- 1Department of Microbial Ecology, Netherlands Institute of Ecology (NIOO-KNAW), Wageningen, Netherlands
- 2Department of Plant and Environmental Sciences, University of Copenhagen, Copenhagen, Denmark
- 3Institute of Biology, Leiden, Netherlands
The phyllosphere, the aboveground part of a plant, is a harsh environment with diverse abiotic and biotic stresses, including oscillating nutrient availability and temperature as well as exposure to UV radiation. Microbial colonization of this dynamic environment requires specific adaptive traits, including tolerance to fluctuating temperatures, the production of secondary metabolites and pigments to successfully compete with other microorganisms and to withstand abiotic stresses. Here, we isolated 175 yeasts, comprising 15 different genera, from the wheat flag leaf and characterized a selection of these for various adaptive traits such as substrate utilization, tolerance to different temperatures, biofilm formation, and antagonism toward the fungal leaf pathogen Fusarium graminearum. Collectively our results revealed that the wheat flag leaf is a rich resource of taxonomically and phenotypically diverse yeast genera that exhibit various traits that can contribute to survival in the harsh phyllosphere environment.
Introduction
The phyllosphere is a reservoir of yet unknown microorganisms with intriguing interactions. Microorganisms colonizing the aboveground plant surfaces and tissues, including floral and vegetative parts, are referred to as the phyllosphere microbiome. The phyllosphere is considered to be a harsh environment due to the microbial exposure to limited nutrient sources, UV radiation, temperature oscillations and toxic compounds (Vorholt, 2012). Phyllosphere microorganisms display a wide range of adaptations and antagonistic activities, which are gaining interest for sustaining plant health (Legein et al., 2020; Kavamura et al., 2021). Among the phyllosphere-inhabiting microorganisms, yeasts are found to be abundant as endo- or epiphytes, reaching on average 103–105 colony forming units (CFU) per gram of leaf (Glushakova and Chernov, 2007). The majority of studies on the phyllosphere microbiome, so far, focuses on bacteria and filamentous fungi, especially plant pathogens. Currently, little is known about the diversity and ecological roles of phyllosphere yeasts (Kavamura et al., 2021).
Previous culture-dependent and independent approaches have shown a predominance of the Basidiomycete yeasts Sporobolomyces, Cryptococcus (often reclassified as Papiliotrema) and Pseudozyma (Nasanit et al., 2015a,c, 2016) in the phyllosphere of rice, corn, and sugarcane. Several biotic and abiotic factors impact the abundance and diversity of yeasts in the phyllosphere. These factors include plant genetics (e.g., plant species, genotype, and developmental stage) and environmental conditions such as UV radiation, moisture, geographic location and fungicides (Sapkota et al., 2015). For example, the abundance of Ascomycete yeasts, such as Metschnikowia and Cryptococcus, generally increased over time, particularly in nectar yielding flowers (Glushakova and Chernov, 2010). Additionally, an increase in nutrients found on damaged berries also increased the number of yeasts in the phyllosphere (Janakiev et al., 2019).
The phyllosphere is considered a resource-poor environment, thus microorganisms compete for nutrients and space. Nutrients leak from leaves or fruits into the environment, mainly consisting of carbohydrates such as fructose, glucose and sucrose, and also amino acids and methanol (Mercier and Lindow, 2000). Yeasts are known for their versatile substrate utilization (Żymańczyk-Duda et al., 2017), they can use different carbon sources (e.g., simple sugars, methanol, and methane) as well as amino acids and nitrogen sources (e.g., methylamine, ammonium salts and nitrate; Moliné et al., 2010; Chi et al., 2015; Shiraishi et al., 2015), allowing them to expand their ecological niche (Deak, 2006).
A key step to elucidate the ecological roles of phyllosphere yeasts lies in determining their ability to withstand harsh environmental conditions and their interactions with other phyllosphere members. A number of mechanisms have been proposed to facilitate the adaptations of yeasts to the phyllosphere environment. For example, the majority of phyllosphere yeasts are present in highly organized multicellular communities called biofilms (Váchová and Palková, 2018), which play a role in stress resilience. More specifically, biofilm formation has been implicated in resistance to fungicides and proposed as a physical barrier on plant surface injuries, preventing fungal hyphae from entering the plant tissue (Villa et al., 2017).
Currently, the majority of studies on environmental yeasts focusses on their biocontrol potential (Schisler et al., 2002a, 2015). Several studies have shown the ability of yeasts to inhibit plant pathogens and to protect against post-harvest diseases via the production of secondary metabolites, cell-wall-degrading enzymes, and so-called killer toxins (Freimoser et al., 2019). The mechanisms underlying these antagonistic activities have been identified only for a few species. For example, the production of pulcherriminic acid by Metschnikowia pulcherrima has been implicated in the growth inhibition of Botrytis cinerea (Sipiczki, 2020), whereas polymers (e.g., pullulan), volatiles (e.g., ethanol, phenylethanol and ethyl acetate) and secondary metabolites (e.g., aureobasidins) produced by A. pullulans have been shown to inhibit the growth of Alternaria alternata and B. cinerea (Contarino et al., 2019; Yalage Don et al., 2020; Di Francesco et al., 2021). For the majority of biocontrol yeasts, however, the mechanisms underlying the antagonistic activity are still unknown (Gore-Lloyd et al., 2019). Investigations of their lifestyles and adaptations to the phyllosphere environment will contribute to a better understanding of the interplay between yeasts and other phyllosphere members.
The present study aimed at investigating the taxonomic diversity of yeasts from wheat flag leaves, i.e., the last leaf before the ear emergence. The flag leaf can contribute up to 40% of the final photosynthetic capacity of wheat plants and therefore has a major impact on yield (Sylvester-Bradley et al., 1990). For a subset of taxonomically different flag leaf yeasts, we assessed their phenotypic and metabolic potential, including biofilm formation, substrate utilization spectrum, growth at different temperatures and antagonism toward the fungal pathogen Fusarium graminearum. Our results provide a first step toward characterizing the yeast diversity specifically found in wheat flag leaves and serve a foundation for further studies on the ecological roles of phyllosphere yeasts.
Materials and Methods
Isolation of Yeasts From the Wheat Flag Leaf
Yeasts were isolated from the flag leaves of wheat (Triticum aestivum) cultivar Elixer. Samples were collected during the spring of 2020 at Taastrup, Denmark (55°38′46″N 12°17′53″E) on a fungicide untreated field plot. Plants were sampled at the flowering stage (growth stage 61–69 according to Zadoks code (Zadoks et al., 1974)). Three different methods were used to isolate both epiphytic and endophytic yeasts. For the first method a washing solution (0.5% Tween80 and 0.9% NaCl) was added to a 15 ml-tube with 33 flag leaves. Samples were vortexed for 1 min, sonicated for 2 min at 45 kHz, and vortexed again for 1 min. Leaves were transferred to a new tube and blended. The blended-washed leaves were stored in 15% (v/v) glycerol. In the second method, the washing solution was centrifuged for 5 min at 5.000 rpm, the supernatant was removed and the pellet was stored in 3 ml of 15% glycerol (v/v). The third method consisted of blending leaves directly (without the washing step) and storing in 15% glycerol (v/v). All samples were stored at −80°C.
For the selective isolation of yeasts, glycerol samples were 10-fold serial diluted (10−1–10−7) with 0.9% NaCl and plated on different media. Suspensions were plated on Malt Extract Agar (MEA), Potato Dextrose Agar (PDA), Sabouraud Agar (SDA), 869 media supplemented with wheat flag leaf extract (Eevers et al., 2015) and Yeast Extract Peptone Dextrose (YEPD), with and without 10% lactic acid to favor yeast growth. Bacterial growth was prevented by adding 50 μl/ml chloramphenicol and 50 μl/ml tetracycline. Plates were incubated for 5–7 days at 25°C. At least two of each yeast colony morphologies were picked and streaked on fresh plates (at least twice) to obtain pure cultures. Isolates were grown in YEPD broth for 2–3 days and stored in 15% glycerol (v/v). Isolates which did not grow well in liquid were streaked on plates and, after 3–5 days, cells were directly resuspended in glycerol. All samples were stored at −80°C for long-term storage.
