
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 30 June 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.926752
This article is part of the Research Topic Somatic Embryogenesis: 60 Years of Research Applied to Plant Cloning to Unravel Plant Totipotency, Volume II View all 12 articles
Plant regeneration occurs when plants repair or replace damaged structures based on the totipotency and pluripotency of their cells. Tissue culture is one of the most widely used regenerative technologies. Recently, a series of breakthroughs were made in the study of plant regeneration. This review summarizes two regenerative pathways in tissue culture: somatic embryogenesis and de novo organogenesis. Furthermore, we review the environmental factors influencing plant regeneration from explant sources, basal culture medium, plant growth regulators, and light/dark treatment. Additionally, we analyse the molecular mechanisms underlying two pathways. This knowledge will promote an understanding of the fundamental principles of plant regeneration from precursor cells and lay a solid foundation for applying plant micropropagation and genetic modification.
An entire plant can be regenerated from an adult tissue or organ, a mass of unorganized calli, or even a single cell in a process referred to as plant regeneration. Plant regeneration refers to the physiological renewal, repair, or replacement of tissue in plants (Ikeuchi et al., 2016). The totipotency or pluripotency of plant cells underlies the ability of plants to regenerate, reflecting the high plasticity of cell fate. Totipotency refers to the ability of a cell to differentiate into a complete individual, whereas pluripotency involves the differentiation of a specific group of tissues or organs from a cell (Verdeil et al., 2007). The concept of tissue culture was proposed as early as a century ago and envisaged the regeneration of whole plants from somatic cells in vitro (Haberlandt, 1902). The tissue culture system has matured since the historical discovery that different concentration ratios of auxin and cytokinin (CK) are critical to regenerating adventitious roots and shoots (Skoog and Miller, 1957). Steward et al. (1958) successfully regenerated new somatic embryos and subsequently developed roots and shoots by using isolated phloem cells from carrot roots, which confirmed the totipotency of plant cells. Since then, tissue culture technology based on regenerative ability has been extensively used in various fields, including basic research, micropropagation, and transgenic breeding.
The ability of plant regeneration is affected by multiple factors, including use of a plant growth regulator (PGR; Çabuk and Özgen, 2016; Gerdakaneh et al., 2020), the composition of basic medium (Sundararajan et al., 2017; Chimdessa, 2020), and explant type (Dhar and Joshi, 2005; Minutolo et al., 2020). Importantly, plant tissue culture presents strong species dependence and genotype specificity. Some plants, such as tobacco (Nicotiana tabacum), Arabidopsis thaliana, and rice (Oryza sativa), can be easily regenerated in vitro, whereas other plants, such as soybean (Glycine Max), wheat (Triticum aestivum), and maize (Zea mays), are more difficult to regenerate. Moreover, Japonica varieties show a higher capacity for callus formation than Indica varieties in rice (Abe and Futsuhara, 1986). The tissue culture capacities of hybrid lines are higher than those of inbred lines in maize (Duncan et al., 1985). Clarifying the regulatory network and genetic control of plant-regeneration ability in tissue culture is helpful to improving plant-regeneration rates and genetic transformation efficiency.
Therefore, this review discusses two pathways of plant regeneration in tissue culture: somatic embryogenesis and de novo organogenesis. We then describe how environmental factors affect plant regeneration from the aspects of explant sources, basal culture medium, PGRs, and light/dark treatment. Importantly, we describe the molecular mechanisms that regulate somatic embryogenesis from three levels: transcription factors, hormone signalling, and epigenetic regulation. Furthermore, we elaborate on the molecular mechanisms underlying pluripotent callus formation, de novo root organogenesis, and de novo shoot organogenesis. This review provides insight into how plants regenerate from explants and important cues for plant micropropagation and genetic modification.
Regeneration pathways in seed plants can be divided into tissue repair, somatic embryogenesis, and de novo organogenesis. The first pathway concerns how young plant tissues, such as root or leaf tips, repair injured parts and is often used in plant-cutting propagation techniques (Xu and Huang, 2014). In tissue culture, plants are regenerated mainly by somatic embryogenesis and de novo organogenesis (Hill and Schaller, 2013).
In somatic embryogenesis, plant somatic cells undergo dedifferentiation into embryonic stem cells and then by way of embryonic development form complete plants, signifying that plant cells are totipotent by virtue of the embryogenic callus (Zimmerman, 1993; Verdeil et al., 2007). Somatic embryogenesis leads to an exchange in cell fate from a somatic cell back into an embryonic stem cell. Dedifferentiation through this pathway is usually accomplished under stress conditions, hormonal induction (e.g., auxin), or gene expression modification (Jiménez and Thomas, 2006; Fehér, 2015; Horstman et al., 2017). Somatic embryos can be directly induced from individual somatic cells or indirectly generated from embryonic callus (Yang and Zhang, 2010; Horstman et al., 2017).
Indirect somatic embryogenesis is the most common pathway, especially in crop plants, and starts with the embryonic callus (an unorganized cell mass; Figure 1A). Embryonic callus induction is followed by the formation of proembryonic masses on the surface or within the callus mass, from which single cells or cell clusters develop into somatic embryos (Toonen et al., 1994). Under appropriate conditions, somatic embryos can develop into shoots and roots (Figure 1A). In the case of maize (Rakshit et al., 2010), embryonic callus can be induced to form from explants, such as immature embryos and shoot tips, in a callus-inducing medium containing a high level of auxin and a low level of CK. When transferred to a shoot-inducing medium (SIM) containing a high level of CK and a low level of auxin, embryonic callus differentiates into shoots. For root regeneration, root-inducing medium containing some auxin without CK is required for incubating embryonic callus.
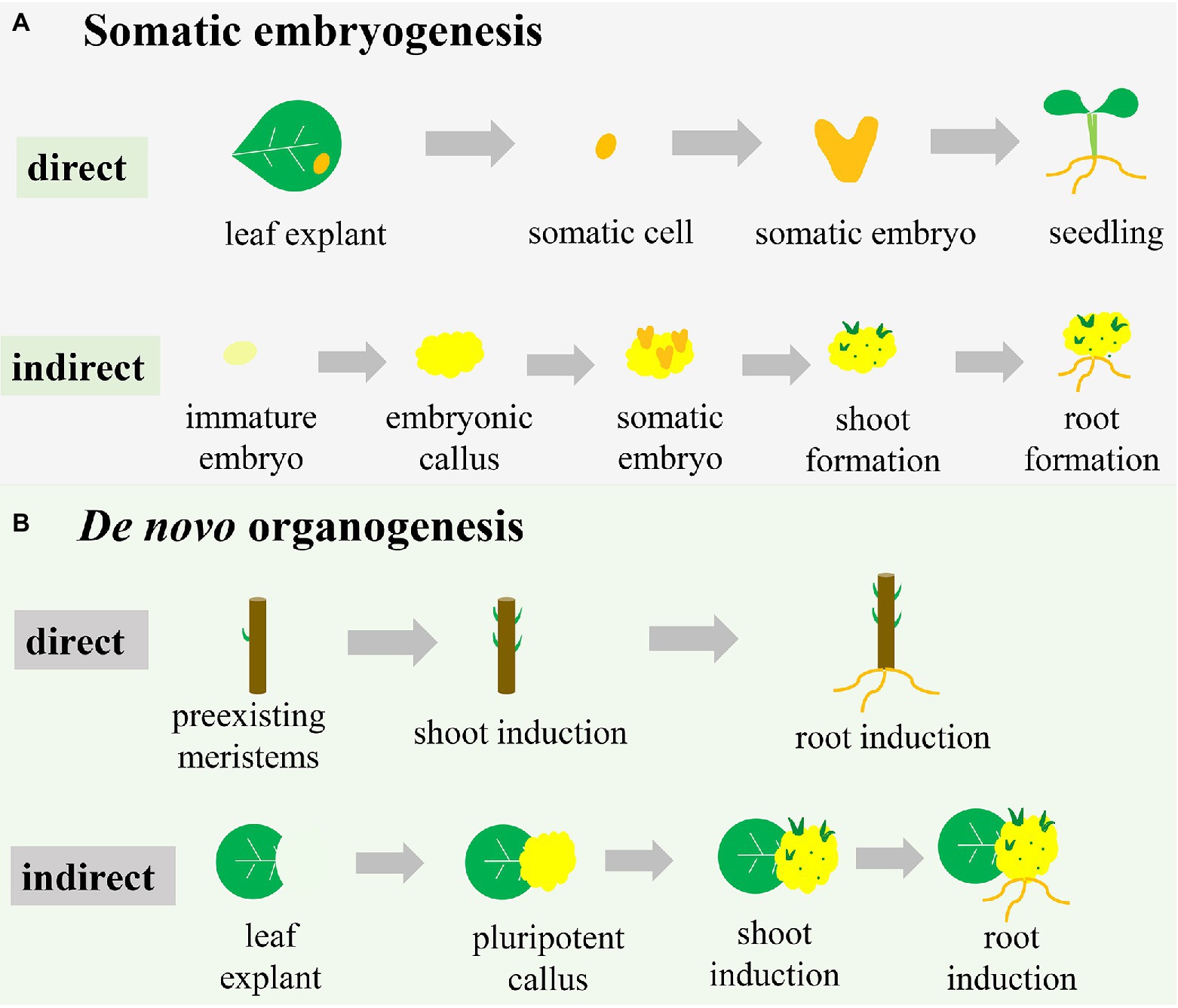
Figure 1. Different pathways of plant regeneration. (A) Somatic embryogenesis. In the direct pathway, the somatic cell originated from explants (e.g., a leaf) is induced to form the somatic embryo, which subsequently drives the development of the whole plant. In the indirect pathway, the explant (e.g., an immature embryo) is induced to initiate the embryonic callus, on which somatic embryos are formed to subsequently develop shoots and roots. (B) De novo organogenesis. In the direct pathway, shoots and roots are induced directly on the stem with pre-existing meristems. In the indirect pathway, pluripotent callus is produced around the wound in a leaf explant, with formation of shoots and roots subsequently induced.
Unlike the formal pathway, direct somatic embryogenesis lacks the callus phase and is less well defined (Figure 1A). In this system, the explant exhibits a more regular compact cell division and is less prolific (Horstman et al., 2017). The individual somatic cell in one or more cell layers divides and bulges under appropriate conditions to develop into a morphologically recognizable new embryo capable of developing into a whole plant (Fitch and Manshardt, 1990; Xu and Huang, 2014; Figure 1A). For example, constitutive expression of the morphogenic transcription factors BABY BOOM (BBM) and WUSCHEL (WUS)2 in maize resulted in rapid formation of abundant somatic embryos on the scutella (Lowe et al., 2018). These somatic embryos could then be directly germinated into plants without the callus phase.
Direct and indirect somatic embryogenesis pathways can occur in the same explant, but the periods of obtaining regenerated plants differ (Zhang et al., 2021). Compared with the direct somatic embryogenesis pathway, the indirect pathway has a longer period to regenerate plants due to the callus-induction process. Therefore, the indirect somatic embryogenesis pathway is frequently associated with somaclonal variation (Miguel and Marum, 2011; Bahmankar et al., 2017). However, the indirect somatic embryogenesis pathway results in more regenerated plantlets than the direct pathway due to the plentiful proliferation of callus (Gaj, 2011). Therefore, if the goal is rapid regeneration of plants, the direct pathway is more efficient than the indirect pathway. However, for species in which explants are difficult to obtain or situations where many regenerated plants are desired, the indirect pathway is the better choice.
De novo organogenesis refers to the regenerative process that does not use a somatic embryo but rather the differentiation of the meristematic centre, reflecting the pluripotency of plant cells (Lardon and Geelen, 2020). Plant-regenerative mechanisms, such as de novo organogenesis, result in regenerating adventitious roots and/or adventitious shoots in vitro or from injured plant organs, with this frequently occurring in nature (Duclercq et al., 2011). The regeneration process of adventitious shoots and roots is referred to as de novo shoot organogenesis and de novo root organogenesis. Like somatic embryogenesis, de novo organogenesis can also be categorized as either a direct or indirect regeneration pathway (Figure 1B). As with somatic embryogenesis, shoots or roots are directly induced from pre-existing meristems or injured organs under advisable conditions (Sang et al., 2018; Figure 1B). Cutting-propagation technology is based on the direct de novo organogenesis used to regenerate organs. During indirect de novo organogenesis, cells undergo dedifferentiation and plant growth regulators stimulate cell division (Sugimoto and Meyerowitz, 2013), after which additional dedifferentiated cells are induced with further culture time and ultimately generate pluripotent callus (Figure 1B). When all conditions are met, the pluripotent callus undergoes physiological and biochemical changes, resulting in different cell-division positions and directions (Wang et al., 2011; de Almeida et al., 2015). De novo shoot organogenesis and de novo root organogenesis are initiated using different combinations of auxin and CK (Street and Henshaw, 1966).
