- 1Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA, United States
- 2Department of Biology, University of Washington, Seattle, WA, United States
The circadian clock represents a critical regulatory network, which allows plants to anticipate environmental changes as inputs and promote plant survival by regulating various physiological outputs. Here, we examine the function of the clock-regulated transcription factor, CYCLING DOF FACTOR 6 (CDF6), during cold stress in Arabidopsis thaliana. We found that the clock gates CDF6 transcript accumulation in the vasculature during cold stress. CDF6 mis-expression results in an altered flowering phenotype during both ambient and cold stress. A genome-wide transcriptome analysis links CDF6 to genes associated with flowering and seed germination during cold and ambient temperatures, respectively. Analysis of key floral regulators indicates that CDF6 alters flowering during cold stress by repressing photoperiodic flowering components, FLOWERING LOCUS T (FT), CONSTANS (CO), and BROTHER OF FT (BFT). Gene ontology enrichment further suggests that CDF6 regulates circadian and developmental-associated genes. These results provide insights into how the clock-controlled CDF6 modulates plant development during moderate cold stress.
Introduction
The circadian clock consists of an expansive regulatory network, which enables eukaryotic organisms to synchronize their metabolism, physiology, and development to daily and seasonal environmental changes (Greenham and McClung, 2015; Creux and Harmer, 2019). Through a coordinated and interconnected series of transcriptional–translational feedback regulations between multiple components, the clock modulates the expression of a large proportion of the transcriptome in plants. For example, the clock regulates 40–50% of genes involved in plant abiotic stress responses (Covington et al., 2008). Several recent transcriptome studies indicate that the time of day impacts the plant transcriptional response to abiotic stimulus (Wilkins et al., 2010; Blair et al., 2019; Grinevich et al., 2019; Bonnot et al., 2021; Markham and Greenham, 2021). The clock is also involved in regulating several critical developmental phenotypes. For example, mis-expression of some clock components (CIRCADIAN CLOCK ASSOCIATED 1/CCA1, EARLY FLOWERING 3/ELF3, PSEUDO RESPONSE REGULATOR 9/PRR9, TIMING OF CAB EXPRESSION 1/TOC1, etc.) results in altered hypocotyl growth and flowering (Nagel and Kay, 2012; Huang and Nusinow, 2016; Nakamichi, 2020).
In Arabidopsis, the clock coordinates aspects of photoperiodic flowering primarily through the regulation of GIGANTEA (GI) (Song et al., 2015). During long-day (16-h light: 8-h dark) conditions, GI forms a complex with FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) to target the degradation of CYCLING DOF FACTORs (CDFs), a small subfamily of the DNA-binding with one finger (DOF) transcription factor (TF) family, which enables CONSTANS (CO) and FLOWERING LOCUS T (FT) to accumulate and promote flowering (Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009; Nohales et al., 2019). However, during short-day conditions, the CDFs bind to the DOF-binding sites (AAAG) of FT and CO and act redundantly to suppress the accumulation of FT and CO mRNAs, resulting in delayed flowering (Imaizumi et al., 2005; Fornara et al., 2009). Recent work indicates that the vasculature-expressed CDF6 negatively regulates FT transcript abundance, resulting in delayed flowering (Krahmer et al., 2019). CDF6 also interacts with GI, consistent with the known mechanism for the GI-FKF1 module degrading CDF family members during long-day conditions (Krahmer et al., 2019).
The CDFs are regulated by several clock components. For example, the day-expressed PRR9, PRR7, and PRR5 sequentially bind to the CDF5 promoter region to repress its accumulation during the day (Martín et al., 2018). PRR7 and PRR5 associate with the promoter region of CDF3 (Nakamichi et al., 2012). Furthermore, PRR9, PRR7, and PRR5 negatively regulate the expression of CDF1, while CCA1 positively regulates the CDF1 expression via regulation of GI (Nakamichi et al., 2007). The expression of both CDF5 and its natural antisense transcript, CDF5 LONG NONCODING RNA (FLORE), is altered in CCA1 over-expression lines (Henriques et al., 2017). In addition, the expression of the lesser characterized CDF6 is also clock-regulated, and PRR9 and LATE ELONGATED HYPOCOTYL (LHY) directly bind to the CDF6 promoter (Liu et al., 2016; Adams et al., 2018; Blair et al., 2019).
Photoperiodic flowering regulators are involved in various abiotic stress responses. GI is required for the drought escape, oxidative stress, and cold stress response (Riboni et al., 2013; Fornara et al., 2015). CDF3 confers tolerance to drought and freezing temperatures, while acting in both GI-dependent and GI-independent abiotic stress response pathways (Corrales et al., 2017; Renau-Morata et al., 2020). Furthermore, plants over-expressing the tomato orthologs of CDF1 and CDF3 show increased tolerance during drought and salinity stress (Corrales et al., 2014). CDF1, 2, 3, 5, and 6 are uniformly upregulated in response to cold stress (Kilian et al., 2007; Calixto et al., 2018; Blair et al., 2019). As such, the CDFs are considered part of the cold-regulated (COR) genes, due to their altered expression in a transcriptome analysis of the central cold regulators, C-REPEAT BINDING FACTORS (CBFs) triple loss-of-function mutants (cbf123) (Shi et al., 2017; Song et al., 2021). Interestingly, both CDF5 and CDF6 were induced over 2-fold times in response to low temperature in parallel with CBF1, CBF2, and CBF3 (Park et al., 2015). While CDF3 confers cold stress tolerance and CDF5 impacts hypocotyl elongation during short days (Corrales et al., 2017; Martín et al., 2020), the CDFs have not been shown to have differential functions in regulating plant growth and development during cold stress.
Here, we use a combination of meta-data, transcriptomic, genetic, and phenotypic approaches to better understand the interplay between the clock regulation and the function of the lesser characterized family member, CDF6, during cold stress. We find that CCA1 modulates CDF6 transcript accumulation in response to moderate cold stress. Through analysis of tissue-specific (SUC2) and mutant genotypes, we find that CDF6 influences photoperiodic flowering in both ambient and cold temperatures, and germination during ambient temperature. Specifically, vasculature-expressed CDF6 represses FT, CO, and BFT to regulate flowering. Finally, we show that CDF6 significantly alters the transcriptome during cold stress including the expression of genes involved in flowering, rhythmic, and metabolic processes.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana seeds were sterilized for 3–4 h (~100 mL of 6% sodium hypochlorite and ~4 mL of concentrated hydrochloric acid), plated on 1X Murashige and Skoog (MS) medium supplemented with 1.5% sucrose (w/v), and stratified in the dark for three nights at 4°C. Seeds were grown at a constant temperature of 22°C with ~90 μmol photons·s−1·m−2, in diurnal (12-h light: 12-h dark; LD) cycles for 8 days. For circadian and time of day experiments, seedlings were transferred to constant light (LL) for 2 days before sampling. Columbia-0 (Col-0) was used as the wild-type (WT) control. Clock genotypes (cca1-1/lhy-21, CCA1-OX (35S::CCA1), SUC2::CCA1 #18, and prr7-3/prr9-1) and CDF mis-expressed line (SUC2::HA-CDF6 #11) were previously characterized (Farré et al., 2005; Pruneda-Paz et al., 2009; Endo et al., 2014; Nagel et al., 2014; Krahmer et al., 2019). The cdf12356 quintuple line was generated using the CRISPR/Cas9-based gene editing constructs in pKIR1.1 plasmids (Tsutsui and Higashiyama, 2017) containing CDF1 (5'- GTTTGGCTGGACAATTACAC-3') and CDF6 (5'-GTCTCAAGTTAGAGATACTC-3') gRNAs using the cdf235 triple T-DNA mutants as a genetic background (Fornara et al., 2009). To generate cdf1 and cdf6 mutations, the cdf1235 quadruple mutant was generated first, and then the cdf6 mutation was induced by gene editing in the cdf1235 quadruple to generate the cdf12356 quintuple mutant (Supplementary Figure 1). The cdf6 single mutant is a SALK T-DNA insertion mutant line (SALK_010734), which was genotyped to confirm homozygosity. The reduced CDF6 expression level was validated via quantitative real-time PCR (qRT-PCR). To generate pCDF6::CDF6-GUS plants, the genomic copy with ~1.2 kb of the promoter and the coding sequence was PCR purified and cloned into pENTR d-TOPO (Invitrogen). LR Clonase II was used to perform the Gateway reaction with pMDC162 to create pCDF6::CDF6-GUS. The sequences were confirmed via Sanger sequencing [Institute for Integrated Genome Biology (IIGB) Genomics Core, University of California, Riverside (UCR)], and the vectors were transformed into WT plants with Agrobacterium-mediated transformation. Two independent T3 lines (Lines A and B) were selected for GUS assays. For cold treatment, seedlings were exposed for 1 h at 10°C and sampled at 4-h intervals (ZT12–ZT36) along with control samples grown at ambient temperature (22°C) after 8 days of entrainment in LD and 2 days in continuous light (LL). For the continuous cold experiments, plants were grown in long-day conditions (16-h light: 8-h dark) for 8 days at 22°C. Then the seedlings were maintained at 22°C or transferred to long-day conditions at 10°C for 2 days before sampling every 4 h.
