- 1MARA Key Laboratory of Soybean Biology (Beijing), Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Life Science and Technology, Harbin Normal University, Harbin, China
Onset of flowering of plants is precisely controlled by extensive environmental factors and internal molecular networks, in which FLOWERING LOCUS T (FT) is a key flowering integrator. In soybean, a typical short-day plant, 11 FT homologues are found in its genome, of which several homologues are functionally diversified in flowering pathways and the others including GmFT3a are yet unknown. In the current study, we characterized GmFT3a, which is located on the same chromosome as the flowering promoters GmFT2a and GmFT5a. Overexpression of GmFT3a significantly promoted flowering of Arabidopsis under the inductive long-day (LD) photoperiod. GmFT3a over-expressed soybean also flowered earlier than the control under LD, but they were not significantly different under inductive short-day (SD) conditions, indicating that GmFT3a acts as a flowering promoter in the non-inductive photoperiod in soybean. Compared with other GmFT homologues, GmFT3a exhibited a slighter effect in flowering promotion than GmFT2a, GmFT5a and GmFT2b under LD conditions. GmFT3a promoted flowering by regulating the expression of downstream flowering-related genes and also affected the expression of other GmFTs. According to the re-sequencing data, the regional distributions of two major haplotypes in 176 soybean varieties were analyzed. The varieties with GmFT3a-Hap2 haplotype matured relatively early, and relative higher expression of GmFT3a was detected in early maturing varieties, implying that Hap2 variation may contribute to the adaptation of soybean to higher latitude regions by increasing expression level of genes in metabolism and signaling pathways. The early flowering germplasm generated by overexpression of GmFT3a has potential to be planted at higher latitudes where non-inductive long day is dominant in the growing season, and GmFT3a can be used to fine-tune soybean flowering and maturity time and improve the geographical adaptation.
Introduction
Precise timing of flowering is critical to the environmental adaptation and productivity of crops (Cockram et al., 2007; Nakamichi, 2015). Understanding the molecular mechanisms underlying reproductive transition is a prerequisite for improving regional adaptability in crop breeding (Blümel et al., 2015; Lin et al., 2021a). In Arabidopsis, photoperiod, vernalization, gibberellin (GA), age and autonomous pathways were confirmed to be involved in the modulation of flowering (Zeevaart, 2008; Blümel et al., 2015). Among the environmental factors, photoperiod acts as a major determinant signal for flowering in many plants (Thomas and Vince-Prue, 1997; He et al., 2020). In the photoperiodic pathway, FLOWERING LOCUS T (FT), a key photoperiod-regulated flowering integrator, encodes florigen which functions as a leaf-derived long-distance mobile signals and promotes floral transition (Corbesier et al., 2007; Wigge, 2011). Homologues of FT are highly conserved and promotive to flowering in diverse species (Tamaki et al., 2007; Pin et al., 2010; Park et al., 2014; Qin et al., 2017; Zhang et al., 2021).
Soybean is a typical short-day plant but is now widely grown in a wide range of latitudes from 53°N to 40°S with diverse daylength (Watanabe et al., 2012; Jia et al., 2014; Liu et al., 2022), resulting from rich varietal diversity in maturity. However, the cultivation area of a given variety is restricted to a narrow range of latitudes because of its sensitivity to photoperiods. In soybean breeding programs, the maturity or growth period which is mainly controlled by the photoperiod and temperature, is one of the most important traits for adaptation to a given environment (Hartwig, 1970; Wu et al., 2015; Song et al., 2019; Lin et al., 2021a).
Soybean maturity ecotypes were classified into a numerical and consecutive system “Maturity groups (MGs)” according to photothermal sensitivity and adaptation to specific environments (Hartwig, 1970; Jia et al., 2014; Mourtzinis and Conley, 2017; Song et al., 2019). The diversity of soybean varieties in MG and adaptation to different environments benefit from the variations and combinations of the genes in the photoperiod pathway (Jiang et al., 2014; Wang et al., 2015; Zhang et al., 2020; Liu et al., 2020a; Lin et al., 2021b). It is important to deeply understand the effect of allelic variations in maturity-related genes on the adaptation of soybean varieties to diverse geographic regions and farming systems. Soybean is a diploid species derived from an ancient tetraploid, and its genome has undergone whole-genome duplications during its long evolutionary history (Wang et al., 2015). Since then, many genes have multiple copies in the genome. For FT homologues, the integrators in the flowering pathway, there are at least 11 homologues in soybean (Kong et al., 2010; Liu et al., 2020b; Lee et al., 2021; Zhang et al., 2021). Among these genes, GmFT2a (Glyma.16 g150700) and GmFT5a (Glyma.16 g044100) play redundant but coordinated roles in flowering promotion by up-regulating the expression of floral determination genes (Kong et al., 2010; Sun et al., 2011; Na et al., 2013; Nan et al., 2014; Takeshima et al., 2019; Lin et al., 2021b). Moreover, GmFT2b, promotes flowering under long-days (LDs; Chen et al., 2020). GmFT2b overexpression plants exhibited early flowering under non-inductive LD conditions, while ft2b mutants flowered later than wildtype (WT) under LD. However, GmFT1a and GmFT4 were identified as inhibiting genes for flowering. Overexpression of GmFT1a in soybean delayed flowering, confirming that it was a flowering repressor in soybean (Liu et al., 2018), and GmFT4 was proven to be a repressor in Arabidopsis (Zhai et al., 2014). GmFT3b acts redundantly in flowering time regulation and may be compensated by other FT homologs in soybean (Su et al., 2022). The existence of multiple soybean GmFT homologues might enhance the adjustability of photoperiodic regulation of flowering (Jiang et al., 2019; Lin et al., 2021b). For instance, the expression of FT-like proteins is not restricted to short-day (SD) conditions (Kobayashi and Weigel, 2007), highlighting the coincident expression of FT-like genes encoding both floral activators and floral inhibitors in the day-neutral species of Solanum lycopersicum (Cao et al., 2016a) and Nicotiana tabacum (Beinecke et al., 2018). However, some important GmFT homologues have yet to be elucidated, which hinders a better understanding of the mechanism of functional allocations of the GmFT family in the photoperiodic flowering pathway. In this study, we focused on the GmFT3a (Glyma.16 g044200) which is highly homologous to GmFT3b, and investigated its function in the flowering control of soybean. These results will deepen the understanding of the functional diversification of GmFT members and provide a new target gene for improving the adaptation of soybean to diverse regions.
Materials and methods
Plant materials and growth conditions
For gene cloning of GmFT3a, Zigongdongdou (ZGDD), a late-maturing (MG IX) and photoperiod-sensitive soybean [Glycine max (L.) Merr.] variety which originated from Sichuan province in the southwest China (Wu et al., 2006) was selected as the plant material. Heihe 27 (HH27), an early maturing (MG 00) and photoperiodic-insensitive and early maturing variety (MG 00) from Heilongjiang province in northeast China, was selected to compare the gene expression pattern with that of ZGDD. For genetic transformation, a medium-maturing (MG III) variety Jack was used as the receptor. Moreover, 12 MG standard soybean varieties were chosen for gene expression analysis of GmFT3a. All of soybean seeds were germinated in pots with soil and vermiculite (1:1) and the plants were grown in growth chambers with constant temperature (28°C) and controlled photoperiods, i.e., LD (16 h light/8 h dark) and SD (12 h light/12 h dark), respectively. A diverse panel of 176 soybean cultivars with consecutive MG groups covering early MG 000 to MG IX, were re-sequenced to investigate the natural variation of GmFT3a. The germplasms were collected from China and the United States (Supplementary Table S2; Zhang et al., 2019; Liu et al., 2020c).
GmFT3a gene cloning
Total RNA was extracted for gene cloning and expression analysis from the first trifoliolate leaf of ZGDD in LD (16 h light/8 h dark) using TransZol UP kit (TransGen Biotech, Beijing, China). The first strand of cDNA was obtained using the TransGene reverse transcription kit (TransGen Biotech, Beijing, China). The corresponding primer sequences are listed in Supplementary Table S1.
Gene expression analysis
ZGDD, HH27 and 12 maturity group (MG) standard varieties (Supplementary Table S2) were treated with LD and SD, respectively. After photoperiodic treatments (LD and SD) for 13 days, various tissues (shoot apex, unifoliolate, trifoliolate, stem, hypocotyl, root and flower) were taken to evaluate the expression level of GmFT3a. The real-time quantitative PCR (qRT-PCR) primers were designed by NCBI Primer Blast online with the amplification products ranging from 100 to 300 bp. Three-step PCR method was set with GmActin as the internal reference on the ABI Prism® 7900HT real-time PCR instrument, and that relative expression of target genes was calculated according to the 2-ΔΔCT algorithm (Pin and Nilsson, 2012).
Subcellular localization of GmFT3a protein
The subcellular localization of GmFT3a protein was conducted by constructing p16318-GmFT3a-GFP fusion expression vector and carrying out plasmolysis isolation, transformation and culture of Arabidopsis protoplasts. The fusion GmFT3a-p16318- GFP subcellular localization vector of the experimental group and the empty p16318 vector of the control group were prepared and transformed into Arabidopsis protoplasts, and the expression position of GFP was observed in confocal laser scanning microscope (Olympus FV31-SD, Olympus Corporation, Japan).