The essential difference between de novo organogenesis and somatic embryogenesis is the absence of somatic embryo formation, whereas both pathways include direct and indirect methods of regeneration (Figure 1). The callus is formed in the two indirect methods, but the characteristics of the callus differ. Somatic embryogenesis leads to embryogenic callus with totipotency and subsequent development into a somatic embryo, whereas de novo organogenesis induces non-embryogenic callus with pluripotency (Yumbla-Orbes et al., 2017; Shin and Seo, 2018; Fehér, 2019). Moreover, indirect de novo organogenesis can result in genetic instability and somaclonal variance similar to somatic embryogenesis (Vitamvas et al., 2019). Organ production directly from explants is a time-saving method but unsuitable for transgenic research due to the production of chimeric shoots containing both transformed and untransformed cells (Firoozabady and Moy, 2004). Many studies have reported the induction of embryonic tissue from immature seeds or embryos of cereal crops, suggesting that somatic embryogenesis is restricted to a certain time of year (Malik et al., 2004; Jones et al., 2019). However, the material used for organogenesis is multiplicative and seasonally flexible. Additionally, for some organs or tissues that are easy to induce de novo organogenesis, it might be difficult to develop somatic embryos. Therefore, two pathways are occasionally combined to enhance the frequency of plantlet regeneration in a giving species for commercial marketplace or scientific research (Abe and Futsuhara, 1986).
Although all plant cells have the totipotential to regenerate entire plants, the ease in expression of that capacity varies in plant species and varieties, even in cells of the same plant (Table 1). For example, only a part of maize stock is capable of plant regeneration in tissue culture. These include a few self-inbred lines, F1 hybrids, and open-pollinated hybrids (Hibberd, 1984; Hodges et al., 2012). A previous study tested 101 maize self-inbred lines to examine the ability of plant regeneration, finding that only 49% were able to regenerate whole plants, with 97% of the hybrids producing callus capable of plant regeneration having at least one regenerable parent (Duncan et al., 1985). Another study evaluated a total of 113 tropical maize inbreds for tissue culture response, revealing that only 42 had the ability of embryonic callus induction (Carvalho et al., 1997). Moreover, the tissue culture capacities of hybrid lines are higher than those of inbred lines, although until recently, it remained difficult to explain this fact. Furthermore, conditions favourable for plant regeneration in one cultivar can sometimes be inadequate to grow plants in another cultivar of the same species (Ali et al., 2007; Satish et al., 2016).

Table 1. Environmental factors and molecular mechanisms affecting plant regeneration in tissue culture.
The age of explants is another factor that affects plant regeneration in tissue culture (Table 1). Although every living cell can regenerate entire plants, most studies use cells or tissues with active growth and vigorous physiological metabolism as explants (Hoque and Mansfield, 2004). Among the explants used in tissue culture, the most widely used are immature embryos, including in maize (Jones et al., 2019), rice (Rakshana et al., 2019), wheat (Kumar et al., 2017), barley (Hinchliffe and Harwood, 2019), and other important cereal crops. Immature inflorescences are also suitable explants for sorghum (Chou et al., 2020), wheat (Mahmood and Razzaq, 2017), and barley (Saeedpour et al., 2021). Moreover, immature cotyledons and hypocotyl segments excised from seedlings are often utilized for medicinal plants, such as Pterocarpus marsupium (Husain et al., 2010), Cassia angustifolia (Parveen and Shahzad, 2014) and Santalum album L. (Akhtar and Shahzad, 2019). Additionally, embryogenic callus was successfully induced from young leaves in wheat (Yu et al., 2012), sorghum (Wernicke et al., 1982), and rye (Haliloglu and Aydin, 2016), and other explants have also been reported, including root tips (Wang et al., 2021), shoot tips (Long et al., 2020), anthers (Han et al., 2021), and pollen (Cho and Zapata, 1988). Regardless of the explant, initial cell division begins at a young part near the cambium and vascular bundles. Explants in the juvenile-development phase are more regenerative and possess higher totipotency than those of adult explants (Lee et al., 2020). For example, a study investigating the frequency of embryonic callus induction among different ages of maize seedlings found a higher frequency of embryonic callus induction for seedlings that were between 2- and 6-cm long than for longer seedlings (Long et al., 2020). Moreover, reports indicate that differences in endogenous hormones and nutrients in various parts of explants may explain the differences in regenerative abilities (Bhaskaran and Smith, 1990; Saeedpour et al., 2021), with variations in endogenous hormones also affecting the demand for exogenous hormones in tissue culture.
Exogenous hormones, especially auxin, CK, and other PGRs, play an important role in plant somatic embryogenesis and de novo organogenesis (Jiménez, 2005; Schwarz and Beaty, 2018; Table 1). Plant regeneration in vitro depends on the addition of exogenous hormones and the response to these hormones during tissue culture (Bernula et al., 2020). Generally, the response of explants to PGRs comprises three stages: (1) cultured explant cells perceive plant hormone signalling to induce subsequent dedifferentiation; (2) due to the influence of plant hormone balance, the differentiation instructions for specific cells in plant tissue are given, laying the foundation for the subsequent differentiation of specific organs; and (3) plant morphogenesis occurs independent of exogenous hormones (Ye et al., 2012). Although somatic embryogenesis is initiated by exogenous auxin, its further occurrence does not require auxin. A possible reason is that exogenous auxin promotes the synthesis of endogenous auxin, with the resulting increases in endogenous auxin promoting regeneration (Michalczuk et al., 1992; Nic-Can and Loyola-Vargas, 2016).
Auxin is the most important determinant of somatic embryogenesis for many species in tissue culture. Exogenous auxin promotes callus formation from cultured materials by inducing the production of endogenous precursors of ethylene synthesis, including 1-aminocyclopropane-1-carboxylic acid (Singla et al., 2007). 2,4-Dichlorophenoxyacetic acid (2,4-D), a synthetic auxin, is widely used in many species, especially cereal crops and medicinal plants. Gaj (2004) reported that in >65% of experiments, 2,4-D was used alone or combined with other hormones. The concentration of 2,4-D affects callus formation, and the optimal concentration varies for different species or tissues. The general principle is that a low concentration promotes embryonic callus formation, whereas a high concentration inhibits its formation. For most Poaceae spp. 2 mg/l of 2,4-D is optimal to induce embryonic callus formation (Wang et al., 2008; Çabuk and Özgen, 2016), and 5–10 μm 2,4-D is suggested for somatic embryos induction in many medicinal plants (Husain et al., 2010; Parveen and Shahzad, 2014). However, there is no need to add 2,4-D to medium after the embryonic callus develops into an embryoid and regenerates seedlings, suggesting that the effect of 2,4-D is promoted during embryogenic callus induction and inhibited during embryogenic callus development into a complete plant (Singla et al., 2007; Parveen and Shahzad, 2014). Additionally, different concentrations of auxins, such as indole-3-acetic acid (IAA) and α-naphthalene acetic acid, also play an important role in promoting the differentiation of adventitious roots in tissue culture (Nissen and Sutter, 1990; El-Sherif, 2018).
CK is the most widely used PGR in adventitious shoot induction and initiation of somatic embryogenesis in tissue culture. De novo shoot regeneration requires cell proliferation involving the activation of cell mitosis. CK affects competent cells in the shoot-regeneration process, leading to cell-mass generation and cell-fate transformation. CK can induce adventitious shoots alone and cooperates with auxin to reinforce proliferation in chosen cells (Cortleven et al., 2019). Skoog and Miller (1957) proposed that a high CK-to-auxin ratio stimulates shoot formation, whereas roots are formed when the ratio is low. To date, the golden hormone-regeneration pattern has been a guiding determinant of the fate of explants in vitro. In addition to inducing shoot regeneration, CK also initiated somatic embryogenesis. It was reported that MS medium containing 6-benzyladenine alone could induce high frequency of somatic embryo differentiation in S. album L. (Akhtar and Shahzad, 2019). Moreover, the effects of PGRs, such as abscisic acid (ABA) and gibberellin (GA), on plant regeneration have also been reported (Nishiwaki et al., 2000). The addition of GA to the medium promotes germination and differentiation of immature embryos, which inhibits somatic embryo development. Ge et al. (2016) reported that the maize transcription factor MYB138 promotes somatic embryogenesis by inhibiting GA signal transduction.
Several types of culture media, including Murashige and Skoog (MS), N6, Woody Plant Medium (WPM), and B5, are used for callus induction and shoot differentiation and significantly influence plant regeneration in tissue culture (Table 1); however, different species or tissues may also require different basal medium. A previous study reported more prolific callus formation and higher shoot differentiation on MS medium than on B5 medium during plant regeneration from Easter lily (Lilium longiflorum L. cv. Ace) ovary tissues (Ramsay et al., 2003). However, N6 medium induced higher percentages of callus and green plants than did MS medium for rice (O. sativa; Sundararajan et al., 2017). For Indian siris (Albizia lebbeck), the WPM medium achieved the highest primary somatic embryoids development, whereas enhanced maturation of primary somatic embryoids occurred on MS medium (Saeed and Shahzad, 2015). During the conversion of somatic embryos into plantlets, a half strength MS medium performed better than other media in many medicinal plants (Sahai et al., 2010; Parveen and Shahzad, 2014; Saeed and Shahzad, 2015). Additionally, the carbon source is a vital component affecting plant regeneration in culture medium (Table 1). Sugar provides energy for the culture and represents the main regulator of the permeation environment, with glucose, sucrose, and maltose most often used in plant tissue culture. Small molecules of sugar can penetrate into living cells and dehydrate somatic embryos, thus promoting somatic embryo maturation (Kaviani, 2011). Moreover, a low sucrose concentration during somatic embryogenesis is advantageous to somatic embryo formation (Yaseen et al., 2013; Long et al., 2020). However, Malik et al. (2017) found that maltose resulted in maximal callusing and regeneration percentage as compared with other carbon sources for mature wheat embryos. Furthermore, compared with glucose and sucrose, maltose may more effectively inhibit the browning of plant cells. Other components, such as mannitol and metal ions, added to the culture medium can also affect the regeneration ability of explants (Simonović et al., 2021).
Under light conditions, phenolic compounds in explants will be oxidized by polyphenol oxidases, and the tissue will turn brown. The oxidation products can darken tissues and inhibit the activity of various proteins, with a potentially adverse effect on the formation of somatic embryos (Bhatia and Bera, 2015). Therefore, callus initiation, maintenance, and maturation require dark conditions in plant for many species. A previous report indicated that light reduces endogenous CK and auxin levels in plants by degrading auxins (Zenser et al., 2001). In this regard, darkness may help maintain a high auxin-to-CK ratio to support callus formation in explants. Additionally, dark conditions can lead to thinner cell walls and lower cell-wall deposits, thereby facilitating the entrance of PGRs into cells (Dai and Castillo, 2007). However, some studies have shown that light can promote callus formation by upregulating the expression of somatic embryogenesis marker genes, such as WUS, BBM, and leafy cotyledon 2 (LEC2; Siddique and Islam, 2015; Yu et al., 2019).
For shoot and root regeneration, a 16-/8-h photoperiod is generally required (Table 1). The frequency and speed of shoot initiation are higher under light conditions for maize regeneration (Li et al., 2002). A previous report showed that light might stimulate apical meristem differentiation by maintaining an optimal CK-to-auxin ratio, with low light intensity (~30–60 μmol m−2 s−1) preferable for shoot and root differentiation (Farhadi et al., 2017). Moreover, a recent study showed that light-emitting diodes (LEDs), which can regulate the level of photomorphogenic radiation necessary for plant morphogenesis, can be excellent substitutes for traditional cool-white fluorescent lamps (Bidabadi and Jain, 2020). Furthermore, LEDs are associated with cellular redox balance and involved in antioxidative metabolic activities during in vitro plant regeneration (Gupta and Karmakar, 2017).
Theoretically, somatic embryogenesis is a typical dedifferentiation process in which differentiated somatic cells are returned to the state of totipotent embryonic stem cells. Dedifferentiation is the basis of totipotency and regeneration in multicellular organisms. Recent research suggests that somatic embryogenesis is a complex process involving transcription factors, hormone signalling pathways, and epigenetic regulation (Figure 2).
