
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 June 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.925744
This article is part of the Research Topic The Role of Stress Proteins in Plants Under Abiotic Stress View all 5 articles
Plants have evolved defense mechanisms to overcome unfavorable climatic conditions. The growth and development of plants are regulated in response to environmental stress. In this study, we investigated the molecular and physiological characteristics of a novel gene PagSAP11 in hybrid poplar (Populus alba × Populus tremula var. glandulosa) under drought stress. PagSAP11, a stress-associated protein (SAP) family gene, encodes a putative protein containing an A20 and AN1 zinc-finger domain at its N- and C-termini, respectively. Knockdown of PagSAP11 transgenic poplars (SAP11-Ri) enhanced their tolerance to drought stress compared with wild type plants. Moreover, the RNAi lines showed increased branching of lateral shoots that led to a gain in fresh weight, even when grown in the living modified organism (LMO) field. In SAP11-Ri transgenic plants, the expression levels of genes involved in axillary bud outgrowth and cell proliferation such as DML10, CYP707A and RAX were increased while the DRM gene which involved in bud dormancy was down-regulated. Taken together, these results indicate that PagSAP11 represents a promising candidate gene for engineering trees with improved stress tolerance and growth during unfavorable conditions.
Plants lead a sedentary life; therefore, they have developed various mechanisms to adapt to changes in the surrounding environment (El-Sappah et al., 2022). Drought is an adverse environmental condition that can affect plant growth and development (El-Sappah and Rather, 2022). Global changes in drought have the potential to trigger widespread die-offs of trees, which could further exacerbate global warming and thus affect ecosystem products and service globally. Forest trees have adapted various defense strategies for drought stress (Lata and Prasad, 2011; Jia et al., 2020; Kim et al., 2020; Jian et al., 2021). For example, plants attempt to minimize water loss by reducing transpiration rate and surface area of the leaves (Estravis-Barcala et al., 2020). At the molecular level, dehydration-responsive-element-binding (DREBs) proteins, WRKY transcription factor and NAC (NAM, ATAF, and CUC) transcription factor regulate the expressions of stress-responsive genes such as LEA (late-embryogenesis) and aquaporin genes (Harfouche et al., 2014; Estravis-Barcala et al., 2020).
One such important family of stress response regulatory proteins, the Stress-Associated Protein (SAP) genes encode a novel class of zinc-finger proteins that are well conserved among various plant species. A distinct feature of SAP protein is the presence of A20/AN1 zinc-finger domains. The A20 zinc finger domain has Cx2-4Cx11Cx2C consensus sequence at the N-terminus whereas AN1 zinc finger domain has two consensus sequences: Cx2Cx9-12Cx1-2Cx4Cx2Hx5HxC and the Cx4Cx9-12Cx1-2Cx4Cx2Hx5HxC at the C-terminus (Jin et al., 2007). The A20 domain of SAPs was first identified as a tumor necrosis factor (TNF) induced gene from the human umbilical cord (Opipari et al., 1990). Furthermore, the AN1 domain was first identified in Xenopus laevis, a protein encoded by animal hemisphere 1 (AN1) maternal RNA (Jin et al., 2007). SAP family genes have been isolated from various plants, including Arabidopsis thaliana, Populus euphratica, Populus trichocarpa, Medicago truncaula, Brassica napus and Zea mays after the identification of the first SAP gene (OsSAP1) in Oryza sativa (Vij and Tyagi, 2006; Jin et al., 2007; Jia et al., 2016).
To date, several SAP genes have been identified that play important roles in the regulation of plant responses to abiotic stress. OsSAP1 improves tolerance to cold, drought, and salinity stresses in transgenic tobacco (Mukhopadhyay et al., 2004). Overexpression of AtSAP5 confers salt, drought and osmotic tolerance in Arabidopsis (Kang et al., 2011). While, GmSAP16-overexpressing plants showed improved resistance to drought and salinity stress and increased abscisic acid (ABA) sensitivity in Arabidopsis (Zhang et al., 2019). Further, overexpression of MdSAP15 in Arabidopsis led to enhanced osmotic and drought stress tolerance (Dong et al., 2018). In addition, silencing of SlSAP4 resulted in reduced resistance to Botrytis cinerea infection in tomato (Liu et al., 2019). In woody plants, it was identified that down-regulation of PagSAP1 enhances tolerance to salt stress in hybrid poplar (Yoon et al., 2018). Li et al. (2019) also reported that overexpression of PtSAP13 increased resistance to salt stress in Arabidopsis. Although several SAPs have been identified in various plants, their exact biological functions in woody plants are not yet fully understood.
Owing to its rapid growth rate, high productivity and extensive adaptation to environment, poplar is used as resource for biofuels and other bioproducts. With the availability of its full genome sequence and the establishment of transgenic techniques, poplar has become a model organism for studying woody plants (Bradshaw et al., 2001; Sannigrahi et al., 2010; Guo et al., 2012). Considering that productivity of poplars closely depends on water availability (Khan et al., 2016), increased drought tolerance is important factor for improving its utility.
In this study, we carried out the genetic and molecular characterization of PagSAP11 in hybrid poplar (Populus alba × Populus tremula var. glandulosa). We generated PagSAP11 transgenic poplar plants to investigate the function of PagSAP11. Downregulated expression of PagSAP11 in transgenic plants results in increased number of branches and leads to improved tolerance to drought stress. Our results suggest that PagSAP11 plays an important role in drought stress response and regulation of plant architecture.
Hybrid poplars [P. alba × P. tremula var. glandulosa, clone Bonghwa (BH)] were used as both WT and transgenic plants in this study. Control and transgenic plants were propagated in vitro prior to acclimation in pots containing commercial horticultural potting soil. The plants were then cultivated in growth chambers (16 h light/8 h dark cycle; light intensity, 150 μM m−2 s−1; 24°C), followed by acclimation in soil and growth for an additional 6 weeks until they reached a plant height of approximately 15 cm. Then, plants were transferred to LMO field and grown there at the Forest Bioresources Department of the National Institute of Forest Science, Republic of Korea (latitude 37.2 N, longitude 126.9E).
For tissue-specific gene expression analysis, leaf, stem, and root tissues were collected from 1-year-old poplar plants whereas flower tissues were collected from 25-year-old hybrid poplar plants.
A cDNA library was constructed as described by Lee et al. (2005) and blasted against previously identified plant SAP genes. The selected cDNA clone (GenBank accession number CX653572.1) was sequenced, and the deduced amino acid sequences and molecular weights of the products were determined using CLC Main Workbench (Qiagen, Aarhus, Denmark). Multiple sequence alignment and phylogenetic tree analysis were performed using the ClustalW (Sievers et al., 2011) and MEGA11 (Tamura et al., 2021) based on previously deduced amino acid sequences of SAP proteins such as from AtSAP3 (NP_001189620.1), AtSAP5 (NP_566429.1), AtSAP7 (NP_192941.1), AtSAP9 (NP_194013.1), and AtSAP10 (NP_194268.1) from A. thaliana; GmSAP5 (XP_003540534.1) from Glycine max; MeSAP5 (XP_021624685.1) from Manihot esculenta; OsSAP1 (XP_015651267), OsSAP11 (XP_015651039.1), and OsSAP15 (XP_015639532.1) from O. sativa; PeSAP5 (XP_011007335.1) from Populus euphratica; PtSAP4 (XP_002298442.1) and PtSAP11 (XP_002314027.1) from P. trichocarpa; PpSAP5 (XP_007218519.1) from Prunus persica; RcSAP5 (XP_002513177.1) from Ricinus communis; VvSAP5 (XP_003633555.1) from Vitis vinifera.
Total RNA was isolated from various tissues of hybrid poplar (clone BH) plants using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Reverse-transcription quantitative PCR (RT-qPCR) primers were designed using the Primer3 program.1 Gene expression levels were determined using the 2−△△Ct method (Livak and Schmittgen, 2001). The first-strand cDNA was synthesized using EcoDry™ premix (Takara, Kyoto, Japan) and RT-qPCR was performed using the SYBR Green PCR Master Mix (Bio-Rad Laboratories, CA, United States) and a CFX96™ Real-Time System (Bio-Rad Laboratories, CA, United States). The cycling conditions for amplification were performed according to the manufacturer’s instructions. All primers used for RT-qPCR are listed in Supplementary Table 1.