Expression vector construction and plant transformation
A fragment with XbaI and KpnI digestion sites was ligated into pCAMBIA1300 and pTF101.1 vector to construct overexpression vectors, and the constructed vectors were transformed into Arabidopsis Columbia-0 by Agrobacterium tumefaciens mediation and inflorescence dipping method (Clough and Bent, 1998). The soybean variety Jack was used for transformation according to a previously published protocol (Chen et al., 2018). The transformed Arabidopsis plants were screened by hygromycin (Hyg), and the soybean plants were screened by applying 160 mg/l glufosinate solution on young leaves, and PCR identification.
Transcriptome sequencing and data analysis
Leaf was sampled at 13 days after emergence (DAE) under SD conditions and 30 DAE under LD conditions. Three biological replicates were analyzed. The total amount of 1.5 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, United States) following manufacturer’s manual. In order to select cDNA fragments of 150 ~ 200 bp in length preferentially, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, United States). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. The raw data have been uploaded to NCBI Sequence Read Archive with the accession number of PRJNA832118.
All the downstream analyses were based on clean data with high quality. Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences); Nt (NCBI non-redundant nucleotide sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KO (KEGG Ortholog database); GO (Gene Ontology). Differential expression analysis of two conditions/groups was performed using the DESeq R package (1.10.1; Li and Dewey, 2011). DESeq provided statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. Genes with an adjusted p value <0.05 found by DESeq were assigned as differentially expressed. Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq R packages-based Wallenius non-central hyper-geometric distribution (Young et al., 2010), which can adjust for gene length bias in DEGs. We used KOBAS (Mao et al., 2005) software to test the statistical enrichment of differential expression genes in KEGG pathways.
Phenotyping and statistical analysis
Transgenic plants were grown in culture rooms under SD and LD conditions, respectively, and the flowering time was determined as the number of days from VE (emergence) to R1 stage (beginning bloom: the first flower appears at any node in the main stem; Fehr and Caviness, 1977). The statistical analysis was carried out by Microsoft Excel, and Student’s t-test was used to assess the significance of difference between the transgenic lines.
Natural variations and haplotype identification
In order to evaluate the effects of GmFT3a natural variations on the adaptability of soybean varieties, the GmFT3a alleles of 176 soybeans germplasm were detected. The genomic data were obtained in our previous investigation (Zhang et al., 2019; Liu et al., 2020c) and the MG classification were carried out according to the standard methodology described by Song et al. (2019).
Results
Bioinformatic characteristics and subcellular localization of GmFT3a
The length of the GmFT3a CDS region is 528 bp in ZGDD, encoding 175 amino acids, which is a hydrophilic non-secretory protein. The region ranging from amino acid 27 to 164 is the PEBP domain (Supplementary Figures S1, S2). The phylogenetic tree divided these proteins into three clear groups based on their protein sequence homology, including (1) FT, (2) MFT (MOTHER of FT and TFL 1) and (3) BFT (BROTHER of FT and TFL 1) and TFL 1 (TERMINAL FLOWER 1) clades. GmFT3a was closely clustered with AtFT (AT1G65480.1; Figure 1A). The tertiary structure of its protein is similar to that of the AtFT protein and GmFT2a protein (Supplementary Figure S3). p16318-GFP in the control group did not show subcellular expression specificity, while fluorescence signals of GmFT3a-GFP were detected in cytoplasm and nucleus (Figure 1B). This result is similar to the subcellular localization of other functional phosphatidylethanolamine-binding proteins in soybean (Wang et al., 2015).
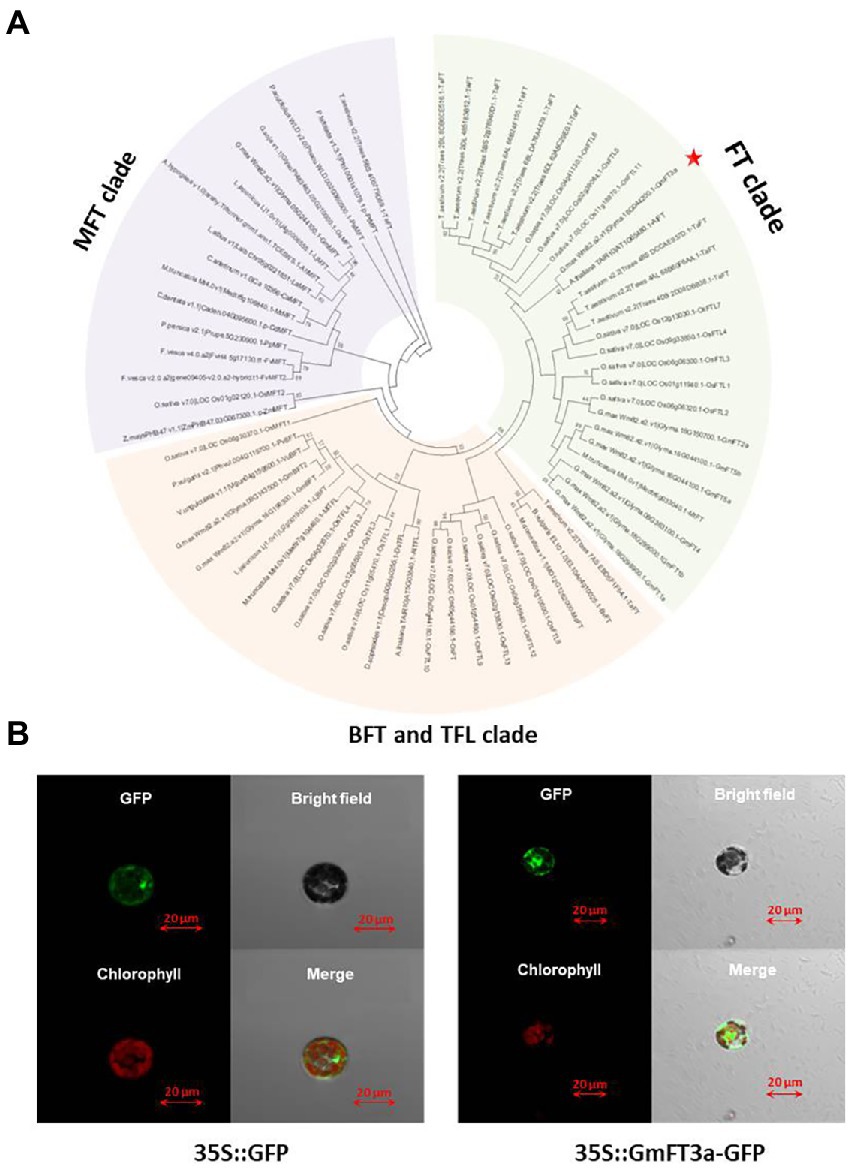
Figure 1. Phylogenetic tree based on protein sequences between GmFT3a and other FT/TFL1 family members from flowering plants (A) and subcellular localization of GmFT3a-GFP fusion protein in Arabidopsis protoplasts. The left four panels are 35S::GmFT3a-GFP constructs, and the right four panels are controls (35S::GFP) (B).
GmFT3a expression was regulated by photoperiods and correlated with maturity groups of varieties
GmFT3a expression was evaluated in different tissues of the photoperiod-sensitive variety ZGDD. Under both SD and LD conditions, relatively higher expression was detected in trifoliolate, followed by the shoot apex and unifoliolate, and extremely lower expression was detected in stem, hypocotyl, root and flower tissues in ZGDD (Supplementary Figure S4a). The expression of GmFT3a was not significantly different among trifoliolate, shoot apex, and unifoliolate under SD, while the expression of trifoliolate was significantly higher than that of shoot apex and unifoliolate under LD and exceeded the average expression under SD (Supplementary Figure S4a).
We also analyzed the diurnal expression patterns of GmFT3a in trifoliolates of both HH27 and ZGDD since 13 DAE. In early variety HH27, and the consecutive 48 h sampling by every 4 h showed two major expression peaks at 4 h and 32 h in SD condition, but relative lower expression under LD, and only one peak on 0/48 h (Supplementary Figure S4b). In late variety ZGDD, the consecutive 48 h sampling every 4 h showed two peaks at 40 h and 4 h under LD, and no significant diurnal rhythm was displayed under SD (Supplementary Figure S4b). Under LD condition, the GmFT3a expression pattern of ZGDD was similar as under SD, but the highest expression was detected in the unifoliolate value in HH27, then followed by the trifoliolate and shoot apex (Supplementary Figure S4a).
Among the varieties belonging to different maturity groups, it was found that the expression of GmFT3a in the trifoliolates of early maturing groups of MG 000, MG 0 and MG VII was higher than that of the late-maturity groups under LD, while MG 000 and MG 00 maintained higher GmFT3a expression than the others under SD conditions (Supplementary Figure S4c).
Overexpression of GmFT3a promoted flowering and regulated down-stream genes in both Arabidopsis and soybean
A total of four independent transgenic lines overexpressing GmFT3a were obtained in Arabidopsis, and an the obvious early flowering phenotype was observed in the T5 homozygous generation under LD treatment (Figure 2A). The average flowering time of the four lines was 3.9 d earlier than that of WT Arabidopsis (p < 0.01, Figure 2B). The average total (rosette and cauline) leaves of WT Arabidopsis were 16.7 ± 0.6, and those of the GmFT3a Arabidopsis overexpressing lines were 11.2 ± 1.0, 12.6 ± 1.3, 10.7 ± 1.4 and 12.5 ± 0.7, respectively. The average total leaf number of four lines was reduced by 29.6% compared with that of the WT (p < 0.01). Overexpression of GmFT3a resulted in a very significant early flowering phenotype (p < 0.01;s Figure 2C) in Arabidopsis. The qRT-PCR results showed that the ectopic GmFT3a caused significantly high expression of AtFT, AtSOC1 and AtCO (p < 0.01, Figure 2D), and the up-regulation of the AtFT and AtCO in the photoperiod pathway might be the underlying reason for early flowering of transgenic GmFT3a Arabidopsis.
