- 1National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2State Key Laboratory of Crop Stress Biology for Arid Areas, College of Life Sciences, Northwest A&F University, Xianyang, China
- 3Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
Transmembrane kinases (TMKs) play important roles in plant growth and signaling cascades of phytohormones. However, its function in the regulation of early leaf senescence (ELS) of plants remains unknown. Here, we report the molecular cloning and functional characterization of the WATER-SOAKED SPOT1 gene which encodes a protein belongs to the TMK family and controls chloroplast development and leaf senescence in rice (Oryza sativa L.). The water-soaked spot1 (oswss1) mutant displays water-soaked spots which subsequently developed into necrotic symptoms at the tillering stage. Moreover, oswss1 exhibits slightly rolled leaves with irregular epidermal cells, decreased chlorophyll contents, and defective stomata and chloroplasts as compared with the wild type. Map-based cloning revealed that OsWSS1 encodes transmembrane kinase TMK1. Genetic complementary experiments verified that a Leu396Pro amino acid substitution, residing in the highly conserved region of leucine-rich repeat (LRR) domain, was responsible for the phenotypes of oswss1. OsWSS1 was constitutively expressed in all tissues and its encoded protein is localized to the plasma membrane. Mutation of OsWSS1 led to hyper-accumulation of reactive oxygen species (ROS), more severe DNA fragmentation, and cell death than that of the wild-type control. In addition, we found that the expression of senescence-associated genes (SAGs) was significantly higher, while the expression of genes associated with chloroplast development and photosynthesis was significantly downregulated in oswss1 as compared with the wild type. Taken together, our results demonstrated that OsWSS1, a member of TMKs, plays a vital role in the regulation of ROS homeostasis, chloroplast development, and leaf senescence in rice.
Introduction
Plant leaf is photosynthetic organ for energy production and nutrient assimilation during plant growth. Plant leaf senescence is the final stage of leaf development and controlled by programmed cell death (PCD) (Lim et al., 2007). During leaf senescence, reactive oxygen species (ROS) accumulate, malondialdehyde (MDA) content increases, the antioxidant enzyme activity decreases, chlorophyll degrades, photosynthesis efficiency reduces, and cell membrane gets damaged (Lim et al., 2007; Woo et al., 2019; Domínguez and Cejudo, 2021). ROS, including singlet oxygen (1O2), superoxide anions (O2–), hydroxyl radicals (OH), and hydrogen peroxide (H2O2), are important signaling molecules that trigger PCD and leaf senescence in plants (Wang et al., 2013; Mittler, 2017). Hyper-accumulation of ROS can disrupt the redox balance in cells and cause severe damage to lipids, proteins, and DNA. In addition, ROS affect the fluidity of biofilm, resulting in enzyme inactivation (Foyer and Noctor, 2005; Saed-Moucheshi et al., 2014). Therefore, production and scavenging of ROS must be tightly controlled by the antioxidant defense system to maintain a dynamic balance of the ROS level in plants (Lee S. et al., 2012; Mittler, 2017). The major antioxidant enzymes are catalase (CAT), superoxide dismutase (SOD), peroxidase (POD), and ascorbate peroxidase (APX) (Saed-Moucheshi et al., 2014).
In agriculture, plant leaves undergo normal leaf senescence positively contributes to crop yield by transferring its photosynthetic products into grains or other crop harvest organs (Wu et al., 2012). Early leaf senescence (ELS) means premature leaf yellowing and/or withering compared to wild type during reproductive growth, which decreases the functional period of leaves, thus affecting crop yield and quality (Lim et al., 2007). Leaf senescence is associated with the expression of specific genes (Woo et al., 2019). More than 185 senescence-associated genes (SAGs) have been identified in rice (Li et al., 2020). These genes are mainly associated with chloroplast development (Yang Y. et al., 2016; Chen et al., 2021), chlorophyll degradation (Kusaba et al., 2007; Morita et al., 2009; Jiang et al., 2011; Yamatani et al., 2013; Cui et al., 2021), hormone signal transduction (Kong et al., 2006; Chen et al., 2013; Liang et al., 2014), protease transport metabolism (Wu et al., 2016; Hong et al., 2018), and energy transport metabolism (Huang et al., 2007; Qiao et al., 2010).
Receptor-like kinases (RLKs) consist in one of the ubiquitous and most abundant gene families in plants. There are more than 1130 RLK genes in rice, twice as many as in Arabidopsis due to gene duplication (Shiu et al., 2004). A typical RLK is composed of a variable extracellular domain to perceive specific ligands, a transmembrane domain to connect the extracellular and intracellular parts for signal transmission, and an intracellular domain to transduce external signals into the cell to activate or inactivate the downstream regulatory components via phosphorylation (Morillo and Tax, 2006; Gish and Clark, 2011). The common structural element of many plant RLKs is an extracellular leucine-rich repeats (LRRs) domain, often referred to as LRR-RLK (Kinoshita et al., 2005). Each LRR is typically 20–30 residues long that has a high portion of leucine and other hydrophobic residues. The number of LRR motifs ranges between 3 and 27, and the distribution interval of LRR varies in different LRR-RLKs (Bella et al., 2008).
The transmembrane kinases (TMKs) of LRR-RLKs family play an important role in regulating plant growth and development (Xu et al., 2014). In Arabidopsis, unique double and triple TMK mutant combinations lead to serious growth defects, such as late flowering and seed sterility (Dai et al., 2013). The Arabidopsis TMK4 acts as a node in auxin and ABA signaling pathways (Wang et al., 2020; Li et al., 2021). In addition, TMKs have vital roles in non-canonical auxin signaling in regulating pavement cell morphogenesis, differential growth of the apical hook, lateral root formation, root gravitropic response, and thermomorphogenesis in Arabidopsis (Lin et al., 2021). Up to the present, only one rice TMK-encoding gene (OsTMK) has been characterized, and it is involved in gibberellin signal transduction (van der Knaap et al., 1999). Whether rice TMKs play a role in regulation of leaf senescence remains unknown.
In this study, we isolated and characterized the rice mutant water-soaked spot1 (oswss1), which exhibits water-soaked spots and necrotic symptoms on leaves. OsWSS1 encodes a member of the TMK subfamily of RLKs. The single amino acid substitution (Leu396Pro) in OsWSS1 results in the water-soaked spots and early leaf senescence phenotypes of the mutant oswss1. These results demonstrated that OsWSS1 encodes a TMK that plays an important role in regulation of rice leaf senescence.
Materials and Methods
Plant Materials and Growth Conditions
The rice early leaf senescence mutant oswss1 was identified from the ethyl methane sulphonate (EMS) mutagenesis library of the indica rice cultivar JG30. The mutant oswss1 was crossed with two japonica cultivars (02428 and Nipponbare) and the indica line JG30 for genetic analysis and gene mapping. The F1 plants were self-pollinated to produce F2 seeds. Unless specified otherwise, all parents, F1 hybrids, F2 populations, and transgenic plants were grown under natural conditions in paddy fields in Beijing and Hainan province, China.
Measurement of Chlorophyll Concentrations and Relative Water Content
For determination of the content of chlorophyll a (Chl a), chlorophyll b (Chl b), and carotenoid (Car), 10–30 mg wild-type leaves and oswss1 green leaves and water-soaked spot leaves were cut into small pieces and chlorophylls were extracted in 95% ethanol solution for 48 h at 4°C in the dark. The absorbance values at wavelengths of 665, 649, and 470 nm were measured using a BioPhotometer plus (Eppendorf, Germany). The 95% ethanol was used as a blank control. The concentrations of Chl a, Chl b, and Car were calculated following a published method (Lichtenthaler, 1987). There are six biological replicates for each group.
For measurement of RWC, the fully expanded leaves of wild type and oswss1 were excised from plants in the field, and the fresh weight was measured and recorded as m1. The leaves were soaked in distilled water in a 50 mL centrifuge tube for 2 h, weighed, and recorded as m2. The leaves were then dried in a 60°C drying oven for 48 h, weighed, and recorded as m3. The percentage of RWC was calculated as (m1 − m3)/(m2 − m3) × 100.
Transmission Electron Microscopy and Scanning Electron Microscopy
Wild-type leaves, oswss1 green leaves, and water-soaked spot leaves were cut into pieces and were immersed in 2.5% pre-cooled glutaraldehyde solution, vacuumed for 60 min, and then kept at 4°C overnight. Subsequent procedures were performed following a published protocol (Zheng et al., 2021). Briefly, the samples were washed with phosphate buffer (0.1 M, pH 7.0) and fixed with 1% osmic acid stationary solution (pH 7.3), then dehydrated using a graded ethanol series. After dehydration, samples were embedded in epoxy resin. Afterward, sections were stained with uranyl acetate and examined with a Hitachi HT7700 transmission electron microscope.
For scanning electron microscopy analysis of the leaf epidermis, the samples were fixed in 2.5% glutaraldehyde solution and washed using phosphate buffer, and then dehydrated through a graded ethanol series. The samples were dried by the critical-point drying method and coated with gold, and then observed using a Hitachi TM-1000 scanning electron microscope (Rao et al., 2015).
Observation of Leaf Cell Morphology
At the tillering stage (87 days after sowing, DAS), leaves of wild type and oswss1 mutant were fixed with formalin/acetic acid/alcohol fixative (FAA, 3.7% formaldehyde, 5% glacial acetic acid, and 50% ethanol) for 24 h, then dehydrated with 40, 60, 80, and 100% alcohol for 50 min, respectively. After that, the leaves were placed in 1.25 g/mL chloral hydrate solution for 60 min at 96°C. The upper epidermal tissues of leaves were scraped, and the lower epidermal cells were examined and photographed using BX43 light microscope (Tokyo, Japan).
For paraffin sectioning, leaves of wild type and oswss1 mutant were fixed in FAA fixative overnight at 4°C. The paraffin sectioning procedures were performed following a published protocol (Ren et al., 2016). In brief, the samples were dehydrated with graded ethanol series, infiltrated with graded xylene series, and embedded in paraffin. The embedded samples were sliced into 10 μm thickness pieces and stained with a solution containing 1% safranine and 1% Fast Green. The sections were observed and photographed using BX43 light microscope (Tokyo, Japan).
Map-Based Cloning of OsWSS1
The phenotypes of F1 plants and the segregation ratio of F2 population were identified for genetic analysis. F2 individuals with mutant phenotype were selected for DNA extraction and gene mapping. The parents and 21 F2 individuals with mutant phenotype from the 02428/oswss1 cross were subjected to initial linkage analysis by genotyping using 189 polymorphic insertion/deletion (InDel) markers covering 12 rice chromosomes. Subsequently, a large F2 population with 1184 mutant individuals were used for fine mapping. New polymorphic primers were developed based on DNA sequence differences between the japonica cultivar Nipponbare (NIP) and indica cultivar JG30 to further narrow down the OsWSS1 locus. The wild type and oswss1 mutants were amplified by PCR using the marker primers (Supplementary Table 2). The results were analyzed using DNAMAN software.
For complementary experiment of oswss1, a 10 kb genomic DNA fragment containing the 6.6 kb wild type OsWSS1 coding sequence (CDS), 2.3 kb upstream and 1.1 kb downstream, was amplified by PCR using primers pWSS1-CF and pWSS1-CR (Supplementary Table 3). The amplified fragment was cloned into the binary vector pCAMBIA1300 using the ClonExpress® II One Step Cloning Kit (Vazyme, China). The constructed vector was transformed into callus cells of oswss1 by Agrobacterium tumefaciens-mediated transformation and the transgenic plants were regenerated.
