
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 July 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.909098
The TGA transcription factors are known to modulate the biosynthesis of secondary metabolites in plants. However, their regulatory function in natural rubber (NR) biosynthesis was not revealed in the rubber tree (Hevea brasiliensis). Here, 14 genes encoding TGA transcription factors (name HbTGA1-HbTGA14) were identified in the rubber tree. HbTGAs were differentially expressed in different tissues. HbTGA1 was expressed at its highest level in latex. We found specific in vitro and in vivo binding of the HbTGA1 protein with promoters of multiple NR biosynthesis genes (HbHMGS2, HbHMGR2, HbCPT6, HbCPT8, and HbSRPP2). The activation of the promoters of HbHMGS2 and HbCPT6 was significantly suppressed by HbTGA1, while the activities of promoters of HbHMGR2, HbCPT8, and HbSRPP2 were increased by HbTGA1. The promoter activities of HbHMGS2, HbHMGR2, HbCPT6, HbCPT8, and HbSRPP2 were significantly increased by HbTGA1 under jasmonate stress, while the promoter activities of HbHMGS2, HbHMGR2, HbCPT6, HbCPT8, and HbSRPP2 were also significantly increased by HbTGA1 under salicylic acid stress. The present study provides insights into the role of TGA transcription factors in regulating the expression of NR biosynthesis genes from H. brasiliensis.
Rubber tree (Hevea brasiliensis Muell. Arg) is an economically essential tropical tree and provides a sole commercial source of natural rubber (NR). NR, cis-1,4-polyisoprene, is an important industrial raw material because of its unique physical properties (Backhaus, 1985; van Beilen and Poirier, 2007). NR is an end-product of a side branch of the plant isoprenoid synthesis pathway (Yamashita and Takahashi, 2020). NR is produced in laticifer cells through a sequential condensation of isopentenyl diphosphates (IPPs) in a cis-1,4 configuration (Cornish, 2001; Chow et al., 2007; Salehi et al., 2021). As a consequence genes involved in IPP synthesis to cis-1,4-polyisoprene are named as NR biosynthesis genes (Tang et al., 2016). The biochemical pathways involving NR biosynthesis are now fully understood (Yamashita and Takahashi, 2020; Salehi et al., 2021), the regulatory mechanism of NR biosynthesis genes has not yet been fully investigated in the rubber tree.
The TGA transcription factors (TFs) are members of the subfamily of basic leucine zipper (bZIP) TFs (Li et al., 2019a; Wang et al., 2019). TGA proteins have the conserved bZIP domain-containing N-x7-R/K-x9-L-x6-L-x6-L motif, and Yx2RL[RQ]ALSS[LS]W motif, which is the unique motif of TGA TFs (Gatz, 2013). TGA TFs regulate gene expression by specific binding of TGACG-motif on gene promoter (Jakoby et al., 2002) and play crucial roles in secondary metabolites biosynthesis (Lv et al., 2019; Han et al., 2020; Huo et al., 2021), plant development (Wang et al., 2019), defense against pathogens (Kesarwani et al., 2007; Shearer et al., 2012), and responses to stress (Gatz, 2013; Li et al., 2019b). Given the potential importance of TGA TFs in plant secondary metabolites biosynthesis, we carried out the analysis of the TGA TFs family in the rubber tree and investigated the roles of TGA TFs in NR biosynthesis.
Ten Arabidopsis TGA protein sequences were downloaded from the NCBI database. These protein sequences were used as query sequences against the local rubber tree genome database (Li et al., 2017) through the BLASTp search (E-value 1.0 × e−5). Then, candidate rubber tree TGAs were further confirmed for conserved domains in the Pfam database (El-Gebali et al., 2018). The final confirmed genes were named HbTGA1-HbTGA14. The molecular weight and isoelectric point of HbTGAs were predicted by the ExPASy server (Gasteiger et al., 2003).
The start and end points of HbTGAs were calculated from the rubber tree annotation GFF3 files, the chromosomal distribution of HbTGAs was displayed using MapChart v2.3.
The exon and intron of HbTGAs were investigated using a web server GSDS (Hu et al., 2015). Multiple sequence alignments for HbTGA and other TGA proteins sequences from Arabidopsis, cassava, and Ricinus communis derived from the NCBI database were carried out using Clustal.1 The phylogenetic tree was constructed based on the HbTGA and other TGA protein sequences from Arabidopsis, cassava, and R. communis using MEGA7.0 software with Neighbor-Joining methods followed by 1,000 bootstrap replicates (Kumar et al., 2016).
The transcriptome data of different tissues, including latex (SRX1554800), bark (SRX1554797), leaf (SRX1554799), root (SRX1554786), female flower (SRX1554813), seed (SRX1554817), and male flower (SRX1554814), were obtained from the NCBI database. The expression levels of HbTGAs were calculated as fragments per kilobase of exon per million fragments mapped (FPKM), and heat-map was drawn based on the log2-transformed FPKM values using MeV 4.9.0 software.
The 3.0 kb upstream sequences of the start codon of NR biosynthesis genes were obtained as the promoter region from the rubber tree local genome database. TGA TF binding sites on the promoters of NR biosynthesis genes were calculated using PlantCARE (Rombauts et al., 1999).
DNA was isolated from the leaves according to method of Allen et al. (2006). RNA was extracted from latex as described previously (Tang et al., 2007).
The coding sequence of HbTGA1 was amplified with PCR primers (see Supplementary Table S1) from latex cDNA, and cloned into the BglII/NcoI restriction site of pCAMBIA1302, resulting in a HbTGA1 fused with green fluorescent protein (GFP) expression vector, which was further introduced into onion epidermal by Agrobacterium-mediated transformation. The transformed onion epidermal were incubated in darkness for 48 h at 25°C and then visualized under fluorescent microscopy.
The promoter fragments of NR biosynthesis genes harboring TGACG-motif were obtained through PCR amplification with primers (see Supplementary Table S1), respectively. The bait vector was constructed by cloning the promoters of NR biosynthesis genes into the site of SacI/SpeI of pHIS2.1 vector (Clontech). The coding sequence of HbTGA1 was inserted into the pGADT7 vector at the site of EcoRI/BamHI, generating prey vectors. Then, yeast Y187 strain was introduced with bait and prey vectors, respectively. The introduced yeast was cultured on SD/−Trp-Leu medium and SD/−Leu/−Trp/-His medium adding corresponding concentrations of 3-AT obtained by screening experiment for 3 days at 30°C.
To determine the binding of the HbTGA1 protein with promoters of NR biosynthesis genes in vitro, electrophoretic mobility shift assay (EMSA) was performed. Firstly, the coding sequence of HbTGA1 was cloned into the pET28a vector at the site of BamHI/EcoRI, resulting in an HbTGA1-His-tagged protein expression vector. The expression vector was introduced into Escherichia coli BL21 (DE3). HbTGA1-His-tagged protein was purified using a HiTrap affinity column (GE Healthcare) in accordance with the instruction manual. Secondly, total 205 bp-long DNA fragments of the NR biosynthesis genes promoter (except HbHMGS2) were PCR-amplified, which containing the predicted TGACG-motifs with 100 bp upstream and 100 bp downstream. There were two TGACG-motifs between in 217 bp, total 417 bp-long DNA fragments of HbHMGS2 promoter was PCR-amplified, from 100 bp upstream of the first TGACG-motif to 100 bp downstream of the second TGACG-motif. The 205 bp-long DNA fragments which no TGACG-motif was PCR-amplified as negative control. The PCR products were purified using purification kit (Foregene, DE-03011). The primers used for EMSA are listed in Supplementary Table S1. Finally, EMSA was implemented according to the protocol of the EMSA kit (Invitrogen, E33075). In brief, the purified HbTGA1 was incubated with double-stranded promoter nucleotides for 30 min at room temperature. The DNA/protein complex samples were subjected to 12% polyacrylamide gel electrophoresis at 120 V for 30 min. Then, gel was examined using SYBR Green EMSA stain.