To determine the subcellular localization of PagSAP11, eGFP was fused in-frame with the C-terminus of PagSAP11. The coding sequence of PagSAP11 was amplified from the genomic DNA of hybrid poplar (clone BH) using gene-specific primers (Supplementary Table 1). The PCR products were cloned into the pENTR/dTOPO Vector (Invitrogen, CA, United States). Correct construction of the clones was confirmed by sequencing. The PagSAP11 fragment was then cloned into the Gateway version of pCsVMV-eGFP-N-999 vector by LR recombination reaction (Kim et al., 2008). Next, CsVMV::PagSAP11-eGFP recombinant plasmid was introduced into poplar protoplasts using the polyethylene glycol method (Abe and Theologis, 1998). The localization of PagSAP11-eGFP was observed using confocal laser scanning microscope (Leica, TCS SP5, Wetzlar, Germany).
PCR amplification of PagSAP11 was performed using a specific primer set, using a forward primer (5′-GAA TCT AGA GGA TCC ACT TTT GAG AAG ATG-3′) and a reverse primer (5′-ATA AAC AAT GAG CTC AGT CCA TGG C-3′). The cycling conditions for amplification were performed according to the manufacturer’s instructions. To construct the RNAi vector, a partial sequence of PagSAP11 was inserted into the pH7GWIWG vector (Karimi et al., 2002) using the Gateway system (Invitrogen, CA, United States). A schematic diagram of vector construction is shown in Figure 1. To construct the vector for PagSAP11-OX, the PCR products were ligated into the pGEM-T Easy Vector (Promega, WI, United States). The vector was digested with XbaI and SacI and inserted into the pBI121 binary vector. Plant transformation was performed as previously described by Choi et al. (2005). The transformed cells were selected on MS medium containing 1 mg/l 2, 4-D, 0.1 mg/l naphthaleneacetic acid (NAA), 0.01 mg/l benzyladenine (BA), and 2 mg/l hygromycin for the RNAi lines, or 50 mg/l kanamycin for the OX lines. Putative transgenic shoots were regenerated in woody plant medium (WPM; Duchefa, Haarlem, Netherlands) containing 1.0 mg/l t-zeatin, 0.1 mg/l BA, and 0.01 mg/l NAA under the same concentrations of antibiotics. The regenerated shoots were transferred to MS medium containing 0.2 mg/l indole-3-butyric acid (IBA) for rooting.
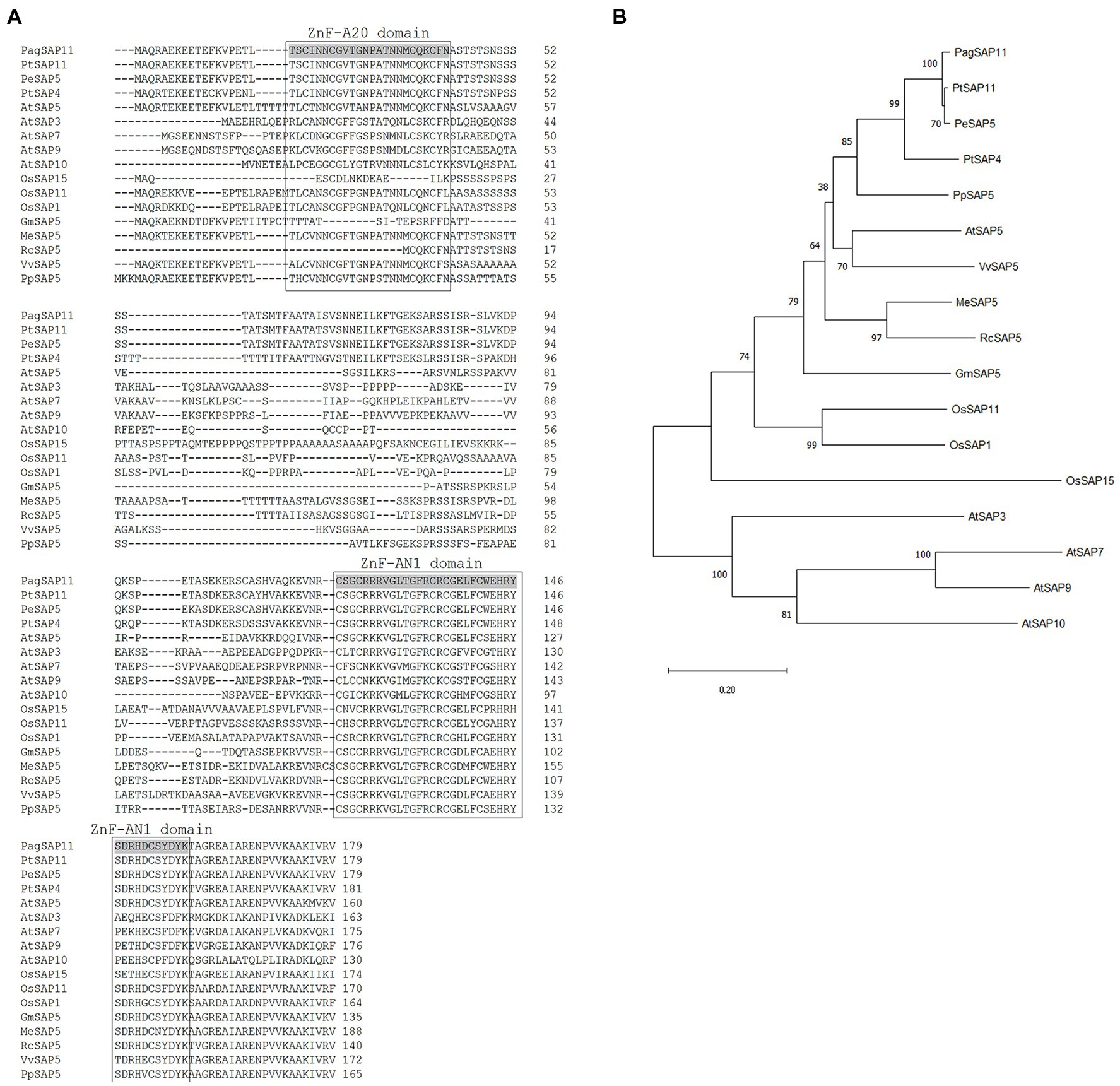
Figure 1. Comparison of the predicted protein sequences of PagSAP11 with other plant SAP proteins. (A) Multiple sequence alignment of PagSAP11 and related proteins from Populus trichocarpa, Populus euphratica, Arabidopsis thaliana, Oryza sativa, Glycine max, Manihot esculenta, Ricinus communis, Vitis vinifera, and Prunus persica. (B) Phylogenetic analysis of proteins from (A).
To estimate expression levels of PagSAP11 under stress conditions, WT poplar (clone BH) plants were grown in soil pots under the normal watering conditions for 8 weeks and then subjected to the following treatments: water (control), 150 mM NaCl and 10% PEG for 24 h. For heat and cold treatments, 2-month-old WT poplars grown in soil under normal conditions were exposed to 40°C or 4°C for 24 h in growth chambers (Vision Scientific, Daejeon, Korea). To investigate response to drought stress, WT and transgenic poplar trees were grown in soil under the normal condition for 8 weeks and then watering was suspended for 10 days. Afterward, the photochemical efficiency of photosystem II (PSII) and chlorophyll contents were measured in the 6-8th leaves from the top using a plant efficiency analyzer (Hansatech Instruments, Morfolk, UK) and ethanol extraction method (Lee et al., 2016; Cho et al., 2021), respectively. To analyze water loss, the detached leaves exposed to dehydration on a clean-bench (Hu et al., 2010). Briefly, five fully expanded leaves from three 8-week-old soil-acclimated WT and transgenic poplar trees were detached and placed on a clean-bench. Leaves were weighed at the indicated times to measure the rate of water loss.
All data represent the mean values (±standard deviation) from at least three independent replicates. Statistical analysis was conducted using the Student T-test, and significance level is indicated by asterisks.
