
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 June 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.922561
This article is part of the Research TopicPhysiological, Biochemical and Molecular Approaches in Response to Abiotic Stresses in PlantsView all 17 articles
Heat shock transcription factor (Hsf) exists widely in eukaryotes and responds to various abiotic stresses by regulating the expression of downstream transcription factors, functional enzymes, and molecular chaperones. In this study, TaHsfA2-13, a heat shock transcription factor belonging to A2 subclass, was cloned from wheat (Triticum aestivum) and its function was analyzed. TaHsfA2-13 encodes a protein containing 368 amino acids and has the basic characteristics of Hsfs. Multiple sequence alignment analysis showed that TaHsfA2-13 protein had the highest similarity with TdHsfA2c-like protein from Triticum dicoccoides, which reached 100%. The analysis of tissue expression characteristics revealed that TaHsfA2-13 was highly expressed in root, shoot, and leaf during the seedling stage of wheat. The expression of TaHsfA2-13 could be upregulated by heat stress, low temperature, H2O2, mannitol, salinity and multiple phytohormones. The TaHsfA2-13 protein was located in the nucleus under the normal growth conditions and showed a transcriptional activation activity in yeast. Further studies found that overexpression of TaHsfA2-13 in Arabidopsis thaliana Col-0 or athsfa2 mutant results in improved tolerance to heat stress, H2O2, SA and mannitol by regulating the expression of multiple heat shock protein (Hsp) genes. In summary, our study identified TaHsfA2-13 from wheat, revealed its regulatory function in varieties of abiotic stresses, and will provide a new target gene to improve stress tolerance for wheat breeding.
Abiotic stresses such as drought, high salt, and high temperature can affect the survival, productivity, and geographical distribution of many important crops worldwidely (Gong et al., 2020). There seem to be significant differences among the various stresses, while they also show overlapping responses. For example, osmotic stress caused by drought and salinity is an important environmental threat, limiting plant growth and agricultural productivity (Chen et al., 2020). Oxidative stress is an integral component of various stress conditions, including excessive light, cold, heat, drought, and this fact largely determines the substantial overlap in plant physiological and molecular responses to various environmental challenges (Savchenko and Tikhonov, 2021). In the process of fighting against these adverse environmental factors, plants have evolved a complex and elaborate regulatory network (Guo et al., 2016). With the development of molecular techniques such as large-scale transcriptome analysis, a large number of abiotic stress associated genes have been discovered in plants (Nakashima et al., 2009; Duan et al., 2019). Genes encoding transcription factors are downstream components of signal transduction and play indispensable roles in the regulation of multiple abiotic stresses. About 7% of the coding sequences in plant genomes are transcription factors, many of which typically belong to large gene families, such as the Hsf family (Baniwal et al., 2004; Udvardi et al., 2007). They are excellent candidates for crop improvement (Baillo et al., 2019). In contrast to the few Hsfs in yeast and mammals, the plant Hsf families contain a large number of Hsf members, forming a complex plant-specific superfamily that is widespread in many species (Guo et al., 2016). There are 21 Hsfs in Arabidopsis, 24 in tomato, 25 in pepper, 41 in bamboo, 52 in soybean, and 82 in wheat (Scharf et al., 2012; Fragkostefanakis et al., 2015; Guo et al., 2015; Duan et al., 2019; Huang et al., 2021). Compared to other plant species, the large number of Hsfs in wheat makes the corresponding research more complicated.
In general, plant Hsf proteins have conserved modular structures, including N-terminal DNA binding domain (DBD) characterized by a central helix-turn-helix motif, an oligomerization domain (OD) with a bipartite heptad pattern of hydrophobic amino acid residues (HR-A/B region), the C-terminal nuclear localization signal (NLS) and nuclear export signal (NES) for regulating Hsf proteins subcellular localization (Baniwal et al., 2004). Plant Hsf were classified into A, B, and C based on the sequence length between DBD and HR-A/B regions and the number of amino acid residues inserted into HR-A/B region (Guo et al., 2016; Liu et al., 2020). Among these three types of Hsf proteins, HsfA class members usually contain an activation domain at the C-terminal, characterized by aromatic, hydrophobic, and acidic amino acid residues (AHA), whereas HsfB and C lack the AHA motif and in many cases do not function as activator (Kotak et al., 2004). The different protein structures of HsfA, HsfB, and HsfC determine the different functions they perform. In Arabidopsis, HsfA1 and HsfA2 have been reported as positive regulators of plant responses to heat stress (HS), whereas two members of Class B Hsf, HsfB1, and HsfB2b, are active repressors of Hsfs and Hsps (Nishizawa et al., 2006; Charng et al., 2007; Ikeda et al., 2011; Liu et al., 2011; Nishizawa-Yokoi et al., 2011).
Current understanding about the individual functions for A2 subclass of Hsf in plant mainly comes from model species such as Arabidopsis and tomato (Heerklotz et al., 2001; Charng et al., 2007; Ogawa et al., 2007; Chan-Schaminet et al., 2009; Fragkostefanakis et al., 2016; Liu et al., 2019). The expression of HsfA2 was not detected in control cell cultures, but was strongly upregulated after HS treatment, indicating an important regulatory role of HsfA2 during HS in Arabidopsis (Nover et al., 2001). HsfA2 transcript level was also strongly induced by HS in tomato (Scharf et al., 1998). In addition, the expression of HsfA2 in tomato anther is higher than that in other flower tissues, and can be further induced under both short and prolonged HS conditions, similar to its expression in leaves (Giorno et al., 2010). Recent studies have found that HsfA2, as a central regulatory factor, is involved in the transgenerational memory to high temperature in plants through complex epigenetic regulation such as histone demethylation (Liu et al., 2019). In addition to heat stress, many other abiotic stresses such as cold, salt, and drought, as well as phytohormones such as abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), and ethylene have also been shown to induce the expression of HsfA2 in plants (Nishizawa et al., 2006; Zhang et al., 2015; Duan et al., 2019). As an important regulatory factor of HS, the Arabidopsis HsfA2 is involved in other abiotic stress, such as salt stress, osmotic stress and anoxia stress (Charng et al., 2007; Banti et al., 2010). Coincidentally, overexpression of OsHsfA2e in Arabidopsis also increased tolerance to high-salinity stress (Yokotani et al., 2008). Additionally, transgenic Arabidopsis plants overexpressing TaHsfA2d showed considerable tolerance to salinity and drought stresses (Chauhan et al., 2013). More recently, TaHsfA2e-5D has been shown to positively regulate heat and drought stress, at least in part by promoting the expression of stress-responsive genes (Bi et al., 2022). It seems that plant Hsfs may act as the intersection of multiple signaling pathways, on the one hand participating in the regulation of abiotic signaling pathways and on the other hand mediating phytohormone signaling (Guo et al., 2016). Indeed, our previous studies have shown that many Hsfs, such as the A2 subclass of Hsfs, are responsive to many abiotic stresses and phytohormones (Duan et al., 2019).
Although basic biological information about Hsfs in wheat is well known, fewer Hsfs have been cloned and identified from wheat than other crops, and the function of most Hsfs are unclear. Our previous studies have demonstrated the expression of TaHsfA2-13 was induced by various abiotic stresses (Duan et al., 2019). In this study, the biological functions and potential mechanisms of wheat TaHsfA2-13 in plant tolerance to multiple abiotic stresses will be further revealed.
Cang6005, a semi-winter heat-tolerant wheat variety, was used for gene cloning and expression analysis. After surface disinfection with 0.1% HgCl2, wheat seeds were cleaned with sterile water and soaked at room temperature for 12 h. When the germ grew to 1–2 cm, seedlings were transplanted and cultivated in Hoagland nutrient solution under day/night conditions of 16 h/8 h (70–100 μmol m−2 S−1), 25°C, and 50–60% relative humidity. The stress treatments were performed as described by previous studies (Duan et al., 2019; Liu et al., 2020; Bi et al., 2022). Seedlings at the two-leaf and one-heart stage were treated with high temperature (37°C), low temperature (4°C), H2O2 (10 mmol L−1), mannitol (15% W/V), NaCl (0.2 mol L−1), MeJA (0.1 mmol L−1), ABA (0.2 mmol L−1) and SA (0.8 mmol L−1) for different time, respectively. The second leaves were collected in each treatment, and then immediately frozen in liquid nitrogen for RNA extraction.
Total RNA of plants was extracted using the RNArose Reagent Systems Kit (Huashun Bioengineering Co., LTD., Shanghai, China), according to the manufacturer’s instructions. RNA was further purified through the PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian) to remove genomic DNA contamination. Then, 2 μg of purified RNA was used as template to synthesize first-strand cDNA. The specific primers were designed using DNAMAN 8.01 software to amplify the TaHsfA2-13 sequence. The PCR amplified products were constructed to pEasy-Blunt cloning vectors (pEasy-Blunt Simple Cloning Kit, TransGen Biotech) and sequenced by Shanghai Shenggong Bioengineering Technology Co., LTD. Primer sequence information is presented in Supplementary Table S1.