To identify if HbTGA1 activates the promoters of NR biosynthesis genes in plant, a dual-luciferase assay system was employed for this purpose. In brief, HbTGA1 was amplified and cloned into a pGreenII62 SK vector, generating an effector vector. The promoter of NR biosynthesis genes was amplified and cloned into a pGreenII0800-luciferase (LUC) vector, as a reporter vector. Then Agrobacterium tumefaciens GV3101 was introduced with effector or reporter vector. The A. tumefaciens containing the reporter and effector were mixed in the ratio of 1:2 (v:v) and then infiltrated into the tobacco following previously described method (Hellens et al., 2005). The infiltrated leaves were collected to measure LUC activities until 72 h after infiltration. For methyl jasmonate (MeJA) and salicylic acid (SA) treatment, tobacco leaves infiltrated with A. tumefaciens were sprayed with 100 μM MeJA, 100 μM SA, or water as control at 48 h after infiltration. The infiltrated leaves were collected and extracted protein for LUC activities assay at 24 h after treatment. The fluorescent values of LUC and the reference Renilla (REN) were measured in accordance with the instruction manual of the Dual-Luciferase Reporter Assay System (Promega, E1910). Experiments were performed in three biological replicates per treatment. Statistical analyses were carried out by one-way ANOVA (SAS6.11). Statistical significance was defined as p < 0.05.
A total of 14 putative genes encoding TGATFs were identified in the rubber tree genome. These genes were named as HbTGA1-HbTGA14. These predicted HbTGAs were distributed across 10 of 18 chromosomes (Figure 1). HbTGA proteins span 361 (HbTGA4) to 536 (HbTGA13) amino acids in length, with molecular weights spanning 40.62 kDa (HbTGA4) to 59.79 kDa (HbTGA13), and exhibit isoelectric points that range from 5.55 (HbTGA3) to 8.38 (HbTGA1; Table 1).
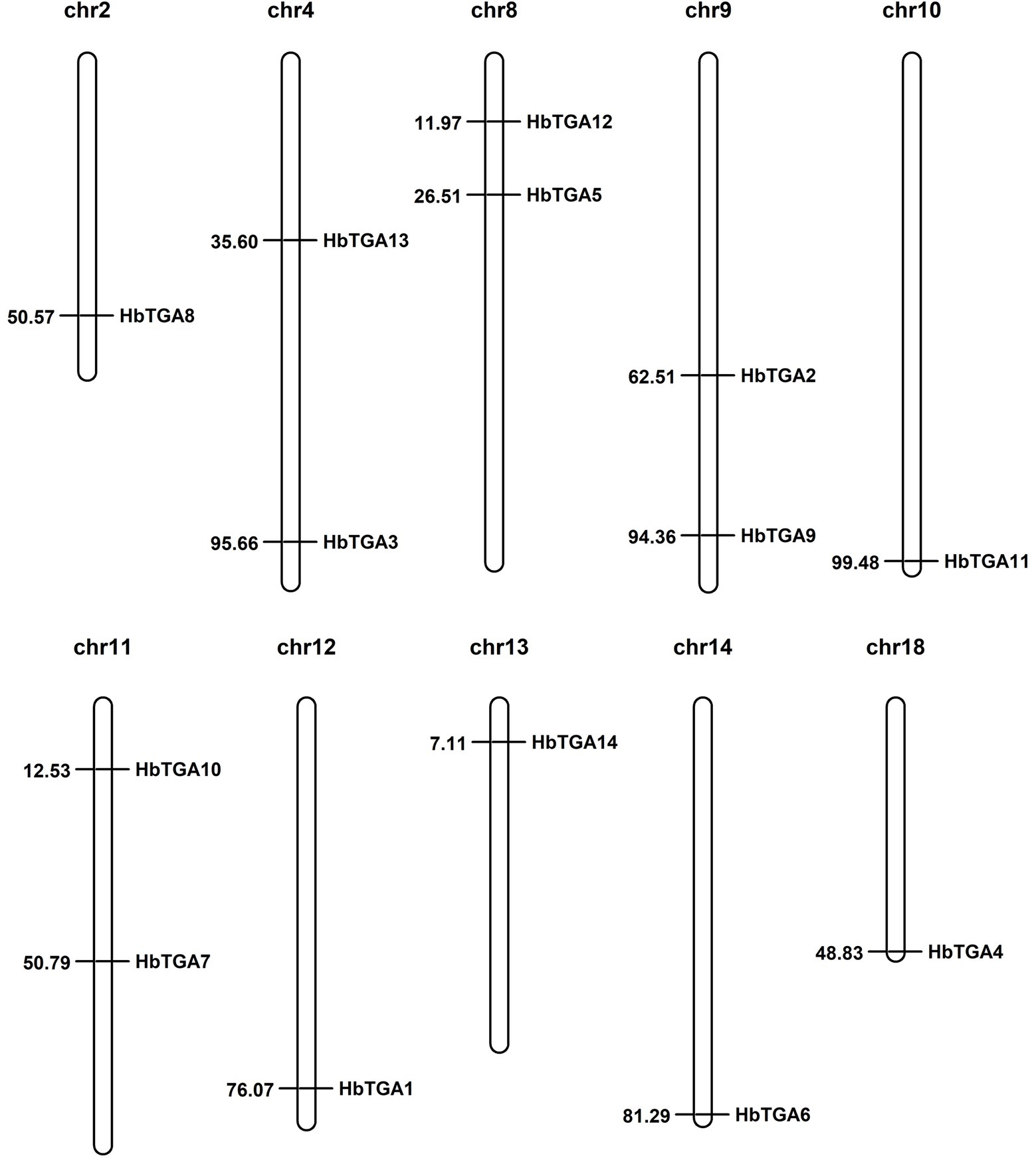
Figure 1. Chromosome location of HbTGAs in Hevea brasiliensis. 14 HbTGAs were localized onto 10 of 18 chromosomes. Map positions (in Mbp) are shown on the left.
To explore the evolutionary relationships between HbTGAs and TGAs from other plants, a phylogenetic tree was constructed by comparing HbTGAs with TGA proteins from Arabidopsis, cassava, and R. communis. As shown in Figure 2, 42 TGA proteins were separated into five clades. Clades II and IV both contain four HbTGAs, while clade I, III, and V have two members of the HbTGAs family.
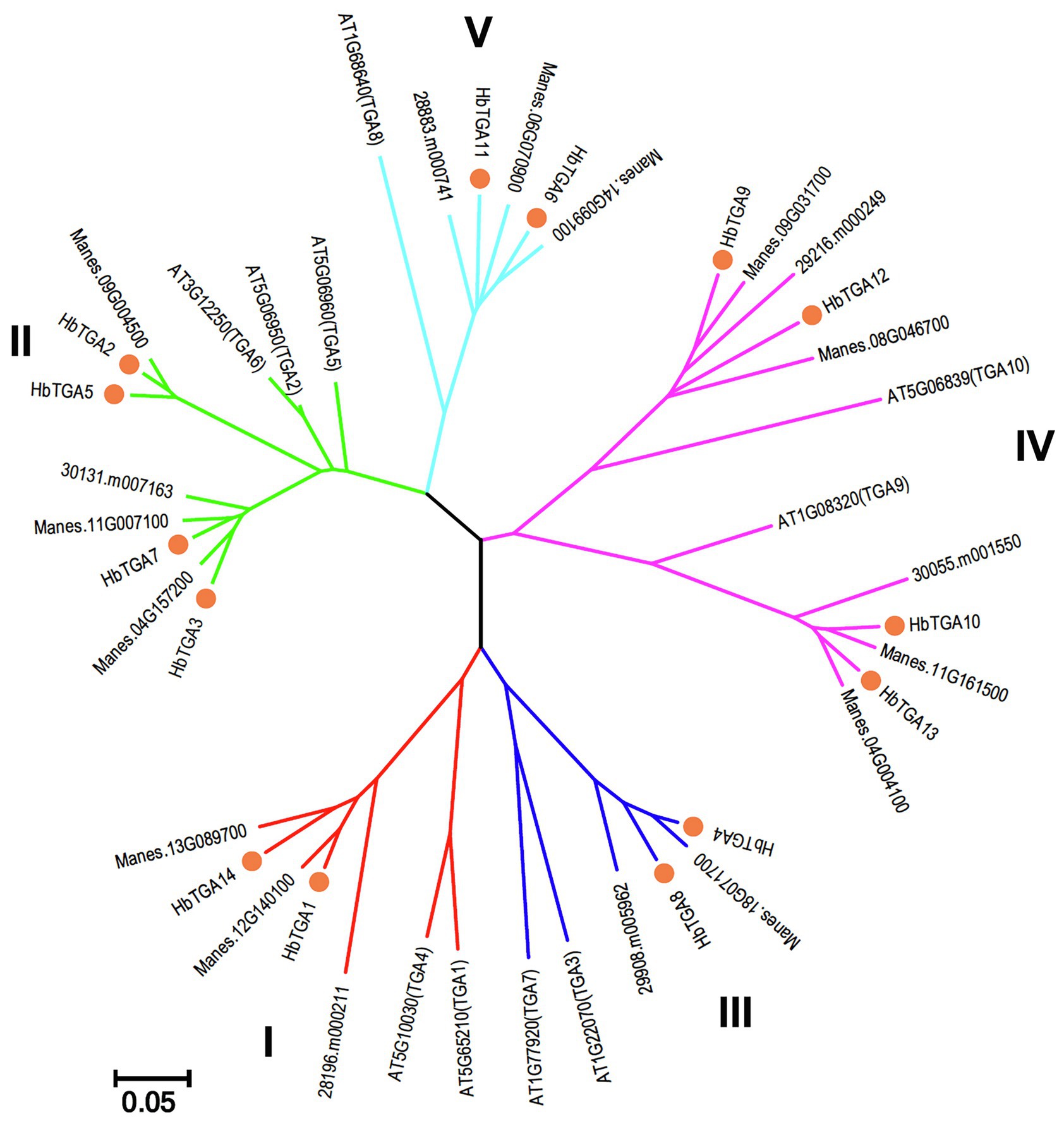
Figure 2. Phylogenetic analysis of TGA proteins from Arabidopsis, cassava, and Ricinus communis with Neighbor-Joining method using 1,000 bootstrap replicates. Each clade is demonstrated by different color.
