Y., et al. (2015). ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate - and ethylene-responsive defence genes. Plant Cell Environ. 38, 2721–2734. doi: 10.1111/pce.12583
Chen, L., Zhang, L., Xiang, S., Chen, Y., Zhang, H., and Yu, D. (2021). The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 72, 1473–1489. doi: 10.1093/jxb/eraa529
Chen, R., He, H., Yang, Y., Qu, Y., Ge, F., and Liu, D. (2017). Functional characterization of a pathogenesis-related protein family 10 gene, LrPR10-5, from Lilium regale Wilson. Australas. Plant Pathol. 46, 251–259. doi: 10.1007/s13313-017-0485-0
Chen, W., Kui, L., Zhang, G., Zhu, S., Zhang, J., Wang, X., et al. (2017). Whole-genome sequencing and analysis of the Chinese herbal plant Panax notoginseng. Mol. Plant. 10, 899–902. doi: 10.1016/j.molp.2017.02.010
Hao, W., Gray, M. A., Forster, H., and Adaskaveg, J. E. (2019). Evaluation of new oomycota fungicides for management of phytophthora root rot of citrus in California. Plant Dis. 103, 619–628. doi: 10.1094/PDIS-07-18-1152-RE
Holsters, M., de Waele, D., Depicker, A., Messens, E., van Montagu, M., and Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163, 181–187. doi: 10.1007/BF00267408
Horsch, R., Rogers, S., and Fraley, R. (1985). Transgenic plant. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437. doi: 10.1101/SQB.1985.050.01.054
Hussain, A., Li, X., Weng, Y., Liu, Z., Ashraf, M. F., Noman, A., et al. (2018). CaWRKY22 acts as a positive regulator in pepper response to Ralstonia solanacearum by constituting networks with CaWRKY6, CaWRKY27, CaWRKY40, and CaWRKY58. Int. J. Mol. Sci. 19:1426. doi: 10.3390/ijms19051426
Ifnan Khan, M., Zhang, Y., Liu, Z., Hu, J., Liu, C., Yang, S., et al. (2018). CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 19:1403. doi: 10.3390/ijms19051403
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., and Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx008
Jiao, Z., Sun, J., Wang, C., Dong, Y., Xiao, S., Gao, X., et al. (2018). Genome-wide characterization, evolutionary analysis of WRKY genes in Cucurbitaceae species and assessment of its roles in resisting to powdery mildew disease. PLoS ONE. 13:e0199851. doi: 10.1371/journal.pone.0199851
Khaliq, A., Alam, S., Khan, I. U., Khan, D., Naz, S., Zhang, Y., et al. (2020). Integrated control of dry root rot of chickpea caused by Rhizoctonia bataticola under the natural field condition. Biotechnol. Rep. 25:e00423. doi: 10.1016/j.btre.2020.e00423
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 16, 319–331. doi: 10.1105/tpc.016980
Li, J., Xiong, Y., Li, Y., Ye, S., Yin, Q., Gao, S., et al. (2019). Comprehensive analysis and functional studies of WRKY transcription factors in Nelumbo nucifera. Int. J. Mol. Sci. 20:5006. doi: 10.3390/ijms20205006
Li, S., Hai, J., Wang, Z., Deng, J., Liang, T., Su, L., et al. (2021a). Lilium regale Wilson WRKY2 regulates chitinase gene expression during the response to the root rot pathogen Fusarium oxysporum. Front. Plant Sci. 12:741463. doi: 10.3389/fpls.2021.741463
Li, S., Liu, G., Pu, L., Liu, X., Wang, Z., Zhao, Q., et al. (2021b). WRKY transcription factors actively respond to Fusarium oxysporum in Lilium regale Wilson. Phytopathology 111, 1625–1637. doi: 10.1094/PHYTO-10-20-0480-R