The protein sequences used for multiple sequence alignment analysis were downloaded from NCBI website.2 GenBank accession numbers of the proteins are as follows: TaHsfA2-13 (T. aestivum, MT 680017), TdHsfA2c-like (T. dicoccoides, XP_037425119), AetHsfA2c (Aegilops tauschii, XP_020147337), BdHsfA2c (Brachypodium distachyon, XP_010234740), ZmHsf30 (Zea mays, NP_001147128), AtHsfA2 (A. thaliana, NP_001324815; Charng et al., 2007). Multiple sequence alignment analysis was carried out by the DNAMAN 8.0 software.3
Real-time PCR was performed with SYBR Green PCR master mix (TaKaRa, Dalian) on a Bio-Rad CFX96 realtime PCR detector. According to the methods of Liu et al. (2020), TaRP15 and Atactin8 were selected as internal reference genes in wheat and Arabidopsis, respectively, to calculate the relative transcription levels of the interest genes. The primers used are listed in Supplemental Table S1. Three replicates were performed for each qRT-PCR reaction, and the results were reported as means and standard deviation. SPSS 19.0 software was used to analyze the significance.
The coding sequence of TaHsfA2-13 without a stop codon was inserted into the pJIT1-hGFP vector through LR recombination reaction to generate TaHsfA2-13-GFP fusion protein. The vectors was transformed into Agrobacterium strain GV3101 and injected into N. benthamiana leaves with a needleless 1 ml syringe. The nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI, 1 mg ml−1) dye before imaging, fluorescence observation was performed under a fluorescence microscope (Zeiss META510).
The full-length and truncated TaHsfA2-13 were cloned into pGBKT7 and then introduced into yeast strain AH109, respectively. The transformants were grown on the SD/-Trp and SD/Trp-/His-/Ade-media for 3 days before observation. The transcriptional activation activities was evaluated according to their growth status and the activity of α-galactosidase.
For the construction of prey, the coding sequence of TaHsfA2-13 was amplified from wheat cDNA and then cloned into pGADT7 vector. For bait construction, the promoters of five AtHsps were amplified from Arabidopsis genomic DNA and then cloned into pHIS2.1 vector according to the methods described by Liu et al. (2020). The bait and prey constructs were co-transformed into yeast strain Y187, and grown on the selective medium with 10 mM 3-aminotriazole (3-AT) or not.
To obtain the plants overexpressing TaHsfA2-13 in Arabidopsis, the coding region of TaHsfA2-13 was amplified by PCR and cloned into pCAMBIA1300. The recombinant plasmid was transformed into Agrobacterium strain GV3101. At the stage of flower-budding, Arabidopsis wild type (Col-0) and athsfa2 mutants were transformed by classical floral dip method (Liu et al., 2020). The T-DNA insertion transgenic seeds were selected on MS-agar plates supplemented with 50 μg·μL−1 of hygromycin and confirmed by semi-quantitative RT-PCR. Atactin8 gene was used for normalizing the gene expression in Arabidopsis.
For heat stress treatment, sterilized seeds of wild-type (or athsfa2) and Arabidopsis homozygous transgenic lines were planted on MS agar plates. Seedlings of 5 days old were subjected to basal thermotolerance (45°C for 50 min, placed at 22°C for 8 days) and acquired thermotolerance treatment (37°C for 1 h, incubated at 22°C for 2 days, 46°C for 50 min, placed at 22°C for 8 days) according to the previous methods (Larkindale et al., 2005; Li et al., 2019). After 8 days, photography was taken and the rosette leaves of different lines were collected to determine chlorophyll content. For oxidative stress, osmotic stress and phytohormone treatments, sterilized wild-type and transgenic seeds were planted on MS agar plates supplemented with H2O2 (10 mmol l−1), mannitol (15% W/V), and SA (0.8 mmol l−1), respectively. After 2 weeks, photography was taken and the rosette leaves of different lines were collected to determine chlorophyll content. The chlorophyll contents were measured as described by Liu et al. (2020). In each experiment, 30 seedlings of each line were treated at least, and three independent experiments were conducted.
The rosette leaves of Arabidopsis seedlings were harvested to measure the physiological indexes after seedlings of 5 days old were subjected to basal thermotolerance (45°C for 50 min, placed at 22°C for 8 days). The activities of superoxide dismutase (SOD) and peroxidase (POD) were analyzed using the nitrotetrazolium blue chloride reduction and guaiacol colorimetric method, respectively (Li et al., 2015). Osmotic potential was determined with a VIESCOR 5100C vapour pressure osmometer (Wesor Inc., Logan, UT, United States of America) as described by Liu et al. (2002).
Our previous RNA-Seq data showed that TaHsfA2-13, a Hsf of subclass A2, was strongly upregulated by heat stress, H2O2 and ABA, suggesting that this gene may be an important node in abiotic stress and phytohormones (Duan et al., 2019). To further clarify the biological function of TaHsfA2-13, we obtained the coding sequence of it from heat-treated wheat leaves, which encoding 369 amino acids. Analysis of amino acid sequence showed that TaHsfA2-13 had the basic characteristics similar to other Hsfs of class A, including DNA binding domain (DBD), oligomerization domain (OD), nuclear localization signal (NLS), nuclear export signal (NES) and activator peptide motif (AHA). Moreover, multiple sequence alignment analysis revealed that TaHsfA2-13 shared the highest similarity with TdHsfA2c-like from T. dicoccoides (Figure 1).
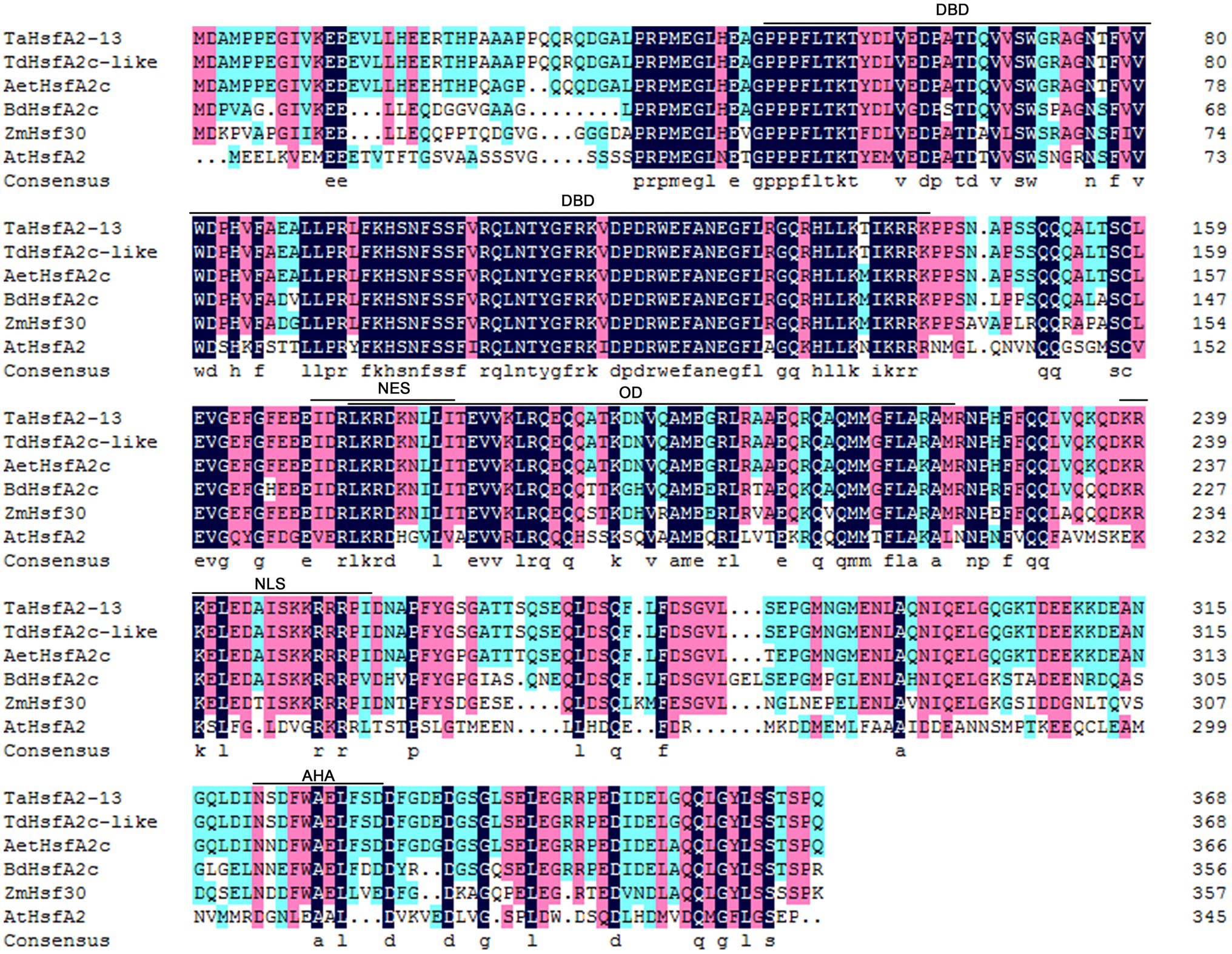
Figure 1. Sequence alignment of TaHsfA2-13 and other plant HsfA2 proteins. Multiple sequence alignment of amino acid sequences from Aegilops tauschii (AetHsfA2c), Arabidopsis thaliana (AtHsfA2), Brachypodium distachyon (BdHsfA2c), Triticum aestivum (TaHsfA2-13), Triticum dicoccoides (TdHsfA2c-like), and Zea mays (ZmHsf30). Conservative domains such as: DNA-binding domain (DBD), Oligomerization domain (OD), Nuclear localization signal (NLS), Nuclear export signal (NES) and Activator peptide motif (AHA) are marked with line segments.