Quantitative Real-Time PCR
Seedlings were prepared and grown as described above. The total mRNA was isolated with the GeneJET Plant RNA Purification Kit (Thermo Fisher Scientific). cDNA was synthesized using 1 μg of total RNA and was reverse-transcribed with the iScript cDNA synthesis kit (Bio-rad). qRT-PCR was performed with SYBR Green Master Mix (Thermo Fisher Scientific) with the CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Three biological and three technical replicates were analyzed. The relative expression was calculated against the housekeeping gene, ISPOENTENYL-DIPOHSPHATE DELTA ISOMERASE II (IPP2), using the ΔΔCq method. See Supplementary Table 1 for gene-specific primer sequences and qRT-PCR conditions used in this study.
GUS Assay
For beta-glucuronidase (GUS) assays, plants were grown in long-day conditions (16-h light: 8-h dark), similar to the flowering time experiments described in the following section, and GUS staining was performed as previously described with the following modifications (Yang et al., 2018). Four seedlings were harvested in 1 mL of cold 90% (v/v) acetone and then vacuumed for 10 min. Seedlings were fixed at room temperature for 60 min. Acetone was replaced with ~500 μl of wash buffer (10 mM EDTA, 50 mM phosphate buffer (pH 7), 0.1% (v/v) Triton X-100, 1 mM potassium ferrocyanide, and 1 mM potassium ferricyanide in 20% methanol) on ice. After vacuuming three times for 10 min, wash buffer was replaced with ~500 μl of staining buffer [10 mM EDTA, 50 mM phosphate buffer (pH 7), 0.1% (v/v) Triton X-100, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 20% methanol, and 2 mM X-Gluc (Carbosynth Ltd, United Kingdom)]. Seeds were vacuum-infiltrated for 40 min and then incubated for 20 min for pCDF6::CDF6-GUS line A and ~16 h for line B at 37°C. After incubation, seedlings were washed and stored in 70% (v/v) ethanol until imaging on a Leica M165 FC stereoscope (Leica Microsystems CMS GmbH, Wetzlar, Germany) using white light.
Flowering Time and Seed Germination Assays
For flowering time assays, seeds were stratified for 2–3 nights after sterilization and grown for ~10 days on 1X MS plates at ~90 μmol photons·s−1·m-2. Seedlings were transferred to soil and grown in long-day (16-h light: 8-h dark) or short-day (8-h light: 16-h dark) conditions at 22°C or 10°C as indicated in figure legends. Flowering time was measured as the mean number of rosette leaves for at least three independent replicates with n≥6 plants per genotype. Germination was defined as the emergence of the radicle and measured for three biological replicates of 75–100 seeds from WT, cdf6, cdf12356, and SUC2::CDF6, using a modified method of Nelson et al. (2009). Briefly, seeds were liquid sterilized with 70% (v/v) ethanol and 0.05% (v/v) Triton X-100 followed by incubations with 70% (v/v) and 95% (v/v) ethanol. Seeds were sprinkled on filter papers placed on 0.8% (w/v) Bacto-agar plates. After plating, seeds were immediately transferred to constant light conditions (μmol photons·s−1·m-2) at 22°C or 10°C.
RNA-Sequencing Setup and Analysis
Three biological replicates of WT, cdf6, and SUC2::CDF6 were grown as described above and sampled with lights on (Zeitgeber Time 0, ZT0) after 1 h of temperature treatment at 10°C along with control seedlings grown at 22°C. About 1 μg of total RNA was isolated and treated with DNAse I (Millipore Sigma). mRNA purification and libraries were prepared as described previously (Blair et al., 2019). Final libraries were purified by Ampure XP beads. Library quality was confirmed via Qubit 2.0 Fluorescence Reader (ThermoFisher Scientific) and Bioanalyzer 2100 (Agilent Genomics). The analysis pipeline was also described previously (Blair et al., 2019). Briefly, the sequencing analysis was performed at the UCR IIGB Genomics Core facility on the NextSeq500 (Illumina), which generated single-end 75-bp sequences. Reads were mapped to the TAIR10 genome using Hisat2, and limma.voom was used to determine differential gene expression (Law et al., 2014; Kim et al., 2015; H Backman and Girke, 2016). Differentially expressed genes (DEGs) were defined as genes with −1 > Log2 fold change > 1 and false discovery rate < 0.05. Gene ontology (GO) terms were assigned with a previously published pipeline, and flowering-related genes were identified using a previously published dataset (Bonnot et al., 2019; Kinoshita and Richter, 2020).
Meta-Data Analysis
The transcription factor binding site motif analysis was conducted by inputting 500 bp upstream of the DOF-TF family members, from the TAIR bulk data download, into Find Individual Motif Occurrences (FIMO) (Grant et al., 2011; Le Hir and Bellini, 2013). The timing of peak expression (phase) was determined with the Phaser tool from the DIURNAL database (Mockler et al., 2007). The list of COR genes was obtained from an analysis of the cbf1, cbf2, and cbf3 transcriptomes (Shi et al., 2017). The authors used Cuffdiff default parameters to normalize, perform statistical analysis, and identify differentially expressed genes from FPKM values; please refer to the “Methods” section of Shi et al. (2017) for additional details. The upstream clock regulators of DOF TFs were compiled from published Chromatin Immunoprecipitation-sequencing (ChIP-seq) datasets (Huang et al., 2012; Liu et al., 2013, 2016; Nagel et al., 2015; Kamioka et al., 2016; Adams et al., 2018). TFs that may bind to CDF6 in vitro were identified using the DNA Affinity Purification (DAP)-sequencing genome browser by selecting TF families, searching for AT1G26790 (CDF6) in the viewer, and checking for peaks that might indicate TF occupancy (O'Malley et al., 2016).
Identifiers of Genes Referenced in This Study
AT5G62430/CDF1, AT5G39660/CDF2, AT3G47500/CDF3, AT2G34140/CDF4, AT1G69570/CDF5, AT1G26790/CDF6, AT1G69572/FLORE, AT2G31230/ERF15, AT1G60960/ATIRT3, AT2G46790/PRR9, AT5G02810/PRR7, AT1G01060/LHY, AT2G46830/CCA1, AT5G15840/CO, AT1G65480/FT, AT1G07887, AT1G53480/MRD1, AT1G53490/HEI10, AT1G76960, AT5G35935, AT5G35940, AT1G60960/ATIRT3, AT1G75945, AT2G06995, AT3G59930, AT5G33355, AT1G68050/FKF1, AT1G68840/ATRAV2, AT1G80340/GA3OX2, AT2G45660/SOC1, AT5G24470/PRR5, AT5G60910/AGL8, and AT5G62040/BFT.
Data Availability
The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database and can be accessed through GEO Series accession GSE197581.
Results
CCA1 Gates CDF6 Expression in Response to Cold Stress
We previously showed that the expression of CDF6 is altered in the cca1lhy clock mutants under diurnal (12-h light: 12-h dark; LD) conditions (Blair et al., 2019). In this study, we found that in constant light (LL) and temperature, CDF6 expression is also altered in the CCA1 over-expressor (35S::CCA1), cca1lhy, and prr7prr9 mutants, which is evident by dampened transcript accumulation and changes in peak expression (Supplementary Figures 2A–C). Similar to the other CDFs, we observed that CDF6 is localized to the vasculature (Figure 1A). We therefore assessed the expression of CDF6 in previously published lines where CCA1 is driven by the phloem companion cell (PCC)-specific promoter of SUCROSE-PROTON SYMPORTER (SUC2::CCA1) (Endo et al., 2014). Interestingly, when CCA1 is expressed in the PCC, CDF6 transcript accumulation is significantly altered relative to WT (Col-0) (Figure 1B) and to a higher extent compared to changes in 35S::CCA1, specifically between ZT20 and ZT24 (Supplementary Figure 2A), the time points where the transcript levels of both CDF6 and CCA1 are at the highest in the WT (Figure 1B; Supplementary Figure 3A). These results indicate that CCA1 regulates CDF6 transcript accumulation in the PCC of the vasculature.
