
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 June 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.918508
This article is part of the Research Topic Genomics and disease resistance in wheat and maize View all 16 articles
 Xiubin Tian1,2
Xiubin Tian1,2 Qifan Chen1,2
Qifan Chen1,2 Chao Ma1,2
Chao Ma1,2 Wenqiang Men1,2
Wenqiang Men1,2 Qianqian Liu1,2
Qianqian Liu1,2 Yue Zhao1,2
Yue Zhao1,2 Jiajun Qian1,2
Jiajun Qian1,2 Ziwei Fan1,2
Ziwei Fan1,2 Jingnan Miao1,2
Jingnan Miao1,2 Jinqiu He1,2
Jinqiu He1,2 Sunish K. Sehgal3
Sunish K. Sehgal3 Huanhuan Li1,2*
Huanhuan Li1,2* Wenxuan Liu1,2*
Wenxuan Liu1,2*Powdery mildew of wheat is a foliar disease that is spread worldwide. Cultivation of resistant varieties is the most effective, economical, and environmentally friendly strategy to curb this disease. Powdery mildew resistance genes (Pm) are the primary resources for resistance breeding, and new Pm genes are in constant demand. Previously, we identified Aegilops longissima chromosome 6Sl#3 as a carrier of powdery mildew resistance and designated the resistance gene as Pm6Sl. Here, we reported the design of 24 markers specific to 6Sl#3 on the basis of the full-length cDNA sequences of 6Sl#3 donor Ae. longissma accession TA1910, and the development of wheat-Ae. longissima 6Sl#3 introgression stocks by ph1b-induced homoeologous recombination. Further, 6Sl#3 introgression lines were identified and characterized by integration analysis of powdery mildew responses, in situ hybridization, and molecular markers and Pm6Sl was mapped to a distal interval of 42.80 Mb between markers Ael58410 and Ael57699 in the long arm of 6Sl#3. Two resistant recombinants, R43 (T6BS.6BL-6Sl#3L) and T27 (Ti6AS.6AL-6Sl#3L-6AL), contained segments harboring Pm6Sl with less than 8% of 6Sl#3 genomic length, and two markers were diagnostic for Pm6Sl. This study broadened powdery mildew resistance gene resources for wheat improvement and provided a fundamental basis for fine mapping and cloning of Pm6Sl to further understand its molecular mechanism of disease resistance.
Bread wheat (Triticum aestivum L., 2n = 6x = 42; AABBDD) is a hexaploid species of the genus Triticum in the grass family Poaceae (or Gramineae). It is the most extensively grown and traded cereal crop with the highest monetary value and the second most-produced cereal after maize. Therefore, the sustainable production of wheat is essential for social progress and stability worldwide. Powdery mildew caused by the fungus Blumeria graminis f. sp. tritici (Bgt) is one of the most damaging foliage diseases of wheat. Severe infection can cause yield loss of 5–30% and decrease grain quality in epidemic years (Fried et al., 1981; Bennett, 1984; Conner et al., 2003; Morgounov et al., 2012). The development and cultivation of resistant varieties is currently the most effective, economical, and environmentally friendly approach to constraining yield and grain quality losses caused by this disease (Wang et al., 2005). As resources for resistance breeding, many powdery mildew resistance genes (Pm) have been identified and introgressed into contemporary wheat varieties from landraces, ancestral species, and other wild relatives. Currently, at least 100 permanently and temporarily designated genes for powdery mildew resistance have been documented (McIntosh et al., 2017; Li et al., 2020; Zhang et al., 2020; He et al., 2021). Fourteen cataloged Pm genes have been cloned, including Pm1, Pm2, Pm3, Pm4b, Pm5e, Pm8, Pm17, Pm21, Pm24, Pm38, Pm40, Pm41, Pm46, and Pm60 (Brunner et al., 2010; Hewitt et al., 2021; Sánchez-Martín et al., 2021; Yang et al., 2021). Only a fraction of the known Pm genes, such as Pm2, Pm4b, Pm8, Pm13, and Pm21, have been widely deployed in wheat production (Yang et al., 2013, 2021; Xing et al., 2018). A large number of Pm genes have been defeated by virulent races, and others could not be deployed in wheat breeding owing to either race-specificity or deleterious gene linkage drag (Klindworth et al., 2013; Tan et al., 2019). Consequently, new Pm genes for continuous improvement of wheat are still in demand.
Cultivated bread wheat has a large genome of 17 giga-base pairs (Gb) comprising three subgenomes, A, B, and D, which were contributed by T. urartu Tumanian ex Gandilyan (2n = 2x = 14, AA), Ae. speltoides Tausch (2n = 2x = 14, SS), and Ae. tauschii Coss. (2n = 2x = 14, DD) (Dubcovsky and Dvorak, 2007; El Baidouri et al., 2017). Wild species of common wheat serve as important gene resources for wheat improvement. Many of these species contain chromosomes that are not homologous to those of wheat, and the genes they bear are difficult to transfer to stable wheat lines. Since those chromosomes are functionally similar or homoeologous to wheat chromosomes, they can often be induced to pair and recombine with wheat chromosomes by suppression of strict bivalent formation, which is controlled by genes Ph1 on chromosome arm 5BL and Ph2 in homoeologous group 3 chromosomes (Riley et al., 1968; Sears, 1977; Qi et al., 2007). Other methods of achieving chromosome transfers include induction of centric fusion of unpaired wheat and foreign chromosomes by double monosomy (breakage and reunion) (Friebe et al., 2005; Liu et al., 2011, 2016) or radiation of seeds or pollen to induce random chromosome breaks and reunions (Sears, 1956; Liu et al., 1999). At least 15 cataloged Pm genes have been transferred into wheat from wild wheat relatives, including Pm13 and Pm66 from Ae. longissima Schweinf. & Muschl. (2n = 2x = 14, SlSl), Pm29 probably from Ae. geniculata Roth. (2n = 4x = 28, UgUgMgMg), Pm57 from Ae. searsii Feldman & Kislev ex Hammer (2n = 2x = 14, SsSs), Pm7, Pm8, Pm17, Pm20, and Pm56 from Secale cereale L. (2n = 2x = 14, RR), Pm21(= Pm31), Pm55, Pm62 and Pm67 from Dasypyrum villosum (L.) P. Candargy (2n = 2x = 14, VV), Pm51 from Thinopyrum elongatum (Host) D. R. Dewey (2n = 2x = 14, EE), and Pm2b from Agropryron cristatum (L.) Gaertn. (2n = 2x = 14, PP). Several of these Pm genes, such as Pm8 on 1RS, and Pm21 on 6VS, have been widely deployed in wheat resistance breeding programs across the world (He et al., 2018; Li et al., 2020; Zhang et al., 2020).
Ae. longissima Schweinf. & Muschl. (2n = 2x = 14, SlSl), an annual grass species native to the eastern Mediterranean basin is one of the five diploid Aegilops species carrying the S, or a modified S genome (Feldman et al., 1995; Friebe et al., 1996). Ae. longissima represents a valuable reservoir of genetic diversity for resistance to stem rust, stripe rust, powdery mildew, Septoria glume blotch, and eyespot (Ceoloni et al., 1992; Sheng et al., 2012; Wang et al., 2018; Xia et al., 2018). However, the potential of Ae. longissima in wheat improvement is far from being fully assessed; only powdery mildew resistance genes Pm13 (3SlS) and Pm66 (4SlS) have been used in wheat breeding programs (Li et al., 2021). Recently, genome sequences of Ae. longissima accession of TL05 (Li et al., 2022) and AEG-6782-2 (Avni et al., 2022) have been published. These genomic resources will accelerate the exploration and utilization of beneficial genes from this species.
Previously, we identified a Chinese Spring (CS)-Ae. longissima 6Sl#3 disomic addition line TA7548 with resistance to wheat powdery mildew (Xia et al., 2018). In this study, we described the development of 6Sl#3 recombinants conferring resistance to powdery mildew and the physical mapping of Pm6Sl in Ae. longissima chromosome 6Sl#3.