Subcellular Localization
To detect the subcellular localization of OsWSS1, the full-length CDS of OsWSS1 without the termination codon was amplified by PCR using primers WSS1-GFP-F and WSS1-GFP-R (Supplementary Table 3) and fused into the N-terminal of enhanced green fluorescent protein (eGFP) in the pCAMBIA1300 vector under the control of the CaMV 35S promoter to generate a p35S:OsWSS1:eGFP vector. The constructs were then transiently expressed or co-expressed with PIP2-mCherry, a plasma membrane marker, in rice protoplasts derived from the wild-type rice seedlings following a published protocol (Rao et al., 2015). In addition, the constructs were transiently expressed or co-expressed in Nicotiana benthamiana leaves by Agrobacterium tumefaciens-mediated infiltration following the methods described previously (Gao et al., 2016). GFP fluorescence signals in rice protoplasts and epidermal cell of N. benthamiana leaves were observed using a Zeiss LSM700 laser scanning confocal microscope.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted using the RNAiso Plus reagent (Takara, Japan) according to the manufacturer’s manual. Total of 1 μg RNA was reversely transcribed into complementary DNA (cDNA) using a FastQuant RT Kit with gDNA remover (Tiangen, China). Quantitative reverse transcription PCR (qRT-PCR) was performed on an ABI 7500 Real-Time PCR system (Life Technologies, Carlsbad, CA, United States) with an SYBR® Premix ExTaq™ II (Takara). Rice OsActin (LOC_Os03g50885) was used as an internal reference gene. The relative quantitative analysis was performed by 2–Δ ΔCT method (Livak and Schmittgen, 2001). Primers used for qRT-PCR are listed in Supplementary Table 4.
Measurement of Hydrogen Peroxide (H2O2) and Malondialdehyde Contents, and Antioxidants Activities
At the tillering stage, fresh leaves (0.3 g) of wild type and oswss1 were collected and ground into homogenate in a precooled mortar with 2 mL 50 mmol L–1 precooled sodium phosphate buffer (pH 7.8). The homogenate was centrifuged at 12, 000 × g at 4°C for 10 min. The supernatant was collected for determination of the content of H2O2 and MDA, the activities of SOD and CAT, using the kits (A064-1-1, A003-1-2, A001-1-2, and A007-1-1) following the manufacturer’s manuals (Nanjing Jiancheng Bioengineering Institute, China).
Histochemical Analysis and TUNEL Assay
The 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining assays were performed as described previously (Thordal-Christensen et al., 1997). Briefly, leaves of wild type and oswss1 were collected at the tillering stage, then soaked in 0.1% (w/v) DAB (Sigma) or 0.05% (w/v) NBT (Duchefa) solution and dyed for more than 8 h under dark conditions. The staining leaves were transferred into 95% ethanol for decolorization. Trypan blue staining was used to detect dead cells according to the method of a published protocol (Zheng et al., 2021). In brief, leaves at the same part were cut off and immersed in trypan blue solution and boiled for 10 min. After being kept in dark overnight, the leaves were transferred to 25 mg/mL chloral hydrate to decolorize for 3 days.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay was performed following a published protocol (Huang et al., 2007). Briefly, leaves of wild type and oswss1 were collected and fixed in FAA solution for 24 h, then embedded in paraffin and cut into thin slices. Slides were hydrated with ethanol series and treated with proteinase K in phosphate buffer (pH 7.4). The TUNEL assay was performed using a Fluorescein In Situ Cell Death Detection Kit (Roche) according to the manufacturer’s instructions.
Bioinformatics Analyses
OsWSS1 protein transmembrane domain prediction was performed using TMHMM server v. 2.01. Conserved domains of the OsWSS1-encoded proteins were analyzed using SMART2. The sequences used for phylogenetic analysis were obtained from the NCBI Blast website3 using OsWSS1 protein sequence (BAF28192) as the query. Alignment of the full-length amino acid sequences was performed using the DNAMAN software, and sequence logos of each protein were generated online with WebLogo3 (Crooks et al., 2004). The phylogenetic trees were constructed using MAGE 7.0 version software using the bootstrap method with 1000 bootstrap replicates. Protein sequence alignment was performed among OsWSS1 and its homologs from Oryza brachyantha (XP_006663422.1), Sorghum bicolor (XP_002450707.1), Setaria italic (XP_004979222.1), Zea mays (XP_008670729.1, PWZ25297.1), Panicum hallii (XP_025827113.1), Triticum dicoccoides (XP_037421068.1), Triticum aestivum (KAF7042763.1), Aegilops tauschii subsp. tauschii (XP_020188972.1), Brachypodium distachyon (XP_003576062.1), Panicum miliaceum (RLM70345.1), Ananas comosus (OAY69618.1), Triticum urartu (EMS63078.1), Triticum turgidum subsp. durum (VAH91395.1), Hordeum vulgare (KAE8766681.1), Musa balbisiana (THU51216.1), Carex littledalei (KAF3334352.1), Phoenix dactylifera (XP_008812470.1), Capsicum annuum (XP_016551399.1), Cocos nucifera (AID55112.1), Cannabis sativa (XP_030507417.1), Vitis vinifera (RVW82026.1), Camellia sinensis (XP_028113257.1), Helianthus annuus (XP_021981103.1), Medicago truncatula (XP_003601704.1), Nelumbo nucifera (XP_010246532.1), and Arabidopsis thaliana (AAP04161.1).
Results
The Oswss1 Mutant Exhibits Water-Soaked Spots and Withered Leaves
The rice oswss1 mutant was isolated from the indica rice cultivar JG30 that was treated with EMS mutagenesis. Under natural conditions of the paddy field, light green water-soaked spots gradually appeared on oswss1 leaves at the initial tillering stage (60 days after sowing, DAS), the water-soaked spot leaves gradually withered, while the wild-type (WT) leaves remain green (Figure 1A and Supplementary Figures 1A–C). An abundance of light green spots appeared on the oswss1 leaves at the booting stage (105 DAS, Supplementary Figure 1D). We measured the chlorophyll (Chl) content of the water-soaked spots and green parts of oswss1 leaves at the initial tillering stage. The results showed no significant difference between the green part of oswss1 and WT leaves, while the contents of Chl a, Chl b, and Car in the water-soaked zone of oswss1 leaves were significantly lower than those in the WT leaves (Figure 1B). At the heading stage (118 DAS), the panicle length of oswss1 was shorter than that of the WT (Figures 1C,D). The lengths of the internodes I, II, III, IV, and V of oswss1 were shortened by 22.6, 30.2, 33.8, 22.5, and 32.8%, respectively, in comparison with WT (Supplementary Figure 2). In addition, the plant height, seed-setting rate, and 1000-grain weight were lower in oswss1 than that of the WT (Figures 1E–G). In short, the water-soaked spots of oswss1 have an obvious negative effect on the agronomic traits.
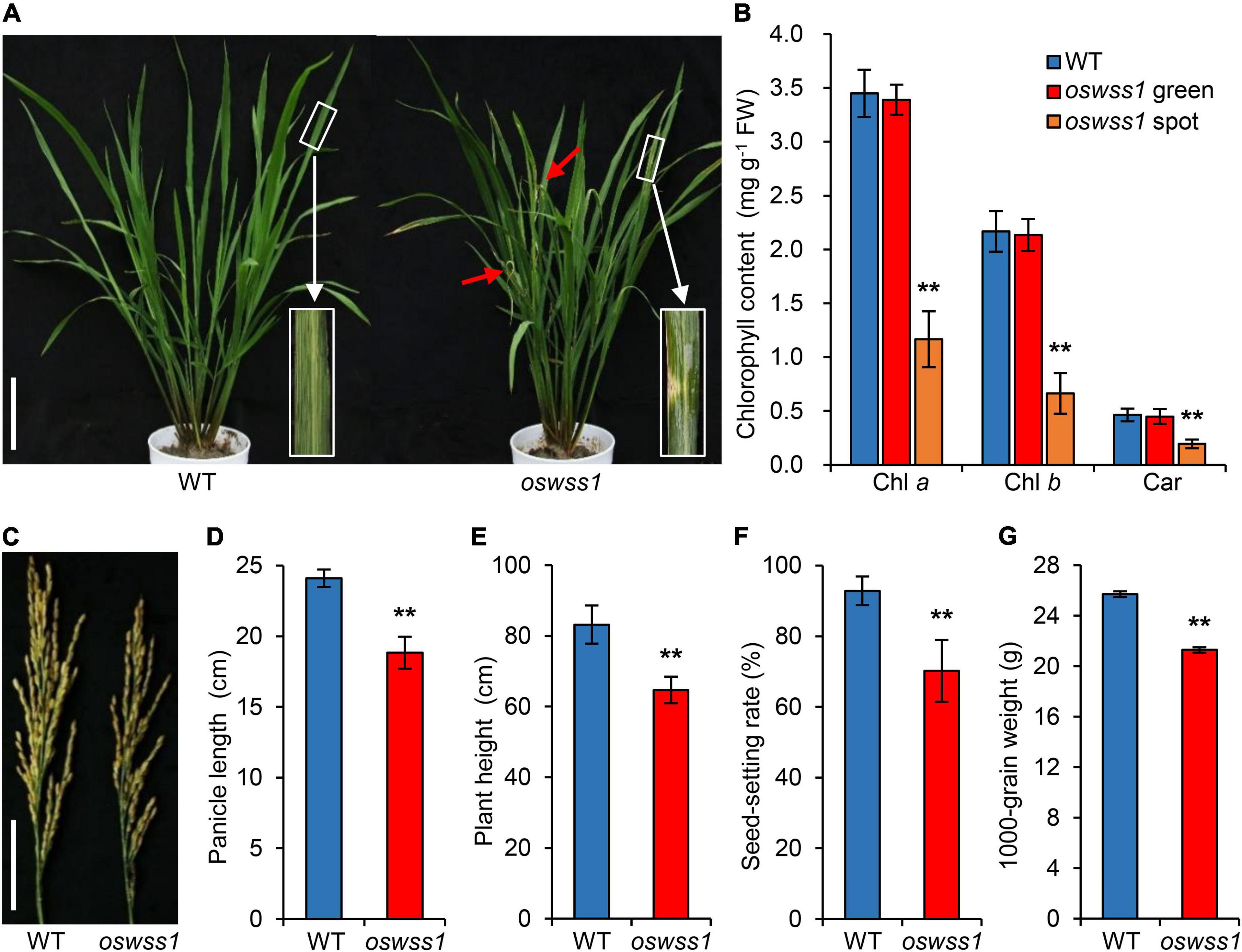
Figure 1. Phenotype characterization and major agronomical traits of the rice oswss1 mutant. (A) Phenotype of wild type (WT) and oswss1 at the initial tillering stage (60 days after sowing, DAS). Inset shows a magnified view of a water-soaked spot of oswss1 leaves. Scale bar = 14 cm. (B) Comparison of chlorophyll content between WT and oswss1 at the initial tillering stage (60 DAS). Values are mean ± SD (n = 6). (C) Panicle morphology comparison between WT and oswss1 at the heading stage (118 DAS). Scale bar = 6 cm. (D–G) Agronomic traits of WT and oswss1 plants. (D) Panicle length; (E) plant height; (F) seed-setting rate; (G) 1,000-grain weight. Values are mean ± SD (n = 10). **P < 0.01 (Student’s t-test).
Irregular Cell Structure and Abnormal Chloroplast Development in the Oswss1 Leaves
At the maximum tillering stage (87 DAS), the water-soaked zone of oswss1 leaves were slightly curled (Figure 2A). Paraffin sectioning assay showed that the mesophyll cells were arranged normally and regularly in the WT leaves (Figure 2B). In contrast, the volume of bulliform cells (BCs) in the water-soaked zone of oswss1 leaves was significantly reduced; the mesophyll cells (MCs) were abnormal, and the cell arrangement was irregular (Figure 2B). The leaf width of the water-soaked zone of oswss1 mutant was narrower than that of WT (Figure 2C). In addition, the relative water content (RWC) of the water-soaked zone of oswss1 leaves was lower, compared with that of WT leaves (Figure 2D). These results implied that the slightly rolled oswss1 leaves may be caused by the atrophied BCs and MCs.