Taxonomic Identification of Yeasts
All 175 yeast isolates were characterized by ITS rRNA gene sequencing. PCR amplifications were conducted using the primers ITS1 (5′-TCCGTAGGT GAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTT ATTGATATGC-3′), synthesized by Integrated DNA Technologies (IDT; White et al., 1990; Lane, 1991). Additionally, the selection of 51 isolates was also characterized by 28S rRNA gene sequencing (also known as D1/D2 sequencing) using the primers LR5 (5′-TCCTGAGG GAAACTTCG-3′) and LROR (5′-ACCCGCTG AACTTAAGC-3′). DNA template for colony PCR was obtained by disrupting the cells by heating in a microwave at 600 W for 60 s, followed by vortexing and again microwaving at 600 W for 60 s in sterile demineralized-water. Samples were centrifuged at 5.000 rpm for 5 min and an aliquot of 3 μl was used as DNA template. The reaction mixture contained Go Taq G2 Hot Start Green Master Mix 2×, 0.4 μm for both primers, DNA template and nuclease-free water. The PCR conditions in the Thermal cycler were: 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 55°C for 30 s, and 72°C for 1 min 30 s, with a final extension at 72°C for 5 min and stored at 12°C. The presence of the PCR products was verified on a 1% agarose gel in TBE buffer. PCR products were purified via ethanol precipitation or using a PCR purification kit (Zymo Research fungal/bacterial DNA Miniprep Kit). Samples were sequenced at BaseClear (Leiden, Netherlands) or Macrogen Inc. (Amsterdam, Netherlands) using the primer ITS1 and LR5. Taxonomic characterization was based on partial internal transcribed spacers (ITS) and the D1/D2 domains of the large subunit of 26S rRNA gene sequences using the NCBI BLAST database.1 For the phylogenetic analysis, sequences were aligned using the MUSCLE algorithm in the MEGA software (version 7.0) and a phylogenetic tree was constructed using Maximum Likelihood method and Tamura-Nei model with a bootstrap of 1.000 replications (Kumar et al., 2016). The tree was visualized using iTOL (version 6). ITS and D1/D2 sequences are deposited at European Nucleotide Archive (ENA) under the project number PRJEB51687.
Culture Conditions
For all in vitro assays, the fungal isolate F. graminearum strain 8/1 (Miedaner et al., 2000) was grown on Potato Dextrose Agar (PDA, pH 7) plates for 5–7 days at 25°C. Yeast cultures were started from glycerol stocks. Isolates were plated on PDA plates and incubated at 25°C for 5–10 days. For all in vitro assays, a loop of the yeast cells was collected and inoculated in 0.9% NaCl. Cells were washed twice by centrifugation at 5.000 rpm for 5 min. An initial cell density (OD600nm) of 0.1 was used for all experiments except the Biolog plate, where an OD600nm of 0.01 was used. All assays were performed at 25°C, which was the temperature used for the isolation of yeasts, unless stated otherwise.
Metabolic Fingerprinting
The Biolog EcoPlate™ system (Biolog Inc., Hayward, CA, United States of America) were used to analyze differential utilization of carbon sources and thereby providing a metabolic fingerprint for each individual strain. Every plate contains 31 different carbon sources grouped into six categories (including amino acids, amines, carbohydrates, carboxylic acids miscellaneous, and polymer compounds) and a water control in three replicates. Yeast cells, described above, were adjusted to OD600nm 0.01 and 100 μl per well was inoculated into the Biolog EcoPlate™. Plates were incubated at 25°C for 10 days at 180 rpm. Metabolism of specific substrates, and consequently growth of the cultures, resulted in a change in the tetrazolium dye. Cell density was measured after 2, 4, 7, and 10 days using a microtiter plate reader (OD590nm), minus values were adjusted to zero. The average well color development (AWCD) was calculated by dividing the total of all values (excluding the water control) by 93. Additionally, the AWCD was calculated for each substrate group. MetaboAnalyst (Version 5.0) was used to visualize the 10 day-inoculation data. A heatmap was created following a Euclidean distance measure and a ward clustering method (Xia et al., 2009).
Growth at Different Temperatures
Yeast cells were collected as described above. The cell density was adjusted to OD600nm 0.1 and 20 μl were added to 180 μl PDB pH 7 in a round-bottom microtiter 96-wells plate. Plates were sealed with plastic wrap and incubated at different temperatures, ranging from 4, 10, 25 to 37°C, at 200 rpm for 7 days. Cell density was measured after 2, 4, and 7 days using a microtiter plate reader (OD600nm).
In vitro Biofilm Formation
Yeast cells were collected from plates and OD600nm was adjusted to 0.1 as described above. A total of 20 μl of this suspension was added to 180 μl PDB in a flat-bottom microtiter plate. Plate was sealed and incubated statically in the dark for 3–4 days at 25°C. After incubation, cells were stained by adding 10 μl of 0.1% crystal violet to each well of the microtiter plate. Plate was incubated at room temperature for 15 min. Cells were washed three times with demineralized-water to remove platonic cells. After that, cells in biofilm were dissolved in 200 μl of 96% ethanol and incubated for 5 min. Cell density was measured using a microtiter plate reader at OD600nm.
Antifungal Activity via Agar-Diffusible and Volatile Compounds
The effect of yeast diffusible compounds on the growth of the fungal pathogen F. graminearum was tested using in vitro dual culture assay. Yeast isolates and the fungal pathogen were previously grown as described above. An aliquot of 10 μl was streaked on one side of a 9-cm Petri dish (ϕ 9 cm) containing PDA at pH 7 (0.5 cm from the edge of the plate). Control treatments were inoculated with 10 μl 0.9% NaCl. A total of four replicates was prepared. Plates were incubated for 24 h at 25°C. After that, a fungal mycelial agar plug (ϕ 5 mm) was placed 24 h later at the opposite side of the Petri dish. Plates were incubated for 6 days at 25°C until the fungus on the control plates reached the edge of the plate. After 6 days, the inhibition zone was determined by measuring the area between the fungal hyphae and the yeast colony (area treatment) using ImageJ (FIJI). The inhibition percentage was calculated as
The effect of volatile organic compounds (VOCs) on the growth of F. graminearum was investigated using two-compartment Petri dishes, which allowed the physical separation between the yeast and the fungal isolates. Yeast and fungal inocula were prepared as described above. An aliquot of 10 μl of the yeast isolates at OD600nm 0.1 was inoculated on one of the compartments of the Petri dish containing PDA (pH 7) and spread evenly. Control treatments were inoculated with 10 μl 0.9% NaCl. A total of four replicates were prepared. Plates were incubated for 24 h at 25°C before inoculation of the fungal pathogen. A mycelium agar plug was placed (ϕ 5 mm) on the other compartment of the Petri dish. Plates were sealed three times with plastic wrap and incubated at 25°C for 4 days. Growth inhibition was calculated by measuring the mycelial growth (area treatment) after VOC exposure. The inhibition of the four replicates was calculated by
Statistical significance was determined with one-way ANOVA, Tukey’s HSD test (p < 0.05).
Scanning Electron Microscopic Analysis
Scanning electron microscopy (JEOL SEM 6400 equipped with Image Convert for windows) was used to visualize the influence of yeasts on the morphological changes in the fungal hyphae of F. graminearum. Samples from the dual culture confrontation assay (described above) were fixed with 1.5% glutaraldehyde in PBS for 1 h while shaking. Then, samples were dehydrated in an increasing percentage of acetone for 20 min each (70, 80, 90, 96, and 100% EtOH) and critical point dried (Baltec CPD-030). Afterward, the samples were sputter coated with platina and palladium to a 20-mm-thickness and stored in a vacuum until use.
Results
Isolation and Phylogenetic Delineation of Phyllosphere Yeasts
A total of 175 yeasts were isolated from the surfaces and internal tissues of wheat flag leaves. ITS and D1/D2-amplicon sequencing revealed a total of 15 genera, representing 25 different species (Figures 1, 2; Supplementary Table S1). Isolation on SDA and PDA media yielded the highest diversity of isolates, 10 and 9 out of 15 different genera, respectively, followed by YEPD (eight genera). The majority of the yeast isolates belonged to the phylum Basidiomycota (145 isolates; 82.9%), including the genera Vishniacozyma (46 isolates; 26.3%), Sporobolomyces (42 isolates; 24.0%) and Papiliotrema (19 isolates; 9.1%). Less frequent genera detected were Pseudozyma, Anthracocystis, Dioszegia, and Rhodotorula. Isolates belonging to the phylum Ascomycota (30 isolates; 17.1%) included Aureobasidium (28 isolates; 16.0%) and Metschnikowia (2 isolates; 1.1%). For further phenotypic and metabolic characterization, we selected 51 yeast isolates based on their phylogenetic delineation, with at least two isolates of the same genus (Figures 1, 2; Supplementary Figure 1).
