
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 22 June 2022
Sec. Plant Systematics and Evolution
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.911702
 Jun-Yi Zhang1,2†
Jun-Yi Zhang1,2† Min Liao1,3†
Min Liao1,3† Yue-Hong Cheng4
Yue-Hong Cheng4 Yu Feng1
Yu Feng1 Wen-Bing Ju1,3
Wen-Bing Ju1,3 Heng-Ning Deng1
Heng-Ning Deng1 Xiong Li1,3
Xiong Li1,3 Andelka Plenković-Moraj5
Andelka Plenković-Moraj5 Bo Xu1,3*
Bo Xu1,3*The species in the genus Cypripedium (Orchidaceae) are considered endangered, mainly distributed in the temperate regions of the Northern Hemisphere, with high ornamental and economic value. Despite previous extensive studies based on both morphology and molecular data, species and sections relationships within Cypripedium remain controversial. Here, we employed two newly generated Cypripedium chloroplast genomes with five other published genomes to elucidate their genomic characteristics. The two genomes were 162,773–207,142 bp in length and contained 128–130 genes, including 82–84 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. We identified 2,192 simple sequence repeats, 786 large repeat sequences, and 7,929 variable loci. The increase of repeat sequences (simple sequence repeats and large repeat sequences) causes a significant amplification in the chloroplast genome size of Cypripedium. The expansion of the IR region led to the pseudogenization or loss of genes in the SSC region. In addition, we identified 12 highly polymorphic loci (Pi > 0.09) suitable for inferring the phylogeny of Cypripedium species. Based on data sets of whole chloroplast genomes (IRa excluded) and protein-coding sequences, a well-supported phylogenetic tree was reconstructed, strongly supporting the five subfamilies of Orchidaceae and the genus Cypripedium as monophyletic taxa. Our findings also supported that C. palangshanense belonged to sect. Palangshanensia rather than sect. Retinervia. This study also enriched the genomic resources of Cypripedium, which may help to promote the conservation efforts of these endangered species.
Orchidaceae is one of the two largest families of angiosperms, consisting of approximately 880 genera and 27,000 species, accounting for 8% of all vascular plant species and growing in a wide range of habitats worldwide (Chase et al., 2003, 2015). The genus Cypripedium L. consisted of approximately 51 species with unique colorful labellum (known as lady’s slipper), which mainly distributed in subtropical to temperate in the Northern Hemisphere (Cribb, 1997; Chen and Cribb, 2005; Chen et al., 2013). More than two-thirds of these species were native to China, especially in Yunnan, Sichuan and Tibet (Chen et al., 2013). In the wild, many species have become rare and endangered due to shrinking natural habitats and over-collection for gardens and herbarium (Nelson, 1994; Farrell, 1999; Szlachetko et al., 2020). Thus, this genus is currently listed on the Convention on International Trade in Endangered Species of Wild Fauna and Flora, CITES Appendix II (Mcgough et al., 2006).
A robust and well-resolved orchid family phylogeny is fundamental to understanding the evolution and diversification of Orchidaceae, including individual traits, species diversification and conservation. Over the past two decades, growing evidence of molecular markers (such as plastid markers, nuclear ribosomal DNA, mitochondrial or low-copy nuclear genes) has greatly advanced our understanding of orchid relationships, clarifying relationships among orchid subfamilies with morphologically confused taxa (Górniak et al., 2010; Guo et al., 2012; Chase et al., 2015; Deng et al., 2015; Givnish et al., 2015; Li et al., 2016). However, the relationship between subfamily Cypripedioideae and Vanilloideae has been controversial. For example, Deng et al. (2015); Givnish et al. (2015); Li et al. (2016) indicated that subfamily Vanilloideae (vs. Cypripedioideae; Cameron et al., 1999; Li et al., 2019) belonged to the second diverged taxa of the orchid family. In addition, recent molecular phylogenetic studies within the genus Cypripedium (nuclear ribosomal ITS, low copy nuclear gene (ACO) and chloroplast genes (matK, rbcL, trnH-psbA, atpI-atpH, trnS-trnfM and trnL-F) supported that the genus was monophyletic and roughly divided into 15 sections (Li et al., 2011; Chen et al., 2013; Szlachetko et al., 2020). But the internal structure is not well resolved, such as the sect. Retinervia and sect. Palangshanensia (Cox et al., 1997; Cribb, 1997; Eccarius, 2009).
Chloroplast is an important organelle that promotes the growth and development of most plants and plays an important role in the biosynthesis of plant carbohydrates, proteins and lipids (Daniell et al., 2016). The chloroplast genome consists of a helical, double-stranded genome with the aptitude to replicate independently of the nuclear genome (Palmer, 1985; Abdullah et al., 2021). Chloroplast genomes of land plants are typically circular DNA molecules with highly conserved regions, gene content, and gene order (Wicke et al., 2011). The average chloroplast genome size of land plants is 151 kb, with most species ranging from 130–170 kb in length, and the average GC content is 36.3% (Guo et al., 2021). A typical chloroplast genome consists of a pair of inverted repeats (IR) regions separated by a large single copy (LSC) region and a small single copy (SSC) region (Sugiura, 1992). In recent years, with the rapid development of next-generation sequencing technology, thousands of complete chloroplast genomes from various land plants have been sequenced (Yu et al., 2017), among which 394 were from Orchidaceae (NCBI1, accessed on April 7, 2021). To date, only five chloroplast genomes have been reported for this genus, Cypripedium japonicum Thunberg (Kim et al., 2015), C. formosanum Hayata (Lin et al., 2015), C. calceolus L. (Zhang et al., 2019), C. subtropicum Chen & Lang (Guo et al., 2021) and C. tibeticum King ex Rolfe (Guo et al., 2021). The lengths of all reported chloroplast genomes of Cypripedium (174,417–212,668 bp) were higher than the average of land plants (151 kb). The chloroplast genome of C. subtropicum was 212,668 bp, which is the largest known genome of orchids and the sixth largest of terrestrial plants (Guo et al., 2021). Chloroplast genome sequences have been widely recognized for phylogenetic and divergence history studies in flowering plants (Tonti-Filippini et al., 2017; Feng et al., 2020). Also, chloroplast genomes can provide unique and substantial information for the analysis of plant systematics and evolutionary relationships with matrilineal inheritance characteristics (Wang et al., 2016). Meanwhile, highly variable loci identified in the chloroplast genome can make significant contributions to future phylogenetic studies of the genus (Chen et al., 2015).
In the present study, we generated two newly sequenced chloroplast genomes of Cypripedium species and performed comparative genomic analysis in combination with five other published chloroplast genomes from this genus. We also included 47 whole plastid genomes and plastid protein-coding genes (CDSs) representing five subfamilies of Orchidaceae for phylogenetic analysis. Our aims were to (1) explore the patterns of long sequence repeats (LSRs) and simple sequence repeats (SSRs) in seven Cypripedium plastid genomes that cause significant expansion and contraction of the genome; (2) identify polymorphic loci for future phylogenetic inference of the genus Cypripedium; and (3) elucidate the molecular evolution and phylogenetic relationships of Cypripedium species and Orchidaceae.
Fresh leaf samples of Cypripedium palangshanense Tang & Wang and C. debile Rchb. were collected from the native environment in Wolong National Nature Reserve, Sichuan, China. The collected leaf samples were kept in silica gel and stored at the Herbarium of Chengdu Institute of Biology (CDBI). Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method (Allen et al., 2006). Sheared low molecular weight DNA fragments were used to construct paired-end (PE 150) libraries according to the protocol of the Illumina manual (Illumina, CA, United States). Completed libraries were pooled and sequenced in the Illumina NovaSeq 6000 platform with 350 bp insert size (Berry Genomics, Beijing, China).