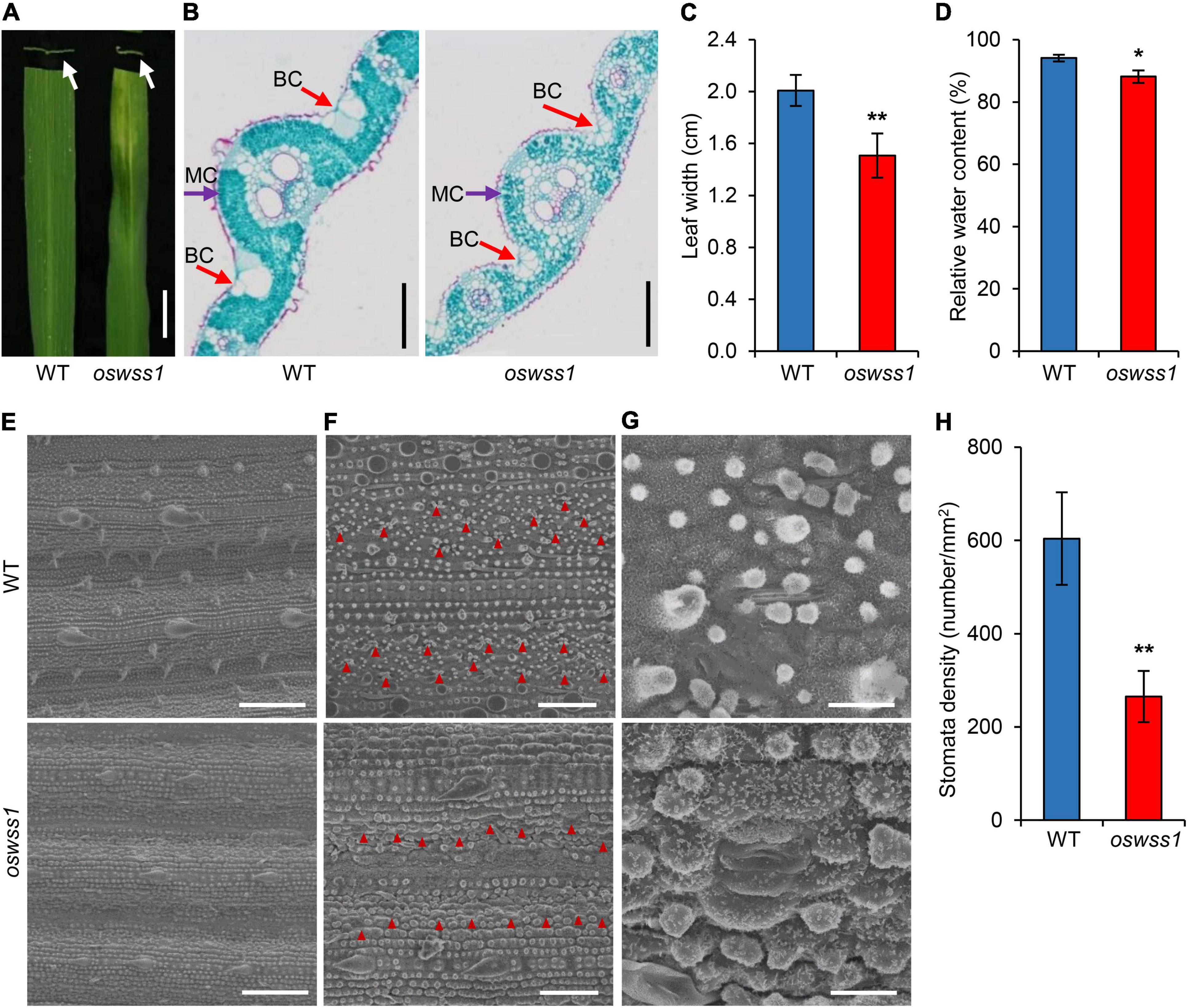
Figure 2. Morphology comparison between wild type (WT) and oswss1 leaves at the tillering stage (87 DAS). (A) Close-up image and cross section of WT and oswss1 mutant leaves. Arrows indicate the cross section of water-soaked spot leaf. Scale bar = 2 cm. (B) Comparison of cell structure between WT and oswss1 leaves. BC, bulliform cell; MC, mesophyll cell. Scale bars = 200 μm. (C) Comparison of leaf width between WT and oswss1 leaves. (D) Comparison of relative water content between WT and oswss1 leaves. (E) Scanning electron microscopy analysis of leaf morphological characteristics between WT and oswss1 leaves. Scale bars = 100 μm. (F) Comparison of stomata density between WT and oswss1 leaves. The red triangles indicate the positions of stomata. Scale bars = 50 μm. (G) Morphological characteristics of stomata between WT and oswss1 leaves. Scale bars = 10 μm. (H) Statistical analysis of stomata density between WT and oswss1. Values are mean ± SD (n = 6). *P < 0.05; **P < 0.01 (Student’s t-test).
Scanning electron microscopy (SEM) assays showed the leaf surface in the water-soaked zone of oswss1 leaves was seriously shrunk, the cuticular papillae were reduced, the stomatal morphology was deformed, and the stomatal density was also significantly reduced compared with that of WT (Figures 2E–H). Furthermore, the lower epidermal cells of the leaves were observed under light microscope. The leaf epidermal cells of WT showed regular rectangular shape, tightly arranged and had a complete cell structure (Supplementary Figure 3A). However, the leaf epidermal cell shape in the water-soaked zone of oswss1 leaves was irregular, the cell arrangement was loose, and the cell structure changed significantly (Supplementary Figure 3B). Overall, these results suggested that OsWSS1 play a role in regulation of leaf development.
The chloroplast ultrastructure of the WT leaves, the green parts and water-soaked zone of oswss1 leaves were compared by transmission electron microscopy (TEM) at the tillering stage (87 DAS). TEM observation revealed that the chloroplasts were normal and structural integrity in the WT and the green part of oswss1, including intact membrane and dense grana lamella (Figures 3A–D). In contrast, in the water-soaked zone of oswss1 leaves, the chloroplast development was impaired, the thylakoid lamellar structure was abnormal, and the grana lamellar was severely reduced, the number of chloroplasts was decreased (Figures 3E,F). The results suggested that OsWSS1 plays an important role in chloroplast development.
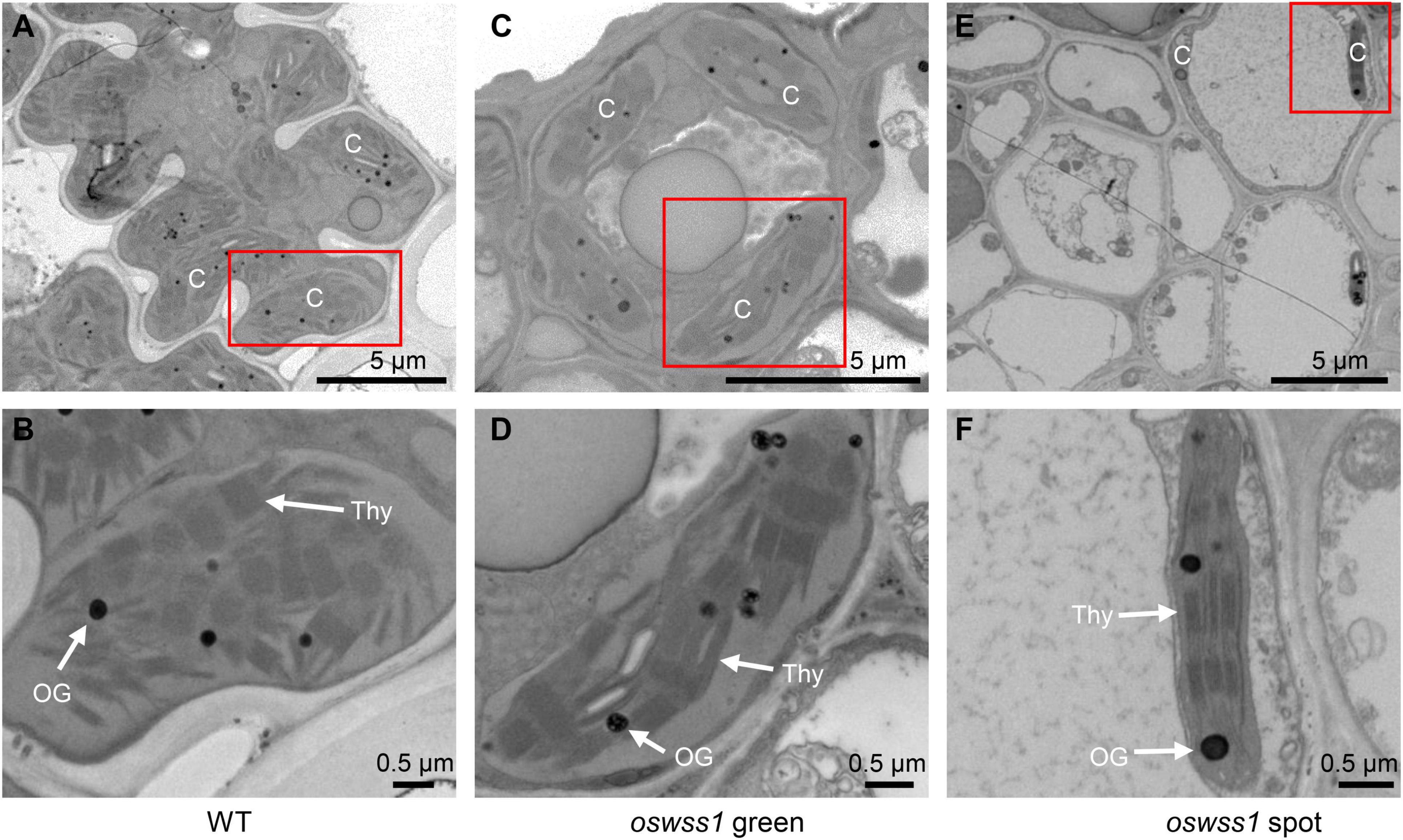
Figure 3. Comparison of chloroplast ultrastructural between wild type (WT) and oswss1 leaves at the tillering stage (87 DAS). Transmission electron microscopy (TEM) was used to detect the chloroplasts ultrastructural in WT leaves (A,B), oswss1 green sectors (C,D) and oswss1 spot sectors (E,F). C, chloroplast; OG, osmiophilic plastoglobuli; Thy, thylakoid. The red box indicates the area magnified.
Map-Based Cloning of OsWSS1
The oswss1 was crossed with two japonica rice cultivars (NIP and 02428) and the indica line JG30, respectively. The leaves of F1 plants were the same as the WT. The phenotype of leaf morphology showed 3:1 (WT: mutant) segregation ratio in the F2 populations (Supplementary Table 1). These results indicated that the water-soaked spot phenotype of oswss1 was controlled by a single recessive gene.
For molecular cloning of OsWSS1 locus, the polymorphism between the parents of oswss1 and 02428 was analyzed, a total of 189 pairs of polymorphic insertion/deletion (InDel) markers were screened. Subsequently, 21 mutant individuals from the 02428/oswss1 F2 populations were used for linkage analysis. The OsWSS1 locus was preliminarily mapped on chromosome 11 between the markers ID11-131 and ID11-12 (Figure 4A). For fine mapping of OsWSS1, nine new InDel markers were developed (Supplementary Table 2). Through genotyping of 1184 mutant individuals from the F2 populations, OsWSS1 was narrowed down to an 89.53 kb region between the markers Ch11-38 and Ch11-42 (Figure 4A). This region contains 13 annotated open reading frames (ORFs). Sequencing of these 13 ORFs revealed a T-to-C single-base substitution in the first exon of LOC_Os11g26130. This T to C change results in an amino acid substitution from leucine (Leu) to proline (Pro) in the 396th residue (Figures 4B,C). Additionally, the mutation of the OsWSS1 gene was verified by a cleaved amplified polymorphic sequence (CAPS) marker which was developed based on this point mutation (Figure 4D).
