- 1College of Biological and Pharmaceutical Engineering, West Anhui University, Lu’an, China
- 2Anhui Engineering Laboratory for Conservation and Sustainable Utilization of Traditional Chinese Medicine Resources, Lu’an, China
- 3College of Life Science, Anhui Agricultural University, Hefei, China
- 4Analytical and Testing Center, West Anhui University, Lu’an, China
Peucedanum praeruptorum Dunn is a commonly used traditional Chinese medicine that is abundant in furano- and dihydropyrano coumarins. When P. praeruptorum reaches the bolting stage, the roots gradually lignified, and the content of coumarins declines rapidly. Non-bolting has always been a decisive factor for harvesting the P. praeruptorum materials. To evaluate the amount of coumarin components in unbolted and bolted P. praeruptorum, the variations of praeruptorin A, praeruptorin B, praeruptorin E, peucedanocoumarin I, and peucedanocoumarin II were determined. Additionally, 336,505 transcripts were obtained from the comparative transcriptome data. Among them, a total of 1,573 differentially expressed genes were screened out. To identify the critical genes involved in coumarin biosynthesis, comparative transcriptomics coupled with co-expression associated analysis was conducted. Finally, coumarin biosynthesis-related eighteen candidate genes were selected for the validation of qPCR. Additionally, a phylogenetic tree and the expression profile of ATP-binding cassette (ABC) transporters were constructed. To clarify the main genes in the regulation of coumarin biosynthesis, the interaction network of the co-expression genes from thirteen modules was constructed. Current results exhibited the significant increment of praeruptorin A, praeruptorin B and praeruptorin E in the bolted P. praeruptorum. Although, peucedanocoumarin I and peucedanocoumarin II were slightly increased. Besides the content of coumarins, the essential genes involved in the coumarin biosynthesis also exhibited an overall downward trend after bolting. Three peroxidases (PRXs) involved in the production of lignin monomers had been demonstrated to be downregulated. PAL, C4H, HCT, COMT, CCoAOMT, and some ABC transporters were dramatically downregulated at the bolting stage. These results indicated that the downregulation of coumarin biosynthetic genes in the bolted P. praeruptorum ultimately reduced the formation of coumarins. However, the mechanism through which bolting indirectly affects the formation of coumarin still needs extra functional verification.
Introduction
Peucedanum praeruptorum is a perennial herb in the Umbelliferae family whose dried roots are commonly used in traditional Chinese medicine (Song et al., 2021). Coumarins, which are rich in P. praeruptorum, have a wide range of applications in the prevention and treatment of cardiovascular and cerebrovascular diseases, anti-inflammatory, reversal of multidrug resistance, anti-cancer, and neuroprotection (Lee et al., 2015; Stelzhammer et al., 2017; Wang et al., 2017; Liu et al., 2020). The medicinal ingredients of P. praeruptorum are mainly furan- and dihydropyran-type coumarins (Zhao et al., 2015; Yu et al., 2020). Furanocoumarins are a class of secondary metabolites derived from structurally simple coumarins (Dugrand-Judek et al., 2015). Structurally, furanocoumarins are available in two isomeric forms: linear and angular, which are considered to originate from the phenylpropanoid pathway (Tian et al., 2017). The upstream genes of the phenylpropanoid pathway were involved in the formation of coumarins (Sui et al., 2019). PAL is the first rate-limiting enzyme in the regulation of coumarin biosynthesis, whose expression was susceptible to multiple abiotic stresses (Shang et al., 2012; Sui et al., 2019). 4CL catalyzed the formation of cinnamoyl-CoA, p-coumaryl-CoA, caffeoyl-CoA, ferulic-acid-CoA, and various hydroxycinnamate-CoA esters (Rastogi et al., 2013; Li and Nair, 2015). From three 4CLs found in P. praeruptorum, Pp4CL1 preferentially takes coumarate and ferulic acid as substrates, although it can also use caffeic acid, cinnamic acid, o-coumaric acid, and other precursors. Pp4CL7/4CL10 lacked catalytic activity for hydroxycinnamic acid (Liu et al., 2017). The ortho-hydroxylation of hydroxycinnamate is an essential step in the biosynthesis of coumarins, especially for the subsequent cyclization of coumarin lactones (Matsumoto et al., 2012). C2′H is required for the formation of umbelliferone. The expression of C2′H was high in the roots of P. praeruptorum and was induced by MeJA and UV-B treatments (Yao et al., 2017). Caffeic acid O-methyltransferase-similar (COMT-S) was found to be responsible for the O-methylation of hydroxycoumarins (Zhao et al., 2019). Cinnamic acid was catalyzed and lactonized by cinnamic acid-4-hydroxylase (C4H), 4-coumaric acid-CoA ligase (4CL), and p-coumaroyl-CoA 2′-hydroxylase (C2′H) to yield umbelliferone (Song et al., 2020). Some CYP450 family members and MDR transporters may participate in the biosynthesis and transportation of coumarins (Zhao et al., 2015). However, the downstream branch and specific coumarin transporters related to dihydropyranocoumarin remain unclear.
The biosynthesis of coumarins still requires some post-modifying enzymes, which are mainly composed of methyltransferase, o-methyltransferase, prenyltransferase, and monooxygenase. Umbelliferone dimethylallyl transferase (UDT) plays an important role in the prenylation of umbelliferone (Munakata et al., 2016). Umbelliferone can be prenylated at the C6 and C8 positions to yield linear and angular furocoumarins, respectively. Psoralen is hydroxylated at the C5 and C8 positions to form xanthotoxol and bergaptol, respectively. The osthenol catalyzed by U8DT satisfies the structural basis for dihydropyranocoumarins. Depending on U6DT, umbelliferone is prenylated to form demethylsuberosin (DMS). Marmesin synthase (MS) catalyzes DMS to form marmesin, which is then converted to psoralen via psoralen synthase (PS) (Jian et al., 2020). Marmesin is hydroxylated by marmesin monooxygenase (MO) to produce bergaptol or xanthol. Bergaptol O-methyltransferase (BMT) is involved in the O-methylation reaction of bergaptol with high substrate specificity (Zhao et al., 2016b). Imperatorin is produced from bergaptol and xanthomol by prenyltransferase (PT), bergaptol-O-methyltransferase (BMT), and xanthomol-O-methyltransferase (XMT) to form imperatorin, isoimperatorin, and other furanocoumarins (Munakata et al., 2020). A portion of the osthenol precursors are added to angelicin, while the residue will form dihydrofuranocoumarins by multi-step reactions (Munakata et al., 2016). Based on the biogenic pathway, lomatin may participate in the biosynthesis of praeruptorin A, praeruptorin B, praeruptorin E, and other dihydrofuranocoumarins through PT, OMT, and CYP450 monooxygenase. The current issue is that the key genes or transcription factors responsible for the biosynthesis and regulation of P. praeruptorum coumarins have not been widely identified and investigated. To some extent, this limits the application of synthetic biology strategies for large-scale production of such active ingredients in heterologous expression systems.
Early bolting has a significant impact on the accumulation of secondary metabolites in traditional Chinese medicine (Zhou et al., 2014). The bolted herbs in the Umbelliferae family, such as Peucedanum, Angelica, Saposhnikovia, Notopterygium, and Glehnia, were generally not harvested (Yu et al., 2019; Song et al., 2021). P. praeruptorum began to lignify once it entered into reproductive growth, and the content of coumarins gradually declined (Guo et al., 2021; Li et al., 2021). During the reproductive growth, a large amount of nutrients are consumed. A lack of carbon sources led to an increment in the secondary xylem area and a decrease in coumarins (Chen et al., 2019; Robe et al., 2021). Our previous study suggested that post-transcriptional modification, signal transduction, and secondary metabolism might play vital roles in coumarin biosynthesis (Song et al., 2021). Among them, the key enzymes of the phenylpropanoid pathway such as ATP-binding cassette (ABC) transporters, apoptosis-related genes, and circadian rhythm-related genes participated in the regulation of coumarin biosynthesis. Despite that, what is the expression pattern of key genes involved in coumarin biosynthesis between the bolted and unbolted P. praeruptorum, and which ABC transporter subfamily may be involved in coumarin transportation and distribution? How do the co-expressed genes involved in coumarin biosynthesis connect and interact? These are still unanswered questions that are worthy of further exploration. Here, the contents of five pyranocoumarins were determined at the bolting stages. The lignification of P. praeruptorum root was determined by phloroglucinol staining. A total of 18 candidate genes were identified, which are involved in the regulation of coumarin biosynthesis. Among them, PAL, C3H, COMT, and HCT in the phenylpropanoid pathway in bolted P. praeruptorum and AS, PS1, and BMT involved in coumarin biosynthesis were dramatically downregulated Three PRXs related to lignin polymerization were also negatively regulated. These findings will contribute to a better understanding of the coumarin biosynthetic pathway and bolting mechanism in P. praeruptorum.