Approximately 19 Gb of clean data for Cypripedium palangshanense and 15 Gb of clean data for C. debile were used to assemble the chloroplast genomes with GetOrganelle v1.7.2 (Jin et al., 2020). Bandage (Wick et al., 2015) was used to identify the circular maps to assess the quality of the assembly. The average coverage for the assembled chloroplast genomes was 770.6 × and 510.1 × for C. palangshanense and C. debile, respectively. The assembled chloroplast genomes were annotated using PGA (Qu et al., 2019) with chloroplast genomes of C. calceolus (NC_045400) and C. japonicum (NC_027227) as reference sequences. To quantify IR boundaries, raw sequencing reads were remapped to the 600-bp surroundings of the IR ends. Manual correction of genes with missing start and stop codons in annotations was performed using Geneious Prime 2021 (Biomatters Ltd., Auckland, New Zealand). The circular chloroplast genome maps were visualized using OGDRAW v1.3.132 (Greiner et al., 2019).
The sequence of Cypripedium calceolus, C. japonicum, C. subtropicum, C. tibeticum and C. formosanum was included in the comparative chloroplast genome analysis. The base content was determined using DNA Baser Sequence Assembler v5.152. To identify highly variable regions, the seven chloroplast genomes were aligned using the MAFFT v7.475 (Katoh and Standley, 2013) with default parameters. The number of polymorphic sites and nucleotide variability (Pi) were evaluated using a sliding window with 200-bp step size and a 600-bp window length implemented in DnaSP v5.10.1 (Librado and Rozas, 2009). Full alignments with annotation were visualized using the mVISTA software (Frazer et al., 2004), and gene arrangement was further analyzed by the Mauve alignment plugin in Geneious Prime 2021 (Biomatters Ltd., Auckland, New Zealand). The junction of chloroplast genomes was analyzed in IRscope (Amiryousefi et al., 2018) to visualize the expansion and contraction of inverted repeats.
Large sequence repeats (LSRs), including forward (F), reverse (R), complement (C) and palindrome (P) sequence repeats, were identified in whole chloroplast genome, LSC, SSC and IR regions using REPuter (Kurtz et al., 2001; Hamming distance = 3 and minimum repeat size of 30 bp). Simple sequence repeats (SSRs) (≥ 10 bp) were detected using MISA (Beier et al., 2017) with the minimum thresholds for mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide were set to 10, 5, 4, 3, 3, and 3, respectively. In addition, tandem repeats were identified with Tandem Repeats Finder v4.09 (Benson, 1999) with default parameters. The overlapped repeats of the results were removed manually.
The whole chloroplast genome sequences and protein-coding sequences of 45 Orchidaceae (Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae, and Epidendroideae) species were downloaded from the National Center for Biotechnology Information Search database (Supplementary Table 1). Phylogenetic relationships within Orchidaceae were reconstructed in combination with the 45 published accessions and the two newly generated Cypripedium chloroplast genomes. Two species of family Iridaceae (Iris dichotoma Pall., MK593157) and Amaryllidaceae (Lycoris sanguinea Maxim., NC_047453) were included as outgroups in the phylogenetic analysis. Before constructing the phylogenetic tree, we manually corrected all inversions using Geneious Prime 2021 to obtain consistent gene and base order. Phylogenetic analyses were performed based on the following two data sets: (1) the complete chloroplast genome sequences (IRa excluded); (2) the extracted sequences representing all coding sequences (CDSs). We used MAFFT v7.475 (Katoh and Standley, 2013) with default parameters to obtain aligned whole chloroplast genomes (IRa excluded) and CDSs, as well as manual adjustments where necessary. Three different methods including Maximum parsimony (MP), Maximum likelihood (ML) and Bayesian inference (BI) were employed in the phylogenetic analysis. The MP analysis based on the concatenated data set was carried out using PAUP v4.0b10 (Swofford, 2003), with a heuristic search with 1,000 random taxon stepwise addition sequences, tree bisection reconnection branch swapping, and 1,000 bootstrap replications. The ML analysis was performed using IQ-TREE v.1.4.241 (Nguyen et al., 2014), the ModelFinder in IQ-TREE tested a total of 286 DNA models and chose TIM + F + R3 as the best-fit nucleotide substitution model for the two data matrices, and branch support was estimated using 2,000 replicates of both SH-like approximate likelihood-ratio test (SH-aLRT) (Guindon et al., 2010) and the ultrafast bootstrapping algorithm (UFboot) (Minh et al., 2013). For BI analysis, the best-fit nucleotide substitution models (GTR + I + G) for the two data matrices were chosen based on the corrected Akaike Information Criterion (AICc) using jModeltest v2.1.6 (Posada, 2008) software. The BI analysis was conducted using MrBayes v3.2.7a (Ronquist and Huelsenbeck, 2003) with two parallel runs (20 million generations). The first 25% of trees from all runs were discarded as burn-in. The results were visualized in Figtree v1.4.43.
We obtained the whole chloroplast genomes of 207,142 bp for Cypripedium palangshanense and 162,773 bp for C. debile (GenBank accession Nos. MW924110 and MW924111, respectively). The chloroplast genomes of these two species showed a typical quadripartite structure containing a pair of IRs separated by an SSC region and an LSC region (Figure 1). The IR boundaries were quantified by the remapping of short reads, which showed above 500 × for the IR ends and surrounding areas (Supplementary Datasheet). The LSC region of C. palangshanense expanded to 128,862 bp, similar to C. subtropicum (129,998 bp), while the LSC region of C. debile was 89,446 bp, the smallest of the published Cypripedium genomes (Table 1). The IR regions of the two species (34,415 and 31,639 bp, respectively) were slightly larger than the other five species, but the SSC regions (9,450 and 10,049 bp, respectively) were smaller than previously sequenced species (Supplementary Table 2). The GC content of the two chloroplast genomes was uneven, with approximately 29.5% for C. palangshanense and 35.4% for C. debile. The GC content of the two chloroplast genomes varied considerably in the LSC region (24.7% and 32.7%, respectively) while similar in the IRs regions (38.6% and 40.1%, respectively) and in the SSC regions (both 29.0%) (Table 1). Accordingly, C. palangshanense had high proportions of A (37.6%) and T (37.8%) nucleotides and low proportions of G (11.6%) and C (12.1%) nucleotides in the LSC region, while the base/nucleotide composition in the SSC and IRs regions were similar to other five species (Supplementary Figure 1 and Supplementary Table 2).
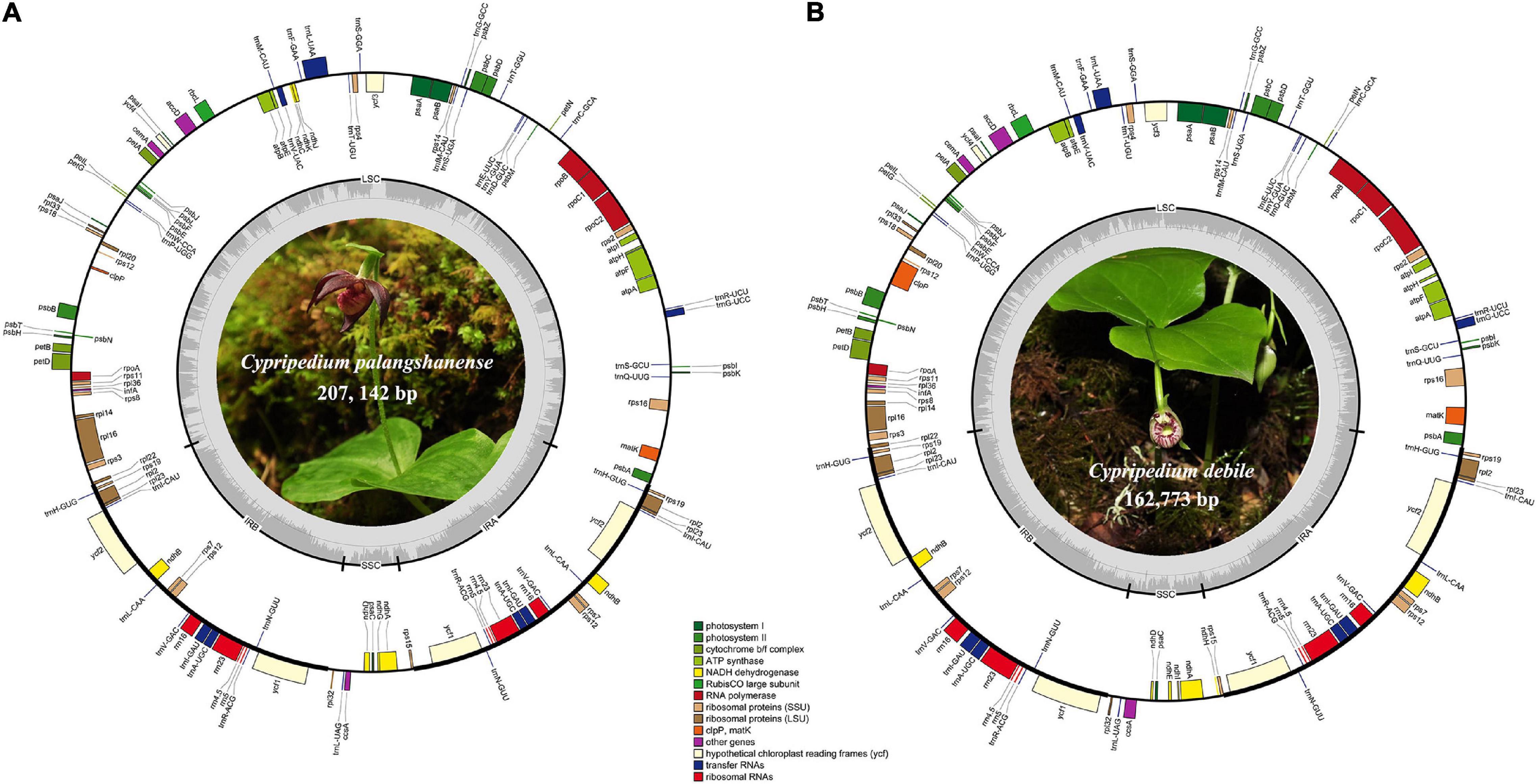
Figure 1. Chloroplast genome maps of C. palangshanense (A) and C. debile (B). Genes of different functional groups are shown in colored bars. The inner circle (dashed gray area) indicates the proportional GC content of the corresponding genes. Regions of the large single-copy (LSC), small single-copy (SSC) and inverted repeats (IRA and IRB) are indicated.