The plant material used in this study included common wheat CS, CS ph1b mutant TA3809 lacking pairing homologous gene Ph1, thereby permitting homoeologous recombination, CS-Ae. longissima 6Sl#3 disomic addition line TA7548, where a pair of chromosomes 6Sl#3 from Ae. longissima was added to CS background, 6Sl#3 donor Ae. longissima accession TA1910, CS nulli-tetrasomic stocks N6AT6B (TA3152), N6BT6D (TA3155), and N6DT6A (TA3156), in which the chromosomes 6A, 6B and 6D pairs were respectively replaced by homoeologous pairs 6B, 6D, and 6A. The number following the chromosome designation (6Sl#3) indicated the origin of the alien chromosome derived from different Ae. longissima accessions (Raupp et al., 1995). All materials were provided by the Wheat Genetics Resource Center (WGRC), Kansas State University, USA, and increased in China at the Experimental Station of Henan Agricultural University.
Two populations segregating for 6Sl#3 were developed. Population 1 produced by crossing CS monosomic 6A (CSM6A, 20W″ + 6A′) with TA7548 (21W″ + 6Sl#3″) was used to develop compensating Robertsonian translocations (RobTs) involving wheat chromosome 6A and 6Sl#3. F1 plants with 42 chromosomes and positive for 6Sl#3-specific markers were selected and self-pollinated.
Population 2 used to develop 6Sl#3 recombinants was derived from the hybrid of CS ph1b mutant TA3809 crossed with TA7548. F2 individuals with homozygous ph1b and monosomic 6Sl#3 were selected using the ph1b-specific marker ABC302.3 (Wang et al., 2002) and 6Sl#3-specific markers. Self-pollinated progenies of these plants were used to screen putative 6Sl#3 recombinants.
Individual plants of both populations were screened by a few 6Sl#3-specific molecular markers to select the plants showing marker-disassociation as putative recombinants involving 6Sl#3 and then verified by cytogenetic and molecular marker analyses.
Preparation, sequencing and alignment of an Ae. longissima full-length cDNA library was performed by BGI Genomics (Shenzhen, China) with the Pacific Bioscience Sequel platform (Pacific Biosciences, Silicon Valley, California, USA). Briefly, the second leaves from Ae. longissima TA1910 were collected at 0, 12, 24, and 48 h post-inoculation with Bgt isolate E26 and equally mixed for total RNA extraction using a mirVana miRNA Isolation Kit (Cat. No. AM1561, Ambion, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's protocol. Total RNA was converted to first-strand cDNA using a SMARTer PCR cDNA Synthesis Kit (Cat. No. 634925, Clontech, Takara Biomedical Technology (Beijing) Co., Ltd., Beijing). After PCR optimization, a large-scale PCR was performed to synthesize second-strand cDNA. After another large-scale PCR, the DNA was ready for library template preparation by PacBio SMRTbell; selected fragments > 4 kb were sequenced by Pacific Biosciences Sequel. Sequencing data were processed by Single-Molecule, Real-Time (SMRT) sequencing analysis through reads of insert, classify, and cluster to obtain consensus full-length isoforms. Isoforms that could not be aligned to any database were predicted by TransDecoder version v3.0.1 (https://transdecoder.github.io).
Fifty-three markers were used in this study, of which 24 were 6Sl#3-specific markers developed from full-length cDNA sequences of TA1910, and the other 29 simple sequence repeat (SSR) markers specific for wheat chromosomes 6A (15), 6B (8), and 6D (6) were obtained from GrainGenes. SSR marker details are listed in Supplementary Table S1. The genomic positions of the markers of 6Sl#3-specificity were ordered using blastn alignment against both the CS reference genomic sequence (Wheat_IWGSC_RefSeq_v2.1) (Zhu et al., 2021) and Ae. longissima accession TL05 reference sequences (Li et al., 2022).
Genomic DNA (gDNA) was isolated from 5 to 10 cm segments of young leaves with a DNeasy Plant Mini Kit (Qiagen, Cat No. 69104) following the instructions. PCR reactions were conducted in a 15-μl volume containing 2.0 μl template gDNA (100 ng/μl), 1.0 μl of each primer (5.0 μmol/l), 7.5 μl Taq MasterMix (CW Bio Inc., China) and 3.5 μl ddH2O. Using 6Sl#3-specific primers, PCR amplifications were performed by Touchdown63 and by F50SSR using SSR markers (Liu et al., 2017). PCR products of 6Sl#3-specific markers and SSR markers were resolved in 1.5% and 3.0% agarose gels and visualized by ethidium bromide staining under UV light.
Collection and nitrous oxide treatment of root tips, squash preparations, and genomic in situ hybridization (GISH) were performed following protocols described by Liu et al. (2017). Ae. longissima gDNA was labeled with fluorescein-12-dUTP. Common wheat CS gDNA was used as blocking. The ratio of gDNA of Ae. longissima to CS was 1:120. Fluorescence in situ hybridization (FISH) was performed with eight oligonucleotide probes, of which six were 6-carboxytetramethylrhodamine (TAMRA)-modified oligonucleotides (pAs1-1, pAs1-3, pAs1-4, pAs1-6, AFA-3, and AFA-4) and displayed red signals, and the other two were 6-carboxyfluorescein (FAM)-modified oligonucleotides (pSc119.2-1 and (GAA)10), which fluoresced green (Du et al., 2017; Huang et al., 2018). After hybridization and slide washing, a drop (25–30 μl) of Vectashield mounting medium containing 1 μg/ml DAPI (Vector Laboratories Inc, Burlingame, CA, USA) was added to each slide and then covered with a 24 × 30 cm glass coverslip. Fluorescent images were observed under a Zeiss Axio Scope A1 fluorescence microscope (Jena, Germany) and captured with an AxioCam MRc5 CCD camera. Images were further processed with Adobe Photoshop CS3 (Version 10.0.1) (Adobe Systems Inc., San Jose, CA, USA).
Thirty single-pustule-derived Bgt isolates were collected from different regions in China and used to test TA7548. Isolate E26, kindly provided by the Nanjing Agricultural University and locally maintained, was used to test the 6Sl#3 derivatives. The remaining 29 isolates were maintained at the University of Yantai, Shandong Province. The preparation of plant materials and Bgt inoculation followed protocols was described by Li et al. (2020). Infection types were recorded on a 0–4 scale at 10 days post inoculation, while conidia were fully developed on the first leaves of susceptible control CS. Plants with ITs of 0–2 were considered to be resistant, whereas those with ITs of 3–4 were susceptible (Shi et al., 1987).
Thirty Bgt isolates were used to test the Bgt response of TA7548 together with CS as a susceptible control when the first leaves were fully expanded. Seedling reactions 10 days post-inoculation indicated that 28 of 30 Bgt isolates were avirulent on TA7548, with ITs ranging from 0 to 2, and the other two isolates were virulent on TA7548 with ITs 3 and 4. CS, the susceptible control, was highly susceptible to all Bgt-isolates, with ITs 3-4 (Table 1). Thus, the gene(s) on chromosome 6Sl#3, designated as Pm6Sl, confers resistance to multiple Bgt-isolates.
Sequenced by the Pacific Bioscience Sequel platform generated 2,136,574,821 bp CCS (reads of insert) data (1,161,885 reads), of which, 685,686 (59.01%) reads were full-length non-chimeric, with a mean length of 1,494.7 bp. After clustering and polishing the consensus with Quiver and removing redundancies, a total of 69,907 high-quality consensus isoforms were identified, covering a total length of 113,954,143 bp, with a mean length of 1,630 bp and N50 of 1,920 bp. The isoforms were further analyzed using TransDecoder software to identify candidate coding regions. The longest open reading frame (ORF) was then selected and blasted to the SwissProt and Hmmscan databases to search for Pfam protein homology sequences for CDS prediction, leading to the prediction of 62,761 (71,013,186 bp) CDS with lengths ranging from 297 to 5,913 bp were predicted.
By blasting the full-length sequences of 62,761 predicted CDS of Ae. longissima to CS reference genomic sequences (IWGSC RefSeq v2.1, IWGSC, 2021), a total of 8,461 (13.48%) CDS were uniquely assigned to homoeologous group 6. One hundred and thirty-four unique CDS with similarities of 80–90% to wheat group 6 genomic sequences were selected for further PCR primer design. Based on a comparison of these CDS sequences with those of CS, 134 PCR primer pairs were designed. The comparison of amplification of CS and CS-Ae. longissima 6Sl#3 disomic addition line TA7548 led to the identification of 24 (17.91%) markers specific to chromosome 6Sl#3 (Table 2).