In order to explore the expression patterns of TaHsfA2-13 in wheat, qRT-PCR was used to detect the expression of TaHsfA2-13 in different tissues at seedling and reproductive stages. Compared with the reproductive stage of wheat, TaHsfA2-13 was more greatly expressed in seedling stage, and the expression level in young shoot or leaf was 2.5 fold higher than that in root under normal growth conditions. Specifically, the expression level of TaHsfA2-13 in embryos was almost undetectable, suggesting that TaHsfA2-13 may not be involved in embryonic development (Figure 2A).
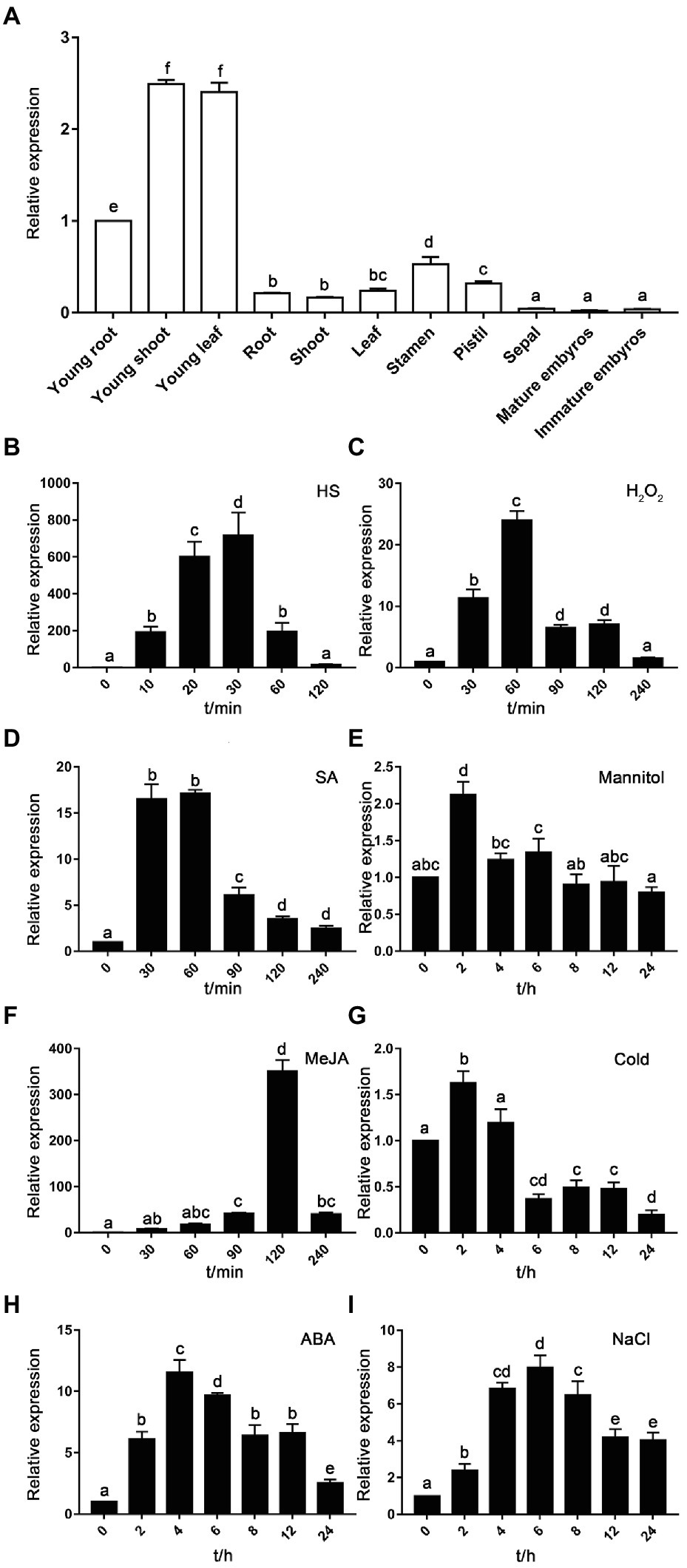
Figure 2. Expression analysis of TaHsfA2-13. (A) Tissue-specific expression patterns of TaHsfA2-13. The expression level in young root was set as 1. (B–I) Expression levels of TaHsfA2-13 in response to abiotic stresses and phytohormones. The expression of untreated samples was set as 1. The wheat TaRP15 gene was used as an internal control. All data represent means ± standard deviation (SD) of three biological replicates. Different lowercase letters above the bars denote significant differences at the p < 0.05 level.
Next, to confirm the induction of TaHsfA2-13 gene expression under various abiotic stresses and phytohormones, we detected the transcript levels of TaHsfA2-13 in wheat leaves which were treated with 37°C HS treatment, 4°C cold treatment, H2O2, mannitol, NaCl, MeJA, ABA and SA, respectively. It was found that the TaHsfA2-13 gene was substantially induced by HS and its highest level was about 700 fold greater than the untreated control (Figure 2B). Under H2O2 treatment, the expression level of TaHsfA2-13 was greatly increased and reached the peak at 60 min (Figure 2C). Following NaCl stress, TaHsfA2-13 expression was induced at 2 h, peaked at 6 h, and then decreased slowly (Figure 2I). However, for mannitol and cold treatments, the responses of TaHsfA2-13 were relatively weak, and its transcript levels slightly increased at 2 h (Figures 2E,G). Strikingly, the responses of TaHsfA2-13 to phytohormones were extremely differently. SA treatment could rapidly induce expression of TaHsfA2-13 to the peak within 30 min (Figure 2D). Compared with the SA treatment, the peak time of TaHsfA2-13 transcription level reached under the MeJA and ABA treatments was delayed, which was 120 min and 2 h, respectively (Figures 2F,H).
As shown in Figure 1, TaHsfA2-13 has an AHA domain at the C-terminal of its protein, suggesting that TaHsfA2-13 may be a transcriptional activator. To verify this, we analyzed the transcriptional activation activity of TaHsfA2-13 in yeast with the GAL4 yeast system. The yeast harboring pGBKT7-TaHsfA2-13 and pGBKT7-TaHsfA2-13301-368 grew well like positive controls, which carrying pGBKT7-P53 on the yeast medium (SD/Trp-/His−/Ade−/X-α-gal), while the yeast harboring pGBKT7-TaHsfA2-131-300 and negative controls barely grew at all (Figure 3A). These results indicate that TaHsfA2-13 has a transcriptional activation activity in yeast.