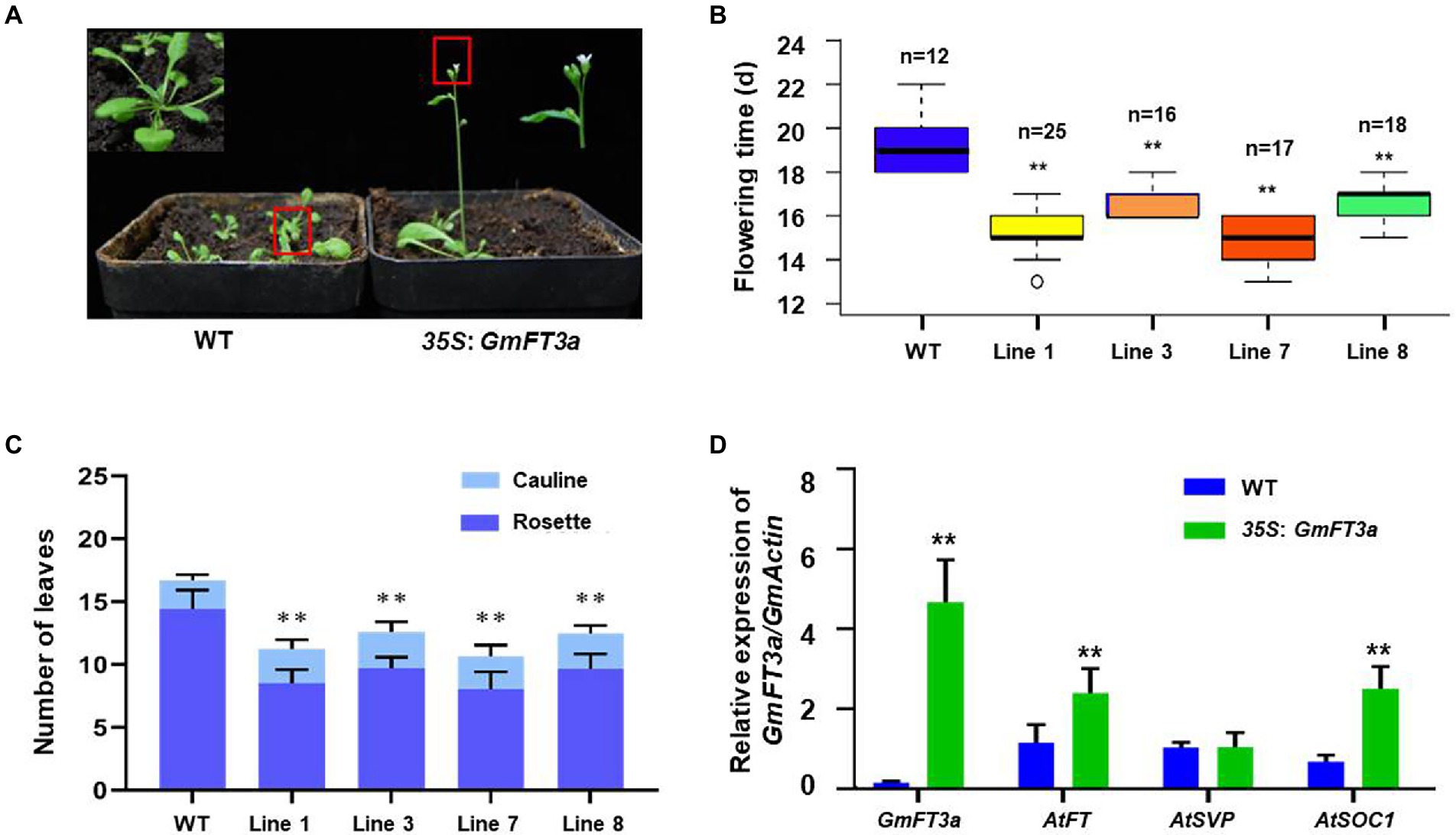
Figure 2. The phenotype (A), flowering time (B), the number of rosette and cauline rosette leaves at flowering (C) and the expression of flowering regulatory genes (D) of the transgenic GmFT3a lines under LD condition in Arabidopsis.
The fusion expression vector of GmFT3a was constructed and the variety Jack was used for Agrobacterium-mediated transformation. According to the flowering time of T1 generation and the number of seeds harvested, we selected three (Line 7, Line 9 and Line 14) from the GmFT3a overexpression lines (Figure 3A). In the T2 generation, the average flowering time of the Jack variety was 23.7 ± 1.5 DAE under SD, and that of the three transgenic GmFT3a lines were 21.7 ± 3.3, 21.5 ± 1.0 and 22.6 ± 3.7 DAE, respectively (Figure 3B). Under LD, the average flowering time of WT was 51.7 ± 4.1 DAE, while the three transgenic GmFT3a lines were 8.9 d, 4.0 d and 5.6 d earlier than that the WT, respectively (Figures 3B,C).
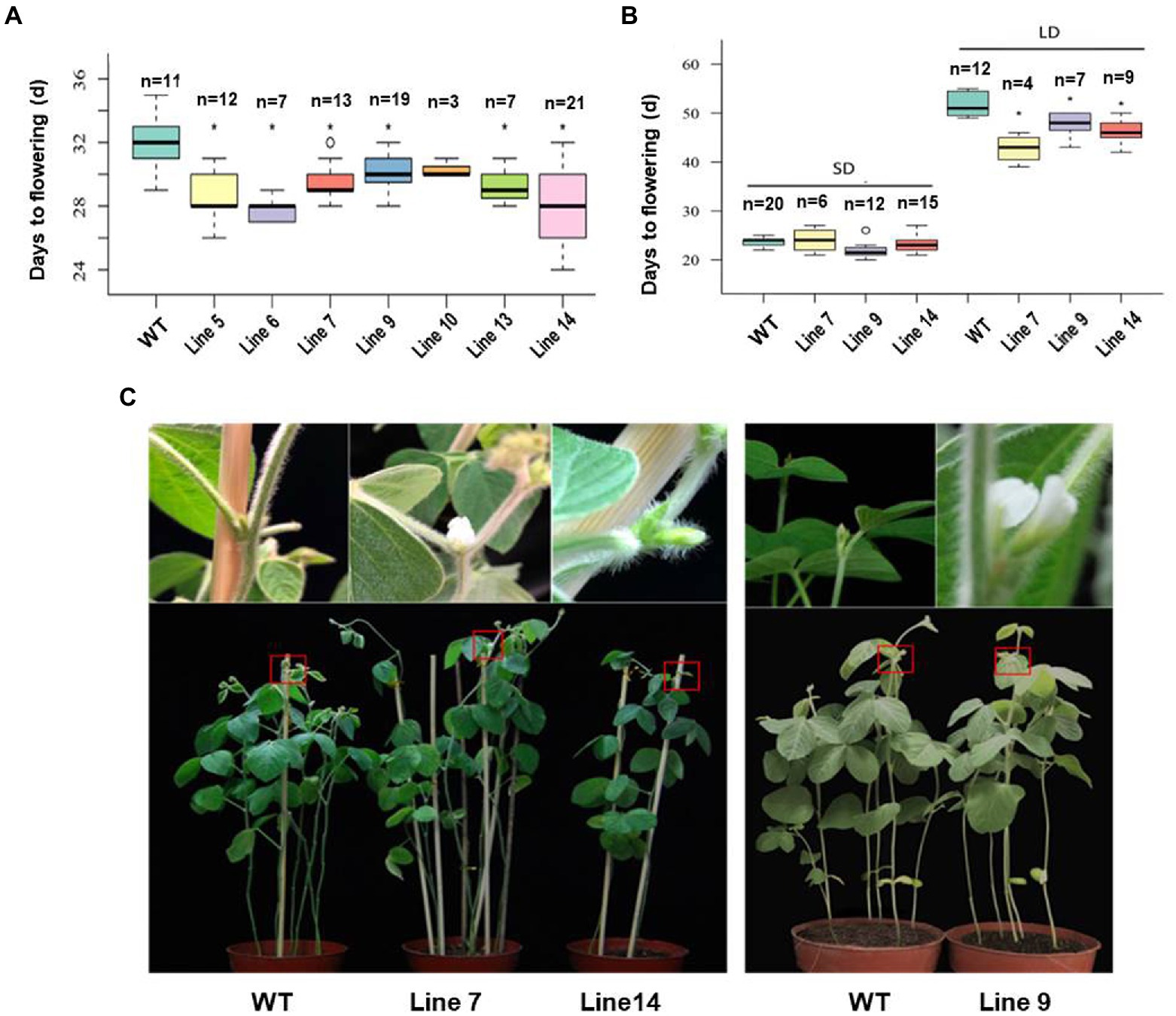
Figure 3. The flowering time of the transgenic GmFT3a soybean in natural (A) and long-day (LD) and short-day (SD) (B) conditions and the flowering phenotypes of transgenic GmFT3a T2 generation in LD condition (C). The seeds sown in Beijing, China (N39°58′, E116°20′) on June 20, 2018. The data represent the mean ± standard deviation, and statistical significance was determined using Student’s t-tests (*p < 0.05, **p < 0.01). DAE: days after emergence.