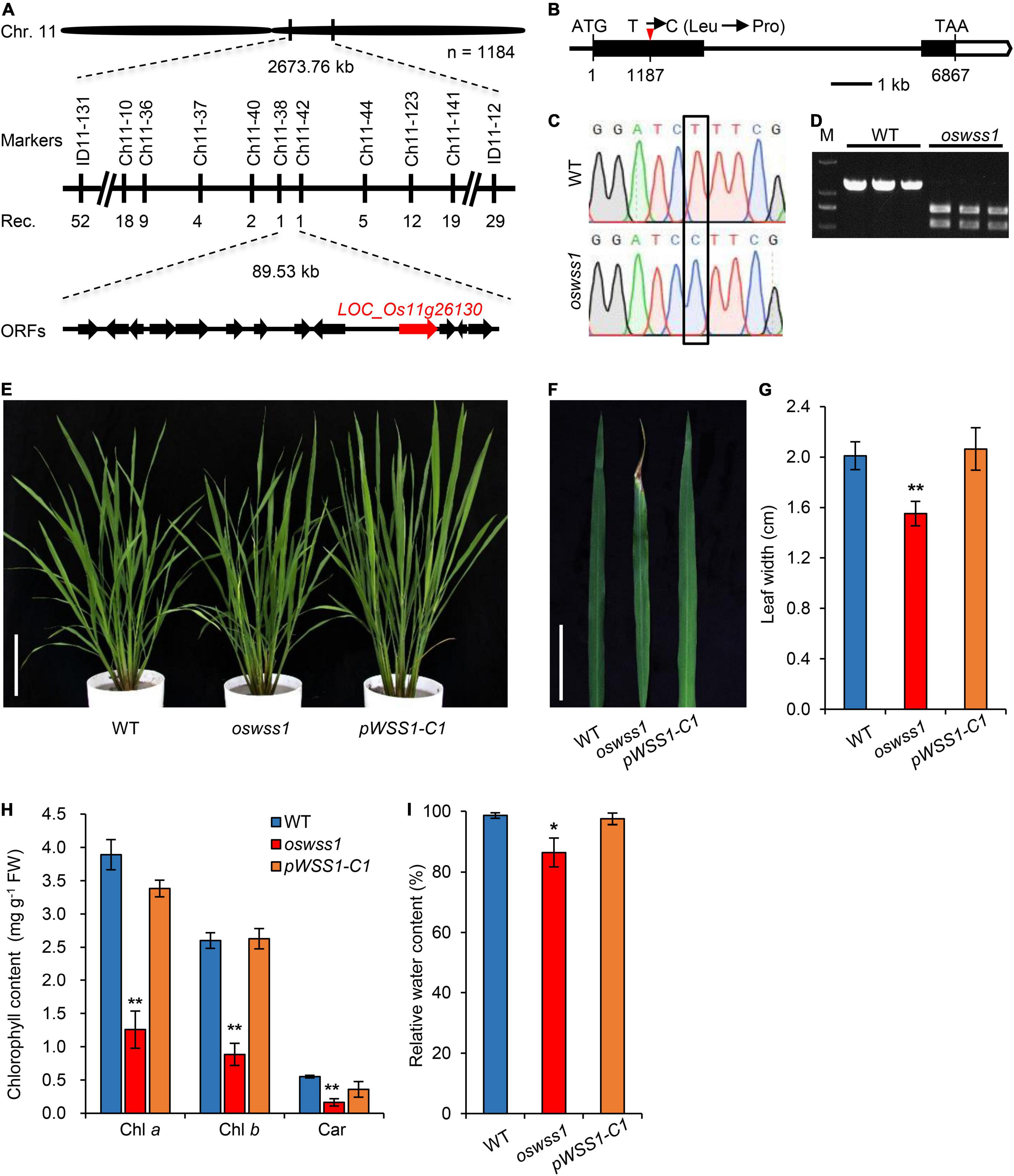
Figure 4. Map-based cloning of OsWSS1 and complementation test. (A) Fine mapping of OsWSS1. The OsWSS1 locus was mapped to an 89.53 kb genomic region on chromosome 11 between markers Ch11-38 and Ch11-42. The molecular markers and recombinant numbers are displayed. (B) The structure and mutation site in OsWSS1. The white boxes, black boxes, and black lines represent untranslated regions, exons, and introns, respectively. (C) DNA sequencing to confirm the point mutation of the OsWSS1 in the oswss1 mutant. The black box indicates the mutation position. (D) Confirmation of the mutation by cleaved amplified polymorphic sequence. PCR products from oswss1 were digested by BamHI while PCR products from wild type (WT) were not digested by BamH1. (E) The plant morphology among WT, oswss1, and complementation transgenic line at the initial tillering stage (60 DAS). Scale bar = 15 cm. (F) Leaf morphology among WT, oswss1 and complementation transgenic line at the tillering stage. Scale bar = 5 cm. (G) Comparison of leaf width of WT, oswss1, and complementation plants. (H) Comparison of chlorophyll content of WT, oswss1, and complementation plants. (I) Relative water content among WT, oswss1, and complementation leaves. Values are mean ± SD (n = 6). *P < 0.05, **P < 0.01 (Student’s t-test).
The genetic complementation was performed to confirm that the mutation in LOC_Os11g26130 is responsible for the oswss1 mutant phenotype. The complementation vector pCAMBIA1300:OsWSS1 with a WT genomic fragment containing the entire coding region of OsWSS1 along with 2.3 kb upstream sequence and 1.1 kb downstream sequence was constructed and transformed into the oswss1-derived callus through Agrobacterium tumefaciens-mediated transformation. A total of 25 transgenic lines were generated, among them, 19 independent and positive complementary lines (including pWSS1-C1) exhibited the WT normal leaf phenotype (Figures 4E,F and Supplementary Figure 4A). Sequencing revealed that the pWSS1-C1 transgenic line was heterozygous for the oswss1 mutation in the T0 transgenic plant (Supplementary Figure 4B). There was no significant difference in the leaf width, chlorophyll content, and RWC between the WT and pWSS1-C1 complementary lines (Figures 4G–I). Furthermore, the plant height, panicle length, 1,000-grain weight, and seed-setting rate of 10 pWSS1-C complementary lines tested were recovered to the WT level (Supplementary Figures 5A–E and Supplementary Table 5). Quantitative real-time PCR (qRT-PCR) analysis revealed that expression of OsWSS1 in oswss1 was similar to that of the WT plants, whereas the relative transcript level of OsWSS1 was markedly increased in the pWSS1-C1 compared with that in the WT and oswss1 plants (Supplementary Figure 5F), indicating that the phenotype of oswss1 is not caused by the difference in OsWSS1 expression. Taken together, these data confirmed that the mutation of LOC_Os11g26130 was responsible for the early leaf senescence phenotype of oswss1.
OsWSS1 Encodes a TMK1 Type LRR-RLK
According to the RAP-DB database4, the coding sequence of OsWSS1 is composed of 2 exons with 2739 bp of cDNA sequence. Using the deduced OsWSS1 protein sequence as a query to search on the NCBI database5, an identical protein which annotated as receptor protein kinase TMK1 was found. TMK1 belongs the LRR-RLK subfamily of the RLK family. SMART (see Text Footnote 2) analysis showed that OsWSS1 contains an N-terminal signal peptide domain (SP), seven LRR domains, a transmembrane domain (TM), and a serine/threonine kinase domain (Figure 5A and Supplementary Figure 6).
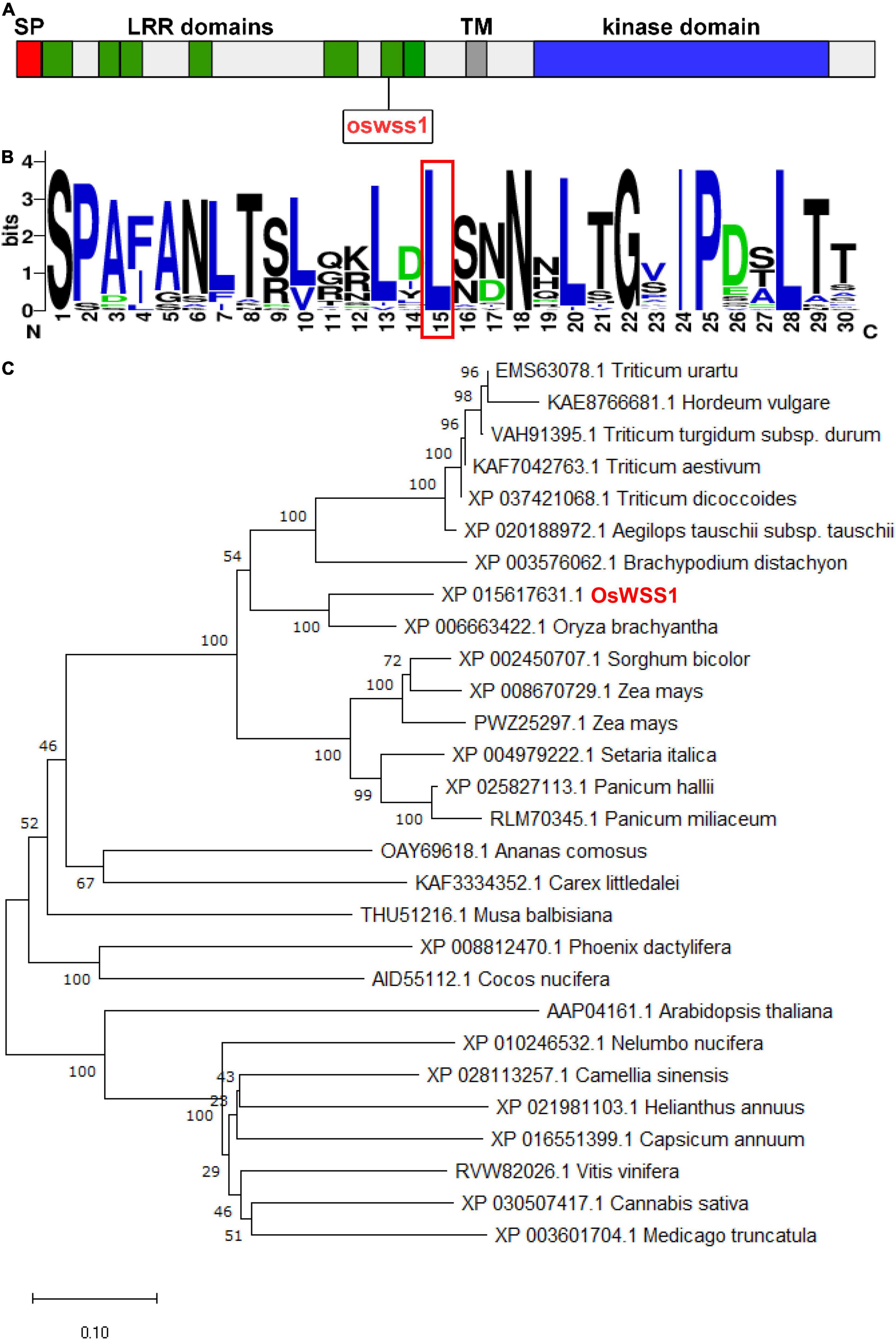
Figure 5. Phylogenic analysis of OsWSS1. (A) Schematic diagram of the OsWSS1 protein. SP, signal peptide; LRR domain, leucine-rich repeat region; TM, transmembrane domain. The rectangle indicates the position of the point mutation of the OsWSS1 in the oswss1 mutant. (B) Conservation analysis of the amino acid substitution region and the frequency of the 50 homologous genes. The red-boxed region indicates the position of the amino acid transition. (C) Phylogenetic tree of the OsWSS1 protein. OsWSS1 is highlighted in red. The phylogenetic tree was constructed using MEGA 7.0 with the neighbor-joining method and 1,000 bootstrap replicates.
A total of 27 OsWSS1 homologous proteins (more than 60% identity with OsWSS1) were identified from different plant species to determine their evolutionary relationship. Phylogenetic analysis showed that OsWSS1 homologs were conserved in both monocots and dicots, and OsWSS1 was closely related to the homologous genes in S. bicolor, Z. mays, and T. aestivum (Figure 5C). In addition, multiple alignment analysis of protein sequences revealed that most OsWSS1-like proteins have a highly conserved serine/threonine kinase domain (Supplementary Figure 7). Multiple sequence alignment and motif analysis revealed that the Leu396Pro amino acid substitution site in the oswss1 mutant was located in a highly conserved LRR region (Figure 5B), suggesting that this site is critical for the function of OsWSS1.
OsWSS1 Is Constitutively Expressed and the Encoded Protein Localizes to the Plasma Membrane
The qRT-PCR was performed to detect the relative expression level of OsWSS1 in different organs of the WT plants. OsWSS1 was constitutively expressed in all tissues at young seedlings stage and heading stage, its expression level was relatively lower in roots, stems, and panicles, and relatively higher in leaves (Figure 6A). To further assess the transcript level of OsWSS1 during leaf senescence, we tested OsWSS1 expression in WT leaves at different developmental stages by qRT-PCR. The OsWSS1 transcript levels were not significantly different in various ages and regions of WT leaves (Supplementary Figure 8), indicating that OsWSS1 is not induced by senescence.