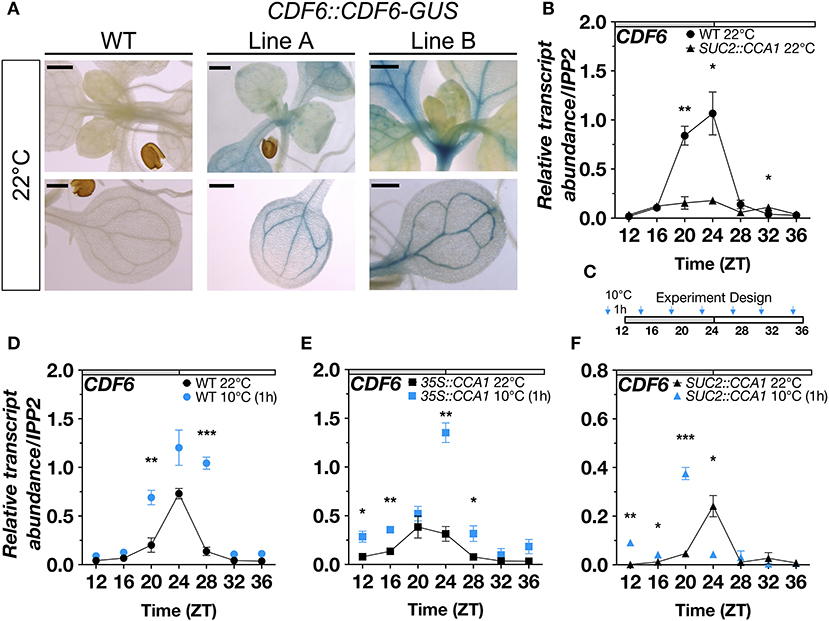
Figure 1. Vasculature-specific CDF6 expression is gated by the clock in response to cold stress. (A) GUS activity in WT and two independent T3 CDF6::CDF6-GUS plants (Lines A and B) indicate the expression pattern of CDF6 at ZT0 in 11-day-old seedlings grown in long-day (16-h light: 8-h dark) conditions. Bars correspond to 500 μm. (B) qRT-PCR of CDF6 transcript abundance in WT (Col-0) and SUC2::CCA1 in ambient temperature. qRT-PCR of CDF6 transcript abundance in (C) continuous 22°C or 1 h of 10°C exposure in (D) WT, (E) 35S::CCA1, and (F) SUC2::CCA1. Seedlings were grown in continuous light (LL) for 2 days after 8 days of entrainment in diurnal (12-h light:12-h dark; LD) cycles. Gray and white bars indicate the subjective night and day periods, respectively. Time (ZT) represents hours. mRNA levels are normalized to IPP2 (mean values ± standard error (SE), n = 3; ***P ≤ 0.001, **P ≤ 0.05, *P ≤ 0.01; unpaired student's t-test).
A subset of DOF-TFs (CDF1,2,3,5,6, ADOF1) is COR genes based on transcriptomic analysis of cbf mutants (Shi et al., 2017). In response to moderate cold stress (10°C), CDFs are mostly upregulated during the morning (Blair et al., 2019). CDF6 is significantly upregulated at ZT1 but not at ZT6, while CDF1 and CDF3 are significantly upregulated at both times of day (Blair et al., 2019). To determine whether CCA1 is responsible for CDF6 mRNA accumulation during cold stress, we exposed WT, 35S::CCA1, SUC2::CCA1, and cca1lhy seedlings to 10°C for 1 h before sampling every 4 h (ZT12–ZT36) and quantifying CDF6 transcript abundance. Seedlings were entrained for 8 days in LD before transfer to continuous light (LL). Of note, both CCA1 and its partially redundant partner, LHY, displayed increased transcript abundance in response to cold depending on the time of the day (Supplementary Figures 3A–D). In WT, we found that CDF6 transcript abundance, similar to CCA1, was significantly elevated in response to cold at the time points surrounding dawn (ZT20 and ZT28) (Figures 1C,D; Supplementary Figure 3A). However, CDF6 displayed higher transcript abundance in response to cold at dawn (ZT24), late morning (ZT28), and dusk (ZT12 and ZT16) in the 35S::CCA1 line (Figure 1E). While in the SUC2::CCA1 line, we observed that CDF6 exhibits higher transcript accumulation across the night period (ZT12–ZT20) but lower at dawn (ZT24) in response to cold (Figure 1F). Furthermore, CDF6 induction in response to cold at ZT24 in WT (though not significant) and 35S::CCA1 was significantly reduced in SUC2::CCA1 but restored in the cca1lhy mutants (Figures 1D–F; Supplementary Figure 2D). However, the induction of CDF6 transcript abundance at ZT28 in WT, 35S::CCA1, and cca1lhy was abolished in SUC2::CCA1 (Figures 1D–F; Supplementary Figure 2D). A comparison between WT, 35::CCA1, and SUC2::CCA1 further supports the temporal mis-regulation of CDF6 in response to cold (Supplementary Figure 2E). Of note, similar to what was observed at 22°C, CDF6::CDF6-GUS lines also showed vasculature protein accumulation at ZT0 after 1 h of exposure to 10°C (Supplementary Figure 3E). Together, these data suggest that the clock via CCA1 gates CDF6 expression which in turn diminishes the normal cold induction of CDF6 during the day and may promote cold induction during the subjective night.
CDF6 Regulates Flowering and Seed Germination
To date, higher orders of multiple mutants of cdf1, cdf2, cdf3, and cdf5 are known to cause early flowering, and a more recent study demonstrated that SUC2::CDF6 results in delayed flowering (Fornara et al., 2009; Krahmer et al., 2019). However, how cold stress impacts CDFs regulation of flowering time is still an open question. To examine further the role of CDF6 in photoperiodic flowering during cold stress, we measured the flowering phenotype of SUC2::CDF6 and a SALK T-DNA insertion line for cdf6 (Supplementary Figures 4A,B), under continuous ambient (22°C) or cold (10°C) temperatures. As the CDFs share high-sequence homology and likely redundant function, we also measured flowering in the cdf quintuple mutant (cdf12356, Supplementary Figure 1). In long-day (16-h light: 8-h dark) conditions, the SUC2::CDF6 plants displayed late flowering relative to WT under both ambient and cold temperatures (Figure 2A). Interestingly, the cdf12356 mutant exhibits significant early flowering under both ambient and cold temperatures; however, this observed early flowering phenotype is notably reduced during cold (Figure 2A). While the cdf6 single mutant did not show a significant flowering phenotype relative to WT at ambient temperature, under cold temperature, cdf6 plants display a modest but significant early flowering phenotypes (Figure 2A). Furthermore, we observe a statistically significant interaction between temperature and genotype (Supplementary Figure 4C). These observations suggest that CDF6 plays a redundant role in the regulation of photoperiodic flowering during ambient temperature and a predominant role during cold temperatures. In short-day conditions (8-h light: 16-h dark), only cdf12356 shows a difference relative to WT with early flowering in both cold and ambient temperatures (Supplementary Figure 4D).
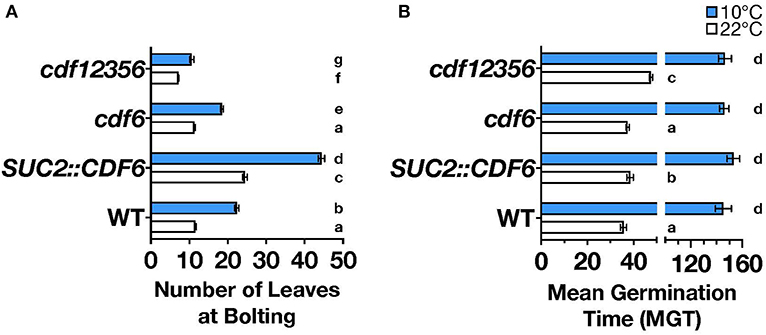
Figure 2. CDF6 mis-expression alters flowering and seed germination. (A) The number of leaves at bolting for WT, SUC2::CDF6, cdf6, and cdf12356 plants grown at constant 22°C (white bars) or 10°C (blue bars) in long-day conditions (mean values ± SE, n = 3+, two-way ANOVA with Tukey's multiple comparison test). (B) Mean germination time (MGT) for WT, SUC2::CDF6, cdf6, and cdf12356 plants grown in constant light (LL) at 10°C or 22°C with lowercase a-c letters indicating statistical significance at 22°C and d at 10°C (mean values ± SE, n = 4, one-way ANOVA with Dunnett's multiple comparison test).
CDF4 and other DOF-TFs, such as DOF AFFECTING GERMINATION1 (DAG1), DAG2, and DOF6, have been shown to play a role in germination (Ruta et al., 2020). To determine whether CDF6 functions similarly to CDF4 and these other DOFs, we measured germination over time in constant light and temperature (22°C or 10°C) and in the SUC2::CDF6, cdf6, and cdf12356 lines. SUC2::CDF6 and cdf12356 seeds have delayed germination relative to WT during ambient temperature but not under cold, whereas there is no significant difference in cdf6 germination at either temperature (Figure 2B). Next, we measured seed dormancy after cold (4°C) and dark treatment of seeds for three nights and found germination synchrony across all genotypes tested (Supplementary Figures 4E,F). Thus, we conclude that during ambient temperature, CDF6 may contribute to the regulation of seed germination rather than dormancy.