The chloroplast genomes of Cypripedium palangshanense and C. debile consisted of 128–130 genes, including 82–84 protein-coding genes, 38 tRNA genes, and 8 rRNA genes (Table 1). Each IR contain four rRNA genes (rrn4.5, rrn5, rrn16, rrn23), eight tRNA genes (trnH-GHG, trnL-CAU, trnL-CAA, trnV-GAC, trnL-GAU, trnA-UGC, trnR-ACG, trnN-GUU) and eight protein-coding genes (rps7, rps12, rps19, rpl2, rpl23, ndhB, ycf1, ycf2; Supplementary Table 3). A total of eleven genes contained one intron, including eight protein-coding genes (rps16, rpl2, rpl16, rpoC1, petB, petD, ndhA, ndhB) and three tRNA genes (trnG-UCC, trnK-UUU, trnA-UGC). Three protein-coding genes (atpF, rps12, clpP1) and three tRNA genes (trnL-UAA, trnV-UAC, trnA-UGC) contained two introns, while ycf3 gene which has four introns (Supplementary Table 3).
The chloroplast genomes of seven Cypripedium species showed a significant difference in genome size (162,773–212,668 bp; Table 1 and Supplementary Figure 2) and a 75-kb inversion (trnG-UCC to trnP-UGG) occurred in the LSC region for three species: C. subtropicum, C. tibeticum and C. formosanum (Supplementary Figure 2). The whole aligned sequences show high variability in the LSC and SSC regions, and high similarities in IRs except the ycf2-ndhB regions’ sequence identities significantly falling below 50% (Supplementary Figure 3). The LSC/IRb (JLB: rpl22) boundary and the LSC/IRa (JLA: rps19 & psbA) boundary were stable among the seven Cypripedium chloroplast genomes. However, the two newly sequenced species show great differences in the locations of the SSC/IRb (JSB) boundary and the SSC/IRa (JSA) boundary compared to previously published species. The JSB boundary was located between ycf1 gene and rpl32 gene in C. debile and exclusively on the rpl32 gene in C. palangshanense, while stable (ycf1 & ndhF) in other five species. The JSA boundary was located on the rpl15 gene in both C. debile and C. palangshanense, while on the ycf1 gene in the other five species (Figure 2).
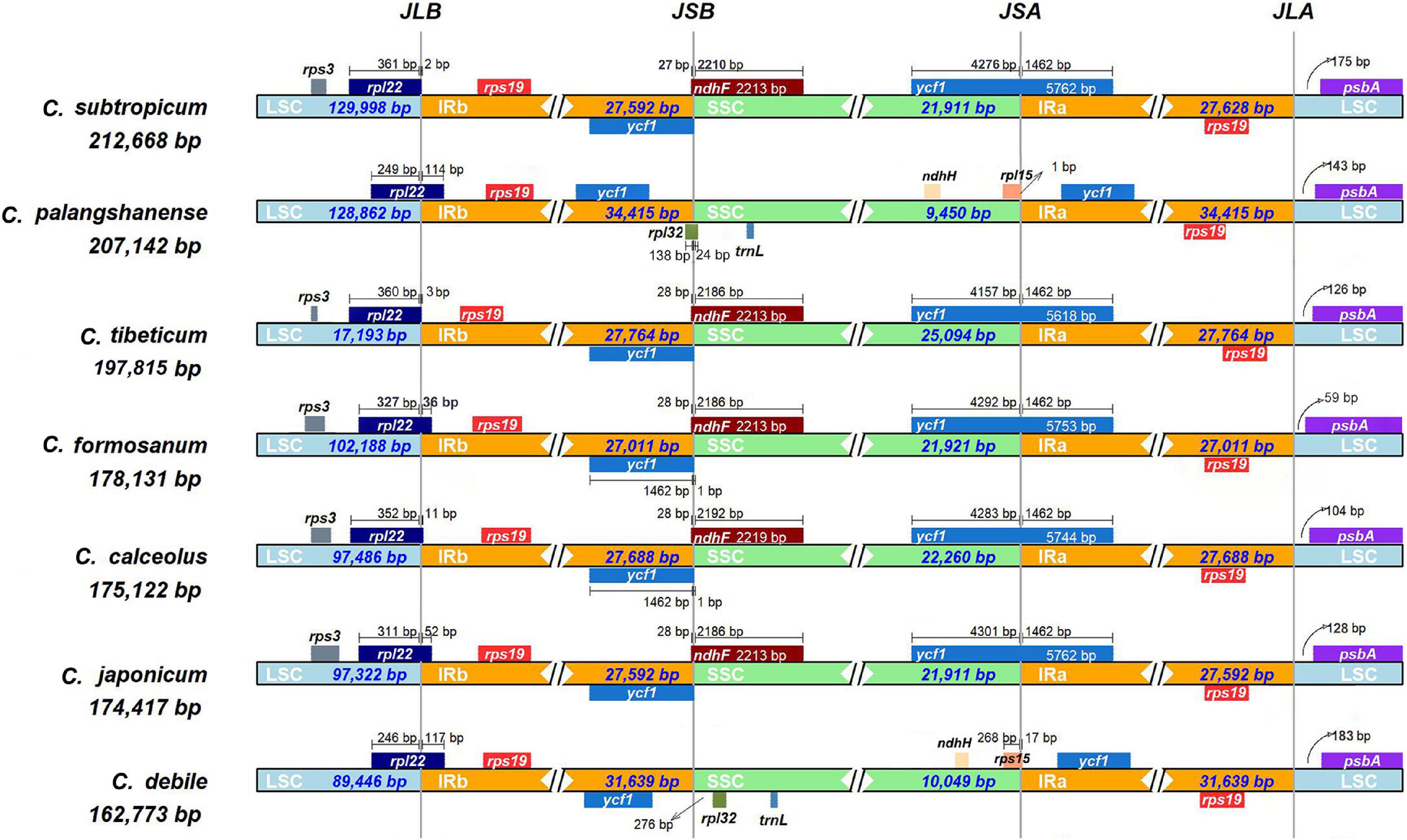
Figure 2. Analyses of expansion and contraction of inverted repeats in the seven Cypripedium chloroplast genomes.