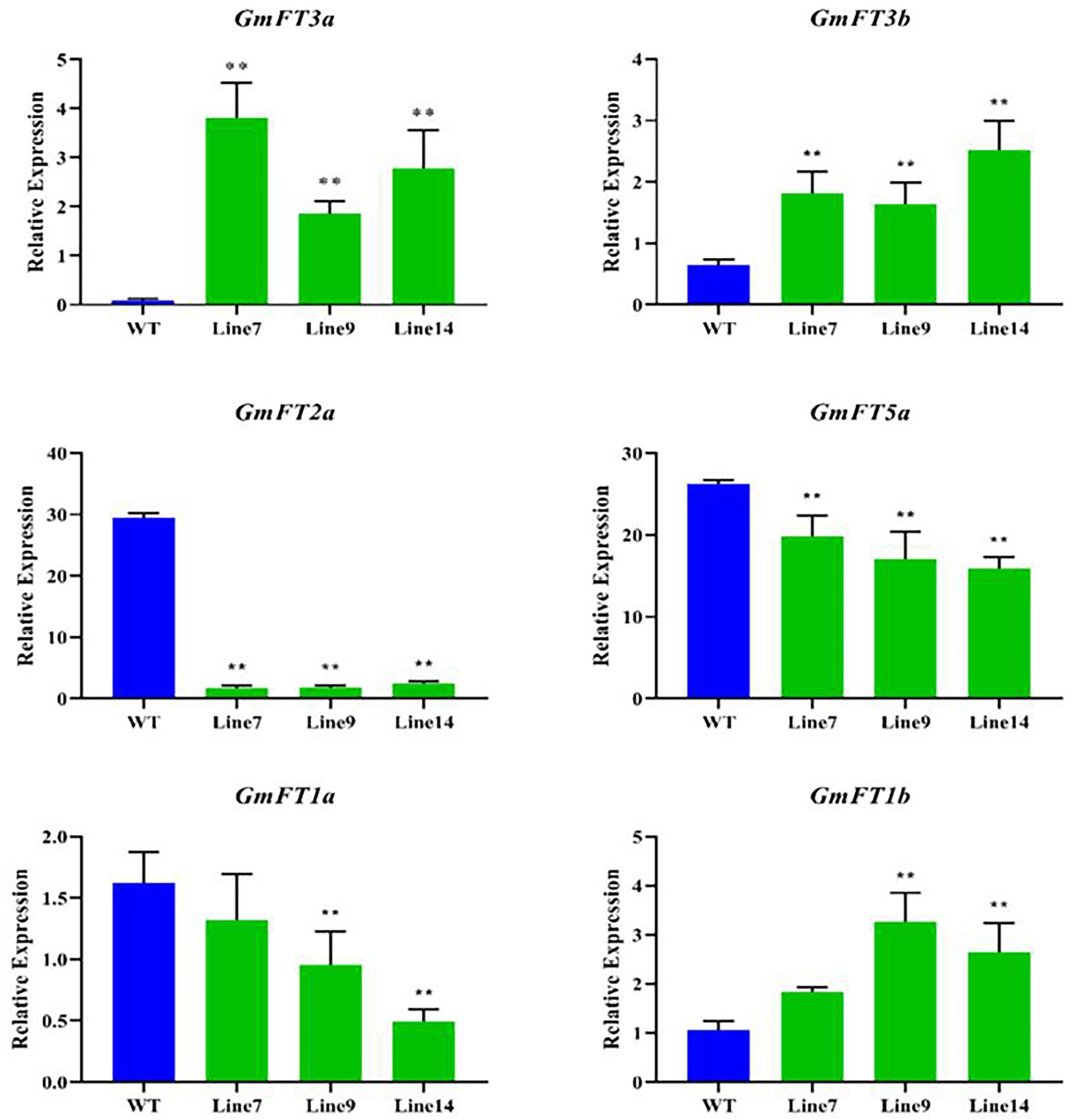
The expression of GmFT homologues was measured in the transgenic GmFT3a soybean under LD condition, and their influence on other GmFT genes was analyzed. The results showed that the expression level of transgenic GmFT3a plants was significantly higher than that of WT (p < 0.01), and the expression level of GmFT3b, which has the highest homology with GmFT3a, was also significantly higher (p < 0.01), while the expression of GmFT2a and GmFT5a decreased significantly in the transgenic GmFT3a soybean (p < 0.01). The declining degree of GmFT2a expression was higher than that of GmFT5a. The expression of GmFT1a significantly decreased, while GmFT1b increased significantly in transgenic Line 9 and Line 14 (p < 0.01; Figure 4).
More genes in metabolism and signaling pathways were enriched under LD than under SD among DEGs in transgenic GmFT3a soybean
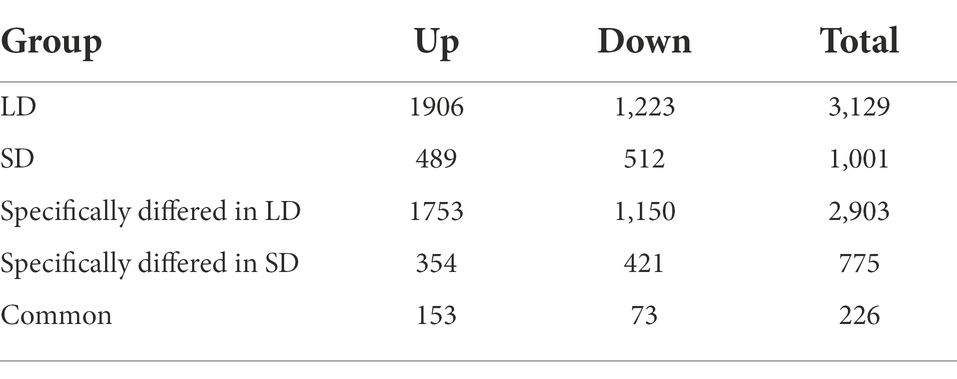
According to the transcriptomic profiles, a total of 3,129 DEGs with 1906 up-regulated and 1,223 down-regulated unigenes were observed under LD, and 1,001 DEGs including 489 up- and 512 down-regulated unigenes were expressed under SD (Table 1; Figure 5). Among these DEGs, there were 226 (153 up- and 73 down-regulated) unigenes under both LD and SD conditions (Table 1; Figure 5). Among the up-regulated unigenes under LD, 558 up-regulated genes were identified in the transgenic GmFT3a soybean (p < 0.05). These genes included F-box protein (a gene for the controlled degradation of cellular protein); disease resistance protein (a resistance protein guard the plant against pathogens); abscisic stress-ripening protein 3 (an abscisic acid-, stress-, and ripening-induced protein); heat shock protein 83 (a gene for promoting maturation), structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction.
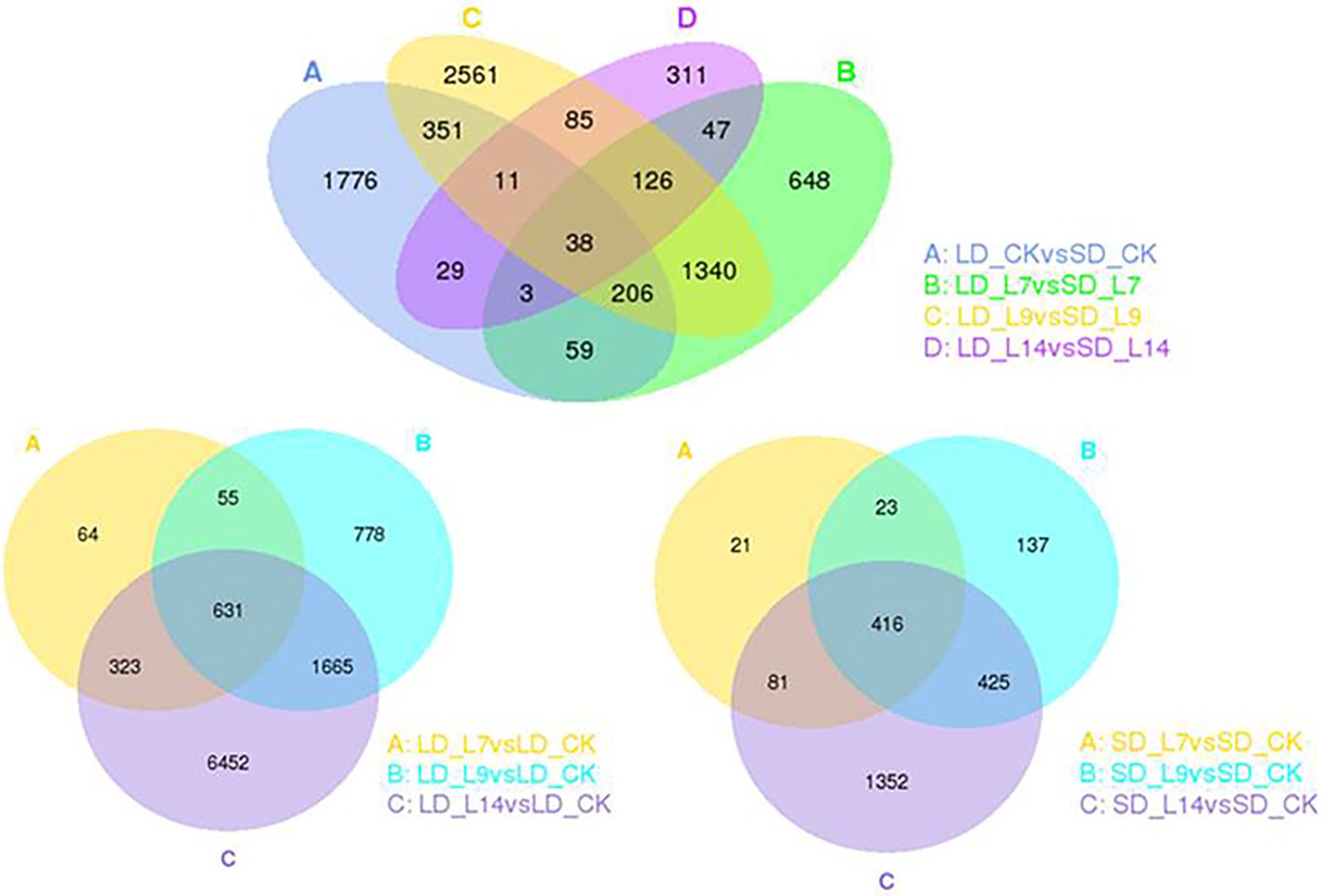
Figure 5. The Venn diagram of DEGs between transgenic GmFT3a soybean and control in LD and SD, respectively.
