
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 28 July 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.903306
 Zheng Ying1†
Zheng Ying1† Muhammad Awais2†
Muhammad Awais2† Reshmi Akter2
Reshmi Akter2 Fengjiao Xu2
Fengjiao Xu2 Sul Baik2
Sul Baik2 Daehyo Jung2
Daehyo Jung2 Deok Chun Yang2
Deok Chun Yang2 Gi-Young Kwak2*
Gi-Young Kwak2* You Wenying1*
You Wenying1*Discrimination of plant species, cultivars, and landraces is challenging because plants have high phenotypic and genotypic resemblance. Panax ginseng is commonly referred to as Korean ginseng, which contains saponins with high efficacy on cells, and has been reported to be worth billions in agroeconomic value. Korean ginseng’s increasing global agroeconomic value includes additional species and cultivars that are not Korean ginseng but have physical characteristics close to it. This almost unidentifiable physical characteristic of Korean ginseng-like species is discriminated via molecular markers. Single nucleotide polymorphism (SNP), found across the plant species in abundance, is a valuable tool in the molecular mapping of genes and distinguishing a plant species from adulterants. Differentiating the composition of genes in species is quite evident, but the varieties and landraces have fewer differences in addition to single nucleotide mismatch. Especially in the exon region, there exist both favorable and adverse effects on species. With the aforementioned ideas in discriminating ginseng based on molecular markers, SNP has proven reliable and convenient, with advanced markers available. This article provides the simplest cost-effective guidelines for experiments in a traditional laboratory setting to get hands-on SNP marker analysis. Hence, the current review provides detailed up-to-date information about the discrimination of Panax ginseng exclusively based on SNP adding with a straightforward method explained which can be followed to perform the analysis.
Ginseng belongs to the genus Panax and the family Araliaceae, commonly known as Korean ginseng, and has excellent medicinal properties. It is widely used in East Asia as an herbal medicinal plant (Wang et al., 2019; Zhang et al., 2020). Local Korean and overseas consumers favor Korean ginseng (Panax ginseng). The favored Korean ginseng has a modern medical science verification of effectiveness in improving blood circulation and brain function (Kennedy and Scholey, 2003; Li et al., 2017), enhancing immune function (Rivera et al., 2005), preventing diabetes (Lai et al., 2006), and improving sexual performance (de Andrade et al., 2007), as well as having anticancer and anti-aging properties (Keum et al., 2000; Li et al., 2017). The other most used species of the genus Panax include P. quinquefolius, P. notoginseng, P. japonicus, P. pseudoginseng, P. vietnamensis, P. omeiensis, P. assamicus, P. shangianus, P. sinensis, P. stipuleanatus, P. trifolius, P. variabilis, P. wangianus, P. bipinnatifidus, P. sokpayensis, and P. zingiberensis. In addition, several cultivars are introduced mainly for enhancing saponins or protecting against root rot diseases in ginseng species. The Korean cultivars include Chunpoong, Yunpoong, Gopoong, Sunpoong, Gumpoong, Cheongsun, Sunhyang, Sunun, Sunone, K-1, G-1, and Kowon. Furthermore, Jilin Huangguo Renshen, Jishen, Fuxing 01, Fuxing 02, Kangmei 01, Xinkaihe 01, Xinkaihe 02, Zhongnong Huangfengshen, and Zhongda Linxiashen belongs to the Chinese cultivars (Zhang et al., 2020). The conclusive presentation of ginseng species and cultivars belonging to Panax ginseng and P. notoginseng is shown in Figure 1.
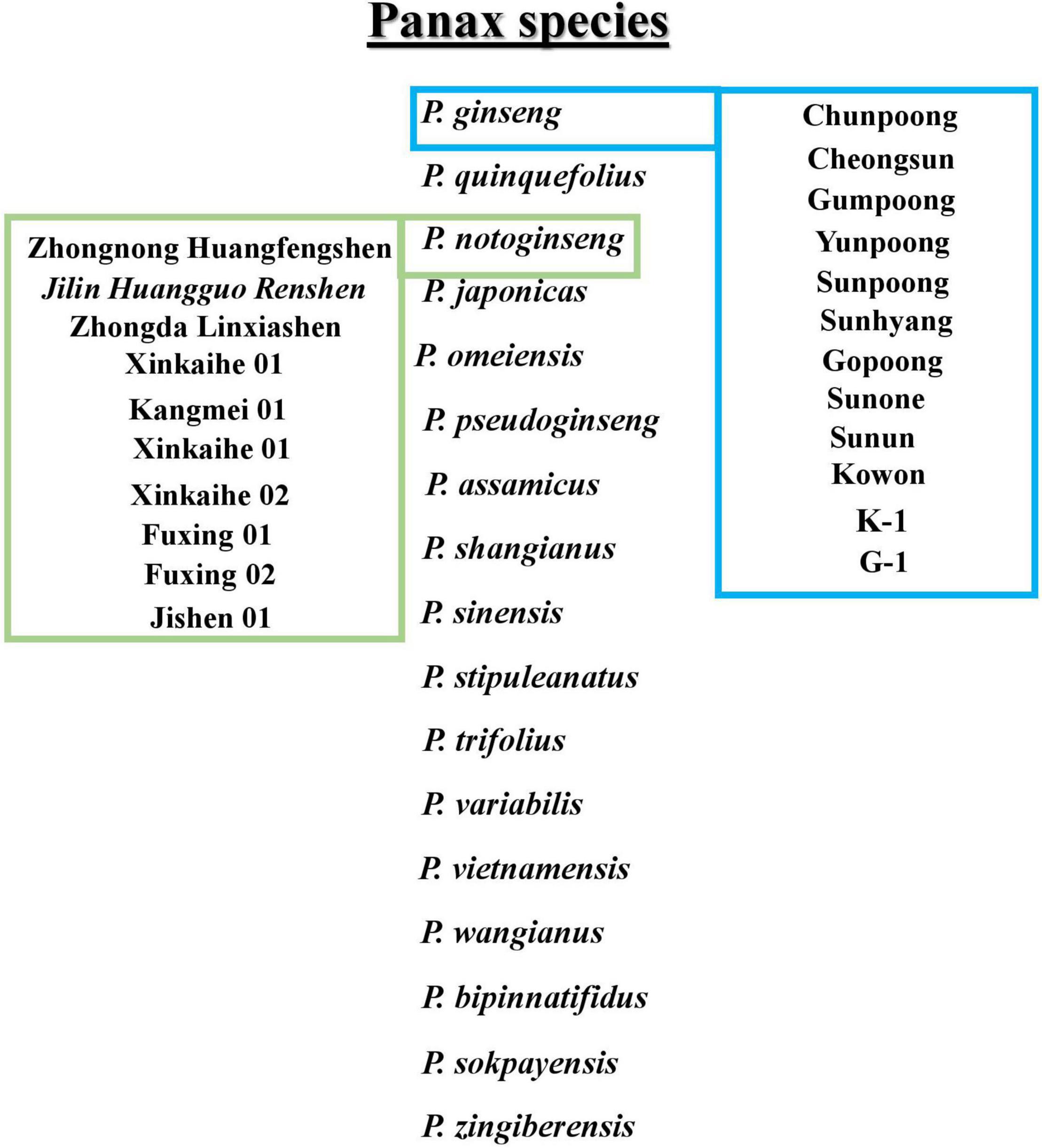
Figure 1. Description of Panax species. In boxes Panax ginseng and Panax notoginseng cultivars are mentioned.