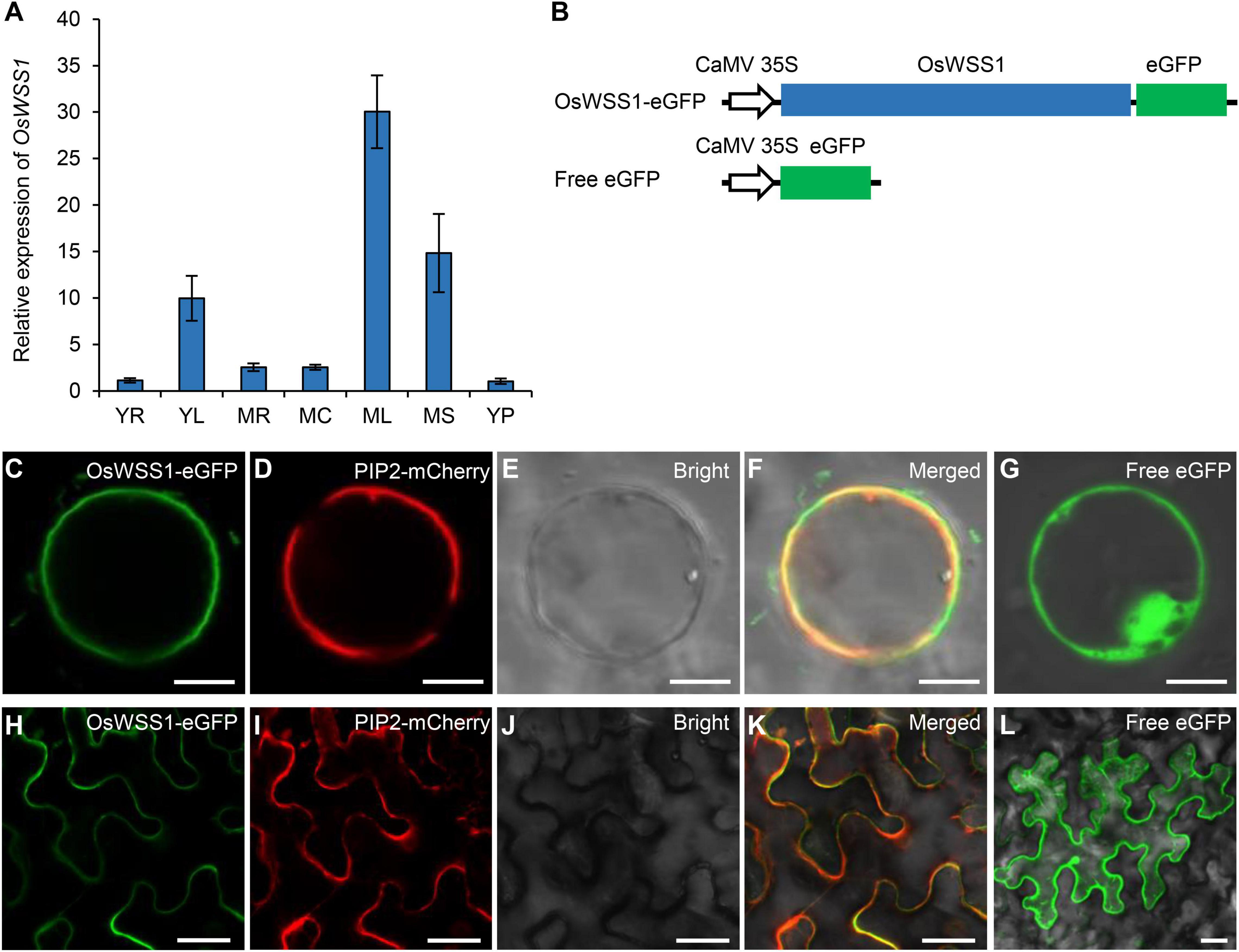
Figure 6. Expression pattern of OsWSS1 and subcellular localization of the OsWSS1-eGFP protein. (A) Expression pattern of OsWSS1 in different tissues of the wild type plants. YR, Young root; YL, young leaf; MR, mature root; MC, mature culm; ML, mature leaf; MS, mature sheath; YP, young panicle. OsWSS1 expression was normalized using the OsActin gene as an internal control. OsWSS1 expression of YR was normalized to 1 after the normalization with the value of OsActin. Values are mean ± SD (n = 3). (B) A schematic diagram of the constructs used for subcellular localization. (C–F) Subcellular localization of OsWSS1 in rice protoplast. The fusion protein (OsWSS1-eGFP) (C) was co-expressed with the plasma membrane marker (PIP2-mCherry) (D). (G) Subcellular localization of the control (Free-eGFP) in rice protoplast. (H–K) Subcellular localization of OsWSS1 in epidermal cell of Nicotiana benthamiana leaves. The fusion protein (OsWSS1-eGFP) (H) was co-expressed with the plasma membrane marker (PIP2.1-mCherry) (I). (L) Subcellular localization of the control (Free-eGFP) epidermal cell of N. benthamiana leaves. Scale bars: 5 μm (C–G); 15 μm (H–L).
Using the Cell-PLoc 2.0 program6, OsWSS1 was predicted to be localized in plasma membrane. To confirm OsWSS1 localization, the expression vector of the OsWSS1 and eGFP fusion was constructed and co-expressed with the plasma membrane marker PIP2-mCherry (Heng et al., 2018) in rice leaf sheath protoplasts (Figure 6B). Confocal laser-scanning microscopy revealed that the green fluorescent signal of OsWSS1-eGFP was indeed colocalized with PIP2 on the plasma membrane, while the negative control, 35S-eGFP, was localized at both the cytoplasm and nucleus (Figures 6C–G). Similar results were obtained in Nicotiana benthamiana leaf epidermal cells (Figures 6H–L). These results demonstrated that OsWSS1 is a plasma membrane protein.
Mutation of OsWSS1 Causes Early Leaf Senescence and Premature Cell Death
Besides the water-soaked spots on leaves, oswss1 mutant displays early leaf senescence (Figure 7A). The oswss1 leaves exhibit water-soaked spots which subsequently developed into necrotic symptoms. Since the expression levels of many SAGs will change during leaf senescence, we determined the expression levels of SAGs in oswss1 and WT plants by qRT-PCR. Results showed that the expression levels of all tested SAGs were significantly upregulated in oswss1 (Figure 7B). In addition, the content of the lipid peroxidation product MDA was increased in oswss1 by 1.4 times as compared with the WT (Figure 7C).
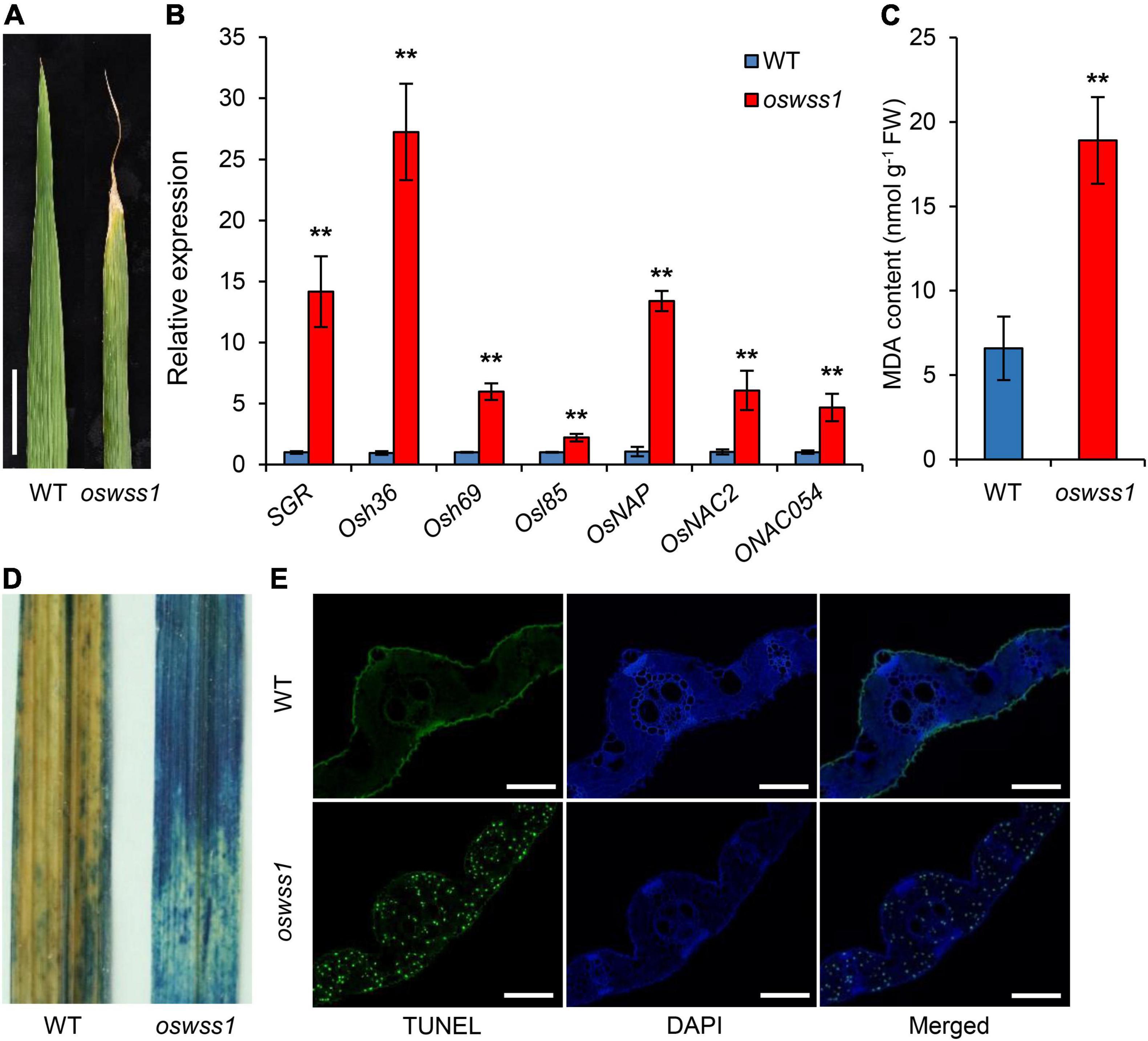
Figure 7. Mutation of OsWSS1 causes early leaf senescence and premature cell death. (A) Leaf morphology between wild type (WT) and oswss1 plants at the heading stage (118 DAS). Scale bar = 4 cm. (B) Transcriptional level analysis of senescence-related genes between WT and oswss1 plants at the tillering stage (87 DAS) by qRT-PCR. OsActin was used as an internal control. The gene expression of WT was normalized to 1 after the normalization with the value of OsActin. Values are mean ± SD (n = 3). **P < 0.01 (Student’s t-test). (C) Comparison of the malondialdehyde (MDA) contents between WT and oswss1 leaves at the tillering stage. Values are mean ± SD (n = 6). **P < 0.01 (Student’s t-test). (D) The trypan blue staining of leaves between WT and oswss1 plants at the tillering stage. (E) TUNEL assay of WT and oswss1 leaves at the tillering stage. Blue signal is 4′,6-diamino-phenylindole (DAPI) staining, green fluorescence represents TUNEL-positive signals. Scale bars = 50 μm.
Trypan blue is an indicator of leaf cell death. After trypan blue staining, large area of the oswss1 leaves was stained blue while only small area of the WT leaves was stained blue (Figure 7D). The TUNEL assay can detect DNA fragmentation that results from apoptotic signaling cascades by labeling the terminal end of nucleic acids. We accordingly used TUNEL assays to determine cell death in the oswss1 mutant. As expected, there were few TUNEL positive signal spots in the WT leaves, whereas there were many strong and high TUNEL positive signals in oswss1 leaves (Figure 7E and Supplementary Figure 9). These data indicated that DNA damage was more serious in oswss1 leaves compared with that of WT leaves. Overall, these results suggested that the oswss1 mutant stimulated premature cell death (PCD) and accelerated the leaf senescence.
Reactive Oxygen Species Is Highly Accumulated in the Oswss1 Leaves
The ROS accumulation usually causes severe damage to most tissues and cells in plant leaves, leading to leaf senescence (Khanna-Chopra, 2012; Xu et al., 2022). To detect the ROS level in the water-soaked spots of the oswss1 mutant, we used DAB and NBT staining to detect H2O2 and O2– accumulation, respectively. The oswss1 leaves showed dark brownish red color while the WT leaves exhibited white color after DAB staining (Figure 8A). NBT stained blue spots were densely distributed in the oswss1 leaves and sparsely distributed in WT leaves (Figure 8B). We also measured the H2O2 concentration in the leaves and results showed the H2O2 content in oswss1 leaves was increased by 40.4% compared with that of WT leaves at the tillering stage (Figure 8C). These data showed that ROS accumulated at a higher level in oswss1 leaves compared with that of WT. To determine whether hyper-accumulation of ROS would activate the antioxidant enzymes, the activities of SOD and CAT were detected. The activities of SOD and CAT of oswss1 leaves were significantly lower compared with that of the WT control (Figures 8D,E). Our findings suggested that mutation of OsWSS1 results in ROS accumulation in rice leaves.