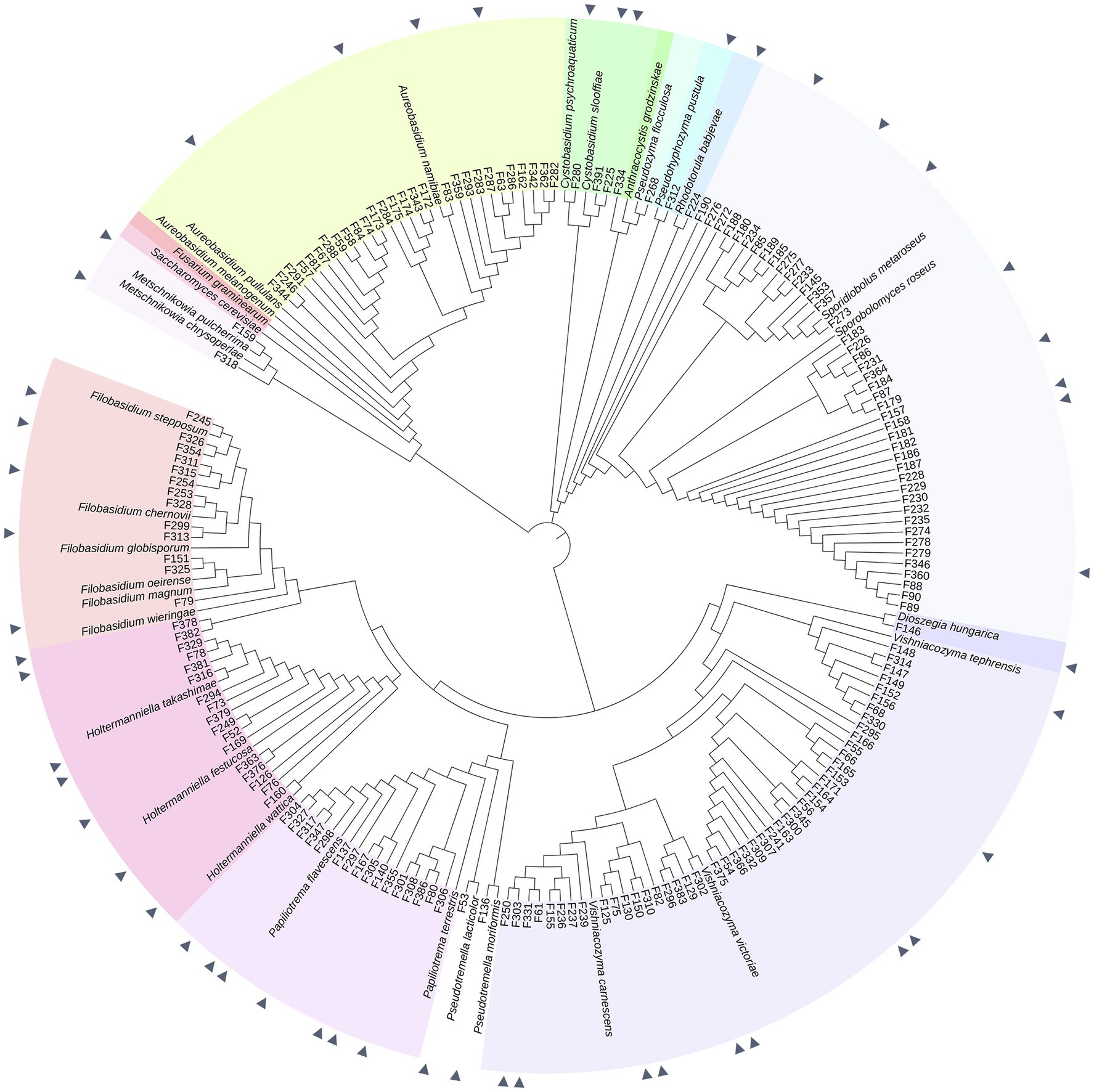
Figure 1. Phylogeny of 175 phyllosphere yeasts isolated from the wheat flag leaf. Phylogeny is based on partial ITS sequences compared to NCBI database. Neighbor Joining tree was constructed with MEGA, MUSCLE for alignment. The iTol software was used to visualize and color-code the tree. Isolates indicated with a triangle are the 51 yeasts selected for the screening of different phenotypic and metabolic traits. Fusarium graminearum strain CBS 131786 was used as an outgroup. Type strains used are described in Supplementary Table S1.

Figure 2. Phenotypic and taxonomic characterization of the selected yeast isolates. Pictures depict 5 to 10-day-old isolates grown on PDA at 25°C. Partial ITS and D1/D2 rRNA gene sequences were compared to the NCBI database for taxonomic identification.
Metabolic Profiling of Phyllosphere Yeasts
To determine the metabolic diversity of the 51 selected phyllosphere yeasts, Biolog EcoPlate was used to screen for their ability to metabolize 31 nutrient sources of which at least 20 are of plant and/or microbial origin. Growth was measured spectrophotometrically, i.e., average well color development values (Supplementary Table S2). The results showed that isolates classified as Vishniacozyma (F300, F345), Aureobasidium (F359, F57) or Papiliotrema (F301) were able to utilize various carbon sources, while none of the Sporobolomyces and Dioszegia isolates grew on any of the substrates tested (Figure 3A). More specifically, Vishniacozyma isolate F345 had the broadest substrate utilization spectrum, followed by Vishniacozyma isolates F75, F300 and F66. Intriguingly, F. graminearum strain 8/1 was able to use 24 out of the 31 different carbon sources, indicating a broad substrate utilization spectrum for this prevalent fungal pathogen of wheat leaves. Among the different types of carbon sources tested, carbohydrates were the preferred substrates followed by polymers and carboxylic acid; none of the 51 yeast isolates were able to use amines under the tested conditions. The most frequently metabolized carbohydrates were D-Xylose, D-Mannitol and N-Acetyl-D-Glucosamine, the polymers were Tween 40 and 80, and the carboxylic acids were D-Galacturonic acid and D-Glucosaminic acid (Figures 3B,C).
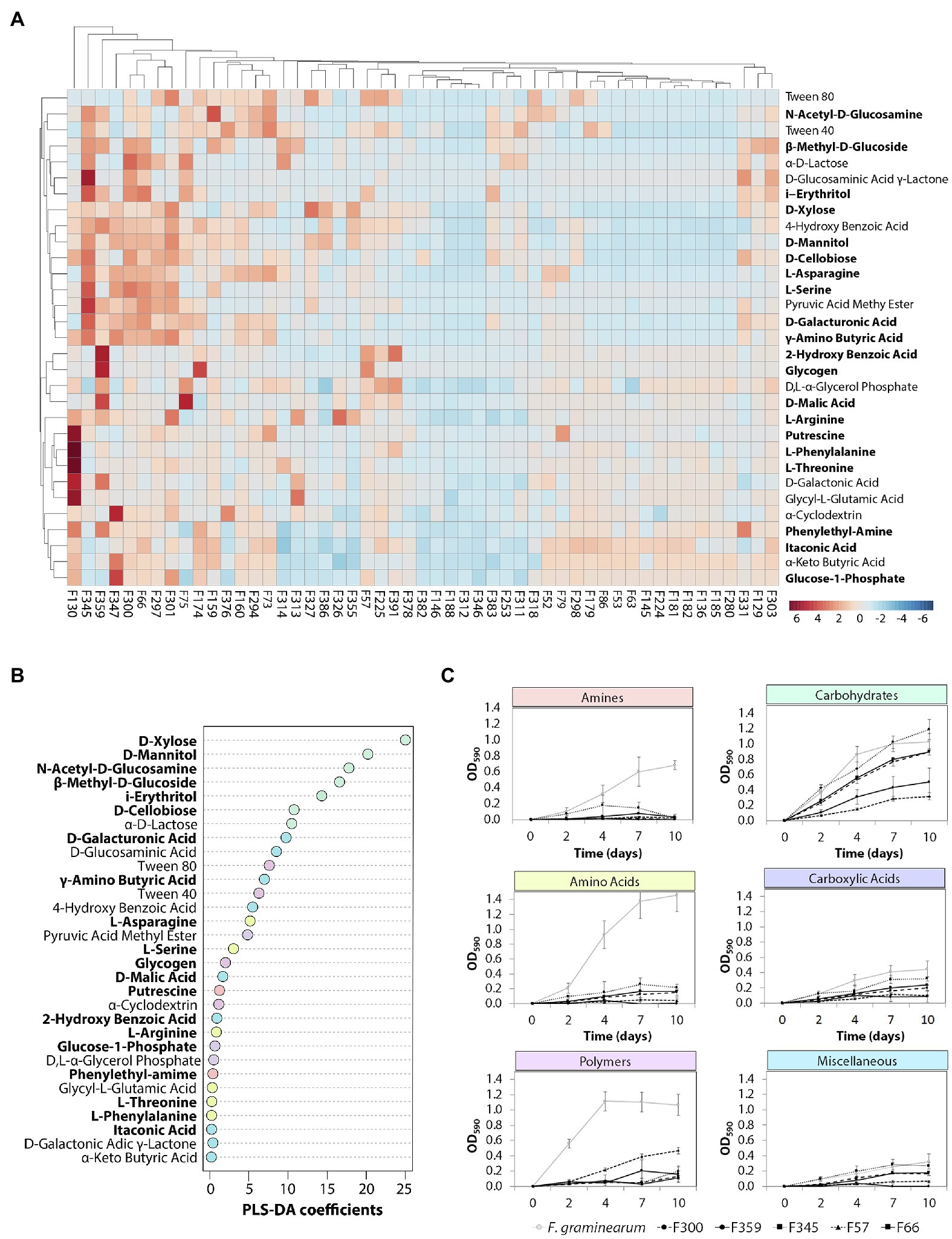
Figure 3. Metabolic profiling of the phyllosphere yeasts. (A) Hierarchical cluster and heat-map analyses of nutrient sources utilized by the selected yeast isolates performed with MetaboAnalyst. Yeast isolates were inoculated (OD600 = 0.01) in the BIOLOG EcoPlates, incubated for 10 days at 25°C and 180 rpm, growth was measured with a plate reader at OD590nm. Columns represent the average of three replicates of each of the 51 isolates. Rows represent the different carbon sources (blue: low abundance, red: high abundance). Compounds of plant and/or microbial origin are displayed in bold. (B) Partial least squares-discriminant analysis (PLS-DA) indicating the importance of each carbon source. (C) The dynamics of the top 5 isolates including F. graminearum with the most diverse metabolic profile per carbon type.