Analysis of the expression profiles under drought stress will provide insights for investigation of candidates; thus, we first analyzed the expression patterns of SAP family genes of the hybrid poplar (P. alba × P. tremula var. glandulosa, clone BH) in response to drought stress using qRT-PCR. We detected SAP genes based on the public database of P. trichocarpa.2 As a result, except for three SAPs (SAP3, −14 and − 18), the expression of other SAPs increased under drought conditions (Supplementary Figure S1). Among them, four SAPs (SAP7, −11, −13 and − 16) exhibited significantly up-regulated expression patterns under drought stress. Notably, according to the previous report, the expression of PtSAP11 increased under salt treatment (Li et al., 2019). Taken together, we selected SAP11 for further investigation of gene function.
Thereafter, we determined the full-length coding sequence (CDS) of PagSAP11. The full-length CDS of PagSAP11 without introns was 540 bp long, and encoded protein was estimated to comprise 179 amino acids, with one AN1 domain and one A20 domain (Figure 2A). The sequence data was submitted to GenBank (Accession number ON519817). The molecular weight of PagSAP11 was predicted 19.65 kDa. We also performed a multiple alignment analysis based on the amino acid sequences of PagSAP11 with genetically closed proteins from other plant species. The amino acid sequence of PagSAP11 shared 98% identity with PtSAP11 and 84% with PtSAP4 from P. trichocarpa, 97% with PeSAP5 from P. euphratica, 67% with PpSAP5 from P. persica, 61% with AtSAP5 from Arabidopsis, 60% with VvSAP5 from V. vinifera, and 52% with OsSAP11 from O. sativa (Figure 2A). To identify the phylogenetic relationships of PagSAP11, we developed an unrooted phylogenetic tree using the protein sequence of PagSAP11 and its orthologs (Figure 2B). As shown in Figure 2B, PagSAP11 was clustered in the same clade as PtSAP11 and PeSAP5. In addition, PagSAP11 was closely associated with AtSAP5, which is involved in conferring resistance towards salt stress, osmotic stress and water deficiency (Kang et al., 2011). These results suggest that PagSAP11 is a suitable candidate gene associated with drought stress response.
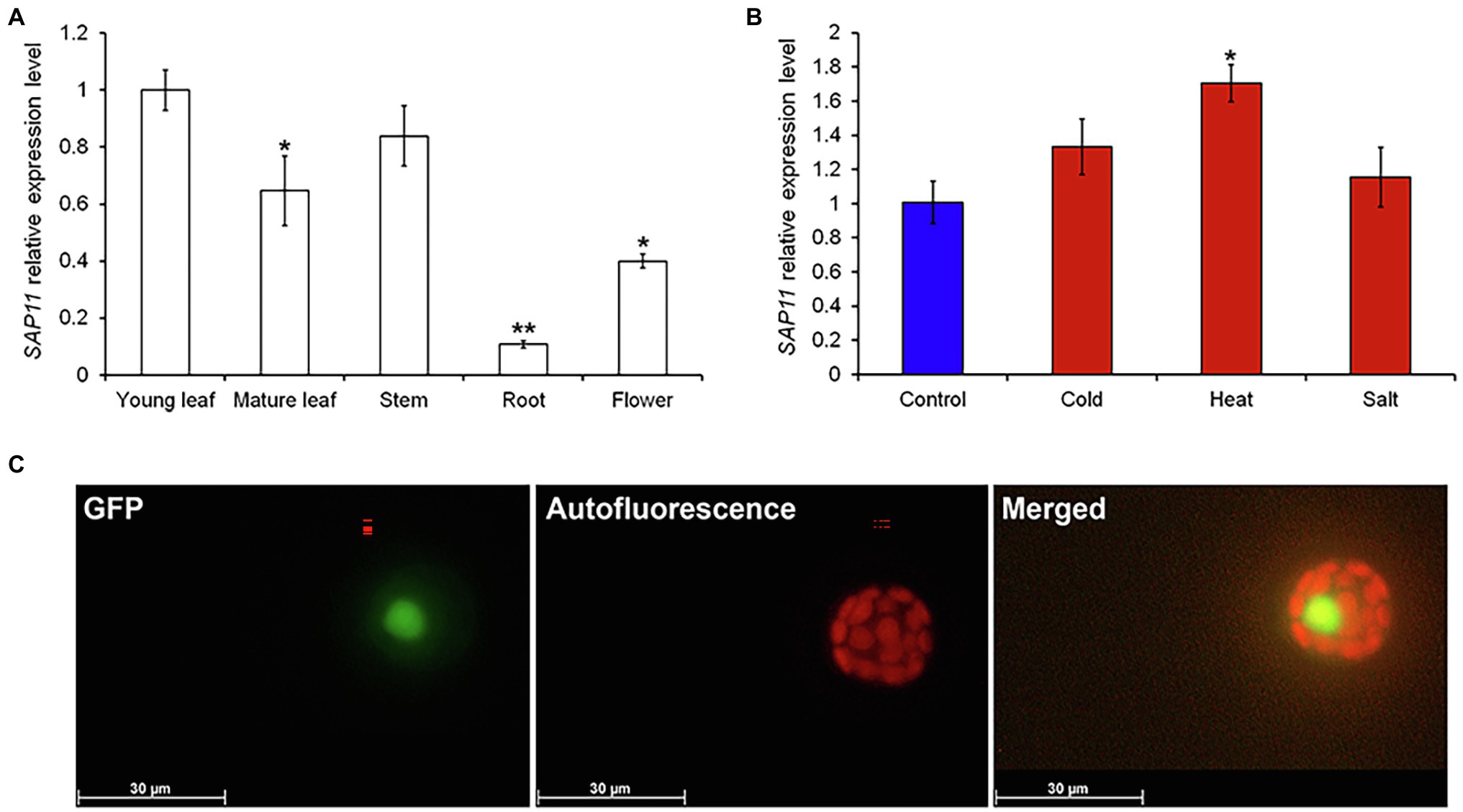
Figure 2. Characterization of PagSAP11. (A) Expression analysis of PagSAP11 gene in poplar tissues by qRT-PCR: young leaf, mature leaf, stem, root and flower. (B) Expression analysis of PagSAP11 under abiotic stress conditions (cold, heat and salt). The values represent the means ± standard deviations (SDs) of three biological replicates. Asterisks indicate significant differences by student t-test (*p < 0.05, **p < 0.005). (C) Subcellular localization of PagSAP11-GFP protein. The green color indicates PagSAP11-GFP expression and red color means autofluorescence. Scale bar = 30 μm.
To elucidate the molecular function of PagSAP11, we estimated expression patterns of PagSAP11 gene in various tissues and under stress conditions. Transcript levels of PagSAP11 were highest in young leaves (Figure 3A). Additionally, expression of PagSAP11 increased under heat as well as drought stress conditions (Figure 3B; Supplementary Figure S1). Further, we analyzed the subcellular localization of PagSAP11 by transient expression of PagSAP11-GFP fusion protein in protoplasts from hybrid poplars (Figure 3C). We observed strong GFP signals in the nucleus (Figure 3C), indicating that PagSAP11 is a nuclear protein.