Korean ginseng has outstanding qualities compared to other species in the genus, primarily due to the higher content of ginsenosides. Korean ginseng is highly valued as it provides a basis for its higher price; however, such quality sometimes causes fraudulent labeling (Jung et al., 2014). Additionally, cultivars have been frequently mixed-cultivated on local farms alongside seeds of ginseng cultivars sold in the market by home seed producers. The complexities involved in Korean ginseng’s growth condition, such as the cultivation process, require a careful monitoring system in its cultivating condition, close to its natural habitat (Jo et al., 2017). If proper measures are not taken when growing the medicinal plants, it hinders the management of ginseng cultivation and affects the quality control of ginseng products (Wang et al., 2016a), safeguarding public health and consumers’ rights (Wang et al., 2011a). Though the medicinal values and economic benefits of different ginseng preparation processes are significantly different, product adulteration and substitution severely affect its quality and causes widespread problems in the ginseng market (Ichim and de Boer, 2020).
To overcome the problem of counterfeits, efficient discrimination methods for ginseng varieties/cultivars are necessary to combat adulterants. For the last few decades, traditional methods of discriminating ginseng cultivars have been based on phenotypic observations, but environmental and developmental factors often affect morphological characteristics. Different ginseng cultivars are highly similar in their morphology, especially in the early developmental stage of ginseng, dried ginseng, seeds, and seedlings. Hence, rendering differentiation is quite tricky and sometimes impossible. Besides, differentiating ginseng cultivars based on their morphological characteristics cannot be used to screen many ginseng samples. Therefore, a simple method of DNA analysis is straightforward and highly desirable (Wang et al., 2010a). Thus, techniques have been introduced in the form of allozymes (Koren et al., 2003), restriction fragment length polymorphism (RFLP) (Ngan et al., 1999), simple sequence repeats (SSR) (Bang et al., 2011), amplified fragment length polymorphism (AFLP) (Ha et al., 2002), random amplified polymorphic DNA (RAPD), polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) (Um et al., 2001), and single nucleotide polymorphism (SNP) (Kim et al., 2016). A comprehensive description of biochemical and DNA markers is provided in Figure 2.
Though several review articles have been published on the discrimination of Ginseng (Jo et al., 2017; Goodwin and Proctor, 2019; Ichim and de Boer, 2020) that cover the overall genetic markers, a concise review article on SNP-based studies has not been published yet. Furthermore, regarding research articles, a high amount of data has been published on SNP in cpDNA, mtDNA, and nuclear DNA. Therefore, there is an immediate need for summarized literature to be published that wholly deals with the studies based on single nucleotide polymorphism (SNP). The current review article explains SNP-based discrimination of the ginseng species and cultivars step-by-step, i.e., starting from the chloroplast to mitochondria and then the nuclear genome. In addition, a simple method to perform the SNP analysis in traditional labs is provided.
Figure 3 Illustration of the three genomes found in a plant cell. Chloroplast and mitochondrial genome are drawn with OGDRAW (Lohse et al., 2007).1
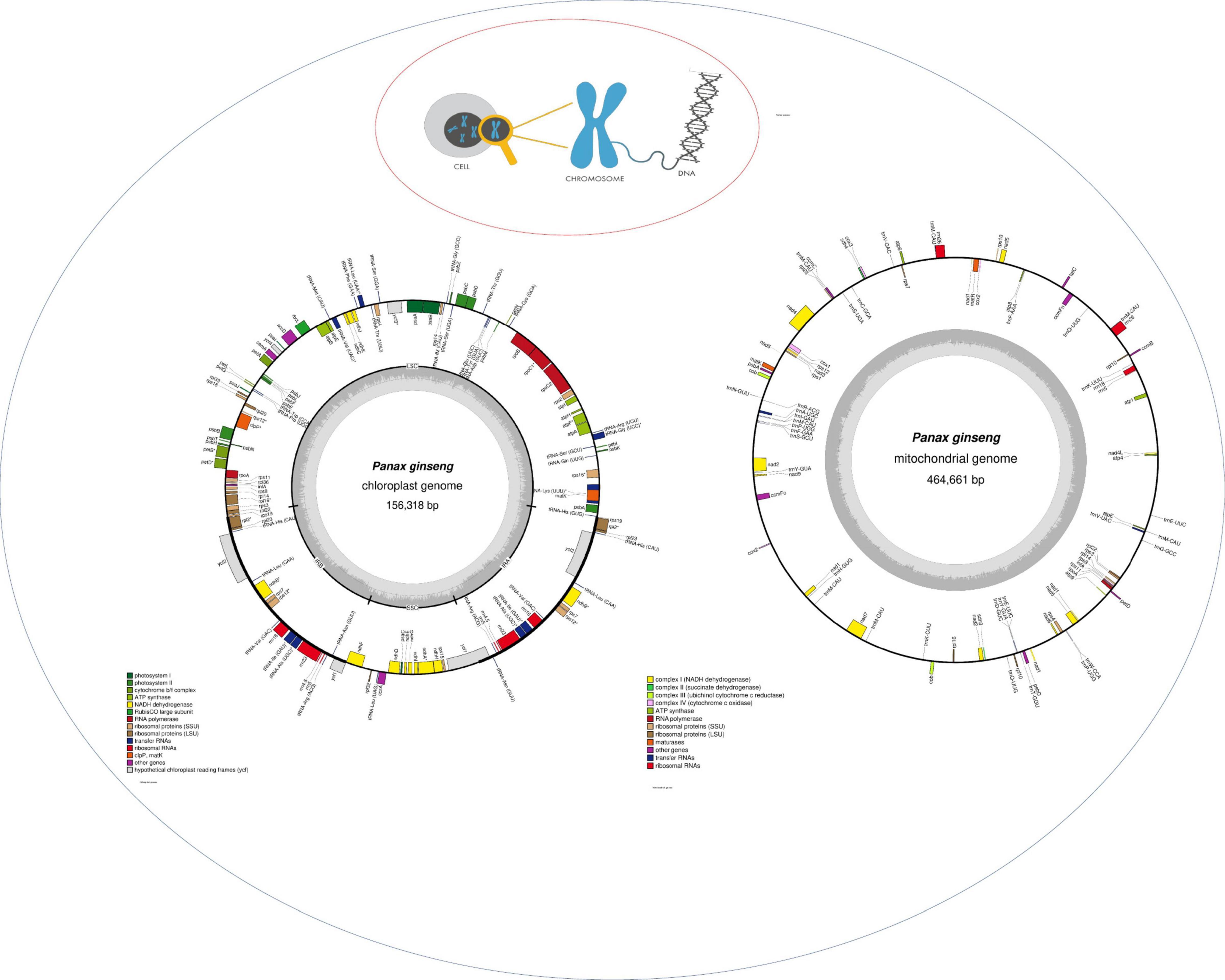
Figure 3. Representation of the typical genomes found in plant cell. The description of genes in cpDNA and mtDNA is highlighted in colors.