Temperature Growth Range of Phyllosphere Yeasts
In the phyllosphere environment, yeasts are exposed to substantial temperature oscillations. To investigate the temperature range for growth, the selected 51 isolates were tested in vitro at 4, 10, 25 and 37°C. All yeast isolates were able to grow at 25°C, although the growth rate ranged considerably between the different isolates (Supplementary Table S3; Supplementary Figure 2). Most of the isolates (94%) grew well within 2 days of incubation at 25°C, whereas Filobasidium isolate F253 and Aureobasidium isolate F63 required up to 7 days of incubation at this temperature. When the temperature was reduced to 10°C, only 27 and 45 isolates were able to grow after 48 and 96 h, respectively. At 4°C, 13 isolates were able to grow after 4 days of incubation, and an additional 31 isolates showed growth with longer incubation. Isolates F225, F280 and F391 classified as Cystobasidium were unable to grow at 4°C, but grew at 10°C after 7 days. None of the 51 selected yeast isolates was able to grow at 37°C.
Biofilm Formation by Phyllosphere Yeasts
Biofilms play a vital role in stress resilience and can protect yeasts against UV radiation and fungicides. Additionally, they can form a physical barrier on leaf surfaces, preventing penetration of the leaf surface by fungal pathogens. In vitro biofilm formation was detected by crystal violet staining of liquid cultures after 7 days of growth at 25°C with a threshold of OD600nm at 0.05 to prevent any false positives due to background signal. A total of 19 yeast isolates (38%) were able to form a visible biofilm in at least 2 of the 3 screenings performed (Supplementary Figure 3). Biofilm formation was detected for all Metschnikowia (F159 and F318) and Papiliotrema (F297, F298, F301, F327, F347, F355, and F386) isolates, whereas no biofilm formation was detected for Filobasidium, Dioszegia, Rhodotorula and Sporobolomyces isolates under the tested conditions.
Interactions Between Phyllosphere Yeasts and Fusarium graminearum
To investigate the potential of phyllosphere yeasts to inhibit the growth of the leaf pathogen F. graminearum, different in vitro assays were performed. Dual-confrontation assays showed that several yeast isolates inhibited mycelial growth of the pathogen through the production of volatile and/or diffusible metabolites (Figure 4). Ten of the 51 isolates (19,6%) exhibited significant volatile-mediated antifungal activity. Two of these antagonistic yeast isolates (F318 and F159) that were classified as Metschnikowia inhibited the fungal growth by 66 and 56%, respectively. Also isolates classified as Aureobasidium, Papiliotrema, Rhodotorula, Sporobolomyces or Vishniacozyma were able to inhibit the growth of this fungal pathogen via volatiles. Two other Papiliotrema isolates and one Metschnikowia isolate also inhibited hyphal growth via agar-diffusible metabolites, by 55, 54 and 21%, respectively. Altogether, these results indicated that various, taxonomically diverse phyllosphere yeasts can inhibit the growth of F. graminearum via volatile and diffusible metabolites.
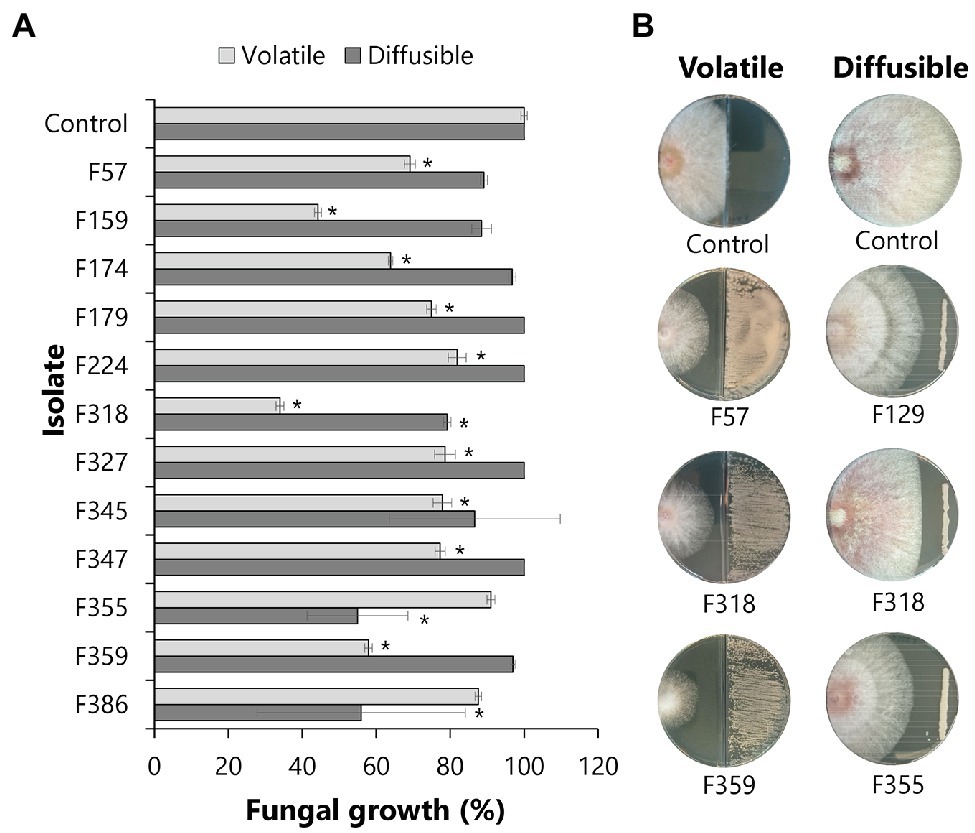
Figure 4. Antagonistic activity of phyllosphere yeasts against F. graminearum. Each isolate was screened against F. graminearum for the production of volatile organic compounds (VOCs) or agar-diffusible compounds. (A) Bars represent the standard deviation of the mean growth percentage of 4 independent replicates. Statistical differences as compared to control (exposed to medium alone) were determined using Student’s t-test (p < 0.05) and are indicated with an asterisk (*). (B) Pictures of the antagonistic activities by representative isolates made after 4 and 6 days of exposure for the volatile and agar-diffusible compounds, respectively.
To visualize the interaction between phyllosphere yeasts and F. graminearum, scanning electron microscopy showed that Metschnikowia isolate F318 and Papiliotrema isolate F386 both altered hyphal morphology compared to the control conditions, where smooth fungal hyphae were observed for F. graminearum (Figure 5). More specifically, direct confrontation between Metschnikowia and F. graminearum resulted in the production of an extracellular matrix (presumably extracellular polysaccharides (EPS)) on the hyphal surface as well as in between the hyphae (Figure 5). Confrontation with Papiliotrema resulted in shrunken and distorted hyphae (Figure 5).