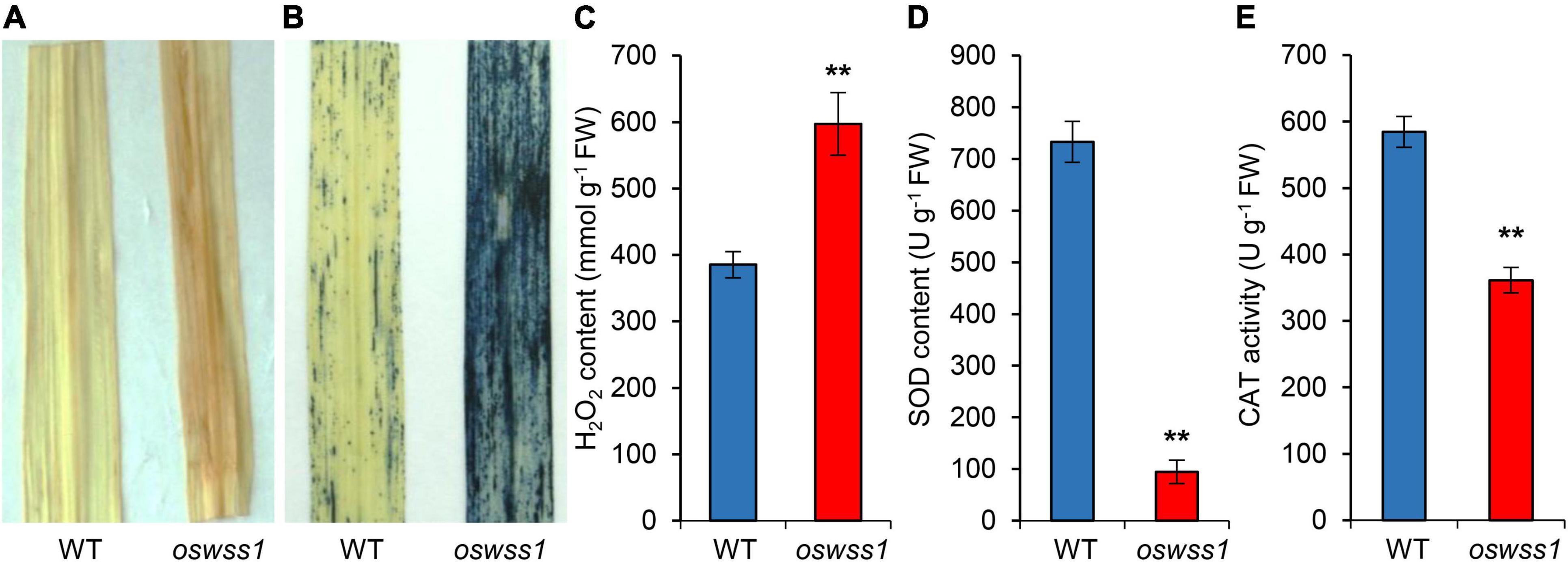
Figure 8. Reactive oxygen species is highly accumulated in the oswss1 leaves at the tillering stage (87 DAS). (A) 3,3′-diaminobenzidine (DAB) staining of leaves between wild type (WT) and oswss1 plants. (B) The nitroblue tetrazolium (NBT) staining of leaves between WT and oswss1 plants. (C–E) Quantification of H2O2 contents (C), SOD activity (D), and CAT activity (E) in leaves between the WT and oswss1 plants. Values are mean ± SD (n = 6). **P < 0.01 (Student’s t-test).
The Expression of Genes Related to Chloroplast Development, Chlorophyll Synthesis and Photosynthesis Altered in Oswss1 Mutant
To investigate the role of OsWSS1 in chloroplast development, the expression of genes related to chloroplast development and photosynthesis (psaA, psbA, rbcL, rbcS, Cab1R, Cab2R, RpoA, and RpoB) (Huang et al., 2020) was compared between WT and oswss1 leaves. The expression of all the tested genes except RpoA was significantly lower in oswss1 as compared with that of the WT (Figure 9A). Since the chlorophyll content of oswss1 decreased (Figure 1B), the expression of 12 chlorophyll biosynthesis-related genes (HEMA, HEMC, HEMD, HEME, OsPORA, OsPORB, OsCAO1, OsCAO2, V1, V2, OsDVR, and OschlH) were compared between WT and oswss1 leaves. The expression of chloroplast biosynthesis-related genes, such as OsPORB (Sakuraba et al., 2013), OsCAO1 (Lee S. et al., 2005), OsCAO2 (Lee S. et al., 2005), and V2 (Sugimoto et al., 2007), were significantly less in oswss1 leaves than that of the WT (Figure 9B). These data suggest the OsWSS1 mutation (Leu396Pro amino acid substitution) affects the expression of genes related to chloroplast development, photosynthesis, and chloroplast biosynthesis. These results also imply that OsWSS1 is involved in chloroplast development in rice.
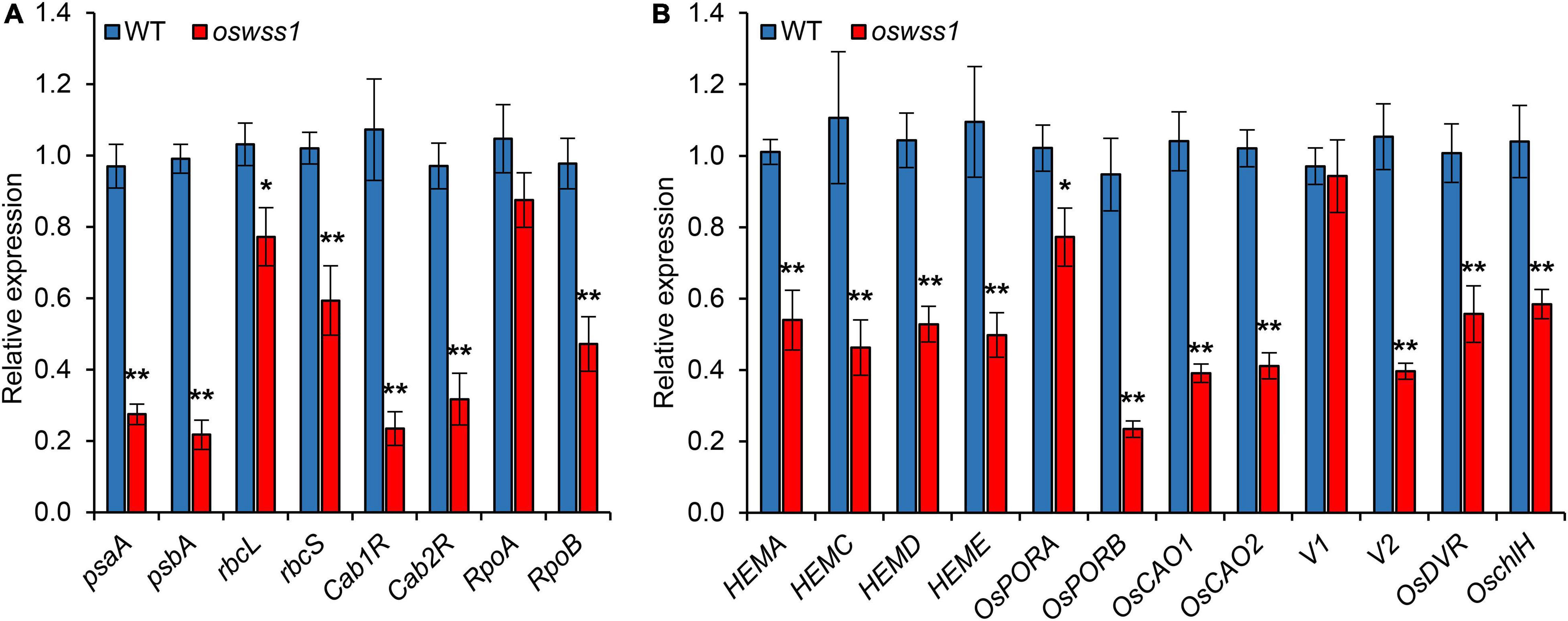
Figure 9. The expression of genes related to chloroplast development, chlorophyll synthesis and photosynthesis altered in oswss1 mutant at the tillering stage (87 DAS) by qRT-PCR. (A) The expression of genes involved in photosynthesis between wild type (WT) and oswss1 mutant. (B) Transcriptional level analysis of expression of genes involved in chlorophyll synthesis and chloroplast development between WT and oswss1 mutant. OsActin was used as an internal control. The gene expression of WT was normalized to 1 after the normalization with the value of OsActin. Values are mean ± SD (n = 3). *P < 0.05, **P < 0.01 (Student’s t-test).
Discussion
OsWSS1 Encodes a Transmembrane Kinases Which Is Essential for Normal Growth and Development of Rice
Despite the extensive studies of some RLKs (including TMK) on regulation of plant growth and development (Clark et al., 1997; Diévart and Clark, 2004; Lee L. Y. et al., 2012; He et al., 2021), up to the present, the function of TMK controlling plant leaf senescence and cell death remains unknown. In this study, we have revealed that rice OsWSS1, a TMK protein, affects plant leaf senescence and cell death. The mutant oswss1 showed reduced chlorophyll content, abnormal chloroplast development, reduced plant height, reduced seed-setting rate, and decreased 1,000-grain weight (Figures 1–3 and Supplementary Figure 1). Unlike rice, there is no significant difference of plant growth and development between Arabidopsis TMK T-DNA insertion single mutant and the wild type (Dai et al., 2013). This is possibly due to the difference of redundancy of TMK genes between Arabidopsis and rice. There are four TMK genes in Arabidopsis (Dai et al., 2013) but only one TMK gene in rice genome. The mutant phenotype of oswss1 was complemented by expressing the wild type OsWSS1 gene (Figure 4 and Supplementary Figure 4), demonstrating that OsWSS1 plays an important role in the growth and development of rice plants.
OsWSS1 Is Involved in the Leaf Epidermal Development and Chloroplast Development in Rice Leaves
The lesion-mimic and necrotic lesion in the leaf suggested that major changes in cell structure have occurred (Qiu et al., 2019; Cui et al., 2021). In this study, we observed changes in leaf cell structure between oswss1 and WT plants. Paraffin section showed atrophy in the BCs and MCs in the water-soaked zone of oswss1 leaves compared to the WT leaves (Figure 2B). Light microscope revealed the abnormal leaf epidermal cell shape in oswss1 and the significantly changed cell structure (Supplementary Figure 3). It has been reported that the defective leaf epidermis is the main contributors to leaf rolling (Li et al., 2017). Thus, the rolled oswss1 leaves may be caused by the defective leaf cell development in the epidermis. Moreover, the surface of water-soaked spots was severely shrunk in oswss1 leaves (Figures 2E–G). The shrunken BCs (Figure 2B) explained the wrinkled leaf epidermal cell shape (Figure 2G and Supplementary Figure 3) observed on the oswss1 mutant. Stomata is a key channel for water and gas exchange, which acts an important role to regulate water evapotranspiration and water utilization (Buckley, 2005). The es1 (early senescence1) (Rao et al., 2015) and ospls1 (Oryza sativa premature leaf senescence 1) (Yang X. et al., 2016) mutants exhibited excessive water loss due to increased stomatal density. However, the oswss1 mutant exhibited lower stomatal density, altered stomatal morphology, and decreased cuticular papillae (Figures 2E–H), indicating that the water evapotranspiration capacity was reduced in oswss1. We speculate that the weakening of water transpiration leads to transpiration force reduction and organic transport obstruction, then would obstruct the leaf normal physiological activities, resulting in water-soaked spots of leaves.