The chloroplast is photosynthetic organelles that provide sustenance to green plants. In angiosperms, most of the chloroplast genomes are circular, having duplex DNA, containing one large single-copy region (LSC), one small single-copy region (SSC), and a pair of inverted repeats (IRs). In chloroplast, gene content and order are highly conserved, but the genome size fluctuates from 120 to 160 kb in length. Most studies are concerned with plant species discrimination; evolutionary studies and phylogenetic relationships are based on cpDNA because of its haploid nature, maternal inheritance, highly conserved gene content, and genome structure (Song et al., 2017). Adding to it, cpDNA is sometimes highlighted as useful to identify species (Kim et al., 2017), though, in the case of P. ginseng, few studies have been performed. With cpDNA, the first-ever study to distinguish ginseng species, cultivars, and landraces with SNP and dCAPS markers (In dCAPS assay, a mismatch/or mismatches in PCR primer is used to create restriction endonuclease (RE)-sensitive polymorphism based on the target mutation) was conducted by Kim et al. (2015) using InDeL regions on tandem repeats. One of the InDeL at intergenic regions of rps16-trnUUG had two variants of tandem repeats, 13 and 33 bp, which identified one inbred Korean cultivar “Sunhyang” and P. quinquefolius, respectively, in comparison to other Korean ginseng cultivars. Another Tandem repeat with a size of 57 bp in the ycf1 gene discriminated Chunpoong and Hwangsook from different P. ginseng cultivars and P. quinquefolius. In addition, the dCAPS method using restriction enzymes in the rpoc2 gene was considered and, on the SNP site, with a cleavage, generated unique bands of Gumpoong and Cheongsun, among other cultivars; furthermore, rpoc1 exon SNP was detectible by dCAPS marker and developed a unique amplicon for Chunpoong.
While continuing with the trend to distinguish ginseng species on the plastid genome, Nguyen et al. (2017) performed a comparative analysis of the five Panax species and developed gcpm1-14 markers that could discriminate the Panax species. Out of 14, 8 markers produced unique amplicon sizes and were specific to different Panax species. Among the 14 markers, 13 markers could not distinguish between the two cultivars of P. ginseng, i.e., Chunpoong and Yunpoong. Still, the gcpm12 discriminated between the two cultivars and produced an amplicon size of 316 and 373, respectively. Furthermore, gcpm3, gcpm8, and gcpm10 markers could distinguish P. notoginseng from the other Panax species, while the gcpm4 marker was specific to P. quinquefolius. The marker gcpm6 discriminated against P. japonicus while gcpm9 and gcpm14 discriminated against P. vietnamensis from the rest of the 4 Panax species. Among all the primer sets, gcpm12 was the most variable and produced amplicon sizes ranging from 202 (P. vietnamensis) to 316 (P. ginseng, Chunpoong).
Kim et al. (2015) and Giang et al. (2020) used dCAPS method for differentiating ginseng. Basically, dCAPS is a modification of CAPS. It requires a restriction enzyme that cleaves the DNA at specific sites; research conducted by Giang et al. (2020), who did a comparative analysis of seven Panax species, showed the complete chloroplast genome was carried out and that 1,128 SNP in 71 coding gene sequences were identified, while the psaJ, psbN, rpl23, psbF, psbL, rps18, and rps7 were identical among the seven species. Species delimitation was performed based on unique polymorphism found in the sequences found upon multiple alignments; 18 dCAPS primer pairs were designed. For the first two primer sets, Pgdm1-2 were designed on the genes rpl20 and ndhK using the restriction enzyme Cla1. The 3rd primer pair was derived from rps15 using the restriction enzyme Sma1 resulting in specificity for P. ginseng compared to other species. The markers Pqdm4-6 were specific to P. quinquefolius and were designed on the rpoC1, ndhA, and ndhK genes, digested by Sal1, Cha1, and Alu1, respectively. Pndm7-9 markers were obtained from the genes rpoC1, rpoC2, and ndhK digested with the help of Sal1 HindIII, and Cla1 made a unique pattern that was specific to P. notoginseng. For discriminating P. japonicus, markers Pjdm10-11 derived from rpoC2 and rpoB sequences were cleaved with Sal1 and Rsa1, respectively. The markers Pvdm12 and 13, Psdm14-16, and Ptdm17 and 18 were able to discriminate the P. vietnamensis, P. stipuleanatus, and P. trifolius from the other Panax species successfully. Figure 4 describes the patterns of the bands via the gel-based electrophoresis. The genes discussed in the current article are summarized in Table 1.
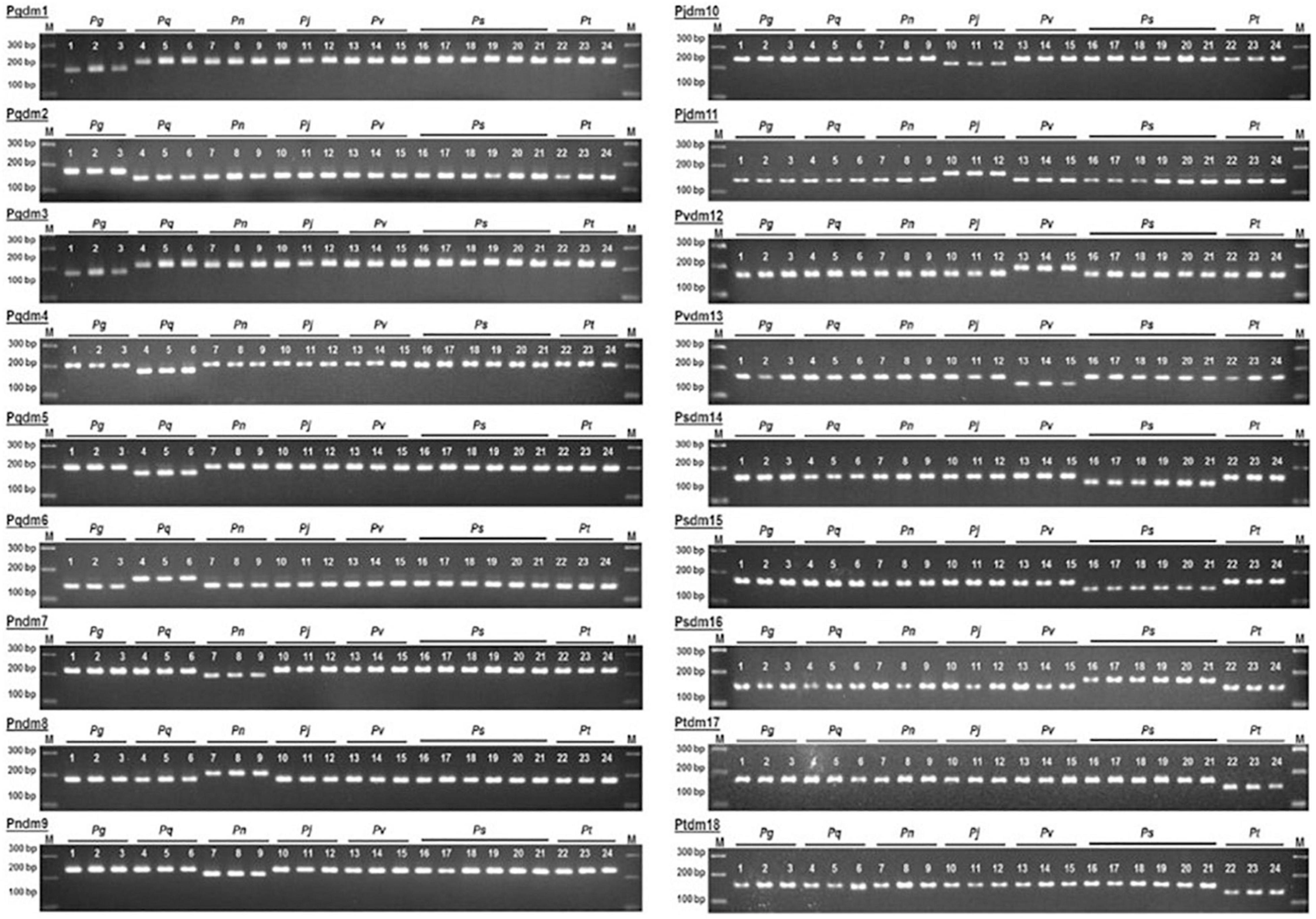
Figure 4. Representation of 18 SNP dCAPS markers for the discrimination of different Panax species. The image is obtained from Giang et al. (2020).