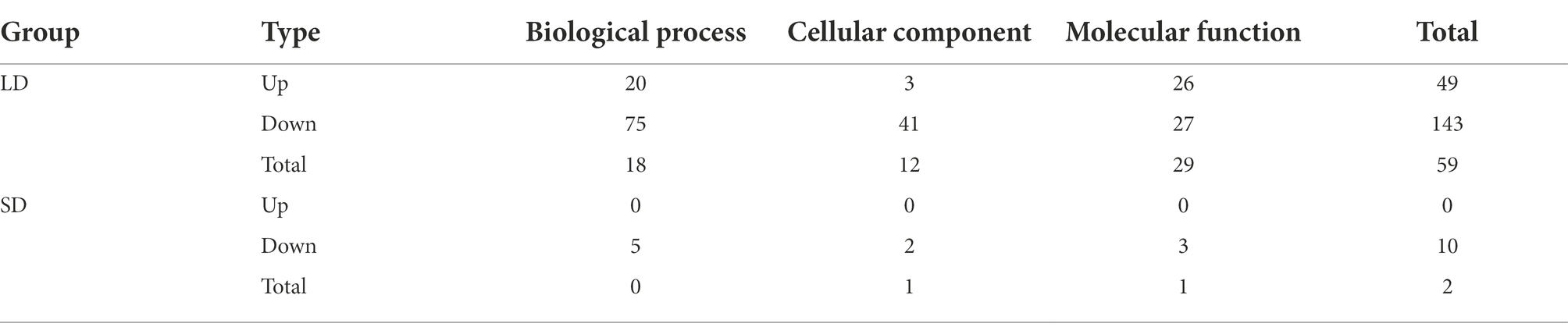
For further identifying the gene function, 49 and 143 GO terms were annotated in the up- and down-regulated DEGs, respectively under LD (Table 2). Under SD condition, only 10 GO terms significantly enriched in the down-regulated DEGs (corrected p < 0.05). Among the 49 up-regulated GO items, the “glucosamine-containing compound metabolic process,” “fatty acid biosynthetic process,” “signal transduction,” and “cellular protein modification process” items were significantly overrepresented in biological process; Other terms, such as “proteinaceous extracellular matrix,” “extracellular matrix,” “extracellular region part” were in cellular component, and “oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor,” “kinase activity” and “ATP binding” were in molecular function. While under SD condition, no GO items were significantly enriched in the up-regulated DEGs and only 10 GO items were enriched in the down-regulated DEGs. The top 20 obviously enriched KEGG pathways are shown in Figure 6. By comparing transgenic GmFT3a group with control, “Plant hormone signal transduction” and “Amino sugar and nucleotide sugar metabolism” pathway enriched the most in the DEGs under LD condition; while only the “Biosynthesis of secondary metabolites” and “Sesquiterpenoid and triterpenoid biosynthesis” pathway were enriched under the SD condition (Figure 6).
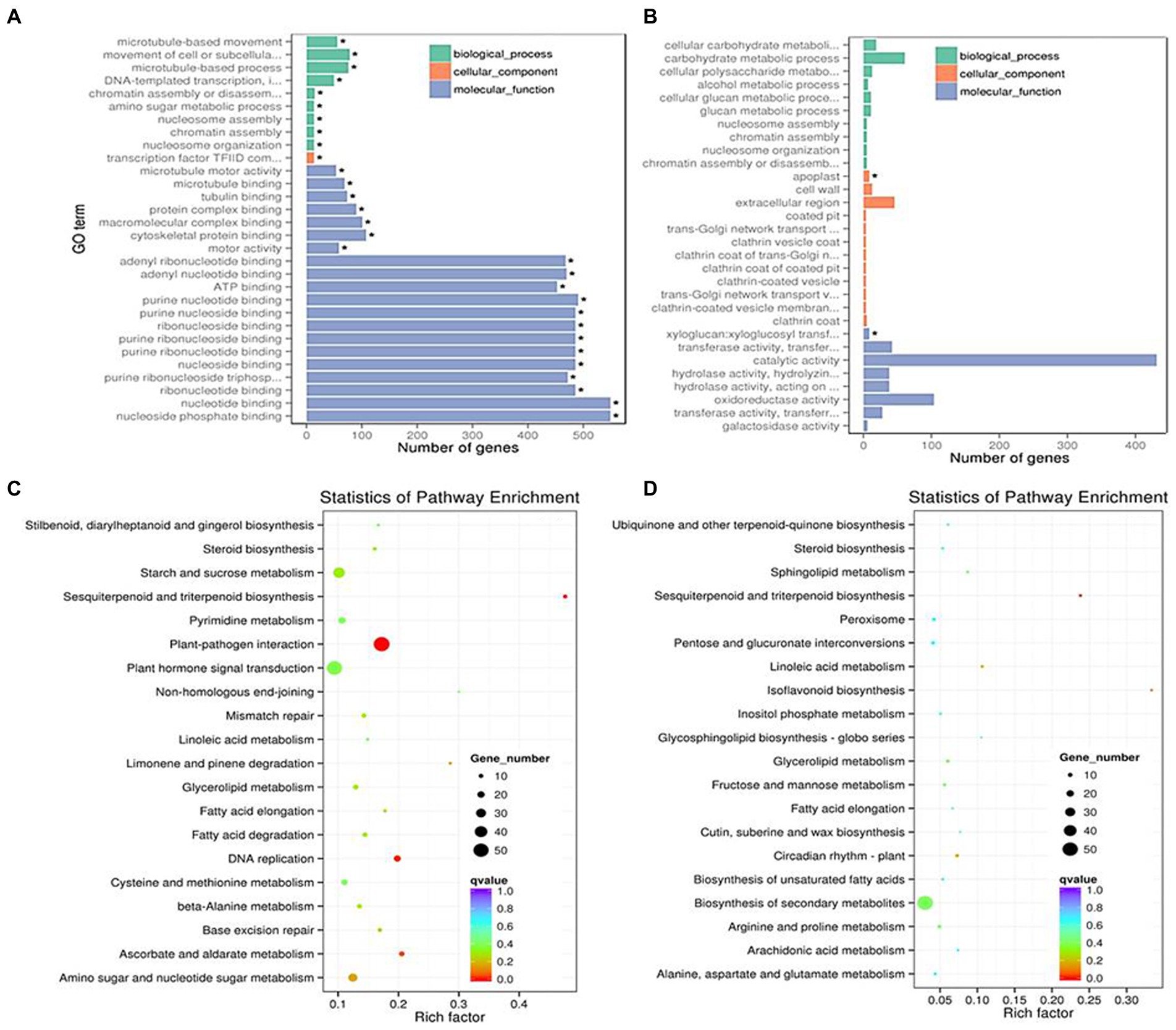
Figure 6. The most enriched GO terms in LD (A) and SD (B) and KEGG pathway terms in LD (C) and SD (D) conditions.
Natural variations of GmFT3a existed in soybean varieties with diverse maturity groups and origins
Based on the re-sequencing data, the natural variations of GmFT3a in 176 soybean varieties with diverse maturity groups and origins were analyzed and a total of 8 SNPs including 3 alleles in the promoter region (Gm16:4160524.0.4162523) and 5 alleles in genomic region (Gm16:4162524.0.4164824) were identified (Figure 7). Among the two major haplotypes, Hap1 was detected in 169 varieties ranging from MG 000 to MG IX, and Hap2 was detected in 7 varieties with MG 00 to MG II (Figure 7; Supplementary Table S2), indicating that Hap2 would be related to early maturity. Almost all Hap2 varieties collected from the high-latitude regions in northeast China, e.g., Heihe 9, Heihe 18, Heihe 38 and Huajiang 4 were bred in Heihe city (approximately 50°N), Heilongjiang Province, and the Tiefeng 18 and Tiefeng 20 varieties were bred in Tieling (approximately 42°N), Liaoning Province in northeast China.