The conserved motifs of all the 14 HbTGAs were found (Figure 3A) using MEME online tool using “searching for 12 motifs.” As shown in Figure 3B, all 14 HbTGAs contained motifs 1, 2, 3, 4, 5, 6, and 8. Motif 1(N-x7-R/K-x9-L-x6-L-x6-L) is a typical bZIP domain in b-zip TFs and motif 2(Yx2RL[R/Q]ALSS [L/S]W) is a unique bZIP-D box in TGA TFs. Except for HbTGA9, 10, 12, and 13, other TGAs contain motif 7. Except for HbTGA2, 3, 5, and 7, 9, 12, other TGAs do not contain motif 9. HbTGA2, 3, 5, and 7 have motif 10. HbTGA10 and HbTGA13 have motif 11. HbTGA6 and HbTGA11 have motif 12.
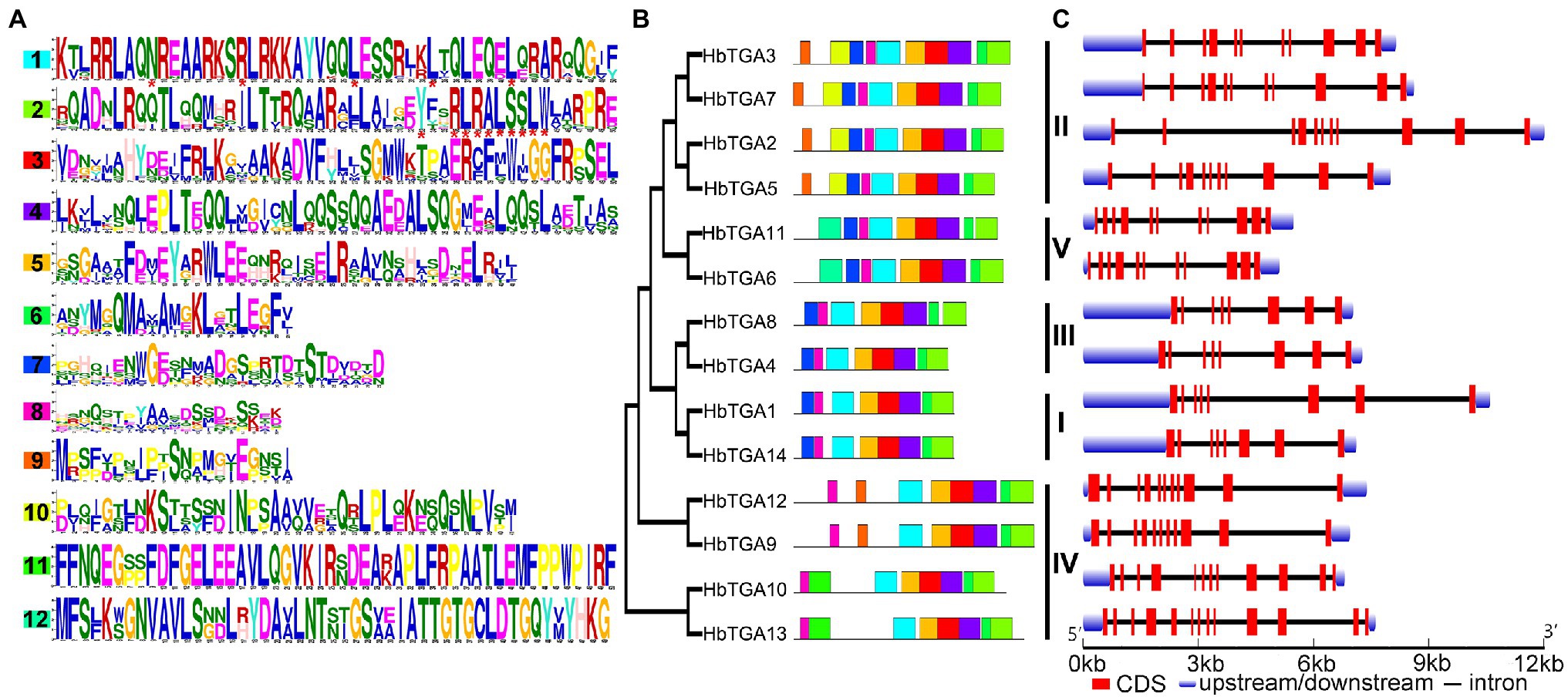
Figure 3. Conserved motifs, phylogenetic tree, and gene structure in HbTGAs. (A) Conserved motifs of the TGA proteins. Each motif is represented by a number in colored box. (B) Phylogenetic relationships. (C) Gene structure in HbTGAs.
The gene structures of 14 HbTGAs were also examined. HbTGAs were interrupted by more than six introns (Figure 3C). HbTGAs in Clades II, IV, and V contain more than 10 introns, while HbTGAs in Clades I and II have seven introns. The same group of HbTGAs has a similar gene structure and the same conserved domain, which may reflect their similar functions.
The expression patterns of HbTGAs were analyzed using the transcriptome data of different rubber tree tissues (Figure 4). All the 14 HbTGAs were expressed in the evaluated tissues, but there were differences in expression patterns. For example, the expression of HbTGA1 showed the highest level in latex. HbTGA2 and HbTGA3 had higher expression levels in all evaluated tissues, while HbTGA6, HbTGA7, and HbTGA11 had lower expression levels in all evaluated tissues. HbTGA12 and HbTGA13 were expressed most strongly in the root, HbTGA2, HbTGA5, HbTGA9, and HbTGA12 were expressed at their highest levels in the seed. In addition, HbTGA10, 11, 12 were not expressed in latex, HbTGA12 was not expressed in leaf, while HbTGA12 was not expressed in the female flower.