Materials and Methods
Samples and Reagents
The samples for the experiment were taken from the Ta-pieh Mountain Medicinal Botanical Garden of West Anhui University in October 2020. All P. praeruptorum samples were grown for more than one year. The whole plants of the unbolted and bolted P. praeruptorum were collected for further experiments. Ten biological replicates were taken at the bolting stages for the determination of coumarins, and three biological replicates were taken for qPCR analysis. By comparing the original plant to the reference medicinal material (Batch No. WKQ-DZYC-01607) from Sichuan Vikeqi Biotechnology Co., Ltd. (Chengdu, China), the authenticity of the plant was confirmed. The fibrous roots of P. praeruptorum were removed. The taproots were preserved and rinsed with sterile water, and the surface water was absorbed and dried naturally.
Peucedanocoumarin I (Shanghai Yuanye Biotechnology Co., Ltd., batch number: B50414, purity > 95.0%). Praeruptorin A (National Institute for Food and Drug Control, purity > 99.4%). Peucedanocoumarin II (Shanghai Yuanye Biotechnology Co., Ltd., batch number: B5041, purity > 97.8%). Praeruptorin B (National Institute for Food and Drug Control, purity > 98.9%). Praeruptorin E (Shanghai Yuanye Biotechnology Co., Ltd., batch number: B20036, purity > 99.9%). Methanol (GR, Shanghai McLean Biochemical Technology Co., Ltd.). Other reagents were of analytical grade.
The Histochemical Staining
Peucedanum praeruptorum taproots were stained with phloroglucinol-hydrochloric acid dye solution by the previous methods (Liljegren, 2010; Chauhan et al., 2015). In brief, 1mL of concentrated hydrochloric acid was dropped on the cross-section of the front beard and left for five minutes. Then, 1mL of the phloroglucinol-alcohol mixture was dropped to dye the lignified cell wall. Finally, the dyed pink area in the xylem was recorded.
Preparation of Coumarin Extraction
The extraction of total coumarin was referred to as the Chinese Pharmacopoeia, followed by the previous method (Hou et al., 2010). The roots of P. praeruptorum were naturally dried in the shade and then pulverized into a coarse powder. 0.5 g of powder was mixed with 25 mL of chloroform and ultrasonically extracted for 10 minutes (250 W, 33 kHz). After cooling the extract, the lost weight was replaced with chloroform. To prepare the test solution, 5 mL of the continuous filtrate was evaporated to dryness and dissolved with proper methanol. All samples were filtered through a 0.45 μm organic-based microporous membrane (ANPEL Laboratory Technologies (Shanghai) Inc.) prior to the HPLC analysis.
Optimization of HPLC Conditions and Methodology
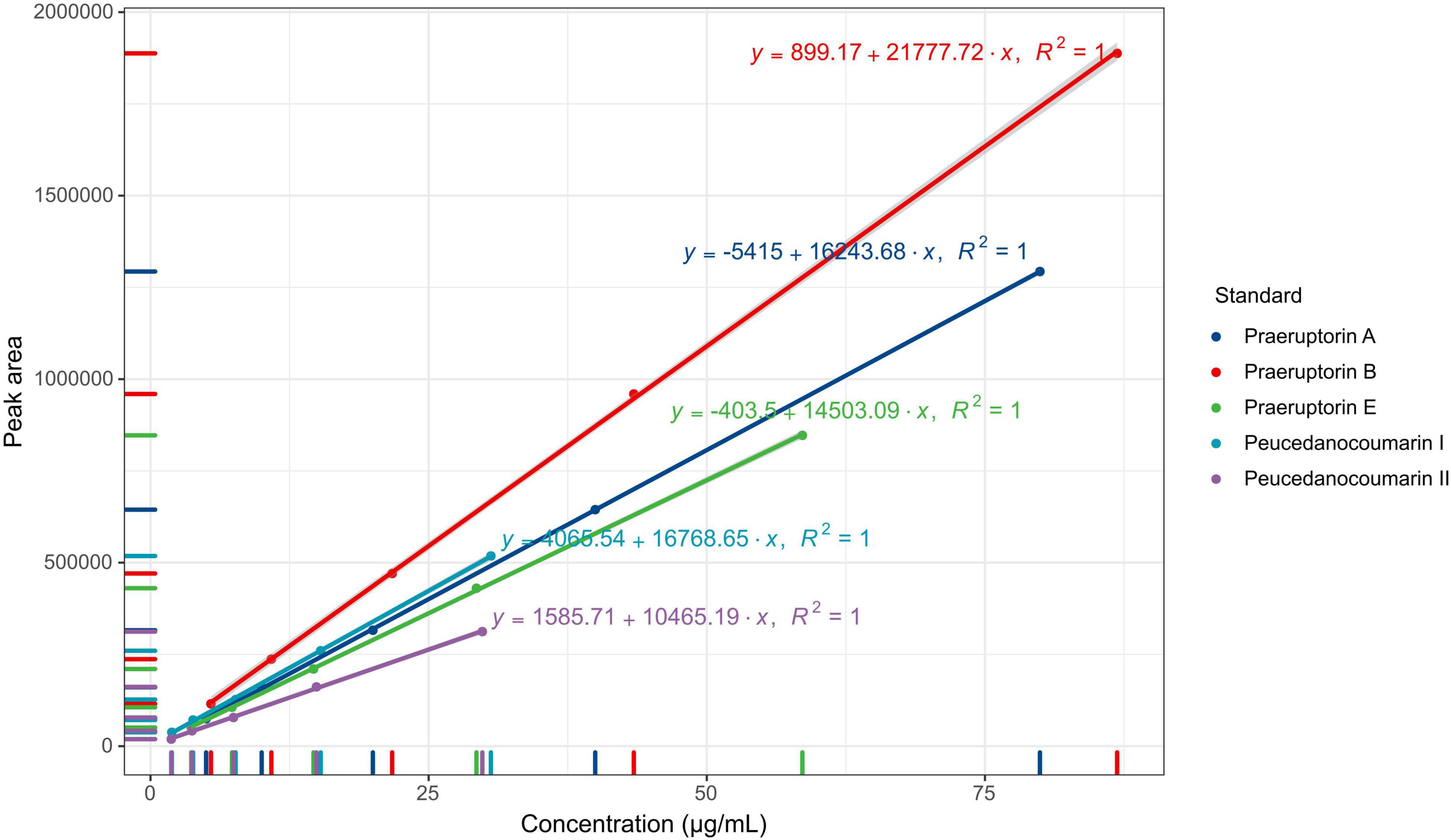
The content of coumarins was determined using LC-2030C high-performance liquid chromatography (Shimadzu, Japan). The chromatographic column used in the test was ZORBAX Eclipse Plus C18 (150 mm x 4.6 mm, 5 μm) (Agilent, United States). Methanol-water (volume ratio of 75:25) was used as the mobile phase. The flow rate was set at 1.0 mL/min. The column temperature was set at 30°C, and the detection wavelength was at 235 nm. The injection volume was 10 μL. To produce the reference substance solutions, the appropriate amounts of the peucedanocoumarin I, praeruptorin A, peucedanocoumarin II, praeruptorin B, and praeruptorin E standards were mixed with methanol, respectively. The concentrations of Peucedanocoumarin I, praeruptorin A, peucedanocoumarin II, praeruptorin B, and praeruptorin E were 30.59 g/mL, 79.94 g/mL, 29.82 g/mL, 86.88 g/mL, and 58.69 g/mL, respectively.
The methodological investigation concluded with the precision test, stability test, repeatability test, and recovery rate test by using previous methods (Tao et al., 2009). The precision test was conducted by injecting 10 μL of test solution under the specified chromatographic conditions and repeating six times. The stability test included the injection of the test solution at 4, 8, 12, 16, 20, and 24 h intervals under the specified chromatographic conditions. The repeatability test evaluates six batches of P. praeruptorum. The test solution was injected at a volume of 10 μL under the specified chromatographic conditions. The recovery rate experiment was conducted to weigh 0.50 g of P. praeruptorum (including 0.49 mg/g of peucedanocoumarin I, 6.04 mg/g of praeruptorin A, 0.46 mg/g of peucedanocoumarin II, 3.43 mg/g of praeruptorin B, and 2.09 mg/g of praeruptorin E). The test was repeated six times. Coumarin standards were added to each batch, and the peak area of each sample was measured. The recovery rate and relative standard deviation (RSD) of each standard were calculated, respectively. The optimized chromatography method was used to determine the five coumarins in the samples, and ten biological replicates were performed for each coumarin.
The Hierarchical Clustering, Phylogenetic Tree and Expression Profile of the Differential Genes
The raw data of the transcriptome used for the analysis of expression profiles were obtained from the Sequence Read Archive (SRA) database1. The BioProject accession was PRJNA714368. To investigate the differential genes at the bolting stages, transcripts from the annual bolted and unbolted P. praeruptorum were selected for subsequent analysis. CDD, KOG, COG, NR, NT, PFAM, Swissprot, and TrEMBL databases were used for the functional annotation (Altschul et al., 1997). The GO function annotation is obtained based on the annotation of the transcript from Swissprot and TrEMBL. The KEGG Automatic Annotation Server (KAAS) was used to obtain KEGG annotation information (Moriya et al., 2007). The transdecoder software2 was used for CDS prediction after blasting the transcripts with databases. Based on the annotation information, all possible unigenes were screened and the blastn (e-value < 10–5) was used for additional validation. By setting the significant difference (p < 0.01), false discovery rate (FDR) correction (q < 0.05), and | fold change| > 2, a total of 1,573 genes with significantly differential expression were obtained (Benjamini and Hochberg, 1995; Anders and Huber, 2010).