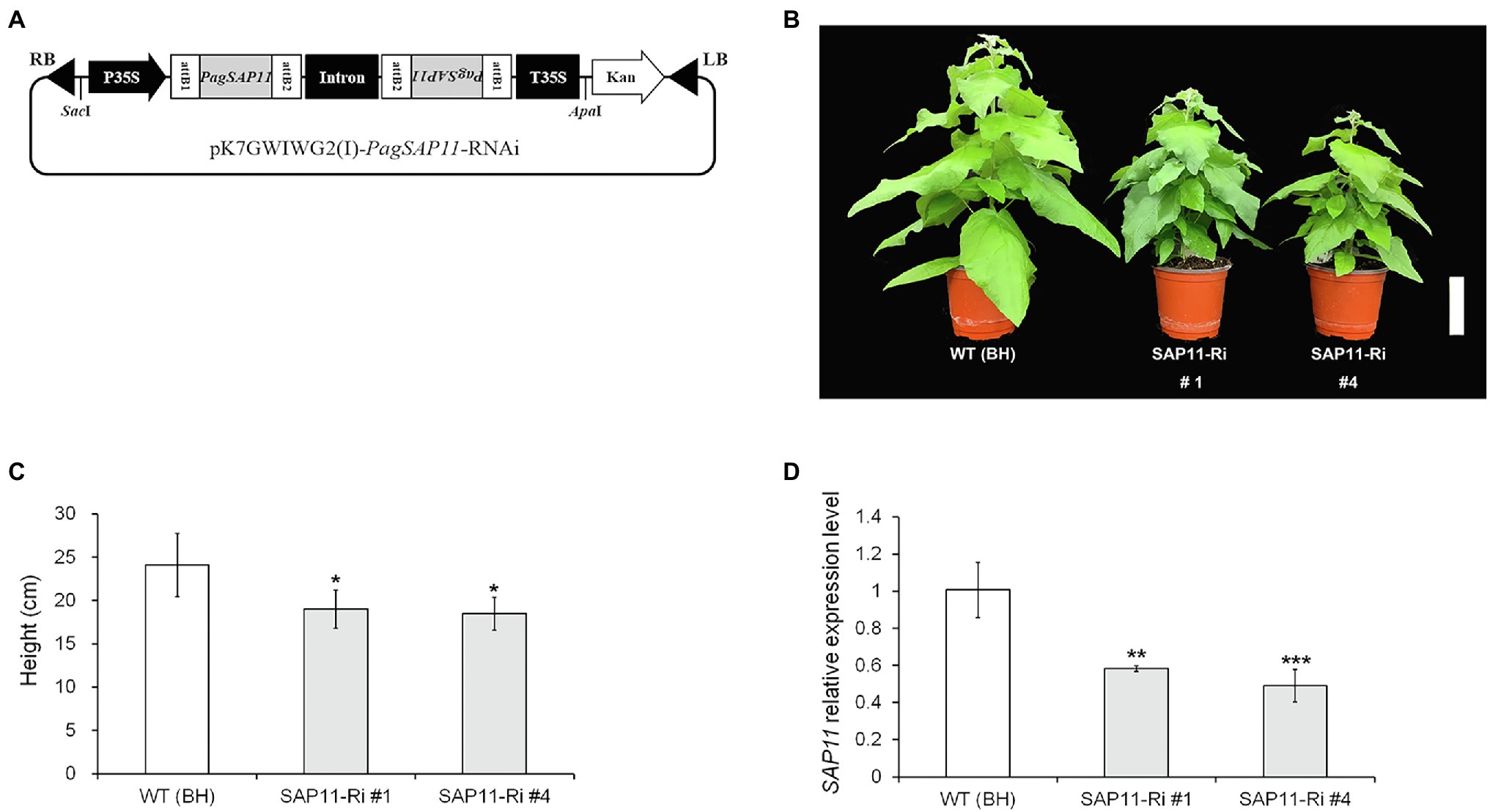
Figure 3. Generation of PagSAP11-RNAi transgenic poplar using Agrobacterium-mediated transformation. (A) Strategy for construction of expression vector pK7GWIWG2(I), containing the PagSAP11-RNAi. (B) Representative image of WT (clone BH) and independent two transgenic (SAP11-Ri #1 and #4) poplar plants grown for 2 months on soil after acclimation (n = 15). (C) Measurement of plant heights from (B). (D) Relative PagSAP11 transcript levels in each transgenic line. The expression levels of transgenic plants were confirmed using real-time qPCR. The values represent the means ± standard deviations (SDs) of three biological replicates. Asterisks indicate significant differences by student t-test (*p < 0.05, **p < 0.005, ***p < 0.001). Scale bar = 10 cm.
To explore the function of PagSAP11, we generated both knockdown (SAP11-Ri) and overexpressing (SAP11-OX) transgenic poplars. Under normal growth conditions, the two independently transformed lines of SAP11-Ri (SAP11-Ri #1 and #4) exhibited reduced shoot height and developed lateral branches (Figures 1B,C). Transcript levels of PagSAP11 were lower in knockdown transgenic poplars than in the WT poplar plants (Figure 1D). In contrast, PagSAP11-overexpressing plants also showed relatively reduced shoot height (Supplementary Figure S2) but exhibited a phenotype similar to that of WT plants. Overall, these results suggested that PagSAP11 is associated with lateral shoot growth.
To analyze the drought stress resistance of SAP11-Ri plants, we suspended the watering of 8-week-old WT and SAP11-Ri plants for 10 days. The plants were then kept in a growth room and maintained at low humidity. As a result, compared to the WT plants, SAP11-Ri plants showed drought stress tolerance even after 10 days without watering (Figure 4A). Thereafter, to evaluate the drought resistance of SAP11-Ri plants, we measured transpirational water loss of detached leaves from WT and transgenic plants. Leaves from SAP11-Ri plants had a significantly reduced water loss, while leaves from WT plants lost about 20% of their FW (Figure 4B). In addition, we also estimated photochemical efficiency of photosystem II (PSII) and chlorophyll contents in detached leaves from WT and SAP11-Ri plants. As shown in Figure 4C, the photochemical efficiency of PSII, a representative physiological marker of leaf senescence (Lee et al., 2016), rapidly declined in WT leaves compared to SAP11-Ri leaves. Consequently, chlorophyll contents of SAP11-Ri leaves were slowly decreased compared to WT leaves (Figure 4D). Moreover, we found that the expression levels of LEA, RD29B (Responsive to Desiccation 29B), ACS11 (1-Aminocyclopropane-1-Carnoxylate Synthase 11) and GolS (Inositol-3-α-Galactosyltransferase) were increased in WT leaves, whereas they were slightly increased or unchanged in SAP11-Ri leaves (Figure 4E). In contrast, SAP11-OX lines showed a drought-sensitive phenotype under drought stress (Supplementary Figure S3). Taken together, these results indicate that the knockdown of PagSAP11 confers drought resistance.