Another typical feature of lesion formation is the change of chloroplast development and the chloroplast degradation (Cui et al., 2021). The chlorophyll content in the water-soaked spots of oswss1 decreased by 30% compared to the WT (Figure 1B). In general, changes in chlorophyll content are associated with chloroplast development in rice (Zhang et al., 2017; Cui et al., 2019; Sun et al., 2019; He et al., 2020). As expected, TEM observation showed that chloroplast development was impaired, chloroplast number was reduced, and there were more degraded chloroplasts in the water-soaked zone of oswss1 compared with that of the WT and the green part of oswss1 plants (Figure 3). The formation of a functional chloroplast requires the coordinated expression of nuclear and chloroplastic genes (He et al., 2018; Chen et al., 2019). This process involves two types of RNA polymerases, nucleus-encoded polymerase (NEP) and plastid-encoded polymerase (PEP) (Jarvis and López-Juez, 2013). PEP transcribes chloroplast genes involved in photosynthesis, such as psaA, psbA, and rbcL; NEP largely transcribes housekeeping genes, such as rpoA and rpoB, which regulate plastid development at the early stages of plant growth (Shiina et al., 2005). In this study, we found the transcripts of PEP-dependent genes were significantly decreased, and the expression of chlorophyll synthesis-associated genes, including HEMA, HEMC, HEMD, HEME, OsPORA, OsPORB, OsCAO1, OsCAO2, V2, OsDVR, and OschlH, were also downregulated in the oswss1 mutant (Figure 9). Overall, these results indicate that OsWSS1 may play a vital role in regulating leaf epidermal development and chloroplast development.
Mutation of OsWSS1 Promotes Reactive Oxygen Species Hyper-Accumulation and Cell Death
Reactive oxygen species are important signaling molecules and able to interact with many kinds of target molecules and metabolites, like DNA, proteins, lipids, and other cellular molecules, ROS accumulation can highly oxidative damage in cell structure and cellular components (Triantaphylidès et al., 2008; Ge et al., 2015). The oswss1 exhibited hyper-accumulation of ROS in the water-soaked spots manifested by NBT and DAB staining, and measurement of H2O2 concentration (Figures 8A–C). ROS is generally in a dynamic balance in plant cells, and its content is determined by the delicate balance between ROS scavenging and ROS generation (Hu et al., 2011). SOD is the first defense line of plant ROS scavenging system to remove O2– in the cell. CAT is another main ROS scavenging enzyme which catalyzes H2O2 to form H2O and O2 (Scandalios, 2002; Mittler et al., 2004). The activities of SOD and CAT were decreased in oswss1 leaves (Figures 8D,E), suggesting that the hyper-accumulation of ROS may be caused by the reduction in the activity of these two key enzymes, thereby resulting in the water-soaked spots phenotype of oswss1. Taken together, these findings suggest that OsWSS1 plays a key role in maintaining ROS homeostasis during rice leaf development and chloroplast development. In addition, ROS can act as a signal to activate defense and offer wide possibilities for broad-spectrum disease resistance (Qi et al., 2017), future studies can examine whether OsWSS1 is involved in the disease resistance in rice.
Reactive oxygen species are also important factors in triggering early leaf senescence and PCD in plants (Loor et al., 2010; Wang et al., 2013; Mittler, 2017; Chen et al., 2021). The oswss1 mutant exhibited remarkable increased amounts of oxidative-stress-induced toxic compound MDA, enhanced PCD (Figures 7C–E and Supplementary Figure 9). During leaf senescence, the expression of some SAGs is promoted (Lim et al., 2007; Yang et al., 2020). The results also showed that the expression levels of SAGs, SGR (Jiang et al., 2011), Osh36, Osh69, Osl85 (Lee et al., 2001), OsNAP (Liang et al., 2014), OsNAC2 (Mao et al., 2017), and ONAC054 (Sakuraba et al., 2020) were significantly upregulated in the oswss1 mutant (Figure 7B). Collectively, we predicted that the early leaf senescence of oswss1 was caused by ROS-mediated PCD. In addition, leaf senescence is also influenced by various phytohormones, including abscisic acid (ABA) and jasmonic acid (JA) (Lim et al., 2007; Liang et al., 2014). The Arabidopsis TMK1 and TMK4 act as key regulators in the signaling cascades of ABA (Li et al., 2021; Yang et al., 2021); the Nicotiana tabacum NtTMK1 was strongly induced by the treatment with JA (Cho and Pai, 2000), thus it is possible that OsWSS1 is involved in the regulation of leaf senescence partially by modulating the signaling responses of phytohormones.
The Putative Molecular Mechanism of OsWSS1 in Regulation of Reactive Oxygen Species Homeostasis
Plant LRR-RLKs are important membrane receptors that regulate plant development by sensing various ligands (Man et al., 2020). The LRR domain binds to ligands and transmits extracellular signals to the downstream (Chen, 2021). There was a point mutation in the 7th LRR domain of OsWSS1, in which the hydrophobic residue Leu396 was replaced by Pro amino acid in oswss1 mutant (Figure 5B and Supplementary Figure 7). The hydrophobic residues play key roles in protein folding, ligand-protein and protein–protein binding, protein-nucleic acid interactions (Southall et al., 2002; Matsushima et al., 2005). Several pieces of evidence suggest that LRR-RLKs play a role in ROS homeostasis (Ouyang et al., 2010; Lin et al., 2020). Overexpression of OsSIK2 in rice reduced ROS accumulation by enhancing the activities of POD, SOD, and CAT (Ouyang et al., 2010). OsSTLK-overexpressing rice plants remarkably reduced ROS concentration by upregulating ROS-scavenging activities (Lin et al., 2020). In this study, the oswss1 mutant exhibits hyper-accumulation of ROS and lower SOD and CAT activities (Figures 8C–E). The expression levels of OsWSS1 were not significantly different between WT and oswss1 mutant (Supplementary Figure 5F), and there was no distinct difference in OsWSS1 expression during leaf senescence (Supplementary Figure 8), suggesting that the expression level of OsWSS1 may not affect the aging process. Based on these results, we speculate that OsWSS1 probably stimulates the activities of the SOD or CAT and regulates ROS homeostasis through phosphorylation and activation of unknown proteins.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
KZ, JX, CW, and ZJ conceived and designed the research. JX, FX, FW, and YZ performed the experiments. TZ, ZJ, YT, and ZW participated in data analysis. JX, KZ, and TZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Science Foundation of China (U20A2035 to KZ), National Natural Science Foundation of China (31801729 to ZJ), and the Innovation Program of Chinese Academy of Agricultural Sciences to KZ and CW.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.918673/full#supplementary-material
Footnotes
- ^ http://www.cbs.dtu.dk/services/TMHMM
- ^ http://smart.embl-heidelberg.de/
- ^ http://www.ebi.ac.uk/Tools/sss/ncbiblast/
- ^ https://rapdb.dna.affrc.go.jp/#
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/
References
Bella, J., Hindle, K. L., Mcewan, P. A., and Lovell, S. C. (2008). The leucine-rich repeat structure. Cell. Mol. Life Sci. 65, 2307–2333. doi: 10.1007/s00018-008-8019-0
Buckley, T. N. (2005). The control of stomata by water balance. New Phytol. 168, 275–292. doi: 10.1111/j.1469-8137.2005.01543.x
Chen, D., Qiu, Z., He, L., Hou, L., Li, M., Zhang, G., et al. (2021). The rice LRR-like1 protein YELLOW AND PREMATURE DWARF 1 is involved in leaf senescence induced by high light. J. Exp. Bot. 72, 1589–1605. doi: 10.1093/jxb/eraa532
Chen, L., Huang, L., Dai, L., Gao, Y., Zou, W., Lu, X., et al. (2019). PALE-GREEN LEAF12 encodes a novel pentatricopeptide repeat protein required for chloroplast development and 16S rRNA processing in rice. Plant Cell Physiol. 60, 587–598. doi: 10.1093/pcp/pcy229
Chen, T. (2021). Identification and characterization of the LRR repeats in plant LRR-RLKs. BMC Mol. Cell Biol. 22:9. doi: 10.1186/s12860-021-00344-y
Chen, Y., Xu, Y., Luo, W., Li, W., Na, C., and Chong, Z. K. (2013). The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 163, 1673–1685. doi: 10.1104/pp.113.224527
Cho, H. S., and Pai, H. S. (2000). Cloning and characterization of NtTMK1 gene encoding a TMK1-homologous receptor-like kinase in tobacco. Mol. Cells 10, 317–324. doi: 10.1016/S0014-5793(00)01678-1
Clark, S. E., Williams, R. W., and Meyerowitz, E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. doi: 10.1016/s0092-8674(00)80239-1
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Cui, X., Wang, Y., Wu, J., Han, X., Gu, X., Lu, T., et al. (2019). The RNA editing factor DUA1 is crucial to chloroplast development at low temperature in rice. New Phytol. 221, 834–849. doi: 10.1111/nph.15448
Cui, Y., Peng, Y., Zhang, Q., Xia, S., Ruan, B., Xu, Q., et al. (2021). Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. 105, 942–956. doi: 10.1111/tpj.15079
Dai, N., Wang, W., Patterson, S. E., and Bleecker, A. B. (2013). The TMK subfamily of receptor-like kinases in Arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLoS One 8:e60990. doi: 10.1371/journal.pone.0060990
Diévart, A., and Clark, S. E. (2004). LRR-containing receptors regulating plant development and defense. Development 131, 251–261. doi: 10.1242/dev.00998
Domínguez, F., and Cejudo, F. J. (2021). Chloroplast dismantling in leaf senescence. J. Exp. Bot. 72, 5905–5918. doi: 10.1093/jxb/erab200
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
Gao, Y., Jia, S., Wang, C., Wang, F., Wang, F., and Zhao, K. (2016). BjMYB1, a transcription factor implicated in plant defence through activating BjCHI1 chitinase expression by binding to a W-box-like element. J. Exp. Bot. 67, 4647–4658. doi: 10.1093/jxb/erw240
Ge, C. W., Zhi-Guo, E., Pan, J. J., Jiang, H., Zhang, X. Q., Zeng, D. L., et al. (2015). Map-based cloning of a spotted-leaf mutant gene OsSL5 in Japonica rice. Plant Growth Regul. 75, 595–603. doi: 10.1007/s10725-014-9962-4
Gish, L. A., and Clark, S. E. (2011). The RLK/Pelle family of kinases. Plant J. 66, 117–127. doi: 10.1111/j.1365-313X.2011.04518.x
He, L., Zhang, S., Qiu, Z., Zhao, J., Nie, W., Lin, H., et al. (2018). FRUCTOKINASE-LIKE PROTEIN 1 interacts with TRXz to regulate chloroplast development in rice. J. Integr. Plant Biol. 60, 94–111. doi: 10.1111/jipb.12631
He, Y., Shi, Y., Zhang, X., Xu, X., Wang, H., Li, L., et al. (2020). The OsABCI7 transporter interacts with OsHCF222 to stabilize the thylakoid membrane in rice. Plant Physiol. 184, 283–299. doi: 10.1104/pp.20.00445
He, Z., Zou, T., Xiao, Q., Yuan, G., Liu, M., Tao, Y., et al. (2021). An L-type lectin receptor-like kinase promotes starch accumulation during rice pollen maturation. Development 148, dev196378. doi: 10.1242/dev.196378
Heng, Y., Wu, C., Long, Y., Luo, S., Ma, J., Chen, J., et al. (2018). OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. Plant Cell 30, 889–906. doi: 10.1105/tpc.17.00998
Hong, Y., Zhang, Y., Sinumporn, S., Yu, N., Zhan, X., Shen, X., et al. (2018). Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. 95, 877–891. doi: 10.1111/tpj.13995
Hu, L., Liang, W., Yin, C., Cui, X., Zong, J., Wang, X., et al. (2011). Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23, 515–533. doi: 10.1105/tpc.110.074369
Huang, L., Sun, Q., Qin, F., Li, C., Zhao, Y., and Zhou, D. X. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144, 1508–1519. doi: 10.1104/pp.107.099473