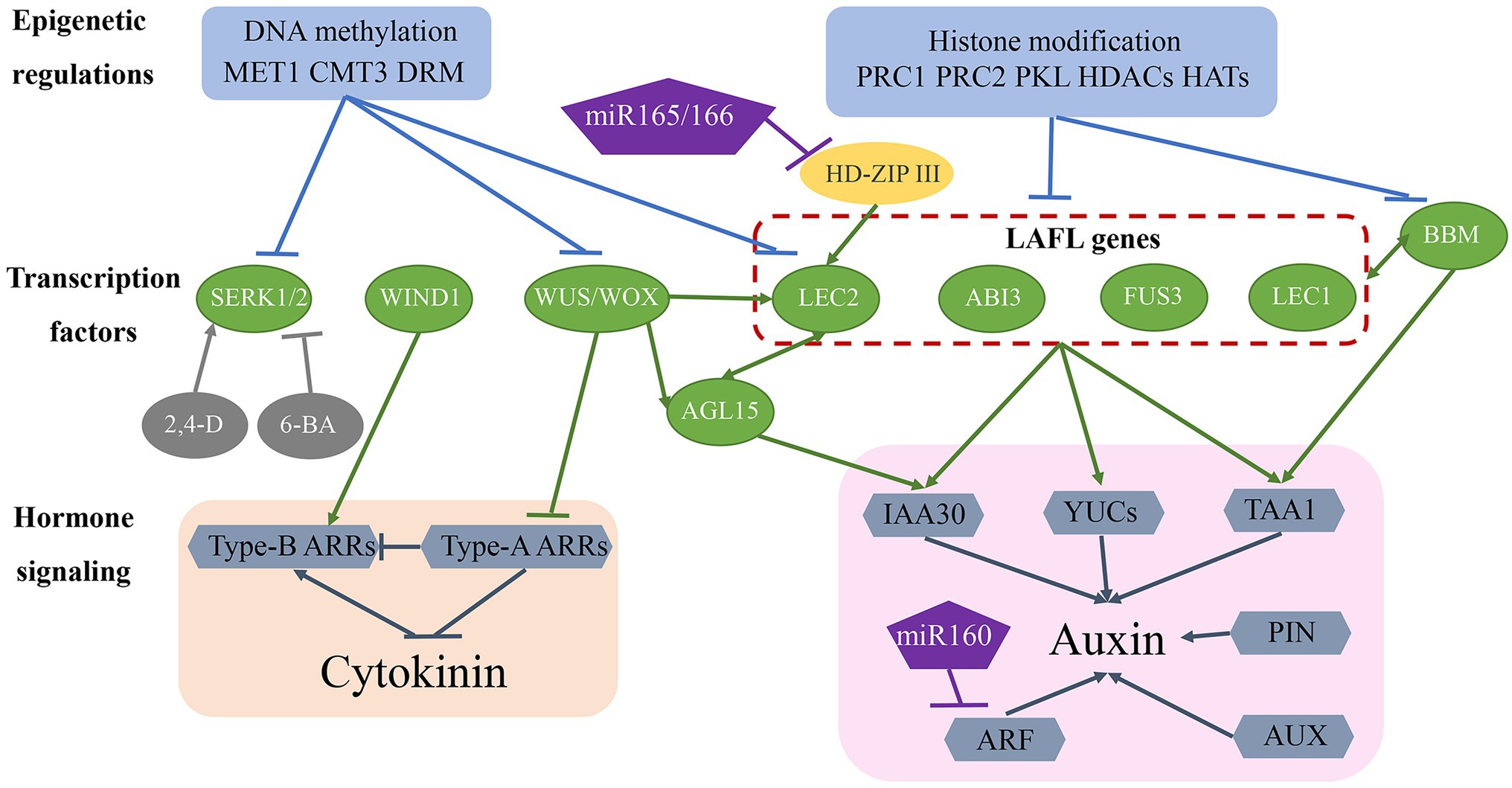
Figure 2. Molecular mechanisms of somatic embryogenesis. The somatic embryogenesis process is influenced by epigenetic regulation, transcription factors, and hormone signalling pathways. Epigenetic regulation, including DNA methylation and histone modifications, repress transcription factor access to gene-promoter regions, thereby inhibiting the expression of genes involved in somatic embryogenesis. Many transcription factors (green ovals) are involved in this regulatory network and also regulate each other and activate downstream auxin and CK signalling pathways. Additionally, miR-160 and miR-165/-166 are involved in regulating somatic embryogenesis.
Several transcription factors have been identified as essential regulators of the somatic embryogenesis process (Figure 2; Table 1). Fusca 3 (FUS3), LEC2, and abscisic acid insensitive 3 (ABI3), encode plant-specific B3-domain-containing proteins that are members of the AFL subfamily of transcription factors (Parcy et al., 1994; Luerßen et al., 1998; Stone et al., 2001). These genes/proteins together with LEC1-encoded CCAAT-binding transcription factors harbouring a HAP3 subunit form LAFL complexes (Lotan et al., 1998; Lepiniec et al., 2018). Overexpression of each of these genes promotes the formation of somatic embryos or embryonic traits in somatic tissues in the absence of additional hormones (Xu and Huang, 2014). The expression of LAFL genes is regulated by epigenetic factors, hormone signalling, and other transcription factors, such as BBM (Salaün et al., 2021). LEC2 and agamous-like 15 (AGL15) encode a MADS-box transcription factor that controls each of their respective expression in a regulatory feedback loop that also regulates the expression of the auxin-responsive protein gene IAA30, a primary factor in auxin signalling (Heck et al., 1995; Sato and Yamamoto, 2008). Additionally, LEC2 induces the expression of other auxin-related genes (IAA1, IAA17, and 1-aminocyclopropane-1-carboxylate synthase 4), as well as those encoding key enzymes involved in auxin biosynthesis, such as tryptophan aminotransferase of Arabidopsis 1 (TAA1) and YUCCA (YUC) genes (YUC1, YUC2, YUC4, and YUC10; Braybrook and Harada, 2008; Wójcikowska et al., 2013; Horstman et al., 2017). The BBM transcription factor upregulates the expression of LAFL genes and AGL15 during the somatic embryogenesis process, and LAFL proteins regulate BBM expression (Horstman et al., 2017), with BBM overexpression promoting callus proliferation and formation of somatic embryos (Salaün et al., 2021).
The WUS homeobox-containing transcription factor is involved in regulating embryonic cell fate by inducing the vegetative-to-embryonic transition (Jha et al., 2020). Overexpression of WUS promotes somatic embryo production without requiring the addition of hormones in Arabidopsis (Zuo et al., 2002) and upregulates LEC1, LEC2, and AGL15 expression during somatic embryogenesis (Jha et al., 2020; Figure 2; Table 1). Wus-related homeobox (WOX) genes encode similar sequences to the WUS homeodomain and a specific WUS box downstream of the homeodomain (Haecker et al., 2004). WOX proteins perform an essential role in early embryonic patterning (Salaün et al., 2021), and overexpression of WOX9 results in improved efficiency in somatic embryogenesis by increasing the levels of AGL15 and AGL8 (Tvorogova et al., 2019). Additionally, WOX5 is regarded as a marker of dedifferentiation based on its significant upregulation from the early stage of somatic embryogenesis (Orłowska and Kępczyńska, 2018).
Wound-induced differentiation 1 (WIND1), which encodes another APETALA2/ethylene-responsive element-binding factor transcription factor, induces the acquisition of regeneration competency (Iwase et al., 2011); however, it is not directly involved in promoting somatic embryo formation, although it does play a role in the induction of callus in the indirect somatic embryogenesis pathway. Similar to WUS, WIND1 acts upstream of LEC2 during regeneration (Iwase et al., 2015). Compared with LAFL proteins, WUS and WIND1 induce somatic embryogenesis through a different hormone pathway (Figure 2; Table 1) and are mainly involved in CK-specific responses rather than auxin biosynthesis and signal transduction (Horstman et al., 2017). Specifically, WUS represses negative regulators [type-A Arabidopsis response regulator (ARR) genes] of CK response, whereas WIND1 stimulates the expression of positive regulators (type-B ARR genes) of CK response (Leibfried et al., 2005; Iwase et al., 2011).
Somatic embryogenesis receptor-like kinase (SERK) belongs to the RLK gene family, and as the first key gene screened in a carrot hypocotyl regeneration study, it regulates the transition from somatic cells to embryonic cells (Schmidt et al., 1997). Studies show that single cells expressing SERK can develop into regenerative somatic embryos, with regenerative somatic cells and zygotic embryos demonstrating the same signal transduction pathway. SERK genes were subsequently cloned from Arabidopsis (Hecht et al., 2001), rice (Hu et al., 2005), wheat (Singla et al., 2008), maize (Zhang et al., 2011), and other plants and showed higher expression levels in the embryogenic callus and maturation stage than in the non-embryogenic callus (Gulzar et al., 2020). In maize, ZmSERK1 and ZmSERK2 exhibit redundant functions in the initiation of embryonic cell formation and division and are regulated by auxin and CK (Zhang et al., 2011). Additionally, 2,4-D enhances ZmSERK1 and ZmSERK2 levels, which promote somatic embryogenesis, whereas the CK 6-benzyladenine reduces their respective expression, thereby inhibiting somatic embryogenesis (Zhang et al., 2011; Méndez-Hernández et al., 2019; Figure 2; Table 1).
Several other transcription factors are also critical for regulating somatic embryogenesis. PGA37/MYB118 and MYB115 promote somatic embryo formation by positively regulating the expression of lec1 (Wang et al., 2009). Additionally, LEC1-like, the most closely related subunit of LEC1, plays an important role in embryogenesis (Kwong et al., 2003). Furthermore, a double mutant of the genes viviparous1/ABI3-like 1 (VAL)1 and VAL2 exhibited embryo-like proliferations, suggesting that VAL1 and VAL2 negatively regulate somatic embryogenesis (Suzuki et al., 2006).
Plant hormones, especially auxins and CKs, are key factors in the somatic embryogenesis pathway. Therefore, genes associated with hormone signalling pathways are likely to play an important role in that process (Figure 2; Table 1). The LAFL protein complex upregulates the expression of auxin-biosynthesis-related genes (TAA1 and YUC genes) and the auxin signalling gene IAA30, and WUS and WIND1 negatively and positively regulate type-A ARR and type-B ARR genes corresponding to CK responses. Additionally, polar auxin transport induces concentration gradients maximal necessary for plant development. Pin-formed (PIN) and AUX proteins achieve differential distributions by controlling auxin efflux and influx, respectively (Petrásek and Friml, 2009). Moreover, differential expression of AUX/IAA genes and auxin response factors (ARFs), the core components of the auxin signalling pathway, is related to induction of somatic embryogenesis (Quintana-Escobar et al., 2019; Wójcik et al., 2020). Furthermore, microRNA (miR)-165/-166 and miR-160 may contribute to auxin-related induction of somatic embryogenesis by targeting the HD-ZIP III family genes phabulosa/phavoluta (PHB/PHV), positive regulators of LEC2 expression, and ARF genes (ARF10, ARF16, and ARF17), respectively (Wójcik et al., 2017; Jin et al., 2020).
Epigenetic regulation is key to maintaining somatic cell identity by suppressing the expression of embryo-specific genes (Figure 2; Table 1). DNA methylation and histone modification play an important role in regulating gene expression and determining cell fate (Méndez-Hernández et al., 2019). During callus formation, DNA methyltransferase activity regulates gene transcription. A previous study showed that mutation in methyltransferase 1 (MET1) results in decreased CG methylation and dysregulated expression of the auxin efflux carrier PIN1 engaged in polar auxin transport during somatic embryogenesis (Wójcikowska et al., 2020). Decreased methylation has been reported in SERK, LEC2, and WUS in the embryogenic callus (Karim et al., 2018). Additionally, studies revealed relatively lower levels of DNA methylation at CG, CHG, and CHH sequence contexts in association with MET1, chromomethylase 3 (CMT3), and domains rearranged methyltransferase 2 (DRM2) activities related to somatic embryogenesis and regeneration ability (Karim et al., 2018; Wójcikowska et al., 2020).
In addition to DNA methylation, histone modifications, including methylation, acetylation, and ubiquitination, also play an important role in regulating somatic embryogenesis. Polycomb repressive complex (PRC)1 and PRC2 are required to establish and maintain stable epigenetic suppression in response to developmental or environmental signals (Mozgova and Hennig, 2015; Figure 2; Table 1). PRC2 exhibits histone 3 lysine 27 trimethylation (H3K27me3) activity, and PRC2 mutation results in incomplete transition from embryo to seedling, disorderly cell division in seedlings, and formation of callus with embryo traits (Xu and Huang, 2014). PRC1 recognizes H3K27me3 alterations and promotes chromatin compaction via histone H2A lysine ubiquitination (Salaün et al., 2021). A recent study showed that PRC1 and PRC2 repress the expression of embryo-specific genes, including LAFL, AGL15, WOX5, BBM, and PIN1 (Duarte-Aké et al., 2019). Additionally, pickle (PKL), a member of the chromodomain helicase DNA-binding protein 3 family of chromatin ATPase remodelers, is another epigenetic factor that plays a key role in preventing somatic cells from producing embryonic traits (Ogas et al., 1999). Similar to PCR1 and PCR2, PKL represses the expression of embryonic genes, including LAFL genes, by promoting H3K27me3 alterations (Dean Rider et al., 2003; Aichinger et al., 2009; Figure 2). Furthermore, histone acetylation regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) plays a critical role in somatic embryogenesis (Tanaka et al., 2008; Figure 2). Trichostatin A, an HDAC inhibitor, upregulates the expression of genes related to embryogenesis, including LEC1, FUS3, and ABI3 (Tanaka et al., 2008).