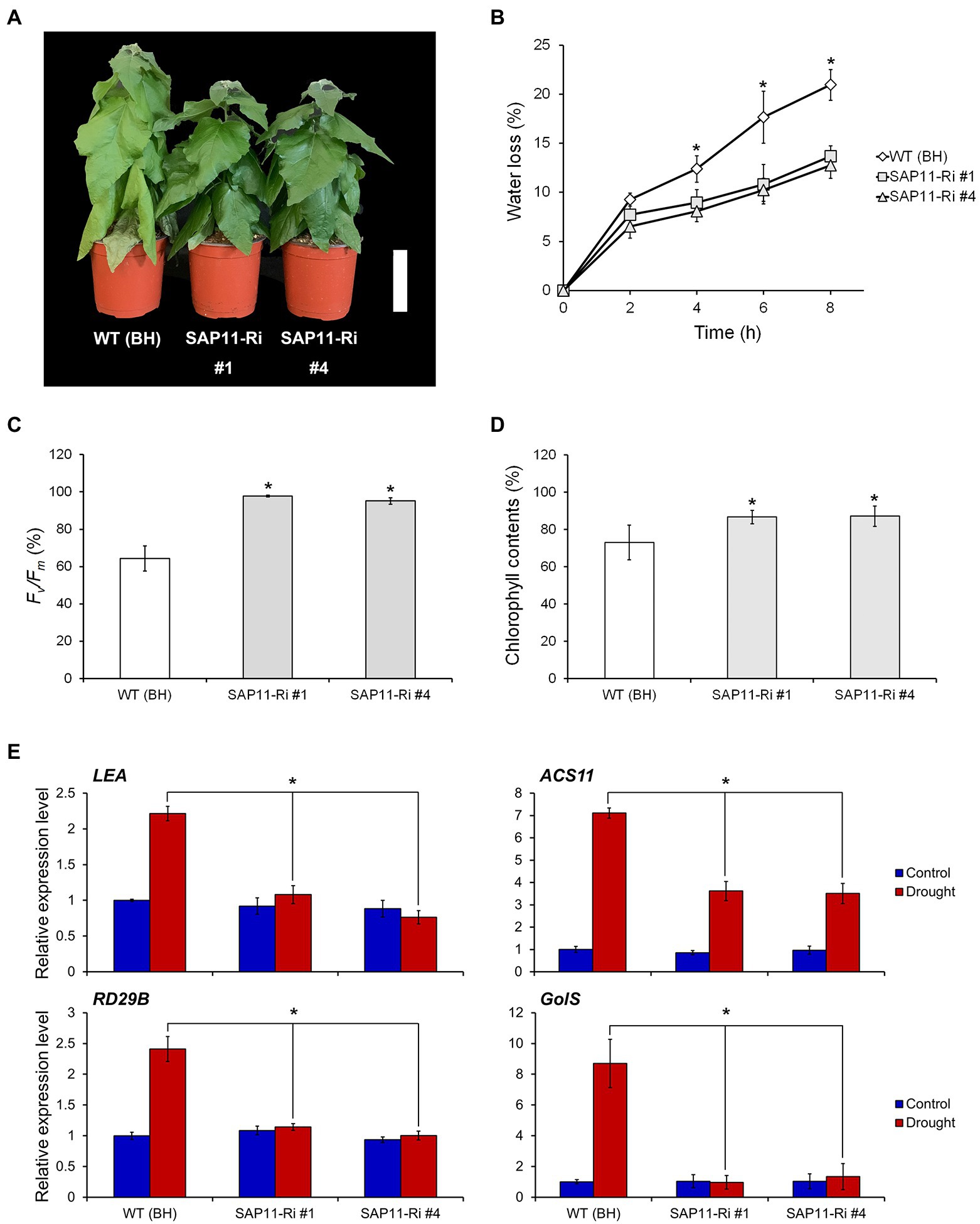
Figure 4. Knockdown of PagSAP11 improves drought tolerance. (A) Phenotype of WT (BH clone) and transgenic poplars (SAP11-Ri #1, #4) after water withdrawal for 10 days. (B-D) Transpirational water loss (B), photochemical efficiency (C), and chlorophyll contents (D) were measured in detached leaves from WT and transgenic plants (n = 5). (E) Expression changes of drought-responsive genes in WT and transgenic poplars after water withdrawal for 3 days. The values represent the means ± standard deviations (SDs) of three biological replicates. Asterisks indicate significant differences by student t-test (*p < 0.05, **p < 0.005). Scale bar = 10 cm.
As described above, SAP11-Ri plants showed reduced shoot height and an increased number of lateral branches compared to WT plants (Figure 1). To estimate association between PagSAP11 and shoot branching, we measured the number of lateral branches in WT and SAP11-Ri plants. As a result, SAP11-Ri plants showed increased branch numbers compared with WT plants (Figures 5A,B), whereas the SAP11-OX plants produced only a few branches (Supplementary Figures S4A,B). Next, we analyzed the expression levels of genes involved in axillary bud formation and bud dormancy. The mRNA abundances of DML10 (DEMETER-like demethylase), CYP707A (Cryptochrome P450 707A family) and RAX (Regulator of Axillary meristems) were increased in SAP11-Ri plants, while the transcript levels of DRM (Dormancy-associated protein 1) were reduced (Figure 5C). In addition, the transcript levels of these genes were unchanged or decreased in SAP11-OX plants compared with WT plants (Supplementary Figure S4C). However, the expression levels of auxin, cytokinin and strigolactone-related genes did not change significantly between SAP11-Ri and SAP11-OX plants compared with WT plants (Supplementary Figure S5). Overall, these results indicate that PagSAP11 is involved in shoot branching via regulating axillary bud formation and bud dormancy.
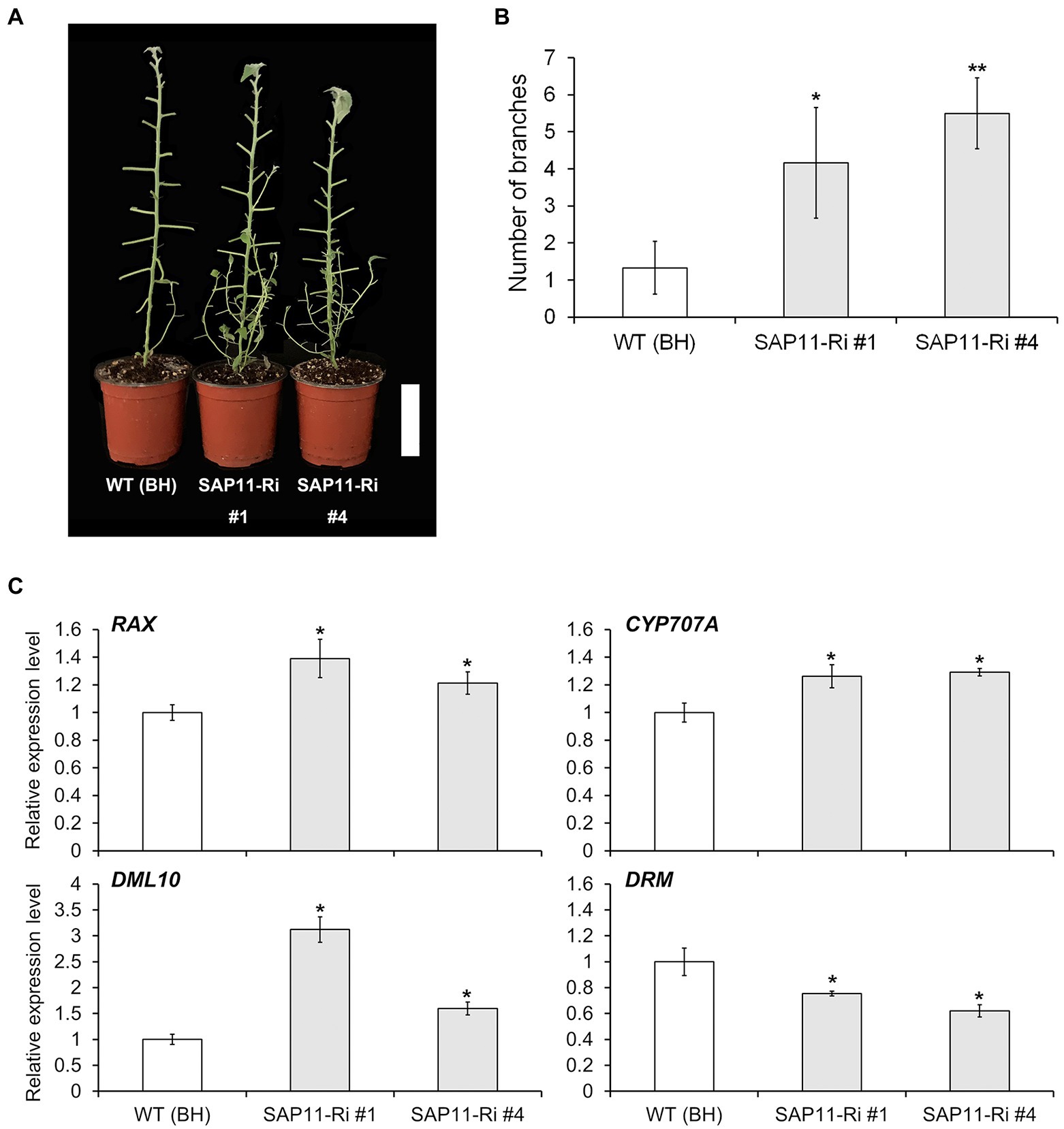
Figure 5. PagSAP11-RNAi hybrid poplars exhibit increased sylleptic branching. (A) Sylleptic branching in representative defoliated WT (clone BH) and transgenic poplars. (B) Sylleptic branch number in WT and transgenic poplars grown for 2 months on the soil after acclimation (n = 5). (C) Relative expression levels of genes involved in bud outgrowth. The values represent the means ± standard deviations (SDs) of three biological replicates. Asterisks indicate significant differences by student t-test (*p < 0.05, **p < 0.005). Scale bar = 10 cm.