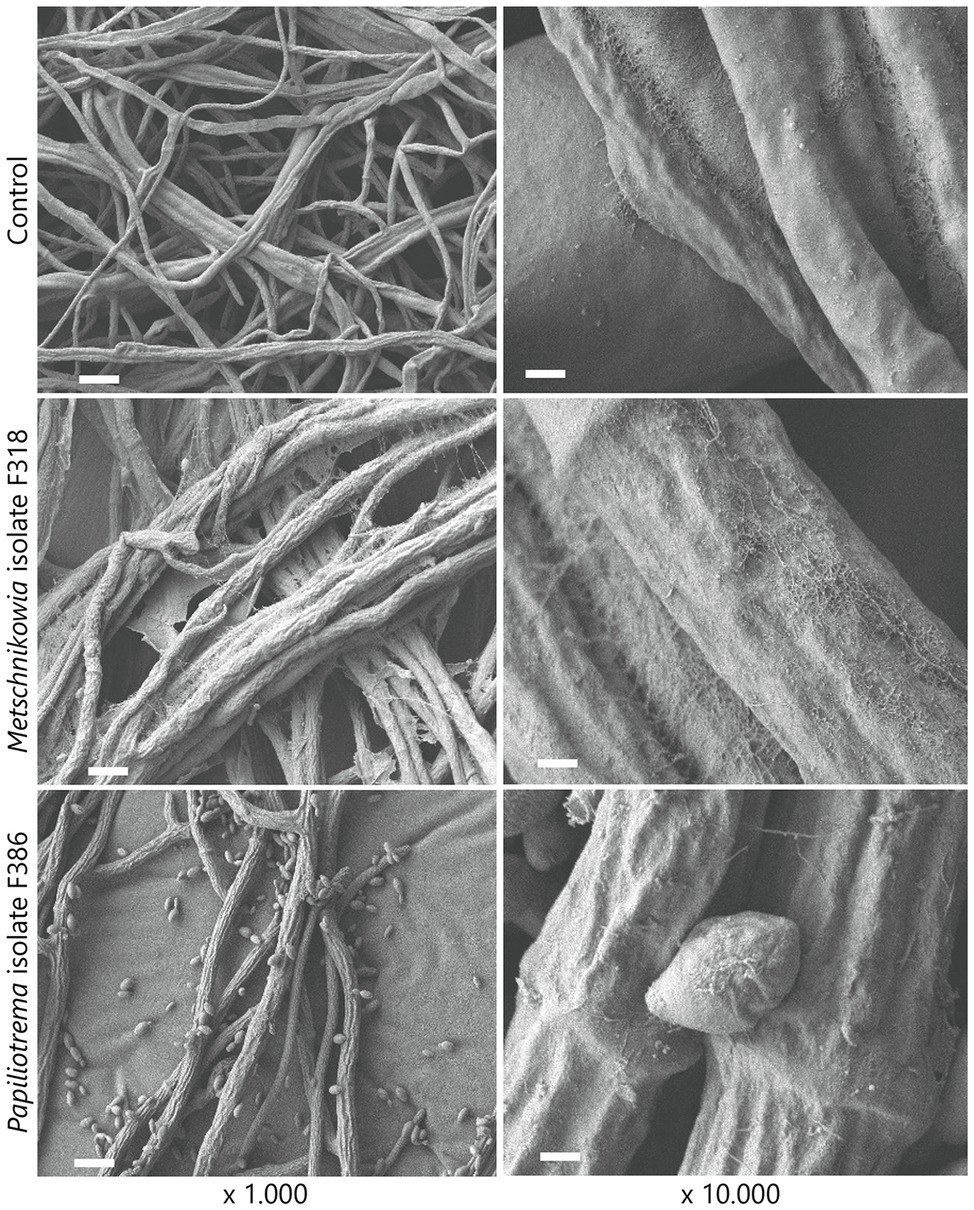
Figure 5. Scanning electron microscopic (SEM) images of the interactions between phyllosphere yeasts and the fungal pathogen F. graminearum. Dual culture assays of F. graminearum growing alone (top panels) and exposed to Metschnikowia (middle panels) and Papiliotrema (bottom panels) isolates. Bars represent 10 μm and 1 μm for ×1.000 and ×10.000 magnification images, respectively.
Discussion
Yeasts are well-known for their biotechnological and medical importance, with Candida and Saccharomyces spp. extensively explored. Plant-associated yeasts have been described for their biocontrol potential, especially in post-harvest disease management but knowledge on yeast ecology and the mechanisms underlying their interactions with other members of the phyllosphere microbiome are still limited. In this study, we examined the diversity of culturable yeasts colonizing wheat flag leaves, their adaptive traits and ability to inhibit the growth of the fungal pathogen F. graminearum (Figure 6).
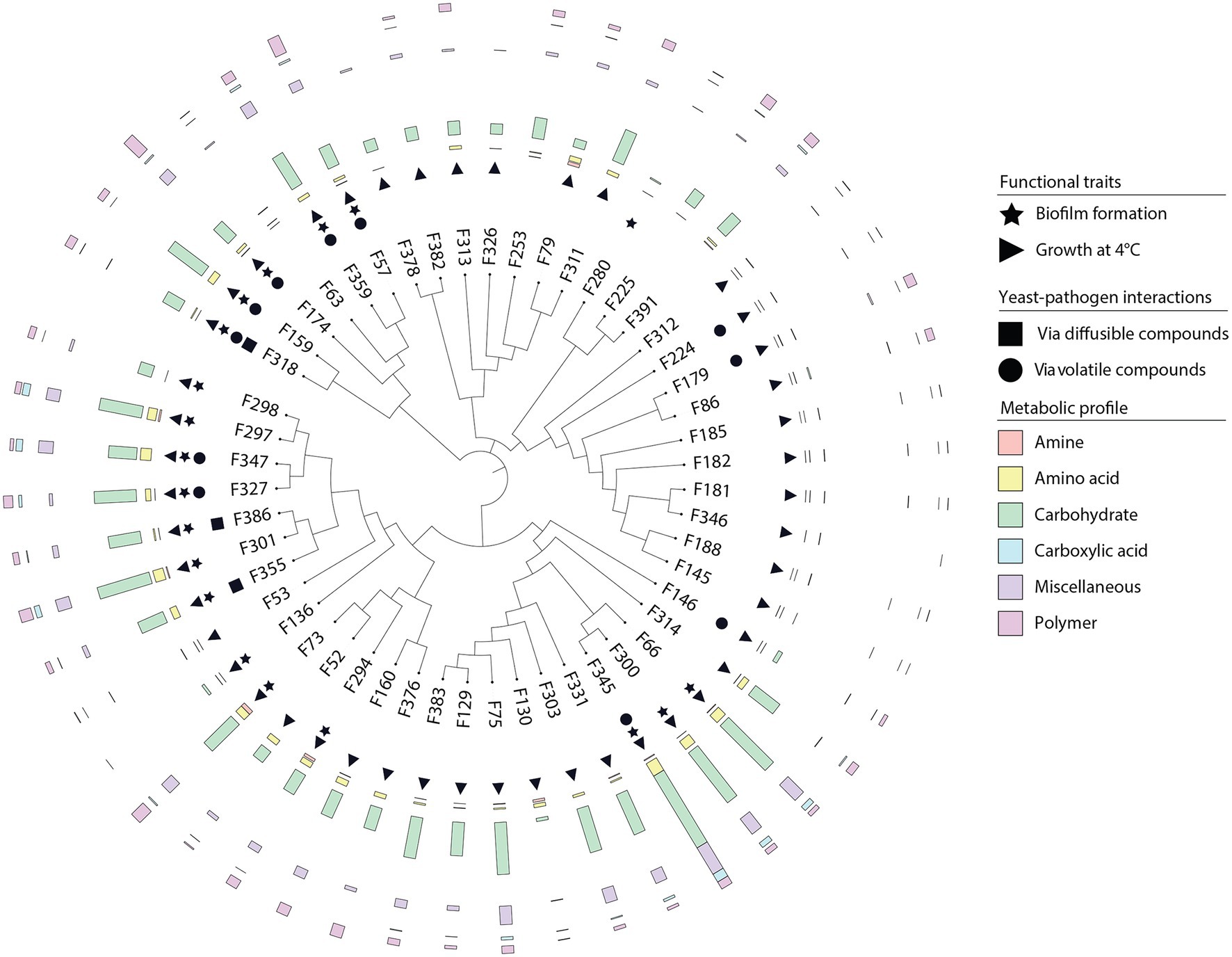
Figure 6. Overview of the adaptive traits of yeasts inhabiting the wheat flag leaf. Neighbor-joining phylogenetic tree based on partial ITS sequences of yeast isolates. Isolates were screened for different traits including biofilm formation, growth at different temperatures, competition with the fungal pathogen F. graminearum via the production of agar-diffusible and volatile compounds, and substrate utilization.
