- College of Plant Science, Jilin University, Changchun, China
The life of higher plants progresses successively through embryonic, juvenile, adult, and reproductive stages. LEAFY COTYLEDON (LEC) transcription factors, first discovered in Arabidopsis thaliana several decades ago, play a key role in regulating plant embryonic development, seed maturation, and subsequent growth. Existing studies have demonstrated that LECs together with other transcription factors form a huge and complex regulatory network to regulate many aspects of plant growth and development and respond to environmental stresses. Here, we focus on the role that has received little attention about the LECs linking different developmental stages and generational cycles in plants. We summarize the current fragmented research progress on the LECs role and molecular mechanism in connecting embryonic and vegetative growth periods and the reproductive stage. Furthermore, the possibility of LECs controlling the maintenance and transition of plant growth stages through epigenetic modifications is discussed.
Introduction
The plant lifecycle goes through embryonic, juvenile, adult, and reproductive stages. Transitions between different developmental stages of plants ensure that plants can complete the lifecycle and maintain normal growth and development and adaptation to environmental stresses.
The embryonic stage, which begins with the formation of a single-celled zygote after the fertilization of an egg by a male gamete, can be divided into morphogenesis, maturation, and dehydration (Cridge et al., 2016). Forward genetics methods have identified a series of the master genes LAFL that regulate embryonic development, including LEAFY COTYLEDON1 (LEC1) and LEC1-LIKE (L1L) of the NF-YB gene family, and ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEC2 (LEAFY COTYLEDON2) of the B3-AFL gene family (Giraudat et al., 1992). LEAFY COTYLEDONs (LEC1, LEC2, and FUSCA3) are the key genes in plant embryonic development. During embryonic morphogenesis, the LEAFY COTYLEDONs (LECs) determine the fate of stalk cells and control cotyledon characteristics. In the later stage of embryonic maturation, ABI3 promotes the accumulation of storage substances and increases the dehydration resistance of embryos (Giraudat et al., 1992; Meinke, 1992; Meinke et al., 1994; West et al., 1994). Similar to LEC1, L1L is expressed primarily during seed development. However, suppression of L1L gene expression induced defects in embryo development that differed from those of lec1 mutants, suggesting that LEC1 and L1L play unique roles in embryogenesis (Kwong et al., 2003). Furthermore, LEC1 and L1L genes act as key regulators to coordinate the expression of fatty acid biosynthetic genes (Mu et al., 2008). Loss of function of each of these genes produced distinct phenotypes, with enhanced phenotypes in double mutants, suggesting that these genes regulate embryonic development through distinct but redundant pathways. Subsequent studies found that the LEC1 encodes a transcriptional activator that is mainly expressed in seeds and encodes a HAP3 subunit belonging to the HAP (NF-Y or CBF) family of transcription factors, which binds the CCAAT-box. The HAP complex includes three distinct subunits: HAP2/NF-YA/CBF-B, HAP3/NF-YB/CBF-A, and HAP5/NF-YC/CBF-C, and HAP3 plays an important role in plant embryonic development, chlorophyll biosynthesis, flowering regulation, etc. (Lotan et al., 1998), whereas LEC2, FUS3, and ABI3 all belong to the AFL subfamily of the LAV family in the B3 transcription factor superfamily; hence, they are called AFL-B3 regulators (Luerssen et al., 1998; Stone et al., 2001). LEC2, FUS3, and ABI3 can activate downstream target gene expression by binding the conserved region of B3 to the RY/Sph conserved sequence (CATGCAT) cis-regulator. Numerous studies have shown that these genes are involved in embryonic development and need to be inhibited in the vegetative phase after seed germination to ensure that the plant can enter the vegetative phase smoothly (Bentsink and Koornneef, 2008). Several repressors, including chromatin remodeling proteins (Henderson et al., 2004; Tang et al., 2008), and DNA-binding transcription factors (Moon et al., 2003) are involved in suppressing the expression of embryonic development genes in vegetative organs. Therefore, plant seeds can smoothly enter the vegetative growth stage after germination. The traditional view is that the embryonic stage is the period when the stem and root meristems are initially formed (Tsukagoshi et al., 2007), whereas the juvenile stage in vegetative growth is when the shoot apical meristem begins to form stems, true leaves, and lateral buds (Poethig, 1990). However, for some subterranean sprouting plants, such as beans and maize, a certain number of leaves are already present during embryogenesis, indicating that the juvenile stage begins during embryonic development. The above results show that there is a close relationship between plant embryonic development and vegetative growth. However, research in this area is relatively fragmented and there is a lack of sufficient research on how LECs regulates vegetative growth. Hence, the underlying molecular mechanisms and the significance of low-level expression of LEAFY COTYLEDONs during vegetative periods remain unclear and are both discussed thoroughly in this study. For the first time, we have systematically discussed the key role of LEAFY COTYLEDONs in regulating embryonic and vegetative stages, in order to provide new ideas for researchers to further study the relationship between embryonic and vegetative stages.
The vegetative growth period includes juvenile and adult stages. The transition of plants from juvenile to adult is called the vegetative phase change (VPC; Huang et al., 2015a), whereas flowering represents the transition from plant adulthood to the reproductive stage. And there are many studies on the relationship between vegetative growth and reproductive growth.
Many perennial overwintering plants undergo vernalization to flower (Xu and Chong, 2018), cycling through embryonic, juvenile, adult, and reproductive stages. In Arabidopsis thaliana, repression of flowering via chromatin modification (lysine trimethylation at position 27 of histone H3–H3K27me3; Bouché et al., 2017; Xu and Chong, 2018). The vernalization-induced FLC silencing is stable in the current generation, but the next generation needs to reactivate the expression of FLC to ensure that the plants are vernalized through cold winters in each generation, thereby preventing the plants from blooming before or during the winter (He and Li, 2018). It has been found that the seed-specific “pioneer” transcription factor LEC1 initiates this reset process after fertilization and cooperates with LEC2 and FUS3 to form a “vernalization memory” embryo reset pathway, thereby activating the expression of FLC (He and Li, 2018) so that the plant can smoothly re-enter the embryonic stage from the reproductive stage for the generation cycle.