For the specific species, the suitability of genetic markers is sometimes measured in interspecific distance to observe the number of variable sites or pairwise distance between sequences. Highlighted methods include the Refined Single Linkage (RESL) algorithm implemented in the Barcode of Life Database (BOLD) (Ratnasingham and Hebert, 2013) or OBITools (Boyer et al., 2016), etc., whereas a more nuanced technique, mPTP, is a tree-based method that enhances the quality of the selection process of the barcoding markers compared to traditional markers. Furthermore, the technique becomes more useful if some regions are carefully selected which highlight specific structural patterns that enable the discrimination of species, as in the case of ginseng species, i.e., P. ginseng, P. quinquefolius, P. bipinnatifidus, and P. stipuleatus whose identification is quite a complex process because of recently evolved plastid genome. Manzanilla et al. (2018) used the mPTP approach toward the determination of all the Panax species. The method includes the selection of suitable markers upon the extraction of SNP density over the whole plastid genomes and retrieved 2052 SNP. With further analysis, three regions were selected for barcoding analysis, each of the regions contain 83 SNP on average within the Panax species, and fifteen markers were designed. Out of 15 analyzed markers, only four (trnC-rps16, trnS-trnG, petB, and trnE-trnM) were able to discriminate most ginseng species. When used together, three markers (trnC-rps16, trnE-trnM, and psbM-trnD) confidently determine the most traded ginseng species, i.e., P. ginseng, P. quinquefolius, and P. vietnamensis. For further directions, confirming the result via gel-based method or real-time PCR, all the Panax samples with multiple accessions per taxon are to be made to observe variation in the selected markers.
In angiosperms, the mitogenome, compared to the chloroplast genome, was not used to analyze the phylogeny over several decades. In the recent past, researchers have been interested in the potential usefulness of mitochondrial data for phylogenetic data (Han et al., 2021). With regard to differentiating between species or cultivars, much work has been published on the mitogenome in ginseng. One of the genes present in Figure 3, mitochondrial NADH dehydrogenase subunit 7 (nad7) intron 3, was identified by Li et al. (2017) to discriminate between Russian wild ginseng, Chinese cultivated ginseng, and P. ginseng cultivars. With the help of a multi aligned sequence, it was identified that nad7 intron three domains have a homologous sequence between Russian wild ginseng, Korean cultivated ginseng, and Chinese cultivated ginseng, except that a single nucleotide polymorphism “G” was detected in Russian wild ginseng at 700 bp in comparison to Chinese cultivated ginseng “T” at the same position. Allele-specific PCR and real-time PCR were performed to obtain an optimized result for distinguishing the plant species. Furthermore, a research article by Wang et al. (2011b) used a multiplex amplification refractory mutation system (MARMS) to distinguish between “Gumpoong” and “Chungsun” from all of the other cultivars of P. ginseng using mitochondrial nicotinamide adenine dinucleotide (NADH) dehydrogenase subunit 7 (nad7) intron three region. Adding to it, (Wang et al., 2009) considered mitochondrial nad7 intron four region to discriminate Chunpoong cultivar by designing a modified allele-specific primer and using multiplex PCR.
Chunpoong has superior quality over other cultivars and gives a high yield and high resistance to ginseng rusty root rot disease. Wang et al. (2010a) discriminated Chunpoong using mitochondrial cox2 intron II region. A total of six primer pairs were used, from which two acted as universal to produce a 2,179-bp fragment and one acted as a control. In the case of Chunpoong, three primer sets, i.e., Forward, Reverse, and Specific, were designed and used to obtain a size of 1,037 bp. In addition to the other eight cultivars, a specific primer pair set was designed and gave an 1,183 bp band in size. To validate the results, a large number of ginseng samples were analyzed, and discrimination of the Chunpoong was obtained with accuracy.
The major ginseng cultivars cultivated in China, such as Damaya, Ermaya, Biaotiao, Changbo, and Huangguo, are mainly distinguished according to their phenotype. Damaya has higher medicinal and commercial value, often mixed with other cultivars for commercial purposes. To address the issue, Wang et al. (2016b) identified (cox2) as a marker to discriminate Damaya from other cultivars, Ermaya, Biantiao, Changbo, and Huangguo. Upon their sequencing alignments, the resulting sequences were utterly identical. The exception of SNIP (variation of single nucleotide at a specific position among the individuals) was found to be on the nucleotide position 386, replacing a nucleotide in cultivar Damaya A to C. Three primer pairs (forward, reverse, and specific) were designed. Allele-specific PCR was performed, which resulted in a 410 bp band specific to Damaya, while the other cultivars generated only a specific band of 771 bp.
As technology progresses, new techniques are being introduced into the scientific community. One such technique is Kompetitive Allele-Specific PCR (KASP), a newly developed method of single nucleotide polymorphism (SNP) genotyping, which requires only a few SNP markers to genotype various samples. The technique is set by the LGC Genomics Ltd., and is an efficient and low-cost genotyping method. The accuracy and convenience of the KASP assay have made it popular in the analysis of corn (Zea mays) and wheat (Triticum aestivum) (Ma et al., 2021). Furthermore, concerning the P. ginseng cultivar, Gumpoong assembly of the whole mitogenome was performed by Jang et al. (2020). In addition, using the technique, 10 SNP markers were designed to evaluate the diversity between 59 Korean ginseng accessions, including ten accessions of P. quinquefolius and P. notoginseng.
The nuclear genome mainly contains functional genes for metabolic pathways, such as auxin repressed genes. These candidate genes can sometimes be used to identify the best cultivar. Accordingly, Wang et al. (2019) discriminated against the P. ginseng cultivar K-1, which is well known for its good root shape and thus suitable for producing red ginseng and productive lateral roots and effective disease resistance. Data mining of 5 pathogenesis-related (PR) proteins was performed; out of five, one gene PR5 (thaumatin-like protein) contained the SNP region at 289th position (A for G). Multiplex PCR and real-time PCR assays were applied to distinguish K-1 from the rest of the P. ginseng cultivars and 4 Chinese ginseng cultivars (Damaya, Ermaya, Biantiao, and Huangguo). With the help of a specific primer set, an exclusive 310 bp band was generated, which was unique to the K-1 cultivar, while a 577 bp common band was generated by using two universal primer pairs; it was used as a positive control to show that PCR conditions are well optimized. To determine whether the base substitution (A for G) in the K-1 cultivar is responsible for its disease resistance, bioinformatics tools were used, revealing that non-synonymous substitution leads to modifying amino acid residue from aspartic acid (D) to asparagine (N). Modification of the amino acid residue increased the instability index and isoelectric point of PR5 of K-1. The base substitution also changed the secondary structure, including the length of the coil and strand. Further, it changed the tertiary structure of the PR5 protein of K-1, which could be the reason for disease resistance in the K-1 cultivar.