Huang, W., Zhang, Y., Shen, L., Fang, Q., Liu, Q., Gong, C., et al. (2020). Accumulation of the RNA polymerase subunit RpoB depends on RNA editing by OsPPR16 and affects chloroplast development during early leaf development in rice. New Phytol. 228, 1401–1416. doi: 10.1111/nph.16769
Jarvis, P., and López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Jiang, H., Chen, Y., Li, M., Xu, X., and Wu, G. (2011). Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J. Integr. Plant Biol. 53, 375–387. doi: 10.1111/j.1744-7909.2011.01037.x
Khanna-Chopra, R. (2012). Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249, 469–481. doi: 10.1007/s00709-011-0308-z
Kinoshita, T., Caño-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., et al. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171. doi: 10.1038/nature03227
Kong, Z., Li, M., Yang, W., Xu, W., and Xue, Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141, 1376–1388. doi: 10.1104/pp.106.082941
Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y., Fujimoto, M., et al. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19, 1362–1375. doi: 10.1105/tpc.106.042911
Lee, S., Seo, P. J., Lee, H. J., and Park, C. M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70, 831–844. doi: 10.1111/j.1365-313X.2012.04932.x
Lee, L. Y., Hou, X., Fang, L., Fan, S., Kumar, P. P., and Yu, H. (2012). STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development 139, 1568–1576. doi: 10.1242/dev.079426
Lee, R. H., Wang, C. H., Huang, L. T., and Chen, S. C. (2001). Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 52, 1117–1121. doi: 10.1093/jexbot/52.358.1117
Lee, S., Kim, J. H., Yoo, E. S., Lee, C. H., Hirochika, H., and An, G. (2005). Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol. 57, 805–818. doi: 10.1007/s11103-005-2066-9
Li, L., Li, B., Zhu, S., Wang, L., Song, L., Chen, J., et al. (2021). TMK4 receptor kinase negatively modulates ABA signaling by phosphorylating ABI2 and enhancing its activity. J. Integr. Plant Biol. 63, 1161–1178. doi: 10.1111/jipb.13096
Li, W. Q., Zhang, M. J., Gan, P. F., Qiao, L., Yang, S. Q., Miao, H., et al. (2017). CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 92, 904–923. doi: 10.1111/tpj.13728
Li, Z., Zhang, Y., Zou, D., Zhao, Y., Wang, H. L., Zhang, Y., et al. (2020). LSD 3.0: a comprehensive resource for the leaf senescence research community. Nucleic Acids Res. 48, D1069–D1075. doi: 10.1093/nar/gkz898
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., and Liu, L. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Method. Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Lin, F., Li, S., Wang, K., Tian, H., Gao, J., Zhao, Q., et al. (2020). A leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci. 296:110465. doi: 10.1016/j.plantsci.2020.110465
Lin, W., Zhou, X., Tang, W., Takahashi, K., Pan, X., Dai, J., et al. (2021). TMK-based cell-surface auxin signalling activates cell-wall acidification. Nature 599, 278–282. doi: 10.1038/s41586-021-03976-4
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loor, G., Kondapalli, J., Schriewer, J. M., Chandel, N. S., Vanden Hoek, T. L., and Schumacker, P. T. (2010). Menadione triggers cell death through ROS-dependent mechanisms involving PARP activation without requiring apoptosis. Free Radic. Biol. Med. 49, 1925–1936. doi: 10.1016/j.freeradbiomed.2010.09.021
Man, J., Gallagher, J. P., and Bartlett, M. (2020). Structural evolution drives diversification of the large LRR-RLK gene family. New Phytol. 226, 1492–1505. doi: 10.1111/nph.16455
Mao, C., Lu, S., Lv, B., Zhang, B., Shen, J., He, J., et al. (2017). A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 174, 1747–1763. doi: 10.1104/pp.17.00542
Matsushima, N., Tachi, N., Kuroki, Y., Enkhbayar, P., Osaki, M., Kamiya, M., et al. (2005). Structural analysis of leucine-rich-repeat variants in proteins associated with human diseases. Cell. Mol. Life Sci. 62, 2771–2791. doi: 10.1007/s00018-005-5187-z
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Morillo, S. A., and Tax, F. E. (2006). Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 9, 460–469. doi: 10.1016/j.pbi.2006.07.009
Morita, R., Sato, Y., Masuda, Y., Nishimura, M., and Kusaba, M. (2009). Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 59, 940–952. doi: 10.1111/j.1365-313X.2009.03919.x
Ouyang, S. Q., Liu, Y. F., Liu, P., Lei, G., He, S. J., Ma, B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329. doi: 10.1111/j.1365-313X.2010.04146.x
Qi, J., Wang, J., Gong, Z., and Zhou, J. M. (2017). Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. doi: 10.1016/j.pbi.2017.04.022
Qiao, Y., Jiang, W., Lee, J., Park, B., Choi, M. S., Piao, R., et al. (2010). SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 185, 258–274. doi: 10.1111/j.1469-8137.2009.03047.x
Qiu, Z., Zhu, L., He, L., Chen, D., Zeng, D., Chen, G., et al. (2019). DNA damage and reactive oxygen species cause cell death in the rice local lesions 1 mutant under high light and high temperature. New Phytol. 222, 349–365. doi: 10.1111/nph.15597
Rao, Y., Yang, Y., Xu, J., Li, X., Leng, Y., Dai, L., et al. (2015). EARLY SENESCENCE 1 encodes a SCAR-like protein2 that affects water loss in rice. Plant Physiol. 169, 1225–1239. doi: 10.1104/pp.15.00991
Ren, D., Rao, Y., Wu, L., Xu, Q., Li, Z., Yu, H., et al. (2016). The pleiotropic ABNORMAL FLOWER AND DWARF1 affects plant height, floral development and grain yield in rice. J. Integr. Plant Biol. 58, 529–539. doi: 10.1111/jipb.12441
Saed-Moucheshi, A., Shekoofa, A., and Pessarakli, M. (2014). Reactive oxygen species (ROS) generation and detoxifying in plants. J. Plant Nutr. 37, 1573–1585. doi: 10.1080/01904167.2013.868483
Sakuraba, Y., Kim, D., Han, S. H., Kim, S. H., Piao, W., Yanagisawa, S., et al. (2020). Multilayered regulation of membrane-bound ONAC054 is essential for abscisic acid-induced leaf senescence in rice. Plant Cell 32, 630–649. doi: 10.1105/tpc.19.00569
Sakuraba, Y., Rahman, M. L., Cho, S. H., Kim, Y. S., Koh, H. J., Yoo, S. C., et al. (2013). The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 74, 122–133. doi: 10.1111/tpj.12110
Scandalios, J. G. (2002). The rise of ROS. Trends Bioche. Sci. 27, 483–486. doi: 10.1016/s0968-0004(02)02170-9
Shiina, T., Tsunoyama, Y., Nakahira, Y., and Khan, M. S. (2005). Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 244, 1–68. doi: 10.1016/S0074-7696(05)44001-2
Shiu, S. H., Karlowski, W. M., Pan, R., Tzeng, Y. H., Mayer, K. F., and Li, W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Southall, N. T., Dill, K. A., and Haymet, A. (2002). A view of the hydrophobic effect. J. Phy. Chem. B 106, 521–533. doi: 10.1021/jp015514e
Sugimoto, H., Kusumi, K., Noguchi, K., Yano, M., Yoshimura, A., and Iba, K. (2007). The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J. 52, 512–527. doi: 10.1111/j.1365-313X.2007.03251.x
Sun, Y., Tian, Y., Cheng, S., Wang, Y., Hao, Y., Zhu, J., et al. (2019). WSL6 encoding an Era-type GTP-binding protein is essential for chloroplast development in rice. Plant Mol. Biol. 100, 635–645. doi: 10.1007/s11103-019-00885-z
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Triantaphylidès, C., Krischke, M., Hoeberichts, F. A., Ksas, B., Gresser, G., Havaux, M., et al. (2008). Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 148, 960–968. doi: 10.1104/pp.108.125690
van der Knaap, E., Song, W. Y., Ruan, D. L., Sauter, M., Ronald, P. C., and Kende, H. (1999). Expression of a gibberellin-induced leucine-rich repeat receptor-like protein kinase in deepwater rice and its interaction with kinase-associated protein phosphatase. Plant Physiol. 120, 559–570. doi: 10.1104/pp.120.2.559
Wang, Q., Qin, G., Cao, M., Chen, R., He, Y., Yang, L., et al. (2020). A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants. Nat. Commun. 11:679. doi: 10.1038/s41467-020-14395-w
Wang, Y., Lin, A., Loake, G. J., and Chu, C. (2013). H2O2-induced leaf cell death and the crosstalk of reactive nitric/oxygen species. J. Integr. Plant Biol. 55, 202–208. doi: 10.1111/jipb.12032
Woo, H. R., Kim, H. J., Lim, P. O., and Nam, H. G. (2019). Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant biol. 70, 347–376. doi: 10.1146/annurev-arplant-050718-095859
Wu, L., Ren, D., Hu, S., Li, G., Dong, G., Jiang, L., et al. (2016). Down-regulation of a nicotinate phosphoribosyl transferase gene, OsNaPRT1, leads to withered leaf tips. Plant Physiol. 171, 1085–1098. doi: 10.1104/pp.15.01898
Wu, X. Y., Kuai, B. K., Jia, J. Z., and Jing, H. C. (2012). Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 54, 936–952. doi: 10.1111/jipb.12005
Xu, J., Pan, C., Lin, H., Ye, H., Wang, S., Lu, T., et al. (2022). A rice XANTHINE DEHYDROGENASE gene regulates leaf senescence and response to abiotic stresses. Crop J. 10, 310–322. doi: 10.1016/j.cj.2021.05.011
Xu, T., Dai, N., Chen, J., Nagawa, S., Cao, M., Li, H., et al. (2014). Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343, 1025–1028. doi: 10.1126/science.1245125
Yamatani, H., Sato, Y., Masuda, Y., Kato, Y., Morita, R., Fukunaga, K., et al. (2013). NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll - protein complexes during leaf senescence. Plant J. 74, 652–662. doi: 10.1111/tpj.12154
Yang, J., He, H., He, Y., Zheng, Q., Li, Q., Feng, X., et al. (2021). TMK1-based auxin signaling regulates abscisic acid responses via phosphorylating ABI1/2 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118:e2102544118. doi: 10.1073/pnas.2102544118
Yang, S., Fang, G., Zhang, A., Ruan, B., Jiang, H., Ding, S., et al. (2020). Rice EARLY SENESCENCE 2, encoding an inositol polyphosphate kinase, is involved in leaf senescence. BMC Plant Biol. 20:393. doi: 10.1186/s12870-020-02610-1
Yang, X., Gong, P., Li, K., Huang, F., Cheng, F., and Pan, G. (2016). A single cytosine deletion in the OsPLS1 gene encoding vacuolar-type H+-ATPase subunit A1 leads to premature leaf senescence and seed dormancy in rice. J. Exp. Bot. 67, 2761–2776. doi: 10.1093/jxb/erw109
Yang, Y., Xu, J., Huang, L., Leng, Y., Dai, L., Rao, Y., et al. (2016). PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 67, 1297–1310. doi: 10.1093/jxb/erv529
Zhang, Z., Cui, X., Wang, Y., Wu, J., Gu, X., and Lu, T. (2017). The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol. Plant 10, 86–98. doi: 10.1016/j.molp.2016.08.009
Keywords: transmembrane kinases, water-soaked spot, leaf senescence, chloroplast development, reactive oxygen species, rice
Citation: Xu J, Ji Z, Wang C, Xu F, Wang F, Zheng Y, Tang Y, Wei Z, Zhao T and Zhao K (2022) WATER-SOAKED SPOT1 Controls Chloroplast Development and Leaf Senescence via Regulating Reactive Oxygen Species Homeostasis in Rice. Front. Plant Sci. 13:918673. doi: 10.3389/fpls.2022.918673
Received: 12 April 2022; Accepted: 26 April 2022;
Published: 26 May 2022.
Edited by:
Lei Wang, Institute of Botany (CAS), ChinaReviewed by:
Yasuhito Sakuraba, The University of Tokyo, JapanZhonghai Li, Beijing Forestry University, China
Copyright © 2022 Xu, Ji, Wang, Xu, Wang, Zheng, Tang, Wei, Zhao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaijun Zhao, emhhb2thaWp1bkBjYWFzLmNu; Tianyong Zhao, dHpoYW8yQG53YWZ1LmVkdS5jbg==
†These authors have contributed equally to this work
 Jiangmin Xu
Jiangmin Xu Zhiyuan Ji
Zhiyuan Ji Chunlian Wang
Chunlian Wang Feifei Xu1
Feifei Xu1 Fujun Wang
Fujun Wang Yuhan Zheng
Yuhan Zheng Yongchao Tang
Yongchao Tang Zheng Wei
Zheng Wei Kaijun Zhao
Kaijun Zhao