Here, we focus on LEAFY COTYLEDONs (LECs) and elucidate the molecular and epigenetic mechanisms of embryonic developmental regulators that play a role in linking different developmental stages to regulate the plant life cycle. In addition, previous studies were mostly limited to the function of LEAFY COTYLEDONs at a specific developmental stage. Hence, little attention has been paid to the role of LEAFY COTYLEDONs in plant phase transition and life cycle, and there is a lack of systematic and in-depth research on its molecular mechanism. We further examine the molecular mechanisms by which LECs regulate plant phase transitions and propose a possible model of the complete life cycle.
Plant phase transition and distinguishable boundaries
The plant cycle goes through the embryonic stage, the vegetative growth period, and the reproductive growth period (also called the flowering period). Dicotyledonous embryogenesis starts from the fertilized egg (zygote), which is divided into several morphologically distinct stages: proembryo, heart-shaped embryo, spherical embryo, torpedo-shaped embryo, and horseshoe-shaped embryo (De Smet et al., 2010). After embryonic morphogenesis, seeds enter a period of desiccation and then dormancy. Previous studies have found that seed development strictly follows a specific order, which is regulated by a series of embryonic development regulators LEAFY COTYLEDONs (such as LEC1, LEC2, FUSCA3, and ABI3) that are activated in sequence (Raz et al., 2001).
After morphogenesis, the embryo differentiation and development need to be arrested in time to ensure that the seeds enter the desiccation phase smoothly and acquire desiccation tolerance. The embryo growth arrest is regulated by the FUS3/LEC type genes to enable the seed to enter the maturity stage, as mutations in these genes cause a continuation of growth of immature embryos (Raz et al., 2001). Furthermore, LEC1, LEC2, FUS3, and ABI3 can cooperate with each other to activate target genes (Santos-Mendoza et al., 2008) and thus participate in the signaling pathway of ABA-mediated seed maturation regulated by ABI3 and ABI5 (Suzuki and McCarty, 2008).
After embryonic development, the radicle and plumule break through the seed coat, and the cotyledons unfold to allow photosynthesis, signifying that the plant is entering the vegetative growth phase. Juvenile and adult stages have easily distinguishable morphological features in the model plant Arabidopsis. Juvenile leaves are small and round, with smooth and non-notched edges, and epidermal hairs on the adaxial surface (Tanaka et al., 2008). Previous studies have found that miR156 is a master control factor in the transition from juvenile to adult stage and is evolutionarily conserved (Wang et al., 2009). The miR156 controls the leaf shape and epidermal hair traits of plants by regulating the SQUAMOSA promoter-binding protein-like9 (SPL9) gene family (Wu et al., 2009). When the vegetative growth enters the final stage, the plant reaches mature/adult state, whereby the expression of a series of genes changes under the stimulation of seasonal and temperature signals. For example, the inhibition of flowering by FLC is relieved (Searle et al., 2006) and the floral meristem-determining gene LFY is upregulated (Blázquez et al., 1997), simultaneously initiating flower buds to begin differentiation, which marks the beginning of the reproductive growth phase (Blázquez et al., 1997). In addition, a series of hormonal regulation and epigenetic modifications are also involved in different stages of this transition from vegetative to reproductive growth.
LEAFY COTYLEDONs are inhibited to ensure that the plant smoothly enters the vegetative growth stage
After entering the vegetative growth phase, the expression of the whole plant genome changes. Some genes that are highly expressed in the embryonic stage, such as LAFL, need to be suppressed to ensure that the plant can enter the vegetative phase smoothly (Jia et al., 2014). Several epigenetic modifications and transcription factors play an important role (Jia et al., 2014).
Chromatin remodeling complexes POLYCOMB REPRESSIVE COMPLEXES (PRC) 1 and 2 are required for establishing and maintaining stable epigenetic repression in response to developmental or environmental signals (Mozgova and Hennig, 2015). Briefly, PRC2 has a histone 3, lysine 27 trimethylation (H3K27me3) activity, and PRC1 can recognize H3K27me3 and lead to chromatin compaction through histone H2A lysine ubiquitination (H2Aub; Bouyer et al., 2011). The LAFL genes are repressed by the Polycomb machinery via its histone-modifying activities, including the histone H3 K27 trimethyltransferase activity of the PRC2 and the histone H2A E3 monoubiquitin ligase activity of the PRC1 during seed germination (Makarevich et al., 2006; Bratzel et al., 2010; Yang et al., 2013; Schneider et al., 2016). The PCR2 is the main factor involved in cell identity to maintain stable repression of embryogenesis program genes (Salaün et al., 2021).
VAL1 and 2 (VIVIPAROUS/ABI3LIKE) are transcriptional regulators containing DNA-and chromatin-binding domains, and the expression of VAL1 and 2 increased in this process as embryo-repressed genes (Yuan et al., 2021). Specifically, VAL proteins recruit histone deacetylase 19 (HDA19) and PRC1 to the chromatin regions, which contain genes involved in the regulation of development and dormancy. Thereby, HDA19 removes histone acetylation marks, whereas PRC1 incorporates monoubiquitinated histone H2A (H2Aub) marks to initiate initial repression of the target gene (Makarevich et al., 2006). Thus, the VAL proteins appear to cause the initial repression of the seed-development and germination-related genes. Later on, this repression is maintained by PRC2-mediated trimethylation at H3K27 (Makarevich et al., 2006). It is critical to note that VAL1 was shown to interact with HDA19 and to repress LAFL gene expression during germination (Zhou et al., 2013).