To evaluate the enhanced lateral bud outgrowth of SAP11-Ri plants under LMO field conditions, we planted SAP11-Ri transgenic and WT poplars in the LMO facility and monitored their growth performance. After 3 months of active growth in the spring to summer seasons of the second year, we measured stem height and diameter of transgenic poplars (Figure 6). Selected trees were photographed in the winter (Figure 6A). SAP11-Ri poplars exhibited decreased shoot height compared to the WT poplars (Figure 6B). Next, we estimated the number of their side stalks and sylleptic branches. As a result, SAP11-Ri poplars showed increased number of side stalks and sylleptic branches (Figures 6C,D). Moreover, SAP11-Ri poplars showed increased dry weight due of an increase in lateral shoot branching (Figure 6E). Therefore, these results suggest that increased sylleptic shoot branching supports maintenance of biomass in SAP11-Ri plants.
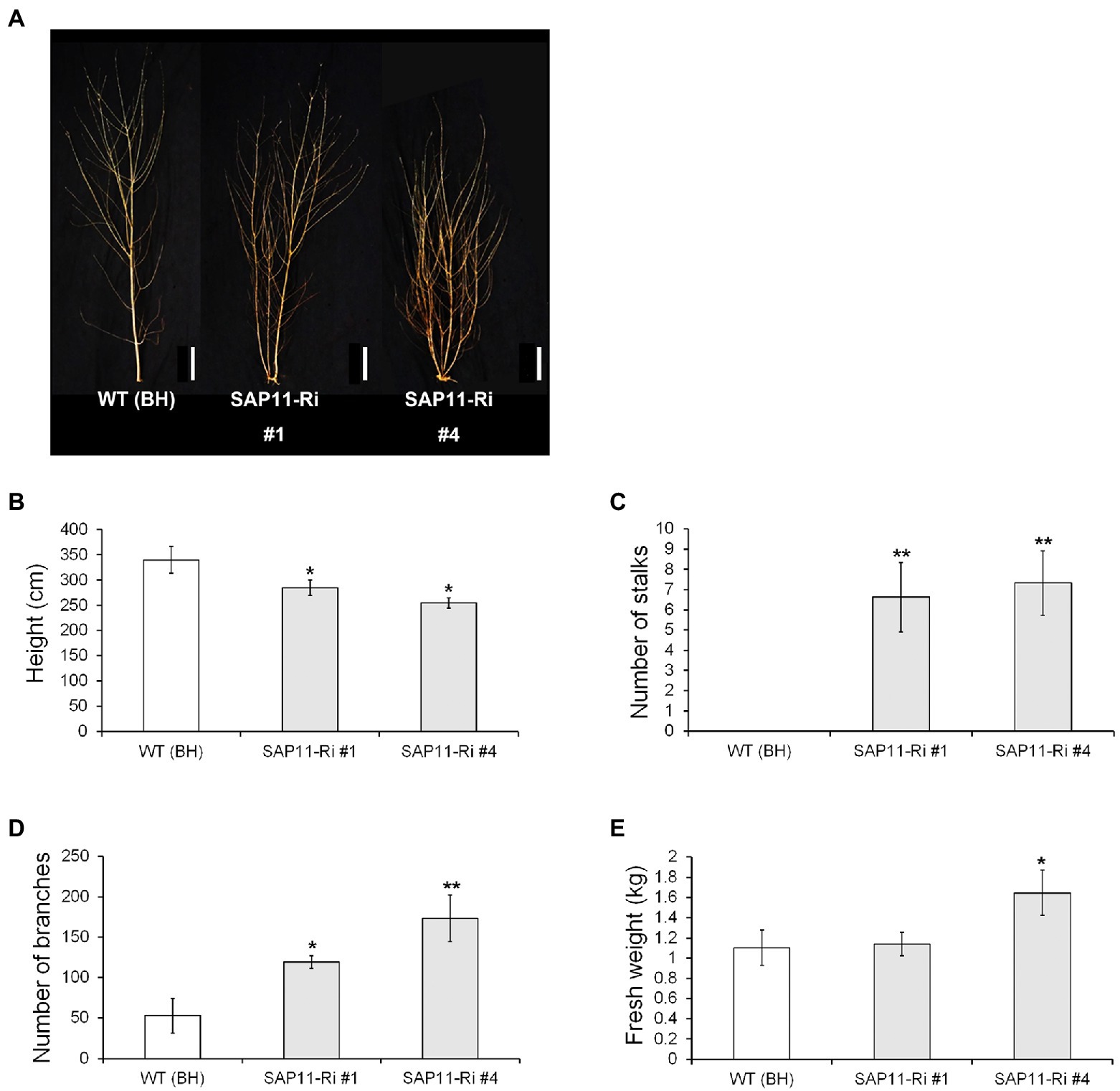
Figure 6. Growth-related data recorded from SAP11-engineered poplars of the field trial during second cultivation cycle. (A) Image of representative WT (clone BH) and transgenic poplar grown in the LMO field. (B-D) Shoot height (B), number of side stalks (C) and number of sylleptic branches (D) in WT and transgenic poplars (n = 10). (E) Aboveground biomass yields of the WT and SAP11-engineered poplars. The values represent the means ± standard deviations (SDs) of three biological replicates. Asterisks indicate significant differences by student t-test (*p < 0.05, **p < 0.005), Scale bar = 30 cm.
Drought stress severely affects plant growth and productivity so that adjusting plant architecture in response to drought stress is important for plant survival (El-Sappah et al., 2021). Lateral shoot branching in plants determines plant architecture, which is complicatedly regulated by genetic factors, phytohormones, nutrition, and the environmental signals. Stress-associated protein (SAP) genes have been reported that are involved in response to abiotic stress in various plant species, including woody plants. In this study, we identified that expression of PagSAP11 was induced under drought stress (Supplementary Figure S1). We used a transgenic approach to silence and upregulate the expression of PagSAP11 to investigate its function. As a result, we explored that knockdown of PagSAP11 induced tolerance to drought stress, whereas overexpressing lines exhibited a drought-sensitive phenotype (Figure 4; Supplementary Figure S3). Hence, we concluded that PagSAP11 provides an important adaptation strategy under drought stress conditions by modulating plant architecture.
Poplar species and their hybrids are considered as important industrial stemwood due to their rapid growth and high productivity (Larchevêque et al., 2011). Among these, hybrid poplar (P. alba × P. tremula var. glandulosa) can tolerate flooding, has extensive root system, and has salt and cold tolerance (Qiu et al., 2019). Additionally, hybrid poplar has been used in many researches due to their high rates of transformation and well-documented genomic resources (Bae et al., 2021).
Amino acid sequence of PagSAP11 shared 98% similarity with that of PtSAP11 (Figure 2). Jin et al. reported that PtSAP11 has one C-repeat (CRT)/dehydration responsive elements (DRE) element in its upstream of the translation start site. A DRE with a 9 bp conserved core sequence (5’-TACCGACAT-3′) was identified in the promoter of the drought-responsive gene rd29A in Arabidopsis (Yamaguchi-Shinozaki and Shinozaki, 1993). CRT with the 5 bp core sequence (5’-CCGAC-3′) was identified in the promoter of cold-inducible genes from Arabidopsis (Baker et al., 1994). DREB proteins have been reported that are important transcription factors which regulate stress signal transduction and modulate gene expression by interacting with a DRE/CRT cis-element present in the promoter region of various abiotic stress-responsive genes (Lata and Prasad, 2011). PagSAP11 also is likely to have a DRE element in its promoter region, suggesting that DREBs can bind to the promoter region of PagSAP11 to induce expression of PagSAP11 in response to drought stress. Structural analysis revealed that PagSAP11 consists of one AN1 domain and one A20 domain (Figure 2). Although the biochemical function of these domains is still largely unknown, OsSAP8 and PagSAP1 are localized in the cytoplasm and presumably function via protein–protein interactions mediated by their A20 and AN1 zinc finger domains (Kanneganti and Gupta, 2008; Yoon et al., 2018). In addition, it was identified that the OsSAP1 could interact with self, its close homolog OsSAP11 and a rice receptor-like cytoplasmic kinase, OsRLCK253 via A20 domain (Giri et al., 2011). AtSAP5, the ortholog of PagSAP11 in Arabidopsis, exhibits E3 ubiquitin ligase activity in vitro (Kang et al., 2011). Moreover, it is generally known that many of the zinc-finger domain proteins act as transcriptional regulators. PagSAP11 protein was localized in the nucleus (Figure 3C), indicating that PagSAP11 may act as a transcriptional regulator in the nucleus.