A total of 7,929 variable (polymorphic) sites were found in 251,932 nucleotide loci, including 5,411 singleton variable sites (SVS) and 2,518 parsimony informative sites (PIS). Three different categories under SVS were observed, 5,133 sites with two variants (SV2V), 275 sites with three variants (SV3V) and 3 sites with four variants (SV4V). Similarly, PIS has three categories: 2,265 sites with two variants (PIS2V), 240 sites with three variants (PIS3V) and 13 sites with four variants (PIS4V; Supplementary Table 4). The window-based nucleotide variability (Pi) values for the alignment of the seven chloroplast genomes ranged from 0 to 0.28833. We identified 12 highly divergent regions (Pi > 0.09) with Pi values ranging from 0.09 to 0.28833, including nine intergenic spacer (IGS) regions (psbL-trnG, trnY-trnT, trnE-trnT, petA-psbJ, clpP1-psbT, psbB-psbT, ycf2-ndhB, trnT-trnL, and trnF-trnV) and three coding sequence regions (ndhD, ndhA and rps16; Figure 3; Supplementary Table 5).
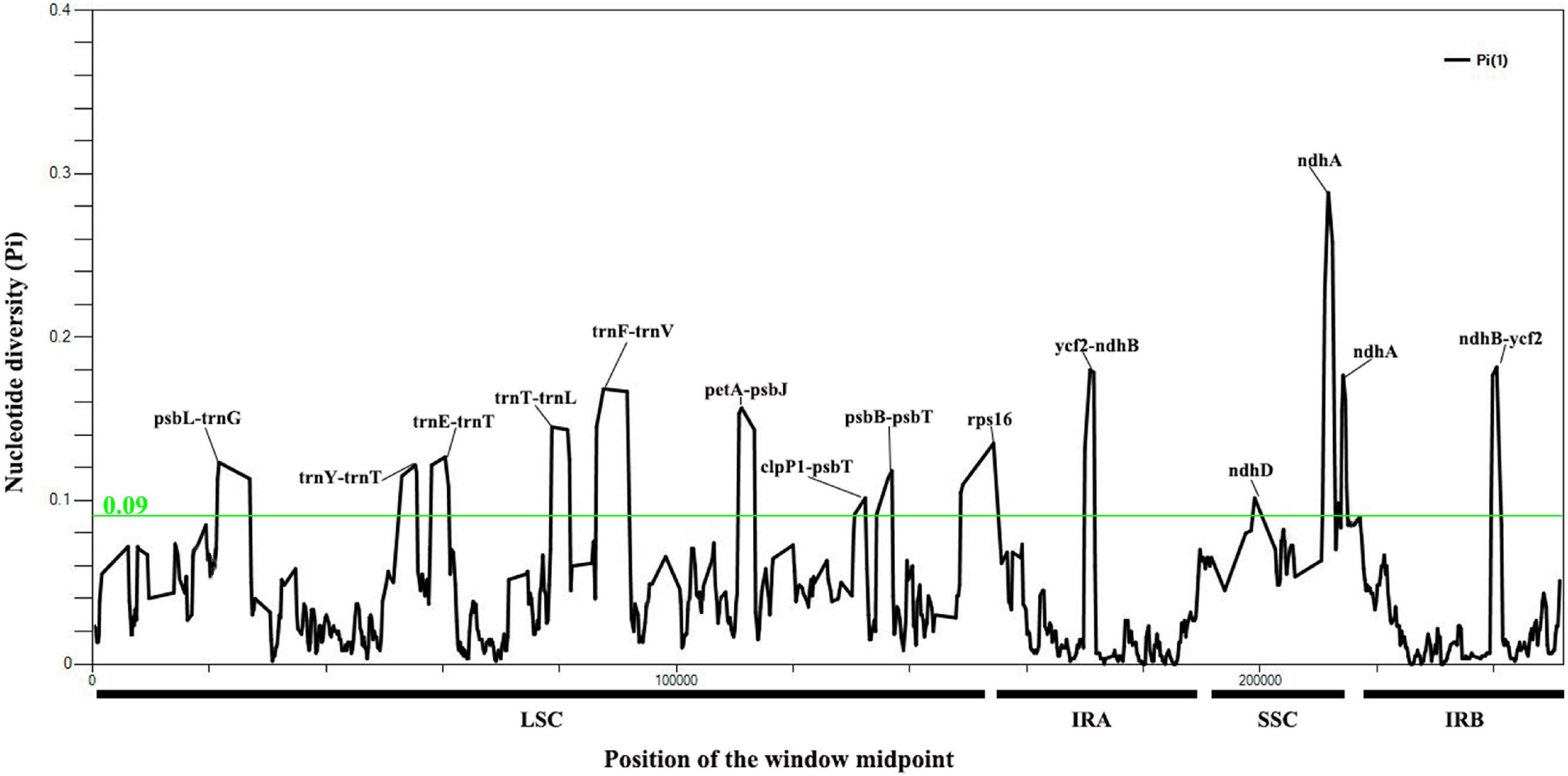
Figure 3. Nucleotide variability values compared between the seven chloroplast genomes of Cypripedium using the window sliding analysis. (X-axis indicates the position of the midpoint of the window, while Y-axis indicates the nucleotide diversity of each window).
A total of 2,192 SSRs were identified in the seven Cypripedium chloroplast genomes (Figure 4A and Supplementary Table 6). The mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide of these SSRs were account for 20.2, 24.7, 25.0, 13.5, 12.4 and 4.2%, respectively. The five dominant motif types in the SSRs were A/T, AT/TA, AAT/ATT, AAAT/ATTT, and AATAT/ATATT. Among 41 different SSR types, C. palangshanense had 3 unique types, C. subtropicum and C. tibeticum had 2 unique types, while C. formosanum had one unique type (Figure 4A and Supplementary Table 6). A total of 786 large sequence repeats (LSRs; ≥ 30 bp and Hamming distance = 3) were identified in the seven Cypripedium chloroplast genomes (Figure 4B and Supplementary Table 7). In general, F repeats (386) were the most common type, while C repeats (54) were the least. Among the seven species, C. japonicum (87) contained the least LSRs, and C. palangshanense contained the most (154; Figure 4B and Supplementary Table 7). In the LSC region, the seven Cypripedium species had similar numbers of LSRs (40–50) but showed significant differences in the SSC (3–50) and IR (4–50) regions (Figures 4C-E and Supplementary Table 8).