Four markers mapped to each arm of 6Sl#3 were used to select putative RobTs involving 6Sl#3 in Population 1. Markers Ael69501 and Ael42958 were distally located at opposite ends of 6Sl#3, whereas Ael67319 and Ael63185 occupied intermediate positions in the short and long arms. Plants lacking either the short arm- or long arm-specific marker(s) were selected as putative 6Sl#3 RobTs. From 608 plants in population 1, 17 (2.80%) showing disassociations of markers were selected as putative RobTs or having telosomes (Telos) for either 6Sl#3S or 6Sl#3L. Seven plants lacked long arm markers and 10 lacked short arm markers (Figure 1). GISH and FISH analyses of the seven plants lacking the long arm markers identified a compensating RobT T6Sl#3S.6AL (1S71), whereas the remaining plants were monotelosomic 6Sl#3S (Figure 2). The 10 plants with only long arm markers were all monotelosomic 6Sl#3L (represented by T42) (Figure 2). A powdery mildew test using Bgt-isolate E26 showed that the RobT T6Sl#3S.6AL and the six monotelosomic 6Sl#3S were susceptible, whereas all 10 monotelosomic 6Sl#3L were resistant with IT of 0 (Figure 3). Thus, Pm6Sl was located in the long arm of chromosome 6Sl#3.
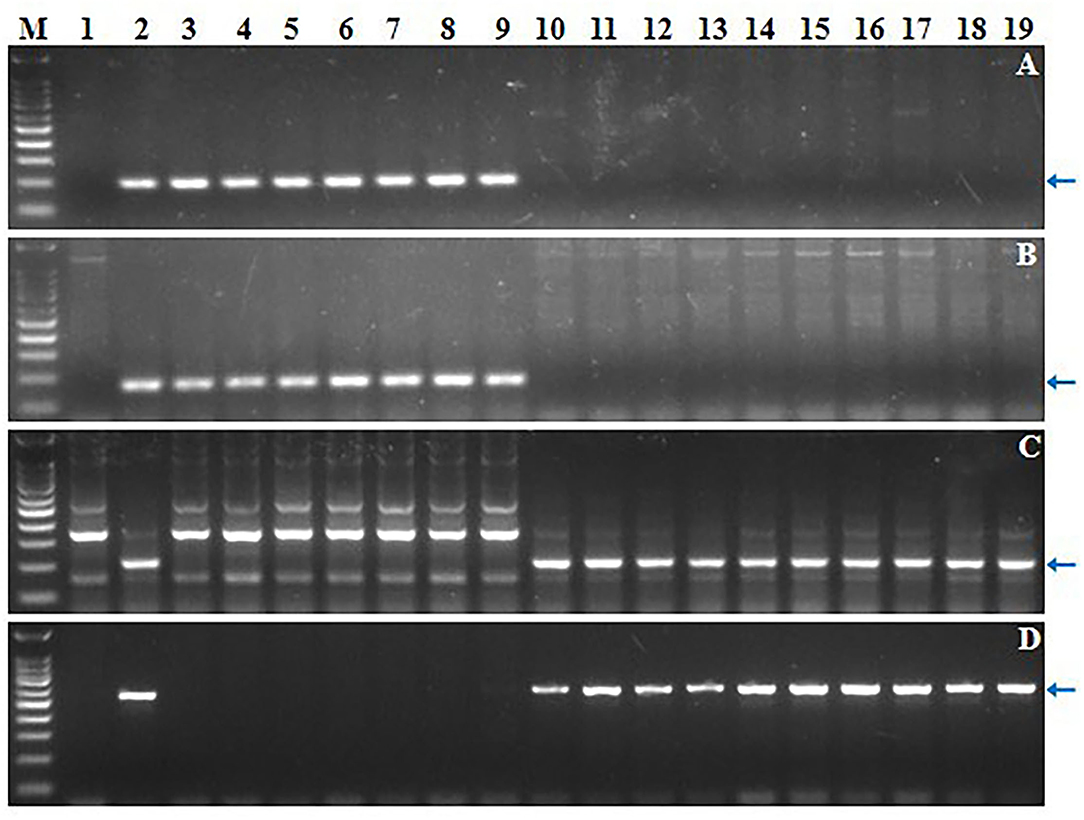
Figure 1. Screening of 17 CS-Ae. longissima 6Sl#3 RobTs by molecular marker analysis. (A–D) Electrophoresis patterns of 6Sl#3-specific markers Ael69501, Ael67319, Ael63185 and Ael42958, respectively. M: 100 bp DNA Ladder Marker; 1: common wheat CS; 2: CS-Ae. longissima 6Sl#3 disomic addition line TA7548; 3-19: the newly developed CS-Ae. longissima 6Sl#3 RobTs and telosomes. Arrows pointed to the polymorphic bands of the chromosome 6Sl#3-specific molecular markers.
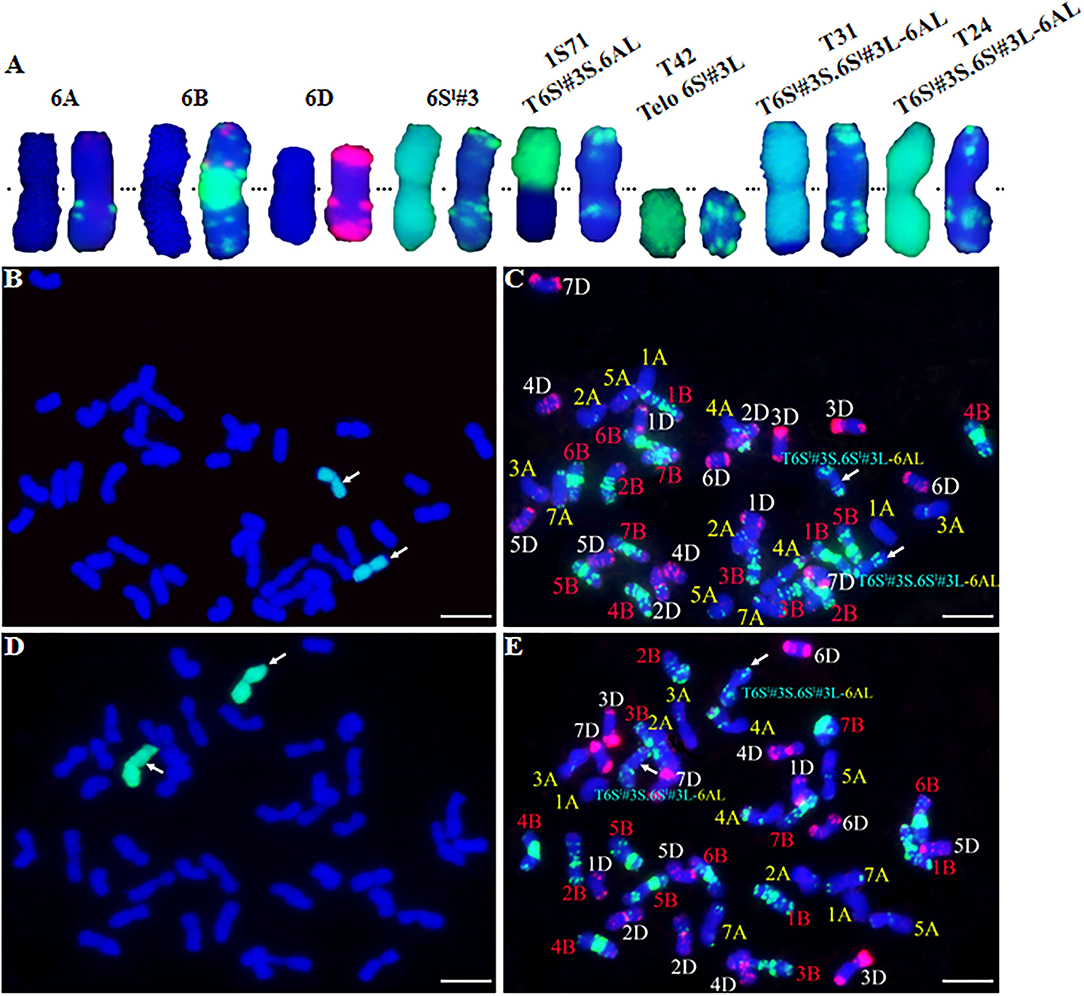
Figure 2. GISH and FISH analysis of CS-Ae. longissima 6Sl#3 introgression lines. (A) From left to right, chromosome pairs of 6A, 6B, 6D, 6Sl#3, T6Sl#3S.6AL (1S71), 6Sl#3L telosome (T42), T6Sl#3S.6Sl#3L-6AL (T31), and T6Sl#3S.6Sl#3L-6AL (T24). The left and right chromosome of each pair is GISH and FISH, respectively. (B–E) GISH and FISH patterns of T31 (B,C) and T24 (D,E). Arrows indicated the recombined chromosomes involving 6Sl#3. Ae. longissima chromatin is visualized by green florescence and wheat chromosomes are counterstained with DAPI and fluoresced blue in GISH. In FISH, TAMRA-modified oligonucleoties (pAs1-1, pAs1-3, pAs1-4, pAs1-6, AFA-3 and AFA-4) are in red color, FAM-modified pSc119.2-1 and (GAA)10 are in green color. Scale bar = 10 μm.