The PICKLE genes (PKL and PICKLE RELATED 1 and 2—PKR1 and PKR2) are also upregulated in the process, and they are members of the CHD3 family of chromatin ATPase remodelers (Han et al., 2015). It was found that PKL acts throughout the seedling, repressing the expression of embryonic traits, and is required for GA-dependent responses in shoots (Henderson et al., 2004; Smolikova et al., 2021). Furthermore, in the pkl and val1 mutants, the chromatin-based repression of LAFL is less important and, consequently, both PKL and VAL have a repressive effect on the expression of LAFL genes during zygotic embryo development (Hurtado et al., 2006; Bouyer et al., 2011; Han et al., 2015; Shen et al., 2015).
miRNAs also play an important role in the process of transitioning from embryonic to the vegetative growth phase. The miR156-SPLs control the transition from juvenile to adult stage and are evolutionary conserved (Morea et al., 2016). The expression of miR156 decreased gradually with age, whereas the expression pattern of SPL was negatively correlated with age (Kim, 2020). The miRNAs were shown to target LEC1, LEC2, and FUS3 directly or indirectly and to suppress their activities in the embryo during embryo development, until they are required (Wu and Poethig, 2006; Willmann et al., 2011). Interestingly, LAFL genes such as FUSCA3 can target miR156A and miR156C to activate miR156, thus delaying the VPC (Lumba et al., 2012; Vashisht and Nodine, 2014). In summary, the expression of miRNA inhibited LAFL to ensure the smooth entry of plants into the vegetative growth phase, and for the transition of the juvenile to adult phase.
Function of LEAFY COTYLEDONs during vegetative growth
LECs have been extensively studied as regulators of embryonic development and seed maturation. However, little is known about how they regulate postembryonic development and vegetative growth.
LEC1 encodes the B subunit of NF-Y that consists of NF-YA, NF-YB, and NF-YC, and often functions as a pioneer transcription factor (Lotan et al., 1998; Wang et al., 2011, 2012; Nardini et al., 2013; Wang and Perry, 2013) and is first activated after pollination and is expressed in both embryo and endosperm. Song et al. found that an embryonic development arrest, caused by loss of LEC1 function in the embryo, can be rescued by normally expressed LEC1 in the endosperm (Fleming et al., 2013). The loss of LEC1 function in the endosperm leads directly to the embryonic development arrest and cannot be rescued by LEC1 in the embryo (Fleming et al., 2013). Therefore, LEAFY COTYLEDONs also play a crucial role in tissues other than embryos in plant growth and development. Hou et al. found that hypocotyl elongation was inhibited in the lec1 mutants relative to wild type in Arabidopsis (Oldfield et al., 2014). LEC1 and PHYTOCHROME-INTERACTING FACTOR4 (PIF4) have a direct interdependent relationship, and can jointly bind to the promoter G-box elements of the hypocotyl elongation-related genes such as INDOLE-3-ACETIC ACID INDUCIBLE 19 (IAA19) and CYTOKININ INDUCED ROOT CURLING 2 (YUC8), and coordinately regulate hypocotyl elongation (Oldfield et al., 2014). In addition, Hou et al. also revealed that LEC1 inhibited the development of cotyledon trichomes by regulating the expression of the trichome development-related genes such as GLABROUS1 (GL1), GL2, and GL3 and recruiting the transcription factor TRICHOMELESS2 (TCL2; Huang et al., 2015a).
AFL-B3 regulators (FUSCA3, LEC2, and ABI3) have also been shown to be associated with root development. In culture medium free of any exogenous hormone supplementation, while adventitious root emergence and growth was prominently observed in wild-type cells, no such features were observed in abi3-6 cells (Sherwood et al., 2014). Expression of auxin-responsive AUX1 and GH3 genes was significantly reduced in abi3-6 cells, indicating that auxin levels or distribution may be altered in absence of ABI3 (Sherwood et al., 2014). In addition, the function of auxin in promoting lateral root formation may be related to FUSCA3 and LEC2, and these two homologous B3 transcription factors have been shown to interact to bind to the auxin biosynthesis gene YUCCA4 (YUC4) and synergistically activate its transcription during lateral root formation (Song et al., 2021). Furthermore, FUSCA3 expression is activated by LEC2 in lateral root initiation. The observations indicate that the FUS3-LEC2 complex functions as a key regulator in auxin-regulated lateral root formation (Song et al., 2021).
These findings suggest that LEAFY COTYLEDONs play an important role in regulating embryonic development and vegetative growth.
LEAFY COTYLEDONs regulate the vegetable phase change
During the transition from juvenile to adult stage, plants are accompanied by multiple physiological and biochemical metabolic processes, such as biomass, reproductive capacity, resistance to pests and diseases, stress resistance, and plant architecture (Chuck et al., 2011; Huang et al., 2015b; Tang et al., 2017; Sengupta and Nag Chaudhuri, 2020). Previous studies have shown that LEAFY COTYLEDONs play an important role in regulating plant stage transitions.
The FUSCA3 is a transcription factor belonging to the B3 family and plays an important role in the determination of embryonic leaf identity and the timing of development (Luerssen et al., 1998). Whole-genome microarray analysis found that the expression of the genes related to ethylene signaling pathway increased in fus3 mutants, including ACS6, ERF1, EDF4, EDF2, etc. (Lumba et al., 2012). The loss of FUSCA3 function resulted in advanced vegetative phase change and cotyledon replacement by leaves (Vashisht and Nodine, 2014). It has been proven that FUSCA3 may partially depend on the ethylene signaling pathway to regulate the vegetative stage transition. In seeds, FUSCA3 binds to and represses AtGA3ox2, reducing levels of bioactive GA in tissues where it is expressed, thus repressing germination (Wang et al., 2012). Dwarf8, a DELLA protein that, when mutated, results in shortening of a delay in VPC, is downregulated along with another DELLA (Wang et al., 2012).