One of the critical genes responsible for the growth and development of the plant is Auxin repressed genes (Park and Han, 2003). Kim et al. (2016) used Auxin repressed protein gene to distinguish the Chunpoong (P. ginseng cultivar) from P. quinquefolius and eight cultivars of Korean ginseng using modified allele-specific primer pair producing a specific 215 bp band. In contrast, on InDel region, a specific primer set for P. quinquefolius was designed, generating a specific band of 489 bp. The universal primer pair produced 609-bp amplicons that were common to all the ginseng cultivars and P. quinquefolius.
Sun et al. (2010) revived the work of Wang et al. (2009) as it was efficient on a limited range of cultivars. Sun et al. (2010) used the “major latex-like protein (MLP-like) gene” expressed highly in 4-year-old Chunpoong. An InDeL and SNP-based marker research was conducted. The InDel marker could identify hybrid F1, which is cross-cultivated between P. ginseng and P. quinquefolius. The SNP-based marker helped in analyzing the Chunpoong population or plant breeding program based on real-time PCR. The genes discussed in the current article are summarized in Table 1.
The worth of P. ginseng is primarily due to ginsenosides, which are saponins. Some researchers like Wen-Ru et al. (2020) considered both the metabolomics and molecular techniques to distinguish the ginseng species and cultivars. In the case of molecular discrimination, functional genes like Dammarenediol synthase (DS), which is the critical functional enzyme in the cyclization of 2, 3-oxidosqualene to dammarenediol II, are the first step of skeleton formation for the most bioactive ginsenoside. Upon the multi align, a sequence of P. ginseng and P. quinquefolius was found, two SNIPS in the intron region, and designed two specific primer sets to discriminate between P. ginseng and P. quinquefolius and gave 571 and 341 bp bands, respectively. Furthermore, the method is reliable and sensitive as it can generate results only with 0.1 ng of DNA. Dammarenediol synthase (DS) enzyme is involved in the biosynthetic pathway of ginsenosides. In addition, ginsenoside’s profile varies within the species and genus with temperature, region, and age. Taking this into consideration, Grazina et al. (2021) hypothesized DS gene as a potential marker and discriminated five major ginseng species, namely P. ginseng, P. quinquefolius, P. notoginseng, P. japonicus, and P. trifolius, using high resolution melting (HRM) analysis. In addition, the applicability of the method developed was effective for commercial ginseng products that include herbal infusions, dried roots, and plant food supplements.
According to the China plant BOL group, the internal transcribed spacer ITS region has higher discriminatory power than the plastid genome (Li et al., 2015). Furthermore, Yang et al. (2012) confirmed that the ITS region provides geographic information about the plant species as a molecular marker. In addition, the ITS region is a good selection for a marker to determine phylogeny at the interspecific and intergeneric levels among flowering plants and other eukaryotes. In contrast, a specific primer set designed in the divergent ITS regions can be used to authenticate the original plants from counterfeits (Feng et al., 2010). Continuing with the trend to identify species based on ITS region, (Osathanunkul and Madesis, 2019) used the combination of a DNA barcode with HRM to discriminate P. ginseng from P. notoginseng. Additionally, Thai ginseng (Talinum paniculatum) and Phytolacca americana were differentiated from Korean ginseng due to their strong resemblance in root shape. Initially, three primer sets were selected, ITS2, rbcL, and trnL, to simultaneously identify the best among them for discrimination of all selected species. rbcL primer pair generated a unique melting curve for Talinum and Panax species, while no distant curves were obtained for Phytolacca species. While in the case of trnl melting curves for P. americana, P. japonica, P. ginseng, and P. notoginseng were nearly identical. ITS2 region, on the other hand, had distant melting curves for all the species and determined all the species successfully. The reference diagram is provided in Supplementary Figure.
Yang et al. (2017) analyzed the 45S region of Korean ginseng cultivar and P. quinquefolius, a few snips were exclusive to G-1 and P. quinquefolius. The Exclusive primer set for G-1 and P. quinquefolius yielded a band in size of 449 bp. In contrast, the specific primer pairs for P. quinquefolius produced a band size of 255 bp. In contrast, the positive control band size for all the ginseng cultivars was 562. The experiments were performed ten times to validate the results. Wang et al. (2010b) adopted the Multiplex PCR method to discriminate the Gumpoong (P. ginseng cultivar) from other cultivars. Also, P. notoginseng, P. quinquefolius, and a few Chinese cultivars such as Biantiao Damaya and Ermaya use the 26S rDNA region.
Over the last 10 years, nucleic acids have been widely altered by substituting the phosphodiester linker or the sugar-phosphodiester backbone with various neutral or charged structures. More than a few of these modified oligonucleotides have developed properties in terms of affinity and binding to DNA and RNA. One of the significant representatives of this new type is peptide nucleic acids (PNAs) (Pellestor and Paulasova, 2004). Lee et al. (2010) used the technology of PNAs to identify between P. ginseng, P. quinquefolius, and P. japonicus using 5 PNA probes designed on 3 SNP in the 5.8S ITS region. Furthermore, a signal intensity comparison between PNA and DNA microarrays was performed to show PNA microarray provides a significantly more stable and specific fluorescent signal intensity than the DNA microarray. With the trend in the identification of P. ginseng based on the ITS region, Kim et al. (2007) identified several SNP in ITS and the 5.8S region of rDNA in Korean ginseng cultivars, and two of the ginseng species, P. quinquefolius, and P. japonicas, that were cleaved by Taq 1 polymerase at specific sites for discrimination of the species and cultivars. This discrimination method provides the breeders with basic information on target-specific cultivars. Figure 5 covers the region from 18S to 26S rDNA. The location of a specific primer pair on the ITS1 region is visualized since specific primer set position could differ among the desirability and regions of SNIP.
The external transcribed spacer (ETS) lies in the intergenic spacer region, which separates the repetitive 18S–5.8S–26S ribosomal gene blocks from each other (Poczai and Hyvönen, 2010). Wang et al. (2011a) upon multi alignment sequence of P. ginseng and P. quinquefolius exploited the two SNP in the ETS region. For both the species, specific primer sets were designed to have an intentional mismatch for more specificity. The remaining two forward and reverse primers were used as universal. A multiplex PCR analysis revealed P. ginseng with 388 bp and P. quinquefolius with 201 bp band sizes. Furthermore, mixed samples of both species and capsules, tea, and processed ginseng were also tested, which were identified efficiently, proving the method was reliable. The Supplementary File contains the Universal primer set position and specific primers pairs position for all the genes discussed in the current article.
Here, we provide the simplest and most reliable method for working with the single nucleotide polymorphism (SNP). SNP are found in plant species across three genomes: the nuclear genome, mitochondrial genome (mtDNA), or chloroplast genome (cpDNA) (Li et al., 2015) explained and discussed the barcodes in genomes for the discrimination of plant species from a basic to newly developed system. But, most researchers themselves decide which regions they want to target and why they choose the specific region, as in the case of Wang et al. (2019) who used targeted functional genes “pathogenesis-related protein 5 gene” or Sun et al. (2010) who used “Major Latex-Like Protein Gene.” Likewise some researchers prefer to use mitochondrial genome or otherwise Chloroplast genome is preferred to discriminate the plant species, cultivars, and varieties.