Figure 3. Powdery mildew resistance evaluation of 6Sl#3 recombinants. 1S71: CS-Ae. longissima RobT T6Sl#3S.6AL; T42: 6Sl#3L telosome line; T31: CS-Ae. longissima T6Sl#3S.6Sl#3L-6AL recombinant; R43: CS-Ae. longissima T6BS.6BL-6Sl#3L recombinant; T27: CS-Ae. longissima Ti6AS.6AL-6Sl#3L-6AL recombinant; T24: CS-Ae. longissima T6Sl#3S.6Sl#3L-6AL recombinant. R: resistant to powdery mildew, S: susceptible to powdery mildew.
In order to develop CS-Ae. longissima recombinants with breakpoints at 6Sl#3L where Pm6Sl resides, four markers specific for 6Sl#3L were used to select putative recombinants in Population 2. Two markers (Ael65131 and Ael56039) were near the centromere of 6Sl#3L, whereas the other two, Ael58410 and Ael42958, were located at the distal region of 6Sl#3L. Fourteen plants (1.76%) displayed disassociations with the four markers and were selected as putative 6Sl#3L recombinants. These putative 6Sl#3L recombinants were further characterized using 24 6Sl#3-specific markers. Based on the presence of different markers, the 14 plants were grouped into four types: T31, T24, R43, and T27 (Figure 4).
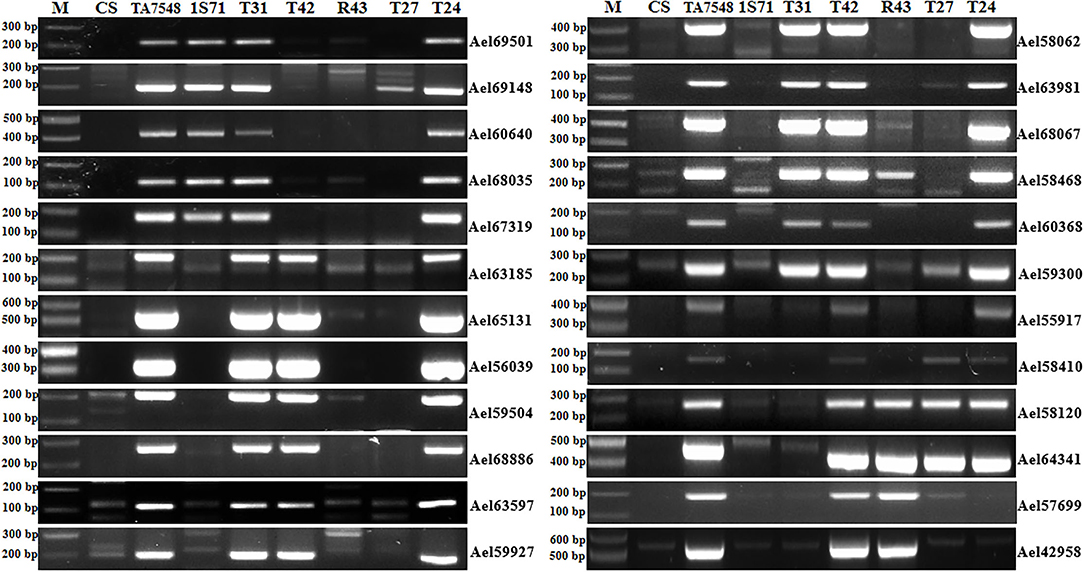
Figure 4. PCR patterns of 6Sl#3 recombinants using 6Sl#3-specific markers. M: 100 bp DNA ladder marker.
T31 type lost six proximal and distal markers of 6Sl#3L (Ael55917, Ael58410, Ael58120, Ael64341, Ael57699, and Ael42958), which covered a distal segment of <81.77 Mb. T24 plants had all 6Sl#3-specific markers except two-terminal markers (Ael57699 and Ael42958), both at 659.64 Mb. The R43 type presented only four distal markers (Ael58120, Ael64341, Ael57699, and Ael42958) of 6Sl#3L covering a segment of <45.37 Mb. T27 was positive for only three distal markers of 6Sl#3L covering a segment of <51.40 Mb; however, both terminal markers of 6Sl#3L (Ael57699 and Ael42958) at 659.64 Mb were absent (Figure 4). Chromosome 6Sl segments in both recombinants R43 and T27 were < 8% of 6Sl genomic length.
GISH and FISH were performed to identify chromosome recombinants involving 6Sl#3L. GISH showed that type T31 contained a recombined 6Sl#3 chromosome with a terminal wheat segment consistent with 6AL (Figures 2B,C). Therefore, the T31 recombinants were identified as T6Sl#3S.6Sl#3L-6AL. Neither GISH nor FISH identified a wheat segment replacing distal 6Sl#3L in the T24 type despite positive indications from marker analysis (Figures 2D,E). The results indicated that those plants had a small distal segment of wheat replacing the 6Sl#3L counterpart. GISH of both R43 and T27 types failed to detect the green color for Ae. longissima chromatin, and FISH also failed to detect a difference from CS. This confirmed that any 6Sl#3 segments in these lines should be small and beyond the resolving power of these methods.
The combined GISH, FISH patterns, and 6Sl#3-specific marker analyses indicated that all putative recombinants contained 6Sl#3L segments of various sizes, whereas the identities of the wheat segments in the other groups except T31 were unresolved. SSR markers were used to further characterize those 6Sl#3L recombinants. Twenty-nine SSR markers specific to chromosomes 6A (15), 6B (8) and 6D (6) were selected to perform PCR amplification (Supplementary Table S1). Compared with CS and TA7548, different PCR products were amplified from T31, T27, and T24 for 6A-SSRs, and R43 for 6B-SSRs.
Analyses of 15 6A-specific SSR markers displayed that recombinants of type T31 lacked all 6A-specific SSR markers except seven distal markers in chromosome bin 6AL8-0.90-1.00 (Figure 5; Supplementary Table S2), indicating that T31 lines had recombined chromosomes containing only small segments of 6AL, thus designated as T6Sl#3S.6Sl#3L-6AL (Supplementary Figure S1). Type T24 presented SSR patterns similar to those of T31 and was also designated as T6Sl#3S.6Sl#3L-6AL, but the wheat segment was smaller with just distal SSR marker Xmwg2053 (Figure 5; Supplementary Table S2). T27 recombinants lacked six distal markers in chromosome bin 6AL8-0.90-1.00, whereas they retained two proximal markers (Xwmc201 and Xgwm570) (Figure 5; Supplementary Table S2). By combining 6Sl#3 specific marker and SSR markers analyses, T27 should be an interstitial recombinant formed by proximal 6Sl#3L segments replaced 6AL counterparts, thus designated as Ti6AS.6AL-6Sl#3L-6AL (Supplementary Figure S1).