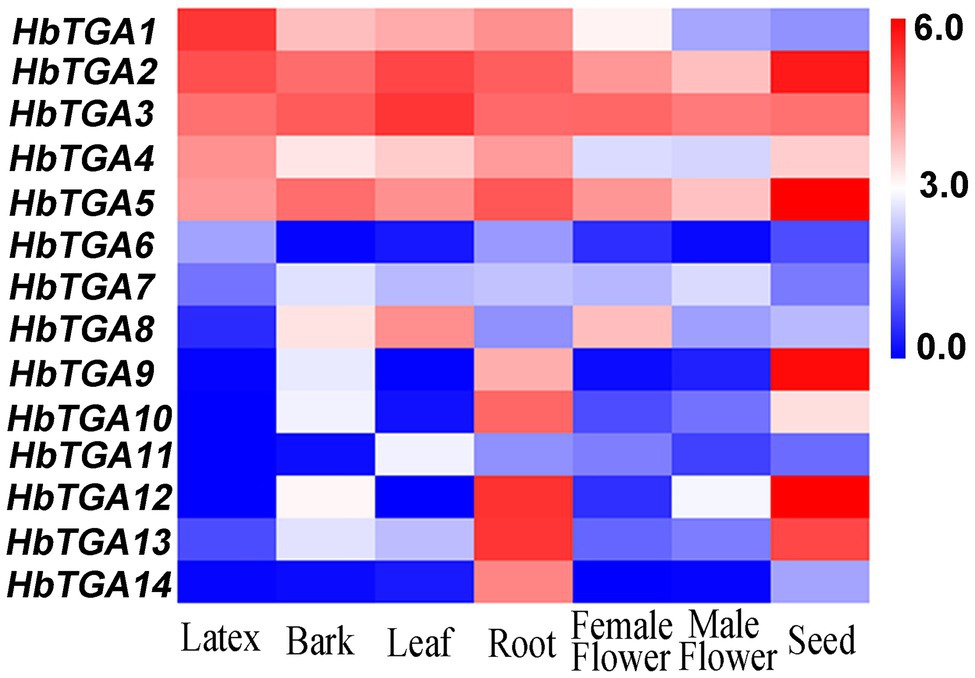
Figure 4. Expression patterns of HbTGAs in different tissues. The heat-map was created using log2-transformed fragments per kilobase of exon per million fragments mapped (FPKM) values from the transcriptome data of different tissues, including latex, bark, leaf, root, female flower, and male flower seed.
The 3 kb upstream fragment of the start codon (ATG) of NR biosynthesis-related gene was isolated as the promoter region. The putative TGA TFs binding site (TGACG-motif) was predicted on the promoters of NR biosynthesis genes. As shown in Figure 5A, the promoters of these natural rubber synthesis genes contain 1–3 TGA TFs recognition sites. The expression of NR biosynthesis genes was analyzed using the transcriptome data of seven rubber tree tissues and organs (Figure 5B). According to the expression abundance of these genes in latex, HbHMGS2, HbHMGR2, HbCPT6, HbCPT8, and HbSRPP2 which were expressed at a higher level in latex were selected to further verify whether TGA TFs interact with promoters of NR biosynthesis genes.
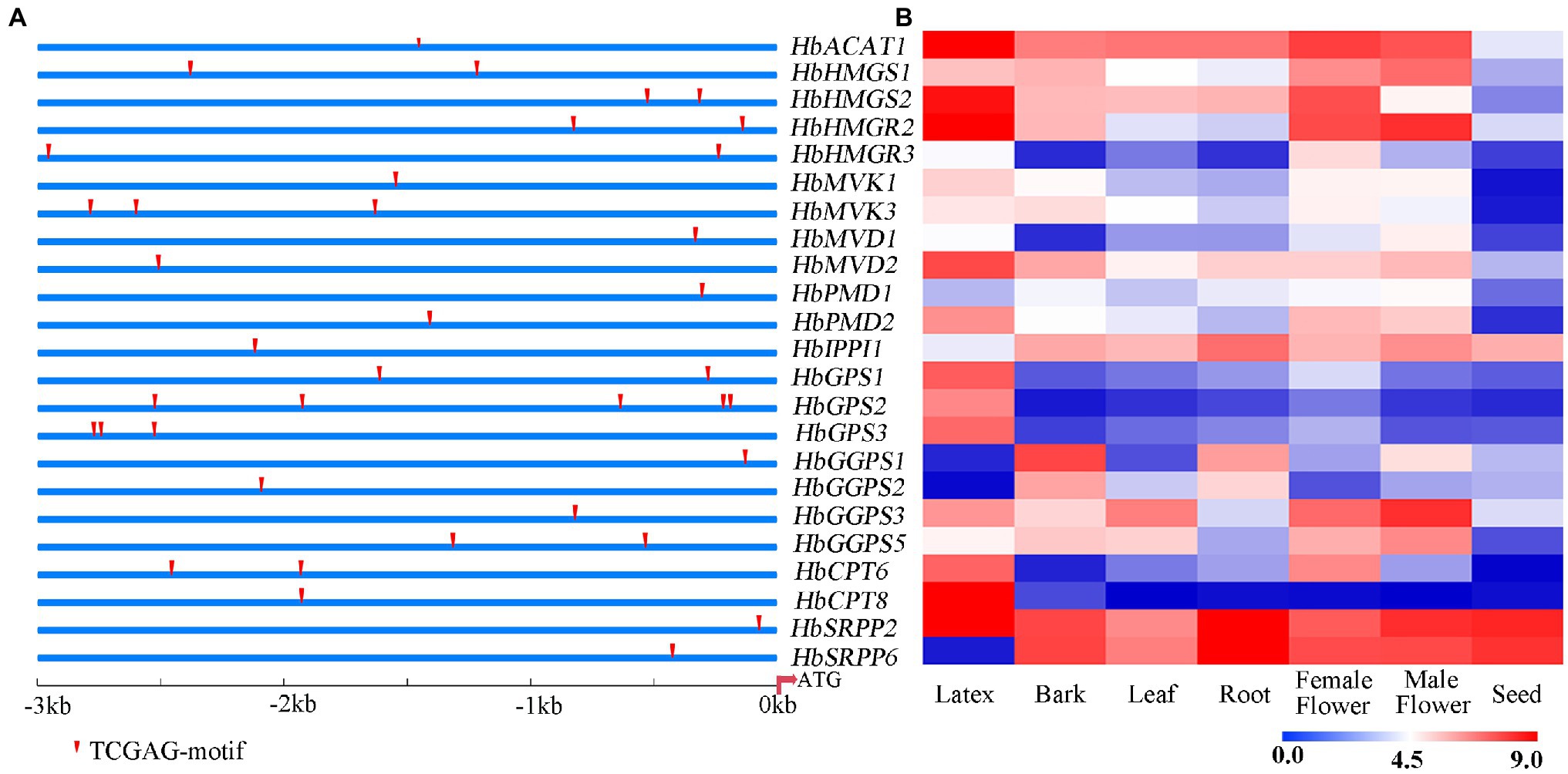
Figure 5. Prediction of binding site of TGA TFs on the promoters of NR biosynthesis genes (A) and expression patterns of NR biosynthesis genes in different tissues (B). The heat-map was created using log2-transformed FPKM values from the transcriptome data of different tissues, including latex, bark, leaf, root, female flower, male flower seed.
The subcellular localization of HbTGA1 was analyzed in transiently transformed onion epidermal cells. As shown in Figure 6, HbTGA1 was located in the nucleus. The binding specificity of these promoters of NR biosynthesis genes with HbTGA1 was determined by the yeast one-hybrid experiment (Figure 7). An in vitro EMSA assay further suggested HbTGA1 bound with promoters of HbHMGS2, HbHMGR2, HbCPT6, HbCPT8, and HbSRPP2 (Figure 8). This finding indicates that HbTGA1 binds to the promoters of these NR biosynthesis genes and activates the transcription in yeast.