Starting with genomic DNA Isolation is mainly done using CTAB (Doyle and Doyle, 1987), modified CTAB (Allen et al., 2006), or the DNA isolation kit method. The quantity and quality had to be ensured as it affects PCR reaction. Most researchers use 5 to 50 ng of DNA to perform PCR analysis. The next step to follow is the PCR with the universal primer set; if the plant cpDNA/mtDNA/nDNA is already available at NCBI2 or other available sites3 the target gene can be taken into the consideration and designing of the universal primer pair is brought about.4 The sequence data can be multi aligned with the help of online software.5 Multiple sequence alignment provides a better view to observe the SNIP or InDeL region between two or more species. A specific primer set design is brought about on the SNP site or InDeL region. Optimizing the Multiplex PCR is tedious as researchers work with 3–6 primer pairs in one reaction to optimize them with a single reaction condition. The schematic representation to work with analysis is shown in Figure 6.
This would be different, however, if a plant is chosen whose online data regarding genomes are not available, as in the case of Terminalia ferdinandiana, an Australian native plant commonly known as Kakadu plum plant found in Australia (Mohanty and Cock, 2012). In such cases, some researchers have already designed universal primer pairs on conserved regions, as Dong et al. (2013) who designed 138 sets of primers on the conserved regions on the plastid genome. Such primer pairs can be used to start identifying the genes along with the heterologous sites between related species. Table 2 provides only the primer sets with an amplification rate of 100%. According to Li et al. (2015), on the one hand, the whole-plastid-barcodes have great success in species distinguishing the closely related taxa and are cost-efficient, while standard technology is not available to most laboratories. However, researchers may have to choose between cost efficiency or particularity. In traditional labs, working with a single gene would be cost-effective and less time-consuming.
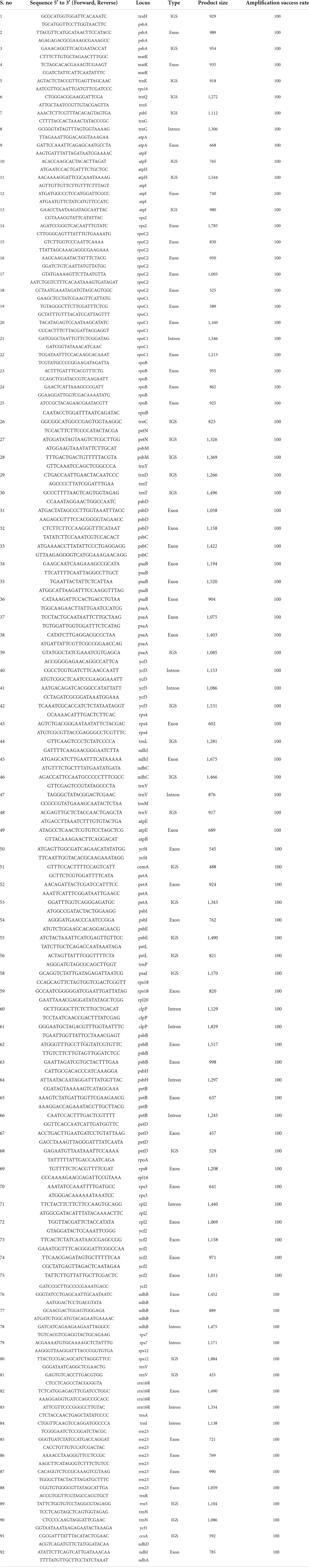
Table 2. Set of universal primer pairs according to Dong et al. (2013).
To detect the polymorphism within the plant species, cultivars, and varieties, a plethora of markers are readily available and used for the discrimination. A low mutation rate can be detected when it comes to SNP analysis. Furthermore, the outcome of the results is much swifter. As technology has been on the rise in the last few decades, the SNP analysis in discrimination has become recognizably important in herbal medicine. In particular, when it comes to P. ginseng, the most valuable medicinal herb, the estimated market worth is $2,084 million (Baeg and So, 2013). As the current article discusses distinguishing between the ginseng species, cultivars, and varieties, few reports discuss functional genes and how one cultivar acts better than the other. Some researchers have outlined a phenomenon known as horizontal plastid genome transfer into the mitochondrial and nuclear genomes (MTPTs) or nuclear genome sequences of plastid origin (NUPTs). These complexities in the genome can cause confusion, false results, and misidentification (Park et al., 2020). Though the work concerning the problem has already started, much more has to be done in the field. Fortunately, there are articles available online for P. ginseng cpDNA, mtDNA, and nDNA (see text footnote 2). So, dealing with MTPTs or NUPTs becomes easier as the researcher does not have to begin from scratch.
As the data above proves, most researchers have focused on nDNA or mtDNA regarding discrimination in ginseng plants. The mtDNA, along with cpDNA, is maternally inherited, and the evolution rate compared with the nuclear genome is much slower. It is well-established that the Mito genome is much larger. In addition, the protein-coding genes are fewer, as mentioned by Cui et al. (2021). In comparison ginseng plastid genome is much smaller which is easy to assemble furthermore, functional or protein-coding genes are higher in number. The above article discusses discrimination of ginseng on the functional genes in mtDNA and nDNA but in case cpDNA the gap is to be filled.
This review article summarizes the work on the discrimination of P. ginseng with the typical three genomes found in plants. It provides a concise work chart on working with SNP as a reliable method. The discrimination of P. ginseng via reliable SNP analysis is suitable for all plant species with convenience and cost-effectiveness that gives a chance to traditional labs to work with confidence in distinguishing plant species.