Vasculature-Expressed CDF6 Regulates the Transcriptome During Cold Stress
To determine a broader role for CDF6 in both development and cold responses, we examined transcriptome changes in WT and CDF6 mis-expression lines (SUC2::CDF6 and cdf6). For this, seedlings were entrained for 8 days in LD and then transferred to constant light (LL) for 2 days. On day 11, seedlings were subjected to 10°C for 1 h and sampled at subjective dawn (ZT0), the time of day CDF6 expression is significantly altered in the cdf6 mutant and SUC2::CDF6 lines (Supplementary Figures 4B, 5F,G). Using a cut-off of −1 > Log2 fold change > 1 and false discovery rate (FDR) < 0.05, we identified 473 differentially expressed genes (DEGs) in the WT (10°C vs. 22°C), and we consider these as the generally cold-responsive DEGs in this study (Figure 3A, Supplementary Dataset 1). Of these generally cold-responsive genes, ~31% were previously identified as COR genes in the cbf1, cbf2, and cbf3 mutants (Supplementary Dataset 2; Shi et al., 2017). Using the Phaser database which provides insight into clock regulation of transcript levels, we identified that ~50% of these DEGs exhibit rhythmic expression with significant enrichment from the afternoon to early evening period; this is consistent with the peak expression for cold-responsive genes and the proportion of the transcriptome that cycles during cold stress (Covington et al., 2008; Grundy et al., 2015; Supplementary Figure 5A; Supplementary Dataset 3).
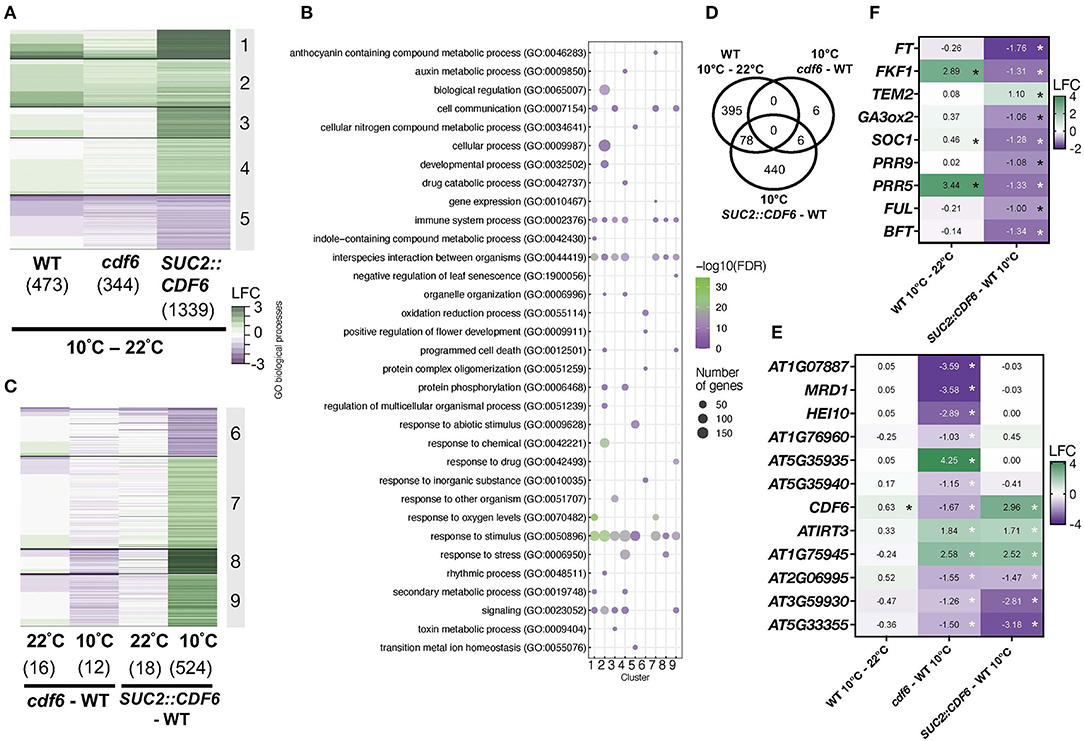
Figure 3. Vasculature-expressed CDF6 regulates cold-responsive and flowering-associated genes in the morning. (A) Temperature and (C) genotype-specific DEGs [defined as −1 > Log2 fold change (LFC) > 1 and false discovery rate (FDR) < 0.05] identified after RNA-sequencing of three replicates of 11-day-old WT, SUC2::CDF6, and cdf6 seedlings. Seedlings were grown in constant light for 2 days at 22°C after entrainment for 8 days in diurnal (LD) conditions before sampling at ZT0 after 1 h at 10°C or continuously at 22°C. (B) Summary of enriched GO biological process terms with indicated false discovery rate (FDR-corrected p-value < 0.05). GO terms selected based on a previously published protocol using a FDR cut-off < 0.05 (Bonnot et al., 2019). (D) DEGs specific to the cold response vs. CDF6 mis-regulation during cold stress. Expression profiles of (E) 12 DEGs mis-expressed in cdf6 and (F) flowering-associated genes identified in SUC2::CDF6 during cold stress; *FDR < 0.05.
Next, we identified 344 genes that are differentially expressed in cdf6 at 10°C compared to 22°C; and ~71% of these are also found in the WT (10°C vs. 22°C) dataset (Figure 3A; Supplementary Figure 5B). However, the effect of SUC2::CDF6 at 10°C compared to 22°C is most striking with 1339 DEGs being detected, and of these, only ~27% overlapped with the generally cold-responsive genes (Figure 3A; Supplementary Figure 5B). We speculate that the other ~73% of DEGs may be specifically regulated by CDF6 in the vasculature in response to cold stress rather than across cell types in response to cold. A gene ontology (GO) analysis reveals enrichment for terms associated with the clock (Cluster 2), responses to abiotic and biotic factors (Clusters 1, 3, 4, and 5), and metabolic processes (Clusters 4 and 5) (Figure 3B). Cluster 2 also contains a number of flowering- and clock-related genes (Figure 3B).
To further dissect the impact of CDF6 mis-expression on the transcriptome during cold stress, we compared each genotype relative to WT at each temperature. First, we observed that there are very few DEGs in cdf6 compared to the WT (16 DEGs at 22°C and 12 DEGs at 10°C (Figure 3C). However, in vasculature-expressed CDF6 (SUC2:CDF6), we observe a greater impact on the transcriptome during cold stress (524 DEGs) compared to ambient (18 DEGs) temperature (Figure 3C). Cluster 6 genes generally show downregulation in SUC2::CDF6 compared to WT at 10°C with minimal expression change in the other comparisons. These genes were enriched for the biological GO term “positive regulation of flowering,” and many genes involved with flowering are found in this cluster, including the florigen molecule FT (Figure 3B). The other clusters show enrichment for terms associated with responses to stimulus/stress, immune system processes, and development (Figure 3B).
We also identified genes that may contribute to the delayed germination phenotype that we previously observed in SUC2::CDF6 lines under ambient temperatures (Figure 2B; Supplementary Figure 5C). For example, IRON REGULATED TRANSPORTER 3 (ATIRT3) transports zinc, which is essential for the development of reproductive organs, and is needed for proper seed development (Lee et al., 2021). IRT3 is upregulated in SUC2::CDF6 at both temperatures, although this upregulation is reduced at 10°C when compared to at 22°C, suggesting that SUC2::CDF6 seeds may have increased zinc transport to support germination during ambient temperature (Supplementary Figure 5C). In addition, ETHYLENE RESPONSE FACTOR15 (ERF15), a positive regulator of ABA, is upregulated in SUC2::CDF6 under both ambient and cold temperatures (Supplementary Figure 5C). Interestingly, ERF15 over-expression results in delayed germination, similar to the phenotype we observed in SUC2::CDF6 line (Lee et al., 2015). This suggests that vasculature-expressed CDF6 may upregulate ERF15 to delay germination and impact ABA responsiveness.