The abundance of transcripts directly reflects the level of expression of a specific gene. In the experiment, the TPM value was used to compare the gene expression between the two groups. Subsequently, the salmon tool was used to calculate the expression levels by using RNA-seq data (Patro et al., 2017). For the replicates in the same group, the expression level is the average of all repeated data. The TPM values of all the unigenes were used for hierarchical clustering and expression profiling of differentially expressed genes. TBtools (v.1.098) was used to compare the gene expression profiles among groups (Chen et al., 2020). MEGA (v.6.0.6) was used to align the target sequences and construct a phylogenetic tree based on the Neighbor-Joining method (Tamura et al., 2013).
Analysis of Coumarin Biosynthesis Genes by Quantitative Real-Time PCR Analysis
Before performing the total RNA extraction, the sampling equipment was disinfected and de-RNAsed. Fresh P. praeruptorum roots were collected, and the surface was quickly cleaned with RNase-free water. The samples were placed in an enzyme-free tube before being quickly frozen in liquid nitrogen. After it had completely frozen, it was transferred to a refrigerator set at –80 degrees Celsius for storage. Each P. praeruptorum sample was ground to 100 mg in liquid nitrogen. Total RNA was isolated from bolted and unbolted P. praeruptorum using the UNIQ-10 column Trizol total RNA extraction kit (Sangon Biotech Ltd., Shanghai). Electrophoresis was used to detect RNA concentration. Subsequently, mRNA was isolated and fragmented. The mixture was centrifuged for 3-5 s after it had been mixed. After 10 min of incubation at 25°C, the reaction was performed at 50°C for 30 min and at 85°C for 5 min to obtain cDNA. GAPDH was selected as a reference gene referred to in this study (Zhao et al., 2016a). Based on the sequences of eighteen candidate genes and the reference gene, Primer Premier 5.0 was used to design the primer sequences (Supplementary Table 1). The qPCR experiment was carried out using the StepOne Plus real-time PCR equipment (ABI, Foster, CA, United States). Relative quantification of target genes was performed by using the SYBR Green I method with 2X SG Fast qPCR Master Mix (High Rox, B639273, BBI, ABI). The 2–ΔΔ Ct method was applied to calculate the relative expression of the target genes after bolting (Livak and Schmittgen, 2001).
The Interaction Network Analysis
Several genes relevant to coumarin biosynthesis were screened out from 13 gene modules of the WGCNA data. Briefly, the WGCNA script was used to create a gene set matrix for the co-expression correlation analysis (Langfelder and Horvath, 2008). The differential expression profile obtained by the transcriptome analysis was used in the WGCNA. Following the selection of an appropriate soft threshold, the co-expressed gene modules were performed to determine the number of genes in each module. The co-expression correlation coefficient between genes was calculated first based on the measured gene expression levels, and then the genes were clustered using euclidean distance by drawing a gene tree. The phenotypic traits were weighted, and the correlation and credibility of each gene module were calculated in relation to them. The core module was selected based on its relevance and significance. Based on the weighted scores of these gene pairs, an intergenic interaction network was visualized through the Cytoscape (v.3.9.0) (Doncheva et al., 2019).
Results and Discussion
Sample Collection and Histochemical Staining of P. praeruptorum Roots
Due to the influence of genetic, ecological, and growth circumstances, P. praeruptorum from the same period may grow at different rates (Liang et al., 2018). To compare the degree of the lignification at the bolting stage, both unbolted and bolted P. praeruptorum roots were collected (Figure 1). In the bolting stage, the taproot of P. praeruptorum is relatively slender and has more fibrous roots. The results from phloroglucinol staining showed that the area of bolted xylem was darker and larger, which implied that the lignin content in the bolted roots was higher (Chen et al., 2019). Early bolting has become a crucial factor affecting the quality of the crude materials. The quality of these materials depends on when they are harvested (Zhao et al., 2011; Li et al., 2020). Currently, there is no conclusive evidence that bolting results in a loss of ability to use medications (Pouteau and Albertini, 2009; Li et al., 2022). The bolted P. praeruptorum conveys large amounts of nutrients to the aerial parts, which may be the possible reason for early bolting (Song et al., 2021).

Figure 1. Sample collection and the phloroglucinol staining of unbolted and bolted P. praeruptorum roots. (A) Unbolted P. praeruptorum, (B) Bolted P. praeruptorum, (C) The root of unbolted P. praeruptorum, (D) The root of bolted P. praeruptorum, (E) Phloroglucinol staining of the cross section of the unbolted taproot, (F) Phloroglucinol staining of the cross section of the bolted taproot.
Extraction of Coumarins and Methodology Investigation
The five coumarins (peucedanocoumarin I, praeruptorin A, peucedanocoumarin II, praeruptorin B, and praeruptorin E) were determined from the methanol extraction of P. praeruptorum roots. The chromatographic conditions were investigated as a priority to optimize the detection of the samples. These results indicated that the five coumarins kept a better linear relationship in the range of concentration (Figure 2). The linear ranges of peucedanocoumarin I, praeruptorin A, peucedanocoumarin II, praeruptorin B, and praeruptorin E were 0.48∼7.65 μg, 1.25∼19.99 μg, 0.47∼7.46 μg, 1.36∼21.72 μg, and 0.92∼14.65 μg, respectively. Six independent injections were performed under the optimized chromatographic conditions. The RSD of the peak areas of peucedanocoumarin I, praeruptorin A, peucedanocoumarin II, praeruptorin B, and praeruptorin E were 0.3, 0.26, 0.27, 0.26, and 0.25%, respectively, which indicated that the precision of the instrument was good. The samples were injected at 4, 8, 12, 16, 20, and 24 h, with the RSDs of the peak areas of 0.97, 0.18, 0.18, 0.06, and 0.26%. Six samples from the same batch were tested, and the RSDs of the five coumarin contents were 3.62, 0.92, 2.68, 2.49, and 1.43%, implying good repeatability. The recoveries of peucedanocoumarin I, praeruptorin A, peucedanocoumarin II, praeruptorin B, and praeruptorin E were 99.97, 99.79, 101.17, 99.73, and 99.95%, respectively. The RSDs of the five standards were 0.28, 0.17, 0.93, 0.23, 0.21%, respectively, which suggested the test method has good accuracy.
Determination of the Coumarins in the Unbolted and Bolted P. praeruptorum
The standards and test samples were determined under the defined chromatographic conditions (Figure 3). Except for praeruptorin A and peucedanocoumarin II, the peaks of peucedanocoumarin I, praeruptorin B, and praeruptorin E had better shape and resolution. The five standards used in the experiment had a common hexacyclic skeleton. However, praeruptorin A and peucedanocoumarin II are isomers and their polarities are closely related to each other, which results in poor separation on a C18 column (Wang et al., 2015). Using the optimized chromatographic method, 10 batches of samples were used to determine the five coumarins (Figure 4).