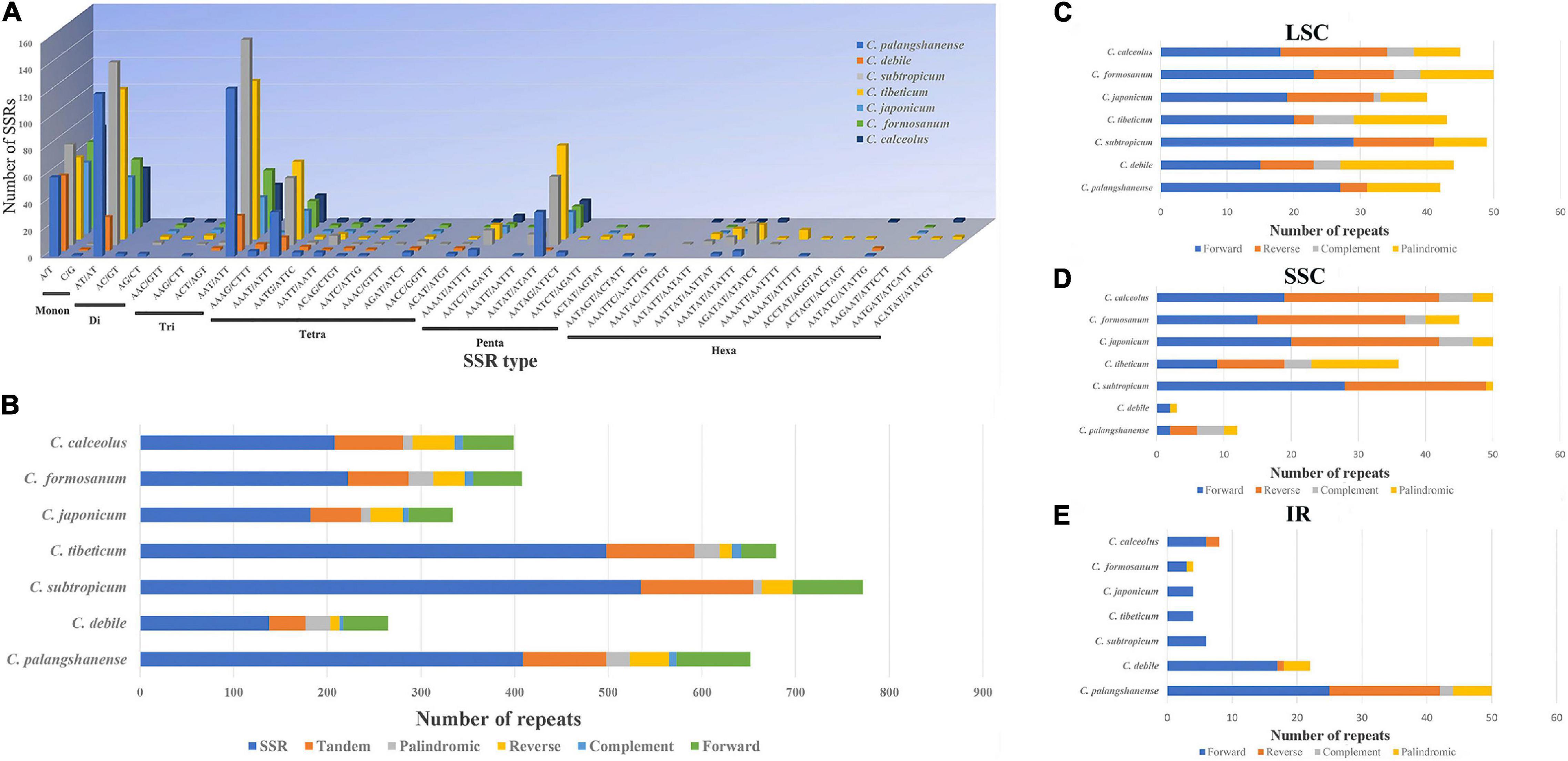
Figure 4. Analyses of repeat sequences in seven chloroplast genomes. (A) Number of different SSRs types. (B) Number of SSRs, tandem repeats and large LSRs. (C) Number of different LSRs types in the LSC regions. (D) Number of different LSRs types in the SSC region. (E) Number of different LSRs types in the IR region.
The phylogenetic analysis using MP, BI and ML methods based on whole chloroplast genomes (IRa excluded) and CDSs yielded identical topologies with high support values (Figure 5 and Supplementary Figures 4–8). Thus, we present the topology resulting from ML analysis based on CDSs, with posterior probability (PP), maximum likelihood (BSML) and maximum parsimony (BSMP) bootstrap values labeled on the tree branches. Our phylogenetic tree indicated that the subfamily Apostasioideae (PP = 1, BSML = 100%, BSMP = 100%) diverged first and was sister to remaining taxa of the orchid family, followed by Vanilloideae (PP = 1, BSML = 100%, BSMP = 100%), which is sister to a strongly supported group (PP = 1, BSML = 100%, BSMP = 98%) comprising Cypripedioideae (PP = 1, BSML = 100%, BSMP = 100%), Orchidoideae (PP = 1, BSML = 100%, BSMP = 100%) and Epidendroideae (PP = 1, BSML = 100%, BSMP = 100%). Within the subfamily Cypripedioidea, we resolved a strongly supported clade consisting of the genus Paphiopedium (PP = 1, BSML = 100%, BSMP = 100%) and Cypripedium (PP = 1, BSML = 100%, BSMP = 100%). Our phylogenetic analyses also strongly supported (PP = 1, BSML ≥ 90%, BSMP ≥ 92%) the monophyly of the sections of genus Cypripedium, sect Cypripedium, Flabellinervia, Subtropica, Retinervia and Palangshanensia.
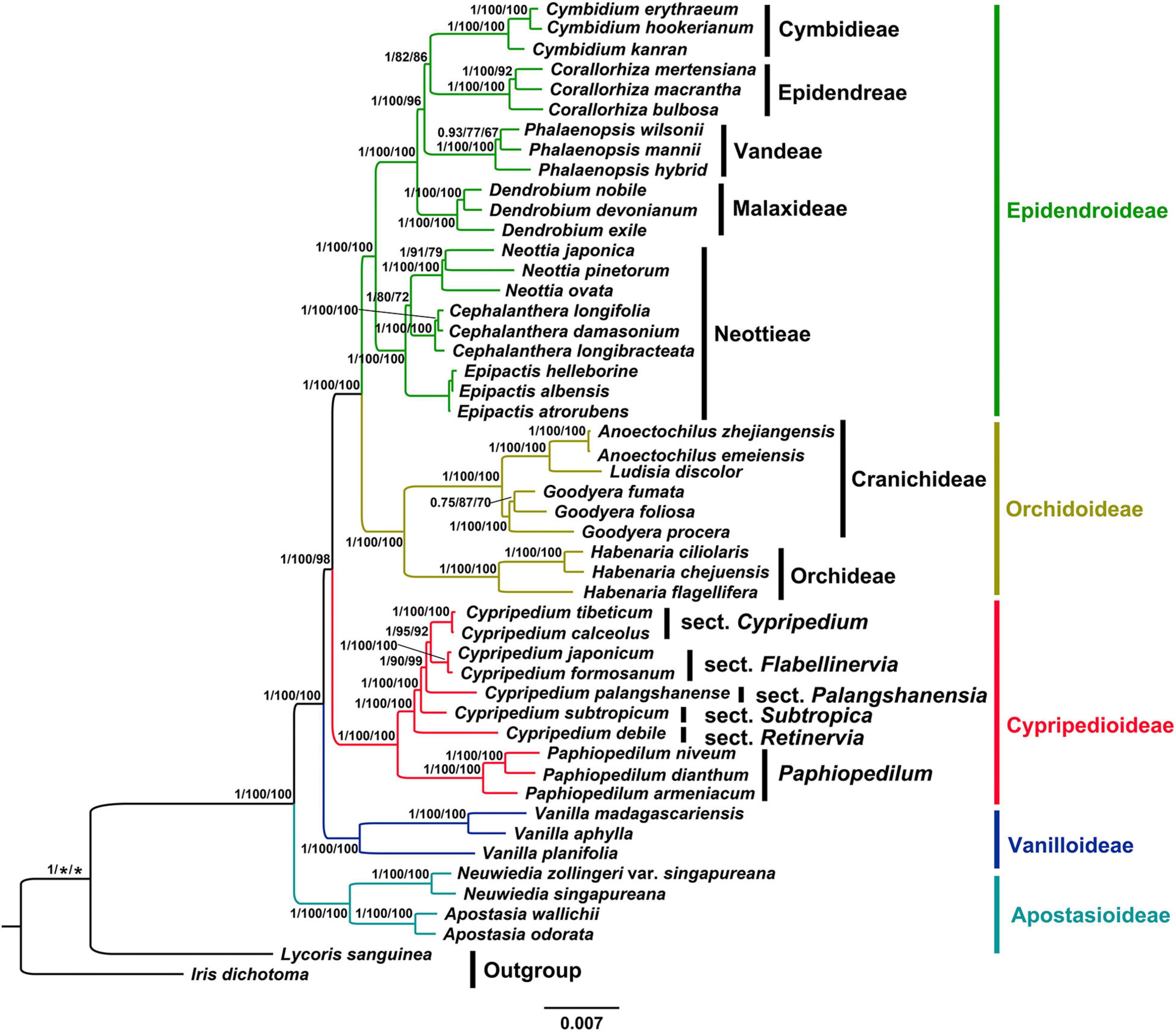
Figure 5. Phylogram depicting the relationships of the plastid CDSs of 47 different Orchidaceae species using ML method. Numbers associated with nodes indicated Bayesian posterior probabilities (PP), maximum likelihood (BSML) and maximum parsimony (BSMP) bootstrap values, respectively. The PP on the left, BSML in the middle, BSMP on the right.