In addition, FUSCA3 can be phosphorylated by SNF-related protein kinases (SnRKs) in Arabidopsis, and exogenous application of JA delays VPC and rejuvenizes leaf five in both epidermal characteristics and gene expression patterns (Wang et al., 2012). Using Genome-Wide Identification of in Vivo Binding Sites for FUS3, it was found that FUSCA3 can directly bind to the promoters of miR156A and miR156C (two major precursors of miR156; Lumba et al., 2012). The miR156-SPL is the master pathway regulating vegetative stage transition. The miR156 inhibits SPL3, SPL4, SPL5, SPL9, SPL10, and SPL13, and targets miR172 (a microRNA that represses the AP2 gene); this pathway mechanism is evolutionary conserved (Schneider et al., 2016). To sum up, FUSCA3 cooperates with ABA/GA during the embryonic stage to regulate the transition from embryonic to the vegetative stage, whereas the function of FUSCA3 in the vegetative stage regulates the transition of the vegetative to reproductive stage by coordinating the regulation of the ethylene signaling pathway and balancing the two hormones JA and GA. Therefore, FUS3 may regulate VPC through two pathways (via hormones and via miR156-SPL). However, it remains unclear whether these two pathways cross and what their molecular mechanisms are.
In Arabidopsis, Polycomb repressive group complex 2 (PRC2) inhibits gene expression by catalyzing the trimethylation of histone H3 at lysine 27 (H3K27me3; Wang et al., 2012). The major seed regulatory genes, including LEC2, FUSCA3, and ABI3, were found to be enriched in the CURLY LEAF (CLF) and SWINGER (SWN) binding regions through ChIP seq analyses to profile the genome-wide distributions of CLF and SWN in Arabidopsis seedlings (Cui et al., 2014).CLF and SWN are the two major H3K27 methyltransferases and are the core components of PRC2, targeting transcripts of MADS-box transcription factors and SPLs, and those of miR156A/C and MIR172, as well as playing roles in VPC (Cui et al., 2014). Furthermore, in contrast to wild type with abaxial trichomes on leaf 7, the swn-3 mutant exhibited the phenotypes with significantly delayed vegetative phase change and phenotypes with later abaxial trichomes on leaf 11 and rounder leaves; by contrast, clf29 had similar trichome production and leaf shape to wild type (Mao et al., 2017). Even though clf-29 did not exhibit a vegetative phase change phenotype, we cannot rule out the possibility that CLF acts redundantly with SWN in influencing vegetative phase change (Mao et al., 2017). Hence, we can hypothesize that LEAFY COTYLEDONs recruit SWN or CLF in miR156 loci to influence VPC. It is worth noting that miR156 has an age-dependent expression pattern because it is highly expressed in young plants and gradually decreases with age. Therefore, the miR156 must have undergone a reset process during the generational change. The latest research shows that LEC2 is involved in regulating the transgenerational reset process of miR156A/C in the late embryo to ensure the smooth lifecycle of plants (Osadchuk et al., 2021). However, the research on the regulation of VPC by LECs is still relatively small, and more attention needs to be devoted to this area.
LEAFY COTYLEDONs participate in the regulation of plant flowering via the vernalization pathway
Many over-wintering plants in temperate climates acquire the competence to flower in spring after experiencing prolonged cold exposure through a process known as vernalization (Bouché et al., 2017; Xu and Chong, 2018). The vernalization pathways shut down the expression of a potent floral repressor and this vernalization-mediated repression or “vernalized state” is maintained in cell divisions during subsequent growth and development when the temperature rises in spring, named epigenetic “memory of winter cold,” enabling plants to flower in spring (Xu and Chong, 2018; Shu et al., 2019). However, the “memory of winter cold” must be reset in the next generation to ensure that each generation or each growth cycle experiences winter cold before flowering and thus flowers at the right season to maximize reproductive success (He and Li, 2018; Shu et al., 2019). In Arabidopsis thaliana, the vernalization pathway blocks the expression of the potent floral repressor FLC to enable spring flowering (Xu and Chong, 2018). Recent studies reveal that shortly after fertilization LEC1 functions as a pioneer transcription factor to initiate FLC resetting/re-activation, and that LEC2 and FUS3 subsequently function together with LEC1 to fully re-activate FLC expression in early embryogenesis by recruiting the plant-specific scaffold protein FRI, which physically associates with the H3K36 methyltransferase Embryonal Fyn-associated substrate (EFS) and other active chromatin modifiers (Xu et al., 2016; Gao et al., 2022). Thus, LEC1, LEC2, and FUSCA3 govern the reprogramming process of vernalized memory. Interestingly, VAL1/2 belongs to the B3 family of transcription factors as FUSCA3 and LEC2, and both can bind to the cold memory element CME of FLC. Therefore, after the plants entered the vegetative phase, the expression of these embryonic developmental genes decreased, and VAL1/2 competed to bind to the FLC promoter motif to shut down FLC expression, allowing plants to flower in warm spring (Xu et al., 2016). Therefore, these three LEC genes reset the parental “memory of winter cold” during early embryogenesis in Arabidopsis and indirectly involved in repressing FLC to allow flowering, and this is of great significance for plants to successfully complete the generational alternation (Andrés and Coupland, 2012; Niu and He, 2019).
LEAFY COTYLEDONs regulate phase transitions and complete the lifecycle through epigenetic modifications
Epigenetic modifications are involved in the phase transition and lifecycle of plants, and LECs play an important role in them.