Pluripotent callus formation is initiated by the division of pericycle cells in the xylem pole in a process similar to lateral root initiation (Atta et al., 2009), with molecular factors participating in lateral root initiation also involved in pluripotent callus formation. During this process, some root meristem marker genes, including WOX5, scarecrow (SCR), short root (SHR), plethora (PLT)1, PLT2, and root clavata-homolog 1 (RCH1), are significantly upregulated (Atta et al., 2009; Figure 3; Table 1). WOX5, SCR, PLT1, and PLT2 are transcriptionally activated by HAT of the GNAT/MYST superfamily 1, which binds directly to their respective promoters to initiate acetylation (Kim et al., 2018). Additionally, the rapid induction of PLT3, PLT5, and PLT7 expression by auxin results in transcriptional regulation of PLT1 and PLT2 (Kareem et al., 2015). Moreover, WOX11 promotes pluripotency acquisition by activating the expression of lateral organ boundaries domain 16 (LBD16), which is activated via ARFs and promotes the expression of WOX5, PLT1, and PLT2 (Xu and Hu, 2020).
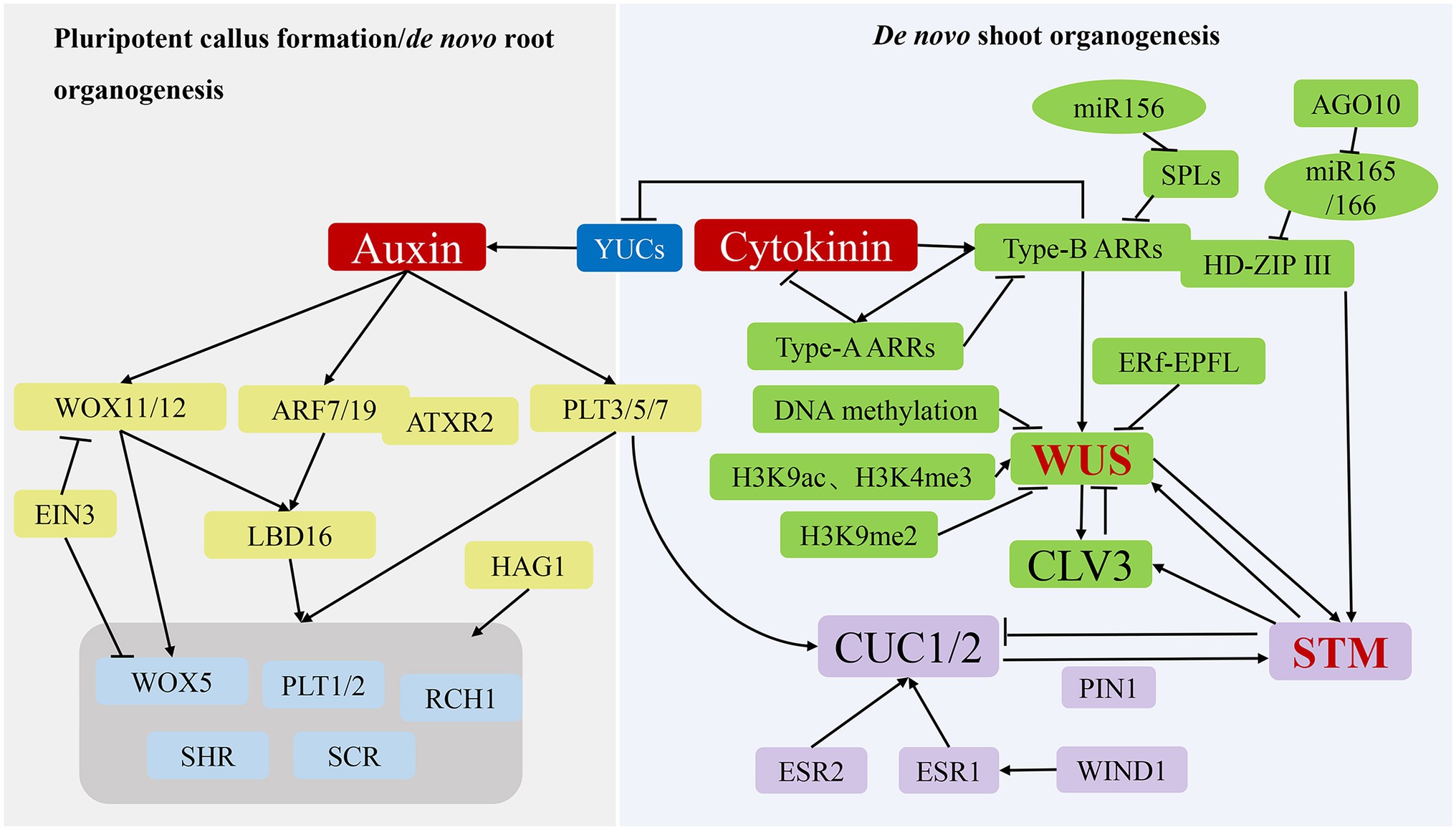
Figure 3. Molecular mechanisms of de novo organogenesis in tissue culture. During the process of pluripotent callus formation or de novo root organogenesis (left panel), YUC-mediated auxin acts as a key regulator to activate WOX11/12, ARF7/19 and PLT3/5/7 expression, after which their translated products directly or indirectly promote the expression of genes, including WOX5, PLT1/2, SHR, SCR, and RCH1, to induce pluripotent callus or root apical meristem formation. During the process of de novo shoot organogenesis (right panel), two pathways (WUS-CLV3 and STM-CUC) establish negative-feedback loops and play critical regulatory roles. The WUS-CLV3 pathway is mainly regulated by DNA methylation, histone modification, and hormone signalling. CK activates the expression of type-B ARRs to stimulate WUS expression, whereas type-B ARRs repress YUC-mediated auxin biosynthesis. In the STM-CUC pathway, STM expression is promoted by CUC1 and CUC2, both of which have their expression upregulated by PLT3/5/7, ESR1, ESR2, WIND1, and PIN1. Moreover, WUS and STM interact directly to activate CLV3 expression, suggesting that the two pathways converge and coordinate to control shoot regeneration.
In tissue culture, de novo root organogenesis is induced by transferring pluripotent callus to root induction medium rich in auxin. In past years, the analysis of transcriptome, epigenome, and cell lineage in pluripotent callus has revealed that the formation of pluripotent callus and de novo root organogenesis share similar genetic pathways (Liu et al., 2014; Xu and Huang, 2014; Figure 3; Table 1). The de novo root organogenesis process can be divided into two steps: the transition of competent cells to root founder cells, which is marked by WOX11, and the switch of root founder cells to root primordium cells, which is marked by WOX5 (Liu et al., 2014). Inhibition of polar auxin transportation blocks the rooting process, suggesting that auxin is the key hormone that regulates de novo root organogenesis (Greenwood et al., 2001). Suppression of YUC genes (YUC1, 2, 4, 5, 6, 8, and 9) mediating auxin biogenesis inhibits the expression of WOX11 and prevents the fate transition of competent cells (Chen et al., 2016; Pan et al., 2019).
In the first step, auxin directly activates the expression of WOX11 and its homolog WOX12 (Sang et al., 2018; Figure 3; Table 1). During the next step, WOX11/12 promotes the expression of WOX5 and LBD16 responsible for activating the expression of WOX5, PLT1, and PLT2 (Sang et al., 2018). It was found that the transcription factor ethylene insensitive 3 (EIN3) strongly decreased the de novo root organogenesis rate by suppressing the transcription of WOX11 and WOX5 (Li et al., 2021; Figure 3; Table 1), and older explants showed increased EIN3 activity, which is in accord with the observation that younger organs possess a higher regeneration ability (Li et al., 2021). As mentioned above, auxin also induces PLT3, PLT5, and PLT7 expression, which subsequently regulate downstream root meristem marker genes. In addition to WOX11/12 and PLTs genes, the auxin response factors ARF7 and ARF19 target and activate the expression of LBD16 (Okushima et al., 2007). The Arabidopsis trithoprax-related 2 (ATXR2) protein can physically interact with ARF7 and ARF19. The complex catalyses H3K36me3 deposition at the promoter of LBD16 to promote its expression in the root regeneration process (Lee et al., 2018).
After culturing on SIM rich in CK, the pluripotent callus continues to divide under CK-mediated actions, and cell populations gradually generate for subsequent differentiation, signifying the construction of the stem cell niche (Ikeuchi et al., 2019). Shoot stem cell homeostasis is maintained by two regulatory pathways: WUS-clavata 3 (CLV3) and shoot meristemless (STM)-cup-shaped cotyledon (CUC; Figure 3; Table 1). As the determining factor in the early stage of stem cell niche construction, WUS expression begins 2 to 3 days after SIM culture (Zhang et al., 2017), with initial expression of WUS marking the establishment of shoot-progenitor cells and representing the most critical molecular event in de novo shoot organogenesis. The regeneration ability of the WUS mutant is completely lost, whereas WUS overexpression results in ectopic formation of shoots, indicating that WUS is necessary for de novo shoot regeneration (Gordon et al., 2007). WUS promotes the expression of CLV3, which encodes a signal peptide, whereas CLV3 inhibits WUS expression in a negative-feedback loop that plays a key role in maintaining the stem cell population (Han et al., 2020). Similarly, STM is expressed throughout the shoot meristem and represses the expression of CUC1 and CUC2, whereas CUC1 and CUC2 activate STM expression to maintain the shoot meristem (Balkunde et al., 2017).
The WUS-CLV3 pathway is regulated by DNA methylation, histone modification, and hormone signalling (Figure 3). Mutations of MET1, CMT3, DRM1, and DRM2 result in loss or reduction in DNA methylation in the regulatory regions of the WUS promoter, which enhances WUS expression and the shoot-regeneration rate (Sugimoto et al., 2019). WUS gene promotes both somatic embryogenesis and de novo organogenesis (Figures 2, 3) so that the lower levels of DNA methylation at CG, CHG, and CHH sequence contexts in association with MET1, CMT3, DRM1, and DRM2 activities are beneficial for two pathways. In somatic embryogenesis pathway, H3K27me3 alterations prevent somatic cells from producing embryonic traits by repressing the expression of WUS gene. However, de novo shoot regeneration involves different histone modification sites at WUS. The abundance of markers of histone 3 lysine 9 acetylation (H3K9ac) and histone 3 lysine 4 trimethylation (H3K4me3) at WUS sites increases, whereas the abundance of inhibitory markers histone 3 lysine 9 di-methylation (H3K9me2) decreases during shoot regeneration (Li et al., 2011). By contrast, kryptonite, an H3K9 methyltransferase, and Jumanji-domain-containing 14, an H3K4 demethylase, are responsible for repressing WUS transcription, which decreases shoot production. However, HAC1, a HAT, and lysine-specific demethylase 1-like 3, an H3K4 demethylase, activated WUS transcription, which increased shoot production (Ishihara et al., 2019).
Additionally, the auxin and CK signalling pathways affect WUS expression. As transcriptional activators of CK signalling, type-B ARRs (ARR1, ARR2, ARR10, and ARR12) directly activate WUS expression following binding to its promoter (Zhang et al., 2017), while also suppressing YUC-mediated auxin accumulation to further promote WUS expression (Meng et al., 2017). Type-A ARRs (ARR5, ARR6, ARR7, and ARR15), as negative regulators of CK signalling, are directly regulated by type-B ARRs and interfere with the function of type-B ARRs, thereby creating a negative-feedback loop (Sugimoto et al., 2019). Furthermore, targeting of squamosa promoter binding protein-like (SPL) mRNA by miR-156 decreases regulation of the activities of type-B ARRs in an age-dependent manner (Zhang et al., 2015). In young explants, miR156 levels are elevated relative to those in adult explants and repress SPL expression, thus increasing type-B ARR activity and shoot-regeneration ability. Moreover, miR-165/-166 inhibits shoot regeneration by splicing and reducing the translation of mRNAs encoding proteins harbouring an HD-ZIP III domain, including PHB, PHV, and REVOLUTA (Shin et al., 2020). Argonaute 10 inhibits shoot regeneration by suppressing miR-165/-166 activity. Another study found that type-B ARRs interact with HD-ZIP III proteins to form transcription complexes that specifically activate WUS expression during the early stage of shoot regeneration (Zhang et al., 2017), and a recent study demonstrated that accurate spatial expression of WUS and CLV3 influences their function (Zhang et al., 2021). Specifically, a signalling pathway comprising ERECTA family receptors and epidermal-pattern factor-like ligands inhibits the expression of WUS and CLV3 in the periphery of the shoot apical meristem, confining them to the centre. These findings demonstrate that WUS expression is determined by multiple regulators in a complicated molecular network.