The chloroplast genome of the newly generated Cypripedium debile in this study is the smallest in the genus, while the previously published genome of C. subtropicum remains the largest in the genus, as well as in the Orchid family. The chloroplast genomes of Cypripedium (162,773–212,668 bp) were larger than the average of sequenced land plants (151 kb), while the GC content (28.2–35.4%) was lower than the average of sequenced land plants (37.6%; Guo et al., 2021). The IR regions of the two newly reported chloroplast genomes (31,639–34,415 bp) were significantly larger than five published Cypripedium (27,579–27,764 bp), while the SSC regions were the opposite (9,450–10,049 bp and 27,414–31,911 bp; Table 1). This indicates the expansion of IR into SSC region has contributed the most to the decrease of SSC region in C. debile and C. palangshanense, which has been reported in Corydalis species (Xu and Wang, 2021).
Among the seven species, three species (Cypripedium tibeticum, C. palangshanense and C. subtropicum, 197,815–212,668 bp) with larger chloroplast genomes had lower GC content (28.2–30.5%), while the other four species (C. debile, C. japonicum, C. calceolus and C. formosanum, 162,773–178,131 bp) with smaller chloroplast genomes had higher GC content (33.9–35.4%). In addition, we find all Cypripedium species had much lower GC content in the LSC and SSC regions (23.7–31.7% and 20.6–29.0%, respectively) than in the IR region (38.6–42.7%) due to the reduction of AT nucleotides in the four rRNA genes (rrn23s, rrn16s, rrn4.5s, and rrn5s), which is also reported in Meng et al. (2018); Hishamuddin et al. (2020). Meanwhile, this study suggested that the chloroplast genomes of Cypripedium showed very high level of A + T content (64.6%–71.8%) and low level of G + C content (28.2%–35.4%), a feature rarely observed in chloroplast genome sequences of many land plants (Bi et al., 2018). The high AT content was caused by repetitive sequences consisting of poly (A), poly (T) or poly (AT) regions in the non-coding regions of the single-copy regions, especially in the LSC region (Kim et al., 2015).
The gene order of the two newly sequenced chloroplast genomes in this study is conserved without gene rearrangement and inversion events, but a long inversion (75-kb) occurred in the LSC region in three published Cypripedium species (C. subtropicum, C. tibeticum and C. formosanum), resulted in very high variability of this region in the whole aligned sequences (Supplementary Figures 2, 3). Interestingly, remarkable divergence was also observed in the SSC region, where no inversion event occurred (Supplementary Figure 3). Meanwhile, we found that four ndh genes (ndhE, ndhF, ndhH, and ndhI) in C. palangshanense disappeared in the SSC region compared to C. tibeticum, C. japonicum, C. calceolus, C. formosanum, and C. subtropicum, as well as three ndh genes (ndhF, ndhG and ndhH) in C. debile were lost in the SSC region (Figure 1). A study by Lin et al. (2015) showed that in C. formosanum, the ndh genes transferred from the chloroplast genome to the mitochondrial genome. Thus, a similar phenomenon may occur in the two newly sequenced species. Also, different degrees of loss or deletion of the ndh genes among species in Cypripedium resulted in significant variability in the SSC region. Furthermore, gene loss or pseudogenization has partially counteracted the increase of chloroplast genomes size in this genus, which showed a similar expansion-contraction mechanism in Corydalis (Xu and Wang, 2021) and Pelargonium (Weng et al., 2017).
Highly variable regions have potential for species identification and wide-range phylogenetic analysis (Zhang et al., 2009). In the present study, the ndhA located in the SSC region had the highest nucleotide variation (Pi = 0.28833; Figure 3), which may be associated with the expansion and contraction of IRs (Goulding et al., 1996) and the transfer of ndh from the chloroplast genome to the mitochondrial genome (Lin et al., 2015). The IR region is relatively conserved except in the region between ycf2 and ndhB (Figure 3 and Supplementary Figure 3). The introns of rps16 and intergenic regions with relatively high divergence values located in the LSC region (psbL-trnG, trnY-trnT, trnE-trnT, trnT-rnL, trnF-trnV, petA-psbJ, clpP1-psbT, and psbB-psbT) were associated with repetitive sequences (Figure 3). For example, a various number of repetitive sequences can be found in the IGS region of psbL-trnG, trnF-trnV, trnT-trnL and petA-psbJ. The highly polymorphic regions identified in this study have potential to be exploited as candidate barcode sequences in the phylogenetic analysis of Cypripedium. Further work is needed to verify whether these markers can be recommended as valid barcodes for species of the genus Cypripedium.
SSRs (≥ 10 bp) are small repetitive units of chloroplast DNA, together with LSRs, have played an important role in the evolution of the chloroplast genome and may contribute to the development of future molecular markers (Zhang et al., 2016; Li et al., 2021). Interestingly, the three larger chloroplast genomes (Cympripedium tibeticum, C. palangshanense, and C. subtropicum; 98–154 LSRs, and 409–535 SSRs; Figure 4B and Supplementary Table 7) had more LSRs and SSRs than the four smaller ones (C. debile, C. japonicum, C. calceolus, and C. formicum; 87–114 LSRs, and 138–222 SSRs). This indicates increase of LSRs and SSRs led to the obvious enlargement of these chloroplast genomes (C. tibeticum, C. palangshanense, and C. subtropicum). Moreover, in the IR region, the number of LSRs of the two newly sequenced chloroplast genomes (22–50) is approximately six times higher than the five published plastids (4–8). At the SSC region, the number of LSRs of the two newly sequenced chloroplast genomes (3–12) are approximately one-fifth of the five published plastids (36–50). Thus, we speculate that the increase of LSRs in the IR regions of C. debile and C. palangshanense led to the marked enlargement of their IR regions, while the decrease of LSRs in the SSC region of C. debile and C. palangshanense that led to the significantly smaller of their SSC region. Furthermore, we find that the coding region of this genus is conserved and that chloroplast genome expansion is closely associated with the proliferation of IGS regions, especially in the LSC region, which is also reported in Guo et al. (2021). And frequent variation in the repeat region also plays an important role in the variation and sequence rearrangement of the chloroplast genome (Zhang et al., 2016; Yuan et al., 2017).
Although chloroplast genome data-set has been considered as a single locus due to their uniparental inheritance, a growing number of studies indicate the implementation of complete chloroplast genome data-set has the potential to resolve the phylogenetic relationships of controversial genus (Wolfe et al., 1987; Green, 2011; Wicke et al., 2011; Schwarz et al., 2017; Gonçalves et al., 2019; Zhang et al., 2020). Our phylogenetic tree based on both whole chloroplast genomes (IRa excluded) and CDSs sequences resolved Cypripedium as a monophyletic taxon with high support values (PP = 1, BSML = 100%, BSMP = 100%), which is consistent with the results from the plastid markers (matK, rbcL, trnH-psbA, trnS-trnfM, atpI-atpH, vspace*-0.5pttextittrnL intron, and trnL-F; Cox et al., 1997; Fatihah et al., 2011; Li et al., 2011) and nuclear ribosomal ITS analyses (Fatihah et al., 2011; Szlachetko et al., 2020). The inter-section relationships of the genus were resolved as: (sect. Retinervia (Subtropica (Palangshanensia (Flabellinervia, Cypripedium)))). However, the systematic position of sect. Retinervia and Palangshanensia has been controversial. The traditional Retinervia (C. debile, C. palangshanense and C. elegans Reichenbach) is described by Cribb (1997) based on the following morphological characteristics: two opposite leaves, situated near the middle of the stem and produce a single-flowered inflorescence terminated by the smallest flower within the genus with tepals distinctly longer than the lip. Recently, Li et al. (2011) firstly showed that C. palangshanense is not included in the sect. Retinervia based on five plastid markers and nuclear gene. Then Chen et al. (2013) first proposed to separate C. palangshanense from the sect. Retinervia to create sect. Palangshanensia based on the phylogenetic results of Li et al. (2011). Finally, both results from ITS/ACO (BS = 84, PP = 1.0; Szlachetko et al., 2020) and our whole chloroplast genomes (IRa excluded) and CDSs analysis support the establishment of the sect. Palangshanensia. Unfortunately, C. elegans of this section was not included in this study, and further studies are needed to verify its systematic position in the future.