Branching or branch number is regarded as an integral trait of tree crown architecture. Since this trait determines leaf area index (LAI), it contributes directly to the biomass production in the short-rotation poplar coppice (Ding et al., 2021). Shoot branching is an orchestrated process involved in a wide range of external environmental, internal, physiological and developmental factors (Leyser, 2009). Previous studies have reported that phytohormone such as auxin, cytokinin and strigolactone are involved in lateral shoot branching as internal factors (Cline and Dong-Il, 2002; Muhr et al., 2016; Barbier et al., 2019; Yang et al., 2020). The role of auxin in shoot branching has been well established through studies in which the removal of the shoot tip of plants stimulates axillary bud outgrowth and this growth could be suppressed by applying the plant hormone auxin to the stump of decapitated plants (Thimann and Skoog, 1933). Moreover, it was recently identified that the apically derived auxin transported in the stem upregulates expression of strigolactone synthesis genes and downregulates cytokinin levels, resulting in the inhibition and promotion of bud outgrowth, respectively (Muhr et al., 2016; Barbier et al., 2019). However, in this study, the expressions of auxin-, cytokinin- and strigolactone-related genes were similar to WT, SAP11-Ri and SAP11-OX plants (Supplementary Figure S5). In contrast, the transcript levels of RAX1, which encodes an MYB transcription factor involved the initiation of axillary meristems (Jia et al., 2020) were increased in SAP11-Ri. Furthermore, the expression levels of DML10, a gene involved in bud break (Conde et al., 2019), and CYP707A, a gene related to bud dormancy (Zhao et al., 2021), were also upregulated in SAP11-Ri. However, the mRNA abundance of DRM, a marker of bud dormancy (Yoon et al., 2017), was decreased in SAP11-Ri compared with the WT plants (Figure 5). On the other hands, knockdown and overexpressing of PagSAP11 resulted in reduced plant height, suggesting that the phenotypes finely tuned and influenced by various factors. For example, in rice, overexpression and knockout of OsCKX9 resulted in retarded shoot growth (Duan et al., 2019). These findings indicate that PagSAP11 regulates bud dormancy different from hormone-regulated pathway.
In this study, we investigated the gene characteristics and functions of PagSAP11 in drought stress response and shoot branching in hybrid poplar. Our results suggest that PagSAP11 negatively regulates response to drought stress and the growth of lateral branches. Downregulation or overexpression of PagSAP11 can increase or decrease drought stress resistance in transgenic plants, respectively. Moreover, reduced expression of PagSAP11 can induce lateral shoot branching in the transgenic poplars, in greenhouses as well as in the LMO field. Overall, our research suggests that PagSAP11 is a potential candidate gene to engineer woody plants that maintain their biomass under drought stress conditions.
The data presented in the study are deposited in the GenBank, accession number ON519817.
SP, HC, S-KY, and H-AJ performed the experiments, analyzed the data, and wrote and revised the manuscript. EK-B, HC, Y-IC and HL designed the study and wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported in part by the National Institute of Forest Science (project no. FG0702-2018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We appreciate K. I. Lee, M. S. Park and S. K. Yang for excellent technical assistance. We apologize to other colleagues whose work has not been cited here because of space restrictions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.925744/full#supplementary-material
Abe, S., and Theologis, A. (1998). Transient gene expression in protoplasts of Arabidopsis thaliana. Methods Mol. Biol. 82, 209–217. doi: 10.1385/0-89603-391-0:209
Bae, E., Choi, H., Choi, J., Lee, H., Kim, S., Ko, J., et al. (2021). Efficient knockout of the phytoene desaturase gene in a hybrid poplar (Populus alba × Populus glandulosa) using the CRISPR/Cas9 system with a single gRNA. Transgenic Res. 30, 837–849. doi: 10.1007/s11248-021-00272-9
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5′-region of Arabidopsis thaliana corl5a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Barbier, F. F., Dun, E. A., Derr, S. C., Chabikwa, T. G., and Beveridge, C. A. (2019). An update on the signals controlling shoot branching. Trends in Plant Sci. 24, 220–236. doi: 10.1016/j.tplants.2018.12.001
Bradshaw, H. D., Ceulemans, R., Davis, J., and Stettler, R. (2001). Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J. Plant Growth Regul. 19, 306–313. doi: 10.1007/s003440000030
Cho, J., Kim, M., Bae, E., Choi, Y., Jeon, H., Han, K., et al. (2021). Field evaluation of transgenic hybrid poplars with desirable wood properties and enhanced growth for biofuel production by bicistronic expression of PdGA20ox1 and PtrMYB3 in wood-forming tissue. Biotechnol. Biofuels 14:177. doi: 10.1186/s13068-021-02029-2
Choi, Y., Noh, E., Han, M., Lee, J., and Choi, K. (2005). An efficient and novel plant selectable marker based on organomercurial resistance. J. Plant. Biol. 48, 351–355. doi: 10.1007/BF03030576
Cline, M. G., and Dong-Il, K. (2002). A preliminary investigation of the role of auxin and cytokinin in sylleptic branching of three hybrid poplar clones exhibiting contrasting degrees of sylleptic branching. Annals of Bot. 90, 417–421. doi: 10.1093/aob/mcf195
Conde, D., Perales, M., Sreedasyam, A., Tuskan, G. A., Lloret, A., Badenes, M. L., et al. (2019). Engineering tree seasonal cycles of growth through chromatin modification. Front. Plant Sci. 10:412. doi: 10.3389/fpls.2019.00412
Ding, C., Wang, N., Huang, Q., Zhang, W., Huang, J., Yan, S., et al. (2021). The importance of proleptic branch traits in biomass production of poplar in high-density plantations. J. For. Res. 33, 463–473. doi: 10.1007/s11676-021-01418-5
Dong, Q., Duan, D., Zhao, S., Xu, B., Luo, J., Wang, Q., et al. (2018). Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic Arabidopsis. Int. J. Mol. Sci. 19:2478. doi: 10.3390/ijms19092478
Duan, J., Yu, H., Yuan, K., Liao, Z., Meng, X., Jing, Y., et al. (2019). Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice. Proc. Natl. Acad. Sci. U. S. A. 116, 14319–14324. doi: 10.1073/pnas.1810980116
El-Sappah, A. H., and Rather, S. A. (eds). (2022). Genomics Approaches to Study Abiotic Stress Tolerance in Plants in Plant Abiotic Stress Physiology. Vol. 2. Burlington: Apple Academic Press.