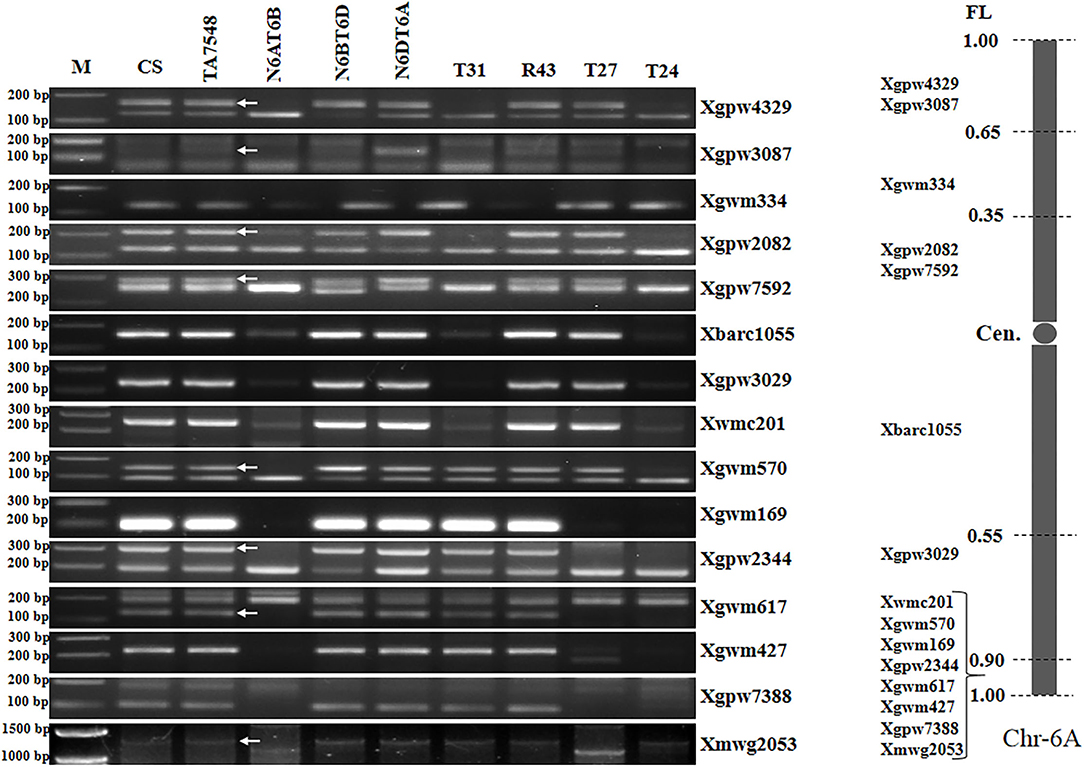
Figure 5. Molecular markers analysis of 6Sl#3 recombinants using wheat 6A SSR markers. Arrows pointed to 6A-specific bands when two or more bands were amplified using a single primer pair. M: 100 bp DNA ladder marker.
PCR patterns of eight 6B-specific SSR markers displayed that type R43 recombinants were negative for two of five markers (Xgwm219 and Xwmc417) in chromosome bin 6BL5-0.40-1.00 but positive for the remaining six markers (Figure 6; Supplementary Table S2). We concluded that the recombined chromosomes in R43 were formed by the 6Sl#3L distal segments substituting for distal counterparts of 6BL. Thus, R43 was designated T6BS.6BL-6Sl#3L (Supplementary Figure S1).
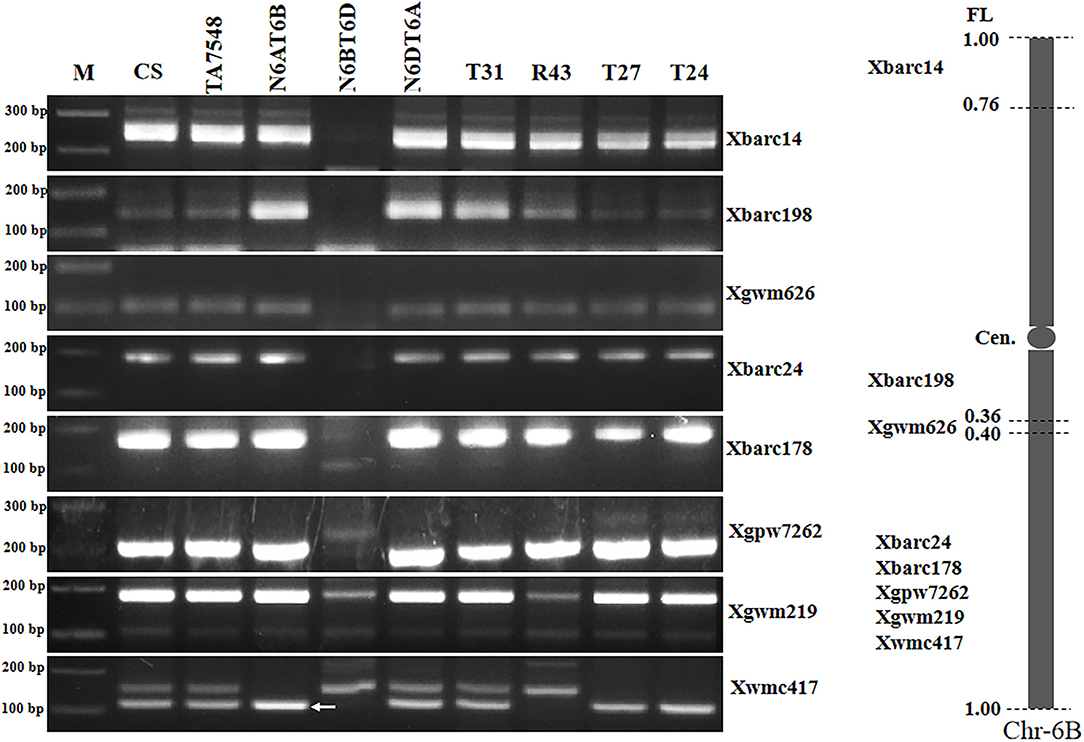
Figure 6. Molecular markers analysis of 6Sl#3 recombinants using wheat 6B SSR markers. Arrows pointed to 6B-specific bands when two or more bands were amplified using a single primer pair. M: 100 bp DNA ladder marker.
The 6Sl#3L recombinants, together with susceptible control CS and resistant control TA7548, were assayed by inoculation of the Bgt-isolate E26. Infection types at 10 days post-inoculation indicated that recombinants of T24, R43, T27, and TA7548 were resistant (IT 0), whereas T31 plants and CS were susceptible (IT 4) (Figure 3). By integration of the Bgt-responsive assay and molecular marker analyses, Pm6Sl was physically mapped to a distal interval between markers Ael58410 and Ael42958 in the long arm of 6Sl#3, which spanned 42.8 Mb in Ae. longissima TL05 reference sequence (Figure 7).