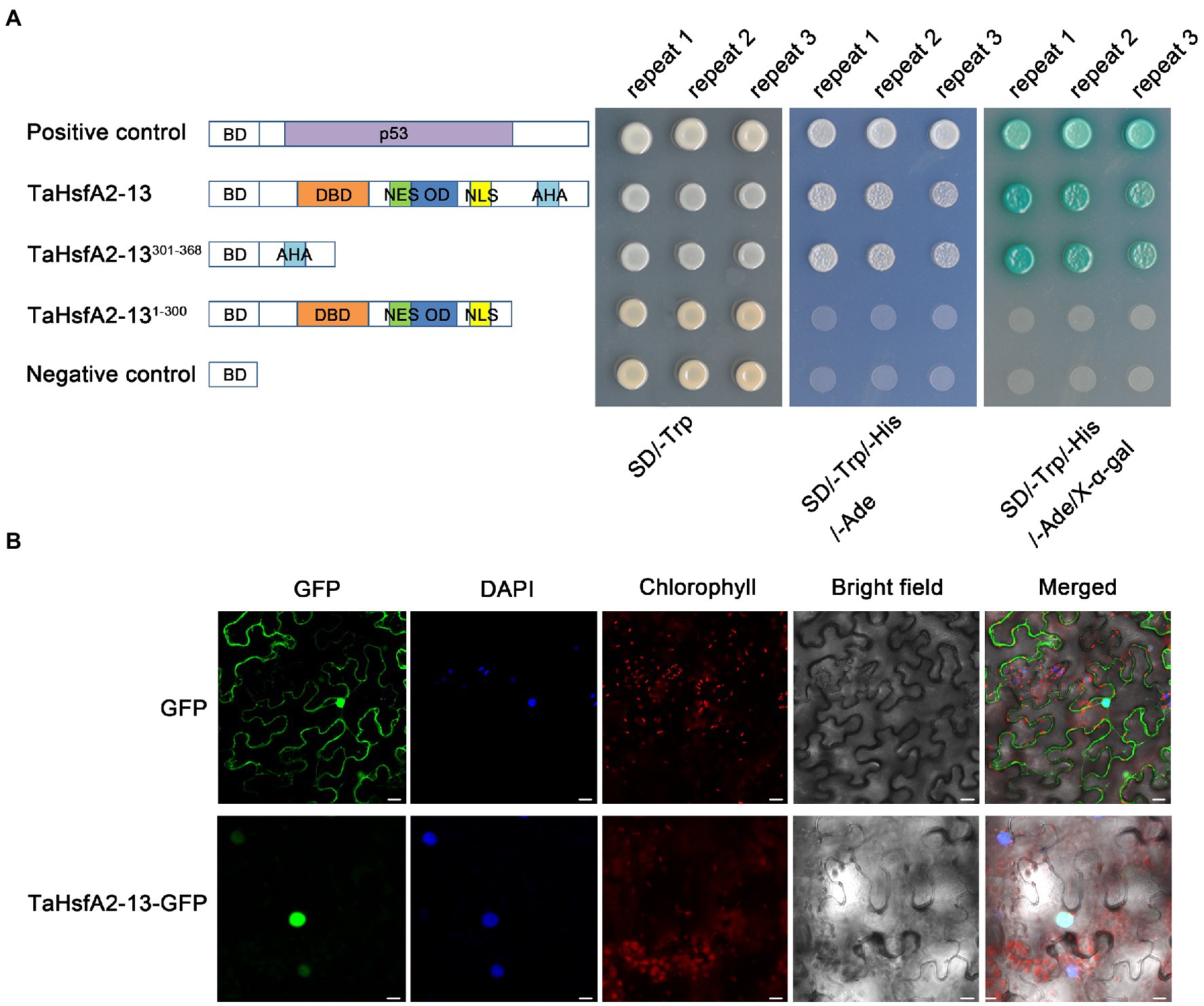
Figure 3. Transcriptional activity and subcellular localization analyses of TaHsfA2-13. (A) Transcriptional activation of TaHsfA2-13 in yeast. Schematic representation of the full-length TaHsfA2-13 and truncated protein constructs in the pGBKT7 vector. Fusion proteins of the GAL4 DNA-binding domain (BD) and full-length TaHsfA2-13 or truncated TaHsfA2-13 with (301–368 aa) or without (1–300 aa) AHA domain were expressed in the yeast strain AH109. Transformants were spotted onto SD/-Trp and SD/Trp-/His-/Ade-media. The plates were incubated at 30°C for 3 days. The pGBKT7 and pGBKT7-P53 vectors were used as negative and positive controls, respectively. (B) Subcellular localization of TaHsfA2-13 in tobacco leaf epidermal cells. Bars = 20 μm.
Analysis of amino acid sequence revealed that TaHsfA2-13 possesses both NLS and NES (Figure 1), so the true localization of its protein is worth considering. The transient expression vector pJIT1-TaHsfA2-13-hGFP was constructed and transformed into tobacco leaf epidermal cells. The subcellular localization of the target protein was revealed by fluorescence microscope. Under normal circumstances, compared to the whole cell distribution of free GFP, the green fluorescence of the target protein is only distributed in the nucleus, which completely overlaps with the fluorescence of the nuclear specific dye DAPI. This indicates that TaHsfA2-13 is a nuclear localization transcription factor (Figure 3B).
To further explore the biological functions of TaHsfA2-13 in plants, transgenic plants overexpressing TaHsfA2-13 in Col-0 (7–28, 8–29 and 18–30) or athsfa2 mutant (6–11, 10–16 and 11–12) were obtained (Supplementary Figure S1). Firstly, the role of TaHsfA2-13 in thermotolerance was analyzed. As shown in Figure 4A; Supplementary Figure S2, under normal growth conditions, there was no significant growth difference between transgenic lines and WT (or athsfa2). However, under heat stress conditions, transgenic plants had fewer withered leaves and better growth than WT (or athsfa2). Consistent with this, the content of chlorophyll in transgenic plants was also significantly higher than that in WT (or athsfa2), indicating a less degree of injury in transgenic lines after HS treatment (Figure 4B). Meanwhile, we also detected the activities of POD, SOD and osmotic potential in transgenic lines and WT under HS treatment or not. The results showed that under normal growth conditions, there was no difference between the transgenic lines and WT. But, when subjected to HS treatment, the activities of POD and SOD were significantly higher in transgenic lines, and osmotic potential of the transgenic lines was significantly lower (Figures 4C–E).
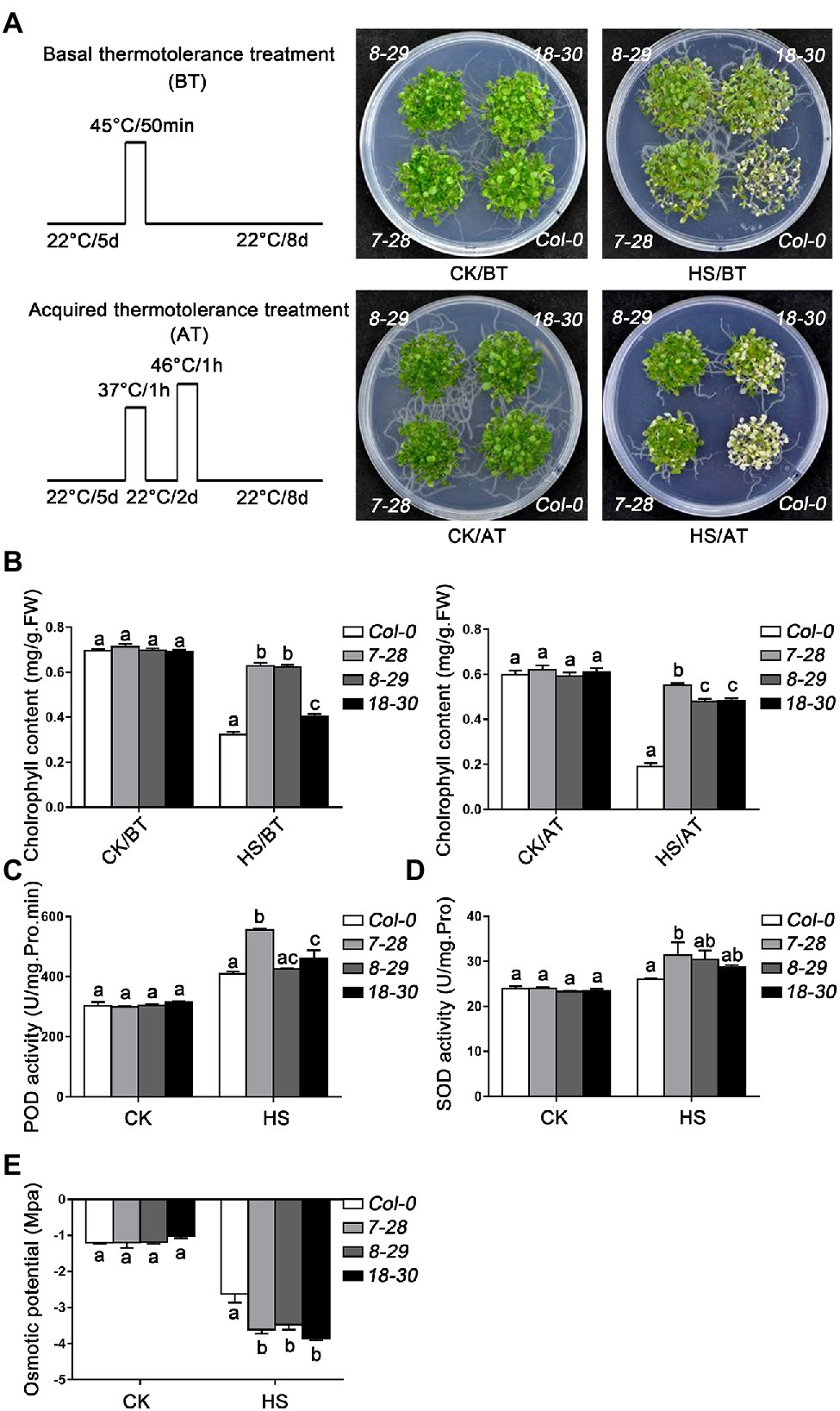
Figure 4. Thermotolerance assays of TaHsfA2-13 overexpressing transgenic Arabidopsis lines. (A) The phenotypes of WT and transgenic seedlings after basal thermotolerance (BT, 45°C for 50 min, placed at 22°C for 8 days) and acquired thermotolerance (AT, 37°C for 1 h, incubated at 22°C for 2 days, 46°C for 1 h, placed at 22°C for 8 days) treatment. The plants were photographed 8 d after different HS. (B) Chlorophyll content of WT and transgenic seedlings treated with different HS. (C–E) Peroxidase (POD), superoxide dismutase (SOD) activities, and osmotic potential of WT and transgenic seedlings under HS treatment or not. For each experiment, at least 30 plants per line were used. Values are means ± SD from three independent measurements. Different lowercase letters above the bars denote significant differences at the p < 0.05 level.