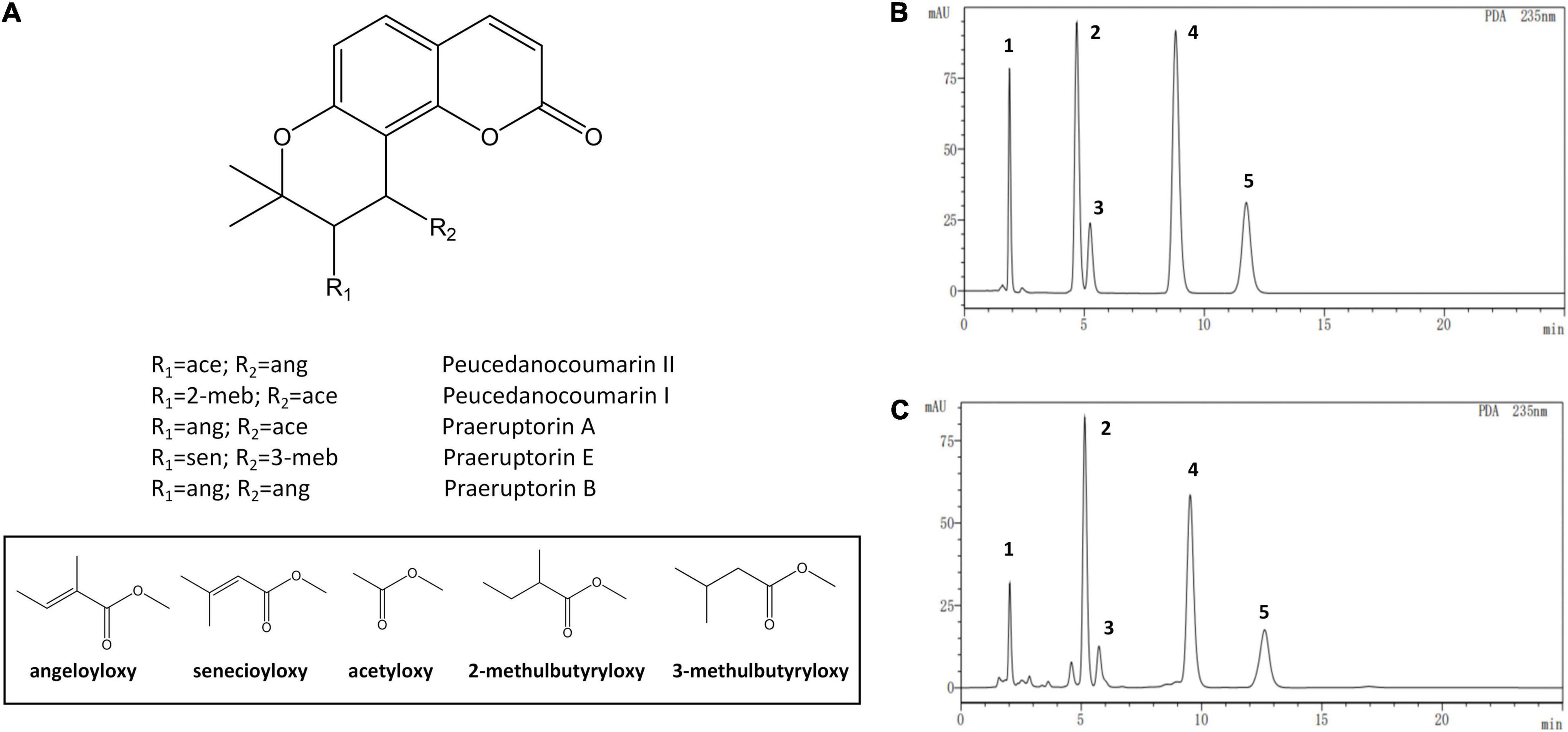
Figure 3. The chemical structure and high performance liquid chromatograms of five coumarin components. (A) The structures of the coumarins, (B) The liquid chromatogram of the standards, (C) The liquid chromatogram of test sample. Numbers one to five depict peucedanocoumarin I, praeruptorin A, peucedanocoumarin II, praeruptorin B, and praeruptorin E, respectively.
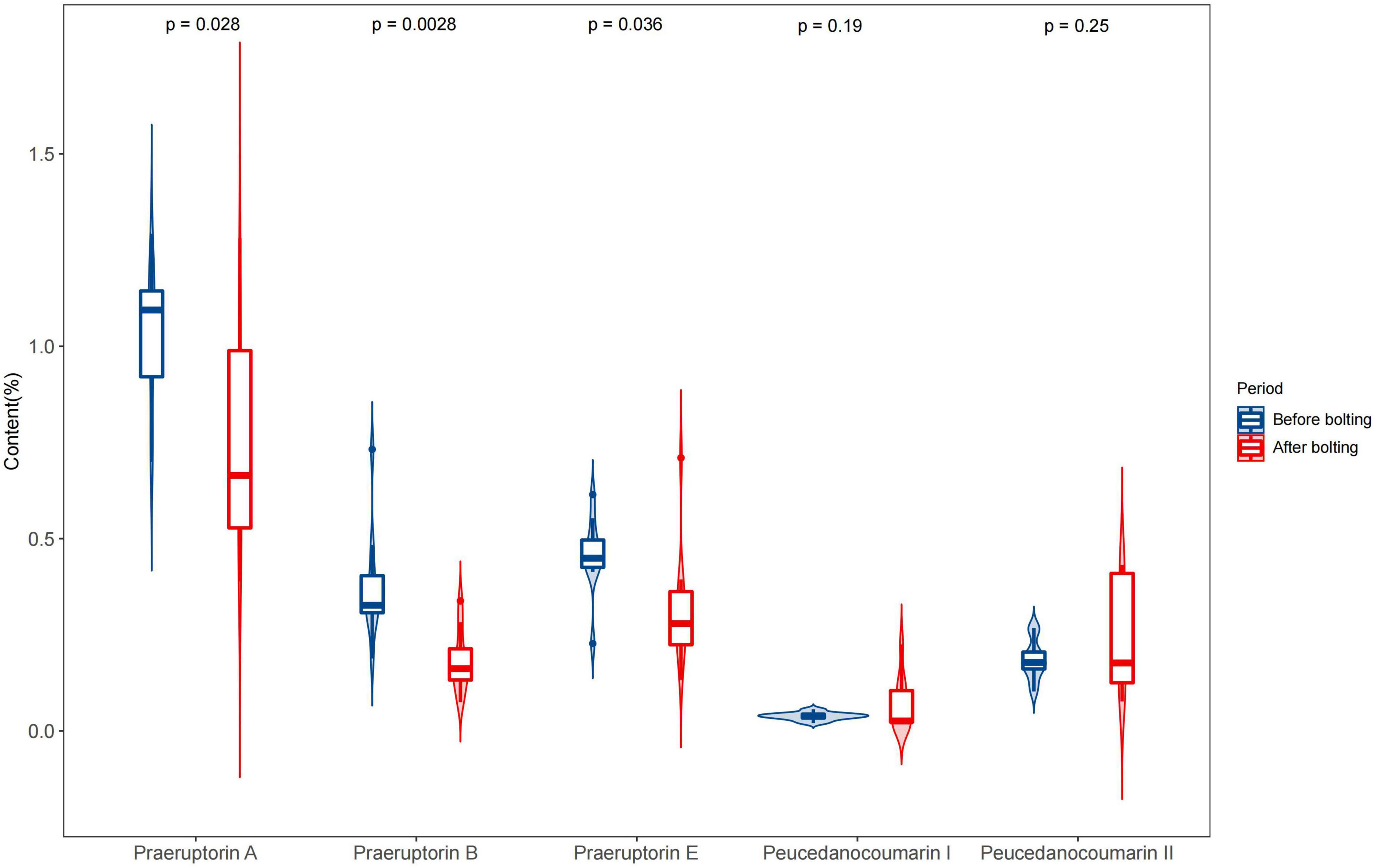
Figure 4. Determination of five coumarins of the unbolted and bolted P. praeruptorum. Ten batches of experiments were repeated for each group. The content of each coumarin was calculated as the mean plus standard error. A significant analysis was carried out on the content of coumarins in unbolted and bolted P. praeruptorum. “p-value < 0.05” means there is a significant difference.
In the bolted P. praeruptorum, the average contents of praeruptorin A, praeruptorin B, and praeruptorin E declined by 26.6, 52.1, and 30.3%, respectively. However, the contents of peucedanocoumarin I and peucedanocoumarin II were raised by 45.7 and 24.8%, respectively. Previous study also demonstrated that the contents of praeruptorin A and praeruptorin B decreased after bolting (Sarkhail et al., 2013). Praeruptorin A and peucedanocoumarin II accumulate in a distinct way depending on the differential expression of their biosynthetic genes, such as prenyltransferase (PT), acetyl-CoA acetyltransferase (AACT), and O-methyltransferase (O-MT) (Song et al., 2014). The major difference between praeruptorin A and praeruptorin B was that the R2 group is linked to acetyloxy or angeloyloxy groups, and praeruptorin B has two angeloyloxy groups (Figure 3). However, the content of praeruptorin B was significantly lower than that of praeruptorin A after bolting, which suggested that the expression level of PT genes was more capable of influencing the biosynthesis of this group of coumarins. In addition, the molecular weight of peucedanocoumarin I had only two hydrogen atoms more than that of praeruptorin A, implying that peucedanocoumarin I was generated from praeruptorin A under the catalysis of some oxidoreductases. The content of peucedanocoumarin I increased, presumably due to the transformation of praeruptorin A. This also explains the decline of praeruptorin A in the bolted P. praeruptorum.
Identification of Coumarin Biosynthetic Genes of Unbolted and Bolted P. praeruptorum
To further investigate the coumarin biosynthesis of P. praeruptorum, we compared and analyzed unbolted and bolted transcriptomic data in the annual plants (Supplementary Table 2). The hierarchical clustering divides these differential genes into two subgroups (Supplementary Figure 1). A total of 1,573 differentially expressed genes were screened out, of which 298 genes were up-regulated and 1,275 genes were down-regulated. Among them, 63 candidates involved in coumarin biosynthesis, including PAL, 4CL, C4H, C3H, HCT, COMT, CCoAOMT, UDT, AS, PS, BMT, SGT, and PRX, were identified from the transcriptomics data (Supplementary Table 3). Compared with the unbolted, 4CL, C4H, PS2, and C3H were significantly upregulated, whereas UDT, COMT, AS, BMT, and PRX were remarkably downregulated in the bolted P. praeruptorum (Figure 5). The expression levels of some homologs involved in coumarin biosynthesis, such as PAL, HCT, AS, PS, SGT, etc., had emerged different expressions. The reduction of the key genes in the umbelliferone branch pathway may result in the decline of coumarin accumulation. Such genes with bidirectional expression patterns suggest that functional divergence and spatiotemporal expression exist in these paralogs (Siwinska et al., 2018).