Next, we considered that a large number of DEGs could be generally cold-responsive rather than differentially expressed as a result of the absence of CDF6. Thus, we compared the 524 DEGs (SUC2::CDF6) and 12 DEGs (cdf6) to our generally cold-responsive DEGs and identified 6 DEGs specific to cdf6 and 440 DEGs specific to SUC2::CDF6 with an additional 6 shared by both SUC2::CDF6 and cdf6 genotypes (Figure 3D). The 12 cdf6 DEGs perform various functions, including defense (AT3G59930, AT5G33355), class 1 crossover (HEI10), methionine biosynthesis (MRD1), and metal transport (IRT3), in addition to a handful of genes with unknown functions (Goto and Naito, 2002; Silverstein et al., 2005; Chelysheva et al., 2012; Lee et al., 2021) (Figure 3E). To further assess the extent of mis-expression due to cold stress rather than the effect of CDF6, we also compared the 473, 344, and 1,339 DEGs (shown in Figure 3A) at 10°C vs. 22°C in the WT, cdf6, and SUC2::CDF6 genotypes, respectively. This analysis identified 61 DEGs that can be considered unique to cdf6 (Supplementary Figure 5D), which are enriched for the following three biological GO terms: cellular response to heat, response to heat, and response to temperature stimulus (Supplementary Dataset 3). While these DEGs are not generally cold-responsive in comparison to our WT dataset, they may be generally temperature responsive. The other CDFs are not differentially expressed in cdf6 or SUC2::CDF6 at either temperature, with the exception of CDF3 downregulation in SUC2::CDF6 at 10°C (Supplementary Figure 5E). The data also confirm a significant depletion of CDF6 mRNA in cdf6 and elevation in SUC2::CDF6, both the response to cold and at ambient temperature, confirming this genotype elevates mRNA abundance in PCCs (Figure 3E; Supplementary Figures 5F,G). Future functional work with additional cdf6 alleles and complementation lines is needed to define the regulatory mechanism of CDF6.
In the 440 SUC2::CDF6-specific DEGs at 10°C, we identified nine genes associated with flowering (FT, FKF1, TEM2, GA3ox2, SOC1, PRR9, PRR5, FUL, and BFT), all of which are downregulated with the exception of TEM2 (Figure 3F). This complements previous reports of reduced expression in SUC2::CDF6 in the evening under ambient temperature conditions (Krahmer et al., 2019). Interestingly, a few clock- or circadian-regulated genes, specifically PRR9, PRR5, and FKF1, are differentially expressed in SUC2::CDF6 compared to WT at 10°C, indicating that there may be feedback regulation between CDF6 and these components (Figure 3F). Many other flowering-related components identified are either TFs or involved in hormone signaling (Kinoshita and Richter, 2020; Supplementary Dataset 4).
CDF6 Alters the Expression of Key Flowering Genes During Cold Under Long-Day Conditions
To better understand how CDF6 regulates photoperiodic flowering during cold, we measured the abundance of the photoperiodic flowering component mRNAs, FT, CO, and BFT, via qRT-PCR over a 24-h period in continuous 10°C or 22°C long-day conditions. In WT and under constant cold, FT transcript accumulation is significantly increased from ZT20 to ZT36 and displays a shift in peak expression to ZT20 compared to its peak expression at ZT16 during ambient temperature (Supplementary Figure 6A). In SUC2::CDF6 lines, we observed a significant reduction in FT transcript abundance relative to WT in the evening (ZT16 and ZT20), at dawn (ZT24), and late afternoon (ZT32, ZT12, and ZT36) (Figure 4A). At 10°C, FT accumulation is reduced relative to WT throughout the 24-h period (Figure 4B). Interestingly, in SUC2::CDF6, FT repression was significantly enhanced during the cold at ZT16, further supporting a distinct role for CDF6 in regulating photoperiodic flowering during cold stress (Figures 2A, 4C).
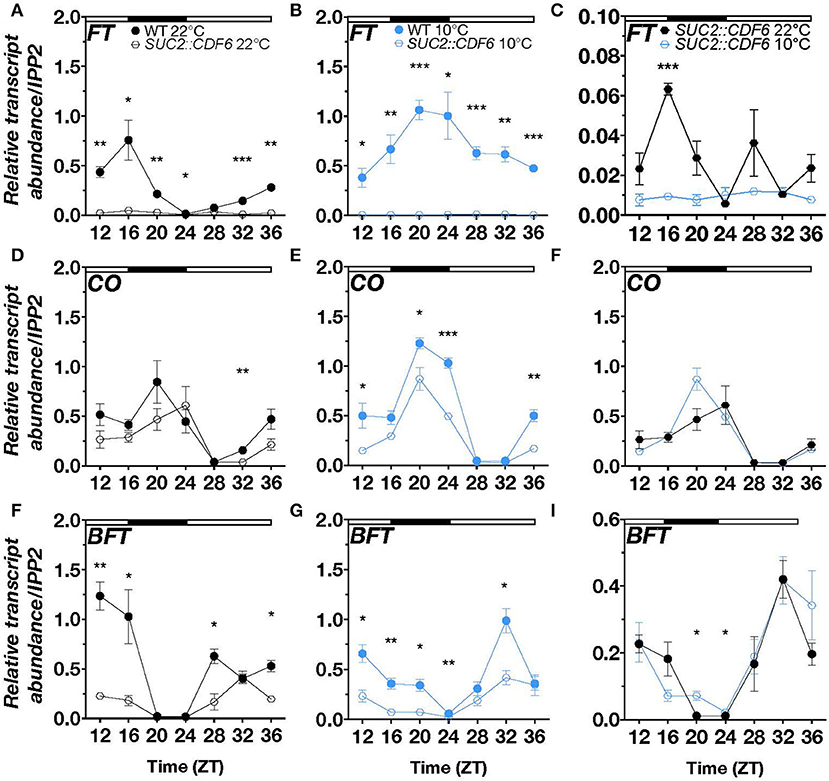
Figure 4. CDF6 alters FT, CO, and BFT expression during cold stress. qRT-PCR of (A–C) FT, (D–F) CO, and (G–I) BFT relative transcript abundance in WT and SUC2::CDF6 seedlings. Plants were grown in long-day conditions at 22°C for 8 days followed by 2 days continuously at 22°C or 10°C. Seedlings were sampled every 4 h starting at 12 h after dawn. (C) FT (F) CO, and (I) BFT transcript abundance in SUC2::CDF6 at 10°C and 22°C from the data shown in A and B, D and E, and G and H, respectively. White and black bars indicate the day and night periods, respectively. mRNA levels are normalized to IPP2 (mean values ± SE, n = 3; ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05; unpaired student's t-test).
Under ambient temperature, CO shows a shift in peak expression from ZT20 to ZT24 in SUC2::CDF6 relative to WT and is significantly downregulated in SUC2::CDF6 at ZT32 (Figure 4D). CO has peak expression at ZT20 under continuous cold conditions, and is significantly repressed in the early morning (ZT20), at dawn (ZT24), and late evening (ZT12 and ZT36) in SUC2::CDF6 compared to WT (Figure 4E). In addition, CO is significantly upregulated in response to cold in WT at ZT24, so it is likely that pSUC2-driven expression of CDF6 prevents CO accumulation during the cold at this time point (Figure 4E, Supplementary Figure 6B). Although the repressive effect of SUC2::CDF6 is not significantly different at 10°C compared to 22°C, our data together indicate that CDF6 expression is required for appropriate accumulation of CO in WT at specific time points under both temperatures (Figure 4F). These findings are consistent with the transcriptome analysis, which shows that FT and CO are strongly downregulated during cold stress when CDF6 is regulated by pSUC2 (Figures 3D, 4). FT and CO are not significantly different in cdf6 compared to the WT at either temperature tested, likely due to redundancy by other CDFs in photoperiodic flowering regulation (Supplementary Figures 6C,D).
Besides FT, CO, and the two clock components PRR9 and PRR5, BFT is specifically localized to the PCC under control conditions (Supplementary Figure 7). In addition, transcriptome analysis revealed BFT to be downregulated in SUC2::CDF6 seedlings during cold stress (Figure 3F). Therefore, we also assessed BFT transcript accumulation during continuous cold under long-day conditions in SUC2::CDF6. At ambient temperature, CDF6 downregulates BFT during the day (ZT12, ZT16, ZT28, and ZT36) (Figure 4G). However, during continuous cold, BFT transcript abundance is significantly reduced in SUC2::CDF6 during both the day and night periods (ZT12-ZT24 and ZT32) (Figure 4H). Together, these data indicate that CDF6 represses BFT independent of temperature, although BFT mRNA accumulation is significantly increased in SUC2::CDF6 in the early morning (ZT20 and ZT24) during continuous cold compared to ambient temperature (Figure 4I).
Discussion
Previous work shows that the clock regulates a range of developmental programs, including germination, flowering, and senescence, to promote optimal survival and reproduction (Lu et al., 2012; Adams et al., 2018; Kim et al., 2018; Zha et al., 2019; Kyung et al., 2021). The clock is also known to regulate responses to abiotic stress. A classic example involves the clock gating of CBF1-3 accumulation during cold stress, which promotes the activation of COR genes to confer increased cold tolerance (Fowler and Thomashow, 2002; Gilmour et al., 2004; Fowler et al., 2005). Additional evidence suggests that the expression of CBFs is regulated by PRR5, PRR7, and PRR9 (Nakamichi et al., 2009).