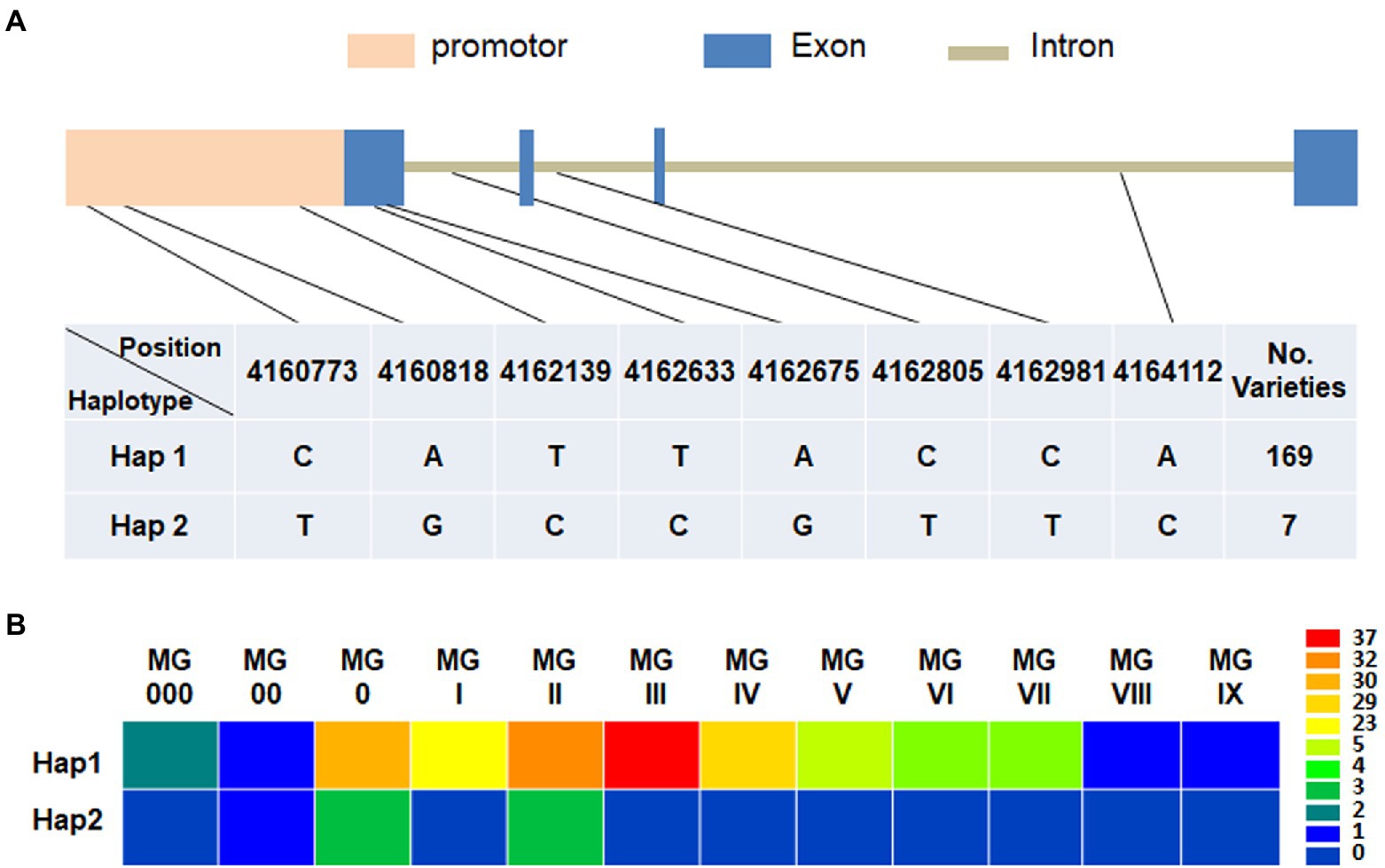
Figure 7. Haplotype identification (A) and corresponding maturity groups (B) of GmFT3a in 176 representative soybean varieties.
Discussion
GmFT3a fine-tunes flowering time of soybean under LD condition
FT belongs to a phosphatidylethanolamine-binding protein family composed of six members in Arabidopsis (Kardailsky et al., 1999; Jin et al., 2021). FT is considered to be a major component of florigen and plays a critical role in the photoperiodic flowering pathway (Corbesier et al., 2007; Wigge, 2011). To date, the photoperiod-dependent flowering pathway was intensively studied and a total of 11 florigen (FT) homologues have been characterized in soybean (Kong et al., 2010; Sun et al., 2011; Liu et al., 2018, 2020b; Zhang et al., 2021; Su et al., 2022). GmFT3a, which is located on the same chromosome as the key flowering promoters GmFT2a and GmFT5a, has yet to be studied for functional identification. In this study, GmFT3a promoted flowering by 3.9 d in Arabidopsis under an inductive LD photoperiod, and the overexpression of GmFT3a in soybean resulted in earlier flowering significantly by 4.0–8.9 d than that of the control under non-inductive LD conditions (Figures 2A, 3C). Compared with other GmFT homologues, GmFT2a exhibited a stronger effect in promoting flowering than GmFT3a. Overexpression of GmFT2a resulted in earlier flowering by approximately 9 d under SD and extremely early flowering phenotypes by approximately 32 d under LD in Jack background (Cai et al., 2020); and under LD, it even drove flowering of the extreme late-maturing variety ZGDD which would have retained its vegetative growth under non-inductive LD conditions (Li et al., 2005; Sun et al., 2011). In addition, GmFT3a seems to have a partially similar but weaker function to GmFT5a and GmFT2b, which held dominant functions under LD condition. The overexpression of GmFT5a and GmFT2b caused earlier flowering by approximately 16 d and 7 d, respectively under LD (Chen et al., 2018; Cai et al., 2020). Besides, GmFT3b, which has the highest homology with GmFT3a, was functionally redundant in regulation of flowering time (Jiang et al., 2019; Su et al., 2022). Collectively, the supporting role of GmFT3a fine-tuned soybean flowering under the non-inductive LD condition.
GmFT3a promoted flowering by regulating the downstream flowering-related genes
The transition from vegetative growth to reproductive growth is due to the balance of flowering activators and flowering inhibitors according to previous models for photoperiodic flowering in soybean (Liu et al., 2018; Chen et al., 2020). The overexpression of GmFT3a unexpectedly down-regulated that of GmFT2a and GmFT5a under LD (Figure 4), which was inconsistent with the up-regulation of GmFT2a and GmFT5a by overexpression of GmFT2b (Chen et al., 2020). Under LD conditions, higher expression levels of flowering activators are required to overcome the enhanced the effects of flowering inhibitors. This implied that the function of GmFT3a might differ from that of GmFT2b in flowering regulation, and the lowered expression of GmFT2a and GmFT5a is probably responsible for the minor/weak effect on promoting the flowering of GmFT3a. GmFT3b was up-regulated under the overexpression of GmFT3a (Figure 4), and a previous study showed that GmFT3a and GmFT3b were both significantly up-regulated in the ft2a and ft5a single soybean mutant, respectively (Li et al., 2021), suggesting that this pair of homologues might share a similar trend under the lack of major promoting effector GmFT2a and GmFT5a. However, there was no significant change in the expression of GmFT3a displayed in both overexpression of GmFT3b and the ft3b mutant (Su et al., 2022). It could be inferred that GmFT3a probably functioned upstream on the GmFT3b, and the hypothesis still needs more molecular evidence and flowering phenotype of the GmFT3a mutant. In addition, higher expression of GmFT3a was detected in the relatively early maturing varieties (MG 000 and MG 00) under SD, suggesting that GmFT3a might correlate with early flowering and function as an effective promoter of flowering in non-inductive photoperiodic conditions (Jia et al., 2014; Cao et al., 2016b; Liu et al., 2020b).
In Arabidopsis, the expression of the AtFT, AtSOC1 and AtCO genes was significantly increased in the overexpression of GmFT3a (p < 0.05, Figure 2D). A similar pattern was detected in the overexpression of GmFT2a Arabidopsis, which directed the up-regulation of AtFT and AtSOC1 (Sun et al., 2011). The increase in AtSOC1 in the age pathway was potentially involved in flowering signal integration (Srikanth and Schmid, 2011; Lv et al., 2021). The RNA transcripts of floral identity genes, including GmAP1, GmSOC1, GmLFY and GmFDL19, were increased in GmFT2a- or GmFT5a-overexpressing soybean lines compared with the wild type (Nan et al., 2014; Takeshima et al., 2019).
Among the transcriptomic DEGs, the large number under LD conditions was more than 3-fold compared to SD conditions (Table 1; Figure 5). The functional patterns of GmFT3a were divided into two main aspects: the vegetative growth accelerator and signal transduction pathway. GmFT3a probably achieved its promotional roles through the enhancement of genes involved in cell division, photosynthesis, carbohydrate metabolism, fatty acid biosynthesis, and ascorbate metabolism. Moreover, the genes encoding signaling regulation and transcription factors, such as ethylene-responsive transcription factor AP2-EREBP, auxin response factor ARF, zinc finger protein COL16, MADS-box family, and GRF4 (GROWTH-REGULATING FACTOR 4), were involved in plant growth and metabolisms (Supplementary Table S3). These differentially expressed transcription factors were significantly correlated with plant signaling and the regulation of flowering development (Ratcliffe and Riechmann, 2002; Matias-Hernandez et al., 2016; Xu et al., 2021a). Furthermore, the KEGG pathway maps provided abundant information on the exploration of signaling and metabolic pathways involved in GmFT3a. Many differentially expressed unigenes were enriched in signaling (Plant hormone signal transduction pathway) under LD conditions and metabolic processes (Biosynthesis of secondary metabolites) under SD conditions, respectively. Transcriptome analysis revealed GmFT3a might promote flowering by regulating reproductive signals and growth regulators. However, the functional complementarity and interactions among multiple FT homologues remain to be further verified (Lee et al., 2021). Actually, the interaction and allocation mechanism among the GmFT family genes will be further certificated confirmed by evaluation of the phenotypes of the Gmft3a mutant and/or the double mutant of different combinations of Gmft members in future studies.