Next, we also explore the role of TaHsfA2-13 in oxidative stress, osmotic stress and response to phytohormone. As expected, following the H2O2, SA and mannitol treatments, the growth of transgenic seedlings was better than that of WT (or athsfa2), and the chlorophyll content of transgenic seedlings was higher, suggesting that TaHsfA2-13 could also enhance the resistance to oxidative stress, osmotic stress and toxicity of excess phytohormone other than improve the basic and acquired thermotolerance of transgenic plants (Figures 5A,B; Supplementary Figure S2).
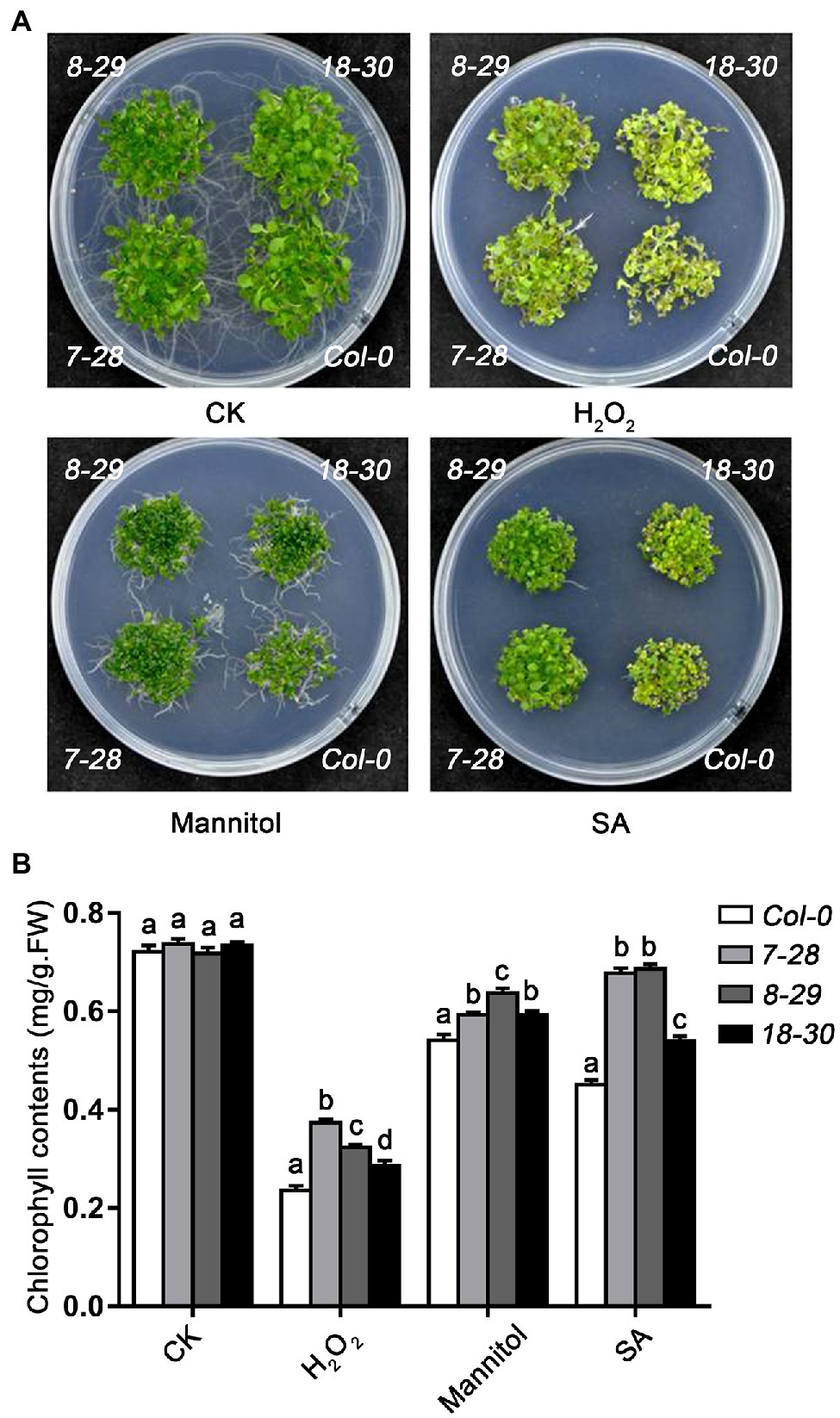
Figure 5. Effect of H2O2, salicylic acid (SA) and mannitol on WT, and transgenic plants. (A) Morphology and (B) Chlorophyll content of WT and transgenic seedlings after H2O2 (10 mmol L−1), mannitol (15% W/V), and SA (0.8 mmol L−1) treatment. The data was recorded after 2 weeks of germination on MS agar plates. For each experiment, at least 30 plants per line were used. Values are means ± SD from three independent measurements. Different lowercase letters above the bars denote significant differences at the p < 0.05 level.
To determine the regulatory role of TaHsfA2-13 on Hsps in Arabidopsis, five Hsp genes, including Hsp21, Hsp70b, Hsp70T, Hsp90, Hsp101, which can be positively regulated by AtHsfA2 were selected in this study (Nishizawa et al., 2006; Ogawa et al., 2007). After mannitol and SA treatments, the expression levels of these Hsp genes were higher in the TaHsfA2-13 overexpressing lines than those in WT. However, not all the expression levels of these Hsp genes were higher in the TaHsfA2-13 overexpressing lines, especially after heat treatment and H2O2 treatment. During basic thermotolerance, the expression level of Hsp90 in transgenic lines was significantly lower than that in WT. The transcript levels of Hsp70T and Hsp90 in transgenic lines were not changed during acquired thermotolerance or H2O2 treatment compared with WT (Figure 6). Further experiments showed that TaHsfA2-13 could interact with promoter sequences of these Hsps, which containing HSE motifs (Figure 7). These results suggest that TaHsfA2-13 enhances the tolerance of transgenic lines to various abiotic stresses, probably mediates by activation of Hsps. Of course, more experiments are needed to explore the specific molecular mechanisms involved in regulating plant responses to various abiotic stresses.
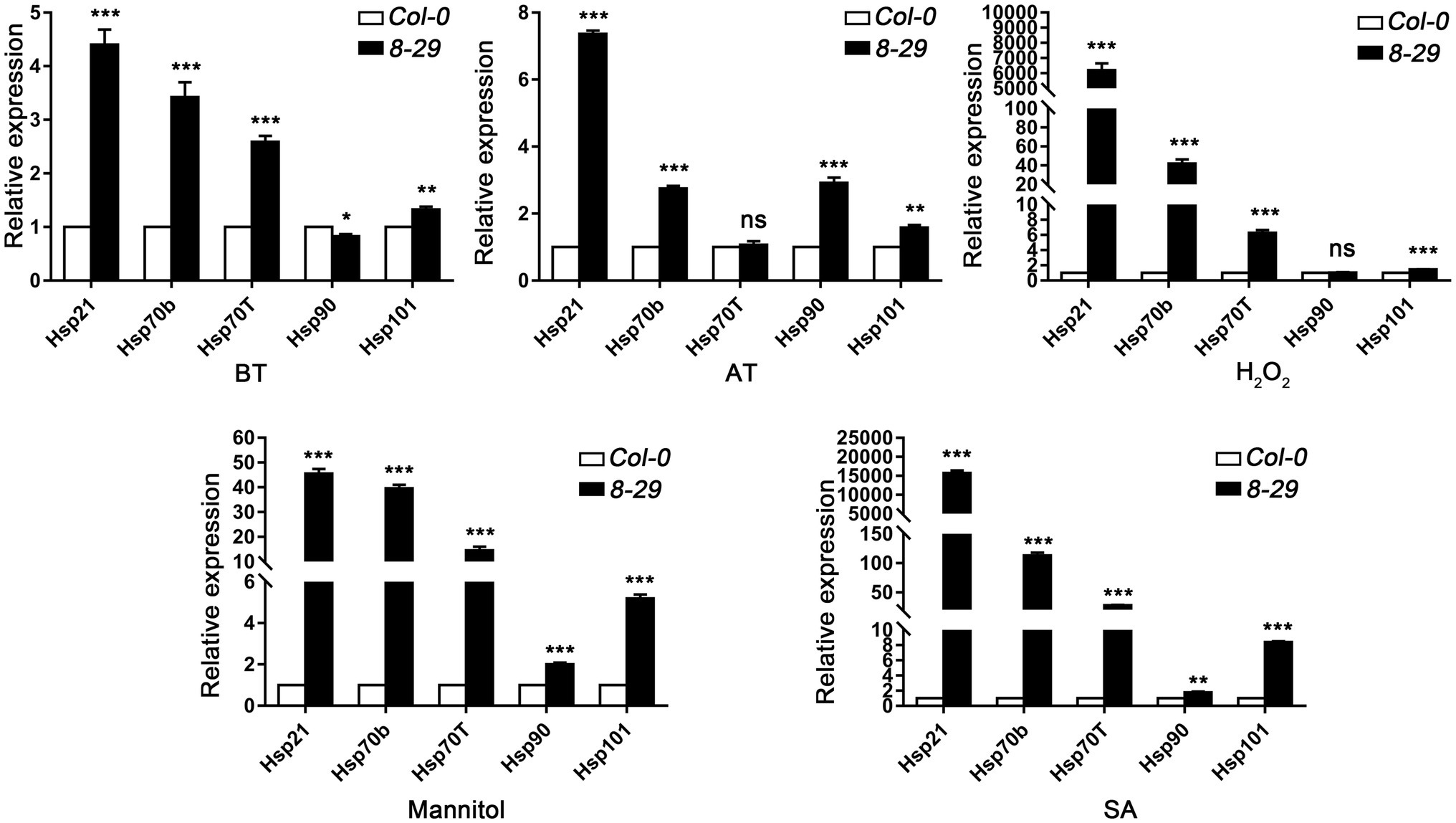
Figure 6. Expression analysis of Hsp genes in WT and transgenic lines after various stresses and phytohormones treatments. For analysis of Hsp genes transcription levels under heat stress, total RNAs were extracted from five-old day seedlings after treatment with slightly modified BT (45°C for 50 min) or AT (37°C for 1 h, incubated at 22°C for 2 days, 46°C for 1 h). For analysis of Hsp genes transcription levels under H2O2, SA and mannitol treatments, total RNAs were extracted from 2-week-old seedlings grown on MS plates with H2O2 (10 mmol L−1), mannitol (15% W/V), and SA (0.8 mmol L−1). The relative expression of Hsp genes were normalized to that of Atactin8, which was used as an internal control gene. All values are presented as the mean ± standard error of values from three independent experiments. Asterisks indicate significant differences compared to WT, and ns means no significant difference (*p < 0.05, **p < 0.01, ***p < 0.005, Student’s t-test).