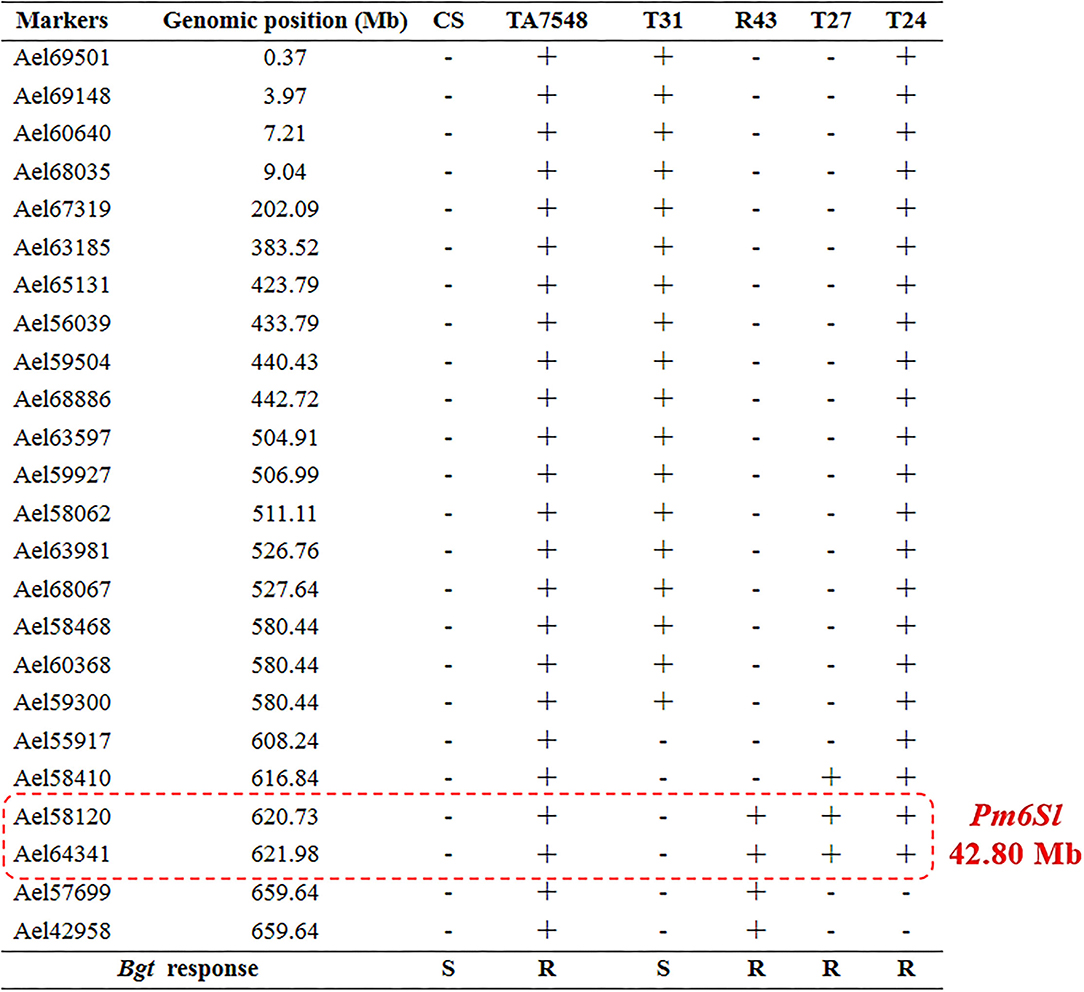
Figure 7. Physical mapping of gene Pm6Sl. “+” indicated the presence of 6Sl#3-specific molecular markers, “–” indicated the absence of 6Sl#3-specific molecular marker. R indicated resistance to powdery mildew, S indicated susceptible to powdery mildew.
Two markers developed in this study, Ael58120 and Ael64341, were close to Pm6Sl. PCR amplification was performed to determine whether they were diagnostic with four Pm6Sl stocks, including CS-Ae. longissima 6Sl#3 disomic addition line TA7548, recombinant R43, T24, and T27, and 23 wheat lines. The target bands were amplified from Pm6Sl stocks but were absent in those wheat varieties not containing Pm6Sl (Figure 8). Thus, both PCR markers can be used to assist the selection of Pm6Sl plants in the hybrid progeny of these Pm6Sl stocks in breeding programs.
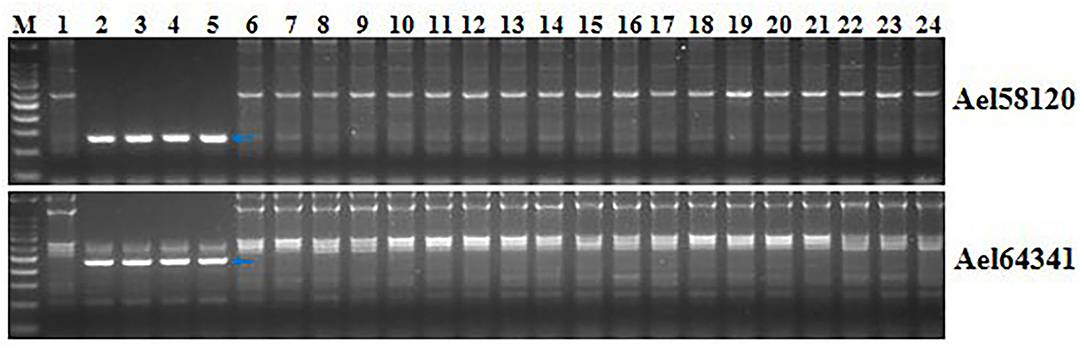
Figure 8. Validation of the usefulness of two markers Ael58120 and Ael64341 closely linked to Pm6Sl. M: 100 bp DNA ladder; 1: CS; 2: CS-Ae. longissima 6Sl#3 disomic addition line TA7548; 3-5: 6Sl#3 recombinants R43, T24 and T27 with resistance to powdery mildew; 6: Bainong 207; 7: Bainong 64; 8: Aikang 58; 9: Zhoumai 16; 10: Zhoumai 18; 11: Zhoumai 22; 12: Zhoumai 28; 13: Pingan 0518; 14: Pingan 602; 15: Pingan 901; 16: Jimai 22; 17: Yanzhan 4110; 18: Shengxuan 6; 19: Yangmai 5; 20: Nannong 9918; 21: Linxuan 101; 22: Hua 5; 23: Ningmai 3; 24: Xinong 979. Arrows pointed to the polymorphic bands of the respective molecular markers linked to Pm6Sl.
in situ hybridization (GISH and FISH) provides a highly efficient approach to resolving alien chromosome segments in wheat (Li et al., 2019). The combined application of both techniques can display the size of both donor and recipient wheat chromosomes and the number and position of breakage points. However, the resolution of GISH is about 25 Mb (Mukai et al., 1993). Similarly, FISH can also fail to resolve chromosomes or segments unless there are presented abundant signal bands. In contrast, transcriptome sequencing can quickly and economically yield numerous sequences to facilitate the rapid and efficient development of molecular markers covering all the genomes of wild wheat relatives. Combined GISH/FISH with chromosome-specific markers has been widely used to accelerate the detection of alien chromatin harboring useful genes in wheat introgression and breeding programs (Wang et al., 2018; Zhang et al., 2019; Li et al., 2020). Ae. longissima belongs to the section Sitopsis in the genus Aegilops. The S or modified S genomes of the section are genetically related to the wheat B and D genomes, thus resolving small segments of Ae. longissima chromatins introgressed into wheat by GISH is challenging. In this study, we successfully identified RobT lines with centric fusion T6Sl#3S.6AL (1S71), telosomes 6Sl#3S and 6Sl#3L, and recombinant T6Sl#3S.6Sl#3L-6AL (T31) by combining GISH and FISH, whereas other recombinants were unresolved. By integrating both 6Sl#3-specific and wheat SSR marker analyses, we identified recombinants T27 (Ti6AS.6AL-6Sl#3L-6AL), T24 (T6Sl#3S.6Sl#3L-6AL), and R43 (T6BS.6BL-6Sl#3L).
Mapping and cloning of genes from wild species still face tough challenges due to Ph gene-suppressed recombination between alien chromosomes and their wheat homoeologous counterparts, lack of reference genomic sequences for those wild species, and insufficient markers specific to alien chromosomes. In this study, we constructed a full-length cDNA sequence database of Ae. longissima accession TA1910, and designed PCR primers based on isoform sequence comparisons with CS reference genomic sequences. About 18% of molecular markers designed in the study gave polymorphic bands to distinguish Ae. longissima 6Sl#3 chromatin from wheat chromatin. This was much more efficient than the EST-PCR, SSR, and mapped-flcDNA-based marker development that we reported previously (Liu et al., 2016). By marker analysis and tests of powdery mildew response, Pm6Sl was located at the distal interval of 42.80 Mb flanked by markers Ael58410 and Ael57699 in the long arm of 6Sl#3. The resistant recombinants with small 6Sl#3 segments and the full-length cDNA sequence database of TA1910 developed in this study will help future fine mapping and cloning of Pm6Sl.
The transfer of favorable genes from wild relatives of common wheat is an effective approach to broadening the genetic base of modern wheat. Homoeologous recombination-based transfer is currently the best way to introgress desirable genes from wild species to common wheat. However, homoeologous recombination between alien chromatin and wheat homoeologous counterparts was restrained in the presence of Ph genes (Gyawali et al., 2019). The deletion mutant (ph1b) of the pairing homologous gene Ph1 at 5BL is mostly deployed to promote homoeologous recombination between wild species and wheat. A lot of alien genes conferring disease and pest resistance have been transferred into wheat and mapped by ph1b-induced homoeologous recombination (Dong et al., 2020; Wan et al., 2020). In this study, 24 CS-Ae. longissima 6Sl#3 recombinants were developed based on ph1b-induced homoeologous recombination. By integration of cytogenetic analyses and powdery mildew resistance evaluation, the novel powdery mildew resistance gene Pm6Sl was mapped to the distal interval of 42.8 Mb in the long arm of 6Sl#3.