BRAHMA(BRM) is a SWI2/SNF2 chromatin remodeling ATPase, and BRM antagonizes the PcG protein complexed with SWN protein influencing the histone methylation status of the MIR156A promoter region (Smolikova et al., 2021). In addition, the brm mutants accelerate VPC and flowering in Arabidopsis. Furthermore, BRM opposes PICKLE at the target loci, whereby BRM maintains active expression and PKL balances active and repressive states of these targets and promotes H3K27me3, leading up to phase change as SWI/SNF chromatin remodelers (Hurtado et al., 2006). In Arabidopsis, the pkl mutants delay VPC and flowering, and PKL cooperatively interacts with SWN or CLF to regulate target genes. In addition, the LAFL network (LEC1, LEC2, ABI3, and FUSCA3), as a negative regulator of seed germination, needs to be suppressed before seedling development. This repressive signal is mediated by PRC1 and PRC2, as well as PKL and PICKLERELATED2 (PKR2) proteins (Tao et al., 2019). However, chromodomain, helicase, and DNA-binding CHD1 protein (CHR5) are involved in establishing the active chromatin state of seed maturation genes. CHR5 is expressed during late embryogenesis, antagonizing PKL in embryo gene expression and seed storage protein accumulation (Tao et al., 2017). In addition, CHR5 was shown to be associated with the promoters of ABI3 and FUS3 and to be required for reducing nucleosome occupancy near the transcriptional start site (Shen et al., 2015). In view of the above, LEAFY COTYLEDONs have distinct spatiotemporal expression patterns and create a complex dynamic regulatory network with other transcription factors and epigenetic modifiers and consequently play a key role in connecting different developmental stages of plants. Based on the above, we propose a possible model (Figure 1) of the molecular mechanism of LEAFY COTYLEDONs involved in regulating plant phase transition from the macro perspective of plant generation cycle.
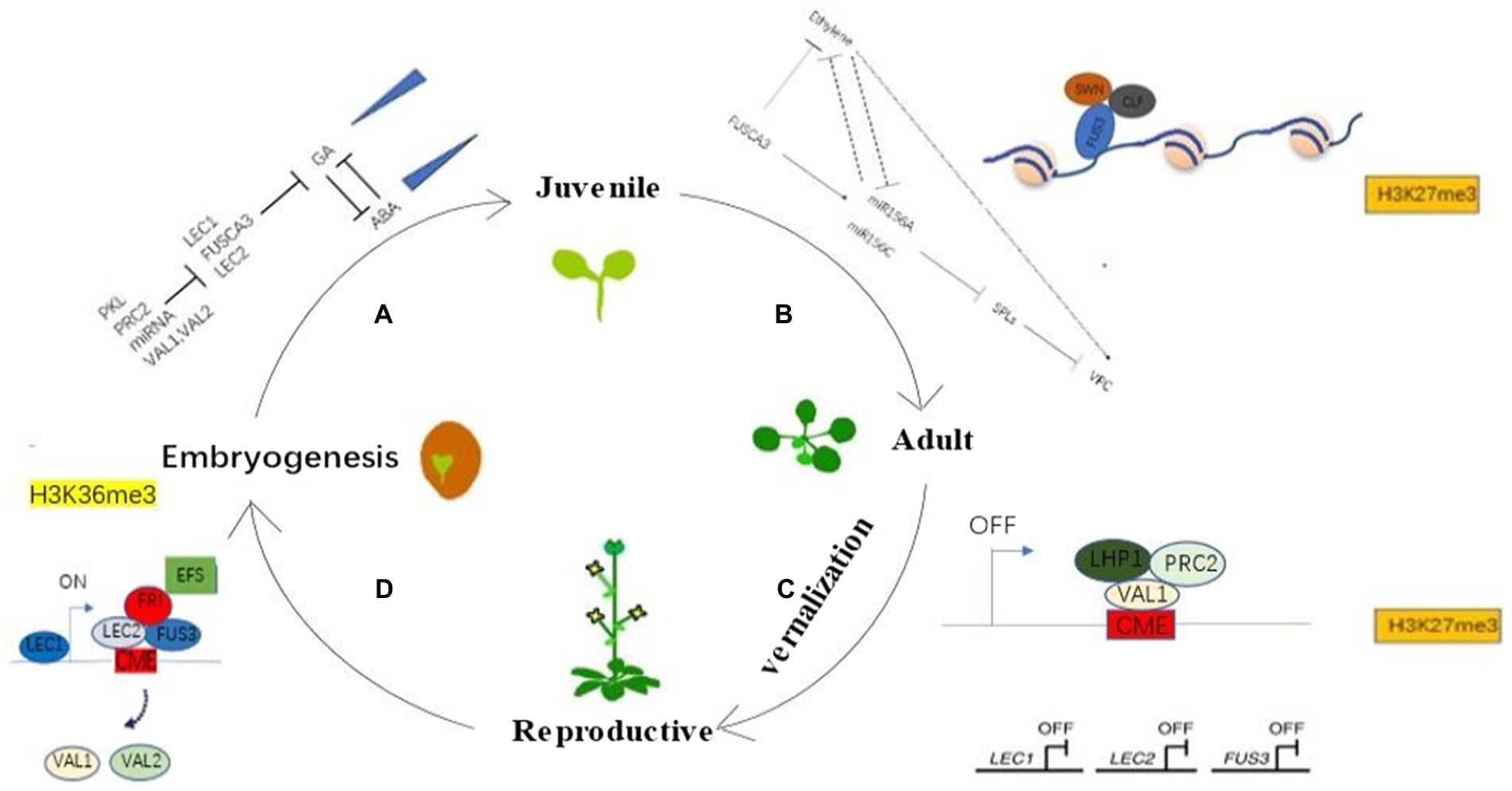
Figure 1. LEAFY COTYLEDONs connect different growth stages of plants and complete the generation cycle through epigenetic modification and mediating phytohormonal signaling. (A) LEAFY COTYLEDONs regulate the transition from embryogenesis to juvenile stage. PKL, PRC2, miRNA, and VAL1/VAL2 inhibit the expression of LEAFY COTYLEDONs to inhibit the GA signaling pathway. GA is antagonistic to ABA, and the expression of GA increases continuously, whereas ABA shows the opposite expression pattern during this stage. (B) LEAFY COTYLEDONs regulate the transition from juvenile to adult stage. There are potentially two pathways of how LEAFY COTYLEDONs regulate the transition from juvenile to adult stage. One is that FUS3 promotes VPC by recruiting chromatin repressors such as SWN and CLF for chromatin modification of miR156 to inhibit the expression of miR156 and then promote the expression of downstream SPLs. Another is that FUS3 regulates VPC by participating in the ethylene signaling pathway, and there may be a cross-talk between these two pathways. (C) Perennials experience vernalization from juvenile to adulthood. VAL1/2 competes with LEAFY COTYLEDONs for binding to the FLC cold memory element CME, and VAL1/2 shuts down FLC expression by recruiting PRC2 and LHP1 for chromatin modification of FLC, as LEAFY COTYLEDONs shut down expression during this period. (D) Plants enter embryonic stage after fertilization. The LEAFY COTYLEDONs are expressed sequentially, and the plant enters a new life cycle by recruiting active chromatin modifiers such as FRI and EFS to activate the expression of FLC and a series of other genes such as miR156. VAL1/2 are suppressed during this period.