In the STM-CUC pathway, the negative-feedback loop between STM and CUC plays a critical role in regulating de novo shoot organogenesis (Figure 3; Table 1). CUC proteins are essential in establishing the shoot promeristem (Aida et al., 1999). Polar localisation of PIN1 induced by CUC determines the location of shoot progenitors, with the polarized upregulation of PIN promoting STM expression in the promeristem (Gordon et al., 2007) Additionally, PLT3, PLT5, and PLT7 upregulate CUC1 and CUC2 expression during shoot regeneration, with these PLT proteins controlling shoot regeneration via a two-step mechanism that first establishes competence by activating PLT1 and PLT2 expression during pluripotent callus formation. Moreover, PLTs regulate CUCs to accomplish regeneration (Kareem et al., 2015). In addition to PLTs, enhancer of shoot regeneration (ESR)1 and ESR2 act as upstream regulators of CUC genes during de novo shoot organogenesis by activating CUC1 expression by directly binding to its promoter (Ikeda et al., 2006; Matsuo et al., 2011). Notably, ESR1 expression is regulated by WIND1, which connects wound signalling to shoot regeneration (Iwase et al., 2017).
Both the WUS-CLV3 and STM-CUC pathways are essential for stem cell development during de novo shoot organogenesis. A recent study reported that the two pathways converge and coordinate through direct interaction between the WUS and STM proteins (Su et al., 2020; Figure 3; Table 1). Specifically, STM directly activates CLV3 expression by binding to its promoter at a different site from that of WUS. Additionally, WUS–STM interactions enhance WUS binding to the CLV3 promoter and activation of CLV3 transcription, suggesting that CLV3 is simultaneously regulated by WUS, STM, and their complex (Su et al., 2020; du and Homeostasis, 2020). Furthermore, STM activity is regulated by WUS activity in the shoot meristem (Lenhard et al., 2002; Su et al., 2020).
Plant-regeneration techniques in tissue culture have been used in many fields, including gene-function research, transgenic breeding, and rapid micropropagation. In gene-function research, multiple methods, including overexpression, gene knockout, and genome editing, rely on genetic transformation in plants. The embryogenic callus is the most widely used genetic transformation receptor in most species. For example, CRISPR-Cas9 promoter editing of maize Arabidopsis CLV3-LIKE genes enhanced grain-yield-related traits (Liu et al., 2021). However, only a few plant lines can establish an efficient transgenic system. Genotype has become the inhibitory factor in genetic transformation and gene-function verification. Therefore, understanding the mechanisms associated with embryogenic callus induction and plant regeneration can facilitate gene-function validation and research.
In addition to gene-function research, transgenic plant breeding is also based on genetic transformation. Compared with traditional breeding, transgenic technology can break the reproductive isolation between species, realize the precision improvement of certain genes, estimate offspring traits, and offer the advantages of accurate targeting and shorter breeding cycles (Gepts, 2002). However, genotype limitations to genetic transformation inhibit the development of transgenic plant breeding. In the case of maize transgenic breeding, most backbone lines used for commercial production are recalcitrant to transformation, resulting in the desirable gene needing to first be introduced into a few good transgenic receptors, followed by import of the desirable gene fragment into the target inbred line through successive backcrosses. Therefore, conventional maize breeding systems must undergo genetic transformation, successive self-pollinations, and backcrosses that require at least 3 to 6 years and greatly extends the maize transgenic breeding cycle. Hence, analysing the mechanism of plant regeneration can create more transgenic receptors, address genotype-specific limitations, and further accelerate the transgenic breeding process.
Micropropagation is among the most important plant tissue culture techniques because of its ability to rapidly multiply a selected plant with a minimal number of starting materials. Compared with conventional propagation by seeds or vegetative methods, micropropagation enables large-scale propagation of multiple plants, resulting in its wide use in research and commerce. Moreover, micropropagation is an efficient technology for preserving gene pools and genetic diversity in plants (Chokheli et al., 2020). Many endangered or rare species have been successfully propagated using micropropagation, including Artemisia hololeuca and Hyssopus angustifolius (Zayova et al., 2018; Chokheli et al., 2020). Furthermore, many high-demand medicinal plants have been mass-developed using micropropagation (Moraes et al., 2021). Efficient regeneration depends on an appropriate micropropagation protocol, including explant types, medium compositions, and culture conditions (Singh, 2015). Therefore, understanding the plant-regeneration mechanism promotes the use of effective protocols for plant micropropagation.
In this review, we discussed how regeneration happens through two different pathways (somatic embryogenesis and de novo organogenesis in tissue culture) and the environmental factors and molecular mechanisms affecting these two pathways. This information offers a reference for scientific research and technology development in this field.
Despite the extent of research and the remarkable advances made as a result, the mechanisms that regulate plant regeneration require further elucidation. Plant regeneration in vitro is a complex developmental process, with only part of this process currently understood and requiring additional study for a comprehensive and integral understanding. First, although the regulatory network involved in plant regeneration has been initially determined, how these players and signalling molecules coordinate the different stages of regeneration remains unclear. Second, although we understand that plant regeneration is regulated by complex networks of gene regulation and influenced by external environmental stimulation, the interaction between external and internal signals to achieve the dynamic balance of growth and development requires further investigation. Specifically, it is unclear how external signals selectively activate internal plant regulators. Therefore, future studies on regeneration mechanisms should explore the interaction between external environmental factors and internal signalling networks. In tissue culture, the traditional way to improve plant regeneration is to change external environmental factors; therefore, combining an understanding of molecular mechanisms with traditional methods to achieve targeted plant regeneration should be a focus of future research. Third, the factors that control plant regeneration have mainly been outlined in Arabidopsis; however, whether other plants exhibit the same molecular mechanisms remains unverified. Rapid micropropagation and genetic transformation of most important crops and medicinal plants remain difficult; therefore, a future developmental direction for plant-regeneration research might involve applying theoretical concepts of plant-regeneration mechanisms to agricultural practice in order to help establish efficient regeneration systems and promote the industrialisation of agricultural biotechnology. Finally, the computer modelling, based on integral understanding, might be a promising research direction in plant tissue culture. In future, we just input the genotype of the species, then select the explant sources and desired regeneration pathway, the computer may automatically design the culture conditions we need, such as the composition of the culture medium and the amount of PGRs. Or we tell computer the genetic information and environmental conditions used for a certain species, it might simulate the entire culture process and the expected outcomes. That would greatly accelerate the research process of plant tissue culture.
YL wrote the initial manuscript. YY prepared these figures. YS and GP reviewed the manuscript and made significant editorial contributions. All authors contributed to the article and approved the submitted version.
This work was supported by the Fundamental Research Funds of China West Normal University (412906).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abe, T., and Futsuhara, Y. (1986). Genotypic variability for callus formation and plant regeneration in Rice (Oryza sativa L.). Theor. Appl. Genet. 72, 3–10. doi: 10.1007/BF00261446
Aichinger, E., Villar, C. B., Farrona, S., Reyes, J. C., Hennig, L., and Köhler, C. (2009). Chd3 proteins and Polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5:e1000605. doi: 10.1371/journal.pgen.1000605
Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and Cotyledon formation During Arabidopsis embryogenesis: interaction among the cup-shaped Cotyledon and shoot Meristemless genes. Development 126, 1563–1570. doi: 10.1242/dev.126.8.1563
Akhtar, R., and Shahzad, A. (2019). Morphology and ontogeny of directly differentiating shoot buds and somatic embryos in Santalum album L. J. For. Res. 30, 1179–1189. doi: 10.1007/s11676-018-0679-5
Ali, G., Hadi, F., Ali, Z., Tariq, M., and Khan, M. A. (2007). Callus induction and in vitro complete plant regeneration of different cultivars of tobacco (Nicotiana tabacum L.) on Media of Different Hormonal Concentrations. Biotechnology 6, 561–566. doi: 10.3923/biotech.2007.561.566
Atta, R., Laurens, L., Boucheron-Dubuisson, E., Guivarc’h, A., Carnero, E., Giraudat-Pautot, V., et al. (2009). Pluripotency of Arabidopsis xylem Pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57, 626–644. doi: 10.1111/j.1365-313X.2008.03715.x
Bahmankar, M., Mortazavian, S. M. M., Tohidfar, M., Sadat Noori, S. A., Izadi Darbandi, A., Corrado, G., et al. (2017). Chemical compositions, somatic embryogenesis, and Somaclonal variation in cumin. Biomed. Res. Int. 2017:e7283806. doi: 10.1155/2017/7283806
Balkunde, R., Kitagawa, M., Xu, X. M., Wang, J., and Jackson, D. (2017). Shoot Meristemless trafficking controls axillary meristem formation, meristem size and organ boundaries in Arabidopsis. Plant J. 90, 435–446. doi: 10.1111/tpj.13504
Bernula, D., Benkő, P., Kaszler, N., Domonkos, I., Valkai, I., Szőllősi, R., et al. (2020). Timely removal of exogenous Cytokinin and the prevention of auxin transport from the shoot to the root affect the regeneration potential of Arabidopsis roots. PCTOC 140, 327–339. doi: 10.1007/s11240-019-01730-3
Bhaskaran, S., and Smith, R. H. (1990). Regeneration in cereal tissue culture: A review. Crop Sci. 30, 1328–1337. doi: 10.2135/cropsci1990.0011183X003000060034x
Bhatia, S., and Bera, T. (2015). “Somatic embryogenesis and organogenesis,” in Modern Applications of Plant Biotechnology in Pharmaceutical Sciences. eds. S. Bhatia, K. Sharma, R. Dahiya, and T. Bera (Boston: Academic Press), 209–230.
Bidabadi, S. S., and Jain, S. M. (2020). Cellular, molecular, and physiological aspects of in vitro plant regeneration. Plan. Theory 9:702. doi: 10.3390/plants9060702
Braybrook, S. A., and Harada, J. J. (2008). Lecs go crazy in embryo development. Trends Plant Sci. 13, 624–630. doi: 10.1016/j.tplants.2008.09.008
Çabuk, B., and Özgen, M. (2016). The Effect of Different 2, 4-D Doses on Callus Induction and Chromosomal Structure in Maize (Zea mays L.). Infection 2, 188–194.
Carvalho, C. H. S., Bohorova, N., Bordallo, P. N., Abreu, L. L., Valicente, F. H., Bressan, W., et al. (1997). Type ii callus production and plant regeneration in tropical maize genotypes. Plant Cell Rep. 17, 73–76. doi: 10.1007/s002990050355
Chen, L., Tong, J., Xiao, L., Ruan, Y., Liu, J., Zeng, M., et al. (2016). Yucca-mediated auxin biogenesis is required for cell fate transition occurring During De novo root organogenesis in Arabidopsis. J. Exp. Bot. 67, 4273–4284. doi: 10.1093/jxb/erw213
Chimdessa, E. (2020). Composition and preparation of plant tissue culture medium. J. Tissue Cult. Bioengin. 3:120. doi: 10.29011/2688-6502.000020
Cho, M., and Zapata, F. (1988). Callus formation and plant regeneration in isolated pollen culture of Rice (Oryza sativa L. Cv. Taipei 309). Plant Sci. 58, 239–244. doi: 10.1016/0168-9452(88)90014-3
Chokheli, V. A., Dmitriev, P. A., Rajput, V. D., Bakulin, S. D., Azarov, A. S., Varduni, T. V., et al. (2020). Recent development in micropropagation techniques for rare plant species. Plants 9:1733. doi: 10.3390/plants9121733
Chou, J., Huang, J., and Huang, Y. (2020). Simple and efficient genetic transformation of Sorghum using immature inflorescences. Acta Physiol. Plant. 42, 1–8. doi: 10.1007/s11738-020-3023-6
Cortleven, A., Leuendorf, J. E., Frank, M., Pezzetta, D., Bolt, S., and Schmülling, T. (2019). Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 42, 998–1018. doi: 10.1111/pce.13494
Dai, W., and Castillo, C. (2007). Factors affecting plant regeneration from leaf tissues of buddleia species. HortScience 42, 1670–1673. doi: 10.21273/HORTSCI.42.7.1670
de Almeida, M., Graner, É. M., Brondani, G. E., de Oliveira, L. S., Artioli, F. A., de Almeida, L. V., et al. (2015). Plant morphogenesis: Theorical bases. Adv. Fore. Sci. 2, 13–22.