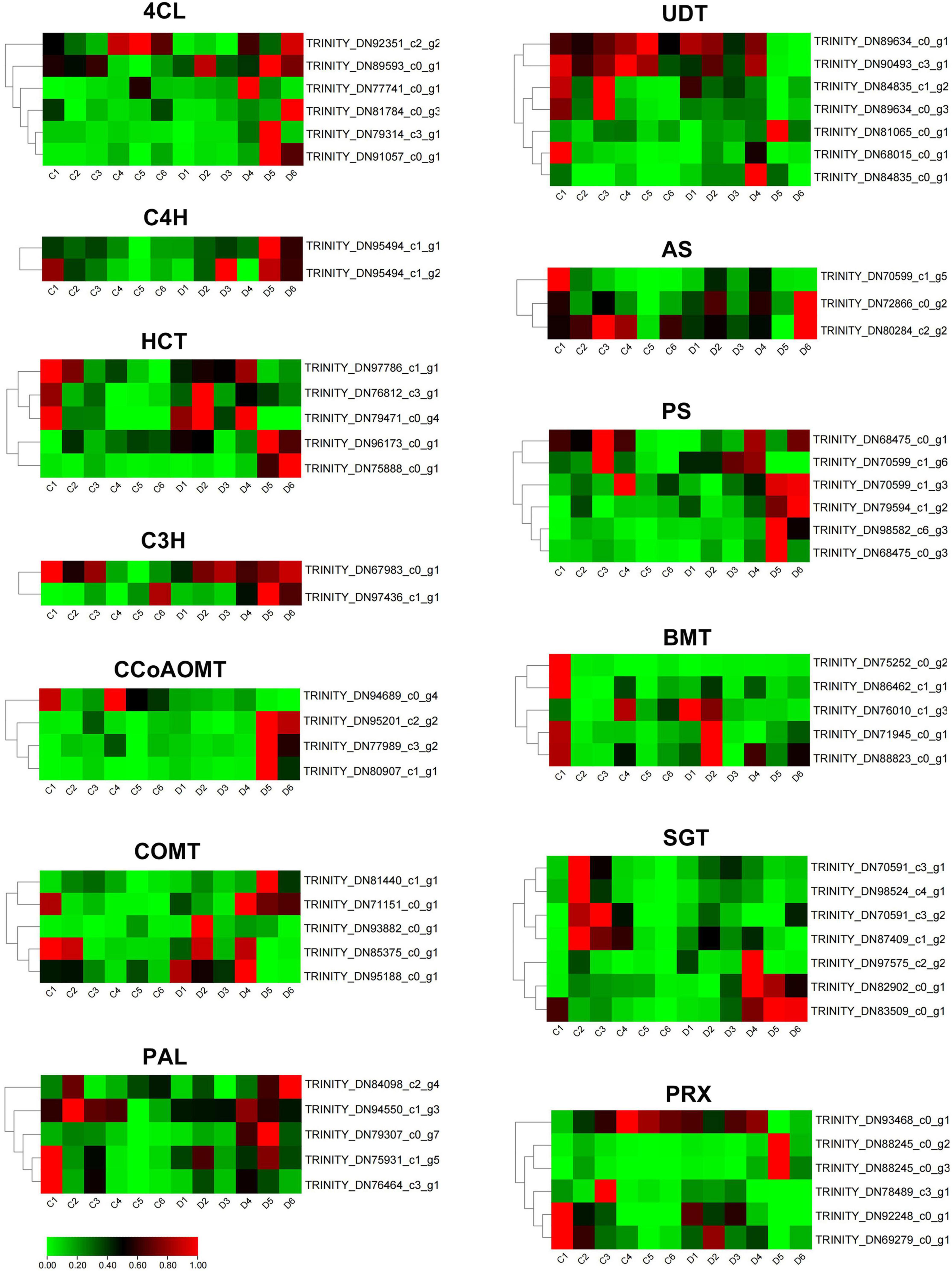
Figure 5. The expression profiles of coumarin biosynthetic genes of the unbolted and bolted P. praeruptorum. TPM values were used to compare the gene expression between the two groups. These values were normalized by the log2 function. The red represents the fold change in expression levels reaching two folds, the green represents the fold change in expression levels reaching one fold, and the black represents the fold change between one and two folds.
The Expression Profile and qPCR Validation of Genes Associated With Coumarin Biosynthesis
To narrow down the biosynthetic genes related to coumarins, eighteen important genes involved in the coumarin biosynthesis pathway were identified from 63 candidates to determine which genes were differentially expressed in the unbolted P. praeruptorum. As shown in Supplementary Figure 2, the coumarin biosynthesis pathway is a branch of the phenylpropanoid metabolism. p-Coumaric acid is situated at the crossover point of this branch. Through multi-step reactions, p-coumaric acid subsequently forms a linear coumarin scepolin under the catalysis of C3H, CA2H, OMT, and SGT. p-Coumaric acid is catalyzed by 4CL, C3H, HCT, COMT, CCoAOMT to form feruloyl-CoA, which will further form G-type lignin monomer. p-Coumaric acid is catalyzed by C’2H and lactionization to yield umbelliferone, the precursor of furano- and dihydropyranocoumarin (Supplementary Figure 2). Compared with the expression profiling of coumarin biosynthetic genes in the bolting stage, the expression levels of almost all genes were not significantly different. Except for PS1 and PRX3, the average expression levels of the other genes were upregulated in the bolted stage. To verify the expression of the candidate genes, we further detected the expression level of the genes by qPCR analysis. The expression of the PAL, C4H1, C3H, HCT, COMT, CCoAOMT, AS, PS1, BMTs, and PRXs were decreased in the bolted P. praeruptorum, whereas the expression of the 4CL1, C4H2, PS2, and SGT genes were increased (Figure 6). These results were inconsistent with the expression profile of coumarin biosynthesis genes. We re-analyzed the differences between the transcriptome and qPCR analysis results. From the measured expression data, it may be that the expressions of the biological replicates of the transcriptome are in a wide range, and the average expression levels of most genes in the bolting stage are not significantly different.
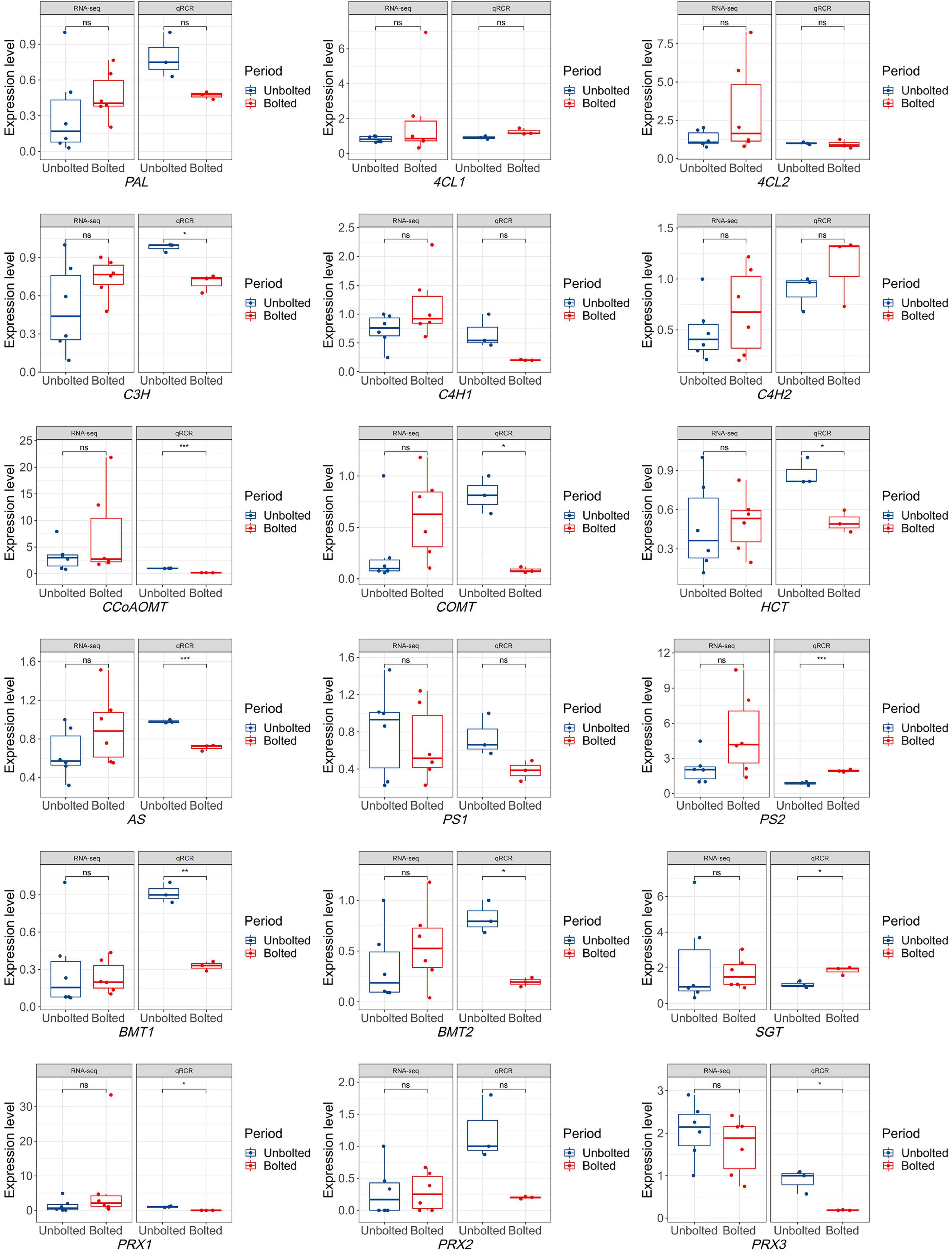
Figure 6. The expression levels of eighteen coumarin biosynthetic genes obtained by RNA-seq and qPCR analysis. The expression level obtained by RNA-seq technology contains 6 biological replicates, while the expression level obtained by qPCR method contains 3 biological replicates. Significant differences in the expression levels of coumarin biosynthesis genes were analyzed in the unbolted and bolted P. praeruptorum. One asterisk indicates significant difference between two periods. Three asterisks indicate extremely significant difference between two periods. “ns” indicates no significance. *denote there is significant difference between two groups (0.01 < P <0.05). **denote there is extremely significant difference between two groups (P < 0.01). ***denote there is extremely significant difference between two groups (P < 0.001).