MA and ZY wrote, drew figures and tables, and collected data of the manuscript. RA, FX, SB, and DJ contributed to collect the data. G-YK performed editing and helped with tables and figures. DY and YW conceptualized the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agri-Food Export Business Model Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (Project No: 320104-03).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.903306/full#supplementary-material
Allen, G. C., Flores-vergara, M., Krasynanski, S., Kumar, S., and Thompson, W. J. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Baeg, I.-H., and So, S.-H. (2013). The world ginseng market and the ginseng (Korea). J. Ginseng Res. 37, 1–7. doi: 10.5142/jgr.2013.37.1
Bang, K.-H., Chung, J.-W., Kim, Y.-C., Lee, J.-W., Jo, I.-H., and Seo, A. (2011). Development of SSR markers for identification of Korean ginseng (Panax ginseng CA Meyer) cultivars. Korean J. Med. Crop Sci. 19, 185–190. doi: 10.7783/KJMCS.2011.19.3.185
Boyer, F., Mercier, C., Bonin, A., Le Bras, Y., Taberlet, P., and Coissac, E. (2016). obitools: a unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182. doi: 10.1111/1755-0998.12428
Cui, H., Ding, Z., Zhu, Q., Wu, Y., Qiu, B., and Gao, P. J. S. R. (2021). Comparative analysis of nuclear, chloroplast, and mitochondrial genomes of watermelon and melon provides evidence of gene transfer. Sci. Rep. 11:1595. doi: 10.1038/s41598-020-80149-9
de Andrade, E., de Mesquita, A. A., de Almeida Claro, J., de Andrade, P. M., Ortiz, V., and Paranhos, M. (2007). Study of the efficacy of Korean Red Ginseng in the treatment of erectile dysfunction. Asian J. Androl. 9, 241–244. doi: 10.1111/j.1745-7262.2007.00210.x
Dong, W., Xu, C., Cheng, T., Lin, K., and Zhou, S. (2013). Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 5, 989–997. doi: 10.1093/gbe/evt063
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Feng, T., Liu, S., and He, X.-J. (2010). Molecular authentication of the traditional Chinese medicinal plant Angelica sinensis based on internal transcribed spacer of nrDNA. Electronic J. Biotechnol. 13, 9–10. doi: 10.2225/vol13-issue1-fulltext-13
Giang, V. N. L., Waminal, N. E., Park, H.-S., Kim, N.-H., Jang, W., and Lee, J. (2020). Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique single nucleotide polymorphism markers. J. Ginseng 44, 135–144. doi: 10.1016/j.jgr.2018.06.003
Goodwin, P., and Proctor, E. (2019). Molecular techniques to assess genetic variation within and between Panax ginseng and Panax quinquefolius. Fitoterapia 138:104343. doi: 10.1016/j.fitote.2019.104343
Grazina, L., Amaral, J. S., Costa, J., and Mafra, I. (2021). Towards authentication of Korean ginseng-containing foods: differentiation of five Panax species by a novel diagnostic tool. LWT 151:112211. doi: 10.1016/j.lwt.2021.112211
Ha, W., Shaw, P., Liu, J., Yau, F. C., and Wang, J. (2002). Authentication of Panax ginseng and Panax quinquefolius using amplified fragment length polymorphism (AFLP) and directed amplification of minisatellite region DNA (DAMD). J. Agric. Food Chem. 50, 1871–1875. doi: 10.1021/jf011365l
Han, E.-K., Cho, W.-B., Tamaki, I., Choi, I.-S., and Lee, J.-H. (2021). Comparative mitogenomic analysis reveals gene and intron dynamics in rubiaceae and intra-specific diversification in damnacanthus indicus. Int. J. Mol. Sci. 22:7237. doi: 10.3390/ijms22137237
Ichim, M. C., and de Boer, H. J. (2020). A review of authenticity and authentication of commercial ginseng herbal medicines and food supplements. Front. Pharmacol. 11:2185. doi: 10.3389/fphar.2020.612071
Jang, W., Lee, H. O., Kim, J. U., Lee, J. W., Hong, C. E., Bang, K. H., et al. (2020). Complete mitochondrial genome and a set of 10 novel Kompetitive allele-specific PCR markers in ginseng (Panax ginseng CA Mey.). Agronomy 10:1868. doi: 10.3390/agronomy10121868
Jo, I. H., Kim, Y. C., Kim, D. H., Kim, K. H., Hyun, T. K., Ryu, H., et al. (2017). Applications of molecular markers in the discrimination of Panax species and Korean ginseng cultivars (Panax ginseng). J. Ginseng Res. 41, 444–449. doi: 10.1016/j.jgr.2016.09.001
Jung, J., Kim, K. H., Yang, K., Bang, K. H., and Yang, T. J. (2014). Practical application of DNA markers for high-throughput authentication of Panax ginseng and Panax quinquefolius from commercial ginseng products. J. Ginseng Res. 38, 123–129. doi: 10.1016/j.jgr.2013.11.017
Kennedy, D. O., and Scholey, A. B. (2003). Ginseng: potential for the enhancement of cognitive performance and mood. Pharmacol. Biochem. Behav. 75, 687–700. doi: 10.1016/S0091-3057(03)00126-6
Keum, Y.-S., Park, K.-K., Lee, J.-M., Chun, K.-S., Park, J. H., and Lee, S. K. (2000). Antioxidant and anti-tumor promoting activities of the methanol extract of heat-processed ginseng. Cancer Lett. 150, 41–48. doi: 10.1016/S0304-3835(99)00369-9
Kim, J.-H., Kim, M.-K., Wang, H., Lee, H.-N., Jin, C.-G., Kwon, W.-S., et al. (2016). Discrimination of Korean ginseng (Panax ginseng Meyer) cultivar Chunpoong and American ginseng (Panax quinquefolius) using the auxin repressed protein gene. J. Ginseng Res. 40, 395–399. doi: 10.1016/j.jgr.2015.12.002
Kim, J. H., Moon, J.-C., Kang, T. S., Kwon, K., and Jang, C. S. (2017). Development of cpDNA markers for discrimination between Cynanchum wilfordii and Cynanchum auriculatum and their application in commercial C. wilfordii food products. Appl. Biol. Chem. 60, 79–86. doi: 10.1007/s13765-017-0252-5
Kim, K., Lee, S.-C., Lee, J., Lee, H. O., Joh, H. J., and Kim, N.-H. (2015). Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One 10:e0117159. doi: 10.1371/journal.pone.0117159
Kim, O. T., Bang, K. H., In, D. S., Lee, J. W., Kim, Y. C., and Shin, Y. S. (2007). Molecular authentication of ginseng cultivars by comparison of internal transcribed spacer and 5.8 S rDNA sequences. Plant Biotechnol. Rep. 1, 163–167. doi: 10.1007/s11816-007-0019-2
Koren, O. G., Potenko, V. V., and Zhuravlev, Y. N. (2003). Inheritance and variation of allozymes in Panax ginseng CA Meyer (Araliaceae). Int. J. Plant Sci. 164, 189–195. doi: 10.1086/344758
Lai, D.-M., Tu, Y.-K., Liu, I.-M., Chen, P.-F., and Cheng, J.-T. (2006). Mediation of β-endorphin by ginsenoside Rh2 to lower plasma glucose in streptozotocin-induced diabetic rats. Planta Med. 72, 9–13. doi: 10.1055/s-2005-916177
Lee, J.-W., Bang, K.-H., Choi, J.-J., Chung, J.-W., Lee, J.-H., and Jo, I.-H. (2010). Development of peptide nucleic acid (PNA) microarray for identification of Panax species based on the nuclear ribosomal internal transcribed spacer (ITS) and 5.8 S rDNA regions. Genes Genomics 32, 463–468. doi: 10.1007/s13258-010-0040-7
Li, G., Cui, Y., Wang, H., Kwon, W.-S., and Yang, D.-C. (2017). Molecular differentiation of Russian wild ginseng using mitochondrial nad7 intron 3 region. J. Ginseng Res. 41, 326–329. doi: 10.1016/j.jgr.2016.06.003
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y., and Chen, S. J. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Lohse, M., Drechsel, O., and Bock, R. J. (2007). OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274. doi: 10.1007/s00294-007-0161-y