Ae. longissima has been reported to contain diverse biotic stress resistance genes. However, currently, very few genes from this species are deployed in wheat breeding programs, except Pm13 (Ceoloni et al., 1992). In the present study, we developed two 6Sl#3 recombinants, T27 (Ti6AS.6AL-6Sl#3L-6AL) and R43 (T6BS.6BL-6Sl#3L) by ph1b-induced homoeologous recombination. Both lines conferred broad-spectrum resistance to powdery mildew and harbored Pm6Sl in an Ae. longissima segment of <8% of the 6Sl genomic length. These resistant recombinants with tiny 6Sl segments will decrease the linkage drags and be potentially useful in wheat disease resistance breeding programs.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
WL and HL conceived the research and wrote the paper. HL and SS performed in situ hybridization. XT, CM, and WM conducted full-length cDNA data analysis and molecular marker development. YZ, QC, and JM conducted molecular marker analysis. WM evaluated responses to Bgt isolates. JH, JQ, and ZF conducted SSR marker analyses. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (Nos. 31971887 and 31801361), the Scientific and Technological Research Project of Henan Province of China (No. 212102110059), and the Topnotch Talents of Henan Agricultural University (30500939).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are thankful to Prof. Aizhong Cao from the College of Agronomy, Nanjing Agricultural University, China, for kindly providing Bgt isolate E26.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.918508/full#supplementary-material
Supplementary Figure S1. Characterization of 6Sl#3 recombinants by wheat SSR markers and 6Sl#3-specific markers. Wheat 6A chromatins were represented by red column, 6B by purple and 6Sl#3 by green. Wheat 6A and 6B SSR markers were in red and purple italic, respectively, ordered by locating chromosome bin and primer-aligned CS reference genomic sequences in the same bin; 6Sl#3-specific markers in green bold, ordered by derived full-length cDNA sequence-aligned Ae. longissima TL05 reference genomic sequences. Bold black numbers on the right of 6A and 6B columns represented chromosome fragment length (FL). Based on analyses of SSR markers and 6Sl#3-specific markers, T31 was designated as T6Sl#3S.6Sl#3L-6AL, R43 as T6BS.6BL-6Sl#3L, T24 as T6Sl#3S.6Sl#3L-6AL, and T27 as Ti6AS.6AL-6Sl#3L-6AL.
Supplementary Table S1. SSR primers used in this study.
Supplementary Table S2. Characterization of 6Sl#3 recombinants using wheat SSR markers.
Avni, R., Lux, T., Minz-Dub, A., Millet, E., Sela, H., Distelfeld, A., et al. (2022). Genome sequences of three Aegilops species of the section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J. 110, 179–192. doi: 10.1111/tpj.15664
Bennett, F. (1984). Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programs. Plant Pathol. 33, 279–300. doi: 10.1111/j.1365-3059.1984.tb01324.x
Brunner, S., Hurni, S., Streckeisen, P., Mayr, G., Albrecht, M., Yahiaoui, N., et al. (2010). Intragenic allele pyramiding combines different specificities of wheat Pm3 resistance alleles. Plant J. 64, 433–445. doi: 10.1111/j.1365-313X.2010.04342.x
Ceoloni, C., Signore, G., Ercoll, L., and Donini, P. (1992). Locating the alien chromatin segment in common wheat-Aegilops longissima mildew resistant transfers. Hereditas 116, 239–245. doi: 10.1111/j.1601-5223.1992.tb00830.x
Conner, R. L., Kuzyk, A. D., and Su, H. (2003). Impact of powdery mildew on the yield of soft white spring wheat cultivars. Can. J. Plant Sci. 83, 725–728. doi: 10.4141/P03-043
Dong, Z. J., Tian, X. B., Ma, C., Xia, Q., Wang, B. L., Chen, Q. F., et al. (2020). Physical mapping of Pm57, a powdery mildew resistance gene derived from Aegilops searsii. Int. J. Mol. Sci. 21, 322. doi: 10.3390/ijms21010322
Du, P., Zhuang, L. F., Wang, Y., Yuan, L., Wang, Q., Wang, D. R., et al. (2017). Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 60, 93–103. doi: 10.1139/gen-2016-0095
Dubcovsky, J., and Dvorak, J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 318, 393. doi: 10.1126/science.1143986
El Baidouri, M., Murat, F., Veyssiere, M., Molinier, M., Flores, R., Burlot, L., et al. (2017). Reconciling the evolutionary origin of bread wheat (Triticum aestivum). New Phytol. 213, 1477–1486. doi: 10.1111/nph.14113
Feldman, M., Lupton, F., and Miller, T. (1995). “Wheats,” in Evolution of Crop Plants, 2nd Edn. Eds J. Smartt and N. W. Simmonds (London: Longman Scientific), 184–192.
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., and Gill, B. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pest: current status. Euphytica 91, 59–87. doi: 10.1007/BF00035277
Friebe, B., Zhang, P., Linc, G., and Gill, B. S. (2005). Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenet. Genome Res. 109, 293–297. doi: 10.1159/000082412
Fried, P., Mackenzie, D., and Nelson, R. (1981). Yield loss caused by Erysiphe graminis f. sp. tritici on single culms of “Chancellor” wheat and four multilines. J. Plant. Dis. Prot. 88, 256–264.
Gyawali, Y., Zhang, W., Chao, S., Xu, S., and Cai, X. W. (2019). Delimitation of wheat ph1b deletion and development of ph1b-specific DNA markers. Theor. Appl. Genet. 132, 195–204. doi: 10.1007/s00122-018-3207-2
He, H. G., Liu, R. K., Ma, P. T., Du, H. N., Zhang, H. H., Wu, Q. H., et al. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
He, H. G., Zhu, S. Y., Zhao, R. H., Jiang, Z. N., Ji, Y. Y., Ji, J., et al. (2018). Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 11, 879–882. doi: 10.1016/j.molp.2018.03.004
Hewitt, T., Muller, M. C., Molnar, I., Mascher, M., Holušová, K., Šimková, H., et al. (2021). A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 229, 2812–2826. doi: 10.1111/nph.17075
Huang, X. Y., Zhu, M. Q., Zhuang, L. F., Zhang, S. Y., Wang, J. J., Chen, X. J., et al. (2018). Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor. Appl. Genet. 131, 1967–1986. doi: 10.1007/s00122-018-3126-2
Klindworth, D. L., Hareland, G. A., Elias, E. M., and Xu, S. S. (2013). Attempted compensation for linkage drag affecting agronomic characteristics of durum wheat 1AS/1DL translocation lines. Crop Sci. 53, 422–429. doi: 10.2135/cropsci2012.05.0310
Li, H. H., Dong, Z. J., Ma, C., Tian, X. B., Qi, Z. J., Wu, N., et al. (2019). Physical mapping of stem rust resistance gene Sr52 from Dasypyrum villosum based on ph1b-induced homoeologous recombination. Int. J. Mol. Sci. 20:4887. doi: 10.3390/ijms20194887
Li, H. H., Dong, Z. J., Ma, C., Xia, Q., Tian, X. B., Sehgal, S., et al. (2020). A spontaneous wheat-Aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor. Appl. Genet. 133, 1149–1159. doi: 10.1007/s00122-020-03538-8
Li, H. H., Tian, X. B., Pei, S. L., Men, W. Q., Ma, C., Sehgal, S., et al. (2021). Development of novel wheat-Aegilops longissima 3Sl translocations conferring powdery mildew resistance and specific molecular markers for chromosome 3Sl. Plant Dis. 105, 2938–2945. doi: 10.1094/PDIS-12-20-2691-RE
Li, L. F., Zhang, Z. B., Wang, Z. H., Li, N., Sha, Y., Wang, X. F., et al. (2022). Genome sequences of five Sitopsis species of Aegilops and the origin of polyploid wheat B subgenome. Mol. Plant. 15, 488–503. doi: 10.1016/j.molp.2021.12.019
Liu, W. X., Chen, P. D., and Liu, D. J. (1999). Development of Triticum aestivum-Leymus racemosus translocation lines by irradiating adult plants at meiosis. Acta. Bot. Sin. 41, 463–467.