Conclusion
The development of higher plants starts from the zygote, and plants already begin to prepare for subsequent vegetative growth, flowering, and reproduction during embryonic development. However, there are many studies on the relationship between vegetative growth and reproductive growth, but few studies on how the embryonic stage affects vegetative growth and reproductive growth. In fact, epigenetic modification in the embryonic stage allows the plant to remember this modification and pass it on to the postembryonic vegetative and reproductive growth stages. We think that LEAFY COTYLEDONs may play a key role in linking different reproductive stages of plants through the establishment of active chromatin during the embryonic stage and the recruitment of chromatin modifiers. The Polycomb repressive group complex (PcG) proteins EMF1, CLF, LHP, and SWN are involved in these processes. FUSCA3 has been shown to perform different functions in embryonic and vegetative stages, synergize with JA, GA, and ABA, and participate in the epigenetic modification process to regulate the stage transition in plants. LEAFY COTYLEDONs form a complex network with histone epigenetic modifiers, miRNAs, and transcription factors specifically expressed at each growth stage, and they also mediate phytohormone signals. However, previous studies were mostly limited to the function of LEAFY COTYLEDONs at a specific developmental stage, with little attention paid to the role of LEAFY COTYLEDONs in plant phase transition and life cycle. However, the progression of the life cycle is critical for perennials. During the cycle of each generation, the reactivation of many genes is required to start a new round of life activities. For example, the flowering suppressor gene FLC is reset by LEAFY COTYLEDONs during embryonic stage, and the age master gene miR156A/C is reset by LEAFY COTYLEDONs during embryonic stage. A growing number of studies have demonstrated the critical role of LEC genes in altering plant generations, but these studies are only in their infancy. We hope this paper will provide new research ideas for researchers to study various physiological activities associated with the alteration of plant generations. LEAFY COTYLEDONs are involved in many physiological and biochemical processes in plants, especially in epigenetic modifications, which we believe play a crucial role in tandem with different developmental stages in plants. We propose a model for the role of LEAFY COTYLEDONs in linking different growth stages and completing the generational cycle in this study (Figure 1). Further exploration of LEAFY COTYLEDONs’ intrinsic molecular mechanism has far-reaching significance for understanding the principle of the plant lifecycle and applying it to agricultural production.
Author contributions
CC and XD contributed to the conception and design of the study. CC wrote the first draft of the manuscript. XD make critical revisions to the work. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andrés, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Bentsink, L., and Koornneef, M. (2008). Seed dormancy and germination. Arabidopsis Book. 6:e0119. doi: 10.1199/tab.0119
Blázquez, M. A., Soowal, L. N., Lee, I., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124, 3835–3844. doi: 10.1242/dev.124.19.3835
Bouché, F., Woods, D. P., and Amasino, R. M. (2017). Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol. 173, 27–35. doi: 10.1104/pp.16.01322
Bouyer, D., Roudier, F., Heese, M., Andersen, E. D., Gey, D., Nowack, M. K., et al. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7:e1002014. doi: 10.1371/journal.pgen.1002014
Bratzel, F., López-Torrejón, G., Koch, M., Del Pozo, J. C., and Calonje, M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859. doi: 10.1016/j.cub.2010.09.046
Chuck, G. S., Tobias, C., Sun, L., Kraemer, F., Li, C., Dibble, D., et al. (2011). Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc. Natl. Acad. Sci. U. S. A. 108, 17550–17555. doi: 10.1073/pnas.1113971108
Cridge, A. G., Dearden, P. K., and Brownfield, L. R. (2016). Convergent occurrence of the developmental hourglass in plant and animal embryogenesis? Ann. Bot. 117, 833–843. doi: 10.1093/aob/mcw024
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., and Lin, H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
De Smet, I., Lau, S., Mayer, U., and Jürgens, G. (2010). Embryogenesis - the humble beginnings of plant life. Plant J. 61, 959–970. doi: 10.1111/j.1365-313X.2010.04143.x
Fleming, J. D., Pavesi, G., Benatti, P., Imbriano, C., Mantovani, R., and Struhl, K. (2013). NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 23, 1195–1209. doi: 10.1101/gr.148080.112
Gao, J., Zhang, K., Cheng, Y. J., Yu, S., Shang, G. D., Wang, F. X., et al. (2022). A robust mechanism for resetting juvenility during each generation in Arabidopsis. Nat. Plants 8, 257–268. doi: 10.1038/s41477-022-01110-4
Giraudat, J., Hauge, B. M., Valon, C., Smalle, J., Parcy, F., and Goodman, H. M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. doi: 10.1105/tpc.4.10.1251