Ma, K. B., Yang, S.-J., Jo, Y.-S., Kang, S. S., and Nam, M. (2021). Development of Kompetitive Allele Specific PCR markers for identification of persimmon varieties using genotyping-by-sequencing. Electronic J. Biotechnol. 49, 72–81. doi: 10.1016/j.ejbt.2020.11.003
Manzanilla, V., Kool, A., Nguyen Nhat, L., Nong Van, H., Le Thi Thu, H., et al. (2018). Phylogenomics and barcoding of Panax: toward the identification of ginseng species. BMC Evolutionary Biol. 18:44. doi: 10.1186/s12862-018-1160-y
Mohanty, S., and Cock, I. E. (2012). The chemotherapeutic potential of Terminalia ferdinandiana: phytochemistry and bioactivity. Pharmacognosy Rev. 6, 29–36. doi: 10.4103/0973-7847.95855
Ngan, F., Shaw, P., But, P., and Wang, J. J. P. (1999). Molecular authentication of Panax species. Phytochemistr 50, 787–791. doi: 10.1016/S0031-9422(98)00606-2
Nguyen, V. B., Park, H.-S., Lee, S.-C., Lee, J., Park, J. Y., and Yang, T.-J. (2017). Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences. J. Agric. Food Chem. 65, 6298–6306. doi: 10.1021/acs.jafc.7b00925
Osathanunkul, M., and Madesis, P. (2019). Bar-HRM: a reliable and fast method for species identification of ginseng (Panax ginseng. Panax notoginseng, Talinum paniculatum and Phytolacca Americana). PeerJ 7:e7660. doi: 10.7717/peerj.7660
Park, H.-S., Jayakodi, M., Lee, S. H., Jeon, J.-H., Lee, H.-O., and Park, J. Y. (2020). Mitochondrial plastid DNA can cause DNA barcoding paradox in plants. Sci. Rep. 10:9599. doi: 10.1038/s41598-020-63233-y
Park, S., and Han, K.-H. (2003). An auxin-repressed gene (RpARP) from black locust (Robinia pseudoacacia) is posttranscriptionally regulated and negatively associated with shoot elongation. Tree Physiol. 23, 815–823. doi: 10.1093/treephys/23.12.815
Pellestor, F., and Paulasova, P. J. (2004). The peptide nucleic acids (PNAs), powerful tools for molecular genetics and cytogenetics. Eur. J. Hum. Genet. 12, 694–700. doi: 10.1038/sj.ejhg.5201226
Poczai, P., and Hyvönen, J. J. (2010). Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Mol. Biol. Rep. 37, 1897–1912. doi: 10.1007/s11033-009-9630-3
Ratnasingham, S., and Hebert, P. D. (2013). A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS One 8:e66213. doi: 10.1371/journal.pone.0066213
Rivera, E., Pettersson, F. E., Inganäs, M., Paulie, S., and Grönvik, K.-O. (2005). The Rb1 fraction of ginseng elicits a balanced Th1 and Th2 immune response. Vaccine 23, 5411–5419. doi: 10.1016/j.vaccine.2005.04.007
Song, Y., Wang, S., Ding, Y., Xu, J., Li, M. F., Zhu, S., et al. (2017). Chloroplast genomic resource of Paris for species discrimination. Sci. Rep. 7:3427. doi: 10.1038/s41598-017-02083-7
Sun, H., Wang, H. T., Kwon, W. S., In, J. G., Lee, B. S., and Yang, D. C. (2010). Development of molecular markers for the determination of the new cultivar ‘Chunpoong’in Panax ginseng CA M EYER associated with a major latex-like protein gene. Biol. Pharm. Bull. 33, 183–187. doi: 10.1248/bpb.33.183
Um, J.-Y., Chung, H.-S., Kim, M.-S., Na, H.-J., Kwon, H.-J., and Kim, J.-J. (2001). Molecular authentication of Panax ginseng species by RAPD analysis and PCR-RFLP. Biol. Pharm. Bull. 24, 872–875. doi: 10.1248/bpb.24.872
Wang, H., Kim, M.-K., Kwon, W.-S., Jin, H., Liang, Z., and Yang, D.-C. (2011a). Molecular authentication of Panax ginseng and ginseng products using robust SNP markers in ribosomal external transcribed spacer region. J. Pharm. Biomed. Anal. 55, 972–976. doi: 10.1016/j.jpba.2011.03.037
Wang, H., Kwon, W.-S., Yang, D.-U., Kim, M.-K., Sathiyamoorthy, S., and Jin, H. (2011b). Development of a multiplex amplification refractory mutation system for simultaneous authentication of Korean ginseng cultivars “Gumpoong” and “Chungsun”. Mitochondrial DNA 22, 35–38. doi: 10.3109/19401736.2011.588216
Wang, H., Wang, J., and Li, G. (2016b). A simple real-time polymerase chain reaction (PCR)-based assay for authentication of the Chinese Panax ginseng cultivar Damaya from a local ginseng population. Genet. Mol. Res. 15:gmr8801. doi: 10.4238/gmr.15028801
Wang, H., Li, G., Kwon, W.-S., and Yang, D.-C. (2016a). Development of EST intron-targeting SNP markers for Panax ginseng and their application to cultivar authentication. Int. J. Mol. Sci. 17:884. doi: 10.3390/ijms17060884
Wang, H., Sun, H., Kwon, W.-S., Jin, H., and Yang, D.-C. (2009). Molecular identification of the Korean ginseng cultivar “Chunpoong” using the mitochondrial nad7 intron 4 region. Mitochondrial DNA 20, 41–45. doi: 10.1080/19401730902856738
Wang, H., Sun, H., Kwon, W.-S., Jin, H., and Yang, D.-C. (2010a). A PCR-based SNP marker for specific authentication of Korean ginseng (Panax ginseng) cultivar “Chunpoong”. Mol. Biol. Rep. 37:1053. doi: 10.1007/s11033-009-9827-5
Wang, H., Sun, H., Kwon, W.-S., Jin, H., and Yang, D.-C. (2010b). A simplified method for identifying the Panax ginseng cultivar gumpoong based on 26S rDNA. Planta Med. 76, 399–401. doi: 10.1055/s-0029-1186161
Wang, H., Xu, F., Wang, X., Kwon, W.-S., and Yang, D.-C. (2019). Molecular discrimination of Panax ginseng cultivar K-1 using pathogenesis-related protein 5 gene. J. Ginseng Res. 43, 482–487. doi: 10.1016/j.jgr.2018.07.001
Wen-Ru, W., Cheng, C.-S., Cheng, Q.-Q., Chi-Chou, L., Hao, C., and Zi-Yu, T. (2020). Novel SNP markers on ginsenosides biosynthesis functional gene for authentication of ginseng herbs and commercial products. Chin. J. Nat. Med. 18, 770–778. doi: 10.1016/S1875-5364(20)60017-6
Yang, D.-U., Kim, M.-K., Mohanan, P., Mathiyalagan, R., Seo, K.-H., and Kwon, W.-S. (2017). Development of a single-nucleotide-polymorphism marker for specific authentication of Korean ginseng (Panax ginseng Meyer) new cultivar “G-1”. J. Ginseng Res. 41, 31–35. doi: 10.1016/j.jgr.2015.12.007
Yang, J. Y., Jang, S. Y., Kim, H.-K., and Park, S. J. (2012). Development of a molecular marker to discriminate Korean Rubus species medicinal plants based on the nuclear ribosomal DNA internal transcribed spacer and chloroplast trnL-F intergenic region sequences. J. Korean Soc. Appl. Biol. Chem. 55, 281–289. doi: 10.1007/s13765-012-1044-6
Keywords: single nucleotide polymorphism, saponins, landraces, cultivars, markers
Citation: Ying Z, Awais M, Akter R, Xu F, Baik S, Jung D, Yang DC, Kwak G-Y and Wenying Y (2022) Discrimination of Panax ginseng from counterfeits using single nucleotide polymorphism: A focused review. Front. Plant Sci. 13:903306. doi: 10.3389/fpls.2022.903306
Received: 24 March 2022; Accepted: 31 May 2022;
Published: 28 July 2022.
Edited by:
Isabel Mafra, University of Porto, PortugalReviewed by:
Panagiotis Madesis, University of Thessaly, GreeceCopyright © 2022 Ying, Awais, Akter, Xu, Baik, Jung, Yang, Kwak and Wenying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: You Wenying, MjU0MDMxNDE3QHFxLmNvbQ==; Gi-Young Kwak, a3dha2dpeW91bmc4QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.