Dean Rider, S. Jr., Henderson, J. T., Jerome, R. E., Edenberg, H. J., Romero-Severson, J., and Ogas, J. (2003). Coordinate repression of regulators of embryonic identity by pickle During germination in Arabidopsis. Plant J. 35, 33–43. doi: 10.1046/j.1365-313X.2003.01783.x
Dhar, U., and Joshi, M. (2005). Efficient plant regeneration protocol through callus for Saussurea Obvallata (dc.) Edgew. (Asteraceae): effect of explant type, age and plant growth regulators. Plant Cell Rep. 24, 195–200. doi: 10.1007/s00299-005-0932-1
du, F., and Homeostasis, S. R. S. S. C. (2020). Integrated Signals Regulate Shoot Stem Cell Homeostasis. Mol. Plant 13:1535. doi: 10.1016/j.molp.2020.09.022
Duarte-Aké, F., Nic-Can, G., and De-la-Peña, C. (2019). “Somatic embryogenesis: polycomb complexes control cell-to-embryo transition,” in Fundamentals and Applications. eds. R. Alvarez-Venegas, C. De-la-Peña, and J. A. Casas-Mollano (United States: Springer), 339–354.
Duclercq, J., Sangwan-Norreel, B., Catterou, M., and Sangwan, R. S. (2011). De novo shoot organogenesis: from art to science. Trends Plant Sci. 16, 597–606. doi: 10.1016/j.tplants.2011.08.004
Duncan, D., Williams, M., Zehr, B., and Widholm, J. (1985). The production of callus capable of plant regeneration from immature embryos of numerous Zea mays genotypes. Planta 165, 322–332. doi: 10.1007/BF00392228
El-Sherif, N. A. (2018). Impact of Plant Tissue Culture on Agricultural Sustainability. Sustainability of Agricultural Environment in Egypt: Part II. United States: Springer. p. 93–107.
Farhadi, N., Panahandeh, J., Azar, A. M., and Salte, S. A. (2017). Effects of explant type, growth regulators and light intensity on callus induction and plant regeneration in four ecotypes of Persian shallot (Allium Hirtifolium). Sci. Hortic. 218, 80–86. doi: 10.1016/j.scienta.2016.11.056
Fehér, A. (2015). Somatic embryogenesis—stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta. 1849, 385–402. doi: 10.1016/j.bbagrm.2014.07.005.
Fehér, A. (2019). Callus, dedifferentiation, Totipotency, somatic embryogenesis: what These terms mean in the era of molecular plant biology? Front. Plant Sci. 10:536. doi: 10.3389/fpls.2019.00536
Firoozabady, E., and Moy, Y. (2004). Regeneration of pineapple plants via somatic embryogenesis and organogenesis. In Vitro Cell. Dev. Biol. 40, 67–74. doi: 10.1079/IVP2003494
Fitch, M. M. M., and Manshardt, R. M. (1990). Somatic embryogenesis and plant regeneration from immature zygotic embryos of papaya (Carica papaya L.). Plant Cell Rep. 9, 320–324. doi: 10.1007/BF00232860
Gaj, M. D. (2004). Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) Heynh. Plant Growth Regul. 43, 27–47. doi: 10.1023/B:GROW.0000038275.29262.fb
Gaj, M. D. (2011). “Somatic embryogenesis and plant regeneration in the culture of Arabidopsis thaliana (L.) Heynh. Immature zygotic embryos,” in Plant Embryo Culture. eds. T. A. Thorpe and E. C. Yeung (Berlin, Heidelberg: Springer), 257–265.
Ge, F., Luo, X., Huang, X., Zhang, Y., He, X., Liu, M., et al. (2016). Genome-wide analysis of transcription factors involved in maize embryonic callus formation. Physiol. Plant. 158, 452–462. doi: 10.1111/ppl.12470
Gepts, P. (2002). A comparison between crop domestication, classical plant breeding, and genetic engineering. Crop Sci. 42, 1780–1790. doi: 10.2135/cropsci2002.1780
Gerdakaneh, M., Badakhshan, H., Mohamadi, M., and Arji, I. (2020). Effect of different media and growth regulators on micropropagation of Gf677. J. Plant Prod. 43, 241–254. doi: 10.22055/PPD.2019.27439.1667
Gordon, S. P., Heisler, M. G., Reddy, G. V., Ohno, C., Das, P., and Meyerowitz, E. M. (2007). Pattern formation During De novo assembly of the Arabidopsis shoot meristem. Development 134, 3539–3548. doi: 10.1242/dev.010298
Greenwood, M. S., Cui, X., and Xu, F. (2001). Response to auxin changes During maturation-related loss of adventitious rooting competence in loblolly pine (Pinus taeda) stem cuttings. Physiol. Plant. 111, 373–380. doi: 10.1034/j.1399-3054.2001.1110315.x
Gulzar, B., Mujib, A., Malik, M. Q., Sayeed, R., Mamgain, J., and Ejaz, B. (2020). Genes, proteins and other networks regulating somatic embryogenesis in plants. J. Gene. Engin. Biotechnol. 18, 1–15. doi: 10.1186/s43141-020-00047-5
Gupta, S. D., and Karmakar, A. (2017). Machine vision based evaluation of impact of light emitting diodes (Leds) on shoot regeneration and the effect of spectral quality on phenolic content and antioxidant capacity in Swertia Chirata. J. Photochem. Photobiol. B Biol. 174, 162–172. doi: 10.1016/j.jphotobiol.2017.07.029
Haberlandt, G. (1902). Cultur Versuche Mit Isolierten Pflanzenzellen Sitz. Ber Mat Nat ki Kais Akad Wiss, Wien 111, 69–92.
Haecker, A., Groß-Hardt, R., Geiges, B., Sarkar, A., Breuninger, H., Herrmann, M., et al. (2004). Expression dynamics ofWOXgenes mark cell fate decisions during early embryonic patterning inArabidopsis thaliana. J. Embryol. Exp. Morphol. 131, 657–668. doi: 10.1242/dev.00963
Haliloglu, K., and Aydin, M. (2016). Efficient regeneration system from Rye Leaf Base segments. Springerplus 5:2005. doi: 10.1186/s40064-016-3689-9
Han, Y., Broughton, S., Liu, L., Zhang, X.-Q., Zeng, J., He, X., et al. (2021). Highly efficient and genotype-independent barley gene editing based on anther culture. Plant commun. 2:100082. doi: 10.1016/j.xplc.2020.100082
Han, H., Liu, X., and Zhou, Y. (2020). Transcriptional circuits in control of shoot stem cell Homeostasis. Curr. Opin. Plant Biol. 53, 50–56. doi: 10.1016/j.pbi.2019.10.004
Hecht, V., Vielle-Calzada, J.-P., Hartog, M. V., Schmidt, E. D., Boutilier, K., Grossniklaus, U., et al. (2001). The Arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816. doi: 10.1104/pp.010324
Heck, G. R., Perry, S. E., Nichols, K. W., and Fernandez, D. E. (1995). Agl15, a mads domain protein expressed in developing embryos. Plant Cell 7, 1271–1282. doi: 10.1105/tpc.7.8.1271
Hibberd, K. A. (1984). Induction, selection, and characterization of mutants in maize cell. Culture 1, 571–576.
Hill, K., and Schaller, G. E. (2013). Enhancing plant regeneration in tissue culture: A molecular approach through manipulation of Cytokinin sensitivity. Plant Signal. Behav. 8:e25709. doi: 10.4161/psb.25709
Hinchliffe, A., and Harwood, W. A. (2019). Agrobacterium-mediated transformation of barley immature embryos. Barley. Humana Press, New York, NY 115–126.
Hodges, T. K., Kamo, K. K., Becwar, M. R., and Schroll, S. (2012). Regeneration of Maize. United States: Elsevier 15–33.
Hoque, M. E., and Mansfield, J. W. (2004). Effect of genotype and explant age on callus induction and subsequent plant regeneration from root-derived callus of Indica Rice genotypes. Plant Cell Tissue Organ Cult. 78, 217–223. doi: 10.1023/B:TICU.0000025640.75168.2d
Horstman, A., Bemer, M., and Boutilier, K. (2017). A transcriptional view on somatic embryogenesis. Regeneration 4, 201–216. doi: 10.1002/reg2.91
Horstman, A., Li, M., Heidmann, I., Weemen, M., Chen, B., Muino, J. M., et al. (2017). The Baby Boom transcription factor activates the Lec1-Abi3-Fus3-Lec2 network to induce somatic embryogenesis. Plant Physiol. 175, 848–857. doi: 10.1104/pp.17.00232
Hu, H., Xiong, L., and Yang, Y. (2005). Rice Serk1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta 222, 107–117. doi: 10.1007/s00425-005-1534-4
Husain, M. K., Anis, M., and Shahzad, A. (2010). Somatic embryogenesis and plant regeneration in Pterocarpus marsupium Roxb. Trees 24, 781–787. doi: 10.1007/s00468-010-0448-3
Ikeda, Y., Banno, H., Niu, Q.-W., Howell, S. H., and Chua, N.-H. (2006). The enhancer of shoot regeneration 2 gene in Arabidopsis regulates cup-shaped Cotyledon 1 at the transcriptional level and controls Cotyledon development. Plant Cell Physiol. 47, 1443–1456. doi: 10.1093/pcp/pcl023
Ikeuchi, M., Favero, D. S., Sakamoto, Y., Iwase, A., Coleman, D., Rymen, B., et al. (2019). Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 70, 377–406. doi: 10.1146/annurev-arplant-050718-100434
Ikeuchi, M., Ogawa, Y., Iwase, A., and Sugimoto, K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143, 1442–1451. doi: 10.1242/dev.134668
Ishihara, H., Sugimoto, K., Tarr, P. T., Temman, H., Kadokura, S., Inui, Y., et al. (2019). Primed histone demethylation regulates shoot regenerative competency. Nat. Commun. 10, 1–15. doi: 10.1038/s41467-019-09386-5
Iwase, A., Harashima, H., Ikeuchi, M., Rymen, B., Ohnuma, M., Komaki, S., et al. (2017). Wind1 promotes shoot regeneration through transcriptional activation of enhancer of shoot Regeneration1 in Arabidopsis. Plant Cell 29, 54–69. doi: 10.1105/tpc.16.00623
Iwase, A., Mita, K., Nonaka, S., Ikeuchi, M., Koizuka, C., Ohnuma, M., et al. (2015). Wind1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 128, 389–397. doi: 10.1007/s10265-015-0714-y
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Kojima, M., Arai, T., et al. (2011). The Ap2/erf transcription factor Wind1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 21, 508–514. doi: 10.1016/j.cub.2011.02.020
Jha, P., Ochatt, S. J., and Kumar, V. (2020). Wuschel: A master regulator in plant growth signaling. Plant Cell Rep. 39, 431–444. doi: 10.1007/s00299-020-02511-5
Jiménez, V. M. (2005). Involvement of plant hormones and plant growth regulators on in vitro somatic embryogenesis. Plant Growth Regul. 47, 91–110. doi: 10.1007/s10725-005-3478-x
Jiménez, V. M., and Thomas, C. (2006). “Participation of plant hormones in determination and progression of somatic embryogenesis,” in Somatic Embryogenesis. eds. A. Mujib and J. Šamaj (Berlin, Heidelberg: Springer), 103–118.
Jin, L., Yarra, R., Zhou, L., Zhao, Z., and Cao, H. (2020). miRNAs as key regulators via targeting the phytohormone signaling pathways during somatic embryogenesis of plants. Biotech 10:495. doi: 10.1007/s13205-020-02487-9
Jones, T., Lowe, K., Hoerster, G., Anand, A., Wu, E., Wang, N., et al. (2019). “Maize transformation using the Morphogenic genes Baby Boom and Wuschel2,” in Transgenic Plants (New York, NY: Humana Press), 81–93.
Kareem, A., Durgaprasad, K., Sugimoto, K., Du, Y., Pulianmackal, A. J., Trivedi, Z. B., et al. (2015). Plethora genes control regeneration by a two-step mechanism. Curr. Biol. 25, 1017–1030. doi: 10.1016/j.cub.2015.02.022
Karim, R., Tan, Y. S., Singh, P., Nuruzzaman, M., Khalid, N., and Harikrishna, J. A. (2018). Expression and DNA methylation of Met1, Cmt3 and Drm2 During in vitro culture of Boesenbergia rotunda (L.) Mansf. Philippine Agri. Sci. 101, 261–270.
Kaviani, B. (2011). Conservation of plant genetic resources by cryopreservation. Aust. J. Crop. Sci. 5, 778–800.