Here, we investigate the clock regulation of a poorly characterized member of the CDF family, CDF6, during ambient and cold temperatures. We find that CCA1 represses CDF6 transcript accumulation during ambient temperature and gates the accumulation of CDF6 during moderate cold stress (Figure 5). To better understand the clock regulation of CDF6, we utilized the FIMO tool to scan the 500 bp upstream of each DOF-TF family member for motifs associated with clock TF binding (Grant et al., 2011). This analysis identified that proximal promoter regions of ~39% of the DOF-TFs contain a full or partial evening element or CCA1-binding site, whereas none of the DOF-TF promoters contain a primary G-box motif, the element which is associated with PRR binding (Supplementary Dataset 5). This may explain the low number of DOF family members in PRR7 (~5%), PRR9 (~11%), and TOC1 (0) ChIP datasets compared to the CCA1 (~25%) and LHY (~14%) ChIP datasets, although it is important to note the CCA1 and LHY datasets also identified many more target genes than the PRR datasets (Huang et al., 2012; Liu et al., 2013, 2016; Nagel et al., 2015; Kamioka et al., 2016; Adams et al., 2018) (Supplementary Dataset 5). We also considered that clock genes may not be the only upstream regulators of CDF6, therefore, we utilized the DAP-sequencing database to identify TFs that may bind to the CDF6 promoter region (O'Malley et al., 2016). This revealed 142 unique gene IDs/TFs that bind in vitro along the CDF6 promoter, gene body, or UTRs (Supplementary Dataset 6). Of these, ~12% were DOF-TFs (including CDF3, CDF4, and CDF5), suggesting that these CDFs may regulate CDF6 or other DOF-TFs. The finding that CDF6 expression is significantly downregulated in 35S::CDF3 lines further supports the conclusion that CDF3 modulates CDF6 transcription (Corrales et al., 2017). Finally, ~20% of the TFs identified in the DAP-sequencing analysis were also significantly mis-regulated in our generally cold-responsive dataset (FDR < 0.05), implying that some of these TFs may also play a role in the cold regulation of CDF6 (Supplementary Dataset 6). Future work to investigate whether these TFs play a role in modulating the cold response of CDF6 and other DOFs is needed.
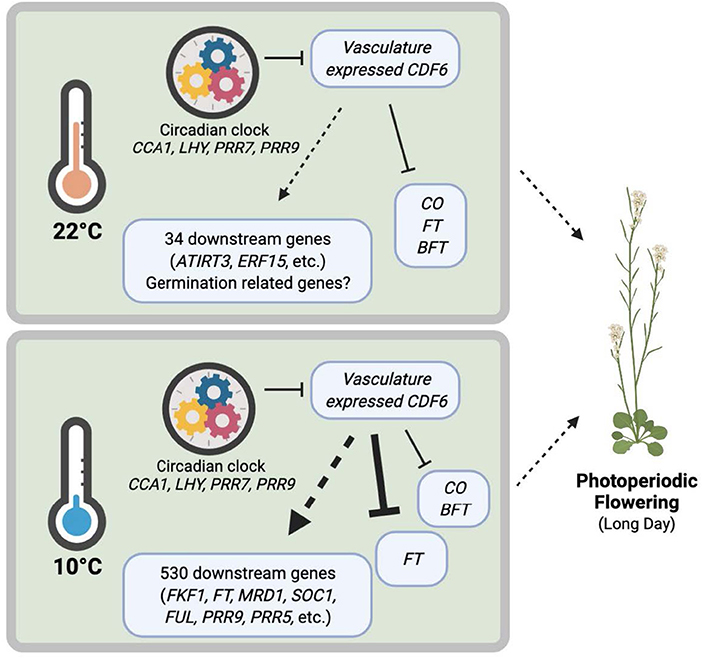
Figure 5. Clock-regulated, cold-induced, and vasculature-expressed CDF6 finetunes plant growth and development under different temperatures. The circadian clock regulates CDF6 transcript accumulation in the vasculature at specific times of the day. During ambient temperature, CDF6 regulates FT, CO, BFT, and at least 34 other downstream genes (16 in cdf6-WT and 18 in SUC2::CDF6-WT), including ATIRT3 and ERF15, which contribute to germination and flowering phenotypes. During cold temperatures, CDF6 regulates 530 downstream genes and represses FT, CO, and BFT in the vasculature/phloem companion cells to control photoperiodic flowering. Model created with BioRender.com.
The results of this study indicate that CDF6 participates in seed germination regulation during ambient temperature and photoperiodic flowering during cold stress (Figure 2). Other DOF-TFs function in vascular system development and germination; however, with the exception of CDF4, the CDFs have not been previously associated with germination (Le Hir and Bellini, 2013). Our transcriptomic analysis revealed that IRT3, ERF15, and 16 other DEGs may contribute to the observed germination phenotype (Figure 5; Supplementary Figure 4). Of note, our dataset was generated under a later developmental stage than when germination was phenotypically assessed, thus future work at the same developmental stage could elucidate any additional DEGs that may contribute to the delayed germination in SUC2::CDF6.
Our study supports a greater role for CDF6 function under cold vs. ambient temperature to broadly control plant growth and development. We identified a number of flowering-associated genes that have altered expression in vasculature-expressed CDF6 during cold stress, four (FT, BFT, PRR9, and PRR5) of which are also expressed in the PCC (Figure 3; Supplementary Figures 7A–D) (Mustroph et al., 2009). Of note, single-cell sequencing data in Arabidopsis shows that CDF6 may also be expressed in phloem parenchyma, xylem, and even mesophyll cells (Mustroph et al., 2009; Kim et al., 2021). We also identified a number of GO terms associated with responses to stress, development, and metabolism (Figure 3). While the GO terms corresponding to the response to stress and development corroborate our earlier findings for CDF6 during cold stress and in relation to both the observed germination and flowering phenotypes, the metabolism GO terms are interesting, as other CDFs have been previously implicated in metabolic regulation. A transcriptomic analysis of 35S::CDF3 indicates the enrichment of metabolism GO terms, and more specifically, a metabolic analysis indicates that CDF3 over-expression impacts the metabolism of sugars and amino acids (Corrales et al., 2017). While the results of the GO analysis for both SUC2::CDF6 and 35S::CDF3 yielded similar enriched terms, only a single gene is shared between the DEGs in the CDF3 (531) and CDF6 (18) datasets. The shared gene, AT4G01390, is significantly downregulated in SUC2::CDF6 and upregulated in 35S::CDF3. Based on public annotation, AT4G01390 contains a MATH [meprin and TRAF (tumor necrosis factor receptor-associated factor) homology] domain which is found in proteins involved in several of the GO terms shared between the two datasets, such as stress responses, plant development, signaling, and metabolism (Oelmuller et al., 2005; Inzé et al., 2012; Qi et al., 2021). Of note, the lack of significant overlap between the two datasets is not surprising given the differences in tissue-specific expression (35S::CDF3 vs. SUC2::CDF6), experimental design, and analysis. Together, this finding indicates that CDF3 and CDF6 may work similarly but independently to integrate temperature signals to alter metabolism and development.
Next, we observed that vasculature-expressed CDF6 results in altered FT, CO, and BFT accumulation across the 24-h period at both 22 and 10°C, but notably during continuous cold on long days (Figure 4). Specifically, we found that FT shows higher accumulation during the cold from ZT20 to ZT36, while CO is higher at ZT24 and lower at ZT32 (Supplementary Figure 6). At first glance, this seems contrary to a previous work, which indicates that lower temperatures decrease FT accumulation and delay flowering (Song et al., 2013). However, the impact of temperature on FT accumulation is highly dependent on the timing of the temperature stress and the photoperiod length (Kinmonth-Schultz et al., 2016; Krahmer et al., 2019). For example, when plants are grown on ambient long days with cool nights, CO has higher transcript accumulation at dawn (ZT0) and no change at midday (ZT8) or dusk (ZT16), while FT is higher at dawn with a decrease or no change at midday and dusk when compared to the constant ambient temperature control conditions (Kinmonth-Schultz et al., 2016). In constant light, FT exhibits increased transcript abundance during cold treatment (Schwartz et al., 2009). FT is dynamically controlled during cold temperature exposure by multiple regulators, including FLC, SHORT VEGETATIVE PHASE (SVP), HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1), miR156, miR172, and sequestration of FT, through its interaction with phospholipid phosphatidylglycerol (PG) at cellular membranes (Song et al., 2013; Susila et al., 2021). Our study shows for the first time that a CDF family member contributes to the differential transcript accumulation of key photoperiod regulators (FT, CO, and BFT) during cold stress. Additionally, ectopic expression of CDF6 with pSUC2 results in the differential expression of 34 and 530 downstream genes under ambient and cold temperatures, respectively (Figure 5). We conclude that CDF6 directly or indirectly regulates the transcription of numerous genes, particularly at low temperatures.