Liu, W. X., Jin, Y., Rouse, M., Friebe, B., Gill, B. S., and Pumphery, M. O. (2011). Development and characterization of wheat-Ae. searsii Robertsonian translocations and a recombinant chromosome conferring resistance to stem rust. Theor. Appl. Genet. 122, 1537–1545. doi: 10.1007/s00122-011-1553-4
Liu, W. X., Koo, D. H., Friebe, B., and Gill, B. S. (2016). A set of Triticum aestivum-Aegilops speltoides Robertsonian translocation lines. Theor. Appl. Genet. 129, 2359–2368. doi: 10.1007/s00122-016-2774-3
Liu, W. X., Koo, D. H., Xia, Q., Li, C. X., Bai, F. Q., Song, Y. L., et al. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 130, 841–848. doi: 10.1007/s00122-017-2855-y
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C., and Xia, X. C. (2017). Catalogue of gene symbols for wheat. 2017. Supplement. Annu. Wheat. Newsl. 53, 107–128.
Morgounov, A., Tufan, H. A., Sharma, R., Akin, B., Bagci, A., Braun, H. J., et al. (2012). Global incidence of wheat rusts and powdery mildew during 1969-2010 and durability of resistance of winter wheat variety Bezostaya 1. Eur. J. Plant. Pathol.132, 323–340. doi: 10.1007/s10658-011-9879-y
Mukai, Y., Friebe, B., Hatchet, J. H., Yamamoto, M., and Gill, B. S. (1993). Molecular cytogentic analysis of radiation-induced wheat-rye terminal and intercalary chromosomal translocations and the detection of rye chromatin specifying resistance to Hessian fly. Chromosoma 102, 88–95. doi: 10.1007/BF00356025
Qi, L. L., Friebe, B., Zhang, P., and Gill, B. S. (2007). Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res. 15, 3–19. doi: 10.1007/s10577-006-1108-8
Raupp, W. J., Friebe, B., and Gill, B. S. (1995). Suggested guidelines for the nomenclature and abbreviation of the genetic stocks of wheat, Triticum aestivum L. em Thell, and its relatives. Wheat. Inf. Serv. 81, 51–55.
Riley, R., Chapman, V., and Johnson, R. (1968). Introduction of yellow rust resistance of Aegilops comosa into wheat by genetically induced homoeologous recombination. Nature 217, 383–384. doi: 10.1038/217383a0
Sánchez-Martín, J., Widrig, V., Herren, G., Wicker, T., Zbinden, H., Gronnier, J., et al. (2021). Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants. 7, 327–341. doi: 10.1038/s41477-021-00869-2
Sears, E. (1956). “The transfer of leaf rust resistance from Aegilops umbellulata to wheat, gentics in plant breeding,” in Brook-haven Symposia in Biology (Upton, NY: Brookhaven National Laboratory), 1–22.
Sears, E. (1977). An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19, 585–593. doi: 10.1139/g77-063
Sheng, H. Y., See, D. R., and Murray, T. D. (2012). Mapping QTL for resistance to eyespot of wheat in Aegilops longissima. Theor. Appl. Genet. 125, 355–366. doi: 10.1007/s00122-012-1838-2
Shi, Q. M., Zhang, X. X., Duan, X. Y., and Sheng, B. Q. (1987). Identification of isolates of Blumeria graminis f. sp. tritici. Sci. Agric. Sin. 20, 64–70.
Tan, C. C., Li, G. Q., Cowger, C., Carver, B. F., and Xu, X. Y. (2019). Characterization of Pm63, a powdery mildew resistance gene in Iranian landrace PI 628024. Theor. Appl. Genet. 132, 1137–1144. doi: 10.1007/s00122-018-3265-5
Wan, W. T., Xiao, J., Li, M. L., Tang, X., Wen, M. X., Cheruiyot, A. K., et al. (2020). Fine mapping of wheat powdery mildew resistance gene Pm6 using 2B/2G homoeologous recombinants induced by the ph1b mutant. Theor. Appl. Genet. 133, 1265–1275. doi: 10.1007/s00122-020-03546-8
Wang, K. Y., Lin, Z. S., Wang, L., Wang, K., Shi, Q. H., Du, L. P., et al. (2018). Development of a set of PCR markers specific to Aegilops longissima chromosome arms and application in breeding a translocation line. Theor. Appl. Genet. 131, 13–25. doi: 10.1007/s00122-017-2982-5
Wang, X. W., Lai, J. R., Liu, G. T., and Chen, F. (2002). Development of a Scar marker for the Ph1 locus in common wheat and its application. Crop Sci. 42, 1365–1368. doi: 10.2135/cropsci2002.1365
Wang, Z. L., Li, L. H., He, Z. H., Duan, X. Y., Zhou, Y. L., Chen, X. M., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463. doi: 10.1094/PD-89-0457
Xia, Q., Mai, Y. N., Dong, Z. J., and Liu, W. X. (2018). Identification of powdery mildew resistance resources from wheat-wild relative disomic addition lines and development of molecular markers of alien chromosome-specialty. J. Henan Agric. Sci. 47, 64–69. doi: 10.15933/j.cnki.1004-3268.2018.06.012
Xing, L. P., Hu, P., Liu, J. Q., Witek, K., Zhou, S., Xu, J. F., et al. (2018). Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant. 11, 874–878. doi: 10.1016/j.molp.2018.02.013
Yang, H., Zhong, S. F., Chen, C., Yang, H., Chen, W., Tan, F. Q., et al. (2021). Identification and cloning of a CC-NBS-NBS-LRR gene as a candidate of Pm40 by integrated analysis of both the available transcriptional data and published linkage mapping. Int. J. Mol. Sci. 22, 10239. doi: 10.3390/ijms221910239
Yang, S. H., Li, J., Zhang, X. H., Zhang, Q. J., Huang, J., Chen, J. Q., et al. (2013). Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl. Acad. Sci. U. S. A. 110, 18572–18577. doi: 10.1073/pnas.1318211110
Zhang, J. P., Zhang, P., Hewitt, T., Li, J. B., Dundas, I., Schnippenkoetter, W., et al. (2019). A strategy for identifying markers linked with stem rust resistance in wheat harboring an alien chromosome introgression from a non-sequenced genome. Theor. Appl. Genet. 132, 125–135. doi: 10.1007/s00122-018-3201-8
Zhang, R. Q., Xiong, C. X., Mu, H. Q., Yao, R. N., Meng, X. R., Kong, L. N., et al. (2020). Pm67, a new powdery mildew resistance gene transferred from Dasypyrum villosum chromosome 1V to common wheat (Triticum aestivum L.). Crop J. 9, 882–888. doi: 10.1016/j.cj.2020.09.012
Keywords: common wheat, Ae. longissima, 6Sl#3 recombinants, powdery mildew resistance, physical mapping
Citation: Tian X, Chen Q, Ma C, Men W, Liu Q, Zhao Y, Qian J, Fan Z, Miao J, He J, Sehgal SK, Li H and Liu W (2022) Development and Characterization of Triticum aestivum-Aegilops longissima 6Sl Recombinants Harboring a Novel Powdery Mildew Resistance Gene Pm6Sl. Front. Plant Sci. 13:918508. doi: 10.3389/fpls.2022.918508
Received: 12 April 2022; Accepted: 02 May 2022;
Published: 02 June 2022.
Edited by:
Cheng Liu, Shandong Academy of Agricultural Sciences, ChinaReviewed by:
Xiangyang Xu, United States Department of Agriculture, United StatesCopyright © 2022 Tian, Chen, Ma, Men, Liu, Zhao, Qian, Fan, Miao, He, Sehgal, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanhuan Li, bGlodWFuaHVhbmhhcHB5QGhlbmF1LmVkdS5jbg==; Wenxuan Liu, bGl1d2VueHVhbkBoZW5hdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.