Liu, D., Zhao, Q., Cui, X., Chen, R., Li, X., Qiu, B., et al. (2019). A transcriptome analysis uncovers Panax notoginseng resistance to Fusarium solani induced by methyl jasmonate. Genes Genomics 41, 1383–1396. doi: 10.1007/s13258-019-00865-z
Liu, X., Li, D., Zhang, S., Xu, Y., and Zhang, Z. (2019). Genome-wide characterization of the rose (Rosa chinensis) WRKY family and role of RcWRKY41 in gray mold resistance. BMC Plant Biol. 19:522. doi: 10.1186/s12870-019-2139-6
Luan, Q., Chen, C., Liu, M., Li, Q., Wang, L., and Ren, Z. (2019). CsWRKY50 mediates defense responses to Pseudoperonospora cubensis infection in Cucumis sativus. Plant Sci. 279, 59–69. doi: 10.1016/j.plantsci.2018.11.002
Luise, H. B., Nina, M. F., Klaus, H., Oliver, K., and Dierk, W. (2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41, 9764–9778. doi: 10.1093/nar/gkt732
Ma, Y. N., Chen, C. J., Li, Q. Q., Xu, F. R., Cheng, Y. X., and Dong, X. (2019). Monitoring antifungal agents of Artemisia annua against Fusarium oxysporum and Fusarium solani, associated with Panax notoginseng root-rot disease. Molecules 24:213. doi: 10.3390/molecules24010213
Qiu, B., Zhang, Y., Wang, Q., Wang, Z., Chen, H., Cui, X., et al. (2020). Panax notoginseng snakin gene increases resistance to Fusarium solani in transgenic tobacco. Ind. Crops Prod. 157:112902. doi: 10.1016/j.indcrop.2020.112902
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schollenberger, M., Muller, H. M., Rufle, M., Suchy, S., Planck, S., and Drochner, W. (2005). Survey of Fusarium toxins of plant origin marketed. Int. J. Food Microbiol. 97, 317–326. doi: 10.1016/j.ijfoodmicro.2004.05.001
Shen, X., Guo, X., Guo, X., Zhao, D., Zhao, W., Chen, J., et al. (2017). PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 112, 302–311. doi: 10.1016/j.plaphy.2017.01.015
Wang, L., Liu, F., Zhang, X., Wang, W., Sun, T., Chen, Y., et al. (2018). Expression characteristics and functional analysis of the ScWRKY3 gene from sugarcane. Int. J. Mol. Sci. 19:4059. doi: 10.3390/ijms19124059
Wang, Q., Qiu, B. L., Li, S., Zhang, Y. P., Cui, X. M., Ge, F., et al. (2019). A methyl jasmonate induced defensin like protein from Panax notoginseng confers resistance against Fusarium solani in transgenic tobacco. Biol. Plant. 63, 797–807. doi: 10.32615/bp.2019.123
Wang, W., Li, T., Chen, Q., Deng, B., Deng, L., and Zeng, K. (2021). Transcription factor CsWRKY65 participates in the establishment of disease resistance of citrus fruits to Penicillium digitatum. J. Agric. Food Chem. 69, 5671–5682. doi: 10.1021/acs.jafc.1c01411
Wang, X., Li, J., Guo, X., Ma, Y., Qiao, Q., and Guo, J. (2019). PlWRKY13: A transcription factor involved in abiotic and biotic stress responses in Paeonia lactiflora. Int. J. Mol. Sci. 20:5953. doi: 10.3390/ijms20235953
Xie, W., Meng, X., Zhai, Y., Zhou, P., Ye, T., Wang, Z., et al. (2018). Panax Notoginseng saponins: a review of its mechanisms of antidepressant or anxiolytic effects and network analysis on phytochemistry and pharmacology. Molecules 23:940. doi: 10.3390/molecules23040940
Xiong, X. P., Sun, S. C., Zhang, X. Y., Li, Y. J., Liu, F., Zhu, Q. H., et al. (2020). GhWRKY70D13 regulates resistance to Verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways. Front. Plant Sci. 11:69. doi: 10.3389/fpls.2020.00069