Taken together, our data suggest that vasculature-expressed CDF6 plays a role in regulating photoperiodic flowering during cold stress, and some of this regulation involves functional redundancy with other CDFs. We provide new insights on the regulatory relationship between CDF6 and the clock, cold stress, and plant development. As climate change continues to cause erratic weather events, the precise regulation of photoperiod flowering components in specific cell types during cold temperatures should be further explored as a tool to combat potential crop losses.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DN conceived the project. EB and DN designed and performed experiments, analyzed the data, and wrote the manuscript. GG, ML, and TI generated and confirmed the plant materials. EB, TI, and DN revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by an NSF Early Career Award (IOS 1942949) to DN and in part by the National Institute of Health (R01GM079712) to TI.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Julia Bailey-Serres, Samantha Cordingley, Dr. Titouan Bonnot, Dr. Jeoffrey George, and Sabrina Gilmour for critical reading of the manuscript and members of the Bailey-Serres lab for helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.919676/full#supplementary-material
References
Adams, S., Grundy, J., Veflingstad, S. R., Dyer, N. P., Hannah, M. A., Ott, S., et al. (2018). Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 220, 893–907 doi: 10.1111/nph.15415
Backman, H. T. W., and Girke, T. (2016). systemPipeR: NGS workflow and report generation environment. BMC Bioinformat. 17, 388. doi: 10.1186/s12859-016-1241-0
Blair, E. J., Bonnot, T., Hummel, M., Hay, E., Marzolino, J. M., Quijada, I. A., et al. (2019). Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Sci. Rep. 9, 4814. doi: 10.1038/s41598-019-41234-w
Bonnot, T., Blair, E. J., Cordingley, S. J., and Nagel, D. H. (2021). Circadian coordination of cellular processes and abiotic stress responses. Curr. Opin. Plant Biol. 64, 102133. doi: 10.1016/j.pbi.2021.102133
Bonnot, T., Gillard, M., and Nagel, D. (2019). A simple protocol for informative visualization of enriched gene ontology terms. BIO-PROTOCOL 9:e3429. doi: 10.21769/BioProtoc.3429
Calixto, C. P. G., Guo, W., James, A. B., Tzioutziou, N. A., Entizne, J. C., Panter, P. E., et al. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell. 30, 1424–1444 doi: 10.1105/tpc.18.00177
Chelysheva, L., Vezon, D., Chambon, A., Gendrot, G., Pereira, L., Lemhemdi, A., et al. (2012). The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799. doi: 10.1371/journal.pgen.1002799
Corrales, A-. R., Carrillo, L., Lasierra, P., Nebauer, S. G., Dominguez-Figueroa, J., Renau-Morata, B., et al. (2017). Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 40, 748–764. doi: 10.1111/pce.12894
Corrales, A-. R., Nebauer, S. G., Carrillo, L., Fernández-Nohales, P., Marqués, J., Renau-Morata, B., et al. (2014). Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65, 995–1012. doi: 10.1093/jxb/ert451
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9, R130. doi: 10.1186/gb-2008-9-8-r130
Creux, N., and Harmer, S. (2019). Circadian rhythms in plants. Cold Spring Harb. Perspect. Biol. 11, a034611 doi: 10.1101/cshperspect.a034611
Endo, M., Shimizu, H., Nohales, M. A., Araki, T., and Kay, S. A. (2014). Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515, 419–422. doi: 10.1038/nature13919
Farré, E. M., Harmer, S. L., Harmon, F. G., Yanovsky, M. J., and Kay, S. A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15, 47–54. doi: 10.1016/j.cub.2004.12.067
Fornara, F., de Montaigu, A., Sánchez-Villarreal, A., Takahashi, Y., Ver Loren van Themaat, E., Huettel, B., et al. (2015). The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 81, 695–706. doi: 10.1111/tpj.12759
Fornara, F., Panigrahi, K. C. S., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Fowler, S. G., Cook, D., and Thomashow, M. F. (2005). Low temperature induction of Arabidopsis CBF1, 2. and 3 is gated by the circadian clock. Plant Physiol. 137, 961–968. doi: 10.1104/pp.104.058354
Gilmour, S. J., Fowler, S. G., and Thomashow, M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4
Goto, D. B., and Naito, S. (2002). AtMRD1 and AtMRU1, two novel genes with altered mRNA levels in the methionine over-accumulating mto1-1 mutant of Arabidopsis thaliana. Plant Cell Physiol. 43, 923–931. doi: 10.1093/pcp/pcf107
Grant, C. E., Bailey, T. L., and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Greenham, K., and McClung, C. R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16, 598–610. doi: 10.1038/nrg3976
Grinevich, D. O., Desai, J. S., Stroup, K. P., Duan, J., Slabaugh, E., Doherty, C. J., et al. (2019). Novel transcriptional responses to heat revealed by turning up the heat at night. Plant Mol. Biol. 101, 1–19 doi: 10.1007/s11103-019-00873-3
Grundy, J., Stoker, C., and Carré, I. A. (2015). Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 6, 648. doi: 10.3389/fpls.2015.00648
Henriques, R., Wang, H., Liu, J., Boix, M., Huang, L-. F., Chua, N-. H., et al. (2017). The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 216, 854–867. doi: 10.1111/nph.14703
Huang, H., and Nusinow, D. A. (2016). Into the evening: complex interactions in the arabidopsis circadian clock. Trends Genet. 32, 674–686. doi: 10.1016/j.tig.2016.08.002
Huang, W., Pérez-García, P., Pokhilko, A., Millar, A. J., Antoshechkin, I., Riechmann, J. L., et al. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79. doi: 10.1126/science.1219075
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Inzé, A., Vanderauwera, S., Hoeberichts, F. A., Vandorpe, M., Van Gaever, T., Van Breusegem, F., et al. (2012). A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 35, 308–320. doi: 10.1111/j.1365-3040.2011.02323.x
Kamioka, M., Takao, S., Suzuki, T., Taki, K., Higashiyama, T., Kinoshita, T., et al. (2016). Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the arabidopsis circadian clock. Plant Cell 28, 696–711. doi: 10.1105/tpc.15.00737
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, H., Kim, H. J., Vu, Q. T., Jung, S., McClung, C. R., Hong, S., et al. (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 115, 8448–8453 doi: 10.1073/pnas.1722407115
Kim, J-. Y., Symeonidi, E., Pang, T. Y., Denyer, T., Weidauer, D., Bezrutczyk, M., et al. (2021). Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell 33, 511–530. doi: 10.1093/plcell/koaa060
Kinmonth-Schultz, H. A., Tong, X., Lee, J., Song, Y. H., Ito, S., Kim, S-. H., et al. (2016). Cool night-time temperatures induce the expression of CONSTANS and FLOWERING LOCUS T to regulate flowering in Arabidopsis. New Phytol. 211, 208–224. doi: 10.1111/nph.13883
Kinoshita, A., and Richter, R. (2020). Genetic and molecular basis of floral induction in Arabidopsis thaliana. J. Exp. Bot. 71, 2490–2504. doi: 10.1093/jxb/eraa057
Krahmer, J., Goralogia, G. S., Kubota, A., Zardilis, A., Johnson, R. S., Song, Y. H., et al. (2019). Time-resolved interaction proteomics of the GIGANTEA protein under diurnal cycles in Arabidopsis. FEBS Lett. 593, 319–338. doi: 10.1002/1873-3468.13311
Kyung, J., Jeon, M., Jeong, G., Shin, Y., Seo, E., Yu, J., et al. (2021). The two clock proteins CCA1 and LHY activate VIN3 transcription during vernalization through the vernalization-responsive cis-element. Plant Cell. 34, 1020–1037 doi: 10.1093/plcell/koab304
Law, C. W., Chen, Y., Shi, W., and Smyth, G. K. (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. doi: 10.1186/gb-2014-15-2-r29
Le Hir, R., and Bellini, C. (2013). The plant-specific dof transcription factors family: new players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 4, 164. doi: 10.3389/fpls.2013.00164
Lee, S., Lee, J., Ricachenevsky, F. K., Punshon, T., Tappero, R., Salt, D. E., et al. (2021). Redundant roles of four ZIP family members in zinc homeostasis and seed development in Arabidopsis thaliana. Plant J. 108, 1162–1173. doi: 10.1111/tpj.15506