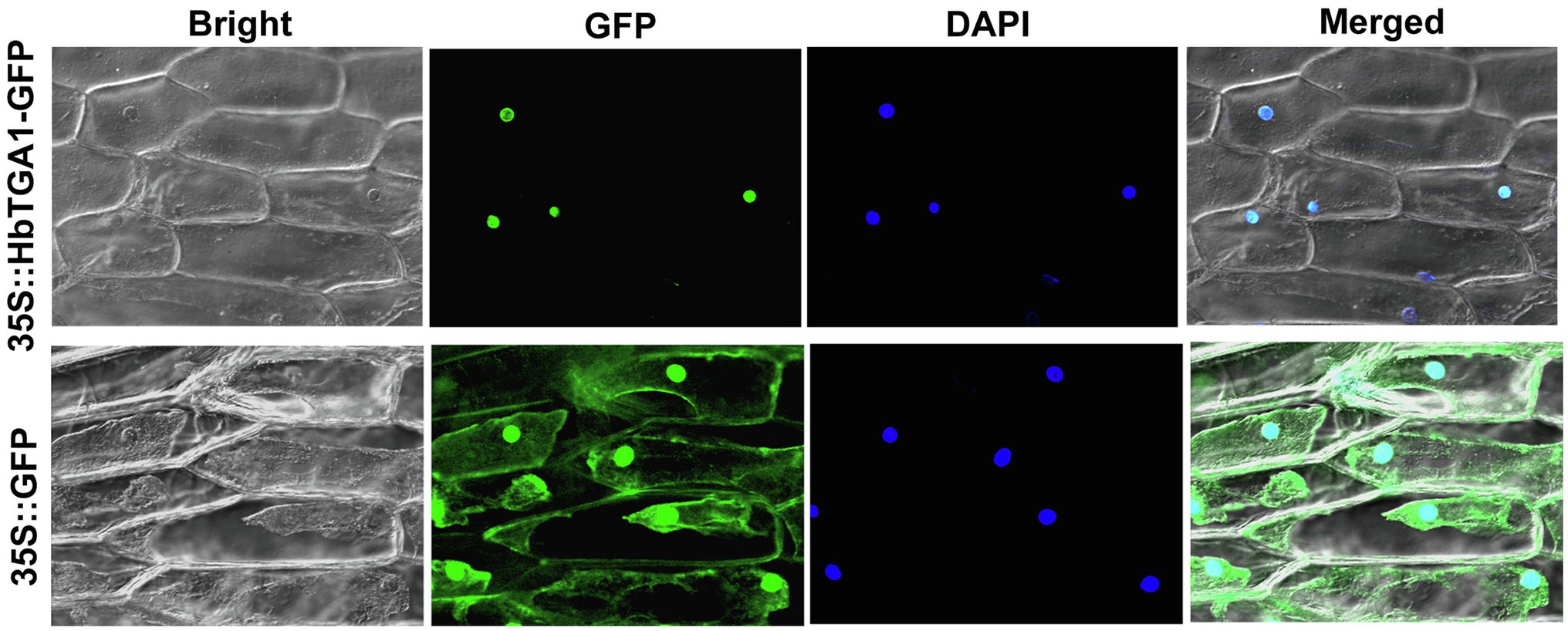
Figure 6. Subcellular localization of HbTGA1. Transient expression of 35S::HbTGA1-GFP (upper layer) and 35S::GFP (under layer, control vector) in onion epidermal cells, showing the nuclear localization of HbTGA1.
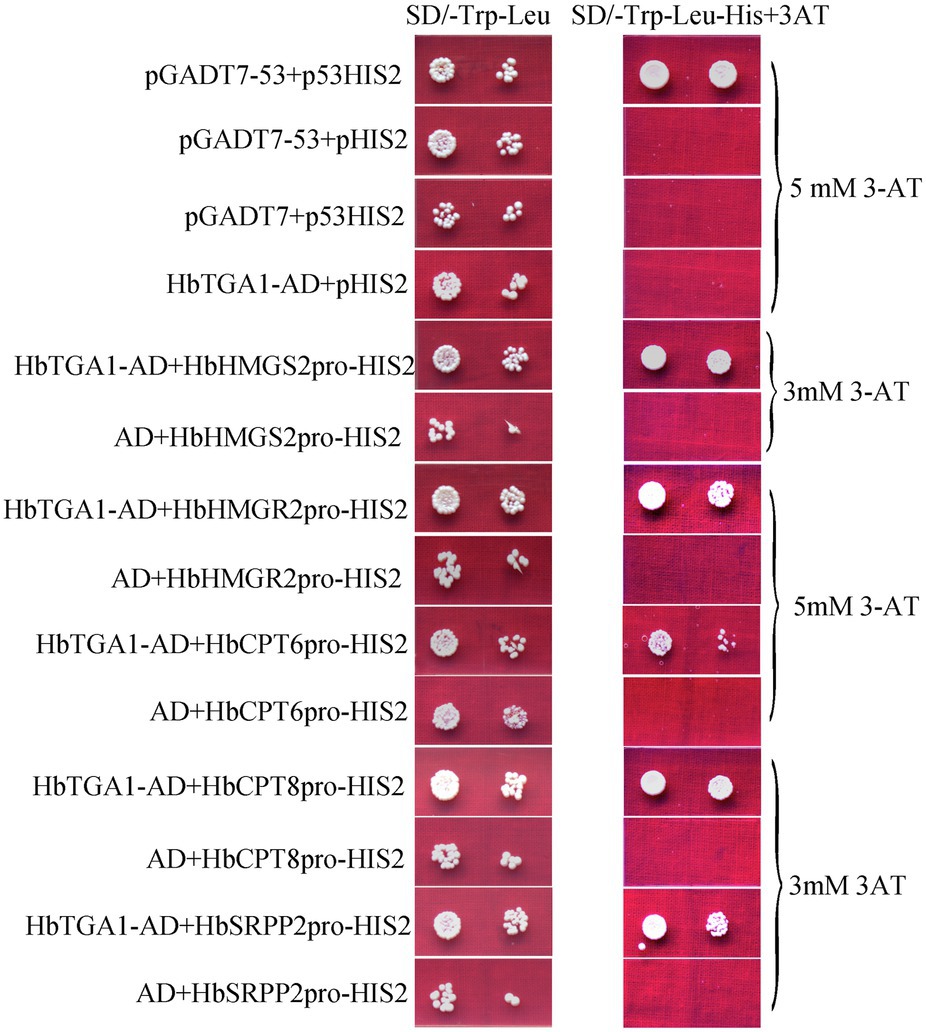
Figure 7. HbTGA1 binding with the promoters of NR biosynthesis genes in yeast. Yeast harboring bait and prey vectors were grown in SD/−Trp-Leu medium and SD/−Leu/−Trp/-His selective medium adding corresponding concentrations of 3-AT obtained by screening experiment for 3 days at 30°C.
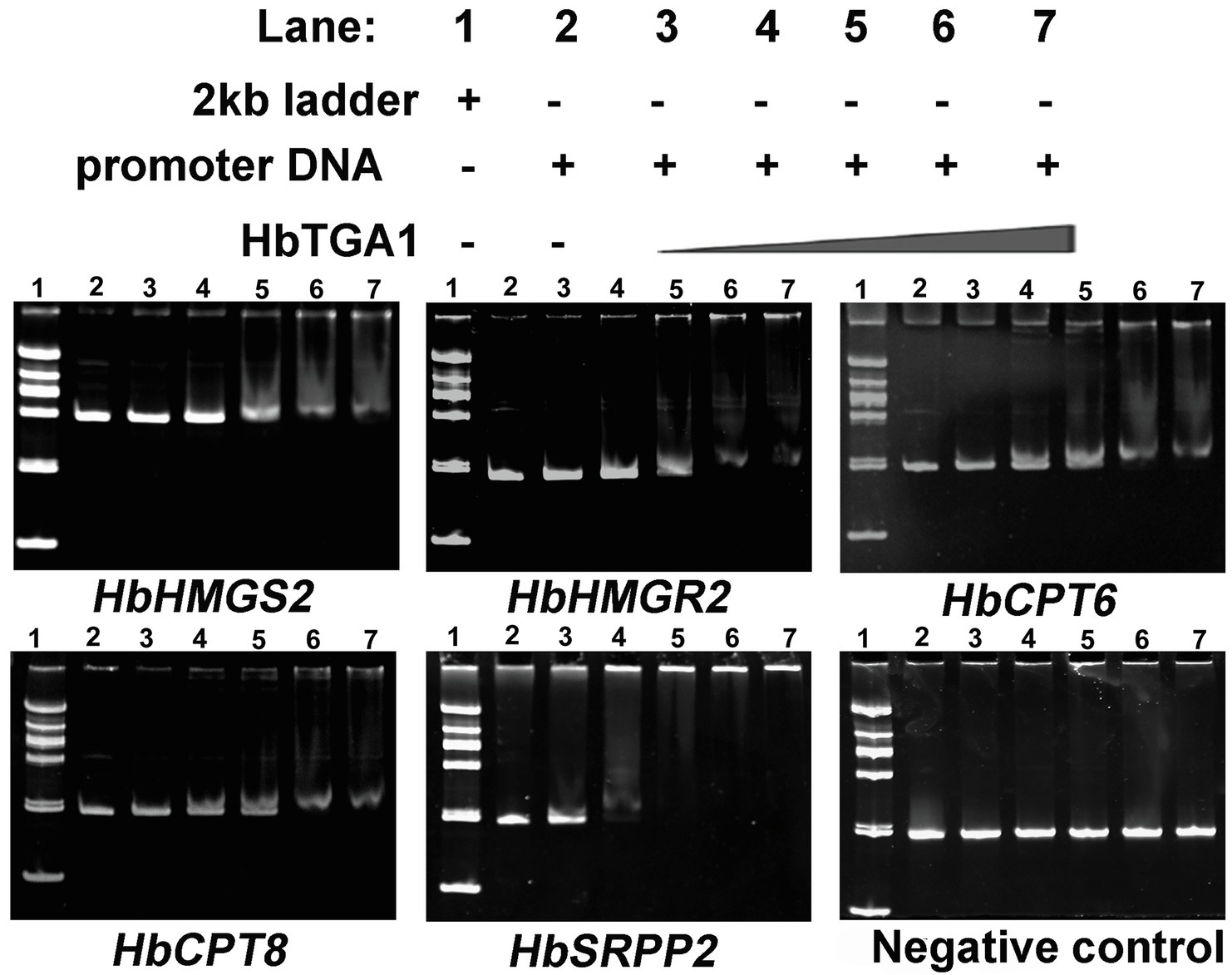
Figure 8. HbTGA1 binding with the promoters of NR biosynthesis genes was analyzed via electrophoretic mobility shift assay (EMSA). Lane 1: DNA 2,000 markers. Lane 2: 50 ng, the promoter of NR biosynthesis genes only. Lanes 3–7: 50 ng, the promoter of NR biosynthesis genes with increasing amounts of HbTGA1 protein (100, 200,300, 400, and 500 ng).