The Wheat Flag Leaf Harbors a Taxonomically Diverse Reservoir of Yeasts
Previous studies have shown that yeasts colonize a wide range of natural environments, including soils, oceans, insects and plants (Boekhout et al., 2021). Plant-associated yeasts are receiving increased attention in the past two decades, in particular those colonizing fruits (Boekhout et al., 2021). In line with previous culture-dependent and independent studies of the wheat phyllosphere (Karlsson et al., 2014; Sapkota et al., 2015; Knorr et al., 2019), we found that Basidiomycetes were the most frequently isolated yeasts. Among these, yeasts belonging to the genera Aureobasidium, Vishniacozyma and Sporobolomyces, also found in phyllosphere of other crops, such as rice, sugarcane and corn (Nasanit et al., 2015b,c, 2016), were the most abundant. Additionally, we also detected Pseudohyphozyma and Pseudotremella species, which were described previously for litter and forest soil (Mašínová et al., 2017) but are rarely isolated from the phyllosphere (Elisashvili et al., 2009; Nguyen et al., 2021).
Here we characterized the yeast isolates based on the variable ITS region as well as the 28S domain (also known as D1/D2) for better taxonomic delineation. Yeast taxonomy has long been based on these phylogenetic markers, but the resolution is limited due to the highly conserved sequences in these regions, especially in basidiomycetous yeasts (Li et al., 2020). Therefore, a large number of yeast species are being reclassified (Yurkov et al., 2021). For example, Cryptococcus species are being reclassified as Papiliotrema and Filobasidium, whereas Candida species can be found in multiple teleomorphic taxa with different generic names. Hence, the yeasts described here will be subjected in the near future to whole genome sequencing for improved taxonomic delineation.
Adaptations of Yeasts to the Harsh Phyllosphere Environment
The phyllosphere is a heterogeneous environment where microorganisms are exposed to different (a)biotic stresses. Here we studied a number of traits that may contribute to the yeast’s adaptation to the phyllosphere, including carbon utilization, growth at different temperatures, and biofilm formation. Nutrients, such as carbon compounds, are important for growth and the synthesis of secondary metabolites (Andrews, 1992). A diverse metabolic profile can offer an advantage to withstand the harsh conditions in the phyllosphere, allowing microorganisms to expand their ecological niche by making better use of the available carbon sources (Deak, 2006). Here we showed that the yeasts from the wheat flag leaf displayed diverse metabolic profiles with varying degradation rates. Vishniacozyma and Aureobasidium isolates, both highly abundant on the wheat flag leaf, displayed the most diverse substrate utilization profiles. Unexpectedly, Sporobolomyces isolates were unable to use any of the carbon sources tested under these conditions, despite their frequent and high abundances in wheat leaves found in the present and previous studies (Bashi and Fokkema, 1977; Sapkota et al., 2017). It is likely that Sporobolomyces species colonize the wheat flag leaves through the utilization of glucose, fructose or sucrose, sugars that are present in the phyllosphere and enable rapid colonization (Leveau and Lindow, 2000) and competition with leaf pathogens such as Cochliobolus sativus, partly by limiting glucose and amino acid availability. Also, Metschnikowia isolates have been shown to inhibit the post-harvest pathogen Colletotrichum gloeosporioides through competition for glucose and fructose (Tian et al., 2018; Sipiczki, 2020). To begin to understand ecological niche overlap or niche differentiation between the phyllosphere yeasts and the fungal pathogen F. graminearum strain 8/1, we also determined the metabolic profile of this pathogen. We found that F. graminearum is not only able to utilize most of the nutrient sources tested, but also utilizes amino acids, amines and polymers that could not be utilized by the phyllosphere yeasts tested. This versatile carbon utilization profile likely contributes to the successful infection of a wide range of cereal crops by this pathogen.
In addition to oscillating nutrient availability, phyllosphere yeasts also need to adapt to oscillating temperatures, which can differ drastically between day and night, winter and spring. Yeasts from the genera Sporobolomyces, Rhodotorula and Vishniacozyma have been proposed to tolerate extreme environmental conditions and to help plants adapting to cold environments (Buzzini et al., 2018; Vujanovic, 2021). Our results showed that all isolates from these genera grew at different temperatures, including low temperatures. Also, biofilms play an important role in stress resilience and protection against harsh environmental conditions (Váchová and Palková, 2018), including temperature oscillations. Additionally, biofilms form a physical barrier on injuries of plant surfaces preventing the invasion by pathogenic fungi. We show that biofilm formation was wide-spread among the wheat phyllosphere yeasts, including all Metschnikowia and Papiliotrema isolates. Previous studies have proposed biofilm formation as one of the antagonistic mechanisms employed by biocontrol Metschnikowia isolates on grape wounds (Sipiczki, 2020). This mode of action is hypothesized to be based on growth inhibition of the pathogen by increasing colonization efficiency (e.g., competition for space). However, other biocontrol mechanisms, e.g., production of secondary metabolites, cannot be excluded to contribute to the observed antagonistic activity. Currently, data on biofilm formation by environmental yeasts is sparse (Cordero-Bueso et al., 2017) and our results suggest this may be a more common trait in the phyllosphere. Future investigations involving microscopy should address if these biofilms occur in planta and if they contribute to the phyllosphere competence of yeasts.
Interactions Between Yeasts and the Fungal Pathogen Fusarium graminearum in the Phyllosphere
We found that the production of secondary diffusible compounds with inhibitory activity against F. graminearum was not a wide-spread mechanism among the yeasts isolated from wheat flag leaves. Only two out of the 51 phyllosphere yeasts tested were able to significantly inhibit hyphal growth. However, it is still possible that these yeast isolates can inhibit the fungal pathogen by competing for nutrients and space, rather than via the production of secondary metabolites. Competition for nutrients and space have been proposed as the primary mechanism by which biocontrol yeasts inhibit pathogens (Freimoser et al., 2019). Isolates F355 and F386, which showed the strongest inhibitory effects against F. graminearum, are closely related to Papiliotrema flavescens, formerly known as Cryptococcus flavescens/C. nodaensis. Earlier studies on P. flavescens OH182.9, isolated from wheat heads, showed no antagonism in in vitro experiments, but this isolate was able to reduce disease incidence caused by F. graminearum by 56% in a bioassay (Khan et al., 2001). Field experiments where different winter wheat cultivars were inoculated with this strain also demonstrated reduced disease severity of F. graminearum by 60% (Schisler et al., 2002b; Khan et al., 2004).
Scanning electron microscopy further showed hyphal morphological changes on the hyphae of F. graminearum during the interaction with specific yeast isolates. The interaction with P. flavescens (isolate F386) not only reduced hyphal growth but also changed the hyphal morphology from smooth to shrunken and distorted. This morphological change has been previously observed for F. graminearum confronted by different Paenibacillus peoriae strains, bacteria with a high potential of biocontrol and plant growth promotion (Ali et al., 2021). Interestingly, the interaction of F. graminearum with Metschnikowia isolate F318, an isolate displaying little reduction of hyphal growth in vitro, led to considerable changes in hyphal morphology. These results suggest that Metschnikowia isolate F318 imposed stress to the fungal pathogen resulting in the production of extracellular polysaccharides (EPS). Biofilm formation by other Fusarium species, more specifically F. oxysporum, has been described to protect the fungal pathogen against several fungicides (Peiqian et al., 2014).
Contrary to diffusible secondary metabolites, volatile compounds with inhibitory effects on F. gramineraum were more frequently produced by the phyllosphere yeasts. These low molecular weight compounds serve as a means of interspecies communication from a distance and are proposed as an effective biological control strategy against multiple pathogens (Tilocca et al., 2020). The most promising volatile-producing antagonists belonged to M. pulcherrima (isolates F159 and F318), Aureobasidium pullulans (isolates F57, F174 and F359) and Papiliotrema flavescens (isolates F327 and F347), inhibiting hyphal growth by up to 65%. Volatile compounds produced by Metschnikowia and Aureobasidium species have been previously described for their biocontrol potential against several fungal pathogens, including A. alternata and B. cinerea (Oro et al., 2018). The volatile compounds ethanol, 2-phenylethanol and ethyl acetate have been proposed to be involved in this inhibitory activity (Oro et al., 2018; Contarino et al., 2019; Sipiczki, 2020; Yalage Don et al., 2020) and hypothesized to disrupt the fungal membranes leading to leakage and deformed hyphae (Yalage Don et al., 2020). To date, however, most of the knowledge on yeast volatiles stems from studies using yeasts of biotechnological importance, e.g., the production of volatiles by yeasts and their effects on wine aroma (Bordet et al., 2020). Further research efforts should be put on characterization of the volatile compounds produced by environmental yeasts and their ecological roles.
Concluding Remarks
The wheat flag leaf is a diverse reservoir of yeasts which can withstand the harsh conditions found in the phyllosphere through different mechanisms. Here we provide a first insight into the genetic and phenotypic diversity of yeasts living on and in the wheat flag leaf. Further research should focus on the molecular mechanisms underlying the interactions between yeasts, including the identification of genes and metabolites involved in antifungal activity. To this end, current approaches and tools already in use for model yeasts can be employed for comparative genomics and functional analysis of the phyllosphere yeasts. Unraveling the mechanisms underlying the antagonistic activities would not only improve our understanding of the ecology of phyllosphere yeasts, but also contribute to the discovery of novel yeast-based biocontrol products.