Lee, S-. B., Lee, S-. J., and Kim, S. Y. (2015). AtERF15 is a positive regulator of ABA response. Plant Cell Rep. 34, 71–81. doi: 10.1007/s00299-014-1688-2
Liu, T., Carlsson, J., Takeuchi, T., Newton, L., and Farré, E. M. (2013). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 76, 101–114. doi: 10.1111/tpj.12276
Liu, T. L., Newton, L., Liu, M-. J., Shiu, S-. H., and Farré, E. M. (2016). A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in arabidopsis. Plant Physiol. 170, 528–539. doi: 10.1104/pp.15.01562
Lu, S. X., Webb, C. J., Knowles, S. M., Kim, S. H. J., Wang, Z., Tobin, E. M., et al. (2012). CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 158, 1079–1088. doi: 10.1104/pp.111.189670
Markham, K. K., and Greenham, K. (2021). Abiotic stress through time. New Phytol. 231, 40–46 doi: 10.1111/nph.17367
Martín, G., Rovira, A., Veciana, N., Soy, J., Toledo-Ortiz, G., Gommers, C. M. M., et al. (2018). Circadian waves of transcriptional repression shape pif-regulated photoperiod-responsive growth in Arabidopsis. Curr. Biol. 28, 311–318.e5. doi: 10.1016/j.cub.2017.12.021
Martín, G., Veciana, N., Boix, M., Rovira, A., Henriques, R., Monte, E., et al. (2020). The photoperiodic response of hypocotyl elongation involves regulation of CDF1 and CDF5 activity. Physiol. Plant. 169, 480–490 doi: 10.1111/ppl.13119
Mockler, T. C., Michael, T. P., Priest, H. D., Shen, R., Sullivan, C. M., Givan, S. A., et al. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72, 353–363. doi: 10.1101/sqb.2007.72.006
Mustroph, A., Zanetti, M. E., Jang, C. J. H., Holtan, H. E., Repetti, P. P., Galbraith, D. W., et al. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 18843–18848. doi: 10.1073/pnas.0906131106
Nagel, D. H., Doherty, C. J., Pruneda-Paz, J. L., Schmitz, R. J., Ecker, J. R., Kay, S. A., et al. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 112, E4802–10. doi: 10.1073/pnas.1513609112
Nagel, D. H., and Kay, S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22, R648–57. doi: 10.1016/j.cub.2012.07.025
Nagel, D. H., Pruneda-Paz, J. L., and Kay, S. A. (2014). FBH1 affects warm temperature responses in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U. S. A. 111, 14595–14600. doi: 10.1073/pnas.1416666111
Nakamichi, N. (2020). The transcriptional network in the arabidopsis circadian clock system. Genes 11, 1284. doi: 10.3390/genes11111284
Nakamichi, N., Kiba, T., Kamioka, M., Suzuki, T., Yamashino, T., Higashiyama, T., et al. (2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U. S. A. 109, 17123–17128. doi: 10.1073/pnas.1205156109
Nakamichi, N., Kita, M., Niinuma, K., Ito, S., Yamashino, T., Mizoguchi, T., et al. (2007). Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 48, 822–832. doi: 10.1093/pcp/pcm056
Nakamichi, N., Kusano, M., Fukushima, A., Kita, M., Ito, S., Yamashino, T., et al. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50, 447–462. doi: 10.1093/pcp/pcp004
Nelson, D. C., Riseborough, J-. A., Flematti, G. R., Stevens, J., Ghisalberti, E. L., Dixon, K. W., et al. (2009). Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149, 863–873. doi: 10.1104/pp.108.131516
Nohales, M. A., Liu, W., Duffy, T., Nozue, K., Sawa, M., Pruneda-Paz, J. L., et al. (2019). Multi-level modulation of light signaling by GIGANTEA regulates both the output and pace of the circadian clock. Dev. Cell. 49, 840–851.e8 doi: 10.1016/j.devcel.2019.04.030
Oelmuller, R., Peskan-Berghofer, T., Shahollari, B., Trebicka, A., Sherameti, I., Varma, A., et al. (2005). MATH domain proteins represent a novel protein family in Arabidopsis thaliana, and at least one member is modified in roots during the course of a plant-microbe interaction. Physiol. Plant. 124, 152–166. doi: 10.1111/j.1399-3054.2005.00505.x
O'Malley, R. C., Huang, S-. S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. doi: 10.1016/j.cell.2016.04.038
Park, S., Lee, C-. M., Doherty, C. J., Gilmour, S. J., Kim, Y., Thomashow, M. F., et al. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82, 193–207. doi: 10.1111/tpj.12796
Pruneda-Paz, J. L., Breton, G., Para, A., and Kay, S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485. doi: 10.1126/science.1167206
Qi, H., Xia, F-. N., Xiao, S., and Li, J. (2021). TRAF proteins as key regulators of plant development and stress responses. J. Integr. Plant Biol. 64, 431–448 doi: 10.1111/jipb.13182
Renau-Morata, B., Carrillo, L., Dominguez-Figueroa, J., Vicente-Carbajosa, J., Molina, R. V., Nebauer, S. G., et al. (2020). CDF transcription factors: plant regulators to deal with extreme environmental conditions. J. Exp. Bot. 71, 3803–3815. doi: 10.1093/jxb/eraa088
Riboni, M., Galbiati, M., Tonelli, C., and Conti, L. (2013). GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol. 162, 1706–1719. doi: 10.1104/pp.113.217729
Ruta, V., Longo, C., Lepri, A., De Angelis, V., Occhigrossi, S., Costantino, P., et al. (2020). The DOF transcription factors in seed and seedling development. Plants. 9, 218. doi: 10.20944/preprints202001.0243.v1
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schwartz, C., Balasubramanian, S., Warthmann, N., Michael, T. P., Lempe, J., Sureshkumar, S., et al. (2009). Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics 183, 723–32. 1SI−7SI. doi: 10.1534/genetics.109.104984
Shi, Y., Huang, J., Sun, T., Wang, X., Zhu, C., Ai, Y., et al. (2017). The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant Biol. 59, 118–133. doi: 10.1111/jipb.12515
Silverstein, K. A. T., Graham, M. A., Paape, T. D., and VandenBosch, K. A. (2005). Genome organization of more than 300 defensin-like genes in Arabidopsis. Plant Physiol. 138, 600–610. doi: 10.1104/pp.105.060079
Song, Y., Zhang, X., Li, M., Yang, H., Fu, D., Lv, J., et al. (2021). The direct targets of CBFs: in cold stress response and beyond. J. Integr. Plant Biol 63, 1874–1887. doi: 10.1111/jipb.13161
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464. doi: 10.1146/annurev-arplant-043014-115555
Susila, H., Jurić, S., Liu, L., Gawarecka, K., Chung, K. S., Jin, S., et al. (2021). Florigen sequestration in cellular membranes modulates temperature-responsive flowering. Science 373, 1137–1142. doi: 10.1126/science.abh4054
Tsutsui, H., and Higashiyama, T. (2017). pKAMA-ITACHI vectors for highly efficient crispr/cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 58, 46–56. doi: 10.1093/pcp/pcw191
Wilkins, O., Bräutigam, K., and Campbell, M. M. (2010). Time of day shapes Arabidopsis drought transcriptomes: Arabidopsis drought transcriptomes. Plant J. 63, 715–727. doi: 10.1111/j.1365-313X.2010.04274.x
Yang, J., Luo, D., Yang, B., Frommer, W. B., and Eom, J. (2018). SWEET 11 and 15 as key players in seed filling in rice. New Phytologist 218, 604–615. doi: 10.1111/nph.15004
Keywords: cold stress, circadian clock, CDF6, transcription factor, vasculature, abiotic stress
Citation: Blair EJ, Goralogia GS, Lincoln MJ, Imaizumi T and Nagel DH (2022) Clock-Controlled and Cold-Induced CYCLING DOF FACTOR6 Alters Growth and Development in Arabidopsis. Front. Plant Sci. 13:919676. doi: 10.3389/fpls.2022.919676
Received: 13 April 2022; Accepted: 23 June 2022;
Published: 26 July 2022.
Edited by:
Wei Huang, South China Agricultural University, ChinaCopyright © 2022 Blair, Goralogia, Lincoln, Imaizumi and Nagel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dawn H. Nagel, ZGF3bm5AdWNyLmVkdQ==
†Present Address: Greg S. Goralogia, Department of Forest Ecosystems and Society, Oregon State University, Corvallis, OR, United States
Matthew J. Lincoln, Institute of Molecular Plant Sciences, University of Edinburgh, Edinburgh, United Kingdom
 Emily J. Blair
Emily J. Blair Greg S. Goralogia
Greg S. Goralogia Matthew J. Lincoln2†
Matthew J. Lincoln2† Takato Imaizumi
Takato Imaizumi Dawn H. Nagel
Dawn H. Nagel