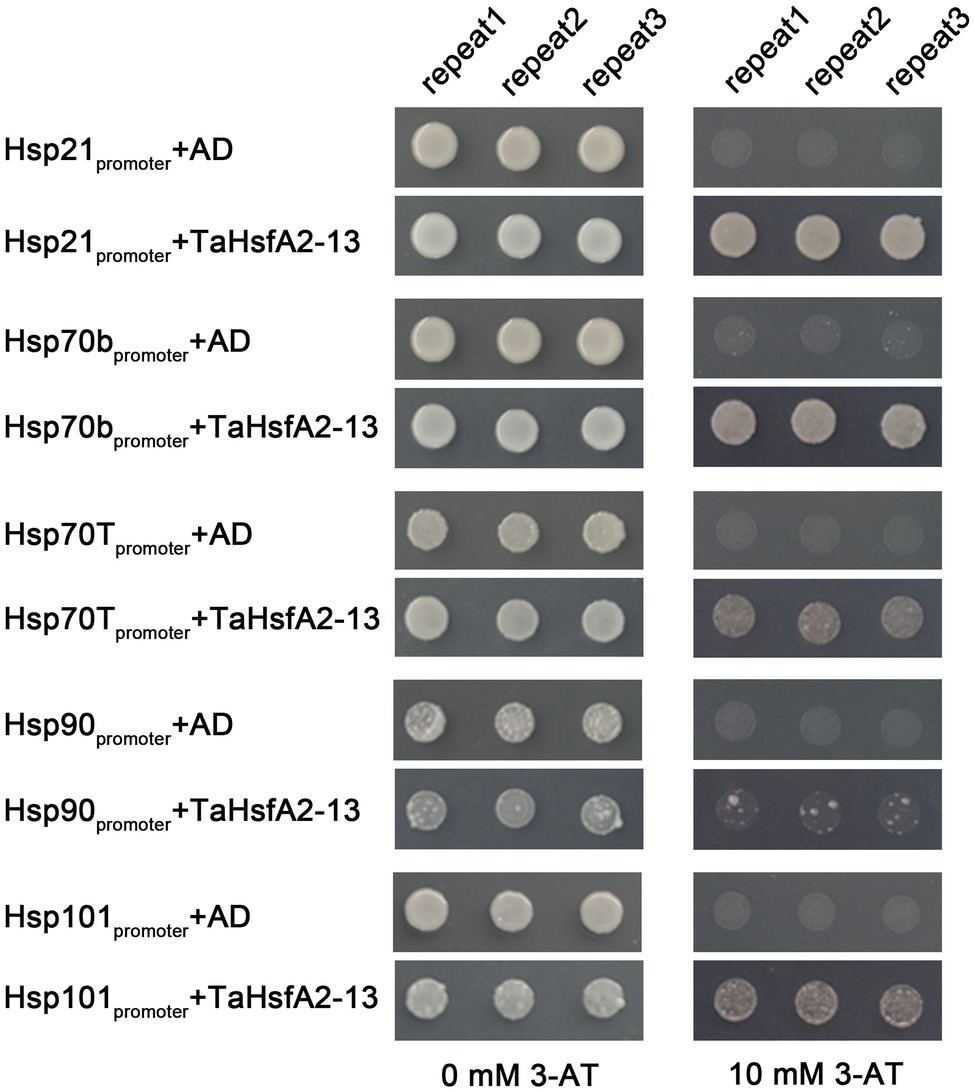
Figure 7. Growth of yeast cells transformed with the bait and prey constructs on SD-Trp-Leu-His supplemented with or without 10 mM 3-AT. The transformation with the empty vector pGADT7 was taken as a negative control. The experiments were repeated three times with the same results.
Wheat is one of the important staple food crops, providing 21% of calories and 20% of protein globally (Lobell and Gourdji, 2012; Shiferaw et al., 2013). It is estimated that the demand for wheat will increase by 60% by 2050, but yields might go down by 29% because of environmental stresses (Manickavelu et al., 2012; Kulkarni et al., 2017). The main objective of agronomic research remains to improve crop productivity under various abiotic stresses (Puranik et al., 2012). Therefore, understanding the molecular mechanisms of plant responses to abiotic stresses is a prerequisite for improving plant stress tolerance and productivity (Guo et al., 2016). Hsfs are downstream components of signal transduction and play important roles in regulating the expression of many genes responding to various environmental stresses (Hrmova and Hussain, 2021). Despite there are large number of Hsfs in wheat, few Hsfs have been cloned and identified due to the complexity of the wheat genome. In this study, we cloned TaHsfA2-13, a member of the A2 subclass, from wheat. Analysis of amino acid sequence showed that TaHsfA2-13 possessed the basic properties of HsfAs (Figure 1). Unlike HsfBs and HsfCs, HsfAs tend to have the AHA domain, which also determines their transcriptional activation activity (Kotak et al., 2004). Indeed, TaHsfA2-13 was found to exhibit transcriptional activation activity in this study (Figure 3A). Similarly, in a recent study, the AHA motif was found to be important for the transcriptional activity of TaHsfA2e-5D (Bi et al., 2022). Although NLS and NES coexist, TaHsfA2-13 is normally located in the nucleus (Figures 1, 3B), which differs its orthologue from Arabidopsis (Evrard et al., 2013). Furthermore, the sequence of TaHsfA2-13 is highly similar to TdHsfA2c-like from diploid T. dicoccoides, indicating that TaHsfA2-13 is highly conserved during the polyploidization (Figure 1). In the A, B and D subgenomes of common bread wheat, approximately 60.1–61.3% of the genes have orthologues in diploid species (International Wheat Genome Sequencing, 2014). It seems that polyploidization has been shown to enhance abiotic stress tolerance in plants by aggregating stress resistance genes (Chao et al., 2013; Yang et al., 2014).
Although there is high sequence similarities among members of the A2 subclass, the expression pattern of each gene is quite different (Duan et al., 2019). In this study, gene expression profiles showed that TaHsfA2-13 transcripts were differentially accumulated in all tested tissues, and the highest expression level was found in young roots, stems, and leaves, suggesting that TaHsfA2-13 might be involved in the growth of wheat seedlings (Figure 2A). However, previous study showed that subclass A2 members including A2b, A2c, and A2e were predominantly expressed in endosperm (Xue et al., 2014). TaHsfA2-1, also belonging to A2 subclass, was mainly expressed in mature leaves (Liu et al., 2020). These results indicate the functional diversity of A2 subclass members in wheat.
In recent years, there have been many reports about Hsfs regulating plant responses to various abiotic stresses and these transcription factors are not only involved in individual stress. For example, overexpression of AtHsfA2 in Arabidopsis increases tolerance to heat, osmosis, and salt stress (Nishizawa et al., 2006). TaHsfA6f can enhance the tolerance of transgenic plants to high temperature, drought and salt stress in wheat (Xue et al., 2015; Bi et al., 2020). Like many Hsfs, TaHsfA2-13 not only improves the basic and acquired thermotolerance of transgenic plants, but also involve in response and tolerance to phytohormones, osmotic, and oxidative stresses (Figures 2, 4, 5; Supplementary Figure S2). Firstly, TaHsfA2-13 was dramatically induced by heat stress (Figure 2B) and transgenic plants overexpressing TaHsfA2-13 in Col-0 or athsfa2 mutant showed improved thermotolerance (Figure 4). These results imply that the improvement of heat tolerance by TaHsfA2-13 is conserved between monocot and dicot plant species. Next, we found that TaHsfA2-13 was upregulated by other abiotic stresses besides heat stress, such as osmotic and oxidative stresses (Figure 2). Indeed, transgenic Arabidopsis lines clearly exhibited better growth status under H2O2 and mannitol treatments, with fewer withered leaves and higher chlorophyll content (Figure 5). Finally, we also found that TaHsfA2-13 responds to SA, ABA and MeJA treatments (Figure 2). A large number of studies have shown that phytohormones play critical roles in helping plants to adapt to various environmental stresses (Verma et al., 2016). For instance, SA alleviates the adverse effects of heat stress on photosynthesis by altering proline production and ethylene formation (Khan et al., 2013). In general, ABA is responsible for plant defense against abiotic stresses such as drought and salinity (Zhang et al., 2006). MeJA operate in parallel, presumably synergistically, to ABA, and are indispensable for osmotic stress tolerance in rice (Tang et al., 2020). Therefore, our future research will focus on the potential molecular mechanism of TaHsfA2-13 regulated by phytohormones in response to various abiotic stresses.