Han, S. K., Wu, M. F., Cui, S., and Wagner, D. (2015). Roles and activities of chromatin remodeling ATPases in plants. Plant J. 83, 62–77. doi: 10.1111/tpj.12877
He, Y., and Li, Z. (2018). Epigenetic environmental memories in plants: establishment, maintenance, and reprogramming. Trends Genet. 34, 856–866. doi: 10.1016/j.tig.2018.07.006
Henderson, J. T., Li, H. C., Rider, D. S., Mordhorst, A. P., Romero-Severson, J., Cheng, J. C., et al. (2004). PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 134, 995–1005. doi: 10.1104/pp.103.030148
Huang, M., Hu, Y., Liu, X., Li, Y., and Hou, X. (2015a). Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front. Plant Sci. 6:955. doi: 10.3389/fpls.2015.00955
Huang, M., Hu, Y., Liu, X., Li, Y., and Hou, X. (2015b). Arabidopsis LEAFY COTYLEDON1 mediates postembryonic development via interacting with PHYTOCHROME-INTERACTING FACTOR4. Plant Cell 27, 3099–3111. doi: 10.1105/tpc.15.00750
Hurtado, L., Farrona, S., and Reyes, J. C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62, 291–304. doi: 10.1007/s11103-006-9021-2
Jia, H., Suzuki, M., and McCarty, D. R. (2014). Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip. Rev. Dev. Biol. 3, 135–145. doi: 10.1002/wdev.126
Kim, D. H. (2020). Current understanding of flowering pathways in plants: focusing on the vernalization pathway in Arabidopsis and several vegetable crop plants. Hortic. Environ. Biotechnol. 61, 209–227. doi: 10.1007/s13580-019-00218-5
Kwong, R. W., Bui, A. Q., Lee, H., Kwong, L. W., Fischer, R. L., Goldberg, R. B., et al. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15, 5–18. doi: 10.1105/tpc.006973
Lotan, T., Ohio, M., Yee, K. M., West, M. A., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/S0092-8674(00)81463-4
Luerssen, H., Kirik, V., and Herrmann, P. (1998). Mise’ra S encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15, 755–764. doi: 10.1046/j.1365-313X.1998.00259.x
Lumba, S., Tsuchiya, Y., Delmas, F., Hezky, J., Provart, N. J., Shi, L. Q., et al. (2012). The embryonic leaf identity gene FUSCA3 regulates vegetative phase transitions by negatively modulating ethylene-regulated gene expression in Arabidopsis. BMC Biol. 10:8. doi: 10.1186/1741-7007-10-8
Makarevich, G., Leroy, O., Akinci, U., Schubert, D., Clarenz, O., Goodrich, J., et al. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7, 947–952. doi: 10.1038/sj.embor.7400760
Mao, Y. B., Liu, Y. Q., Chen, D. Y., Chen, F. Y., Fang, X., Hong, G. J., et al. (2017). Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 8:13925. doi: 10.1038/ncomms13925
Meinke, D. (1992). A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258, 1647–1650. doi: 10.1126/science.258.5088.1647
Meinke, D., Franzmann, L. H., Nickle, T., and Yeung, E. C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell 6, 1049–1064. doi: 10.2307/3869884
Moon, Y. H., Chen, L., Pan, R. L., Chang, H. S., Zhu, T., Maffeo, D. M., et al. (2003). EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15, 681–693. doi: 10.1105/tpc.007831
Morea, E. G., da Silva, E. M., Valente, G. T., Barrera Rojas, C. H., Vincentz, M., and Nogueira, F. T. (2016). Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol. 16:153. doi: 10.1186/s12870-016-0716-5
Mozgova, I., and Hennig, L. (2015). The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296. doi: 10.1146/annurev-arplant-043014-115627
Mu, J., Tan, H., Zheng, Q., Fu, F., Liang, Y., Zhang, J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148, 1042–1054. doi: 10.1104/pp.108.126342
Nardini, M., Gnesutta, N., Donati, G., Gatta, R., Forni, C., Fossati, A., et al. (2013). Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152, 132–143. doi: 10.1016/j.cell.2012.11.047
Niu, D., and He, Y. (2019). LEAFY COTYLEDONs: old genes with new roles beyond seed development. F1000Res 8:F1000 Faculty Rev-2144. doi: 10.12688/f1000research.21180.1
Oldfield, A. J., Yang, P., Conway, A. E., Cinghu, S., Freudenberg, J. M., Yellaboina, S., et al. (2014). Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 55, 708–722. doi: 10.1016/j.molcel.2014.07.005
Osadchuk, K., Cheng, C. L., and Irish, E. E. (2021). The integration of leaf-derived signals sets the timing of vegetative phase change in maize, a process coordinated by epigenetic remodeling. Plant Sci. 312:111035. doi: 10.1016/j.plantsci.2021.111035
Poethig, R. S. (1990). Phase change and the regulation of shoot morphogenesis in plants. Science 250, 923–930. doi: 10.1126/science.250.4983.923
Raz, V., Bergervoet, J. H., and Koornneef, M. (2001). Sequential steps for developmental arrest in Arabidopsis seeds. Development 128, 243–252. doi: 10.1242/dev.128.2.243
Salaün, C., Lepiniec, L., and Dubreucq, B. (2021). Genetic and molecular control of somatic embryogenesis. Plants 10:1467. doi: 10.3390/plants10071467
Santos-Mendoza, M., Dubreucq, B., Baud, S., Parcy, F., Caboche, M., and Lepiniec, L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54, 608–620. doi: 10.1111/j.1365-313X.2008.03461.x
Schneider, A., Aghamirzaie, D., Elmarakeby, H., Poudel, A. N., Koo, A. J., Heath, L. S., et al. (2016). Potential targets of VIVIPAROUS1/ABI3-LIKE1 (VAL1) repression in developing Arabidopsis thaliana embryos. Plant J. 85, 305–319. doi: 10.1111/tpj.13106