Since NR is synthesized in laticifers, genes highly expressed in laticifers may participate in NR biosynthesis (Oh et al., 1999). The expression level of HbTGA1 is the most highest among all HbTGAs. To investigate whether HbTGA1 involves in regulating NR biosynthesis-related genes, we transiently expressed HbTGA1 along with a LUC controlled by the promoters of NR biosynthesis genes in tobacco. The schematic diagrams of the effectors and reporters used in dual-luciferase assay system are shown in Figure 9A. Compared to the control, the expression of HbTGA1 enabled to change the level of the LUC activity controlled by the promoters of NR biosynthesis genes. As shown in Figure 9B, the activation of the promoter of HbHMGS2, and HbCPT6 were significantly suppressed by HbTGA1, decreased by 27 and 55%, respectively; while the activation of promoters of HbHMGR2, HbCPT8, and HbSRPP2, increased to 2.5-, 3.3-, and 1.4-fold, respectively (Figure 9B). Taken together, HbTGA1 takes part in regulating the expression of multiple genes in the NR synthesis pathway.
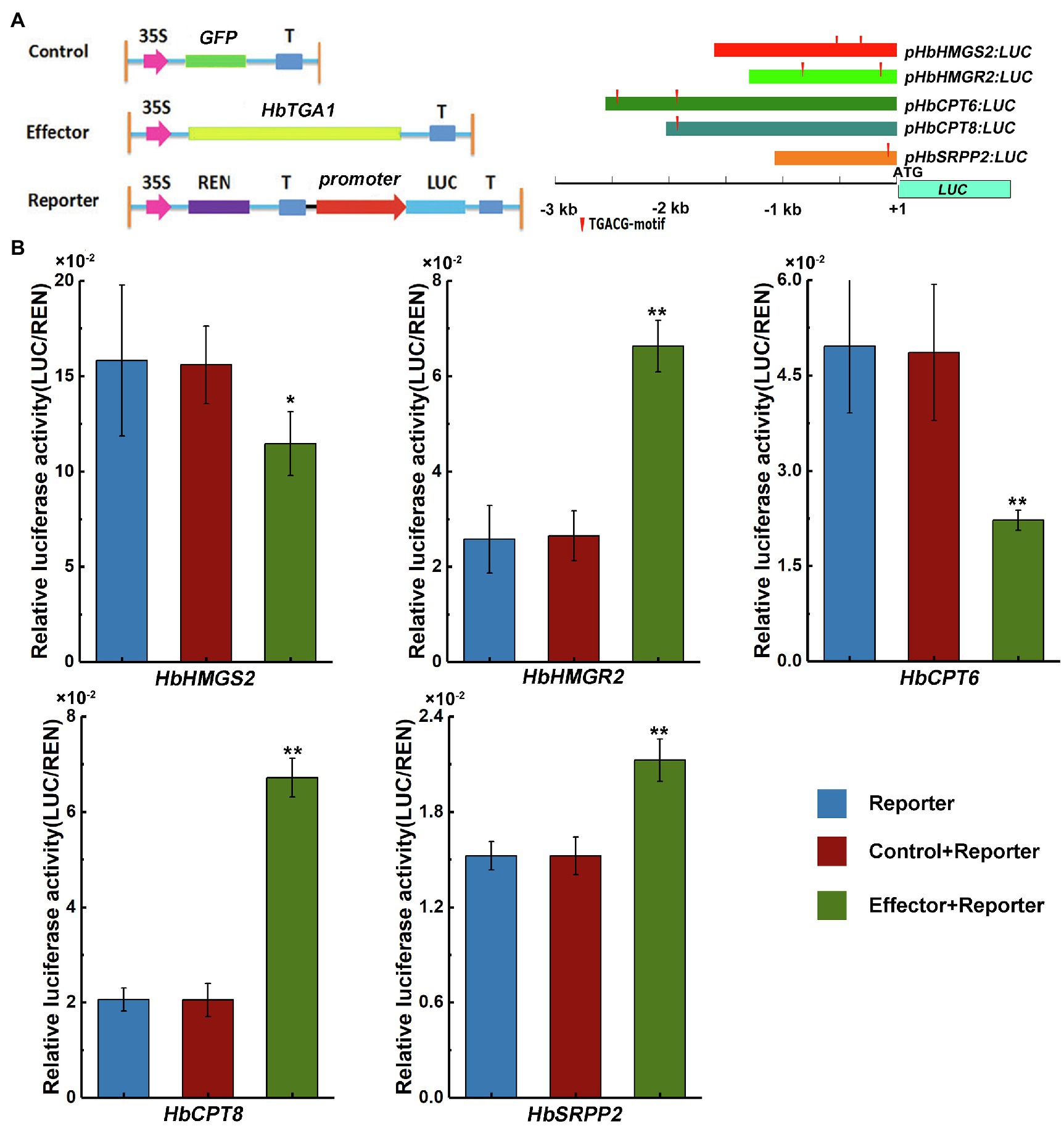
Figure 9. Effect of HbTGA1 on the activation of the promoter of NR biosynthesis genes. Schematic maps of e-effector and reporter (A). The promoter activity was determined by a transient dual-luciferase (LUC) assay (B). The relative LUC activities (LUC/REN) were normalized to the reference Renilla (REN) LUC. Error bars represent SD of three technical replicates. *p < 0.05, **p < 0.01.
Subsequently, we transiently expressed HbTGA1 along with a LUC controlled by the promoters of NR biosynthesis genes in tobacco under MeJA and SA treatment conditions (Figure 10). Compared to the control, the promoter activities of HbHMGS2 and HbCPT6, HbCPT8, and HbSRPP2 were significantly increased by HbTGA1 under MeJA stress, increased to 1.5-, 1.5-, 1.3-, and 1.4-fold, respectively; the promoter activities of HbHMGS2, HbHMGR2, HbCPT6, HbCPT8, and HbSRPP2 were also significantly increased by HbTGA1 under SA stress, increased to 1.3-, 2.0-, 1.6-, 1.4-, and 1.9-fold, respectively. The result revealed that HbTGA1 might participate in the MeJA- and SA-inducible NR biosynthesis in rubber tree.