El-Sappah, A. H., Rather, S. A., Wani, S. H., Elrys, A. S., Bilal, M., Huang, Q., et al. (2022). Heat stress-mediated constraints in maize (Zea mays) production: challenges and solutions. Front. Plant Sci. 13:879366. doi: 10.3389/fpls.2022.879366
El-Sappah, A. H., Yan, K., Huang, Q., Islam, M. M., Li, Q., Wang, Y., et al. (2021). Comprehensive mechanism of gene silencing and its role in plants growth and development. Front. Plant Sci. 12:705249. doi: 10.3389/fpls.2021.705249
Estravis-Barcala, M., Mattera, M. G., Soliani, C., Bellora, N., Opgenoorth, L., Heer, K., et al. (2020). Molecular bases of responses to abiotic stress in trees. J. Exp. Bot. 71, 3765–3779. doi: 10.1093/jxb/erz532
Giri, J., Vij, S., Dansana, P. K., and Tyagi, A. K. (2011). Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 191, 721–732. doi: 10.1111/j.1469-8137.2011.03740.x
Guo, J., Morrell-Falvey, J. L., Labbé, J. L., Muchero, W., Kalluri, U. C., Tuskan, G. A., et al. (2012). Highly efficient isolation of Populus mesophyll protoplasts and its application in transient expression assays. PLoS One 7:e44908. doi: 10.1371/journal.pone.0044908
Harfouche, A., Meilan, R., and Altman, A. (2014). Molecular and physiological responses to abiotic stress in forest trees and their relevance to tree improvement. Tree Physiol. 34, 1181–1198. doi: 10.1093/treephys/tpu012
Hu, L., Wang, Z., Du, H., and Huang, B. (2010). Differential accumulation of dehydrins in response to water stress for hybrid and common bermudagrass genotypes differing in drought tolerance. J. Plant Physiol. 167, 103–109. doi: 10.1016/j.jplph.2009.07.008
Jia, H., Li, J., Zhang, J., Ren, Y., Hu, J., and Lu, M. (2016). Genome-wide survey and expression analysis of the stress-associated protein gene family in desert poplar. Populus euphratica. Tree Genet. Genomes. 12:78. doi: 10.1007/s11295-016-1033-8
Jia, T., Zhang, K., Li, F., Huang, Y., Fan, M., and Huang, T. (2020). The AtMYB2 inhibits the formation of axillary meristem in Arabidopsis by repressing RAX1 gene under environmental stresses. Plant Cell Rep. 39, 1755–1765. doi: 10.1007/s00299-020-02602-3
Jian, W., Zheng, Y., Yo, T., Cao, H., Chen, Y., Cui, Q., et al. (2021). SlNAC6, a NAC transcription factor, is involved in drought stress response and reproductive process in tomato. J. Plant Physiol. 264:153483. doi: 10.1016/j.jplph.2021.153483
Jin, Y., Wang, M., Fu, J., Xuan, N., Zhu, Y., Lian, Y., et al. (2007). Phylogenetic and expression analysis of ZnF-AN1 genes in plants. Genomics 90, 265–275. doi: 10.1016/j.ygeno.2007.03.019
Kang, M., Fokar, M., Abdelmageed, H., and Allen, R. D. (2011). Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 75, 451–466. doi: 10.1007/s11103-011-9748-2
Kanneganti, V., and Gupta, A. K. (2008). Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 66, 445–462. doi: 10.1007/s11103-007-9284-2
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/s1360-1385(02)02251-3
Khan, Z., Rho, H., Firrincieli, A., Hung, S. H., Luna, V., Masciarelli, O., et al. (2016). Growth enhancement and drought tolerance of hybrid poplar upon inoculation with endophyte consortia. Curr. Plant. Biol. 6, 38–47. doi: 10.1016/j.cpb.2016.08.001
Kim, M., Cho, J., Park, E., Lee, H., Choi, Y., Bae, E., et al. (2020). Overexpression of a poplar RING-H2 zinc finger, Ptxerico, confers enhanced drought tolerance reduced water loss and ion leakage in Populus. Int. J. of Mol. Sci. 21:9454. doi: 10.3390/ijms21249454
Kim, J., Kim, U., Yeom, M., Kim, J., and Nam, H. (2008). FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20, 307–319. doi: 10.1105/tpc.107.055715
Larchevêque, M., Maurel, M., Desrochers, A., and Larocque, G. R. (2011). How does drought tolerance compare between two improved hybrids of balsam poplar and an unimproved native species? Tree Physiol. 31, 240–249. doi: 10.1093/treephys/tpr011
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lee, I., Lee, I., Kim, J., Kim, J., Chung, E., Kim, H., et al. (2016). NORE1/SAUL1 integrates temperature-dependent defense programs involving SGT1b and PAD4 pathways and leaf senescence in Arabidopsis. Physiol. Plant. 158, 180–199. doi: 10.1111/ppl.12434
Lee, H., Lee, J., Noh, E., Bae, E., Choi, Y., and Han, M. (2005). Generation and analysis of expressed sequence tags from poplar (Populus alba × P. tremula var. glandulosa) suspension cells. Plant Sci. 169, 1118–1124. doi: 10.1016/j.plantsci.2005.07.013
Leyser, O. (2009). The control of shoot branching: an example of plant information processing. Plant Cell Environ. 32, 694–703. doi: 10.1111/j.1365-3040.2009.01930.x
Li, J., Sun, P., Xia, Y., Zheng, G., Sun, J., and Jia, H. (2019). A stress-associated protein, PtSAP13, from Populus trichocarpa provides tolerance to salt stress. Int. J. Mol. Sci. 20:5782. doi: 10.3390/ijms20225782
Liu, S., Yuan, X., Wang, Y., Wang, H., Wang, J., Shen, Z., et al. (2019). Tomato stress-associated protein 4 contributes positively to immunity against necrotrophic fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 32, 566–582. doi: 10.1094/MPMI-04-18-0097-R
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Muhr, M., Prüfer, N., Paulat, M., and Teichmann, T. (2016). Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED1 expression and shoot architecture. New Phytol. 212, 613–626. doi: 10.1111/nph.14076
Mukhopadhyay, A., Vij, S., and Tyagi, A. K. (2004). Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U. S. A. 101, 6309–6314. doi: 10.1073/pnas.0401572101
Opipari, A. W., Boguski, M. S., and Dixit, V. M. (1990). The A20 cDNA induced by tumor necrosis factor α encodes a novel type of zinc finger protein. J. Biol. Chem. 265, 14705–14708. doi: 10.1016/s0021-9258(18)77165-2
Qiu, D., Bai, S., Ma, J., Zhang, L., Shao, F., Zhang, K., et al. (2019). The genome of Populus alba × Populus tremula var. glandulosa clone 84K. DNA Res. 26, 423–431. doi: 10.1093/dnares/dsz020
Sannigrahi, P., Ragauskas, A. J., and Tuskan, G. A. (2010). Poplar as a feedstock for biofuels: A review of compositional characteristics. Biofuels Bioprod. Biorefin. 4, 209–226. doi: 10.1002/bbb.206
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thimann, K. V., and Skoog, F. (1933). Studies on the growth hormone of plants. III. The inhibiting action of the growth substance on bud development. Proc. Natl. Acad. Sci. U. S. A. 19, 714–716. doi: 10.1073/pnas.19.7.714
Vij, S., and Tyagi, A. K. (2006). Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Gen. Genomics. 276, 565–575. doi: 10.1007/s00438-006-0165-1
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 236, 331–340. doi: 10.1007/BF00277130
Yang, T., Lian, Y., Kang, J., Bian, Z., Xuan, L., Gao, Z., et al. (2020). The SUPPRESSOR of MAX2 1 (SMAX1)-Like SMXL6, SMXL7 and SMXL8 act as negative regulators in response to drought stress in Arabidopsis. Plant Cell Physiol. 61, 1477–1492. doi: 10.1093/pcp/pcaa066
Yoon, S., Bae, E., Choi, H., Choi, Y., and Lee, H. (2017). Isolation and expression of dormancy-associated protein 1 (DRM1) in poplar (Populus alba × P. glandulosa). J. Plant Biotechnol. 44, 69–75. doi: 10.5010/JPB.2017.44.1.069
Yoon, S., Bae, E., Lee, H., Choi, Y., Han, M., Choi, H., et al. (2018). Downregulation of stress-associated protein 1 (PagSAP1) increases slat stress tolerance in poplar (Populus alba × P. glandulosa). Trees 32, 823–833. doi: 10.1007/s00468-018-1675-2
Zhang, X., Zheng, W., Cao, X., Cui, X., Zhao, S., Yu, T., et al. (2019). Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and soybean. Front. Plant Sci. 10:1453. doi: 10.3389/fpls.2019.01453
Keywords: stress-associated protein, PagSAP11, drought tolerance, lateral shoot branching, bud dormancy, hybrid poplar
Citation: Park SJ, Bae E-K, Choi H, Yoon S-K, Jang H-A, Choi Y-I and Lee H (2022) Knockdown of PagSAP11 Confers Drought Resistance and Promotes Lateral Shoot Growth in Hybrid Poplar (Populus alba × Populus tremula var. glandulosa). Front. Plant Sci. 13:925744. doi: 10.3389/fpls.2022.925744
Received: 21 April 2022; Accepted: 30 May 2022;
Published: 24 June 2022.
Edited by:
Longxing Hu, Hunan Agricultural University, ChinaReviewed by:
Ahmed H. El-Sappah, Zagazig University, EgyptCopyright © 2022 Park, Bae, Choi, Yoon, Jang, Choi and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyoshin Lee, aHlvc2hpbmxlZUBrb3JlYS5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.