Plant Hsps, as chaperone proteins, play key roles in biotic and abiotic stress tolerance of plants by maintaining the functional conformation of plant proteins and preventing the aggregation of non-native proteins (Ul Haq et al., 2019). Numerous studies have shown that the regulation of hsps by hsfs plays an important role in plant response to various environmental stresses (Guo et al., 2008; Xue et al., 2014; Driedonks et al., 2015). For example, the overexpression of GmHsfA1 improved the thermotolerance of transgenic soybeans, possibly due to the activation of downstream genes GmHsp70, GmHsp22 and other GmHsps (Zhu et al., 2009). In Arabidopsis, Hsp90 is regulated by HsfA2 and enhances tolerance to oxidative stress (Nishizawa-Yokoi et al., 2010). During seedling stage of wheat, TaHsfA6e was reported to regulate the expression of TaHsps genes, such as Hsp17, Hsp70 and Hsp90 in response to high temperature and drought (Kumar et al., 2018). Additionally, TaHsfA6f could improve thermotolerance in wheat, which depended on the function of downstream target genes, including TaHsps (TaHSP16.8, TaHSP17, TaHSP17.3 and TaHSP90.1-A1), TaRof1, galactinol synthase, and glutathione-S-transferase (Xue et al., 2015). In this study, with a few exceptions, almost all tested Hsps were strongly upregulated in TaHsfA2-13 overexpressing transgenic lines after various stresses and phytohormones treatments (Figure 6). Previous studies have found that HSPs genes (sHsp., Hsp70, Hsp90 and DnaJ) and Hsfs may be closely related to the structural integrity and function of chloroplast (Wu et al., 2018). Here, due to the protection of high level of Hsps, TaHsfA2-13 overexpressing transgenic lines grew better and had higher chlorophyll content than WT (or athsfa2; Figures 4B, 5B; Supplementary Figure S2). Taken together, our results suggest that TaHsfA2-13 is a positive regulator of abiotic stresses, including heat, osmotic and oxidative stress, and involved in response to phytohormones treatments, possibly by upregulation of stress-related response genes, such as Hsps.
In summary, we clarified important roles of wheat TaHsfA2-13 gene in response to HS, H2O2, mannitol, and SA in Arabidopsis. TaHsfA2-13 exerts stress tolerance functions by regulating the expression of downstream genes such as Hsps. It indicates that different signal pathways share the same signal molecules under stress conditions. Our study elevates the mechanistic understanding of individual gene involved in various stress and cross-talk tolerance in plant and provides genetic materials for crop stress tolerance breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XM, XG, and GL designed this study. XM performed bioinformatics analysis, generated and analyzed data, and wrote the manuscript. BZ, ML, RL, and QR performed the experiments. XG and GL edited this manuscript and contributed with valuable discussions. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of Hebei Province (no. C2021301024); HAAFs Science and Technology Innovation Special Project (no. 2022KJCXZX-SSS-2); Talents Construction Project of Science and Technology Innovation of HAAFs (no. C22R1301); Basic Research Fund of Hebei Academy of Agriculture and Forestry Sciences (no. 2021110204).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Yee-Yung Charng (Agricultural Biotechnology Research Center, Academia Sinica, Taipei) for providing athsfa2 mutant seeds.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.922561/full#supplementary-material
Supplementary Figure S1 | Identification of transgenic plants. The mRNA levels of TaHsfA2-13 overexpressing lines and WT (or athsfa2) were examined by semi-quantitative RT-PCR analysis, and the expression of Atactin8 was analyzed as a loading control.
Supplementary Figure S2 | Comparison of wild type (WT), athsfa2 and TaHsfA2-13 overexpressing transgenic plants in response to various abiotic stresses and phytohormones. (A) Morphology and (B) Chlorophyll content of WT, athsfa2 and transgenic seedlings after various abiotic stresses and phytohormones. For each experiment, at least 30 plants per line were used. Values are means ± SD from three independent measurements. Different lowercase letters above the bars denote significant differences (p < 0.05).
Hsf, Heat shock transcription factor; HS, Heat shock; SA, Salicylic acid; WT, Wild type; Hsp, Heat shock protein; HSE, HS responsive element; DBD, DNA-binding domain; OD, Oligomerization domain; NLS, Nuclear localization signal; NES, Nuclear export signal; AHA, Activator peptide motif.
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes (Basel) 10:771. doi: 10.3390/genes10100771
Baniwal, S. K., Bharti, K., Chan, K. Y., Fauth, M., Ganguli, A., Kotak, S., et al. (2004). Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487. doi: 10.1007/BF02712120
Banti, V., Mafessoni, F., Loreti, E., Alpi, A., and Perata, P. (2010). The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 152, 1471–1483. doi: 10.1104/pp.109.149815
Bi, H., Miao, J., He, J., Chen, Q., Qian, J., Li, H., et al. (2022). Characterization of the wheat heat shock factor TaHsfA2e-5D conferring heat and drought tolerance in Arabidopsis. Int. J. Mol. Sci. 23, 2784. doi: 10.3390/ijms23052784
Bi, H., Zhao, Y., Li, H., and Liu, W. (2020). Wheat heat shock factor TaHsfA6f increases ABA levels and enhances tolerance to multiple abiotic stresses in transgenic plants. Int. J. Mol. Sci. 21, 3121. doi: 10.3390/ijms21093121
Chan-Schaminet, K. Y., Baniwal, S. K., Bublak, D., Nover, L., and Scharf, K. D. (2009). Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 284, 20848–20857. doi: 10.1074/jbc.M109.007336
Chao, D. Y., Dilkes, B., Luo, H., Douglas, A., Yakubova, E., Lahner, B., et al. (2013). Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341, 658–659. doi: 10.1126/science.1240561
Charng, Y. Y., Liu, H. C., Liu, N. Y., Chi, W. T., Wang, C. N., Chang, S. H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. doi: 10.1104/pp.106.091322
Chauhan, H., Khurana, N., Agarwal, P., Khurana, J. P., and Khurana, P. (2013). A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS One 8:e79577. doi: 10.1371/journal.pone.0079577
Chen, K., Gao, J. H., Sun, S. J., Zhang, Z. J., Yu, B., Li, J., et al. (2020). BONZAI proteins control global osmotic stress responses in plants. Curr. Biol. 30, 4815–4825.e4. doi: 10.1016/j.cub.2020.09.016
Driedonks, N., Xu, J., Peters, J. L., Park, S., and Rieu, I. (2015). Multi-level interactions between heat shock factors, heat shock proteins, and the rRedox system regulate acclimation to heat. Front. Plant Sci. 6:999. doi: 10.3389/fpls.2015.00999
Duan, S., Liu, B., Zhang, Y., Li, G., and Guo, X. (2019). Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genomics 20:257. doi: 10.1186/s12864-019-5617-1
Evrard, A., Kumar, M., Lecourieux, D., Lucks, J., von Koskull-Doring, P., and Hirt, H. (2013). Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ. 1:e59. doi: 10.7717/peerj.59
Fragkostefanakis, S., Mesihovic, A., Simm, S., Paupiere, M. J., Hu, Y., Paul, P., et al. (2016). HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in yomato male reproductive tissues. Plant Physiol. 170, 2461–2477. doi: 10.1104/pp.15.01913
Fragkostefanakis, S., Roth, S., Schleiff, E., and Scharf, K. D. (2015). Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 38, 1881–1895. doi: 10.1111/pce.12396
Giorno, F., Wolters-Arts, M., Grillo, S., Scharf, K. D., Vriezen, W. H., and Mariani, C. (2010). Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J. Exp. Bot. 61, 453–462. doi: 10.1093/jxb/erp316
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Guo, M., Liu, J. H., Ma, X., Luo, D. X., Gong, Z. H., and Lu, M. H. (2016). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7:114. doi: 10.3389/fpls.2016.00114
Guo, M., Lu, J. P., Zhai, Y. F., Chai, W. G., Gong, Z. H., and Lu, M. H. (2015). Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 15:151. doi: 10.1186/s12870-015-0512-7
Guo, J., Wu, J., Ji, Q., Wang, C., Luo, L., Yuan, Y., et al. (2008). Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genomics 35, 105–118. doi: 10.1016/S1673-8527(08)60016-8
Heerklotz, D., Doring, P., Bonzelius, F., Winkelhaus, S., and Nover, L. (2001). The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 21, 1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001