Xu, H., Watanabe, K. A., Zhang, L., and Shen, Q. J. (2016). WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. 23, 311–323. doi: 10.1093/dnares/dsw025
Xu, Y. P., Xu, H., Wang, B., and Su, X. D. (2020). Crystal structures of N-terminal WRKY transcription factors and DNA complexes. Protein Cell. 11, 208–213. doi: 10.1007/s13238-019-00670-0
Yang, Y., Liu, J., Zhou, X., Liu, S., and Zhuang, Y. (2020). Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ. 8:e8777. doi: 10.7717/peerj.8777
Yousefi, R., Jevdokimenko, K., Kluever, V., Pacheu-Grau, D., and Fornasiero, E. F. (2021). Influence of subcellular localization and functional state on protein turnover. Cells 10:1747. doi: 10.3390/cells10071747
Zhang, X., Liu, J., Wu, L., Wang, Z., and Zhang, S. (2019). GbWRKY1, a member of the WRKY transcription factor family identified from Gossypium barbadense, is involved in resistance to Verticillium wilt. Biotechnol. Biotechnol. Equip. 33, 1354–1364. doi: 10.1080/13102818.2019.1667873
Zhao, L., Zhang, W., Song, Q., Xuan, Y., Li, K., Cheng, L., et al. (2020). A WRKY transcription factor, TaWRKY40-D, promotes leaf senescence associated with jasmonic acid and abscisic acid pathways in wheat. Plant Biol. 22, 1072–1085. doi: 10.1111/plb.13155
Zhao, X. Y., Qi, C. H., Jiang, H., Zhong, M. S., You, C. X., Li, Y. Y., et al. (2019a). MdHIR4 transcription and translation levels associated with disease in apple are regulated by MdWRKY31. Plant Mol. Biol. 101, 149–162. doi: 10.1007/s11103-019-00898-8
Zhao, X. Y., Qi, C. H., Jiang, H., Zhong, M. S., Zhao, Q., You, C. X., et al. (2019b). MdWRKY46-enhanced apple resistance to Botryosphaeria dothidea by activating the expression of MdPBS3.1 in the salicylic acid signaling pathway. Mol. Plant Microbe Interact. 32, 1391–1401. doi: 10.1094/MPMI-03-19-0089-R
Zhao, Y., Yang, Z., Ding, Y., Liu, L., Han, X., Zhan, J., et al. (2019). Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 286, 28–36. doi: 10.1016/j.plantsci.2019.05.021
Zheng, J., Liu, F., Zhu, C., Li, X., Dai, X., Yang, B., et al. (2019). Identification, expression, alternative splicing and functional analysis of pepper WRKY gene family in response to biotic and abiotic stresses. PLoS ONE. 14:e0219775. doi: 10.1371/journal.pone.0219775
Zhou, M. L., and Memelink, J. (2016). Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 34, 441–449. doi: 10.1016/j.biotechadv.2016.02.004
Zhou, T., Yang, X., Wang, G., and Cao, F. (2021). Molecular cloning and expression analysis of a WRKY transcription factor gene, GbWRKY20, from Ginkgo biloba. Plant Signal. Behav. 16:1930442. doi: 10.1080/15592324.2021.1930442
Keywords: jasmonic acid, Fusarium solani, WRKY transcription factors, transgenic tobacco, transcriptional activation
Citation: Zheng L, Qiu B, Su L, Wang H, Cui X, Ge F and Liu D (2022) Panax notoginseng WRKY Transcription Factor 9 Is a Positive Regulator in Responding to Root Rot Pathogen Fusarium solani. Front. Plant Sci. 13:930644. doi: 10.3389/fpls.2022.930644
Received: 28 April 2022; Accepted: 23 June 2022;
Published: 14 July 2022.
Edited by:
Laurence Veronique Bindschedler, University of London, United KingdomReviewed by:
Huamin Chen, Institute of Plant Protection (CAAS), ChinaCopyright © 2022 Zheng, Qiu, Su, Wang, Cui, Ge and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diqiu Liu, RGlxaXVsaXVAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.