The Phylogenetic Analysis and Expression Profile of ABC Transporters in P. praeruptorum
ATP-binding cassette (ABC) transporters are a wide and ancestral transmembrane protein family found in many natural species, which have received a lot of attention because of their multiple biological functions (Caña-Bozada et al., 2019; Huang et al., 2021). Here, 188 tentative ABC transporter genes or fragments were screened out from the transcriptome data (Supplementary Table 4). By constructing a phylogenetic tree with 129 ABC transporters from A. thaliala, only a limited number of transcripts could be clustered with the ABC family genes from P. praeruptorum (Supplementary Figure 3). It was conceivable that these genes or segments included only one or a few domains inside the transmembrane domains (TMDs) and nucleotide-binding domains (NBDs) domains and had little homology with the Arabidopsis ABC transporter family. Most of the PpABC genes have high homology with the AtABCA/ABCD/ABCE/ABCG/ABCI subfamilies. TRINITY_DN96014_c1_g1 shares a high homology with AtABCG22. TRINITY DN96811_c2_g1 and TRINITY_DN96361_c1_g2 have a high homology with AtABCG37. TRINITY_DN94634_c1_g6, TRINITY_DN78862_c0_g1, and TRINITY_DN181606_c0_g1 all have high homology with the AtABCG2 family. TRINITY_DN63312_c0_g1 has high homology with AtABCC10. TRINITY_DN80626_c2_g1, TRINITY_DN102046_c0_g1 have high homology with ABCA family. TRINITY_DN82877_c1_g2 has high homology with AtABCB5. We further analyzed the expression profile of 103 PpABC genes (Supplementary Table 5). The results indicated that the expression levels of these genes had several patterns. The first group of genes had higher expression in both unbolted and bolted P. praeruptorum, such as TRINITY_DN94350_c2_g4, TRINITY_DN96988_c1_g4 and TRINITY_DN88597_c0_g3. TRINITY_DN94350_c2_g4 shared more homology with AtABCE2, whereas the other two genes shared more with AtABCF1. The second group of genes had low expression in the unbolted P. praeruptorum and increased expression in the bolted, such as TRINITY_DN86296_c0_g1, TRINITY_DN749574350_c0_g4, and TRINITY_DN94728_c2_g1. TRINITY_DN86296_c0_g1 and TRINITY_DN7495_c0_g4 were both classified as members of the AtABCG subfamily, and TRINITY_DN94728_c2_g1 was found to be related to AtABCB20. The third group of genes had higher expression in the unbolted P. praeruptorum and lower expression in the bolted, such as TRINITY_DN96014_c1_g1 and TRINITY_DN91059_c0_g2. TRINITY_DN96014_c1_g1 and TRINITY_DN91059_c0_g2 shared more homology with AtABCG22 and AtABCG4, respectively. The remaining genes were expressed at lower levels both in unbolted and bolted P. praeruptorum (Figure 7). ABC transporters play multiple roles in trafficking ions, carbohydrates, lipids, xenobiotics, antibiotics, medicines, and heavy metals (Gadsby et al., 2006; Khan et al., 2020). AtABCB1 and AtABCB2 are known as auxin transporters. Overexpression of AtABCB1 promoted hypocotyl cell elongation (Lewis et al., 2007). A. thaliala, Z. mays, and O. sativa also contain ABCC subfamily members that are responsible for phytate trafficking (Nagy et al., 2009; Badone et al., 2010; Tagashira et al., 2015). Both AtABCC1 and AtABCC2 mediate tolerance to cadmium (Cd) and mercury via vacuolar sequestration (Park et al., 2012). AtABCF3 was involved in root development and growth (Kato et al., 2009). ABCG subfamily members have been associated with cuticle development and mental impairment (Bessire et al., 2011; Fourcroy et al., 2014; Lefèvre et al., 2018). AtABCG36 stimulated Cd uptake in root epidermal cells and was induced by Cd treatment (Kim et al., 2007). AtABCG37 was primarily responsible for delivering highly oxygenated coumarins to root exudation (Ziegler et al., 2017). The ABCG subfamily might be the main regulators participating in the extracellular secretion and transport of coumarin.

Figure 7. The expression profile of the possible ABC transporter genes in the unbolted and bolted P. praeruptorum. The TPM values are normalized by the log2 function. The expression patterns of PpABC genes can be divided into four groups: the first group has higher levels of expression both at the bolted and unbolted stages. The second group displayed high expressions prior to bolting and low expressions after bolting. The third group is the absolute opposite of the second group. The fourth group had lower expression both at the bolted and unbolted stages.
The Interaction Network of Coumarin Biosynthetic Genes
To clarify which genes were involved in the regulation of coumarin biosynthesis, the WGCNA data from the previous study was deeply mined (Song et al., 2021). Among thirteen gene modules, three modules relevant to coumarin biosynthesis were screened out by a gene-module correlation analysis (Supplementary Table 6). A total of 210 gene pairs were associated with the genes related to coumarins biosynthesis (Supplementary Table 7). TRINITY_DN77989_c3_g2 (CCoAOMT), TRINITY_DN68475_c0_g1 (PS1), TRINITY_ DN95494_c1_g1 (C4H1), TRINITY_DN79594_c1_g2 (PS2), and TRINITY_DN90493_c3_g1 (UDT) interacted strongly with some genes in the turquoise module. PS1 and C4H1 have strong interactions with TRINITY_DN97736_c4_g6 (UPL3), implying that UPL3 is involved in the ubiquitination of the two proteins. TRINITY_DN77619_c2_g1 (HSL1) interacts with UDT. TRINITY_DN74096_c0_g1 (ABCB1) interacts strongly with TRINITY_DN98338_c2_g1 (LOX2), TRINITY_DN95583_c1_g4 (MPK3), and TRINITY_DN79029_c0_g2 (Scarecrow-like TF). TRINITY_DN75913_c1_g5 (PAL) interacts with TRINITY_DN84719_c0_g2 (aquaporin 1) and TRINITY_DN92098_c0_g7 (EARLI 1) in the yellow-green module (Supplementary Figure 4).
Conclusion
Early bolting seriously affects the yield and quality of P. praeruptorum. The underlying mechanisms of the ineffectiveness of the bolted P. praeruptorum for medicinal purposes remain unclear. We found that lignification was more severe in the root of bolted P. praeruptorum. The contents of praeruptorin A, B, and E were lower in the bolted P. praeruptorum. We further compared the transcriptome at each bolting stage and found that some genes on the phenylpropanoid pathway branch were involved in coumarin biosynthesis, such as PAL, C4H, HCT, COMT, and CCoAOMT. Additionally, we screened several ABC transporters implicated in coumarin transport, including those belonging to the ABCA/D/E/G/I subfamilies. The ABCG subfamily may also play a role in the transportation of coumarin during the bolting stage. The network of co-expressed genes indicated that PS1 and C4H1 both have strong interactions with UPL3. Bolting may have a negative effect on the accumulation of coumarins in P. praeruptorum and the regulation of associated biosynthetic genes. Our results provide some scientific references for the quality evaluation of P. praeruptorum drugs. However, more evidence is needed to figure out how bolting changes the endogenous signaling cascades and how this influences the activation of downstream genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
BH and CS discussed the writing plan. CS, XL, and BJ drafted the manuscript. CS, XL, LL, PW, MM, CC, and FW edited the manuscript. XL, BJ, BL, and GW conduct the experiment. BH acquired the funding. All authors have read, reviewed, and approved the submitted version.