Hrmova, M., and Hussain, S. S. (2021). Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 22:5662. doi: 10.3390/ijms22115662
Huang, B., Huang, Z., Ma, R., Chen, J., Zhang, Z., and Yrjala, K. (2021). Genome-wide identification and analysis of the heat shock transcription factor family in moso bamboo (Phyllostachys edulis). Sci. Rep. 11:16492. doi: 10.1038/s41598-021-95899-3
Ikeda, M., Mitsuda, N., and Ohme-Takagi, M. (2011). Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254. doi: 10.1104/pp.111.179036
International Wheat Genome Sequencing, C. (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. doi: 10.1126/science.1251788
Khan, M. I. R., Iqbal, N., Masood, A., Per, T. S., and Khan, N. A. (2013). Salicylic acid alleviates adverse effects of heat stress on photosynthesis through changes in proline production and ethylene formation. Plant Signal. Behav. 8:e26374. doi: 10.4161/psb.26374
Kotak, S., Port, M., Ganguli, A., Bicker, F., and von Koskull-Doring, P. (2004). Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 39, 98–112. doi: 10.1111/j.1365-313X.2004.02111.x
Kulkarni, M., Soolanayakanahally, R., Ogawa, S., Uga, Y. G., Selvaraj, M., and Kagale, S. (2017). Drought response in wheat: key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front. Chem. 5:106. doi: 10.3389/fchem.2017.00106
Kumar, R. R., Goswami, S., Singh, K., Dubey, K., Rai, G. K., Singh, B., et al. (2018). Characterization of novel heat-responsive transcription factor (TaHSFA6e) gene involved in regulation of heat shock proteins (HSPs)—a key member of heat stress-tolerance network of wheat. J. Biotechnol. 279, 1–12. doi: 10.1016/j.jbiotec.2018.05.008
Larkindale, J., Hall, J. D., Knight, M. R., and Vierling, E. (2005). Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 138, 882–897. doi: 10.1104/pp.105.062257
Li, H. C., Zhang, H. N., Li, G. L., Liu, Z. H., Zhang, Y. M., Zhang, H. M., et al. (2015). Expression of maize heat shock transcription factor gene ZmHsf06 enhances the thermotolerance and drought-stress tolerance of transgenic Arabidopsis. Funct. Plant Biol. 42, 1080–1091. doi: 10.1071/FP15080
Li, G. L., Zhang, H. N., Shao, H., Wang, G. Y., Zhang, Y. Y., Zhang, Y. J., et al. (2019). ZmHsf05, a new heat shock transcription factor from Zea mays L. improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant. Plant Sci. 283, 375–384. doi: 10.1016/j.plantsci.2019.03.002
Liu, G. R., Chen, X. Z., and Duan, W. Q. (2002). The relationship between wheat leaf osmotic adjustment ability and varietal drought resistance under water stress [in Chinese]. J. Agric. Univ. Hebei 25, 1–3. doi: 10.3969/j.issn.1000-1573.2002.02.001
Liu, J., Feng, L., Gu, X., Deng, X., Qiu, Q., Li, Q., et al. (2019). An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 29, 379–390. doi: 10.1038/s41422-019-0145-8
Liu, Z., Li, G., Zhang, H., Zhang, Y., Zhang, Y., Duan, S., et al. (2020). TaHsfA2-1, a new gene for thermotolerance in wheat seedlings: characterization and functional roles. J. Plant Physiol. 246-247:153135. doi: 10.1016/j.jplph.2020.153135
Liu, H. C., Liao, H. T., and Charng, Y. Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34, 738–751. doi: 10.1111/j.1365-3040.2011.02278.x
Lobell, D. B., and Gourdji, S. M. (2012). The influence of climate change on global crop productivity. Plant Physiol. 160, 1686–1697. doi: 10.1104/pp.112.208298
Manickavelu, A., Kawaura, K., Oishi, K., Shin-I, T., Kohara, Y., Yahiaoui, N., et al. (2012). Comprehensive functional analyses of expressed sequence tags in common wheat (Triticum aestivum). DNA Res. 19, 165–177. doi: 10.1093/dnares/dss001
Nakashima, K., Ito, Y., and Yamaguchi-Shinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., and Shigeoka, S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. doi: 10.1111/j.1365-313X.2006.02889.x
Nishizawa-Yokoi, A., Nosaka, R., Hayashi, H., Tainaka, H., Maruta, T., Tamoi, M., et al. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52, 933–945. doi: 10.1093/pcp/pcr045
Nishizawa-Yokoi, A., Tainaka, H., Yoshida, E., Tamoi, M., Yabuta, Y., and Shigeoka, S. (2010). The 26S proteasome function and Hsp90 activity involved in the regulation of HsfA2 expression in response to oxidative stress. Plant Cell Physiol. 51, 486–496. doi: 10.1093/pcp/pcq015
Nover, L., Bharti, K., Döring, P., Mishra, S. K., Ganguli, A., and Scharf, K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2
Ogawa, D., Yamaguchi, K., and Nishiuchi, T. (2007). High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 58, 3373–3383. doi: 10.1093/jxb/erm184
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2016.08.015
Savchenko, T., and Tikhonov, K. (2021). Oxidative stress-induced alteration of plant central metabolism. Life (Basel). 11:304. doi: 10.3390/life11040304
Scharf, K. D., Berberich, T., Ebersberger, I., and Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Scharf, K. D., Heider, H., Hohfeld, I., Lyck, R., Schmidt, E., and Nover, L. (1998). The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 18, 2240–2251. doi: 10.1128/MCB.18.4.2240
Shiferaw, B., Smale, M., Braun, H. J., Duveiller, E., Reynolds, M., and Muricho, G. (2013). Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 5, 291–317. doi: 10.1007/s12571-013-0263-y
Tang, G. L., Ma, J. N., Hause, B., Nick, P., and Riemann, M. (2020). Jasmonate is required for the response to osmotic stress in rice. Environ. Exp. Bot. 175:104047. doi: 10.1016/j.envexpbot.2020.104047
Udvardi, M. K., Kakar, K., Wandrey, M., Montanari, O., Murray, J., Andriankaja, A., et al. (2007). Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol. 144, 538–549. doi: 10.1104/pp.107.098061
Ul Haq, S., Khan, A., Ali, M., Khattak, A. M., Gai, W. X., Zhang, H. X., et al. (2019). Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 20:5321. doi: 10.3390/ijms20215321
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wu, H., Shi, N., An, X., Liu, C., Fu, H., Cao, L., et al. (2018). Candidate genes for yellow leaf color in common wheat (Triticum aestivum L.) and major related metabolic pathways according to transcriptome profiling. Int. J. Mol. Sci. 19:1594. doi: 10.3390/ijms19061594
Xue, G. P., Drenth, J., and McIntyre, C. L. (2015). TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 66, 1025–1039. doi: 10.1093/jxb/eru462
Xue, G. P., Sadat, S., Drenth, J., and McIntyre, C. L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557. doi: 10.1093/jxb/ert399
Yang, C., Zhao, L., Zhang, H., Yang, Z., Wang, H., Wen, S., et al. (2014). Evolution of physiological responses to salt stress in hexaploid wheat. Proc. Natl. Acad. Sci. U. S. A. 111, 11882–11887. doi: 10.1073/pnas.1412839111
Yokotani, N., Ichikawa, T., Kondou, Y., Matsui, M., Hirochika, H., Iwabuchi, M., et al. (2008). Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 227, 957–967. doi: 10.1007/s00425-007-0670-4
Zhang, J. H., Jia, W. S., Yang, J. C., and Ismail, A. M. (2006). Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 97, 111–119. doi: 10.1016/j.fcr.2005.08.018
Zhang, J., Li, Y., Jia, H. X., Li, J. B., Huang, J., Lu, M. Z., et al. (2015). The heat shock factor gene family in Salix suchowensis: a genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 6:748. doi: 10.3389/fpls.2015.00748
Zhu, Y., Wang, Z., Jing, Y., Wang, L., Liu, X., Liu, Y., et al. (2009). Ectopic over-expression of BhHsf1, a heat shock factor from the resurrection plant Boea hygrometrica, leads to increased thermotolerance and retarded growth in transgenic Arabidopsis and tobacco. Plant Mol. Biol. 71, 451–467. doi: 10.1007/s11103-009-9538-2
Keywords: wheat, Hsf, TaHsfA2-13, abiotic stresses, transgenic, Arabidopsis
Citation: Meng X, Zhao B, Li M, Liu R, Ren Q, Li G and Guo X (2022) Characteristics and Regulating Roles of Wheat TaHsfA2-13 in Abiotic Stresses. Front. Plant Sci. 13:922561. doi: 10.3389/fpls.2022.922561
Received: 18 April 2022; Accepted: 10 June 2022;
Published: 27 June 2022.
Edited by:
Caroline Müller, Universidade Federal da Fronteira Sul, BrazilReviewed by:
Shabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaCopyright © 2022 Meng, Zhao, Li, Liu, Ren, Li and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiulin Guo, bXloZjIwMDJAMTYzLmNvbQ==; Guoliang Li, Z3VvbGlhbmdsaWxpQGhhYWZzLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.