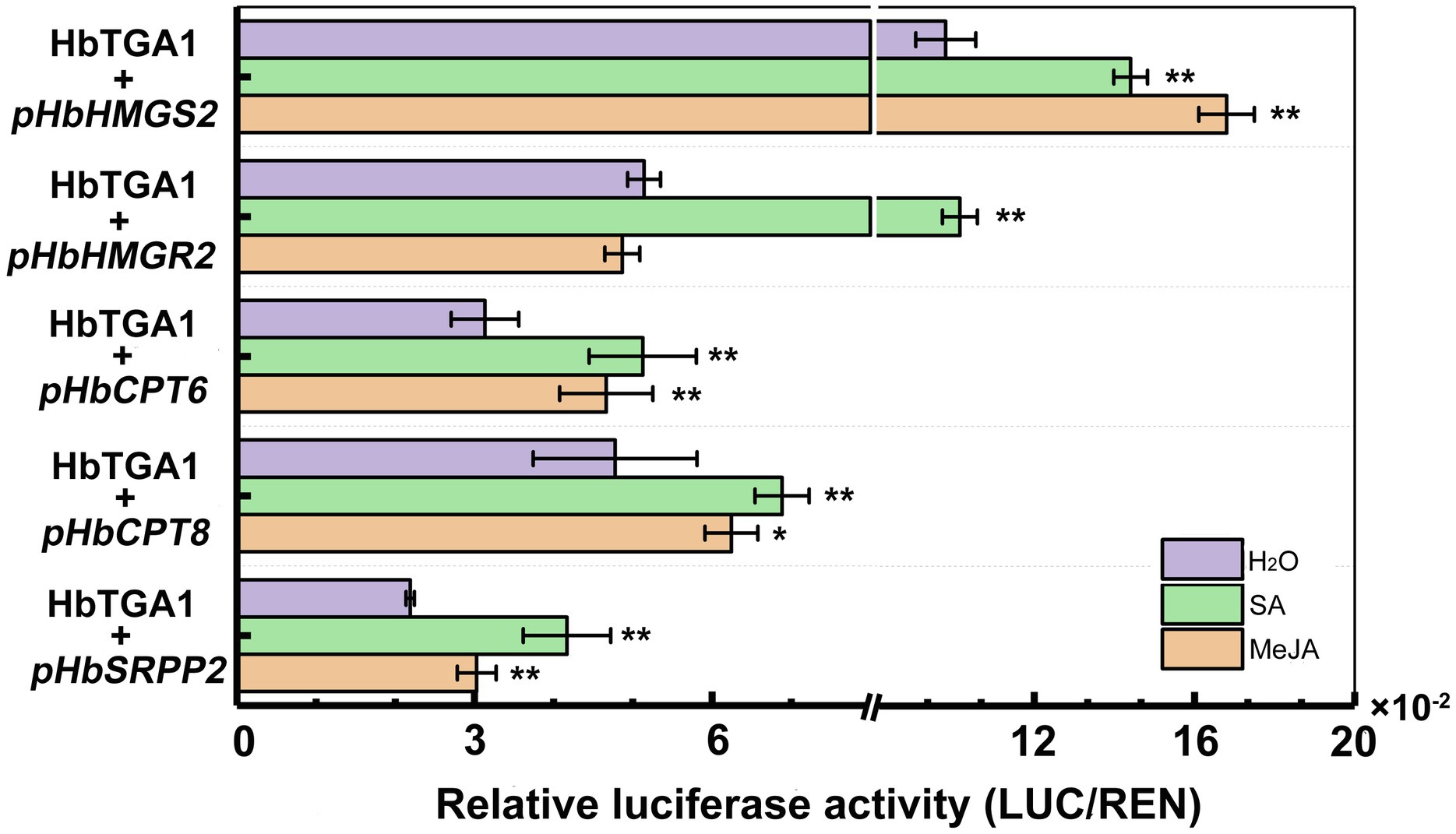
Figure 10. Activation of the promoter of NR biosynthesis genes by HbTGA1 under JA and SA. Error bars represent SD of three technical replicates. *p < 0.05, **p < 0.01.
A number of investigates have proved the involvement of TGA TFs in several biological processes (Shearer et al., 2012; Gatz, 2013; Canales et al., 2017; Sun et al., 2017; Lv et al., 2019; Wang et al., 2019; Han et al., 2020; Huo et al., 2021). However, the TGA TFs family and their role in the rubber tree were scarcely understood. Here, we identified 14 TGA TFs members in the rubber tree. The HbTGAs could be clustered together with TGAs from Arabidopsis, cassava, and R. communis in the same clade, suggesting that the evolution of TGA genes is conserved.
Transcription factors regulate gene expression and play key roles in biological processes. In rubber tree, a few TFs take part in regulation of the expression of NR biosynthesis genes. For example, three TFs, including HbWRKY1, HbWRKY14, and HbMADS4, downregulated the expression of HbSRPP (Wang et al., 2013; Li et al., 2016, 2020). HbCZF1 upregulates the expression of hmg1 (Guo et al., 2015). HblMYB19, HblMYB44, and HbWRKY27 upregulated the expression of HbFPS1 (Wang et al., 2017; Qu et al., 2020). HbRZFP1 downregulated HRT2 expression (Guo et al., 2018). HbMYC2b activates HbSRPP expression (Guo et al., 2019). These data showed TFs play critical roles in NR biosynthesis. It was found that TGA TFs regulate plant secondary metabolites biosynthesis. In Artemisia annua, AaTGA6 regulates artemisinin content by directly binding to the promoter of AaERF1. In Tripterygium wilfordii, TwTGA1 binds with promoters of PMT and MPO1 and activates their expressions, and modulates secondary metabolites biosynthesis (Han et al., 2020). In our study, Y1H and EMSA assays showed that HbTGA1 bound to the promoter of NR biosynthesis genes. The activation of promoters of multiple NR biosynthesis genes was regulated by HbTGA1. These results suggested HbTGA1 might modulate the expression of NR biosynthesis genes and participate in NR synthesis in rubber tree. The functions of HbTGA1 participating in NR synthesis in rubber tree needs to be explored in rubber tree in future. Additionally, TGA factors have been shown to interact with other TFs to modulate their activity (Gatz, 2013), the potential regulators of HbTGA1 also needs to further be investigated.
In plant TGA, TFs have key roles in secondary metabolites biosynthesis through SA and JA signaling pathways (Lv et al., 2019; Huo et al., 2021). For example, TwTGA1 was reported to increase the MeJA-inducible triptolide synthesis by upregulating the expression of TwTPS27a/b in T. wilfordii (Han et al., 2020; Huo et al., 2021). AaTGA6 was reported to modulate SA-inducible artemisinin synthesis in A. annua (Lv et al., 2019). In this study, the activation of promoters of multiple NR biosynthesis genes was significantly increased by HbTGA1 under SA and JA. In addition, SA could also induce a transient increase in NR yield in rubber tree (Tungngoen et al., 2011), suggesting SA signaling pathway might play role in regulating NR biosynthesis. However, there is still a lack of direct evidence to show SA signaling regulatory involvement in NR biosynthesis and the core module of SA signaling needs to further clarify in rubber tree. JA signaling has been reported to modulate NR biosynthesis in rubber tree (Deng et al., 2018). Being the target of SA signaling, HbTGA1 can connect the SA pathway with the JA pathway (Gatz, 2013). Thus, it will be of great interest to further study the regulatory mechanisms of HbTGA1 modulating MeJA- and SA-inducible NR biosynthesis. HbTGA1 might become a biotechnological tool in rubber tree breeding.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
S-QP: conceptualization. DG, H-LL, J-HZ, and YW: investigation. DG and S-QP: writing—original draft and writing—review and editing. All authors contributed to the article and approved the submitted version.