Data Availability Statement
Data was deposited at the ENA database under accession number PRJEB51687 (secondary accession ERP136341), available from June 19th, 2022.
Author Contributions
LG and VC designed the study, processed and interpreted the data, and drafted the manuscript. LG, CV, and VC performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
All authors are supported by the Novo Nordisk Foundation (Grant NNF19SA0059348).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Alex Gobbi (Copenhagen University) for processing the wheat flag leaves. We thank Wilhelm Schäfer (Hamburg University) for providing the Fusarium graminearum strain 8/1. We also thank Joost Willemse (Leiden University) for the assistance with the scanning electron microscopy. This manuscript is publication number 7454 of the Netherlands Institute of Ecology (NIOO-KNAW).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.908628/full#supplementary-material
Footnotes
References
Ali, M. A., Lou, Y., Hafeez, R., Li, X., Hossain, A., Xie, T., et al. (2021). Functional analysis and genome mining reveal high potential of biocontrol and plant growth promotion in nodule-inhabiting bacteria within Paenibacillus polymyxa complex. Front. Microbiol. 11:618601. doi: 10.3389/fmicb.2020.618601
Andrews, J. H. (1992). Biological control in the phyllosphere. Annu. Rev. Phytopathol 30, 603–635. doi: 10.1146/annurev.py.30.090192.003131
Bashi, E., and Fokkema, N. J. (1977). Environmental factors limiting growth of Sporobolomyces roseus, an antagonist of Cochliobolus sativus, on wheat leaves. Trans. Br. Mycol. Soc. 68, 17–25. doi: 10.1016/s0007-1,536(77)80146-0
Boekhout, T., Amend, A. S., el Baidouri, F., Gabaldón, T., Geml, J., Mittelbach, M., et al. (2021). Trends in yeast diversity discovery. Fungal Divers. 2021, 1–47. doi: 10.1007/S13225-021-00494-6
Bordet, F., Joran, A., Klein, G., Roullier-Gall, C., and Alexandre, H. (2020). Yeast-yeast interactions: mechanisms, methodologies and impact on composition. Microorganisms 8, 1–33. doi: 10.3390/microorganisms8040600
Buzzini, P., Turchetti, B., and Yurkov, A. (2018). Extremophilic yeasts: the toughest yeasts around? Yeast 35, 487–497. doi: 10.1002/YEA.3314
Chi, M., Li, G., Liu, Y., Liu, G., Li, M., Zhang, X., et al. (2015). Increase in antioxidant enzyme activity, stress tolerance and biocontrol efficacy of Pichia kudriavzevii with the transition from a yeast-like to biofilm morphology. Biol. Control 90, 113–119. doi: 10.1016/J.BIOCONTROL.2015.06.006
Contarino, R., Brighina, S., Fallico, B., Cirvilleri, G., Parafati, L., and Restuccia, C. (2019). Volatile organic compounds (VOCs) produced by biocontrol yeasts. Food Microbiol. 82, 70–74. doi: 10.1016/J.FM.2019.01.008
Cordero-Bueso, G., Mangieri, N., Maghradze, D., Foschino, R., Valdetara, F., Cantoral, J. M., et al. (2017). Wild grape-associated yeasts as promising biocontrol agents against Vitis vinifera fungal pathogens. Front. Microbiol. 8:2025. doi: 10.3389/fmicb.2017.02025
Deak, T. (2006). “Environmental factors influencing yeasts,” in The Yeast Handbook. eds. C. A. Rosa and G. Péter (Berlin, Heidelberg: Springer), 155–174.
Di Francesco, A., Di Foggia, M., Corbetta, M., Baldo, D., Ratti, C., and Baraldi, E. (2021). Biocontrol activity and plant growth promotion exerted by Aureobasidium pullulans strains. J. Plant Growth Regul. 40:244. doi: 10.1007/S00344-020-10,184-3/FIGURES/5
Eevers, N., Gielen, M., Sánchez-López, A., Jaspers, S., White, J. C., Vangronsveld, J., et al. (2015). Optimization of isolation and cultivation of bacterial endophytes through addition of plant extract to nutrient media. Microbial Biotechnology 8, 707–715. doi: 10.1111/1751-7915.12291
Elisashvili, V., Kachlishvili, E., Tsiklauri, N., Metreveli, E., Khardziani, T., and Agathos, S. N. (2009). Lignocellulose-degrading enzyme production by white-rot Basidiomycetes isolated from the forests of Georgia. World Journal of Microb. Biotechnol. 25, 331–339. doi: 10.1007/s11274-008-9,897-x
Freimoser, F. M., Rueda-Mejia, M. P., Tilocca, B., and Migheli, Q. (2019). Biocontrol yeasts: mechanisms and applications. World J. Microbiol. Biotechnol. 35:154. doi: 10.1007/s11274-019-2,728-4
Glushakova, A. M., and Chernov, I. Y. (2007). Seasonal dynamic of the numbers of epiphytic yeasts. Microbiology (N Y) 76, 590–595. doi: 10.1134/S0026261707050128
Glushakova, A. M., and Chernov, I. Y. (2010). Seasonal dynamics of the structure of epiphytic yeast communities. Microbiology (N Y) 79, 830–839. doi: 10.1134/S0026261710060160
Gore-Lloyd, D., Sumann, I., Brachmann, A. O., Schneeberger, K., Ortiz-Merino, R. A., Moreno-Beltrán, M., et al. (2019). Snf2 controls pulcherriminic acid biosynthesis and antifungal activity of the biocontrol yeast Metschnikowia pulcherrima. Mol. Microbiol. 112, 317–332. doi: 10.1111/MMI.14272
Janakiev, T., Dimkić, I., Unković, N., Ljaljević Grbić, M., Opsenica, D., Gašić, U., et al. (2019). Phyllosphere fungal communities of plum and antifungal activity of indigenous phenazine-producing Pseudomonas synxantha against Monilinia laxa. Front. Microbiol. 10, 1–20. doi: 10.3389/fmicb.2019.02287
Karlsson, I., Friberg, H., Steinberg, C., and Persson, P. (2014). Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS ONE 9:e111786. doi: 10.1371/JOURNAL.PONE.0111786
Kavamura, V. N., Mendes, R., Bargaz, A., and Mauchline, T. H. (2021). Defining the wheat microbiome: Towards microbiome-facilitated crop production. Comput. Struct. Biotechnol. J. 19, 1200–1213. doi: 10.1016/j.csbj.2021.01.045
Khan, N. I., Schisler, D. A., Boehm, M. J., Lipps, P. E., and Slininger, P. J. (2004). Field testing of antagonists of Fusarium head blight incited by Gibberella zeae. Biological Control 29, 245–255. doi: 10.1016/S1049-9644(03)00157-9
Khan, N. I., Schisler, D. A., Boehm, M. J., Slininger, P. J., and Bothast, R. J. (2001). Selection and evaluation of microorganisms for biocontrol of Fusarium head blight of wheat incited by Gibberella zeae. Plant Dis. 85, 1253–1258. doi: 10.1094/PDIS.2001.85.12.1253
Knorr, K., Jørgensen, L. N., and Nicolaisen, M. (2019). Fungicides have complex effects on the wheat phyllosphere mycobiome. PLOS ONE 14:e0213176. doi: 10.1371/journal.pone.0213176
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics. eds. E. Stackebrandt and M. Goodfellow (Chichester: John Wiley and Sons), 115–175.
Legein, M., Smets, W., Vandenheuvel, D., Eilers, T., Muyshondt, B., Prinsen, E., et al. (2020). Modes of action of microbial biocontrol in the phyllosphere. Front. Microbiol. 11:1619. doi: 10.3389/fmicb.2020.01619
Leveau, J. H. J., and Lindow, S. E. (2000). Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. 98, 3446–3453. doi: 10.1073/pnas.061629598
Li, A. H., Yuan, F. X., Groenewald, M., Bensch, K., Yurkov, A. M., Li, K., et al. (2020). Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: Proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud. Mycol. 96, 17–140. doi: 10.1016/j.simyco.2020.01.002
Mašínová, T., Bahnmann, B. D., Větrovský, T., Tomšovský, M., Merunková, K., and Baldrian, P. (2017). Drivers of yeast community composition in the litter and soil of a temperate forest. FEMS Microbiol. Ecol. 93:223. doi: 10.1093/FEMSEC/FIW223
Mercier, J., and Lindow, S. E. (2000). Role of Leaf Surface Sugars in Colonization of Plants by Bacterial Epiphytes. Appl. Environ Microbiol. 66, 369–374. doi: 10.1128/AEM.66.1.369-374.2000
Miedaner, T., Reinbrecht, C., and Schilling, A. G. (2000). Association among aggressiveness, fungal colonization, and mycotoxin production of 26 isolates of Fusarium graminearum in winter rye head blight. Zeitschrift fur Pflanzenkrankheiten und Pflanzenschutz 107, 124–134.