The natural variations of GmFT3a are related to the high-latitude adaptability of soybean
The adaptation of soybean varieties to different latitudes or daylengths was mainly attributed to the combinations and functional diversifications among the photoperiod-related genes (Liu et al., 2020b; Lin et al., 2021b). The wide distribution of soybean varieties results from rich natural variations and different combinations of genes and QTLs controlling flowering time (Watanabe et al., 2012; Liu et al., 2022). The early maturity of soybeans in Northeast China was largely attributed to local exotic migration and selection against positive alleles causing new recombination, while only few new allele emergence/mutation occurred (Fu et al., 2020). Therefore, allelic variations in flowering time-related genes could be applied as an effective means for breeding (Cao et al., 2016b; Khan et al., 2022). There were natural variations within the GmFT family genes that contribute to soybean adaptation to various growing areas (Jiang et al., 2019). Some natural variations in GmFT2a caused significantly late flowering, which is suitable for soybean accessions which adapted to low-latitude areas (Li et al., 2021). In contrast, some single-nucleotide polymorphisms (SNPs) in GmFT5a result in early flowering, which was preferred in high-latitude areas (Takeshima et al., 2016). For GmFT3a, multiple SNP alleles in both promoter and genomic regions suggested that genomic variations and regulation of gene expression of GmFT3a both contributed to the flowering and maturity diversities (Figure 7). Notably, the rare Hap2 of GmFT3a showed relatively early maturity and most of these varieties were bred for high-latitude regions in China, suggesting that GmFT3a-Hap2 may have contributed to the adaptation of soybean to higher latitude areas. For soybean breeding, it is an effective way to broaden the adaptive region of soybean varieties through recombining and modifying genes in photoperiod pathway, e.g., various combinations of mutations at the E loci (E1, E2, E3 and E4) provide considerable genetic plasticity that contributes to soybean cultivation at diverse latitudes (Jiang et al., 2014; Liu et al., 2020c, 2022); J (the major gene for adaptation to short-day and high temperature conditions at low latitudes; Yue et al., 2016; Lu et al., 2017), GmPRR37 (Wang et al., 2020), the ft2aft5a double mutants showed late flowering and produced more pods and seeds under SD conditions (Cai et al., 2020). When using these genes with large impacts on flowering time and maturity, it might be difficult to maintain the original yield and quality traits of elite varieties. However, fine-tuning florigen expression is a promising strategy for high yield (Xu et al., 2021b). In this study, we found that the overexpression of GmFT3a promoted flowering by approximately 4.0–8.9 d under LD conditions, suggesting that GmFT3a provides an opportunity to lightly modify the flowering time and maturity of soybean varieties, which is helpful to retain yield and agronomic traits of an elite variety with extension of its adaptive regions (Qi et al., 2021).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA832118.
Author contributions
TH and LY conceived the study. YW and CZ performed the Arabidopsis transformation experiment. LC and WH performed the soybean transformation. JW and LZ recorded the phenotype of transgenic soybean. SS and CW provided and maintained the MG standard varieties. TW and BJ analyzed the re-sequencing data. YC and FC performed the gene expression experiment. SY, XX, and WS analyzed the transcriptomic data. SY and YW drafted the manuscripts. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from National Natural Science Foundation of China (32001566), the Central Public-interest Scientific Institution Basal Research Fund (No. Y2022GH08), and the China Agriculture Research System (CARS-04).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.929747/full#supplementary-material
References
Beinecke, F. A., Grundmann, L., Wiedmann, D. R., Schmidt, F. J., Caesar, A. S., Zimmermann, M., et al. (2018). The FT/FD-dependent initiation of flowering under long-day conditions in the day-neutral species Nicotiana tabacum originates from the facultative short-day ancestor Nicotiana tomentosiformis. Plant J. 96, 329–342. doi: 10.1111/tpj.14033
Blümel, M., Dally, N., and Jung, C. (2015). Flowering time regulation in crops-what did we learn from Arabidopsis? Curr. Opin. Plant Biol. 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Cai, Y., Wang, L., Chen, L., Wu, T., Liu, L., Sun, S., et al. (2020). Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 18, 298–309. doi: 10.1111/pbi.13199
Cao, K., Cui, L., Zhou, X., Ye, L., Zou, Z., and Deng, S. (2016a). Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front. Plant Sci. 6:1213. doi: 10.3389/fpls.2015.01213
Cao, D., Takeshima, R., Zhao, C., Liu, B., Jun, A., and Kong, F. (2016b). Molecular mechanisms of flowering under long days and stem growth habit in soybean. J. Exp. Bot. 68, erw1873–erw1884. doi: 10.1093/jxb/erw394
Chen, L., Cai, Y., Liu, X., Yao, W., Guo, C., Sun, S., et al. (2018). Improvement of soybean Agrobacterium-mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int. J. Mol. Sci. 19, 3039. doi: 10.3390/ijms19103039
Chen, L., Cai, Y., Qu, M., Wang, L., Sun, H., Jiang, B., et al. (2020). Soybean adaption to high-latitude regions is associated with natural variations of GmFT2b, an ortholog of FLOWERING LOCUS T. Plant Cell Environ. 43, 934–944. doi: 10.1111/pce.13695
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cockram, J., Jones, H., Leigh, F. J., O'Sullivan, D., Powell, W., Laurie, D. A., et al. (2007). Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot. 58, 1231–1244. doi: 10.1093/jxb/erm042
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q. Z., Searle, L., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Fehr, W. R., and Caviness, C. E. (1977). “Stages of soybean development,” in Cooperative Extension Service, Agriculture and Home Economics Experiment Station. eds. C. E. Donhowe (Ames, IA, USA: Iowa State University of Science and Technology).
Fu, M., Wang, Y., Ren, H., Du, W., Wang, D., Bao, R., et al. (2020). Genetic dynamics of earlier maturity group emergence in south-to-north extension of Northeast China soybeans. Theor. Appl. Genet. 133, 1839–1857. doi: 10.1007/s00122-020-03558-4
Hartwig, E. E. (1970). Growth and reproductive characteristics of soybeans (Glycine max (L.) Merr.) grown under short-day conditions. Trop. Sci. 12, 47–53.
He, Y., Chen, T., and Zeng, X. (2020). Genetic and epigenetic understanding of seasonal timing of flowering. Plant Commun. 1, 13. doi: 10.1016/j.xplc.2019.100008
Jia, H., Jiang, B., Wu, C., Hou, W., Sun, S., Yan, H., et al. (2014). Maturity group classification and maturity locus genotyping of early-maturing soybean varieties from high-latitude cold regions. PLoS One 9:e94139. doi: 10.1371/journal.pone.0094139
Jiang, B., Nan, H., Gao, Y., Tang, L., Yue, Y., Lu, S., et al. (2014). Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS One 9:e106042. doi: 10.1371/journal.pone.0106042
Jiang, B., Zhang, S., Song, W., Abdul, M., Khan, A., Sun, S., et al. (2019). Natural variations of FT family genes in soybean varieties covering a wide range of maturity groups. BMC Genomics 20, 230. doi: 10.1186/s12864-019-5577-5
Jin, S., Nasim, Z., Susila, H., and Ahn, J. H. (2021). Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin. Cell Dev. Biol. 109, 20–30. doi: 10.1016/j.semcdb.2020.05.007
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
Khan, M. A. A., Zhang, S. W., Emon, R. M., Chen, F., Song, W., Wu, T., et al. (2022). CONSTANS polymorphism modulates flowering time and maturity in soybean. Front. Plant Sci. 13:817544. doi: 10.3389/fpls.2022.817544
Kobayashi, Y., and Weigel, D. (2007). Move on up, it’s time for change mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21, 2371–2384. doi: 10.1101/gad.1589007
Kong, F., Liu, B., Xia, Z., Sato, S., Kim, B. M., Watanabe, S., et al. (2010). Two coordinately regulated homologues of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231. doi: 10.1104/pp.110.160796
Lee, S. H., Choi, C. W., Park, K. M., Jung, W. H., Chun, H. J., Baek, D., et al. (2021). Diversification in functions and expressions of soybean FLOWERING LOCUS T genes fine-tunes seasonal flowering. Front. Plant Sci. 12:613675. doi: 10.3389/fpls.2021.613675
Li, B., and Dewey, C. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 93–99. doi: 10.1186/1471-2105-12-323
Li, X., Fang, C., Yang, Y., Lv, T., Su, T., Chen, L., et al. (2021). Overcoming the genetic compensation response of soybean florigens to improve adaptation and yield at low latitudes. Curr. Biol. 31, 3755–3767.e4. doi: 10.1016/j.cub.2021.06.037