This study was supported by Hainan Provincial Natural Science Foundation of China (No. 322MS125), National Natural Science Foundation of China (No. 31970620), and Central Public-Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630052022009).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.909098/full#supplementary-material
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Canales, J., Contreras-López, O., Álvarez, J. M., and Gutiérrez, R. A. (2017). Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. Plant J. 92, 305–316. doi: 10.1111/tpj.13656
Chow, K. S., Wan, K. L., Isa, M. N., Bahari, A., Tan, S. H., Harikrishna, K., et al. (2007). Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J. Exp. Bot. 58, 2429–2440. doi: 10.1093/jxb/erm093
Cornish, K. (2001). Similarities and differences in rubber biochemistry among plant species. Phytochemistry 57, 1123–1134. doi: 10.1016/S0031-9422(01)00097-8
Deng, X., Guo, D., Yang, S., Shi, M., Chao, J., Li, H., et al. (2018). Jasmonate signalling in the regulation of rubber biosynthesis in laticifer cells of rubber tree, Hevea brasiliensis. J. Exp. Bot. 69, 3559–3571. doi: 10.1093/jxb/ery169
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2018). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gatz, C. (2013). From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant-Microbe Interact. 26, 151–159. doi: 10.1094/MPMI-04-12-0078-IA
Guo, D., Li, H. L., Wang, Y., Zhu, J. H., and Peng, S. Q. (2019). A myelocytomatosis transcription factor from Hevea brasiliensis positively regulates the expression of the small rubber particle protein gene. Ind. Crop. Prod. 133, 90–97. doi: 10.1016/j.indcrop.2019.01.052
Guo, D., Yang, Z. P., Li, H. L., Wang, Y., Zhu, J. H., and Peng, S. Q. (2018). The 14-3-3 protein HbGF14a interacts with a RING zinc finger protein to regulate expression of the rubber transferase gene in Hevea brasiliensis. J. Exp. Bot. 69, 1903–1912. doi: 10.1093/jxb/ery049
Guo, D., Yi, H. Y., Li, H. L., Liu, C., Yang, Z. P., and Peng, S. Q. (2015). Molecular characterization of HbCZF1, a Hevea brasiliensis CCCH-type zinc finger protein that regulates hmg1. Plant Cell Rep. 34, 1569–1578. doi: 10.1007/s00299-015-1809-6
Han, J., Liu, H. T., Wang, S. C., Wang, C. R., and Miao, G. P. (2020). A class I TGA transcription factor from Tripterygium wilfordii Hook. f. Modulates the biosynthesis of secondary metabolites in both native and heterologous hosts. Plant Sci. 290:110293. doi: 10.1016/j.plantsci.2019.110293
Hellens, R. G., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13. doi: 10.1186/1746-4811-1-13
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huo, Y., Zhang, B., Chen, L., Zhang, J., Zhang, X., and Zhu, C. (2021). Isolation and functional characterization of the promoters of miltiradiene synthase genes, TwTPS27a and TwTPS27b, and interaction analysis with the transcription factor TwTGA1 from Tripterygium wilfordii. Plan. Theory 10:418. doi: 10.3390/plants10020418
Jakoby, M., Weisshaar, B., DrÖge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. doi: 10.1104/pp.106.095299
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, H. L., Guo, D., Zhu, J. H., Wang, Y., and Peng, S. Q. (2017). Identification and expression analysis of genes involved in histone acetylation in Hevea brasiliensis. Tree Genet. Genomes 13:98. doi: 10.1007/s11295-017-1178-0
Li, B., Liu, Y., Cui, X. Y., Fu, J. D., Zhou, Y. B., Zheng, W. J., et al. (2019b). Genome-wide characterization and expression analysis of soybean TGA transcription factors identified a novel TGA gene involved in drought and salt tolerance. Front. Plant Sci. 10:549. doi: 10.3389/fpls.2019.00549
Li, N., Muthreich, M., Huang, L. J., Thurow, C., Sun, T., Zhang, Y., et al. (2019a). TGACG-BINDING FACTORs (TGAs) and TGA-interacting CC-type glutaredoxins modulate hyponastic growth in Arabidopsis thaliana. New Phytol. 221, 1906–1918. doi: 10.1111/nph.15496
Li, H. L., Qu, L., Guo, D., Zhu, J. H., Wang, Y., and Peng, S. Q. (2020). Histone deacetylase interacts with a WRKY transcription factor to regulate the expression of the small rubber particle protein gene from Hevea brasiliensis. Ind. Crop. Prod. 145:111989. doi: 10.1016/j.indcrop.2019.111989
Li, H. L., Wei, L. R., Guo, D., Wang, Y., Zhu, J. H., Chen, X. T., et al. (2016). HbMADS4, a MADS-box transcription factor from Hevea brasiliensis, negatively regulates HbSRPP. Front. Plant Sci. 7:1709. doi: 10.3389/fpls.2016.01709
Lv, Z., Guo, Z., Zhang, L., Zhang, F., Jiang, W., Shen, Q., et al. (2019). Interaction of BZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in Artemisia Annua. J. Exp. Bot. 70, 3969–3979. doi: 10.1093/jxb/erz166
Oh, S. K., Kang, H., Shin, D. H., Yang, J., Chow, K. S., Yeang, H. Y., et al. (1999). Isolation, characterisation and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J. Biol. Chem. 274, 17132–17138. doi: 10.1074/jbc.274.24.17132
Qu, L., Li, H. L., Guo, D., Zhu, J. H., Wang, Y., Yin, L. Y., et al. (2020). HbWRKY27, a group IIe WRKY transcription factor, positively regulates HbFPS1 expression in Hevea brasiliensis. Sci. Rep. 10:20639. doi: 10.1038/s41598-020-77805-5
Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Salehi, M., Cornish, K., Bahmankar, M., and Naghavi, M. R. (2021). Natural rubber-producing sources, systems, and perspectives for breeding and biotechnology studies of Taraxacum kok-saghyz. Ind. Crop. Prod. 170:113667. doi: 10.1016/j.indcrop.2021.113667
Shearer, H. L., Cheng, Y. T., Wang, L., Liu, J., Boyle, P., Despres, C., et al. (2012). Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol. Plant-Microbe Interact. 25, 1459–1468. doi: 10.1094/MPMI-09-11-0256
Sun, T., Busta, L., Zhang, Q., Ding, P., Jetter, R., and Zhang, Y. (2017). TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 217, 344–354. doi: 10.1111/nph.14780
Tang, C., Qi, G., Li, H., Zhang, C., and Wang, Y. (2007). A convenient and efficient protocol for isolating high-quality RNA from latex of Hevea brasiliensis (Para rubber tree). J. Biochem. Biophys. Methods 70, 749–754. doi: 10.1016/j.jbbm.2007.04.002
Tang, C., Yang, M., Fang, Y., Luo, Y., Gao, S., Xiao, X., et al. (2016). The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2:16073. doi: 10.1038/nplants.2016.73
Tungngoen, K., Viboonjun, U., Kongsawadworakul, P., Katsuharad, M., Julien, J. L., Sakre, S., et al. (2011). Hormonal treatment of the bark of rubber trees (Hevea brasiliensis) increases latex yield through latex dilution in relation with the differential expression of two aquaporin genes. J. Plant Physiol. 168, 253–262. doi: 10.1016/j.jplph.2010.06.009
van Beilen, J. B., and Poirier, Y. (2007). Establishment of new crops for the production of natural rubber. Trends Biotechnol. 25, 522–529. doi: 10.1016/j.tibtech.2007.08.009
Wang, Y., Guo, D., Li, H. L., and Peng, S. Q. (2013). Characterization of HbWRKY1, a WRKY transcription factor from Hevea brasiliensis that negatively regulates HbSRPP. Plant Physiol. Biochem. 71, 283–289. doi: 10.1016/j.plaphy.2013.07.020
Wang, Y., Salasini, B. C., Khan, M., Devi, B., Bush, M., Subramaniam, R., et al. (2019). Clade I TGACG-motif binding basic leucine zipper transcription factors mediate BLADE-ON-PETIOLE-dependent regulation of development. Plant Physiol. 180, 937–951. doi: 10.1104/pp.18.00805
Wang, Y., Zhan, D. F., Li, H. L., Guo, D., Zhu, J. H., and Peng, S. Q. (2017). Transcriptome-wide identification and characterization of MYB transcription factor genes in the laticifer cells of Hevea brasiliensis. Front. Plant Sci. 8:1974. doi: 10.3389/fpls.2017.01974
Keywords: Hevea brasiliensis, TGA transcription factor, biosynthesis of natural rubber, gene, promoter
Citation: Guo D, Li H-L, Zhu J-H, Wang Y and Peng S-Q (2022) HbTGA1, a TGA Transcription Factor From Hevea brasiliensis, Regulates the Expression of Multiple Natural Rubber Biosynthesis Genes. Front. Plant Sci. 13:909098. doi: 10.3389/fpls.2022.909098
Received: 31 March 2022; Accepted: 13 June 2022;
Published: 06 July 2022.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Stephen Beungtae Ryu, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South KoreaCopyright © 2022 Guo, Li, Zhu, Wang and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-Qing Peng, c2hxcGVuZ0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.