Moliné, M., Flores, M. R., Libkind, D., Del Carmen Diéguez, M., Farías, M. E., and Van Broock, M. (2010). Photoprotection by carotenoid pigments in the yeast Rhodotorula mucilaginosa: the role of torularhodin. Photochem. Photobiol. Sci. 9, 1145–1151. doi: 10.1039/c0pp00009d
Nasanit, R., Jaibangyang, S., Tantirungkij, M., and Limtong, S. (2016). Yeast diversity and novel yeast D1/D2 sequences from corn phylloplane obtained by a culture-independent approach. Antonie Van Leeuwenhoek 109, 1615–1634. doi: 10.1007/S10482-016-0762-X
Nasanit, R., Krataithong, K., Tantirungkij, M., and Limtong, S. (2015a). Assessment of epiphytic yeast diversity in rice (Oryza sativa) phyllosphere in Thailand by a culture-independent approach. Antonie Van Leeuwenhoek 107, 1475–1490. doi: 10.1007/S10482-015-0442-2
Nasanit, R., Krataithong, K., Tantirungkij, M., and Limtong, S. (2015b). Assessment of epiphytic yeast diversity in rice (O. sativa) phyllosphere in Thailand by a culture-independent approach. Antonie Van Leeuwenhoek 107, 1475–1490. doi: 10.1007/S10482-015-0442-2/TABLES/3
Nasanit, R., Tangwong-o-thai, A., Tantirungkij, M., and Limtong, S. (2015c). The assessment of epiphytic yeast diversity in sugarcane phyllosphere in Thailand by culture-independent method. Fungal Biol. 119, 1145–1157. doi: 10.1016/J.FUNBIO.2015.08.021
Nguyen, M. P., Lehosmaa, K., Martz, F., Koskimäki, J. J., Pirttilä, A. M., and Häggman, H. (2021). Host species shape the community structure of culturable endophytes in fruits of wild berry species (Vaccinium myrtillus L., Empetrum nigrum L. And Vaccinium vitis-idaea L.). FEMS Microbiol. Ecol. 97, 1–13. doi: 10.1093/femsec/fiab097
Oro, L., Feliziani, E., Ciani, M., Romanazzi, G., and Comitini, F. (2018). Volatile organic compounds from Wickerhamomyces anomalus, Metschnikowia pulcherrima and Saccharomyces cerevisiae inhibit growth of decay causing fungi and control postharvest diseases of strawberries. Int. J. Food Microbiol. 265, 18–22. doi: 10.1016/j.ijfoodmicro.2017.10.027
Peiqian, L., Xiaoming, P., Huifang, S., Jingxin, Z., Ning, H., and Birun, L. (2014). Biofilm formation by Fusarium oxysporum f. sp. cucumerinum and susceptibility to environmental stress. FEMS Microbiol. Lett. 350, 138–145. doi: 10.1111/1574-6968.12310
Sapkota, R., Jørgensen, L. N., and Nicolaisen, M. (2017). Spatiotemporal variation and networks in the mycobiome of the wheat canopy. Front. Plant Sci. 8, 1–10. doi: 10.3389/fpls.2017.01357
Sapkota, R., Knorr, K., Jørgensen, L. N., O’Hanlon, K. A., and Nicolaisen, M. (2015). Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytologist 207, 1134–1144. doi: 10.1111/NPH.13418
Schisler, D. A., Boehm, M. J., Paul, P. A., Rooney, A. P., and Dunlap, C. A. (2015). Reduction of Fusarium head blight using prothioconazole and prothioconazole-tolerant variants of the Fusarium head blight antagonist Cryptococcus flavescens OH 182.9. Biol. Control 86, 36–45. doi: 10.1016/J.BIOCONTROL.2015.04.002
Schisler, D. A., Khan, N. I., Boehm, M. J., and Slininger, P. J. (2002a). Greenhouse and field evaluation of biological control of Fusarium head blight on durum wheat. Plant Dis. 86, 1350–1356. doi: 10.1094/PDIS.2002.86.12.1350
Schisler, D. A., Khan, N. I., Boehm, M. J., and Slininger, P. J. (2002b). Greenhouse and field evaluation of biological control of Fusarium head blight on durum wheat. Plant Dis. 86, 1350–1356. doi: 10.1094/PDIS.2002.86.12.1350
Shiraishi, K., Oku, M., Uchida, D., Yurimoto, H., and Sakai, Y. (2015). Regulation of nitrate and methylamine metabolism by multiple nitrogen sources in the methylotrophic yeast Candida boidinii. FEMS Yeast Res. 15, 1–9. doi: 10.1093/femsyr/fov084
Sipiczki, M. (2020). Metschnikowia pulcherrima and related pulcherrimin-producing yeasts: Fuzzy species boundaries and complex antimicrobial antagonism. Microorganisms 8, 1–19. doi: 10.3390/microorganisms8071029
Sylvester-Bradley, R., Scott, R. K., and Wright, C. E. (1990). Physiology in the production and improvement of cereals. Res. Rev. 18, 1–156.
Tian, Y. Q., Li, W., Jiang, Z. t., Jing, M. m., and Shao, Y. z. (2018). The preservation effect of Metschnikowia pulcherrima yeast on anthracnose of postharvest mango fruits and the possible mechanism. Food Sci. Biotechnol. 27, 95–105. doi: 10.1007/s10068-017-0213-0
Tilocca, B., Cao, A., and Migheli, Q. (2020). Scent of a killer: microbial volatilome and its role in the biological control of plant pathogens. Front. Microbiol. 11:41. doi: 10.3389/FMICB.2020.00041
Váchová, L., and Palková, Z. (2018). How structured yeast multicellular communities live, age and die? FEMS Yeast Res. 18, 1–9. doi: 10.1093/femsyr/foy033
Villa, F., Cappitelli, F., Cortesi, P., and Kunova, A. (2017). Fungal biofilms: targets for the development of novel strategies in plant disease management. Front. Microbiol 8:654. doi: 10.3389/fmicb.2017.00654
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Vujanovic, V. (2021). Tremellomycetes yeasts in kernel ecological niche: early indicators of enhanced competitiveness of endophytic and mycoparasitic symbionts against wheat pathobiota. Plants 10, 1–11. doi: 10.3390/plants10050905
White, T. J., Bruns, T. D., Lee, S. B., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics,” in PCR – Protocols and Applications – A Laboratory Manual (New York: Academic Press), 315–322.
Xia, J., Psychogios, N., Young, N., and Wishart, D. S. (2009). MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660. doi: 10.1093/NAR/GKP356
Yalage Don, S. M., Schmidtke, L. M., Gambetta, J. M., and Steel, C. C. (2020). Aureobasidium pullulans volatilome identified by a novel, quantitative approach employing SPME-GC–MS, suppressed Botrytis cinerea and A. alternata in vitro. Scientific Reports 10:4498. doi: 10.1038/s41598-020-61,471-8
Yurkov, A., Alves, A., Bai, F. Y., Boundy-Mills, K., Buzzini, P., Čadež, N., et al. (2021). Nomenclatural issues concerning cultured yeasts and other fungi: why it is important to avoid unneeded name changes. IMA Fungus 12:18. doi: 10.1186/s43008-021-00067-x
Zadoks, J. C., Chang, T. T., and Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Research 14, 415–421. doi: 10.1111/J.1365-3180.1974.TB01084.X
Keywords: yeast ecology, phyllosphere, (a)biotic stresses, antagonism, culturomics, functional characterization, biofilm formation, carbon utilization
Citation: Gouka L, Vogels C, Hansen LH, Raaijmakers JM and Cordovez V (2022) Genetic, Phenotypic and Metabolic Diversity of Yeasts From Wheat Flag Leaves. Front. Plant Sci. 13:908628. doi: 10.3389/fpls.2022.908628
Edited by:
Tomislav Cernava, Graz University of Technology, AustriaReviewed by:
Maged M. Saad, King Abdullah University of Science and Technology, Saudi ArabiaZachary Albert Noel, Auburn University, United States
Copyright © 2022 Gouka, Vogels, Hansen, Raaijmakers and Cordovez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Viviane Cordovez, di5jb3Jkb3ZlekBuaW9vLmtuYXcubmw=
 Linda Gouka
Linda Gouka Caroline Vogels1
Caroline Vogels1 Lars H. Hansen
Lars H. Hansen Jos M. Raaijmakers
Jos M. Raaijmakers Viviane Cordovez
Viviane Cordovez