Li, X., Wu, C., Ma, Q., Zhang, S., Li, C., Zhang, X., et al. (2005). Morphology and anatomy of the differentiation of flower bud and the process of flowering reversion in soybean cv. Zigongdongdou. Acta Agron. Sin. 31, 1437–1442. doi: 10.3321/j.issn:0496-3490.2005.11.008
Lin, X., Fang, C., Liu, B., and Kong, F. (2021a). Natural variation and artificial selection of photoperiodic flowering genes and their applications in crop adaptation. aBIOTECH 2, 156. doi: 10.1007/s42994-021-00039-0
Lin, X., Liu, B., Weller, J. L., Abe, J., and Kong, F. (2021b). Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 63, 981–994. doi: 10.1111/jipb.13021
Liu, L., Gao, L., Zhang, L., Cai, Y., Song, W., Chen, L., et al. (2022). Co-silencing E1 and its homologs in an extremely late-maturing soybean cultivar confers super-early maturity and adaptation to high-latitude short-season regions. J. Integr. Agric. 21, 326–335. doi: 10.1016/S2095-3119(20)63391-3
Liu, X., He, J., Wang, Y., Xing, G., Li, Y., Yang, S., et al. (2020a). Geographic differentiation and phylogeographic relationships among world soybean populations. Crop J. 8, 260–272. doi: 10.1016/j.cj.2019.09.010
Liu, W., Jiang, B., Ma, L., Zhang, S., Zhai, H., Xu, X., et al. (2018). Functional diversification of FLOWERING LOCUS T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 217, 1335–1345. doi: 10.1111/nph.14884
Liu, L., Song, W., Wang, L., Sun, X., Qi, Y., Wu, T., et al. (2020c). Allele combinations of maturity genes E1-E4 affect adaptation of soybean to diverse geographic regions and farming systems in China. PLoS One 15:e0235397. doi: 10.1371/journal.pone.0235397
Liu, L., Xuan, L., Jiang, Y., and Yu, H. (2020b). Regulation by FLOWERING LOCUS T and TERMINAL FLOWER 1 in flowering time and plant architecture. Small Struct. 2:2000125. doi: 10.1002/sstr.202000125
Lu, S., Zhao, X., Hu, Y., Liu, S., Nan, H., Li, X., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. doi: 10.1038/ng.3819
Lv, X., Zeng, X., Hu, H., Chen, L., Zhang, F., Liu, R., et al. (2021). Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex. Plant Cell 33, 1182–1195. doi: 10.1093/plcell/koab016
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.2307/1592215
Matias-Hernandez, L., Aguilar-Jaramillo, A. E., Cigliano, R. A., Sanseverino, W., and Pelaz, S. (2016). Flowering and trichome development share hormonal and transcription factor regulation. J. Exp. Bot. 67, 1209–1219. doi: 10.1093/jxb/erv534
Mourtzinis, S., and Conley, S. P. (2017). Delineating soybean maturity groups across the United States. Agron. J. 109, 1397–1403. doi: 10.2134/agronj2016.10.0581
Na, X., Jian, B., Yao, W., Wu, C., Hou, W., Jiang, B., et al. (2013). Cloning and functional analysis of the flowering gene GmSOC1-like, a putative SUPPRESSOR OF OVEREXPRESSION CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in soybean. Plant Cell Rep. 32, 1219–1229. doi: 10.1007/s00299-013-1419-0
Nakamichi, N. (2015). Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol. 56, 594–604. doi: 10.1093/pcp/pcu181
Nan, H., Cao, D., Zhang, D., Li, Y., Lu, S., Tang, L., et al. (2014). GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS One 9:e97669. doi: 10.1371/journal.pone.0097669
Park, S. J., Jiang, K., Tal, L., Yichie, Y., Gar, O., Zamir, D., et al. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46, 1337–1342. doi: 10.1038/ng.3131
Pin, P. A., Benlloch, R., Bonnet, D., Wremerth-Weich, E., Kraft, T., Gielen, J., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400. doi: 10.1126/science.1197004
Pin, P. A., and Nilsson, O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x
Qi, X., Jiang, B., Wu, T., Sun, S., Wang, C., Song, W., et al. (2021). Genomic dissection of widely planted soybean cultivars leads to a new breeding strategy of crops in the post-genomic era. Crop J. 9, 1079–1087. doi: 10.1016/j.cj.2021.01.001
Qin, Z., Wu, J., Geng, S., Feng, N., Chen, F., Kong, X., et al. (2017). Regulation of FT splicing by an endogenous cue in temperate grasses. Nat. Commun. 8, 14320. doi: 10.1038/ncomms14320
Ratcliffe, O. J., and Riechmann, J. L. (2002). Arabidopsis transcription factors and the regulation of flowering time: a genomic perspective. Curr. Issues Mol. Biol. 296, 285–289. doi: 10.1126/science.296.5566.285
Song, W., Sun, S., Ibrahim, S. E., Xu, Z., Wu, H., Hu, X., et al. (2019). Standard cultivar selection and digital quantification for precise classification of maturity groups in soybean. Crop Sci. 59, 1997–2006. doi: 10.2135/cropsci2019.02.0095
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Su, Q., Chen, L., Cai, Y., Chen, Y., Yuan, S., Li, M., et al. (2022). Functional redundancy of FLOWERING LOCUS T 3b in soybean flowering time regulation. Int. J. Mol. Sci. 23, 2497. doi: 10.3390/ijms23052497
Sun, H., Jia, Z., Cao, D., Jiang, B., Wu, C., Hou, W., et al. (2011). GmFT2a, a soybean homologue of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS One 6:e29238. doi: 10.1371/journal.pone.0029238
Takeshima, R., Hayashi, T., Zhu, J., Zhao, C., Xu, M., Yamaguchi, N., et al. (2016). A soybean quantitative trait locus that promotes FLOWERING under long days is identified as FT5a, a FLOWERING LOCUS T ortholog. J. Exp. Bot. 67, 5247–5258. doi: 10.1093/jxb/erw283
Takeshima, R., Nan, H., Harigai, K., Dong, L., and Abe, J. (2019). Functional divergence between soybean FLOWERING LOCUS T orthologues, FT2a. J. Exp. Bot. 70, 3941–3953. doi: 10.1093/jxb/erz199
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Wang, L., Sun, S., Wu, T., Liu, L., Sun, X., Cai, Y., et al. (2020). Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 18, 1869–1881. doi: 10.1111/pbi.13346
Wang, Z., Zhou, Z., Liu, Y., Liu, T., Li, Q., Ji, Y., et al. (2015). Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 27, 323–336. doi: 10.1105/tpc.114.135103
Watanabe, S., Harada, K., and Abe, J. (2012). Genetic and molecular bases of photoperiod responses of flowering in soybean. Breed. Sci. 61, 531–543. doi: 10.1270/jsbbs.61.531
Wigge, P. (2011). FT, A mobile developmental signal in plants. Curr. Biol. 21, R374–R378. doi: 10.1016/j.cub.2011.03.038
Wu, T., Li, J., Wu, C., Sun, S., Mao, T., Jiang, B., et al. (2015). Analysis of the independent-and interactive-photo-thermal effects on soybean flowering. J. Integr. Agric. 14, 622–632. doi: 10.1016/S2095-3119(14)60856-X
Wu, C., Ma, Q., Yam, K. M., Cheung, M. Y., Xu, Y., Han, T., et al. (2006). In situ expression of the GmNMH7 gene is photoperiod-dependent in a unique soybean (Glycine max [L.] Merr.) flowering reversion system. Planta 223, 725–735. doi: 10.1007/s00425-005-0130-y
Xu, X., Zhang, L., Cao, X., Liu, L., Jiang, B., Zhang, C., et al. (2021a). Cotyledons facilitate the adaptation of early maturing soybean varieties to high latitude long day environments. Plant Cell Environ. 44, 2551–2564. doi: 10.1111/pce.14120
Xu, K., Zhang, X. M., Chen, H., Zhang, C., Zhu, J., Cheng, Z., et al. (2021b). Fine-tuning florigen increases field yield through improving photosynthesis in soybean. Front. Plant Sci. 12:710754. doi: 10.3389/fpls.2021.710754
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Yue, Y., Liu, N., Jiang, B., Li, M., Wang, H., Jiang, Z., et al. (2016). A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol. Plant 10, 656–658. doi: 10.1016/j.molp.2016.12.004
Zeevaart, J. A. (2008). Leaf-produced floral signals. Curr. Opin. Plant Biol. 11, 541–547. doi: 10.1016/j.pbi.2008.06.009
Zhai, H., Lü, S., Liang, S., Wu, H., Zhang, X., Liu, B., et al. (2014). GmFT4, a homologue of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS One 9:e89030. doi: 10.1371/journal.pone.0089030
Zhang, L., Liu, W., Mesfin, T., Xu, X., Qi, Y., Sapey, E., et al. (2020). Principles and practices of the photo-thermal adaptability improvement in soybean. J. Integr. Agric. 19, 295–310. doi: 10.1016/S2095-3119(19)62850-9
Zhang, S., Singh, M. B., and Bhalla, P. L. (2021). Molecular characterization of a soybean FT homologue, GMFT7. Sci. Rep. 11, 3651. doi: 10.1038/s41598-021-83305-x
Keywords: soybean, GmFT3a, flowering time, photoperiod, adaptation
Citation: Yuan S, Wang Y, Wang J, Zhang C, Zhang L, Jiang B, Wu T, Chen L, Xu X, Cai Y, Sun S, Chen F, Song W, Wu C, Hou W, Yu L and Han T (2022) GmFT3a fine-tunes flowering time and improves adaptation of soybean to higher latitudes. Front. Plant Sci. 13:929747. doi: 10.3389/fpls.2022.929747
Edited by:
Fanjiang Kong, Guangzhou University, ChinaReviewed by:
José Antonio Abelenda, National Institute of Agricultural and Food Research and Technology, SpainLin Zhao, Northeast Agricultural University, China
Copyright © 2022 Yuan, Wang, Wang, Zhang, Zhang, Jiang, Wu, Chen, Xu, Cai, Sun, Chen, Song, Wu, Hou, Yu and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianfu Han, aGFudGlhbmZ1QGNhYXMuY24=; Lijie Yu, eXVsaWppZTE5NjFAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Shan Yuan
Shan Yuan Yining Wang1,2†
Yining Wang1,2† Tingting Wu
Tingting Wu Li Chen
Li Chen Yupeng Cai
Yupeng Cai Shi Sun
Shi Sun Fulu Chen
Fulu Chen Wenwen Song
Wenwen Song Cunxiang Wu
Cunxiang Wu Wensheng Hou
Wensheng Hou Tianfu Han
Tianfu Han