The results of the phylogenetic inference based on whole chloroplast genomes (IRa excluded) and CDSs support the relationships between the five subfamilies of the orchid family: Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae, and Epidendroideae, which is consistent with previous studies (Cameron et al., 1999; Guo et al., 2012; Chase et al., 2015; Deng et al., 2015; Givnish et al., 2015; Li et al., 2016, 2019). In the present study, the subfamily Apostasioideae (including Neuwiedia and Apostasia) diverged firstly and is sister to all the other subfamilies in Orchidaceae, which is congruent with morphological characters (Pridgeon et al., 1999). Subfamily Vanilloideae diverged secondly, followed by subfamily Cypripedioideae, which is consistent with previous studies (Guo et al., 2012; Chase et al., 2015; Deng et al., 2015; Givnish et al., 2015; Li et al., 2016). The subfamily Orchidoideae, consisting of approximately 190 genera and 3600 species, is the second largest subfamily within Orchidaceae (Li et al., 2016). The researchers reached a consensus on the four orchid genera sampled in this study (Cameron et al., 1999; Li et al., 2016), which were divided into two clades, Cranichideae and Orchidaceae. The subfamily Epidendroideae is the most complicated taxon, representing a greater number of genera (ca. 650 genera) and species (ca. 18,000 species) than the total number of the other four subfamilies (Li et al., 2016). In this study, we selected several genera of key nodes in the subfamily and revealed their phylogenetic relationships. Our phylogenetic tree indicated that the tribe Neottieae diverged as sister to the remaining Epidendroideae, followed by the tribe Malaxideae, which is sister to three well-supported tribes comprising Vandeae, Epidendreae and Cymbidieae. However, the relationship among these three tribes has been controversial as well. Chase et al. (2015); Li et al. (2016) supported the topology (Cymbidieae (Vandeae, Epidendreae)) based on low-copy nuclear gene (Xdh) and plastid markers (rbcL, matK, psaB, ycf1). Givnish et al. (2015); Li et al. (2019) supported another topology (Epidendreae (Vandeae, Cymbidieae)) based on plastid and mitochondrial genomes. Thus, future studies with extensive taxon sampling and molecular and/or morphological evidence are needed to provide a higher resolution of the relationships among three tribes in this subfamily.
In the present study, we reported the complete chloroplast genomes of two newly sequenced Cypripedium species for comparative genomic analysis with five other published species. We revealed the mechanism of significant genome amplification of this genus and discussed that expansion of the IR region leads to gene pseudogenization or loss in the SSC region. Also, identification of polymorphic loci and molecular markers was performed, which will be useful for species identification and determination of phylogenetic relationships in the future. We made efforts to enrich the genomic resources of Cypripedium, which may help to promote the conservation of these endangered species. Meanwhile, the data-sets of the whole chloroplast genomes (IRa excluded) and CDSs sequences provided new insights in addressing the phylogeny of Orchidaceae, as well as genetic resources for further phylogenetic studies for this family. Future studies should be complemented by larger sample sizes to elucidate the phylogenetic relationship of these species.
The chloroplast genome sequences of Cypripedium palangshanense and C. debile were submitted to the National Center for Biotechnology Information (NCBI) and the accession numbers were: MW924110 and MW924111, respectively. Raw reads were submitted to the NCBI database under the BioProject number: PRJNA838021.
BX and JZ designed the study. YC, WJ, and HD collected the plant materials. JZ, YF, AP-M, and XL performed the data analysis. JZ and ML drafted the manuscript. BX, ML, YF, and AP-M revised the manuscript. All authors reviewed and approved the final manuscript.
This work was supported by the National Key Research and Development Program of China (Grant No. 2020YFE0203200), the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (Grant No. 2019QZKK0502), and Wild Plants Sharing and Service Platform of Sichuan Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Qi Yu for help with sample collection.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.911702/full#supplementary-material
Abdullah, Henriquez, C. L., Mehmood, F., Hayat, A., Sammad, A., Waseem, S., et al. (2021). Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae). Genomics 113, 183–192. doi: 10.1016/j.ygeno.2020.12.016
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Amiryousefi, A., Hyvönen, J., and Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Beier, S., Thiel, T., Münch, T., Scholz, U., and Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bi, Y., Zhang, M. F., Xue, J., Dong, R., Du, Y. P., and Zhang, X. H. (2018). Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci. Rep. 8:1184. doi: 10.1038/s41598-018-19591-9
Cameron, K. M., Chase, M. W., Whitten, W. M., Kores, P. J., Jarrell, D. C., Albert, V. A., et al. (1999). A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am. J. Bot. 86, 208–224. doi: 10.2307/2656938
Chase, M. W., Cameron, K. M., Barrett, R. L., and Freudenstein, J. V. (2003). “DNA data and Orchidaceae systematics: a new phylogenetic classification,” in Orchid Conservation, eds K. W. Dixon, S. P. Kell, R. L. Barrett, and P. J. Cribb (Kota Kinabalu: Natural History Publications), 69–89.
Chase, M. W., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., van-den-Berg, C., et al. (2015). An updated classification of Orchidaceae. Bot. J. Linn. Soc. 177, 151–174. doi: 10.1111/boj.12234
Chen, J., Hao, Z. D., Xu, H. B., Yang, L. M., Liu, G. X., Sheng, Y., et al. (2015). The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng. Front. Plant Sci. 6:447. doi: 10.3389/fpls.2015.00447
Chen, S. C., and Cribb, P. J. (2005). “Cypripedium,” in Flora of China, eds Z. Y. Wu, P. H. Raven, and D. Y. Hong (Beijing: Science Press), 22–33.
Chen, S. C., Liu, Z. J., Chen, L. J., and Li, L. Q. (2013). The Genus Cypripedium in China. Beijing: Science Press, 10–12.
Cox, A. V., Pridgeon, A. M., Albert, V. A., and Chase, M. W. (1997). Phylogenetics of the slipper orchids (Cypripedioideae, Orchidaceae): nuclear rDNA ITS sequences. Plant Syst. Evol. 208, 197–223. doi: 10.1007/BF00985442
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17:134. doi: 10.1186/s13059-016-1004-2
Deng, H., Zhang, G. Q., Lin, M., Wang, Y., and Liu, Z. J. (2015). Mining from transcriptomes: 315 single-copy orthologous genes concatenated for the phylogenetic analyses of Orchidaceae. Ecol. Evol. 5, 3800–3807. doi: 10.1002/ece3.1642
Farrell, L. (1999). Cypripedium calceolus L. (Orchidaceae): lady’s slipper orchid. Br. Red Data Books.1 Vascular Plants 1:114.
Fatihah, N. H. N., Fay, M. F., and Maxted, N. (2011). Molecular phylogenetics of Cypripedium L. (Cypripedioideae: Orchidaceae) based on plastid and nuclear DNA sequences. J. Agrobiotechnol. 2, 35–51.