Kim, J. Y., Yang, W., Forner, J., Lohmann, J. U., Noh, B., and Noh, Y. S. (2018). Epigenetic reprogramming by histone acetyltransferase Hag1/Atgcn5 is required for pluripotency Acquisition in Arabidopsis. EMBO J. 37:e98726. doi: 10.15252/embj.201798726
Kumar, R., Mamrutha, H. M., Kaur, A., Venkatesh, K., Grewal, A., Kumar, R., et al. (2017). Development of an efficient and reproducible regeneration system in wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plants 23, 945–954. doi: 10.1007/s12298-017-0463-6
Kwong, R. W., Bui, A. Q., Lee, H., Kwong, L. W., Fischer, R. L., Goldberg, R. B., et al. (2003). Leafy Cotyledon1-Like defines a class of regulators essential for embryo development. Plant Cell 15, 5–18. doi: 10.1105/tpc.006973
Lardon, R., and Geelen, D. (2020). Natural variation in plant pluripotency and regeneration. Plants 9:1261. doi: 10.3390/plants9101261
Lee, M. H., Lee, J., Jie, E. Y., Choi, S. H., Jiang, L., Ahn, W. S., et al. (2020). Temporal and Spatial Expression Analysis of Shoot-Regeneration Regulatory Genes During the Adventitious Shoot Formation in Hypocotyl and Cotyledon Explants of Tomato (Cv. Micro-Tom). Int. J. Mol. Sci. 21:5309. doi: 10.3390/ijms21155309
Lee, K., Park, O.-S., and Seo, P. J. (2018). Atxr2 as a Core regulator of De novo root organogenesis. Plant Signal. Behav. 13:e1449543. doi: 10.1080/15592324.2018.1449543
Leibfried, A., To, J. P., Busch, W., Stehling, S., Kehle, A., Demar, M., et al. (2005). Wuschel controls meristem function by direct regulation of Cytokinin-inducible response regulators. Nature 438, 1172–1175. doi: 10.1038/nature04270
Lenhard, M., Jürgens, G., and Laux, T. (2002). The Wuschel and Shootmeristemless genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206. doi: 10.1242/dev.129.13.3195
Lepiniec, L., Devic, M., Roscoe, T., Bouyer, D., Zhou, D.-X., Boulard, C., et al. (2018). Molecular and epigenetic regulations and functions of the Lafl transcriptional regulators That control seed development. Plant Reprod. 31, 291–307. doi: 10.1007/s00497-018-0337-2
Li, W., Liu, H., Cheng, Z. J., Su, Y. H., Han, H. N., Zhang, Y., et al. (2011). DNA methylation and histone modifications regulate De novo shoot regeneration in Arabidopsis by modulating Wuschel expression and auxin signaling. PLoS Genet. 7:e1002243. doi: 10.1371/journal.pgen.1002243
Li, W., Masilamany, P., Kasha, K. J., and Pauls, K. P. (2002). Developmental, Tissue culture, and genotypic factors affecting plant regeneration from shoot apical meristems of germinated Zea mays L. seedlings. In Vitro Cel. Dev. Biol. 38, 285–292. doi: 10.1079/IVP2002291
Li, H., Yao, L., Sun, L., and Zhu, Z. (2021). Correction: Ethylene insensitive 3 suppresses plant De novo root regeneration from leaf explants and mediates age-regulated regeneration decline. Development 148:dev199635. doi: 10.1242/dev.199635
Liu, L., Gallagher, J., Arevalo, E. D., Chen, R., Skopelitis, T., Wu, Q., et al. (2021). Enhancing grain-yield-related traits by Crispr–Cas9 promoter editing of maize Cle genes. Nat. Plants 7, 287–294. doi: 10.1038/s41477-021-00858-5
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014). Wox11 and 12 are involved in the first-step cell fate transition During De novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093. doi: 10.1105/tpc.114.122887
Long, Y., Yang, Y., Ge, F., Pan, G., and Shen, Y. (2020). Establishment of a maize callus regeneration system from haploid shoot tips. PCTOC 141, 583–592. doi: 10.1007/s11240-020-01817-2
Lotan, T., Ohto, M.-a., Yee, K. M., West, M. A., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis leafy Cotyledon1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/S0092-8674(00)81463-4
Lowe, K., La Rota, M., Hoerster, G., Hastings, C., Wang, N., Chamberlin, M., et al. (2018). Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev. Biol. Plant 54, 240–252. doi: 10.1007/s11627-018-9905-2
Luerßen, H., Kirik, V., Herrmann, P., and Miséra, S. (1998). Fusca3 encodes a protein with a conserved Vp1/Abi3-Like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15, 755–764. doi: 10.1046/j.1365-313X.1998.00259.x
Mahmood, I., and Razzaq, A. (2017). Responses of explant type of wheat (Triticum aestivum L.) genotypes to different tissue culture media. J. Natl. Sci. Found. 45:265. doi: 10.4038/jnsfsr.v45i3.8191
Malik, K., Birla, D., Yadav, H., Sainger, M., Chaudhary, D., and Jaiwal, P. K. (2017). Evaluation of carbon sources, gelling agents, growth hormones and additives for efficient callus induction and plant regeneration in Indian wheat (Triticum aestivum L.) genotypes using mature embryos. J. Crop. Sci. Biotechnol. 20, 185–192. doi: 10.1007/s12892-017-0046-0
Malik, S. I., Rashid, H., Yasmin, T., and Minhas, N. M. (2004). Plant regeneration by somatic embryogenesis from callus of mature seed explants of bread wheat (Triticum aestivum L.). Pak. J. Bot. 36, 629–634.
Matsuo, N., Makino, M., and Banno, H. (2011). Arabidopsis enhancer of shoot regeneration (Esr) 1 and Esr2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci. 181, 39–46. doi: 10.1016/j.plantsci.2011.03.007
Méndez-Hernández, H. A., Ledezma-Rodríguez, M., Avilez-Montalvo, R. N., Juárez-Gómez, Y. L., Skeete, A., Avilez-Montalvo, J., et al. (2019). Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 10:77. doi: 10.3389/fpls.2019.00077
Meng, W. J., Cheng, Z. J., Sang, Y. L., Zhang, M. M., Rong, X. F., Wang, Z. W., et al. (2017). Type-B Arabidopsis response regulators specify the shoot stem cell niche by dual regulation of Wuschel. Plant Cell 29, 1357–1372. doi: 10.1105/tpc.16.00640
Michalczuk, L., Cooke, T. J., and Cohen, J. D. (1992). Auxin levels at different stages of carrot somatic embryogenesis. Phytochemistry 31, 1097–1103. doi: 10.1016/0031-9422(92)80241-6
Miguel, C., and Marum, L. (2011). An epigenetic view of plant cells cultured in vitro: Somaclonal variation and Beyond. J. Exp. Bot. 62, 3713–3725. doi: 10.1093/jxb/err155
Minutolo, M., Chiaiese, P., Di Matteo, A., Errico, A., and Corrado, G. (2020). Accumulation of ascorbic acid in tomato cell culture: influence of the genotype, source explant and time of in vitro cultivation. Antioxidants 9:222. doi: 10.3390/antiox9030222
Moraes, R. M., Cerdeira, A. L., and Lourenço, M. V. (2021). Using micropropagation to develop medicinal plants into crops. Molecules 26:1752. doi: 10.3390/molecules26061752
Mozgova, I., and Hennig, L. (2015). The Polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296. doi: 10.1146/annurev-arplant-043014-115627
Nic-Can, G. I., and Loyola-Vargas, V. M. (2016). “The role of the auxins during somatic embryogenesis,” in Somatic Embryogenesis: Fundamental Aspects and Applications. eds. V. Loyola-Vargas and N. Ochoa-Alejo (Switzerland: Springer), 171–182.
Nishiwaki, M., Fujino, K., Koda, Y., Masuda, K., and Kikuta, Y. (2000). Somatic embryogenesis induced by the simple application of Abscisic acid to carrot (Daucus carota L.) seedlings in culture. Planta 211, 756–759. doi: 10.1007/s004250000387
Nissen, S. J., and Sutter, E. G. (1990). Stability of Iaa and Iba in nutrient medium to several tissue culture procedures. HortScience 25, 800–802. doi: 10.21273/HORTSCI.25.7.800
Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). Pickle is a Chd3 chromatin-remodeling factor That regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. 96, 13839–13844. doi: 10.1073/pnas.96.24.13839
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). Arf7 and Arf19 regulate lateral root formation via direct activation of Lbd/Asl genes in Arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Orłowska, A., and Kępczyńska, E. (2018). Identification of Polycomb repressive Complex1, Trithorax group genes and their simultaneous expression with Wuschel, Wuschel-related Homeobox5 and shoot Meristemless During the induction phase of somatic embryogenesis in Medicago truncatula Gaertn. PCTOC 134, 345–356. doi: 10.1007/s11240-018-1425-6
Pan, J., Zhao, F., Zhang, G., Pan, Y., Sun, L., Bao, N., et al. (2019). Control of De novo root regeneration efficiency by developmental status of Arabidopsis leaf explants. J. Genet. Genomics 46, 133–140. doi: 10.1016/j.jgg.2019.03.001
Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during arabidopsis seed development: roles of the Abi3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582. doi: 10.1105/tpc.6.11.1567
Parveen, S., and Shahzad, A. (2014). Somatic embryogenesis and plantlet regeneration of Cassia angustifolia from immature Cotyledon-derived callus. Biol. Plant. 58, 411–418. doi: 10.1007/s10535-014-0409-6
Petrásek, J., and Friml, J. (2009). Auxin transport routes in plant development. Development 136, 2675–2688. doi: 10.1242/dev.030353
Quintana-Escobar, A. O., Nic-Can, G. I., Galaz Avalos, R. M., Loyola-Vargas, V. M., and Gongora-Castillo, E. (2019). Transcriptome analysis of the induction of somatic embryogenesis in Coffea Canephora and the participation of Arf and aux/Iaa genes. PeerJ 7:e7752. doi: 10.7717/peerj.7752
Rakshana, P., Valarmathi, R., and Raveendran, M. (2019). Optimization of tissue culture protocol for rapid regeneration of traditional therapeutic Rice genotype ‘Kavuni’. Elect. J. Plant Breed. 10, 334–340. doi: 10.5958/0975-928X.2019.00043.7
Rakshit, S., Rashid, Z., Sekhar, J., Fatma, T., and Dass, S. (2010). Callus induction and whole plant regeneration in elite Indian maize (Zea mays L.) Inbreds. PCTOC 100, 31–37. doi: 10.1007/s11240-009-9613-z
Ramsay, J. L., Galitz, D. S., and Lee, C. W. (2003). Basal medium and sucrose concentration influence regeneration of Easter lily in ovary culture. HortScience 38, 404–406. doi: 10.21273/HORTSCI.38.3.404
Saeed, T., and Shahzad, A. (2015). High frequency plant regeneration in Indian Siris via cyclic somatic embryogenesis with biochemical, histological and Sem investigations. Ind. Crop. Prod. 76, 623–637. doi: 10.1016/j.indcrop.2015.07.060
Saeedpour, A., Jahanbakhsh Godehkahriz, S., Lohrasebi, T., Esfahani, K., Salmanian, A. H., and Razavi, K. (2021). The effect of endogenous hormones, Total antioxidant and Total phenol changes on regeneration of barley cultivars. Iran. J. Biotechnol. 19, 30–39. doi: 10.30498/IJB.2021.2838
Sahai, A., Shahzad, A., and Sharma, S. (2010). Histology of organogenesis and somatic embryogenesis in excised root cultures of an endangered species Tylophora Indica (Asclepiadaceae). Aust. J. Bot. 58, 198–205. doi: 10.1071/BT09220
Salaün, C., Lepiniec, L., and Dubreucq, B. (2021). Genetic and molecular control of somatic embryogenesis. Plan. Theory 10:1467.
Sang, Y. L., Cheng, Z. J., and Zhang, X. S. (2018). Plant Stem Cells and De Novo Organogenesis. New Phytol. 218, 1334–1339. doi: 10.1111/nph.15106
Satish, L., Rathinapriya, P., Ceasar, S. A., Rency, A. S., Pandian, S., Rameshkumar, R., et al. (2016). Effects of Cefotaxime, amino acids and carbon source on somatic embryogenesis and plant regeneration in four Indian genotypes of foxtail millet (Setaria italica L.). In Vitro Cell. Dev. Biol. 52, 140–153. doi: 10.1007/s11627-015-9724-7
Sato, A., and Yamamoto, K. T. (2008). Overexpression of the non-canonical aux/Iaa genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol. Plant. 133, 397–405. doi: 10.1111/j.1399-3054.2008.01055.x
Schmidt, E., Guzzo, F., Toonen, M., and De Vries, S. (1997). A leucine-rich repeat containing receptor-Like kinase Marks somatic plant cells competent to form embryos. Development 124, 2049–2062. doi: 10.1242/dev.124.10.2049
Schwarz, O. J., and Beaty, R. M. (2018). “Organogenesis,” in Plant Tissue Culture Concepts and Laboratory Exercises (Oxford, UK: Routledge), 125–138.