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Sengupta, S., and Nag Chaudhuri, R. (2020). ABI3 plays a role in de-novo root regeneration from Arabidopsis thaliana callus cells. Plant Signal. Behav. 15:1794147. doi: 10.1080/15592324.2020.1794147
Shen, Y., Devic, M., Lepiniec, L., and Zhou, D. X. (2015). Chromodomain, helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes. Plant Biotechnol. J. 13, 811–820. doi: 10.1111/pbi.12315
Sherwood, R. I., Hashimoto, T., O'Donnell, C. W., Lewis, S., Barkal, A. A., van Hoff, J. P., et al. (2014). Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 32, 171–178. doi: 10.1038/nbt.2798
Shu, J., Chen, C., Thapa, R. K., Bian, S., Nguyen, V., Yu, K., et al. (2019). Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings. Plant Direct 3:e00100. doi: 10.1002/pld3.100
Smolikova, G., Strygina, K., Krylova, E., Leonova, T., Frolov, A., Khlestkina, E., et al. (2021). Transition from seeds to seedlings: hormonal and epigenetic aspects. Plants 10:1884. doi: 10.3390/plants10091884
Song, J., Xie, X., Chen, C., Shu, J., Thapa, R. K., Nguyen, V., et al. (2021). LEAFY COTYLEDON1 expression in the endosperm enables embryo maturation in Arabidopsis. Nat. Commun. 12:3963. doi: 10.1038/s41467-021-24234-1
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFYCOTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U. S. A. 98, 11806–11811. doi: 10.1073/pnas.201413498
Suzuki, M., and McCarty, D. R. (2008). Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 11, 548–553. doi: 10.1016/j.pbi.2008.06.015
Tanaka, M., Kikuchi, A., and Kamada, H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146, 149–161. doi: 10.1104/pp.107.111674
Tang, X., Hou, A., Babu, M., Nguyen, V., Hurtado, L., Lu, Q., et al. (2008). The Arabidopsis BRAHMA chromatin remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 147, 1143–1157. doi: 10.1104/pp.108.121996
Tang, L. P., Zhou, C., Wang, S. S., Yuan, J., Zhang, X. S., and Su, Y. H. (2017). FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 213, 1740–1754. doi: 10.1111/nph.14313
Tao, Z., Hu, H., Luo, X., Jia, B., Du, J., and He, Y. (2019). Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nat. Plants 5, 424–435. doi: 10.1038/s41477-019-0402-3
Tao, Z., Shen, L., Gu, X., Wang, Y., Yu, H., and He, Y. (2017). Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551, 124–128. doi: 10.1038/nature24300
Tsukagoshi, H., Morikami, A., and Nakamura, K. (2007). Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. U. S. A. 104, 2543–2547. doi: 10.1073/pnas.0607940104
Vashisht, D., and Nodine, M. D. (2014). MicroRNA functions in plant embryos. Biochem. Soc. Trans. 42, 352–357. doi: 10.1042/BST20130252
Wang, J. W., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, J. W., Park, M. Y., Wang, L. J., Koo, Y., Chen, X. Y., Weigel, D., et al. (2011). miRNA control of vegetative phase change in trees. PLoS Genet. 7:e1002012. doi: 10.1371/journal.pgen.1002012
Wang, F., and Perry, S. E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161, 1251–1264. doi: 10.1104/pp.112.212282
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape, and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
West, M., Yee, K. M., Danao, J., Zimmerman, J. L., Fischer, R. L., Goldberg, R. B., et al. (1994). LEAFYCOTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6, 1731–1745. doi: 10.2307/3869904
Willmann, M. R., Mehalick, A. J., Packer, R. L., and Jenik, P. D. (2011). MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 155, 1871–1884. doi: 10.1104/pp.110.171355
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Wu, G., and Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. doi: 10.1242/dev.02521
Xu, S., and Chong, K. (2018). Remembering winter through vernalisation. Nat. Plants 4, 997–1009. doi: 10.1038/s41477-018-0301-z
Xu, Y., Guo, C., Zhou, B., Li, C., Wang, H., Zheng, B., et al. (2016). Regulation of vegetative phase change by SWI2/SNF2 chromatin remodeling ATPase BRAHMA. Plant Physiol. 172, 2416–2428. doi: 10.1104/pp.16.01588
Yang, C., Bratzel, F., Hohmann, N., Koch, M., Turck, F., and Calonje, M. (2013). VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23, 1324–1329. doi: 10.1016/j.cub.2013.05.050
Yuan, L., Song, X., Zhang, L., Yu, Y., Liang, Z., Lei, Y., et al. (2021). The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide Polycomb silencing in Arabidopsis. Nucleic Acids Res. 49, 98–113. doi: 10.1093/nar/gkaa1129
Keywords: LEC1, LEC2, FUS3, ABI3, phase transition, flowering, epigenetic modification
Citation: Chen C and Du X (2022) LEAFY COTYLEDONs: Connecting different stages of plant development. Front. Plant Sci. 13:916831. doi: 10.3389/fpls.2022.916831
Edited by:
Ranjan Swarup, University of Nottingham, United KingdomReviewed by:
David Horvath, Agricultural Research Service (USDA), United StatesYuhai Cui, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2022 Chen, Du and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinglin Du, ZHV4aW5nbGluMjAwNEAxNjMuY29t
 Chen Chen
Chen Chen Xinglin Du*
Xinglin Du*