Funding
This work was supported by China Agricultural Research System of MOF and MARA (CARS-21), National level major increase and decrease projects (2060302), and Anhui Outstanding Youth Fund (1808085J17).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.899819/full#supplementary-material
Footnotes
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anders, S., and Huber, W. (2010). Differential expression and sequence-specific interaction of karyopherin α with nuclear localization sequences. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Badone, F. C., Cassani, E., Landoni, M., Doria, E., Panzeri, D., Lago, C., et al. (2010). The low phytic acid1-241 (lpa1-241) maize mutation alters the accumulation of anthocyanin pigment in the kernel. Planta 231, 1189–1199. doi: 10.1007/s00425-010-1123-z
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Statist. Soc. Series B 57, 289–300. doi: 10.2307/2346101
Bessire, M., Borel, S., Fabre, G., Carrac, L., Efremova, N., Yephremov, A., et al. (2011). A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23, 1958–1970. doi: 10.1105/tpc.111.083121
Caña-Bozada, V., Morales-Serna, F. N., García-Gasca, A., Llera-Herrera, R., and Fajer-Ávila, E. J. (2019). Genome-wide identification of ABC transporters in monogeneans. Mol. Biochem. Parasitol. 234:111234. doi: 10.1016/j.molbiopara.2019.111234
Chauhan, S. K., Upadhyaya, S. K., Uniyal, M. R., and Rana, S. (2015). Development of pharmacognostic profile of Alpinia galanga, Willd. (Zingiberaceae). Int. J. Pharmacol. Res. 5, 57–61. doi: 10.7439/ijpr
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Chu, S., Zhang, L., Xie, J., Dai, M., Wu, X., et al. (2019). Tissue-specific metabolite profiling on the different parts of bolting and unbolting Peucedanum praeruptorum dunn (Qianhu) by laser microdissection combined with UPLC-Q/TOF–MS and HPLC–DAD. Molecules 24:1439. doi: 10.3390/molecules24071439
Doncheva, N. T., Morris, J. H., Gorodkin, J., and Jensen, L. J. (2019). Cytoscape stringapp: network analysis and visualization of proteomics data. J. Proteome Res. 18, 623–632. doi: 10.1021/acs.jproteome.8b00702
Dugrand-Judek, A., Olry, A., Hehn, A., Costantino, G., Ollitrault, P., Froelicher, Y., et al. (2015). The distribution of coumarins and furanocoumarins in Citrus species closely matches Citrus phylogeny and reflects the organization of biosynthetic pathways. PLoS One 10:e0142757. doi: 10.1371/journal.pone.0142757
Fourcroy, P., Sisó-Terraza, P., Sudre, D., Savirón, M., Reyt, G., Gaymard, F., et al. (2014). Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 201, 155–167. doi: 10.1111/nph.12471
Gadsby, D. C., Vergani, P., and Csanády, L. (2006). The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature 440, 477–483. doi: 10.1038/nature04712
Guo, F. X., Xiao, W. J., Chen, Y., Zhang, Y. J., Chen, Y. Z., Liu, L. L., et al. (2021). Initiation of early bolting by pre-enhancing anthocyanin and catalase activity in Angelica sinensis tender leaf during medicine formation cultivation year. Russ. J. Plant Physiol. 68, 763–773. doi: 10.1134/S1021443721040063
Hou, Z., Luo, J., Wang, J., and Kong, L. (2010). Separation of minor coumarins from Peucedanum praeruptorum using HSCCC and preparative HPLC guided by HPLC/MS. Sep. Purif. Technol. 75, 132–137.
Huang, J., Li, X., Chen, X., Guo, Y., Liang, W., and Wang, H. (2021). Genome-wide identification of soybean abc transporters relate to aluminum toxicity. Int. J. Mol. Sci. 22:6556. doi: 10.3390/ijms22126556
Jian, X., Zhao, Y., Wang, Z., Li, S., Li, L., Luo, J., et al. (2020). Two CYP71AJ enzymes function as psoralen synthase and angelicin synthase in the biosynthesis of furanocoumarins in Peucedanum praeruptorum Dunn. Plant Mol. Biol. 104, 327–337. doi: 10.1007/s11103-020-01045-4
Kato, T., Tabata, S., and Sato, S. (2009). Analyses of expression and phenotypes of knockout lines for Arabidopsis ABCF subfamily members. Plant Biotechnol. 26, 409–414. doi: 10.5511/plantbiotechnology.26.409
Khan, N., You, F. M., Datla, R., Ravichandran, S., Jia, B., and Cloutier, S. (2020). Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.). BMC Genomics 21:722. doi: 10.1186/s12864-020-07121-9
Kim, D. Y., Bovet, L., Maeshima, M., Martinoia, E., and Lee, Y. (2007). The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218. doi: 10.1111/j.1365-313X.2007.03044.x
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lee, J., Lee, Y. J., Kim, J., and Bang, O. S. (2015). Pyranocoumarins from root extracts of Peucedanum praeruptorum Dunn with multidrug resistance reversal and anti-inflammatory activities. Molecules 20, 20967–20978. doi: 10.3390/molecules201219738
Lefèvre, F., Fourmeau, J., Pottier, M., Baijot, A., Cornet, T., Abadía, J., et al. (2018). The Nicotiana tabacum ABC transporter NtPDR3 secretes O-methylated coumarins in response to iron deficiency. J. Exp. Bot. 69, 4419–4431. doi: 10.1093/jxb/ery221
Lewis, D. R., Miller, N. D., Splitt, B. L., Wu, G., and Spalding, E. P. (2007). Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell 19, 1838–1850. doi: 10.1105/tpc.107.051599
Li, M., Cui, X., Jin, L., Li, M., and Wei, J. (2022). Bolting reduces ferulic acid and flavonoid biosynthesis and induces root lignification in Angelica sinensis. Plant Physiol. Biochem. 170, 171–179. doi: 10.1016/j.plaphy.2021.12.005
Li, M., Li, J., Wei, J., and Paré, P. W. (2021). Transcriptional controls for early bolting and flowering in Angelica sinensis. Plants 10:1931 doi: 10.3390/plants10091931
Li, Y., Kong, D., Fu, Y., Sussman, M. R., and Wu, H. (2020). The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 148, 80–89. doi: 10.1016/j.plaphy.2020.01.006
Li, Z., and Nair, S. K. (2015). Structural basis for specificity and flexibility in a plant 4-coumarate:CoA Ligase. Structure 23, 2032–2042. doi: 10.1016/j.str.2015.08.012
Liang, W. H., Chang, T. W., and Charng, Y. C. (2018). Influence of harvest stage on the pharmacological effect of Angelica dahurica. Bot. Stud 59:14. doi: 10.1186/s40529-018-0230-1
Liljegren, S. (2010). Phloroglucinol stain for lignin. Cold Spring Harb. Protoc. 2010:4954. doi: 10.1101/pdb.prot4954
Liu, C.-M., Shen, H., Lin, Y., and Yu, Y.-L. (2020). Antiproliferative and antimetastatic effects of praeruptorin C on human non–small cell lung cancer through inactivating ERK/CTSD signalling pathways. Molecules 25:1625. doi: 10.3390/molecules25071625
Liu, T., Yao, R., Zhao, Y., Xu, S., Huang, C., Luo, J., et al. (2017). Cloning, functional characterization and site-directed mutagenesis of 4-Coumarate: coenzyme a ligase (4CL) involved in coumarin biosynthesis in peucedanum praeruptorum Dunn. Front. Plant Sci. 8:4. doi: 10.3389/fpls.2017.00004
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Matsumoto, S., Mizutani, M., Sakata, K., and Shimizu, B. I. (2012). Molecular cloning and functional analysis of the ortho-hydroxylases of p-coumaroyl coenzyme A/feruloyl coenzyme A involved in formation of umbelliferone and scopoletin in sweet potato, Ipomoea batatas (L.) Lam. Phytochemistry 74, 49–57. doi: 10.1016/j.phytochem.2011.11.009
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, 182–185. doi: 10.1093/nar/gkm321
Munakata, R., Kitajima, S., Nuttens, A., Tatsumi, K., Takemura, T., Ichino, T., et al. (2020). Convergent evolution of the UbiA prenyltransferase family underlies the independent acquisition of furanocoumarins in plants. New Phytol. 225, 2166–2182. doi: 10.1111/nph.16277
Munakata, R., Olry, A., Karamat, F., Courdavault, V., Sugiyama, A., Date, Y., et al. (2016). Molecular evolution of parsnip (Pastinaca sativa) membrane-bound prenyltransferases for linear and/or angular furanocoumarin biosynthesis. New Phytol. 211, 332–344. doi: 10.1111/nph.13899
Nagy, R., Grob, H., Weder, B., Green, P., Klein, M., Frelet-Barrand, A., et al. (2009). The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporterinvolved in guard cell signaling and phytate storage. J. Biol. Chem. 284, 33614–33622. doi: 10.1074/jbc.M109.030247
Park, J., Song, W. Y., Ko, D., Eom, Y., Hansen, T. H., Schiller, M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288. doi: 10.1111/j.1365-313X.2011.04789.x
Patro, R., Duggal, G., Love, M. I, Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Pouteau, S., and Albertini, C. (2009). The significance of bolting and floral transitions as indicators of reproductive phase change in Arabidopsis. J. Exp. Bot. 60, 3367–3377. doi: 10.1093/jxb/erp173
Rastogi, S., Kumar, R., Chanotiya, C. S., Shanker, K., Gupta, M. M., Nagegowda, D. A., et al. (2013). 4-Coumarate: CoA ligase partitions metabolites for eugenol biosynthesis. Plant Cell Physiol. 54, 1238–1252. doi: 10.1093/pcp/pct073
Robe, K., Conejero, G., Gao, F., Lefebvre-Legendre, L., Sylvestre-Gonon, E., Rofidal, V., et al. (2021). Coumarin accumulation and trafficking in Arabidopsis thaliana: a complex and dynamic process. New Phytol. 229, 2062–2079. doi: 10.1111/nph.17090
Sarkhail, P., Shafiee, A., and Sarkheil, P. (2013). Biological activities and pharmacokinetics of praeruptorins from peucedanum species: a systematic review. Biomed. Res. Int. 2013:343808. doi: 10.1155/2013/343808
Shang, Q. M., Li, L., and Dong, C. J. (2012). Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 236, 1093–1105. doi: 10.1007/s00425-012-1659-1
Siwinska, J., Siatkowska, K., Olry, A., Grosjean, J., Hehn, A., Bourgaud, F., et al. (2018). Scopoletin 8-hydroxylase: a novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis. J. Exp. Bot. 69, 1735–1748. doi: 10.1093/jxb/ery005
Song, C., Li, X., Jia, B., Liu, L., Ou, J., and Han, B. (2021). De novo transcriptome sequencing coupled with co-expression analysis reveal the transcriptional regulation of key genes involved in the formation of active ingredients in peucedanum praeruptorum dunn under bolting period. Front. Genet. 12:683037. doi: 10.3389/fgene.2021.683037
Song, J., Luo, H., Xu, Z., Zhang, Y., and Xin, H. (2020). Mining genes associated with furanocoumarin biosynthesis in an endangered medicinal plant, Glehnia littoralis. J. Genet. 99, 1–11.