Shin, J., Bae, S., and Seo, P. J. (2020). De novo shoot organogenesis During plant regeneration. J. Exp. Bot. 71, 63–72. doi: 10.1093/jxb/erz395
Shin, J., and Seo, P. J. (2018). Varying auxin levels induce distinct pluripotent states in callus cells. Front. Plant Sci. 9:1653. doi: 10.3389/fpls.2018.01653
Siddique, A. B., and Islam, S. S. (2015). Effect of light and dark on callus induction and regeneration in tobacco (Nicotiana tabacum L.). Bangladesh J. Bot. 44, 643–651. doi: 10.3329/bjb.v44i4.38636
Simonović, A. D., Trifunović-Momčilov, M., Filipović, B. K., Marković, M. P., Bogdanović, M. D., and Subotić, A. R. (2021). Somatic embryogenesis in Centaurium erythraea Rafn—current status and perspectives: a review. Plan. Theory 10:70. doi: 10.3390/plants10010070
Singh, A. (2015). “Micropropagation of Plants,” in Plant Biology and Biotechnology. eds. B. Bahadur, M. Venkat Rajam, L. Sahijram, and K. V. Krishnamurthy (United States: Springer), 329–346.
Singla, B., Khurana, J. P., and Khurana, P. (2008). Characterization of three somatic embryogenesis receptor kinase genes from wheat, Triticum aestivum. Plant Cell Rep. 27, 833–843. doi: 10.1007/s00299-008-0505-1
Singla, B., Tyagi, A. K., Khurana, J. P., and Khurana, P. (2007). Analysis of expression profile of selected genes expressed During auxin-induced somatic embryogenesis in Leaf Base system of wheat (Triticum aestivum) and their possible interactions. Plant Mol. Biol. 65, 677–692. doi: 10.1007/s11103-007-9234-z
Skoog, F., and Miller, C. (1957). Chemical regulation of growth and organ formation in plant tissues cultured. Vitro Symp. Soc. Exp. Biol. 11, 118–130.
Steward, F. C., Mapes, M. O., and Smith, J. (1958). Growth and organized development of cultured cells. I. Growth and division of freely suspended cells. Am. J. Bot. 45, 693–703. doi: 10.1002/j.1537-2197.1958.tb12224.x
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). Leafy Cotyledon2 encodes a B3 domain transcription factor That induces embryo development. Proc. Natl. Acad. Sci. 98, 11806–11811. doi: 10.1073/pnas.201413498
Street, H. E., and Henshaw, G. G. (1966). “Introduction and methods employed in plant tissue culture” in Cells and Tissues in Culture Methods, Biology and Physiology. ed. E. N. Willmer (London, NY: Academic Press), 459–532.
Su, Y. H., Zhou, C., Li, Y. J., Yu, Y., Tang, L. P., Zhang, W. J., et al. (2020). Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. 117, 22561–22571. doi: 10.1073/pnas.2015248117
Sugimoto, K., and Meyerowitz, E. M. (2013).Regeneration in Arabidopsis tissue culture. Plant Organogenesis. ed. Ive SmetDe Totowa, NJ: Springer p. 265–275.
Sugimoto, K., Temman, H., Kadokura, S., and Matsunaga, S. (2019). To regenerate or not to regenerate: factors That drive plant regeneration. Curr. Opin. Plant Biol. 47, 138–150. doi: 10.1016/j.pbi.2018.12.002
Sundararajan, S., Sivaraman, B., Rajendran, V., and Ramalingam, S. (2017). Tissue culture and agrobacterium-mediated genetic transformation studies in four commercially important Indica Rice cultivars. J. Crop. Sci. Biotechnol. 20, 175–183. doi: 10.1007/s12892-017-0045-0
Suzuki, M., Wang, H. H.-Y., and McCarty, D. R. (2006). Repression of the leafy Cotyledon 1/B3 regulatory network in plant embryo development by Vp1/Abscisic acid insensitive 3-Like B3 genes. Plant Physiol. 143, 902–911. doi: 10.1104/pp.106.092320
Tanaka, M., Kikuchi, A., and Kamada, H. (2008). The Arabidopsis histone deacetylases Hda6 and Hda19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146, 149–161. doi: 10.1104/pp.107.111674
Toonen, M. A., Hendriks, T., Schmidt, E. D., Verhoeven, H. A., van Kammen, A., and de Vries, S. C. (1994). Description of somatic-embryo-forming single cells in carrot suspension cultures employing video cell tracking. Planta 194, 565–572. doi: 10.1007/BF00714471
Tvorogova, V. E., Fedorova, Y. A., Potsenkovskaya, E. A., Kudriashov, A. A., Efremova, E. P., Kvitkovskaya, V. A., et al. (2019). The Wuschel-related Homeobox transcription factor Mtwox9-1 stimulates somatic embryogenesis in Medicago truncatula. PCTOC 138, 517–527. doi: 10.1007/s11240-019-01648-w
Verdeil, J.-L., Alemanno, L., Niemenak, N., and Tranbarger, T. J. (2007). Pluripotent versus totipotent plant stem cells: dependence versus autonomy? Trends Plant Sci. 12, 245–252. doi: 10.1016/j.tplants.2007.04.002
Vitamvas, J., Viehmannova, I., Cepkova, P. H., Mrhalova, H., and Eliasova, K. (2019). Assessment of Somaclonal variation in indirect morphogenesis-derived plants of Arracacia xanthorrhiza. Pesq. Agrop. Brasileira 54:e00301. doi: 10.1590/s1678-3921.pab2019.v54.00301
Wang, C., Ma, H., Zhu, W., Zhang, J., Zhao, X., and Li, X. (2021). Seedling-derived leaf and root tip as alternative explants for callus induction and plant regeneration in maize. Physiol. Plant. 172, 1570–1581. doi: 10.1111/ppl.13347
Wang, X., Niu, Q.-W., Teng, C., Li, C., Mu, J., Chua, N.-H., et al. (2009). Overexpression of Pga37/Myb118 and Myb115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 19, 224–235. doi: 10.1038/cr.2008.276
Wang, X.-D., Nolan, K. E., Irwanto, R. R., Sheahan, M. B., and Rose, R. J. (2011). Ontogeny of embryogenic callus in Medicago truncatula: The fate of the pluripotent and totipotent stem cells. Ann. Bot. 107, 599–609. doi: 10.1093/aob/mcq269
Wang, W., Zhao, X., Zhuang, G., Wang, S., and Chen, F. (2008). Simple hormonal regulation of somatic embryogenesis and/or shoot organogenesis in caryopsis cultures of Pogonatherum Paniceum (Poaceae). Plant Cell Tissue Organ Cult. 95, 57–67. doi: 10.1007/s11240-008-9414-9
Wernicke, W., Potrykus, I., and Thomas, E. (1982). Morphogenesis from cultured leaf tissue of Sorghum bicolor—the morphogenetic pathways. Protoplasma 111, 53–62. doi: 10.1007/BF01287646
Wójcik, A. M., Nodine, M. D., and Gaj, M. D. (2017). Mir160 and Mir166/165 contribute to the Lec2-mediated auxin response involved in the somatic embryogenesis induction in Arabidopsis. Front. Plant Sci. 8:2024. doi: 10.3389/fpls.2017.02024
Wójcik, A. M., Wójcikowska, B., and Gaj, M. D. (2020). Current perspectives on the auxin-mediated genetic network That controls the induction of somatic embryogenesis in plants. Int. J. Mol. Sci. 21:1333. doi: 10.3390/ijms21041333
Wójcikowska, B., Jaskóła, K., Gąsiorek, P., Meus, M., Nowak, K., and Gaj, M. D. (2013). Leafy Cotyledon2 (Lec2) promotes embryogenic induction in somatic tissues of Arabidopsis, Via Yucca-Mediated Auxin Biosynthesis. Planta 238, 425–440. doi: 10.1007/s00425-013-1892-2
Wójcikowska, B., Wójcik, A. M., and Gaj, M. D. (2020). Epigenetic regulation of auxin-induced somatic embryogenesis in plants. Int. J. Mol. Sci. 21:2307. doi: 10.3390/ijms21072307
Xu, C., and Hu, Y. (2020). The molecular regulation of cell pluripotency in plants. aBIOTECH 1, 169–177. doi: 10.1007/s42994-020-00028-9
Xu, L., and Huang, H. (2014). Genetic and epigenetic controls of plant regeneration. Curr. Top. Dev. Biol. 108, 1–33. doi: 10.1016/B978-0-12-391498-9.00009-7
Yang, X., and Zhang, X. (2010). Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 29, 36–57. doi: 10.1080/07352680903436291
Yaseen, M., Ahmad, T., Sablok, G., Standardi, A., and Hafiz, I. A. (2013). Role of carbon sources for in vitro plant growth and development. Mol. Biol. Rep. 40, 2837–2849. doi: 10.1007/s11033-012-2299-z
Ye, X., She, M., Wang, K., and Xu, H. (2012). Identification, cloning, and potential application of genes related to somatic embryogenesis in plant tissue culture. Acta Agron. Sin. 38, 191–201. doi: 10.3724/SP.J.1006.2012.00191
Yu, Y., Qin, W., Li, Y., Zhang, C., Wang, Y., Yang, Z., et al. (2019). Red light promotes cotton embryogenic callus formation by influencing endogenous hormones, polyamines and Antioxidative enzyme activities. Plant Growth Regul. 87, 187–199. doi: 10.1007/s10725-018-0461-x
Yu, H., Wang, W., Wang, Y., and Hou, B. (2012). High frequency wheat regeneration from leaf tissue explants of regenerated plantlets. Adv. Biosci. Biotechnol. 3, 46–50. doi: 10.4236/abb.2012.31008
Yumbla-Orbes, M., da Cruz, A. C. F., Pinheiro, M. V. M., Rocha, D. I., Batista, D. S., Koehler, A. D., et al. (2017). Somatic embryogenesis and De novo shoot organogenesis Can be alternatively induced by reactivating Pericycle cells in Lisianthus (Eustoma grandiflorum (Raf.) Shinners) root explants. In Vitro Cell. Dev. Biol. 53, 209–218. doi: 10.1007/s11627-017-9800-2
Zayova, E., Geneva, M., Stancheva, I., Dimitrova, L., Petrova, M., Hristozkova, M., et al. (2018). Evaluation of the antioxidant potential of in vitro propagated hyssop (Hyssopus officinalis L.) with different plant growth regulators. Med. Plan. Int. J. Phytomed. Rela. Indus. 10, 295–304. doi: 10.5958/0975-6892.2018.00044.8
Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of aux/Iaa proteins. Proc. Natl. Acad. Sci. 98, 11795–11800. doi: 10.1073/pnas.211312798
Zhang, L., DeGennaro, D., Lin, G., Chai, J., and Shpak, E. D. (2021). Erecta family signaling constrains Clavata3 and Wuschel to the center of the shoot apical meristem. Development 148:dev189753.
Zhang, T.-Q., Lian, H., Tang, H., Dolezal, K., Zhou, C.-M., Yu, S., et al. (2015). An intrinsic Microrna timer regulates progressive decline in shoot regenerative capacity in plants. Plant Cell 27, 349–360. doi: 10.1105/tpc.114.135186
Zhang, T.-Q., Lian, H., Zhou, C.-M., Xu, L., Jiao, Y., and Wang, J.-W. (2017). A two-step model for De novo activation of Wuschel During plant shoot regeneration. Plant Cell 29, 1073–1087. doi: 10.1105/tpc.16.00863
Zhang, S., Liu, X., Lin, Y., Xie, G., Fu, F., Liu, H., et al. (2011). Characterization of a Zmserk gene and its relationship to somatic embryogenesis in a maize culture. PCTOC 105, 29–37. doi: 10.1007/s11240-010-9834-1
Zhang, M., Wang, A., Qin, M., Qin, X., Yang, S., Su, S., et al. (2021). Direct and indirect somatic embryogenesis induction in Camellia Oleifera Abel. Front. Plant Sci. 12:451. doi: 10.3389/fpls.2021.644389
Zimmerman, J. L. (1993). Somatic embryogenesis: A model for early development in higher plants. Plant Cell 5:1411. doi: 10.2307/3869792
Keywords: somatic embryogenesis, de novo organogenesis, environmental factors, molecular mechanisms, plant regeneration
Citation: Long Y, Yang Y, Pan G and Shen Y (2022) New Insights Into Tissue Culture Plant-Regeneration Mechanisms. Front. Plant Sci. 13:926752. doi: 10.3389/fpls.2022.926752
Received: 23 April 2022; Accepted: 31 May 2022;
Published: 30 June 2022.
Edited by:
Paloma Moncaleán, Neiker Tecnalia, SpainReviewed by:
Piotr Tomasz Bednarek, Plant Breeding and Acclimatization Institute, PolandCopyright © 2022 Long, Yang, Pan and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Long, d29haWN1b2N1b2RhQDE2My5jb20=; Yaou Shen, c2hlbnlhb3VAc2ljYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.