Feng, Y., Comes, H. P., and Qiu, Y. X. (2020). Phylogenomic insights into the temporal-spatial divergence history, evolution of leaf habit and hybridization in Stachyurus (Stachyuraceae). Mol. Phylogenet. Evol. 150:106878. doi: 10.1016/j.ympev.2020.106878
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279. doi: 10.1093/nar/gkh458
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B Biol. Sci. 282:20151553. doi: 10.1098/rspb.2015.1553
Gonçalves, D. J. P., Simpson, B. B., Ortiz, E. M., Shimizu, G. H., and Jansen, R. K. (2019). Incongruence between gene trees and species trees and phylogenetic signal variation in plastid genes. Mol. Phylogenet. Evol. 138, 219–232. doi: 10.1016/j.ympev.2019.05.022
Górniak, M., Paun, O., and Chase, M. W. (2010). Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol. Phylogenet. Evol. 56, 784–795. doi: 10.1016/j.ympev.2010.03.003
Goulding, S. E., Wolfe, K. H., Olmstead, R. G., and Morden, C. W. (1996). Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 252, 195–206. doi: 10.1007/BF02173220
Green, B. R. (2011). Chloroplast genomes of photosynthetic eukaryotes. Plant J. 66, 34–44. doi: 10.1111/j.1365-313X.2011.04541.x
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guo, Y. Y., Luo, Y. B., Liu, Z. J., and Wang, X. Q. (2012). Evolution and biogeography of the slipper orchids: eocene vicariance of the conduplicate genera in the Old and New World Tropics. PLoS One 7:e38788. doi: 10.1371/journal.pone.0038788
Guo, Y. Y., Yang, J. X., Li, H. K., and Zhao, H. S. (2021). Chloroplast genomes of two species of Cypripedium: expanded genome size and proliferation of AT-biased repeat sequences. Front. Plant Sci. 12:142. doi: 10.3389/fpls.2021.609729
Hishamuddin, M. S., Lee, S. Y., Ng, W. L., Ramlee, S. I., Lamasudin, D. U., and Mohamed, R. (2020). Comparison of eight complete chloroplast genomes of the endangered Aquilaria tree species (Thymelaeaceae) and their phylogenetic relationships. Sci. Rep. 10:13034. doi: 10.1038/s41598-020-70030-0
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., Depamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241. doi: 10.1186/s13059-020-02154-5
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, J. S., Kim, H. T., and Kim, J. H. (2015). The largest plastid genome of monocots: a novel genome type containing AT residue repeats in the slipper orchid Cypripedium japonicum. Plant Mol. Biol. Rep. 33, 1210–1220. doi: 10.1007/s11105-014-0833-y
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Li, J. h, Liu, Z. J., Salazar, G. A., Bernhardt, P., Perner, H., Tomohisa, Y., et al. (2011). Molecular phylogeny of Cypripedium (Orchidaceae: Cypripedioideae) inferred from multiple nuclear and chloroplast regions. Mol. Phylogenet. Evol. 61, 308–320. doi: 10.1016/j.ympev.2011.06.006
Li, L., Hu, Y., He, M., Zhang, B., Wu, W., Cai, P., et al. (2021). Comparative chloroplast genomes: insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genomics 22:138. doi: 10.1186/s12864-021-07427-2
Li, M. H., Zhang, G. Q., Lan, S. R., and Liu, Z. J. (2016). A molecular phylogeny of Chinese orchids. J. Syst. Evol. 54, 349–362. doi: 10.1111/jse.12187
Li, Y. X., Li, Z. H., Schuiteman, A., Chase, M. W., Li, J. W., Huang, W. C., et al. (2019). Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol. Phylogenet. Evol. 139:106540. doi: 10.1016/j.ympev.2019.106540
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, C. S., Chen, J. J. W., Huang, Y. T., Chan, M. T., Daniell, H., Chang, W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 5:9040. doi: 10.1038/srep09040
Mcgough, H. N., Roberts, D. L., Brodie, C., and Kowalczyk, J. (2006). CITES and Slipper Orchids: an Introduction to Slipper Orchids Covered by the Convention on Internal Trade in Endangered Species. Kew: The Board of Trustees, Royal Botanic Gardens.
Meng, J., Li, X., Li, H., Yang, J., Wang, H., and He, J. (2018). Comparative analysis of the complete chloroplast genomes of four Aconitum medicinal species. Molecules 23:1015. doi: 10.3390/molecules23051015
Minh, B. Q., Nguyen, M. A. T., and von-Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Nelson, E. C. (1994). Historical data from specimens in the herbarium, National Botanic Gardens, Glasnevin, Dublin (DBN), especially on Cypripedium calceolus. BSBI News 67, 21–22.
Nguyen, L. T., Schmidt, H. A., von-Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-Likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Palmer, J. D. (1985). Comparative organization of chloroplast genomes. Annual Review of Genetics 19, 325–354. doi: 10.1146/annurev.ge.19.120185.001545
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Pridgeon, A. M., Cribb, P. J., Chase, M. W., and Rasmussen, F. N. (1999). Genera Orchidacearum Vol. 1. General Introduction, Apostasioideae, Cypipedioideae. Oxford: Oxford University Press.
Qu, X. J., Moore, M. J., Li, D. Z., and Yi, T. S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15:50. doi: 10.1186/s13007-019-0435-7
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Schwarz, E. N., Ruhlman, T. A., Weng, M. L., Khiyami, M. A., Sabir, J. S. M., Hajarah, N. H., et al. (2017). Plastome-wide nucleotide substitution rates reveal accelerated rates in Papilionoideae and correlations with genome features across Legume subfamilies. J. Mol. Evol. 84, 187–203. doi: 10.1007/s00239-017-9792-x
Sugiura, M. (1992). The chloroplast genome. Plant Mol. Biol. 19, 149–168. doi: 10.1007/978-94-011-2656-4_10
Swofford, D. (2003). PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), Version 4.0 b10. Sunderland: Sinauer Associates.
Szlachetko, D. L., Górniak, M., Kowalkowska, A. K., Kolanowska, M., Jurczak-Kurek, A., and Morales, F. A. (2020). The natural history of the genus Cypripedium (Orchidaceae). Plant Biosyst. Int. J. Deal. Aspects Plant Biol. 155, 772–796. doi: 10.1080/11263504.2020.1785963
Tonti-Filippini, J., Nevill, P., Dixon, K., and Small, I. (2017). What can we do with 1000 plastid genomes? Plant J. Cell Mol. Biol. 90, 808–818. doi: 10.1111/tpj.13491
Wang, Y., Zhan, D. F., Jia, X., Mei, W. L., Dai, H. F., Chen, X. T., et al. (2016). Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) Gilg and evolution analysis within the malvales order. Front. Plant Sci. 7:280. doi: 10.3389/fpls.2016.00280
Weng, M. L., Ruhlman, T. A., and Jansen, R. K. (2017). Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 214, 842–851. doi: 10.1111/nph.14375
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Wicke, S., Schneeweiss, G. M., dePamphilis, C. W., Müller, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. 84:9054. doi: 10.1073/pnas.84.24.9054
Xu, X., and Wang, D. (2021). Comparative chloroplast genomics of Corydalis species (Papaveraceae): evolutionary perspectives on their unusual large scale rearrangements. Front. Plant Sci. 11:600354. doi: 10.3389/fpls.2020.600354
Yu, X. Q., Drew, B. T., Yang, J. B., Gao, L. M., and Li, D. Z. (2017). Comparative chloroplast genomes of eleven Schima (Theaceae) species: insights into DNA barcoding and phylogeny. PLoS One 12:e0178026. doi: 10.1371/journal.pone.0178026
Yuan, C., Zhong, W. J., Mou, F. S., Gong, Y. Y., Pu, D. Q., Ji, P. C., et al. (2017). The complete chloroplast genome sequence and phylogenetic analysis of Chuanminshen (Chuanminshen violaceum Sheh et Shan). Physiol. Mol. Biol. Plants 23, 35–41. doi: 10.1007/s12298-016-0395-6
Zhang, J., Wang, J., Xia, T., and Zhou, S. (2009). DNA barcoding: species delimitation in tree peonies. Sci. China Series C Life Sci. 52, 568–578. doi: 10.1007/s11427-009-0069-5
Zhang, L. J., Ding, R., Meng, W. W., Hu, H. L., Chen, X. H., and Wu, H. H. (2019). The complete chloroplast genome sequence of the threatened Cypripedium calceolus (Orchidaceae). Mitochondrial DNA Part B 4, 4220–4222. doi: 10.1080/23802359.2019.1693933
Zhang, X., Sun, Y., Landis, J. B., Lv, Z., Shen, J., Zhang, H., et al. (2020). Plastome phylogenomic study of Gentianeae (Gentianaceae): widespread gene tree discordance and its association with evolutionary rate heterogeneity of plastid genes. BMC Plant Biol. 20:340. doi: 10.1186/s12870-020-02518-w
Keywords: Cypripedium, chloroplast genome, IR expansion, molecular markers, Orchidaceae, phylogenomics
Citation: Zhang J-Y, Liao M, Cheng Y-H, Feng Y, Ju W-B, Deng H-N, Li X, Plenković-Moraj A and Xu B (2022) Comparative Chloroplast Genomics of Seven Endangered Cypripedium Species and Phylogenetic Relationships of Orchidaceae. Front. Plant Sci. 13:911702. doi: 10.3389/fpls.2022.911702
Received: 03 April 2022; Accepted: 25 May 2022;
Published: 22 June 2022.
Edited by:
Guo-Qiang Zhang, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 Zhang, Liao, Cheng, Feng, Ju, Deng, Li, Plenković-Moraj and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Xu, eHVib0BjaWIuYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.