Song, Y. L., Jing, W. H., Du, G., Yang, F. Q., Yan, R., and Wang, Y. T. (2014). Qualitative analysis and enantiospecific determination of angular-type pyranocoumarins in Peucedani Radix using achiral and chiral liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr. A 1338, 24–37. doi: 10.1016/j.chroma.2014.01.078
Stelzhammer, V., Ozcan, S., Michael, G., Steeb, H., Hodes, G. E., Guest, C., et al. (2017). Reduced allergic lung inflammation by root extracts from two species of Peucedanum through inhibition of Th2 cell activation. J. Ethnopharmacol. 196, 75–83. doi: 10.1016/j.dineu.2015.08.001
Sui, Z., Luo, J., Yao, R., Huang, C., Zhao, Y., and Kong, L. (2019). Functional characterization and correlation analysis of phenylalanine ammonia-lyase (PAL) in coumarin biosynthesis from Peucedanum praeruptorum Dunn. -Phytochemistry 158, 35–45. doi: 10.1016/j.phytochem.2018.11.006
Tagashira, Y., Shimizu, T., Miyamoto, M., Nishida, S., and Yoshida, K. T. (2015). Overexpression of a gene involved in phytic acid biosynthesis substantially increases phytic acid and total phosphorus in rice seeds. Plants 4, 196–208. doi: 10.3390/plants4020196
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tao, Y., Luo, J., Lu, Y., Xu, D., Hou, Z., and Kong, L. (2009). Rapid identification of two species of peucedanum by high-performance liquid chromatography-diode array detection-electrospray ionization tandem mass spectrometry. Nat. Prod. Commun. 4, 1079–1084. doi: 10.1177/1934578X0900400812
Tian, Y., Shi, R., Gao, M., Wang, H., Du, Y., Zhang, L., et al. (2017). Differentiation of furanocoumarin isomers with ratio of relative abundance of characteristic fragment ions and application in Angelicae dahuricae radix. Chromatographia 80, 1401–1410. doi: 10.1007/s10337-017-3348-5
Wang, L., Wang, J., Yang, L., Zhou, S. M., Guan, S. Y., Yang, L. K., et al. (2017). Effect of Praeruptorin C on 3-nitropropionic acid induced Huntington’s disease-like symptoms in mice. Biomed. Pharmacother. 86, 81–87. doi: 10.1016/j.biopha.2016.11.111
Wang, X. Y., Li, J. F., Jian, Y. M., Wu, Z., Fang, M. J., and Qiu, Y. K. (2015). On-line comprehensive two-dimensional normal-phase liquid chromatography×reversed-phase liquid chromatography for preparative isolation of Peucedanum praeruptorum. J. Chromatogr. A 1387, 60–68. doi: 10.1016/j.chroma.2015.02.003
Yao, R., Zhao, Y., Liu, T., Huang, C., Xu, S., Sui, Z., et al. (2017). Identification and functional characterization of a p-coumaroyl CoA 2′-hydroxylase involved in the biosynthesis of coumarin skeleton from Peucedanum praeruptorum Dunn. Plant Mol. Biol. 95, 199–213. doi: 10.1007/s11103-017-0650-4
Yu, C. L., Yu, Y. L., Yang, S. F., Hsu, C. E., Lin, C. L., Hsieh, Y. H., et al. (2020). Praeruptorin A reduces metastasis of human hepatocellular carcinoma cells by targeting ERK/MMP1 signaling pathway. Environ. Toxicol. 36, 540–549. doi: 10.1002/tox.23059
Yu, G., Zhou, Y., Yu, J., Hu, X., Tang, Y., Yan, H., et al. (2019). Transcriptome and digital gene expression analysis unravels the novel mechanism of early flowering in Angelica sinensis. Sci. Rep. 9:10035. doi: 10.1038/s41598-019-46414-2
Zhao, Y., Liu, T., Luo, J., Zhang, Q., Xu, S., Han, C., et al. (2015). Integration of a decrescent transcriptome and metabolomics dataset of Peucedanum praeruptorum to investigate the CYP450 and MDR genes involved in coumarins biosynthesis and transport. Front. Plant Sci. 6:996. doi: 10.3389/fpls.2015.00996
Zhao, Y., Luo, J., Xu, S., Wang, W., Liu, T., Han, C., et al. (2016a). Selection of reference genes for gene expression normalization in Peucedanum praeruptorum Dunn under abiotic stresses, hormone treatments and different tissues. PLoS One 11:e0152356. doi: 10.1371/journal.pone.0152356
Zhao, Y., Wang, N., Zeng, Z., Xu, S., Huang, C., Wang, W., et al. (2016b). Cloning, functional characterization, and catalytic mechanism of a Bergaptol O-Methyltransferase from Peucedanum praeruptorum dunn. Front. Plant Sci. 7:722. doi: 10.3389/fpls.2016.00722
Zhao, Y., Wang, N., Sui, Z., Huang, C., Zeng, Z., and Kong, L. (2019). The molecular and structural basis of o-methylation reaction in coumarin biosynthesis in Peucedanum praeruptorum dunn. Int. J. Mol. Sci. 20, 1533. doi: 10.3390/ijms20071533
Zhao, Z., Liang, Z., and Ping, G. (2011). Macroscopic identification of Chinese medicinal materials: traditional experiences and modern understanding. J. Ethnopharmacol. 134, 556–564. doi: 10.1016/j.jep.2011.01.018
Zhou, J., Wang, W., Liu, M., and Liu, Z. (2014). Molecular authentication of the traditional medicinal plant Peucedanum praeruptorum and its substitutes and adulterants by dna - Barcoding technique. Pharmacogn. Mag. 10, 385–390. doi: 10.4103/0973-1296.141754
Keywords: bolting, lignification, coumarin biosynthesis, transcriptional regulation, ABC transporters
Citation: Song C, Li X, Jia B, Liu L, Wei P, Manzoor MA, Wang F, Li BY, Wang G, Chen C and Han B (2022) Comparative Transcriptomics Unveil the Crucial Genes Involved in Coumarin Biosynthesis in Peucedanum praeruptorum Dunn. Front. Plant Sci. 13:899819. doi: 10.3389/fpls.2022.899819
Received: 19 March 2022; Accepted: 29 April 2022;
Published: 17 May 2022.
Edited by:
Md Abdur Rahim, Sher-e-Bangla Agricultural University, BangladeshReviewed by:
Md Atikur Rahman, National Institute of Animal Science, Rural Development Administration, South KoreaMd. Mizanur Rahman, Islamic University, Bangladesh
Copyright © 2022 Song, Li, Jia, Liu, Wei, Manzoor, Wang, Li, Wang, Chen and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bangxing Han, hanbx1978@sina.com
†These authors have contributed equally to this work
 Cheng Song
Cheng Song